Abstract
The endoplasmic reticulum (ER) is divided into rough and smooth domains (RER and SER). The two domains share most proteins, but RER is enriched in some membrane proteins by an unknown mechanism. We studied RER protein targeting by expressing fluorescent protein fusions to ER membrane proteins in Caenorhabditis elegans. In several cell types RER and general ER proteins colocalized, but in neurons RER proteins were concentrated in the cell body, whereas general ER proteins were also found in neurites. Surprisingly RER membrane proteins diffused rapidly within the cell body, indicating they are not localized by immobilization. Ribosomes were also concentrated in the cell body, suggesting they may be in part responsible for targeting RER membrane proteins.
INTRODUCTION
The endoplasmic reticulum (ER) is an extensive intracellular membrane system. It is important for a number of cellular functions including translocation of secretory proteins across the membrane, insertion of membrane proteins, lipid synthesis, calcium storage and signaling, and separation of nucleoplasm from cytoplasm. Its structure varies depending on cell type. Often two domains, rough and smooth ER (RER and SER), can be distinguished. Although this distinction has been noted for many years, nothing is known about how proteins are targeted to the two domains.
In animal cells the ER forms a network that extends throughout the cell, and in several different cell types this network has been shown to be continuous. In one kind of experiment, green fluorescent protein (GFP) fused to a membrane protein that was localized in the ER, or GFP targeted to the lumen of the ER, could be bleached from the entire cell by repeatedly exposing a part of the cell to intense laser light (Cole et al., 1996; Subramanian and Meyer, 1997; Dayel et al., 1999). The rapidity of bleaching suggested that the proteins are freely diffusible in a continuous membrane network. In a different kind of experiment, fluorescent dye from an oil droplet diffused from directly contacted membranes into a continuous membrane network, which extended throughout both sea urchin eggs and Purkinje neurons, and is most likely the ER (Terasaki and Jaffe, 1991; Terasaki et al., 1994). In view of this continuity it is interesting to understand how domains within the ER might be established.
SER and RER were initially identified by electron microscopy; the RER is decorated with ribosomes, whereas the SER is not. Although the membranes often look quite different, they were classified as domains of the same organelle because connections between the two types of membrane were observed (cf. Fawcett, 1981). RER must be present in all cells because in all cells nascent proteins are inserted into the membrane from ER-bound ribosomes. SER is prominent in certain cell types, such as liver, steroid-synthesizing cells, muscle, and neurons. The relationship between SER and RER composition has been best studied in liver tissue, where the two types of membranes can be separated by biochemical fractionation. Subsequent analysis of their enzyme activities and protein composition indicated that most proteins present in one domain are also found in the other (Depierre and Dallner, 1975; Kreibich et al., 1978). The major exception to the generalization that RER and SER have the same protein composition is the enrichment of several membrane proteins in the RER (Kreibich et al., 1978). ER membrane proteins can thus be divided between those that are concentrated in the RER, RER membrane proteins, and those that are not, general ER proteins. By fractionation of liver cells, ribophorins I and II (components of the oligosaccharyl transferase) were found to be enriched in the RER (Kreibich et al., 1978), as was a subunit of signal peptidase and TRAPα (SSRα; Vogel et al., 1990) and Sec61α (Meyer et al., 2000). The common feature of these proteins is that they are involved either in translocation of proteins across the ER membrane (Rapoport et al., 1996) or in their modification during translocation. Several studies have suggested that another membrane protein involved in the translocation process, the SRP-receptor, is not restricted to the RER (Tajima et al., 1986; Vogel et al., 1990). Thus, some, but perhaps not all, membrane proteins involved in translocation of newly synthesized proteins across the ER membrane are highly concentrated in the RER.
The basic questions about RER protein localization are unresolved, in part because most experiments have been performed with fractionated liver. It is not known how evolutionarily conserved the targeting of RER membrane proteins within the ER might be. Nor is it known how general the phenomenon is between cell types: are RER membrane proteins targeted to a subregion of the ER membrane only in liver and a few other cell types, or is their localization a general feature of all cells? It is also not clear whether RER membrane proteins are in fact localized in live cells or only in mechanically disrupted cells. In two cases RER membrane proteins have been observed by immunoelectron microscopy to be localized within the ER (Hortsch et al., 1985; Vogel et al., 1990), but again the cells were severely perturbed before examination. The mechanism by which RER membrane proteins are localized also remains unknown. Several models have been suggested but not tested. For example RER membrane proteins have been proposed to be interconnected by a filamentous network that would allow them to segregate into portions of the ER (Kreibich et al., 1978; Ivessa et al., 1992). It has also been proposed that the linkage of translocation proteins to the ribosome would restrict their diffusion and allow them to be localized (Vogel et al., 1990). Alternatively a selective diffusion barrier could exist between RER and SER.
Protein targeting to the nuclear envelope (NE), a different ER domain, has been studied, and may be instructive for thinking about RER membrane protein localization (for a review of ER domains, see Baumann and Walz, 2001). The NE is a double-membrane structure in which the outer membrane is connected to the peripheral ER and the inner membrane is connected to the outer at the nuclear pore. In animal cells it is distinguished from the rest of the ER by nuclear pores and a set of membrane proteins enriched in the inner NE. For the lamin B receptor (LBR) the NE targeting domain was shown to be present in the nucleoplasmic portion of the protein (Soullam and Worman, 1993). The same region of the protein contains determinants for binding lamins (Worman et al., 1988; Ye and Worman, 1994). These observations led to the proposal that LBR is synthesized in the peripheral ER like other membrane proteins and then diffuses throughout the ER until entering the inner NE, where it binds to lamins. The binding of LBR to nuclear proteins would serve to concentrate it in the inner NE (Soullam and Worman, 1993, 1995). This model has since gained support from studies of the diffusional mobility of NE membrane proteins. In contrast to general ER membrane proteins which diffuse very rapidly, NE membrane proteins are essentially immobile (Ellenberg et al., 1997; Östlund et al., 1999; Rolls et al., 1999), most likely because of the binding interaction which is responsible for concentrating them in the NE. RER membrane proteins could be similarly immobilized and localized by a binding partner.
In this study we establish a system to examine RER membrane protein localization in live cells. We expressed fluorescent protein (FP, variants of GFP) fusions to membrane proteins in Caenorhabditis elegans and observed their localization in a variety of cell types in live worms. In several cell types tagged RER and general ER proteins colocalized. However in neurons RER markers, and ribosomes, were concentrated in the cell body while general ER markers were present in both the cell body and neurites. We found that the mobility of RER membrane proteins in the cell body was high compared with NE membrane proteins, indicating that RER membrane proteins are not localized by immobilization. We consider models for RER membrane protein localization in light of the unexpected mobility of these proteins.
MATERIALS AND METHODS
C. elegans Culture and Transgenic Lines
C. elegans were grown according to standard methods (Lewis and Fleming, 1995). Transgenic worm lines were constructed by injecting DNA into the gonad of young adult worms (Mello and Fire, 1995). For all worm lines 4 μg/ml of each expression plasmid, generally a cyan FP (CFP)-encoding plasmid and a yellow FP (YFP)-encoding one, were mixed in water with 92 μg/ml pRF4 DNA, which was used as an array marker (Mello and Fire, 1995). Roller worms were maintained by repeated selection of the phenotype. Several transgenic lines were made for each construct, and the one expressing a low amount of protein was chosen for analysis to minimize mislocalization due to overexpression.
Plasmid Construction
A set of vectors to make C- and N- terminal CFP and YFP protein fusions was created from the Fire lab vectors (www.ciwemb.edu). The parent plasmid was pPD122.13. Nuclear localization signals were excised from this vector with KpnI. Next the GFP was replaced with CFP, PCR amplified from pPD136.61 or YFP from pPD136.64 using the KpnI and NheI sites. During this PCR step, polylinkers were added at the N- or C-terminus of the CFP or YFP coding sequence. To make fusion proteins with the FP at the N terminus of the protein, the FP was amplified with CTAAA before the start codon of the FP. At the 3′ end of the FP coding sequence the stop codon was omitted, and the following sequence was added in frame with the FP coding sequence: 5′GGCGGGGGACTCGACACGCGTATGCATCCCGGGAGATCTGGCGCGCC3′. This sequence adds a flexible linker containing several glycines as well as restriction sites in which to insert coding sequences. The two vectors generated were called pCN and pYN (for C/YFP at the N-terminus). Vectors to fuse the FP to the C-terminus of proteins were also created. The following sequence was added upstream of the FP start codon: 5′GGCGCGCCATGCATAGATCTCCCGGGACGCGTGGCGGGG-GACTCGACGGC3′. Again cloning sites and a flexible linker were added. In this case the FP coding region was amplified with the stop codon intact. The vectors generated were pYC and pCC.
To these basic vectors, promoters were added into the polylinker present from the starting vector. The rpl-28 promoter was PCR amplified from Fire lab vector pPD129.57. The myo-3 promoter was PCR amplified from pPD136.61 The glr-1 promoter was PCR amplified from plasmid pKP6 (Hart et al., 1995). The dpy-7 promoter was PCR amplified from genomic DNA based on Gilleard et al. (1997); the amplified region corresponded to nucleotides 567–781 in locus CEDPY7. Each promoter was cloned into the four fusion vectors so that there were sets of vectors, for example, pgYC, pgYN, pgCC, pgCN, all of which contained a particular promoter, in this case glr-1.
To make plasmids that expressed FP-fusion proteins in worms, genomic coding regions, amplified from cosmids or genomic DNA, of predicted proteins were inserted into vectors like pgYC. Complete coding regions were used except for Golgi markers. The sequence names for the predicted ER proteins are listed in Figure 3D. One of the ER proteins was not predicted in WormBase, but a sequence very similar to TRAPγ was present on cosmid Y69A2. The location of the tag is also indicated in Figure 3D. For the NE marker, full-length emerin (M01D7.6) was PCR amplified and inserted into the pYC series of vectors such that the FP would be fused to the C-terminus of the protein. Similarly the Golgi marker, mans (for mannosidase-short) was tagged at the C-terminus with the FP. In this case, however, only a short region of the coding sequence was PCR amplified. The fusion protein is predicted to contain 82 amino acids from the sequence F58H1.1 fused to YFP. The plasma membrane marker YFP-GPI was constructed by inserting a signal sequence upstream of YFP and a GPI-anchoring sequence downstream. The signal sequence and GPI anchor sequence were kindly provided by Joachim Füllekrug (Max-Planck-Institute of Molecular Cell Biology and Genetics).
Figure 3.
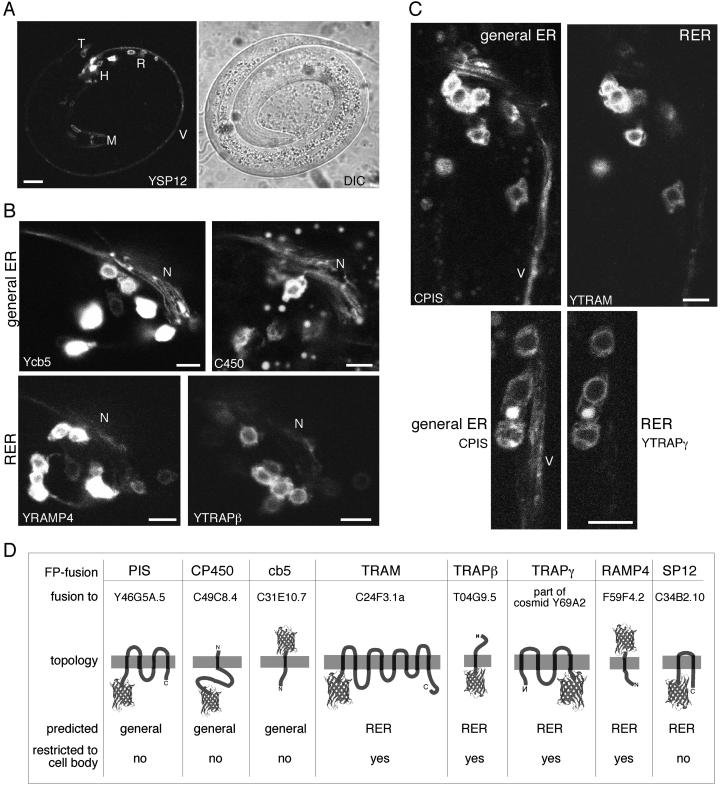
The localization of different ER membrane proteins in neurons is distinct. (A) YFP-SP12 (YSP12) was expressed under control of the glr-1 promoter. Muscle cells in the nose of the worm (M, cells at the tip of the nose on either side) as well as some other head cells express the FP. Fluorescence is also seen in neurons in several head ganglia (H) near the nerve ring, in the retrovesicular ganglion (R), in tail ganglia (T), and in neurites that project along the ventral nerve cord (V). (B) Nerve rings of worms expressing different predicted FP-ER markers were imaged. The predicted general ER markers shown are YFP-cytochrome b5 (Ycb5) and CFP-cytochrome P450 (C450), and predicted RER membrane proteins shown are YFP-RAMP4 (YRAMP4) and YFP-TRAPβ (YTRAPβ). Neurites sweeping across the nerve ring are marked N. (C) CFP-PIS (PIS) was coexpressed with YFP-TRAM (top panel) and YFP-TRAPγ (bottom panel). Neurites in the ventral nerve cord are labeled V. (D) Summary of predicted FP-ER membrane proteins tested. FP fusions were to the full-length genomic region of the gene listed. The FP is represented by the barrel structure. The predicted topologies are shown with the lumen at the top of the diagram. Scale bars are 5 μm, except that in A, which is 10 μm.
Confocal Microscopy
For observation, C. elegans were mounted on 2% agarose pads on glass slides in 10 μl 0.1% tetramisole/1% tricaine in M9. A coverslip was placed on top, excess agarose was cut away, and the coverslip was sealed with nail polish. Worms were observed between 10 and 60 min after mounting. Generally L2 or L3 worms were analyzed.
Microscopy was performed using the Compact Confocal Camera (CCC) at the European Molecular Biology Laboratory. CFP was excited with a 430-nm laser, and YFP with a 514-nm laser as described in White et al. (1999). Frame interlace collection was used for most images, except Figure 2B for which line interlace collection was used. Images were taken using a 63× 1.4 NA Plan-Apochromat DIC objective (Carl Zeiss, Thornwood, NY), and processed using NIH Image 1.62 and Canvas 6 (Deneba Systems, Inc., Miami, FL).
Figure 2.
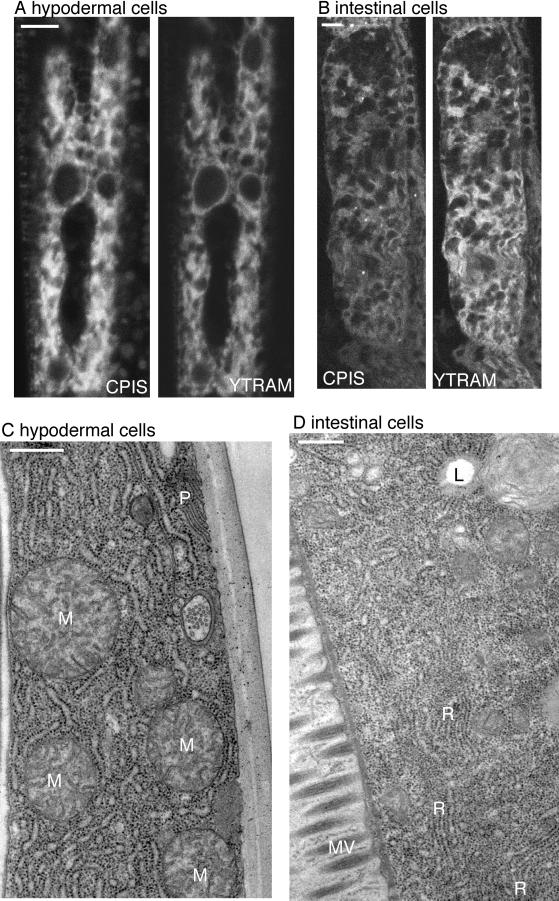
Distribution of ER membrane proteins, and the ER itself, in hypodermal and intestinal cells. (A) CFP-PIS and YFP-TRAM were expressed in hypodermal cells and imaged by confocal microscopy. (B) CFP-PIS (CPIS) and YFP-TRAM (YTRAM) were imaged in intestinal cells by confocal microscopy. Several regions of the cell appear wavy because of movement of the worm during imaging. (C) A transverse section of a worm was examined by electron microscopy after immersion fixation. A portion of a hypodermal cell is shown. The cytoplasm is filled with RER and free ribosomes. M, mitochondria; P, an infolding of the plasma membrane. The hypodermal cell is bounded on the right by cuticle. (D) A portion of an intestinal cell from a transverse section of a worm viewed by electron microscopy and fixed as in C is shown. MV, microvilli in the lumen of the intestine; L, a probable lipid droplet; and R, stacked regions of RER. Scale bars: A and B, 5 μm; C and D, 0.5 μm.
Photobleaching
Fluorescence recovery after photobleaching (FRAP) experiments were also performed using the CCC set up as described for imaging. A single bleach scan at full laser power and integration time of 250 μs was used for most experiments. For YFP bleaching either a 20/80 universal beamsplitter or a specific CFP/YFP beamsplitter was used with similar results. For CFP bleaching the CFP/YFP beamsplitter was used and integration time was 100 μs. Images of the whole cell were collected before the bleach, immediately after, and every 10 s thereafter. Quantitation was performed using NIH Image 1.62. Total pixel intensity was summed in a background region of the image, part of the bleached region of the cell, and part of the unbleached region for each time point. Background was subtracted from both bleached and unbleached values, and then the ratio of bleached to unbleached was taken for each time point and divided by the initial prebleach ratio to correct for difference in intensity between regions of the cell. Simulations of FRAP experiments were performed using Virtual Cell (Schaff et al., 2000; http://www.nrcam.uchc.edu/). A computer program that analyzes diffusion in complex structures (Siggia et al., 2000) was also used to determine diffusion coefficients for some data sets.
Electron Microscopy
General TEM methods have been described previously (Hall, 1995). To accentuate the staining of ribosomes within neuronal cytoplasm, several fast freezing methods were explored, followed by freeze substitution and plastic embedment (cf. McDonald, 1998; Williams-Masson et al., 1998). Briefly, we used either metal mirror fixation or high-pressure freezing to quickly immobilize live animals on a piece of filter paper, inside a sealed piece of flexible dialysis tubing, or in a slurry of yeast or Escherichia coli. The frozen samples were then slowly exposed to a primary fixative of osmium tetroxide in methanol or acetone and, while still kept very cold, dehydrated through solvents, and infiltrated with plastic resin. After curing, the animals were thin-sectioned and poststained for TEM by conventional means. Microscopy was done using a Philips CM10 electron microscope (Mahwah, NJ).
RESULTS
C. elegans Can Be Used to Study ER Proteins in Live Differentiated Cells
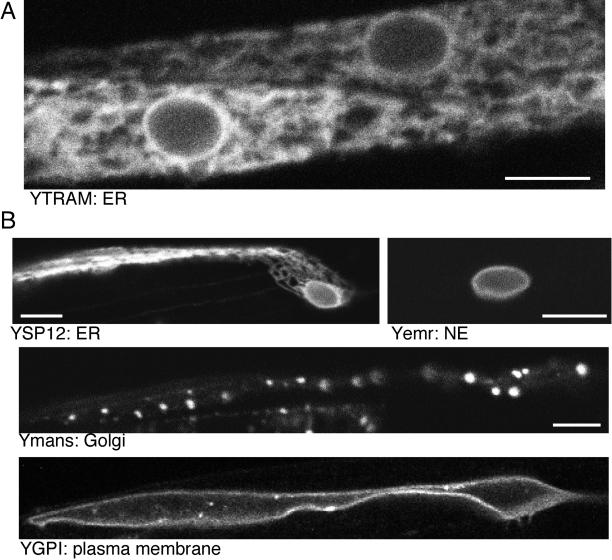
We wanted to establish a system in which to study ER protein localization in multiple differentiated cell types. C. elegans cells can be observed while the organism is alive and intact. To visualize the ER in different C. elegans cell types, the coding sequence of GFP variants was fused to the genomic coding region of predicted ER membrane proteins. These fusions were expressed under the control of cell type–specific promoters. In various cell types, for example, body wall muscle (Figure 1A), the FP-ER fusion was localized to a reticular intracellular network that appeared similar to the ER in many other types of cells. In head muscle cells the distributions of predicted ER markers were compared with those of FP-fusions to proteins predicted to be targeted to other intracellular organelles. Again FP-ER markers, for example, the signal peptidase 12-kDa subunit (SP12), were localized to a reticular network (Figure 1B). In contrast a FP-fusion to a NE protein, worm emerin (Lee et al., 2000), was observed exclusively at the nuclear rim (Figure 1B). FP-fusions to the stalk and transmembrane regions of two predicted Golgi resident enzymes were targeted to spots scattered throughout the cell (Figure 1B and our unpublished results), a pattern consistent with localization of Golgi proteins in other invertebrates (for example, Drosophila [Stanley et al., 1997] and mosquito [Rolls et al., 1997]). The plasma membrane was labeled with GPI anchored yellow FP (YFP) and was clearly distinct from intracellular membranes (Figure 1B). Thus FP-fusions to worm proteins can be constructed based on analogy with mammalian homologues, correctly targeted, and visualized in live cells.
Figure 1.
FP fusions to predicted ER membrane proteins can be localized to the ER in C. elegans. (A) YFP-TRAM (YTRAM) was expressed in body wall muscle. A single confocal plane is shown, and the NE and surrounding reticulum are visible. (B) In head muscle markers for different cellular membranes are easily distinguishable. YFP-SP12 (YSPR), YFP-mannosidase transmembrane and stalk (Ymans), YFP-emerin (Yemr), and YGPI were expressed under the control of the glr-1 promoter and imaged in head muscles. In the Ymans and YSP12 panels the nucleus is at the right, and the anterior contractile region of the cell is on the left. Scale bars, 5 μm.
In Several Cell Types RER and General ER Proteins Colocalize
To determine whether RER membrane proteins are localized to specific regions of the ER in different C. elegans cell types, spectrally distinct variants of GFP were fused to ER proteins and imaged in the same cells. Predicted RER membrane proteins were chosen based on sequence similarity to mammalian proteins involved in translocation across the ER membrane. General ER proteins were considered to be all those ER proteins involved in functions other than translocation across the membrane, for example, lipid synthesis.
FP fusions to predicted RER and general ER proteins were expressed in hypodermal cells using the dpy-7 promoter, and in intestinal cells using the general promoter rpl-28. In both cell types phosphatidylinositol synthase (PIS, a general ER protein) and TRAM (a protein involved in translocation across the ER membrane) were present in the same reticular membranes (Figure 2, A and B). In intestinal cells we occasionally saw patches of membranes enriched only in PIS but they were most obvious in deteriorating worms. Overall RER membrane markers were not restricted to a region of the ER.
Ultrastructural analysis of C. elegans hypodermal and intestinal cells suggested an explanation for the colocalization of FP fusions to predicted RER and general ER proteins. Both cell types were filled with ribosomes and contained abundant RER (Figure 2, C and D). We did not see any evidence of SER, and if it is present in these cells, it must account for only a small portion of the total ER. Thus, it is likely that the colocalization of RER and general ER markers in these cells is due to the paucity of SER.
In Neurons RER Membrane Proteins Are Localized to a Subregion of the ER
Because neurons in other organisms contain SER, neurons were chosen as a candidate cell type in which RER membrane proteins might be spatially segregated. Neuronal membranes have been best studied in mammals in highly polarized neurons with axons and dendrites. C. elegans neurons generally project only one or two unbranched neurites, which are often both pre- and postsynaptic, and so are very different from these mammalian neurons.
To visualize the ER in C. elegans neurons, FP-SP12 was expressed under control of the glr-1 promoter (Figure 3A). The glr-1 promoter drives expression in different classes of motorneurons and interneurons (Hart et al., 1995), quite a few of which send neurites into the ventral nerve cord. Most of the cell bodies are located in the ganglia near the nerve ring, although a few are in the retrovesicular ganglion posterior to the nerve ring, and some are in the tail ganglia. FP-SP12 fluorescence was visible continuously in the ventral nerve cord (Figure 3A, labeled V), indicating that the neurites contain ER. In addition to neuronal expression, the vectors with the glr-1 promoter gave some expression in several head muscles (see Figure 1B).
To determine whether any difference in localization of RER and general ER membrane proteins could be detected in neurons, FP-fusions to the two classes of predicted proteins were expressed under the glr-1 promoter. The predicted general ER proteins were present in neurites as well as the cell body, whereas most of the predicted RER membrane proteins were concentrated in the cell body. Representative confocal images of nerve rings from these two groups are shown in Figure 3B. Both yellow FP (YFP)-cytochrome b5 and cyan FP (CFP)-cytochrome P450, general ER markers, can be seen in neurites that sweep across the nerve ring. In contrast the cell bodies of neurons expressing the predicted RER markers YFP-TRAM and YFP-TRAPβ are brightly fluorescent, while the neurites are barely visible. Slight fluorescence in the neurites is probably due to overexpression.
More rigorous comparisons between different classes of ER membrane proteins were made in worms expressing two ER proteins in the same cells. The expression patterns of pairs of ER membrane proteins were compared by making transgenic worm lines coexpressing CFP and YFP fusion proteins. With the microscopy setup used, cross-talk between the two channels is negligible (White et al., 1999). Although CFP-PIS was present in the cell body and neurites, YFP-TRAM was much more concentrated in the cell body than the neurites (Figure 3C). Similar observations were made for the CFP-PIS/YFP-TRAPγ pair: the general ER protein, CFP-PIS, was observed throughout the cell, whereas the translocation protein, YFP-TRAPγ was strongly enriched in the cell body (Figure 3C).
Because fusions to GFP variants can sometimes cause mistargeting of proteins and because the ER has not been studied in worms, a number of different FP-ER fusions were tested. The correlation between the predicted category of the protein and observed localization (Figure 3D) strengthened the validity of the fusions as markers for different classes of ER proteins. Of three predicted general ER proteins tested, all localized to both the cell body and neurites, consistent with the hypothesis that they incorporate into, and distribute throughout, the ER membrane. Of the five homologues of translocation proteins tested, four were enriched in the cell body. The FP-fusion to the 12-kDa subunit of signal peptidase (SP12) was distributed throughout the neurons like the general ER membrane proteins. The signal peptidase complex has been reported to fractionate with rough membranes (Vogel et al., 1990) so this divergence in behavior from other translocation proteins is likely to be caused by fusion to the FP.
Although the major difference in localization of ER membrane proteins in neurons was between those that were concentrated in the cell body and those that were not, several other differences were also observed. Relative to the other FP-ER proteins studied, little FP-PIS was present in the NE, and at the other extreme YFP-cytochrome b5 was particularly abundant in the NE. At present we have no explanation for these differences in individual proteins so we have focused on the broader distinction between general ER proteins and those involved in translocation.
All Predicted ER Markers Are Localized to the ER, and Observed Differences in Localization Are Independent of the Imaging Conditions
Conclusions about localization of ER membrane proteins in neurons from these experiments require that the FP-tagged membrane proteins are stably localized to the ER. An alternate explanation for the difference in distribution between general ER markers and RER markers is that the general ER markers escaped to the plasma membrane. We consider this explanation unlikely. C. elegans neurons are small, and so it is difficult to see a reticular ER structure in them; however, all markers were observed in intracellular membranes. A plasma membrane marker expressed in neurons, YFP-GPI (see Figure 1B), was distinguishable from ER markers in the cell body (unpublished results). Because the glr-1 promoter also drove expression in head muscle cells, all ER markers were examined in these cells as well. For each ER marker, a reticular pattern was observed in head muscle cells (for example, see Figure 1B). In these cells, and in the larger cells (e.g., intestine, hypodermis, and body wall muscle) we examined, no evidence of plasma membrane fluorescence was seen (Figures 1 and 2, A and B).
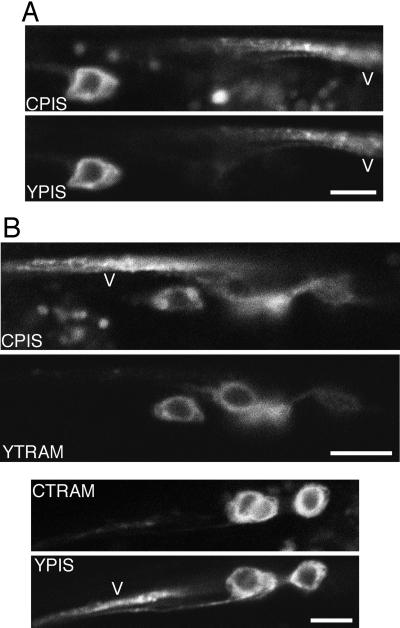
We tested the imaging conditions to make sure they were robust enough to reliably detect fluorescence in neurites and were not influenced by differences in the properties of YFP and CFP. First we fused CFP and YFP to the same membrane protein, PIS, and imaged the two fusions in the same neurons. Both color tags gave the same result: PIS is present in both the cell body and neurite (Figure 4A). Autofluorescence is always higher in the CFP channel; the blobs that are seen in many CFP images are autofluorescence and are unrelated to the fusion protein being expressed. The second test we performed was to make reciprocally tagged pairs of fusion proteins and image both pairs. We expressed CFP-PIS and YFP-TRAM in the same worm and compared the result to YFP-PIS and CFP-TRAM expressed in a different worm. Both pairs yielded the same conclusion: FP-TRAM is concentrated in the cell body, whereas FP-PIS is present in both the cell body and neurites (Figure 4B). Therefore, the results obtained were independent of which protein was tagged with a particular GFP variant.
Figure 4.
Differences in the distribution of ER markers in neurons are independent of the imaging conditions used. (A) CFP-PIS (CPIS) and YFP-PIS (YPIS) were coexpressed under the glr-1 promoter and imaged in a neuron in the retrovesicular ganglion with the ventral nerve cord (V) nearby. (B) In all panels cells in the retrovesicular ganglion were imaged with the ventral nerve cord (V) passing close by. In the top pair of panels CFP-PIS and YFP-TRAM (YTRAM) were imaged in the same cells; in the bottom panels the tags were switched so the TRAM was labeled with CFP and PIS was labeled with YFP. Scale bars, 5 μm.
Localization of RER Membrane Proteins Correlates with Ultrastructural Observations of RER
Because most of the predicted RER membrane markers were concentrated in the cell body, we tested whether morphologically recognizable RER was also localized there. Electron micrographs of C. elegans neurons show abundant free ribosomes, ribosomes on the outer nuclear membrane, and ribosomes on stretches of membrane in the rest of the cell body, which in some sections are seen to be continuous with the nuclear envelope (Figure 5A). In contrast membranes studded with ribosomes are not seen in the neurites. In regions of synaptic contact many intracellular membranes are present within the neurite, most identifiable as synaptic vesicles. However, even in regions of the neurite that do not make synaptic contact, an intracellular membrane profile is often seen (Figure 5B). This membrane profile is smooth (ribosome-free) and often visible in many consecutive sections. It varies in dimensions from section to section, shows irregular swellings along its length, and is always larger than the microtubules. The appearance of this membrane is consistent with it being SER.
Figure 5.
At the ultrastructural level RER is observed in the cell body of C. elegans neurons, and smooth membranes are present in neurites. (A) The cell body of a neuron in which the ER membranes have become slightly distended during fixation is shown. The distention of the ER makes it clear that the membranes are tightly covered with ribosomes, and highlights the connection of the peripheral ER with the NE. Free ribosomes are also present. (B) A cross section of neurites in the ventral nerve cord is shown. Small regular circles are microtubules. Larger irregular profiles are smooth membranes (arrowheads). Scale bars, 0.5 μm.
Ribosomes Are Concentrated and Immobilized in the Neuron Cell Body
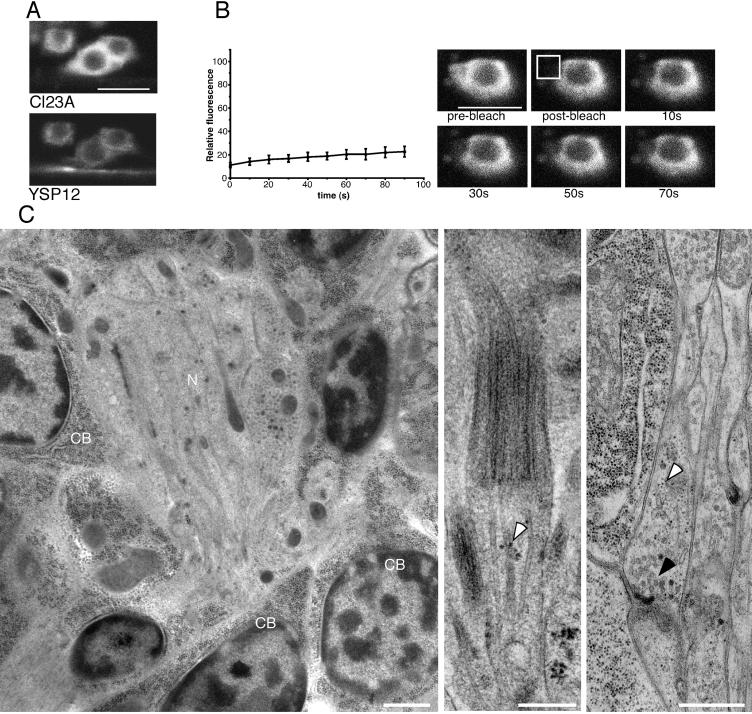
Because RER membrane proteins often associate with ribosomes, we tested whether ribosomes were also concentrated in the cell body. By light microscopy a CFP-fusion to a ribosomal subunit, L23A, was examined. L23A was chosen to visualize ribosomes because a GFP-tagged version of the yeast homolog can rescue an L23A knockout in yeast (Hurt et al., 1999). In C. elegans neurons CFP-L23A was concentrated in the cell body (Figure 6A). Localization to a region of the neuron was consistent with the tagged protein being incorporated into ribosomes.
Figure 6.
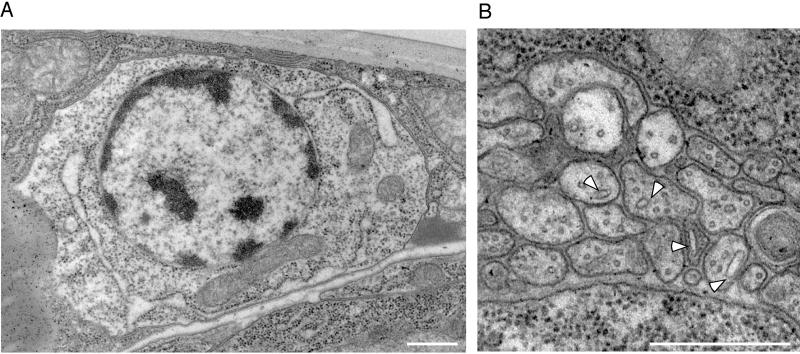
Ribosomes are highly concentrated in the cell bodies of neurons. (A) CFP-L23A (CL23A) and YFP-SP12 (YSP12) were coexpressed under control of the glr-1 promoter. Neurons of the retrovesicular ganglion and the nearby ventral nerve cord were imaged. (B) Photobleaching experiments were performed on cell bodies of neurons expressing CFP-L23A. A region of the cell was bleached (boxed area in example at the right), and images were acquired immediately after the bleach and then every 10 s. After background fluorescence was subtracted, the ratio of fluorescence in the bleached and an unbleached area of the cell was calculated (relative fluorescence). The average of six experiments and the 90% confidence interval are plotted on the graph. (C) In the electron micrograph in the left panel ribosomes (electron-dense pepper-like spots) can be seen filling neuronal cell bodies (CB) and are absent from a bundle of neurites (N). In the middle panel the ending of a ciliated sensory neuron in the nose is shown. A cluster of ribosomes near a microtubule is identified by an arrowhead. In the right panel a lengthwise section of part of the ventral nerve cord is shown with adjacent cells at the left of the panel. In the adjacent cells ribosomes appear as very distinct electron dense dots. A few similar dots are present in a presynaptic nerve terminal (white arrowhead). Synaptic vesicles are also present in this nerve terminal (black arrowhead) and below the synaptic vesicles is a dyad synapse onto the neurite below and the muscle cell to the left. A few electron dense dots that are probably ribosomes are also present in the postsynaptic neurite. Scale bars, A and B: 5 μm. Scale bars, C, left and right panels: 0.5 μm; C, middle panel: 0.2 μm.
To test the incorporation of CFP-L23A into the ribosome, we performed photobleaching experiments. Large objects diffuse more slowly than small ones, and in cytoplasm several groups have found this effect is much greater than predicted based on theoretical considerations, suggesting particles the size of ribosomes may be virtually immobile (reviewed in Luby-Phelps, 2000). The mobility of ribosomes themselves has not, however, been studied. Photobleaching was performed by exposing a portion of a neuron expressing CFP-L23A briefly to intense laser light. The cell was then imaged over time to detect the change in fluorescence in the bleached region relative to the fluorescence in the rest of the cell. Over more than a minute very little fluorescence equilibration was seen between the bleached and unbleached regions of the cell (Figure 6B). In contrast, soluble YFP equilibrated so quickly that under similar conditions the bleached region could not be detected because recovery was complete by the first postbleach image. The differences in mobility are not due to bleaching CFP and YFP: in other experiments the recovery of CFP and YFP versions of the same protein was indistinguishable (our unpublished results). The low mobility of CFP-L23A was consistent with it being incorporated into ribosomes. It also confirms that ribosomes diffuse little in the cytoplasm. In some worms CFP-L23A fluorescence was detectable at lower levels in the neurites, but when it was bleached it recovered much more quickly than the fluorescence in the cell body (unpublished results). Thus the fluorescence in the neurite was probably free CFP-L23A that had escaped being incorporated into ribosomes.
To examine ribosome distribution at higher resolution, electron micrographs of C. elegans from a variety of fixation regimes were analyzed. Ribosomes were identifiable in all preparations but were particularly prominent in high pressure frozen worms because of enhanced contrast (Figure 6C, left panel). In all worms ribosomes were abundant in neuron cell bodies and rare or absent in neurites. Occasionally small processes in nerve cords contained abundant ribosomes but most could be attributed to fingers of hypodermal cells. One case in which ribosomes were identified in neurites was a cluster of ribosomes in the ciliated sensory ending of a dendrite in the nose (Figure 6C, middle panel). Small clusters of ribosomes were also occasionally noted within neurites in regions of synaptic neuropil (Figure 6C, right panel), either presynaptically, postsynaptically, or as small clusters close to a microtubule bundle. Most appeared to be free ribosomes but rarely may have been associated with membrane. Thus, at both the light microscope and ultrastructural level, ribosomes were abundant in the cell body and rare in neurites, similar to RER membrane protein localization.
RER Membrane Proteins Diffuse Rapidly
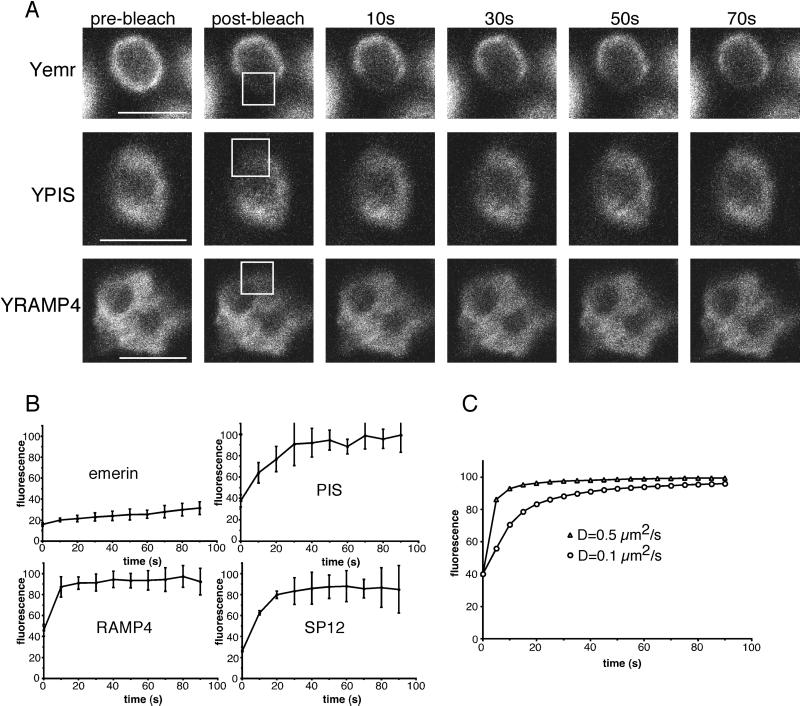
To test whether RER membrane proteins are immobilized like NE membrane proteins, we compared the diffusion of RER and general ER membrane proteins. To compare the two classes of proteins, photobleaching experiments (FRAP) were performed on worm neurons expressing FP-ER membrane proteins. These experiments were technically difficult because of slight movements of the worms, low fluorescence signal, the small size of the neurons, and their localization inside the body. To demonstrate that the bleaching protocol used can detect differences in mobility, we compared the fluorescence recovery of a NE protein with that of general ER proteins. The NE protein used was a YFP fusion to C. elegans emerin, which resides in the NE (Lee et al., 2000). The mammalian homolog has a low diffusional mobility relative to general ER proteins (Östlund et al., 1999; Rolls et al., 1999), and we found that the C. elegans protein also diffused very slowly. The bleached area remained obvious throughout the time course of the experiment (Figure 7A). Quantitation confirmed that there was little equilibration between bleached and unbleached areas of the cell (Figure 7B). In contrast, when a general ER protein was tested, fluorescence equilibrated between the bleached and unbleached regions of cell bodies in less than 30 s (see Figure 7A for examples). Quantitation of the equilibration confirmed that it was very rapid (Figure 7B).
Figure 7.
Concentration of RER membrane markers in the cell body does not require immobilization. (A) Photobleaching experiments were performed as in Figure 6. Bleached regions are indicated with boxes. The examples shown are YFP-emerin (Yemr), YFP-PIS (YPIS), and YFP-RAMP4 (YRAMP4). The bleached region of PIS and RAMP4 is less distinct because they are mobile and proteins move in and out of the region during bleaching. (B) Quantitation of bleaching experiments was performed as in Figure 6, except different numbers of experiments were used for the different proteins. For emerin, six experiments were averaged, for PIS and RAMP4 three experiments, and for SP12 two experiments. (C) Simulations of FRAP in two dimensions were performed using Virtual Cell (http://www.nrcam.uchc.edu/). Diffusion coefficients of 0.5 μm2/s (which is a standard value for membrane proteins in the ER; Nehls et al., 2000) and 0.1 μm2/s were used to model recovery at the center of a 3 × 3-μm2 bleach area. The total computational area was 20 × 20 μm2, and calculations were performed using a 0.4-μm mesh size and a 0.1-s time step. Scale bars, 5 μm.
Next we compared the mobility of RER and general ER proteins (Figure 7A). In both cases, recovery was rapid (Figure 7B). Like the general ER membrane proteins, fluorescence of RER membrane proteins had equilibrated between bleached and unbleached regions of the cells by 30 s. Thus, RER membrane proteins diffuse rapidly, and their mobility is more comparable to that of general ER membrane proteins than NE membrane proteins.
DISCUSSION
We have established C. elegans as a system in which to study ER structure and membrane protein localization in live, differentiated cells. It has not previously been possible to compare the distribution of RER and general ER membrane proteins easily in different cell types. By visualizing FP fusions to ER membrane proteins we have been able to compare localization of different classes of ER proteins in various cell types. In several cell types the distribution of FP fusions to RER and general ER membrane proteins overlapped at the light microscope level. In contrast, in neurons RER membrane markers, ribosomes and RER itself, were restricted to the cell body, whereas general ER markers were present throughout the cell body and neurites.
SER Is a Specialized Domain Abundant Only in Some Cell Types
In several different C. elegans cell types, including intestinal and hypodermal cells, predicted general ER and RER membrane proteins colocalized. An explanation for this lack of separation comes from the ultrastructural observation that RER is very abundant in these cell types and SER is rare or absent. Thus, there is simply no, or little, membrane from which RER membrane proteins would be expected to be excluded. Our observations support the idea that SER is present only in cells in which ER functions other than protein translocation across the membrane are very important. We found general ER markers along the entire length of neurites, consistent with the idea that calcium regulation is an important function of neuronal ER (Takei et al., 1992). Although the distribution of general ER proteins has been relatively well studied in neurons, that of RER proteins has not. Concentration of RER membrane proteins to a region of the neuronal ER has only been reported with one antibody to a single protein (Krijnse-Locker et al., 1995). In this case one cannot exclude lack of detection of the RER membrane protein by the antibody in narrow axons and dendrites.
Segregation of RER Membrane Proteins
In all cases where it has been tested, the ER is a continuous membrane system (Baumann and Walz, 2001). Thus, the lowest energy state for ER proteins is to be evenly distributed throughout the membrane. Within the ER there are at least two well-established classes of membrane proteins that are not distributed throughout the whole membrane system: inner NE membrane proteins and RER membrane proteins. C. elegans has recently been shown to contain an NE protein analogous to vertebrate emerin (Lee et al., 2000), and we show that its behavior at the NE is similar to that of mammalian NE membrane proteins. The localization of RER membrane proteins to a region of the ER has previously been observed only in vertebrates. By demonstrating that segregation of RER membrane proteins occurs in C. elegans, we show that it is likely to be an evolutionarily ancient process.
For inner NE membrane proteins, specific regions of the protein contain targeting signals that are responsible for localization. Our data suggest that RER membrane proteins contain positive targeting signals that localize them to the cell body in neurons. Although most RER markers expressed in C. elegans neurons were concentrated in the cell body, one subunit of an established RER protein complex, signal peptidase, was present in neurites when tagged with YFP. The YFP perhaps interfered with assembly of the subunit into the complex or disrupted an RER targeting signal within the subunit itself. In either case, localization of this protein throughout the neuron indicates that this pattern is the default one; no targeting signal is required to gain access to SER in the neurite. At high expression levels other RER membrane proteins were observed in neurites (not shown). Again this suggests that a signal is not required to enter the neurite. It also indicates that the targeting mechanism to the cell body is saturable.
Several basic mechanisms could be responsible for localization of RER membrane proteins in neurons. One mechanism that has been proposed (see Introduction), is a network of interactions between RER proteins. This model predicts random localization of RER in patches throughout the cell and cannot explain why in neurons the RER is always found in the cell body. At least three other basic mechanisms could account for this localization. One type of mechanism is binding of RER membrane proteins to a component present only in the cell body. A second mechanism is a selective diffusion barrier that does not allow RER membrane proteins to gain access to the neurite. A third mechanism is active retrieval from the neurite to the cell body.
Binding to a localized partner is conceptually the simplest model and is analogous to the mechanism used to target proteins to the NE. Ribosomes are the best candidate binding partner. Many RER membrane proteins associate directly or indirectly with ribosomes (Görlich and Rapoport, 1993), and so targeting could occur by either direct binding to ribosomes or by lateral interactions with other RER membrane proteins associated with ribosomes. Additionally in C. elegans neurons the distribution of ribosomes and RER membrane proteins is the same. Is this simple model consistent with the behavior of RER and SER membrane proteins that is observed in worm neurons?
Binding of RER membrane proteins to ribosomes would be expected to affect both the localization and diffusion of RER membrane proteins because ribosomes are virtually immobile and are localized to a region of the cell. In the case in which the binding reaction is rapid relative to diffusion, which is likely to be the case for RER membrane proteins and ribosomes, the degree of localization is directly related to the observed diffusion in a FRAP experiment (Cowan et al., 1997). For example a 10-fold concentration of an RER membrane protein in the cell body would correspond to a 10-fold reduction in effective diffusion coefficient (Deff).
Therefore FRAP experiments like those in Figure 7 should provide a test of this model. The FRAP data did not show large differences between proteins that would not be expected to bind ribosomes, general ER proteins, and RER membrane proteins, which would be expected to interact with ribosomes (Figure 7B). These data do not, however, rule out more subtle differences in Deff. Simulations from the Virtual Cell program (Schaff et al., 2000; http://www.nrcam.uchc.edu/) show that fluorescence recovery plots of proteins with fivefold differences in Deff are not grossly different (Figure 7C).
To determine whether subtle differences in Deff were present in our results, we processed sets of data collected at 5-s intervals with a program designed to analyze diffusion in complex structures like the ER (Siggia et al., 2000). Several data sets were analyzed reasonably well by the program and yielded Deff ranging from 0.1 to 0.5 μm2/s, expected values for ER proteins. Four of the data sets represented RER membrane proteins and had Deff ranging from 0.1 to 0.2 μm2/s. Two of the data sets were from general ER membrane proteins and had Deff 0.3 and 0.5 μm2/s. It is impossible to draw strong conclusions from so few samples and from a system in which FRAP experiments are very difficult to perform, but the results are consistent with a two- to fivefold slower mobility of RER proteins. They could thus explain up to a fivefold concentration of RER membrane proteins in the cell body by immobilized ribosomes. To test this model further, more precise FRAP experiments, preferably using larger cells in which RER membrane protein targeting can also be observed, will be necessary. In addition, the C. elegans system established here could be used to perform a screen to identify factors required for RER membrane protein targeting. These factors may be involved in targeting via ribosomes or may suggest additional mechanisms by which RER membrane proteins are localized.
Targeting of Ribosomes within C. elegans Neurons
Protein synthesis in neurites is believed to be important for synaptic modulation, so it is interesting to compare the distribution of ribosomes in C. elegans with that in other organisms. In mammalian neurons, ribosomes are most concentrated in the cell body and base of the dendrites. They are also present in distal dendrites, but are largely excluded from axons (Deitch and Banker, 1993; Knowles et al., 1996). The axonal exclusion is not complete (Koenig et al., 2000), but relative to dendrites few ribosomes are present in axons. In invertebrates no compartment analogous to the mammalian axon, in which ribosomes are very scarce, has been described. This is, in part, due to lack of study. Where it has been examined, protein synthetic machinery is present in invertebrate neurites. In one of the simplest organisms with neurons, hydra, ribosomes appear in the neurites (Lentz, 1966). In squid and mollusks ribosomes are abundant in giant axons (Sotelo et al., 1999; Spencer et al., 2000), and in the mollusk Aplysia presynaptic protein synthesis can be important for synaptic plasticity (Martin et al., 1997). It has been suggested that ribosomes are not excluded from axons in invertebrates because the neurons are not as strictly polarized as vertebrate CNS neurons (Martin et al., 2000).
In C. elegans we found ribosomes were concentrated in the cell body of neurons, and very rare in neurites. To our knowledge this is the first description of an invertebrate neuronal compartment in which ribosomes are rare, comparable to mammalian axons. It is likely that similar compartments exist in other invertebrates but have just not been described in detail. Because most C. elegans neurites are both pre- and postsynaptic, strict axonal polarity is not required for exclusion of ribosomes from neurites. Another implication of this observation is that localization of ribosomes to particular neuronal compartments may be ancestral to the divergence of nematodes, mollusks, and vertebrates.
The mechanism by which ribosomes are excluded from some neuronal compartments is not known. In other cell types, one possibility that has been proposed to localize ribosomes, at least to regions of the ER (Bergmann and Fusco, 1990), is binding to RER membrane proteins. Considering the high mobility of RER membrane proteins relative to ribosomes this is very unlikely. Another possibility is that narrow processes exclude ribosomes based on their large size. However, mitochondria are present in nematode neurites, and other processes do not exclude ribosomes. For example, fingers of hypodermal cells, which are similar in diameter to neurites, often invade the ventral nerve cord in C. elegans but are filled with ribosomes. It is therefore probable that a more specific mechanism exists to exclude ribosomes from neurites. Although ribosomes are rare in both mammalian axons and C. elegans neurites, some are present (Koenig et al., 2000 and Figure 6C). In both cases the ribosomes seem to be localized to specific regions of the neurite, indicating they may be specifically targeted there. It will be interesting to determine whether they are accompanied by particular mRNAs, as they are in mammalian dendrites (reviewed in Kiebler and DesGroseillers, 2000). C. elegans neurons may provide a useful system for understanding how ribosomes are excluded from some neuronal compartments and whether ribosomes that escape exclusion are important for synaptic modulation.
ACKNOWLEDGMENTS
The authors are grateful to the following: Jamie White for help with the light microscopy; James Jonkman and Stephen Grill, for writing the bleaching macro for the CCC; the CCC development team; the Analytical Ultrastructure Center at AECOM and the laboratory of Stan Erlandsen at the University of Minnesota, for providing specialized facilities; Frank Macaluso and Ya Chen for expert help in fast freezing and freeze substitution methods; Kent McDonald for help in discussing these protocols; Gloria and Tylon Stephney for excellent help in electron microscopy; Yang Shi and the Hyman lab for providing worm support. They are also grateful to Anne Hart for providing the glr-1 promoter; the Fire lab at the Carnegie Institution for vectors; and Alan Coulson at the Sanger Center for cosmids. The ideas in this article were helped along by discussions with many people, in particular Ed Hedgecock, Mark Terasaki, Boris Slepchenko, and Reinhart Heinrich. The authors also thank Eric Siggia for allowing them to use his diffusion analysis program; Erik Snapp for indispensable help with the program; and Will Prinz and particularly David Sabatini for comments on the manuscript. This work was supported by National Institutes of Health grant RR 12596 to D.H.H. and GM58012 to Y.S. in whose lab M.V. works. M.M.R. was a Howard Hughes Predoctoral Fellow in the Biological Sciences. T.A.R. is an investigator of the Howard Hughes Medical Institute.
Abbreviations used:
- Deff
effective diffusion coefficient
- ER
endoplasmic reticulum
- SER
smooth endoplasmic reticulum
- RER
rough endoplasmic reticulum
- NE
nuclear envelope
- FP
fluorescent protein
- FRAP
fluorescence recovery after photobleaching
- GFP
green fluorescent protein
- CFP
cyan fluorescent protein
- YFP
yellow fluorescent protein
- LBR
lamin B receptor
- SP12
signal peptidase 12-kDa subunit
- PIS
phosphatidylinositol synthase
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–10–0514. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–10–0514.
REFERENCES
- Baumann O, Walz B. Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int Rev Cytol. 2001;205:149–214. doi: 10.1016/s0074-7696(01)05004-5. [DOI] [PubMed] [Google Scholar]
- Bergmann JE, Fusco PJ. The G protein of vesicular stomatitis virus has free access into and egress from the smooth endoplasmic reticulum of UT-1 cells. J Cell Biol. 1990;110:625–635. doi: 10.1083/jcb.110.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole NB, Smith CL, Sciaky N, Terasaki M, Edidin M, Lippincott-Schwartz J. Diffusional mobility of Golgi proteins in membranes of living cells. Science. 1996;273:797–801. doi: 10.1126/science.273.5276.797. [DOI] [PubMed] [Google Scholar]
- Cowan AE, Nakhimovsky L, Myles DG, Koppel DE. Barriers to diffusion of plasma membrane proteins form early during guinea pig spermiogenesis. Biophys J. 1997;73:507–516. doi: 10.1016/S0006-3495(97)78089-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayel MJ, Hom EF, Verkman AS. Diffusion of green fluorescent protein in the aqueous-phase lumen of the endoplasmic reticulum. Biophys J. 1999;76:2843–2851. doi: 10.1016/S0006-3495(99)77438-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitch JS, Banker GA. An electron microscopic analysis of hippocampal neurons developing in culture: early stages in the emergence of polarity. J Neurosci. 1993;13:4301–4315. doi: 10.1523/JNEUROSCI.13-10-04301.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depierre JW, Dallner G. Structural aspects of the membrane of the endoplasmic reticulum. Biochim Biophys Acta. 1975;415:411–472. doi: 10.1016/0304-4157(75)90006-4. [DOI] [PubMed] [Google Scholar]
- Ellenberg J, Siggia ED, Moreira JE, Smith CL, Presley JF, Worman HJ, Lippincott-Schwartz J. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J Cell Biol. 1997;138:1193–1206. doi: 10.1083/jcb.138.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett DW. The Cell. Philadelphia: W.B. Saunders Company; 1981. [Google Scholar]
- Gilleard JS, Barry JD, Johnstone IL. cis regulatory requirements for hypodermal cell-specific expression of the Caenorhabditis elegans cuticle collagen gene dpy-7. Mol Cell Biol. 1997;17:2301–2311. doi: 10.1128/mcb.17.4.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Rapoport TA. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane [see comments] Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- Hall, D.H. (1995). Electron microscopy and 3D image reconstruction. In: C. elegans: Modern Biological Analysis of an Organism, ed. H.F. Epstein, D.C. Shakes, Methods in Cell Biology, San Diego, CA: Academic Press, Vol 48, 396–436.
- Hart AC, Sims S, Kaplan JM. Synaptic code for sensory modalities revealed by C. elegans GLR-1 glutamate receptor. Nature. 1995;378:82–85. doi: 10.1038/378082a0. [DOI] [PubMed] [Google Scholar]
- Hortsch M, Griffiths G, Meyer DI. Restriction of docking protein to the rough endoplasmic reticulum: immunocytochemical localization in rat liver. Eur J Cell Biol. 1985;38:271–279. [PubMed] [Google Scholar]
- Hurt E, Hannus S, Schmelzl B, Lau D, Tollervey D, Simos G. A novel in vivo assay reveals inhibition of ribosomal nuclear export in Ran-cycle and nucleoporin mutants. J Cell Biol. 1999;144:389–401. doi: 10.1083/jcb.144.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa NE, De Lemos-Chiarandini C, Tsao YS, Takatsuki A, Adesnik M, Sabatini D, Kreibich G. O-Glycosylation of intact and truncated ribophorins in brefeldin A-treated cells: newly synthesized intact ribophorins are only transiently accessible to the relocated glycosyltransferases. J Cell Biol. 1992;117:949–958. doi: 10.1083/jcb.117.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler MA, DesGroseillers L. Molecular insights into mRNA transport and local translation in the mammalian nervous system. Neuron. 2000;25:19–28. doi: 10.1016/s0896-6273(00)80868-5. [DOI] [PubMed] [Google Scholar]
- Knowles RB, Sabry JH, Martone ME, Deerinck TJ, Ellisman MH, Bassell GJ, Kosik KS. Translocation of RNA granules in living neurons. J Neurosci. 1996;16:7812–7820. doi: 10.1523/JNEUROSCI.16-24-07812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig E, Martin R, Titmus M, Sotelo-Silveira JR. Cryptic peripheral ribosomal domains distributed intermittently along mammalian myelinated axons. J Neurosci. 2000;20:8390–8400. doi: 10.1523/JNEUROSCI.20-22-08390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreibich G, Ulrich BL, Sabatini DD. Proteins of rough microsomal membranes related to ribosome binding: 1. Identification of ribophorins I and II, membrane proteins characteristic of rough microsomes. J Cell Biol. 1978;77:464–487. doi: 10.1083/jcb.77.2.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijnse-Locker J, Parton RG, Fuller SD, Griffiths G, Dotti CG. The organization of the endoplasmic reticulum and the intermediate compartment in cultured rat hippocampal neurons. Mol Biol Cell. 1995;6:1315–1332. doi: 10.1091/mbc.6.10.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Gruenbaum Y, Spann P, Liu J, Wilson KL. C. elegans nuclear envelope proteins emerin, MAN1, lamin, and nucleoporins reveal unique timing of nuclear envelope breakdown during mitosis. Mol Biol Cell. 2000;11:3089–3099. doi: 10.1091/mbc.11.9.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentz TH. The Cell Biology of Hydra. New York: John Wiley & Sons, Inc; 1966. [Google Scholar]
- Lewis JA, Fleming JT. In: Basic culture methods. In Caenorhabditis elegans: modern biological analysis of an organism. Epstein HF, Shakes DC, editors. 1995. Methods in Cell Biology, San Diego, CA: Academic Press, Vol 48, 3–29. [Google Scholar]
- Luby-Phelps K. Cytoarchitecture and physical properties of cytoplasm: volume, viscosity, diffusion, intracellular surface area. Int Rev Cytol. 2000;192:189–221. doi: 10.1016/s0074-7696(08)60527-6. [DOI] [PubMed] [Google Scholar]
- Martin KC, Barad M, Kandel ER. Local protein synthesis and its role in synapse-specific plasticity. Curr Opin Neurobiol. 2000;10:587–592. doi: 10.1016/s0959-4388(00)00128-8. [DOI] [PubMed] [Google Scholar]
- Martin KC, Casadio A, Zhu H, E Y, Rose JC, Chen M, Bailey CH, Kandel ER. Synapse-specific, long-term facilitation of aplysia sensory to motor synapses: a function for local protein synthesis in memory storage. Cell. 1997;91:927–938. doi: 10.1016/s0092-8674(00)80484-5. [DOI] [PubMed] [Google Scholar]
- McDonald K. High pressure freezing for preservation of high resolution fine structure and antigenicity for immunolabelling. Methods Mol Biol. 1998;117:77–97. doi: 10.1385/1-59259-201-5:77. [DOI] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA transformation. In Caenorhabditis elegans: modern biological analysis of an organism. H.F. Epstein and D.C. Shakes, editors. Methods in Cell Biology. San Diego, CA: Academic Press, Vol; 1995. p. 48. 451–482. [Google Scholar]
- Meyer HA, Grau H, Kraft R, Kostka S, Prehn S, Kalies KU, Hartmann E. Mammalian Sec61 is associated with Sec62 and Sec63. J Biol Chem. 2000;275:14550–14557. doi: 10.1074/jbc.275.19.14550. [DOI] [PubMed] [Google Scholar]
- Nehls S, Snapp EL, Cole NB, Zaal KJ, Kenworthy AK, Roberts TH, Ellenberg J, Presley JF, Siggia E, Lippincott-Schwartz J. Dynamics and retention of misfolded proteins in native ER membranes. Nat Cell Biol. 2000;2:288–295. doi: 10.1038/35010558. [DOI] [PubMed] [Google Scholar]
- Östlund C, Ellenberg J, Hallberg E, Lippincott-Schwartz J, Worman HJ. Intracellular trafficking of emerin, the Emery-Dreifuss muscular dystrophy protein. J Cell Sci. 1999;112:1709–1719. doi: 10.1242/jcs.112.11.1709. [DOI] [PubMed] [Google Scholar]
- Rapoport TA, Jungnickel B, Kutay U. Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu Rev Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- Rolls MM, Marquardt MT, Kielian M, Machamer CE. Cholesterol-independent targeting of Golgi membrane proteins in insect cells. Mol Biol Cell. 1997;8:2111–2118. doi: 10.1091/mbc.8.11.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls MM, Stein PS, Taylor SS, Ha E, McKeon F, Rapoport TA. A visual screen of a GFP-fusion library identifies a new type of nuclear envelope membrane protein. J Cell Biol. 1999;146:29–43. doi: 10.1083/jcb.146.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaff JC, Slepchenko BM, Loew LM. Physiological modeling with virtual cell framework. Methods Enzymol. 2000;321:1–23. doi: 10.1016/s0076-6879(00)21184-1. [DOI] [PubMed] [Google Scholar]
- Siggia ED, Lippincott-Schwartz J, Bekiranov S. Diffusion in inhomogeneous media: theory and simulations applied to whole cell photobleach recovery. Biophys J. 2000;79:1761–1770. doi: 10.1016/S0006-3495(00)76428-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotelo JR, Kun A, Benech JC, Giuditta A, Morillas J, Benech CR. Ribosomes and polyribosomes are present in the squid giant axon: an immunocytochemical study. Neuroscience. 1999;90:705–715. doi: 10.1016/s0306-4522(98)00587-9. [DOI] [PubMed] [Google Scholar]
- Soullam B, Worman HJ. The amino-terminal domain of the lamin B receptor is a nuclear envelope targeting signal. J Cell Biol. 1993;120:1093–1100. doi: 10.1083/jcb.120.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soullam B, Worman HJ. Signals and structural features involved in integral membrane protein targeting to the inner nuclear envelope. J Cell Biol. 1995;130:15–27. doi: 10.1083/jcb.130.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer GE, Syed NI, van Kesteren E, Lukowiak K, Geraerts WP, van Minnen J. Synthesis and functional integration of a neurotransmitter receptor in isolated invertebrate axons. J Neurobiol. 2000;44:72–81. doi: 10.1002/1097-4695(200007)44:1<72::aid-neu7>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Stanley H, Botas J, Malhotra V. The mechanism of Golgi segregation during mitosis is cell type-specific. Proc Natl Acad Sci USA. 1997;94:14467–14470. doi: 10.1073/pnas.94.26.14467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian K, Meyer T. Calcium-induced restructuring of nuclear envelope and endoplasmic reticulum calcium stores. Cell. 1997;89:963–971. doi: 10.1016/s0092-8674(00)80281-0. [DOI] [PubMed] [Google Scholar]
- Tajima S, Lauffer L, Rath VL, Walter P. The signal recognition particle receptor is a complex that contains two distinct polypeptide chains. J Cell Biol. 1986;103:1167–1178. doi: 10.1083/jcb.103.4.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei K, Stukenbrok H, Metcalf A, Mignery GA, Sudhof TC, Volpe P, De Camilli P. Ca2+ stores in Purkinje neurons: endoplasmic reticulum subcompartments demonstrated by the heterogeneous distribution of the InsP3 receptor, Ca(2+)-ATPase, and calsequestrin. J Neurosci. 1992;12:489–505. doi: 10.1523/JNEUROSCI.12-02-00489.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaki M, Jaffe LA. Organization of the sea urchin egg endoplasmic reticulum and its reorganization at fertilization. J Cell Biol. 1991;114:929–940. doi: 10.1083/jcb.114.5.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaki M, Slater NT, Fein A, Schmidek A, Reese TS. Continuous network of endoplasmic reticulum in cerebellar Purkinje neurons. Proc Natl Acad Sci USA. 1994;91:7510–7514. doi: 10.1073/pnas.91.16.7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel F, Hartmann E, Görlich D, Rapoport TA. Segregation of the signal sequence receptor protein in the rough endoplasmic reticulum membrane. Eur J Cell Biol. 1990;53:197–202. [PubMed] [Google Scholar]
- White J, Johannes L, Mallard F, Girod A, Grill S, Reinsch S, Keller P, Tzschaschel B, Echard A, Goud B, Stelzer EH. Rab6 coordinates a novel Golgi to ER retrograde transport pathway in live cells. J Cell Biol. 1999;147:743–760. doi: 10.1083/jcb.147.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Masson EM, Heid PJ, Lavin CA, Hardin J. The cellular mechanism of epithelial rearrangement during morphogenesis of the Caenorhabditis elegans dorsal hypodermis. Dev Biol. 1998;204:263–276. doi: 10.1006/dbio.1998.9048. [DOI] [PubMed] [Google Scholar]
- Worman HJ, Yuan J, Blobel G, Georgatos SD. A lamin B receptor in the nuclear envelope. Proc Natl Acad Sci USA. 1988;85:8531–8534. doi: 10.1073/pnas.85.22.8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Worman HJ. Primary structure analysis and lamin B and DNA binding of human LBR, and integral protein of the nuclear envelope inner membrane. J Biol Chem. 1994;269:11306–11311. [PubMed] [Google Scholar]