Abstract
Resistance to therapy commonly develops in patients with high-grade serous ovarian carcinoma (HGSC) and triple-negative breast cancer (TNBC), urging the search for improved therapeutic combinations and their predictive biomarkers. Starting from a CRISPR knockout screen, we identified that loss of RB1 in TNBC or HGSC cells generates a synthetic lethal dependency on casein kinase 2 (CK2) for surviving the treatment with replication-perturbing therapeutics such as carboplatin, gemcitabine, or PARP inhibitors. CK2 inhibition in RB1-deficient cells resulted in the degradation of another RB family cell cycle regulator, p130, which led to S phase accumulation, micronuclei formation, and accelerated PARP inhibition–induced aneuploidy and mitotic cell death. CK2 inhibition was also effective in primary patient-derived cells. It selectively prevented the regrowth of RB1-deficient patient HGSC organoids after treatment with carboplatin or niraparib. As about 25% of HGSCs and 40% of TNBCs have lost RB1 expression, CK2 inhibition is a promising approach to overcome resistance to standard therapeutics in large strata of patients.
Casein kinase 2 inhibition overcomes chemoresistance in RB1-deficient cancer cells.
INTRODUCTION
High-grade serous ovarian carcinoma (HGSC) and triple-negative breast cancer (TNBC) are malignancies with low patient survival due to the frequent development of therapy resistance (1, 2). Standard-of-care treatment for both cancer types includes DNA-damaging agents, such as platinum drugs (alone or combined with taxanes) or gemcitabine. Because of similar molecular characteristics [high genomic instability, high prevalence of BRCA mutations, and functional homologous recombination (HR) deficiency] (3), poly(ADP-ribose) polymerase (PARP) inhibitors have been applied to treat HGSC and TNBC. Maintenance therapy with PARP inhibitors (PARPis) extends the progression-free survival of patients with BRCA mutant tumors (4, 5). However, despite the initial success of the stratified approach, patients in this “better prognosis” cohort still often develop resistance to both chemotherapy and PARPis (6–8), leading to disease relapses. There is, therefore, still a need for the development of strategies to improve therapeutic responses in patients with HGSC and TNBC.
Therapeutics used for HGSC and TNBC generally intervene with the progression of DNA replication by blocking DNA polymerase by masked chain termination (gemcitabine) (9) or by cross-linking the neighboring nucleotides (carboplatin and cisplatin) (6, 10). PARPis have a dual effect: They block DNA repair signaling by inhibiting PARylation, while in the absence of auto-PARylation, PARP itself remains trapped onto the chromatin, blocking the replication fork progression (11, 12). Upon misregulation of cell cycle checkpoints, the replication-born DNA lesions persist until mitosis and lead to aneuploidy and mitotic cell death (13–16). Hence, the replicative DNA lesions become particularly detrimental to tumor cells lacking cell cycle checkpoint functionality, which is explored as a therapeutic target for single agent and combinatorial treatments [reviewed in (17)].
Effective cancer treatments often rely on synergistically acting therapeutic agents. Synergy is defined as a cytotoxic effect of independent pharmacological perturbations that exceeds the reduction of cancer cell viability caused by either of the perturbations alone, and it can be assessed using methods such as Bliss, Loewe, highest single agent (HSA), and others (18, 19). The extreme case of a synergistic interaction is synthetic lethality, where the combination of two nonlethal perturbations becomes fully lethal (such that 0 + 0 = 1). In cancer therapy, this is commonly applied in cases where one cancer-specific genetic alteration causes a de novo dependence on another targetable protein or cellular signal. The therapeutic advantage of these types of interactions is that they become cancer cell specific, while normal nonmalignant cells are unaffected by the drug addition. The synthetic lethal interaction between BRCA mutations and sensitivity to PARP inhibition is a key example of a targetable synthetic lethality that led to effective, stratified, and even curative treatments for patients with ovarian and breast cancer (2, 20). In the search for other context-specific therapeutic synergistic and synthetic lethal combinations, the application of high-throughput chemical screens and functional genomics (CRISPR-based loss- or gain-of-function screens) has become an important tool (21–25).
Casein kinase 2, a constitutively active protein kinase, has been considered a relevant cancer therapeutic target because of its frequent overexpression in cancer cells and its multiple cellular functions, including proliferation, cell cycle control, protection from apoptosis, and DNA repair response (26–29). However, because of the lack of strategies for stratified use, the use of CK2 inhibitors in cancer therapy has yet to prove clinically beneficial. Using a CRISPR loss-of-function screening, we identified a cytoprotective role for CK2 catalytic subunit α′ for carboplatin treatment survival in HGSC cells deficient for the retinoblastoma (RB1) tumor suppressor. Validation studies revealed that under the pressure of replication-perturbing drugs, RB1-deficient cells have a synthetic lethal dependency on CK2 activity for controlling S phase progression and mitotic fidelity. CK2 inhibitors, therefore, specifically sensitized RB1-deficient TNBC and HGSC cells to carboplatin or PARPi treatment. Our study provides a rationale for exploring CK2 inhibitors together with standard therapeutics to treat RB1-deficient triple-negative breast and high-grade ovarian cancers.
RESULTS
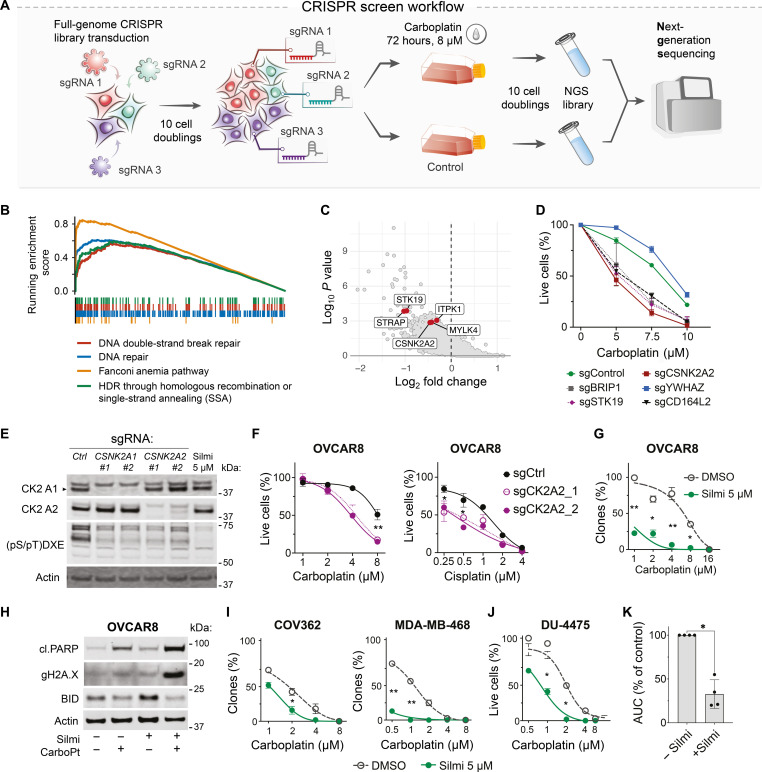
A genome-wide CRISPR screen identifies modulators of carboplatin sensitivity
To identify genes whose ablation increases the carboplatin sensitivity in HGSC cells, we performed a pooled genome-wide CRISPR loss-of-function screen (Fig. 1A) in the OVCAR8, a cell line derived from an ovarian tumor that appeared resistant to high-dose carboplatin treatment (30) and bearing mutant TP53 (ubiquitous in HGSC) (31, 32) and a functional loss of HR-mediated DNA repair (33).
Fig. 1. A CRISPR-Cas9 knockout screen identifies CK2 subunit α′ among carboplatin-sensitizing factors.
(A) Schematic representation of the CRISPR-Cas9 screen experiment. Puromycin-selected cells were propagated for 10 divisions to eliminate the cells with fitness gene knockouts. (B) Gene Ontology analysis with Enrichr (90) reveals significant enrichment in DNA repair factors among carboplatin response-essential genes. (C) Volcano plot, the hit genes encoding for kinases are highlighted. (D) Viability response to carboplatin in OVCAR8 cells with the knockouts of screening hit genes. Image cytometry–based counts of live and dead cells. Hoechst/CellTox green staining, n = 2; data points represent mean ± SD. (E) Immunoblotting for CK2 kinase catalytic subunits and phosphorylation motif in OVCAR8 cells upon CSNK2A1/2 knockout or silmitasertib (5 μM, Silmi) treatment for 4 hours. (F) Viability of OVCAR8 cells with CK2 a′ subunit CRISPR knockout after 7 days of drug exposure. (G) Quantification of the clonogenic survival assay. Treatment: carboplatin ± 5 μM silmitasertib or vehicle (5 days), drug-free regrowth (9 days). Mean ± SD, n = 2, N = 2 for each cell line. (H) Immunoblotting for apoptotic markers in OVCAR8 cells treated for 5 days with 5 μM silmitasertib, 8 μM carboplatin, or their combination. (I) Quantification of the clonogenic survival assay, performed as in (G). (J) Quantification of the survival assay for DU-4475 cells. Treatment: carboplatin ± 5 μM silmitasertib or vehicle (5 days), drug-free regrowth (9 days). Mean ± SD, n = 2, N = 2. Because of suspension growth, DU4475 were counted using Trypan blue exclusion assay and Countess II counter. (K) Difference in the area under the curve (AUC) for carboplatin response curves in OVCAR8, COV362, MDA-MB-468, and DU-4475 in the presence of silmitasertib. *P < 0.05 and **P < 0.01; two-way ANOVA with Tukey posttest for (D), (F), (G), (I), and (J) and Mann-Whitney test for (K). NGS, next-generation sequencing; HDR, homology-directed repair.
As expected, the analysis of the single guide RNA (sgRNA) abundances upon DNA-damaging carboplatin treatment identified multiple DNA repair and cell cycle checkpoint factors as statistically significantly enriched among hit genes (Fig. 1B and tables S1 and S2). We further focused on druggable carboplatin-sensitizing genes, such as genes encoding kinases (CSNK2A2, STK19, ITPK1, MYLK4, and STRAP) or receptor molecules (CD164L2 and HTR5A) from the top 100 genes ranked by MAGeCK robust rank aggregation (RRA) score (34) as genes that promote survival after the carboplatin treatment (Fig. 1C and table S1). To validate the screen results, we retested several hit genes with different RRA scores using individual sgRNAs. In agreement with the genome-wide CRISPR screen, the targeted knockouts of sensitizer genes (CSNK2A2, STK19, CD164L2, and top 1 hit BRIP) or resistance genes (YWHAZ) reduced or improved the viability of OVCAR8 cells in response to carboplatin, respectively (Fig. 1D).
We chose to explore the carboplatin-sensitizing effect of the knockout of CSNK2A2 encoding the α′ catalytic subunit of the serine-threonine kinase CK2, which can be targeted with the selective inhibitor silmitasertib (35, 36) and has a documented role in DNA damage response [reviewed in (37)]. We modeled the loss of CK2 activity by knocking out catalytic subunit-encoding genes CSNK2A1 (α) and CSNK2A2 (α′). Knockouts of α and α′ subunits only partly decreased the levels of CK2 phosphorylated substrates [(pS/pT)DXE; Fig. 1E], suggesting that the CK2 catalytic domain consisting of a-a or a′-a′ homodimers are still capable of maintaining partial CK2 activity in OVCAR8 cells. Chemical inhibition with silmitasertib, a selective CK2 inhibitor, efficiently inhibited CK2 substrate phosphorylation levels (Fig. 1E). Both genetic and chemical inhibition of CK2 activity (Fig. 1E) reduced the viability of OVCAR8 cells in response to platinum drugs (Fig. 1, F and G). Consistent with its greater inhibitory effect on CK2-mediated phosphorylation, silmitasertib had a carboplatin-sensitizing effect both in short-term viability assays and in long-term clonogenic survival tests (Fig. 1G). OVCAR8 treatment with carboplatin and silmitasertib induced apoptotic markers, PARP cleavage, γH2AX accumulation, and BH3 interacting-domain death agonist (BID) degradation (Fig. 1H), indicating the cytotoxic effect of the combination.
We tested the carboplatin-sensitizing effect of silmitasertib in a broader panel of cell models (N = 14; table S4), representing cancers treated with platinum drugs and derived from both chemo-resistant and chemo-naive tumors. In addition to eight HGSC cell lines, we tested the cervix cancer cell line HeLa and five TNBC cell lines. We observed sensitization to carboplatin by silmitasertib in 6 of 14 tested cell lines (Fig. 1, I to K, and fig. S1), which indicated a stratified effect of the drug combination.
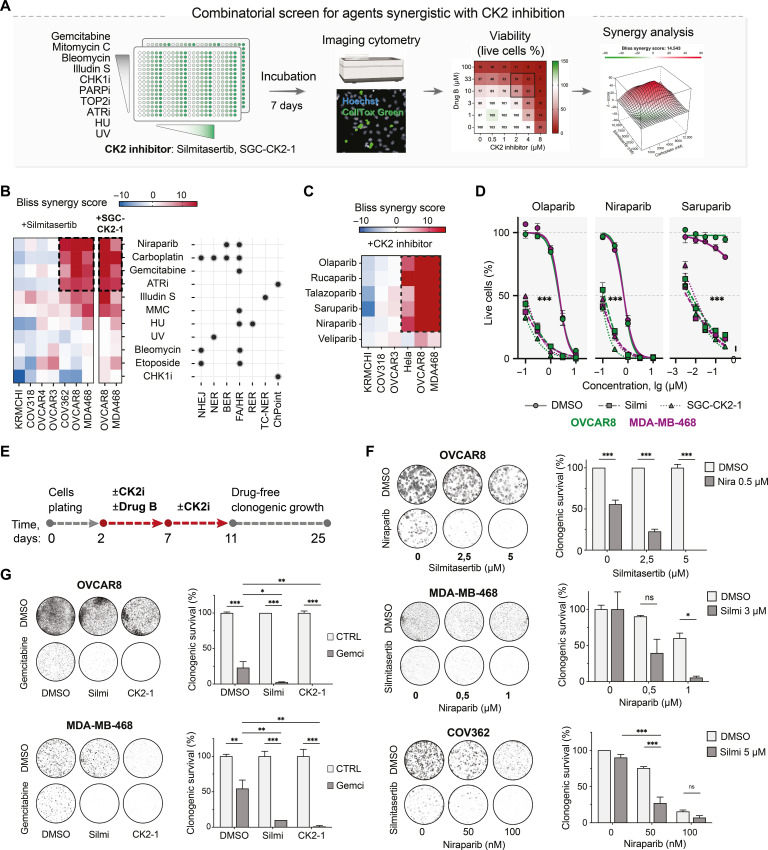
CK2 inhibition sensitizes cancer cells to PARPis and gemcitabine
Carboplatin kills cancer cells by inducing multiple types of DNA damage and cell cycle defects, including GpG intrastrand and interstrand DNA cross-links that lead to replication fork stalling and double-strand breaks that can be repaired following the active G1/S and G2/M checkpoints (6). To determine what types of carboplatin-induced DNA lesions or cell cycle defects cause cytotoxicity upon CK2 inhibition, we performed an imaging cytometry–based combinatorial drug screen testing the sensitizing interaction between silmitasertib and 10 other agents that induce DNA damage via different mechanisms relevant to carboplatin cytotoxicity using a physiologically relevant range of the drug concentrations (Fig. 2A and table S3). We compared the drug combination responses in three cell lines that displayed a sensitizing carboplatin-silmitasertib interaction (OVCAR8, COV362, and MDA-MB-468; Fig. 1, G and I) and four cell lines where CK2 inhibition did not have a sensitizing effect with carboplatin (OVCAR3, COV318, OVCAR4, and KURAMOCHI; fig. S1, A to D). The drug screen showed that CK2 inhibition enhanced the toxicity of PARP and ataxia telangiectasia and Rad3-related protein (ATR) inhibitors and gemcitabine (similar to the effect of CK2 inhibition on carboplatin) in OVCAR8, COV362, and MDA-MB-468 cells but not in the other tested models (Fig. 2B and figs. S2 and S3). These agents induce replication-associated DNA damage (38). Illudin S, an inducer of transcription-associated DNA damage, and mitomycin C, an inducer of DNA cross-links, showed an additive effect with silmitasertib. CK2 inhibition failed to potentiate the toxicity of double-strand break inducers etoposide and bleomycin, deoxynucleoside triphosphate–depleting hydroxyurea, checkpoint kinase 1 (CHK1) inhibitor prexasertib, or ultraviolet irradiation (Fig. 2B and figs. S2 and S3).
Fig. 2. Interactions between silmitasertib and replication-perturbing drugs in HGSC and TNBC cells.
(A) Schematic depiction of the combinatorial drug screening approach. The cell lines (OVCAR8, OVCAR3, OVCAR4, COV362, COV318, and MDA-MB-468) were plated to multiwell plates with single drugs or drug combinations predispensed at a range of doses. After 7 days of incubation, the number of live and dead cells for each treatment was measured by image cytometry using Hoechst and CellTox Green dyes. Viability was calculated as a fraction of the live cells normalized to DMSO-treated controls. (B) Bliss synergy scores heatmaps for drug-drug interactions (mean of two independent experiments for each cell line). NHEJ, nonhomologous end joining; NER, nucleotide excision repair; BER, base excision repair; FA/HR, Fanconi anemia/HR repair pathway; RER, ribonucleotide excision repair; TC-NER, transcription-coupled nucleotide excision repair. The dots mark the published evidence for the contribution of the pathways to the repair of the lesions by the tested drugs. (C) Bliss synergy scores heatmap for PARPi-CK2i interactions in the tested cell lines (mean of two independent experiments for each cell line). (D) PARPi dose response of OVCAR8 and MDA-MB-468 cells in the presence of 5 μM silmitasertib or 1 μM SGC-CK2-1 for 7 days. Image cytometry–based counts of live and dead cells. Hoechst/CellTox green staining, n = 3; data points represent mean ± SD. (E) Schematic timeline of the clonogenic survival test for screen results validation. (F) Clonogenic survival of the indicated cell lines treated with niraparib and silmitasertib as in (E). Mean ± SD (n = 2). (G) Clonogenic survival of OVCAR8 and MDA-MB-468 treated with gemcitabine and CK2 inhibitors silmitasertib or SGC-CK2-1. Bars represent the gemcitabine-surviving fraction of clones normalized to DMSO control or CK2 inhibitor–only condition, respectively. Mean ± SD (n = 2). In (F) and (G), the experiments were performed twice. *P < 0.05, **P < 0.01, and ***P < 0.001, two-way ANOVA with Tukey posttest. UV, ultraviolet; ns, not significant.
We further investigated the interaction between the CK2 and PARPis (N = 6) in the dose matrix drug combination screen (setup as in Fig. 2A). Inhibition of CK2 with silmitasertib or the more selective CK2 inhibitor SGC-CK2-1 (39) combined with any tested PARPi, except least potent PARP trapper veliparib (40), led to synergistic inhibition of the viability in OVCAR8, MDA-MB-468, and HeLa cells but not in OVCAR3, COV318, or KURAMOCHI (Fig. 2, C and D). Notably, CK2 inhibition markedly increased the sensitivity of OVCAR8 and MDA-MB-468 to the PARP1-selective inhibitor saruparib (Fig. 2D) (41). In the long-term validation experiments (Fig. 2E), CK2 and PARP coinhibition reduced the survival of OVCAR8, COV362, MDA-MB-468, DU-4475, and HeLa cells (Fig. 2F and fig. S4, A to C). Similarly, CRISPR-mediated knockout of CSNK2A2 led to a moderate but significant decrease in the clonogenic survival of OVCAR8 cells upon niraparib treatment (fig. S4D). In addition, we validated the enhanced toxicity of gemcitabine in OVCAR8 and MDA-MB-468 cells upon CK2 inhibition (Fig. 2F). Collectively, the results demonstrated that CK2 inhibition enhanced the toxicity of agents that induce replication-associated lesions, such as platinum drugs, PARPis, and gemcitabine in breast, ovarian, and cervical cancer cell lines.
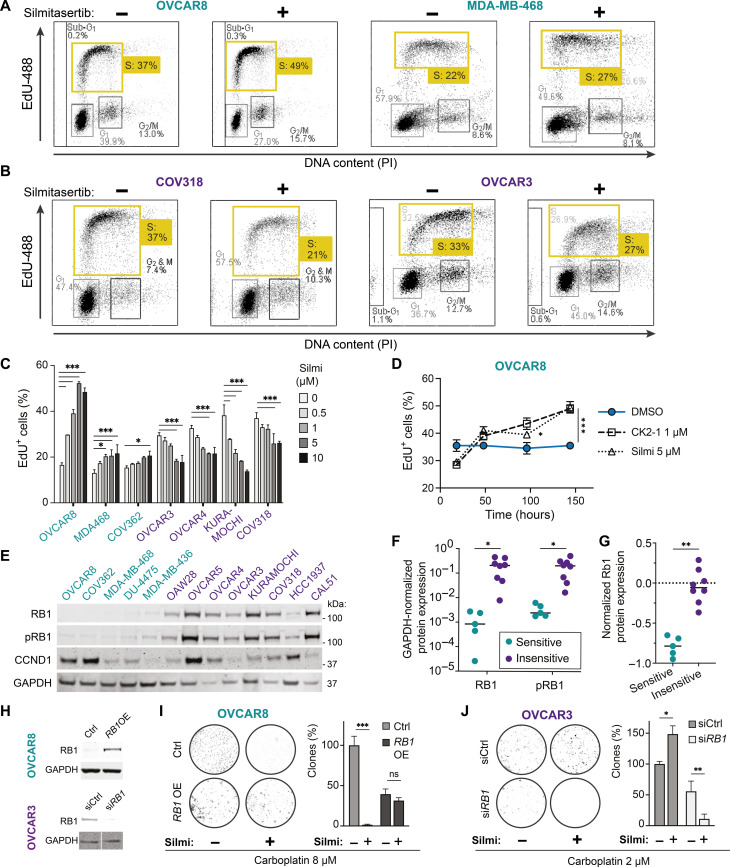
CK2 inhibition deregulates S phase progression in RB1-deficient cells
On the basis of the drug screening data, we hypothesized that CK2 differentially influenced cell cycle S phase progression in the cell lines vulnerable to the combinations of CK2 inhibitors with replication-affecting agents. To test this, we analyzed the cell cycle progression in CK2-inhibited cells by 5-ethynyl-2’-deoxyuridine (EdU) incorporation assay and mitotic index quantification. CK2 inhibition with silmitasertib or SGC-CK2-1 increased the EdU incorporation rate and the S phase cell fraction in OVCAR8, MDA-MB-468, and COV362 lines in flow cytometry and imaging assays [Fig. 3A; cerulean-shaded in Fig. 3C and fig. S4 (E to G)] and increased the mitotic index of OVCAR8 cells (fig. S4H). In contrast, in OVCAR3, COV318, OVCAR4, and KURAMOCHI, CK2 inhibition had a cytostatic effect reflected by a decrease in the fraction of EdU-incorporating cells and the intensity of EdU incorporation (Fig. 3B; violet-shaded in Fig. 3C and fig. S4E) and by a dose-dependent reduction in the mitotic index (fig. S4H). In the drug combination–sensitive OVCAR8 cell line, CK2 inhibition led to a time-dependent increase in the EdU-incorporating cell fraction, with the strongest difference at 144 hours of drug exposure (Fig. 3D); however, no substantial increase in the markers of replicative DNA damage [accumulation of γH2AX, nuclear replication protein A (RPA) loading, and Fanconi anemia group D2 protein (FANCD2) foci] (42, 43) was detected (figs. S5 and S6). These data indicated that CK2 kinase inhibition accelerated the S phase onset in the drug combination–sensitive cell lines (cerulean-shaded in Fig. 3), while it hindered the G1/S transition in the insensitive cell lines (violet-shaded in Fig. 3).
Fig. 3. The CK2 inhibition–dependent sensitization to carboplatin or niraparib in HGSC and TNBC cell lines depends on the loss of RB1 expression.
Cell lines were classified as sensitive (cerulean-shaded) or insensitive (violet-shaded) if they presented or did not present sensitization to carboplatin by silmitasertib (Figs. 1 and 2). (A and B) Flow cytometry analysis of the EdU incorporation. The cell lines were pretreated with silmitasertib or vehicle (72 hours), pulse-labeled with EdU (5 μM, 30 min), and immediately fixed and processed using Click-iT assay. (C) Quantification of the EdU incorporation imaging in the cell lines pretreated with silmitasertib for 72 hours. (D) Quantification of the EdU incorporation imaging in OVCAR8 treated with silmitasertib or SGC-CK2-1 for the indicated time. (E) Immunoblotting for RB1, phospho-RB1 (T608), and cyclin D1 expression in the cell lines. (F) Quantification of RB1 and phospho-RB1 immunoblotting in (E). (G) RPPA-based quantification of RB1 protein expression in cell lines from the CCLE study (44). Classification of the cell lines as in (E). (H) Immunoblotting analysis of RB1 expression in OVCAR8 cells transduced with TFORF1844 lentivector (top) and OVCAR3 transfected with 30 nM RB1-targeting siRNA (bottom). (I and J) Clonogenic survival of RB1-overexpressing OVCAR8 and RB1-depleted OVCAR3 cells. The cells were treated with indicated concetrations of carboplatin ± silmitasertib (5 or 2 μM for OVCAR8 or OVCAR3, respectively) or their combination for 48 hours and replated to six-well plates in drug-free media. The number of colonies in carboplatin-treated control transfectants is taken as 100%. For (C), (D), (I), and (J), the bars represent mean ± SD, n = 2, N = 2. *P < 0.05, **P < 0.01, and ***P < 0.001, two-way ANOVA with Tukey posttest. For (F) and (G), *P < 0.05 and **P < 0.01, Mann-Whitney test. PI, propidium iodide; GAPDH, glyceraldehyde phosphate dehydrogenase.
To identify molecular factors contributing to the differential effect of CK2 inhibition on cell cycle and therapeutics sensitivity, we analyzed the expression of G1/S and S phase regulators in HGSC and TNBC cell lines in publicly available gene expression datasets [Cancer Cell Line Encyclopedia (CCLE)] (44). We found differential expression of RB1 pathway genes stratifying the cell lines (fig. S4I), with a loss of RB1 expression characteristic for the cell lines vulnerable to the combination of CK2 inhibitors with replication-affecting agents. Protein expression analysis confirmed the low-to-absent RB1 protein level in the cell lines where CK2 inhibitors acted synergistically with carboplatin or PARPis (cerulean-shaded, Fig. 3, E and F). CCLE reverse-phase protein array (RPPA) data on RB1 expression confirmed our Western blot findings (Fig. 3G). No association with CK2 inhibition–mediated sensitization was observed for cyclin D1, another commonly deregulated G1/S transition mediator (Fig. 3E).
We assessed whether the expression of RB1 is sufficient to block the hypersensitivity to carboplatin or olaparib imparted by CK2 inhibition in RB1-deficient cells. Lentiviral delivery of RB1 open reading frame (ORF) to OVCAR8 and MDA-MB-468 line resulted in elevated RB1 protein level (Fig. 3H) and reverted the silmitasertib-mediated sensitization to carboplatin (Fig. 3I and fig. S4, J and K) and olaparib (fig. S4J). In RB1-proficient cell lines, RB1 mRNA depletion or heterozygous gene knockout increased the sensitivity to the combinations of CK2 inhibitors with carboplatin or olaparib (Fig. 3, H and J, and fig. S7, A to E), while the controls transfectants remained protected by the cytostatic effect of silmitasertib (fig. S7, C and E). Notably, depletion of BRCA2, a well-recognized determinant of PARPi and platinum drugs sensitivity, increased the sensitivity to niraparib but failed to deepen the response of RB1-proficient OVCAR3 and OVCAR4 lines to the combination of niraparib with silmitasertib (fig. S7G). Similarly, we observed no sensitization to the combinations in the HCC1937 breast cancer cell line (figs. S1H and S7H), which carries mutant breast cancer 1 gene (BRCA1) (table S4) but retains low level of RB1 protein expression (Fig. 3E). The findings indicated that the loss of RB1 in cancer cells created a specific context for a synthetic lethal dependence on CK2 in response to PARPis or carboplatin.
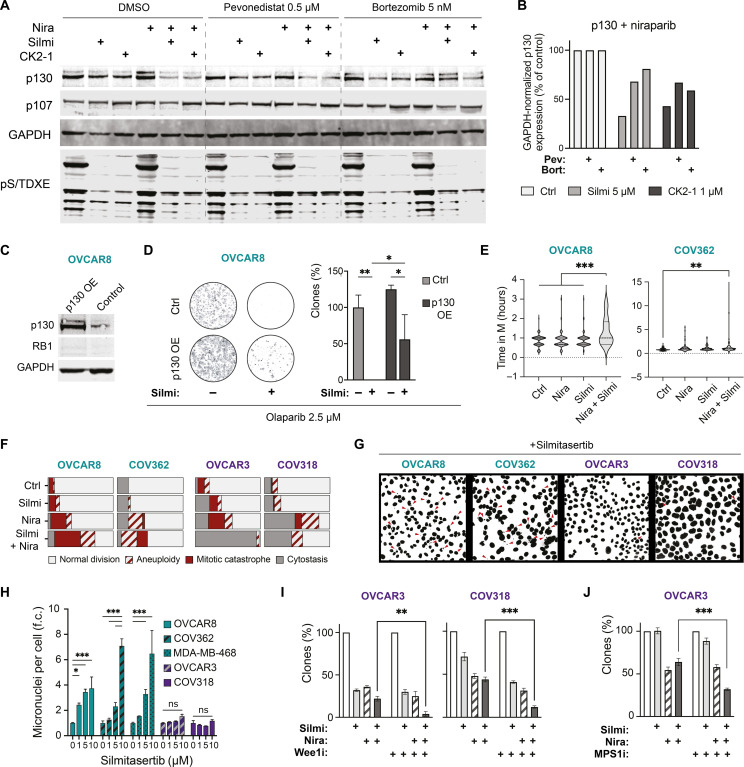
The RB1 family proteins, p107 and p130, act as redundant mediators of cell cycle control in RB1-deficient cells (45–47). Therefore, we analyzed how CK2 inhibition affected the p107 and p130 protein abundance in RB1-deficient OVCAR8 and RB1-proficient OVCAR3 cells upon PARP inhibition. We observed a 40 to 60% reduction in the level of p130 in the cells concurrently treated with niraparib and CK2 inhibitors in both cell lines (Fig. 4A and fig. S7F). At the same time, the p107 abundance remained unchanged in the niraparib-treated OVCAR8 (Fig. 4A). The inhibition of protein degradation by proteasome or neddylation inhibitors, bortezomib or pevonedistat, partly reverted the drop in the p130 protein level (Fig. 4, A and B). We concluded that CK2 inhibition accelerated p130 degradation following the PARP inhibition, hence contributing to further deregulation of the G1/S transition in RB1-deficient cells.
Fig. 4. CK2 inhibition triggers p130 degradation, mitotic cell death, and postmitotic aneuploidy in RB1-deficient HGSC cells.
(A) Immunoblotting for p107 and p130 abundance in OVCAR8 cells. Treatment: 1 μM niraparib ± 5 μM silmitasertib or 1 μM SGC-CK2-1 for 48 hours and DMSO, bortezomib, or pevonedistat for an additional 24 hours. pS/TDXE, CK2 phosphorylation motif. Image-wide nonlinear adjustments of the brightness of the Western blot membrane scan were applied for presentation clarity. (B) Quantification of p130 protein levels in niraparib-treated cells in (A). (C) Immunoblotting for p130 expression in TFORF1500 lentivector-expressing OVCAR8. (D) Clonogenic survival of p130-overexpressing (p130 OE) OVCAR8. Treatment: Olaparib ± 5 μM silmitasertib for 48 hours and then drug-free media for 7 days. The number of colonies in olaparib-treated control transfectants is taken as 100%. (E) Time from prometaphase until cytokinesis for the mitotic division resulting in two daughter cells. Treatment: Niraparib ± 5 μM silmitasertib, 48 hours before the start of the time-lapse imaging (every 15 min for 96 hours in total). Asynchronous cells. (F) Outcomes of mitoses in RB1-deficient (cerulean-shaded) and RB1-proficient (violet-shaded) HGSC cell lines in (E). Cells were considered cytostatic if no mitotic rounding and division were observed during 48 hours. N = 2, 50 to 60 cells per condition. (G) Representative images of Hoechst-stained nuclei of the indicated cell lines. Treatment: Silmitasertib, 120 hours. Red arrowheads indicate micronuclei. (H) Quantification of the imaging experiment in (G) presented as fold-change (f.c.) of the controls. At least 500 nuclei were counted for each condition. N = 2. (I and J) Clonogenic survival of RB1-proficient cell lines treated with silmitasertib and niraparib for 72 hours. Wee1 inhibitor adavosertib (0.3 μM) or MPS1 inhibitor AZ3146 (0.5 μM) was added for the last 18 hours of the drug treatment, followed by drug-free growth for 7 days. N = 2. For (D), (F), and (G), *P < 0.05, **P < 0.01, and ***P < 0.001, two-way ANOVA with Tukey posttest.
We explored whether the p130 expression changes had functional consequences for the sensitivity to the combinatorial PARP and CK2 inhibition. Ectopic expression of p130 inhibited the olaparib-sensitizing effect of CK2 inhibitors in a clonogenic survival test in RB1-deficient OVCAR8 cells (Fig. 4, C and D). Depletion of RBL2 transcript resulted in the up-regulation of RB1 protein in RB1-proficient cells and failed to sensitize to the combination (fig. S7, I and J). These data indicate that p130 functions to counteract the carboplatin- or PARPi-induced cytotoxicity upon CK2 inhibition in the absence of RB1.
CK2 inhibition exacerbates aneuploidy and mitotic cell death in niraparib-treated RB1-deficient cells
To determine whether concurrent CK2 and PARP inhibition triggers cell death in a particular cell cycle phase, we followed the cell division in RB1-deficient and RB1-proficient cells using time-lapse microscopy (movies S1 to S16). Exposure to niraparib alone for 48 hours elevated the incidence of mitotic cell death and aneuploidy in all tested cell lines (Fig. 4F and fig. S8), in agreement with the previous reports (48–50). In the presence of both silmitasertib and niraparib, RB1-deficient cells displayed an increase in the number of mitotic catastrophes and aneuploid divisions (Fig. 4F and fig. S8), indicating that mitotic aberrancies contribute to the increased cytotoxicity of the drug combination. Notably, we observed extended mitosis duration even for normal divisions resulting in two daughter cells in combination-treated RB1-deficient lines (Fig. 4E). The CK2 inhibition alone did not affect the mitotic outcome. However, it induced a dose-dependent accumulation of postmitotic micronuclei in RB1-deficient, but not in RB1-proficient, cell lines (Fig. 4, G and H). Similarly, CK2 inhibition exacerbated the micronucleation upon short-term carboplatin treatment in RB1-deficient cells (fig. S6C). Together, the data suggest that CK2 plays a role in maintaining cell division fidelity that, in the absence of RB1, becomes critical for surviving replication errors introduced by therapeutic agents.
To assess the role of cell cycle checkpoints in response to the CK2 and PARPi combination in RB1-proficient cells, we tested the sensitivity in the presence of cell cycle checkpoint inhibitors. Additional inhibition of Wee1 kinase following the silmitasertib + niraparib treatment sensitized RB1-proficient cell lines COV318 and OVCAR3 (Fig. 4I). The inhibition of spindle assembly checkpoint kinase MPS1 had a similar effect in OVCAR3 (Fig. 4J). We concluded that the activity of G2/M and M checkpoint kinases contributed to the survival of RB1-proficient cells upon concurrent CK2 and PARP inhibition.
Silmitasertib potentiates the long-term efficacy of carboplatin in patient-derived RB1-deficient HGSC organoids
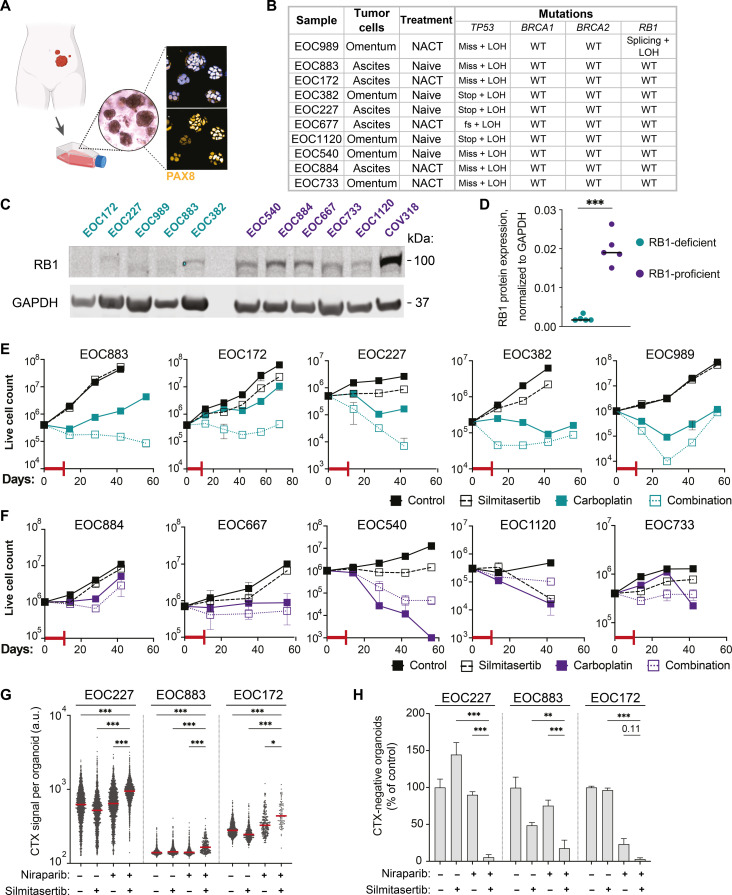
Testing drug responses in long-term HGSC ex vivo three-dimensional cultures, or organoids, is a powerful functional assay to predict the therapeutic efficacy in patients (51–53). To evaluate the translational potential of the carboplatin-silmitasertib combination, we assessed its efficacy in a set of 10 previously established, clinically relevant, BRCA wild-type and TP53-mutant HGSC patient–derived organoid cultures derived from ascites (N = 5) or omentum (N = 5) and collected at different stages of the treatment (Fig. 5, A and B) (54). First, we determined the RB1, BRCA1, and BRCA2 protein expression in actively growing, asynchronous organoids and classified them into RB1-deficient and RB1-proficient groups based on the relative RB1 protein expression (Fig. 5, C and D, and fig. S9, A and B). Next, we assessed the long-term survival and regrowth of the organoids after treatment with carboplatin ± silmitasertib. The combination of silmitasertib with carboplatin substantially attenuated the survival of RB1-deficient patient-derived organoids as compared to carboplatin alone (Fig. 5E). For three of five RB1-deficient patient samples, CK2 inhibition had a carboplatin-sensitizing effect during the whole experiment timeline, while for the remaining two samples, the effect was time point dependent (EOC989 and EOC382), reflecting sample-specific differences in carboplatin sensitivity. In contrast, silmitasertib did not enhance the carboplatin cytotoxicity in RB1-proficient samples, and, in one sample, it even protected the cells from the carboplatin cytotoxicity (Fig. 5F). Coherent with the genomic BRCA status of the organoids, BRCA1/2 protein expression did not significantly differ between responder and nonresponder cultures (fig. S9, A to C).
Fig. 5. Efficacy of silmitasertib in combination with carboplatin or niraparib in HGSC patient–derived organoids.
(A) Schematics of the establishment of the HGSC organoids (created with BioRender). Immunofluorescent imaging of PAX8, HGSC histologic marker (91), in organoids. (B) Characteristics of the selected set of organoids. NACT, neoadjuvant platinum-based chemotherapy. Mutations: fs, frameshift; stop, stop gain; miss, missense mutation; splicing, splicing isoform mutation; LOH, loss of heterozygosity; WT, wild type. (C) Immunoblotting analysis of RB1 protein expression in the set of organoids. (D) Quantification of the immunoblot in (C). (E and F) Long-term survival of RB1-deficient (cerulean-shaded) and RB1-proficient (violet-shaded) organoids. The organoids were exposed to the drugs alone or in combination for 7 days, followed by additional 4 days in the presence of a vehicle or silmitasertib alone (red T-line, drug treatment, 11 days in total). After the drug treatment, the organoids were passaged at the density of 5 × 105 live cells per 200 μl of the gel for 6 to 8 weeks. N = 2, mean 土 SD. (G) Quantification of the CellTox Green fluorescent signal per organoid. N = 2, n of organoids (30 to 300 per well) varied depending on the treatment. (H) Viability of RB1-deficient organoids in (H), the number of CellTox Green-negative (live) organoids expressed as the % of DMSO control. Plots present mean ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001, Mann-Whitney test for (C) and two-way ANOVA with Tukey posttest for (G) and (H). a.u., arbitrary units.
We further tested whether silmitasertib potentiated the cytotoxicity of niraparib in RB1-deficient patient-derived samples EOC172, EOC227, and EOC883. Confocal imaging–based quantification of live and dead organoids discriminated by CellTox Green staining showed a significantly lower survival after treatment with niraparib in the presence of silmitasertib (Fig. 5, G and H, and fig. S10), indicating that CK2 inhibition enhances the efficacy of the PARP inhibition in RB1-deficient patient-derived HGSC models. These data indicate that CK2 inhibition combined with standard therapeutics is a promising treatment strategy for RB1-deficient HGSC.
DISCUSSION
CRISPR screening offers an effective approach to systematically identify gene-drug interaction and for the identification of drivers of sensitivity or resistance to cancer therapeutics (23–25, 55–57). Our CRISPR screen study identified CSNK2A2 as a carboplatin sensitizer gene, refining the previous findings on the interaction between CK2 inhibitors and platinum drugs (29, 58, 59). Chemical inhibition of CK2 activity with two selective inhibitors confirmed the role of CK2 in carboplatin sensitivity, validating the phenotype identified in the screen. The combination of carboplatin with silmitasertib was effective in producing long-term responses in models that lacked RB1 tumor suppressor protein expression. Therefore, the identified specific interaction between the drug combination and RB1 deficiency can be classified as a context-specific synthetic lethality (19). In addition to CK2, we identified four kinases (STK19, STRAP, MYLK4, and ITPK1; Fig. 1 and table S1) that have not been implicated in carboplatin response before, therefore expanding the knowledge on targetable regulators of platinum resistance (24, 38, 60). STK19 was reported as a regulator of transcription-coupled nucleotide excision repair (38, 61), a mechanism of platinum-DNA adduct removal (62, 63). As STK19 can be targeted by the recently developed compound ZT-12-037-01 (64), our data also warrant the evaluation of the STK19 inhibitor as a carboplatin sensitizer in HGSC models in future studies.
Our study demonstrates that, in addition to chemotherapy, CK2 inhibition enhances the efficacy of the PARPis in RB1-deficient cell line models of breast, ovarian, and cervical cancer (Figs. 2 and 3 and figs. S4, A to D, and S7, A to E), while ectopic expression of RB1 reverted this phenotype (Fig. 3, H to J, and fig. S4, J and K). A remarkable synergy was seen between CK2 inhibitors and the PARP1-selective inhibitor saruparib (41) in RB1-deficient models, which warrants further investigation of the mechanistic role of CK2 in PARP1 activity regulation. In addition to PARPis and platinum drugs, we found CK2 activity critical for RB1-deficient cancer cell survival after treatment with replication poisons, such as illudin S or ATR kinase inhibitors (Fig. 3). It suggests a shared mechanism of toxicity for those drug combinations, which may involve recently reported interaction between CK2 and ATR at replication forks (65). CK2 inhibition did not affect the responses to hydroxyurea (HU) or CHK1 inhibitor, suggesting the differential impact of CK2 activity on different stages of G1/S and S phase checkpoint activation and stalled replication forks restart and requires further investigation. The lack of synergistic interaction with the several tested agents argues against the general sensitization to apoptosis by CK2 inhibition.
Our data illuminate a mechanism of regulation of RB1-deficient cell cycle progression and division outcomes by CK2 via preventing the proteasome-mediated degradation of another RB family protein, p130 (summarized on Fig. 6). Pronounced S phase deregulation upon CK2 inhibition in RB1-deficient cells evidenced by dose- and time-dependent accumulation of S phase (Fig. 3, A, C, and D) likely led to the dose-dependent formation of micronuclei (Fig. 4, E and F), indicative of the defects resulting from the mitotic entry upon incomplete replication (66–69). When RB1-deficient cells were challenged by PARP inhibition and trapping to chromatin, the concurrent inhibition of CK2 resulted in exacerbated mitotic infidelity as evidenced by extended mitosis, aneuploidy, and mitotic cell death (Fig. 4 and movies S1 to S16). The cells lacking both RB1 and p130 enter the S phase prematurely and have extended replication time (70). According to previous studies, the targeted degradation of p130 during S phase progression can be mediated by CK2-mediated cyclin F E2 ubiquitin ligase binding (71), while CK2 phosphorylates cyclin F and modifies its target-binding activity in a context-dependent manner (72). The observed mitotic phenotypes in RB1-deficient cells upon CK2 inhibition align with reports of replication stress-triggered mitotic cell death and postmitotic aberrations in cells lacking cell cycle checkpoints and/or DNA repair activity (13–15). Furthermore, inhibition of G2/M checkpoint kinase Wee1 (73) sensitized RB1-proficient ovarian cancer cells to the combination of silmitasertib with carboplatin or niraparib (Fig. 4G), indicating that intact cell cycle checkpoints activity can counteract the synergistic action of CK2 and PARPi combination.
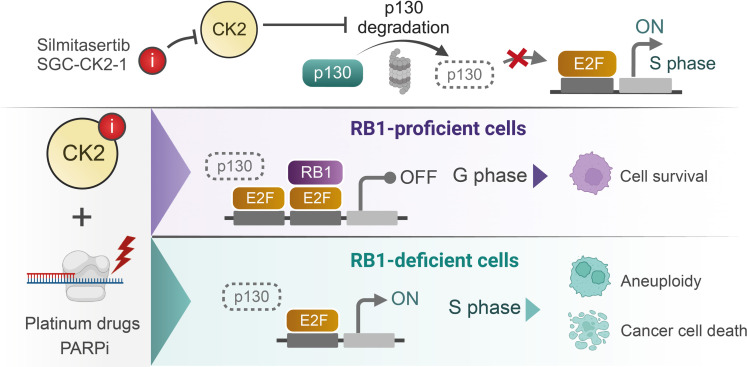
Fig. 6. A schematic putative mechanism of action of the CK2 inhibitors combined with replication-perturbing therapeutics.
Inhibition of CK2 kinase results in degradation of RB1 paralog p130, which contributes to deregulation of the S phase progression and triggers postmitotic aneuploidy and mitotic cell death in RB1-deficient cancer cells exposed to the carboplatin or PARPis (created with BioRender).
RB1 inactivation by deletions or mutations occurs in 15 to 25% of HGSCs (31, 32), 20 to 40% of basal-like breast cancers (3) (a gene expression–defined subtype closely corresponding to histologically defined TNBC), 37% of metastatic prostate cancers (74), and more than 90% of small cell lung cancers (75). In addition to genomic inactivation, human papillomavirus type 16 (HPV16) oncoprotein E7 inactivates RB1 protein, extending the functional RB1 deficiency to 50% of cervical cancers that are HPV16 positive (76). Hence, the CK2 inhibitor drug combination could be relevant for a substantial fraction of patients with cancer. To our knowledge, no molecular patient stratification has been done in clinical studies of silmitasertib, either as a single agent (clinical trials NCT04663737, NCT03897036, NCT03904862, NCT01199718, NCT00891280, NCT04663737, and NCT04668209) or in combination with chemotherapy (NCT02128282). Profiling RB1 deficiency in the tumors (either by DNA sequencing, transcriptomics, or immunohistochemistry) offers a promising strategy to stratify patients in future clinical studies of CK2 inhibitor combinatorial treatments and help retrospectively reevaluate the outcomes of earlier studies performed in nonstratified patient populations. RB1 deficiency in patient-derived ex vivo HGSC organoids stratifies the responses to silmitasertib combinations with platinum or niraparib. The tumor-derived organoids usually recapitulate the clinical responses of the tumor of origin (51, 53, 54, 77), and the effect of CK2 inhibition on RB1-deficient organoids suggests that the respective tumors could have shown sensitivity as well. Notably, two of the “responder” organoid cultures were established from chemotherapy-persisting tumor cells from tumor samples collected at interval debulking surgery after the patients had received neoadjuvant chemotherapy, highlighting that the combination could also benefit the treatment of platinum-resistant disease.
Silmitasertib is an orally administered drug candidate that has been tested in combination with platinum and gemcitabine in patients with metastatic cholangiocarcinoma with reported manageable toxicity (78). Given their tolerability in vivo (35, 78, 79), these drug combinations may be promising for overcoming chemotherapy resistance and producing durable responses in the subset of patients with RB1-deficient tumors. The safety of the CK2 inhibitor + PARPi combination has not been studied in clinical setting. However, a very recent study reported regression of mouse xenografts established from two RB1-deficient, BRCA wild-type cell lines and treated with silmitasertib combined with olaparib (80), suggesting tolerability and efficacy of the combination in vivo. The very fact that most healthy cells in the body have functional RB1 and therefore are expected to be insensitive to the silmitasertib therapeutic combinations argues in favor of a mild toxicity profile of such treatment. Collectively, our data suggest that the combination of a CK2 inhibitor with platinum drugs or PARPis should be considered for clinical testing as treatment of RB1-deficient tumors, which opens a possibility to expand the use of PARPis beyond BRCA-mutant cases. The example of CK2 inhibition potentiating the efficacy of the standard-of-care therapeutics, specifically in the context of RB1 deficiency, highlights the pressing need for better accounting for the tumor molecular landscape in drug combination research, biomarker discovery, and clinical study design.
MATERIALS AND METHODS
Cell lines
Cell lines and culture conditions are listed in table S4. The cells were maintained in the appropriate culture medium at 37°C with 5% CO2 in a humidified incubator. The identities of the cell lines were confirmed using GenePrint10 System (Promega) at the Institute for Molecular Medicine Finland (FIMM) Sequencing service. Cells were routinely tested for mycoplasma negativity using a MycoAlert kit (Lonza, #11600271).
CRISPR library production
LentiCas9-Blast vector was a gift from F. Zhang (Addgene, #52962). Human Brunello CRISPR knockout pooled library was a gift from D. Root and J. Doench (Addgene, #73178). The library was transformed into electrocompetent Lucigen Endura Escherichia coli (Lucigen; catalog no. 60242-2) using a Bio-Rad MicroPulser Electroporator (#1652100), program EC1 according to the manufacturer’s protocol. The electroporated bacteria were plated onto 10 15-cm LB agar plates with ampicillin (100 μg/ml). After overnight incubation at 32°C, the DNA plasmid was extracted using a NucleoBond Xtra Midi kit (MACHEREY-NAGEL, #740410.50). The transformation efficiency was controlled by plating 0.01% of the transformation reaction to a 15-cm LB agar plate with ampicillin (100 μg/ml).
Virus production
The human embryonic kidney 293-FT cells were seeded at ~105 cells/cm2 16 hours before transfection. Transfection of lenti-Guide-Puro plasmid with CRISPR library, packaging plasmids VSV-G and psPAX2 (Addgene, #14888 and #12260) was done using Lipofectamine 2000 transfection reagent according to the manufacturer’s protocol. The viral supernatant was collected 48 hours posttransfection, and the titer was assessed according to Stewart et al. (81).
Lentiviral transduction
Cells were incubated with the lentiviral supernatant for 24 hours in the presence of polybrene (8 μg/μl; Sigma-Aldrich) at a multiplicity of infection > 5 unless otherwise stated.
CRISPR screening
Cas9-expressing OVCAR8 cells were transduced with Brunello lentiviral knockout sgRNA library (4 sgRNAs per gene) (82) at a multiplicity of infection of 0.3. After puromycin selection, the transduced cells were propagated for 10 divisions to allow for the elimination of fitness gene-targeting sgRNAs. Next, the cell pool was divided to control and treatment pools; carboplatin at 8 μM was applied to the treatment pool for 72 hours, then the drug was washed off, and both control and carboplatin-treated pools were expanded for 10 cell doublings. A total of 8 × 106 cells were collected 3 days after infection (T0), 10 divisions after infection (T1), and 10 divisions after carboplatin treatment started (T2) for genomic DNA extraction and sgRNA amplification for sequencing.
Next-generation sequencing
For CRISPR screening, the genomic DNA was extracted using a NucleoSpin Tissue kit (MACHEREY-NAGEL, #740952.50) according to the manufacturer’s protocol. sgRNA cassettes were amplified from 2.5 μg of the template in 50-μl reactions using OneTaq DNA Polymerase (New England Biolabs, #M0480) and LG.Lib.ampl1.F and LG.Lib.ampl1.R primers (table S5). Illumina sequencing primer binding sites were added by polymerase chain reaction (PCR) amplification of sgRNA amplicons with primer mix WS Stager Mix and LG.gRNA.Ampl.NGS.R (table S5). Illumina indices and adapters for sample multiplexing were added by PCR amplification with Illumina_indX_F and Illumina_indX_R primers using NEBNext Ultra II Q5 Master Mix (New England Biolabs, #M0544). Samples were purified using AMPure XP beads (Beckman Coulter, #A63880). The library was sequenced with a NextSeq500 Illumina sequencer using the PE100 protocol (with 10% PhiX spike-in). Genomic profiling of the patient-derived HGSC organoids, the NGS, mutation, and copy number variation (CNV) calling were performed earlier and described by Senkowski and coauthors (54).
CRISPR screen analysis
Samples were demultiplexed using Illumina bcl2fastq to generate FASTQ files. Individual sgRNA counts were extracted using the count_spacers.py script (83). Positively selected genes were identified using the MAGeCK tool (34) and DESeq2(Wald) (84) using simplified routines provided by DEBRA R package (85). The overrepresentation and gene set enrichment analysis for GO-BP (Gene Ontology–biological process) and GO-CC (cellular component) terms were performed with clusterProfiler R package (86) using the top 150 genes with the following parameters pAdjustMethod = “BH,” pvalueCutoff = 0.25, qvalueCutoff = 0.25.
Clonogenic survival assay
Cells (3 to 5 × 104 per well) were plated in six-well plates in duplicates for each treatment. After 24 hours, the drugs at the indicated concentrations were added, and the cells were incubated for 5 days, followed by additional growth in a drug-free medium or in the presence of a CK2 inhibitor or vehicle for 3 days. After that, the cells were trypsinized, and 10, 25, or 50% of the sample was replated to the fresh drug-free medium in new plates for clonogenic growth for 10 more days, during which the medium was replenished twice. For the quantification, cells were fixed with ice-cold methanol:acetic acid mix (7:1) and stained with 0.1% crystal violet. The bright-field whole-well images of the wells were taken using Cytation5 imager (BioTek) and analyzed automatically using Gen5 software (BioTek) to count the number of clones per well.
Drug sensitivity screening
Drugs diluted in dimethyl sulfoxide (DMSO) or water were dispensed at 30-nl volume to 384-well black plates (Corning, #3864) using an Echo 550 acoustic liquid handler (Labcyte). The compounds were plated at five different concentrations in threefold dilutions covering a concentration range relevant for each drug. Cell-killing benzethonium chloride (BzCl, 100 μM) and compound vehicle (DMSO, 0.1%) were used as positive and negative controls, respectively. Cells were diluted to medium at the desired concentration, and the suspension was dispensed to the predrugged plates at 30 μl using a MultiFlo dispenser (BioTek). After 7 days of incubation at 37°C, 10 μl of phosphate-buffered saline (PBS) containing Hoechst (4 μg/ml) and 1/10,000 CellTox Green Dye (Promega) was dispensed per well 1 hour before imaging at a Cytation5 image cytometer or an Opera Phenix (PerkinElmer) confocal screening microscope.
Organoid cultures
Previously established long-term HGSC organoid cultures (54) were expanded in BME-2 Cultrex gel for at least two passages before drug sensitivity testing to get actively proliferating cultures. The media, passaging, and dissociation were done as described (54).
Drug sensitivity testing on organoids
For long-term drug survival assay, dissociated organoid cells (5 × 105 to 106) were seeded to BME-2 gel (Cultrex, Bio-Techne) at the density of 2.5 to 5 × 103 cells/μl and allowed to take up for 4 days in the presence of 5 μM Rho-associated protein kinase (ROCK) kinase inhibitor. After ROCK kinase inhibitor was removed, the organoids were exposed to carboplatin or silmitasertib alone or in combination for 7 days, followed by additional 4 days in the presence of vehicle or silmitasertib alone. After drug treatment, the organoids were dissociated, and live cells were counted using the Trypan blue exclusion method and the Countess II cell counter (Invitrogen). Live cells (3 to 6 × 105 or all surviving cells) were passaged to fresh BME-2 droplets at the same density. The passaging and live cell counting were repeated in a similar manner biweekly for 6 to 8 weeks. The experiment was performed in duplicates.
For short-term drug sensitivity assay, dissociated organoid cells (2 to 5 × 104) were seeded in 10 μl of BME-2 gel droplets (Cultrex, Bio-Techne), one droplet per well in 96-well Cell Carrier Ultra plates (PerkinElmer) and allowed to take up for 4 days in media containing 5 μM ROCK kinase inhibitor. Next, the medium was changed to 200 μl of medium containing niraparib, silmitasertib, or the combination. Medium and drugs were replenished after 4 days. After 7 days since drug addition, the medium was changed to drug-free, and after another 7 days of incubation at 37°C, 20 μl of PBS containing Hoechst 33342 (10 μg/ml) and 1/4000 CellTox Green Dye (Promega) was dispensed per well 8 hours before imaging at an Opera Phenix (PerkinElmer) confocal screening microscope. Image analysis was performed on the maximum projection image of 10 whole-well z-planes of the confocal imaging using Harmony software (PerkinElmer).
Drug combination analysis
To assess the effect of the drug combination treatments, we applied the Bliss independence model (87). Bliss scores were calculated for each drug combination based on the 4 × 4 dose-response matrix, and the Bliss synergy score values in the most synergistic area were calculated by SynergyFinder 2.0 (88), with values higher than 8 considered as synergy and negative values considered as antagonism.
Gene editing
Parental Cas9 cell lines were generated by viral infection with lentiCas9-Blast (Addgene, #52962), followed by blasticidin selection for 7 days. sgRNAs for targeting CSNK2A1, CSNK2A2, and RB1 (table S6) were cloned to lentiGuide-Puro vector (Addgene, #52963) and delivered by lentiviral transduction to the Cas9-expressing cells followed by puromycin selection (1 μg/ml) for 5 days. The efficiency of the knockouts was validated by immunoblotting.
RB1 and RBL2 overexpression
Lentiviral vectors TORF1844 and TORF1500, engineered to carry RB1 and RBL2 ORFs in the pLX_TRC317 backbone (89), were gift from F. Zhang and ordered via Addgene (#144313 and #143960, respectively). OVCAR8 and MDA-MB-468 cells (105 in 1 ml of medium) were infected with 0.1 ml of the lentiviral supernatant of each construct in the presence of polybrene (8 μg/ml) overnight, and the selection of the transduced cells was done using puromycin (1 μg/ml) for 5 days. Selected clones were pooled, expanded, and tested for the expression of the targets.
RNA interference
Small interfering RNAs (siRNAs) against RBL2 (SI02664473 and SI02664480), RB1 (SI00007091 and SI02653819), and AllStarNegative control siRNA (QIAGEN) were transfected at the final concentration of 30 nM using Lipofectamine RNAiMAX (Thermo Fisher Scientific) according to the manufacturer’s instructions. After 24 hours, the medium was changed, and the transfectants were treated as stated in the figure legends.
Flow cytometry cell cycle analysis
EdU incorporation assay was done using a Click-IT kit (Thermo Fisher Scientific, #C10337) according to the manufacturer’s instructions. Cells were labeled with 5 μM EdU for 30 min. For flow cytometry, DNA staining was done with propidium iodide in the presence of ribonuclease A (100 μg/ml) for 1 hour at 37°C. At least 10,000 events per sample were analyzed on an Accuri C6 Plus (Becton Dickinson). Data were analyzed using Accuri C6 Plus Sampler software.
Immunostaining and confocal imaging
Antibodies used for immunostaining are listed in table S7. Cells were preextracted with 0.35% Triton X-100 in PBS on ice, fixed with 4% paraformaldehyde, and, after three washes with PBS, incubated in the blocking buffer (PBS, 0.5% bovine serum albumin, 20 mM glycine, and 0.05% Triton X-100) for 1 hour at room temperature. For immunostaining combined with EdU detection, the click reaction was performed according to a Cilck-IT kit (Thermo Fisher Scientific, #C10337) instruction, after which the wells were washed with blocking buffer and processed for antibody incubation. Immunostaining was performed overnight at 4°C. After three washes with the blocking buffer, the secondary antibodies were applied for 1 hour. After washing off the secondary antibodies, counterstaining with Hoechst 33342 was done at 10 μg/ml in PBS for 5 min. The cells were imaged at an Opera Phenix confocal screening microscope (PerkinElmer) with the 40× water immersion objective.
Mitotic timing and outcomes analysis
Cells were seeded to 12-well plates and incubated with the indicated drugs for 48 hours. After that, the plates were placed in the Incucyte ZOOM equipped with a 10× objective. The plates were imaged with 15- to 20-min intervals in the presence of the drugs for an additional consecutive 48 hours. The individual cells’ fates were traced from the image time-lapse sequences. The mitotic time was counted from characteristic cell rounding in anaphase till separation of two daughter cells; or, in case of aneuploidy, in spreading of one bi-/multinuclear cell without division; or, in case of cell death, in the mitotic cell vacuolization, membrane blebbing, detachment, followed by disappearance from the field of view.
Immunoblotting
Cells were lysed in radioimmunoprecipitation assay buffer without EDTA supplemented with Pierce protease and phosphatase inhibitors cocktail (Thermo Fisher Scientific) and benzonase (20 U/ml; Millipore) on ice for 30 min. The samples were centrifuged for 15 min at 17,000×g at 4°C, and the supernatants were processed for loading to bis-tris 4 to 12% gradient Bolt polyacrylamide gels using Bolt loading dye solution and reducing agents and electrophoresis in MES running buffer (all from Thermo Fisher Scientific) according to the manufacturer’s instructions. Protein transfer to the nitrocellulose membranes was done overnight in Towbin transfer buffer, after which the membranes were stained with Ponceau Red to detect the protein transfer efficiency, washed with tris-buffered saline–0.05% Tween-20 (TBS-T), and blocked in 5% nonfat milk in TBS-T. The membranes were incubated with primary antibodies listed in table S8 at the indicated dilutions in 5% nonfat milk in TBS-T overnight at 4°C except for glyceraldehyde phosphate dehydrogenase and β-actin antibodies (1 hour at room temperature). After three washes in TBS-T, the membranes were incubated with the secondary fluorophore-conjugated antibodies. Primary and secondary antibodies and their working dilutions used for immunoblotting are listed in table S7. The membranes were scanned in a LiCor Odyssey imager, and the fluorescence intensities of bands were quantified in ImageLite software. Immunoblotting experiments were repeated twice. Representative images are shown.
Statistics and reproducibility
Statistical analyses were conducted using GraphPad Prism. For the nonparametric comparison of two groups of data, the Mann-Whitney test was applied. For multiple comparisons, statistical significance (adjusted P values) was calculated using the two-way analysis of variance (ANOVA) with the Tukey multiple comparisons test. Results are reported as nonsignificant at P > 0.05 and with increasing degrees of significance: *0.01 < P ≤ 0.05, **0.001 < P ≤ 0.01, and ***P ≤ 0.001.
Acknowledgments
We thank the staff of the Biomedicum Helsinki Flow Cytometry Unit, High-Content Analysis Unit, High-Throughput Biomedicine Unit, and Genomics Unit at the FIMM for providing research infrastructure and excellent technical help. We thank the HERCULES consortium for providing access to molecularly characterized HGSC patient samples. We thank C. Sørensen and his team (University of Copenhagen) for fruitful discussions that shaped the experimental design and helped data interpretation. Figures 5A and 6 were created with BioRender.
Funding: This work was supported by European Union’s Horizon 2020 research and innovation program grant 667403 (HERCULES) to D.B., Y.A., J.O., S.H., J.H., T.A., and K.W.; European Union’s Horizon 2020 research and innovation program grant 965193 (DECIDER) to S.H. and J.H.; European Union’s Horizon 2020 research and innovation program grant 845045 (RESIST3D) to W.S.; Danish Cancer Society grant R204-A12322 to W.S.; Danish Cancer Society grant R302-A17398 to K.W.; Novo Nordisk Foundation Center for Stem Cell Biology grant NNF17CC0027852 to D.B. and K.W.; Novo Nordisk Foundation Infrastructure grant number NNF20OC0061734 to K.W.; Novo Nordisk Foundation Interdisciplinary Synergy Programme 2021 grant NNF21OC0070381 to K.W. and T.A.; Innovation Fund Denmark/ERA PerMed JTC2020 grant 0204-00005B (PARIS) to K.W.; Cancer Society of Finland to T.A.; Sigrid Jusélius Foundation to T.A.; and Academy of Finland (grants 326238, 340141, 345803, and 344698) to T.A.
Author contributions: Conceptualization: D.B., Y.A., T.A., and K.W. Methodology: D.B., W.S., Y.A., T.A., K.W., and S.H. Investigation: D.B., W.S., Y.A., J.O., L.G.-M., M.E., S.T., and J.H. Visualization: D.B. Software: Y.A. Formal analysis: D.B., Y.A., J.O., and T.A. Data curation: J.O. and J.H. Supervision: T.A., K.W., and S.H. Validation: D.B. and S.H. Writing—original draft: D.B. Writing—review and editing: D.B., Y.A., W.S., L.G.-M., S.T., M.E., J.O., J.H., S.H., T.A., and K.W. Resources: W.S., D.B., T.A., K.W., S.H., and J.H. Funding acquisition: W.S., D.B., T.A., K.W., and J.H. Project administration: K.W.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Legends for movies S1 to S16
Legend for data file S1
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S16
Data file S1
REFERENCES AND NOTES
- 1.Bowtell D. D., Böhm S., Ahmed A. A., Aspuria P.-J., Bast R. C., Beral V., Berek J. S., Birrer M. J., Blagden S., Bookman M. A., Brenton J. D., Chiappinelli K. B., Martins F. C., Coukos G., Drapkin R., Edmondson R., Fotopoulou C., Gabra H., Galon J., Gourley C., Heong V., Huntsman D. G., Iwanicki M., Karlan B. Y., Kaye A., Lengyel E., Levine D. A., Lu K. H., McNeish I. A., Menon U., Narod S. A., Nelson B. H., Nephew K. P., Pharoah P., Powell D. J., Ramos P., Romero I. L., Scott C. L., Sood A. K., Stronach E. A., Balkwill F. R., Rethinking ovarian cancer II: Reducing mortality from high-grade serous ovarian cancer. Nat. Rev. Cancer 15, 668–679 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bianchini G., De Angelis C., Licata L., Gianni L., Treatment landscape of triple-negative breast cancer —expanded options, evolving needs. Nat. Rev. Clin. Oncol. 19, 91–113 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Network , Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tutt A., Robson M., Garber J. E., Domchek S. M., Audeh M. W., Weitzel J. N., Friedlander M., Arun B., Loman N., Schmutzler R. K., Wardley A., Mitchell G., Earl H., Wickens M., Carmichael J., Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: A proof-of-concept trial. Lancet 376, 235–244 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Audeh M. W., Audeh M. W., Carmichael J., Penson R. T., Friedlander M., Powell B., Bell-McGuinn K. M., Scott C., Weitzel J. N., Oaknin A., Loman N., Lu K., Schmutzler R. K., Matulonis U., Wickens M., Tutt A., Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: A proof-of-concept trial. Lancet 376, 245–251 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Rottenberg S., Disler C., Perego P., The rediscovery of platinum-based cancer therapy. Nat. Rev. Cancer 21, 37–50 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Annunziato S., Barazas M., Rottenberg S., Jonkers J., Genetic dissection of cancer development, therapy response, and resistance in mouse models of breast cancer. Cold Spring Harb. Symp. Quant. Biol. 81, 141–150 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Ang J. E., Gourley C., Powell C. B., High H., Shapira-Frommer R., Castonguay V., De Greve J., Atkinson T., Yap T. A., Sandhu S., Banerjee S., Chen L.-M., Friedlander M. L., Kaufman B., Oza A. M., Matulonis U., Barber L. J., Kozarewa I., Fenwick K., Assiotis I., Campbell J., Chen L., de Bono J. S., Gore M. E., Lord C. J., Ashworth A., Kaye S. B., Efficacy of chemotherapy in BRCA1/2 mutation carrier ovarian cancer in the setting of PARP inhibitor resistance: A multi-institutional study. Clin. Cancer Res. 19, 5485–5493 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Plunkett W., Huang P., Gandhi V., Preclinical characteristics of gemcitabine. Anticancer Drugs 6 (Suppl 6), 7–13 (1995). [DOI] [PubMed] [Google Scholar]
- 10.Rabik C. A., Eileen Dolan M., Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat. Rev. 33, 9–23 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pommier Y., O’Connor M. J., de Bono J., Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci. Transl. Med. 8, 362ps17 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Valabrega G., Scotto G., Tuninetti V., Pani A., Scaglione F., Differences in PARP inhibitors for the treatment of ovarian cancer: Mechanisms of action, pharmacology, safety, and efficacy. Int. J. Mol. Sci. 22, 4203 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schoonen P. M., Talens F., Stok C., Gogola E., Heijink A. M., Bouwman P., Foijer F., Tarsounas M., Blatter S., Jonkers J., Rottenberg S., van Vugt M. A. T. M., Progression through mitosis promotes PARP inhibitor-induced cytotoxicity in homologous recombination-deficient cancer cells. Nat. Commun. 8, 15981 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim H., George E., Ragland R., Rafail S., Zhang R., Krepler C., Morgan M., Herlyn M., Brown E., Simpkins F., Targeting the ATR/CHK1 axis with PARP inhibition results in tumor regression inbrca-mutant ovarian cancer models. Clin. Cancer Res. 23, 3097–3108 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang Y., McGrail D. J., Sun C., Labrie M., Chen X., Zhang D., Ju Z., Vellano C. P., Lu Y., Li Y., Jeong K. J., Ding Z., Liang J., Wang S. W., Dai H., Lee S., Sahni N., Mercado-Uribe I., Kim T.-B., Chen K., Lin S.-Y., Peng G., Westin S. N., Liu J., O’Connor M. J., Yap T. A., Mills G. B., Sequential therapy with PARP and WEE1 inhibitors minimizes toxicity while maintaining efficacy. Cancer Cell 35, 851–867.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elbæk C. R., Petrosius V., Sørensen C. S., WEE1 kinase limits CDK activities to safeguard DNA replication and mitotic entry. Mutat. Res. 819-820, 111694 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Li S., Wang L., Wang Y., Zhang C., Hong Z., Han Z., The synthetic lethality of targeting cell cycle checkpoints and PARPs in cancer treatment. J. Hematol. Oncol. 15, 147 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaelin W. G., The concept of synthetic lethality in the context of anticancer therapy. Nat. Rev. Cancer 5, 689–698 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Akimov Y., Aittokallio T., Re-defining synthetic lethality by phenotypic profiling for precision oncology. Cell Chem. Biol. 28, 246–256 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Mirza M. R., Coleman R. L., González-Martín A., Moore K. N., Colombo N., Ray-Coquard I., Pignata S., The forefront of ovarian cancer therapy: Update on PARP inhibitors. Ann. Oncol. 31, 1148–1159 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Han K., Jeng E. E., Hess G. T., Morgens D. W., Li A., Bassik M. C., Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat. Biotechnol. 35, 463–474 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Behan F. M., Iorio F., Picco G., Gonçalves E., Beaver C. M., Migliardi G., Santos R., Rao Y., Sassi F., Pinnelli M., Ansari R., Harper S., Jackson D. A., McRae R., Pooley R., Wilkinson P., van der Meer D., Dow D., Buser-Doepner C., Bertotti A., Trusolino L., Stronach E. A., Saez-Rodriguez J., Yusa K., Garnett M. J., Prioritization of cancer therapeutic targets using CRISPR–Cas9 screens. Nature 568, 511–516 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Dai M., Yan G., Wang N., Daliah G., Edick A. M., Poulet S., Boudreault J., Ali S., Burgos S. A., Lebrun J.-J., In vivo genome-wide CRISPR screen reveals breast cancer vulnerabilities and synergistic mTOR/Hippo targeted combination therapy. Nat. Commun. 12, 3055 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He Y. J., Meghani K., Caron M.-C., Yang C., Ronato D. A., Bian J., Sharma A., Moore J., Niraj J., Detappe A., Doench J. G., Legube G., Root D. E., D’Andrea A. D., Drané P., De S., Konstantinopoulos P. A., Masson J.-Y., Chowdhury D., DYNLL1 binds to MRE11 to limit DNA end resection in BRCA1-deficient cells. Nature 563, 522–526 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y., Yang C., Liu Z., Du S., Can S., Zhang H., Zhang L., Huang X., Xiao Z., Li X., Fang J., Qin W., Sun C., Wang C., Chen J., Chen H., Integrative analysis of CRISPR screening data uncovers new opportunities for optimizing cancer immunotherapy. Mol. Cancer 21, 2 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borgo C., D’Amore C., Sarno S., Salvi M., Ruzzene M., Protein kinase CK2: A potential therapeutic target for diverse human diseases. Signal Transduct. Target. Ther. 6, 183 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kulbe H., Iorio F., Chakravarty P., Milagre C. S., Moore R., Thompson R. G., Everitt G., Canosa M., Montoya A., Drygin D., Braicu I., Sehouli J., Saez-Rodriguez J., Cutillas P. R., Balkwill F. R., Integrated transcriptomic and proteomic analysis identifies protein kinase CK2 as a key signaling node in an inflammatory cytokine network in ovarian cancer cells. Oncotarget 7, 15648–15661 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hessenauer A., Schneider C. C., Götz C., Montenarh M., CK2 inhibition induces apoptosis via the ER stress response. Cell. Signal. 23, 145–151 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Siddiqui-Jain A., Bliesath J., Macalino D., Omori M., Huser N., Streiner N., Ho C. B., Anderes K., Proffitt C., O’Brien S. E., Lim J. K. C., Von Hoff D. D., Ryckman D. M., Rice W. G., Drygin D., CK2 inhibitor CX-4945 suppresses DNA repair response triggered by DNA-targeted anticancer drugs and augments efficacy: Mechanistic rationale for drug combination therapy. Mol. Cancer Ther. 11, 994–1005 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Schilder R. J., Hall L., Monks A., Handel L. M., Fornace A. J. Jr., Ozols R. F., Fojo A. T., Hamilton T. C., Metallothionein gene expression and resistance to cisplatin in human ovarian cancer. Int. J. Cancer 45, 416–422 (1990). [DOI] [PubMed] [Google Scholar]
- 31.Patch A.-M., Christie E. L., Etemadmoghadam D., Garsed D. W., George J., Fereday S., Nones K., Cowin P., Alsop K., Bailey P. J., Kassahn K. S., Newell F., Quinn M. C. J., Kazakoff S., Quek K., Wilhelm-Benartzi C., Curry E., Leong H. S.; The Australian Ovarian Cancer Study Group, Hamilton A., Mileshkin L., Au-Yeung G., Kennedy C., Hung J., Chiew Y.-E., Harnett P., Friedlander M., Quinn M., Pyman J., Cordner S., O’Brien P., Leditschke J., Young G., Strachan K., Waring P., Azar W., Mitchell C., Traficante N., Hendley J., Thorne H., Shackleton M., Miller D. K., Arnau G. M., Tothill R. W., Holloway T. P., Semple T., Harliwong I., Nourse C., Nourbakhsh E., Manning S., Idrisoglu S., Bruxner T. J. C., Christ A. N., Poudel B., Holmes O., Anderson M., Leonard C., Lonie A., Hall N., Wood S., Taylor D. F., Xu Q., Fink J. L., Waddell N., Drapkin R., Stronach E., Gabra H., Brown R., Jewell A., Nagaraj S. H., Markham E., Wilson P. J., Ellul J., McNally O., Doyle M. A., Vedururu R., Stewart C., Lengyel E., Pearson J. V., Waddell N., deFazio A., Grimmond S. M., Bowtell D. D. L., Whole–genome characterization of chemoresistant ovarian cancer. Nature 521, 489–494 (2015). [DOI] [PubMed] [Google Scholar]
- 32.The Cancer Genome Atlas Research Network , Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tumiati M., Hietanen S., Hynninen J., Pietilä E., Färkkilä A., Kaipio K., Roering P., Huhtinen K., Alkodsi A., Li Y., Lehtonen R., Erkan E. P., Tuominen M. M., Lehti K., Hautaniemi S. K., Vähärautio A., Grénman S., Carpén O., Kauppi L., A functional homologous recombination assay predicts primary chemotherapy response and long-term survival in ovarian cancer patients. Clin. Cancer Res. 24, 4482–4493 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Li W., Xu H., Xiao T., Cong L., Love M. I., Zhang F., Irizarry R. A., Liu J. S., Brown M., Liu X. S., MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pierre F., Chua P. C., O’Brien S. E., Siddiqui-Jain A., Bourbon P., Haddach M., Michaux J., Nagasawa J., Schwaebe M. K., Stefan E., Vialettes A., Whitten J. P., Chen T. K., Darjania L., Stansfield R., Anderes K., Bliesath J., Drygin D., Ho C., Omori M., Proffitt C., Streiner N., Trent K., Rice W. G., Ryckman D. M., Discovery and SAR of 5-(3-chlorophenylamino)benzo[c][2,6]naphthyridine-8-carboxylic acid (CX-4945), the first clinical stage inhibitor of protein kinase CK2 for the treatment of cancer. J. Med. Chem. 54, 635–654 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Lertsuwan J., Sawasdichai A., Tasnawijitwong N., Gaston K., Jayaraman P.-S., Satayavivad J., Sequence of CX-4945 and cisplatin administration determines the effectiveness of drug combination and cellular response in cholangiocarcinoma cells in vitro. Anticancer Res 41, 6155–6167 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Borgo C., Ruzzene M., Role of protein kinase CK2 in antitumor drug resistance. J. Exp. Clin. Cancer Res. 38, 287 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olivieri M., Cho T., Álvarez-Quilón A., Li K., Schellenberg M. J., Zimmermann M., Hustedt N., Rossi S. E., Adam S., Melo H., Heijink A. M., Sastre-Moreno G., Moatti N., Szilard R. K., McEwan A., Ling A. K., Serrano-Benitez A., Ubhi T., Feng S., Pawling J., Delgado-Sainz I., Ferguson M. W., Dennis J. W., Brown G. W., Cortés-Ledesma F., Williams R. S., Martin A., Xu D., Durocher D., A genetic map of the response to DNA damage in human cells. Cell 182, 481–496.e21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wells C. I., Drewry D. H., Pickett J. E., Tjaden A., Krämer A., Müller S., Gyenis L., Menyhart D., Litchfield D. W., Knapp S., Axtman A. D., Development of a potent and selective chemical probe for the pleiotropic kinase CK2. Cell Chem. Biol. 28, 546–558.e10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murai J., Huang S.-Y. N., Das B. B., Renaud A., Zhang Y., Doroshow J. H., Ji J., Takeda S., Pommier Y., Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 72, 5588–5599 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Illuzzi G., Staniszewska A. D., Gill S. J., Pike A., McWilliams L., Critchlow S. E., Cronin A., Fawell S., Hawthorne G., Jamal K., Johannes J., Leonard E., Macdonald R., Maglennon G., Nikkilä J., O’Connor M. J., Smith A., Southgate H., Wilson J., Yates J., Cosulich S., Leo E., Preclinical characterization of AZD5305, A next-generation, highly selective PARP1 inhibitor and trapper. Clin. Cancer Res. 28, 4724–4736 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toledo L., Neelsen K. J., Lukas J., Replication catastrophe: When a checkpoint fails because of exhaustion. Mol. Cell 66, 735–749 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Schlacher K., Wu H., Jasin M., A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 22, 106–116 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghandi M., Huang F. W., Jané-Valbuena J., Kryukov G. V., Lo C. C., McDonald E. R., Barretina J., Gelfand E. T., Bielski C. M., Li H., Hu K., Andreev-Drakhlin A. Y., Kim J., Hess J. M., Haas B. J., Aguet F., Weir B. A., Rothberg M. V., Paolella B. R., Lawrence M. S., Akbani R., Lu Y., Tiv H. L., Gokhale P. C., de Weck A., Mansour A. A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J. M., Porter D. A., Jones M. D., Golji J., Caponigro G., Taylor J. E., Dunning C. M., Creech A. L., Warren A. C., McFarland J. M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y. E., Cherniack A. D., Tsherniak A., Vazquez F., Jaffe J. D., Lane A. A., Weinstock D. M., Johannessen C. M., Morrissey M. P., Stegmeier F., Schlegel R., Hahn W. C., Getz G., Mills G. B., Boehm J. S., Golub T. R., Garraway L. A., Sellers W. R., Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sage J., Mulligan G. J., Attardi L. D., Miller A., Chen S., Williams B., Theodorou E., Jacks T., Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev. 14, 3037–3050 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Witkiewicz A. K., Kumarasamy V., Sanidas I., Knudsen E. S., Cancer cell cycle dystopia: Heterogeneity, plasticity, and therapy. Trends Cancer 8, 711–725 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burkhart D. L., Sage J., Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat. Rev. Cancer 8, 671–682 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dale Rein I., Solberg Landsverk K., Micci F., Patzke S., Stokke T., Replication-induced DNA damage after PARP inhibition causes G2 delay, and cell line-dependent apoptosis, necrosis and multinucleation. Cell Cycle 14, 3248–3260 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schoonen P. M., Kok Y. P., Wierenga E., Bakker B., Foijer F., Spierings D. C. J., van Vugt M. A. T. M., Premature mitotic entry induced by ATR inhibition potentiates olaparib inhibition-mediated genomic instability, inflammatory signaling, and cytotoxicity in BRCA2-deficient cancer cells. Mol. Oncol. 13, 2422–2440 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slade D., Mitotic functions of poly(ADP-ribose) polymerases. Biochem. Pharmacol. 167, 33–43 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hill S. J., Decker B., Roberts E. A., Horowitz N. S., Muto M. G., Worley M. J., Feltmate C. M., Nucci M. R., Swisher E. M., Nguyen H., Yang C., Morizane R., Kochupurakkal B. S., Do K. T., Konstantinopoulos P. A., Liu J. F., Bonventre J. V., Matulonis U. A., Shapiro G. I., Berkowitz R. S., Crum C. P., D’Andrea A. D., Prediction of DNA repair inhibitor response in short-term patient-derived ovarian cancer organoids. Cancer Discov. 8, 1404–1421 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ooft S. N., Weeber F., Dijkstra K. K., McLean C. M., Kaing S., van Werkhoven E., Schipper L., Hoes L., Vis D. J., van de Haar J., Prevoo W., Snaebjornsson P., van der Velden D., Klein M., Chalabi M., Boot H., van Leerdam M., Bloemendal H. J., Beerepoot L. V., Wessels L., Cuppen E., Clevers H., Voest E. E., Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci. Transl. Med. 11, eaay2574 (2019). [DOI] [PubMed] [Google Scholar]
- 53.de Witte C. J., Espejo Valle-Inclan J., Hami N., Lõhmussaar K., Kopper O., Vreuls C. P. H., Jonges G. N., van Diest P., Nguyen L., Clevers H., Kloosterman W. P., Cuppen E., Snippert H. J. G., Zweemer R. P., Witteveen P. O., Stelloo E., Patient-derived ovarian cancer organoids mimic clinical response and exhibit heterogeneous inter- and intrapatient drug responses. Cell Rep. 31, 107762 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Senkowski W., Gall-Mas L., Falco M. M., Li Y., Lavikka K., Kriegbaum M. C., Oikkonen J., Bulanova D., Pietras E. J., Voßgröne K., Chen Y. J., Erkan E. P., Dai J., Lundgren A., Mk G. H., Larsen I. M., Lamminen T., Kaipio K., Huvila J., Virtanen A., Engelholm L., Christiansen P., Santoni-Rugiu E., Huhtinen K., Carpén O., Hynninen J., Hautaniemi S., Vähärautio A., Wennerberg K., A platform for efficient establishment and drug-response profiling of high-grade serous ovarian cancer organoids. Dev. Cell 58, 1106–1121.e7 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Palit S. A. L., van Dorp J., Vis D., Lieftink C., Linder S., Beijersbergen R., Bergman A. M., Zwart W., van der Heijden M. S., A kinome-centered CRISPR-Cas9 screen identifies activated BRAF to modulate enzalutamide resistance with potential therapeutic implications in BRAF-mutated prostate cancer. Sci. Rep. 11, 13683 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soares F., Chen B., Lee J. B., Ahmed M., Ly D., Tin E., Kang H., Zeng Y., Akhtar N., Minden M. D., He H. H., Zhang L., CRISPR screen identifies genes that sensitize AML cells to double-negative T-cell therapy. Blood 137, 2171–2181 (2021). [DOI] [PubMed] [Google Scholar]
- 57.Xu S., Zhan M., Jiang C., He M., Yang L., Shen H., Huang S., Huang X., Lin R., Shi Y., Liu Q., Chen W., Mohan M., Wang J., Genome-wide CRISPR screen identifies ELP5 as a determinant of gemcitabine sensitivity in gallbladder cancer. Nat. Commun. 10, 5492 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zakharia K., Miyabe K., Wang Y., Wu D., Moser C. D., Borad M. J., Roberts L. R., Preclinical in vitro and in vivo evidence of an antitumor effect of CX-4945, a casein kinase II inhibitor in cholangiocarcinoma. Transl. Oncol. 12, 143–153 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trembley J. H., Li B., Kren B. T., Gravely A. A., Caicedo-Granados E., Klein M. A., Ahmed K., CX-4945 and siRNA-mediated knockdown of CK2 improves cisplatin response in HPV(+) and HPV(−) HNSCC cell lines. Biomedicine 9, 571 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nagel R., Avelar A. T., Aben N., Proost N., van de Ven M., van der Vliet J., Cozijnsen M., de Vries H., Wessels L. F. A., Berns A., Inhibition of the replication stress response is a synthetic vulnerability in SCLC that acts synergistically in combination with cisplatin. Mol. Cancer Ther. 18, 762–770 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boeing S., Williamson L., Encheva V., Gori I., Saunders R. E., Instrell R., Aygün O., Rodriguez-Martinez M., Weems J. C., Kelly G. P., Conaway J. W., Conaway R. C., Stewart A., Howell M., Snijders A. P., Svejstrup J. Q., Multiomic analysis of the UV-induced DNA damage response. Cell Rep. 15, 1597–1610 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zamble D. B., Mu D., Reardon J. T., Sancar A., Lippard S. J., Repair of cisplatin−DNA adducts by the mammalian excision nuclease. Biochemistry 35, 10004–10013 (1996). [DOI] [PubMed] [Google Scholar]
- 63.Reardon J. T., Vaisman A., Chaney S. G., Sancar A., Efficient nucleotide excision repair of cisplatin, oxaliplatin, and Bis-aceto-ammine-dichloro-cyclohexylamine-platinum(IV) (JM216) platinum intrastrand DNA diadducts. Cancer Res. 59, 3968–3971 (1999). [PubMed] [Google Scholar]
- 64.Yin C., Zhu B., Zhang T., Liu T., Chen S., Liu Y., Li X., Miao X., Li S., Mi X., Zhang J., Li L., Wei G., Xu Z. X., Gao X., Huang C., Wei Z., Goding C. R., Wang P., Deng X., Cui R., Pharmacological targeting of STK19 inhibits oncogenic NRAS-driven melanomagenesis. Cell 176, 1113–1127.e16 (2019). [DOI] [PubMed] [Google Scholar]
- 65.Guerra B., Doktor T. K., Frederiksen S. B., Somyajit K., Andresen B. S., Essential role of CK2α for the interaction and stability of replication fork factors during DNA synthesis and activation of the S-phase checkpoint. Cell. Mol. Life Sci. 79, 339 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Burrell R. A., McClelland S. E., Endesfelder D., Groth P., Weller M.-C., Shaikh N., Domingo E., Kanu N., Dewhurst S. M., Gronroos E., Chew S. K., Rowan A. J., Schenk A., Sheffer M., Howell M., Kschischo M., Behrens A., Helleday T., Bartek J., Tomlinson I. P., Swanton C., Replication stress links structural and numerical cancer chromosomal instability. Nature 494, 492–496 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Feng W., Jasin M., BRCA2 suppresses replication stress-induced mitotic and G1 abnormalities through homologous recombination. Nat. Commun. 8, 525 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wilhelm T., Magdalou I., Barascu A., Técher H., Debatisse M., Lopez B. S., Spontaneous slow replication fork progression elicits mitosis alterations in homologous recombination-deficient mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 111, 763–768 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Palmerola K. L., Amrane S., De Los Angeles A., Xu S., Wang N., de Pinho J., Zuccaro M. V., Taglialatela A., Massey D. J., Turocy J., Robles A., Subbiah A., Prosser B., Lobo R., Ciccia A., Koren A., Baslan T., Egli D., Replication stress impairs chromosome segregation and preimplantation development in human embryos. Cell 185, 2988–3007.e20 (2022). [DOI] [PubMed] [Google Scholar]
- 70.Salama S., Gorka C., Mulloy R., Braun P., Harlow E., Combinatorial roles for pRB, p107, and p130 in E2F-mediated cell cycle control. Proc. Natl. Acad. Sci. U.S.A. 97, 10820 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Enrico T. P., Stallaert W., Wick E. T., Ngoi P., Wang X., Rubin S. M., Brown N. G., Purvis J. E., Emanuele M. J., Cyclin F drives proliferation through SCF-dependent degradation of the retinoblastoma-like tumor suppressor p130/RBL2. eLife 10, e70691 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Salvi J. S., Kang J., Kim S., Colville A. J., de Morrée A., Billeskov T. B., Larsen M. C., Kanugovi A., van Velthoven C. T. J., Cimprich K. A., Rando T. A., ATR activity controls stem cell quiescence via the cyclin F–SCF complex. Proc. Natl. Acad. Sci. U.S.A. 119, e2115638119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.di Rorà A. G. L., Cerchione C., Martinelli G., Simonetti G., A WEE1 family business: Regulation of mitosis, cancer progression, and therapeutic target. J. Hematol. Oncol. 13, 126 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Taylor B. S., Schultz N., Hieronymus H., Gopalan A., Xiao Y., Carver B. S., Arora V. K., Kaushik P., Cerami E., Reva B., Antipin Y., Mitsiades N., Landers T., Dolgalev I., Major J. E., Wilson M., Socci N. D., Lash A. E., Heguy A., Eastham J. A., Scher H. I., Reuter V. E., Scardino P. T., Sander C., Sawyers C. L., Gerald W. L., Integrative genomic profiling of human prostate cancer. Cancer Cell 18, 11–22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.George J., Lim J. S., Jang S. J., Cun Y., Ozretić L., Kong G., Leenders F., Lu X., Fernández-Cuesta L., Bosco G., Müller C., Dahmen I., Jahchan N. S., Park K.-S., Yang D., Karnezis A. N., Vaka D., Torres A., Wang M. S., Korbel J. O., Menon R., Chun S.-M., Kim D., Wilkerson M., Hayes N., Engelmann D., Pützer B., Bos M., Michels S., Vlasic I., Seidel D., Pinther B., Schaub P., Becker C., Altmüller J., Yokota J., Kohno T., Iwakawa R., Tsuta K., Noguchi M., Muley T., Hoffmann H., Schnabel P. A., Petersen I., Chen Y., Soltermann A., Tischler V., Choi C.-M., Kim Y.-H., Massion P. P., Zou Y., Jovanovic D., Kontic M., Wright G. M., Russell P. A., Solomon B., Koch I., Lindner M., Muscarella L. A., la Torre A., Field J. K., Jakopovic M., Knezevic J., Castaños-Vélez E., Roz L., Pastorino U., Brustugun O.-T., Lund-Iversen M., Thunnissen E., Köhler J., Schuler M., Botling J., Sandelin M., Sanchez-Cespedes M., Salvesen H. B., Achter V., Lang U., Bogus M., Schneider P. M., Zander T., Ansén S., Hallek M., Wolf J., Vingron M., Yatabe Y., Travis W. D., Nürnberg P., Reinhardt C., Perner S., Heukamp L., Büttner R., Haas S. A., Brambilla E., Peifer M., Sage J., Thomas R. K., Comprehensive genomic profiles of small cell lung cancer. Nature 524, 47–53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dyson N., Howley P. M., Münger K., Harlow E., The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243, 934–937 (1989). [DOI] [PubMed] [Google Scholar]
- 77.Maenhoudt N., Defraye C., Boretto M., Jan Z., Heremans R., Boeckx B., Hermans F., Arijs I., Cox B., Van Nieuwenhuysen E., Vergote I., Van Rompuy A.-S., Lambrechts D., Timmerman D., Vankelecom H., Developing organoids from ovarian cancer as experimental and preclinical models. Stem Cell Rep. 14, 717–729 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Borad M. J., Bai L.-Y., Chen M.-H., Hubbard J. M., Mody K., Rha S. Y., Richards D. A., Lindsey Davis S., Soong J., Huang C.-E. C.-E., Tse E., Ahn D. H., Chang H.-M., Yen C.-J., Oh D.-Y., Park J. O., Hsu C., Becerra C. R., Chen J.-S., Chen Y.-Y., Silmitasertib (CX-4945) in combination with gemcitabine and cisplatin as first-line treatment for patients with locally advanced or metastatic cholangiocarcinoma: A phase Ib/II study. J. Clin. Oncol. 39, 312 (2021). [Google Scholar]
- 79.Battistutta R., Cozza G., Pierre F., Papinutto E., Lolli G., Sarno S., O’Brien S. E., Siddiqui-Jain A., Haddach M., Anderes K., Ryckman D. M., Meggio F., Pinna L. A., Unprecedented selectivity and structural determinants of a new class of protein kinase CK2 inhibitors in clinical trials for the treatment of cancer. Biochemistry 50, 8478–8488 (2011). [DOI] [PubMed] [Google Scholar]
- 80.Wang C., Tian L., He Q., Lin S., Wu Y., Qiao Y., Zhu B., Li D., Chen G., Targeting CK2-mediated phosphorylation of p53R2 sensitizes BRCA-proficient cancer cells to PARP inhibitors. Oncogene 42, 2971–2984 (2023). [DOI] [PubMed] [Google Scholar]
- 81.Stewart S. A., Dykxhoorn D. M., Palliser D., Mizuno H., Yu E. Y., An D. S., Sabatini D. M., Chen I. S., Hahn W. C., Sharp P. A., Weinberg R. A., Novina C. D., Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9, 493–501 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Doench J. G., Fusi N., Sullender M., Hegde M., Vaimberg E. W., Donovan K. F., Smith I., Tothova Z., Wilen C., Orchard R., Virgin H. W., Listgarten J., Root D. E., Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Joung J., Konermann S., Gootenberg J. S., Abudayyeh O. O., Platt R. J., Brigham M. D., Sanjana N. E., Zhang F., Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat. Protoc. 12, 828–863 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Akimov Y., Bulanova D., Timonen S., Wennerberg K., Aittokallio T., Improved detection of differentially represented DNA barcodes for high-throughput clonal phenomics. Mol. Syst. Biol. 16, e9195 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu G., Wang L.-G., Han Y., He Q.-Y., clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bansal M., Yang J., Karan C., Menden M. P., Costello J. C., Tang H., Xiao G., Li Y., Allen J., Zhong R., Chen B., Kim M., Wang T., Heiser L. M., Realubit R., Mattioli M., Alvarez M. J., Shen Y., Gallahan D., Singer D., Saez-Rodriguez J., Xie Y., Stolovitzky G., Califano A., A community computational challenge to predict the activity of pairs of compounds. Nat. Biotechnol. 32, 1213–1222 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ianevski A., Giri A. K., Aittokallio T., SynergyFinder 2.0: Visual analytics of multi-drug combination synergies. Nucleic Acids Res. 48, W488–W493 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Joung J., Ma S., Tay T., Geiger-Schuller K. R., Kirchgatterer P. C., Verdine V. K., Guo B., Arias-Garcia M. A., Allen W. E., Singh A., Kuksenko O., Abudayyeh O. O., Gootenberg J. S., Fu Z., Macrae R. K., Buenrostro J. D., Regev A., Zhang F., A transcription factor atlas of directed differentiation. Cell 186, 209–229.e26 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xie Z., Bailey A., Kuleshov M. V., Clarke D. J. B., Evangelista J. E., Jenkins S. L., Lachmann A., Wojciechowicz M. L., Kropiwnicki E., Jagodnik K. M., Jeon M., Ma’ayan A., Gene set knowledge discovery with enrichr. Curr. Protoc. 1, e90 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mhawech-Fauceglia P., Wang D., Samrao D., Godoy H., Ough F., Liu S., Pejovic T., Lele S., Pair Box 8 (PAX8) protein expression in high grade, late stage (stages III and IV) ovarian serous carcinoma. Gynecol. Oncol. 127, 198–201 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Legends for movies S1 to S16
Legend for data file S1
Movies S1 to S16
Data file S1