Abstract
Blastomere biopsy is used in preimplantation genetic diagnosis; however, the long-term implications on the offspring are poorly characterized. We previously reported a high risk of memory defects in adult biopsied mice. Here, we assessed nervous function of aged biopsied mice and further investigated the mechanism of neural impairment after biopsy. We found that aged biopsied mice had poorer spatial learning ability, increased neuron degeneration, and altered expression of proteins involved in neural degeneration or dysfunction in the brain compared to aged control mice. Furthermore, the MeDIP assay indicated a genome-wide low methylation in the brains of adult biopsied mice when compared to the controls, and most of the genes containing differentially methylated loci in promoter regions were associated with neural disorders. When we further compared the genomic DNA methylation profiles of 7.5-days postconception (dpc) embryos between the biopsy and control group, we found the whole genome low methylation in the biopsied group, suggesting that blastomere biopsy was an obstacle to de novo methylation during early embryo development. Further analysis on mRNA profiles of 4.5-dpc embryos indicated that reduced expression of de novo methylation genes in biopsied embryos may impact de novo methylation. In conclusion, we demonstrate an abnormal neural development and function in mice generated after blastomere biopsy. The impaired epigenetic reprogramming during early embryo development may be the latent mechanism contributing to the impairment of the nervous system in the biopsied mice, which results in a hypomethylation status in their brains.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-013-1466-2) contains supplementary material, which is available to authorized users.
Keywords: Blastomere biopsy, Neurodegenerative disorders, Epigenetic reprogramming, DNA methylation, Neuron degeneration, Assisted reproductive technology (ART)
Introduction
Assisted reproductive technologies (ARTs) have been widely used in the treatment of human infertility. However, some processes involved in ART-mediated conception are very different from natural conception, such as ovarian hyperstimulation and exposure of the embryo to tissue culture medium. Both clinical investigations and animal studies have indicated an increased incidence of developmental abnormalities in ART offspring [1–4]. Moreover, the non-physiological manipulations used during ART are thought to contribute to the risk of developmental abnormalities [3, 4].
Preimplantation genetic diagnosis (PGD) has been one of the major clinical components of assisted reproductive technologies (ARTs) since 1990 [5]. PGD is used for two distinct purposes: for aneuploidy screening to enhance the success of in vitro fertilization, and to detect genetic disorders if family members are known to have a disease or are carriers [6, 7]. Compared to other ARTs, the protocol required for PGD includes not only superovulation and in vitro embryo culture but also the more invasive biopsy procedure of removing one or two blastomeres from the embryo. At present, evaluation of the influence of blastomere biopsy on offspring development is less. Some clinical investigations have assessed the biopsy risk by comparing the health outcomes of PGD offspring and ICSI/IVF offspring, which obtained different conclusions [5, 8–13]. Some researchers reported that blastomere biopsy does not have a negative effect on embryo viability or neonatal health [5, 8, 9]. Others think it influences early embryonic development, lowers the live birth rate, and results in lower neurologic scores of 2-year-old children [10–13]. These clinical investigations only assessed embryonic development and young children’s health due to the age restriction of PGD offspring, and could not perform mechanism research. In recent years, animal models were gradually used for studying potential effects of ART technology on its offspring, which can perform mechanism research and enable long-term safety assessments of ART because of the short life span of the animals. Studies on mouse models have revealed that embryo culture and transplant operation cause an abnormal development in their offspring [1, 4], but it has not been used for the assessment of biopsy risk.
In our previous study, we revealed that blastomere biopsy led to a high risk of defective memory function in the resulting adult mice [14], which is consistent with a recent result of population-based study that 2-year-old PGS children had lower neurologic optimality scores than control IVF children [13]. These results imply that development of the nervous system is sensitive to the embryo biopsy procedure. To ascertain the harmful effects of blastomere biopsy on the nervous system of the offspring, in this study we further evaluated the nervous system function of aged control and biopsied mice to determine whether degenerative neural disorders were more frequent in the biopsied group. Moreover, the potential mechanism that contributes to neural impairment in biopsied mice was investigated. From this study, we hope to provide valuable information on the safety and possible procedure improvements of PGD.
Materials and methods
Mouse model
ICR mice in the biopsied or control groups were derived from biopsied embryos or in vitro cultured embryos without biopsy, respectively. Cleavage-stage biopsy and embryo transfer were performed according to previously published procedures [14]. All animal experiments were approved by the ethical board of Nanjing Medical University.
Spatial learning assay and pole climbing test
Spatial learning ability was assayed using the acquisition trials of the Morris water maze test, as previously described [1, 15]. Thirty-seven 84-week-old mice in the biopsied group and fifteen 84-week-old mice in the control group were tested. A total of 18 trials were performed over three consecutive days with six trials per day. Data was analyzed using a tracking system (PolyTrak, San Diego Instruments, San Diego, CA, USA).
The pole climbing test was conducted as previously described [16]. Two parameters were recorded: the time until the mice turned their heads downwards, and the time until the mice touched the floor with their front claws. Each mouse (83 weeks old, biopsy n = 7, control n = 10) was tested three more times individually, with 1-min intervals between tests.
Tissue collection and morphological analysis
The brain tissues from 86-week-old mice were divided into three and fixed in formaldehyde solution for immunohistochemical examination, glutaraldehyde solution for ultrastructural examination, or liquid nitrogen for Western-blot analysis. For ultrastructural examination, the tissue blocks were postfixed with 2 % OsO4 and embedded in Araldite (Zhongxing Bary Technology Co., Beijing, China). Ultrathin sections were stained with uranyl acetate and lead citrate and examined by electron microscopy (JEM.1010, JEOL, Japan). The brain tissues of 10-week-old mice were frozen in liquid nitrogen and used for DNA and protein extraction.
Immunohistochemistry
Paraffin-embedded sections were immunostained as previously described [17] by incubation overnight at 4 °C with primary antibody against MBP (1:200, Santa Cruz Biotechnology, sc-13914) followed by HRP-conjugated secondary antibody (ZSGB-BIO, ZB-2306). Immunoreactivity was visualized using diaminobenzidine, viewed by bright field microscopy (Axioskop 2 plus; Zeiss, Germany), and quantified using ImageJ software (a free Java image processing program, http://imagej.software.informer.com). MBP expression was calculated as the ratio of the intensity of the positive signal/area of striatum.
2-DE, gel image analysis and protein identification
Proteins were extracted from the brains of three 86-week-old mice from each group and separated by two-dimensional gel electrophoresis (2-DE), as previously described [14]. Gels were silver stained, scanned, and analyzed using ImageMaster 2D Platinum software (Version 5.0, GE Healthcare, San Francisco, CA, USA). The relative volume of each spot (% vol) was calculated; mean ± standard deviation values were compared using ImageMaster 2D Platinum software.
Differently expressed spots (≥1.5-fold and p < 0.05, Student’s t test) were excised, denatured, alkylated, trypsin-digested, and analyzed using an Ultraflex II MALDI-TOF-TOF mass spectrometer and FlexControl 2.4 software (Bruker Daltonics GmbH, Bremen, Germany) using previously described analysis and search conditions [14].
Methylated DNA immunoprecipitation (MeDIP) assay
The MeDIP assay and analysis was performed according to the NimbleGen’s DNA Methylation user’s guide and performed at CapitalBio Corporation (Beijing, China). Briefly, genomic DNA was extracted from the brain tissues of 10-week-old adult mice or from 7.5-days postconception (dpc) embryos (three samples in the biopsied group and three samples in the control group) using the DNeasy Blood and Tissue Kit (QIAGEN Inc., Valencia, CA, USA). For MeDIP assay, one sample required more than 5 μg of purified DNA, which was extracted from one 10-week-old mouse or from eighty 7.5-dpc embryos. Then, DNA was sonicated to create random 500-bp fragments, and the methylated DNA was immunoprecipitated using mouse monoclonal antibodies against 5-methylcytidine and anti-mouse-coupled IgG Biomag magnetic beads, eluted, and purified by phenol chloroform extraction and ethanol precipitation. The input and immunoprecipitated DNA were labeled with Cy3- and Cy5-labeled random 9-mers, respectively, hybridized to NimbleGen MM8 CpG Promoter arrays (single arrays containing all known CpG Islands annotated by UCSC and all well-characterized RefSeq promoter regions, from approximately −1,300 to +500 bp of the TSSs, covered by ~385,000 probes) and scanned using the Axon GenePix 4000B microarray scanner.
For each probe, the IP/Input log2 ratio is computed and scaled to center the ratio data around zero. Regions of significant positive enrichment in ChIP-based methylation microarray data were identified using a modified ACME algorithm for peak identification [18]. Briefly, from the scaled log2-ratio data, a fixed-length window (750 bp) was placed around each consecutive probe and the one-sided Kolmogorov–Smirnov (KS) test was applied to determine whether the probes were drawn from a significantly more positive distribution of intensity log-ratios than those in the rest of the array. The resulting score for each probe was the –log 10 p value from the windowed KS test around that probe. Regions with more than two probes scoring above 2.0 were defined as MeDIP peaks, and peaks were mapped to genes and CpG islands.
To identify the differentially methylated loci between the biopsied and control groups, the log2-ratio values of probes locating in the same region of the chip were compared between the two groups. If the values of at least two adjacent probes had significant differences between the biopsied and control groups (p < 0.05, Student’s t test), the corresponding MeDIP peaks were defined as differentially methylated loci.
The methylation patterns of genomic DNA were visualized by hierarchical cluster (HC) analysis, using Cluster analysis 3.0 software (an open source software, http://cluster.updatestar.com). HC analysis was used for all samples, using the log2-ratio values of probes. The differentially methylated loci were then grouped by a hierarchical clustering algorithm in terms of the Pearson correlation coefficient of the sample and the average linkage method.
Bisulfite sequencing
Genomic DNA extracted from 10-week-old brain tissues or 7.5-dpc embryos was subjected to bisulfite treatment (Epitect Bisulfite Kit, QIAGEN Inc.), and differentially methylated regions were amplified by PCR using HotStarTaq Plus Master Mix Kit (QIAGEN Inc.). The PCR conditions were 95 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 52–58 °C for 30 s, and 72 °C for 30 s, with a final extension at 72 °C for 7 min. Primer sequences and annealing temperatures are presented in Supplementary Table S4. The products were electrophoresed, purified using the QIAprep Spin Miniprep Kit, (QIAGEN Inc.), subjected to TA cloning, and sequenced to determine their methylation status.
Quantitative real-time PCR analysis
Equal amounts of RNA (200 ng) were reverse-transcribed into cDNA. Each sample was assayed in duplicate (1 μl cDNA, 8 pmol primers, 10 μl SYBR Premix EX Taq II, 0.4 μl Rox Reference Dye in 20-μl volumes) using the ABI Prism 7300 Sequence Detection System (Perkin-Elmer Applied Biosystems, MA, USA) at 94 °C for 30 s, followed by 40 cycles of 94 °C for 5 s, 53–60 °C for 30 s, and 72 °C for 30 s. Primer sequences and annealing temperatures are presented in Supplementary Table S4. Melting curve analysis was performed to confirm the real-time quantitative PCR products.
Western blotting
Western blotting of brain tissue lysates was performed as previously described [17] using anti-MBP (1:200; sc-13914, Santa Cruz Biotech, CA, USA), anti-SOD1 (1:1,000; ab16831, Abcam, Cambridge, UK), anti-NEFL (1:500; ap7332c, Abgent Inc, CA, USA), anti-ALDH2 (1:500; ap1432d, Abgent Inc.), anti-DPYSL2 (1:500; ap13939c, Abgent Inc.), anti-Myelin PLP (1:500; ab83032, Abcam), and anti-GAPDH (1:1,000; KC-5G5, Kangchen Biotech, Shanghai, China).
Microarray analysis
RNA was isolated from 4.5-dpc embryos (three samples in biopsied group and three samples in control group; each sample was extracted from 70 embryos) and microarray analysis was performed by the Bioassay Laboratory of CapitalBio Corporation (Beijing, China) using GeneChip Mouse 430 2.0 Arrays (Affymetrix, Santa Clara, CA, USA), according to the manufacturer’s protocol [19]. The chips were scanned with a GeneChip 3000 laser confocal slide scanner (Affymetrix) and the data was analyzed using the Microarray Suite 5.0 Analysis software (Affymetrix). Differentially expressed transcripts were selected using a q value <0.05 (false discovery rate when the gene was evaluated for genetic variations) and fold change ≥2 or ≤ −2.
Results
Neural impairments in aged biopsied mice
Behavioral test
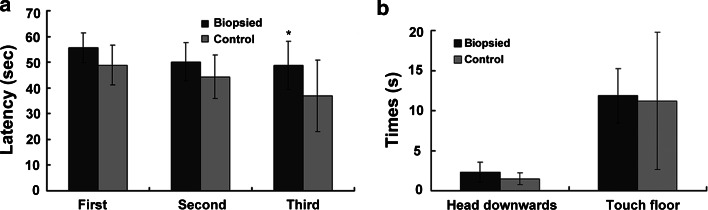
Spatial learning was assayed using the hidden platform version of the Morris water maze test. In the acquisition trials, the mice were expected to find the hidden platform located in the fourth quadrant as quickly as possible within 60 s. The aged control group displayed evident improvements over the 3 days of testing (comparing with the first or second acquisition time, the third time decreased significantly, p < 0.05, Student’s t test), whereas the aged biopsied mice made no progress (the third acquisition time had no obvious difference with the first or second time, p > 0.05, Student’s t test). By comparing the first, second, or third acquisition time, respectively, between the two groups, we found that the third acquisition time differed significantly in the aged control and biopsied mice (p < 0.01, Student’s t test; Fig. 1a). These results preliminarily proved the impaired spatial learning ability in biopsied mice.
Fig. 1.
Nervous system function behavioral test. a Morris water maze acquisition trials on 84-week-old mice. Average time to find a hidden platform was measured, indicating the third acquisition time differed significantly in the control and biopsied mice (biopsy n = 37, control n = 15, *p < 0.01). b Pole climbing test on 83-week-old mice. Times until the mouse turned its head downwards and touched the floor with its front claw were measured; no significant differences were observed between the biopsied and control groups (biopsy n = 7, control n = 10, p > 0.05)
The pole climbing test was conducted to evaluate the muscle endurance and locomotor coordination of mice. In the pole climbing test, the length of time until the mouse turned its head downwards and the length of time until the mouse touched the floor with its front claw were compared. No significant differences were observed between the aged biopsied and control groups (p > 0.05, Student’s t test; Fig. 1b), confirming that the biopsied mice had similar muscle coordination as control mice. This result excluded the locomotor influence on the acquisition time. Combined the results of Morris water maze test and pole climbing test, we demonstrated that blastomere biopsy significantly altered the spatial learning ability of the resulting aged animals.
In our previous study of adult biopsied and control mice, no differences between groups were observed in the acquisition trials, although the poorer memory function of the biopsied mice was revealed by the probe trials [14]. Unfortunately, the aged biopsied and control mice in this study failed to complete the probe trials and reversal trials of the Morris water maze test due to their poor health. Thus, the memory function of the aged offspring could not be evaluated.
Brain morphological analysis
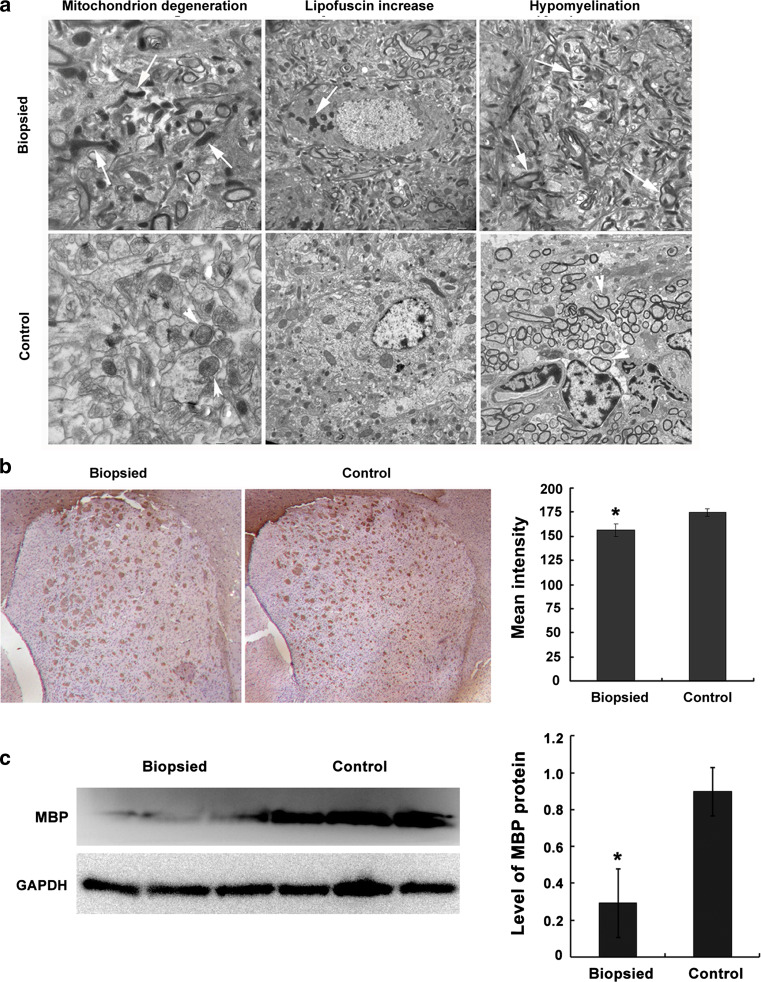
The ultrastructure of the hippocampus from three aged biopsy mice and three aged control mice was examined. A series of neuronal regression phenotypes, including mitochondrial degeneration, increased lipofuscin, synaptic loss, and hypomyelination were observed more frequently in the aged biopsy group compared to control mice (Fig. 2a). Immunostaining verified the lower expression of myelin basic protein (MBP) in the striatum of aged biopsy mice, compared to the control group (p < 0.05; Fig. 2b), in accordance with Western-blot analysis of MBP expression in brain tissues from aged mice (p < 0.05; Fig. 2c).
Fig. 2.
Morphological analysis of brain tissues in aged biopsied and control mice (86 weeks old; n = 3 each). a Ultrastructural analysis of the hippocampus. Increased neuronal regression, mitochondrial degeneration, increased lipofuscin, and hypomyelination (arrows) were observed in the biopsied group; arrowheads indicate normal mitochondria and myelin sheath in control group. b MBP immunostaining in the striatum (*p < 0.05). c Western blotting of MBP in brain tissues (*p < 0.05)
Analysis of differential protein expression in the brain
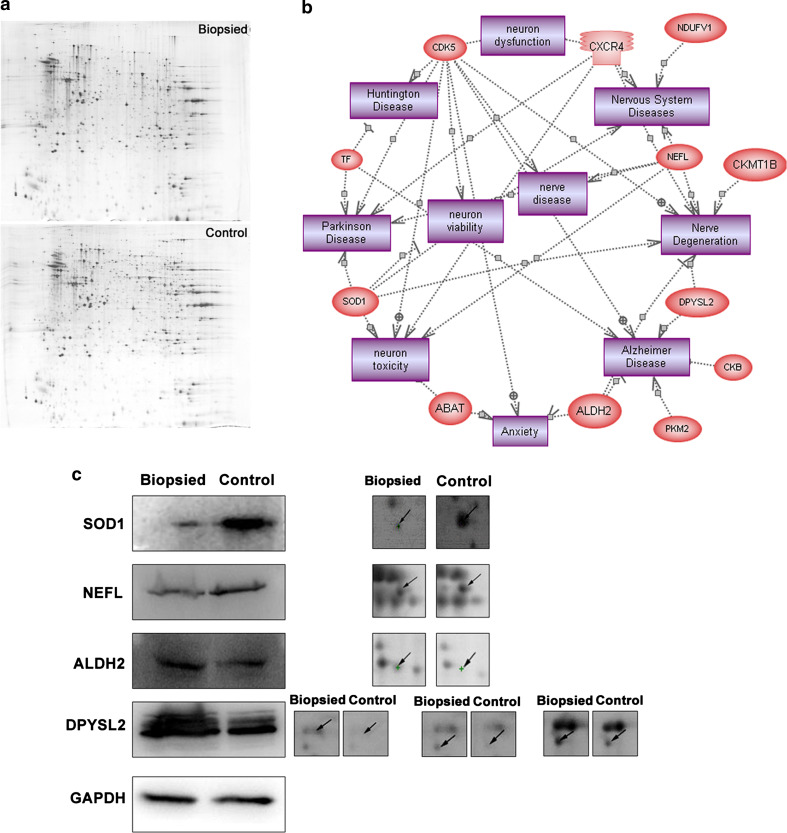
The protein expression changes in the brain of biopsied aged mice were analyzed by two-dimensional gel electrophoresis (2-DE) and matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF-TOF). Quadruple 2-DE maps of the brain proteins from the aged control and biopsied groups are presented in Fig. 3a. By analyzing the 2-DE gels using ImageMaster 2D Platinum software, we identified 58 significantly differentially expressed protein spots in the biopsied group compared to the control group; 44 of these spots were successfully identified, corresponding to 38 known proteins (further detailed information is listed in Table S1). PathwayStudio analysis indicated that 12/38 proteins were involved in cellular processes linked to neural degeneration or dysfunction (Fig. 3b). The expression of four proteins involved in neural degeneration: superoxide dismutase 1 (SOD1), neurofilament protein (NEFL), aldehyde dehydrogenase 2 (ALDH2), and dihydropyrimidinase-like 2 (DPYSL2), was further examined in the brain tissues of aged biopsied and control mice by Western blotting. Consistent with the ImageMaster 2D Platinum software analysis, the levels of SOD1 and NEFL were decreased, and the levels of ALDH2 and DPYSL2 were increased in the aged brains of biopsied mice, compared to the control group (Fig. 3c).
Fig. 3.
Protein alterations in the brain of aged biopsied mice (86 weeks old). a 2-DE maps of brain proteins. b PathwayStudio analysis; 12/38 differentially expressed proteins (ovals) were involved in cellular processes (cubes) related to neural degeneration or dysfunction. c Western blotting confirmation of 2-DE ImageMaster 2D Platinum software analysis
Epigenetic alterations are observed in the brains of biopsied mice
MeDIP assay analysis of genomic methylation
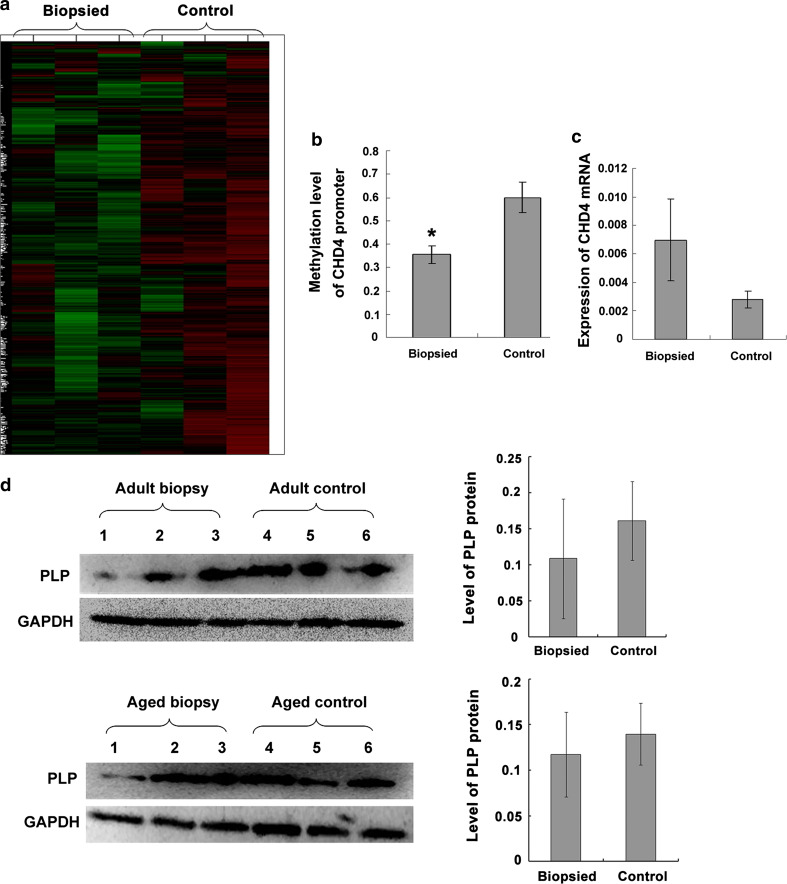
Methylation was analyzed by immunoprecipitating sheared genomic DNA using an anti-5-methylcytosine antibody and hybridization to NimbleGen MM8 Meth 385 K CpG plus Promoter microarrays. Cluster analysis of the hybridization signals was performed to characterize the specific methylation patterns of genomic DNA in the adult biopsied and control brains (brain samples from three biopsy mice and three control mice, respectively). Two clusters corresponding to two distinct methylation patterns were identified. The three biopsied samples grouped in the same cluster which showed low methylation (green), while the three control samples grouped in another cluster which showed high methylation (red; Fig. 4a). These results suggested that genome-wide low methylation was induced in the brain of the adult biopsied mice.
Fig. 4.
Analysis of genomic methylation in the brain of adult biopsied and control mice (10 weeks old; n = 3 each). a MeDIP profiling of global methylation. b Bisulfite sequencing of the CHD4 promoter (*p < 0.01). c Real-time PCR analysis of CDH4. d Western-blot analysis of PLP, a gene regulated by the CHD4-EGR2-NAB complex
We also investigated the differentially methylated loci in the adult biopsied and control brains by examining the distribution of the signals on the arrays. Eleven differentially methylated loci (corresponding to the promoter regions of 11 genes) were identified (1.5-fold difference over at least two adjacent probes, p < 0.05). Six of these genes were found to be associated with neural disorders (Table 1).
Table 1.
The genes containing differentially methylated loci in promoter regions in adult biopsied and control mice
| Gene name | Annotation | Methylation pattern in biopsied mice |
|---|---|---|
| Ireb2 | Alzheimer’s disease; resembling Parkinson’s disease | Hypomethylation |
| Acsm1 | Schizophrenia | Hypermethylation |
| Chd4 | Peripheral nerve myelination | Hypomethylation |
| Kcne2 | Neuronal excitability; schizophrenia | Hypermethylation |
| Gm239 | Atrophin-1 is the protein product of the dentatorubral-pallidoluysian atrophy (DRPLA) gene. DRPLA OMIM:125370 is a progressive neurodegenerative disorder. | Hypermethylation |
| Wbscr22 | Williams-Beuren syndrome (WBS) (mental retardation, unique cognitive and personal profiles) | Hypermethylation |
| Plunc | Mycoplasma infection and allergic inflammation | Hypomethylation |
| Tslp | Inflammation and gastrointestinal allergy | Hypermethylation |
| Dnd1 | Germ cell development | Hypermethylation |
| Prm1 | Spermatogenesis and spermatid development | Hypermethylation |
| Cma2 | Generation and degradation of Ang II | Hypomethylation |
Validation of methylation status and preliminary functional analysis of the CHD4 gene
We selected chromodomain helicase DNA-binding protein 4 (CHD4) for further analysis. Bisulfite sequencing was performed to validate the differential methylation of the CHD4 promoter region. Fifteen positive clones from each sample (six brain samples from three adult biopsy mice and three adult control mice, respectively) were picked for sequencing and the methylation rate was calculated. The biopsied mice displayed hypomethylation of the CHD4 promoter region compared to the control group (p < 0.01; Fig. 4b), consistent with the results of the MeDIP assay. To determine the effects of abnormal methylation of the CHD4 promoter, real-time PCR analysis was performed to examine CHD4 transcription. Increased CHD4 mRNA expression was observed in the brains of the adult mice from the biopsied group compared to the control group; however this difference was not statistically significant (p > 0.05; Fig. 4c).
Myelin proteolipid protein (PLP), a protein necessary for myelination, is a target gene regulated by the CHD4—early growth response-2 (EGR2)—NGFI-A/Egr-binding protein (NAB) complex. To investigate the effect of CHD4 on neural degeneration, we quantified the expression of PLP by Western blotting. Compared with control mice, the expression of PLP was markedly decreased in the brain tissues of some of the adult and aged biopsied mice (Fig. 4d).
Genome-wide low methylation in biopsied 7.5-dpc embryos
MeDIP assay and bioinformatic analysis
To elucidate the potential mechanism contributing to epigenetic alterations and neural impairment in the brain of biopsied mice, we profiled the global methylation level in 7.5-dpc embryos from the biopsied and control groups (n = 3 each group) using the MeDIP assay. In the clustering analysis, the biopsy group replicates sub-clustered together, showing low methylation (green). In contrast, the control group replicates clustered closely together, showing high methylation (red; Fig. 5a). We further examined the overall distribution of signals on the arrays. After data standardization, we identified significantly differentially methylated loci in the biopsied group: 3,468 low methylated loci and 1,308 high methylated loci, compared with the control group (69 of these low methylated loci and two of the high methylated loci were more than 1.8-fold differentially methylated, p < 0.05; Table 2).
Fig. 5.
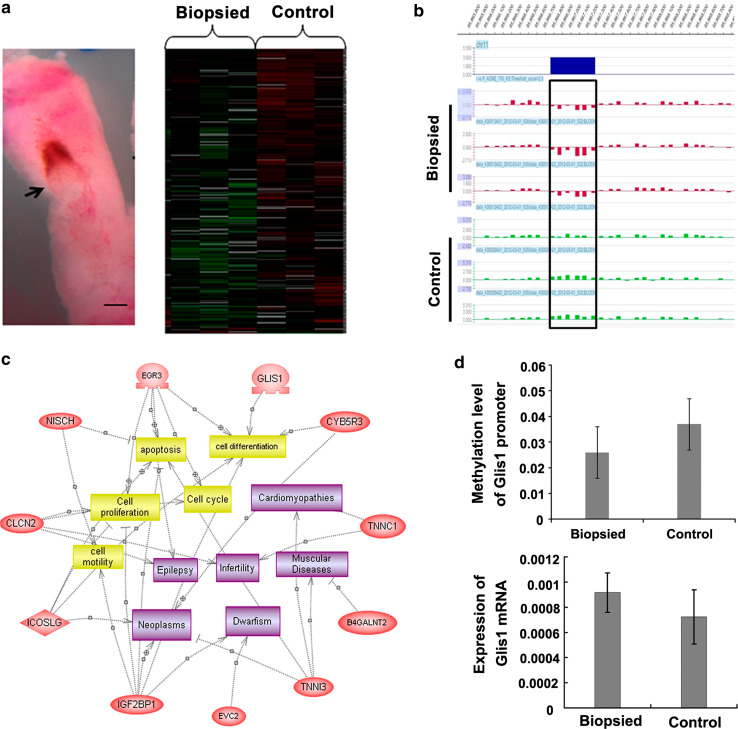
Analysis of genomic methylation in biopsied and control 7.5-dpc embryos (n = 3 each). a MeDIP assay of global methylation. The left lane shows the morphology of embryo and surrounding deciduas at 7.5-dpc (the arrow indicates the position of the embryo). The right lane indicates the result of cluster analysis. b Example of differentially methylated loci (black rectangle; >1.8-fold difference for at least two adjacent probes). c PathwayStudio analysis of differentially methylated genes (ovals); 11 genes were involved in cellular processes (cubes) related to embryo development. d Bisulfite sequencing and real-time PCR validation for Glis
Table 2.
Differentially methylated loci between the biopsied and control 7.5-dpc embryos
| Differentially methylated loci (biopsied vs. control) | Total | >2.0-fold difference | 1.8–2.0-fold difference | 1.5–1.8-fold difference |
|---|---|---|---|---|
| Low methylated loci | 3,468 | 29 | 40 | 326 |
| High methylated loci | 1,308 | 0 | 2 | 7 |
The low methylated loci in the biopsied group were selected for further analysis. Nineteen differentially methylated loci (corresponding to the promoter regions of 20 genes) were screened out on the basis that the regions with differentially methylated peaks for least two adjacent probes were more than 1.8-fold different (Fig. 5b; Table S2). The 20 genes with differentially methylated promoter regions were further analyzed by PathwayStudio software, which indicated that 11 genes were involved in embryo development. Some of these genes participate in important events during embryo development, including cell proliferation, differentiation, and apoptosis; others are involved in development-related diseases (Fig. 5c).
Validation of differential methylation of the Glis1 promoter
The transcription factor Gli-similar 1 (Glis1) regulates the expression of a variety of genes involved in embryo development [20]. Bisulfite sequencing was performed to confirm the differential methylation of the Glis1 promoter. At least 15 positive clones from each sample (three examples from biopsied 7.5-dpc embryos and three examples from control 7.5-dpc embryos) were picked for sequencing and the methylation rate was calculated. Consistent with the MeDIP assay, the biopsied embryos displayed hypomethylation of the Glis1 promoter compared to the control group; however, this difference was not statistically significant (Fig. 5d). Accordingly, real-time PCR analysis confirmed increased transcription of Glis1 in the biopsied embryos compared to the control group; however, this difference was also not statistically significant (Fig. 5d).
Reduced expression of genes involved in methylation in biopsied 4.5-dpc embryos
Microarray analysis
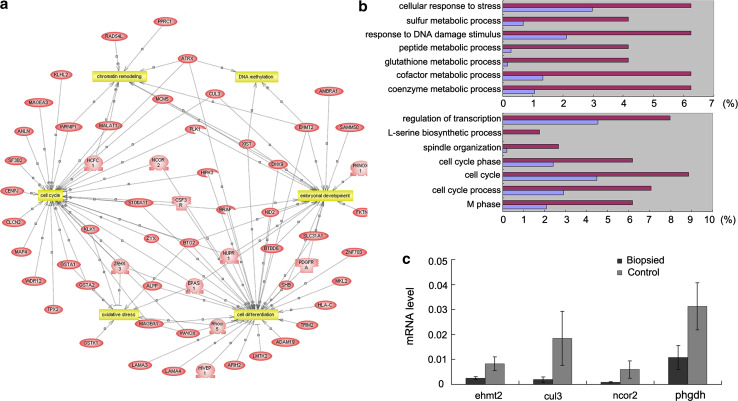
Differential gene expression in mRNA samples derived from 4.5-dpc embryos (blastocysts) of the biopsied group (n = 3) and control group (n = 3) were identified using Affymetrix Mouse Genome 430 v2.0 GeneChips. We identified 183 differentially expressed genes (>1.5-fold difference); 54 genes were upregulated and 129 genes were downregulated in the biopsied embryos (Table S3). PathwayStudio analysis indicated that 58 of these 183 genes (31.7 %) participated in a variety of biological processes, including DNA methylation, chromatin remodeling, the cell cycle, oxidative stress, embryo development, and cell differentiation (Fig. 6a).
Fig. 6.
Differential gene expression in biopsied and control 4.5-dpc embryos (n = 3 each). a Microarray analysis. Differentially expressed genes mainly participate in DNA methylation, chromatin remodeling, cell cycle, oxidative stress, embryo development, and cell differentiation. b DAVID analysis indicated that the transcripts upregulated in the biopsied embryos mainly related to “metabolic process” and the “response” to various stimuli (upper), and the downregulated transcripts in the biopsied embryos mainly associated with cell cycle events and transcriptional regulation (nether). c Real-time PCR confirmation (*p < 0.05)
Database for Annotation, Visualization and Integrated Discovery (DAVID) analysis was also performed for the 183 differentially expressed genes to identify enriched biological themes. Seven gene ontology (GO) terms for biological processes were overrepresented in the transcripts upregulated in the biopsied embryos, mainly related to “metabolic process” and the “response” to various stimuli, such as stress and DNA damage. Seven overrepresented biological process terms were identified for the transcripts that were downregulated in the biopsied embryos, mainly associated with cell cycle events and transcriptional regulation (Fig. 6b).
Real-time PCR verification
Two differentially expressed genes (Ehmt2 and Cul3), which participate in de novo methylation, and two differentially expressed genes (Ncor2 and Phgdh) involved in cell differentiation were selected for further analysis. Real-time PCR confirmed that these transcripts were expressed at lower levels in the biopsied 4.5-dpc embryos, compared to the control group (p < 0.05; Fig. 6c).
Discussion
PGD is currently widely applied in clinical practice, yet concerns regarding the safety of this method are growing [14, 21]. At present, only some clinical investigations have assessed the effect of biopsy procedure on embryonic development and young children’s health, but these have obtained different conclusions [5, 8–13]. The long-term health implications of blastomere biopsy on offspring have not yet been systematically assessed. Our recent study revealed that adult biopsied mice had poorer memory compared to control mice, indicating that development of the nervous system is sensitive to the blastomere biopsy procedure, and that biopsied mice may have a potentially high risk of neurodegenerative diseases [14]. It is common knowledge that aging is a major risk factor for neurodegenerative diseases, and aging is usually accompanied by increased dysfunction in the nervous system [22]. To ascertain the harmful effects of blastomere biopsy on the nervous system of the resulting offspring, in the present study we further evaluated the nervous system function of aged mice to determine whether degenerative neural disorders are more frequent in the biopsied group. Moreover, the potential mechanism contributing to these neural impairments was investigated.
Behavioral activity, brain histology, and molecular alterations were examined to assess nervous system function in the aged mice. The acquisition trials of the Morris water maze test and pole climbing test indicated that the blastomere biopsy procedure significantly affected the spatial learning ability of the resulting aged animals (Fig. 1). In our previous study of adult mice, no difference in spatial learning ability was observed between the biopsied and control groups, although compromised memory function was detected in the biopsied mice [14]. The above results indicated that the adverse effects of biopsy procedure on the offspring’s nervous system become more serious with age. When the biopsied mice at adult stage, the neural impairment is still not sufficient to affect spatial learning; when mice get old, the neural impairment becomes more serious, which results in poorer learning ability. Unfortunately, the aged biopsied and control mice only completed the acquisition trials, and could not continue completing the probe trials and reversal trials of the Morris water maze test because they were old and feeble. Thus, the memory function of the aged offspring could not be evaluated.
Previous studies in rodents have indicated that the hippocampus is crucial for spatial learning [23, 24]. Neuron degeneration in the hippocampus is considered a key contributing factor to the decline in learning and memory function [25]. In this study, a series of neuron regression phenotypes were more frequently observed in the hippocampus of aged biopsy mice compared to control mice (Fig. 2a). The structural alterations to hippocampal neurons were in accordance with the reduced learning ability of the aged biopsied mice. Our previous study of adult mice showed that blastomere biopsy could impair the structure of the myelinated nerve fibers in the corpora striatum, which was confirmed by decreased expression of the myelination-related protein MBP [14]. In this study, immunostaining of the striatum and Western-blotting analysis of brain tissues verified the lower expression of MBP in the brains of the aged biopsy mice (Fig. 2b, c). These results indicate that blastomere biopsy leads to more serious neuron degeneration during aging in the brains of the resulting offspring.
Neuronal impairment in the brain tissues of the aged biopsied mice was also confirmed at the molecular level. Proteomic analysis indicated that the expression of many proteins involved in neural degeneration or dysfunction were altered in the brain tissues of aged biopsied mice compared to control aged mice (Fig. 3b). These changes may have contributed to neuron degeneration. For example, the protein expression levels of SOD1 and NEFL were significantly reduced in the brains of aged biopsied mice. Reduced expression of SOD1 is reported to contribute to mitochondrial dysfunction and neuron degeneration [26, 27], and mutation of NEFL results in severe, early onset axonal neuropathy [28]. Conversely, DPYSL2 and ALDH2 are involved in neuronal repair, regeneration, and development when neuron degeneration or dysfunction occur [29, 30]. Increased expression of DPYSL2 and ALDH2 was observed in the brains of aged biopsied mice compared to controls, further implying that neuronal impairment occurred in the aged biopsied mice.
Taken together, our results suggest that blastomere biopsy affects neural development and function in the resulting offspring, and that the biopsied mice have a higher risk of neurodegenerative diseases in old age. This is consistent with a recent study that reported that 2-year-old PGS children had lower neurologic optimality scores than control IVF/ICSI children [13]. Thus, a large-scale investigation of neural function needs to be performed in adult and even aged PGD offspring.
Currently, the influence of ART procedures on the health of the offspring is gradually emerging [2, 13, 21]. However, the mechanisms contributing to these adverse effects are still not clear. In recent years, epigenetic deregulation has received increasing attention as a possible cause of adverse ART outcomes, as the incidence of imprinting disorders is reported to be increased in ART offspring [31, 32]. Additionally, evidence for the epigenetic effects of ART has been obtained in mice. Both superovulation and embryo culture have been reported to alter imprinted methylation in the resulting embryos, which can influence embryo development and even physiological parameters at the adult age [33, 34]. At present, studies on the epigenetic effects of ART have mainly focused on imprinted genes, and the effects of a limited range of ART procedures on imprinted methylation in the resulting embryos have been studied. The definitive answers to many questions about the adverse effects of ART await genome-wide epigenetic profiling of the offspring of various ART techniques [35].
Here, we characterized the genomic DNA methylation patterns in the brains of adult biopsy and control mice using the MeDIP assay. Genome-wide low methylation was observed in the biopsy group when compared to the control group (Fig. 4a), which might be associated with the neural impairment observed in biopsy mice. On the other hand, most of the genes containing differentially methylated loci in promoter regions were found to be associated with neural disorders (Table 1). We selected a gene showing low methylation in the biopsied group, CHD4, for further investigation. The increased expression of CHD4 gene in the brains of the biopsied mice was found, which might due to the reduced methylation of the CHD4 promoter (Fig. 4c). It has been demonstrated that NAB2 represses transcription by interacting with CHD4, and the expression of many genes is directly down-regulated by the CHD4-Egr2-NAB complex in vivo [36, 37]. Thus, increased expression of CHD4 in the brains of the biopsied group may influence the function of the nervous system by repressing the expression of genes regulated by the CHD4-Egr2-NAB complex. For example, PLP, a target gene repressed by CHD4-EGR2-NAB, is necessary for myelination [36, 38, 39]. We observed that the expression of PLP was markedly decreased in some biopsied mice, compared to controls (Fig. 4d), which could possibly contribute to neuron degeneration of these mice.
Based on these results, we suggest that hypomethylation in the brain is involved in the neural impairment observed in biopsied mice. It is known that an important epigenetic reprogramming event, i.e., whole genomic demethylation and then remethylation (defined as de novo methylation), occurs during early embryo development. Demethylation is initiated immediately after fertilization, and genome hypomethylation is observed until the blastocyst stage [40]. DNA methylation begins to be reestablished during or after implantation at 4.5 dpc, and the remethylation of almost all genes has been completed, and corresponds to the characteristic methylation pattern of adult somatic tissues, by 6.5 dpc [41]. All PGD-related manipulations are performed during the demethylation stage. Since DNA methylation and demethylation are both sensitive to environmental factors, we investigated whether blastomere biopsy disturbed epigenetic reprogramming during early embryo development, and if this could result in epigenetic alterations in the tissues of adult mice.
To address this issue, the MeDIP assay was performed to compare the genomic DNA methylation profiles of 7.5-dpc embryos in the biopsy and control group. Whole-genome low methylation was observed in the biopsied embryos (Fig. 5a), suggesting that blastomere biopsy presented an obstacle to de novo methylation during early embryo development (from 4.5 to 6.5 dpc). This impaired remethylation during early embryo development may have resulted in the hypomethylated status in the brain of the resulting adult mice (Fig. 4a). The genes showing low methylation in promoter regions were then selected for further analysis. It is worth noting that most of these differentially methylated genes identified in 7.5-dpc biopsied embryos are involved in cell proliferation, differentiation, and apoptosis (Fig. 5c). Thus, the altered expression level of these genes may influence the development of various tissues, which in turn may affect organ function. Further evaluations of the physiological functions of other organs in PGD offspring are required.
Profiling of mRNA expression in 4.5-dpc embryos (blastocyst stage) helped to understand why blastomere biopsy influenced remethylation during early embryo development. A total of 183 differently expressed genes were identified between the biopsy and control embryos. Most differently expressed genes (70.5 %) were downregulated in the biopsied embryos; these downregulated genes were mainly associated with cell cycle events and transcriptional regulation. By further analyzing this group of genes, we found that several genes participated in de novo methylation. For example, the protein encoded by Ehmt2 can interact directly with DNA methyltransferases and mediate de novo methylation [42]. Cul3 also participates in de novo methylation by interacting with the DNA methyltransferase DNMT3b [43]. Thus, decreased expression of these genes may disturb de novo methylation at the blastocyst stage (4.5-dpc embryo), which may contribute to the genome hypomethylation observed in biopsied embryos at 7.5 dpc. In addition, it was found that many downregulated genes were involved in cell differentiation, including Ncor2 and Phgdh, which participate in neuron differentiation and neurogenesis [44, 45]. Decreased expression of these genes may influence early embryo development and affect the structure and function of specific organs, such as the nervous system impairment observed in this study.
A small number of differentially genes were upregulated in the biopsied embryos at 4.5 dpc. The enriched biological themes for these upregulated genes were mainly “response to stress or DNA damage” and “metabolic process.” This may reflect a defense response of the embryos to the biopsy procedure, which may increase the expression of metabolism-related genes (including peptides, cofactors, coenzyme metabolism, etc.) in order to promote embryo survival and growth.
It obviously, only some differentially genes were involved DNA methylation. Other genes participate in several biological processes, including cell differentiation, cell cycle, etc. (Fig 6a). These results indicated that the biopsy procedure influenced early embryo development from several aspects. The exact role of methylation-related genes as well as other differentially genes needs to be investigated further.
In conclusion, we demonstrated abnormal neural development and function in mice generated after blastomere biopsy, and showed that biopsied mice may have a higher risk of neurodegenerative disorders in old age. Moreover, we suggest that impaired epigenetic reprogramming during early embryo development may be the latent mechanism that contributes to nervous system impairment in biopsied mice. This study first revealed the negative impacts of biopsy procedure on the offspring’s later development and explored the possible mechanism that will provide valuable information on procedure improvements of PGD. More studies should be performed to address the possible adverse effects of blastomere biopsy on the development of offspring. Unfortunately, the sample size is relative small in this study, and some methods used also have technical limits, which may weaken the comprehensiveness of our results. The more large-scale and depth research needs to be performed in the future. Anyway, this study provides feasibility and foundation for further large sample population study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank Ming Xiao for his technical support during the experiment of brain morphological analysis. We thank Xuejiang Guo for help with the proteomics data analysis. This study was supported by grants from the Major State Basic Research Development Program of China (973 Program) (No. 2012CB944902) and from the National Science Foundation of China (No.31271604).
Footnotes
Y. Wu, Z. Lv, and Y. Yang contributed equally to this work.
Contributor Information
Hui Zhu, Phone: +86-25-86862038, FAX: +86-25-86862908, Email: njzhuhui@njmu.edu.cn.
Qi Zhou, Phone: +86-10-64807299, FAX: +86-10-64807299, Email: qzhou@ioz.ac.cn.
References
- 1.Ecker DJ, Stein P, Xu Z, Williams CJ, Kopf GS, Bilker WB, Abel T, Schultz RM. Long-term effects of culture of preimplantation mouse embryos on behavior. Proc Natl Acad Sci USA. 2004;101(6):1595–1600. doi: 10.1073/pnas.0306846101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson CL, Fisher JR, Hammarberg K, Amor DJ, Halliday JL. Looking downstream: a review of the literature on physical and psychosocial health outcomes in adolescents and young adults who were conceived by ART. Hum Reprod. 2011;26(5):1209–1219. doi: 10.1093/humrep/der041. [DOI] [PubMed] [Google Scholar]
- 3.Maher ER. Imprinting and assisted reproductive technology. Hum Mol Genet. 2005;14:R133–R138. doi: 10.1093/hmg/ddi107. [DOI] [PubMed] [Google Scholar]
- 4.Watkins AJ, Platt D, Papenbrock T, Wilkins A, Eckert JJ, Kwong WY, Osmond C, Hanson M, Fleming TP. From the cover: mouse embryo culture induces changes in postnatal phenotype including raised systolic blood pressure. Proc Natl Acad Sci USA. 2007;104(13):5449–5454. doi: 10.1073/pnas.0610317104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardy K, Martin KL, Leese HJ, Winston RM, Handyside AH. Human preimplantation development in vitro is not adversely affected by biopsy at the 8-cell stage. Hum Reprod. 1990;5(6):708–714. doi: 10.1093/oxfordjournals.humrep.a137173. [DOI] [PubMed] [Google Scholar]
- 6.Sermon K, Van Steirteghem A, Liebaers I. Preimplantation genetic diagnosis. Lancet. 2004;363(9421):1633–1641. doi: 10.1016/S0140-6736(04)16209-0. [DOI] [PubMed] [Google Scholar]
- 7.Moutou C, Viville S. Preimplantation genetic diagnosis of monogenic diseases. Ann Biol Clin (Paris) 2003;61(5):521–532. [PubMed] [Google Scholar]
- 8.Desmyttere S, Bonduelle M, Nekkebroeck J, Roelants M, Liebaers I, De Schepper J. Growth and health outcome of 102 2-year-old children conceived after preimplantation genetic diagnosis or screening. Early Hum Dev. 2009;85(12):755–759. doi: 10.1016/j.earlhumdev.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Nekkebroeck J, Bonduelle M, Desmyttere S, Van den Broeck W, Ponjaert-Kristoffersen I. Mental and psychomotor development of 2-year-old children born after preimplantation genetic diagnosis/screening. Hum Reprod. 2008;23(7):1560–1566. doi: 10.1093/humrep/den033. [DOI] [PubMed] [Google Scholar]
- 10.Banerjee I, Shevlin M, Taranissi M, Thornhill A, Abdalla H, Ozturk O, Barnes J, Sutcliffe A. Health of children conceived after preimplantation genetic diagnosis: a preliminary outcome study. Reprod Biomed Online. 2008;16(3):376–381. doi: 10.1016/S1472-6483(10)60599-8. [DOI] [PubMed] [Google Scholar]
- 11.Mastenbroek S, Twisk M, van der Veen F, Repping S. Preimplantation genetic screening: a systematic review and meta-analysis of RCTs. Hum Reprod Update. 2011;17(4):454–466. doi: 10.1093/humupd/dmr003. [DOI] [PubMed] [Google Scholar]
- 12.Cho YJ, Kim JY, Song IO, Lee HS, Lim CK, Koong MK, Kang IS. Does blastomere biopsy in preimplantation genetic diagnosis affect early serum beta-hCG levels? Clin Exp Reprod Med. 2011;38(1):31–36. doi: 10.5653/cerm.2011.38.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Middelburg KJ, van der Heide M, Houtzager B, Jongbloed-Pereboom M, Fidler V, Bos AF, Kok J, Hadders-Algra M, PGS Follow-up Study Group Mental, psychomotor, neurologic, and behavioral outcomes of 2-year-old children born after preimplantation genetic screening: follow-up of a randomized controlled trial. Fertil Steril. 2011;96(1):165–169. doi: 10.1016/j.fertnstert.2011.04.081. [DOI] [PubMed] [Google Scholar]
- 14.Yu Y, Wu J, Fan Y, Lv Z, Guo X, Zhao C, Zhou R, Zhang Z, Wang F, Xiao M, Chen L, Zhu H, Chen W, Lin M, Liu J, Zhou Z, Wang L, Huo R, Zhou Q, Sha J. Evaluation of blastomere biopsy using a mouse model indicates the potential high risk of neurodegenerative disorders in the offspring. Mol Cell Proteomics. 2009;8(7):1490–1500. doi: 10.1074/mcp.M800273-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vorhees CV, Williams MT. Morris water maze: procedures for assessing spatial and related forms of learning and memory. Nat Protoc. 2006;1(2):848–858. doi: 10.1038/nprot.2006.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogawa N, Hirose Y, Ohara S, Ono T, Watanabe Y. A simple quantitative bradykinesia test in MPTP-treated mice. Res Commun Chem Pathol Pharmacol. 1985;50(3):435–441. [PubMed] [Google Scholar]
- 17.Zhu YF, Cui YG, Guo XJ, Wang L, Bi Y, Hu YQ, Zhao X, Liu Q, Huo R, Lin M, Zhou ZM, Sha JH. Proteomic analysis of effect of hyperthermia on spermatogenesis in adult male mice. J Proteome Res. 2006;5(9):2217–2225. doi: 10.1021/pr0600733. [DOI] [PubMed] [Google Scholar]
- 18.Scacheri P, Crawford GE, Davis S. Statistics for ChIP-chip and DNase hypersensitivity experiments on NimbleGen arrays. Methods Enzymol. 2006;411:270–282. doi: 10.1016/S0076-6879(06)11014-9. [DOI] [PubMed] [Google Scholar]
- 19.Han SY, Kim SH, Heasley LE. Differential gene regulation by specific gain-of-function JNK1 proteins expressed in Swiss 3T3 fibroblasts. J Biol Chem. 2002;277(49):47167–47174. doi: 10.1074/jbc.M204270200. [DOI] [PubMed] [Google Scholar]
- 20.Maekawa M, Yamaguchi K, Nakamura T, Shibukawa R, Kodanaka I, Ichisaka T, Kawamura Y, Mochizuki H, Goshima N, Yamanaka S. Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature. 2011;474(7350):225–229. doi: 10.1038/nature10106. [DOI] [PubMed] [Google Scholar]
- 21.Winston RM, Hardy K. Are we ignoring potential dangers of in vitro fertilization and related treatments? Nat Cell Biol. 2002;4:s14–s18. doi: 10.1038/ncb-nm-fertilityS14. [DOI] [PubMed] [Google Scholar]
- 22.von Bernhardi R, Tichauer JE, Eugenin J. Aging-dependent changes of microglial cells and their relevance for neurodegenerative disorders. J Neurochem. 2010;112(5):1099–1114. doi: 10.1111/j.1471-4159.2009.06537.x. [DOI] [PubMed] [Google Scholar]
- 23.Logue SF, Paylor R, Wehner JM. Hippocampal lesions cause learning deficits in inbred mice in the Morris water maze and conditioned-fear task. Behav Neurosci. 1997;111(1):104–113. doi: 10.1037/0735-7044.111.1.104. [DOI] [PubMed] [Google Scholar]
- 24.Morris RG, Garrud P, Rawlins JN, O’Keefe J. Place navigation impaired in rats with hippocampal lesions. Nature. 1982;297(5868):681–683. doi: 10.1038/297681a0. [DOI] [PubMed] [Google Scholar]
- 25.Scheff SW, Price DA, Schmitt FA, Mufson EJ. Hippocampal synaptic loss in early Alzheimer’s disease and mild cognitive impairment. Neurobiol Aging. 2006;27(10):1372–1384. doi: 10.1016/j.neurobiolaging.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 26.Niwa J, Yamada S, Ishigaki S, Sone J, Takahashi M, Katsuno M, Tanaka F, Doyu M, Sobue G. Disulfide bond mediates aggregation, toxicity, and ubiquitylation of familial amyotrophic lateral sclerosis-linked mutant SOD1. J Biol Chem. 2007;282(38):28087–28095. doi: 10.1074/jbc.M704465200. [DOI] [PubMed] [Google Scholar]
- 27.Magrane J, Hervias I, Henning MS, Damiano M, Kawamata H, Manfredi G. Mutant SOD1 in neuronal mitochondria causes toxicity and mitochondrial dynamics abnormalities. Hum Mol Genet. 2009;18(23):4552–4564. doi: 10.1093/hmg/ddp421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yum SW, Zhang J, Mo K, Li J, Scherer SS. A novel recessive Nefl mutation causes a severe, early-onset axonal neuropathy. Ann Neurol. 2009;66(6):759–770. doi: 10.1002/ana.21728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki Y, Nakagomi S, Namikawa K, Kiryu-Seo S, Inagaki N, Kaibuchi K, Aizawa H, Kikuchi K, Kiyama H. Collapsin response mediator protein-2 accelerates axon regeneration of nerve-injured motor neurons of rat. J Neurochem. 2003;86(4):1042–1050. doi: 10.1046/j.1471-4159.2003.01920.x. [DOI] [PubMed] [Google Scholar]
- 30.Picklo MJ, Olson SJ, Markesbery WR, Montine TJ. Expression and activities of aldo-keto oxidoreductases in Alzheimer disease. J Neuropathol Exp Neurol. 2001;60(7):686–695. doi: 10.1093/jnen/60.7.686. [DOI] [PubMed] [Google Scholar]
- 31.Manipalviratn S, DeCherney A, Segars J. Imprinting disorders and assisted reproductive technology. Fertil Steril. 2009;91(2):305–315. doi: 10.1016/j.fertnstert.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amor DJ, Halliday J. A review of known imprinting syndromes and their association with assisted reproduction technologies. Hum Reprod. 2008;23(12):2826–2834. doi: 10.1093/humrep/den310. [DOI] [PubMed] [Google Scholar]
- 33.Market-Velker BA, Zhang L, Magri LS, Bonvissuto AC, Mann MR. Dual effects of superovulation: loss of maternal and paternal imprinted methylation in a dose-dependent manner. Hum Mol Genet. 2010;19(1):36–51. doi: 10.1093/hmg/ddp465. [DOI] [PubMed] [Google Scholar]
- 34.Market-Velker BA, Fernandes AD, Mann MR. Side-by-side comparison of five commercial media systems in a mouse model: suboptimal in vitro culture interferes with imprint maintenance. Biol Reprod. 2010;83(6):938–950. doi: 10.1095/biolreprod.110.085480. [DOI] [PubMed] [Google Scholar]
- 35.van Montfoort AP, Hanssen LL, de Sutter P, Viville S, Geraedts JP, de Boer P. Assisted reproduction treatment and epigenetic inheritance. Hum Reprod Update. 2012;18(2):171–197. doi: 10.1093/humupd/dmr047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mager GM, Ward RM, Srinivasan R, Jang SW, Wrabetz L, Svaren J. Active gene repression by the Egr2.NAB complex during peripheral nerve myelination. J Biol Chem. 2008;283(26):18187–18197. doi: 10.1074/jbc.M803330200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Srinivasan R, Mager GM, Ward RM, Mayer J, Svaren J. NAB2 represses transcription by interacting with the CHD4 subunit of the nucleosome remodeling and deacetylase (NuRD) complex. J Biol Chem. 2006;281(22):15129–15137. doi: 10.1074/jbc.M600775200. [DOI] [PubMed] [Google Scholar]
- 38.Griffiths IR, Scott I, McCulloch MC, Barrie JA, McPhilemy K, Cattanach BM. Rumpshaker mouse: a new X-linked mutation affecting myelination: evidence for a defect in PLP expression. J Neurocytol. 1990;19(2):273–283. doi: 10.1007/BF01217305. [DOI] [PubMed] [Google Scholar]
- 39.Al-Saktawi K, McLaughlin M, Klugmann M, Schneider A, Barrie JA, McCulloch MC, Montague P, Kirkham D, Nave KA, Griffiths IR. Genetic background determines phenotypic severity of the Plp rumpshaker mutation. J Neurosci Res. 2003;72(1):12–24. doi: 10.1002/jnr.10561. [DOI] [PubMed] [Google Scholar]
- 40.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241(1):172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 41.Borgel J, Guibert S, Li Y, Chiba H, Schübeler D, Sasaki H, Forné T, Weber M. Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet. 2010;42(12):1093–1100. doi: 10.1038/ng.708. [DOI] [PubMed] [Google Scholar]
- 42.Leung DC, Dong KB, Maksakova IA, Goyal P, Appanah R, Lee S, Tachibana M, Shinkai Y, Lehnertz B, Mager DL, Rossi F, Lorincz MC. Lysine methyltransferase G9a is required for de novo DNA methylation and the establishment, but not the maintenance, of proviral silencing. Proc Natl Acad Sci USA. 2011;108(14):5718–5723. doi: 10.1073/pnas.1014660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shamay M, Greenway M, Liao G, Ambinder RF, Hayward SD. De novo DNA methyltransferase DNMT3b interacts with NEDD8-modified proteins. J Biol Chem. 2010;285(47):36377–36386. doi: 10.1074/jbc.M110.155721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jepsen K, Solum D, Zhou T, McEvilly RJ, Kim HJ, Glass CK, Hermanson O, Rosenfeld MG. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature. 2007;450(7168):415–419. doi: 10.1038/nature06270. [DOI] [PubMed] [Google Scholar]
- 45.Kawakami Y, Yoshida K, Yang JH, Suzuki T, Azuma N, Sakai K, Hashikawa T, Watanabe M, Yasuda K, Kuhara S, Hirabayashi Y, Furuya S. Impaired neurogenesis in embryonic spinal cord of Phgdh knockout mice, a serine deficiency disorder model. Neurosci Res. 2009;63(3):184–193. doi: 10.1016/j.neures.2008.12.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.