Abstract
Exposure of plants to adverse environmental conditions leads to extensive transcriptional changes. Genome-wide approaches and gene function studies have revealed the importance of chromatin-level control in the regulation of stress-responsive gene expression. Advances in understanding chromatin modifications implicated in plant stress response and identifying proteins involved in chromatin-mediated transcriptional responses to stress are briefly presented in this review. We then highlight how chromatin-mediated gene expression changes can be coupled to the metabolic status of the cell, since many of the chromatin-modifying proteins involved in transcriptional regulation depend on cofactors and metabolites that are shared with enzymes in basic metabolism. Lastly, we discuss the stability and heritability of stress-induced chromatin changes and the potential of chromatin-based strategies for increasing stress tolerance of crops.
Keywords: Abiotic stress, Energy, Acclimation, Memory, Transgenerational, Epigenetic, Chromatin remodeling, Histone modifications, Methylation, PARP, Sirtuin, Arabidopsis, Crop improvement
Introduction
Plants are constantly exposed to environmental stresses, which adversely affect their growth and development and may ultimately result in death of the plant. Abiotic stresses such as drought, temperature extremes, salinity, soil mineral deficiency and toxicity, account for the majority of crop losses and hence constitute a major limitation to agricultural production. Besides, climate change is expected to increase the frequency and intensity of extreme environmental events, further affecting yields [1, 2]. In this context, the survival of plants exposed to acute stress depends on their ability to perceive environmental changes and trigger defence responses that include the rapid activation of stress defence genes. An appropriate and efficient response to lasting adverse environmental conditions is likely to require the precise reprogramming of gene expression to rebalance growth, development and survival. During the last 15 years, the accumulation of genome-scale gene expression data and their comprehensive analyses has allowed the identification of stress-responsive genes and transcription factors that could be responsible for orchestrating the specific transcriptional response to different environmental stresses [3–5]. However, the mechanisms that change a quiescent gene into an actively transcribed gene in response to stress are still poorly understood.
In eukaryotes, the expression of nuclear genes is influenced by chromatin the structure of the chromatin, the basic repeating unit of which is the nucleosome. Each nucleosome consists of 147 bp of DNA wrapped around a histone protein octamer containing two molecules each of histones H2A, H2B, H3, and H4. In addition, histone H1 associates with linker DNA, packing adjacent nucleosomes together to reach a higher degree of condensation. Condensed chromatin hinders access of the basal transcription machinery (transcription factors, RNA polymerases) to DNA, and results in the silencing of underlying sequences. Stress signals can alter chromatin structure, in a process known as chromatin remodeling, so that the transcription machinery can access previously condensed DNA. This alteration can be accomplished with the help of chromatin remodelers. Changes in DNA methylation, in histone modifications and exchange of histone variants also influence the stress-mediated transcriptional response. Furthermore, evidence is emerging that points to stable and heritable (aka epigenetic) gene expression changes in response to stress.
Here, we first provide an updated overview of the recent advances made in understanding chromatin-mediated changes in gene expression involved in plant stress responses and in transient and stable stress memory. In this context, we then highlight the interconnections between chromatin-modifying enzymes and plant metabolism, and discuss chromatin modification-based strategies to enhance stress tolerance in crops. Chromatin modification and remodeling mechanisms that control developmental processes (e.g., flowering time) or other whole-plant non-stress-related processes in plants have been reviewed elsewhere [6, 7].
Chromatin modification and remodeling in plant stress responses
The main players of plant stress responses at the chromatin level include DNA methylation and demethylation enzymes, histone modification enzymes, histone variants, chromatin remodeling complexes and other chromatin-associated factors. Their function and molecular mode of action are reviewed below in relation to plant stress response.
DNA methylation and demethylation
DNA methylation is a covalent modification that has a profound impact on transcription and genome stability. In plants, it refers to the formation of 5-methylcytosine from cytosine through the action of a DNA methyltransferase. In Arabidopsis, DNA is methylated in three sequence contexts: CG, CHG and CHH, where H is any nucleotide, except guanine. The methyltransferase MET1 is required for CG methylation maintenance and is mainly involved in the silencing of transposons, repetitive elements and some imprinted genes. CMT2, CMT3 and DRM2 methyltransferases collaborate to control non-CG methylation [8–11]. In addition, asymmetric CHH methylation, and also CG and CHG methylations, can be established de novo by RNA-directed DNA methylation (RdDM), which involves the participation of small interfering RNAs (siRNAs) [12]. siRNAs produced by Dicer-like (DCL) nucleases bind to Argonaute (AGO) proteins and recruit DRM2 as well as histone-modifying enzymes to DNA targets (reviewed in [13]).
Cytosine DNA methylation is considered to be a stable mark; nevertheless, active demethylation has been observed in both plants and animals [14]. In plants, DNA demethylation is based on base excision repair and involves the DNA glycosylases of the DEMETER (DME) family such as REPRESSOR OF SILENCING (ROS1) in Arabidopsis [15–17]. By creating a chromatin environment that allows DNA glycosylases to function, the histone acetyltransferase IDM1 also affects DNA demethylation [18].
DNA methylation has traditionally been associated with gene silencing, as exemplified by the high level of methylation of silent heterochromatin DNA. Since DNA methylation is linked to histone modifications (see next section), one way it may affect transcription is by changing chromatin structure. Unlike in heterochromatin, the relation between DNA methylation and transcription is less clear in euchromatin. Heavy methylation in promoter regions often correlates with gene silencing similar to the situation in heterochromatin. In contrast, DNA methylation within gene bodies is compatible with transcription in Arabidopsis [19], although it may impede transcript elongation [20].
A number of studies implicate DNA methylation in the modulation of gene expression in response to biotic and abiotic stresses [21–23]. Furthermore, mutations in the Arabidopsis DNA methyltransferases MET1, CMT3, DRM1 and DMR2 lead to genome-wide hypomethylation, upregulation of defense-related genes, and increased resistance to pathogen attack, in addition to pleiotropic growth and developmental defects such as dwarfism [21]. Remarkably, in this latter study, the flanking regions of defense-related genes were found to be enriched in transposable elements (TEs). Moreover, the methylation state of the TE correlated with the expression of both the TE and the neighboring protein-coding gene, suggesting that dynamic alteration in methylation at TEs may drive stress-induced changes in expression of the TE and, in some cases, the proximal gene [21]. Since TEs insertions are mostly stochastic, it is likely that considerable natural variation in transcriptional gene silencing of defense genes exists [24]. Similarly, genome-wide hypomethylation induced by the DNA methylation inhibitor 5-azacytidine resulted in induced expression of oxidative stress-related genes, and increased tolerance to salinity in rice [25]. Both the demethylation level and the activity of stress-responsive genes were higher in the salt-tolerant than in the salt-sensitive cultivars analyzed in this study. Very similar results were obtained from the study of DNA methylation polymorphisms in response to drought stress in rice. Under drought conditions, hypermethylation was predominant in drought-susceptible genotypes while drought-tolerant genotypes presented hypomethylation [26]. In Arabidopsis, natural variation in CMT2 is associated with genome-wide methylation changes and temperature adaptation and cmt2 mutants have increased tolerance to heat stress [27]. Although this suggests a role for CHH methylation in adaptation to temperature stress, the mechanism(s) linking CMT2, CHH methylation and heat tolerance remain to be identified. The association of stress-induced DNA hypomethylation with transcriptional upregulation of stress-responsive genes suggests that at least some stress genes are kept in a repressed state by DNA methylation under optimal growth conditions. This mechanism would minimize their burden on growth and development under optimal conditions, and allow their activation under stress conditions. However, there is currently no conclusive evidence for widespread dynamic and reversible DNA methylation changes for the regulation of stress-responsive genes in response to abiotic stress.
Histone modifications
The N-terminal tails and to some extent the cores and C-termini of histones are subject to a multitude of covalent post-translational modifications that are catalyzed by histone-modifying enzymes. Post-translational modifications of histones include methylation (lysine and arginine residues) [28], acetylation (lysine) [29], phosphorylation (serine and threonine), monoubiquitination [30], ADP-ribosylation, as well as other poorly studied or yet unknown modifications [31]. Recently, 67 previously undescribed histone modifications were identified, increasing the number of known histone marks by about 70 % and further complicating the understanding of the histone code [32]. To date, the most studied histone modifications are methylation, acetylation and phosphorylation. Some of these histone modifications are thought to be quite stable (e.g., lysine methylation), whereas others may be highly dynamic (e.g., threonine and serine phosphorylation) [33]. A wide range of enzymes specifically catalyzes these chemical modifications, some of them working together. Histone methyltransferases (HMT) catalyze histone methylation and histone acetyltransferases (HAT) are responsible for histone acetylation, while histone demethylases (HDM) and histone deacetylases (HDAC) catalyze the demethylation and deacetylation of histones, respectively. Plant genomes encode many proteins for each of these enzyme families (e.g., 16 HDAC and 12 HAT genes in Arabidopsis [34]); some are highly conserved in all eukaryotes while others appear to be specific to plants [34, 35].
The nature, position and combination of histone modifications affect the accessibility of DNA for transcription. Some of these modifications are associated with transcriptional repression and others with the activation of gene expression. In general, histone hyperacetylation is associated with actively expressed genes and histone hypoacetylation with silent DNA regions. Depending on their targets, histone methylation, phosphorylation and ubiquitination can occur on either active or inactive genes [36]. Lysine and arginine residues of histones can be mono-, di-, or trimethylated, further extending the complexity of histone modification-dependent gene regulation. In plants, the best investigated examples of histone marks associated with actively transcribed genes are H3K4me3, H3K9me3, H3K36me3, H3K4ac and H2Bub1 [37]. In contrast, H3K27me3 is associated with repressed/lowly expressed genes, and H3K9me2 and H3K27me1 are associated with repressed sequences, methylated DNA and transposons.
Similarly to DNA methylation, histone modifications are affected by environmental cues. Firstly, dynamic changes in both histone methylation and acetylation marks and the enzymes that catalyze them have been implicated in the plant response to a range of abiotic stresses, including heat and cold stresses [38, 39], drought and salt stress [40–42]. For instance, AtGCN5, a member of the GNAT/MYST subfamily of HAT enzymes, which associates with the transcriptional co-activator proteins ADA2a and ADA2b, positively regulates the expression of stress-induced genes by maintaining the proper histone acetylation level in response to cold and salt stress. Both the gcn5-1 and ada2b-1 mutant lines are hypersensitive to stress [43–45]. The activities of several HDAC enzymes, including AtHD2C, AtHOS15, and AtHDA6, have also been implicated in controlling the plant stress response at the chromatin level [39, 46, 47]. Mutants of these HDAC genes often display phenotypes similar to those of the HAT mutants, i.e., a hypersensitivity to one or several abiotic stress (salt, drought, cold, and/or treatment with the stress hormone abscisic acid). This is particularly intriguing because HAT and HDAC enzymes have opposing actions on the acetylation of stress-responsive gene loci.
Histone-modifying enzymes are known to interact with other proteins that allow the establishment of a structural link with the histones and the DNA. One of these proteins, named Histone Deacetylation Complex1 (HDC1), was recently functionally characterized in Arabidopsis [48]. HDC1 interacts with at least two deacetylases (HDA6 and HDA19), promotes histone deacetylation, and attenuates the de-repression of genes under moderate drought. Interestingly, and in contrast to the developmental defects often observed when manipulating the expression of HDACs [49], overexpression of HDC1 enhanced plant growth, both under normal and moderate salt and drought stress conditions [48].
Histone variants
Another chromatin feature that can control gene expression is the presence of histone variants in the nucleosome. Amino acid sequence differences between histone variants and the canonical histones can affect the stability of the nucleosome, internucleosomal contacts and interactions with chromatin-binding proteins or histone modifiers [50]. Like in most organisms, the Arabidopsis genome contains multiple copies of genes encoding histone variants (for review see [51]). The large number of histone variants raises questions about the number of different nucleosome structures that exist and whether structural alterations may explain differences in function. The functional diversity of plant histone variants and their potential role in gene expression regulation during development or in response to stress are not currently well understood.
The first insights into a role for histone variants in stress-mediated transcriptional regulation arose from the discovery that plants unable to incorporate the histone variant H2A.Z into nucleosomes phenocopy plants grown at elevated temperature and display a constitutive high-temperature transcriptome [52]. Hence, H2A.Z is proposed to be evicted from nucleosomes when the temperature increases, and to thus play a key role in temperature sensing and integration at the chromatin level. H2A.Z may also serve as “thermosensor” in budding yeast, suggesting that this mechanism is evolutionarily conserved [52]. In line with these findings, H2A.Z deposition is enriched in the bodies of genes associated with response to environmental and developmental stimuli, and correlates negatively with DNA methylation [53]. It has recently been proposed that a major function of DNA methylation is to exclude H2A.Z from constitutively expressed genes [54]. Apart from three genes that encode H2A.Z subtype variants (HTA8/9/11), the Arabidopsis genome contains four genes that encode canonical H2A proteins (HTA1/2/10/13), four genes that encode less well-categorized variants (HTA4/6/7/12), and two genes that encode H2A.X subtype variants (HTA3/5). In mammals, the H2A.X variants have been implicated in DNA repair after DNA damage following replication–transcription collisions, oxidative stress and various environmental stress conditions [55, 56]. However, in plants the H2A variants associated with stress response and acclimation processes remain to be determined.
H3 histone variants can also play a role in regulating transcription. Genome-wide profiling in Arabidopsis has revealed H3.1 enrichment in heterochromatin and H3.3 enrichment in transcriptionally active regions, similarly to what has been reported in animals. This suggests that H3.1 acts as the canonical histone that is incorporated during DNA replication, whereas H3.3 could be incorporated during chromatin remodeling processes [57, 58]. Interestingly, Arabidopsis H3.3 is enriched in the promoters of many strongly regulated genes suggesting that it may function in transcriptional regulation [59]. It is tempting to speculate that the large number and the diversity of genes encoding H3.3 variants in plant genomes reflect special functions in the adaptation of land plants to variable environments [60]. Direct evidence for a role of H3.3 variants in adaptive responses is still missing. However, the recent characterisation of Arabidopsis lines mutated in the HIRA complex, which is involved in H3.3 deposition into chromatin in animals, has shown that the plant HIRA complex is required for the appropriate transcription of genes responsive to biotic and abiotic factors [61].
A striking feature of histone gene expression in response to abiotic stress is the family of minor, “drought-inducible” H1 variants present in all angiosperms [62]. The Arabidopsis HIS1-3 gene, for instance, is induced by drought, high salinity stress and abscisic acid [63]. Similarly, the tomato linker histone variant HIS1-S and the rice linker histone HON702 are also induced by water deficit and drought stress [64, 65]. The tools and techniques are available to test whether H1 distribution changes upon stress and whether H1 variants function in stress tolerance or adaptation.
Chromatin remodeling enzymes and other chromatin-associated factors
Chromatin remodeling can also occur through the disruption of the nucleosome structure and loosening of histone–DNA contacts. These changes in higher-order structure of chromatin contribute to gene expression regulation by facilitating the binding of transcription factors. The process is catalyzed by ATP-dependent chromatin remodelers, multiprotein complexes (encoded by over 40 genes in Arabidopsis) that can be divided in at least five families defined by their ATPase subunits: the SWI/SNF, the ISWI, the NURD/Mi-2/CHD, the INO80, and the SWR1 group [66]. Several of these families also mediate the exchange of histone variants (reviewed in [67]) or are involved in the transcriptional response to stress. For instance, Arabidopsis plants lacking an active PIE1 complex (homolog of SWR1 in yeast), which incorporates H2A.Z into nucleosomes, exhibit misregulation of genes involved in the plant innate immune and temperature responses [52, 68]. Exposure of plants to adverse environmental conditions generally leads to a temporary growth arrest that allows adaptation [69]. Recently, it was shown that the BRAHMA ATPase AtCHR2 of the SWI/SNF chromatin remodeling complex, which acts as a positive regulator of gibberellin-mediated responses, plays an important role in the growth arrest response during drought and heat stress in Arabidopsis [70–72]. Another example of a SWI/SNF class chromatin remodeler involved in the transcriptional response to stress is SPLAYED (SYD). SYD regulates gene expression in pathogen stress signaling pathways, and loss of its activity results in increased susceptibility to the fungal pathogen Botrytis cinerea, but not to the bacterial pathogen Pseudomonas syringae. These results demonstrate that reduced stress tolerance in a chromatin-remodeling mutant can be stress-specific, and is not the due to decreased overall fitness and global, non-specific, deregulation of gene expression [73].
Deregulation of gene expression in response to heat stress is also associated with changes in nucleosome occupancy and the decondensation of heterochromatin. While decondensation persisted after stress release, the loss of DNA-bound nucleosomes and activation of certain TEs were only transient [74]. The nucleosome occupancy-mediated resilencing of TEs was delayed in mutants with impaired CHROMATIN ASSEMBLY FACTOR 1 (CAF-1) activity, suggesting a role for this complex in reloading nucleosomes upon recovery from stress [74].
Another group of chromatin-associated proteins that affect gene expression by changing chromatin structure are the high mobility group (HMG) proteins, which can be grouped into the HMGA and HMGB subfamilies in plants [75]. HMGB proteins are involved in several DNA-related processes including the activation and repression of transcription (reviewed in [76, 77]. Lack of HMGB1 in Arabidopsis, as well as HMGB2 overexpression, affect plant growth and stress tolerance. There is a marked impact on the transcriptome, with an enrichment of stress-responsive genes among the downregulated genes in the hmgb1 mutant [78, 79]. Chromatin structure is also affected by topoisomerases and evidence is accumulating that these enzymes may play an important role in chromatin remodeling during signal-dependent activation or repression of gene transcription. Ju et al. [80] provided the first in vivo molecular evidence for such a role by showing that the human Topo IIβ can generate a transient dsDNA strand break that is required for a nucleosome-specific histone H1–HMGB protein exchange, which in turn leads to local changes of chromatin architecture and the transcriptional activation of target genes. More recently, the plant topoisomerase VI was shown to act as an integrator of multiple signals generated by reactive oxygen species (ROS) and to regulate the expression of oxidative stress-responsive genes [81]. Furthermore, the constitutive expression of the rice topoisomerase VI in Arabidopsis was shown to increase the expression of stress-responsive genes and to confer abiotic stress tolerance [82]. Whether this role involves a molecular mode of action similar to that of Topo IIβ and/or other chromatin remodeling changes remains to be elucidated.
Metabolite control of stress-induced chromatin changes
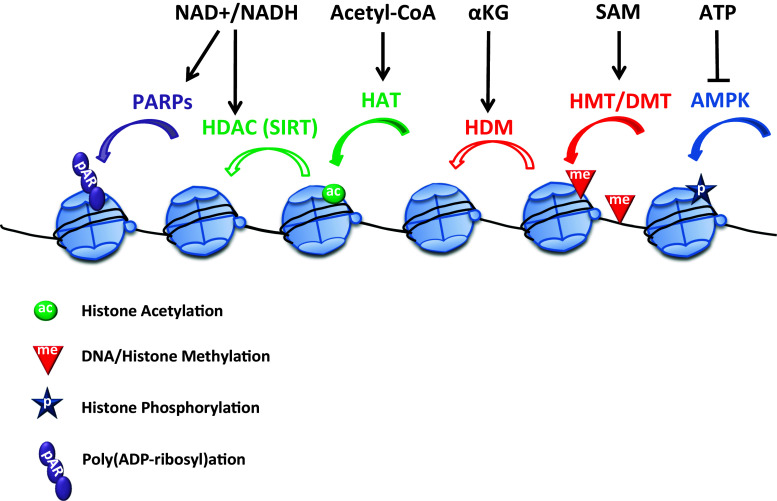
Simultaneously with the fine-tuning of gene expression programs, environmental stress and nutrient availability cues lead to changes in the energy metabolism status of the cell. Interestingly, many of the chromatin factors involved in transcriptional regulation employ essential metabolites such as adenosine triphosphate (ATP), nicotinamide adenine dinucleotide (NAD), the methyl donor S-adenosylmethionine (SAM), and the acetyl donor acetyl coenzyme A (acetyl-CoA) for their function. These small molecules may thus provide a way to couple chromatin-mediated gene expression changes with the metabolic status of the cell, and their availability is likely to be critical in the transcriptional response to stress. This idea is supported by work in yeast and animal systems (reviewed in [83, 84]) (Fig. 1). For instance, increased glucose metabolism results in excess acetyl-CoA production, increased levels of histone acetylation, and increased global gene expression in mammalian cells [85]. This interconnection could involve the sirtuin enzymes, whose study has provided the first evidence for the metabolic regulation of chromatin. Sirtuins are class III HDACs that are named after their homology to the Saccharomyces cerevisiae gene silent information regulator 2 (Sir2), and require NAD+ as a cofactor to deacetylate proteins including histones and transcriptional regulators. Through this activity, yeast and animal sirtuins are known to regulate important biological processes linked to nutrient availability and cell energy status (NAD+ level and NAD+/NADH ratio), including DNA damage/repair, cell apoptosis, longevity, and oxidative stress tolerance (reviewed in [86]). Evidence suggests that these histone-modifying enzymes (encoded by two genes in Arabidopsis) may also play important roles as sensors of the energy status and stress signals as well as transcriptional regulators of stress response in plants [87, 88].
Fig. 1.
Crosstalk between metabolism and chromatin. Deposition and removal of chromatin marks is influenced by metabolites, which function as cofactors or substrates for the chromatin modification enzymes. First, NAD+ concentration and NAD+/NADH ratio regulate the activity of the sirtuin NAD+-dependent histone deacetylases (SIRT) and poly(ADP-ribose) polymerases (PARPs). These two enzymes are involved in DNA damage repair and replication as well as in the regulation of chromatin compaction and gene transcription in eukaryotes. Low ATP/AMP ratios can activate AMPK, a kinase that phosphorylates histones. Acetyl-CoA is a donor for HAT-mediated histone acetylation, and α-ketoglutarate (αKG) is a cofactor for histone demethylation by Jumonji C-domain-containing histone lysine demethylases (HDM). Finally, S-adenosylmethionine (SAM), which is synthesized from the amino acid methionine, is the substrate for histone and DNA methyltransferases (HMT and DMT, respectively). Under stress conditions energy consumption increases while energy production decreases. A decrease in the energy metabolite ATP and a higher NAD+/NADH ratio are expected to lead to increased AMPK and SIRT activities, respectively. Depletion of the citrate cycle intermediates acetyl-CoA, and αKG is likely to affect the acetylation and demethylation of histones
The actions of sirtuins in these processes are likely to be interconnected with those of poly(ADP-ribose) polymerases (PARPs) ([89], Fig. 1). PARPs work in coordination with poly(ADP-ribose) glycohydrolases (PARGs) to regulate transcriptional activity by the addition and removal of ADP-ribose polymers, respectively [90, 91]. Targets of PARP enzymatic activities include histones, the poly(ADP-ribosyl)ation of which induces local relaxation of the chromatin structure. PARP activity is induced by DNA damage such as that caused by ROS formation upon stress exposure. Active PARPs synthesize polymers of ADP-ribose using NAD+ as substrate, causing ATP consumption and NAD+ depletion, and hence leading to a reduction of sirtuin activity [92]. A growing body of evidence suggests that stress tolerance in eukaryotes can be improved by maintaining energy homeostasis under stress conditions, notably via an increase in the NAD+/NADH ratio and sirtuin activity, or conversely a reduction of PARP activity. In fact, it has been reported that plants with reduced PARP activity are more tolerant to multiple stresses [93].
Changes in the pool of another key energy metabolite, ATP, is also likely to affect chromatin modifications via the adenosine monophosphate-activated protein kinase (AMPK), a highly conserved protein kinase in eukaryotes acting as a central regulator of energy homeostasis (Fig. 1). Recently, a critical role has been identified for AMPK in the phosphorylation of histone H2B during stress adaptation in mammals [94]. Another example of a link between chromatin modification and metabolism is the role of SAM on histone and DNA methylation (Fig. 1). SAM is generated by SAM synthetases (also known as methionine adenosyltransferase, MAT). One of these enzymes, the mouse MATII-α isoform, has been found in a chromatin-bound state and was shown to interact with several histone remodelers, including components of the SWI/SNF chromatin remodelers, the NuRD (nucleosome remodeling and histone deacetylase complex) nucleosome remodelers, PARP-1 [poly(ADP-ribose) polymerase], and the PcG (Polycomb group) proteins [95]. In rice, knockdown of genes encoding SAM synthetases results in altered DNA and histone methylation and late flowering [96], thereby providing evidence for a key role of SAM as methyl donor for histone and DNA methyltransferases in plants. Methyltransferases convert SAM to S-adenosylhomocysteine (SAH), which is a potent inhibitor of both histone and DNA methyltransferase activities. The hydrolysis of SAH to homocysteine is therefore essential for preventing the inhibition of trans-methylation reactions and for the recycling of methionine and SAM. In Arabidopsis, the SAH hydrolase 1 SAHH1, originally called HOG1 for HOMOLOGY-DEPENDENT GENE SILENCING 1, is involved in maintaining transcriptional gene silencing at numerous targets [97–100]. A second isoform, SAHH2, seems to have no role in silencing or DNA methylation [98]. Reduced SAH hydrolase activity in weak hog1 alleles relieves transcriptional gene silencing and results in genome-wide demethylation, poor growth, low fertility and reduced seed germination [98]. Although no changes in histone methylation levels were initially detected in a weak hog1 allele [99], the characterization of a stronger allele, hog1-7, has confirmed the effects of SAM depletion on histone methylation [101]. Since homocysteine can also serve as a precursor for the synthesis of glutathione (GSH), an essential redox buffer of the cell that is depleted by increased ROS levels, exposure to oxidative stress can be expected to lead to decreased SAM and hence reduced methyltransferase activity.
In yeast, the myo-inositol derivatives inositol polyphosphates modulate the activity of several ATP-dependent chromatin remodeling complexes [102]. The recent functional characterisation of a myo-inositol phosphate synthase (MIPS1) with a dual function as a metabolic enzyme and a transcriptional regulator demonstrate that myo-inositol is also involved in chromatin remodeling in plants [103]. The authors showed that Arabidopsis MIPS1 stimulates its own expression by binding to its own promoter and locally inhibiting the activity of ATXR5 and ATXR6, two histone methyltransferases responsible for the spreading of heterochromatin marks. This inhibition is released upon pathogen attack, which leads to MIPS1 repression and suggests a function for MIPS1 in pathogen defence. Finally, the citrate cycle intermediate α-ketoglutarate (αKG) is a co-substrate of Jumonji C-domain-containing histone lysine demethylases in eukaryotes, including plants [104].
The integration of metabolic profiling and modeling with studies on plant chromatin function is now needed to unravel the full extent of the functional links between metabolism and chromatin regulation.
Transient and stable stress effects
Acclimation to stress and epigenetic stress memory have emerged as important areas of research in recent years. Three main types of stress-induced gene expression changes may exist: (1) transient changes, which are reversed upon stress release; (2) stable changes that are maintained from several days up to the whole life of the plant after the stress has ceased, and which may be mitotically heritable and contribute to acclimation processes; and (3) stable changes that are inherited by subsequent generations and which might contribute to plant adaptation and evolution. In the context of transgenerationally inherited changes, two possible effects are discussed in the literature. First, an increased stress tolerance in the progeny of stressed plants may result from a directed effect on specific stress-defense genes. Second, an increased genetic or epigenetic variability in the progeny of stressed plants may be the consequence of undirected effects, for instance stochastic changes in DNA methylation induced by stress [105].
Whereas many examples of transient chromatin changes and a few examples of long-lasting chromatin changes exist (reviewed in [6, 105]), unambiguous evidence for directed transgenerational epigenetic inheritance is lacking in plants [105]. In Arabidopsis, this apparent absence of transgenerational transmission of environmentally induced epigenetic states has been notably attributed to the two chromatin regulators DDM1 (Decrease in DNA methylation1) and MOM1 (Morpheus’ Molecule 1). These regulators rapidly erase stress memories and reset the chromatin status to its pre-stress state, therefore preventing mitotic propagation and the transgenerational inheritance of stress-induced epigenetic states [106]. Furthermore, evidence that epigenetic changes induced by abiotic stress have an adaptive advantage for stress tolerance is lacking. Studies of natural occurring epialleles (i.e., alleles that differ in DNA methylation or chromatin state, but not in DNA sequence), such as those carried out by Silveira et al. [107], may represent a promising strategy to test this hypothesis, especially if this variation can be linked to the history of the growth conditions of the natural variant. However, because epialleles can frequently revert, it is unclear at this time whether they play a significant role in evolution [108]. Epiallele stability appears to be strongly sequence context-dependent and much more research is required to understand the kinetics of epiallele emergence and reversion, as well as the potential effects of epialleles on gene expression.
Until recently, reported cases of durable stress ‘memory’ in plants mostly involved transposon activation, homologous recombination, DNA methylation or small non-coding RNAs (reviewed in [6]; [109–113]). More knowledge about the dynamics of chromatin marks is needed to better understand the potential of DNA methylation and histone modifications for mediating lasting stress-induced effects on gene expression. Histone modifications are generally thought to be much less stable than DNA methylation, but positive feedback loops such as recently proposed for plant PcG proteins [114] may establish a dynamic stability that is sufficient for maintaining stress effects long into the recovery period [115].
Stable stress-induced changes do not always involve a lasting maintenance of modified gene activity. Often, a primary stress episode sensitizes plants to subsequent stresses resulting in faster or stronger changes in gene expression upon repeated exposure (Fig. 2). This sensitized gene expression can contribute to the physiological priming syndrome, i.e., improved tolerance to repeated stresses, a phenomenon which is best characterized in plant–pathogen interactions. Stress-responsive genes are kept in a repressed state under optimal conditions, but can be sensitized by even moderate or short stress incidents (Fig. 2). These changes may involve the deposition of activating chromatin marks, the removal of repressive marks or alterations in nucleosome occupancy. Sensitized genes are often inactive but may also show some basal transcription. When a mild stress is repeated, the sensitized genes become active. However, exposure to a more acute or long-lasting stress can directly establish an activated state. Repeated severe stresses may also establish a hyperactivated state, leading to an even more efficient stress response. Upon stress release, genes will be reverted either to the sensitized state, or to the repressed state. Although priming by substances such as β-aminobutyric acid or beneficial microorganisms has large economic potential, the underlying mechanisms are only just starting to emerge. Studies by Jaskiewicz et al. [116] and Ding et al. [117] provide evidence that the sensitization of gene expression via histone modifications or stalled RNA polymerase II contributes to priming to biotic stress. Interestingly, transcriptional memory responses that occur during repeated exposures to stress appear to be different from those that take place in response to a single exposure [118].
Fig. 2.
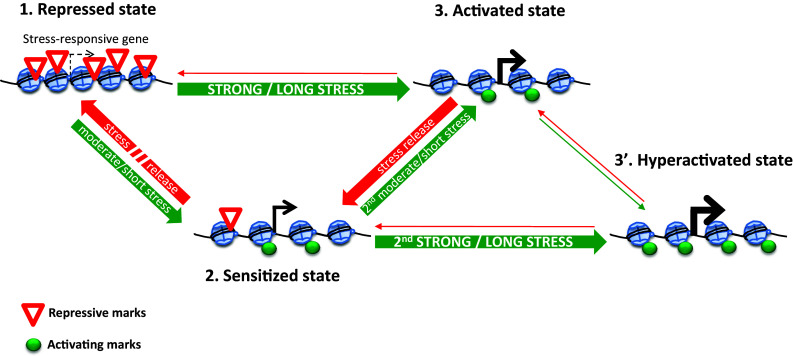
Model for chromatin-level regulation of the response and acclimation of plants to abiotic stress. Under optimal growth conditions, a stress-responsive gene is kept in a repressed (silenced) state (1). Upon moderate or short stress exposure, the deposition of chromatin activating marks and/or the removal of repressive marks establish a sensitized state (2). Genes in this state may be weakly transcribed or continue to be inactive. Longer stress duration or higher intensity induces an activated state (3). This can be achieved directly or via a transient sensitized state. Upon stress release, the gene rapidly reverts to the sensitized state (2) and then slowly to the repressed state (1). As long as the repressed state has not been reestablished, a second moderate or brief stress is sufficient to reactivate the gene. Note that the acclimated/sensitized state does not necessarily depend on prior full activation. For some genes, a second intense or long stress will not only establish the activated but also a hyperactivated state (3′) reflected by a further increase in the rate of transcription
Chromatin modification-based strategies for enhancing stress tolerance
A general aim in crop improvement is to increase productivity under stress conditions, while preserving growth performance under non-stress conditions. Several of the studies presented in this review show that stress results in dynamic changes in DNA methylation and/or histone modifications that result in the transcriptional activation of stress-responsive genes which leads to increased stress tolerance. Therefore, modulating the activities of DNA and histone-modifying enzymes represents an interesting approach towards more stress-tolerant crops. However, constitutive changes in these enzymes often results in growth and developmental defects, which will impinge on any potential agronomical implication. Increased stress tolerance established by chromatin modifiers might hence be unavoidably associated with reduced plant vigor. An interesting exception is HDC1, because plants overexpressing HDC1 outperform wild-type plants under both standard and drought stress conditions [48]. This suggests that specific chromatin modifiers can indeed be engineered to enhance stress tolerance in crops.
Gene expression changes in response to environmental cues also involve histone variants. The histone variant H2A.Z is an important thermosensor that coordinates the sensitivity of temperate plants to increased temperature during grain development. Perturbing H2A.Z occupancy, through higher temperature or genetic modification, strongly reduces yield in grasses [119]. These findings may be useful for obtaining new varieties that are more resilient to thermal stress. More work is needed to unleash the full potential of histone variants or chromatin modifiers to create stress-tolerant crops.
Strategies to enhance stress tolerance in crops based on transgenerational memory are particularly exciting. Although not successfully applied in the context of stress physiology, energy use efficiency has been increased by selection in genetically identical canola (Brassica napus) lines, suggesting that existing or novel epigenetic variability can be stably inherited and selected for increased yield in crops [120]. Remarkably, the epigenetic variants obtained were stable for over eight generations with respect to their phenotypes, DNA methylation patterns, and histone modifications (methylation and acetylation).
Conclusion and perspectives
Storage of DNA in nuclei of eukaryotic cells requires that it is tightly compacted in the form of chromatin. However, the structure of chromatin also serves a second very important role, by conferring the capacity to precisely control gene expression. In particular, chromatin-mediated regulation of gene expression plays a central role in various stress responses.
Optimal plant performance requires the rapid and tailored expression of particular genes in response to specific stresses. Different stresses often share some effects on cellular processes and, therefore, lead to the activation of common genes in addition to stress-specific genes. Understanding the mechanisms that control the level and the specificity of chromatin-mediated regulation of gene expression is now a key focus for future research. Evidence suggests that it is the balance of several modifications that controls the strength of gene expression. For instance, genes with H3K9ac alone are usually highly expressed, whereas genes also modified with H3K27me3 or DNA methylation are less highly expressed [121]. In addition, fine modulation of gene expression may involve interactions between histone-modifying enzymes and transcription factors (e.g., [122]). Identification of the protein complexes that associate with active or repressed genes in normal and stress conditions will help to uncover the basis for specificity.
Studies of plant responses to stress at the chromatin level have been very useful for revealing the level of complexity. Now it is essential to rigorously establish causality and to shift from a static to a dynamic analysis in order to reach reliable models of transcriptional stress responses that have predictive power. However, to date relatively few chromatin-modifying enzymes and remodelers have been functionally characterized and linked to stress responses. Although much of this progress was possible because of reverse genetic approaches, we are convinced that the power of mapping-by-sequencing together with well-designed modifier or suppressor screens will make forward genetics a powerful tool for the identification of new chromatin factors involved in stress responses. In addition, genome-wide chromatin immunoprecipitation, proteomic, and transcriptome analyses under both steady state and stress conditions are required to establish epigenome profiles, identify protein complexes and reveal their genomic targets. Understanding the metabolic control of chromatin changes will also constitute another important area of research.
Finally, it will be of particular interest to determine the stability of stress-induced chromatin changes and to establish whether they are able to confer long-lasting stress memories to allow enhanced tolerance to future stress exposure in the same or even subsequent generations. In striking contrast with the large number of reviews on the topic, few undisputable examples of transgenerational stress memory exist in the literature, and their potential adaptive significance remains unknown to date. However, unlike the limited data available for transgenerational effects, a growing body of evidence now clearly implicates chromatin regulation in the memory of stress, acclimation and priming within a single generation. Here lies great potential for crop improvement.
Acknowledgments
Work in the authors’ laboratories is supported by the French National Research Agency (Grant ANR 2010 JCJC 1205 01 to CL), the Swedish research councils VR and FORMAS and the Knut-and-Alice-Wallenberg foundation (to LH). We thank Dr. Ben Field and the anonymous reviewers for critical reading and helpful comments on the manuscript.
References
- 1.Tubiello FN, Soussana J-F, Howden SM. Crop and pasture response to climate change. Proc Natl Acad Sci USA. 2007;104:19686–19690. doi: 10.1073/pnas.0701728104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Challinor AJ, Watson J, Lobell DB, et al. A meta-analysis of crop yield under climate change and adaptation. Nat Clim Chang. 2014;4:287–291. [Google Scholar]
- 3.Gadjev I, Vanderauwera S, Gechev TS, et al. Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 2006;141:436–445. doi: 10.1104/pp.106.078717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naika M, Shameer K, Mathew OK, et al. STIFDB2: an updated version of plant stress-responsive transcription factor database with additional stress signals, stress-responsive transcription factor binding sites and stress-responsive genes in Arabidopsis and rice. Plant Cell Physiol. 2013;54:e8. doi: 10.1093/pcp/pcs185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borkotoky S, Saravanan V, Jaiswal A, et al. The arabidopsis stress responsive gene database. Int J Plant Genomics. 2013 doi: 10.1155/2013/949564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hauser MT, Aufsatz W, Jonak C, Luschnig C. Transgenerational epigenetic inheritance in plants. Biochim Biophys Acta Gene Regul Mech. 2011;1809:459–468. doi: 10.1016/j.bbagrm.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gentry M, Hennig L. Remodelling chromatin to shape development of plants. Exp Cell Res. 2013;321:1–7. doi: 10.1016/j.yexcr.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 8.Stroud H, Do T, Du J, et al. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol. 2014;21:64–72. doi: 10.1038/nsmb.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zemach A, Kim MY, Hsieh PH, et al. The arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153:193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindroth AM, Cao X, Jackson JP, et al. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292:2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- 11.Du J, Zhong X, Bernatavichute YV, et al. Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell. 2012;151:167–180. doi: 10.1016/j.cell.2012.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saze H, Tsugane K, Kanno T, Nishimura T. DNA methylation in plants: relationship to small RNAs and histone modifications, and functions in transposon inactivation. Plant Cell Physiol. 2012;53:766–784. doi: 10.1093/pcp/pcs008. [DOI] [PubMed] [Google Scholar]
- 14.Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agius F, Kapoor A, Zhu J-K. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc Natl Acad Sci USA. 2006;103:11796–11801. doi: 10.1073/pnas.0603563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gehring M, Huh JH, Hsieh TF, et al. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Penterman J, Zilberman D, Huh JH, et al. DNA demethylation in the Arabidopsis genome. Proc Natl Acad Sci USA. 2007;104:6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian W, Miki D, Zhang H, et al. A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science. 2012;336:1445–1448. doi: 10.1126/science.1219416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, Yazaki J, Sundaresan A, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Zilberman D, Gehring M, Tran RK, et al. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 21.Dowen RH, Pelizzola M, Schmitz RJ, et al. Widespread dynamic DNA methylation in response to biotic stress. Proc Natl Acad Sci. 2012;109:E2183–E2191. doi: 10.1073/pnas.1209329109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song Y, Ji D, Li S, et al. The dynamic changes of DNA methylation and histone modifications of salt responsive transcription factor genes in soybean. PLoS One. 2012;7:e41274. doi: 10.1371/journal.pone.0041274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colaneri AC, Jones AM. Genome-wide quantitative identification of DNA differentially methylated sites in Arabidopsis seedlings growing at different water potential. PLoS One. 2013;8:e59878. doi: 10.1371/journal.pone.0059878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X. Dynamic differential methylation facilitates pathogen stress response in Arabidopsis. Proc Natl Acad Sci USA. 2012;109:12842–12843. doi: 10.1073/pnas.1210292109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong L, Xu YH, Wang JB. The effect of 5-azacytidine on wheat seedlings responses to NaCl stress. Biol Plant. 2010;54:753–756. [Google Scholar]
- 26.Gayacharan, Joel aJ. Epigenetic responses to drought stress in rice (Oryza sativa L.) Physiol Mol Biol Plants. 2013;19:379–387. doi: 10.1007/s12298-013-0176-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen X, De Jonge J, Forsberg SKG et al (2014) Natural CMT2 variation is associated with genome-wide methylation changes and temperature seasonality. PLoS Genet 10:e1004842. doi:10.1371/journal.pgen.1004842 [DOI] [PMC free article] [PubMed]
- 28.Liu C, Lu F, Cui X, Cao X. Histone methylation in higher plants. Annu Rev Plant Biol. 2010;61:395–420. doi: 10.1146/annurev.arplant.043008.091939. [DOI] [PubMed] [Google Scholar]
- 29.Waterborg JH. Plant histone acetylation: in the beginning. Biochim Biophys Acta. 2011;1809:353–359. doi: 10.1016/j.bbagrm.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Feng J, Shen W-H. Dynamic regulation and function of histone monoubiquitination in plants. Front Plant Sci. 2014;5:83. doi: 10.3389/fpls.2014.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 32.Tan M, Luo H, Lee S, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee BM, Mahadevan LC. Stability of histone modifications across mammalian genomes: implications for “epigenetic” marking. J Cell Biochem. 2009;108:22–34. doi: 10.1002/jcb.22250. [DOI] [PubMed] [Google Scholar]
- 34.Pandey R, Müller A, Napoli CA, et al. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002;30:5036–5055. doi: 10.1093/nar/gkf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo M, Liu X, Singh P, et al. Chromatin modifications and remodeling in plant abiotic stress responses. Biochim Biophys Acta Gene Regul Mech. 2012;1819:129–136. doi: 10.1016/j.bbagrm.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 36.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roudier F, Ahmed I, Bérard C, et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 2011;30:1928–1938. doi: 10.1038/emboj.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwon CS, Lee D, Choi G, Chung W-I. Histone occupancy-dependent and -independent removal of H3K27 trimethylation at cold-responsive genes in Arabidopsis. Plant J. 2009;60:112–121. doi: 10.1111/j.1365-313X.2009.03938.x. [DOI] [PubMed] [Google Scholar]
- 39.Zhu J, Jeong JC, Zhu Y, et al. Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance. Proc Natl Acad Sci USA. 2008;105:4945–4950. doi: 10.1073/pnas.0801029105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sokol A, Kwiatkowska A, Jerzmanowski A, Prymakowska-Bosak M. Up-regulation of stress-inducible genes in tobacco and Arabidopsis cells in response to abiotic stresses and ABA treatment correlates with dynamic changes in histone H3 and H4 modifications. Planta. 2007;227:245–254. doi: 10.1007/s00425-007-0612-1. [DOI] [PubMed] [Google Scholar]
- 41.Kim J-M, To TK, Ishida J, et al. Transition of chromatin status during the process of recovery from drought stress in Arabidopsis thaliana. Plant Cell Physiol. 2012;53:847–856. doi: 10.1093/pcp/pcs053. [DOI] [PubMed] [Google Scholar]
- 42.Zong W, Zhong X, You J, Xiong L. Genome-wide profiling of histone H3K4-tri-methylation and gene expression in rice under drought stress. Plant Mol Biol. 2013;81:175–188. doi: 10.1007/s11103-012-9990-2. [DOI] [PubMed] [Google Scholar]
- 43.Hark AT, Vlachonasios KE, Pavangadkar KA, et al. Two Arabidopsis orthologs of the transcriptional coactivator ADA2 have distinct biological functions. Biochim Biophys Acta Gene Regul Mech. 2009;1789:117–124. doi: 10.1016/j.bbagrm.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 44.Kaldis A, Tsementzi D, Tanriverdi O, Vlachonasios KE. Arabidopsis thaliana transcriptional co-activators ADA2b and SGF29a are implicated in salt stress responses. Planta. 2011;233:749–762. doi: 10.1007/s00425-010-1337-0. [DOI] [PubMed] [Google Scholar]
- 45.Pavangadkar K, Thomashow MF, Triezenberg SJ. Histone dynamics and roles of histone acetyltransferases during cold-induced gene regulation in Arabidopsis. Plant Mol Biol. 2010;74:183–200. doi: 10.1007/s11103-010-9665-9. [DOI] [PubMed] [Google Scholar]
- 46.Sridha S, Wu K. Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J. 2006;46:124–133. doi: 10.1111/j.1365-313X.2006.02678.x. [DOI] [PubMed] [Google Scholar]
- 47.Chen L-T, Luo M, Wang Y-Y, Wu K. Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J Exp Bot. 2010;61:3345–3353. doi: 10.1093/jxb/erq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perrella G, Lopez-Vernaza MA, Carr C, et al. Histone deacetylase complex1 expression level titrates plant growth and abscisic acid sensitivity in Arabidopsis. Plant Cell. 2013;25:3491–3505. doi: 10.1105/tpc.113.114835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanaka M, Kikuchi A, Kamada H. The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol. 2008;146:149–161. doi: 10.1104/pp.107.111674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weber CM, Henikoff S. Histone variants: dynamic punctuation in transcription. Genes Dev. 2014;28:672–682. doi: 10.1101/gad.238873.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu Y, Dong A, Shen W-H. Histone variants and chromatin assembly in plant abiotic stress responses. Biochim Biophys Acta. 2012;1819:343–348. doi: 10.1016/j.bbagrm.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 52.Kumar SV, PA Wigge. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 2010;140:136–147. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 53.Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–129. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coleman-Derr D, Zilberman D. Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genet. 2012;8:e1002988. doi: 10.1371/journal.pgen.1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dickey JS, Redon CE, Nakamura AJ, et al. H2AX: functional roles and potential applications. Chromosoma. 2009;118:683–692. doi: 10.1007/s00412-009-0234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seo J, Kim K, Chang D-Y, et al. Genome-wide reorganization of histone H2AX toward particular fragile sites on cell activation. Nucleic Acids Res. 2014;42:1016–1025. doi: 10.1093/nar/gkt951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stroud H, Otero S, Desvoyes B, et al. Genome-wide analysis of histone H3.1 and H3.3 variants in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2012;109:5370–5375. doi: 10.1073/pnas.1203145109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wollmann H, Holec S, Alden K, et al. Dynamic deposition of histone variant H3.3 accompanies developmental remodeling of the Arabidopsis transcriptome. PLoS Genet. 2012;8:e1002658. doi: 10.1371/journal.pgen.1002658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shu H, Nakamura M, Siretskiy A, et al. Arabidopsis replacement histone variant H3.3 occupies promoters of regulated genes. Genome Biol. 2014;15:R62. doi: 10.1186/gb-2014-15-4-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ingouff M, Berger F. Histone3 variants in plants. Chromosoma. 2010;119:27–33. doi: 10.1007/s00412-009-0237-1. [DOI] [PubMed] [Google Scholar]
- 61.Nie X, Wang H, Li J, et al. The HIRA complex that deposits the histone H3.3 is conserved in Arabidopsis and facilitates transcriptional dynamics. Biol Open. 2014;3:794–802. doi: 10.1242/bio.20148680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jerzmanowski A, Przewloka M, Grasser KD. Linker histones and HMG1 proteins of higher plants. Plant Biol. 2000;2:586–597. [Google Scholar]
- 63.Ascenzi R, Gantt JS. A drought-stress-inducible histone gene in Arabidopsis thaliana is a member of a distinct class of plant linker histone variants. Plant Mol Biol. 1997;34:629–641. doi: 10.1023/a:1005886011722. [DOI] [PubMed] [Google Scholar]
- 64.Scippa GS, Griffiths A, Chiatante D, Bray EA. The H1 histone variant of tomato, H1-S, is targeted to the nucleus and accumulates in chromatin in response to water-deficit stress. Planta. 2000;211:173–181. doi: 10.1007/s004250000278. [DOI] [PubMed] [Google Scholar]
- 65.Hruz T, Laule O, Szabo G, et al. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 67.Billon P, Côté J. Precise deposition of histone H2A.Z in chromatin for genome expression and maintenance. Biochim Biophys Acta. 2012;1819:290–302. doi: 10.1016/j.bbagrm.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 68.March-Díaz R, García-Domínguez M, Lozano-Juste J, et al. Histone H2A.Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. Plant J. 2008;53:475–487. doi: 10.1111/j.1365-313X.2007.03361.x. [DOI] [PubMed] [Google Scholar]
- 69.Achard P, Cheng H, De Grauwe L, et al. Integration of plant responses to environmentally activated phytohormonal signals. Science. 2006;311:91–94. doi: 10.1126/science.1118642. [DOI] [PubMed] [Google Scholar]
- 70.Mlynárová L, Nap J-P, Bisseling T. The SWI/SNF chromatin-remodeling gene AtCHR12 mediates temporary growth arrest in Arabidopsis thaliana upon perceiving environmental stress. Plant J. 2007;51:874–885. doi: 10.1111/j.1365-313X.2007.03185.x. [DOI] [PubMed] [Google Scholar]
- 71.Archacki R, Buszewicz D, Sarnowski TJ, et al. BRAHMA ATPase of the SWI/SNF chromatin remodeling complex acts as a positive regulator of gibberellin-mediated responses in arabidopsis. PLoS One. 2013;8:e58588. doi: 10.1371/journal.pone.0058588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Han S-K, Sang Y, Rodrigues A, et al. The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in Arabidopsis. Plant Cell. 2012;24:4892–4906. doi: 10.1105/tpc.112.105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Walley JW, Rowe HC, Xiao Y, et al. The chromatin remodeler SPLAYED regulates specific stress signaling pathways. PLoS Pathog. 2008;4:e1000237. doi: 10.1371/journal.ppat.1000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pecinka A, Dinh HQ, Baubec T, et al. Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell. 2010;22:3118–3129. doi: 10.1105/tpc.110.078493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grasser KD. Chromatin-associated HMGA and HMGB proteins: versatile co-regulators of DNA-dependent processes. Plant Mol Biol. 2003;53:281–295. doi: 10.1023/b:plan.0000007002.99408.ba. [DOI] [PubMed] [Google Scholar]
- 76.Ueda T, Yoshida M. HMGB proteins and transcriptional regulation. Biochim Biophys Acta. 2010;1799:114–118. doi: 10.1016/j.bbagrm.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 77.Stros M. HMGB proteins: interactions with DNA and chromatin. Biochim Biophys Acta. 2010;1799:101–113. doi: 10.1016/j.bbagrm.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 78.Kwak KJ, Kim JY, Kim YO, Kang H. Characterization of transgenic Arabidopsis plants overexpressing high mobility group B proteins under high salinity, drought or cold stress. Plant Cell Physiol. 2007;48:221–231. doi: 10.1093/pcp/pcl057. [DOI] [PubMed] [Google Scholar]
- 79.Lildballe DL, Pedersen DS, Kalamajka R, et al. The expression level of the chromatin-associated HMGB1 protein influences growth, stress tolerance, and transcriptome in Arabidopsis. J Mol Biol. 2008;384:9–21. doi: 10.1016/j.jmb.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 80.Ju B-G, Lunyak VV, Perissi V, et al. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 81.Simková K, Moreau F, Pawlak P, et al. Integration of stress-related and reactive oxygen species-mediated signals by Topoisomerase VI in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2012;109:16360–16365. doi: 10.1073/pnas.1202041109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jain M, Tyagi AK, Khurana JP. Overexpression of putative topoisomerase 6 genes from rice confers stress tolerance in transgenic Arabidopsis plants. FEBS J. 2006;273:5245–5260. doi: 10.1111/j.1742-4658.2006.05518.x. [DOI] [PubMed] [Google Scholar]
- 83.Lu C, Thompson CB. Metabolic regulation of epigenetics. Cell Metab. 2012;16:9–17. doi: 10.1016/j.cmet.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu J, Kim J, Oberdoerffer P. Metabolic modulation of chromatin: implications for DNA repair and genomic integrity. Front Genet. 2013;4:182. doi: 10.3389/fgene.2013.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wellen KE, Hatzivassiliou G, Sachdeva UM, et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rajendran R, Garva R, Krstic-Demonacos M, Demonacos C. Sirtuins: molecular traffic lights in the crossroad of oxidative stress, chromatin remodeling, and transcription. J Biomed Biotechnol. 2011;2011:368276. doi: 10.1155/2011/368276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huang L, Sun Q, Qin F, et al. Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol. 2007;144:1508–1519. doi: 10.1104/pp.107.099473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang C, Gao F, Wu J, et al. Arabidopsis putative deacetylase AtSRT2 regulates basal defense by suppressing PAD4, EDS5 and SID2 expression. Plant Cell Physiol. 2010;51:1291–1299. doi: 10.1093/pcp/pcq087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cantó C, Sauve AA, Bai P. Molecular aspects of medicine crosstalk between poly (ADP-ribose) polymerase and sirtuin enzymes. Mol Aspects Med. 2013;34:1168–1201. doi: 10.1016/j.mam.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Briggs AG, Bent AF. Poly(ADP-ribosyl)ation in plants. Trends Plant Sci. 2011;16:372–380. doi: 10.1016/j.tplants.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 91.Lamb RS, Citarelli M, Teotia S. Functions of the poly(ADP-ribose) polymerase superfamily in plants. Cell Mol Life Sci. 2012;69:175–189. doi: 10.1007/s00018-011-0793-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Luo X, Kraus WL. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012;26:417–432. doi: 10.1101/gad.183509.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.De Block M, Verduyn C, De Brouwer D, Cornelissen M. Poly(ADP-ribose) polymerase in plants affects energy homeostasis, cell death and stress tolerance. Plant J. 2005;41:95–106. doi: 10.1111/j.1365-313X.2004.02277.x. [DOI] [PubMed] [Google Scholar]
- 94.Bungard D, Fuerth BJ, Zeng P-Y, et al. Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science. 2010;329:1201–1205. doi: 10.1126/science.1191241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Katoh Y, Ikura T, Hoshikawa Y, et al. Methionine adenosyltransferase II serves as a transcriptional corepressor of Maf oncoprotein. Mol Cell. 2011;41:554–566. doi: 10.1016/j.molcel.2011.02.018. [DOI] [PubMed] [Google Scholar]
- 96.Li W, Han Y, Tao F, Chong K. Knockdown of SAMS genes encoding S-adenosyl-l-methionine synthetases causes methylation alterations of DNAs and histones and leads to late flowering in rice. J Plant Physiol. 2011;168:1837–1843. doi: 10.1016/j.jplph.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 97.Furner I, Sheikh M, Collett C. Gene silencing and homology-dependent gene silencing in Arabidopsis: genetic modifiers and DNA methylation. Genetics. 1998;149:651–662. doi: 10.1093/genetics/149.2.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rocha PSCF, Sheikh M, Melchiorre R, et al. The Arabidopsis HOMOLOGY-DEPENDENT GENE SILENCING1 gene codes for an S-adenosyl-L-homocysteine hydrolase required for DNA methylation-dependent gene silencing. Plant Cell. 2005;17:404–417. doi: 10.1105/tpc.104.028332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mull L, Ebbs ML, Bender J. A histone methylation-dependent DNA methylation pathway is uniquely impaired by deficiency in Arabidopsis S-adenosylhomocysteine hydrolase. Genetics. 2006;174:1161–1171. doi: 10.1534/genetics.106.063974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jordan ND, West JP, Bottley A, et al. Transcript profiling of the hypomethylated hog1 mutant of Arabidopsis. Plant Mol Biol. 2007;65:571–586. doi: 10.1007/s11103-007-9221-4. [DOI] [PubMed] [Google Scholar]
- 101.Baubec T, Dinh HQ, Pecinka A, et al. Cooperation of multiple chromatin modifications can generate unanticipated stability of epigenetic States in Arabidopsis. Plant Cell. 2010;22:34–47. doi: 10.1105/tpc.109.072819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shen X, Xiao H, Ranallo R, et al. Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science. 2003;299:112–114. doi: 10.1126/science.1078068. [DOI] [PubMed] [Google Scholar]
- 103.Latrasse D, Jégu T, Meng P-H, et al. Dual function of MIPS1 as a metabolic enzyme and transcriptional regulator. Nucleic Acids Res. 2013;41:2907–2917. doi: 10.1093/nar/gks1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang W, Jiang D, Jiang J, He Y. A plant-specific histone H3 lysine 4 demethylase represses the floral transition in Arabidopsis. Plant J. 2010;62:663–673. doi: 10.1111/j.1365-313X.2010.04182.x. [DOI] [PubMed] [Google Scholar]
- 105.Pecinka A, Mittelsten Scheid O. Stress-induced chromatin changes: a critical view on their heritability. Plant Cell Physiol. 2012;53:801–808. doi: 10.1093/pcp/pcs044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Iwasaki M, Paszkowski J. Identification of genes preventing transgenerational transmission of stress-induced epigenetic states. Proc Natl Acad Sci USA. 2014;111:8547–8552. doi: 10.1073/pnas.1402275111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Silveira AB, Trontin C, Cortijo S, et al. Extensive natural epigenetic variation at a de novo originated gene. PLoS Genet. 2013;9:e1003437. doi: 10.1371/journal.pgen.1003437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Becker C, Hagmann J, Müller J, et al. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature. 2011;480:245–249. doi: 10.1038/nature10555. [DOI] [PubMed] [Google Scholar]
- 109.Boyko A, Kovalchuk I. Transgenerational response to stress in Arabidopsis thaliana. Plant Signal Behav. 2010;5:995–998. doi: 10.4161/psb.5.8.12227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Verhoeven KJF, Jansen JJ, van Dijk PJ, Biere A. Stress-induced DNA methylation changes and their heritability in asexual dandelions. New Phytol. 2010;185:1108–1118. doi: 10.1111/j.1469-8137.2009.03121.x. [DOI] [PubMed] [Google Scholar]
- 111.Bilichak A, Ilnystkyy Y, Hollunder J, Kovalchuk I. The progeny of Arabidopsis thaliana plants exposed to salt exhibit changes in DNA methylation, histone modifications and gene expression. PLoS One. 2012;7:e30515. doi: 10.1371/journal.pone.0030515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhong S-H, Liu J-Z, Jin H, et al. Warm temperatures induce transgenerational epigenetic release of RNA silencing by inhibiting siRNA biogenesis in Arabidopsis. Proc Natl Acad Sci USA. 2013;110:9171–9176. doi: 10.1073/pnas.1219655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rasmann S, De Vos M, Casteel CL, et al. Herbivory in the previous generation primes plants for enhanced insect resistance. Plant Physiol. 2012;158:854–863. doi: 10.1104/pp.111.187831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Derkacheva M, Steinbach Y, Wildhaber T, et al. Arabidopsis MSI1 connects LHP1 to PRC2 complexes. EMBO J. 2013;32:2073–2085. doi: 10.1038/emboj.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kleinmanns JA, Schubert D. Polycomb and Trithorax group protein-mediated control of stress responses in plants. Biol Chem. 2014;395:1291–1300. doi: 10.1515/hsz-2014-0197. [DOI] [PubMed] [Google Scholar]
- 116.Jaskiewicz M, Conrath U, Peterhänsel C. Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep. 2011;12:50–55. doi: 10.1038/embor.2010.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ding Y, Fromm M, Avramova Z. Multiple exposures to drought “train” transcriptional responses in Arabidopsis. Nat Commun. 2012;3:740. doi: 10.1038/ncomms1732. [DOI] [PubMed] [Google Scholar]
- 118.Ding Y, Liu N, Virlouvet L, et al. Four distinct types of dehydration stress memory genes in Arabidopsis thaliana. BMC Plant Biol. 2013;13:229. doi: 10.1186/1471-2229-13-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Boden SA, Kavanová M, Finnegan EJ, Wigge PA. Thermal stress effects on grain yield in Brachypodium distachyon occur via H2A.Z-nucleosomes. Genome Biol. 2013;14:R65. doi: 10.1186/gb-2013-14-6-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hauben M, Haesendonckx B, Standaert E, et al. Energy use efficiency is characterized by an epigenetic component that can be directed through artificial selection to increase yield. Proc Natl Acad Sci USA. 2009;106:20109–20114. doi: 10.1073/pnas.0908755106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhou J, Wang X, He K, et al. Genome-wide profiling of histone H3 lysine 9 acetylation and dimethylation in Arabidopsis reveals correlation between multiple histone marks and gene expression. Plant Mol Biol. 2010;72:585–595. doi: 10.1007/s11103-009-9594-7. [DOI] [PubMed] [Google Scholar]
- 122.Kim K-C, Lai Z, Fan B, Chen Z. Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell. 2008;20:2357–2371. doi: 10.1105/tpc.107.055566. [DOI] [PMC free article] [PubMed] [Google Scholar]