Abstract
Enhancers are positive DNA regulatory sequences controlling temporal and tissue-specific gene expression. These elements act independently of their orientation and distance relative to the promoters of target genes. Enhancers act through a variety of transcription factors that ensure their correct match with target promoters and consequent gene activation. There is a growing body of evidence on association of enhancers with transcription factors, co-activators, histone chromatin marks, and lncRNAs. Alterations in enhancers lead to misregulation of gene expression, causing a number of human diseases. In this review, we focus on the common characteristics of enhancers required for transcription stimulation.
Keywords: Chromatin loop, Gene regulation, Epigenetics, Transcription, Regulatory elements
Introduction
Cells establish individual patterns of gene expression during differentiation and development. The spatiotemporal control of transcription by RNA polymerase II (RNAPII) depends on enhancers, DNA regulatory elements that activate gene transcription [1, 2]. The first known enhancer was identified in 1981 [3, 4] in the SV40 virus genome, but subsequently such sequences have been found to be widespread among higher eukaryotes.
Enhancers are modular elements that lack stereotyped sequence composition and are located at a distance from transcription start sites of genes. The distance from an enhancer to its target promoter can vary from a few kb to 1 Mb. For example, this distance in human CD4+ T cells is ~50 kb [5]. Enhancers can be located in intra- and intergenic regions, introns, and even exons of genes [6, 7]. Enhancers usually activate transcription independently of their orientation and position relative to the target gene. Multiple enhancers can control the activity of a certain gene or group of genes [8, 9].
Enhancers are usually several hundred base pairs long and contain clusters of different 4- to 8-bp degenerate DNA sequences that are recognized by multiple transcription factors [10]. As a consequence of transcription factors binding to the enhancer DNA, these elements are characterized by low nucleosome occupancy and could be detected by their hypersensitivity to DNaseI [11–13]. Nucleosome destabilization at enhancers is facilitated by the presence of a highly dynamic H3.3/H2A.Z combination of histone variants [14–16] that were shown to form less stable contacts with DNA [17].
Binding of transcription factors to enhancers leads to a subsequent activation of transcription by recruitment of co-activators, releasing RNAPII pausing and stimulation of elongation. The activity of each enhancer is restricted to a definite spatiotemporal window by a specific set of DNA-binding transcription factors that control their specificity to promoters [10, 18–20]; by enhancer-blocking elements named insulators [21–24] and by chromosome separators termed TADs (Topologically Associated Domains) boundaries [25, 26].
In recent years, our knowledge of enhancers and their features has greatly expanded due to application of genome-wide technologies. Different features of enhancers, including transcription factors, co-factors, histone marks, were mapped throughout the genomes and used to predict novel enhancers [27, 28]. These data have changed our views on the prevalence of enhancers. For example, it has been predicted that the human genome contains approximately one million enhancers [29, 30]. Notwithstanding this progress, there are a lot of gaps and challenges in identification of features significant for enhancer activity.
In the present review, we summarize common features of enhancers that are important for stimulation of transcription: basic steps of enhancer–protein complex formation, factors essential for the interaction of enhancer with the promoter, enhancer-associated histone modifications, and implications of lncRNAs in enhancer activity. We discuss the recent studies that provide evidence for the relationship between mutations in enhancers and various human diseases.
Principles of enhancer-dependent transcription
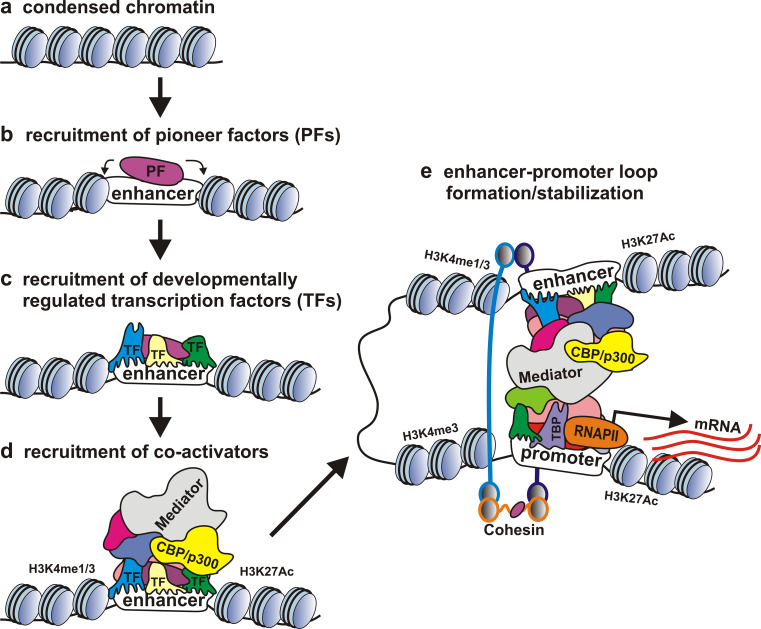
Enhancers act through protein factors that are assembled on their DNA. At the first step of enhancer complex formation, so-called pioneer factors are recruited (Fig. 1a, b) [31, 32]. These factors control the accessibility of enhancer DNA by displacing nucleosomes and opening chromatin locally at inactive enhancers, thereby facilitating the binding of developmentally regulated transcription factors [10, 32, 33]. The recruitment of pioneer factors is accompanied by DNA demethylation [34–36] and can be followed by active histone modification [37, 38]. However, the binding of pioneer factor is usually insufficient to allow enhancers to stimulate transcription [36, 37, 39, 40]. Then a set of developmentally regulated transcription factors, whose activity is restricted to certain spatiotemporal windows, bind to each enhancer sequence in a specific pattern, usually in cooperation with each other. Several transcription factors may compete for the binding to the same or partially overlapping binding sites.
Fig. 1.
Crucial steps of enhancer complex formation and long-range interaction of enhancer with promoter. Condensed chromatin at inactive enhancers opens up upon recruitment of pioneer factors (PFs) capable to displace nucleosomes. PFs facilitate the binding of developmentally regulated transcription factors (TFs) to the enhancer DNA, and they, in turn, recruit co-activators of transcription. The Mediator complex binds to transcription factors at enhancers and to general transcription factors, RNAPII at promoters, and links the enhancer and promoter together. The cohesin complex supports enhancer–promoter contact, forming a ring-like structure around DNA
The binding of developmentally regulated factors (Fig. 1c) is followed by the recruitment of co-activators (Fig. 1d) that lack the DNA-binding capacity, resulting in the formation of an active enhancer complex. These co-activators can mediate contacts between enhancers and general transcription factors at the promoters and/or function as chromatin remodelers (Fig. 1e).
Mediator is a large multiprotein co-activator complex conserved from yeast to humans. In mammals, Mediator consists of about 30 polypeptides (named MED1–MED31), CDK8, and cyclin C [41]. Mediator subunits associate with enhancers and active promoters; knockdown of Mediator subunits reduces the transcription of enhancer-controlled genes [42–45]. Mediator interacts with transcription factors, including general transcription factors bound to promoters, as well as with RNAPII and elongation factors [46, 47]. Mediator transfers the activating signal from enhancer to promoter, stimulating preinitiation complex assembly, activating paused RNAPII, and regulating transcription elongation [48, 49].
As shown recently, Mediator occupies in murine ESC large enhancer domains, or super-enhancers that have an average size of about 8.5 kb, and are enriched in the key transcription factors (master regulators) Oct4, Sox2, Nanog, Klf4, and Esrrb that control the pluripotent state of ESC cells [45]. A significant proportion of human super-enhancers and their target genes are tissue- and cell type-specific [50]. Several super-enhancers correspond to previously identified locus control regions (LCRs): a long cis-regulatory elements consisting of multiple functional enhancers [50].
Other common enhancer co-activators include two histone acetyltransferases: the CREB-binding protein (CBP) and the related E1A-interacting 300-kDa protein p300 [51, 52]. In mammals, these proteins are paralogs that have more than 90 % sequence identity in the HAT domain [53]; therefore, they are often referred to as p300/CBP. The majority of p300/CBP-bound genome regions overlap [54, 55] and tend to localize to active enhancers and promoters [56–59]. Drosophila has only the CBP homologue (named dCBP, or nejire), which also binds to active enhancers and promoters [60, 61]. Several studies suggest that the p300/CBP enrichment on DNA accurately predicts enhancers [56–59, 62, 63].
The p300/CBP protein has at least 400 interacting protein partners. The lysine 27 of histone 3 is one of its main targets in vivo [64, 65]. Genome-wide studies have shown that active enhancers associate with histone 3 acetylated at lysine 27 (H3K27ac) [62, 66, 67]. Similar to p300/CBP, H3K27ac marks both enhancers and active promoters [16]. In addition to histones, p300/CBP acetylates more than 70 non-histone proteins [53], including GATA3/4 [68, 69] and PU.1 [70] enhancer pioneer factors.
Recently, Krebs et al. [71] reported the association of an ATAC histone acetyltransferase complex with enhancers and promoters in two human cell lines. Furthermore, the ATAC complex was found to bind to a group of enhancers deprived of p300/CBP. This is evidence of a novel class of p300/CBP-independent enhancers that waits to be studied.
Current models suggest that the activity of histone acetyltransferases and ATP-dependent chromatin remodelers reduces the affinity of histones to enhancer DNAs and leads to chromatin decompaction, facilitating the binding of transcription factors. However, several studies indicate that acetylation of non-histone proteins may also be important for enhancer-dependent activation of transcription [68, 69].
Long-distance contacts between enhancers and promoters
A critical step required for enhancer-dependent transcription is the establishment of functional contacts between enhancers and target promoters. The dominant “looping” model suggests that active enhancers form direct physical contacts with promoters, while the intervening DNA is looped out (Fig. 1e). Data obtained by 3C and derivative technologies support this model [2, 19, 72, 73]. The first 3C study in mammals was performed on the β-globin LCR located 40–60 kb away from the globin genes [74]. Interactions between LCR and the target promoter were observed in fetal liver cells expressing β-globin but not in brain cells, in which β-globin gene is inactive, suggesting that enhancer–promoter interactions are important for promoter stimulation.
To date, the existence of enhancer–promoter loops has been confirmed for various enhancers [23, 72, 75–77]. Moreover, enhancer–promoter interactions have proved to differ between cell types and correlate with target gene transcription [78, 79]. There is some evidence that enhancer–promoter contacts are not constant but can be established prior to gene activation.
In particular, GR [80], FOXO3 [81] and TNFα [82] dependent enhancers interact with their target promoters prior to signaling. Likewise, long-range interactions involving Oct4 enhancer are established only in a subpopulation of cells prior to activation of Oct4 gene [83]. Specialized proteins might be responsible for the establishment of enhancer–promoter contacts prior to activation. Insulator proteins binding to the promoter regions are possible candidates for this role [84].
The physical contacts between the enhancer and promoter are sensitive to the loss of several DNA-binding transcription factors. For example, EKLF and GATA-1 are sufficient for the establishment of the enhancer–promoter contact at the β-globin locus [85, 86], and Oct4 factor is required for the enhancer–promoter contact at the Nanog locus [87].
The enhancer–promoter loops are further controlled by co-factors such as Mediator and cohesin complexes. The cohesin complex is composed of four core subunits: Smc1(A/B), Smc3, Scc1, and Scc3 (SA1/SA2) [88, 89]. The long coiled-coil polypeptides Smc1(A/B) and Smc3 interact with each other through the hinge domain and are additionally connected through the Scc1 subunit, forming a ring-like structure around DNA, and Scc3 (SA1/SA2) links to the central part of Scc1 [88, 89].
Cohesin acts synergistically with Mediator: these complexes could be co-purified [42, 90, 91]. Cohesin is enriched at Mediator-bound enhancers and promoters [42, 50, 92], and, as in the case of Mediator, knockdown of cohesin reduces the transcription of the enhancer-controlled genes [42]. Depletion of cohesin influences the RNAPII occupancy at predicted enhancers and promoters [93]. Depletion of Mediator [42, 43, 94] and cohesin subunits [42, 91, 94–96] results in a decreased frequency of interaction between enhancers and promoters, suggesting their direct contribution to enhancer–promoter communication.
Mediator proteins may mediate contacts between enhancer- and promoter-bound transcription factors, while cohesin supports chromatin looping by forming a ring-like structure around the interacting elements [42].
Histone marks and enhancers
Histones are subject to covalent modifications (such as acetylation, methylation, phosphorylation and ubiquitination) that occur mainly at their N-terminal tails and may correlate with the transcriptional status of genes. The existence of distinct histone modifications provided a basis for the ‘histone code’ hypothesis, according to which specific histone modification patterns affect binding of proteins to chromatin and determine the active and inactive regions of the genome [97]. For example, H3K27ac (see above) is associated with active gene transcription sites [62, 66, 67], and histone H3 mono-, di-, or trimethylated at lysine 4 (H3K4Me1, H3K4me2, and H3K4Me3, respectively) marks active chromatin [14, 16, 56, 98, 99].
The results of most studies suggest that both active enhancers and promoters are marked by nucleosomes containing H3K4me2 [14, 56, 100], while data concerning the distribution of H3K4Me1 and H3K4Me3 between active enhancers and promoters are contradictory. Some authors report that in human cells (e.g., HeLa, K562, and GM06990) H3K4Me1 is a hallmark of active enhancers, while H3K4Me3 is associated with active promoters [56, 62, 98, 99]. Indeed, the DNA sequences predicted as enhancers by enrichment of H3K4me1 and p300 and depletion of H3K4me3 gives over 75 % of positives in a functional test [62, 101].
Other researchers were not able to detect any significant difference between the presence of H3K4Me3 and H3K4Me1 at active enhancers and promoters in human CD4+ T cells [14, 16]. Moreover, Pekowska et al. [102] reported that H3K4me3 is enriched at active enhancers, while H3K4me1 is distributed independently of enhancer activity. The level of H3K4me3 at enhancers is lower than at promoters. Likewise, experiments with Drosophila embryos have shown that H3K4me1 modification takes place regardless of the functional activity of mesodermal enhancers [103].
The supposed enhancer-specific histone modifications may involve a limited recruitment of specific methyltransferases to enhancers. Therefore, additional information can be extracted from the distribution of methyltransferase proteins. For example, mammalian MLL3/MLL4 proteins—the main regulators of H3K4me1 [104]—are enriched at enhancer regions [104–106]. Similar results have been obtained in Drosophila: Trr (a homologue of mammalian MML3/MML4) and Trx, which are responsible for bulk H3K4me1 [105, 107], are associated with enhancers and co-localize with H3K4me1 and dCBP [107]. However, the distribution of the main H3K4 trimethylases Set1a and Set1b (dSet1) proteins [108–111] relative to enhancers has not yet been analyzed.
To date, the analysis of alternative histone modifications in mammals and Drosophila has failed to reveal their correlation with the majority of enhancers, suggesting that enhancers are heterogeneous [16, 28, 103]. It has also been found that over 20 % of human enhancers are associated with acetylation of histone H3 lysine 18 (H3K18Ac) chromatin hallmark [16]; in Drosophila, enhancers also tend to associate with H3K18Ac [112]; and trimethylated histone H3 lysine 79 (H3K79me3) marks about 15 % of intergenic enhancers [103].
Thus, currently available information about the relationship between histone modifications and enhancer activity is rather limited. The analysis of additional histone marks and an accurate comparison of different signatures in individual cell types are necessary to capture a complete picture of active enhancers.
Long non-coding RNAs and enhancers
Early evidence that transcription could be associated with enhancers came from studies of the human beta-globin locus where a non-coding RNA (ncRNA) is transcribed from the HS2 enhancer within the LCR [113] only in cells where enhancer is active [114, 115]. This ncRNA is transcribed mainly in one direction and from multiple sites of the enhancer [116]. The generated ncRNA were polyadenylated and spliced; however it did not appear to contain the normal cap-structure at 5′-ends [117]. HS2 was found to be associated with RNAPII [118] that seems to be bound to enhancer independently of the promoter-bound RNAPII [118, 119].
Further systematic analysis indicated that a major portion of the genome is being transcribed and that the bulk of genome transcripts account for long (>200 bp) non-protein-coding RNAs (lncRNAs) [120–123], which can have a positive or negative effect on gene transcription [124].
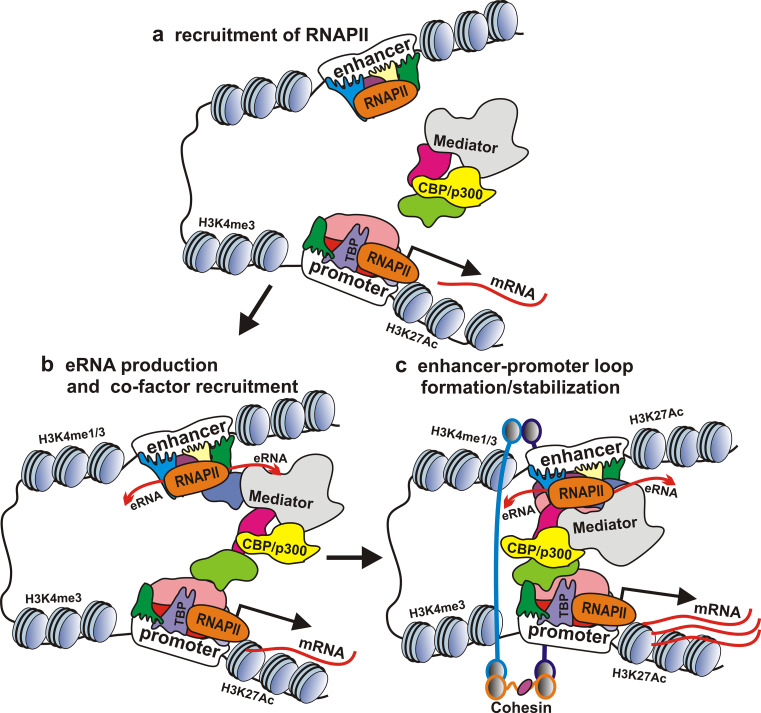
LncRNAs originating from enhancers were named enhancer RNAs (eRNAs) (Fig. 2). Based on the genome-wide studies, they are ~0.5- to 5-kb ncRNAs derived from DNA regions that share enhancer-associated features [57, 120, 125–128]. The eRNAs could be transcribed uni- or bidirectionally; they may contain or lack poly(A) tails [57, 120, 125–127, 129, 130].
Fig. 2.
The hypothesized positive role of eRNAs in the activity of transcribed enhancers. Enhancer activation by developmentally regulated transcription factors leads to RNAPII recruitment and to production of eRNAs that help to recruit co-activators and facilitate enhancer–promoter loop formation
For instance, Kim and colleagues [57] have found in mouse neuronal cells ~12,000 of CBP-bound regions enriched in H3K4Me1 and located distally from known TSSs of protein-coding genes. Among these regions, 25 % recruited RNAPII, and 16.7 % were transcribed, resulting in the production of RNAs with a length of <2 kb. Most of transcripts identified in this study were transcribed bidirectionally and were non-poly(A)+ [57].
Several studies have revealed eRNAs derived from extragenic RNAPII bound sites [125, 127] and from activator binding sites: estrogen receptor [126, 130], p53 [131], and MYOD1 [132].
Orom et al. [128] performed a search of enhancer-associated transcripts based on the functional test. They selected ≈0.1- to 9-kb lncRNAs from intergenic regions whose knockdown by siRNAs resulted in down-regulation of nearby protein-coding genes. The selected transcripts were predominantly spliced and poly(A)+. Unlike in the majority of eRNAs, H3K4me3 was present at their 5′-ends, and H3K36me3 marked their bodies [128]. Based on this difference, the authors classified them into a distinct group of lncRNAs, named ncRNA-a (ncRNA-activating). However, it is difficult to discriminate some of them from unidirectionally transcribed and poly(A)+ eRNAs. Indeed, due to nonexclusive conditions of search for eRNAs and ncRNA-a, transcripts of both groups may overlap.
A number of studies indicate a positive role of eRNAs in the enhancer function. For example, transcription of eRNAs positively correlates with the expression of nearby genes [57, 125, 133, 134], and targeted degradation of a major part of eRNAs leads to reduction of the expression of nearby protein-coding genes, as in the case of ncRNA-a [131–133, 135, 136]. Furthermore, enhancer activation upon stimulation correlates with eRNA production [136, 137]. The expression of eRNAs enhances transcription of the reporter gene in an RNA tethering assay [131, 136]. However, the selected eRNAs were ineffective in trans-stimulation experiments [136].
The role of eRNAs in enhancer–promoter loop formation is unclear. The eRNAs level was found to be higher at enhancers that interact with promoters [78, 136, 138]. However, looping between enhancers and target genes remained intact after inhibition of eRNA transcription [126]. The establishment of enhancer–promoter contact prior to eRNAs synthesis was further supported by the fact that eRNAs were lacking at the enhancer of arc gene in case of promoter deletion [57]. However, ncRNA-a depletion by siRNA reduced chromosomal looping between the ncRNA-a expressing region and the target gene loci. Furthermore, the tested ncRNA-as were found to associate with Mediator implicated in the loop formation [43], and several authors reported association of lncRNAs with enhancer-bound activators [139–141].
Recent studies suggest that transcription of long non-coding RNAs through enhancer-containing regulatory regions correlates with decrease in target gene transcription. For example, Gummalla et al. [142] reported that the 90-kb iab-8 ncRNA is transcribed through the regulatory region of the abd-A gene and participate in its repression in Drosophila. The authors propose that this repression is established in two ways: (1) the iab-8 precursor produces a micro-RNA, which targets the abdominal-A mRNA, and (2) iab-8 transcription directly interferes with the expression of abdominal-A, which lies just downstream of the iab-8 ncRNA poly(A) site [142]. In a previous study, Petruk et al. [143] found that lncRNA transcribed through an enhancer-containing regulatory region interferes with the Ubx gene promoter. However, likewise interfering with gene promoter these transcripts can directly affect enhancers located in transcribed regulatory regions.
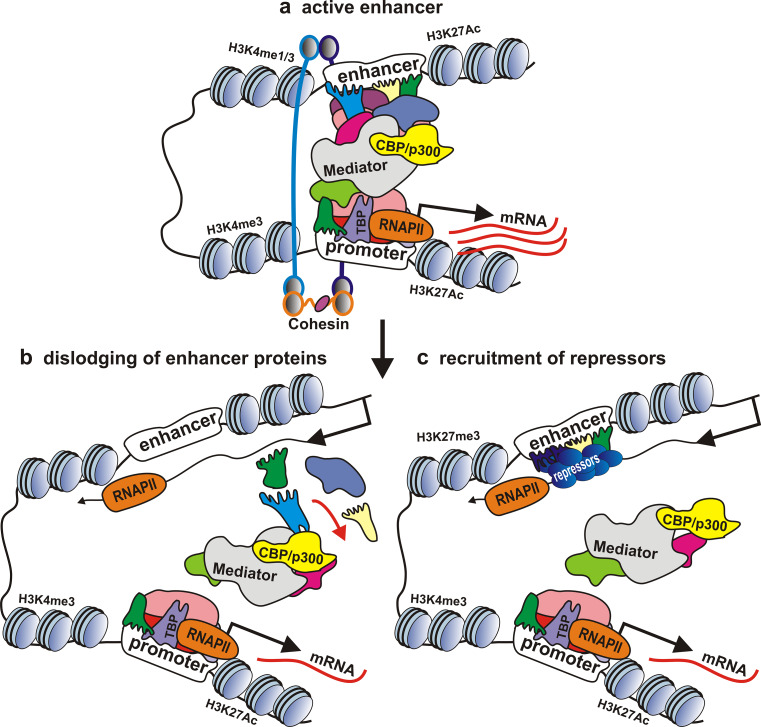
Using an assay in transgenic lines, we found that transcription leads to suppression of enhancers from the regulatory regions of yellow and white genes [144]. Transcription through the enhancer of the white gene resulted in dislodging of Zeste, a protein important for enhancer–promoter communication, suggesting a role for the mechanism of ‘transcriptional interference’ (Fig. 3b). This mechanism probably acts on intergenic enhancers and controls their maximum activity by a negative feedback loop, with excessive activation of transcription inhibiting the enhancer activity. Similar positive enhancer-associated transcription from one enhancer can negatively affect the activity of nearby enhancers that should be inactive in a given tissue or a group of cells.
Fig. 3.
The potential mechanisms of enhancer inactivation by lncRNAs. Transcription through the enhancer from upstream promoter leads to dislodging of the enhancer-bound protein complex from DNA or/and to the recruitment of repressor proteins onto DNA, resulting in enhancer inactivation
Enhancer inactivation by lncRNAs can possibly involve the recruitment of Polycomb group repressors (Fig. 3c) [145]. For example, over 20 % of lincRNAs expressed in various cell types are bound by the PRC2 Polycomb group complex [146].
More information comes from studies on enhancers in human embryonic stem cells (hESCs) [66, 147]. Active enhancers in hESCs cells show canonical enrichment in H3K4me1 and H3K27ac histone modifications and associate with the p300 protein [66, 147]. The authors identified a class of “poised enhancers” that have features of both active and inactive chromatin but are linked to inactive genes. Being also enriched in H3K4me1 and p300, they are distinguished by the absence of H3K27ac and enrichment in H3K27me3 [66, 147] and are bound by Polycomb group repressors [148]. Analysis of PcG and other repressor proteins for association with inactive enhancers and linkage to ongoing transcription at enhancers may provide new insights into enhancer function.
Diseases and enhancers
Great efforts are made to understand the genetic basis of human diseases. In addition to mutations in the coding part of genes, disruption of gene regulatory regions is a major type of disease-associated changes in DNA. Below, we briefly consider only a few examples of enhancer-related diseases.
The first evidence in humans comes from studies of the β-globin locus linked to β-thalassemia, a transfusion-dependent anemia. Several types of thalassemia are characterized by hematological symptoms observed in the absence of β-globin protein, although the β-globin gene in the patients is intact [149–151]. These thalassemias are associated with deletions of DNA regulatory regions. For example, patients with Dutch (γδβ°) thalassemia have a 100-kb deletion that removes the LCR and almost all sequences upstream of the β-globin gene [151]. A deletion of ~30 kb found in the Hispanic δβ-thalassemia likewise removes the LCR and affects β-globin gene expression [150].
Another example concerns limb abnormalities in humans, mice, cats, and chickens with single-point mutations in ZRS, a highly conserved ≈800-bp limb-specific enhancer located 1 Mb from the target sonic hedgehog (SHH) gene. These mutations affect long-range SHH signaling, which plays a central role in patterning numerous embryonic tissues [152].
Hirschsprung (HSCR) disease is a complex genetic disorder attributed to a failure of the enteric neural crest cells to form ganglia in the hindgut. The risk for HSCR is associated with single-nucleotide polymorphism (SNP) in the RET enhancer [153, 154].
Van Buchem (VB) disease is an autosomal skeletal dysplasia characterized by bone overgrowth. This disease is associated with a 52-kb deletion in the regulatory region 35 kb downstream of the SOST gene [155, 156]. The affected region normally contains the ECR5 enhancer [157], deletion of which results in the phenotypes observed in VB disease [158].
Facioscapulohumeral muscular dystrophy (FSHD) is a dominant neuromuscular disease with a prevalence of 1 in 20,000, which leads to weakness and atrophy of specific groups of muscles in the face, shoulder girdle, and lower extremities [159]. FSHD is associated with the subtelomeric region 4q35 containing an array of 3.3-kb-long macrosatellite repeats (D4Z4) [160]. The length of this array varies from 35 to 300 kb in healthy subjects but is consistently lower than 35 kb in FSHD patients [161]. Each D4Z4 repeat contains a potent transcriptional enhancer [162–164], an open reading frame for the double homeobox gene DUX4 [165, 166], and a number of regulatory elements (for review, see [167]). The maintenance of pathological FSHD phenotype is due to the expression of D4Z4-proximal genes that include DUX4, DUX4c, FRG1, FRG2, and ANT1. All these genes are upregulated in FSHD (for review, see [167, 168]). The D4Z4 enhancer interacts with the Krüppel-like transcription factor 15 (KLF15) in FSHD patients, thereby activating the DUX4c and FRG2 genes [169]. Recent studies indicate that lncRNA and miRNA are also implicated in transcriptional regulation in FSHD [170, 171].
Aniridia is a panocular malformation associated with haploinsufficiency of PAX6 transcription factor. As shown by Bhatia et al. [172], aniridia can be caused by point mutation in the conserved SIMO enhancer located 150 kb from PAX6 gene. Another disease causing blindness, nonsyndromic congenital retinal nonattachment (NCRNA), is linked to a deletion of an enhancer 20 kb upstream from the ATOH7 transcription factor gene that is required for retinal ganglion cell and optic nerve development [173].
Alterations in enhancer-containing regulatory regions are also responsible for other development disorders such as Leri–Weill dyschondrosteosis syndrome [174], Axenfeld–Rieger syndrome (ARS) [175], coronary artery disease [176], prostate cancer [177], and MonoMAC syndrome [178].
Many lymphomas, including Bukitt lymphoma (BL), mantle cell lymphoma and follicular lymphoma are caused by translocations that position a strong immunoglobulin heavy chain µ enhancer in a relative proximity (100–1000 kbp) to proto-oncogenes c-myc, Cyclin D1 (CCNDD1) or bcl2, respectively [179]. The µ enhancer is thought to directly activate the proto-oncogenes [180], although this notion has been challenged in a recent study, where the authors show that activation of CCND1 and CMYC is accomplished by proximal nucleolin-dependent enhancers, following the relocalization of the translocated regions to the proximity of the nucleolus [181]. BL is linked to two human viruses, Epstein–Barr’s virus and human immunodeficiency virus [182], while the role of viral enhancers in BL is not known yet; the mutations in the murine Moloney murine leukemia virus enhancer were shown to affect cancer development in mice [183].
Globally, genome-wide association studies (GWAS) localize the majority of disease-associated SNPs to noncoding sequences [29, 184, 185], particularly to enhancers [185–188]. The enhancer-associated SNPs are linked to cancer, diabetes, rheumatoid arthritis, systemic lupus erythematosus, multiple sclerosis, Crohn’s disease, celiac disease, Alzheimer’s disease, etc. [50, 186, 189, 190]. The above examples are only a small part of long list of enhancer-associated diseases. The relationships between changes in lncRNAs transcription and diseases are also currently under a careful study [191].
Individual genome-wide sequencing and analysis of enhancer-associated SNPs will certainly become an integral part of tests for timely detection of malformations and other abnormalities.
Conclusions and outlook
Genome-wide studies indicate that enhancers are enriched in Mediator and cohesin complexes. A significant proportion of enhancers associate with p300/CBP, ATAC, eRNAs, RNAPII, and active histone marks H3K4me, H3K27Ac, H3K18Ac, and H3K79me3.
However, enhancers show an extreme variability in their DNA-binding transcription factors and most of the known enhancer features are not necessarily required for enhancer activity. Such variability appears to be significant for the plasticity and accuracy of gene expression control. At the same time, we suggest that there should be some general principles and elegant mechanisms of enhancer-dependent gene activation. The mechanisms may also vary depending on the chromosomal context and nuclear compartmentalization.
Many more questions need to be answered. How the connection of cofactors with different combinations of activators is achieved? How the collaboration between diverse cofactors is established? What positive signals are translated from activators to promoter-bound factors: protein modifications, changes of protein conformation, chromatin structure, etc.? What defines lncRNA to be positive/negative? What restricts enhancer activity to certain cells? The answers to these questions will certainly provide a deeper insight into the principles of enhancer action and genetic control.
Acknowledgments
We are grateful to N.A. Gorgolyuk for his help in preparing the manuscript. This study was supported by RFBR 15-04-04208-a to D.C., RFBR 15-04-03973-a to M.E., RFBR 13-04-93106-CNRS_a to P.G., and the MEGAFSHD grant from the Association Française contre les Myopathies (AFM) to Y.V.
References
- 1.Buecker C, Wysocka J. Enhancers as information integration hubs in development: lessons from genomics. Trends Genet. 2012;28(6):276–284. doi: 10.1016/j.tig.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144(3):327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- 4.Benoist C, Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981;290(5804):304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- 5.Chepelev I, Wei G, Wangsa D, Tang Q, Zhao K. Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res. 2012;22(3):490–503. doi: 10.1038/cr.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birnbaum RY, Clowney EJ, Agamy O, Kim MJ, Zhao J, Yamanaka T, Pappalardo Z, Clarke SL, Wenger AM, Nguyen L, Gurrieri F, Everman DB, Schwartz CE, Birk OS, Bejerano G, Lomvardas S, Ahituv N. Coding exons function as tissue-specific enhancers of nearby genes. Genome Res. 2012;22(6):1059–1068. doi: 10.1101/gr.133546.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ritter DI, Dong Z, Guo S, Chuang JH. Transcriptional enhancers in protein-coding exons of vertebrate developmental genes. PLoS ONE. 2012;7(5):e35202. doi: 10.1371/journal.pone.0035202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim A, Dean A. Chromatin loop formation in the beta-globin locus and its role in globin gene transcription. Mol Cells. 2012;34(1):1–5. doi: 10.1007/s10059-012-0048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith E, Shilatifard A. Enhancer biology and enhanceropathies. Nat Struct Mol Biol. 2014;21(3):210–219. doi: 10.1038/nsmb.2784. [DOI] [PubMed] [Google Scholar]
- 10.Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13(9):613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 11.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, Garg K, John S, Sandstrom R, Bates D, Boatman L, Canfield TK, Diegel M, Dunn D, Ebersol AK, Frum T, Giste E, Johnson AK, Johnson EM, Kutyavin T, Lajoie B, Lee BK, Lee K, London D, Lotakis D, Neph S, Neri F, Nguyen ED, Qu H, Reynolds AP, Roach V, Safi A, Sanchez ME, Sanyal A, Shafer A, Simon JM, Song L, Vong S, Weaver M, Yan Y, Zhang Z, Zhang Z, Lenhard B, Tewari M, Dorschner MO, Hansen RS, Navas PA, Stamatoyannopoulos G, Iyer VR, Lieb JD, Sunyaev SR, Akey JM, Sabo PJ, Kaul R, Furey TS, Dekker J, Crawford GE, Stamatoyannopoulos JA. The accessible chromatin landscape of the human genome. Nature. 2012;489(7414):75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu C, Wong YC, Elgin SC. The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell. 1979;16(4):807–814. doi: 10.1016/0092-8674(79)90096-5. [DOI] [PubMed] [Google Scholar]
- 13.Xi H, Shulha HP, Lin JM, Vales TR, Fu Y, Bodine DM, McKay RD, Chenoweth JG, Tesar PJ, Furey TS, Ren B, Weng Z, Crawford GE. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 2007;3(8):e136. doi: 10.1371/journal.pgen.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet. 2009;41(8):941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40(7):897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21(12):1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butler JE, Kadonaga JT. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 2001;15(19):2515–2519. doi: 10.1101/gad.924301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Arensbergen J, van Steensel B, Bussemaker HJ. In search of the determinants of enhancer-promoter interaction specificity. Trends Cell Biol. 2014 doi: 10.1016/j.tcb.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zabidi MA, Arnold CD, Schernhuber K, Pagani M, Rath M, Frank O, Stark A. Enhancer–core-promoter specificity separates developmental and housekeeping gene regulation. Nature. 2014 doi: 10.1038/nature13994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chetverina D, Aoki T, Erokhin M, Georgiev P, Schedl P. Making connections: insulators organize eukaryotic chromosomes into independent cis-regulatory networks. BioEssays. 2014;36(2):163–172. doi: 10.1002/bies.201300125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Core LJ, Lis JT. Paused Pol II captures enhancer activity and acts as a potent insulator. Genes Dev. 2009;23(14):1606–1612. doi: 10.1101/gad.1827709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorkin DU, Leung D, Ren B. The 3D genome in transcriptional regulation and pluripotency. Cell Stem Cell. 2014;14(6):762–775. doi: 10.1016/j.stem.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kyrchanova O, Georgiev P. Chromatin insulators and long-distance interactions in Drosophila. FEBS Lett. 2014;588(1):8–14. doi: 10.1016/j.febslet.2013.10.039. [DOI] [PubMed] [Google Scholar]
- 25.Ciabrelli F, Cavalli G. Chromatin-driven behavior of topologically associating domains. J Mol Biol. 2014 doi: 10.1016/j.jmb.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 26.Schwarzer W, Spitz F. The architecture of gene expression: integrating dispersed cis-regulatory modules into coherent regulatory domains. Curr Opin Genet Dev. 2014;27:74–82. doi: 10.1016/j.gde.2014.03.014. [DOI] [PubMed] [Google Scholar]
- 27.Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 2014;15(4):272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 28.Wang C, Zhang MQ, Zhang Z. Computational identification of active enhancers in model organisms. Genomics Proteomics Bioinf. 2013;11(3):142–150. doi: 10.1016/j.gpb.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Consortium EP An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heintzman ND, Ren B. Finding distal regulatory elements in the human genome. Curr Opin Genet Dev. 2009;19(6):541–549. doi: 10.1016/j.gde.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smale ST, Tarakhovsky A, Natoli G. Chromatin contributions to the regulation of innate immunity. Annu Rev Immunol. 2014;32:489–511. doi: 10.1146/annurev-immunol-031210-101303. [DOI] [PubMed] [Google Scholar]
- 32.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25(21):2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell. 2013;49(5):825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Serandour AA, Avner S, Percevault F, Demay F, Bizot M, Lucchetti-Miganeh C, Barloy-Hubler F, Brown M, Lupien M, Metivier R, Salbert G, Eeckhoute J. Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res. 2011;21(4):555–565. doi: 10.1101/gr.111534.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu J, Pope SD, Jazirehi AR, Attema JL, Papathanasiou P, Watts JA, Zaret KS, Weissman IL, Smale ST. Pioneer factor interactions and unmethylated CpG dinucleotides mark silent tissue-specific enhancers in embryonic stem cells. Proc Natl Acad Sci USA. 2007;104(30):12377–12382. doi: 10.1073/pnas.0704579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu J, Watts JA, Pope SD, Gadue P, Kamps M, Plath K, Zaret KS, Smale ST. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 2009;23(24):2824–2838. doi: 10.1101/gad.1861209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, Ragoussis J, Natoli G. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32(3):317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 38.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoogenkamp M, Lichtinger M, Krysinska H, Lancrin C, Clarke D, Williamson A, Mazzarella L, Ingram R, Jorgensen H, Fisher A, Tenen DG, Kouskoff V, Lacaud G, Bonifer C. Early chromatin unfolding by RUNX1: a molecular explanation for differential requirements during specification versus maintenance of the hematopoietic gene expression program. Blood. 2009;114(2):299–309. doi: 10.1182/blood-2008-11-191890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liber D, Domaschenz R, Holmqvist PH, Mazzarella L, Georgiou A, Leleu M, Fisher AG, Labosky PA, Dillon N. Epigenetic priming of a pre-B cell-specific enhancer through binding of Sox2 and Foxd3 at the ESC stage. Cell Stem Cell. 2010;7(1):114–126. doi: 10.1016/j.stem.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 41.Yin JW, Wang G. The Mediator complex: a master coordinator of transcription and cell lineage development. Development. 2014;141(5):977–987. doi: 10.1242/dev.098392. [DOI] [PubMed] [Google Scholar]
- 42.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467(7314):430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494(7438):497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park SW, Li G, Lin YP, Barrero MJ, Ge K, Roeder RG, Wei LN. Thyroid hormone-induced juxtaposition of regulatory elements/factors and chromatin remodeling of Crabp1 dependent on MED1/TRAP220. Mol Cell. 2005;19(5):643–653. doi: 10.1016/j.molcel.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 45.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153(2):307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ansari SA, Morse RH. Mechanisms of Mediator complex action in transcriptional activation. Cell Mol Life Sci. 2013;70(15):2743–2756. doi: 10.1007/s00018-013-1265-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11(11):761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poss ZC, Ebmeier CC, Taatjes DJ. The Mediator complex and transcription regulation. Crit Rev Biochem Mol Biol. 2013;48(6):575–608. doi: 10.3109/10409238.2013.840259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szutorisz H, Dillon N, Tora L. The role of enhancers as centres for general transcription factor recruitment. Trends Biochem Sci. 2005;30(11):593–599. doi: 10.1016/j.tibs.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 50.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155(4):934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bedford DC, Kasper LH, Fukuyama T, Brindle PK. Target gene context influences the transcriptional requirement for the KAT3 family of CBP and p300 histone acetyltransferases. Epigenetics. 2010;5(1):9–15. doi: 10.4161/epi.5.1.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holmqvist PH, Mannervik M. Genomic occupancy of the transcriptional co-activators p300 and CBP. Transcription. 2013;4(1):18–23. doi: 10.4161/trns.22601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang L, Tang Y, Cole PA, Marmorstein R. Structure and chemistry of the p300/CBP and Rtt109 histone acetyltransferases: implications for histone acetyltransferase evolution and function. Curr Opin Struct Biol. 2008;18(6):741–747. doi: 10.1016/j.sbi.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramos YF, Hestand MS, Verlaan M, Krabbendam E, Ariyurek Y, van Galen M, van Dam H, van Ommen GJ, den Dunnen JT, Zantema A, t Hoen PA. Genome-wide assessment of differential roles for p300 and CBP in transcription regulation. Nucleic Acids Res. 2010;38(16):5396–5408. doi: 10.1093/nar/gkq184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138(5):1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 57.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465(7295):182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.May D, Blow MJ, Kaplan T, McCulley DJ, Jensen BC, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Afzal V, Simpson PC, Rubin EM, Black BL, Bristow J, Pennacchio LA, Visel A. Large-scale discovery of enhancers from human heart tissue. Nat Genet. 2012;44(1):89–93. doi: 10.1038/ng.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457(7231):854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holmqvist PH, Boija A, Philip P, Crona F, Stenberg P, Mannervik M. Preferential genome targeting of the CBP co-activator by Rel and Smad proteins in early Drosophila melanogaster embryos. PLoS Genet. 2012;8(6):e1002769. doi: 10.1371/journal.pgen.1002769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Negre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, Li Z, Ishii H, Spokony RF, Chen J, Hwang L, Cheng C, Auburn RP, Davis MB, Domanus M, Shah PK, Morrison CA, Zieba J, Suchy S, Senderowicz L, Victorsen A, Bild NA, Grundstad AJ, Hanley D, MacAlpine DM, Mannervik M, Venken K, Bellen H, White R, Gerstein M, Russell S, Grossman RL, Ren B, Posakony JW, Kellis M, White KP. A cis-regulatory map of the Drosophila genome. Nature. 2011;471(7339):527–531. doi: 10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459(7243):108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCord RP, Zhou VW, Yuh T, Bulyk ML. Distant cis-regulatory elements in human skeletal muscle differentiation. Genomics. 2011;98(6):401–411. doi: 10.1016/j.ygeno.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SY, Ge K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30(2):249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, Harte PJ. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136(18):3131–3141. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107(50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nord AS, Blow MJ, Attanasio C, Akiyama JA, Holt A, Hosseini R, Phouanenavong S, Plajzer-Frick I, Shoukry M, Afzal V, Rubenstein JL, Rubin EM, Pennacchio LA, Visel A. Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell. 2013;155(7):1521–1531. doi: 10.1016/j.cell.2013.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kawamura T, Ono K, Morimoto T, Wada H, Hirai M, Hidaka K, Morisaki T, Heike T, Nakahata T, Kita T, Hasegawa K. Acetylation of GATA-4 is involved in the differentiation of embryonic stem cells into cardiac myocytes. J Biol Chem. 2005;280(20):19682–19688. doi: 10.1074/jbc.M412428200. [DOI] [PubMed] [Google Scholar]
- 69.Yamagata T, Mitani K, Oda H, Suzuki T, Honda H, Asai T, Maki K, Nakamoto T, Hirai H. Acetylation of GATA-3 affects T-cell survival and homing to secondary lymphoid organs. EMBO J. 2000;19(17):4676–4687. doi: 10.1093/emboj/19.17.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bai Y, Srinivasan L, Perkins L, Atchison ML. Protein acetylation regulates both PU.1 transactivation and Ig kappa 3′ enhancer activity. J Immunol. 2005;175(8):5160–5169. doi: 10.4049/jimmunol.175.8.5160. [DOI] [PubMed] [Google Scholar]
- 71.Krebs AR, Karmodiya K, Lindahl-Allen M, Struhl K, Tora L. SAGA and ATAC histone acetyl transferase complexes regulate distinct sets of genes and ATAC defines a class of p300-independent enhancers. Mol Cell. 2011;44(3):410–423. doi: 10.1016/j.molcel.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gibcus JH, Dekker J. The hierarchy of the 3D genome. Mol Cell. 2013;49(5):773–782. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smallwood A, Ren B. Genome organization and long-range regulation of gene expression by enhancers. Curr Opin Cell Biol. 2013;25(3):387–394. doi: 10.1016/j.ceb.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10(6):1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 75.de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 2012;26(1):11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ghavi-Helm Y, Klein FA, Pakozdi T, Ciglar L, Noordermeer D, Huber W, Furlong EE. Enhancer loops appear stable during development and are associated with paused polymerase. Nature. 2014;512(7512):96–100. doi: 10.1038/nature13417. [DOI] [PubMed] [Google Scholar]
- 77.Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, Aiden EL. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159(7):1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489(7414):109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kieffer-Kwon KR, Tang Z, Mathe E, Qian J, Sung MH, Li G, Resch W, Baek S, Pruett N, Grontved L, Vian L, Nelson S, Zare H, Hakim O, Reyon D, Yamane A, Nakahashi H, Kovalchuk AL, Zou J, Joung JK, Sartorelli V, Wei CL, Ruan X, Hager GL, Ruan Y, Casellas R. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell. 2013;155(7):1507–1520. doi: 10.1016/j.cell.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hakim O, Sung MH, Voss TC, Splinter E, John S, Sabo PJ, Thurman RE, Stamatoyannopoulos JA, de Laat W, Hager GL. Diverse gene reprogramming events occur in the same spatial clusters of distal regulatory elements. Genome Res. 2011;21(5):697–706. doi: 10.1101/gr.111153.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Eijkelenboom A, Mokry M, de Wit E, Smits LM, Polderman PE, van Triest MH, van Boxtel R, Schulze A, de Laat W, Cuppen E, Burgering BM. Genome-wide analysis of FOXO3 mediated transcription regulation through RNA polymerase II profiling. Mol Syst Biol. 2013;9:638. doi: 10.1038/msb.2012.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen CA, Schmitt AD, Espinoza CA, Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503(7475):290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wei Z, Gao F, Kim S, Yang H, Lyu J, An W, Wang K, Lu W. Klf4 organizes long-range chromosomal interactions with the oct4 locus in reprogramming and pluripotency. Cell Stem Cell. 2013;13(1):36–47. doi: 10.1016/j.stem.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 84.Negre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, Henikoff JG, Feng X, Ahmad K, Russell S, White RA, Stein L, Henikoff S, Kellis M, White KP. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010;6(1):e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev. 2004;18(20):2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol Cell. 2005;17(3):453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 87.Levasseur DN, Wang J, Dorschner MO, Stamatoyannopoulos JA, Orkin SH. Oct4 dependence of chromatin structure within the extended Nanog locus in ES cells. Genes Dev. 2008;22(5):575–580. doi: 10.1101/gad.1606308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dorsett D. Cohesin: genomic insights into controlling gene transcription and development. Curr Opin Genet Dev. 2011;21(2):199–206. doi: 10.1016/j.gde.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Remeseiro S, Cuadrado A, Losada A. Cohesin in development and disease. Development. 2013;140(18):3715–3718. doi: 10.1242/dev.090605. [DOI] [PubMed] [Google Scholar]
- 90.Ebmeier CC, Taatjes DJ. Activator-Mediator binding regulates Mediator-cofactor interactions. Proc Natl Acad Sci USA. 2010;107(25):11283–11288. doi: 10.1073/pnas.0914215107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang H, Jiao W, Sun L, Fan J, Chen M, Wang H, Xu X, Shen A, Li T, Niu B, Ge S, Li W, Cui J, Wang G, Sun J, Fan X, Hu X, Mrsny RJ, Hoffman AR, Hu JF. Intrachromosomal looping is required for activation of endogenous pluripotency genes during reprogramming. Cell Stem Cell. 2013;13(1):30–35. doi: 10.1016/j.stem.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 92.Yan J, Enge M, Whitington T, Dave K, Liu J, Sur I, Schmierer B, Jolma A, Kivioja T, Taipale M, Taipale J. Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell. 2013;154(4):801–813. doi: 10.1016/j.cell.2013.07.034. [DOI] [PubMed] [Google Scholar]
- 93.Schaaf CA, Kwak H, Koenig A, Misulovin Z, Gohara DW, Watson A, Zhou Y, Lis JT, Dorsett D. Genome-wide control of RNA polymerase II activity by cohesin. PLoS Genet. 2013;9(3):e1003382. doi: 10.1371/journal.pgen.1003382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Apostolou E, Ferrari F, Walsh RM, Bar-Nur O, Stadtfeld M, Cheloufi S, Stuart HT, Polo JM, Ohsumi TK, Borowsky ML, Kharchenko PV, Park PJ, Hochedlinger K. Genome-wide chromatin interactions of the Nanog locus in pluripotency, differentiation, and reprogramming. Cell Stem Cell. 2013;12(6):699–712. doi: 10.1016/j.stem.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chien R, Zeng W, Kawauchi S, Bender MA, Santos R, Gregson HC, Schmiesing JA, Newkirk DA, Kong X, Ball AR, Jr, Calof AL, Lander AD, Groudine MT, Yokomori K. Cohesin mediates chromatin interactions that regulate mammalian beta-globin expression. J Biol Chem. 2011;286(20):17870–17878. doi: 10.1074/jbc.M110.207365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Seitan VC, Hao B, Tachibana-Konwalski K, Lavagnolli T, Mira-Bontenbal H, Brown KE, Teng G, Carroll T, Terry A, Horan K, Marks H, Adams DJ, Schatz DG, Aragon L, Fisher AG, Krangel MS, Nasmyth K, Merkenschlager M. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature. 2011;476(7361):467–471. doi: 10.1038/nature10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 98.Consortium EP, Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H, Giresi PG, Goldy J, Hawrylycz M, Haydock A, Humbert R, James KD, Johnson BE, Johnson EM, Frum TT, Rosenzweig ER, Karnani N, Lee K, Lefebvre GC, Navas PA, Neri F, Parker SC, Sabo PJ, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins FS, Dekker J, Lieb JD, Tullius TD, Crawford GE, Sunyaev S, Noble WS, Dunham I, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker IL, Baertsch R, Keefe D, Dike S, Cheng J, Hirsch HA, Sekinger EA, Lagarde J, Abril JF, Shahab A, Flamm C, Fried C, Hackermuller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen JS, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson MC, Thomas DJ, Weirauch MT, Gilbert J, Drenkow J, Bell I, Zhao X, Srinivasan KG, Sung WK, Ooi HS, Chiu KP, Foissac S, Alioto T, Brent M, Pachter L, Tress ML, Valencia A, Choo SW, Choo CY, Ucla C, Manzano C, Wyss C, Cheung E, Clark TG, Brown JB, Ganesh M, Patel S, Tammana H, Chrast J, Henrichsen CN, Kai C, Kawai J, Nagalakshmi U, Wu J, Lian Z, Lian J, Newburger P, Zhang X, Bickel P, Mattick JS, Carninci P, Hayashizaki Y, Weissman S, Hubbard T, Myers RM, Rogers J, Stadler PF, Lowe TM, Wei CL, Ruan Y, Struhl K, Gerstein M, Antonarakis SE, Fu Y, Green ED, Karaoz U, Siepel A, Taylor J, Liefer LA, Wetterstrand KA, Good PJ, Feingold EA, Guyer MS, Cooper GM, Asimenos G, Dewey CN, Hou M, Nikolaev S, Montoya-Burgos JI, Loytynoja A, Whelan S, Pardi F, Massingham T, Huang H, Zhang NR, Holmes I, Mullikin JC, Ureta-Vidal A, Paten B, Seringhaus M, Church D, Rosenbloom K, Kent WJ, Stone EA, Program NCS, Baylor College of Medicine Human Genome Sequencing C, Washington University Genome Sequencing C, Broad I, Children’s Hospital Oakland Research I, Batzoglou S, Goldman N, Hardison RC, Haussler D, Miller W, Sidow A, Trinklein ND, Zhang ZD, Barrera L, Stuart R, King DC, Ameur A, Enroth S, Bieda MC, Kim J, Bhinge AA, Jiang N, Liu J, Yao F, Vega VB, Lee CW, Ng P, Shahab A, Yang A, Moqtaderi Z, Zhu Z, Xu X, Squazzo S, Oberley MJ, Inman D, Singer MA, Richmond TA, Munn KJ, Rada-Iglesias A, Wallerman O, Komorowski J, Fowler JC, Couttet P, Bruce AW, Dovey OM, Ellis PD, Langford CF, Nix DA, Euskirchen G, Hartman S, Urban AE, Kraus P, Van Calcar S, Heintzman N, Kim TH, Wang K, Qu C, Hon G, Luna R, Glass CK, Rosenfeld MG, Aldred SF, Cooper SJ, Halees A, Lin JM, Shulha HP, Zhang X, Xu M, Haidar JN, Yu Y, Ruan Y, Iyer VR, Green RD, Wadelius C, Farnham PJ, Ren B, Harte RA, Hinrichs AS, Trumbower H, Clawson H, Hillman-Jackson J, Zweig AS, Smith K, Thakkapallayil A, Barber G, Kuhn RM, Karolchik D, Armengol L, Bird CP, de Bakker PI, Kern AD, Lopez-Bigas N, Martin JD, Stranger BE, Woodroffe A, Davydov E, Dimas A, Eyras E, Hallgrimsdottir IB, Huppert J, Zody MC, Abecasis GR, Estivill X, Bouffard GG, Guan X, Hansen NF, Idol JR, Maduro VV, Maskeri B, McDowell JC, Park M, Thomas PJ, Young AC, Blakesley RW, Muzny DM, Sodergren E, Wheeler DA, Worley KC, Jiang H, Weinstock GM, Gibbs RA, Graves T, Fulton R, Mardis ER, Wilson RK, Clamp M, Cuff J, Gnerre S, Jaffe DB, Chang JL, Lindblad-Toh K, Lander ES, Koriabine M, Nefedov M, Osoegawa K, Yoshinaga Y, Zhu B, de Jong PJ Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Koch CM, Andrews RM, Flicek P, Dillon SC, Karaoz U, Clelland GK, Wilcox S, Beare DM, Fowler JC, Couttet P, James KD, Lefebvre GC, Bruce AW, Dovey OM, Ellis PD, Dhami P, Langford CF, Weng Z, Birney E, Carter NP, Vetrie D, Dunham I. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007;17(6):691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, Mieczkowski P, Lieb JD, Zhao K, Brown M, Liu XS. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42(4):343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, Ren B. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488(7409):116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pekowska A, Benoukraf T, Zacarias-Cabeza J, Belhocine M, Koch F, Holota H, Imbert J, Andrau JC, Ferrier P, Spicuglia S. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J. 2011;30(20):4198–4210. doi: 10.1038/emboj.2011.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczynski B, Riddell A, Furlong EE. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet. 2012;44(2):148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- 104.Hu D, Gao X, Morgan MA, Herz HM, Smith ER, Shilatifard A. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol Cell Biol. 2013;33(23):4745–4754. doi: 10.1128/MCB.01181-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Herz HM, Mohan M, Garruss AS, Liang K, Takahashi YH, Mickey K, Voets O, Verrijzer CP, Shilatifard A. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev. 2012;26(23):2604–2620. doi: 10.1101/gad.201327.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee JE, Wang C, Xu S, Cho YW, Wang L, Feng X, Baldridge A, Sartorelli V, Zhuang L, Peng W, Ge K (2013) H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. eLife 2:e01503. doi:10.7554/eLife.01503 [DOI] [PMC free article] [PubMed]
- 107.Tie F, Banerjee R, Saiakhova AR, Howard B, Monteith KE, Scacheri PC, Cosgrove MS, Harte PJ. Trithorax monomethylates histone H3K4 and interacts directly with CBP to promote H3K27 acetylation and antagonize Polycomb silencing. Development. 2014;141(5):1129–1139. doi: 10.1242/dev.102392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ardehali MB, Mei A, Zobeck KL, Caron M, Lis JT, Kusch T. Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription. EMBO J. 2011;30(14):2817–2828. doi: 10.1038/emboj.2011.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hallson G, Hollebakken RE, Li T, Syrzycka M, Kim I, Cotsworth S, Fitzpatrick KA, Sinclair DA, Honda BM. dSet1 is the main H3K4 di- and tri-methyltransferase throughout Drosophila development. Genetics. 2012;190(1):91–100. doi: 10.1534/genetics.111.135863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mohan M, Herz HM, Smith ER, Zhang Y, Jackson J, Washburn MP, Florens L, Eissenberg JC, Shilatifard A. The COMPASS family of H3K4 methylases in Drosophila. Mol Cell Biol. 2011;31(21):4310–4318. doi: 10.1128/MCB.06092-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, Shilatifard A. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol. 2008;28(24):7337–7344. doi: 10.1128/MCB.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, Linder-Basso D, Plachetka A, Shanower G, Tolstorukov MY, Luquette LJ, Xi R, Jung YL, Park RW, Bishop EP, Canfield TK, Sandstrom R, Thurman RE, MacAlpine DM, Stamatoyannopoulos JA, Kellis M, Elgin SC, Kuroda MI, Pirrotta V, Karpen GH, Park PJ. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471(7339):480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Collis P, Antoniou M, Grosveld F. Definition of the minimal requirements within the human beta-globin gene and the dominant control region for high level expression. EMBO J. 1990;9(1):233–240. doi: 10.1002/j.1460-2075.1990.tb08100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ashe HL, Monks J, Wijgerde M, Fraser P, Proudfoot NJ. Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev. 1997;11(19):2494–2509. doi: 10.1101/gad.11.19.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tuan D, Kong S, Hu K. Transcription of the hypersensitive site HS2 enhancer in erythroid cells. Proc Natl Acad Sci USA. 1992;89(23):11219–11223. doi: 10.1073/pnas.89.23.11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kong S, Bohl D, Li C, Tuan D. Transcription of the HS2 enhancer toward a cis-linked gene is independent of the orientation, position, and distance of the enhancer relative to the gene. Mol Cell Biol. 1997;17(7):3955–3965. doi: 10.1128/mcb.17.7.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ling J, Baibakov B, Pi W, Emerson BM, Tuan D. The HS2 enhancer of the beta-globin locus control region initiates synthesis of non-coding, polyadenylated RNAs independent of a cis-linked globin promoter. J Mol Biol. 2005;350(5):883–896. doi: 10.1016/j.jmb.2005.05.039. [DOI] [PubMed] [Google Scholar]
- 118.Johnson KD, Christensen HM, Zhao B, Bresnick EH. Distinct mechanisms control RNA polymerase II recruitment to a tissue-specific locus control region and a downstream promoter. Mol Cell. 2001;8(2):465–471. doi: 10.1016/s1097-2765(01)00309-4. [DOI] [PubMed] [Google Scholar]
- 119.Johnson KD, Grass JA, Park C, Im H, Choi K, Bresnick EH. Highly restricted localization of RNA polymerase II within a locus control region of a tissue-specific chromatin domain. Mol Cell Biol. 2003;23(18):6484–6493. doi: 10.1128/MCB.23.18.6484-6493.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigo R, Gingeras TR. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316(5830):1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 123.Zhang Y, Yang L, Chen LL. Life without A tail: New formats of long noncoding RNAs. Int J Biochem Cell Biol. 2013 doi: 10.1016/j.biocel.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 124.Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS (2011) lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res 39 (Database issue):D146–D151. doi:10.1093/nar/gkq1138 [DOI] [PMC free article] [PubMed]
- 125.De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8(5):e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hah N, Murakami S, Nagari A, Danko CG, Kraus WL. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013;23(8):1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Koch F, Fenouil R, Gut M, Cauchy P, Albert TK, Zacarias-Cabeza J, Spicuglia S, de la Chapelle AL, Heidemann M, Hintermair C, Eick D, Gut I, Ferrier P, Andrau JC. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat Struct Mol Biol. 2011;18(8):956–963. doi: 10.1038/nsmb.2085. [DOI] [PubMed] [Google Scholar]
- 128.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shiekhattar R. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143(1):46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Core LJ, Martins AL, Danko CG, Waters CT, Siepel A, Lis JT. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet. 2014;46(12):1311–1320. doi: 10.1038/ng.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, Kraus WL. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145(4):622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Oude Vrielink JA, Elkon R, Melo SA, Leveille N, Kalluri R, de Laat W, Agami R. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell. 2013;49(3):524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 132.Mousavi K, Zare H, Dell’orso S, Grontved L, Gutierrez-Cruz G, Derfoul A, Hager GL, Sartorelli V. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell. 2013;51(5):606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Onoguchi M, Hirabayashi Y, Koseki H, Gotoh Y. A noncoding RNA regulates the neurogenin1 gene locus during mouse neocortical development. Proc Natl Acad Sci USA. 2012;109(42):16939–16944. doi: 10.1073/pnas.1202956109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009;5(8):e1000617. doi: 10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lam MT, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, Lee CY, Watt A, Grossman TR, Rosenfeld MG, Evans RM, Glass CK. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498(7455):511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, Oh S, Kim HS, Glass CK, Rosenfeld MG. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498(7455):516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, Glass CK, Rosenfeld MG, Fu XD. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474(7351):390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lin YC, Benner C, Mansson R, Heinz S, Miyazaki K, Miyazaki M, Chandra V, Bossen C, Glass CK, Murre C. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat Immunol. 2012;13(12):1196–1204. doi: 10.1038/ni.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bertani S, Sauer S, Bolotin E, Sauer F. The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol Cell. 2011;43(6):1040–1046. doi: 10.1016/j.molcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 140.Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell. 2013;152(4):743–754. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, Chang HY. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gummalla M, Maeda RK, Castro Alvarez JJ, Gyurkovics H, Singari S, Edwards KA, Karch F, Bender W. abd-A regulation by the iab-8 noncoding RNA. PLoS Genet. 2012;8(5):e1002720. doi: 10.1371/journal.pgen.1002720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Petruk S, Sedkov Y, Riley KM, Hodgson J, Schweisguth F, Hirose S, Jaynes JB, Brock HW, Mazo A. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell. 2006;127(6):1209–1221. doi: 10.1016/j.cell.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Erokhin M, Davydova A, Parshikov A, Studitsky VM, Georgiev P, Chetverina D. Transcription through enhancers suppresses their activity in Drosophila. Epigenetics Chromatin. 2013;6(1):31. doi: 10.1186/1756-8935-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Saxena A, Carninci P. Long non-coding RNA modifies chromatin: epigenetic silencing by long non-coding RNAs. BioEssays. 2011;33(11):830–839. doi: 10.1002/bies.201100084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106(28):11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470(7333):279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21(8):1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Curtin P, Pirastu M, Kan YW, Gobert-Jones JA, Stephens AD, Lehmann H. A distant gene deletion affects beta-globin gene function in an atypical gamma delta beta-thalassemia. J Clin Investig. 1985;76(4):1554–1558. doi: 10.1172/JCI112136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Driscoll MC, Dobkin CS, Alter BP. Gamma delta beta-thalassemia due to a de novo mutation deleting the 5′ beta-globin gene activation-region hypersensitive sites. Proc Natl Acad Sci USA. 1989;86(19):7470–7474. doi: 10.1073/pnas.86.19.7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kioussis D, Vanin E, deLange T, Flavell RA, Grosveld FG. Beta-globin gene inactivation by DNA translocation in gamma beta-thalassaemia. Nature. 1983;306(5944):662–666. doi: 10.1038/306662a0. [DOI] [PubMed] [Google Scholar]
- 152.Anderson E, Peluso S, Lettice LA, Hill RE. Human limb abnormalities caused by disruption of hedgehog signaling. Trends Genet. 2012;28(8):364–373. doi: 10.1016/j.tig.2012.03.012. [DOI] [PubMed] [Google Scholar]
- 153.Emison ES, McCallion AS, Kashuk CS, Bush RT, Grice E, Lin S, Portnoy ME, Cutler DJ, Green ED, Chakravarti A. A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature. 2005;434(7035):857–863. doi: 10.1038/nature03467. [DOI] [PubMed] [Google Scholar]
- 154.Fernandez RM, Bleda M, Luzon-Toro B, Garcia-Alonso L, Arnold S, Sribudiani Y, Besmond C, Lantieri F, Doan B, Ceccherini I, Lyonnet S, Hofstra RM, Chakravarti A, Antinolo G, Dopazo J, Borrego S. Pathways systematically associated to Hirschsprung’s disease. Orphanet J Rare Dis. 2013;8:187. doi: 10.1186/1750-1172-8-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Balemans W, Patel N, Ebeling M, Van Hul E, Wuyts W, Lacza C, Dioszegi M, Dikkers FG, Hildering P, Willems PJ, Verheij JB, Lindpaintner K, Vickery B, Foernzler D, Van Hul W. Identification of a 52 kb deletion downstream of the SOST gene in patients with van Buchem disease. J Med Genet. 2002;39(2):91–97. doi: 10.1136/jmg.39.2.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Staehling-Hampton K, Proll S, Paeper BW, Zhao L, Charmley P, Brown A, Gardner JC, Galas D, Schatzman RC, Beighton P, Papapoulos S, Hamersma H, Brunkow ME. A 52-kb deletion in the SOST-MEOX1 intergenic region on 17q12-q21 is associated with van Buchem disease in the Dutch population. Am J Med Genet. 2002;110(2):144–152. doi: 10.1002/ajmg.10401. [DOI] [PubMed] [Google Scholar]
- 157.Loots GG, Kneissel M, Keller H, Baptist M, Chang J, Collette NM, Ovcharenko D, Plajzer-Frick I, Rubin EM. Genomic deletion of a long-range bone enhancer misregulates sclerostin in Van Buchem disease. Genome Res. 2005;15(7):928–935. doi: 10.1101/gr.3437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Collette NM, Genetos DC, Economides AN, Xie L, Shahnazari M, Yao W, Lane NE, Harland RM, Loots GG. Targeted deletion of Sost distal enhancer increases bone formation and bone mass. Proc Natl Acad Sci USA. 2012;109(35):14092–14097. doi: 10.1073/pnas.1207188109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tawil R. Facioscapulohumeral muscular dystrophy. Neurotherapeutics. 2008;5(4):601–606. doi: 10.1016/j.nurt.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wijmenga C, Hewitt JE, Sandkuijl LA, Clark LN, Wright TJ, Dauwerse HG, Gruter AM, Hofker MH, Moerer P, Williamson R, et al. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat Genet. 1992;2(1):26–30. doi: 10.1038/ng0992-26. [DOI] [PubMed] [Google Scholar]
- 161.van Deutekom JC, Wijmenga C, van Tienhoven EA, Gruter AM, Hewitt JE, Padberg GW, van Ommen GJ, Hofker MH, Frants RR. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum Mol Genet. 1993;2(12):2037–2042. doi: 10.1093/hmg/2.12.2037. [DOI] [PubMed] [Google Scholar]
- 162.Petrov A, Pirozhkova I, Laoudj D, Carnac G, Lipinski M, Vassetzky YS. Chromatin loop domain organization within the 4q35 locus in facioscapulohumeral dystrophy patients versus normal human myoblasts. Proc Natl Acad Sci USA. 2006;103:6982–6987. doi: 10.1073/pnas.0511235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Petrov AP, Laoudj D, Vassetzky YS. Genetics and epigenetics of progressive fascioscapulohumeral (landouzy-dejerine) muscular dystrophy. Genetics (Moscow) 2003;39(2):147–151. [PubMed] [Google Scholar]
- 164.Petrov AV, Allinne J, Pirozhkova IV, Laoudj D, Lipinski M, Vassetzky YS. A nuclear matrix attachment site in the 4q35 locus has an enhancer-blocking activity in vivo: implications for the facio-scapulo-humeral dystrophy. Genome Res. 2008;18(1):39–45. doi: 10.1101/gr.6620908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gabriels J, Beckers MC, Ding H, De Vriese A, Plaisance S, van der Maarel SM, Padberg GW, Frants RR, Hewitt JE, Collen D, Belayew A. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene. 1999;236(1):25–32. doi: 10.1016/s0378-1119(99)00267-x. [DOI] [PubMed] [Google Scholar]
- 166.van Geel M, Heather LJ, Lyle R, Hewitt JE, Frants RR, de Jong PJ. The FSHD region on human chromosome 4q35 contains potential coding regions among pseudogenes and a high density of repeat elements. Genomics. 1999;61(1):55–65. doi: 10.1006/geno.1999.5942. [DOI] [PubMed] [Google Scholar]
- 167.Dmitriev P, Lipinski M, Vassetzky YS. Pearls in the junk: dissecting the molecular pathogenesis of facioscapulohumeral muscular dystrophy. Neuromuscul Disord. 2009;19:17–20. doi: 10.1016/j.nmd.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 168.Tawil R, van der Maarel SM, Tapscott SJ. Facioscapulohumeral dystrophy: the path to consensus on pathophysiology. Skelet Muscle. 2014;4:12. doi: 10.1186/2044-5040-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Dmitriev P, Petrov A, Ansseau E, Charron S, Coppée F, Belayew A, Carnac G, Turki A, Laoudj D, Lipinski M, Vassetzky YS (2011) The Krüppel-like factor KLF15 is a key actor in upregulation of the 4q35 genes DUX4c and FRG2 in FSHD muscles. J Biol Chem. doi:10.1074/jbc.M1111.254052
- 170.Cabianca DS, Casa V, Bodega B, Xynos A, Ginelli E, Tanaka Y, Gabellini D. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell. 2012;149(4):819–831. doi: 10.1016/j.cell.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Dmitriev P, Stankevicins L, Ansseau E, Petrov A, Barat A, Dessen P, Robert T, Turki A, Lazar V, Labourer E, Belayew A, Carnac G, Laoudj-Chenivesse D, Lipinski M, Vassetzky YS. Defective regulation of microRNA target genes in myoblasts from facioscapulohumeral dystrophy patients. J Biol Chem. 2013;288(49):34989–35002. doi: 10.1074/jbc.M113.504522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Bhatia S, Bengani H, Fish M, Brown A, Divizia MT, de Marco R, Damante G, Grainger R, van Heyningen V, Kleinjan DA. Disruption of autoregulatory feedback by a mutation in a remote, ultraconserved PAX6 enhancer causes aniridia. Am J Hum Genet. 2013;93(6):1126–1134. doi: 10.1016/j.ajhg.2013.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ghiasvand NM, Rudolph DD, Mashayekhi M, Brzezinski JAt, Goldman D, Glaser T. Deletion of a remote enhancer near ATOH7 disrupts retinal neurogenesis, causing NCRNA disease. Nat Neurosci. 2011;14(5):578–586. doi: 10.1038/nn.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Sabherwal N, Bangs F, Roth R, Weiss B, Jantz K, Tiecke E, Hinkel GK, Spaich C, Hauffa BP, van der Kamp H, Kapeller J, Tickle C, Rappold G. Long-range conserved non-coding SHOX sequences regulate expression in developing chicken limb and are associated with short stature phenotypes in human patients. Hum Mol Genet. 2007;16(2):210–222. doi: 10.1093/hmg/ddl470. [DOI] [PubMed] [Google Scholar]
- 175.Volkmann BA, Zinkevich NS, Mustonen A, Schilter KF, Bosenko DV, Reis LM, Broeckel U, Link BA, Semina EV. Potential novel mechanism for Axenfeld–Rieger syndrome: deletion of a distant region containing regulatory elements of PITX2. Invest Ophthalmol Vis Sci. 2011;52(3):1450–1459. doi: 10.1167/iovs.10-6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Harismendy O, Notani D, Song X, Rahim NG, Tanasa B, Heintzman N, Ren B, Fu XD, Topol EJ, Rosenfeld MG, Frazer KA. 9p21 DNA variants associated with coronary artery disease impair interferon-gamma signalling response. Nature. 2011;470(7333):264–268. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhang X, Cowper-Sal-lari R, Bailey SD, Moore JH, Lupien M. Integrative functional genomics identifies an enhancer looping to the SOX9 gene disrupted by the 17q24.3 prostate cancer risk locus. Genome Res. 2012;22(8):1437–1446. doi: 10.1101/gr.135665.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hsu AP, Johnson KD, Falcone EL, Sanalkumar R, Sanchez L, Hickstein DD, Cuellar-Rodriguez J, Lemieux JE, Zerbe CS, Bresnick EH, Holland SM (2013) GATA2 haploinsufficiency caused by mutations in a conserved intronic element leads to MonoMAC syndrome. Blood 121(19):3830–3837, S3831–3837. doi:10.1182/blood-2012-08-452763 [DOI] [PMC free article] [PubMed]
- 179.Sklyar I, Iarovaia OI, Lipinski M, Vassetzky YS. Translocations affecting human immunoglobulin heavy chain locus. Biopolym Cell. 2014;30(2):91–95. [Google Scholar]
- 180.Gostissa M, Yan CT, Bianco JM, Cogne M, Pinaud E, Alt FW. Long-range oncogenic activation of Igh-c-myc translocations by the Igh 3′ regulatory region. Nature. 2009;462(7274):803–807. doi: 10.1038/nature08633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Allinne J, Pichugin A, Iarovaia O, Klibi M, Barat A, Zlotek-Zlotkiewicz E, Markozashvili D, Petrova N, Camara-Clayette V, Ioudinkova E, Wiels J, Razin SV, Ribrag V, Lipinski M, Vassetzky YS. Perinucleolar relocalization and nucleolin as crucial events in the transcriptional activation of key genes in mantle cell lymphoma. Blood. 2014;123(13):2044–2053. doi: 10.1182/blood-2013-06-510511. [DOI] [PubMed] [Google Scholar]
- 182.Tsfasman T, Klibi M, Pichugin A, Lipinski M, Vassetzky YS. HIV: implication in Burkitt lymphoma. Biopolym Cell. 2012;28:285–288. [Google Scholar]
- 183.Speck NA, Renjifo B, Golemis E, Fredrickson TN, Hartley JW, Hopkins N. Mutation of the core or adjacent LVb elements of the Moloney murine leukemia virus enhancer alters disease specificity. Genes Dev. 1990;4(2):233–242. doi: 10.1101/gad.4.2.233. [DOI] [PubMed] [Google Scholar]
- 184.Genomes Project C. Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, Shafer A, Neri F, Lee K, Kutyavin T, Stehling-Sun S, Johnson AK, Canfield TK, Giste E, Diegel M, Bates D, Hansen RS, Neph S, Sabo PJ, Heimfeld S, Raubitschek A, Ziegler S, Cotsapas C, Sotoodehnia N, Glass I, Sunyaev SR, Kaul R, Stamatoyannopoulos JA. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337(6099):1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, Ntini E, Arner E, Valen E, Li K, Schwarzfischer L, Glatz D, Raithel J, Lilje B, Rapin N, Bagger FO, Jorgensen M, Andersen PR, Bertin N, Rackham O, Burroughs AM, Baillie JK, Ishizu Y, Shimizu Y, Furuhata E, Maeda S, Negishi Y, Mungall CJ, Meehan TF, Lassmann T, Itoh M, Kawaji H, Kondo N, Kawai J, Lennartsson A, Daub CO, Heutink P, Hume DA, Jensen TH, Suzuki H, Hayashizaki Y, Muller F, Consortium F, Forrest AR, Carninci P, Rehli M, Sandelin A (2014) An atlas of active enhancers across human cell types and tissues. Nature 507(7493):455–461. doi:10.1038/nature12787 [DOI] [PMC free article] [PubMed]
- 187.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473(7345):43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Trynka G, Sandor C, Han B, Xu H, Stranger BE, Liu XS, Raychaudhuri S. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat Genet. 2013;45(2):124–130. doi: 10.1038/ng.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Akhtar-Zaidi B, Cowper-Sal-lari R, Corradin O, Saiakhova A, Bartels CF, Balasubramanian D, Myeroff L, Lutterbaugh J, Jarrar A, Kalady MF, Willis J, Moore JH, Tesar PJ, Laframboise T, Markowitz S, Lupien M, Scacheri PC. Epigenomic enhancer profiling defines a signature of colon cancer. Science. 2012;336(6082):736–739. doi: 10.1126/science.1217277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Corradin O, Saiakhova A, Akhtar-Zaidi B, Myeroff L, Willis J, Cowper-Sal-lari R, Lupien M, Markowitz S, Scacheri PC. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 2014;24(1):1–13. doi: 10.1101/gr.164079.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]