Abstract
The mammalian kidney develops from the ureteric bud and the metanephric mesenchyme. In mice, the ureteric bud invades the metanephric mesenchyme at day E10.5 and begins to branch. The tips of the ureteric bud induce the metanephric mesenchyme to condense and form the cap mesenchyme. Some cells of this cap mesenchyme undergo a mesenchymal-to-epithelial transition and differentiate into renal vesicles, which further develop into nephrons. The developing kidney expresses Fibroblast growth factor (Fgf)1, 7, 8, 9, 10, 12 and 20 and Fgf receptors Fgfr1 and Fgfr2. Fgf7 and Fgf10, mainly secreted by the metanephric mesenchyme, bind to Fgfr2b of the ureteric bud and induce branching. Fgfr1 and Fgfr2c are required for formation of the metanephric mesenchyme, however the two receptors can substitute for one another. Fgf8, secreted by renal vesicles, binds to Fgfr1 and supports survival of cells in the nascent nephrons. Fgf9 and Fgf20, expressed in the metanephric mesenchyme, are necessary to maintain survival of progenitor cells in the cortical region of the kidney. FgfrL1 is a novel member of the Fgfr family that lacks the intracellular tyrosine kinase domain. It is expressed in the ureteric bud and all nephrogenic structures. Targeted deletion of FgfrL1 leads to severe kidney dysgenesis due to the lack of renal vesicles. FgfrL1 is known to interact mainly with Fgf8. It is therefore conceivable that FgfrL1 restricts signaling of Fgf8 to the precise location of the nascent nephrons. It might also promote tight adhesion of cells in the condensed metanephric mesenchyme as required for the mesenchymal-to-epithelial transition.
Keywords: Kidney development, Metanephric mesenchyme, Ureteric bud, Fibroblast growth factor (Fgf), Fibroblast growth factor receptor (Fgfr), FgfrL1
Introduction to early kidney development
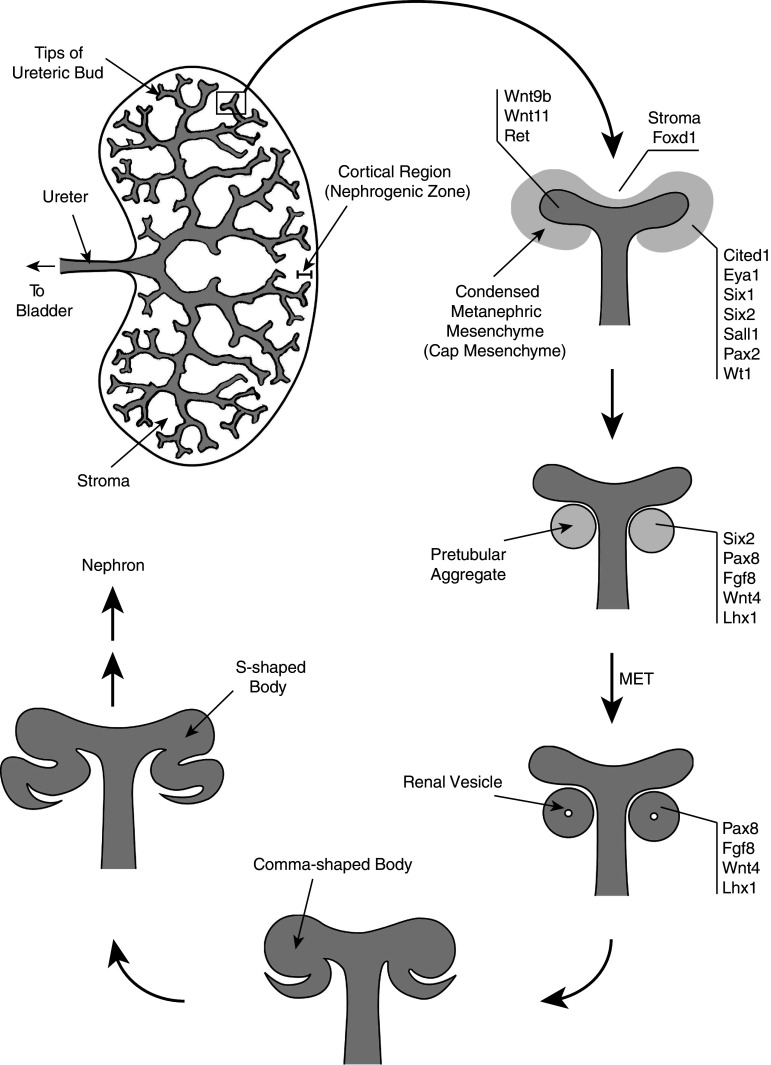
The metanephric (permanent) kidney of higher vertebrates is a complex organ comprising thousands of nephrons that are connected by a branched collecting duct system. The nephrons represent the functional units of the kidney. They filter the blood through a basement membrane and drain the filtrate via tubules and collecting ducts to the bladder. During embryonic development, the metanephric kidney develops from two different tissues, both of which are derived from the intermediate mesoderm, the ureteric bud and the metanephric mesenchyme [1]. In the mouse, development of the permanent kidney is initiated at embryonic day E10.5 when a region of the nephric (Wolffian) duct near the hindlimbs bulges out and gives rise to the ureteric bud. The metanephric mesenchyme sends out signals that induce the ureteric bud to elongate and to invade the metanephric mesenchyme, where it subsequently branches in a stereotypical fashion to form the highly branched collecting duct system. In turn, the ureteric bud releases signals that induce the metanephric mesenchyme to condense around the tips of the bud and to form the cap mesenchyme. Some cells of the cap mesenchyme develop into pretubular aggregates that undergo a mesenchymal-to-epithelial transition and form epithelial renal vesicles (see Fig. 1). At the stage of the late renal vesicle, the distal parts of the nascent nephrons fuse with the ureteric tips to form a contiguous drainage system [2]. Subsequently, the vesicles elongate and form comma-shaped bodies, which then develop into S-shaped bodies. The proximal parts of the S-shaped bodies differentiate into podocytes and the Bowman’s capsule and eventually become vascularized, while the more distal parts differentiate into the tubules of the nephron. These tubules become further segmented into a proximal tubular domain, the loop of Henle and a distal tubular domain.
Fig. 1.
Schematic drawing of early kidney development. Nephron formation takes place in the cortical region of the developing kidney. Reciprocal signaling between the tips of the ureteric bud and the metanephric mesenchyme leads to branching of the ureteric tips and to condensation of the mesenchyme around these tips. Some cells of the condensed mesenchyme form pretubular aggregates that undergo a mesenchymal-to-epithelial transition (MET) into renal vesicles. Subsequently, the vesicles form comma-shaped bodies that further develop into S-shaped bodies. The proximal parts of the S-shaped bodies differentiate to glomeruli, while the distal parts differentiate to tubules. Some of the marker genes that are specifically expressed in ureteric buds, stroma, cap mesenchyme, pretubular aggregates and renal vesicles, respectively, are indicated. The adult mouse kidney comprises approximately 12,000 nephrons
Nephron induction and maturation is a continuous process that takes place in the cortical region of the developing kidney. In the mouse, this process continues until about day P10 after birth and requires nephron progenitor cells that reside in the peripheral cortical zone of the kidney. The cortical zone also contains stromal cells that surround and support the ureteric tips and the nephrogenic mesenchyme (Fig. 1).
Functional regions of the developing kidney can be distinguished by the expression pattern of specific marker genes [1, 3]. The epithelium of the ureteric bud expresses Wnt9b, Wnt11 and Ret. Nephrogenic precursor cells in the cortical region of the developing kidney express Cited1, whereas stromal precursor cells express Foxd1. The uninduced metanephric mesenchyme expresses, amongst other marker genes, Gdnf, Six1, Eya1, Six2, Sall1, Pax2 and Wt1. Results obtained with knockout mice suggest a precise molecular hierarchy, in which one factor acts upstream of the other to induce a molecular signaling cascade: Eya1 > Six1 > Six2 > Sall1 > Pax2. Finally, renal vesicles express Pax8, Fgf8, Wnt4 and Lhx1 (Fig. 1).
Nephron development is a tightly controlled process that is regulated by various growth factors and morphogens. These proteins act in different signaling pathways, including the Fgf/Fgfr, the Wnt/β-catenin and the Notch/Presenilin pathway. To initiate nephron formation, Wnt9b is secreted from the ureteric bud into the metanephric mesenchyme where it activates the canonical β-catenin pathway [4]. In response, the metanephric mesenchyme secretes Fgf8 [5, 6] and Wnt4 [7]. Fgf8 is required for cell maintenance and survival during different stages of nephrogenesis (see also below). Wnt4 induces cells from the cap mesenchyme to undergo the mesenchymal-to-epithelial transition, which finally leads to the formation of renal vesicles. Signaling by Notch and Presenilin is subsequently required to pattern the tubules of the nascent nephrons [8, 9].
The Fgf/Fgfr signaling system
The Fgf/Fgfr signaling pathway will be described here in more detail because it involves FgfrL1, the major topic of this review. The Fgf/Fgfr system comprises a diverse palette of ligands, receptors, coreceptors and modulating proteins. Humans and mice possess 22 different Fgfs that can be grouped according to sequence and specificity in 7 distinct subfamilies [10]: the Fgf1 subfamily (Fgfs 1, 2), the Fgf4 subfamily (Fgfs 4, 5, 6), the Fgf7 subfamily (Fgfs 3, 7, 10, 22), the Fgf8 subfamily (Fgfs 8, 17, 18), the Fgf9 subfamily (Fgfs 9, 16, 20), the endocrine Fgf subfamily (Fgfs 15/19, 21, 23; Fgf19 is the human orthologue of mouse Fgf15), and the subfamily of Fgf homologous factors (Fgfs 11, 12, 13, 14). Fgf homologous factors appear to serve an unrelated, intracellular function and will not be considered further here. All Fgfs are monomeric proteins that interact with heparan sulfate proteoglycans, which act as coreceptors and increase their activity. Together with the heparan sulfate, they bind to four different receptors (Fgfr1–Fgfr4), induce their dimerization and trigger downstream signaling events [11]. A ternary complex consisting of two Fgfrs, two Fgf ligands and two heparan sulfate chains appears to be the active complex at the cell membrane. Once activated, the receptors signal by various pathways to the interior of the cells, including the MAPK pathway, the PI3K/Akt pathway, the Jak/Stat pathway and the PLCγ pathway [12].
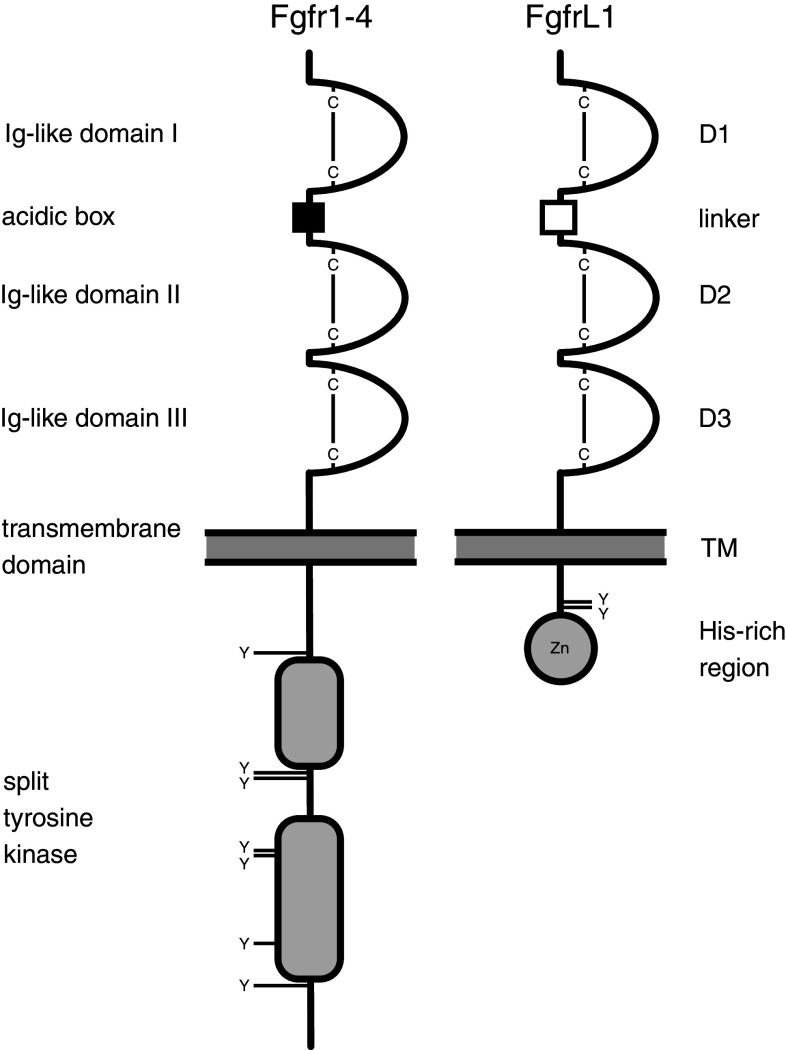
All Fgfrs display the same overall structure [11]. They contain a signal peptide that is cleaved off after synthesis, three extracellular immunoglobulin (Ig)-like domains D1–D3, a single transmembrane domain and a split intracellular tyrosine kinase domain (Fig. 2). The first Ig-domain is separated from the second by a stretch of acidic residues that has been termed the “acidic box”. In the case of Fgfr1, the best-studied Fgf receptor, the intracellular domain contains seven tyrosine residues that become phosphorylated after ligand binding in a sequential and precisely ordered fashion (Y-653, Y-583, Y-463, Y-766, Y-585, Y-654, Y-730) [13]. Phosphorylation leads to binding of adapter proteins, such as Frs2 and Shc, and to downstream signaling, in the case of the MAPK pathway via the Frs2/Grb2/Sos/Ras/Raf/Mek/Erk cascade. Moreover, tyrosine residue Y-766 is a docking site for PLCγ. Binding of PLCγ to the receptor leads to activation of the phospholipase and to cleavage of phosphatidylinositol into diacylglycerol and inositol triphosphate, both of which act as second messengers. Several proteins have been identified that modulate Fgf signaling at different levels. Positive regulation is mediated by anosmin and Flrt3, while negative regulation is mediated by Dusp and several Spry/Spred proteins.
Fig. 2.
Domain structure of the Fgf receptors and comparison with FgfrL1. All proteins have an extracellular part with three Ig-like domains (D1–D3), the first being separated from the second by a linker comprising many hydrophilic residues. This linker has been termed “acidic box” in the case of the classical receptors. Each Ig-like domain is stabilized by a disulfide bond (“C–C”). Fgfr1–Fgfr4 contain a split tyrosine kinase domain in their intracellular part. FgfrL1 is lacking this kinase domain but instead contains a histidine-rich region that interacts with zinc ions. Fgfr1 harbors seven tyrosine residues (indicated by “Y”) that become phosphorylated after ligand binding. These residues are fully conserved in Fgfr2, but only partially conserved in Fgfr3 and Fgfr4. FgfrL1 contains two tyrosine residues in its intracellular domain that are involved in sorting of the receptor to the late endosomes and lysosomes. There is no evidence that these residues become phosphorylated in FgfrL1
A further level of regulation is achieved by alternative splicing of the mRNA precursors for the Fgfs and Fgfrs [11]. In the case of the receptors, the third Ig domain D3 is subject to alternative splicing. In the genes for Fgfr1, Fgfr2 and Fgr3, this domain is encoded by three exons, exons IIIa, IIIb and IIIc. All isoforms contain exon IIIa, but exons IIIb and IIIc are spliced in a mutually exclusive manner to generate two different splice variants, the b- and the c-variant. No similar alternative exon is found in the Fgfr4 gene. Thus, the four different receptor genes can be transcribed and translated into seven different receptor proteins, Fgfr1b, Fgfr1c, Fgfr2b, Fgfr2c, Fgfr3b, Fgfr3c and Fgfr4. The b-variants are usually expressed in epithelial tissues, whereas the c-variants are mainly found in mesenchymal tissues. The Fgf ligands bind to a pocket of the receptor that is formed by Ig domains D2 and D3. Alternative splicing of the D3 domain, as observed in the b- and c-variants, will therefore affect the ligand binding properties of the receptors.
FgfrL1, the fifth Fgf receptor
A decade ago, we discovered a fifth Fgfr and termed it FgfrL1 (fibroblast growth factor receptor-like 1) [14, 15]. This receptor is found in most multicellular animals from Cnidarians to vertebrates [15, 16]. Similar to the classical receptors, it contains a signal peptide and three extracellular Ig-like domains, the first being separated from the second by a stretch of hydrophilic residues comparable to the “acidic box” of the other receptors (Fig. 2). The extracellular part shares about 40 % sequence similarity with receptors Fgfr1–Fgfr4. However, the intracellular domain of FgfrL1 is considerably shorter than those of the other receptors and does not share much similarity with any other protein. This region also reveals very low sequence conservation among different animal species with the exception of a stretch of 20 residues at the very C-terminal end. This end comprises 6–10 dipeptide repeats, where His residues alternate with Thr or Ser residues. We found that this His-rich region binds zinc ions [17] and interacts with proteins of the Spry/Spred family [18]. Preceding the His-rich region, there are two conserved tyrosine motifs that appear to act as sorting signals. These motifs target the receptor to the late endosomes and lysosomes and are therefore involved in the turnover of FgfrL1 [19].
The mode of action of FgfrL1 is not yet known. FgfrL1 binds to Fgf ligands, with relatively high affinity to Fgfs 3, 4, 8, 10 and 22 and with somewhat weaker affinity to Fgfs 2, 5, 17, 18 and 23 [20]. However, it does not appear to bind to Fgf1, 6, 7, 9, 12, 16, 18, 19, 20 and 21. FgfrL1 also interacts with heparin and heparan sulfate containing proteoglycans [21, 22]. The interacting proteins on the surface of HEK293 cells have been identified as glypican-4 and glypican-6 [23]. While binding to ligands and heparin appear to follow the common scheme of the classical receptors, signaling does not. FgfrL1 does not contain any protein tyrosine kinase activity at its intracellular domain and consequently cannot signal by trans-autophosphorylation. Therefore it has been suggested that FgfrL1 might act as a decoy receptor, which sequesters Fgf ligands away from the signaling receptors [20, 24]. Such a decoy activity is particularly likely for the receptor after shedding. We have demonstrated that a certain percentage of FgfrL1 is shed from the cell surface by an as yet unidentified protease [20]. Shedding of FgfrL1 appears to be minimal in HEK293 and COS-1 cells, but nearly complete in C2C12 cells when these cells differentiate into myotubes. The shed receptor appears to bind and neutralize some Fgf ligands in the extracellular space. Another possibility includes that FgfrL1 might dimerize with the classical receptors and block transphosphorylation at the intracellular domain. However, this possibility does not seem likely because FgfrL1 forms constitutive homodimers at the plasma membrane as demonstrated by FRET measurements and co-precipitation studies [22]. A third possibility would be that it accelerates the turnover of the classical receptors. To do so, FgfrL1 would have to interact in some way with the conventional Fgfrs, but so far there is no evidence for such an interaction. Yet a fourth possibility would be that FgfrL1 recruits signaling molecules, such as inhibitors and tyrosine phosphatases, to the sites where the other receptors are expressed. The interaction of FgfrL1 with members of the Spry/Spred family might point into this direction [18]. We have found that FgfrL1 recruits Spred1 to the plasma membrane via its His-rich domain. Spred/Spry proteins are known as negative regulators of tyrosine kinase receptors. In fact, a negative effect on cell proliferation was observed when FgfrL1 constructs were transfected into MG63 cells [21]. Furthermore, FgfrL1 blocked Fgf signaling when its mRNA was injected into Xenopus oocytes [20].
The most astonishing function of FgfrL1, however, is its effect on cell–cell fusion. We observed that FgfrL1 accumulated at sites of cell–cell contact [22]. When coated on plastic surfaces, it promoted adhesion of various cell types, including HEK293, 3T3 and CHO. Furthermore, when over-expressed in CHO cells, it induced fusion of these cells into large syncytia comprising several hundred nuclei [23]. FgfrL1 was also able to fuse heterologous cells, such as HeLa, MG63, COS7, C2C12, with CHO cells. For the fusogenic activity, only the third Ig domain and the transmembrane domain were required. The other domains (D1, D2, intracellular domain) were not needed and appeared to modulate or even repress the fusogenic activity. It is worth mentioning in this context that the D3 domain of the classical receptors Fgfr1–Fgfr4 do not exhibit any cell fusion activity [23].
Cell–cell fusion plays a fundamental role during embryonic development of most animals [25, 26]. It is observed during fertilization when sperm and egg fuse, later in the formation of bones when macrophages fuse to osteoclasts, then in skeletal muscle when myoblasts fuse to myotubes and finally in the placenta where the trophoblasts fuse to the trophoblast layer. While the fusion of viruses with the host plasma membrane has been studied in detail, fusion of entire cells to syncytia is poorly understood. Nevertheless, it has been demonstrated that “fusogens”, proteins that actively merge the two cell membranes, play a decisive role. To date only a few fusogens have been described, such as EFF-1 and AFF-1 from C. elegans and Duf, Rst, Sns and Hbs from Drosophila. Interestingly, EFF-1, AFF-1, Duf, Rst, Sns and Hbs are all members of the Ig-domain superfamily, suggesting that Ig-domains are involved in the fusion process [25, 26]. FgfrL1 is the first mammalian protein that has been shown to trigger cell–cell fusion in vitro.
Fgfs in early kidney development
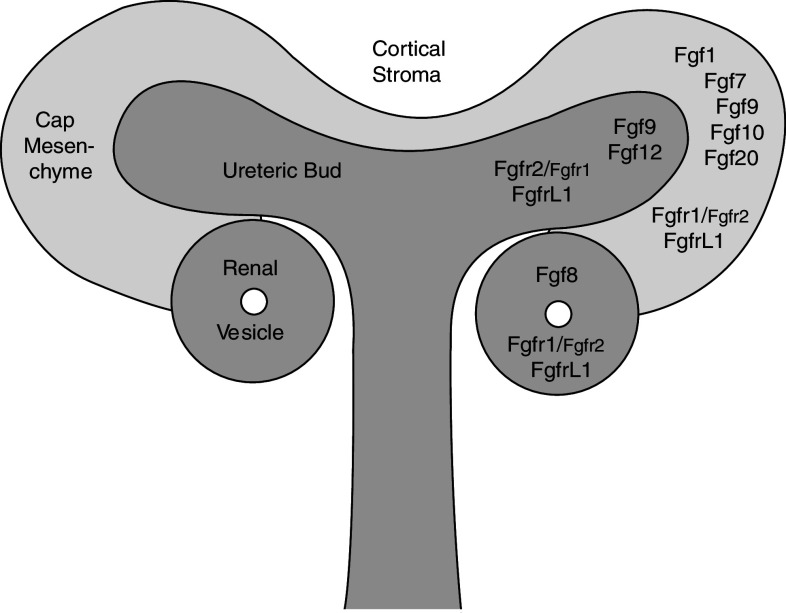
Initial RT-PCR studies demonstrated that most Fgfs are expressed in the developing rat kidney (Fgfs 1, 2, 3, 4, 5, 7, 8, 9, 10) [27]. A recent analysis of the mouse transcriptome provided by the GUDMAP database showed expression of Fgfs 1, 7, 8, 9, 10, 12 and 20 in the nephrogenic zone of developing kidneys [28]. Among these, Fgf1, 7, 10 and 20 are expressed primarily in the cap mesenchyme, Fgf8 in the renal vesicle, Fgf9 in the cap mesenchyme as well as the ureteric epithelium and Fgf12 only in the ureteric bud (Fig. 3).
Fig. 3.
Schematic diagram of a ureteric tip with locations of Fgf and Fgfr expression. A branching ureteric tip surrounded by cap mesenchyme is depicted. Some cells of the cap mesenchyme have aggregated and undergone a transition to epithelial renal vesicles. Expression of Fgf ligands and Fgf receptors is indicated in specific compartments. Differences in the expression levels between Fgfr1 and Fgfr2 are indicated by the relative size of the fonts
Early reports demonstrated that some Fgfs have profound effects on growth and differentiation of cells and tissues derived from metanephric mesenchyme and ureteric bud. When ureteric bud cells were cultivated in vitro, they secreted several soluble factors that stimulated growth of uninduced mesenchymal tissue and prevented apoptosis. One of these factors was identified as Fgf2 (basic Fgf) [29]. When added to mesenchymal tissues cultivated in vitro, Fgf2 promoted mesenchymal condensation, although it did not induce conversion of the mesenchyme to epithelial structures [30]. In cooperation with other soluble factors (TGFβ, LIF), however, it was able to convert kidney mesenchyme into tubules [31, 32].
When tested on isolated ureteric buds cultivated in vitro, several factors, including Fgf1, Fgf2, Fgf7 and Fgf10, were found to have a positive effect on proliferation and branching morphogenesis [33]. Fgf7, if overexpressed in hepatocytes of transgenic mice, induced morphological abnormalities of several organs [34]. The most striking aberrations were hyperplasia and cystic dilation of the collecting duct system in the kidney, suggesting that tight regulation of Fgf7 expression is critical for normal kidney development.
Inactivation studies performed with knockout mice demonstrated altered renal phenotypes for Fgf7, Fgf8, Fgf9, Fgf10 and Fgf20 deficient mice (Table 1). In contrast, deletion of Fgf1, Fgf2 or compound deletion of Fgf1 and Fgf2 resulted in viable, fertile mice that displayed very mild phenotypes and no renal defects [35]. Targeted disruption of Fgf7 and Fgf10, ligands known to bind to Fgfr2b, led to defects in the collecting duct system. Fgf7 null mice were viable and did not show severe abnormalities, although the animal’s fur exhibited a matted appearance due to a defect in the hair shaft [36]. Morphometric analyses of kidneys from such Fgf7 deficient mice revealed that the collecting duct system was smaller than that of control mice. Adult mice had 30 % fewer nephrons than wild-type mice [37].
Table 1.
Effects of targeted disruption of Fgf ligand and Fgf receptor genes on early kidney development in the mouse
| Gene | Targeting strategy (promoter of cre) | Effect relative to wild-type kidney | Ref. |
|---|---|---|---|
| Fgf ligands | |||
| Fgf1−/− | General ko | Normal kidneys | [35] |
| Fgf2−/− | General ko | Normal kidneys | [35] |
| Fgf1−/−; Fgf2−/− | Compound deletion | Normal kidneys | [35] |
| Fgf7−/− | General ko | Mild, smaller kidneys, fewer ureteric tips | [37] |
| Fgf10−/− | General ko | Smaller kidneys, fewer ureteric tips (lung agenesis) | [40] |
| Fgf8MM−/− | Conditional ko in MM (Pax3) | Renal hypoplasia, arrest at renal vesicle stage | [5] |
| Fgf8MM−/− | Conditional ko in mesoderm (T) | Renal hypoplasia, arrest at renal vesicle stage | [6] |
| Fgf9−/− | General ko | Normal kidneys (malformations of lung and testis) | [45] |
| Fgf20−/− | General ko | Reduced kidney size | [46] |
| Fgf9−/−; Fgf20−/− | Compound deletion | Kidney agenesis | [43] |
| Fgf9−/−; Fgf20+/− | Compound deletion | Normal kidneys | [43] |
| Fgf9+/−; Fgf20−/− | Compound deletion | Hypoplastic kidneys (15–60 % reduced size) | [43] |
| Fgf receptors | |||
| Fgfr2b−/− | General ko | Smaller kidneys with fewer nephrons (lung agenesis) | [51] |
| Fgfr1UB−/− | Conditional ko in UB (Hoxb7) | Normal | [56] |
| Fgfr2UB−/− | Conditional ko in UB (Hoxb7) | Smaller kidneys, aberrant branching, fewer nephrons | [56] |
| Fgfr1MM−/− | Conditional ko in MM (Pax3) | Normal | [60] |
| Fgfr2MM−/− | Conditional ko in MM (Pax3) | Normal, sometimes two ureteric buds | [61] |
| Fgfr1MM−/−; Fgfr2MM−/− | Conditional ko in MM (Pax3) | Kidney aplasia, little MM, unbranched UB | [60] |
| Fgfr1MM−/−; Fgfr2c−/− | Conditional ko in MM (Pax3) | Similar to Fgfr1MM−/−; Fgfr2MM−/− but more MM | [62] |
| Fgfr3−/− | General ko | Normal kidneys | [53] |
| Fgfr4−/− | General ko | Normal kidneys | [54] |
| Fgfr3−/−; Fgfr4−/− | Compound deletion | Normal kidneys | [54] |
| FgfrL1−/− (Fgfr5−/−) | General ko | Severe renal dysgenesis/agenesis | [64] |
ko knockout, UB ureteric bud, MM metanephric mesenchyme
Fgf10 knockout mice were generated in two different laboratories [38, 39]. Fgf10 null mice lacked all limbs and died at birth due to complete absence of the lungs, demonstrating a role for Fgf10 in lung and limb morphogenesis. Similar to Fgf7, knockout of Fgf10 resulted in smaller kidneys with fewer renal tubules and collecting ducts compared to wild-type mice [40]. These defects resemble the phenotype of Fgfr2b deficient mice (see below), suggesting that Fgf10 acts as the major ligand for Fgfr2b.
Recently, Michos et al. [41] found that Fgf10 is needed for branching of the ureteric bud in mice that lack Gdnf/Ret and Spry. In the absence of the negative regulator Spry, Gdnf/Ret signaling is no longer required for extensive kidney development. In this case, the removal of only one copy of the Fgf10 gene caused a complete failure of ureter and kidney development. The authors speculated that Gdnf/Ret signaling is the major trigger for growth of the ureteric tips and that in its absence, Fgf10/Fgfr2b signaling might be able to substitute to a large extent [41].
Fgf8 is essential for gastrulation [42]. Consequently, a conditional knockout strategy had to be used to study the effects of Fgf8 on kidney development. Two different groups generated conditional Fgf8 knockout mice. Grieshammer et al. [5] used the Pax3 promoter to drive expression of Cre recombinase in the metanephric mesenchyme and to delete a floxed Fgf8 allele. Perantoni et al. [6] used the Brachyury T promoter to drive Cre recombinase in all mesodermal cells and to delete a floxed Fgf8 allele. Both groups found that mutant offspring died shortly after birth and displayed abnormally small kidneys. Fgf8 is usually expressed in pretubular aggregates, renal vesicles, comma- and S-shaped bodies, but not in the ureteric bud or the interstitial stroma (Fig. 3). Kidneys of Fgf8 mutant mice still contained some renal vesicles but these did not progress to the stage of the comma- and S-shaped bodies. The mutant kidneys also showed reduced branching of the ureteric bud. Cell death occurred at significantly increased rate in regions where the mesenchymal-to-epithelial transition takes place in wildtype animals as well as in the cortical zone where nephrogenic precursor cells reside. Expression of Wnt4 and Lhx1 was absent, an observation that explains the failure of the mutant kidneys to convert the metanephric mesenchyme into nephron epithelia. Rescue experiments proved that Fgf8 as well as Wnt4 were required to sustain Lhx1 expression and tubule formation. Fgf8 is therefore required to maintain survival of cells in S-shaped bodies and to support survival of progenitor cells in the peripheral zone of the developing kidney [5, 6].
The roles of Fgf9 and Fgf20 in kidney formation were recently described [43]. The authors demonstrated that these growth factors are necessary and sufficient to maintain progenitor stemness in the cortical zone of the kidney. The adult kidney does not possess any stem cells for renewal but relies on a surplus of nephrons generated during development from an embryonic pool of progenitor cells. These progenitor cells are located distal to the ureteric bud in the cap mesenchyme and express Six2 and Sall1.
Global disruption of the Fgf9 gene resulted in mice that died at birth due to severe lung hypoplasia [44]. The Fgf9 deficient mice showed aberrant development of the male reproductive tract, but the kidneys developed normally [45]. The Fgf20 gene was targeted by insertion of a β-Galactosidase cDNA into the first exon [46]. Fgf20 null mice were viable and fertile, although they suffered from hearing loss and showed slight reduction in the size of the kidney. It was only when the researchers generated compound mutant animals that they found severe alterations in the kidney [43]. Compound null embryos (Fgf9−/−; Fgf20−/−) showed complete kidney agenesis. In contrast, compound heterozygous animals (Fgf9+/−; Fgf20+/−) did not display any overt abnormalities. Animals lacking both alleles of Fgf20 and one allele of Fgf9 (Fgf9+/−; Fgf20−/−) exhibited hypoplastic kidneys with defects ranging from mild to very severe renal dysgenesis. Depending on the level of Fgf9 and Fgf20, the mutant kidneys exhibited reduced numbers of Six2-positive progenitor cells and also more mature nephrons distal to the ureteric tips. When the metanephric mesenchyme was grown as explant culture in serum free media, addition of Fgf9 or Fgf20 was sufficient to promote survival and proliferation of some progenitor cells. These findings indicate that Fgf9 and Fgf20 act redundantly to maintain the nephrogenic zone. Reduction in the level of these growth factors leads to premature differentiation of the progenitor cells within the niche. Thus, the function of Fgf9 and Fgf20 is to maintain self-renewal of renal progenitors in the condensed metanephric mesenchyme.
Fgfrs in early kidney development
All Fgfrs are expressed in the developing rat kidney as demonstrated by RT-PCR and by in situ hybridization [27]. Fgfr1 was found to be widely distributed in the metanephric mesenchyme and epithelia, Fgfr2 in the ureteric bud and in nephron epithelia, Fgfr3 in mesenchymal condensates, nephron elements and medullary interstitium and Fgfr4 mainly in maturing nephrons. Knockout mice were generated to study the function of these receptors in early development (Table 1). Mice with a targeted disruption of the Fgfr1 gene died between embryonic day E7.5 and E9.5 [47, 48] well before the metanephric kidneys started to develop. Similarly, knockout of Fgfr2 (both isoforms Fgfr2b and Fgfr2c) resulted in embryonic lethality after implantation of the embryo in the uterus [49, 50]. Knockout mice with a targeted deletion of only the Fgfr2b isoform died at birth due to lung agenesis [51]. These animals lacked all limbs and had smaller kidneys, reminiscent of the Fgf10 knockout mice described above [40], indicating that Fgf10 is the major ligand for Fgfr2b. Transgenic mice, which overexpressed the extracellular domain of Fgfr2b in soluble form, displayed severe kidney dysgenesis in addition to multiple abnormalities of the lung, skull, limbs, skin and eyes [52]. The soluble protein appeared to act as a dominant negative receptor for Fgf ligands, sequestering ligands away from the endogenous receptors. Mice with targeted disruptions of Fgfr3 [53], Fgfr4 [54] or compound disruption of Fgfr3 and Fgfr4 [54] did not reveal any overt kidney phenotype. It is therefore unlikely that these receptors play a major role during early kidney development.
To overcome the problem of embryonic lethality of Fgfr1 and Fgfr2 knockout mice, one research group used a conditional targeting approach and studied the phenotypes of animals harboring conditional deletions of the two receptors either in the ureteric bud or the metanephric mesenchyme [55]. The authors used the Hoxb7 promoter to drive expression of Cre recombinase specifically in the ureteric bud of their mice [56]. These mice were crossed with animals carrying lox-p sites flanking critical exons of the Fgfr1 and Fgfr2 genes. Fgfr1 is expressed at low levels in the ureteric bud and consequently, the Fgfr1UB−/− animals did not reveal any overt renal abnormalities. In contrast, absence of Fgfr2 from the ureteric bud resulted in Fgfr2UB−/− mice with abnormalities in ureteric bud branching. These mice exhibited thinner ureteric bud stalks and fewer ureteric bud tips than control animals. The ureteric tips revealed regions of reduced proliferation and inappropriate apoptosis. Hybridization with a probe for Foxd1 demonstrated abnormal thickening of the stroma in the cortical region. However, the animals developed normal nephrons and were vital, although their kidneys displayed an abnormal shape and contained fewer glomeruli.
Since many effects of Fgfr signaling are mediated by the intracellular docking protein Frs2α, the authors also studied the influence of this protein [57]. Utilizing the same strategy as described above, they deleted a critical region of the Frs2α gene specifically in the ureteric bud. Kidneys from Frs2αUB−/− animals were found to be smaller than those from control animals but the reduction in size was less pronounced than that of the Fgfr2UB−/− mice. To further clarify the relationship between Fgfr2 and Frs2α, they generated mice with two point mutations in Fgfr2 in the binding site for Frs2α (L424A and R426A). These point mutations are known to efficiently block the ability of Frs2α to bind to Fgfr2 [58]. Unexpectedly, the Fgfr2LR/LR mice did not show any abnormalities in ureteric branching or in kidney size. This result suggests that Fgfr2 signals in the ureteric epithelium via other adapter molecules than Frs2α. Finally, the researchers generated mice with compound deletions of Fgfr2 and Frs2α [59]. They found that Fgfr2UB−/−;Frs2αUB−/− kidneys were more severely affected than either Fgfr2UB−/− kidneys or Frs2αUB−/− kidneys. Together with the observation gained with the point mutations, these results suggest that Fgfr2 and Frs2α act separately and additively during branching morphogenesis of the kidney. One possibility is that Frs2α is used primarily by the tyrosine kinase Ret, and not by Fgfr2, for signaling in the ureteric bud. Fgfr2 in turn might utilize another adaptor protein (such as PLCγ) for downstream signaling in the ureteric epithelium.
The same research group also analyzed the roles of Fgfr1 and Fgfr2 in the metanephric mesenchyme (Table 1). The Pax3 promoter was used to drive expression of Cre recombinase specifically in the metanephric mesenchyme [60]. The Cre mice were crossed as above with animals carrying lox-p sites flanking critical exons of the Fgfr1 and Fgfr2 genes. Although Fgfr1 and Fgfr2 are both expressed in the metanephric mesenchyme (Fig. 3), Fgfr1MM−/− and Fgfr2MM−/− mice developed normal kidneys. Occasionally, the Fgfr2MM−/− mice had two ureteric buds, suggesting that Fgfr2 is critical for ensuring that a single ureteric bud is formed from the Wolffian duct. Thus, Fgfr2 appears to be involved in the regulation of the ureteric bud induction site [61]. When the researchers deleted both Fgfr1 and Fgfr2 in the metanephric mesenchyme, they observed complete renal aplasia [60]. At E10.5 the compound null mice showed no condensed metanephric mesenchyme, but invasion of the ureteric bud into the mesenchyme occurred normally. At E11.5 the mutant mice had ureteric buds that failed to elongate and to branch. The metanephric mesenchyme still expressed the marker genes Eya1 and Six1, but there was no expression of the downstream genes Six2, Sall1 and Pax2. It is therefore likely that Fgfr1 and Fgfr2 act downstream of Eya1 and Six1 in the metanephric mesenchyme, but upstream of Six2, Sall1 and Pax2. The mesenchyme showed abnormally high rates of apoptosis and relatively low proliferation in regions that would usually develop into condensed metanephric mesenchyme. Together, Fgfr1 and Fgfr2 are therefore required for normal formation of the metanephric mesenchyme but some redundancy exists and each receptor can substitute for the other.
The researchers also studied the roles of the two splice variants Fgfr2b and Fgfr2c in renal development [62]. They found that Fgfr2b was mainly expressed in the epithelium of the Wolffian duct and ureteric tree, but to some extent also in the cap mesenchyme. Deletion of Fgfr2c did not affect kidney development. However, deletion of Fgfr2c together with conditional deletion of Fgfr1 in the metanephric mesenchyme led to renal dysgenesis. The phenotype was somewhat milder than that of the mice with compound deletion of Fgfr1MM−/− and Fgfr2MM−/− (see above). In particular, some metanephric mesenchyme that expressed Six2 and Pax2 was identified at early stages, but the Fgfr1MM−/−;Fgfr2c−/− mice had no renal tissue beyond E12.5. Thus, Fgfr1 and Fgfr2c act redundantly and at least one of the two receptors is required in the metanephric mesenchyme for normal kidney development.
Role of FgfrL1 in early kidney development
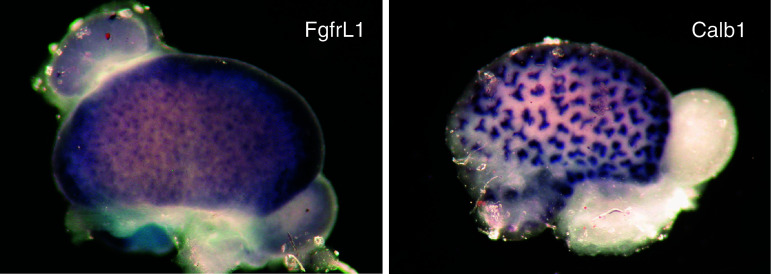
To investigate the function of the fifth receptor, our group has generated mice with a targeted disruption of the FgfrL1 gene. The promoter region and the first two exons of this gene were replaced by a neo cassette [63]. FgfrL1 is normally expressed in all nephrogenic structures of the cortical zone, including pretubular aggregates, renal vesicles, comma- and S-shaped bodies [64, 65] (Fig. 3). It is also found in tubules of the nascent nephrons and in the ureteric bud epithelium, but it is not expressed in the cortical and medullary stroma or in fully differentiated glomeruli. Staining of an E15.5 kidney by whole mount in situ hybridization is shown in Fig. 4. FgfrL1 knockout mice were born alive but died immediately after birth due to respiratory problems. These problems were explained by a rudimentary diaphragm muscle that was not strong enough to inflate the lungs after birth. However, the major alteration of our knockout mice was the nearly complete absence of the metanephric kidneys [64]. The FgfrL1 null mice contained only a small nodule at the end of the ureter that was barely thicker than the ureter itself.
Fig. 4.
Expression pattern of FgfrL1 in mouse kidneys. Distribution of the FgfrL1 mRNA is shown at E15.5 by whole mount in situ hybridization with a probe for mouse FgfrL1 (left) and mouse calbindin (control, right). Expression of FgfrL1 is widely observed in the nephrogenic zone of the developing kidney but barely in the adrenal gland or the gonads. Expression of calbindin is restricted to the ureteric buds. See [64, 65] for details on the experimental procedures
At E11.5, the ureter had begun to invade the metanephric mesenchyme of the mutant kidneys and to branch. At E12.5, the mesenchyme had condensed around the ureteric tips to form the cap mesenchyme but then the condensed mesenchyme did not undergo the mesenchymal-to-epithelial transition. Consequently, no epithelial renal vesicles formed and the ureteric tips ceased to branch. At E14.5 increased apoptosis was observed in the cortical zone of the developing kidney and reduced proliferation was noted in the metanephric mesenchyme. The expression of marker genes was investigated by RT-PCR, by in situ hybridization and by immunohistochemistry [64]. Expression of the nephrogenic markers Wnt4, Lhx1 and Fgf8 was significantly reduced. However, markers of the uninduced mesenchyme (Six2, Eya1, Wt1) or markers of the ureteric bud (Wnt9b, Wnt11) were barely affected. Organ cultures performed with kidney rudiments dissected from mutant mice at E11.5 confirmed these observations [64]. The cultures showed no differentiation of the mesenchyme and severely reduced branching of the ureter. Taken together, our in vivo and in vitro studies suggest that the ureteric bud of FgfrL1 deficient animals received the correct signals to invade the metanephric mesenchyme, where it branched several times and sent out inductive signals to the mesenchyme. However, the metanephric mesenchyme was not responsive to these signals and failed to differentiate into epithelial structures. As a consequence, branching of the ureteric tips stopped and the uninduced cells started to decay by apoptosis.
To identify genes that may act downstream of FgfrL1 in the regulatory hierarchy of genes required for early nephron development, we used DNA microarray profiling [65]. When the gene expression profiles of kidney rudiments from wild-type and knockout mice were compared at E12.5, we detected 56 differentially expressed transcripts with 2-fold or greater reduction in the mutant kidneys, including Fgf8, Wnt4 and Lhx1. The best hits with the most significant changes in expression levels are listed in Table 2. Most of these hits are normally expressed in the renal vesicles and/or the ureteric bud as indicated by the gene expression data provided by GUDMAP [66]. An interesting finding of our microarray experiments was that the lack of FgfrL1 expression was barely compensated for by the up-regulation of other genes. In particular, the classical receptors Fgfr1–Fgfr4 did not show any altered expression in the mutant kidneys. The fact that no signature of increased Fgf signaling could be detected raises the intriguing possibility that FgfrL1 might also be involved in pathways or activities other than Fgf signaling.
Table 2.
Differential gene expression between wild-type and FgfrL1 deficient kidneys at E12.5
| Gene name | Fold change wt/ko | Protein accession | Amino acids | Subcellular localization | Function | Location of expression | |
|---|---|---|---|---|---|---|---|
| Array | qPCR | ||||||
| Svopl | 9.5 | 9.4 | Q6PDF3 | 494 | Membrane | Transporter | RV |
| Krt23 | 7.4 | 17.4 | Q99PS0 | 422 | Cytoskeleton | Intermediate filament | UB |
| Slc32a1 | 7.0 | >20 | O35633 | 525 | Membrane | Transporter | UB, RV |
| Wnt4 | 6.2 | 13.3 | P22724 | 351 | Secreted | Growth factor | RV |
| Dkk1 | 5.4 | 3.0 | O54908 | 272 | Secreted | Antagonist of Wnt | UB, RV |
| Egr1 | 4.9 | 4.6 | P08046 | 533 | Nucleus | Transcription factor | UB, mesenchyme |
| Pcp4 | 4.4 | 17.0 | P63054 | 62 | Cytosol | Neuronal differentiation | Bladder |
| Fgf8 | 4.4 | 9.2 | P37237 | 268 | Secreted | Growth factor | RV |
| Lhx1 | 4.1 | 7.7 | P63006 | 406 | Nucleus | Transcription factor | UB, RV |
| Sp5 | 3.7 | Not done | Q9JHX2 | 398 | Nucleus | Transcription factor | RV |
| Aldh1a1 | 3.5 | 4.0 | P24549 | 501 | Cytosol | Redox enzyme | Ovary, testis |
| Clec18a | 3.4 | 2.3 | Q7TSQ1 | 534 | Secreted | C-type lectin | RV |
| FgfrL1 | 3.1 | 6.8 | Q91V87 | 529 | Membrane | Growth factor receptor | UB, RV |
RV renal vesicle, UB ureteric bud
Mechanism of FgfrL1 action
The molecular mechanisms, by which FgfrL1 controls nephron formation, are not yet clear. Together with Wnt4, FgfrL1 is one of the unique genes whose deletion specifically interferes with kidney formation, without grossly affecting other organs of the animal. We have reported that FgfrL1 knockout mice have a hypoplastic diaphragm muscle but that all other muscles appear to be normal [63]. We also observed subtle alterations in the skeleton, in particular in the skull [19]. Another group of researchers, who generated FgfrL1 deficient mice by deletion of exons 3–7, described hypoplasia of several skeletal elements (shortened bones, malformed vertebrae), heart defects (thickened ventricular valves, septation anomalies) and transient fetal anemia in mutant mice, but they did not report any data about the missing kidneys [67]. We have evidence that most of these relatively mild alterations are secondary effects due to the absence of the kidneys. Bilateral kidney agenesis causes oligohydramnios, a deficiency of amniotic fluid, also termed Potter sequence or Potter syndrome [68]. Furthermore, the lack of the kidneys will affect the renin–angiotensin system of the growing fetus because renin is produced in the arterioles of the kidneys [69]. When renin is absent from the blood circulation, angiotensinogen cannot be converted to angiotensin and the fetus will be subjected to a life-threatening hypotension. Together with the deficiency of the amniotic fluid, this hypotension will cause a dramatic compression of the embryo. In fact, human fetuses with Potter syndrome show typical signs of intrauterine compression, such as facial dysmorphia, lung hypoplasia and skull ossification defects with large sutures [68]. Skull ossification defects are a common consequence of the low blood pressure since nascent bones need a high oxygen tension for normal growth. Oligohydramnios and hypotension are also observed in mice lacking the metanephric kidneys [69, 70]. Most of the alterations in the skeleton of our knockout mice can therefore be explained by the missing kidneys. Our conclusion is further supported by findings from a third group of researchers who developed FgfrL1 deficient mice and published the results as a patent application [71]. These authors reported that all their homozygous FgfrL1−/− mice died around birth and exhibited a fully penetrant kidney phenotype with bilateral kidney dysgenesis/agenesis as observed with our mice. At embryonic stage E10.5, the ureteric bud of their FgfrL1 deficient mice still invaded the metanephric mesenchyme, but then failed to branch and form the characteristic T-shaped structure. Expression of Pax2 was severely reduced in the metanephric mesenchyme as in our mice. It is therefore likely that kidney dysgenesis/agenesis is a general feature of FgfrL1 knock-out mice.
The FgfrL1 gene occurs in most animal classes, from sea anemones to fish and mammals [15, 16]. In contrast, metanephric kidneys are only found in amniotes, that is reptiles, birds and mammals. Fish and amphibians have pronephros and mesonephros but no metanephric kidneys. Therefore FgfrL1 must have taken over a completely novel function during the evolution of higher vertebrates. Interestingly, pronephros and mesonephros, which also develop in mammals, do not appear to be affected in our knockout animals [64]. Curiously enough, FgfrL1 expression is relatively low in the developing kidneys, while it is relatively high in the skeleton, especially in cartilage [14]. To our surprise, cartilaginous elements did not appear to be affected in our FgfrL1 deficient mice.
The kidney alterations observed in FgfrL1 deficient mice appear to be highly similar to the phenotypes of mice that harbor specific deletions of Fgf8, Wnt4 or Lhx1. Animals with a conditional deletion of Fgf8 in the metanephric mesenchyme initiate nephrogenesis, but nephron development is arrested at the stage of the renal vesicle [5, 6]. Mice with a global deletion of Wnt4 fail to form renal vesicles, however other aspects of mesenchymal and ureteric development are barely affected [7]. Mice with a conditional deletion of Lhx1 have kidneys with renal vesicles that do not develop beyond the S-shaped body stage [74]. Nascent nephrons from kidneys that lack Wnt4 or Lhx1 still express Fgf8 [5, 6]. In our FgfrL1 null mice, nephron development appears to be affected at an earlier stage because the condensed mesenchyme fails to undergo the mesenchymal-to-epithelial transition and because no renal vesicles can be observed [64]. Kidney development is blocked even at an earlier stage in mice with a compound deletion of the receptors Fgfr1 and Fgfr2 that lack a proper metanephric mesenchyme [60]. Although a linear hierarchy of genes might be too simplistic and will have to be taken with a lot of precaution, we can tentatively draw a cascade of genes required for early nephron development in the metanephric mesenchyme as follows: Fgfr1/Fgfr2 > FgfrL1 > Fgf8 > Wnt4, Lhx1. This order is consistent with the fact that expression of Fgf8, Wnt4 and Lhx1 is missing in our FgfrL1 deficient mice.
It is worthwhile to mention in this context that Fgf signaling often acts in concert with Wnt signaling to evoke a complete response [72]. Fgf8 for example is required to initiate Wnt4 expression in pretubular aggregates of the metanephric mesenchyme and, in turn, Wnt4 signaling reinforces Fgf8 expression in nascent renal vesicles [5, 6]. Generally, it appears that the two pathways have opposing effects on the transcription of co-regulated genes, at least in the zebrafish tailbud [73]. During development of the mouse kidney, Wnt9b is secreted from the ureteric bud into the metanephric mesenchyme, where it triggers the canonical β-catenin pathway and initiates nephron formation [4]. However, the corresponding Fgf pathway has not yet been described. Given the strikingly similar phenotypes of Wnt9b deficient kidneys and FgfrL1 deficient kidneys and the fact that Wnt9b is still expressed in the FgfrL1 deficient epithelium, it is tempting to speculate that FgfrL1 represents the mediator of the Fgf signal that acts in concert with Wnt9b to initiate nephron formation.
Currently, we envisage two possible molecular mechanisms how FgfrL1 might be involved in nephron formation. We have demonstrated that FgfrL1 binds Fgf ligands and that it can act as a decoy receptor, especially when it is shed from the plasma membrane [20]. It could therefore sequester Fgfs away from the signaling receptors Fgfr1/Fgfr2. Of all Fgfs that are expressed in the metanephric mesenchyme (Fgf1, 7, 8, 9, 10, 12, 20) only Fgf8 appears to bind to FgfrL1 with high affinity, whereas Fgf1, 7, 9, 12 and 20 do not interact at all [20]. Fgf8 might therefore represent the major ligand for FgfrL1 in the kidney. This notion is consistent with the observation that FgfrL1 is usually expressed in tissues that also express Fgf8. In fact, the two proteins have been reported to form a synexpression group in Xenopus laevis [75]. If FgfrL1 binds Fgf8 it may control the diffusion range of this morphogen and restrict it to the location where the renal vesicle has to form. Indeed, Fgf8 has been found to act as an important tissue organizer in other organs. During development of the eye, Fgf8 is part of the “FGF signaling center” that triggers differentiation of retinal ganglion cells [76]. During formation of the encephalon, Fgf8 is involved in the establishment of the isthmic organizer that controls development of the midbrain and hindbrain [77].
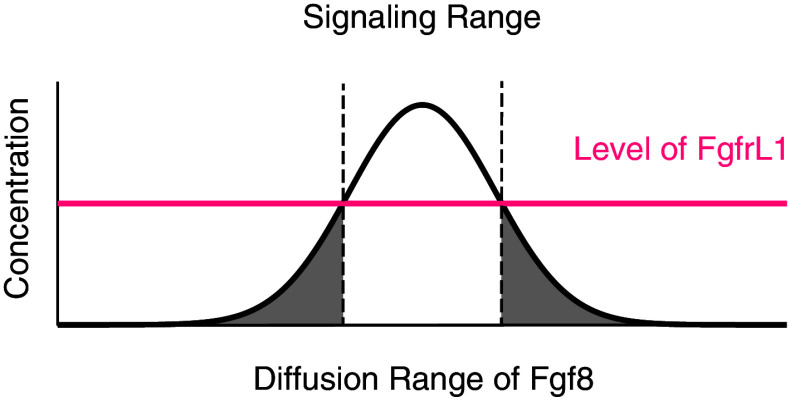
It is of interest to note in this context that the homologous receptor FgfrL1 from the flat worm Dugesia japonica, which was termed nou-darake (“brains everywhere”), might function in a similar way [78]. Nou-darake is specifically expressed in the head region of the flat worms. Depletion of the receptor by RNAi resulted in formation of ectopic brain tissue throughout the body of the animals. These results suggest that nou-darake might bind and neutralize a brain-inducing factor (probably an Fgf) and restrict the diffusion radius of this factor to the planarian head. It is conceivable that FgfrL1 acts in a similar way in the developing mouse kidney. FgfrL1 is expressed at relatively low levels in the renal vesicle. It might therefore be able to neutralize Fgf8 only in regions where the levels of this morphogen are low, that is in the distal region of the secreting source (Fig. 5). In the proximal regions where the Fgf8 levels are high and exceed the levels of FgfrL1, Fgf8 cannot be neutralized and consequently, signaling via its receptors Fgfr1/Fgfr2 may take place despite the presence of FgfrL1. In this way, FgfrL1 would confine Fgf signaling to the precise location of the renal vesicle (Fig. 5).
Fig. 5.
Restriction of Fgf8/Fgfr1 signaling to nephrogenic structures by FgfrL1. FgfrL1, which is widely expressed in the nephrogenic zone of the developing kidney, binds and neutralizes Fgf8, which is secreted by nascent nephrons. If the concentration of FgfrL1 is lower than that of Fgfr1 and if the affinity of FgfrL1/Fgf8 is higher than that of Fgfr1/Fgf8, FgfrL1 could neutralize Fgf8 in regions that have low levels of Fgf8, but not in regions that have high levels. FgfrL1 would therefore restrict the signaling range of Fgf8 to the region of the secreting source, the renal vesicle
A second possibility would be that FgfrL1 acts as a major cell adhesion protein. We have demonstrated that it efficiently promotes fusion of CHO cells in vitro [23]. To our knowledge, there is no evidence for any cell–cell fusion event occurring during nephron development. However, cell fusion might just be the most extreme result of very tight cell adhesion that in the case of CHO cells might end up in membrane fusion. Under normal conditions, FgfrL1 might only bring together the cell membranes into intimate contact. In fact, FgfrL1 is able to induce tight adhesion of various cell types if coated on plastic plates [22]. Furthermore, FgfrL1 is found at the contact sites of cells in the renal vesicles [64, 65]. The receptor could therefore function, together with other cell adhesion proteins, in the condensation of the metanephric mesenchyme around the ureteric tips. If FgfrL1 is absent as observed in our knockout mice, the other adhesion molecules might not be able to bring together the mesenchymal cells into close contact and the mesenchymal-to-epithelial conversion might then not take place.
Further studies will be required to discriminate between the two possibilities outlined above. It is also conceivable that the two possibilities are not mutually exclusive and that FgfrL1 acts as a modulator to restrict the Fgf8 diffusion range as well as an adhesion protein to properly align mesenchymal cells for epithelial conversion.
Acknowledgments
Research in the laboratory of the authors was supported by grants from the Swiss National Science Foundation (31003A-143350), from the Helmut Horten Foundation and from the Swiss Foundation for Research on Muscular Diseases.
References
- 1.Costantini F, Kopan R. Patterning a complex organ: branching morphogenesis and nephron segmentation in kidney development. Dev Cell. 2010;18:698–712. doi: 10.1016/j.devcel.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Georgas K, Rumballe B, Valerius MT, Chiu HS, Thiagarajan RD, Lesieur E, Aronow BJ, Brunskill EW, Combes AN, Tang D, Taylor D, Grimmond SM, Potter SS, McMahon AP, Little MH. Analysis of early nephron patterning reveals a role for distal RV proliferation in fusion to the ureteric tip via a cap mesenchyme-derived connecting segment. Dev Biol. 2009;332:273–286. doi: 10.1016/j.ydbio.2009.05.578. [DOI] [PubMed] [Google Scholar]
- 3.Hendry C, Rumballe B, Moritz K, Little MH. Defining and redefining the nephron progenitor population. Pediatr Nephrol. 2011;26:1395–1406. doi: 10.1007/s00467-010-1750-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park JS, Valerius MT, McMahon AP. Wnt/beta-catenin signaling regulates nephron induction during mouse kidney development. Development. 2007;134:2533–2539. doi: 10.1242/dev.006155. [DOI] [PubMed] [Google Scholar]
- 5.Grieshammer U, Cebrian C, Ilagan R, Meyers E, Herzlinger D, Martin GR. FGF8 is required for cell survival at distinct stages of nephrogenesis and for regulation of gene expression in nascent nephrons. Development. 2005;132:3847–3857. doi: 10.1242/dev.01944. [DOI] [PubMed] [Google Scholar]
- 6.Perantoni AO, Timofeeva O, Naillat F, Richman C, Pajni-Underwood S, Wilson C, Vainio S, Dove LF, Lewandoski M. Inactivation of FGF8 in early mesoderm reveals an essential role in kidney development. Development. 2005;132:3859–3871. doi: 10.1242/dev.01945. [DOI] [PubMed] [Google Scholar]
- 7.Stark K, Vainio S, Vassileva G, McMahon AP. Epithelial transformation of metanephric mesenchyme in the developing kidney regulated by Wnt-4. Nature. 1994;372:679–683. doi: 10.1038/372679a0. [DOI] [PubMed] [Google Scholar]
- 8.Wang P, Pereira FA, Beasley D, Zheng H. Presenilins are required for the formation of comma- and s-shaped bodies during nephrogenesis. Development. 2003;130:5019–5029. doi: 10.1242/dev.00682. [DOI] [PubMed] [Google Scholar]
- 9.Cheng HT, Kim M, Valerius MT, Surendran K, Schuster-Gossler K, Gossler A, McMahon AP, Kopan R. Notch2, but not Notch1, is required for proximal fate acquisition in the mammalian nephron. Development. 2007;134:801–811. doi: 10.1242/dev.02773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Itoh N. The Fgf families in humans, mice, and zebrafish: their evolutional processes and roles in development, metabolism, and disease. Biol Pharm Bull. 2007;30:1819–1825. doi: 10.1248/bpb.30.1819. [DOI] [PubMed] [Google Scholar]
- 11.Beenken A, Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nat Rev Drug Discov. 2009;8:235–253. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dailey L, Ambrosetti D, Mansukhani A, Basilico C. Mechanisms underlying differential responses to FGF signaling. Cytokine Growth Factor Rev. 2005;16:233–247. doi: 10.1016/j.cytogfr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 13.Lew ED, Furdui CM, Anderson KS, Schlessinger J. The precise sequence of FGF receptor autophosphorylation is kinetically driven and is disrupted by oncogenic mutations. Sci Signal. 2009;2:ra6. doi: 10.1126/scisignal.2000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiedemann M, Trueb B. Characterization of a novel protein (FGFRL1) from human cartilage related to FGF receptors. Genomics. 2000;69:275–279. doi: 10.1006/geno.2000.6332. [DOI] [PubMed] [Google Scholar]
- 15.Trueb B. Biology of FGFRL1, the fifth fibroblast growth factor receptor. Cell Mol Life Sci. 2011;68:951–964. doi: 10.1007/s00018-010-0576-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertrand S, Somorjai I, Garcia-Fernandez J, Lamonerie T, Escriva H. FGFRL1 is a neglected putative actor of the FGF signalling pathway present in all major metazoan phyla. BMC Evol Biol. 2009;9:226. doi: 10.1186/1471-2148-9-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhuang L, Karotki AV, Bruecker P, Trueb B. Comparison of the receptor FGFRL1 from sea urchins and humans illustrates evolution of a zinc binding motif in the intracellular domain. BMC Biochem. 2009;10:33. doi: 10.1186/1471-2091-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhuang L, Villiger P, Trueb B. Interaction of the receptor FGFRL1 with the negative regulator Spred1. Cell Signal. 2011;23:1496–1504. doi: 10.1016/j.cellsig.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 19.Rieckmann T, Zhuang L, Flück CE, Trueb B. Characterization of the first FGFRL1 mutation identified in a craniosynostosis patient. Biochim Biophys Acta. 2009;1792:112–121. doi: 10.1016/j.bbadis.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 20.Steinberg F, Zhuang L, Beyeler M, Kalin RE, Mullis PE, Brandli AW, Trueb B. The FGFRL1 receptor is shed from cell membranes, binds FGFs and antagonizes FGF signaling in Xenopus embryos. J Biol Chem. 2010;285:2193–2202. doi: 10.1074/jbc.M109.058248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trueb B, Zhuang L, Taeschler S, Wiedemann M. Characterization of FGFRL1, a novel FGF receptor preferentially expressed in skeletal tissues. J Biol Chem. 2003;278:33857–33865. doi: 10.1074/jbc.M300281200. [DOI] [PubMed] [Google Scholar]
- 22.Rieckmann T, Kotevic I, Trueb B. The cell surface receptor FGFRL1 forms constitutive dimers that promote cell adhesion. Exp Cell Res. 2008;314:1071–1081. doi: 10.1016/j.yexcr.2007.10.029. [DOI] [PubMed] [Google Scholar]
- 23.Steinberg F, Gerber S, Rieckmann T, Trueb B. Rapid fusion and syncytium formation of heterologous cells upon expression of the FGFRL1 receptor. J Biol Chem. 2010;285:37704–37715. doi: 10.1074/jbc.M110.140517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sleeman M, Fraser J, McDonald M, Yuan S, White D, Grandison P, Kumble K, Watson JD, Murison JG. Identification of a new fibroblast growth factor receptor, FGFR5. Gene. 2001;271:171–182. doi: 10.1016/S0378-1119(01)00518-2. [DOI] [PubMed] [Google Scholar]
- 25.Chen EH, Grote E, Mohler W, Vignery A. Cell-cell fusion. FEBS Lett. 2007;581:2181–2193. doi: 10.1016/j.febslet.2007.03.033. [DOI] [PubMed] [Google Scholar]
- 26.Oren-Suissa M, Podbilewicz B. Evolution of programmed cell fusion: common mechanisms and distinct functions. Dev Dyn. 2010;239:1515–1528. doi: 10.1002/dvdy.22284. [DOI] [PubMed] [Google Scholar]
- 27.Cancilla B, Ford-Perriss MD, Bertram JF. Expression and localization of fibroblast growth factors and fibroblast growth factor receptors in the developing rat kidney. Kidney Int. 1999;56:2025–2039. doi: 10.1046/j.1523-1755.1999.00781.x. [DOI] [PubMed] [Google Scholar]
- 28.Brown AC, Adams D, de Caestecker M, Yang X, Friesel R, Oxburgh L. FGF/EGF signaling regulates the renewal of early nephron progenitors during embryonic development. Development. 2011;138:5099–5112. doi: 10.1242/dev.065995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barasch J, Qiao J, McWilliams G, Chen D, Oliver JA, Herzlinger D. Ureteric bud cells secrete multiple factors, including bFGF, which rescue renal progenitors from apoptosis. Am J Physiol. 1997;273:F757–F767. doi: 10.1152/ajprenal.1997.273.5.F757. [DOI] [PubMed] [Google Scholar]
- 30.Perantoni AO, Dove LF, Karavanova I. Basic fibroblast growth factor can mediate the early inductive events in renal development. Proc Natl Acad Sci USA. 1995;92:4696–4700. doi: 10.1073/pnas.92.10.4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barasch J, Yang J, Ware CB, Taga T, Yoshida K, Erdjument-Bromage H, Tempst P, Parravicini E, Malach S, Aranoff T, Oliver JA. Mesenchymal to epithelial conversion in rat metanephros is induced by LIF. Cell. 1999;99:377–386. doi: 10.1016/S0092-8674(00)81524-X. [DOI] [PubMed] [Google Scholar]
- 32.Plisov SY, Yoshino K, Dove LF, Higinbotham KG, Rubin JS, Perantoni AO. TGF beta 2, LIF and FGF2 cooperate to induce nephrogenesis. Development. 2001;128:1045–1057. doi: 10.1242/dev.128.7.1045. [DOI] [PubMed] [Google Scholar]
- 33.Qiao J, Bush KT, Steer DL, Stuart RO, Sakurai H, Wachsman W, Nigam SK. Multiple fibroblast growth factors support growth of the ureteric bud but have different effects on branching morphogenesis. Mech Dev. 2001;109:123–135. doi: 10.1016/S0925-4773(01)00592-5. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen HQ, Danilenko DM, Bucay N, DeRose ML, Van GY, Thomason A, Simonet WS. Expression of keratinocyte growth factor in embryonic liver of transgenic mice causes changes in epithelial growth and differentiation resulting in polycystic kidneys and other organ malformations. Oncogene. 1996;12:2109–2119. [PubMed] [Google Scholar]
- 35.Miller DL, Ortega S, Bashayan O, Basch R, Basilico C. Compensation by fibroblast growth factor 1 (FGF1) does not account for the mild phenotypic defects observed in FGF2 null mice. Mol Cell Biol. 2000;20:2260–2268. doi: 10.1128/MCB.20.6.2260-2268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo L, Degenstein L, Fuchs E. Keratinocyte growth factor is required for hair development but not for wound healing. Genes Dev. 1996;10:165–175. doi: 10.1101/gad.10.2.165. [DOI] [PubMed] [Google Scholar]
- 37.Qiao J, Uzzo R, Obara-Ishihara T, Degenstein L, Fuchs E, Herzlinger D. FGF-7 modulates ureteric bud growth and nephron number in the developing kidney. Development. 1999;126:547–554. doi: 10.1242/dev.126.3.547. [DOI] [PubMed] [Google Scholar]
- 38.Min H, Danilenko DM, Scully SA, Bolon B, Ring BD, Tarpley JE, DeRose M, Simonet WS. Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev. 1998;12:3156–3161. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, Yagishita N, Matsui D, Koga Y, Itoh N, Kato S. Fgf10 is essential for limb and lung formation. Nat Genet. 1999;21:138–141. doi: 10.1038/5096. [DOI] [PubMed] [Google Scholar]
- 40.Ohuchi H, Hori Y, Yamasaki M, Harada H, Sekine K, Kato S, Itoh N. FGF10 acts as a major ligand for FGF receptor 2 IIIb in mouse multi-organ development. Biochem Biophys Res Commun. 2000;277:643–649. doi: 10.1006/bbrc.2000.3721. [DOI] [PubMed] [Google Scholar]
- 41.Michos O, Cebrian C, Hyink D, Grieshammer U, Williams L, D’Agati V, Licht JD, Martin GR, Costantini F. Kidney development in the absence of Gdnf and Spry1 requires Fgf10. PLoS Genet. 2010;6:e1000809. doi: 10.1371/journal.pgen.1000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun X, Meyers EN, Lewandoski M, Martin GR. Targeted disruption of Fgf8 causes failure of cell migration in the gastrulating mouse embryo. Genes Dev. 1999;13:1834–1846. doi: 10.1101/gad.13.14.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barak H, Huh SH, Chen S, Jeanpierre C, Martinovic J, Parisot M, Bole-Feysot C, Nitschke P, Salomon R, Antignac C, et al. FGF9 and FGF20 maintain the stemness of nephron progenitors in mice and man. Dev Cell. 2012;22:1191–1207. doi: 10.1016/j.devcel.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Colvin JS, White AC, Pratt SJ, Ornitz DM. Lung hypoplasia and neonatal death in Fgf9-null mice identify this gene as an essential regulator of lung mesenchyme. Development. 2001;128:2095–2106. doi: 10.1242/dev.128.11.2095. [DOI] [PubMed] [Google Scholar]
- 45.Colvin JS, Green RP, Schmahl J, Capel B, Ornitz DM. Male-to-female sex reversal in mice lacking fibroblast growth factor 9. Cell. 2001;104:875–889. doi: 10.1016/S0092-8674(01)00284-7. [DOI] [PubMed] [Google Scholar]
- 46.Huh SH, Jones J, Warchol ME, Ornitz DM. Differentiation of the lateral compartment of the cochlea requires a temporally restricted FGF20 signal. PLoS Biol. 2012;10:e1001231. doi: 10.1371/journal.pbio.1001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deng CX, Wynshaw-Boris A, Shen MM, Daugherty C, Ornitz DM, Leder P. Murine FGFR-1 is required for early postimplantation growth and axial organization. Genes Dev. 1994;8:3045–3057. doi: 10.1101/gad.8.24.3045. [DOI] [PubMed] [Google Scholar]
- 48.Yamaguchi TP, Harpal K, Henkemeyer M, Rossant J. Fgfr-1 is required for embryonic growth and mesodermal patterning during mouse gastrulation. Genes Dev. 1994;8:3032–3044. doi: 10.1101/gad.8.24.3032. [DOI] [PubMed] [Google Scholar]
- 49.Xu X, Weinstein M, Li C, Naski M, Cohen RI, Ornitz DM, Leder P, Deng C. Fibroblast growth factor receptor 2 (FGFR2)-mediated reciprocal regulation loop between FGF8 and FGF10 is essential for limb induction. Development. 1998;125:753–765. doi: 10.1242/dev.125.4.753. [DOI] [PubMed] [Google Scholar]
- 50.Arman E, Haffner-Krausz R, Chen Y, Heath JK, Lonai P. Targeted disruption of fibroblast growth factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proc Natl Acad Sci USA. 1998;95:5082–5087. doi: 10.1073/pnas.95.9.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Revest JM, Spencer-Dene B, Kerr K, De Moerlooze L, Rosewell I, Dickson C. Fibroblast growth factor receptor 2-IIIb acts upstream of Shh and Fgf4 and is required for limb bud maintenance but not for the induction of Fgf8, Fgf10, Msx1, or Bmp4. Dev Biol. 2001;231:47–62. doi: 10.1006/dbio.2000.0144. [DOI] [PubMed] [Google Scholar]
- 52.Celli G, LaRochelle WJ, Mackem S, Sharp R, Merlino G. Soluble dominant-negative receptor uncovers essential roles for fibroblast growth factors in multi-organ induction and patterning. EMBO J. 1998;17:1642–1655. doi: 10.1093/emboj/17.6.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Colvin JS, Bohne BA, Harding GW, McEwen DG, Ornitz DM. Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nat Genet. 1996;12:390–397. doi: 10.1038/ng0496-390. [DOI] [PubMed] [Google Scholar]
- 54.Weinstein M, Xu X, Ohyama K, Deng CX. FGFR-3 and FGFR-4 function cooperatively to direct alveogenesis in the murine lung. Development. 1998;125:3615–3623. doi: 10.1242/dev.125.18.3615. [DOI] [PubMed] [Google Scholar]
- 55.Bates CM. Role of fibroblast growth factor receptor signaling in kidney development. Am J Physiol Renal Physiol. 2011;301:F245–F251. doi: 10.1152/ajprenal.00186.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao H, Kegg H, Grady S, Truong HT, Robinson ML, Baum M, Bates CM. Role of fibroblast growth factor receptors 1 and 2 in the ureteric bud. Dev Biol. 2004;276:403–415. doi: 10.1016/j.ydbio.2004.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sims-Lucas S, Cullen-McEwen L, Eswarakumar VP, Hains D, Kish K, Becknell B, Zhang J, Bertram JF, Wang F, Bates CM. Deletion of Frs2α from the ureteric epithelium causes renal hypoplasia. Am J Physiol Renal Physiol. 2009;297:F1208–F1219. doi: 10.1152/ajprenal.00262.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eswarakumar VP, Ozcan F, Lew ED, Bae JH, Tome F, Booth CJ, Adams DJ, Lax I, Schlessinger J. Attenuation of signaling pathways stimulated by pathologically activated FGF-receptor 2 mutants prevents craniosynostosis. Proc Natl Acad Sci USA. 2006;103:18603–18608. doi: 10.1073/pnas.0609157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sims-Lucas S, Cusack B, Eswarakumar VP, Zhang J, Wang F, Bates CM. Independent roles of Fgfr2 and Frs2α in ureteric epithelium. Development. 2011;138:1275–1280. doi: 10.1242/dev.062158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poladia DP, Kish K, Kutay B, Hains D, Kegg H, Zhao H, Bates CM. Role of fibroblast growth factor receptors 1 and 2 in the metanephric mesenchyme. Dev Biol. 2006;291:325–339. doi: 10.1016/j.ydbio.2005.12.034. [DOI] [PubMed] [Google Scholar]
- 61.Hains D, Sims-Lucas S, Kish K, Saha M, McHugh K, Bates CM. Role of fibroblast growth factor receptor 2 in kidney mesenchyme. Pediatr Res. 2008;64:592–598. doi: 10.1203/PDR.0b013e318187cc12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sims-Lucas S, Cusack B, Baust J, Eswarakumar VP, Masatoshi H, Takeuchi A, Bates CM. Fgfr1 and the IIIc isoform of Fgfr2 play critical roles in the metanephric mesenchyme mediating early inductive events in kidney development. Dev Dyn. 2011;240:240–249. doi: 10.1002/dvdy.22501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baertschi S, Zhuang L, Trueb B. Mice with a targeted disruption of the FgfrL1 gene die at birth due to alterations in the diaphragm. FEBS J. 2007;274:6241–6253. doi: 10.1111/j.1742-4658.2007.06143.x. [DOI] [PubMed] [Google Scholar]
- 64.Gerber SD, Steinberg F, Beyeler M, Villiger PM, Trueb B. The murine FgfrL1 receptor is essential for the development of the metanephric kidney. Dev Biol. 2009;335:106–119. doi: 10.1016/j.ydbio.2009.08.019. [DOI] [PubMed] [Google Scholar]
- 65.Gerber SD, Amann R, Wyder S, Trueb B. Comparison of the gene expression profiles from normal and FgfrL1 deficient mouse kidneys reveals downstream targets of FgfrL1 signaling. PLoS ONE. 2012;7:e33457. doi: 10.1371/journal.pone.0033457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brunskill EW, Aronow BJ, Georgas K, Rumballe B, Valerius MT, Aronow J, Kaimal V, Jegga AG, Yu J, Grimmond S, McMahon AP, Patterson LT, Little MH, Potter SS. Atlas of gene expression in the developing kidney at microanatomic resolution. Dev Cell. 2008;15:781–791. doi: 10.1016/j.devcel.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Catela C, Bilbao-Cortes D, Slonimsky E, Kratsios P, Rosenthal N, Te Welscher P. Multiple congenital malformations of Wolf-Hirschhorn syndrome are recapitulated in FgfrL1 null mice. Dis Model Mech. 2009;2:283–294. doi: 10.1242/dmm.002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Potter EL. Pathology of the fetus and infant. 2. Chicago: Year Book Medical Publishers; 1962. [Google Scholar]
- 69.Gubler MC, Antignac C. Renin-angiotensin system in kidney development: renal tubular dysgenesis. Kidney Int. 2010;77:400–406. doi: 10.1038/ki.2009.423. [DOI] [PubMed] [Google Scholar]
- 70.Smith NP, Losty PD, Connell MG, Mayer U, Jesudason EC. Abnormal lung development precedes oligohydramnios in a transgenic murine model of renal dysgenesis. J Urol. 2006;175:783–786. doi: 10.1016/S0022-5347(05)00169-2. [DOI] [PubMed] [Google Scholar]
- 71.Anderson SJ, Brennan J, de Sauvage FJ, Ding Z, Edwards J, Fikes NA, Huang W, Ouyang W, Rangel C, Sangha M, Shi Z–Z, Sparks MJ, Trackey J, Vetter M, Wang C-Y, Woodings J (2007) Novel gene disruptions, compositions and methods relating thereto. United States Patent Application 20070292438
- 72.Katoh M, Katoh M. Cross-talk of WNT and FGF signaling pathways at GSK3beta to regulate beta-catenin and SNAIL signaling cascades. Cancer Biol Ther. 2006;5:1059–1064. doi: 10.4161/cbt.5.9.3151. [DOI] [PubMed] [Google Scholar]
- 73.Stulberg MJ, Lin A, Zhao H, Holley SA. Crosstalk between Fgf and Wnt signaling in the zebrafish tailbud. Dev Biol. 2012;369:298–307. doi: 10.1016/j.ydbio.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kobayashi A, Kwan KM, Carroll TJ, McMahon AP, Mendelsohn CL, Behringer RR. Distinct and sequential tissue-specific activities of the LIM-class homeobox gene Lim1 for tubular morphogenesis during kidney development. Development. 2005;132:2809–2823. doi: 10.1242/dev.01858. [DOI] [PubMed] [Google Scholar]
- 75.Hayashi S, Itoh M, Taira S, Agata K, Taira M. Expression patterns of Xenopus FGF receptor-like 1/nou-darake in early Xenopus development resemble those of planarian nou-darake and Xenopus FGF8. Dev Dyn. 2004;230:700–707. doi: 10.1002/dvdy.20040. [DOI] [PubMed] [Google Scholar]
- 76.Martinez-Morales JR, Del Bene F, Nica G, Hammerschmidt M, Bovolenta P, Wittbrodt J. Differentiation of the vertebrate retina is coordinated by an FGF signaling center. Dev Cell. 2005;8:565–574. doi: 10.1016/j.devcel.2005.01.022. [DOI] [PubMed] [Google Scholar]
- 77.Chi CL, Martinez S, Wurst W, Martin GR. The isthmic organizer signal FGF8 is required for cell survival in the prospective midbrain and cerebellum. Development. 2003;130:2633–2644. doi: 10.1242/dev.00487. [DOI] [PubMed] [Google Scholar]
- 78.Cebria F, Kobayashi C, Umesono Y, Nakazawa M, Mineta K, Ikeo K, Gojobori T, Itoh M, Taira M, Sanchez Alvarado A, Agata K. FGFR-related gene nou-darake restricts brain tissues to the head region of planarians. Nature. 2002;419:620–624. doi: 10.1038/nature01042. [DOI] [PubMed] [Google Scholar]