Abstract
The RAF family of kinases are key components acting downstream of receptor tyrosine kinases and cells employ several distinct mechanisms to strictly control their activity. RAF transitions from an inactive state, where the N-terminal regulatory region binds intramolecularly to the C-terminal kinase domain, to an open state capable of executing the phosphoryl transfer reaction. This transition involves changes both within and between the protein domains in RAF. Many different proteins regulate the transition between inactive and active states of RAF, including RAS and KSR, which are arguably the two most prominent regulators of RAF function. Recent developments have added several new twists to our understanding of RAF regulation. Among others, dimerization of the RAF kinase domain is emerging as a crucial step in the RAF activation process. The multitude of regulatory protein–protein interactions involving RAF remains a largely untapped area for therapeutic applications.
Keywords: RAF, KSR, Kinase, Scaffold, Protein–protein interaction, Allosteric regulation
Introduction
In metazoans, the archetypical signal transduction pathway is the RTK–RAS–MAPK module (RTK, receptor tyrosine kinase; MAPK, mitogen-activated protein kinase), and it has garnered much attention due to its deregulation in many human cancers. Upon activation, RTKs recruit the guanine nucleotide exchange factor (GEF) SOS via the adaptor proteins Shc and Grb2, and SOS then converts plasma membrane localized RAS from its inactive, GDP-bound state to its active, GTP-bound state. Activated RAS in turn transmits the signal to a three-tiered kinase cascade consisting of RAF, MEK, and ERK/MAPK. The most complex step during signal transduction through the RTK–RAS–MAPK pathway is the activation of RAF, and this involves several events including membrane recruitment, phosphorylation, and protein oligomerization. A critical regulator of RTK- and RAS-dependent RAF activation is KSR (kinase suppressor of RAS).
Since the discovery of the RAF proteins in the mid-1980s and KSR about a decade later, the interconnections between them have become ever more intricate. Indeed, they are structurally related and likely evolved from a common ancestral gene. Recent advances in our knowledge of RAF and KSR have indicated that KSR is not only structurally similar to and acts as a scaffold for RAF signaling, but that KSR plays a more operative role in the RAF activation process.
The founding member of the RAF family is C-RAF/RAF-1 (hereafter referred to as C-RAF), which was identified as the human homolog of a viral oncoprotein [1]. Subsequently, two C-RAF paralogs were identified in vertebrates, namely A-RAF and B-RAF [2–4]. KSR was originally identified in the fruitfly Drosophila melanogaster and the nematode Caenorhabditis elegans as a protein required for signaling downstream of activated RAS [5–7]. In vertebrates, there are two KSR proteins, KSR1 and KSR2 [7, 8].
In this review, we focus on recent developments concerning the RAF and KSR proteins that have come to light over the past few years. For a broader perspective, the reader is directed to several excellent reviews [9–13]. In particular, we will highlight advances made in our understanding of the requirements for RTK-dependent RAF activation, including membrane recruitment and relief of autoinhibition, the role of scaffold proteins in RAF activation, phosphorylation and dephosphorylation events, as well as allosteric regulation. Although we will discuss these issues separately, in many ways they are interdependent. We will also briefly discuss kinase-independent functions of C-RAF. A common theme throughout is the role of protein–protein interactions in regulating signal transduction.
RAF and KSR proteins
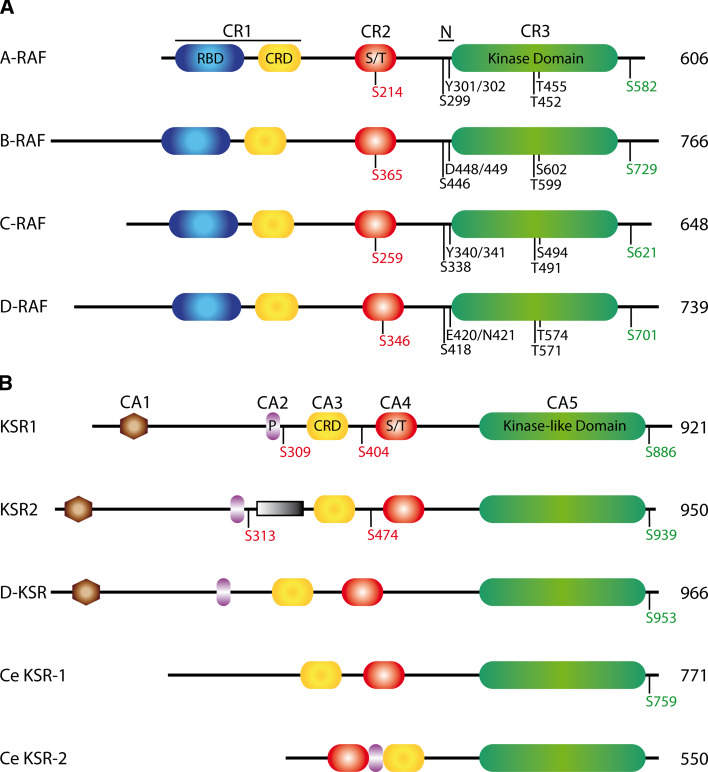
The domain organization of the RAF proteins is shown in Fig. 1a. All RAF proteins contain three conserved regions (CR1–CR3), with Drosophila RAF being most closely related to BRAF at the amino acid level. CR1 contains the RAS-binding domain (RBD) that engages the effector loop of GTP-bound RAS, in addition to a cysteine-rich domain (CRD) that is involved in membrane association and also contributes to RAS binding [14–16]. BRAF and its Drosophila ortholog also contain an extended N-terminus which is believed to enhance the interaction with RAS [17, 18], and has also been reported to mediate calcium-dependent RAF dimerization [19]. CR2 is a Ser/Thr-rich region containing a conserved Ser residue (Ser259 in C-RAF) that has an important regulatory function. Phosphorylation of Ser259 creates a binding site for 14-3-3 proteins, which stabilizes the inactive state of RAF and prevents plasma membrane recruitment [20–24]. Together, CR1 and CR2 comprise a regulatory region that inhibits kinase activity via an intramolecular interaction with the kinase domain of CR3 [20, 25, 26]. The interaction between the regulatory region and kinase domain is crucial for preventing inappropriate activation of RAF, and is therefore exquisitely regulated. Immediately N-terminal to CR3 is the negative charge region (N-region), which is thought to modulate the conformation of the kinase domain and the interaction with the regulatory region [9, 12, 26, 27]. A second 14-3-3 binding site (centered on Ser621 of C-RAF) of considerable importance is located C-terminal to the kinase domain.
Fig. 1.
The domain organization of RAF (a) and KSR (b) proteins. Conserved regions (CR) and conserved areas (CA) are indicated at the top of each panel, as is the negative charge region (N) in the RAF proteins. Conserved phosphorylation sites are indicated underneath each isoform. Phosphorylation sites known or suspected of mediating binding to 14-3-3 proteins are colored red (negative regulation) or green (positive regulation). The size of each isoform is noted at the right. D-RAF and D-KSR are the Drosophila paralogs and Ce denotes C. elegans proteins. All other proteins depicted are from human. S309 and S404 in human KSR1 are the equivalent of S297 and S392 in mouse KSR1 (see text). The unique region of KSR2 is represented as a shaded box. CRD Cysteine-rich domain, P proline-rich, RBD RAS-binding domain, S/T serine/threonine-rich
The domain organization of the KSR proteins is shown in Fig. 1b. Vertebrate and fly KSR proteins share five conserved areas called CA1–CA5. The CA1 domain in Drosophila and mammals has been shown to participate in the formation of a ternary KSR–MEK–RAF complex that facilitates MEK phosphorylation [28, 29]. The C. elegans proteins lack the CA1 domain, and so it is not required for their function or they have evolved a means to compensate for its absence. The CA2 domain is a proline-rich region of unknown function. The CA3 domain is similar to the CRD of RAF and is required for membrane recruitment of KSR1 [30]. However, it does not bind RAS and it cannot substitute for the CRD of RAF [15, 30]. CA4 is a Ser/Thr-rich region similar to CR2 of RAF. CA4 contains an FXFP motif that constitutes a stimulus-dependent ERK docking site [28, 31–33]. CA5 is highly homologous to the kinase domain (CR3) of RAF proteins. However, KSR1 and KSR2 lack the invariant lysine residue in subdomain II of the kinase domain that is necessary for catalytic activity. Interestingly, C. elegans KSR-1 and Drosophila KSR contain the invariant lysine in subdomain II, which would suggest that they are catalytically competent. While this remains to be shown, there is compelling evidence to suggest that KSR proteins do not require catalytic activity for their function. Mutation of the invariant lysine residue in Drosophila KSR does not compromise its function [29], and in C. elegans, ksr-1 transgenes with mutations predicted to nullify catalytic activity are able to complement loss of function alleles [34]. The lack of demonstrable kinase activity has led some to refer to CA5 as a kinase-like or pseudo-kinase domain. The CA5 domain binds constitutively to MEK, and to RAF following stimulation [29, 32, 33, 35, 36]. A study of KSR1 knockout mice indicates that mammalian KSR1 plays an important role in growth factor- and RAS-dependent RAF activation [37, 38]. In addition to, apparently, regulating growth factor- and RAS-dependent RAF activation, KSR2 has several functions that are distinct from KSR1 [39, 40]. KSR2 contains a region of 63 amino acids between CA2 and CA3 that is not present in the other KSR proteins, and this mediates KSR2-specific binding to calcineurin and AMPK [39, 40]. The calcineurin interaction allows KSR2 to activate ERK signaling following an increase in intracellular calcium [40]. KSR2 regulates cellular energy metabolism in part through its interaction with AMPK, and this contributes to the obese phenotype of KSR2 knockout mice [39, 41].
RAS-dependent RAF activation
In the inactive state, RAF exists in the cytosol in a closed conformation with the regulatory region bound to the kinase domain, and this state is stabilized by the binding of 14-3-3 proteins [20, 22, 25, 26, 42, 43]. The first step in the RAF activation process is plasma membrane recruitment by RAS-GTP [21, 44–47]. RAS-GTP transitions RAF from a closed to an open state by relieving the interaction between the regulatory region and kinase domain [20, 26, 42]. Membrane recruitment is associated with dephosphorylation of Ser259, dissociation of 14-3-3 from CR2, and stabilization of the open conformation [21, 23, 42, 48, 49]. Membrane recruitment ultimately brings RAF into the vicinity of other proteins that will complete the activation process, which includes phosphorylation events that stabilize the open, active conformation of RAF.
A recent development in RAS-dependent activation of RAF concerns RAS signaling from plasma membrane microdomains. Upon activation, the mobility of RAS-GTP within the inner leaflet of the membrane is observed to decrease [50]. This decreased mobility results from sequestration of active RAS in small, densely packed nanoclusters [50–57]. Although not fully understood, the formation of these nanoclusters depends on scaffold proteins of the galectin family and possibly also Shoc2/SUR-8 (hereafter referred to as SUR-8) [50, 55, 57–59]. By manipulating RAS nanocluster formation, it has been shown that RAF plasma membrane recruitment and activation occurs almost exclusively in nanoclusters, and the amount of nanocluster formation correlates with the level of ERK activation [50, 53, 55, 56, 58–60]. The different RAS isoforms (H-, K-, and N-RAS) segregate into distinct nanoclusters due to their having different C-terminal membrane anchors [52, 53, 55, 57]. These isoform-specific nanoclusters have different lipid and protein contents, which is likely responsible for generating some or all of the signaling diversity of the RAS isoforms. Artificial tethering of H-RAS to different membrane microdomains indicates that RAS membrane localization can regulate ERK substrate specificity [61].
Several models have been proposed to explain the ability of nanoclusters to facilitate signaling. SOS has an allosteric binding site for RAS-GTP, creating a positive feedback loop between RAS and SOS, and the high concentration of RAS-GTP in nanoclusters may allow SOS to remain bound at the plasma membrane even after the stimulus is removed [62, 63]. RAS nanoclusters have been proposed to function as a low threshold switch, generating the same amount of activated ERK over a wide range of RAF activity [60, 64, 65]. RAS nanoclusters could accumulate a high local concentration of RAF and KSR–MEK–ERK, and protect MEK and ERK from negatively acting cytosolic phosphatases, thus enhancing pathway activation [60, 64]. One interesting consequence of this switch-like behavior noted by Tian et al. [60] is that RAF activation need not be maximal for productive signaling to occur. They suggest that the complex mechanisms involved in RAF activation instead generate levels of redundancy that ensures that at least some of the individual RAF molecules are activated to a sufficient extent [60]. Of course, the downside to this is that a relatively minor perturbation in any one of the mechanisms that regulate RAF activity could lead to inappropriate pathway activation, such as can occur in cancer.
Several important questions remain regarding RAS-dependent RAF activation. It is not known if the different RAF paralogs have similar affinities for isoform-specific nanoclusters, although it has been reported that C-RAF is predominantly recruited to K-RAS nanoclusters following growth factor stimulation [53]. The extent to which RAS can activate RAF on intracellular membranes is also not known. K-RAS appears to localize to and signal exclusively from the plasma membrane [66–68], while active H-RAS and N-RAS have been observed on intracellular membranes, including the Golgi apparatus, endoplasmic reticulum, and endosomes [66, 68–71]. Currently, it has not been conclusively demonstrated that endogenous, active RAF is associated with active RAS on these membranes. Another outstanding question concerns how RAF travels to the membrane. Does this occur by passive diffusion or active transport? If active transport is required then uncoupling RAF from the transport machinery could be explored as an additional avenue for therapeutic intervention in cancer. What role does RAF dimerization play in membrane translocation? RAF inhibitors that induce dimerization also promote membrane translocation in a RAS-dependent manner [72], suggesting that RAF dimerization may precede and facilitate RAS binding.
Scaffold proteins involved in RTK-dependent RAF signaling
A common feature of MAPK cascades is the utilization of scaffold proteins to regulate signaling both spatially and temporally. Scaffold proteins facilitate signaling by coordinating the interaction of two or more components of the cascade, and they also prevent inappropriate pathway cross-talk by insulating MAPK cascades from each other. As we shall see, certain proteins can also inhibit signal transduction. A myriad of scaffold proteins are believed to regulate ERK activation under various circumstances, but we restrict our discussion to those scaffolds involved in regulating RTK-dependent RAF/MEK/ERK signaling.
KSR is the prototypical metazoan scaffold protein for the ERK/MAPK cascade, and the simplistic view of KSR is that of a scaffold protein that facilitates MAPK signaling by colocalizing RAF, MEK, and ERK. Although this is partially true, it is far from the whole story. KSR is constitutively bound to MEK, while binding to RAF and ERK occurs after stimulation [28, 32, 33, 35]. In resting cells, KSR1 is localized to the cytoplasm in a high molecular weight complex containing MEK, the kinase C-TAK1, the catalytic and structural subunits of PP2A, a dimer of 14-3-3 proteins bound to phosphorylated serine residues (Ser297 and Ser392 of mouse KSR1), and several chaperone proteins which are required for the stability of KSR1 [31, 34, 35, 38, 40]. C-TAK1 constitutively associates with mouse KSR1 and phosphorylates Ser392 [32]. Upon stimulation, a regulatory subunit of PP2A is recruited to the complex, allowing PP2A to dephosphorylate Ser392 and release 14-3-3 from this site [35]. The release of 14-3-3 from this site exposes the CA3 domain to mediate membrane recruitment, and may also reveal the ERK docking site in CA4 [32, 35]. Once 14-3-3 is released from Ser392, KSR1 is able to participate in RAF activation.
An additional scaffold/adaptor protein involved in RAF activation is CNK (connector enhancer of KSR). In Drosophila, CNK associates with KSR and the two cooperate in activating RAF [73–76]. The interaction between CNK and KSR depends on the adaptor protein HYP (Hyphen, also known as Aveugle), which binds to CNK via heterodimerization of their sterile alpha motif (SAM) domains [74, 75, 77]. SAM domain heterodimerization may create a composite binding site for KSR, or alternatively it could alter the conformation of CNK and reveal the KSR binding site [77]. CNK also binds to RAF and has opposing roles in RAF regulation [76, 78, 79]. In unstimulated cells, CNK maintains RAF in the inactive state through binding of RAF to the RAF-inhibitory region (RIR) in the C-terminus of CNK [78]. Upon stimulation, repression is relieved and CNK converts to a potent activator of RAF [78, 79]. Compared to Drosophila, relatively little is known about mammalian CNK proteins. CNK1 and CNK2 have been shown to regulate ERK activation in a concentration-dependent manner, which is characteristic of scaffold proteins [80, 81]. However, mammalian CNK proteins lack a RIR, and if they repress RAF in resting cells they might do so by a mechanism different than Drosophila CNK. CNK1 interacts with B- and C-RAF, and contributes to C-RAF activation by promoting Src-dependent phosphorylation of Tyr340 and Tyr341 [81]. CNK2 also interacts with B- and C-RAF, but the same report failed to detect an interaction between CNK2 and KSR1 or MEK [80]. Much work remains to be done to understand the role of mammalian CNK in RAF activation, and further studies may reveal additional similarities between mammalian and Drosophila CNK proteins.
SUR-8 is a leucine-rich repeat protein originally identified in C. elegans that acts downstream of RAS, but upstream of RAF in the RTK–RAS–MAPK pathway [82]. Several mechanisms have been proposed to account for the positive effect of SUR-8 on RAF activation. It has been reported that SUR-8 forms a ternary complex with RAS and RAF, and thus acts as a scaffold to facilitate RAS-dependent RAF activation [83, 84]. However, another group failed to detect an interaction between SUR-8 and C-RAF, or SUR-8 and H-, K-, and N-RAS [85]. Instead, these authors report that SUR-8 binds to M-RAS and recruits the catalytic subunit of the PP1 phosphatase, which then dephosphorylates inhibitory sites on RAF that is bound to other molecules of RAS [85]. As noted previously, SUR-8 may participate in the formation of RAS nanoclusters [50, 55], and a recent report indicating that SUR-8 increases the rate of RAS binding to RAF following EGF treatment is consistent with this [86]. Using a synthetic and inducible RAS-specific GEF, Yoshiki et al. [87] have shown that RAS activation alone is insufficient to achieve maximal ERK activation. They found that plasma membrane recruitment of C-RAF and maximal ERK activation also requires calcium signals, and this was dependent on SUR-8 [87]. These observations lead to the intriguing possibility that calcium signals, acting through SUR-8, play a role in the formation of RAS nanoclusters. Regardless of the mechanism, the importance of SUR-8 in RAF activation is underscored by the discovery of SUR-8 mutations in a Noonan-like syndrome [88]. In Noonan-like syndromes, mutations are frequently observed that deregulate the RAS-MAPK pathway, and the mutations identified in SUR-8 result in constitutive membrane recruitment of SUR-8 and enhanced ERK activation [88].
Two additional scaffold proteins implicated in RTK-dependent ERK signaling are IQGAP1 and paxillin. IQGAP1 binds to all three kinase components of the ERK/MAPK cascade, and IQGAP1 regulates ERK activation in a concentration-dependent manner, which suggests that it is a bona fide ERK pathway scaffold [89–92]. Paxillin associates with RAF, MEK, and ERK following growth factor stimulation, and colocalizes with active ERK [93, 94]. However, its role as a scaffold for the ERK/MAPK pathway remains to be formally tested.
RKIP (RAF kinase inhibitory protein) presents an interesting case of a protein that can bind to two components of the MAPK cascade, C-RAF and MEK. However, RKIP is not a scaffold, but instead uncouples signaling between C-RAF and MEK. RKIP behaves as a competitive inhibitor of MEK phosphorylation by disrupting the association between C-RAF and MEK [95, 96]. RKIP binds to C-RAF and MEK in a mutually exclusive manner [95, 96]. However, the binding sites for RKIP and MEK on C-RAF do not overlap, nor do the binding sites for RKIP and C-RAF on MEK, indicating that RKIP is not a direct competitor of the C-RAF/MEK interaction [95, 96]. This suggests that RKIP induces a conformational change in C-RAF and/or MEK that prevents their interaction. It is not clear if C-RAF is the only RAF isoform targeted by RKIP. One report showed that RKIP could bind to B-RAF, but did not inhibit its kinase activity [97]. Another report showed that RKIP could inhibit growth of melanoma cells with activated B-RAF, and B-RAF-induced differentiation of PC12 cells [98]. However, in the latter case, it is possible that RKIP was actually inhibiting C-RAF that had been transactivated by B-RAF. Several studies have mapped the RKIP binding site on C-RAF to the N-region, and the introduction of a negative charge in this region by phosphorylation or aspartic acid substitutions reduced RKIP binding [95, 97, 99, 100]. B-RAF bears a constitutive negative charge in the N-region, and is likely refractory to inhibition by RKIP. RKIP activity is inhibited by phosphorylation, including PKC-dependent phosphorylation of Ser153 and ERK-dependent feedback phosphorylation of Ser99 [101, 102]. Additional studies of RKIP are warranted, as this could aid the development of inhibitors of the RAF/MEK interaction with clinical benefit.
Phosphorylation of RAF
RAF phosphorylation sites are shown in Fig. 1a. Unless specified otherwise, we will refer to these sites based on the numbering of C-RAF. Phosphorylation of Ser259 mediates binding to 14-3-3 proteins, and this has a negative regulatory role because substitution of alanine at this position increases basal RAF activity [21–24, 103, 104]. Binding of 14-3-3 at this site maintains RAF in the closed conformation, and may also mask the CRD and prevent spurious interactions with RAS at the plasma membrane [20, 22, 25, 26, 42, 43, 105, 106]. The impact of this site on RAF activity is highlighted by the discovery of gain-of-function C-RAF mutations in Noonan syndrome that decrease phosphorylation of Ser259 and binding to 14-3-3 [107, 108]. Two kinases have been reported to phosphorylate Ser259, PKA and Akt. There is strong evidence that PKA phosphorylates Ser259 directly [22, 103], while the effects of Akt could be direct or indirect [109–111]. Upon growth factor stimulation Ser259 dephosphorylation is mediated by either PP2A or PP1 [21, 35, 85, 112].
Ser621 is conserved in all RAF proteins and is also a docking site for 14-3-3 proteins [113, 114]. Binding of 14-3-3 to phospho-Ser621 appears to have multiple roles in RAF regulation. In resting cells, dimerization of 14-3-3 monomers bound to phosphorylated Ser259 and Ser621 is thought to stabilize the closed, inactive conformation of RAF [20, 42, 114]. However, 14-3-3 binding to Ser621 is required for RAF activation following stimulation, because mutants with an alanine substitution at this position are inactive [113–115]. Several observations have been made regarding the requirement of Ser621 for RAF activity. Phosphorylation of Ser621 increases the stability of RAF by preventing proteasomal degradation [116], binding of 14-3-3 to Ser621 enhances dimerization of RAF [117–121], and RAF requires binding of 14-3-3 to Ser621 in order for the catalytic domain to bind ATP [122]. Ser621 phosphorylation is largely mediated by autophosphorylation [116, 122]. However, kinase-inactive RAF retains some Ser621 phosphorylation in vivo [122], indicating that other kinases can phosphorylate this residue. The phosphatase(s) responsible for dephosphorylating Ser621 are currently unknown.
Several additional phosphorylation sites contribute to RAF activation. These include Ser338 and Tyr340/341 in the N-region, as well as two putative sites in the activation segment of the kinase domain. Growth factor-stimulated phosphorylation of Ser338 is RAS-dependent and required for maximal C-RAF activation [123–126]. Substitution of Ser338 with alanine or dephosphorylation of Ser338 by PP5 results in C-RAF inactivation [123–127]. The kinase that mediates RTK-induced phosphorylation of Ser338 remains elusive. Members of the PAK family have been identified as Ser338 kinases [128–131], but several observations make it unlikely that they are involved in RTK-dependent Ser338 phosphorylation. PAK-mediated Ser338 phosphorylation is independent of RAS and neither dominant-negative PAK3 nor PAK1 siRNA blocks RTK-dependent Ser338 phosphorylation [132, 133]. In mammalian cells, Ser338 phosphorylation requires KSR1-mediated recruitment of CK2 [134], but in Drosophila, CK2 is dispensable for KSR-dependent RAF activation [118]. A recent report suggests that Ser338 phosphorylation occurs as a result of autophosphorylation, and this requires homodimerization of C-RAF or heterodimerization of C-RAF and B-RAF [133]. Ser338 phosphorylation appears to synergize with phosphorylation of Tyr340/341. Growth factor-stimulated phosphorylation of Tyr340/341 requires RAS-dependent membrane recruitment and Src family tyrosine kinases [46, 47, 126, 135], and phosphorylation of Tyr340/341 and Ser338 is required for maximal RAF activity [27, 123, 125, 126]. Mutation of Tyr340/341 to aspartic acid increases basal Ser338 phosphorylation and activity of C-RAF [133]. Phosphorylation of the equivalent residues in A-RAF appears to play a similar role, but for reasons unknown, inducible kinase activity of A-RAF is much lower than C-RAF [26, 47]. In contrast, B-RAF contains aspartic acid residues at the equivalent sites of Tyr340/341 (Asp448/449), and B-RAF is constitutively phosphorylated on Ser446 [26, 126]. This is believed to account for the increased basal activity of B-RAF. Phosphorylation of the N-region cooperates with RAS-GTP in disrupting the inhibitory interaction between the regulatory region and kinase domain [26, 27]. Phosphorylation of the N-region is necessary for maximal RAF activation but not sufficient, which is illustrated by the fact that B-RAF responds to RAS activation [26, 47]. Maximal activation of RAF is also thought to require phosphorylation within the activation segment of the kinase domain [74, 123, 125, 133, 136]. Crystallographic studies of B-RAF indicate that, in the inactive state, the kinase domain adopts a conformation in which an intramolecular interaction between the P loop and the DFG motif prevents catalysis, and it is possible that phosphorylation of the activation segment disrupts this interaction [137].
RAF is also subject to ERK-mediated feedback phosphorylation, which was proposed to have both positive and negative effects on C-RAF kinase activity [104, 138]. However, several recent studies have convincingly demonstrated that ERK-mediated feedback phosphorylation of C-RAF and B-RAF has a negative impact on kinase activity. Five residues in C-RAF (Ser29, Ser289, Ser296, Ser301, and Ser642) are phosphorylated by ERK following growth factor stimulation [104, 138]. Phosphorylation of a sixth residue (Ser43) is also ERK-dependent, but it does not match the consensus for ERK phosphorylation sites and so it is unclear whether ERK phosphorylates Ser43 directly [104]. Phosphorylation of these sites on C-RAF, which we refer to as hyperphosphorylated C-RAF, has a negative regulatory role because mutation of the six serine residues to alanine results in sustained growth factor-stimulated C-RAF activity, membrane localization, and Ser338 phosphorylation [104]. The timing of hyperphosphorylation is delayed relative to the activating phosphorylation on Ser338, and correlates with low C-RAF activity that cannot be restimulated by an additional challenge with growth factor [104]. It was also shown that hyperphosphorylated C-RAF is unable to bind RAS and relocalizes to the cytoplasm where it is recycled for activation by PP2A and the Pin1 prolyl isomerase [104]. In mammalian cells, B-RAF and KSR1 have also been shown to be subject to growth factor-stimulated, ERK-dependent feedback phosphorylation [28, 31, 121]. B-RAF is phosphorylated on four sites (Ser151, Thr401, Ser750, and Thr753) as is KSR1 (Thr260, Thr274, Ser320, and Ser443), and phosphorylation of both B-RAF and KSR1 requires the ERK docking site in KSR1 [28]. Hyperphosphorylation of B-RAF prevents binding to activated RAS, and has also been shown to disrupt the B-RAF/KSR1 interaction and relocalize B-RAF and KSR1 to the cytoplasm [28, 121]. Like C-RAF, B-RAF is recycled for activation by PP2A and Pin1 [121]. Although it has not been demonstrated that KSR1 is recycled by the same mechanism as RAF, given the similarity between the two this is likely the case. ERK-dependent feedback phosphorylation has also been shown to disrupt heterodimerization of B-RAF and C-RAF [119, 121].
While there is a considerable wealth of knowledge regarding phosphorylation of the RAF proteins, much remains to be learned. With the exception of ERK and Src, we know little about the kinases that phosphorylate RAF. Several candidates have been proposed to phosphorylate Ser259 and Ser338 (and their equivalents in A-RAF and B-RAF), but their requirement in vivo has not been rigorously tested, nor is it clear how these kinases are themselves regulated with respect to phosphorylation of RAF. In addition, individual phosphorylation sites are not insulated from each other, yet our understanding of the spatial and temporal coordination of these phosphorylation events is limited. Phosphorylation of Ser259 and Ser338 on C-RAF is thought to be mutually exclusive. In contrast, B-RAF is constitutively phosphorylated on Ser446, but remains sensitive to the phosphorylation state of Ser365 (the equivalent of Ser259). Existing data would also suggest that Ser338 phosphorylation occurs at the plasma membrane subsequent to phosphorylation of Tyr340/341, yet Ser338 phosphorylation can occur in the cytosol in the absence of phosphorylation at Tyr340/341. The ability to track multiple phosphorylation events simultaneously in living cells would go a long way towards resolving these issues.
Allosteric regulation of RAF activity
In recent years, it has become increasingly apparent that allosteric mechanisms play a crucial role in RAF activation, and we have already noted several instances where protein–protein interactions can impinge on RAF to modulate its catalytic activity. Here, we will focus on the role of dimerization in the allosteric regulation of RAF activity. Earlier studies reported the intriguing phenomenon that the physical juxtaposition of RAF proteins with each other could stimulate their kinase activity [120, 139–142]. Indeed, subsequent experiments showed that even a catalytically compromised B-RAF was capable of switching on C-RAF activity in trans in a manner dependent on a physical interaction between B-RAF and C-RAF, suggesting that the underlying mechanism is independent of a simple transautophosphorylation route [119, 137]. Most recently, several groups have demonstrated that ATP-competitive B-RAF inhibitors can induce B-RAF/C-RAF complex formation and the resultant stimulation of C-RAF activity in trans, underscoring the relevance of RAF/RAF interactions in modulating kinase activity [72, 143, 144].
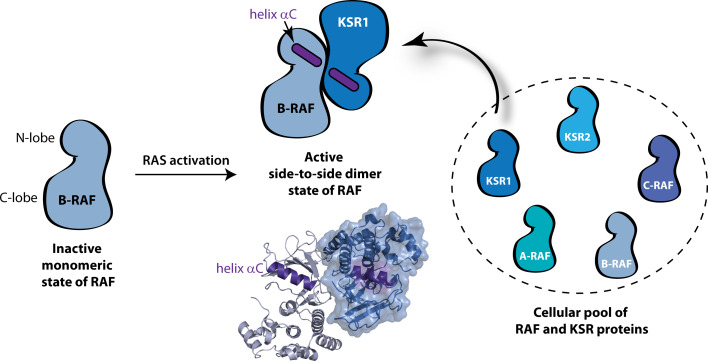
The precise mechanism by which the juxtaposition of RAF molecules stimulates kinase activity was recently elucidated [118]. Activation of the kinase domain of RAF is controlled principally by an allosteric interaction between two kinase domains in a specific side-to-side dimer configuration (Fig. 2). The dimer state is thought to position a critical helix in the kinase domain (helix αC) in a productive conformation necessary for catalytic activity. Most notably, the kinase domain in KSR can also serve the role of an allosteric activator of RAF by forming KSR/RAF side-to-side heterodimers [118]. The intrinsic ability of KSR to allosterically modulate RAF activity is a departure from the conventional dogma that KSR is yet another passive scaffold in RAF signaling.
Fig. 2.
An allosteric mechanism for activation of the kinase domain of RAF. RAF exists in an inactive conformation characterized by the monomeric state of its kinase domain. This monomeric state represents a kinase domain isolated from neighbouring kinase domains, but maintains other regulatory interactions that stabilize the inactive state as described in the text (for simplicity, only the C-terminal kinase domain of RAF is diagrammed; the N- and C-terminal lobes of the kinase domain are indicated). Upon RAS activation, the kinase domain of RAF transitions from an inactive monomer to an active side-to-side dimer state characterized structurally by the juxtaposition of a critical helix (helix αC) in the N-lobe that is thought to lock the kinase domain in a productive conformation competent for catalytic activity. A ribbons representation of the side-to-side dimer is shown (based on PDB ID 1UWH; ref. [137]). The cellular pool of RAF/KSR proteins in higher-order metazoa comprises multiple isoforms and the mechanism by which the cell selectively drives dimer formation between two given isoforms remains unknown
The question still remains as to how RAF dimerization is regulated. Removal of the N-terminal regulatory region causes constitutive activation of RAF, suggesting that, in addition to maintaining inactive RAF in a closed state inaccessible to MEK, the regulatory region may also prevent side-to-side dimer formation. Interestingly, RAS has also been reported to form dimers [145], raising the intriguing possibility that RAS activates RAF by relieving autoinhibition and directly promoting RAF dimerization. Additional proteins may also participate in RAF dimerization. The aforementioned 14-3-3 proteins are candidates for promoting RAF side-to-side dimers [118]. Two kinases, MLK3 and DGKη, have both been shown to facilitate B-RAF/C-RAF complex formation independently of their kinase activity [146, 147]. However, whether this complex formation is as a result of direct B-RAF/C-RAF side-to-side dimerization was not specifically demonstrated in either case, and it would be interesting to see if MLK3 and DGKη regulate dimerization of the RAF kinase domain.
Equally intriguing is the question of how the cell regulates specific dimer formation between the multiple isoforms of RAF and KSR proteins that are present in higher-order metazoa. Given that all isoforms of RAF and KSR share a nearly identical dimer interface, the cell must have a separate mechanism in place for selectively driving dimer formation between any two specific isoforms. What is the relative activity of the different dimer combinations? The heterodimers comprising KSR/RAF are thought to be more active by virtue of KSR’s specific capacity to function as both an allosteric activator of RAF and to simultaneously recruit and present the RAF substrate MEK [118]. If promotion of side-to-side dimers activates RAF, then it follows that the disruption of the dimers must lead to inactivation. While the exact mechanisms of disrupting the specific dimers might be diverse, a common theme likely involves the triggering of negative-feedback loops [121, 148].
Kinase-independent functions of C-RAF
Characterization of C-RAF knockout mice showed that C-RAF protects against apoptosis [149, 150]. However, MEK/ERK activation is normal in these mice, presumably due to B-RAF activity, and kinase activity is not required for C-RAF to confer protection against apoptosis [149, 150]. It is now known that C-RAF can negatively regulate the activity of several proapoptotic kinases, including Rok-α, MST2, and ASK1. While kinase activity of C-RAF is dispensable for this regulation, a direct interaction between C-RAF and the target kinase is requisite. In the case of Rok-α, growth factor stimulation induces an intermolecular interaction between the regulatory region of C-RAF and the kinase domain of Rok-α, resulting in inhibition of Rok-α kinase activity [151]. Inhibition of Rok-α promotes cell migration, reduces sensitivity to Fas-induced apoptosis, and is required for RAS-induced tumorigenesis in the epidermis [152–154]. In the case of MST2, C-RAF binds to MST2 in resting cells and this correlates with reduced oligomerization of MST2 and dephosphorylation at activating sites [155]. Stress signals and mitogens disrupt the C-RAF/MST2 interaction, with the former promoting apoptosis and the latter rendering cells permissive to apoptotic stimuli [155, 156]. Interestingly, there appears to be a reciprocal relationship between C-RAF and MST2, with MST2 being required for full activation of C-RAF following growth factor stimulation [157]. MST2 is required to maintain the expression level of the catalytic subunit of PP2A, and depletion of MST2 increases Ser259 phosphorylation on C-RAF [157]. C-RAF also binds to ASK1, and by an unknown mechanism inhibits its kinase activity and protects cells from ASK1-dependent apoptosis [158–160]. These kinase-independent functions of C-RAF again illustrate the potential therapeutic benefits of targeting protein–protein interactions.
Conclusions and future perspectives
The past several years have seen exciting advances in our understanding of the molecular mechanisms that operate during activation of RAF kinases and signaling events regulated by the KSR proteins. In particular, we note the prominence of protein–protein interactions in all facets of signal transduction involving RAF and KSR. We expect that continued molecular modeling and biophysical studies will elucidate the architecture of these protein binding interfaces, and future studies may reveal novel protein–protein interactions in which RAF or KSR participate. Targeting protein interactions related to RAF and KSR would aid the treatment of many human ailments, including cancer, developmental disorders, and obesity. While the development of protein interaction inhibitors is technically more challenging than inhibitors of enzyme-catalyzed reactions, we feel this is offset by the flexibility and specificity afforded by the multitude of potential targets.
Acknowledgments
This work was supported by funding from the Canadian Institutes for Health Research and from the Canadian Cancer Society to M.T. and F.S. T.R. is a Research Fellow of The Terry Fox Foundation. F.S. holds a Canada Research Chair in Structural Biology of Signal Transduction, and M.T. is a Canada Research Chair (Tier II) in Intracellular Signaling.
References
- 1.Bonner TI, Kerby SB, Sutrave P, Gunnell MA, Mark G, Rapp UR. Structure and biological activity of human homologs of the raf/mil oncogene. Mol Cell Biol. 1985;5:1400–1407. doi: 10.1128/mcb.5.6.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck TW, Huleihel M, Gunnell M, Bonner TI, Rapp UR. The complete coding sequence of the human A-raf-1 oncogene and transforming activity of a human A-raf carrying retrovirus. Nucleic Acids Res. 1987;15:595–609. doi: 10.1093/nar/15.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huebner K, ar-Rushdi A, Griffin CA, Isobe M, Kozak C, Emanuel BS, Nagarajan L, Cleveland JL, Bonner TI, Goldsborough MD, Croce CM, Rapp U. Actively transcribed genes in the raf oncogene group, located on the X chromosome in mouse and human. Proc Natl Acad Sci USA. 1986;83:3934–3938. doi: 10.1073/pnas.83.11.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ikawa S, Fukui M, Ueyama Y, Tamaoki N, Yamamoto T, Toyoshima K. B-raf, a new member of the raf family, is activated by DNA rearrangement. Mol Cell Biol. 1988;8:2651–2654. doi: 10.1128/mcb.8.6.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kornfeld K, Hom DB, Horvitz HR. The ksr-1 gene encodes a novel protein kinase involved in Ras-mediated signaling in C. elegans . Cell. 1995;83:903–913. doi: 10.1016/0092-8674(95)90206-6. [DOI] [PubMed] [Google Scholar]
- 6.Sundaram M, Han M. The C. elegans ksr-1 gene encodes a novel Raf-related kinase involved in Ras-mediated signal transduction. Cell. 1995;83:889–901. doi: 10.1016/0092-8674(95)90205-8. [DOI] [PubMed] [Google Scholar]
- 7.Therrien M, Chang HC, Solomon NM, Karim FD, Wassarman DA, Rubin GM. KSR, a novel protein kinase required for RAS signal transduction. Cell. 1995;83:879–888. doi: 10.1016/0092-8674(95)90204-X. [DOI] [PubMed] [Google Scholar]
- 8.Channavajhala PL, Wu L, Cuozzo JW, Hall JP, Liu W, Lin LL, Zhang Y. Identification of a novel human kinase supporter of Ras (hKSR-2) that functions as a negative regulator of Cot (Tpl2) signaling. J Biol Chem. 2003;278:47089–47097. doi: 10.1074/jbc.M306002200. [DOI] [PubMed] [Google Scholar]
- 9.Claperon A, Therrien M. KSR and CNK: two scaffolds regulating RAS-mediated RAF activation. Oncogene. 2007;26:3143–3158. doi: 10.1038/sj.onc.1210408. [DOI] [PubMed] [Google Scholar]
- 10.Kolch W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat Rev Mol Cell Biol. 2005;6:827–837. doi: 10.1038/nrm1743. [DOI] [PubMed] [Google Scholar]
- 11.McKay MM, Morrison DK. Integrating signals from RTKs to ERK/MAPK. Oncogene. 2007;26:3113–3121. doi: 10.1038/sj.onc.1210394. [DOI] [PubMed] [Google Scholar]
- 12.Wellbrock C, Karasarides M, Marais R. The RAF proteins take centre stage. Nat Rev Mol Cell Biol. 2004;5:875–885. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- 13.Zebisch A, Troppmair J. Back to the roots: the remarkable RAF oncogene story. Cell Mol Life Sci. 2006;63:1314–1330. doi: 10.1007/s00018-006-6005-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo Z, Diaz B, Marshall MS, Avruch J. An intact Raf zinc finger is required for optimal binding to processed Ras and for ras-dependent Raf activation in situ. Mol Cell Biol. 1997;17:46–53. doi: 10.1128/mcb.17.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou M, Horita DA, Waugh DS, Byrd RA, Morrison DK. Solution structure and functional analysis of the cysteine-rich C1 domain of kinase suppressor of Ras (KSR) J Mol Biol. 2002;315:435–446. doi: 10.1006/jmbi.2001.5263. [DOI] [PubMed] [Google Scholar]
- 16.Bondeva T, Balla A, Varnai P, Balla T. Structural determinants of Ras–Raf interaction analyzed in live cells. Mol Biol Cell. 2002;13:2323–2333. doi: 10.1091/mbc.E02-01-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding J, Tchaicheeyan O, Ambrosio L. Drosophila Raf’s N terminus contains a novel conserved region and can contribute to torso RTK signaling. Genetics. 2010;184:717–729. doi: 10.1534/genetics.109.111344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer A, Hekman M, Kuhlmann J, Rubio I, Wiese S, Rapp UR. B- and C-RAF display essential differences in their binding to Ras: the isotype-specific N terminus of B-RAF facilitates Ras binding. J Biol Chem. 2007;282:26503–26516. doi: 10.1074/jbc.M607458200. [DOI] [PubMed] [Google Scholar]
- 19.Terai K, Matsuda M. The amino-terminal B-Raf-specific region mediates calcium-dependent homo- and hetero-dimerization of Raf. EMBO J. 2006;25:3556–3564. doi: 10.1038/sj.emboj.7601241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terai K, Matsuda M. Ras binding opens c-Raf to expose the docking site for mitogen-activated protein kinase kinase. EMBO Rep. 2005;6:251–255. doi: 10.1038/sj.embor.7400349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kubicek M, Pacher M, Abraham D, Podar K, Eulitz M, Baccarini M. Dephosphorylation of Ser-259 regulates Raf-1 membrane association. J Biol Chem. 2002;277:7913–7919. doi: 10.1074/jbc.M108733200. [DOI] [PubMed] [Google Scholar]
- 22.Dumaz N, Marais R. Protein kinase A blocks Raf-1 activity by stimulating 14-3-3 binding and blocking Raf-1 interaction with Ras. J Biol Chem. 2003;278:29819–29823. doi: 10.1074/jbc.C300182200. [DOI] [PubMed] [Google Scholar]
- 23.Light Y, Paterson H, Marais R. 14-3-3 antagonizes Ras-mediated Raf-1 recruitment to the plasma membrane to maintain signaling fidelity. Mol Cell Biol. 2002;22:4984–4996. doi: 10.1128/MCB.22.14.4984-4996.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rommel C, Radziwill G, Moelling K, Hafen E. Negative regulation of Raf activity by binding of 14-3-3 to the amino terminus of Raf in vivo. Mech Dev. 1997;64:95–104. doi: 10.1016/S0925-4773(97)00052-X. [DOI] [PubMed] [Google Scholar]
- 25.Cutler RE, Jr, Stephens RM, Saracino MR, Morrison DK. Autoregulation of the Raf-1 serine/threonine kinase. Proc Natl Acad Sci USA. 1998;95:9214–9219. doi: 10.1073/pnas.95.16.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tran NH, Wu X, Frost JA. B-Raf and Raf-1 are regulated by distinct autoregulatory mechanisms. J Biol Chem. 2005;280:16244–16253. doi: 10.1074/jbc.M501185200. [DOI] [PubMed] [Google Scholar]
- 27.Tran NH, Frost JA. Phosphorylation of Raf-1 by p21-activated kinase 1 and Src regulates Raf-1 autoinhibition. J Biol Chem. 2003;278:11221–11226. doi: 10.1074/jbc.M210318200. [DOI] [PubMed] [Google Scholar]
- 28.McKay MM, Ritt DA, Morrison DK. Signaling dynamics of the KSR1 scaffold complex. Proc Natl Acad Sci USA. 2009;106:11022–11027. doi: 10.1073/pnas.0901590106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roy F, Laberge G, Douziech M, Ferland-McCollough D, Therrien M. KSR is a scaffold required for activation of the ERK/MAPK module. Genes Dev. 2002;16:427–438. doi: 10.1101/gad.962902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michaud NR, Therrien M, Cacace A, Edsall LC, Spiegel S, Rubin GM, Morrison DK. KSR stimulates Raf-1 activity in a kinase-independent manner. Proc Natl Acad Sci USA. 1997;94:12792–12796. doi: 10.1073/pnas.94.24.12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cacace AM, Michaud NR, Therrien M, Mathes K, Copeland T, Rubin GM, Morrison DK. Identification of constitutive and ras-inducible phosphorylation sites of KSR: implications for 14-3-3 binding, mitogen-activated protein kinase binding, and KSR overexpression. Mol Cell Biol. 1999;19:229–240. doi: 10.1128/mcb.19.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller J, Ory S, Copeland T, Piwnica-Worms H, Morrison DK. C-TAK1 regulates Ras signaling by phosphorylating the MAPK scaffold, KSR1. Mol Cell. 2001;8:983–993. doi: 10.1016/S1097-2765(01)00383-5. [DOI] [PubMed] [Google Scholar]
- 33.Therrien M, Michaud NR, Rubin GM, Morrison DK. KSR modulates signal propagation within the MAPK cascade. Genes Dev. 1996;10:2684–2695. doi: 10.1101/gad.10.21.2684. [DOI] [PubMed] [Google Scholar]
- 34.Stewart S, Sundaram M, Zhang Y, Lee J, Han M, Guan KL. Kinase suppressor of Ras forms a multiprotein signaling complex and modulates MEK localization. Mol Cell Biol. 1999;19:5523–5534. doi: 10.1128/mcb.19.8.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ory S, Zhou M, Conrads TP, Veenstra TD, Morrison DK. Protein phosphatase 2A positively regulates Ras signaling by dephosphorylating KSR1 and Raf-1 on critical 14-3-3 binding sites. Curr Biol. 2003;13:1356–1364. doi: 10.1016/S0960-9822(03)00535-9. [DOI] [PubMed] [Google Scholar]
- 36.Yu W, Fantl WJ, Harrowe G, Williams LT. Regulation of the MAP kinase pathway by mammalian Ksr through direct interaction with MEK and ERK. Curr Biol. 1998;8:56–64. doi: 10.1016/S0960-9822(98)70020-X. [DOI] [PubMed] [Google Scholar]
- 37.Lozano J, Xing R, Cai Z, Jensen HL, Trempus C, Mark W, Cannon R, Kolesnick R. Deficiency of kinase suppressor of Ras1 prevents oncogenic ras signaling in mice. Cancer Res. 2003;63:4232–4238. [PubMed] [Google Scholar]
- 38.Nguyen A, Burack WR, Stock JL, Kortum R, Chaika OV, Afkarian M, Muller WJ, Murphy KM, Morrison DK, Lewis RE, McNeish J, Shaw AS. Kinase suppressor of Ras (KSR) is a scaffold which facilitates mitogen-activated protein kinase activation in vivo. Mol Cell Biol. 2002;22:3035–3045. doi: 10.1128/MCB.22.9.3035-3045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Costanzo-Garvey DL, Pfluger PT, Dougherty MK, Stock JL, Boehm M, Chaika O, Fernandez MR, Fisher K, Kortum RL, Hong EG, Jun JY, Ko HJ, Schreiner A, Volle DJ, Treece T, Swift AL, Winer M, Chen D, Wu M, Leon LR, Shaw AS, McNeish J, Kim JK, Morrison DK, Tschop MH, Lewis RE. KSR2 is an essential regulator of AMP kinase, energy expenditure, and insulin sensitivity. Cell Metab. 2009;10:366–378. doi: 10.1016/j.cmet.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dougherty MK, Ritt DA, Zhou M, Specht SI, Monson DM, Veenstra TD, Morrison DK. KSR2 is a calcineurin substrate that promotes ERK cascade activation in response to calcium signals. Mol Cell. 2009;34:652–662. doi: 10.1016/j.molcel.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brommage R, Desai U, Revelli JP, Donoviel DB, Fontenot GK, Dacosta CM, Smith DD, Kirkpatrick LL, Coker KJ, Donoviel MS, Eberhart DE, Holt KH, Kelly MR, Paradee WJ, Philips AV, Platt KA, Suwanichkul A, Hansen GM, Sands AT, Zambrowicz BP, Powell DR. High-throughput screening of mouse knockout lines identifies true lean and obese phenotypes. Obesity. 2008;16:2362–2367. doi: 10.1038/oby.2008.361. [DOI] [PubMed] [Google Scholar]
- 42.Hibino K, Shibata T, Yanagida T, Sako Y. A RasGTP-induced conformational change in C-RAF is essential for accurate molecular recognition. Biophys J. 2009;97:1277–1287. doi: 10.1016/j.bpj.2009.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rommel C, Radziwill G, Lovric J, Noeldeke J, Heinicke T, Jones D, Aitken A, Moelling K. Activated Ras displaces 14-3-3 protein from the amino terminus of c-Raf-1. Oncogene. 1996;12:609–619. [PubMed] [Google Scholar]
- 44.Leevers SJ, Paterson HF, Marshall CJ. Requirement for Ras in Raf activation is overcome by targeting Raf to the plasma membrane. Nature. 1994;369:411–414. doi: 10.1038/369411a0. [DOI] [PubMed] [Google Scholar]
- 45.Stokoe D, Macdonald SG, Cadwallader K, Symons M, Hancock JF. Activation of Raf as a result of recruitment to the plasma membrane. Science. 1994;264:1463–1467. doi: 10.1126/science.7811320. [DOI] [PubMed] [Google Scholar]
- 46.Marais R, Light Y, Paterson HF, Marshall CJ. Ras recruits Raf-1 to the plasma membrane for activation by tyrosine phosphorylation. EMBO J. 1995;14:3136–3145. doi: 10.1002/j.1460-2075.1995.tb07316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marais R, Light Y, Paterson HF, Mason CS, Marshall CJ. Differential regulation of Raf-1, A-Raf, and B-Raf by oncogenic ras and tyrosine kinases. J Biol Chem. 1997;272:4378–4383. doi: 10.1074/jbc.272.7.4378. [DOI] [PubMed] [Google Scholar]
- 48.Roy S, McPherson RA, Apolloni A, Yan J, Lane A, Clyde-Smith J, Hancock JF. 14-3-3 facilitates Ras-dependent Raf-1 activation in vitro and in vivo. Mol Cell Biol. 1998;18:3947–3955. doi: 10.1128/mcb.18.7.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brummer T, Martin P, Herzog S, Misawa Y, Daly RJ, Reth M. Functional analysis of the regulatory requirements of B-Raf and the B-Raf(V600E) oncoprotein. Oncogene. 2006;25:6262–6276. doi: 10.1038/sj.onc.1209640. [DOI] [PubMed] [Google Scholar]
- 50.Murakoshi H, Iino R, Kobayashi T, Fujiwara T, Ohshima C, Yoshimura A, Kusumi A. Single-molecule imaging analysis of Ras activation in living cells. Proc Natl Acad Sci USA. 2004;101:7317–7322. doi: 10.1073/pnas.0401354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eisenberg S, Shvartsman DE, Ehrlich M, Henis YI. Clustering of raft-associated proteins in the external membrane leaflet modulates internal leaflet H-ras diffusion and signaling. Mol Cell Biol. 2006;26:7190–7200. doi: 10.1128/MCB.01059-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Niv H, Gutman O, Kloog Y, Henis YI. Activated K-Ras and H-Ras display different interactions with saturable nonraft sites at the surface of live cells. J Cell Biol. 2002;157:865–872. doi: 10.1083/jcb.200202009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Plowman SJ, Ariotti N, Goodall A, Parton RG, Hancock JF. Electrostatic interactions positively regulate K-Ras nanocluster formation and function. Mol Cell Biol. 2008;28:4377–4385. doi: 10.1128/MCB.00050-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prior IA, Harding A, Yan J, Sluimer J, Parton RG, Hancock JF. GTP-dependent segregation of H-ras from lipid rafts is required for biological activity. Nat Cell Biol. 2001;3:368–375. doi: 10.1038/35070050. [DOI] [PubMed] [Google Scholar]
- 55.Hancock JF, Parton RG. Ras plasma membrane signalling platforms. Biochem J. 2005;389:1–11. doi: 10.1042/BJ20050231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Plowman SJ, Muncke C, Parton RG, Hancock JF. H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc Natl Acad Sci USA. 2005;102:15500–15505. doi: 10.1073/pnas.0504114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prior IA, Muncke C, Parton RG, Hancock JF. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J Cell Biol. 2003;160:165–170. doi: 10.1083/jcb.200209091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Belanis L, Plowman SJ, Rotblat B, Hancock JF, Kloog Y. Galectin-1 is a novel structural component and a major regulator of h-ras nanoclusters. Mol Biol Cell. 2008;19:1404–1414. doi: 10.1091/mbc.E07-10-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shalom-Feuerstein R, Plowman SJ, Rotblat B, Ariotti N, Tian T, Hancock JF, Kloog Y. K-ras nanoclustering is subverted by overexpression of the scaffold protein galectin-3. Cancer Res. 2008;68:6608–6616. doi: 10.1158/0008-5472.CAN-08-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tian T, Harding A, Inder K, Plowman S, Parton RG, Hancock JF. Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nat Cell Biol. 2007;9:905–914. doi: 10.1038/ncb1615. [DOI] [PubMed] [Google Scholar]
- 61.Casar B, Arozarena I, Sanz-Moreno V, Pinto A, Agudo-Ibanez L, Marais R, Lewis RE, Berciano MT, Crespo P. Ras subcellular localization defines extracellular signal-regulated kinase 1 and 2 substrate specificity through distinct utilization of scaffold proteins. Mol Cell Biol. 2009;29:1338–1353. doi: 10.1128/MCB.01359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Das J, Ho M, Zikherman J, Govern C, Yang M, Weiss A, Chakraborty AK, Roose JP. Digital signaling and hysteresis characterize ras activation in lymphoid cells. Cell. 2009;136:337–351. doi: 10.1016/j.cell.2008.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Groves JT, Kuriyan J. Molecular mechanisms in signal transduction at the membrane. Nat Struct Mol Biol. 2010;17:659–665. doi: 10.1038/nsmb.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harding A, Tian T, Westbury E, Frische E, Hancock JF. Subcellular localization determines MAP kinase signal output. Curr Biol. 2005;15:869–873. doi: 10.1016/j.cub.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 65.Inder K, Harding A, Plowman SJ, Philips MR, Parton RG, Hancock JF. Activation of the MAPK module from different spatial locations generates distinct system outputs. Mol Biol Cell. 2008;19:4776–4784. doi: 10.1091/mbc.E08-04-0407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gomez GA, Daniotti JL. H-Ras dynamically interacts with recycling endosomes in CHO-K1 cells: involvement of Rab5 and Rab11 in the trafficking of H-Ras to this pericentriolar endocytic compartment. J Biol Chem. 2005;280:34997–35010. doi: 10.1074/jbc.M506256200. [DOI] [PubMed] [Google Scholar]
- 67.Roy S, Wyse B, Hancock JF. H-Ras signaling and K-Ras signaling are differentially dependent on endocytosis. Mol Cell Biol. 2002;22:5128–5140. doi: 10.1128/MCB.22.14.5128-5140.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang X, Sorkin A. Coordinated traffic of Grb2 and Ras during epidermal growth factor receptor endocytosis visualized in living cells. Mol Biol Cell. 2002;13:1522–1535. doi: 10.1091/mbc.01-11-0552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bivona TG, Perez De Castro I, Ahearn IM, Grana TM, Chiu VK, Lockyer PJ, Cullen PJ, Pellicer A, Cox AD, Philips MR. Phospholipase Cgamma activates Ras on the Golgi apparatus by means of RasGRP1. Nature. 2003;424:694–698. doi: 10.1038/nature01806. [DOI] [PubMed] [Google Scholar]
- 70.Chiu VK, Bivona T, Hach A, Sajous JB, Silletti J, Wiener H, Johnson RL, 2nd, Cox AD, Philips MR. Ras signalling on the endoplasmic reticulum and the Golgi. Nat Cell Biol. 2002;4:343–350. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- 71.Perez de Castro I, Bivona TG, Philips MR, Pellicer A. Ras activation in Jurkat T cells following low-grade stimulation of the T-cell receptor is specific to N-Ras and occurs only on the Golgi apparatus. Mol Cell Biol. 2004;24:3485–3496. doi: 10.1128/MCB.24.8.3485-3496.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, Ludlam MJ, Stokoe D, Gloor SL, Vigers G, Morales T, Aliagas I, Liu B, Sideris S, Hoeflich KP, Jaiswal BS, Seshagiri S, Koeppen H, Belvin M, Friedman LS, Malek S. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 73.Anselmo AN, Bumeister R, Thomas JM, White MA. Critical contribution of linker proteins to Raf kinase activation. J Biol Chem. 2002;277:5940–5943. doi: 10.1074/jbc.M110498200. [DOI] [PubMed] [Google Scholar]
- 74.Douziech M, Sahmi M, Laberge G, Therrien M. A KSR/CNK complex mediated by HYP, a novel SAM domain-containing protein, regulates RAS-dependent RAF activation in Drosophila . Genes Dev. 2006;20:807–819. doi: 10.1101/gad.1390406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roignant JY, Hamel S, Janody F, Treisman JE. The novel SAM domain protein Aveugle is required for Raf activation in the Drosophila EGF receptor signaling pathway. Genes Dev. 2006;20:795–806. doi: 10.1101/gad.1390506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Therrien M, Wong AM, Rubin GM. CNK, a RAF-binding multidomain protein required for RAS signaling. Cell. 1998;95:343–353. doi: 10.1016/S0092-8674(00)81766-3. [DOI] [PubMed] [Google Scholar]
- 77.Rajakulendran T, Sahmi M, Kurinov I, Tyers M, Therrien M, Sicheri F. CNK and HYP form a discrete dimer by their SAM domains to mediate RAF kinase signaling. Proc Natl Acad Sci USA. 2008;105:2836–2841. doi: 10.1073/pnas.0709705105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Douziech M, Roy F, Laberge G, Lefrancois M, Armengod AV, Therrien M. Bimodal regulation of RAF by CNK in Drosophila . EMBO J. 2003;22:5068–5078. doi: 10.1093/emboj/cdg506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Laberge G, Douziech M, Therrien M. Src42 binding activity regulates Drosophila RAF by a novel CNK-dependent derepression mechanism. EMBO J. 2005;24:487–498. doi: 10.1038/sj.emboj.7600558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lanigan TM, Liu A, Huang YZ, Mei L, Margolis B, Guan KL. Human homologue of Drosophila CNK interacts with Ras effector proteins Raf and Rlf. FASEB J. 2003;17:2048–2060. doi: 10.1096/fj.02-1096com. [DOI] [PubMed] [Google Scholar]
- 81.Ziogas A, Moelling K, Radziwill G. CNK1 is a scaffold protein that regulates Src-mediated Raf-1 activation. J Biol Chem. 2005;280:24205–24211. doi: 10.1074/jbc.M413327200. [DOI] [PubMed] [Google Scholar]
- 82.Sieburth DS, Sun Q, Han M. SUR-8, a conserved Ras-binding protein with leucine-rich repeats, positively regulates Ras-mediated signaling in C. elegans . Cell. 1998;94:119–130. doi: 10.1016/S0092-8674(00)81227-1. [DOI] [PubMed] [Google Scholar]
- 83.Dai P, Xiong WC, Mei L. Erbin inhibits RAF activation by disrupting the sur-8-Ras-Raf complex. J Biol Chem. 2006;281:927–933. doi: 10.1074/jbc.M507360200. [DOI] [PubMed] [Google Scholar]
- 84.Li W, Han M, Guan KL. The leucine-rich repeat protein SUR-8 enhances MAP kinase activation and forms a complex with Ras and Raf. Genes Dev. 2000;14:895–900. [PMC free article] [PubMed] [Google Scholar]
- 85.Rodriguez-Viciana P, Oses-Prieto J, Burlingame A, Fried M, McCormick F. A phosphatase holoenzyme comprised of Shoc2/Sur8 and the catalytic subunit of PP1 functions as an M-Ras effector to modulate Raf activity. Mol Cell. 2006;22:217–230. doi: 10.1016/j.molcel.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 86.Matsunaga-Udagawa R, Fujita Y, Yoshiki S, Terai K, Kamioka Y, Kiyokawa E, Yugi K, Aoki K, Matsuda M. The scaffold protein Shoc2/SUR-8 accelerates the interaction of Ras and Raf. J Biol Chem. 2010;285:7818–7826. doi: 10.1074/jbc.M109.053975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yoshiki S, Matsunaga-Udagawa R, Aoki K, Kamioka Y, Kiyokawa E, Matsuda M. Ras and calcium signaling pathways converge at Raf1 via the Shoc2 scaffold protein. Mol Biol Cell. 2010;21:1088–1096. doi: 10.1091/mbc.E09-06-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cordeddu V, Di Schiavi E, Pennacchio LA, Ma’ayan A, Sarkozy A, Fodale V, Cecchetti S, Cardinale A, Martin J, Schackwitz W, Lipzen A, Zampino G, Mazzanti L, Digilio MC, Martinelli S, Flex E, Lepri F, Bartholdi D, Kutsche K, Ferrero GB, Anichini C, Selicorni A, Rossi C, Tenconi R, Zenker M, Merlo D, Dallapiccola B, Iyengar R, Bazzicalupo P, Gelb BD, Tartaglia M. Mutation of SHOC2 promotes aberrant protein N-myristoylation and causes Noonan-like syndrome with loose anagen hair. Nat Genet. 2009;41:1022–1026. doi: 10.1038/ng.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Casar B, Pinto A, Crespo P. Essential role of ERK dimers in the activation of cytoplasmic but not nuclear substrates by ERK-scaffold complexes. Mol Cell. 2008;31:708–721. doi: 10.1016/j.molcel.2008.07.024. [DOI] [PubMed] [Google Scholar]
- 90.Ren JG, Li Z, Sacks DB. IQGAP1 modulates activation of B-Raf. Proc Natl Acad Sci USA. 2007;104:10465–10469. doi: 10.1073/pnas.0611308104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Roy M, Li Z, Sacks DB. IQGAP1 binds ERK2 and modulates its activity. J Biol Chem. 2004;279:17329–17337. doi: 10.1074/jbc.M308405200. [DOI] [PubMed] [Google Scholar]
- 92.Roy M, Li Z, Sacks DB. IQGAP1 is a scaffold for mitogen-activated protein kinase signaling. Mol Cell Biol. 2005;25:7940–7952. doi: 10.1128/MCB.25.18.7940-7952.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ishibe S, Joly D, Liu ZX, Cantley LG. Paxillin serves as an ERK-regulated scaffold for coordinating FAK and Rac activation in epithelial morphogenesis. Mol Cell. 2004;16:257–267. doi: 10.1016/j.molcel.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 94.Ishibe S, Joly D, Zhu X, Cantley LG. Phosphorylation-dependent paxillin-ERK association mediates hepatocyte growth factor-stimulated epithelial morphogenesis. Mol Cell. 2003;12:1275–1285. doi: 10.1016/S1097-2765(03)00406-4. [DOI] [PubMed] [Google Scholar]
- 95.Yeung K, Janosch P, McFerran B, Rose DW, Mischak H, Sedivy JM, Kolch W. Mechanism of suppression of the Raf/MEK/extracellular signal-regulated kinase pathway by the raf kinase inhibitor protein. Mol Cell Biol. 2000;20:3079–3085. doi: 10.1128/MCB.20.9.3079-3085.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yeung K, Seitz T, Li S, Janosch P, McFerran B, Kaiser C, Fee F, Katsanakis KD, Rose DW, Mischak H, Sedivy JM, Kolch W. Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature. 1999;401:173–177. doi: 10.1038/43686. [DOI] [PubMed] [Google Scholar]
- 97.Trakul N, Menard RE, Schade GR, Qian Z, Rosner MR. Raf kinase inhibitory protein regulates Raf-1 but not B-Raf kinase activation. J Biol Chem. 2005;280:24931–24940. doi: 10.1074/jbc.M413929200. [DOI] [PubMed] [Google Scholar]
- 98.Park S, Yeung ML, Beach S, Shields JM, Yeung KC. RKIP downregulates B-Raf kinase activity in melanoma cancer cells. Oncogene. 2005;24:3535–3540. doi: 10.1038/sj.onc.1208435. [DOI] [PubMed] [Google Scholar]
- 99.Park S, Rath O, Beach S, Xiang X, Kelly SM, Luo Z, Kolch W, Yeung KC. Regulation of RKIP binding to the N-region of the Raf-1 kinase. FEBS Lett. 2006;580:6405–6412. doi: 10.1016/j.febslet.2006.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rath O, Park S, Tang HH, Banfield MJ, Brady RL, Lee YC, Dignam JD, Sedivy JM, Kolch W, Yeung KC. The RKIP (Raf-1 Kinase Inhibitor Protein) conserved pocket binds to the phosphorylated N-region of Raf-1 and inhibits the Raf-1-mediated activated phosphorylation of MEK. Cell Signal. 2008;20:935–941. doi: 10.1016/j.cellsig.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 101.Corbit KC, Trakul N, Eves EM, Diaz B, Marshall M, Rosner MR. Activation of Raf-1 signaling by protein kinase C through a mechanism involving Raf kinase inhibitory protein. J Biol Chem. 2003;278:13061–13068. doi: 10.1074/jbc.M210015200. [DOI] [PubMed] [Google Scholar]
- 102.Shin SY, Rath O, Choo SM, Fee F, McFerran B, Kolch W, Cho KH. Positive- and negative-feedback regulations coordinate the dynamic behavior of the Ras-Raf-MEK-ERK signal transduction pathway. J Cell Sci. 2009;122:425–435. doi: 10.1242/jcs.036319. [DOI] [PubMed] [Google Scholar]
- 103.Dhillon AS, Pollock C, Steen H, Shaw PE, Mischak H, Kolch W. Cyclic AMP-dependent kinase regulates Raf-1 kinase mainly by phosphorylation of serine 259. Mol Cell Biol. 2002;22:3237–3246. doi: 10.1128/MCB.22.10.3237-3246.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dougherty MK, Muller J, Ritt DA, Zhou M, Zhou XZ, Copeland TD, Conrads TP, Veenstra TD, Lu KP, Morrison DK. Regulation of Raf-1 by direct feedback phosphorylation. Mol Cell. 2005;17:215–224. doi: 10.1016/j.molcel.2004.11.055. [DOI] [PubMed] [Google Scholar]
- 105.Michaud NR, Fabian JR, Mathes KD, Morrison DK. 14-3-3 is not essential for Raf-1 function: identification of Raf-1 proteins that are biologically activated in a 14-3-3- and Ras-independent manner. Mol Cell Biol. 1995;15:3390–3397. doi: 10.1128/mcb.15.6.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Clark GJ, Drugan JK, Rossman KL, Carpenter JW, Rogers-Graham K, Fu H, Der CJ, Campbell SL. 14-3-3 zeta negatively regulates raf-1 activity by interactions with the Raf-1 cysteine-rich domain. J Biol Chem. 1997;272:20990–20993. doi: 10.1074/jbc.272.34.20990. [DOI] [PubMed] [Google Scholar]
- 107.Kobayashi T, Aoki Y, Niihori T, Cave H, Verloes A, Okamoto N, Kawame H, Fujiwara I, Takada F, Ohata T, Sakazume S, Ando T, Nakagawa N, Lapunzina P, Meneses AG, Gillessen-Kaesbach G, Wieczorek D, Kurosawa K, Mizuno S, Ohashi H, David A, Philip N, Guliyeva A, Narumi Y, Kure S, Tsuchiya S, Matsubara Y. Molecular and clinical analysis of RAF1 in Noonan syndrome and related disorders: dephosphorylation of serine 259 as the essential mechanism for mutant activation. Hum Mutat. 2010;31:284–294. doi: 10.1002/humu.21187. [DOI] [PubMed] [Google Scholar]
- 108.Pandit B, Sarkozy A, Pennacchio LA, Carta C, Oishi K, Martinelli S, Pogna EA, Schackwitz W, Ustaszewska A, Landstrom A, Bos JM, Ommen SR, Esposito G, Lepri F, Faul C, Mundel P, Lopez Siguero JP, Tenconi R, Selicorni A, Rossi C, Mazzanti L, Torrente I, Marino B, Digilio MC, Zampino G, Ackerman MJ, Dallapiccola B, Tartaglia M, Gelb BD. Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nat Genet. 2007;39:1007–1012. doi: 10.1038/ng2073. [DOI] [PubMed] [Google Scholar]
- 109.Guan KL, Figueroa C, Brtva TR, Zhu T, Taylor J, Barber TD, Vojtek AB. Negative regulation of the serine/threonine kinase B-Raf by Akt. J Biol Chem. 2000;275:27354–27359. doi: 10.1074/jbc.M004371200. [DOI] [PubMed] [Google Scholar]
- 110.Rommel C, Clarke BA, Zimmermann S, Nunez L, Rossman R, Reid K, Moelling K, Yancopoulos GD, Glass DJ. Differentiation stage-specific inhibition of the Raf-MEK-ERK pathway by Akt. Science. 1999;286:1738–1741. doi: 10.1126/science.286.5445.1738. [DOI] [PubMed] [Google Scholar]
- 111.Zimmermann S, Moelling K. Phosphorylation and regulation of Raf by Akt (protein kinase B) Science. 1999;286:1741–1744. doi: 10.1126/science.286.5445.1741. [DOI] [PubMed] [Google Scholar]
- 112.Jaumot M, Hancock JF. Protein phosphatases 1 and 2A promote Raf-1 activation by regulating 14-3-3 interactions. Oncogene. 2001;20:3949–3958. doi: 10.1038/sj.onc.1204526. [DOI] [PubMed] [Google Scholar]
- 113.Yip-Schneider MT, Miao W, Lin A, Barnard DS, Tzivion G, Marshall MS. Regulation of the Raf-1 kinase domain by phosphorylation and 14-3-3 association. Biochem J. 2000;351:151–159. doi: 10.1042/0264-6021:3510151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tzivion G, Luo Z, Avruch J. A dimeric 14-3-3 protein is an essential cofactor for Raf kinase activity. Nature. 1998;394:88–92. doi: 10.1038/27938. [DOI] [PubMed] [Google Scholar]
- 115.Mischak H, Seitz T, Janosch P, Eulitz M, Steen H, Schellerer M, Philipp A, Kolch W. Negative regulation of Raf-1 by phosphorylation of serine 621. Mol Cell Biol. 1996;16:5409–5418. doi: 10.1128/mcb.16.10.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Noble C, Mercer K, Hussain J, Carragher L, Giblett S, Hayward R, Patterson C, Marais R, Pritchard CA. CRAF autophosphorylation of serine 621 is required to prevent its proteasome-mediated degradation. Mol Cell. 2008;31:862–872. doi: 10.1016/j.molcel.2008.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Garnett MJ, Rana S, Paterson H, Barford D, Marais R. Wild-type and mutant B-RAF activate C-RAF through distinct mechanisms involving heterodimerization. Mol Cell. 2005;20:963–969. doi: 10.1016/j.molcel.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 118.Rajakulendran T, Sahmi M, Lefrancois M, Sicheri F, Therrien M. A dimerization-dependent mechanism drives RAF catalytic activation. Nature. 2009;461:542–545. doi: 10.1038/nature08314. [DOI] [PubMed] [Google Scholar]
- 119.Rushworth LK, Hindley AD, O’Neill E, Kolch W. Regulation and role of Raf-1/B-Raf heterodimerization. Mol Cell Biol. 2006;26:2262–2272. doi: 10.1128/MCB.26.6.2262-2272.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Weber CK, Slupsky JR, Kalmes HA, Rapp UR. Active Ras induces heterodimerization of cRaf and BRaf. Cancer Res. 2001;61:3595–3598. [PubMed] [Google Scholar]
- 121.Ritt DA, Monson DM, Specht SI, Morrison DK. Impact of feedback phosphorylation and Raf heterodimerization on normal and mutant B-Raf signaling. Mol Cell Biol. 2010;30:806–819. doi: 10.1128/MCB.00569-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dhillon AS, Yip YY, Grindlay GJ, Pakay JL, Dangers M, Hillmann M, Clark W, Pitt A, Mischak H, Kolch W. The C-terminus of Raf-1 acts as a 14-3-3-dependent activation switch. Cell Signal. 2009;21:1645–1651. doi: 10.1016/j.cellsig.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 123.Chong H, Lee J, Guan KL. Positive and negative regulation of Raf kinase activity and function by phosphorylation. EMBO J. 2001;20:3716–3727. doi: 10.1093/emboj/20.14.3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Diaz B, Barnard D, Filson A, MacDonald S, King A, Marshall M. Phosphorylation of Raf-1 serine 338-serine 339 is an essential regulatory event for Ras-dependent activation and biological signaling. Mol Cell Biol. 1997;17:4509–4516. doi: 10.1128/mcb.17.8.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Goetz CA, O’Neil JJ, Farrar MA. Membrane localization, oligomerization, and phosphorylation are required for optimal raf activation. J Biol Chem. 2003;278:51184–51189. doi: 10.1074/jbc.M309183200. [DOI] [PubMed] [Google Scholar]
- 126.Mason CS, Springer CJ, Cooper RG, Superti-Furga G, Marshall CJ, Marais R. Serine and tyrosine phosphorylations cooperate in Raf-1, but not B-Raf activation. EMBO J. 1999;18:2137–2148. doi: 10.1093/emboj/18.8.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.von Kriegsheim A, Pitt A, Grindlay GJ, Kolch W, Dhillon AS. Regulation of the Raf-MEK-ERK pathway by protein phosphatase 5. Nat Cell Biol. 2006;8:1011–1016. doi: 10.1038/ncb1465. [DOI] [PubMed] [Google Scholar]
- 128.Chaudhary A, King WG, Mattaliano MD, Frost JA, Diaz B, Morrison DK, Cobb MH, Marshall MS, Brugge JS. Phosphatidylinositol 3-kinase regulates Raf1 through Pak phosphorylation of serine 338. Curr Biol. 2000;10:551–554. doi: 10.1016/S0960-9822(00)00475-9. [DOI] [PubMed] [Google Scholar]
- 129.King AJ, Sun H, Diaz B, Barnard D, Miao W, Bagrodia S, Marshall MS. The protein kinase Pak3 positively regulates Raf-1 activity through phosphorylation of serine 338. Nature. 1998;396:180–183. doi: 10.1038/24184. [DOI] [PubMed] [Google Scholar]
- 130.Wu X, Carr HS, Dan I, Ruvolo PP, Frost JA. p21 activated kinase 5 activates Raf-1 and targets it to mitochondria. J Cell Biochem. 2008;105:167–175. doi: 10.1002/jcb.21809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zang M, Hayne C, Luo Z. Interaction between active Pak1 and Raf-1 is necessary for phosphorylation and activation of Raf-1. J Biol Chem. 2002;277:4395–4405. doi: 10.1074/jbc.M110000200. [DOI] [PubMed] [Google Scholar]
- 132.Chiloeches A, Mason CS, Marais R. S338 phosphorylation of Raf-1 is independent of phosphatidylinositol 3-kinase and Pak3. Mol Cell Biol. 2001;21:2423–2434. doi: 10.1128/MCB.21.7.2423-2434.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zang M, Gong J, Luo L, Zhou J, Xiang X, Huang W, Huang Q, Luo X, Olbrot M, Peng Y, Chen C, Luo Z. Characterization of Ser338 phosphorylation for Raf-1 activation. J Biol Chem. 2008;283:31429–31437. doi: 10.1074/jbc.M802855200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ritt DA, Zhou M, Conrads TP, Veenstra TD, Copeland TD, Morrison DK. CK2 is a component of the KSR1 scaffold complex that contributes to Raf kinase activation. Curr Biol. 2007;17:179–184. doi: 10.1016/j.cub.2006.11.061. [DOI] [PubMed] [Google Scholar]
- 135.Fabian JR, Daar IO, Morrison DK. Critical tyrosine residues regulate the enzymatic and biological activity of Raf-1 kinase. Mol Cell Biol. 1993;13:7170–7179. doi: 10.1128/mcb.13.11.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang BH, Guan KL. Activation of B-Raf kinase requires phosphorylation of the conserved residues Thr598 and Ser601. EMBO J. 2000;19:5429–5439. doi: 10.1093/emboj/19.20.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, Jones CM, Marshall CJ, Springer CJ, Barford D, Marais R. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/S0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 138.Balan V, Leicht DT, Zhu J, Balan K, Kaplun A, Singh-Gupta V, Qin J, Ruan H, Comb MJ, Tzivion G. Identification of novel in vivo Raf-1 phosphorylation sites mediating positive feedback Raf-1 regulation by extracellular signal-regulated kinase. Mol Biol Cell. 2006;17:1141–1153. doi: 10.1091/mbc.E04-12-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Farrar MA, Alberol I, Perlmutter RM. Activation of the Raf-1 kinase cascade by coumermycin-induced dimerization. Nature. 1996;383:178–181. doi: 10.1038/383178a0. [DOI] [PubMed] [Google Scholar]
- 140.Luo Z, Tzivion G, Belshaw PJ, Vavvas D, Marshall M, Avruch J. Oligomerization activates c-Raf-1 through a Ras-dependent mechanism. Nature. 1996;383:181–185. doi: 10.1038/383181a0. [DOI] [PubMed] [Google Scholar]
- 141.Mizutani S, Inouye K, Koide H, Kaziro Y. Involvement of B-Raf in Ras-induced Raf-1 activation. FEBS Lett. 2001;507:295–298. doi: 10.1016/S0014-5793(01)02992-1. [DOI] [PubMed] [Google Scholar]
- 142.Wojnowski L, Stancato LF, Larner AC, Rapp UR, Zimmer A. Overlapping and specific functions of Braf and Craf-1 proto-oncogenes during mouse embryogenesis. Mech Dev. 2000;91:97–104. doi: 10.1016/S0925-4773(99)00276-2. [DOI] [PubMed] [Google Scholar]
- 143.Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, Hussain J, Reis-Filho JS, Springer CJ, Pritchard C, Marais R. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Inouye K, Mizutani S, Koide H, Kaziro Y. Formation of the Ras dimer is essential for Raf-1 activation. J Biol Chem. 2000;275:3737–3740. doi: 10.1074/jbc.275.6.3737. [DOI] [PubMed] [Google Scholar]
- 146.Chadee DN, Xu D, Hung G, Andalibi A, Lim DJ, Luo Z, Gutmann DH, Kyriakis JM. Mixed-lineage kinase 3 regulates B-Raf through maintenance of the B-Raf/Raf-1 complex and inhibition by the NF2 tumor suppressor protein. Proc Natl Acad Sci USA. 2006;103:4463–4468. doi: 10.1073/pnas.0510651103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yasuda S, Kai M, Imai S, Takeishi K, Taketomi A, Toyota M, Kanoh H, Sakane F. Diacylglycerol kinase eta augments C-Raf activity and B-Raf/C-Raf heterodimerization. J Biol Chem. 2009;284:29559–29570. doi: 10.1074/jbc.M109.043604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Karreth FA, DeNicola GM, Winter SP, Tuveson DA. C-Raf inhibits MAPK activation and transformation by B-Raf(V600E) Mol Cell. 2009;36:477–486. doi: 10.1016/j.molcel.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 149.Huser M, Luckett J, Chiloeches A, Mercer K, Iwobi M, Giblett S, Sun XM, Brown J, Marais R, Pritchard C. MEK kinase activity is not necessary for Raf-1 function. EMBO J. 2001;20:1940–1951. doi: 10.1093/emboj/20.8.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mikula M, Schreiber M, Husak Z, Kucerova L, Ruth J, Wieser R, Zatloukal K, Beug H, Wagner EF, Baccarini M. Embryonic lethality and fetal liver apoptosis in mice lacking the c-raf-1 gene. EMBO J. 2001;20:1952–1962. doi: 10.1093/emboj/20.8.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Niault T, Sobczak I, Meissl K, Weitsman G, Piazzolla D, Maurer G, Kern F, Ehrenreiter K, Hamerl M, Moarefi I, Leung T, Carugo O, Ng T, Baccarini M. From autoinhibition to inhibition in trans: the Raf-1 regulatory domain inhibits Rok-alpha kinase activity. J Cell Biol. 2009;187:335–342. doi: 10.1083/jcb.200906178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ehrenreiter K, Piazzolla D, Velamoor V, Sobczak I, Small JV, Takeda J, Leung T, Baccarini M. Raf-1 regulates Rho signaling and cell migration. J Cell Biol. 2005;168:955–964. doi: 10.1083/jcb.200409162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Piazzolla D, Meissl K, Kucerova L, Rubiolo C, Baccarini M. Raf-1 sets the threshold of Fas sensitivity by modulating Rok-alpha signaling. J Cell Biol. 2005;171:1013–1022. doi: 10.1083/jcb.200504137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ehrenreiter K, Kern F, Velamoor V, Meissl K, Galabova-Kovacs G, Sibilia M, Baccarini M. Raf-1 addiction in Ras-induced skin carcinogenesis. Cancer Cell. 2009;16:149–160. doi: 10.1016/j.ccr.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 155.O’Neill E, Rushworth L, Baccarini M, Kolch W. Role of the kinase MST2 in suppression of apoptosis by the proto-oncogene product Raf-1. Science. 2004;306:2267–2270. doi: 10.1126/science.1103233. [DOI] [PubMed] [Google Scholar]
- 156.O’Neill E, Kolch W. Taming the Hippo: Raf-1 controls apoptosis by suppressing MST2/Hippo. Cell Cycle. 2005;4:365–367. doi: 10.4161/cc.4.3.1531. [DOI] [PubMed] [Google Scholar]
- 157.Kilili GK, Kyriakis JM. Mammalian Ste20-like kinase (Mst2) indirectly supports Raf-1/ERK pathway activity via maintenance of protein phosphatase-2A catalytic subunit levels and consequent suppression of inhibitory Raf-1 phosphorylation. J Biol Chem. 2010;285:15076–15087. doi: 10.1074/jbc.M109.078915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Alavi AS, Acevedo L, Min W, Cheresh DA. Chemoresistance of endothelial cells induced by basic fibroblast growth factor depends on Raf-1-mediated inhibition of the proapoptotic kinase, ASK1. Cancer Res. 2007;67:2766–2772. doi: 10.1158/0008-5472.CAN-06-3648. [DOI] [PubMed] [Google Scholar]
- 159.Chen J, Fujii K, Zhang L, Roberts T, Fu H. Raf-1 promotes cell survival by antagonizing apoptosis signal-regulating kinase 1 through a MEK-ERK independent mechanism. Proc Natl Acad Sci USA. 2001;98:7783–7788. doi: 10.1073/pnas.141224398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yamaguchi O, Watanabe T, Nishida K, Kashiwase K, Higuchi Y, Takeda T, Hikoso S, Hirotani S, Asahi M, Taniike M, Nakai A, Tsujimoto I, Matsumura Y, Miyazaki J, Chien KR, Matsuzawa A, Sadamitsu C, Ichijo H, Baccarini M, Hori M, Otsu K. Cardiac-specific disruption of the c-raf-1 gene induces cardiac dysfunction and apoptosis. J Clin Invest. 2004;114:937–943. doi: 10.1172/JCI20317. [DOI] [PMC free article] [PubMed] [Google Scholar]