Abstract
8-hydroxy-4-methyl-9-nitrobenzo(g)chromen-2-one (NBC) has been found to be a fairly potent ATP site-directed inhibitor of protein kinase CK2 (Ki = 0.22 μM). Here, we show that NBC also inhibits PIM kinases, especially PIM1 and PIM3, the latter as potently as CK2. Upon removal of the nitro group, to give 8-hydroxy-4-methyl-benzo(g)chromen-2-one (here referred to as “denitro NBC”, dNBC), the inhibitory power toward CK2 is almost entirely lost (IC50 > 30 μM) whereas that toward PIM1 and PIM3 is maintained; in addition, dNBC is a potent inhibitor of a number of other kinases that are weakly inhibited or unaffected by NBC, with special reference to DYRK1A whose IC50 values with NBC and dNBC are 15 and 0.60 μM, respectively. Therefore, the observation that NBC, unlike dNBC, is a potent inducer of apoptosis is consistent with the notion that this effect is mediated by inhibition of endogenous CK2. The structural features underlying NBC selectivity have been revealed by inspecting its 3D structure in complex with the catalytic subunit of Z. mays CK2. The crucial role of the nitro group is exerted both through a direct electrostatic interaction with the side chain of Lys68 and, indirectly, by enhancing the acidic dissociation constant of the adjacent hydroxyl group which interacts with a conserved water molecule in the deepest part of the cavity. By contrast, the very same nitro group is deleterious for the binding to the active site of DYRK1A, as disclosed by molecular docking. This provides the rationale for preferential inhibition of DYRK1A by dNBC.
Keywords: Protein kinase CK2, Kinase inhibitors, PIM kinases, DYRK1A, PKB
Introduction
The increasing realization that most human diseases are related to “communication disorders”, resulting from alterations in signalling pathways, accounts for the observation that the largest individual component of the “druggable genome” is provided by the family of protein kinases, collectively referred to as the human “kinome” [1, 2]. As a general rule, protein kinases are signalling molecules par excellence, inactive under basal conditions, whose activity is evoked by specific stimuli. Mutations in protein kinases that result in inappropriate activity, such as gain of function, often underlie global pathologies, as exemplified by the large proportion of the kinome related to human diseases, currently including more than 150 members (http://www.cellsignal.com/reference/kinase_disease.html). In particular the “onco-kinome” is composed of a variety of kinases whose abnormal activity, promoted by genetic alterations or overexpression, is a cause of neoplasia.
Such a paradigm does not apply to the most pleiotropic member of the kinome, protein kinase CK2 (an acronym derived from the misnomer “casein kinase-2”), which is individually responsible for the generation of a large proportion of the human phosphoproteome [3, 4]. The catalytic subunits of CK2 (α and/or α’), which are very similar but encoded by two distinct genes, are constitutively active either by themselves or when they form a heterodimer with the non-catalytic β-subunit. Moreover, mutations causing gain of function of CK2 have never been reported, arguing against the concept that CK2 might be an oncogene in the strict sense of the term. Nevertheless, CK2 is clearly implicated in many of the phenomena associated with cancer, as reviewed elsewhere [5–8], and cancer cells have often been shown to be more dependent on abnormally high CK2 levels than their normal cellular counterparts (e.g. [9–11]). In an attempt to reconcile the undeniable cancer driving potential of CK2 with its lack of features that are typical of kinases encoded by oncogenes, it has recently been proposed [8] that the over-reliance of the tumour phenotype on high levels of CK2 is an example of “non-oncogene addiction” [12], a mechanism which applies to situations where a protein that is not subjected to genetic alterations becomes over-represented in the cell to such a degree that it causes an undue reliance on itself. This would match perfectly with the recurrent observation that CK2 levels are invariably more elevated in cancer cells compared to their “normal” counterparts [13].
Given these premises, it is not surprising that many efforts have been made to develop potent, selective and, most important, cell permeable inhibitors of CK2 (reviewed in [14–17]). Some of these are now available commercially and have proved useful in dissecting cellular functions affected by CK2 and in validating putative endogenous targets of this extremely pleiotropic kinase. Treating cancer cells with these compounds often results in accelerated apoptosis, and one of them, CX-4945, developed by Cylene Pharmaceuticals, has been approved by the Food and Drug Administration (FDA) to enter phase I clinical trials for the treatment of advanced solid tumours, Castelman’s disease and multiple myeloma (http://www.cylenepharma.com/cylene/cx_4945). CX-4945, as well as nearly all the other cell permeable CK2 inhibitors, are competitive with respect to the co-substrate ATP. The 3D structures of some of these compounds in complex with CK2 have been solved (reviewed by Battistutta [18]), providing useful information for the development of even more potent and selective compounds. Given the large number of protein kinases (>500) and the high conservation of their ATP binding sites, achieving selectivity is a formidable task, especially in the case of inhibitors that target the ATP binding site. The CK2 inhibitors most frequently used for cellular studies belong to the class of polyhalogenated benzimidazole (or triazole or pyrazole) derivatives, notably DRB, TBB, TBI and DMAT, whose selectivity, once they started to be profiled against large panels of protein kinases (>50), proved to be not as narrow as previously believed [19]. In particular, it became clear that some classes of protein kinases not previously included in these panels, with special reference to DYRKs, PIMs, HIPK2 and ERK8, tend to be inhibited by these compounds as drastically as CK2. On the other hand, less attention has been devoted to another class of CK2 inhibitors, i.e. condensed polyphenolic compounds, with special reference to those derived from the scaffold of coumarin [20]. One of these, NBC (8-hydroxy-4-methyl-9-nitrobenzo(g)chromen-2-one), was previously shown to be a fairly potent and selective inhibitor of CK2 when tested on a panel of 33 protein kinases [21]. Here, we confirm the remarkable selectivity of NBC by profiling it on a panel of 125 kinases, and we provide both biochemical and structural evidence that such a selectivity is critically reliant on the nitro group of NBC, whose removal gives rise to dNBC, a potent inhibitor of kinases DYRK1A, ERK8 and GSK3β, which is nearly inactive toward CK2.
Materials and methods
Inhibitors
The structure of NBC and dNBC, also previously termed C12 and C11, respectively, dealt with by [20, 21] is shown in Fig. 1.
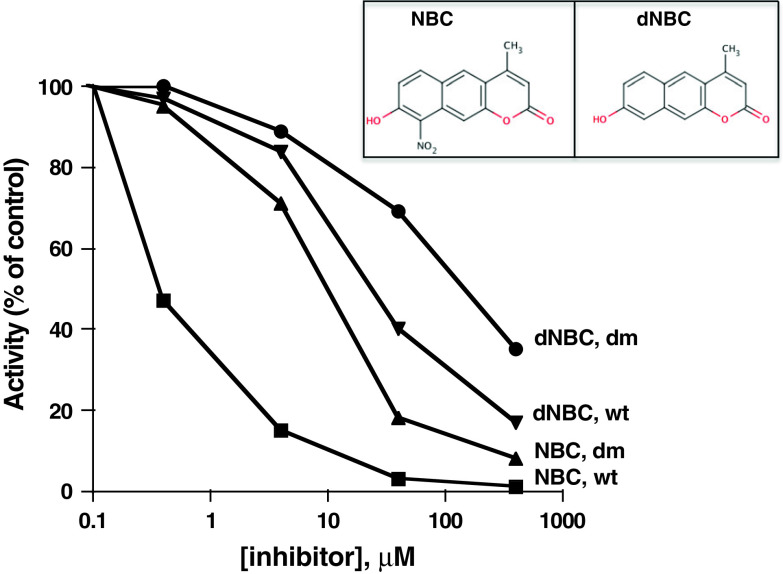
Fig. 1.
Dose-dependent inhibition of recombinant CK2 w.t. by NBC and dNBC and CK2α V66I174A by NBC CK2 activity was assayed either in absence or in the presence of increasing concentration of the inhibitors (whose structure is shown in the inset), as indicated. dm refers to the CK2α double mutant in which Val66 and Ile174 were replaced by alanine. Phosphorylation conditions are described in the text
Source and purification of protein kinases
Native CK2 was purified from rat liver [22]. Human recombinant α and β subunits of CK2 were expressed in Escherichia coli, and the holoenzyme was reconstituted and purified as described previously [23]. A double mutant of CK2α subunit in which V66 and I174 were replaced by alanine was generated as reported in [24]. The expression plasmid of GST-DYRK1A (residues 1–499), dual specificity tyrosine-phosphorylated and regulated kinase, was kindly provided by Dr. W. Becker (Inst. fuer Pharmakologie und Toxikologie, Aachen, Germany). Expression of Glutathione S-transferase (GST) fusion protein was induced with 0.5 μM isopropyl-1-thio-β-D-galactopyranoside in E. coli BL21(DE3) for 5 h at 37°C. Bacteria pellets were resuspended in PBS buffer (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4) and sonicated (6 × 20 s) in ice. Bacteria extract was loaded on a glutathione-Sepharose 4B as described by the manufacter (Pharmacia) and an overnight on-column cleavage with thrombin was performed. All the fractions containing DYRK1A were pooled and loaded on Superdex 75 Hi Load 26/60 (Pharmacia) in 0.5 M NaCl PBS buffer. The fractions in the OD peaks were assayed for DYRK1A activity, pooled, concentrated to 8 mg/ml and stored at −80°C. The source of all the other protein kinases used for specificity assays is as referenced in [25].
Phosphorylation assays
Native CK2 purified from rat liver and recombinat CK2 αw.t. and αV66I174A (0.5–1 pmol) were tested in a final volume of 25 μl containing 50 mM Tris/HCl (pH 7.5), 100 mM NaCl, 12 mM MgCl2, 100 μM synthetic peptide substrate RRRADDSDDDDD and 0.02 mM [γ−33P]ATP (500–1,000 c.p.m./pmol), and incubated for 10 min at 37°C. Assays were stopped by addition of 5 μl of 0.5 M orthophosphoric acid before spotting 20-μl aliquots onto phosphocellulose filters. Activity of DYRK1A (50 ng) was tested on 100 μM DYRKtide [26] for 10 min at 30°C in phosphorylation buffer containing 50 mM Tris, pH 7.5, 12 mM MgCl2 and 0.02 mM ATP [γ −33P]ATP (500–1,000 c.p.m./pmol) in a final volume of 30 μl. Assays were stopped by spotting 25 μl onto phosphocellulose filters. Filters were washed in 75 mM phosphoric acid (5–10 ml each) four times then once in methanol and dried before counting. Conditions for the activity assays of other protein kinases tested in selectivity experiments and in the determination of IC50 values of ERK8, GSK3β, PKBα and PKBβ were as described or referenced in [25].
Kinetic determinations
Initial velocities were determined at each of the substrate concentration tested. Km values were calculated either in the absence or in the presence of increasing concentrations of inhibitor, from Lineweaver–Burk double-reciprocal plots of the data. Inhibition constants were then calculated by linear regression analysis of Km/Vmax against inhibitor concentration plots.
Promiscuity score and Gini coefficient
Promiscuity score [19] and Gini coefficient [27] were determined as described elsewhere.
Cell viability assays
The human leukemia Jurkat and lymphoblastoid CEM T-cell lines were maintained in RPMI-1640, supplemented with 10% (v/v) foetal calf serum, 2 mM l-glutamine, 100 units/ml penicillin and 100 μg/ml streptomycin. For the treatment, cells were suspended at a density of 106 cells/ml in a medium containing 1% (v/v) foetal calf serum, then incubated at 37°C, in the presence of the compounds at the indicated concentrations. Control cells were treated with equal amounts of solvent (0.5% v/v DMSO). Cell viability was assessed in Jurkat cells by means of 3-(4,5-dimethylthiazol-2-yl)-3,5-diphenyltriazolium bromide (MTT) reagent, while apoptosis/necrosis were evaluated in CEM cells by means of the Cell Detection Elisa kit (Roche), based on the quantification of nucleosomes released in the cytosol or in the medium by apoptotic or necrotic cells, respectively. Ten thousand cells were used for each determination, and the manufacturer’s instructions were followed.
Crystal preparation and data collection
The recombinant CK2α-subunit from Zea mays was expressed in Escherichia coli, isolated and purified according to a previously described method [28]. Crystals of the CK2 complex with the inhibitor NBC were obtained by co-crystallisation with the sitting drop vapour-diffusion technique. An 8 mg/ml protein stock solution (in 25 mM Tris–Cl, 500 mM NaCl, 7 mM 2-mercaptoethanol, pH 8.5) was preincubated with 30 mM inhibitor solution (100% DMSO) in the proper amount to have an inhibitor–protein molar ratio of about 3:1, and not to exceed a 5% DMSO concentration in the final protein solution. Crystallisation drops were prepared by mixing a 1-μl drop of protein/NBC solution with 2 μl of water and 1 μl of precipitant solution (10–20% PEG 4000, sodium acetate 0.2 M, Tris 0.1 M pH 8). The drop was equilibrated against 500 μl of the same precipitant solution (20% PEG 4000). Crystals grew in a few days at 293 K.
X-ray diffraction data were collected at ESRF beamline ID13 in Grenoble, at a temperature of 100 K. Crystals were cryo-protected by immersion oil type B (Hampton Research). Data were indexed with MOSFLM v.6.2.6 [29] and then scaled with SCALA [30] from the CCP4 software package [31].
Structure determination and refinement
The CK2-NBC complex crystallises in space group C2, with one molecule in the asymmetric unit. To find the best position inside the unit cell, an initial rigid body transformation on the model of the apo-enzyme (PDB code 1JAM) was adequate. Refinement was carried out using Phenix v.1.6.4 [32], alternating automated cycles and manual inspection steps using the graphic program Coot v 0.6.2 [33]. Data collection and final model statistics are reported in Table 3 (below).
Table 3.
Data collection and final model statistics
| Space group | C2 |
|---|---|
| Data collection | |
| Cell dimensions | |
| a, b, c (Å) | 142.41, 60.96, 45.66 |
| α, β, γ (°) | 90.0, 100.19, 90.0 |
| Total number of observations | 32,616 (4,776) |
| Total number unique | 16,400 (2,491) |
| Resolution (Å) | 55.90–2.20 (2.32–2.20) |
| Rsym (%) | 12.4 (60.0) |
| Mean I/σ(I) | 4.4 (1.6) |
| Completeness (%) | 83.9 (87.4) |
| Multiplicity | 2.0 (1.9) |
| Bwilson (Å2) | 30.8 |
| Solvent content (%) | 54.6 |
| Refinement | |
| Resolution (Å) | 41.3–2.20 (2.34–2.20) |
| No. of reflections (reference set) | 16,393 |
| Rcrys/Rfree | 16.9/23.0 (22.1/29.4) |
| No. atoms | |
| All atoms | 2,863 |
| Protein | 2,728 |
| Ligands: NBC | 20 |
| Water | 115 |
| Root mean square deviations from ideality | |
| Bond lengths | 0.008 |
| Bond Angles | 1.125 |
| Ramachandran plot statistics (%) (excl. Gly, Pro) | |
| Most favored regions | 89.9 |
| Additionally allowed regions | 9.8 |
| Generously allowed regions | 0 |
| Disallowed regions | 0.3 |
Numbers in parentheses refer to the highest resolution bin
Data deposition
The coordinates for the model of CK2 in complex with NBC have been deposited at the RCSB Protein Data Bank with ID code 3PWD.
Molecular modelling
The crystal structure of human DYRK1A was retrieved from the PDB (PDB code 2VX3) and processed in order to remove the ligands and water molecules. Hydrogen atoms were added to the protein structure using standard geometries with the MOE program. To minimise contacts between hydrogens, the structures were subjected to Amber99 force-field minimisation until the rms (root mean square) of conjugate gradient was <0.1 kcal mol−1 Å−1 (1 Å = 0.1 nm) keeping the heavy atoms fixed at their crystallographic positions. To strictly validate the model generated and to calibrate the high-throughput docking protocol, a small database of known DYRK1A inhibitors was built and a set of docking runs was performed. After the calibration phase, flexible ligand-docking steps with three different programs, MOEDock, Glide and Gold, were performed essentially as described previously [34].
Results
Mode of inhibition of CK2 by NBC: relevance of the nitro group
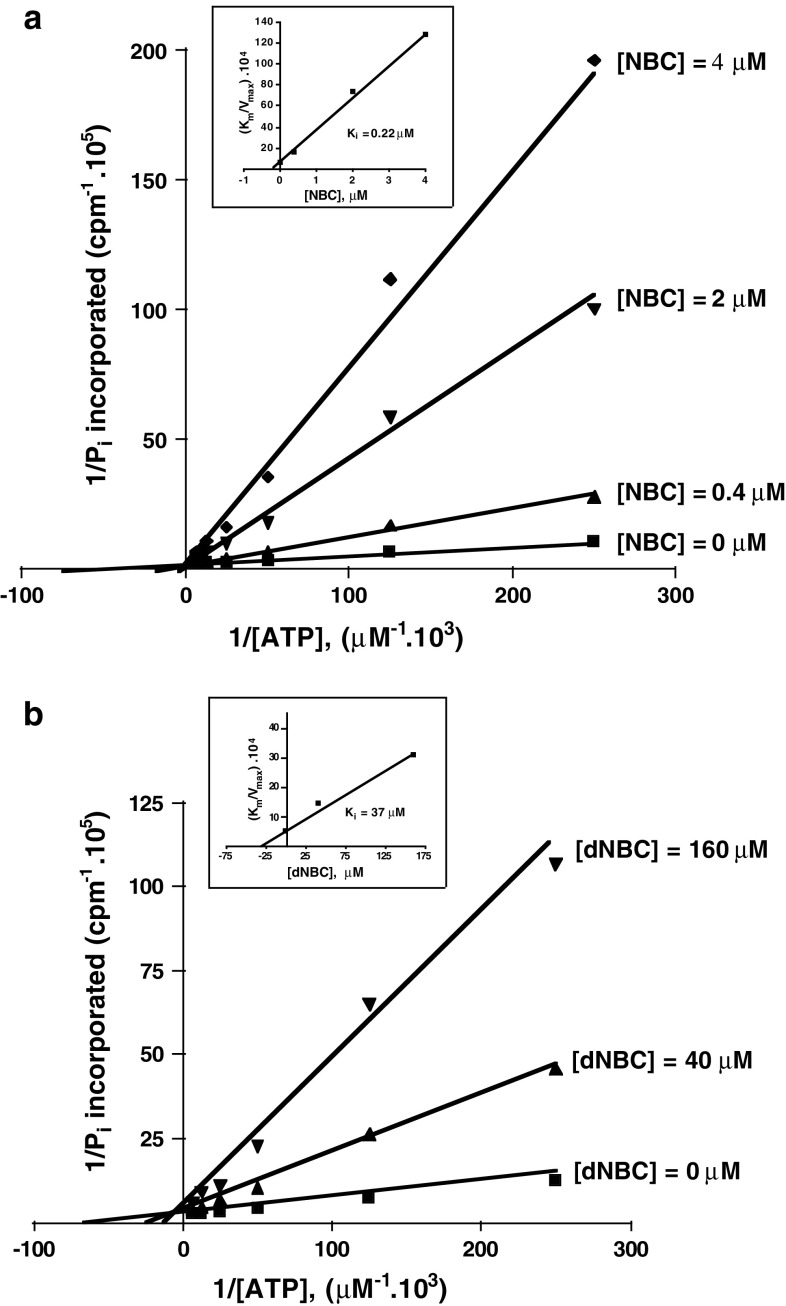
CK2 inhibition by increasing concentration of NBC (8-hydroxy-4-methyl-9-nitrobenzo(g) chromen-2-one) is shown in Fig. 1. From this experiment, an IC50 value of around 0.22 μM can be calculated. As shown by the kinetics reported in Fig. 2, the mode of inhibition is typically competitive with respect to the co-substrate ATP. Figure 1 also shows that inhibition by NBC is critically dependent on the presence of two hydrophobic residues, Val-66 and Ile-174, whose mutation to alanine causes an almost 20-fold increase in IC50. Val-66 is invariably replaced by alanine in all the Ser/Thr kinases other than CK2 and, together with Ile-174, it contributes to make the hydrophobic pocket adjacent to the ATP binding site of CK2 smaller than it is in most other kinases [28]. Inhibition is also critically dependent on the nitro group of NBC, since its removal gives rise to a derivative (8-hydroxy-4-methyl-benzo(g)chromen-2-one, dNBC) whose ability to inhibit CK2 is drastically reduced, as judged by its high IC50 and Ki value (see Figs. 1 and 2b).
Fig. 2.
Kinetic analysis of CK2 inhibition by NBC and dNBC. Inhibition of CK2 by NBC (a) and dNBC (b) is competitive with respect to ATP. Native CK2 activity was determined as described in the text, in the presence of the indicated concentrations of inhibitor. Ki was calculated from the Km/Vmax against [i] (replot shown in the insets). Results are means of experiments run in triplicate, with the SEM never exceeding 10%
Selectivity of NBC and dNBC
The selectivity of NBC and dNBC was compared at a concentration of 10 μM on a panel of more than 115 kinases (Table 1A and B), where the residual activities in the presence of inhibitor are expressed as per cent of control values, and listed in order of decreasing inhibition. As shown in Table 1A, NBC almost entirely suppresses the activity of CK2 (6% residual activity), with similar inhibition of PIM3 (6%) and PIM1 (11%). Also noteworthy is its efficacy toward PKBβ (Akt2) (14% residual activity), especially considering that the other isoform of PKB (α or Akt1) is entirely unaffected (98% residual activity). Only a few additional kinases are inhibited by 50% or more at 10 μM NBC (e.g. HIPK2, DYRK1A, ERK8 and PIM2), the great majority being relatively insensitive to this inhibitor. This accounts for its relatively low promiscuity score (21.8) and rather high Gini coefficient (see below). The IC50 values of NBC with CK2, PKBβ, PIM1 and PIM3 were calculated (see Table 2); they reveal that, although at 10 μM these kinases are inhibited to a similar extent, PIM1 is significantly less susceptible to NBC inhibition than the other three kinases (IC50 3.10 as compared to 0.37, 0.34 and 0.78, respectively).
Table 1.
Selectively profile of NBC (A) and dNBC (B)
| Kinase | Residual activity (% of control) |
|---|---|
| A | |
| CK2 | 6 |
| PIM3 | 6 |
| PIM1 | 11 |
| PKBb | 14 |
| HIPK2 | 21 |
| DYRK1A | 24 |
| VEG-FR | 24 |
| DAPK1 | 26 |
| CAMK1 | 28 |
| ERK8 | 29 |
| PIM2 | 36 |
| SRPK1 | 38 |
| MNK1 | 43 |
| BTK | 45 |
| GCK | 48 |
| FGF-R1 | 49 |
| MINK1 | 50 |
| IR | 52 |
| MLK3 | 52 |
| DYRK3 | 54 |
| RSK2 | 55 |
| MAPKAP-K3 | 56 |
| SGK1 | 58 |
| HIPK3 | 61 |
| Aurora A | 61 |
| MST2 | 64 |
| PRK2 | 65 |
| IRR | 66 |
| ERK1 | 68 |
| CK1 | 68 |
| PAK4 | 70 |
| DYRK2 | 70 |
| IRAK4 | 70 |
| NEK6 | 71 |
| TTK | 71 |
| IKKb | 72 |
| MNK2 | 73 |
| AMPK | 75 |
| MARK4 | 76 |
| GSK3b | 76 |
| MARK2 | 76 |
| JNK1 | 77 |
| p38 g MAPK | 77 |
| NUAK1 | 78 |
| CLK2 | 78 |
| p38a MAPK | 78 |
| S6K1 | 79 |
| PHK | 79 |
| CDK2-Cyc A | 80 |
| MKK1 | 80 |
| SmMLCK | 80 |
| EF2 K | 80 |
| HER4 | 80 |
| IKKe | 81 |
| IGF-1R | 81 |
| EPH-A2 | 81 |
| YES1 | 83 |
| MARK1 | 83 |
| MKK2 | 84 |
| PKCa | 84 |
| CHK2 | 85 |
| MPSK1 | 85 |
| EPH-B3 | 87 |
| Lck | 87 |
| PKD1 | 87 |
| JNK2 | 88 |
| MAPKAP-K2 | 88 |
| BRSK2 | 88 |
| SYK | 89 |
| PKCy | 89 |
| MARK3 | 90 |
| MEKK1 | 91 |
| STK33 | 91 |
| JNK3 | 91 |
| MSK1 | 92 |
| RIPK2 | 93 |
| CAMKKb | 93 |
| Aurora B | 93 |
| TAOl | 93 |
| ASK1 | 94 |
| LKB1 | 94 |
| TrkA | 95 |
| PAK5 | 96 |
| CHK1 | 98 |
| PKBa | 98 |
| p38b MAPK | 98 |
| RSK1 | 99 |
| MLK1 | 99 |
| PRAK | 100 |
| MELK | 100 |
| BRK | 101 |
| MKK6 | 101 |
| PLK1 | 101 |
| CSK | 101 |
| BRSK1 | 101 |
| PDK1 | 101 |
| EPH-A4 | 101 |
| ROCK 2 | 102 |
| p38d MAPK | 104 |
| PKCz | 104 |
| TIE2 | 104 |
| HIPK1 | 105 |
| TAK1 | 105 |
| ZAP70 | 105 |
| PAK2 | 106 |
| ERK2 | 106 |
| TBK1 | 107 |
| Src | 107 |
| NEK2a | 108 |
| ABL | 109 |
| JAK2 | 110 |
| EPH-B2 | 112 |
| PKA | 113 |
| EPH-B4 | 114 |
| EPH-B1 | 118 |
| PAK6 | 120 |
| M5T4 | 125 |
| B | |
| DYRK1A | 3 |
| ERK8 | 4 |
| GSK3b | 6 |
| PIM1 | 10 |
| TAO1 | 12 |
| PIM3 | 12 |
| PRK2 | 13 |
| CLK2 | 18 |
| DYRK3 | 19 |
| AMPK | 20 |
| MKK1 | 21 |
| HIPK2 | 22 |
| SmMLCK | 23 |
| BRSK2 | 24 |
| CDK2-Cyc A | 27 |
| IRR | 29 |
| RSK1 | 29 |
| MKK2 | 31 |
| DYRK2 | 32 |
| MNK2 | 38 |
| MNK1 | 39 |
| DAPK1 | 41 |
| ASK1 | 43 |
| GCK | 45 |
| BTK | 46 |
| VEG-FR | 49 |
| IRAK4 | 49 |
| LKB1 | 49 |
| PRAK | 50 |
| EF2 K | 51 |
| MEKK1 | 51 |
| TTK | 52 |
| RSK2 | 52 |
| PIM2 | 53 |
| S6K1 | 55 |
| CK2 | 55 |
| MLK3 | 56 |
| MST2 | 58 |
| NUAK1 | 59 |
| SGK1 | 60 |
| PAK4 | 61 |
| CK1 | 61 |
| ERK1 | 62 |
| MINK1 | 62 |
| PKD1 | 63 |
| CAMKKb | 64 |
| Aurora B | 64 |
| JNK1 | 65 |
| PKCz | 66 |
| PHK | 67 |
| HIPK3 | 67 |
| CAMK1 | 68 |
| RIPK2 | 68 |
| MELK | 68 |
| SRPK1 | 68 |
| EPH-A2 | 68 |
| PKBb | 69 |
| HIPK1 | 70 |
| CHK1 | 70 |
| IKKe | 71 |
| EPH-B3 | 71 |
| HER4 | 72 |
| MSK1 | 72 |
| TBK1 | 73 |
| FGF-R1 | 74 |
| ROCK 2 | 74 |
| CHK2 | 74 |
| IR | 74 |
| MARK4 | 75 |
| MPSK1 | 75 |
| IKKb | 76 |
| p38a MAPK | 77 |
| STK33 | 78 |
| p38 g MAPK | 78 |
| TAK1 | 78 |
| NEK6 | 78 |
| YES1 | 79 |
| MAPKAP-K2 | 80 |
| Lck | 80 |
| PAK2 | 81 |
| IGF-1R | 81 |
| MLK1 | 81 |
| MARK2 | 81 |
| ERK2 | 82 |
| PAK5 | 83 |
| EPH-B4 | 84 |
| TrkA | 85 |
| JNK2 | 86 |
| MAPKAP-K3 | 87 |
| ABL | 87 |
| BRSK1 | 87 |
| Aurora A | 87 |
| MARK3 | 88 |
| MARK1 | 89 |
| p38d MAPK | 90 |
| EPH-A4 | 90 |
| Src | 91 |
| JNK3 | 92 |
| MKK6 | 92 |
| JAK2 | 92 |
| SYK | 93 |
| PKCa | 93 |
| PLK1 | 95 |
| ZAP70 | 97 |
| PKA | 97 |
| NEK2a | 98 |
| p38b MAPK | 101 |
| PKCγ | 101 |
| MST4 | 103 |
| BRK | 103 |
| CSK | 103 |
| TIE2 | 104 |
| PAK6 | 105 |
| PDK1 | 106 |
| EPH-B2 | 109 |
| PKBa | 111 |
| EPH-B1 | 114 |
Inhibition assays were performed at a concentration of 10 μM of the indicated inhibitor under conditions referenced in the text. The residual activity are sorted in increasing order
Table 2.
IC50 values of NBC and dNBC for selected protein kinases
| IC50 (μM) | ||
|---|---|---|
| Kinase | NBC | dNBC |
| CK2 | 0.37 | 32.34 |
| DYRK1A | 15.00 | 0.60 |
| PIM1 | 3.10 | 3.90 |
| PIM2 | 4.17 | 33.35 |
| PIM3 | 0.34 | 2.05 |
| PKBα | >50.00 | >50.00 |
| PKBβ | 0.78 | >30.00 |
| GSK3β | >30.00 | 0.70 |
| ERK8 | 3.90 | 0.30 |
Conditions for the activity assays of protein kinases tested in the determination of IC50 values were described in the text or referenced in [24]. The values are the means of the results obtained in triplicate with SEM never exceeding 15%
A comparison with dNBC (Table 1B) revealed that loss of the nitro group strikingly alters the selectivity profile of the inhibitor. For example, CK2 inhibition becomes negligible (>50% residual activity), as already mentioned above (Fig. 1), and the same also applies to PKBβ (69% residual activity). In contrast, inhibition of PIM1 persists and a number of protein kinases which are only marginally affected by NBC are now drastically inhibited by dNBC. This is particularly the case for DYRK1A (3% residual activity) ERK8 (4% residual activity), and GSK3β (6% residual activity). The IC50 values of dNBC with these kinases were all in the submicromolar range, i.e. even below that of PIM1 (see Table 2).
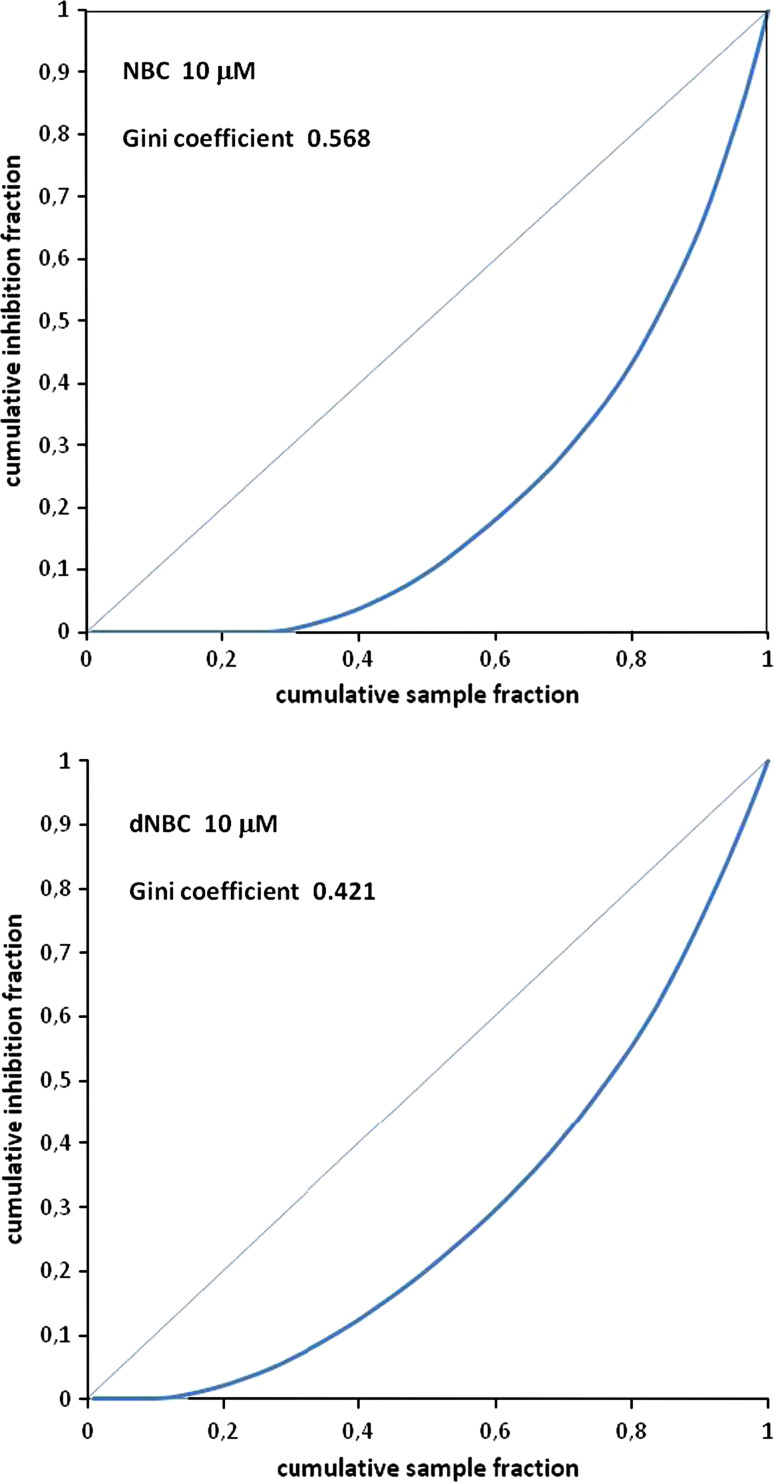
To gain additional information about the selectivity of NBC and dNBC advantage has been taken of the “Gini coefficient”, a parameter originally developed by economists to measure income inequality and recently successfully exploited to evaluate differences in selectivity between protein kinase inhibitors [27]. By following the procedure described in [27], the Lorenz curves were drawn from the data in Table 1A and B: as shown in Fig. 3, this led to the calculation of a Gini coefficient markedly higher in the case of NBC (0.548) as compared to dNBC (0.421). As shown in Table 2, the only kinase inhibited by NBC with similar potency to CK2 and PIM3 was PKBβ, which was hardly affected by dNBC (see Table 1B). Also noteworthy in this connection is the ability of NBC to discriminate between the two isoforms of PKB: PKBβ is inhibited with an IC50 value of 0.78 μM, but the α isoform (Akt1) is only marginally affected (IC50 > 50 μM) (see Table 2).
Fig. 3.
Lorenz curves for NBC and dNBC. Lorenz curves were constructed and Gini coefficients [27] calculated for NBC and dNBC at 10 μM concentration using the inhibition data reported in Table 1A and B, and applying a properly programmed Microsoft Excel spreadsheet (available in the Supporting Information in [27])
Cytotoxicity of NBC versus dNBC
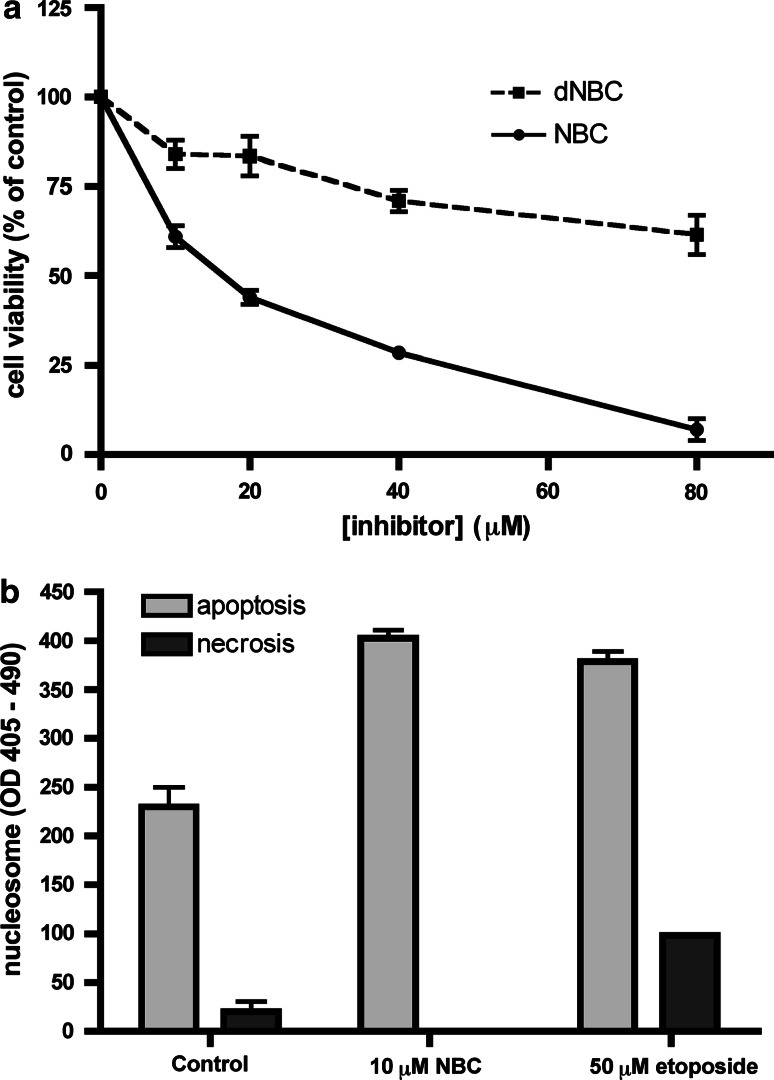
Treatment of Jurkat cells with increasing concentrations of either NBC or dNBC promotes sharply different effects on cell viability (see Fig. 4a). While NBC is highly cytotoxic with a DC50 value (the inhibitor concentration which induces 50% cell death in 24 h) of around 15 μM, treatment with dNBC up to 80 μM is still compatible with >60% cell survival. This rules out the possibility that the cytotoxic effect of NBC is mediated by kinases inhibited by both NBC and dNBC, e.g. PIMs, and supports the concept that the kinase whose inhibition by NBC stimulates apoptosis in these cells is indeed CK2. Also of note in this respect is that quinalizarin, a potent and selective inhibitor of CK2 which, unlike NBC, does not inhibit PKBβ [35], promotes a cellular effect very similar to that shown here for NBC. Indeed, the ratio between DC50 and IC50 for these two inhibitors is almost identical, being 38.5 for quinalizarin [35] and 37.8 for NBC (drawn from Fig. 4a and Table 2). Incidentally, this approach highlights the usefulness of employing different inhibitors of the same kinase with non-overlapping selectivity profiles to implicate a particular kinase in a given cellular event. The cytotoxic effect of NBC was also confirmed using a Cell Death Detection Elisa kit (Roche) which allows discrimination between apoptosis and necrosis. By this approach, it turned out that the effect of NBC, like that of quinalizarin [35], is entirely accounted for by apoptosis (Fig. 4b).
Fig. 4.
Effect of NBC and dNBC on cell viability. a Cells were treated for 24 h with increasing concentration of the indicated compounds. Cell viability was assessed by the MTT method, assigning 100% value to the control cells, treated with the solvent. Reported values represent the means ± SEM from four separate experiments. b Cells were treated with 10 μM NBC for 4 h. Apoptosis and necrosis were evaluated by quantification of nucleosomes present in the cytosol (apoptotic cells) and in extracellular medium (released by necrotic cells), using the Cell Detection Elisa kit (Roche). Treatment with 50 μM etoposide was used as a positive control for apoptosis induction. Reported values represent the means ± SEM from two separate experiments. For details, see text
Structural insights
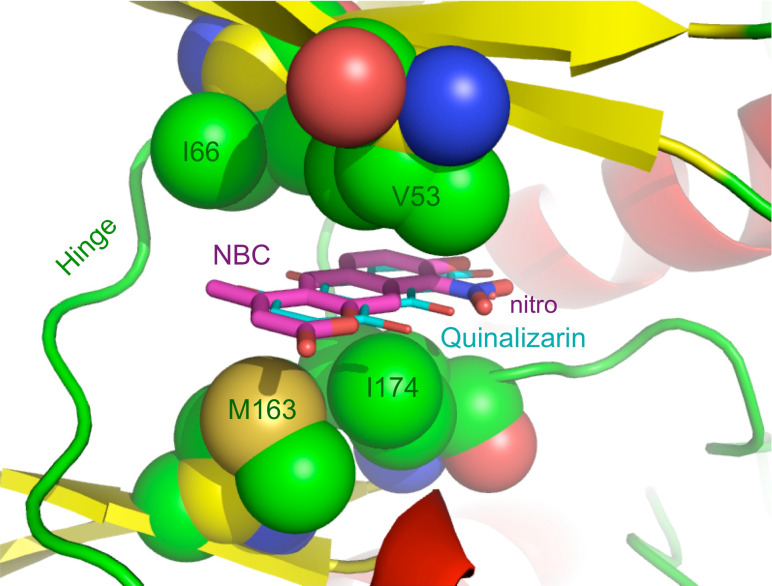
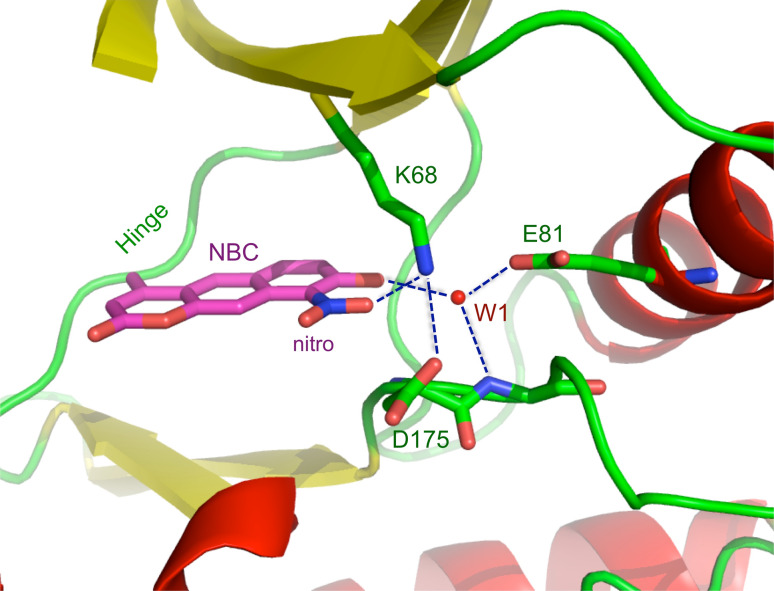
To explore the structural basis for NBC activity and, in particular, the role of the nitro function in binding of the inhibitor, we determined the crystal structure of maize CK2α in complex with NBC at 2.2 Å resolution (statistics on data collection and refinement are reported in Table 3). As suggested by the biochemical characterisation, NBC is an ATP-competitive inhibitor that binds to the active site of protein kinase CK2. NBC is sandwiched between hydrophobic residues, Val53 and Ile66 on one side, Met163 and Ile174 on the other side, in a position similar to that of the inhibitor quinalizarin [35] (see Fig. 5). The importance of the interactions of the ligand with these hydrophobic residues is corroborated by the increase in the IC50 value upon mutation of V66 (equivalent to I-66 in maize CK2) and I174 to alanine (see above, Fig. 1). In comparison with the structurally related quinalizarin and other anthraquinones such as emodin, 1,8-dihydroxy-4-nitro-anthraquinone (MNA), 1,8-dihydroxy-4-nitro-xanthen-9-one (MNX) and 1,4-diamino-5,8-dihydroxy-anthraquinone (DAA), the different nature and position of the substituents in the three condensed six-membered rings scaffold determine the dissimilar orientation of NBC in the binding pocket. In the case of NBC, the presence of a nitro function at position 9 and of an adjacent hydroxyl function at position 8 is essential in positioning the ligand near the positive area in the proximity of Lys68. As illustrated in Fig. 6, the nitro group interacts with Lys68, at 2.61 Å, while the hydroxyl group in ortho interacts with the conserved water W1, at 2.76 Å. Therefore, the nitro function is important for binding, not only for the possibility of establishing a direct electrostatic interaction with Lys68 but also for its role in lowering the pKa of the adjacent OH, allowing it to interact more strongly with water W1 in the positive area. This indirect effect on the phenolic hydroxyl moiety has been noticed and described previously for other inhibitors; for example, for the anthraquinones MNA and MNX that also carry a nitro function in a phenolic ring [36]. The fundamental role of the nitro group in the binding of NBC to CK2 was confirmed by the dramatic decrease in affinity seen for dNBC and by the loss of selectivity observed for this derivative.
Fig. 5.
Inhibitor NBC (magenta carbon atoms) bound to maize CK2α (ribbon coloured according to the secondary structure). NBC is sandwiched between hydrophobic residues Val53 and Ile66 on one side, and Met163 and Ile174 on the other side (all shown as solid spheres). A comparison with the similar binding mode of anthraquinone inhibitor quinalizarin [35] (cyan carbon atoms) is shown
Fig. 6.
Polar interactions between NBC and surrounding residues in maize CK2α. The nitro function is at 2.61 Å from Lys68, while the hydroxyl group in ortho interacts with the conserved water W1, at a distance of 2.76 Å
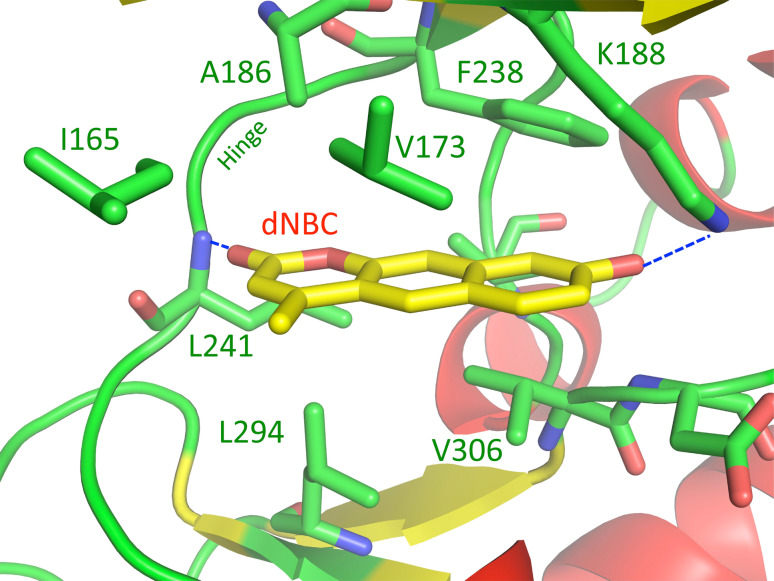
NBC is a fairly potent ATP site-directed inhibitor of protein kinase CK2, poorly effective against DYRK1A (IC50 = 15.00 μM); by contrast, the structurally related compound dNBC is quite effective against DYRK1A (IC50 = 0.6 μM), and only affects CK2 marginally (IC50 = 32.34 μM). Since the two compounds differ only in the presence or absence of a nitro group at position 3, we have exploited a molecular docking approach to try to understand how dNBC and NBC bind to DYRK1A. The docking mode of dNBC suggests that the compound is buried more deeply inside the binding cleft of DYRK1A (PDB code: 2VX3) as compared to NBC in CK2 (Fig. 7). This could be explained by the presence of an alanine residue (Ala186) which in CK2 is replaced by Val66. In this way, dNBC can interact simultaneously with both the hinge region and the phosphate binding region of DYRK1A. In particular. the carbonyl group at position 2 is hydrogen bonded to the backbone amino group of Leu241 in the hinge region, while the phenolic oxygen at position 8 interacts with Lys188 in a similar way to that seen for NBC bound to CK2. The planar aromatic system forms a hydrophobic interaction with the ATP binding zone thanks to the interactions with Phe238, Val173, Ile165 (at the top of the cleft), and Val306, Leu294 and Leu241 (at the bottom of the cleft). A key role in this interaction network is played by the methyl group at position 4, which is oriented in the opposite direction to the way it is positioned in the NBC–CK2 complex. It is embedded between Ile165 and Val173 located in the glycine-rich loop and is therefore protected from the solvent. This type of binding does not allow the insertion of a bulky nitro group (present in NBC) at position 9 due to the presence of Phe238 (Fig. 7) and may explain why NBC fails to inhibit DYRK1A as drastically as dNBC.
Fig. 7.
Molecular docking of dNBC bound to the active site of DYRK1A. Molecular docking was performed as described in the text. The most important polar interactions are highlighted
Discussion
The most remarkable outcome of this study is the finding that by just adding or removing a nitro group to the scaffold of a benzocoumarin derivative it is possible to generate inhibitors with strikingly different selectivity toward diverse classes of protein kinases. While in fact it is very frequent and in a way expectable that a single modification of its scaffold may cause either a loss or an enhancement of the potency of an inhibitor toward an individual kinase, a simultaneous drop and increase of inhibitory efficiency toward different kinases driven by the same structural alteration is an uncommon and noteworthy event. In our case, the addition of the nitro group confers selectivity toward CK2 and to a lesser extent PKBβ, while its removal generates a compound which potently inhibits DYRK1A, ERK8 and GSK3β, but has lost the ability to inhibit CK2 and PKBβ. Both compounds are fairly good inhibitors of PIM1, consistent with the notion that the ATP binding site of this kinase is quite flexible and amenable to accommodating a wide variety of ligands.
The determination of the crystal structure of the nitro compound (NBC) in complex with the CK2 catalytic subunit discloses the essential role of the nitro group at position 9 within the benzocoumarine scaffold, by showing that it both makes a direct electrostatic interaction with the side chain of Lys68 and also enhances the potential of the adjacent OH to interact with the constitutive water (W1) located in the positive area of the CK2 active site. Such an interaction is dependent on the dissociation of the phenolic OH whose pKa is markedly decreased by an adjacent nitro function, as previously shown in the case of the antraquinone derivatives MNA and MNX [36].
On the other hand, a molecular docking approach performed using the crystal structure of DYRK1A accounts very well for the observation that the nitro group, whose removal is detrimental for high affinity binding to CK2, also needs to be removed to ensure high affinity inhibition of DYRK1A. Given the paucity of selective DYRK1A inhibitors available, with the notable exception of harmine [25], advantage could be taken of dNBC to develop new selective inhibitors of this kinase, which plays important roles in neurodegenerative diseases [37]. For the time being, it should be born in mind that, in addition to DYRK1A, dNBC also drastically inhibits ERK8 and GSK3β, exhibiting an overall promiscuity higher than that of NBC, which is reflected in a Gini coefficient of 0.421 as compared to 0.568 for NBC (see Fig. 3).
The different selectivities of NBC and dNBC towards different kinases, despite their structural similarity, provides a new approach for dissecting the biological functions of CK2 since only the former is a potent inhibitor of CK2, the latter being instead a potent inhibitor of DYRK1A, ERK8, GSK3β and TAO1, none of which is susceptible to NBC inhibition. Thus, the ability of NBC, but not of dNBC, to readily induce apoptosis of Jurkat cells is consistent with the concept that CK2 plays a key role in counteracting programmed cell death [38]. Although in fact the cytotoxic effect of NBC could also be mediated by PKBβ, its DC50 to IC50 (with CK2) ratio and its proapoptotic efficacy (Fig. 4) are strikingly similar to those of quinalizarin, a CK2 inhibitor having no effect on PKBβ [35]. Also worthy of note is the ability of NBC to discriminate between the two isoforms of PKB/Akt, only one of which (PKBβ/Akt2) is readily inhibited, and of dNBC to inhibit PIM1 and PIM3 much more efficiently than the third isoform of the family, PIM2.
Acknowledgments
The work was supported by grants to L.A.P. from the European Commission (Prokinaseresearch 503467) and Associazione Italiana per la Ricerca sul Cancro (AIRC), by funds from the Italian Ministry of Education, University and Research (PRIN Project 2008) to L.A.P. and R.B. and by a Strategic Award from the UK Medical Research Council to support the National Centre for Protein Kinase Profiling (http://www.kinase-screen.mrc.ac.uk). We are grateful to Professor Sir Philip Cohen (MRC Protein Phosphorylation Unit, University of Dundee, Dundee, Scotland, U.K.) for a critical reading of the manuscript. Authors thank the staff at beamline ID13 of the ESRF (Grenoble) for on-site assistance in data collection. The Molecular Modelling Section (MMS) coordinated by Professor S. Moro (Padova, Italy) is gratefully acknowledged.
Abbreviations
- PIM
Provirus integration site for Moloney murine leukaemia virus
- GSK
Glycogen synthase kinase
- ERK
Extracellular-signal-regulated kinase
- DYRK
Dual-specificity tyrosine-phosphorylated and regulated kinase
- DRB
5,6- dichloro-1-β-D-ribofuranosylbenzimidazole
- TBB
4,5,6,7-tetrabromo-1H-benzotriazole
- TBI
4,5,6,7-tetrabromo-1H-benzimidazole
- DMAT
4,5,6,7-tetrabromo-2-(dimethylamino)benzimidazole
- HIPK
Homeodomain-interacting protein kinase
- PKB
Protein kinase B
- PRK
Protein kinase C-related kinase
Contributor Information
Roberto Battistutta, Phone: +39-049-923236, FAX: +39-049-7923250, Email: roberto.battistutta@unipd.it.
Lorenzo A. Pinna, Phone: +39-049-8276109, FAX: +39-049-8073310, Email: lorenzo.pinna@unipd.it
References
- 1.Hopkins LA, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 2.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 3.Salvi M, Sarno S, Cesaro L, Nakamura H, Pinna LA. Extraordinary pleiotropy of protein kinase CK2 revealed by weblogo phosphoproteome analysis. Biochim Biophys Acta. 2009;1793:847–859. doi: 10.1016/j.bbamcr.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 4.Salvi M, Cesaro L, Pinna LA. Variable contribution of protein kinases to the generation of the human phosphoproteome: a global weblogo analysis. BioMol Concept. 2010;1:185–195. doi: 10.1515/BMC.2010.013. [DOI] [PubMed] [Google Scholar]
- 5.Dominguez I, Sonenshein GE, Seldin DC. Protein kinase CK2 in health and disease: CK2 and its role in Wnt and NF-kB signalling: linking development and cancer. Cell Mol Life Sci. 2009;66:1850–1857. doi: 10.1007/s00018-009-9153-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guerra B, Issinger O-G. Protein kinase CK2 in human diseases. Curr Med Chem. 2008;15:1870–1886. doi: 10.2174/092986708785132933. [DOI] [PubMed] [Google Scholar]
- 7.Sarno S, Pinna LA. Protein kinase CK2 as a druggable target. Mol Biosyst. 2008;4:889–894. doi: 10.1039/b805534c. [DOI] [PubMed] [Google Scholar]
- 8.Ruzzene M, Pinna LA. Addiction to protein kinase CK2: a common denominator of diverse cancer cells? Biochim Biophys Acta. 2010;1804:499–504. doi: 10.1016/j.bbapap.2009.07.018. [DOI] [PubMed] [Google Scholar]
- 9.Silva A, Yunes JA, Cardoso BA, Martins LR, Jotta PY, Abecasis M, Nowill AE, Leslie NR, Cardoso AA, Barata JT. PTEN posttranslational inactivation and hyperactivation of the PI3 K/Akt pathway sustain primary T cell leukaemia viability. J Clin Invest. 2008;118:3762–3764. doi: 10.1172/JCI34616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piazza FA, Ruzzene M, Gurrieri C, Montini B, Bonanni L, Chioetto G, Di Maira G, Barbon F, Cabrelle A, Zambello R, Adami F, Trentin L, Pinna LA, Semenzato G. Multiple myeloma cell survival relies on high activity of protein kinase CK2. Blood. 2006;108:1698–1707. doi: 10.1182/blood-2005-11-013672. [DOI] [PubMed] [Google Scholar]
- 11.Kim JS, Eom JI, Cheong JW, Lee JK, Yang WI, Min YM. Protein kinase CK2alpha as an unfavourable prognostic marker and novel therapeutic target in acute myeloid leukaemia. Clin Cancer Res. 2007;13:1019–1028. doi: 10.1158/1078-0432.CCR-06-1602. [DOI] [PubMed] [Google Scholar]
- 12.Solimini NL, Luo J, Elledge SJ. Non-oncogene addiction and the stress phenotype of cancer cells. Cell. 2007;130:986–988. doi: 10.1016/j.cell.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 13.Trembley JH, Chen Z, Unger G, Slaton J, Kren BT, Van Waes C, Ahmed K. Emergence of protein kinase CK2 as a key target in cancer therapy. Biofactors. 2010;36:187–195. doi: 10.1002/biof.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarno S, Salvi M, Battistutta R, Zanotti G, Pinna LA. Features and potentials of ATP-site directed CK2 inhibitors. Biochim Biophys Acta. 2005;1754:263–270. doi: 10.1016/j.bbapap.2005.07.043. [DOI] [PubMed] [Google Scholar]
- 15.Duncan JS, Litchfield DW. Too much of a good thing: the role of protein kinase CK2 in tumorigenesis and prospects for therapeutic inhibition of CK2. Biochim Biophys Acta. 2008;1784:33–47. doi: 10.1016/j.bbapap.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 16.Sarno S, Papinutto E, Franchin C, Bain J, Elliott M, Meggio F, Kazimierczuk Z, Orzeszko A, Zanotti G, Battistutta R, Pinna LA (2010) ATP site-directed inhibitors of protein kinase CK2: an update. Curr Top Med Chem (in press) [DOI] [PubMed]
- 17.Mazzorana M, Pinna LA, Battistutta R. A structural insight into CK2 inhibition. Mol Cell Biochem. 2008;316:57–62. doi: 10.1007/s11010-008-9822-5. [DOI] [PubMed] [Google Scholar]
- 18.Battistutta R. Protein kinase CK2 in health and disease: Structural bases of protein kinase CK2 inhibition. Cell Mol Life Sci. 2009;66:1868–1889. doi: 10.1007/s00018-009-9155-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pagano MA, Bain J, Kazimierczuk Z, Sarno S, Ruzzene M, Di Maira G, Elliott M, Orzeszko A, Cozza G, Meggio F, Pinna LA. The selectivity of inhibitors of protein kinase CK2: an update. Biochem J. 2008;415:353–365. doi: 10.1042/BJ20080309. [DOI] [PubMed] [Google Scholar]
- 20.Chilin A, Battistutta R, Bortolato A, Cozza G, Zanatta S, Poletto G, Mazzorana M, Zagotto G, Uriarte E, Guiotto A, Pinna LA, Meggio F, Moro S. Coumarin as attractive casein kinase 2 (CK2) inhibitor scaffold: an integrate approach to elucidate the putative binding motif and explain structure-activity relationships. J Med Chem. 2008;51:752–759. doi: 10.1021/jm070909t. [DOI] [PubMed] [Google Scholar]
- 21.Meggio F, Pagano MA, Moro S, Zagotto G, Ruzzene M, Sarno S, Cozza G, Bain J, Elliott M, Donella Deana A, Brunati AM, Pinna LA. Inhibition of protein kinase CK2 by condensed polyphenolic derivatives. An in vitro and in vivo study. Biochemistry. 2004;43:12931–12936. doi: 10.1021/bi048999g. [DOI] [PubMed] [Google Scholar]
- 22.Meggio F, Donella Deana A, Pinna LA. Endogenous phosphate acceptor proteins for rat liver cytosolic casein kinases. J Biol Chem. 1981;256:11958–11961. [PubMed] [Google Scholar]
- 23.Sarno S, Vaglio P, Meggio F, Issinger O-G, Pinna LA. Protein kinase CK2 mutants defective in substrate recognition: purification and kinetic analysis. J Biol Chem. 1996;271:10595–10601. doi: 10.1074/jbc.271.18.10595. [DOI] [PubMed] [Google Scholar]
- 24.Pagano MA, Andrzejewska M, Ruzzene M, Sarno S, Cesaro L, Bain J, Elliott M, Meggio F, Kazimierczuk Z, Pinna LA. Optimization of protein kinase CK2 inhibitors derived from 4, 5, 6, 7-tetrabromobenzimidazole. J Med Chem. 2004;47:6239–6247. doi: 10.1021/jm049854a. [DOI] [PubMed] [Google Scholar]
- 25.Bain J, Plater L, Elliott M, Shpiro N, Hastie J, McLauchlan H, Klevernic I, Arthur S, Alessi D, Cohen P. The selectivity of protein kinase inhibitors; a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Himpel S, Tegge W, Frank R, Leder S, Joost H-G, Becker W. Specificity determinants of substrate recognition by the protein kinase DYRK1A. J Biol Chem. 2000;275:2431–2438. doi: 10.1074/jbc.275.4.2431. [DOI] [PubMed] [Google Scholar]
- 27.Graczyk PP. Gini coefficient: a new way to express selectivity of kinase inhibitors against a family of kinases. J Med Chem. 2007;50:5773–5779. doi: 10.1021/jm070562u. [DOI] [PubMed] [Google Scholar]
- 28.Battistutta R, De Moliner E, Sarno S, Zanotti G, Pinna LA. Structural features underlying selective inhibition of protein kinase CK2 by ATP site-directed tetrabromo-2-benzotriazole. Protein Sci. 2001;10:2200–2206. doi: 10.1110/ps.19601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leslie AGW (1991) Crystallographic Computing V. In: Moras D, Podjarny AD, Thierry, JP (eds) Crystallographic Computing 5, From Chemistry to Biology. Oxford University Press, Oxford, pp 27–38
- 30.Evans PR “Data reduction”, Proceedings of CCP4 Study Weekend, 1993, on Data Collection & Processing, pp 114–122
- 31.Collaborative Computational Project, Number 4 (1994) The CCP4 Suite: Programs for Protein Crystallography. Acta Cryst D50: 760–763 [DOI] [PubMed]
- 32.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emsley P, Lohkamp B, Scott W, Cowtan K. Features and Development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cozza G, Bonvini P, Zorzi E, Poletto G, Pagano MA, Sarno S, Donella-Deana A, Zagotto G, Rosolen A, Pinna LA. Identification of ellagic acid as potent inhibitor of protein kinase CK2: a successful example of a virtual screening application. J Med Chem. 2006;49:2363–2366. doi: 10.1021/jm060112m. [DOI] [PubMed] [Google Scholar]
- 35.Cozza G, Mazzorana M, Papinutto E, Bain J, Elliott M, Di Maira G, Gianoncelli A, Pagano MA, Sarno S, Ruzzene M, Battistutta R, Meggio F, Moro S, Zagotto G, Pinna LA. Quinalizarin as a potent, selective and cell-permeable inhibitor of protein kinase CK2. Biochem J. 2009;421:387–395. doi: 10.1042/BJ20090069. [DOI] [PubMed] [Google Scholar]
- 36.De Moliner E, Moro S, Sarno S, Zagotto G, Zanotti G, Pinna LA, Battistutta R. Inhibition of Protein Kinase CK2 by Anthraquinone-related Compounds. J Biol Chem. 2003;278:1831–1836. doi: 10.1074/jbc.M209367200. [DOI] [PubMed] [Google Scholar]
- 37.Ryu YS, Park SY, Jung M-S, Yoon S-H, Kwen M-Y, Lee S-Y, Choi S-H, Radnaabazar C, Kim M-K, Kim H, Kim K, Song W-J, Chung S-H. Dyrk1A-mediated phosphorylation of prresenilin 1: a functional link between Down syndrome and Alzheimer’s disease. J Neurochem. 2010;115:574–584. doi: 10.1111/j.1471-4159.2010.06769.x. [DOI] [PubMed] [Google Scholar]
- 38.Ahmad KA, Wang G, Unger G, Slaton J, Ahmed K. Protein Kinase CK2 - A Key Suppressor of Apoptosis. Adv Enzyme Regul. 2008;48:179–187. doi: 10.1016/j.advenzreg.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]