Abstract
Liver receptor homologue-1 (LRH-1) is a member of the nuclear receptor superfamily. We characterized two functional nuclear localization signals (NLSs) in LRH-1. NLS1 (residues 117–168) overlaps the second zinc finger in the DNA binding domain. Mutagenesis showed that the zinc finger structure and two basic clusters on either side of the zinc finger loop are critical for nuclear import of NLS1. NLS2 (residues 169–204) is located in the Ftz-F1 box that contains a bipartite signal. In full-length LRH-1, mutation of either NLS1 or NLS2 had no effect on nuclear localization, but disruption of both NLS1 and NLS2 resulted in the cytoplasmic accumulation of LRH-1. Either NLS1 or NLS2 alone was sufficient to target LRH-1 to the nucleus. Both NLS1 and NLS2 mediate nuclear transport by a mechanism involving importin α/β. Finally, we showed that three crucial basic clusters in the NLSs are involved in the DNA binding and transcriptional activities of LRH-1.
Keywords: Liver receptor homologue-1 (LRH-1), Nuclear localization signal, Zinc finger, Importin, Nuclear receptor, Nuclear import
Introduction
Liver receptor homologue-1 (LRH-1; NR5A2), a member of the nuclear receptor NR5A subfamily, is predominantly expressed in the liver, intestine, pancreas and ovaries [1, 2]. Homozygous LRH-1-deficient mice die around embryonic day 7.5, indicating that it plays a critical role in embryogenesis [3]. LRH-1 is highly related to steroidogenic factor-1 (SF-1; NR5A1), also known as adrenal 4 binding protein (Ad4BP), which governs the development and function of the adrenal glands, gonads, ventromedial hypothalamus and pituitary [4–6]. LRH-1 and SF-1 recognize the same DNA sequence and bind as monomers to activate the transcription of target genes [7]. A number of target genes of LRH-1 have been identified and are involved in reverse cholesterol transport, bile acid synthesis, steroidogenesis and cell proliferation [8–11].
LRH-1 has the typical structure of a nuclear hormone receptor, including an N-terminal activation function-1 domain, a zinc-finger-containing DNA binding domain (DBD), a hinge region, and a C-terminal ligand-binding domain (LBD) possessing a ligand-dependent activation domain (AF-2) [2, 7]. Members of the NR5A subfamily contain a unique 30-amino acid Ftz-F1 box located after the second zinc finger motif [12]. Nuclear hormone receptors are typically ligand-activated transcription factors. Unliganded nuclear receptors can be found in both cytosol and nucleus. However, the ligand-bound receptors become localized to the nucleus and bind to specific sites in regulatory regions to regulate the transcription of target genes. Multiple coregulators have been shown to interact with nuclear receptors and modulate the transcriptional activity of receptors. LRH-1 and SF-1 are orphan receptors whose natural ligands have yet to be defined. Unlike most nuclear receptors, LRH-1 molecules bind to DNA as monomers and are constitutively active when expressed in cells [7, 13].
LRH-1 is always localized to the nucleus in cells [14]. As a nuclear protein, LRH-1 must be imported from the cytoplasm into the nucleus via the nuclear pore complex (NPC). Small molecules (less than 40 kDa) can pass directly through the NPC by passive diffusion. However, transport of large molecules through the NPC is an active process that is mediated by a family of transport factors known as importins and exportins [15]. Protein import into the nucleus usually requires the presence of nuclear localization signal (NLS). The classical NLS is characterized by a short stretch of basic amino acids (monopartite NLS) or two stretches of basic amino acids separated by a spacer of 10–12 residues (bipartite NLS) [16, 17]. Importin β-mediated transport through the NPC is the most commonly utilized nuclear import process. In the classical pathway, the NLS of the cargo proteins is recognized by an adaptor protein, importin α, that forms a complex with importin β [15, 18]. However, importin α may not be required for the nuclear import of target proteins that can interact directly with importin β [19, 20].
Multiple NLSs are usually present in nuclear proteins. Like most DNA-binding proteins, the predominant NLS of nuclear receptors is located in the DBD [21]. The glucocorticoid receptor (GR) is reported to have a second NLS (NLS2) in the LBD that lacks a recognizable basic motif [22, 23]. The androgen receptor (AR) also contains an agonist-specific NLS in the LBD and a ligand-independent NLS in the N-terminal transactivation domain (NTD) [24, 25]. Estrogen receptor α and progesterone receptor has three proto-NLSs located in the DBD and hinge region, but none of them is sufficient for nuclear targeting of protein [26]. These NLSs can mediate the translocation of receptors to the nucleus by different mechanisms. The NLS1 of GR interacts with importin α, importin 7 and importin 8, and GR can be imported by importin α/β and importin 7 [27]. The NLS2 also interacts with importin 7 and importin 8, but is unable to bind importin α. Importin α/β is involved in interacting with the NLS of AR DBD, whereas the NLSs of the NTD and LBD are importin α/β-independent [28].
Nucleocytoplasmic transport is an important mode of regulation of the transcription factor function of nuclear receptors. The transport signals or factors involved in regulating nuclear import of LRH-1 have not been empirically determined. In this report, we describe two functional NLSs in mouse LRH-1. The presence of either NLS is sufficient to introduce LRH-1 into the nucleus. We further show that both NLSs interact with importin α and importin β and that nuclear accumulation of GFP-NLSs was abolished by energy depletion, suggesting that the nuclear transport of LRH-1 works via the importin α/β system.
Materials and methods
Plasmids
The plasmids pGFP-LRH1 and pMyc-LRH1 have been described previously [14]. After plasmid pGFP-LRH1 was digested with KpnI/BamHI or ClaI/BamHI, the ends were blunted with Klenow and then self-ligated to generate pGFP-LRH11–116 and pGFP-LRH11–169. Restriction enzymes ClaI, BsrGI and BsrDI were cut at positions corresponding to codons 169, 193 and 226 of LRH-1, respectively. The C-terminal fragments of LRH-1 were released by digestion with appropriate restriction enzymes and then inserted in-frame after EGFP in the pEGFP-C1 vector (Clontech) to produce pGFP-LRH1169–560, pGFP-LRH1193–560, and pGFP-LRH1226–560. pGFP-LRH1 was digested with KpnI and ClaI, blunted with Klenow and self-ligated to generate pGFP-LRH1Δ116–168. Plasmid pGFP-LRH1Δ170–193 was constructed by PCR amplification using relevant primers incorporating a ClaI site and a BamHI site, and the resulting fragment was inserted into ClaI/BamHI-digested pGFP-LRH1. Plasmid pGFP-LRH1Δ118–205 was constructed by PCR amplification using relevant primers incorporating a KpnI site and a BlpI site, and the resulting fragment was inserted into KpnI/BlpI-digested pGFP-LRH1. The ClaI-KpnI fragment from pGFP-LRH1 was inserted into pEGFP-C2 (Clontech) to generate pGFP-LRH1117–168. Plasmids pGFP-LRH1126–142 and pGFP-LRH1137–160 were generated by ligating a double-stranded oligomer containing residues 126–142 or 137–160 with HindIII and BamHI ends into the HindIII/BamHI sites of pEGFP-C2. Plasmid pGFP-LRH1151–169 was produced by ligating a double-stranded oligomer containing residues 151–169 with BsrGI and NotI ends into the BsrGI/NotI sites of pEGFP-N1 (Clontech). The plasmid pGFP-LRH1169–193 was made by digestion of pGFP-LRH1169–560 with BsrGI and XbaI, blunted with Klenow and self-ligated. Plasmid pGFP-LRH1169–204 was constructed by PCR amplification and the resulting fragment was inserted into SmaI/BamHI digested pEGFP-C1.
Alanine-substitution mutants and C143G mutation were generated based on pMyc-LRH1, pGFP-LRH1117–168, or pGFP-LRH1169–204 by PCR-based site-directed mutagenesis [29] or overlap extension PCR [30]. All mutations were verified by DNA sequencing. For generation of mutants with mutations in disjoint areas, the ClaI fragment from pMyc-LRH1-R181A/R183A was inserted into the same site of pMyc-LRH1-K132A/R133A or pMyc-LRH1-K165A/K166A to produce the mutant pMyc-LRH1-K132A/R133A + R181A/R183A or pMyc-LRH1-K165A/K166A + R181A/R183A. The ClaI fragment from pMyc-LRH1-K194A/R195A was inserted into the same site of the pMyc-LRH1-K132A/R133A or pMyc-LRH1-K165A/K166A to produce the mutant pMyc-LRH1-K132A/R133A + K194A/R195A or pMyc-LRH1-K165A/K166A + K194A/R195A. Two complementary primers (5′-tcgagatgactgctccaaagaagaagcgtaaga-3′ and 5′-agcttcttacgcttcttctttggagcagtcatc-3′) containing an eight amino acid sequence of SV40 NLS were inserted into the XhoI/HindIII sites of pEGFP-C3 to generate pEGFP-NLSSV40.
The luciferase reporter pSCC2.3-Luc was produced by cloning the upstream regions (−2,300 to +55) of human CYP11A1 into pGL3-Basic vector (Promega). Plasmids for fusion proteins GST-importin α and GST-importin β were as previously described [31, 32] and generously provided by Y. Yoneda (Department of Anatomy and Cell Biology, Osaka University Medical School, Osaka, Japan). The GFP-fusion protein constructs for the transcription/translation system were produced by inserting the cDNA into the NheI/SmaI sites of pCl-neo vector (Promega).
Fluorescence microscopy and ATP depletion
COS-7 cells were maintained in DMEM/high glucose (3%), supplemented with 10% fetal bovine serum. Cells were subcultured onto 24-well plates and transfected the following day with 1 μg of plasmid DNA using TurboFect (Fermentas) according to the manufacturer’s protocol. After 24 h, cells were fixed in 4% paraformaldehyde for 10 min and then mounted. For immunocytochemistry, fixed cells were treated with 0.2% Triton X-100 in PBS for 10 min and then incubated with anti-Myc antibody (Upstate Biotechnology) overnight at 4°C. After washing with PBS, the cells were incubated with the Alexa fluor 568 anti-mouse IgG secondary antibody (Molecular Probes) for 1 h. Nuclear DNA was stained with DAPI for 30 min. Images were obtained with a Leica TCS SP5 confocal microscope. For energy-depletion experiments, cells were cultured with 10 mM sodium azide and 6 mM 2-deoxy-d-glucose (Sigma) in glucose-free DMEM (Invitrogen) containing 10% fetal bovine serum. Cells were then examined by fluorescence microscopy after 30 or 60 min incubation in the energy-depletion medium. The cells were stained with MitoTrack Red (Molecular Probes) to monitor mitochondrial function.
Luciferase assays
Cells were subcultured 24 h before transfection onto 24-well plates at a density of 105 cells/well. Cells were transfected with 200 ng of LRH-1 expression plasmids, 100 ng of reporter pSCC2.3-Luc and 1 ng of control reporter phRLuc using Lipofectamine 2000 (Invitrogen). After 24 h, cells were harvested and luciferase activities were determined using the Dual-Glo Luciferase Assay System (Promega). The results were normalized to internal Renilla luciferase activities. Data were obtained from six independent experiments and are presented as means±SEM. The significance of differences between group means was determined using one-way ANOVA.
Western blotting
Western blot analyses were performed as previously described [33]. The anti-Myc antibody was used to assess the expression of Myc-LRH-1, while anti-β-actin antibody determination of β-actin levels (Upstate Biotechnology) was used as a loading control. Immunoblotting levels were determined using the Immobilon Western HRP system (Millipore).
GST pull-down
GST fusion proteins were expressed in Escherichia coli BL21(DE3) cells by induction with 1 mM isopropyl-β-d-thiogalactopyranoside for 4 h at 28°C. Cells were pelleted, frozen at −80°C and then resuspended in extraction buffer (2 mM EDTA, 10 mM dithiothreitol, 2 mM PMSF and 10 μg/ml leupeptin). Cells were lysed by addition of 2 mg/ml lysozyme and 0.1 mg/ml DNase with incubation for 1 h on ice. After sonication and centrifugation, the GST fusion proteins in the supernatant were incubated with glutathione-Sepharose beads (Amersham Biosciences) for 3 h at 4°C. After three washes with PBS containing 0.1% Triton X-100, bead-bound GST proteins were incubated with [35S]methionine-labelled proteins produced by in vitro translation using the TNT T7 Quick Coupled Transcription/Translation System (Promega) according to the manufacturer’s protocol. The reactions were carried out for 2 h at 4°C. The bound proteins were washed with PBS/Triton X-100 and then resolved on 12% SDS-polyacrylamide gels. The gels were dried and signals were detected using a Typhoon 8600 Phosphor Imager (GE Healthcare Life Sciences).
Electrophoretic mobility shift assay
Oligonucleotides containing the LRH-1/SF-1 binding site from the human CYP11A1 promoter were end-labelled with [γ-32P]ATP and T4 polynucleotide kinase (NEB), annealed and purified by Amicon Ultra-0.5 (Millipore). The sense strand sequences of the probes were 5′-TCGACTTCTGGTATGGCCTTGAGCTGGTAG and 5′-CAGCTTCTGGTATCACCGTGAGCTGGTAGTT (mutant probe) [34], where the consensus sequences are underlined. Wild-type LRH-1 and its mutants were translated in vitro using the TNT T3 Quick Coupled Transcription/Translation System (Promega). These proteins were then incubated with radiolabelled probes in 20 μl of binding buffer, comprising 10 mM HEPES, pH 7.5, 1 mM EDTA, 1 mM dithiothreitol, 50 mM KCl, 0.1% bovine serum albumin and 0.5 μg of poly(dI-dC), at room temperature for 30 min. For competition experiments, a 20-fold molar excess of cold oligonucleotide was preincubated with proteins for 30 min. When antibody was included in the reaction, in vitro-translated proteins were incubated with anti-Myc antibody (1 μg) for 120 min at 4°C before addition of the labelled probe. The binding products were resolved on 4% non-denaturing polyacrylamide gels at 4°C. The gels were dried and signals were detected using a Typhoon 8600 Phosphor Imager (GE Healthcare Life Sciences).
Results
Identification of two putative nuclear localization domains in LRH-1
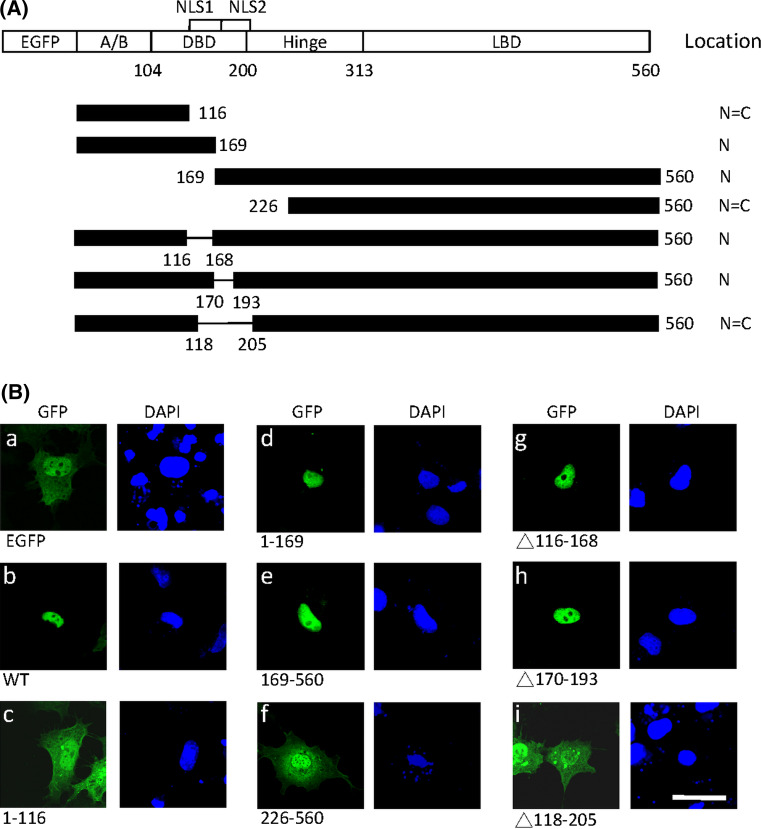
To determine the region responsible for nuclear localization, full-length and various deletion fragments of LRH-1 were fused to the C-terminus of enhanced green fluorescence protein (GFP) (Fig. 1a). As shown in Fig. 1ba) GFP protein (approximately 27 kDa) was dispersed in the nucleus and cytoplasm in COS-7 cells because small molecules can cross the nucleus pore complex by passive diffusion [35]. The full-length LRH-1 fused to GFP was exclusively localized to the nucleus, indicating the presence of an active NLS (Fig. 1bb). The N-terminal 1–169 fragment localized GFP to the nucleus (Fig. 1bd), whereas the 1–116 fragment had no effect on nuclear localization (Fig. 1bc), suggesting that residues 117–169 contained a functional NLS. However, removal of this region (construct 169–560) did not abolish the protein transport to the nucleus (Fig. 1be). The N-terminal deletion construct 193–560 was partially localized in the cytoplasm (data not shown) and the 226–560 fragment was distributed throughout the cell just like GFP alone (Fig. 1bf). These results implied that the 169–226 region contains the second NLS. Deletion of residues 116–168 or 170–193 did not affect nuclear localization (Fig. 1bg, h), whereas deletion of these two nuclear localization motifs (construct Δ118–205) abolished nuclear localization (Fig. 1bi). The results suggested that these two NLSs can target protein to the nucleus independently and one of them is required for nuclear transport of LRH-1.
Fig. 1.
Identification of two nuclear localization domains in LRH-1. a Schematic representation of LRH-1 deletion fragments fused to C-terminal EGFP tag. The full-length LRH-1 contains the A/B domain, DNA binding domain (DBD), hinge region and ligand binding domain (LBD). Two putative NLSs are indicated above. The subcellular localization of LRH-1 mutants is summarized on the right (N nuclear, C cytoplasmic). b GFP-LRH-1 wild-type or deletion mutants were transfected into COS-7 cells. After 24 h, cells were fixed and nuclei counterstained with DAPI. Fusion proteins were visualized by confocal microscopy. Scale bar 25 μm
Identification of key residues for nuclear import in NLS1 and NLS2
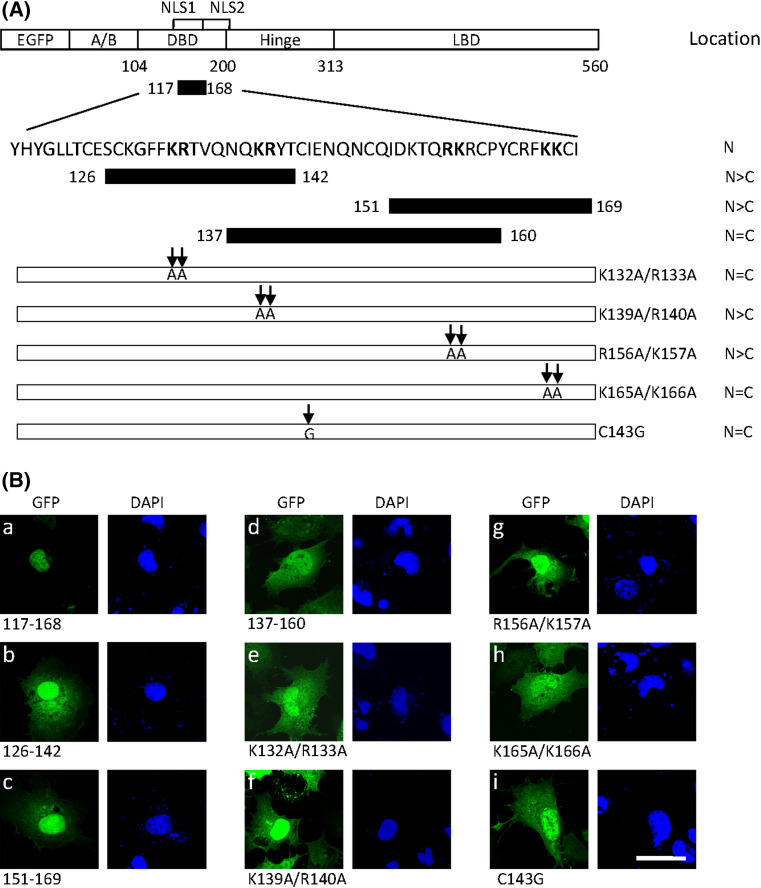
To confirm that residues 117–168 contain the nuclear localization activity, this fragment was fused to GFP to examine the subcellular distribution in COS-7 cells. As shown in Fig. 2ba, this fusion protein was exclusively found in the nucleus, indicating that an active NLS was included in this region. Three smaller fragments were created and individually fused to GFP to define the NLS in this region (Fig. 2a). The fusion protein with the fragment containing residues 137–160 was present in both the nucleus and cytoplasm (Fig. 2bd). Although fusion proteins with residues 126–142 and 151–169 were predominantly nuclear, significant cytoplasmic localization was observed in most cells (Fig. 2bb,c). Thus, the entire domain (117–168, referred to NLS1) appeared to be necessary to drive GFP completely into the nucleus. Four clusters consisting of two adjacent positively charged residues, typically present in a NLS, were identified in this region. To determine which particular positively charged residues are critical for the nuclear import of NLS1, four mutants were generated by mutation of the basic arginine and lysine into the neutral amino acid alanine in each cluster (Fig. 2a). The mutation K139A/R140A or R156A/K157A produced partial cytoplasmic distribution of the fusion protein (Fig. 2bf,g). However, mutation of K132A/R133A or K165A/K166A completely abolished the nuclear targeting of NLS1 (Fig. 2be,h). These results suggest that basic amino acids in cluster 1 (K132 and R133) and cluster 4 (K165 and K166) are necessary for efficient nuclear import of NLS1.
Fig. 2.
Detection of key basic residues in NLS1 for nuclear import. a Sequence of the putative NLS1 is shown. The full-fragment (residues 117–168) and three segments within this region (residues 126–142, 137–160 and 151–169) were fused to EGFP. The basic amino acids lysine and arginine (in boldface) in the 117–168 region were substituted by alanines as indicated by arrows. The subcellular localization of LRH-1 mutants is summarized on the right (N nuclear, C cytoplasmic). b Wild-type and mutant forms were transfected into COS-7 cells. After 24 h, cells were fixed and the nuclei counterstained with DAPI. Fusion proteins were visualized by confocal microscopy. Scale bar 25 μm
The NLS1 is located in the DBD encompassing amino acids 117–168, which contains an intact zinc finger motif (Fig. 7b). To determine whether zinc finger structure is crucial for the nuclear import of NLS1, the first zinc-chelating cysteine was changed into glycine to destabilize the zinc finger structure. As shown in Fig. 2bi, the mutant protein C143G failed to target to the nucleus, suggesting that the zinc finger tertiary structure is required for the nuclear transport of NLS1.
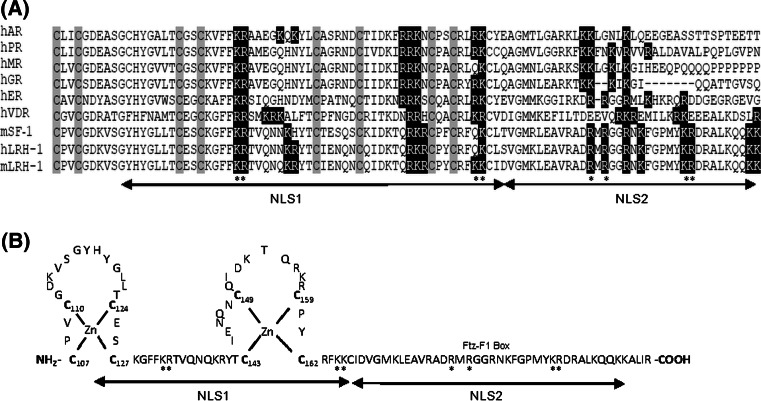
Fig. 7.
Comparison with other nuclear receptors and the structure of LRH-1 NLS domain. a Sequence alignment of the zinc-finger domain and the adjacent region of several nuclear receptors. The cysteine residues that coordinate zinc binding are highlighted by grey boxes. The nuclear localization domains NLS1 and NLS2 of LRH-1 are indicated. The conserved basic amino acids that were mutated in this study are highlighted by dark boxes. The key basic amino acids identified for nuclear import of NLS1 and NLS2 are indicated by asterisks. b Diagram of the zinc binding loops of LRH-1. The unique Ftz-F1 box for NR5A subfamily members is indicated
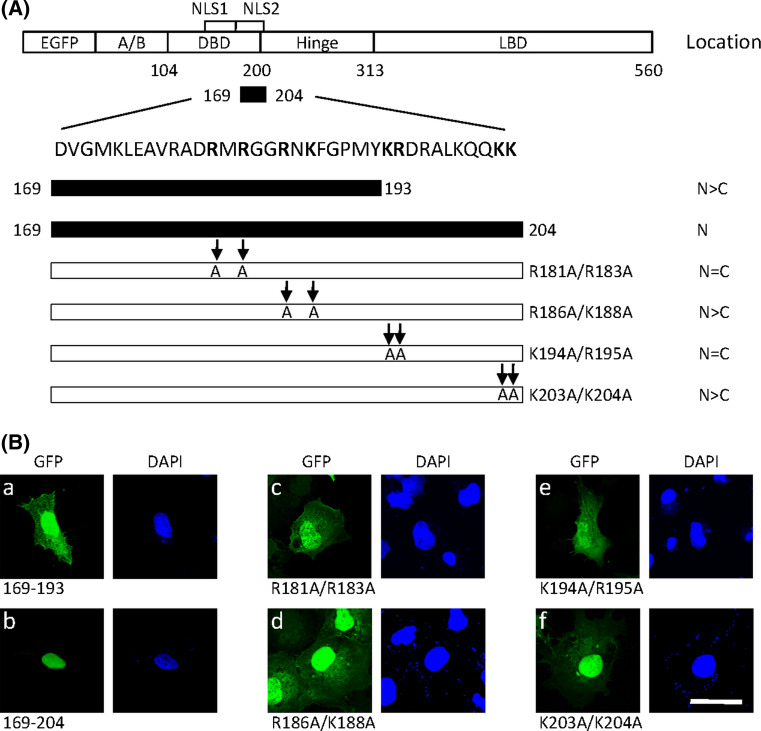
To further delimit the second NLS in the residues 169–226, two fragments 169–193 and 169–204 were separately fused to GFP protein (Fig. 3a). As shown in Fig. 3ba,b, fusion with fragment 169–204, but not fragment 169–193, resulted in complete transport of GFP into the nucleus. This suggests that residues 169–204 contain a functional NLS (referred to as NLS2). Four clusters of positively charged residues were also found in the NLS2 sequence. Mutation of R186A/K188A or K203A/K204A had little effect on nuclear localization of NLS2 (Fig. 3bd,f). However, the mutations R181A/R183A or K194A/R195A inhibited the nuclear localization of NLS2 (Fig. 3bc,e). These results indicate that basic amino acids in cluster 1 (R181 and R183) and cluster 3 (K194 and R195) are necessary for efficient nuclear import of NLS2.
Fig. 3.
Detection of key basic residues in NLS2 for nuclear import. a Sequence of the putative NLS2 is shown. Two segments of either residues 169–193 or residues 169–204 were fused to EGFP. The basic amino acids lysine and arginine (in boldface) in residues 169–204 were substituted by alanines as indicated by arrows. The subcellular localization of LRH-1 mutants is summarized on the right (N nuclear, C cytoplasmic). b Wild-type and mutant forms were transfected into COS-7 cells. After 24 h, cells were fixed and the nuclei counterstained with DAPI. Fusion proteins were visualized by confocal microscopy. Scale bar 25 μm
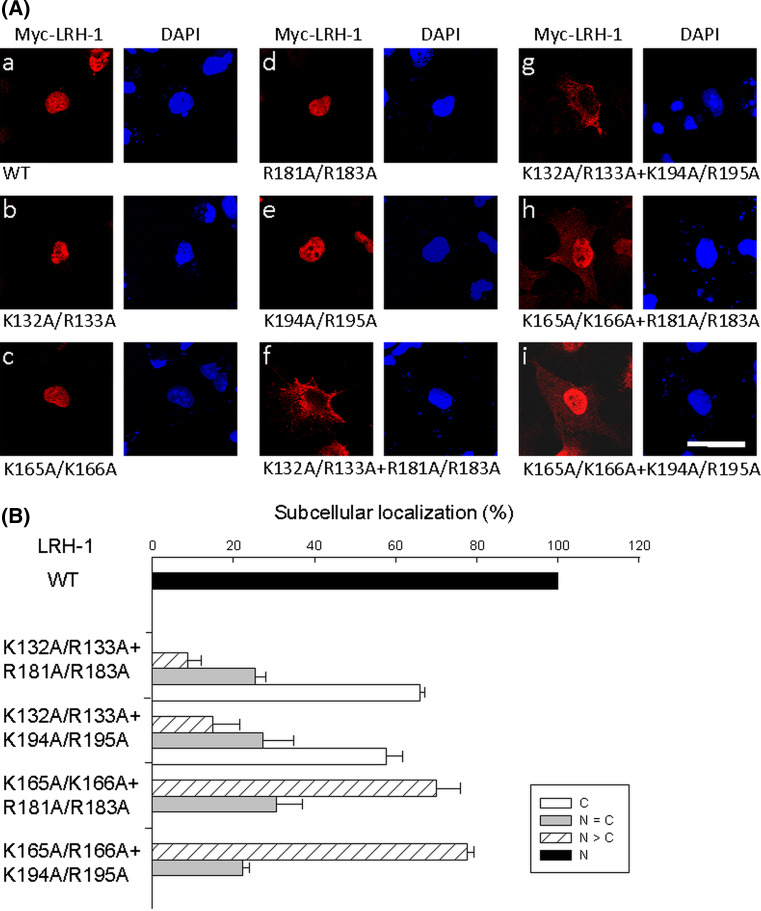
To determine the relative contribution of NLS1 and NLS2 to the nuclear import of LRH-1, alanine mutations were introduced into the full-length LRH-1 protein. As shown in Fig. 4ab–e, mutation of an individual basic cluster in NLS1 (K132A/R133A or K165A/K166A) or NLS2 (R181A/R183A or K194A/R195A) did not change the nuclear localization of LRH-1. In addition, the C143G mutation in the context of the whole protein also failed to affect the nuclear localization of LRH-1 (data not shown). However, the exclusive localization of LRH-1 in the nucleus was abolished by simultaneous mutation of basic clusters in both NLS1 and NLS2. The double mutations K132A/R133A + R181A/R183A and K132A/R133A + K194A/R195A significantly inhibited nuclear import, leading to approximately 60% of cells with cytoplasmic distribution of mutant LRH-1 (Fig. 4af,g). Although the distribution of the double mutants K165A/K166A + R181A/R183A and K165A/K166A + 194A/R195A was predominantly nuclear, minor amounts of the mutant proteins were also observed in the cytoplasm (Fig. 4ah,i). These results indicate that either NLS1 or NLS2 is sufficient to mediate nuclear import of LRH-1. In addition, these data confirm that residues K132/R133 from NLS1 and residues R181/R183 and K194/R195 from NLS2 are crucial for nuclear import.
Fig. 4.
Simultaneous disruption of NLS1 and NLS2 abolishes the nuclear localization of LRH-1. a Basic amino acids within either or both NLSs were mutated to alanines in the full-length LRH-1 tagged with Myc. These constructs were transfected into COS-7 cells. After 24 h, cells were fixed and immunostained with anti-Myc antibody. The nuclei were counterstained with DAPI. Fusion proteins were visualized by confocal microscopy. Wild-type (a) and single cluster mutants in NLS1 (b, c) or NLS2 (d, e) are exclusively localized to the nucleus. The subcellular localization of double cluster mutants in representative cells is shown in f–i. Scale bar 25 μm. b Statistics of subcellular distribution of wild-type (a in a) and double cluster mutants (f–i in a) are summarized. A minimum of 100 cells were counted for each experiment and the means ± SEM three independent experiments are presented
Nuclear import mechanisms of LRH-1 NLS
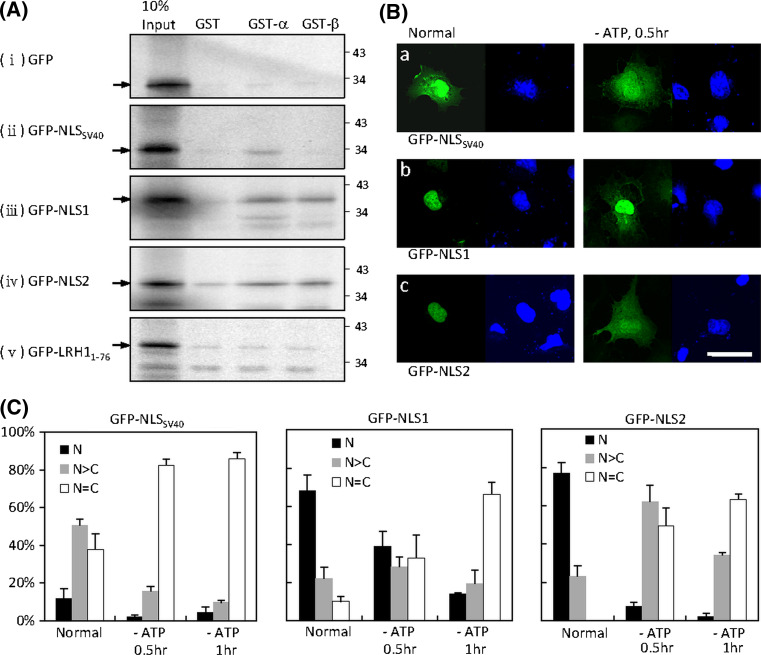
Classical NLSs are recognized by importin α. In addition, some NLSs are known to directly interact with importin β. GST pull-down assays were performed to determine the mechanisms of LRH-1 nuclear import. The [35S]-labelled GFP and GFP-NLS fusion proteins were translated in vitro and assayed for interaction with GST-importin α or GST-importin β, or with GST alone as a negative control. As expected, the classical NLS of simian virus 40 (SV40) interacted with importin α, but not with importin β (Fig. 5a). Both NLS1 and NLS2 exhibited interaction with importin α and importin β, whereas the N-terminal construct (1–76) and GFP alone did not show interaction in GST assays.
Fig. 5.
Nuclear import mechanisms of LRH-1 NLS. a Residues 117–168 (NLS1) or 169–204 (NLS2) of LRH-1 were fused to GFP tag. The classical NLS of SV40 fused to GFP was used as a positive control. The N-terminal residues 1–76 fused to GFP and GFP alone were used as negative controls. [35S]-labelled GFP or fusion proteins were synthesized in vitro and subjected to GST pull-down assays using GST, GST-importin α or GST-importin β. The molecular mass in kilodaltons is indicated on the right-hand side. b The GFP-NLS fusion constructs were transfected into COS-7 cells. After 24 h, cells were incubated in energy-depletion medium for 30 or 60 min, fixed and then counterstained with the nuclear dye DAPI. Fusion proteins were visualized by confocal microscopy. Scale bar 25 μm. c Statistics of subcellular distribution of GFP-NLS fusion proteins in b are summarized. A minimum of 100 cells were counted for each experiment and the means ± SEM of three independent experiments are presented
The nuclear accumulation of small molecules requires constitutive nuclear import, nuclear retention or both [36, 37]. Importin-mediated nuclear transport is an active process, so we then looked for changes the subcellular distribution of NLS-containing proteins in response to energy depletion. Cells expressing GFP-NLS were incubated with sodium azide and 2-deoxy-d-glucose in glucose-free medium to inhibit active nuclear import [36]. The GFP-NLSSV40 was predominantly nuclear in most cells. As expected, a redistribution of GFP-NLSSV40 protein throughout the nucleus and cytoplasm was observed after 30 min incubation in energy-depletion medium (Fig. 5b, c). However, the uniform distribution of GFP or GFP-LRH11–76 (37 kDa) in the nucleus and cytoplasm was not changed under these energy-depleting conditions (data not shown). The energy depletion treatment also led to the redistribution of GFP-NLS1 (34 kDa) and GFP-NLS2 (32 kDa) from the nucleus to the cytoplasm (Fig. 5b, c). This effect was more prominent for GFP-NLS2 protein, in which the exclusively nuclear localization of GFP-NLS2 was almost abolished within 30 min of treatment. These results suggest that NLS1 and NLS2-mediated nuclear import is an energy-dependent mechanism.
Effect of NLS mutation on DNA binding and transactivity of LRH-1
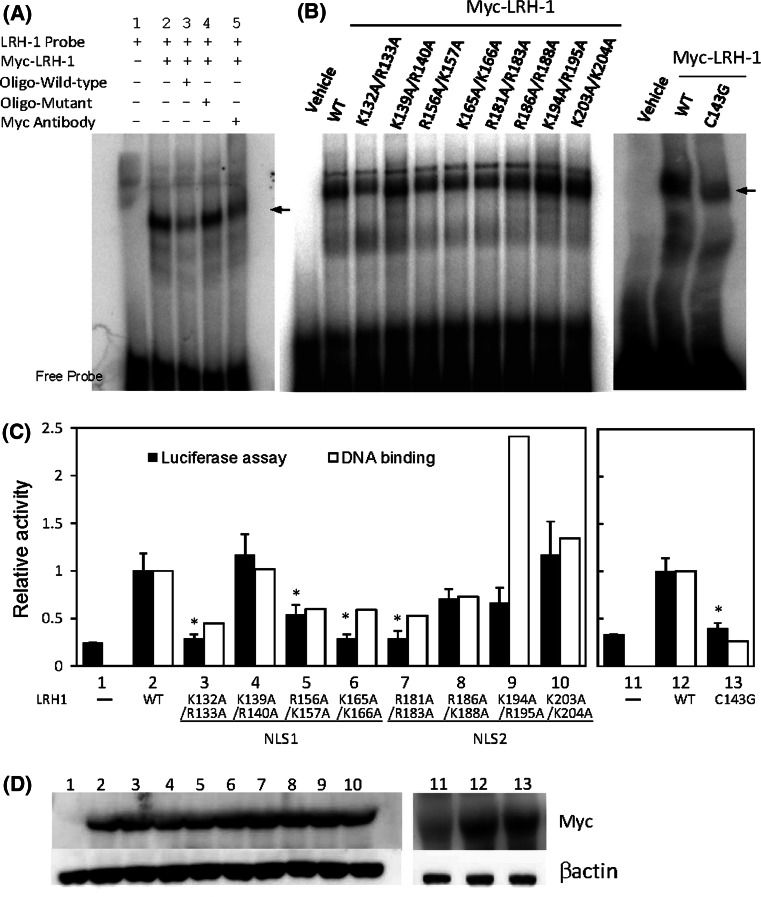
As both NLS1 and NLS2 are located in the DBD, we asked whether the positively charged residues within an NLS would affect the DNA binding and activity of LRH-1. A synthetic oligonucleotide containing a putative LRH-1 binding sequence upstream of the human CYP11A1 gene was used in an electrophoretic mobility shift assay (EMSA). A shifted complex was observed upon addition of in vitro-synthesized Myc-tagged LRH-1 (Fig. 6a, lane 2). The binding of protein to the probe was blocked by anti-Myc antibody (lane 5) and excess unlabelled wild-type (lane 3) but not mutated (lane 4) oligonucleotides, indicating specific binding to Myc-LRH-1. Five out of the eight alanine-substitution mutants, K132A/R133A, R156A/K157A, K165A/K166A, R181A/R183A and R186A/K188A, had reduced DNA binding activity compared to wild-type protein (Fig. 6b, c), whereas mutant K194A/R195A had an increased ability to bind DNA. The transactivity of mutant proteins was further examined by transfection into COS-7 cells with CYP11A1 promoter-linked luciferase. Western blot analysis demonstrated that mutant and wild-type proteins were expressed at similar levels (Fig. 6d). Mutants with reduced DNA binding activity showed lower levels of transcriptional activity, though the decrease with the mutant R186A/K188A was not statistically significant (Fig. 6c). An almost complete loss of transcriptional activity was found in mutants K132A/R133A, K165A/K166A and R181A/R183A, in which the mutated residues were those also required for nuclear import in NLS1 or NLS2. However, although residues K194/R195 were crucial for nuclear import in NLS2, they were not essential for DNA binding and transactivity of LRH-1. The zinc finger is a part of the DBD. Mutation of C143G with consequent defective structure of the second finger not only impaired the DNA binding but also the transcriptional activity of LRH-1 (Fig. 6b, c), indicating that the cysteine residue in the second zinc finger is crucial for DNA binding and functional activity of LRH-1.
Fig. 6.
DNA binding and transcriptional activity of wild-type and mutant LRH-1. a Myc-tagged LRH-1 was synthesized in vitro. EMSA was performed using a [32P]-labelled probe containing a LRH-1 binding site in the absence (lane 2) or presence of a 20-fold molar excess of unlabelled wild-type (lane 3) or mutant (lane 4) oligonucleotide or anti-Myc antibody (lane 5). Migration of DNA–protein complexes is indicated by the arrow. b Myc-tagged wild-type (WT) and mutated LRH-1 were synthesized in vitro and subjected to EMSA using a [32P]-labelled probe containing a LRH-1 binding site. Densitometric quantification of EMSA data are presented as the values relative to the wild-type in c (open boxes). c The CYP11A1 promoter–luciferase construct was cotransfected in COS-7 cells with an empty expression vector (−) or expression vectors for wild-type (WT) or mutated LRH-1 as indicated. Cells were lysed and assayed for luciferase activity after transfection for 24 h. The relative luciferase activity was compared to that with the wild-type construct. The data presented are the means ± SEM (closed boxes) of six independent experiments; *p < 0.05 compared to wild-type. d Expression of Myc-LRH-1 proteins in COS-7 cells was confirmed by immunoblotting with an anti-Myc antibody
Discussion
LRH-1 is an orphan nuclear receptor that is constitutively expressed in the nucleus as a transcriptional activator [7, 13]. In this study, we identified two NLS regions in residues 117–168 (NLS1) and 169–204 (NLS2) that can actively target LRH-1 to the nucleus. The GFP fusion experiments demonstrated that either NLS alone was sufficient to transport heterologous protein to the nucleus (Figs. 2 and 3). Disruption of either NLS1 or NLS2 did not affect the accumulation of LRH-1 in the nucleus, whereas simultaneous mutations of NLS1 and NLS2 abolished the nuclear localization of full-length LRH-1 (Fig 4). Our results indicate that NLS1 and NLS2 independently mediate the nuclear translocation of LRH-1 and the presence of either NLS is sufficient to target LRH-1 to the nucleus.
Like other nuclear receptors, the NLSs of LRH-1 are located in the DBD and the adjacent hinge region. However, two functional NLSs which are closely associated in this region have not been reported previously. NLS1 maps to a 52 amino acid fragment that contains the second zinc finger loop (Fig. 7b). Unlike the entire fragment, three short segments, residues 126–142, 151–169 and 137–160, were not alone sufficient to efficiently target the GFP protein to the nucleus (Fig. 2). Mutation of two basic clusters K132A/R133A or K165A/K166A significantly impaired nuclear targeting, suggesting that these basic amino acids are crucial for the nuclear import of NLS1. The DBD, comprising two zinc fingers, is the most conserved domain of the nuclear receptor family. Comparison of the amino acid sequences of the zinc finger region reveals that these two basic domains essential for NLS1 function are highly conserved between species and in several nuclear receptor family members (Fig. 7a), indicating the functional importance of the sequence. The two basic clusters in NLS1 are separated by a 31-residue spacer that covers the second zinc finger loop. When the zinc finger tertiary structure was changed by mutating the first zinc-chelating cysteine, the mutant NLS1 could not direct GFP protein to the nucleus, suggesting that the zinc finger structure is necessary for nuclear import. Similar results have also been obtained for other zinc finger proteins, where mutations that disrupt the zinc finger structure abolish the nuclear localization of NGFI-A [38]. NLS1 does not resemble the classical bipartite sequence with a 10–12 amino acid spacer between two basic clusters [17, 39]. However, the two basic clusters could be brought into close proximity once the zinc finger module has folded to form a compact loop structure (Fig. 7b). Therefore, it is possible that the spacer region is much narrower in this arrangement and the two basic domains can then function as a bipartite signal. Taken together, our data suggest that the function of NLS1 is dependent on both the presence of basic residues and the zinc finger structure.
The second NLS of LRH-1 is located near the C-terminus of the zinc finger motif. This is a highly basic region that is unique to the NR5A subfamily and is designated the Ftz-F1 (fushi tarazu factor 1) box [12]. Sequence alignment shows that LRH-1 and SF-1 share extensive homology within this region and are only about 10–30% identical to other steroid receptors (Fig. 7a). Mutation of R181A/R183A or K194A/R195A abolished the nuclear localization, indicating that these basic amino acids are essential for the nuclear transport of NLS2. The NLS2 region is characterized by two clusters of basic amino acids separated by a ten-residue spacer (RMRGGRNKFGPMYKR) that is similar to the typical bipartite NLS sequence. Li et al. [40] have also demonstrated that the conserved sequence in this domain of SF-1 is capable of targeting GFP protein to the nucleus.
Single mutations of the crucial basic cluster in NLS1 or NLS2 did not affect the nuclear localization of full-length LRH-1. However, the double mutations K132A/R133A in NLS1 and R181A/R183A or K194A/R195A in NLS2 led to the cytoplasmic accumulation of LRH-1 (Fig. 4). In addition, the amino acid fragments 1–116 and 226–560 failed to direct GFP to the nucleus. Taken together, these results demonstrate that no additional NLS exists in the N-terminal A/B region or C-terminal LBD, and that the presence of one NLS, either NLS1 or NLS2, is sufficient for the nuclear import of LRH-1. Hormone-activated nuclear receptors such as GR and AR usually contain a ligand-dependent NLS in the LBD [22–24]. It is assumed that the conformational changes in LBD induced by ligand binding make the nuclear localization domain accessible to enable nuclear transport [22]. However, we did not detect significant NLS activity in the corresponding regions of LRH-1. The absence of a NLS in the LBD has also been reported for another orphan receptor Rev-erb [41]. Structural and biochemical analysis showed that the activation of mouse LRH-1 is ligand-independent, whereas the binding of phosphatidyl inositol can regulate the activity of human LRH-1 and mouse and human SF-1 [13, 42]. Therefore, whether a masked NLS is present in the LBD of LRH-1 requires further investigation.
In a classical nuclear import process, the NLS of cargo proteins is recognized by importin α to form a trimeric complex with importin β. The importin β then mediates translocation of the complex into the nucleus in an energy-dependent manner [18]. However, the adaptor importin α may not be required for certain proteins whose NLS can directly interact with importin β [19, 20]. Our data showed that both NLS1 and NLS2 of LRH-1 could directly interact with importin α and importin β in vitro (Fig. 5a), suggesting that LRH-1 is able to utilize the importin α/β mechanism for its nuclear transport. Recently, a number of proteins have been found to enter the nucleus utilizing the nonclassical pathway through an interaction with importin β [19, 20]. Both NLSs of LRH-1 can bind to both types of importins, suggesting that multiple pathways may be involved to ensure the translocation of LRH-1 into the nucleus to execute its nuclear activities. The nuclear accumulation of small molecules requires constitutively active import, nuclear retention or both [36, 37]. When active import was prevented by incubation in energy-depletion medium, GFP-NLS1 and GFP-NLS2 were redistributed to the cytoplasm (Fig. 5), indicating that the nuclear import mediated by NLS1 or NLS2 is an energy-dependent pathway. The relocalization of GFP-NLS2 was promptly observed, whereas the majority of GFP-NLS1 was retained in the nucleus within 30 min of incubation, implying that NLS1 may interact with other components of the nucleus. That an NLS sequence is able to mediate nuclear retention has been demonstrated for other proteins. For instance, the NLS of bovine herpesvirus 1 tegument protein VP22 can bind to histones to promote its nuclear accumulation [43]. The NLSs of many transcription factors are located within the DNA-binding domain that is considered to be a potential nuclear retention signal [21]. Although NLS1 contains only one zinc-finger loop, it may have partial DNA-binding capability to enhance the nuclear accumulation of NLS1-attached cargoes.
The transcriptional activity of transcription factors is modulated by their subcellular localization. The presence of two NLSs allows a potential mechanism whereby the nuclear transport of LRH-1 could be differentially regulated. LRH-1 plays a crucial role in embryonic development. Mice deficient in LRH-1 die at early embryonic stages [3]. LRH-1 regulates the expression of Oct4 to maintain pluripotency in embryonic stem cells and early mouse embryos [44]. Furthermore, LRH-1 can replace Oct4 in the reversion of mouse somatic cells to pluripotent cells [45]. This reprogramming capacity of LRH-1 can be enhanced by mutations in SUMO-conjugated lysine residues, which prevents the localization of LRH-1 to transcriptionally inactive nuclear bodies and increases its transactivity [14]. The present study revealed the nuclear transport mechanism of LRH-1 and implied that mutation in residues essential for NLS function could inhibit the nuclear localization of LRH-1 and consequently may influence embryonic development.
In summary, we identified two structurally different types of NLSs that can independently mediate the nuclear localization of LRH-1. Two basic clusters are required for full nuclear import in NLS1 and NLS2, respectively. The NLS1 contains a zinc finger domain whose tertiary structure is important for the function of NLS1. Both NLS1 and NLS2 mediate the nuclear transport process by an importin α/β mechanism. Three crucial basic clusters in the NLSs are involved in the DNA binding and transcriptional activities of LRH-1. Nuclear localization is important for normal function of LRH-1, and this study provides an insight into the mechanisms of LRH-1 nuclear import.
Acknowledgments
We are grateful to Dr. Y. Yoneda for providing the GST-importin plasmids. This work was supported by NSC96-2320-B-002-006 from the National Science Council, Taiwan.
References
- 1.Boerboom D, Pilon N, Behdjani R, Silversides DW, Sirois J. Expression and regulation of transcripts encoding two members of the NR5A nuclear receptor subfamily of orphan nuclear receptors, steroidogenic factor-1 and NR5A2, in equine ovarian cells during the ovulatory process. Endocrinology. 2000;141:4647–4656. doi: 10.1210/en.141.12.4647. [DOI] [PubMed] [Google Scholar]
- 2.Fayard E, Auwerx J, Schoonjans K. LRH-1: an orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol. 2004;14:250–260. doi: 10.1016/j.tcb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Pare JF, Malenfant D, Courtemanche C, Jacob-Wagner M, Roy S, Allard D, Belanger L. The fetoprotein transcription factor (FTF) gene is essential to embryogenesis and cholesterol homeostasis and is regulated by a DR4 element. J Biol Chem. 2004;279:21206–21216. doi: 10.1074/jbc.M401523200. [DOI] [PubMed] [Google Scholar]
- 4.Luo X, Ikeda Y, Parker KL. A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell. 1994;77:481–490. doi: 10.1016/0092-8674(94)90211-9. [DOI] [PubMed] [Google Scholar]
- 5.Morohashi KI, Omura T. Ad4BP/SF-1, a transcription factor essential for the transcription of steroidogenic cytochrome P450 genes and for the establishment of the reproductive function. FASEB J. 1996;10:1569–1577. doi: 10.1096/fasebj.10.14.9002548. [DOI] [PubMed] [Google Scholar]
- 6.Shinoda K, Lei H, Yoshii H, Nomura M, Nagano M, Shiba H, Sasaki H, Osawa Y, Ninomiya Y, Niwa O. Developmental defects of the ventromedial hypothalamic nucleus and pituitary gonadotroph in the Ftz-F1 disrupted mice. Dev Dyn. 1995;204:22–29. doi: 10.1002/aja.1002040104. [DOI] [PubMed] [Google Scholar]
- 7.Galarneau L, Pare JF, Allard D, Hamel D, Levesque L, Tugwood JD, Green S, Belanger L. The alpha1-fetoprotein locus is activated by a nuclear receptor of the Drosophila FTZ-F1 family. Mol Cell Biol. 1996;16:3853–3865. doi: 10.1128/mcb.16.7.3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo Y, Liang CP, Tall AR. The orphan nuclear receptor LRH-1 potentiates the sterol-mediated induction of the human CETP gene by liver X receptor. J Biol Chem. 2001;276:24767–24773. doi: 10.1074/jbc.M100912200. [DOI] [PubMed] [Google Scholar]
- 9.Nitta M, Ku S, Brown C, Okamoto AY, Shan B. CPF: an orphan nuclear receptor that regulates liver-specific expression of the human cholesterol 7alpha-hydroxylase gene. Proc Natl Acad Sci U S A. 1999;96:6660–6665. doi: 10.1073/pnas.96.12.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clyne CD, Speed CJ, Zhou J, Simpson ER. Liver receptor homologue-1 (LRH-1) regulates expression of aromatase in preadipocytes. J Biol Chem. 2002;277:20591–20597. doi: 10.1074/jbc.M201117200. [DOI] [PubMed] [Google Scholar]
- 11.Botrugno OA, Fayard E, Annicotte JS, Haby C, Brennan T, Wendling O, Tanaka T, Kodama T, Thomas W, Auwerx J, Schoonjans K. Synergy between LRH-1 and beta-catenin induces G1 cyclin-mediated cell proliferation. Mol Cell. 2004;15:499–509. doi: 10.1016/j.molcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 12.Ueda H, Sun GC, Murata T, Hirose S. A novel DNA-binding motif abuts the zinc finger domain of insect nuclear hormone receptor FTZ-F1 and mouse embryonal long terminal repeat-binding protein. Mol Cell Biol. 1992;12:5667–5672. doi: 10.1128/mcb.12.12.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sablin EP, Krylova IN, Fletterick RJ, Ingraham HA. Structural basis for ligand-independent activation of the orphan nuclear receptor LRH-1. Mol Cell. 2003;11:1575–1585. doi: 10.1016/S1097-2765(03)00236-3. [DOI] [PubMed] [Google Scholar]
- 14.Yang FM, Pan CT, Tsai HM, Chiu TW, Wu ML, Hu MC. Liver receptor homolog-1 localization in the nuclear body is regulated by sumoylation and cAMP signaling in rat granulosa cells. FEBS J. 2009;276:425–436. doi: 10.1111/j.1742-4658.2008.06785.x. [DOI] [PubMed] [Google Scholar]
- 15.Sorokin AV, Kim ER, Ovchinnikov LP. Nucleocytoplasmic transport of proteins. Biochemistry (Mosc) 2007;72:1439–1457. doi: 10.1134/S0006297907130032. [DOI] [PubMed] [Google Scholar]
- 16.Kalderon D, Richardson WD, Markham AF, Smith AE. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature. 1984;311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- 17.Robbins J, Dilworth SM, Laskey RA, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-T. [DOI] [PubMed] [Google Scholar]
- 18.Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmeri D, Malim MH. Importin beta can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin alpha. Mol Cell Biol. 1999;19:1218–1225. doi: 10.1128/mcb.19.2.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Philips AS, Kwok JC, Chong BH. Analysis of the signals and mechanisms mediating nuclear trafficking of GATA-4. Loss of DNA binding is associated with localization in intranuclear speckles. J Biol Chem. 2007;282:24915–24927. doi: 10.1074/jbc.M701789200. [DOI] [PubMed] [Google Scholar]
- 21.LaCasse EC, Lefebvre YA. Nuclear localization signals overlap DNA- or RNA-binding domains in nucleic acid-binding proteins. Nucleic Acids Res. 1995;23:1647–1656. doi: 10.1093/nar/23.10.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Picard D, Yamamoto KR. Two signals mediate hormone-dependent nuclear localization of the glucocorticoid receptor. EMBO J. 1987;6:3333–3340. doi: 10.1002/j.1460-2075.1987.tb02654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Savory JG, Hsu B, Laquian IR, Giffin W, Reich T, Hache RJ, Lefebvre YA. Discrimination between NL1- and NL2-mediated nuclear localization of the glucocorticoid receptor. Mol Cell Biol. 1999;19:1025–1037. doi: 10.1128/mcb.19.2.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poukka H, Karvonen U, Yoshikawa N, Tanaka H, Palvimo JJ, Janne OA. The RING finger protein SNURF modulates nuclear trafficking of the androgen receptor. J Cell Sci. 2000;113(Pt 17):2991–3001. doi: 10.1242/jcs.113.17.2991. [DOI] [PubMed] [Google Scholar]
- 25.Zhou ZX, Sar M, Simental JA, Lane MV, Wilson EM. A ligand-dependent bipartite nuclear targeting signal in the human androgen receptor. Requirement for the DNA-binding domain and modulation by NH2-terminal and carboxyl-terminal sequences. J Biol Chem. 1994;269:13115–13123. [PubMed] [Google Scholar]
- 26.Ylikomi T, Bocquel MT, Berry M, Gronemeyer H, Chambon P. Cooperation of proto-signals for nuclear accumulation of estrogen and progesterone receptors. EMBO J. 1992;11:3681–3694. doi: 10.1002/j.1460-2075.1992.tb05453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freedman ND, Yamamoto KR. Importin 7 and importin alpha/importin beta are nuclear import receptors for the glucocorticoid receptor. Mol Biol Cell. 2004;15:2276–2286. doi: 10.1091/mbc.E03-11-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaku N, Matsuda K, Tsujimura A, Kawata M. Characterization of nuclear import of the domain-specific androgen receptor in association with the importin alpha/beta and Ran-guanosine 5′-triphosphate systems. Endocrinology. 2008;149:3960–3969. doi: 10.1210/en.2008-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fisher CL, Pei GK. Modification of a PCR-based site-directed mutagenesis method. Biotechniques. 1997;23:570–574. doi: 10.2144/97234bm01. [DOI] [PubMed] [Google Scholar]
- 30.Higuchi R, Krummel B, Saiki RK. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imamoto N, Shimamoto T, Takao T, Tachibana T, Kose S, Matsubae M, Sekimoto T, Shimonishi Y, Yoneda Y. In vivo evidence for involvement of a 58 kDa component of nuclear pore-targeting complex in nuclear protein import. EMBO J. 1995;14:3617–3626. doi: 10.1002/j.1460-2075.1995.tb00031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kose S, Imamoto N, Tachibana T, Shimamoto T, Yoneda Y. Ran-unassisted nuclear migration of a 97-kD component of nuclear pore-targeting complex. J Cell Biol. 1997;139:841–849. doi: 10.1083/jcb.139.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsieh HT, Wang CH, Wu ML, Yang FM, Tai YC, Hu MC. PIASy inhibits LRH-1-dependent CYP11A1 expression by competing for SRC-1 binding. Biochem J. 2009;419:201–209. doi: 10.1042/BJ20081402. [DOI] [PubMed] [Google Scholar]
- 34.Hu MC, Hsu NC, Pai CI, Wang CK, Chung B. Functions of the upstream and proximal steroidogenic factor 1 (SF-1)-binding sites in the CYP11A1 promoter in basal transcription and hormonal response. Mol Endocrinol. 2001;15:812–818. doi: 10.1210/me.15.5.812. [DOI] [PubMed] [Google Scholar]
- 35.Wei X, Henke VG, Strubing C, Brown EB, Clapham DE. Real-time imaging of nuclear permeation by EGFP in single intact cells. Biophys J. 2003;84:1317–1327. doi: 10.1016/S0006-3495(03)74947-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cardarelli F, Serresi M, Bizzarri R, Giacca M, Beltram F. In vivo study of HIV-1 Tat arginine-rich motif unveils its transport properties. Mol Ther. 2007;15:1313–1322. doi: 10.1038/sj.mt.6300172. [DOI] [PubMed] [Google Scholar]
- 37.Stochaj U, Rassadi R, Chiu J. Stress-mediated inhibition of the classical nuclear protein import pathway and nuclear accumulation of the small GTPase Gsp1p. FASEB J. 2000;14:2130–2132. doi: 10.1096/fj.99-0751fje. [DOI] [PubMed] [Google Scholar]
- 38.Matheny C, Day ML, Milbrandt J. The nuclear localization signal of NGFI-A is located within the zinc finger DNA binding domain. J Biol Chem. 1994;269:8176–8181. [PubMed] [Google Scholar]
- 39.Jans DA, Xiao CY, Lam MH. Nuclear targeting signal recognition: a key control point in nuclear transport? Bioessays. 2000;22:532–544. doi: 10.1002/(SICI)1521-1878(200006)22:6<532::AID-BIES6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 40.Li LA, Chiang EF, Chen JC, Hsu NC, Chen YJ, Chung BC. Function of steroidogenic factor 1 domains in nuclear localization, transactivation, and interaction with transcription factor TFIIB and c-Jun. Mol Endocrinol. 1999;13:1588–1598. doi: 10.1210/me.13.9.1588. [DOI] [PubMed] [Google Scholar]
- 41.Chopin-Delannoy S, Thenot S, Delaunay F, Buisine E, Begue A, Duterque-Coquillaud M, Laudet V. A specific and unusual nuclear localization signal in the DNA binding domain of the Rev-erb orphan receptors. J Mol Endocrinol. 2003;30:197–211. doi: 10.1677/jme.0.0300197. [DOI] [PubMed] [Google Scholar]
- 42.Krylova IN, Sablin EP, Moore J, Xu RX, Waitt GM, MacKay JA, Juzumiene D, Bynum JM, Madauss K, Montana V, Lebedeva L, Suzawa M, Williams JD, Williams SP, Guy RK, Thornton JW, Fletterick RJ, Willson TM, Ingraham HA. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell. 2005;120:343–355. doi: 10.1016/j.cell.2005.01.024. [DOI] [PubMed] [Google Scholar]
- 43.Zhu J, Qiu Z, Wiese C, Ishii Y, Friedrichsen J, Rajashekara G, Splitter GA. Nuclear and mitochondrial localization signals overlap within bovine herpesvirus 1 tegument protein VP22. J Biol Chem. 2005;280:16038–16044. doi: 10.1074/jbc.M500054200. [DOI] [PubMed] [Google Scholar]
- 44.Gu P, Goodwin B, Chung AC, Xu X, Wheeler DA, Price RR, Galardi C, Peng L, Latour AM, Koller BH, Gossen J, Kliewer SA, Cooney AJ. Orphan nuclear receptor LRH-1 is required to maintain Oct4 expression at the epiblast stage of embryonic development. Mol Cell Biol. 2005;25:3492–3505. doi: 10.1128/MCB.25.9.3492-3505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heng JC, Feng B, Han J, Jiang J, Kraus P, Ng JH, Orlov YL, Huss M, Yang L, Lufkin T, Lufkin T, Lim B, Ng HH. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell. 2010;6:167–174. doi: 10.1016/j.stem.2009.12.009. [DOI] [PubMed] [Google Scholar]