Abstract
HIF-1α is the regulated subunit of the HIF-1 transcription factor, which induces transcription of a number of genes involved in the cellular response to hypoxia. The HIF-1α protein is rapidly degraded in cells supplied with adequate oxygen but is stabilized in hypoxic cells. Using polysome profile analysis, we found that translation of HIF-1α mRNA in NIH3T3 cells is spared the general reduction in translation rate that occurs during hypoxia. To assess whether the 5′UTR of the HIF-1α mRNA contains an internal ribosome entry site (IRES), we constructed a dicistronic reporter with the HIF-1α 5′UTR inserted between two reporter coding regions. We found that the HIF-1α 5′UTR promoted translation of the downstream reporter, indicating the presence of an IRES. The IRES had activity comparable to that of the well-characterized c-myc IRES. IRES activity was not affected by hypoxic conditions that caused a reduction in cap-dependent translation, and IRES activity was less affected by serum-starvation than was cap-dependent translation. These data indicate that the presence of an IRES in the HIF-1α 5′UTR allows translation to be maintained under conditions that are inhibitory to cap-dependent translation.
INTRODUCTION
When vertebrate tissues are deprived of oxygen, cellular metabolic responses are initiated to conserve energy and enhance survival. These changes include arrest in the cell cycle (Amellem and Pettersen, 1991; Schmaltz et al., 1998), suppression of protein synthesis (Kraggerud et al., 1995; Tinton and Buc-Calderon, 1999), and increased expression of a number of genes involved in energy metabolism. Hypoxia also causes the induction of growth factors that lead to enhancement of the oxygen supply: vascular endothelial growth factor (VEGF) initiates angiogenesis and enhances blood vessel permeability (Shweiki et al., 1992; Ladoux and Frelin, 1993), while erythropoietin production increases the level of circulating red blood cells.
The transcriptional induction of many genes that respond to hypoxia is mediated by the transcription factor hypoxia-inducible factor-1 (HIF-1). HIF-1 was originally identified as a factor that induces transcription of the erythropoietin gene under hypoxic conditions but has subsequently been found to participate in the hypoxic induction of numerous genes, including VEGF (Shweiki et al., 1992; Ladoux and Frelin, 1993), tyrosine hydroxylase (Norris and Millhorn, 1995), phosphoglycerate kinase 1, lactate dehydrogenase (Firth et al., 1994), aldolase A, pyruvate kinase M (Semenza et al., 1994), and the glucose transporters 1 and 3 (Ebert et al., 1995).
Both subunits of the heterodimeric HIF-1 protein belong to the basic helix-loop-helix family of transcription factors and contain the Per-Arnt-Sim (PAS) region of homology. HIF-1 activity is regulated primarily by regulation of the availability of the HIF-1α subunit. The HIF-1β subunit, also known as the aryl hydrocarbon nuclear translocator (ARNT) protein, is expressed at relatively constant level and has been shown to act as a dimerization partner for a number of other proteins including the aryl hydrocarbon receptor (Wang et al., 1995). HIF-1α mRNA is expressed constitutively, but the HIF-1α protein is almost undetectable in normal oxygen conditions because it is rapidly degraded by the ubiquitin/proteasome system, with a half-life <5 min (Huang et al., 1996; Salceda and Caro, 1997). During hypoxia the HIF-1α protein is stabilized, and the level of HIF-1 activity rapidly increases. Because the HIF-1α level is very low before the onset of hypoxia, the accumulation of HIF-1 activity requires de novo synthesis of HIF-1α protein, during conditions that may not be favorable to maximal rates of protein synthesis. This raises the question of whether the HIF-1α mRNA has features that favor its translation during hypoxia.
Translation of most eukaryotic mRNAs involves interaction of the mRNA 5′ m7GpppN cap with the eIF4E subunit of the eIF4F translation initiation complex. Once the eIF4F complex has assembled on the mRNA the small ribosomal subunit is recruited to the mRNA and scanning for a favorable AUG initiation codon commences. The amount of eIF4E can be limiting for translation and its availability is dependent upon the phosphorylation state of its inhibitory binding partners, 4EBP1 and 4EBP2 (Pause et al., 1994). Hypoxia has been shown to reduce the availability of eIF4E by increasing its association with 4EBP1, although it is suspected that other mechanisms contribute to the suppression of protein synthesis (Tinton and Buc-Calderon, 1999).
An alternative mode of translation initiation, that does not require eIF4E and the 5′ cap, involves recruitment of the translation initiation complex by an internal ribosome entry site (IRES). Translation by internal ribosome entry was first identified in picornaviruses, but a number of cellular mRNAs have subsequently been found to contain an IRES, including basic fibroblast growth factor (Vagner et al., 1995), vascular endothelial growth factor (Akiri et al., 1998; Miller et al., 1998; Stein et al., 1998), c-myc (Stoneley et al., 1998), immunoglobulin heavy-chain binding protein (BiP; Macejak and Sarnow, 1991), and NF-κB–repressing factor (Oumard et al., 2000).
Although the advantages for viruses to contain an IRES are quite clear, the advantages for a cellular IRES were not immediately understood. Expression of all the IRES containing cellular genes reported thus far appear to be tightly regulated for normal cellular function and/or required under conditions where cap-dependent translation is suppressed. Because HIF-1α is expressed during hypoxia and because the 5′UTR is long and G + C rich, we investigated whether HIF-1α is efficiently translated during hypoxia. We show that the translation of endogenous HIF-1α mRNA in NIH3T3 cells is not compromised under hypoxic conditions, in agreement with a recent study of the effect of hypoxia on HIF1-α translation in HeLaS3 cells (Gorlach et al., 2000). Using dicistronic assays we demonstrate that the HIF-1α 5′UTR contains an IRES that has activity similar to that of the IRES in c-myc. IRES activity was maintained during hypoxia, suggesting that the HIF-1α IRES is likely to be utilized in conditions where cap-dependent translation is suppressed.
MATERIALS AND METHODS
Cell Culture and Transfection
NIH3T3, HEK293, and HeLa cells were cultivated in DMEM (CSL, Melbourne, Australia) containing 10% heat-inactivated fetal calf serum (CSL), 100 U/ml penicillin, 10 μg/ml streptomycin, 2 mM glutamine, and 10 mM HEPES (CSL). All lines were kept at 37°C in a humidified atmosphere containing 5% CO2. For hypoxic treatment, cells were placed into a hypoxic chamber (Edwards Instrument Co., Wilmington, MA) with a regulated environment of 1% O2, 5% CO2, 94% N2. For transient transfections, 1 × 105 cells were plated into 24-well plates the day before transfection. Cells were transfected using LipoFectamine 2000 (GIBCO-BRL, Rockville, MD) as per the manufacturer's recommendations and were incubated for 24 h as described above. Cells were harvested 24 h posttransfection, washed with 1× PBS, and lysed in 100 μl passive reporter lysis buffer (Promega, Madison, WI). Lysates were stored at −20°C until assay.
Plasmid Constructs
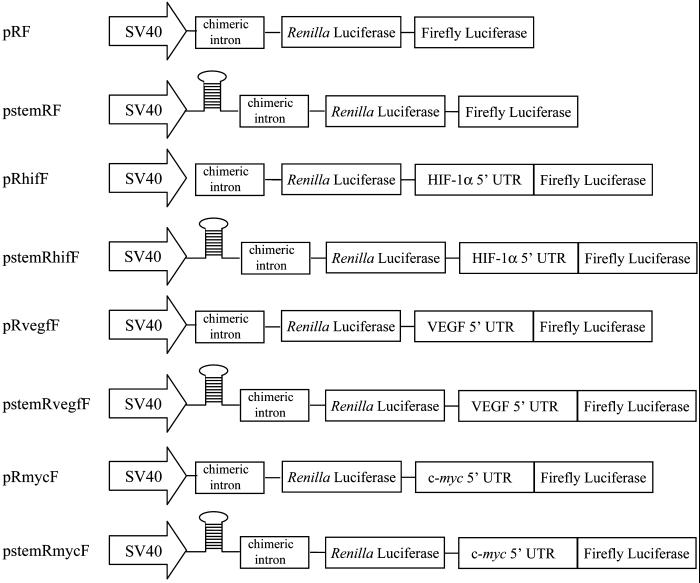
The dicistronic constructs used in this study are shown schematically in Figure 1. The plasmids, pRF (formerly pGL3R) and pstemRmycF (formerly pGL3utrH), have been described previously (Stoneley et al., 1998; a kind gift from Dr. A. Willis, University of Leicester). Briefly, pRF is derived from pGL3 (Promega), with the Renilla Luciferase coding region inserted upstream of the firefly luciferase coding region. pstemRmycF was constructed from pRF, with the c-myc 5′UTR inserted into the linking region between the Renilla and firefly luciferase coding regions, such that firefly luciferase is driven from the putative IRES. Upstream of the Renilla coding region, a stable stem-loop was inserted to suppress translation of the first cistron by preventing ribosome binding.
Figure 1.
Schematic illustration of dicistronic reporter gene constructs. The control dicistronic reporter gene, in plasmid pRF, contains cDNAs encoding sea pansy (Renilla reniformis) luciferase and firefly luciferase, separated by a short linker sequence. Expression is driven by the SV40 early promoter. pstemRF is similar to pRF, but with the addition of a stem-loop at the 5′ end of the transcript to suppress cap-dependent translation. Plasmids pRhifF, pRmycF, and pRvegfF contain the murine HIF-1α, c-myc, and VEGF 5′ UTRs, respectively, inserted between the two reporter reading frames. Plasmids pstemRhifF, pstemRmycF, and pstemRvegfF are identical to their corresponding vectors described above, except they also contain the stable stem-loop at the 5′ end of the transcript.
Plasmid pstemRF was constructed by excising the c-myc 5′ UTR from pstemRmycF by digesting with PvuII/NcoI (NEB), blunt-ending with T4 DNA polymerase (NEB) as per manufacturer's recommendations, and ligating shut. The mouse HIF-1α 5′ UTR was cloned by 5′-RACE using a kit (Roche Laboratories, Nutley, NJ) and gene-specific oligonucleotides 5′-GCCTTCCATGGCGAATCG-3′, 5′-AACGCCGGCTCTGTCCGCG-3′ and 5′-AAGAGACAAGTCCAGAGGCG-3′ as first, second, and third primer, respectively. The PCR product was digested with SalI, subcloned into the SalI site of pKSBluescriptII., cut with NheI and NotI, and ligated to the NheI- and NotI-digested mouse HIF-1α cDNA that was amplified by PCR as described (Kappel et al., 1999) to generate pKSHIFIRES. The dicistronic reporter containing the HIF-1α 5′UTR was made by excising the HIF-1α 5′ UTR from pKSHIFIRES by digesting with ApaLI, blunt-ending, and digesting with NcoI. The resulting 286-base pair fragment was isolated and ligated into pRF digested with PvuII/NcoI and pstemRhifF with SpeI, blunt-ended, and then digested with NcoI, to create pRhifF and pstemRhifF, respectively. Dual reporter VEGF plasmids were generated by digesting pBSV5 (Miller et al., 1998) with HindIII and blunt-ended, followed by digestion with NcoI to excise the 1013-base pair murine VEGF 5′ UTR. This fragment was ligated into pRF digested with PvuII/NcoI and pstemRF digested with SpeI, blunt-ended, and then digested with NcoI to generate plasmids pRvegfF and pstemRvegfF, respectively. pRmycF was constructed by digesting pstemRmycF with SpeI/NcoI and ligating the 396-base pair 5′ UTR into similarly digested pRF.
To construct plasmid pGemRhifF, a 624-nt DNA fragment was PCR amplified from pRhifF using primers 5′-CGGGATCCGCAAGAAGATGCACCTGATG-3′ and 5′-CCCAAGCTTGCGTATCTCTTCATAGCCT-3′, digested with BamHI and HindIII, and ligated into pGEM4Z (Promega).
Reporter Assays
Firefly luciferase and Renilla luciferase activities were measured in a luminometer (Model TD 20/20; Turner Designs, Mountain View, CA) using the reagents provided with the dual luciferase reporter kit (Promega). The assay was carried out using one quarter of the manufacturer's recommendations per assay. Ratios of firefly to Renilla luciferase were not altered when compared with assays performed under conditions recommended by the manufacturer. Transfection efficiency and extract preparations were corrected by normalizing the data to the corresponding Renilla luciferase activity for each construct.
35S-Methionine Incorporation
NIH3T3 cells were plated at a density of 5 × 105 cells per 24-well dish and incubated for 24 h, followed by a further 24-h incubation under either normoxic or hypoxic conditions. Cells were then washed with PBS and incubated in methionine-free DMEM (Life Technologies-BRL, Rockville, MD), but containing 30 μCi/ml l-35S-methionine/cysteine (New England Nuclear, Boston, MA). Cells were subsequently washed twice with PBS, four times with TCA, and twice with ethanol, air dried, and dissolved in 0.4 ml of 0.3 M NaOH. After neutralization with 80 μl of 1.5 M HCl, one half of the sample was subjected to liquid scintillation counting (United Technology, Packard Bell 2000, La Jolla, CA), and the other half was used for measurement of total protein concentration using Bradford Reagent (Bio-Rad, Richmond, CA).
Polysome Analysis
Polysome analysis was performed essentially as described (Savant Bhonsale and Cleveland, 1992). Briefly, three 100-mm dishes of NIH3T3 cells at just subconfluent density were used for each polysome distribution analysis. Cells were washed three times with ice-cold PBS containing 100 μg/ml cycloheximide, removed using a cell scraper, and lysed in polysome lysis buffer (100 mM KCl, 5 mM MgCl2, 10 mM HEPES [pH 7.4], 100 μg/ml cycloheximide, 0.5% NP-40, 20 U/ml RNasin ([Promega]). Nuclei were pelleted by centrifugation at 12,000 × g at 4°C for 5 min. Total RNA in the resulting supernatant was determined by measuring OD260, and equivalent amounts of RNA was loaded onto each 15–50% (wt/vol) linear sucrose gradient. Gradients were centrifuged at 38,000 rpm for 2 h at 4°C in a Beckman SW41Ti rotor (Fullerton, CA). Gradients were analyzed by passing the contents through a Pharmacia single-path UV-1 optical unit to monitor continual OD260. Total RNA from each fraction was extracted with phenol/chloroform, followed by chloroform alone after incubation with SDS/proteinase K. The identities of the 40S and 60S ribosome subunit peaks were confirmed by comparison to profiles from extracts to which EDTA was added before centrifugation. HIF-1α, β-actin, and GAPDH transcripts were quantified by RNase protection assay using complementary RNA probes synthesized with T7 RNA polymerase from XmnI-restricted pKSHIFIRES, SP6 RNA polymerase from PvuII-restricted pβ-actin (Ponte et al., 1983), and T7 RNA polymerase from DdeI-restricted pGAPM, respectively, essentially as described (Lagnado et al., 1994). Gels were scanned using a PhosphorImager and quantified using ImageQuant (Molecular Dynamics, Sunnyvale, CA).
RNase Protection Assay
The RNase protection probe for assessing transcripts from cells transfected with pRhifF was synthesized with T7 RNA polymerase from BstXI–digested pGemRhifF as previously described (Lagnado et al., 1994). Poly(A)+ RNA was isolated from cells lysed in SDS/proteinase K as described previously (Hogg et al., 1997), and 2 μg was subjected to RNase protection assay as described (Lagnado et al., 1994).
Northern Analysis
Poly(A)+ RNA was isolated as described previously (Hogg et al., 1997), and 4 μg was denatured in formaldehyde and separated on a 1% formaldehyde-agarose gel. RNAs were transferred to a nylon membrane by capillary blotting. Filters were UV cross-linked, and hybridizations were performed in ExpressHyb Hybridization solution (CLONTECH, Palo Alto, CA) as recommended by the manufacturer. The hybridization probes were Renilla luciferase (988-base pair SpeI/EcoRV cDNA fragment) and firefly luciferase (1656-base pair NcoI/XbaI cDNA fragment).
RESULTS
Total Translation Is Reduced During Hypoxia But Translation of HIF-1α mRNA Is Not Affected
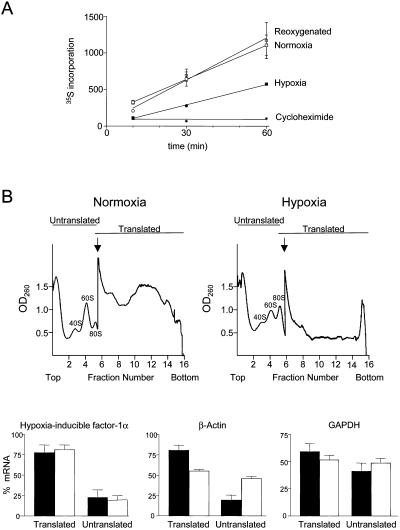
To determine the extent to which translation in NIH3T3 cells is suppressed by hypoxia, we compared the rate of [35S]methionine incorporation into cells that had been exposed to hypoxia for 24 h or cultured under normoxic conditions. Hypoxia caused a 50% decrease in the rate of methionine incorporation (Figure 2). In cells exposed to hypoxia for 23 h but returned to normoxia for 1 h before measurement of methionine incorporation, the incorporation rate returned to that of the control cells that had not been exposed to hypoxia, indicating that the effect of hypoxia on translation was reversible.
Figure 2.
Effect of hypoxia on protein synthesis. (A) Time course of incorporation of 35S-methionine/cysteine into TCA-precipitable protein. Cells were preincubated for 24 h under normoxia or hypoxia (1% O2) or incubated for 23 h under hypoxia, followed by 1 h of normoxia. Also shown is the incorporation into normoxic cells in the presence of 10 μg/ml cycloheximide. Each measurement was normalized with respect to total protein as determined by Bradford assay. (B) Effect of hypoxia on the translation of HIF-1α, β-actin, and GAPDH mRNAs determined by polysome profile analysis. NIH3T3 cells were incubated for 24 h in normoxia or hypoxia (1% O2), and cytoplasmic extracts were fractionated by 15–50% sucrose gradient centrifugation. Examples of polysome profiles showing the location of 80S ribosomes and free ribosome subunits are shown. The arrow indicates where the sensitivity of the UV monitor was increased fivefold. Transcripts that were being translated at the time of extract preparation are polysome-associated and sediment in fractions 7–16, with the most actively translated transcripts toward the bottom of the gradient. Cytoplasmic extracts from cells grown under normoxic or hypoxic conditions were fractionated, RNA was prepared from each fraction, and the HIF-1α, β-actin, and GAPDH mRNAs were detected by RNase protection assay. Each mRNA from the RNase protection assay was quantitated by phosphorimager and calculated as a percentage of the total for that mRNA. Data for fractions 1–6 and 7–16 were pooled and represented as translated or untranslated regions of the gradient, respectively. Black bars show data for normoxic cells, and white bars show data for hypoxic cells. The data represent the mean (±SEM) from three independent experiments.
To investigate whether the translation rate of HIF-1α mRNA is reduced under hypoxia or whether there might be a mechanism that spares HIF-1α from the overall reduction in translation, we examined polysome profiles for HIF-1α under normoxic and hypoxic growth conditions and compared the HIF-1α profiles with those of β-actin, a protein that is not induced during hypoxia, and GAPDH, which, although often considered a housekeeping gene, is induced during hypoxia. Cytoplasmic extracts from NIH3T3 cells grown under normoxic or hypoxic conditions were sedimented through sucrose gradients. The location of the HIF-1α, β-actin, and GAPDH mRNAs within the gradient was determined by RNase protection assay of fractions from the gradient and the proportion of mRNA that was associated with polysomes was quantitated (Figure 2). In cells grown under normoxic conditions the majority of the β-actin mRNA was found in the polysomal fraction, with only 23% of the mRNA in the fractions corresponding to untranslated mRNA. However, the proportion of untranslated β-actin mRNA was significantly increased in cells grown under hypoxia, with 46% of the mRNA located in the nonpolysomal region of the gradient. In contrast, the proportion of HIF-1α mRNA associated with polysomes was unaffected by hypoxia, indicating that the translation rate of HIF-1α was maintained. The proportion of GAPDH mRNA within the polysome region of the gradient was also largely unaffected by hypoxia, although the GAPDH mRNA was less efficiently translated than either HIF-1α or β-actin under both growth conditions, with only 50–60% of the mRNA sedimenting within the polysome region. These results indicate that translation of the HIF-1α and GAPDH mRNAs is not suppressed by hypoxia. We have not further investigated the features of the GAPDH mRNA that allow translation to be maintained during hypoxia. Unlike HIF1-α the GAPDH 5′UTR is of average length and base composition, although it contains a 5′ terminal polypyrimidine tract.
The HIF-1α 5′UTR Contains an Internal Ribosome Entry Site
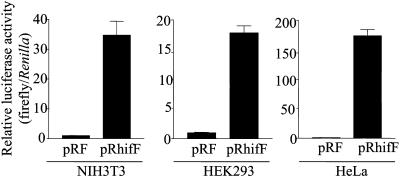
Because the 5′UTR of HIF-1α mRNA is rather long (286 nt) and GC-rich (72%) but is nevertheless translated efficiently under both normoxic and hypoxic conditions, we suspected it may contain an internal ribosome entry site. To test this, we inserted the HIF-1α 5′UTR into a dicistronic reporter gene, between the cDNAs encoding two distinct luciferase enzymes (Renilla luciferase and firefly luciferase), and examined the effect the HIF-1α segment had on the relative expression of activity from the downstream firefly luciferase cistron. The plasmids pRF, which contains a short linker sequence between the two luciferase cistrons, and pRhifF, which contains the HIF-1α 5′UTR inserted between the cistrons (Figure 1), were each transiently transfected into NIH3T3, HEK293, and HeLa cells. The presence of the HIF-1α 5′UTR increased the expression of the downstream firefly luciferase relative to Renilla luciferase by 30-fold in NIH3T3 cells, 18-fold in HEK293 cells, and 180-fold in HeLa cells (Figure 3), suggesting that the HIF-1α 5′UTR can indeed promote translation by internal ribosome entry.
Figure 3.
Relative enhancement of expression of the downstream reporter enzyme by the HIF-1α 5′UTR in various cell types. NIH3T3, HEK293, and HeLa cells were transiently transfected with pRF or pRhifF (Figure 1) as indicated. Renilla and firefly luciferase activities were determined 24 h posttransfection, and IRES activities represented as ratios of firefly to Renilla luciferase. The ratios for each cell line are graphed relative to pRF, which was given a value of 1. Data presented are the mean (±SEM) of triplicate samples from three independent experiments.
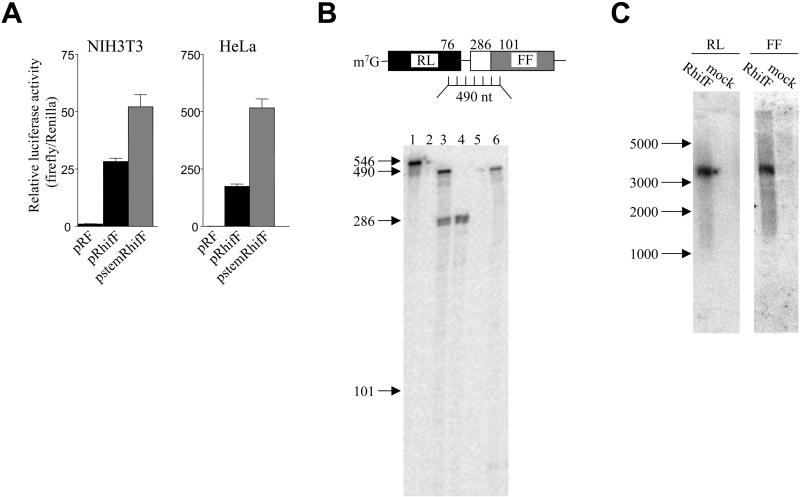
To check that the firefly luciferase was translated by internal ribosome entry and was not translated as a result of reinitiation of ribosomes that failed to dissociate from the mRNA after encountering the stop codon of the upstream Renilla luciferase, we inserted a 60-base pair palindrome sequence at the transcription start site, so that the transcript would have a strong stem-loop structure adjacent to the 5′cap, as described previously (Stoneley et al., 1998). This stem-loop was previously shown to inhibit cap-dependent translation (Stoneley et al., 1998). The presence of the stem-loop in stemRhifF reduced expression of Renilla luciferase without affecting expression of firefly luciferase, resulting in an increase in the relative firefly luciferase activity in both NIH3T3 and HeLa cells (Figure 4). This result demonstrates that the translation of the firefly luciferase does not result primarily from failure of ribosomes to dissociate after translation of the upstream cistron.
Figure 4.
The HIF-1α 5′UTR contains an IRES. (A) Effect of a 5′ stem-loop on relative expression of the downstream reporter enzyme. NIH3T3 and HeLa cells were transfected with pRF, pRhifF, or pstemRhifF (described in Figure 1) as indicated. IRES activities are represented as ratios of firefly luciferase to Renilla luciferase. All luciferase ratios are graphed relative to pRF, which was given a value of 1. Data are presented as mean (±SEM) of triplicate samples from three independent experiments. (B) Ribonuclease protection mapping of the intercistronic region of the dicistronic mRNA. A schematic showing the dicistronic mRNA containing the HIF-1α 5′ UTR hybridized to the antisense RNA probe used for RNase protection analysis is shown. The lengths of probe regions protected by the Renilla luciferase coding region, HIF1-α 5′UTR, and firefly luciferase coding region are indicated. The RNase protection gel is shown below, with sizes relative to molecular weight markers indicated. Lane 1, undigested RNA probe. Lane 2, 2 μg yeast tRNA. Lane 3, Poly(A)+ mRNA from NIH3T3 cells transfected with pRhifF. Lane 4, Poly(A)+ mRNA from mock transfected NIH3T3 cells. Lane 5, Poly(A)+ mRNA from mock transfected HeLa cells. Lane 6, Poly(A)+ mRNA from HeLa cells transfected with pRhifF. (C) Northern analysis of pRhifF mRNA in NIH3T3 cells. Poly(A)+ RNA was isolated from NIH3T3 cells either transiently transfected with pRhifF, or mock transfected, as indicated. Northern analysis was performed using 32P-labeled DNA probes for the Renilla (RL) or firefly (FF) luciferase regions as indicated. The migration of RNA size standards is indicated.
To verify that the firefly luciferase protein was synthesized by translation of the downstream cistron of an intact dicistronic transcript, rather than from a functionally monocistronic mRNA generated by splicing or by use of a cryptic promoter within the dicistronic gene, we performed an RNase protection mapping of the region spanning the end of the Renilla luciferase open reading frame into the firefly luciferase open reading frame. Assay of RNA from transfected NIH3T3 cells produced a major band of 490 nt, which is the size expected from intact dicistronic mRNA, and a second band of 286 nt, which is the size expected from protection by endogenous HIF1-α mRNA (Figure 4). When RNA from mock-transfected NIH3T3 cells was assayed, only the endogenous HIF1-α product was seen, verifying the identity of this band. Assay of RNA from transfected HeLa cells produced a single major product of the size expected from protection by intact dicistronic mRNA. No endogenous HIF1-α from HeLa cells was seen because the probe only detects mouse HIF1-α mRNA. No product of a size that would indicate the presence of a cryptic promoter or alternative splicing event was seen either in NIH3T3 or HeLa cells. Importantly, apart from the endogenous HIF-1α product in NIH3T3 cells, there was no product of a size intermediate between the 490-nt fragment protected by intact dicistronic transcript and 101 nt, which is the minimal size product required from a transcript that encodes firefly luciferase open reading frame. Hence, the increased expression of firefly luciferase that results from insertion of the HIF-1α 5′UTR must result from translation of intact dicistronic mRNA. To further demonstrate the firefly luciferase expression was from an intact dicistronic mRNA, Northern analysis was performed on RNA isolated from NIH3T3 cells transfected with pRhifF. Probes for both Renilla luciferase and firefly luciferase detected a single RNA species corresponding to the size expected of the dicistronic mRNA (3.2 kb) in transfected cells but not in mock-transfected cells (Figure 4C).
Relative Efficiency of the HIF-1α IRES Compared with Other Cellular IRESs
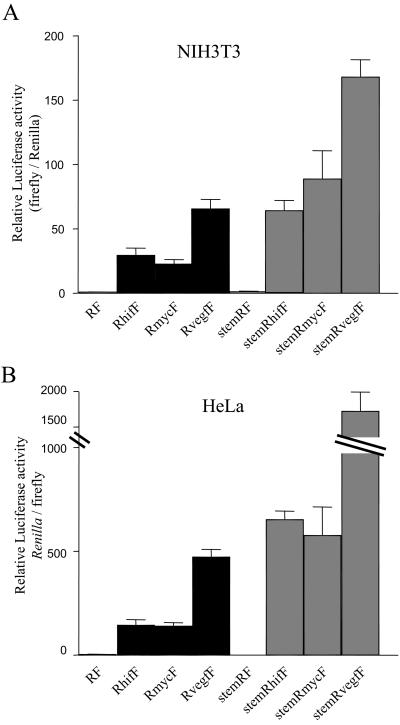
A number of mRNAs that encode proteins involved in regulation of cell growth or response to stress have been found to contain IRESs. These include eIF4G, X-linked inhibitor-of-apoptosis (XIAP), NF-κB repressing factor (NRF), c-myc, and vascular endothelial growth factor (VEGF; Gan and Rhoads, 1996; Akiri et al., 1998; Miller et al., 1998; Stein et al., 1998; Stoneley et al., 1998; Holcik et al., 1999; Oumard et al., 2000). To compare the activity of the HIF-1α IRES with other cellular IRESs we transfected NIH3T3 and HeLa cells with dicistronic reporters containing either the HIF-1α, c-myc, or VEGF 5′UTR (Figure 5). In both cell types the HIF-1α IRES had activity comparable to the c-myc IRES, whereas the VEGF 5′UTR was two- to threefold more active. All three IRESs were considerably more active in HeLa cells than in NIH3T3 cells. A similar relationship between the activities of the different IRESs was seen when a stem-loop was inserted upstream of the Renilla luciferase cistron to inhibit cap-dependent translation. The relative ratio of firefly to Renilla luciferase was increased two- to threefold for each IRES and in both cell types.
Figure 5.
Comparison of the activities of the HIF-1α, c-myc, and VEGF IRESs. NIH3T3 and HeLa cells were transiently transfected with the various dicistronic reporter constructs described in Figure 1. Renilla and firefly luciferase activities were determined 24 h posttransfection, and IRES activities were represented as ratios of firefly to Renilla luciferase, relative to the ratio obtained with pRF, which was given a value of 1. Data are presented as the mean (±SEM) of triplicate samples from three independent experiments.
IRES Activity During Hypoxia and Serum Deprivation
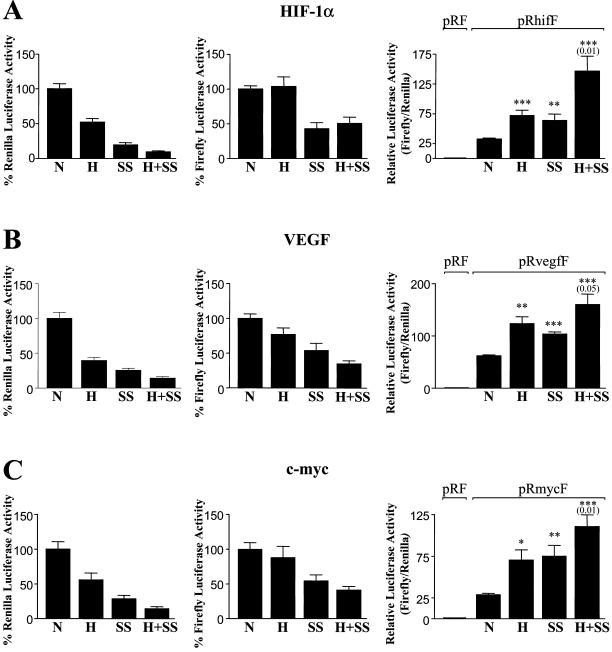
The function of the IRES in the HIF-1α 5′UTR might be to allow efficient translation during hypoxia. If this were the case, we would expect translation of firefly luciferase from the dicistronic RhifF mRNA to be relatively unaffected by hypoxia. To test this, we subjected NIH3T3 cells transfected with the dicistronic RhifF gene to hypoxia for 24 h and then measured the relative luciferase activities (Figure 6). Under hypoxia Renilla luciferase activity was reduced to 50% of the level seen in normoxic cells, consistent with the effect of hypoxia on total translation rate measured by incorporation of [35S]methionine (Figure 2). In contrast firefly luciferase activity was the same in normoxic and hypoxic cells, indicating that IRES-mediated translation is unaffected by hypoxia. To examine whether the HIF-1α IRES also conferred a translational advantage during other forms of cellular stress, we also subjected cells to serum starvation for 24 h. Serum starvation reduced expression of both Renilla luciferase and firefly luciferase, but the Renilla luciferase activity was the more affected, suggesting that IRES-dependent translation was less sensitive than cap-dependent translation to the serum starvation. When serum-starved cells were subjected to hypoxia, the Renilla luciferase activity was further reduced by 50%, but firefly luciferase activity was maintained at the same level as in normoxic serum-starved cells. Thus, the ratio of firefly luciferase to Renilla luciferase was greatest in cells subjected to deprivation of both serum and oxygen. These results are consistent with the function of the IRES being to maintain efficient translation of HIF-1α during hypoxia.
Figure 6.
IRES activity is maintained during hypoxia. NIH3T3 cells were transiently transfected with pRhifF (A), pRvegfF (B), or pRmycF (C). Four hours posttransfection, cells were subjected to either hypoxia, serum starvation, or serum starvation and hypoxia together or were left under control conditions for a further 24 h before harvesting and assay for Renilla and firefly luciferase. Each luciferase activity is shown relative to the activity under normoxia, which is expressed as 100%. The histograms at left show Renilla luciferase activity, the middle histograms show firefly luciferase, and the histograms at right show the ratio of firefly to Renilla. The ratios are graphed relative to the ratio obtained with pRF under normoxia, which was given a value of 1. The data shown are means (±SEM) of triplicate samples from each of five independent experiments. Statistical significance of the difference between normoxic and treatment was tested using the two-tailed Student's t test (*p < 0.01; **p < 0.001; ***p < 0.0001). The significance level for the effect of hypoxia on serum-starved cells (H+SS vs. SS) is shown in brackets.
The property of the HIF-1α IRES of protecting translation from suppression by hypoxia might be a specific feature of the HIF-1α 5′UTR or could be a general property of cellular IRESs. To investigate whether this property might be shared by other cellular IRESs, we compared the effect of hypoxia on the HIF-1α IRES to the effect on the activity of the IRES from VEGF, which also functions during hypoxia (Stein et al., 1998), and from c-myc, which is not known to play a role in the response to hypoxia. Firefly luciferase activity from dicistronic reporters containing either the c-myc or the VEGF IRES was only marginally reduced by hypoxia, whereas Renilla activity was significantly reduced (Figure 6). The VEGF and c-myc IRESs also resembled the HIF-1α in being less sensitive to serum starvation than was the cap-dependent translation of Renilla luciferase, indicated by the approximately twofold increase in the ratio of firefly to Renilla luciferase in serum-starved cells. The differential sensitivity to hypoxia was maintained in cells starved of serum. Thus, the maintenance of activity during hypoxia may be a general property of IRES-mediated initiation of translation.
DISCUSSION
We find that protein synthesis in NIH3T3 cells is reduced when cells are cultured under hypoxic (1% O2) conditions. Total protein synthesis measured by methionine plus cysteine incorporation was reduced by 50% after 24-h exposure to hypoxia, and a similar reduction was seen in the level of cap-dependent luciferase reporter in transfected cells subjected to hypoxia. These observations are consistent with previous reports that hypoxia causes a suppression of protein synthesis by 55% in rat astrocytes (Stein et al., 1998), 70–85% in rat hepatocytes (Tinton and Buc-Calderon, 1999), and 60–70% in the human cervical cell line NHIK 3025 (Kraggerud et al., 1995). Using polysome profile analysis to assess the effect of hypoxia on the proportion of mRNA associated with translationally active polyribosomes, we found that the proportion of β-globin mRNA that cosedimented with polysomes was reduced by hypoxia, consistent with the reduction in overall protein synthesis. However, the association of HIF-1α mRNA with polysomes was unaffected by the hypoxia, suggesting the HIF-1α mRNA is translated equally well under normoxia and hypoxia. This finding is consistent with a recent report of the effect of hypoxia on polysome profiles in HeLaS3 cells (Gorlach et al., 2000), who found that the translational activities of β-actin and the ribosomal protein L28 were reduced by hypoxia but HIF-1α was unaffected.
We found using a dicistronic reporter that the HIF-1α mRNA 5′UTR can function as an IRES and that the IRES activity is not affected by hypoxia. Internal ribosome entry was first recognized as an alternative mechanism of initiation of translation in studies of poliovirus and encephalomyocarditis virus, which have uncapped mRNAs and thus require a cap-independent mechanism of initiation (Jang et al., 1988; Pelletier and Sonenberg, 1988). The viruses make use of this cap-independent mechanism to allow preferential synthesis of viral proteins while translation of host cell proteins is shut off, in part because of cleavage of the translation initiation factor eIF4G by a viral protease. When the first examples of cellular mRNAs that contain IRESs were found it was not obvious what advantage there would be in having this alternative mechanism of translation initiation for an mRNA that has a 5′ cap. However, it is now evident that a number of proteins that function during periods of cellular stress are translated from mRNAs that contain an IRES (Vagner et al., 1996; Holcik et al., 1999; Stoneley et al., 2000), and it is reasonable to postulate that these mRNAs contain IRESs to ensure their translation is not compromised during the stress condition. This has been proposed to be the function of the IRES in VEGF mRNA, which like HIF-1α has an important function during hypoxia and like the HIF-1α mRNA, contains an IRES (Stein et al., 1998). Although additional regulatory mechanisms contribute to the expression of both HIF-1α and VEGF during hypoxia, polysome profile analysis indicates that both transcripts remain efficiently translated during hypoxia (Stein et al., 1998; Gorlach et al., 2000; this report).
The maintenance of rate of translation during hypoxia is not restricted to hypoxia-induced genes, because the c-myc IRES also has this property. Furthermore, we also found that translation from the HIF-1α, VEGF, and c-myc IRESs was less inhibited by serum starvation than was the translation of a cap-dependent reporter, and a similar observation has recently been made with the IRES from the X-linked inhibitor of apoptosis (XIAP) mRNA (Holcik et al., 1999). Thus, the preservation of translational efficiency during cellular stress may be a common feature of cellular IRESs.
Another pattern that is emerging from studies of cellular IRESs relates to the large size and GC-richness of the 5′UTRs of mRNAs that contain IRESs. We were first alerted to the possibility that HIF-1α contains an IRES by this property, and we suggest that any gene found to have a 5′UTR that is GC-rich and longer than ∼250 bases is a good candidate for having an IRES. Whether there are more specific features that are universally essential to IRES activity remains to be determined, but the likely negative effect on cap-dependent translation of a large GC-rich 5′UTR, combined with the frequency with which genes containing a long, GC-rich 5′UTR have been found to contain IRESs, makes it a good indicator. Predicting the relative strengths of cellular IRESs does not appear possible at this stage, because only a few comparisons have been made, and no feature that correlates well with IRES strength is evident. We found that the HIF-1α IRES has a similar strength in the dicistronic assay to the c-myc IRES and about half the activity of the VEGF 5′UTR in both a mouse and a human cell line. The greater activity of the VEGF 5′UTR does not appear to relate to its greater size or to the fact that the VEGF 5′UTR contains two IRESs (Huez et al., 1998), because deletion of a considerable portion of the VEGF 5′UTR can produce an even more active IRES (Stein et al., 1998).
Several observations suggest that the relative activity of an IRES may depend on the cell type. We found that the activity of the HIF-1α, c-myc, and VEGF IRESs is greater in HeLa cells than in NIH3T3 mouse fibroblasts. The c-myc IRES has also been found to be more active in HeLa cells than in a number of other cell lines, including BALB/c 3T3 mouse fibroblasts, and COS7 cells (Stoneley et al., 1998). Presumably a factor that participates in IRES function is present at limiting levels in some cells. Although we found IRES activity to be considerably greater in HeLa than in NIH3T3 cells, there was good concordance between the relative activities of the different IRESs in the two cell lines.
Our findings contribute to an emerging pattern of IRESs being present in cellular mRNAs whose translation is required at times when protein synthesis in general is submaximal. Downregulation of translation is achieved in most situations by further limiting the availability of eIF4E, the cap-binding protein. Thus, any mRNAs that can initiate translation by a cap-independent method are protected against this form of suppression of translation. Situations where protein synthesis is downregulated include mitosis, amino acid starvation, glucose starvation, heat shock, oxygen deprivation, and apoptosis. Most of the cellular mRNAs known to contain an IRES have a function during one or more of these special circumstances. In the case of HIF-1α, the protein plays a key role in adaptation to hypoxia, and the IRES in the 5′UTR is likely to contribute to ensuring it is adequately expressed during hypoxia.
ACKNOWLEDGMENTS
The authors thank A. Willis for gifts of the plasmids, pRF (formerly pGL3R) and pstemRmycF (formerly pGL3utrH), B. May for use of the luminometer, T. Gonda and B. Wattenberg for critical reading of the manuscript, and all members of the Goodall lab for helpful discussion. This work was supported by grants from the National Heart Foundation of Australia and the Anti-Cancer Foundation of South Australia. K.L. is the recipient of a Dawes Scholarship from the Royal Adelaide Hospital.
Abbreviations used:
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HEPES
N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid
- HIF
hypoxia-inducible factor
- IRES
internal ribosome entry site
- PBS
phosphate-buffered saline
- UTR
untranslated region
- VEGF vascular endothelial growth factor.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.02–02–0017. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.02–02–0017.
REFERENCES
- Akiri G, Nahari D, Finkelstein Y, Le SY, Elroy-Stein O, Levi BZ. Regulation of vascular endothelial growth factor (VEGF) expression is mediated by internal initiation of translation and alternative initiation of transcription. Oncogene. 1998;17:227–236. doi: 10.1038/sj.onc.1202019. [DOI] [PubMed] [Google Scholar]
- Amellem O, Pettersen EO. Cell inactivation and cell cycle inhibition as induced by extreme hypoxia: the possible role of cell cycle arrest as a protection against hypoxia-induced lethal damage. Cell Prolif. 1991;24:127–141. doi: 10.1111/j.1365-2184.1991.tb01144.x. [DOI] [PubMed] [Google Scholar]
- Ebert BL, Firth JD, Ratcliffe PJ. Hypoxia and mitochondrial inhibitors regulate expression of glucose transporter-1 via distinct Cis-acting sequences. J Biol Chem. 1995;270:29083–29089. doi: 10.1074/jbc.270.49.29083. [DOI] [PubMed] [Google Scholar]
- Firth JD, Ebert BL, Pugh CW, Ratcliffe PJ. Oxygen-regulated control elements in the phosphoglycerate kinase 1 and lactate dehydrogenase A genes: similarities with the erythropoietin 3′ enhancer. Proc Natl Acad Sci USA. 1994;91:6496–6500. doi: 10.1073/pnas.91.14.6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan W, Rhoads RE. Internal initiation of translation directed by the 5′- untranslated region of the mRNA for eIF4G, a factor involved in the picornavirus- induced switch from cap-dependent to internal initiation. J Biol Chem. 1996;271:623–626. doi: 10.1074/jbc.271.2.623. [DOI] [PubMed] [Google Scholar]
- Gorlach A, Camenisch G, Kvietikova I, Vogt L, Wenger RH, Gassmann M. Efficient translation of mouse hypoxia-inducible factor-1alpha under normoxic and hypoxic conditions. Biochim Biophys Acta. 2000;1493:125–134. doi: 10.1016/s0167-4781(00)00172-x. [DOI] [PubMed] [Google Scholar]
- Hogg A, Schirm S, Nakagoshi H, Bartley P, Ishii S, Bishop JM, Gonda TJ. Inactivation of a c-Myb/estrogen receptor fusion protein in transformed primary cells leads to granulocyte/macrophage differentiation and down regulation of c-kit but not c-myc or cdc2. Oncogene. 1997;15:2885–2898. doi: 10.1038/sj.onc.1201472. [DOI] [PubMed] [Google Scholar]
- Holcik M, Lefebvre C, Yeh C, Chow T, Korneluk RG. A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nat Cell Biol. 1999;1:190–192. doi: 10.1038/11109. [DOI] [PubMed] [Google Scholar]
- Huang LE, Arany Z, Livingston DM, Bunn HF. Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J Biol Chem. 1996;271:32253–32259. doi: 10.1074/jbc.271.50.32253. [DOI] [PubMed] [Google Scholar]
- Huez I, Creancier L, Audigier S, Gensac MC, Prats AC, Prats H. Two independent internal ribosome entry sites are involved in translation initiation of vascular endothelial growth factor mRNA. Mol Cell Biol. 1998;18:6178–6190. doi: 10.1128/mcb.18.11.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappel A, Ronicke V, Damert A, Flamme I, Risau W, Breier G. Identification of vascular endothelial growth factor (VEGF) receptor-2 (Flk-1) promoter/enhancer sequences sufficient for angioblast and endothelial cell-specific transcription in transgenic mice. Blood. 1999;93:4284–4292. [PubMed] [Google Scholar]
- Kraggerud SM, Sandvik JA, Pettersen EO. Regulation of protein synthesis in human cells exposed to extreme hypoxia. Anticancer Res. 1995;15:683–686. [PubMed] [Google Scholar]
- Ladoux A, Frelin C. Hypoxia is a strong inducer of vascular endothelial growth factor mRNA expression in the heart. Biochem Biophys Res Commun. 1993;195:1005–1010. doi: 10.1006/bbrc.1993.2144. [DOI] [PubMed] [Google Scholar]
- Lagnado CA, Brown CY, Goodall GJ. AUUUA is not sufficient to promote poly(A) shortening and degradation of an mRNA: the functional sequence within AU-rich elements may be UUAUUUA(U/A)(U/A) Mol Cell Biol. 1994;14:7984–7995. doi: 10.1128/mcb.14.12.7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macejak DG, Sarnow P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature. 1991;353:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- Miller DL, Dibbens JA, Damert A, Risau W, Vadas MA, Goodall GJ. The vascular endothelial growth factor mRNA contains an internal ribosome entry site. FEBS Lett. 1998;434:417–420. doi: 10.1016/s0014-5793(98)01025-4. [DOI] [PubMed] [Google Scholar]
- Norris ML, Millhorn DE. Hypoxia-induced protein binding to O2-responsive sequences on the tyrosine hydroxylase gene. J Biol Chem. 1995;270:23774–23779. doi: 10.1074/jbc.270.40.23774. [DOI] [PubMed] [Google Scholar]
- Oumard A, Hennecke M, Hauser H, Nourbakhsh M. Translation of NRF mRNA is mediated by highly efficient internal ribosome entry. Mol Cell Biol. 2000;20:2755–2759. doi: 10.1128/mcb.20.8.2755-2759.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- Ponte P, Gunning P, Blau H, Kedes L. Human actin genes are single copy for alpha-skeletal and alpha-cardiac actin but multicopy for beta- and gamma-cytoskeletal genes: 3′ untranslated regions are isotype specific but are conserved in evolution. Mol Cell Biol. 1983;3:1783–1791. doi: 10.1128/mcb.3.10.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salceda S, Caro J. Hypoxia-inducible factor 1alpha (HIF-1alpha) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions. Its stabilization by hypoxia depends on redox-induced changes. J Biol Chem. 1997;272:22642–22647. doi: 10.1074/jbc.272.36.22642. [DOI] [PubMed] [Google Scholar]
- Savant Bhonsale S, Cleveland DW. Evidence for instability of mRNAs containing AUUUA motifs mediated through translation-dependent assembly of a >20S degradation complex. Genes Dev. 1992;6:1927–1939. doi: 10.1101/gad.6.10.1927. [DOI] [PubMed] [Google Scholar]
- Schmaltz C, Hardenbergh PH, Wells A, Fisher DE. Regulation of proliferation-survival decisions during tumor cell hypoxia. Mol Cell Biol. 1998;18:2845–2854. doi: 10.1128/mcb.18.5.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL, Roth PH, Fang HM, Wang GL. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J Biol Chem. 1994;269:23757–23763. [PubMed] [Google Scholar]
- Shweiki D, Itin A, Soffer D, Keshet E. Vascular endothelial growth factor induced by hypoxia may mediate hypoxia-initiated angiogenesis. Nature. 1992;359:843–845. doi: 10.1038/359843a0. [DOI] [PubMed] [Google Scholar]
- Stein I, Itin A, Einat P, Skaliter R, Grossman Z, Keshet E. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol Cell Biol. 1998;18:3112–3119. doi: 10.1128/mcb.18.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneley M, Chappell SA, Jopling CL, Dickens M, MacFarlane M, Willis AE. c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Mol Cell Biol. 2000;20:1162–1169. doi: 10.1128/mcb.20.4.1162-1169.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneley M, Paulin FE, Le Quesne JP, Chappell SA, Willis AE. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- Tinton SA, Buc-Calderon PM. Hypoxia increases the association of 4E-binding protein 1 with the initiation factor 4E in isolated rat hepatocytes. FEBS Lett. 1999;446:55–59. doi: 10.1016/s0014-5793(99)00185-4. [DOI] [PubMed] [Google Scholar]
- Vagner S, Gensac MC, Maret A, Bayard F, Amalric F, Prats H, Prats AC. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol Cell Biol. 1995;15:35–44. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner S, Touriol C, Galy B, Audigier S, Gensac MC, Amalric F, Bayard F, Prats H, Prats AC. Translation of CUG- but not AUG-initiated forms of human fibroblast growth factor 2 is activated in transformed and stressed cells. J Cell Biol. 1996;135:1391–1402. doi: 10.1083/jcb.135.5.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]