Abstract
α-Amylase (EC 3.2.1.1) represents the best known amylolytic enzyme. It catalyzes the hydrolysis of α-1,4-glucosidic bonds in starch and related α-glucans. In general, the α-amylase is an enzyme with a broad substrate preference and product specificity. In the sequence-based classification system of all carbohydrate-active enzymes, it is one of the most frequently occurring glycoside hydrolases (GH). α-Amylase is the main representative of family GH13, but it is probably also present in the families GH57 and GH119, and possibly even in GH126. Family GH13, known generally as the main α-amylase family, forms clan GH-H together with families GH70 and GH77 that, however, contain no α-amylase. Within the family GH13, the α-amylase specificity is currently present in several subfamilies, such as GH13_1, 5, 6, 7, 15, 24, 27, 28, 36, 37, and, possibly in a few more that are not yet defined. The α-amylases classified in family GH13 employ a reaction mechanism giving retention of configuration, share 4–7 conserved sequence regions (CSRs) and catalytic machinery, and adopt the (β/α)8-barrel catalytic domain. Although the family GH57 α-amylases also employ the retaining reaction mechanism, they possess their own five CSRs and catalytic machinery, and adopt a (β/α)7-barrel fold. These family GH57 attributes are likely to be characteristic of α-amylases from the family GH119, too. With regard to family GH126, confirmation of the unambiguous presence of the α-amylase specificity may need more biochemical investigation because of an obvious, but unexpected, homology with inverting β-glucan-active hydrolases.
Keywords: α-Amylase; Glycoside hydrolase families; GH13, GH57, GH119, GH126; Conserved sequence regions; Catalytic machinery; Evolutionary relationships
Introduction
α-Amylase represents the best known and most intensively studied amylolytic enzyme. Amylolytic enzymes are enzymes degrading starch and starchy substrates and are applied widely in various branches of the food, pharmaceutical, and chemical industries. In a broader sense, the designation “amylolytic enzymes” has been used for the variety of starch hydrolases and related enzymes that are active—in terms of hydrolysis, transglycosylation, and isomerization—towards the α-glucosidic bonds present in starch and related poly- and oligo-saccharides.
As our understanding of the details of protein primary and tertiary structure of individual amylolytic enzymes increased in the last couple of decades, it has become clear that these enzymes, which are very closely related by their function on starch, e.g., α-amylase, β-amylase, and glucoamylase, are not closely related in terms of structure and reaction mechanism. It has turned out to be more appropriate to base a classification of amylolytic enzymes on similarities in their amino acid sequences and three-dimensional structures, reaction mechanisms, and catalytic machineries, all reflecting evolutionary relatedness, than on specificity. Such an approach, however, opens the door to ideas that enzymes closely related in function will classify separately, but also that similar reactions can be catalyzed by structurally different and thus evolutionarily unrelated proteins.
This appears to be the situation for the α-amylase enzymes that are the topic of the present review. The main goal is to describe the most recent knowledge of α-amylases with regard to the existence of structurally different enzymes apparently possessing the same α-amylase-type activity. Since the α-amylase is an enzyme with a broad substrate preference and product specificity, emphasis is also given to subtle but unique structural features discriminating between closely related α-amylases, taking into account, for example, their taxonomical origin.
CAZy classification system and α-amylases
α-Amylases are glycoside hydrolases (GHs) and have therefore become a part of the sequence-based classification system of enzymes active towards various saccharides. This system was first developed some 20 years ago for GHs [1] and later updated [2, 3]. Now, the entire system exists online (since 1998) for all Carbohydrate-Active enZymes (CAZy), as the so-called CAZy database (or CAZy server; http://www.cazy.org/) covering: (1) catalytic modules involved in degradation, creation, and modification of glycosidic linkages of saccharides; and (2) associated carbohydrate-binding modules (CBM) responsible for adhesion to saccharides [4]. The classification of catalytic modules includes, in addition to GHs, glycosyl transferases, polysaccharide lyases, and carbohydrate esterases.
The CAZy classification system based on comparison of amino acid sequences was established in an effort to overcome the inability of classical IUB Enzyme Nomenclature (for GHs: EC 3.2.1.x) to reflect structural features and evolutionary aspects of GHs [1]. Within the CAZy system, individual enzymes are classified into sequence-based families. Currently, more than 130 such families (designated with Arabic numerals, e.g., GH13) have been created chronologically since 1991. The enzymes (proteins) belonging to the same GH family should, in principle, exhibit sequence similarities (usually with conserved sequence regions; CSRs), share catalytic machinery (the same catalytic residues located on corresponding structural elements), employ the same reaction mechanism (either retaining or inverting) and adopt the same type of catalytic domain fold [1]. There are two additional levels of hierarchical classification in the CAZy database, i.e., clans as a higher level and subfamilies as a lower level of hierarchy [4, 5]. A GH clan groups together families sharing catalytic machinery and adopting the same structural fold of the catalytic domain, but with significant difference in overall sequence. A clan is designated by a letter preceded by a dash (e.g., GH-H) attributed in the alphabetical order of their appearance, with every GH clan creation being based on tertiary structure information [4]. A GH subfamily is a group of members of one family that shares more sequence/structural and functional characteristics than applicable for the entire family, i.e., a subfamily members should share a more recent evolutionary ancestor. Subfamilies of a given GH family are designated by a numeral suffix preceded by underscore (e.g., GH13_7). In the scientific literature proposals have been made to split various GH families into subfamilies, and the α-amylase family GH13 was the first to have been officially divided in this way, in this case into 37 subfamilies, by the CAZy curators [6]. In addition to GH13, of all 131 GH families, official subfamilies have been defined only for the families GH5 [7] and GH30 [8]. Moreover, the entries for the families GH21, GH40, GH41, GH60, and GH69 were deleted from the GH system (CAZy; [4]).
Currently, the entire CAZy database covers the sequence data from more than 2,100 genomes of Bacteria, almost 150 genomes of Archaea and around 70 of Eukaryotes (CAZy; [4]). The encyclopedic project CAZypedia (http://www.cazypedia.org/) was created about 5 years ago and, being written and curated by scientists directly involved in studying such enzymes, it constitutes a very comprehensive and complementary source of information.
α-Amylase (EC 3.2.1.1), the most widely studied amylolytic enzyme, catalyzes the hydrolysis, with retention of configuration, of the internal α-1,4-glucosidic bonds in starch and related poly- and oligo-saccharides and is an endo-acting enzyme [9]. Some exo-acting enzymes can be considered as α-amylases, but are not addressed in detail here. The α-amylases, however, exhibit quite varied product profiles, depending on their origin [10]. In addition to the main hydrolytic reaction, many α-amylases are also able to catalyze transglycosylation, e.g., those from Aspergillus oryzae [11], Alteromonas haloplanktis [12], human saliva [13], and human pancreas [14], and even a chimera from Bacillus amyloliquefaciens and Bacillus licheniformis [15]. Without a serious biochemical characterization, the α-amylase specificity should not be ascribed unambiguously to an enzyme displaying simply amylolytic (i.e., a starch-hydrolyzing) activity [16]. Any experimental data, on the other hand, can now be supported by reliable in silico analyses of primary structure and predictions.
Currently, the α-amylase enzyme specificity within the CAZy database can be found unambiguously in the families GH13, GH57 and GH119, moreover it has been suggested to also be present in family GH126 (CAZy; [4, 17]. Of these, the family GH13 can be considered to be the main α-amylase family first proposed in the CAZy system in 1991 [1], with family GH57 as the second and smaller α-amylase family established in 1996 [3] and the families GH119 and GH126 defined in 2006 and 2011, respectively (CAZy; [4]).
α-Amylase family GH13
α-Amylase family GH13 was established in 1991 when the classification of GHs, i.e., the CAZy database [4], was first published [1]. At that time, the family GH13 was (due to classification criteria) defined as a polyspecific enzyme family covering, in addition to α-amylase, enzymes such as α-glucosidase, dextran glucosidase, isoamylase, pullulanase, amylopullulanase, neopullulanase, cyclodextrin glucanotransferase, and some exo-acting amylases [1]. The classification reflected the predictions made previously that various starch hydrolases and related amylolytic enzymes would exhibit mutual sequence similarities and share catalytic residues and folds [18–21]. Currently, the family GH13 contains approximately 30 different enzyme specificities from three EC groups, i.e., hydrolases, transferases, and isomerases [4, 22], plus non-enzymatic members represented by the heavy-chains of heteromeric amino acid transporters and 4F2 antigens [23–25]. With regard to the number of sequences belonging to GH13, this family ranks among the largest in CAZy with more than 13,500 members in March 2013 (CAZy; [4]) originating predominantly from Bacteria (~11,500), with fewer from Eukaryotes (~1,800) and Archaea (~200).
In general, the GH13 α-amylases and other family GH13 members are three-domain proteins (Fig. 1) consisting of the main catalytic (β/α)8-barrel domain (domain A) with a small domain B protruding out of the barrel as a longer loop between the strand β3 and helix α3 and succeeded at the C-terminal end by domain C, adopting an antiparallel β-sandwich fold [10, 26–30]. This domain organization was first determined for Taka-amylase A [31], i.e., the α-amylase from A. oryzae. The domain of the (β/α)8-barrel is composed of eight inner parallel β-strands surrounded by eight α-helices and, because it was first recognized in the structure of triose-phosphate isomerase (TIM; [32]), it is often called the TIM-barrel [33]. Individual GH13 members and sometimes all members with a given specificity may contain additional domains on either terminus of their polypeptide chain. Although their functions have still not been completely understood, such domains are often involved in binding starch, glycogen, and other related saccharides [34–38]. Typical starch-binding domains (SBDs) have also been classified within the CAZy database as the so-called CBM families [4] with CBM20 as a representative of the C-terminal SBD that was first recognized [39–41]. At present, ten CBM families are considered as SBD families: CBM20, CBM21, CBM25, CBM26, CBM34, CBM41, CBM45, CBM48, CBM53, and CBM58 [42–45].
Fig. 1.
Tertiary structures of representative family GH13 α-amylases. The structures from following subfamilies and origins are shown: a GH13_1, Aspergillus oryzae (PDB code: 2TAA; [31]); b GH13_5, Bacillus licheniformis (PDB code: 1BLI; [249]); c GH13_6, Hordeum vulgare—barley isozyme AMY-1 (PDB code: 1P6W; [129]); d GH13_37, uncultured bacterium AmyP (modeled structure; residues Leu6-Thr491) obtained from the Phyre server [245] based on the Flavobacterium sp. 92 GH13 cyclomaltodextrinase (PDB code: 3EDE; [205]) as template; e unclassified, A. haloplanktis (PDB code: 1G94; [12]); f unclassified, Halothermothrix orenii AmyB (PDB code: 3BC9; [70]); g unclassified, Bacteroides thetaiotaomicron (PDB code: 3K8L; [38]). The individual domains are colored as follows: catalytic (β/α)8-barrel blue, domain B green, domain C red, N-terminal domain cyan; starch-binding domain of CBM58 family magenta. The GH13 catalytic triad, i.e., catalytic nucleophile—Asp (top), proton donor—Glu (left) and transition-state stabilizer—Asp (right)—is highlighted in the (β/α)8-barrel domain in each structure in a similar position. Saccharide molecules are colored yellow to emphasize: c an additional surface binding site as “a pair of sugar tongs” with the side-chain of tyrosine (in black) involved in binding; e a heptasaccharide occupying the subsites from −4 to +3 as a transglycosylation product from acarbose (a pseudotetrasaccharide); and g a maltopentaose bound by the SBD of the family CBM58 inserted within the domain B. The structures were retrieved from the Protein Data Bank (PDB; [250]) and visualized with the program WebLabViewerLite (Molecular Simulations, Inc.)
History
All the family GH13 members should obey several distinct criteria [10, 46]: (1) employing the retaining mechanism of α-glycosidic bond cleavage; (2) adopting a (β/α)8-barrel as the catalytic domain; (3) exhibiting 4–7 CSRs positioned mostly on β-strands of the barrel (Fig. 2); and (4) sharing the catalytic machinery, consisting of strand β4-aspartic acid (catalytic nucleophile), β5-glutamic acid (proton donor), and β7-aspartic acid (transition-state stabilizer)—here referred to as the catalytic triad. These criteria were defined explicitly by [47] in a more stringent version that reflected the situation 20 years ago when a much lower number of sequences was available.
Fig. 2.
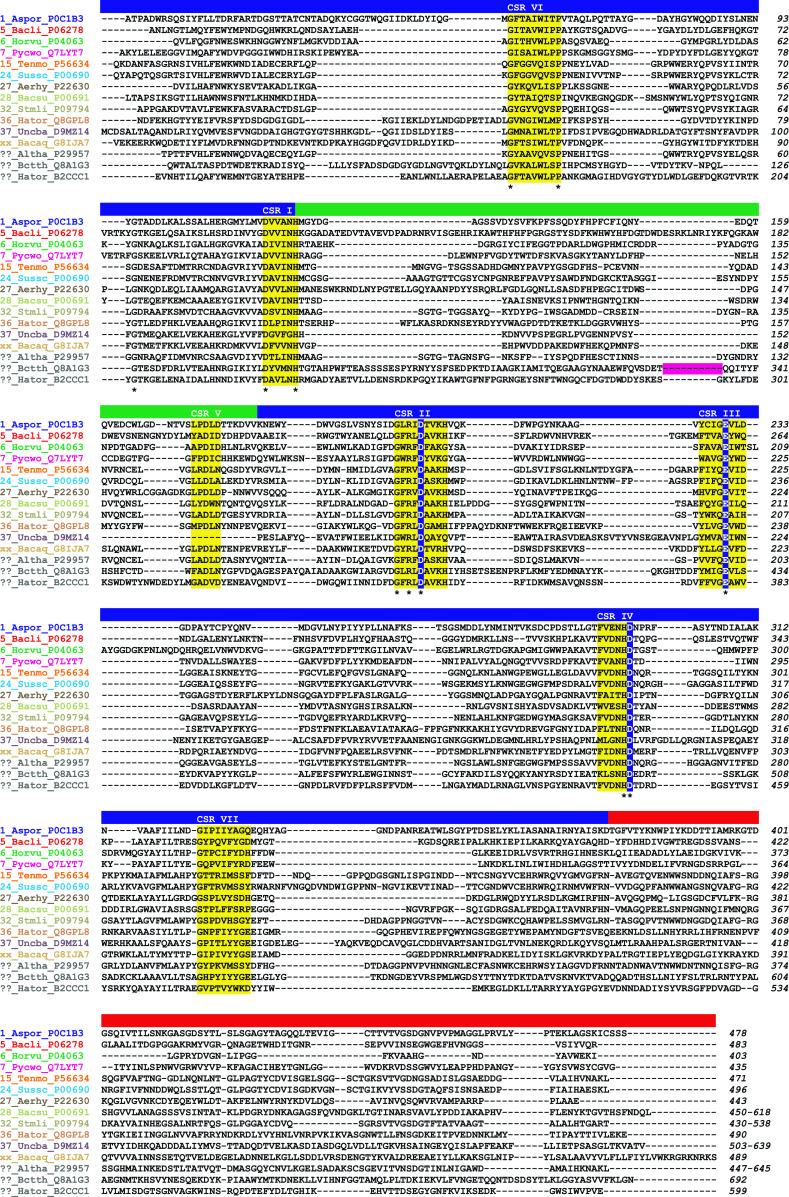
Amino acid sequence alignment of GH13 α-amylases representing the individual α-amylase subfamilies. The aligned sequences span the typical GH13 α-amylase’s domain arrangement consisting of catalytic (β/α)8-barrel with domain B (inserted between the strand β3 and helix α3) and domain C (succeeding the TIM-barrel). The name of an enzyme is composed of the GH13 subfamily number (“xx” for the recently described α-amylase from Bacillus aquimaris and “??” for α-amylases from A. haloplanktis, Bacteroides thetaiotaomicron, and Halothermothrix orenii AmyB that currently are still not assigned any GH13 subfamily in CAZy) followed by the abbreviation of the source (organism) and the UniProt accession number. Note there are two α-amylases from Halothermothrix orenii, the AmyA assigned to subfamily GH13_36 (UniProt Q8GPL8) and the AmyB not assigned as yet to any GH13 subfamily (UniProt B2CCC1). The organisms are abbreviated as follows: Aspor, Aspergillus oryzae; Bacli, Bacillus licheniformis; Horvu, Hordeum vulgare; Pycwo, Pyrococcus woesei; Tenmo, Tenebrio molitor; Sussc, Sus scrofa (pancreas); Aerhy, Aeromonas hydrophila; Bacsu, Bacillus subtilis; Stmli, Streptomyces limosus; Hator, Halothermothrix orenii; Uncba, uncultured bacterium; Bacaq, Bacillus aquimaris; Altha, A. haloplanktis; and Bctth, Bacteroides thetaiotaomicron. The seven CSRs typical for the family GH13 [46] and the catalytic triad are highlighted in yellow and blue, respectively. The residues conserved invariantly are marked by an asterisk below the alignment. The individual α-amylase family GH13 domains are indicated as a colored lane above the alignment: blue catalytic domain A, green domain B, red C-terminal domain C. The positions corresponding to the deleted SBD of the family CBM58 in ??_Bctth_Q8A1G3 (SusG α-amylase) are signified by a magenta lane. All sequences were retrieved from the UniProt knowledge database [251]. The sequence alignments were done using the program Clustal-W2 [252] and then manually tuned in order to maximize sequence similarities
Throughout family GH13, sequence identity is extremely low and only the catalytic triad, i.e., Asp206, Glu230, and Asp297 (Taka-amylase A numbering; [31]), plus the arginine (Arg204) positioned two residues before the catalytic nucleophile (Fig. 2) are conserved invariantly [46]. This is, however, not applicable for the non-enzymatic GH13 members, that, depending on their taxonomic origin, may not contain the catalytic residues [23, 25].
In general, however, most functionally important and other conserved residues for any GH13 family member are found in the 4–7 CSRs [46] (Fig. 2). The four best known regions, i.e., CSRs I, II, III, and IV, were well established in 1986 by comparison of 11 α-amylases originating from microorganisms, plants and animals [48]. It is worth mentioning that the CSRs I, II, and IV were initially proposed by Toda et al. [49] who pointed out these regions for Taka-amylase A and pig pancreatic α-amylase, then by Friedberg [50] who added the regions in the B. amyloliquefaciens α-amylase and finally by Rogers [51] who completed the picture by describing them in the barley α-amylase. The three additional regions, i.e., CSRs V, VI, and VII, were identified later [52, 53]. The first four CSRs are located at or near the C-termini of strands β3, β4, β5 and β7 of the catalytic (β/α)8-barrel domain and include the catalytic triad. The three additional CSRs, positioned near the C-terminus of domain B and at or near the C-termini of the barrel strands β2 and β8, contain residues that may be used to distinguish the GH13 specificities from each other.
Clan GH-H
Nowadays, enzymes related structurally to α-amylase are represented by the CAZy clan GH-H consisting of three families, i.e., the families GH70 and GH77 in addition to the main family GH13 [4, 10].
The family GH70 contains glucosyltransferases having a circularly permuted version of the α-amylase-type (β/α)8-barrel catalytic domain, first predicted by MacGregor et al. [54] and confirmed by solving the three-dimensional structure of Lactobacillus reuteri glucansucrase [55] followed by the structures of a few closely related enzymes [56–58].
The members of family GH77 are 4-α-glucanotransferases with a regular catalytic (β/α)8-barrel, but lacking the domain C that follows the barrel in family GH13 [59–61]. Moreover, in several GH77 4-α-glucanotransferases from Borrelia, the above-mentioned conserved arginine that is situated two residues preceding the catalytic nucleophile, in CSR II, is substituted by a lysine [62]. This means that, when the enzymes of clan GH-H are considered as a whole, only the catalytic triad is truly invariantly conserved [63].
Despite the observed differences between the individual GH families of clan GH-H, there is no doubt that all the members of this clan containing α-amylases (i.e., GH13, GH70, and GH77) share a common ancestor [64, 65] and may be readily discriminated from the remotely homologous family GH31 of α-glucosidases [66].
Subfamilies in GH13
The many specificities, large number of sequences, and obvious subgroups of enzymes, e.g., the so-called oligo-1,6-glucosidase and neopullulanase subfamilies [67], suggested the need for further subdivision of GH13. A major breakthrough to describe the family members at a lower level of hierarchy came in 2006 when GH13 was broken up into 35 subfamilies by CAZy curators [6]. This basically reflects the idea that there are groups of enzymes in family GH13 that exhibit, within such a subfamily, a substantially higher degree of similarity in sequence, taxonomy, and/or specificity than in family GH13 as a whole. Currently, 37 subfamilies of GH13 have been defined (CAZy; [4]), but several sequences and characterized enzymes are not yet assigned to a subfamily [38, 68–71].
Subfamily GH13_1
The subfamily GH13_1 covers the eukaryotic α-amylases from fungi and yeast only [6] with no α-amylase of a different taxonomic origin (CAZy; [4]). The number “1” for this GH13 subfamily may well reflect the fact that the α-amylase from A. oryzae (i.e., Taka-amylase A; [49]) is classified here, which was the first α-amylase with its three-dimensional structure solved (Fig. 1; [31]). Tertiary structures are available also for two additional closely related α-amylases from Aspergillus niger; one being the so-called acid-stable α-amylase [72] and the other one exhibiting 100 % sequence identity to Taka-amylase A [73]. There are two additional Taka-amylase A tertiary structures exhibiting an increased thermostability, one with chemically modified Asp197 [74] and the other as a heavy-atom derivative [75].
Although there is no structure as yet for GH13_1 α-amylase from a yeast (CAZy; [4]), several yeast α-amylases were demonstrated to possess SBDs of families CBM20, e.g., the enzymes from Cryptococcus sp. S-2 [76] and Cryptococcus flavus [77], or CBM21, e.g., the enzymes from Lipomyces kononenkoae [78] and Lipomyces starkeyi [79]. Some show an ability to degrade raw starch without a distinct SBD, for example the α-amylase of Saccharomycopsis fibuligera KZ [80]. This may reflect the presence of surface (i.e., secondary) binding sites, which is a general feature of CAZy [81]. Approximately half of the known surface binding sites have been found within several GH13 subfamilies [82], but they are best known from barley and other plant α-amylases of subfamily GH13_6 [83]. In the subfamily GH13_1, a surface binding site was seen, e.g., outside the active site in the A. niger α-amylase structure [73]. While SBDs may be present also in some fungal α-amylases, e.g., from Aspergillus kawachii [84], it is not clear why some of these α-amylases contain an SBD whereas others do not [40, 41, 85].
Interestingly, the GH13_1 members exhibit similarities in their CSR VI (strand β2) to cyclodextrin glucanotransferases from GH13_2 and in CSR VII (strand β8) to α-glucosidases from GH13_31, respectively [46]. This relatedness was also seen in the evolutionary tree of the entire family GH13 [6], where subfamilies GH13_1 and GH13_2 are adjacent to each other, and the GH13_31 α-glucosidases are on the neighboring branch as a part of the so-called oligo-1,6-glucosidase subfamily cluster covering several GH13 enzyme specificities [67]. Structural details can be found in various three-dimensional structures of the subfamily GH13_2 cyclodextrin glucanotransferases [86, 87] and subfamily GH13_31 that contains oligo-1,6-glucosidases [88, 89], α-glucosidase [90], dextran glucosidases [91, 92], and sucrose isomerases [93–95]. The presence of a glutamic acid residue in the position i-3 from the transition-state stabilizer, i.e., the invariant aspartic acid residue at the strand β7 in CSR IV (Fig. 2) may be considered as another characteristic sequence feature of most subfamily GH13_1 members.
Subfamilies GH13_5 and GH13_28
The subfamilies GH13_5 and GH13_28 include bacterial liquefying and saccharifying α-amylases, respectively. Tertiary structures are available for three typical representatives of the liquefying α-amylases of GH13_5, i.e., from B. licheniformis (Fig. 1; [96, 97]), Bacillus stearothermophilus [98] and B. amyloliquefaciens [99] and one saccharifying α-amylase of GH13_28 from Bacillus subtilis [100]. Structures have been determined for additional GH13_5 α-amylases, e.g., that from Bacillus halmapalus [101], for which also secondary (surface) binding sites were observed [102], two others from Bacillus strains [103, 104], and even an exo-amylase from Bacillus sp. 707 [105], i.e., a maltohexaose-producing amylase [106] also classified in subfamily GH13_5 (CAZy; [4]). Interestingly, from the evolutionary point of view, the unclassified polyextremophilic α-amylase AmyB from Halothermothrix orenii [70] that possesses an N-terminal domain preceding the canonical (β/α)8-barrel (Fig. 1), seems to be closely related to GH13_5 subfamily enzymes (Fig. 3).
Fig. 3.

Evolutionary trees of GH13 α-amylases representing the individual α-amylase subfamilies. The trees are based on the alignment of: a catalytic (β/α)8-barrel including domain B with succeeding domain C (653 positions; aligned in Fig. 2); and b seven GH13 characteristic CSRs (52 residues; highlighted in Fig. 2). The names of the α-amylases are explained in the legend to Fig. 2. The evolutionary trees were calculated as a Phylip-tree type using the neighbor-joining clustering [253] and the bootstrapping procedure [254] (the number of bootstrap trials used was 1,000) implemented in the ClustalX package [252], and then displayed with the program TreeView [255]
Although the enzymes of these two subfamilies possess quite different sequences, e.g., one of the most obvious differences is the length and sequence of domain B [23], both these α-amylase subfamilies belong to the so-called “amylase” part in the evolutionary tree of the entire family GH13 covering subfamilies 5, 6, 7, 15, 24, 27, 28, and 32 [6].
Later, some fungal α-amylases produced intracellularly were added to the subfamily GH13_5 [107]. These enzymes in some cases, e.g., from Histoplasma capsulatum [108] and Paracoccidioides brasiliensis [109], seem to be associated with virulence by their involvement in the production of an outer α-(1,3)-glucan layer. Recently, this subfamily was increased by addition of a few potential α-amylases from Archaea (CAZy; [4]).
In subfamily GH13_28, the presence of the SBD from family CBM26 occurring as repeated motifs in α-amylases from lactobacilli [110, 111] is of a special interest [112, 113]. It should be pointed out that the α-amylases from GH13_5 are closely related to plant and archaeal α-amylases from subfamilies GH13_6 and GH13_7 (Fig. 3), whereas those from GH13_28 are closer to animal α-amylases from subfamilies GH13_15 and GH13_24 [80, 107, 114, 115].
Subfamilies GH13_6 and GH13_7
The subfamilies GH13_6 and GH13_7 represent mainly α-amylases from plants and Archaea, respectively, which were originally revealed as constituting a group of closely related α-amylases [114, 116]. Such a close relatedness (Fig. 3) is remarkable not only due to a long taxonomical distance between prokaryotic archaebacteria and eukaryotic plants, but also because of, for example, marked differences in thermostability of archaeal and plant α-amylases. While plant α-amylases do not have exceptional thermostability, the GH13_7 α-amylases are, in general, produced by hyperthermophilic archaeons from various strains of two genera, Pyrococcus and Thermococcus [117], and these enzymes tend to be active and stable at temperatures around 80 °C and higher.
The best characterized representatives of the GH13_7 subfamily are the α-amylases from Thermococcus hydrothermalis [118], Thermococcus onnurineus [119], Pyrococcus furiosus [120, 121], and Pyrococcus woesei [122], for which also the three-dimensional structure has already been determined [123]. Recently, a few hypothetical α-amylases from Bacteria were assigned to the GH13_7 subfamily (CAZy; [4]) that was for a long period considered to be solely the archaeal α-amylase subfamily [107, 114, 115, 124]. All of these hypothetical enzymes originate interestingly from flavobacteria.
On the other hand, the subfamily GH13_6 basically covers higher plant α-amylases and those from green algae, but the bacterium Saccharophagus degradans was found to contain in its genome a plant-like copy of α-amylase in addition to a bacterial one, indicating a horizontal gene transfer event [115]. At present, there may be four GH13_6 α-amylases from Bacteria, the one from S. degradans, two from strains of Spirochaeta thermophila and one from Stigmatella aurantiaca (CAZy; [4]). With regard to the best characterized plant α-amylases, these are the two barley isozymes AMY1, a low pI isozyme [125] and AMY2, a high pI isozyme [126], with solved three-dimensional structures (Fig. 1) determined both in free forms and as complexes [127–130]. α-Amylases from other higher plants, such as wheat [131], rice [132], maize [133], kidney bean [134], apple [135], Arabidopsis thaliana [136], banana [137], and others are also included in GH13_6 (CAZy; [4]). Interestingly, the plant α-amylases belong to the GH13 α-amylases having the shortest polypeptide chain (Fig. 2).
Surface binding sites were observed among the representatives of both subfamilies. In addition to four such sites seen in the structure of the archaeal GH13_7 α-amylase from P. woesei [123], the surface binding sites have been best studied in two barley α-amylase isozymes [129, 130, 138–140], especially the one located in domain C of the low pI isozyme AMY1 called “a pair of sugar tongs” [129] that shows no equivalent binding in the counterpart high pI isozyme AMY2 [141].
Both GH13_6 and GH13_7 subfamilies were originally revealed as two closely related groups positioned in the evolutionary tree on adjacent branches [114]. More recent studies have indicated [107, 115, 142, 143], however, that these two together with the subfamily GH13_5 belong to one evolutionary cluster of α-amylases (Fig. 3), both when the phylogenetic tree is based on the alignment of CSRs only and when a substantial part of amino acid sequences, typically the catalytic domain, are included in the calculation (e.g., Fig. 2). This applies even in the evolutionary tree of the entire family GH13 in the so-called “amylase” part [6]. The subfamilies GH13_6 and GH13_7 share several unique sequence features that discriminate them from the α-amylases in the remaining GH13 subfamilies [114]. One of the most notable features may be the Gly202 (T. hydrothermalis α-amylase numbering; [118]) at the end of the CSR II at the strand β4 of the catalytic (β/α)8-barrel (Fig. 2) the carbonyl group of which serves as a specific ligand for the calcium ion in the complex structure of barley α-amylase (high pI isozyme AMY2) with acarbose [128] and could play the same role in the archaeal counterpart, i.e., the P. woesei α-amylase [123].
Subfamilies GH13_15, GH13_24 and GH13_32
The α-amylase subfamilies GH13_15, GH13_24 and GH13_32 represent mainly the α-amylases from insects, animals (including mammals) and Actinobacteria, respectively. Their mutual relatedness (Fig. 3) was revealed in 1994 [53] and confirmed in many subsequent studies [107, 115, 142–144] including the comparison made when the entire family GH13 was officially divided into subfamilies [6].
A special case is represented by the α-amylase from the Antarctic psychrophile A. haloplanktis [145] that was demonstrated to exhibit close similarity to animal α-amylases [53, 115, 146], but up to now has not been assigned to any GH13 subfamily (CAZy; [4]). Its three-dimensional structure is available [68, 69] and based on structural studies [12] this α-amylase was also found to perform a transglycosylation reaction (Fig. 1e). Together with related animal α-amylases (Fig. 3) it belongs to a group of the so-called chloride-activated α-amylases [147, 148] and is a useful model for comparative studies [149].
Among insects, the α-amylases belong to the subfamily GH13_15, and the α-amylases from a model organism Drosophila melanogaster [150] and related fruit flies have been the subject of many evolutionarily oriented studies [151–153]. The tertiary structure is available for the α-amylase from a yellow meal worm Tenebrio molitor [154, 155].
In the subfamily GH13_24, covering animals from, for example, molluscs [156, 157] and arthropods [158] to vertebrates (i.e., frogs, fishes, birds, and mammals including human beings; [159]), the best known representatives with tertiary structures available are pig pancreatic α-amylase [160–163] plus those from human saliva [164] and pancreas [165]. Recently the structure of the α-amylase of the fish Oryzias latipes was solved [166]. The surface binding sites were also well recognized in tertiary structures of several subfamily GH13_24 members, e.g., in the α-amylases from pig pancreas [163, 167–169] and both human saliva [13] and pancreas [170].
The third subfamily GH13_32 interestingly contains bacterial α-amylases mostly from actinomycetes. Some of these amylases may exhibit a maltotriohydrolase specificity (EC 3.2.1.116), e.g., those from Thermobifida fusca [171] and Brachybacterium sp. [172], both belonging to Actinobacteria. Such α-amylases from Actinobacteria are often called “animal-type” α-amylases [115]. In this subfamily, the halophilic α-amylase from Kocuria varians has been described recently [173] as possessing, C-terminal to the catalytic domain, two tandem copies of an SBD of the family CBM25 [174, 175]. A similar domain arrangement was found previously in the α-amylase from Bacillus sp. No. 195 [176] that also belongs to the subfamily GH13_32 (CAZy; [4]). Typical GH13_32 α-amylases from streptomycetes [177, 178] may also contain an SBD, usually of the family CBM20 [41] at their C-terminus. A very recent evolutionary study has identified the subfamily GH13_32 type of α-amylase in basidiomycetes of fungi, i.e., Eucarya [179] and the authors, in agreement with another study [180], concluded that the gene donor may have originated from Actinobacteria.
Subfamily GH13_27
The subfamily GH13_27 covers a small group (~50 sequences) of bacterial α-amylases (Fig. 3) recognized as homologous before the CAZy classification was established [181]. In most studies [53, 80, 115] this subfamily is usually represented by two experimentally characterized α-amylases from Aeromonas hydrophila [182] and Xanthomonas campestris [181]. Although no three-dimensional structure is available for any GH13_27 α-amylase, there is no doubt—based on sequence comparison—these enzymes are typical 3-domain GH13 α-amylases (i.e., without additional domains such as SBDs). In addition to the two representatives from A. hydrophila and X. campestris, two α-amylases from Pseudomonas sp. KFCC 10818 have been cloned, expressed, sequenced, and characterized [183, 184].
Subfamily GH13_36
The subfamily GH13_36 seems to contain a group of α-amylases possessing related GH13 enzyme specificities [185]. This subfamily was originally defined as the group of amylolytic enzymes with an “intermediary” position in the evolutionary tree between the polyspecific subfamilies of oligo-1,6-glucosidase and neopullulanase [67]. That proposal was mainly based on a specific sequence in the CSR V—typically QPDLN and MPKLN for oligo-1,6-glucosidases and neopullulanases, respectively, the “intermediary” group being characterized by MPDLN (Fig. 2). Nowadays, the oligo-1,6-glucosidase and neopullulanase subfamilies cover several CAZy-curated GH13 subfamilies [6]. The “intermediary” group was, however, assigned to the subfamily GH13_36 only recently (CAZy; [4]). Only 11 GH13_36 members have been biochemically characterized to any extent, although in some cases, these enzymes were described just as an “amylase” [185], e.g., the enzymes from Dictyoglomus thermophilum AmyC [186] and Bacillus megaterium [187]. The activity towards α-1,6-branched glucans together with a transferase ability were, however, demonstrated for the amylolytic enzyme from B. megaterium [188, 189] as well as for the periplasmic one from X. campestris [190]. Furthermore, the GH13_36 “amylase” from Paenibacillus polymyxa was able to release panose from pullulan [191, 192], the one from Bacillus clarkii exhibited predominating activity toward γ-cyclodextrins [193] and the lipoprotein amylase from Anaerobranca gottschalkii showed both cyclodextrin-hydrolyzing and transglycosylating activities [194]. No additional activity was observed for the other lipoprotein α-amylase from Thermotoga maritima [195] or for the halothermophilic α-amylase from H. orenii [196], for which the three-dimensional structure is available as the only representative of the subfamily GH13_36 [197].
Recently established subfamily GH13_37
The subfamily GH13_37 is the most recently established α-amylase subfamily in CAZy (CAZy; [4]). It was created based on isolation and phylogenetic analysis of a novel α-amylase designated AmyP from a marine metagenomic library [198]. The enzyme was later shown to exhibit the ability to degrade raw starch [199]. Currently this subfamily contains, in addition to the AmyP, <20 hypothetical members that come mostly from marine bacteria (CAZy; [4]). These α-amylases are most closely related to maltohexaose-producing amylases from the subfamily GH13_19 [198]. If only α-amylases are considered, however, the subfamily GH13_37 members share the branch of the evolutionary tree with those from the subfamily GH13_36 (Fig. 3). Of special interest is the fact that they may lack domain B (Fig. 1) since the loop connecting the strand β3 to helix α3 of their catalytic (β/α)8-barrel seems to be too short (i.e., <25 residues; Fig. 2) to form a domain B typical of the family GH13. The situation will be clearer when the announced three-dimensional structure of the novel α-amylase AmyP [200] will be solved and described in detail.
As yet non-defined GH13 subfamily
An additional GH13 α-amylase subfamily that may be defined in CAZy in the near future could be based on the α-amylase BaqA from Bacillus aquimaris described very recently by Puspasari et al. [71]. The fact that the source is again a marine bacterium and the enzyme is also a raw starch-degrading α-amylase [201] deserves attention. However, this new group of α-amylases seems to be most closely related to those from subfamily GH13_1, especially if only the GH13 characteristic CSRs I–VII are considered (Fig. 3). The α-amylases from GH13_37, together with the above-mentioned GH13_36 intermediary α-amylases and the GH13_19 maltohexaose-producing amylases, were found to occupy the adjacent cluster in the evolutionary tree [71]. The exclusive sequence feature of this newly proposed GH13 α-amylase subfamily is the presence of two consecutive tryptophans (Trp201 and Trp202, B. aquimaris α-amylase numbering), located at helix α3 (Fig. 2) preceding strand β4 (i.e., the CSR II) of the catalytic (β/α)8-barrel domain, that may indicate a surface binding site [71]. The same feature is present in α-amylase homologues from Geobacillus thermoleovorans [202] and Anoxybacillus sp. SK3–4 and sp. DT3–1 [203]. The recently solved three-dimensional structure of the G. thermoleovorans α-amylase has shown [204] that the above-mentioned tryptophan pair is positioned in a region with additional aromatic residues exhibiting a structural resemblance to the neopullulanase subfamily GH13_20, especially to the cyclomaltodextrinase from Flavobacterium sp. 92 [205]. In the latter enzyme the aromatic region interacts with domain N, which is not present in α-amylases. Moreover, the second tryptophan of the exclusive tryptophan pair (W205, G. thermoleovorans α-amylase numbering) is not exposed to the solvent but buried [204]. Further, it was pointed out that the G. thermoleovorans α-amylase resembles in structure, not only GH13_1 and GH13_20 enzymes but also some from the GH13_2 subfamily [204] that contains cyclodextrin glucanotransferases [6].
Notably there is another interesting and yet unclassified α-amylase with three-dimensional structure already solved—the SusG protein from Bacteroides thetaiotaomicron [38]. It is related to subfamilies GH13_36 and GH13_37 and, in a wider sense also to the currently unclassified group represented by marine B. aquimaris α-amylase mentioned above (Fig. 3). The α-amylase SusG is encoded by a member of the sus (starch utilization system) locus of the human gut symbiont B. thetaiotaomicron [206–208] and represents the only GH13 α-amylase with an SBD inserted in domain B (Fig. 1); in this particular case the SBD is of family CBM58 [38].
Family GH57
A new family, GH57, containing α-amylases was created in 1996 [3] after long efforts to find the family GH13 sequence features in two supposed “α-amylases”, one from the thermophilic bacterium D. thermophilum [209] and the other from the hyperthermophilic archaeon P. furiosus [210]. Even after the family GH57 was established, the possibility of a relationship between the two families GH13 and GH57 was not definitively eliminated, although it had proved impossible to align convincingly the two above-mentioned GH57 enzymes with representatives of GH13 α-amylases [211]. The first GH57 crystal structure, however, solved for the 4-α-glucanotransferase from Thermococcus litoralis in 2003, unambiguously confirmed that the GH57 family cannot belong to the GH-H clan [212].
Nowadays the family GH57 represents a second and smaller α-amylase family [213]. It contains ~900 members all originating from prokaryotes with the approximate Archaea:Bacteria ratio 3:1 and, like the main α-amylase family GH13, the family GH57 is a polyspecific family (Fig. 4a; CAZy; [4]). Interestingly, the two fundamental family members, which were originally considered to be α-amylases [209, 210], now are both recognized as 4-α-glucanotransferases: the enzyme from D. thermophilum was re-evaluated as a glucanotransferase in 2004 [214], whereas a transferase activity for the one from P. furiosus was identified immediately [215]. There are five well-established enzyme specificities in the family GH57 [216, 217], i.e., α-amylase [218], 4-α-glucanotransferase [209, 210, 219–221], amylopullulanase [222–225], branching enzyme [226–228], and even α-galactosidase [229]. Noticeably, two additional amylolytic specificities in the family GH57 may be defined by two partially characterized GH57 members: the PF0870 open reading frame from the P. furiosus genome [230] and a non-specified amylase of an uncultured bacterium isolated from a hydrothermal vent in the Atlantic Ocean [231]. Recently, a GH57 enzyme was described possessing dual specificity: the amylopullulanase from the archaeon Staphylothermus marinus also exhibits cyclomaltodextrinase activity [232]. Dual specificity may also be found for the AmyC enzyme from T. maritima, described originally as an α-amylase [226], but its sequence-structural features clearly indicate it may have also branching enzyme activity [217].
Fig. 4.
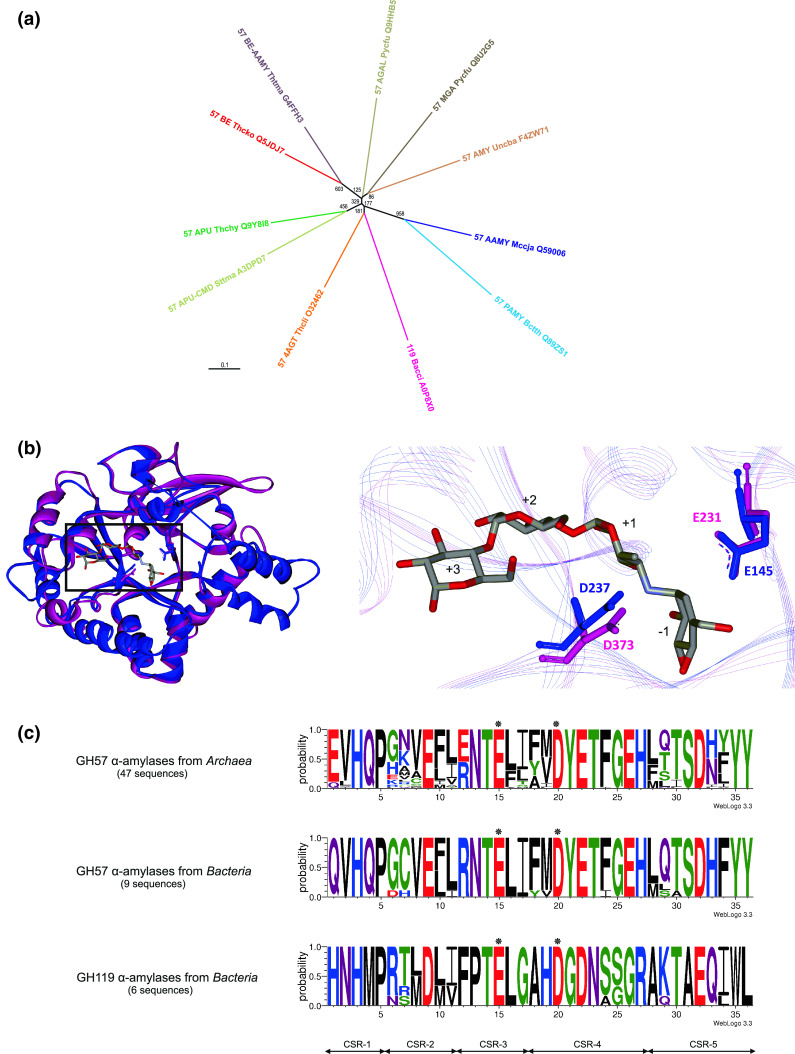
a Evolutionary tree showing the α-amylase representative of the family GH57 among the other GH57 enzyme specificities together with the α-amylase representative of the family GH119. The tree is based on the alignment of the five GH57 characteristic CSRs (36 residues). The name of an enzyme is composed of the GH57 or GH119 family number followed by the abbreviation of specificity (in capitals), abbreviation of source (organism) and the UniProt accession number. All sequences were retrieved from the UniProt knowledge database [251]. The specificities are abbreviated as follows: AAMY α-amylase, PAMY putative α-amylase-like protein, MGA maltogenic amylase, AMY unspecified amylase, APU amylopullulanase, APU-CMD amylopullulanase-cyclomaltodextrinase, BE branching enzyme, BE-AAMY α-amylase-branching enzyme, 4AGT 4-α-glucanotransferase, AGAL α-galactosidase. The organisms are abbreviated as follows: Bacci, Bacillus circulans; Bccth, Bacteroides thetaiotaomicron; Mccja, Methanocaldococcus jannaschii; Pycfu, Pyrococcus furiosus; Sttma, Staphylothermus marinus; Thchy, Thermococcus hydrothermalis; Thcko, Thermococcus kodakaraensis; Thcli, Thermococcus litoralis; Thtma, Thermotoga maritima; Uncba, uncultured bacterium. The evolutionary tree was calculated as a Phylip-tree type using the neighbor-joining clustering [253] and the bootstrapping procedure [254] (the number of bootstrap trials used was 1,000) implemented in the ClustalX package [252], and then displayed with the program TreeView [255]. b Structural models of α-amylases from families GH57 and GH119. Left superimposed modeled structures of the GH57 Methanocaldococcus jannaschii α-amylase (blue) and GH119 Bacillus circulans α-amylase (magenta). The α-helical bundle succeeding the catalytic (β/α)7-barrel was modeled only in the GH57 α-amylase. The rectangle indicates a detailed view on the right. Right a close-up focused on predicted catalytic residues of the α-amylases from GH57 (Glu145 and Asp237) and GH119 (Glu231 and Asp373). Both models were superimposed with the real structure of GH57 Thermococcus litoralis 4-α-glucanotransferase (PDB code: 1K1Y; [212]; not shown). The structural models of α-amylases were obtained from the Phyre server [245] based on the GH57 4-α-glucanotransferase template as follows: residues Met1-Tyr356 of the GH57 α-amylase [216] and Thr121-Asp429 of the GH119 α-amylase [242]. The superimposed part covers 218 Cα-atoms with a 0.75 Å root-mean square deviation; the superimposition was done using the MultiProt server [256]. Acarbose-occupying subsites −1 through +3 [257] from the complex with GH57 4-α-glucanotransferase structure [212] is shown. The structures were visualized with the program WebLabViewerLite (Molecular Simulations, Inc.). c Sequence logos of α-amylases from families GH57 from Archaea and Bacteria and GH119. CSR-1, residues 1–5; CSR-2, residues 6–11; CSR-3, residues 12–17; CSR-4, residues 18–27; CSR-5, residues 28–36. Asterisks signify the catalytic nucleophile (glutamic acid) in position 15 (in CSR-3) and proton donor (aspartic acid) in position 20 (in CSR-4). The logos are based on identifying the CSRs in both families [217, 242] as follows: for 59 α-amylase sequences (47 from Archaea and nine from Bacteria) from family GH57 and for six bacterial GH119 α-amylases. Sequence logos were created using the WebLogo 3.0 server [258]
Although family GH57 shares the retaining reaction mechanism with the main α-amylase family GH13 [87, 228], a (β/α)7-barrel (i.e., an incomplete TIM barrel) is adopted as the fold bearing the catalytic residues [212, 228, 233–235]. Any GH57 member can be characterized by five CSRs—identified originally by Zona et al. [236] and refined recently by Blesak and Janecek [217]—that are clearly different from those characteristic for family GH13 [46]. The catalytic machinery also discriminates the families GH57 and GH13 from each other, because a glutamic acid at strand β4 (Glu123 in T. litoralis 4-α-glucanotransferase) and aspartic acid at strand β7 (Asp214) of the (β/α)7-barrel act as the catalytic nucleophile and proton donor, respectively [212, 228]. Remarkably, in GH57 enzymes, there is no third catalytic amino acid residue like the transition-state stabilizer necessary in family GH13 [10, 87, 237]. The (β/α)7-barrel domain is succeeded in every GH57 member by a helical segment consisting of a bundle of 3–4 α-helices [216, 217]. Since the CSR-5 positioned in this α-helical bundle was revealed to contain functionally essential residues [228], it was proposed that both the barrel and the helical bundle together form the catalytic area of enzymes in the family GH57 [217].
α-Amylase and its putative homologues
It should be pointed out that although the family GH57 is considered to be an α-amylase family, the true α-amylase (EC 3.2.1.1) enzyme specificity has still not been confirmed unambiguously. The only evidence comes from the study by Kim et al. [218] who showed that the α-amylase from the methanogenic archaeon Methanococcus jannaschii degrades soluble starch. The uncertainty concerning the exact specificity has arisen from the fact the α-amylase was also able to degrade pullulan at a relative rate of 82 % of that determined for starch [218, 238]. Thus the potential α-amylase, identified first as a hypothetical MJ1611 open reading frame in the M. jannaschii genome [239], is the leading family GH57 member warranting the “α-amylase” designation [240].
What is, however, more interesting is the observation that family GH57 contains members, i.e., the so-called α-amylase-like proteins, exhibiting clear sequence features of the α-amylase from M. jannaschii, but simultaneously lacking one or both catalytic residues [216]. With regard to their origin, GH57 α-amylases come predominantly from Archaea (~80 %), whereas the GH57 α-amylase-like proteins originate mostly from Bacteria (~85 %). Both of these groups contain proteins approximately 500 amino acid residues long that exhibit a high degree of sequence identity to each other, especially in the CSRs that have been suggested to be sequence fingerprints of the individual family GH57 enzyme specificities [217]. The most substantial difference discriminating the α-amylases and α-amylase-like proteins of family GH57 from each other is the presence of both characteristic catalytic residues in α-amylases (Fig. 4b) and the lack of one or both of these residues in their homologues [216]. The catalytic nucleophile located at the strand β4 of the catalytic (β/α)7-barrel in CSR-3 (Glu145 in M. jannaschii α-amylase) and proton donor at the strand β7 in CSR-4 (Asp237) are most frequently substituted by a serine and glutamic acid, respectively, in amylase-like proteins that are found mainly in two genera Bacteroides and Prevotella, both from the order Bacteroidales of Bacteria.
The family GH57 enzymes with specificities of amylopullulanase, branching enzyme and 4-α-glucanotransferase contain some sequence-structural segments at their C-termini additional to catalytic (β/α)7-barrel with succeeding three-helix bundle [212, 228, 234, 235], while both α-amylases and α-amylase-like proteins seem to be composed of only the barrel and helical bundle [217]. A similar two-domain arrangement is also found in α-galactosidases, but the domain covering the α-helical region in α-amylases seems usually to be ~50–100 residues longer than that present in the α-galactosidases [217]. With regard to the N-terminus, there is probably no signal peptide in α-amylases and α-amylase-like proteins since the CSR-1 is almost exclusively located very close to the N-terminus [216] and no information is available on a signal peptide for the only characterized α-amylase from M. jannaschii [218].
The unique sequence features that discriminate both α-amylases and α-amylase-like proteins from the remaining family GH57 enzyme specificities as well as from each other are seen in their CSRs and best represented by their sequence logos. They were suggested to define the so-called sequence fingerprints of a given enzyme specificity [217]. Based on a detailed bioinformatics analysis of 367 sequences resulting in creating the sequence logos for five GH57 enzyme specificities, the positions 1, 12, 13, 21, 27, and 35–36 were revealed to be specifically characteristic for α-amylase as follows (Fig. 4c): (1) positions 1 (CSR-1) and 12 (CSR-3) contain mostly glutamic acid or glutamine and arginine or glutamic acid, respectively, with all four remaining well-established GH57 specificities being characterized by a histidine and tryptophan in the corresponding positions; (2) positions 13 (CSR-3) and 21 (CSR-4) are exclusively occupied by invariant asparagine and tyrosine, respectively, the other specificities possessing different residues there and not so strictly conserved; (3) the position 27 (CSR-4) is an invariant histidine, but this position is not so unique for α-amylases since a corresponding histidine can also be found in amylopullulanases; and (4) positions 35–36 at the end of logo (CSR-5) consist of two adjacent tyrosines and may represent the most unambiguous GH57 α-amylase feature because a tyrosine residue has not been seen at these positions in any GH57 sequences ascribed to a specificity other than that of α-amylase. It should be emphasized here that these last two positions in a GH57 sequence logo (positions 35–36; CSR-5) represent a sequence fingerprint that most reliably distinguishes the individual enzyme specificities from each other [217].
Sequence logos reveal also specific differences between α-amylases and their α-amylase-like protein counterparts [216]. For example: (1) the invariant proline in position 5 (the end of CSR-1) in α-amylases is often substituted by an isoleucine in bacterial α-amylase-like proteins; (2) the position 7 (CSR-2) is frequently occupied by a cysteine if either of the two, i.e., an α-amylase and/or an α-amylase-like protein, originates from Bacteria (for the α-amylase-like proteins from the genus Bacteroides, there are two adjacent cysteines in the CSR-2); (3) highly specific for α-amylases is the presence of three aromatic residues in positions 18, 21 and 24 (CSR-4), whereas only the position 18 may be considered as aromatic one in the α-amylase-like proteins; and (4) the last three positions (34–36; CSR-5) of the α-amylase sequence logo are mostly aromatic residues, too, the middle position 35 being replaced by an arginine in the α-amylase-like proteins. These specific differences between the enzymatically active α-amylases and their most probably inactive α-amylase-like counterparts from the family GH57 are, however, still smaller than the differences between the α-amylases and the other well-established enzymes specificities within GH57, as indicated by the evolutionary tree (Fig. 4a).
Family GH119
The family GH119 was established in 2006 following the study by Watanabe et al. [241] who described a novel α-amylase as a product of the gene IgtZ from Bacillus circulans AM7 and found no obvious sequence similarity to any α-amylase from either family GH13 or GH57. The specificity of α-amylase was ascribed to the IgtZ protein based on production of glucose and maltooligosaccharides up to maltopentaose from maltooligosaccharides longer than four glucose units, amylose and soluble starch [241]. The α-amylase IgtZ also contains SBDs of families CBM20 (1 copy) and CBM25 (2 copies). The family GH119 may thus be considered as the third CAZy α-amylase family, but it is worth mentioning that currently, i.e., 7 years after it was created in the CAZy database, the family is still very small since only five hypothetical bacterial proteins obtained from genome sequencing projects have been added to the α-amylase IgtZ described originally (CAZy; [4]). Until recently, when a close relatedness of the family GH119 to GH57 was revealed (Fig. 4b; [242]), information on family GH119 concerning sequence-structural details (e.g., catalytic machinery and fold) was, in fact, lacking [243].
A relatedness to the family GH57
Based on a detailed in silico analysis that involved comparison of amino acid sequences of all family GH119 members (CAZy; [4]) in combination with the BLAST tool [244] and tertiary structure modeling at the Phyre-2 [245] and SwissModel [246] servers, an unambiguous evolutionary relatedness was revealed between families GH119 and GH57 [242]. The five CSRs characteristic of the family GH57 [217, 236] were identified in the sequences of all six GH119 members. It was thus possible to predict the GH119 catalytic residues, i.e., Glu231 and Asp373 in the sequence of the α-amylase IgtZ from B. circulans, which are conserved invariantly in GH119 (Fig. 4c). This prediction is further supported by three-dimensional structure modeling indicating that the family GH119 shares with the family GH57 both the catalytic (β/α)7-barrel fold and catalytic machinery (a glutamic acid as the catalytic nucleophile and an aspartic acid as the proton donor at the strands β4 and β7 of the barrel; Fig. 4b). Despite the clear similarity, the family GH119 retains its own identity in the evolutionary tree common for both families (Fig. 4a) reflecting its characteristic sequence features also in the CSRs [242]. As suggested, the creation of a novel CAZy GH clan of families GH57 and GH119 is thus highly probable but this needs to be confirmed by solving the tertiary structure and identifying experimentally the catalytic residues in a family GH119 representative [242].
Family GH126
The last CAZy GH family supposedly containing the specificity of α-amylase is the family GH126, created on the basis of a report by Ficko-Blean et al. [17]. They assessed the CPF_2247 protein product from the Clostridium perfringens ATCC 13124 genome as an α-amylase since the enzyme showed activity on maltooligosaccharides, amylose and glycogen, but no attempt was made to show that anomeric configuration is conserved. Remarkably, although the CPF_2247 defines its own family GH126, which currently also contains approximately 60 hypothetical proteins from Bacteria (CAZy; [4]), it exhibits ~20 % sequence identity to endo-β-1,4-glucanases from the family GH8 [17]. Moreover, the CPF_2247 is structurally most closely related to GH8 endoglucanase CelA [247] and GH48 cellobiohydrolase CelS [248] both from Clostridium thermocellum. These two families form together the clan GH-M (CAZy; [4]). The enzymes from the clan GH-M employ, however, an inverting mechanism of glucosidic bond cleavage and the CPF_2247 “α-amylase” seems to share with them, in addition to a catalytic (α/α)6-barrel fold, its catalytic nucleophile (Glu84) plus a few other invariantly conserved and functionally important residues (Fig. 5; [17]). These facts are not consistent with the general definition of α-amylase enzyme specificity [9, 87] employing the retaining mechanism, that has been confirmed by crystal structures of various α-amylases from family GH13 [11, 12, 14, 73, 100, 101, 123, 128, 130, 148, 162] and representatives from family GH57 [212, 228].
Fig. 5.
Structure of the “α-amylase” of family GH126. a GH126 structure of the α-amylase from Clostridium perfringens (PDB code: 3REN; [17]) showing the (α/α)6-barrel with highlighted residues (colored by element) involved in catalysis: Glu84 (general acid), Asp136 (general base), and Tyr194 (contributing to catalysis). b Superimposition of Clostridium perfringens GH126 α-amylase (blue) with Clostridium thermocellum GH8 endoglucanase CelA (red PDB code: 1KWF; [247]). The superimposed part covers 193 Cα-atoms with a 1.93 Å root-mean square deviation; the superimposition was done using the MultiProt server [256]. The detailed view focuses on the catalytic residues in the structure of GH8 endoglucanase CelA (Glu95 and Asp278 with Tyr215) and the proposed residues in the GH126 α-amylase (Glu84 and Asp136 with Tyr194). Cellopentaose occupying subsites −3 through +2 [257] in complex with GH8 endoglucanase CelA is shown. A comparison with the family GH15 glucoamylase, i.e., an α-glucan-active enzyme with (α/α)6-barrel catalytic fold employing the inverting mechanism [259], reveals less similarity since the superimposed part between the GH126 α-amylase and Aspergillus niger GH15 glucoamylase [260] covered only 146 Cα-atoms with a 1.97 Å root-mean square deviation. The structures were retrieved from the PDB [250] and visualized with the program WebLabViewerLite (Molecular Simulations, Inc.)
Despite these apparent inconsistencies, monitoring the depolymerization of various polysaccharides by the CPF_2247 using thin-layer chromatography revealed clear activity on amylose and glycogen (with production of a mixture of maltooligosaccharides from maltose to maltoheptaose), but no activity on pullulan and cellulose [17]. Furthermore, reaction products from the series of maltotriose to maltoheptaose tested by high-performance anion exchange chromatography showed that maltopentaose is the minimal oligosaccharide substrate for CPF_2247, from which the enzyme removes a single glucose residue [17]. This means that, as well as the difficulty of understanding how an α-amylase could share the catalytic mechanism with inverting β-glucanases, there is also some uncertainty concerning the endo/exo fashion of action of this remarkable GH126 amylolytic enzyme CPF_2247 from C. perfringens.
Conclusions
As documented in the present review, α-amylase ranks among the most frequently occurring CAZy. It is likely that α-amylases are present in families GH57 and GH119, and possibly even in GH126, but α-amylase is considered the main representative of family GH13—known generally as the main α-amylase family. This GH family forms a clan together with families GH70 and GH77, but in the latter two, no α-amylase specificity has been found. Within the family GH13, however, the α-amylases define several GH13 subfamilies, the members of which exhibit closer sequence-structural similarities than observed for the family as a whole. Nevertheless, despite more than 13,500 sequences classified currently in the family GH13, all the members share 4–7 CSRs, the α-amylase-type of (β/α)8-barrel catalytic domain, catalytic machinery, and retaining reaction mechanism. The α-amylase classified in family GH57 employs the same retaining mechanism as GH13 α-amylase. All the family GH57 members differ, however, from those of family GH13 in that they possess their own five CSRs and adopt a (β/α)7-barrel fold (i.e., an incomplete TIM-barrel) bearing catalytic machinery different from that used in the family GH13. All of these GH57 characteristics are likely to be shared by family GH119, which is the third GH family containing α-amylase specificity. The last GH family that has been indicated to contain the specificity of an α-amylase, i.e., the family GH126, is of special interest and the situation may be clarified when further biochemical characterization and structure determination have been carried out, since proteins of the family GH126 exhibit an unambiguous homology to β-glucan-active hydrolases of families GH8 and GH48 that employ an inverting reaction mechanism.
Future discoveries relating to α-amylases may show wider, but related, specificities and new structures, giving new families within the CAZy system, but thorough biochemical characterization and structure determination is required before new and surprising conclusions can be accepted.
Acknowledgments
SJ thanks the Slovak Research and Development Agency for financial support under contract No. LPP-0417-09 and the Slovak Grant Agency VEGA for Grant No. 2/0148/11. BS thanks the Danish Research Council for Independent Research | Natural Sciences (FNU) for financial support under Grant No. 09-072151.
Abbreviations
- CAZy
Carbohydrate-Active enZymes
- CBM
Carbohydrate-binding module
- CSR
Conserved sequence region
- GH
Glycoside hydrolase
- SBD
Starch-binding domain
- TIM
Triose-phosphate isomerase
References
- 1.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henrissat B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henrissat B, Bairoch A. Updating the sequence-based classification of glycosyl hydrolases. Biochem J. 1996;316:695–696. doi: 10.1042/bj3160695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henrissat B, Davies GJ. Structural and sequence-based classification of glycoside hydrolases. Curr Opin Struct Biol. 1997;7:637–644. doi: 10.1016/s0959-440x(97)80072-3. [DOI] [PubMed] [Google Scholar]
- 6.Stam MR, Danchin EG, Rancurel C, Coutinho PM, Henrissat B. Dividing the large glycoside hydrolase family 13 into subfamilies: towards improved functional annotations of α-amylase-related proteins. Protein Eng Des Sel. 2006;19:555–562. doi: 10.1093/protein/gzl044. [DOI] [PubMed] [Google Scholar]
- 7.Aspeborg H, Coutinho PM, Wang Y, Brumer H, 3rd, Henrissat B. Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5) BMC Evol Biol. 2012;12:186. doi: 10.1186/1471-2148-12-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.St John FJ, Gonzalez JM, Pozharski E. Consolidation of glycosyl hydrolase family 30: a dual domain 4/7 hydrolase family consisting of two structurally distinct groups. FEBS Lett. 2010;584:4435–4441. doi: 10.1016/j.febslet.2010.09.051. [DOI] [PubMed] [Google Scholar]
- 9.MacGregor EA. α-Amylase structure and activity. J Protein Chem. 1988;7:399–415. doi: 10.1007/BF01024888. [DOI] [PubMed] [Google Scholar]
- 10.MacGregor EA, Janecek S, Svensson B. Relationship of sequence and structure to specificity in the α-amylase family of enzymes. Biochim Biophys Acta. 2001;1546:1–20. doi: 10.1016/s0167-4838(00)00302-2. [DOI] [PubMed] [Google Scholar]
- 11.Brzozowski AM, Davies GJ. Structure of the Aspergillus oryzae α-amylase complexed with the inhibitor acarbose at 2.0 Å resolution. Biochemistry. 1997;36:10837–10845. doi: 10.1021/bi970539i. [DOI] [PubMed] [Google Scholar]
- 12.Aghajari N, Roth M, Haser R. Crystallographic evidence of a transglycosylation reaction: ternary complexes of a psychrophilic α-amylase. Biochemistry. 2002;41:4273–4280. doi: 10.1021/bi0160516. [DOI] [PubMed] [Google Scholar]
- 13.Ramasubbu N, Ragunath C, Mishra PJ. Probing the role of a mobile loop in substrate binding and enzyme activity of human salivary amylase. J Mol Biol. 2003;325:1061–1076. doi: 10.1016/s0022-2836(02)01326-8. [DOI] [PubMed] [Google Scholar]
- 14.Li C, Begum A, Numao S, Park KH, Withers SG, Brayer GD. Acarbose rearrangement mechanism implied by the kinetic and structural analysis of human pancreatic α-amylase in complex with analogues and their elongated counterparts. Biochemistry. 2005;44:3347–3357. doi: 10.1021/bi048334e. [DOI] [PubMed] [Google Scholar]
- 15.Brzozowski AM, Lawson DM, Turkenburg JP, Bisgaard-Frantzen H, Svendsen A, Borchert TV, Dauter Z, Wilson KS, Davies GJ. Structural analysis of a chimeric bacterial α-amylase. High-resolution analysis of native and ligand complexes. Biochemistry. 2000;39:9099–9107. doi: 10.1021/bi0000317. [DOI] [PubMed] [Google Scholar]
- 16.Janecek S, Svensson B, MacGregor EA. Characteristic differences in the primary structure allow discrimination of cyclodextrin glucanotransferases from α-amylases. Biochem J. 1995;305:685–686. doi: 10.1042/bj3050685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ficko-Blean E, Stuart CP, Boraston AB. Structural analysis of CPF_2247, a novel α-amylase from Clostridium perfringens . Proteins. 2011;79:2771–2777. doi: 10.1002/prot.23116. [DOI] [PubMed] [Google Scholar]
- 18.Svensson B. Regional distant sequence homology between amylases, α-glucosidases and transglucanosylases. FEBS Lett. 1988;230:72–76. doi: 10.1016/0014-5793(88)80644-6. [DOI] [PubMed] [Google Scholar]
- 19.Kuriki T, Imanaka T. Nucleotide sequence of the neopullulanase gene from Bacillus stearothermophilus . J Gen Microbiol. 1989;135:1521–1528. doi: 10.1099/00221287-135-6-1521. [DOI] [PubMed] [Google Scholar]
- 20.MacGregor EA, Svensson B. A super-secondary structure predicted to be common to several α-1,4-d-glucan-cleaving enzymes. Biochem J. 1989;259:145–152. doi: 10.1042/bj2590145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jespersen HM, MacGregor EA, Sierks MR, Svensson B. Comparison of the domain-level organization of starch hydrolases and related enzymes. Biochem J. 1991;280:51–55. doi: 10.1042/bj2800051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Svensson B, Janecek S (2013) Glycoside hydrolase family 13. CAZypedia. http://www.cazypedia.org/. Accessed 18 Mar 2013
- 23.Janecek S, Svensson B, Henrissat B. Domain evolution in the α-amylase family. J Mol Evol. 1997;45:322–331. doi: 10.1007/pl00006236. [DOI] [PubMed] [Google Scholar]
- 24.Fort J, de la Ballina LR, Burghardt HE, Ferrer-Costa C, Turnay J, Ferrer-Orta C, Uson I, Zorzano A, Fernandez-Recio J, Orozco M, Lizarbe MA, Fita I, Palacin M. The structure of human 4F2hc ectodomain provides a model for homodimerization and electrostatic interaction with plasma membrane. J Biol Chem. 2007;282:31444–31452. doi: 10.1074/jbc.M704524200. [DOI] [PubMed] [Google Scholar]
- 25.Gabrisko M, Janecek S. Looking for the ancestry of the heavy-chain subunits of heteromeric amino acid transporters rBAT and 4F2hc within the GH13 α-amylase family. FEBS J. 2009;276:7265–7278. doi: 10.1111/j.1742-4658.2009.07434.x. [DOI] [PubMed] [Google Scholar]
- 26.MacGregor EA. Relationships between structure and activity in the α-amylase family of starch-metabolising enzymes. Starch/Staerke. 1993;45:232–237. [Google Scholar]
- 27.Janecek S. Parallel β/α-barrels of α-amylase, cyclodextrin glycosyltransferase and oligo-1,6-glucosidase versus the barrel of β-amylase: evolutionary distance is a reflection of unrelated sequences. FEBS Lett. 1994;353:119–123. doi: 10.1016/0014-5793(94)01019-6. [DOI] [PubMed] [Google Scholar]
- 28.Svensson B. Protein engineering in the α-amylase family: catalytic mechanism, substrate specificity, and stability. Plant Mol Biol. 1994;25:141–157. doi: 10.1007/BF00023233. [DOI] [PubMed] [Google Scholar]
- 29.Kuriki T, Imanaka T. The concept of the α-amylase family: structural similarity and common catalytic mechanism. J Biosci Bioeng. 1999;87:557–565. doi: 10.1016/s1389-1723(99)80114-5. [DOI] [PubMed] [Google Scholar]
- 30.van der Maarel MJ, van der Veen B, Uitdehaag JC, Leemhuis H, Dijkhuizen L. Properties and applications of starch-converting enzymes of the α-amylase family. J Biotechnol. 2002;94:137–155. doi: 10.1016/s0168-1656(01)00407-2. [DOI] [PubMed] [Google Scholar]
- 31.Matsuura Y, Kusunoki M, Harada W, Kakudo M. Structure and possible catalytic residues of Taka-amylase A. J Biochem. 1984;95:697–702. doi: 10.1093/oxfordjournals.jbchem.a134659. [DOI] [PubMed] [Google Scholar]
- 32.Banner DW, Bloomer AC, Petsko GA, Phillips DC, Pogson CI, Wilson IA, Corran PH, Furth AJ, Milman JD, Offord RE, Priddle JD, Waley SG. Structure of chicken muscle triose phosphate isomerase determined crystallographically at 2.5 angstrom resolution using amino acid sequence data. Nature. 1975;255:609–614. doi: 10.1038/255609a0. [DOI] [PubMed] [Google Scholar]
- 33.Farber GK, Petsko GA. The evolution of α/β barrel enzymes. Trends Biochem Sci. 1990;15:228–234. doi: 10.1016/0968-0004(90)90035-a. [DOI] [PubMed] [Google Scholar]
- 34.Penninga D, van der Veen BA, Knegtel RM, van Hijum SA, Rozeboom HJ, Kalk KH, Dijkstra BW, Dijkhuizen L. The raw starch binding domain of cyclodextrin glycosyltransferase from Bacillus circulans strain 251. J Biol Chem. 1996;271:32777–32784. doi: 10.1074/jbc.271.51.32777. [DOI] [PubMed] [Google Scholar]
- 35.Abe A, Tonozuka T, Sakano Y, Kamitori S. Complex structures of Thermoactinomyces vulgaris R-47 α-amylase 1 with malto-oligosaccharides demonstrate the role of domain N acting as a starch-binding domain. J Mol Biol. 2004;335:811–822. doi: 10.1016/j.jmb.2003.10.078. [DOI] [PubMed] [Google Scholar]
- 36.Boraston AB, Healey M, Klassen J, Ficko-Blean E, Lammerts van Bueren A, Law V. A structural and functional analysis of α-glucan recognition by family 25 and 26 carbohydrate-binding modules reveals a conserved mode of starch recognition. J Biol Chem. 2006;281:587–598. doi: 10.1074/jbc.M509958200. [DOI] [PubMed] [Google Scholar]
- 37.van Bueren AL, Boraston AB. The structural basis of α-glucan recognition by a family 41 carbohydrate-binding module from Thermotoga maritima . J Mol Biol. 2007;365:555–560. doi: 10.1016/j.jmb.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 38.Koropatkin NM, Smith TJ. SusG: a unique cell-membrane-associated α-amylase from a prominent human gut symbiont targets complex starch molecules. Structure. 2010;18:200–215. doi: 10.1016/j.str.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 39.Svensson B, Jespersen H, Sierks MR, MacGregor EA. Sequence homology between putative raw-starch binding domains from different starch-degrading enzymes. Biochem J. 1989;264:309–311. doi: 10.1042/bj2640309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Janecek S, Sevcik J. The evolution of starch-binding domain. FEBS Lett. 1999;456:119–125. doi: 10.1016/s0014-5793(99)00919-9. [DOI] [PubMed] [Google Scholar]
- 41.Janecek S, Svensson B, MacGregor EA. Relation between domain evolution, specificity, and taxonomy of the α-amylase family members containing a C-terminal starch-binding domain. Eur J Biochem. 2003;270:635–645. doi: 10.1046/j.1432-1033.2003.03404.x. [DOI] [PubMed] [Google Scholar]
- 42.Rodriguez-Sanoja R, Oviedo N, Sanchez S. Microbial starch-binding domain. Curr Opin Microbiol. 2005;8:260–267. doi: 10.1016/j.mib.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 43.Machovic M, Janecek S. Starch-binding domains in the post-genome era. Cell Mol Life Sci. 2006;63:2710–2724. doi: 10.1007/s00018-006-6246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Christiansen C, Abou Hachem M, Janecek S, Viksø-Nielsen A, Blennow A, Svensson B. The carbohydrate-binding module family 20—diversity, structure, and function. FEBS J. 2009;276:5006–5029. doi: 10.1111/j.1742-4658.2009.07221.x. [DOI] [PubMed] [Google Scholar]
- 45.Janecek S, Svensson B, MacGregor EA. Structural and evolutionary aspects of two families of non-catalytic domains present in starch and glycogen binding proteins from microbes, plants and animals. Enzyme Microb Technol. 2011;49:429–440. doi: 10.1016/j.enzmictec.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 46.Janecek S. How many conserved sequence regions are there in the α-amylase family? Biologia. 2002;57(Suppl 11):29–41. [Google Scholar]
- 47.Takata H, Kuriki T, Okada S, Takesada Y, Iizuka M, Minamiura N, Imanaka T. Action of neopullulanase. Neopullulanase catalyzes both hydrolysis and transglycosylation at α-(1,4)- and α-(1,6)-glucosidic linkages. J Biol Chem. 1992;267:18447–18452. [PubMed] [Google Scholar]
- 48.Nakajima R, Imanaka T, Aiba S. Comparison of amino acid sequences of eleven different α-amylases. Appl Microbiol Biotechnol. 1986;23:355–360. [Google Scholar]
- 49.Toda H, Kondo K, Narita K. The complete amino acid sequence of Taka-amylase A. Proc Jpn Acad. 1982;B58:208–212. [Google Scholar]
- 50.Friedberg F. On the primary structure of amylases. FEBS Lett. 1983;152:139–140. doi: 10.1016/0014-5793(83)80365-2. [DOI] [PubMed] [Google Scholar]
- 51.Rogers JC. Conserved amino acid sequence domains in α-amylases from plants, mammals, and bacteria. Biochem Biophys Res Commun. 1985;128:470–476. doi: 10.1016/0006-291x(85)91702-4. [DOI] [PubMed] [Google Scholar]
- 52.Janecek S. New conserved amino acid region of α-amylases in the third loop of their (β/α)8-barrel domains. Biochem J. 1992;288:1069–1070. doi: 10.1042/bj2881069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Janecek S. Sequence similarities and evolutionary relationships of microbial, plant and animal α-amylases. Eur J Biochem. 1994;224:519–524. doi: 10.1111/j.1432-1033.1994.00519.x. [DOI] [PubMed] [Google Scholar]
- 54.MacGregor EA, Jespersen HM, Svensson B. A circularly permuted α-amylase-type α/β-barrel structure in glucan-synthesizing glucosyltransferases. FEBS Lett. 1996;378:263–266. doi: 10.1016/0014-5793(95)01428-4. [DOI] [PubMed] [Google Scholar]
- 55.Vujicic-Zagar A, Pijning T, Kralj S, Lopez CA, Eeuwema W, Dijkhuizen L, Dijkstra BW. Crystal structure of a 117-kDa glucansucrase fragment provides insight into evolution and product specificity of GH70 enzymes. Proc Natl Acad Sci USA. 2010;107:21406–21411. doi: 10.1073/pnas.1007531107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ito K, Ito S, Shimamura T, Weyand S, Kawarasaki Y, Misaka T, Abe K, Kobayashi T, Cameron AD, Iwata S. Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans . J Mol Biol. 2011;408:177–186. doi: 10.1016/j.jmb.2011.02.028. [DOI] [PubMed] [Google Scholar]
- 57.Brison Y, Pijning T, Malbert Y, Fabre E, Mourey L, Morel S, Potocki-Veronese G, Monsan P, Tranier S, Remaud-Simeon M, Dijkstra BW. Functional and structural characterization of α-(1,2) branching sucrase derived from DSR-E glucansucrase. J Biol Chem. 2012;287:7915–7924. doi: 10.1074/jbc.M111.305078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pijning T, Vujicic-Zagar A, Kralj S, Dijkhuizen L, Dijkstra BW. Structure of the α-1,6/α-1,4-specific glucansucrase GTFA from Lactobacillus reuteri 121. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012;68:1448–1454. doi: 10.1107/S1744309112044168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Przylas I, Tomoo K, Terada Y, Takaha T, Fujii K, Saenger W, Sträter N. Crystal structure of amylomaltase from Thermus aquaticus, a glycosyltransferase catalysing the production of large cyclic glucans. J Mol Biol. 2000;296:873–886. doi: 10.1006/jmbi.1999.3503. [DOI] [PubMed] [Google Scholar]
- 60.Barends TR, Bultema JB, Kaper T, van der Maarel MJ, Dijkhuizen L, Dijkstra BW. Three-way stabilization of the covalent intermediate in amylomaltase, an α-amylase-like transglycosylase. J Biol Chem. 2007;282:17242–17249. doi: 10.1074/jbc.M701444200. [DOI] [PubMed] [Google Scholar]
- 61.Jung JH, Jung TY, Seo DH, Yoon SM, Choi HC, Park BC, Park CS, Woo EJ. Structural and functional analysis of substrate recognition by the 250s loop in amylomaltase from Thermus brockianus . Proteins. 2011;79:633–644. doi: 10.1002/prot.22911. [DOI] [PubMed] [Google Scholar]
- 62.Godany A, Vidova B, Janecek S. The unique glycoside hydrolase family 77 amylomaltase from Borrelia burgdorferi with only catalytic triad conserved. FEMS Microbiol Lett. 2008;284:84–91. doi: 10.1111/j.1574-6968.2008.01191.x. [DOI] [PubMed] [Google Scholar]
- 63.Machovic M, Janecek S. The invariant residues in the α-amylase family: just the catalytic triad. Biologia. 2003;58:1127–1132. [Google Scholar]
- 64.Jespersen HM, MacGregor EA, Henrissat B, Sierks MR, Svensson B. Starch- and glycogen-debranching and branching enzymes: prediction of structural features of the catalytic (β/α)8-barrel domain and evolutionary relationship to other amylolytic enzymes. J Protein Chem. 1993;12:791–805. doi: 10.1007/BF01024938. [DOI] [PubMed] [Google Scholar]
- 65.Janecek S. α-Amylase family: molecular biology and evolution. Prog Biophys Mol Biol. 1997;67:67–97. doi: 10.1016/s0079-6107(97)00015-1. [DOI] [PubMed] [Google Scholar]
- 66.Janecek S, Svensson B, MacGregor EA. A remote but significant sequence homology between glycoside hydrolase clan GH-H and family GH31. FEBS Lett. 2007;581:1261–1268. doi: 10.1016/j.febslet.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 67.Oslancova A, Janecek S. Oligo-1,6-glucosidase and neopullulanase enzyme subfamilies from the α-amylase family defined by the fifth conserved sequence region. Cell Mol Life Sci. 2002;59:1945–1959. doi: 10.1007/PL00012517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aghajari N, Feller G, Gerday C, Haser R. Crystal structures of the psychrophilic α-amylase from Alteromonas haloplanctis in its native form and complexed with an inhibitor. Protein Sci. 1998;7:564–572. doi: 10.1002/pro.5560070304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aghajari N, Feller G, Gerday C, Haser R. Structures of the psychrophilic Alteromonas haloplanctis α-amylase give insights into cold adaptation at a molecular level. Structure. 1998;6:1503–1516. doi: 10.1016/s0969-2126(98)00149-x. [DOI] [PubMed] [Google Scholar]
- 70.Tan TC, Mijts BN, Swaminathan K, Patel BK, Divne C. Crystal structure of the polyextremophilic α-amylase AmyB from Halothermothrix orenii: details of a productive enzyme–substrate complex and an N domain with a role in binding raw starch. J Mol Biol. 2008;378:852–870. doi: 10.1016/j.jmb.2008.02.041. [DOI] [PubMed] [Google Scholar]
- 71.Puspasari F, Radjasa OK, Noer AS, Nurachman Z, Syah YM, van der Maarel M, Dijkhuizen L, Janecek S, Natalia D. Raw starch-degrading α-amylase from Bacillus aquimaris MKSC 6.2: isolation and expression of the gene, bioinformatics and biochemical characterization of the recombinant enzyme. J Appl Microbiol. 2013;114:108–120. doi: 10.1111/jam.12025. [DOI] [PubMed] [Google Scholar]
- 72.Boel E, Brady L, Brzozowski AM, Derewenda Z, Dodson GG, Jensen VJ, Petersen SB, Swift H, Thim L, Woldike HF. Calcium binding in α-amylases: an X-ray diffraction study at 2.1-Å resolution of two enzymes from Aspergillus . Biochemistry. 1990;29:6244–6249. doi: 10.1021/bi00478a019. [DOI] [PubMed] [Google Scholar]
- 73.Vujicic-Zagar A, Dijkstra BW. Monoclinic crystal form of Aspergillus niger α-amylase in complex with maltose at 1.8 Å resolution. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62:716–721. doi: 10.1107/S1744309106024729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Siddiqui KS, Poljak A, De Francisci D, Guerriero G, Pilak O, Burg D, Raftery MJ, Parkin DM, Trewhella J, Cavicchioli R. A chemically modified α-amylase with a molten-globule state has entropically driven enhanced thermal stability. Protein Eng Des Sel. 2010;23:769–780. doi: 10.1093/protein/gzq051. [DOI] [PubMed] [Google Scholar]
- 75.Sugahara M, Takehira M, Yutani K. Effect of heavy atoms on the thermal stability of α-amylase from Aspergillus oryzae . PLoS ONE. 2013;8:e57432. doi: 10.1371/journal.pone.0057432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Iefuji H, Chino M, Kato M, Iimura Y. Raw-starch-digesting and thermostable α-amylase from the yeast Cryptococcus sp. S-2: purification, characterization, cloning and sequencing. Biochem J. 1996;318:989–996. doi: 10.1042/bj3180989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Galdino AS, Ulhoa CJ, Moraes LM, Prates MV, Bloch C, Jr, Torres FA. Cloning, molecular characterization and heterologous expression of AMY1, an α-amylase gene from Cryptococcus flavus . FEMS Microbiol Lett. 2008;280:189–194. doi: 10.1111/j.1574-6968.2007.01059.x. [DOI] [PubMed] [Google Scholar]
- 78.Steyn AJ, Marmur J, Pretorius IS. Cloning, sequence analysis and expression in yeasts of a cDNA containing a Lipomyces kononenkoae α-amylase-encoding gene. Gene. 1995;166:65–71. doi: 10.1016/0378-1119(95)00633-0. [DOI] [PubMed] [Google Scholar]
- 79.Kang HK, Lee JH, Kim D, Day DF, Robyt JF, Park KH, Moon TW. Cloning and expression of Lipomyces starkeyi α-amylase in Escherichia coli and determination of some of its properties. FEMS Microbiol Lett. 2004;233:53–64. doi: 10.1016/j.femsle.2004.01.036. [DOI] [PubMed] [Google Scholar]
- 80.Hostinova E, Janecek S, Gasperik J. Gene sequence, bioinformatics and enzymatic characterization of α-amylase from Saccharomycopsis fibuligera KZ. Protein J. 2010;29:355–364. doi: 10.1007/s10930-010-9260-6. [DOI] [PubMed] [Google Scholar]
- 81.Cuyvers S, Dornez E, Delcour JA, Courtin CM. Occurrence and functional significance of secondary carbohydrate binding sites in glycoside hydrolases. Crit Rev Biotechnol. 2012;32:93–107. doi: 10.3109/07388551.2011.561537. [DOI] [PubMed] [Google Scholar]
- 82.Cockburn D, Svensson B (2013) Surface binding sites in carbohydrate active enzymes: an emerging picture of structural and functional diversity. In: Lindhorst TK, Rauter AP (eds) SPR carbohydrate chemistry—chemical and biological approaches, vol 39. Royal Society of Chemistry, Cambridge. doi:10.1039/9781849737173-00204
- 83.Møller MS, Cockburn D, Nielsen JW, Jensen JM, Vester-Christensen MB, Nielsen MM, Andersen JM, Wilkens C, Rannes J, Hägglund P, Henriksen A, Abou Hachem M, Willemoës M, Svensson B (2013) Surface binding sites (SBSs), mechanism and regulation of enzymes degrading amylopectin and α-limit dextrins. J Appl Glycosci. doi:10.5458/jag.jag.JAG-2012_023
- 84.Kaneko A, Sudo S, Sakamoto Y, Tamura G, Ishikawa T, Ohba T. Molecular-cloning and determination of the nucleotide-sequence of a gene encoding an acid-stable α-amylase from Aspergillus-kawachii . J Ferment Bioeng. 1996;81:292–298. [Google Scholar]
- 85.Machovic M, Janecek S. The evolution of putative starch-binding domains. FEBS Lett. 2006;580:6349–6356. doi: 10.1016/j.febslet.2006.10.041. [DOI] [PubMed] [Google Scholar]
- 86.Klein C, Schulz GE. Structure of cyclodextrin glycosyltransferase refined at 2.0 Å resolution. J Mol Biol. 1991;217:737–750. doi: 10.1016/0022-2836(91)90530-j. [DOI] [PubMed] [Google Scholar]
- 87.Uitdehaag JC, Mosi R, Kalk KH, van der Veen BA, Dijkhuizen L, Withers SG, Dijkstra BW. X-ray structures along the reaction pathway of cyclodextrin glycosyltransferase elucidate catalysis in the α-amylase family. Nat Struct Biol. 1999;6:432–436. doi: 10.1038/8235. [DOI] [PubMed] [Google Scholar]
- 88.Watanabe K, Hata Y, Kizaki H, Katsube Y, Suzuki Y. The refined crystal structure of Bacillus cereus oligo-1,6-glucosidase at 2.0 Å resolution: structural characterization of proline-substitution sites for protein thermostabilization. J Mol Biol. 1997;269:142–153. doi: 10.1006/jmbi.1997.1018. [DOI] [PubMed] [Google Scholar]
- 89.Yamamoto K, Miyake H, Kusunoki M, Osaki S. Crystal structures of isomaltase from Saccharomyces cerevisiae and in complex with its competitive inhibitor maltose. FEBS J. 2010;277:4205–4214. doi: 10.1111/j.1742-4658.2010.07810.x. [DOI] [PubMed] [Google Scholar]
- 90.Shirai T, Hung VS, Morinaka K, Kobayashi T, Ito S. Crystal structure of GH13 α-glucosidase GSJ from one of the deepest sea bacteria. Proteins. 2008;73:126–133. doi: 10.1002/prot.22044. [DOI] [PubMed] [Google Scholar]
- 91.Hondoh H, Saburi W, Mori H, Okuyama M, Nakada T, Matsuura Y, Kimura A. Substrate recognition mechanism of α-1,6-glucosidic linkage hydrolyzing enzyme, dextran glucosidase from Streptococcus mutans . J Mol Biol. 2008;378:913–922. doi: 10.1016/j.jmb.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 92.Møller MS, Fredslund F, Majumder A, Nakai H, Poulsen JC, Lo Leggio L, Svensson B, Abou Hachem M. Enzymology and structure of the GH13_31 glucan 1,6-α-glucosidase that confers isomaltooligosaccharide utilization in the probiotic Lactobacillus acidophilus NCFM. J Bacteriol. 2012;194:4249–4259. doi: 10.1128/JB.00622-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang D, Li N, Lok SM, Zhang LH, Swaminathan K. Isomaltulose synthase (PalI) of Klebsiella sp. LX3. Crystal structure and implication of mechanism. J Biol Chem. 2003;278:35428–35434. doi: 10.1074/jbc.M302616200. [DOI] [PubMed] [Google Scholar]
- 94.Ravaud S, Robert X, Watzlawick H, Haser R, Mattes R, Aghajari N. Trehalulose synthase native and carbohydrate complexed structures provide insights into sucrose isomerization. J Biol Chem. 2007;282:28126–28136. doi: 10.1074/jbc.M704515200. [DOI] [PubMed] [Google Scholar]
- 95.Ravaud S, Robert X, Watzlawick H, Haser R, Mattes R, Aghajari N. Structural determinants of product specificity of sucrose isomerases. FEBS Lett. 2009;583:1964–1968. doi: 10.1016/j.febslet.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 96.Machius M, Wiegand G, Huber R. Crystal structure of calcium-depleted Bacillus licheniformis α-amylase at 2.2 Å resolution. J Mol Biol. 1995;246:545–559. doi: 10.1006/jmbi.1994.0106. [DOI] [PubMed] [Google Scholar]
- 97.Hwang KY, Song HK, Chang C, Lee J, Lee SY, Kim KK, Choe S, Sweet RM, Suh SW. Crystal structure of thermostable α-amylase from Bacillus licheniformis refined at 1.7 Å resolution. Mol Cells. 1997;7:251–258. [PubMed] [Google Scholar]
- 98.Suvd D, Fujimoto Z, Takase K, Matsumura M, Mizuno H. Crystal structure of Bacillus stearothermophilus α-amylase: possible factors determining the thermostability. J Biochem. 2001;129:461–468. doi: 10.1093/oxfordjournals.jbchem.a002878. [DOI] [PubMed] [Google Scholar]
- 99.Alikhajeh J, Khajeh K, Ranjbar B, Naderi-Manesh H, Lin YH, Liu E, Guan HH, Hsieh YC, Chuankhayan P, Huang YC, Jeyaraman J, Liu MY, Chen CJ. Structure of Bacillus amyloliquefaciens α-amylase at high resolution: implications for thermal stability. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:121–129. doi: 10.1107/S1744309109051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fujimoto Z, Takase K, Doui N, Momma M, Matsumoto T, Mizuno H. Crystal structure of a catalytic-site mutant α-amylase from Bacillus subtilis complexed with maltopentaose. J Mol Biol. 1998;277:393–407. doi: 10.1006/jmbi.1997.1599. [DOI] [PubMed] [Google Scholar]
- 101.Davies GJ, Brzozowski AM, Dauter Z, Rasmussen MD, Borchert TV, Wilson KS. Structure of a Bacillus halmapalus family 13 α-amylase, BHA, in complex with an acarbose-derived nonasaccharide at 2.1 Å resolution. Acta Crystallogr D Biol Crystallogr. 2005;61:190–193. doi: 10.1107/S0907444904027118. [DOI] [PubMed] [Google Scholar]
- 102.Lyhne-Iversen L, Hobley TJ, Kaasgaard SG, Harris P. Structure of Bacillus halmapalus α-amylase crystallized with and without the substrate analogue acarbose and maltose. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62:849–854. doi: 10.1107/S174430910603096X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nonaka T, Fujihashi M, Kita A, Hagihara H, Ozaki K, Ito S, Miki K. Crystal structure of calcium-free α-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites. J Biol Chem. 2003;278:24818–24824. doi: 10.1074/jbc.M212763200. [DOI] [PubMed] [Google Scholar]
- 104.Shirai T, Igarashi K, Ozawa T, Hagihara H, Kobayashi T, Ozaki K, Ito S. Ancestral sequence evolutionary trace and crystal structure analyses of alkaline α-amylase from Bacillus sp. KSM-1378 to clarify the alkaline adaptation process of proteins. Proteins. 2007;66:600–610. doi: 10.1002/prot.21255. [DOI] [PubMed] [Google Scholar]
- 105.Kanai R, Haga K, Akiba T, Yamane K, Harata K. Biochemical and crystallographic analyses of maltohexaose-producing amylase from alkalophilic Bacillus sp. 707. Biochemistry. 2004;43:14047–14056. doi: 10.1021/bi048489m. [DOI] [PubMed] [Google Scholar]
- 106.Tsukamoto A, Kimura K, Ishii Y, Takano T, Yamane K. Nucleotide sequence of the maltohexaose-producing amylase gene from an alkalophilic Bacillus sp. #707 and structural similarity to liquefying type α-amylases. Biochem Biophys Res Commun. 1988;151:25–31. doi: 10.1016/0006-291x(88)90554-2. [DOI] [PubMed] [Google Scholar]
- 107.van der Kaaij RM, Janecek S, van der Maarel MJ, Dijkhuizen L. Phylogenetic and biochemical characterization of a novel cluster of intracellular fungal α-amylase enzymes. Microbiology. 2007;153:4003–4015. doi: 10.1099/mic.0.2007/008607-0. [DOI] [PubMed] [Google Scholar]
- 108.Marion CL, Rappleye CA, Engle JT, Goldman WE. An α-(1,4)-amylase is essential for α-(1,3)-glucan production and virulence in Histoplasma capsulatum . Mol Microbiol. 2006;62:970–983. doi: 10.1111/j.1365-2958.2006.05436.x. [DOI] [PubMed] [Google Scholar]
- 109.Camacho E, Sepulveda VE, Goldman WE, San-Blas G, Nino-Vega GA. Expression of Paracoccidioides brasiliensis AMY1 in a Histoplasma capsulatum amy1 mutant, relates an α-(1,4)-amylase to cell wall α-(1,3)-glucan synthesis. PLoS ONE. 2012;7:e50201. doi: 10.1371/journal.pone.0050201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rodriguez Sanoja R, Morlon-Guyot J, Jore J, Pintado J, Juge N, Guyot JP. Comparative characterization of complete and truncated forms of Lactobacillus amylovorus α-amylase and role of the C-terminal direct repeats in raw-starch binding. Appl Environ Microbiol. 2000;66:3350–3356. doi: 10.1128/aem.66.8.3350-3356.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rodriguez-Sanoja R, Ruiz B, Guyot JP, Sanchez S. Starch-binding domain affects catalysis in two Lactobacillus α-amylases. Appl Environ Microbiol. 2005;71:297–302. doi: 10.1128/AEM.71.1.297-302.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Guillen D, Santiago M, Linares L, Perez R, Morlon J, Ruiz B, Sanchez S, Rodriguez-Sanoja R. α-Amylase starch binding domains: cooperative effects of binding to starch granules of multiple tandemly arranged domains. Appl Environ Microbiol. 2007;73:3833–3837. doi: 10.1128/AEM.02628-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rodriguez-Sanoja R, Oviedo N, Escalante L, Ruiz B, Sanchez S. A single residue mutation abolishes attachment of the CBM26 starch-binding domain from Lactobacillus amylovorus α-amylase. J Ind Microbiol Biotechnol. 2009;36:341–346. doi: 10.1007/s10295-008-0502-y. [DOI] [PubMed] [Google Scholar]
- 114.Janecek S, Leveque E, Belarbi A, Haye B. Close evolutionary relatedness of α-amylases from Archaea and plants. J Mol Evol. 1999;48:421–426. doi: 10.1007/pl00006486. [DOI] [PubMed] [Google Scholar]
- 115.Da Lage JL, Feller G, Janecek S. Horizontal gene transfer from Eukarya to bacteria and domain shuffling: the α-amylase model. Cell Mol Life Sci. 2004;61:97–109. doi: 10.1007/s00018-003-3334-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jones RA, Jermiin LS, Easteal S, Patel BK, Beacham IR. Amylase and 16S rRNA genes from a hyperthermophilic archaebacterium. J Appl Microbiol. 1999;86:93–107. doi: 10.1046/j.1365-2672.1999.00642.x. [DOI] [PubMed] [Google Scholar]
- 117.Leveque E, Janecek S, Belarbi A, Haye B. Thermophilic archaeal amylolytic enzymes. Enzyme Microb Technol. 2000;26:2–13. [Google Scholar]
- 118.Leveque E, Haye B, Belarbi A. Cloning and expression of an α-amylase encoding gene from the hyperthermophilic archaebacterium Thermococcus hydrothermalis and biochemical characterisation of the recombinant enzyme. FEMS Microbiol Lett. 2000;186:67–71. doi: 10.1111/j.1574-6968.2000.tb09083.x. [DOI] [PubMed] [Google Scholar]
- 119.Lim JK, Lee HS, Kim YJ, Bae SS, Jeon JH, Kang SG, Lee JH. Critical factors to high thermostability of an α-amylase from hyperthermophilic archaeon Thermococcus onnurineus NA1. J Microbiol Biotechnol. 2007;17:1242–1248. [PubMed] [Google Scholar]
- 120.Dong G, Vieille C, Savchenko A, Zeikus JG. Cloning, sequencing, and expression of the gene encoding extracellular α-amylase from Pyrococcus furiosus and biochemical characterization of the recombinant enzyme. Appl Environ Microbiol. 1997;63:3569–3576. doi: 10.1128/aem.63.9.3569-3576.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jorgensen S, Vorgias CE, Antranikian G. Cloning, sequencing, characterization, and expression of an extracellular α-amylase from the hyperthermophilic archaeon Pyrococcus furiosus in Escherichia coli and Bacillus subtilis . J Biol Chem. 1997;272:16335–16342. doi: 10.1074/jbc.272.26.16335. [DOI] [PubMed] [Google Scholar]
- 122.Frillingos S, Linden A, Niehaus F, Vargas C, Nieto JJ, Ventosa A, Antranikian G, Drainas C. Cloning and expression of α-amylase from the hyperthermophilic archaeon Pyrococcus woesei in the moderately halophilic bacterium Halomonas elongata . J Appl Microbiol. 2000;88:495–503. doi: 10.1046/j.1365-2672.2000.00988.x. [DOI] [PubMed] [Google Scholar]
- 123.Linden A, Mayans O, Meyer-Klaucke W, Antranikian G, Wilmanns M. Differential regulation of a hyperthermophilic α-amylase with a novel (Ca,Zn) two-metal center by zinc. J Biol Chem. 2003;278:9875–9884. doi: 10.1074/jbc.M211339200. [DOI] [PubMed] [Google Scholar]
- 124.Linden A, Wilmanns M. Adaptation of class-13 α-amylases to diverse living conditions. ChemBioChem. 2004;5:231–239. doi: 10.1002/cbic.200300734. [DOI] [PubMed] [Google Scholar]
- 125.Rogers JC, Milliman C. Isolation and sequence analysis of a barley α-amylase cDNA clone. J Biol Chem. 1983;258:8169–8174. [PubMed] [Google Scholar]
- 126.Rogers JC. Two barley α-amylase gene families are regulated differently in aleurone cells. J Biol Chem. 1985;260:3731–3738. [PubMed] [Google Scholar]
- 127.Kadziola A, Abe J, Svensson B, Haser R. Crystal and molecular structure of barley α-amylase. J Mol Biol. 1994;239:104–121. doi: 10.1006/jmbi.1994.1354. [DOI] [PubMed] [Google Scholar]
- 128.Kadziola A, Søgaard M, Svensson B, Haser R. Molecular structure of a barley α-amylase–inhibitor complex: implications for starch binding and catalysis. J Mol Biol. 1998;278:205–217. doi: 10.1006/jmbi.1998.1683. [DOI] [PubMed] [Google Scholar]
- 129.Robert X, Haser R, Gottschalk TE, Ratajczak F, Driguez H, Svensson B, Aghajari N. The structure of barley α-amylase isozyme 1 reveals a novel role of domain C in substrate recognition and binding: a pair of sugar tongs. Structure. 2003;11:973–984. doi: 10.1016/s0969-2126(03)00151-5. [DOI] [PubMed] [Google Scholar]
- 130.Robert X, Haser R, Mori H, Svensson B, Aghajari N. Oligosaccharide binding to barley α-amylase 1. J Biol Chem. 2005;280:32968–32978. doi: 10.1074/jbc.M505515200. [DOI] [PubMed] [Google Scholar]
- 131.Baulcombe DC, Huttly AK, Martienssen RA, Barker RF, Jarvis MG. A novel wheat α-amylase gene (α-Amy3) Mol Gen Genet. 1987;209:33–40. doi: 10.1007/BF00329833. [DOI] [PubMed] [Google Scholar]
- 132.Huang N, Sutliff TD, Litts JC, Rodriguez RL. Classification and characterization of the rice α-amylase multigene family. Plant Mol Biol. 1990;14:655–668. doi: 10.1007/BF00016499. [DOI] [PubMed] [Google Scholar]
- 133.Young TE, DeMason DA, Close TJ. Cloning of an α-amylase cDNA from aleurone tissue of germinating maize seed. Plant Physiol. 1994;105:759–760. doi: 10.1104/pp.105.2.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mori H, Kobayashi T, Tonokawa T, Tatematsu A, Matsui H, Kimura A, Chiba S. Molecular cloning of an α-amylase cDNA from germinating cotyledons of kidney bean (Phaseolus vulgaris L. cv. Toramame) J Appl Glycosci. 1997;45:261–267. [Google Scholar]
- 135.Wegrzyn T, Reilly K, Cipriani G, Murphy P, Newcomb R, Gardner R, MacRae E. A novel α-amylase gene is transiently upregulated during low temperature exposure in apple fruit. Eur J Biochem. 2000;267:1313–1322. doi: 10.1046/j.1432-1327.2000.01087.x. [DOI] [PubMed] [Google Scholar]
- 136.Stanley D, Fitzgerald AM, Farnden KJA, MacRae EA. Characterisation of putative α-amylases from apple (Malus domestica) and Arabidopsis thaliana . Biologia. 2002;57(Suppl 11):137–148. [Google Scholar]
- 137.Junior AV, do Nascimento JR, Lajolo FM. Molecular cloning and characterization of an α-amylase occurring in the pulp of ripening bananas and its expression in Pichia pastoris . J Agric Food Chem. 2006;54:8222–8228. doi: 10.1021/jf060805b. [DOI] [PubMed] [Google Scholar]
- 138.Bozonnet S, Jensen MT, Nielsen MM, Aghajari N, Jensen MH, Kramhøft B, Willemoës M, Tranier S, Haser R, Svensson B. The ‘pair of sugar tongs’ site on the non-catalytic domain C of barley α-amylase participates in substrate binding and activity. FEBS J. 2007;274:5055–5067. doi: 10.1111/j.1742-4658.2007.06024.x. [DOI] [PubMed] [Google Scholar]
- 139.Nielsen MM, Bozonnet S, Seo ES, Motyan JA, Andersen JM, Dilokpimol A, Abou Hachem M, Gyemant G, Naested H, Kandra L, Sigurskjold BW, Svensson B. Two secondary carbohydrate binding sites on the surface of barley α-amylase 1 have distinct functions and display synergy in hydrolysis of starch granules. Biochemistry. 2009;48:7686–7697. doi: 10.1021/bi900795a. [DOI] [PubMed] [Google Scholar]
- 140.Nielsen JW, Kramhøft B, Bozonnet S, Abou Hachem M, Stipp SL, Svensson B, Willemoës M. Degradation of the starch components amylopectin and amylose by barley α-amylase 1: role of surface binding site 2. Arch Biochem Biophys. 2012;528:1–6. doi: 10.1016/j.abb.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 141.Tranier S, Deville K, Robert X, Bozonnet S, Haser R, Svensson B, Aghajari N. Insights into the “pair of sugar tongs” surface binding site in barley α-amylase isozymes and crystallization of appropriate sugar tongs mutants. Biologia. 2005;60(Suppl 16):37–46. [Google Scholar]
- 142.Pujadas G, Palau J. Evolution of α-amylases: architectural features and key residues in the stabilization of the (β/α)8 scaffold. Mol Biol Evol. 2001;18:38–54. doi: 10.1093/oxfordjournals.molbev.a003718. [DOI] [PubMed] [Google Scholar]
- 143.Godany A, Majzlova K, Horvathova V, Vidova B, Janecek S. Tyrosine 39 of GH13 α-amylase from Thermococcus hydrothermalis contributes to its thermostability. Biologia. 2010;65:408–415. [Google Scholar]
- 144.Da Lage JL, Danchin EG, Casane D. Where do animal α-amylases come from? An interkingdom trip. FEBS Lett. 2007;581:3927–3935. doi: 10.1016/j.febslet.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 145.Feller G, Lonhienne T, Deroanne C, Libioulle C, Van Beeumen J, Gerday C. Purification, characterization, and nucleotide sequence of the thermolabile α-amylase from the Antarctic psychrotroph Alteromonas haloplanctis A23. J Biol Chem. 1992;267:5217–5221. [PubMed] [Google Scholar]
- 146.Feller G, Payan F, Theys F, Qian M, Haser R, Gerday C. Stability and structural analysis of α-amylase from the Antarctic psychrophile Alteromonas haloplanctis A23. Eur J Biochem. 1994;222:441–447. doi: 10.1111/j.1432-1033.1994.tb18883.x. [DOI] [PubMed] [Google Scholar]
- 147.D’Amico S, Gerday C, Feller G. Structural similarities and evolutionary relationships in chloride-dependent α-amylases. Gene. 2000;253:95–105. doi: 10.1016/s0378-1119(00)00229-8. [DOI] [PubMed] [Google Scholar]
- 148.Aghajari N, Feller G, Gerday C, Haser R. Structural basis of α-amylase activation by chloride. Protein Sci. 2002;11:1435–1441. doi: 10.1110/ps.0202602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cipolla A, Delbrassine F, Da Lage JL, Feller G. Temperature adaptations in psychrophilic, mesophilic and thermophilic chloride-dependent α-amylases. Biochimie. 2012;94:1943–1950. doi: 10.1016/j.biochi.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 150.Boer PH, Hickey DA. The α-amylase gene in Drosophila melanogaster: nucleotide sequence, gene structure and expression motifs. Nucleic Acids Res. 1986;14:8399–8411. doi: 10.1093/nar/14.21.8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Da Lage JL, Wegnez M, Cariou ML. Distribution and evolution of introns in Drosophila amylase genes. J Mol Evol. 1996;43:334–347. doi: 10.1007/BF02339008. [DOI] [PubMed] [Google Scholar]
- 152.Da Lage JL, Renard E, Chartois F, Lemeunier F, Cariou ML. Amyrel, a paralogous gene of the amylase gene family in Drosophila melanogaster and the Sophophora subgenus. Proc Natl Acad Sci USA. 1998;95:6848–6853. doi: 10.1073/pnas.95.12.6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Da Lage JL, Maisonhaute C, Maczkowiak F, Cariou ML. A nested α-amylase gene in Drosophila ananassae . J Mol Evol. 2003;57:355–362. doi: 10.1007/s00239-003-2488-4. [DOI] [PubMed] [Google Scholar]
- 154.Strobl S, Gomis-Ruth FX, Maskos K, Frank G, Huber R, Glockshuber R. The α-amylase from the yellow meal worm: complete primary structure, crystallization and preliminary X-ray analysis. FEBS Lett. 1997;409:109–114. doi: 10.1016/s0014-5793(97)00451-1. [DOI] [PubMed] [Google Scholar]
- 155.Strobl S, Maskos K, Betz M, Wiegand G, Huber R, Gomis-Ruth FX, Glockshuber R. Crystal structure of yellow meal worm α-amylase at 1.64 Å resolution. J Mol Biol. 1998;278:617–628. doi: 10.1006/jmbi.1998.1667. [DOI] [PubMed] [Google Scholar]
- 156.Le Moine S, Sellos D, Moal J, Daniel JY, San Juan Serrano F, Samain JF, Van Wormhoudt A. Amylase on Pecten maximus (Mollusca, bivalves): protein and cDNA characterization; quantification of the expression in the digestive gland. Mol Mar Biol Biotechnol. 1997;6:228–237. [PubMed] [Google Scholar]
- 157.Nikapitiya C, Oh C, Whang I, Kim CG, Lee YH, Kim SJ, Lee J. Molecular characterization, gene expression analysis and biochemical properties of α-amylase from the disk abalone, Haliotis discus discus . Comp Biochem Physiol B Biochem Mol Biol. 2009;152:271–281. doi: 10.1016/j.cbpb.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 158.Van Wormhoudt A, Sellos D. Cloning and sequencing analysis of three amylase cDNAs in the shrimp Penaeus vannamei (Crustacea decapoda): evolutionary aspects. J Mol Evol. 1996;42:543–551. doi: 10.1007/BF02352284. [DOI] [PubMed] [Google Scholar]
- 159.Da Lage JL, Maczkowiak F, Cariou ML. Phylogenetic distribution of intron positions in α-amylase genes of bilateria suggests numerous gains and losses. PLoS ONE. 2011;6:e19673. doi: 10.1371/journal.pone.0019673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Buisson G, Duee E, Haser R, Payan F. Three-dimensional structure of porcine pancreatic α-amylase at 2.9 Å resolution. Role of calcium in structure and activity. EMBO J. 1987;6:3909–3916. doi: 10.1002/j.1460-2075.1987.tb02731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Qian M, Haser R, Payan F. Structure and molecular model refinement of pig pancreatic α-amylase at 2.1 Å resolution. J Mol Biol. 1993;231:785–799. doi: 10.1006/jmbi.1993.1326. [DOI] [PubMed] [Google Scholar]
- 162.Qian M, Haser R, Buisson G, Duee E, Payan F. The active center of a mammalian α-amylase. Structure of the complex of a pancreatic α-amylase with a carbohydrate inhibitor refined to 2.2-Å resolution. Biochemistry. 1994;33:6284–6294. doi: 10.1021/bi00186a031. [DOI] [PubMed] [Google Scholar]
- 163.Gilles C, Astier JP, Marchis-Mouren G, Cambillau C, Payan F. Crystal structure of pig pancreatic α-amylase isoenzyme II, in complex with the carbohydrate inhibitor acarbose. Eur J Biochem. 1996;238:561–569. doi: 10.1111/j.1432-1033.1996.0561z.x. [DOI] [PubMed] [Google Scholar]
- 164.Ramasubbu N, Paloth V, Luo Y, Brayer GD, Levine MJ. Structure of human salivary α-amylase at 1.6 Angstrom resolution: implications for its role in the oral cavity. Acta Crystallogr D. 1996;52:435–446. doi: 10.1107/S0907444995014119. [DOI] [PubMed] [Google Scholar]
- 165.Brayer GD, Luo Y, Withers SG. The structure of human pancreatic α-amylase at 1.8 Å resolution and comparisons with related enzymes. Protein Sci. 1995;4:1730–1742. doi: 10.1002/pro.5560040908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mizutani K, Toyoda M, Otake Y, Yoshioka S, Takahashi N, Mikami B. Structural and functional characterization of recombinant medaka fish α-amylase expressed in yeast Pichia pastoris . Biochim Biophys Acta. 2012;1824:954–962. doi: 10.1016/j.bbapap.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 167.Machius M, Vertesy L, Huber R, Wiegand G. Carbohydrate and protein-based inhibitors of porcine pancreatic α-amylase: structure analysis and comparison of their binding characteristics. J Mol Biol. 1996;260:409–421. doi: 10.1006/jmbi.1996.0410. [DOI] [PubMed] [Google Scholar]
- 168.Payan F, Qian M. Crystal structure of the pig pancreatic α-amylase complexed with malto-oligosaccharides. J Protein Chem. 2003;22:275–284. doi: 10.1023/a:1025072520607. [DOI] [PubMed] [Google Scholar]
- 169.Larson SB, Day JS, McPherson A. X-ray crystallographic analyses of pig pancreatic α-amylase with limit dextrin, oligosaccharide, and α-cyclodextrin. Biochemistry. 2010;49:3101–3115. doi: 10.1021/bi902183w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Qin X, Ren L, Yang X, Bai F, Wang L, Geng P, Bai G, Shen Y. Structures of human pancreatic α-amylase in complex with acarviostatins: implications for drug design against type II diabetes. J Struct Biol. 2011;174:196–202. doi: 10.1016/j.jsb.2010.11.020. [DOI] [PubMed] [Google Scholar]
- 171.Yang CH, Liu WH. Cloning and characterization of a maltotriose-producing α-amylase gene from Thermobifida fusca . J Ind Microbiol Biotechnol. 2007;34:325–330. doi: 10.1007/s10295-006-0200-6. [DOI] [PubMed] [Google Scholar]
- 172.Doukyu N, Yamagishi W, Kuwahara H, Ogino H. A maltooligosaccharide-forming amylase gene from Brachybacterium sp. strain LB25: cloning and expression in Escherichia coli . Biosci Biotechnol Biochem. 2008;72:2444–2447. doi: 10.1271/bbb.80207. [DOI] [PubMed] [Google Scholar]
- 173.Yamaguchi R, Tokunaga H, Ishibashi M, Arakawa T, Tokunaga M. Salt-dependent thermo-reversible α-amylase: cloning and characterization of halophilic α-amylase from moderately halophilic bacterium, Kocuria varians . Appl Microbiol Biotechnol. 2011;89:673–684. doi: 10.1007/s00253-010-2882-y. [DOI] [PubMed] [Google Scholar]
- 174.Yamaguchi R, Arakawa T, Tokunaga H, Ishibashi M, Tokunaga M. Distinct characteristics of single starch-binding domain SBD1 derived from tandem domains SBD1–SBD2 of halophilic Kocuria varians α-amylase. Protein J. 2012;31:250–258. doi: 10.1007/s10930-012-9400-2. [DOI] [PubMed] [Google Scholar]
- 175.Yamaguchi R, Inoue Y, Tokunaga H, Ishibashi M, Arakawa T, Sumitani J, Kawaguchi T, Tokunaga M. Halophilic characterization of starch-binding domain from Kocuria varians α-amylase. Int J Biol Macromol. 2012;50:95–102. doi: 10.1016/j.ijbiomac.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 176.Sumitani J, Tottori T, Kawaguchi T, Arai M. New type of starch-binding domain: the direct repeat motif in the C-terminal region of Bacillus sp. no. 195 α-amylase contributes to starch binding and raw starch degrading. Biochem J. 2000;350:477–484. [PMC free article] [PubMed] [Google Scholar]
- 177.Virolle MJ, Long CM, Chang S, Bibb MJ. Cloning, characterisation and regulation of an α-amylase gene from Streptomyces venezuelae . Gene. 1988;74:321–334. doi: 10.1016/0378-1119(88)90166-7. [DOI] [PubMed] [Google Scholar]
- 178.Vigal T, Gil JA, Daza A, Garcia-Gonzalez MD, Martin JF. Cloning, characterization and expression of an α-amylase gene from Streptomyces griseus IMRU3570. Mol Gen Genet. 1991;225:278–288. doi: 10.1007/BF00269860. [DOI] [PubMed] [Google Scholar]
- 179.Da Lage JL, Binder M, Hua-Van A, Janecek S, Casane D. Gene make-up: rapid and massive intron gains after horizontal transfer of a bacterial α-amylase gene to Basidiomycetes. BMC Evol Biol. 2013;13:40. doi: 10.1186/1471-2148-13-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Chen W, Xie T, Shao Y, Chen F. Phylogenomic relationships between amylolytic enzymes from 85 strains of fungi. PLoS ONE. 2012;7:e49679. doi: 10.1371/journal.pone.0049679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hu NT, Hung MN, Huang AM, Tsai HF, Yang BY, Chow TY, Tseng YH. Molecular cloning, characterization and nucleotide sequence of the gene for secreted α-amylase from Xanthomonas campestris pv. campestris . J Gen Microbiol. 1992;138:1647–1655. doi: 10.1099/00221287-138-8-1647. [DOI] [PubMed] [Google Scholar]
- 182.Gobius KS, Pemberton JM. Molecular cloning, characterization, and nucleotide sequence of an extracellular amylase gene from Aeromonas hydrophila . J Bacteriol. 1988;170:1325–1332. doi: 10.1128/jb.170.3.1325-1332.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Na HK, Kim ES, Lee HH, Yoo OJ, Jhon DY. Cloning and nucleotide sequence of the α-amylase gene from alkalophilic Pseudomonas sp. KFCC 10818. Mol Cells. 1996;2:203–208. [Google Scholar]
- 184.Kang EJ, Kim ES, Lee JE, Jhon DY. Cloning, sequencing, characterization, and expression of a new α-amylase isozyme gene (amy3) from Pseudomonas sp. Biotechnol Lett. 2001;23:811–816. [Google Scholar]
- 185.Majzlova K, Pukajova Z, Janecek S. Tracing the evolution of the α-amylase subfamily GH13_36 covering the amylolytic enzymes intermediate between oligo-1,6-glucosidases and neopullulanases. Carbohydr Res. 2013;367:48–57. doi: 10.1016/j.carres.2012.11.022. [DOI] [PubMed] [Google Scholar]
- 186.Horinouchi S, Fukusumi S, Ohshima T, Beppu T. Cloning and expression in Escherichia coli of two additional amylase genes of a strictly anaerobic thermophile, Dictyoglomus thermophilum, and their nucleotide sequences with extremely low guanine-plus-cytosine contents. Eur J Biochem. 1988;176:243–253. doi: 10.1111/j.1432-1033.1988.tb14275.x. [DOI] [PubMed] [Google Scholar]
- 187.Metz RJ, Allen LN, Cao TM, Zeman NW. Nucleotide sequence of an amylase gene from Bacillus megaterium . Nucleic Acids Res. 1988;16:5203. doi: 10.1093/nar/16.11.5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Brumm PJ, Hebeda RE, Teague WM. Purification and characterization of the commercialized, cloned Bacillus megaterium α-amylase. Part I: purification and hydrolytic properties. Starch/Staerke. 1991;43:315–319. [Google Scholar]
- 189.Brumm PJ, Hebeda RE, Teague WM. Purification and characterization of the commercialized, cloned Bacillus megaterium α-amylase. Part II: transferase properties. Starch/Staerke. 1991;43:319–323. [Google Scholar]
- 190.Abe J, Shibata Y, Fujisue M, Hizukuri S. Expression of periplasmic α-amylase of Xanthomonas campestris K-11151 in Escherichia coli and its action on maltose. Microbiology. 1996;142:1505–1512. doi: 10.1099/13500872-142-6-1505. [DOI] [PubMed] [Google Scholar]
- 191.Yebra MJ, Arroyo J, Sanz P, Priet JA. Characterization of novel neopullulanase from Bacillus polymyxa . Appl Biochem Biotechnol. 1997;68:113–120. [Google Scholar]
- 192.Yebra MJ, Blasco A, Sanz P. Expression and secretion of Bacillus polymyxa neopullulanase in Saccharomyces cerevisiae . FEMS Microbiol Lett. 1999;170:41–49. doi: 10.1111/j.1574-6968.1999.tb13353.x. [DOI] [PubMed] [Google Scholar]
- 193.Nakagawa Y, Saburi W, Takada M, Hatada Y, Horikoshi K. Gene cloning and enzymatic characteristics of a novel γ-cyclodextrin-specific cyclodextrinase from alkalophilic Bacillus clarkii 7364. Biochim Biophys Acta. 2008;1784:2004–2011. doi: 10.1016/j.bbapap.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 194.Ballschmiter M, Armbrecht M, Ivanova K, Antranikian G, Liebl W. AmyA, an α-amylase with β-cyclodextrin-forming activity, and AmyB from the thermoalkaliphilic organism Anaerobranca gottschalkii: two α-amylases adapted to their different cellular localizations. Appl Environ Microbiol. 2005;71:3709–3715. doi: 10.1128/AEM.71.7.3709-3715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Liebl W, Stemplinger I, Ruile P. Properties and gene structure of the Thermotoga maritima α-amylase AmyA, a putative lipoprotein of a hyperthermophilic bacterium. J Bacteriol. 1997;179:941–948. doi: 10.1128/jb.179.3.941-948.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Mijts BN, Patel BK. Cloning, sequencing and expression of an α-amylase gene, amyA, from the thermophilic halophile Halothermothrix orenii and purification and biochemical characterization of the recombinant enzyme. Microbiology. 2002;148:2343–2349. doi: 10.1099/00221287-148-8-2343. [DOI] [PubMed] [Google Scholar]
- 197.Sivakumar N, Li N, Tang JW, Patel BK, Swaminathan K. Crystal structure of AmyA lacks acidic surface and provide insights into protein stability at poly-extreme condition. FEBS Lett. 2006;580:2646–2652. doi: 10.1016/j.febslet.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 198.Liu Y, Lei Y, Zhang X, Gao Y, Xiao Y, Peng H. Identification and phylogenetic characterization of a new subfamily of α-amylase enzymes from marine microorganisms. Mar Biotechnol. 2012;14:253–260. doi: 10.1007/s10126-011-9414-3. [DOI] [PubMed] [Google Scholar]
- 199.Lei Y, Peng H, Wang Y, Liu Y, Han F, Xiao Y, Gao Y. Preferential and rapid degradation of raw rice starch by an α-amylase of glycoside hydrolase subfamily GH13_37. Appl Microbiol Biotechnol. 2012;94:1577–1584. doi: 10.1007/s00253-012-4114-0. [DOI] [PubMed] [Google Scholar]
- 200.Yu J, Wang C, Hu Y, Dong Y, Wang Y, Tu X, Peng H, Zhang X. Purification, crystallization and preliminary crystallographic analysis of the marine α-amylase AmyP. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013;69:263–266. doi: 10.1107/S1744309113001693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Puspasari F, Nurachman Z, Noer AS, Radjasa OK, van der Maarel MJEC, Natalia D. Characteristics of raw starch degrading α-amylase from Bacillus aquimaris MKSC 6.2 associated with soft coral Sinularia sp. Starch/Staerke. 2011;63:461–467. [Google Scholar]
- 202.Finore I, Kasavi C, Poli A, Romano I, Toksoy Oner E, Kirdar B, Dipasquale L, Nicolaus B, Lama L. Purification, biochemical characterization and gene sequencing of a thermostable raw starch digesting α-amylase from Geobacillus thermoleovorans subsp. stromboliensis subsp. nov. World J Microbiol Biotechnol. 2011;27:2425–2433. [Google Scholar]
- 203.Chai YY, Rahman RN, Illias RM, Goh KM. Cloning and characterization of two new thermostable and alkalitolerant α-amylases from the Anoxybacillus species that produce high levels of maltose. J Ind Microbiol Biotechnol. 2012;39:731–741. doi: 10.1007/s10295-011-1074-9. [DOI] [PubMed] [Google Scholar]
- 204.Mok SC, Teh AH, Saito JA, Najimudin N, Alam M. Crystal structure of a compact α-amylase from Geobacillus thermoleovorans . Enzyme Microb Technol. 2013;53:46–54. doi: 10.1016/j.enzmictec.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 205.Buedenbender S, Schulz GE. Structural base for enzymatic cyclodextrin hydrolysis. J Mol Biol. 2009;385:606–617. doi: 10.1016/j.jmb.2008.10.085. [DOI] [PubMed] [Google Scholar]
- 206.Shipman JA, Cho KH, Siegel HA, Salyers AA. Physiological characterization of SusG, an outer membrane protein essential for starch utilization by Bacteroides thetaiotaomicron . J Bacteriol. 1999;181:7206–7211. doi: 10.1128/jb.181.23.7206-7211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Martens EC, Koropatkin NM, Smith TJ, Gordon JI. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J Biol Chem. 2009;284:24673–24677. doi: 10.1074/jbc.R109.022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Cameron EA, Maynard MA, Smith CJ, Smith TJ, Koropatkin NM, Martens EC. Multidomain carbohydrate-binding proteins involved in Bacteroides thetaiotaomicron starch metabolism. J Biol Chem. 2012;287:34614–34625. doi: 10.1074/jbc.M112.397380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Fukusumi S, Kamizono A, Horinouchi S, Beppu T. Cloning and nucleotide sequence of a heat-stable amylase gene from an anaerobic thermophile, Dictyoglomus thermophilum . Eur J Biochem. 1988;174:15–21. doi: 10.1111/j.1432-1033.1988.tb14056.x. [DOI] [PubMed] [Google Scholar]
- 210.Laderman KA, Asada K, Uemori T, Mukai H, Taguchi Y, Kato I, Anfinsen CB. α-Amylase from the hyperthermophilic archaebacterium Pyrococcus furiosus. Cloning and sequencing of the gene and expression in Escherichia coli . J Biol Chem. 1993;268:24402–24407. [PubMed] [Google Scholar]
- 211.Janecek S. Sequence of archaeal Methanococcus jannaschii α-amylase contains features of families 13 and 57 of glycosyl hydrolases: a trace of their common ancestor? Folia Microbiol. 1998;43:123–128. doi: 10.1007/BF02816496. [DOI] [PubMed] [Google Scholar]
- 212.Imamura H, Fushinobu S, Yamamoto M, Kumasaka T, Jeon BS, Wakagi T, Matsuzawa H. Crystal structures of 4-α-glucanotransferase from Thermococcus litoralis and its complex with an inhibitor. J Biol Chem. 2003;278:19378–19386. doi: 10.1074/jbc.M213134200. [DOI] [PubMed] [Google Scholar]
- 213.Janecek S. Amylolytic families of glycoside hydrolases: focus on the family GH-57. Biologia. 2005;60(Suppl 16):177–184. [Google Scholar]
- 214.Nakajima M, Imamura H, Shoun H, Horinouchi S, Wakagi T. Transglycosylation activity of Dictyoglomus thermophilum amylase A. Biosci Biotechnol Biochem. 2004;68:2369–2373. doi: 10.1271/bbb.68.2369. [DOI] [PubMed] [Google Scholar]
- 215.Laderman KA, Davis BR, Krutzsch HC, Lewis MS, Griko YV, Privalov PL, Anfinsen CB. The purification and characterization of an extremely thermostable α-amylase from the hyperthermophilic archaebacterium Pyrococcus furiosus . J Biol Chem. 1993;268:24394–24401. [PubMed] [Google Scholar]
- 216.Janecek S, Blesak K. Sequence-structural features and evolutionary relationships of family GH57 α-amylases and their putative α-amylase-like homologues. Protein J. 2011;30:429–435. doi: 10.1007/s10930-011-9348-7. [DOI] [PubMed] [Google Scholar]
- 217.Blesak K, Janecek S. Sequence fingerprints of enzyme specificities from the glycoside hydrolase family GH57. Extremophiles. 2012;16:497–506. doi: 10.1007/s00792-012-0449-9. [DOI] [PubMed] [Google Scholar]
- 218.Kim JW, Flowers LO, Whiteley M, Peeples TL. Biochemical confirmation and characterization of the family-57-like α-amylase of Methanococcus jannaschii . Folia Microbiol. 2001;46:467–473. doi: 10.1007/BF02817988. [DOI] [PubMed] [Google Scholar]
- 219.Jeon BS, Taguchi H, Sakai H, Ohshima T, Wakagi T, Matsuzawa H. 4-α-Glucanotransferase from the hyperthermophilic archaeon Thermococcus litoralis. Enzyme purification and characterization, and gene cloning, sequencing and expression in Escherichia coli . Eur J Biochem. 1997;248:171–178. doi: 10.1111/j.1432-1033.1997.00171.x. [DOI] [PubMed] [Google Scholar]
- 220.Tachibana Y, Fujiwara S, Takagi M, Imanaka T. Cloning and expression of the 4-α-glucanotransferase gene from the hyperthermophilic archaeon Pyrococcus sp. KOD1, and characterization of the enzyme. J Ferment Bioeng. 1997;83:540–548. [Google Scholar]
- 221.Labes A, Schonheit P. Unusual starch degradation pathway via cyclodextrins in the hyperthermophilic sulfate-reducing archaeon Archaeoglobus fulgidus strain 7324. J Bacteriol. 2007;189:8901–8913. doi: 10.1128/JB.01136-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Dong G, Vieille C, Zeikus JG. Cloning, sequencing, and expression of the gene encoding amylopullulanase from Pyrococcus furiosus and biochemical characterization of the recombinant enzyme. Appl Environ Microbiol. 1997;63:3577–3584. doi: 10.1128/aem.63.9.3577-3584.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Erra-Pujada M, Debeire P, Duchiron F, O’Donohue MJ. The type II pullulanase of Thermococcus hydrothermalis: molecular characterization of the gene and expression of the catalytic domain. J Bacteriol. 1999;181:3284–3287. doi: 10.1128/jb.181.10.3284-3287.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Imamura H, Jeon BS, Wakagi T. Molecular evolution of the ATPase subunit of three archaeal sugar ABC transporters. Biochem Biophys Res Commun. 2004;319:230–234. doi: 10.1016/j.bbrc.2004.04.174. [DOI] [PubMed] [Google Scholar]
- 225.Jiao YL, Wang SJ, Lv MS, Xu JL, Fang YW, Liu S. A GH57 family amylopullulanase from deep-sea Thermococcus siculi: expression of the gene and characterization of the recombinant enzyme. Curr Microbiol. 2011;62:222–228. doi: 10.1007/s00284-010-9690-6. [DOI] [PubMed] [Google Scholar]
- 226.Ballschmiter M, Fütterer O, Liebl W. Identification and characterization of a novel intracellular alkaline α-amylase from the hyperthermophilic bacterium Thermotoga maritima MSB8. Appl Environ Microbiol. 2006;72:2206–2211. doi: 10.1128/AEM.72.3.2206-2211.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Murakami T, Kanai T, Takata H, Kuriki T, Imanaka T. A novel branching enzyme of the GH-57 family in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J Bacteriol. 2006;188:5915–5924. doi: 10.1128/JB.00390-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Palomo M, Pijning T, Booiman T, Dobruchowska JM, van der Vlist J, Kralj S, Planas A, Loos K, Kamerling JP, Dijkstra BW, van der Maarel MJ, Dijkhuizen L, Leemhuis H. Thermus thermophilus glycoside hydrolase family 57 branching enzyme: crystal structure, mechanism of action, and products formed. J Biol Chem. 2011;286:3520–3530. doi: 10.1074/jbc.M110.179515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.van Lieshout JFT, Verhees CH, van der Oost J, de Vos WM, Ettema TJG, van der Sar S, Imamura H, Matsuzawa H. Identification and molecular characterization of a novel type of α-galactosidase from Pyrococcus furiosus . Biocatal Biotransform. 2003;21:243–252. [Google Scholar]
- 230.Comfort DA, Chou CJ, Conners SB, VanFossen AL, Kelly RM. Functional-genomics-based identification and characterization of open reading frames encoding α-glucoside-processing enzymes in the hyperthermophilic archaeon Pyrococcus furiosus . Appl Environ Microbiol. 2008;74:1281–1283. doi: 10.1128/AEM.01920-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Wang H, Gong Y, Xie W, Xiao W, Wang J, Zheng Y, Hu J, Liu Z. Identification and characterization of a novel thermostable gh-57 gene from metagenomic fosmid library of the Juan de Fuca Ridge hydrothermal vent. Appl Biochem Biotechnol. 2011;164:1323–1338. doi: 10.1007/s12010-011-9215-1. [DOI] [PubMed] [Google Scholar]
- 232.Li D, Li X, Park KH. An extremely thermostable amylopullulanase from Staphylothermus marinus displays both pullulan- and cyclodextrin-degrading activities. Appl Microbiol Biotechnol. 2013;97:5359–5369. doi: 10.1007/s00253-012-4397-1. [DOI] [PubMed] [Google Scholar]
- 233.Imamura H, Fushinobu S, Jeon BS, Wakagi T, Matsuzawa H. Identification of the catalytic residue of Thermococcus litoralis 4-α-glucanotransferase through mechanism-based labeling. Biochemistry. 2001;40:12400–12406. doi: 10.1021/bi011017c. [DOI] [PubMed] [Google Scholar]
- 234.Dickmanns A, Ballschmiter M, Liebl W, Ficner R. Structure of the novel α-amylase AmyC from Thermotoga maritima . Acta Crystallogr D Biol Crystallogr. 2006;62:262–270. doi: 10.1107/S0907444905041363. [DOI] [PubMed] [Google Scholar]
- 235.Santos CR, Tonoli CC, Trindade DM, Betzel C, Takata H, Kuriki T, Kanai T, Imanaka T, Arni RK, Murakami MT. Structural basis for branching-enzyme activity of glycoside hydrolase family 57: structure and stability studies of a novel branching enzyme from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. Proteins. 2011;79:547–557. doi: 10.1002/prot.22902. [DOI] [PubMed] [Google Scholar]
- 236.Zona R, Chang-Pi-Hin F, O’Donohue MJ, Janecek S. Bioinformatics of the glycoside hydrolase family 57 and identification of catalytic residues in amylopullulanase from Thermococcus hydrothermalis . Eur J Biochem. 2004;271:2863–2872. doi: 10.1111/j.1432-1033.2004.04144.x. [DOI] [PubMed] [Google Scholar]
- 237.Matsuura Y. A possible mechanism of catalysis involving three essential residues in the enzymes of α-amylase family. Biologia. 2002;57(Suppl 11):21–27. [Google Scholar]
- 238.Li M, Peeples TL. Purification of hyperthermophilic archaeal amylolytic enzyme (MJA1) using thermoseparating aqueous two-phase systems. J Chromatogr B. 2004;807:69–74. doi: 10.1016/j.jchromb.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 239.Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, Blake JA, FitzGerald LM, Clayton RA, Gocayne JD, Kerlavage AR, Dougherty BA, Tomb JF, Adams MD, Reich CI, Overbeek R, Kirkness EF, Weinstock KG, Merrick JM, Glodek A, Scott JL, Geoghagen NS, Venter JC. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii . Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 240.Janecek S (2013) Glycoside hydrolase family 57. CAZypedia. http://www.cazypedia.org/. Accessed 18 Mar 2013
- 241.Watanabe H, Nishimoto T, Kubota M, Chaen H, Fukuda S. Cloning, sequencing, and expression of the genes encoding an isocyclomaltooligosaccharide glucanotransferase and an α-amylase from a Bacillus circulans strain. Biosci Biotechnol Biochem. 2006;70:2690–2702. doi: 10.1271/bbb.60294. [DOI] [PubMed] [Google Scholar]
- 242.Janecek S, Kuchtova A. In silico identification of catalytic residues and domain fold of the family GH119 sharing the catalytic machinery with the α-amylase family GH57. FEBS Lett. 2012;586:3360–3366. doi: 10.1016/j.febslet.2012.07.020. [DOI] [PubMed] [Google Scholar]
- 243.Naumoff DG. Hierarchical classification of glycoside hydrolases. Biochemistry (Moscow) 2011;76:622–635. doi: 10.1134/S0006297911060022. [DOI] [PubMed] [Google Scholar]
- 244.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 245.Kelley LA, Sternberg MJE. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 246.Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL repository and associated resources. Nucleic Acids Res. 2009;37:D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Guerin DM, Lascombe MB, Costabel M, Souchon H, Lamzin V, Beguin P, Alzari PM. Atomic (0.94 Å) resolution structure of an inverting glycosidase in complex with substrate. J Mol Biol. 2002;316:1061–1069. doi: 10.1006/jmbi.2001.5404. [DOI] [PubMed] [Google Scholar]
- 248.Guimaraes BG, Souchon H, Lytle BL, David Wu JH, Alzari PM. The crystal structure and catalytic mechanism of cellobiohydrolase CelS, the major enzymatic component of the Clostridium thermocellum cellulosome. J Mol Biol. 2002;320:587–596. doi: 10.1016/s0022-2836(02)00497-7. [DOI] [PubMed] [Google Scholar]
- 249.Machius M, Declerck N, Huber R, Wiegand G. Activation of Bacillus licheniformis α-amylase through a disorder→order transition of the substrate-binding site mediated by a calcium–sodium–calcium metal triad. Structure. 1998;6:281–292. doi: 10.1016/s0969-2126(98)00032-x. [DOI] [PubMed] [Google Scholar]
- 250.Deshpande N, Addess KJ, Bluhm WF, Merino-Ott JC, Townsend-Merino W, Zhang Q, Knezevich C, Xie L, Chen L, Feng Z, Green RK, Flippen-Anderson JL, Westbrook J, Berman HM, Bourne PE. The RCSB Protein Data Bank: a redesigned query system and relational database based on the mmCIF schema. Nucleic Acids Res. 2005;3:D233–D237. doi: 10.1093/nar/gki057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.UniProt Consortium Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res. 2013;41:D43–D47. doi: 10.1093/nar/gks1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 253.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 254.Felsenstein J. Confidence-limits on phylogenies—an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 255.Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 256.Shatsky M, Nussinov R, Wolfson HJ. A method for simultaneous alignment of multiple protein structures. Proteins. 2004;56:143–156. doi: 10.1002/prot.10628. [DOI] [PubMed] [Google Scholar]
- 257.Davies GJ, Wilson KS, Henrissat B. Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem J. 1997;321:557–559. doi: 10.1042/bj3210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Sauer J, Sigurskjold BW, Christensen U, Frandsen TP, Mirgorodskaya E, Harrison M, Roepstorff P, Svensson B. Glucoamylase: structure/function relationships, and protein engineering. Biochim Biophys Acta. 2000;1543:275–293. doi: 10.1016/s0167-4838(00)00232-6. [DOI] [PubMed] [Google Scholar]
- 260.Aleshin AE, Firsov LM, Honzatko RB. Refined structure for the complex of acarbose with glucoamylase from Aspergillus awamori var. X100 to 2.4-Å resolution. J Biol Chem. 1994;269:15631–15639. [PubMed] [Google Scholar]




