Abstract
O-GlcNAcylation is a dynamic post-translational modification that takes place on ser/thr residues of nucleocytoplasmic proteins. O-GlcNAcylation regulates almost all cellular events as a nutrient sensor, a transcriptional and translational regulator, and a disease-related factor. Although the role of O-GlcNAcylation in insulin signaling and metabolism are well established, the relationship between O-GlcNAcylation and autophagy is largely unknown. Here, we manipulated O-GlcNAcylation in Drosophila and found that it regulates autophagy through Akt/dFOXO signaling. We demonstrate that O-GlcNAcylation and the levels of O-GlcNAc transferase (OGT) are increased during starvation. Furthermore, Atg proteins and autolysosomes are increased in OGT-reduced flies without fasting. Atg proteins and autophagosomes are reduced in OGT-overexpressing flies. Our results suggest that not only autophagy gene expression but also autophagic structures are regulated by OGT through Akt and dFOXO. These data imply that O-GlcNAcylation is important in modulating autophagy as well as insulin signaling in Drosophila.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-015-1889-z) contains supplementary material, which is available to authorized users.
Keywords: O-GlcNAc modification, OGT, Autophagy, Drosophila melanogaster, dFOXO
Introduction
O-GlcNAc modification of proteins occurs on serine or threonine residues, the same amino acids as phosphorylation, resulting in the regulation of cell signaling [1]. Two genes encode the enzymes for O-GlcNAcylation. O-GlcNAc transferase (OGT) catalyzes the addition of UDP-GlcNAc to target proteins, whereas O-GlcNAcase (OGA) removes the GlcNAc moiety from proteins [2]. Because UDP-GlcNAc and protein O-GlcNAc modification are influenced by glucose, fatty acid, uridine, and glutamine, O-GlcNAcylation is regarded as a nutrient sensor [3].
Autophagy is a mechanism for the degradation of cellular components and organelles in response to starvation, cellular stress, and development [4–6]. It is reported that beta cells of diabetic db/db mice have increased autophagosomes, and beta-cell-specific Atg7 knockout mice show hypoinsulinemia and hyperglycemia, indicating that autophagy is important for beta cell function [7]. Although autophagy is closely related to diabetes and the hyperglycemic condition, very little is known about the connection between O-GlcNAc and autophagy, even though it is well known that protein O-GlcNAcylation is increased and affects proteins’ function in diabetes and hyperglycemia [8–10]. Beclin1 and Bcl-2 have been identified as O-GlcNAcylated proteins; the level of beclin1 is decreased in the hearts of db/db mice and GlcNAc-treated cardiomyocytes [11]. LGG-1-GFP, a C. elegans homolog of Atg8, and its active form, PE-LGG-1-GFP, are increased in ogt-1 and oga-1 mutants. Although these two enzymes have opposite functions, the loss of OGT or OGA enhances the level of autophagy marker proteins [12]. Recently, it has been reported that transient overexpression of OGA led to an increase in autophagic flux by enhancing the formation of autolysosome in Neuro2A cells [13]. Previous studies on the expression of autophagy proteins yielded controversial results as to whether O-GlcNAcylation of the autophagy pathway increased or decreased the autophagy response. These discrepancies may be attributed to differences in cell type, species, or experimental design. Moreover, the function of OGT during the autophagic process in vivo and in vitro has not yet been investigated. Here, we study the modulation of OGT and the resulting effects on autophagy; we determined that basal autophagy is reduced with OGT overexpression and is enhanced with OGT RNAi. However, starvation-induced autophagy did not occur in OGT RNAi flies. These changes are mediated, at least part, by modulation of the activity of Akt and dFOXO.
Results
Altered OGT levels lead to changes in autophagic structures
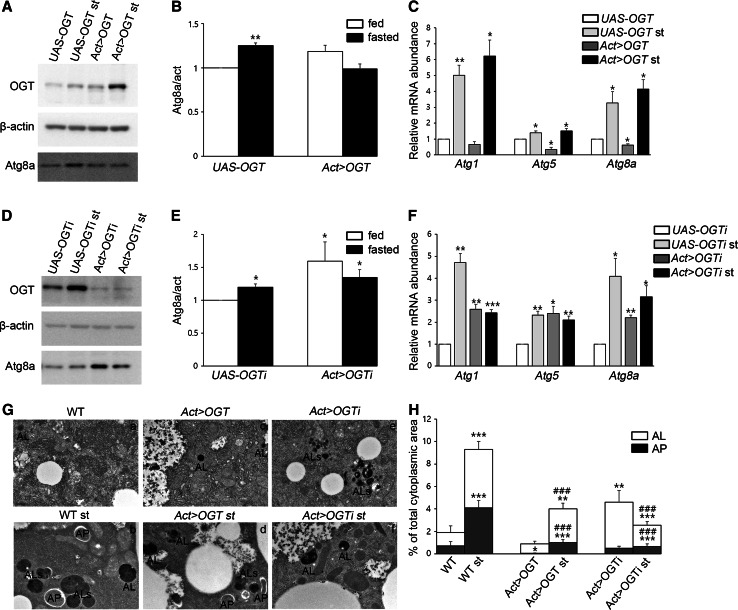
To understand the role of O-GlcNAc modification in Drosophila autophagy, we manipulated the ogt gene using UAS-OGT RNAiII [14] or UAS-OGT [15] lines to either knockdown or overexpress OGT, respectively. We knocked down OGT using UAS-OGT RNAiII by crossing with flies carrying the actin-gal4 driver line (hereafter Act > OGTi). Next, we sought to increase the extent of O-GlcNAcylation in whole cells by overexpressing Drosophila OGT (hereafter Act > OGT) (Figures S1A and S1B). We first assayed the levels of Atg8a, an autophagic marker protein, by Western blot from early third-instar larval extracts [16]. The level of Atg8a protein and mRNA was increased in fasting larvae from either UAS-OGT or UAS-OGTi (Fig. 1A–F). However, there was no difference in Atg8a protein between feeding and fasting when OGT was overexpressed (Fig. 1A, B). On the other hand, Atg8a mRNA is decreased in OGT-overexpressed larvae (Fig. 1C), and transient overexpression of OGT in Drosophila S2 cells resulted in a reduction of Atg8a protein (Figures S2A and S2B). Reduction of OGT protein by RNAi led to a dramatic increase in the level of Atg8a without starvation (Fig. 1D–F). We also checked mRNA levels of other Atgs, including Atg1 and Atg5. mRNA expression of Atg1 and Atg5 is upregulated in OGT knock-down and Atg5 mRNA is downregulated in OGT overexpressed larvae (Fig. 1C, F). We concluded that O-GlcNAc levels affect expression of autophagy-related genes.
Fig. 1.
The effect of O-GlcNAcylation on autophagosomes and autolysosomes. A Lysates from UAS-OGT and Act > OGT larvae with or without fasting were analyzed by Western blotting using OGT, β-actin, and Atg8a antibodies. B Quantification of Atg8a in (A) (**p < 0.01, Student’s t test). The data are represented as mean ± SE for four independent experiments. C Relative mRNA levels based on real-time quantitative PCR with standard error. mRNAs of Atg5 and Atg8a show a decrease in OGT-overexpressed larvae (*p < 0.05, **p < 0.01, Student’s t test). D Larvae lysates used for immunoblotting from UAS-OGTi and Act > OGTi with or without starvation. E Quantification was performed by using Multi Gauge V3.1, mean ± SE, n = 4 (*p < 0.05, Student’s t test). F Relative mRNA levels on real-time RT-PCR with standard error. All of Atg mRNA levels are upregulated in OGT knock-down larvae. Four independent experiments were carried out in triplicate each (*p < 0.05, **p < 0.01, ***p < 0.001, Student’s t test). G TEM images reveal abundant autophagosomes (AP) and autolysosomes (AL, ALs) in WT (Oregon R)-fasted larval fat bodies (Gb) but not OGT-overexpressing larval fat bodies (Gc, Gd) or fat bodies lacking OGT (Gf). The number of AL is greatly increased with OGT reduction (Ge). The graph in (H) shows AP and AL area ratios calculated form transmission electron micrographs of three larvae per genotype. Error bars show SE from the mean (*p < 0.05, **p < 0.01, ***p < 0.001 compared to feeding control, ### p < 0.001 compared to feeding siblings, Student’s t test). Bar represents 500 nm
To further confirm these findings, we used transmission electron microscopy (TEM). Ultrastructural analysis showed that numerous autophagosomes and autolysosomes were present in the wild-type (WT) larval fat body cells following 3 h of starvation (Fig. 1Ga, Gb, H). Overexpression of OGT resulted in a strong reduction of autophagosomes compared to WT. Following starvation, many autophagosomes and autolysosomes were observed in OGT-overexpressing larvae; however, both autophagic structures were reduced in OGT-overexpressing larvae compare to WT starved larvae (Fig. 1Gb, Gd, H). Surprisingly, the cumulative area of autolysosomes was significantly increased in cross sections of OGT RNAi fat body cells during feeding. After 3 h of starvation, the cumulative area of autolysosomes exhibited a strong reduction in OGT RNAi cells (Fig. 1Ge, Gf, H). Taken together, these results indicate that alteration of O-GlcNAc modification changes autophagic structures not only during basal autophagy but also during starvation-induced autophagy.
Changing the level of OGT alters the expression of Atg proteins
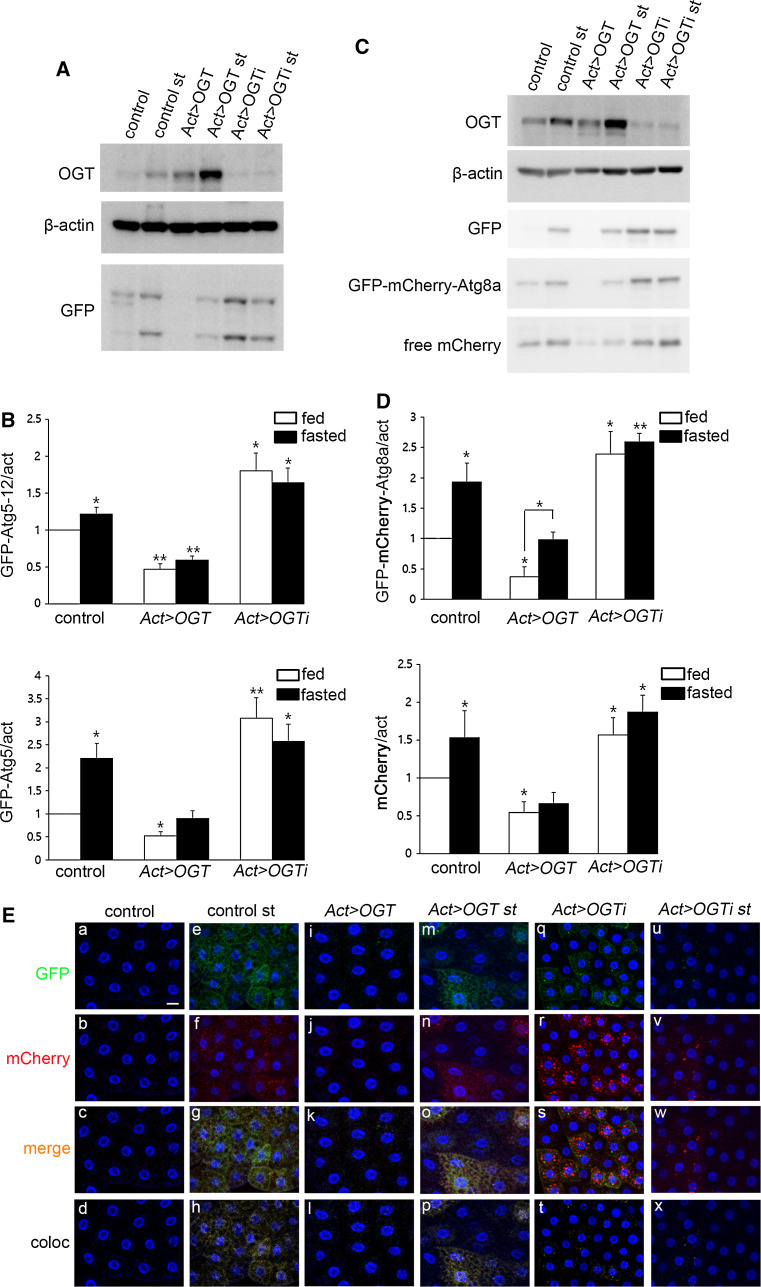
To visualize early structures of autophagy in OGT-overexpressing or OGT RNAi flies, we used GFP-Atg5 [6]. Atg5 conjugates with Atg12 and acts as an E3-like enzyme, essential for formation of phosphatidylethanolamine (PE)-Atg8 [17]. The protein levels of GFP-Atg5 (lower band) and GFP-Atg5-conjugated with Atg12 (GFP-Atg5-Atg12, upper band) were increased during amino acid starvation and decreased in OGT-overexpressing larvae. On the contrary, protein levels of GFP-Atg5 and GFP-Atg5-Atg12 were dramatically increased in OGT RNAi larvae during feeding and starvation does not induce expression of these proteins (Fig. 2A, B).
Fig. 2.
O-GlcNAc modification changes autophagy pathway protein expression. A Larval lysates from UAS-OGT and Act > OGT with or without fasting were analyzed by immunoblot with OGT, β-actin, and GFP antibodies. Because we used GFP-Atg5 to detect early structures of autophagy, we checked Atg5 expression by GFP antibody. B Quantification of GFP (*p < 0.05, **p < 0.01, Student’s t test). The data are represented as mean ± SE for four independent experiments. C Lysates from control, Act > OGT, and Act > OGTi larvae with or without fasting were analyzed by Western blot using OGT, β-actin, GFP, and mCherry antibodies. D Quantification of mCherry bands were performed by using Multi Gauge V3.1, mean ± SE, n = 4 (*p < 0.05, **p < 0.01, Student’s t test). E Early third-instar larval fat body images of each GFP-mCherry-Atg8a-expressing line. Practically no GFP-mcherry-Atg8a puncta are seen in fat body cells of well-fed control larvae (a–d) and OGT-overexpressing larvae (i–l). Dots positive for both mCherry and GFP appear in control cells (e–h) and OGT-overexpressing cells (m–p) upon fasting. Reduction of OGT results in the formation of mostly mCherry-positive dots during feeding (q–t), while the number of mCherry-positive puncta was reduced during fasting (u–x). a, e, i, m, q, u GFP channels (green). b, f, j, n, r, v mCherry channels (red). c, g, k, o, s, w Merged images. d, h, l, p, t, x Colocalized images. DAPI was used for nuclei staining (blue). Scale bar represents 20 μm
To determine if increased GFP-Atg5 in OGT RNAi larvae is due to increased autophagic flux or decreased lysosomal activity, we used tandem tagged (GFP-mCherry) Atg8a [18]. Because of the different pH sensitivity of GFP and mCherry, a non-acidified autophagic compartment such as the phagophore and autophagosome are co-labeled with GFP and mCherry, whereas the autolysosome will show only mCherry signal. Therefore, this marker can be an indicator of autophagic flux. First, we analyzed autophagic flux by Western blot using mCherry and GFP antibodies. During amino acid starvation, the protein levels of GFP-mCherry-Atg8a and free mCherry were increased; because Atg8a itself is rapidly broken down in the autolysosome, free mCherry or GFP was generated. OGT-overexpressing larvae showed a large decrease in both GFP-mCherry-Atg8a and autolysosomal conversion of GFP-mCherry-Atg8a to free mCherry. Starvation-induced autophagy occurred in OGT-overexpressing larvae but the amount of GFP-mCherry-Atg8a and free mCherry was less than in control fasted larvae. On the other hand, OGT RNAi larvae showed increased levels of GFP-mCherry-Atg8a and free mCherry and starvation did not induce additional levels of these proteins (Fig. 2C, D).
Autophagic flux can also be analyzed by immunofluorescence assay using confocal microscopy. We observed no punctate structures in fat body cells during feeding (Fig. 2Ea–Ed, Ei–El), but numerous puncta, including GFP-mCherry double positive and mCherry positive, were seen after amino acid removal (Fig. 2Ee–Eh). Overexpression of OGT led to a decrease in GFP-mCherry double-positive and mCherry-positive puncta in fasted fat body cells compare to fasted control cells (Fig. 2Ee–Eh, Em–Ep). Surprisingly, reduction of OGT increased GFP-mCherry double-positive (51.4 % of the fat body cells) and significantly increased mCherry-positive puncta (60 % of the fat body cells), while these puncta were dramatically reduced in fasted fat bodies of OGT RNAi cells (Fig. 2Eq–Ex). Our observation of GFP-mCherry-Atg8a in the autolysosome using both Western blots and confocal microscopy suggests that increased O-GlcNAcylation reduces autophagy, and decreased O-GlcNAcylation enhances autophagic flux; however, starvation-induced autophagy is impaired in OGT RNAi larvae.
Starvation reduces Akt signaling and O-GlcNAc modification enhances Akt signaling
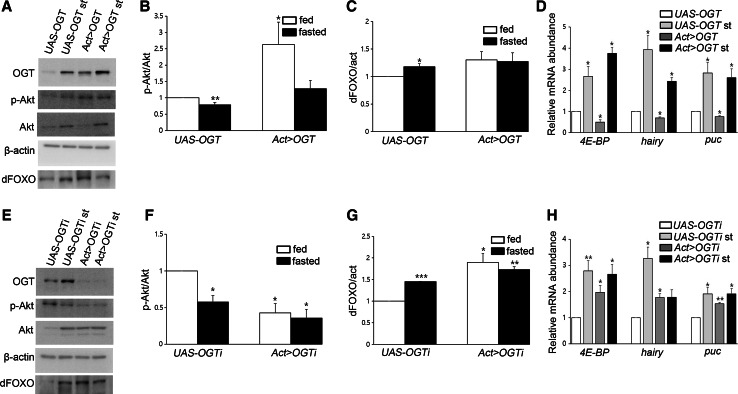
We have previously shown that increased O-GlcNAcylation following OGA knockdown enhances not only O-GlcNAcylation of Akt but also its phosphorylation, and that these two modifications enhance Akt activity [14]. Furthermore, phosphorylation of Akt and dFOXO is decreased in fasted WT L3 larvae [19] and fasted mice [20]. Therefore, we reasoned that OGT modulation would affect autophagy through Akt and its downstream signaling. To measure Akt activity during fluctuating OGT levels, we examined Akt phosphorylation in feeding and fasting larvae. Western-blot analysis showed that phosphorylation of Akt at ser505 was reduced in starved UAS-controls (Fig. 3a, b, e, f). OGT overexpression increased phosphorylation on Akt, as expected, whereas phosphorylation of Akt in the fasted sibling was strongly reduced (Fig. 3a, b). Conversely, OGT RNAi larvae had a dramatic decrease in Akt phosphorylation in the feeding state; this reduced level was less than in starved UAS-OGTi larvae (Fig. 3e, f). Thus, Akt phosphorylation is decreased by reduction of OGT as much as during starvation.
Fig. 3.
Akt signaling changes depending on the level of OGT. a Lysates from UAS-OGT and Act > OGT larvae with or without starvation were analyzed by Western blot using OGT, phosphor-Akt, Akt, dFOXO, and β-actin antibodies. b Quantification of p-Akt in (a) (*p < 0.05, **p < 0.01, Student’s t test). c Quantification of dFOXO in (a) (*p < 0.05, Student’s t test). The data are represented as mean ± SE for four independent experiments. d The mRNA levels of three dFOXO target genes were measured by real-time RT-PCR in UAS-control and OGT overexpressing larvae under feeding and fasting condition (*p < 0.05, Student’s t test). e Larval lysates from UAS-OGTi and Act > OGTi with or without fasting analyzed by immunoblot. f, g Quantifications of p-Akt and dFOXO bands in (e) by using Multi Gauge V3.1, mean ± SE, n = 4 (*p < 0.05, **p < 0.01, ***p < 0.001 Student’s t test). h 4E-BP, hairy, and puc mRNA levels was determined by Student’s t test (*p < 0.05, **p < 0.01). Total RNAs of UAS-OGTi and Act > OGTi in feeding and fasting were prepared and analyzed by real-time RT-PCR. Four independent experiments were carried out in triplicate each
O-GlcNAc modification of dFOXO affects its activity
We have previously reported on the nuclear distribution of dFOXO in OGA or OGT knockdown larval fat bodies; OGT RNAi showed a strong nuclear distribution of dFOXO, while increased O-GlcNAcylation by reduction of OGA inhibited nuclear localization of dFOXO in fat body cells [14]. Therefore, we studied the levels of dFOXO protein under feeding and fasting conditions to determine whether modulation of OGT could also affect dFOXO levels. We determined that levels of dFOXO protein are increased in fasted UAS-controls (Fig. 3a, c, e, g) and that there were no significant differences in dFOXO levels between feeding and fasting in either OGT overexpressing (Fig. 3a, c) or OGT knockdown larvae. However, the levels of dFOXO were upregulated in OGT RNAi larvae compared to UAS-control larvae (Fig. 3e, g). To test whether level of OGT influences transcriptional activity of dFOXO, we measured mRNA levels of dFOXO target genes [21]. In starvation, autophagy genes are upregulated in UAS controls (Fig. 3d, h). dFOXO target mRNAs that were assessed by real-time RT-PCR are reduced in OGT-overexpressing larvae (Fig. 3d). On the contrary, reduction of O-GlcNAc modification stimulates the expression of dFOXO target genes (Fig. 3h). Taken together, we may conclude that transcriptional activity of dFOXO is changed depending on OGT levels.
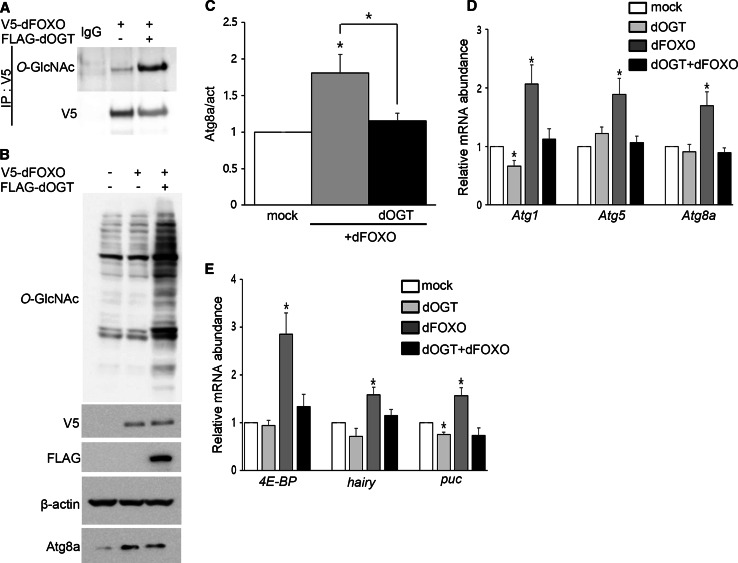
It has been reported that mammalian FoxO1 is O-GlcNAcylated and that this modification regulates FoxO1 activity in response to glucose [22]. Based on findings in mammals and Drosophila, we predicted that dFOXO would be a candidate for O-GlcNAcylation and that O-GlcNAcylation of dFOXO might affect regulation of autophagy. In Drosophila, a null mutation of foxo strongly inhibited starvation-induced autophagy, and dFOXO expression was sufficient to induce autophagy [23]. It has been shown that the levels of both dFOXO protein and mRNA increase during fasting [24]; furthermore, overexpression of FoxO1 or FoxO3 induces autophagy through activation of autophagy genes including LC3, Gabarap1, and Atg12 in cardiomyocytes [20]. Nevertheless, the function of dFOXO as a transcriptional regulator of autophagy pathway genes has not been demonstrated in Drosophila. Therefore, we examined the role of dFOXO in regulating autophagy and the effects of O-GlcNAcylation of dFOXO using Drosophila larvae and S2 cells. V5-tagged dfoxo was modified with O-GlcNAc and we observed a substantial increase in O-GlcNAcylation after OGT overexpression (Fig. 4a). In addition, endogenous dFOXO was pulled down by succinylated wheat germ agglutinin (sWGA) (Figure S3A), a modified lectin that is able to bind O-GlcNAc on proteins [25]. To determine whether dFOXO is able to induce autophagy pathway genes, Drosophila S2 cells were transfected with V5-dfoxo with or without FLAG-dogt. The induction of autophagy pathway genes was determined by Western blots and real-time RT-PCR. Atg8a proteins were significantly increased in dFOXO-transfected cells, and co-expression of dOGT with dFOXO resulted in a reduction of Atg8a protein (Fig. 4b, c). Overexpression of dFOXO induces expression of its target genes as well as autophagy-related genes, while co-expression of dOGT with dFOXO suppresses induction of these genes (Fig. 4d, e). Taken together, our results suggest that dFOXO is O-GlcNAcylated and that this modification diminishes its transcriptional activity.
Fig. 4.
O-GlcNAcylation on dFOXO affects its function as a transcription factor. a V5-tagged dfoxo was expressed in S2 cells either alone or with dogt. To demonstrate O-GlcNAcylation on dFOXO, dFOXO was immunoprecipitated using V5-agarose followed by Western blotting with anti-O-GlcNAc antibody, RL2. As a control, the same immunoprecipitates were blotted with anti-V5 antibody. b S2 cells were transiently transfected with V5-dfoxo alone or in combination with FLAG-dogt. Lysates were analyzed by RL2, V5, FLAG, β-actin, and Atg8a antibodies. c Quantification of Atg8a in (b) (*p < 0.05, Student’s t test). Real-time RT-PCR showed activation of autophagy-related genes (d) and dFOXO target genes (e) in S2 cells after co-transfection with V5-dfoxo alone or FLAG-dogt alone or in combination of two (*p < 0.05, Student’s t test). Five independent experiments were carried out in triplicate each
Lastly, we tested whether other proteins in the autophagy pathway were modified with O-GlcNAc in Drosophila and mammals. Among the autophagy pathway proteins tested in mouse brain lysates, Atg7 and LC3-I were pulled down by sWGA. We confirmed that O-GlcNAcylation on Atg7 by immune-precipitation experiment using O-GlcNAc-specific antibody (Figure S3C). Total O-GlcNAcylation was dramatically increased in the brain lysate of thiamet-G-injected mice (Figure S3B). Atg5 in mouse brain lysate was not modified with O-GlcNAc (Figure S3C). In addition, Drosophila Atg5, Atg8a, and GFP-mCherry-Atg8a were not pulled down by sWGA (Figure S3D and S3E). These results imply that O-GlcNAcylation of other autophagy pathway protein, including Atg7 may affect autophagic signaling in mammal.
Discussion
Earlier reports have implicated O-GlcNAc modification in the regulation of autophagy acting at several levels. For example, levels of beclin1 and LC3-II protein were shown to be diminished in hearts of diabetic mice and beclin1 was O-GlcNAcylated [11]. In C. elegans, mutants in OGT and OGA show increased levels of the C. elegans homolog of the Atg8-reporter during feeding and fasting [12]. However, the role of O-GlcNAcylation on beclin1 and Bcl-2 has not been identified, and it is not clear why OGT and OGA mutants increased the Atg8-reporter in C. elegans, given that these two enzymes have opposite function. To understand the function of OGT in basal autophagy and starvation-induced autophagy, we used Drosophila as an in vivo model. Here, we demonstrated the function of OGT in regulating autophagy in S2 cells as well as in Drosophila under basal and starvation conditions. When larvae are deprived of nutrients, especially amino acids, autophagy is induced in the fat body. Starvation-induced autophagy is regulated by PI3K signaling [26]. We hypothesized that O-GlcNAcylation of proteins may influence autophagy through PI3K signaling; thus, we examined the protein levels of dFOXO and the phosphorylation of Akt during feeding and fasting. When global O-GlcNAcylation is increased, Akt is more active and dFOXO is inactivated in the cytoplasm. By contrast, phosphorylation on Akt is reduced and protein levels of dFOXO are increased with reduction of OGT. Based on our findings, we conclude that O-GlcNAc modification enhances Akt signaling and negatively regulates autophagy.
Developmentally programmed autophagy and subsequent cell death occur in tissue remodeling during metamorphosis [27, 28]. Because programmed autophagy is known to be initiated 108 h after egg laying [6], we used small-feeding early L3 larvae (72 h after egg laying) for our experiments to rule out programmed basal autophagy. Under basal conditions, a low level of autophagy is required to remove and recycle long-lived or damaged proteins and organelles to maintain homeostasis [29]. Interestingly, the number of autophagic structures and expression of autophagy pathway genes are altered depending the level of OGT. Autophagy is downregulated, whereas the number of lysosome is greatly increased in OGT overexpressing larvae (Figure S2C). Furthermore, the level of autophagy pathway proteins and autolysosomes are significantly increased in OGT-reduced larvae in their early third-instar stage. These transgenic larvae showed an early lethal pupae phenotype with increased apoptosis [14]. Therefore, we assumed that uncontrolled autophagy would also affect the viability of OGT RNAi larvae. Based on the phenotype of OGT overexpressing and OGT knockdown larvae, we conclude that proper O-GlcNAc modification is important for maintaining cellular degradation pathways including autophagy and the lysosome.
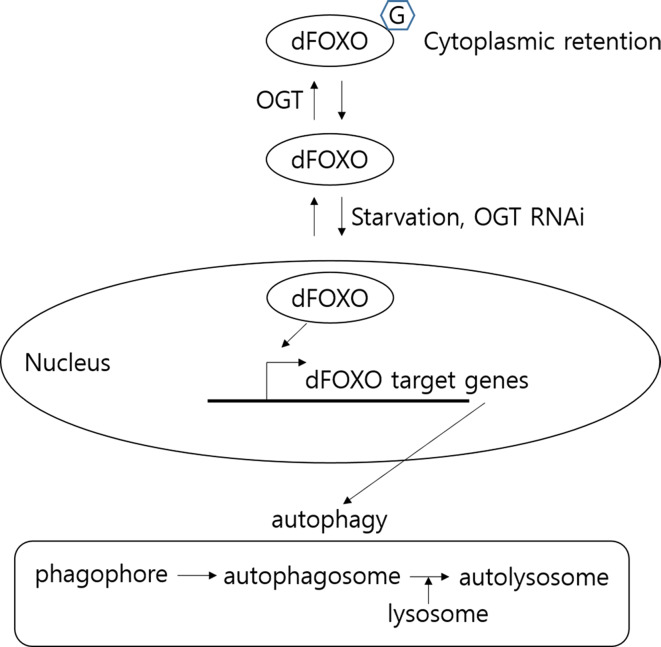
To determine whether decreased cellular O-GlcNAcylation enhances dFOXO activity related to the increase in Atg proteins, and how O-GlcNAcylation on dFOXO affects its activity in a cell autonomous manner, we overexpressed dFOXO with or without OGT in S2 cells. Overexpression of dFOXO induced to increase the level of Atgs, while overexpression of dFOXO with OGT does not induce Atg mRNAs, and dFOXO was modified with O-GlcNAc. Furthermore, when dFOXO was overexpressed along with OGT, O-GlcNAc modification of dFOXO was increased and the protein level of Atg8a as well as several Atg mRNAs was substantially decreased. Taken together, our results show that dFOXO increases expression of autophagy-related genes and that O-GlcNAcylation of dFOXO reduces its activity in vitro, which correlates with a change in autophagy in vivo. Under conditions of reduced O-GlcNAcylation, dFOXO is upregulated and predominantly localizes to the nucleus, where dFOXO activates autophagy pathway gene expression. As a result, the number of autophagic structures is dramatically increased in OGT knockdown flies. dFOXO is equally distributed in the nucleus and cytoplasm in OGT-overexpressing larvae, resulting in reduction of autophagy pathway gene expression and a decrease of autophagosomes (Fig. 5).
Fig. 5.
Working model of regulation of autophagy by OGT. Overexpression of ogt leads to reduced basal autophagy, whereas loss of ogt increases autophagy. The findings suggest O-GlcNAc modification influences autophagy pathway gene induction and autophagic flux
Materials and methods
Fly strains and maintenance
Flies were raised at 25 °C on a standard diet. UAS-OGT RNAi and UAS-OGT flies were described previously [14, 15]. Actin-Gal4, GFP-Atg5 (50877), and GFP-mCherry-Atg8a (37749) were from the Bloomington Drosophila Stock Center.
Cell culture
Drosophila S2 cells were purchased from Invitrogen and cultured in Schneider’s Insect Medium (Welgene) supplemented with 10 % FBS (Gibco) and 1 % Penicillin–Streptomycin (Gibco). PP242 was purchased from Selleckchem. Thiamet-G was synthesized according the original paper [31].
Plasmids and transfection
The pMT-FLAG-dogt and pMT-dfoxo-V5 plasmids were described previously [14, 32]. S2 cells were transfected using effectene reagent following the manufacturer’s protocol (Qiagen). Recombinant proteins under the control of the pMT promoter were induced by adding 500 μM CuSO4 to the medium.
Animals and intraperitoneal (IP) injection of thiamet-G
B6SJL wild-type (WT) mice were purchased from SLC and used for the experiments. All procedures were approved by the Committee on Animal Investigations of the Yonsei University. We used 40 mg/kg thiamet-G dissolved in saline solution to inhibit O-GlcNAcase. Both male and female mice were used in the experiments. Thiamet-G or the vehicle (saline solution) was injected into the abdominal cavity of 8-week-old mice every day for 7 days.
Immunoblots and immunoprecipitation
Early third-instar larvae were homogenized in lysis buffer [50 mM Tris–HCl (pH 7.5), 150 mM sodium chloride, 1 % Nonidet P40, 0.5 % sodium deoxycholate, and one tablet of complete Mini Protein Inhibitor Cocktail/10 ml]. Transfected S2 cells, and tissue lysates from control and thiamet-G injected mice were collected and homogenized in the same cold lysis buffer. Homogenates were centrifuged at 4 °C for 20 min at 19,320 × g. Protein concentrations were determined with the Bradford assay (Bio-Rad). Supernatants containing 25 μg of protein were mixed with Laemmli buffer and boiled for 5 min, then separated on a polyacrylamide-SDS gel and transferred to a nitrocellulose (NC) membrane (Amersham). The membrane was incubated in TBST containing 5 % nonfat dry milk at room temperature for 1 h. The membrane was then incubated with primary antibody at 4 °C overnight. The primary antibodies used were anti-RL2 (Thermo Scientific, MA1-072), anti-CTD 110.6 (Covance, MMS-248R), anti-OGT (Santa Cruz, H300), anti-β-actin (Cell Signaling, 4967), anti-Atg8a (MBL, PM037), anti-Flag (Sigma, F1804), anti-GFP (Santa Cruz, B2), anti-mCherry (abcam, ab125096), anti-dFOXO [30], anti-dFOXO (Cosmobio, CAC-THU-A-DFOXO), phospho-Akt (Cell Signaling, 4054), Akt (Cell Signaling, 9272), anti-LC3 (MBL, PM036), anti-V5 (Invitrogen, 46-0705), Atg7 (Cell Signaling, 8558), Atg5 (Cell Signaling, 2011), and anti-dAtg5 (abcam, ab64147). Membranes were washed in TBST and peroxidase-based detection was performed with enhanced chemiluminescence (ECL) detection reagent (Santa Cruz, 2048) followed by imaging (LAS-3000, Fujifilm, Tokyo, Japan) and quantification of protein bands (Multigauge V3.1, Fujifilm). Data were expressed as mean ± SE. The two-tailed Student’s t test was used for statistical analysis (*p < 0.05, **p < 0.01 and ***p < 0.001).
For co-immunoprecipitation, the supernatants were preabsorbed with V5-agarose (Invitrogen, A7345) or RL2 antibody-bounded protein A/G PLUS-Agarose (Santa Cruz, 2003) for at least 2 h at 4 °C. Lysates were then added to each agarose suspension and incubated overnight at 4 °C. The beads were incubated and washed five times for 5 min each with washing buffer [50 mM Tris–HCl (pH 7.5), 500 mM sodium chloride, 0.1 % Nonidet P40, 0.05 % sodium deoxycholate] and then with the washing buffer without sodium chloride. The agarose beads were then suspended in Laemmli buffer and boiled for 5 min. sWGA pull-down was performed by adding sWGA-agarose (L-1020S, Vector Laboratories) to the protein extracts; bound material was analyzed by immunoblotting.
Immunohistochemistry
Immunohistochemistry was performed as described previously [32]. After fixing the tissue, the samples were mounted in Prolong Gold plus DAPI (Invitrogen) and observed with a LSM 700 (Carl Zeiss) using a Neo-Fluar 10 X NA (0.3) objective lens at room temperature. Colocalization channels were calculated using Meta Imaging SeriesR MetaMorph software (Meta Series Software 7.7.0; Molecular Devices) and appear in white in all figures. Illustrations were generated using Photoshop CS6.
Transmission EM
Tissues were dissected and fixed for 2 h at RT in 2 % PFA (freshly prepared from paraformaldehyde, Merck) and 0.5 % glutaraldehyde (vacuum distilled, Polysciences) in 0.1 M sodium cacodylate buffer, pH 7.2. To stop fixation, free aldehyde groups were blocked by soaking the tissues in 50 mM NH4Cl in buffer for 1 h. The tissues were rinsed in buffer and postfixed with 1 % osmium tetroxide (Electron Microscopy Science) in distilled water for 1 h, followed by en bloc staining with 1 % aqueous uranyl acetate for 1 h. After dehydration in a series of graded ethanol washes, embedding in Epon-Araldite (Fluka) was performed according to standard protocol. Ultrathin sections (80 nm thickness) were prepared on copper slot grids, stained with uranyl acetate and lead citrate and then observed with a Hitachi H-7650 electron microscope at 80 kV. Electron micrographs were taken with an 11-megapixel CCD XR611-M digital camera (Advanced Microscopy Techniques). For statistical analysis, six images were taken randomly at 3000× magnification per section, from three larvae per genotype (18 pictures). The area of autophagosomes, autolysosomes, lysosomes, and total cytoplasm was calculated using Photoshop CS6, by manually encircling relevant structures. Relative cytoplasmic area values and two-tailed, two-sample unequal variance Student’s t tests were calculated using Excel.
Quantitative RT-PCR
Total RNA was extracted from 10 whole flies in TRIzol reagent (Invitrogen, Grand Island, NY, USA). DNase-treated total RNA was quantified with a Nano Drop ND-1000. 1 μg total RNA from each genotype was reverse-transcribed into cDNA using ReverTra Ace qPCR RT Master Mixe (Toyobo, Japan). Expression levels were analyzed by quantitative real-time PCR (qRT-PCR) with SYBR Premix Ex Taq II (Takara, Japan) and measured on an ABI prism 7300 Sequence (Applied Biosystems, USA) using primer sets for target genes. mRNA abundance of each gene was normalized relative to ribosomal protein L32 (RpL32, also known as rp49) by the method of comparative CT. Primer sequences are Atg1F: GAGTATTGCAATGGCGGCGACT, Atg1R: CAGGAATCGCGC AAACCCAA, Atg5F: GCACGCACGGCATTGATCTACA, Atg5R: GCCCTGGGATTTGCTGGAAT, Atg8aF: GCAAATATCCAGACCGTGTGCC, Atg8aR: AGCCCATGGTAGCCGATGTT, rp49F: CAGTCGGATCGATAT GCTAAGCTGT, rp49R: TAACCGATGTTGGGCATCAGATACT, hairyF: AAGGCCGACATTCTGGAGAAG ACA, hairyR: GGCGAATCCGGCCTTGAATTTGTT, pucF ATCGCATTGGTGCCAATCAGAAGG, pucR: GC CGTTCATTGCCGTTGATGATGA, 4ebpF: CCATGATCACCAGGAAGGTTGTCA, 4ebpR: AGCCCGCTCG TAGATAAGTTTGGT.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 Figure S1. The effects of increased OGT or OGT knockdown on overall O-GlcNAc levels. (A) Larval lysates from UAS-OGT and Act>OGT in feeding and fasting were analyzed by immunoblot with RL2 antibody. (B) Larvae lysates used for immunoblotting from UAS-OGTi and Act>OGTi in feeding and fasting to detect overall O-GlcNAc levels (TIFF 7877 kb)
Supplementary material 2 Figure S2. The effect of O-GlcNAcylation on protein levels of Atg8a and the number of lysosomes. (A) S2 cells were transfected with pMT-FLAG-dogt for 48 h and harvested for immunoblotting. Anti-RL2, anti-FLAG, anti-β-actin, and anti-Atg8a antibodies are used. (B) Quantification of the amount of Atg8a in (A) (*p<0.05, student’s t-test). (C) A morphometric evaluation of EM images (***p<0.001, student’s t-test). TEM reveals many lysosomes in OGT-overexpressing larval fat bodies (TIFF 10380 kb)
Supplementary material 3 Figure S3. O-GlcNAcylated proteins of the autophagy pathway. (A) S2 cells were treated with 10μM PP242 for 4 h to induce autophagy. Representative western blots showing β-actin and Atg8a in control and PP242-treated S2 cells. Lysates were prepared and analyzed either directly by immunoblotting (Input) or following pull-down with sWGA-agarose to detect O-GlcNAc modification on dFOXO. (B) WT mice were treated with the O-GlcNAcase inhibitor, thiamet-G (TG) for 7 days to increase total O-GlcNAcylation. Total O-GlcNAcylation of control and thiamet-G injected mouse brain. (C) To demonstrate O-GlcNAcylation on Atg7, Atg7 was immunoprecipitated using specific O-GlcNAc antibody followed by Western blotting with anti-Atg7 antibody. Brain lysates were prepared and analyzed either directly by immunoblotting or following pull-down with sWGA agarose to detect O-GlcNAcylation on Atg7, LC3, and Atg5. β-actin was used as a loading control. (D) Control and PP242-treated S2 cell lysates were prepared and analyzed either directly by immunoblotting as input or following pull-down with sWGA agarose to detect O-GlcNAcylation on dAtg5 and Atg8a. β-actin was used as a loading control. (E) Whole cell lysate of GFP-mCherry-Atg8a-expressing larvae with feeding or fasting. Larval lysates were prepared and analyzed either directly by immunoblotting (input) or following pull-down with sWGA agarose to detect O-GlcNAcylation on Atg8a using mCherry and GFP antibodies. Upper arrow is mCherry-GFP-Atg8a and lower arrow is free mCherry. β-actin was used as a loading control (TIFF 173286 kb)
Acknowledgments
We thank K. M. Choe, and K. Yu for fly stocks and DNA clones, and O. Puig and K. Yu for providing the anti dFOXO antibody. We are also grateful to other members of the Cho and Choe laboratories for helpful discussions. This work is partly supported by the Graduate School of Yonsei University. This work was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (NRF-2013R1A2A1A01008067) to J. W. C.
Conflict of interest
The authors declare that they have no conflicts of interest to declare.
Footnotes
J. Roth and J. W. Cho are co-corresponding authors.
Contributor Information
Jürgen Roth, Phone: 82-2-2123-4083, Email: jurgen.roth@bluewin.ch.
Jin Won Cho, Phone: 82-2-363-4083, Email: chojw311@yonsei.ac.kr.
References
- 1.Dias WB, Cheung WD, Hart GW. O-GlcNAcylation of kinases. Biochem Biophys Res Commun. 2012;422:224–228. doi: 10.1016/j.bbrc.2012.04.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruan HB, Singh JP, Li MD, Wu J, Yang X. Cracking the O-GlcNAc code in metabolism. Trends Endocrinol Metab. 2013;24:301–309. doi: 10.1016/j.tem.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zachara NE, Hart GW. O-GlcNAc a sensor of cellular state: the role of nucleocytoplasmic glycosylation in modulating cellular function in response to nutrition and stress. Biochim Biophys Acta. 2004;1673:13–28. doi: 10.1016/j.bbagen.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 4.Mehrpour M, Esclatine A, Beau I, Codogno P. Autophagy in health and disease. 1. Regulation and significance of autophagy: an overview. Am J Physiol Cell Physiol. 2010;298:C776–C785. doi: 10.1152/ajpcell.00507.2009. [DOI] [PubMed] [Google Scholar]
- 5.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 6.Rusten TE, Lindmo K, Juhasz G, Sass M, Seglen PO, Brech A, Stenmark H. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell. 2004;7:179–192. doi: 10.1016/j.devcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Fujitani Y, Ebato C, Uchida T, Kawamori R, Watada H. Beta-cell autophagy: a novel mechanism regulating beta-cell function and mass: lessons from beta-cell-specific Atg7-deficient mice. Islets. 2009;1:151–153. doi: 10.4161/isl.1.2.9057. [DOI] [PubMed] [Google Scholar]
- 8.Erickson JR, Pereira L, Wang L, Han G, Ferguson A, Dao K, Copeland RJ, Despa F, Hart GW, Ripplinger CM, et al. Diabetic hyperglycaemia activates CaMKII and arrhythmias by O-linked glycosylation. Nature. 2013;502:372–376. doi: 10.1038/nature12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu K, Paterson AJ, Chin E, Kudlow JE. Glucose stimulates protein modification by O-linked GlcNAc in pancreatic beta cells: linkage of O-linked GlcNAc to beta cell death. Proc Natl Acad Sci USA. 2000;97:2820–2825. doi: 10.1073/pnas.97.6.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma J, Hart GW. Protein O-GlcNAcylation in diabetes and diabetic complications. Expert Rev Proteomics. 2013;10:365–380. doi: 10.1586/14789450.2013.820536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marsh SA, Powell PC, Dell’italia LJ, Chatham JC. Cardiac O-GlcNAcylation blunts autophagic signaling in the diabetic heart. Life Sci. 2013;92(11):648–656. doi: 10.1016/j.lfs.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang P, Lazarus BD, Forsythe ME, Love DC, Krause MW, Hanover JA. O-GlcNAc cycling mutants modulate proteotoxicity in Caenorhabditis elegans models of human neurodegenerative diseases. Proc Natl Acad Sci USA. 2012;109:17669–17674. doi: 10.1073/pnas.1205748109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar A, Singh PK, Parihar R, Dwivedi V, Lakhotia SC, Ganesh S. Decreased O-linked GlcNAcylation protects from cytotoxicity mediated by huntingtin exon1 protein fragment. J Biol Chem. 2014;289:13543–13553. doi: 10.1074/jbc.M114.553321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park S, Park SH, Baek JY, Jy YJ, Kim KS, Roth J, Cho JW, Choe KM. Protein O-GlcNAcylation regulates Drosophila growth through the insulin signaling pathway. Cell Mol Life Sci. 2011;68:3377–3384. doi: 10.1007/s00018-011-0640-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinclair DA, Syrzycka M, Macauley MS, Rastgardani T, Komljenovic I, Vocadlo DJ, Brock HW, Honda BM. Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc) Proc Natl Acad Sci USA. 2009;106:13427–13432. doi: 10.1073/pnas.0904638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta VK, Scheunemann L, Eisenberg T, Mertel S, Bhukel A, Koemans TS, Kramer JM, Liu KS, Schroeder S, Stunnenberg HG, et al. Restoring polyamines protects from age-induced memory impairment in an autophagy-dependent manner. Nat Neurosci. 2013;16:1453–1460. doi: 10.1038/nn.3512. [DOI] [PubMed] [Google Scholar]
- 17.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 18.Nezis IP, Shravage BV, Sagona AP, Lamark T, Bjorkoy G, Johansen T, Rusten TE, Brech A, Baehrecke EH, Stenmark H. Autophagic degradation of dBruce controls DNA fragmentation in nurse cells during late Drosophila melanogaster oogenesis. J Cell Biol. 2010;190:523–531. doi: 10.1083/jcb.201002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang B, Moya N, Niessen S, Hoover H, Mihaylova MM, Shaw RJ, Yates JR, 3rd, Fischer WH, Thomas JB, Montminy M. A hormone-dependent module regulating energy balance. Cell. 2011;145:596–606. doi: 10.1016/j.cell.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sengupta A, Molkentin JD, Yutzey KE. FoxO transcription factors promote autophagy in cardiomyocytes. J Biol Chem. 2009;284:28319–28331. doi: 10.1074/jbc.M109.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bai Hua, Kang Ping, Hernandez Ana Maria, Tatar Marc. Activin signaling targeted by insulin/dFOXO regulates aging and muscle proteostasis in Drosophila . PLoS Genet. 2013;9(11):e1003941. doi: 10.1371/journal.pgen.1003941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Housley MP, Rodgers JT, Udeshi ND, Kelly TJ, Shabanowitz J, Hunt DF, Puigserver P, Hart GW. O-GlcNAc regulates FoxO activation in response to glucose. J Biol Chem. 2008;283:16283–16292. doi: 10.1074/jbc.M802240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juhasz G, Puskas LG, Komonyi O, Erdi B, Maroy P, Neufeld TP, Sass M. Gene expression profiling identifies FKBP39 as an inhibitor of autophagy in larval Drosophila fat body. Cell Death Differ. 2007;14:1181–1190. doi: 10.1038/sj.cdd.4402123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Villa-Cuesta E, Sage BT, Tatar M. A role for Drosophila dFoxO and dFoxO 5′UTR internal ribosomal entry sites during fasting. PloS One. 2010;5(7):e11521. doi: 10.1371/journal.pone.0011521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim EY, Jeong EH, Park S, Jeong HJ, Edery I, Cho JW. A role for O-GlcNAcylation in setting circadian clock speed. Genes Dev. 2012;26:490–502. doi: 10.1101/gad.182378.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 27.Juhasz G, Csikos G, Sinka R, Erdelyi M, Sass M. The Drosophila homolog of Aut1 is essential for autophagy and development. FEBS Lett. 2003;543:154–158. doi: 10.1016/S0014-5793(03)00431-9. [DOI] [PubMed] [Google Scholar]
- 28.Thummel CS. Steroid-triggered death by autophagy. Bioessays. 2001;23:677–682. doi: 10.1002/bies.1096. [DOI] [PubMed] [Google Scholar]
- 29.Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010;90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 30.Yuzwa SA, Macauley MS, Heinonen JE, Shan X, Dennis RJ, He Y, Whitworth GE, Stubbs KA, McEachern EJ, Davies GJ, et al. A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat Chem Biol. 2008;4:483–490. doi: 10.1038/nchembio.96. [DOI] [PubMed] [Google Scholar]
- 31.Puig O, Marr MT, Ruhf ML, Tjian R. Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 2003;17:2006–2020. doi: 10.1101/gad.1098703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee KS, Kwon OY, Lee JH, Kwon K, Min KJ, Jung SA, Kim AK, You KH, Tatar M, Yu K. Drosophila short neuropeptide F signalling regulates growth by ERK-mediated insulin signalling. Nat Cell Biol. 2008;10(4):468–475. doi: 10.1038/ncb1710. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1 Figure S1. The effects of increased OGT or OGT knockdown on overall O-GlcNAc levels. (A) Larval lysates from UAS-OGT and Act>OGT in feeding and fasting were analyzed by immunoblot with RL2 antibody. (B) Larvae lysates used for immunoblotting from UAS-OGTi and Act>OGTi in feeding and fasting to detect overall O-GlcNAc levels (TIFF 7877 kb)
Supplementary material 2 Figure S2. The effect of O-GlcNAcylation on protein levels of Atg8a and the number of lysosomes. (A) S2 cells were transfected with pMT-FLAG-dogt for 48 h and harvested for immunoblotting. Anti-RL2, anti-FLAG, anti-β-actin, and anti-Atg8a antibodies are used. (B) Quantification of the amount of Atg8a in (A) (*p<0.05, student’s t-test). (C) A morphometric evaluation of EM images (***p<0.001, student’s t-test). TEM reveals many lysosomes in OGT-overexpressing larval fat bodies (TIFF 10380 kb)
Supplementary material 3 Figure S3. O-GlcNAcylated proteins of the autophagy pathway. (A) S2 cells were treated with 10μM PP242 for 4 h to induce autophagy. Representative western blots showing β-actin and Atg8a in control and PP242-treated S2 cells. Lysates were prepared and analyzed either directly by immunoblotting (Input) or following pull-down with sWGA-agarose to detect O-GlcNAc modification on dFOXO. (B) WT mice were treated with the O-GlcNAcase inhibitor, thiamet-G (TG) for 7 days to increase total O-GlcNAcylation. Total O-GlcNAcylation of control and thiamet-G injected mouse brain. (C) To demonstrate O-GlcNAcylation on Atg7, Atg7 was immunoprecipitated using specific O-GlcNAc antibody followed by Western blotting with anti-Atg7 antibody. Brain lysates were prepared and analyzed either directly by immunoblotting or following pull-down with sWGA agarose to detect O-GlcNAcylation on Atg7, LC3, and Atg5. β-actin was used as a loading control. (D) Control and PP242-treated S2 cell lysates were prepared and analyzed either directly by immunoblotting as input or following pull-down with sWGA agarose to detect O-GlcNAcylation on dAtg5 and Atg8a. β-actin was used as a loading control. (E) Whole cell lysate of GFP-mCherry-Atg8a-expressing larvae with feeding or fasting. Larval lysates were prepared and analyzed either directly by immunoblotting (input) or following pull-down with sWGA agarose to detect O-GlcNAcylation on Atg8a using mCherry and GFP antibodies. Upper arrow is mCherry-GFP-Atg8a and lower arrow is free mCherry. β-actin was used as a loading control (TIFF 173286 kb)