Abstract
Centriolar satellites are small, microscopically visible granules that cluster around centrosomes. These structures, which contain numerous proteins directly involved in centrosome maintenance, ciliogenesis, and neurogenesis, have traditionally been viewed as vehicles for protein trafficking towards the centrosome. However, the recent identification of several new centriolar satellite components suggests that this model offers only an incomplete picture of their cellular functions. While the mechanisms controlling centriolar satellite status and function are not yet understood in detail, emerging evidence points to these structures as important hubs for dynamic, multi-faceted regulation in response to a variety of cues. In this review, we summarize the current knowledge of the roles of centriolar satellites in regulating centrosome functions, ciliogenesis, and neurogenesis. We also highlight newly discovered regulatory mechanisms targeting centriolar satellites and their functional status, and we discuss how defects in centriolar satellite components are intimately linked to a wide spectrum of human diseases.
Keywords: Centriolar satellites, Centrosome, Ciliogenesis, Neurogenesis, PCM1, Protein transport
Introduction
Centriolar satellites (CS) are small, spherical granules with a diameter of approximately 70–100 nm that cluster in the vicinity of the centrosome (Fig. 1a, b). They can be visualized as small, electron-dense entities by electron microscopy and as small punctate structures by fluorescence microscopy [1–4]. These structures were first observed during studies of the ultrastructure of the centrosome using electron microscopy, but until the mid-1990s they received little attention [1, 2, 5]. Indeed, to this day these structures remain poorly studied.
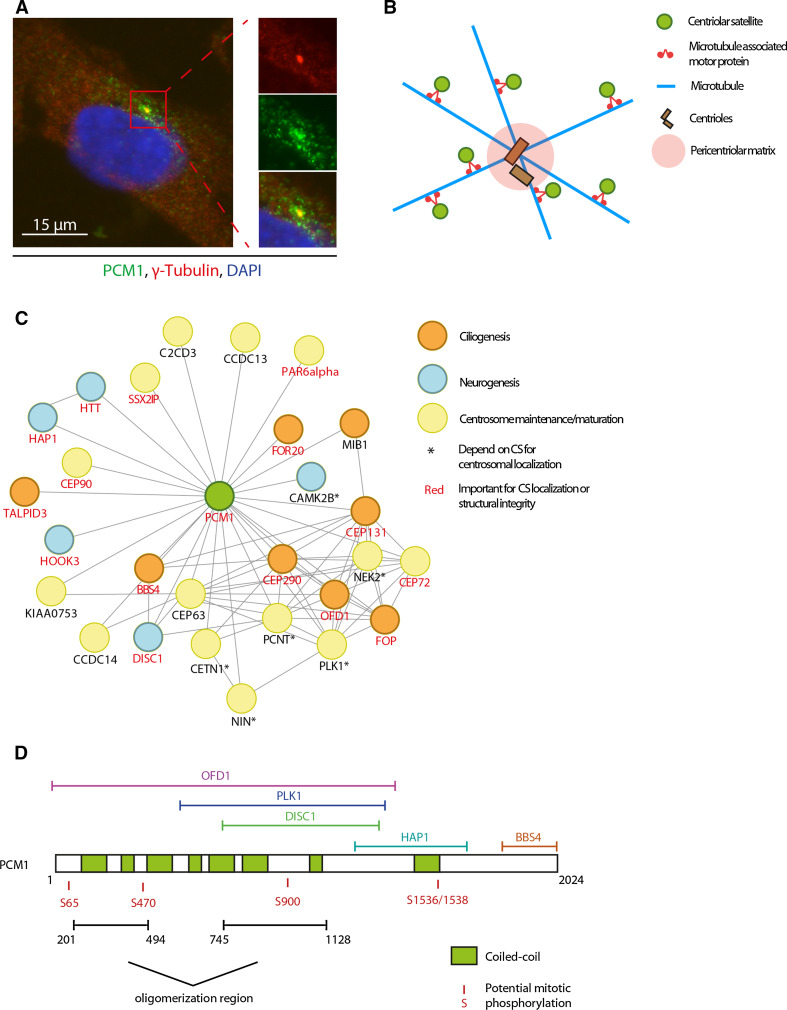
Fig. 1.
Structure and composition of centriolar satellites. a Immunofluorescence staining of centriolar satellites in U2OS cells. CS are visualized by PCM1 staining (green), γ-tubulin (red) is used as a marker for the centrosome. b Schematic representation of CS in interphase cells. CS cluster around the centrosome and are, to a lesser extent, distributed throughout the cytoplasm. CS are anchored to and move across the microtubules through microtubule-associated motor proteins such as dynein and kinesin. c Representation of known and predicted protein–protein interactions among proteins localizing to or interacting with CS. Proteins marked with an asterisk are, at least partially, dependent on CS for their centrosomal localization, proteins highlighted in red are important for the structural integrity or subcellular localization of satellites in general. Network was created using the STRING database (version 9.1) [101]. Interactions between PCM1 and SSX2IP, CEP63, CEP90, CAMK2B, C2CD3, Par6α, CCDC13, CCDC14, KIAA0753, and TALPID3 were added manually based on the literature. d Schematic representation of the PCM1 protein showing known protein interaction regions, functional domains, and potential mitotic phosphorylation sites
The centrosome in mammalian cells is best known for its capability to nucleate microtubules and its function as a microtubule-organizing center (MTOC). However, it is now firmly established that the function of the centrosome extends far beyond its role as a MTOC. Thus, it acts as a multifunctional platform for numerous signaling processes and is functionally implicated in processes such as cell cycle progression, cell polarity, migration, mitosis, and ciliogenesis (extensively reviewed in [6] and [7]). In contrast to CS, the centrosome is an extensively studied cellular organelle with a well-defined structure. It consists of a mother and daughter centriole, which can be visualized by electron microscopy as two perpendicularly organized tubulin-containing and barrel-shaped entities of ~200 × 500 nm with a distinct ninefold radial symmetry. A cloud of fibrous material, termed the pericentriolar matrix, surrounds the two centrioles. The pericentriolar matrix consists of high concentrations of centrosomal proteins involved in the cellular processes mentioned above. A range of proteins bind directly to the centrioles and are, depending on their localization, referred to as subdistal or distal appendages.
In addition to their striking enrichment around the centrosome, early observations described CS as clustering in the vicinity of the basal body of ciliated cells [1–4]. This basal body, which contains a differentiated form of the centrosome, can be formed when cells exit the cell cycle and the centrioles migrate to the apical side of the cell. Once the basal body is correctly situated at the plasma membrane, it is able to promote nucleation and maintenance of axonemal microtubules from the mother centriole, forming the ciliary backbone [8]. Although CS are detectable in almost all mammalian cell types, their size, molecular composition, abundance, and localization can vary considerably [3]. They are only detectable in interphase cells and undergo dissolution during mitosis [3]. While CS are concentrated around the centrosome, and distributed to a lesser extent throughout the cytoplasm in interphase cells, they coalesce at the centrosome upon mitotic entry after which they gradually disappear. Upon completion of cytokinesis they reassemble and regain their original localization [3].
While dedicated studies of CS remain scarce, they are generally thought to be microtubule-anchored protein complexes that are transported towards the centrosome, and which may play important regulatory roles in centrosome biology [9]. PCM1, the first CS protein discovered, has traditionally been viewed as a fundamental component, and is often used as a marker to identify these structures [1–4]. In agreement with this, PCM1 is required for CS assembly, and depleting PCM1 from cells is a commonly used means of perturbing their cellular functions. At the molecular level, PCM1 is thought to function as a scaffold for other proteins that reside within the satellites (Fig. 1c). PCM1 contains 8 predicted coiled-coil motifs, most of which are present in the N-terminus of the protein (Fig. 1d). Moreover, based on the few available mapping studies of PCM1-associated proteins, these coiled-coil domains appear to mediate the majority of its protein–protein interactions [10–13]. To date, approximately 30 proteins have been identified as bona fide CS components, and with the notable exception of PCM1, all of these also localize to the centrosome and/or have reported functions at either the centrosome or the basal body. In particular, CS have been shown to harbor many proteins essential for ciliogenesis [10, 14], microtubule organization [4], mitotic spindle pole maintenance [15, 16], centrosome duplication [17, 18], dendritic patterning [19] and neuronal development [11, 20].
Centriolar satellites as regulators of centrosomal proteostasis
The centrosome is involved in a variety of processes and signaling pathways that require timely recruitment and exchange of proteins. CS harbor a range of proteins that are also present at the centrosome, and a prevalent view of CS function is that they ensure the stable supply of such factors to centrosomes, thus regulating centrosomal proteostasis [9]. CS may regulate protein composition of the centrosome and basal body by either microtubule- and dynein/dynactin-dependent active transport, or by sequestering centrosomal proteins by retaining them at the satellites.
One of the earliest examples of dynein- and CS-mediated transport is the centrosomal recruitment of ninein, centrin, and pericentrin, three proteins involved in microtubule anchorage at the centrosome [4, 21–23]. Ablation of CS by knockdown of PCM1 leads to reduced concentrations of these proteins at the centrosome and results in failure to anchor microtubules and to establish a radial microtubule network [4, 22, 23]. CS depend on an intact microtubule network and require a direct interaction with dynein/dynactin motor proteins for correct localization and cargo delivery [2, 24–26]. This physical coupling between the satellites and microtubule-associated motor proteins can be mediated by several CS proteins such as BBS4, Par6α, and CEP290 [24–26]. Hence, satellite transport can be suppressed by depolymerization of microtubules (e.g. by colcemide or vanadate treatment), inhibition of dynein motor proteins, or ablation of dynactin, which mediates contacts between dynein and CS components [4].
Similar to the case of ninein, centrin, and pericentrin, additional components such as Nek2A and CaMKIIβ depend on CS-mediated transport for optimal enrichment at the centrosome [19, 27]. However, there seems to be no universal requirement of CS for targeting proteins to the centrosome, as the centrosomal levels of several other factors, such as OFD1 and CEP290, are largely unaffected by PCM1 status [10, 26]. Moreover, disruption of CS has been suggested to increase the centrosomal localization of several proteins that are normally present at the satellites, including CEP72 and CEP90 [15, 28, 29]. These proteins instead appear to be recruited to centrosomes via mechanisms other than satellite-mediated transport, such as intrinsic centrosomal targeting domains (e.g. LisH or coiled-coil domains). The notion that the presence of CS reduces the concentration of the latter proteins at the centrosome suggests that these structures can serve as temporary storage sites, thereby enabling fine-tuning of the protein composition at the centrosome. This aspect of CS function may be important for ensuring centrosome protein homeostasis in several settings. During mitotic entry, for example, the duplicated centrosomes must be separated for the spindle poles to form [7, 30–33]. Several CS proteins, including CEP72, CEP90, CEP131 (also known as AZI1) and the recently identified factors SSX2IP and CCDC13 have been implicated in centrosome and spindle pole maintenance in mitosis [15, 16, 18, 34, 35]. An increased influx of these proteins resulting from CS dissolution upon mitotic entry might be required to ensure an adequate concentration of these proteins at the centrosome during the chromosome segregation process [33]. Alternatively, it has been proposed that CS may protect some of these factors from degradation, serve as chaperones, or shield them from engaging in unwanted protein–protein interactions [36, 37].
Centriolar satellites in centrosome maintenance and maturation
Several CS proteins play roles in centrosome duplication, separation, and spindle pole assembly. These include CEP72 and CEP90, which localize to satellites and have been shown to interact with one or more CS components [15, 29, 38]. CEP90 and CEP72 are required for maintaining spindle pole stability and chromosome alignment during metaphase through the recruitment of Kizuna and γ-tubulin [15, 28, 39]. Knockdown of CEP72 or CEP90 causes spindle pole fragmentation, resulting in detrimental mono- or multipolar spindles. Concurrently, the spindle checkpoint is not activated as a direct effect of chromosome misalignment and spindle pole fragmentation, which leads to an increase of apoptotic cells or mitotic arrest [15, 28].
CEP131 and CCDC13 are two other CS factors required for faithful chromosome segregation [34, 35]. Both proteins physically interact with PCM1 and localize to satellites as well as to the centrosome [34, 35]. Cells depleted of CEP131 or CCDC13 display similar phenotypes that include an increased frequency of multipolar spindle poles and formation of micronuclei and chromatin bridges [34]. In addition, cells that manage to complete mitosis display elevated levels of DNA damage markers such as phosphorylated H2AX (γH2AX), phospho-ATM and phospho-Chk2, most likely reflecting post-mitotic DNA damage resulting from chromosome missegregation [34].
These observations highlight the involvement of CS factors in mitotic centrosome function and faithful chromosome segregation. However, CEP72, CEP90, Kizuna, CEP131, and CCDC13 are not dependent on the satellites for their centrosomal localization. In addition, a study of mitotic catastrophe and cell death resulting from depletion of a number of centrosomal proteins did not observe significant detrimental consequences of PCM1 knockdown [40]. Hence, while the precise role(s) of CS in spindle pole maintenance is uncertain, the above observations raise the possibility that CS proteins may have specific functions at satellites in addition to their established roles at the centrosome.
Centriolar satellites contain many proteins involved in the recruitment of core centriolar factors and have been implicated on a number of occasions in centrosome maturation and duplication [18, 41, 42]. Nek2A is a cell cycle-regulated kinase that facilitates entry into mitosis, centrosome separation, and spindle pole formation [43]. A study on the spatiotemporal dynamics of Nek2A localization revealed a supporting role of CS in recruiting Nek2A and its substrate C-Nap1 to the centrosome [27]. Another example is the centrosome maturation factor C2CD3 that localizes to CS in a PCM1-dependent manner. Lack of centrosomal C2CD3 results in perturbed recruitment of centriolar distal appendage proteins, manifesting as severe defects during ciliogenesis [42].
Similar to DNA, the centrosome must be duplicated once and only once during each cell cycle. Indeed, DNA replication and centrosome duplication occur synchronously and these processes are controlled by the same cyclin-dependent kinases (CDKs) [44, 45]. Centriole duplication is a hierarchical and tightly regulated process that requires the timely recruitment of numerous specific factors [31]. Interestingly, CEP63, one such factor required for centriole duplication, has recently been identified as a component of CS through immunofluorescence and proximity interaction screens [46, 47]. While the centrosomal targeting of CEP63 is dependent on its interaction with CEP152, the factors CCDC14 and KIAA0753 also exert control over this localization. Thus, CCDC14 limits the availability of CEP63 to the centrosome, a process that is antagonized by KIAA0753. Strikingly, both CCDC14 and KIAA0753 also localize to CS, suggesting that in this setting the satellites might function as temporary storage containers, as described previously [15, 47]. Further supporting the alleged roles of CS in centrosome biogenesis and maintenance, disruption of CS integrity, for example by PCM1 knock down, can lead to a marked increase in fragmentation or even loss of centrosomes [17, 48]. In response to such perturbations, the stress responsive p38 mitogen-activated protein kinase (p38MAPK) is activated, phosphorylating p53 and p21 to promote cell cycle arrest through CDK2 inhibition [48]. Arrested cells with dysfunctional centrosomes are generally unable to differentiate or form functional cilia [48]. In further support of a role for CS in centrosome maintenance, PCM1 depletion can also manifest as a G1 arrest through the activation of the p38-p53 signaling axis, similar to the response to centrosome fragmentation [17, 48]. Consequently, upon depletion of PCM1, both DNA replication and cell cycle progression can be restored by deletion of p53 or chemical inhibition of the p38 kinase, although these cells display defective spindle poles and aneuploidy as a consequence of dysfunctional centrosomes.
Centriolar satellites and ciliogenesis
Primary cilia are antenna-like structures that originate from the centrosome and protrude from the cell body for up to a few micrometers. The central scaffold is composed of a rigid microtubule network that extends away from the cell surface but is still covered by the cell membrane. Primary cilia serve as “sensing antennae” to detect various extracellular signals and perform specialized sensory tasks (reviewed in [8] and [49]). To this end, cilia are richly decorated with various receptors (e.g. odorant receptors and rhodopsin) to establish olfaction and vision, and are fundamental for key signal transduction responses including the Wnt and hedgehog signaling pathways. Additionally, nodal cilia, which generate the leftward flow of extra-embryonic fluid, are required to establish correct left–right asymmetry during embryonic development in vertebrates [50]. CS are recognized as key regulators of ciliogenesis, most likely by controlling licensing of centriole maturation and integrating signals from the cilium, the cell interior, and the cell cycle machinery (Fig. 2) [51, 52]. As mentioned above, CS factors such as C2CD3 facilitate the recruitment of distal appendage proteins such as Ttkb2, CEP164, Ift88 and Ift52 during ciliogenesis [42]. More specifically, Ttkb2 is required for removal of CP110, which serves as an antagonizer of ciliogenesis, while CEP164 is a key factor for establishing the primary cilium as discovered in a microscopy-based siRNA screen [14, 53]. Ift88 and Ift52 are crucial proteins involved in intra-flagellar transport (IFT), which mediates cargo transport inside the cilium [54].
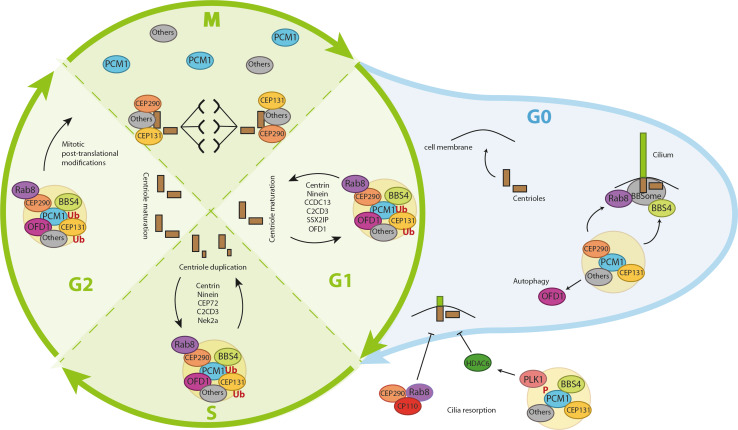
Fig. 2.
Dynamic regulation of centriolar satellites through the cell cycle. CS are only detected in interphase cells and are involved in maintaining centrosomal proteostasis by storing or actively transporting centrosomal proteins throughout interphase. Consequently, CS are involved in the maturation and duplication of the centrosome. In addition, CS are centrally involved in ciliogenesis. Upon cell cycle exit, MIB1-mediated ubiquitylation is suppressed and CS facilitate the recruitment of various factors to the ciliary transition zone. Upon cell cycle re-entry, the primary cilium is disassembled. CS facilitate this process through two separate pathways, one involving PCM1-dependent PLK1 loading to CS and activation of HDAC6, and another causing inhibition of CEP290 and Rab8a transport by CP110. Subsequently, upon mitotic entry, centriolar satellites are dissolved followed by redistribution of numerous centriolar satellite factors to the centrosome
The role of CS in ciliogenesis is perhaps best underscored by the crucial role of the BBSome in the process. The BBSome is a stable multi-protein complex localizing to the ciliary transition zone, which consists of at least seven core components (BBS1, BBS2, BBS4, BBS5, BBS7, BBS8, and BBS9). Current models posit a specific role for the BBSome in protrusion of the microtubule network from the ciliary base by recruitment of internal cell membrane proteins to the growing structure [36, 55]. Mouse models with targeted disruption of BBS components have shown that loss of the BBSome perturbs trafficking of cilia membrane precursors and compromises cilium formation [56–59]. In humans, mutations in any of the BBS proteins are causative of the Bardet–Biedl Syndrome (BBS-OMIM209900). BBS patients display a high frequency of renal failure/abnormalities, obesity, retinal dystrophy, mental retardation, and limb malformations (e.g. polydactyly). It should be noted, however, that BBS and other ciliopathies are genetically heterogenous disorders, and linking clinical phenotypes to specific mutations has, therefore, proven to be challenging [60, 61].
BBSome assembly occurs in a highly hierarchical manner, with BBS4 being the last added component [62]. Interestingly, BBS4 is the only BBSome factor that stably localizes to CS through interactions with PCM1, CEP131, and CEP290 [37, 63]. BBS4 binds the extreme C-terminus of PCM1 and localizes to satellites, the centrosome, and the basal body in ciliated cells [10, 13, 24]. BBS4 was also shown to impact CS localization by bridging the interaction between PCM1 and dynein/dynactin, but more recently it has been established that BBS4 itself depends on CS for recruitment to the basal body and subsequent BBSome formation [13, 24, 37, 63]. In 2007, Nachury and colleagues suggested a model in which CS could spatially limit BBSome activation to the basal body by acting as a chaperone for BBS4 during cytoplasmic transport [36]. This hypothesis has recently been strengthened through a study on CEP131. CEP131 not only binds PCM1 and BBS4 at CS, but it also inhibits BBS4 recruitment to the basal body, and cells depleted of CEP131 display increased accumulation of BBS4 and the BBSome at the ciliary transition zone [37]. Furthermore, zebrafish morphants lacking CEP131 are visually impaired and display delayed melanosome trafficking, perturbed Kupffer’s vesicles, and randomized left–right asymmetry in embryos, phenotypes that are all related to defects in ciliogenesis [37, 64]. In addition, the Drosophila CEP131 orthologue, DILA, localizes to the basal body in ciliating cells and is required for ciliogenesis in sensory neurons and germ cells [65]. Together, these results suggest that CEP131 and CS are important mediators of ciliogenesis through regulation of BBSome accumulation at cilia [37]. However, as CEP131 also localizes to the centrosome independently of CS, it is equally possible that the observed phenotypes can be attributed to a centrosome-specific defect rather than perturbed CS functionality. Interestingly, whereas cells transiently depleted of CEP131 are clearly impaired in ciliogenesis in a number of cell culture models and lower organisms, possibly as a consequence of perturbed BBSome trafficking, a recent study demonstrated, unexpectedly, that CEP131 knockout mice are both viable and proficient in ciliogenesis [66]. This suggests that a compensatory pathway exists in higher eukaryotes and that alternative mechanisms may further regulate BBSome assembly. Indeed, no CEP131 mutations have so far been associated with ciliopathies in humans. The molecular mechanisms underlying the dynamic exchange of BBS4 from satellites and its integration into the BBSome upon ciliogenesis remain largely unexplored but may be regulated through post-translational modifications. One potential regulator could be the ubiquitin E3 ligase MIB1, which has been recently identified as a bona fide component of CS, and which was shown to ubiquitylate PCM1 and CEP131 in cycling cells [67, 68]. Strikingly, a marked attenuation of MIB1 activity and decreased ubiquitylation of PCM1 and CEP131 was observed upon serum starvation, which is a common cue for inducing ciliogenesis in human fibroblasts [68]. In addition, depletion of MIB1 was accompanied by activation of the ciliation program in cycling cells. These observations suggest that MIB1 may inhibit ciliogenesis during the cell cycle through ubiquitylation of CS factors, including PCM1 and CEP131 [68, 69].
In addition to the inhibitory role of MIB1, CS were recently found to antagonize ciliogenesis in a surprising manner. OFD1 was shown to serve as yet another ciliogenesis inhibitor functioning at CS [70]. In addition to its association with CS, OFD1 also localizes to centrioles and it has long been suggested to play roles in centrosome maintenance and primary cilia assembly [10, 14, 71–73]. OFD1 facilitates plasma membrane docking of the centrosome and recruitment of Ift88 to distal appendages of the centrioles. Moreover, embryonic fibroblasts from OFD1 knockout mice display abnormally long centrioles compared to their wild-type counterparts [72]. Accordingly, mutations in OFD1 in humans manifest as primary cilia dysfunction in disorders such as oral–facial–digital syndrome, Joubert syndrome, and nephronophthisis-related ciliopathies [10, 74]. Tang and colleagues demonstrated that OFD1 removal from satellites through the autophagy pathway stimulates ciliogenesis [70]. Thus, autophagy-deficient cells demonstrated elevated OFD1 levels at CS, reduced BBS4 accumulation around the ciliary transition zone, and perturbed ciliogenesis. Reduction of the satellite-associated OFD1 pool by partial knockdown (which leaves the centrosomal fraction intact) rescued the ciliogenetic potential of these cells [70]. Since the centriolar pool of OFD1 seems to be a prerequisite for centrosome maintenance and primary cilia formation, while CS-associated OFD1 seems to inhibit this process, it is attractive to speculate that many of the proteins mentioned above may have different functions depending on their localization to the satellites, centrosome, or basal body. Interestingly, a different link between autophagy and ciliogenesis was reported by Cuervo and colleagues [75]. In contrast to a stimulatory role of autophagy in ciliogenesis, these authors observed an inhibitory role of autophagy mediated by the degradation of IFT20. This protein is required for cargo transport inside the cilium and is implicated in the nucleation and maintenance of this structure. Upon serum starvation, IFT20 is no longer targeted by the autophagy pathway and localizes to the basal body to promote ciliogenesis [75]. Conversely, upon ciliogenesis highly localized autophagosome formation near the basal body can be observed, suggesting that there are two distinct phases of autophagy governing ciliogenesis [75]. While basal autophagy negatively controls ciliogenesis through IFT20 degradation, centrosomal or CS-localized OFD1 is not efficiently targeted by this system. Upon ciliogenesis, the combination of CS accumulation at the basal body and increased formation of cilia-associated autophagosomes might offer a permissive environment for OFD1 removal from CS to further stimulate ciliogenesis [70].
CEP290 and TALPID3/KIAA0586 are two other CS factors with prominent roles in ciliogenesis. Both proteins localize to the ciliary transition zone and further stimulate ciliogenesis by recruiting Rab8a to the BBSome [26, 52, 76–78]. Rab8a and its GTP exchange factor, Rabin8, physically interact with BBS1 and are both required for ciliary vesicle trafficking [36, 79, 80]. In addition, TALPID3 is required for disassembly of CS at the ciliary transition zone prior to ciliary vesicle formation [78]. Knockdown of PCM1, CEP290, or TALPID3 leads to reduction of Rab8a at the cilium and impaired ciliogenesis. In line with the role of CS and CEP290 in Rab8a transport, dysfunction or absence of CEP290 perturbs the ciliary influx and localization of specific photo-transducers and odorant signaling molecules such as Golf and Gy in the cilia of olfactory receptor neurons [26, 76, 81]. Consistent with the above, mutations in CEP290 are linked to ciliopathies such as Joubert Syndrome, Meckel Syndrome, Leber Congenital Amaurosis, and even BBS. Moreover, both physical and genetic interactions between CEP290 and BBS4 have recently been demonstrated in mouse models, potentially offering an explanation for the overlapping spectrum of ciliopathies caused by mutations in CEP290 and BBS4 [63]. CS-mediated transport of Rab8 supported by CEP290 seems to be antagonized by CP110, a centrosomal protein involved in cell cycle progression and centrosome duplication [52, 82–84]. CP110-mediated cilia resorption is dependent on its interaction with Rab8a and CEP290, but CP110 has so far not been detected in CS. Consistent with the notion that ciliogenesis is suppressed in cycling cells, CP110 expression is strongly induced when cells re-enter the cell cycle from quiescence [82].
A second CS-mediated pathway involved in cilia resorption involves PCM1 and PLK1 (Polo-like kinase 1) [12]. Microtubules in cilia are highly glutamylated, acetylated, and glycosylated, ensuring their structural integrity [80]. PLK1 activates HDAC6 (histone deacetylase 6), which is the deacetylase responsible for removing acetyl groups from axonemal microtubules during cilium disassembly [12, 85]. PLK1 localization to the pericentriolar matrix was shown to be dependent on a direct interaction with PCM1, the presence of CS, and active microtubule-mediated transport. Interestingly, the interaction between PCM1 and PLK1 is fully dependent on CDK1-mediated phosphorylation of Thr-703 on PCM1 [12]. Similar to CP110, the activity of CDK1 is suppressed throughout G1 phase and peaks in early mitosis, suggesting that CS are subject to strong cell cycle-dependent regulation (Fig. 2).
The involvement of CS in ciliogenesis is, however, not limited to the mechanisms described above. Other CS proteins such as SSX2IP, CCDC13, FOP, FOR20, and CEP72 have also been implicated in ciliogenesis [18, 28, 29, 35, 86, 87]. Despite the molecular details of how these proteins facilitate ciliogenesis remain obscure, perturbed centrosome maturation or failure to target CEP290 and Rab8a to the basal body are common phenotypes resulting from ablation of these factors [16, 18, 88]. However, similar to several other CS proteins, neither SSX2IP, CCDC13, FOP, FOR20, nor CEP72 require CS for their centrosomal localization. Additionally, it remains to be established whether the phenotypes caused by ablation of these factors are a consequence of centrosome/basal body dysfunction, perturbed CS functionality, or both.
Centriolar satellites and neurogenesis
Besides controlling the localization of proteins involved in centrosome maturation and ciliogenesis, CS have also been implicated in neurogenesis. Functional interactions between PCM1 and several proteins involved in neurogenesis, such as Huntingtin (HTT), HAP1 (Huntingtin associated protein 1), DISC1 (disrupted in schizophrenia 1), Hook3, and CaMKIIβ have been reported [11, 13, 19, 20, 89]. These proteins can all be detected at the centrosome, which plays a pivotal role in various neuro-developmental processes. DISC1, BBS4, and PCM1 have been shown to operate synergistically to mediate centrosomal trafficking of CS [13]. As its name suggests, the DISC1 gene has been found to be disrupted in patients suffering from schizophrenia and is recognized as a risk factor for general psychiatric illness, including recurring depression and bipolar disorder [90, 91]. Disrupting the DISC1-BBS4-PCM1 complex by depleting any of these factors impairs neuronal migration during cortical development [13]. Comparable to the role of CS in ciliogenesis and the organization of microtubule arrays, it was suggested that these proteins are dependent on each other and function cooperatively to recruit specific proteins involved in neurogenesis to the centrosome [10, 13].
CaMKIIβ is a Calcium/Calmodulin-dependent kinase that functions at the centrosome and is responsible for dendrite retraction. Knockdown or mislocalization of CaMKIIβ leads to excessive growth of dendrites, a consequence of which is defective formation of dendrite patterns and neuronal impairment [19, 92]. Interestingly, the centrosomal localization of CaMKIIβ is almost completely dependent on its interaction with PCM1 and the presence of CS [19]. Of note, interaction studies have identified associations between CEP131, Calmodulin, and a majority of the Calmodulin kinase isoforms, indicating additional regulatory roles of CS components in neurogenesis [68].
Further evidence for the implications of CS in neurogenesis comes from the functional interactions between PCM1, HAP1, and HTT [11, 89]. Although the role of HAP1 and HTT at CS remains elusive, there is evidence that they are required for centrosomal protein transport, which is to some extent dependent on PCM1 [89]. Depletion of HTT or HAP1 does not only lead to PCM1 dispersal but also negatively affects ciliogenesis. Abnormally long polyglutamine stretches in the HTT protein are causative of the devastating neurodegenerative disorder Huntington’s disease, and olfactory abnormalities have been reported in several cases [93]. These studies make it tempting to speculate that impaired ciliogenesis in Huntington’s disease models can to some extent be attributed to CS dysregulation [89, 94].
Dynamic regulation of centriolar satellites
As alluded to in the previous paragraphs, disruption of CS is causative of a range of cellular defects, including an unorganized microtubule network, dysfunctional ciliogenesis, centrosome fragmentation, cell cycle arrest, and impaired neurogenesis [4, 10, 14, 17, 19, 20, 29, 48]. As the satellites seem to affect such a wide range of processes, it is likely that they are subject to tight regulation at different levels. Indeed, evidence that CS are dynamically regulated in response to changing cellular conditions or perturbations has recently emerged. Early CS studies established that these structures are only present in interphase cells and gradually dissolve when cells enter mitosis, becoming completely absent during metaphase and anaphase (Fig. 3) [1, 2, 4]. During this window of the cell cycle, a subset of CS proteins localize to the centrosome until the satellites are reassembled after mitosis. The molecular mechanisms behind the assembly and disassembly of CS are largely unknown, but as PCM1 protein levels remain stable throughout the cell cycle, mitotic satellite dissolution is likely to be regulated by post-translational modifications of one or more CS components [95]. Quantitative phosphoproteomic screens have identified multiple residues in PCM1 that are phosphorylated in mitosis and consistently, PCM1 displays a characteristic gel-mobility shift in extracts from mitotic cells which is dependent on CDK1 activity (Fig. 1d) [12, 96]. Thus far, only CDK1-dependent phosphorylation of Thr-703 on PCM1 has been confirmed, however, this specific phosphorylation event does not seem to mediate CS dissolution but is required for recruitment of PLK1 to CS and cilia resorption [12].
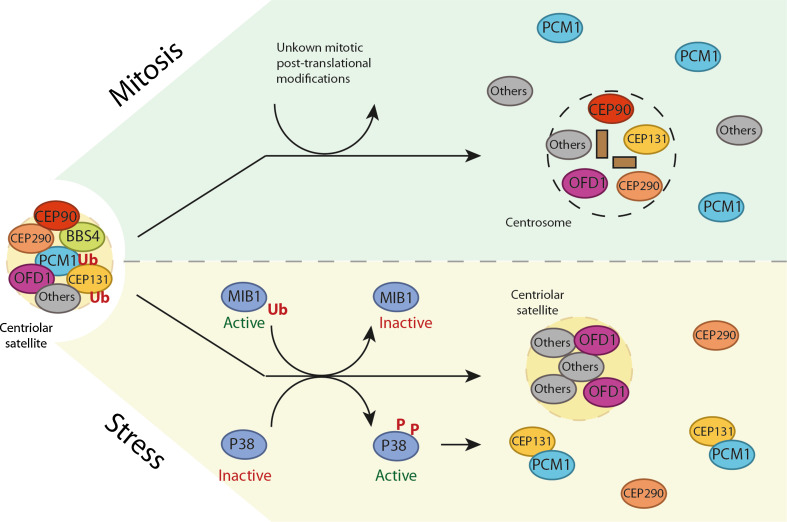
Fig. 3.
Centriolar satellite remodeling in mitosis and in response to cell stress. Centriolar satellites undergo extensive remodeling upon mitotic entry and in response to a range of cellular stresses, including proteotoxic stress and transcriptional blocks. Mitotic CS dissolution results in the redistribution of various satellite proteins to either the centrosome or cytoplasm and is likely to be regulated by post-translational protein modifications. Stress-induced CS remodeling is fully dependent on p38MAPK activity and coincides with MIB1 inactivation, leading to stimulation of ciliogenesis and AZI1-PCM1 complex formation
A recent study demonstrated that CS are also exquisitely sensitive to a broad range of cellular stresses such as UV radiation and heat shock, which induce rapid removal of a subset of CS factors including CEP131, PCM1, and CEP290 (Fig. 3) [68]. Stress-induced CS reorganization is fully dependent on p38 activity and coincides with inactivation of the ubiquitin E3 ligase MIB1. Similar to CS dissolution in mitosis, the mechanistic details on how p38 activation leads to CS reorganization are currently lacking. Perhaps somewhat counterintuitive, UV- and p38-dependent CS remodelling is accompanied by a marked increase in PCM1–CEP131 interaction, a conundrum which could be key to understand this process [68]. Even though it is unclear what the long-term fate of these PCM1-, CEP131-, and CEP290-deprived satellites are, these observations suggest that PCM1 might not be an essential scaffold for satellites per se, as has long been thought [2, 9].
While PCM1 is essential for long-term maintenance of CS, it is not the only factor responsible for the structural integrity of these structures. Depletion of several other CS components also causes loss or mislocalization of satellites (Fig. 1c, proteins highlighted in red). In addition, overexpression of several CS factors such as BBS4, CEP290, and PCM1 leads to formation of large aggregates that stain positive for other satellite markers, suggesting that these proteins are themselves able to nucleate CS formation [24, 26, 41]. Based on these observations it appears that the integrity of CS depends on a number of factors that all contribute to a stable environment in these structures.
Apart from the molecular composition of CS, the dynamics and trafficking activity of these structures also appear to be subject to tight regulation. Both BBS4, Par6α, and CEP290 have been implicated in directly regulating the subcellular localization of CS by functionally linking them to microtubule-associated motor proteins [24–26, 34, 76]. BBS4 and Par6α are both able to bind p150glued, which physically links dynein motor proteins and their respective cargo. Accordingly, depletion of BBS4 or Par6α results in the loss of CS in the vicinity of the centrosome [24, 25]. CEP290 associates with both dynein and kinesin motor proteins, suggesting that this factor can impact on both anterograde (away from the centrosome) and retrograde (towards the centrosome) transport of satellites along the microtubule network. Indeed, depletion of CEP290 results in an even tighter clustering of CS around the centrosome [26, 76]. Together, these observations suggest an attractive model in which the combined effects of CEP290, BBS4, and Par6α binding to dynein and/or kinesin motor proteins regulate the dynamic shuttling of CS along microtubules.
Similar to our limited understanding of the dynamic regulation of CS, the process of CS biogenesis remains largely unexplored. Early studies of satellite assembly mainly focused on PCM1, a protein capable of self-interaction and aggregation, dependent on two non-overlapping regions (201–494 and 745–1128) (Fig. 1d) [3, 95]. PCM1 truncations encompassing these regions are able to form large aggregates that retain the ability to interact with other CS markers including endogenous PCM1 [3]. A different mechanism of CS assembly that is linked to the formation of supernumerary centrosomes in S phase-arrested cells has been proposed [97]. CS contain a number of proteins required for centrosome biogenesis (e.g. centrin and pericentrin) and have been suggested to impact on centrosome duplication and stability [17, 18, 48]. Two studies independently demonstrated that prior to the formation of supernumerary centrosomes during a prolonged S phase, a remarkable increase in nuclear centrin granules could be observed, which was dependent on CDK2 and Chk1 [97, 98]. Following nuclear export, these centrin granules appeared to coalesce with PCM1 and showed remarkable similarities to CS as judged by electron microscopy. While suggesting an interesting link between CS and centrosome over-duplication, which is a common hallmark of cancer cells, it is not clear if these observations actually reflect a molecular mechanism underlying CS biogenesis in unperturbed interphase cells [99, 100].
Concluding remarks
Centriolar satellites harbor a wide range of proteins that have specialized functions at the centrosome, centrioles, and/or basal body. Although the exact functions of CS are still not fully understood, it is generally accepted that they contribute to the trafficking of protein cargo to the pericentriolar matrix. CS facilitate this through active microtubule-dependent transport and by acting as temporary storage containers [9]. In addition, it is clear that several proteins present in the satellites are directly responsible for maintaining the structural integrity and/or localization of these structures [10, 13, 66]. Besides their importance in the recruitment of centrosomal proteins, CS are also directly involved in assembly of the BBSome and the timely disassembly of cilia, and thus seem to play a central role in centrosome biology (Fig. 4). Hence, perturbed CS function leads to various centrosomal defects such as a failure to maintain interphase and mitotic microtubules, and impaired ciliogenesis and neurogenesis [4, 19, 27].
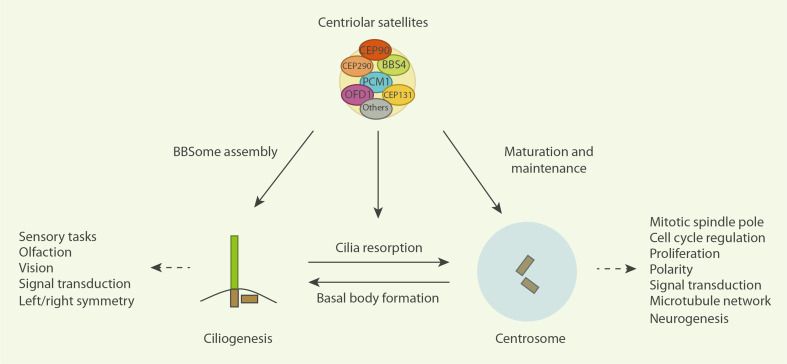
Fig. 4.
Centriolar satellites as central modulators of centrosome biology. CS contain numerous proteins directly implicated in centrosome biogenesis and are involved in numerous centrosome-dependent processes such as neurogenesis, ciliogenesis, and establishment of an organized microtubule network. Through their transporting and storage capabilities CS are able to directly modulate these processes. As CS are dynamic structures and many of the indicated centrosomal processes are under strict, cell cycle-regulated control, it is likely the CS themselves are also subject to several levels of regulatory control to ensure faithful centrosome functionality
Despite a complete inventory of CS factors is lacking, research over the last decade has pinpointed approximately 30 proteins that localize to these structures or directly interact with PCM1 (Fig. 1c). The identification and functional studies of these proteins have greatly improved our understanding of the multi-faceted roles of CS. While the original view of these structures as protein transporting vehicles still seems valid, a considerable number of CS factors can localize to the centrosome independently of CS integrity, and in many cases it is still unclear whether these proteins have additional functions directly at the satellites. Recent reports have partially addressed this issue by demonstrating that some proteins indeed have CS-specific functions, with roles of CEP131 in regulating BBS4 release from satellites and of CS-localized OFD1 in inhibiting ciliogenesis serving as prime examples [37, 70].
It has also become clear that CS are dynamic structures that change their molecular composition throughout the cell cycle and during conditions of cell stress (Figs. 2, 3) [12, 37, 68, 70]. In the latter case, CS dissolution is mediated by the stress responsive kinase p38, possibly stimulating ciliogenesis [68]. The mechanism and functional consequences of mitotic CS dissolution remain unexplored, but it has been proposed that mitosis-specific post-translational modifications, such as CDK1-mediated phosphorylation, govern this process [95, 96].
It seems likely that CS harbor many additional proteins that are involved in centrosome biogenesis, ciliogenesis, and/or neurogenesis. Future studies will undoubtedly uncover such factors, thus expanding the presently known repertoire of cellular proteins localizing to CS and increasing our understanding of the biology and functions of these fascinating structures. Further exploration of the mechanisms governing the regulation and spatiotemporal dynamics of CS organization will also be important for shedding more light on these topics. The fact that several kinases and an E3 ubiquitin ligase have been shown to function at CS naturally raises the question of what other regulatory mechanisms control CS status under different conditions. In combination, future studies along these lines are likely to offer exciting new insights into the biology and mechanistic underpinnings of CS function at the crossroads of many critical cellular processes and human diseases such as ciliopathies and neurological disorders.
Acknowledgments
Work in the laboratory of the authors is funded by the Novo Nordisk Foundation, Danish Medical Research Council, the Danish Cancer Society, and the Lundbeck Foundation.
Conflict of interest
The authors declare no conflict of interest.
Contributor Information
Niels Mailand, Phone: +45 35 32 50 23, Email: niels.mailand@cpr.ku.dk.
Simon Bekker-Jensen, Phone: +45 35 25 50 24, Email: simon.bekker-jensen@cpr.ku.dk.
References
- 1.Balczon R, Bao L, Zimmer WE. PCM-1, A 228-kD centrosome autoantigen with a distinct cell cycle distribution. J Cell Biol. 1994;124:783–793. doi: 10.1083/jcb.124.5.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kubo A, Sasaki H, Yuba-Kubo A, et al. Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J Cell Biol. 1999;147:969–980. doi: 10.1083/jcb.147.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kubo A, Tsukita S. Non-membranous granular organelle consisting of PCM-1: subcellular distribution and cell-cycle-dependent assembly/disassembly. J Cell Sci. 2003;116:919–928. doi: 10.1242/jcs.00282. [DOI] [PubMed] [Google Scholar]
- 4.Dammermann A, Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol. 2002;159:255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernhard W, de Harven E (1960) L’ultrastructure du centriole et d’autres éléments de l’appareil achromatique. In: Bargmann W, Peters D, Wolpers C (eds) Verhandlungen Band II/Biologisch-Medizinischer Teil. Springer Berlin Heidelberg, pp 217–227
- 6.Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 7.Doxsey S, Zimmerman W, Mikule K. Centrosome control of the cell cycle. Trends Cell Biol. 2005;15:303–311. doi: 10.1016/j.tcb.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Ishikawa H, Marshall WF. Ciliogenesis: building the cell’s antenna. Nat Rev Mol Cell Biol. 2011;12:222–234. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- 9.Bärenz F, Mayilo D, Gruss OJ. Centriolar satellites: busy orbits around the centrosome. Eur J Cell Biol. 2011;90:983–989. doi: 10.1016/j.ejcb.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 10.Lopes CAM, Prosser SL, Romio L, et al. Centriolar satellites are assembly points for proteins implicated in human ciliopathies, including oral-facial-digital syndrome 1. J Cell Sci. 2011;124:600–612. doi: 10.1242/jcs.077156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engelender S, Sharp AH, Colomer V, et al. Huntingtin-associated protein 1 (HAP1) interacts with the p150Glued subunit of dynactin. Hum Mol Genet. 1997;6:2205–2212. doi: 10.1093/hmg/6.13.2205. [DOI] [PubMed] [Google Scholar]
- 12.Wang G, Chen Q, Zhang X, et al. PCM1 recruits Plk1 to the pericentriolar matrix to promote primary cilia disassembly before mitotic entry. J Cell Sci. 2013;126:1355–1365. doi: 10.1242/jcs.114918. [DOI] [PubMed] [Google Scholar]
- 13.Kamiya A, Tan PL, Kubo K-I, et al. Recruitment of PCM1 to the centrosome by the cooperative action of DISC1 and BBS4: a candidate for psychiatric illnesses. Arch Gen Psychiatry. 2008;65:996–1006. doi: 10.1001/archpsyc.65.9.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graser S, Stierhof Y-D, Lavoie SB, et al. Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol. 2007;179:321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim K, Rhee K. The pericentriolar satellite protein CEP90 is crucial for integrity of the mitotic spindle pole. J Cell Sci. 2011;124:338–347. doi: 10.1242/jcs.078329. [DOI] [PubMed] [Google Scholar]
- 16.Hori A, Ikebe C, Tada M, Toda T. Msd1/SSX2IP-dependent microtubule anchorage ensures spindle orientation and primary cilia formation. EMBO Rep. 2014 doi: 10.1002/embr.201337929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srsen V, Gnadt N, Dammermann A, Merdes A. Inhibition of centrosome protein assembly leads to p53-dependent exit from the cell cycle. J Cell Biol. 2006;174:625–630. doi: 10.1083/jcb.200606051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bärenz F, Inoue D, Yokoyama H, et al. The centriolar satellite protein SSX2IP promotes centrosome maturation. J Cell Biol. 2013;202:81–95. doi: 10.1083/jcb.201302122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puram SV, Kim AH, Ikeuchi Y, et al. A CaMKIIβ signaling pathway at the centrosome regulates dendrite patterning in the brain. Nat Neurosci. 2011;14:973–983. doi: 10.1038/nn.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ge XX, Frank CLC, de Anda FFC, Tsai L-HL. Hook3 interacts with PCM1 to regulate pericentriolar material assembly and the timing of neurogenesis. Neuron. 2010;65:13. doi: 10.1016/j.neuron.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piel M, Meyer P, Khodjakov A, et al. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J Cell Biol. 2000;149:317–330. doi: 10.1083/jcb.149.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Q, Hansen D, Killilea A, et al. Kendrin/pericentrin-B, a centrosome protein with homology to pericentrin that complexes with PCM-1. J Cell Sci. 2001;114:797–809. doi: 10.1242/jcs.114.4.797. [DOI] [PubMed] [Google Scholar]
- 23.Young A, Dictenberg JB, Purohit A, et al. Cytoplasmic dynein-mediated assembly of pericentrin and gamma tubulin onto centrosomes. Mol Biol Cell. 2000;11:2047–2056. doi: 10.1091/mbc.11.6.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim JC, Badano JL, Sibold S, et al. The Bardet-Biedl protein BBS4 targets cargo to the pericentriolar region and is required for microtubule anchoring and cell cycle progression. Nat Genet. 2004;36:462–470. doi: 10.1038/ng1352. [DOI] [PubMed] [Google Scholar]
- 25.Kodani A, Tonthat V, Wu B, Sütterlin C. Par6 alpha interacts with the dynactin subunit p150 Glued and is a critical regulator of centrosomal protein recruitment. Mol Biol Cell. 2010;21:3376–3385. doi: 10.1091/mbc.E10-05-0430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J, Krishnaswami SR, Gleeson JG. CEP290 interacts with the centriolar satellite component PCM-1 and is required for Rab8 localization to the primary cilium. Hum Mol Genet. 2008;17:3796–3805. doi: 10.1093/hmg/ddn277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hames RSR, Crookes RER, Straatman KRK, et al. Dynamic recruitment of Nek2 kinase to the centrosome involves microtubules, PCM-1, and localized proteasomal degradation. Mol Biol Cell. 2005;16:1711–1724. doi: 10.1091/mbc.E04-08-0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oshimori N, Li X, Ohsugi M, Yamamoto T. Cep72 regulates the localization of key centrosomal proteins and proper bipolar spindle formation. EMBO J. 2009;28:2066–2076. doi: 10.1038/emboj.2009.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stowe TR, Wilkinson CJ, Iqbal A, Stearns T. The centriolar satellite proteins Cep72 and Cep290 interact and are required for recruitment of BBS proteins to the cilium. Mol Biol Cell. 2012;23:3322–3335. doi: 10.1091/mbc.E12-02-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piehl M, Tulu US, Wadsworth P, Cassimeris L. Centrosome maturation: measurement of microtubule nucleation throughout the cell cycle by using GFP-tagged EB1. Proc Natl Acad Sci USA. 2004;101:1584–1588. doi: 10.1073/pnas.0308205100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Avidor-Reiss T, Gopalakrishnan J. Building a centriole. Curr Opin Cell Biol. 2013;25:72–77. doi: 10.1016/j.ceb.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doxsey S. Re-evaluating centrosome function. Nat Rev Mol Cell Biol. 2001;2:688–698. doi: 10.1038/35089575. [DOI] [PubMed] [Google Scholar]
- 33.Blagden SP, Glover DM. Polar expeditions–provisioning the centrosome for mitosis. Nat Cell Biol. 2003;5:505–511. doi: 10.1038/ncb0603-505. [DOI] [PubMed] [Google Scholar]
- 34.Staples CJ, Myers KN, Beveridge RDD, et al. The centriolar satellite protein Cep131 is important for genome stability. J Cell Sci. 2012;125:4770–4779. doi: 10.1242/jcs.104059. [DOI] [PubMed] [Google Scholar]
- 35.Staples CJ, Myers KN, Beveridge RDD, et al. Ccdc13; a novel human centriolar satellite protein required for ciliogenesis and genome stability. J Cell Sci. 2014 doi: 10.1242/jcs.147785. [DOI] [PubMed] [Google Scholar]
- 36.Nachury MV, Loktev AV, Zhang Q, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 37.Chamling X, Seo S, Searby CC, et al. The centriolar satellite protein AZI1 interacts with BBS4 and regulates ciliary trafficking of the BBSome. PLoS Genet. 2014;10:e1004083. doi: 10.1371/journal.pgen.1004083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim K, Lee K, Rhee K. CEP90 is required for the assembly and centrosomal accumulation of centriolar satellites, which is essential for primary cilia formation. PLoS One. 2012;7:e48196. doi: 10.1371/journal.pone.0048196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oshimori N, Ohsugi M, Yamamoto T. The Plk1 target Kizuna stabilizes mitotic centrosomes to ensure spindle bipolarity. Nat Cell Biol. 2006;8:1095–1101. doi: 10.1038/ncb1474. [DOI] [PubMed] [Google Scholar]
- 40.Kimura M, Yoshioka T, Saio M, et al. Mitotic catastrophe and cell death induced by depletion of centrosomal proteins. Cell Death Dis. 2013;4:e603. doi: 10.1038/cddis.2013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vladar EK, Stearns T. Molecular characterization of centriole assembly in ciliated epithelial cells. J Cell Biol. 2007;178:31–42. doi: 10.1083/jcb.200703064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye X, Zeng H, Ning G, et al. C2cd3 is critical for centriolar distal appendage assembly and ciliary vesicle docking in mammals. Proc Natl Acad Sci USA. 2014 doi: 10.1073/pnas.1318737111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hames RS, Fry AM. Alternative splice variants of the human centrosome kinase Nek2 exhibit distinct patterns of expression in mitosis. Biochem J. 2002;361:77–85. doi: 10.1042/0264-6021:3610077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lacey KR, Jackson PK, Stearns T. Cyclin-dependent kinase control of centrosome duplication. Proc Natl Acad Sci USA. 1999;96:2817–2822. doi: 10.1073/pnas.96.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krämer A, Mailand N, Lukas C, et al. Centrosome-associated Chk1 prevents premature activation of cyclin-B-Cdk1 kinase. Nat Cell Biol. 2004;6:884–891. doi: 10.1038/ncb1165. [DOI] [PubMed] [Google Scholar]
- 46.Brown NJ, Marjanović M, Lüders J, et al. Cep63 and cep152 cooperate to ensure centriole duplication. PLoS One. 2013;8:e69986. doi: 10.1371/journal.pone.0069986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Firat-Karalar EN, Rauniyar N, Yates JR, Stearns T. Proximity interactions among centrosome components identify regulators of centriole duplication. Curr Biol. 2014;24:664–670. doi: 10.1016/j.cub.2014.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mikule K, Delaval B, Kaldis P, et al. Loss of centrosome integrity induces p38-p53-p21-dependent G1-S arrest. Nat Cell Biol. 2007;9:160–170. doi: 10.1038/ncb1529. [DOI] [PubMed] [Google Scholar]
- 49.Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Babu D, Roy S. Left-right asymmetry: cilia stir up new surprises in the node. Open Biol. 2013;3:130052. doi: 10.1098/rsob.130052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ansley SJ, Badano JL, Blacque OE, et al. Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature. 2003;425:628–633. doi: 10.1038/nature02030. [DOI] [PubMed] [Google Scholar]
- 52.Tsang WY, Bossard C, Khanna H, et al. CP110 suppresses primary cilia formation through its interaction with CEP290, a protein deficient in human ciliary disease. Dev Cell. 2008;15:187–197. doi: 10.1016/j.devcel.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goetz SC, Liem KF, Anderson KV. The spinocerebellar ataxia-associated gene Tau tubulin kinase 2 controls the initiation of ciliogenesis. Cell. 2012;151:847–858. doi: 10.1016/j.cell.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pedersen LB, Rosenbaum JL. Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Curr Top Dev Biol. 2008;85:23–61. doi: 10.1016/S0070-2153(08)00802-8. [DOI] [PubMed] [Google Scholar]
- 55.Jin H, White SR, Shida T, et al. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 2010;141:1208–1219. doi: 10.1016/j.cell.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tadenev ALD, Kulaga HM, May-Simera HL, et al. Loss of Bardet-Biedl syndrome protein-8 (BBS8) perturbs olfactory function, protein localization, and axon targeting. Proc Natl Acad Sci USA. 2011;108:10320–10325. doi: 10.1073/pnas.1016531108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tan PL, Barr T, Inglis PN, et al. Loss of Bardet Biedl syndrome proteins causes defects in peripheral sensory innervation and function. Proc Natl Acad Sci USA. 2007;104:17524–17529. doi: 10.1073/pnas.0706618104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kulaga HM, Leitch CC, Eichers ER, et al. Loss of BBS proteins causes anosmia in humans and defects in olfactory cilia structure and function in the mouse. Nat Genet. 2004;36:994–998. doi: 10.1038/ng1418. [DOI] [PubMed] [Google Scholar]
- 59.Eichers ER, Abd-El-Barr MM, Paylor R, et al. Phenotypic characterization of Bbs4 null mice reveals age-dependent penetrance and variable expressivity. Hum Genet. 2006;120:211–226. doi: 10.1007/s00439-006-0197-y. [DOI] [PubMed] [Google Scholar]
- 60.Coppieters F, Lefever S, Leroy BP, De Baere E. CEP290, a gene with many faces: mutation overview and presentation of CEP290base. Hum Mutat. 2010;31:1097–1108. doi: 10.1002/humu.21337. [DOI] [PubMed] [Google Scholar]
- 61.Fattahi Z, Rostami P, Najmabadi A, et al. Mutation profile of BBS genes in Iranian patients with Bardet-Biedl syndrome: genetic characterization and report of nine novel mutations in five BBS genes. J Hum Genet. 2014 doi: 10.1038/jhg.2014.28. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Q, Yu D, Seo S, et al. Intrinsic protein–protein interaction-mediated and chaperonin-assisted sequential assembly of stable Bardet-Biedl syndrome protein complex, the BBSome. J Biol Chem. 2012;287:20625–20635. doi: 10.1074/jbc.M112.341487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y, Seo S, Bhattarai S, et al. BBS mutations modify phenotypic expression of CEP290-related ciliopathies. Hum Mol Genet. 2014;23:40–51. doi: 10.1093/hmg/ddt394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilkinson CJ, Carl M, Harris WA. Cep70 and Cep131 contribute to ciliogenesis in zebrafish embryos. BMC Cell Biol. 2009;10:17. doi: 10.1186/1471-2121-10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ma L, Jarman AP. Dilatory is a drosophila protein related to AZI1 (CEP131) that is located at the ciliary base and required for cilium formation. J Cell Sci. 2011;124:2622–2630. doi: 10.1242/jcs.084798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hall EA, Keighren M, Ford MJ, et al. Acute versus chronic loss of mammalian azi1/cep131 results in distinct ciliary phenotypes. PLoS Genet. 2013;9:e1003928. doi: 10.1371/journal.pgen.1003928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Akimov V, Rigbolt KTG, Nielsen MM, Blagoev B. Characterization of ubiquitination dependent dynamics in growth factor receptor signaling by quantitative proteomics. Mol BioSyst. 2011;7:3223–3233. doi: 10.1039/c1mb05185g. [DOI] [PubMed] [Google Scholar]
- 68.Villumsen BH, Danielsen JR, Povlsen L, et al. A new cellular stress response that triggers centriolar satellite reorganization and ciliogenesis. EMBO J. 2013 doi: 10.1038/emboj.2013.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chavali PL, Gergely F. Cilia born out of shock and stress. EMBO J. 2013;32:3011–3013. doi: 10.1038/emboj.2013.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tang Z, Lin MG, Stowe TR, et al. Autophagy promotes primary ciliogenesis by removing OFD1 from centriolar satellites. Nature. 2013 doi: 10.1038/nature12606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Romio L, Fry AM, Winyard PJD, et al. OFD1 is a centrosomal/basal body protein expressed during mesenchymal-epithelial transition in human nephrogenesis. J Am Soc Nephrol. 2004;15:2556–2568. doi: 10.1097/01.ASN.0000140220.46477.5C. [DOI] [PubMed] [Google Scholar]
- 72.Singla V, Romaguera-Ros M, Garcia-Verdugo JM, Reiter JF. Ofd1, a human disease gene, regulates the length and distal structure of centrioles. Dev Cell. 2010;18:410–424. doi: 10.1016/j.devcel.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ferrante MI, Zullo A, Barra A, et al. Oral-facial-digital type I protein is required for primary cilia formation and left-right axis specification. Nat Genet. 2006;38:112–117. doi: 10.1038/ng1684. [DOI] [PubMed] [Google Scholar]
- 74.Coene KLM, Roepman R, Doherty D, et al. OFD1 is mutated in X-linked Joubert syndrome and interacts with LCA5-encoded lebercilin. Am J Hum Genet. 2009;85:465–481. doi: 10.1016/j.ajhg.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pampliega O, Orhon I, Patel B, et al. Functional interaction between autophagy and ciliogenesis. Nature. 2013;502:194–200. doi: 10.1038/nature12639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McEwen DP, Koenekoop RK, Khanna H, et al. Hypomorphic CEP290/NPHP6 mutations result in anosmia caused by the selective loss of G proteins in cilia of olfactory sensory neurons. Proc Natl Acad Sci USA. 2007;104:15917–15922. doi: 10.1073/pnas.0704140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Craige B, Tsao C-C, Diener DR, et al. CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J Cell Biol. 2010;190:927–940. doi: 10.1083/jcb.201006105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kobayashi T, Kim S, Lin Y-C, et al. The CP110-interacting proteins Talpid3 and Cep290 play overlapping and distinct roles in cilia assembly. J Cell Biol. 2014 doi: 10.1083/jcb.201304153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 80.Nachury MV, Seeley ES, Jin H. Trafficking to the ciliary membrane: how to get across the periciliary diffusion barrier? Annu Rev Cell Dev Biol. 2010;26:59–87. doi: 10.1146/annurev.cellbio.042308.113337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chang BB, Khanna HH, Hawes NN, et al. In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum Mol Genet. 2006;15:1847–1857. doi: 10.1093/hmg/ddl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen Z, Indjeian VB, McManus M, et al. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev Cell. 2002;3:339–350. doi: 10.1016/S1534-5807(02)00258-7. [DOI] [PubMed] [Google Scholar]
- 83.Spektor A, Tsang WY, Khoo D, Dynlacht BD. Cep97 and CP110 suppress a cilia assembly program. Cell. 2007;130:678–690. doi: 10.1016/j.cell.2007.06.027. [DOI] [PubMed] [Google Scholar]
- 84.Tsang WY, Dynlacht BD. CP110 and its network of partners coordinately regulate cilia assembly. Cilia. 2013;2:9. doi: 10.1186/2046-2530-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pugacheva EN, Jablonski SA, Hartman TR, et al. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 2007;129:1351–1363. doi: 10.1016/j.cell.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee JY, Stearns T. FOP is a centriolar satellite protein involved in ciliogenesis. PLoS One. 2013;8:e58589. doi: 10.1371/journal.pone.0058589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sedjaï F, Acquaviva C, Chevrier V, et al. Control of ciliogenesis by FOR20, a novel centrosome and pericentriolar satellite protein. J Cell Sci. 2010;123:2391–2401. doi: 10.1242/jcs.065045. [DOI] [PubMed] [Google Scholar]
- 88.Klinger M, Wang W, Kuhns S, et al. The novel centriolar satellite protein SSX2IP targets Cep290 to the ciliary transition zone. Mol Biol Cell. 2013 doi: 10.1091/mbc.E13-09-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Keryer G, Pineda JR, Liot G, et al. Ciliogenesis is regulated by a huntingtin-HAP1-PCM1 pathway and is altered in Huntington disease. J Clin Invest. 2011;121:4372–4382. doi: 10.1172/JCI57552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Porteous DJ, Thomson P, Brandon NJ, Millar JK. The genetics and biology of DISC1–an emerging role in psychosis and cognition. Biol Psychiatry. 2006;60:123–131. doi: 10.1016/j.biopsych.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 91.Park N, Juo SH, Cheng R, et al. Linkage analysis of psychosis in bipolar pedigrees suggests novel putative loci for bipolar disorder and shared susceptibility with schizophrenia. Mol Psychiatry. 2004;9:1091–1099. doi: 10.1038/sj.mp.4001541. [DOI] [PubMed] [Google Scholar]
- 92.Puram SV, Riccio A, Koirala S, et al. A TRPC5-regulated calcium signaling pathway controls dendrite patterning in the mammalian brain. Genes Dev. 2011;25:2659–2673. doi: 10.1101/gad.174060.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lazic SE, Goodman AOG, Grote HE, et al. Olfactory abnormalities in Huntington’s disease: decreased plasticity in the primary olfactory cortex of R6/1 transgenic mice and reduced olfactory discrimination in patients. Brain Res. 2007;1151:219–226. doi: 10.1016/j.brainres.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 94.Liu J-P, Zeitlin SO. The long and the short of aberrant ciliogenesis in Huntington disease. J Clin Invest. 2011;121:4237–4241. doi: 10.1172/JCI60243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Spalluto C, Wilson DI, Hearn T. Evidence for centriolar satellite localization of CDK1 and cyclin B2. Cell Cycle. 2013;12:1802–1803. doi: 10.4161/cc.24840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Olsen JV, Vermeulen M, Santamaria A, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 97.Prosser SL, Straatman KR, Fry AM. Molecular dissection of the centrosome overduplication pathway in S-phase-arrested cells. Mol Cell Biol. 2009;29:1760–1773. doi: 10.1128/MCB.01124-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Löffler H, Fechter A, Liu FY, et al. DNA damage-induced centrosome amplification occurs via excessive formation of centriolar satellites. Oncogene. 2012 doi: 10.1038/onc.2012.310. [DOI] [PubMed] [Google Scholar]
- 99.Pihan GA, Wallace J, Zhou Y, Doxsey SJ. Centrosome abnormalities and chromosome instability occur together in pre-invasive carcinomas. Cancer Res. 2003;63:1398–1404. [PubMed] [Google Scholar]
- 100.Dodson H, Bourke E, Jeffers LJ, et al. Centrosome amplification induced by DNA damage occurs during a prolonged G2 phase and involves ATM. EMBO J. 2004;23:3864–3873. doi: 10.1038/sj.emboj.7600393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein–protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]