Abstract
Coxsackievirus B3 (CVB3) is the primary causal agent of viral myocarditis. During infection, it hijacks host genes to favour its own replication. However, the underlying mechanism is still unclear. Although the viral receptor is an important factor for viral infectivity, other factors such as microRNAs (miRNA) may also play an essential role in its replication after host cell entry. miRNAs are post-transcriptional gene regulators involved in various fundamental biological processes as well as in diseases. To identify miRNAs involved in CVB3 pathogenesis, we performed microarray analysis of miRNAs using CVB3-infected murine hearts and identified miR-203 as one of the most upregulated candidates. We found that miR-203 upregulation is through the activation of protein kinase C/transcription factor AP-1 pathway. We further identified zinc finger protein-148 (ZFP-148), a transcription factor, as a novel target of miR-203. Ectopic expression of miR-203 downregulated ZFP-148 translation, increased cell viability and subsequently enhanced CVB3 replication. Silencing of ZFP-148 by siRNA showed similar effects on CVB3 replication. Finally, analyses of the signalling cascade downstream of ZFP-148 revealed that miR-203-induced suppression of ZFP-148 differentially regulated the expression of prosurvival and proapoptotic genes of the Bcl-2 family proteins as well as the cell cycle regulators. This altered gene expression promoted cell survival and growth, which provided a favourable environment for CVB3 replication, contributing to the further damage of the infected cells. Taken together, this study identified a novel target of miR-203 and revealed, for the first time, the molecular link between miR-203/ZFP-148 and the pathogenesis of CVB3.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-012-1104-4) contains supplementary material, which is available to authorized users.
Keywords: MicroRNA-203, ZFP-148, Coxsackievirus B3, Myocarditis
Introduction
Coxsackievirus B3 (CVB3), a positive single-stranded RNA virus in the family Picornaviridae, can infect multiple organs of humans and cause different diseases, such as myocarditis, pancreatitis, meningitis, etc. However, the most common disease is myocarditis, particularly in infants, children and pregnant women [1]. Viral myocarditis often enters its late phase, dilated cardiomyopathy (DCM), leading to heart failure [2]. CVB3 infects these organs largely depending on the expression of viral receptor. However, the expression levels of this receptor do not always correlate well with the viral infectivity in certain organs including the heart [3, 4], suggesting that other factors such as microRNA (miRNA) may also play important roles in viral infection and replication.
MiRNAs are short RNA molecules approximately 22 nucleotides (nts) in length. They are gene regulators at the post-transcriptional level through suppression of translation and regulation of RNA stability [5]. The 5′ end sequence of the miRNA (particularly bases 2–8) is called the seed region and is mainly responsible for hybridization with its complimentary sequence (seed match sequence) in the 3′ untranslated region (3′UTR) of the target mRNA. This complementary binding usually results in the silencing of the target genes [5]. Mutations within the seed match region often inhibit the miRNA activity [6]. Previous studies reported that a single miRNA may regulate a large number of genes, but the extent of regulation of protein levels usually does not exceed two to three folds [7].
Human genome has been so far found to encode more than 1,000 miRNAs [8] which regulate fundamental biological processes, such as cell proliferation, stress response, differentiation, and apoptosis [9]. They are also known to play important roles in pathogenesis of a variety of diseases, such as cardiovascular diseases, cancers, and infectious diseases [10, 11]. For viral infections, miRNAs have been found to regulate the host and virus interaction network. These include the following: (1) cellular miRNAs suppress or promote viral gene expression [12]; (2) cellular miRNAs suppress other cellular gene expression that regulates viral replication [13]; and (3) virus-encoded miRNAs regulate viral or possibly cellular genes [14, 15]. To date, a number of studies on the role of miRNAs in cardiovascular diseases have been published [16]; however, the function of coxsackievirus-induced alternations of miRNA expression in myocarditis has not yet been reported. The viral miRNAs known so far are all encoded by big nuclear DNA viruses. RNA viruses, such as CVB3 and other picornaviruses that replicate themselves in cytoplasm, likely do not encode miRNAs [6]. Thus, we focused our investigation on the identification of cellular miRNAs regulating CVB3 replication. In this study, we performed microarray analyses of miRNA expression profiles in CVB3-nfected A/J mouse hearts. We found that a number of the miRNA expressions were altered and that miR-203 was one of the most upregulated candidates. We further found that CVB3-induced upregulation of miR-203 is through the activation of protein kinase C (PKC)/transcription factor AP-1 pathway. Further, we used HeLa cells and HL-1 cardiomyocytes to carry out functional characterization of miR-203, because these cell lines have a very low level of endogenous miR-203 expression and are suitable for ectopic expression by transfection. In addition, these cell lines are CVB3 receptor-positive cells and a well-established model system for study of viral replication and pathogenesis. By luciferase reporter assay, we found that zinc finger protein 148 (ZFP-148), a transcription factor (also known as ZBP-89, BFCOL1, and BERF1) [17], is a novel target of miR-203. Silencing of ZFP-148 by miR-203 differentially regulated expression of its downstream pro-survival and pro-apoptotic genes. This led to the enhanced cell growth, which created favourable conditions for CVB3 replication and thus damaged the target cells. To our best knowledge, this is the first report to link the miR-203-induced suppression of ZFP-148 to the underlying mechanism of CVB3 pathogenesis.
Materials and methods
An expanded Materials and methods section is available in the Online Resource.
Virus, cell culture, animals and infection
CVB3 was kindly provided by Charles J. Gauntt (University of Texas, Austin, USA). The virus was propagated in HeLa cells (ATCC) and titred routinely by plaque assay. The HL-1 cell line, a murine cardiac cell line established from an AT-1 mouse atrial cardiomyocyte tumor lineage, was a generous gift from Dr. William C. Claycomb (Louisiana State University Medical Center, USA). Animal experimental protocol was approved by the Animal Care Committee of Faculty of Medicine, University of British Columbia. Four-week-old A/J male mice (Jackson Laboratory) were infected intraperitoneally with 5 × 104 plaque-forming unit (pfu) of CVB3 or sham-infected with phosphate buffer saline (PBS).
H&E staining and echocardiography
The hearts were collected at day 4 post infection (pi). The evaluation of myocarditis based on H&E staining and heart functional measurement by echocardiography were conducted by the methods described previously [18].
RNA extraction
Total RNAs were extracted from mouse hearts, HL-1 cells or HeLa cells using the RNeasy® Mini Kit (Qiagen). Briefly, 30 mg of the basal portion of ventricular tissues were homogenized in a RLT buffer. Culture cells (1 × 106) were lysed also in the RLT buffer. Total cellular RNAs or viral RNAs were isolated from the supernatants of these samples following the manufacturer’s instructions. Measurement of RNA concentration was done using a NanoDrop ND-1000 Spectrophotometer (Thermo Scientific, MA, USA).
MicroRNA microarray
Total RNA was extracted as described above from CVB3- or sham-infected mouse hearts at day 4 pi and submitted to the LC Sciences (Houston, TX, USA) for microarray analyses of miRNA expression profiles as described earlier [19]. (See Online Resource 1).
Cell culture transfection
HeLa cells and HL-1 cells were cultured in the DMEM and Claycomb medium (JRH Biosciences), respectively. Cells were transfected with either pre-miR-203, pre-miR-CL (control) (Applied Biosystems), miRNA inhibitor or siRNA (Invitrogen) using Oligofectamine™ as described previously [20]. Cells were then infected with CVB3 (see online Data Supplement).
Quantitative real-time RT-PCR
Q-RT-PCRs were performed by using the TaqMan® MicroRNA Assays (Applied BioSystems) according to the instructions of the kit and as previously described [21]. The internal controls used to normalize the data were SnoR-202 for measuring miRNA-203 levels in heart tissues and HL-1 cells and U6 RNA for determining miRNA-203 maturation in HeLa cells transfected with pre-miRNA-203 for 48 h. For quantification of CVB3 RNA in miR-203-expressing cells infected with CVB3, oligonucleotide primers targeting CVB3 2A gene were synthesized (see online Table 1). The primers for determining miRNA-203 expression levels were purchased from Applied BioSystems (see Online Resource).
Live-cell confocal imaging
Live-cell confocal imaging was performed according to a published method [22] with some modifications. Briefly, HeLa cells were plated in 35-mm glass-bottom culture dishes (MatTek Culture Ware, Ashland, MA, USA) for 24 h and then transfected with either the pre-miR-203 or pre-miR-CL using Oligofectamine (Life Technology). The microscope incubator was maintained at 37 °C, and the CO2 concentration was adjusted to 5 %. Observation of cell morphology was performed at 24 and 48 h post-transfection under a Leica confocal microscope (TCS SP2) using differential interference contrast.
Cell viability assay
HeLa cells growing in 24-well plates at approximately 50 % confluence were transfected with either pre-miR-203 or pre-miR-CL as described above. At 24 and 48 h post-transfection, cell viability was measured by using a 3-(4, 5-dimethylthiazol-2-yl) -5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium salt (MTS) assay kit (Promega) as described previously [20].
MiRNA target prediction
The mi-203 target genes were predicted by using the TargetScan program (release 4.1) (http://www.targetscan.org/) [8]. We also used the MicroRNA.org (http://www.microrna.org), a comprehensive resource of miRNA target predictions (released August 2010) to confirm the results.
Western blot analysis
Western blotting was carried out according to the standard protocol described previously [20]. The primary antibodies used in this study are as follows: mouse monoclonal antibodies against CVB3 capsid protein VP-1 (DAKO), beta-actin (Sigma), GAPDH, p21waf1, p27kip1, Rb, cyclin D1 and cyclin E (Santa Cruz); rabbit polyclonal antibodies against ZFP-148 (Aviva Systems Biology), Bcl-X (BD Pharmingen™), as well as phosphorylated protein kinase c (p-PKC), p-cJun and JunB (Cell Signalling). Secondary antibodies used were either goat anti-mouse or goat anti-rabbit IgG (Santa Cruz) depending on the first antibody used. PKC inhibitors GF109203X was from Calbiochem, Germany (see Online Resource).
Viral plaque assay
The virus titre was determined by plaque assay according to the standard protocol described previously [20]. The plaques were counted, and the virus titre was calculated as the pfu/mL.
Construction of luciferase reporter plasmids and its mutants
The partial 3′UTR of ZFP-148 was amplified by RT-PCR from HeLa cell total RNAs as described previously [23] and cloned into the pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega) to produce the wild-type (Wt) reporter. Two mutant reporters were generated by site-directed mutagenesis within the miR-203 seed match region. The primers for site-directed mutagenesis are listed in online Table 1 (see Online Resource).
Luciferase reporter analysis
HeLa cells were co-transfected with the ZFP-148-3′UTR reporter construct or its mutant and pre-miR-203 using lipofectamine™ 2000 (Invitrogen). Forty-eight hours post-transfection, firefly and Renilla luciferase activities were detected on an ELISA reader (TECAN) using the Dual-Glo® Luciferase Assay System (Promega) following the method described previously [23].
Statistical analysis
Statistical significance was evaluated using the Student’s t test for paired comparisons. All values are expressed as mean ± SD. A P value of less than 0.05 was considered statistically significant.
Results
CVB3 infection upregulated miR-203 in HL-1 cardiomyocytes and mouse heart
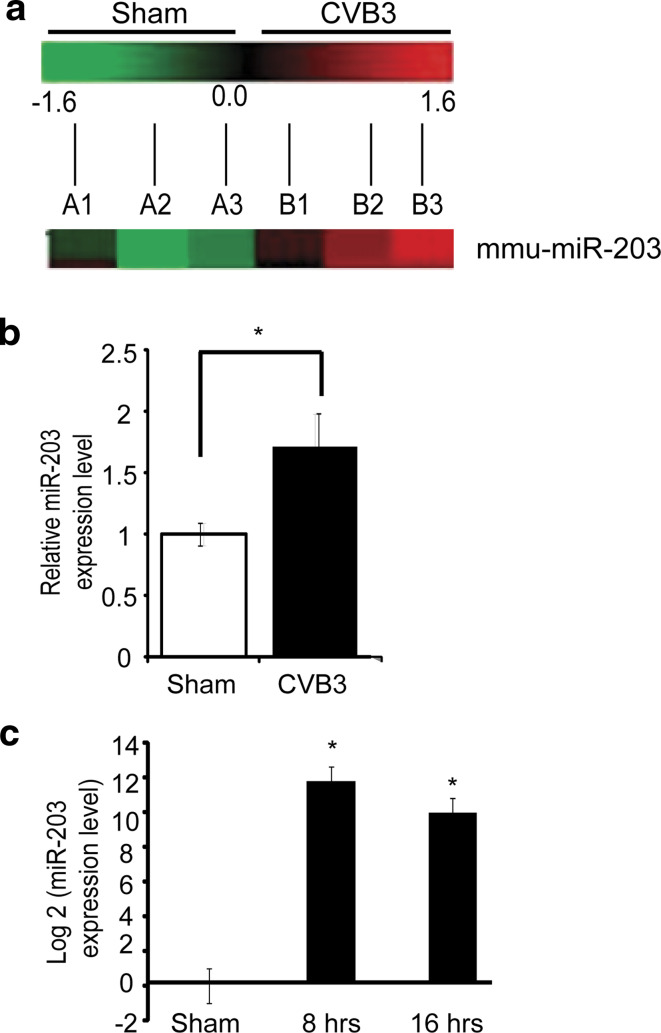
To determine whether CVB3 infection can alter the host miRNA expression profiles, A/J mice were infected with CVB3 or sham-infected with PBS. These animals at day 4 pi showed cardiac dysfunction, which was confirmed by echocardiography (supplement Fig. S1a). Histopathologically, the heart showed massive mononuclear infiltration (supplement Fig. S1b). Total RNAs were then isolated from the hearts and the miRNA expression profiles were analyzed by using the microarray service (LC Science). This analysis demonstrated significant alterations in miRNA expression profiles in CVB3-infected mouse hearts as compared to the sham-infected group. We selected candidate miRNAs from the profiles according to the following criteria: signal intensity > 500, P < 0.05 and fold change ≥ 1.5. The results revealed that, at day 4 pi, 22 miRNAs were downregulated and 4 were upregulated (data not shown). Among these miRNA candidates, miR-203 (whose function in the heart or in CVB3 infection has not been reported) showed a 1.6-fold upregulation in CVB3-infected mouse hearts (P = 0.020) as compared to the control (Fig. 1a). The upregulation of miR-203 in these heart tissues as well as in CVB3-infected HL-1 cardiomyocytes was further confirmed by real time PCR (Fig. 1b, c). MiR-203 was then selected for further study.
Fig. 1.
CVB3 infection upregulates miR-203 in HL-1 cardiomyocytes and mouse heart. a Partial heat map of the mouse miR-203 expression profile. A1–A3 are the three control mice sham-infected with PBS and B1–B3 are mice infected with CVB3. The heat map was generated with the log2 value of each CVB3/sham pair of microarray signal at day 4 pi. Red upregulation, green downregulation, black no changes. MiR-203 had a 1.6-fold upregulation as compared to the control group, P < 0.05. b Q-RT-PCR measurement of miR-203 expression. RNAs were extracted from the hearts at day 4 pi. Endogenous small RNA (SnoR-202) control was used to normalize the data, P = 0.014. c miR-203 expression levels in HL-1 cells at 8 and 16 h pi (10 MOI for 8 and 16 h) were determined by Q-RT-PCR
Upregulation of miR-203 is through the activation of PKC/AP-1 cascade
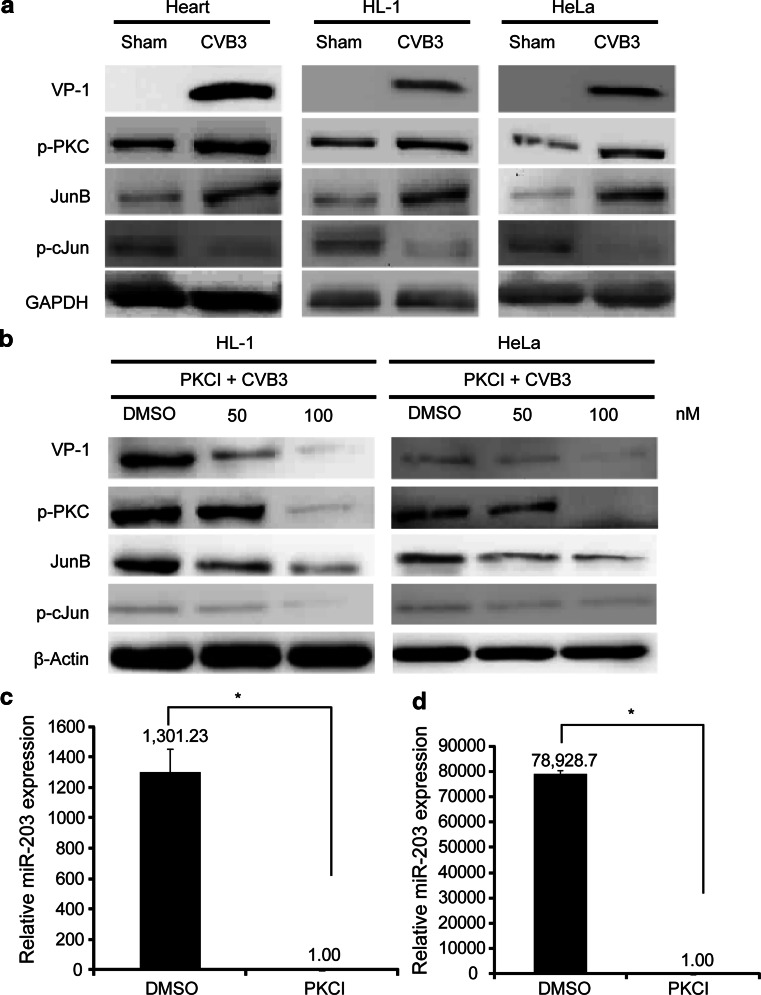
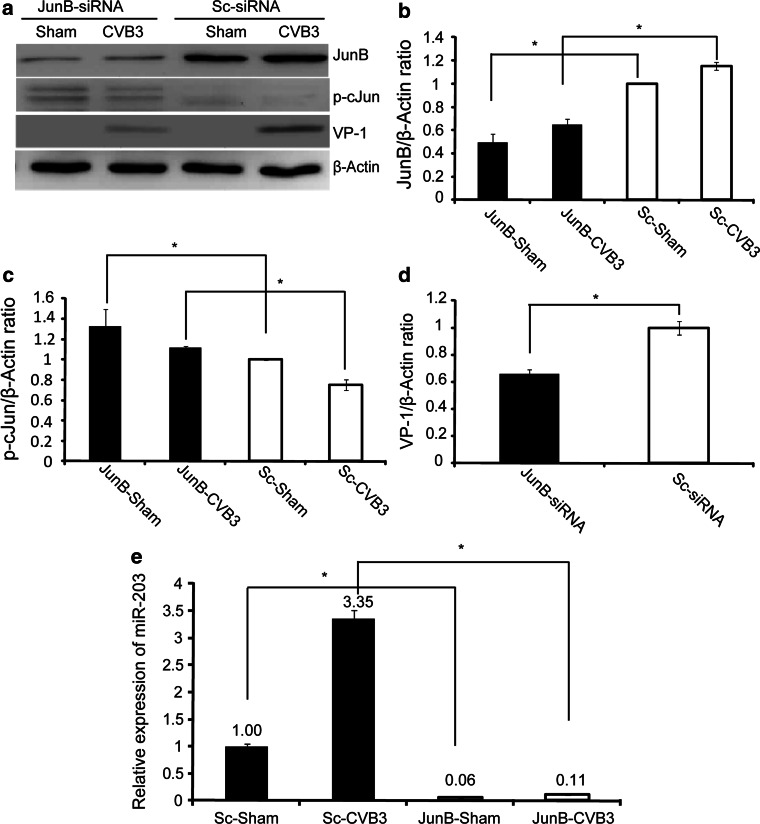
To explore the signalling pathway leading to the upregulation of miR-203 in CVB3-infected heart, we first examined the activation of PKC and its downstream transcription factor AP-1 family proteins c-Jun and JunB, since this cascade activation is essential for miR-203 upregulation in human keratinocytes during differentiation [24]. Our western blot analyses using cell lysates from CVB3-infected murine heart tissues, HeLa cells and HL-1 cardiomyocytes demonstrated that CVB3 infection induced upregulation of p-PKC and further upregulated JunB and downregulated p-cJun as compared to the sham-infected controls (Fig. 2a). This data was further solidified by the experiments in which addition of PKC inhibitor GF109203X significantly suppressed expression of both AP-1 (JunB, p-cJun) and CVB3 protein VP-1 in a dose-dependent manner (Fig. 2b). As JunB and cJun are positive and negative regulators of miR-203 expression, respectively, in human kerantinocytes [24], our data suggest that miR-203 upregulation in CVB3-infected mouse heart depends on activation of PKC/AP-1 cascade. To support this notion, we further measured the miRNA-203 expression by Q-RT-PCR and showed that inhibition of PKC with a specific inhibitor reduced AP-1-regulated expression of miR-203 in HeLa cell (Fig. 2c) and HL-1 cardiomyocytes (Fig. 2d). To further confirm the involvement of JunB in the upregulation of miR-203 during CVB3 infection, we used specific siRNA to knockdown JunB and found that this siRNA markedly reduced JunB and CVB3 VP-1 expression (Fig. 3a, b, d). Interestingly, p-cJun was found to be upregulaed (Fig. 3a, c). We also checked the effect of JunB-siRNA on miR-203 expression levels and found a marked reduction (at least threefold) (Fig. 3e). These data confirmed that CVB3 infection induces miR-203 expression through the PKC/AP-1 cascade.
Fig. 2.
CVB3 induces upregulation of miR-203 through activation of PKC and differential regulation of downstream AP-1 family transcription factors JunB and c-Jun. a Cell lysates from CVB3-infected or sham-infected mouse hearts, HL-1 cardiomyocytes and HeLa cells were used to conduct western blot analysis. b Suppression of PKC with specific inhibitor GF109203X blocked the PKC/AP-1activation. Both HeLa cells and HL-1 cells were treated with either the PKC inhibitor (PKCI) or DMSO and then infected with CVB3. AP-1 (JunB and c-Jun) proteins were detected by western blot analysis. MiR-203 expression in CVB3-infected HeLa cells (c) and HL-1 cells (d) was measured by Q-RT-PCR
Fig. 3.
Silencing of JunB with siRNAs markedly reduces miR-203 expression. a HeLa cells were transfected with JunB siRNAs or control siRNAs and then infected with CVB3. Cell lysates were used for western blot analysis of JunB, p-cJun and CVB3VP-1. β-actin was used as a loading control. b–d Densitometry analyses of expression levels of JunB, p-cJun, and VP-1 (*P < 0.05). e Q-RT-PCR to detect downregulation of miR-203 in cells transfected with JunB siRNA and then infected with CVB3 as described in (a)
ZFP-148 is a novel target gene of miR-203
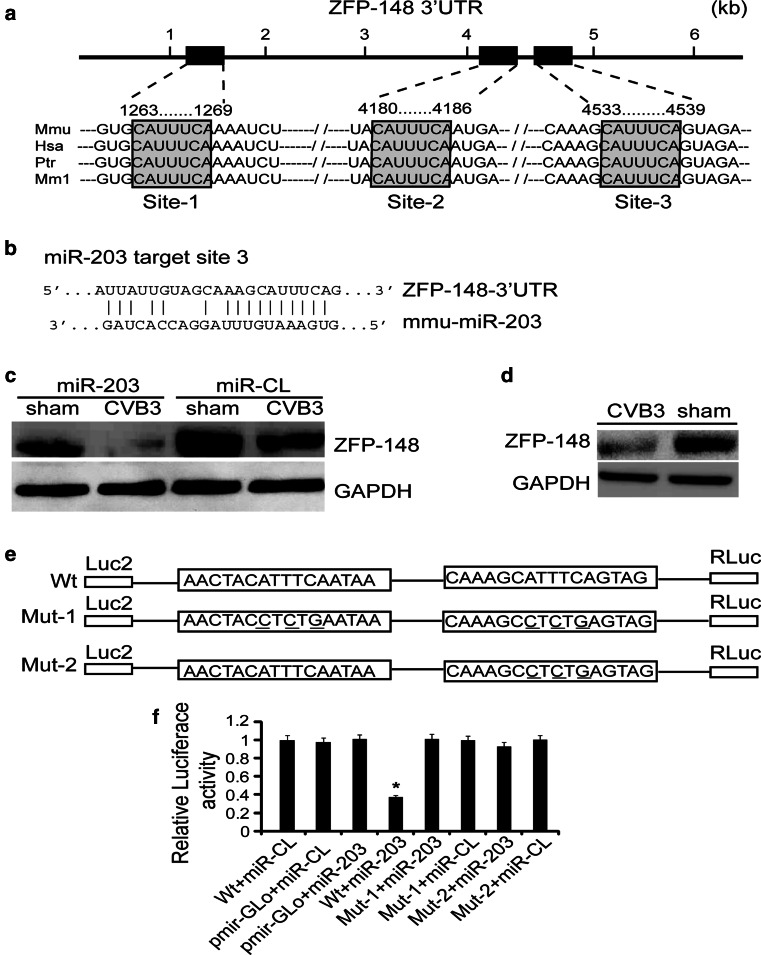
To investigate the signal pathways contributed to the miR-203 effect on CVB3 replication, we sought to identify its target genes. Bioinformatic analyses using TargetScan (http://www.targetscan.org/) program showed that ZFP-148 is one of the top predicted targets, which also include p63, suppressor of cytokine signalling-3 (SOCS3) and V-abl Abelson murine leukemia viral oncogene homolog-1 (ABL-1). As p63, SOCS3 and ABL-1 have already been reported to be targets of miRNA-203 [25], we selected ZFP-148 for further study. Sequence analysis revealed that ZFP-148 has three target sites within its 3′UTR, located at nts 1,263–1,269, 4,180–4,186, and 4,533–4,539 (named site-1, -2, and -3, respectively) (Fig. 4a). Site-1 is closed to the 5′ end region of the 3′UTR while the site-2 and site-3 are close to each other and located near the 3′ end region. Sequence alignment shows that these three sites are highly conserved among human, mice, chimpanzee and monkey. Figure 4b shows the hybridization of the miR-203 to site-3, one representative target of the three, in the ZFP-148 3′UTR. To confirm the targeting of ZFP-148 by miR-203 experimentally, we detected ZFP-148 expression by western blot analysis after transfection of HeLa cells with pre-miR-203. Figure 4c shows marked downregulation of ZFP-148 expression in the miR-203-transfected cells as compared to the control. Meanwhile, we confirmed significant downregulation of ZFP-148 in mouse heart at day 4 pi (Fig. 4d), correlating to the upregulation of miR-203 showing in Fig. 1b.
Fig. 4.
ZFP-148 is a novel target of miR-203. a Physical map of three binding sites of miR-203 within the 3′UTR of ZFP-148 and the sequence alignment of these binding sites within mouse, human, chimpanzee, and Rhesus monkey. The numbers above the sequences indicate the nucleotide positions of these sites. The seed match sequences are highlighted and boxed. b Partial base pairing of miR-203 with binding site-3 in the 3′UTR of ZFP-148. c miR-203 expression suppressed ZFP-148 translation. Total proteins were prepared from HeLa cells transfected with either pre-miR-203 or control (pre-miR-CL) and then infected with CVB3 or sham-infected with PBS. ZFP-148 expression was detected using a specific antibody. GAPDH expression was a loading control. d ZFP-148 expression was inhibited in CVB3-infected mouse heart at day 4 pi. ZFP-148 was detected by western blot analysis. e Diagram of luciferase reporter plasmids. A fragment of the 3′UTR of ZFP-148 carrying miR-203 binding site-2 and -3 was inserted downstream of the Renilla luciferase gene. The ZFP-148-3′UTR mutant-1 (Mut-1) construct was designed to have several point mutations, which are underlined, in both binding sites; while the Mut-2 was constructed by point mutations at only binding site-3. f Luciferase reporter assay. HeLa cells were transfected with different combinations of the ZFP-148-3′UTR (Wt) or mutant (Mut) plasmid or vector only (pmir-GLo) and miR-203 as indicated. Luciferase reporter assay was conducted and the relative luciferase activity was calculated after normalizing the data against that of HeLa cells cotransfected with Wt + miR-CL, which was set as 1.0. Data are mean ± SD. *P < 0.05
To validate ZFP-148 as a true target of miR-203, a luciferase reporter plasmid, carrying a 3′UTR fragment of ZFP-148 (nts 7,061–7,504) containing the downstream two sites (site-2 and site-3) of miR-203 (Fig. 4a), was constructed. Two mutant constructs, ZFP-148-3′UTR-Mut-1 and ZFP-148-3′UTR-Mut-2, were also generated by site-directed mutagenesis of the miR-203 seed match sequence (Fig. 4e). The luciferase reporter constructs were co-transfected with pre-miR-203 or pre-miR-CL into HeLa cells. The luciferase assay data revealed that HeLa cells co-transfected with Wt ZFP-148-3′UTR (containing two binding sites) and miR-203 (Wt + miR-203) showed an up to 70 % reduction of the relative luciferase activity as compared to the control cells co-transfected with ZFP-148-3′UTR and pre-miR-CL (Wt + miR-CL) (Fig. 4f). In contrast, when one site or both sites together were mutated, miR-203 showed no significant targeting effect on reporter gene expression, indicating that ZFP-148 is a novel and specific target gene of miR-203.
miR-203 promotes CVB3 replication in HeLa cells
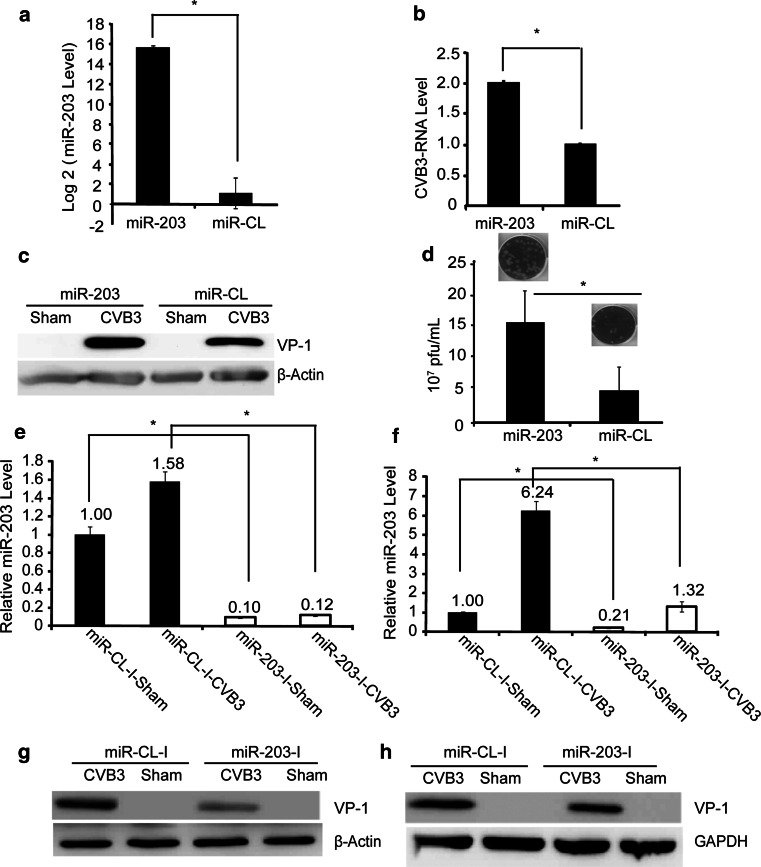
To study the positive effect of miR-203 upregulation on CVB3 replication, we transfected HeLa cells with either pre-miR-203 or pre-miR-CL for 48 h and then infected with CVB3 at a multiplicity of infection (MOI) of 10 or sham-infected with PBS for 6 h. Q-RT-PCR showed that pre-miR-203-transfected cells had an approximately 15-log2 increase in miR-203 expression as compared to the control cells (Fig. 5a). The enhanced CVB3 replication efficiency was evaluated by Q-RT-PCR to detect viral RNA transcription (Fig. 5b) and western blot to detect CVB3 VP-1 protein production (Fig. 5c), respectively. This miR-203-promoted CVB3 replication was further confirmed by viral plaque assay, showing a significant increase in viral particle release in miR-203-expressing cells as compared to the control (Fig. 5d). Meanwhile, inhibition of the endogenous levels of miR-203 in both HeLa and HL-1 cells using specific inhibitors resulted in a marked reduction of miR-203 expression (Fig. 5e, f), respectively, and also caused significant downregulation of CVB3-VP-1 expression in both cell lines (Fig. 5g, h).
Fig. 5.
MiR-203 expression promotes CVB3 replication in HeLa cells. a Q-RT-PCR to detect mature miR-203 levels in HeLa cells transfected with pre-miR-203 or pre-miR-CL. U6 was used as an endogenous control to normalize the results. b Q-RT-PCR to measure CVB3 RNA expression. HeLa cells were transfected as described above and then infected with CVB3. RNAs extracted from the cell culture supernatants were used to measure the CVB3 RNA using primers targeting viral 2A gene. The data of the control was set as 1.0. c Western blot to detect CVB3 VP-1 expression. HeLa cells were transfected and then infected with CVB3 as described above. The protein lysates were collected for western blot analysis. β-actin served as a loading control. d Viral plaque assay. HeLa cell culture supernatants described in (b) were employed to determine CVB3 particle formation by plaque assay on HeLa cell monolayers. e, f Q-RT-PCR to detect miRNA-203 expression levels after treatment of HeLa cells (e) and HL-1 cells (f) with miR-203 inhibitors for 48 h and then infection with 10 MOI of CVB3 for 5 and 8 h, respectively. U6 and snoR-203 as endogenous controls used to normalize our data for HeLa cells and HL-1 cells respectively. g, h Western blot to detect VP-1 in HeLa (g) and HL-1(h) cells transfected with miR-203 inhibitors or control inhibitors and then infected with CVB3
Silencing of ZFP-148 with specific siRNAs benefits CVB3 replication
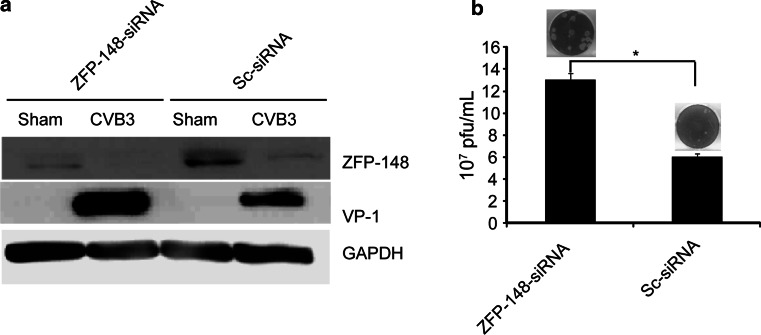
To further verify the effect of ZFP-148 downregulation on CVB3 replication, we transfected HeLa cells with specific siRNAs to silence the ZFP-148 expression and then infected the cells with CVB3 for 6 h. The suppression of the ZFP-148 expression and the effect on CVB3 replication were detected by western blot analysis and viral plaque assay, respectively. Figure 6a demonstrated that ZFP-148 expression was dramatically suppressed and CVB3 VP-1 production was significantly increased in cells transfected with ZFP-148 siRNAs as compared to the control transfected with scrambled siRNAs. These data were further confirmed by plaque assay, showing that silencing of the ZFP-148 resulted in a significant increase of CVB3 progeny release (Fig. 6b).
Fig. 6.
Silencing of ZFP-148 with specific siRNA promotes CVB3 replication. a HeLa cells were transfected with ZFP-148-specific siRNA or scrambled siRNA and then infected with CVB3 for 6 h or sham-infected with PBS. ZFP-148 and CVB3 VP-1 expression was detected by western blot analysis. b Viral particle releases. Cell culture supernatants collected from HeLa cell culture described in (a) above were used for plaque assay to detect CVB3 particle release. Values shown here are means of ±SD of three independent experiments, *P < 0.05
Ectopic expression of miR-203 promotes cell survival
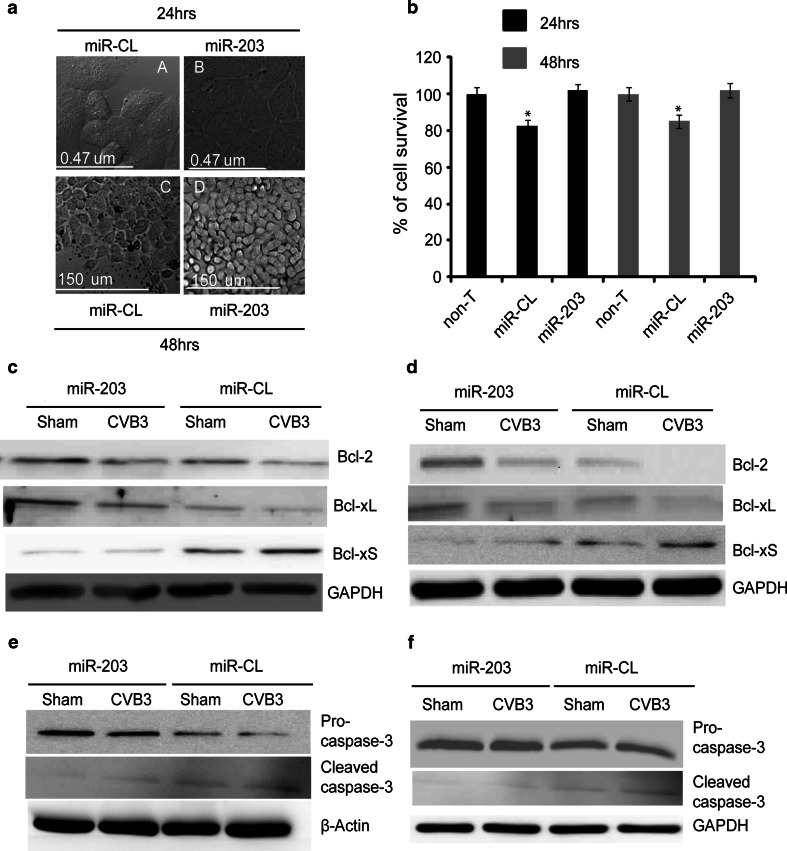
In further elucidating the mechanism by which miR-203 promotes CVB3 replication, we speculated that miR-203 expression might promote host cell growth and thus provide favourable conditions for CVB3 replication. To test this hypothesis, we first examined the morphological changes in miR-203-expressing HeLa cells by live cell confocal imaging. Figure 7a shows that miR-203 expression (panels B and D) promoted cell proliferation at both 24 and 48 h post-transfection as compared to the control cells transfected with pre-miR-CL (panels A and C). This miR-203-promoted cell growth was further verified by cell viability assay at 24 and 48 h post-transfection. Figure 7b demonstrates that cells transfected with pre-miR-203 had a 20 % increase in cell survival as compared to the control samples at both 24 and 48 h post-transfection. This is likely because miR-203 (but not the miR-CL) expression can counteract the unfavourable effects created by transfection reagents and resume the normal environment for cell growth. Further analysis by western blot revealed the upregulation of the survival markers (Bcl-2 and Bcl-xL) as well as the downregulation of the apoptotic marker (Bcl-xS) in pre-miR-203-transfected HeLa cells (Figs. 7c; S2a, c, e) and HL-1 cardiomyocytes (Figs. 7d; S2b, d, f) that were either sham- or CVB3-infected for 6 h. In addition, we also detected the activation of caspase-3 and demonstrated that miR-203 expression suppressed the cleavage of pro-caspase-3 in HeLa cells (Figs. 7e, S3a, b) and HL-1 cells (Fig. 7f, S3c, d). Here, it is worth mentioning that the image of cleavage product of caspase-3 is weak. This is probably due to ubiquitin-proteasome-mediated protein degradation caused by viral infection or other cellular stresses [26].
Fig. 7.
MiR-203 expression enhances HeLa cell growth. a Morphological changes. HeLa cells were transfected as described above and cell morphology was analyzed by live cell confocal imaging at 24 and 48 h post-transfection. Note that HeLa cells transfected with pre-miR-203 (B and D) have more cells than that of the control cells (A and C) at both time points. b MTS cell viability assay. HeLa cells were transfected as described above and MTS assay was conducted at 24 and 48 h post-transfection. The absorbance of non-treated (non-T) cells was defined as 100 % survival (control). Values shown here are means of ±SD of three independent experiments, *P < 0.05. c, d Western blot analyses of Bcl-2 family proteins in pre-miR-203 or pre-miR-CL transfected HeLa cells (c) and HL-1 cardiomyocytes (d), respectively, at 6 h pi. e, f Western blot analyses of pro-caspase-3 cleavage in pre-miR-203 or pre-miR-CL-transfected HeLa cells and HL-1 cells, respectively, at 6 h pi
Suppression of ZFP-148 by miR-203 leads to the differential expression of cell cycle regulators
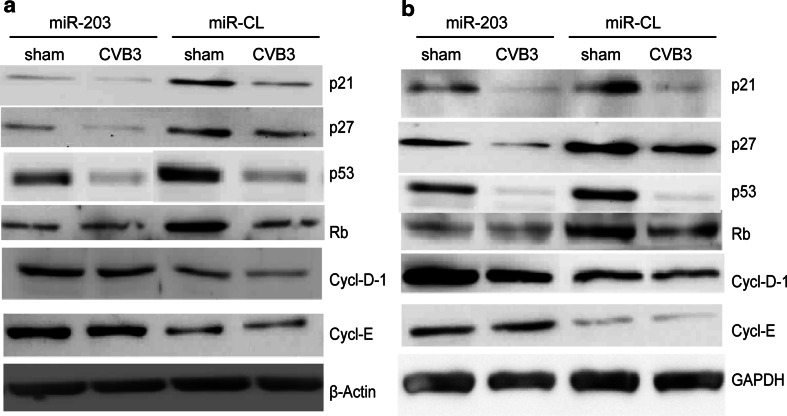
The data presented in Figs. 5, 6 and 7 above suggest that miR-203 enhances CVB3 replication, likely through promoting host cell viability. Thus, we next investigated the signalling pathways leading to the enhanced cell survival/growth in the conditions of suppression of ZFP-148 expression by miR-203. We detected the expression levels of the downstream genes of ZFP-148 involved in cell cycle regulation, which include cyclin-dependent kinase inhibitors (p21Waf1 and, p27Kip1), tumor suppressors [retinoblastoma (Rb) and p53] and cell cycle progression genes (cyclin E and cyclin D1) [27, 28], in pre-miR-203-transfected HeLa cells infected with CVB3 or sham-infected with PBS. Figures 8a and S4 demonstrate that these cell cycle arrest genes and tumor suppressor genes were markedly downregulated, while the cell cycle progression genes (cyclin E and cyclin D1) were significantly upregulated in the miR-203 expressing cells as compared to the control. These data were further confirmed by experiments using HL-1 cardiomyocytes (Figs. 8b; S5). It is worth noting that, in CVB3-infected samples, the changes of expression levels of certain responsive genes such as p21, p53 and Rb were not dramatic as compared to that in sham-infected cells. This is probably due to the shut-down of host gene expression by CVB3 infection and thus masked the effect of miRNA-203. The effect of ZFP-148 suppression by miR-203 on downstream gene expression was further verified in HeLa cells by silencing ZFP-148 with siRNA, which showed the downregulation of p53, p21Waf1 and p27Kip1 (Fig. S6). Finally, we also demonstrated the downregulation of p53, p21Waf1 and p27Kip1 as well as the upregulation of DCM gene makers GATA4 and Mef2a in CVB3-infected mouse hearts used for microarray analysis of miRNA expression profiles (Fig. S7).
Fig. 8.
MiR-203 expression induces differential expression of downstream responsive genes of ZFP-148. HeLa cells (a) and HL-1 cell (b) were transfected with either the pre-miR-203 or control miRNA and then infected with CVB3 as described above. Western blot analyses were conducted to detect the expression of cell cycle regulators or tumour suppressors using indicated antibodies
Discussion
CVB3 is a cardiotropic virus and can reach a high viral load and cause massive damage to the heart in a very short duration after infection. This infection is initiated by entry of host cells through viral receptor, which then hijacks the host gene expression machinery to benefit its own replication [29]. However, the underlying mechanism has not been precisely defined. miRNAs are a group of newly identified regulators of gene expression at the post-transcriptional level and are involved in many human disorders, including cardiovascular diseases [11]. To identify the miRNA candidates regulating CVB3 infectivity and replication, we performed miRNA microarray analyses and identified a number of differentially expressed miRNAs in CVB3-infected heart of A/J mice, an established viral myocarditis mouse model. One of the significantly upregulated candidate miRNAs was miR-203 and its expression level was increased at least 1.6-fold as compared to the sham-infected control mice, implying its role in gene regulation during CVB3 infection. This upregulation was further validated by Q-RT-PCR. To explore the signalling pathway leading to miR-203 upregulation, we focused on the analyses of the PKC/AP-1 cascade, as AP-1, a transcription factor consisting of homodimers or heterodimers of the Jun and Fos families of nuclear proteins [30], is known to be involved in cell differentiation, proliferation and apoptosis [31]. Although it has been reported that miR-203 upregulation is modulated through PKC/AP-1 activation during keratinocyte differentiation [24], there is no report on the transcriptional regulation of miR-203 in a virus-infected cardiomyocyte model system. Our data demonstrated that CVB3 infection of mouse heart or HL-1 cardiomyocytes activated PKC and further upregulated JunB, a positive regulator of miR-203, and meanwhile downregulated c-Jun, a negative regulator of miR-203 [24]. These data were further solidified by using siRNAs targeting JunB, which suppressed JunB, upregulated p-cJun, and subsequently downregulated miR-203 expression. These data suggest that miR-203 upregulation in CVB3-infected heart is through the activation of the PKC/AP-1 cascade.
miR-203 has been previously described as one of the skin-abundant miRNAs which promotes differentiation by repressing ‘stemness’ [32]. It was also found to be overexpressed in pancreatic adenocarcinoma and rheumatoid arthritis [33]. To elucidate the biological function of miR-203 upregulation during CVB3 infection, we identified the target genes of miR-203. We conducted bioinformatic prediction and found a number of target candidates for miR-203, which included ZFP-148 and several known targets, such as p63, SOCS3, ABL-1, etc. [24, 32]. We chose ZFP-148 as a candidate for further study not only because it is an unknown new target of miR-203 but also for the following reasons: (1) ZFP-148 is a transcription factor that regulates the expression of many cell cycle regulatory genes, such as p21WAF1 and p27KIP1, as well as the genes for apoptosis induction pathways [19, 34]; (2) ZFPs act as antiviral agents for many viruses and inhibit viral replication through different mechanisms inducing inhibition of the viral RNA synthesis and induction of host cell apoptosis [35, 36]; (3) ZFP-148 was reported to be involved in the pathogenesis of many diseases, such as colon cancer development, neuroectodermal tumours, and myocardial infarction [37, 38,] and (4) bioinformatics prediction showed that ZFP-148 has three miR-203 target sites (with high scores) and two of these sites are close to each other and located near the 3′ end region of the 3′UTR (nts 4,180–4,186 and 4,533–4,539, respectively) [39]. This structural organization supports current knowledge that one given miRNA usually has more than one binding site within the 3′UTR of the target gene and presents in a cluster manner [40]. This selection of ZFP-148, as the target gene of miR-203 was finally confirmed to be correct by the following findings: (1) ectopic expression of miR-203 suppressed ZFP-148 translation and this suppression was verified by western blot analysis using mouse heart tissue with CVB3-induced upregulation of miR-203 expression; and (2) luciferase assay by co-transfection of different combinations of reporter plasmids (Wt or mutant) and miR-203 demonstrated that only the co-transfection with Wt reporter and pre-miR-203 could dramatically reduce the luciferase activity. It is worth mentioning that, in the luciferase reporter assays to verify the binding sites, we only selected site-2 and site-3 for the following reasons: (1) the effective sites preferentially reside near both end regions of the 3′UTR, and (2) closely spaced sites often act synergistically [41]. Our data support the previous finding [41] and indicate that ZFP-148 is a true target gene of miR-203.
To study the effect of suppression of ZFP-148 by miR-203 on cellular signal networks associating with CVB3 replication, we first demonstrated that ectopic expression of miR-203 promoted cell growth and enhanced CVB3 replication, which is supported by the increased cell viability and decreased activation of caspase-3. Similar results were obtained by transfection of cells with siRNAs targeting ZFP-148, showing that, upon suppression of ZFP-148, CVB3 replication was enhanced. These data provide evidence to support the fact that miR-203 benefits CVB3 replication by targeting ZFP-148, which is involved in the regulation of cell growth [28, 42, 43]. To further elucidate the mechanism by which miR-203 promotes CVB3 replication by targeting ZFP-148, we next performed experiments to characterize the expression of downstream responsive genes of ZFP-148 in the signal transduction pathway leading to cell survival and growth. ZFP-148 is known as a potent repressor of the Bcl-xL, one of the pro-survival proteins in the BcL-2 family [44]. We found that Bcl-xL was upregulated while Bcl-xS (a proapoptotic marker in the BcL-2 family) was downregulated in pre-miR-203-transfected HeLa and HL-1 cells. ZFP-148 is also a transcription factor that governs a number of cell cycle regulatory genes, such as p21Waf1 and p27Kip1, through binding to the GC-rich region of its promoter [45]. The p21Waf1 is a cyclin-dependent kinase inhibitor (CDKI) which arrests cell cycle progression at the G1 phase in response to various stress factors [46]. Previous studies have also reported that p21Waf1 was involved in the inhibition of cardiac hypertrophy and DCM occurrence through inhibiting ROCK kinases [47]. Our data demonstrated the downregulation of p21Waf1 in the pre-miR-203-transfected cells as compared to the control group. This downregulation of p21Waf1 may contribute to the CVB3-induced DCM, a heart disease characterized by heart enlargement, chamber dilation and increased ventricular wall thickness. This speculation was further supported by the downregulation of p21Waf1 as well as the upregulation of DCM markers, Mef2a and GATA4, in CVB3-infected mouse hearts at day 4 pi; however, the remodelling of the heart is not obvious at this early time point (data not shown). p27Kip1, another CDKI involved in the regulation of the cell cycle [48], was also downregulated in pre-miR-203-transfected cells. This downregulation is believed to further contribute to the cell growth. We further demonstrated the upregulation of cell cycle progression genes (cyclin D-1 and cyclin E) and the downregulation of the tumour suppressor p53 and Rb in miRNA-203-expressing HeLa cells and HL-1 cells. Based on all these data, we proposed a signal transduction model wherein CVB3-induced upregulation of miR-203 expression inhibits ZFP-148 translation, which results in differential regulation of downstream pro-survival and pro-apoptotic genes, leading to enhanced cell growth and creating an optimum survival environment to support CVB3 infection and replication (Fig. 9). This signal pathway produces a positive feedback loop at the early stage of CVB3 infection to continue the promotion of CVB3 replication. However, at late stage of infection, when viruses reach a high titre, they will induce cell apoptosis and damage the cardiomyocytes, which is likely the underlying mechanism of viral pathogenesis.
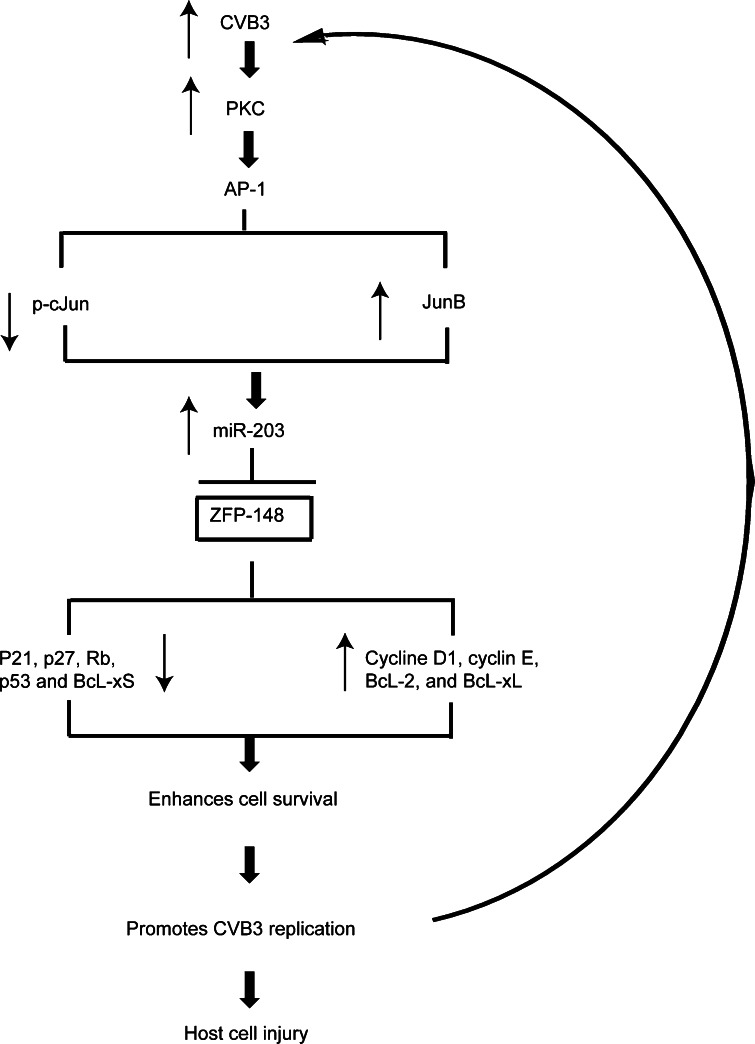
Fig. 9.
A putative model of miR-203 action leading to enhanced CVB3 replication. CVB3 infection induces upregulation of miR-203 via the PKC-AP-1 signalling cascade, which in turn suppresses its target gene ZFP-148 expression. The suppression of ZFP-148 translation results in downregulation of pro-apoptotic/cell cycle arrest genes and upregulation of prosurvival/cell growth genes, leading to cell survival/growth, which benefits CVB3 replication. Persistent infection of CVB3 triggers a positive feedback loop for viral infection and replication, which subsequently causes target cell injury and disease. The thin arrows indicate up- or downregulation of gene expression
In summary, our data demonstrated that CVB3 infection caused upregulation of miR-203 expression in mouse hearts at early stage (day 4 pi) of infection. MiR-203 expression downregulated ZFP-148 mRNA translation by specifically targeting its 3′UTR, which resulted in differential expression of its downstream target genes, such as cell cycle regulators, tumour suppressors and Bcl-2 family proteins, which in turn promote cell survival to foster CVB3 replication. To our knowledge, this is the first report on ZFP-148 as a novel target of miR-203 as well as the connection between the miR-203 targeting of ZFP-148 and the pathogenesis of CVB3-induced myocarditis. Therefore, our finding may provide a potential strategy in developing anti-CVB3 interventions.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
Dr. Maged Gomaa Hemida is a recipient of the CIHR-IMPACT postdoctoral training fellowship and the Heart and Stroke foundation of Canada postdoctoral training fellowship. Xin Ye is supported by a UGF Award from the University of British Columbia. We would like to thank Claire Smith and Lubos Bohunek for their help with animal experiments. We also want to thank Dr. Thomas Ibraham for his help with the confocal microscopy.
Abbreviations
- CVB3
Coxsackievirus B3
- DCM
Dilated cardiomyopathy
- DMEM
Dulbecco’s modified Eagle’s medium
- miRNA
microRNA
- MOI
Multiplicity of infection
- PKC
Protein kinase C
- pi
Post-infection
- pfu
Plaque-forming unit
- UTR
Untranslated region
- ZFP
Zinc finger protein
Footnotes
Maged Gomaa Hemida and Xin Ye contributed equally to this work.
References
- 1.Esfandiarei M, McManus BM. Molecular biology and pathogenesis of viral myocarditis. Annu Rev Pathol. 2008;3:127–155. doi: 10.1146/annurev.pathmechdis.3.121806.151534. [DOI] [PubMed] [Google Scholar]
- 2.Kuhl U, Pauschinger M, Seeberg B, Lassner D, Noutsias M, Poller W, Schultheiss HP. Viral persistence in the myocardium is associated with progressive cardiac dysfunction. Circulation. 2005;112(13):1965–1970. doi: 10.1161/CIRCULATIONAHA.105.548156. [DOI] [PubMed] [Google Scholar]
- 3.Cheung PK, Yuan J, Zhang HM, Chau D, Yanagawa B, Suarez A, McManus B, Yang D. Specific interactions of mouse organ proteins with the 5′ untranslated region of Coxsackievirus B3: potential determinants of viral tissue tropism. J Med Virol. 2005;77(3):414–424. doi: 10.1002/jmv.20470. [DOI] [PubMed] [Google Scholar]
- 4.Bergelson JM, Cunningham JA, Droguett G, Kurt-Jones EA, Krithivas A, Hong JS, Horwitz MS, Crowell RL, Finberg RW. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275(5304):1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cullen BR. Five questions about viruses and microRNAs. PLoS Pathog. 2010;6(2):e1000787. doi: 10.1371/journal.ppat.1000787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 8.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 9.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 10.Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31(5):766–776. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- 11.Ikeda S, Pu WT. Expression and function of microRNAs in heart disease. Curr Drug Targets. 2010;11(8):913–925. doi: 10.2174/138945010791591304. [DOI] [PubMed] [Google Scholar]
- 12.Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449(7164):919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ziegelbauer JM, Sullivan CS, Ganem D. Tandem array-based expression screens identify host mRNA targets of virus-encoded microRNAs. Nat Genet. 2009;41(1):130–134. doi: 10.1038/ng.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choy EY, Siu KL, Kok KH, Lung RW, Tsang CM, To KF, Kwong DL, Tsao SW, Jin DY. An Epstein–Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med. 2008;205(11):2551–2560. doi: 10.1084/jem.20072581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435(7042):682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 16.Voellenkle C, van Rooij J, Cappuzzello C, Greco S, Arcelli D, Di Vito L, Melillo G, Rigolini R, Costa E, Crea F, Capogrossi MC, Napolitano M, Martelli F. MicroRNA signatures in peripheral blood mononuclear cells of chronic heart failure patients. Physiol Genomics. 2010;42(3):420–426. doi: 10.1152/physiolgenomics.00211.2009. [DOI] [PubMed] [Google Scholar]
- 17.Cheng PY, Kagawa N, Takahashi Y, Waterman MR. Three zinc finger nuclear proteins, Sp1, Sp3, and a ZBP-89 homologue, bind to the cyclic adenosine monophosphate-responsive sequence of the bovine adrenodoxin gene and regulate transcription. Biochemistry. 2000;39(15):4347–4357. doi: 10.1021/bi992298f. [DOI] [PubMed] [Google Scholar]
- 18.Yuan J, Liu Z, Lim T, Zhang H, He J, Walker E, Shier C, Wang Y, Su Y, Sall A, McManus B, Yang D. CXCL10 inhibits viral replication through recruitment of natural killer cells in Coxsackievirus B3-induced myocarditis. Circ Res. 2009;104(5):628–638. doi: 10.1161/CIRCRESAHA.108.192179. [DOI] [PubMed] [Google Scholar]
- 19.Luo L, Ye L, Liu G, Shao G, Zheng R, Ren Z, Zuo B, Xu D, Lei M, Jiang S, Deng C, Xiong Y, Li F. Microarray-based approach identifies differentially expressed microRNAs in porcine sexually immature and mature testes. PLoS One. 2010;5(8):e11744. doi: 10.1371/journal.pone.0011744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan J, Stein DA, Lim T, Qiu D, Coughlin S, Liu Z, Wang Y, Blouch R, Moulton HM, Iversen PL, Yang D. Inhibition of Coxsackievirus B3 in cell cultures and in mice by peptide-conjugated morpholino oligomers targeting the internal ribosome entry site. J Virol. 2006;80(23):11510–11519. doi: 10.1128/JVI.00900-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mestdagh P, Feys T, Bernard N, Guenther S, Chen C, Speleman F, Vandesompele J. High-throughput stem-loop RT-qPCR miRNA expression profiling using minute amounts of input RNA. Nucleic Acids Res. 2008;36(21):e143. doi: 10.1093/nar/gkn725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paloheimo O, Ihalainen TO, Tauriainen S, Valilehto O, Kirjavainen S, Niskanen EA, Laakkonen JP, Hyoty H, Vihinen-Ranta M. Coxsackievirus B3-induced cellular protrusions: structural characteristics and functional competence. J Virol. 2011;85(13):6714–6724. doi: 10.1128/JVI.00247-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye X, Liu Z, Hemida MG, Yang D. Targeted delivery of mutant tolerant anti-coxsackievirus artificial microRNAs using folate conjugated bacteriophage Phi29 pRNA. PLoS One. 2011;6(6):e21215. doi: 10.1371/journal.pone.0021215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sonkoly E, Wei T, Pavez Lorie E, Suzuki H, Kato M, Torma H, Stahle M, Pivarcsi A. Protein kinase C-dependent upregulation of miR-203 induces the differentiation of human keratinocytes. J Invest Dermatol. 2010;130(1):124–134. doi: 10.1038/jid.2009.294. [DOI] [PubMed] [Google Scholar]
- 25.Moffatt CE, Lamont RJ. Porphyromonas gingivalis induction of microRNA-203 expression controls suppressor of cytokine signaling 3 in gingival epithelial cells. Infect Immun. 2011;79(7):2632–2637. doi: 10.1128/IAI.00082-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Si X, Gao G, Wong J, Wang Y, Zhang J, Luo H. Ubiquitination is required for effective replication of Coxsackievirus B3. PLoS One. 2008;3(7):e2585. doi: 10.1371/journal.pone.0002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lowe SW, Sherr CJ. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr Opin Genet Dev. 2003;13(1):77–83. doi: 10.1016/S0959-437X(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 28.Zhang CZ, Chen GG, Lai PB. Transcription factor ZBP-89 in cancer growth and apoptosis. Biochim Biophys Acta. 2010;1806(1):36–41. doi: 10.1016/j.bbcan.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 29.Shi Y, Chen C, Lisewski U, Wrackmeyer U, Radke M, Westermann D, Sauter M, Tschope C, Poller W, Klingel K, Gotthardt M. Cardiac deletion of the Coxsackievirus-adenovirus receptor abolishes Coxsackievirus B3 infection and prevents myocarditis in vivo. J Am Coll Cardiol. 2009;53(14):1219–1226. doi: 10.1016/j.jacc.2008.10.064. [DOI] [PubMed] [Google Scholar]
- 30.Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J Cell Sci. 2004;117(Pt 25):5965–5973. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- 31.Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001;20(19):2390–2400. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- 32.Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature. 2008;452(7184):225–229. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stanczyk J, Ospelt C, Karouzakis E, Filer A, Raza K, Kolling C, Gay R, Buckley CD, Tak PP, Gay S, Kyburz D. Altered expression of microRNA-203 in rheumatoid arthritis synovial fibroblasts and its role in fibroblast activation. Arthr Rheum. 2011;63(2):373–381. doi: 10.1002/art.30115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmittgen TD, Jiang J, Liu Q, Yang L. A high-throughput method to monitor the expression of microRNA precursors. Nucleic Acids Res. 2004;32(4):e43. doi: 10.1093/nar/gnh040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sera T. Inhibition of virus DNA replication by artificial zinc finger proteins. J Virol. 2005;79(4):2614–2619. doi: 10.1128/JVI.79.4.2614-2619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zimmerman KA, Fischer KP, Joyce MA, Tyrrell DL. Zinc finger proteins designed to specifically target duck hepatitis B virus covalently closed circular DNA inhibit viral transcription in tissue culture. J Virol. 2008;82(16):8013–8021. doi: 10.1128/JVI.00366-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Law DJ, Labut EM, Merchant JL. Intestinal overexpression of ZNF148 suppresses ApcMin/+ neoplasia. Mamm Genome. 2006;17(10):999–1004. doi: 10.1007/s00335-006-0052-4. [DOI] [PubMed] [Google Scholar]
- 38.Yuan J, Cheung PK, Zhang H, Chau D, Yanagawa B, Cheung C, Luo H, Wang Y, Suarez A, McManus BM, Yang D. A phosphorothioate antisense oligodeoxynucleotide specifically inhibits Coxsackievirus B3 replication in cardiomyocytes and mouse hearts. Lab Invest. 2004;84(6):703–714. doi: 10.1038/labinvest.3700083. [DOI] [PubMed] [Google Scholar]
- 39.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38(Suppl):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 40.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 41.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27(1):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng Y, Wang X, Xu L, Pan H, Zhu S, Liang Q, Huang B, Lu J. The transcription factor ZBP-89 suppresses p16 expression through a histone modification mechanism to affect cell senescence. FEBS J. 2009;276(15):4197–4206. doi: 10.1111/j.1742-4658.2009.07128.x. [DOI] [PubMed] [Google Scholar]
- 43.Salmon M, Owens GK, Zehner ZE. Over-expression of the transcription factor, ZBP-89, leads to enhancement of the C2C12 myogenic program. Biochim Biophys Acta. 2009;1793(7):1144–1155. doi: 10.1016/j.bbamcr.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bai L, Yoon SO, King PD, Merchant JL. ZBP-89-induced apoptosis is p53-independent and requires JNK. Cell Death Differ. 2004;11(6):663–673. doi: 10.1038/sj.cdd.4401393. [DOI] [PubMed] [Google Scholar]
- 45.Merchant JL, Bai L, Okada M. ZBP-89 mediates butyrate regulation of gene expression. J Nutr. 2003;133(7 Suppl):2456S–2460S. doi: 10.1093/jn/133.7.2456S. [DOI] [PubMed] [Google Scholar]
- 46.Klopfleisch R, Gruber AD. Differential expression of cell cycle regulators p21, p27 and p53 in metastasizing canine mammary adenocarcinomas versus normal mammary glands. Res Vet Sci. 2009;87(1):91–96. doi: 10.1016/j.rvsc.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 47.Hauck L, Harms C, Grothe D, An J, Gertz K, Kronenberg G, Dietz R, Endres M, von Harsdorf R. Critical role for FoxO3a-dependent regulation of p21CIP1/WAF1 in response to statin signaling in cardiac myocytes. Circ Res. 2007;100(1):50–60. doi: 10.1161/01.RES.0000254704.92532.b9. [DOI] [PubMed] [Google Scholar]
- 48.Chiarle R, Pagano M, Inghirami G. The cyclin dependent kinase inhibitor p27 and its prognostic role in breast cancer. Breast Cancer Res. 2001;3(2):91–94. doi: 10.1186/bcr277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.