Abstract
Antimicrobial peptides are a vital component of the innate immune system of all eukaryotic organisms and many of these peptides have potent antifungal activity. They have potential application in the control of fungal pathogens that are a serious threat to both human health and food security. Development of antifungal peptides as therapeutics requires an understanding of their mechanism of action on fungal cells. To date, most research on antimicrobial peptides has focused on their activity against bacteria. Several antimicrobial peptides specifically target fungal cells and are not active against bacteria. Others with broader specificity often have different mechanisms of action against bacteria and fungi. This review focuses on the mechanism of action of naturally occurring antifungal peptides from a diverse range of sources including plants, mammals, amphibians, insects, crabs, spiders, and fungi. While antimicrobial peptides were originally proposed to act via membrane permeabilization, the mechanism of antifungal activity for these peptides is generally more complex and often involves entry of the peptide into the cell.
Keywords: Antifungal peptides, Antimicrobial peptide
Introduction
Innate immune systems have evolved in all kingdoms of eukaryotes to protect against infection by bacteria, viruses, and eukaryotic pathogens such as fungi and parasites. The components of these immune systems consist of small-molecule secondary metabolites as well as small proteins and peptides with a broad spectrum of activities against pathogens. The number of identified peptides now exceeds 1,700 and is continually increasing [1]. Although they often possess common attributes such as small size, an overall positive charge and amphipathicity, they fall into a number of diverse and distinct groups. These include α-helical peptides, β-sheet peptides, those with mixed α-helical and β-sheet structures, extended peptides and peptides enriched in specific amino acids. Whereas the antibacterial activities of a number of peptides have been described in detail, less is known about their antifungal effects, which will be the main focus of this review. Gene encoded peptides are of particular interest as the genes can be cloned and expressed recombinantly for pharmaceutical applications or employed in the generation of transgenic plants for agricultural applications. This review focuses on antifungal peptides with activity in the low micromolar range whose mechanism of action has been investigated.
Bacteria and viruses generally get more public attention than fungi because they spread rapidly and have drastic effects on human health and well-being but a number of fungal species are also serious pathogens [2, 3]. Advances in medicine, particularly the treatment of HIV/AIDS and cancer and the increased success rates in organ transplantation, has led to an increase in the number of people with compromised immune systems. These people are highly susceptible to fungal infection. The fourth most common cause of nosocomial infection is now the major human fungal pathogen Candida albicans [4]. Mortality rates associated with systemic fungal infection are close to 50 %, with rates reaching 100 % for some fungal pathogens in the developing world [5]. Antifungal treatments that are currently employed in the clinic are being rendered insufficient due to issues with toxic side effects, poor efficacy, and the development of resistance in pathogens. These factors all contribute to the need to investigate the potential use of innate immunity peptides as novel therapeutics for maintenance of human health. Other fungal species, particularly filamentous fungi, are pathogenic to plant species and cause crop losses of major economic significance each year [3, 6]. Expression of certain antifungal proteins in transgenic plants has increased resistance to disease [7–9]. For these reasons, the development of novel antifungal molecules is of great interest for both human and plant protection. An understanding of the mechanisms of action of naturally occurring antifungal peptides will be essential in achieving these goals.
Fungal cell architecture
The cell wall
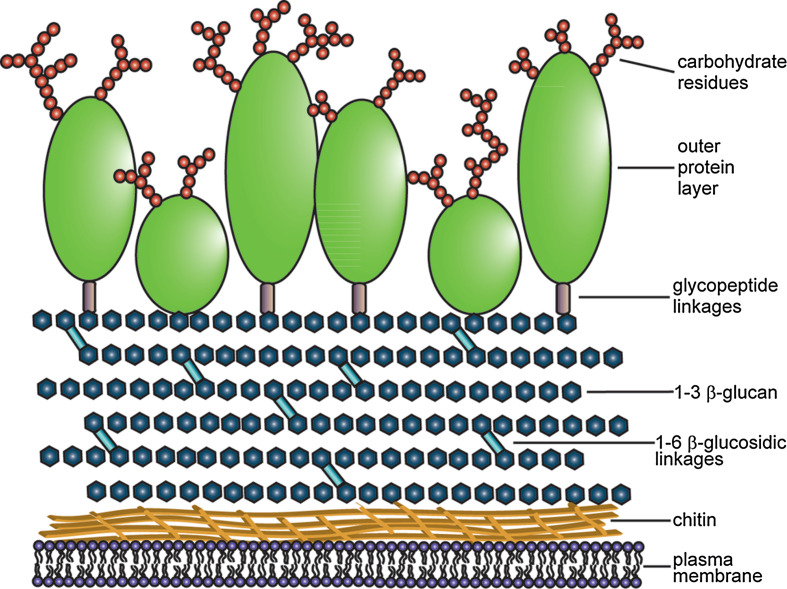
Fungal cells differ from other eukaryotic cells in many ways. The cell wall acts as a protective barrier, limiting the access of molecules to the plasma membrane, as well as being involved in cell adhesion, pathogenesis, and cell signaling [10]. Yeast and filamentous fungi share similar cell wall architecture (Fig. 1). The chitin layer sits adjacent to the plasma membrane and, while it is the least abundant cell wall constituent (2–10 %), its crystalline structure plays a large role in cell wall stability [11]. The β-glucan network consists largely of (1 → 3)-β-glucans with (1 → 6)-β-branches. However, in yeast, (1 → 6)-β-glucans are also present [11, 12]. This network represents 50–60 % of the cell wall by weight [13]. The outermost layer of the cell wall is comprised of glycosylated proteins and constitutes between 20 and 60 % of cell wall mass [11, 13]. Glycosylated proteins on the yeast cell surface are decorated by mannose residues while those in filamentous fungi can also include galactose, glucose, and uronic acids [13]. A large degree of intermolecular disulfide bridging also occurs between proteins in the outer layer. These disulfide linkages, as well as the carbohydrate moieties extending from these proteins, play a major role in limiting the porosity of the wall [14]. Apart from being cross-linked to each other, cell wall proteins are also covalently linked to the β-glucan network in two ways. The first involves a GPI-anchor that links the protein to the (1 → 3)-β-glucan via a (1 → 6)-β-glucosidic linkage. These proteins are trafficked to the plasma membrane before part of the GPI-anchor is removed and the β-glucan is attached [15]. The second linkage involves direct attachment of the protein to the (1 → 3)-β-glucan via an alkali-sensitive bond [16].
Fig. 1.
Fungal cell wall composition. Schematic representation of the fungal cell wall showing the outer layer of glycosylated proteins (green) with the linked carbohydrate residues (red circles), the (1–3)-β-glucan network (dark blue) with (1–6)-β-glucosidic linkages (light blue), and the chitin layer (yellow) that lies adjacent to the plasma membrane of the cell (purple)
Plasma membrane composition
Plasma membranes are composed of three main lipids: phospholipids, sphingolipids, and sterols. The plasma membranes of mammalian cells typically include zwitterionic phospholipids such as phosphatidylcholine, in contrast to bacteria and fungi, which are richer in anionic phospholipids. This difference in lipid composition is proposed to contribute to the selectivity of some antimicrobial peptides [17]. In bacteria, the common anionic lipids are phosphatidylglycerol and cardiolipin, whereas phosphatidylserine and phosphatidylinositol are more common in fungal membranes [18]. The plasma membranes of eukaryotic cells also contain sterols. In animal cells, this is generally cholesterol, while lower eukaryotes, including fungi, contain ergosterol [19]. This difference in sterol content is exploited in the mechanisms of antifungal drugs including amphotericin B and the azoles [20, 21].
Sterols and sphingolipids are often associated in “lipid rafts” with GPI-anchored proteins [22]. Disruption of either ergosterol or sphingolipid synthesis prevents raft formation in yeast and also impairs delivery of GPI-anchored proteins to the cell membrane [23]. Since cell wall proteins are also trafficked via a GPI anchor, it is likely that delivery of these proteins is also affected by disruption of lipid rafts. Sphingolipids are potentially specific targets for antifungal molecules due to structural differences between fungal, plant, and mammalian sphingolipids such as 9-methyl group branching of the sphingoid base and different degrees of unsaturation in fungal sphingolipids [24].
Antimicrobial peptides
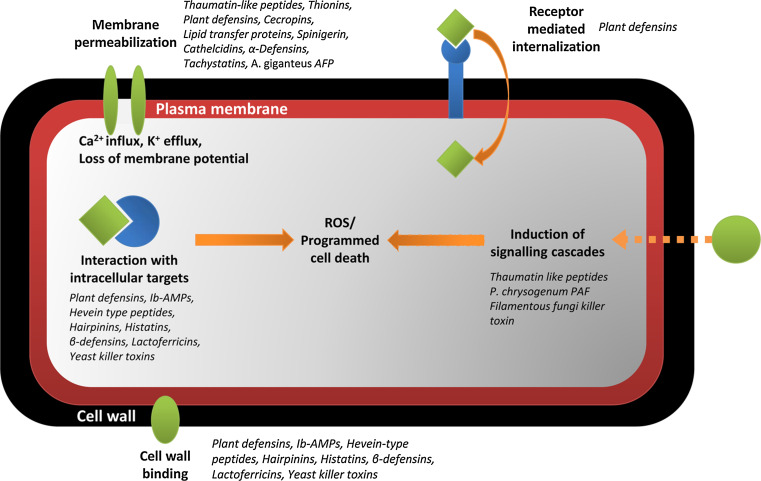
The survival of all higher organisms is dependent upon their ability to protect themselves from attack by pathogens. In many instances, this involves the production of a myriad of antimicrobial peptides. The main focus for discovery of antimicrobial peptides has long been restricted to those with antibacterial activity. As such, antibacterial peptides are the best characterized. Relatively little is known about the mechanism of action of antifungal peptides. Table 1 lists the antifungal peptides described in this review. Some of these peptides act specifically against fungi, while others have broader activity. Figure 2 summarizes the range of mechanisms of action that have been described for these peptides.
Table 1.
Naturally occurring antifungal peptides
| Peptide | Source | Structural class | Target(s) |
|---|---|---|---|
| Thaumatin-like proteins | Plants | β-sheets | Filamentous fungi, yeast |
| Thionins | Plants | Mixed αβ | Filamentous fungi, yeast, bacteria, mammalian cells |
| Plant defensins | Plants | Cysteine-stabilized αβ motif | Filamentous fungi, yeast |
| Lipid transfer proteins | Plants | α-helical bundle | Filamentous fungi, bacteria |
| IbAMPs | Plants | β-hairpin | Filamentous fungi, less active against yeast |
| Snakins | Plants | Not reported | Filamentous fungi, bacteria |
| Hevein-type peptides | Plants | Mixed αβ | Filamentous fungi, yeast, bacteria |
| Knottin-type peptides | Plants | β-sheet (cystein-knot) | Filamentous fungi, some Gram positive bacteria |
| 2S albumin peptides | Plants | α-helical | Filamentous fungi |
| Hairpinins | Plants | α-helical | Filamentous fungi |
| Histatins | Humans | α-helical | Yeast |
| Cathelicidins | Mammals | Variable | Bacteria, yeast, less active against filamentous fungi |
| Mammalian defensins | Mammals | Mixed αβ | Yeast |
| Lactoferrin-derived | Mammals | Variety of structures | Bacteria, yeast, filamentous fungi |
| Temporins | Insects | α-helical | Bacteria, yeast, filamentous fungi |
| Brevinin | Insects | Mixed αβ | Yeast, bacteria |
| Insect defensins | Insects | Cysteine-stabilized αβ motif | Filamentous fungi, yeast, some active against bacteria |
| Thanatin | Insects | β-hairpin | Filamentous fungi, bacteria |
| Glycine-rich peptides | Insects | Not reported | Yeast |
| Cecropins | Insects | α-helical | Filamentous fungi, bacteria |
| Spinigerin | Insects | α-helical | Filamentous fungi, yeast, bacteria |
| Insect knottin-type peptides | Insects | β-sheet (cystein-knot) | Yeast |
| Penaeidins | Shrimp | α-helical, random coil | Filamentous fungi, Gram positive bacteria |
| Hemocyanin-derived peptides | Shrimp | Not reported | Filamentous fungi |
| Crab β-hairpin peptides | Crab | β-hairpin | Yeast, bacteria |
| Tachystatins | Crab | β-sheet (cystein-knot) | Yeast, bacteria |
| Big defensin | Crab | Mixed αβ | Yeast, bacteria |
| Cenchritis muricatus peptides | Mollusk | α-helical | Filamentous fungi, yeast |
| Gomesin | Spiders | β-hairpin | Filamentous fungi, yeast |
| Yeast killer toxins | Yeast | Mixed | Yeast |
| Peptides from filamentous fungi | Fungi | β-sheet (barrel-like) | Filamentous fungi |
Fig. 2.
Mechanisms of action of antifungal peptides. Schematic representation of the various mechanisms of action proposed for antifungal peptides described in this review
Antifungal peptides from plants
Due to the constant threat of attack from fungal pathogens and their lack of an adaptive immune response, plants express a large number of antifungal proteins for protection against fungal disease. These proteins either play a role in the plant’s constitutive immunity or can be induced upon attack by pathogens. The inducible, pathogen-related (PR) proteins are generally expressed not only at the site of infection but also systemically. Seventeen families of PR proteins have been described to date. Many of these are enzymes including (1 → 3)-β-glucanases (PR-2), chitinases (PR-3, -4, -8 and -11), proteinases (PR-7) peroxidases (PR-9), and oxalate oxidases (PR-16 and -17). Their activities are relatively well understood and have been reviewed previously so they will not be discussed in detail here [see review by 18]. An ever-increasing number of small, disulfide-rich proteins also exist including thaumatin-like proteins, thionins, lipid transfer proteins (LTP), plant defensins, hevein- and knottin-type proteins, Ib-AMPs and snakins. Some members of these families have been classified into PR groups such as defensins (PR-12), thionins (PR-13), and LTPs (PR-14) [25]. Since only pathogen-induced proteins can be placed in these groups, many family members are excluded so this nomenclature will not be used here.
Thaumatin-like proteins
Thaumatin, from the African shrub Thaumatococcus daniellii, is a 22-kDa extremely sweet tasting protein that displays antifungal activity. Similar thaumatin-like (TL) proteins with antifungal activity have been isolated from other plant species including tobacco, maize, barley, winter wheat, and black nightshade [26–30]. The structures of several of these proteins have been solved [31–34] and they display a conserved fold consisting of three domains. Domain I is an 11-strand flattened β-sandwich that forms the core of the molecule, from which a number of disulfide-stabilized loops extend (domains II and III, Fig. 3a) [31]. Another conserved feature is the presence of a cleft between domains I and II. This cleft has an overall basic charge in thaumatin but is acidic in the other family members [26, 28, 32]. This cleft is believed to be involved in binding of the TL protein osmotin, from Nicotiana tabacum, to fungal cell wall components including β-1,3 glucans [28, 32, 35]. Some TL proteins also exhibit glucanase activity [36].
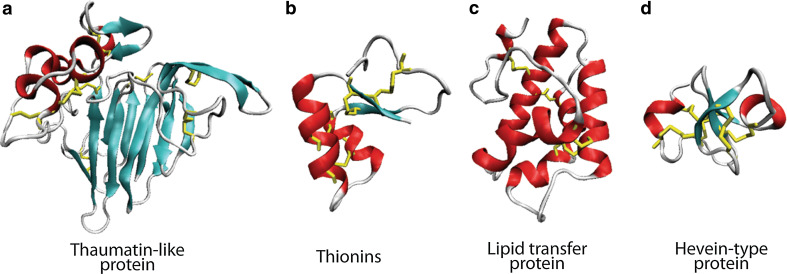
Fig. 3.
Plant antifungal protein structures. a Thaumatin-like protein-domain I is the 11-stranded flattened β-sandwich from which domain II and III extend. b Thionin-Г (gamma) shaped fold with two α-helices forming the long arm and two β-strands forming the short arm. c Lipid transfer protein-bundle of four α-helices joined by loop regions. d Hevein type protein-three strand antiparallel β-sheet with α-helices on either side
Osmotin is the best characterized TL protein. It is produced in response to osmotic stress and accumulates to high concentrations in the vacuole. It is growth inhibitory toward a number of fungi but does not retain the sweet-tasting characteristic of thaumatin [29]. Osmotin, along with zeamatin, another TL protein from maize, permeabilizes the membranes of susceptible fungi such as Fusarium oxysporum [30, 37]. It is unlikely that this results from direct interaction of the protein with the membrane because TL proteins do not exhibit any of the structural characteristics of membrane permeabilizing peptides such as amphipathicity. However, permeabilization by zeamatin occurs readily at 4 °C, indicating that involvement of an enzymatic activity in the antifungal mechanism is unlikely [30]. Zeamatin is known to inhibit α-amylase and trypsin [38] but such inhibitory activities has not been linked to antifungal activity or membrane permeabilization. In contrast, a flax seed TL protein permeabilizes artificial liposomes [39], which suggests direct peptide–lipid interaction. TL proteins demonstrate variable activity against a spectrum of fungal species, even among different strains of the same species. This specificity is predicted to result, in part, from differences in the fungal cell wall. S. cerevisiae, for example, is resistant to osmotin and this resistance has been attributed to the presence of three Pir proteins (Pir 1–3) in the cell wall [40]. Disruption of the gene responsible for targeting of these proteins to the cell wall (SSD1) renders the yeast sensitive to osmotin [41]. Furthermore, the expression of one of these S. cerevisiae Pir proteins in the osmotin sensitive fungus Fusarium oxysporum leads to osmotin resistance [42]. While the presence of Pir proteins leads to osmotin resistance, other cell wall proteins, in particular the carbohydrate moieties on these proteins, are required for osmotin sensitivity [43]. Disruption of the enzymes (mannosyltransferase) responsible for the transfer of phosphomannans to glycosylated proteins leads to a decrease in osmotin sensitivity [43]. This is postulated to result from the decrease in negative charges on the cell surface and the associated decrease in binding of the positively charged osmotin.
Another interesting feature of osmotin is its ability to induce kinase signaling cascades. Osmotin activates the pheromone-response MAP kinase signal pathway in S. cerevisiae, leading to modifications in the cell wall that increase the susceptibility of the cell to the protein [44]. It also activates a RAS2/cAMP stress response pathway that induces apoptosis. This activation is mediated through a G-protein coupled receptor (GPCR)-like integral membrane protein [45]. This involvement of proteins on the plasma membrane demonstrates that antifungal molecules may have more than one distinct target on a single fungal species. Taken together, these observations suggest that the mode of action of osmotin is complex, and does not simply involve permeabilization of membranes as once thought. Synergistic activity between TL proteins suggests that they act on different fungal targets. For example, when induced to express in grape vines, osmotin and TL protein display synergistic antifungal activity towards Uncinula necator and Phomopsis viticola in infected leaves and berries [46]. Constitutive expression of the rice TL protein in banana plants leads to increased resistance to wilt caused by F. oxysporum sp. Cubensec [47] demonstrating the utility of small antifungal peptides from this class in agricultural biotechnology.
Cysteine-rich peptides
Thionins
The antimicrobial activity of thionins was recorded long before they were isolated from plant tissue. In the late 19th century, brewers noticed that barley seeds contained a substance that was toxic to yeast. In 1942, this substance was isolated and named purothionin [48]. A number of thionins have since been purified from various plant species and tissues. They are small proteins (~5 kDa) that are stabilized by three to four disulfide bonds. They generally carry an overall positive charge of seven or ten depending on which group they belong to (groups I–III). One group (group IV), carries no charge [49]. Despite a high degree of sequence variability, thionins share a common structural fold (Fig. 3b). The structure can be represented by the Greek capital letter Γ (gamma), with two α-helices forming the long arm and two β-strands forming the short arm [49]. A groove between these two regions contains the five amino acids that are most highly conserved between family members. One amino acid in this region, tyrosine 13, is fully conserved among all toxic thionins and iodination of this residue leads to a loss in activity [49].
Thionins display a broad range of toxicity against bacterial, fungal, mammalian, and insect cells. This lack of specificity and the ability of thionins to permeabilize the membranes of target cells [50, 51] implies a direct interaction of thionins with lipids. In support of this, thionins are known to bind to lipids during protein extraction and the addition of phospholipids inhibits their antimicrobial effects [48]. The membrane permeabilizing activity appears to occur via a specific interaction with anionic phospholipids at the conserved cleft that results in solubilization of these lipids and extraction from the membrane [52, 53]. Membrane permeabilization by thionins is inhibited by mono and di-valent cations, a property which is likely to restrict the utility of thionins as antimicrobial agents unless variants can be generated with decreased sensitivity to cations [54].
Plant defensins
Plant defensins are small, cysteine-rich proteins (45–54 amino acids) that have been isolated from many plant species and tissues [55]. A variety of functions have been attributed to plant defensins. While many have antifungal activity, plant defensins have also been described with functions in antibacterial activity, zinc tolerance, and blocking of ion channels [reviewed in 56], as well as inhibition of protein translation machinery, α-amylases and proteases [57–60]. Plant defensins with antifungal activity have shown promise for use in both agricultural and therapeutic settings. Potatoes expressing the alfalfa defensin (MsDef1, previously known as alfAFP) showed significant resistance against the fungal pathogen Verticillium dahlia in the field compared to non-transformed controls [61]. Expression of a Dahlia defensin (DmAMP1) in rice was sufficient to provide protection against two major rice pathogens, Magnaporthe oryzae and Rhizoctonia solani [62]. In addition, treatment with the radish defensin, RsAFP2 led to protection against Candidiasis in a mouse model [63]. A review of the use of plant defensins to engineer fungal resistance of crops can be found in [64] including a table of fungal pathogens for which resistance has been generated through expression of transgenic defensin.
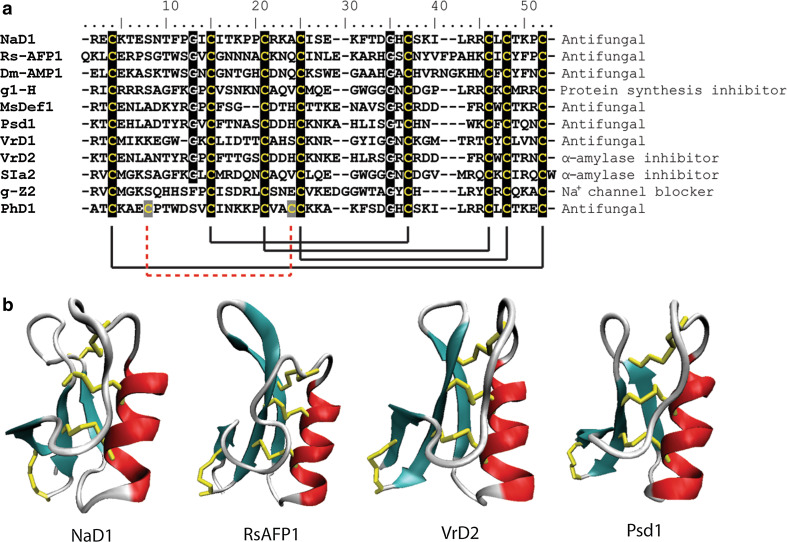
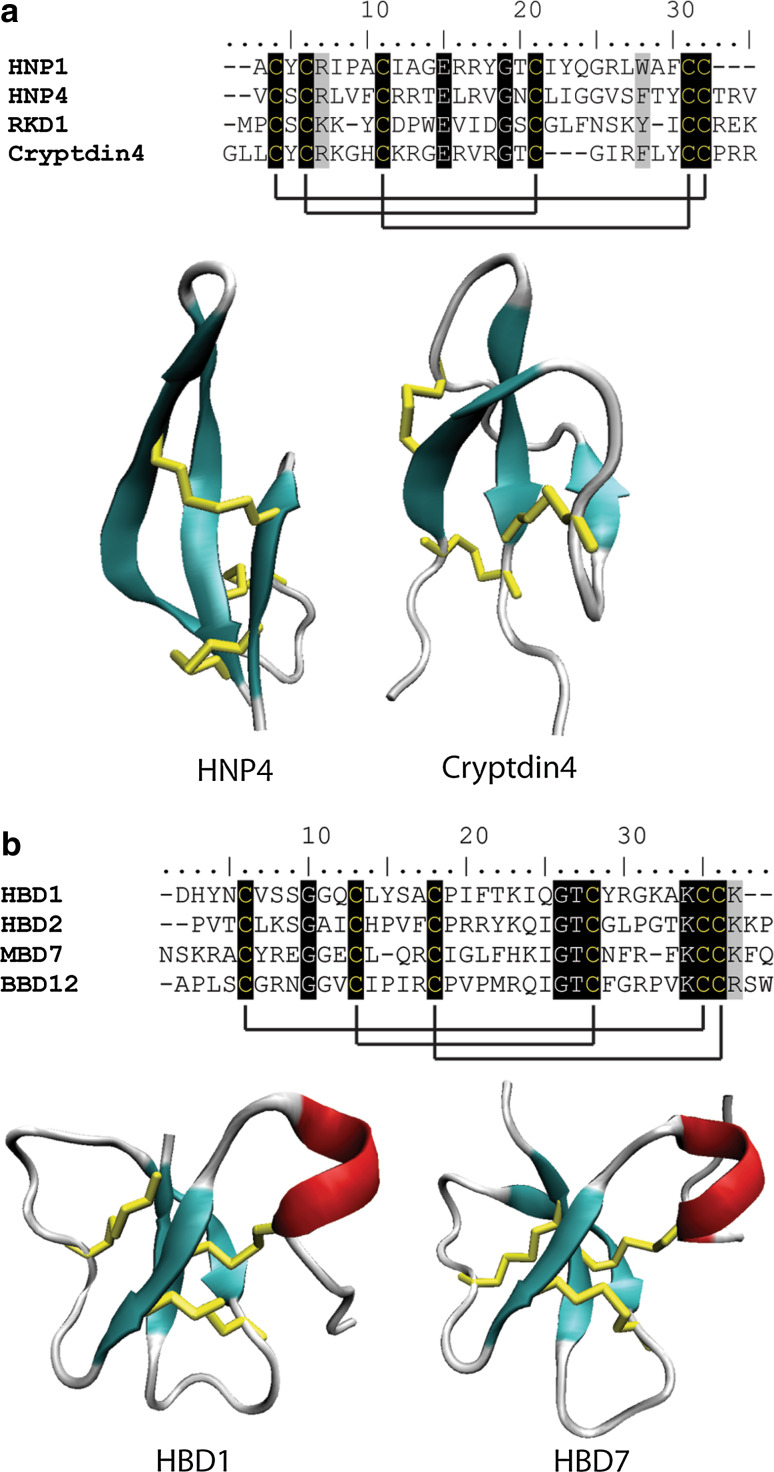
The structures of many defensins have been solved. They display a common fold consisting of a triple-stranded, anti-parallel β-sheet connected to an α-helix by three disulfide bonds forming a cysteine-stabilized αβ motif (CSαβ, Fig. 4b). A fourth disulfide joins the N- and C-termini creating an extremely stable protein [65–69]. Despite this common fold, the level of sequence identity between defensins is very low (Fig. 4a). The eight cysteine residues are invariant, and the two glycine residues (positions 13 and 34), an aromatic residue (position 11) and a glutamate (position 29) are highly conserved (numbering relative to RsAFP2) [70]. Two defensins have also been isolated that contain a fifth disulfide bond [71]. These conserved residues are thought to maintain the structural confirmation of the core defensin fold while variation in the reactive surface loops is the root of the variable specificity with respect to target species and the diversity of functions carried out by plant defensins [60].
Fig. 4.
Sequence alignment and structural comparison of plant defensins. a Sequence alignment of a variety of plant defensins. The eight conserved cysteines that form the four characteristic disulfide bonds (with the extra disulfide in PhD1 as a dotted red line) are highlighted as are the conserved glycine residues. b The conserved cysteine stabilized αβ (CS αβ) fold consisting of a triple-stranded, anti-parallel β-sheet connected to an α-helix by three disulfide bonds. Beta-strands are represented in cyan, random coils and turns are in white, and disulfide bonds are in yellow
Observations to date on the activity of various plant defensins suggest that while those with similar sequences may act via similar mechanisms, those with a low level of sequence identity are likely to act via differing mechanisms. A plant defensin from radish (RsAFP2) and one from dahlia (DmAMP1) cause Ca2+ influx and K+ efflux in Neurospora crassa hyphae as well as alkalinization of the growth media [72]. The sequence identity between RsAFP2 and DmAMP1 is 50 % as calculated using the BLASTp suite-2 sequences (NCBI). Alkalinization initiated by RsAFP2 is inhibited by G-protein inhibitors, but the alkalinization caused by DmAMP1 is not [72]. This suggests that these defensins may induce similar responses via different mechanisms. The presence of high-affinity binding sites for plant defensins on target cells [73, 74] has led to the suggestion that these peptides may undergo receptor-mediated insertion into membranes [74]. This is consistent with the observation that both RsAFP2 and DmAMP1 permeabilize fungal membranes [75] but not artificial bilayers [72, 76].
Sphingolipids have been reported as binding sites for both RsAFP2 and DmAMP1. Disruption of the biosynthetic pathway for the sphingolipid mannosediinositolphosphorylceramide (M(IP)2C) in S. cerevisiae results in resistance to DmAMP1 [74]. DmAMP1 binds to purified M(IP)2C and this binding is enhanced in the presence of ergosterol [77]. RsAFP2 binds to glucosylceramide (GlcCer), another sphingolipid in P. pastoris and C. albicans; [78]. Strains that do not contain this lipid are resistant to RsAFP2-induced permeabilization and growth inhibition but are susceptible to DmAMP1, confirming that RsAFP2 and DmAMP1 bind to different lipid receptors. Interestingly, an RsAFP2 variant (Y38G) that lacks antifungal activity still binds to GlcCer [78]. Also, RsAFP2 is unable to permeabilize artificial liposomes containing GlcCer indicating that binding alone is not sufficient for membrane permeabilization. RsAFP2 does not bind to human or soybean GlcCer and this may determine the protein’s spectrum of activity. Treatment of C. albicans with RsAFP2 leads to generation of reactive oxygen species (ROS) [76] and induction of programmed cell death [79]. The defensin from Heuchera sanguinea also induces ROS and programmed cell death but does not rely on an interaction with sphingolipids and is hypothesized to interact with another, yet to be identified, membrane component [80]. Screening of a C. albicans deletion collection revealed a role for genes with functions in cell wall integrity and hyphal growth/septin ring formation in RsAFP2 tolerance [81]. This work continued to provide evidence for interaction between the defensin and glucosylceramides in the cell wall. RsAFP2 induced morphological changes in the cell wall and altered septum formation leading to activation of the cell wall integrity pathway [81].
Like RsAFP2, MsDef1, a defensin from Medicago sativa, is unable to inhibit the growth of F. graminearum variants lacking GlcCer [82] suggesting a commonality in the mechanism involving GlcCer binding. However, while MsDef1 interacts with, and blocks mammalian L-type calcium channels, RsAFP2 does not [83]. This indicates overlapping but different mechanisms of action for these two peptides. MsDef1 activates two of the three MAP kinase signaling cascades present in Fusarium graminearum (Gpmk1 and Mgv1) and deletions of genes in these pathways leads to increased sensitivity of the fungus to MsDef1, RsAFP2, and MtDef2 (from Medicago truncatula) [84]. Interestingly, a second M. truncatula defensin, MtDef4, does not activate these pathways and does not display enhanced activity toward the deletion mutants. This indicates that signaling cascades are involved in mediating adaptive resistance to some but not all plant defensins, for example, through cell wall modifications.
Some plant defensins enter fungal cells and interact with intracellular targets. NaD1, a defensin expressed at high levels in the floral tissue of Nicotiana alata, interacts with the fungal cell wall and permeabilizes the plasma membrane before traversing into the cytoplasm [85]. Interaction with the cell wall is critical for activity and the kinetics of permeabilization differ significantly to that of other AMPs that act through membrane permeabilization [86]. Permeabilization is also saturable, suggesting that interaction with a receptor may be involved. Whether interaction with specific intracellular targets is also required for activity is as yet unknown. PsD1 (from Pisum sativum) localizes to the nucleus of treated N. crassa cells, interacts with a cell cycle control protein, cyclin F, and halts the cell cycle [87]. Overall, the studies carried out to date indicate the sequence divergence of plant defensins may be indicative of abundant, variable mechanisms of antifungal activity.
Lipid transfer proteins
Lipid transfer proteins (LTPs) are slightly larger than thionins and defensins (~10 kDa) and contain four disulfide bonds. Members of this family share an overall sequence identity of about 30 %, including the eight cysteine residues. While LTPs were originally identified for their ability to transfer lipids from mitochondria to artificial liposomes [88], their function as intracellular lipid traffickers is unlikely, as they are not present in the cytosol and are targeted to the cell wall [89]. Furthermore, an LTP from onion seeds (AceAMP1) is unable to transfer phospholipids from liposomes to mitochondria, indicating not all LTPs function in the same manner [90]. The structure of LTPs consists of a bundle of four α-helices joined by loop regions [91] (Fig. 3C). The four α-helices form a hydrophobic pocket that can accommodate a fatty-acyl chain.
Antifungal and antibacterial activities have been reported for a number of LTPs, including AceAMP1 [91]. As yet, the mechanism of antifungal activity of LTPs is not well understood, although a recent study into the activity of Ha-AP10 from Helianthus annuus reveals that it permeabilizes fungal spores as well as artificial liposomes composed of the anionic lipid phosphatidylglycerol but not the zwitterionic lipid phosphatidylcholine [92]. The mechanism behind this permeabilization and whether it is representative of the activity of other LTPs is still unknown. Lipid transfer proteins isolated from Coffea canephora and Capsicum annuum have also been shown to inhibit mammalian α-amylases [93, 94]
Ib-AMPs
Four small (20-mer) peptides have been isolated from the seeds of Impatiens balsamina (Ib-AMP1-4). They are the smallest antifungal peptides identified in plants, and are produced through processing of a single polypeptide precursor [95]. The structure of Ib-AMP1 has been solved by NMR and reveals a well-defined loop structure stabilized by two disulfide bonds [96]. The peptides inhibit the growth of bacteria and fungi, and are 2–20 times more active against filamentous fungi than single-celled yeast [97]. This selectivity may be explained by binding of IbAMP1 to chitin. Filamentous fungi have about five times more chitin than yeast and it is likely that the higher concentration of chitin on the surface of filamentous fungi increases the binding of Ib-AMP to the cell. The peptides bind to the hyphal walls and are not thought to act via pore formation because they do not significantly disrupt artificial liposomes [98]. IbAMP1 enters the cytoplasm of C. albicans cells [97] but the mechanism of this internalization and its role in growth inhibition is not understood. Variants containing all d-amino acids are just as active as the native peptide [97], suggesting that interaction with a specific receptor is not required for activity. In addition, linear analogues of IbAMP1 have improved activity against Staphylococcus aureus, probably due to their enhanced ability to depolarize the plasma membrane [99].
Snakins
Snakins are 63–66 amino acid peptides (~7 kDa) that were first isolated from potatoes [100]. Homologous cDNAs have been obtained from many other plant species [101]. All snakins have 12 conserved cysteine residues and six disulfide bonds [100]. Snakin-1 (SN-1) is expressed constitutively in the outer layers of the potato tuber, stems, axillary buds, and flower petals. Expression of the SN-1 gene is not induced by activation of hormone-induced stress response pathways as exposure to methyl jasmonate, ethylene, abscisic acid, salicylic acid, isonicotinic acid, or indolacetic did not result in induction of SN-1 mRNA and challenge by bacterial and fungal pathogens also failed to induce expression of the SN-1 gene in potato leaves [100]. The activity of SN-1 varies against different fungal species and, while SN-1 exhibits synergy with potato tuber defensin (PTH1) against some fungal strains, the combined effect is merely additive against other species [100]. This indicates that combinations of antimicrobial peptides can work together against pathogens and that the mechanism of activity of a single antifungal molecule may vary against different pathogens. Snakin-2 (SN-2) is also basally expressed in potato tissue and exhibits only 38 % amino acid sequence identity with SN-1. However, SN-2 is induced upon wounding and in response to pathogen infection [101]. Interestingly, despite their high level of sequence variation, SN-1 and SN-2 display very similar activity spectra [101]. Snakins are not predicted to act via membrane permeabilization and have no effect on artificial liposomes [100].
Hevein-type peptides
Hevein, a protein from the rubber tree, and related peptides range in size from 30 to 43 amino acids and contain three to five disulfide bonds. Their structure consists of a triple-stranded, antiparallel β-sheet with α-helices on either side (Fig. 3d) [102]. Generally, hevein-type peptides exhibit chitin-binding properties and consequently were thought to inhibit fungal growth by interfering with cell wall biosynthesis [103]. However, hevein-type peptides from Pharbitis nil (Pn-AMP1) and Eucommia ulmoides (EAFP2) have recently been demonstrated to have activity against fungal species lacking chitin [102, 104]. Pn-AMP1 does not enter fungal hyphae but it does cause actin depolarization leading to growth arrest [104]. As reported for osmotin, the sensitivity of cells to Pn-AMP1 is mediated by a mannosyltransferase indicating cell wall binding to carbohydrate moieties may be essential [104]. Hevein-type peptides from wasabi (WjAMP-1), cycad (Cy-AMP1) and spindle tree (Ee-CBP) are also active against bacteria [105–107]. Interestingly, Cy-AMP1 requires its chitin-binding activity for antifungal, but not for antibacterial activity [107]. Hevein-type proteins from the wheat Triticum kiharae have recently been described to play a role in salinity stress as well as in fungal resistance, expanding the function of hevein-type proteins beyond the antifungal response [108].
Knottin-type peptides
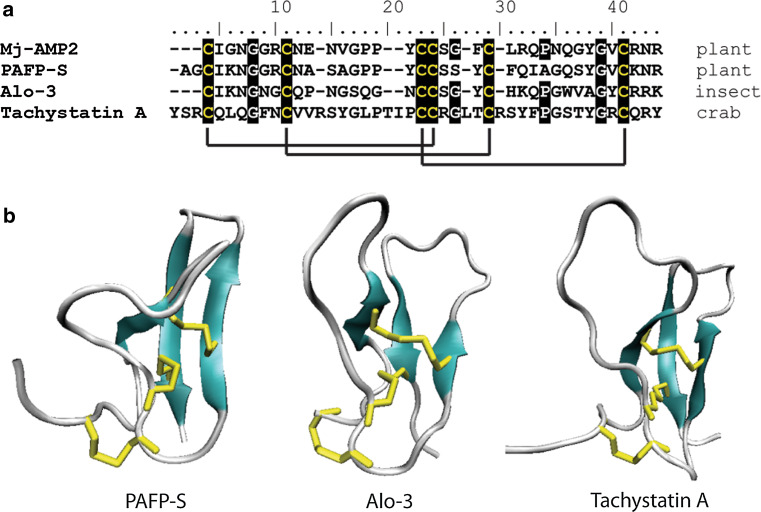
The knottin-type peptides are a family of proteins with, as the name suggests, a cysteine-stabilized, “knotted” topology, defined by two parallel disulfide bonds threaded by a third disulfide bond (Fig. 5) [109]. Proteins from diverse sources and with diverse functions are members of this group, including spider neurotoxins, protease inhibitors and antifungal peptides from insects and crabs. A comprehensive database of knottin-type peptides can be found at http://knottin.cbs.cnrs.fr. Knottin-type, antifungal peptides have been isolated from the plants Mirabilis jalapa L. (Mj-AMP1 and 2) [110] and Phytolacca americana (PAFP-S) [111]. The structure of PAFP-S consists of a triple-stranded, anti-parallel, β-sheet with a long loop region connecting β-strands 1 and 2.
Fig. 5.
Sequence alignment and structural comparison of knottin-type peptides. a Sequence conservation within knottin-type peptides. The six conserved cysteines that form the three characteristic disulfide bonds are highlighted as are two conserved glycine residues and a conserved proline. Cysteine resides are highlighted in black with solid black lines denoting disulfide bonds. b The conserved structure of knottin-type peptides including PAFP-S (pdb code 1DKC), alo-3 (pdb code 1Q3J), and tachystatin A (pdb code 1CIX) include a triple-stranded antiparallel β-sheet with two parallel disulfide bonds threaded by a third. Beta-strands are represented in cyan, random coils and turns are in white, and disulfide bonds are in yellow
The two peptides, Mj-AMP1 and Mj-AMP2, differ by only four amino acids and yet Mj-AMP2 has ten-fold higher activity against most fungi [110]. Three of these differences occur in the C-terminal hairpin domain, thus, this region may be essential for activity. Interestingly, this C-terminal domain represents a β-hairpin motif, referred to as the γ-core, which is conserved among many classes of antimicrobial peptides and is often implicated as essential for the activity of these peptides [112, 113]. Despite their similarity to spider neurotoxins, these peptides have no effect on insect neuronal cells [110]. However, a knottin-like peptide from garden pea (PA1b) has recently been found to act as an insecticide through inhibition of vacuolar ATPase, a property not previously described for peptide inhibitors [114].
2S albumin proteins
Proteins in the 2S family are storage proteins in the seeds of both monocots and dicots. They have been investigated mostly as allergens [115, 116] but have the characteristic molecular weight, cationic residues, and disulfide bonds of antimicrobial peptides. Antifungal activity has been demonstrated for 2S albumin proteins, which are heterodimeric, from the seeds of Malva parviflora [117], Passiflora edulis f. flavicarpa [118], and Raphanus sativus [119]. The mechanism by which 2S proteins inhibit fungal growth is not very well understood. Incubation of various fungal species with the R. sativus proteins resulted in growth inhibition but not a loss in viability [119]. The P. edulis 2S protein prevents acidification of media by F. oxysporum leading to a proposed inhibition of H+-ATPases or increased permeability of the membrane to protons [118].
Hairpinins
A novel class of antifungal peptides has recently been identified in the seeds of Echinochloa crus-galli [120] and Fagopyrum esculentum [121]. These peptides have the unique structure among plant defense peptides of two helices linked by two disulfide bonds. The mechanism of EcAMP1 from E. crus-galli was shown to occur via a two-step process whereby EcAMP1 first binds to the cell surface of Fusarium solani and is then internalized, without disruption of the membrane, and accumulates in vesicular structures [120].
Antifungal peptides from vertebrates
Vertebrates boast an adaptive immune system that responds to pathogenic attack and customizes the organism’s defense based on previous exposure to pathogens. While this response is highly effective, the delay involved and the enormous level of pathogenic challenge faced requires a faster, more general, first line of defense. Part of this “innate” immune response involves the production or release of antimicrobial peptides.
Mammalian peptides
The site of antimicrobial peptide expression in mammals is most often the cells of the epithelial layers and neutrophils. While these peptides are most often antibacterial, reflecting the increased level of threat faced from these microbes, a number also possess antifungal activity, particularly against yeast. The major groups of peptides are the histatins, cathelicidins, defensins, and lactoferricins. The defensins probably represent an ancient defense system as they are conserved among all higher eukaryotes and even fungi [122]. The cathelicidins have only been identified in mammals while histatins are only found in humans and closely related primates. The antimicrobial activity of these peptides was identified some time ago, however, it is now becoming clear that the role of these peptides in vivo is probably much more complex and that a functional interrelationship exists between the innate and adaptive immune response [reviewed by 123].
Histatins
The histatins are a group of histidine-rich peptides from in human saliva that exhibit antifungal activity against a number of Candida species. Histatin 1 and 3 are gene products and the remaining ten members of the family are proteolytic cleavage products of these peptides [124].
Histatin 5, a cleavage product of histatin 3, is the most potent of these molecules and as such, the most well studied. Histatin 5 is a 24-amino-acid peptide containing seven histidines, four arginines, and three lysines [125]. In an aqueous environment, it adopts a random coil structure, however, in a non-aqueous environment the peptide adopts an α-helical conformation [125]. The heat shock protein Ssa2p, a 70-kDa cell wall protein in C. albicans, is the binding site for histatin 5 [126]. The presence of extracellular Ca2+ prevents binding of histatin 5 to C. albicans [127], presumably by disrupting the interaction between histatin 5 and Ssa2p. The presence of the Ssa2p is required for susceptibility of C. albicans to histatin 5 and the internalization of histatin 5 into cells [128]. Uptake of histatin 5 into C. albicans cells is dependent on the presence of two polyamine transporters, Dur3 and Dur31 [129], which usually function in spermidine uptake. Internalization must occur by translocation, not endocytosis, for histatin to act as an antifungal molecule against C. albicans [130]. Upon internalization, histatin 5 is localized to the mitochondria if respiration is underway and causes a loss of mitochondrial membrane integrity [131].
Movement of histatin 5 to mitochondria suggests it does not act simply via membrane disruption and, consistent with this, it is unable to cause substantial release of calcein from C. albicans cells or affect the cytoplasmic transmembrane potential [132]. Histatin 5 does not display strong amphipathicity, which is common in other pore-forming molecules [132]. Interestingly, mutagenesis of histatin 5 to increase its amphipathicity generates the ability to dissipate the cytoplasmic transmembrane potential of treated cells [133]. Although it does not appear to lyse cells, histatin 5 does induce propidium iodide (PI) uptake and ATP release [131, 134] and this ATP release correlates with candidacidal activity [134]. Time lapse confocal microscopy has shown that there is a delay between histatin 5 internalization and PI uptake, confirming that histatin 5 does not directly permeabilize fungal cell membranes [130]. It appears that extracellular activity of ATP that is responsible for cell death rather than ATP release itself, since anaerobically grown cells are able to release ATP upon histatin 5 treatment but are not killed [135]. This extracellular activity appears to be mediated by P2X receptors, which are nucleotide-binding receptors on the plasma membrane that are involved in extracellular signaling by ATP. P2 agonists that activate these receptors kill aerobically, but not anaerobically, grown C. albicans cells and P2 antagonists prevent histatin 5-induced cell death [135]. This suggests that histatin 5 causes the release of ATP that subsequently binds to P2X receptors on the cell surface, thus inducing signaling cascades leading to cell death. The release of ATP in response to histatin 5 is mediated by the K+ channel, Trk1p, and this release can be inhibited by anion channel blockers [136]. C. albicans mutants with decreased levels of Trk1p exhibit a normal amount of histatin 5 uptake but a marked decrease in histatin 5-induced cell death [136]. The production of ROS in response to histatin 5 has also been reported [133, 137]. However, the relevance of this has been debated and ROS is not believed to be directly involved in histatin 5-mediated killing [138]. In addition, histatin 5 does not induce programmed cell death pathways [137].
Cathelicidins
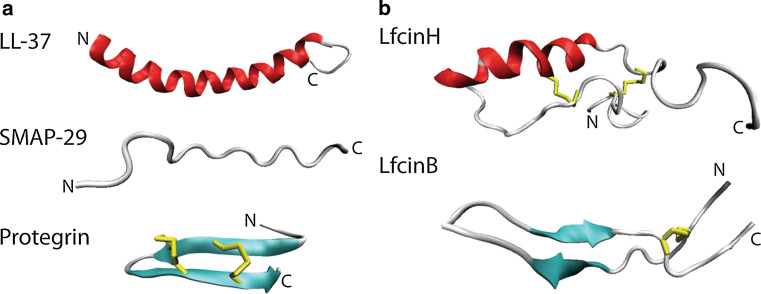
Antimicrobial peptides belonging to the cathelicidin family are extremely diverse in both sequence and structure; however their similarity lies in the N-terminal prosequence they all possess. This sequence of approximately 100 amino acids is also related to cathelin, a cystatin-like protein. Cathelicidins are expressed predominantly in neutrophils although they are also present in epithelial layers in humans [139]. Cathelicidins can form α-helical, β-hairpin, or extended conformations (Fig. 6a). They predominantly exhibit antibacterial activity although several are active against fungal species [139]. The recent sequencing of marsupial genomes has led to the discovery of new cathelicidin genes, one of which has shown to have potent activity towards multidrug resistant strains of bacteria [140].
Fig. 6.
NMR structures of mammalian antifungal peptides from the (a) cathelicidin and (b) lactoferrin-derived families. a Peptides in the cathelicidin family can take α helical (LL37, pdb code 2K60), extended (SMAP29, pdb code 1FRY), or β hairpin conformations (protegrin, pdb code 1PG1). b Lactoferrin-derived peptides with α-helix and extended loops (LfcinH, pdb code 1Z6V) and antiparallel β-sheet (LfcinB, pdb code 1LFC) conformations. Beta-strands are represented in cyan, α-helices are in red, random coils and turns are in white, and disulfide bonds are in yellow
Cathelicidins have superior activity against yeast compared to filamentous fungi [141], which may be an evolutionary response to the greater threat posed by yeast to mammals compared to filamentous fungi. Cathelicidins, despite their structural divergence, appear to kill cells via perturbation of the plasma membrane. A number of antifungal cathelicidins from sheep (SMAP-29) and cow (BMAP-27, BMAP-28) form amphipathic α-helices in a hydrophobic environment. They have C-terminal hydrophobic domains, with strong membrane permeabilization activities [141, 142]. SMAP-29 concentrates on the plasma membrane of treated cells and causes propidium iodide uptake provided the cells are metabolically active [143]. In a hydrophobic environment, PMAP-23 (from pigs) forms two short α-helices joined by a flexible region [144]. This peptide binds to the plasma membrane of treated cells and is active against C. albicans protoplasts indicating interaction with the cell wall is not required for inhibitory activity [145]. The β-hairpin peptide protegrin and the extended, tryptophan-rich peptide indolicidin (both from pigs), also exhibit candidacidal activity through membrane permeabilization [141, 146].
So far, only one cathelicidin, LL-37, has been identified in humans [139]. It forms an amphipathic α-helix [147] (Fig. 6) and binds to the cell wall and plasma membrane of treated cells [148]. It disrupts the C. albicans cell membrane completely and allows leakage of proteins of up to 40 kDa into the medium [148]. The kinetics of permeabilization are very rapid with complete lysis occurring within 5 min, supporting the idea that membrane disruption is the sole mechanism of cathelicidin activity. Insertion of LL-37 into membranes is equally dependent on hydrophobic interactions between the peptide and acyl chains of the membrane lipids as it is on electrostatic interactions with lipid head groups [149]. Sterols decrease the membrane association of LL-37 with cholesterol having a stronger effect than ergosterol [150] leading to the hypothesis that protection of the host cell against secreted AMPs may have been a significant selective pressure in the evolution of cholesterol. LL-37 and the mouse equivalent, mCRAMP, are believed to act in vivo by creating a barrier on the skin for protection against C. albicans invasion [151]. Furthermore LL-37 is secreted into human sweat and processed to the more active peptides, RK-31 and KS-30. Importantly, these peptides retain their activity even in the high salt conditions present in human sweat. These truncated variants are also able to enter the cytoplasm of C. albicans cells suggesting that their increased activity may result from access to intracellular targets [152]. The expression of mCRAMP is also induced in mouse skin upon C. albicans infection [151]. One disadvantage of cathelicidins as novel antifungal peptides is the toxicity often demonstrated toward erythrocytes. However, some synthetic variants exhibit decreased erythrocytic activity compared to antimicrobial activity [146]. A comprehensive review of LL-37 as an antimicrobial peptide along with other roles the peptide plays in innate immunity can be found in [153].
Defensins
Mammalian defensins represent a large group of peptides with an important role in the host’s immune system. Deficiencies in various defensins have been implicated in some diseases such as inflammatory bowel disease [154]. Mammalian defensins share structural and functional similarities with defensins from plants, insects and fungi. They can be divided into the α-defensins and the β-defensins based on their structural characteristics and cysteine spacing pattern (Fig. 7). The α-defensins are 29–35 amino acid peptides that share six conserved cysteine residues forming three disulfide bonds. Their structure is comprised of three antiparallel β-strands that form an amphipathic β-sheet. The β-defensins are longer than their α-counterparts, ranging from 34 to 42 residues in length. They contain the signature triple-stranded, antiparallel β-sheet as well as a short α-helix.
Fig. 7.
Sequence alignment and structural comparison of mammalian (a) α-defensins and (b) β-defensins. a Six cysteine residues which form three disulfide bonds are conserved in the three stranded β-sheet fold characteristic of mammalian α-defensins. Cysteine resides are highlighted in black with solid black lines denoting disulfide bonds. Residues highlighted in grey are highly conserved among the sequences HNP4 (pdb code 1ZMM) and cryptdin4 (pdb code 2GW9) form a triple-stranded anti-parallel β-sheet. b β-defensins contain six conserved cysteines that form three disulfide bonds although the connectivity of these disulfides differs to that of α-defensins. β-defensins also contain an additional short α-helix (HBD1, pdb code 1E4S and HBD2, pdb code 1E4Q). β-strands are represented in cyan, α-helices are in red, random coils and turns are in white, and disulfide bonds are in yellow
The α-defensins
These defensins were first identified in rabbits and have since been found in guinea pigs, rats, hamsters, macaques and humans [155]. In humans, a number of these proteins are expressed in neutrophils [human neutrophil defensins 1–4 (HNP 1–4)] and their combined concentration can be up to 1.5 mM [155]. Three of these (HNP 1–3) are identical apart from one N-terminal amino acid. Interestingly, only HNP-1 and HNP-2 display candidacidal activity while HNP-3, which has an acidic amino acid at the N-terminus, is completely inactive on C. albicans [156]. HNP-4 is also toxic to C. albicans cells [157]. The antibacterial activities of α-defensins have been attributed to their ability to permeabilize membranes [155, 158]. The candidacidal activity of the rabbit neutrophil defensin (NP-1) is also attributed to this ability [159]. HNP-1, like histatin 5, causes C. albicans to release ATP but does not lyse cells [160]. Additionally, the antifungal activity of HNP-1, in contrast to NP-1, is dependent on the metabolic activity of the target cell [156], supporting a different mechanism of action for the human and rabbit α-defensins. While C. albicans cells treated with HNP-1 remain metabolically active after 2 h, they are not able to form colonies [160], which indicates a fungistatic mode of action.
The β-defensins
While only a few mammalian β-defensins have been characterized in any detail, they are mainly found in the epithelial layers, consistent with a role in host defense. The antibacterial activity of β-defensins, as with α-defensins, is attributed to their membrane permeabilizing properties. Studies on the antibacterial activity along with other general features of human β-defensin 2 (hBD-2) and hBD-3 have been reviewed in [161] and [162]. In addition to their antibacterial activity, hBD-2 and hBD-3 are potent inhibitors of Candida species [163] and hBD-2 is up-regulated in response to exposure to C. albicans and Trycophyton rubrum in keratinocytes [164] and in response to Aspergillus fumigatus in lung epithelial cells [165]. Exposure of lung epithelial cells to A. fumigatus also induces expression of hBD9. The mechanism underlying the antifungal activity of hBD2 and hBD3 has only recently been elucidated and, similar to human α-defensins, does not appear to result from simple membrane disruption. Both peptides require the presence of Ssa2p, the histatin 5 binding protein in the fungal cell wall, for activity [166]. However, their activity is not dependent on Trk1p, the second mediator of histatin 5 activity, indicating that they act via related but different mechanisms to histatin 5 [166]. Deletion of genes involved in the high osmolarity glycerol (HOG) pathway in C. albicans increased sensitivity to hBD-2 and hBD-3 [167]. This pathway has also been implicated in histatin 5 tolerance [168]. This is likely to be indicative of the role of the HOG pathway (for review see [169]) in a wide variety of stress responses as opposed to a common mechanism between histatin 5 and the human β defensins. The exact mechanisms by which hBD-2 and hBD-3 act remain unknown. However, evidence from calcein release assays suggests that antifungal activity is unlikely to involve membrane disruption. Additionally, beta defensin activity was found to require target cells to be metabolically active [170]. As a strong positive charge is a feature common to many antifungal molecules the presence of high concentrations of cations often decreases efficacy. The antibacterial activity of hBD-3 is salt insensitive, however, its antifungal activity is abolished by low concentrations of both monovalent and divalent cations [170], pointing to differences between the antibacterial and antifungal mechanisms of action.
Lactoferricins
Lactoferrin is a multifunctional, 80-kDa protein, first isolated from bovine milk and later identified in a number of species including humans, pigs, and mice. Lactoferrin is a member of the transferrin family of proteins all of which share characteristic iron binding properties. The antimicrobial activity of lactoferrin was originally attributed to the high affinity iron binding function [171] as iron is an essential nutrient for virtually all organisms and sequestering iron from microorganisms would inhibit their growth. Additional functions for lactoferrin were revealed when proteolytic cleavages of lactoferrin produced several peptides with antifungal activity equivalent to or better than the whole protein [172–176]. Further studies on the antifungal activity of intact human lactoferrin revealed that the protein causes slight K+ efflux from C. albicans cells, but does not allow Na+ release and does not disrupt the membrane [177]. This indicates that non-specific membrane permeabilization is not the mechanism of action. Spheroplasts are more resistant to killing by lactoferrin and the peptide is known to induce changes in the cell wall of C. albicans, suggesting that cell wall interactions are probably central to the mechanism of action. However, lactoferrin also depolarizes the plasma membrane and leads to acidification of the cytoplasm, suggesting additional targets beyond the fungal cell wall. Cellular respiration is also required for its cytotoxic activity [177], which again supports the notion that lactoferrin does not act via non-specific membrane permeabilization.
Aside from the activity of lactoferrin as a full protein, peptides from the N-terminus of the protein have antibacterial and antifungal activity. One of these is lactoferricin B (LfcinB), which comprises the region spanning residues 17 to 40 of the bovine lactoferrin protein [178]. Peptides spanning the equivalent region of the N-termini of the human [172], murine [179] and porcine [176] proteins have also been assayed for antimicrobial activity; revealing the bovine peptide as the most active of any species tested [180]. Variations in antimicrobial activity are not surprising as there is only 41 % sequence identity between the human and bovine peptides. Structures of both the bovine (LfcinB) and human (LfcinH) derived peptides have been solved (Fig. 6b) and they adopt different folding patterns which also likely contributes to differences in activity. LfcinB adopts an antiparallel β-sheet [181] while LfcinH, which is 17 amino acids longer, forms an α-helix followed by an extended region [182]. Both peptides are stabilized by disulfide bonds but these disulfide bonds are not essential for antimicrobial activity [183].
Neither LfcinB nor LfcinH are likely to act by non-specific permeabilization of membranes as they do not lyse bacteria or cause calcein release from artificial liposomes [184, 185]. However, both peptides dissipate the proton gradient across the plasma membrane suggesting some interaction with the membrane [172]. Antibacterial activity of LfcinB involves binding to lipopolysaccharide (LPS) on the surface of Gram-negative bacteria [186]. The peptide is also found at a high concentration inside treated E. coli cells [187]. The slow killing kinetics displayed by this peptide supports the role of an intracellular target [188]. Phosphorylation of response regulators in the two-component system in E. coli is inhibited by LfcinB, which in turn inhibits growth [189]. Lfpep is a truncated version of LfcinH from the equivalent region that LfcinB is derived from full-length lactoferrin. The mechanism of Lfpep differs from Lfcins as it causes propidium iodide uptake and almost total release of intracellular K+ from treated C. albicans cells indicating that membrane permeabilization is a component of antimicrobial activity [174]. Lfpep is also able to permeabilize artificial liposomes [185].
Truncated versions of the LfcinB have been produced synthetically to identify the region of the molecule responsible for antifungal activity. The smallest active peptide was a hexapeptide comprising amino acids 20–25 of bovine lactoferrin (RRWQWR–NH2) [175]. This peptide inhibited the growth of filamentous fungi but had no activity against E. coli or S. cerevisiae [175]. This may be explained by the loss of ability to translocate into the cytoplasm. A slightly longer peptide (residues 17–31) was more active against filamentous fungi and was active against E. coli and S. cerevisiae [175]. Interestingly, while both peptides permeabilized the membrane of Penicillium digitatum hyphae, the hexapeptide was fungicidal while the longer peptide was not. Variability indicates that the two peptides act by are different mechanisms and that each peptide has different mechanisms against bacteria as well as different types of fungi.
Amphibian peptides
Many diverse antimicrobial peptides have been isolated from non-mammalian vertebrates such as reptiles, amphibians, and fish. These creatures often inhabit environments that are rich in pathogens yet they are generally disease free. The skin secretions of various frog and toad species are one of the richest sources of antimicrobial peptides found to date [reviewed by 190]. The antibacterial properties of antimicrobial peptides from these species remain the main focus and it is only relatively recently that their antifungal activities have been investigated in any detail. Fungi are significant pathogens of many amphibious species and some fungi have even been associated with a decline in worldwide amphibian numbers [191].
Temporins
The temporins are 10–14-amino-acid peptides originally identified in the skin secretions of the European red frog, Rana temporaria [192]. While they were initially thought to be limited to this species, they have since been isolated from several other frog species as well as wasp venom [191]. Their antifungal properties have been harnessed for the production of transgenic potato plants resistant to late blight and pink rot, caused by Phytophthora species [9]. Temporins are not as basic as other cationic antimicrobial peptides although in the most potent, temporin A, the one basic residue is essential for activity [193]. The peptides are also amidated at their C-termini, increasing the overall positive charge by one [192].
The mechanism underlying the antifungal activity of temporins has not been investigated, although the antibacterial activity has been attributed to efficient permeabilization of bacterial membranes [193]. Permeabilization alone may not be sufficient for cell death as permeabilization of E. coli was observed at sub-lethal concentrations of temporin L [194]. However, an interaction between temporins and specific biological molecules on the cell surface is unlikely as temporins permeabilize artificial liposomes with the size of molecules released increasing with increased concentration of temporin [195]. Discrimination between anionic and zwitterionic membranes was not observed for permeabilization by temporins. However, temporins do not lyse human erythrocytes, which suggests there are additional factors involved in the mechanism of action on different cell types [194].
Brevinin-1 family
Another class of proteins isolated from frog skin secretions belong to the brevinin-1 family. Brevinin-1BYa, from the foothill yellow-legged frog, is the most effective member of this class at inhibiting the growth of Candida species [126]. This 24-amino-acid peptide carries a charge of +4 and adopts an α-helical conformation in a hydrophobic environment [126]. The C-terminal region contains two cysteine residues that form a single disulfide bond and create a loop of five amino acids. A variant containing a single amino acid change (Phe12 → Leu) has four-fold less activity against Candida, whereas activity against the Gram-positive bacterium, Staphylococcus aureus decreased only two-fold and its activity against the Gram-negative bacterium, E. coli, was not altered [126]. In contrast, substituting the two cysteines with serines abolished the antifungal activity and greatly reduced the activity against E. coli but did not affect the activity against S. aureus [196]. This implies that not only does the peptide’s mechanism of action differ between bacteria and fungi, it also differs between Gram-positive and Gram-negative species of bacteria.
Antifungal peptides from invertebrates
Invertebrates, like plants, lack an adaptive immune response and as such rely heavily on antimicrobial proteins for protection. Invertebrate species are thus a rich source of antimicrobial peptides. Again, the focus has been on discovery of antibacterial peptides, although many with antifungal activity, and some exclusively so, have been reported. Some of these, such as insect defensins, share sequence identity with peptides from other organisms, while others have no obvious homologues.
Insect peptides
Antimicrobial peptides from insects are expressed in the fat body (equivalent of liver), and secreted into the hemolymph (equivalent of blood) in response to bacterial or fungal infections [197]. In Drosophila, two separate pathways are involved in this response: Gram-negative bacteria initiate the Immune-deficiency (IMD) pathway, while Gram-positive bacteria and fungi activate the Toll-receptor pathway [198]. Antimicrobial peptides from insects are often isolated after bacterial challenge, this may explain why fewer antifungal molecules have been identified.
Insect defensins
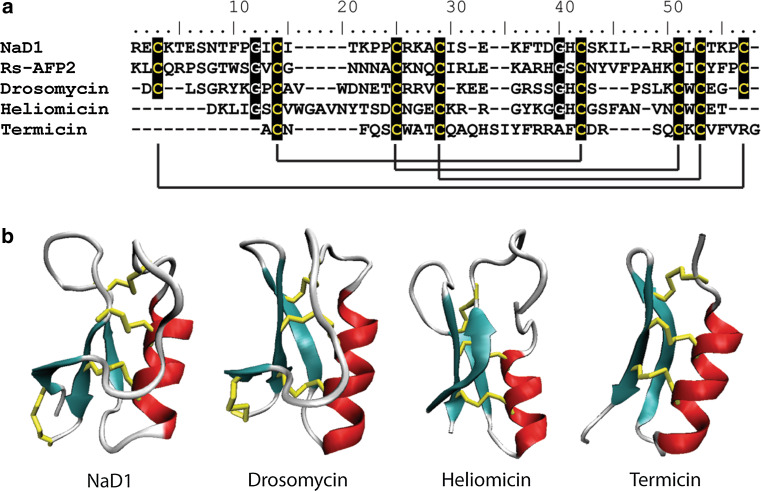
Defensins have been identified in every insect for which they have been screened. Three of these, heliomicin, from Heliothis virescens [199], termicin, from Pseudacanthotermes spiniger [200] and drosomycin, from Drosophila melanogaster [201], display potent antifungal activity. Drosomycin is a 44-residue protein with a high degree of similarity to plant defensins. Interestingly, it also shares the same cysteine spacing pattern; one of the major determinants for classification of these families of proteins (Fig. 8). Drosomycin also displays a similar three-dimensional structure, comprised of a triple-stranded anti-parallel β-sheet tethered to an α-helix (CSαβ motif, Fig. 8) [201]. Heliomicin (44 amino acids) shares a similar structure to drosomycin and plant defensins, although it lacks the fourth disulfide bond that joins the N- and C- termini [202]. Termicin (36 amino acids) is smaller than drosomycin and heliomicin, lacking the first β-strand of the antiparallel β-sheet [203]. As such, it is more similar to the antibacterial insect defensins and a defensin from mussel (MGD-1). Termicin is expressed constitutively and is believed to play a role in protecting the termite against invasion by the symbiotic fungus that lives on fecal pellets in termite nests and predigests lignocellulose to aid food digestion in termites [200].
Fig. 8.
Sequence alignment and structural comparison of insect defensins with the plant defensin NaD1. a Six cysteines that form three disulfide bonds are conserved in the insect defensins Heliomycin and Termicin. Drosomycin has two additional cysteine residues that form an extra disulfide bond with the same connectivity as observed in plant defensins such as NaD1. Cysteine residues are highlighted in black with solid black lines denoting disulfide bonds. b The insect defensins drosomycin (pdb code 1MYN), heliomicin (pdb code 1I2U) and termicin (pdb code 1MM0) share the common CSαβ-fold with the plant defensins (NaD1, pdb code 1MR4). Beta-strands are represented in cyan, α-helices are in red, random coils and turns are in white, and disulfide bonds are in yellow
The range of target pathogens towards which specific insect defensins are active varies considerably. Drosomycin is active against filamentous fungi, but not against yeast or bacteria. Heliomicin is active against filamentous fungi and yeast, but not bacteria [199] and termicin is most active toward filamentous fungi, although it also inhibits the growth of yeast at moderate concentrations and bacteria at high concentrations [200]. The effects of these insect defensins on their target cells has not been well characterized, although structure–function studies have been undertaken to identify regions of the molecules involved in activity. Mutation of the four N-terminal amino acids of heliomicin to alanine resulted in a 15-fold reduction in antifungal activity [202]. Intriguingly, only a two-fold reduction in antifungal activity occurred when the two basic residues at the beginning of β-strand two were substituted with leucines, which are present in antibacterial insect defensins. Heliomycin with these substitutions has an overall charge of −1 yet gains antibacterial activity. Site-directed mutagenesis of drosomycin led to identification of five cationic (R6, K8, R20, R21, and K38) and two anionic (D1 and E25) residues located in different secondary structures that are involved in antifungal activity [204]. It was also suggested that the location of these charged residues, as opposed to the overall charge of the protein, is important for activity, possibly through an electrostatic interaction with target molecules. Although specific residues do not seem to be conserved between the antifungal defensins, one common feature is the presence of a basic residue buried in a hydrophobic patch [205]. This may be essential for activity.
Thanatin
Thanatin is a peptide that is induced upon challenge by microbial pathogens from the spined soldier bug Podisus maculiventris. It inhibits the growth of bacteria and filamentous fungi, but is not active against yeast species and is not hemolytic [206]. It is similar in sequence to brevinin-1 although the N-terminus is seven amino acids shorter and structural analysis revealed that it lacks the α-helix formed by the N-terminus of the brevinin-1 peptide [206, 207]. Rather, the N-terminus forms a long extended arm (residues 1–7). The remainder of the peptide forms a cysteine-stabilized β-sheet loop as observed in brevenins [207].
Thanatin does not permeabilize bacterial membranes but instead causes rapid agglutination of bacterial cells. An all D-enantiomer of the peptide has no antibacterial activity yet activity against filamentous fungi is unchanged [206] implying that, like many of the peptides discussed thus far, the peptide differs in its mode of action against bacteria and fungi. While the peptide is fungicidal to spores at a high concentration [206], the effect on hyphae has not been investigated. Thanatin also has a basic residue (Arg) in the center of a hydrophobic patch [207] as reported for drosomycin and other antifungal peptides although its significance has not been investigated. Transgenic rice expressing thanatin is resistant to the rice blast fungus Magnaporthe oryzae [208].
Glycine-rich peptides
Three glycine-rich peptides with antifungal activity have been isolated from insects, namely, AFP, holotricin-3 and tenecin-3 [209–211]. Of these, only tenecin-3 has been studied in detail. This peptide is 78 residues long and Gly, His and Glu residues represent 80 % of its amino acid composition [211]. The peptide has no cysteine residues and it adopts a random structure even in a hydrophobic environment . Non-specific membrane permeabilization is probably not responsible for the antifungal activity of tenecin-3 as it does not cause calcein release from artificial liposomes [212]. Furthermore, uptake of tenecin-3 into the cytoplasm of treated C. albicans cells is required for cell death indicating involvement of an intracellular target may be involved. Interestingly, only C. albicans cells in the log phase of growth were able to internalize tenecin-3 [212]. Uptake requires cellular processes and is inhibited at 0 °C and by the presence of the oxidative phosphorylation inhibitor, sodium azide.
Cecropins
Cecropins are 35–39-amino-acid peptides that were originally isolated from the cecropin moth (Hyalophora cecropia) and have since been found in other insect species, including Drosophila. They form two α-helices separated by a flexible hinge region in a hydrophobic environment [213]. In Drosophila, cecropins are induced in response to both bacterial and fungal pathogens and may reach concentrations between 25 and 100 μM in the hemolymph, [213]. Cecropin A is lethal to germinating and non-germinating Fusarium but only to germinating Aspergillus. Furthermore, fluorescently labeled cecropin A only bound to particular hyphae and this binding resulted in cell death. That is, cecropin A bound to germinating Aspergillus hyphae to induce death whereas binding and cell death was not observed with non-germinating hyphae [214]. This selectivity relates to changes in the cell wall composition that occur at various stages of growth. The mode of action of cecropins against fungi has been reported to involve disruption of the plasma membrane [215] although this has not been studied in great detail.
Spinigerin
A 25-amino-acid peptide with both antibacterial and antifungal properties is expressed constitutively and stored in hemocyte granules of the termite P. spiniger [200]. This peptide adopts a random coil structure in an aqueous environment but forms a stable, amphipathic α-helix in a hydrophobic environment [216]. The α-helix is slightly bent and shares characteristics with magainin 2, an antimicrobial peptide from Xenopus [202]. These similarities have led to the hypothesis that spinigerin acts via membrane permeabilization in a similar manner to magainin 2 [217], although this hypothesis has not been tested.
Insect knottin-type peptides
Three peptides from the insect Acrocinus longimanus have activity against Candida glabrata. They are members of the knottin-type family [218] and the most potent of these peptides, Alo-3, is very similar in sequence to the other two peptides (Fig. 4). It has two additional basic amino acids at its C-terminus that form part of a cationic pole at the base of the molecule [218]. The three peptides share sequence similarity with the plant peptide MjAMP; but the mechanism of action for these peptides has not yet been investigated.
Moricin-like peptides
Screening of hemolymph from immune-stimulated Galleria mellonella revealed a number of novel peptides similar to the antibacterial moricin peptides from lepidopterans [219]. These peptides are activity against the plant pathogen F. graminearum, but not fungi that infect insects. Moricins adopt a helical structure with an amphipathic N-terminal region that is thought to be crucial for antibacterial activity [220].
Marine invertebrates
Marine invertebrates have also proved to be a source of many novel antimicrobial peptides. Unlike in insects, where peptides are expressed in the fat body in response to infection, antimicrobial peptides in marine invertebrates are produced constitutively in hemocytes. The peptides are stored in granules and released via exocytosis upon bacterial or fungal infection.
Penaeidins
The penaeidins are 47–63 residue peptides first isolated from the tropical shrimp Penaeus vannamei and have since been found in a number of other shrimp species [221, 222]. The N-terminal domain of the peptide is proline-rich and does not have a well-defined structure. The C-terminal domain forms an α-helix with loop regions either side connected by three disulfide bonds [223, 224]. Penaedin-3 and -4 bind chitin in vitro, however, this is believed to be related to their ability to bind to the gill cuticle, which is composed largely chitin and not to their antifungal activity [225, 226]. The N-terminal proline rich domain of penaeidin-4, but not penaeidin-3, has similar antifungal activity to that of the whole protein [225, 227] further supporting the notion that chitin binding by the C-terminal domain is for localization in the gill cuticle as opposed to binding to fungal chitin. The effect of penaeidins on filamentous fungi has been examined by microscopy, which revealed increased hyphal branching and a lack of sporulation after exposure to the penaeidins [221]. Penaeidins and other shrimp antimicrobial peptides are of particular interest to aquaculture, a billion dollar industry, because of potential control of microbial infections which can have a significant impact on production [228].
Crab β-hairpin peptides
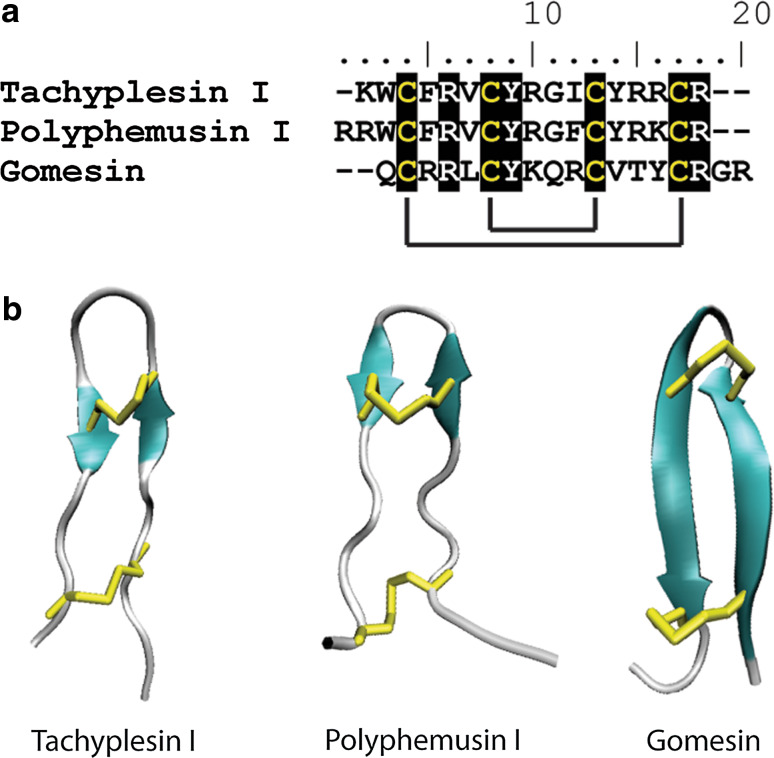
Several small (17–18 amino acid) β-hairpin peptides known as tachyplesins and polyphemusins have been isolated from Japanese and American horseshoe crabs, respectively (Fig. 9) [229]. Tachyplesin II and polyphemusins I and II, are active against fungi while tachyplesins I and III are not. The three antifungal peptides have an arginine at their N-terminus while the two without antifungal activity have a lysine at this position [230]. In fact, this amino acid change is the only difference between tachyplesins II and III. Once again, the mechanism of action for their antifungal activity has not been investigated; although it is known that they enter the cytoplasm of treated E. coli cells [231].
Fig. 9.
Sequence alignment and structural comparison of β-hairpin peptides. a Conservation of the four cysteine residues that form two conserved disulfide bonds in β-hairpin peptides. Cysteine resides are highlighted in black with solid black lines denoting disulfide bonds. Conserved arginine and tyrosine residues are also highlighted. b The β-hairpin structure of tachyplesin I (pdb code 1MA2), polyphemusin I (pdb code 1RKK) and Gomesin (pdb code 1KFP) is stabilized by the two conserved disulfide bonds. β-strands are represented in cyan with disulfide bonds in yellow
Tachystatins
The tachystatins are another group of antifungal peptides from the hemocytes of the Japanese horseshoe crab. They are 41–44 amino acid peptides and are members of the knottin-type family of proteins with sequence and structural similarities to neurotoxins from spider venom, as well as plant and insect antifungal peptides [232]. The structure of tachystatin A has been solved by NMR and consists of a triple-stranded, anti-parallel β-sheet with a long loop connecting β-strands 1 and 2 [233] (Fig. 4). Tachystatin A, along with tachystatin B and C, bind chitin, and a causal relationship exists between this binding and antifungal activity [232]. Tachystatin C binds to the periphery of Pichia pastoris cells and is able to lyse these cells [232], indicating the antifungal activity of these peptides may result from membrane disruption.
Big defensin
A 79-amino-acid peptide produced by horseshoe crab hemocytes is active against Gram-negative and Gram-positive bacteria as well as fungi [234]. The protein is composed of two domains, an N-terminal hydrophobic domain of 42 amino acids and defensin-like domain corresponding to amino acids 43 to 79 with homology to rat neutrophil defensin 2 (NP-2). The N-terminal, hydrophobic domain is toxic to Gram-positive bacteria while the C-terminal, defensin-like domain is active against Gram-negative strains [234]. The activity of these individual domains on fungi and the mechanism of action of the intact protein have not been investigated.
Hemocyanin-derived peptides
Peptides from the C-terminal region of hemocyanin, a shrimp respiratory protein, have broad-spectrum antifungal activity [235]. They accumulate in the hemolymph relative to intact hemocyanin in response to fungal immune challenge, suggesting a novel mechanism for production of antimicrobial peptides upon infection in invertebrates. A number of other antimicrobial peptides are proteolytic cleavage products of larger proteins, including the lactoferrin-derived peptides discussed previously. Another novel feature of these peptides is their acidic nature; they carry a negative charge of between −2 and −8. [235]. They do not share sequence identity with any other antimicrobial peptides identified to date, and their structure has not been elucidated; thus, no hypothesis regarding their mechanism of action has been put forward.
Cenchritis muricatus peptides
Cm-p1, a small hydrophillic peptide from the marine snail C. muricatus, is active against a broad spectrum of fungal species but has no activity against bacteria or mammalian cells. Modeling studies demonstrated that this peptide could form a helical conformation with exposed basic residues and a hydrophobic region essential for antifungal activity [236].
Spiders
Gomesin
In the context of evolution, spiders are not-too-distant relatives of crabs. This is evident in the similarity of the previously described crab tachystatins with spider venom toxins. It is not surprising, therefore, that an antifungal peptide that is closely related to the crab β-hairpin peptides, tachyplesin and polyphemusin, has been identified in the spider Acanthoscurria gomesiana [237]. This 18-amino-acid peptide, gomesin, has the same cysteine spacing as the crab β-hairpin peptides (Fig. 9) and forms the common β-hairpin structure as determined by NMR [238].
Gomesin is expressed constitutively in spider hemocytes [239] and inhibits the growth of filamentous fungi and yeasts at low concentrations. The mechanism underlying its activity has not been elucidated. However, it displays an amphipathic nature with positively charged poles and a hydrophobic patch in the center. Mutagenesis of this hydrophobic region leads to substantial reduction in activity [240]. This is consistent with the ability of gomesin to lyse giant unilamellar vesicles [241] and supports membrane permeabilization as a mechanism of action. Studies on the antitumor activity of gomesin revealed a mechanism that involved an increase in cellular calcium levels, induction of MAPK/ERK, PKC, and PI3 K signal transduction pathways, and generation of reactive oxygen species [242].
Antifungal peptides from fungi
Filamentous fungi and yeast secrete antifungal peptides into the extracellular environment, presumably to provide themselves with an advantage over competing species. Most of these peptides are not related to antifungal peptides from other species, reflecting the ancient divergence of these organisms from the other eukaryotes. However, a number of fungal defensins have been isolated recently, indicating that this family of proteins may predate this divergence [122, 243].
Peptides from filamentous fungi
AFP and PAF are 51- and 55-amino-acid peptides secreted by Aspergillus giganteus and Penicillium chrysogenum, respectively. They share 47 % amino acid sequence identity. The structure of AFP has been solved and displays a β-barrel-like fold, a conformation that is not shared by any other antimicrobial peptides [244]. The modes of action of both of these peptides have been investigated. When the rice blast fungus Magnaporthe grisea is treated with AFP, the peptide permeabilizes the membrane before entering the cell and moving to the nucleus [245]. AFP has an oligonucleotide-binding fold and the peptide binds to DNA and causes DNA condensation in vitro [246]. This property could be responsible for the toxicity of the peptide. AFP is expressed as an inactive precursor with six additional amino acids at the N-terminus that are cleaved following secretion [246]. This process of activation after secretion prevents interaction with DNA and hence toxicity to the host cell.
Despite 47 % sequence similarity with AFP, PAF appears to act via a different mechanism. PAF also causes membrane permeabilization [247] and enters hyphae [248], but it does not travel to the nucleus. PAF induces hallmarks of apoptotis-like cell death in treated hyphae, which include production of reactive oxygen species, exposure of phosphatidylserine on the outer leaflet of the plasma membrane, and disintegration of subcellular organelles [249]. PAF uptake is an active process and is blocked by the oxidative phosphorylation inhibitor sodium azide and by the oxidative phosphorylation uncoupler carbonyl cyanide m-chlorophenyl hydrazone (CCCP). Inhibition of endocytosis blocks uptake of PAF [248]. Resistance to PAF is imparted by the inactivation of G-protein signaling cascades [249].
Killer toxins
Yeast killer toxins
Yeast killer toxins are an interesting class of antifungal molecules because they are often not encoded by the host genome, but rather by dsRNA viruses that infect yeast species. As a consequence of this, the genes required for immunity are also virally encoded and the toxins are, therefore, active against non-infected cells of the same species [250]. Virally encoded proteins are produced as preproproteins containing a signal domain, followed by α-, γ- and β-domains. During folding, the α- and β-domains are brought together and linked via a disulfide bond. The γ-domain is then cleaved at its N- and C- termini to release the mature protein for secretion [251]. The mechanism that protects the toxin-producing cells from the secreted toxins is not well understood but the γ-domain is presumed to be involved.
Two toxins produced by S. cerevisiae, K1 and K28, have been studied in detail and exert their effects via different mechanisms. One common feature is that the initial interaction between the toxin and the target cell involves binding to the cell wall. For K1, this interaction is mediated through (1 → 6)-β-glucan binding, while K28 binds to mannoproteins [251]. Following cell wall binding, K1 is transferred to the plasma membrane where it forms cation-selective ion channels. The mechanism of this cation-channel formation is not understood. Following binding to the cell wall, the K28 toxin is endocytosed and transported to the cytoplasm via a retrograde transport pathway [252]. Once in the cytoplasm, the protein diffuses into the nucleus and interacts with proteins to block the cell cycle.
A killer toxin WmKT, from the yeast Williopsis mrakii (formerly known as HM1 and Hansenula mrakii, respectively) is chromosomally encoded and expressed as a preproprotein with a signal domain and an N-terminal propeptide that is cleaved to release the mature 88-amino-acid peptide [253]. This peptide forms a Greek key β-barrel structure that displays strong similarities with the plant antifungal peptide, MiAMP1, from macadamia. WmKT binds to the cell wall, probably via interaction with (1 → 6)-β-glucan [253]. The peptide then inhibits the activity of (1 → 3)-β-glucan synthase, an enzyme responsible for synthesis of the β-glucan network of the cell wall, and leads to cell lysis [253–255].
Filamentous fungi killer toxin
The killer toxin, KP4, is one of three produced by the only filamentous fungus that has been reported to express killer toxins to date. It is expressed by Ustilago maydis cells after infection with a dsRNA virus that encodes the toxin, as described for the S. cerevisiae toxins [256]. The KP4 peptide is 105 amino acids in length and displays an α/β sandwich structure. Unlike other killer toxins, KP4 does not seem to require cell wall binding since spheroplasts are equally susceptible to growth inhibition as intact cells. It inhibits Ca2+ uptake into cells by blocking Ca2+ channels, leading to inhibition of Ca2+-regulated pathways required for cell growth and division. KP4 is fungistatic rather than fungicidal [256] and transgenic expression of KP4 in maize provided protection against Ustilago maydis infection [257].
Biotechnological potential for antifungal peptides
Emerging resistance to conventional antifungal strategies has created a requirement for novel methods for protection against fungal pathogens in both agriculture and medicine. Antifungal peptides from natural sources are an important resource for the control of fungal infections. Transgenic expression of a number of AFPs in a variety of crops has provided protection against fungal pathogens with no reported effect on the plant or crop yield [258, 259, 260, 261]. This has been limited to a research environment but the potential for use in agriculture has been established. AFPs have also been shown to control mammalian fungal infections in a laboratory setting [262, 263]. Based on promising data a number of AFPs have undergone pre-clinical and clinical trials yet none has yet gained acceptance for clinical use. As our understanding of the biology and biochemistry of AFPs increases it is anticipated that the potential for AFPs in agriculture and medicine will be achieved.
Concluding remarks
As the number of antifungal peptides identified increases and more information regarding their activities becomes available, one thing is becoming clear. These peptides have evolved to act via a number of different mechanisms, and a single peptide is often capable of more than one mode of action, depending on the target cell type. Very few peptides that demonstrate both antibacterial and antifungal activity appear to exert these effects via the same mechanism and for this reason the antifungal activities of peptides cannot be inferred from studies on their antibacterial activities.
Particularly in relation to antifungal peptides from plants, the multitudinous activities demonstrated by various members of these peptide groups indicate that their structural similarities may merely represent advantageous, stable scaffolds which plants use for a variety of activities. In addition, the similarities seen between antimicrobial peptides from diverse species indicates these peptides developed early in the evolutionary process. It is also possible that some peptides have evolved to display similar characteristics via convergent evolution. The wide variety of features and activities identified for various peptides makes them extremely promising for use as therapeutics and for crop protection in agriculture. In order for these peptides to be used to their full potential, their modes of action must be understood as well as the mechanisms employed by resistant strains to avoid or counteract their toxic effects.
References
- 1.Brahmachary M, et al. ANTIMIC: a database of antimicrobial sequences. Nucleic Acids Res. 2004;32(database issue):D586–D589. doi: 10.1093/nar/gkh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Selitrennikoff CP. Antifungal proteins. Appl Environ Microbiol. 2001;67(7):2883–2894. doi: 10.1128/AEM.67.7.2883-2894.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fisher MC, et al. Emerging fungal threats to animal, plant and ecosystem health. Nature. 2012;484(7393):186–194. doi: 10.1038/nature10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wisplinghoff H, et al. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis (an official publication of the Infectious Diseases Society of America) 2004;39(3):309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 5.Husain S, et al. Opportunistic mycelial fungal infections in organ transplant recipients: emerging importance of non-Aspergillus mycelial fungi. Clin Infect Dis (an official publication of the Infectious Diseases Society of America) 2003;37(2):221–229. doi: 10.1086/375822. [DOI] [PubMed] [Google Scholar]
- 6.Pennisi E. The push to pit genomics against fungal pathogens. Science. 2001;292(5525):2273–2274. doi: 10.1126/science.292.5525.2273. [DOI] [PubMed] [Google Scholar]
- 7.Coca M, et al. Transgenic rice plants expressing the antifungal AFP protein from Aspergillus giganteus show enhanced resistance to the rice blast fungus Magnaporthe grisea . Plant Mol Biol. 2004;54(2):245–259. doi: 10.1023/B:PLAN.0000028791.34706.80. [DOI] [PubMed] [Google Scholar]
- 8.Anand A, et al. Greenhouse and field testing of transgenic wheat plants stably expressing genes for thaumatin-like protein, chitinase and glucanase against Fusarium graminearum . J Exp Bot. 2003;54(384):1101–1111. doi: 10.1093/jxb/erg110. [DOI] [PubMed] [Google Scholar]
- 9.Osusky M, et al. Transgenic potatoes expressing a novel cationic peptide are resistant to late blight and pink rot. Transgenic Res. 2004;13(2):181–190. doi: 10.1023/B:TRAG.0000026076.72779.60. [DOI] [PubMed] [Google Scholar]
- 10.Bowman SM, Free SJ. The structure and synthesis of the fungal cell wall. BioEssays (News and Reviews in Molecular, Cellular and Developmental Biology. 2006;28(8):799–808. doi: 10.1002/bies.20441. [DOI] [PubMed] [Google Scholar]
- 11.Bowman SM, Free SJ. The structure and synthesis of the fungal cell wall. BioEssays. 2006;28(8):799–808. doi: 10.1002/bies.20441. [DOI] [PubMed] [Google Scholar]
- 12.Fontaine T, et al. Molecular organization of the alkali-insoluble fraction of Aspergillus fumigatus cell wall. J Biol Chem. 2000;275(36):27594–27607. doi: 10.1074/jbc.M909975199. [DOI] [PubMed] [Google Scholar]
- 13.Schoffelmeer EA, et al. The cell wall of Fusarium oxysporum . Fungal Genet Biol. 1999;27(2–3):275–282. doi: 10.1006/fgbi.1999.1153. [DOI] [PubMed] [Google Scholar]
- 14.Zlotnik H, et al. Saccharomyces cerevisiae mannoproteins form an external cell wall layer that determines wall porosity. J Bacteriol. 1984;159(3):1018–1026. doi: 10.1128/jb.159.3.1018-1026.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brul S, et al. The incorporation of mannoproteins in the cell wall of S. cerevisiae and filamentous Ascomycetes. Antonie Van Leeuwenhoek. 1997;72(3):229–237. doi: 10.1023/A:1000429208049. [DOI] [PubMed] [Google Scholar]
- 16.Klis FM, et al. Dynamics of cell wall structure in Saccharomyces cerevisiae . FEMS Microbiol Rev. 2002;26(3):239–256. doi: 10.1111/j.1574-6976.2002.tb00613.x. [DOI] [PubMed] [Google Scholar]
- 17.Matsuzaki K. Why and how are peptide-lipid interactions utilized for self-defense? Magainins and tachyplesins as archetypes. Biochim Biophys Acta. 1999;1462(1–2):1–10. doi: 10.1016/s0005-2736(99)00197-2. [DOI] [PubMed] [Google Scholar]
- 18.Theis T, Stahl U. Antifungal proteins: targets, mechanisms and prospective applications. Cell Mol Life Sci. 2004;61(4):437–455. doi: 10.1007/s00018-003-3231-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bloch KE. Sterol structure and membrane function. CRC Crit Rev Biochem. 1983;14(1):47–92. doi: 10.3109/10409238309102790. [DOI] [PubMed] [Google Scholar]
- 20.Brajtburg J, et al. Amphotericin B: current understanding of mechanisms of action. Antimicrob Agents Chemother. 1990;34(2):183–188. doi: 10.1128/AAC.34.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lupetti A, et al. Molecular basis of resistance to azole antifungals. Trends Mol Med. 2002;8(2):76–81. doi: 10.1016/S1471-4914(02)02280-3. [DOI] [PubMed] [Google Scholar]
- 22.Wang TY, Leventis R, Silvius JR. Fluorescence-based evaluation of the partitioning of lipids and lipidated peptides into liquid-ordered lipid microdomains: a model for molecular partitioning into “lipid rafts”. Biophys J . 2000;79(2):919–933. doi: 10.1016/S0006-3495(00)76347-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bagnat M, et al. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc Natl Acad Sci USA. 2000;97(7):3254–3259. doi: 10.1073/pnas.97.7.3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thevissen K, et al. Fungal sphingolipids as targets for the development of selective antifungal therapeutics. Curr Drug Targets. 2005;6(8):923–928. doi: 10.2174/138945005774912771. [DOI] [PubMed] [Google Scholar]
- 25.Loon LCV, Strien EAV. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant Pathol. 1999;55:85–97. doi: 10.1006/pmpp.1999.0213. [DOI] [Google Scholar]
- 26.Jami SK, et al. Molecular, biochemical and structural characterization of osmotin-like protein from black nightshade (Solanum nigrum) J Plant Physiol. 2006;164(3):238–252. doi: 10.1016/j.jplph.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Kuwabara C, et al. Abscisic acid- and cold-induced thaumatin-like protein in winter wheat has an antifungal activity against snow mould Microdochium nivale . Physiol Plant. 2002;115(1):101–110. doi: 10.1034/j.1399-3054.2002.1150112.x. [DOI] [PubMed] [Google Scholar]
- 28.Osmond RI, et al. Binding interactions between barley thaumatin-like proteins and (1,3)-beta-d-glucans. Kinetics, specificity, structural analysis and biological implications. Eur J Biochem. 2001;268(15):4190–4199. doi: 10.1046/j.1432-1327.2001.02331.x. [DOI] [PubMed] [Google Scholar]
- 29.Singh NK, et al. Characterization of osmotin: a thaumatin-like protein associated with osmotic adaptation in plant cells. Plant Physiol. 1987;85(2):529–536. doi: 10.1104/pp.85.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts W, Selitrennikoff C. Zeamatin, an antifungal protein from maize with membrane-permeabilizing activity. J Gen Microbiol. 1990;136:1771–1778. doi: 10.1099/00221287-136-9-1771. [DOI] [Google Scholar]
- 31.de Vos AM, et al. Three-dimensional structure of thaumatin I, an intensely sweet protein. Proc Natl Acad Sci USA. 1985;82(5):1406–1409. doi: 10.1073/pnas.82.5.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Min K, et al. Crystal structure of osmotin, a plant antifungal protein. Proteins. 2004;54(1):170–173. doi: 10.1002/prot.10571. [DOI] [PubMed] [Google Scholar]
- 33.Batalia MA, et al. The crystal structure of the antifungal protein zeamatin, a member of the thaumatin-like, PR-5 protein family. Nat Struct Biol. 1996;3(1):19–23. doi: 10.1038/nsb0196-19. [DOI] [PubMed] [Google Scholar]
- 34.Koiwa H, et al. Crystal structure of tobacco PR-5d protein at 1.8 Å resolution reveals a conserved acidic cleft structure in antifungal thaumatin-like proteins. J Mol Biol. 1999;286(4):1137–1145. doi: 10.1006/jmbi.1998.2540. [DOI] [PubMed] [Google Scholar]
- 35.Salzman RA, et al. Inorganic cations mediate plant PR5 protein antifungal activity through fungal Mnn1- and Mnn4-regulated cell surface glycans. Mol Plant Microbe Interact. 2004;17(7):780–788. doi: 10.1094/MPMI.2004.17.7.780. [DOI] [PubMed] [Google Scholar]
- 36.Ghosh R, Chakrabarti C. Crystal structure analysis of NP24-I: a thaumatin-like protein. Planta. 2008;228(5):883–890. doi: 10.1007/s00425-008-0790-5. [DOI] [PubMed] [Google Scholar]
- 37.Abad LR, et al. Antifungal activity of tobacco osmotin has specificity and involves plasma membrane permeabilization. Plant Sci. 1996;118(1):11–23. doi: 10.1016/0168-9452(96)04420-2. [DOI] [Google Scholar]
- 38.Schimoler-O’Rourke R, Richardson M, Selitrennikoff CP. Zeamatin inhibits trypsin and alpha-amylase activities. Appl Environ Microbiol. 2001;67(5):2365–2366. doi: 10.1128/AEM.67.5.2365-2366.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anzlovar S, et al. Membrane permeabilizing activity of pathogenesis-related protein linusitin from flax seed. Mol Plant Microbe Interact. 1998;11(7):610–617. doi: 10.1094/MPMI.1998.11.7.610. [DOI] [Google Scholar]
- 40.Yun DJ, et al. Stress proteins on the yeast cell surface determine resistance to osmotin, a plant antifungal protein. Proc Natl Acad Sci USA. 1997;94(13):7082–7087. doi: 10.1073/pnas.94.13.7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ibeas JI, et al. Resistance to the plant PR-5 protein osmotin in the model fungus Saccharomyces cerevisiae is mediated by the regulatory effects of SSD1 on cell wall composition. Plant J. 2001;25(3):271–280. doi: 10.1046/j.1365-313x.2001.00967.x. [DOI] [PubMed] [Google Scholar]
- 42.Narasimhan ML, et al. Overexpression of a cell wall glycoprotein in Fusarium oxysporum increases virulence and resistance to a plant PR-5 protein. Plant J. 2003;36(3):390–400. doi: 10.1046/j.1365-313X.2003.01886.x. [DOI] [PubMed] [Google Scholar]
- 43.Ibeas JI, et al. Fungal cell wall phosphomannans facilitate the toxic activity of a plant PR-5 protein. Plant J. 2000;23(3):375–383. doi: 10.1046/j.1365-313x.2000.00792.x. [DOI] [PubMed] [Google Scholar]
- 44.Yun DJ, et al. Osmotin, a plant antifungal protein, subverts signal transduction to enhance fungal cell susceptibility. Mol Cell. 1998;1(6):807–817. doi: 10.1016/S1097-2765(00)80080-5. [DOI] [PubMed] [Google Scholar]
- 45.Narasimhan ML, et al. Osmotin is a homolog of mammalian adiponectin and controls apoptosis in yeast through a homolog of mammalian adiponectin receptor. Mol Cell. 2005;17(2):171–180. doi: 10.1016/j.molcel.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 46.Monteiro S, et al. Osmotin and thaumatin from grape: a putative general defense mechanism against pathogenic fungi. Phytopathology. 2003;93(12):1505–1512. doi: 10.1094/PHYTO.2003.93.12.1505. [DOI] [PubMed] [Google Scholar]
- 47.Mahdavi F, Sariah M, Maziah M. Expression of rice thaumatin-like protein gene in transgenic banana plants enhances resistance to fusarium wilt. Appl Biochem Biotechnol. 2012;166(4):1008–1019. doi: 10.1007/s12010-011-9489-3. [DOI] [PubMed] [Google Scholar]
- 48.Florack DE, Stiekema WJ. Thionins: properties, possible biological roles and mechanisms of action. Plant Mol Biol. 1994;26(1):25–37. doi: 10.1007/BF00039517. [DOI] [PubMed] [Google Scholar]
- 49.Stec B. Plant thionins-the structural perspective. Cell Mol Life Sci. 2006;63(12):1370–1385. doi: 10.1007/s00018-005-5574-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hughes P, et al. The cytotoxic plant protein, beta-purothionin, forms ion channels in lipid membranes. J Biol Chem. 2000;275(2):823–827. doi: 10.1074/jbc.275.2.823. [DOI] [PubMed] [Google Scholar]
- 51.Carrasco L, et al. Thionins: plant peptides that modify membrane permeability in cultured mammalian cells. Eur J Biochem. 1981;116(1):185–189. doi: 10.1111/j.1432-1033.1981.tb05317.x. [DOI] [PubMed] [Google Scholar]
- 52.Stec B, et al. Proposal for molecular mechanism of thionins deduced from physico-chemical studies of plant toxins. J Pept Res. 2004;64(6):210–224. doi: 10.1111/j.1399-3011.2004.00187.x. [DOI] [PubMed] [Google Scholar]
- 53.Majewski J, Stec B. X-ray scattering studies of model lipid membrane interacting with purothionin provide support for a previously proposed mechanism of membrane lysis. Eur Biophys J. 2010;39(8):1155–1165. doi: 10.1007/s00249-009-0568-0. [DOI] [PubMed] [Google Scholar]
- 54.Oard S, Karki B, Enright F. Is there a difference in metal ion-based inhibition between members of thionin family: molecular dynamics simulation study. Biophys Chem. 2007;130(1–2):65–75. doi: 10.1016/j.bpc.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 55.Lay FT, Anderson MA. Defensins-components of the innate immune system in plants. Curr Protein Pept Sci. 2005;6(1):85–101. doi: 10.2174/1389203053027575. [DOI] [PubMed] [Google Scholar]
- 56.Carvalho Ade O, Gomes VM. Plant defensins—prospects for the biological functions and biotechnological properties. Peptides. 2009;30(5):1007–1020. doi: 10.1016/j.peptides.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 57.Colilla FJ, Rocher A, Mendez E. Gamma-purothionins: amino acid sequence of two polypeptides of a new family of thionins from wheat endosperm. FEBS Lett. 1990;270(1–2):191–194. doi: 10.1016/0014-5793(90)81265-P. [DOI] [PubMed] [Google Scholar]
- 58.Bloch C, Jr, Richardson M. A new family of small (5 kDa) protein inhibitors of insect alpha-amylases from seeds or sorghum (Sorghum bicolor (L) Moench) have sequence homologies with wheat gamma-purothionins. FEBS Lett. 1991;279(1):101–104. doi: 10.1016/0014-5793(91)80261-Z. [DOI] [PubMed] [Google Scholar]
- 59.Thomma BP, Cammue BP, Thevissen K. Plant defensins. Planta. 2002;216(2):193–202. doi: 10.1007/s00425-002-0902-6. [DOI] [PubMed] [Google Scholar]
- 60.van der Weerden NL, Anderson MA (2012) Plant defensins: common fold, multiple functions. Fungal Biol Rev
- 61.Gao AG, et al. Fungal pathogen protection in potato by expression of a plant defensin peptide. Nat Biotechnol. 2000;18(12):1307–1310. doi: 10.1038/82436. [DOI] [PubMed] [Google Scholar]
- 62.Jha S, et al. Expression of Dm-AMP1 in rice confers resistance to Magnaporthe oryzae and Rhizoctonia solani . Transgenic Res. 2009;18(1):59–69. doi: 10.1007/s11248-008-9196-1. [DOI] [PubMed] [Google Scholar]
- 63.Tavares PM, et al. In vitro activity of the antifungal plant defensin RsAFP2 against Candida isolates and its in vivo efficacy in prophylactic murine models of candidiasis. Antimicrob Agents Chemother. 2008;52(12):4522–4525. doi: 10.1128/AAC.00448-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaur J, Sagaram US, Shah D. Can plant defensins be used to engineer durable commercially useful fungal resistance in crop plants? Fungal Biol Rev. 2011;25(3):128–135. doi: 10.1016/j.fbr.2011.07.004. [DOI] [Google Scholar]
- 65.Almeida MS, et al. Solution structure of Pisum sativum defensin 1 by high resolution NMR: plant defensins, identical backbone with different mechanisms of action. J Mol Biol. 2002;315(4):749–757. doi: 10.1006/jmbi.2001.5252. [DOI] [PubMed] [Google Scholar]
- 66.Lay FT, et al. The three-dimensional solution structure of NaD1, a new floral defensin from Nicotiana alata and its application to a homology model of the crop defense protein alfAFP. J Mol Biol. 2003;325(1):175–188. doi: 10.1016/S0022-2836(02)01103-8. [DOI] [PubMed] [Google Scholar]
- 67.Liu YJ, et al. Solution structure of the plant defensin VrD1 from mung bean and its possible role in insecticidal activity against bruchids. Proteins. 2006;63(4):777–786. doi: 10.1002/prot.20962. [DOI] [PubMed] [Google Scholar]
- 68.Fant F, Vranken WF, Borremans FA. The three-dimensional solution structure of Aesculus hippocastanum antimicrobial protein 1 determined by 1H nuclear magnetic resonance. Proteins. 1999;37(3):388–403. doi: 10.1002/(SICI)1097-0134(19991115)37:3<388::AID-PROT7>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 69.Fant F, et al. Determination of the three-dimensional solution structure of Raphanus sativus antifungal protein 1 by 1H NMR. J Mol Biol. 1998;279(1):257–270. doi: 10.1006/jmbi.1998.1767. [DOI] [PubMed] [Google Scholar]
- 70.Broekaert WF, et al. Plant defensins: novel antimicrobial peptides as components of the host defense system. Plant Physiol. 1995;108(4):1353–1358. doi: 10.1104/pp.108.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lay FT, Brugliera F, Anderson MA. Isolation and properties of floral defensins from ornamental tobacco and petunia. Plant Physiol. 2003;131(3):1283–1293. doi: 10.1104/pp.102.016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thevissen K, et al. Fungal membrane responses induced by plant defensins and thionins. J Biol Chem. 1996;271(25):15018–15025. doi: 10.1074/jbc.271.25.15018. [DOI] [PubMed] [Google Scholar]
- 73.Thevissen K, et al. Specific, high affinity binding sites for an antifungal plant defensin on Neurospora crassa hyphae and microsomal membranes. J Biol Chem. 1997;272(51):32176–32181. doi: 10.1074/jbc.272.51.32176. [DOI] [PubMed] [Google Scholar]
- 74.Thevissen K, et al. Specific binding sites for an antifungal plant defensin from Dahlia (Dahlia merckii) on fungal cells are required for antifungal activity. Mol Plant Microbe Interact. 2000;13(1):54–61. doi: 10.1094/MPMI.2000.13.1.54. [DOI] [PubMed] [Google Scholar]
- 75.Thevissen K, Terras FR, Broekaert WF. Permeabilization of fungal membranes by plant defensins inhibits fungal growth. Appl Environ Microbiol. 1999;65(12):5451–5458. doi: 10.1128/aem.65.12.5451-5458.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aerts AM et al. (2012) The antifungal activity of RsAFP2, a plant defensin from Raphanus sativus, involves the induction of reactive oxygen species in Candida albicans. J Mol Microbiol Biotechnol (in press) [DOI] [PubMed]
- 77.Thevissen K, et al. DmAMP1, an antifungal plant defensin from dahlia (Dahlia merckii), interacts with sphingolipids from Saccharomyces cerevisiae . FEMS Microbiol Lett. 2003;226(1):169–173. doi: 10.1016/S0378-1097(03)00590-1. [DOI] [PubMed] [Google Scholar]
- 78.Thevissen K, et al. Defensins from insects and plants interact with fungal glucosylceramides. J Biol Chem. 2004;279(6):3900–3905. doi: 10.1074/jbc.M311165200. [DOI] [PubMed] [Google Scholar]
- 79.Aerts AM, et al. The antifungal plant defensin RsAFP2 from radish induces apoptosis in a metacaspase independent way in Candida albicans . FEBS Lett. 2009;583(15):2513–2516. doi: 10.1016/j.febslet.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 80.Aerts AM, et al. The antifungal plant defensin HsAFP1 from Heuchera sanguinea induces apoptosis in Candida albicans . Front Microbiol. 2011;2:47. doi: 10.3389/fmicb.2011.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thevissen K, et al. The plant defensin RsAFP2 induces cell wall stress, septin mislocalization and accumulation of ceramides in Candida albicans . Mol Microbiol. 2012;84(1):166–180. doi: 10.1111/j.1365-2958.2012.08017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ramamoorthy V, et al. Glucosylceramide synthase is essential for alfalfa defensin-mediated growth inhibition but not for pathogenicity of Fusarium graminearum . Mol Microbiol. 2007;66(3):771–786. doi: 10.1111/j.1365-2958.2007.05955.x. [DOI] [PubMed] [Google Scholar]
- 83.Spelbrink RG, et al. Differential antifungal and calcium channel-blocking activity among structurally related plant defensins. Plant Physiol. 2004;135(4):2055–2067. doi: 10.1104/pp.104.040873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ramamoorthy V, et al. Two mitogen-activated protein kinase signalling cascades mediate basal resistance to antifungal plant defensins in Fusarium graminearum . Cell Microbiol. 2007;9(6):1491–1506. doi: 10.1111/j.1462-5822.2006.00887.x. [DOI] [PubMed] [Google Scholar]
- 85.van der Weerden NL, Lay FT, Anderson MA. The plant defensin, NaD1, enters the cytoplasm of Fusarium oxysporum hyphae. J Biol Chem. 2008;283(21):14445–14452. doi: 10.1074/jbc.M709867200. [DOI] [PubMed] [Google Scholar]
- 86.van der Weerden NL, Hancock RE, Anderson MA. Permeabilization of fungal hyphae by the plant defensin NaD1 occurs through a cell wall-dependent process. J Biol Chem. 2010;285(48):37513–37520. doi: 10.1074/jbc.M110.134882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lobo DS, et al. Antifungal Pisum sativum defensin 1 interacts with Neurospora crassa cyclin F related to the cell cycle. Biochemistry. 2007;46(4):987–996. doi: 10.1021/bi061441j. [DOI] [PubMed] [Google Scholar]
- 88.Kader JC. Lipid-transfer proteins in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:627–654. doi: 10.1146/annurev.arplant.47.1.627. [DOI] [PubMed] [Google Scholar]
- 89.Thoma S, Kaneko Y, Somerville C. A non-specific lipid transfer protein from Arabidopsis is a cell wall protein. Plant J. 1993;3(3):427–436. doi: 10.1046/j.1365-313X.1993.t01-25-00999.x. [DOI] [PubMed] [Google Scholar]
- 90.Cammue BP, et al. A potent antimicrobial protein from onion seeds showing sequence homology to plant lipid transfer proteins. Plant Physiol. 1995;109(2):445–455. doi: 10.1104/pp.109.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cheng CS, et al. Binding mechanism of nonspecific lipid transfer proteins and their role in plant defense. Biochemistry. 2004;43(43):13628–13636. doi: 10.1021/bi048873j. [DOI] [PubMed] [Google Scholar]
- 92.Regente MC, et al. The cytotoxic properties of a plant lipid transfer protein involve membrane permeabilization of target cells. Lett Appl Microbiol. 2005;40(3):183–189. doi: 10.1111/j.1472-765X.2004.01647.x. [DOI] [PubMed] [Google Scholar]
- 93.Zottich U, et al. Purification, biochemical characterization and antifungal activity of a new lipid transfer protein (LTP) from Coffea canephora seeds with alpha-amylase inhibitor properties. Biochim Biophys Acta. 2011;1810(4):375–383. doi: 10.1016/j.bbagen.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 94.Diz MS, et al. Characterisation, immunolocalisation and antifungal activity of a lipid transfer protein from chili pepper (Capsicum annuum) seeds with novel alpha-amylase inhibitory properties. Physiol Plant. 2011;142(3):233–246. doi: 10.1111/j.1399-3054.2011.01464.x. [DOI] [PubMed] [Google Scholar]
- 95.Tailor RH, et al. A novel family of small cysteine-rich antimicrobial peptides from seed of Impatiens balsamina is derived from a single precursor protein. J Biol Chem. 1997;272(39):24480–24487. doi: 10.1074/jbc.272.39.24480. [DOI] [PubMed] [Google Scholar]
- 96.Patel SU, et al. Structural studies of Impatiens balsamina antimicrobial protein (Ib-AMP1) Biochemistry. 1998;37(4):983–990. doi: 10.1021/bi971747d. [DOI] [PubMed] [Google Scholar]
- 97.Thevissen K, et al. Antifungal activity of synthetic peptides derived from Impatiens balsamina antimicrobial peptides Ib-AMP1 and Ib-AMP4. Peptides. 2005;26(7):1113–1119. doi: 10.1016/j.peptides.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 98.Lee DG, et al. Antifungal mechanism of a cysteine-rich antimicrobial peptide, Ib-AMP1, from Impatiens balsamina against Candida albicans . Biotechnol Lett. 1999;21:1047–1050. doi: 10.1023/A:1005636610512. [DOI] [Google Scholar]
- 99.Wang P, et al. Antimicrobial specificity and mechanism of action of disulfide-removed linear analogs of the plant-derived Cys-rich antimicrobial peptide Ib-AMP1. Peptides. 2009;30(12):2144–2149. doi: 10.1016/j.peptides.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 100.Segura A, et al. Snakin-1, a peptide from potato that is active against plant pathogens. Mol Plant Microbe Interact. 1999;12(1):16–23. doi: 10.1094/MPMI.1999.12.1.16. [DOI] [PubMed] [Google Scholar]
- 101.Berrocal-Lobo M, et al. Snakin-2, an antimicrobial peptide from potato whose gene is locally induced by wounding and responds to pathogen infection. Plant Physiol. 2002;128(3):951–961. doi: 10.1104/pp.010685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang RH, et al. Solution structure of Eucommia antifungal peptide: a novel structural model distinct with a five-disulfide motif. Biochemistry. 2004;43(20):6005–6012. doi: 10.1021/bi036263y. [DOI] [PubMed] [Google Scholar]
- 103.Broekaert WF, et al. Antimicrobial peptides from Amaranthus caudatus seeds with sequence homology to the cysteine/glycine-rich domain of chitin-binding proteins. Biochemistry. 1992;31(17):4308–4314. doi: 10.1021/bi00132a023. [DOI] [PubMed] [Google Scholar]
- 104.Koo JC, et al. Pn-AMP1, a plant defense protein, induces actin depolarization in yeasts. Plant Cell Physiol. 2004;45(11):1669–1680. doi: 10.1093/pcp/pch189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kiba A, et al. C-terminal domain of a hevein-like protein from Wasabia japonica has potent antimicrobial activity. Plant Cell Physiol. 2003;44(3):296–303. doi: 10.1093/pcp/pcg035. [DOI] [PubMed] [Google Scholar]
- 106.Van den Bergh KP, et al. Five disulfide bridges stabilize a hevein-type antimicrobial peptide from the bark of spindle tree (Euonymus europaeus L.) FEBS Lett. 2002;530(1–3):181–185. doi: 10.1016/S0014-5793(02)03474-9. [DOI] [PubMed] [Google Scholar]
- 107.Yokoyama S, et al. The chitin-binding capability of Cy-AMP1 from cycad is essential to antifungal activity. J Pept Sci. 2009;15(7):492–497. doi: 10.1002/psc.1147. [DOI] [PubMed] [Google Scholar]
- 108.Andreev YA, et al. Genes encoding hevein-like defense peptides in wheat: distribution, evolution, and role in stress response. Biochimie. 2012;94(4):1009–1016. doi: 10.1016/j.biochi.2011.12.023. [DOI] [PubMed] [Google Scholar]
- 109.Rees DC, Lipscomb WN. Refined crystal structure of the potato inhibitor complex of carboxypeptidase A at 2.5 Å resolution. J Mol Biol. 1982;160(3):475–498. doi: 10.1016/0022-2836(82)90309-6. [DOI] [PubMed] [Google Scholar]
- 110.Cammue BP, et al. Isolation and characterization of a novel class of plant antimicrobial peptides form Mirabilis jalapa L. seeds. J Biol Chem. 1992;267(4):2228–2233. [PubMed] [Google Scholar]
- 111.Gao GH, et al. Solution structure of PAFP-S: a new knottin-type antifungal peptide from the seeds of Phytolacca americana . Biochemistry. 2001;40(37):10973–10978. doi: 10.1021/bi010167k. [DOI] [PubMed] [Google Scholar]
- 112.Yount NY, Yeaman MR. Multidimensional signatures in antimicrobial peptides. Proc Natl Acad Sci USA. 2004;101(19):7363–7368. doi: 10.1073/pnas.0401567101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yeaman MR, Yount NY. Unifying themes in host defence effector polypeptides. Nat Rev Microbiol. 2007;5(9):727–740. doi: 10.1038/nrmicro1744. [DOI] [PubMed] [Google Scholar]
- 114.Chouabe C, et al. New mode of action for a knottin protein bioinsecticide: pea albumin 1 subunit b (PA1b) is the first peptidic inhibitor of V-ATPase. J Biol Chem. 2011;286(42):36291–36296. doi: 10.1074/jbc.M111.281055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Robotham JM, et al. Ana o 3, an important cashew nut (Anacardium occidentale L.) allergen of the 2S albumin family. J Allergy Clin Immunol. 2005;115(6):1284–1290. doi: 10.1016/j.jaci.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 116.Puumalainen TJ, et al. Napins, 2S albumins, are major allergens in oilseed rape and turnip rape. J Allergy Clin Immunol. 2006;117(2):426–432. doi: 10.1016/j.jaci.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 117.Wang X, Bunkers GJ. Potent heterologous antifungal proteins from cheeseweed (Malva parviflora) Biochem Biophys Res Commun. 2000;279(2):669–673. doi: 10.1006/bbrc.2000.3997. [DOI] [PubMed] [Google Scholar]
- 118.Agizzio AP, et al. A 2S albumin-homologous protein from passion fruit seeds inhibits the fungal growth and acidification of the medium by Fusarium oxysporum . Arch Biochem Biophys. 2003;416(2):188–195. doi: 10.1016/S0003-9861(03)00313-8. [DOI] [PubMed] [Google Scholar]
- 119.Terras FR, et al. Analysis of two novel classes of plant antifungal proteins from radish (Raphanus sativus L.) seeds. J Biol Chem. 1992;267(22):15301–15309. [PubMed] [Google Scholar]
- 120.Nolde SB, et al. Disulfide-stabilized helical hairpin structure and activity of a novel antifungal peptide EcAMP1 from seeds of barnyard grass (Echinochloa crus-galli) J Biol Chem. 2011;286(28):25145–25153. doi: 10.1074/jbc.M110.200378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Oparin PB, et al. Buckwheat trypsin inhibitor with helical hairpin structure belongs to a new family of plant defence peptides. Biochem J. 2012;446(1):69–77. doi: 10.1042/BJ20120548. [DOI] [PubMed] [Google Scholar]
- 122.Zhu S. Discovery of six families of fungal defensin-like peptides provides insights into origin and evolution of the CSalphabeta defensins. Mol Immunol. 2007;45(3):828–838. doi: 10.1016/j.molimm.2007.06.354. [DOI] [PubMed] [Google Scholar]
- 123.Yang D, Chertov O, Oppenheim JJ. The role of mammalian antimicrobial peptides and proteins in awakening of innate host defenses and adaptive immunity. Cell Mol Life Sci. 2001;58(7):978–989. doi: 10.1007/PL00000914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fitzgerald DH, Coleman DC, O’Connell BC. Binding, internalisation and degradation of histatin 3 in histatin-resistant derivatives of Candida albicans . FEMS Microbiol Lett. 2003;220(2):247–253. doi: 10.1016/S0378-1097(03)00121-6. [DOI] [PubMed] [Google Scholar]
- 125.Raj PA, Marcus E, Sukumaran DK. Structure of human salivary histatin 5 in aqueous and nonaqueous solutions. Biopolymers. 1998;45(1):51–67. doi: 10.1002/(SICI)1097-0282(199801)45:1<51::AID-BIP5>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 126.Conlon JM, et al. Isolation of peptides of the brevinin-1 family with potent candidacidal activity from the skin secretions of the frog Rana boylii . J Pept Res. 2003;62(5):207–213. doi: 10.1034/j.1399-3011.2003.00090.x. [DOI] [PubMed] [Google Scholar]
- 127.Dong J, et al. Calcium blocks fungicidal activity of human salivary histatin 5 through disruption of binding with Candida albicans . J Dent Res. 2003;82(9):748–752. doi: 10.1177/154405910308200917. [DOI] [PubMed] [Google Scholar]
- 128.Li XS, et al. Candida albicans cell wall ssa proteins bind and facilitate import of salivary histatin 5 required for toxicity. J Biol Chem. 2006;281(32):22453–22463. doi: 10.1074/jbc.M604064200. [DOI] [PubMed] [Google Scholar]
- 129.Kumar R, et al. Histatin 5 uptake by Candida albicans utilizes polyamine transporters Dur3 and Dur31 proteins. J Biol Chem. 2011;286(51):43748–43758. doi: 10.1074/jbc.M111.311175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jang WS, et al. Salivary histatin 5 internalization by translocation, but not endocytosis, is required for fungicidal activity in Candida albicans . Mol Microbiol. 2010;77(2):354–370. doi: 10.1111/j.1365-2958.2010.07210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Helmerhorst EJ, et al. The cellular target of histatin 5 on Candida albicans is the energized mitochondrion. J Biol Chem. 1999;274(11):7286–7291. doi: 10.1074/jbc.274.11.7286. [DOI] [PubMed] [Google Scholar]
- 132.Edgerton M, et al. Candidacidal activity of salivary histatins. Identification of a histatin 5-binding protein on Candida albicans . J Biol Chem. 1998;273(32):20438–20447. doi: 10.1074/jbc.273.32.20438. [DOI] [PubMed] [Google Scholar]
- 133.Helmerhorst EJ, Troxler RF, Oppenheim FG. The human salivary peptide histatin 5 exerts its antifungal activity through the formation of reactive oxygen species. Proc Natl Acad Sci USA. 2001;98(25):14637–14642. doi: 10.1073/pnas.141366998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Koshlukova SE, et al. Salivary histatin 5 induces non-lytic release of ATP from Candida albicans leading to cell death. J Biol Chem. 1999;274(27):18872–18889. doi: 10.1074/jbc.274.27.18872. [DOI] [PubMed] [Google Scholar]
- 135.Koshlukova SE, et al. Released ATP is an extracellular cytotoxic mediator in salivary histatin 5-induced killing of Candida albicans . Infect Immun. 2000;68(12):6848–6856. doi: 10.1128/IAI.68.12.6848-6856.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Baev D, et al. The TRK1 potassium transporter is the critical effector for killing of Candida albicans by the cationic protein, histatin 5. J Biol Chem. 2004;279(53):55060–55072. doi: 10.1074/jbc.M411031200. [DOI] [PubMed] [Google Scholar]
- 137.Wunder D, et al. Human salivary histatin 5 fungicidal action does not induce programmed cell death pathways in Candida albicans . Antimicrob Agents Chemother. 2004;48(1):110–115. doi: 10.1128/AAC.48.1.110-115.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Veerman EC, et al. Reactive oxygen species play no role in the candidacidal activity of the salivary antimicrobial peptide histatin 5. Biochem J. 2004;381(Pt 2):447–452. doi: 10.1042/BJ20040208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gennaro R, Zanetti M. Structural features and biological activities of the cathelicidin-derived antimicrobial peptides. Biopolymers. 2000;55(1):31–49. doi: 10.1002/1097-0282(2000)55:1<31::AID-BIP40>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 140.Wang J, et al. Ancient antimicrobial peptides kill antibiotic-resistant pathogens: Australian mammals provide new options. PLoS ONE. 2011;6(8):e24030. doi: 10.1371/journal.pone.0024030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Benincasa M, et al. Fungicidal activity of five cathelicidin peptides against clinically isolated yeasts. J Antimicrob Chemother. 2006;58(5):950–959. doi: 10.1093/jac/dkl382. [DOI] [PubMed] [Google Scholar]
- 142.Skerlavaj B, et al. SMAP-29: a potent antibacterial and antifungal peptide from sheep leukocytes. FEBS Lett. 1999;463(1–2):58–62. doi: 10.1016/S0014-5793(99)01600-2. [DOI] [PubMed] [Google Scholar]
- 143.Lee DG, et al. Antifungal mechanism of SMAP-29 (1–18) isolated from sheep myeloid mRNA against Trichosporon beigelii . Biochem Biophys Res Commun. 2002;295(3):591–596. doi: 10.1016/S0006-291X(02)00717-9. [DOI] [PubMed] [Google Scholar]
- 144.Park K, et al. Structural studies of porcine myeloid antibacterial peptide PMAP-23 and its analogues in DPC micelles by NMR spectroscopy. Biochem Biophys Res Commun. 2002;290(1):204–212. doi: 10.1006/bbrc.2001.6173. [DOI] [PubMed] [Google Scholar]
- 145.Lee DG, et al. Fungicidal effect of antimicrobial peptide, PMAP-23, isolated from porcine myeloid against Candida albicans . Biochem Biophys Res Commun. 2001;282(2):570–574. doi: 10.1006/bbrc.2001.4602. [DOI] [PubMed] [Google Scholar]
- 146.Falla TJ, Hancock RE. Improved activity of a synthetic indolicidin analog. Antimicrob Agents Chemother. 1997;41(4):771–775. doi: 10.1128/aac.41.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Porcelli F, et al. NMR structure of the cathelicidin-derived human antimicrobial peptide LL-37 in dodecylphosphocholine micelles. Biochemistry. 2008;47(20):5565–5572. doi: 10.1021/bi702036s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.den Hertog AL, et al. Candidacidal effects of two antimicrobial peptides: histatin 5 causes small membrane defects, but LL-37 causes massive disruption of the cell membrane. Biochem J. 2005;388(Pt 2):689–695. doi: 10.1042/BJ20042099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Henzler-Wildman KA, et al. Perturbation of the hydrophobic core of lipid bilayers by the human antimicrobial peptide LL-37. Biochemistry. 2004;43(26):8459–8469. doi: 10.1021/bi036284s. [DOI] [PubMed] [Google Scholar]
- 150.Sood R, Kinnunen PK. Cholesterol, lanosterol, and ergosterol attenuate the membrane association of LL-37(W27F) and temporin L. Biochim Biophys Acta. 2008;1778(6):1460–1466. doi: 10.1016/j.bbamem.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 151.Lopez-Garcia B, et al. Anti-fungal activity of cathelicidins and their potential role in Candida albicans skin infection. J Invest Dermatol. 2005;125(1):108–115. doi: 10.1111/j.0022-202X.2005.23713.x. [DOI] [PubMed] [Google Scholar]
- 152.den Hertog AL, et al. The human cathelicidin peptide LL-37 and truncated variants induce segregation of lipids and proteins in the plasma membrane of Candida albicans . Biol Chem. 2006;387(10–11):1495–1502. doi: 10.1515/BC.2006.187. [DOI] [PubMed] [Google Scholar]
- 153.Durr UH, Sudheendra US, Ramamoorthy A. LL-37, the only human member of the cathelicidin family of antimicrobial peptides. Biochim Biophys Acta. 2006;1758(9):1408–1425. doi: 10.1016/j.bbamem.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 154.Fellermann K, et al. Crohn’s disease: a defensin deficiency syndrome? Eur J Gastroenterol Hepatol. 2003;15(6):627–634. doi: 10.1097/00042737-200306000-00008. [DOI] [PubMed] [Google Scholar]
- 155.Raj PA, Dentino AR. Current status of defensins and their role in innate and adaptive immunity. FEMS Microbiol Lett. 2002;206(1):9–18. doi: 10.1111/j.1574-6968.2002.tb10979.x. [DOI] [PubMed] [Google Scholar]
- 156.Lehrer RI, et al. Modulation of the in vitro candidacidal activity of human neutrophil defensins by target cell metabolism and divalent cations. J Clin Invest. 1988;81(6):1829–1835. doi: 10.1172/JCI113527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wilde CG, et al. Purification and characterization of human neutrophil peptide 4, a novel member of the defensin family. J Biol Chem. 1989;264(19):11200–11203. [PubMed] [Google Scholar]
- 158.Ganz T, Lehrer RI. Defensins. Pharmacol Ther. 1995;66(2):191–205. doi: 10.1016/0163-7258(94)00076-F. [DOI] [PubMed] [Google Scholar]
- 159.Lehrer RI, et al. Correlation of binding of rabbit granulocyte peptides to Candida albicans with candidacidal activity. Infect Immun. 1985;49(1):207–211. doi: 10.1128/iai.49.1.207-211.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Edgerton M, et al. Salivary histatin 5 and human neutrophil defensin 1 kill Candida albicans via shared pathways. Antimicrob Agents Chemother. 2000;44(12):3310–3316. doi: 10.1128/AAC.44.12.3310-3316.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Schroder JM, Harder J. Human beta-defensin-2. Int J Biochem Cell Biol. 1999;31(6):645–651. doi: 10.1016/S1357-2725(99)00013-8. [DOI] [PubMed] [Google Scholar]
- 162.Dhople V, Krukemeyer A, Ramamoorthy A. The human beta-defensin-3, an antibacterial peptide with multiple biological functions. Biochim Biophys Acta. 2006;1758(9):1499–1512. doi: 10.1016/j.bbamem.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 163.Joly S, et al. Human beta-defensins 2 and 3 demonstrate strain-selective activity against oral microorganisms. J Clin Microbiol. 2004;42(3):1024–1029. doi: 10.1128/JCM.42.3.1024-1029.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ishikawa H, Bae S, Katayama I. Human beta defensin-2 expression by keratinocytes is induces by co culture with Trycophyton rubrum through Toll-like receptors 2 and 4. Open Dermatol J. 2009;3:81–85. doi: 10.2174/1874372200903010081. [DOI] [Google Scholar]
- 165.Alekseeva L, et al. Inducible expression of beta defensins by human respiratory epithelial cells exposed to Aspergillus fumigatus organisms. BMC Microbiol. 2009;9:33. doi: 10.1186/1471-2180-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Vylkova S, et al. Distinct antifungal mechanisms: beta-defensins require Candida albicans Ssa1 protein, while Trk1p mediates activity of cysteine-free cationic peptides. Antimicrob Agents Chemother. 2006;50(1):324–331. doi: 10.1128/AAC.50.1.324-331.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Argimon S, et al. Interaction between the Candida albicans high-osmolarity glycerol (HOG) pathway and the response to human beta-defensins 2 and 3. Eukaryot Cell. 2011;10(2):272–275. doi: 10.1128/EC.00133-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Vylkova S, et al. Histatin 5 initiates osmotic stress response in Candida albicans via activation of the Hog1 mitogen-activated protein kinase pathway. Eukaryot Cell. 2007;6(10):1876–1888. doi: 10.1128/EC.00039-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.O’Rourke SM, Herskowitz I, O’Shea EK. Yeast go the whole HOG for the hyperosmotic response. Trends Genet. 2002;18(8):405–412. doi: 10.1016/S0168-9525(02)02723-3. [DOI] [PubMed] [Google Scholar]
- 170.Vylkova S, et al. Human beta-defensins kill Candida albicans in an energy-dependent and salt-sensitive manner without causing membrane disruption. Antimicrob Agents Chemother. 2007;51(1):154–161. doi: 10.1128/AAC.00478-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Farnaud S, Evans RW. Lactoferrin—a multifunctional protein with antimicrobial properties. Mol Immunol. 2003;40(7):395–405. doi: 10.1016/S0161-5890(03)00152-4. [DOI] [PubMed] [Google Scholar]
- 172.Lupetti A, et al. Candidacidal activities of human lactoferrin peptides derived from the N terminus. Antimicrob Agents Chemother. 2000;44(12):3257–3263. doi: 10.1128/AAC.44.12.3257-3263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Gifford JL, Hunter HN, Vogel HJ. Lactoferricin: a lactoferrin-derived peptide with antimicrobial, antiviral, antitumor and immunological properties. Cell Mol Life Sci. 2005;62(22):2588–2598. doi: 10.1007/s00018-005-5373-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Viejo-Diaz M, Andres MT, Fierro JF. Different anti-Candida activities of two human lactoferrin-derived peptides, Lfpep and kaliocin-1. Antimicrob Agents Chemother. 2005;49(7):2583–2588. doi: 10.1128/AAC.49.7.2583-2588.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Munoz A, Marcos JF. Activity and mode of action against fungal phytopathogens of bovine lactoferricin-derived peptides. J Appl Microbiol. 2006;101(6):1199–1207. doi: 10.1111/j.1365-2672.2006.03089.x. [DOI] [PubMed] [Google Scholar]
- 176.Chen HL, et al. Synthetic porcine lactoferricin with a 20-residue peptide exhibits antimicrobial activity against Escherichia coli, Staphylococcus aureus, and Candida albicans . J Agric Food Chem. 2006;54(9):3277–3282. doi: 10.1021/jf053031s. [DOI] [PubMed] [Google Scholar]
- 177.Viejo-Diaz M, Andres MT, Fierro JF. Effects of human lactoferrin on the cytoplasmic membrane of Candida albicans cells related with its candidacidal activity. FEMS Immunol Med Microbiol. 2004;42(2):181–185. doi: 10.1016/j.femsim.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 178.Yamauchi K, et al. Antibacterial activity of lactoferrin and a pepsin-derived lactoferrin peptide fragment. Infect Immun. 1993;61(2):719–728. doi: 10.1128/iai.61.2.719-728.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Strom MB, et al. Increased antibacterial activity of 15-residue murine lactoferricin derivatives. J Pept Res. 2001;57(2):127–139. doi: 10.1034/j.1399-3011.2001.00806.x. [DOI] [PubMed] [Google Scholar]
- 180.Vorland LH, et al. Lactoferricin of bovine origin is more active than lactoferricins of human, murine and caprine origin. Scand J Infect Dis. 1998;30(5):513–517. doi: 10.1080/00365549850161557. [DOI] [PubMed] [Google Scholar]
- 181.Hwang PM, et al. Three-dimensional solution structure of lactoferricin B, an antimicrobial peptide derived from bovine lactoferrin. Biochemistry. 1998;37(12):4288–4298. doi: 10.1021/bi972323m. [DOI] [PubMed] [Google Scholar]
- 182.Hunter HN, et al. Human lactoferricin is partially folded in aqueous solution and is better stabilized in a membrane mimetic solvent. Antimicrob Agents Chemother. 2005;49(8):3387–3395. doi: 10.1128/AAC.49.8.3387-3395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hoek KS, et al. Antibacterial activity in bovine lactoferrin-derived peptides. Antimicrob Agents Chemother. 1997;41(1):54–59. doi: 10.1128/aac.41.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Nguyen LT, Schibli DJ, Vogel HJ. Structural studies and model membrane interactions of two peptides derived from bovine lactoferricin. J Pept Sci. 2005;11(7):379–389. doi: 10.1002/psc.629. [DOI] [PubMed] [Google Scholar]
- 185.Viejo-Diaz M, et al. Potassium efflux induced by a new lactoferrin-derived peptide mimicking the effect of native human lactoferrin on the bacterial cytoplasmic membrane. Biochemistry (Mosc) 2003;68(2):217–227. doi: 10.1023/A:1022657630698. [DOI] [PubMed] [Google Scholar]
- 186.Farnaud S, et al. Interactions of lactoferricin-derived peptides with LPS and antimicrobial activity. FEMS Microbiol Lett. 2004;233(2):193–199. doi: 10.1111/j.1574-6968.2004.tb09482.x. [DOI] [PubMed] [Google Scholar]
- 187.Haukland HH, et al. The antimicrobial peptides lactoferricin B and magainin 2 cross over the bacterial cytoplasmic membrane and reside in the cytoplasm. FEBS Lett. 2001;508(3):389–393. doi: 10.1016/S0014-5793(01)03100-3. [DOI] [PubMed] [Google Scholar]
- 188.Ulvatne H, Vorland LH. Bactericidal kinetics of 3 lactoferricins against Staphylococcus aureus and Escherichia coli . Scand J Infect Dis. 2001;33(7):507–511. doi: 10.1080/00365540110026692. [DOI] [PubMed] [Google Scholar]
- 189.Ho YH, Sung TC, Chen CS. Lactoferricin B inhibits the phosphorylation of the two-component system response regulators BasR and CreB. Mol Cell Proteomics. 2012;11(4):M111.014720. doi: 10.1074/mcp.M111.014720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Rinaldi AC. Antimicrobial peptides from amphibian skin: an expanding scenario. Curr Opin Chem Biol. 2002;6(6):799–804. doi: 10.1016/S1367-5931(02)00401-5. [DOI] [PubMed] [Google Scholar]
- 191.Rollins-Smith LA, et al. Activities of temporin family peptides against the chytrid fungus (Batrachochytrium dendrobatidis) associated with global amphibian declines. Antimicrob Agents Chemother. 2003;47(3):1157–1160. doi: 10.1128/AAC.47.3.1157-1160.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Simmaco M, et al. Temporins, antimicrobial peptides from the European red frog Rana temporaria . Eur J Biochem. 1996;242(3):788–792. doi: 10.1111/j.1432-1033.1996.0788r.x. [DOI] [PubMed] [Google Scholar]
- 193.Mangoni ML, et al. Structure-function relationships of temporins, small antimicrobial peptides from amphibian skin. Eur J Biochem. 2000;267(5):1447–1454. doi: 10.1046/j.1432-1327.2000.01143.x. [DOI] [PubMed] [Google Scholar]
- 194.Mangoni ML. Temporins, anti-infective peptides with expanding properties. Cell Mol Life Sci. 2006;63(9):1060–1069. doi: 10.1007/s00018-005-5536-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Rinaldi AC, et al. Effects of temporins on molecular dynamics and membrane permeabilization in lipid vesicles. J Pept Res. 2001;58(3):213–220. doi: 10.1034/j.1399-3011.2001.00896.x. [DOI] [PubMed] [Google Scholar]
- 196.Pal T, et al. Brevinin-1BYa: a naturally occurring peptide from frog skin with broad-spectrum antibacterial and antifungal properties. Int J Antimicrob Agents. 2006;27(6):525–529. doi: 10.1016/j.ijantimicag.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 197.Bulet P, Stocklin R. Insect antimicrobial peptides: structures, properties and gene regulation. Protein Pept Lett. 2005;12(1):3–11. doi: 10.2174/0929866053406011. [DOI] [PubMed] [Google Scholar]
- 198.Jiggins FM, Kim KW. The evolution of antifungal peptides in Drosophila . Genetics. 2005;171(4):1847–1859. doi: 10.1534/genetics.105.045435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lamberty M, et al. Insect immunity. Isolation from the lepidopteran Heliothis virescens of a novel insect defensin with potent antifungal activity. J Biol Chem. 1999;274(14):9320–9326. doi: 10.1074/jbc.274.14.9320. [DOI] [PubMed] [Google Scholar]
- 200.Lamberty M, et al. Insect immunity. Constitutive expression of a cysteine-rich antifungal and a linear antibacterial peptide in a termite insect. J Biol Chem. 2001;276(6):4085–4092. doi: 10.1074/jbc.M002998200. [DOI] [PubMed] [Google Scholar]
- 201.Landon C, et al. Solution structure of drosomycin, the first inducible antifungal protein from insects. Protein Sci. 1997;6(9):1878–1884. doi: 10.1002/pro.5560060908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Lamberty M, et al. Solution structures of the antifungal heliomicin and a selected variant with both antibacterial and antifungal activities. Biochemistry. 2001;40(40):11995–12003. doi: 10.1021/bi0103563. [DOI] [PubMed] [Google Scholar]
- 203.Da Silva P, et al. Solution structure of termicin, an antimicrobial peptide from the termite Pseudacanthotermes spiniger . Protein Sci. 2003;12(3):438–446. doi: 10.1110/ps.0228303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zhang Z, Zhu S. Functional role of charged residues in drosomycin, a Drosophila antifungal peptide. Dev Comp Immunol. 2010;34(9):953–958. doi: 10.1016/j.dci.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 205.Landon C, et al. The active site of drosomycin, a small insect antifungal protein, delineated by comparison with the modeled structure of Rs-AFP2, a plant antifungal protein. J Pept Res. 2000;56(4):231–238. doi: 10.1034/j.1399-3011.2000.00757.x. [DOI] [PubMed] [Google Scholar]
- 206.Fehlbaum P, et al. Structure-activity analysis of thanatin, a 21-residue inducible insect defense peptide with sequence homology to frog skin antimicrobial peptides. Proc Natl Acad Sci USA. 1996;93(3):1221–1225. doi: 10.1073/pnas.93.3.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Mandard N, et al. Solution structure of thanatin, a potent bactericidal and fungicidal insect peptide, determined from proton two-dimensional nuclear magnetic resonance data. Eur J Biochem. 1998;256(2):404–410. doi: 10.1046/j.1432-1327.1998.2560404.x. [DOI] [PubMed] [Google Scholar]
- 208.Imamura T, et al. Acquired resistance to the rice blast in transgenic rice accumulating the antimicrobial peptide thanatin. Transgenic Res. 2010;19(3):415–424. doi: 10.1007/s11248-009-9320-x. [DOI] [PubMed] [Google Scholar]
- 209.Iijima R, Kurata S, Natori S. Purification, characterization, and cDNA cloning of an antifungal protein from the hemolymph of Sarcophaga peregrina (flesh fly) larvae. J Biol Chem. 1993;268(16):12055–12061. [PubMed] [Google Scholar]
- 210.Kim DH, et al. Bacterial expression of tenecin 3, an insect antifungal protein isolated from Tenebrio molitor, and its efficient purification. Mol Cells. 1998;8(6):786–789. [PubMed] [Google Scholar]
- 211.Lee SY, et al. Purification and cDNA cloning of an antifungal protein from the hemolymph of Holotrichia diomphalia larvae. Biol Pharm Bull. 1995;18(8):1049–1052. doi: 10.1248/bpb.18.1049. [DOI] [PubMed] [Google Scholar]
- 212.Kim DH, et al. Internalization of tenecin 3 by a fungal cellular process is essential for its fungicidal effect on Candida albicans . Eur J Biochem. 2001;268(16):4449–4458. doi: 10.1046/j.1432-1327.2001.02364.x. [DOI] [PubMed] [Google Scholar]
- 213.Ekengren S, Hultmark D. Drosophila cecropin as an antifungal agent. Insect Biochem Mol Biol. 1999;29(11):965–972. doi: 10.1016/S0965-1748(99)00071-5. [DOI] [PubMed] [Google Scholar]
- 214.DeLucca AJ, et al. Fungicidal activity of cecropin A. Antimicrob Agents Chemother. 1997;41(2):481–483. doi: 10.1128/aac.41.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.De Lucca AJ, et al. d-cecropin B: proteolytic resistance, lethality for pathogenic fungi and binding properties. Med Mycol. 2000;38(4):301–308. doi: 10.1080/mmy.38.4.301.308. [DOI] [PubMed] [Google Scholar]
- 216.Landon C, et al. Solution structures of stomoxyn and spinigerin, two insect antimicrobial peptides with an alpha-helical conformation. Biopolymers. 2006;81(2):92–103. doi: 10.1002/bip.20370. [DOI] [PubMed] [Google Scholar]
- 217.Zasloff M. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc Natl Acad Sci USA. 1987;84(15):5449–5453. doi: 10.1073/pnas.84.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Barbault F, et al. Solution structure of Alo-3: a new knottin-type antifungal peptide from the insect Acrocinus longimanus . Biochemistry. 2003;42(49):14434–14442. doi: 10.1021/bi035400o. [DOI] [PubMed] [Google Scholar]
- 219.Brown SE, et al. The discovery and analysis of a diverged family of novel antifungal moricin-like peptides in the wax moth Galleria mellonella . Insect Biochem Mol Biol. 2008;38(2):201–212. doi: 10.1016/j.ibmb.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 220.Hemmi H, et al. Solution structure of moricin, an antibacterial peptide, isolated from the silkworm Bombyx mori . FEBS Lett. 2002;518(1–3):33–38. doi: 10.1016/S0014-5793(02)02637-6. [DOI] [PubMed] [Google Scholar]
- 221.Destoumieux D, et al. Recombinant expression and range of activity of penaeidins, antimicrobial peptides from penaeid shrimp. Eur J Biochem. 1999;266(2):335–346. doi: 10.1046/j.1432-1327.1999.00855.x. [DOI] [PubMed] [Google Scholar]
- 222.Cuthbertson BJ, et al. Diversity of the penaeidin antimicrobial peptides in two shrimp species. Immunogenetics. 2002;54(6):442–445. doi: 10.1007/s00251-002-0487-z. [DOI] [PubMed] [Google Scholar]
- 223.Cuthbertson BJ, et al. Solution structure of synthetic penaeidin-4 with structural and functional comparisons with penaeidin-3. J Biol Chem. 2005;280(16):16009–16018. doi: 10.1074/jbc.M412420200. [DOI] [PubMed] [Google Scholar]
- 224.Yang Y, et al. Solution structure of the recombinant penaeidin-3, a shrimp antimicrobial peptide. J Biol Chem. 2003;278(38):36859–36867. doi: 10.1074/jbc.M305450200. [DOI] [PubMed] [Google Scholar]
- 225.Cuthbertson BJ, et al. A new class (penaeidin class 4) of antimicrobial peptides from the Atlantic white shrimp (Litopenaeus setiferus) exhibits target specificity and an independent proline-rich-domain function. Biochem J. 2004;381(Pt 1):79–86. doi: 10.1042/BJ20040330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Destoumieux D, et al. Penaeidins, antimicrobial peptides with chitin-binding activity, are produced and stored in shrimp granulocytes and released after microbial challenge. J Cell Sci. 2000;113(Pt 3):461–469. doi: 10.1242/jcs.113.3.461. [DOI] [PubMed] [Google Scholar]
- 227.Cuthbertson BJ, Bullesbach EE, Gross PS. Discovery of synthetic penaeidin activity against antibiotic-resistant fungi. Chem Biol Drug Des. 2006;68(2):120–127. doi: 10.1111/j.1747-0285.2006.00417.x. [DOI] [PubMed] [Google Scholar]
- 228.Tassanakajon A, et al. Cationic antimicrobial peptides in penaeid shrimp. Mar Biotechnol. 2011;13(4):639–657. doi: 10.1007/s10126-011-9381-8. [DOI] [PubMed] [Google Scholar]
- 229.Ohta M, et al. Mechanisms of antibacterial action of tachyplesins and polyphemusins, a group of antimicrobial peptides isolated from horseshoe crab hemocytes. Antimicrob Agents Chemother. 1992;36(7):1460–1465. doi: 10.1128/AAC.36.7.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Tincu JA, Taylor SW. Antimicrobial peptides from marine invertebrates. Antimicrob Agents Chemother. 2004;48(10):3645–3654. doi: 10.1128/AAC.48.10.3645-3654.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Powers JP, et al. The antimicrobial peptide polyphemusin localizes to the cytoplasm of Escherichia coli following treatment. Antimicrob Agents Chemother. 2006;50(4):1522–1524. doi: 10.1128/AAC.50.4.1522-1524.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Osaki T, et al. Horseshoe crab hemocyte-derived antimicrobial polypeptides, tachystatins, with sequence similarity to spider neurotoxins. J Biol Chem. 1999;274(37):26172–26178. doi: 10.1074/jbc.274.37.26172. [DOI] [PubMed] [Google Scholar]
- 233.Fujitani N, et al. Structure of the antimicrobial peptide tachystatin A. J Biol Chem. 2002;277(26):23651–23657. doi: 10.1074/jbc.M111120200. [DOI] [PubMed] [Google Scholar]
- 234.Saito T, et al. A novel big defensin identified in horseshoe crab hemocytes: isolation, amino acid sequence, and antibacterial activity. J Biochem (Tokyo) 1995;117(5):1131–1137. doi: 10.1093/oxfordjournals.jbchem.a124818. [DOI] [PubMed] [Google Scholar]
- 235.Destoumieux-Garzon D, et al. Crustacean immunity. Antifungal peptides are generated from the C terminus of shrimp hemocyanin in response to microbial challenge. J Biol Chem. 2001;276(50):47070–47077. doi: 10.1074/jbc.M103817200. [DOI] [PubMed] [Google Scholar]
- 236.Lopez-Abarrategui C, et al. Functional characterization of a synthetic hydrophilic antifungal peptide derived from the marine snail Cenchritis muricatus . Biochimie. 2012;94(4):968–974. doi: 10.1016/j.biochi.2011.12.016. [DOI] [PubMed] [Google Scholar]
- 237.Silva PI, Jr, Daffre S, Bulet P. Isolation and characterization of gomesin, an 18-residue cysteine-rich defense peptide from the spider Acanthoscurria gomesiana hemocytes with sequence similarities to horseshoe crab antimicrobial peptides of the tachyplesin family. J Biol Chem. 2000;275(43):33464–33470. doi: 10.1074/jbc.M001491200. [DOI] [PubMed] [Google Scholar]
- 238.Mandard N, et al. The solution structure of gomesin, an antimicrobial cysteine-rich peptide from the spider. Eur J Biochem. 2002;269(4):1190–1198. doi: 10.1046/j.0014-2956.2002.02760.x. [DOI] [PubMed] [Google Scholar]
- 239.Lorenzini DM, et al. Molecular cloning, expression analysis and cellular localization of gomesin, an anti-microbial peptide from hemocytes of the spider Acanthoscurria gomesiana . Insect Biochem Mol Biol. 2003;33(10):1011–1016. doi: 10.1016/S0965-1748(03)00115-2. [DOI] [PubMed] [Google Scholar]
- 240.Fazio MA, et al. Biological and structural characterization of new linear gomesin analogues with improved therapeutic indices. Biopolymers. 2006;88(3):386–400. doi: 10.1002/bip.20660. [DOI] [PubMed] [Google Scholar]
- 241.Domingues TM, Riske KA, Miranda A. Revealing the lytic mechanism of the antimicrobial peptide gomesin by observing giant unilamellar vesicles. Langmuir (ACS J Surf Colloids) 2010;26(13):11077–11084. doi: 10.1021/la100662a. [DOI] [PubMed] [Google Scholar]
- 242.Soletti RC, et al. Peptide gomesin triggers cell death through l-type channel calcium influx, MAPK/ERK, PKC and PI3K signaling and generation of reactive oxygen species. Chem Biol Interact. 2010;186(2):135–143. doi: 10.1016/j.cbi.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 243.Mygind PH, et al. Plectasin is a peptide antibiotic with therapeutic potential from a saprophytic fungus. Nature. 2005;437(7061):975–980. doi: 10.1038/nature04051. [DOI] [PubMed] [Google Scholar]
- 244.Campos-Olivas R, et al. NMR solution structure of the antifungal protein from Aspergillus giganteus: evidence for cysteine pairing isomerism. Biochemistry. 1995;34(9):3009–3021. doi: 10.1021/bi00009a032. [DOI] [PubMed] [Google Scholar]
- 245.Moreno AB, Del Pozo Martinez A, San Segundo B. Biotechnologically relevant enzymes and proteins. Antifungal mechanism of the Aspergillus giganteus AFP against the rice blast fungus Magnaporthe grisea . Appl Microbiol Biotechnol. 2006;72(5):883–895. doi: 10.1007/s00253-006-0362-1. [DOI] [PubMed] [Google Scholar]
- 246.Martinez Del Pozo A, et al. The antifungal protein AFP of Aspergillus giganteus is an oligonucleotide/oligosaccharide binding (OB) fold-containing protein that produces condensation of DNA. J Biol Chem. 2002;277(48):46179–46183. doi: 10.1074/jbc.M207472200. [DOI] [PubMed] [Google Scholar]
- 247.Kaiserer L, et al. Characterization of the Penicillium chrysogenum antifungal protein PAF. Arch Microbiol. 2003;180(3):204–210. doi: 10.1007/s00203-003-0578-8. [DOI] [PubMed] [Google Scholar]
- 248.Oberparleiter C, et al. Active internalization of the Penicillium chrysogenum antifungal protein PAF in sensitive Aspergilli. Antimicrob Agents Chemother. 2003;47(11):3598–3601. doi: 10.1128/AAC.47.11.3598-3601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Leiter E, et al. Antifungal protein PAF severely affects the integrity of the plasma membrane of Aspergillus nidulans and induces an apoptosis-like phenotype. Antimicrob Agents Chemother. 2005;49(6):2445–2453. doi: 10.1128/AAC.49.6.2445-2453.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Schmitt MJ, Breinig F. Yeast viral killer toxins: lethality and self-protection. Nat Rev Microbiol. 2006;4(3):212–221. doi: 10.1038/nrmicro1347. [DOI] [PubMed] [Google Scholar]
- 251.Breinig F, Tipper DJ, Schmitt MJ. Kre1p, the plasma membrane receptor for the yeast K1 viral toxin. Cell. 2002;108(3):395–405. doi: 10.1016/S0092-8674(02)00634-7. [DOI] [PubMed] [Google Scholar]
- 252.Eisfeld K, et al. Endocytotic uptake and retrograde transport of a virally encoded killer toxin in yeast. Mol Microbiol. 2000;37(4):926–940. doi: 10.1046/j.1365-2958.2000.02063.x. [DOI] [PubMed] [Google Scholar]
- 253.Kasahara S, et al. Involvement of cell wall beta-glucan in the action of HM-1 killer toxin. FEBS Lett. 1994;348(1):27–32. doi: 10.1016/0014-5793(94)00575-3. [DOI] [PubMed] [Google Scholar]
- 254.Guyard C, et al. Characterization of a Williopsis saturnus var. mrakii high molecular weight secreted killer toxin with broad-spectrum antimicrobial activity. Antimicrob Agents Chemother. 2002;49(6):961–971. doi: 10.1093/jac/dkf040. [DOI] [PubMed] [Google Scholar]
- 255.Takasuka T, et al. Cell wall synthesis specific cytocidal effect of Hansenula mrakii toxin-1 on Saccharomyces cerevisiae . Cell Mol Biol Res. 1995;41(6):575–581. [PubMed] [Google Scholar]
- 256.Gage MJ, et al. KP4 fungal toxin inhibits growth in Ustilago maydis by blocking calcium uptake. Mol Microbiol. 2001;41(4):775–785. doi: 10.1046/j.1365-2958.2001.02554.x. [DOI] [PubMed] [Google Scholar]
- 257.Allen A, et al. Transgenic maize plants expressing the Totivirus antifungal protein, KP4, are highly resistant to corn smut. Plant Biotechnol J. 2011;9(8):857–864. doi: 10.1111/j.1467-7652.2011.00590.x. [DOI] [PubMed] [Google Scholar]
- 258.Osusky M, et al. Transgenic plants expressing cationic peptide chimeras exhibit broad-spectrum resistance to phytopathogens. Nat Biotechnol. 2000;18(11):1162–1166. doi: 10.1038/81145. [DOI] [PubMed] [Google Scholar]
- 259.Jach G, et al. Enhanced quantitative resistance against fungal disease by combinatorial expression of different barley antifungal proteins in transgenic tobacco. Plant J (Cell Mol Biol) 1995;8(1):97–109. doi: 10.1046/j.1365-313X.1995.08010097.x. [DOI] [PubMed] [Google Scholar]
- 260.Muramoto N, et al. Transgenic sweet potato expressing thionin from barley gives resistance to black rot disease caused by Ceratocystis fimbriata in leaves and storage roots. Plant Cell Rep. 2012;31(6):987–997. doi: 10.1007/s00299-011-1217-5. [DOI] [PubMed] [Google Scholar]
- 261.Li Z, et al. Expression of a radish defensin in transgenic wheat confers increased resistance to Fusarium graminearum and Rhizoctonia cerealis . Funct Integr Genomics. 2011;11(1):63–70. doi: 10.1007/s10142-011-0211-x. [DOI] [PubMed] [Google Scholar]
- 262.Rossi DC, et al. Therapeutic use of a cationic antimicrobial peptide from the spider Acanthoscurria gomesiana in the control of experimental candidiasis. BMC Microbiol. 2012;12:28. doi: 10.1186/1471-2180-12-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Kondori N, et al. Fungicidal activity of human lactoferrin-derived peptides based on the antimicrobial alphabeta region. Int J Antimicrob Agents. 2011;37(1):51–57. doi: 10.1016/j.ijantimicag.2010.08.020. [DOI] [PubMed] [Google Scholar]