Abstract
Rho GTPases are a class of evolutionarily conserved proteins comprising 20 members, which are predominantly known for their role in regulating the actin cytoskeleton. They are primarily regulated by binding of GTP/GDP, which is again controlled by regulators like GEFs, GAPs, and RhoGDIs. Rho GTPases are thus far well known for their role in the regulation of actin cytoskeleton and migration. Here we present an overview on the role of Rho GTPases in regulating cell shape and plasticity of cell migration. Finally, we discuss the emerging roles of ubiquitination and sumoylation in regulating Rho GTPases and cell migration.
Keywords: Rac1, RhoA, Cdc42, Rho GTPases, Cell shape, Cell migration, Plasticity, Amoeboid, Mesenchymal, Metastases
Introduction
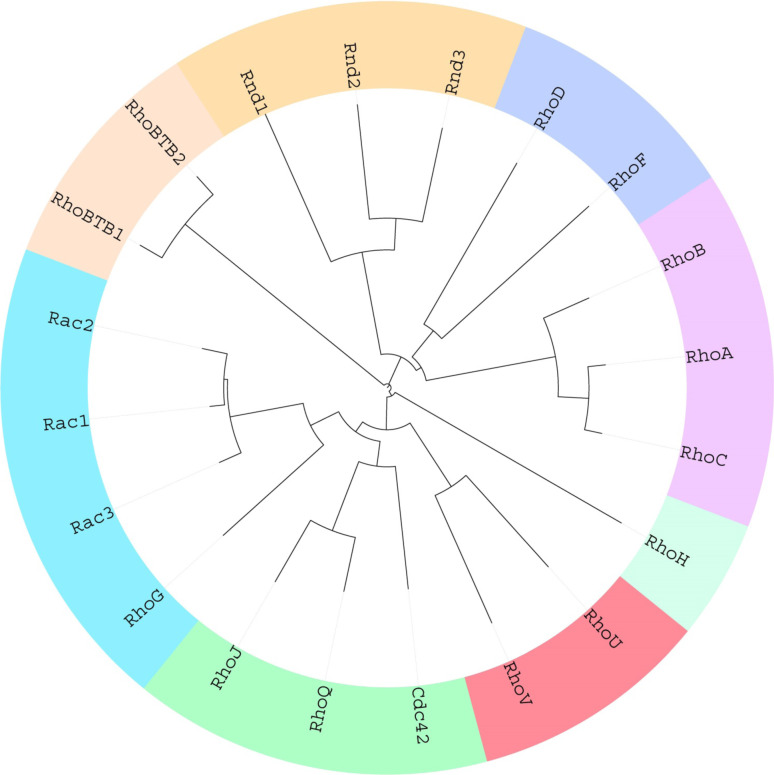
Rho GTPases are small (21–25 kDa), evolutionarily conserved proteins and constitute a unique subfamily within the Ras superfamily of small GTPases. Each subfamily in the Ras protein superfamily shares a common G domain core that is responsible for GTPase activity as well as for nucleotide exchange. The Rho family differs from the other Ras sub families by the presence of a “Rho insert domain” within the 5th β strand and the 4th α helix in the small GTPase domain [1]. The Rho family of GTPases comprises of 20 proteins (Fig. 1), which are further divided into eight sub-groups based on their primary amino acid sequence identity, structural motifs, and biological function—Rac proteins (Rac1, Rac2, Rac3, and RhoG), Rho proteins (RhoA, RhoB and RhoC), Cdc42 proteins (Cdc42, RhoQ, and RhoJ), Rnd proteins (Rnd1, Rnd2/RhoN, and Rnd3/RhoE), RhoBTB proteins (RhoBTB1, RhoBTB2), RhoH, RhoUV (RhoU (Wrch) and RhoV (Chp)), and the RhoF subfamily (RhoD and RhoF) (Figs. 1, 2).
Fig. 1.
Phylogenetic tree of the Rho GTPases. Multiple sequence alignment was performed with amino acids 5–173 of Rac1 as the basis using Clustal Omega. By employing the data generated in this process, a phylogenetic tree was created and visualized with iTOL [195]. The 20 existing Rho GTPases are divided into eight distinct sub families with the lines representing the degree of divergence between each member of the family
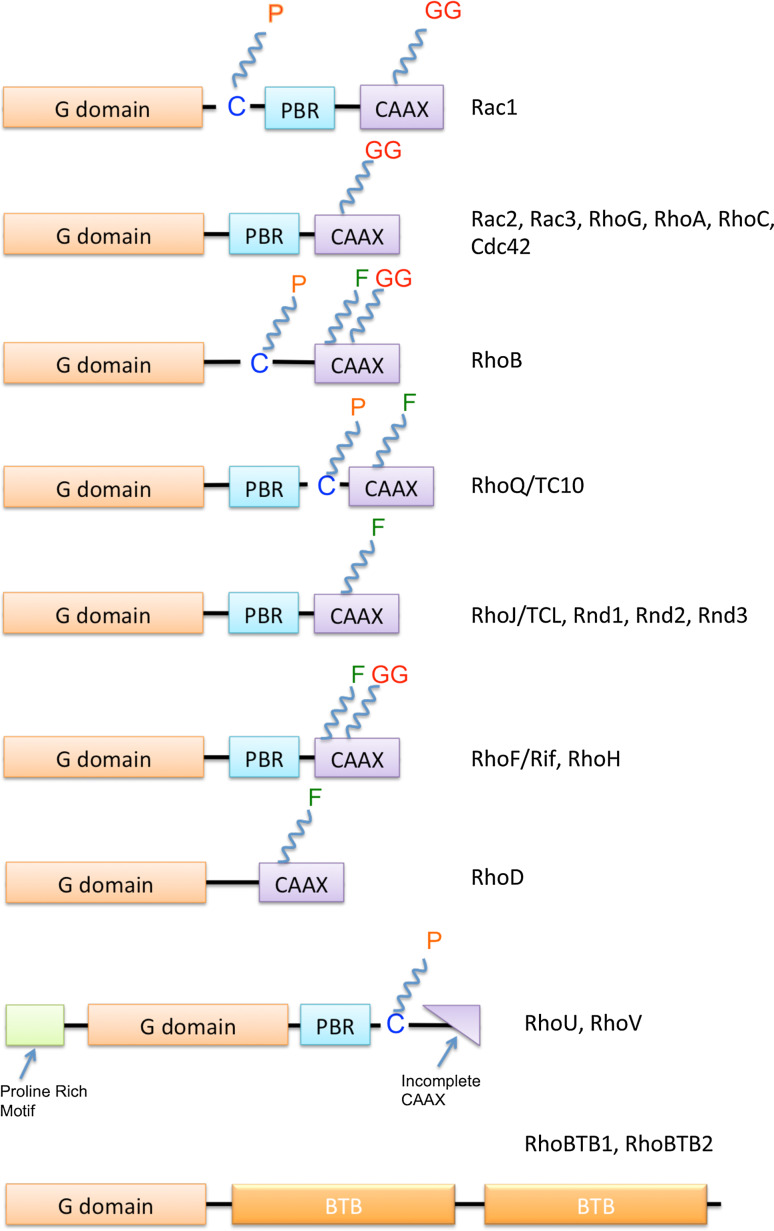
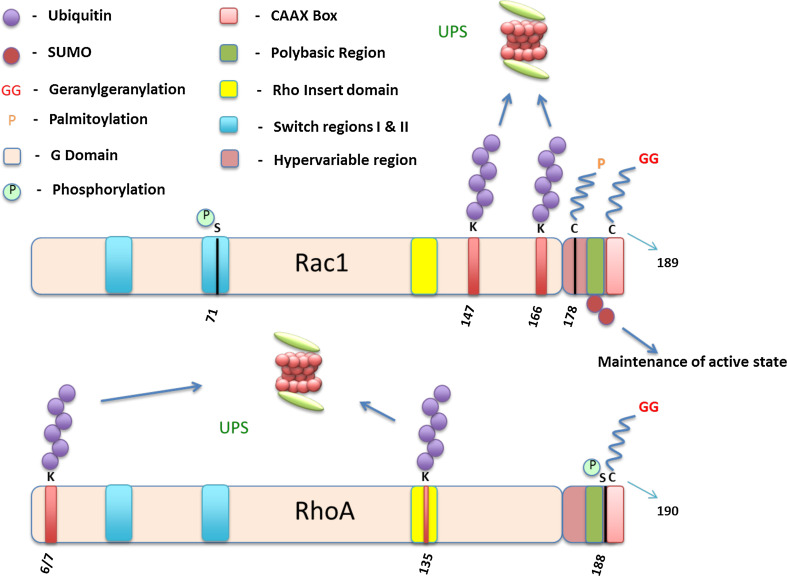
Fig. 2.
Rho GTPases—domain organization and post-translational modifications: All Rho GTPases contain a G domain, though Rnd1-3, RhoH, and the Rho BTB proteins have modifications in the GTP/GDP binding region that make them lack GTPase activity. The Polybasic Region (PBR) domain is present in most Rho GTPases, while RhoU and RhoV contain an N-terminal proline-rich motif as well as an incomplete CAAX box. Due to the importance of the CAAX box in post-translational modifications like geranylgeranylation (GG) and farnesylation (F), RhoU and RhoV are mainly palmitoylated (P). All the other proteins are also subject to various post-translational modifications including farnesylation (F), geranylgeranylation (GG), as well as palmitoylation (P)
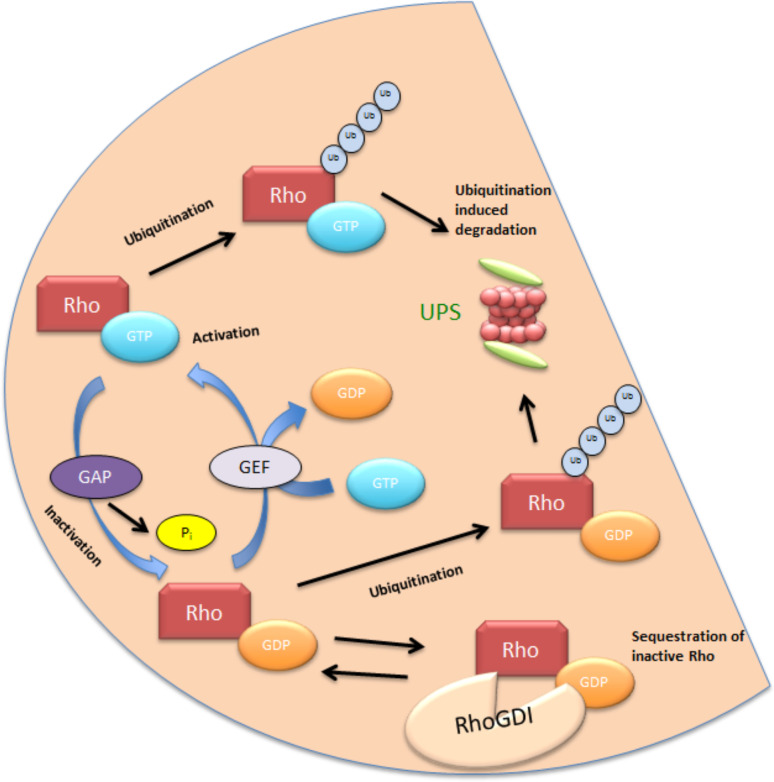
Like other GTPases, most members of the Rho family also cycle between an inactive GDP bound state and an active GTP bound state [2]. The conversion from the GDP bound form to the GTP bound form is promoted by guanine nucleotide exchange factors (GEFs), which release bound GDP and help in subsequent binding of GTP, which is more abundant in the cell. The GTPases, as the name indicates, have an intrinsic but weak GTPase activity, which hydrolyses GTP to GDP and is enhanced by GTPase activating proteins (GAPs) (Fig. 3). Another mechanism of inactivating the Rho GTPases is through Rho guanine nucleotide dissociation inhibitors (RhoGDIs), which bind the cytosolic GTPases and sequester them, thus preventing any access to downstream targets (Fig. 3). While Rho proteins are believed to persist in their inactive GDP-bound state in resting cells, a recent qualitative analysis of the molecular switch functions of the family suggested that RhoD and RhoF exist in a GTP bound state, due to relatively higher GDP dissociation as well as low GTP hydrolysis [3]. Other ways of regulating this switch include several post-translational modifications including but not limited to phosphorylation, oxidation and the well-known C-terminal isoprenylation that mediates the targeting of GTPases to the plasma membrane (Figs. 2, 3) or other intra cellular membranes as well as in pathological conditions when the cell is hijacked by pathogens. Some “atypical” members of the Rho family including Rnd1-3, RhoH, RhoBTB1, and RhoBTB2 do not possess the ability to hydrolyze GTP and lack a GDP binding site, and are thus permanently bound to GTP [4–6]. In these cases, regulation is achieved through modulation of gene expression [7, 8], phosphorylation, and protein degradation [5].
Fig. 3.
A general scheme for regulation of Rho GTPases: most Rho GTPases cycle between an inactive GDP bound conformation and an active GTP bound conformation. This process is overseen by GEFs and GAPs. GEFs remove the GDP bound to the Rho GTPase and GTP binds to the protein due to the higher levels of GTP in the cytosol, while GAPs promote the hydrolysis of GTP and inactivate the protein. Sequestering of the protein by Rho GDI in the cytosol constitutes another layer of regulation. Finally, ubiquitination and subsequent proteasomal degradation of the proteins ensures protein homeostasis in cells. While proteins involved in ubiquitination are known for a few Rho GTPases, namely RhoA and Rac1, not much is known about ubiquitin dependent regulation of the other Rho GTPases
Actin cytoskeleton gives the cell shape, structure, and polarity, and undergoes constant remodeling. Lamellipodia, filopodia, stress fibers, and focal adhesions constitute the structural framework of the actin cytoskeleton, which is crucially regulated by Rho GTPases. Activation of Rho GTPases leads to interaction with numerous effectors which trigger various downstream pathways that influence cellular processes including cell division, proliferation, gene expression [9] and transcriptional regulation, actin and microtubule organization, cell adhesion, migration, invasion, metastasis, wound healing [4], cell mobility, and cell shape, of which the last two will be discussed in greater detail in this review. Amongst the sub-families, the best-studied proteins are Rac1, RhoA, and Cdc42. A variety of insights into the in vivo functions of these three proteins have been provided by loss of function studies in Drosophila melanogaster, Caenorhabditis elegans, and in mice.
Rac subfamily
Four Rho GTPases, Rac1, Rac2, Rac3, and RhoG constitute the Rac subfamily [10] (Fig. 1). Sharing over 80 % sequence homology, these proteins are involved in the formation of lamellipodia and membrane ruffles, via the WAVE complex [11] as well as in phagocytosis [9]. Rac1 is the most well studied member of this family and has been implicated in various cellular functions such as intercellular adhesion [12], establishment of cell polarity, cell proliferation, tumorigenesis, migration [12], endocytosis, phagocytosis [13, 14], and in various stages of the cell cycle and cytokinesis. More recently, the development of a photoactivatable Rac has also made it possible to study the spatio-temporal dynamics of Rac1 activation in moving cells [15]. The studies with the photoactivatable Rac1 showed that localized activation of Rac1 immediately led to an inhibition of RhoA activity in a spatially controlled manner [16]. In the border cells of the Drosophila ovary, it was also shown that focalized activation of Rac1 led to a response in other cells of the cluster, suggesting that these cells sense the direction as a group [15, 17].
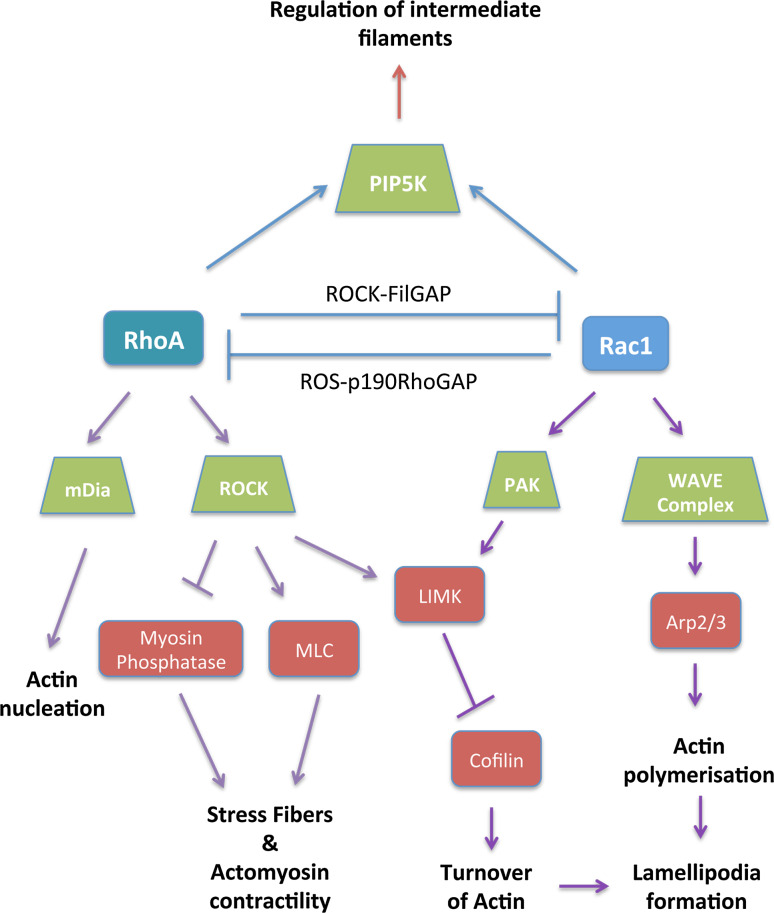
Rac1 has been further shown to regulate tumor cell function through various important proteins like JNK [18], cAMP [19], and phospholipase Cγ1 (PLCγ1) [20]. It promotes cytoskeletal reorganization through two main pathways (Fig. 4). The first one involves the WAVE (WASP-family verprolin-homologous protein) complex and its activation which in turn leads to activation of Arp2/3, a seven-subunit protein complex that serves as a nucleation site for new actin filaments. It binds to existing actin filaments and initiates growth of new ones at a 70° angle to the parental filament [21]. The second mechanism by which Rac1 regulates cytoskeletal organization is via the p21-activated kinase (Pak). The Paks, a family of six serine/threonine kinases bind to both Rac1 as well as Cdc42 via an N-terminal GTPase binding domain. They primarily modulate cytoskeletal reorganization by phosphorylating various proteins, including myosin light chain kinase (MLC), LIM kinase, as well as the Arpc1b subunit of the Arp2/3 complex (Fig. 4) [22–24].
Fig. 4.
RhoA, Rac1, and their effectors: two of the best-studied Rho GTPases, RhoA and Rac1, often share an antagonistic relationship, with both Rho GTPases known to inhibit the activity of the other. Rac1 induces actin polymerization by activating the Arp2/3 complex. Rac1 also activates PAK, which phosphorylates LIM kinase. LIMK in turn phosphorylates and inhibits cofilin, thus regulating actin turnover. RhoA activates ROCK leading to stress fiber formation via activation of myosin light chain or inhibition of myosin phosphatase. RhoA also activates mDia resulting in actin nucleation
Pak1 is the best studied amongst the Paks and has been shown to be highly upregulated in ovarian, breast, and bladder cancers [25]. It is also crucial in the context of cell motility, cell survival, and cell cycle progression [26]. Despite their high sequence homology, studies in knockout mice suggest that the functions of Rac family members are non-redundant. Rac1 knockout results in embryonic lethality [27] and conditional knockout mice have been studied in great detail to learn more about the physiological tissue-specific roles of Rac1 [28–30]. Rac2, Rac3, and RhoG knockout mice do not show significant developmental defects but display cell type-specific defects [31, 32].
Recent studies have identified a recurrent somatic mutation in Rac1, present in melanomas [33, 34]. Both studies suggested that UV induced a P29S mutation because of the characteristic C > T transition as well as its presence in sun-exposed melanomas. This gain-of-function mutation is shown to increase proliferation and migration, possibly due to increased binding to Pak1 and induced Rac1 accumulation in ruffling membranes, a hallmark of Rac1 activation [33, 34]. This study also opens up the possibility of therapeutic targeting of Rac1 or its downstream effectors in treating melanomas.
Rac1b is a splice variant of Rac1, which was initially observed to be upregulated in colon cancers [35]. It contains a 19-residue insert in the C-terminal region close to the GTP-binding region. Due to its inability to interact with RhoGDI and a reduced intrinsic GTPase activity, it is predominantly present in a GTP-bound active state [36]. Rac1b, while defective in activating several Rac1-regulated signaling pathways, is known to stimulate the NF-κB pathway in certain cell types contributing to cell cycle progression and survival [37, 38]. Rac1b is also less susceptible to ubiquitin-dependent proteasomal degradation [38].
While Rac1 is ubiquitously expressed, Rac2 expression is limited to hematopoietic cells, most likely because of silencing of the Rac2 gene locus in non-hematopoietic cells from DNA methylation [39]. Rac3 is highly upregulated in the brain and in some human cell lines [40]. Rac2 is expressed at various levels with Rac1 in hematopoietic cells depending on the stage of differentiation and maturation [41]. Rac2 deficiency in particular affects the function of neutrophils and macrophages [42–44]. Rac2 regulates the activation of NADPH oxidase [45] and the generation of reactive oxygen species (ROS) in hematopoietic cells [46]. Further, in macrophages, Rac2 is required for phagocytosis [44]. Rac3 was found to be constitutively active and/or deregulated in human breast cancer-derived epithelial cell lines and is highly localized to membranes [47, 48], while it is also up-regulated when fibroblasts are stimulated with serum [40].
RhoG has been implicated in cell migration and is known to stimulate lamellipodium formation by activating Rac GEFs [49]. Pathogenic bacteria including Salmonella and Yersinia are known to exploit RhoG function of host cells during their infection cycle [50, 51]. RhoG has also been shown to regulate EGF-induced cell migration in epithelial cells as well as early EGF receptor internalization processes [52]. RhoG was also involved in the indirect activation of PI3K, along with Rac1 and Cdc42 [53]. However, compared to Rac1, our knowledge of RhoG regulation and its role in cell motility remains obscure.
Rho subfamily
The Rho subfamily of Rho GTPases consists of three highly conserved proteins—RhoA, RhoB, and RhoC. While all three members induce the formation of stress fibers and focal adhesion complexes, RhoA and RhoC are frequently upregulated in human tumors, while RhoB seems to act as a tumor suppressor [54] and is observed to have pro-apoptotic functions [55].
Despite significant homology between the three Rho isoforms, they are each preferentially bound by their respective regulating enzymes and all three members have their own roles in cancer cell migration [56]. The functional divergence is a consequence of differences at the C-terminal end where a set of 15 amino acids plays an important role in mediating the interaction with their effector molecules. RhoA, the best-studied isoform, has been implicated in most stages of cancer progression, while in normal epithelia it helps to generate epithelial polarity as well as junction assembly and function [57]. The three major downstream effectors of RhoA are ROCK, PIP5K, and mDia (Fig. 4). RhoA contributes to both amoeboid and mesenchymal mode of migration. Induction of actomyosin contractility via RhoA-ROCK signaling leads to blebbing, thus causing amoeboid migration, while it also regulates tail retraction in mesenchymal migration [58]. The kinds of migration and the roles each Rho GTPase play are discussed in greater detail later in the text.
RhoC, unlike RhoA, functions mostly in metastatic processes. While there have been reports of RhoC playing a role in cell proliferation as well, most studies indicate that RhoC is essential for invasion, although dispensable for tumor initiation [59, 60]. It is observed that there is a reciprocal regulation between RhoA and RhoC. RhoC is upregulated during EMT while RhoA levels are reduced [61].
RhoB is located primarily in endosomes or at the plasma membrane and regulates endosomal trafficking [62, 63]. The difference in localization (RhoA and RhoC localize to the plasma membrane when not bound to RhoGDI) is due to the C-terminal lipid modifications in RhoB as well as the fact that RhoB does not bind to RhoGDI. RhoB knockout mice develop normally but are susceptible to carcinogen-induced skin tumor formation, further emphasizing the role of RhoB as a tumor suppressor [64].
RhoA and RhoC can both bind to the Rho-associated kinases ROCK I and ROCK II, which leads to the activation of LIMK, which in turn phosphorylates cofilin. Cofilin then works in tandem with the Arp2/3 complex in the reorganization of actin filaments. A reduction in the levels of RhoA or RhoC usually corresponds to a higher level of RhoB [65, 66], whereas the reverse does not hold true, i.e., a reduced expression of RhoB does not correspond to increased expression of RhoA and RhoC. Phosphorylation of RhoA at Serine-188 by various kinases like PKA, PKG, and SLK [67–69] weakens the association of the protein with the membrane surface and due to RhoGDI’s higher affinity for phosphorylated RhoA, finally dissociates from the plasma membrane (Fig. 7). The presence of valine or isoleucine at position 43 is predicted to play a crucial role in Rho activation. In RhoA and RhoB, this residue is valine, while in RhoC it is isoleucine. This is the only point of divergence in the first two-thirds of the sequence and has an impact on basal as well as GEF-stimulated activity of these proteins [70].
Fig. 7.
Rac1, RhoA, and a detailed representation of various post-translational modifications on both proteins. The Rho insert domain, the Polybasic region, as well as the switch regions are shown in both Rho GTPases
Cdc42 subfamily
The Cdc42 subfamily of proteins consists of three proteins: Cdc42, RhoJ, and RhoQ (Fig. 1). Of the three, Cdc42 is by far the most studied protein and the members of this family are known to stimulate filopodia formation and in maintaining cell polarity. It has been implicated in a variety of processes in the nervous system including cell motility, cytoskeletal reorganization, establishment of neuronal polarity, morphology, and cell cycle progression. It is essential for axon generation in hippocampal neurons [71], axon myelination in glial cells [72], and also plays a critical role in innate immunity [73]. Complete germline deletion of Cdc42 is embryonic lethal, and conditional knockouts have been used to elucidate the roles of Cdc42.
The role of Cdc42 in filopodia formation is ambiguous as Cdc42-null mouse embryonic fibroblasts (MEFs) completely lack filopodia activity [74], Cdc42-null neurons show reduced filopodia [71], while fibroblastoid cells derived from Cdc42-null embryonic stem cells form normal filopodia [75]. It is hypothesized that other Rho GTPases, including the RhoUV subclass, and the RhoF subclass are also involved in this process [8].
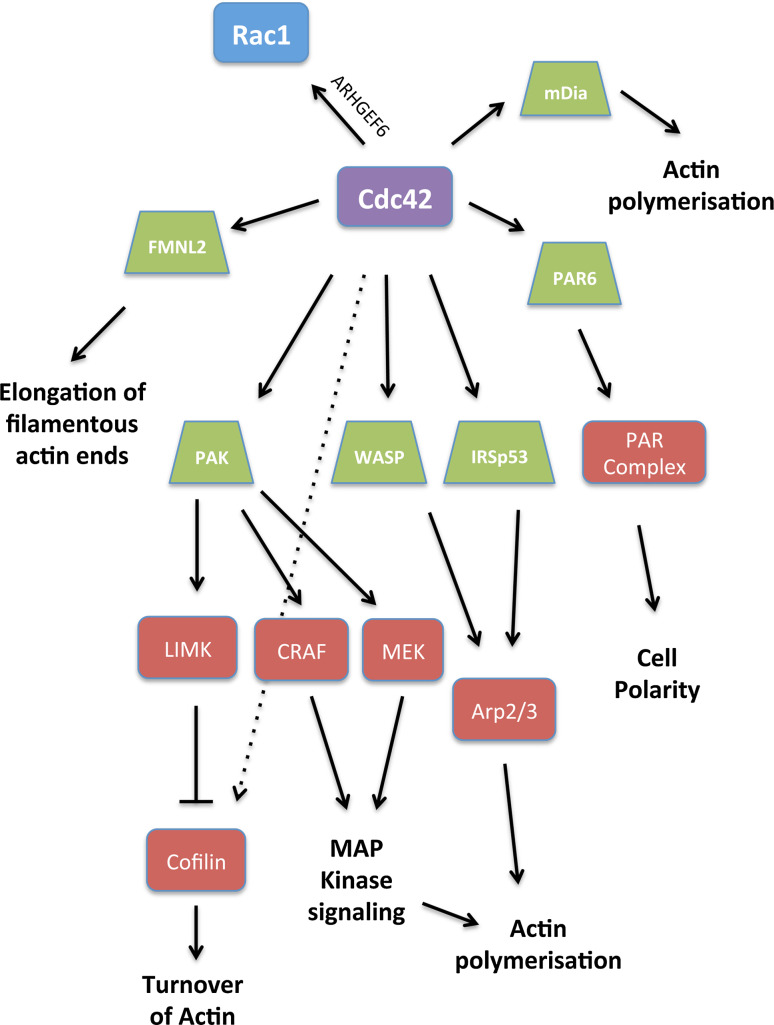
Cdc42 and Rac1 work in tandem to regulate signals during tumor progression. The main effectors of Cdc42 are WASP, PAK, and PAR (partitioning-defective) (Fig. 5). Cell polarity in particular is established and mediated by Cdc42 via formation of the PAR complex (PAR3-PAR6-aPKC) [31].
Fig. 5.
Cdc42 and its effectors: Cdc42 induces actin polymerization by binding to WASP, or through IRSp53. PAK is a downstream effector of Cdc42 that activates LIM kinase (LIMK) leading to inhibition of cofilin. PAK phosphorylates CRAF and MEK activating the MAP kinase pathway and indirectly actin polymerization. FMNL2 was recently characterized as a downstream effector of Cdc42 that modulates actin polymerization at the lamellipodium tip. Cdc42 also contributes to cell polarity by interacting with the PAR complex
RhoQ, also called TC-10, is primarily involved in insulin-stimulated glucose uptake where it plays a role in insulin-regulated translocation of glucose transporter 4 (GLUT4) [76]. GTP hydrolysis of RhoQ is also seen to promote exocytic fusion and when in the vicinity of the plasma membrane, was seen to be Rac1 dependent [77]. RhoJ, also called TCL (TC10-like) for its close similarity to RhoQ, plays a role in early endocytosis, adipocyte differentiation in vitro, as well as in regulating the actin cytoskeleton [78].
Cell mobility and Rho GTPases
Cell mobility refers to the process of controlled movement of cells and is a fundamental feature of cellular existence. Cytoskeletal reorganization, formation of distinct actin-based membrane protrusions such as lamellipodia and filopodia, the control of intercellular adhesion and regulation of cell polarity are all empirical for cell mobility. As discussed in earlier sections, Rho GTPases are crucial for the aforementioned processes and hence it is natural to argue that Rho GTPases play an integral role in regulating cell mobility.
Movement within an extracellular matrix (ECM) is a multistep process that requires changes in cytoskeletal structure, adhesions, and is dependent on the constituents of the ECM. While the study of cell migration primarily started with two-dimensional (2D) cell culture models, recent developments have propelled further research towards studying both one-dimensional (1D) and three-dimensional (3D) cell culture models. The seminal paper by Lauffenburger et al. in 1996 discussed 2D cell migration by dividing the process into five steps: Lamellipodium extension, formation and stabilization of focal adhesion complexes, proteolysis of the ECM using surface proteases, contraction of the cell body, and finally, the retraction of the tail.
Imaging of cells in 3D models of the ECM as well as in vivo revealed that lamellipodia-based motility is only one among multiple migration strategies employed by cells. While ECM composition and stiffness affect migration regardless of dimensionality, factors such as ECM porosity, crosslinking, and topography become important factors in 3D migration [79]. 3D models also reveal that cells not only form different protrusions at the leading edge but also otherwise, resulting from distinct signaling pathways and actin-regulatory proteins. Apart from lamellipodia and filopodia, cells can form lobopodia, which are blunt, cylindrical protrusions formed due to intracellular pressure and were discovered in migrating fibroblasts [80]. Increased intracellular pressure can also trigger the formation of membrane blebs, which help in amoeboid form of movement [81]. Cells also form invadopodia and podosomes, actin-rich structures that proteolytically degrade the ECM for 3D migration [82].
One-dimensional models were designed to understand the role of topography in aligned 3D ECMs. Aligned topography in 3D ECMs results in a faster migration rate and this is observed in in vitro models, as well as mouse tumor and in vivo metastasis models [83, 84]. In 2D migration, ECM molecules are presented as a flat sheet of globular molecules without any appreciable fibrillar structure; 3D migration results in a fibrillar topography which prevents lateral spreading. As a consequence, apical-basal polarity is not imposed on the actin cytoskeleton in 3D migration unlike in 2D models. The review by Baker et al. [85] describes in detail the ways in which 3D migration differs drastically from 2D migration and they are not further discussed in this review.
Cell migration and modes of migration
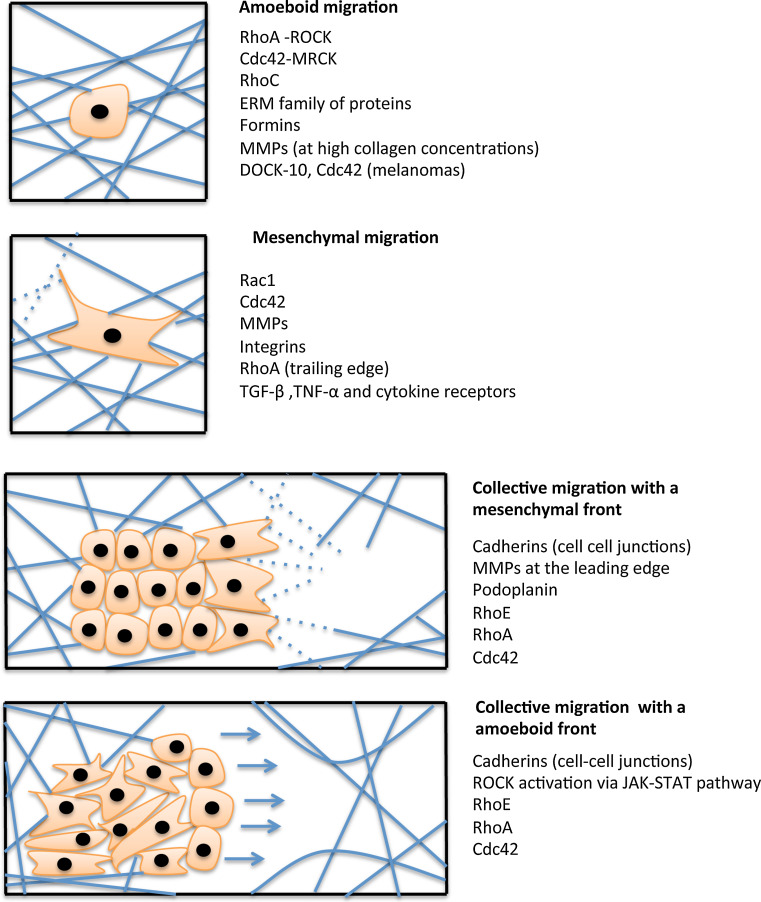
Migration is a key underlying phenomenon for tissue formation and maintenance, as well as for regeneration of tissues and for invasion of tumor cells. In general, modes of migration can be classified into two major types based on whether the cells migrate as single cells or in a group, aptly termed single cell migration and collective cell migration, respectively (Fig. 6). The kind of migration exerted is cell type-dependent and distinct signaling pathways are activated in response to various extracellular cues. Based on various factors such as ECM density, stiffness, and orientation as well as cell-specific parameters such as cell–cell, cell–matrix adhesion, extracellular protease activities and cytoskeletal polarity, the migration mode of cells is determined. Table 1 summarizes the different kinds of migration and how activation or inactivation of various Rho GTPases influences the type of migration employed by cells.
Fig. 6.
Modes of cell migration. Cells can migrate as single cells or in a group. Single cell migration can either be amoeboid or mesenchymal in nature while collective migration involves cells moving in groups where the position of a cell within a cluster decides its function. The factors responsible for each kind of migration are mentioned next to each figure. See text for detailed description
Table 1.
Different migration modes and the contribution of each Rho GTPase and the interacting proteins were depicted
| Type of migration | Subtype | Rho GTPase | Mechanism of action | Other proteins/factors involved | Functional relevance |
|---|---|---|---|---|---|
| Single cell migration | Mesenchymal | Rac1 | Recruitment to the leading edge | PIP3 | Various cell types [91] |
| Activation | JNK pathway, MMPs | Fibroblasts [99] | |||
| Activation leading to MMP-1 expression | Fibroblasts [98] | ||||
| Required for MMP-9 generation | Chondrocytes [98] | ||||
| Activation | DOCK3, NEDD9 | Melanoma [88] | |||
| Activation | P130Cas, MMP14 | Bone marrow myeloid progenitors [97] | |||
| RhoA | Active, but downstream signaling is blocked | NEDD9, Src | [89] | ||
| Spatial activation | Various cell types [91], leukocytes [94] | ||||
| Block | MT1-MMP, PI3K, MMP-2 | SMECs (rat endothelial cells) [189] | |||
| Cdc42 | Directionality | Rac1 | Various cell types [91, 90] | ||
| Activation | FMNL2 | Melanoma [93] | |||
| Activation | MT1-MMP, PI3K, MMP-2 | SMECs (rat endothelial cells) [189] | |||
| Amoeboid | RhoA | Activation | PDK1, ROCK | Melanoma [112] | |
| Activation | FilGAP, Rac1 | Carcinoma [190] | |||
| Activation | Hic-5 | Murine melanoma [191] | |||
| Activation | EphA2 | Prostate carcinoma [192] | |||
| Cdc42 | Activation | DOCK-10 | Melanoma [117] | ||
| Activation | Dip, mDia2 | [113] | |||
| RhoC | Activation | FMNL2 | Melanoma [193] | ||
| Rac1 | Inactivation | EphA2 | Prostate carcinoma [192] | ||
| Collective migration | RhoE | Inhibits Rho/ROCK | Par3, Par6, DDR1 | Carcinoma [129] | |
| RhoA | Activation | α5-integrin | Fibroblasts [135] | ||
| Local regulation of activity | Myosin-IXA | Human bronchial epithelial cells [194] | |||
| Activation | GP130-IL6ST-JAK1-STAT3 | Melanoma [136] | |||
| Cdc42 | Regulation of MLC | Carcinoma [135] |
Single cell migration
As the name indicates, here the cells tend to move individually rather than in strands or sheets as is seen in collective cell migration. This is witnessed during embryonic development when neural crest cells leave the neural tube or in migrating limb muscle precursor cells. This usually involves the loss of epithelial polarity, thus leading to the gain of a mesenchymal morphology. Single cell migration is also seen in tumors when loss of E-cadherin leads to loss of intercellular adhesion, thus leading to the migration of detached cells. Individual cell movement is essential for tumor cells to cross basement membranes and enter blood vessels, thus leading to metastasis [86].
Based on the morphology and interplay with the ECM, single cell movement can be further classified into mesenchymal and amoeboid movement (Fig. 6).
Mesenchymal movement
This type of single cell movement is characterized by elongated cell morphology, focalized cell–matrix interactions with high attachment to the ECM, high contractility as well as established cell polarity and the secretion of proteases to degrade the ECM to move in a fibroblast-like manner. This form of movement exhibits polarity where the production of actin assembly driven protrusions at the front co-ordinate with the actomyosin contractility driven retraction at the tail to facilitate movement. Here the cells move at a relatively low speed (around 1 μm/min) [87].
Rac1 is highly implied in driving the mesenchymal mode of migration. DOCK3, a Rac-specific GEF, has been identified as being crucial in cells undergoing the mesenchymal form of migration. DOCK-3 complexes with NEDD9 [88], a signal transduction molecule whose levels are elevated in various cancers as well as in metastasis, to activate Rac1 in migration of melanoma cells [88]. NEDD9 also blocks Rho/ROCKII signaling by Src-dependent phosphorylation of the negative regulatory Tyr722 site on ROCKII. Src has been shown to play a key role in maintaining mesenchymal movement and suppression of amoeboid, rounded form of movement [89].
Cdc42 activity serves to maintain the directionality of the cell and, in tandem with Rac1, promotes actin polymerization at the leading edge of the cell [90, 91]. In the osteosarcoma cell line LM8, Cdc42 was responsible for the fibroblastic morphology that led to pulmonary metastasis of the cells via a mesenchymal mode of migration [92]. FMNL2, a downstream effector of Cdc42, promotes mesenchymal movement by modulating actin polymerization at the lamellipodium tip (Fig. 5) [93]. The role of RhoA is complex, as it is present at both the retracting edge as well as the leading edge. The latter is consistent with the role of Rho/ROCK signaling in tail retraction in leukocytes [94]. It is seen in Madin–Darby canine kidney (MDCK) cells that there is no RhoA activity at the rear end of the cell [95], suggesting that the role of the Rho/ROCK pathway in cell migration is also cell type-dependent.
Mesenchymal movement also uses secreted proteases to remodel the extracellular matrix by proteolysis. Matrix metalloproteases (MMPs) are produced as pro-enzymes in cells and are usually cleaved either by other MMPs or by proteases such as plasmin. Rho GTPases have been implicated in the regulation of the activity of these MMPs. Inhibition of protease function causes a rounded Rho-dependent morphology that suggests promotion of Rac1 activity by proteases, for example the MMP-14-dependent recruitment of p130Cas, an activator of Rac1 [96, 97]. Activated Rac1 can induce the expression of MMP-1 in fibroblasts while Rac1 is needed for shear stress-induced MMP-9 generation in chondrocytes [98, 99]. It is also necessary for MMP-13 production in the osteosarcoma cell line, SaOS-2 [100]. Activation of Rac1 also induces activation of the JNK pathway, which regulates the transcription of MMP genes among others [99].
The Rho/ROCK pathway plays a critical role in lysophosphatidic acid (LPA, a biolipid that has been implicated in ovarian cancer initiation and progression)-induced ovarian cancer progression. A recent study showed that ROCK inhibition reduced the levels of MMP-9 and reduced the invasive capabilities of these cells [101]. Cdc42 and RhoA are observed to play opposing roles in regulating cell surface localization of MT1 (membrane type 1)-MMP and MMP-2 activation; constitutive activation of Cdc42 as well as ROCK inhibition caused the same effect, namely the increased cell surface localization of MT1-MMP and also increased PI3K activity, which is required for MMP-2 activation. Proteases such as MMP-2, MMP-9, and membrane type MMP (MT–MMP), ADAMs, plasmin, as well as cathepsins are involved in this form of migration. Various receptors have also been shown to trigger mesenchymal mode of movement. These include the TGF-β and TNF-α receptors, GPCRs, integrins, and cytokine receptors.
Amoeboid movement
This movement is usually seen in cells with a rounded morphology, such as leukocytes, hematopoietic stem cells, and cancer cells. This type of movement is largely independent from any matrix contact and proteolytic degradation is avoided, as the cells tend to squeeze through gaps in the extracellular matrix rather than degrading it (Fig. 6). This results in a faster form of migration with speeds up to 10 μm/min [102] as seen in carcinoma cells showing amoeboid morphology. Several studies correlate amoeboid motility with blood-borne metastasis [103–105].
Amoeboid migration can further be classified into two types. The first features blebby migration of cells that do not adhere to the matrix and use a propulsive, pushing mode of migration [106], where the site of the blebbing usually decides the direction of migration [107]. The second type of amoeboid migration occurs in slightly more elongated amoeboid cells where there are poor matrix adhesions and the absence of focal adhesive complexes [108]. Amoeboid migration in cancer cells is different from the amoeboid migration in leukocytes or Dictyostelium, where there is a lot more adhesion to the substrate, as well as formation of protrusions in the direction of movement and a uropod at the cell rear.
Actomyosin contractility through myosin activation driven by Rho/ROCK signaling is characteristic of amoeboid movement [58, 109, 110]. Contractility of the cortical actin network is important to propel the nucleus of dendritic cells (DCs), B-lymphocytes, and granulocytes through small gaps [111]. Pinner et al. [112] showed that phosphoinositide-dependent protein kinase-1 (PDK1) is required to localize ROCK to the plasma membrane where it directly competes with RhoE to bind ROCK-I, thus preventing RhoE from inhibiting the Rho/ROCK pathway. It is also observed that Dip, an interaction partner of mDia2, promotes blebbing, thus indicating a role for formins in amoeboid migration [113]. Leukocytes remodel the membrane physically to substitute for proteolysis and Rabodzey et al. [114] showed that some leukocytes appear to induce endothelial cells to displace the basement membrane. While proteolysis is not a feature of amoeboid migration, it is observed that cells utilize MMP-14 at high concentrations of collagen I [115, 116].
In contrast to mesenchymal movement, where Rac1 is localized to the leading edge of cells, it has been shown in melanoma cells that DOCK-10-mediated Cdc42 activation is essential for amoeboid migration of cells [117]. Recent work on melanoma cells has brought up an interesting observation which questions the notion that amoeboid single cell migration is not polarized. A375 melanoma cells exhibit multiple blebs and this would theoretically suggest their inability to move directionally. This led the authors to prove the existence of an ezrin-rich uropod-like structure that inhibits blebbing at the rear of the cell and drives cell invasion into a 3D matrix. Thus, in these cells, cell movement is decided by the rear end of the cell and not the front end [118].
Collective cell migration
Single cell migration has provided insights into the molecular mechanisms as well as the cellular framework in place especially in tumor cells that migrate constitutively as single cells. However, for many cancer types, histomorphologically, the most frequently observed invasion unit is a group of cells which together define a malignant function [119, 120]. This form of migration is also seen in various developmental morphogenic processes. It is observed in the movement of cells of the inner blastocyst, dorsal surface closure, as well as trachea morphogenesis in Drosophila embryo, vascular sprouts during angiogenesis, and also in keratinocytes migrating across the wound matrix [121]. This is different from multicellular streaming where directed migration of single cells occurs in response to a chemokine or an extracellular cue, thus forming a multicellular stream of migrating cells each of which form protrusions and generate their own traction force on the matrix [122]. In this form of motility, cells migrate in strands, sheets, clusters, and tubes, while maintaining cell–cell junctions. Proteins like cadherins and adhesion receptors of the immunoglobulin superfamily provide cell–cell adhesion in the body of migrating cells [123]. This sort of migration is seen in various cancer types, including melanomas, epithelial prostate cancer, and most commonly in squamous cell carcinomas [124].
For a multicellular migrating group, it is observed that the function of cells varies depending on the position in the cluster. Collective invasion models usually hypothesize the presence of a mesenchymal mode of migration amongst the leading layer of cells, which ensures actin-based protrusions to drive the cell forward as well as matrix proteolysis using local expression of MMPs to guide the group along the membrane. In contrast to single cell migration where tail retraction is seen, in collective cell migration, it is the pulling forces exerted on neighboring cells that drive the cells forward. There are exceptions, however, such as in mammary morphogenesis [125], where the absence of actin-rich protrusions at the front most likely implies that the cells push forward by force generated in a layer behind the leading edge [125, 126].
Podoplanin is a cell surface glycoprotein that is highly expressed along the leading cell layer of migrating clusters in various cancer types. Forced expression of podoplanin in MCF7 breast cancer cells and human keratinocytes induces cell migration with an increase in the formation of filopodia-like membrane protrusions and a decrease in stress fibres [127, 128].
Due to the fact that only the leading edge generates actomyosin-based traction force, the cells inside the migrating group may not interact with the ECM and are restricted to cell–cell contacts and any intercellular matrix contact. In order to maintain cell–cell contact, it is also necessary that actomyosin contractility be kept at low levels near the adhesions to maintain cohesion [129, 130]. The inhibition of the Rho/ROCK pathway is accomplished in a RhoE-dependent manner by the polarity proteins Par3 and Par6, in interaction with discoidin domain receptor 1 (DDR1) [129], which also upregulates N-cadherin levels, which in turn inhibits membrane protrusions and Rac1 activity at cell–cell contacts in collectively migrating neural crest cells [131–133]. The morphological organization of these clusters varies based on factors such as specific cell–cell and cell–matrix adhesion as well as proteolysis off the leading edge.
The role of stromal cells in collective cell migration has also been discussed. Stromal cells were identified at the migration front and are highly polarized along with the formation of lamellipodia [134]. E-cadherin-mediated junctions connect them to their neighbors that are dragged along. Co-culture experiments between carcinoma cells and stromal fibroblasts also showed that fibroblasts prepare migratory tracks for the carcinoma cells by taking the lead and proteolytically digesting as well as force remodeling the matrix via α5-integrin-mediated RhoA activation leading to subsequent activation of MLC [135]. The carcinoma cells trailing behind the fibroblasts follow the generated track via Cdc42-mediated regulation of MLC. In experiments involving melanomas, it was observed that the invasive front consists of rounded cells following an amoeboid form of movement, though the tumor body consisted of elongated cells, leading to the suggestion that melanoma cells are using the amoeboid form of cell movement while migrating collectively [136]. Cytokine signaling via the GP130-IL6ST-JAK1-STAT3 pathway was found to influence this rounded form of migration in melanomas via activation of ROCK, while this pathway is not required in squamous cell carcinoma cells [136].
Another form of cell migration is scaffold cell-dependent migration, which depends on cell–cell adhesions. This form of migration is detected during normal brain development, when immature neurons move along radial glial fibers, which cover most of the neuronal migration route [137]. N-cadherin is required for intercellular adhesion under these conditions [138, 139].
Plasticity of cell migration
Cells preferentially employ one of the above-described migration types to move in their natural environment. For example, leukocytes prefer amoeboid movement while fibroblasts and stromal cells in general prefer mesenchymal migration. While attempts have been made to identify mechanisms of migration using these cell types as models, the quest to find one dominant pathway influencing cell migration, or determining the rate-limiting step of the invasion process has not yet yielded any straightforward results.
It is becoming increasingly evident that changes to the microenvironment or intrinsic cell properties result in cells adapting to the new conditions by genomic and epigenetic alterations. Modulating various proteins, receptor expression levels and the presence of different cytoskeletal regulators responsible for cell–cell and cell–matrix adhesion ultimately leads to a change in the mode of migration of the cell. This ability of cells to adapt to their environment and change their migration styles instead of completely abandoning them is referred to as cell plasticity.
There are various ways in which tumor cells could change their form of migration depending on the kind of environment encountered. One such transition is the epithelial-to-mesenchymal transition (EMT). EMT is involved in many developmental processes as well as in invasive tumors, where immotile epithelial cells lose their cell–cell contacts, polarity, and acquire mesenchymal traits like motility and invasiveness, thus allowing them to move away from their location [140]. Another characteristic feature of EMT is the loss of E-cadherin. EMT is characterized by an increase in signals from receptors such as Wnt, TGF-β, FGF, and EGF, which lead to the activation of transcriptional repressors such as Snail1 and 2, Twist, and ZEB1-2 [141, 142]. These changes often lead to inhibition of E-cadherin and a corresponding increase in the mesenchymal morphology-associated N-cadherin, thus weakening cell–cell junctions. This weakening causes disruption of cell anchoring to the basement membrane as well as a disturbance in apicobasal polarity, allowing cells to acquire a mesenchymal phenotype [143]. Exogenous addition of MMPs such as MMP-2, -3, and -9 is seen to facilitate EMT by cleavage of E-cadherin, and the consequent activation of Rac1 [144]. EMT is also a route for cancer cells to intravasate and metastasize. EMT allows them to spread to distant organs and then undergo an MET (mesenchymal-to-epithelial transition) to proliferate in the newly adapted microenvironment. Loss of the Rac GEF Tiam1 is required for the induction of EMT in HGF-induced epithelial cells [145, 146].
Single cell migration strategies such as amoeboid and mesenchymal movement are also readily interchangeable in response to various cues. Suppression or enhancement of particular molecular pathways readily causes a switch between the two types of movement. These transitions are called amoeboid-to-mesenchymal transition (AMT) or mesenchymal-to-amoeboid transition (MAT).
The regulation of Rac1 activity appears to be a key switch in AMT. In melanomas, DOCK3, a RacGEF, in complex with NEDD9, activates Rac1, leading to the mesenchymal form of migration, while the switch to amoeboid signaling is facilitated by ARHGAP22, which on activation by the Rho pathway suppresses Rac1, thus facilitating amoeboid movement [88]. NEDD9 also drives cells into a mesenchymal mode by inhibiting ROCKII in a Src-dependent manner [147]. EphA2 is an indirect RhoA activator that also leads to amoeboid movement [148]. Reduction of WAVE2 levels leads to cells switching to the amoeboid form from the mesenchymal form (MAT) [88, 149]. Other processes influencing MAT include interference of Rab5-mediated endocytosis, thus disrupting Rac1 recycling to cell protrusions [150], as well as inhibition of Smurf1, an E3 ligase of RhoA [151]. Recent studies revealed that loss of inhibitors of apoptosis (IAPs) leads to stabilization of Rac1 and AMT in many cell types [152, 153].
Cdc42 signaling via PAK2 or N-WASP is also involved in AMTs and Gadea et al. [117] showed recently that DOCK10-mediated regulation of Cdc42 maintains amoeboid movement in A375M2 melanoma cells. Integrin availability is another factor that determines the mode of movement. Loss of p53 leads to enhanced integrin turnover, thus converting cells to the amoeboid mode [154].
Microtubule stability also influences the mode of migration. Experiments in HT-1080 cells showed that stathmin, a microtubule destabilizing protein [155], enhances migration in a rounded form in sarcomas [156], suggesting that stathmin might contribute to MAT. Siva1, a molecule involved in apoptosis regulation, was shown to inhibit stathmin, thus leading to a block in EMT [157]. The role of stathmin in these transitions is still not clear, but it has been suggested as an important biomarker as it is highly upregulated in many cancers [158].
Therapeutic inhibition of MMPs was considered a novel avenue to inhibit cells from migrating in a mesenchymal fashion. This was until the discovery of a protease-independent mode of invasiveness [159]. It was observed in fibrosarcoma and melanoma cells that inhibition of MMP function led to cells switching immediately into the amoeboid form of migration [148, 159, 160]. However, this finding was disputed later by Sabeh et al. [115], when they suggested that the protease-independent mechanisms of cell migration are only plausible when the collagen network is devoid of the covalent cross-links that are characteristic of normal tissues. They suggested that employing 3D ECM models may not faithfully replicate conditions the cells would face in vivo and thus it was not an accurate indicator of whether reverting to amoeboid form of migration was relevant in vivo [115].
RhoE, a known ROCK inhibitor, has been associated with increased invasiveness in fibroblasts. Inhibition of RhoE is associated with increased metastatic potential [161]. It is also downregulated in hepatocellular carcinoma and this is associated with poor prognosis and tumor progression [162, 163]. siRNA-mediated knockdown of RhoE induced a loss of E-cadherin at cell–cell junctions. This was linked to EMT via the upregulation of ZEB2, the zinc-finger E-box binding homeobox protein, and the down regulation of mir-200b and mir-200c [162]. Conversely, in gastric cancer cells, RhoE is up-regulated under hypoxic conditions by hypoxia inducing factor 1 (HIF-1), thus leading to EMT [164].
Recent studies in primary fibroblasts observed the formation of lamellipodia, with Rac1 and Cdc42 targeted to the leading edge in 3D collagen while cells formed blunt, cylindrical protrusions called lobopodia and a nonpolarized localized concentration of Rac1 and Cdc42 in cells migrating inside dermal explants. These cells in turn converted back to a lamellipodia form of movement upon inhibition of RhoA, ROCK, or myosin II activity. This study also reported that the elasticity of the matrix influenced the type of migration as much as the interplay between Rac1, Cdc42, and RhoA [165] (Table 1).
Other categories of invasion include collective-to-mesenchymal transitions as well as collective-to-amoeboid form of migration triggered either by Rac1 or RhoA, respectively. One trigger for collectively migrating cells to detach from the rest and migrate on their own is a local expression of Rac1, which allows abnormal cell tip behavior, increase in substrate cell interactions, and eventually cell detachment in an E-cadherin-dependent manner [166]. An alternative method is the release of cells from the tissue in an amoeboid manner. This depends on the regulation of actomyosin contractility as well a dependence on integrin-mediated adhesion [88, 167]. Detached tumor cells from the main epithelial mass in breast cancer lesions move in an amoeboid manner and are guided by EGF, which is secreted by activated macrophages [168].
In conclusion, while amoeboid and mesenchymal forms of movement are discussed as two distinct types, it is prudent to see them as the two extremes in a spectrum modulated by various signaling pathways as well as environmental cues, all of which influence the decision of the cell to switch between one mode to the other. Thus, plasticity of cell migration needs to be considered duly while designing inhibitors targeting one particular form of migration. Utilizing combinations of inhibitors as well as identifying potential master switches in the signaling pathways regulating migration remains an aim for the future.
Regulation of Rho GTPases by ubiquitination and SUMOylation
Apart from GEFs, GAPs, and RhoGDIs, Rho GTPases are regulated by various post-translational modifications which include, but are not restricted to, phosphorylation, isoprenylation, ubiquitination, and SUMOylation. Recent studies [152, 169] revealed that ubiquitin-dependent regulation as well as other post-translational modifications on Rho GTPases can affect their stability and consequently influence the plasticity of cell migration.
Ubiquitination and degradation of Rho GTPases was first witnessed during host–pathogen interactions. Cytotoxic necrotizing factor (CNF1), a toxin secreted by uropathogenic Escherichia coli, constitutively activated Rac1 and Cdc42 by deamidating the glutamine at the 61st position, while similarly affecting RhoA at the 63rd residue [170, 171]. This triggered the degradation of the proteins in a proteasome-mediated manner [172] in various cell types.
Consequently, studies to identify E3 ligases of these proteins gained steam and the HECT domain-containing Smurf1 was identified as a specific E3 ligase for RhoA by ubiquitinating the 6/7 lysine residues, thus targeting it for proteasomal degradation (Fig. 7) [173, 174]. Correspondingly, an over-expression in Smurf1 led to a loss of stress fibers as well as reduced cell motility in epithelial cells [151, 174, 175]. aPKCζ recruits Smurf1 to membrane protrusions resulting in degradation of RhoA and ensures cell polarity [174]. CRL3BACURD was the second E3 ligase shown to ubiquitinate RhoA. It belongs to the Cullin-RING multi-subunit ubiquitin ligase family (CRL). To ubiquitinate RhoA, the CUL3 protein associates with the evolutionary conserved BTB domain-containing proteins BACURD1/2 that function as the receptors for RhoA [176]. CRL3BACURD targets inactive GDP bound RhoA for degradation unlike Smurf1 [177]. Skp1-Cul1-F-box (SCF) FBXL19 (SCFFBXL19) was recently identified as another E3 ligase for RhoA in lung epithelial cells. Unlike Smurf1, it ubiquitinates lysine-135 and targets RhoA for proteasomal degradation in an Erk2-dependent manner (Fig. 7) [178].
Two studies in 2011–2012 unveiled the E3 ubiquitin ligases of Rac1 [152, 179]. Oberoi et al. discovered that both XIAP and cIAP-1 directly bind to Rac1 and directly conjugate polyubiquitin chains to lysine-147 and target it for degradation (Fig. 7). Thus, loss of these two IAPs led to enhanced migration with elongated cell morphology. It is also shown that depletion of XIAP and cIAP-1 prevents CNF-1 toxin-mediated degradation of Rac1 or loss of Rac1 by RhoGDI depletion [152]. Hace1 was another HECT domain-containing protein that was identified to be an E3 ligase of Rac1 [179]. The key difference between Hace1 and the IAPs was that while Hace1 specifically interacted with active Rac1, the IAPs were indiscriminate about the activation status of Rac1. SCFFBXL19 was identified as a novel E3 ligase for Rac1 in mouse epithelial cells where it was shown that the F-Box protein polyubiquitinates Rac1 at lysine-166 (Fig. 7) and targets it for proteasomal degradation in an Akt-dependent manner [169].
Relatively little is known about the regulation of Cdc42 via proteasomal degradation and the identity of an E3 ligase for Cdc42 is still being investigated. Cdc42 does play a role in the degradation of RhoA though, as it binds to the aPKCζ-Par complex, which controls Smurf1 [174]. This suggests a spatio-temporal regulation for control of RhoA levels at the leading edge during protrusion-dependent movements by Cdc42.
SUMOylation is a process in which small-ubiquitin-like modifier (SUMO) proteins 1, 2, and 3 are covalently conjugated to specific lysine residues on target proteins. It is a reversible modification catalyzed by a family of SUMO-specific proteases called SENPs. Angeliki Malliri’s group first showed that Rho GTPases are also SUMOylated when they demonstrated that Rac1, primarily in its activated form, interacts with PIAS3. PIAS family proteins are known to function as SUMO E3 ligases and this study showed that HGF treatment led to co-localization of PIAS3 and Rac1-GTP at the cell membrane. Depletion of PIAS3 also led to reduced Rac1 activity, resulting in impaired lamellipodia-membrane ruffle formation. Ultimately, the study showed that SUMOylation is required to maintain sustained activation of Rac1 and consequently for optimal cell migration (Fig. 7) [180]. They also reported that Cdc42-GTP levels were not influenced by PIAS3 depletion. Due to the conserved PBR domains in the Rho GTPase family, this leads to speculation whether any of the other Rho GTPases are also targeted for SUMOylation.
Another recent study reported that RhoGDI1 SUMOylation at lysine-138 increases its binding to Rho GTPases, thus preventing cell migration. They also reported that XIAP binds to RhoGDI1 in a RING-dependent manner, thus preventing SUMOylation at lysine-138 [181, 182]. RhoGDIs add another dimension of regulation to the picture. There are only three genes encoding for RhoGDIs in mammals [183]. RhoGDI1 is the most abundant and the best-characterized member of the family and interacts with several Rho GTPases including Rac1, Rac2, Cdc42, RhoA, and RhoC [184, 185]. RhoGDIs bind to the cytosolic Rho pool and shield the isoprenoid moiety from water. Absence of RhoGDI1 results in the proteasome-dependent degradation of the unstable cytosolic pool of Rho GTPases [186, 187]. Intriguingly, XIAP is required for degradation of Rac1 upon RhoGDI depletion.
Finally, another mode of regulation of Rho GTPase levels is post-transcriptional modifications of Rho GTPase mRNAs by microRNAs. This is discussed in detail in the recent review by Liu and colleagues [188] and is not discussed in this review.
Concluding remarks
In recent years, the regulation of Rho GTPases and their role in controlling various cellular processes have been studied in great detail though many such studies are pursued using overexpression of these proteins in tumor cells. Further, the role of many Rho GTPases in regulating physiological forms of migration is understudied as most of the studies pertain to tumor cells. Numerous in vitro 2D/3D cell culture models mimicking the scenario in tissue have been continuously pursued albeit with limitations. The development of two-photon/multiphoton microscopes and other in vivo imaging techniques have enabled us to gain first-hand insights into the dynamics of cell shape and migration under physiological settings. Apart from Rac1, RhoA, and Cdc42, the role of other Rho GTPases is still unclear, and further studies are clearly warranted. Increasing evidence argues for the role for ubiquitination and SUMOylation in the regulation of Rho GTPases and deciphering the cross-talk between these various post-translational modifications remains a challenging issue. In the same lines, development of novel tools has provided vital inputs into the current understanding of the spatiotemporal dynamics of Rho GTPase activation in migrating cells. The explicit role of Rho GTPases in regulating tumorigenesis, and tumor cell migration in particular, has triggered enormous interest in targeting Rho GTPases or the downstream signaling components for tumor therapeutics. Further understanding of the pathophysiological relevance of Rho GTPases will assist in adroitly targeting some of these molecules and to adopt them for clinical trials.
Acknowledgments
The work in KR’s lab was funded through ENP programme grant RA1739/1-1 from the DFG and a PLUS3 grant from Boehringer Ingelheim Stiftung to KR. We apologize to colleagues whose work could not be cited due to space constraints.
References
- 1.Valencia A, Chardin P, Wittinghofer A, Sander C. The Ras protein family: evolutionary tree and role of conserved amino acids. Biochemistry. 1991;30:4637–4648. doi: 10.1021/bi00233a001. [DOI] [PubMed] [Google Scholar]
- 2.Sahai E, Marshall CJ. RHO-GTPases and cancer. Nat Rev Cancer. 2002;2:133–142. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- 3.Jaiswal M, Fansa EK, Dvorsky R, Ahmadian MR. New insight into the molecular switch mechanism of human Rho family proteins: shifting a paradigm. Biol Chem. 2013;394:89–95. doi: 10.1515/hsz-2012-0207. [DOI] [PubMed] [Google Scholar]
- 4.Aspenstrom P, Ruusala A, Pacholsky D. Taking Rho GTPases to the next level: the cellular functions of atypical Rho GTPases. Exp Cell Res. 2007;313:3673–3679. doi: 10.1016/j.yexcr.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 5.Chardin P. Function and regulation of Rnd proteins. Nat Rev Mol Cell Biol. 2006;7:54–62. doi: 10.1038/nrm1788. [DOI] [PubMed] [Google Scholar]
- 6.Wherlock M, Mellor H. The Rho GTPase family: a Racs to Wrchs story. J Cell Sci. 2002;115:239–240. doi: 10.1242/jcs.115.2.239. [DOI] [PubMed] [Google Scholar]
- 7.Ongusaha PP, Kim HG, Boswell SA, Ridley AJ, Der CJ, Dotto GP, Kim YB, Aaronson SA, Lee SW. RhoE is a pro-survival p53 target gene that inhibits ROCK I-mediated apoptosis in response to genotoxic stress. Curr Biol. 2006;16:2466–2472. doi: 10.1016/j.cub.2006.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Tao W, Pennica D, Xu L, Kalejta RF, Levine AJ. Wrch-1, a novel member of the Rho gene family that is regulated by Wnt-1. Genes Dev. 2001;15:1796–1807. doi: 10.1101/gad.894301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 10.Boureux A, Vignal E, Faure S, Fort P. Evolution of the Rho family of Ras-like GTPases in eukaryotes. Mol Biol Evol. 2007;24:203–216. doi: 10.1093/molbev/msl145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature. 2002;418:790–793. doi: 10.1038/nature00859. [DOI] [PubMed] [Google Scholar]
- 12.Machacek M, Hodgson L, Welch C, Elliott H, Pertz O, Nalbant P, Abell A, Johnson GL, Hahn KM, Danuser G. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thiel DA, Reeder MK, Pfaff A, Coleman TR, Sells MA, Chernoff J. Cell cycle-regulated phosphorylation of p21-activated kinase 1. Curr Biol. 2002;12:1227–1232. doi: 10.1016/s0960-9822(02)00931-4. [DOI] [PubMed] [Google Scholar]
- 14.Wells CM, Walmsley M, Ooi S, Tybulewicz V, Ridley AJ. Rac1-deficient macrophages exhibit defects in cell spreading and membrane ruffling but not migration. J Cell Sci. 2004;117:1259–1268. doi: 10.1242/jcs.00997. [DOI] [PubMed] [Google Scholar]
- 15.Wu YI, Wang X, He L, Montell D, Hahn KM. Spatiotemporal control of small GTPases with light using the LOV domain. Methods Enzymol. 2011;497:393–407. doi: 10.1016/B978-0-12-385075-1.00016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, He L, Wu YI, Hahn KM, Montell DJ. Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat Cell Biol. 2010;12:591–597. doi: 10.1038/ncb2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Rivera Rosado LA, Moon SY, Zhang B. Silencing of D4-GDI inhibits growth and invasive behavior in MDA-MB-231 cells by activation of Rac-dependent p38 and JNK signaling. J Biol Chem. 2009;284:12956–12965. doi: 10.1074/jbc.M807845200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L, Zhang JJ, Huang XY. cAMP inhibits cell migration by interfering with Rac-induced lamellipodium formation. J Biol Chem. 2008;283:13799–13805. doi: 10.1074/jbc.M800555200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sala G, Dituri F, Raimondi C, Previdi S, Maffucci T, Mazzoletti M, Rossi C, Iezzi M, Lattanzio R, Piantelli M, Iacobelli S, Broggini M, Falasca M. Phospholipase Cgamma1 is required for metastasis development and progression. Cancer Res. 2008;68:10187–10196. doi: 10.1158/0008-5472.CAN-08-1181. [DOI] [PubMed] [Google Scholar]
- 21.Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci USA. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bokoch GM. Regulation of cell function by Rho family GTPases. Immunol Res. 2000;21:139–148. doi: 10.1385/IR:21:2-3:139. [DOI] [PubMed] [Google Scholar]
- 23.Edwards DC, Sanders LC, Bokoch GM, Gill GN. Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nat Cell Biol. 1999;1:253–259. doi: 10.1038/12963. [DOI] [PubMed] [Google Scholar]
- 24.Vadlamudi RK, Li F, Barnes CJ, Bagheri-Yarmand R, Kumar R. p41-Arc subunit of human Arp2/3 complex is a p21-activated kinase-1-interacting substrate. EMBO Rep. 2004;5:154–160. doi: 10.1038/sj.embor.7400079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ong CC, Jubb AM, Zhou W, Haverty PM, Harris AL, Belvin M, Friedman LS, Koeppen H, Hoeflich KP. p21-activated kinase 1: PAK’ed with potential. Oncotarget. 2011;2:491–496. doi: 10.18632/oncotarget.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rayala SK, Molli PR, Kumar R. Nuclear p21-activated kinase 1 in breast cancer packs off tamoxifen sensitivity. Cancer Res. 2006;66:5985–5988. doi: 10.1158/0008-5472.CAN-06-0978. [DOI] [PubMed] [Google Scholar]
- 27.Sugihara K, Nakatsuji N, Nakamura K, Nakao K, Hashimoto R, Otani H, Sakagami H, Kondo H, Nozawa S, Aiba A, Katsuki M. Rac1 is required for the formation of three germ layers during gastrulation. Oncogene. 1998;17:3427–3433. doi: 10.1038/sj.onc.1202595. [DOI] [PubMed] [Google Scholar]
- 28.Benninger Y, Thurnherr T, Pereira JA, Krause S, Wu X, Chrostek-Grashoff A, Herzog D, Nave KA, Franklin RJ, Meijer D, Brakebusch C, Suter U, Relvas JB. Essential and distinct roles for cdc42 and rac1 in the regulation of Schwann cell biology during peripheral nervous system development. J Cell Biol. 2007;177:1051–1061. doi: 10.1083/jcb.200610108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo F, Cancelas JA, Hildeman D, Williams DA, Zheng Y. Rac GTPase isoforms Rac1 and Rac2 play a redundant and crucial role in T-cell development. Blood. 2008;112:1767–1775. doi: 10.1182/blood-2008-01-132068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walmsley MJ, Ooi SK, Reynolds LF, Smith SH, Ruf S, Mathiot A, Vanes L, Williams DA, Cancro MP, Tybulewicz VL. Critical roles for Rac1 and Rac2 GTPases in B cell development and signaling. Science. 2003;302:459–462. doi: 10.1126/science.1089709. [DOI] [PubMed] [Google Scholar]
- 31.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 32.Corbetta S, Gualdoni S, Albertinazzi C, Paris S, Croci L, Consalez GG, de Curtis I. Generation and characterization of Rac3 knockout mice. Mol Cell Biol. 2005;25:5763–5776. doi: 10.1128/MCB.25.13.5763-5776.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, Dicara D, Ramos AH, Lawrence MS, Cibulskis K, Sivachenko A, Voet D, Saksena G, Stransky N, Onofrio RC, Winckler W, Ardlie K, Wagle N, Wargo J, Chong K, Morton DL, Stemke-Hale K, Chen G, Noble M, Meyerson M, Ladbury JE, Davies MA, Gershenwald JE, Wagner SN, Hoon DS, Schadendorf D, Lander ES, Gabriel SB, Getz G, Garraway LA, Chin L. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, Cheng E, Davis MJ, Goh G, Choi M, Ariyan S, Narayan D, Dutton-Regester K, Capatana A, Holman EC, Bosenberg M, Sznol M, Kluger HM, Brash DE, Stern DF, Materin MA, Lo RS, Mane S, Ma S, Kidd KK, Hayward NK, Lifton RP, Schlessinger J, Boggon TJ, Halaban R. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jordan P, Brazao R, Boavida MG, Gespach C, Chastre E. Cloning of a novel human Rac1b splice variant with increased expression in colorectal tumors. Oncogene. 1999;18:6835–6839. doi: 10.1038/sj.onc.1203233. [DOI] [PubMed] [Google Scholar]
- 36.Matos P, Collard JG, Jordan P. Tumor-related alternatively spliced Rac1b is not regulated by Rho-GDP dissociation inhibitors and exhibits selective downstream signaling. J Biol Chem. 2003;278:50442–50448. doi: 10.1074/jbc.M308215200. [DOI] [PubMed] [Google Scholar]
- 37.Singh A, Karnoub AE, Palmby TR, Lengyel E, Sondek J, Der CJ. Rac1b, a tumor associated, constitutively active Rac1 splice variant, promotes cellular transformation. Oncogene. 2004;23:9369–9380. doi: 10.1038/sj.onc.1208182. [DOI] [PubMed] [Google Scholar]
- 38.Visvikis O, Lores P, Boyer L, Chardin P, Lemichez E, Gacon G. Activated Rac1, but not the tumorigenic variant Rac1b, is ubiquitinated on Lys 147 through a JNK-regulated process. FEBS J. 2008;275:386–396. doi: 10.1111/j.1742-4658.2007.06209.x. [DOI] [PubMed] [Google Scholar]
- 39.Ladd PD, Butler JS, Skalnik DG. Identification of a genomic fragment that directs hematopoietic-specific expression of Rac2 and analysis of the DNA methylation profile of the gene locus. Gene. 2004;341:323–333. doi: 10.1016/j.gene.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 40.Haataja L, Groffen J, Heisterkamp N. Characterization of RAC3, a novel member of the Rho family. J Biol Chem. 1997;272:20384–20388. doi: 10.1074/jbc.272.33.20384. [DOI] [PubMed] [Google Scholar]
- 41.Van Hennik PB, Hordijk PL. Rho GTPases in hematopoietic cells. Antioxid Redox Signal. 2005;7:1440–1455. doi: 10.1089/ars.2005.7.1440. [DOI] [PubMed] [Google Scholar]
- 42.Roberts AW, Kim C, Zhen L, Lowe JB, Kapur R, Petryniak B, Spaetti A, Pollock JD, Borneo JB, Bradford GB, Atkinson SJ, Dinauer MC, Williams DA. Deficiency of the hematopoietic cell-specific Rho family GTPase Rac2 is characterized by abnormalities in neutrophil function and host defense. Immunity. 1999;10:183–196. doi: 10.1016/s1074-7613(00)80019-9. [DOI] [PubMed] [Google Scholar]
- 43.Williams DA, Tao W, Yang F, Kim C, Gu Y, Mansfield P, Levine JE, Petryniak B, Derrow CW, Harris C, Jia B, Zheng Y, Ambruso DR, Lowe JB, Atkinson SJ, Dinauer MC, Boxer L. Dominant negative mutation of the hematopoietic-specific Rho GTPase, Rac2, is associated with a human phagocyte immunodeficiency. Blood. 2000;96:1646–1654. [PubMed] [Google Scholar]
- 44.Yamauchi A, Kim C, Li S, Marchal CC, Towe J, Atkinson SJ, Dinauer MC. Rac2-deficient murine macrophages have selective defects in superoxide production and phagocytosis of opsonized particles. J Immunol. 2004;173:5971–5979. doi: 10.4049/jimmunol.173.10.5971. [DOI] [PubMed] [Google Scholar]
- 45.Dinauer MC. Regulation of neutrophil function by Rac GTPases. Curr Opin Hematol. 2003;10:8–15. doi: 10.1097/00062752-200301000-00003. [DOI] [PubMed] [Google Scholar]
- 46.Werner E. GTPases and reactive oxygen species: switches for killing and signaling. J Cell Sci. 2004;117:143–153. doi: 10.1242/jcs.00937. [DOI] [PubMed] [Google Scholar]
- 47.Mira JP, Benard V, Groffen J, Sanders LC, Knaus UG. Endogenous, hyperactive Rac3 controls proliferation of breast cancer cells by a p21-activated kinase-dependent pathway. Proc Natl Acad Sci USA. 2000;97:185–189. doi: 10.1073/pnas.97.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morris CM, Haataja L, McDonald M, Gough S, Markie D, Groffen J, Heisterkamp N. The small GTPase RAC3 gene is located within chromosome band 17q25.3 outside and telomeric of a region commonly deleted in breast and ovarian tumours. Cytogenet Cell Genet. 2000;89:18–23. doi: 10.1159/000015583. [DOI] [PubMed] [Google Scholar]
- 49.Katoh H, Hiramoto K, Negishi M. Activation of Rac1 by RhoG regulates cell migration. J Cell Sci. 2006;119:56–65. doi: 10.1242/jcs.02720. [DOI] [PubMed] [Google Scholar]
- 50.Patel JC, Galan JE. Investigating the function of Rho family GTPases during Salmonella/host cell interactions. Methods Enzymol. 2008;439:145–158. doi: 10.1016/S0076-6879(07)00411-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roppenser B, Roder A, Hentschke M, Ruckdeschel K, Aepfelbacher M. Yersinia enterocolitica differentially modulates RhoG activity in host cells. J Cell Sci. 2009;122:696–705. doi: 10.1242/jcs.040345. [DOI] [PubMed] [Google Scholar]
- 52.Samson T, Welch C, Monaghan-Benson E, Hahn KM, Burridge K. Endogenous RhoG is rapidly activated after epidermal growth factor stimulation through multiple guanine-nucleotide exchange factors. Mol Biol Cell. 2010;21:1629–1642. doi: 10.1091/mbc.E09-09-0809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang HW, Shin MG, Lee S, Kim JR, Park WS, Cho KH, Meyer T, Do Heo W. Cooperative activation of PI3K by Ras and Rho family small GTPases. Mol Cell. 2012;47:281–290. doi: 10.1016/j.molcel.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gomez del Pulgar T, Benitah SA, Valeron PF, Espina C, Lacal JC. Rho GTPase expression in tumourigenesis: evidence for a significant link. BioEssays. 2005;27:602–613. doi: 10.1002/bies.20238. [DOI] [PubMed] [Google Scholar]
- 55.Huang M, Prendergast GC. RhoB in cancer suppression. Histol Histopathol. 2006;21:213–218. doi: 10.14670/HH-21.213. [DOI] [PubMed] [Google Scholar]
- 56.Ridley AJ. RhoA, RhoB and RhoC have different roles in cancer cell migration. J Microsc. 2013;251:242–249. doi: 10.1111/jmi.12025. [DOI] [PubMed] [Google Scholar]
- 57.Braga VM, Yap AS. The challenges of abundance: epithelial junctions and small GTPase signalling. Curr Opin Cell Biol. 2005;17:466–474. doi: 10.1016/j.ceb.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 58.Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer. 2003;3:362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- 59.Merajver SD, Usmani SZ. Multifaceted role of Rho proteins in angiogenesis. J Mammary Gland Biol Neoplasia. 2005;10:291–298. doi: 10.1007/s10911-006-9002-8. [DOI] [PubMed] [Google Scholar]
- 60.Wang W, Wu F, Fang F, Tao Y, Yang L. RhoC is essential for angiogenesis induced by hepatocellular carcinoma cells via regulation of endothelial cell organization. Cancer Sci. 2008;99:2012–2018. doi: 10.1111/j.1349-7006.2008.00902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dietrich KA, Schwarz R, Liska M, Grass S, Menke A, Meister M, Kierschke G, Langle C, Genze F, Giehl K. Specific induction of migration and invasion of pancreatic carcinoma cells by RhoC, which differs from RhoA in its localisation and activity. Biol Chem. 2009;390:1063–1077. doi: 10.1515/BC.2009.110. [DOI] [PubMed] [Google Scholar]
- 62.Ellis S, Mellor H. Regulation of endocytic traffic by rho family GTPases. Trends Cell Biol. 2000;10:85–88. doi: 10.1016/s0962-8924(99)01710-9. [DOI] [PubMed] [Google Scholar]
- 63.Ridley AJ. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 2006;16:522–529. doi: 10.1016/j.tcb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 64.Liu AX, Rane N, Liu JP, Prendergast GC. RhoB is dispensable for mouse development, but it modifies susceptibility to tumor formation as well as cell adhesion and growth factor signaling in transformed cells. Mol Cell Biol. 2001;21:6906–6912. doi: 10.1128/MCB.21.20.6906-6912.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ho TT, Merajver SD, Lapiere CM, Nusgens BV, Deroanne CF. RhoA-GDP regulates RhoB protein stability. Potential involvement of RhoGDIalpha. J Biol Chem. 2008;283:21588–21598. doi: 10.1074/jbc.M710033200. [DOI] [PubMed] [Google Scholar]
- 66.Vega FM, Fruhwirth G, Ng T, Ridley AJ. RhoA and RhoC have distinct roles in migration and invasion by acting through different targets. J Cell Biol. 2011;193:655–665. doi: 10.1083/jcb.201011038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lang P, Gesbert F, Delespine-Carmagnat M, Stancou R, Pouchelet M, Bertoglio J. Protein kinase A phosphorylation of RhoA mediates the morphological and functional effects of cyclic AMP in cytotoxic lymphocytes. EMBO J. 1996;15:510–519. [PMC free article] [PubMed] [Google Scholar]
- 68.Sauzeau V, Le Jeune H, Cario-Toumaniantz C, Smolenski A, Lohmann SM, Bertoglio J, Chardin P, Pacaud P, Loirand G. Cyclic GMP-dependent protein kinase signaling pathway inhibits RhoA-induced Ca2+ sensitization of contraction in vascular smooth muscle. J Biol Chem. 2000;275:21722–21729. doi: 10.1074/jbc.M000753200. [DOI] [PubMed] [Google Scholar]
- 69.Savoia C, Tabet F, Yao G, Schiffrin EL, Touyz RM. Negative regulation of RhoA/Rho kinase by angiotensin II type 2 receptor in vascular smooth muscle cells: role in angiotensin II-induced vasodilation in stroke-prone spontaneously hypertensive rats. J Hypertens. 2005;23:1037–1045. doi: 10.1097/01.hjh.0000166845.49850.39. [DOI] [PubMed] [Google Scholar]
- 70.Sloan CM, Quinn CV, Peters JP, Farley J, Goetzinger C, Wernli M, DeMali KA, Ellerbroek SM. Divergence of Rho residue 43 impacts GEF activity. Small GTPases. 2012;3:15–22. doi: 10.4161/sgtp.19557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garvalov BK, Flynn KC, Neukirchen D, Meyn L, Teusch N, Wu X, Brakebusch C, Bamburg JR, Bradke F. Cdc42 regulates cofilin during the establishment of neuronal polarity. J Neurosci. 2007;27:13117–13129. doi: 10.1523/JNEUROSCI.3322-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thurnherr T, Benninger Y, Wu X, Chrostek A, Krause SM, Nave KA, Franklin RJ, Brakebusch C, Suter U, Relvas JB. Cdc42 and Rac1 signaling are both required for and act synergistically in the correct formation of myelin sheaths in the CNS. J Neurosci. 2006;26:10110–10119. doi: 10.1523/JNEUROSCI.2158-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee K, Boyd KL, Parekh DV, Kehl-Fie TE, Baldwin HS, Brakebusch C, Skaar EP, Boothby M, Zent R. Cdc42 promotes host defenses against fatal infection. Infect Immun. 2013;81:2714–2723. doi: 10.1128/IAI.01114-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang L, Wang L, Zheng Y. Gene targeting of Cdc42 and Cdc42GAP affirms the critical involvement of Cdc42 in filopodia induction, directed migration, and proliferation in primary mouse embryonic fibroblasts. Mol Biol Cell. 2006;17:4675–4685. doi: 10.1091/mbc.E06-05-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Czuchra A, Wu X, Meyer H, van Hengel J, Schroeder T, Geffers R, Rottner K, Brakebusch C. Cdc42 is not essential for filopodium formation, directed migration, cell polarization, and mitosis in fibroblastoid cells. Mol Biol Cell. 2005;16:4473–4484. doi: 10.1091/mbc.E05-01-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chiang SH, Baumann CA, Kanzaki M, Thurmond DC, Watson RT, Neudauer CL, Macara IG, Pessin JE, Saltiel AR. Insulin-stimulated GLUT4 translocation requires the CAP-dependent activation of TC10. Nature. 2001;410:944–948. doi: 10.1038/35073608. [DOI] [PubMed] [Google Scholar]
- 77.Kawase K, Nakamura T, Takaya A, Aoki K, Namikawa K, Kiyama H, Inagaki S, Takemoto H, Saltiel AR, Matsuda M. GTP hydrolysis by the Rho family GTPase TC10 promotes exocytic vesicle fusion. Dev Cell. 2006;11:411–421. doi: 10.1016/j.devcel.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 78.Abe T, Kato M, Miki H, Takenawa T, Endo T. Small GTPase Tc10 and its homologue RhoT induce N-WASP-mediated long process formation and neurite outgrowth. J Cell Sci. 2003;116:155–168. doi: 10.1242/jcs.00208. [DOI] [PubMed] [Google Scholar]
- 79.Doyle AD, Petrie RJ, Kutys ML, Yamada KM. Dimensions in cell migration. Curr Opin Cell Biol. 2013;25:642–649. doi: 10.1016/j.ceb.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Petrie RJ, Yamada KM. At the leading edge of three-dimensional cell migration. J Cell Sci. 2012;125:5917–5926. doi: 10.1242/jcs.093732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Charras GT, Yarrow JC, Horton MA, Mahadevan L, Mitchison TJ. Non-equilibration of hydrostatic pressure in blebbing cells. Nature. 2005;435:365–369. doi: 10.1038/nature03550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Linder S, Wiesner C, Himmel M. Degrading devices: invadosomes in proteolytic cell invasion. Annu Rev Cell Dev Biol. 2011;27:185–211. doi: 10.1146/annurev-cellbio-092910-154216. [DOI] [PubMed] [Google Scholar]
- 83.Provenzano PP, Inman DR, Eliceiri KW, Trier SM, Keely PJ. Contact guidance mediated three-dimensional cell migration is regulated by Rho/ROCK-dependent matrix reorganization. Biophys J. 2008;95:5374–5384. doi: 10.1529/biophysj.108.133116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sidani M, Wyckoff J, Xue C, Segall JE, Condeelis J. Probing the microenvironment of mammary tumors using multiphoton microscopy. J Mammary Gland Biol Neoplasia. 2006;11:151–163. doi: 10.1007/s10911-006-9021-5. [DOI] [PubMed] [Google Scholar]
- 85.Baker BM, Chen CS. Deconstructing the third dimension: how 3D culture microenvironments alter cellular cues. J Cell Sci. 2012;125:3015–3024. doi: 10.1242/jcs.079509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Giampieri S, Manning C, Hooper S, Jones L, Hill CS, Sahai E. Localized and reversible TGFbeta signalling switches breast cancer cells from cohesive to single cell motility. Nat Cell Biol. 2009;11:1287–1296. doi: 10.1038/ncb1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Grinnell F. Fibroblast mechanics in three-dimensional collagen matrices. J Bodyw Mov Ther. 2008;12:191–193. doi: 10.1016/j.jbmt.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sanz-Moreno V, Gadea G, Ahn J, Paterson H, Marra P, Pinner S, Sahai E, Marshall CJ. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135:510–523. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 89.Lee HH, Tien SC, Jou TS, Chang YC, Jhong JG, Chang ZF. Src-dependent phosphorylation of ROCK participates in regulation of focal adhesion dynamics. J Cell Sci. 2010;123:3368–3377. doi: 10.1242/jcs.071555. [DOI] [PubMed] [Google Scholar]
- 90.Etienne-Manneville S, Hall A. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCzeta. Cell. 2001;106:489–498. doi: 10.1016/s0092-8674(01)00471-8. [DOI] [PubMed] [Google Scholar]
- 91.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 92.Yui Y, Itoh K, Yoshioka K, Naka N, Watanabe M, Hiraumi Y, Matsubara H, Watanabe K, Sano K, Nakahata T, Adachi S. Mesenchymal mode of migration participates in pulmonary metastasis of mouse osteosarcoma LM8. Clin Exp Metastasis. 2010;27:619–630. doi: 10.1007/s10585-010-9352-x. [DOI] [PubMed] [Google Scholar]
- 93.Block J, Breitsprecher D, Kuhn S, Winterhoff M, Kage F, Geffers R, Duwe P, Rohn JL, Baum B, Brakebusch C, Geyer M, Stradal TE, Faix J, Rottner K. FMNL2 drives actin-based protrusion and migration downstream of Cdc42. Curr Biol. 2012;22:1005–1012. doi: 10.1016/j.cub.2012.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Alblas J, Ulfman L, Hordijk P, Koenderman L. Activation of Rhoa and ROCK are essential for detachment of migrating leukocytes. Mol Biol Cell. 2001;12:2137–2145. doi: 10.1091/mbc.12.7.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kurokawa K, Matsuda M. Localized RhoA activation as a requirement for the induction of membrane ruffling. Mol Biol Cell. 2005;16:4294–4303. doi: 10.1091/mbc.E04-12-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bachy S, Letourneur F, Rousselle P. Syndecan-1 interaction with the LG4/5 domain in laminin-332 is essential for keratinocyte migration. J Cell Physiol. 2008;214:238–249. doi: 10.1002/jcp.21184. [DOI] [PubMed] [Google Scholar]
- 97.Gonzalo P, Guadamillas MC, Hernandez-Riquer MV, Pollan A, Grande-Garcia A, Bartolome RA, Vasanji A, Ambrogio C, Chiarle R, Teixido J, Risteli J, Apte SS, del Pozo MA, Arroyo AG. MT1-MMP is required for myeloid cell fusion via regulation of Rac1 signaling. Dev Cell. 2010;18:77–89. doi: 10.1016/j.devcel.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kheradmand F, Werner E, Tremble P, Symons M, Werb Z. Role of Rac1 and oxygen radicals in collagenase-1 expression induced by cell shape change. Science. 1998;280:898–902. doi: 10.1126/science.280.5365.898. [DOI] [PubMed] [Google Scholar]
- 99.Westermarck J, Kahari VM. Regulation of matrix metalloproteinase expression in tumor invasion. Faseb J. 1999;13:781–792. [PubMed] [Google Scholar]
- 100.Yamagata K, Li X, Ikegaki S, Oneyama C, Okada M, Nishita M, Minami Y. Dissection of Wnt5a-Ror2 signaling leading to matrix metalloproteinase (MMP-13) expression. J Biol Chem. 2012;287:1588–1599. doi: 10.1074/jbc.M111.315127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jeong KJ, Park SY, Cho KH, Sohn JS, Lee J, Kim YK, Kang J, Park CG, Han JW, Lee HY. The Rho/ROCK pathway for lysophosphatidic acid-induced proteolytic enzyme expression and ovarian cancer cell invasion. Oncogene. 2012;31:4279–4289. doi: 10.1038/onc.2011.595. [DOI] [PubMed] [Google Scholar]
- 102.Wyckoff JB, Jones JG, Condeelis JS, Segall JE. A critical step in metastasis: in vivo analysis of intravasation at the primary tumor. Cancer Res. 2000;60:2504–2511. [PubMed] [Google Scholar]
- 103.Philippar U, Roussos ET, Oser M, Yamaguchi H, Kim HD, Giampieri S, Wang Y, Goswami S, Wyckoff JB, Lauffenburger DA, Sahai E, Condeelis JS, Gertler FB. A Mena invasion isoform potentiates EGF-induced carcinoma cell invasion and metastasis. Dev Cell. 2008;15:813–828. doi: 10.1016/j.devcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sahai E, Wyckoff J, Philippar U, Segall JE, Gertler F, Condeelis J. Simultaneous imaging of GFP, CFP and collagen in tumors in vivo using multiphoton microscopy. BMC Biotechnol. 2005;5:14. doi: 10.1186/1472-6750-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xue C, Wyckoff J, Liang F, Sidani M, Violini S, Tsai KL, Zhang ZY, Sahai E, Condeelis J, Segall JE. Epidermal growth factor receptor overexpression results in increased tumor cell motility in vivo coordinately with enhanced intravasation and metastasis. Cancer Res. 2006;66:192–197. doi: 10.1158/0008-5472.CAN-05-1242. [DOI] [PubMed] [Google Scholar]
- 106.Fackler OT, Grosse R. Cell motility through plasma membrane blebbing. J Cell Biol. 2008;181:879–884. doi: 10.1083/jcb.200802081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Keller HU, Bebie H. Protrusive activity quantitatively determines the rate and direction of cell locomotion. Cell Motil Cytoskelet. 1996;33:241–251. doi: 10.1002/(SICI)1097-0169(1996)33:4<241::AID-CM1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 108.Yoshida K, Soldati T. Dissection of amoeboid movement into two mechanically distinct modes. J Cell Sci. 2006;119:3833–3844. doi: 10.1242/jcs.03152. [DOI] [PubMed] [Google Scholar]
- 109.Friedl P, Wolf K. Plasticity of cell migration: a multiscale tuning model. J Cell Biol. 2010;188:11–19. doi: 10.1083/jcb.200909003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sahai E, Marshall CJ. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat Cell Biol. 2003;5:711–719. doi: 10.1038/ncb1019. [DOI] [PubMed] [Google Scholar]
- 111.Lammermann T, Bader BL, Monkley SJ, Worbs T, Wedlich-Soldner R, Hirsch K, Keller M, Forster R, Critchley DR, Fassler R, Sixt M. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature. 2008;453:51–55. doi: 10.1038/nature06887. [DOI] [PubMed] [Google Scholar]
- 112.Pinner S, Sahai E. PDK1 regulates cancer cell motility by antagonising inhibition of ROCK1 by RhoE. Nat Cell Biol. 2008;10:127–137. doi: 10.1038/ncb1675. [DOI] [PubMed] [Google Scholar]
- 113.Eisenmann KM, Harris ES, Kitchen SM, Holman HA, Higgs HN, Alberts AS. Dia-interacting protein modulates formin-mediated actin assembly at the cell cortex. Curr Biol. 2007;17:579–591. doi: 10.1016/j.cub.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 114.Rabodzey A, Alcaide P, Luscinskas FW, Ladoux B. Mechanical forces induced by the transendothelial migration of human neutrophils. Biophys J. 2008;95:1428–1438. doi: 10.1529/biophysj.107.119156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sabeh F, Shimizu-Hirota R, Weiss SJ. Protease-dependent versus -independent cancer cell invasion programs: three-dimensional amoeboid movement revisited. J Cell Biol. 2009;185:11–19. doi: 10.1083/jcb.200807195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wolf K, Wu YI, Liu Y, Geiger J, Tam E, Overall C, Stack MS, Friedl P. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat Cell Biol. 2007;9:893–904. doi: 10.1038/ncb1616. [DOI] [PubMed] [Google Scholar]
- 117.Gadea G, Sanz-Moreno V, Self A, Godi A, Marshall CJ. DOCK10-mediated Cdc42 activation is necessary for amoeboid invasion of melanoma cells. Curr Biol. 2008;18:1456–1465. doi: 10.1016/j.cub.2008.08.053. [DOI] [PubMed] [Google Scholar]
- 118.Lorentzen A, Bamber J, Sadok A, Elson-Schwab I, Marshall CJ. An ezrin-rich, rigid uropod-like structure directs movement of amoeboid blebbing cells. J Cell Sci. 2011;124:1256–1267. doi: 10.1242/jcs.074849. [DOI] [PubMed] [Google Scholar]
- 119.Friedl P, Hegerfeldt Y, Tusch M. Collective cell migration in morphogenesis and cancer. Int J Dev Biol. 2004;48:441–449. doi: 10.1387/ijdb.041821pf. [DOI] [PubMed] [Google Scholar]
- 120.Friedl P, Noble PB, Walton PA, Laird DW, Chauvin PJ, Tabah RJ, Black M, Zanker KS. Migration of coordinated cell clusters in mesenchymal and epithelial cancer explants in vitro. Cancer Res. 1995;55:4557–4560. [PubMed] [Google Scholar]
- 121.Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10:445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 122.Garcia GL, Rericha EC, Heger CD, Goldsmith PK, Parent CA. The group migration of Dictyostelium cells is regulated by extracellular chemoattractant degradation. Mol Biol Cell. 2009;20:3295–3304. doi: 10.1091/mbc.E09-03-0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rieger S, Senghaas N, Walch A, Koster RW. Cadherin-2 controls directional chain migration of cerebellar granule neurons. PLoS Biol. 2009;7:e1000240. doi: 10.1371/journal.pbio.1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Parri M, Chiarugi P. Rac and Rho GTPases in cancer cell motility control. Cell Commun Signal. 2010;8:23. doi: 10.1186/1478-811X-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ewald AJ, Brenot A, Duong M, Chan BS, Werb Z. Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Dev Cell. 2008;14:570–581. doi: 10.1016/j.devcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gray RS, Cheung KJ, Ewald AJ. Cellular mechanisms regulating epithelial morphogenesis and cancer invasion. Curr Opin Cell Biol. 2010;22:640–650. doi: 10.1016/j.ceb.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Scholl FG, Gamallo C, Vilaro S, Quintanilla M. Identification of PA2.26 antigen as a novel cell-surface mucin-type glycoprotein that induces plasma membrane extensions and increased motility in keratinocytes. J Cell Sci. 1999;112(Pt 24):4601–4613. doi: 10.1242/jcs.112.24.4601. [DOI] [PubMed] [Google Scholar]
- 128.Wicki A, Lehembre F, Wick N, Hantusch B, Kerjaschki D, Christofori G. Tumor invasion in the absence of epithelial–mesenchymal transition: podoplanin-mediated remodeling of the actin cytoskeleton. Cancer Cell. 2006;9:261–272. doi: 10.1016/j.ccr.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 129.Hidalgo-Carcedo C, Hooper S, Chaudhry SI, Williamson P, Harrington K, Leitinger B, Sahai E. Collective cell migration requires suppression of actomyosin at cell–cell contacts mediated by DDR1 and the cell polarity regulators Par3 and Par6. Nat Cell Biol. 2011;13:49–58. doi: 10.1038/ncb2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Levayer R, Pelissier-Monier A, Lecuit T. Spatial regulation of Dia and Myosin-II by RhoGEF2 controls initiation of E-cadherin endocytosis during epithelial morphogenesis. Nat Cell Biol. 2011;13:529–540. doi: 10.1038/ncb2224. [DOI] [PubMed] [Google Scholar]
- 131.Shintani Y, Fukumoto Y, Chaika N, Svoboda R, Wheelock MJ, Johnson KR. Collagen I-mediated up-regulation of N-cadherin requires cooperative signals from integrins and discoidin domain receptor 1. J Cell Biol. 2008;180:1277–1289. doi: 10.1083/jcb.200708137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Theveneau E, Marchant L, Kuriyama S, Gull M, Moepps B, Parsons M, Mayor R. Collective chemotaxis requires contact-dependent cell polarity. Dev Cell. 2010;19:39–53. doi: 10.1016/j.devcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Theveneau E, Mayor R. Integrating chemotaxis and contact-inhibition during collective cell migration: small GTPases at work. Small GTPases. 2010;1:113–117. doi: 10.4161/sgtp.1.2.13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Poujade M, Grasland-Mongrain E, Hertzog A, Jouanneau J, Chavrier P, Ladoux B, Buguin A, Silberzan P. Collective migration of an epithelial monolayer in response to a model wound. Proc Natl Acad Sci USA. 2007;104:15988–15993. doi: 10.1073/pnas.0705062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gaggioli C, Hooper S, Hidalgo-Carcedo C, Grosse R, Marshall JF, Harrington K, Sahai E. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat Cell Biol. 2007;9:1392–1400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- 136.Sanz-Moreno V, Gaggioli C, Yeo M, Albrengues J, Wallberg F, Viros A, Hooper S, Mitter R, Feral CC, Cook M, Larkin J, Marais R, Meneguzzi G, Sahai E, Marshall CJ. ROCK and JAK1 signaling cooperate to control actomyosin contractility in tumor cells and stroma. Cancer Cell. 2011;20:229–245. doi: 10.1016/j.ccr.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 137.Rakic P. Mode of cell migration to the superficial layers of fetal monkey neocortex. J Comp Neurol. 1972;145:61–83. doi: 10.1002/cne.901450105. [DOI] [PubMed] [Google Scholar]
- 138.Kawauchi T, Sekine K, Shikanai M, Chihama K, Tomita K, Kubo K, Nakajima K, Nabeshima Y, Hoshino M. Rab GTPases-dependent endocytic pathways regulate neuronal migration and maturation through N-cadherin trafficking. Neuron. 2010;67:588–602. doi: 10.1016/j.neuron.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 139.Shikanai M, Nakajima K, Kawauchi T. N-cadherin regulates radial glial fiber-dependent migration of cortical locomoting neurons. Commun Integr Biol. 2011;4:326–330. doi: 10.4161/cib.4.3.14886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kalluri R. EMT: when epithelial cells decide to become mesenchymal-like cells. J Clin Invest. 2009;119:1417–1419. doi: 10.1172/JCI39675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Spaderna S, Schmalhofer O, Wahlbuhl M, Dimmler A, Bauer K, Sultan A, Hlubek F, Jung A, Strand D, Eger A, Kirchner T, Behrens J, Brabletz T. The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res. 2008;68:537–544. doi: 10.1158/0008-5472.CAN-07-5682. [DOI] [PubMed] [Google Scholar]
- 142.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 143.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial–mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 144.Radisky DC, Levy DD, Littlepage LE, Liu H, Nelson CM, Fata JE, Leake D, Godden EL, Albertson DG, Nieto MA, Werb Z, Bissell MJ. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature. 2005;436:123–127. doi: 10.1038/nature03688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hordijk PL, ten Klooster JP, van der Kammen RA, Michiels F, Oomen LC, Collard JG. Inhibition of invasion of epithelial cells by Tiam1-Rac signaling. Science. 1997;278:1464–1466. doi: 10.1126/science.278.5342.1464. [DOI] [PubMed] [Google Scholar]
- 146.Malliri A, van Es S, Huveneers S, Collard JG. The Rac exchange factor Tiam1 is required for the establishment and maintenance of cadherin-based adhesions. J Biol Chem. 2004;279:30092–30098. doi: 10.1074/jbc.M401192200. [DOI] [PubMed] [Google Scholar]
- 147.Ahn J, Sanz-Moreno V, Marshall CJ. The metastasis gene NEDD9 product acts through integrin beta3 and Src to promote mesenchymal motility and inhibit amoeboid motility. J Cell Sci. 2012;125:1814–1826. doi: 10.1242/jcs.101444. [DOI] [PubMed] [Google Scholar]
- 148.Parri M, Taddei ML, Bianchini F, Calorini L, Chiarugi P. EphA2 reexpression prompts invasion of melanoma cells shifting from mesenchymal to amoeboid-like motility style. Cancer Res. 2009;69:2072–2081. doi: 10.1158/0008-5472.CAN-08-1845. [DOI] [PubMed] [Google Scholar]
- 149.Yamazaki D, Kurisu S, Takenawa T. Involvement of Rac and Rho signaling in cancer cell motility in 3D substrates. Oncogene. 2009;28:1570–1583. doi: 10.1038/onc.2009.2. [DOI] [PubMed] [Google Scholar]
- 150.Palamidessi A, Frittoli E, Garre M, Faretta M, Mione M, Testa I, Diaspro A, Lanzetti L, Scita G, Di Fiore PP. Endocytic trafficking of Rac is required for the spatial restriction of signaling in cell migration. Cell. 2008;134:135–147. doi: 10.1016/j.cell.2008.05.034. [DOI] [PubMed] [Google Scholar]
- 151.Sahai E, Garcia-Medina R, Pouyssegur J, Vial E. Smurf1 regulates tumor cell plasticity and motility through degradation of RhoA leading to localized inhibition of contractility. J Cell Biol. 2007;176:35–42. doi: 10.1083/jcb.200605135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Oberoi TK, Dogan T, Hocking JC, Scholz RP, Mooz J, Anderson CL, Karreman C, Meyer zu Heringdorf D, Schmidt G, Ruonala M, Namikawa K, Harms GS, Carpy A, Macek B, Koster RW, Rajalingam K. IAPs regulate the plasticity of cell migration by directly targeting Rac1 for degradation. Embo J. 2012;31:14–28. doi: 10.1038/emboj.2011.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Oberoi-Khanuja TK, Rajalingam K. IAPs as E3 ligases of Rac1: shaping the move. Small GTPases. 2012;3:131–136. doi: 10.4161/sgtp.19988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, Lukashchuk N, Gillespie DA, Ludwig RL, Gosselin P, Cromer A, Brugge JS, Sansom OJ, Norman JC, Vousden KH. Mutant p53 drives invasion by promoting integrin recycling. Cell. 2009;139:1327–1341. doi: 10.1016/j.cell.2009.11.026. [DOI] [PubMed] [Google Scholar]
- 155.Curmi PA, Gavet O, Charbaut E, Ozon S, Lachkar-Colmerauer S, Manceau V, Siavoshian S, Maucuer A, Sobel A. Stathmin and its phosphoprotein family: general properties, biochemical and functional interaction with tubulin. Cell Struct Funct. 1999;24:345–357. doi: 10.1247/csf.24.345. [DOI] [PubMed] [Google Scholar]
- 156.Belletti B, Nicoloso MS, Schiappacassi M, Berton S, Lovat F, Wolf K, Canzonieri V, D’Andrea S, Zucchetto A, Friedl P, Colombatti A, Baldassarre G. Stathmin activity influences sarcoma cell shape, motility, and metastatic potential. Mol Biol Cell. 2008;19:2003–2013. doi: 10.1091/mbc.E07-09-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Li N, Jiang P, Du W, Wu Z, Li C, Qiao M, Yang X, Wu M. Siva1 suppresses epithelial-mesenchymal transition and metastasis of tumor cells by inhibiting stathmin and stabilizing microtubules. Proc Natl Acad Sci USA. 2011;108:12851–12856. doi: 10.1073/pnas.1017372108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Belletti B, Baldassarre G. Stathmin: a protein with many tasks. New biomarker and potential target in cancer. Expert Opin Ther Targets. 2011;15:1249–1266. doi: 10.1517/14728222.2011.620951. [DOI] [PubMed] [Google Scholar]
- 159.Wolf K, Mazo I, Leung H, Engelke K, von Andrian UH, Deryugina EI, Strongin AY, Brocker EB, Friedl P. Compensation mechanism in tumor cell migration: mesenchymal–amoeboid transition after blocking of pericellular proteolysis. J Cell Biol. 2003;160:267–277. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Friedl P. Prespecification and plasticity: shifting mechanisms of cell migration. Curr Opin Cell Biol. 2004;16:14–23. doi: 10.1016/j.ceb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 161.Belgiovine C, Frapolli R, Bonezzi K, Chiodi I, Favero F, Mello-Grand M, Dei Tos AP, Giulotto E, Taraboletti G, D’Incalci M, Mondello C. Reduced expression of the ROCK inhibitor Rnd3 is associated with increased invasiveness and metastatic potential in mesenchymal tumor cells. PLoS ONE. 2010;5:e14154. doi: 10.1371/journal.pone.0014154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Grise F, Sena S, Bidaud-Meynard A, Baud J, Hiriart JB, Makki K, Dugot-Senant N, Staedel C, Bioulac-Sage P, Zucman-Rossi J, Rosenbaum J, Moreau V. Rnd3/RhoE Is down-regulated in hepatocellular carcinoma and controls cellular invasion. Hepatology. 2012;55:1766–1775. doi: 10.1002/hep.25568. [DOI] [PubMed] [Google Scholar]
- 163.Luo H, Dong Z, Zou J, Zeng Q, Wu D, Liu L. Down-regulation of RhoE is associated with progression and poor prognosis in hepatocellular carcinoma. J Surg Oncol. 2012;105:699–704. doi: 10.1002/jso.23019. [DOI] [PubMed] [Google Scholar]
- 164.Zhou J, Li K, Gu Y, Feng B, Ren G, Zhang L, Wang Y, Nie Y, Fan D. Transcriptional up-regulation of RhoE by hypoxia-inducible factor (HIF)-1 promotes epithelial to mesenchymal transition of gastric cancer cells during hypoxia. Biochem Biophys Res Commun. 2011;415:348–354. doi: 10.1016/j.bbrc.2011.10.065. [DOI] [PubMed] [Google Scholar]
- 165.Petrie RJ, Gavara N, Chadwick RS, Yamada KM. Nonpolarized signaling reveals two distinct modes of 3D cell migration. J Cell Biol. 2012;197:439–455. doi: 10.1083/jcb.201201124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Friedl P, Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147:992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 167.Hegerfeldt Y, Tusch M, Brocker EB, Friedl P. Collective cell movement in primary melanoma explants: plasticity of cell–cell interaction, beta1-integrin function, and migration strategies. Cancer Res. 2002;62:2125–2130. [PubMed] [Google Scholar]
- 168.Wyckoff J, Wang W, Lin EY, Wang Y, Pixley F, Stanley ER, Graf T, Pollard JW, Segall J, Condeelis J. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 2004;64:7022–7029. doi: 10.1158/0008-5472.CAN-04-1449. [DOI] [PubMed] [Google Scholar]
- 169.Zhao J, Mialki RK, Wei J, Coon TA, Zou C, Chen BB, Mallampalli RK, Zhao Y. SCF E3 ligase F-box protein complex SCF(FBXL19) regulates cell migration by mediating Rac1 ubiquitination and degradation. Faseb J. 2013;27:2611–2619. doi: 10.1096/fj.12-223099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Flatau G, Lemichez E, Gauthier M, Chardin P, Paris S, Fiorentini C, Boquet P. Toxin-induced activation of the G protein p21 Rho by deamidation of glutamine. Nature. 1997;387:729–733. doi: 10.1038/42743. [DOI] [PubMed] [Google Scholar]
- 171.Schmidt G, Sehr P, Wilm M, Selzer J, Mann M, Aktories K. Gln 63 of Rho is deamidated by Escherichia coli cytotoxic necrotizing factor-1. Nature. 1997;387:725–729. doi: 10.1038/42735. [DOI] [PubMed] [Google Scholar]
- 172.Lerm M, Pop M, Fritz G, Aktories K, Schmidt G. Proteasomal degradation of cytotoxic necrotizing factor 1-activated rac. Infect Immun. 2002;70:4053–4058. doi: 10.1128/IAI.70.8.4053-4058.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ozdamar B, Bose R, Barrios-Rodiles M, Wang HR, Zhang Y, Wrana JL. Regulation of the polarity protein Par6 by TGFbeta receptors controls epithelial cell plasticity. Science. 2005;307:1603–1609. doi: 10.1126/science.1105718. [DOI] [PubMed] [Google Scholar]
- 174.Wang HR, Zhang Y, Ozdamar B, Ogunjimi AA, Alexandrova E, Thomsen GH, Wrana JL. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science. 2003;302:1775–1779. doi: 10.1126/science.1090772. [DOI] [PubMed] [Google Scholar]
- 175.Bryan B, Cai Y, Wrighton K, Wu G, Feng XH, Liu M. Ubiquitination of RhoA by Smurf1 promotes neurite outgrowth. FEBS Lett. 2005;579:1015–1019. doi: 10.1016/j.febslet.2004.12.074. [DOI] [PubMed] [Google Scholar]
- 176.Zimmerman ES, Schulman BA, Zheng N. Structural assembly of cullin-RING ubiquitin ligase complexes. Curr Opin Struct Biol. 2010;20:714–721. doi: 10.1016/j.sbi.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Chen Y, Yang Z, Meng M, Zhao Y, Dong N, Yan H, Liu L, Ding M, Peng HB, Shao F. Cullin mediates degradation of RhoA through evolutionarily conserved BTB adaptors to control actin cytoskeleton structure and cell movement. Mol Cell. 2009;35:841–855. doi: 10.1016/j.molcel.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 178.Wei J, Mialki RK, Dong S, Khoo A, Mallampalli RK, Zhao Y, Zhao J. A new mechanism of RhoA ubiquitination and degradation: roles of SCF E3 ligase and Erk2. Biochim Biophys Acta. 2013;1833:2757–2764. doi: 10.1016/j.bbamcr.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Torrino S, Visvikis O, Doye A, Boyer L, Stefani C, Munro P, Bertoglio J, Gacon G, Mettouchi A, Lemichez E. The E3 ubiquitin-ligase HACE1 catalyzes the ubiquitylation of active Rac1. Dev Cell. 2011;21:959–965. doi: 10.1016/j.devcel.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 180.Castillo-Lluva S, Tatham MH, Jones RC, Jaffray EG, Edmondson RD, Hay RT, Malliri A. SUMOylation of the GTPase Rac1 is required for optimal cell migration. Nat Cell Biol. 2010;12:1078–1085. doi: 10.1038/ncb2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yu J, Zhang D, Liu J, Li J, Yu Y, Wu XR, Huang C. RhoGDI SUMOylation at Lys-138 increases its binding activity to Rho GTPase and its inhibiting cancer cell motility. J Biol Chem. 2012;287:13752–13760. doi: 10.1074/jbc.M111.337469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Liu J, Zhang D, Luo W, Yu Y, Yu J, Li J, Zhang X, Zhang B, Chen J, Wu XR, Rosas-Acosta G, Huang C. X-linked inhibitor of apoptosis protein (XIAP) mediates cancer cell motility via Rho GDP dissociation inhibitor (RhoGDI)-dependent regulation of the cytoskeleton. J Biol Chem. 2011;286:15630–15640. doi: 10.1074/jbc.M110.176982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.DerMardirossian C, Bokoch GM. GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 2005;15:356–363. doi: 10.1016/j.tcb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 184.Fukumoto Y, Kaibuchi K, Hori Y, Fujioka H, Araki S, Ueda T, Kikuchi A, Takai Y. Molecular cloning and characterization of a novel type of regulatory protein (GDI) for the rho proteins, ras p21-like small GTP-binding proteins. Oncogene. 1990;5:1321–1328. [PubMed] [Google Scholar]
- 185.Leonard D, Hart MJ, Platko JV, Eva A, Henzel W, Evans T, Cerione RA. The identification and characterization of a GDP-dissociation inhibitor (GDI) for the CDC42Hs protein. J Biol Chem. 1992;267:22860–22868. [PubMed] [Google Scholar]
- 186.Boulter E, Garcia-Mata R, Guilluy C, Dubash A, Rossi G, Brennwald PJ, Burridge K. Regulation of Rho GTPase crosstalk, degradation and activity by RhoGDI1. Nat Cell Biol. 2010;12:477–483. doi: 10.1038/ncb2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Bielek H, Anselmo A, Dermardirossian C. Morphological and proliferative abnormalities in renal mesangial cells lacking RhoGDI. Cell Signal. 2009;21:1974–1983. doi: 10.1016/j.cellsig.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Liu M, Bi F, Zhou X, Zheng Y. Rho GTPase regulation by miRNAs and covalent modifications. Trends Cell Biol. 2012;22:365–373. doi: 10.1016/j.tcb.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ispanovic E, Serio D, Haas TL. Cdc42 and RhoA have opposing roles in regulating membrane type 1-matrix metalloproteinase localization and matrix metalloproteinase-2 activation. Am J Physiol Cell Physiol. 2008;295:C600–C610. doi: 10.1152/ajpcell.00460.2007. [DOI] [PubMed] [Google Scholar]
- 190.Saito K, Ozawa Y, Hibino K, Ohta Y. FilGAP, a Rho/Rho-associated protein kinase-regulated GTPase-activating protein for Rac, controls tumor cell migration. Mol Biol Cell. 2012;23:4739–4750. doi: 10.1091/mbc.E12-04-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Noguchi F, Inui S, Nakajima T, Itami S. Hic-5 affects proliferation, migration and invasion of B16 murine melanoma cells. Pigment Cell Melanoma Res. 2012;25:773–782. doi: 10.1111/pcmr.12005. [DOI] [PubMed] [Google Scholar]
- 192.Taddei ML, Parri M, Angelucci A, Bianchini F, Marconi C, Giannoni E, Raugei G, Bologna M, Calorini L, Chiarugi P. EphA2 induces metastatic growth regulating amoeboid motility and clonogenic potential in prostate carcinoma cells. Mol Cancer Res. 2011;9:149–160. doi: 10.1158/1541-7786.MCR-10-0298. [DOI] [PubMed] [Google Scholar]
- 193.Kitzing TM, Wang Y, Pertz O, Copeland JW, Grosse R. Formin-like 2 drives amoeboid invasive cell motility downstream of RhoC. Oncogene. 2010;29:2441–2448. doi: 10.1038/onc.2009.515. [DOI] [PubMed] [Google Scholar]
- 194.Omelchenko T, Hall A. Myosin-IXA regulates collective epithelial cell migration by targeting RhoGAP activity to cell–cell junctions. Curr Biol. 2012;22:278–288. doi: 10.1016/j.cub.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]