Abstract
We have cloned and sequenced a cDNA that encodes for a nuclear protein of 238 kDa in the dipteran Chironomus tentans. This protein, that we call p2D10, is structurally similar to the α subunit of the general transcription factor TFIIIC. Using immunoelectron microscopy we have shown that a fraction of p2D10 is located at sites of transcription, which is consistent with a possible role of this protein in transcription initiation. We have also found that a large fraction of p2D10 is located in the nucleoplasm and in the nuclear pore complexes. Using gel filtration chromatography and coimmunoprecipitation methods, we have identified and characterized two p2D10-containing complexes that differ in molecular mass and composition. The heavy p2D10-containing complex contains at least one other component of the TFIIIC complex, TFIIIC-ε. Based on its molecular mass and composition, the heavy p2D10-containing complex may be the Pol III holoenzyme. The light p2D10-containing complex contains RNA together with at least two proteins that are thought to be involved in mRNA trafficking, RAE1 and hrp65. The observations reported here suggest that this new TFIIIC-α-like protein is involved in posttranscriptional steps of premRNA metabolism in Chironomus tentans.
INTRODUCTION
Protein-coding RNAs are synthesized as precursors (premRNAs), packed into ribonucleoprotein (premRNP) complexes, and processed inside the cell nucleus. Nuclear mRNA export factors are recruited to the mRNPs, which are then transported to the cytoplasm (reviewed by Görlich and Kutay, 1999; Nakielny and Dreyfuss, 1999). Several proteins required for mRNA export have been identified, including TAP/Mex67, RAE1/Gle2, Gle1, Dbp5, REF/Aly/Yra1, and UAP56/HEL/Sub2p. The functions of these proteins are not fully understood, but it is known that they promote transport of mRNA to the cytoplasm, presumably by mediating the interactions between the mRNP export cargo and the nuclear pore complex (NPC; reviewed by Cullen, 2000; Cole, 2000; Conti and Izaurralde, 2001; Linder and Stutz, 2001).
The role that nuclear structure plays in mRNP export is not clear, and it is still controversial whether the mRNPs are free to diffuse from the gene to the NPC or whether they interact with nuclear structures. Studies in several experimental systems (such as those carried out by Politz et al., 1998, 1999; Singh et al., 1999) have shown that mRNPs move randomly throughout the nucleus, and a diffusion-based model has been proposed to explain how mature mRNPs are preferentially exported to the cytoplasm (reviewed by Politz and Pederson, 2000). However, there is also evidence for intranuclear retention of mRNPs. For instance, by analyzing in vivo the export of premRNAs that are defective in splicing and/or 3′ end formation, Custodio et al. (1999) showed that unprocessed premRNAs accumulate at the sites of transcription and proposed that such a retention mechanism might be crucial to prevent the export of aberrant mRNAs (reviewed by Carmo-Fonseca et al., 1999).
The Balbiani ring (BR) genes of the dipteran Chironomus tentans and their transcripts, the BR premRNP particles, have been used as a model system to analyze different aspects of gene expression in eukaryotes (reviewed by Wieslander 1994; Daneholt, 2001). In the salivary gland cells of C. tentans, the chromatin is organized into giant polytene chromosomes leaving the interchromatin space, or nucleoplasm, free of chromatin fibers and thus amenable to structural analysis. Moreover, in the C. tentans salivary gland cells, the assembly, processing, and transport of the BR premRNP particles can be directly visualized using transmission electron microscopy (EM; Skoglund et al., 1983). In a previous study, we applied electron tomography to analyze in situ whether BR mRNPs bind to nucleoplasmic structures (Miralles et al., 2000). We found that approximately one third of the nucleoplasmic BR particles were in contact with a fibrous network. We identified one of the proteins located in such fibers, an evolutionarily conserved protein called hrp65 in C. tentans, NonA in Drosophila melanogaster, and PSF and p54nrb in human (Miralles et al., 2000 and references therein). Our observations showed that mRNPs are not always freely diffusible in the nucleoplasm. However, the tomography analysis could not determine whether the interaction of the mRNPs with nucleoplasmic fibers promotes movement of the mRNP toward the NPC or restricts the movement of the mRNP, maybe as part of a mechanism for sorting and retaining unprocessed premRNPs. To further characterize the nucleoplasmic structures identified by Miralles et al. (2000), we sought to identify additional nucleoplasmic proteins using an immunological method that we present here. The large dimensions of the salivary gland cells allowed us to isolate the nucleoplasm by manual microdissection, and we subsequently raised monoclonal antibodies (mAbs) against nucleoplasmic proteins. Two of the mAbs obtained in this study, mAbs 2D10 and 1F2, recognize a nuclear protein that is predominantly located in the nucleoplasm and in the nuclear envelope. We have called this protein p2D10. Immunoscreening of cDNA expression libraries and sequencing of positive cDNA clones revealed that p2D10 is structurally similar to a general transcription factor, TFIIIC-α. We also found that p2D10 is associated in vivo with at least one other component of the TFIIIC2 complex, TFIIIC-ε, which suggests that p2D10 might be the C. tentans ortholog of TFIIIC-α. Moreover, a significant fraction of p2D10 is associated with RAE1 and hrp65, two proteins that are thought to be involved in mRNP trafficking, which suggests that p2D10 is involved in posttranscriptional events of the RNA pol II gene expression pathway.
MATERIALS AND METHODS
Animals and Cell Culture
C. tentans were raised under the conditions described by Lezzi et al. (1981). Fourth instar larvae were used for the experiments. For drug treatments, larvae were grown in the presence of either 10 μg/ml actinomycin D (Calbiochem, La Jolla, CA) or 85 μg/ml DRB (Sigma Chemical, St. Louis, MO) for 120 min before dissection and fixation of the salivary glands.
C. tentans cells were cultivated as described by Wyss (1982).
Monoclonal Antibody Production
We prepared nucleoplasm by microdissection of salivary gland cells of C. tentans and used this as the antigen for immunization. Salivary glands were fixed on ice with ethanol-acetic acid 3:1 for ∼30 min, washed with cold 70% ethanol, transferred to ethanol-glycerol 1:1, and mounted in a dissection chamber with paraffin oil. Nucleoplasmic material was pooled and stored at −20°C until use. The immunization was carried out following standard procedures described by Harlow and Lane (1988). BALB/c mice were injected intraperitoneally with the nucleoplasm from ∼100 cells in Freund's complete adjuvant. Two intraperitoneal booster injections were administered in Freund's incomplete adjuvant at intervals of 2 weeks. One week after the second booster, the serum was tested by Western blotting for the presence of antibodies against C. tentans nuclear proteins. Three weeks after this test, the immunized mouse was given a final intravenous injection of nucleoplasmic material in PBS. Two days later, the spleen was extracted and macerated, and the spleen cells were fused to mouse myeloma Sp20 cells following standard procedures. Hybridoma selection was done in 96-well plates using a hypoxanthine-, aminopterin-, and thymidine-selective medium as described by Harlow and Lane (1988). The resulting hybridomas were screened by Western blot against nuclear extracts. Hybridomas that were positive in this screening were subsequently analyzed by immunocytology on salivary gland sections. Selected hybridomas were cloned by limiting dilution at least three times. All mAbs obtained were IgM. MAbs 2D10 and 1F2 were used in the present study.
Cloning and Sequencing of Full-Length p2D10
MAb 2D10 was used to screen a random-primed λgt11 cDNA library from salivary gland cells of C. tentans following standard procedures (Sambrook et al., 1989). Approximately 7.5 × 105 clones were screened, and one antibody-specific clone was isolated, λ-2D10–1. The insert of λ-2D10–1 was amplified by PCR with λgt11 forward and reverse primers, PCR-labeled with digoxigenin (Roche, Mannheim, Germany) and used as a probe to screen an oligo(dT)-primed λZAP cDNA library from tissue culture cells of C. tentans as previously described (Visa et al., 1996a). Approximately 7.3 × 105 clones were screened and three positives were isolated: λ-2D10-A, λ-2D10-B, and λ-2D10-D. The inserts of all three clones were PCR-amplified with SK and T7 primers and sequenced at least three times using walking primers. Sequence analysis was performed with the University of Wisconsin Genetics Computer Group Sequence Analysis Program and EGCC extensions to the Wisconsin Package Sequence Analysis Program. The sequence of p2D10 (AJ313507) was aligned with the sequences of TFIIICα proteins from Drosophila (Q9VJY7), rat (Q63505), and human (Q12789) using the PileUp program. The sequence was divided into stretches of amino acid of length ten, and the percentages of identical residues in at least 3 out of 4 species were calculated manually.
Subcloning and Expression of Recombinant p2D10
To produce 6xHis-tagged recombinant protein, the cDNA that encodes p2D10 (residues 960-2043) was subcloned into the NdeI-XhoI sites of pET-21b (Novagen, Madison, WI). The resulting plasmid was introduced into Escherichia coli (DE3). After induction with isopropyl-β-d-thiogalactopyranoside (IPTG), the recombinant protein was purified by NTA-agarose affinity chromatography (QIAGEN, Hilden, Germany) and analyzed by SDS-PAGE and by Western blotting.
Antibodies
Pep-I and Pep-II antibodies were obtained by immunizing rabbits with two peptides VNLDKDRTATIDGDP(C) and LNDNGEIPGDRKGAG(C) conjugated to keyhole limpet hemocyanin. These peptides corresponded to amino acids 1714–1728 and 990-1005 of p2D10. Antisera were purified on protein G in a HiTrap affinity column (Pharmacia Biotechnology, Uppsala, Sweden) and by peptide-affinity chromatography on Sulfolink columns (Pierce, Rockford, IL). MAb Y1D2 against the PEP protein of D. melanogaster (Amero et al., 1991) was kindly provided by S. Amero; this mAb cross-reacts with the PEP homolog in C. tentans, referred to as hrp120 (N. Visa and B. Daneholt, unpublished results). MAbs 4E9 against hrp65 (Miralles et al., 2000), 2E4 against hrp45 (Wurtz et al., 1996), 4F9 against hrp36 (Wurtz et al., 1996), and 1D3 against hrp23 (Sun et al., 1998) were kindly provided by B. Daneholt. MAb Y12 against the Sm epitope of snRNP core proteins was a gift from J. Steitz. Rabbit polyclonal antisera against RAE1 and CBP20 were kindly provided by E. Izaurralde (Sabri and Visa, 2000; Izaurralde et al., 1995). A rabbit serum against TFIIIC-ε was a gift from R.G. Roeder (Hsieh et al., 1999). The antibody against His-tag was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-mouse IgM immunoglobulins used for immunoprecipitation and anti-IgM antibodies conjugated with alkaline phosphatase (AP) or horseradish peroxidase (HRP) used for Western blot analysis were purchased from Southern Biotechnology Association (Birmingham, AL). Other secondary antibodies conjugated with AP or HRP for Western blotting were purchased from Dako (Glostrup, Denmark). Secondary antibodies conjugated to 6 nm colloidal gold used for immuno-LM and immuno-EM were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA).
Preparation of Cytoplasmic and Nuclear Extracts from C. tentans Tissue Culture Cells
C. tentans tissue culture cells were homogenized in PBS (137 mM NaCl, 3 mM KCl, 8 mM Na2HPO4, 2 mM NaH2PO4, pH 7.2) containing 0.2% Nonident P40 (NP-40) and 100 μg/ml PMSF using a glass homogenizer (tight pestle). The homogenate was centrifuged at 2000 × g for 10 min at 4°C. The supernatant was the cytoplasmic fraction. The pellet containing the nuclei was resuspended in PBS containing 100 μg/ml PMSF, sonicated three times for 4–5 s each time, and centrifuged as above. The resulting supernatant was the soluble nuclear extract, and it was used as input for the immunoprecipitations, or it was fractionated by gel filtration. For electrophoretic analysis, proteins were precipitated by the addition of 6 volumes of cold acetone and stored at −20°C. For screening of hybridoma supernatants by Western blotting, the nuclear pellet was prepared as described above, resuspended in PBS, sonicated six times for 10 s, and precipitated with acetone. When indicated, the following protease inhibitors were added to all buffers: aprotinin (1 μg/ml), leupeptin (1 μg/ml), and pepstatin A (1 μg/ml). For partial digestion of soluble nuclear extracts with trypsin, the nuclear pellet was resuspended in 10 mM Tris-HCl, pH 7.5, 5 mM KCl, in the absence of protease inhibitors, before sonication and centrifugation. The supernatant was supplemented with CaCl2 to give a final concentration of 10 mM and subsequently incubated with different concentrations (0–50 μg/ml) of trypsin (Sigma Chemical) for 10 min at room temperature.
SDS-PAGE and Western Blot Analysis
Proteins were separated by SDS-PAGE and stained with Coomassie blue following standard procedures. For Western blotting, proteins were transferred to polyvinylidenefluoride membranes (PVDF; Millipore, Bedford, MA) in Tris-glycine buffer supplemented with 0.02% SDS and 4 M urea using a semidry electophoretic transfer cell (Bio-Rad, Cambridge, MA). Membranes were blocked with 10% nonfat dry milk in PBS and probed with antibodies. Both primary and secondary antibodies were diluted in PBS containing 1% milk and 0.05% Tween 20. The secondary antibodies were conjugated to AP or HRP. Detection of AP conjugates was performed using an NBT/BCIP system. HRP conjugates were detected by chemiluminescence (Amersham Pharmacia Biotechnology, Amersham, UK).
Immunoprecipitation
Immunoprecipitations with mAbs were carried out in two steps. First, the mAb was added to either crude nuclear extract or selected fractions from gel filtration chromatography to give a final concentration of ∼10 μg/ml in the presence of 0.1% NP-40. The specimen was then incubated for 90 min at 4°C with gentle rotation. Anti-mouse IgM immunoglobulins covalently coupled to protein G-agarose (Zymed Laboratories, South San Francisco, CA) were added, and the rotation was continued for an additional 90 min at 4°C. The agarose beads were washed twice with PBS containing 0.1% NP-40 and once with PBS. In some cases, the beads were washed with PBS supplemented with increasing concentrations of NaCl or Na-deoxycholate (DOC) for 20 min at 4°C and then washed as above. The proteins were eluted with 0.5% SDS at room temperature. The eluted proteins were precipitated with acetone and subsequently analyzed by SDS-PAGE and Western blotting. When peptide-specific antibodies were used for immunoprecipitation, the affinity-purified antibodies were covalently cross-linked to protein A-agarose and incubated with protein extracts as indicated above.
In-Gel Digestion and Mass Spectrometry Analysis
The Coomassie-stained bands were excised from the SDS-PAGE gel and subjected to in-gel digestion as described (Hellman, 2000). The peptides were extracted and analyzed by matrix-assisted laser desorption ionization time of flight (MALDI-TOF) mass spectrometry, using a Bruker Autoflex instrument (Bremen, Germany), equipped with delayed extraction and reflector. The instrument was optimized for peptide analysis and alfa-cyano-4-hydroxy cinnamic acid was used as matrix. Spectra were internally calibrated using autolytic fragments from trypsin. The peptide mass fingerprinting analysis was done using ProFound over the internet and GPMAW software locally.
Gel Filtration
Nuclear extracts were prepared in PBS containing 0.2% NP-40, 100 μg/ml PMSF, 1 μg/ml aprotinin, 1 μg/ml leupeptins, and 1 μg/ml pepstatin A. Extract (200 μl), prepared from ∼100 ml C. tentans cell culture, was fractionated on a Superose HR6 column (Amersham Pharmacia Biotechnology) previously equilibrated with the same buffer. The sample was fractionated at a flow rate of 0.2 ml/min, and fractions of 0.6 ml were collected. The proteins in the fractions were precipitated with acetone and analyzed by Coomassie staining or by Western blotting. For RNase treatment, the nuclear extract was incubated with 25 μg/ml RNase A (Roche) at room temperature for 20 min before fractionation.
Immunostaining of Salivary Gland Cryosections (Light Microscopy)
Immunostaining of semithin cryosections was performed as previously described (Visa et al., 1996a). Salivary glands from fourth instar larvae were fixed for 60 min at room temperature in 0.1 M cacodylate buffer at pH 7.2 containing 4% paraformaldehyde, cryoprotected with 2.3 M sucrose, and frozen by immersion in liquid nitrogen. Sections of 0.5-μm thickness were cut on a cryoultramicrotome (Ultracut S/FC S; Leica, Heerbrugg, Switzerland) and mounted on glass slides. Before immunolabeling, the sections were incubated in PBS containing 0.1 M glycine and 2% BSA. The first antibody was hybridoma culture medium diluted 1:5 in PBS containing 0.5% BSA. The secondary antibody was an anti-mouse IgM conjugated to 6 nm gold particles (Jackson Immuno Research Laboratories) diluted 1:50 in PBS containing 0.5% BSA. The immunogold labeling was silver-enhanced with IntenSE (Amersham Life Science, Arlington Heights, IL). In negative controls, the primary antibody was replaced by either PBS or an unrelated IgM.
Immuno-EM
Immuno-EM was carried out essentially according to the method described by Tokuyasu (1980) with some modifications (Visa et al., 1996a). The specimens were prepared as described above for light microscopy, except that fixation was carried out for 20–25 min in 4% paraformaldehyde and 0.1% glutaraldehyde. The cryosections, 70–80 nm thick, were picked up on drops of 2.3 M sucrose and mounted onto nickel grids coated with formvar and carbon. The grids were floated onto drops of PBS containing 0.1 M glycine and 10% fetal calf serum before incubation with the antibody solutions. Hybridoma cell culture media from 2D10 and 1F2 were diluted 1:2 and 1:10, respectively, in PBS containing 5% fetal calf serum. Two types of negative controls were carried out by replacing the primary antibody either by PBS or by an unrelated IgM. The secondary antibody against mouse IgM, conjugated to 6 nm gold particles (Jackson ImmunoResearch Laboratories), was diluted 1:50 in PBS containing 5% fetal calf serum. After immunolabeling, the sections were stained with 2% aqueous uranyl acetate for 3–5 min and embedded in polyvinyl alcohol (9–10 kDa; Aldrich Chemical, Milwaukee, WI). The specimens were examined and photographed in a Zeiss CEM 902 electron microscope (Thornwood, NY) at 80 KV.
RESULTS
MAbs 2D10 and 1F2 Recognize a Nucleoplasmic Protein in C. tentans
We have used an immunological method to identify new nucleoplasmic proteins in C. tentans. Nucleoplasm from fixed salivary glands was manually microdissected, and the isolated nucleoplasmic material was used to immunize mice and to raise mAbs following standard procedures (see MATERIALS AND METHODS). The resulting hybridomas were screened by Western blotting to demonstrate antibody reaction to nuclear proteins prepared from C. tentans tissue culture cells. Clones that scored positive in the Western blotting were further screened by immunocytology on sections of salivary gland cells.
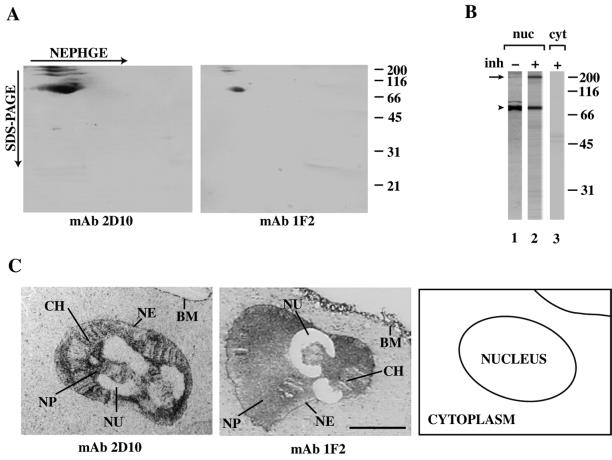
Two antibodies, mAbs 2D10 and 1F2, showed very similar patterns in the immunochemical assays. When nuclear extracts of C. tentans were analyzed by two-dimensional Western blotting, both mAbs recognized a prominent spot of molecular mass 75–80 kDa and several weaker spots up to ∼220 kDa (Figure 1A). The high-molecular-mass spots became stronger when a cocktail of broad-range protease inhibitors was used during extract preparation, as shown for mAb 2D10 in Figure 1B (compare lanes 1 and 2). In cytoplasmic extracts, mAbs 2D10 and 1F2 did not recognize any proteins (lane 3 in Figure 1B and our unpublished results). When nuclear extracts were partially digested with trypsin, mAbs 2D10 and 1F2 showed different patterns of recognition of the degradation products (our unpublished results), which suggests that the epitopes that are recognized by the two mAbs lie on different parts of the protein.
Figure 1.
MAbs 2D10 and 1F2 recognize a nucleoplasmic protein of C. tentans. (A) Nuclear proteins were separated by nonequilibrium pH gradient gel electrophoresis (NEPHGE) using ampholines in the pH 3–10 range. The second dimension was conventional SDS-PAGE. Proteins were analyzed by Western blot with mAbs 2D10 and 1F2. (B) Western blot of proteins in nuclear (nuc) and cytoplasmic (cyt) extracts of C. tentans tissue culture cells probed with mAb 2D10. Protein extraction was performed without (lane1) or with (lanes 2 and 3) a cocktail of broad-range protease inhibitors. In nuclear extracts, an intense band at 75–80 kDa (arrowhead) and a weak band at ∼220 kDa (arrow) were observed (lanes 1 and 2). In A and B, the mobilities of molecular mass standards in SDS-PAGE are shown on the right. (C) Semithin cryosections of salivary gland cells stained with mAbs 2D10 and 1F2. The antibody-binding sites were detected with a gold-conjugated secondary antibody and visual-ized after silver enhancement. The results were analyzed and photographed under bright field microscopy. A schematic interpretation of the micrographs is provided on the right. BM, basal membrane; CH, chromosomes; NE, nuclear envelope; NP, nucleoplasm; NU, nucleolus. Bar, 20 μm.
To study the cellular localization of the protein that was recognized by mAbs 2D10 and 1F2, semithin cryosections of salivary gland cells were stained with hybridoma supernatants. The large dimensions of the salivary gland cells of C. tentans and their highly organized polytene nucleus allow three distinct nuclear compartments to be identified in the preparations: the polytene chromosomes, the nucleolus, and the nucleoplasm or interchromosome space. MAbs 2D10 and 1F2 labeled extensively the cell nucleus (Figure 1C). The nucleoplasm was the most intensely stained area, and the nuclear envelope was also labeled. The chromosomes displayed a weak, banded staining. A weak labeling was also observed in the cytoplasm. In the negative control, no labeling could be detected in either the nucleus or the cytoplasm of the salivary gland cells (not shown).
In summary, the immunocytology and Western blotting results indicated that the mAbs 2D10 and 1F2 recognized a nuclear protein with molecular mass of ∼220 kDa in C. tentans that we called p2D10. Both mAbs also recognized bands of molecular mass 75–80 kDa, which are proteolytic fragments of p2D10 (see below).
Cloning and Sequencing of a cDNA Encoding p2D10, a TFIIIC-α-like Protein
To characterize p2D10 at the molecular level, mAb 2D10 was used to screen cDNA expression libraries of C. tentans. Three different clones were isolated and subsequently sequenced (designated λ-2D10-A, λ-2D10-B, and λ-2D10-D). The clone λ-2D10-D, which had a length of 6300 base pairs, turned out to harbor a 5′ UTR of length 73 nt, an ORF encoding 2043 amino acids, and a 3′ UTR of length 98 nt. The sequences of the other two clones, λ-2D10-A and λ-2D10-B, were included in λ-2D10-D.
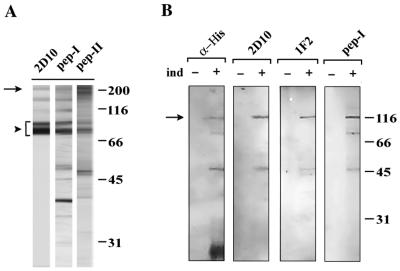
To confirm that the ORF encoded by λ-2D10-D was indeed the protein recognized by mAbs 2D10 and 1F2 in nuclear extracts, two polyclonal antibodies were generated against synthetic peptides that encompassed amino acid residues 1714–1728 and 990-1005 of the λ-2D10-D ORF. These two antibodies were designated pep-I and pep-II, respectively. In Western blot assays against nuclear proteins of C. tentans, pep-I and pep-II recognized the same bands that had been observed with mAbs 2D10 and 1F2, which confirmed the specificity of the positive clone. Pep-I and pep-II recognized the bands of molecular mass 75–80 kDa (arrowhead in Figure 2A) as well as high-molecular-mass bands up to 220 kDa. The predicted size of the protein encoded by λ-2D10-D was 238 kDa, similar to the apparent mobility of 220 kDa observed in SDS-PAGE.
Figure 2.
The protein encoded by λ-2D10-D is recognized by mAbs 2D10 and 1F2. (A) Nuclear extracts of C. tentans cells were analyzed by Western blot with either mAb 2D10 or two independent peptide-specific antibodies (pep-I and pep-II) raised against sequences encoded in λ-2D10-D. Pep-I, pep-II, and 2D10 recognized bands of molecular mass 75–80 kDa (arrowhead) and also high-molecular-mass bands up to ∼220 kDa (arrow). Additional bands, partially due to degradation, were also revealed by pep-I and pep-II. (B) The C-terminal half of λ-2D10-D ORF was expressed in E. coli with a His tag at the C terminus and purified on NTA-agarose beads. The purified recombinant protein (arrow) was recognized by antibodies against His, 2D10, 1F2, and pep-I. Extracts prepared before (−ind) or after (+ind) induction of expression were analyzed in parallel. The mobilities of molecular standards expressed in kDa are shown on the right.
The fact that all four antibodies against p2D10, including pep-I and pep-II, recognized not only the 220 kDa band but also the 75–80 kDa bands strongly suggested that the 75–80 kDa bands corresponded to proteolytic fragments of p2D10. To confirm this point, the 220 and 75–80 kDa band were analyzed by peptide mass fingerprinting on MALDI-TOF-MS. For this purpose, the sample was immunoprecipitated from a nuclear extract using the pep-II antibody, resolved by SDS-PAGE, and the 75–80 and 220 kDa bands were cut out of the gel and in situ digested with trypsin before mass spectrometry analysis. A total of 21 and 28 peptides were recovered from the 220 and 75–80 kDa bands, respectively, and 12 peptide masses were found in common between the two samples, which suggests the existence of common peptides in the two bands. However, peptide mass analyses of large proteins are usually limited by a high frequency of random matches. For this reason, and in order to further confirm the significance of our mass comparisons, the MH+ values from the 75–80 kDa band were matched against the amino acid sequences predicted from the p2D10 cDNA and against two other unrelated proteins of similar size, Tpr (267 kDa, Q99968) and myosin (222 kDa, P13535). Disregarding partial digestion products and considering MH+ values below 1500, eight sequence matches were found in p2D10 whereas only two and three possible matches were found in myosin and Tpr, respectively. This result shows that in spite of the high background levels, the frequency of observed matches between the 75–80 kDa band and p2D10 is two- to threefold higher than expected from random hits. Furthermore, the most intensive peptide mass peaks in the spectra are those that are common between the 220 and 75–80 kDa bands, supporting the conclusion that these peptides are identical. Furthermore, those peptide mass peaks do not belong to any common contaminating protein such as various keratins. Taken together, the present mass spectrometry analysis and the antibody results reported above support the conclusion that the 75–80 kDa bands are proteolytic fragments of p2D10.
The C-terminal half of the ORF encoded by λ-2D10-D, from residue 960 to residue 2043 (128 kDa), was expressed in E. coli with a His tag at the C terminus. The resulting recombinant protein was purified and analyzed by Western blotting. As expected, mAbs 2D10 and 1F2 recognized the recombinant, His-tagged protein (Figure 2B).
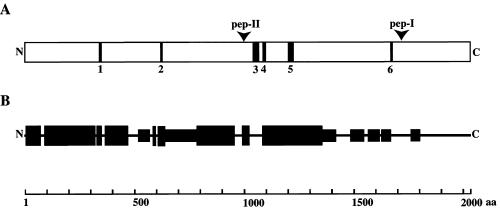
The predicted pI of the protein encoded by λ-2D10-D was 8.7. This protein had several putative phosphorylation and glycosylation sites. It also contained several unusual charged stretches (Figure 3A). Two acidic stretches consisting of 8 acidic residues each were found starting from amino acid positions 338 and 1671, respectively (Figure 3A, motifs 1 and 6). Two stretches of basic amino acids were found at amino acid positions 1037–1067 (motif 3) and 1082–1098 (motif 4), containing 14 and 7 lysines, respectively. The ORF of λ-2D10-D also contained a stretch of hydrophobic amino acids (7 serines or leucines in a stretch of length 9 amino acids) starting from residue 625 (motif 2). Except for a short coiled-coil between positions 1203 and 1231 (motif 5), no obvious structural motifs were identified in the λ-2D10-D ORF.
Figure 3.
Primary structure of p2D10 (A) The positions of sequence motifs (motifs 1–6) found in p2D10 are shown schematically. The data were obtained with an integrated search in Prosit, Pfam, and Prints at http://www.expasy.ch and Psort II at http://psort.nibb.ac.jp. Motif 1: Asp/Glu-rich region encompassing residues 338–351. Motif 2: Leu/Ser-rich region at 625–633. Motif 3: Lys-rich region at 1037–1067. Motif 4: Lys-rich region at 1082–1098. Motif 5: coiled-coil corresponding to residues 1203–1231 Motif 6: Asp/Glu-rich region at 1671–1682. The arrowheads show the positions of peptides used to raise the specific antibodies, pep-I and pep-II. (B) Structural conservation of TFIIIC-α. The sequence of p2D10 (AJ313507) was aligned with the sequences of TFIIIC-α proteins from Drosophila (Q9VJY7), rat (Q63505), and human (Q12789) with the PileUp program of the GCG package, and the percentage of identity was analyzed as described in MATERIALS AND METHODS. Large and small boxes are correspond to regions with >28% and 14–28% identity, respectively.
A search of nonredundant databases was performed with BLAST and FASTA. The best hits were the general transcription factors TFIIIC-α from human (hTFIIIC-α) and rat (rTFIIIC-α) with 28% identity. A similar level of homology was also found for the product of the CG7099 gene of D. melanogaster (Q9VJY7). The product Q9VJY7 of D. melanogaster has not been characterized, but it appears to be the Drosophila ortholog of TFIIIC-α, as indicated by database searches that compared the hTFIIIC-α against the D. melanogaster genome database. No other related proteins were found in the databases.
Comparing the primary structure of the TFIIIC-α proteins of human, rat, and Drosophila showed that the most conserved sequences were located within the N-terminal two-thirds of the protein, whereas the C-terminal third was much less conserved. A higher degree of conservation at the N-terminus was also found when p2D10 was compared with known TFIIIC-α proteins (Figure 3B).
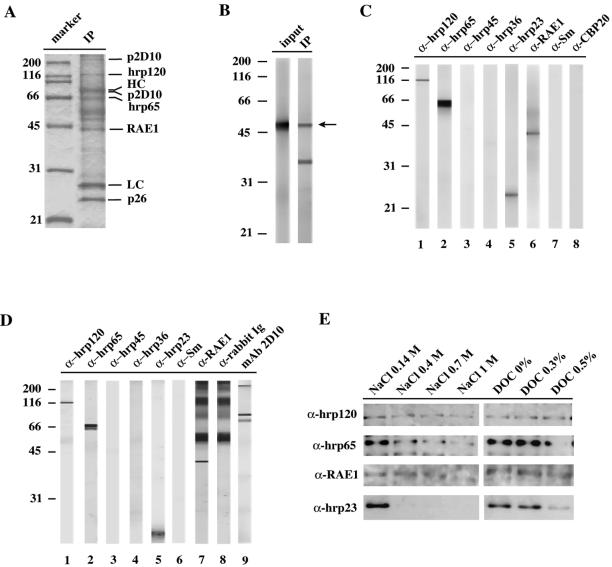
We wanted to determine whether p2D10 was associated in vivo with components of the TFIIIC complex. Human TFIIIC-α is the largest subunit of the TFIIIC2 factor, which is one of the components of the Pol III transcription machinery (reviewed by Geiduschek and Kassavetis, 2001; Huang and Maraia, 2001). One of the TFIIIC2 subunits, TFIIIC-ε, is conserved (Hsieh et al., 1999), and this allowed us to use antibodies against hTFIIIC-ε for Western blot analyses in C. tentans. A nuclear extract was prepared from C. tentans tissue culture cells, and mAb 2D10 was used to immunoprecipitate p2D10. As controls, either the antibody or the nuclear extract were excluded from the immunoprecipitation. The immunoprecipitated proteins were resolved by SDS-PAGE and either stained with Coomassie or analyzed by Western blotting. Coomassie-staining revealed that at least 10 polypeptides were coimmunoprecipitated with p2D10 (Figure 4A). A polyclonal antibody against hTFIIIC-ε reacted with a major band of approximate molecular mass 50 kDa when the immunoprecipitated material was analyzed by Western blotting (Figure 4B). The molecular mass of the hTFIIIC-ε is ∼63 kDa (Hsieh et al., 1999), whereas the protein that is believed to be the TFIIIC-ε homolog in Caenorhabditis elegans (GenBank accession no. Z35603) is 50 kDa (Hsieh et al., 1999). Therefore, the apparent molecular mass of the protein recognized by antihTFIIIC-ε in C. tentans (50 kDa) agrees with the size of TFIIIC-ε in invertebrates, and we suggest that this protein is the counterpart in C. tentans of TFIIIC-ε (ctTFIIIC-ε).
Figure 4.
Identification of proteins coimmunoprecipitated with p2D10. (A) Immunoprecipitation on nuclear extract from tissue culture cells of C. tentans with mAb 2D10. The eluted proteins were separated by SDS-PAGE and stained with Coomassie blue. Bands corresponding to identified proteins are indicated on the right. HC and LC correspond to the heavy and light chains, respectively, of mAb 2D10. The p26 protein is a C. tentans protein similar to Drosophila Q9VIX5 (Kiesler, Sabri and Visa, unpublished observations). (B) The presence of TFIIIC-ε was analyzed by immunoblotting on either nuclear extract (input) or immunoprecipitated (IP) proteins using antihTFIIIC-ε antibody. A main band of 50 kDa (arrow) corresponding to TFIIIC-ε was observed in both the input and the immunoprecipitated material. (C) Proteins immunoprecipitated by mAb 2D10 were analyzed by Western blot using antibodies against different proteins, as indicated above the lanes. (D) Proteins immunoprecipitated with pep-II were analyzed by Western blot as in C. The additional bands observed with the anti-RAE1 antibody (lane 7) are due to cross-reactivity of the secondary antibody with pep-II, as shown by the control with only secondary antibody shown in lane 8. (E) Im-munoprecipitations were performed as in A, and the bound proteins were washed with either PBS or PBS containing increasing concentrations of NaCl or DOC for 20 min before elution. The eluted proteins were subsequently analyzed by Western blot with antibodies against hrp120, hrp65, hrp23, and RAE1.
Coprecipitation of other TFIIIC2 factors could not be tested because of lack of evolutionary conservation.
In summary, the p2D10 protein is structurally similar to TFIIIC-α and is associated in vivo with at least one other component of the TFIIIC2 complex. Although it remains to be proven whether p2D10 is active as transcription factor, our present results suggest that p2D10 is the C. tentans ortholog of TFIIIC-α (ctTFIIIC-α).
Association of p2D10 with Nucleoplasmic Proteins
The location of p2D10 predominantly in the nucleoplasm of the salivary gland cells and the significant labeling observed at the nuclear envelope (Figure 1C) led us to analyze whether p2D10 is associated with other nuclear proteins previously characterized in C. tentans. We carried out immunoprecipitation experiments with mAb 2D10 as described above, and the immunoprecipitated proteins were eluted, separated by SDS-PAGE and probed with a panel of antibodies against nuclear proteins (Figure 4C). All of the available antibodies against p2D10 (mAb 2D10, mAb 1F2, pep-I, and pep-II) were positive against the immunoprecipitated material, as expected (our unpublished results). The following antibodies against other nuclear proteins were tested: mAb Y1D2 against hrp120/Pep (Amero et al., 1991), mAb 4E9 against hrp65 (Miralles et al., 2000), mAb 2E4 against hrp45 (Alzhanova-Ericsson et al., 1996), mAb 4F9 against hrp36 (Visa et al., 1996a), mAb 1D3 against hrp23 (Sun et al., 1998), anti-hRAE1 polyclonal antibody recognizing C. tentans RAE1 (Sabri and Visa, 2000), mAb Y12 against the Sm epitope of snRNP core proteins (Pettersson et al., 1984), and a polyclonal antibody against the cap-binding protein CBP20 (Visa et al., 1996b). The hrp45 and hrp36 proteins were highly abundant in the extracts that were used as input for the immunoprecipitations, but they were not detected in the eluate (Figure 4C, lanes 3 and 4, respectively). The Sm-core proteins and the CBP20 were not coimmunoprecipitated either (lanes 7 and 8, respectively). Instead, four other proteins, hrp120, hrp65, hrp23 and RAE1, coimmunopurified with p2D10 in a very reproducible manner (Figure 4C, lanes 1, 2, 5, 6).
The coimmunoprecipitated experiments were also carried out with mAb 1F2 and pep-I (Figure 4D). Identical results were obtained in all cases irrespective of the antibody used for immunoprecipitation, which supports the specificity of the coimmunoprecipitation results.
To assess the stability of the interactions detected, the immunoprecipitations were followed by highly stringent washes with buffers containing up to 1 M NaCl or 0.5% DOC (Figure 4E). Hrp120, hrp65, and RAE1 were associated with p2D10 in the presence of either NaCl or DOC. In contrast, hrp23 could not be detected in association with p2D10 at high ionic strength (≥0.4 M NaCl).
In summary, our results indicate that p2D10 is in a complex with ctTFIIIC-ε (Figure 4B) and that p2D10 is associated with hrp120, hrp65, hrp23, and RAE1 (Figure 4, C and D). CtTFIIIC-ε is a component of the Pol III transcription machinery, whereas hrp120, hrp65, hrp23, and RAE1 are involved in processing and/or transport of premRNAs. This means that our results suggest that p2D10 functions in both Pol III and Pol II gene expression pathways.
p2D10 Is Associated with hrp65 and RAE1 in a Complex that Contains RNA
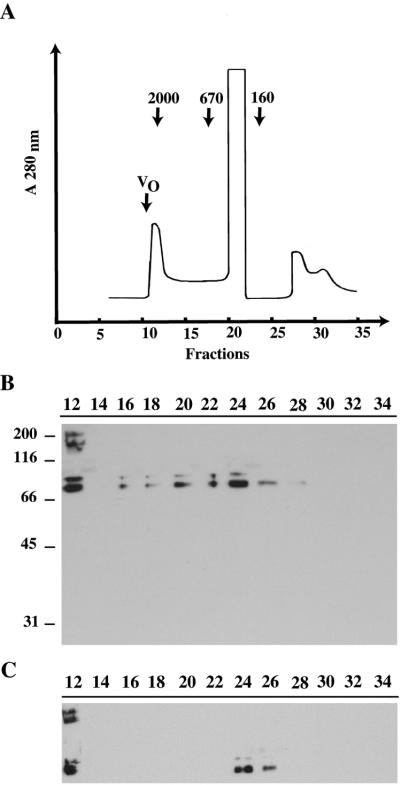
The results described above suggest that p2D10 has several functions, and this led us to expect that the p2D10 protein would be found in different complexes of defined composition. To check this hypothesis, nuclear extracts were fractionated in a size-exclusion Superose HR6 chromatography column. Proteins in each fraction were analyzed by Western blotting using mAb 2D10. As shown in Figure 5, p2D10 was detected in a heavy peak with a molecular mass of 1800–2000 kDa (fractions 12–13) and in a second much broader peak extending from ∼1100 to <160 kDa (fractions 16–27). The 220-kDa band recognized by mAb 2D10 was visible in the heavy peak but not in the other 2D10-positive fractions, which contained only the 75–80 kDa proteolytic fragments.
Figure 5.
Chromatographic analysis of TFIIIC-α–containing complexes. Nuclear proteins from tissue culture cells of C. tentans were fractionated on a gel filtration Superose HR6 column. (A) The chromatogram, showing the fractionation of some molecular mass standards, in kDa, used for calibration. VO: void volume. (B) Fractions were pooled two by two (for example, lane 12 contains fractions 12 and 13), separated by SDS-PAGE, and analyzed by immunoblotting using mAb 2D10. The mobilities of molecular mass standards in SDS-PAGE are shown on the left. (C) Nuclear extract was preincubated with 25 μg/ml RNase A for 20 min before chromatography and Western blot analysis as in B.
To check whether any of the p2D10 complexes contained RNA, a nuclear extract was treated with RNase A, fractionated, and analyzed as above. The result is shown in Figure 5C. The same amount of nuclear extracts was loaded in Figure 5, B and C. The heavy peak remained unchanged after RNase digestion. The light peak, however, became much narrower and was restricted to fractions 24–27, corresponding to a mass of <160 kDa. We concluded that the light peak was a ribonucleoprotein complex that contained both protein and RNA.
The finding that a TFIIIC-α-like protein was present in two complexes of different sizes and compositions encouraged us to analyze in more detail the association of p2D10 with proteins involved in either transcription or posttranscriptional events in the two different complexes. For this purpose, nuclear extracts were fractionated as described above, and selected fractions were used as the input to immunoprecipitation assays with mAb 2D10. The fractions were pooled in four groups, fractions 12–14, 16–18, 20–22, and 24–26. All immunoprecipitations were performed under identical conditions. The eluted proteins were analyzed by Western blotting with antibodies against the general transcription factor ctTFIIIC-ε and the putative RNA export factor RAE1. As shown in Figure 6, the association of p2D10 with ctTFIIIC-ε was restricted to fractions 12–14, whereas p2D10 was associated with RAE1 in fractions 20–22. The same type of analysis was carried out with antibodies against hrp65, which was also preferentially detected in fractions 20–22 (our unpublished results). On the other hand, hrp120 was associated with the heavy p2D10-containing complex (unpublished observation). The association of hrp23 with the p2D10 complexes could not be determined after gel filtration, probably because of the lower affinity of the interaction.
Figure 6.
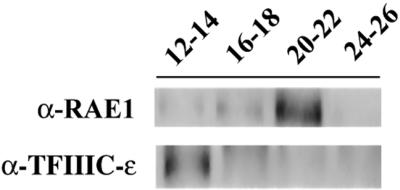
Coimmunoprecipitation of TFIIIC-α–associated proteins from selected gel filtration fractions. After fractionation of the nuclear extract on a Superose HR6 column, selected fractions were pooled in four groups including fractions 12–14, 16–18, 20–22, and 24–26 and used for immunoprecipitation with mAb 2D10. The eluted proteins were analyzed by Western blot using antibodies against RAE1 and TFIIIC-ε.
In summary, p2D10 is found in vivo in at least two different complexes. One of the complexes, referred to as the heavy p2D10 complex, has a mass of about 1800 kDa, is not sensitive to RNase digestion and contains at least one transcription factor, ctTFIIIC-ε. The second p2D10 complex, which is called the light p2D10 complex, contains RNA and at least two other proteins, RAE1 and hrp65, involved in premRNA metabolism.
p2D10 Is Located in the Nucleoplasm and at Nuclear Pores
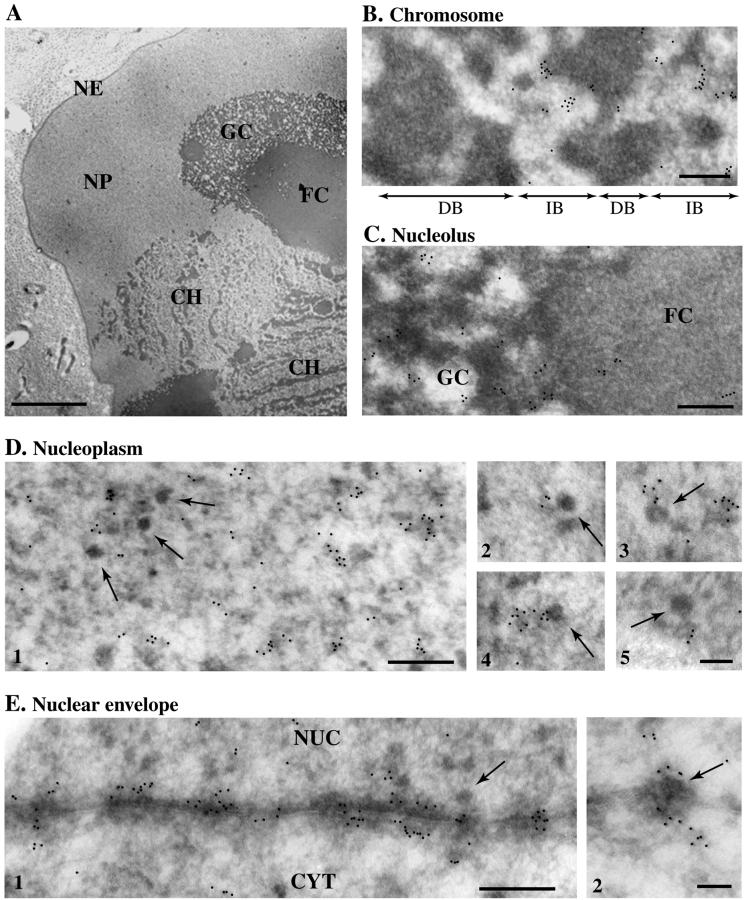
We also determined the subcellular location of the p2D10 protein in the salivary gland cells of C. tentans by immuno-EM. Thin cryo-sections of salivary glands were immunolabeled with either mAb 2D10 or mAb 1F2, with an unrelated IgM being used as a negative control. Labeling was visualized with a gold-conjugated secondary antibody. The same distribution of labeling was obtained with both mAbs. However, labeling with mAb 2D10 was weak in the immuno-EM assays, and for this reason the results shown were obtained with mAb 1F2. The distribution of p2D10 in the different nuclear compartments is illustrated in Figure 7. In the chromosomes, the dense chromatin was not labeled, whereas some interbands were labeled (Figure 7B). The nucleolus, in particular the granular component, was also weakly labeled (Figure 7C). The highest density of labeling was found in the nucleoplasm and at the nuclear envelope, which agrees with the distribution seen in the light microscope, shown in Figure 1C. The nucleoplasmic BR particles of the salivary gland cells, which are known to contain a premRNA molecule associated with hnRNP proteins (reviewed by Daneholt, 2001), were not labeled (Figure 7D1). The nucleoplasmic labeling was often observed on fibrillar structures located close to the BR particles (Figure 7D, 2–5). These labeled structures correspond to connecting fibers (CFs) and fibrogranular clusters (FGCs) described previously (Miralles et al., 2000). Interestingly, the CFs also contain hrp65 (Miralles et al., 2000).
Figure 7.
Immuno-EM analysis of p2D10 in salivary gland cells of C. tentans. Thin cryosections of salivary glands were stained with mAb 1F2 and with a secondary antibody coupled to colloidal gold. (A) Overview of a salivary gland cell as seen in the EM at low magnification. CH, chromosome; FC, fibrillar component of the nucleolus; GC, granular component of the nucleolus; NE, nuclear envelope; NP, nucleoplasm. Bar, 5 μm. (B) In the chromosomes, labeling was concentrated in the interbands (IB), whereas the dense chromatin bands (DB) were not labeled. Bar, 300 nm. (C) In the nucleolus, labeling was observed mainly in the granular component (GC). Bar, 300 nm. (D) Examples of labeling in the nucleoplasm. The arrows point at BR RNP particles located in the nucleoplasm. Bar, D1: 300 nm; D2–5: 100 nm. (E) Labeling along the nuclear envelope. The arrows point at BR RNP particles located at the NPC. The nuclear (NUC) and cytoplasmic (CYT) compartments are indicated. Bars, 300 and 100 nm in E1 and E2, respectively. A black dot was pasted onto each of the original gold markers to facilitate the visualization of the results.
Most of the nuclear pore complexes (NPCs) were intensely labeled (Figure 7E), with gold particles decorating both the nuclear and the cytoplasmic sides of the nuclear envelope. This labeling was highly specific, and a total absence of gold was observed in the negative controls (our unpublished results). The BR particles in transit through the NPC were not significantly labeled; the NPC itself was labeled, but not the exported mRNP cargo (Figure 7E2).
The location of RAE1 at the NPC depends on ongoing RNA pol II transcription (Pritchard et al., 1999) and is related to the presence of an export cargo at the NPC (Sabri and Visa, 2000). To determine whether the association of p2D10 with the NPC shows the same behavior, fourth instar larvae were treated with transcription inhibitors, actinomycin D or DRB, before dissection of salivary glands and immuno-EM analysis. Neither actinomycin D nor DRB had any effect on the location of p2D10 at the NPC (our unpublished results).
In summary, the in situ studies demonstrate that p2D10 is located in the chromosomes but also present in other nuclear compartments that contain hrp65 and RAE1, such as the nucleoplasm and the nuclear envelope. Moreover, the location of p2D10 at the NPC does not depend on Pol II transcription.
DISCUSSION
We report here the molecular cloning of a cDNA that encodes for p2D10, a 238-kDa nuclear protein that shares many properties with TFIIIC-α, the largest subunit of the TFIIIC2 factor. The assembly of transcription complexes on Pol III promoters requires at least two multimeric factors, TFIIIB and TFIIIC, in addition to Pol III itself (reviewed by Geiduschek and Kassavetis, 2001; Huang and Maraia, 2001). In mammals, TFIIIC can be resolved into two complexes of specific compositions and functions, designated TFIIIC1 and TFIIIC2. The TFIIIC2 complex, which is required for promoter recognition (Dean and Berk, 1988), consists of five subunits of molecular mass of ∼220, 110, 102, 90, and 63 kDa (Yoshinaga et al., 1989; Sinn et al., 1995; Wang and Roeder, 1996). We have shown that p2D10 coimmunoprecipitates with at least one other component of TFIIIC2, TFIIIC-ε. We have also shown by immuno-EM in the salivary gland cells of C. tentans that a fraction of p2D10 is located in the nucleoplasm and at the nuclear envelope. Moreover, our results show that p2D10 is associated with hrp65 and with RAE1, two RNA-binding proteins that are thought to be involved in intranuclear transport and/or retention of premRNA. Our data suggest that this new TFIIIC-α-like protein may be involved in posttranscriptional steps of premRNA metabolism.
p2D10, a TFIIIC-α-like Protein in C. tentans
Three experiments support the conclusion that mAbs 2D10 and 1F2 recognize p2D10, a TFIIICα-like protein in C. tentans. First, two independent peptide-specific antibodies raised against sequences that are encoded in the p2D10 cDNA gave the same labeling pattern as mAbs 2D10 and 1F2 in Western blotting. Second, recombinant p2D10 expressed in E. coli was recognized by mAbs 2D10 and 1F2. Third, the mAb 2D10 could immunoprecipitate another component of the TFIIIC2 complex, ctTFIIIC-ε, from crude nuclear extracts. Although the function of p2D10 in transcription remains to be demonstrated, the observations summarized above suggest that the protein that is recognized by mAbs 2D10 and 1F2 is not only structurally similar but also functionally related to TFIIIC-α.
Four different antibodies against p2D10, two mAbs and two peptide-specific antibodies, have been used in the present study. In Western blots of C. tentans proteins, all four antibodies recognized prominent, very intense bands of 75–80 kDa in addition to the full-length p2D10 that migrates to ∼220 kDa. Short p2D10 isoforms may exist in C. tentans, but a comprehensive screening of cDNA expression libraries failed to reveal any other related proteins. Based on this observation, on the specificity of our antibodies, and on mass spectrometry data, we conclude that the 75–80 kDa proteins detected in our Western blots correspond to proteolytic fragments of p2D10.
Complexes that Contain p2D10
The human Pol III holoenzyme has an apparent molecular mass of ∼1500 kDa (Wang et al., 1997). We have identified a complex of similar mass in C. tentans. This complex contains p2D10 and ctTFIIIC-ε, and we refer to it as the heavy p2D10 complex. The Pol III machinery in insects has not been characterized. However, the mass of the heavy p2D10 complex, and the fact that it contains both p2D10 and ctTFIIIC-ε, suggest that the heavy p2D10 complex might be the Pol III holoenzyme of C. tentans. Our immuno-EM experiments have shown that a fraction of p2D10 is located in the chromosomes and specifically in the interbands, where actively transcribed genes are known to be located (Sass and Bautz, 1982). This distribution is consistent with p2D10 having a role in transcription.
We have also detected another complex that contains p2D10. This complex has a lower molecular mass than the first and is more abundant in the nuclear extracts as judged from the intensity of labeling in Western blots. We have termed this complex the light p2D10 complex. Two other proteins, RAE1 and hrp65, were found in the light p2D10 complex. Hrp65 belongs to a family of RNA-binding proteins that includes PSF and p54nrb/NonO in mammals (Dong et al., 1993; Patton et al., 1993), and Bj6/NonA in Drosophila (von Besser et al., 1990). The function of these proteins is still controversial, but they appear to play several roles in the expression of class II genes. The human PSF was initially identified as a premRNA splicing factor (Patton et al., 1993), and more recent reports have suggested a role for both PSF and p54nrb/NonO in transcription regulation (Mathur et al., 2001 and references therein) and nuclear retention of hyper-edited RNA (Zhang and Carmichael, 2001). In C. tentans, hrp65 was identified as a component of the nucleoplasmic fibers that are associated with premRNP particles in transit from the gene to the NPC (Miralles et al., 2000). The RAE1 protein, also called Gle2p or mrnp41, is highly conserved in eukaryotes and is involved in mRNA export (reviewed by Görlich and Kutay, 1999; Nakielny and Dreyfuss, 1999; Cullen, 2000). A recent report has suggested that RAE1 plays a role in mitotic regulation (Wang et al., 2001). RAE1 is located at the NPC through direct binding to a nucleoporin, Nup116p/NUP98, and it is thought to bridge the interaction between the mRNA and the NPC (Bailer et al., 1998; Pritchard et al., 1999; Sabri and Visa, 2000).
The light p2D10 complex eluted in a broad peak, with molecular masses ranging from 1100 to <160 kDa. The molecular mass of the complex following RNase digestion was <160 kDa. Thus, we conclude that the light TFIIIC-α complex in native, nondigested extracts is a heterogeneous complex that contains both protein and RNA. The drastic reduction in size caused by RNase digestion suggests that the RNA contained in the light p2D10 complex is relatively long, maybe rRNA or mRNA. We favor the hypothesis that the light p2D10 complex contains mRNA (or premRNA), because the other proteins detected in this complex, RAE1 and hrp65, have been found in association with mRNA (Bharathi et al., 1997; Kraemer and Blobel, 1997; Miralles et al., 2000).
We could not detect the full-length p2D10 protein in the light p2D10 complex. This may be due to proteolytic degradation during our experiments, or it may suggest that a specific cleavage product of p2D10 binds, directly or indirectly, to hrp65 and RAE1 in a ribonucleoprotein complex in vivo. Interestingly, the human TFIIICα protein is specifically cleaved both in vitro and in vivo by the poliovirus 3C protease, which results in stable proteolytic fragments of ∼90 and 125 kDa (Shen et al., 1996). It will be interesting to determine whether the 75–80 kDa bands observed in the case of p2D10 also originate from specific cleavage events mediated by viral or cellular proteases.
Subcellular Location and Posttranscriptional Role of p2D10
Our coimmunoprecipitation experiments show that p2D10 and hrp65 are associated, as are p2D10 and RAE1, but we have not demonstrated the existence in vivo of a ternary complex containing all three proteins. The light p2D10 complex is likely to be heterogeneous and to include different subcomplexes. Indeed, the fact that the proteins identified in this complex are found in several subcellular locations suggests that the interactions between these proteins are dynamic and are different in different cell compartments.
The immuno-EM analysis presented in Figure 7 reveals that the nucleoplasmic fraction of p2D10 is widely distributed in connecting fibers (CFs) and fibrogranular clusters (FGCs) that are in contact with the BR particles (Miralles et al., 2000). The hrp65 protein is also found in the CFs, but it is confined to the proximal part of the fibers, close to the BR particles (Miralles et al., 2000). Thus, hrp65 and p2D10 might interact with each other in the CFs, maybe bridging the binding of the BR particles to the FGCs. It is interesting to point out that hrp23, which coimmunoprecipitates with both p2D10 and hrp65, is one of the major RNA-binding proteins in the BR particles (Sun et al., 1998). Thus, it is tempting to speculate that the binding of BR particles to the CFs is maintained by the interaction between hrp23 and hrp65, and, in turn, the CFs are associated with the large FGCs by interactions between hrp65 and p2D10.
The RAE1 protein is predominantly located at the nuclear envelope, which suggests that the interaction between p2D10 and RAE1 occurs in a later step of the gene expression pathway. We have shown that p2D10 remains associated with the NPC after transcription inhibition, whereas RAE1 is not found at the nuclear envelope if transcription is inhibited (Pritchard et al., 1999). These findings indicate that the binding of p2D10 to the NPC cannot be mediated by RAE1.
To our surprise, a fraction of p2D10 was located in the granular compartment of the nucleolus, as shown by immuno-EM. The hrp23 displays a very similar nucleolar localization (Sun et al., 1998), which suggests that p2D10 localizes to the nucleolus through its association with hrp23.
The spatial relationships of p2D10 with BR particles observed in the immuno-EM experiments suggests that the p2D10 protein functions in the intranuclear transport or retention of mRNP particles, maybe participating in the delivery of the mRNP export cargoes to the nuclear pore. The further characterization of direct protein–protein and protein–RNA interactions in the light p2D10 complex remains as goal for the future and will contribute to our understanding of the role of this complex in mRNA biogenesis.
ACKNOWLEDGMENTS
The authors thank S. Amero, B. Daneholt, E. Izaurralde, J. Steitz, and R.G. Roeder for the gift of antibodies; L. Fjelkestam and M. Tibäck for technical assistance; and L. Wieslander for the gift of C. tentans cDNA libraries; and George Farrants for language editing. This work was supported by grants from the Swedish Natural Science Research Council (NFR), Marcus and Marianne Wallenberg Foundation, Lars Hierta Foundation, Åke Wiberg Foundation, and Carl Trygger Foundation. N.S. is supported by a scholarship from the Ministry of Culture and Higher Education of Iran.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–09–0436. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–09–0436.
REFERENCES
- Alzhanova-Ericsson AT, Sun X, Visa N, Kiseleva E, Wurtz T, Daneholt B. A protein of the SR family of splicing factors binds extensively to exonic Balbiani ring pre-mRNA and accompanies the RNA from the gene to the nuclear pore. Genes Dev. 1996;10:2881–2893. doi: 10.1101/gad.10.22.2881. [DOI] [PubMed] [Google Scholar]
- Amero SA, Elgin SC, Beyer AL. A unique zinc finger protein is associated preferentially with active ecdysone-responsive loci in Drosophila. Genes Dev. 1991;5:188–200. doi: 10.1101/gad.5.2.188. [DOI] [PubMed] [Google Scholar]
- Bailer SM, Siniossoglou S, Podtelejnikov A, Hellwig A, Mann M, Hurt E. Nup116p and nup100p are interchangeable through a conserved motif that constitutes a docking site for the mRNA transport factor gle2p. EMBO J. 1998;17:1107–1119. doi: 10.1093/emboj/17.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharathi A, Ghosh A, Whalen WA, Yoon JH, Pu R, Dasso M, Dhar R. The human RAE1 gene is a functional homologue of Schizosaccharomyces pombe rae1 gene involved in nuclear export of Poly(A)+ RNA. Gene. 1997;198:251–258. doi: 10.1016/s0378-1119(97)00322-3. [DOI] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Custodio N, Calado A. Intranuclear trafficking of messenger RNA. Crit Rev Eukaryot Gene Expr. 1999;9:213–219. doi: 10.1615/critreveukargeneexpr.v9.i3-4.60. [DOI] [PubMed] [Google Scholar]
- Cole CN. mRNA export. the long and winding road. Nat Cell Biol. 2000;2:E55–E58. doi: 10.1038/35008681. [DOI] [PubMed] [Google Scholar]
- Conti E, Izaurralde E. Nucleocytoplasmic transport enters the atomic age. Curr Opin Cell Biol. 2001;13:310–319. doi: 10.1016/s0955-0674(00)00213-1. [DOI] [PubMed] [Google Scholar]
- Cullen BR. Nuclear RNA export pathways. Mol Cell Biol. 2000;20:4181–4187. doi: 10.1128/mcb.20.12.4181-4187.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custodio N, Carmo-Fonseca M, Geraghty F, Pereira HS, Grosveld F, Antoniou M. Inefficient processing impairs release of RNA from the site of transcription. EMBO J. 1999;18:2855–2866. doi: 10.1093/emboj/18.10.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneholt B. Assembly and transport of a premessenger RNP particle. Proc Natl Acad Sci USA. 2001;98:7012–7017. doi: 10.1073/pnas.111145498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean N, Berk AJ. Ordering promoter binding of class III transcription factors TFIIIC1 and TFIIIC2. Mol Cell Biol. 1988;8:3017–3025. doi: 10.1128/mcb.8.8.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B, Horowitz DS, Kobayashi R, Krainer AR. Purification and cDNA cloning of HeLa cell p54nrb, a nuclear protein with two RNA recognition motifs and extensive homology to human splicing factor PSF and Drosophila NONA/BJ6. Nucleic Acids Res. 1993;21:4085–4092. doi: 10.1093/nar/21.17.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiduschek EP, Kassavetis GA. The RNA polymerase III transcription apparatus. J Mol Biol. 2001;310:1–26. doi: 10.1006/jmbi.2001.4732. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Hellman U. Sample preparation by SDS-PAGE and in-gel digestion. EXS. 2000;88:43–54. doi: 10.1007/978-3-0348-8458-7_3. [DOI] [PubMed] [Google Scholar]
- Hsieh YJ, Wang Z, Kovelman R, Roeder RG. Cloning and characterization of two evolutionarily conserved subunits (TFIIIC102 and TFIIIC63) of human TFIIIC and their involvement in functional interactions with TFIIIB and RNA polymerase III. Mol Cell Biol. 1999;19:4944–4952. doi: 10.1128/mcb.19.7.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Maraia RJ. Comparison of the RNA polymerase III transcription machinery in S. pombe, S. cerevisiae, and human. Nucleic Acids Res. 2001;29:2675–2690. doi: 10.1093/nar/29.13.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Lewis J, Gamberi C, Jarmolowski A, McGuigan C, Mattaj IW. A cap-binding protein complex mediating U snRNA export. Nature. 1995;376:709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- Kraemer D, Blobel G. mRNA binding protein mrnp 41 localizes to both nucleus and cytoplasm. Proc Natl Acad Sci USA. 1997;94:9119–9124. doi: 10.1073/pnas.94.17.9119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezzi M, Meyer B, Mahr R. Heat shock phenomena in Chironomus tentans I. In vivo effects of heat, overheat, and quenching on salivary chromosome puffing. Chromosoma. 1981;83:327–339. doi: 10.1007/BF00327356. [DOI] [PubMed] [Google Scholar]
- Linder P, Stutz F. Traveling with DEAD box proteins. Curr Biol. 2001;11:R961–R963. doi: 10.1016/s0960-9822(01)00574-7. [DOI] [PubMed] [Google Scholar]
- Mathur M, Tucker PW, Samuels HH. PSF is a novel corepressor that mediates its effect through Sin3A and the DNA binding domain of nuclear hormone receptors. Mol Cell Biol. 2001;21:2298–2311. doi: 10.1128/MCB.21.7.2298-2311.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles F, Öfverstedt LG, Sabri N, Aissouni Y, Hellman U, Skoglund U, Visa N. Electron tomography reveals post-transcriptional binding of pre-mRNPs to specific fibers in the nucleoplasm. J Cell Biol. 2000;148:271–282. doi: 10.1083/jcb.148.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S, Dreyfuss G. Transport of proteins and RNAs in and out of the nucleus. Cell. 1999;99:677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- Patton JG, Porro EB, Galceran J, Tempst P, Nadal-Ginard B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 1993;7:393–406. doi: 10.1101/gad.7.3.393. [DOI] [PubMed] [Google Scholar]
- Pettersson I, Hinterberger M, Mimori T, Gottlieb E, Steitz JA. The structure of mammalian small nuclear ribonucleoproteins. Identification of multiple protein components reactive with anti-(U1)ribonucleoprotein and anti-Sm autoantibodies. J Biol Chem. 1984;259:5907–5914. [PubMed] [Google Scholar]
- Politz JC, Browne ES, Wolf DE, Pederson T. Intranuclear diffusion and hybridization state of oligonucleotides measured by fluorescence correlation spectroscopy in living cells. Proc Natl Acad Sci USA. 1998;95:6043–6048. doi: 10.1073/pnas.95.11.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz JC, Pederson T. Review: movement of mRNA from transcription site to nuclear pores. J Struct Biol. 2000;129:252–257. doi: 10.1006/jsbi.2000.4227. [DOI] [PubMed] [Google Scholar]
- Politz JC, Tuft RA, Pederson T, Singer RH. Movement of nuclear poly(A) RNA throughout the interchromatin space in living cells. Curr Biol. 1999;9:258–291. doi: 10.1016/s0960-9822(99)80136-5. [DOI] [PubMed] [Google Scholar]
- Pritchard CE, Fornerod M, Kasper LH, van Deursen JM. RAE1 is a shuttling mRNA export factor that binds to a GLEBS-like NUP98 motif at the nuclear pore complex through multiple domains. J Cell Biol. 1999;145:237–254. doi: 10.1083/jcb.145.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabri N, Visa N. The Ct-RAE1 protein interacts with Balbiani ring RNP particles at the nuclear pore. RNA. 2000;6:1597–1609. doi: 10.1017/s1355838200001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sass H, Bautz EK. Immunoelectron microscopic localization of RNA polymerase B on isolated polytene chromosomes of Chironomus tentans. Chromosoma. 1982;85:633–642. doi: 10.1007/BF00330777. [DOI] [PubMed] [Google Scholar]
- Shen Y, Igo M, Yalamanchili P, Berk AJ, Dasgupta A. DNA binding domain and subunit interactions of transcription factor IIIC revealed by dissection with poliovirus 3C protease. Mol Cell Biol. 1996;16:4163–4171. doi: 10.1128/mcb.16.8.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh OP, Björkroth B, Masich S, Wieslander L, Daneholt B. The intranuclear movement of Balbiani ring premessenger ribonucleoprotein particles. Exp Cell Res. 1999;251:135–146. doi: 10.1006/excr.1999.4490. [DOI] [PubMed] [Google Scholar]
- Sinn E, Wang Z, Kovelman R, Roeder RG. Cloning and characterization of a TFIIIC2 subunit (TFIIIC beta) whose presence correlates with activation of RNA polymerase III-mediated transcription by adenovirus E1A expression and serum factors. Genes Dev. 1995;9:675–685. doi: 10.1101/gad.9.6.675. [DOI] [PubMed] [Google Scholar]
- Skoglund U, Andersson K, Björkroth B, Lamb MM, Daneholt B. Visualization of the formation and transport of a specific hnRNP particle. Cell. 1983;34:847–855. doi: 10.1016/0092-8674(83)90542-1. [DOI] [PubMed] [Google Scholar]
- Sun X, Alzhanova-Ericsson AT, Visa N, Aissouni Y, Zhao J, Daneholt B. The hrp23 protein in the Balbiani ring pre-mRNP particles is released just before or at the binding of the particles to the nuclear pore complex. J Cell Biol. 1998;142:1181–1193. doi: 10.1083/jcb.142.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyasu KT. Immunochemistry on ultrathin frozen sections. Histochem J. 1980;12:381–403. doi: 10.1007/BF01011956. [DOI] [PubMed] [Google Scholar]
- Visa N, Alzhanova-Ericsson AT, Sun X, Kiseleva E, Björkroth B, Wurtz T, Daneholt B. A pre-mRNA binding protein accompanies the RNA from the gene through the nuclear pores and into polysomes. Cell. 1996a;84:253–264. doi: 10.1016/s0092-8674(00)80980-0. [DOI] [PubMed] [Google Scholar]
- Visa N, Izaurralde E, Ferreira J, Daneholt B, Mattaj IW. A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J Cell Biol. 1996b;133:5–14. doi: 10.1083/jcb.133.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Besser H, Schnabel P, Wieland C, Fritz E, Stanewsky R, Saumweber H. The puff-specific Drosophila protein Bj6, encoded by the gene no-on transient A, shows homology to RNA-binding proteins. Chromosoma. 1990;100:37–47. doi: 10.1007/BF00337601. [DOI] [PubMed] [Google Scholar]
- Wang X, Babu JR, Harden JM, Jablonski SA, Gazi MH, Lingle WL, de Groen PC, Yen TJ, van Deursen JM. The mitotic checkpoint protein hBUB3 and the mRNA export factor hRAE1 interact with GLEBS-containing proteins. J Biol Chem. 2001;276:26559–26567. doi: 10.1074/jbc.M101083200. [DOI] [PubMed] [Google Scholar]
- Wang Z, Luo T, Roeder RG. Identification of an autonomously initiating RNA polymerase III holoenzyme containing a novel factor that is selectively inactivated during protein synthesis inhibition. Genes Dev. 1997;11:2371–2382. doi: 10.1101/gad.11.18.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Roeder RG. TFIIIC1 acts through a downstream region to stabilize TFIIIC2 binding to RNA polymerase III promoters. Mol Cell Biol. 1996;16:6841–6850. doi: 10.1128/mcb.16.12.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieslander L. The Balbiani ring multigene family: coding repetitive sequences and evolution of a tissue-specific cell function. Prog Nucleic Acid Res. 1994;48:275–313. doi: 10.1016/s0079-6603(08)60858-2. [DOI] [PubMed] [Google Scholar]
- Wurtz T, Kiseleva E, Nacheva G, Alzhanova-Ericcson A, Rosen A, Daneholt B. Identification of two RNA-binding proteins in Balbiani ring premessenger ribonucleoprotein granules and presence of these proteins in specific subsets of heterogeneous nuclear ribonucleoprotein particles. Mol Cell Biol. 1996;16:1425–1435. doi: 10.1128/mcb.16.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss C. Chironomus tentans epithelial cell lines sensitive to ecdysteroids, juvenile hormone, insulin and heat shock. Exp Cell Res. 1982;139:309–319. doi: 10.1016/0014-4827(82)90255-5. [DOI] [PubMed] [Google Scholar]
- Yoshinaga SK, L'Etoile ND, Berk AJ. Purification and characterization of transcription factor IIIC2. J Biol Chem. 1989;264:10726–10731. [PubMed] [Google Scholar]
- Zhang Z, Carmichael GG. The fate of double-stranded RNA in the nucleus: a multiprotein complex containing the inosine-specific RNA-binding protein p54nrb mediates the nuclear retention of A-to-I edited RNAs. Cell. 2001;106:465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]