Abstract
Identification and characterization of the normal epithelial lineages in the mammary gland is a fundamental step in understanding both development and cellular origin of cancer. In contrast to other tissues where lineage tracing has been widely accepted as a method of choice for dissecting the stem cell hierarchy, mammary gland has long remained a challenge due to its unique developmental and topological features. Recent advances in high-resolution single-cell imaging, combined with the use of inducible Cre-recombinase and in situ cell ablation, have provided unprecedented insight into mammary epithelial cell composition and function. Here, we briefly summarize and compare different mammary gland lineage tracing strategies, examine associated caveats and discuss future challenges and opportunities.
Keywords: Breast, Lineages, Fate mapping, In situ cell ablation, Cell-of-origin, Genetically engineered mouse models, Topological context
Introduction
One of the central unresolved issues in mammary gland biology is the relationship between the normal cellular developmental hierarchy and different subtypes of breast cancer. The following are some of the key questions: do different tumor subtypes originate from a distinct cell-of-origin? What is the nature of the relationship between normal and cancer stem cells? Does cell fate influence susceptibility to malignant transformation? What degree of cell fate plasticity do different cell types exhibit during normal development and tumorigenesis? The prerequisite for resolving these questions is an in depth understanding of the cellular hierarchies in the normal breast tissue.
The mammary gland consists of hollow, bi-layered epithelial tubes that are organized into elaborate tree-like structures embedded in fatty stroma. The gland undergoes intense remodeling during three distinct developmental stages. In puberty, the network of primary and secondary branches is formed by proliferative expansion of terminal end buds (TEBs), bifurcation and side branching. In adult gland, tertiary branches and small alveolar buds are formed alongside primary and secondary ducts during each estrus cycle. In pregnancy, the alveolar epithelium undergoes major expansion to yield milk-producing alveoli. Upon each lactating period, the gland undergoes massive remodeling and involution. It is widely accepted that such profound regenerative capacity is sustained by stem cells; however, their nature and location remain a subject of intense debate.
Transplantation assay
For over half a century, transplantation assay has been the gold standard method for evaluation of stem and progenitor cell activity in mammary epithelium. The first evidence of the presence of stem cells in the mammary gland came from DeOme and Faulkin [1] who showed that transplantation of small fragments of mouse mammary epithelium into de-epithelialized mammary fat pads gives rise to entire ductal epithelial tree. The progeny of the transplanted cells could be serially transplanted confirming that mammary epithelium contains cells with self-renewing properties and multi-lineage potential. Subsequent studies demonstrated that transplantation of dissociated mammary epithelial cells (MECs) at limiting dilutions gives rise to three types of outgrowths: ductal, alveolar (lobular) and complete glands, suggesting the presence of lineage-restricted progenitors [2, 3].
More recently, transplantation assays have mostly been used for characterization of prospectively isolated mouse MEC subpopulations obtained using different combinations of markers [4–15] and other parameters such as cell size [16] or label retention [17]. In addition, lentiviral genomic barcoding has been employed for analysis of clonal dynamics within serially transplanted cells [18]. Collectively, these studies demonstrated significant heterogeneity in both luminal and basal/myoepithelial layers (reviewed in [19, 20]). Markers specific for stem or progenitor cells have not been identified to date, the existing combinations merely enrich for these subpopulations.
One of the key findings that emerged from these analyses is that MECs within the basal/myoepithelial layer have stem cell-like properties [4, 5]. It is unclear, however, whether all myoepithelial cells can potentially function as stem cells in transplantation assays, or whether there is a specific subset that has intrinsic stem cell-like properties. A recent report by Prater et al. [21] suggests that a high proportion of myoepithelial cells can acquire multi-lineage potential in transplantation assays when cultured in vitro in the presence of agents that disrupt actin–myosin interactions.
Lineage tracing
There is accumulating evidence that transplanted MECs do not always reflect their physiological developmental fate [21–25], emphasizing the need to revisit and refine traditional hierarchy models using methodologies that preserve tissue architecture. Lineage tracing, a technique originally developed to analyze early embryos, is one of the most powerful methods available for identifying stem and progenitor cells and analyzing their function in physiological developmental context. The method is based on permanent labeling of a single cell or a group of cells and subsequent tracking of their fate and the fate of their progeny in vivo. This approach is also essential for identification and characterization of topologically and functionally specialized microenvironments such as the stem cell niche. Genetic lineage tracing (also called genetic fate mapping) is a critical tool for analysis of tissue development, homeostasis and cellular origin of cancer [26].
In situ lineage tracing
The first in situ fate mapping in the mammary gland was done over a decade ago by the Smith laboratory [27]. The term “in situ” refers here to the procedure consisting of labeling and tracking of the cells in an intact mammary gland of the host animal. Wagner et al. [27] identified a MEC subpopulation called “parity-induced” mammary epithelial cells (PI-MECs), using transgenic mice carrying Cre-recombinase under control of the pregnancy-specific promoter for whey acidic protein (WAP).
In recent years, a combination of inducible Cre-recombinase and high-resolution imaging has provided extraordinary insight into the mouse MEC composition and function. Several conditional genetic labeling strategies have been employed to track mouse MECs in situ, using either knock-in or transgenic mice. They include doxycycline-inducible TetO-Cre system, Tamoxifen-inducible Cre-recombinase (CreER, CreERT2) and orthotopic adenoviral delivery of lineage-specific Cre-recombinases (AdCre) [21–24, 28–31]. All methods are based on temporal and cell-type-specific regulation of Cre-recombinase activity, resulting in permanent expression of the reporter allele in target cells.
Each approach has some general, as well as mammary specific, advantages and disadvantages. Generation of new transgenic or knock-in strains and their subsequent breeding is time consuming and expensive. Intraductal injection of lineage-specific AdCre is not only faster and cheaper, but also enables spatial control of Cre-recombinase activity, which is especially useful for downstream applications such as modeling breast cancer, where systemic expression of Cre-recombinase could have deleterious effects. Tamoxifen can temporarily delay ductal development and skew the relative distribution and proliferation rate of MEC subpopulations in a dose-dependent manner; thus caution should be used with CreER system when interpreting short-term in situ tracing studies of a quantitative nature [32]. Doxycycline-inducible Tet-strains contain constructs consisting of endogenous gene promoter sequence and tTA or rtTA cassette. In transgenic Tet-strains, these constructs are randomly inserted into the genome and thus may not faithfully reflect endogenous gene expression due to the chromosomal position effects or uncontrolled copy number. Therefore, the lack of positive cells in lineage tracing studies using transgenic Tet-strains may not reflect their true absence in a tissue and should thus be interpreted with caution. The same caveat applies to transgenic strains carrying Cre-recombinase.
Various gene promoters have been used to drive Cre expression in specific MEC subpopulations (summarized in Table 1). They include components of the developmental signaling pathways (Wnt, Notch), structural genes (cytokeratins, actin) and mammary-related genes (WAP, Elf5). The choice of the drivers has mostly been based on information obtained in ex vivo analyses of various MEC subpopulations, or stem cell markers characterized in other tissues, such as the skin and intestine.
Table 1.
Mouse strains used for in situ lineage tracing in the mammary gland
| Driver | Inducible system | Transgenic (T) Knock-in (K) | References |
|---|---|---|---|
| K5 | CreER | K | Van Keymeulen et al. [22] |
| CreERT2 | T | Rios et al. [33] | |
| rtTA/TetO-Cre | T | Rios et al. [33] | |
| K8 | CreER | T | Van Keymeulen et al. [22] |
| K14 | No | T | Van Keymeulen et al. [22] |
| rtTA/TetO-Cre | T | Van Keymeulen et al. [22] | |
| CreERT2 | T | Rios et al. [33] | |
| K18 | CreER | T | Van Keymeulen et al. [22] |
| Acta2 | CreERT2 | T | Prater et al. [21] |
| Lgr5 | CreERT2 | K | Van Keymeulen et al. [22], de Visser et al. [24], Rios et al. [33] |
| Procr | CreERT2 | K | Wang et al. [31] |
| Axin2 | CreERT2 | K | van Amerongen et al. [23] |
| Notch2 | CreERT2 | K | Sale et al. [28] |
| Notch3 | CreERT2 | K | Lafkas et al. [30] |
| Elf5 | rtTA/TetO-Cre | T | Rios et al. [33] |
| Wap | No | T | Wagner et al. [27], Chang et al. [25] |
One important consideration when interpreting lineage tracing data is that a gene promoter chosen to drive Cre expression, can be switched on or off in a developmental stage-dependent manner in a particular MEC subset. Conversely, a single reporter gene may label two or more MEC subpopulations at different developmental stages. Thus, a single marker does not necessarily identify the same and unique MEC subpopulation at different stages of morphogenesis.
3D visualization
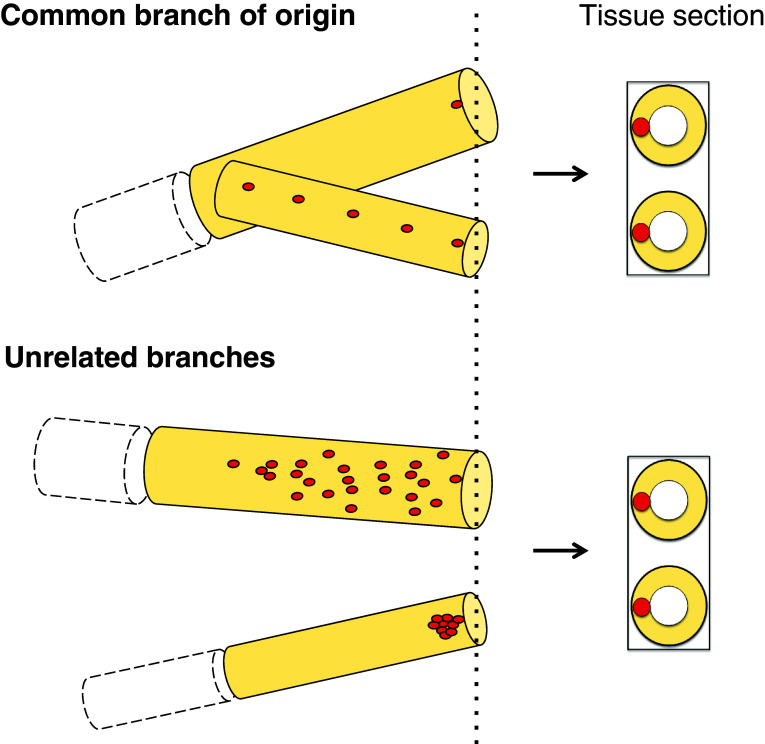
Mammary gland consists of spatially complex network of branches that undergo intense remodeling in puberty, pregnancy, lactation and involution. This presents some unique challenges for visualization of labeled cells and interpretation of long-term lineage tracing data. Standard two-dimensional tissue sections are inadequate for quantitative clonal analysis and can also be misleading in cases where traced cells are rare or exhibit complex spatial patterns (Fig. 1). Analysis of the whole-mount specimens in different focal planes is essential in such cases, however, traditional protocols for visualization of LacZ-positive cells are not suitable for high-resolution imaging at the cellular level.
Fig. 1.
Two-dimensional tissue sections do not reveal important topological and quantitative information in lineage tracing experiments. Schematic presentation of mammary ducts harboring genetically labeled cells and respective tissue sections. Dotted line represents the plane of the section. Complex topological patterns, the branch of origin and quantitative information, cannot be discerned from tissue sections
To visualize individual cells within their intact three-dimensional context, Sale and colleagues developed an improved x-gal labeling protocol that enables high-resolution, single-cell imaging of mammary whole-mounts [28]. Using Notch2 paralogue as a genetic marker, they discovered and functionally characterized two previously unrecognized mammary epithelial cell types: S and L cells [28]. S cells exhibit a unique combination of morphological and topological features: small size (S cell diameter is approximately one-third of the luminal cell diameter), formation of strings around single large Notch2-negative cell and a distinct reiterative spatial placement relative to the longitudinal axis and the circumference of the duct. During the estrous cycle, tertiary branches are formed directly above the S cell strings. These complex topological arrangements would be very difficult, if not impossible, to extrapolate from serial sections.
An alternative approach to whole-mount analysis of x-gal labeled samples is 3D confocal reconstruction of optical sections from fluorescently labeled specimens. This method is particularly useful in conjunction with multicolor reporter system for analysis of multiple clones in close proximity [31, 33].
In situ cell ablation
A major drawback of transplantation assays is disruption of the physiological developmental and topological context, which may induce non-physiological lineage commitment in transplanted cells. Moreover, in some cases, transplantation assay may not reveal important functional features of the target cells. For example, S and L cells do not give rise to outgrowths in transplantation assays, nor do they form clones in situ after long-term lineage tracing [28]. To gain more insight into their functional properties, Sale and colleagues used conditional in situ cell ablation, a powerful method for the characterization of cells in their physiological developmental and spatial context. This technique is based on conditional expression of diphtheria toxin receptor (iDTR) in Cre-positive cells [34]. Upon administration of diphtheria toxin, Cre-expressing cells are depleted whereas other cells remain intact. Ablation of S and L cells during active ductal morphogenesis revealed that their function indeed reflects their topological context: S cells regulate spatial placement of tertiary branches alongside the ductal tree, whereas L cells orchestrate formation of alveolar lumen and spatial organization of alveolar clusters [28].
A recent report by Wang et al. [31] described an alternative approach based on conditional expression of Diphtheria toxin fragment A (DTA) in Cre-positive cells. This method does not require administration of diphtheria toxin; however, it may require multiple Tamoxifen injections in a short period of time to achieve optimal ablation efficiency [31]. DTA-based ablation of Procr (protein C receptor)-positive MECs during puberty largely prevented ductal growth, thus corroborating lineage tracing data, which indicated that this population of cells exhibits stem cell properties. The authors note that the role of Procr-positive stromal cells should also be taken into account when interpreting these experiments, since they were also affected by the ablation [31].
Lineage tracing and MEC hierarchy models
Traditional depictions of the mammary epithelial cell hierarchy conform to a classical Waddington’s landscape model [35] and are based on the premise that MECs comprise two major lineages, luminal and basal/myoepithelial. It is becoming increasingly evident, however, that mammary epithelium is much more complex than previously thought, both developmentally and functionally, and exhibits a considerable degree of cell fate plasticity.
The first evidence that mammary epithelium consists of several distinct morphotypes, comes from the ultrastructural studies done by Chepko and Smith [36]. They divided MECs into five subtypes: classical luminal and myoepithelial cells, undifferentiated and differentiated large light cells (ULLCs, DLLCs) and small light cells (SLCs). Sale and colleagues provide the first genetic evidence that MECs constitute at least four independent lineages: classical luminal and myoepithelial lineage, L-cell lineage and S-cell lineage [28]. There are marked morphological and topological similarities between the light cells (small and large) identified by electron microscopy and S and L cells, respectively, identified by genetic labeling. Whether these populations overlap remains to be demonstrated experimentally. These studies highlight the importance of defining distinct MEC subsets based on multiple variables that, in addition to classical marker profile, include morphology, topological placement, function and developmental context.
Alveologenesis is another example where fate mapping has recently revealed previously unrecognized developmental complexity of the mammary epithelium. Two independent groups have demonstrated using different genetic labeling strategies that each alveolus consists of at least two distinct luminal alveolar lineages with independent progenitors [25, 28]. Using Notch2-CreERT2 knock-in mice, Sale and colleagues have identified single and paired Notch2-positive L-alveolar cells in each alveolus, whereas classical secretory alveolar cells remained unlabeled. In situ depletion of L-alveolar progenitors prior to pregnancy did not impair proliferation of secretory alveolar cells, instead they formed outgrowths resembling alveoli but with no lumen [28]. Chang et al. [25] used Wap-Cre transgenics to label PI-MECs and found an inverse labeling pattern: whereas all luminal estrogen receptor (ER) negative cells in an alveolus could be derived from PI-MECs, single or paired alveolar ER-positive cells were unlabeled, indicating that they are derived from a different lineage. Importantly, in both studies, basal alveolar cells remained unlabeled indicating they have a different origin. This result is consistent with the report by van Amerongen and colleagues who demonstrated using Axin2-CreERT2 knock-in mice that a subset of MECs positive for Axin2 in prepubescent animals gives rise to myoepithelial but not luminal alveolar cells [23]. Different origin of basal and luminal alveolar cells in individual alveoli was also demonstrated by Rios et al. [33] using K5-rtTA/TetO-cre and Elf5-rtTA/TetO-cre transgenics.
In addition to classical secretory alveolar cells and L cells, Notch2 lineage tracing has revealed a third, possibly functionally distinct subset of luminal alveolar lineage, called “transient alveolar cells” [28]. They fill in the lumen of actively growing tertiary branches and sprouting alveolar buds in early pregnancy and are cleared as soon as these structures are formed. Transit alveolar cells are Notch2 positive, thus indicating that they are not direct descendants of the secretory alveolar lineage.
Taken together, these studies imply that individual alveoli are not clonal outgrowths, but are formed by spatially and temporally coordinated growth of different cell lineages.
One of the most controversial issues arising from in situ lineage tracing studies is the identity and the role of stem cells in the adult mammary gland homeostasis. Two opposing stem cell hierarchy models have been proposed based on data obtained with partially overlapping set of drivers (Table 1) [22, 33]. One model suggests that the luminal and basal compartment is maintained by lineage-restricted, unipotent stem cells [22]; while the other proposes that adult mammary epithelium is maintained by bi-potent stem cells [33]. Discrepancies between the results obtained using the same set of drivers could be attributed to several factors discussed in detail above (see pages 4–6). These include differences in imaging techniques (2D versus 3D), inherent differences between the mouse models (knock-in versus transgenic strains, Tamoxifen versus doxycycline induction) and heterogeneity within the stem cell compartment.
The evidence for the latter comes from Wang and colleagues [31], who demonstrated the existence of at least two distinct subsets of basal stem cells in transplantation assays: Procrpositive/K5low/K14low and Procrnegative/K5high/K14high population. It is unclear, however, whether both subsets contribute to the maintenance of the basal lineage under physiological conditions. While multi-potency of Procrpositive basal cells was confirmed by in situ lineage tracing, developmental potential of Procrnegative basal cells was not assessed in situ, because they presently lack a unique marker suitable for such analysis.
To address the issue of lineage identity and developmental plasticity, Granit and colleagues have recently proposed an alternative to a traditional MEC hierarchy model [37]. They suggest a multidimensional classification of MECs along several distinct axes, including stem cell versus differentiated identity, basal versus luminal identity and mesenchymal versus epithelial identity. Such multidimensional models allow description of intermediate and mixed differentiation states and can be applied to both normal mammary gland and breast cancer.
Breast cancer cell-of-origin
In situ lineage tracing combined with gain- and loss-of-function analyses will be instrumental for elucidating the cell-of-origin of different breast cancer subtypes. So far, the Cre-lox-based murine models of breast cancer have not been used for classical in situ fate mapping, wherein specific MEC subsets are transformed, labeled and traced in an intact mammary gland of the host animal.
A distinctive genetic labeling strategy based on ubiquitous (i.e. non cell type-specific) labeling has recently been utilized for intravital analysis of clonal dynamics in MMTV-PyMT model of breast cancer [38]. Four-week-old MMTV-PyMT transgenic mice develop mammary hyperplasia, which gradually progresses to invasive cancer [39]. Zomer et al. [38] have generated compound MMTV-PyMT/R26-CreERT2/R26R-Confetti mice and treated them with Tamoxifen at different time points, to induce random expression of one of the four confetti colors in each mouse cell. They then surgically implanted mammary optical window to visualize fluorescently labeled mammary tumor cells and to track the fate of their progeny in live animals. These analyses have revealed high degree of plasticity in putative cancer stem cells [38].
Going forward, a critical factor in tracing the cellular origin of breast cancer will be the use of specific drivers that allow targeting of non-overlapping MEC subpopulations at different stages of development. Simultaneous transformation and labeling of specific MEC subsets will enable in situ visualization of individual cells in initial stages of tumorigenesis, clonal analysis and fate mapping in fully formed tumors, ex vivo characterization of labeled cells and evaluation of therapeutic efficacy of anticancer drugs.
Concluding remarks
Lineage tracing has recently provided some exciting and unexpected new insights into mammary gland biology and has emerged as an indispensable tool for dissecting MEC hierarchy. Many questions still remain, in particular, those regarding the location and the nature of the stem cell niche. A major challenge facing the field will be to reconcile data obtained using different tracers and developmental time points. The extent of the overlap between various MEC subpopulations identified using prospective isolation and fate mapping, remains to be determined.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Deome KB, Faulkin LJ, Jr, Bern HA, Blair PB. Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland-free mammary fat pads of female C3H mice. Cancer Res. 1959;19(5):515–520. [PubMed] [Google Scholar]
- 2.Smith GH. Experimental mammary epithelial morphogenesis in an in vivo model: evidence for distinct cellular progenitors of the ductal and lobular phenotype. Breast Cancer Res Treat. 1996;39(1):21–31. doi: 10.1007/BF01806075. [DOI] [PubMed] [Google Scholar]
- 3.Kordon EC, Smith GH. An entire functional mammary gland may comprise the progeny from a single cell. Development. 1998;125(10):1921–1930. doi: 10.1242/dev.125.10.1921. [DOI] [PubMed] [Google Scholar]
- 4.Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, Wu L, Lindeman GJ, Visvader JE. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439(7072):84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 5.Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, Eaves CJ. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439(7079):993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 6.Sleeman KE, Kendrick H, Robertson D, Isacke CM, Ashworth A, Smalley MJ. Dissociation of estrogen receptor expression and in vivo stem cell activity in the mammary gland. J Cell Biol. 2007;176(1):19–26. doi: 10.1083/jcb.200604065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shehata M, Teschendorff A, Sharp G, Novcic N, Russell IA, Avril S, Prater M, Eirew P, Caldas C, Watson CJ, et al. Phenotypic and functional characterisation of the luminal cell hierarchy of the mammary gland. Breast Cancer Res BCR. 2012;14(5):R134. doi: 10.1186/bcr3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sleeman KE, Kendrick H, Ashworth A, Isacke CM, Smalley MJ. CD24 staining of mouse mammary gland cells defines luminal epithelial, myoepithelial/basal and non-epithelial cells. Breast cancer Res BCR. 2006;8(1):R7. doi: 10.1186/bcr1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spike BT, Engle DD, Lin JC, Cheung SK, La J, Wahl GM. A mammary stem cell population identified and characterized in late embryogenesis reveals similarities to human breast cancer. Cell Stem Cell. 2012;10(2):183–197. doi: 10.1016/j.stem.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smalley MJ, Kendrick H, Sheridan JM, Regan JL, Prater MD, Lindeman GJ, Watson CJ, Visvader JE, Stingl J. Isolation of mouse mammary epithelial subpopulations: a comparison of leading methods. J Mammary Gland Biol Neoplasia. 2012;17(2):91–97. doi: 10.1007/s10911-012-9257-1. [DOI] [PubMed] [Google Scholar]
- 11.Bai L, Rohrschneider LR. s-SHIP promoter expression marks activated stem cells in developing mouse mammary tissue. Genes Dev. 2010;24(17):1882–1892. doi: 10.1101/gad.1932810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plaks V, Brenot A, Lawson DA, Linnemann JR, Van Kappel EC, Wong KC, de Sauvage F, Klein OD, Werb Z. Lgr5-expressing cells are sufficient and necessary for postnatal mammary gland organogenesis. Cell Rep. 2013;3(1):70–78. doi: 10.1016/j.celrep.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makarem M, Kannan N, Nguyen LV, Knapp DJ, Balani S, Prater MD, Stingl J, Raouf A, Nemirovsky O, Eirew P, et al. Developmental changes in the in vitro activated regenerative activity of primitive mammary epithelial cells. PLoS Biol. 2013;11(8):e1001630. doi: 10.1371/journal.pbio.1001630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Regan JL, Kendrick H, Magnay FA, Vafaizadeh V, Groner B, Smalley MJ. c-Kit is required for growth and survival of the cells of origin of Brca1-mutation-associated breast cancer. Oncogene. 2012;31(7):869–883. doi: 10.1038/onc.2011.289. [DOI] [PubMed] [Google Scholar]
- 15.Zeng YA, Nusse R. Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture. Cell Stem Cell. 2010;6(6):568–577. doi: 10.1016/j.stem.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Machado HL, Kittrell FS, Edwards D, White AN, Atkinson RL, Rosen JM, Medina D, Lewis MT. Separation by cell size enriches for mammary stem cell repopulation activity. Stem Cells Transl Med. 2013;2(3):199–203. doi: 10.5966/sctm.2012-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaanta AS, Virtanen C, Selfors LM, Brugge JS, Neel BG. Evidence for a multipotent mammary progenitor with pregnancy-specific activity. Breast Cancer Res BCR. 2013;15(4):R65. doi: 10.1186/bcr3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen LV, Makarem M, Carles A, Moksa M, Kannan N, Pandoh P, Eirew P, Osako T, Kardel M, Cheung AM, et al. Clonal analysis via barcoding reveals diverse growth and differentiation of transplanted mouse and human mammary stem cells. Cell Stem Cell. 2014;14(2):253–263. doi: 10.1016/j.stem.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 19.Oakes SR, Gallego-Ortega D, Ormandy CJ. The mammary cellular hierarchy and breast cancer. Cell Mol Life Sci CMLS. 2014;71:4301–4324. doi: 10.1007/s00018-014-1674-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Visvader JE, Stingl J. Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev. 2014;28(11):1143–1158. doi: 10.1101/gad.242511.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prater MD, Petit V, Alasdair Russell I, Giraddi RR, Shehata M, Menon S, Schulte R, Kalajzic I, Rath N, Olson MF et al (2014) Mammary stem cells have myoepithelial cell properties. Nat Cell Biol 16(10):942–950, 941–947 [DOI] [PMC free article] [PubMed]
- 22.Van Keymeulen A, Rocha AS, Ousset M, Beck B, Bouvencourt G, Rock J, Sharma N, Dekoninck S, Blanpain C. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479(7372):189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- 23.van Amerongen R, Bowman AN, Nusse R. Developmental stage and time dictate the fate of Wnt/beta-catenin-responsive stem cells in the mammary gland. Cell Stem Cell. 2012;11(3):387–400. doi: 10.1016/j.stem.2012.05.023. [DOI] [PubMed] [Google Scholar]
- 24.de Visser KE, Ciampricotti M, Michalak EM, Tan DW, Speksnijder EN, Hau CS, Clevers H, Barker N, Jonkers J. Developmental stage-specific contribution of LGR5(+) cells to basal and luminal epithelial lineages in the postnatal mammary gland. J Pathol. 2012;228(3):300–309. doi: 10.1002/path.4096. [DOI] [PubMed] [Google Scholar]
- 25.Chang TH, Kunasegaran K, Tarulli GA, De Silva D, Voorhoeve PM, Pietersen AM. New insights into lineage restriction of mammary gland epithelium using parity-identified mammary epithelial cells. Breast Cancer Res BCR. 2014;16(1):R1. doi: 10.1186/bcr3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kretzschmar K, Watt FM. Lineage tracing. Cell. 2012;148(1–2):33–45. doi: 10.1016/j.cell.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Wagner KU, Boulanger CA, Henry MD, Sgagias M, Hennighausen L, Smith GH. An adjunct mammary epithelial cell population in parous females: its role in functional adaptation and tissue renewal. Development. 2002;129(6):1377–1386. doi: 10.1242/dev.129.6.1377. [DOI] [PubMed] [Google Scholar]
- 28.Sale S, Lafkas D, Artavanis-Tsakonas S. Notch2 genetic fate mapping reveals two previously unrecognized mammary epithelial lineages. Nat Cell Biol. 2013;15(5):451–460. doi: 10.1038/ncb2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tao L, van Bragt MP, Laudadio E, Li Z. Lineage tracing of mammary epithelial cells using cell-type-specific cre-expressing adenoviruses. Stem Cell Reports. 2014;2(6):770–779. doi: 10.1016/j.stemcr.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lafkas D, Rodilla V, Huyghe M, Mourao L, Kiaris H, Fre S. Notch3 marks clonogenic mammary luminal progenitor cells in vivo. J Cell Biol. 2013;203(1):47–56. doi: 10.1083/jcb.201307046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang D, Cai C, Dong X, Yu QC, Zhang XO, Yang L, Zeng YA. Identification of multipotent mammary stem cells by protein C receptor expression. Nature. 2014;517:81–84. doi: 10.1038/nature13851. [DOI] [PubMed] [Google Scholar]
- 32.Shehata M, van Amerongen R, Zeeman AL, Giraddi RR, Stingl J. The influence of tamoxifen on normal mouse mammary gland homeostasis. Breast Cancer Res BCR. 2014;16(4):411. doi: 10.1186/s13058-014-0411-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rios AC, Fu NY, Lindeman GJ, Visvader JE. In situ identification of bipotent stem cells in the mammary gland. Nature. 2014;506(7488):322–327. doi: 10.1038/nature12948. [DOI] [PubMed] [Google Scholar]
- 34.Saito M, Iwawaki T, Taya C, Yonekawa H, Noda M, Inui Y, Mekada E, Kimata Y, Tsuru A, Kohno K. Diphtheria toxin receptor-mediated conditional and targeted cell ablation in transgenic mice. Nat Biotechnol. 2001;19(8):746–750. doi: 10.1038/90795. [DOI] [PubMed] [Google Scholar]
- 35.Waddington CH. The strategy of the genes; a discussion of some aspects of theoretical biology. London: Allen & Unwin; 1957. [Google Scholar]
- 36.Chepko G, Smith GH. Three division-competent, structurally-distinct cell populations contribute to murine mammary epithelial renewal. Tissue Cell. 1997;29(2):239–253. doi: 10.1016/S0040-8166(97)80024-9. [DOI] [PubMed] [Google Scholar]
- 37.Granit RZ, Slyper M, Ben-Porath I. Axes of differentiation in breast cancer: untangling stemness, lineage identity, and the epithelial to mesenchymal transition. Wiley Interdiscip Rev Syst Biol Med. 2014;6(1):93–106. doi: 10.1002/wsbm.1252. [DOI] [PubMed] [Google Scholar]
- 38.Zomer A, Ellenbroek SI, Ritsma L, Beerling E, Vrisekoop N, Van Rheenen J. Intravital imaging of cancer stem cell plasticity in mammary tumors. Stem Cells. 2013;31(3):602–606. doi: 10.1002/stem.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, Pollard JW. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol. 2003;163(5):2113–2126. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]