Abstract
In mammals, pronucleus formation, a landmark event for egg activation and fertilization, is critical for embryonic development. However, the mechanisms underlying pronucleus formation remain unclear. Increasing evidence has shown that the transition from a mature egg to a developing embryo and the early steps of development are driven by the control of maternal cytoplasmic factors. Herein, a two-dimensional-electrophoresis-based proteomic approach was used in metaphase II and parthenogenetically activated mouse eggs to search for maternal proteins involved in egg activation, one of which was poly(rC)-binding protein 1 (PCBP1). Phosphoprotein staining indicated that PCBP1 displayed dephosphorylation in parthenogenetically activated egg, which possibly boosts its ability to bind to mRNAs. We identified 75 mRNAs expressed in mouse eggs that contained the characteristic PCBP1-binding CU-rich sequence in the 3′-UTR. Among them, we focused on H2a.x mRNA, as it was closely related to pronucleus formation in Xenopus oocytes. Further studies suggested that PCBP1 could bind to H2a.x mRNA and enhance its stability, thus promoting mouse pronucleus formation during parthenogenetic activation of murine eggs, while the inhibition of PCBP1 evidently retarded pronucleus formation. In summary, these data propose that PCBP1 may serve as a novel maternal factor that is required for determining the normal timing of pronucleus formation.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-015-1905-3) contains supplementary material, which is available to authorized users.
Keywords: hnRNP E1, mRNA stabilization, Pronuclear development, Microinjection
Introduction
At the end of oogenesis, mature oocytes are held in a developmentally quiescent state until an appropriate trigger initiates development. The process whereby the oocyte undergoes the final transition from this arrested state to a new cellular state that can support embryogenesis is called egg activation [1, 2]. Egg activation involves a stepwise series of events, including cortical granule exocytosis, release of oocyte meiotic arrest, and formation of haploid female pronucleus that is capable of combining with the haploid male pronucleus. Maternal mRNAs and proteins underwent dynamic changes during egg activation, including recruitment and regulated degradation of maternal mRNAs, translation, and post-translational modifications. These changes during egg activation provide necessary support to the zygote’s growth and development [1–4].
Pronucleus formation, a landmark event for egg activation and fertilization, is critical for embryonic development. The female pronucleus is assembled upon egg activation from a condensed organization of chromatin and relies on contributions from cytoplasmic components. Whilst male pronucleus development generally requires egg activation in most mammalian species, once the second polar body extrusion and female pronucleus construction begin, the sperm nuclei undergo rapid morphological and biochemical transformations. These changes include disassembly of the nuclear lamina, decondensation of chromatin, replacement of protamines by maternal histones, reassembly of a nuclear envelope, nuclear swelling, and formation of a male pronucleus [5–8]. Fusion of female and male pronuclei allows their DNA to combine, resulting in a diploid zygote and the completion of fertilization.
Increasing evidence has shown that the transition from a mature egg to a developing embryo and the early steps of development are driven by the control of maternal cytoplasmic factors (mRNAs and proteins), that are produced during oogenesis and stored in the mature egg. Upon egg activation and fertilization, these maternal factors initiate development cascades that carry out the subsequent embryonic development program [2, 9]. Furthermore, a wealth of evidence has demonstrated the existence of maternal factors that affect various nuclear functions including chromatin assembly and the structure and establishment of components of the nuclear structure, such as nucleoplasmin, histones, and nuclear lamins [10].
Despite the crucial role of maternal factors in egg activation, only a few factors have been identified. Poly(rC)-binding protein 1 (PCBP1), also referred to as hnRNP E1 or αCP1, is a maternal protein that was identified in the protein profile of mouse metaphase II (MII) eggs [11, 12]. PCBP1 contains highly conserved triple repeats of the KH domain and belongs to the superfamily of nucleic acid-binding proteins, which are involved in a variety of mRNA processing steps, including transcription [13], nucleocytoplasmic mRNA shuttling [14], mRNA stability [15–19], mRNA translation [20–22], and protein–protein interactions [23, 24]. In our previous work [25], we showed that PCBP1 plays a key role in maintaining the transcriptionally silent state in the fully grown oocyte, which is a prerequisite for preovulatory oocytes to eventually achieve meiotic maturation, fertilization, and early embryonic development.
In this study, we examined differentially expressed proteins in normal MII eggs compared with parthenogenetically activated mouse eggs through two-dimensional gel electrophoresis (2-DE). PCBP1 was identified as a differentially expressed protein and found to be required for determining the normal timing of pronucleus formation during egg activation.
Materials and methods
Reagents and animals
All reagents were from Sigma Chemicals (St Louis, MO, USA), unless stated otherwise. ICR white mice were maintained under a controlled environment of 20–22 °C, 12/12-h light/dark cycle, and 50–70 % humidity, with food and water provided ad libitum. Animal care was conducted in accordance with the Animal Research Committee guidelines of Nanjing Medical University.
Egg collection and culture
Mature cumulus-free eggs were collected from superovulated 6- to 8-week-old ICR female mice as previously described [11]. Parthenogenetically activated eggs were obtained by culturing cumulus-free eggs in Ca2+-free CZB medium supplemented with 5.56 mM d-glucose containing both 10 mM SrCl2 and 5 μg/mL cytochalasin B for 1 h at 37 °C in a humidified atmosphere of 5 % CO2. The zona pellucida was then removed by treating the eggs for 3 min in acid PBS (Obiogene, Carlsbad, CA, USA) followed by mechanical shearing. Both denuded MII eggs and parthenogenetically activated eggs were extracted in lysis buffer as previously described [11] and used for proteomic studies.
Two-dimensional gel electrophoresis (2-DE) and protein identification
Lysates from MII eggs and parthenogenetically activated eggs were used for 2-DE to compare the protein expression profiles [11]. A spot was regarded as differentially expressed between groups if the difference in spot intensity was greater than twofold. The differential spots were excised and identified as previously described [26].
Phosphoprotein separation and detection
For phosphoprotein detection, MII egg protein extracts (30 μg) were separated by 2-DE and stained with Pro-Q Diamond phosphoprotein gel stain (Molecular Probes, Eugene, OR, USA) [11].
Bioinformatics analysis
PCBP1 binds to a pyrimidine-rich cluster conforming to the consensus sequence (C/U)CCANxCCC(U/A)PyxUC(C/U)CC [15, 16, 18, 19, 27] in the 3′-UTR of mRNAs. We searched for this consensus sequence in the 3′-UTR of the mRNAs expressed in mouse oocytes in the UTResource database (http://www.ba.itb.cnr.it/UTR/) [28]. An mRNA was determined to be expressed in oocytes if there was corresponding expressed sequence tags (ESTs) in the library of the Eppig Hampl Solter fully grown mouse oocyte (Library ID: dbEST:18552) [29].
RNA immunoprecipitation (RIP) assay
RNA immunoprecipitation was performed using the EZ-Magna RIP kit (Millipore, Billerica, MA, USA) following the manufacturer’s protocol. Ovaries from 10 wild-type ICR mice were fully ground in liquid nitrogen and then lysed in complete RIP lysis buffer. The ovary protein extracts were then incubated with RIP buffer containing magnetic beads conjugated with a goat anti-PCBP1 antibody (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) or negative control normal goat IgG (Santa Cruz Biotechnology Inc.). Samples were incubated with Protein K with shaking to digest the protein and then immunoprecipitated RNA was isolated. Purified RNA was subjected to qRT-PCR analysis to evaluate the level of H2a.x mRNA, and the primer sequences targeting the binding region were as follows: H2a.x: (sense) 5′-CCGGCCTGTGGACAAGAGT-3′ and (antisense) 5′-GGTGTTTGGGTTGTAGTTGAGT-3′. AR (androgen receptor), a well-known target that binds with PCBP1 [30], was amplified as a positive control, and Smarca4, which lacks the PCBP1-binding motif in its sequence, was also analyzed in the immunoprecipitated RNA. The primer sequences were as follows: AR (sense) 5′-CCCACCTTGTTCCCTTTCCA-3′ and (antisense) 5′-AGCCACAATACGCAGCAGAT-3′; Smarca4 (sense) 5′-CAACCACCCCTACATGTTCCA-3′ and (antisense) 5′-TTCGTTGCACGGAGTTTGG-3′.
RNA electrophoretic mobility shift (RNA-EMSA) assay
Ovary extract proteins (50 μg) were incubated with 50 pM biotin-labeled H2a.x RNA probes in RNA-EMSA buffer (50 mM KCl, 5 % (vol/vol) glycerol, 0.1 % (vol/vol) NP-40, 1 mM MgCl2, 1 mM dithiothreitol, 10 mM Tris–HCl, 0.05 mg/mL heparin, 10 units of RNase inhibitor and 0.5 mg/mL yeast tRNA) for 20 min at 20 °C. Samples were subjected to electrophoresis on a 5 % polyacrylamide 0.5× TBE gels, and transferred to positive-charge nylon membranes. The signals of RNA-EMSA reaction were detected by LightShift chemiluminescent kit (Pierce Chemical, Rockford, IL, USA). For competition assays, excess unlabeled RNA probes (1.25 mM) or antibodies (1 μg) were added directly to binding reactions. All the synthesized mRNAs used in this experiment were obtained from Invitrogen [27], and the oligonucleotide sequences were as follows: wild-type H2a.x probe (WT) 5′-GCCAUCCAUCCCCUCUUCCCCAGC-3′, mutated H2a.x probe (Mut) 5′-GCCAUCCAUCCCCUCUUGCCAGC-3′ and irrelevant sequence (IS) 5′-AAGGGUGGAGCCAAAAGGGUCAUC-3′.
Western blot analysis
The proteins extracted from mouse ovaries were separated by SDS-PAGE and then electrically transferred to a nitrocellulose membrane (GE Healthcare, San Francisco, CA, USA) as described previously [26]. Following transfer, the membranes were blocked in Tris-buffered saline (TBS) containing 5 % skimmed milk for 1 h at room temperature and then incubated for 2 h at 37 °C with a goat anti-PCBP1 antibody and a polyclonal rabbit anti-H2A.X antibody (Cell signaling technology Inc., Danvers, MA, USA) diluted 1:100 and 1:500, respectively, in TBST (TBS containing 0.1 % Tween-20) with 5 % skimmed milk. After washing three times in TBST for 10 min each, the membrane was incubated for 1 h at 37 °C with horseradish peroxidase-conjugated rabbit anti-goat or goat anti-rabbit IgGs (Santa Cruz Biotechnology Inc.) diluted 1:1000 in TBST. The membrane was then washed three times in TBST for 10 min each, and processed using an enhanced chemiluminescence detection system (Alpha Innotech, San Leandro, CA, USA).
Microinjection of anti-H2A.X antibody or anti-PCBP1 antibody into MII eggs
Rabbit anti-H2A.X antibody or goat anti-PCBP1 antibody (0.2 mg/mL in PBS, pH 7.4), whose specificities were verified by western blots (Supplemental Fig. 1), was microinjected into the cytoplasm of MII eggs as described by Dai et al. [31]. The experiment was repeated six times. A Nikon Diaphot ECLIPSE TE 300 inverting microscope (Nikon UK Ltd, Kingston upon Thames, Surrey, UK) equipped with Narishige MM0-202N hydraulic three-dimensional micromanipulators (Narishige Inc., Sea Cliff, NY, USA) was used for these experiments. All microinjections were completed in 40 min. A microinjection volume of about 5–7 pl per egg was used in all experiments. The same amount of rabbit or goat IgG diluted in PBS was microinjected as a control. After microinjection, MII eggs were washed thoroughly and cultured in CZB medium for 3 h at 37 °C in 5 % CO2 and then parthenogenetically activated in CZB medium supplemented with 10 mM SrCl2 and 5 μg/mL cytochalasin B for 1 h, after which they were washed thoroughly and cultured in CZB medium for further observation.
siRNA microinjection into mouse eggs
Two pairs of siRNA duplexes, which have been described previously with proven inhibition efficiency [25], were microinjected into MII eggs. Scrambled siRNA nucleotides were used as a negative control. MII eggs from three groups (Pcbp1 siRNA injected, negative control siRNA injected, and non-injected control) were cultured in CZB medium for 3 h after microinjection. Some of the injected samples were collected at this point to further confirm the knockdown efficiency of siRNA by real-time RT-PCR. The GAPDH gene was used as an endogenous control. The primer sequences and the expected sizes of PCR products were as follows: Pcbp1 (sense) 5′-CCAGCTCGCCAGTCATCT-3′ and (antisense) 5′-TGCCCAATAGCCTTTCACC-3′ (253 bp); Gapdh (sense) 5′-TCCCGTAGACAAAATGGT-3′ and (antisense) 5′-TCCTGGAAGATGGTGATG-3′ (242 bp). RNAi-treated eggs were activated in Ca2+-free CZB medium supplemented with 5.56 mM d-glucose containing both 10 mM SrCl2 and 5 μg/mL cytochalasin B for 1 h at 37 °C under 5 % CO2. Eggs were washed and cultured in CZB medium after parthenogenetic activation, and the number of pronuclei in each of the three groups was counted using live cell imaging and analysis.
Construction of Pcbp1 plasmid and in vitro transcription of RNA
The plasmid-containing full-length mouse Pcbp1 was purchased from OriGene (MD, USA), and the Pcbp1 coding sequence was amplified and cloned at FseI and AscI of pCS2+ vector. The pCS2+ vector, which has a myc tag, allows in vitro transcription of polyadenylated mRNA from the SP6 promoter.
In vitro synthesis of capped RNAs was performed using linearized plasmids with the SP6 Message Machine kit (Ambion, Austin, TX, USA) and the mRNAs were purified using RNeasy cleanup kit (Qiagen, Valencia, CA, USA). Synthesized RNA was portioned into aliquots and stored at −80 °C.
Assessment of mRNA levels after PCBP1 antibody or Pcbp1 mRNA microinjection
Goat anti-PCBP1 antibody or normal Goat IgG (0.5 mg/mL in PBS, pH 7.4) was microinjected into the cytoplasm of MII eggs as previously described. After microinjection, MII eggs were washed thoroughly and cultured in CZB medium for 3 h at 37 °C in 5 % CO2 and then parthenogenetically activated in CZB medium supplemented with 10 mM SrCl2 and 5 μg/mL cytochalasin B for 1 h, after which they were washed thoroughly and cultured in CZB medium for 3 h. Total RNA was extracted from the two groups using an Absolutely RNA Nanoprep Kit (Qiagen) according to the manufacturer’s protocol. Real-time RT-PCR analysis was carried out using an Invitrogen Real-time PCR Kit and GAPDH or 18 s rRNA was used as an endogenous control. Real-time quantitative RT-PCR data were calculated using the method.
To evaluate the effect of PCBP1 overexpression, in vitro transcribed myc-Pcbp1 mRNA (10 ng/μL) was injected into the cytoplasm of MII eggs using the same procedure as the antibody microinjection. The same amount of Rnase-free PBS was injected as control. Overexpression of exogenous myc-PCBP1 protein was confirmed by immunofluorescence of anti-myc Tag antibody (Abcam, MA, USA). The mRNA levels of the genes detected after antibody microinjection were compared between the myc-Pcbp1 mRNA microinjection group and the control group.
Live cell imaging and analysis
Antibody/IgG/siRNA microinjected eggs that had been parthenogenetically activated using SrCl2 for 1 h were moved to gridded coverglass dishes (MatTek, CatNo. P35G-1.5-7-C-grid) before imaging. Images were acquired automatically at multiple locations on the coverslip using a Nikon TE2000E inverting microscope fitted with an X20 Nikon Plan Fluor objective, a linearly encoded stage (Proscan, Prior) and a Hamamatsu Orca-ER CCD camera. The microscope was controlled using Simple PCI (Compix). The microscope was housed in a custom-designed 37 °C chamber with a secondary internal chamber that delivered humidified 5 % CO2. Differential interference contrast images were obtained every 5 min for a period of 5 h.
Statistical analysis
Each experiment was repeated at least three times. Independent samples Student’s t tests were performed to test the differences between antibody and IgG-injected groups, as well as those between siRNA or exogenous mRNA and control groups. Before independent samples Student’s t tests were conducted, the ratios were arcsine-root transformed to assure approximation of normality. The data were given as mean ± STDEV. Numbers of embryos examined are indicated (n). P < 0.05 was considered statistically significant.
Results
Comparison between normal MII eggs and parthenogenetically activated eggs
To systematically investigate proteins that might be involved in egg activation, we compared the total protein expression profiles of mature mouse MII eggs and parthenogenetically activated mouse eggs without zona pellucida. We performed 2-DE gel electrophoresis of the two groups over a pH range of 3–10. The volume of the stained spots was used as a measure of protein abundance. A spot was regarded differentially expressed between groups if the spot intensity showed a greater than twofold difference, and these differentially expressed spots were selected for mass spectrum (MS) analysis.
Among the identified proteins, PCBP1 exhibited differential expression patterns in MII eggs and parthenogenetically activated eggs. MS identification showed four spots to represent the PCBP1 protein. All spots had the same molecular weight but different isoelectric points (pIs). Among the four spots, spots 1 and 2 (pI values 5.57 and 5.78, respectively) were downregulated, whereas spots 3 and 4 (pI values 6.02 and 6.31) were upregulated after parthenogenetic activation (Fig. 1a, b). The matched-peptides, sequence coverage rates and Mascot Scores of four spots from MS analysis are shown in Supplemental Table 1. These results suggest that during egg activation, PCBP1 protein undergoes post-translational modification, perhaps phosphorylation.
Fig. 1.
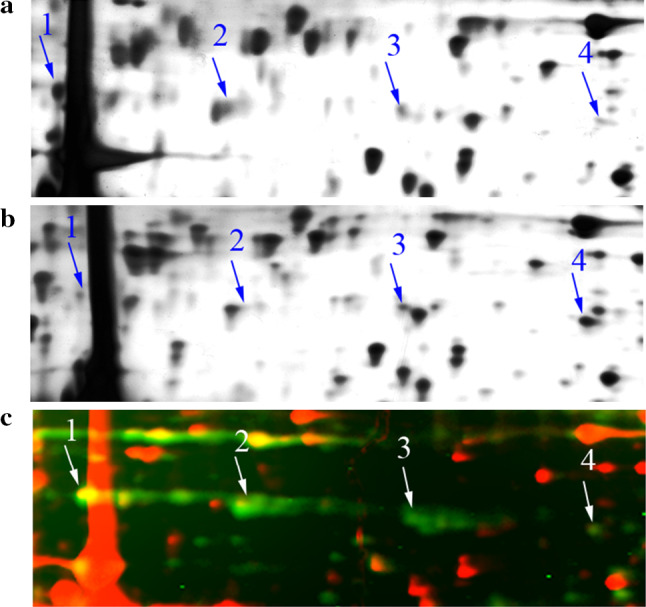
Comparison of normal MII eggs and parthenogenetically activated eggs. Four spots (arrows 1–4) were identified as PCBP1 in gels from both normal MII mouse eggs (a) and parthenogenetically activated mouse eggs (b) by MALDI-TOF. The intensity of spots 1 and 2 in the activated gel was downregulated by more than twofold; in contrast, spots 3 and 4 were upregulated by more than twofold. c Cropped phosphoprotein gel image of normal MII mouse eggs. Four spots (arrows 1–4) correspond to PCBP1. The phosphorylated forms of proteins stained by Pro-Q diamond dye showed green fluorescence; red fluorescence represents silver staining proteins
Phosphorylation analysis of PCBP1 protein in MII eggs
We stained a 2-DE gel of normal MII egg lysates with a fluorescently labeled Pro-Q Diamond dye that recognizes phosphorylated proteins. The four spots of PCBP1 were all labeled with fluorescence (Fig. 1c). This result suggested that PCBP1 was phosphorylated to varying degrees in normal MII eggs.
PCBP1 might be involved in mRNA stability in mouse eggs
In eukaryotic cells, PCBP1 mostly plays an important role in mRNA stability and/or translation by interacting with a pyrimidine-rich sequence cluster conforming to the consensus sequence (C/U)CCANxCCC(U/A)PyxUC(C/U)CC in the 3′-UTR of mRNAs [15, 16, 18, 19, 27]. Thus, we searched for the consensus sequence in the 3′-UTR of the mRNAs expressed in the mouse oocytes as described in the Methods section. We found that 75 sequences in the 3′-UTR of mRNAs (Supplemental Table 2) expressed in mouse oocytes matched the consensus sequence, suggesting these mRNAs could be potential binding targets of PCBP1 protein. To determine whether PCBP1 was involved in regulating the stability of these mRNAs, we evaluated the levels of 11 randomly selected mRNAs by real-time quantitative RT-PCR during egg activation when PCBP1 was blocked or overexpressed. We also randomly evaluated levels of four presumably unrelated mRNAs.
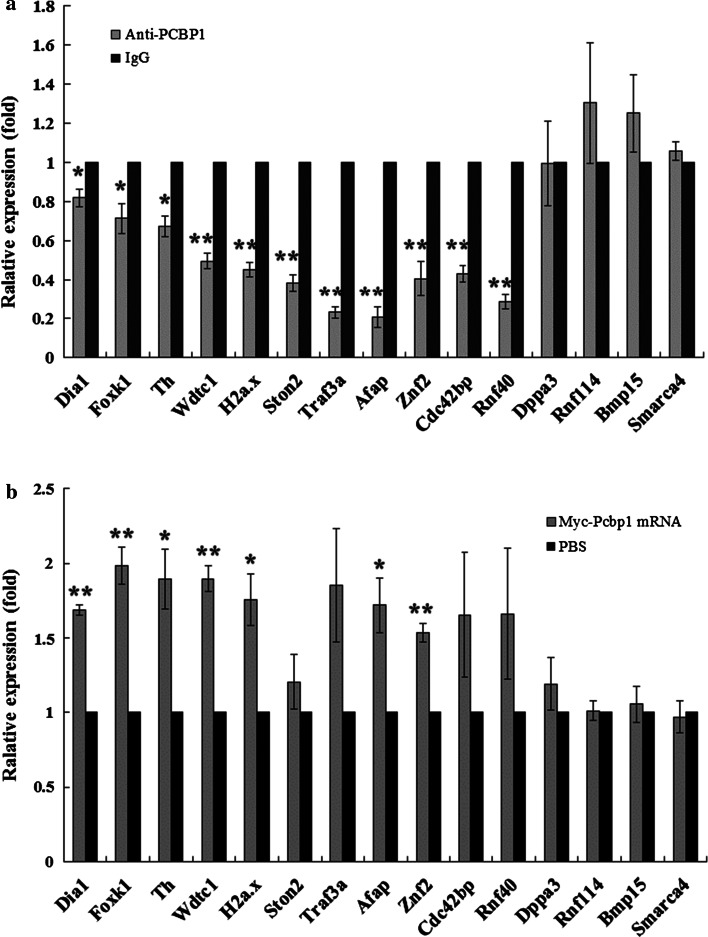
Under conditions in which the PCBP1 protein was blocked during egg activation, the expression levels of the 11 putative PCBP1-binding mRNAs were all significantly decreased (Fig. 2a). We also overexpressed PCBP1 through exogenous myc-pcbp1 mRNA microinjection and confirmed efficient overexpression by immunofluorescence with anti-myc antibody (Supplemental Fig. 2). Almost all of the putative PCBP1-binding mRNAs were elevated to varying degrees after PCBP1 overexpression, though several showed no statistical significance (Fig. 2b). Notably, the expression levels of the four mRNAs lacking the PCBP1-binding sequence showed no changes regardless of PCBP1 blocking or overexpression (Fig. 2).
Fig. 2.
Involvement of PCBP1 in mRNA stability in mouse eggs. a Blockade of PCBP1 by antibody microinjection led to a significant reduction in the levels of 11 putative PCBP1-binding mRNAs (including Dia1, Foxk1, Th, Wdtc1, H2a.x, Ston2, Traf3a, Afap, Znf2, Cdc42 bp and Rnf40) during egg parthenogenetic activation. In comparison, the expression levels of putative non-PCBP1-binding mRNAs (including Dppa3, Rnf114, Bmp15, and Smarca4) remained unchanged significantly. b Overexpression of PCBP1 through exogenous pcbp1 mRNA microinjection caused elevation of expression levels of the 11 putative PCBP1-binding mRNAs, although the changes of Ston2, Traf3a, Cdc42 bp, and Rnf40 had no statistical significance. The expression levels of the four other putative non-PCBP1-binding mRNAs were also unchanged. The y-axis represents the relative quantification of target genes normalized to GAPDH or 18sRNA as calculated by method in the real-time PCR assays. *P < 0.05, **P < 0.01
H2a.x mRNA is a target of PCBP1
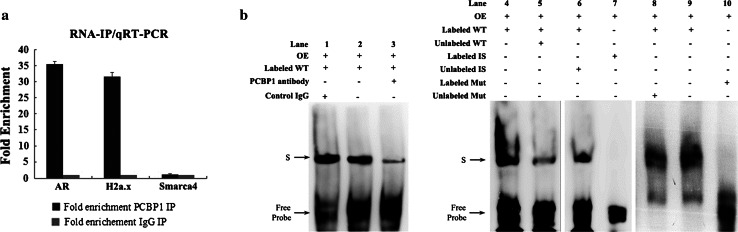
H2a.x was one of the mRNAs containing the PCBP1-binding sequence in its 3′-UTR that showed statistically significant differences in expression upon PCBP1 blocking or overexpression. To confirm that H2a.x mRNA was a target of PCBP1, RIP assays were performed on ovarian tissue extracts with an antibody against PCBP1, and RNA levels in immunoprecipitates were determined by qRT-PCR. The results showed that AR, as a well-known binding target of PCBP1, was preferentially enriched (about 35-fold) in PCBP1-containing mRNPs relative to control IgG. Notably, H2a.x mRNA was also detected at a level of about 32-fold greater than that of control IgG. The level of Samarca4, one of the mRNAs lacking the PCBP1-binding motif examined above, was comparable between the anti-PCBP1 antibody group and the control IgG group (Fig. 3a). These results indicate that PCBP1 protein could bind to H2a.x mRNA.
Fig. 3.
H2a.x mRNA is a binding target for PCBP1. a RNA immunoprecipitation (RIP) experiment using anti-PCBP1 or control IgG antibodies. RNA levels in immunoprecipitates were determined by qRT-PCR. AR and H2a.x mRNAs were enriched by 35- and 32-fold, respectively, in PCBP1-containing mRNPs relative to control IgG. But the level of Samarca4 was comparable between the anti-PCBP1 antibody group and the control IgG group. b RNA-EMSA assays were performed to verify the interaction of PCBP1 and H2a.x mRNA. Components for every reaction are indicated above each lane. Protein extracts for mouse ovaries were bound to a biotin-labeled oligonucleotide probe encompassing the PCBP1-binding site in the H2a.x mRNA (lane 2, 4, and 9). Addition of anti-PCBP1 antibody reduced the assembly (lane 3) but IgG had no effect (lane 1). Irrelevant probe (lane 7) or mutant H2a.x probe (lane 10) failed to form the RNA–protein complex. The prominent band was competed with the unlabeled H2a.x probe (lane 5) but not the irrelevant probe (lane 6) or mutated H2a.x probe (lane 8). Arrowheads specific RNA–protein complexes (S) and free probes. OE, ovary protein extracts; WT, H2a.x probe encompassing the PCBP1-binding site; Mut, mutant H2a.x probe; IS, irrelevant sequence
RNA-EMSA was further performed using ovarian protein extracts and biotin-labeled oligonucleotide probes encompassing the PCBP1-binding motif in the 3′-UTR of H2a.x mRNA. The results confirmed the presence of the H2a.x mRNA–protein complex, while the mutant H2a.x probe or irrelevant probe failed to form the RNA–protein complex. Addition of an anti-PCBP1 antibody reduced the complex formation, but IgG had no effect. The complexes were effectively competed by the unlabeled H2a.x probe but were not competed with an irrelevant probe or mutated H2a.x probe (Fig. 3b), further confirming the specificity of this binding.
Microinjection of MII eggs with H2A.X antibody retards pronucleus formation
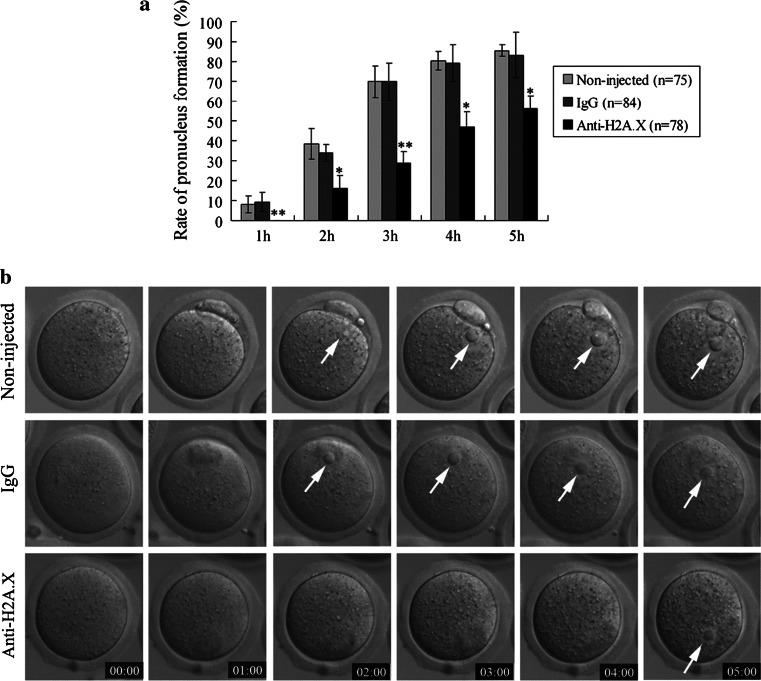
The H2A.X protein is required for pronucleus formation in Xenopus oocytes. To examine whether H2A.X also plays a similar role in mice, mouse MII eggs were microinjected with an H2A.X antibody. The percentage of pronuclei formed in the H2A.X antibody-injected group was markedly lower than that of the IgG-injected group and the non-injected group at different time points from 1 to 5 h after parthenogenetic activation (P < 0.05). In contrast, IgG injection did not affect the rate of pronucleus formation (Fig. 4).
Fig. 4.
Blockade of H2A.X by H2A.X antibody microinjection leads to delay in pronucleus formation. a A significant time-dependent inhibitory effect on pronucleus formation was observed in the H2A.X antibody-injected group compared with the non-injected group and the IgG-injected group. Data are presented as mean values ± SD; *P < 0.05, **P < 0.01 versus control. b The H2A.X antibody-injected group displayed an obvious delay in pronucleus formation (pronuclei indicated by white arrows). Time stamp is h:min after parthenogenetic activation
Inhibition of maternal PCBP1 protein by antibody microinjection retards pronucleus formation
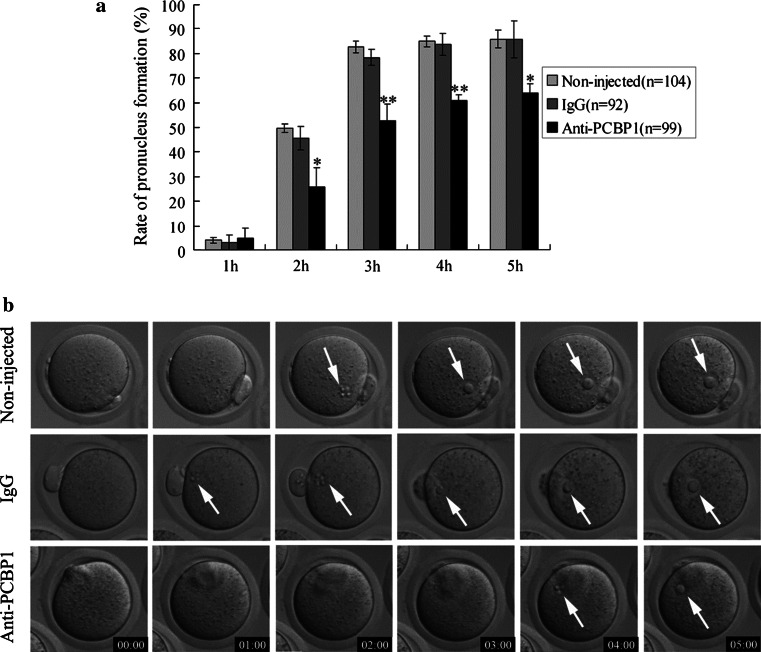
To clarify the role of PCBP1 in pronucleus formation, we observed this process dynamically after microinjection of PCBP1 antibody. The percentage of pronuclei formed in the PCBP1 antibody-injected group was markedly lower than that in the IgG-injected group and the non-injected group at different time points from 2 to 5 h after parthenogenetic activation (P < 0.05). In contrast, IgG injection did not affect the rate of pronucleus formation (Fig. 5; Supplemental movie 1–3).
Fig. 5.
Blockade of PCBP1 by PCBP1 antibody microinjection leads to delay in pronucleus formation. a A significant time-dependent inhibitory effect on pronucleus formation was observed in the PCBP1 antibody-injected group compared with the non-injected group and the IgG-injected group. Data are presented as mean values ± SD; *P < 0.05, **P < 0.01 versus control. b Dynamic observation of pronucleus formation in the non-injected group, IgG-injected group and PCBP1 antibody-injected group by live cell imaging (pronuclei indicated by white arrows). PCBP1 antibody-injected eggs displayed obvious delay in pronucleus formation. Time stamp is h:min after parthenogenetic activation. Selected frames from Supplementary Movies 1, 2 and 3
Depletion of maternal Pcbp1 mRNA by stealth siRNA microinjection leads to pronucleus formation delay
Two Pcbp1-specific siRNAs, whose knockdown efficiencies were verified by real-time PCR (Fig. 6a), were microinjected into mouse MII eggs. The rate of pronucleus formation in the Pcbp1 siRNA-injected group was notably lower than that of the negative control siRNA injected or non-injected group from 2 to 4 h after parthenogenetic activation, but the pronucleus formation rate recovered to a similar extent at 5 h (Fig. 6b). These results also indicated a delay in pronucleus formation after microinjection of MII eggs with stealth siRNAs targeting Pcbp1.
Fig. 6.
Knockdown of Pcbp1 by Pcbp1-siRNAs microinjection retarded pronucleus formation. a The knockdown efficiency of Pcbp1-specific siRNAs was confirmed by real-time PCR. The samples were collected from MII oocytes cultured in CZB medium for 3 h after siRNA microinjection. b The rate of pronucleus formation of Pcbp1 siRNA (upper panel, Pcbp1-siRNA-1#; lower panel, Pcbp1-siRNA-2#) microinjected eggs was evidently lower than that of the two other groups at 2–4 h after injection and parthenogenetic activation. Data are presented as mean values ± SD; *P < 0.05, **P < 0.01 versus control
Discussion
Pronucleus formation is a landmark event for egg activation and fertilization. However, little is known about the maternal factors involved in pronucleus formation. In this study, we compared the protein profiles of parthenogenetically activated eggs to normal MII mouse eggs with the aim of searching for maternal factors facilitating egg activation. This approach makes no assumptions about the involvement of known or unknown molecules, allowing the process to be independent of any presupposed hypotheses. Our results showed that PCBP1 was one of the most significantly altered maternal proteins.
Analysis using 2-DE identified four spots in a line as PCBP1 in gels from both normal and parthenogenetically activated eggs, showing that these proteins had the same molecular weight but different pIs and suggesting that there might be some post-translational modification of PCBP1. Bioinformatics techniques have demonstrated that PCBP1 has many conserved phosphorylation sites. Thus, we anticipated that PCBP1 may be phosphorylated in mouse eggs. To confirm this hypothesis, we carried out phosphoprotein staining of 2-DE gels from normal MII eggs with a fluorescent-labeled Pro-Q Diamond dye and visualized the putative phosphorylated proteins [32]. The image verified that all four spots representing PCBP1 in the 2-DE gel from normal MII eggs displayed phosphorylation. Given that the two spots in the partial acid position were downregulated and the two spots in the partial alkaline position were upregulated in the gel from activated eggs, it is not difficult to conclude that PCBP1 might undergo dephosphorylation after parthenogenetic activation of mouse eggs, and the occurrence of such evident post-translational modification of PCBP1 also suggests that it may play an important role in egg activation. Furthermore, previous studies have shown that phosphorylation of PCBP1 regulated its binding affinity to target consensus sequences. PAK1 kinase-mediated phosphorylation of cytoplasmic PCBP1 was shown to reduce its RNA-binding activity [33], while another study showed that dephosphorylation of PCBP1 could boost its ability to bind to mRNAs [34].
Bioinformatics analysis identified 75 mRNAs expressed in mouse oocyte that had the consensus PCBP1-binding CU-rich motif in their 3′-UTR, which implied they may be potential binding targets of PCBP1. Eleven mRNAs were randomly chosen for further investigation, and we found that their mRNA levels were significantly decreased when PCBP1 was blocked during egg activation; on the contrary, their levels were elevated to varying degrees when PCBP1 was overexpressed. However, the expressional levels of the putative non-PCBP1-binding mRNAs remained unchanged significantly even after PCBP1 was blocked or was overexpressed. The results suggested that PCBP1 protein might specifically bind the target mRNAs and be involved in regulating the stability of these mRNAs during egg activation. Among these 11 putative PCBP1-binding mRNAs, we focused on H2a.x.
H2A.X is a variant of H2A that is highly conserved from Saccharomyces cerevisiae to humans and has a major role in chromatin metabolism [35]. H2A.X plays an important role in nucleosome assembly [36], chromatin organization [37], and DNA repair [38], in addition, H2A.X has been reported to be involved in chromatin decondensation and pronucleus formation in Xenopus laevis [36, 39]. RIP was performed to investigate whether PCBP1 could bind with H2a.x mRNA. This technique has been demonstrated to be an effective approach to identify putative targets of mRNA-binding proteins using antibodies against RNA-binding proteins [40]. As expected, the results showed that H2a.x mRNA was mainly enriched in the mRNP pellets immunoprecipitated by the anti-PCBP1 antibody compared with IgG, indicating it was indeed a binding target of PCBP1. In addition, RNA-EMSA further verified that PCBP1 and H2a.x mRNA could form an RNA–protein complex, while irrelevant or mutant H2a.x mRNAs failed to form the RNA–protein complex. Assembly of H2a.x into mRNPs was reduced by the anti-PCBP1 antibody or competed with the unlabeled self-probe but not with the mutated probe or the irrelevant probe. Thus, we concluded from the above results that PCBP1 could undoubtedly and specifically bind to H2a.x mRNA. Because of the limitations of materials, while the interaction of PCBP1 protein with H2a.x mRNA was verified in the ovary, direct evidence that PCBP1 actually binds to H2a.x in oocytes remains lacking. However, the obvious change in the expression level of H2a.x mRNA when PCBP1 was blocked or overexpressed during egg activation implied that PCBP1 could bind to maternal H2a.x mRNA in activated eggs and enhance its stability. H2A.X incorporation into chromatin was observed during the pronucleus formation in Xenopus laevis [39], however, in mammals, the direct role of H2A.X in pronucleus formation has not been proven, despite the deposition of H2A.X in the parent nuclei in mouse zygotes [41, 42]. Thus, we performed H2A.X antibody microinjection in mouse eggs, and the significantly retarded pronucleus formation in parthenogenetically activated mouse eggs after H2A.X antibody microinjection suggested that H2A.X is also involved in pronucleus formation in mice. We speculated that PCBP1 protein might contribute to pronucleus formation through its regulation on the stability of H2a.x mRNA.
Pronucleus formation delay was expectedly observed after inhibition of PCBP1 at the protein level or mRNA level by antibody or specific siRNAs microinjection into mouse eggs followed by parthenogenetic activation. These results indicated that PCBP1 determines the normal timing of pronucleus formation. Studies in different species (including mice, cattle and humans) have shown a positive correlation between the timing of the first cell cycle of embryo and subsequent developmental potential. Early onset of first cleavage is associated with increased blastocyst formation, higher implantation and pregnancy rates [43–45]. The appearance of pronuclei marks entry into G1 phase of the 1-cell stage of development [45], and delay in pronucleus formation was always observed in aged animals and considered to be one of the main factors causing the preimplantation wastage of embryos [46]. These observations suggest that the timing of pronuclear formation may be an important determinant of embryo development.
In summary, here we have demonstrated, using proteomic techniques, that PCBP1 is one of the most significantly altered maternal proteins during egg activation. Our results suggest that maternal PCBP1 displays dephosphorylation in parthenogenetically activated eggs, boosting its ability to bind to H2a.x mRNA and enhance its stability, thus determining the normal timing of pronucleus formation in mouse eggs. These findings improve our understanding of the molecular mechanisms regulating pronucleus formation and may aid in the search for the causes of failure of early embryonic development and in nuclear transplant embryonic development. In the previous work, we found PCBP1 also played an important role in the regulation of global transcriptional silencing in fully grown oocytes. Together with this work, we speculate that PCBP1 is a multifunctional protein in mouse oocytes, and might perform diverse roles according to different post-translational modifications, cellular localization, and physiological events. Further studies are required to fully investigate the roles and mechanisms of PCBP1.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Table 1. The identification information of PCBP1 protein. (XLSX 12 kb)
Supplemental Table 2. Maternal mRNAs suggested to be potential binding targets for PCBP1. (XLSX 19 kb)
Supplemental Fig. 1. Only one band with predicted molecular weight was present on western blots of ovary protein extracts using anti-PCBP1 (a) or anti-H2A.X (b) antibody confirmed the specificity of these antibodies. (TIFF 2153 kb)
Supplemental Fig. 2. RNase-free PBS (control group) or exogenous myc-Pcbp1 mRNA (overexpression group) was microinjected into MII eggs and then immunofluorescence detecting with anti-myc Tag antibody indicated that myc-PCBP1 protein was efficiently overexpressed. (TIFF 10699 kb)
Supplemental movie 1. The process of pronucleus formation in non-injected group. (MOV 1011 kb)
Supplemental movie 2. The process of pronucleus formation in IgG-injected group. (MOV 859 kb)
Supplemental movie 3. The process of pronucleus formation in anti-PCBP1 antibody-injected group. (MOV 1222 kb)
Acknowledgments
We are grateful to Prof. Qiang Wang (Nanjing Medical University) for providing anti-myc antibody. We gratefully acknowledge Liwen Bianji for editing the article. This work is supported by the China 973 Program (2012CB944704) and the National Science Foundation of China (30700275).
Footnotes
Z. Shi, C. Zhao, and Y. Yang contributed equally to this work.
Contributor Information
Ran Huo, Phone: 86-25-86862038, Email: huoran@njmu.edu.cn.
Qi Zhou, Phone: 86-10-64807299, Email: qzhou@ioz.ac.cn.
References
- 1.Horner VL, Wolfner MF. Transitioning from egg to embryo: triggers and mechanisms of egg activation. Dev Dyn. 2008;237:527–544. doi: 10.1002/dvdy.21454. [DOI] [PubMed] [Google Scholar]
- 2.Krauchunas AR, Wolfner MF. Molecular changes during egg activation. Curr Top Dev Biol. 2013;102:267–292. doi: 10.1016/B978-0-12-416024-8.00010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malcuit C, Kurokawa M, Fissore RA. Calcium oscillations and mammalian egg activation. J Cell Physiol. 2006;206:565–573. doi: 10.1002/jcp.20471. [DOI] [PubMed] [Google Scholar]
- 4.Stitzel ML, Seydoux G. Regulation of the oocyte-to-zygote transition. Science. 2007;316:407–408. doi: 10.1126/science.1138236. [DOI] [PubMed] [Google Scholar]
- 5.Collas P. Cytoplasmic control of nuclear assembly. Reprod Fertil Dev. 1998;10:581–592. doi: 10.1071/RD98049. [DOI] [PubMed] [Google Scholar]
- 6.Poccia D, Collas P. Nuclear envelope dynamics during male pronuclear development. Dev Growth Differ. 1997;39:541–550. doi: 10.1046/j.1440-169X.1997.t01-4-00001.x. [DOI] [PubMed] [Google Scholar]
- 7.Perreault SD, Naish SJ, Zirkin BR. The timing of hamster sperm nuclear decondensation and male pronucleus formation is related to sperm nuclear disulfide bond content. Biol Reprod. 1987;36:239–244. doi: 10.1095/biolreprod36.1.239. [DOI] [PubMed] [Google Scholar]
- 8.Clift D, Schuh M. Restarting life: fertilization and the transition from meiosis to mitosis. Nat Rev Mol Cell Biol. 2013;14:549–562. doi: 10.1038/nrm3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li L, Zheng P, Dean J. Maternal control of early mouse development. Development. 2010;137:859–870. doi: 10.1242/dev.039487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakamura H, Wu C, Kuang J, Larabell C, Etkin LD. XCS-1, a maternally expressed gene product involved in regulating mitosis in Xenopus . J Cell Sci. 2000;113(Pt 13):2497–2505. doi: 10.1242/jcs.113.13.2497. [DOI] [PubMed] [Google Scholar]
- 11.Ma M, Guo X, Wang F, Zhao C, Liu Z, Shi Z, Wang Y, Zhang P, Zhang K, Wang N, Lin M, Zhou Z, Liu J, Li Q, Wang L, Huo R, Sha J, Zhou Q. Protein expression profile of the mouse metaphase-II oocyte. J Proteome Res. 2008;7:4821–4830. doi: 10.1021/pr800392s. [DOI] [PubMed] [Google Scholar]
- 12.Wang S, Kou Z, Jing Z, Zhang Y, Guo X, Dong M, Wilmut I, Gao S. Proteome of mouse oocytes at different developmental stages. Proc Natl Acad Sci USA. 2010;107:17639–17644. doi: 10.1073/pnas.1013185107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ko JL, Loh HH. Poly C binding protein, a single-stranded DNA binding protein, regulates mouse mu-opioid receptor gene expression. J Neurochem. 2005;93:749–761. doi: 10.1111/j.1471-4159.2005.03089.x. [DOI] [PubMed] [Google Scholar]
- 14.Kim JH, Hahm B, Kim YK, Choi M, Jang SK. Protein-protein interaction among hnRNPs shuttling between nucleus and cytoplasm. J Mol Biol. 2000;298:395–405. doi: 10.1006/jmbi.2000.3687. [DOI] [PubMed] [Google Scholar]
- 15.Weiss IM, Liebhaber SA. Erythroid cell-specific determinants of alpha-globin mRNA stability. Mol Cell Biol. 1994;14:8123–8132. doi: 10.1128/mcb.14.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiss IM, Liebhaber SA. Erythroid cell-specific mRNA stability elements in the alpha 2-globin 3′ nontranslated region. Mol Cell Biol. 1995;15:2457–2465. doi: 10.1128/mcb.15.5.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thiele BJ, Doller A, Kahne T, Pregla R, Hetzer R, Regitz-Zagrosek V. RNA-binding proteins heterogeneous nuclear ribonucleoprotein A1, E1, and K are involved in post-transcriptional control of collagen I and III synthesis. Circ Res. 2004;95:1058–1066. doi: 10.1161/01.RES.0000149166.33833.08. [DOI] [PubMed] [Google Scholar]
- 18.Paulding WR, Czyzyk-Krzeska MF. Regulation of tyrosine hydroxylase mRNA stability by protein-binding, pyrimidine-rich sequence in the 3′-untranslated region. J Biol Chem. 1999;274:2532–2538. doi: 10.1074/jbc.274.4.2532. [DOI] [PubMed] [Google Scholar]
- 19.Czyzyk-Krzeska MF, Bendixen AC. Identification of the poly(C) binding protein in the complex associated with the 3′ untranslated region of erythropoietin messenger RNA. Blood. 1999;93:2111–2120. [PubMed] [Google Scholar]
- 20.Evans JR, Mitchell SA, Spriggs KA, Ostrowski J, Bomsztyk K, Ostarek D, Willis AE. Members of the poly (rC) binding protein family stimulate the activity of the c-myc internal ribosome entry segment in vitro and in vivo. Oncogene. 2003;22:8012–8020. doi: 10.1038/sj.onc.1206645. [DOI] [PubMed] [Google Scholar]
- 21.Pickering BM, Mitchell SA, Spriggs KA, Stoneley M, Willis AE. Bag-1 internal ribosome entry segment activity is promoted by structural changes mediated by poly(rC) binding protein 1 and recruitment of polypyrimidine tract binding protein 1. Mol Cell Biol. 2004;24:5595–5605. doi: 10.1128/MCB.24.12.5595-5605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pickering BM, Mitchell SA, Evans JR, Willis AE. Polypyrimidine tract binding protein and poly r(C) binding protein 1 interact with the BAG-1 IRES and stimulate its activity in vitro and in vivo. Nucleic Acids Res. 2003;31:639–646. doi: 10.1093/nar/gkg146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong N, Radu G, Ju W, Brown WT. Novel progerin-interactive partner proteins hnRNP E1, EGF, Mel 18, and UBC9 interact with lamin A/C. Biochem Biophys Res Commun. 2005;338:855–861. doi: 10.1016/j.bbrc.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 24.Lim J, Hao T, Shaw C, Patel AJ, Szabo G, Rual JF, Fisk CJ, Li N, Smolyar A, Hill DE, Barabasi AL, Vidal M, Zoghbi HY. A protein–protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 25.Xia M, He H, Wang Y, Liu M, Zhou T, Lin M, Zhou Z, Huo R, Zhou Q, Sha J. PCBP1 is required for maintenance of the transcriptionally silent state in fully grown mouse oocytes. Cell Cycle. 2012;11:2833–2842. doi: 10.4161/cc.21169. [DOI] [PubMed] [Google Scholar]
- 26.Huang XY, Guo XJ, Shen J, Wang YF, Chen L, Xie J, Wang NL, Wang FQ, Zhao C, Huo R, Lin M, Wang X, Zhou ZM, Sha JH. Construction of a proteome profile and functional analysis of the proteins involved in the initiation of mouse spermatogenesis. J Proteome Res. 2008;7:3435–3446. doi: 10.1021/pr800179h. [DOI] [PubMed] [Google Scholar]
- 27.Holcik M, Liebhaber SA. Four highly stable eukaryotic mRNAs assemble 3′ untranslated region RNA-protein complexes sharing cis and trans components. Proc Natl Acad Sci USA. 1997;94:2410–2414. doi: 10.1073/pnas.94.6.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mignone F, Grillo G, Licciulli F, Iacono M, Liuni S, Kersey PJ, Duarte J, Saccone C, Pesole G. UTRdb and UTRsite: a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 2005;33:D141–D146. doi: 10.1093/nar/gki021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evsikov AV, Graber JH, Brockman JM, Hampl A, Holbrook AE, Singh P, Eppig JJ, Solter D, Knowles BB. Cracking the egg: molecular dynamics and evolutionary aspects of the transition from the fully grown oocyte to embryo. Genes Dev. 2006;20:2713–2727. doi: 10.1101/gad.1471006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeap BB, Voon DC, Vivian JP, McCulloch RK, Thomson AM, Giles KM, Czyzyk-Krzeska MF, Furneaux H, Wilce MC, Wilce JA, Leedman PJ. Novel binding of HuR and poly(C)-binding protein to a conserved UC-rich motif within the 3′-untranslated region of the androgen receptor messenger RNA. J Biol Chem. 2002;277:27183–27192. doi: 10.1074/jbc.M202883200. [DOI] [PubMed] [Google Scholar]
- 31.Dai Y, Lee C, Hutchings A, Sun Y, Moor R. Selective requirement for Cdc25C protein synthesis during meiotic progression in porcine oocytes. Biol Reprod. 2000;62:519–532. doi: 10.1095/biolreprod62.3.519. [DOI] [PubMed] [Google Scholar]
- 32.Agrawal GK, Thelen JJ. Development of a simplified, economical polyacrylamide gel staining protocol for phosphoproteins. Proteomics. 2005;5:4684–4688. doi: 10.1002/pmic.200500021. [DOI] [PubMed] [Google Scholar]
- 33.Meng Q, Rayala SK, Gururaj AE, Talukder AH, O’Malley BW, Kumar R. Signaling-dependent and coordinated regulation of transcription, splicing, and translation resides in a single coregulator, PCBP1. Proc Natl Acad Sci USA. 2007;104:5866–5871. doi: 10.1073/pnas.0701065104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leffers H, Dejgaard K, Celis JE. Characterisation of two major cellular poly(rC)-binding human proteins, each containing three K-homologous (KH) domains. Eur J Biochem. 1995;230:447–453. doi: 10.1111/j.1432-1033.1995.tb20581.x. [DOI] [PubMed] [Google Scholar]
- 35.Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W. Histone H2A variants H2AX and H2AZ. Curr Opin Genet Dev. 2002;12:162–169. doi: 10.1016/S0959-437X(02)00282-4. [DOI] [PubMed] [Google Scholar]
- 36.Kleinschmidt JA, Steinbeisser H. DNA-dependent phosphorylation of histone H2A.X during nucleosome assembly in Xenopus laevis oocytes: involvement of protein phosphorylation in nucleosome spacing. EMBO J. 1991;10:3043–3050. doi: 10.1002/j.1460-2075.1991.tb07855.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuccotti M, Garagna S, Merico V, Monti M, Alberto Redi C. Chromatin organisation and nuclear architecture in growing mouse oocytes. Mol Cell Endocrinol. 2005;234:11–17. doi: 10.1016/j.mce.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 38.Tuteja N, Singh MB, Misra MK, Bhalla PL, Tuteja R. Molecular mechanisms of DNA damage and repair: progress in plants. Crit Rev Biochem Mol Biol. 2001;36:337–397. doi: 10.1080/20014091074219. [DOI] [PubMed] [Google Scholar]
- 39.Dimitrov S, Dasso MC, Wolffe AP. Remodeling sperm chromatin in Xenopus laevis egg extracts: the role of core histone phosphorylation and linker histone B4 in chromatin assembly. J Cell Biol. 1994;126:591–601. doi: 10.1083/jcb.126.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Racki WJ, Richter JD. CPEB controls oocyte growth and follicle development in the mouse. Development. 2006;133:4527–4537. doi: 10.1242/dev.02651. [DOI] [PubMed] [Google Scholar]
- 41.Inoue A, Zhang Y. Nucleosome assembly is required for nuclear pore complex assembly in mouse zygotes. Nat Struct Mol Biol. 2014;21:609–616. doi: 10.1038/nsmb.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu BJ, Dong FL, Ma XS, Wang XG, Lin F, Liu HL. Localization and expression of histone H2A variants during mouse oogenesis and preimplantation embryo development. Genet Mol Res. 2014;13:5929–5939. doi: 10.4238/2014.August.7.8. [DOI] [PubMed] [Google Scholar]
- 43.McLaren A, Bowman P. Genetic effects on the timing of early development in the mouse. J Embryol Exp Morphol. 1973;30:491–498. [PubMed] [Google Scholar]
- 44.Lonergan P, Khatir H, Piumi F, Rieger D, Humblot P, Boland MP. Effect of time interval from insemination to first cleavage on the developmental characteristics, sex ratio and pregnancy rate after transfer of bovine embryos. J Reprod Fertil. 1999;117:159–167. doi: 10.1530/jrf.0.1170159. [DOI] [PubMed] [Google Scholar]
- 45.Fenwick J, Platteau P, Murdoch AP, Herbert M. Time from insemination to first cleavage predicts developmental competence of human preimplantation embryos in vitro. Hum Reprod. 2002;17:407–412. doi: 10.1093/humrep/17.2.407. [DOI] [PubMed] [Google Scholar]
- 46.Koyama K, Kang SS, Huang W, Yanagawa Y, Takahashi Y, Nagano M. Aging-related changes in in vitro-matured bovine oocytes: oxidative stress, mitochondrial activity and ATP content after nuclear maturation. J Reprod Dev. 2014;60:136–142. doi: 10.1262/jrd.2013-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. The identification information of PCBP1 protein. (XLSX 12 kb)
Supplemental Table 2. Maternal mRNAs suggested to be potential binding targets for PCBP1. (XLSX 19 kb)
Supplemental Fig. 1. Only one band with predicted molecular weight was present on western blots of ovary protein extracts using anti-PCBP1 (a) or anti-H2A.X (b) antibody confirmed the specificity of these antibodies. (TIFF 2153 kb)
Supplemental Fig. 2. RNase-free PBS (control group) or exogenous myc-Pcbp1 mRNA (overexpression group) was microinjected into MII eggs and then immunofluorescence detecting with anti-myc Tag antibody indicated that myc-PCBP1 protein was efficiently overexpressed. (TIFF 10699 kb)
Supplemental movie 1. The process of pronucleus formation in non-injected group. (MOV 1011 kb)
Supplemental movie 2. The process of pronucleus formation in IgG-injected group. (MOV 859 kb)
Supplemental movie 3. The process of pronucleus formation in anti-PCBP1 antibody-injected group. (MOV 1222 kb)