Abstract
Gene expression can be modulated depending on physiological and developmental requirements. A multitude of regulatory genes, which are organized in interdependent networks, guide development and eventually generate specific phenotypes. Transcription factors (TF) are a key element in the regulatory cascade controlling cell fate and effector functions. In this review, we discuss recent data on the diversity of TF that determine natural killer (NK) cell fate and NK cell function.
Keywords: Transcription factor, NK cell differentiation, Migration, Effector function
Transcriptional regulation of NK cell development
The basis for the transcriptional regulation of NK cell development is just beginning to come into view. Compared to T and B cells for which an extensive transcriptional program is well established, NK cells are the Cinderellas of the lymphocyte family. The contribution of individual transcription factors to the development of conventional NK cells development is outlined below.
Ets1
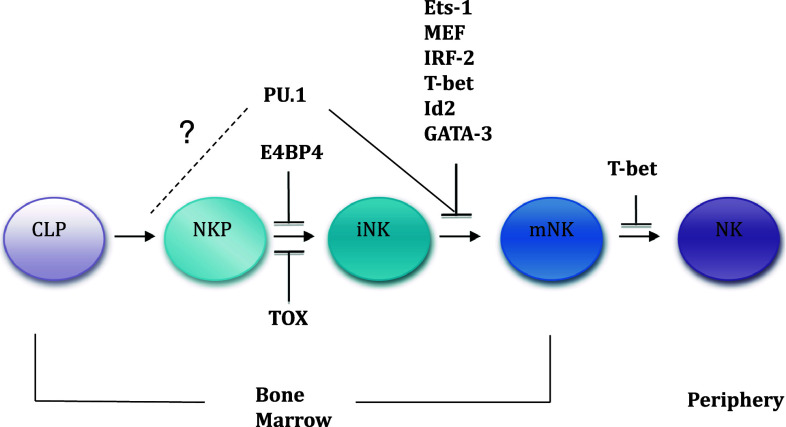
E-twenty-six (Ets)1 is a member of the large Ets family of transcription factors that have a winged helix-turn-helix DNA binding domain and are highly expressed in lymphoid cells with multiple roles in B and T cells, particularly in regulating cytokine expression [1]. Ets1 −/− mice have about threefold less NK cells in the spleen than wild-type littermates, and the remaining NK cells show both impaired cytotoxicity and cytokine release [2]. These data suggest that Ets1 has a role in the production of mature NK cells (mNK). However, functional mNK cells are still produced in the absence of Ets1 which indicates that the action of Ets1 itself is not essential and may overlap that of other Ets family transcription factors such as PU.1 or MEF, both of which have roles in NK cell development (see below).
PU.1
PU.1 is involved in the development of multi-potent lymphoid progenitor cells and, for example, is required for B cell production [3]. SPI1 −/− mice lack the expression of the PU.1 protein and die in utero, though survive long enough to develop a fetal liver that is a rich source of hematopoietic stem cells. Chimeric mice can then be produced by transferring fetal liver cells into irradiated Rag −/− Il2rg −/− recipients [4]. All the lymphocytes in the chimeras lack PU.1 and this results in a complete loss of B or T cells but NK cells still develop. However, the number of mNK cells produced is reduced four- to tenfold (depending on the number of fetal liver cells transplanted). A decrease in the number of Spi1 −/−-derived NK cell precursors (NKP) in the bone marrow (BM) is also found. This effect on NKP is not necessarily a NK cell intrinsic effect as the lack of PU.1 could also severely impair the function of the lymphoid precursors that may be reflected in the loss of B and T cells. Mature Spi1 −/− NK cells are poorly activated by IL-2 and IL-12, but their cytotoxicity is apparently unaffected. The latter clearly differentiates PU.1 from Ets1 in NK cell effector function.
MEF
The myeloid ELF1-like (MEF) factor is a member of the Ets family of transcription factors that can influence NK cell development. Similar to other Ets members, Mef −/− mice have only 40% of the normal number of splenic mNK cells [5] and the remaining NK cells do not proliferate normally in response to IL-2.
T-bet
The T-box expressed in T cells (also known as Tbx21) is a member of the T-box transcription factor family and is well characterized with respect to its role in regulating the Th1 lineage specification of CD4+ T cells [6]. Tbx21 −/− mice have approximately only one-third the normal number of mature splenic NK cells but increased numbers of NK cells within their BM [7]. Long-term BM chimeras where Tbx21 −/− BM was transferred into Tbx21 +/+ or Tbx21 −/− hosts, and mixed BM chimera where Tbx21 +/+ and Tbx21 −/− BM were co-transferred into irradiated host, showed that, compared to Tbx21 +/+ BM, Tbx21 −/− BM gave rise to a reduced but detectable number of NK1.1+ TCRβ− NK cells [7]. These results led the authors to conclude that the defects in NK cell development were entirely stem cell intrinsic. Nonetheless, several lines of evidence support the idea that expression of T-bet in NK cells may not be sufficient to complete late stages of peripheral NK cell differentiation, such as the CD11bhiCD27hi to CD11bhiCD27low transition (A.M.F., unpublished data).
GATA-3
GATA-3 is a zinc finger transcription factor and has a well-characterized role in T cell development [8]. Due to the embryonic lethality of Gata3 −/− mice, studies on GATA-3’s role in NK cells have relied upon the generation of hematopoietic chimeras by transferring fetal liver cells into irradiated Rag −/− Il2rg −/− recipients as for PU.1. Splenic NK cell numbers are not affected by GATA-3 loss nor is their cytotoxic potential, although the number of hepatic NK cells is reduced in the chimeric mice [9]. Gata3 −/− NK cells have a more immature phenotype and more restricted Ly49 repertoire than normal, suggesting a role for GATA-3 in the immature NK (iNK) to mNK cell transition [9]. GATA-3 is, however, essential for the production of thymic (t) NK cells, a distinct NK cell population found in the thymus with a characteristic elevated expression of IL-7Rα [10].
IRF-2
The interferon regulatory factor (IRF)-2 is a member of the IFN regulatory transcription factor (IRF) family which regulates expression of the IFN-α and IFN-β genes. Irf2 −/− mice have a large reduction in mNK cells, but normal levels of iNK cells, in the spleen and liver [11, 12].
Id2
Id2 is a transcription factor that acts to repress transcriptional activity of the basic helix-loop-helix E-box family of transcription factors such as E2A, E2-2 and HEB. The role of Id2 and E-box factors has been extensively studied in both B and T cells [13]. Initial observations indicated decreased numbers of NK cells in Id2 −/− mice [14, 15]. Further analysis of Id2 −/− mice demonstrates a tenfold decrease in mNK cells in the BM and periphery but no effect on the numbers of iNK cells [16]. NKP cells are also present at normal levels in the BM of Id2−/− mice. Since Id2 acts via its suppression of E2A, Id2 −/− x E2a −/− mice were generated and show a restoration of mNK cell numbers within BM but do not rescue them in spleen. However, splenic NK cells from Id2 −/− x E2a −/− mice produce more IFN-γ after stimulation much better than do Id2 −/− splenic NK cells.
These data demonstrate that the additional loss of E2A is not sufficient to restore migration of Id2 −/− NK cells out of the BM, suggesting that at least one other E-box protein is active during NK cell development. Even though Id2 is highly expressed in BM, NKP numbers are normal in Id2 −/− mice so it is suggested that Id3 may act to replace Id2 function resulting in normal NKP levels [17]. Recent data have shown that over-expression of Id2 in human thymic progenitor cells in vitro strongly favors NK cell production so providing further evidence for the central role of Id2 in NK cell development [18].
E4bp4
E4bp4 (also known as Nfil3) is a basic leucine zipper transcription factor with significantly higher expression in NK cells than in B or T cells. E4bp4 −/− mice have virtually no detectable mNK cells in the periphery and no NK cell-mediated cytotoxicity [19]. There are very few mature or iNK cells in the BM of E4bp4 −/− although NKP cell numbers are unchanged. This is directly reflected in the relative expression level of E4bp4 that is low in NKP cells but eight- to tenfold higher in iNK and mNK cells. IL-15 is a crucial factor promoting NK cell development and survival [19]. Over-expression of E4bp4 in either hematopoietic progenitors from Il15ra −/− mice or in progenitors cultured without IL-15 rescued NK cell production, suggesting that E4bp4 acts downstream of IL-15. Gene expression analysis demonstrated that expression of both Id2 and Gata-3 is reduced in the absence of E4bp4. Over-expression of E4bp4 in wild-type progenitors resulted in induction of Gata-3 and Id2 expression indicating that both should act downstream of E4bp4. Retroviral transduction of E4bp4 −/− hematopoietic progenitors by Id2 rescues NK cell development. Retroviral transduction of wild-type hematopoietic progenitors with E4bp4 greatly enhances the number of NK cells produced [19]. These data identify E4bp4 as an essential transcription factor for development of the NK cell lineage that acts downstream of IL-15 and Id2.
Bcl11b
Bcl11b is a Kruppel-like transcriptional repressor that plays a major role in early T cell development [20]. T cell development in the thymus of Bcl11b −/− mice is blocked at the DN2 stage, but these thymic progenitors can be kept in continuous cell culture and have superior proliferative capacity to wild-type cells [21]. These cells have also elevated expression of genes such as E4bp4 and Id2 that are associated with NK cell development. Therefore, Bcl11b is required to attenuate the proliferative capacity of thymic progenitors and enhance T cell development by repressing the expression of genes that would induce commitment to alternative lineages. Bcl11b−/− T cells from all developmental stages have gained the ability to grow in response to IL-2 and exhibit a transcriptional profile that is very similar to normal activated NK cells. These Bcl11b −/− cells are referred to as induced T-to-NK (ITNK) cells [21]. The ITNK cells are very similar to conventional NK cells morphologically, in terms of transcriptional profile, ability to kill tumor cells in vitro and preventing tumor metastasis in vivo. It is possible that, due to their strong proliferative ability, ITNK cells could represent a potential source of transferable NK cell activity for therapeutic application [22].
Tox
Tox is a member of the diverse transcription factor family with a high mobility group (HMG) box and involved in regulating chromatin accessibility for other transcription factors. Tox has already been characterized in T cell development where it has an essential role during T cell selection [23]. mNK cells are almost completely absent in the spleen of Tox −/− mice [24]. Similarly, mNK cells are virtually absent from the BM, and iNK cells are reduced by 50% whereas the NKP cell numbers are unchanged. The relative expression level of Tox increases in iNK and mNK cells in BM relative to NKP cells in an analogous manner to that seen for E4bp4, while Id2 expression appears to be Tox dependent. However, retroviral transduction of Tox −/− hematopoietic progenitors by Id2 does not rescue NK cell development [24]. Tox seems to act in a similar fashion to E4bp4, but the Id2 rescue data suggest that E4bp4 and Tox act via separate downstream pathways. Retroviral transduction of Tox −/− hematopoietic progenitors by E4bp4 (and vice versa) will help to clarify the relationship of these TF for NK cell development.
NK cell migration and effector functions
As indicated above, NK cells develop primarily in the BM, thymus, and lymph nodes and are distributed in lymphoid tissues, and in many organs throughout the body, including uterus, liver, lung, intestine, and peritoneum. Resident and recruited NK cells play important roles in immunological surveillance [25] and homeostasis of mucosal surfaces [26]. Mobilization to inflamed tissues and lymphoid organs requires a dedicated network of surface molecules expressed by NK cells that allow interaction with endothelial cells whereas NK cell effector functions involve the coordinated action of multiple proteins such as cytokines, chemokines, and proteases. In this section, we review emerging data on the contribution of TF to modulate NK cell traffic and effector functions.
NK cell migration to peripheral lymphoid organs and inflamed tissues
A number of TF that were originally described in the context of CD4+ T cell development and effector function [27] have now also been shown to have important roles in migration. For example, Tbx21 −/− mice are resistant to a wide range of autoimmune disorders, including type I diabetes, inflammatory colitis and arthritis, lupus nephritis, and experimental autoimmune encephalomyelitis [28]. This resistance to inflammatory diseases is characterized by a lack of T cell infiltration at pathological sites, due to modulation of molecules involved in rolling and adhesion to inflamed endothelium, such as selectins and chemokine receptors [29]. In contrast to T cells, the contribution of T-bet to NK cell migration remains poorly characterized. For example, the observation that Tbx21 −/− NK cells migrate to reactive lymph nodes as efficiently as wild-type NK cells (A.M.F., unpublished data) suggests that the absence of T-bet does not affect rolling and adhesion to high endothelial venules. In line with this idea, a recent study addressed the role of T-bet expressed by NK cells in controlling melanoma metastasis to the lung, to conclude that T-bet does not seem to modulate NK cell migration to pathological tissues [30]. In the latter study, Werneck and colleagues co-transferred wild-type and T-bet-deficient NK cells labeled with different dyes and showed no differences in the recovery of NK cells from lungs, demonstrating that homing of Tbx21 −/− NK cells is not distinguishable from Tbx21 +/+ NK cells. Thus, the failure of Tbx21 −/− NK cells to protect mice after melanoma B16.F10 challenge was attributed to a defect in survival and inefficient lysis of tumour cells rather than a defective mobilization.
The chemokine receptor CXCR3 is critically involved in targeting effector T cells to inflamed tissues [31]. Current literature supports a role for T-bet in promoting CXCR3 expression by in vitro-polarized CD4+ Th1 cells [29]. Indeed, in the absence of T-bet, levels of CXCR3 transcripts and surface expression of CXCR3 protein are minimal compared to Tbx21 +/+ CD4+ T cells activated under Th1-polarizing conditions. In addition, the functional absence of CXCR3 was demonstrated by an impaired response to CXCR3 ligands in vitro that was rescued when Tbx21 −/− CD4+ T cells were retrovirally transduced with T-bet, indicating that T-bet can drive functional expression of CXCR3. Migration of NK cells to inflamed lymph nodes [32] and to tumor sites [33] seems to be exclusively dependent upon the expression of CXCR3 on NK cells and of CXCR3 ligands CXCL-9, CXCL-10, and CXCL-11 in tissues. Nevertheless, Tbx21 −/− NK cells express levels of CXCR3 similar to Tbx21 +/+ counterparts and migrate efficiently to inflamed lymph nodes (A.M.F., unpublished data). Thus, the different role of T-bet in modulating CXCR3 expression and function in NK cells and in antigen-primed CD4+ T cells may reflect tissue-specific differences similar to those reported for other genes. For example, T-bet has been shown to repress TNF expression in dendritic cells (DCs) with dramatic consequences in gut homeostasis [34, 35], whereas T-bet activates TNF in CD4+ T cells [36].
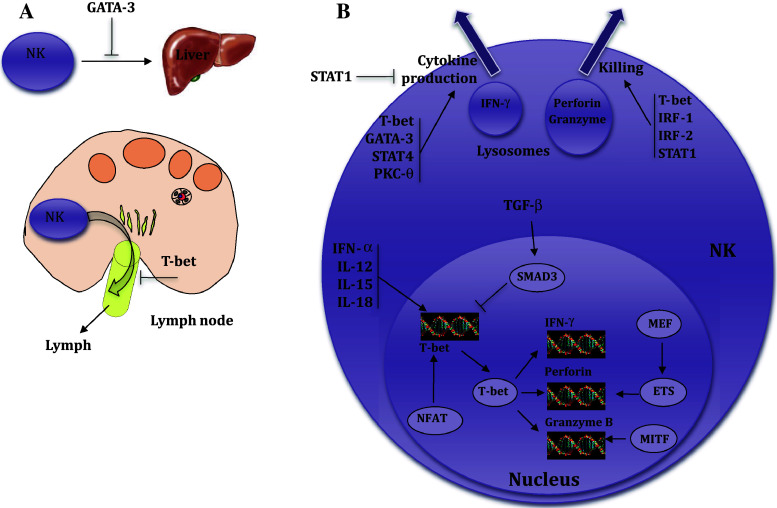
The role of the TF GATA-3 in NK cell migration has been addressed in hematopoietic chimeras generated by injecting GATA-3-deficient (Gata3 −/−) embryonic hematopoietic stem cells into alymphoid recipient hosts (Rag −/− Il2rg −/−9). These studies revealed a critical contribution of GATA-3 to modulating NK cell migration specifically to the liver. The analysis of adhesion molecules expressed on NK cells from control and Gata3 −/− chimeras showed that the expression levels of CD11c and CD49b were reduced in Gata3 −/− NK cells from BM, spleen, and liver, whereas CD62L expression was increased. Furthermore, the absolute numbers of NK cells in the BM of Gata3 −/− chimeras were increased relative to controls, consistent with the hypothesis that Gata3 −/− NK cells might be unable to home normally from the BM to the liver. Adoptive transfer experiments where CFSE-labeled BM from Gata3 −/− or control chimeras were grafted into NK-deficient Rag −/− Il2rg −/− hosts show similar absolute numbers of Gata3 −/− and Gata3 +/+ NK cells recovered from the spleen of recipients. In marked contrast, NK cell homing to the liver was clearly defective in the absence of GATA-3. Together, these data identify a critical role for GATA-3 in promoting NK cell migration to the liver. Nevertheless, the molecular events downstream of GATA-3 that modulate entry into the liver remain unknown.
NK cell exit from lymphoid organs
In contrast to the multiple steps required for migration into lymphoid organs and inflammatory sites, only one requirement has been identified so far for egress from lymphoid organs that involves G protein-coupled receptors [37]. The immunosuppressive drug FTY720 was initially thought to cause lymphopenia by accelerating the homing of lymphocytes to secondary lymphoid organs [38]. Today, it is widely accepted that FTY720-mediated lymphocyte depletion from peripheral blood is more consistent with a block in egress from peripheral organs. Indeed, histological studies have shown that lymph node medullary sinuses of FTY720-treated mice are devoid of lymphocytes, which suggests that cells cannot enter efferent lymphatics [39]. A molecular mechanism is suggested by the finding that phosphorylated FTY720 becomes a potent agonist of G protein-coupled receptors specific for the lysophospholipid sphingosine-1-phosphate (S1P), like S1P1, S1P3, S1P4, and S1P5 [37]. Furthermore, intravenous administration of S1P also causes depletion of circulating lymphocytes [39], supporting the hypothesis that FTY720 blocks exit through S1P receptors. Recently, it has been shown that the percentage of NK cells in the blood, spleen, and lungs is much lower in S1P5-deficient than in wild-type mice [40]. Conversely, the percentages of NK cells in BM and lymph nodes are twice as high in S1P5-deficient mice as they are in wild-type mice. By using genetic and functional models, this study has provided compelling evidence for a role of S1P5 in NK cell trafficking.
An ethylnitrosourea-induced mutation in mice affecting blood NK cells has recently been reported, in which NK cells were reduced in blood and spleen but increased in lymph nodes and BM [41], somewhat mimicking the FTY720 phenotype. The accumulation of NK cells in lymph nodes reflected a decreased ability to exit into lymph. Interestingly enough, this strain designated Duane carries a point mutation within the Tbx21 gene, which generates a defective T-bet protein. Remarkably, Duane NK cells have a 30-fold reduced transcript levels of S1P5, and chromatin immunoprecipitation confirmed binding of T-bet to the S1p5 locus. Together, these findings identify S1P5 as a T-bet-induced gene that is required for NK cell egress from lymphoid tissues.
Transcription factors regulating NK cell effector functions
NK cells sense and respond to a wide variety of stimuli either directly through engagement of Toll-like receptors (TLR) and activating receptors, or indirectly through cytokines released by other cells in the inflammatory milieu [25]. How nuclear TF integrate this information to transmit downstream signals that regulate gene transcription is discussed below.
T-bet
T-bet mRNA is induced in primary human NK cells following activation with immobilized human IgG and IL-12 [42]. Also, NK cells purified from Tbx21 −/− mice produce significantly reduced amounts of IFN-γ following co-activation via CD16 and IL-12 when compared with wild-type animals, both at the protein and mRNA levels [42]. Up-regulation of T-bet at the mRNA level has also been detected in wild-type NK cells stimulated with various combinations of cytokines and an agonistic antibody against the activating receptor Ly49D [7]. It has been shown that IL-12 and IL-15 strongly induces T-bet expression. Other cytokines such as IL-18, IFN-α, and IFN-γ did not cause significant T-bet induction. In addition, ligation of the Ly49D receptor modestly induced T-bet. In particular, IL-12 together with IL-18, a powerful inducer of IFN-γ production by NK cells, was the most potent inducer of T-bet expression observed. Consistent with the potent induction of T-bet expression by IL-12 and IL-18, Stat4 −/− NK cells have a significant defect in T-bet induction upon treatment with these cytokines. T-bet expression also increased when the Ly49D receptor was stimulated in combination with IL-12 or IFN-α as compared with ligation of the receptor or cytokine alone. In contrast to previous studies that show an IFN-γ-dependent up-regulation of T-bet in CD4+ Th1 T cells, monocytes and DCs, T-bet expression in NK cells seems to be independent of IFN-γ, as Ifng −/− NK cells have no impairment in T-bet expression. These findings reinforce the idea that tissue-specific restrictions may account for the different outcomes observed [28].
JNK, ERK
A specific signalling pathway has recently been identified that operates downstream of the immunoreceptor tyrosine-based activation motif (ITAM)-coupled NK-cell receptors NK1.1, Ly49D, Ly49H, and NKG2D [43]. Using primary NK cells from Bcl10-deficient mice, a key role for Bcl10 signalosomes was demonstrated in the activation of canonical NF-κB signaling upon NK-cell triggering. These Bcl10-dependent cascades selectively control cytokine and chemokine production, but seemingly without affecting NK-cell differentiation or killing [43]. Within the ITAM pathway, distinct signaling intermediates are variably involved in cytotoxicity and/or IFN-γ secretion. Engagement of NK cell receptors that signal through ITAMs results in the rapid activation of protein kinase C-θ (PKC-θ). Analyses of Prkcz-deficient NK cells from PKC-θ-deficient mice indicated that PKC-θ is required for ITAM-mediated IFN-γ secretion, whereas it has no marked influence on the release of cytolytic mediators [44]. In the same study, it was found that PKC-θ deficiency preferentially impairs sustained extracellular-regulated kinase (ERK) signaling as well as activation of c-Jun N-terminal kinase (JNK) and the transcription factors AP-1 and NFAT, but it does not affect activation of NF-κB. Whether PKC-θ is required for IFN-γ secretion in response to IL-12 [45], or a combination of IL-12 with IL-18 [44], is debated.
C/EBPγ
Much of the available information on how TF orchestrate NK cell lytic responses has been obtained from TF knockout models that result in defective expression of lysosomal proteases such as perforin and granzymes, and the use of the chromatin immunoprecipitation (ChIP) technique that allows the analysis of promoters that are directly and specifically bound by a TF. Using ChIP in IL-2-activated NK cells from wild-type and Tbx21 −/− mice, it was shown that, together with IFN-γ, T-bet binds the promoters of perforin and granzyme B [7]. Furthermore, real-time RT-PCR of perforin and granzyme B mRNA shows up-regulation of these genes in the presence of a combination of IL-12 and IL-18. Perforin transcription has also been shown to be indirectly modulated by MEF [5]. Using electrophoretic mobility shift assays (EMSA), it was shown that MEF binds to Ets sites that, in turn, activate the perforin promoter in NK cells. Mice lacking the CCAAT/enhancer binding protein gamma chain (C/EBPγ) also show defects in perforin-mediated NK cell killing [46]. Although C/EBPγ does not contain transcription-activating domains, it can interact with other transcription factors through the leucine zipper domain and function as a dominant negative form [47]. In some cases, C/EBPγ can augment the DNA binding ability of other transcription factors [48]. C/EBPγ BM chimeras generated by transferring C/EBPγ marrow into irradiated Rag2 −/− mice show that perforin expression and cytolytic activity of spleen NK cells is reduced in the absence of C/EBPγ TF [46]. Similarly, reduced granzyme B expression has been shown in microphthalmia transcription factor (MITF)-deficient NK cells, and low level expression of granzyme B and perforin at the protein level was consistent with a significant reduction of NK cell activity [49].
Irf1
The expression of a number of genes pivotal in CD4+ Th1 differentiation is affected by IRF-1, which has therefore been called a master TF for Th1 cell differentiation [50]. Although the number and phenotype of spleen NK cells of Irf1 −/− mice is similar to that of wild-type animals, IL-12-augmented lysis of the prototypical NK cell target YAC-1 is defective in Irf1-deficient mice [51, 52]. Accordingly, in vivo elimination of class-I-deficient tumor RMA-S cells and rejection of parental BM transplants are impaired in Irf1 −/− mice [51]. Similar reduced cytolytic activity has been reported in Irf2 −/− NK cells [11], consistent with the idea that IRF-2 can act as a functional agonist rather than antagonist of IRF-1.
Gata3
Samson and colleagues found that splenic NK cells from control and Gata3 −/− chimeras (described above) showed similar cytolytic capacity, which was boosted by IL-12. These observations suggest that NK cells developing in the absence of GATA-3 have normal cell-mediated killing, and express functional receptors for IL-12 [9].
T-bet is responsible for direct transactivation of the IFN-γ gene in CD4+ Th1 T cells [27]. Both Tbx21 +/+ and Tbx21 −/− NK cells stimulated with IL-12 and IL-18 rapidly secrete large quantities of IFN-γ. Interestingly, after 24 h of stimulation, Tbx21 −/− cells produce significantly lower levels of IFN-γ, an observation consistent with the higher rate of cell death as determined by annexin V staining. GATA-3 plays an essential role in T cell differentiation, whereby it promotes the generation of CD4+ T cells specialized in the production of T helper 2 (Th2)-type cytokines [53]. Somewhat surprisingly, Gata3 −/− NK cells are poor IFN-γ producers compared to controls, likely due to a much lower expression levels of T-bet suggesting that GATA-3 may act upstream of T-bet in regulating IFN-γ expression. Furthermore, the cross-linking of cell surface receptors, including CD11b and 2B4, was found to induce IFN-γ production by control but not by Gata3 −/− NK cells. As a result, Gata3 −/− mice were unable to provide early protection in vivo against infection with Listeria monocytogenes. These data may indicate that a complex network of interacting TF govern the outcome of NK cell activation. This possibility is reinforced by recent data suggesting that T-bet and GATA-3 occupy many of the same genes, including those that are differentially expressed between Th1 and Th2 CD4+ T cells [36]. The latter study indicates that a particular phenotype, such as the choice between Th1 and Th2 lineage commitment, might be the result of the opposing action of these two TF at a set of shared target genes.
Additional signals may contribute to IFN-γ production by NK cells, raising the level of complexity of TF networks. Indeed, IFN-γ release is attenuated by TGFβ [54]. This might be a direct effect of TGFβ, or might result indirectly from cell–cell contact between NK cells and regulatory T cells producing this cytokine. In support of a direct effect, TGFβ can suppress IFN-γ production through SMAD3 [42], a transcription factor downstream from TGFβ-induced signaling, which results in suppression of T-bet.
NFAT
The Ca2+-dependent transcription factor family known as nuclear factor of activated T cells (NFAT) has been shown to be important in T-cell immune responses [55]. Because NFAT proteins have a weak DNA-binding capacity, they cooperate with other transcription factors at composite sites within the promoters of target genes. Recently, NFAT was shown to also be important for the induction of specific genetic programs that guide the differentiation and effector or regulatory activities of CD4+ T helper subsets via the transcriptional regulation of their lineage-specific transcription factors, specifically T-bet and GATA-3, therefore opening the possibility for similar regulatory networks in NK cells [44].
STAT proteins
There are a total of seven signal transducers and activators of transcription (STAT) proteins. Once activated by phosphorylation, the STATs function as dimers to induce the expression of genes with complementary promoters. Receptor binding of type 1 IFN elicits STAT1 and STAT2 phosphorylation, leading to the formation of transcriptionally active STAT1–STAT2 heterodimers or STAT1–STAT1 homodimers. In addition, STAT1 is required for type 1-induced IFN induction of NK cell cytotoxicity [56].The STAT4 molecule is crucial for IL-12 induction of NK cell IFN-γ production [56]. Conversely, STAT1 functions as a negative regulator of NK cell IFN-γ production, and cells conditioned by exposure to type 1 IFN, are reduced in their responsiveness to IL-12 [57].
Endogenous and fluorescently tagged IFN-γ and TNF have been used in freshly isolated primary NK cells and in live and fixed NK-92 cells to study cytokine and perforin secretion in granules, leading to the conclusion that these soluble mediators are released via different pathways [58]. Whether distinct TF selectively modulate NK cell killing versus cytokine release, and whether the polarization of cytokine- and perforin-containing granules to the NK cell–target interface is controlled by TF remain unknown.
Concluding remarks
While our knowledge of the TF network in NK cells is limited, the emerging picture is that these networks provide insights into a regulatory core of TF that drive all network interconnectivity. The information gathered from genetic models correlates NK cell phenotypes to the absence of TF. Nonetheless, mice deficient for certain TF (T-bet, IRF-1, IRF-2, MEF) display reduced numbers of NK cells, or NK cells showing an immature phenotype (T-bet, GATA-3), therefore making it difficult to directly link NK cell function to specific TF. Yet, NK cells remain the Cinderella of the lymphocyte family in terms of understanding the genetic pathway that leads to their differentiation and function. However, NK cells are getting ready to attend the Ball. The recent burst of activity defining transcriptional regulation in the NK cell lineage is quite dramatic and, as Figs. 1 and 2 show, a real genetic pathway for NK cell differentiation and effector functions is beginning to appear. If this progress continues and can allow improved means of NK cell production for use in NK cell-based immunotherapy, then this Cinderella story will also have a very happy ending.
Fig. 1.
Indication of points at which loss of individual transcription factors blocks NK cell development in BM or egress to the periphery. CLP Common lymphoid progenitor, NKP NK cell precursor, iNK immature NK cell, mNK mature NK cell
Fig. 2.
Schematic representation of key events modulating NK cell migration (a) and effector functions (b) by transcription factors
Acknowledgments
The authors acknowledge the support of the Swiss National Science Foundation (31-109832 to AMF) and the Medical Research Council-UK (G0802068 to G.M.L. and A.M.F., and G0901737 to H.J.B.). The authors also acknowledge financial support from the Department of Health via the National Institute for Health research (NIHR) comprehensive Biomedical Research Centre award to Guy’s & St Thomas’ NHS Foundation Trust in partnership with King’s College London and King’s College Hospital NHS Foundation Trust.
Conflict of interest
The authors have no conflicting financial interests.
References
- 1.Russell L, Garrett-Sinha LA. Transcription factor Ets-1 in cytokine and chemokine gene regulation. Cytokine. 2010;51:217–226. doi: 10.1016/j.cyto.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Barton K, et al. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity. 1998;9:555–563. doi: 10.1016/S1074-7613(00)80638-X. [DOI] [PubMed] [Google Scholar]
- 3.Medina KL, et al. Assembling a gene regulatory network for specification of the B cell fate. Dev Cell. 2004;7:607–617. doi: 10.1016/j.devcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Colucci F, et al. Differential requirement for the transcription factor PU.1 in the generation of natural killer cells versus B and T cells. Blood. 2001;97:2625–2632. doi: 10.1182/blood.V97.9.2625. [DOI] [PubMed] [Google Scholar]
- 5.Lacorazza HD, et al. The ETS protein MEF plays a critical role in perforin gene expression and the development of natural killer and NK-T cells. Immunity. 2002;17:437–449. doi: 10.1016/S1074-7613(02)00422-3. [DOI] [PubMed] [Google Scholar]
- 6.Szabo SJ, et al. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science. 2002;295:338–342. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- 7.Townsend MJ, et al. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20:477–494. doi: 10.1016/S1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 8.Hosoya T, Maillard I, Engel JD. From the cradle to the grave: activities of GATA-3 throughout T-cell development and differentiation. Immunol Rev. 2010;238:110–125. doi: 10.1111/j.1600-065X.2010.00954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samson SI, et al. GATA-3 promotes maturation, IFN-gamma production, and liver-specific homing of NK cells. Immunity. 2003;19:701–711. doi: 10.1016/S1074-7613(03)00294-2. [DOI] [PubMed] [Google Scholar]
- 10.Vosshenrich CA, et al. A thymic pathway of mouse natural killer cell development characterized by expression of GATA-3 and CD127. Nat Immunol. 2006;7:1217–1224. doi: 10.1038/ni1395. [DOI] [PubMed] [Google Scholar]
- 11.Lohoff M, et al. Deficiency in the transcription factor interferon regulatory factor (IRF)-2 leads to severely compromised development of natural killer and T helper type 1 cells. J Exp Med. 2000;192:325–336. doi: 10.1084/jem.192.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taki S, Nakajima S, Ichikawa E, Saito T, Hida S. IFN regulatory factor-2 deficiency revealed a novel checkpoint critical for the generation of peripheral NK cells. J Immunol. 2005;174:6005–6012. doi: 10.4049/jimmunol.174.10.6005. [DOI] [PubMed] [Google Scholar]
- 13.Quong MW, Romanow WJ, Murre C. E protein function in lymphocyte development. Annu Rev Immunol. 2002;20:301–322. doi: 10.1146/annurev.immunol.20.092501.162048. [DOI] [PubMed] [Google Scholar]
- 14.Yokota Y, et al. Development of peripheral lymphoid organs and natural killer cells depends on the helix–loop–helix inhibitor Id2. Nature. 1999;397:702–706. doi: 10.1038/17812. [DOI] [PubMed] [Google Scholar]
- 15.Ikawa T, Fujimoto S, Kawamoto H, Katsura Y, Yokota Y. Commitment to natural killer cells requires the helix–loop–helix inhibitor Id2. Proc Natl Acad Sci USA. 2001;98:5164–5169. doi: 10.1073/pnas.091537598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boos MD, Yokota Y, Eberl G, Kee BL. Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of E protein activity. J Exp Med. 2007;204:1119–1130. doi: 10.1084/jem.20061959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramirez K, Kee BL. Multiple hats for natural killers. Curr Opin Immunol. 2010;22:193–198. doi: 10.1016/j.coi.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schotte R, et al. Synergy between IL-15 and Id2 promotes the expansion of human NK progenitor cells, which can be counteracted by the E protein HEB required to drive T cell development. J Immunol. 2010;184:6670–6679. doi: 10.4049/jimmunol.0901508. [DOI] [PubMed] [Google Scholar]
- 19.Gascoyne DM, et al. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat Immunol. 2009;10:1118–1124. doi: 10.1038/ni.1787. [DOI] [PubMed] [Google Scholar]
- 20.Liu P, et al. Bcl11a is essential for normal lymphoid development. Nat Immunol. 2003;4:525–532. doi: 10.1038/ni925. [DOI] [PubMed] [Google Scholar]
- 21.Li P, et al. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science. 2010;329:85–89. doi: 10.1126/science.1188063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Santo JP. Immunology. A guardian of T cell fate. Science. 2010;329:44–45. doi: 10.1126/science.1191664. [DOI] [PubMed] [Google Scholar]
- 23.Wilkinson B, et al. TOX: an HMG box protein implicated in the regulation of thymocyte selection. Nat Immunol. 2002;3:272–280. doi: 10.1038/ni767. [DOI] [PubMed] [Google Scholar]
- 24.Aliahmad P, de la Torre B, Kaye J. Shared dependence on the DNA-binding factor TOX for the development of lymphoid tissue-inducer cell and NK cell lineages. Nat Immunol. 2010;11:945–952. doi: 10.1038/ni.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9:503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 26.Colonna M. Interleukin-22-producing natural killer cells and lymphoid tissue inducer-like cells in mucosal immunity. Immunity. 2009;31:15–23. doi: 10.1016/j.immuni.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 27.Szabo SJ, et al. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/S0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 28.Glimcher LH, Townsend MJ, Sullivan BM, Lord GM. Recent developments in the transcriptional regulation of cytolytic effector cells. Nat Rev Immunol. 2004;4:900–911. doi: 10.1038/nri1490. [DOI] [PubMed] [Google Scholar]
- 29.Lord GM, et al. T-bet is required for optimal proinflammatory CD4+T-cell trafficking. Blood. 2005;106:3432–3439. doi: 10.1182/blood-2005-04-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Werneck MB, Lugo-Villarino G, Hwang ES, Cantor H, Glimcher LH. T-bet plays a key role in NK-mediated control of melanoma metastatic disease. J Immunol. 2008;180:8004–8010. doi: 10.4049/jimmunol.180.12.8004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rot A, von Andrian UH. Chemokines in innate and adaptive host defense: basic chemokinese grammar for immune cells. Annu Rev Immunol. 2004;22:891–928. doi: 10.1146/annurev.immunol.22.012703.104543. [DOI] [PubMed] [Google Scholar]
- 32.Martin-Fontecha A, et al. Induced recruitment of NK cells to lymph nodes provides IFN-gamma for T(H)1 priming. Nat Immunol. 2004;5:1260–1265. doi: 10.1038/ni1138. [DOI] [PubMed] [Google Scholar]
- 33.Wendel M, Galani IE, Suri-Payer E, Cerwenka A. Natural killer cell accumulation in tumors is dependent on IFN-gamma and CXCR3 ligands. Cancer Res. 2008;68:8437–8445. doi: 10.1158/0008-5472.CAN-08-1440. [DOI] [PubMed] [Google Scholar]
- 34.Garrett WS, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garrett WS, et al. Colitis-associated colorectal cancer driven by T-bet deficiency in dendritic cells. Cancer Cell. 2009;16:208–219. doi: 10.1016/j.ccr.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jenner RG, et al. The transcription factors T-bet and GATA-3 control alternative pathways of T-cell differentiation through a shared set of target genes. Proc Natl Acad Sci USA. 2009;106:17876–17881. doi: 10.1073/pnas.0909357106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cyster JG. Chemokines, sphingosine-1-phosphate, and cell migration in secondary lymphoid organs. Annu Rev Immunol. 2005;23:127–159. doi: 10.1146/annurev.immunol.23.021704.115628. [DOI] [PubMed] [Google Scholar]
- 38.Chiba K, et al. FTY720, a novel immunosuppressant, induces sequestration of circulating mature lymphocytes by acceleration of lymphocyte homing in rats. I. FTY720 selectively decreases the number of circulating mature lymphocytes by acceleration of lymphocyte homing. J Immunol. 1998;160:5037–5044. [PubMed] [Google Scholar]
- 39.Mandala S, et al. Alteration of lymphocyte trafficking by sphingosine-1-phosphate receptor agonists. Science. 2002;296:346–349. doi: 10.1126/science.1070238. [DOI] [PubMed] [Google Scholar]
- 40.Walzer T, et al. Natural killer cell trafficking in vivo requires a dedicated sphingosine 1-phosphate receptor. Nat Immunol. 2007;8:1337–1344. doi: 10.1038/ni1523. [DOI] [PubMed] [Google Scholar]
- 41.Jenne CN, et al. T-bet-dependent S1P5 expression in NK cells promotes egress from lymph nodes and bone marrow. J Exp Med. 2009;206:2469–2481. doi: 10.1084/jem.20090525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trotta R, et al. TGF-beta utilizes SMAD3 to inhibit CD16-mediated IFN-gamma production and antibody-dependent cellular cytotoxicity in human NK cells. J Immunol. 2008;181:3784–3792. doi: 10.4049/jimmunol.181.6.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gross O, et al. Multiple ITAM-coupled NK-cell receptors engage the Bcl10/Malt1 complex via Carma1 for NF-kappaB and MAPK activation to selectively control cytokine production. Blood. 2008;112:2421–2428. doi: 10.1182/blood-2007-11-123513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tassi I, et al. NK cell-activating receptors require PKC-theta for sustained signaling, transcriptional activation, and IFN-gamma secretion. Blood. 2008;112:4109–4116. doi: 10.1182/blood-2008-02-139527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Page KM, Chaudhary D, Goldman SJ, Kasaian MT. Natural killer cells from protein kinase C theta-/- mice stimulated with interleukin-12 are deficient in production of interferon-gamma. J Leukoc Biol. 2008;83:1267–1276. doi: 10.1189/jlb.1107745. [DOI] [PubMed] [Google Scholar]
- 46.Kaisho T, et al. Impairment of natural killer cytotoxic activity and interferon gamma production in CCAAT/enhancer binding protein gamma-deficient mice. J Exp Med. 1999;190:1573–1582. doi: 10.1084/jem.190.11.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cooper C, Henderson A, Artandi S, Avitahl N, Calame K. Ig/EBP (C/EBP gamma) is a transdominant negative inhibitor of C/EBP family transcriptional activators. Nucleic Acids Res. 1995;23:4371–4377. doi: 10.1093/nar/23.21.4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davydov IV, Bohmann D, Krammer PH, Li-Weber M. Cloning of the cDNA encoding human C/EBP gamma, a protein binding to the PRE-I enhancer element of the human interleukin-4 promoter. Gene. 1995;161:271–275. doi: 10.1016/0378-1119(95)00271-7. [DOI] [PubMed] [Google Scholar]
- 49.Ito A, et al. Inhibitory effect on natural killer activity of microphthalmia transcription factor encoded by the mutant mi allele of mice. Blood. 2001;97:2075–2083. doi: 10.1182/blood.V97.7.2075. [DOI] [PubMed] [Google Scholar]
- 50.Lohoff M, et al. Interferon regulatory factor-1 is required for a T helper 1 immune response in vivo. Immunity. 1997;6:681–689. doi: 10.1016/S1074-7613(00)80444-6. [DOI] [PubMed] [Google Scholar]
- 51.Duncan GS, Mittrucker HW, Kagi D, Matsuyama T, Mak TW. The transcription factor interferon regulatory factor-1 is essential for natural killer cell function in vivo. J Exp Med. 1996;184:2043–2048. doi: 10.1084/jem.184.5.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taki S, et al. Multistage regulation of Th1-type immune responses by the transcription factor IRF-1. Immunity. 1997;6:673–679. doi: 10.1016/S1074-7613(00)80443-4. [DOI] [PubMed] [Google Scholar]
- 53.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/S0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 54.Wahl SM, Wen J, Moutsopoulos NM. The kiss of death: interrupted by NK-cell close encounters of another kind. Trends Immunol. 2006;27:161–164. doi: 10.1016/j.it.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 55.Macian F. NFAT proteins: key regulators of T-cell development and function. Nat Rev Immunol. 2005;5:472–484. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]
- 56.Nguyen KB, et al. Coordinated and distinct roles for IFN-alpha beta, IL-12, and IL-15 regulation of NK cell responses to viral infection. J Immunol. 2002;169:4279–4287. doi: 10.4049/jimmunol.169.8.4279. [DOI] [PubMed] [Google Scholar]
- 57.Nguyen KB, et al. Interferon alpha/beta-mediated inhibition and promotion of interferon gamma: STAT1 resolves a paradox. Nat Immunol. 2000;1:70–76. doi: 10.1038/76940. [DOI] [PubMed] [Google Scholar]
- 58.Reefman E, et al. Cytokine secretion is distinct from secretion of cytotoxic granules in NK cells. J Immunol. 2010;184:4852–4862. doi: 10.4049/jimmunol.0803954. [DOI] [PubMed] [Google Scholar]