Abstract
Affinity proteins based on small scaffolds are currently emerging as alternatives to antibodies for therapy. Similarly to antibodies, they can be engineered to have high affinity for specific proteins. A potential problem with small proteins and peptides is their short in vivo circulation time, which might limit the therapeutic efficacy. To circumvent this issue, we have engineered bispecificity into an albumin-binding domain (ABD) derived from streptococcal Protein G. The inherent albumin binding was preserved while the opposite side of the molecule was randomized for selection of high-affinity binders. Here we present novel ABD variants with the ability to bind to the epidermal growth factor receptor 3 (ErbB3). Isolated candidates were shown to have an extraordinary thermal stability and affinity for ErbB3 in the nanomolar range. Importantly, they were also shown to retain their affinity to albumin, hence demonstrating that the intended strategy to engineer bispecific single-domain proteins against a tumor-associated receptor was successful. Moreover, competition assays revealed that the new binders could block the natural ligand Neuregulin-1 from binding to ErbB3, indicating a potential anti-proliferative effect. These new binders thus represent promising candidates for further development into ErbB3-signaling inhibitors, where the albumin interaction could result in prolonged in vivo half-life.
Keywords: Albumin-binding domain, ABD, Phage display, ErbB3, HSA, Bispecific
Introduction
During the past 15 years, antibodies have been established as the primary affinity proteins used in cancer therapy [1, 2]. More recently, promising alternative scaffolds of both immunoglobulin and non-immunoglobulin origins have gained increased interest [3, 4]. Several of these alternative molecular formats demonstrate an improved stability and solubility, more cost-efficient production, and ease of manipulation, as compared to antibodies [3–5]. Antibodies, as well as other large proteins, have limited vascular permeability and tumor-penetrating capabilities [6]. Smaller binding molecules often have a better tissue penetration, but their small size also results in a rapid renal clearance and, thus, a shorter half-life [6, 7]. Elimination of molecules smaller than the glomerular filtration cutoff can be avoided through association or conjugation to larger serum proteins such as albumin or by chemical modifications that increase the hydrodynamic radius, for example by PEGylation [8, 9]. IgG and albumin have much longer residence times in the circulation than other proteins of similar size. This is a result of active rescue from degradation by binding to the neonatal Fc-receptor (FcRn) [8, 10]. Therefore, association to albumin or Fc can improve the half-life of small proteins and peptides by utilization of this FcRn-mediated recycling [8, 10]. Several studies have demonstrated the utility of gene fusions to obtain multifunctional proteins that in addition to the affinity for a therapeutically relevant target protein also endow it with affinity for either albumin or IgG to improve the pharmacokinetic properties [11–17]. In addition, albumin itself is known to accumulate in tumors [18–20] and has therefore been explored as a delivery vehicle for chemotherapy [21] and as a fusion partner to protein-based drugs [22–24].
To engineer a small single-domain affinity protein with a long half-life, we have utilized an albumin-binding domain (ABD) as a scaffold for selection of bispecific affinity molecules [25, 26]. ABD is one of three albumin-binding motifs present in streptococcal Protein G and shows high affinity to albumin of several species [27]. The small size of only 46 amino acids puts this novel scaffold among the smallest of all structured cysteine-free engineered affinity proteins described to date [3, 4]. In addition to the prolonged half-life obtained in animal models, it has also been shown that ABD does not interfere with the FcRn-albumin-binding kinetics [17]. Recently, an ABD-based affinity protein to interferon gamma has been developed by another group [28]. In that work, the albumin-binding region was modified and replaced by a novel binding surface and the binder was hence monospecific. We have also constructed a combinatorial library for selection of ABD-based binders by phage display. However, our approach has instead been to randomize 11 surface-exposed residues on the surface of the domain that is formed by the first and third helix [25]. Since the surface area responsible for the albumin-binding is mainly localized to the second helix [29–31], the intention is hence to retain albumin affinity and engineer bispecific binders (Fig. 1a). We have previously utilized this approach for the development of bispecific binders against a domain from Protein A and TNF-alpha, respectively [25, 26]. The affinity domains showed retained affinity for albumin, demonstrating that bispecificity could be achieved. These different utilizations of ABD show that the domain has a high tolerance for amino acid substitutions and hence supports the potential of our efforts in this study.
Fig. 1.
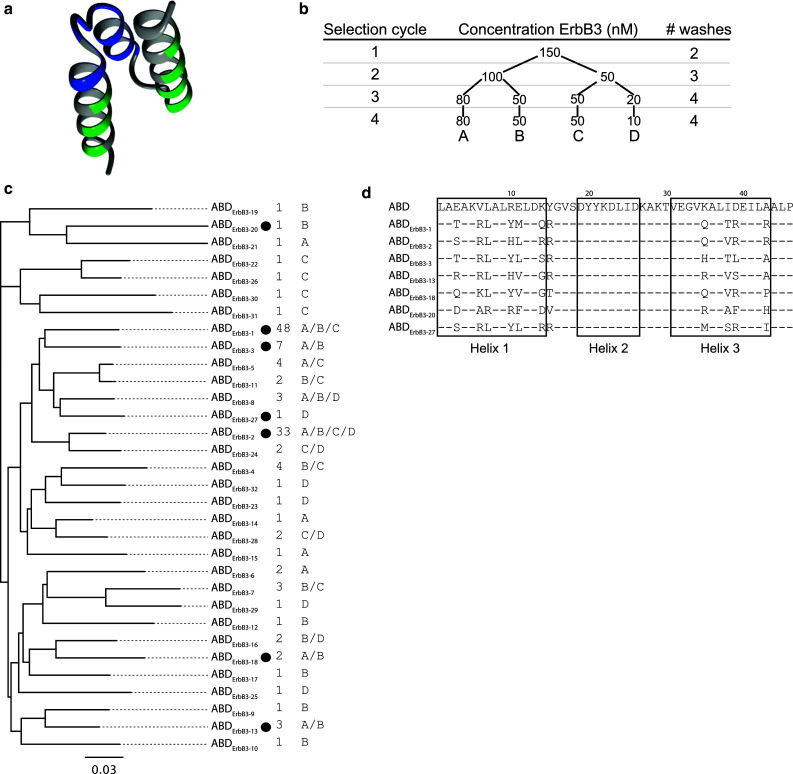
Phage display selection of hErbB3-binding ABD molecules. a Molecular representation of ABD (PDB-file: 1GJS). Residues colored in green represent randomized parts of the protein domain. The blue parts indicate residues binding to HSA (according to Linhult et al. [29]). b Four cycles of phage display selection were performed with increasing selection pressure in each round. This was achieved by decreasing the ErbB3-concentration in parallel tracks and increasing the number of washes prior to elution. c Multi-alignment tree of all ABD sequences recovered from the phage display selection. Variants chosen for expression and further analysis are indicated (filled circle). The number of times a clone was observed during sequencing as well as track origin is displayed to the right. The scale bar indicates the number of substitutions per residue, i.e., a longer distance corresponds to larger differences between the sequences. The sequence similarity tree was constructed using the Geneious software v4.8 (Biomatters, Auckland, New Zealand). d Amino-acid sequences of ABD variants chosen for further analysis. The boxes indicate the three helices in non-randomized ABD and bars represent residues that are common with the scaffold sequence
Here, we have developed bispecific, single-domain binders targeted towards the human epidermal growth factor receptor 3 (ErbB3 or HER3). ErbB3 is a member of the EGFR tyrosine kinase family, which plays important roles in the regulation of many cellular functions such as proliferation, differentiation, and migration, and is often dysregulated in cancer [32–36]. Binding of any of several identified ligands, e.g., Neuregulin-1 (NRG-β1) or Neuregulin-2, to ErbB3 facilitates heterodimerization, which then triggers intracellular signaling by transphosphorylation of the cytoplasmatic domains [34]. ErbB3 lacks intrinsic tyrosine kinase activity [37], but is phosphorylated and activated upon heterodimerization with for example epidermal growth factor receptor 1 (EGFR, ErbB1) or ErbB2. This results in activation of the mitogen-activated protein kinase (MAPK) and phosphoinositide 3-kinase (PI3 K)-Akt pathways and contributes to the suppression of apoptosis and increased proliferation [34]. In this receptor family, EGFR and ErbB2 have been explored extensively as drug targets. However, ErbB3 has recently gained attention because it is the preferred dimerization partner of ErbB2 and since it is also involved in resistance to ErbB2-targeted therapies [34, 38, 39]. Consequently, inhibition of ErbB3 activity is an interesting approach to address some of the limitations of the drugs currently used to target EGFR or ErbB2 [40–43]. Importantly, a therapeutic effect that is mediated by inhibition of ErbB3 signaling would not be dependent on Fc function and a small ligand-blocking affinity protein is hence expected to have at least the same therapeutic potential as a large monoclonal antibody. Using an alternative to a full-length IgG would also avoid potential Fc-mediated adverse effects in healthy tissues with a normal ErbB3 expression.
The results from our study validate the earlier shown strategy [25, 26] to combine small size, intrinsic albumin-binding and, in this case, ability of specific high-affinity binding to a tumor target in a single domain. In summary, the isolated candidates from phage display selections were shown to bind to both recombinant ErbB3 and the native receptor on a cancer cell line. Furthermore, the new binders were shown to compete with the natural ligand NRG-β1 for binding to the receptor, and these novel molecules hence represent promising candidates for further development into potent inhibitors of ErbB3 signaling.
Materials and methods
Library construction
A phage display library has previously been constructed by randomizing 11 surface-exposed residues in helices one and three of ABD. The randomized residues are located at the opposite side of the albumin-binding surface to potentially retain the affinity for HSA and enable selection of single-domain bispecific binders. The randomization was done by using the degenerate NNK codons (see Fig. 1a). The design and assembly of the library has been described in detail elsewhere [25].
Phage display selection
The Escherichia coli RR1∆M15 strain [44] that carries the phagemid library [25] was cultured by inoculating 500 ml of tryptic soy broth (TSB), supplemented with 2 % (w/v) glucose and 100 μg ml−1 ampicillin, with 100 μl bacterial stock with a cell density of 2.2 × 1010 cfu ml−1, which corresponds to approximately a 100-fold excess of bacteria compared to the experimentally determined library size [25]. In cycles 3 and 4, the culture volume was reduced to 100 ml. The number of cells used for inoculation was always at least a 104-fold excess compared to the number of phages eluted from the previous cycle to ensure retained coverage. A 15-fold excess of M13K07 helper-phage (New England Biolabs (NEB), Ipswich, MA, USA) was allowed to infect an 8-ml culture (4 ml in cycle 3 and 4) aliquot during 2 h at 37 °C. Cells were harvested by centrifugation and used to inoculate 500 ml of TSB medium (100 ml for cycles 3 and 4) supplemented with 5 % (w/v) yeast extract (TSB+Y), 100 μg ml−1 ampicillin, 100 μg ml−1 kanamycin and 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG, Apollo Scientific, Derbyshire, UK) followed by overnight incubation at 30 °C to produce library-expressing phages. A phage-stock was prepared by two successive precipitation steps using polyethylene glycol (PEG)/NaCl followed by resuspension in 1 ml of phosphate buffered saline (PBS) pH 7.4 supplemented with 3 % (w/v) BSA and 0.05 % (v/v) Tween 20 (3 % PBSTB). As a negative pre-selection step, the phages were incubated for 30 min with 100 nM of purified human polyclonal Fc derived from IgG (Bethyl Laboratories, Montgomery, TX, USA) and 0.6 mg of Dynabeads® Protein A magnetic beads (Invitrogen, Carlsbad, CA, USA) that had been washed twice with 500 μl of PBS supplemented with 0.1 % Tween 20 (PBST) and blocked for 20 min with 500 μl PBS supplemented with 0.1 % (v/v) Tween 20 and 5 % (w/v) BSA (5 % PBSTB). Fc was not included during pre-selection for the first selection cycle. The entire phage-stock (1011 phages) was used in cycle 1 and a 104-fold excess compared to the number of previously eluted phages was used in subsequent cycles. The unbound phages were recovered in the supernatant, transferred to a new tube, and incubated for 2 h with recombinant human ErbB3–Fc chimera (R&D Systems, Minneapolis, MN, USA) before being transferred to 1.5 mg of washed Dynabeads Protein A (0.6 mg in cycle 3 and 4). Beads from both pre-selection and selection were washed with PBST according to Fig. 1b and the bound phages were eluted from the beads by incubation with 500 μl 50 mM glycine–HCl pH 2.7 for 10 min. The eluate was immediately neutralized upon transfer to a new tube containing an equal volume of PBS with 10 % (v/v) 1 M Tris–HCl pH 8.0. The eluate was used to infect a fresh culture of RRIΔM15 grown to OD600 ≈ 0.5 and thereafter incubated at 37 °C for 30 min. Infected cells were harvested by centrifugation, resuspended in TSB+Y and spread on tryptone yeast extract (TYE) agar plates with 100 μg ml−1 ampicillin and 2 % (w/v) glucose and incubated at 37 °C over night. Colonies were harvested and used to produce phages for the next round of selection. During selection, samples were collected from the last washes and eluates of both negative and positive selection and used to infect RR1ΔM15 to determine phage titers. A total of four cycles of selection were performed with decreasing concentration of target and increasing number of washes according to Fig. 1b. All micro-centrifuge tubes were blocked with 500 μl of 5 % PBSTB end over end for at least 1 h. All steps were performed at room temperature and incubations were made in a Rotamixer.
DNA sequencing and sub-cloning
ABD inserts in colonies originating from the fourth cycle of selection were amplified by polymerase chain reaction (PCR), sequenced by Sanger sequencing and analyzed on an ABI Prism 3700 DNA analyzer (Applied Biosystems, Foster City, CA, USA). Phagemids from colonies of interest were purified from small-scale cultivations using a plasmid purification kit (Qiagen, Venlo, Netherlands) and used as templates for PCR-amplification of the ABD genes. PCR fragments were restricted with EcoRI and XhoI (NEB) and ligated into an expression vector with a T7 promoter and an N-terminal His6-tag that had been treated with the same enzymes, dephosphorylated with alkaline phosphatase (NEB), and purified from a 1 % agarose gel with a DNA gel extraction kit (Qiagen). For production of fusion proteins consisting of ABD variants connected via a G(S3G)2-linker to the IgG-binding Z-domain from staphylococcal Protein A (ABD-Z), DNA constructs were assembled from PCR products using specific primers. The fragments were sub-cloned into the same expression vector as described above using restriction sites EcoRI and AscI (NEB). The linker was introduced by primers including a BamHI (NEB) restriction site. Ligated vectors were transformed to RRIΔM15 and grown on TYE agar supplemented with 50 μg ml−1 kanamycin. Single colonies were picked for PCR-amplification and sequence verification. Plasmids carrying the correct constructs were purified and transformed to Rosetta (DE3) E. coli (Novagen, Madison, WI, USA) for protein expression.
Protein expression and purification
Single colonies of E. coli Rosetta (DE3) with the ABD variants cloned into the expression vector were used to inoculate overnight cultures of TSB supplemented with 50 μg ml−1 kanamycin and 20 μg ml−1 chloramphenicol. One milliliter of each culture was used to inoculate 100 ml of TSB+Y supplemented with the same antibiotics and cultured at 37 °C to OD600 ≈ 0.5. Protein expression was induced by the addition of IPTG to a final concentration of 1 mM and cultures were incubated overnight at 25 °C. Cells were harvested by centrifugation and resuspended in Tris-buffered saline (TST; 25 mM Tris–HCl, 200 mM NaCl, 1 mM EDTA, 0.05 % (v/v) Tween 20, pH 8). Cells were lysed by sonication using a Vibra-Cell (Sonics & Materials Inc., Newtown, CT, USA) and cell debris was removed by an additional centrifugation. The supernatants were filtered prior to loading onto human serum albumin (HSA)-Sepharose resin, with column volumes (CV) of 7.5 ml, pre-equilibrated with TST. Following sample loading, the columns were washed with 10 CV of TST followed by 7 CV of 5 mM NH4Ac pH 5.5 and the proteins were eluted in fractions of 1 ml in 0.5 M HAc pH 2.8. Protein-containing fractions (as determined by absorbance at 280 nm) were evaporated overnight using a Savant AES2010 SpeedVac system (Thermo Scientific, Rockford, IL, USA). Proteins were dissolved in PBST and analyzed by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) and mass spectrometry on a 6520 Accurate Q-TOF LC/MS (Agilent, Santa Clara, CA, USA). Protein concentrations were determined from amino acid sequences and amino acid analysis (Aminosyraanalyscentralen Uppsala University, Uppsala, Sweden). ABD-Z proteins were expressed and purified as described earlier and an additional purification on IgG-Sepharose utilizing the Z-domain was included. The IgG-Sepharose purification was performed using essentially the same protocol as for HSA-Sepharose. The proteins were characterized by SDS-PAGE, mass spectrometry and binding to ErbB3 was confirmed by surface plasmon resonance (SPR) as described below.
Analysis of binding kinetics
Binding kinetics for ErbB3-binding ABD variants to albumin and ErbB3 were evaluated by SPR using a ProteOn XPR36 protein interaction array system (Bio-Rad, Hercules, CA, USA) and compared to non-randomized ABD. Recombinant human ErbB3–Fc chimera (R&D Systems), His6-tagged human ErbB3 (Sino Biological, Beijing, China), His6-tagged murine ErbB3 (mErbB3; Sino Biological), human serum albumin and murine serum albumin (Sigma-Aldrich, St. Louis, MO, USA) were immobilized to 2,000–4,000 RU in separate flow cells of general layer medium (GLM) sensor chips using standard amine coupling according to the manufacturer’s recommendations. One ligand surface was left blank in all experiments and all analytes were injected with flow rates of 50 μl min−1 at 25 °C. Samples were injected for 252 or 300 s and dissociation was monitored for 600 or 1,000 s. The surfaces were regenerated between injections with 10 mM NaOH (ErbB3) or 20 mM HCl (albumin). Each analyte was serially diluted to five different concentrations in running buffer (PBST pH 7.4) and injected with a simultaneous blank injection (PBST). All response levels were double referenced against the blank ligand channel and the buffer injection. Kinetic constants were calculated from experimental data by curve fitting to a 1:1 Langmuir binding isotherm using the ProteOn Manager software version 3.1.0.6 (Bio-Rad). All kinetic measurements were performed at least in duplicates and human polyclonal Fc (Bethyl Laboratories, Montgomery, TX, USA) and Fc-fused human ErbB2 (R&D Systems) were immobilized as negative controls in one of the experiments.
Secondary structure and thermal stability measurements
The secondary structure content and thermal stability of selected ABD molecules were assessed by circular dichroism (CD) using a Jasco J-810 Spectropolarimeter (Jasco, Essex, UK). Proteins were buffer exchanged to PBS, using NAP-5 gel filtration columns (GE Healthcare, Uppsala, Sweden) according to the manufacturer’s recommendations, and diluted to 0.4 mg ml−1. Measurements were carried out in three steps: first, secondary structure content was assessed by measuring the degree of ellipticity from 250 to 195 nm at 25 °C. Second, the ellipticity at 221 nm was measured during heating of the sample from 25 to 90 °C to determine the melting temperature (T m). Lastly, spectra were again recorded from 250 to 195 nm at 25 °C to evaluate refolding of the proteins.
Competition biosensor assay
Binding competition between NRG-β1 and ABD variants for human ErbB3 (hErbB3) was monitored by SPR using a ProteOn XPR36 protein interaction array system. NRG-β1 extracellular domain (ECD, S2-K246) or NRG-β1 epidermal growth factor-like domain (EGF, T176-K246) (R&D Systems) was immobilized by amine coupling to 1,000–2,000 RU on a GLM chip. ErbB3 (Fc-chimera) was pre-incubated with varying concentrations of ABD (12.5–200 nM with 5 nM ErbB3) for 1 h at room temperature in PBST before being injected (50 μl min−1 at 25 °C) over the chip for 200 s. Surfaces were regenerated with 10 mM NaOH. All response levels were double referenced as described above and response levels towards the end of injection (at 160 s) were extracted. Responses from the pre-incubated samples were normalized against reference injections with only ErbB3. As a negative control, the original ABD scaffold was included in the experiment. Positive controls consisted of the NRG-β1 EGF domain that competed with itself and an ErbB3-binding Affibody molecule (Z05417) that is known to bind to the same site on ErbB3 as NRG-β1 [45].
Biosensor analysis of ErbB3 binding in the presence of albumin
The ErbB3-binding of four selected ABD variants and the negative control ABD in the presence of HSA was measured in a biosensor assay using a ProteOn XPR36 protein interaction array system to (1) monitor ErbB3-binding behavior in increasing concentrations of albumin and (2) assess simultaneous binding capacity. Human (Fc-fused, R&D Systems) and murine (His6-tagged, Sino Biological) ErbB3 were immobilized to 1,000–4,000 RU on separate surfaces on a GLM-chip. 200 nM ABD was incubated with varying concentrations of HSA (0–2,000 nM) before being injected over the chip (50 μl min−1 at 25 °C) and the association to ErbB3 was monitored during 200 s. Dissociation was subsequently measured during 600 s. Differences between responses measured during association and dissociation were normalized against corresponding values obtained from reference injections without albumin present. The same experiment was also performed with ABD pre-incubated with MSA prior to injection over immobilized mErbB3.
Flow-cytometric evaluation of ErbB3-binding on human cells
To test for binding to human ErbB3-expressing cells by flow cytometry, two selected ErbB3-binding ABD variants and the control protein ABD were genetically fused with a C-terminal IgG-binding Z-domain (the fusion proteins are denoted ABD-Z) derived from domain B of Protein A [46]. The Z-domain was introduced to facilitate detection of bound ABD variants in the flow cytometer. Human ErbB3-expressing AU565-cells (American Type Culture Collection (ATCC), Manassas, VA, USA) were cultured to 80–90 % confluence in RPMI 1640 medium (Sigma-Aldrich) supplemented with 10 % fetal bovine serum (FBS) (Sigma-Aldrich). As a control, SKOV-3 cells (ErbB3low; ATCC) cultured in McCoy’s 5A modified medium (Sigma-Aldrich), were also treated and analyzed in the same manner. Cells were harvested by incubation in PBS supplemented with 5 mM EDTA and 0.1 mg ml−1 trypsin (Sigma-Aldrich) for 5 min at 37 °C and, following centrifugation, resuspended in fresh medium. Between 2 × 105 and 7 × 105 cells were incubated with 500 nM ABD-Z at room temperature for 1 h. Binding to the cell surface was detected through incubation with biotinylated polyclonal human IgG, followed by incubation with streptavidin R-phycoerythrin conjugate (SAPE, Invitrogen). The cells were washed with 3 % PBSTB between incubations and prior to flow-cytometric analysis. The median fluorescence intensities of the analyzed cells were compared to the auto-fluorescence of non-labeled cells and cells incubated with detection reagents only (background). Non-randomized ABD, fused to Z, was used as a negative control and all analyses were performed at least in triplicates.
To identify if the novel binders are able to compete with NRG-β1 also on cell surface expressed ErbB3, between 2 × 105 and 7 × 105 AU565 cells were, in separate experiments, incubated with 50 nM NRG-β1 EGF (R&D Systems) for 30 min at room temperature followed by incubation with 500 nM ABD-Z for 30 min. Bound ABD variants were detected using biotinylated human IgG and SAPE as described above. The cells were washed with 3 % PBSTB between each incubation step and prior to flow-cytometric analysis. All flow-cytometric data were analyzed using Kaluza flow analysis software version 1.0 (Beckman Coulter, Brea, CA, USA).
Results
Phage display selection, expression, and purification of bispecific ErbB3-binding ABD variants
ErbB3-binding molecules were selected by phage display from a combinatorial library based on an albumin-binding domain where 11 surface-exposed residues not directly involved in albumin-binding had been randomized (Fig. 1a). Four rounds of selection were performed using a recombinant form of the extracellular human receptor fused to an Fc-region, derived from human IgG1, as a target (Fig. 1b). Phages were pre-incubated with a polyclonal human Fc in a negative selection step prior to positive selection against ErbB3. After four rounds of selection, 32 unique ABD variants with an overall high sequence similarity, as indicated by a sequence clustering analysis (Fig. 1c), were observed among the 136 clones that were sequenced. Two clones were observed more frequently, ABDErbB3-1 was found 48 times and ABDErbB3-2 33 times in multiple selection tracks, whereas the remaining sequences appeared 1–7 times each in the data set (Fig. 1c). Multi-alignment of all sequences revealed similarities and patterns of conservation for a number of the residues that were randomized in the library. Several candidates in the data set contained similar sequence patterns in the first helix, for example a positively charged amino acid, lysine or arginine, was preferred in position six. Moreover, the leucine residue in position seven was conserved in most sequences and is also present in ABD before randomization, which might indicate that this amino acid is structurally important. The differences were mainly located in the third helix, which suggests that some of the residues in the first helix are especially important for strong binding. Seven individual clones (Fig. 1d and highlighted with dots in Fig. 1c) were sub-cloned to an expression vector and expressed in E. coli for further characterization. All expressed proteins were successfully purified by affinity chromatography using HSA as affinity ligand, which confirmed that the selected clones had retained their binding to HSA. High purity was obtained, as demonstrated by SDS-PAGE, and expected molecular weights were confirmed for all purified proteins by mass spectrometry (data not shown).
Analysis of binding kinetics
SPR-measurements of the interactions with ErbB3 and serum albumin of human and murine origin (hErbB3, mErbB3, murine serum albumin (MSA) and HSA) were performed to investigate the binding properties of the selected ABD variants and compare them with the parental ABD scaffold. The kinetic data, at least duplicate experiments for each interaction, on seven selected variants (Fig. 1d) and ABD are summarized in Table 1. The binding to ErbB3 was generally characterized by a fast on-rate whereas the off-rate varied more between the variants (Table 1). ABDErbB3-3 and ABDErbB3-27 (Fig. 2a) were shown to have somewhat slower off-rates, and thereby possessed the lowest K D-values among the candidates (10 and 12 nM for hErbB3, respectively). Data from immobilized hErbB3-Fc and hErbB3-His6 correlated well and data from both versions of the receptor are therefore presented together. In addition, the affinities for mErbB3 and hErbB3 were almost identical (Table 1), which suggests that the binders target an epitope on ErbB3 with a high degree of species conservation. As expected, no binding of ABD to ErbB3 was detected, whereas all tested phage-selected variants bound both human and mouse ErbB3 with affinities ranging from 10 to 100 nM. None of the ErbB3-binding ABD variants gave any signal on control surfaces immobilized with polyclonal human Fc or the related hErbB2 receptor (data not shown).
Table 1.
Measured affinity constants for ErbB3-binding ABD variants
| ABD variant | hErbB3 | mErbB3 | HSA | MSA | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| k a (M−1s−1) | k d (s−1) | K D (nM) | k a (M−1s−1) | k d (s−1) | K D (nM) | k a (M−1s−1) | k d (s−1) | K D (nM) | k a (M−1s−1) | k d (s−1) | K D (nM) | T m (°C) | |
| ABDErbB3-1 | 7.5 (± 0.2) × 105 | 5.6 (± 0.3) × 10−2 | 75 [4] | 9.1 (± 0.5) × 105 | 6.3 (± 0.3) × 10−2 | 69 [2] | 4.5 (± 1.9) × 105 | 5.7 (± 1.8) × 10−5 | 0.1 [6] | 2.6 (± 1.2) × 105 | 5.5 (± 0.3) × 10−4 | 2.1 [4] | ≥ 75 |
| ABDErbB3-2 | 1.0 (± 0.2) × 106 | 2.7 (± 0.3) × 10−2 | 26 [4] | 9.3 (± 0.3) × 105 | 3.1 (± 0.04) × 10−2 | 34 [2] | 6.6 (± 3.4) × 105 | 1.7 (± 0.4) × 10−4 | 0.3 [6] | 3.6 (± 1.9) × 105 | 1.2 (± 0.1) × 10−3 | 3.4 [4] | ≥ 80 |
| ABDErbB3-3 | 7.8 (± 0.6) × 105 | 7.6 (± 0.7) × 10−3 | 10 [4] | 8.3 (± 0.2) × 105 | 9.4 (± 0.06) × 10−3 | 11 [2] | 2.7 (± 0.4) × 105 | 1.1 (± 0.3) × 10−4 | 0.4 [6] | 1.8 (± 0.2) × 105 | 5.3 (± 0.3) × 10−4 | 3.0 [4] | ≥ 80 |
| ABDErbB3-13 | 6.1 (± 0.6) × 105 | 2.4 (± 0.1) × 10−2 | 39 [4] | 4.9 (± 0.01) × 105 | 1.8 (± 0.1) × 10−2 | 37 [2] | 2.2 (± 0.4) × 105 | 1.8 (± 0.6) × 10−4 | 0.8 [6] | 1.6 (± 0.4) × 105 | 1.4 (± 0.1) × 10−3 | 8.9 [4] | ≥ 75 |
| ABDErbB3-18 | 2.0 (± 0.2) × 105 | 1.8 (± 0.2) × 10−2 | 94 [4] | 1.8 (± 0.02) × 105 | 1.8 (± 0.1) × 10−2 | 97 [2] | 1.3 (± 0.3) × 105 | 1.1 (± 0.3) × 10−2 | 81 [6] | 5.6 (± 5.0) × 104 | 2.0 (± 1.3) × 10−2 | 367 [4] | ≥ 60 |
| ABDErbB3-20 | 2.7 (± 0.3) × 105 | 1.6 (± 0.1) × 10−2 | 60 [4] | 2.0 (± 0.1) × 105 | 1.7 (± 0.2) × 10−2 | 86 [2] | 1.6 (± 0.1) × 105 | 2.7 (± 0.3) × 10−4 | 1.7 [6] | 1.1 (± 0.04) × 105 | 1.2 (± 0.1) × 10−3 | 11 [4] | ≥ 60 |
| ABDErbB3-27 | 6.2 (± 1.4) × 105 | 7.4 (± 2.5) × 10−3 | 12 [4] | 2.4 (± 0.8) × 106 | 8.6 (± 4.6) × 10−3 | 4 [2] | 9.2 (± 0.6) × 104 | 1.7 (± 0.4) × 10−4 | 1.8 [3] | 7.1 (± 0.3) × 104 | 8.3 (± 0.4) × 10−4 | 12 [2] | ≥ 60 |
| ABD | – | – | – | – | – | – | 7.1 (± 1.2) × 104 | 5.8 (± 0.5) × 10−4 | 8.2 [9] | 6.7 (± 1.2) × 104 | 3.1 (± 0.2) × 10−3 | 46 [6] | ≥ 8 |
Affinities were determined by SPR analysis from multiple experiments. Constants are represented as mean values (± standard deviation) from the number of interactions indicated within brackets. K D values were calculated by k d/k a. Good reproducibility between replicate experiments of all interactions was achieved, as determined by the measured standard deviations and curve-fitting parameters (χ2 and residual plot, data not shown)
Fig. 2.
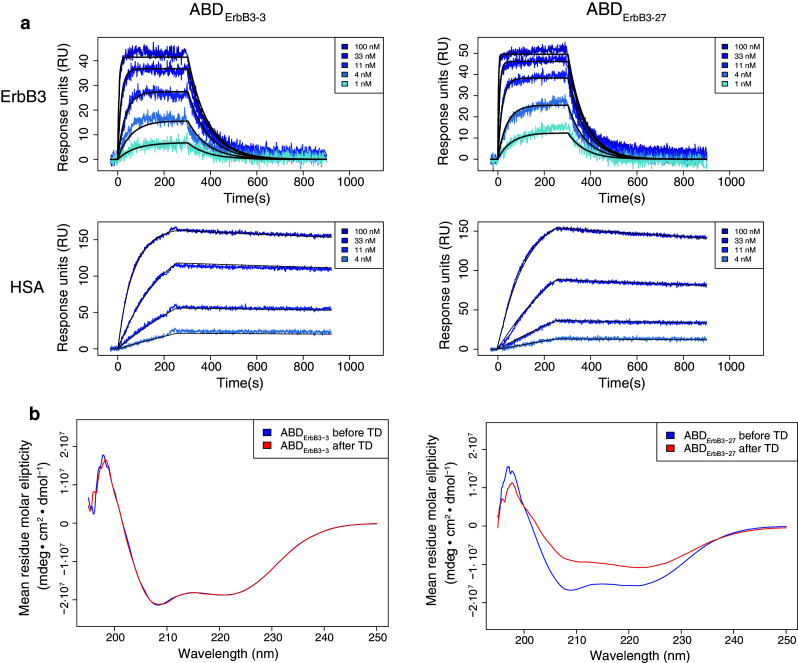
Representative sensorgrams and circular dichroism spectra. a ABDErbB3-3 and ABDErbB3-27 binding to hErbB3 and human serum albumin (HSA). b Overlays of circular dichroism spectra for ABDErbB3-3 and ABDErbB3-27 before (blue) and after (red) thermal denaturation (TD)
Affinities to HSA were in the nanomolar region, ranging from 0.1 nM for ABDErbB3-3 and up to 80 nM for ABDErbB3-18 (Table 1). As seen in Fig. 2a, the analyzed ErbB3-binding ABD variants had slow dissociation from HSA with off-rates (k d) around 10−4 s−1. For all analyzed ABD variants, the affinity for MSA was approximately tenfold lower than for HSA, mainly due to faster dissociation. This correlates with what has been shown previously for ABD [47].
Secondary structure and thermal stability measurements
CD measurements were performed to verify that the selected variants had retained a secondary structure and stability comparable to the ABD scaffold. All analyzed variants had similar spectra with two local inflection points at around 208 and 221 nm, comparable to what has been shown earlier for ABD [29, 30, 48]. For the majority of the analyzed protein variants, the spectra before and after heat treatment overlapped well, which indicated that the proteins were able to refold after heat-induced denaturation. The only binder that did not completely refold after heating was ABDErbB3-27, which is shown in Fig. 2b. Interestingly, this binder exhibited a different denaturation profile with an intermediate plateau at 70 °C followed by an increasing CD signal (data not shown). Complete unfolding was not observed for any of the protein domains, even after heating to 90 °C. Hence, the T m values could not be determined from the thermal denaturation profiles at 221 nm. However, the values were estimated to exceed 60 °C for all of the variants and to even exceed 80 °C for three of them. The melting point for the parental ABD domain was also estimated to exceed 80 °C Table 1.
Competition assay for ErbB3-binding
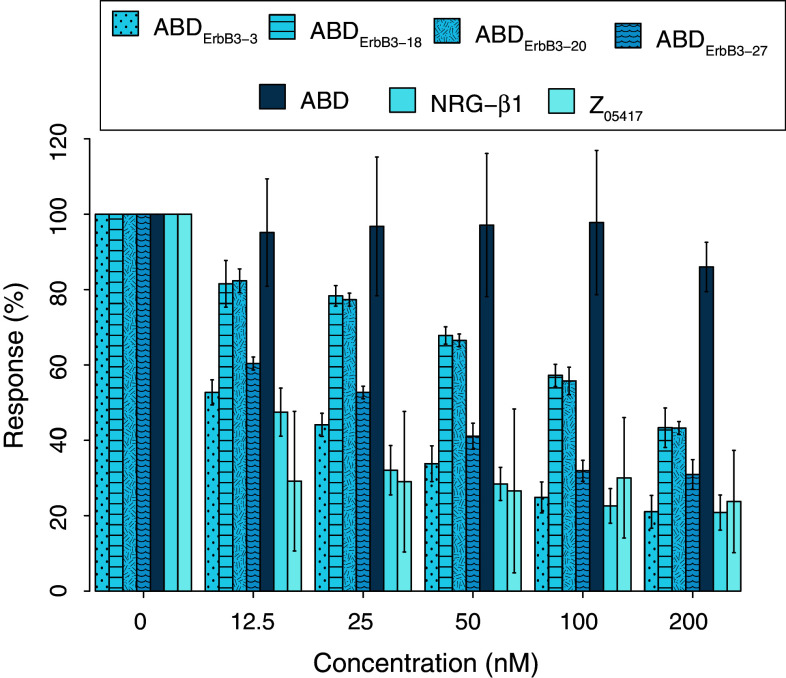
A biosensor assay was employed to investigate if the selected ABD variants recognized the same region as the natural ligand NRG-β1. Four representative ErbB3-binding ABD variants, including the strongest binders ABDErbB3-3 and ABDErbB3-27, were used to assess if the interaction between soluble ErbB3 and immobilized NRG-β1 was affected by the binding of the ABD variants. Two versions of NRG-β1 were used although the same results were expected from both. NRG-β1 ECD consists of the full-length extracellular domain (26.9 kDa) while NRG-β1 EGF contains only the receptor-binding part (7.5 kDa). A constant concentration (5 nM) of ErbB3 was mixed with a series of different concentrations of the respective ABD variants. The mixtures were incubated for 1 h to reach equilibrium, which would theoretically take between 15 and 30 min as calculated from the affinities [49], prior to injection over NRG-β1 immobilized on a sensor chip. To compensate for variations in immobilization level, reference injections with ErbB3 alone were used to normalize the signals in all other injections. The observed binding of ErbB3 to the chip surface was found to decrease with increasing concentration of ABD variant for all tested ErbB3-binding ABD domains (Fig. 3). However, no influence on binding was observed for the parental ABD, which does not have any measurable affinity for ErbB3 Table 1. This inhibitory effect correlates well to the affinities for ErbB3 of the evaluated ABD molecules Table 1. These data indicate that all analyzed ABD variants, which have similar sequences but still represent somewhat different sequence clusters (Fig. 1c), have epitopes on ErbB3 that overlap with the binding-site for NRG-β1. The same competing behavior was observed with immobilized NRG-β1 EGF (data not shown).
Fig. 3.
ErbB3-binding ABD molecules disrupt binding of NRG-β1 to ErbB3. 5 nM hErbB3 was incubated with indicated concentrations of binder and injected over immobilized NRG-β1 ECD (entire extracellular domain). Obtained max responses were normalized against a corresponding injection with untreated hErbB3. Bars represent mean values with indicated standard deviations from two injections over three surfaces (n = 6). Control injection NRG-β1 EGF (receptor-binding part) was performed only once (n = 3). Z05417, an ErbB3-binding Affibody molecule [45], was used as positive control
Investigation of ErbB3-binding in the presence of albumin
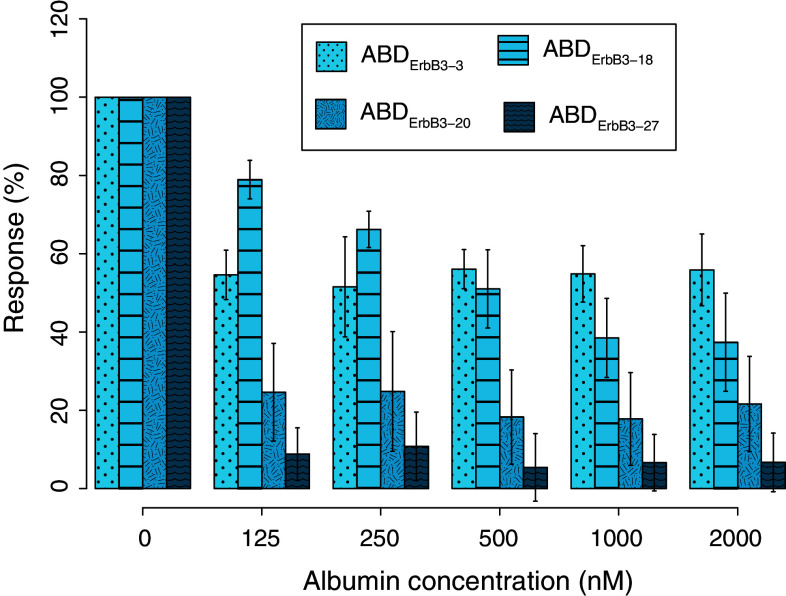
Next, SPR was used to assess the ability of the new bispecific protein domains to bind ErbB3 in the presence of increasing concentrations of albumin. While the ABD concentration was kept constant (at 200 nM), the HSA concentrations were varied between 0 and 2 μM. After mixing and incubating, the binding to ErbB3 was detected by injection over a biosensor surface with immobilized ErbB3 (Fig. 4). ABDErbB3-18, ABDErbB3-20, and ABDErbB3-27 gradually lost their ability to bind to hErbB3 when the concentration of albumin was increased. However, the excess of albumin required to achieve this effect was different for each variant. ABDErbB3-27, for example, lost the ability to bind hErbB3 with HSA present at close to a 1:1 molar ratio. Interestingly, ABDErbB3-3, which has similar kinetic properties for ErbB3 binding as ABDErbB3-27 but binds albumin with higher affinity (Table 1), showed a significant ErbB3-binding even in the presence of a tenfold molar excess of albumin. The same patterns were observed when the ABD molecules were incubated with MSA prior to injection over immobilized mErbB3 for all candidates (data not shown).
Fig. 4.
ABD binding to hErbB3 in the presence of albumin. ABD variants were incubated with varying concentrations of albumin for at least 1 h at room temperature and subsequently injected over a sensor chip with immobilized hErbB3. 200 nM ABD variant was used in all incubations, which were injected over two surfaces with hErbB3. All observed responses were normalized against reference injections without albumin. Data bars represent mean values from four interactions in total (error bars ± standard deviation)
Flow-cytometric evaluation of ErbB3-binding on cells
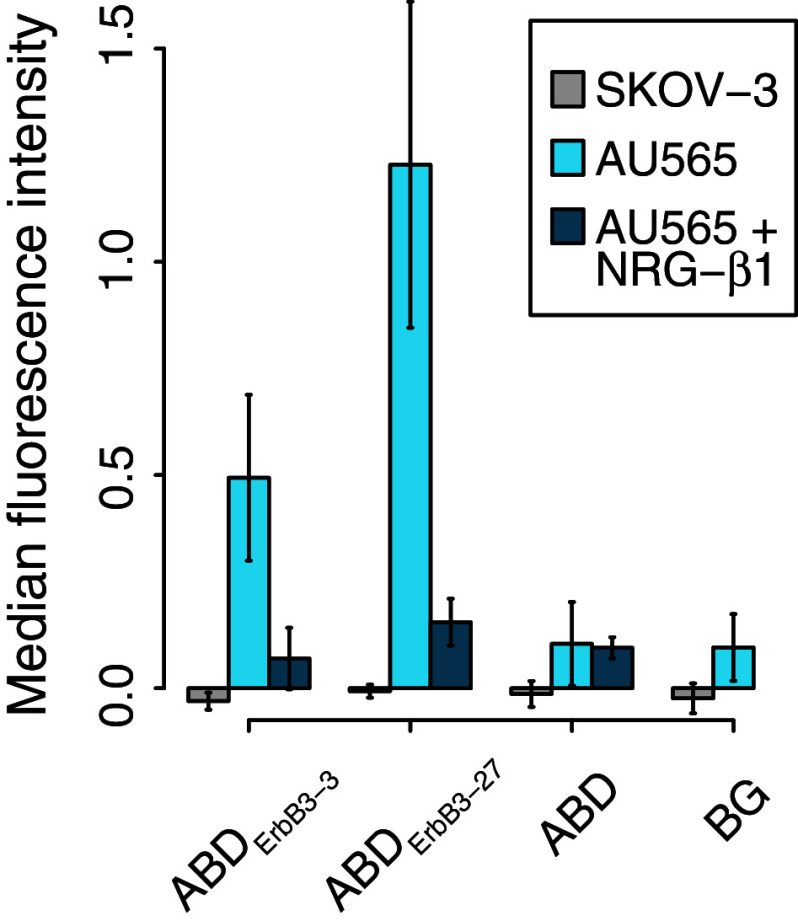
Since the selection and initial characterizations were performed using recombinant ErbB3, we wanted to investigate if the new binders were able to target the native receptors on mammalian cells as well. Consequently, binding of the two candidates with highest ErbB3-affinities, ABDErbB3-3 and ABDErbB3-27, to the ErbB3-expressing human breast cancer cell line AU565 was analyzed by flow cytometry. ABD variants fused to a C-terminal Z-domain were incubated with the cells and detected with biotinylated IgG and a streptavidin R-phycoerythrin conjugate. ABDErbB3-3 and ABDErbB3-27 bound to the AU565 cells, while no binding was detected for the ABD control (Fig. 5). The signal from ABDErbB3-27 compared to ABDErbB3-3 was higher than expected from their respective affinities for ErbB3. Furthermore, no detectable binding was seen for either variant to the ErbB3low-expressing SKOV-3 cell line.
Fig. 5.
ErbB3-binding ABD molecules can bind to hErbB3-expressing cells in vitro. ABD molecules were genetically fused N-terminally of an IgG-binding Z-domain used for detection. 2 × 105−7 × 105 AU565 (ErbB3+, n = 8) or SKOV-3 (ErbB3low, n = 3) cells were incubated with 500 nM ABD-Z and detected with biotinylated polyclonal human IgG and streptavidin–phycoerythrin. To further confirm the earlier shown competition between NRG-β1 and the ABD variants, AU565 cells were pre-incubated with 100 nM NRG-β1 EGF [n = 3 (ABDErbB3-3), n = 2 (ABDErbB3-27 and ABD)]. Bars represent mean values from multiple measurements performed on different days (error bars ± standard deviation). The background was measured from cells treated with detection reagents only. All median fluorescence intensity values were normalized by subtracting the median autofluorescent intensity measured from cells treated with 3 % PBSTB only
The potential competition of ABDErbB3-3 and ABDErbB3-27 with NRG-β1 for ErbB3-binding was further assessed by pre-incubating the AU565 cells with NRG-β1 EGF. The cells were subsequently incubated with 500 nM of the ABD variants fused to a C-terminal Z-domain and detected as described above. The fluorescence intensity from cells that had been pre-incubated with NRG-β1 EGF was close to or equal to the background intensity (Fig. 5). Both ABDErbB3-3 and ABDErbB3-27 were thus blocked from binding to the cells by NRG-β1 EGF, which demonstrate that the binders indeed target an epitope on ErbB3 that is overlapping with the binding site of NRG-β1 EGF. The results thus confirmed the results from the earlier-described SPR-based competition experiments. Moreover, the data from this competition assay clearly demonstrated that the binding to the cells was ErbB3-specific. Since ErbB3 signaling is dependent on the binding of a growth factor, the ABD variants represent promising candidates for the development of ErbB3 signaling inhibitors.
Discussion
Binding proteins that are engineered from alternative, non-antibody scaffolds constitute an interesting class of affinity reagents. Such engineered proteins are promising alternatives to antibodies for different therapy applications and often have several advantages, such as high stability, ease of production and modification, and effective penetration into tissues [5, 50]. A major challenge is to achieve a long in vivo half-life without compromising the small size of the binder. In this study, we demonstrate the selection of ErbB3-specific binding proteins based on a small helical scaffold that is derived from an albumin-binding domain of streptococcal Protein G, which has inherent affinity for albumin. Eleven residues in ABD have earlier been randomized to enable selection of bispecific binders with affinity for two different target proteins, i.e., introducing one novel specificity on a surface of the domain that faces in one direction while preserving the residues known to contribute to albumin binding on the opposite side of the domain [25].
Using this library for phage display selections against hErbB3 we identified 32 unique ABD variants. Seven candidates, selected from sequence data and cluster analysis (Fig. 1c, d), were effectively purified by affinity chromatography through their inherent binding to albumin. Biophysical characterization demonstrated an extraordinary thermal stability with melting temperatures above 80 °C for some of the selected candidates (Table 1). Binding analysis to hErbB3, mErbB3, HSA, and MSA identified two candidates, ABDErbB3-3 and ABDErbB3-27, that demonstrated the strongest affinities for hErbB3, showing equilibrium dissociation constants (K D) of 10 and 12 nM, respectively (Table 1; Fig. 2a). Interestingly, all variants except ABDErbB3-18 had higher affinities for HSA compared to the parental ABD domain. This property might be a consequence of the accumulated positive charge in the selected protein domains (Fig. 1d). Arginine was frequently found in the sequences as one of the most commonly observed residues in the randomized positions. Since albumin has a negative net charge (pI 5.7 for HSA) at the pH used for selection and characterization (pH 7.4), the positive charge of the non-albumin-binding side of the ABD variants might influence the association rate constant (k a) in the interaction. This was indeed observed in the kinetic data for most ABD variants, although minor differences in dissociation rate (k d) were also seen (Table 1). The accumulation of positively charged residues in the selected sequences might be due to the negative net charge of hErbB3 (pI 6.5) during the selection. Furthermore, since the affinities for hErbB3 and mErbB3 (which share 91 % sequence homology) were similar, the data indicated that binding occurred at an epitope that has been conserved between species.
An affinity protein that is able to inhibit natural ligand binding might have an anti-proliferative effect on ErbB3-expressing cancer cells since this ligand plays a central role in the activation of downstream signaling. Such an inhibitory effect might be useful for cancer therapy. Hence, we wanted to investigate whether the novel affinity proteins were able to compete for binding to ErBb3 with the natural ligand NRG-β1. A competition assay was therefore used to test for ABD-mediated blocking of ErbB3 binding to immobilized NRGβ1 (Fig. 3). The results clearly showed that all four analyzed ABD variants were capable of competing for ErbB3 binding with NRG-β1. Furthermore, their efficacies to block NRG-β1 correlated with their respective affinities and the performance of ABDErbB3-3 was comparable to both NRG-β1 blocking itself and an ErbB3-specific Affibody molecule (Z05417). Interestingly, recent encouraging results with this Affibody molecule have demonstrated an anti-proliferative effect on breast cancer cell lines mediated by inhibition of the NRG-β1-induced phosphorylation of ErbB3 [51]. In addition, the same accumulation of arginines and lysines that was observed for the ABD variants is also seen in the ErbB3-specific Affibody molecule, which further indicates that they are targeting a similar epitope [45].
Although the new candidates were clearly bispecific, i.e., demonstrating specific binding to two different targets, we also wanted to investigate if they might be able to interact with ErbB3 and albumin simultaneously. In the crystal complex between the homologous GA-module (an HSA-binding domain with similar structure and binding surface as ABD) and HSA, the surface on GA that is corresponding to the randomized part of ABD faces in the opposite direction from the bound albumin [52], which at least in theory may allow simultaneous binding to occur. To investigate this, ErbB3-binding by four of the bispecific ABD variants (ABDErbB3-3, ABDErbB3-18, ABDErbB3-20, and ABDErbB3-27) in the presence of increasing concentrations of albumin was measured using an SPR assay (Fig. 4). The results showed that ErbB3-binding decreased with increasing concentration of albumin. However, for three of the candidates, ErbB3-binding activity was detected also at the highest albumin concentration (2 μM). The experiments hence indicated that the bispecific domains were able to bind both HSA and ErbB3, but not simultaneously (Fig. 4). The same result has been observed earlier for bispecific binders to TNF-alpha that were based on the same scaffold [26]. Important to mention is that it is not evident that simultaneous binding is preferred in all applications. Moreover, how the different binding modes influence for example biodistribution and biological efficacy have to be investigated in a case-by-case manner in future in vivo studies. However, if desired, simultaneous binding based on the ABD scaffold may still be feasible, since this will depend on the availability of epitopes on the target that can bind ABD without interfering with a bound albumin molecule. It is also important to note that the selections have not been performed under conditions that favor simultaneous binding.
To demonstrate targeting of the native receptor expressed on human cancer cells, two of the ABD molecules that showed the highest affinity for ErbB3 were incubated with ErbB3-positive AU565 cells and analyzed by flow cytometry. ABDErbB3-3 and ABDErbB3-27 were both able to bind to the ErbB3-overexpressing cells (Fig. 5), showing that the epitope is also available for binding when the receptor is anchored in the cell membrane. An ErbB3low-expressing cell line (SKOV-3) was included as a control and showed no detectable binding, which confirmed that the ABD variants were targeting the ErbB3 receptor. The capability of the ABD variants to compete with NRG-β1 was further evaluated by measuring binding to AU565 cells after pre-incubation with NRG-β1. Binding was completely inhibited by the natural ligand, which suggests specific binding to an overlapping region of ErbB3 also in the native form on the surface of cancer cells.
Altogether, here we show the selection of bispecific albumin-binding domains with high affinity for ErbB3 and ability to bind to a biologically relevant epitope. Several promising protein fusion strategies based on association with albumin or immunoglobulins for half-life improvement have been described earlier; for example, ABD has been fused to single-chain diabodies (scDb) [11] and also to an Affibody molecule targeting ErbB2 [16]. Moreover, fusions of a single-chain fragment variable (scFv) or (scDb) to domains of Protein A, Protein G, or Protein L [13, 14], albumin-binding human VH-domain antibodies [53] and albumin-binding peptides [54] have been made. In contrast to these examples, the bispecific ABD molecules presented here can bind albumin without compromising their small size, and existing data indicates that the two binding surfaces may be independently tunable by combinatorial protein engineering [26]. In addition, previous reports have demonstrated that even an ABD with a weak affinity for albumin can have a large impact on half-life in vivo [12]. The strategy of using bispecific ABD molecules therefore represent a unique complement to existing approaches and may also provide a novel bi-functional fusion partner to expand the use of other protein drugs.
Acknowledgments
This work was supported by the Swedish Research Council (VR), the Knut and Alice Wallenberg foundation (KAW), and the Royal Swedish Academy of Sciences (KVA). Prof. Per-Åke Nygren is acknowledged for scientific advice and helpful discussion.
Conflict of interest
The authors declare that they have no conflicts of interest.
Footnotes
Johan Nilvebrant and Mikael Astrand contributed equally to this study.
References
- 1.Weiner LM, Surana R, Wang S. Monoclonal antibodies: versatile platforms for cancer immunotherapy. Nat Rev Immunol. 2010;10(5):317–327. doi: 10.1038/nri2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scott AM, Wolchok JD, Old LJ. Antibody therapy of cancer. Nat Rev Cancer. 2012;12(4):278–287. doi: 10.1038/nrc3236. [DOI] [PubMed] [Google Scholar]
- 3.Lofblom J, Frejd FY, Stahl S. Non-immunoglobulin based protein scaffolds. Curr Opin Biotechnol. 2011;22(6):843–848. doi: 10.1016/j.copbio.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Gebauer M, Skerra A. Engineered protein scaffolds as next-generation antibody therapeutics. Curr Opin Chem Biol. 2009;13(3):245–255. doi: 10.1016/j.cbpa.2009.04.627. [DOI] [PubMed] [Google Scholar]
- 5.Gilbreth RN, Koide S. Structural insights for engineering binding proteins based on non-antibody scaffolds. Curr Opin Struct Biol. 2012;22(4):413–420. doi: 10.1016/j.sbi.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zahnd C, Kawe M, Stumpp MT, de Pasquale C, Tamaskovic R, Nagy-Davidescu G, Dreier B, Schibli R, Binz HK, Waibel R, Pluckthun A. Efficient tumor targeting with high-affinity designed ankyrin repeat proteins: effects of affinity and molecular size. Cancer Res. 2010;70(4):1595–1605. doi: 10.1158/0008-5472.CAN-09-2724. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt MM, Wittrup KD. A modeling analysis of the effects of molecular size and binding affinity on tumor targeting. Mol Cancer Ther. 2009;8(10):2861–2871. doi: 10.1158/1535-7163.MCT-09-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kontermann RE. Strategies to extend plasma half-lives of recombinant antibodies. BioDrugs. 2009;23(2):93–109. doi: 10.2165/00063030-200923020-00003. [DOI] [PubMed] [Google Scholar]
- 9.Kratz F, Elsadek B. Clinical impact of serum proteins on drug delivery. J Control Release. 2012;161(2):429–445. doi: 10.1016/j.jconrel.2011.11.028. [DOI] [PubMed] [Google Scholar]
- 10.Anderson CL, Chaudhury C, Kim J, Bronson CL, Wani MA, Mohanty S. Perspective–FcRn transports albumin: relevance to immunology and medicine. Trends Immunol. 2006;27(7):343–348. doi: 10.1016/j.it.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Stork R, Campigna E, Robert B, Muller D, Kontermann RE. Biodistribution of a bispecific single-chain diabody and its half-life extended derivatives. J Biol Chem. 2009;284(38):25612–25619. doi: 10.1074/jbc.M109.027078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hopp J, Hornig N, Zettlitz KA, Schwarz A, Fuss N, Muller D, Kontermann RE. The effects of affinity and valency of an albumin-binding domain (ABD) on the half-life of a single-chain diabody-ABD fusion protein. Protein Eng Des Sel. 2010;23(11):827–834. doi: 10.1093/protein/gzq058. [DOI] [PubMed] [Google Scholar]
- 13.Hutt M, Farber-Schwarz A, Unverdorben F, Richter F, Kontermann RE. Plasma half-life extension of small recombinant antibodies by fusion to immunoglobulin-binding domains. J Biol Chem. 2012;287(7):4462–4469. doi: 10.1074/jbc.M111.311522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Unverdorben F, Färber-Schwarz A, Richter F, Hutt M, Kontermann RE. Half-life extension of a single-chain diabody by fusion to domain B of staphylococcal Protein A. Protein Eng Des Sel. 2012;25(2):81–88. doi: 10.1093/protein/gzr061. [DOI] [PubMed] [Google Scholar]
- 15.Dennis MS, Jin H, Dugger D, Yang R, McFarland L, Ogasawara A, Williams S, Cole MJ, Ross S, Schwall R. Imaging tumors with an albumin-binding Fab, a novel tumor-targeting agent. Cancer Res. 2007;67(1):254–261. doi: 10.1158/0008-5472.CAN-06-2531. [DOI] [PubMed] [Google Scholar]
- 16.Tolmachev V, Orlova A, Pehrson R, Galli J, Baastrup B, Andersson K, Sandstrom M, Rosik D, Carlsson J, Lundqvist H, Wennborg A, Nilsson FY. Radionuclide therapy of HER2-positive microxenografts using a 177Lu-labeled HER2-specific Affibody molecule. Cancer Res. 2007;67(6):2773–2782. doi: 10.1158/0008-5472.CAN-06-1630. [DOI] [PubMed] [Google Scholar]
- 17.Andersen JT, Pehrson R, Tolmachev V, Daba MB, Abrahmsen L, Ekblad C. Extending half-life by indirect targeting of the neonatal Fc receptor (FcRn) using a minimal albumin-binding domain. J Biol Chem. 2011;286(7):5234–5241. doi: 10.1074/jbc.M110.164848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wunder A, Stehle G, Schrenk HH, Hartung G, Heene DL, Maier-Borst W, Sinn H. Antitumor activity of methotrexate-albumin conjugates in rats bearing a Walker-256 carcinoma. Int J Cancer. 1998;76(6):884–890. doi: 10.1002/(SICI)1097-0215(19980610)76:6<884::AID-IJC19>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 19.Stehle G, Sinn H, Wunder A, Schrenk HH, Stewart JC, Hartung G, Maier-Borst W, Heene DL. Plasma protein (albumin) catabolism by the tumor itself-implications for tumor metabolism and the genesis of cachexia. Crit Rev Oncol Hematol. 1997;26(2):77–100. doi: 10.1016/S1040-8428(97)00015-2. [DOI] [PubMed] [Google Scholar]
- 20.Jain RK. Determinants of tumor blood flow: a review. Cancer Res. 1988;48(10):2641–2658. [PubMed] [Google Scholar]
- 21.Burger AM, Hartung G, Stehle G, Sinn H, Fiebig HH. Pre-clinical evaluation of a methotrexate–albumin conjugate (MTX-HSA) in human tumor xenografts in vivo. Int J Cancer. 2001;92(5):718–724. doi: 10.1002/1097-0215(20010601)92:5<718::AID-IJC1257>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 22.Subramanian GM, Fiscella M, Lamouse-Smith A, Zeuzem S, McHutchison JG. Albinterferon alpha-2b: a genetic fusion protein for the treatment of chronic hepatitis C. Nat Biotechnol. 2007;25(12):1411–1419. doi: 10.1038/nbt1364. [DOI] [PubMed] [Google Scholar]
- 23.Metzner HJ, Weimer T, Kronthaler U, Lang W, Schulte S. Genetic fusion to albumin improves the pharmacokinetic properties of factor IX. Thromb Haemost. 2009;102(4):634–644. doi: 10.1160/TH09-04-0255. [DOI] [PubMed] [Google Scholar]
- 24.Flisiak R, Flisiak I. Albinterferon-alpha 2b: a new treatment option for hepatitis C. Expert Opin Biol Ther. 2010;10(10):1509–1515. doi: 10.1517/14712598.2010.521494. [DOI] [PubMed] [Google Scholar]
- 25.Alm T, Yderland L, Nilvebrant J, Halldin A, Hober S. A small bispecific protein selected for orthogonal affinity purification. Biotechnol J. 2010;5(6):605–617. doi: 10.1002/biot.201000041. [DOI] [PubMed] [Google Scholar]
- 26.Nilvebrant J, Alm T, Hober S, Lofblom J. Engineering bispecificity into a single albumin-binding domain. PLoS ONE. 2011;6(10):e25791. doi: 10.1371/journal.pone.0025791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makrides SC, Nygren PA, Andrews B, Ford PJ, Evans KS, Hayman EG, Adari H, Uhlen M, Toth CA. Extended in vivo half-life of human soluble complement receptor type 1 fused to a serum albumin-binding receptor. J Pharmacol Exp Ther. 1996;277(1):534–542. [PubMed] [Google Scholar]
- 28.Ahmad JN, Li J, Biedermannova L, Kuchar M, Sipova H, Semeradtova A, Cerny J, Petrokova H, Mikulecky P, Polinek J, Stanek O, Vondrasek J, Homola J, Maly J, Osicka R, Sebo P, Maly P. Novel high-affinity binders of human interferon gamma derived from albumin-binding domain of Protein G. Proteins. 2012;80(3):774–789. doi: 10.1002/prot.23234. [DOI] [PubMed] [Google Scholar]
- 29.Linhult M, Binz HK, Uhlen M, Hober S. Mutational analysis of the interaction between albumin-binding domain from streptococcal Protein G and human serum albumin. Protein Sci. 2002;11(2):206–213. doi: 10.1110/ps.02802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kraulis PJ, Jonasson P, Nygren PA, Uhlen M, Jendeberg L, Nilsson B, Kordel J. The serum albumin-binding domain of streptococcal Protein G is a three-helical bundle: a heteronuclear NMR study. FEBS Lett. 1996;378(2):190–194. doi: 10.1016/0014-5793(95)01452-7. [DOI] [PubMed] [Google Scholar]
- 31.Cramer JF, Nordberg PA, Hajdu J, Lejon S. Crystal structure of a bacterial albumin-binding domain at 1.4 A resolution. FEBS Lett. 2007;581(17):3178–3182. doi: 10.1016/j.febslet.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 32.Citri A, Skaria KB, Yarden Y. The deaf and the dumb: the biology of ErbB-2 and ErbB-3. Exp Cell Res. 2003;284(1):54–65. doi: 10.1016/S0014-4827(02)00101-5. [DOI] [PubMed] [Google Scholar]
- 33.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2(2):127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 34.Baselga J, Swain SM. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer. 2009;9(7):463–475. doi: 10.1038/nrc2656. [DOI] [PubMed] [Google Scholar]
- 35.Schoeberl B, Pace EA, Fitzgerald JB, Harms BD, Xu L, Nie L, Linggi B, Kalra A, Paragas V, Bukhalid R, Grantcharova V, Kohli N, West KA, Leszczyniecka M, Feldhaus MJ, Kudla AJ, Nielsen UB (2009) Therapeutically targeting ErbB3: a key node in ligand-induced activation of the ErbB receptor-PI3 K axis. Sci Signal 2 (77): ra31 doi: 10.1126/scisignal.2000352 [DOI] [PubMed]
- 36.Carlsson J. Potential for clinical radionuclide-based imaging and therapy of common cancers expressing EGFR-family receptors. Tumor Biol. 2012;33(3):653–659. doi: 10.1007/s13277-011-0307-x. [DOI] [PubMed] [Google Scholar]
- 37.Linggi B, Carpenter G. ErbB-4 s80 intracellular domain abrogates ETO2-dependent transcriptional repression. J Biol Chem. 2006;281(35):25373–25380. doi: 10.1074/jbc.M603998200. [DOI] [PubMed] [Google Scholar]
- 38.Huang X, Gao L, Wang S, McManaman JL, Thor AD, Yang X, Esteva FJ, Liu B. Heterotrimerization of the growth factor receptors erbB2, erbB3, and insulin-like growth factor-i receptor in breast cancer cells resistant to herceptin. Cancer Res. 2010;70(3):1204–1214. doi: 10.1158/0008-5472.CAN-09-3321. [DOI] [PubMed] [Google Scholar]
- 39.Sergina NV, Rausch M, Wang D, Blair J, Hann B, Shokat KM, Moasser MM. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature. 2007;445(7126):437–441. doi: 10.1038/nature05474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Foreman PK, Gore M, Kobel PA, Xu L, Yee H, Hannum C, Ho H, Wang SM, Tran HV, Horowitz M, Horowitz L, Bhatt RR. ErbB3 inhibitory surrobodies inhibit tumor cell proliferation in vitro and in vivo. Mol Cancer Ther. 2012;11(7):1411–1420. doi: 10.1158/1535-7163.MCT-12-0068. [DOI] [PubMed] [Google Scholar]
- 41.Grovdal LM, Kim J, Holst MR, Knudsen SL, Grandal MV, van Deurs B. EGF receptor inhibitors increase ErbB3 mRNA and protein levels in breast cancer cells. Cell Signal. 2012;24(1):296–301. doi: 10.1016/j.cellsig.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 42.Vaught DB, Stanford JC, Young C, Hicks DJ, Wheeler F, Rinehart C, Sanchez V, Koland J, Muller WJ, Arteaga CL, Cook RS. HER3 is required for HER2-induced preneoplastic changes to the breast epithelium and tumor formation. Cancer Res. 2012;72(10):2672–2682. doi: 10.1158/0008-5472.CAN-11-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amin DN, Sergina N, Lim L, Goga A, Moasser MM. HER3 signalling is regulated through a multitude of redundant mechanisms in HER2-driven tumour cells. Biochem J. 2012;447(3):417–425. doi: 10.1042/BJ20120724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruther U. pUR 250 allows rapid chemical sequencing of both DNA strands of its inserts. Nucleic Acids Res. 1982;10(19):5765–5772. doi: 10.1093/nar/10.19.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kronqvist N, Malm M, Gostring L, Gunneriusson E, Nilsson M, Hoiden Guthenberg I, Gedda L, Frejd FY, Stahl S, Lofblom J. Combining phage and staphylococcal surface display for generation of ErbB3-specific affibody molecules. Protein Eng Des Sel. 2011;24(4):385–396. doi: 10.1093/protein/gzq118. [DOI] [PubMed] [Google Scholar]
- 46.Nilsson B, Moks T, Jansson B, Abrahmsen L, Elmblad A, Holmgren E, Henrichson C, Jones TA, Uhlen M. A synthetic IgG-binding domain based on staphylococcal Protein A. Protein Eng. 1987;1(2):107–113. doi: 10.1093/protein/1.2.107. [DOI] [PubMed] [Google Scholar]
- 47.Johansson MU, Frick IM, Nilsson H, Kraulis PJ, Hober S, Jonasson P, Linhult M, Nygren PA, Uhlen M, Bjorck L, Drakenberg T, Forsen S, Wikstrom M. Structure, specificity, and mode of interaction for bacterial albumin-binding modules. J Biol Chem. 2002;277(10):8114–8120. doi: 10.1074/jbc.M109943200. [DOI] [PubMed] [Google Scholar]
- 48.Gulich S, Linhult M, Nygren P, Uhlen M, Hober S. Stability towards alkaline conditions can be engineered into a protein ligand. J Biotechnol. 2000;80(2):169–178. doi: 10.1016/S0168-1656(00)00259-5. [DOI] [PubMed] [Google Scholar]
- 49.Hulme EC, Trevethick MA. Ligand binding assays at equilibrium: validation and interpretation. Br J Pharmacol. 2010;161(6):1219–1237. doi: 10.1111/j.1476-5381.2009.00604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hosse RJ, Rothe A, Power BE. A new generation of protein display scaffolds for molecular recognition. Protein Sci. 2006;15(1):14–27. doi: 10.1110/ps.051817606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gostring L, Malm M, Hoiden-Guthenberg I, Frejd FY, Stahl S, Lofblom J, Gedda L. Cellular effects of HER3-specific affibody molecules. PLoS ONE. 2012;7(6):e40023. doi: 10.1371/journal.pone.0040023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lejon S, Frick IM, Bjorck L, Wikstrom M, Svensson S. Crystal structure and biological implications of a bacterial albumin-binding module in complex with human serum albumin. J Biol Chem. 2004;279(41):42924–42928. doi: 10.1074/jbc.M406957200. [DOI] [PubMed] [Google Scholar]
- 53.Holt LJ, Basran A, Jones K, Chorlton J, Jespers LS, Brewis ND, Tomlinson IM. Anti-serum albumin domain antibodies for extending the half-lives of short-lived drugs. Protein Eng Des Sel. 2008;21(5):283–288. doi: 10.1093/protein/gzm067. [DOI] [PubMed] [Google Scholar]
- 54.Dennis MS, Zhang M, Meng YG, Kadkhodayan M, Kirchhofer D, Combs D, Damico LA. Albumin binding as a general strategy for improving the pharmacokinetics of proteins. J Biol Chem. 2002;277(38):35035–35043. doi: 10.1074/jbc.M205854200. [DOI] [PubMed] [Google Scholar]