Abstract
Internal ribosomal entry site (IRES)-mediated translation initiation is constitutively activated during stress conditions such as tumorigenesis and hypoxia. The RNA editing enzyme ADAR1 plays an important role in physiology and pathology. Initially, we found that the ADAR1 p150 or p110 transcript levels were decreased in glioma cells compared with normal astrocyte cells. In contrast, protein levels of ADAR1 p110 were significantly upregulated in glioma tissues and cells. This expression pattern indicated translationally controlled regulation. We identified an 885-nt sequence that was located between AUG1 and AUG2 within the ADAR1 mRNA that exhibited IRES-like activity. Furthermore, we confirmed that the translational mode of ADAR1 p110 was mediated by PTBP1 in glioma cells. The protein levels of PTBP1 and ADAR1 were cooperatively expressed in glioma tissues and cells. Knocking down ADAR1 p110 significantly decreased cell proliferation in three types of glioma cells (T98G, U87MG and A172). The removal of a minimal IRES-like sequence in a p150-overexpression construct could effectively abolish p110 induction and resulted in the slight suppression of cell proliferation compared with ADAR1-p150 overexpression in siPTBP1-treated T98G cells. In summary, our study revealed a mechanism whereby ADAR1 p110 can be activated by PTBP1 through an IRES-like element in glioma cells, and ADAR1 is essential for the maintenance of gliomagenesis.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-015-1938-7) contains supplementary material, which is available to authorized users.
Keywords: ADAR1, PTBP1, IRES-like, Cell proliferation, Gliomas
Introduction
The initiation of translation in eukaryotic cells can occur by at least two distinct mechanisms: cap-dependent scanning and internal ribosome entry [1]. IRES-dependent translation is critical for physiological and pathological progress and can drive the expression of proteins under conditions where cap-dependent translation is suppressed, which indicates a translation-regulated response to stress environments such as hypoxia, apoptosis, inflammation and tumorigenesis [2–5]. IRES trans-acting factors (ITAFs) are known to contribute to IRES-mediated translation progress, and a subset of RNA-binding proteins can function as ITAFs, such as polypyrimidine tract binding protein (PTB, also known as PTBP1), upstream of N-ras (UNR, also known as CSDE1) and poly(C)-binding protein (PCBP) [6–8].
A-to-I RNA editing, which is catalyzed by adenosine deaminases that act on RNA (ADAR), is an important posttranscriptional process [9–12]. ADAR1 (also known as ADAR) is widely expressed, especially in liver and brain. ADAR1 is found as two isoforms: a long, 150-kDa protein (ADAR1 p150) that is located in the cytoplasm and nucleus and is interferon inducible and a 110-kDa protein (ADAR1 p110) that is constitutively expressed in the nucleus [13, 14]. The two isoforms are encoded from two different AUGs that are located in exons 1A or 2, and the p150 isoform is N-terminally extended (295 amino acids) compared to p110. ADAR1 plays important roles in physiology and pathology. Disrupted RNA editing levels or abnormal ADAR expression is associated with many diseases, such as cancer. In recent years, several studies have revealed the relationship between ADAR and malignant tumors, mainly in leukemia and glioblastoma (GBM) [15–20]. Over-expressed ADAR2 (also known as ADARB1) can inhibit glioma proliferation and the cell cycle through CDC-14B editing [19]; however, the expression and biological role of ADAR1 in gliomas remain unclear.
Here, we show that ADAR1 mRNA contains an IRES-like element, and PTBP1 might contribute by regulating the coordinated expression of ADAR1 p110 in glioma cells.
Materials and methods
Cell culture and tumor tissues
Human astrocyte cells (NHA, HA, HA-c and HA-sp), glioma cell lines (T98G, U87MG, A172, U251, HS-683, SF-126, SF-763, SF-767, SHG-44, CCF-STGG1, H4, LN-18, LN-229 and U118MG) and human 293ET cells were purchased from the American Type Culture Collection (ATCC) and grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10 % fetal calf serum (Invitrogen) plus antibiotics.
We included 29 glioma tissue samples and 6 normal brain tissues (from the Department of Neurosurgery, Beijing Tiantan Hospital). Malignancy grade (8 samples were grade II, 8 samples were grade III and 13 samples were grade IV) was defined according to the guidelines of the World Health Organization (WHO).
RNA extraction and quantitative real-time PCR
Total RNA was isolated from treated cells using Trizol reagent (Invitrogen) and was reverse transcribed using a reverse transcription system to generate a cDNA template according to the manufacturer’s instructions. Quantitative real-time (qRT) PCR was performed using an SYBR-green-containing PCR kit (Takara) and the IQ5 sequence detection system (Applied Biosystems) according to the manufacturer’s instructions. To detect relative gene expression, the RNA input was normalized to levels of human GAPDH, 18S rRNA mRNA. The primer sequences that were used for RT-PCR and qRT-PCR are listed in Supplementary Information Table 1.
Western blotting
Tissues or cells were harvested using TNTE lysis buffer (pH 7.4) containing 50 mM Tris–HCl, 150 mM NaCl, 1 % NP-40, 0.1 % SDS and protease inhibitor cocktail (Roche, Mannheim, Germany).
Proteins for immunoblotting were resolved in 8–15 % SDS-PAGE gels (Invitrogen) and transferred to nitrocellulose (NC) membranes. The NC membranes were blocked with 5 % skim milk in TBS-T (20 mM Tris–HCl, pH 7.4, 150 mM NaCl, 0.1 % Tween 20) for at least 1 h. Antibodies were added in TBS-T containing 5 % skim milk, and the blots were washed with TBS-T. Immunoblot signals were developed using an enhanced chemiluminescence reagent (GE Healthcare Bio-Sciences Corp.). The following antibodies were used in this study: mouse anti-human ADAR1 (cat. ab88574, Abcam, USA), rabbit anti-human HIF-1α (cat. ab2185, Abcam, USA), rabbit anti-human PCBP2 (Aviva Systems Biology, USA), rabbit anti-human PTBP1 (provided by our lab) and goat anti-human UNR (cat. sc-79292, Santa Cruz, USA).
Plasmid constructs and luciferase assays
Human ADAR (NM_001111) cDNA clone plasmids were obtained from Origene. Fragments (1–885 nt) of ADAR1 and the 5′UTR of UNR were PCR amplified and cloned into the pGL3-basic and pHRF vectors. The primer sequences that were used for PCR amplification are listed in Supplementary Information Table 1. Twenty-four hours before transfection, 293ET and T98G cells were plated into a 24-well plate. For the pGL-3 vector transfection system, pGL-3 plasmids (800 ng) and pRL-TK (100 ng) were transfected into 293ET cells in one well using Lipofectamine™ 2000 (Invitrogen Life Technology Inc.). For the pHRF vector transfection system, the β-galactosidase reporter plasmid (100 ng) and pHRF vectors (600 ng) were co-transfected into cells in one well using Lipofectamine™ 2000 (for 293ET cells) or vigorous transfection reagent (for T98G cells). After 48 h, luciferase assays were performed using the Dual Luciferase Reporter Assay System (Promega, USA). The β-galactosidase activity was assayed using a β-galactosidase enzyme assay system (Beyotime, China). For each experimental trial, cells were transfected with the same plasmids in quadruplicate, and each well was assayed. These experiments were repeated more than three times, and the P value was calculated using a two-tailed t test.
RNAi duplexes and transfection
RNAi duplexes for ADAR1 were designed and synthesized by Genepharma (China). SiADAR1 (5′-CGCAGAGUUCCUCACCUGUATT-3′ and 5′-UACAGGUGAGGAACUCUGCGCGTT-3′) was synthesized [21] and bound the coding region of the human ADAR1 mRNA sequence (GenBank accession no. NM_001111.4). Stealth RNAi™ siRNA for PTBP1 (5′-AGAAGGACCGCAAGAUGGCACUGAU-3′ and 5′-AUCAGUGCCAUCUUGCGGUCCUUCU-3′) was designed and synthesized by Invitrogen. SiNC duplexes were also synthesized. Cells were seeded in DMEM containing 10 % fetal bovine serum without antibiotics. Twenty-four hours later, the cells were transfected with siRNA or siNC (final concentration of 100 nM) using Lipofectamine™ 2000, and progress was followed using the manufacturer’s protocol for Lipofectamine™ 2000-based transfections.
Biotin pulldown assay
ADAR1-885 and EV71 5′UTR RNA probes were prepared from pGEM-3zf(+) constructs. The reaction mixture contained 200 mg of the cell extracts and 12.5 pmol of each biotinylated RNA probe. The RNA-binding buffer (buffer I) contained 10 mM HEPES (pH 7.1), 50 mM KCl, 1.5 mM MgCl2 and 0.5 % NP-40. When using the RNA-binding buffer (buffer II), we added 2 mM DTT, 1 U RNasin, 1 mM EDTA, 0.2 % vanadyl-ribonucleoside complexes (VRC), 20 μM protease inhibitor cocktail and 100 μg/ml tRNA in a final volume of 4 mL, which contained 3.6 mL of buffer I. The final system included 100 μL of T98G extract, 700 μL of buffer II and 10 μL of mRNA probe. The mixture was incubated for 30 min at 30 °C and then added to 500 mL of Streptavidin MagneSphere Paramagnetic Particles (Promega) for 10 min at room temperature to allow binding. The protein–RNA complexes were washed three times with buffer I. After the final wash, 30 μL of DEPC-PBS and 20 μL of protein loading buffer were added to the beads and incubated for 10 min at 98 °C to dissociate the proteins from the RNA. The samples containing the eluted proteins were then subjected to 10 % SDS-PAGE and visualized by using western blotting.
Recombinant adenovirus packaging and infection
Adenoviruses (AD-shNC and AD-shADAR1-1#/2#) were packaged by GeneChem Co., Ltd, Shanghai, China. Before infection, the cell medium was replaced with fresh medium without FBS. Adenoviruses were added at a final concentration of 1010 pfu/mL of medium, and the cells were incubated and shaken gently every 15 min. Two hours later, the medium was replaced with complete medium.
MTT and colony formation assay
Glioblastoma cells (T98G, U87MG and A172) were seeded at a density of 3000 cells per well containing 100 μL of medium in a 96-well plate for 24 h and then transfected with siNC/siRNA for ADAR1 or AD-shNC/AD-shADAR1-2#. Every 24 h after transfection, the surviving cell number was determined as described previously [22]. For colony formation assays, transfected or infected cells were plated in six-well plates at 1000 cells per well. Approximately 14 days later, the cell colonies were stained and counted.
Results
Differential expression patterns of ADAR1 in glioma tissues and cells
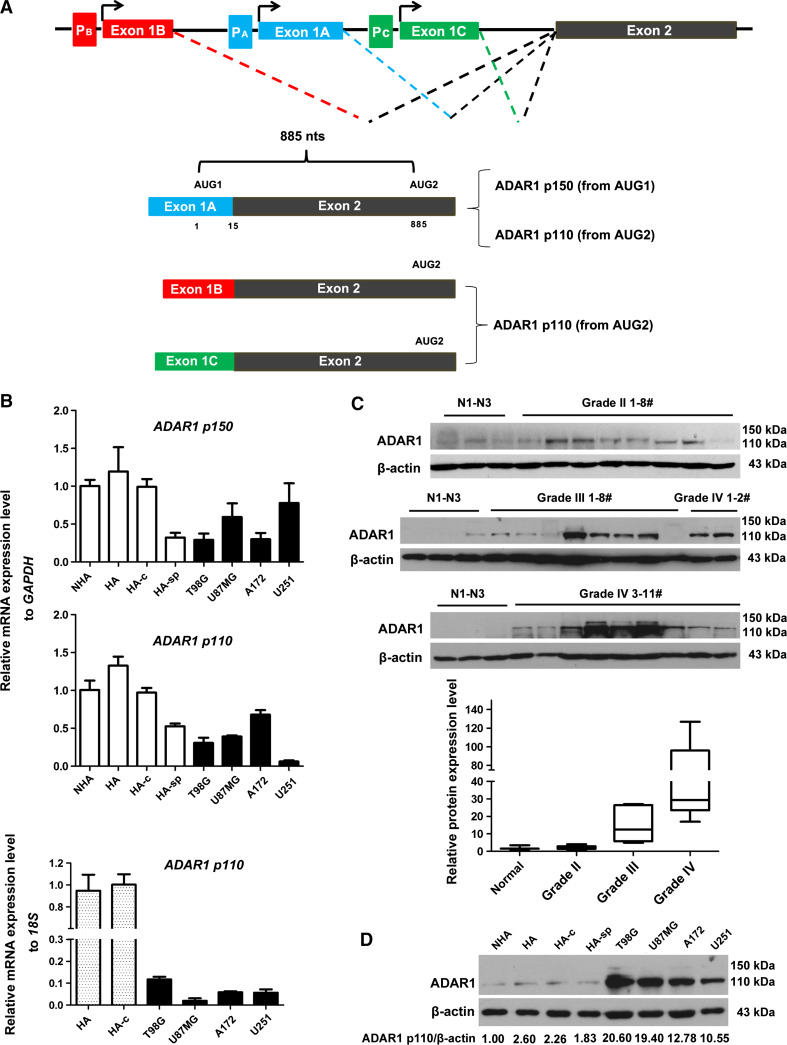
It has been reported that three different promoters control the transcriptional levels of the human ADAR1 gene [23–26] (Fig. 1a). The PA promoter, which is interferon (IFN)-induced, leads to the production of a p150 mRNA that contains the p150-specific exon 1A. In contrast, the two other promoters (PB and PC), which are constitutively active, lead to the production of p110 mRNAs. Through complicated alternative splicing, three transcript variants with mutually exclusive first exons (exon 1A, 1B and 1C) exist in the human transcriptome. The 150-kDa and 110-kDa isoforms have different N-terminal extensions and are expressed either from AUG1- or AUG2-containing mRNAs.
Fig. 1.
The expression levels of ADAR1 in glioma tissue and cells. a Schematic map of the promoter region of the ADAR1 gene. The three promoters (PA, PB and PC) initiate transcription from their corresponding ADAR1 gene, each of which contains a specific exon 1. Exons and introns are depicted as boxes and lines, respectively. b The mRNA levels of ADAR1 (p150- or p110-specific oligonucleotide products, respectively) in glioma and astrocyte cells. The mRNA levels of p110 relative to p150 in glioma cells are shown. The RNA input was normalized to GAPDH or 18S rRNA, and the relative mRNA levels of ADAR1 in NHA cells or HA-c cells were normalized to 1. Expression data are presented as the mean ± standard deviation (SD) of triplicate samples. c The protein levels of ADAR1 in glioma tissues (grade II, 1–9#; grade III, 1–9#; grade IV, 1–11#) compared to normal brain samples (N1–N3). d The protein level of ADAR1 in glioma cells (T98G, U87MG, U251 and A172) compared to normal astrocyte cell lines. Quantification of western blot bands was performed using Quantity One (4.6.2 basic), and the volume values for each band were normalized to corresponding values for β-actin
To investigate the expression of ADAR1 in gliomas, qRT-PCR assays were carried out to analyze the mRNA levels of four glioma cell lines (T98G, U87MG, U251 and A172) and four astrocyte cell lines (NHA, HA, HA-c and HA-sp) (Fig. 1b). The qRT-PCR primers for p150 or p110 (depicted as numbers 1 and 2 in Supplementary Fig. 1A) were designed as described (Ma et al. [15]); the p150 qRT-PCR products contained the first start codon (AUG1), whereas the p110 qRT-PCR products spanned exon 1B and exon 2. We also introduced another pair of primers (designed for the p150 and p110 transcripts, depicted as number 7 in Supplementary Fig. 1A) whose 62-bp product spanned the exon 7–8 boundary (Supplementary Fig. 1B). Specific oligonucleotides for different ADAR1 transcripts showed that mRNA levels were downregulated in glioma cells when compared with normal astrocyte cells. To rule out the possibility that some housekeeping genes such as GAPDH may change their expression profile during glioma genesis, we performed qRT-PCR assays (Fig. 1b) to detect ADAR1 p110 mRNA levels using 18S rRNA as loading controls. In general, ADAR1 p110 levels also demonstrated a reduction in glioma cells. However, p110 levels exhibited moderate differences from one reference to another; this may be because the two loading references exhibit different expression profiles in gliomas compared with normal glia cells.
A 136-nt alternative intron was identified within the ADAR1 p150 transcripts between AUG1 and AUG2 in a previous study [26] that could generate other isoforms (approximately, 144 kDa). Splice variants of a large number of genes could exhibit abnormal expression in GBM cells, such as GLI1 [27]. To examine the impact of alternative splicing on the mRNA levels of ADAR1, we designed corresponding primers [identified as 3–6 in Supplementary Fig. 1A, with the forward primer complementary to the specific first exon (1A or 1B) and the reverse primer matching either the exon junction (without the 136-nt intron) or the intron] and performed qRT-PCR assays (Supplementary Fig. 1C). The results indicated a decreased abundance of the transcripts without the 136-nt intron in glioma cells. Notably, ADAR1 p110 transcripts without the 136-nt intron were significantly downregulated in four types of glioma cells compared to HNA cells. Additionally, the Cancer Genome Atlas (TCGA) database [28] indicated that the mutation rate of the ADAR1 gene in glioma tissues was 0.8 %, and no splice or frameshift mutations were detected in the sequenced glioma samples (Supplementary Fig. 2A and 2B). Bioinformatic analysis (The Data Browser, http://tcga-portal.nci.nih.gov/tcga-portal/AnomalySearch.jsp) also revealed that, of the 424 GBM specimens, 17 (4 %) were significantly upregulated (1.4-fold, mRNA level) when compared with normal brain tissue (AgilentG4502A_07 log2 tumor/normal ratio ≥0.5), and only 1 sample was elevated by twofold (<1 %) (log2 tumor/normal ratio ≥1). Approximately, 67 % of samples were unchanged (Supplementary Fig. 2C and 3A). The TCGA analysis also supported the evidence that the mRNA levels of ADAR1 decreased in GBM tissues.
We then found that the protein levels of ADAR1 p110 were remarkably enhanced in most of the 29 primary glioma samples (8 samples were WHO grade II, 8 were grade III, and 13 were grade IV) when compared with three normal brain tissues (Fig. 1c), and a similar phenomenon was observed in four types of glioblastoma cell lines when compared to four normal human astrocyte cell lines (Fig. 1d). However, the protein levels of ADAR1 p150 were slightly increased in glioma tissue and cells. All of these data demonstrated that, unlike the concomitant decrease in mRNA levels, the protein levels of ADAR1 p110 were over-expressed in glioma tissues and cells when compared with normal brain tissue and astrocyte cell lines. These findings strongly suggest that the upregulation of ADAR1 p110 in glioblastoma is posttranscriptionally or translationally controlled.
The 885-nt sequence between the two AUGs within the ADAR1 p150 transcript exhibits IRES-like activity
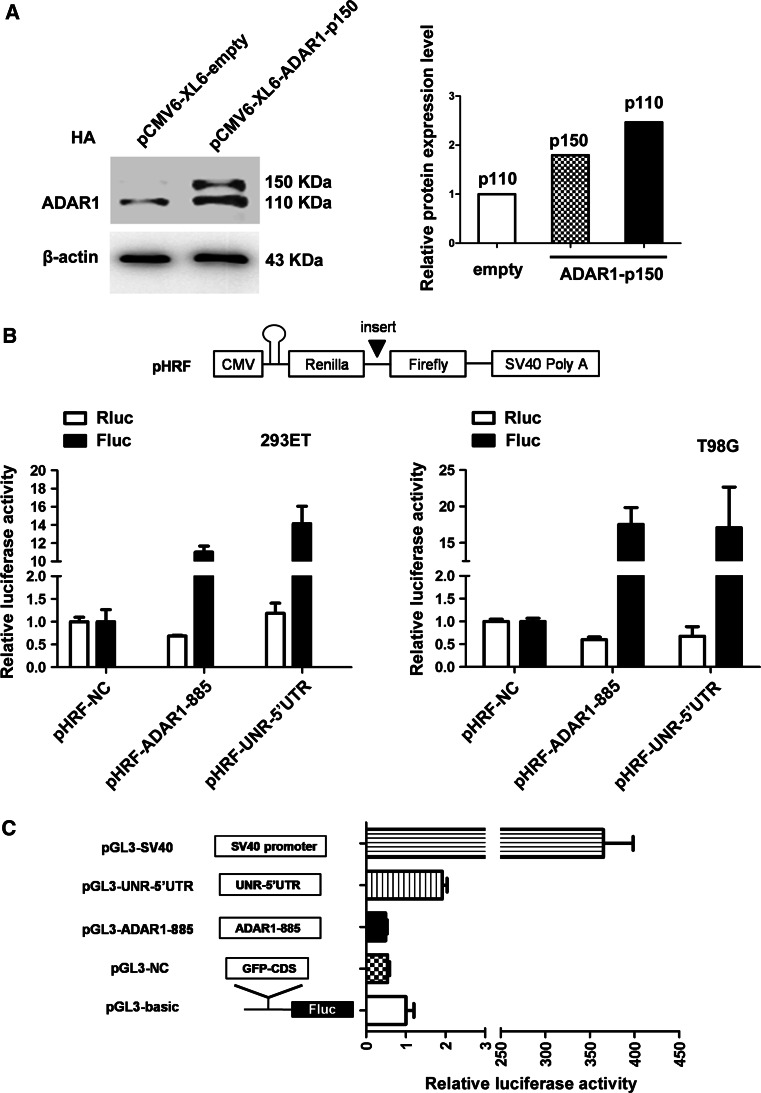
An inconsistent expression pattern in mRNA and protein levels of ADAR1 in GBMs was observed in our study, and cap-independent translational regulation might account for this contradiction. Wang et al. speculated that a cryptic internal ribosome entry site (IRES)-like element existed in the ADAR1 p150 mRNA based on the fact that the p110 isoform could be produced from p150 cDNA vectors in ADAR1 knockout cells [29]. We also transfected the pCMV6-XL6–ADAR1-p150 plasmid into HA cells in which ADAR1 is expressed at lower levels and then performed a western blotting assay. As shown in Fig. 2a, the p110 protein was also produced from p150 mRNA; the relative protein expression of p110 in the empty group was set to 1, and this value was 2.5-fold in the ADAR1 p150 over-expressed group, which may represent the activity of the IRES. To determine whether the 885-nt sequence (ADAR1-885) between AUG1 and AUG2 can function as an IRES element, the 885-nt sequence from ADAR1 was introduced into bicistronic vectors to detect possible IRES-like activity. As negative/positive controls, the GFP-CDS and UNR-5′UTR sequences were cloned, respectively (Fig. 2b). UNR had been reported to contain a functional IRES structure in its 5′UTR region [30]. The pHRF plasmid contains an artificial hairpin stem loop that effectively prevents the translation of Renilla luciferase (Rluc), and the translation of Firefly luciferase (Fluc) could be enhanced if an IRES sequence was inserted. 293ET and T98G cells that were transiently cotransfected with pHRF–NC, pHRF–ADAR1-885 or pHRF–UNR-5′UTR in combination with a β-Gal transfection control exhibited significantly higher relative Fluc activity in the presence of ADAR1-885. The results also demonstrated that Fluc activity for pHRF–ADAR1-885 was similar to that of pHRF–UNR-5′UTR. To exclude the possibility of a promoter-like function of the element, we introduced GFP-CDS/ADAR1-885/UNR-5′UTR into the promoterless pGL3-basic vector. Insertion of ADAR1-885 did not enhance Fluc activity in transiently transfected 293ET cells compared to the pGL3-basic or pGL3-NC vectors. In contrast, the positive control containing an SV40 promoter exhibited strong relative luciferase activity (~350-fold) compared to the promoterless vector (Fig. 2c). These data suggested that ADAR1-885 oligonucleotides did not demonstrate marked promoter activity.
Fig. 2.
The ADAR1-885 sequence between AUG1 and AUG2 exhibits IRES-like activity, and the enhanced expression of Fluc is not induced by cryptic promoter activity. a 293ET cells were cotransfected with the indicated reporter constructs and the TK plasmid. Forty-eight hours after transfection, Fluc activity was measured and normalized to Rluc activity. The expression data are presented as the mean ± SD of triplicate samples. b Forty-eight hours after transfection with the pCMV6-XL6–empty or pCMV6-XL6–ADAR1 vector, HA cells were collected, and total protein was extracted using lysis buffer for western blotting to determine the expression of ADAR1. Quantification of western blot bands was performed using Quantity One (4.6.2 basic), and the volume values for each band were normalized to corresponding values for β-actin. c Schematic representation of the pHRF bicistronic constructs. Within the pHRF plasmid, a hairpin stem loop that impairs ribosome scanning effectively blocks cap-dependent translation. The GFP-CDS, ADAR1-885 and UNR-5′UTR sequences were introduced into the intercistronic region between the Rluc and Fluc genes. Bicistronic reporter plasmids were co-transfected with a β-galactosidase plasmid into 293ET and T98G cells. Forty-eight hours after transfection, Rluc and Fluc activities were measured and normalized to β-galactosidase activity. Values presented are normalized against the fluorescence intensity produced from the pHRF–NC plasmid, which was set to 1. The expression data are presented as the mean ± SD of triplicate samples
These findings suggested that the 885-nt sequence between two AUGs within the ADAR1 p150 transcript exhibit IRES-like activity, but not cryptic promoter activity.
ADAR1-301–600 exhibits necessary, core IRES-like activity
To map the core sequence that was responsible for the putative IRES-like activity, we generated a series of pHRF constructs containing different lengths of DNA (marked as, e.g., ADAR1-1–300, 301–600, 601–885) (Fig. 3a). Deletion analysis indicated that the fragment 301–600 represented the minimal IRES-like active element in 293ET cells (Fig. 3b), and pHRF–ADAR1-301–885 demonstrated full IRES-like activity.
Fig. 3.
The ADAR1-301–600 sequence exhibits minimal IRES-like properties. a The indicated fragments were inserted into the region between the Rluc and Fluc reporter genes in the pHRF vector. b Bicistronic vectors were transfected into 293ET cells and Fluc/Rluc activity was determined. The values are expressed relative to that of the pHRF–NC vector. Expression data are presented as the mean ± SD of triplicate samples. c RNAdraw (v1.1) software was used to predict the secondary structure of the human ADAR1 nt 301–600 region
In some cases, an IRES is a special mRNA motif that can form a typical clover leaf-like structure with a length of 150–250 bp [4, 31, 32]. Therefore, we employed the bioinformatics tools RNAdraw and RNA fold (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) to predict the stable secondary structures of the selected sequences (Fig. 3c and Supplementary Fig. 4). As expected, a typical cloverleaf-like style was exhibited when we input the human ADAR1-301–600 sequence.
PTBP1 can bind the ADAR1-885 domain and regulates ADAR1 p110 expression in glioma cells
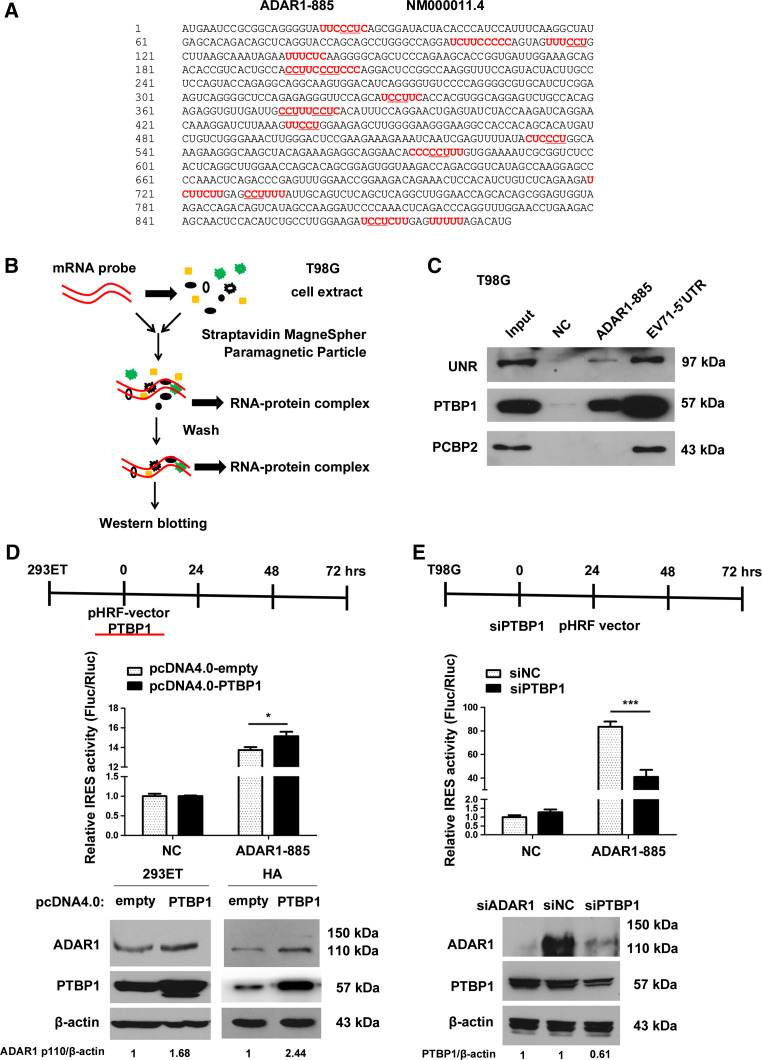
Given that the ADAR1-885 transcript exhibited IRES-like activity, we speculated that IRES trans-acting factors (ITAF) could be independently recruited and facilitate the translation process. It has been reported that PTBP1 can bind to CCUn at polypyrimidine-rich regions of RNA [33, 34]. We further confirmed several motifs (CCU) within the ADAR1-885 fragment by prediction (Fig. 4a). Then, RNA pulldown assays were performed on T98G cells (Fig. 4b), and the EV71-5′UTR, which contained IRES (741 bp), was used as a positive control [35].
Fig. 4.
PTBP1 can bind to the ADAR1-885 sequence and mediates the expression of ADAR1-p110. a Potential PTBP1 binding regions within the ADAR1-885 sequence are indicated by underlining (CCU) and the pyrimidine-rich region is marked in bold red. b Outline of the RNA pulldown assay in T98G cells. The protocol was modified by Lin et al. [35]. c The specific association between UNR/PTBP1 and ADAR1-885 was confirmed by western blot analysis. A negative control (NC) and EV71-5′UTR were also used. d Schematic design for the over-expression of PTBP1 in 293ET cell lines. 293ET cells were transferred to 24- or 6-well plates; the cells in the 24-well plate were co-transfected with the pHRF and pcDNA4.0-PTBP1 vectors, and Fluc/Rluc luciferase activity was examined 48 h after transfection. The IRES activities of the samples containing the pHRF–NC and pcDNA-4.0-empty vectors were set to 1. The protein extracts of the cells in the six-well plates were harvested 48 h after transfection with either pcDNA4.0-empty or pcDNA4.0–PTBP1 plasmid, and western blotting analysis for ADAR1 p110 was performed in the 293ET and HA cell lines and is shown below. Quantification of western blot bands was performed using Quantity One (4.6.2 basic), and the volume values for each band were normalized to corresponding values for β-actin. Finally, the normalized values of ADAR1 p110 in cells transfected with the pcDNA4.0 vector were set to 1. e Schematic design for the knockdown of PTBP1 in the T98G cell lines. T98G cells were transferred to a 24- or 6-well plate. The 24-well plate was then transfected with siPTBP1 or siNC, and, 24 h later, the pHRF vectors were transfected. Final Fluc/Rluc activity was measured 48 h after transfection with the pHRF vectors. The IRES activities of the samples with pHRF–NC and siNC were set to 1. The protein extracts of the cells in the six-well plate were harvested, and western blotting analysis was performed 72 h after transfection with either siNC or siADAR1/PTBP1, as shown below. Quantification of western blot bands was performed using Quantity One (4.6.2 basic), and the volume values for each band were normalized to their corresponding β-actin values. Finally, the normalized values of PTBP1 of the siNC group were set to 1. All of the values are represented as the mean ± SD of four parallel wells, and asterisks indicate as follows: *P < 0.05, **P < 0.01, ***P < 0.001
As shown in Fig. 4c, UNR and PTBP1 could specifically bind to ADAR1-885, but PCBP2 could not. We further investigated the relationship between PTBP1 and ADAR1 p110 in 293ET and T98G cells. We introduced pcDNA4.0-PTBP1 into 293ET cells, and pcDNA4.0 empty plasmid was used as a negative control. An approximately 10 % enhancement in IRES activity with the pHRF–ADAR1-885 construct (significant, P < 0.05) was observed after over-expression of PTBP1 (Fig. 4d). In contrast, the ratio of pHRF–NC was not influenced. Western blotting analysis also confirmed that the protein levels of ADAR1 p110 were improved when cells were transfected with the PTBP1 expression vector. Given that 293ET already exhibits high levels of PTBP1 expression, the over-expression experiment was also performed with HA cell lines, and the results showed a significant increase in ADAR1 p110 protein levels after transfection with pCDNA4.0–PTBP1. We then knocked down PTBP1 and analyzed the Fluc/Rluc ratio of pHRF–ADAR1-885 and the protein levels of ADAR1 p110 in T98G cells (Fig. 4e). Surprisingly, the relative IRES activity of ADAR1-885 was decreased by approximately 50 % after knocking down PTBP1 by approximately 40 %, and the protein levels of ADAR1 p110 were significantly decreased. However, knocking down ADAR1 could not impair PTBP1 in T98G cells, which indicated that ADAR1 was a downstream target of PTBP1. The data above demonstrated that PTBP1 could positively regulate ADAR1 p110 expression via its IRES-like domain in glioma cells.
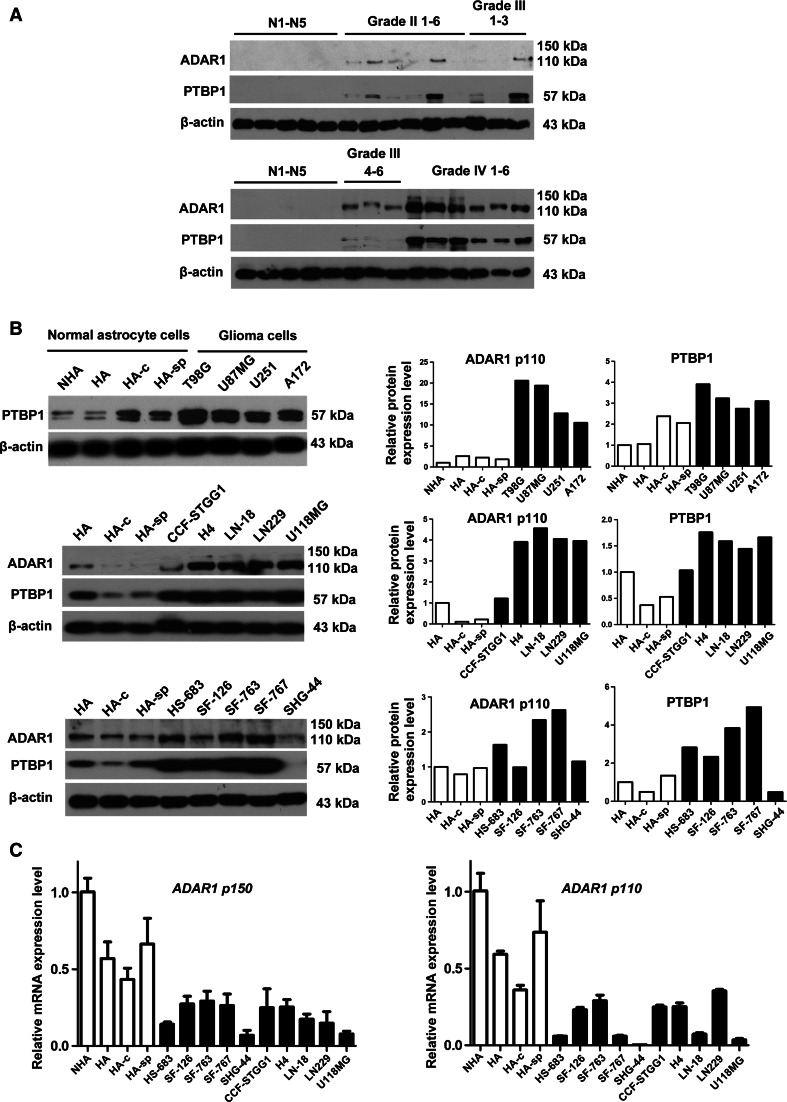
The protein expression of PTBP1 is consistent with ADAR1 p110 in glioma tissue and cells
PTBP1 is known to be aberrantly over-expressed in gliomas [36]. Therefore, we also detected the expression of PTBP1 in glioma tissues and cells. As shown in Fig. 5a, western blotting analysis showed that PTBP1 protein levels increased with malignancy grade (5 normal brain tissues, 6 WHO grade II samples, 6 WHO grade III and 6 WHO grade IV), and ADAR1 p110 exhibited the same pattern in glioma tissues. In addition, we analyzed the RNAseq RPKM values of ADAR1 and PTBP1 from 241 glioma patients (the data were supported by Beijing Tiantan Hospital, as shown in Supplementary Information Table 2). As shown in Supplementary Fig. 5A, the mRNA levels of PTBP1 positively correlated with the malignancy grade of the GBM tumors (83 samples were WHO grade II, 70 were grade III and 88 were grade IV). In the TCGA database, the mRNA levels of PTBP1 were upregulated in glioma tissues (~99 % of samples were higher by 1.4-fold and ~88 % increased twofold over that of normal brain tissues), which implied that the transcriptional and translational levels of PTBP1 were higher in the glioma samples (Supplementary Figs. 2C and 3B). In contrast, the mRNA levels of ADAR1 remained unchanged among the different grades (no significant difference for multiple comparisons, one-way ANOVA with Bonferroni correction). Elevated protein levels of ADAR1 and PTBP1 were obtained when glioma cell lines (HS-683, SF-126, SF-763, SF-767, SHG-44, CCF-STGG1, H4, LN-18, LN-229 and U118MG) were assayed and compared to three astrocyte cell lines (HA, HA-c and HA-sp) (Fig. 5b). Moreover, we showed that the mRNA levels of ADAR1 were downregulated in nearly all tested glioma cells when compared with normal glia cells (Fig. 5c). These findings provide novel information regarding the relevance of PTBP1 and ADAR1 p110 in gliomas.
Fig. 5.
The protein expression of PTBP1 is consistent with that of ADAR1 p110 in glioma tissue and cells. a The protein levels of ADAR1 and PTBP1 in selected glioma tissues (grade II, 1–6#; grade III, 1–6#; grade IV, 1–6#) compared to normal brain samples (N1–N5). b The protein levels of PTBP1 in four types of glioma cells (T98G, U87MG, U251 and A172) compared to normal astrocyte cell lines (upper). ADAR1 protein abundance in four types of glioma cells (T98G, U87MG, U251 and A172) compared to normal astrocyte cell lines is shown in Fig. 1d. The protein levels of ADAR1 p110 and PTBP1 in ten other types of glioma cells (HS-683, SF-126, SF-763, SF-767, SHG-44, CCF-STGG1, H4, LN-18, LN-229 and U118MG) compared with normal astrocyte cell lines (middle and lower). Quantification of western blot bands was performed using Quantity One (4.6.2 basic), and the volume values for each band were normalized to their corresponding β-actin values. Finally, the normalized values of PTBP1 or ADAR1 p110 in NHA cells or HA cells were set to 1. c The mRNA levels of ADAR1 were assayed in the cells mentioned in Fig. 5b. RNA input was normalized to GAPDH mRNA, and the relative mRNA levels of ADAR1 in NHA cells were normalized to 1. Expression data are presented as the mean ± SD of triplicate samples
Above all, PTBP1 is a potential functional RNA-binding protein that is over-expressed in gliomas and results in aberrant upregulation of ADAR1 p110 at the translational, but not transcriptional level.
The biological effects of ADAR1 on cell proliferation in gliomas
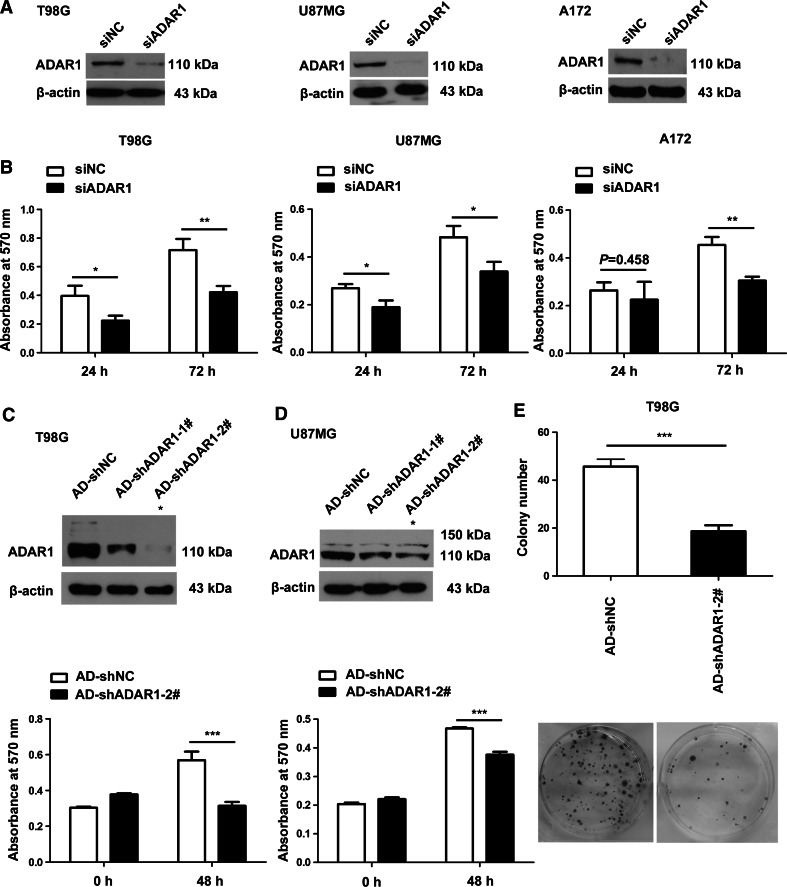
To examine the biological role of ADAR1 in glioma cells, we synthesized siADAR1 duplexes as described [21]. After transfection with siNC or siADAR1 for 72 h, the protein expression of ADAR1 was examined in the T98G, U87MG and A172 cell lines by western blotting. The results showed that the levels of ADAR1 in the siADAR1 group were significantly reduced compared to the siNC group (Fig. 6a), and a dimethyl thiazolyl diphenyl tetrazolium (MTT) assay showed that the reduction of ADAR1 decreased the growth of glioma cells (Fig. 6b). To avoid off-target effects, recombinant adenovirus vectors (AD-shADAR1-1# and 2#) that could bind different sites on ADAR1 mRNA were designed and synthesized. As shown in Fig. 6c, d, knocking down ADAR1 with AD-shADAR1-2# suppressed the endogenous levels of ADAR1 more effectively than AD-shADAR1-1# in glioma cells. Thus, AD-shADAR1-2# was used for subsequent experiments. We also found that knocking down ADAR1 with AD-shADAR1-2# inhibited the growth and survival of T98G and U87MG cells after transfection for 48 h [the glioma cell growth was strongly inhibited when transfected with adenovirus (AD-shNC or AD-shADAR1-2#) for 72 h]. Moreover, the reduction of ADAR1 decreased the colony-forming ability of T98G cells (Fig. 6e).
Fig. 6.
ADAR1 knockdown inhibits the growth and survival of glioma cells. a Seventy-two hours after transfection with siNC or siADAR1, glioma cells were collected, total protein was extracted using lysis buffer and western blotting was used to determine the expression of ADAR1 in 3 glioma cell lines (T98G, U87MG and A172). b MTT assays show the proliferation of T98G, U87MG and A172 cell lines after transfection with siRNA against ADAR1 or control siRNA. c, d Forty-eight hours after transfection with adenovirus (AD-shNC and AD-shADAR1-1#/2#), glioma cells were collected, total protein was extracted using lysis buffer and western blotting was performed to determine the expression of ADAR1 in T98G and U87MG cells. The asterisks indicate that AD-shADAR1-2# could suppress the endogenous levels of ADAR1 effectively and could be used for subsequent experiments. MTT assays were also performed to detect cell growth and cell survival. e The effects of knocking down ADAR1 with AD-shADAR1-2# on the colony-forming abilities of T98G cells. Cell colonies were counted and plotted. The expression data are presented as the mean ± SD of triplicate samples, and asterisks indicate as follows: *P < 0.05, **P < 0.01, ***P < 0.001
Deletion of the IRES-like element resulted in a reduction in p110 induction/cell proliferation in T98G cells
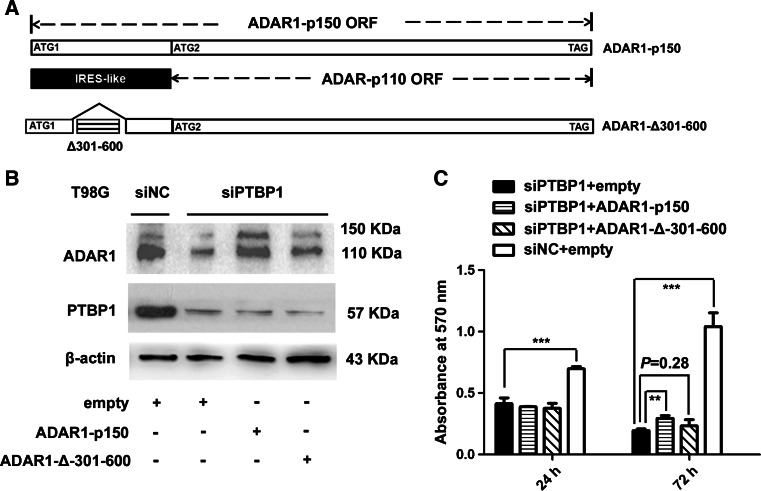
To determine the function of the IRES-like element on p110 induction/cell proliferation, we first generated an IRES-deletion construct, marked as pCMV6-XL6–ADAR1-Δ301–600 (Fig. 7a). Due to ADAR1 p110 expression at high levels in T98G cells, we then performed transfections with empty or ADAR1-p150 or ADAR1-Δ-301–600 plasmid in siPTBP1-treated T98G cells. As shown in Fig. 7b, the ADAR1 p110 levels were abolished in the ADAR1-Δ-301–600 group compared to the ADAR1-p150 group. MTT assays showed that siRNA target PTBP1 could suppress cell proliferation (P < 0.001, siNC group with empty vector versus siPTBP1 group with empty vector) (Fig. 7c) and also decrease the protein levels of ADAR1 p110 in T98G cells (Fig. 7b). However, over-expressing the ADAR1-p150 vector led to the slight rescue of cell proliferation in siPTBP1-treated T98G cells (P < 0.01, siPTBP1 group with ADAR1-p150 vector versus siPTBP1 group with empty vector). The results demonstrated that ADAR1 p110 is essential for the maintenance of gliomagenesis.
Fig. 7.
The deletion of the IRES-like element could eliminate p110 induction, and over-expression of the ADAR1-p150 vector resulted in the slight rescue of cell proliferation in siPTBP1-treated T98G cells. a A schematic representation of the ADAR1 IRES-deletion cDNAs used in western blotting or MTT assays is shown. In the Δ301–600 vector, the ADAR1-301–600 sequence was removed. b T98G cells were transferred to a 12-well plate. The 12-well plate was then transfected with siPTBP1 or siNC, and, 24 h later, the pCMV6-XL6–empty/ADAR1-p150/ADAR1-Δ301–600 vectors were transfected. Western blotting analysis was performed 60 h after transfection with either siNC or siPTBP1. c MTT assays show the proliferation of the siNC/siPTBP1-treated T98G cell lines after transfection with pCMV6-XL6–ADAR1-p150, pCMV6-XL6–ADAR1-Δ301–600 or the empty vector. The expression data are presented as the mean ± SD of triplicate samples, and asterisks indicate as follows: *P < 0.05, **P < 0.01, ***P < 0.001
Based on our findings, we propose that PTBP1 is expressed at background levels in normal astrocyte cells, and once malignant transformation occurs, PTBP1 is significantly enhanced and can upregulate ADAR1 p110 expression via its IRES-like domain in glioblastoma cell lines. In this study, our results showed that knocking down ADAR1 could suppress cell proliferation in glioma cells. The deletion of the IRES-like element could abolish p110 induction, and over-expression of the ADAR1-p150 vector resulted in the slight rescue of cell proliferation in siPTBP1-treated T98G cells.
Discussion
Downregulated ADAR1 mRNA levels in gliomas compared with normal brain tissue were demonstrated in our study and have been confirmed by other groups [37–39]. For instance, only 1 GBM sample exhibited higher expression of ADAR1, whereas 24 other glioma samples demonstrated decreased mRNA levels by 50–80 % relative to that of normal brain (NB) [38]. Next, the reason for deficient mRNA transcripts generating abundant protein products in glioma cells was determined to be due to an IRES-like element. A previous report confirmed that an IRES sequence was located between the AUG1 and AUG2 sites in protein 4.1 R (EPB41) transcripts [40], and another report demonstrated that the increase in c-Jun protein accumulation in glioblastoma cells was activated by an IRES mechanism with no corresponding increase in c-Jun mRNA [5]. However, a core IRES-like element (ADAR1-301–600) is present in all three transcripts (within exons 1A, 1B and 1C, respectively), which indicates that the p110 isoform can be produced by all three transcripts through IRES-like dependent translation initiation. We showed that multiple mechanisms (transcriptional or translational regulation or alternative splicing) could control the expression of ADAR1 p110, but in glioma cells cryptic IRES-like translation plays a leading role.
Like many other IRES, the ADAR1-IRES-like element required PTBP1 for its activity (Fig. 4). Interestingly, we found similar expression patterns for ADAR1 p110 and PTBP1 in glioma cell lines (Fig. 5). PTBP1 is a multi-functional, RNA-binding protein that is aberrantly overexpressed in gliomas [36]. Our data revealed that ADAR1 p110 was directly regulated by PTBP1 in gliomas. In addition, other ITAFs, such as UNR, could also bind to ADAR1-885 (Fig. 4c), and the protein expression levels of UNR were also consistent with that of ADAR1 in glioma cells (Supplementary Fig. 5B). In the TCGA database, the mRNA levels of UNR were unchanged in glioma tissues (6 % of samples were higher by 1.4-fold and 1 % of samples were increased by twofold over normal brain tissues) (Supplementary Figs. 2C and 3C). Whether UNR expression is enhanced by IRES-mediated translation in gliomas requires further study.
In some cases, an mRNA sequence containing an IRES element is expressed under stress or conditions where cap-dependent translation is decreased without corresponding changes in transcript abundance [41]. Furthermore, PTBP1 plays a stimulatory role in the IRES-mediated translation of HIF-1α when the oxygen supply is limited [2]. We found that the protein levels of ADAR1 p110 exhibited a time-dependent increase, similar to HIF-1α (as a positive control) during hypoxia with 1 % O2 or DFO treatment (Supplementary Figs. 6A, 6B and 6C). The mRNA levels of ADAR1 were also examined, and the findings indicated that the p150 and p110 mRNAs exhibited time-dependent decreases after DFO treatment in glioma cells (Supplementary Fig. 6D). Interestingly, protein levels of PTBP1 were induced after oxygen deficiency in U87MG cells (Supplementary Fig. 6A), which suggested that the upregulation of PTBP1 and ADAR1 p110 was consistent under hypoxic conditions.
A-to-I RNA editing is involved in many types of cancers [42–45]. ADAR1 is upregulated in acute lymphoblastic leukemias (ALL), GBM, human hepatocellular carcinoma (HCC) and esophageal squamous cell carcinoma (ESCC) [15, 18, 46, 47]. ADAR1 has a role as an oncogene in HCC and ESCC. Similarly, our study indicated that knocking down ADAR1 could decrease cell proliferation in glioma cells. Another report demonstrated that ADAR1 could mediate microRNA processing by forming a complex with Dicer [48], which suggests that the role of microRNAs in GBM is mediated by ADAR1. A variety of studies in mouse models, C. elegans and cell culture have demonstrated that ADAR1 protects against apoptosis and promotes cell survival [49–52], and our data also supported the notion that knocking down ADAR1 could suppress the proliferation of glioma cells (Fig. 6).The consequences of ADAR1 upregulation on editing activity must be further examined. We also transfected HA cells with empty, ADAR1-p150 or ADAR1-Δ-301–600 plasmids. ADAR1-p110 induction/cell proliferation was improved in the ADAR1-p150 group and was less enhanced in the ADAR1-Δ-301–600 group (all compared with the empty group, data not shown). The data provided supplemental information regarding the function of the ADAR1 in normal glia cells.
Taken together, our study results revealed a model in which an IRES-like element that is located between two AUGs within the mRNA sequence of ADAR1 p150 allows for the use of an internal second AUG site and, therefore, synthesis of 110-kDa isoforms. Moreover, IRES-mediated expression of ADAR1 p110, which is mediated by PTBP1, was upregulated in glioma cells and exerted functions by affecting cell proliferation.
Author contributions
B. Yang, J. Yuan, B. Qiang, and X. Peng designed research; B. Yang performed research; P. Hu and X. Lin provided assistance in research; B. Yang, W. Han, L. Zhu, X. Tan, F. Ye, G. Wang, F. Wu, B. Yin, Z. Bao, T. Jiang, J. Yuan, B. Qiang, and X. Peng contributed new reagents/analytic tools; B. Yang analyzed data; and B. Yang and X. Peng wrote the paper.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by the “973” Project, grants 2011CBA01104, 2009CB825403 and 2007CB946902; the National Science Foundation of China, grants 30825203 and 31071203; the Program for New Century Excellent Talents, grant NCET-07-0505; a “111” project grant; the International Scientific Cooperation Project of China, grant 2011DFB30370; and the National High Technology Research and Development Program of China, grant 2012AA020205.
Contributor Information
Boqin Qiang, Email: chiangbq@imicams.ac.cn.
Xiaozhong Peng, Phone: +86 10 69156411, Email: pengxiaozhong@pumc.edu.cn.
References
- 1.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schepens B, Tinton SA, Bruynooghe Y, Beyaert R, Cornelis S. The polypyrimidine tract-binding protein stimulates HIF-1alpha IRES-mediated translation during hypoxia. Nucleic Acids Res. 2005;33:6884–6894. doi: 10.1093/nar/gki1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bushell M, Stoneley M, Kong YW, Hamilton TL, Spriggs KA, Dobbyn HC, et al. Polypyrimidine tract binding protein regulates IRES-mediated gene expression during apoptosis. Mol Cell. 2006;23:401–412. doi: 10.1016/j.molcel.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 4.Rubsamen D, Blees JS, Schulz K, Doring C, Hansmann ML, Heide H, et al. IRES-dependent translation of egr2 is induced under inflammatory conditions. RNA. 2012;18:1910–1920. doi: 10.1261/rna.033019.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blau L, Knirsh R, Ben-Dror I, Oren S, Kuphal S, Hau P, et al. Aberrant expression of c-Jun in glioblastoma by internal ribosome entry site (IRES)-mediated translational activation. Proc Natl Acad Sci USA. 2012;109:E2875–E2884. doi: 10.1073/pnas.1203659109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grover R, Ray PS, Das S. Polypyrimidine tract binding protein regulates IRES-mediated translation of p53 isoforms. Cell Cycle. 2008;7:2189–2198. doi: 10.4161/cc.7.14.6271. [DOI] [PubMed] [Google Scholar]
- 7.Evans JR, Mitchell SA, Spriggs KA, Ostrowski J, Bomsztyk K, Ostarek D, et al. Members of the poly (rC) binding protein family stimulate the activity of the c-myc internal ribosome entry segment in vitro and in vivo. Oncogene. 2003;22:8012–8020. doi: 10.1038/sj.onc.1206645. [DOI] [PubMed] [Google Scholar]
- 8.Schepens B, Tinton SA, Bruynooghe Y, Parthoens E, Haegman M, Beyaert R, et al. A role for hnRNP C1/C2 and Unr in internal initiation of translation during mitosis. EMBO J. 2007;26:158–169. doi: 10.1038/sj.emboj.7601468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc Natl Acad Sci USA. 1994;91:11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerber A, O’Connell MA, Keller W. Two forms of human double-stranded RNA-specific editase 1 (hRED1) generated by the insertion of an Alu cassette. RNA. 1997;3:453–463. [PMC free article] [PubMed] [Google Scholar]
- 11.Melcher T, Maas S, Herb A, Sprengel R, Higuchi M, Seeburg PH. RED2, a brain-specific member of the RNA-specific adenosine deaminase family. J Biol Chem. 1996;271:31795–31798. doi: 10.1074/jbc.271.50.31795. [DOI] [PubMed] [Google Scholar]
- 12.Chen CX, Cho DS, Wang Q, Lai F, Carter KC, Nishikura K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA. 2000;6:755–767. doi: 10.1017/S1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patterson JB, Samuel CE. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol Cell Biol. 1995;15:5376–5388. doi: 10.1128/MCB.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desterro JM, Keegan LP, Lafarga M, Berciano MT, O’Connell M, Carmo-Fonseca M. Dynamic association of RNA-editing enzymes with the nucleolus. J Cell Sci. 2003;116:1805–1818. doi: 10.1242/jcs.00371. [DOI] [PubMed] [Google Scholar]
- 15.Ma CH, Chong JH, Guo Y, Zeng HM, Liu SY, Xu LL, et al. Abnormal expression of ADAR1 isoforms in Chinese pediatric acute leukemias. Biochem Biophys Res Commun. 2011;406:245–251. doi: 10.1016/j.bbrc.2011.02.025. [DOI] [PubMed] [Google Scholar]
- 16.Steinman RA, Yang Q, Gasparetto M, Robinson LJ, Liu X, Lenzner DE, et al. Deletion of the RNA-editing enzyme ADAR1 causes regression of established chronic myelogenous leukemia in mice. Int J Cancer. 2013;132:1741–1750. doi: 10.1002/ijc.27851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang Q, Crews LA, Barrett CL, Chun HJ, Court AC, Isquith JM, et al. ADAR1 promotes malignant progenitor reprogramming in chronic myeloid leukemia. Proc Natl Acad Sci USA. 2013;110:1041–1046. doi: 10.1073/pnas.1213021110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cenci C, Barzotti R, Galeano F, Corbelli S, Rota R, Massimi L, et al. Down-regulation of RNA editing in pediatric astrocytomas: ADAR2 editing activity inhibits cell migration and proliferation. J Biol Chem. 2008;283:7251–7260. doi: 10.1074/jbc.M708316200. [DOI] [PubMed] [Google Scholar]
- 19.Galeano F, Rossetti C, Tomaselli S, Cifaldi L, Lezzerini M, et al. ADAR2-editing activity inhibits glioblastoma growth through the modulation of the CDC14B/Skp2/p21/p27 axis. Oncogene. 2013;32:998–1009. doi: 10.1038/onc.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maas S, Patt S, Schrey M, Rich A. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc Natl Acad Sci USA. 2001;98:14687–14692. doi: 10.1073/pnas.251531398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jayan GC, Casey JL. Inhibition of hepatitis delta virus RNA editing by short inhibitory RNA-mediated knockdown of ADAR1 but not ADAR2 expression. J Virol. 2002;76:12399–12404. doi: 10.1128/JVI.76.23.12399-12404.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Chao T, Li R, Liu W, Chen Y, Yan X, et al. MicroRNA-128 inhibits glioma cells proliferation by targeting transcription factor E2F3a. J Mol Med. 2008;87:43–51. doi: 10.1007/s00109-008-0403-6. [DOI] [PubMed] [Google Scholar]
- 23.George CX, Samuel CE. Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc Natl Acad Sci USA. 1999;96:4621–4626. doi: 10.1073/pnas.96.8.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.George CX, Samuel CE. Characterization of the 5′-flanking region of the human RNA-specific adenosine deaminase ADAR1 gene and identification of an interferon-inducible ADAR1 promoter. Gene. 1999;229:203–213. doi: 10.1016/S0378-1119(99)00017-7. [DOI] [PubMed] [Google Scholar]
- 25.Kawakubo K, Samuel CE. Human RNA-specific adenosine deaminase (ADAR1) gene specifies transcripts that initiate from a constitutively active alternative promoter. Gene. 2000;258:165–172. doi: 10.1016/S0378-1119(00)00368-1. [DOI] [PubMed] [Google Scholar]
- 26.Lykke-Andersen S, Pinol-Roma S, Kjems J. Alternative splicing of the ADAR1 transcript in a region that functions either as a 5′-UTR or an ORF. RNA. 2007;13:1732–1744. doi: 10.1261/rna.567807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lo HW, Zhu H, Cao X, Aldrich A, Ali-Osman F. A novel splice variant of GLI1 that promotes glioblastoma cell migration and invasion. Cancer Res. 2009;69:6790–6798. doi: 10.1158/0008-5472.CAN-09-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steinman RA, Wang Q. ADAR1 isoform involvement in embryonic lethality. Proc Nat Acad Sci USA. 2011;108:E199. doi: 10.1073/pnas.1105004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cornelis S, Tinton SA, Schepens B, Bruynooghe Y, Beyaert R. UNR translation can be driven by an IRES element that is negatively regulated by polypyrimidine tract binding protein. Nucleic Acids Res. 2005;33:3095–3108. doi: 10.1093/nar/gki611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boutsma E, Noback S, van Lohuizen M. The Polycomb protein and E3 ubiquitin ligase Ring1B harbors an IRES in its highly conserved 5′ UTR. PLoS ONE. 2008;3:e2322. doi: 10.1371/journal.pone.0002322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allam H, Ali N. Initiation factor eIF2-independent mode of c-Src mRNA translation occurs via an internal ribosome entry site. J Biol Chem. 2010;285:5713–5725. doi: 10.1074/jbc.M109.029462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell SA, Spriggs KA, Bushell M, Evans JR, Stoneley M, Le Quesne JP, et al. Identification of a motif that mediates polypyrimidine tract-binding protein-dependent internal ribosome entry. Genes Dev. 2005;19:1556–1571. doi: 10.1101/gad.339105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spriggs KA, Cobbold LC, Ridley SH, Coldwell M, Bottley A, Bushell M, et al. The human insulin receptor mRNA contains a functional internal ribosome entry segment. Nucleic Acids Res. 2009;37:5881–5893. doi: 10.1093/nar/gkp623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin JY, Li ML, Huang PN, Chien KY, Horng JT, Shih SR. Heterogeneous nuclear ribonuclear protein K interacts with the enterovirus 71 5′ untranslated region and participates in virus replication. J Gen Virol. 2008;89:2540–2549. doi: 10.1099/vir.0.2008/003673-0. [DOI] [PubMed] [Google Scholar]
- 36.Cheung HC, Hai T, Zhu W, Baggerly KA, Tsavachidis S, Krahe R, et al. Splicing factors PTBP1 and PTBP2 promote proliferation and migration of glioma cell lines. Brain J Neurol. 2009;132:2277–2288. doi: 10.1093/brain/awp153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paz N, Levanon EY, Amariglio N, Heimberger AB, Ram Z, Constantini S, et al. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 2007;17:1586–1595. doi: 10.1101/gr.6493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choudhury Y, Tay FC, Lam DH, Sandanaraj E, Tang C, Ang BT, et al. Attenuated adenosine-to-inosine editing of microRNA-376a* promotes invasiveness of glioblastoma cells. J Clin Invest. 2012;122:4059–4076. doi: 10.1172/JCI62925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dominissini D, Amariglio N, Rechavi G. Micro-editing mistake translates into a devastating brain tumor. J Clin Invest. 2012;122:3842–3845. doi: 10.1172/JCI66178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lospitao E, Perez-Ferreiro CM, Gosalbez A, Alonso MA, Correas I. An internal ribosome entry site element directs the synthesis of the 80 kDa isoforms of protein 4.1R. BMC Biol. 2008;6:51. doi: 10.1186/1741-7007-6-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson SR. So you want to know if your message has an IRES? Wiley Interdiscip Rev RNA. 2012;3:697–705. doi: 10.1002/wrna.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maas S, Kawahara Y, Tamburro KM, Nishikura K. A-to-I RNA editing and human disease. RNA Biol. 2006;3:1–9. doi: 10.4161/rna.3.1.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishikura K. Editor meets silencer: crosstalk between RNA editing and RNA interference. Nat Rev Mol Cell Biol. 2006;7:919–931. doi: 10.1038/nrm2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gallo A, Galardi S. A-to-I RNA editing and cancer: from pathology to basic science. RNA Biol. 2008;5:135–139. doi: 10.4161/rna.5.3.6739. [DOI] [PubMed] [Google Scholar]
- 45.Galeano F, Tomaselli S, Locatelli F, Gallo A. A-to-I RNA editing: the “ADAR” side of human cancer. Semin Cell Dev Biol. 2012;23:244–250. doi: 10.1016/j.semcdb.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 46.Chen L, Li Y, Lin CH, Chan TH, Chow RK, Song Y, et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat Med. 2013;19:209–216. doi: 10.1038/nm.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qin YR, Qiao JJ, Chan TH, Zhu YH, Li FF, Liu H, et al. Adenosine-to-inosine RNA editing mediated by ADARs in esophageal squamous cell carcinoma. Cancer Res. 2014;74:840–851. doi: 10.1158/0008-5472.CAN-13-2545. [DOI] [PubMed] [Google Scholar]
- 48.Ota H, Sakurai M, Gupta R, Valente L, Wulff BE, Ariyoshi K, et al. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell. 2013;153:575–589. doi: 10.1016/j.cell.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Q, Miyakoda M, Yang W, Khillan J, Stachura DL, Weiss MJ, et al. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J Biol Chem. 2004;279:4952–4961. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- 50.Hartner JC, Walkley CR, Lu J, Orkin SH. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat Immunol. 2008;10:109–115. doi: 10.1038/ni.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.XuFeng R, Boyer MJ, Shen H, Li Y, Yu H, Gao Y, et al. ADAR1 is required for hematopoietic progenitor cell survival via RNA editing. Proc Natl Acad Sci USA. 2009;106:17763–17768. doi: 10.1073/pnas.0903324106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sebastiani P, Montano M, Puca A, Solovieff N, Kojima T, Wang MC, et al. RNA editing genes associated with extreme old age in humans and with lifespan in C. elegans . PLoS One. 2009;4:e8210. doi: 10.1371/journal.pone.0008210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.