Abstract
More and more studies have shown chromatin remodelers and histone modifiers play essential roles in regulating developmental patterns by organizing specific chromosomal architecture to establish programmed transcriptional profiles, with implications that histone chaperones execute a coordinating role in these processes. Chromatin assembly factor-1 (CAF-1), an evolutionarily conserved three-subunit protein complex, was identified as a histone chaperone coupled with DNA replication and repair in cultured mammalian cells and yeasts. Interestingly, recent findings indicate CAF-1 may have important regulatory roles during development by interacting with specific transcription factors and epigenetic regulators. In this review, we focus on the essential roles of CAF-1 in regulating heterochromatin organization, asymmetric cell division, and specific signal transduction through epigenetic modulations of the chromatin. In the end, we aim at providing a current image of facets of CAF-1 as a histone chaperone to orchestrate cell proliferation and differentiation during multi-cellular organism development.
Keywords: Heterochromatin, Signal transduction, Cell proliferation, Cell differentiation, Epigenetic regulation, Genome stability
Introduction
In multi-cellular organisms, development is a delicately orchestrated process controlled by programmed cell proliferation and differentiation. Temporally and spatially organized epigenetic profiles play key roles in determining cell differentiation by modulating transcriptional outputs at the chromatin level. In the past two decades, studies have shown epigenetic regulators, including histone modifiers and chromatin remodelers, play essential roles in mediating developmental programs [1]. On one hand, these epigenetic regulators help to direct particular signal transduction by cooperating with specific transcriptional machineries. For example, nucleosome remodeling factor (NURF) is reported to be involved in T cell maturation in mice by physically interacting with DNA binding factor serum response factor (Srf) to regulate Egr1 gene expression, which is important for thymocyte development [2]. In Drosophila, NURF was reported to regulate Wg signaling by regulating transcriptional affinity at the target gene enhancers [3]. On the other hand, it was also revealed histone modifiers and chromatin remodelers help to generate global epigenetic landscapes during multi-cellular organism development [4, 5]. Interestingly, recent findings indicate that during the epigenetic regulation of developmental processes, histone chaperones also appear to execute more and more important functions. For example, histone chaperone anti-silencing function 1 (Asf1) has been reported to interact with a repressive transcription complex to regulate Notch target gene expression during Drosophila development [6, 7]. Recent studies indicate that histone cell cycle regulation defective homolog A (HIRA), the histone H3.3 chaperone, is required for gastrulation and nuclear transfer-induced transcriptional reprogramming in Xenopus [8, 9]. In the same line as described above, both histone chaperone HIRA and nucleosome assembly protein 1 (NAP-1) are found to accumulate in mouse primordial germ cells (PGCs) during reprogramming [10]. In this review, we will focus on the developmental roles of another histone chaperone, chromatin assembly factor 1 (CAF-1).
Over two decades ago, CAF-1 was first purified from human cells and shown to be capable of depositing histone H3–H4 onto newly synthesized DNA [11]. Subsequent studies in yeast and multi-cellular organisms further implicated its role as an evolutionarily conserved histone chaperone in eukaryotes (Table 1). In addition, biochemists and cell biologists have shown CAF-1 plays pivotal roles in controlling not only chromatin restoration after DNA synthesis and cell cycle progression, but also the formation and maintenance of heterochromatin [12–17]. More recently, developmental biologists and geneticists have more closely investigated the in vivo roles of CAF-1, and have suggested CAF-1 is also an important regulator of developmental pathways, a determinant of epigenetic memories, and a key factor controlling asymmetric cell division [18–21]. In this review, we first briefly summarize the biochemical functions of CAF-1 that mainly based on studies in purified in vitro systems, cell lines, and yeast before we focus on the more intriguing functions of CAF-1 in multi-cellular organism development. Finally, we propose CAF-1 to be a protein complex that may also orchestrate cell proliferation and differentiation during multi-cellular organism development.
Table 1.
Evolutionarily conserved subunits of CAF-1 complex
| Species | Large subunit | Middle subunit | Small subunit |
|---|---|---|---|
| H. sapiens | p150 | p60 | p48 |
| M. musculus | p150 | p60 | p48 |
| D. melanogaster | p180 | p105 | p55 |
| S. mediterranea | p150 | p60 | p48 |
| C. elegans | Chaf1 | Chaf2 | Rba1 |
| S. cerevisiae | Cac1 | Cac2 | Cac3 |
| A. thaliana | FAS1 | FAS2 | MSII |
A key player in chromatin duplication and restoration
Biologists have been studying on the biochemical and cellular functions of CAF-1 using in vitro systems and cultured cell lines since it was first isolated more than 20 years ago [11]. As a histone chaperone, CAF-1 acts in the following major functions in both yeast and mammalian cells: (1) a histone carrier for DNA replication-coupled nucleosome assembly; (2) a player in heterochromatin duplication; and (3) a chromatin restorer after DNA repair.
DNA replication-coupled nucleosome assembly
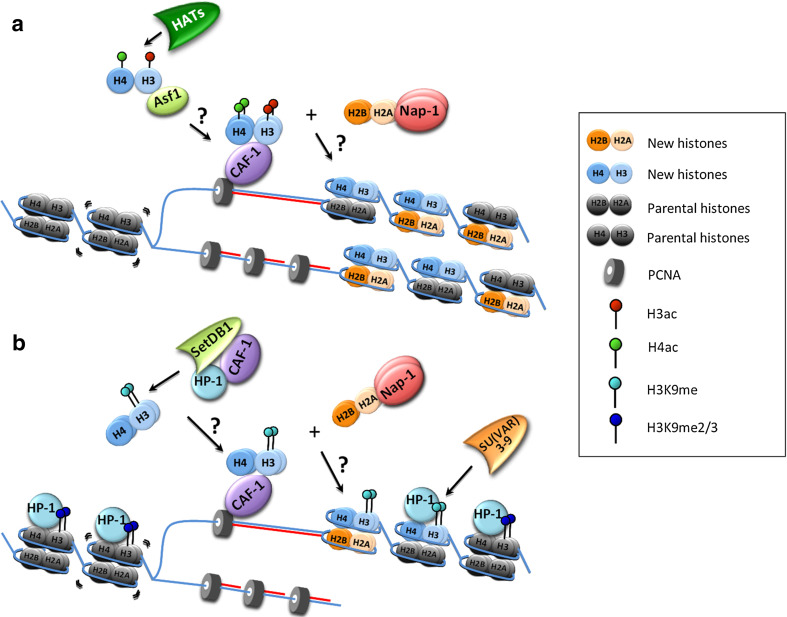
In the process of eukaryotic DNA replication, CAF-1 functions as a histone chaperone that deposits histone H3–H4 heterodimers onto newly synthesized DNA to facilitate nucleosome assembly behind the replication fork [22]. On the newly assembled chromatin, a fraction (perhaps half) of the histones are inherited from the parental chromatin and the rest of the histones are newly synthesized. Although the mechanism of how parental histones are transferred to the newly synthesized DNA for replication-coupled nucleosome assembly remains poorly understood, it is known that newly synthesized histones H3 and H4 are usually acetylated and methylated before they are loaded into the new-born nucleosomes [22]. The K56 residue of histone H3 is reported to be acetylated by CBP or Gcn5 in humans and by Rtt109 in yeasts [23–26] (Fig. 1a). Acetylated H3K56 increases the binding affinity of CAF-1 with H3–H4 and facilitates CAF-1-mediated nucleosome assembly in the yeast model [23, 27]. Newly synthesized histone H4 is also shown to be di-acetylated at the K5 and K12 residues, a conserved feature in single cell eukaryotes (Tetrahymena), flies and humans [28–30]. HAT1 in yeast and human cells is reported to be responsible for the acetylation of H4K5 and H4K12 [31, 32]. However, unlike that the acetylation of H3K56 increases CAF-1’s binding affinity to H3–H4 in vitro [23, 27], the acetylation of H4K5 or K12 reduces the association of H3–H4 with CAF-1 in mammalian cells [32]. Furthermore, studies in yeast show that mutations of H4K5 and K12 have no impact on nucleosome deposition [33]. Based on these assessments, the role of acetylation on H4K5 and K12 remains to be accurately determined. Collectively, a possible scenario is: histone H3–H4 dimers with acetylated H3K56 residues are first captured by histone chaperone Asf1 before transfer to CAF-1, a process that is mediated by the direct interaction between Asf1 and the middle subunit of the CAF-1 complex [22, 23, 34]. Finally, CAF-1 loads the histone H3–H4 tetramers onto naked DNA by interacting with proliferating cell nuclear antigen (PCNA) to fulfill the chromatin assembly flux (Fig. 1a) [22, 35, 36].
Fig. 1.
Roles of CAF-1 in DNA replication-coupled assembly of euchromatin and heterochromatin. a During euchromatic DNA replication, PCNA is loaded at the fork. Newly synthesized histone H3–H4 dimers, acetylated by specific histone acetyltransferases (HATs), are brought by Asf1 to the nascent DNA site, then transferred onto the CAF-1 complex, forming a H3–H4 tetramer. Together with the H2A–H2B tetramer brought by Nap1, these acetylated H3–H4 tetramers are assembled into nucleosomes, which consist of mixed histones, either parental or newly synthesized. b During de novo heterochromatin formation, K9 residues on histone H3 tails are mono-methylated by SetDB1 of the CAF-1/HP-1/SetDB1 complex. SU(VAR)3–9 is responsible for the di- and tri-methylation of H3K9 (H3K9me2/3) based on mono-methylated H3K9 (H3k9me1)
Heterochromatin duplication
Heterochromatin is a special chromatin domain with densely packed DNA, histones, and other associated proteins, and is less accessible than euchromatin to transcription and replication machineries. In multi-cellular organisms, a hallmark of constitutive heterochromatin is the accumulation of heterochromatic protein HP1 and repressive histone modifications H3K9me2/3 and H4K20me3 [37], which provide an epigenetic memory for the heterochromatic architecture from mother cells to daughter cells, particularly during heterochromatin duplication. The roles of CAF-1 in regulating heterochromatin are highly conserved in eukaryotes. In Saccharomyces cerevisiae, CAF-1 (Cac1, Cac2, and Cac3 in yeasts) was found to mediate telomeric heterochromatin silencing by contributing to the spread of Sir proteins [38–41]. In multi-cellular organisms, CAF-1 regulates heterochromatin by interacting with HP1 to facilitate the establishment of epigenetic markers at the heterochromatic regions [16, 20, 42] (Fig. 1b). During replication-coupled heterochromatin formation in late S phase, the K9 residue of free histone H3 is likely mono-methylated by SetDB1, a component of the HP1-CAF-1-SetDB1 complex [43]. The mono-methylated H3K9 is delivered onto DNA via CAF-1 and is further methylated to become H3K9me2/3 by SU(VAR)3–9, which then recruits HP1 to mature the duplicated heterochromatin (Fig. 1b). However, the precise molecular functions of CAF-1 in this process remain obscure.
Chromatin restoration after DNA repair
Genomic stability and integrity are constantly challenged by both environmental stimuli and metabolic products that can induce DNA damages [44]. When DNA lesions occur, DNA damage response (DDR) is initiated at the damaged DNA loci regardless of the timing of cell cycle progression. In eukaryotic organisms, DDR is a series of coordinated events to not only fix the DNA lesions but also to restore the altered chromatin status [22]. A variety of epigenetic regulators have been shown to be involved in this chromatin restoration process. In yeast, INO80 and HDACs are likely responsible for the clearance of H2A-variant and histone de-acetylations at the repaired double strand break (DSB) sites, respectively [45, 46]. A recent discovery in mammalian cells reveals that histone chaperone HIRA directs H3.3–H4-mediated transcriptional recovery after DNA repair [17]. CAF-1 has been shown to participate in UV-induced nucleotide excision repair (NER) and is necessary for repair-coupled chromatin formation in both yeast and mammalian cells [13, 41, 47]. Moreover, CAF-1 is also crucial for successful DSB repair [48]. CAF-1 has been found to cooperate in different DNA repair processes with multiple proteins, including replication clamp protein PCNA [13, 35], DNA helicases BLM and WRN [49, 50], and of interest, heterochromatin protein HP1 [15]. In addition to its function in the recruitment of HP1 during heterochromatin formation (Fig. 1b), studies in mouse cells show CAF-1 is also required for de novo accumulation of HP1 at both euchromatic and heterochromatic DNA damage sites to promote homologous recombination (HR) at the early stage of DDR [15], suggesting CAF-1 may function as a general HP1 loader in different cellular contexts. Based on findings described above, CAF-1 may recruit a variety of repair-associated proteins at the damaged chromatin regions via its role as a histone chaperone and/or a protein platform, contributing to both DNA repair and chromatin restoration at different stages of DDR.
A protein complex required for integrating epigenetic information during model organism development
Epigenetic regulations play essential roles in fine-tuning the transcriptional output coordinating multi-cellular organism development. In addition to its activity as histone chaperone, recent studies of CAF-1 in model organisms have provided evidence that it may also function as a protein platform integrating epigenetic information, either repressive heterochromatic markers or active histone modifications for signal transduction, both of which are required for proper tissue development [19, 20, 42].
Roles of CAF-1 in integrating heterochromatin information during development
Organization of heterochromatin in the nucleus is highly dynamic, which is required for the regulation of specific nuclear architecture and gene silencing during early stages of development [51]. In mouse embryos, pericentric heterochromatin visualized by staining of DAPI and HP1 is progressively assembled into high-order chromatin structures during early developmental stages, starting from the 2-cell stage until the 32-cell stage [42]. Houlard et al. [42] first reported CAF-1-p150, the largest subunit of CAF-1, is involved in determining heterochromatin organization during early embryo development in mice. In homozygous CAF-1-p150 mutant embryos, DAPI and HP1 signals are not enriched to form detectable chromocenters, but rather display diffused patterns in the nucleus, which is evidence of abnormal organization of heterochromatin. Furthermore, by knocking down CAF-1-p150 in cultured embryonic stem (ES) cells, Houlard and colleagues also show epigenetic markers of pericentric heterochromatin, H3K9me3 and H4K20me3, are completely abolished.
After the first 13 rapid cycles of nuclear division, Drosophila early embryos transit to the 14th cycle, initiating the early events of morphogenesis, a process called mid-blastula transition (MBT), which is associated with significant prolongation of the S phase and late replication of the heterochromatic regions [52]. During this process, progressive accumulation of HP1 occurs following the late replication events [52]; thus both HP1 and H3K9me2/3 are detectable at cycle 14 of Drosophila embryogenesis [20]. Consistent with the results observed in early mouse embryos, Huang and colleagues reported a reduction of HP1 and H3K9me2/3 signals at cycle 14 after depletion of dCAF-1-p180 [20]. Together, these reports indicate a conserved role for CAF-1 in recruiting epigenetic signals for the establishment of heterochromatin during early development in model organisms (Fig. 1b).
Early embryo development is a particular stage for de novo establishment of pericentric heterochromatin [42, 52]. It would be interesting to know whether CAF-1 regulates heterochromatin formation and/or maintenance in more cellular contexts than just ES or early embryonic cells. Studies in Drosophila provide evidence that depletion of dCAF-1-p180 suppresses gene silencing as assessed by a variety of PEV models [20], indicating the role of CAF-1 in regulating heterochromatin is not restricted in early-stage animal development. Drosophila polytene chromosomes are a wonderful system to study heterochromatin formation and maintenance and endo-replication [53–55]. Knockdown of dCAF-1-p180 does not apparently affect endo-replication or the overall development of salivary glands [20, 56], while HP1 accumulation and repressive histone modifications, namely H3K9me2/3 and H4K20me3, are significantly reduced at the chromocenter [20], indicating CAF-1 functions as well in salivary gland cells to regulate heterochromatin formation and/or maintenance. Furthermore, the physical interaction between CAF-1 and HP1 detected in both cultured cells [15, 43] and developmental processes [20] supports a general role for CAF-1 in regulating heterochromatin organization. The telomere is another well-studied heterochromatic domain in various model systems. Dysfunction of telomere maintenance is a major cause of many severe diseases and aging [57]. In yeast, it was reported that CAF-1 mutants display telomere shortening and reduced telomeric gene silencing, likely resultant from a failure of Sir spreading [38, 40]. In Arabidopsis, disruption of CAF-1 function also results in telomere shortening [58], the mechanism of which remains to be elucidated. Collectively, these studies suggest essential roles for CAF-1 in both establishing and maintaining pericentric and telomeric heterochromatin during development.
Roles of CAF-1 in integrating epigenetic information required for signal transduction during development
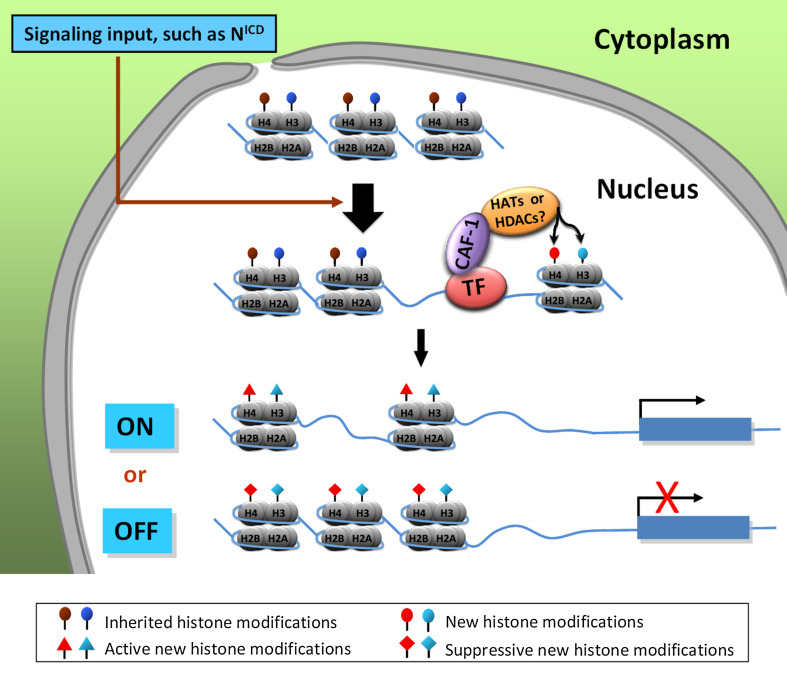
Signal transduction plays a key role in orchestrating tissue development by translating environmental and intrinsic signals into transcriptional outputs. During this process, epigenetic regulation at the chromatin level controls the accessibility of the transcriptional machinery. Chromatin remodelers and histone modifiers have been observed to be involved in this epigenetic regulatory process during model organism development through organizing different kinds of epigenetic information. In Drosophila for example, chromatin remodeling complex NURF was shown to regulate the Wg pathway by directly interacting with the transcription factor Armadillo [3]. Other studies report Trithorax-related (Trr), a histone H3K4 methyltransferase, interacts with Ecdysone receptor (EcR) to modulate its target’s expression [59]. In addition, histone chaperones Asf1 and Nap1 have been observed to specifically mediate Notch target gene repression in association with LAF (LID-associated factor) and RLAF (RPD3-LID-associated factor) complexes [6]. Of interest, our recent studies indicate the CAF-1 complex functions specifically to activate Notch target genes’ expression through a different epigenetic mechanism. Knocking down each CAF-1 subunit in developing wing imaginal discs reduces the expression of Notch targets cut and wingless, resulting in a notched wing phenotype. CAF-1 was found to physically interact with Su(H) (Suppressor of Hairless) in vivo, regulating the enrichment of active epigenetic modifications, such as histone H4 acetylation (H4ac) [19]. It remains an open question as to why and how different histone chaperones are involved in mediating different histone modifications, leading to different transcriptional outputs of either gene repression or gene activation in one specific signal transduction pathway (Fig. 2).
Fig. 2.
Possible roles of CAF-1 in signal transduction governing multi-cellular organism development, a hypothetic model. During development, specific signals, such as the Notch ligand, initiate a signaling cascade from outside of the cell, generating signals that enter the nucleus, e.g., NICD, to interact with specific transcription factors (TF), such as Su(H), recruiting CAF-1 to a specific chromatin region. In this context, CAF-1 regulates the local histone modifications (such as acetylation level) likely by interacting with specific histone modifiers such as a histone acetyltransferase (HAT) or a histone deacetylase (HDAC), which consequently result in either turning-on or shutting-down specific target genes, depending on the newly modified histone marks (active or repressive). However, it is unknown whether in this process CAF-1’s function is dependent on its histone chaperone activity
Roles of CAF-1 in maintaining genome stability during development
Extensive studies demonstrate CAF-1 is critical both for single-strand damage repair through the NER or base excision repair (BER) pathways and for DSB repair through the non-homologous end joining (NHEJ) pathway in in vitro assays and cultured cells [13, 41, 48–50, 60]. Work in plants and mouse cells also shows CAF-1 mediates homologous recombination (HR)-mediated DNA repair [15, 61, 62]. Using an in vivo model, Song and colleagues reported that reduction of dCAF-1-p180 activity directly compromises the efficiency of HR-mediated gap repair during Drosophila development [63], suggesting that during development, CAF-1 may act as a guardian of genome stability to ensure the correct passage of an integrate genome. In Arabidopsis, HR is a process that is used for precise DNA repair in somatic cells and redistribution of genetic information during meiosis [64]. T-DNA integration, a widely used tool for genetic engineering and mutagenesis, is thought to use the NHEJ mechanism [65]. Interestingly, depletion of CAF-1 strongly increases the frequency of somatic HR and T-DNA integration, which is likely due to the increased presence of DNA DSBs [62]. Mutations in yeast linker histone HHO1 result in an increased HR frequency, as is the case for the depletion of histone H4 [66, 67]. As CAF-1 is required for histone H3–H4 assembly during DNA replication processes, it is possible that CAF-1 and H3–H4 promote HR via one pathway. However, considering that CAF-1 is also required for the recruitment of DNA repair proteins during different stages of DDR in cultured mammalian cells [13, 15, 17, 22], suggesting that CAF-1 also functions in another pathway that involves its interaction with DDR proteins rather than histones. The question arises how the two pathways may integrate and/or cross talk with each other. The answer to this question will help us to understand better molecular functions of CAF-1 in maintaining genome stability.
A histone chaperone orchestrating both cell proliferation and differentiation during development
Cell proliferation and differentiation are two fundamental events determining multi-cellular organism development, leading to tissue/organ growth and specification. However, how the two processes are coordinated in different developmental contexts is still poorly understood. Previous studies in cultured cells have long suggested the major function of CAF-1 is to facilitate cell cycle progression by regulating S-phase specific chromatin duplication [22]. More recent studies, however, challenge and modify our previous idea of CAF-1 as just one of the DNA replication regulators needed for cell proliferation, suggesting that CAF-1 may also play essential roles in cell differentiation during multi-cellular organism development.
CAF-1 is essential for viability only in multi-cellular organisms
Studies have shown that CAF-1 is essential for higher eukaryotic animal development [19, 42, 56, 63, 68, 69]. Absence of CAF-1-p150, the largest subunit of CAF-1, in mice causes development to cease at the early embryonic stage [42]. In Drosophila, knocking out any of the CAF-1 subunits, namely dCAF-1-p180, dCAF-1-p105, or dCAF-1-p55, results in a similar lethal larval phenotype, indicating the CAF-1 complex is indispensable for Drosophila development [19, 56, 63, 69]. However, in uni-cellular budding yeast, cells in which each of the CAF-1 subunits are depleted are viable and do not show apparent growth defects under normal growth conditions [41]. Two possibilities may explain this viability difference between yeast cells and multi-cellular animals following CAF-1 depletion: (1) existence of redundant factor(s) in yeast cells, but not in multi-cellular organisms, which can help the cells overcome the replicative defects caused by depletion of CAF-1; or (2) the lethality in multi-cellular organisms upon the depletion of CAF-1 is not due to the defective function of CAF-1 in DNA replication-related cell proliferation, but the defective function of CAF-1 in cell differentiation during multi-cellular development.
Roles of CAF-1 in cell proliferation
It is known that, in cultured cells, CAF-1 functions as a histone chaperone during replication-coupled chromatin assembly to facilitate S-phase progression. The depletion of CAF-1 results in Chk1 activation and S-phase arrest [14, 70]. During animal development, CAF-1 is also required for rapid cell divisions in the early stages of Xenopus laevis development [71] and for S-phase progression during Danio rerio retinal development [68], which supports the in vivo roles of CAF-1 in DNA replication. However, studies in mouse ES cells show that when CAF-1-p150 is depleted by RNAi, DNA replication is still active as revealed by incorporation of bromodeoxyuridine (BrdU) and distribution of PCNA at the replication foci [42]. This observation of unaffected cell cycle upon depletion of CAF-1 seems to be consistent with the observation that yeast cells mutant for CAF-1 grow well under normal culture conditions [41] and with the observation that during Arabidopsis development, FAS1 and FAS2 (the two large subunits of CAF-1 in plants) mutants are viable although they display developmental defects [72]. Interestingly, genetic analyses in yeasts reveal that CAF-1 and Asf1 are partially redundant during cell cycle progression [73], Asf1 mutation partially rescues the synthetic lethality of cac1 and Hir double mutants [74], and triple mutant analyses show that Swi/Snf family protein Rad54 compensates for the functions of CAF-1 and Asf1 [74]. Collectively, the above observations in both uni-cellular and multi-cellular organisms suggest CAF-1 has a function in controlling cell proliferation, which is more likely the case in differentiated cells than undifferentiated cells. It will be intriguing to examine whether the counterparts of yeast Asf1 or Rad54 in higher organisms can compensate for the roles of CAF-1 in cell proliferation during multi-cellular organism development.
Roles of CAF-1 in transcriptional regulation during development
Cell differentiation is a process of cell type specification, which is controlled by the regulation of specific gene expression during development. Signaling cascades play pivotal roles in transducing extrinsic/environmental information to the nucleus, initiating specific transcription program(s) that control cell size, shape and/or metabolic activity, etc. [75]. It is known that histone modifications and chromatin remodeling control the accessibility of transcriptional apparatus and help to implant specific epigenetic “memories”; however, whether replication-coupled chromatin assembly factors are involved in this process is largely unknown. Genome-wide transcriptional analyses in Arabidopsis first show that all the subunits of CAF-1 regulate gene expression during development and are particularly involved in genes that are important for chromatin dynamics and cellular organization, as judged by the significant upregulation of these genes in mutants of different CAF-1 subunits [76]. Moreover, studies in Arabidopsis embryogenesis indicate that disruption of FAS2 and MSI1 located on the chromosomes of maternal origin affects the transcription of genes that are on the chromosomes of paternal origin [77], supporting a regulatory role of CAF-1 in transcriptional regulation. The influence of CAF-1 on the genome-wide profile of transcription during plant development may be specific because only a fraction of genes encoded by the entire genome are affected in the plants mutant for CAF-1. However, the molecular basis for how CAF-1 specifically regulates gene expression has been poorly understood.
Our recent study in Drosophila has started to provide insights into the mechanisms of how CAF-1 specifically regulates gene expression through epigenetic modifications of local chromatin during development [19]. For the first time, a connection between CAF-1 and Notch signaling has been established. Yu et al. show CAF-1 physically interacts with the Notch pathway regulator, Notch intra cellular domain (NICD), and the transcriptional activator Su(H), altering the epigenetic status of the local chromatin in the promoter regions of Notch pathway target genes. Since CAF-1 has been shown to regulate S-phase progression in several cellular contexts as discussed above, it would be interesting to assess whether the regulation of CAF-1 in Notch signaling is dependent on its function in replication-coupled nucleosome assembly. Studies both in mammalian cell lines and Drosophila have revealed Notch pathway is highly sensitive to chromatin regulators [6, 78, 79], including proteins that function in DNA replication, such as TOP2B, RFC1 and some of the MCM (mini-chromosome maintenance) components [78]. Hence, it is tempting for us to propose that transcriptional regulation mediated by CAF-1 is coordinated with DNA replication during development, but direct evidence needs to be displayed.
Roles of CAF-1 in cell differentiation
It has long been proposed that cell differentiation and chromatin duplication are coupled processes [80]. This idea is supported by emerging evidence from studies carried out using cellular reprogramming and the developing nematode nervous system [18, 81]. Embryonic stem cell (ESC) reprogramming is a robust system for investigating the roles of DNA replication-associated cell differentiation [82]. Relevant studies in ESCs reveal that successful reprogramming is associated with nucleotide incorporation during S phase, and the pluripotent conversion is blocked by DNA polymerase inhibitor [81], supporting the idea that DNA synthesis is essential for cell fusion-mediated reprogramming.
Planarians can regenerate any of the missing parts of their body from a population of pluripotent adult stem cells (ASCs) called neoblasts [83, 84]. Upon injury, neoblasts undergo extensive cell division to form the regenerating blastoma that differentiates into specific cell types [85, 86]. However, the genes involved in this regeneration process and how these genes are coordinated are not fully understood. By a systematic RNAi screen on epigenetic regulators, Zeng and colleagues found that replication-related chromatin regulators, such as the CAF-1 complex (p150, p60, and p48) and MCM 2–7, are required for the regeneration of the missing body parts, providing evidence that replication-coupled nucleosome assembly may facilitate differentiation of neoblasts [21].
In addition, it was shown by Nakano et al. [18] that in C. elegans, bilateral asymmetric cell division in the nervous system is dependent on nucleosome assembly mediated by CAF-1 and PCNA. Histone H3 carrying a small deletion (H3ΔQ125−A135) affects CAF-1-mediated nucleosome assembly and asymmetric cell division because the deletion disrupts H3 dimerization and subsequent H3–H4 tetramer formation. Moreover, whereas the core components of replication-coupled nucleosome assembly such as CAF-1 and PCNA are found to be required for this nervous cell-specific asymmetric division, Asf1 is found to be dispensable. Therefore, it requires further investigations to discover the underlying mechanism(s) how CAF-1 specifically regulates cell differentiation.
A histone chaperone versus a “protein platform” during development
Extensive in vitro studies have highlighted the idea that the chaperone activity of CAF-1 during chromatin assembly is responsible for depositing histone H3–H4 onto replicated DNA [35, 47, 87–89]. Other experiments also suggest CAF-1 is involved in new histone incorporation without immediate interruption of nascent DNA synthesis at the damaged loci [13, 17], indicating a consistent role for CAF-1 in histone deposition in vivo. Combining in vitro nucleosome assembly assay with functional genetic analysis, Nakano and colleagues showed for the first time that the chaperone activity of CAF-1 is critical for neuronal asymmetric cell fate determination [18]. To this end, it would be reasonable to hypothesize that the chaperone activity of CAF-1 is essential for all its functions involved in multi-cellular organism development.
However, as discussed in the previous sections of this review: in addition to functions that definitely require its chaperone activity, such as DNA synthesis-coupled processes, CAF-1 has also been shown to interact with epigenetic regulators and transcriptional factors in the processes of heterochromatin formation and signaling transduction [12, 19, 20, 23, 43]. In mouse ES cells, depletion of CAF-1-p150 results in severe alterations of constitutive heterochromatin hallmarks including HP1 and H3K9me3, but does not apparently affect global DNA replication and nuclear architecture, as revealed by proper BrdU incorporation and PML (promyelocytic leukemia) protein distribution [42]. Although the requirement of chaperone activity for CAF-1’s involvement in heterochromatin formation remains to be verified, an alternative mechanism is that CAF-1 functions as a protein “platform”, interacting with HP1 to facilitate the heterochromatin formation [12, 20, 42]. Recently, our lab has observed that CAF-1, as a protein complex, is required for Notch signal transduction via interaction with the Notch pathway core transcriptional factor Su(H), specifying wing margins during Drosophila development [19]. In this case, it is also unknown whether the chaperone activity of CAF-1 is required or it simply provides a “platform” to interact with specific transcription factors and epigenetic modifiers, organizing local chromatin structure so that specific cell differentiation genes can be transcribed. To distinguish these possibilities, it would be interesting to assess: (1) whether other DNA replication components, such as PCNA, are also involved in cell fate determination via the Notch pathway; and (2) whether mutations that disable H3 dimerization affect Notch pathway output differently in the presence and absence of CAF-1. Meanwhile, more detailed domain dissection that determines the CAF-1 and Su(H) physical interaction, and identification of a possible super transcriptional complex, will also help to elucidate the precise mechanism by which CAF-1 is involved in regulating the Notch signal transduction during development.
Concluding remarks and perspectives
For more than two decades, biologists have been working on elucidating the roles of histone chaperone CAF-1 in the processes of DNA replication-coupled nucleosome assembly and DNA repair. Recently, emerging studies have shown CAF-1 also plays essential roles in multi-cellular organism development by regulating heterochromatin formation, signal transduction, and transcription. In this review, we focus on the developmental roles of CAF-1 in multi-cellular organisms. Despite the lack of detailed mechanistic studies, we propose CAF-1 functions not only as a histone chaperone facilitating replication-coupled cell cycle progression, but also as a “protein platform” integrating epigenetic information by recruiting heterochromatic proteins and/or interacting with transcriptional factors to orchestrate cell proliferation and cell differentiation in specific cellular contexts. However, it is not known whether these kinds of interactions are independent of histones. In addition, it is also important to bear in mind whether a particular function of CAF-1 is dependent on the integrity of CAF-1 as a protein complex, i.e., whether the function is a specific function of a particular subunit of CAF-1. Furthermore, to get a complete picture of the roles of CAF-1 in cell proliferation, cell differentiation, and transcriptional regulation, specific questions remain to be answered in different developmental contexts. Genome-wide analyses of CAF-1 mutants should be carried out with different cell types during development. Construction of a delicate protein interactome for CAF-1 would help to address how CAF-1 links transcriptional events to replication-coupled nucleosome assembly processes. At the cellular level, high-resolution imaging systems may be critical for observing the exact behaviors of both CAF-1 and histones at the replication fork, the key to understanding epigenetic inheritance and asymmetric cell division. New technologies based on TALEN and CRISPR/Cas9 [90] may help to systematize investigation into CAF-1’s developmental functions in model organisms, such as planarians, which are the best subjects in which to study cell proliferation and differentiation through genetic manipulations.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (NSFC) [Nos. 31271573, 31228015, 31201007, 81470846] and the 973 Program [2012CB825504]. We are grateful to Gabriel Calvin and Jen Kennedy for critical reading of the manuscript. We thank the anonymous reviewers for constructive suggestions.
Footnotes
Z. Yu and J. Liu contributed equally to this work.
References
- 1.Eccleston A, Cesari F, Skipper M. Transcription and epigenetics. Nature. 2013;502(7472):461. doi: 10.1038/502461a. [DOI] [PubMed] [Google Scholar]
- 2.Landry JW, Banerjee S, Taylor B, Aplan PD, Singer A, Wu C. Chromatin remodeling complex NURF regulates thymocyte maturation. Genes Dev. 2011;25(3):275–286. doi: 10.1101/gad.2007311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song H, Spichiger-Haeusermann C, Basler K. The ISWI-containing NURF complex regulates the output of the canonical Wingless pathway. EMBO Rep. 2009;10(10):1140–1146. doi: 10.1038/embor.2009.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saitou M, Kagiwada S, Kurimoto K. Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development. 2012;139(1):15–31. doi: 10.1242/dev.050849. [DOI] [PubMed] [Google Scholar]
- 5.Cantone I, Fisher AG. Epigenetic programming and reprogramming during development. Nat Struct Mol Biol. 2013;20(3):282–289. doi: 10.1038/nsmb.2489. [DOI] [PubMed] [Google Scholar]
- 6.Moshkin YM, Kan TW, Goodfellow H, Bezstarosti K, Maeda RK, Pilyugin M, Karch F, Bray SJ, Demmers JA, Verrijzer CP. Histone chaperones ASF1 and NAP1 differentially modulate removal of active histone marks by LID-RPD3 complexes during NOTCH silencing. Mol Cell. 2009;35(6):782–793. doi: 10.1016/j.molcel.2009.07.020. [DOI] [PubMed] [Google Scholar]
- 7.Goodfellow H, Krejci A, Moshkin Y, Verrijzer CP, Karch F, Bray SJ. Gene-specific targeting of the histone chaperone asf1 to mediate silencing. Dev Cell. 2007;13(4):593–600. doi: 10.1016/j.devcel.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 8.Jullien J, Astrand C, Szenker E, Garrett N, Almouzni G, Gurdon JB. HIRA dependent H3.3 deposition is required for transcriptional reprogramming following nuclear transfer to Xenopus oocytes. Epigenet Chromatin. 2012;5(1):17. doi: 10.1186/1756-8935-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szenker E, Lacoste N, Almouzni G. A developmental requirement for HIRA-dependent H3.3 deposition revealed at gastrulation in Xenopus. Cell reports. 2012;1(6):730–740. doi: 10.1016/j.celrep.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 10.Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, Lee C, Almouzni G, Schneider R, Surani MA. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452(7189):877–881. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58(1):15–25. doi: 10.1016/0092-8674(89)90398-X. [DOI] [PubMed] [Google Scholar]
- 12.Quivy JP, Gerard A, Cook AJ, Roche D, Almouzni G. The HP1-p150/CAF-1 interaction is required for pericentric heterochromatin replication and S-phase progression in mouse cells. Nat Struct Mol Biol. 2008;15(9):972–979. doi: 10.1038/nsmb.1470. [DOI] [PubMed] [Google Scholar]
- 13.Polo SE, Roche D, Almouzni G. New histone incorporation marks sites of UV repair in human cells. Cell. 2006;127(3):481–493. doi: 10.1016/j.cell.2006.08.049. [DOI] [PubMed] [Google Scholar]
- 14.Ye X, Franco AA, Santos H, Nelson DM, Kaufman PD, Adams PD. Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol Cell. 2003;11(2):341–351. doi: 10.1016/S1097-2765(03)00037-6. [DOI] [PubMed] [Google Scholar]
- 15.Baldeyron C, Soria G, Roche D, Cook AJ, Almouzni G. HP1alpha recruitment to DNA damage by p150CAF-1 promotes homologous recombination repair. J Cell Biol. 2011;193(1):81–95. doi: 10.1083/jcb.201101030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quivy JP, Roche D, Kirschner D, Tagami H, Nakatani Y, Almouzni G. A CAF-1 dependent pool of HP1 during heterochromatin duplication. EMBO J. 2004;23(17):3516–3526. doi: 10.1038/sj.emboj.7600362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adam S, Polo SE, Almouzni G. Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell. 2013;155(1):94–106. doi: 10.1016/j.cell.2013.08.029. [DOI] [PubMed] [Google Scholar]
- 18.Nakano S, Stillman B, Horvitz HR. Replication-coupled chromatin assembly generates a neuronal bilateral asymmetry in C. elegans . Cell. 2011;147(7):1525–1536. doi: 10.1016/j.cell.2011.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu Z, Wu H, Chen H, Wang R, Liang X, Liu J, Li C, Deng WM, Jiao R. CAF-1 promotes Notch signaling through epigenetic control of target gene expression during Drosophila development. Development. 2013;140(17):3635–3644. doi: 10.1242/dev.094599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang H, Yu Z, Zhang S, Liang X, Chen J, Li C, Ma J, Jiao R. Drosophila CAF-1 regulates HP1-mediated epigenetic silencing and pericentric heterochromatin stability. J Cell Sci. 2010;123(Pt 16):2853–2861. doi: 10.1242/jcs.063610. [DOI] [PubMed] [Google Scholar]
- 21.Zeng A, Li YQ, Wang C, Han XS, Li G, Wang JY, Li DS, Qin YW, Shi Y, Brewer G, Jing Q. Heterochromatin protein 1 promotes self-renewal and triggers regenerative proliferation in adult stem cells. J Cell Biol. 2013;201(3):409–425. doi: 10.1083/jcb.201207172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128(4):721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 23.Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008;134(2):244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459(7243):113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tjeertes JV, Miller KM, Jackson SP. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J. 2009;28(13):1878–1889. doi: 10.1038/emboj.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vempati RK, Jayani RS, Notani D, Sengupta A, Galande S, Haldar D. p300-mediated acetylation of histone H3 lysine 56 functions in DNA damage response in mammals. J Biol Chem. 2010;285(37):28553–28564. doi: 10.1074/jbc.M110.149393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burgess RJ, Zhou H, Han J, Zhang Z. A role for Gcn5 in replication-coupled nucleosome assembly. Mol Cell. 2010;37(4):469–480. doi: 10.1016/j.molcel.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allis CD, Chicoine LG, Richman R, Schulman IG. Deposition-related histone acetylation in micronuclei of conjugating Tetrahymena. Proc Natl Acad Sci USA. 1985;82(23):8048–8052. doi: 10.1073/pnas.82.23.8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sobel RE, Cook RG, Allis CD. Non-random acetylation of histone H4 by a cytoplasmic histone acetyltransferase as determined by novel methodology. J Biol Chem. 1994;269(28):18576–18582. [PubMed] [Google Scholar]
- 30.Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci USA. 1995;92(4):1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87(1):95–104. doi: 10.1016/S0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 32.Zhang H, Han J, Kang B, Burgess R, Zhang Z. Human histone acetyltransferase 1 protein preferentially acetylates H4 histone molecules in H3.1-H4 over H3.3-H4. J Biol Chem. 2012;287(9):6573–6581. doi: 10.1074/jbc.M111.312637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma XJ, Wu J, Altheim BA, Schultz MC, Grunstein M. Deposition-related sites K5/K12 in histone H4 are not required for nucleosome deposition in yeast. Proc Natl Acad Sci USA. 1998;95(12):6693–6698. doi: 10.1073/pnas.95.12.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mello JA, Sillje HH, Roche DM, Kirschner DB, Nigg EA, Almouzni G. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep. 2002;3(4):329–334. doi: 10.1093/embo-reports/kvf068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96(4):575–585. doi: 10.1016/S0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- 36.Winkler DD, Zhou H, Dar MA, Zhang Z, Luger K. Yeast CAF-1 assembles histone (H3-H4)2 tetramers prior to DNA deposition. Nucleic Acids Res. 2012;40(20):10139–10149. doi: 10.1093/nar/gks812. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Maison C, Almouzni G. HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol. 2004;5(4):296–304. doi: 10.1038/nrm1355. [DOI] [PubMed] [Google Scholar]
- 38.Huang S, Zhou H, Tarara J, Zhang Z. A novel role for histone chaperones CAF-1 and Rtt106p in heterochromatin silencing. EMBO J. 2007;26(9):2274–2283. doi: 10.1038/sj.emboj.7601670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Enomoto S, Berman J. Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev. 1998;12(2):219–232. doi: 10.1101/gad.12.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Enomoto S, McCune-Zierath PD, Gerami-Nejad M, Sanders MA, Berman J. RLF2, a subunit of yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes Dev. 1997;11(3):358–370. doi: 10.1101/gad.11.3.358. [DOI] [PubMed] [Google Scholar]
- 41.Kaufman PD, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 1997;11(3):345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 42.Houlard M, Berlivet S, Probst AV, Quivy JP, Hery P, Almouzni G, Gerard M. CAF-1 is essential for heterochromatin organization in pluripotent embryonic cells. PLoS Genet. 2006;2(11):e181. doi: 10.1371/journal.pgen.0020181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loyola A, Tagami H, Bonaldi T, Roche D, Quivy JP, Imhof A, Nakatani Y, Dent SY, Almouzni G. The HP1alpha-CAF1-SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin. EMBO Rep. 2009;10(7):769–775. doi: 10.1038/embor.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamburini BA, Tyler JK. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol. 2005;25(12):4903–4913. doi: 10.1128/MCB.25.12.4903-4913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Papamichos-Chronakis M, Krebs JE, Peterson CL. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev. 2006;20(17):2437–2449. doi: 10.1101/gad.1440206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gaillard PH, Martini EM, Kaufman PD, Stillman B, Moustacchi E, Almouzni G. Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell. 1996;86(6):887–896. doi: 10.1016/S0092-8674(00)80164-6. [DOI] [PubMed] [Google Scholar]
- 48.Nabatiyan A, Szuts D, Krude T. Induction of CAF-1 expression in response to DNA strand breaks in quiescent human cells. Mol Cell Biol. 2006;26(5):1839–1849. doi: 10.1128/MCB.26.5.1839-1849.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiao R, Harrigan JA, Shevelev I, Dietschy T, Selak N, Indig FE, Piotrowski J, Janscak P, Bohr VA, Stagljar I. The Werner syndrome protein is required for recruitment of chromatin assembly factor 1 following DNA damage. Oncogene. 2007;26(26):3811–3822. doi: 10.1038/sj.onc.1210150. [DOI] [PubMed] [Google Scholar]
- 50.Jiao R, Bachrati CZ, Pedrazzi G, Kuster P, Petkovic M, Li JL, Egli D, Hickson ID, Stagljar I. Physical and functional interaction between the Bloom’s syndrome gene product and the largest subunit of chromatin assembly factor 1. Mol Cell Biol. 2004;24(11):4710–4719. doi: 10.1128/MCB.24.11.4710-4719.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fussner E, Djuric U, Strauss M, Hotta A, Perez-Iratxeta C, Lanner F, Dilworth FJ, Ellis J, Bazett-Jones DP. Constitutive heterochromatin reorganization during somatic cell reprogramming. EMBO J. 2011;30(9):1778–1789. doi: 10.1038/emboj.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shermoen AW, McCleland ML, O’Farrell PH. Developmental control of late replication and S phase length. Curr Biol. 2010;20(23):2067–2077. doi: 10.1016/j.cub.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lis JT. Imaging Drosophila gene activation and polymerase pausing in vivo. Nature. 2007;450(7167):198–202. doi: 10.1038/nature06324. [DOI] [PubMed] [Google Scholar]
- 54.Ebert A, Lein S, Schotta G, Reuter G. Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res. 2006;14(4):377–392. doi: 10.1007/s10577-006-1066-1. [DOI] [PubMed] [Google Scholar]
- 55.Zielke N, Kim KJ, Tran V, Shibutani ST, Bravo MJ, Nagarajan S, van Straaten M, Woods B, von Dassow G, Rottig C, Lehner CF, Grewal SS, Duronio RJ, Edgar BA. Control of Drosophila endocycles by E2F and CRL4(CDT2) Nature. 2011;480(7375):123–127. doi: 10.1038/nature10579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klapholz B, Dietrich BH, Schaffner C, Heredia F, Quivy JP, Almouzni G, Dostatni N. CAF-1 is required for efficient replication of euchromatic DNA in Drosophila larval endocycling cells. Chromosoma. 2009;118(2):235–248. doi: 10.1007/s00412-008-0192-2. [DOI] [PubMed] [Google Scholar]
- 57.Calado R, Young N. Telomeres in disease. F1000 Med Rep. 2012;4:8. doi: 10.3410/M4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mozgova I, Mokros P, Fajkus J. Dysfunction of chromatin assembly factor 1 induces shortening of telomeres and loss of 45S rDNA in Arabidopsis thaliana . Plant Cell. 2010;22(8):2768–2780. doi: 10.1105/tpc.110.076182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sedkov Y, Cho E, Petruk S, Cherbas L, Smith ST, Jones RS, Cherbas P, Canaani E, Jaynes JB, Mazo A. Methylation at lysine 4 of histone H3 in ecdysone-dependent development of Drosophila. Nature. 2003;426(6962):78–83. doi: 10.1038/nature02080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Linger J, Tyler JK. The yeast histone chaperone chromatin assembly factor 1 protects against double-strand DNA-damaging agents. Genetics. 2005;171(4):1513–1522. doi: 10.1534/genetics.105.043000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kirik A, Pecinka A, Wendeler E, Reiss B. The chromatin assembly factor subunit FASCIATA1 is involved in homologous recombination in plants. Plant Cell. 2006;18(10):2431–2442. doi: 10.1105/tpc.106.045088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Endo M, Ishikawa Y, Osakabe K, Nakayama S, Kaya H, Araki T, Shibahara K, Abe K, Ichikawa H, Valentine L, Hohn B, Toki S. Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. The EMBO journal. 2006;25(23):5579–5590. doi: 10.1038/sj.emboj.7601434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Song Y, He F, Xie G, Guo X, Xu Y, Chen Y, Liang X, Stagljar I, Egli D, Ma J, Jiao R. CAF-1 is essential for Drosophila development and involved in the maintenance of epigenetic memory. Dev Biol. 2007;311(1):213–222. doi: 10.1016/j.ydbio.2007.08.039. [DOI] [PubMed] [Google Scholar]
- 64.Schuermann D, Molinier J, Fritsch O, Hohn B. The dual nature of homologous recombination in plants. Trends Genet. 2005;21(3):172–181. doi: 10.1016/j.tig.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 65.Friesner J, Britt AB. Ku80- and DNA ligase IV-deficient plants are sensitive to ionizing radiation and defective in T-DNA integration. Plant J. 2003;34(4):427–440. doi: 10.1046/j.1365-313X.2003.01738.x. [DOI] [PubMed] [Google Scholar]
- 66.Downs JA, Kosmidou E, Morgan A, Jackson SP. Suppression of homologous recombination by the Saccharomyces cerevisiae linker histone. Mol Cell. 2003;11(6):1685–1692. doi: 10.1016/S1097-2765(03)00197-7. [DOI] [PubMed] [Google Scholar]
- 67.Prado F, Aguilera A. Partial depletion of histone H4 increases homologous recombination-mediated genetic instability. Mol Cell Biol. 2005;25(4):1526–1536. doi: 10.1128/MCB.25.4.1526-1536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fischer S, Prykhozhij S, Rau MJ, Neumann CJ. Mutation of zebrafish caf-1b results in S phase arrest, defective differentiation, and p53-mediated apoptosis during organogenesis. Cell Cycle. 2007;6(23):2962–2969. doi: 10.4161/cc.6.23.4950. [DOI] [PubMed] [Google Scholar]
- 69.Wen P, Quan Z, Xi R. The biological function of the WD40 repeat-containing protein p55/Caf1 in Drosophila. Dev Dyn. 2012;241(3):455–464. doi: 10.1002/dvdy.23730. [DOI] [PubMed] [Google Scholar]
- 70.Hoek M, Stillman B. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc Natl Acad Sci USA. 2003;100(21):12183–12188. doi: 10.1073/pnas.1635158100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Quivy JP, Grandi P, Almouzni G. Dimerization of the largest subunit of chromatin assembly factor 1: importance in vitro and during Xenopus early development. EMBO J. 2001;20(8):2015–2027. doi: 10.1093/emboj/20.8.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaya H, Shibahara KI, Taoka KI, Iwabuchi M, Stillman B, Araki T. FASCIATA genes for chromatin assembly factor-1 in arabidopsis maintain the cellular organization of apical meristems. Cell. 2001;104(1):131–142. doi: 10.1016/S0092-8674(01)00197-0. [DOI] [PubMed] [Google Scholar]
- 73.Kats ES, Albuquerque CP, Zhou H, Kolodner RD. Checkpoint functions are required for normal S-phase progression in Saccharomyces cerevisiae RCAF- and CAF-I-defective mutants. Proc Natl Acad Sci USA. 2006;103(10):3710–3715. doi: 10.1073/pnas.0511102103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haber JE, Braberg H, Wu Q, Alexander R, Haase J, Ryan C, Lipkin-Moore Z, Franks-Skiba KE, Johnson T, Shales M, Lenstra TL, Holstege FC, Johnson JR, Bloom K, Krogan NJ. Systematic triple-mutant analysis uncovers functional connectivity between pathways involved in chromosome regulation. Cell reports. 2013;3(6):2168–2178. doi: 10.1016/j.celrep.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pires-daSilva A, Sommer RJ. The evolution of signalling pathways in animal development. Nat Rev Genet. 2003;4(1):39–49. doi: 10.1038/nrg977. [DOI] [PubMed] [Google Scholar]
- 76.Schonrock N, Exner V, Probst A, Gruissem W, Hennig L. Functional genomic analysis of CAF-1 mutants in Arabidopsis thaliana. J Biol Chem. 2006;281(14):9560–9568. doi: 10.1074/jbc.M513426200. [DOI] [PubMed] [Google Scholar]
- 77.Autran D, Baroux C, Raissig MT, Lenormand T, Wittig M, Grob S, Steimer A, Barann M, Klostermeier UC, Leblanc O, Vielle-Calzada JP, Rosenstiel P, Grimanelli D, Grossniklaus U. Maternal epigenetic pathways control parental contributions to Arabidopsis early embryogenesis. Cell. 2011;145(5):707–719. doi: 10.1016/j.cell.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 78.Yatim A, Benne C, Sobhian B, Laurent-Chabalier S, Deas O, Judde JG, Lelievre JD, Levy Y, Benkirane M. NOTCH1 nuclear interactome reveals key regulators of its transcriptional activity and oncogenic function. Mol Cell. 2012;48(3):445–458. doi: 10.1016/j.molcel.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mourikis P, Lake RJ, Firnhaber CB, DeDecker BS. Modifiers of notch transcriptional activity identified by genome-wide RNAi. BMC Dev Biol. 2010;10:107. doi: 10.1186/1471-213X-10-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mueller GC, Kajiwara K, Kim UH, Graham J. Proposed coupling of chromatin replication, hormone action, and cell differentiation. Cancer Res. 1978;38(11 Pt 2):4041–4045. [PubMed] [Google Scholar]
- 81.Tsubouchi T, Soza-Ried J, Brown K, Piccolo FM, Cantone I, Landeira D, Bagci H, Hochegger H, Merkenschlager M, Fisher AG. DNA synthesis is required for reprogramming mediated by stem cell fusion. Cell. 2013;152(4):873–883. doi: 10.1016/j.cell.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chia G, Egli D. Connecting the cell cycle with cellular identity. Cell Reprogram. 2013;15(5):356–366. doi: 10.1089/cell.2013.0041. [DOI] [PubMed] [Google Scholar]
- 83.Newmark PA, Sanchez Alvarado A. Not your father’s planarian: a classic model enters the era of functional genomics. Nat Rev Genet. 2002;3(3):210–219. doi: 10.1038/nrg759. [DOI] [PubMed] [Google Scholar]
- 84.Wagner DE, Wang IE, Reddien PW. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science. 2011;332(6031):811–816. doi: 10.1126/science.1203983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Newmark PA, Sanchez Alvarado A. Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev Biol. 2000;220(2):142–153. doi: 10.1006/dbio.2000.9645. [DOI] [PubMed] [Google Scholar]
- 86.Wenemoser D, Lapan SW, Wilkinson AW, Bell GW, Reddien PW. A molecular wound response program associated with regeneration initiation in planarians. Genes Dev. 2012;26(9):988–1002. doi: 10.1101/gad.187377.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kaufman PD, Kobayashi R, Kessler N, Stillman B. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell. 1995;81(7):1105–1114. doi: 10.1016/S0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- 88.Smith S, Stillman B. Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 1991;10(4):971–980. doi: 10.1002/j.1460-2075.1991.tb08031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tyler JK, Collins KA, Prasad-Sinha J, Amiott E, Bulger M, Harte PJ, Kobayashi R, Kadonaga JT. Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol Cell Biol. 2001;21(19):6574–6584. doi: 10.1128/MCB.21.19.6574-6584.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wei C, Liu J, Yu Z, Zhang B, Gao G, Jiao R. TALEN or Cas9, rapid, efficient and specific choices for genome modifications. J Genet Genom. 2013;40(6):281–289. doi: 10.1016/j.jgg.2013.03.013. [DOI] [PubMed] [Google Scholar]