Abstract
The NFE2 transcription factor was identified over 25 years ago. The NFE2 protein forms heterodimers with small MAF proteins, and the resulting complex binds to regulatory elements in a large number of target genes. In contrast to other CNC transcription family members including NFE2L1 (NRF1), NFE2L2 (NRF2) and NFE2L3 (NRF3), which are widely expressed, earlier studies had suggested that the major sites of NFE2 expression are hematopoietic cells. Based on cell culture studies it was proposed that this protein acts as a critical regulator of globin gene expression. However, the knockout mouse model displayed only mild erythroid abnormalities, while the major phenotype was a defect in megakaryocyte biogenesis. Indeed, absence of NFE2 led to severely impaired platelet production. A series of recent data, also summarized here, shed new light on the various functional roles of NFE2 and the regulation of its activity. NFE2 is part of a complex regulatory network, including transcription factors such as GATA1 and RUNX1, controlling megakaryocytic and/or erythroid cell function. Surprisingly, it was recently found that NFE2 also has a role in non-hematopoietic tissues, such as the trophoblast, in which it is also expressed, as well as the bone, opening the door to new research areas for this transcription factor. Additional data showed that NFE2 function is controlled by a series of posttranslational modifications. Important strides have been made with respect to the clinical significance of NFE2, linking this transcription factor to hematological disorders such as polycythemias.
Keywords: NFE2, CNC transcription factor, Megakaryocytes, Erythroid cells, Trophoblast cells, Gene regulation, Hematopoiesis, Polycythemia
Introduction
History and nomenclature
The NFE2 transcription factor was first identified in the late 1980s as one of two DNA binding activities in extracts of erythroid cells [1]. The first protein was named NF-E1 for nuclear factor-erythroid 1, and this protein is now known as the GATA1 transcription factor [2]. The second protein called NF-E2 for nuclear factor-erythroid 2 was characterized as a protein recognizing a sequence comprising an activator protein 1 (AP-1) core motif, but was clearly different from AP-1 factors, as it needed residues outside of the core sequence for efficient binding [1, 3, 4]. The NF-E2 binding activity originally referred to the heterodimer of the large p45 NFE2 (~45 kD) and the small MAF (~18 kD) subunits [5]. In accordance with the current HUGO and MGI nomenclatures, we use in this review the terms NFE2 and Nfe2, the official human and mouse gene names respectively, to refer to the gene coding for the larger p45 kD subunit only.
The transcription factor NFE2
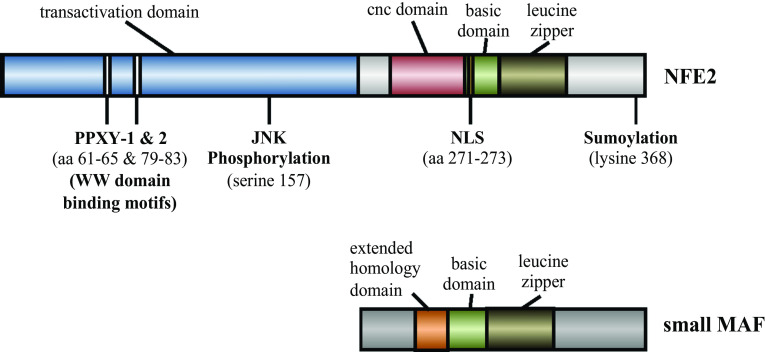
The NFE2 protein (Fig. 1), comprising 373 amino acids, is a member of the Cap’n’Collar (CNC) family of transcription factors [3], a classification that arose due to their homology to the Drosophila CNC1 [6] and C. elegans Skn-1 [7], implicated in development. In vertebrates, other members include NFE2L1 (nuclear factor, erythroid 2-like 1), also called NRF1, NFE2L2/NRF2 (nuclear factor, erythroid 2-like 2), NFE2L3/NRF3 (nuclear factor, erythroid 2-like 3), BACH1 (BTB and CNC homolog 1) and BACH2 (BTB and CNC homolog 2) [8]. The gene encoding the transcription factor NFE2 localizes to chromosome 12 in human [4] and to chromosome 15 in mice [9] and its cloning was reported over 20 years ago [3, 4]. Two transcripts are present with differences in the 5′ non-coding region, one of them being more abundant in adult hematopoietic sites, while the other one dominates in the fetal liver [10]. The binding partners of NFE2 have been identified as small MAF proteins [11, 12], members of a family of proto-oncogenes ubiquitously expressed related to the viral v-maf [13], coding for the MAFF, MAFG and MAFK (formerly also called p18) proteins, all approximately 18 kD in size [14–21] (Fig. 1). The basic leucine zipper bZIP domains of NFE2 and small MAFs mediate DNA binding and dimerization. DNA binding occurs through hydrogen bonds and hydrophobic interactions between the basic residues and the bases in the major groove. The region comprising the leucines consists of a coiled-coil domain with two parallel α-helices mediating protein–protein interactions by providing a hydrophobic interface [4, 11, 12, 22, 23]. The sequence recognized by the NFE2/small MAF heterodimer is an extended AP-1 motif, which is (T/C)GCTGA(C/G)TCA(T/C) [11]. The small MAF proteins are devoid of an obvious transactivation domain, but can act as repressors when homodimerizing between themselves or as activators when heterodimerizing with members of the CNC transcription factors [12, 14, 15, 24]. NFE2 has been found to interact with all three of the small MAFs [11, 12]. Evidence suggests the major interaction in megakaryocytes occurs primarily with MAFG and MAFF [16] while in erythroid cells NFE2 interacts with MAFG [15] and MAFK [18]. The small MAFs are ubiquitous proteins, which are expressed in many different cell types and tissues, although at different levels [19, 20]. In contrast to small MAFs and the other members of the CNC transcription factor family, NFE2 exhibits tissue restriction. The major sites of NFE2 expression are hematopoietic cells, such as erythroid cells, megakaryocytes and mast cells [3, 4, 25]. Expression of NFE2 transcripts in human peripheral granulocytes, consisting primarily of neutrophils, was also reported [26]. In mice it has been shown that NFE2 expression is restricted to hematopoietic tissues, including fetal liver and adult bone marrow or spleen [11]. Intriguingly, more recently NFE2 has been detected in trophoblast cells, challenging the notion that it is a hematopoietic-restricted factor [27].
Fig. 1.
Structure of human NFE2 and small MAF. The main domains and posttranslational modification sites of NFE2. The domains of its dimerization partners, the small MAFs are also shown. PPXY WW domain binding motif, JNK c-JUN N-terminal kinase, NLS nuclear localization signal, cnc cap’n’collar
Control of NFE2 activity
Localization
To exert its transcriptional activity NFE2 needs to be transported into the nucleus. This has been shown to be an active process necessitating an intact monopartite nuclear localization signal (NLS) and importin-7. Mutations in the NLS reduced its nuclear translocation and transcriptional activity, leading to impaired platelet production in megakaryocytes [28]. Within the nucleus NFE2 is located within euchromatin, while its dimerization partner MAFK is present in heterochromatin regions in mouse erythroleukemia MEL cells. Upon differentiation the small MAF subunit relocalizes to euchromatin, allowing its binding to NFE2 to exert its transactivation function [29]. The molecular mechanisms of this subnuclear compartmentalization have yet to be defined. Also, it is not known, whether nuclear export of NFE2 is regulated as well, as has been shown for example for the CNC family member NFE2L2 [30].
Posttranslational modifications
In recent years, posttranslational modifications have emerged as important aspects in the function of transcription factors, since they can influence such aspects as their subcellular localization and their stability, interactions with other proteins or their binding to DNA. NFE2 is subject to various posttranslational modifications that modulate its transcriptional activity (Fig. 1). In human erythroleukemia K562 cells NFE2 has been shown to be SUMOylated at lysine 368, a modification that enhanced its DNA binding affinity as well as transactivation and was essential for the transcription of the β-globin gene [31]. In contrast, ubiquitination of NFE2 by the itchy E3 ubiquitin protein ligase (ITCH) diminishes its transactivating capacity by retaining it in the cytoplasm and not by targeting it for degradation [32]. The latter result was obtained by overexpressing ITCH, hence it would be of interest to determine whether knockdown of the E3 ubiquitin ligase would result in increased nuclear localization of NFE2. NFE2 is also phosphorylated on serine 157 by activated c-JUN N-terminal kinase (phospho-JNK) in uninduced MEL cells, a modification that induces its ubiquitination and targeted degradation. This process was reversed in MEL cells induced to differentiate with DMSO when JNK was inactive [33]. It has been reported that NFE2/small MAF DNA binding complex formation was impaired in cAMP-dependent protein kinase A (PKA)-deficient MEL cells [34]. Although PKA was shown to phosphorylate NFE2 and MAFK in vitro this did not alter the binding of the NFE2/small MAF complex nor transactivation activity, suggesting an indirect mode of regulation. Possibly PKA regulates the interaction of NFE2 with downstream effectors [34, 35]. More recently, it has been demonstrated in vitro using purified proteins that SUMOylation of NFE2/small MAF heterodimer was enhanced by PKA and ERK1. It was proposed that heterodimerization may serve as a physical switch for posttranslational modification and thus control the transcriptional activity of the complex [36]. In addition, the transcriptional activity of NFE2 was increased by the acetylation of its binding partner MAFG mediated by CREB-binding protein [37]. From these data, it is clear that posttranslational modifications play a crucial role in the control of NFE2 activity, but the detailed molecular mechanisms, the interplay between signaling pathways and their importance in vivo still need to be elucidated. Moreover, the posttranslational modifications of NFE2 represent an interesting avenue as potential targets for treatments that would modulate NFE2 levels in hematological disorders, which are presented in a later section.
Regulatory motifs
The NFE2 protein contains within its transactivation domain motifs termed PPXY that bind to WW domains of proteins, one of them being necessary for transactivation [38, 39]. More precisely, this PPXY motif in NFE2 is required for the protein to induce hyperacetylation at histone H3, methylation of K4 at histone H3 and recruitment of RNA polymerase II at the site of the β-globin genes [40]. NFE2 comprises also two potential heme regulatory motifs (HRMs), which have been shown to be dispensable for globin gene induction in MEL cells. Nevertheless, induction of β-globin gene expression by hemin requires the presence of NFE2 in these cells. These data suggested that heme controls NFE2 activity indirectly through other proteins [41]. Indeed, the CNC protein BACH1 possesses six HRMs and heme is negatively regulating its DNA binding, by displacing it from its DNA bound complexes with small MAFs. This leads to dynamic interactions, as for instance, it has been shown, that at the β-globin locus in MEL cells, MAFK forms a repressive complex with BACH1, but switches partners, by interacting with NFE2 upon induction of differentiation to form a complex that drives globin activation [18, 42, 43]. This heme-dependent regulation has also been shown for the α-globin locus in K562 cells [44]. With respect to the molecular mechanism involved, it was found that heme triggers CRM1-dependent nuclear export of BACH1 [45]. Hence, these cell culture based studies provide a framework of how heme and globin synthesis are coordinately regulated.
Regulation of NFE2 expression
Early studies revealed the presence of two alternative promoters of the NFE2 gene resulting in the expression of two mRNA isoforms [10, 26, 46]. Expression and activity of NFE2 are controlled by a variety of reagents and proteins. Induction of differentiation in MEL cells following dimethylsulfoxide (DMSO) treatment leads to the increase of NFE2 transcript as well as protein levels [33, 47, 48]. On the other side, nuclear overexpression of phospholipase C (PLC) β1 in MEL cells, has been associated with inhibition of DMSO-induced differentiation, one of the postulated mechanisms being diminished levels of the NFE2 [49]. Expression levels of NFE2 are also regulated by cytokines or growth factors. In the myelogenous leukemia cell line K562 induced to differentiate with activin A, NFE2 expression and activity were shown to be downregulated by interferon-γ, a negative regulator of erythroid differentiation [50]. In megakaryocytes, interleukin-4 has been shown to downregulate NFE2 mRNA and protein expression [51], while interleukin-1β was shown to upregulate its transcript and protein levels in those cells [52, 53]. In addition, platelet-derived growth factor (PDGF) induced NFE2 transcripts and protein in megakaryocytic cells [54]. NFE2 expression is also controlled by other transcription factors. A major contributor to the transcriptional activity and function of NFE2 in megakaryocytes is GATA1, which has been shown in an elegant hierarchical model to act upstream. Using transgenic mouse models, it was demonstrated that GATA-1-dependent control of the Nfe2 gene significantly contributes to the function of NFE2 in megakaryocyte biogenesis [55]. Human erythroleukemia cells transfected with NF-κB display a decrease in NFE2, indicating that this transcription factor exerts a negative regulation on NFE2, consistent with the high levels of NF-κB in early erythroid progenitors, which display low levels of NFE2 [56]. Another transcription factor that negatively affects NFE2 activity in erythroid cells is c-Jun, which forms a heterocomplex with the small MAF subunit of NFE2 that is inactive and inhibits globin genes transcription [57]. In megakaryocytes, NFE2 was shown to be a target of the transcription factor RUNX1, a key regulator of this cell type [58]. Clearly, NFE2 is embedded in a complex network of signaling proteins and transcription factors, allowing the fine-tuning of its activity in a cell type- and exogenous signal-specific manner.
Role of NFE2 in megakaryocytes
NFE2 is a crucial regulator of megakaryocyte biogenesis and function
The generation of knockout mice has been highly valuable in determining the function of the NFE2 transcription factor in vivo. The various phenotypes of Nfe2 −/− mice are summarized (Table 1 ). When mice deficient in NFE2 were generated, their most striking phenotype was the absence of platelets, which lead to the death of over 90 % of pups by hemorrhage shortly after birth. It resulted from an arrest in maturation in the late-stages of megakaryocyte development, including reduced formation of cytoplasmic granules, but the cells still proliferated normally in response to thrombopoietin [59]. Interestingly surviving adults do not die of hemorrhage or display signs of bleeding in spite of an almost complete lack of platelets, indicating that other elements of the coagulation system play crucial roles. Those mice were also shown to lack production of proplatelets, which are filamentous extensions of mature megakaryocytes, identified as a mechanism of release of platelets [60]. It has also been reported that the proportion of colony forming units-megakaryocytes (CFU-Mks) versus total cells was lower in fetal livers and adult spleens of knockout mice [61]. Despite the presence of thrombocytopenia in Nfe2 −/− mice, the animals did not show an increase in serum thrombopoietin (TPO) levels. Experiments using labeled 125I-TPO showed that the cytokine is bound to hematopoietic tissues, in particular bone marrow and spleen, associated with megakaryocytes (bone marrow) and platelet-like particles (spleen), thus possibly explaining its absence in the circulation [62]. On the contrary, overexpression of NFE2 in murine bone marrow cells, gave rise to increased number of CFU-Mks, enhanced megakaryocyte maturation and resulted in a higher release of platelets, suggesting a role for this transcription factor not only in terminal megakaryocyte development, but also in its initial steps [63]. In primary murine megakaryocytes, the small MAF protein MAFG is a critical binding partner of NFE2 [16]. MAFG not only mediates DNA binding through its bZIP domain, but also targets the NFE2/MAFG heterodimer to a specific subnuclear localization, since deletion of the C-terminal domain of the small MAF factor does not prevent DNA binding, but recapitulates the platelet phenotype observed in Nfe2 −/− mice. This data suggest that the MAFG C-terminus is essential for proplatelet formation and platelet gene activation [64]. Overexpression of NFE2 and small MAF protein in mouse and human fibroblast cells resulted in the generation of megakaryocytes that were able to release platelets [65]. Recently, it was shown that blocking Rho kinase (ROCK) results in increased polyploidization, promotion of demarcation membrane and proplatelet formation as well as in the release of platelets in umbilical cord blood-derived megakaryocytes. ROCK inhibitor treatments in mature megakaryocytes lead also to the downregulation of MYC, NFE2, MAFG and MAFK transcript levels, suggesting that lowering the levels of these transcription factors is a prerequisite to drive the late stages of megakaryocyte maturation [66]. In addition to a well-established role in platelet biogenesis a more recent report suggests that absence of NFE2 results in impaired platelet function. This study showed that transgenic mice expressing a hypomorphic NFE2 mutant exhibit a hypofunction of platelets. This leads to an inhibition of lung metastasis following injection of melanoma cells in this mouse model, since platelet activation is required for the metastatic process. This data opens an interesting new chapter for NFE2 with regards to a possible role in cancer progression [67].
Table 1.
Phenotype of Nfe2 −/− mice
| Tissue type | Phenotype | References |
|---|---|---|
| Hematopoietic | Neonatal death of 90 % of the mice due to hemorrhage, severe thrombocytopenia | [59] |
| Lack of proplatelets | [60] | |
| Reduced colony-forming units megakaryocytes | [61] | |
| Severe anemia in neonates, mild anemia in surviving adults, splenomegaly, extensive reticulocytosis | [109] | |
| Increased sensitivity of red blood cells to phenylhydrazine treatment | [107] | |
| Increased serum EPO, altered erythroid precursor distribution in the spleen and bone marrow | [110] | |
| Non-hematopoietic | Increased bone mass and bone formation | [125, 126] |
| Intrauterine growth restriction, increased syncytiotrophoblast formation, impaired placental vascularization | [27, 128] |
Targets in megakaryocytes
The major role of NFE2 in megakaryocytes is transcriptional regulation of genes involved in their maturation and in the biogenesis of platelets. The first direct target of NFE2 in megakaryocytes to be identified has been the gene coding for thromboxane synthase [68]. The NFE2 protein is also recruited to the promoter of RAB27B, a small GTPase localized in granules and implicated in platelet synthesis [69]. The megakaryocyte-specific β1-tubulin (TUBB1) has also been identified as a target of NFE2, as its expression was recovered upon restoration of this transcription factor [70, 71]. In addition, it has been shown that activation of integrin alpha2b/integrin beta 3 (ITGA2B/ITGB3), also called alphaIIbbeta3, an integrin complex found on the surface of platelets, is defective in NFE2−/− megakaryocytes [72]. Screening for genes regulated by NFE2 that control ITGA2B/ITGB3 activation led to the identification of Caspase-12 as a gene to be downregulated in NFE2 deficient megakaryocytes [73].
A key enzyme for the synthesis of estrogen, 3beta-hydroxysteroid dehydrogenase (HSD3B1) is also a target of NFE2. Of interest, 3beta-HSD has been shown to rescue proplatelet formation in NFE2 null megakaryocytes [74]. Furthermore, the adaptor protein LIM and senescent cell antigen-like domains 1 (LIMS1), implicated in integrin signaling and cell motility is another target of NFE2, as demonstrated by chromatin immunoprecipitation (ChIP) and transactivation experiments [75]. A more recent study combined whole genome-wide analysis by chromatin immunoprecipitation-sequencing (ChIP-seq) and microarrays in primary megakaryocytes [67]. This approach showed a typical NFE2 binding site, A/G TGA C/G TCA GC, to be overrepresented at the occupancy sites. Among 844 genes comprising ChIP-seq peaks for NFE2, they identified 49 candidate genes that can be activated by NFE2 directly, but also 10 candidate genes that can be repressed. Of note, 15 of the genes that are candidates for direct activation have established roles in platelet function during thrombogenesis. The gene encoding P-selectin (Selp), which plays a crucial role in platelet function, and Myosin, light polypeptide 9 (Myl9), which is important for proplatelet formation, have been identified as direct targets of NFE2 [67]. In addition to transcriptional regulation of platelet genes, NFE2 contributes to megakaryocyte maturation by promoting the accumulation of intracellular reactive oxygen species (ROS). The elimination of ROS is largely accomplished by induction of NFE2L2 (NRF2) targets and since NFE2 shows very similar binding specificity, it can compete with NFE2L2 for induction of proteins that eliminate ROS. There is an increase in ROS during megakaryocyte maturation, suggesting that they play an important role in signaling [76]. Definitely, NFE2 plays a crucial role in the generation of platelets as well as in platelet function. Although a series of its targets in this lineage are now known, the coordination between NFE2 controlled pathways and the regulation of megakaryopoiesis by other transcription factors, including EVI1, GATA1, FLI1, RUNX1, SRF and TAL1, as well as miRNAs, epigenetic and posttranslational mechanisms [77], still needs to be elucidated.
Function of NFE2 in the erythroid lineage
A role for NFE2 as a factor promoting erythroid maturation has been suggested by experiments where enforced overexpression of this transcription factor in the monoblastoid M1 cell line yields erythroid and megakaryocytic colonies, while its overexpression in hematopoietic progenitors from fetal liver increases erythroid colonies [78]. As illustrated in the following paragraphs, binding sites for NFE2 have been identified in the regulatory sequences of multiple erythroid genes.
NFE2 and heme biosynthesis
The NFE2 transcription factor has been initially identified as a protein binding to the promoter region of the porphobilinogen deaminase (PBGD) gene, coding for the third enzyme of the heme biosynthesis pathway [1]. Subsequent analysis showed that mutation in the NFE2 binding site abolishes inducibility of the PBDG gene, suggesting that NFE2 is required for proper regulation of this promoter in erythroid cells [79]. The gene of another enzyme, ferrochelatase, catalyzing the last step of heme synthesis, possesses a NFE2 recognition site in its promoter [80], and DNase I footprinting analysis suggested binding of NFE2 to its cognate element in K562 cells [81]. The gene coding for erythroid aminolevulinate synthase 2 (ALAS2) also contains a potential NFE2 element [82], but later studies showed that this site is not essential for expression of the gene, and overexpression of NFE2 and MAFF failed to increase transcriptional activity as assessed by a luciferase reporter assay [83]. A recent study using ChIP has shown that NFE2 binds to regulatory regions of other heme biosynthesis genes, including the ones coding for uroporphyrinogen III synthase (UROS), uroporphyrinogen III decarboxylase (UROD), coproporphyrinogen oxidase (CPOX), but not aminolevulinic acid dehydratase (ALAD). Additional loss-of-function experiments in human erythroleukemia (HEL) cells showed that NFE2 contributes to UROS, CPOX and UROD expression. Furthermore, transfection of NFE2 and MAFG in HEK293 cells resulted in the induction of a luciferase reporter comprising UROS regulatory sequences [84]. The gene coding for the enzyme heme oxygenase 1 (HO1) responsible for the catabolism of heme also has a potential binding sequence for NFE2 [85]. Of interest, heme, the end product of the heme biosynthesis pathway, is able to induce the activity of the NFE2/small MAF heterodimer in K562 cells, as shown by reporter studies [86]. Earlier data have also shown that downregulation of ALAS2 using antisense technology leads to a decrease of NFE2 transcript levels, but does not affect the expression of MAFK, its binding partner [87]. Thus, NFE2 plays an important role at different steps of heme synthesis, but given its tissue restriction and the fact that virtually all cells synthesize heme, it would be of interest to identify the proteins that can replace its function in cell types that are devoid of it.
Regulation of globin chain synthesis and other erythroid targets
NFE2 is required for the production of globin proteins in erythroleukemia cell lines. An erythroleukemia cell line, termed CB3, which is devoid of NFE2, due to the integration of Friend virus in one allele and the loss of the other allele, has served as a valuable model. These cells fail to express high levels of α- and β-globin upon induction of differentiation, while reintroduction of NFE2 or a tethered NFE2/small MAF dimer rescues globin expression [14, 15, 88]. In addition to adult globin chains, NFE2 has been shown to be necessary for the induction of the fetal γ-globin [89]. Transcript and protein levels as well as transactivation and DNA-binding of NFE2 are induced following differentiation of MEL cells with DMSO [48]. Furthermore, hemin (iron protoporphyrin IX) induces globin expression in human erythroleukemia K562 cells and induces the transcription of a reporter containing NFE2 binding sites [90]. The regions necessary for NFE2 transactivation consist of two proline rich sequences within its transactivation and CNC domains [91]. It has been demonstrated that NFE2 in addition to GATA1 is necessary for the formation of DNAse I hypersensitive site 4 present in the enhancer region of β-globin, which allows the chromatin to assume a more “open” conformation [92]. Similarly, NFE2 sites are also necessary for transactivation mediated by DNAse I hypersensitive sites 2 and 3 in the β-globin locus control region [93]. In fact, NFE2 exerts a chromatin remodelling action on the DNAse I hypersensitive site 2 (HS2) of β-globin, by disrupting nucleosomes in an ATP-dependent fashion and allowing subsequent binding of GATA1 [94]. There is evidence that the enhancer HS2 interacts with the promoter at the β-globin locus, since NFE2 binds to the HS2 and is also interacting through its N-terminal domain with a protein present at the promoter site, namely TAFII130, which is associated with TATA binding protein [95]. A confirmation of in vivo binding of NFE2 to the HS2 site came from a study analyzing erythroleukemia cells and cells from mouse fetal liver using ChIP [96]. It has also been reported that NFE2 binds to the DNAse I HS2 site of β-globin, when the chromatin is still in the repressive state, before remodelling occurs [97]. However, in MEL cells the recruitment of NFE2 to the locus control region (LCR) and promoter of β-globin as shown by ChIP is greatly increased upon induction of differentiation. Interestingly, the promoter does not contain NFE2 binding sites, so its presence could be explained through binding to proteins associated with the promoter, or through DNA looping of regions of the LCR to the promoter [98]. In the human K562 cell line NFE2 has been shown to bind to all four HS sites of the β-globin LCR [99]. In human multipotent progenitors NFE2 is recruited to the LCR and promoter regions, in a fashion that is dependent on the erythroid krüppel-like factor (EKLF) and that includes also the recruitment of cofactors TATA-binding protein (TBP), CREB-binding protein (CBP) and Brahma-related gene-1 (BRG1) [100]. However, in contrast to GATA1 and EKLF, NFE2 appears to be dispensable for the formation of the active chromatin hub (ACH), a structure in which the distant LCR loops towards regions where globin genes are present. In Nfe2 knockout mice there is increased binding of NFE2L2 (NRF2) at the LCR, but not to the promoter, suggesting a possible compensation through this transcription factor [101]. The transcription factor NFE2 also binds to and recruits the methyltransferase G9a at the β-globin locus, which plays an activating role for adult β-major chains, while repressing embryonic chains [102]. The recruitment of polymerase II to the promoter of the adult β-globin gene has been demonstrated to necessitate NFE2 and upstream stimulatory factor (USF), a ubiquitous transcription factor shown to interact with NFE2 [103]. There has also been a demonstration of binding of NFE2 to the promoter of alpha-spectrin, a protein that is found in the membrane of erythrocytes, as shown by reporter assays [104]. In addition, NFE2 has been found to positively regulate the expression of the alpha-hemoglobin stabilizing protein (AHSP), a chaperone for α-globin [105]. With respect to erythroid-specific gene transcription, a recent ChIP based study supports the notion that NFE2L2 (NRF2), which is able to bind to NFE2 recognition sites, may also play a role in heme metabolism and erythropoiesis, in particular in response to cellular stress [106]. A potential role for NFE2 in erythroid cells in the protection against oxidative stress has been proposed. At such, red blood cells from Nfe2 −/− mice have been shown to express higher levels of ROS at baseline, as well as markedly increased ROS when treated with H2O2 compared with those from wild-type mice. Those results translated in vivo into a higher sensitivity to the oxidative stress inducer phenylhydrazine, which caused a more severe drop in hematocrit and increased reticulocytosis in Nfe2 knockout mice compared to wild-type animals. One postulated mechanism is the reduced levels of catalase in red blood cells in Nfe2 knockout mice [107]. Recent data have identified the miR-199b-5p as a key regulator of erythropoiesis in humans. The expression of this microRNA is induced during the erythroid differentiation of K562 cells. The upregulation was controlled by the binding of GATA-1 and NFE2 to the miR-199b-5p locus in hemin-treated K562 cells [108].
Erythroid phenotype of Nfe2−/− mice
In vivo, the importance of NFE2 in erythroid cells is illustrated by the fact that mice generated with a deficiency in NFE2 display erythroid abnormalities and anemia, although milder than expected. Erythroid abnormalities are more pronounced in neonates, of which more than 90 % die of hemorrhage due to the lack of platelets. However, surviving adult mice still display hypochromia and presence of target cells, as well as slightly lower hematocrit and hemoglobin. It has been speculated that increased bleeding due to the lack of platelets is causing this phenotype. These mice also display extensive reticulocytosis and splenomegaly, possibly signs of increased compensatory erythropoiesis [59, 109]. More recently compensatory splenic erythropoiesis was confirmed by the observation of an increase in earlier erythroid precursors and enhanced EPO levels in the serum. This can be attributed to a partial block in the differentiation of erythroid precursors in the bone marrow between the basophilic and late basophilic/polychromatic erythroblast stage in Nfe2 −/− mice [110]. The relatively mild erythroid effect cannot be attributed to functional compensation by the related factor NFE2L2, since mice deficient in both of these transcription factors do not display more severe erythroid abnormalities than those deficient in NFE2 alone [111, 112]. Also, transcript levels of neither NFE2L1 nor NFE2L2 are altered in Nfe2 −/− mice, and gel shift assays did not reveal a new binding activity in erythroid nuclear extracts from Nfe2 −/− animals [109]. Hence, the molecular basis for the discrepancy between cell culture based studies, showing a role for NFE2 in globin gene regulation and the absence of an obvious role in vivo in the erythroid compartment in the knockout mouse model still needs to be elucidated. In addition, the absence of NFE2 is an important step in the progression of Friend virus induced erythroleukemia in mice, since heterozygous mice infected with the virus present higher tumor incidence and increased tumor size compared to their wild-type counterparts [113].
Linking NFE2 to hematological disorders
In a clinical setting, overexpression of NFE2 has been identified in polycythemia vera (PV) patients, a disorder characterized by overproduction of erythroid cells and sometimes megakaryocytes and platelets. The severity of the symptoms correlated with the degree of upregulation of NFE2 [114]. Overexpression of NFE2 in CD34+ cells also results in a delay in early erythroid maturation leading to erythroid progenitor expansion and consequently an increased number of erythrocytes derived from one CD34+ cell [115]. In addition, overexpression of NFE2 in CD34+ cells from healthy donors recapitulated the PV phenotype of increased EPO-independent erythroid differentiation and expansion of HSCs/CMPs, while silencing of NFE2 in cells from PV patients had the opposite effect [116]. In addition to PV, increased expression of NFE2 has also been observed in patients with other myeloproliferative neoplasms (MPNs), such as essential thrombocythemia and primary myelofibrosis, through a mechanism that may involve the transcriptional upregulation of NFE2 mediated by Runt-related transcription factor-1 (RUNX1) [117]. Although a large majority of MPN patients harbors the JAK2 (V617F) mutation, there was no correlation between the JAK2 mutational status and the overexpression of both RUNX1 and NFE2 [117]. Furthermore, insertion and deletion mutations in NFE2 in MPN patients were identified. The truncated versions, although not able to bind to DNA nor to transactivate on their own, enhanced the activity of wild-type NFE2 in transfection studies and resulted in erythrocytosis, thrombocytosis and neutrophilia in a mouse model [118]. As NFE2 is mislocalized in primary myelofibrosis (PMF), it was also shown that immunohistochemical staining of NFE2 reliably allows a differential diagnosis between PMF and essential thrombocytopenia [119]. A novel transgenic mouse model generated by overexpressing NFE2 in hematopoietic cells recapitulates many features of MPN, including thrombocytosis, leukocytosis, formation of colonies independent of EPO, typical bone marrow histology, as well as the increase of stem and progenitor cell compartments and spontaneous transformation to acute myeloid leukemia. This in vivo model should help investigating the role of NFE2 in MPNs and identifying new targets for therapeutic intervention [120]. Based on this model, it was hypothesized that NFE2 may have a crucial role in maintaining chronic inflammation and driving clonal evolution and mutagenesis in MPNs [121]. Of interest, treatment of JAK2V617F cells with the histone deacetylase (HDAC) inhibitor Givinostat (GVS) results in the downregulation of NFE2 and C-MYB mRNA and protein levels. Although GVS also blocked JAK2 signal transducer and activator 5-extracellular signal-regulated kinase 1/2 phosphorylation, the control of NFE2 and C-MYB occurred independently of JAK2. GVS acts directly on the NFE2 promoter and the downregulation of NFE2 in CD34+ cells derived from MPN patients lead to inhibition of cell proliferation and erythroid differentiation [122]. Treatment with the HDAC inhibitor Vorinostat also results in the decrease of NFE2 transcript levels in JAK2V617F HEL cells. In a Jak2 V617F knock-in mouse model, treatment with Vorinostat leads to an improvement of peripheral blood counts and a reduction of splenomegaly, underlining its therapeutic potential [123].
Recently, it was shown that upregulation of RUNX1 and its target NFE2 is not specific for PV and other MPNs, as their transcript levels were also substantially increased in polycythemias with higher hypoxia-inducible factor activity, whose progenitors exhibited increased erythropoietin (EPO) sensitivity. In contrast, RUNX1 and NFE2 overexpression was not observed in patients harboring EPO receptor gain-of-function mutations. It was concluded that elevated levels of RUNX1 and NFE2 are not typically present in all primary polycythemias such as primary familial and congenital polycythemia (PFCP), but are rather a characteristic of primary polycythemias with increased HIF signaling [124]. Collectively, these clinically relevant studies suggest that modulation of NFE2 and associated pathways may have therapeutic potential for myeloproliferative disorders.
NFE2 and non-hematopoietic cells
Nfe2 −/− mice also exhibit an increase in trabecular bone volume and in the rate of bone formation, accompanied by augmented serum osteocalcin levels. Intriguingly, cells of the osteoblast lineage do not express Nfe2 transcripts [125]. It was shown that the increased bone phenotype could be transferred to irradiated wild-type animals by using spleen cells derived from Nfe2 −/− animals, suggesting the involvement of hematopoietic cells with respect to the bone phenotype. Experiments of co-culturing osteoblasts with megakaryocytes from NFE2-deficient mice revealed that increased osteoblast proliferation required cell-to-cell contact. It was suggested that the increased bone mass in Nfe2 −/− mice might be due to a megakaryocyte-osteoblast interaction that is anabolic for bone [125, 126]. NFE2-deficient mice also display a substantial increase in cortical bone area and cortical thickness, as well as higher bone mineral density [127]. Hence, elucidation of the molecular mechanisms involved in this regulation by NFE2 may be of interest to identify therapeutic targets for bone disease characterized by a decrease in bone mass and density, such as osteoporosis.
The absence of NFE2 in mice also leads to intrauterine growth restriction (IUGR) and a clue with respect to this phenotype has been recently revealed by the finding that this transcription factor is expressed in cells of the trophoblast. NFE2 has been found to be essential for normal syncytiotrophoblast formation, vascularisation of the placenta as well as embryonic growth. Loss of NFE2 leads to increased expression of the transcription factor glial cells missing homolog 1 (GCM1) and its target genes, resulting in increased syncytiotrophoblast formation. It was found that absence of NFE2 augments acetylation of the GCM1 protein, and in addition enhances acetylation of histone H4 within the GCM1 promoter. Promoting acetylation with HDAC inhibitors in wild-type embryos phenocopies the alterations detected in Nfe2 −/− embryos, whereas blocking of acetylation with the histone acetyltransferase inhibitor curcumin corrects the changes in NFE2 deficient embryos [27]. An additional study showed that the mechanism involves repression by NFE2 of the binding of the JUND transcription factor to the promoter of GCM1 [128]. Although previous thinking confined NFE2 expression exclusively to blood cells, these recent studies underline the importance of NFE2 in non-hematopoietic lineages, opening up new interesting avenues of investigation for this transcription factor.
Conclusions and outlook
The NFE2 transcription factor functions as an important regulator of megakaryocyte maturation. Its vital role in the biogenesis of platelets has been well characterized, especially using the knockout mouse model, which presents severe thrombocytopenia. In contrast, many aspects of its role in the erythroid lineage remain elusive. One of the major issues is the divergence between its vital function in globin expression in mouse erythroleukemia cells and its relatively mild erythroid phenotype in Nfe2 −/− mice. It has been suggested that other members of the CNC transcription factors could be compensating for the role of NFE2 in mice erythroid cells. However, the analysis of Nfe2 and Nfe2l2 compound knockout mice did not reveal any obvious additional anomalies when compared to Nfe2 single knock out animals [111], and we obtained similar results with respect to compound Nfe2 −/− and Nfe2l3 −/− mice (unpublished data). Nfe2l1 −/− mice are embryonic lethal and display anemia [129], making this CNC member a likely factor to compensate. Hence, study of the role of NFE2L1 in the compensation would require a conditional targeting approach. In addition, future studies should address the function of NFE2 in mast cells, as these hematopoietic cells also express this transcription factor [3]. An earlier study suggested that NFE2 is involved in the control of the gene coding for l-histidine decarboxylase (HDC), an important player in the differentiation of mast cells [130]. However, further studies are lacking on the role of NFE2 in this lineage. Based on data linking NFE2 to polycythemias, the transcription factor and/or associated pathways may be valuable therapeutic targets to decrease the number of circulating red blood cells in patients with myeloproliferative disorders. In the future, in-depth analyses of its roles in hematopoietic cells will provide further intriguing insights into the network of NFE2 dependent gene regulation. For a long time considered to be a protein restricted to hematopoietic lineages, and studied in particular in erythroid and megakaryocytic cells, more recent data have shown a function for NFE2 in non-hematopoietic tissues. These initial studies, revealing a role for this transcription factor in trophoblast cells, in bone formation and cancer metastasis, have opened a door to new fascinating studies on the NFE2 transcription factor in a variety of fields.
Acknowledgments
This work was supported by a McGill University Faculty of Medicine internal studentship and a joint fellowship from the CIHR and the Thalassemia Foundation of Canada to JG. We would also like to acknowledge grants from the Thalassemia Foundation of Canada as well as CIHR (MOP-79361 and MOP-97932) to VB.
References
- 1.Mignotte V, Wall L, deBoer E, Grosveld F, Romeo PH. Two tissue-specific factors bind the erythroid promoter of the human porphobilinogen deaminase gene. Nucleic Acids Res. 1989;17(1):37–54. doi: 10.1093/nar/17.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whitelaw E, Tsai SF, Hogben P, Orkin SH. Regulated expression of globin chains and the erythroid transcription factor GATA-1 during erythropoiesis in the developing mouse. Mol Cell Biol. 1990;10(12):6596–6606. doi: 10.1128/mcb.10.12.6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrews NC, Erdjument-Bromage H, Davidson MB, Tempst P, Orkin SH. Erythroid transcription factor NF-E2 is a haematopoietic-specific basic-leucine zipper protein. Nature. 1993;362(6422):722–728. doi: 10.1038/362722a0. [DOI] [PubMed] [Google Scholar]
- 4.Ney PA, Andrews NC, Jane SM, Safer B, Purucker ME, Weremowicz S, Morton CC, Goff SC, Orkin SH, Nienhuis AW. Purification of the human NF-E2 complex: cDNA cloning of the hematopoietic cell-specific subunit and evidence for an associated partner. Mol Cell Biol. 1993;13(9):5604–5612. doi: 10.1128/mcb.13.9.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrews NC. The NF-E2 transcription factor. Int J Biochem Cell Biol. 1998;30(4):429–432. doi: 10.1016/S1357-2725(97)00135-0. [DOI] [PubMed] [Google Scholar]
- 6.Mohler J, Vani K, Leung S, Epstein A. Segmentally restricted, cephalic expression of a leucine zipper gene during Drosophila embryogenesis. Mech Dev. 1991;34(1):3–9. doi: 10.1016/0925-4773(91)90086-L. [DOI] [PubMed] [Google Scholar]
- 7.Bowerman B, Eaton BA, Priess JR. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell. 1992;68(6):1061–1075. doi: 10.1016/0092-8674(92)90078-Q. [DOI] [PubMed] [Google Scholar]
- 8.Motohashi H, O’Connor T, Katsuoka F, Engel J, Yamamoto M. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene. 2002;294(1–2):1–12. doi: 10.1016/S0378-1119(02)00788-6. [DOI] [PubMed] [Google Scholar]
- 9.Peters LL, Andrews NC, Eicher EM, Davidson MB, Orkin SH, Lux SE. Mouse microcytic anaemia caused by a defect in the gene encoding the globin enhancer-binding protein NF-E2. Nature. 1993;362(6422):768–770. doi: 10.1038/362768a0. [DOI] [PubMed] [Google Scholar]
- 10.Pischedda C, Cocco S, Melis A, Marini MG, Kan YW, Cao A, Moi P. Isolation of a differentially regulated splicing isoform of human NF-E2. Proc Natl Acad Sci USA. 1995;92(8):3511–3515. doi: 10.1073/pnas.92.8.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andrews NC, Kotkow KJ, Ney PA, Erdjument-Bromage H, Tempst P, Orkin SH. The ubiquitous subunit of erythroid transcription factor NF-E2 is a small basic-leucine zipper protein related to the v-maf oncogene. Proc Natl Acad Sci USA. 1993;90(24):11488–11492. doi: 10.1073/pnas.90.24.11488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Igarashi K, Kataoka K, Itoh K, Hayashi N, Nishizawa M, Yamamoto M. Regulation of transcription by dimerization of erythroid factor NF-E2 p45 with small Maf proteins (see comments) Nature. 1994;367(6463):568–572. doi: 10.1038/367568a0. [DOI] [PubMed] [Google Scholar]
- 13.Nishizawa M, Kataoka K, Goto N, Fujiwara KT, Kawai S. v-maf, a viral oncogene that encodes a “leucine zipper” motif. Proc Natl Acad Sci USA. 1989;86(20):7711–7715. doi: 10.1073/pnas.86.20.7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kotkow KJ, Orkin SH. Dependence of globin gene expression in mouse erythroleukemia cells on the NF-E2 heterodimer. Mol Cell Biol. 1995;15(8):4640–4647. doi: 10.1128/mcb.15.8.4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blank V, Kim MJ, Andrews NC. Human MafG is a functional partner for p45 NF-E2 in activating globin gene expression. Blood. 1997;89(11):3925–3935. [PubMed] [Google Scholar]
- 16.Lecine P, Blank V, Shivdasani R. Characterization of the hematopoietic transcription factor NF-E2 in primary murine megakaryocytes. J Biol Chem. 1998;273(13):7572–7578. doi: 10.1074/jbc.273.13.7572. [DOI] [PubMed] [Google Scholar]
- 17.Marini MG, Asunis I, Chan K, Chan JY, Kan YW, Porcu L, Cao A, Moi P. Cloning MafF by recognition site screening with the NF-E2 tandem repeat of HS2: analysis of its role in globin and GCSI genes regulation. Blood Cells Mol Dis. 2002;29(2):145–148. doi: 10.1006/bcmd.2002.0550. [DOI] [PubMed] [Google Scholar]
- 18.Brand M, Ranish JA, Kummer NT, Hamilton J, Igarashi K, Francastel C, Chi TC, Crabtree GR, Aebersold R, Groudine M. Dynamic changes in transcription factor complexes during erythroid differentiation revealed by quantitative proteomics. Nat Struct Mol Biol. 2004;11(1):73–80. doi: 10.1038/nsmb713. [DOI] [PubMed] [Google Scholar]
- 19.Blank V, Andrews NC. The Maf transcription factors: regulators of differentiation. Trends Biochem Sci. 1997;22(11):437–441. doi: 10.1016/S0968-0004(97)01105-5. [DOI] [PubMed] [Google Scholar]
- 20.Blank V. Small Maf proteins in mammalian gene control: mere dimerization partners or dynamic transcriptional regulators? J Mol Biol. 2008;376:913–925. doi: 10.1016/j.jmb.2007.11.074. [DOI] [PubMed] [Google Scholar]
- 21.Kannan MB, Solovieva V. Blank V (2012) The small MAF transcription factors MAFF, MAFG and MAFK: current knowledge and perspectives. Biochim Biophys Acta. 1823;10:1841–1846. doi: 10.1016/j.bbamcr.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 22.Chan JY, Han XL, Kan YW. Isolation of cDNA encoding the human NF-E2 protein. Proc Natl Acad Sci USA. 1993;90(23):11366–11370. doi: 10.1073/pnas.90.23.11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toki T, Itoh J, Kitazawa J, Arai K, Hatakeyama K, Akasaka J, Igarashi K, Nomura N, Yokoyama M, Yamamoto M, Ito E. Human small Maf proteins form heterodimers with CNC family transcription factors and recognize the NF-E2 motif. Oncogene. 1997;14(16):1901–1910. doi: 10.1038/sj.onc.1201024. [DOI] [PubMed] [Google Scholar]
- 24.Kataoka K, Igarashi K, Itoh K, Fujiwara KT, Noda M, Yamamoto M, Nishizawa M. Small Maf proteins heterodimerize with Fos and may act as competitive repressors of the NF-E2 transcription factor (published erratum appears in Mol Cell Biol 1995 Jun; 15(6):3461) Mol Cell Biol. 1995;15(4):2180–2190. doi: 10.1128/mcb.15.4.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romeo PH, Prandini MH, Joulin V, Mignotte V, Prenant M, Vainchenker W, Marguerie G, Uzan G. Megakaryocytic and erythrocytic lineages share specific transcription factors. Nature. 1990;344(6265):447–449. doi: 10.1038/344447a0. [DOI] [PubMed] [Google Scholar]
- 26.Toki T, Itoh J, Arai K, Kitazawa J, Yokoyama M, Igarashi K, Yamamoto M, Ito E. Abundant expression of erythroid transcription factor P45 NF-E2 mRNA in human peripheral granurocytes. Biochem Biophys Res Commun. 1996;219(3):760–765. doi: 10.1006/bbrc.1996.0307. [DOI] [PubMed] [Google Scholar]
- 27.Kashif M, Hellwig A, Kolleker A, Shahzad K, Wang H, Lang S, Wolter J, Thati M, Vinnikov I, Bierhaus A, Nawroth PP, Isermann B. p45NF-E2 represses Gcm1 in trophoblast cells to regulate syncytium formation, placental vascularization and embryonic growth. Development. 2011;138(11):2235–2247. doi: 10.1242/dev.059105. [DOI] [PubMed] [Google Scholar]
- 28.Perdomo J, Fock EL, Kaur G, Yan F, Khachigian LM, Jans DA, Chong BH. A monopartite sequence is essential for p45 NF-E2 nuclear translocation, transcriptional activity and platelet production. J Thromb Haemost. 2010;8(11):2542–2553. doi: 10.1111/j.1538-7836.2010.04058.x. [DOI] [PubMed] [Google Scholar]
- 29.Francastel C, Magis W, Groudine M. Nuclear relocation of a transactivator subunit precedes target gene activation. Proc Natl Acad Sci USA. 2001;98(21):12120–12125. doi: 10.1073/pnas.211444898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jain AK, Bloom DA, Jaiswal AK. Nuclear import and export signals in control of Nrf2. J Biol Chem. 2005;280(32):29158–29168. doi: 10.1074/jbc.M502083200. [DOI] [PubMed] [Google Scholar]
- 31.Shyu YC, Lee TL, Ting CY, Wen SC, Hsieh LJ, Li YC, Hwang JL, Lin CC, Shen CK. Sumoylation of p45/NF-E2: nuclear positioning and transcriptional activation of the mammanlian beta-like globin gene locus. Mol Cell Biol. 2005;25(23):10365–10378. doi: 10.1128/MCB.25.23.10365-10378.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee TL, Shyu YC, Hsu TY, Shen CK. Itch regulates p45/NF-E2 in vivo by Lys63-linked ubiquitination. Biochem Biophys Res Commun. 2008;375(3):326–330. doi: 10.1016/j.bbrc.2008.07.164. [DOI] [PubMed] [Google Scholar]
- 33.Lee TL, Shyu YC, Hsu PH, Chang CW, Wen SC, Hsiao WY, Tsai MD, Shen CK. JNK-mediated turnover and stabilization of the transcription factor p45/NF-E2 during differentiation of murine erythroleukemia cells. Proc Natl Acad Sci USA. 2010;107(1):52–57. doi: 10.1073/pnas.0909153107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garingo AD, Suhasini M, Andrews NC, Pilz RB. cAMP-dependent protein kinase is necessary for increased NF-E2.DNA complex formation during erythroleukemia cell differentiation. J Biol Chem. 1995;270(16):9169–9177. doi: 10.1074/jbc.270.16.9169. [DOI] [PubMed] [Google Scholar]
- 35.Casteel D, Suhasini M, Gudi T, Naima R, Pilz RB. Regulation of the erythroid transcription factor NF-E2 by cyclic adenosine monophosphate-dependent protein kinase. Blood. 1998;91(9):3193–3201. [PubMed] [Google Scholar]
- 36.Su YF, Shyu YC, Shen CK, Hwang J. Phosphorylation-dependent SUMOylation of the transcription factor NF-E2. PLoS One. 2012;7(9):e44608. doi: 10.1371/journal.pone.0044608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hung HL, Kim AY, Hong W, Rakowski C, Blobel GA. Stimulation of NF-E2 DNA binding by CREB-binding protein (CBP)-mediated acetylation. J Biol Chem. 2001;276(14):10715–10721. doi: 10.1074/jbc.M007846200. [DOI] [PubMed] [Google Scholar]
- 38.Gavva NR, Gavva R, Ermekova K, Sudol M, Shen CJ. Interaction of WW domains with hematopoietic transcription factor p45/NF-E2 and RNA polymerase II. J Biol Chem. 1997;272(39):24105–24108. doi: 10.1074/jbc.272.39.24105. [DOI] [PubMed] [Google Scholar]
- 39.Mosser EA, Kasanov JD, Forsberg EC, Kay BK, Ney PA, Bresnick EH. Physical and functional interactions between the transactivation domain of the hematopoietic transcription factor NF-E2 and WW domains. Biochemistry. 1998;37(39):13686–13695. doi: 10.1021/bi981310l. [DOI] [PubMed] [Google Scholar]
- 40.Kiekhaefer CM, Boyer ME, Johnson KD, Bresnick EH. A WW domain-binding motif within the activation domain of the hematopoietic transcription factor NF-E2 is essential for establishment of a tissue-specific histone modification pattern. J Biol Chem. 2004;279(9):7456–7461. doi: 10.1074/jbc.M309750200. [DOI] [PubMed] [Google Scholar]
- 41.Moore A, Boudia MM, Lehalle D, Massrieh W, Derjuga A, Blank V. Regulation of globin gene transcription by heme in erythroleukemia cells: analysis of putative heme regulatory motifs in the p45 NF-E2 transcription factor. Antioxid Redox Signal. 2006;8(1–2):68–75. doi: 10.1089/ars.2006.8.68. [DOI] [PubMed] [Google Scholar]
- 42.Tahara T, Sun J, Nakanishi K, Yamamoto M, Mori H, Saito T, Fujita H, Igarashi K, Taketani S. Heme positively regulates the expression of b-globin at the locus control region via the transcriptional factor Bach1 in erythroid cells. J Biol Chem. 2004;279(7):5480–5487. doi: 10.1074/jbc.M302733200. [DOI] [PubMed] [Google Scholar]
- 43.Sun J, Brand M, Zenke Y, Tashiro S, Groudine M, Igarashi K. Heme regulates the dynamic exchange of Bach1 and NF-E2-related factors in the Maf transcription factor network. Proc Natl Acad Sci USA. 2004;101(6):1461–1466. doi: 10.1073/pnas.0308083100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tahara T, Sun J, Igarashi K, Taketani S. Heme-dependent up-regulation of the alpha-globin gene expression by transcriptional repressor Bach1 in erythroid cells. Biochem Biophys Res Commun. 2004;324(1):77–85. doi: 10.1016/j.bbrc.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki H, Tashiro S, Hira S, Sun J, Yamazaki C, Zenke Y, Ikeda-Saito M, Yoshida M, Igarashi K. Heme regulates gene expression by triggering Crm1-dependent nuclear export of Bach1. EMBO J. 2004;23(13):2544–2553. doi: 10.1038/sj.emboj.7600248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toki T, Arai K, Terui K, Komatsu N, Yokoyama M, Katsuoka F, Yamamoto M, Ito E. Functional characterization of the two alternative promoters of human p45 NF-E2 gene. Exp Hematol. 2000;28(10):1113–1119. doi: 10.1016/S0301-472X(00)00523-3. [DOI] [PubMed] [Google Scholar]
- 47.Francastel C, Poindessous-Jazat V, Augery-Bourget Y, Robert-Lezenes J. NF-E2p18/mafK is required in DMSO-induced differentiation of Friend erythroleukemia cells by enhancing NF-E2 activity. Leukemia. 1997;11(2):273–280. doi: 10.1038/sj.leu.2400552. [DOI] [PubMed] [Google Scholar]
- 48.Nagai T, Igarashi K, Akasaka J, Furuyama K, Fujita H, Hayashi N, Yamamoto M, Sassa S. Regulation of NF-E2 activity in erythroleukemia cell differentiation. J Biol Chem. 1998;273(9):5358–5365. doi: 10.1074/jbc.273.9.5358. [DOI] [PubMed] [Google Scholar]
- 49.Faenza I, Matteucci A, Bavelloni A, Marmiroli S, Martelli AM, Gilmour RS, Suh PG, Manzoli L, Cocco L. Nuclear PLCbeta(1) acts as a negative regulator of p45/NF-E2 expression levels in Friend erythroleukemia cells. Biochim Biophys Acta. 2002;1589(3):305–310. doi: 10.1016/S0167-4889(02)00192-1. [DOI] [PubMed] [Google Scholar]
- 50.Lee WH, Chung MH, Tsai YH, Chang JL, Huang HM. Interferon-γ suppresses activin A/NF-E2 induction of erythroid gene expression through the NF-κB/c-Jun pathway. Am J Physiol Cell Physiol. 2014;306(4):C407–C414. doi: 10.1152/ajpcell.00312.2013. [DOI] [PubMed] [Google Scholar]
- 51.Catani L, Amabile M, Luatti S, Valdre L, Vianelli N, Martinelli G, Tura S. Interleukin-4 downregulates nuclear factor-erythroid 2 (NF-E2) expression in primary megakaryocytes and in megakaryoblastic cell lines. Stem Cells. 2001;19(4):339–347. doi: 10.1634/stemcells.19-4-339. [DOI] [PubMed] [Google Scholar]
- 52.Chuen CK, Li K, Yang M, Fok TF, Li CK, Chui CM, YP M. Interleukin-1beta up-regulates the expression of thrombopoietin and transcription factors c-Jun, c-Fos, GATA-1, and NF-E2 in megakaryocytic cells. J Lab Clin Med. 2004;143(2):75–88. doi: 10.1016/j.lab.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 53.Yang M, Li K, Chui CM, Yuen PM, Chan PK, Chuen CK, Li CK, Fok TF. Expression of interleukin (IL) 1 type I and type II receptors in megakaryocytic cells and enhancing effects of IL-1beta on megakaryocytopoiesis and NF-E2 expression. Br J Haematol. 2000;111(1):371–380. doi: 10.1046/j.1365-2141.2000.02340.x. [DOI] [PubMed] [Google Scholar]
- 54.Chui CM, Li K, Yang M, Chuen CK, Fok TF, Li CK, Yuen PM. Platelet-derived growth factor up-regulates the expression of transcription factors NF-E2, GATA-1 and c-Fos in megakaryocytic cell lines. Cytokine. 2003;21(2):51–64. doi: 10.1016/S1043-4666(02)00499-4. [DOI] [PubMed] [Google Scholar]
- 55.Takayama M, Fujita R, Suzuki M, Okuyama R, Aiba S, Motohashi H, Yamamoto M. Genetic analysis of hierarchical regulation for Gata1 and NF-E2 p45 gene expression in megakaryopoiesis. Mol Cell Biol. 2010;30(11):2668–2680. doi: 10.1128/MCB.01304-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu JJ, Hou SC, Shen CK. Erythroid gene suppression by NF-kappa B. J Biol Chem. 2003;278(21):19534–19540. doi: 10.1074/jbc.M212278200. [DOI] [PubMed] [Google Scholar]
- 57.Francastel C, Augery-Bourget Y, Prenant M, Walters M, Martin DI, Robert-Lezenes J. c-Jun inhibits NF-E2 transcriptional activity in association with p18/maf in Friend erythroleukemia cells. Oncogene. 1997;14(7):873–877. doi: 10.1038/sj.onc.1200902. [DOI] [PubMed] [Google Scholar]
- 58.Glembotsky A, Bluteau D, Espasandin Y, Goette N, Marta R, Marin Oyarzun C, Korin L, Lev P, Laguens R, Molinas F, Raslova H, Heller P. Mechanisms underlying platelet function defect in a pedigree with FPD/AML: potential role for candidate RUNX1-targets. J Thromb Haemost. 2014;12(5):761–772. doi: 10.1111/jth.12550. [DOI] [PubMed] [Google Scholar]
- 59.Shivdasani RA, Rosenblatt MF, Zucker-Franklin D, Jackson CW, Hunt P, Saris CJ, Orkin SH. Transcription factor NF-E2 is required for platelet formation independent of the actions of thrombopoietin/MGDF in megakaryocyte development. Cell. 1995;81(5):695–704. doi: 10.1016/0092-8674(95)90531-6. [DOI] [PubMed] [Google Scholar]
- 60.Lecine P, Villeval JL, Vyas P, Swencki B, Xu Y, Shivdasani RA. Mice lacking transcription factor NF-E2 provide in vivo validation of the proplatelet model of thrombocytopoiesis and show a platelet production defect that is intrinsic to megakaryocytes. Blood. 1998;92(5):1608–1616. [PubMed] [Google Scholar]
- 61.Levin J, Peng JP, Baker GR, Villeval JL, Lecine P, Burstein SA, Shivdasani RA. Pathophysiology of thrombocytopenia and anemia in mice lacking transcription factor NF-E2. Blood. 1999;94(9):3037–3047. [PubMed] [Google Scholar]
- 62.Shivdasani RA, Fielder P, Keller GA, Orkin SH, de Sauvage FJ. Regulation of the serum concentration of thrombopoietin in thrombocytopenic NF-E2 knockout mice. Blood. 1997;90(5):1821–1827. [PubMed] [Google Scholar]
- 63.Fock EL, Yan F, Pan S, Chong BH. NF-E2-mediated enhancement of megakaryocytic differentiation and platelet production in vitro and in vivo. Exp Hematol. 2008;36(1):78–92. doi: 10.1016/j.exphem.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 64.Motohashi H, Fujita R, Takayama M, Inoue A, Katsuoka F, Bresnick EH, Yamamoto M. Molecular determinants for small Maf protein control of platelet production. Mol Cell Biol. 2011;31(1):151–162. doi: 10.1128/MCB.00798-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ono Y, Wang Y, Suzuki H, Okamoto S, Ikeda Y, Murata M, Poncz M, Matsubara Y. Induction of functional platelets from mouse and human fibroblasts by p45NF-E2/Maf. Blood. 2012;120(18):3812–3821. doi: 10.1182/blood-2012-02-413617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Avanzi MP, Goldberg F, Davila J, Langhi D, Chiattone C, Mitchell WB. Rho kinase inhibition drives megakaryocyte polyploidization and proplatelet formation through MYC and NFE2 downregulation. Br J Haematol. 2014;164(6):867–876. doi: 10.1111/bjh.12709. [DOI] [PubMed] [Google Scholar]
- 67.Fujita R, Takayama-Tsujimoto M, Satoh H, Gutierrez L, Aburatani H, Fujii S, Sarai A, Bresnick EH, Yamamoto M, Motohashi H. NF-E2 p45 is important for establishing normal function of platelets. Mol Cell Biol. 2013;33(14):2659–2670. doi: 10.1128/MCB.01274-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deveaux S, Cohen-Kaminsky S, Shivdasani RA, Andrews NC, Filipe A, Kuzniak I, Orkin SH, Romeo PH, Mignotte V. p45 NF-E2 regulates expression of thromboxane synthase in megakaryocytes. EMBO J. 1997;16(18):5654–5661. doi: 10.1093/emboj/16.18.5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tiwari S, Italiano JEJ, Barral DC, Mules EH, Novak EK, Swank RT, Seabra MC, Shivdasani RA. A role for Rab27b in NF-E2-dependent pathways of platelet formation. Blood. 2003;102(12):3970–3979. doi: 10.1182/blood-2003-03-0977. [DOI] [PubMed] [Google Scholar]
- 70.Lecine P, Italiano JEJ, Kim SW, Villeval JL, Shivdasani RA. Hematopoietic-specific beta 1 tubulin participates in a pathway of platelet biogenesis dependent on the transcription factor NF-E2. Blood. 2000;96(4):1366–1373. [PubMed] [Google Scholar]
- 71.Schwer HD, Lecine P, Tiwari S, Italiano JE, Jr, Hartwig JH, Shivdasani RA. A lineage-restricted and divergent beta-tubulin isoform is essential for the biogenesis, structure and function of blood platelets. Curr Biol. 2001;11(8):579–586. doi: 10.1016/S0960-9822(01)00153-1. [DOI] [PubMed] [Google Scholar]
- 72.Shiraga M, Ritchie A, Aidoudi S, Baron V, Wilcox D, White G, Ybarrondo B, Murphy G, Leavitt A, Shattil S. Primary megakaryocytes reveal a role for transcription factor NF-E2 in integrin alpha IIb beta 3 signaling. J Cell Biol. 1999;147(7):1419–1430. doi: 10.1083/jcb.147.7.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kerrigan SW, Gaur M, Murphy RP, Shattil SJ, Leavitt AD. Caspase-12: a developmental link between G-protein-coupled receptors and integrin alphaIIbbeta3 activation. Blood. 2004;104(5):1327–1334. doi: 10.1182/blood-2003-10-3633. [DOI] [PubMed] [Google Scholar]
- 74.Nagata Y, Yoshikawa J, Hashimoto A, Yamamoto M, Payne AH, Todokoro K. Proplatelet formation of megakaryocytes is triggered by autocrine-synthesized estradiol. Genes Dev. 2003;17(23):2864–2869. doi: 10.1101/gad.1128003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen Z, Hu M, Shivdasani RA. Expression analysis of primary mouse megakaryocyte differentiation and its application in identifying stage-specific molecular markers and a novel transcriptional target of NF-E2. Blood. 2007;109(4):1451–1459. doi: 10.1182/blood-2006-08-038901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Motohashi H, Kimura M, Fujita R, Inoue A, Pan X, Takayama M, Katsuoka F, Aburatani H, Bresnick EH, Yamamoto M. NF-E2 domination over Nrf2 promotes ROS accumulation and megakaryocytic maturation. Blood. 2010;115(3):677–686. doi: 10.1182/blood-2009-05-223107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tijssen MR, Ghevaert C. Transcription factors in late megakaryopoiesis and related platelet disorders. J Thromb Haemost. 2013;11(4):593–604. doi: 10.1111/jth.12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sayer MS, Tilbrook PA, Spadaccini A, Ingley E, Sarna MK, Williams JH, Andrews NC, Klinken SP. Ectopic expression of transcription factor NF-E2 alters the phenotype of erythroid and monoblastoid cells. J Biol Chem. 2000;275(33):25292–25298. doi: 10.1074/jbc.M908695199. [DOI] [PubMed] [Google Scholar]
- 79.Mignotte V, Eleouet JF, Raich N, Romeo PH. Cis- and trans-acting elements involved in the regulation of the erythroid promoter of the human porphobilinogen deaminase gene. Proc Natl Acad Sci USA. 1989;86(17):6548–6552. doi: 10.1073/pnas.86.17.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Taketani S, Inazawa J, Nakahashi Y, Abe T, Tokunaga R. Structure of the human ferrochelatase gene. Exon/intron gene organization and location of the gene to chromosome 18. Eur J Biochem. 1992;205(1):217–222. doi: 10.1111/j.1432-1033.1992.tb16771.x. [DOI] [PubMed] [Google Scholar]
- 81.Tugores A, Magness ST, Brenner DA. A single promoter directs both housekeeping and erythroid preferential expression of the human ferrochelatase gene. J Biol Chem. 1994;269(49):30789–30797. [PubMed] [Google Scholar]
- 82.Cox TC, Bawden MJ, Martin A, May BK. Human erythroid 5-aminolevulinate synthase: promoter analysis and identification of an iron-responsive element in the mRNA. EMBO J. 1991;10(7):1891–1902. doi: 10.1002/j.1460-2075.1991.tb07715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Surinya KH, Cox TC, May BK. Transcriptional regulation of the human erythroid 5-aminolevulinate synthase gene. Identification of promoter elements and role of regulatory proteins. J Biol Chem. 1997;272(42):26585–26594. doi: 10.1074/jbc.272.42.26585. [DOI] [PubMed] [Google Scholar]
- 84.Rheinemann L, Seeger TS, Wehrle J, Pahl HL. NFE2 regulates transcription of multiple enzymes in the heme biosynthesis pathway. Haematologica. 2014;99(10):e208–e210. doi: 10.3324/haematol.2014.106393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Inamdar NM, Ahn YI, Alam J. The heme-responsive element of the mouse heme oxygenase-1 gene is an extended AP-1 binding site that resembles the recognition sequences for MAF and NF-E2 transcription factors. Biochem Biophys Res Commun. 1996;221(3):570–576. doi: 10.1006/bbrc.1996.0637. [DOI] [PubMed] [Google Scholar]
- 86.Solomon WB, Lin CH, Palma J, Gao XY, Wu S. Suppression of a cellular differentiation program by phorbol esters coincides with inhibition of binding of a cell-specific transcription factor (NF-E2) to an enhancer element required for expression of an erythroid-specific gene. J Biol Chem. 1993;268(7):5089–5096. [PubMed] [Google Scholar]
- 87.Meguro K, Igarashi K, Yamamoto M, Fujita H, Sassa S. The role of the erythroid-specific delta-aminolevulinate synthase gene expression in erythroid heme synthesis. Blood. 1995;86(3):940–948. [PubMed] [Google Scholar]
- 88.Lu SJ, Rowan S, Bani MR, Ben-David Y. Retroviral integration within the Fli-2 locus results in inactivation of the erythroid transcription factor NF-E2 in Friend erythroleukemias: evidence that NF-E2 is essential for globin expression. Proc Natl Acad Sci USA. 1994;91(18):8398–8402. doi: 10.1073/pnas.91.18.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Woon Kim Y, Kim S, Geun Kim C, Kim A. The distinctive roles of erythroid specific activator GATA-1 and NF-E2 in transcription of the human fetal γ-globin genes. Nucleic Acids Res. 2011;39(16):6944–6955. doi: 10.1093/nar/gkr253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Palma JF, Gao X, Lin CH, Wu S, Solomon WB. Iron protoporphyrin IX (hemin) but not tin or zinc protoporphyrin IX can stimulate gene expression in K562 cells from enhancer elements containing binding sites for NF-E2. Blood. 1994;84(4):1288–1297. [PubMed] [Google Scholar]
- 91.Bean TL, Ney PA. Multiple regions of p45 NF-E2 are required for beta-globin gene expression in erythroid cells. Nucleic Acids Res. 1997;25(12):2509–2515. doi: 10.1093/nar/25.12.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stamatoyannopoulos JA, Goodwin A, Joyce T, Lowrey CH. NF-E2 and GATA binding motifs are required for the formation of DNase I hypersensitive site 4 of the human beta-globin locus control region. EMBO J. 1995;14(1):106–116. doi: 10.1002/j.1460-2075.1995.tb06980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pomerantz O, Goodwin AJ, Joyce T, Lowrey CH. Conserved elements containing NF-E2 and tandem GATA binding sites are required for erythroid-specific chromatin structure reorganization within the human beta-globin locus control region. Nucleic Acids Res. 1998;26(24):5684–5691. doi: 10.1093/nar/26.24.5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Armstrong JA, Emerson BM. NF-E2 disrupts chromatin structure at human beta-globin locus control region hypersensitive site 2 in vitro. Mol Cell Biol. 1996;16(10):5634–5644. doi: 10.1128/mcb.16.10.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Amrolia PJ, Ramamurthy L, Saluja D, Tanese N, Jane SM, Cunningham JM. The activation domain of the enhancer binding protein p45NF-E2 interacts with TAFII130 and mediates long-range activation of the alpha- and beta-globin gene loci in an erythroid cell line. Proc Natl Acad Sci USA. 1997;94(19):10051–10056. doi: 10.1073/pnas.94.19.10051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Forsberg EC, Downs KM, Bresnick EH. Direct interaction of NF-E2 with hypersensitive site 2 of the beta-globin locus control region in living cells. Blood. 2000;96(1):334–339. [PubMed] [Google Scholar]
- 97.Onishi Y, Kiyama R. Interaction of NF-E2 in the human beta-globin locus control region before chromatin remodeling. J Biol Chem. 2003;278(10):8163–8171. doi: 10.1074/jbc.M209612200. [DOI] [PubMed] [Google Scholar]
- 98.Sawado T, Igarashi K, Groudine M. Activation of beta-major globin gene transcription is associated with recruitment of NF-E2 to the beta-globin LCR and gene promoter. Proc Natl Acad Sci USA. 2001;98(18):10226–10231. doi: 10.1073/pnas.181344198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Daftari P, Gawa NR, Shen CK. Distinction between AP1 and NF-E2 factor-binding at specific chromatin regions in mammalian cells. Oncogene. 1999;18(39):5482–5486. doi: 10.1038/sj.onc.1202916. [DOI] [PubMed] [Google Scholar]
- 100.Bottardi S, Ross J, Pierre-Charles N, Blank V, Milot E. Lineage-specific activators affect beta-globin locus chromatin in multipotent hematopoietic progenitors. EMBO J. 2006;25(15):3586–3595. doi: 10.1038/sj.emboj.7601232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kooren J, Palstra RJ, Klous P, Splinter E, von Lindern M, Grosveld F, de Laat W. Beta-globin active chromatin Hub formation in differentiating erythroid cells and in p45 NF-E2 knock-out mice. J Biol Chem. 2007;282(22):16544–16552. doi: 10.1074/jbc.M701159200. [DOI] [PubMed] [Google Scholar]
- 102.Chaturvedi CP, Hosey AM, Palii C, Perez-Iratxeta C, Nakatani Y, Ranish JA, Dilworth FJ, Brand M. Dual role for the methyltransferase G9a in the maintenance of beta-globin gene transcription in adult erythroid cells. Proc Natl Acad Sci USA. 2009;106(43):18303–18308. doi: 10.1073/pnas.0906769106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhou Z, Li X, Deng C, Ney PA, Huang S, Bungert J. USF and NF-E2 cooperate to regulate the recruitment and activity of RNA polymerase II in the beta-globin gene locus. J Biol Chem. 2010;285(21):15894–15905. doi: 10.1074/jbc.M109.098376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Boulanger L, Sabatino DE, Wong EY, Cline AP, Garrett LJ, Garbarz M, Dhermy D, Bodine DM, Gallagher PG. Erythroid expression of the human alpha-spectrin gene promoter is mediated by GATA-1- and NF-E2-binding proteins. J Biol Chem. 2002;277(44):41563–41570. doi: 10.1074/jbc.M208184200. [DOI] [PubMed] [Google Scholar]
- 105.Guo-wei Z, Rui-feng Y, Xiang L, Mitchell WJ, De-pei L, Chih-chuan L. NF-E2: a novel regulator of alpha-hemoglobin stabilizing protein gene expression. Chin Med Sci J. 2010;25(4):193–198. doi: 10.1016/S1001-9294(11)60001-1. [DOI] [PubMed] [Google Scholar]
- 106.Campbell MR, Karaca M, Adamski KN, Chorley BN, Wang X, Bell DA. Novel hematopoietic target genes in the NRF2-mediated transcriptional pathway. Oxid Med Cell Longev. 2013;2013:120305. doi: 10.1155/2013/120305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chan JY, Kwong M, Lo M, Emerson R, Kuypers FA. Reduced oxidative-stress response in red blood cells from p45NFE2-deficient mice. Blood. 2001;97(7):2151–2158. doi: 10.1182/blood.V97.7.2151. [DOI] [PubMed] [Google Scholar]
- 108.Li Y, Bai H, Zhang Z, Li W, Dong L, Wei X, Ma Y, Zhang J, Yu J, Sun G, Wang F. The up-regulation of miR-199b-5p in erythroid differentiation is associated with GATA-1 and NF-E2. Mol Cells. 2014;37(3):213–219. doi: 10.14348/molcells.2014.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shivdasani RA, Orkin SH. Erythropoiesis and globin gene expression in mice lacking the transcription factor NF-E2. Proc Natl Acad Sci USA. 1995;92(19):8690–8694. doi: 10.1073/pnas.92.19.8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gasiorek JJ, Nouhi Z, Blank V. Abnormal differentiation of erythroid precursors in p45 NF-E2(−/−) mice. Exp Hematol. 2012;40(5):393–400. doi: 10.1016/j.exphem.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 111.Martin F, van Deursen JM, Shivdasani RA, Jackson CW, Troutman AG, Ney PA. Erythroid maturation and globin gene expression in mice with combined deficiency of NF-E2 and nrf-2. Blood. 1998;91(9):3459–3466. [PubMed] [Google Scholar]
- 112.Kuroha T, Takahashi S, Komeno T, Itoh K, Nagasawa T, Yamamoto M. Ablation of Nrf2 function does not increase the erythroid or megakaryocytic cell lineage dysfunction caused by p45 NF-E2 gene disruption. J Biochem (Tokyo) 1998;123(3):376–379. doi: 10.1093/oxfordjournals.jbchem.a021947. [DOI] [PubMed] [Google Scholar]
- 113.Li YJ, Higgins RR, Pak BJ, Shivdasani RA, Ney PA, Archer M, Ben-David Y. p45(NFE2) is a negative regulator of erythroid proliferation which contributes to the progression of Friend virus-induced erythroleukemias. Mol Cell Biol. 2001;21(1):73–80. doi: 10.1128/MCB.21.1.73-80.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Goerttler PS, Kreutz C, Donauer J, Faller D, Maiwald T, Marz E, Rumberger B, Sparna T, Schmitt-Graff A, Wilpert J, Timmer J, Walz G, Pahl HL. Gene expression profiling in polycythaemia vera: overexpression of transcription factor NF-E2. Br J Haematol. 2005;129(1):138–150. doi: 10.1111/j.1365-2141.2005.05416.x. [DOI] [PubMed] [Google Scholar]
- 115.Mutschler M, Magin AS, Buerge M, Roelz R, Schanne DH, Will B, Pilz IH, Migliaccio AR, Pahl HL. NF-E2 overexpression delays erythroid maturation and increases erythrocyte production. Br J Haematol. 2009;146(2):203–217. doi: 10.1111/j.1365-2141.2009.07742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bogeska R, Pahl HL. Elevated nuclear factor erythroid-2 levels promote epo-independent erythroid maturation and recapitulate the hematopoietic stem cell and common myeloid progenitor expansion observed in polycythemia vera patients. Stem Cells Transl Med. 2013;2(2):112–117. doi: 10.5966/sctm.2012-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang W, Schwemmers S, Hexner EO, Pahl HL. AML1 is overexpressed in patients with myeloproliferative neoplasms and mediates JAK2V617F-independent overexpression of NF-E2. Blood. 2010;116(2):254–266. doi: 10.1182/blood-2009-11-254664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jutzi JS, Bogeska R, Nikoloski G, Schmid CA, Seeger TS, Stegelmann F, Schwemmers S, Grunder A, Peeken JC, Gothwal M, Wehrle J, Aumann K, Hamdi K, Dierks C, Kamar Wang W, Dohner K, Jansen JH, Pahl HL. MPN patients harbor recurrent truncating mutations in transcription factor NF-E2. J Exp Med. 2013;210(5):1003–1019. doi: 10.1084/jem.20120521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Aumann K, Frey AV, May AM, Hauschke D, Kreutz C, Marx JP, Timmer J, Werner M, Pahl HL. Subcellular mislocalization of the transcription factor NF-E2 in erythroid cells discriminates prefibrotic primary myelofibrosis from essential thrombocythemia. Blood. 2013;122(1):93–99. doi: 10.1182/blood-2012-11-463257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kaufmann KB, Grunder A, Hadlich T, Wehrle J, Gothwal M, Bogeska R, Seeger TS, Kayser S, Pham KB, Jutzi JS, Ganzenmuller L, Steinemann D, Schlegelberger B, Wagner JM, Jung M, Will B, Steidl U, Aumann K, Werner M, Gunther T, Schule R, Rambaldi A, Pahl HL. A novel murine model of myeloproliferative disorders generated by overexpression of the transcription factor NF-E2. J Exp Med. 2012;209(1):35–50. doi: 10.1084/jem.20110540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hasselbalch HC. A role of NF-E2 in chronic inflammation and clonal evolution in essential thrombocythemia, polycythemia vera and myelofibrosis? Leuk Res. 2014;38(2):263–266. doi: 10.1016/j.leukres.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 122.Amaru Calzada A, Todoerti K, Donadoni L, Pellicioli A, Tuana G, Gatta R, Neri A, Finazzi G, Mantovani R, Rambaldi A, Introna M, Lombardi L, Golay J, Investigators A. The HDAC inhibitor Givinostat modulates the hematopoietic transcription factors NFE2 and C-MYB in JAK2(V617F) myeloproliferative neoplasm cells. Exp Hematol. 2012;40(8):634–645. doi: 10.1016/j.exphem.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 123.Akada H, Akada S, Gajra A, Bair A, Graziano S, Hutchison RE, Mohi G. Efficacy of vorinostat in a murine model of polycythemia vera. Blood. 2012;119(16):3779–3789. doi: 10.1182/blood-2011-02-336743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kapralova K, Lanikova L, Lorenzo F, Song J, Horvathova M, Divoky V, Prchal JT. RUNX1 and NF-E2 upregulation is not specific for MPNs, but is seen in polycythemic disorders with augmented HIF signaling. Blood. 2014;123(3):391–394. doi: 10.1182/blood-2013-10-534222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kacena MA, Gundberg CM, Nelson T, Horowitz MC. Loss of the transcription factor p45 NF-E2 results in a developmental arrest of megakaryocyte differentiation and the onset of a high bone mass phenotype. Bone. 2005;36(2):215–223. doi: 10.1016/j.bone.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 126.Kacena MA, Shivdasani RA, Wilson K, Xi Y, Troiano N, Nazarian A, Gundberg CM, Bouxsein ML, Lorenzo JA, Horowitz MC. Megakaryocyte-osteoblast interaction revealed in mice deficient in transcription factors GATA-1 and NF-E2. J Bone Miner Res. 2004;19(4):652–660. doi: 10.1359/JBMR.0301254. [DOI] [PubMed] [Google Scholar]
- 127.Kacena MA, Gundberg CM, Kacena WJ, 3rd, Landis WJ, Boskey AL, Bouxsein ML, Horowitz MC. The effects of GATA-1 and NF-E2 deficiency on bone biomechanical, biochemical, and mineral properties. J Cell Physiol. 2013;228(7):1594–1600. doi: 10.1002/jcp.24322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kashif M, Hellwig A, Hashemolhosseini S, Kumar V, Bock F, Wang H, Shahzad K, Ranjan S, Wolter J, Madhusudhan T, Bierhaus A, Nawroth P, Isermann B. Nuclear factor erythroid-derived 2 (Nfe2) regulates JunD DNA-binding activity via acetylation: a novel mechanism regulating trophoblast differentiation. J Biol Chem. 2012;287(8):5400–5411. doi: 10.1074/jbc.M111.289801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chan JY, Kwong M, Lu R, Chang J, Wang B, Yen TS, Kan YW. Targeted disruption of the ubiquitous CNC-bZIP transcription factor, Nrf-1, results in anemia and embryonic lethality in mice. EMBO J. 1998;17(6):1779–1787. doi: 10.1093/emboj/17.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ohtsu H, Kuramasu A, Suzuki S, Igarashi K, Ohuchi Y, Sato M, Tanaka S, Nakagawa S, Shirato K, Yamamoto M, Ichikawa A, Watanabe T. Histidine decarboxylase expression in mouse mast cell line P815 is induced by mouse peritoneal cavity incubation. J Biol Chem. 1996;271(45):28439–28444. doi: 10.1074/jbc.271.45.28439. [DOI] [PubMed] [Google Scholar]