Abstract
Macroautophagy, the process by which cytosolic components and organelles are engulfed and degraded by a double-membrane structure, could be viewed as a specialized, multistep membrane transport process. As such, it intersects with the exocytic and endocytic membrane trafficking pathways. A number of Rab GTPases which regulate secretory and endocytic membrane traffic have been shown to play either critical or accessory roles in autophagy. The biogenesis of the pre-autophagosomal isolation membrane (or phagophore) is dependent on the functionality of Rab1. A non-canonical, Atg5/Atg7-independent mode of autophagosome generation from the trans-Golgi or endosome requires Rab9. Other Rabs, such as Rab5, Rab24, Rab33, and Rab7 have all been shown to be required, or involved at various stages of autophagosomal genesis and maturation. Another small GTPase, RalB, was very recently demonstrated to induce isolation membrane formation and maturation via its engagement of the exocyst complex, a known Rab effector. We summarize here what is now known about the involvement of Rabs in autophagy, and discuss plausible mechanisms with future perspectives.
Keywords: Autophagy, Autophagosome, Parkinson’s disease, Rab, RalB
Introduction
Macroautophagy is an evolutionarily conserved process in which cytoplasmic contents (including membranous organelles like mitochondria and peroxisomes) are engulfed by a double-membrane autophagosome, which eventually has its content degraded at the vacuole or lysosome [1, 2]. Autophagy is induced under conditions of nutrient or growth factor deprivation in the cell, and during various conditions of stress [3]. It plays important roles in cellular homeostasis during development, and modulates cell survival. Its impairment is often associated with disease conditions such as cancer and neurodegeneration [4–9]. The autophagic process entails the formation of the immature autophagosome, or the autophagic vacuole, and its subsequent maturation, or fusion with the late-endosome/lysosome to form autolysosomes. Through work over the years, aided by the identification of the autophagy (Atg) genes in Saccharomyces cerevisiae [10], a large set of molecular players involved in autophagosome formation and various stages of maturation are now known. However, several aspects of autophagy have remained unclear. In particular, details concerning the initial steps of pre-autophagosomal structure (phagophore) and isolation membrane (IM) formation, elongation, and eventual closure to form the immature autophagosome, have been a subject of intense investigation and debate.
A set of molecules is now known to be responsible for the induction and execution of autophagy, and the following paragraphs broadly outline their roles. In yeast, autophagosome formation likely arises from a single phagophore assembly site (PAS), with evidence of membrane input from an Atg9-containing compartment [11]. Existence of an equivalent of the PAS is unclear in mammalian cells. Instead, multiple autophagosomal assembly sites, including the ER, Golgi, endosome, and mitochondria have been recognized and described [12, 13]. A main site of autophagosome generation appears to be ER-associated punctuated compartments enriched in phosphatidylinositol 3-phosphate (PI3P) and the protein double FYVE-domain containing protein 1 (DFCP1), named omegasomes [14]. These are likely to be either part of, or the same as, the ER–IM complexes, which have been documented with particular clarity by ultrastructural analysis, showing membranous interconnectivity between the ER and IM [15, 16]. In spite of the proximity and membranous connection between the omegasome and the ER, the nature of membrane traffic or relationship between the two is still unclear. Recent findings, such as the transient labeling of autophagosomal markers on mitochondrial membranes, also indicate that at least in some cases, the outer membrane of the mitochondria could supply membranes for autophagosome biogenesis [17].
Over 30 Atg proteins are known in yeast, and many of these have orthologues in invertebrates and mammals. These form various subcomplexes, and their functional hierarchy during autophagy have been intensively investigated, but several details are still unclear [18–21]. During nutrient starvation, the unc51-like kinase 1 (ULK1)/Atg1-KIAA0652/Atg13-FIP200/Atg17-Atg101-containing complex appears to be an upstream effector of autophagic signaling that connects to the nutrient-sensing pathways, controlled by molecules such as the mammalian target of rapamycin (mTOR) and AMP-dependent kinase (AMPK), which are involved in cell growth regulation and metabolism. ULK1 could be directly phosphorylated by both mTOR and AMPK, with recent findings indicating that during nutrient starvation, AMPK promotes autophagy by directly activating Ulk1 through phosphorylation of Ser 317 and Ser 777 [22]. During nutrient starvation, high mTOR activity prevents Ulk1 activation by phosphorylating Ulk1 Ser 757, thus disrupting the interaction between Ulk1 and AMPK. Details of its activities are incomplete, but the ULK1/Atg1-containing complex appears to serve in recruiting other Atg proteins, including Atg9, the only membrane component, to the pre-autophagosome assembly site [23–26]. Another bunch of molecules involved in the early stages of autophagy is the PI3-kinase complex. PI3P production is vital for autophagosome formation [27], and this is mediated by class III PI3 kinase complexes consisting of beclin1/Atg6, Vps34, Vps15/p150, and Atg14. The omegasome marker DFCP1, which is normally localized to the ER and Golgi under nutrient-rich conditions, becomes localized to PI3P on the omegasome under starvation conditions, and the PI3-kinase complex positively regulates the DFCP1-positive omegasome formation.
The two complexes above likely initiated the formation of the IM, which occurs inside the ring of the "Ω"-shaped omegasome. IM then undergoes expansion to enclose cytoplasmic contents, but the membrane source for this expansion is controversial, ranging from ER to a post-Golgi source [28]. In yeast, the expansion and eventual closure of the IM to form the double membrane autophagic vesicle is classically known to be dependent on the formation of LC3II, the microtubule-associated protein light chain 3 (LC3), or Atg8, lipidated with phosphoethanolamine (PE). This process involves two ubiquitin-like conjugation pathways. The first of these, involving the E1-like Atg7 and E2-like Atg10, leads to the conjugation of Atg5 and Atg12 and subsequent formation of the Atg16L complex (Atg5–Atg12–Atg16L). The latter is localized to the IM and acts like an E3-enzyme in promoting the recruitment of Atg3-LC3 to PE on the membrane [29]. Interestingly, in mammalian cells, IM expansion is not drastically affected by blocking LC3II formation.
A recently described non-canonical, Atg5–Atg7-independent autophagy process also does not involve the generation of LC3II [30]. In this non-canonical autophagic process, the membrane material for IM expansion appears to be derived from vesicular transport vesicles from trans-Golgi and endosome. Another component that may be critical to IM expansion is Bax-interacting factor 1 (Bif-1), or Endophilin B1. Bif-1 forms a complex with beclin1, and may act in driving membrane curvature in the process of IM extension and closure [31].
Nascent autophagic vesicles (AVs) appear to be able to fuse with multiple compartments in the endocytic pathway, including early endosome, late endosome, and multivesicular bodies (MVBs) [19, 32, 33]. Interestingly, depletion of endosomal COPI subunits results in an inhibition of early endosome maturation and causes an accumulation of AV. This suggests that fusion of AVs with functional early endosomes is a necessary step for its subsequent maturation [34]. Effective autophagic degradation also requires functional MVBs, and in some cases AVs fuses with MVBs to form a hybrid organelle that has been termed "amphisome", which then fuses with lysosomes for degradation of enclosed material [35].
It is clear from the above discussions that autophagosome formation and maturation intersects, and could indeed be dependent on, multiple steps of the exocytic and endocytic pathway. This point is further elaborated below.
Autophagy involves components that mediate secretory and endocytic traffic
Given that autophagosome formation and maturation is basically part of the eukaryotic cellular membrane dynamics, one would think that it might involve known molecular players of the secretory and endocytic machinery. However, the yeast genetic screen for autophagy-defective mutants appeared to have identified a distinct set of core autophagic components that is different from the other components of membrane transport such as Rabs and SNAREs [10]. However, it is also clear that known molecules functioning in vesicular membrane traffic are important, or essential, for the completion of autophagy.
Pertaining to the early secretory pathway, it has been shown that a specific combination of the coat protein II (COPII) (which is responsible for secretory vesicle budding from the ER) components, including Sec12p and Sec24p, is required for autophagy [36]. Sar1, the small GTPase required for the nucleation of the COPII coat, has also recently been shown to be important for autophagy [37]. As discussed above, perturbation of COPI components affects AV maturation [34]. However, COPI components may have a more indirect upstream role in autophagosome formation, as ADP ribosylation factor 1 (Arf1), the small GTPase responsible for COPI assembly, affects the activity of mTORC1, the mTOR complex involved in promoting cell growth [38]. The Rab GTPase regulating ER–Golgi transport, Rab1, and the yeast orthologue Ypt1, has also recently been shown to be essential for autophagy, which shall be elaborated upon below.
Other than ER–Golgi transport, Golgi and post-Golgi trafficking processes and their components also affect autophagy, either directly or indirectly. Two Golgi-localized Rabs, Rab24 [39] and Rab33 [40], have distinctive roles in autophagy. The Golgi syntaxin 5 containing SNARE complex is important in proper transport of lysosomal proteins, and reduction in syntaxin-5 complex components results in the accumulation of autophagosomes as a result of lysosomal dysfunction [41]. As mentioned earlier, Rab9, which mediates late endosome to Golgi transport, is critical for autophagosome formation in the Atg5–Atg7 independent non-canonical autophagic process [30]. The roles of Rab9, Rab24, and Rab33 in autophagy shall be elaborated in further detail below.
With regards to the endocytic pathway, components of the endocytic machinery that affect autophagy include the early endosomal Rab5, which, other than affecting mTORC1 activation [38], also plays a role in autophagosome formation through the beclin1-Vps34-containing complex [42]. Late endosomal Rab7 is required for fusion of autophagosomes with late endosome [43, 44], while Rab11, which is associated with recycling endosomes, is involved in the fusion between AVs and MVBs [35, 45]. Two v-SNAREs, VAMP3 and VAMP7/TI-VAMP, which are implicated in both endo- and exocytosis, have been shown to mediate AV-MVB fusion [35, 45]. In yeast, N-ethylmaleimide sensitive factor (NSF)/Sec18p and the SNARE Vti1 are required for AV fusion with the vacuole [36]. In mammalian cells, a SNARE complex containing VAMP8 and Vti1b is responsible for fusion of autophagosome with lysosomes [46]. In the plant Arabidopsis, a Rab GTPase, RabG3b, plays a role in the differentiation of tracheary elements of the xylem [47].
As evidenced above, Rab GTPases, which have been established to be key regulators of exocytic and endocytic vesicular transport, appear to also have an involvement in autophagy. In considering the intersections and interactions between the exocytic and endocytic pathways and autophagy, the paragraphs below shall focus on new findings pertaining to the involvement of Rab family GTPases in autophagy, particularly in autophagosome formation.
Rab1/Ypt1 and autophagosome formation at the ER
Rab1/Ypt1p regulates ER–Golgi and Golgi transport [48], and has been suspected to be involved in autophagy. An early indication of this notion is a study using the mutant Ypt1(Q67L)p, which is GTPase-deficient and therefore is constitutively active, alongside a background of Ypt1-GAP gene deletion. These cells exhibited growth defects at typically permissive temperatures, as well as various morphological alterations that resembled autophagy [49]. Two recent reports have now confirmed that Rab1/Ypt1p is essential for autophagy in yeast and mammalian cells [37, 50].
The yeast Ypt1p is activated by a multimeric guanine nucleotide exchange factor [51, 52], the TRAnsport Protein Particle (TRAPP) complex [53]. Two forms of the TRAPP complexes have been identified earlier, with TRAPPI functioning in ER–Golgi traffic [54] and a larger TRAPPII complex mediating Golgi traffic [55]. Previously, one of the nonessential subunits of the TRAPP1 complex, Trs85, was shown to be required in yeast for both general macroautophagy [56] and the more specific cytoplasm-to-vacuole targeting (Cvt) pathway [57], in which biosynthetic cargo is sequestered from the cytoplasm by a double-membrane vesicle that eventually fuses with a vacuole. Lynch-Day et al. [50] have recently identified a novel, Ypt1p-activating, Trs85-containing TRAPP complex (known as TRAPPIII)50. The authors also found that ypt1 temperature-sensitive mutants showed some degree of defects at both permissive and non-permissive temperatures for the nitrogen starvation-induced vacuolar transport of a mutant vacuolar alkaline phosphatase Pho8Δ60. These mutants also show varying severity in impairment of Atg8 targeting to the PAS and vacuole. On the other hand, a constitutively active mutant of Ypt1 was able to suppress autophagic defects exhibited by the trs85 deletion mutant. Imaging studies indicate that Ypt1p tend to colocalize with Trs85p at the PAS at a much higher rate than Trs65p and Trs130p, which are exclusively found in the TRAPPII complex. This colocalization is enhanced in an atg1 deletion background, in which autophagic proteins accumulate at the PAS because of the defect in autophagosome formation. The results thus indicate that the Trs85 targets a Ypt1p-specific GEF activity in the form of TRAPPIII to the PAS, although the exact role of Ypt1p in subsequent steps of autophagosome genesis remains to be elucidated.
The early secretory pathway of mammalian cells shares some similar features with that of yeast, but also portrays substantial difference. For one, while ER-derived COPII-coated vesicles in yeast fuse with Golgi membranes by heterotypic fusion (i.e., the fusion of membranes originating from different cellular compartments), mammalian COPII vesicles appear to congregate homotypically into a pre-Golgi intermediate structure, termed by some as vesicular tubular clusters (VTCs) [58]. Although there appears to exist only a single type of stable TRAPP complex in mammalian cells, a recent proteomic analysis of the autophagy interaction network in human cells identified a possible link with mammalian Trs85, or KIAA1012 [59]. The mammalian Ypt1 orthologue Rab1 exists in two closely related isoforms, Rab1a and Rab1b. A recent report by Zoppino et al. [37] showed that both the wild-type and a constitutively active mutant of Rab1b were observed to some degree in autophagic structures labeled with RFP-LC337. This Rab1b-LC3 colocalization could be enhanced by subjecting the cells to starvation and bafilomycin treatment, which promote the accumulation of autophagosomes by blocking their degradation. Over-expression of Rab1b increased the number of LC3-positive puncta, while both the dominant-negative form of Rab1b, Rab1bN121I, and Rab1b silencing reduced the number of autophagosomes. Interestingly, although the authors showed that both Sar1 and Rab1b are important for autophagy, the process is not inhibited by brefeldin A (BFA), which inhibits Arf GEFs and COP1-mediated retrograde processes in the early secretory pathway. BFA-treated cells are still able to elevate the number of LC3-labeled autophagosomes in response to cell starvation, which suggests that autophagosome formation is dependent on an intact trafficking pathway between the ER and Golgi, but perhaps not the retrograde transport process in the early secretory pathway.
In another report, Rab1 is shown to be important for antibacterial autophagy, a parallel mechanism of autophagy used by the cell for targeting intracellular pathogens [60]. Both GFP-tagged Rab1a and Rab1b could be found on LC3-positive autophagosomes of the intracellular pathogen Salmonella typhimurium, and this localization is confirmed by antibody against endogenous Rab1b. These authors also found that Rab1 activity is important for S. typhimurium autophagy and the latter is diminished by Rab1 silencing and dominant negative mutant. Interestingly, the importance of Rab1 is also extended to the autophagy of peroxisomes. In agreement with Zoppino et al., BFA was shown not to inhibit antibacterial autophagy. However, the authors also found, in contrast to earlier reports in yeast [36], that disruption of COPII function did not significantly affect autophagy of S. typhimurium. Whether this reflects a different requirement for different autophagic substrates, or a species difference in mechanistic details, remains to be confirmed. In spite of the advances noted above, the exact role of Rab1b in autophagosome generation remains undefined. Interestingly, another finding indicates that Rab1b’s close paralogue Rab1a may influence the localization of Atg9, the only membrane protein amongst the Atg family. Rab1a knockdown and α-synuclein over-expression (which impairs Rab1a activity) caused Atg9 to relocate from its normal perinuclear TGN location, to a diffuse distribution in cells [61]. Both of these manipulations also impaired autophagy. In this report, however, the authors’ silencing of Rab1b resulted in an increase in LC3II-labeled structures, which is in contrast with the results of the other reports. In fact, Rab1a’s role in cellular membrane traffic appears to be fairly elaborate and not restricted to the ER–Golgi region, and it has been implicated in transcytosis [62] and endocytosis [63]. Whether Rab1a and Rab1b regulate different steps of, or acts to mobilize materials from different membrane sources to effect autophagosome formation, is unclear at the moment.
Rab9 and autophagosome formation at the Golgi/TGN in non-canonical autophagy
Other than the conventional autophagic process, another non-canonical, Atg5/Atg7-independent autophagic process has been described [30]. In examining for the possibility of autophagic process in Atg5 −/− mouse embryonic fibroblasts, the authors found that in spite of being poorly induced by rapamycin, which is known to be a potent inducer of autophagy by inactivating mTOR, other cytotoxic stressors such as etoposide and starvation could still induce autophagosome formation in these cells. These autophagosomes could eventually mature to autolysosomes and are functionally active in terms of protein degradation. This Atg5-independent form of autophagy, as well as that induced in Atg7 −/− cells, does not involve lipidation of LC3 to form LC3II. However, these can still be inhibited by PI3 kinase inhibitors, and is still dependent on Ulk1 and beclin1. Importantly, the authors observed that this Atg5/Atg7-independent form of autophagy is in operation in vivo during embryonic development, and has an apparent function in clearing mitochondria during erythroid maturation, when erythrocytes undergo organelle clearance during terminal differentiation.
Compared to conventional autophagy, the alternative form of autophagy has some peculiar features. Unlike the predominantly ER association, these alternative autophagic vacuoles were localized near the Golgi apparatus, with some isolation membranes seen to have extended from the Golgi membranes. Closure of some isolation membranes appears to occur by fusion with vesicles with thicker membranes that may be derived from the trans-Golgi, or TGN. Most prominently, this alternative form of autophagy, unlike the conventional form, is inhibited by BFA, suggesting that it could be attenuated by inhibition of Golgi retrograde transport. The authors found that etoposide alters the localization pattern of the TGN marker TGN38, and the TGN-late endosome marker mannose 6-phosphate receptor, which suggests that Rab9 might play a role as the latter has long been known to be involved in late endosome–Golgi transport [64]. Prompted by the above observations to check the possible involvement of Rab9, the authors found significant association of Rab9 with Lamp2-positive autolysosomes in cells with induction of alternative autophagy. Constitutively active Rab9 has elevated, while dominant-negative mutant has reduced co-localization of Lamp2 in Atg5−/− cells. Rab9 silencing, while having no effect on conventional autophagy, appeared to decrease the number of etoposide-induced autophagic vacuoles in Atg5−/− cells, but also induced an accumulation of isolation membranes (which is not observed when Ulk1 was silenced). These observations suggest that a Rab9-mediated trafficking process may be required for autophagosome maturation in the alternative mode of autophagy.
The Golgi Rab24 and Rab33’s roles in autophagy
Two mammalian Golgi-associated Rabs have been implicated in the canonical autophagy process. Rab24, whose function in conventional exocytic traffic has been undefined, has a membrane localization that concentrates around the perinuclear region, which partially overlaps with both the ER and the Golgi. Starvation of cells caused a drastic change to the labeling pattern, with Rab24 redistributed to punctate spots, larger dots, ring-shaped small vesicles, as well as some elongated tubular-like structures [39]. These changes could be inhibited by both N-ethylmaleimide, which inhibits vesicular transport, and Wortmannin, which inhibits PI3 kinase activity. Rab24 colocalizes with autophagosome markers such as LC3, and the degree of colocalization increased upon starvation. Further, over-expression of Rab24 appeared to increase the number of autophagic vacuoles. These early observations indicate that Rab24 has a role in autophagy, perhaps in ways that are analogous to Rab1 and Rab9 described above. Another finding revealed an increase in Rab24 mRNA in nerve-injured hypoglossal motor neurons of rats [65]. Induction of LC3 mRNA and accumulation of LC3-II were also observed, with partial co-localization of Rab24 and LC3 observed in the injured neurons. Nerve injury could therefore upregulate autophagy events, and Rab24 appears to be part of this autophagic injury response.
Rab33A and Rab33B are two Golgi Rabs sharing a high degree of homology. Unlike the brain-specific Rab33A [66], Rab33B is ubiquitously expressed [67], and has been implicated in mediating intra-Golgi trafficking and retrograde Golgi–ER trafficking [68, 69]. Itoh et al. showed that both Rab33A and Rab33B interact with Atg16L in a GTP-dependent manner, and are involved in autophagy [40, 70]. Constitutively active, GTPase-deficient mutant of Rab33B enhanced LC3 lipidation, even under nutrient-rich conditions. Interestingly, this form of Rab33B appeared to inhibit basal or constitutive autophagy, as suggested by an accumulation of p62, an autophagic substrate. However, upon starvation, p62 is efficiently degraded. The implication of this observation is not yet clear, but suggests that basal and nutrient starvation-induced autophagy may differ slightly in their component requirements. Conversely, expression of the Rab33B-binding domain of Atg16L (to interfere with the endogenous Rab33B and Atg16L interaction) or silencing of Rab33B increased Atg12- and LC3-positive punctae. Rab33B’s specific interaction with Atg16L indicates that its role in autophagy may be distinct from that of Rab24 and the other Rabs.
Endocytic Rabs and the maturation of autophagic vesicles
The autophagic vesicle, once formed, matures via fusion with endocytic compartments [19], and several endosomal Rabs are expected to function in this regard. Surprisingly, the early endosomal Rab5 has been shown to act at an early stage of autophagosome formation, and its over-expression attenuated the toxicity induced by mutant huntingtin in cell culture and Drosophila models by ameliorating the buildup of toxic aggregates [42]. Rab5’s role in autophagic vesicle fusion to the early endosome compartment, if any, is not particularly clear. On the other hand, Rab11’s role in the autophagic process during erythrocyte maturation has been well documented by Fader et al. [45, 71, 72], using the erythroleukemia cell line K562 as a model. In these cells, Rab11 is found to label MVB membranes. Nutrient starvation and rapamycin caused an enlargement of the structures decorated with GFP-Rab11 and its colocalization with LC3, which was abrogated by a Rab11 dominant negative mutant [72].
As the functional Rab long recognized to be essential for processes involving late-endosomal/lysosomal fusion, Rab7 is expectedly involved in the late stages of autophagosome maturation [43, 44]. Rab7 is also important for the Group A streptococcus (GAS)-containing autophagosome-like vacuoles in infected cells [73]. The exact function of Rab7 in autophagosome maturation is hinted at by a recent report identifying a novel FYVE and coiled-coil (CC) domain-containing protein, FYCO1, as an LC3-interacting protein and Rab7 effector [74]. FYCO1 interacts with and decorates the autophagosome, and the endosomal-lysosome compartments. Recruited to the membrane by Rab7 and its FYVE domain interaction with PI3P, the Rab7–FYCO1 complex promotes microtubule plus end-directed transport of autophagic vesicles. Silencing of FYCO1 resulted in the accumulation of LC3-positive autophagosomal clusters at the perinuclear region, suggesting an inhibition of their transport. Rab7 could therefore function through its effector FYCO1 to promote microtubule based transport and eventual fusion of autophagosome with lysosomes.
RalB activation of autophagosome formation
The Ras-like GTPases RalA and RalB are prominent cancer-related proteins [75]. An interesting connection between the Rals and some Rab proteins is that they share an effector, a membrane tethering complex known as the exocyst complex [76]. The exocyst is an evolutionarily conserved, multimeric protein complex. Its principle function is to mediate the tethering of post-Golgi secretory vesicles to the plasma membrane, and is important for surface exocytosis in polarized cell types [77]. In a yeast interaction screen, Bodemann et al. identified modulators of autophagy, such as the beclin1 interacting proteins Atg14L and Rubicon [78], as well as FIP200, in the first-degree interaction neighborhood of the exocyst subunit Sec3 [79]. Considering the known association between the exocyst and RalA/B, the authors were prompted to check if these are involved in autophagy. Expression of an interaction disrupting minimal Ral-binding domain of a Ral effector suppressed formation of LC3-labeled punctae. Interestingly, siRNA-mediated depletion of RalB, but not RalA, in a stable GFP-LC3 expressing cell line significantly impaired GFP-LC3 signal accumulation and punctae formation. Consistent with this finding, RalB, but not RalA, is activated (i.e., becomes GTP-bound) by nutrient starvation. Ectopic expression of RalB was also sufficient to induce the accumulation of LC3 punctae.
The authors also found that beclin1 is recruited to distinct exocyst subcomplexes under different conditions of nutrient availability. In HEK293 cells, there is an abundance of Sec5/beclin1 complexes under nutrient-rich growth conditions. Nutrient starvation induced the assembly of Exo84/Beclin1 complexes, while Sec5/beclin1 complexes were disassembled within 90 min of nutrient deprivation. This phenomenon could be reversed by the addition of non-essential amino acids. On the other hand, Sec8/beclin1 complexes appeared to be constantly present under both nutrient-rich and nutrient-starved conditions. Vps34 associates with the above exocyst subunits in a manner that is reminiscent of that of beclin1. Expression of constitutively active RalB(G23V) could recapitulate the above observations in the absence of nutrient starvation. Furthermore, either nutrient starvation or RalB(G23V) expression was sufficient to induce assembly of ULK1/Exo84 complexes. Importantly, although ULK1 was present in both Exo84 and Sec5 complexes under nutrient-poor conditions, only Exo84-associated ULK1 displayed significant catalytic activity. Functioning in conjunction with the exocyst subunit Exo84 upon nutrient starvation, RalB therefore appears to induce the nucleation of both the catalytically active ULK1 complex and the beclin1-VPS34 complexes on the exocyst, thus promoting IM formation and maturation. The signaling pathways leading from nutrient starvation to RalB activation by specific RalB GEFs are not yet clear. Different subunits and subcomplexes of the exocyst also interact with members of the Rab and Rho family [80], potential crosstalk or cooperation between these GTPases in the autophagy process would provide interesting courses of investigation.
Small GTPases and autophagy in pathological conditions
Autophagy and its dysregulation and impairment have been extensively implicated in pathological conditions such as neurodegeneration [5, 6] and cancer [8, 9]. Autophagy is a key mechanism for clearing toxic cellular aggregates, and its impairment or an overwhelming of its clearance capacity underlies the pathology of several neurodegenerative diseases. A particularly prominent example is the involvement of α-synuclein in Parkinson’s disease (PD) [81, 82]. A protein of yet unclear endogenous function, α-synuclein gene mutations cause juvenile onset forms of PD, and insoluble aggregates of the protein is the major component of Lewy bodies, a distinct pathological feature of PD and other synucleinopathies [83]. Excessive amounts of α-synuclein or its mutants result in neural toxicity through the formation of oligomers and fibrillar aggregates that perturb critical cellular processes such as axonal transport. In the context of Rab’s regulation of autophagy, it has been shown that Rab1 appeared to be able to rescue neuronal loss in Parkinson’s disease models associated with α-synuclein toxicity [84]. A similar suppressive activity of α-synuclein toxicity has also been demonstrated for Rab3a and Rab8a [85].
The basis of how Rab1 over-expression could effectively attenuate impairments incurred by toxic α-synuclein oligomers has recently been highlighted by the finding that α-synuclein impairs neuronal autophagy, specifically affecting omegasome formation via Atg9 [61]. This effect of α-synuclein is likely through its inhibition of Rab1a activity, as both Rab1a silencing and α-synuclein overexpression changes Atg9 distribution. Interestingly, although the number of omegasomes is above that of normal basal condition in α-synuclein-expressing cells could be enhanced by Rab1a expression, the latter was unable to increase LC3 punctae in the presence of α-synuclein. This observation, in agreement with others, suggests that Rab1 functions early in autophagosome formation.
Dysregulation of autophagy has also been implicated in lysosomal storage disorders, which are metabolic disorders that result from defective lysosomal function. A typical example is Niemann–Pick Type C disease (NPC), which is characterized by an accumulation of cholesterol and sphingolipids in late endosomes and lysosomes. Patients with NPC develop progressive neurodegeneration, and are presented with ataxia, dystonia, and dementia. Mutations in npc1 or npc2, which are associated with cholesterol metabolism, have been implicated in the disease [86, 87]. Evidence suggests that the neurodegeneration observed in NPC is associated with dysregulated autophagy. Ko et al. have observed autophagic vacuoles and increased levels of LC3-II in Purkinje cells of mutant npc1 mice [88]. It is likely that the dysregulation of lipid trafficking triggers autophagy, which in turn results in cellular stress when the cell is unable to keep up with the demand on the autophagic system.
A link between Rabs, autophagy, and NPC comes with the observation that overexpression of certain Rabs rescue lipid trafficking defects in npc mutant fibroblasts [89, 90]. For example, Rab9 appears to have a protective effect in npc1 mutant mice, as evidenced by a decrease in ganglioside accumulation in the brain and an extended lifespan in the animals [91]. It appears that elevated endosomal cholesterol levels perturb the Rab activation cycle and result in trafficking defects that exacerbate the condition. Given the role of Rab9 in late endosome-to-Golgi retrograde transport, over-expression of Rab9 could speculatively restores the transport of cholesterol to the ER, where it is esterified, thus reducing the trapping of lipids in the late endosomes.
RalB’s now-recognized role in autophagy also opens up a new range of possible investigations to further understand the role of autophagy in cancer development. Although many details have yet to be worked out, it appears that RalB activation could functionally couple a variety of nutrient-signaling processes to autophagy. RalA and RalB could be activated by a family of GEFs, for which individual specificity and functional connection between them is not yet clearly established. Interestingly, RalA and RalB have been known to act antagonistically in terms of cancer cell migration [92]. This could likewise be the case for autophagy. RalA and its activator Ral guanine nucleotide dissociation stimulator (RalGDS) have been shown to be indispensable for activation of mammalian target of rapamycin complex 1 (mTORC1) [93], which inactivates Ulk1. The role of autophagy in cancer appears to be more complex than in the case of neurodegenerative diseases with toxic aggregates. Autophagic processes promote cell survival, and could be associated with tumorigenesis. On the other hand, autophagic cell death is a recognized mode of programmed cell death, and could, in the right context, acts as a tumor suppressive mechanism. RalB’s activation and its engagement of the exocyst affect multiple aspects of tumor cell behavior, particularly in the aspects of anchorage-dependent growth, migration, and invasion. It would be interesting to see how its roles in autophagy induction feature in synchrony with its other oncogenic activities.
Concluding remarks
In this short overview, we discussed our current knowledge with regards to the involvement of classical components of the eukaryotic membrane trafficking machinery in autophagy, with a focus on ER and Golgi Rabs (Fig. 1). The recent discovery of RalB and the exocyst’s roles in autophagy has added to the complexity of autophagy induction. While it is clear that these small GTPases are critical for the various stages of autophagosome formation and maturation, a clear molecular picture of how these regulate or interact with the core autophagic complexes is still lacking. Adding these molecular details, particularly pertaining to the hierarchical and temporal ordering of activities and functions, would be important pursuits in the immediate future. Further, it remains perplexing that multiple membrane trafficking components that are spread throughout the secretory pathway appear critical for autophagosome formation, which brings us back to the old, but yet unanswered question of the exact source of autophagosome membrane. It would appear that the cell could initiate IM formation from multiple places at the vicinity of the ER, Golgi, and mitochondria. A full understanding of the role of membrane flow mediated by Rabs in this process would be of both academic and translational importance.
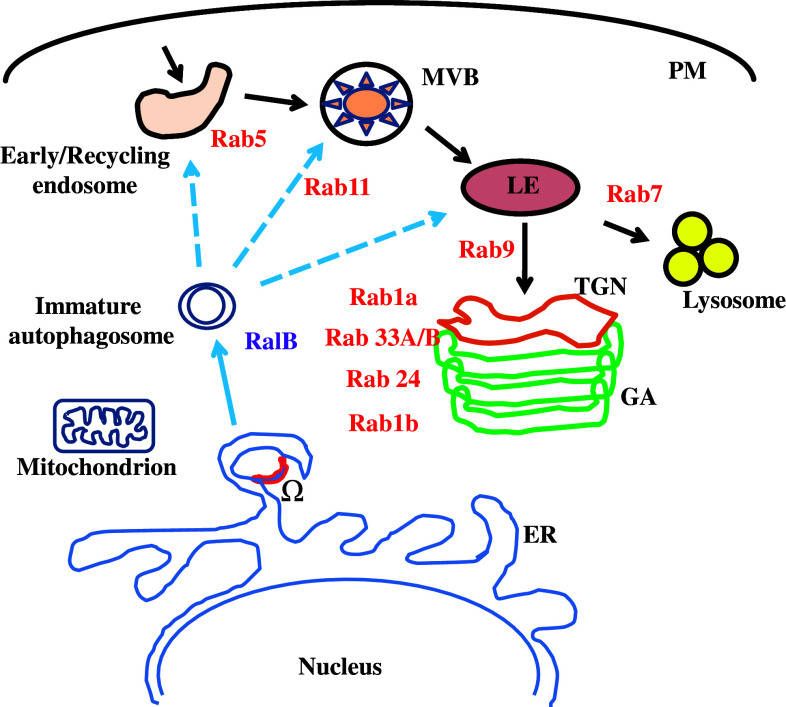
Fig. 1.
Schematic diagram illustrating the membrane compartments in the exocytic and endoscytic machinery that is connected to autophagosome formation and maturation. As depicted, the omegasome (Ω) is associated with the ER, but IM membranes could have a source in ER, Golgi, TGN, and the mitochondria. Also shown are the Rabs with a demonstrated involvement in autophagy, as well as RalB. The blue arrow indicates the generation of a double membrane enclosed immature autophagosome, or autophagic vesicle, from the ER. Blue dotted arrows point to potential endocytic compartments this vesicle could fuse with during its subsequent maturation. Black arrows indicate membrane traffic flow between the endocytic compartments. PM plasma membrane, MVB multivesicular bodies, LE late endosome, TGN trans-Golgi network, GA Golgi apparatus
Acknowledgements
Rab-associated work in BLT’s laboratory was supported by a grant from the Biomedical Research Council (08/1/21/19/533). SNARE-associated work was funded by an AcRF tier 1 grant from the Ministry of Education of Singapore (Project no. T13-0802-P13). CEC is a recipient of the NUS Graduate School of Integrative Sciences and Engineering (NGS) scholarship. The authors declare no financial conflicts of interest.
References
- 1.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mariño G, Madeo F, Kroemer G. Autophagy for tissue homeostasis and neuroprotection. Curr Opin Cell Biol. 2010;23:198–206. doi: 10.1016/j.ceb.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Banerjee R, Beal MF, Thomas B. Autophagy in neurodegenerative disorders: pathogenic roles and therapeutic implications. Trends Neurosci. 2010;33:541–549. doi: 10.1016/j.tins.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xilouri M, Stefanis L. Autophagy in the central nervous system: implications for neurodegenerative disorders. CNS Neurol Disord Drug Targets. 2010;9:701–719. doi: 10.2174/187152710793237421. [DOI] [PubMed] [Google Scholar]
- 7.Beau I, Mehrpour M, Codogno P. Autophagosomes and human diseases. Int J Biochem Cell Biol. 2011;43:460–464. doi: 10.1016/j.biocel.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Mathew R, White E. Autophagy in tumorigenesis and energy metabolism: friend by day, foe by night. Curr Opin Genet Dev. 2011;21:113–119. doi: 10.1016/j.gde.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenfeldt MT, Ryan KM (2011) The multiple roles of autophagy in cancer. Carcinogenesis [DOI] [PMC free article] [PubMed]
- 10.Reggiori F, Klionsky DJ. Autophagy in the eukaryotic cell. Eukaryotic Cell. 2002;1:11–21. doi: 10.1128/EC.01.1.11-21.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol. 2010;190:1005–1022. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamasaki M, Yoshimori T. Where do they come from? Insights into autophagosome formation. FEBS Lett. 2010;584:1296–1301. doi: 10.1016/j.febslet.2010.02.061. [DOI] [PubMed] [Google Scholar]
- 13.Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol. 2010;12:831–835. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- 14.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 16.Ylä-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 17.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Longatti A, Tooze SA. Vesicular trafficking and autophagosome formation. Cell Death Differ. 2009;16:956–965. doi: 10.1038/cdd.2009.39. [DOI] [PubMed] [Google Scholar]
- 19.Noda T, Fujita N, Yoshimori T. The late stages of autophagy: how does the end begin? Cell Death Differ. 2009;16:984–990. doi: 10.1038/cdd.2009.54. [DOI] [PubMed] [Google Scholar]
- 20.Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6:764–776. doi: 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6:79–90. doi: 10.1016/S1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 24.Young ARJ, Chan EYW, Hu XW, Köchl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- 25.Cheong H, Nair U, Geng J, Klionsky DJ. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae . Mol Biol Cell. 2008;19:668–681. doi: 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sekito T, Kawamata T, Ichikawa R, Suzuki K, Ohsumi Y. Atg17 recruits Atg9 to organize the pre-autophagosomal structure. Genes Cells Devot Mol Cell Mech. 2009;14:525–538. doi: 10.1111/j.1365-2443.2009.01299.x. [DOI] [PubMed] [Google Scholar]
- 27.Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol. 2009;186:773–782. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Vaart A, Griffith J, Reggiori F. Exit from the Golgi is required for the expansion of the autophagosomal phagophore in yeast Saccharomyces cerevisiae . Mol Biol Cell. 2010;21:2270–2284. doi: 10.1091/mbc.E09-04-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y, Shimizu S. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi Y, Meyerkord CL, Wang HG. Bif-1/endophilin B1: a candidate for crescent driving force in autophagy. Cell Death Differ. 2009;16:947–955. doi: 10.1038/cdd.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orsi A, Polson HEJ, Tooze SA. Membrane trafficking events that partake in autophagy. Curr Opin Cell Biol. 2010;22:150–156. doi: 10.1016/j.ceb.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 33.Metcalf D, Isaacs AM. The role of ESCRT proteins in fusion events involving lysosomes, endosomes and autophagosomes. Biochem Soc Trans. 2010;38:1469–1473. doi: 10.1042/BST0381469. [DOI] [PubMed] [Google Scholar]
- 34.Razi M, Chan EYW, Tooze SA. Early endosomes and endosomal coatomer are required for autophagy. J Cell Biol. 2009;185:305–321. doi: 10.1083/jcb.200810098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fader CM, Colombo MI. Autophagy and multivesicular bodies: two closely related partners. Cell Death Differ. 2009;16:70–78. doi: 10.1038/cdd.2008.168. [DOI] [PubMed] [Google Scholar]
- 36.Ishihara N, Hamasaki M, Yokota S, Suzuki K, Kamada Y, Kihara A, Yoshimori T, Noda T, Ohsumi Y. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol Biol Cell. 2001;12:3690–3702. doi: 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zoppino FCM, Militello RD, Slavin I, Alvarez C, Colombo MI. Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic. 2010;11:1246–1261. doi: 10.1111/j.1600-0854.2010.01086.x. [DOI] [PubMed] [Google Scholar]
- 38.Li L, Kim E, Yuan H, Inoki K, Goraksha-Hicks P, Schiesher RL, Neufeld TP, Guan KL. Regulation of mTORC1 by the Rab and Arf GTPases. J Biol Chem. 2010;285:19705–19709. doi: 10.1074/jbc.C110.102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Munafó DB, Colombo MI. Induction of autophagy causes dramatic changes in the subcellular distribution of GFP-Rab24. Traffic. 2002;3:472–482. doi: 10.1034/j.1600-0854.2002.30704.x. [DOI] [PubMed] [Google Scholar]
- 40.Itoh T, Fujita N, Kanno E, Yamamoto A, Yoshimori T, Fukuda M. Golgi-resident small GTPase Rab33B interacts with Atg16L and modulates autophagosome formation. Mol Biol Cell. 2008;19:2916–2925. doi: 10.1091/mbc.E07-12-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Renna M, Schaffner C, Winslow AR, Menzies FM, Peden AA, Floto RA, Rubinsztein DC. Autophagic substrate clearance requires activity of the syntaxin-5 SNARE complex. J Cell Sci. 2011;124:469–482. doi: 10.1242/jcs.076489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ravikumar B, Imarisio S, Sarkar S, O’Kane CJ, Rubinsztein DC. Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J Cell Sci. 2008;121:1649–1660. doi: 10.1242/jcs.025726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gutierrez MG, Munafó DB, Berón W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- 44.Jäger S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, Eskelinen EL. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 45.Fader CM, Sánchez DG, Mestre MB, Colombo MI. TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim Biophys Acta. 2009;1793:1901–1916. doi: 10.1016/j.bbamcr.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 46.Furuta N, Fujita N, Noda T, Yoshimori T, Amano A. Combinational soluble N-ethylmaleimide-sensitive factor attachment protein receptor proteins VAMP8 and Vti1b mediate fusion of antimicrobial and canonical autophagosomes with lysosomes. Mol Biol Cell. 2010;21:1001–1010. doi: 10.1091/mbc.E09-08-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwon SI, Cho HJ, Jung JH, Yoshimoto K, Shirasu K, Park OK. The Rab GTPase RabG3b functions in autophagy and contributes to tracheary element differentiation in Arabidopsis. Plant J Cell Mol Biol. 2010;64:151–164. doi: 10.1111/j.1365-313X.2010.04315.x. [DOI] [PubMed] [Google Scholar]
- 48.Plutner H, Cox AD, Pind S, Khosravi-Far R, Bourne JR, Schwaninger R, Der CJ, Balch WE. Rab1b regulates vesicular transport between the endoplasmic reticulum and successive Golgi compartments. J Cell Biol. 1991;115:31–43. doi: 10.1083/jcb.115.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Antoni A, Schmitzová J, Trepte HH, Gallwitz D, Albert S. Significance of GTP hydrolysis in Ypt1p-regulated endoplasmic reticulum to Golgi transport revealed by the analysis of two novel Ypt1-GAPs. J Biol Chem. 2002;277:41023–41031. doi: 10.1074/jbc.M205783200. [DOI] [PubMed] [Google Scholar]
- 50.Lynch-Day MA, Bhandari D, Menon S, Huang J, Cai H, Bartholomew CR, Brumell JH, Ferro-Novick S, Klionsky DJ. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci USA. 2010;107:7811–7816. doi: 10.1073/pnas.1000063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jones S, Newman C, Liu F, Segev N. The TRAPP complex is a nucleotide exchanger for Ypt1 and Ypt31/32. Mol Biol Cell. 2000;11:4403–4411. doi: 10.1091/mbc.11.12.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang W, Sacher M, Ferro-Novick S. TRAPP stimulates guanine nucleotide exchange on Ypt1p. J Cell Biol. 2000;151:289–296. doi: 10.1083/jcb.151.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barrowman J, Bhandari D, Reinisch K, Ferro-Novick S. TRAPP complexes in membrane traffic: convergence through a common Rab. Nature reviews. Mol Cell Biol. 2010;11:759–763. doi: 10.1038/nrm2999. [DOI] [PubMed] [Google Scholar]
- 54.Sacher M, Barrowman J, Wang W, Horecka J, Zhang Y, Pypaert M, Ferro-Novick S. TRAPP I implicated in the specificity of tethering in ER-to-Golgi transport. Mol Cell. 2001;7:433–442. doi: 10.1016/S1097-2765(01)00190-3. [DOI] [PubMed] [Google Scholar]
- 55.Yamasaki A, Menon S, Yu S, Barrowman J, Meerloo T, Oorschot V, Klumperman J, Satoh A, Ferro-Novick S. mTrs130 is a component of a mammalian TRAPPII complex, a Rab1 GEF that binds to COPI-coated vesicles. Mol Biol Cell. 2009;20:4205–4215. doi: 10.1091/mbc.E09-05-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meiling-Wesse K, Epple UD, Krick R, Barth H, Appelles A, Voss C, Eskelinen EL, Thumm M. Trs85 (Gsg1), a component of the TRAPP complexes, is required for the organization of the preautophagosomal structure during selective autophagy via the Cvt pathway. J Biol Chem. 2005;280:33669–33678. doi: 10.1074/jbc.M501701200. [DOI] [PubMed] [Google Scholar]
- 57.Nazarko TY, Huang J, Nicaud JM, Klionsky DJ, Sibirny AA. Trs85 is required for macroautophagy, pexophagy and cytoplasm to vacuole targeting in Yarrowia lipolytica and Saccharomyces cerevisiae . Autophagy. 2005;1:37–45. doi: 10.4161/auto.1.1.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bannykh SI, Nishimura N, Balch WE. Getting into the Golgi. Trends Cell Biol. 1998;8:21–25. doi: 10.1016/S0962-8924(97)01184-7. [DOI] [PubMed] [Google Scholar]
- 59.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang J, Birmingham CL, Shahnazari S, Shiu J, Zheng YT, Smith AC, Campellone KG, Heo WD, Gruenheid S, Meyer T, Welch MD, Ktistakis NT, Kim PK, Klionsky DJ, Brumell JH. Antibacterial autophagy occurs at PI(3)P-enriched domains of the endoplasmic reticulum and requires Rab1 GTPase. Autophagy. 2011;7:17–26. doi: 10.4161/auto.7.1.13840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winslow AR, Chen CW, Corrochano S, Acevedo-Arozena A, Gordon DE, Peden AA, Lichtenberg M, Menzies FM, Ravikumar B, Imarisio S, Brown S, O’Kane CJ, Rubinsztein DC. α-Synuclein impairs macroautophagy: implications for Parkinson’s disease. J Cell Biol. 2010;190:1023–1037. doi: 10.1083/jcb.201003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jin M, Saucan L, Farquhar MG, Palade GE. Rab1a and multiple other Rab proteins are associated with the transcytotic pathway in rat liver. J Biol Chem. 1996;271:30105–30113. doi: 10.1074/jbc.271.47.30158. [DOI] [PubMed] [Google Scholar]
- 63.Mukhopadhyay A, Nieves E, Che FY, Wang J, Jin L, Murray JW, Gordon K, Angeletti RH, Wolkoff AW. Proteomic analysis of endocytic vesicles: Rab1a regulates motility of early endocytic vesicles. J Cell Sci. 2011;124:765–775. doi: 10.1242/jcs.079020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barbero P, Bittova L, Pfeffer SR. Visualization of Rab9-mediated vesicle transport from endosomes to the trans-Golgi in living cells. J Cell Biol. 2002;156:511–518. doi: 10.1083/jcb.200109030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Egami Y, Kiryu-Seo S, Yoshimori T, Kiyama H. Induced expressions of Rab24 GTPase and LC3 in nerve-injured motor neurons. Biochem Biophys Res Commun. 2005;337:1206–1213. doi: 10.1016/j.bbrc.2005.09.171. [DOI] [PubMed] [Google Scholar]
- 66.Zheng JY, Koda T, Arimura Y, Kishi M, Kakinuma M. Structure and expression of the mouse S10 gene. Biochim Biophys Acta. 1997;1351:47–50. doi: 10.1016/s0167-4781(97)00022-5. [DOI] [PubMed] [Google Scholar]
- 67.Zheng JY, Koda T, Fujiwara T, Kishi M, Ikehara Y, Kakinuma M. A novel Rab GTPase, Rab33B, is ubiquitously expressed and localized to the medial Golgi cisternae. J Cell Sci. 1998;111(Pt 8):1061–1069. doi: 10.1242/jcs.111.8.1061. [DOI] [PubMed] [Google Scholar]
- 68.Valsdottir R, Hashimoto H, Ashman K, Koda T, Storrie B, Nilsson T. Identification of rabaptin-5, rabex-5, and GM130 as putative effectors of rab33b, a regulator of retrograde traffic between the Golgi apparatus and ER. FEBS Lett. 2001;508:201–209. doi: 10.1016/S0014-5793(01)02993-3. [DOI] [PubMed] [Google Scholar]
- 69.Starr T, Sun Y, Wilkins N, Storrie B. Rab33b and Rab6 are functionally overlapping regulators of Golgi homeostasis and trafficking. Traffic. 2010;11:626–636. doi: 10.1111/j.1600-0854.2010.01051.x. [DOI] [PubMed] [Google Scholar]
- 70.Fukuda M, Itoh T. Direct link between Atg protein and small GTPase Rab: Atg16L functions as a potential Rab33 effector in mammals. Autophagy. 2008;4:824–826. doi: 10.4161/auto.6542. [DOI] [PubMed] [Google Scholar]
- 71.Fader CM, Savina A, Sánchez D, Colombo MI. Exosome secretion and red cell maturation: Exploring molecular components involved in the docking and fusion of multivesicular bodies in K562 cells. Blood Cells Mol Dis. 2005;35:153–157. doi: 10.1016/j.bcmd.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 72.Fader CM, Sánchez D, Furlán M, Colombo MI. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic. 2008;9:230–250. doi: 10.1111/j.1600-0854.2007.00677.x. [DOI] [PubMed] [Google Scholar]
- 73.Yamaguchi H, Nakagawa I, Yamamoto A, Amano A, Noda T, Yoshimori T. An initial step of GAS-containing autophagosome-like vacuoles formation requires Rab7. PLoS pathogens. 2009;5:e1000670. doi: 10.1371/journal.ppat.1000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pankiv S, Alemu EA, Brech A, Bruun JA, Lamark T, Overvatn A, Bjørkøy G, Johansen T. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol. 2010;188:253–269. doi: 10.1083/jcb.200907015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bodemann BO, White MA. Ral GTPases and cancer: linchpin support of the tumorigenic platform. Nature reviews. Cancer. 2008;8:133–140. doi: 10.1038/nrc2296. [DOI] [PubMed] [Google Scholar]
- 76.Camonis JH, White MA. Ral GTPases: corrupting the exocyst in cancer cells. Trends Cell Biol. 2005;15:327–332. doi: 10.1016/j.tcb.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 77.He B, Guo W. The exocyst complex in polarized exocytosis. Curr Opin Cell Biol. 2009;21:537–542. doi: 10.1016/j.ceb.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, Akira S, Noda T, Yoshimori T. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 79.Bodemann BO, Orvedahl A, Cheng T, Ram RR, Ou YH, Formstecher E, Maiti M, Hazelett CC, Wauson EM, Balakireva M, Camonis JH, Yeaman C, Levine B, White MA. RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell. 2011;144:253–267. doi: 10.1016/j.cell.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Novick P, Guo W. Ras family therapy: Rab, Rho and Ral talk to the exocyst. Trends Cell Biol. 2002;12:247–249. doi: 10.1016/S0962-8924(02)02293-6. [DOI] [PubMed] [Google Scholar]
- 81.Auluck PK, Caraveo G, Lindquist S. α-Synuclein: membrane interactions and toxicity in Parkinson’s disease. Annu Rev Cell Dev Biol. 2010;26:211–233. doi: 10.1146/annurev.cellbio.042308.113313. [DOI] [PubMed] [Google Scholar]
- 82.Chua CEL, Tang BL (2011) Rabs, SNAREs and α-synuclein—membrane trafficking defects in synucleinopathies. Brain Res Rev [DOI] [PubMed]
- 83.Uversky VN. Alpha-synuclein misfolding and neurodegenerative diseases. Curr Protein Pept Sci. 2008;9:507–540. doi: 10.2174/138920308785915218. [DOI] [PubMed] [Google Scholar]
- 84.Cooper AA, Gitler AD, Cashikar A, Haynes CM, Hill KJ, Bhullar B, Liu K, Xu K, Strathearn KE, Liu F, Cao S, Caldwell KA, Caldwell GA, Marsischky G, Kolodner RD, Labaer J, Rochet JC, Bonini NM, Lindquist S. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson’s models. Science. 2006;313:324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gitler AD, Bevis BJ, Shorter J, Strathearn KE, Hamamichi S, Su LJ, Caldwell KA, Caldwell GA, Rochet JC, McCaffery JM, Barlowe C, Lindquist S. The Parkinson’s disease protein alpha-synuclein disrupts cellular Rab homeostasis. Proc Natl Acad Sci USA. 2008;105:145–150. doi: 10.1073/pnas.0710685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Loftus SK, Morris JA, Carstea ED, Gu JZ, Cummings C, Brown A, Ellison J, Ohno K, Rosenfeld MA, Tagle DA, Pentchev PG, Pavan WJ. Murine model of Niemann–Pick C disease: mutation in a cholesterol homeostasis gene. Science. 1997;277:232–235. doi: 10.1126/science.277.5323.232. [DOI] [PubMed] [Google Scholar]
- 87.Naureckiene S, Sleat DE, Lackland H, Fensom A, Vanier MT, Wattiaux R, Jadot M, Lobel P. Identification of HE1 as the second gene of Niemann–Pick C disease. Science. 2000;290:2298–2301. doi: 10.1126/science.290.5500.2298. [DOI] [PubMed] [Google Scholar]
- 88.Ko DC, Milenkovic L, Beier SM, Manuel H, Buchanan J, Scott MP. Cell-autonomous death of cerebellar purkinje neurons with autophagy in Niemann–Pick type C disease. PLoS Genet. 2005;1:81–95. doi: 10.1371/journal.pgen.0010081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Narita K, Choudhury A, Dobrenis K, Sharma DK, Holicky EL, Marks DL, Walkley SU, Pagano RE. Protein transduction of Rab9 in Niemann–Pick C cells reduces cholesterol storage. FASEB J Off Publ Fed Am Soc Exp Biol. 2005;19:1558–1560. doi: 10.1096/fj.04-2714fje. [DOI] [PubMed] [Google Scholar]
- 90.Linder MD, Uronen RL, Hölttä-Vuori M, van der Sluijs P, Peränen J, Ikonen E. Rab8-dependent recycling promotes endosomal cholesterol removal in normal and sphingolipidosis cells. Mol Biol Cell. 2007;18:47–56. doi: 10.1091/mbc.E06-07-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kaptzan T, West SA, Holicky EL, Wheatley CL, Marks DL, Wang T, Peake KB, Vance J, Walkley SU, Pagano RE. Development of a Rab9 transgenic mouse and its ability to increase the lifespan of a murine model of Niemann–Pick type C disease. Am J Pathol. 2009;174:14–20. doi: 10.2353/ajpath.2009.080660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Oxford G, Owens CR, Titus BJ, Foreman TL, Herlevsen MC, Smith SC, Theodorescu D. RalA and RalB: antagonistic relatives in cancer cell migration. Cancer Res. 2005;65:7111–7120. doi: 10.1158/0008-5472.CAN-04-1957. [DOI] [PubMed] [Google Scholar]
- 93.Maehama T, Tanaka M, Nishina H, Murakami M, Kanaho Y, Hanada K. RalA functions as an indispensable signal mediator for the nutrient-sensing system. J Biol Chem. 2008;283:35053–35059. doi: 10.1074/jbc.M805822200. [DOI] [PMC free article] [PubMed] [Google Scholar]