Abstract
Fibroblast growth factors (FGFs) deliver extracellular signals that govern many developmental and regenerative processes, but the mechanisms regulating FGF signaling remain incompletely understood. Here, we explored the relationship between intrinsic stability of FGF proteins and their biological activity for all 18 members of the FGF family. We report that FGF1, FGF3, FGF4, FGF6, FGF8, FGF9, FGF10, FGF16, FGF17, FGF18, FGF20, and FGF22 exist as unstable proteins, which are rapidly degraded in cell cultivation media. Biological activity of FGF1, FGF3, FGF4, FGF6, FGF8, FGF10, FGF16, FGF17, and FGF20 is limited by their instability, manifesting as failure to activate FGF receptor signal transduction over long periods of time, and influence specific cell behavior in vitro and in vivo. Stabilization via exogenous heparin binding, introduction of stabilizing mutations or lowering the cell cultivation temperature rescues signaling of unstable FGFs. Thus, the intrinsic ligand instability is an important elementary level of regulation in the FGF signaling system.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-015-1856-8) contains supplementary material, which is available to authorized users.
Keywords: Fibroblast growth factor, FGF, Unstable, Proteoglycan, Regulation
Introduction
Maintenance of tissue homeostasis depends on complex growth factor signaling networks that govern many basic cell functions. The family of fibroblast growth factors (FGF) represents one of the fundamental tools of such cell-to-cell communication. Eighteen FGFs act as tissue growth factors or metabolic hormones to regulate many important processes throughout development, life, and disease [1]. FGFs signal to varied distances in organism, ranging from relatively short distance auto- and paracrine tissue signaling typical for most of the FGF growth factors, to long-range communication among organs in case of metabolic FGF hormones FGF19, FGF21, and FGF23 [2]. To perform their functions, the FGFs must penetrate easily through the extracellular environment and also remain stable enough to reach the recipient cells in proper conformation to bind and activate its cognate receptors at the cell surface.
Several lines of evidence suggest that limited stability affects FGF signaling, evidenced mostly by research carried-out on FGF1 as a prototypic member of FGF family. FGF1 is unstable protein with limited half-life in vivo. At physiological temperatures, almost 50 % of FGF1 exists in an unfolded state, which is prone to proteolytic degradation [3–6]. Increasing FGF1’s thermal stability by mutagenesis or heparin binding enhanced its biological activity [3, 7, 8]. Limited information exists about stability of other FGFs. It is known that both FGF7 and FGF10 are unstable and, similar to FGF1, their stability can be markedly improved upon heparin binding [9]. A recent study demonstrated that among 17 FGFs tested, only FGF2, FGF4, FGF6, and FGF9 activate FGF receptor (FGFR) signaling in human embryonic stem cells (hESC). Moreover, normal FGFR activation was observed in hESC stimulated by stable FGF1 mutant but not its wild-type (wt) counterpart, suggesting that failure to signal in hESC may stem from instability in as many as 13 different members of FGF family [10].
At present, the stability seems to play a critical role in pathological signaling of at least one FGF, the FGF23. FGF23 acts as metabolic hormone with ‘phosphatonin’ activity, which is produced by mineralized tissues and regulates phosphate levels via inhibiting its reabsorption in kidney proximal tubules [2]. Hypophosphatemic rickets (ADHR) is a human condition caused by aberrant renal phosphate wasting, and characterized by hypophosphatemia, defective bone and cartilage mineralization, short stature, and dental abscesses [11]. ADHR is caused by missense mutations in FGF23 that lead to R176Q, R179Q, or R179W amino acid substitutions which stabilize full-length FGF23 by disrupting the 176RHTR179 motif that serves as a cleavage site for subtilisin-like proprotein convertase or PHEX endopeptidase. Correspondingly, mutated FGF23 is resistant to intracellular proteolytic processing and circulates as a full-length 32-kDa variant only, in contrast to wt FGF23 that is released from cells as a mixture of full-length and truncated 12-kDa variants [12]. Exactly the opposite situation exists in familial tumoral calcinosis (FTC), which is metabolic disorder caused by progressive deposition of basic calcium phosphate crystals in periarticular spaces in hip, elbow, and shoulder. FTC is caused by inactivating mutations in GALNT3 glycosylase which glycosylates Thr178 of FGF23, localized within the 176RHTR179 motif. This glycosylation prevents FGF23 degradation by stabilizing its structure [13, 14].
Although most of the experimental work addressing the effect of stability on FGF signaling was carried-out on FGF1, the physiological functions of FGF1 appear limited in vivo, as demonstrated by the absence of the phenotype in the fgf1 −/− mice [1]. Other FGFs in contrast regulate important developmental processes in mammalians, such as blastocyst formation (FGF4), gastrulation (FGF8), epithelial–mesenchymal interactions necessary for development of epithelial (FGF10) or mesenchymal (FGF9, FGF18) components of multiple organs, and heart and brain morphogenesis (FGF15, FGF16, FGF17) [15–21]. These processes rely on FGF-mediated communication between distant cell populations within the embryo, and therefore demand the FGF structure to be stable enough to fulfill the required spatiotemporal signaling needs.
Altogether, the structural stability of given FGF (i.e., resistance to proteolysis or to thermally induced unfolding) may, when limited, constitute an important basic regulatory level of its signaling. With exception of FGF1, the regulatory role of structural stability has never been systematically evaluated among the remaining 17 members of the FGF family. The present study was undertaken to determine the levels of structural stability among the entire human FGF family (FGF1–10/16–23), and establish to what extent this stability regulates FGF signaling in vitro and in vivo.
Materials and methods
Cell culture and FGF culture medium stability experiments: RCS and MCF7 cells were propagated in DMEM medium supplemented with 10 % FBS and antibiotics (Invitrogene, Carlsbad, CA). Recombinant FGFs were purchased from R&D Systems (Minneapolis, MN), except for FGF18, which was obtained from Life Technologies (Grand Island, NY). Heparin was obtained from Invitrogene. 1 × 104 cells/well were seeded in 24-well plates (Costar, Cambridge, MA) in 0.5 ml of culture medium, treated with FGFs for 72 h, and counted using an automated cell counter (Beckman Coulter, Brea, CA). For the culture medium stability experiments, 100 ng of recombinant FGF was added to 100 μl of cell-free DMEM medium pre-conditioned by a confluent RCS culture (grown in 14 ml of medium in T75 tissue culture flasks) for 12–24 h, and then was incubated in 24-well plates (Costar) in a tissue culture incubator (36.5 °C, 5 % CO2). To avoid the possibility of FGF binding to the plate plastic, samples were harvested by direct addition of 100 μl of SDS-PAGE loading buffer (125 mM Tris–HCl pH 6.8, 20 % glycerol, 4 % SDS, 0.005 % bromophenol blue, 5 % β-mercaptoethanol).
Western blotting (WB), FGFR plasmid generation, and cell transfection: Cells were lysed in buffer containing 50 mM Tris–HCl pH 7.4, 150 mM NaCl, 0.5 % NP-40, 1 mM EDTA, and 25 mM NaF. Protein samples were resolved by SDS-PAGE, transferred onto a PVDF membrane and visualized by chemiluminescence (Thermo Scientific, Rockford, IL). The following antibodies were used: ERK1/2, pERK1/2T202/Y204 (Cell Signaling, Beverly, MA); FGF1, FGF3, FGF4, FGF6–10, FGF16, FGF19–21, ACTIN, FGFR1–4 (Santa Cruz Biotechnology, Santa Cruz, CA); FGF5, FGF17, FGF18, FGF22, FGF23 (R&D Systems); FGF2 (Sigma-Aldrich, St. Louis, MO). To generate transgenic controls for FGFR expression, plasmids expressing FGFR1–4 were created by cloning human full-length FGFR1–4 cDNAs into a pcDNA3.1. vector according to the manufacturer’s protocol (Invitrogene). Cells were transfected using the FuGENE6 reagent (Roche, Indianapolis, IN).
Fluorescence-based thermal shift assay and CD spectroscopy: For the fluorescence-based thermal shift assay, 4 μg of recombinant FGF was mixed with 0.2 μl of SYPRO Orange (Sigma-Aldrich) and added to 96-well plates (Roche) to a total volume of 20 μl. Heparin was obtained from Sigma-Aldrich. Samples were heated from 25 to 90 °C in 0.06 °C increments using a LightCycler480 instrument (Roche). Ten fluorescence readings were taken every 1 °C using excitation and emission wavelengths at 465 and 580 nm, respectively. CD spectra were recorded at 20 °C using a Chirascan CD spectrometer (Applied Photophysics, Leatherhead, UK). Data were collected from 200 to 260 nm with a scan rate of 100 nm/min, 1 s response time, and 2 nm bandwidth using a 0.1-cm quartz cuvette containing recombinant FGFs obtained from R&D Systems, except for FGF1 and FGF2, which were produced as described below. FGFs, lyophilized from various buffer solutions (Table 1), were dissolved in sterile 50 mM phosphate buffer (pH 7.5) to a final concentration of 0.20–0.35 mg/ml. The measured CD spectra were presented as an average of five individual scans and corrected for absorbance due to the buffer. Data were expressed in terms of the mean residue ellipticity (Θ MRE) using the following equation:
where Θ obs is the observed ellipticity in degrees, M w is the protein molecular weight, n is number of residues, l is the cell path length (0.1 cm), c is the protein concentration, and the factor 100 originates from the conversion of the molecular weight to mg/dmol. Thermal unfolding of FGF proteins was followed by monitoring the CD spectra at a heating rate of 1 °C/min using a Chirascan CD spectrometer equipped with a Peltier set-up (Applied Photophysics). Spectra were scanned at 1 °C intervals over a temperature range of 20–80 °C. The resulting thermal denaturation curves at selected wavelengths were roughly normalized to represent a signal change between approximately 1 and 0 and fitted to sigmoidal curves using Origin 8 software (OriginLab, Massachusetts, MA). The melting temperature (T m), i.e., temperature at which half of the protein was in an unfolded state, was derived from the midpoint of the normalized thermal transition.
Table 1.
Formulation of lyophilized FGF proteins. PBS—phosphate buffer saline (10 mM Na2HPO4·2H20, 2 mM K2HPO4, 137 mM NaCl, 2.7 mM KCl)
| FGF | Buffer composition |
|---|---|
| FGF1 | 20 mM Tris, 2 M NaCl, 1 mM EDTA, 1 mM DTT (pH 7.4) |
| FGF2 | 20 mM Tris, 2 M NaCl, 1 mM EDTA, 1 mM DTT (pH 7.4) |
| FGF3 | 20 mM MOPS, 50 mM Na2SO4, 0.5 mM EDTA (pH 7.2) |
| FGF4 | PBS (pH 7.4) |
| FGF5 | 10 mM MOPS, 50 mM Na2SO4, 0.5 mM EDTA, 0.5 mM DTT (pH 7.0) |
| FGF6 | PBS, 0.05 % (w/v) CHAPS (pH 7.4) |
| FGF7 | PBS (pH 7.4) |
| FGF8 | 10 mM MOPS, 50 mM Na2SO4, 0.2 mM EDTA, 0.2 mM DTT (pH 7.0) |
| FGF9 | PBS (pH 7.4) |
| FGF10 | 20 mM MOPS, 50 mM Na2SO4, 0.5 mM EDTA, 1 mM DTT (pH 7.2) |
| FGF16 | 20 mM MOPS, 100 mM Na2SO4, 0.5 mM EDTA, 0.5 mM DTT (pH 7.0) |
| FGF17 | PBS (pH 7.4) |
| FGF19 | PBS (pH 7.4) |
FGF mutant production and analysis: For production of recombinant proteins, the pET-3c vector containing human FGF1 (residues 21-154) and pDEST15 vector containing N-terminally GST-tagged human FGF2 (residues 1-154) were used. Mutagenesis and protein expression were performed as described previously [5, 7, 22]. Recombinant proteins were expressed in E. coli Bl21(DE3)pLysS or Bl21(DE3)-RIL strains and purified on a heparin-Sepharose column (GE Healthcare, Piscataway, NJ). Purified FGF2-GST fusion proteins were cleaved by rTEV protease and subjected to tandem GSH-Sepharose HiTrap and heparin-Sepharose HiTrap chromatography using an Äkta Prime FPLC system (GE Healthcare). Protein purity was confirmed by SDS-PAGE, and molecular masses were verified by mass spectrometry. Thermodynamic properties of the recombinant FGFs were probed by CD and fluorescence spectrometry. Denaturation data were collected using cuvettes with a 10-mm path length and a scan rate of 0.25 °C/min. Temperature-induced ellipticity changes at 227 nm were followed using a slit width of 2 nm and 8 s response time. Fluorescence measurements monitored changes in the emission signal at 353 nm of the single tryptophan residue excited at 280 nm (Trp107 in FGF1 and Trp114 in FGF2). Both excitation and emission slits were 5-nm wide. The protein concentration was approximately 2 × 10−6 M in all thermodynamic experiments. Data were analyzed using PeakFit software (Jandel Scientific, San Rafael, CA) assuming a two-state reversible equilibrium transition. The fraction of protein in the native state at 36.5 °C was calculated from denaturation curves.
Bead implantation experiments: Fertilized chicken eggs (ISA brown) were purchased from Integra (Zabcice, Czech Republic). Eggs were incubated in a humidified forced air incubator at 37.8 °C and embryos were staged according to Hamburger and Hamilton [23]. All procedures were conducted following a protocol approved by the Ethical Committee of the University of Veterinary and Pharmaceutical Sciences (Brno, Czech Republic). Affigel Blue beads (Bio-Rad Laboratories, Hercules, Canada) of 200-μm diameter were soaked in FGF proteins and implanted into the right wing bud at stage HH20–22. Embryos were collected after 10–12 days of incubation and stained with alizarin red/alcian blue solution as previously described [24]. The following activity score was used to compare the strength of the observed phenotypes: the maximal phenotype scored one point for each of the individual skeletal element (humerus, radius, ulna), whereas if only partial morphological changes occurred, half a point was scored for each of the analyzed bones. Thus, the highest total score of 3 indicated the maximal phenotype, whereas a score of 0 indicated normal skeletal development.
Results and discussion
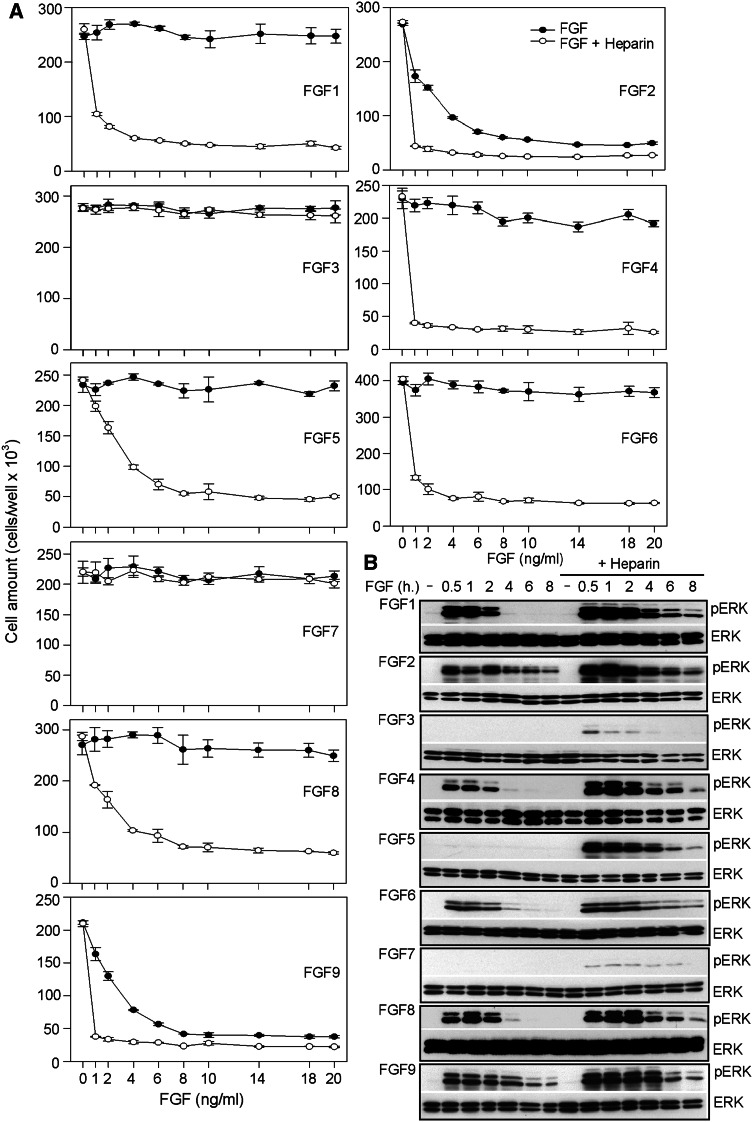
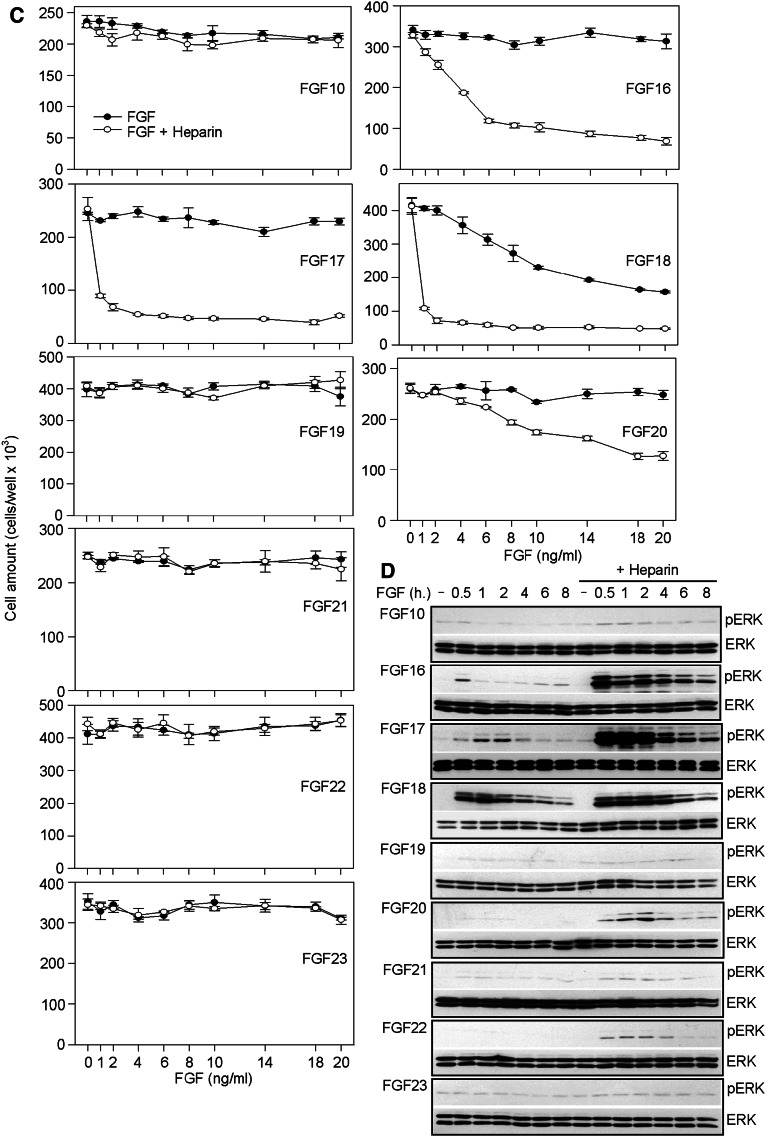
To assess FGF biological activity, we compared their effect on proliferation of rat chondrosarcoma (RCS) chondrocytes which responds to addition of exogenous FGF with potent growth arrest and characteristic changes in cellular morphology [25, 26]. We obtained growth curves of cells treated with 1–20 ng/ml of all 18 members of the FGF family. A total of 11 FGFs were found to inhibit RCS proliferation and to induce changes in cell morphology typically accompanying the growth arrest (Figs. 1; S1). These FGFs belonged to FGF4 (FGF4/6), FGF5 (FGF1/2/5), FGF8 (FGF8/17/18), and FGF9 (FGF9/16/20) subfamilies. In contrast, FGF3 together with FGFs belonging to FGF7 (FGF7/10/22) and FGF19 (FGF19/21/23) subfamilies showed no effect (Figs. 1a, c; S1). The results are in good agreement with the known ligand–receptor affinities within the FGF system. RCS cells express the so-called ‘IIIc’ variants of FGFRs, and therefore can only respond to FGFs belonging to FGF4, FGF5, FGF8, and FGF9 subfamilies but not FGF3 or members of the FGF7 subfamily that require the ‘IIIb’ variants of FGFRs [27–29]. On the other hand, FGF19/21/23 require the βKLOTHO transmembrane co-receptor for FGFR activation, and thus may not inhibit RCS proliferation due to the lack of βKLOTHO expression [30–32].
Fig. 1.
FGF inhibition of RCS cell proliferation. a, c Cells were treated as indicated for 72 h and then counted. Only FGF2/9/18-induced RCS growth arrest alone, whereas FGF1/4/5/6/8/17/20 required heparin to achieve this effect. No effect was observed for FGF3 and members of the FGF7 subfamily (FGF7/10/22) or for the FGF hormones (FGF19/21/23). b, d Cells were treated with 20 ng/ml of FGF for the indicated times and analyzed for ERK MAP kinase phosphorylation (pERK) by Western blotting (WB). Total ERK levels served as a loading control. Prolonged FGF-mediated ERK activation has been shown to lead to the growth arrest phenotype [26]; only FGF2/9/18 caused prolonged ERK activation in the absence of heparin
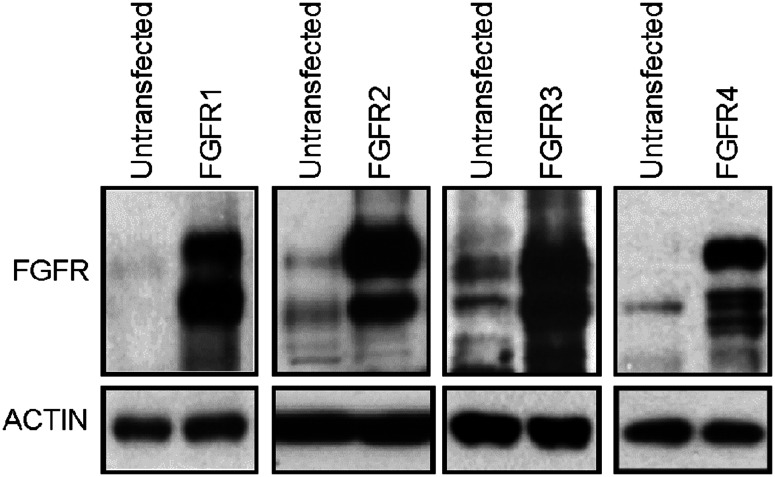
Among the 11 FGFs that inhibited RCS proliferation, only FGF2/9/18 achieved this phenotype when used alone; FGF1/4/5/6/8/16/17/20 required addition of exogenous heparin to inhibit RCS proliferation (Fig. 1a, c). It is well established that FGFs cause RCS growth arrest via sustained activation of the ERK MAP kinase pathway [27, 33]. We detected prolonged ERK activation only in cells treated with FGF2/9/18 alone, whereas FGF1/4/5/6/8/16/17/20 needed heparin to induce strong and prolonged ERK activation (Fig. 1b, d). The surprising dependence of FGF-mediated RCS cell growth arrest on exogenously added heparin observed for FGF1/4/5/6/8/16/17/20 is unlikely to be caused by the lack of cognate FGFRs or an absence of appropriate low-affinity FGF co-receptors at the cell surface. RCS cells express at least two FGFRs at their surface, i.e., FGFR2 and FGFR3 (Fig. 2), which serve as high-affinity receptors for FGF1/4/5/6/8/16–17/20. In fact, both FGF1 and FGF16 have been shown to exhibit higher affinity for FGFR2 and FGFR3 than FGF2 [28, 29], and yet we found that they failed to signal in the absence of heparin when compared to FGF2 (Fig. 1). Moreover, RCS cells produce abundant extracellular matrix rich in sulfated proteoglycans, which serve as low-affinity FGF co-receptors [34, 35].
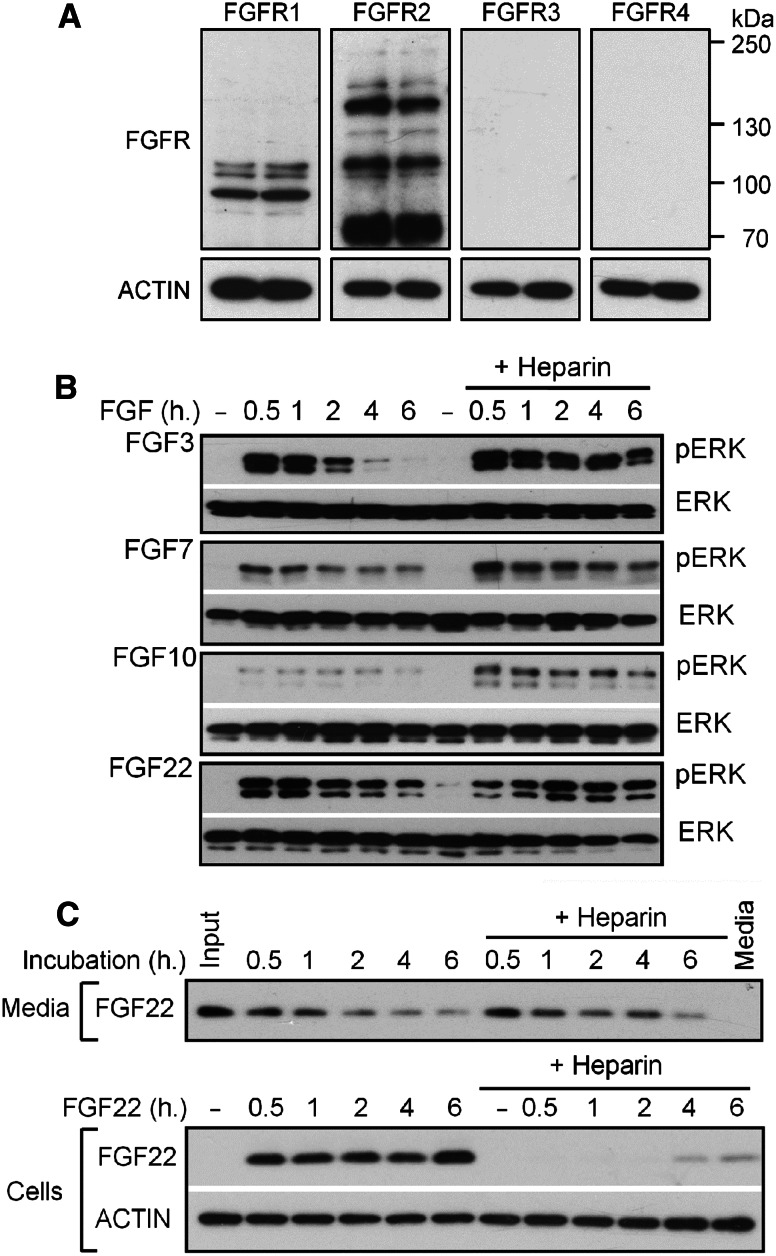
Fig. 2.
FGFR expression in RCS cells. Cells were transfected with plasmids expressing each of the four FGFR receptors (FGFR1–4) and the WB signal generated by each transgenic FGFR was compared with endogenous expression. Note the expression of endogenous FGFR2 and FGFR3 in untransfected cells
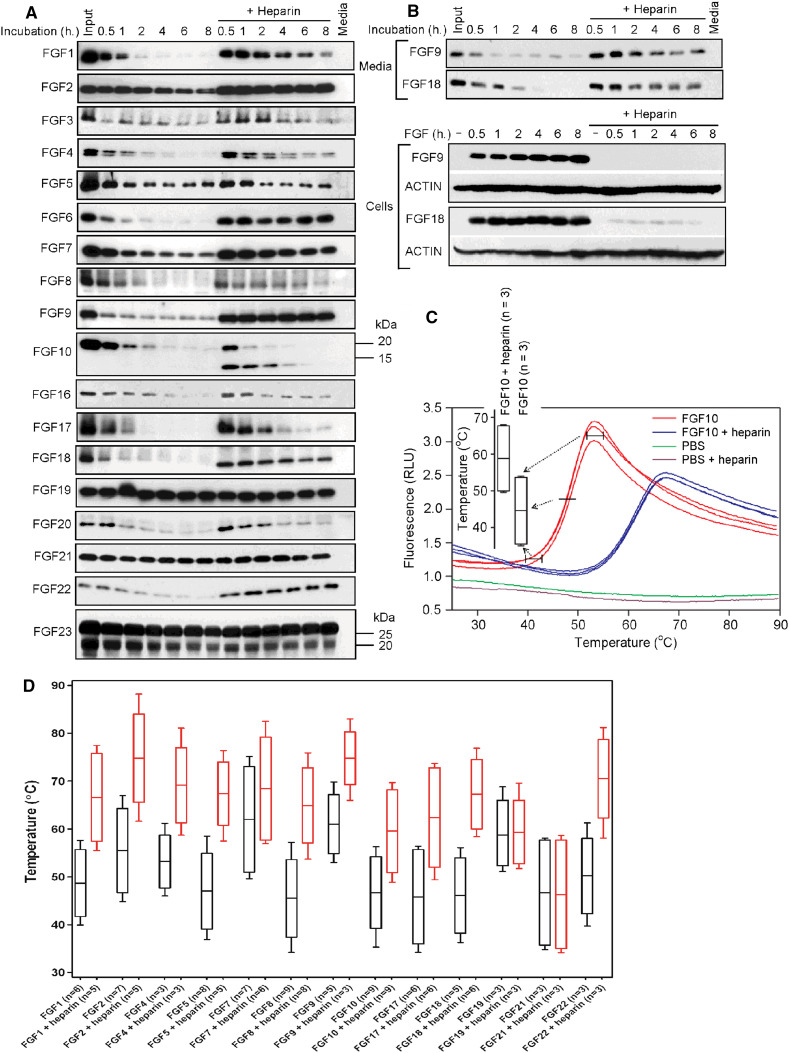
To test whether limited FGF stability accounts for their failure to inhibit RCS proliferation, we determined the rate of degradation of individual FGFs in cell-free tissue culture medium pre-conditioned by RCS cells for 12–24 h. Figure 3a shows that as many as 12 of the 18 tested FGFs (FGF1/3/4/6/8–10/16–18/20/22) were significantly degraded within the first 2 h in the media. Only partial degradation took place in medium containing FGF2/5/7, with ~20–50 % of input protein remaining for the entire 8 h incubation period. Finally, FGF19/21/23 remained exceptionally stable in the conditioned media, with virtually no degradation detected for the entire incubation period (Fig. 3a). This subfamily includes FGF hormones that regulate phosphate wasting (FGF23), metabolic adaptation to starvation (FGF21), and bile acid synthesis and glucose homeostasis (FGF19) [2, 36]. As the metabolic action of FGF hormones is rather slow, it requires FGF19/21/23 to exist in the circulation for prolonged periods of time [31, 36]. Thus the current understanding of FGF19/21/23 biology explains their superior stability in RCS-conditioned media.
Fig. 3.
FGF degradation in cell-conditioned culture media. a Recombinant FGFs were incubated in cell-free medium conditioned by RCS cells, with the amounts of FGF determined by WB. Note the degradation of FGF1/3/4/6/8–10/16–18/20/22, which was inhibited by exogenous heparin addition. Also note the persistence of intact FGF hormones FGF19/21/23. b RCS cells were treated with FGF9/18 for the indicated times and then both medium and cells were harvested and analyzed for FGF9/18 presence by WB. Note the FGF9/18 stabilization upon cell binding in cells treated with FGF9/18 alone, in contrast to heparin-mediated stabilization of soluble FGFs in cells treated with FGF9/18 in the presence of heparin. ACTIN served as a loading control. c Recombinant FGF10 was mixed with SYPRO Orange in phosphate-buffered saline (PBS), and thermal denaturation profiles were generated by heating the sample from 25 to 90 °C. The graph shows melting curves used to generate the box whisker plots presented in (d). FGF thermal denaturation profiles generated by fluorescence-based thermal shift assay (d). Note the stabilizing effect of heparin addition (red) in all but two (FGF19/21) of the tested FGFs. n number of independent experiments
Addition of heparin had a protective effect on the majority of the 15 paracrine FGFs (FGF1–4/6–9/16–18/20/22), with two exceptions. FGF10 was rapidly degraded regardless of heparin presence, whereas FGF5 remained largely intact throughout the entire incubation period (Fig. 3a). Correlation of the rate of FGF degradation with their activity in the RCS growth arrest experiments clearly demonstrated that a prolonged presence in the culture medium allowed FGF2 to inhibit RCS proliferation in the absence of heparin (Figs. 1, 3a). On the other hand, the failure to inhibit RCS proliferation observed for FGF1/4/6/8/16/17/20 stemmed from their rapid degradation in medium without added heparin. The only exceptions were FGF9/18, which were rapidly degraded in cell-free culture medium in the absence of heparin, yet activated ERK and inhibited RCS proliferation to an extent comparable to the relatively ‘stable’ FGF2 (Figs. 1, 3a). We showed that FGF9/18 are stabilized at the cell surface, possibly via interaction with cell-bound proteoglycans, thus explaining the inhibitory effect of FGF9/18 on RCS proliferation in the absence of heparin (Fig. 3b).
As RCS cells did not respond to FGF3, FGF7, FGF10, and FGF22 (Fig. 1), we used FGFR1 and FGFR2-expressing epithelial cell line MCF7 to analyze the effect of the latter FGFs on ERK activation (Fig. 4a). We found prolonged ERK activation in MCF7 cells treated FGF3 and FGF10 only in the presence of heparin (Fig. 4b), corresponding to their instability in the culture media (Fig. 3a). The significant ERK activation caused by FGF7 in the absence of heparin may be explained by its relative stability in the culture media. On the other hand, FGF22 was found unstable in cell-conditioned culture media (Fig. 3a), yet it caused strong ERK activation even in the absence of heparin (Figs. 3a, 4b). Similar to FGF9/18, we found the FGF22 stabilization at the cell surface (Fig. 4c), explaining its ability to cause strong ERK activation in the absence of heparin.
Fig. 4.
FGF3, FGF7, FGF10, and FGF22 effect on MCF7 cells. a Expression of endogenous FGFR1–4 in two independent protein samples extracted from MCF7 cells as determined by WB. Note that only FGFR1 and FGFR2 are expressed. ACTIN served as loading control. b MCF7 cells were cultivated in media conditioned by RCS cells for 12 h, treated with 20 ng/ml of FGF for the indicated times, and analyzed for ERK MAP kinase phosphorylation (pERK) by WB. Total ERK levels served as a loading control. c MCF7 cells were treated with FGF22 for the indicated times and then both medium and cells were harvested and analyzed for FGF22 presence by WB. Note the FGF22 stabilization upon cell binding in cells treated with FGF22 alone, in contrast to heparin-mediated stabilization of soluble FGF22 in cells treated with FGF22 in the presence of heparin. ACTIN served as a loading control
Rapid proteolytic degradation is one of several possible ways to limit FGF activity. It is known that some FGFs, such as FGF1, possess unstable tertiary structures. Thus, thermal instability of the proper fold may render a given FGF inactive before its proteolytic degradation takes place. Interaction with heparin restores the optimal FGF1 folding, allowing for FGFR binding and activation [22, 37]. Thermal instability may explain the failure of FGF5 to signal despite only partial degradation in RCS-conditioned medium, which is fully rescued by addition of exogenous heparin (Figs. 1, 3a; S1). We used a fluorescence-based thermal shift assay, in which SYPRO Orange fluorescence emitted during its interaction with hydrophobic patches exposed by denaturing proteins was measured, to monitor the stability of FGF tertiary structure [38]. Melting curves showing thermal denaturation of FGFs exposed to a temperature range of 30–90 °C were obtained for 13 FGFs that generated a signal in the fluorescence-based thermal shift assay (Fig. 3c). Heparin markedly improved the thermal stability of all FGFs except for FGF7, which was only weakly stabilized by heparin, and FGF19/21, for which heparin provided no improvement in stability (Fig. 3d). The latter finding corresponded well with the known poor affinity of FGF19/21 for heparin [31].
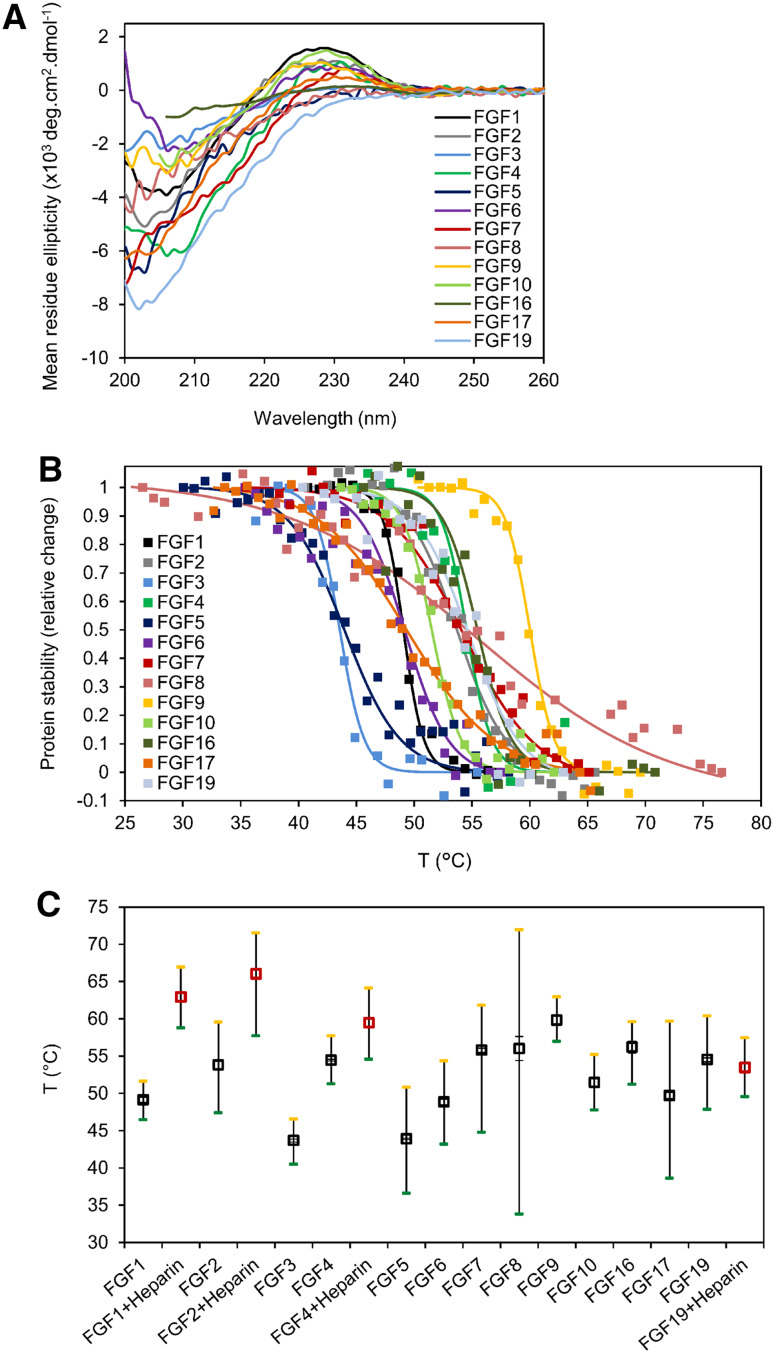
The fluorescence-based thermal shift assay data suggested substantial differences in the thermal stability across the 13 analyzed FGFs, although the extent of the interaction of SYPRO Orange with FGF may have been affected by the different distribution of hydrophobic residues unique to each studied FGF. Moreover, we were unable to determine thermal stability profiles for FGF3/6/20/23, possibly due to their precipitation in the carrier-free phosphate buffer solvent used in the assay. We therefore complemented the fluorescence-based thermal shift assay data with direct determination of FGF thermal stability profiles. Circular dichroism (CD) spectroscopy is a label-free method suitable for monitoring the secondary structure and conformational changes of proteins in solution [39]. Using CD, we probed native conformations and determined the thermal denaturation profiles of FGF1–10/16/17/19. Although the FGFs were analyzed in aqueous buffers of different composition (Table 1), they all exhibited CD spectra with similar patterns: a minimum at around 204–206 nm and a broad maximum centered near 227 nm (Fig. 5a), characteristic of β-rich proteins of β-II type [7, 39]. The studied FGF proteins could be ordered according to their thermal stability as follows: FGF3 ~ FGF5 < FGF8 ~ FGF18 ~ FGF21 < FGF6 ~ FGF1 ~ FGF17 ~ FGF22 < FGF10 < FGF2 ~ FGF4 ~ FGF19 < FGF7 ~ FGF16 < FGF9 (Table 2). The FGF thermal stability profiles determined by CD spectroscopy were in good agreement with those obtained by the fluorescence-based thermal shift assay; the only exception was FGF8, whose CD melting temperature was 10.6 °C lower than melting temperature determined by the fluorescence-based thermal shift assay (Table 2). Further, FGF8 exhibited the broadest thermal stability profile, suggesting that the protein undergoes conformational change at low temperatures (Fig. 5b, c). Similar to the fluorescence-based thermal shift assay, addition of heparin significantly improved the melting temperatures of all but one (FGF19) of the tested FGFs (Fig. 5c).
Fig. 5.
FGF CD spectra and thermal denaturation profiles. a Far-UV CD spectra of FGFs measured in aqueous buffers of various compositions (Table 1) at 20 °C. b Temperature-dependent unfolding of FGFs expressed as a relative change of protein stability, where fully folded and unfolded protein is represented by a value of 1 and 0, respectively. Squares represent relative changes in ellipticity at 227 nm derived from CD spectra of a particular FGF protein at different temperatures. Solid lines represent sigmoidal fits to the experimental data. c Thermal denaturation profiles of FGFs determined by CD spectroscopy. Black squares represent the determined melting temperature of a particular FGF, defined as the temperature at which half of the protein is in an unfolded state. Data represent the average of at least three independent measurements. The green and yellow marks represent temperatures at which 95 and 5 % of a particular protein is folded, respectively. Solid black lines represent the temperature range at which a particular protein undergoes unfolding. The short line indicates a sharp temperature-induced conformational transition of the protein. The red squares represent melting temperatures for particular FGF proteins in the presence of heparin
Table 2.
Comparison of FGF melting temperatures (T m) determined by fluorescence-based thermal shift assay (T 1m) and CD spectroscopy (T 2m)
| FGF | T 1m (°C) | T 2m (°C) | ΔT 1−2m (°C) |
|---|---|---|---|
| FGF1 | 48.7 ± 1.8 | 49.1 ± 0.3 | −0.4 |
| FGF2 | 55.5 ± 1.5 | 53.8 ± 0.5 | 1.7 |
| FGF3 | n.d. | 43.7 ± 0.2 | – |
| FGF4 | 53.2 ± 2.1 | 54.5 ± 0.1 | −1.3 |
| FGF5 | 47.0 ± 1.9 | 43.9 ± 0.1 | 3.1 |
| FGF6 | n.d. | 48.8 ± 0.4 | – |
| FGF7 | 62.0 ± 1.7 | 55.8 ± 0.3 | 6.2 |
| FGF8 | 45.5 ± 1.9 | 56.0 ± 1.6 | −10.5 |
| FGF9 | 61.0 ± 2.2 | 59.8 ± 0.5 | 1.2 |
| FGF10 | 46.7 ± 2.7 | 51.4 ± 0.6 | −4.7 |
| FGF16 | n.d. | 56.2 ± 0.8 | – |
| FGF17 | 45.8 ± 1.2 | 49.7 ± 0.4 | −3.9 |
| FGF18 | 46.1 ± 1.7 | n.d. | – |
| FGF19 | 58.7 ± 2.0 | 54.5 ± 0.3 | 4.2 |
| FGF21 | 46.7 ± 0.6 | n.d. | – |
| FGF22 | 50.2 ± 0.6 | n.d. | – |
n.d. not determined
We altered the FGF stability to experimentally evaluate its effect on FGF signaling. For a gain-of-function approach, we selected the ‘unstable’ FGF1 (Fig. 3a), taking advantage of the published stable FGF1 mutant (FGF1Q40P/S47I/H93G). The combination of Q40P/S47I/H93G substitutions improved the thermal stability of FGF1 by over 21 °C compared to the wild-type (wt) FGF1, without altering other properties such as interactions with FGFR and heparin [7]. FGF1Q40P/S47I/H93G was less than 0.001 % denatured (biologically inactive and highly susceptible to protease action) at physiological temperature, in contrast to wt FGF1, for which ~25–30 % of the molecules were unfolded under such conditions (Table 3). In an opposite approach, we used point-directed mutagenesis to destabilize the ‘stable’ FGF, FGF2 (Fig. 3a). Two FGF2 mutants, FGF2L140A and FGF2R109A/K110A, were generated with thermal stability lowered by 4.6 or 3.3 °C, respectively, when compared to wt FGF2 (Table 3).
Table 3.
Thermal stability of FGF1/2 mutants
| Method | CD | Fluorescence | ||||||
|---|---|---|---|---|---|---|---|---|
| FGF | T m (°C) | ΔT m (°C) | Native fraction at 36.5 °C (%) | Native fraction at 34 °C (%) | T m (°C) | ΔT m (°C) | Native fraction at 36.5 °C (%) | Native fraction at 34 °C (%) |
| FGF1 | 39.9 ± 0.1 | – | 75.5 ± 0.2 | 87.9 ± 0.1 | 39.2 ± 0.1 | – | 70.6 ± 0.8 | 84.8 ± 0.8 |
| FGF1Q40P/S471/H93G | 61.4 ± 0.4 | 21.6 | 100.0 ± 0.0 | 100.0 ± 0.0 | 60.1 ± 0.4 | 21.0 | 100.0 ± 0.0 | 100.0 ± 0.0 |
| FGF2 | 43.4 ± 0.3 | – | 93.2 ± 1.1 | 97.3 ± 0.5 | 43.8 ± 0.3 | – | 92.8 ± 1.3 | 97.0 ± 0.7 |
| FGF2L140A | 38.8 ± 0.1 | −4.6 | 68.3 ± 0.4 | 83.5 ± 0.6 | 39.2 ± 0.4 | −4.6 | 72.7 ± 2.5 | 86.9 ± 1.5 |
| FGF2R109A/K110A | 40.2 ± 0.4 | −3.3 | 78.8 ± 2.3 | 90.2 ± 1.3 | 40.2 ± 0.2 | −3.6 | 78.9 ± 1.5 | 90.3 ± 0.9 |
Measurements were performed by CD at 227 nm and fluorescence at 353 nm due to the single tryptophan residue excited at 280 nm (Trp107 in FGF1, Trp114 in FGF2), in the presence of 0.7 M guanidinium chloride (GdmCl) to prevent protein aggregation and accumulation of folding intermediates [5, 22]. Data represent the average of three independent denaturation experiments with the indicated range. Fractions of proteins in the native state at 36.5 °C are indicated. Note the wt FGF1/2 T m differences when compared with Table 2, which are due to the GdmCl use. ΔT m = T m (mutant) − T m (wt)
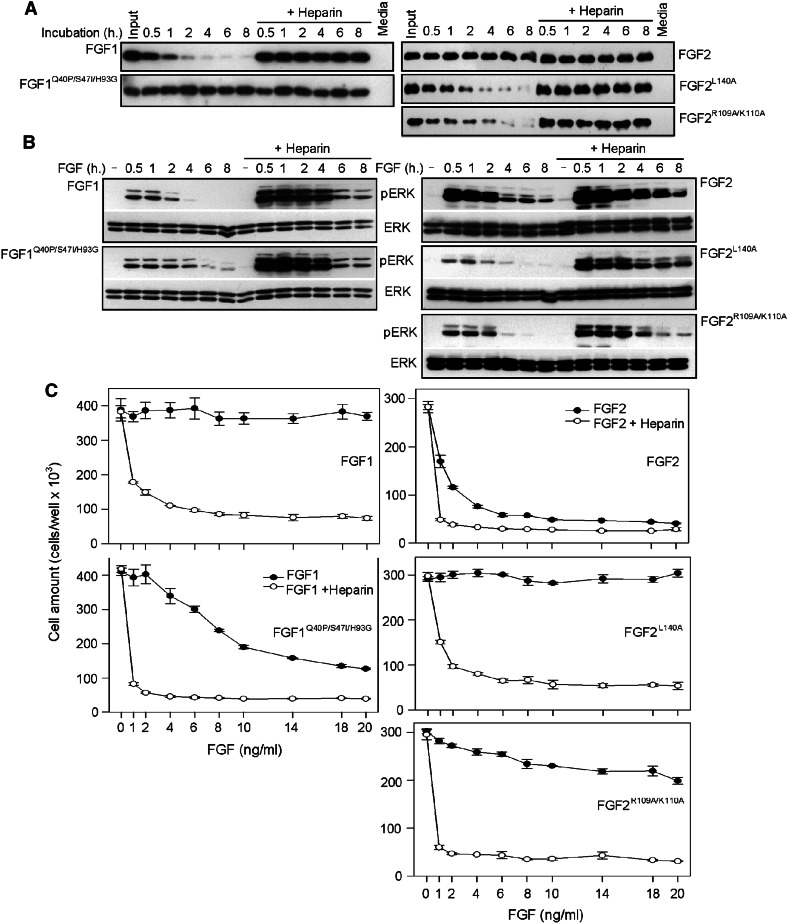
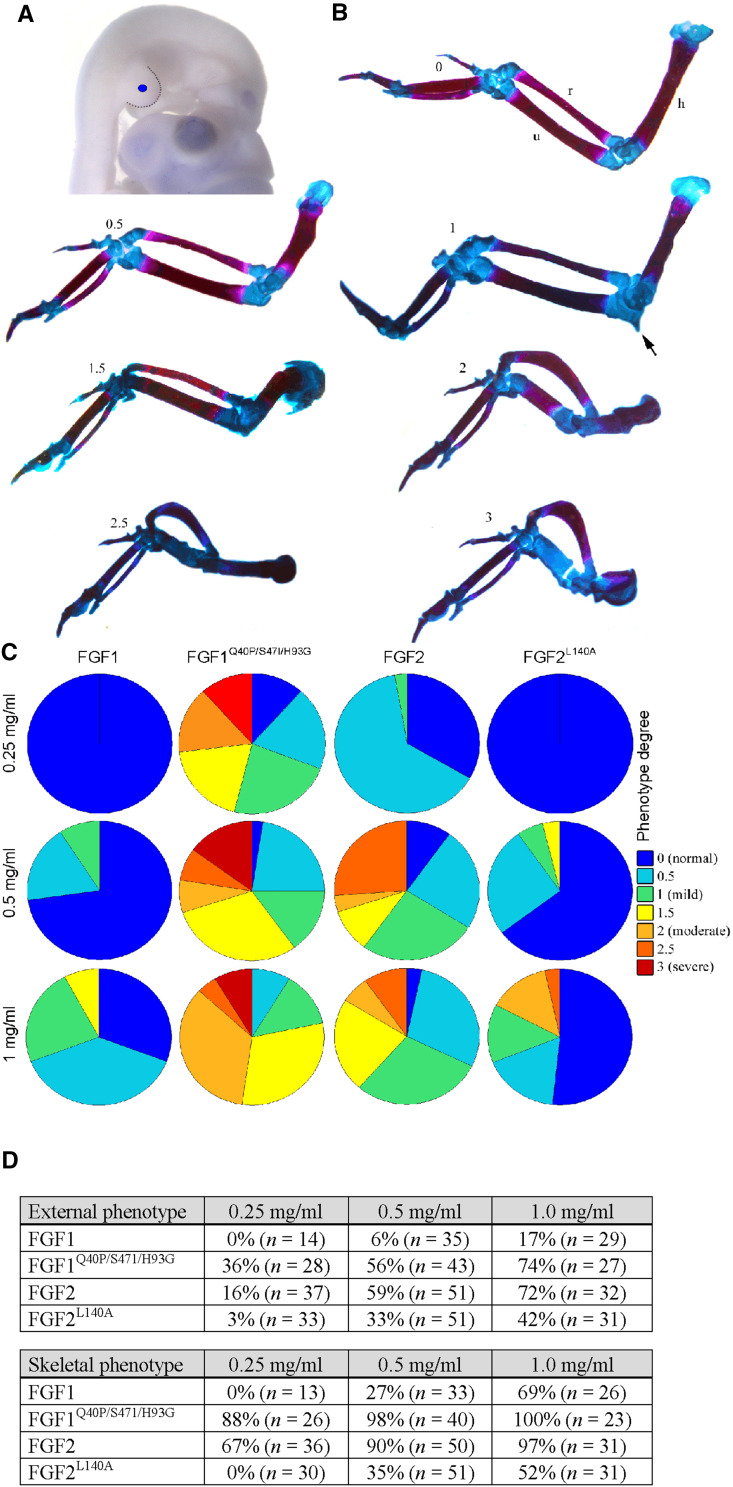
FGF1 stabilization rescued its failure to inhibit RCS proliferation in the absence of heparin (Fig. 6a–c). Both FGF2L140A and FGF2R109A/K110A showed significantly lower resistance to degradation in cell-conditioned medium compared to wt FGF2 (Fig. 6a), with corresponding loss of activity in the growth arrest experiment (Fig. 6b, c). Since cartilage represents a well-characterized tissue system with which to study the effects FGF signaling [40], we used the process of FGF/FGFR-mediated inhibition of skeletal growth for in vivo evaluation of biological activity of FGF1 and FGF2 mutants. Affigel Blue beads soaked with FGFs were implanted into the basis of chicken wing buds at embryonic stages 20–22 and the FGF effect on skeletal development was determined 10–12 days later (Fig. 7a). FGF implantation resulted in skeletal malformations resembling aberrant activation of FGFR/ERK MAP kinase signaling in the growth plate cartilage [40]. These included hypoplasia in both the humerus and ulna accompanied by ossification delay and ectopic cartilaginous outgrowths on the humerus or proximal antebrachium (Fig. 7b). The embryos implanted with the stable FGF1Q40P/S47I/H93G mutant showed a significantly higher percentage of skeletal abnormalities when compared to wt FGF1, at all three concentrations used. Exactly the opposite effect was observed for FGF2, with more abnormal embryos among those implanted with wt FGF2 (97 %, n = 31, at 1.0 mg/ml) compared to the less stable FGF2L140A (52 %, n = 31) (Fig. 7c, d).
Fig. 6.
Cell culture medium degradation and biological activity of FGF1/2 mutants. a Recombinant FGF1/2 and their mutants were incubated in cell-free medium conditioned by RCS, and levels of both proteins were determined by WB. When compared to wt FGF1, the stable FGF1Q40P/S47I/H93G mutant showed increased phosphorylation of ERK MAP kinase over a long period of time and inhibition of RCS proliferation even in the absence of heparin (b, c). In contrast, FGF2 destabilization by L140A or R109A/K110A substitutions resulted in increased degradation and lack of activity in the RCS growth arrest assay (a–c)
Fig. 7.
Effect of FGF1/2 on chicken wing development. a Wing buds of chicken embryos (stage 20–22) were implanted as indicated with beads soaked by various concentrations of wt FGF1, FGF2, stabilized FGF1 mutant (FGF1Q40P/S47I/H93G), and destabilized FGF2 mutant (FGF2L140A). Embryos were left to develop for about 12 days before skeletal preparations were made (alcian blue and alizarin red staining for cartilage and bone, respectively) (b). Note the different severity of wing bone malformations following treatment with FGF1Q40P/S47I/H93G (divided according to a 0.5–3 scale) compared to normal wing skeleton (scale 0). The arrow indicates an ectopic cartilaginous process on the antebrachium. The percentages of actual malformations obtained for each FGF concentration are represented by pie charts (c), and summarized in (d). A higher percentage of abnormal embryos was observed when stabilizing FGF1Q40P/S47I/H93G was implanted compared to ‘unstable’ wt FGF1. In contrast, implantation of ‘unstable’ FGF2L140A resulted in a lower percentage of abnormal bones when compared to ‘stable’ wt FGF2. d Summary of external and skeletal malformations in chicken limb buds implanted with FGF1/2. The selected FGF concentration range was 0.25 mg/ml (no phenotype observed in embryos implanted with wt FGF1 or FGF2L140A) to 1.0 mg/ml (100 % embryos affected when implanted with FGF1Q40P/S47I/H93G). Embryos were first analyzed for external phenotype before skeletal preparations were made. The tables show the percentage of abnormal embryos in each group; n number of implanted embryos
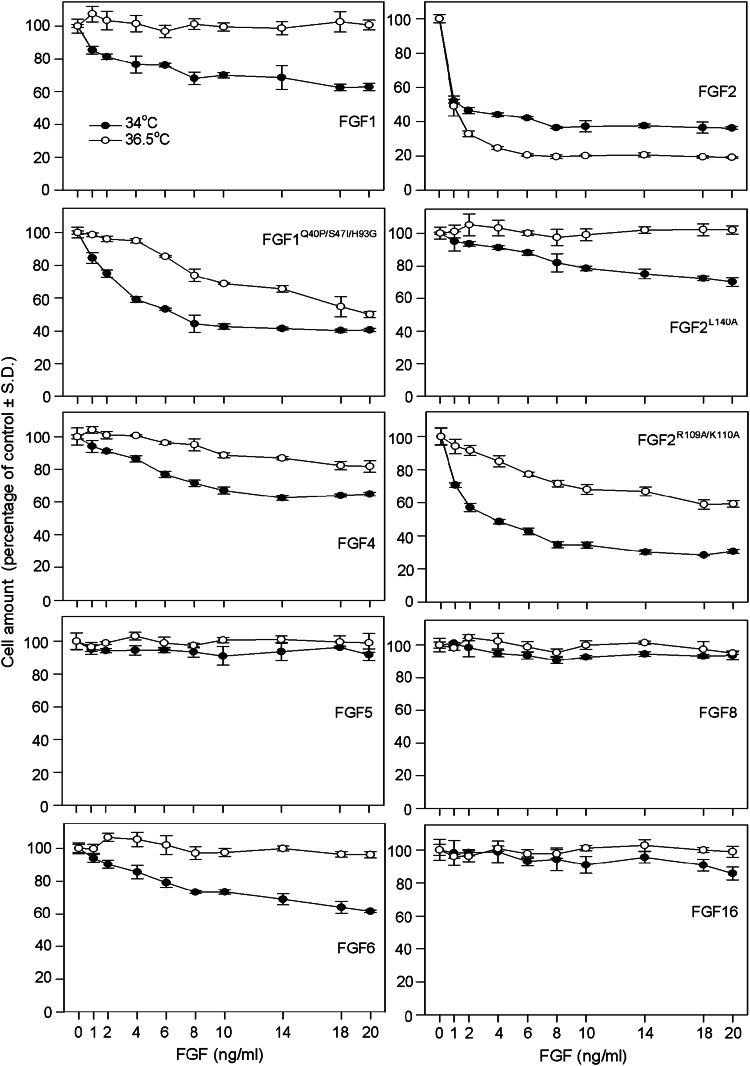
Lowering the RCS cell cultivation temperature by 2.5 °C (34 vs. 36.5 °C) enhanced the growth-inhibitory activities of the stable FGF1Q40P/S47I/H93G mutant and both FGF2L140A and FGF2R109A/K110A mutants (Fig. 8). In addition, wt FGF1 inhibited the proliferation of cells grown at 34 °C in the absence of heparin, compared to 36.5 °C where it had no effect (Figs. 1a, 6c, 8). We next determined the effect of lower cultivation temperature on other unstable FGFs that required heparin-mediated stabilization to inhibit RCS proliferation at 36.5 °C. Among the tested FGFs (FGF4/5/6/8/16), FGF4/6 were found to inhibit RCS proliferation at 34 °C (Fig. 8).
Fig. 8.
Temperature effect on FGF-mediated inhibition of RCS proliferation. RCS cells were treated with the indicated FGFs for 72 h and counted. Graphs compare cell growth at standard (36.5 °C) and lower (34 °C) temperature. Data are expressed as a percentage of the control to compensate for the lower proliferation rate of cells grown at 34 °C. Note the more potent inhibition of RCS proliferation mediated by wt FGF1, FGF1Q40P/S47I/H93G, FGF2L140A, FGF2R109A/K110A, FGF4, and FGF6 at 34 °C
We present evidence that the majority of the 15 human paracrine FGFs display intrinsically low stability. This low stability attenuates biological activity of at least 10 different FGFs, i.e., FGF1/3/4/5/6/8/10/16/17/20. Experimental stabilization via exogenous heparin binding, introduction of stabilizing mutations or lowering the cell cultivation temperature rescues the biological activity of unstable FGFs in vitro and in vivo. In this study, we correlated the FGF stability and their biological functions in only two cell types, thus overlooking the possible specific effects of cell environment on particular FGF stability. This represents one limitation on the conclusions obtained here, since it is likely that the proteolytic stability of the given FGF depends, in vivo, on the cellular and tissue context, for instance on the proteinases produced by the particular cell types involved. As FGFs are expressed at different levels during development, a high production of an individual FGF may partially counteract its instability in vivo, allowing for prolonged signaling.
Another important regulatory factor is an extent of proteoglycan-mediated FGF stabilization, which varies significantly among the individual FGFs (Fig. 3a, d). Although all FGFs carry a proteoglycan binding domain, none of the residues involved in proteoglycan interaction appears completely conserved throughout the FGF family [41, 42]. Basic information on mechanism of FGF interaction with proteoglycans comes from co-crystal structures of FGF1-heparan sulfate glycosaminoglycan (HSGAG), FGF2-HSGAG, FGF2-FGFR1-HSGAG, and FGF1-FGFR2-HSGAG. Analysis of FGF–HSGAG binding interfaces demonstrated that specific topology of the HSGAG-binding loops and the spatial arrangement of basic amino acids in these loops impose distinctive structural requirements on the HSGAG chain for different FGF family members [43]. Thus, the differences in spatial arrangement of basic amino acids in the HSGAG-binding loops account for differences in interaction energies between individual FGFs and the HSGAG.
In summary, our findings demonstrate that the biological activity of many FGFs may be critically regulated by their half-life, reflecting both stability and degradation. This has several implications for our understanding of the FGF system. First, differences in stability inherent to the individual FGFs may constitute an elementary level of regulation of FGF activity in developmental processes that are co-regulated by several FGF ligands. Second, the majority of paracrine FGFs may signal for a prolonged period of time only when stabilized by interaction with proteoglycans, which are predominantly cell-bound in vivo. This may represent a critical factor in the establishment of FGF gradient in developing tissues, as well as constitute an important physiological barrier to ectopic FGF signaling. Third, the thermal stabilization/destabilization may play a role in pathological conditions involving FGF signaling, such as tumor lesions that grow at lower than physiological temperatures or maternal hyperthermia, where multiple developmental malformations result from prolonged fetal exposure to high body temperatures [44, 45].
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank M. Pokorna and K. Tvaruzkova for assistance with CD measurements, J. Medalova and T. Spoustova for assistance with manuscript preparation, and P. B. Mekikian for technical support. The study was supported by the Ministry of Education, Youth and Sports of the Czech Republic (KONTAKT LH12004, CZ.1.07/2.3.00/30.0053, LO1214), Grant Agency of Masaryk University (0071-2013), Czech Science Foundation (14-31540S, P207/12/0775, GBP302/12/G157), European Regional Development Fund (FNUSA-ICRC No.CZ.1.05/1.1.00/02.0123), European Union (ICRC-ERA-HumanBridge No.316345), Netherlands Organization for Scientific Research (700.59.426) and Polish National Science Centre (NCN, 2011/02/A/NZ1/00066).
References
- 1.Itoh N, Ornitz DM. Functional evolutionary history of the mouse Fgf gene family. Dev Dyn. 2008;237:18–27. doi: 10.1002/dvdy.21388. [DOI] [PubMed] [Google Scholar]
- 2.Itoh N. Hormone-like (endocrine) Fgfs: their evolutionary history and roles in development, metabolism, and disease. Cell Tissue Res. 2010;342:1–11. doi: 10.1007/s00441-010-1024-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Copeland RA, Ji H, Halfpenny AJ, Williams RW, Thompson KC, Herber WK, Thomas KA, Bruner MW, Ryan JA, Marquis-Omer D. The structure of human acidic fibroblast growth factor and its interaction with heparin. Arch Biochem Biophys. 1991;289(1):53–61. doi: 10.1016/0003-9861(91)90441-K. [DOI] [PubMed] [Google Scholar]
- 4.Culajay JF, Blaber SI, Khurana A, Blaber M. Thermodynamic characterization of mutants of human fibroblast growth factor 1 with an increased physiological half-life. Biochemistry. 2000;39:7153–7158. doi: 10.1021/bi9927742. [DOI] [PubMed] [Google Scholar]
- 5.Zakrzewska M, Krowarsch D, Wiedlocha A, Otlewski J. Design of fully active FGF-1 variants with increased stability. Protein Eng Des Sel. 2004;17:603–611. doi: 10.1093/protein/gzh076. [DOI] [PubMed] [Google Scholar]
- 6.Buczek O, Krowarsch D, Otlewski J. Thermodynamics of single peptide bond cleavage in bovine pancreatic trypsin inhibitor (BPTI) Protein Sci. 2002;11:924–932. doi: 10.1110/ps.4460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zakrzewska M, Krowarsch D, Wiedlocha A, Olsnes S, Otlewski J. Highly stable mutants of human fibroblast growth factor-1 exhibit prolonged biological action. J Mol Biol. 2005;352:860–875. doi: 10.1016/j.jmb.2005.07.066. [DOI] [PubMed] [Google Scholar]
- 8.Damon DH, Lobb RR, D’Amore PA, Wagner JA. Heparin potentiates the action of acidic fibroblast growth factor by prolonging its biological half-life. J Cell Physiol. 1989;138:221–226. doi: 10.1002/jcp.1041380202. [DOI] [PubMed] [Google Scholar]
- 9.Derrick T, Grillo AO, Vitharana SN, Jones L, Rexroad J, Shah A, Perkins, Spitznagel TM, Middaugh CR. Effect of polyanions on the structure and stability of repifermin (keratinocyte growth factor-2) J Pharm Sci. 2007;96:761–776. doi: 10.1002/jps.20797. [DOI] [PubMed] [Google Scholar]
- 10.Chen G, Gulbranson DR, Yu P, Hou Z, Thomson JA. Thermal stability of fibroblast growth factor protein is a determinant factor in regulating self-renewal, differentiation, and reprogramming in human pluripotent stem cells. Stem Cells. 2012;30:623–630. doi: 10.1002/stem.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Econs MJ, McEnery PT. Autosomal dominant hypophosphatemic rickets/osteomalacia: clinical characterization of a novel renal phosphate-wasting disorder. J Clin Endocrinol Metab. 1997;82(2):674–681. doi: 10.1210/jcem.82.2.3765. [DOI] [PubMed] [Google Scholar]
- 12.White KE, Carn G, Lorenz-Depiereux B, Benet-Pages A, Strom TM, Econs MJ. Autosomal-dominant hypophosphatemic rickets (ADHR) mutations stabilize FGF-23. Kidney Int. 2001;60(6):2079–2086. doi: 10.1046/j.1523-1755.2001.00064.x. [DOI] [PubMed] [Google Scholar]
- 13.Kato K, Jeanneau C, Tarp MA, Benet-Pagès A, Lorenz-Depiereux B, Bennett EP, Mandel U, Strom TM, Clausen H. Polypeptide GalNAc-transferase T3 and familial tumoral calcinosis. Secretion of fibroblast growth factor 23 requires O-glycosylation. J Biol Chem. 2006;281(27):18370–18377. doi: 10.1074/jbc.M602469200. [DOI] [PubMed] [Google Scholar]
- 14.Fukumoto S, Yamashita T. FGF23 is a hormone-regulating phosphate metabolism-unique biological characteristics of FGF23. Bone. 2007;40(5):1190–1195. doi: 10.1016/j.bone.2006.12.062. [DOI] [PubMed] [Google Scholar]
- 15.Colvin JS, White AC, Pratt SJ, Ornitz DM. Lung hypoplasia and neonatal death in Fgf9-null mice identify this gene as an essential regulator of lung mesenchyme. Development. 2001;128(11):2095–2106. doi: 10.1242/dev.128.11.2095. [DOI] [PubMed] [Google Scholar]
- 16.Ohbayashi N, Shibayama M, Kurotaki Y, Imanishi M, Fujimori T, Itoh N, Takada S. FGF18 is required for normal cell proliferation and differentiation during osteogenesis and chondrogenesis. Genes Dev. 2002;16(7):870–879. doi: 10.1101/gad.965702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Usui H, Shibayama M, Ohbayashi N, Konishi M, Takada S, Itoh N. Fgf18 is required for embryonic lung alveolar development. Biochem Biophys Res Commun. 2004;322(3):887–892. doi: 10.1016/j.bbrc.2004.07.198. [DOI] [PubMed] [Google Scholar]
- 18.Vincentz JW, McWhirter JR, Murre C, Baldini A, Furuta Y. Fgf15 is required for proper morphogenesis of the mouse cardiac outflow tract. Genesis. 2005;41(4):192–201. doi: 10.1002/gene.20114. [DOI] [PubMed] [Google Scholar]
- 19.Lu SY, Sheikh F, Sheppard PC, Fresnoza A, Duckworth ML, Detillieux KA, Cattini PA. FGF-16 is required for embryonic heart development. Biochem Biophys Res Commun. 2008;373(2):270–274. doi: 10.1016/j.bbrc.2008.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cholfin JA, Rubenstein JL. Patterning of frontal cortex subdivisions by Fgf17. Proc Natl Acad Sci USA. 2007;104(18):7652–7657. doi: 10.1073/pnas.0702225104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, Yagishita N, Matsui D, Koga Y, Itoh N, Kato S. Fgf10 is essential for limb and lung formation. Nat Genet. 1999;21(1):138–141. doi: 10.1038/5096. [DOI] [PubMed] [Google Scholar]
- 22.Zakrzewska M, Wiedlocha A, Szlachcic A, Krowarsch D, Otlewski J, Olsnes S. Increased protein stability of FGF1 can compensate for its reduced affinity for heparin. J Biol Chem. 2009;284:25388–25403. doi: 10.1074/jbc.M109.001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. J Morphol. 1951;88:49–92. doi: 10.1002/jmor.1050880104. [DOI] [PubMed] [Google Scholar]
- 24.Plant MR, MacDonald ME, Grad LI, Ritchie SJ, Richman JM. Locally released retinoic acid repatterns the first branchial arch cartilages in vivo. Dev Biol. 2000;222:12–26. doi: 10.1006/dbio.2000.9706. [DOI] [PubMed] [Google Scholar]
- 25.Krejci P, Prochazkova J, Smutny J, Chlebova K, Lin P, Aklian A, Bryja V, Kozubik A, Wilcox WR. FGFR3 signaling induces a reversible senescence phenotype in chondrocytes similar to oncogene-induced premature senescence. Bone. 2010;47:102–110. doi: 10.1016/j.bone.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krejci P, Salazar L, Goodridge HS, Kashiwada TA, Schibler MJ, Jelinkova P, Thompson LM, Wilcox WR. STAT1 and STAT3 do not participate in FGF-mediated growth arrest in chondrocytes. J Cell Sci. 2008;121:272–281. doi: 10.1242/jcs.017160. [DOI] [PubMed] [Google Scholar]
- 27.Krejci P, Bryja V, Pachernik J, Hampl A, Pogue R, Mekikian P, Wilcox WR. FGF2 inhibits proliferation and alters the cartilage-like phenotype of RCS cells. Exp Cell Res. 2004;297:152–164. doi: 10.1016/j.yexcr.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 28.Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, Gao G, Goldfarb M. Receptor specificity of the fibroblast growth factor family. J Biol Chem. 1996;27:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J Biol Chem. 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu X, Ge H, Gupte J, Weiszmann J, Shimamoto G, Stevens J, Hawkins N, Lemon B, Shen W, Xu J, Veniant MM, Li YS, Lindberg R, Chen JL, Tian H, Li Y. Co-receptor requirements for fibroblast growth factor-19 signaling. J Biol Chem. 2007;282:29069–29072. doi: 10.1074/jbc.C700130200. [DOI] [PubMed] [Google Scholar]
- 31.Goetz R, Beenken A, Ibrahimi OA, Kalinina J, Olsen SK, Eliseenkova AV, Xu C, Neubert TA, Zhang F, Linhardt RJ, Yu X, White KE, Inagaki T, Kliewer SA, Yamamoto M, Kurosu H, Ogawa Y, Kuro-o M, Lanske B, Razzaque MS, Mohammadi M. Molecular insights into the klotho-dependent, endocrine mode of action of fibroblast growth factor 19 subfamily members. Mol Cell Biol. 2007;27:3417–3428. doi: 10.1128/MCB.02249-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krejci P, Krakow D, Mekikian PB, Wilcox WR. Fibroblast growth factors 1, 2, 17, and 19 are the predominant FGF ligands expressed in human fetal growth plate cartilage. Pediatr Res. 2007;61:267–272. doi: 10.1203/pdr.0b013e318030d157. [DOI] [PubMed] [Google Scholar]
- 33.Raucci A, Laplantine E, Mansukhani A, Basilico C. Activation of the ERK1/2 and p38 mitogen-activated protein kinase pathways mediates fibroblast growth factor-induced growth arrest of chondrocytes. J Biol Chem. 2004;279:1747–1756. doi: 10.1074/jbc.M310384200. [DOI] [PubMed] [Google Scholar]
- 34.Krejci P, Masri B, Fontaine V, Mekikian PB, Weis M, Prats H, Wilcox WR. Interaction of fibroblast growth factor and C-natriuretic peptide signaling in regulation of chondrocyte proliferation and extracellular matrix homeostasis. J Cell Sci. 2005;118:5089–5100. doi: 10.1242/jcs.02618. [DOI] [PubMed] [Google Scholar]
- 35.Yayon A, Klagsbrun M, Esko JD, Leder P, Ornitz DM. Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell. 1991;64:841–848. doi: 10.1016/0092-8674(91)90512-W. [DOI] [PubMed] [Google Scholar]
- 36.Angelin B, Larsson TE, Rudling M. Circulating fibroblast growth factors as metabolic regulators-a critical appraisal. Cell Metab. 2012;16:693–705. doi: 10.1016/j.cmet.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 37.Burke CJ, Volkin DB, Mach H, Middaugh CR. Effect of polyanions on the unfolding of acidic fibroblast growth factor. Biochemistry. 1993;32:6419–6426. doi: 10.1021/bi00076a015. [DOI] [PubMed] [Google Scholar]
- 38.Lavinder JJ, Hari SB, Sullivan BJ, Magliery TJ. High-throughput thermal scanning: a general, rapid dye-binding thermal shift screen for protein engineering. J Am Chem Soc. 2009;131:3794–3795. doi: 10.1021/ja8049063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fasman GD. Circular dichroism and conformational analysis of biomolecules. New York: Plenum Press; 1996. [Google Scholar]
- 40.Foldynova-Trantirkova S, Wilcox WR, Krejci P. Sixteen years and counting: the current understanding of fibroblast growth factor receptor 3 (FGFR3) signaling in skeletal dysplasias. Hum Mutat. 2012;33:29–41. doi: 10.1002/humu.21636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol. 2001;2:REVIEWS3005. doi: 10.1186/gb-2001-2-3-reviews3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Faham S, Lindardt RJ, Rees DC. Diversity does make a difference: fibroblast growth factor-heparin interactions. Curr Opin Struct Biol. 1998;8:578–586. doi: 10.1016/S0959-440X(98)80147-4. [DOI] [PubMed] [Google Scholar]
- 43.Raman R, Venkataraman G, Ernst S, Sasisekharan V, Sasisekharan R. Structural specifity of heparin binding in the fibroblast growth factor family of proteins. Proc Natl Acad Sci USA. 2003;100:2357–2362. doi: 10.1073/pnas.0437842100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jayasundar R, Singh VP. In vivo temperature measurements in brain tumors using proton MR spectroscopy. Neurol India. 2002;50:436–439. [PubMed] [Google Scholar]
- 45.Chambers CD, Johnson KA, Dick LM, Felix RJ, Jones KL. Maternal fever and birth outcome: a prospective study. Teratology. 1998;58:251–257. doi: 10.1002/(SICI)1096-9926(199812)58:6<251::AID-TERA6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.