Abstract
Capsulin and Musculin are basic helix-loop-helix transcription factors, but their biophysiological roles in zebrafish cranial myogenesis are unclear. Expressions of endogenous capsulin transcripts are detected at the central- (~24-hpf) and at dorsal- and ventral-mesoderm cores (~30–72 hpf) of branchial arches. In contrast, musculin transcripts are expressed as a two-phase manner: early phase (20–22 hpf) expressions of musculin are detected at the head mesoderm, whereas late-phase (36–72 hpf) are detected at all presumptive head-muscle precursors. Knockdown of either capsulin or musculin leads to loss of all cranial muscles without affecting trunk muscle development. The defective phenotypes of Capsulin- and Musculin-morphant can be rescued by co-injection of mRNA of each other. Both myf5 and myod transcripts are down-regulated in the Capsulin-morphant while myod transcripts are up-regulated in the Musculin-morphant. Therefore, we propose a putative regulatory network to understand how capsulin/musculin regulate distinctly either myf5 or myod during zebrafish craniofacial myogenesis.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-011-0637-2) contains supplementary material, which is available to authorized users.
Keywords: Capsulin, Mesoderm core, Morpholino, Musculin, Myogenesis, Zebrafish
Introduction
In vertebrates, head muscles are derived from multiple cell lineages, including prechordal mesoderm anterior to the first somite and paraxial mesodermal precursors that migrate into the branchial arches to form a central mesoderm core. Subsequently, the central mesoderm core is further subdivided into dorsal and ventral regions, surrounded by cranial neural crest (CNC) cells and that mesoderm cores will contribute to epaxial and hypaxial muscles of the head [1, 2]. Although these head-muscle precursor cells carry some positional information from their origins, they receive many environmental cues from the surrounding cells, especially from pharyngeal ectoderm and CNC cells. For example, CNC cells can regulate the migration, patterning, and differentiation of head-muscle precursors. In the absence of CNC cells, accumulated myoblasts are kept in a proliferative state, which leads to abnormalities in both differentiation and subsequent myofibril organization in the head [3]. These observations highlight the importance of CNC cells and mesoderm core during cranial muscle development.
Genetically, skeletal muscle development in vertebrates is controlled by myogenic basic helix-loop-helix (bHLH) transcription factors: Myod, Myf5, Myogenin, and MRF4 [4, 5]. These myogenic genes control a developmental program shared by all skeletal muscles, including head muscles. In other words, head-muscle precursor cells in mesoderm core receive environmental cues, activate these myogenic genes, and undergo myogenic lineage specification. Inactivation of Myf5 and/or Myod leading to malformation of different populations of cranial muscles indicates distinct functions of Myf5 and Myod during craniofacial myogenesis [6]. However, the developmental control genes expressed in the mesoderm core, which are responsible for the formation of Myf5, Myod, and all head muscles, are still little known.
Capsulin/Epicardin/Pod-1/Tcf21 and MyoR/Musculin are bHLH transcription factors involved in the regulation of cell differentiation. They have been isolated from human, mouse, rat, bovine, chicken, and Xenopus [7–12]. In mice and as well as in chicken, capsulin is transiently expressed in lung, spleen, and kidney. Both capsulin and musculin are co-expressed in the developing branchiomeric muscles derived from the non-segmented head mesoderm [7, 9, 12]. Gene knock-out experiment revealed that mice lacking Capsulin are not viable, suffering from the lack of lung alveoli, cardiac defects, hypoblastic kidneys, arrest of development and apoptosis of their spleen and from defective gonad development but no overt skeletal muscle abnormalities [13–15]. In mice lacking Capsulin and Musculin, in addition to the Capsulin-dependent phenotypes, most of the branchiomeric muscles in the head were lost [13].
To uncover the roles of capsulin and musculin in craniofacial myogenesis, it is worthy to analyze comparatively capsulin and musculin genes across species. We have recently reported that Capsulin in zebrafish was 55% α-helical, and shared very high sequence identity with similar proteins from other species (72–82%) [16]. Here, we examined the spatiotemporal expression of capsulin and musculin in zebrafish embryo, a well-established model for head and trunk skeletal myogenesis. We also investigated the biological functions of capsulin and musculin in vivo, especially in cranial myogenesis, using the morpholino knock-down approach.
Materials and methods
Cloning of zebrafish musculin and bioinformation
For amplifying musculin cDNA, Musculin-F (5′-ACTACCACACTGCGGAGCTGA-3′) and Musculin-R (5′-AGGTGCCAGATGTTTATGCTGTAGCT-3′) were designed according to an ensemble contig (Ensemble Gene ID: ENSDARG00000045353) encoding of a putative zebrafish Musculin. Amplified DNA fragments were subcloned and sequenced. The presumptive Capsulin and Musculin amino acid sequences were determined using the Wisconsin Sequence Analysis Package v.10.0 (GCG). The Gap program of that package was used for pair comparisons, and the Pileup and Prettybox programs were used for multiple comparisons. The Clustalw molecular evolution genetic program was used for our phylogenic tree analysis (http://www.ebi.ac.uk/clustalw/).
Fish embryos, whole-mount in situ hybridization, antibody labeling, and cryosection
The procedures for zebrafish culture, embryo collection, fluorescent observation, whole-mount in situ hybridization, antibody labeling, and cryosection have been described previously [17, 18], except that capsulin (NM_011545) [16], musculin (this study), myf5 (AF270789) [19], myod (NM_131262) [20], myogenin (AF202639) [21], α-actin (NM_131591) [22], dlx2a (NM_131311) [23], foxd3 (NM_131290) [24], edn1 (NM_131519) [25] and tbx1 (NM_183339) were used as probes. They were digoxigenin-labeled after their partial DNA fragments were cloned. The designation of developmental stages of zebrafish followed those of Kimmel et al. [26].
Preparation and microinjection of morpholino
Capsulin-MO1 (5′-GGACCCGGTGGACATGTTGGCTGGA-3′; Gene Tools) was established according to zebrafish capsulin cDNA sequence for blocking translation. Capsulin-MO2 (5′-GTGTCTCACCAGGTTGACGGATGT-3′) was established according to the correspondent splicing donor/acceptor sites for specifically disturbing splicing of capsulin. Musculin-MO1 (5′-TACTGAACCCGTGGACATCACTTCA-3′) and Musculin-MO2 (ACTCACTCATACCAGGTTGGCAGGG) were employed to block translation and splicing of musculin, respectively. Myf5-MO (5′-TCTGGGATGTGGAGAATACGTCCAT-3′) and Myod-MO (5′-ATATCCGACAACTCCATCTTTTTTG-3′) were synthesized according to previous studies [6, 27]. Negative control MO (5′-CCTCTTACCTCAGTTACAATTTATA-3′) was designed according to the random nucleotide sequences (Gene Tools). All of the above were prepared at stocking concentrations of 1 mM and diluted with double-distilled water to the proper concentrations (4.5 ng per 2.3 nl).
RNA isolation and quantitative reverse transcription-polymerase chain reaction
One hundred embryos derived from uninjected (control) or experimental groups (Capsulin- and Musculin-morphant) at 24 hpf were collected and their total RNA was isolated using a standard procedure as described previously [19]. Around 25 μg of total RNA from each group was used for cDNA synthesis; 1% of cDNA was used for each quantitative PCR reaction. Each assay was run on an Applied Biosystems 7300 Real-Time PCR system in triplicates and expression fold-changes were derived using the comparative CT method (https://products.appliedbiosystems.com). Wang et al. [28, 29] described the procedures of quantitative PCR. capsulin and musculin were selected as targets (Supplementary Table S1). β-actin was used as endogenous control for relative quantification.
Capped mRNA preparation for rescue experiments
Capped mRNAs of capsulin, musculin, myf5, myod, and gfp were synthesized according to the protocol of the manufacturer (Ambion). The resultant mRNAs were diluted to desired concentrations prior to each 2.3-nl injection.
Chemical treatment
For cyclopamine (Sigma-Aldrich) treatment, zebrafish embryos at 6 hpf were collected, randomly divided into several groups of 30 embryos each, and exposed to either water (vehicle-control) or water containing 50 μM of cyclopamine. All embryos were cultivated in 9-cm cell culture plates until embryos developed at 24 and 30 hpf. For SU5402 (Calbiochem) treatment, SU5402 was dissolved in dimethyl sulfoxide at 25 mM as a stock solution and stored at −20°C. Embryos at 18 hpf were dechorionated manually and cultured at 28.5°C for appropriate periods in the presence of the inhibitor at 75 μM in 1/3 Ringer’s solution. After treatment, SU5402 was removed by extensive washing with 1/3 Ringer’s solution, and embryos were fixed immediately after removal of the reagent, or further cultured at 28.5°C until appropriate stages.
Microscopy
All embryos were observed at specific stages under a microscope (DM 2500, Leica) equipped with Nomarski differential interference contrast optics and a fluorescent module having a GFP filter cube (Kramer Scientific). Photographs of embryos at specific stages were taken with a CCD (DFC490, Leica).
Statistical analysis
The two-sample t test with a significance level of 0.05 was employed to identify significant fold-changes between the treatment and control samples in the comparative CT method for capsulin and musculin genes.
Results
Cloning of zebrafish musculin cDNA and comparison of deduced amino acid sequences
A full-length zebrafish musculin cDNA was cloned using a combination of RT-PCR, 5′-RACE and 3′-RACE. A zebrafish musculin gene is composed of two exons and one intron. The deduced zebrafish Musculin amino acid sequence consists of a 160-amino acid polypeptide containing a putative basic helix-loop-helix (basic domain, amino acids positions 63Q to 75M; helix 1 76R to 89L; loop, 90P to 99S; helix 2, 100K to 114L numbered according to the zebrafish Musculin (Supplementary Fig. S1). The zebrafish Musculin polypeptide shares sequence identities of 56, 57, 58, 57, and 62% with the reported Musculin of human, cattle, rat, mouse, and Xenopus, respectively (Supplementary Fig. S1). We used the Clustalw program to determine the phylogenic similarities between zebrafish Musculin and human, cattle, rat, mouse, and Xenopus Musculin. The phylogenic tree generated by the program showed that zebrafish Capsulin/Musculin possesses a unique pattern, different from those of higher vertebrates (Supplementary Fig. S2).
Zebrafish capsulin transiently expressed in mesoderm core of branchial arches
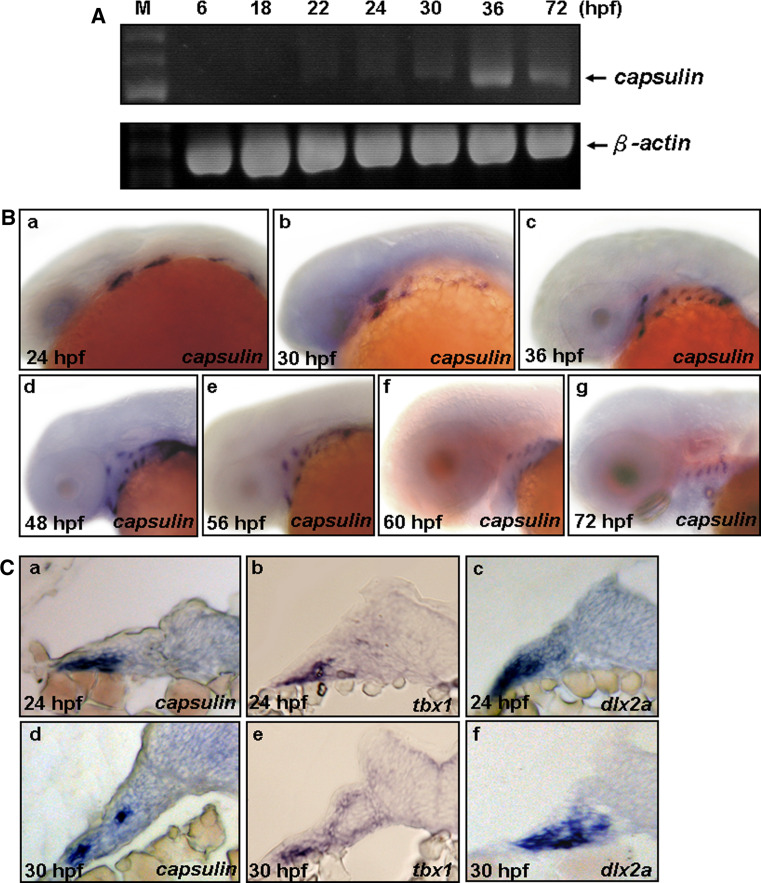
To determine the spatiotemporal expression patterns of capsulin during early development, we performed whole-mount in situ hybridization and RT-PCR experiments to examine the mRNA expression. RT-PCR experiments showed that endogenous zebrafish capsulin were detected at 18 and 22 hpf in a very faint manner, and became more evident at 24–72 hpf (Fig. 1A). Whole-mount in situ hybridization showed that capsulin expression in the head periphery was not detected until 24 hpf. At this stage, four small bilateral groups of mesenchymal cells adjacent to the midbrain and hindbrain express capsulin, and appear with one capsulin-expressing cluster of cells in each pharyngeal arch (Fig. 1B-a). Our inspections of capsulin-expressing cluster show that endogenous capsulin is expressed at the lateral plate mesoderm (Fig. 1C-a). Later (~30 hpf), the pharyngeal arch is further subdivided into dorsal and ventral regions, and surrounded with neural crest-derived cells that will contribute to epaxial and hypaxial muscles of the head. By this time, capsulin-expressing cluster of cells appear in the mesoderm core of the divided dorsal and ventral regions of each branchial arch (Fig. 1B-b; C-d). Expression persists in mesoderm core at 36, 48, 56, 60, and 72 hpf (Fig. 1B-c-g). After 72 hpf, no capsulin signals were observed in the head region, but the expression persisted in the heart field and pectoral fin buds (data not shown). On the other hand, because the early expression domains of capsulin (24 and 30 hpf) are very similar to the expression domains of a T-box transcription factor, tbx1 (a mesoderm core marker), and a homeobox transcription factor, dlx2a, a known marker, labels the positions of post-migratory cranial neural crest (CNC) cells and pharyngeal arch mesenchyme [23]. We also examined the expression patterns of tbx1 and dlx2a by whole-mount in situ hybridization followed by cryosection. By 24 hpf, the expression domains of tbx1 and dlx2a are overlapped with that of capsulin in pharyngeal arch mesenchyme (Fig. 1C-a-c). By 30 hpf, only tbx1 signals but not dlx2a are overlapped with capsulin signals at the ventral mesoderm core (Fig. 1C-d-f). These observations suggest transient expression of capsulin in mesoderm core of branchial arches.
Fig. 1.
capsulin, tbx1, and dlx2a expression during early embryonic stages. A RT-PCR analysis of the zebrafish capsulin and β-actin gene transcripts, using total RNA extracted from different developmental stages. B Whole-mount in situ hybridization of zebrafish capsulin expressions at different embryonic stages. a At 24 h postfertilization (hpf), lateral view. b At 30 hpf, lateral view. c At 36 hpf, lateral view. d At 48 hpf, lateral view. e At 56 hpf, lateral view. f At 60 hpf, lateral view. g At 72 hpf, lateral view. C Cross sections of capsulin, tbx1 and dlx2a expressions of 24- (a–c) and 30-hpf (d–f) embryos
Two-phase expressions of zebrafish musculin
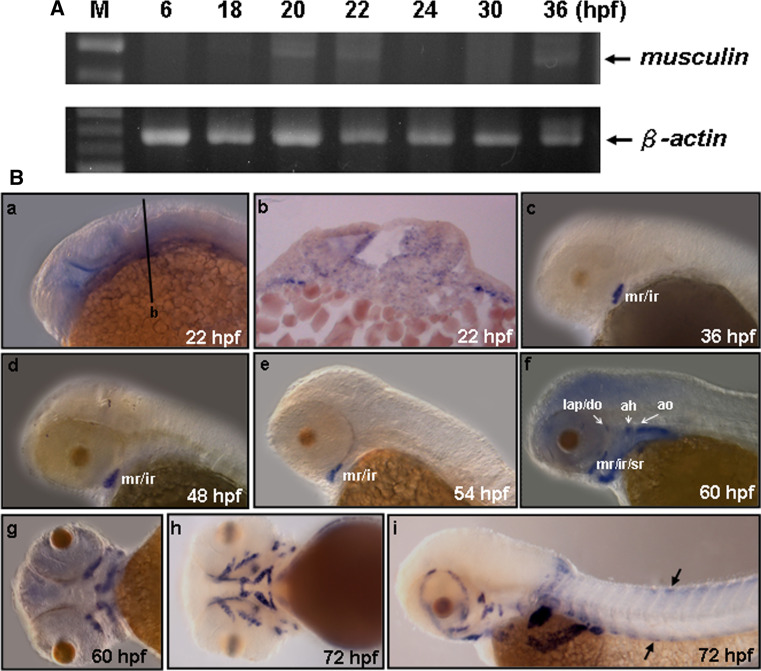
RT-PCR experiments showed that endogenous zebrafish musculin exhibited two phases of expressions. The first phase of zebrafish musculin transcripts were detected at 18–24 hpf, whereas the second phase began its expression at 36 hpf (Fig. 2A). Whole-mount in situ hybridization revealed that musculin transcripts were faintly observed in the head mesoderm of the 18-hpf embryos (data not shown), extended their expression to the mesoderm core of the first arch by 22 hpf (Fig. 2B-a, -b), and were down-regulated to an undetectable level between 24 and 36 hpf. At 36–54 hpf, the second phase of musculin expression was observed at the medial rectus (mr) and inferior rectus (ir) (Fig. 2B-c-e), and then expanded to the superior rectus (sr), levator arcus palatine (lap), adductor operculi (ao), dilator operculi (do) and adductor hyoideu (ah) at 60 hpf (Fig. 2B-f, -g). Later (~72 hpf), musculin expression was observed around all cranial muscles, including ah, adductor mandibulae (am), ao, do, hyohyoideus (hh), interhyoideus (ih), intermandibularis anterior (ima), intermandibularis posterior (imp), lap, inferior oblique (io), ir, lateral rectus (lr), mr, superior oblique (so), sr, and sternohyoideus (sh) (Fig. 2B-h, -i). In addition, we also found that musculin signals were detected at the trunk muscle progenitors by 72 hpf (Fig. 2B-i). These observations correspond with the late-phase expressions of mouse Musculin whose signals are observed in cranial muscles as well as in trunk muscles [13].
Fig. 2.
musculin expression during early embryonic stages. A RT-PCR analysis of the zebrafish musculin and β-actin gene transcripts, using total RNA extracted from different developmental stages. B Whole-mount in situ hybridization of zebrafish musculin expressions at different embryonic stages. a At 22 h postfertilization (hpf), lateral view. b Cross section along the plane indicated by line b in a. c At 36 hpf, lateral view. d At 48 hpf, lateral view. e At 54 hpf, lateral view. f At 60 hpf, lateral view, and g ventral view. h At 72 hpf, ventral view, and i lateral view. Arrows indicate the positions of trunk muscle progenitors. mr medial rectus, ir inferior rectus, lap levator arcus palatine, do dilator operculi, ah adductor hyoideu, ao adductor operculi
Targeting efficiencies of Capsulin- and Musculin-MOs
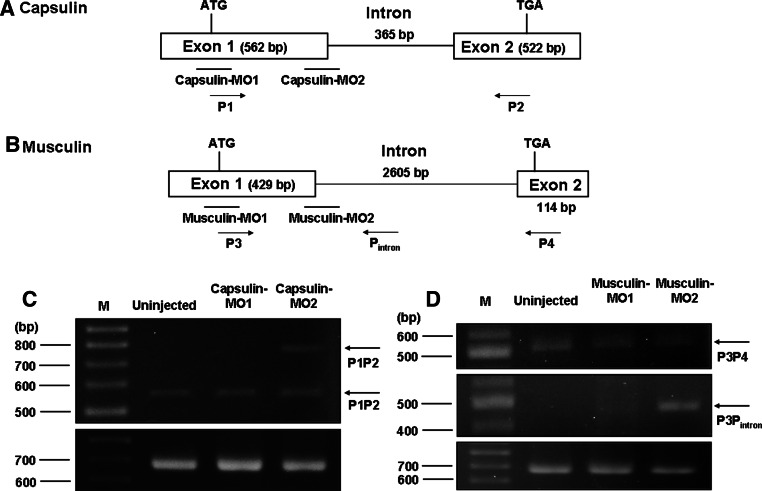
To characterize the function of Capsulin and Musculin in vivo, we used morpholino (MO) to knock down endogenous capsulin and/or musculin expression specifically. According to their genomic structure, we designed two Capsulin-MOs (Capsulin-MO1, Capsulin-MO2) and two Musculin-MOs (Musculin-MO1, Musculin-MO2) in order to specifically block translation and disturb splicing of capsulin and musculin (Fig. 3a, b). Since MO2s were designed for disruption of splicing, we carried out RT-PCR experiments in order to test the targeting efficiency of Capsulin-MO2 and Musculin-MO2. As shown in Fig. 3c, a 531-bp RT-PCR product was detected in uninjected-, Capsulin-MO1- and Capsulin-MO2-injected groups but a longer, ~ 896-bp DNA fragment was observed in the Capsulin-MO2-injected group because a 365-bp intron fragment of capsulin gene was not spliced out. Similar results were also observed in the Musculin-MO2-injected group, which clearly indicated that a 469-bp exon-intron junction of musculin gene fragment was amplified by primers P3 and Pintron (Fig. 3d). These data clearly demonstrated that endogenous capsulin and musculin expressions were inactivated by injecting Capsulin-MO2 or Musculin-MO2.
Fig. 3.
Target sites of Capsulin- and Musculin-morpholinos (MO). a, b Both capsulin and musculin gene possess 2 exons and 1 intron. The locations of sequence-specific Capsulin-MO1, Capsulin-MO2, Musculin-MO1, and Musculin-MO2 are listed below. MO1s are translation-blocking MOs, whereas MO2s are splice-blocking MOs. c RT-PCR analysis to test the targeting efficiency for each MO, using total RNA extracted from different injection groups. The primers P1 and P2 were used for detecting splicing variants of embryos derived from Uninjected group and Capsulin-morphants; d whereas primers P3, P4 and Pintron were used for detecting splicing variants of embryos derived from Uninjected group and Musculin-morphants
Knockdown of capsulin and/or musculin led to reduction of muscle precursors
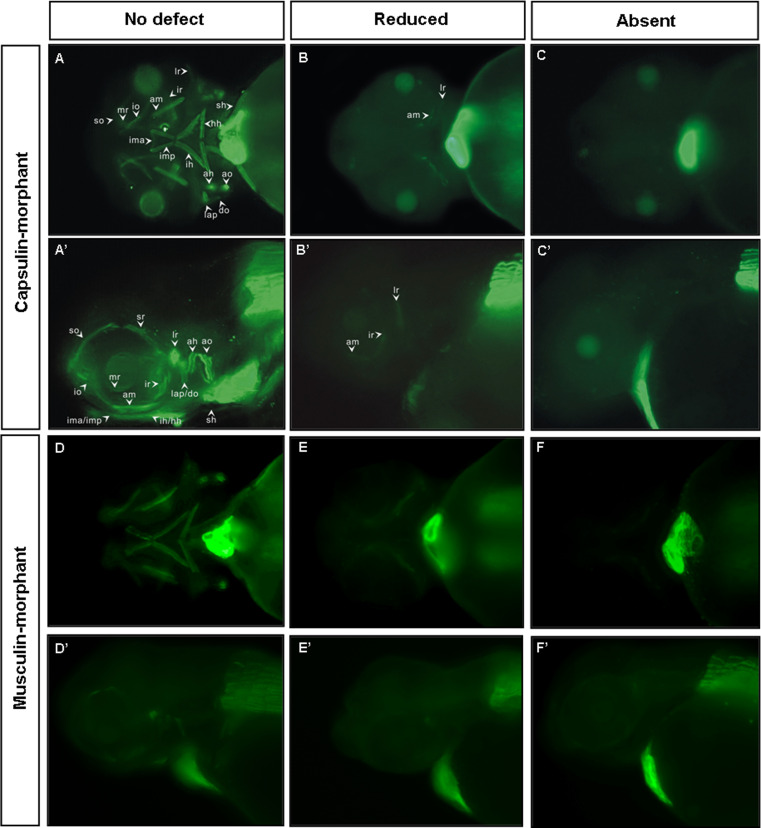
Since capsulin is expressed in the mesoderm core of the pharyngeal arch, it is expected to be highly associated with cranial myogenesis. To address this issue, embryos injected with Capsulin-MOs (MO1 and MO2; Capsulin-morphants) were collected and stained with slow muscle-specific monoclonal antibody F59 to visualize muscle malformation phenotypes. In uninjected embryos, all cranial muscles, including ah, am, ao, do, hh, ih, ima, imp, lap, io, ir, lr, mr, so, sr and sh were labeled with F59 (no defect; Fig. 4a, a′, d, d′). However, cranial muscle labeled with F59 was significantly reduced or absent in the head regions after injection with Capsulin-MO1 (Fig. 4b, b′, c, c′) or Capsulin-MO2 (data not shown). Similar effects were also observed in the Musculin-MO1- (data not shown) or Musculin-MO2-injected embryos (Musculin-morphants; Fig. 4e, e′, f, f′), suggesting that Capsulin and Musculin play important role in cranial myogenesis.
Fig. 4.
Loss-of-function analyses using antisense morpholinos demonstrate that both Capsulin and Musculin protein is required for head-muscle development. Embryos were injected with optimal amounts of Capsulin-MO1 (a–c, a′–c′) and or Musculin-MO2 (d–f, d′–f′) at the 1–2 cell stage to inhibit gene function, and head-muscle development was visualized with F59 antibody staining at 72 hpf. a–f Ventral view, and a′–f′ lateral view
Dose-dependent Capsulin- and Musculin-morphant abnormalities
Our results show that the frequencies of phenotypic abnormalities in consequence of Capsulin-MO1 injection followed a dose-dependent manner (Table 1). At 0.5 ng, 27.6% (total cases in triplicate experiments, n = 36) of the surviving embryos (n = 130) displayed defective head-muscle phenotypes (reduced plus absent); and at 1.5 ng, the percentage increased to 79.2% (n = 72). Similar results were also observed in the Capsulin-MO2-injected embryos. At 0.5 ng, 26.5% (n = 42) of the surviving embryos (n = 158) displayed defective head-muscle phenotypes; and at 1.5 ng, the percentage increased to 67.6% (n = 69). In contrast, no defective head-muscle phenotype was observed in the negative control MO-injected embryos (Table 1). The data suggest that the defective head-muscle phenotypes were the evident phenomena of capsulin-knockdown phenotypes. Furthermore, we co-injected Capsulin-MO2 and capped capsulin mRNA into embryos to further prove that the defective morphant phenotype was the result of the endogenous capsulin-knockdown. As shown in Table 1, the incidences of defective head-muscle phenotypes were reduced when embryos were co-injected with various dosages of capped capsulin mRNA and 1.5 ng of Capsulin-MO1. In the low-dosage-injection group (80 pg/embryo), the abnormality rates of defective head-muscle phenotypes decreased from 79.2% (n = 72) to 65.4% (n = 47); while in the high-dosage-injection group (160 pg/embryo), the abnormality rate decreased significantly to 31.4% (n = 26). In contrast, the defective head-muscle phenotypes were not rescued when embryos were co-injected with various dosages of capped gfp mRNA and 1.5 ng of Capsulin-MO1 (Table 1). According to the data, the effect of Capsulin-MOs on knockdown of capsulin was both distinct and consistent.
Table 1.
Morphological phenotypes of zebrafish embryos derived from fertilized eggs injected with capsulin-MO and other materials
| Injection group | Injection dose (per embryo) | Survival embryosa (Survival rates %)b | No defect embryos (%)* | Embryos with defective head-muscle phenotype (%)* |
|---|---|---|---|---|
| Negative control MO | 4 ng | 304 (86.9%) | 304 (100%) | 0 (0%) |
| Capsulin-MO1 | 0.5 ng | 130 (29.7%) | 94 (72.4%) | 36 (27.6%) |
| 1.5 ng | 91 (22.3%) | 19 (20.8%) | 72 (79.2%) | |
| 4.0 ng | 36 (17.7%) | 18 (50.0%) | 18 (50.0%) | |
| Capsulin-MO2 | 0.5 ng | 158 (35.3%) | 116 (73.5%) | 42 (26.5%) |
| 1.5 ng | 102 (28.7%) | 33 (32.4%) | 69 (67.6%) | |
| Capsulin-MO1 + capsulin mRNA | 1.5 ng + 80 pg | 72 (19.3%) | 25 (34.6%) | 47 (65.4%) |
| 1.5 ng + 160 pg | 83 (21.4%) | 57 (68.6%) | 26 (31.4%) | |
| Capsulin-MO2 + capsulin mRNA | 1.5 ng + 80 pg | 96 (24.0%) | 27 (28.1%) | 69 (71.9%) |
| 1.5 ng + 160 pg | 107 (22.4%) | 76 (70.8%) | 31 (29.2%) | |
| Capsulin-MO1 + musculin mRNA | 1.5 ng + 30 pg | 64 (23.6%) | 50 (78.4%) | 14 (21.6%) |
| 1.5 ng + 60 pg | 77 (17.8%) | 70 (90.7%) | 7 (9.3%) | |
| Capsulin-MO1 + myf5 mRNA | 1.5 ng + 50 pg | 125 (24.5%) | 67 (53.5%) | 58 (46.5%) |
| 1.5 ng + 120 pg | 32 (10.2%) | 27 (84.1%) | 5 (15.9%) | |
| Capsulin-MO1 + myod mRNA | 1.5 ng + 70 pg | 188 (26.4%) | 73 (38.8%) | 115 (61.2%) |
| 1.5 ng + 140 pg | 267 (22.7%) | 130 (48.7%) | 137 (51.3%) | |
| Capsulin-MO1 + gfp mRNA | 1.5 ng + 60 pg | 124 (31.4%) | 36 (29.1%) | 88 (70.9%) |
| 1.5 ng + 120 pg | 155 (26.5%) | 44 (28.4%) | 111 (71.6%) |
aTotal numbers of the survival embryos at 72 hpf in triplicate experiments. b Survival rates indicate the numbers of survival embryos among the numbers of injected embryos at 72 hpf. * Percentages indicate the numbers of no defect or defective head-muscle embryos among the numbers of surviving embryos at 72 hpf. hpf hours post-fertilization
On the other hand, after 1.0-ng Musculin-MO1 injection, 75.1% (n = 431) of embryos survived by 72 hpf, and 20.0% (n = 86) of the surviving embryos displayed defective head-muscle phenotypes (reduced plus absent). However, no embryos survived after being injected with 3-ng Musculin-MO1, suggesting that Musculin might be required for cell survival (Table 2). Similar results were also observed in the Musculin-MO2 injection groups (Table 2). Again, the data suggest that the defective head-muscle phenotypes were the evident phenomena of musculin-knockdown phenotypes. Furthermore, we co-injected Musculin-morpholinos (MO1 or MO2) and capped musculin mRNA into embryos and found that defective head-muscle phenotypes were rescued by capped musculin mRNA (Table 2). Taken together, the results suggested that defective head-muscle phenotypes were the consequences of inactivation of either capsulin or musculin.
Table 2.
Morphological phenotypes of zebrafish embryos derived from fertilized eggs injected with Musculin-MO and other materials
| Injection group | Injection dose (per embryo) | Survival embryosa (Survival rates %)b | No defect embryos (%)* | Embryos with defective head-muscle phenotype (%)* |
|---|---|---|---|---|
| Musculin-MO1 | 1.0 ng | 431 (75.1%) | 345 (80.0%) | 86 (20.0%) |
| 1.5 ng | 161 (51.1%) | 140 (86.9%) | 21 (13.1%) | |
| 3 ng | 0 (0%) | 0 (0%) | 0 (0%) | |
| Musculin-MO2 | 1.0 ng | 303 (64.6%) | 169 (55.7%) | 134 (44.3%) |
| 3 ng | 80 (8.1%) | 63 (78.7%) | 17 (21.3%) | |
| Musculin-MO1 + musculin mRNA | 1.0 ng + 10 pg | 296 (63.4%) | 244 (82.4%) | 52 (17.6%) |
| 1.0 ng + 25 pg | 229 (54.5%) | 209 (91.3%) | 20 (8.7%) | |
| Musculin-MO2 + musculin mRNA | 1.0 ng + 10 pg | 256 (66.7%) | 176 (68.7%) | 80 (31.3%) |
| 1.0 ng + 25 pg | 278 (57.7%) | 236 (84.9%) | 42 (15.1%) | |
| Musculin-MO2 + capsulin mRNA | 1.0 ng + 15 pg | 263 (62.6%) | 197 (74.9%) | 66 (25.1%) |
| 1.0 ng + 30 pg | 225 (76.3%) | 189 (84.0%) | 36 (16.0%) | |
| Musculin-MO2 + myf5 mRNA | 1.0 ng + 50 pg | 342 (56.3%) | 202 (59.0%) | 140 (41.0%) |
| 1.0 ng + 120 pg | 297 (49.7%) | 223 (75.1%) | 74 (24.9%) | |
| Musculin-MO2 + myod mRNA | 1.0 ng + 70 pg | 269 (66.7%) | 165 (61.3%) | 104 (38.7%) |
| 1.0 ng + 140 pg | 220 (52.8%) | 151 (68.6%) | 69 (31.4%) |
aTotal numbers of the survival embryos at 72 hpf in triplicate experiments. b Survival rates indicate the numbers of survival embryos among the numbers of injected embryos at 72 hpf. * Percentages indicate the numbers of no defect or defective head-muscle embryos among the numbers of surviving embryos at 72 hpf. hpf hours post-fertilization
Capsulin and Musculin function distinctly in regulation of Myf5/Myod during cranial myogenesis
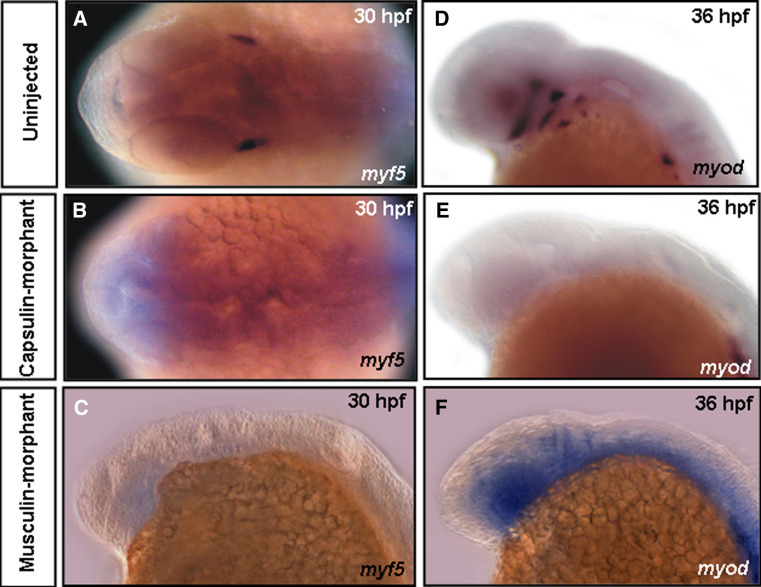
To investigate whether molecular mechanisms lead to defective head-muscle phenotypes, we detected expression patterns of myogenic regulatory factors (MRFs), such as myf5, myod, myogenin, and muscle structure gene α-actin in Capsulin-morphants (injected with Capsulin-MO1), Musculin-morphants (injected with Musculin-MO2) to determine whether down-regulation of these genes resulted in loss of cranial muscles. In uninjected embryos, myf5 transcripts were detected in the mesoderm core of 1st pharyngeal arch/lr at 30 hpf (Fig. 5a). Meanwhile, myod was expressed in the craniofacial muscles mr/ir, sr, lr, mp, im, chd, and chv at 36 hpf (Fig. 5d). myogenin and α-actin were expressed in all craniofacial muscles at 48 or 56 hpf (Supplementary Fig. S3). However, myf5 signals were absent in the Capsulin-morphants at 30 hpf (Fig. 5b); myod transcripts were down-regulated at 36 hpf (myogenin at 48 hpf; α-actin at 56 hpf) in the head region but maintained strong expression at trunk (Fig. 5e, Supplementary Fig. S3). In contrast, myf5 signals were absent in the Musculin-morphants (Fig. 5c); myod signals were still observed with little changes, but myod-positive cells were in a dispersed manner in comparison with those of uninjected embryos (Fig. 5f vs. d). These results indicate that capsulin knockdown leading to loss of all cranial muscles should be due to the absence of myf5- and myod-positive muscle precursor cells, whereas musculin knockdown might have different results. Therefore, we hypothesized that Capsulin and Musculin function distinctly in the regulation of Myf5/Myod during cranial myogenesis.
Fig. 5.
Myogenic genes were regulated by capsulin and/or musculin. The transcripts of myf5 (a–c) and myod (d–f) in Uninjected embryos, Capsulin- and Musculin-morphants at the embryonic stages (hpf) indicated were analyzed by whole-mount in situ hybridization
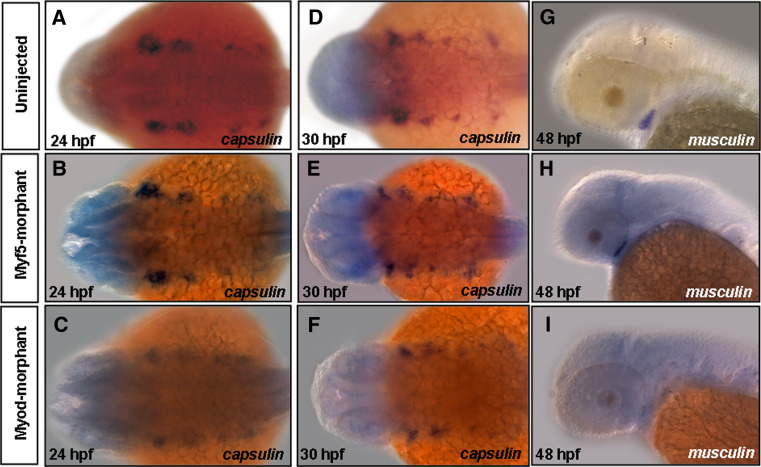
To test this hypothesis, we further examined and compared the capsulin signals between uninjected embryos and embryos derived from either Myf5- or Myod-morphants. Our data show that capsulin signals show no significant differences in 24- and 30-hpf embryos derived from uninjected, Myf5- or Myod-morphants (Fig. 6a–f). Meanwhile, experiments on co-injection of Capsulin-MO2 with capped mRNA of either myf5 or myod were carried out. As shown in Table 1, the incidences of defective head-muscle phenotypes were reduced when embryos were co-injected with various dosages of either capped myf5 or myod mRNA and 1.5 ng of Capsulin-MO1. In the (Capsulin-MO1 + myf5 mRNA) co-injection group (120 pg/embryo), the abnormality rate of defective head-muscle phenotypes decreased from 79.2% (n = 72) to 15.9% (n = 5); and in the (Capsulin-MO1 + myod mRNA) co-injection group (140 pg/embryo), the abnormality rate decreased to 51.3% (n = 137). These results clearly demonstrated that defective head-muscle phenotypes in Capsulin-morphants could be rescued by either capped myf5 or myod mRNA. In contrast, there was little change in musculin signals between uninjected embryos and Myf5-morphants (Fig. 6g, h), but musculin signals were down-regulated in Myod-morphants (Fig. 6i). Again, in the (Musculin-MO2 + myf5 mRNA) co-injection group (120 pg/embryo), the abnormality rate of defective head-muscle phenotypes decreased from 44.3% (n = 134) to 24.9% (n = 74); and in the (Musculin-MO2 + myod mRNA) co-injection group (140 pg/embryo), the abnormality rate decreased slightly to 31.4% (n = 69; Table 2), suggesting that the capped myod mRNA was less efficient than myf5 in rescuing Musculin-MO2-induced defective head-muscle phenotypes. On the basis of these evidences, we proposed that Capsulin and Musculin are both upstream activators of Myf5, but only Capsulin is the upstream activator of Myod during cranial myogenesis.
Fig. 6.
capsulin and musculin expression patterns in the Myf5- and Myod-morphants. The transcripts of capsulin (a–f) and musculin (g–i) in Uninjected embryos, Capsulin- and Musculin-morphants at the embryonic stages (hpf) indicated were analyzed by whole-mount in situ hybridization
Endogenous capsulin expression was down-regulated in Musculin-morphants and vice versa
Lu et al. [13] found that inactivation of neither Capsulin nor Musculin in mice had little effect on cranial muscle development, suggesting that Capsulin and Musculin might compensate each other. Our data showed that knockdown of either Capsulin or Musculin led to defective head-muscle phenotypes in zebrafish (Fig. 4). To understand further their functions, we injected capped musculin mRNA into Capsulin-morphants and capped capsulin mRNA into Musculin-morphants. Results showed that the percentages of defective head-muscle phenotypes were decreased from 79.2% (Capsulin-morphant) to 9.3% (Capsulin-morphant plus capped musculin mRNA) (Table 1), and from 44.7% (Musculin-morphant) to 16.0% (Musculin-morphant plus capped capsulin mRNA) (Table 2). Moreover, quantitative reverse transcription-polymerase chain reaction (qRT-PCR) showed that the expression level of musculin in Capsulin-morphants was significantly decreased to 0.47 (p < 0.0001) fold, in comparison with uninjected embryos (Table 3). On the other hand, the expression level of capsulin (p < 0.0001) in Musculin-morphants was significantly decreased to 0.21 fold (Table 3). These results suggested that Capsulin and Musculin might be able to activate each other, and both of them are essential for cranial myogenesis.
Table 3.
Relative quantification of capsulin and musculin expression levels using the comparative CT method
| Group | Target average CT | β-actin Average CT | ΔCT Target-(β-actin) | ΔΔCT ΔCT(experimental)−ΔCT(control) | Relative folds to control group# |
|---|---|---|---|---|---|
| Capsulin | |||||
| Uninjected | 25.47 ± 0.03 | 18.61 ± 0.03 | 6.85 | 0.00 | 1.00 |
| Musculin-morphant | 25.08 ± 0.02 | 17.15 ± 0.02 | 7.93 | 1.07 | 0.47* |
| Musculin | |||||
| Uninjected | 26.37 ± 0.02 | 18.61 ± 0.03 | 7.75 | 0.00 | 1.00 |
| Capsulin-morphant | 27.84 ± 0.05 | 17.87 ± 0.05 | 9.97 | 2.22 | 0.21* |
C T cycles of qPCR
# Relative folds to control group = 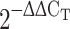
* Star indicates that the control (uninjected) and experimental (Capsulin-, Musculin-morphant) groups are significantly different
Mesoderm cores are unaffected in both Capsulin- and Musculin-morphants
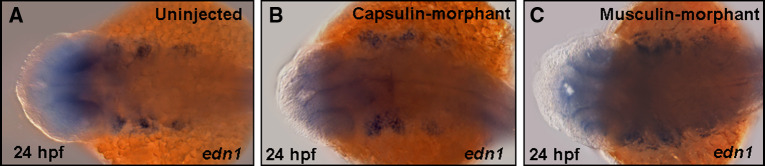
It was reported that mesoderm core played an essential role during cranial myogenesis [1]. To further investigate whether the mesoderm core is affected in either Capsulin- or Musculin-morphant, we used a known mesoderm core marker, edn1, as a riboprobe. As shown in Fig. 7, edn1 labeled three distinct streams of ventral mesoderm core cells in the embryos derived from the uninjected group (Fig. 7a). In contrast, edn1 signals changed little in either Capsulin- (Fig. 7b) or Musculin-morphant (Fig. 7c), indicating that ventral mesoderm cores are unaffected by knocking down either capsulin or musculin.
Fig. 7.
Mesoderm cores are unaffected in the Capsulin- and/or Musculin-morphants. The transcripts of edn1 in a Uninjected embryos, b Capsulin- and c Musculin-morphants at 24 hpf
Neural crest cells change little in Capsulin-morphants
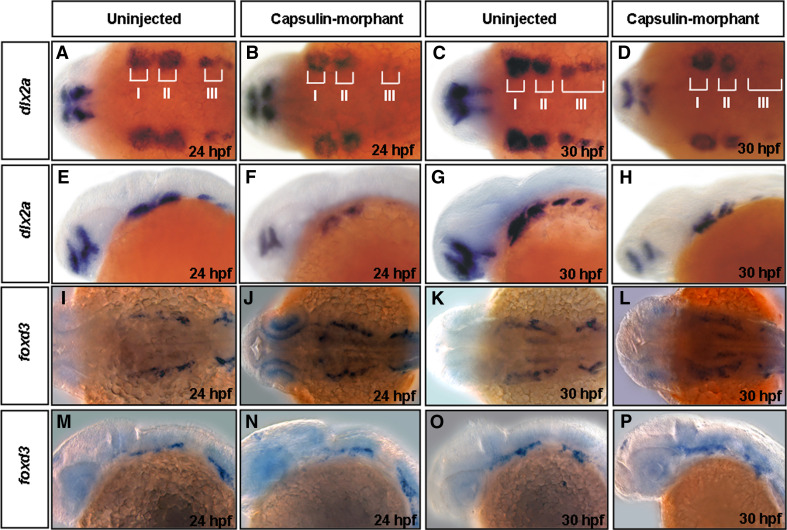
Defects in neural crest cell migration might lead to the loss or malformation of craniofacial elements [30]. We used dlx2a (a post-migratory cell marker) [1] and foxd3 (a pan-neural crest cell marker) [24] probes in whole-mount in situ hybridization to determine whether cranial neural crest cell migration occurs normally in Capsulin-morphants. In 24-hpf (Fig. 8a, e) and 30-hpf (Fig. 8c, g) uninjected embryos, the expression of dlx2a was in three distinct streams of neural crest cells that migrate to the mandibular (I), hyoid (II), and branchial arch (III). In 24-hpf (Fig. 8b, f) and 30-hpf (Fig. 8d, h) Capsulin-MO-injected embryos, dlx2a expression was slightly reduced in hyoid arches (II) but was greatly reduced in the mandibular (I) and branchial arches (III). In contrast, foxd3 signals change little in the embryos derived from both uninjected embryos and Capsulin-morphants (Fig. 8i, m, k, o vs. j, n, l, p), suggesting no significant decrease in total amounts of cranial neural crest cells.
Fig. 8.
Expression patterns of neural crest cell’s genes in the Capsulin-morphants. The transcripts of dlx2a (a–h) and foxd3 (i–p) in Uninjected embryos, and Capsulin-morphants at the embryonic stages (hpf) indicated were analyzed by whole-mount in situ hybridization. I, mandibular arch; II, hyoid arch; III, branchial arch
Extracellular signals regulate capsulin expression
It was reported that the protein Sonic hedgehog (Shh), secreted partly via activation of Fgf8, is essential for cell survival and tissue outgrowth of the developing first pharyngeal arch [31]. To test whether Hh and Fgf signaling play a role in capsulin activation, we detected the capsulin signals of cyclopamine- and SU5402-treated embryos. Results showed that capsulin signals were down-regulated to undetected level in SU5402-treated embryos, whereas capsulin signals were dispersed in head regions of cyclopamine-treated embryos (Supplementary Fig. S4). These observations suggested that Fgf but not Shh might be able to activate capsulin expression.
Discussion
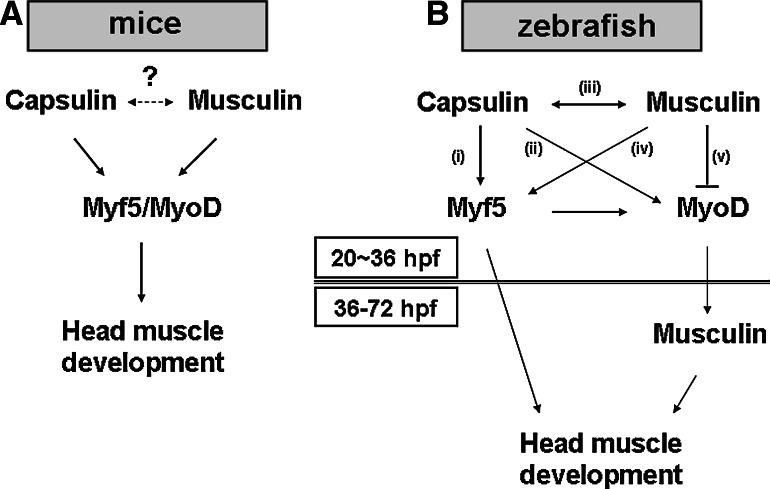
Using an animal model where endogenous Capsulin/Musculin activities can be disrupted enables us to study the physiological roles of Capsulin/Musculin in vivo more precisely. In the mice model, it has been reported that Capsulin and Musculin function redundantly during mice head-muscle development (Fig. 9a) [13]. Although embryonic death was noted in either capsulin or musculin knockout mice embryos, defects in cranial myogenesis have not been reported. However, most of the branchiomeric muscles in the head were lost in mice lacking both Capsulin and Musculin [13, 32]. These observations suggested that Capsulin and Musculin may compensate each other during mice cranial myogenesis. In this study, our data showed that zebrafish embryos treated with either Capsulin- or Musculin-morpholino showed developmental defects, such as lethal effect of embryos, and especially cranial myogenesis defects (Fig. 4). Moreover, Capsulin- and/or Musculin-morphant can be rescued by co-injection with capped musculin and/or capsulin mRNA (Fig. 4, Table 1); endogenous capsulin expression was down-regulated in the Musculin-morphant and vice versa (Table 3). Thus, we proposed that Capsulin and Musculin are mutually-dependent during head-muscle development of zebrafish; and that both Capsulin and Musculin are required for cranial myogenesis. These inconsistencies between zebrafish and mice may be due to the following two reasons. One is that the morpholino knockdown approach does not alter chromosome structure of zebrafish; and the other is that Capsulin and/or Musculin perform different biological functions in fish and mammalian systems.
Fig. 9.
Models of the Capsulin and Musculin regulation network during cranial myogenesis in mice and zebrafish. a Myf5/Myod function downstream of Capsulin/Musculin [13]. b Early phase (20–36 hpf) and late-phase (36–72 hpf) expression of Musculin may have different functions
In this study, we proposed a regulatory network for explaining the complex pathways that involve Capsulin/Musculin during zebrafish head-muscle development. As shown in Fig. 9b, Capsulin might promote head-muscle development through (i) activation of myf5, (ii) activation of myod, and (iii) activation of musculin. Both myod and myf5 mRNAs can rescue capsulin-morphant; however, excess myod mRNA is less efficient in rescuing capsulin-morphant than myf5 mRNA (Table 1). Thus, we suggested that myf5 is more important than myod, and that myf5 and myod play different roles in cranial myogenesis. This observation was further supported by a three-pathway-model proposed by Lin et al. [6], who demonstrated that myf5 and myod control different subsets of head-muscle development. Furthermore, capsulin is a putative down-stream target of Tbx1 transcription factor. Lin et al. [27] reported that myf5- and myod-regulated cranial myogenesis were through tbx1-dependent and tbx1-independent (six1a-pax3) pathways, respectively.
In mice, although Musculin (Myod repressor, MyoR) is known to block myogenesis and activate E-box-dependent muscle genes [32], it can also inhibit differentiation of non-myogenic cells and regulate functions of kidney side population cells [33, 34]. These observations indicate that Musculin might have multiple functions. In zebrafish, musculin expressions are detected at two phases, 20–22 hpf, and 36–72 hpf (Fig. 2). Our data suggested that early phase expression (20–22 hpf) of musculin might activate capsulin and myf5 but inhibit myod (Fig. 9b-iv, -v). At a later stage (older than 36 hpf), musculin transcripts were undetectable in the myod-morphant, suggesting that Myod is a potent activator for the later-phase expression of musculin (Fig. 6l). Notably, the two phases of Musculin expressions are functionally different. Moreover, two-phase expressions of musculin are also reported in mice model [32, 35]. Taken together, our results revealed that the expression patterns of Musculin between mice and zebrafish are similar, and the biological functions of early and/or late-phase expression of Musculin merit further investigation.
Head muscle develops in two distinct populations: branchiomeric and nonbranchiomeric [30, 36]. Branchiomeric muscles include the muscles of mastication, derived from the first or mandibular arch; the muscles of facial expression, derived from the second or hyoid arch; and the muscles of the pharynx and larynx, derived from more caudal arches. Non-branchiomeric head muscles include extraocular muscles and tongue muscles [36, 37]. During zebrafish cranial muscle development, mesoderm cores of the first two arches (arches I and II) are subdivided into dorsal and ventral mesoderm cores. We previously demonstrated capsulin expression at the dorsal and ventral mesoderm cores (Fig. 1). Under the premise of this observation, only branchiomeric muscles would be disrupted in the Capsulin-morphant. However, our data showed that non-branchiomeric head muscles as well as extra-ocular muscles are also affected in the Capsulin-morphant (Fig. 4). Why does endogenous capsulin express neither at non-branchiomeric head muscles nor extra-ocular muscles, which are both affected by capsulin? Because endogenous musculin expression was largely reduced in the Capsulin-morphant (Table 3), we suggest that musculin (especially later-phase expression of musculin) might play an essential role during head-muscle development.
Many genetic studies have revealed that endothelin 1 (edn1) and sonic hedgehog (shh) are involved in ventral arch patterning and first arch development, respectively [25, 31]. edn1 is expressed throughout the pharyngeal endoderm, mesoderm core and surface ectoderm, and has been taken as a good marker for the ventral mesoderm core [38]. Our data showed that (a) edn1 expression was unaffected in neither Capsulin- nor Musculin-morphant (Fig. 7); and (b) inhibition of shh expression had little effect on capsulin expression (Fig. S4). Thus, we proposed that capsulin might not play a role in formation of mesoderm core and the 1st arch. In other words, although Capsulin-morphant embryos developed defective head-muscle phenotypes, both their mesoderm cores and first arches were normal. In summary, this study uncovered a complicated regulatory mechanism showing that Capsulin and Musculin are involved during zebrafish cranial myogenesis. This could provide novel insights which are worthy of further tests in higher vertebrates.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1. Comparison of the deduced amino acid sequence of zebrafish Musculin with those of other known species. The information was obtained from the GenBank nucleotide sequence database with the following accession numbers: human (NP_005089.2), cattle (NM_001192146), rat (XP_001063707.1), mouse (NP_034957.1), and Xenopus (NP_001096235.1) Musculin. Amino acid residues similar to those of the zebrafish Musculin are presented in gray. (TIFF 535 kb)
Fig. S2. Molecular phylogenic analysis of zebrafish Capsulin and Musculin with those of other known species. Data were obtained from GenBank nucleotide sequence database with the following accession numbers: human (Capsulin: NM_003206; Musculin: NP_005089.2), cattle (NM_001014899; NM_001192146), rat (XM_419734; XP_001063707.1), mouse (NM_011545; NP_034957.1), and Xenopus (AY660871; NP_001096235.1). (TIFF 161 kb)
Fig. S3. Myogenic genes, myogenin and α-actin, were affected in the Capsulin-morphant. The transcripts of myogenin (A-B) and α-actin (C-D) in Uninjected embryos, and Capsulin-morphants at the embryonic stages (hpf) indicated were analyzed by whole-mount in situ hybridization. (TIFF 353 kb)
Fig. S4. Upstream signals regulate capsulin expression. The expression patterns of capsulin in (A) vehicle-treated, (B) cyclopamine-treated and (C) SU5402-treated embryos at 24 and 30 hpf. (TIFF 849 kb)
Acknowledgments
This work was supported by the National Science Council, Republic of China, under grant number of NSC 97-2313-B-032-001-MY3. We are also grateful to the Zebrafish Core in Academia Sinica (ZCAS) for providing tbx1 plasmid.
Footnotes
G.-H. Lee and M.-Y. Chang contributed equally to this work.
References
- 1.Yelick PC, Schilling TF. Molecular dissection of craniofacial development using zebrafish. Crit Rev Oral Biol Med. 2002;13:308–322. doi: 10.1177/154411130201300402. [DOI] [PubMed] [Google Scholar]
- 2.Noden DM, Trainor PA. Relations and interactions between cranial mesoderm and neural crest populations. J Anat. 2005;207:575–601. doi: 10.1111/j.1469-7580.2005.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rinon A, Lazar S, Marshall H, Büchmann-Møller S, Neufeld A, Elhanany-Tamir H, Taketo MM, Sommer L, Krumlauf R, Tzahor E. Cranial neural crest cells regulate head muscle patterning and differentiation during vertebrate embryogenesis. Development. 2007;134:3065–3075. doi: 10.1242/dev.002501. [DOI] [PubMed] [Google Scholar]
- 4.Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol. 2005;16:585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 5.Chen YH, Tsai HJ. Myogenic regulatory factors Myf5 and Mrf4 of fish: current status and perspective. J Fish Biol. 2008;73:1872–1890. doi: 10.1111/j.1095-8649.2008.01991.x. [DOI] [Google Scholar]
- 6.Lin CY, Yung RF, Lee HC, Chen WT, Chen YH, Tsai HJ. Myogenic regulatory factor Myf5 and MyoD function distinctly during craniofacial myogenesis of zebrafish. Dev Biol. 2006;299:594–608. doi: 10.1016/j.ydbio.2006.08.042. [DOI] [PubMed] [Google Scholar]
- 7.Lu J, Richardson JA, Olson EN. Capsulin: a novel bHLH transcription factor expressed in epicardial progenitors and mesenchyme of visceral organs. Mech Dev. 1998;73:23–32. doi: 10.1016/S0925-4773(98)00030-6. [DOI] [PubMed] [Google Scholar]
- 8.Quaggin SE, Vanden Heuvel GB, Igarashi P. Pod-1, a mesoderm-specific basic-helix-loop-helix protein expressed in mesenchymal and glomerular epithelial cells in the developing kidney. Mech Dev. 1998;71:37–48. doi: 10.1016/S0925-4773(97)00201-3. [DOI] [PubMed] [Google Scholar]
- 9.Robb L, Mifsud L, Hartley L, Biben C, Copeland NG, Gilbert DJ, Jenkins NA, Harvey RP. Epicardin: A novel basic helix-loop-helix transcription factor gene expressed in epicardium, branchial arch myoblasts, and mesenchyme of developing lung, gut, kidney, and gonads. Dev Dyn. 1998;213:105–113. doi: 10.1002/(SICI)1097-0177(199809)213:1<105::AID-AJA10>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 10.Miyagishi M, Nakajima T, Fukamizu A. Molecular characterization of mesoderm-restricted basic helix-loop-helix protein, POD-1/Capsulin. Int J Mol Med. 2000;5:27–31. doi: 10.3892/ijmm.5.1.27. [DOI] [PubMed] [Google Scholar]
- 11.Klein SL, Strausberg RL, Wagner L, Pontius J, Clifton SW, Richardson P. Genetic and genomic tools for Xenopus research: The NIH Xenopus initiative. Dev Dyn. 2002;225:384–391. doi: 10.1002/dvdy.10174. [DOI] [PubMed] [Google Scholar]
- 12.von Scheven G, Bothe I, Ahmed MU, Alvares LE, Dietrich S. Protein and genomic organization of vertebrate MyoR and Capsulin genes and their expression during avian development. Gene Expr Patterns. 2006;6:383–393. doi: 10.1016/j.modgep.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Lu JR, Bassel-Duby R, Hawkins A, Chang P, Valdez R, Wu H, Gan L, Shelton JM, Richardson JA, Olson EN. Control of facial muscle development by MyoR and capsulin. Science. 2002;298:2378–2381. doi: 10.1126/science.1078273. [DOI] [PubMed] [Google Scholar]
- 14.Cui S, Schwartz L, Quaggin SE. Pod1 is required in stromal cells for glomerulogenesis. Dev Dyn. 2003;226:512–522. doi: 10.1002/dvdy.10244. [DOI] [PubMed] [Google Scholar]
- 15.Cui S, Ross A, Stallings N, Parker KL, Capel B, Quaggin SE. Disrupted gonadogenesis and male-to-female sex reversal in Pod1 knockout mice. Development. 2004;131:4095–4105. doi: 10.1242/dev.01266. [DOI] [PubMed] [Google Scholar]
- 16.Chou CY, Hsu CH, Wang YH, Chang MY, Chen LC, Cheng SC, Chen YH. Biochemical and structural properties of zebrafish Capsulin produced by Escherichia coli . Protein Expr Purif. 2011;75:21–27. doi: 10.1016/j.pep.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Chen YH, Wang YH, Chang MY, Lin CY, Weng CW, Westerfield M, Tsai HJ. Multiple upstream modules regulate zebrafish myf5 expression. BMC Dev Biol. 2007;7:1. doi: 10.1186/1471-213X-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen YH, Lin YT, Lee GH. Novel and unexpected functions of zebrafish CCAAT box binding transcription factor (NF-Y) B subunit during cartilages development. Bone. 2009;44:777–784. doi: 10.1016/j.bone.2009.01.374. [DOI] [PubMed] [Google Scholar]
- 19.Chen YH, Lee WC, Liu CF, Tsai HJ. Molecular structure, dynamic expression and promoter analysis of zebrafish (Danio rerio) myf-5 gene. Genesis. 2001;29:22–35. doi: 10.1002/1526-968X(200101)29:1<22::AID-GENE1002>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 20.Weinberg ES, Allende ML, Kelly CS, Abdelhamid A, Murakami T, Andermann P, Doerre OG, Grunwald DJ, Riggleman B. Developmental regulation of zebrafish MyoD in wild-type, no-tail and spadetail embryos. Development. 1996;122:2711–2780. doi: 10.1242/dev.122.1.271. [DOI] [PubMed] [Google Scholar]
- 21.Chen YH, Lee WC, Cheng CH, Tsai HJ. Muscle regulatory factor gene: zebrafish (Danio rerio) myogenin cDNA. Comp Biochem Physiol B: Biochem Mol Biol. 2000;127:97–103. doi: 10.1016/S0305-0491(00)00242-X. [DOI] [PubMed] [Google Scholar]
- 22.Xu YF, He JY, Wang XK, Lim TM, Gong ZY. Asynchronous activation of 10 muscle-specific protein (MSP) genes during zebrafish somitogenesis. Dev Dyn. 2000;219:201–215. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1043>3.3.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 23.Akimenko MA, Ekker M, Wegner J, Lin W, Westerfield M. Combinatorial expression of three zebrafish genes related to distal-less: part of a homeobox gene code for the head. J Neurosci. 1994;14:3475–3486. doi: 10.1523/JNEUROSCI.14-06-03475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Odenthal J, Nusslein-Volhard C. Fork head domain genes in zebrafish. Dev Genes Evol. 1998;208:245–258. doi: 10.1007/s004270050179. [DOI] [PubMed] [Google Scholar]
- 25.Kimmel CB, Ullmann B, Walker M, Miller CT, Crump JG. Endothelin 1-mediated regulation of pharyngeal bone development in zebrafish. Development. 2003;130:1339–1351. doi: 10.1242/dev.00338. [DOI] [PubMed] [Google Scholar]
- 26.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 27.Lin CY, Chen WT, Lee HC, Yang PH, Yang HJ, Tsai HJ. The transcription factor Six1a plays an essential role in the craniofacial myogenesis of zebrafish. Dev Biol. 2009;331:152–166. doi: 10.1016/j.ydbio.2009.04.029. [DOI] [PubMed] [Google Scholar]
- 28.Wang YH, Wen CC, Yang ZS, Cheng CC, Tsai JN, Ku CC, Wu HJ, Chen YH. Development of a whole-organism model to screen new compounds for sun protection. Mar Biotechnol. 2009;11:419–429. doi: 10.1007/s10126-008-9159-9. [DOI] [PubMed] [Google Scholar]
- 29.Wang YH, Cheng CC, Lee WJ, Chiou ML, Pai CW, Wen CC, Chen WL, Chen YH. A novel phenotype-based approach for systematically screening antiproliferation metallodrugs. Chem-Biol Interact. 2009;182:84–91. doi: 10.1016/j.cbi.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 30.Noden DM. The role of the neural crest in patterning of avian cranial skeletal, connective, and muscle tissues. Dev Biol. 1983;96:144–165. doi: 10.1016/0012-1606(83)90318-4. [DOI] [PubMed] [Google Scholar]
- 31.Yamagishi C, Yamagishi H, Maeda J, Tsuchihashi T, Ivey K, Hu T, Srivastava S. Sonic hedgehog is essential for first pharyngeal arch development. Pediatr Res. 2006;59:349–354. doi: 10.1203/01.pdr.0000199911.17287.3e. [DOI] [PubMed] [Google Scholar]
- 32.Lu J, Webb R, Richardson JA, Olson EN. MyoR: a muscle restricted basic helix–loop–helix transcription factor that antagonizes the actions of MyoD. Proc Natl Acad Sci USA. 1999;96:552–557. doi: 10.1073/pnas.96.2.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu L, Mikloocich J, Sangster N, Perez A, McCormick PJ. MyoR is expressed in nonmyogenic cells and can inhibit their differentiation. Exp Cell Res. 2003;289:162–173. doi: 10.1016/S0014-4827(03)00252-0. [DOI] [PubMed] [Google Scholar]
- 34.Hishikawa K, Marumo T, Miura S, Nakanishi A, Matsuzaki Y, Shibata K, Ichiyanagi T, Kohike H, Komori T, Takahashi I, Takase O, Imai N, Yoshikawa M, Inowa T, Hayashi M, Nakaki T, Nakauchi H, Okano H, Fujita T. Musculin/MyoR is expressed in kidney side population cells and can regulate their function. J Cell Biol. 2005;169:921–928. doi: 10.1083/jcb.200412167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu L, Sangster N, Perez A, McCormick PJ. The bHLH protein MyoR inhibits the differentiation of early embryonic endoderm. Differentiation. 2004;72:341–347. doi: 10.1111/j.1432-0436.2004.07207005.x. [DOI] [PubMed] [Google Scholar]
- 36.Noden DM, Francis-West P. The differentiation and morphogenesis of craniofacial muscles. Dev Dyn. 2006;235:1194–1218. doi: 10.1002/dvdy.20697. [DOI] [PubMed] [Google Scholar]
- 37.Mackenzie S, Walsh FS, Graham A. Migration of hypoglossal myoblast precursors. Dev Dyn. 1998;213:349–358. doi: 10.1002/(SICI)1097-0177(199812)213:4<349::AID-AJA1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 38.Miller CT, Schilling TF, Lee K, Parker J, Kimmel CB. sucker encodes a zebrafish endothelin-1 required for ventral pharyngeal arch development. Development. 2000;127:3815–3828. doi: 10.1242/dev.127.17.3815. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Comparison of the deduced amino acid sequence of zebrafish Musculin with those of other known species. The information was obtained from the GenBank nucleotide sequence database with the following accession numbers: human (NP_005089.2), cattle (NM_001192146), rat (XP_001063707.1), mouse (NP_034957.1), and Xenopus (NP_001096235.1) Musculin. Amino acid residues similar to those of the zebrafish Musculin are presented in gray. (TIFF 535 kb)
Fig. S2. Molecular phylogenic analysis of zebrafish Capsulin and Musculin with those of other known species. Data were obtained from GenBank nucleotide sequence database with the following accession numbers: human (Capsulin: NM_003206; Musculin: NP_005089.2), cattle (NM_001014899; NM_001192146), rat (XM_419734; XP_001063707.1), mouse (NM_011545; NP_034957.1), and Xenopus (AY660871; NP_001096235.1). (TIFF 161 kb)
Fig. S3. Myogenic genes, myogenin and α-actin, were affected in the Capsulin-morphant. The transcripts of myogenin (A-B) and α-actin (C-D) in Uninjected embryos, and Capsulin-morphants at the embryonic stages (hpf) indicated were analyzed by whole-mount in situ hybridization. (TIFF 353 kb)
Fig. S4. Upstream signals regulate capsulin expression. The expression patterns of capsulin in (A) vehicle-treated, (B) cyclopamine-treated and (C) SU5402-treated embryos at 24 and 30 hpf. (TIFF 849 kb)