Abstract
The prion protein (PrPC) when mis-folded is causally linked with a group of fatal neurodegenerative diseases called transmissible spongiform encephalopathies or prion diseases. PrPC normal function is still incompletely defined with such investigations complicated by PrPC post-translational modifications, such as internal cleavage, which feasibly could change, activate, or deactivate the function of this protein. Oxidative stress induces β-cleavage and the N-terminal product of this cleavage event, N2, demonstrates a cellular protective response against oxidative stress. The mechanisms by which N2 mediates cellular antioxidant protection were investigated within an in vitro cell model. N2 protection was regulated by copper binding to the octarepeat domain, directing the route of internalisation, which stimulated MEK1 signalling. Precise membrane interactions of N2, determined by copper saturation, and involving both the copper-co-ordinating octarepeat region and the structure conferred upon the N-terminal polybasic region by the proline motif, were essential for the correct engagement of this pathway. The phenomenon of PrPC post-translational modification, such as cleavage and copper co-ordination, as a molecular “switch” for activation or deactivation of certain functions provides new insight into the apparent multi-functionality of PrPC.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-014-1777-y) contains supplementary material, which is available to authorized users.
Keywords: CJD, N-terminus, PrP23-89, Beta-cleavage, Reactive oxygen species, Signal transduction, Mitogen-activated protein kinase
Introduction
Prion diseases, fatal and transmissible neurodegenerative diseases of humans and animals, are causally linked with the prion protein (PrP). The disease-associated isoforms of the prion protein (PrPSc) are conformationally altered forms of a ‘normal’ cellular protein (PrPC). The predominant central nervous system function of PrPC is as yet undefined; however, various studies have reported a neuroprotective role [1–7]. PrPC undergoes at least two internal cleavage events, termed α- and β-cleavage, producing N1/C1 and N2/C2 fragment pairs, respectively [8, 9]. Diversity in functionality of the different PrP fragments, may contribute to the difficulties in assigning one single function to PrPC with each fragment potentially exerting unique effects on the cell. Hence, the possibility that PrPC cleavage acts as a post-translational molecular “switch” has been suggested [10, 11].
The products of PrPC β-cleavage are increased in human prion-diseased brains and mouse models of prion disease, concurrent with the appearance of PrPSc, and therefore, PrPC β-cleavage has been mainly considered a likely pathogenic event [9, 12, 13]. However, inability to undergo β-cleavage has been shown to render cells in culture more vulnerable during times of oxidative stress, when an imbalance between reactive oxygen species (ROS) production and detoxification causes cell damage leading to death [14]. During the course of prion infection, oxidative damage occurs in the brain paralleling increased deposition of PrPRes (a protease-resistant species of PrP considered a biochemical marker for PrPSc; [12]) and cellular compensation responses are essential for maintaining homoeostasis and viability in chronically infected cultured cells [15, 16]. Significantly, the β-cleavage event, itself, is caused by ROS [17] and, therefore, in contrast to being a toxic event of prion disease, this endoproteolysis may form part of a cellular protective response against heightened oxidative stress.
Recently the N-terminus of PrP has received considerable attention with diverse findings demonstrating that it is integral to PrPSc-mediated toxicity [18, 19], influences conversion of PrPC to PrPSc [20–22], exerts a neuroprotective function [23, 24] and binds to soluble oligomeric amyloid-β (Aβ) peptides [25, 26] mediating the toxic signalling of oligomeric Aβ in Alzheimer’s disease, unless PrPC undergoes α-cleavage, whereby the N1 fragment is reported to be protective against Aβ toxicity [27, 28].
Our previous work has shown that the N-terminal β-cleavage fragment (N2) is able to reduce intra-cellular ROS induced by serum deprivation, a reaction requiring copper saturation of the eight amino acid repeat (octarepeat) domain [24]. When the regions of N2 that are important for the transduction of the protective signal were investigated, it was found that amino acid residues 23–50 alone were sufficient to initiate the reaction, indicating that the octameric repeat region (residues 51–89) played a restraining role with inhibition of signalling when present in its apo form. Transduction of the protective effect additionally required cell surface proteins, the glycosaminoglycan (GAG) heparan sulphate (HS) and intact lipid-raft domains, and was dependent upon a proline motif within the far N-terminal polybasic region. To investigate the mechanism of neuroprotective signal transduction by N2 from the exterior of the cell, this study followed N2 trafficking and assessed candidate signal transduction pathways. Our results show that MEK1 signalling is the primary mediating pathway, which is dependent upon intra-cellular trafficking, a process highly influenced by N2 copper saturation.
Materials and methods
Generation of PrP peptide sequences
The synthetic peptides derived from the mouse PrP23-89 sequence have been described previously [24] and were made as described in Karas et al. [29].
Cell culture
CF10 (PrP null, mouse neuronal; [30]), Neuro2a (N2a; mouse neuroblastoma) and OBL-21 (mouse olfactory bulb) cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Lonza, AUS) supplemented with 10 % (v/v) foetal bovine serum (Lonza) and 50 U/ml penicillin/50 μg/ml streptomycin solution (Sigma, Melbourne, VIC, AUS). Cell cultures were maintained at 37 °C with 5 % CO2 in a humidified incubator.
DCFDA assay
The DCFDA assay using 5 µM 5-(and-6)-chloromethyl-2′,7′-dichlorodihydro-fluorescein diacetate, acetyl ester (CM-H2-DCFDA; Invitrogen, VIC, AUS) was performed as described previously [24, 31].
MTS viability/metabolism assay
Five microlitres of one solution MTS reagent (Promega; VIC AUS) per 100 μl media was added to test and medium-only background control conditions, and incubated under normal culture conditions for 90 min. Reaction product was quantified using absorbance at 462 nm in a Fluostar Optima (BMG Labtech, VIC, AUS).
CyQUANT DNA assay
The CyQUANT assay (Invitrogen) was carried out as per the manufacturer’s product protocol for CyQUANT® NF.
Cytosolic redox potential
Cells were incubated in Opti-MEM®I reduced serum culture medium (phenol red free; Invitrogen) containing 5 µM RedoxSensor™ Red CC-1 probe (Invitrogen) and 1 µM MitoTracker® Green (Invitrogen) for 10 min. Media was removed and cells were incubated in fresh media with or without serum and test peptides.
Fluorescence and confocal microscopy
Fluorescence microscope images were visualised with a 60× oil immersion lens and captured using a Nikon Eclipse TE2000-E epi-fluorescence microscope (Nikon-Roper Scientific).
Confocal images were collected on a Leica SP8 using a 60× oil immersion lens. For quantification, all image parameters were maintained throughout an experiment, across all conditions, and the channels of probes sensitive to photo-oxidation were collected before other channels. For imaging data, the ‘n’ represents independent experiments, which each included capturing sufficient fields to analyse >50 cells per experiment. Cell intensity and co-localisation analyses were performed using Nikon NIS-elements 3.0 software with regions of interest used to gate individual cells.
Indirect immunofluorescence
Cells were grown on 13 mm glass coverslips in standard media with immunofluorescence (IF) performed when cells were 80 % confluent. Plates were incubated in the dark at all light-sensitive stages during this protocol. Cells were fixed in 4 % (v/v) paraformaldehyde/PBS for 20 min and then permeabilised in immunodiluent (ID; 1 % w/v BSA in PBS) with 0.1 % (v/v) Triton-X 100 for 5 min. Coverslips were blocked with ID containing 10 % FBS (v/v) for 30 min. Primary antibodies against PrP Saf32 (1 in 200; Cayman Chemical, Sapphire Biosciences, AUS) and 8B4 (1 in 200; Alicon Switzerland), LAMP-1 (1 in 100; Abcam, AUS), SOD2 (1 in 100; Abcam), and pMEK1 (1 in 100; Abcam) were diluted in ID and applied overnight at 4 °C. Alexafluor488/647-conjugated goat anti-rabbit/mouse secondary antibodies (Invitrogen) diluted 1 in 250 in ID were applied for 1–2 h at room temperature. Coverslips were mounted onto glass slides with mowiol or Pro-Long anti-fade (Invitrogen) mounting media.
Laurdan live cell imaging assay
Cells were incubated with a 5 μM solution of laurdan probe (Invitrogen) in normal media for 30 min prior to beginning the assay. At time 0, normal media was replaced with serum-free, phenol red-free OptiMEM. Peptide was added to a final concentration of 10 μM at the start of imaging and images were collected every minute for 10 min using a near UV excitation and blue emission filter set.
MitoSOX assay
Cells were labelled with 5 μM MitoSOX fluorescent indicator probe (Invitrogen) in standard media for 10 min under routine incubator conditions (as described above). Media was then replaced with fresh, phenol red-free OptiMEM for the duration of the experiment.
Pull-down assays
Dynabeads amine (Invitrogen) were resuspended by brief vortexing. Two hundred and fifty microlitres of bead suspension was transferred into microfuge tubes. Beads were washed twice, by magnetic separation, in conjugation buffer (0.1 M MES, 0.5 M NaCl, pH 6) and resuspended in 200 μl of conjugation buffer. One hundred and fifty micrograms of each peptide was resuspended in 800 μl conjugation buffer and added to the beads, with brief vortex mixing. Ten milligrams EDC and 15 mg NHS (Sigma-Aldrich) were dissolved in 1 ml cold deionised H2O immediately prior to use and 15 μl EDC/NHS solution per reaction added with brief vortexing. Samples were incubated for 2 h at room temperature with slow tilt rotation. Hydroxylamine, to a final concentration of 10 mM, was added to quench the reaction and incubated for 15 min at room temperature with slow tilt rotation. Beads were washed three times with PBS-BSA followed by a final PBS only wash. Coated beads were resuspended in 250 μl PBS. Cells were lysed either mechanically in PBS (by freeze-fracture followed by needle aspiration) or using detergent (0.1 % Triton-X-100 in PBS). One milligram total cell lysate (protein quantification was achieved using BCA assay—Pierce) was diluted into 950 μl PBS and added to the coated beads. Lysates and beads were incubated at room temperature with slow tilting rotation for 1 h. Beads and supernatants were separated magnetically and supernatants were kept. Beads were washed three times with PBS and once in a detergent wash (0.1 % Triton-X-100 in PBS) before elution. Elutions were carried out in the order of deceasing pH followed by increasing salt and finally the beads were boiled to remove anything still bound. For each elution, 100 μl of elution buffer was added to beads, which were briefly vortexed then incubated at room temperature for 2 min of gentle mixing before collecting. The elution buffers used were 0.01 M MES, pH 6 (1.95 g/L MES, 8.75 g/L NaCl); acetate buffer pH 5 (0.6 g/L acetic acid, 8.75 g/L NaCl); acetate buffer pH 4 (0.6 g/L acetic acid, 8.75 g/L NaCl); 0.1 M citrate buffer, pH 3 (21 g/L citric acid); 300 mM NaCl; 1 M NaCl.
Dot blotting
Two microlitres per spot was dotted onto dry nitrocellulose and spots dried for 1 h at 37 °C. Membranes were then blotted as per the western blot procedures using 1 in 100,000 CTXb-HRP (Sigma-Aldrich) and 1 in 1,000 10E4 primary antibody (Jomar Bioscience, SA) with 1 in 2,000 anti-mouse-HRP secondary antibody.
Western blotting
Cells were lysed in RIPA buffer [50 mM Tris–HCl pH 7.4, 150 mM NaCl, 0.1 % (w/v) SDS, 0.5 % (w/v) sodium deoxycholate, 1 % (v/v) NP-40] with 1 in 2000 benzonase (Sigma) at 37 °C for 20 min. Samples were electrophoresed using the NuPAGE system (Invitrogen), transferred onto PVDF membranes (millipore) and blocked as described previously [32]. Protein of interest detection was made using the following dilutions of primary antibodies; 1 in 1,000 37/67 kDa laminin receptor (Abcam), pMEK1 (Abcam), pMEK2 (Abcam), pAKT (Abcam), pERK1/2 (Cell Signalling Technologies, Genesearch, AUS), 1 in 400 p-p38 (R&D Systems, Sapphire Biosciences); and 1 in 5,000 Actin (Sigma-Aldrich) in phosphate buffered saline with 0.1 % (v/v) Tween-20 (PBS-t). Secondary antibodies (Dako, Melbourne, VIC, AUS) were used at 1 in 2,000 for the signalling proteins and 1 in 5,000 for actin. ECL-advance (GE-Healthcare) was used to visualise bands with images collected using a Las-3000 intelligent dark box (FujiFilm, Berthold, Victoria AUS). Following blotting, total protein levels on all gels were assessed to ensure even loading by Coomassie brilliant blue staining [0.1 % (w/v) Coomassie Brilliant Blue in 50 % (v/v) methanol and 7 % (v/v) acetic acid] for 2 min at room temperature with agitation. Membranes were rinsed briefly in destain [50 % (v/v) methanol and 7 % (v/v) acetic acid], then washed in de-stain for 10 min before drying for image capture.
Statistical analyses
Statistical analyses were carried out using GraphPad Prism 5 statistical software. Data are represented as mean ± SEM. The numerical value for each independent experiment (‘n’) is the average of the technical replicates. Primary statistical tests are stated in the text and tested with 95 % confidence intervals. For ANOVA analysis Tukey’s (comparisons of all conditions) or Dunnett’s (comparisons with control values) secondary tests were applied.
Results
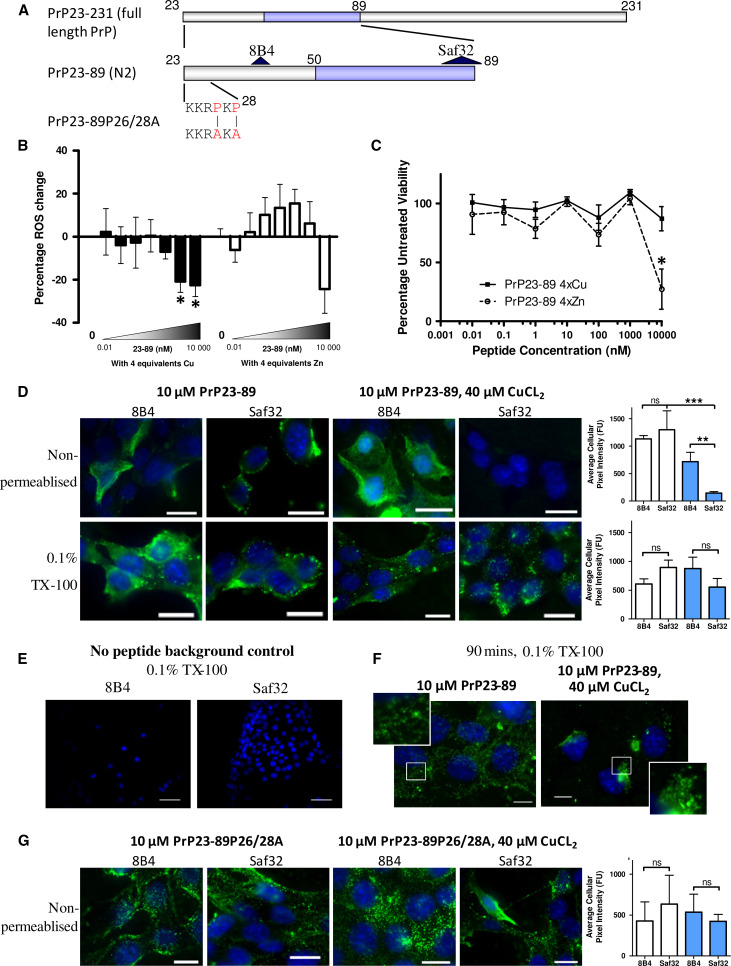
We have previously shown that a synthetic peptide equivalent to the PrPC N2 fragment (PrP23-89 and fragments thereof, Fig. 1a) can reduce intra-cellular ROS burden in response to the stress of serum deprivation [24]. Copper coordination was important for the action of full-length PrP23-89. Serum deprivation was used as a mild insult that induces intracellular ROS and changes cellular metabolism but does not cause acute cell death (Sup Fig. 1 [24]). To extend our previous work, we performed a dose–response following the activity of copper and zinc-saturated (four molar equivalents) PrP23-89 on the intracellular ROS production in CF10 cells (prnp gene ablated [30]; these cells are used throughout unless otherwise stated). The response showed significant ROS reduction by the copper-saturated PrP23-89 fragment at 1 and 10 μM (Fig. 1b; One-way ANOVA, F = 5.327, p = 0.019, n = 3; copper-only responses are shown in Sup Fig. 2), but no significant decrease was observed when zinc loaded (One-way ANOVA, F = 2.26, p = 0.07, n = 4). A dose–response curve of the apo PrP23-89 ROS response has previously been published [24]. Further analysis of the metabolism of these cells showed that the ROS reduction seen when copper-bound is not due to a loss of cellular viability; however, viability is reduced when zinc-bound (Fig. 1c; 2-way ANOVA, F = 6.833, p < 0.001, n = 3). No zinc toxicity was observed in the absence of the N2 peptide (Sup Fig. 2).
Fig. 1.
Copper saturation of N2 influences its anti-oxidant function, cell surface interactions and trafficking. a Schematic of the PrP N2 cleavage fragment (residues 23–89) with antibody binding sites and the P26/28A mutation shown. The octarepeat (copper-binding) region is indicated in blue. b Log10 dose–response of the intracellular ROS reduction activity displayed by copper-saturated (filled bars) and zinc-saturated (hollow bars) PrP23-89 following serum deprivation. c Measurement of MTS metabolism 24 h after serum deprivation and peptide addition. d Antibody detection of the PrP23-89 fragment at the cell surface (non-permeabilised cells) and inside the cell 15 min after peptide addition with or without copper saturation. Antibody signal is shown in green and blue staining indicates DAPI within the nucleus. Graphs show quantification of the detected PrP23-89 signal with white bars indicating apo-PrP23-89 and blue bars indicating copper-saturated PrP23-89 staining. e Staining of CF10 cells in the absence of peptide addition to control for non-specific antibody binding. f Immunofluorescent detection of PrP23-89 inside of cells 90 min post-peptide addition with or without copper. g Immunofluorescent detection of PrP23-89P26/28A at the cell surface, with and without copper saturation, 15 min after peptide addition. Scale bars = 25 µm. *p < 0.05, **p < 0.01, ***p < 0.001
The peptides were applied exogenously, therefore, the cell surface interactions of the apo- and copper-saturated PrP23-89 were investigated by IF localisation. Peptides were added to the serum-free media and cells incubated for 15 min before fixing. Two antibodies, 8B4 and Saf32 (recognising different epitopes, Fig. 1a) were used to probe peptide binding to the cell surface in unpermeabilised cells and throughout the cell, using Triton-X-100 permeabilisation after fixing. The cell surface (unpermeabilised) staining with the 8B4 antibody showed no difference in the pattern of staining or total intensity between the copper-loaded and apo-PrP23-89 applications (Fig. 1d). However, the PrP23-89 showed altered cell surface interactions when detection was performed with the Saf32 antibody (directed against the octameric repeat region). Here, when the copper-loaded PrP23-89 peptide was examined, slides were almost completely devoid of antibody signal (Fig. 1d; One-way ANOVA, F = 10.27, p = 0.0003). In permeabilised cells, no significant differences in the intensity of cell staining were seen across any condition (Fig. 1d; One-way ANOVA, F = 1.391, p = 0.276, n = 3). Control staining of permeabilised CF10 cells showed signal is specific for PrP23-89 (Fig. 1e). When cells were exposed to PrP23-89 with and without copper-loading for a longer time period, 90 min, the intra-cellular staining pattern was visibly different between the two conditions; the apo-PrP23-89 appeared more diffuse with greater cell surface staining and the copper-loaded PrP23-89 showed a punctuate, vesicular, and sometimes perinuclear staining pattern (Fig. 1f).
To explore whether the lack of Saf32 immuno-reactivity with the copper-loaded PrP23-89 was due to copper co-ordination changing epitope availability, PrP23-89P26/28A, a peptide with the two proline residues at positions 26 and 28 mutated to alanine (Fig. 1a), was incubated with the CF10 cells under the same apo- and copper-saturated conditions. This mutant peptide, previously shown to co-ordinate copper in an identical way to PrP23-89 [24], was clearly detectable at the cell surface with Saf32 in both the apo- and copper-saturated states with signal no longer significantly different from 8B4 detection or the apo-peptide (Fig. 1g; One-way ANOVA, F = 0.1595, p = 0.9209, n = 3).
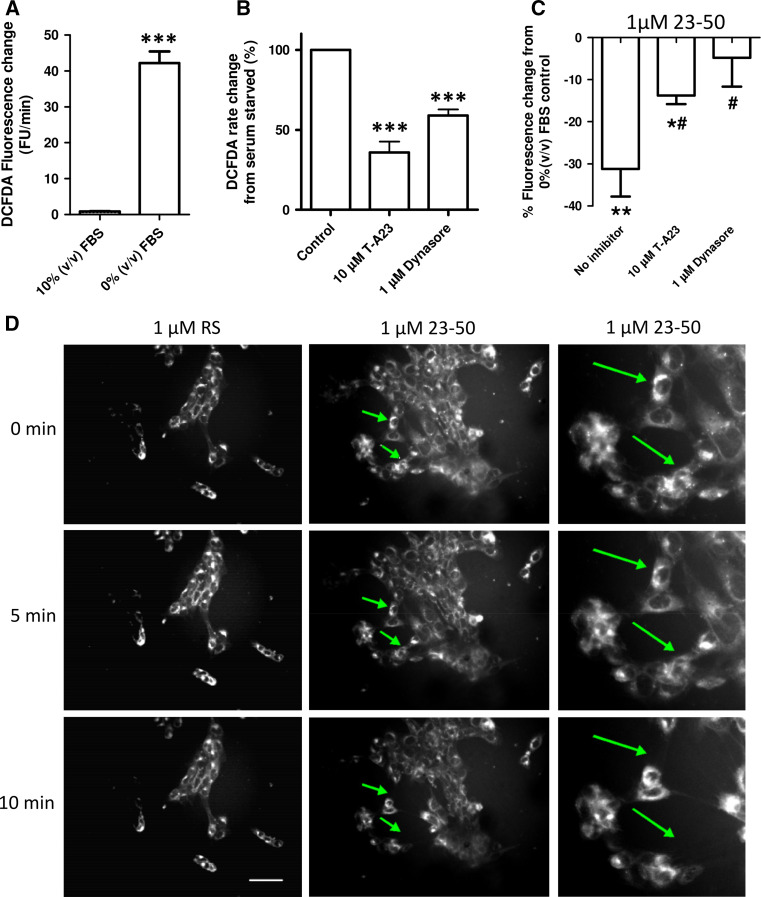
As the intracellular trafficking and sub-cellular localisation of the PrP23-89 peptide was influenced by its copper saturation, we investigated whether altering trafficking pathways would alter antioxidant activity. First, we assessed the influence of internalisation inhibition on intra-cellular ROS production using the DCFDA assay. The results showed that serum deprivation causes a large increase in intracellular ROS production (student’s t test; t = 12.7, p < 0.001, n = 3; Fig. 2a; Sup Fig. 1F) and this was significantly reduced when internalisation was impeded using inhibitors of clathrin-mediated (T-A23) and dynamin-mediated (Dynasore) pathways (Fig. 2b; One-way ANOVA; F = 52.7, p < 0.001, n = 4). For all internalisation (and signalling) pathway inhibition studies, a dose–response curve of the inhibiting compound for both ROS production and cellular viability was tested (Sup Fig. 3) from which a concentration of compound that might influence the pathway, without causing gross cell death, was determined.
Fig. 2.
Inhibition of internalisation reduces stress-induced intracellular ROS and inhibits the protective action of PrP23-50. a DCFDA assay for the detection of intracellular ROS produced in response to serum deprivation. b Cells were stressed by serum deprivation with or without the internalisation inhibitors T-A23 and Dynasore. Changes are shown normalised to 0 % (v/v) serum control values as 100 % ROS production. c Measurement of the percentage ROS reduction exerted by PrP23-50 with and without internalisation inhibition. Results are shown as percentage reduction from the serum-deprived ∓ inhibitor control. Asterisks denote significantly different from no peptide control (*p < 0.05, **p < 0.01, ***p < 0.001) and hash shows significantly different from the no inhibitor action of PrP23-50 (# < 0.05). d Incorporation of the lipophilic dye laurdan into cell membranes before treatment with PrP23-50 (or a RS peptide control) in serum-free media
To assess the involvement of internalisation pathways in PrP N-terminally mediated intra-cellular ROS reduction response, a C-terminally truncated fragment of PrP23-89, PrP23-50, was used. The PrP23-50 fragment was previously shown to reduce intracellular ROS in the absence of the octarepeat region [24], thus not requiring copper and the use of this peptide, therefore, eliminates any confounding effects of adding redox-active copper to these reactions. Results are shown relative to the internalisation inhibitor action on serum-deprived cells; a decrease is indicative of the inhibitor and PrP23-50 effects being additive (suggesting different pathway engagement) and where no change or an attenuated response is seen the effects of the peptide may be competing with the internalisation inhibitor for engagement of the same pathway or mechanism, with overall negation of any additional effect of the PrP23-50. Inhibition of internalisation significantly reduced the ROS mitigating action of PrP23-50 with both T-A23 and dynasore-treated cells showing significantly different responses from the PrP23-50 action without inhibitors, and for the dynasore-treated cells, ROS reduction was no longer significantly different to that of the serum-deprived cells alone (Fig. 2c; One-way ANOVA; F = 4.908, p = 0.0407, n = 3). We have previously detected PrP23-50 in cell lysates harvested after exogenous application [24], indicating that the peptide is taken up by cells. As such we considered if cell membrane changes were occurring in real time. Laurdan fluorescent membrane probe was loaded into cells before addition of PrP23-50, or a random sequence (RS) peptide to control for random peptide uptake effects on membranes, and a time course of images to 10 min were collected. Cells exposed to PrP23-50 showed increased movement and retraction of their membranes into a smaller area, which was not seen for the RS peptide.
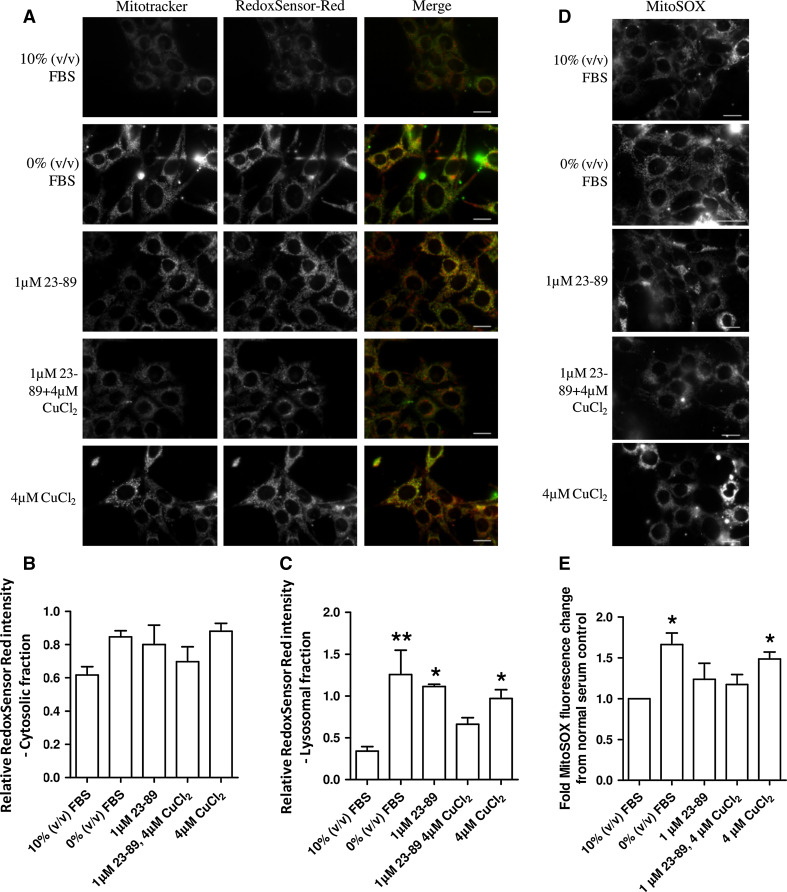
Elucidating the localisation of ROS production was considered important for identifying the site of PrP23-89 activity. To determine the site(s) of ROS production within the cell following serum deprivation and whether this site/these sites were the target of N2 action, intracellular ROS-localisation probes RedoxSensor Red (cytosolic and lysosomal ROS [33]) and mitoSOX (mitochondrial superoxide) were incubated within live cells throughout serum deprivation and peptide treatments. Increased ROS production upon serum deprivation was detected by RedoxSensor Red. RedoxSensor Red partitions into mitochondria if it is oxidised in the cytosol otherwise it is transported to lysosomes where it is oxidised. We analysed the contributions of cytosolic and lysosomal ROS to oxidation of this probe using co-localisation with MitoTracker [33] (Fig. 3a), which indicated the significant increase was not contributed by cytosolic ROS (Fig. 3b; One-way ANOVA; F = 2.395, p = 0.0937, n = 4) but instead by lysosomes (Fig. 3c; One-way ANOVA; F = 5.31, p = 0.0072, n = 4). Copper-loaded N2 significantly reduced the ROS produced in the lysosomes. Mitochondrial superoxide production was also significantly increased in response to serum deprivation and PrP23-89 was able to reduce superoxide production back to insignificance against that produced by cells incubated in serum-containing media both when applied apo and copper-loaded (Fig. 3d, e). The same ROS reduction response was reproduced by the PrP23-50 peptide for both the RedoxSensor Red and MitoSOX probes (Sup Fig. 6 and 7). Despite being sites of ROS production neither lysosomal nor mitochondrial function, as determined by acidity and ATP levels, respectively, were altered (Sup Fig. 4). However, incubation with the PrP23-89 peptide (either with or without copper saturation) did increase cell viability at 24 h post-exposure (Sup Fig. 5).
Fig. 3.
Copper-saturated PrP23-89 reduces the lysosomal and mitochondrial ROS caused by serum deprivation. a Representative plates of RedoxSensor Red changes due to serum deprivation and as influenced by PrP23-89 with and without copper saturation. RedoxSensor Red fluorescence was co-localised with MitoTracker green to indicate compartmental partitioning. Scale bars = 25 µM. Quantification of the cytosolic [co-localised with MitoTracker; (b)] and lysosomal (c) ROS fractions shown relative to 10 % (v/v) serum control. d Representative plates of mitoSOX fluorescence production in response to serum deprivation. Scale bars = 25 µM. e Quantification of mitoSOX fluorescence intensity. *p < 0.05, **p < 0.01
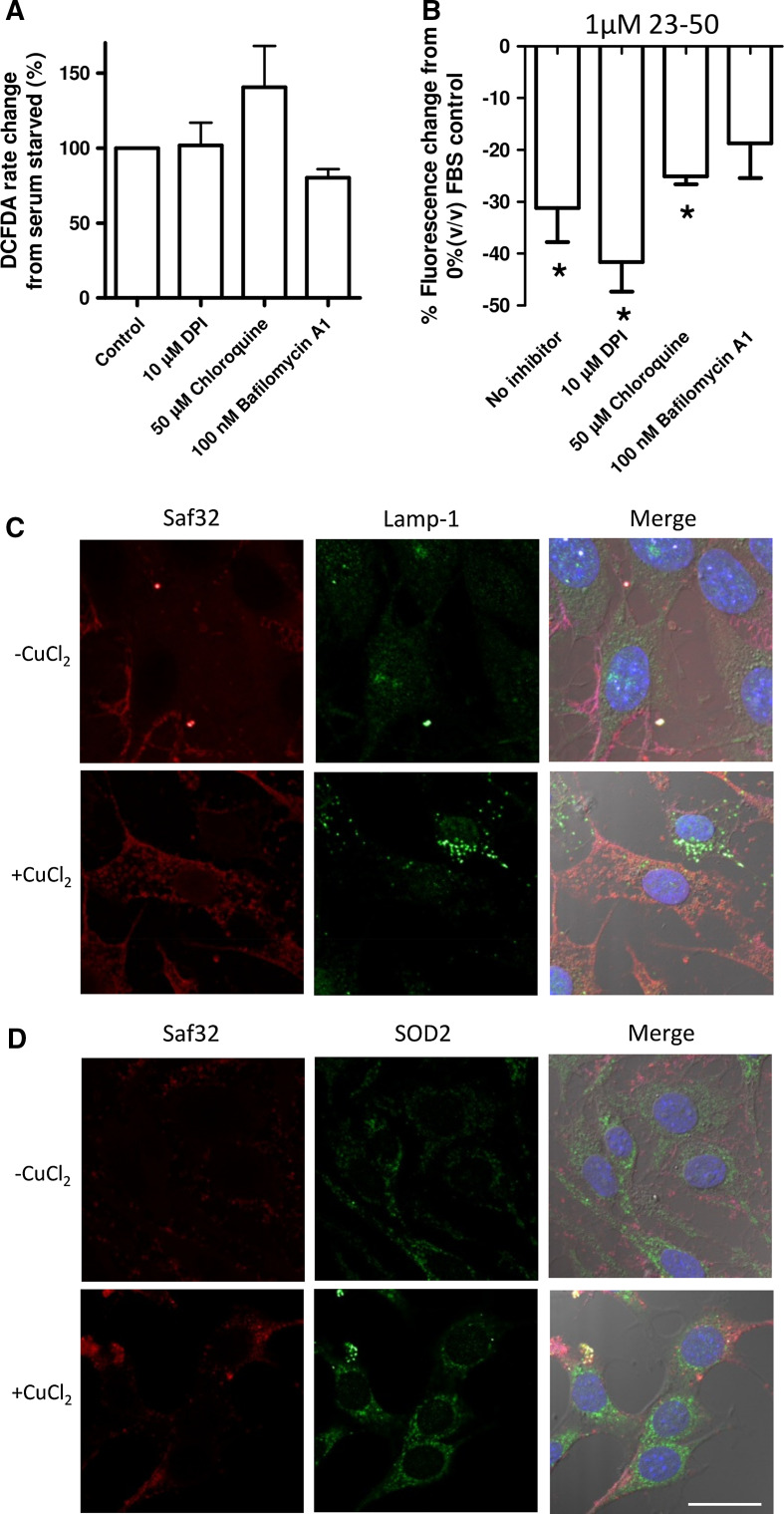
To test if the effects on cellular ROS producing systems were directly due to the actions of the N-terminal peptides (the peptides themselves have no intrinsic anti-oxidant activity [24] ), cytosolic ROS and lysosomal ROS production were investigated using inhibition of NADPH oxidase (NOX; a cytosolic signalling protein that generates superoxide to signal) and inhibitors of lysosomal acidity/acidification. The inhibitors were incubated with the serum-deprived cells and ROS production measured by DCFDA assay. No significant changes in ROS response to serum deprivation were seen with any of these inhibitors (Fig. 4a; One-way ANOVA; F = 2.528, p = 0.0941, n = 5). The model PrP23-50 peptide was used to look at capacity to reduce the serum deprivation-induced ROS in the presence of the NOX and lysosomal acidity/acidification inhibitors. PrP23-50 treatment of the serum-deprived cells treated with the NOX and lysosomal inhibitors showed no significant difference from the serum-deprived no inhibitor control (One-way ANOVA; F = 2.202, p = 0.1453, n = 3); however, the bafilomycin-treated cell response was variable and no longer significantly different from the no peptide control serum-deprived ROS production (Fig. 4b; student’s t test; t = 2.794, p = 0.0682, n = 4). We additionally investigated the co-localisation of PrP23-89 when applied both copper-saturated and in the apo form with the lysosomal marker Lamp 1 and the mitochondrially localised superoxide dismutase (SOD)-2. The peptides were incubated with the cells for 90 min before fixing to allow significant internalisation to occur. Whilst a small amount of co-staining was seen for copper-bound PrP23-89 with SOD2, the staining did not suggest a pronounced co-localisation of PrP23-89 with either of these organelle markers (Fig. 4c, d).
Fig. 4.
Downstream targeting of lysosomes does not play a prominent role in 23–50 ROS reduction and PrP23-89 is not trafficked to this destination. a Effect of the inhibition of NADPH oxidase ROS signalling and lysosomal acidity on serum-deprived ROS production as assayed by DCFDA fluorescence. Data are plotted as a percentage of the serum-deprived control. b 23–50 ROS reduction response after inhibition of NADPH oxidases or lysosomal acidification. Results are shown as percentage reduction from the serum-deprived ∓ inhibitor control. *p < 0.05. Co-localisation of 1 µM PrP23-89 (∓ copper saturation) with lamp-1 lysosomal marker (c) and SOD2 mitochondrial marker (d). Blue = DAPI nuclei staining. Scale bar = 20 µm
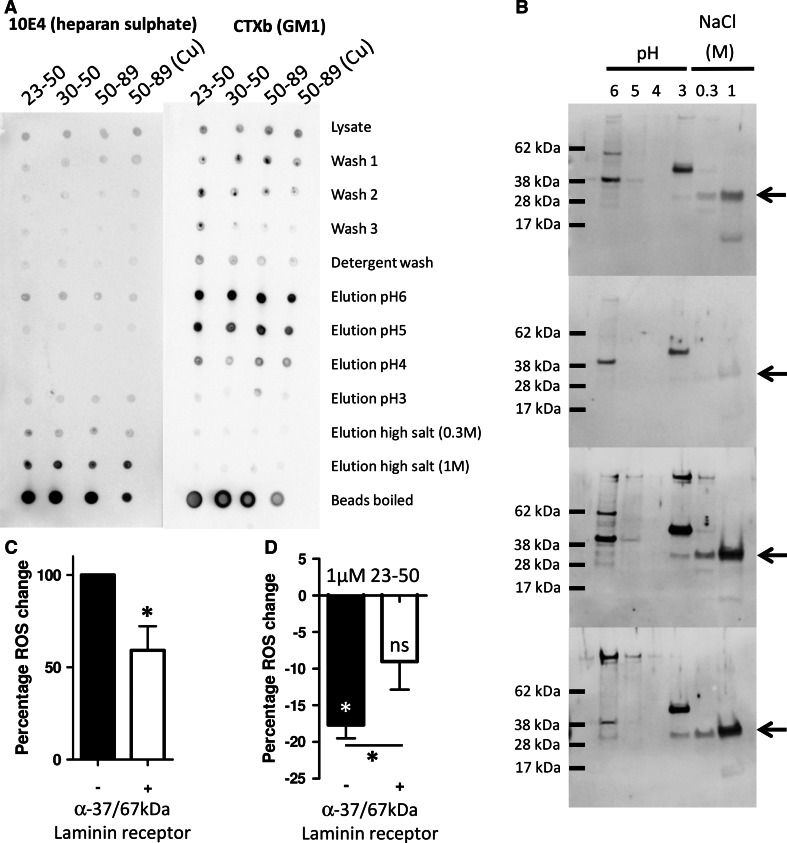
We next considered if we could detect a putative receptor for N-terminal PrP that might be involved in mediating the N2 protective signalling. To look for N-terminal binding partners, we conjugated PrP23-50, PrP30-50 (lacking the N-terminal polybasic domain), and PrP51-89 to magnetic beads. The beads were incubated with whole cell lysate prepared by mechanical disruption or detergent lysis. Lysate was separated from the beads and beads were washed four times. Elution of bound proteins was done using decreasing pH (6, 5, 4, 3), followed by two high salt washes (0.3 and 1 M NaCl—to disrupt GAG-binding interactions). Both HS and lipid rafts were previously identified as essential for PrP23-50 ROS reduction; therefore, we determined if either had been pulled down by spotting 2 μl of each wash and elution onto nitrocellulose and blotting for HS content (using 10E4 antibody) and for the lipid-raft marker GM1. For the mechanically disrupted cell lysates, both HS and GM1 were detected in the elutions but eluted under different conditions with HS eluting at high salt concentrations and GM1 eluting as pH dropped (Fig. 5a). No HS or GM1 pull-down was observed when lipid domains were disrupted by detergent lysis (Sup Fig. 8).
Fig. 5.
The PrP N-terminus pulls down a complex containing heparan sulphate, lipid rafts and the 37/67 kDa laminin receptor. a Dot blots for HS and GM1 of pull downs from cell lysate using PrP N-terminal fragments. b The fractions were western blotted for the 37/67kDA laminin receptor (expected size of the 37/67 kDa laminin receptor band is indicated by arrows). c DCFDA assay of the effect of blocking the 37/67 kDa laminin receptor with antibodies on the induction of ROS during serum deprivation. d Assessment of the activity of PrP23-50 in reducing intracellular ROS produced in response to serum deprivation following targeting of the 37/67 kDa laminin receptor with antibodies. *p < 0.05
The 37/67 kDa laminin receptor has previously been shown to bind to the N-terminus of PrP in an interaction that also requires GAGs. Therefore, samples from the elutions were electrophoresed and western blotted for the 37/67 kDa laminin receptor. This was found most strongly in the elutions from beads conjugated to PrP51-89 but was also found for PrP23-50 and very weakly for PrP30-50, which lacks the charged region thought to bind GAGs (Fig. 5b). Other bands of differing molecular weight are seen in the blots and whilst these are most likely non-specific it cannot be ruled out that they are modified species of the laminin receptor. To test if the 37/67 kDa laminin receptor might be involved in the ROS reduction action of the PrP N-terminus we used anti-37/67 kDa antibodies to sterically block interaction with the cell surface laminin receptor. Blocking the receptor reduced the ROS produced in response to serum deprivation (Fig. 5c; student’s t test, t = 4.006, p = 0.016, n = 5) as well as reducing the efficiency of the PrP23-50 ROS reduction response (Fig. 5d; student’s t test, t = 2.953, p = 0.042, n = 5).
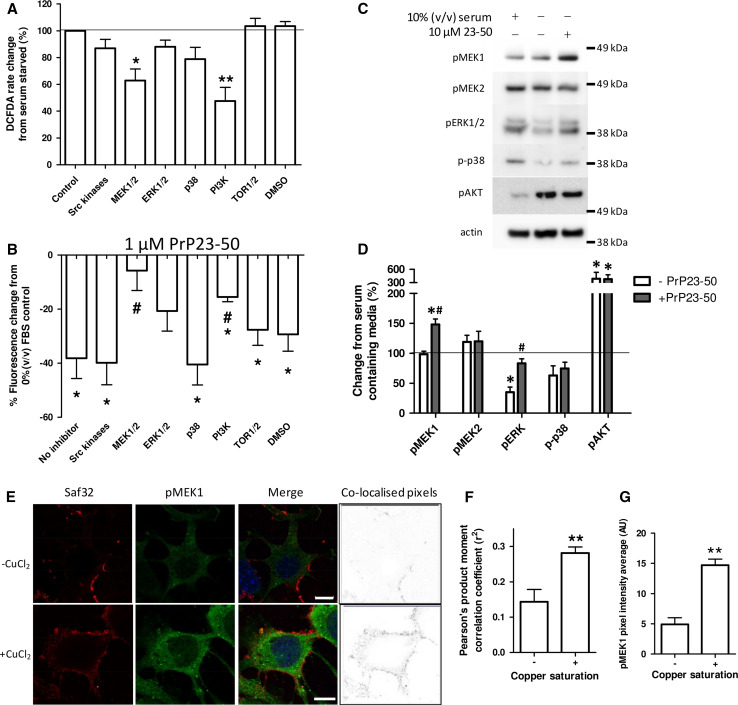
To investigate candidate signal transduction intermediates that might be recruited into the pathway by which copper-saturated PrP23-89 exerts its influence, we first considered the effect of signalling intermediate inhibition on the production of ROS in response to serum deprivation. Inhibitors were directed against key signalling intermediates from well-defined pathways within the cell. Only inhibition of MEK1/2 and PI3K were observed to change the ROS produced in response to serum deprivation, with both reducing the ROS produced (Fig. 6a; One-way ANOVA; F = 5.677, p < 0.001, n = 3). Since these experiments were undertaken in PrP knock-out cells for the purpose of excluding confounding effects from endogenous PrP, the cellular responses may be influenced by their lack of PrP expression. Therefore, select inhibitor assays were also performed with two cell lines, N2a and OBL-21, which express detectable levels of endogenous PrP [32]. Significant differences were seen between the CF10 and N2a/OBL-21 cells in the intra-cellular ROS production upon serum starvation only when NADPH oxidases were inhibited with DPI (Sup Fig. 9). The identified candidate pathways for copper-loaded PrP23-89 function responded consistently across the cell lines regardless of PrP expression. When the ROS-reducing action of PrP23-50 was assessed in the presence of these inhibitors, MEK1/2 and PI3K inhibitors suppressed the action of PrP23-50 significantly compared with the control, no inhibitor condition, and MEK1/2 and ERK1/2 inhibition reduced the ROS reduction such that it was no longer significantly different from the control serum-deprived condition (Fig. 6b; Two-way ANOVA, F = 18.69, p < 0.001, n = 3).
Fig. 6.
PrP23-50 influences MEK-1, ERK1/2, and PI3K signalling pathways and copper-saturated N2 shows increased co-localisation with pMEK1. a Inhibitors of cellular signalling pathways were assayed for their ability to interfere with the ROS produced in response to serum deprivation. The inhibitors used were as follows; Src kinases—500 nM PP2; MEK—10 µM U0126; ERK—10 µM 3-(2-Aminoethyl)-5-((4-ethoxyphenyl)methylene)-2,4-thiazolidinedione hydrochloride; p38—10 µM SB203580; PI3K—1 µM wortmannin; TOR1/2—1 µM rapamycin; DMSO control—0.1 % (v/v); the target protein is indicated in the graph; DCFDA assay results are shown relative to the serum-deprived 100 % control value. b PrP23-50 ROS reduction was assessed by DCFDA fluorescence and shown as percentage reduction from the serum-deprived ± inhibitor control. *p < 0.05 from the no peptide control, # p < 0.05 from the no inhibitor action of PrP23-50. c Western blots and d corresponding densitometry of changes in phosphorylation (activation) of central signal transduction intermediates in response to serum deprivation with and without PrP23-50. Graphical data are presented relative to serum control. e Co-localisation of PrP23-89 with pMEK1. PrP23-89, as detected by saf32, is shown in red and pMEK1 staining is shown in green, with DAPI staining of the nuclei in blue. The heat map (right plates) shows co-localising pixels. Scale bars = 10 µM. f Pearson’s product moment correlation co-efficient analysis of the apo- and copper-saturated PrP23-89 co-localisation. **p < 0.01. g Quantification of pixel intensity for the pMEK1 staining. **p < 0.01
To confirm the involvement of the identified pathways, cells were incubated with serum and without serum plus or minus PrP23-50 for 15 min, then western blotted for central signal protein phosphorylation changes. PrP23-50 inclusion in the serum-free media induced phosphorylation of MEK1 (but not MEK2) and ERK1/2. AKT showed significantly increased phosphorylation in the serum-deprived cells, but PrP23-50 did not appear to additionally influence this pathway (Fig. 6c, d). Co-localisation of the apo- or copper-loaded PrP23-89 peptides with phosphorylated MEK1 (pMEK1) were conducted. Greater co-localisation of copper-saturated PrP23-89 is seen compared with the apo-peptide (Fig. 6e, f; student’s t test, t = 3.989, p = 0.0013, n = 3). Additionally, concordant with the PrP23-50 responses, intensity quantification of the pMEK1 IF demonstrated significantly increased staining in the cells treated with copper-loaded PrP23-89 as compared with the apo-PrP23-89 (Fig. 6g; student’s t test, t = 6.58, p = 0.001, n = 3).
Discussion
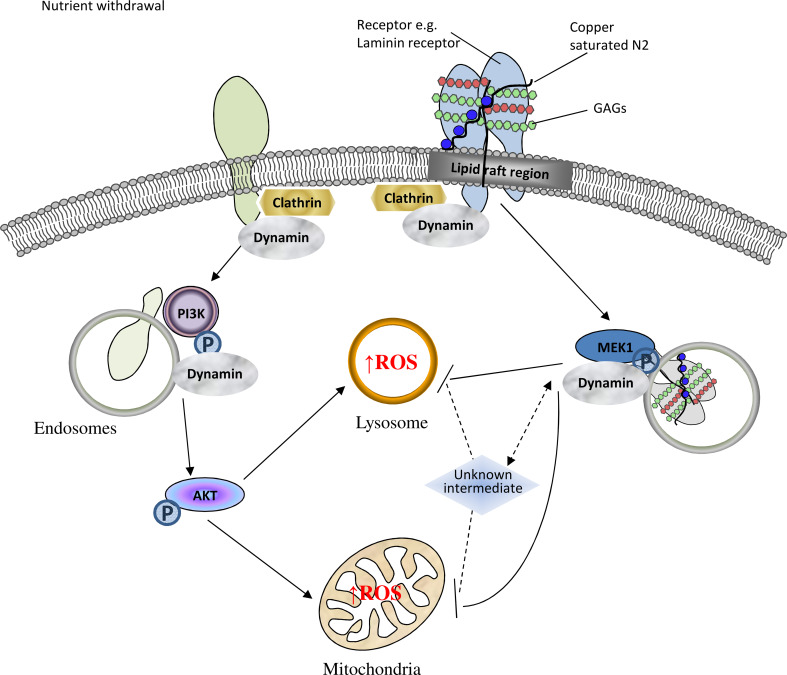
Conditions of heightened ROS cause β-cleavage of PrPC at the cell surface [14, 17] and the β-cleavage event and its N-terminal cleavage product, N2, have been shown to be protective against cellular stress [14, 24, 34, 35]. Herein, we have shown that the copper-saturated N2 β-cleavage product initiates a cellular protective response through binding to a lipid-raft membrane complex containing the 37/67 kDa laminin receptor, engaging its internalisation and stimulating MEK1 signal transduction pathways. The precise membrane interactions of N2, within lipid rafts and with its GAG-binding partners, and also involving both the copper-co-ordinating octarepeat region and the structure conferred upon the N-terminal polybasic region by the proline motif, were also shown essential for the correct engagement of this pathway. Figure 7 provides a schematic overview of the postulated membrane binding, internalisation mechanism and protective modulation pathway for copper-saturated N2.
Fig. 7.
Schematic representation of intracellular signalling pathways influenced by N2 during cellular protection against oxidative stress. Upon serum withdrawal, an intracellular ROS increase is stimulated within mitochondria and lysosomes. The ROS reaction is signalled through internalisation/trafficking of membrane complexes and involves central intermediates of the AKT and MAPK signal transduction pathways. Correct engagement of a lipid-raft membrane complex containing glycosaminoglycans and the 37/67 kDa laminin receptor by copper-loaded the PrP N2 fragment modulates intracellular trafficking and activates MEK1 signalling to oppose ROS production
A critical role for the PrP N-terminus in cellular protection is becoming well established [14, 22–24, 27, 28, 34]. The N-terminus is also required for PrPC internalisation [36–38]. A requirement for PrP internalisation as part of ERK1/2 signalling by full-length PrPC has previously been reported for stress signalling through STI-1 [39, 40]. Mutation of the N-terminal polybasic region, which abolishes internalisation [38], also eliminated the associated signal transduction [40]. Whilst an interaction with STI-1 is unlikely to be involved in the N2 ROS reduction pathway presented in the current study, as STI-1 binds PrP at residues 113–128 [41] beyond the N2 cleavage site, our finding that the N-terminus is able to instigate such pathways on its own suggests that PrPC cleavage at the cell surface may be an important and deliberate event for initiating protective cellular signalling pathways. Earlier studies have also shown that copper stimulates full-length PrPC internalisation [42, 43] and that copper-induced internalisation of full-length PrP requires the N-terminus and at least two octarepeats and is lost when the α-cleavage site is mutated [37, 44]. Copper has been linked with PrPC α-cleavage and signal transduction in the context of an altered membrane environment [32]. Therefore, it is probable that copper-binding interactions modulate the specific function(s) of the PrP23-89 peptide.
Zinc is also known to co-ordinate to the octameric repeat domain [45] and, therefore, could have a functional influence on the N2 fragment. However, in the presented experiments we only observed heightened toxicity of zinc when bound to N2 rather than a functional outcome in the context of these experiments. During prion disease, brain metal levels are known to be altered [46, 47], and this may have significant implications for cellular functions that require copper as a molecular “switch”. To the best of our knowledge, this study is the first to show that the N2 fragment intracellular ROS protection response requires copper-induced internalisation to effectively engage its downstream signalling pathways.
Whilst this study identified the 37/67 kDa laminin receptor as part of a complex that is pulled down by N-terminal fragments of PrP, we hypothesise that this is one part of a much larger complex and we cannot state from the data presented herein that it is the definitive receptor for N2. However, there is significant evidence in the literature to support a functional role for the 37/67 kDa laminin receptor in the PrP N2 interaction; the 37/67 kDa laminin receptor binds to PrPC through GAG interactions with the N2 region of PrPC [48, 49], in a pathway that is also linked with internalisation of PrPSc during disease [50–53]. Other reported PrP receptors were not investigated in the current study but include low-density lipoprotein receptor-related protein 1, with binding in conjunction with GAGs [54], Glypican-1, an HS containing proteoglycan that co-internalises with PrPC [55, 56], and α7 nicotinic receptors [57]. The latter have been shown to form a functional complex with PrPC that mediates STI-1 signalling in a pathway which is known to require internalisation for signal transduction [57]. Potentially N2 may be modulating any or all of these pathways depending upon its membrane environment and copper saturation.
Our presented data indicated that peptide-partner binding interactions might be masking the octarepeat epitope at the cell surface, thus preventing antibody binding. Further, the differing binding pattern of the PrP23-89P26/28A peptide when compared with the wild-type PrP23-89 showed that the cell surface interaction of the octameric repeat domain is influenced by the proline motif within the N-terminal polybasic region. The observations might be due to interactions with the signalling complex through which the N2 fragment mediates its function or, alternatively, as the N-terminal amino-acids alone were able to instigate the same ROS reduction response to serum deprivation, the function of this region may be orientation of the interaction within/at the cell membrane. Our previous findings have shown the importance of the octarepeat for membrane engagement under specific conditions [58], and therefore, the context of the cell membrane lipid binding interactions may also modulate correct pathway engagement for signal transduction in the present study. The rapid retraction of the cell membrane that was shown on addition of PrP23-50 could indicate that changes are occurring at the cell membrane and could have occurred due to a rapid formation of membrane domains required for internalisation. Furthermore, the PrP N-terminus has also been shown to bind to the cytoskeletal protein tubulin [59]. The observed rapid rearrangement of the cell membrane could in theory be due to cytoskeletal restructuring either through direct N2-tubulin interaction or via N2 activation of MEK1 signalling, in similarity with pathways previously reported for actin cytoskeletal rearrangements [60].
Herein, two signalling pathways were shown to be especially involved in modulating the intracellular ROS produced in response to serum deprivation, the PI3K/AKT and the MEK1/ERK1/2 pathways, with the latter influenced by PrP23-50 to act against the ROS increase. Our results showed that direct inhibition of ERK did not produce the same cellular responses as MEK1/2 inhibition and so MEK1 might be acting to stimulate further downstream targets to exert its full protective function. The vesicular internalisation pattern presented for copper-co-ordinating PrP23-89, co-localising with pMEK1, shows this peptide is found in close proximity to the signalling intermediates in the pathways with which it interacts; therefore, copper-loaded PrP23-89 may be part of an internalised complex that acts directly to influence the activity of specific signalling pathways, such as MEK1.
Whilst the ability to engage signalling pathways appears important for N2 function, the properties of this amino acid sequence also affect full-length PrP. Studies using cells expressing full-length PrP have shown that PrP expression is regulated by signalling through the MEK1 pathway [61] and PrP expression is increased in response to conditions of oxidative stress [62]. An environmental stress-induced increase in paracrine N2 signalling could be able to initiate these events even in cells with low endogenous PrP expression. β-cleavage may also prevent signalling of full-length PrP or permit signalling of the C-terminal cleavage fragments. Previous studies have found that PrP can signal through NADPH oxidase [63], which was not seen for the truncated N2 fragment in the current study, and that the C2 fragment is able to initiate signalling through p38 and ERK1/2 in astrocytes and microglia [64]. This highlights a difficulty of attributing functions to full-length PrP as many cells and tissue display more cleaved PrP than full length [65, 66], and further supports the role of cleavage as a functional event for controlling PrP activity.
Our findings are consistent with interactions at the cell surface, in the first phases of internalisation being able to influence the downstream ROS production sites, supporting the importance of correct engagement of a receptor complex at the cell surface by the N2 fragment. We showed that engagement of the N2 peptide at the cell surface was determined by the synergy between the polybasic proline motif and the copper-co-ordinating octarepeats. A predominant PrPC function has remained elusive despite many years of intensive investigation, with many studies convincingly suggesting the likelihood of many different cellular roles. Whilst the choice of model used in each such study may lead to differing results and, therefore, add to the contention around this issue, post-translational modification to PrPC itself is unlikely to represent an irrelevant epiphenomenon in all systems. The hypothesis that PrPC post-translational modification occurs as a molecular switch for activation or deactivation of certain functions facilitates the beginning of our understanding of its multi-functionality. The apparent contextual dependence of the protective effects offered by the N-terminal cleavage products, as shown in our data, demonstrates the finely balanced nature of certain functions. The role of such protective functions and their potential to be corrupted during disease processes will provide new insights into both prion disease pathogenesis and other stresses deleteriously affecting the brain.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors would like to thank Dr Victoria Lawson for her support and helpful discussions. The CF10 cells were a kind gift to Dr Victoria Lawson from Dr Suzette Priola (National Institute of Health, USA). This work was supported by an NH&MRC program grant (#628946) and SJC is supported by an NH&MRC Practitioner Fellowship (#APP100581).
Contributor Information
C. L. Haigh, Email: chaigh@unimelb.edu.au
S. J. Collins, Email: stevenjc@unimelb.edu.au
References
- 1.Haigh CL, Brown DR. Prion protein reduces both oxidative and non-oxidative copper toxicity. J Neurochem. 2006;98(3):677–689. doi: 10.1111/j.1471-4159.2006.03906.x. [DOI] [PubMed] [Google Scholar]
- 2.Brown DR, Schulz-Schaeffer WJ, Schmidt B, Kretzschmar HA. Prion protein-deficient cells show altered response to oxidative stress due to decreased SOD-1 activity. Exp Neurol. 1997;146(1):104–112. doi: 10.1006/exnr.1997.6505. [DOI] [PubMed] [Google Scholar]
- 3.Klamt F, Dal-Pizzol F, Conte da Frota ML, Jr, Walz R, Andrades ME, da Silva EG, Brentani RR, Izquierdo I, Fonseca Moreira JC. Imbalance of antioxidant defense in mice lacking cellular prion protein. Free Radic Biol Med. 2001;30(10):1137–1144. doi: 10.1016/S0891-5849(01)00512-3. [DOI] [PubMed] [Google Scholar]
- 4.Rachidi W, Vilette D, Guiraud P, Arlotto M, Riondel J, Laude H, Lehmann S, Favier A. Expression of prion protein increases cellular copper binding and antioxidant enzyme activities but not copper delivery. J Biol Chem. 2003;278(11):9064–9072. doi: 10.1074/jbc.M211830200. [DOI] [PubMed] [Google Scholar]
- 5.Senator A, Rachidi W, Lehmann S, Favier A, Benboubetra M. Prion protein protects against DNA damage induced by paraquat in cultured cells. Free Radic Biol Med. 2004;37(8):1224–1230. doi: 10.1016/j.freeradbiomed.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Watt NT, Routledge MN, Wild CP, Hooper NM. Cellular prion protein protects against reactive-oxygen-species-induced DNA damage. Free Radic Biol Med. 2007;43(6):959–967. doi: 10.1016/j.freeradbiomed.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Chiarini LB, Freitas AR, Zanata SM, Brentani RR, Martins VR, Linden R. Cellular prion protein transduces neuroprotective signals. EMBO J. 2002;21(13):3317–3326. doi: 10.1093/emboj/cdf324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris DA, Huber MT, van Dijken P, Shyng SL, Chait BT, Wang R. Processing of a cellular prion protein: identification of N- and C-terminal cleavage sites. Biochemistry. 1993;32(4):1009–1016. doi: 10.1021/bi00055a003. [DOI] [PubMed] [Google Scholar]
- 9.Chen SG, Teplow DB, Parchi P, Teller JK, Gambetti P, Autilio-Gambetti L. Truncated forms of the human prion protein in normal brain and in prion diseases. J Biol Chem. 1995;270(32):19173–19180. doi: 10.1074/jbc.270.32.19173. [DOI] [PubMed] [Google Scholar]
- 10.Haigh CL, Marom SY, Collins SJ. Copper, endoproteolytic processing of the prion protein and cell signalling. Front Biosci (Landmark Ed) 2010;15:1086–1104. doi: 10.2741/3663. [DOI] [PubMed] [Google Scholar]
- 11.McMahon HE. Prion processing: a double-edged sword? Biochem Soc Trans. 2012;40(4):735–738. doi: 10.1042/BST20120031. [DOI] [PubMed] [Google Scholar]
- 12.Brazier MW, Lewis V, Ciccotosto GD, Klug GM, Lawson VA, Cappai R, Ironside JW, Masters CL, Hill AF, White AR, Collins S. Correlative studies support lipid peroxidation is linked to PrP(res) propagation as an early primary pathogenic event in prion disease. Brain Res Bull. 2006;68(5):346–354. doi: 10.1016/j.brainresbull.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 13.Yadavalli R, Guttmann RP, Seward T, Centers AP, Williamson RA, Telling GC. Calpain-dependent endoproteolytic cleavage of PrPSc modulates scrapie prion propagation. J Biol Chem. 2004;279(21):21948–21956. doi: 10.1074/jbc.M400793200. [DOI] [PubMed] [Google Scholar]
- 14.Watt NT, Taylor DR, Gillott A, Thomas DA, Perera WS, Hooper NM. Reactive oxygen species-mediated beta-cleavage of the prion protein in the cellular response to oxidative stress. J Biol Chem. 2005;280(43):35914–35921. doi: 10.1074/jbc.M507327200. [DOI] [PubMed] [Google Scholar]
- 15.Haigh CL, McGlade AR, Lewis V, Masters CL, Lawson VA, Collins SJ. Acute exposure to prion infection induces transient oxidative stress progressing to be cumulatively deleterious with chronic propagation in vitro. Free Radic Biol Med. 2011;51(3):594–608. doi: 10.1016/j.freeradbiomed.2011.03.035. [DOI] [PubMed] [Google Scholar]
- 16.Sinclair L, Lewis V, Collins SJ, Haigh CL. Cytosolic caspases mediate mislocalised SOD2 depletion in an in vitro model of chronic prion infection. Dis Model Mech. 2013;6(4):952–963. doi: 10.1242/dmm.010678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McMahon HE, Mange A, Nishida N, Creminon C, Casanova D, Lehmann S. Cleavage of the amino terminus of the prion protein by reactive oxygen species. J Biol Chem. 2001;276(3):2286–2291. doi: 10.1074/jbc.M007243200. [DOI] [PubMed] [Google Scholar]
- 18.Westergard L, Turnbaugh JA, Harris DA. A nine amino acid domain is essential for mutant prion protein toxicity. J Neurosci. 2011;31(39):14005–14017. doi: 10.1523/JNEUROSCI.1243-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sonati T, Reimann RR, Falsig J, Baral PK, O’Connor T, Hornemann S, Yaganoglu S, Li B, Herrmann US, Wieland B, Swayampakula M, Rahman MH, Das D, Kav N, Riek R, Liberski PP, James MN, Aguzzi A. The toxicity of antiprion antibodies is mediated by the flexible tail of the prion protein. Nature. 2013;501(7465):102–106. doi: 10.1038/nature12402. [DOI] [PubMed] [Google Scholar]
- 20.Lawson VA, Priola SA, Meade-White K, Lawson M, Chesebro B. Flexible N-terminal region of prion protein influences conformation of protease-resistant prion protein isoforms associated with cross-species scrapie infection in vivo and in vitro. J Biol Chem. 2004;279(14):13689–13695. doi: 10.1074/jbc.M303697200. [DOI] [PubMed] [Google Scholar]
- 21.Lawson VA, Priola SA, Wehrly K, Chesebro B. N-terminal truncation of prion protein affects both formation and conformation of abnormal protease-resistant prion protein generated in vitro. J Biol Chem. 2001;276(38):35265–35271. doi: 10.1074/jbc.M103799200. [DOI] [PubMed] [Google Scholar]
- 22.Turnbaugh JA, Unterberger U, Saa P, Massignan T, Fluharty BR, Bowman FP, Miller MB, Supattapone S, Biasini E, Harris DA. The N-terminal, polybasic region of PrP(C) dictates the efficiency of prion propagation by binding to PrP(Sc) J Neurosci. 2012;32(26):8817–8830. doi: 10.1523/JNEUROSCI.1103-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guillot-Sestier MV, Sunyach C, Druon C, Scarzello S, Checler F. The alpha-secretase-derived N-terminal product of cellular prion, N1, displays neuroprotective function in vitro and in vivo. J Biol Chem. 2009;284(51):35973–35986. doi: 10.1074/jbc.M109.051086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haigh CL, Drew SC, Boland MP, Masters CL, Barnham KJ, Lawson VA, Collins SJ. Dominant roles of the polybasic proline motif and copper in the PrP23-89-mediated stress protection response. J Cell Sci. 2009;122(Pt 10):1518–1528. doi: 10.1242/jcs.043604. [DOI] [PubMed] [Google Scholar]
- 25.Chen S, Yadav SP, Surewicz WK. Interaction between human prion protein and amyloid-beta (Abeta) oligomers: role OF N-terminal residues. J Biol Chem. 2010;285(34):26377–26383. doi: 10.1074/jbc.M110.145516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Younan ND, Sarell CJ, Davies P, Brown DR, Viles JH. The cellular prion protein traps Alzheimer’s Abeta in an oligomeric form and disassembles amyloid fibers. FASEB J. 2013;27(5):1847–1858. doi: 10.1096/fj.12-222588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guillot-Sestier MV, Sunyach C, Ferreira ST, Marzolo MP, Bauer C, Thevenet A, Checler F. Alpha-Secretase-derived fragment of cellular prion, N1, protects against monomeric and oligomeric amyloid beta (Abeta)-associated cell death. J Biol Chem. 2012;287(7):5021–5032. doi: 10.1074/jbc.M111.323626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fluharty BR, Biasini E, Stravalaci M, Sclip A, Diomede L, Balducci C, La Vitola P, Messa M, Colombo L, Forloni G, Borsello T, Gobbi M, Harris DA. An N-terminal fragment of the prion protein binds to amyloid-beta oligomers and inhibits their neurotoxicity in vivo. J Biol Chem. 2013;288(11):7857–7866. doi: 10.1074/jbc.M112.423954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karas JA, Boland M, Haigh C, Johanssen V, Hill A, Barnham K, Collins S, Scanlon D. Microwave synthesis of prion protein fragments up to 111 amino acids in length generates biologically active peptides. Int J Pept Res Ther. 2012;18(1):21–29. doi: 10.1007/s10989-011-9275-7. [DOI] [Google Scholar]
- 30.Greil CS, Vorberg IM, Ward AE, Meade-White KD, Harris DA, Priola SA. Acute cellular uptake of abnormal prion protein is cell type and scrapie-strain independent. Virology. 2008;379(2):284–293. doi: 10.1016/j.virol.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haigh CL, Brown DR (2008) Investigation of PrPC metabolism and function in live cells : methods for studying individual cells and cell populations. Methods Mol Biol (Clifton, NJ) 459:21–34. doi:10.1007/978-1-59745-234-2_2 [DOI] [PubMed]
- 32.Haigh CL, Lewis VA, Vella LJ, Masters CL, Hill AF, Lawson VA, Collins SJ. PrPC-related signal transduction is influenced by copper, membrane integrity and the alpha cleavage site. Cell Res. 2009;19(9):1062–1078. doi: 10.1038/cr.2009.86. [DOI] [PubMed] [Google Scholar]
- 33.Chen CS, Gee KR. Redox-dependent trafficking of 2,3,4,5, 6-pentafluorodihydrotetramethylrosamine, a novel fluorogenic indicator of cellular oxidative activity. Free Radic Biol Med. 2000;28(8):1266–1278. doi: 10.1016/S0891-5849(00)00265-3. [DOI] [PubMed] [Google Scholar]
- 34.Zeng F, Watt NT, Walmsley AR, Hooper NM. Tethering the N-terminus of the prion protein compromises the cellular response to oxidative stress. J Neurochem. 2003;84(3):480–490. doi: 10.1046/j.1471-4159.2003.01529.x. [DOI] [PubMed] [Google Scholar]
- 35.Dupiereux I, Falisse-Poirrier N, Zorzi W, Watt NT, Thellin O, Zorzi D, Pierard O, Hooper NM, Heinen E, Elmoualij B. Protective effect of prion protein via the N-terminal region in mediating a protective effect on paraquat-induced oxidative injury in neuronal cells. J Neurosci Res. 2008;86(3):653–659. doi: 10.1002/jnr.21506. [DOI] [PubMed] [Google Scholar]
- 36.Nunziante M, Gilch S, Schatzl HM. Essential role of the prion protein N terminus in subcellular trafficking and half-life of cellular prion protein. J Biol Chem. 2003;278(6):3726–3734. doi: 10.1074/jbc.M206313200. [DOI] [PubMed] [Google Scholar]
- 37.Taylor DR, Watt NT, Perera WS, Hooper NM. Assigning functions to distinct regions of the N-terminus of the prion protein that are involved in its copper-stimulated, clathrin-dependent endocytosis. J Cell Sci. 2005;118(Pt 21):5141–5153. doi: 10.1242/jcs.02627. [DOI] [PubMed] [Google Scholar]
- 38.Sunyach C, Jen A, Deng J, Fitzgerald KT, Frobert Y, Grassi J, McCaffrey MW, Morris R. The mechanism of internalization of glycosylphosphatidylinositol-anchored prion protein. EMBO J. 2003;22(14):3591–3601. doi: 10.1093/emboj/cdg344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Americo TA, Chiarini LB, Linden R. Signaling induced by hop/STI-1 depends on endocytosis. Biochem Biophys Res Commun. 2007;358(2):620–625. doi: 10.1016/j.bbrc.2007.04.202. [DOI] [PubMed] [Google Scholar]
- 40.Caetano FA, Lopes MH, Hajj GN, Machado CF, Pinto Arantes C, Magalhaes AC, Vieira Mde P, Americo TA, Massensini AR, Priola SA, Vorberg I, Gomez MV, Linden R, Prado VF, Martins VR, Prado MA. Endocytosis of prion protein is required for ERK1/2 signaling induced by stress-inducible protein 1. J Neurosci. 2008;28(26):6691–6702. doi: 10.1523/JNEUROSCI.1701-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zanata SM, Lopes MH, Mercadante AF, Hajj GN, Chiarini LB, Nomizo R, Freitas AR, Cabral AL, Lee KS, Juliano MA, de Oliveira E, Jachieri SG, Burlingame A, Huang L, Linden R, Brentani RR, Martins VR. Stress-inducible protein 1 is a cell surface ligand for cellular prion that triggers neuroprotection. EMBO J. 2002;21(13):3307–3316. doi: 10.1093/emboj/cdf325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pauly PC, Harris DA. Copper stimulates endocytosis of the prion protein. J Biol Chem. 1998;273(50):33107–33110. doi: 10.1074/jbc.273.50.33107. [DOI] [PubMed] [Google Scholar]
- 43.Perera WS, Hooper NM. Ablation of the metal ion-induced endocytosis of the prion protein by disease-associated mutation of the octarepeat region. Curr Biol. 2001;11(7):519–523. doi: 10.1016/S0960-9822(01)00147-6. [DOI] [PubMed] [Google Scholar]
- 44.Haigh CL, Edwards K, Brown DR. Copper binding is the governing determinant of prion protein turnover. Mol Cell Neurosci. 2005;30(2):186–196. doi: 10.1016/j.mcn.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 45.Jackson GS, Murray I, Hosszu LL, Gibbs N, Waltho JP, Clarke AR, Collinge J. Location and properties of metal-binding sites on the human prion protein. Proc Natl Acad Sci USA. 2001;98(15):8531–8535. doi: 10.1073/pnas.151038498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hesketh S, Sassoon J, Knight R, Brown DR. Elevated manganese levels in blood and CNS in human prion disease. Mol Cell Neurosci. 2008;37(3):590–598. doi: 10.1016/j.mcn.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 47.Hesketh S, Sassoon J, Knight R, Hopkins J, Brown DR. Elevated manganese levels in blood and central nervous system occur before onset of clinical signs in scrapie and bovine spongiform encephalopathy. J Anim Sci. 2007;85(6):1596–1609. doi: 10.2527/jas.2006-714. [DOI] [PubMed] [Google Scholar]
- 48.Gauczynski S, Peyrin JM, Haik S, Leucht C, Hundt C, Rieger R, Krasemann S, Deslys JP, Dormont D, Lasmezas CI, Weiss S. The 37-kDa/67-kDa laminin receptor acts as the cell-surface receptor for the cellular prion protein. EMBO J. 2001;20(21):5863–5875. doi: 10.1093/emboj/20.21.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hundt C, Peyrin JM, Haik S, Gauczynski S, Leucht C, Rieger R, Riley ML, Deslys JP, Dormont D, Lasmezas CI, Weiss S. Identification of interaction domains of the prion protein with its 37-kDa/67-kDa laminin receptor. EMBO J. 2001;20(21):5876–5886. doi: 10.1093/emboj/20.21.5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gauczynski S, Nikles D, El-Gogo S, Papy-Garcia D, Rey C, Alban S, Barritault D, Lasmezas CI, Weiss S. The 37-kDa/67-kDa laminin receptor acts as a receptor for infectious prions and is inhibited by polysulfated glycanes. J Infect Dis. 2006;194(5):702–709. doi: 10.1086/505914. [DOI] [PubMed] [Google Scholar]
- 51.Morel E, Andrieu T, Casagrande F, Gauczynski S, Weiss S, Grassi J, Rousset M, Dormont D, Chambaz J. Bovine prion is endocytosed by human enterocytes via the 37 kDa/67 kDa laminin receptor. Am J Pathol. 2005;167(4):1033–1042. doi: 10.1016/S0002-9440(10)61192-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kolodziejczak D, Da Costa Dias B, Zuber C, Jovanovic K, Omar A, Beck J, Vana K, Mbazima V, Richt J, Brenig B, Weiss SF. Prion interaction with the 37-kDa/67-kDa laminin receptor on enterocytes as a cellular model for intestinal uptake of prions. J Mol Biol. 2010;402(2):293–300. doi: 10.1016/j.jmb.2010.06.055. [DOI] [PubMed] [Google Scholar]
- 53.Pflanz H, Vana K, Mitteregger G, Renner-Muller I, Pace C, Kuchenhoff H, Kretzschmar HA, Wolf E, Weiss S. Scrapie-infected transgenic mice expressing a laminin receptor decoy mutant reveal a prolonged incubation time associated with low levels of PrPRes . J Mol Biol. 2009;388(4):721–729. doi: 10.1016/j.jmb.2009.03.045. [DOI] [PubMed] [Google Scholar]
- 54.Parkyn CJ, Vermeulen EG, Mootoosamy RC, Sunyach C, Jacobsen C, Oxvig C, Moestrup S, Liu Q, Bu G, Jen A, Morris RJ. LRP1 controls biosynthetic and endocytic trafficking of neuronal prion protein. J Cell Sci. 2008;121(Pt 6):773–783. doi: 10.1242/jcs.021816. [DOI] [PubMed] [Google Scholar]
- 55.Cheng F, Lindqvist J, Haigh CL, Brown DR, Mani K. Copper-dependent co-internalization of the prion protein and glypican-1. J Neurochem. 2006;98(5):1445–1457. doi: 10.1111/j.1471-4159.2006.03981.x. [DOI] [PubMed] [Google Scholar]
- 56.Taylor DR, Whitehouse IJ, Hooper NM. Glypican-1 mediates both prion protein lipid raft association and disease isoform formation. PLoS Pathog. 2009;5(11):e1000666. doi: 10.1371/journal.ppat.1000666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beraldo FH, Arantes CP, Santos TG, Queiroz NG, Young K, Rylett RJ, Markus RP, Prado MA, Martins VR. Role of alpha7 nicotinic acetylcholine receptor in calcium signaling induced by prion protein interaction with stress-inducible protein 1. J Biol Chem. 2010;285(47):36542–36550. doi: 10.1074/jbc.M110.157263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boland MP, Hatty CR, Separovic F, Hill AF, Tew DJ, Barnham KJ, Haigh CL, James M, Masters CL, Collins SJ. Anionic phospholipid interactions of the prion protein N terminus are minimally perturbing and not driven solely by the octapeptide repeat domain. J Biol Chem. 2010;285(42):32282–32292. doi: 10.1074/jbc.M110.123398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Osiecka KM, Nieznanska H, Skowronek KJ, Karolczak J, Schneider G, Nieznanski K. Prion protein region 23-32 interacts with tubulin and inhibits microtubule assembly. Proteins. 2009;77(2):279–296. doi: 10.1002/prot.22435. [DOI] [PubMed] [Google Scholar]
- 60.Barros JC, Marshall CJ. Activation of either ERK1/2 or ERK5 MAP kinase pathways can lead to disruption of the actin cytoskeleton. J Cell Sci. 2005;118(Pt 8):1663–1671. doi: 10.1242/jcs.02308. [DOI] [PubMed] [Google Scholar]
- 61.Zawlik I, Witusik M, Hulas-Bigoszewska K, Piaskowski S, Szybka M, Golanska E, Liberski PP, Rieske P. Regulation of PrPC expression: nerve growth factor (NGF) activates the prion gene promoter through the MEK1 pathway in PC12 cells. Neurosci Lett. 2006;400(1–2):58–62. doi: 10.1016/j.neulet.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 62.Cichon AC, Brown DR (2014) Nrf-2 regulation of prion protein expression is independent of oxidative stress. Mol Cell Neurosci 63c:31–37. doi:10.1016/j.mcn.2014.09.001 [DOI] [PubMed]
- 63.Schneider B, Mutel V, Pietri M, Ermonval M, Mouillet-Richard S, Kellermann O. NADPH oxidase and extracellular regulated kinases 1/2 are targets of prion protein signaling in neuronal and nonneuronal cells. Proc Natl Acad Sci USA. 2003;100(23):13326–13331. doi: 10.1073/pnas.2235648100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thellung S, Villa V, Corsaro A, Pellistri F, Venezia V, Russo C, Aceto A, Robello M, Florio T. ERK1/2 and p38 MAP kinases control prion protein fragment 90-231-induced astrocyte proliferation and microglia activation. Glia. 2007;55(14):1469–1485. doi: 10.1002/glia.20559. [DOI] [PubMed] [Google Scholar]
- 65.Lewis V, Haigh CL, Masters CL, Hill AF, Lawson VA, Collins SJ. Prion subcellular fractionation reveals infectivity spectrum, with a high titre-low PrPRes level disparity. Mol Neurodegener. 2012;7:18. doi: 10.1186/1750-1326-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dron M, Moudjou M, Chapuis J, Salamat MK, Bernard J, Cronier S, Langevin C, Laude H. Endogenous proteolytic cleavage of disease-associated prion protein to produce C2 fragments is strongly cell- and tissue-dependent. J Biol Chem. 2010;285(14):10252–10264. doi: 10.1074/jbc.M109.083857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.