Abstract
The innate immune system relies on its capability to detect invading microbes, tissue damage, or stress via evolutionarily conserved receptors. The nucleotide-binding domain leucine-rich repeat (NLR)-containing family of pattern recognition receptors includes several proteins that drive inflammation in response to a wide variety of molecular patterns. In particular, the NLRs that participate in the formation of a molecular scaffold termed the “inflammasome” have been intensively studied in past years. Inflammasome activation by multiple types of tissue damage or by pathogen-associated signatures results in the autocatalytic cleavage of caspase-1 and ultimately leads to the processing and thus secretion of pro-inflammatory cytokines, most importantly interleukin (IL)-1β and IL-18. Here, we review the current knowledge of mechanisms leading to the activation of inflammasomes. In particular, we focus on the controversial molecular mechanisms that regulate NLRP3 signaling and highlight recent advancements in DNA sensing by the inflammasome receptor AIM2.
Keywords: NLR, NLRP3, AIM2, Caspase-1, IL-1β, AIM2
Introduction
In recent years, considerable progress has been made in our understanding of how microbial pathogens are sensed by the innate immune pathways that form the first line of defense. In addition, much research has been conducted on how these pathways translate into the signal transduction and transcription of antimicrobial effector molecules and also on the mechanisms that license and promote adaptive immune responses. A limited number of well-conserved microbial structures, which are known as pathogen-associated molecular patterns (PAMPs), are recognized by germline-encoded pattern recognition receptors (PRRs), which can be found both in “classical” immune and nonimmune cells. Interestingly, some of these receptors are also able to sense various endogenous signals that arise during tissue or cell damage. These signals are commonly referred to as danger-associated molecular patterns (DAMPs).
Our innate immune system has evolved at least four major PRR families that cooperatively operate to recognize microbial pathogens or endogenous danger signals: the toll-like receptor system (TLRs), the RIG-I-like receptors (RLRs), the C-type lectin receptors (CLRs), and the nucleotide-binding domain leucine-rich repeats (NLRs).
As TLRs were the first family of PRRs identified, this group of receptors has been studied the most intensively. TLRs are localized on the cell surface and endosomal compartments and are able to sense a plethora of PAMPs and DAMPs of different physiochemical natures. These include peptides, lipopeptides, glycopeptides, glycolipids, and nucleic acids. On the other hand, cell-membrane-associated CLRs recognize a rather specific repertoire of glycopeptides and polysaccharides. Unlike TLRs and CLRs, RLRs and NLRs are cytosolic proteins that survey for the presence of intracellular microbial molecules or danger signals. The RLRs RIG-I and MDA-5 sense viral RNA and induce an antiviral response via their shared mitochondrial adaptor protein MAVS. Moreover, the two CARD-containing NLRs, NOD1 and NOD2, recognize glycopeptides and signal via their common adaptor molecule RIP2K. While all these PRRs have signal pathways that converge in the transcription of cytokines, chemokines, or proteins involved in antigen presentation and processing, a prominent subgroup of NLRs instead triggers the activation of pro-inflammatory cytokines such as interleukin (IL)-1β through a proteolytic process that is controlled by caspase-1. For PRRs involved in caspase-1 activation in particular, emerging evidence reveals that these sensors, which were initially identified to specifically recognize a broad variety of foreign molecules, also sense endogenous molecules of similar structure under certain conditions. For instance, during tissue damage, cells or nuclei lose their structural integrity, and endogenous DAMPs that are sequestered under normal conditions, such as lipids, proteins, or nucleic acids, gain access to cytosolic innate immune receptors. Here we review these caspase-1-activating proteolytic platforms, the so-called inflammasomes.
The NLR family
In the human system, 22 NLR genes have been identified so far (Fig. 1). NLRs display a tripartite structural organization consisting of C-terminal leucine-rich repeats (LRRs), a central NACHT domain (NACHT stands for domain present in NAIP, CIITA, HET-E, and TP-1), which is commonly associated with an additional domain that is known as NACHT-associated domain (NAD), and an N-terminal effector domain that initiates signaling. In NLR proteins, the NACHT domain and its associated domain NAD are often summarized as nucleotide-binding domains (NBD).
Fig. 1.
Members of the human NLR family. The known human NLR proteins are depicted and grouped according to their N-terminal domains. Grouping NLRs according to their N-terminal domain, five distinct families can be identified: NLRA [A stands for AD (acidic activation domain)], NLRB [B stands for BIR (baculovirus inhibitor of apoptosis) domain], NLRC [C stands for CARD (caspase recruitment domain)], NLRP (P stands for pyrin domain), and NLRX (X stands for domain with no strong homology to the N-terminal domain of any other NLR subfamily member)
LRR domains are very diverse and found in many proteins with a wide variety of different cell biological functions [1]. Despite these functional differences, LRR domains share a common structural framework consisting of a motif of 20–30 amino acids with a characteristic sequence pattern rich in the hydrophobic amino acid leucine that makes them suitable for protein–protein interactions. The architecture of the LRR domain also appears to be well suited for ligand binding [1]. In analogy to TLRs that sense PAMPs via their LRR domain, LRRs in NLRs are believed to be potential ligand-binding domains that directly detect microbial products or intermediate signaling molecules in the cytosol. Nevertheless, a clear receptor-ligand interaction has not been demonstrated for any NLR yet.
The NBD domain shares similarity with the NB-ARC motif of the apoptotic mediator apoptotic protease-activating factor 1 (APAF1) and belongs to the signal transduction ATPases with numerous domains (STAND) family of NTPase domains. Like other NTPase domains, NACHT domains are thought to oligomerize upon activation in the presence of ATP. The NACHT domain seems to be a pivotal module involved in the activation of NLR proteins, since mutations in the ATP-binding region (Walker’s A box) or the Mg-binding region (Walker’s B box) abrogate signaling [2].
NLR subfamilies differ in the N-terminal effector domains, which they require for signal transduction. The majority of NLRs harbor a death-fold domain (DD) at their N terminus, which can be either a caspase recruitment domain (CARD) or a pyrin (PYD) domain. Based on similarity interactions, death-fold domains usually form dimers or multimers with other members of the same subfamily. An interaction across the subfamilies is very uncommon, which is somewhat surprising given the structural similarities among the different subfamilies. For NLRs, an N-terminal PYD recruits a PYD-containing molecule and an N-terminal CARD recruits a CARD-containing molecule to bridge NLRs to other signaling or effector proteins. In addition to NLRs with an N-terminal CARD or PYD domain, there are also NLRs with an N-terminal BIR (baculovirus inhibitor of apoptosis) domain, an N-terminal acidic activation domain (AD), or a further, undefined domain. Based on the molecular structure of the N-terminal domain, a recent nomenclature committee has distinguished the following NLR subfamily designations [3]: NLRA (NLRs with an acidic activation domain), NLRBs (NLR with a BIR domain), NLRCs (NLRs with a CARD domain), NLRPs (NLRs with a pyrin domain), and NLRXs (NLR family with no strong homology to the N-terminal domain of any other NLR subfamily member).
Whereas the two intensively studied NLRCs, NOD1 and NOD2, initiate NF-κB signaling and lead to the transcription of pro-inflammatory genes, several NLRCs and NLRPs are involved in the posttranslational activation of inflammatory caspases. A central paradigm to both caspase-1 activation and NF-κB activation by NLRs is the assembly of oligomeric complexes. These large molecular platforms activate their effectors via proximity-induced autoactivation. In allusion to the word “inflammation” and the Greek suffix “soma” meaning body, the high-molecular-weight complexes activating caspase-1 were termed “inflammasomes” [4]. Within the NLR family, we distinguish the NLRP1 inflammasome that assembles in response to MDP [5] and anthrax lethal toxin [6], the flagellin-sensing NLRC4 inflammasome [7], and the NLRP3 inflammasome, which is engaged by various diverse stimuli such as crystalline material, pore-forming toxins, bacteria, and viruses [8, 9]. Recently, we and others described the protein AIM2, which does not belong in the group of the NLR family, yet also forms an inflammasome in response to cytosolic DNA [10–13] (Fig. 2).
Fig. 2.
Known inflammasomes. The four known inflammasomes are depicted. NLRP1, NLRP3, and AIM2 all indirectly recruit and thereby activate caspase-1 through the bridging molecule ASC. IPAF has been shown to activate caspase-1 independent of ASC, yet there are several reports that stress the requirement for ASC in this process. Apart from this controversy, the exact role of NOD2 and caspase-5 in the NLRP1 inflammasome has to be elucidated and the interaction of NAIP5 with NLRC4 also needs additional clarification
Inflammasomes activate interleukin-1β
Caspases are generally thought to be apoptotic initiators or effectors. A subclass of caspases, known as the inflammatory caspases, is also involved in pro-inflammatory responses. Caspase-1 is the prime example of this subfamily. The inflammasome functions to convert inactive pro-caspase-1 to active caspase-1, which then cleaves inactive cytokine precursors to a secreted and active form. The known substrates that are processed by caspase-1 are the precursors of IL-1β, IL-18, IL-33, and IL-1F7 [14].
It is generally assumed that the respective inflammasome components oligomerize upon engagement of a direct or indirect ligand to recruit pro-caspase-1 via homotypic CARD–CARD interactions. The inflammasome initiators NLRP1, NLRP3, and AIM2 harbor an N-terminal PYD domain through which they cannot activate caspase-1 directly. Instead, they recruit the adaptor molecule apoptosis-associated speck-like protein (ASC) via their PYD domain. The CARD domain of ASC then in turn interacts with the CARD of pro-caspase-1 (Fig. 2). The formation of an inflammasome initiates proximity-induced autoactivation through autoproteolytic cleavage of the 45 kDa pro-enzyme of caspase-1 into active fragments. Active caspase-1 is an oligomeric enzyme of two subunits, 10 kDa (p10) and 20 kDa (p20), forming tetramers in solution. The cysteine protease caspase-1 was formerly known as IL-1β converting enzyme (ICE) due to its ability to process the 33 kDa inactive precursor of interleukin-1β (pro-IL-1β) to the carboxyl-terminal active fragment by proteolytic processing between the amino acid residues Asp116 and Ala117 [15]. Bioactive interleukin-1β is best known as endogenous pyrogen because of its ability to cause fever in experimental settings. Moreover, it has been shown to fulfill multiple functions in mediating inflammatory responses, e.g., by directly activating lymphocytes and epithelial cells. IL-1β is also a potent inducer of intercellular and endothelial adhesion molecule expression and thus promotes the infiltration of inflammatory and immunocompetent cells from the circulation into the extravascular space. IL-1β production has also been closely linked to a special type of cell death known as pyroptosis [16].
Due to the detrimental effects caused by overproduction of bioactive IL-1β, its activation and secretion are regulated by a tightly controlled process. Three steps are involved to avoid the accidental release of IL-1β. First, unlike caspase-1, which is constitutively expressed in resting cells, expression of IL-1β precursor mRNA needs to be induced by NF-κB-activating pathways. Although nearly all microbes induce IL-1β via TLR, CLR, or RLR ligands, IL-1β can additionally induce expression of itself via activating MyD88 through IL-1R binding. Second, inactive proIL-1β that accumulates in the cytosol is proteolytically processed by the inflammasome, which itself is tightly regulated. Third, IL-1β lacks a secretory signal peptide and does not take the classical exocytotic route of secretion. In fact, the nonclassical secretion pathway utilized by IL-1β is only partially understood. However, a process by which phospholipase C and A2 control externalization by specialized secretory lysosomes [17] as well as shedding of plasma membrane microvesicles [18] and direct release via transporters or multivesicular bodies containing exosomes have all been reported [19].
Caspase-1 can also cleave other members of the IL-1 family, namely IL-18 and IL-33. Unlike IL-1β, IL-18 is constitutively expressed in human PBMCs and other cell types and thus does not require a priming process for precursor induction [20]. Cleaved IL-18 activates NK cells and is a potent interferon gamma-inducing cytokine [21]. IL-33 binds to the orphan receptor T1/ST2 and drives Th2 responses [22]. In addition to its role in activating cell surface receptors, IL-33 acts as an “intracellular cytokine” by binding to nuclear DNA to regulate gene transcription [23], and in contrast to IL-1β and IL-18, IL-33 is not activated but inactivated through a proteolytic process involving caspase-1 [24–26]. Lastly, the cytokine IL-1F7 has been reported to be an anti-inflammatory member of the IL-1 family. It is structurally related to IL-18 but lacks true agonist activity. Inhibition of innate immune responses by IL-1F7 may be due to direct interactions with IL-18BP or IL-1Rα. However, it is still unclear whether cleavage of IL-1F7 plays a role in its in vivo function [14].
The NLRP1 inflammasome
Human NLRP1 differs from other NLRPs in its domain architecture. In addition to the N-terminal PYD and the centrally located NACHT and LRR domain, it also has a C-terminal extension consisting of a domain with unknown function (FIIND) and a CARD domain. The other known NLRPs do not have a comparable C-terminal extension (Fig. 2). The composition of two signal transduction domains (PYD and CARD) suggests that human NLRP1 may have multiple signaling roles, and it is worth noting that murine NLRP1 lacks the N-terminal PYD. In addition, unlike humans, who have a single NLRP1 gene, mice have several paralogs located on chromosome 11 (NLRP1a, NLRP1b, NLRP1c), and moreover five different strain-specific alleles exist for NLRP1b [6]. It was even hypothesized that NLRP1 could have a different role in mouse and man and that murine NLRP1 could rather fulfill CARD8 (CARDINAL)-like functions in mice [27]. The regulatory CARD-containing protein CARD8 shares high homology with the CARD of caspase-1 and the C-terminal region of NLRP1 [28], yet CARD8 is not present in the mouse genome. CARD8 has initially been shown to interact with NLRP3 thereby facilitating its inflammasome formation [29, 30], however, recent studies indicated that it might be dispensable for NLRP3 inflammasome activation [9].
NLRP1 was the first NLR family member characterized with respect to inflammasome assembly and caspase-1 activation. It was identified to be the central component of an inducible high-molecular-weight complex of approximately 700 kDa triggering caspase-1 activation. The term “inflammasome,” which had previously only been used for the NLRP1 multiprotein complex, was subsequently broadened for the description of further caspase-1 recruitment platforms. The human NLRP1 inflammasome was initially described to comprise NLRP1, ASC, caspase-1, and caspase-5. The NLRP1–caspase-5 interaction induces processing of caspase-5, whereas the adaptor molecule ASC is required to mediate interaction and subsequent processing of caspase-1 [4]. In recent work, the elicitors for NLRP1 signaling have been suggested to be the peptidoglycan component muramyl dipeptide (MDP) of bacterial cell walls and anthrax lethal factor (LF) of the Bacillus anthracis lethal toxin (LeTx) [5, 6].
The molecular mechanism of NLRP1 activation was mainly elucidated by Reed and colleagues employing cell-free human in vitro reconstitution models [5]. They defined the minimal protein components for NLRP1 inflammasome assembly as NLRP1 and caspase-1, which interact via CARD–CARD interactions. MDP-dependent NLRP1 activation was sufficient for caspase-1 cleavage, but robust NLRP1 inflammasome activation required the addition of small amounts of the adaptor molecule ASC. A two-step mechanism was suggested for NLRP1 activation. First, MDP binds to or alters the LRR of NLRP1 to induce a conformational change, which in turn allows subsequent binding of NTPs and self oligomerization via the NACHT domain [5]. This model is supported by previous reports stating that deletion of LRRs renders NLRP1 able to bind ATP and constitutively active [4]. Oligomers of NLRP1 form double ring structure in dimensions with striking similarity to the central hub of the APAF-1 apoptosome suggesting that APAF-1 and NLRP1 organize in a similar higher order structure by using their oligomerization domain as a central hub [5]. The anti-apoptotic proteins BCL-2 and BCL-xl directly bind and potently inhibit NLRP1 inflammasome activation and subsequent caspase-1 cleavage, possibly by inhibition of ATP binding. The inhibitory effect could be mapped to a 10-mer loop peptide, which binds to and inhibits NLRP1 with high affinity. Moreover, MDP-mediated IL-1β production was elevated in mice deficient for BCL-2, providing evidence that the inhibitory effect may also account for the murine system [31, 32]. Even though the bacterial cell wall component MDP strongly activates caspase-1, direct binding of MDP to NLRP1 was not sufficiently demonstrated and raises the possibility that MDP-triggered NLRP1 activation may be indirect. In fact, recent work suggests a surprising role for NOD2 (NLRC2)—another NLR member—in the assembly of the NLRP1 inflammasome in response to both MDP and anthrax lethal toxin [33]. NOD2 serves as an intracellular sensor for MDP and contains two N-terminal CARDs and initiates activation of NF-κB and MAPK kinases via RIP2 [34]. Even though in a cell-free system NLRP1 is sufficient to activate caspase-1 in response to MDP, NOD2 is needed for in vivo sensing of both MDP and LT. It was shown in this study that NOD2 could directly interact with caspase-1 and NLRP1 but not with NLRP3. It seems that NOD2 could generate both signals required for secretion of bioactive IL-1β, NF-κB-dependent induction of pro-IL-1β expression and second, binding and activation of caspase-1 through its N-terminal CARD to process the pro-cytokine to its active form [33, 35].
The second well-characterized stimulus for NLRP1 is cytosolic anthrax lethal factor derived from the spore-forming bacterium B. anthracis, the causative agent of anthrax disease. B. anthracis secretes lethal toxin (LT), which is composed of two protein subunits, protective antigen (PA) and lethal factor (LF). LF can be transported into the cytosol of many cell types by PA. In mouse macrophages, LF causes rapid necrosis that is central to the pathology of systemic B. anthracis infections. LF is a Zn2+-dependent endoprotease, cleaving the N terminus of MAPK kinases to alter signaling pathways. Inbred mouse strains display variable sensitivity to LT-induced macrophage necrosis [36]. Genetic studies have mapped this trait difference to variations in the polymorphic gene NLRP1b on chromosome 11. Moreover, the NLRP1 paralog NLRP1b has been identified to be the primary mediator of mouse macrophage susceptibility for LT. It has been reported that LT induced macrophage death does indeed require caspase-1, which is activated in susceptible but not resistant macrophages after in vitro treatment with LT [6, 37]. Furthermore, activation of caspase-1 by LT requires binding, uptake, and endosome acidification to mediate translocation of LF into the host’s cytosol. Catalytically active LF cleaves cytosolic substrates by a mechanism involving proteasome activity and Ca2+ ion flux. Catalytically inactive mutant LF, which shares a structure virtually identical with active LF, fails to activate caspase-1, making a model of direct recognition of LF as a ligand for NLRP1 unlikely. LF may instead cleave additional, as yet unknown targets, leading to the degradation of NLRP1 inhibitors or the production of activating factors that trigger NLRP1 inflammasome activation [38–40].
Undoubtedly, further work is needed to elucidate the exact function of NLRP1 and NOD2 in response to anthrax lethal toxin and MDP. It is currently unclear whether MDP and anthrax LF are sensed directly by NLRP1 or together with NOD2. Up to now, the role of MDP in NLRP1 inflammasome activation has been exclusively addressed in human cells by loss of function experiments, whereas the requirement of NOD2 has been concluded from mouse studies [33]. Thus, it cannot be formally excluded that NLRP1 may be an actual MDP receptor. Another issue that needs to be clarified is the exact role of ASC in the NLRP1 inflammasome. Since murine NLRP1 lacks an N-terminal PYD, a direct interaction of murine ASC and NLRP1 seems to be unlikely.
Interestingly, DNA sequence variants in the human genomic NLRP1 region confer risk of several autoimmune diseases, such as generalized vitiligo, vitiligo-associated type I diabetes, and Addison’s disease [41, 42]. Generalized vitiligo is a multifactorial disease in which loss of melanocytes results in spotty depigmentation of skin, hair, and mucous membranes. Serum levels of IL-1β are elevated in patients with generalized vitiligo demonstrating the involvement of the IL-1β-processing pathway in the pathogenesis of this disease [43]. A single nucleotide polymorphism (SNP) in the coding region of NLRP1 has provided evidence of an association with vitiligo. The leucine to histidine substitution at amino acid 155 is located between the N-terminal PYD and the NACHT domain of human NLRP1. Even though this region does not code for a known motif, it is highly conserved suggesting that this region is critical for protein function and in fact, it has been implicated as a region for NOD2–NLRP1 interaction [44]. Other disease-linked SNPs are located in the promoter region and could affect transcription-factor-binding sites [41, 42].
The NLRC4 inflammasome
NLRC4 (also known as IPAF, Card12) has been shown to regulate caspase-1 activation and IL-1β processing in response to infection with various gram-negative bacteria. Activation of NLRC4 pathways is closely related to the subsequent cell death of infected cells. NLRC4 contains an N-terminal CARD, a central NBD, and a C-terminal LRR domain. The deletion of the LRR domain of NLRC4 results in a constitutively active form of the molecule suggesting that NLRC4 activation occurs in a similar manner to other NLRs that also gain activity upon deletion of their LRR. NLRC4 was shown to associate directly and specifically with the CARD domain of pro-caspase-1 through CARD–CARD interactions [45]. Nevertheless, some studies have demonstrated that ASC is functionally involved in NLRC4-mediated caspase-1 activation, which makes the requirement of ASC a subject of controversy [46]. ASC may stabilize or facilitate caspase-1 recruitment to NLRC4. Alternatively, NLRC4 may cooperate with a yet-to-be-defined PYD-containing NLRP protein to recruit ASC via PYD–PYD interaction, which in turn interacts with caspase-1.
NLRC4-deficient macrophages show a markedly reduced activation of caspase-1, release of bioactive IL-1β, and pyroptosis after infection with the gram-negative bacteria Salmonella typhimurium [46], Legionella pneumophila [47], Shigella flexneri [48], and Pseudomonas aeruginosa [49]. These bacteria have in common that they possess either a type III (T3SS) or type IV secretion system. It was initially found that S. typhimurium and L. pneumophila strains deficient in flagellin were defective in their ability to activate caspase-1 in macrophages [47, 50]. In addition, delivery of purified flagellin into the host cell cytosol was capable of activating caspase-1 in an NLRC4-dependent manner. Notably, caspase-1 activation by means of NLRC4 is independent of TLR5 [7]. These results led to the presumption that flagellin is the primary PAMP activating NLRC4.
Polymers of the protein flagellin are structural compounds of flagella, a structure anchored to the bacterial cell wall that enables bacterial motility. Flagellin is the prime example of a PAMP activating host innate immune responses since it is an evolutionarily conserved element and vital for mobile bacteria in their natural environment. However, only flagellin from specific bacteria induces inflammasome activation via NLRC4. Flagellin of Escherichia coli fails to trigger cytosolic inflammasome activation, even if delivered to the host cytosol [51]. Thus the ability of NLRC4 to distinguish different sources of flagellin might be a host strategy to distinguish pathogenic from commensal bacteria. In addition, the expression of flagellin alone is not sufficient to trigger NLRC4 activation. A specific virulence factor transport system is additionally needed for cytosolic delivery. Salmonella requires SipB, a translocase of the salmonella SPI1 T3SS, to induce IL-1β secretion in infected cells. SipB is a component of the transmembrane pore inserted into the eukaryotic cell membrane that delivers proteins to the eukaryotic cell cytosol. The flagellated gram-positive bacterium Listeria monocytogenes, on the other hand, can also trigger NLRC4 activation, even though these bacteria do not express a secretion system. In this case, flagellin can gain access to the cytosol and thus NLRC4 when Listeria escape the phagolysosome to replicate in the cytosol [52].
L. pneumophila is unique amongst flagellated bacteria activating NLRC4 in that NAIP5 in addition to NLRC4 is critically involved in susceptibility to caspase-1 cleavage. NAIP5 and NLRC4 interact physically, and a C-terminal region of Legionella flagellin spanning 35 amino acids was identified to specifically activate this inflammasome. Of note, NAIP5 was dispensable if full-length flagellin from Legionella was delivered to the cytosol. These conflicting results were discussed to be due to overexpression artifacts, since flagellin-dependent activation of the inflammasome is strictly dependent on NAIP5 after natural L. pneumophila infection [53]. Other reports suggest that NAIP5 may have additional functions beyond its interaction with NLRC4. NAIP5 signaling may render macrophages permissive to L. pneumophila infection. Nunez and colleagues proposed an essential role for NAIP5 signaling to promote phagosome–lysosome fusion. The restriction of Legionella growth within macrophages and different bacterial loads of macrophages could explain the results seen with NAIP5-deficient cells regarding flagellin-dependent activation of caspase-1 [54].
The NLRC4 inflammasome can also be activated independent of flagellin, yet this still requires a functional SPI1 T3SS [50]. The nonflagellated bacterium S. flexneri is capable of activating caspase-1 in an NLRC4-dependent manner [48]. In addition, the P. aeruginosa mutant PAK (Delta) fliC, which is deficient in flagellin, is still capable of activating caspase-1 in an NLRC4-dependent manner [55]. Yet it should be mentioned that other groups did not observe caspase-1 activation upon infection with P. aeruginosa deficient in flagellin [49, 56]. A recent publication can explain many of these puzzling findings. Miao et al. showed that NLRC4 can indeed detect the basal body rod component of the T3SS apparatus (rod protein) of several gram-negative bacteria. These rod proteins share a sequence motif that is similar to the C-terminal part of flagellin and essential for the detection by NLRC4 [57]. In summary, these data imply that NLRC4 senses not only cytosolic flagellin but also conserved structural features of virulence factors shared by gram-negative pathogens.
Although it has been sufficiently demonstrated that bacterial flagellin triggers activation of caspase-1 via NLRC4, the molecular mechanism rendering NLRC4 active is rather ambiguous. One possibility is direct binding of flagellin to NLRC4. Binding to NLRC4 might also require the cooperation of a yet-to-be-defined host factor, possibly another NLR or NLRC4 might simply act as a signaling adaptor downstream of the flagellin recognition machinery. Finally, flagellin itself may not be recognized by NLRC4, but instead may induce a cellular response factor that is sensed by NLRC4. So far there is no proof for direct interaction of NLRC4 and flagellin [7]. Since the interaction may only be transient, this does not formally exclude that NLRC4 is an actual receptor.
The NLRP3 inflammasome
The NLRP3 inflammasome has received particular attention due to its diverse and prominent function in sterile inflammatory responses, antimicrobial responses, adjuvanticity, and hereditary autoinflammatory syndromes [58]. NLRP3 is the most extensively studied NLR family member and thought to be a general sensor of DAMP signals. NLRP3 (also known as NALP3, cryopyrin, PYPAF1) contains an N-terminal PYD, a central NBD, and a C-terminal LRR domain. NLRP3 recruits the bridging molecule ASC via homotypic PYD–PYD interactions to activate NF-κB [59] and caspase-1 [29, 30]. The oligomerization of NLRP3 in response to a stimulus requires binding of ATP to the central nucleotide-binding element [2]. In addition, CARD8 has been implicated as a facilitator of NLRP3 inflammasome assembly.
NLRP3 activation can be triggered in response to a broad number of diverse stimuli including those of microbial origin (bacteria and viruses), endogenous origin (endogenous danger signals), and exogenous nonmicrobial origin (crystalline particles). It is still unclear whether these stimuli converge in a common pathway to activate NLRP3.
Stimuli activating NLRP3
Microbes that can activate the NLRP3 pathway include Sendai virus [60], influenza A virus [9, 61, 62], adenoviruses [63], Staphylococcus aureus [64], L. monocytogenes, [52, 64, 65] E. coli, Mycobacterium marinum, S. flexneri, Neisseria gonorrhoe [66], and Candida albicans [67–69]. In several studies, NLRP3 inflammasome activation by bacteria could be pinpointed to a specific pore-forming toxin by either showing that the respective purified pore-forming toxins activate NLRP3 or by showing that bacteria deficient for specific pore-forming toxins lost their ability to activate NLRP3 (Table 1). Moreover, for influenza A virus, it could be demonstrated that the virus-encoded M2 ion channel protein was responsible for NLRP3 inflammasome activation [70]. Nevertheless, it remains still unclear for many of the live microbes whether inflammasome activation is caused by a single PAMP, a combination of multiple pathogenicity factors, or damaging side effects caused by the mode of infection.
Table 1.
Defined molecular structures activating inflammasome pathways
| Inflammasome | Stimulus | Reference |
|---|---|---|
| NLRP1 | Anthrax lethal toxin | [6] |
| MDP | [32] | |
| NLRP3 | Pore-forming agents | |
| Listerolysin O | [64] | |
| Streptolysin | [136] | |
| Nigericin | [64] | |
| Maitotoxin | [137] | |
| Saponin | [116] | |
| Hemolysins | [138] | |
| Ion channels or their activators | ||
| P2X7/ATP | [64] | |
| Influenza virus M2 channel protein | [70] | |
| Phagocytosed materials (crystals, crystalline material, particles, protein aggregates) | ||
| Monosodium uric acid crystals | [73] | |
| Asbestos fibers | [76] | |
| Silica | [75, 76] | |
| Aluminum salts | [75, 78, 116] | |
| Hemozoin crystals | [80, 82] | |
| Cholesterol crystals | [99] | |
| Poly lactide-co-glycolide and polystyrene microparticles | [139] | |
| Amyloid beta | [84] | |
| Hyaluronan | [140] | |
| NLRC4 | Flagellin | [7] |
| Components of the T3SS | [57] | |
| AIM2 | Double-stranded DNA | [10–13, 130, 131] |
For clarity, only studies that have reported a molecularly defined activating signal in a loss-of-function or knock-out experiment for the respective inflammasome component were considered. In this regard, NLRP3-activating signals that are only defined for their priming component (signal I) but not their activating component (signal II) are not listed
A high concentration of extracellular ATP, one of the first activators described to induce NLRP3 inflammasome formation, is ascribed to the group of endogenous DAMPs. The ubiquitous energy source ATP is found at high intracellular levels. On the contrary, the concentration is low in the extracellular space due to the ubiquitous activity of ATPases. A low level of extracellular ATP correlates with the presence of live cells, and transient increases are physiologically used for signaling, best known in the nervous system. However, a large increase in extracellular ATP is often seen during cell death. Binding of extracellular ATP to the purinergic receptor P2X7 leads to NLRP3 activation via ion flux effects mediated by the P2X7-receptor-associated hemi-channel pannexin-1 [71]. While this is an appealing model of necrotic cell sensing by the NLRP3 inflammasome, it is notable that in vitro high ATP concentrations in the millimolar range are required to activate NLRP3. Even though ATP can be measured in the extracellular space during cell death, the levels that are found here are far lower.
Nevertheless, a common denominator of extracellular ATP and bacterial toxins activating NLRP3 is the ability to form membrane pores that induce damage of membrane integrity or cause perturbation of the intracellular ion concentration. An involvement of potassium ion fluxes in NLRP3 activation in response to ATP and pore-forming toxins is supported by the fact that high extracellular potassium levels that decrease the concentration gradient between the extra- and intracellular compartment prevent NLRP3 activation [72].
Phagosomal material (crystals, crystalline material, particles, protein aggregates) is another class of stimuli activating NLRP3. The first crystal-induced inflammatory diseases linked to the NLRP3 inflammasome were gout and pseudogout. The demonstration of NLRP3 activation and IL-1β release by monosodium urate (MSU) crystals and calcium pyrophosphate dehydrate (CPPD) crystals was a milestone in inflammasome research [73]. Uric acid, which is normally soluble under physiologic conditions, precipitates in the joints of patients with hyperuricemia to form MSU crystals. Although it has been known for a long time that IL-1β is activated in monocytes by MSU crystals in vitro, IL-1β was only recently identified as the pivotal cytokine in gouty inflammation [74].
In analogy to the effect caused by gouty crystals, subsequent studies have described several other environmental pollutants consisting of inorganic crystalline material and also endogenous aggregates that trigger NLRP3 activation. Inhalation of crystalline silica and asbestos is known to cause pneumonitis and the progressive pulmonary fibrotic disorders silicosis and asbestosis, respectively. This process was shown to occur in an NLRP3-dependent manner. Macrophages deficient in components of the NLRP3 inflammasome were incapable of secreting the pro-inflammatory cytokines IL-1β and IL-18 in response to silica or asbestos fibers [75–77]. A similar mechanism seems to be engaged by aluminum salts that are commonly used as adjuvants in vaccines [75, 78, 79]. Alum, the most commonly used adjuvant, triggers caspase-1 cleavage via NLRP3. Whether general engagement of NLRP3 is the crucial step used by adjuvants for the induction of an adaptive immune response is somewhat controversial and discussed separately in this review.
Sensing of crystalline particles by NLRP3 seems to be a general phenomenon and not restricted to exogenous inorganic material. For example, malarial hemozoin is also sensed by NLRP3 due to its particulate nature. Hemozoin is an insoluble heme crystal formed by Plasmodium spp. to protect themselves from oxidative damage that results from the presence of free heme [80]. However, whether processing of pro-inflammatory cytokines by the inflammasome is critically involved in animal models of experimental malaria is controversial [80–82]. It has also been hypothesized that inflammasome activation by hemozoin could be indirect via the release of uric acid [83].
In addition to particles formed by pathogens, endogenous peptide aggregates can cause NLRP3 activation. Accumulation of extracellular amyloid-beta in senile plaques is a morphologic feature occurring in the pathogenesis of Alzheimer’s disease. These fibrillar aggregates are sensed by microglia cells via NLRP3 in the central nervous system and lead to the secretion of pro-inflammatory cytokines, nitric oxide, and to subsequent cell death of neuronal bystander cells [84]. Elevated levels of IL-1β were found in brain tissue sections of patients suffering from Alzheimer’s disease [85]. Further animal studies are required to address whether inflammasome activation plays a role under more physiologic conditions in the pathogenesis and progression of Alzheimer’s disease (for an overview of NLRP3 activating structures, see Table 1).
The NLRP3 inflammasome is tightly regulated by a transcriptional step
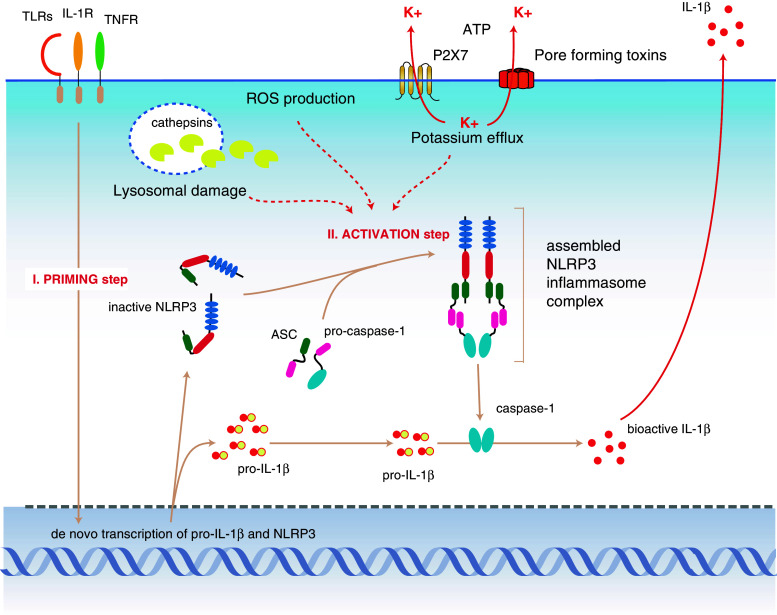
Numerous microbial components, in particular LPS, single-stranded RNA, double-stranded RNA, peptidoglycans, and CpG DNA, have been reported to activate the NLRP3 inflammasome [86, 87]. A direct entry via pores and cytosolic recognition was favored for those stimuli [86], yet it remained unclear how one sensor could bind to all the reported ligands given the differences in their structure and shape. However, two recent studies have indicated that the spectrum of ligands activating NLRP3 is much less broad from a mechanistic point of view [8, 88]. Comprehensive studies using macrophages deficient in TLRs, TLR adaptor proteins, and NLRs showed that the contribution to inflammasome activation of ligands described above is strictly dependent on TLRs and TLR signaling. These TLR-engaging PAMPs lead to an enhanced transcription of NLRP3 via the transcription factor NF-κB. The level of NLRP3 expression is critical for inflammasome activation by NLRP3-activating stimuli, and normal baseline expression is not sufficient for caspase-1 cleavage in unprimed macrophages. However, macrophages that are modified by retroviral gene transfer to constitutively express elevated levels of NLRP3 do not require a priming step via TLRs or NF-κB-activating cytokine receptors for caspase-1 cleavage in response to ATP or nigericin [8]. Therefore, it makes sense to categorize stimuli involved in NLRP3 activation as NLRP3 “primers” and NLRP3 “activators” (Fig. 3). Stimuli priming NLRP3 include all ligands for TLRs, RLRs, NLRs, and cytokine receptors that lead to enhanced NLRP3 expression. Actual NLRP3 activators induce caspase-1 cleavage in a rapid process that is independent of de novo transcription. At this point, at least three technically distinct pathways have been implicated as NLRP3 activators: K+-efflux-inducing stimuli (e.g., pore-forming toxins, ion channels), stimuli inducing lysosomal disintegration (e.g., crystals, protein aggregates, bacteria, viruses) or ROS-inducing stimuli.
Fig. 3.
The NLRP3 inflammasome. NLRP3 expression is the limiting factor in cells that can principally respond to NLRP3 activation. Stimulation of PRRs or cytokine receptors leads to the upregulation of NLRP3 expression and also induces pro-IL-1β expression (priming step). The activation of NLRP3 itself is distinct from this initial priming step. Three presumably distinct pathways have been postulated that can lead to the activation of NLRP3 (activation step). Efflux of potassium, which is initiated by pore-forming toxins, membrane disruption, or ligand-triggered channels, leads to NLRP3 inflammasome assembly by a yet-unknown mechanism. On the other hand, it has been shown that lysosomal disintegration, which leads to the leakage of lysosomal enzymes into the cytosol, can also trigger NLRP3 activation. The lysosomal protease cathepsin B plays a major role in this pathway, yet the direct link between cathepsins and NLRP3 activation hasn’t been elucidated. Lastly, it has been demonstrated that production of reactive oxygen species (ROS), which can be triggered by numerous means, leads to the indirect activation of NLRP3 through the release of TXNIP from thioredoxin. TXNIP can then bind to NLRP3
The fact that priming is a necessary step for NLRP3 inflammasome assembly suggests that macrophages need to acquire a signal that indicates either the presence of infection (via activation of pattern recognition receptors by microbial products) or the presence of other stimulated cells (via the presence of pro-inflammatory cytokines) to commit to sense danger signals in their immediate environment via the activation of the NLRP3 inflammasome. This dual stimulation requirement may operate to prevent accidental or uncontrolled NLRP3 activation, which can have devastating consequences for the host. Priming, as a critical necessity for NLRP3 inflammasome activation, has been overlooked for a long time. This is due to the fact that priming stimuli were usually included in NLRP3 studies to induce pro-IL-1β production, which is often studied as a read out for NLRP3 activation instead of caspase-1. However, pro-IL-1β and NLRP3 expression are not necessarily linked, and NLRP3 activation also exerts biological effects in the absence of caspase-1 or pro-IL-1β. This implies that dissecting pro-IL-1β from NLRP3 priming is not only important from a mechanistic point of view but also relevant with regard to the various biological outcomes downstream of NLRP3 activation.
Another paradigm that has been confusing the field is the observation that TLR stimulation is known to induce secretion of bioactive IL-1β in human PBMCs or monocytes without the need of a second exogenous NLRP3 stimulus such as ATP. Thus, it was hypothesized that the dual step mechanism of NLRP3 activation described above does not account for the human system. However, two observations argue against this theory: Firstly, extracellular release of ATP—a potent NLRP3 activator—is observed in human monocytes upon TLR stimulation and may explain the ability of TLR ligands to induce IL-1β maturation in the presumed absence of a second stimulus [89, 90]. Secondly, a very low concentration of TLR ligands that is still sufficient to induce pro-IL-1β does not lead to NLRP3 activation and IL-1β release [8]. Hence, the requirement of two signals for inflammasome activation is also applicable to human PBMCs.
In summary, macrophages need to acquire a licensing signal provided by transcriptionally active signaling receptors that enables them to respond to NLRP3 activators. The licensing or priming step via NF-κB activation induces the expression of both pro-IL-1β and NLRP3. Live bacteria often provide both signals required for IL-1β secretion, induction of IL-1β and NLRP3 via NF-κB-activating PAMPs, and NLRP3 activation via bacterial toxins or ion channels.
Molecular mechanism of NLRP3 activation
As already mentioned above, the actual event leading to NLRP3 activation remains elusive. It has been shown that potassium efflux is required for NLRP3 activation after various stimuli [72, 91]. These observations stem from experiments in which high extracelluar potassium concentrations are used to inhibit its efflux from cells. However, additional studies have indicated that not only NLRP3, but also NLRP1 [39], NLRC4 [92], and AIM2 [63, 93] are sensitive to high, albeit different, extracellular potassium concentrations. Therefore, it remains unclear whether potassium efflux alone represents a NLRP3-activating stimulus itself or whether low intracellular potassium acts as an additional requirement for inflammasome assembly per se. It is possible that formation of the NLRP3 inflammasome is highly favored at low intracellular potassium concentrations, which may be required, yet not be sufficient for activation.
Because of the C-terminal LRR, NLRP3 was implicated in a model where it directly senses a ligand by functioning as a cytosolic receptor. LRRs are utilized by other receptors to directly bind microbial products. The ligand-receptor interaction could result in a conformational change allowing the recruitment of ASC. Since NLRP3 is activated by a broad range of stimuli—and it seems unlikely that one sensor could bind to all the diverse activators—this hypothesis was abandoned to favor a model where NLRP3 detects an intermediate signaling molecule induced by NLRP3 activators. Another theory deals with a posttranslational modification of NLRP3 induced by an upstream event leading to activation. It is even possible that NLRP3 is solely a signaling protein downstream of the actual receptor or employs other co-receptors such as NLRC4 to cooperatively sense its ligands. In favor of the latter hypothesis, the production of reactive oxygen species (ROS) has been suggested to act as a common event upstream of NLRP3 induced by extracellular ATP, MSU crystals, asbestos, and silica [76]. It is well known that phagocytosis of particulate matter in macrophages results in generation of reactive oxygen species (ROS) [94], and ATP-induced inflammasome activation has been linked to ROS, as well [95]. ROS production is catalyzed by the family of NADPH oxidases (NOX) that include the membrane-bound NOX2 system, which is predominant in phagocytes, but also the NOX1, NOX3, NOX4, NOX5, DUOX1, and DUOX2 complexes. [96]. In addition, ROS can also derive from other cellular sources, such as mitochondria or xanthine oxidase. ROS inhibition by N-acetyl-l-cysteine (NAC) and (2R,4R)-4-aminopyrrolidine-2,4,-dicarboxylate (APDC) can reduce the amount of IL-1β secretion after stimulation with ATP and crystals [72, 76, 95]. Even H2O2 is sufficient to trigger the processing of caspase-1 through activation of NLRP3. Furthermore, ROS production and NLRP3 activation is reduced upon treatment with diphenylene iodonium and apocynin, which inhibit the NADPH oxidase. The knock down of the NADPH oxidase subunit p22phox, which is critically required for the activity of NOX1–4, also diminishes IL-1β secretion [76]. In line with a model predicting NLRP3 inflammasome activation by NADPH oxidase-produced ROS, RNAi-mediated knock down of the ROS-detoxifying protein thioredoxin (TRX) in macrophages shows increased IL-1β secretion upon NLRP3 activation [76]. In addition, Tschopp and colleagues have demonstrated a direct interaction between NLRP3 and thioredoxin-interacting protein (TXNIP), a protein linked to insulin resistance. Inflammasome activators such as uric acid crystals induce the dissociation of TXNIP from thioredoxin in a ROS-sensitive manner and allow binding of TXNIP to NLRP3. TXNIP deficiency incompletely impaired activation of the NLRP3 inflammasome for crystals and ATP, and mice deficient for TXNIP have a reduced influx of neutrophils after intraperitoneal injection of MSU [97].
However, macrophages from mice deficient for NOX2 (also known as gp91phox) respond normally to ATP or crystals [75], although mice deficient in this subunit lack phagocyte superoxide production and are hypersusceptible to various pathogens [98]. In addition, macrophages from chronic granulomatous disease (CGD) patients deficient in any of the phagocyte oxidase subunits p22phox, p47phox, or gp91phox respond normally to established NLRP3 ligands. In fact, these patients even display symptoms of sterile inflammation while they are defective in ROS production. In line with these observations, it was recently reported that ROS negatively regulates caspase-1 activity. In this respect, it was shown that mice deficient in the reactive superoxide species detoxifying enzyme SOD1 display a markedly decreased caspase-1 activity. The reason for these contradictory results is currently still unknown, yet it is possible that differences in species or the experimental systems account for these stark contrasts.
Partially conflicting with this model, where ROS induced by NLRP3 activators release TXNIP from thioredoxin to bind and activate NLRP3, an alternative hypothesis favors a mechanism in which lysosomal rupture in response to crystals activates NLRP3. NLRP3 activation by particles requires binding and uptake by phagocytosis. Macrophages rapidly engulf crystals into intracellular compartments, a process inhibited by the actin-destabilizing drug cytochalasin D. Crystal uptake then leads to lysosomal swelling and destabilization resulting in the leakage of lysosomal contents into the cytosol. Lysosomes contain a plethora of proteolytic enzymes, many of which are activated by acidification of lysosomal pathways, and blocking the H+-ATPase system, which is required for acidification of lysosomal compartments by bafilomycin A, inhibits NLRP3 activation upon crystal treatment whereas stimulation with ATP proceeds normally. Inhibition of the lysosomal protease cathepsin B as well as cathepsin B deficiency leads to greatly reduced caspase-1 activation in response to silica or MSU crystals [75, 84, 99]. This suggests a pivotal function for lysosomes in crystal-mediated NLRP3 activation. Moreover, a crystal-independent induction of lysosomal damage by the molecule Leu-Leu-OMe, which is converted into a membrane-disrupting compound by a lysosomal peptidase, can trigger the NLRP3 inflammasome. Of note, cathepsin B inhibition only down-modulates NLRP3 activators that require phagocytosis, whereas stimuli that activate from the cell surface (e.g., ATP) are not affected.
These data suggest a common mechanism of crystal-induced activation of the NLRP3 inflammasome whereby lysosomal perturbation, not the crystal structure itself, is sensed. According to this model, particles such as silica, MSU, or amyloid-beta induce lysosomal damage and leakage, which is perceived as an endogenous danger signal by the innate immune system. Cathepsin B may gain access to the cytosol to induce the cleavage of a yet-unidentified substrate, which in turn leads to NLRP3 activation [75, 84]. It seems likely that other lysosomal proteases are involved as well because cathepsin B-deficient macrophages still display NLRP3 activation triggered by phagosomal activators, when stimulated at very high concentrations [84, 99]. In this regard, the potent inhibition of NLRP3 activation by pharmacological inhibition of cathepsin B using CA-074-Me implies that other enzymes might be targeted by this small molecule as well.
While these theories for different modes of NLRP3 inflammasome activation are seemingly different for crystal-induced NLRP3 activation, they are not mutually exclusive. It is possible that ROS generation contributes to lysosomal destabilization. On the other hand, the release of lysosomal protease could influence ROS production. Moreover, neither hypothesis can fully explain the general need for lowering the intracellular potassium levels for the activation of NLRP3 and other inflammasome pathways. Further studies are needed to address how these particular aspects of NLRP3 activation integrate into an overall picture.
Hereditary NLRP3-associated syndromes
Hereditary or de novo mutations in the coding region of NLRP3 are responsible for a group of disorders known as cryopyrin-associated periodic syndromes (CAPS, also known as cryopyrinopathies). These hereditary periodic fever syndromes differ in the severity of symptoms and encompass a spectrum from mild to severe disease. Patients with familial cold autoinflammatory syndrome (FCAS), Muckle-Wells syndrome (MWS), and neonatal-onset multisystem inflammatory disease (NOMID, also known as chronic infantile neurologic cutaneous articular syndrome) present with cold-induced fevers, urticaria-like rash, constitutional syndromes, arthritis, aseptic meningitis, and other types of localized inflammation [100]. The gene and protein product responsible for these symptoms was initially denoted as CIAS1 or cryopyrin because of its role in cold-induced fevers. These mutations were discovered long before the molecular function of cryopyrin was known in any detail [58, 101, 102]. However, in the current nomenclature, the structurally derived name NLRP3 has now replaced the functionally descriptive name cryopyrin [3].
PBMCs of mutation-positive MWS and NOMID patients secrete higher levels of IL-1β in response to LPS compared with healthy controls [103], suggesting that these mutations lead to constitutively activated NLRP3 molecules. Indeed, the genomic sequence coding for the NACHT domain of NLRP3 has been confirmed to be the major locus of CAPS-associated mutations, and some disease-associated NLRP3 variants exhibit increased oligomerization of ASC, thereby facilitating caspase-1 activation [104]. Sequence variants may confer ATP binding to the ATP-binding motif of NLRP3 independent of stimulation by exogenous PAMPs or DAMPs, rendering the molecule autoactive, and disease-associated forms of NLRP3 still require nucleotide binding for its activation [2].
However, approximately one-third of patients with classic presentation lack mutations in the coding region of NLRP3. Sequence variants in the promoter region were found in a portion of these mutation-negative patients, possibly leading to enhanced gene transcription [105]. In addition, a single nucleotide polymorphism in the 3′ UTR of NLRP3 has been shown to be linked to disease [106]. Thus, these data also provide a genetic indication that the expression level of NLRP3 is a critical determinant of NLRP3 inflammasome activity. This is in line with the finding that NLRP3 critically requires priming for its activation.
Therapies based on the current understanding of NLRP3 mechanisms are remarkably effective. Even treatment of the most severe cryopyrinopathy, NOMID, with interleukin-1 receptor antagonist anakinra markedly improves clinical and laboratory manifestations. Of note, the therapy responsiveness of patients with NOMID is independent of the prevalence of NLRP3 mutations [107]. The aforementioned findings were recently verified by the use of NLRP3 CAPS mutant knock-in mouse strains. Mice bearing the mutant allele have poor growth and systemic inflammation similar to human CAPS patients and demonstrate early mortality that is primarily mediated by myeloid cells. The disease phenotype requires the inflammasome components ASC and caspase-1 but is only partially dependent on IL-1β and independent of adaptive immune responses.
Role of NLRP3 in endotoxic shock
Mice treated with high doses of lipopolysaccharides (LPS) produce a plethora of pro-inflammatory cytokines accompanied by high lethality—a phenomenon also known as endotoxic shock. Many genetically modified mouse models lacking key molecules involved in LPS-mediated signaling are protected from endotoxic shock, and, interestingly, ASC-deficient and caspase-1 deficient mice have also been shown to have an increased resistance to high levels of LPS, possibly mediated by a decreased production of IL-18 and IL-1β [108, 109]. In contrast to ASC-deficient mice, no significantly enhanced survival was observed for NLRP3-deficient mice treated with high dose LPS, despite reduced serum levels of IL-1β. However, the protection from endotoxic shock was partial for low doses of LPS [64, 110]. Hence, NLRP3 seems to play a partial role in protection from endotoxic shock, although to a far lesser degree than ASC and caspase-1. Another ASC-interacting pyrin-domain-containing protein could account for the partial phenotype observed in NLRP3-deficient mice.
NLRP3 and adaptive immune responses
For almost one century, aluminum hydroxide (alum) has been the only vaccine adjuvant approved worldwide. Despite the fact that it has been injected in billions of people, the molecular mechanisms responsible for its induction of adaptive immune responses are still not fully understood [111]. It is well known that alum absorption increases antigen uptake by dendritic cells [112]. However, in addition to functioning as an antigen delivery system, alum also has immunostimulatory activity in vivo and leads to recruitment of monocytes to the site of injection. Subsequently, dendritic cell migration into lymph nodes and the spleen is implicated in priming and expansion of antigen-specific immunity [113]. Notably, the strong alum-induced Th2 response is independent of TLR signaling [114, 115]. Recently, several reports have demonstrated that aluminum hydroxide is sensed by the NLRP3 inflammasome in vitro in analogy to other crystalline particles [75, 78, 79, 116–118].
While several reports agree on the molecular mechanism of NLRP3 activation in vitro, there is conflicting data on the contribution of NLRP3 activation to alum adjuvanticity in vivo. Li et al. [116] and Eisenbarth et al. [78] show that the NLRP3 inflammasome components NLRP3, ASC, and caspase-1 are required for the induction of a robust antibody response after subcutaneous administration of the model antigen ovalbumin together with alum. In contrast, Franchi et al. [118] demonstrate that NLRP3 is not essentially required for an alum-induced antibody response to intraperitoneally administered human serum albumin. Of note, these studies differ in many variables including vaccine formulation, choice of the model antigen, route of administration, immunization schedule, and also in the methods applied for measuring adaptive immune responses. Consequently, it is difficult to compare these data. Additional discrepancies may be due to the multiple mechanisms of action employed by alum. On the one hand, it functions as an antigen delivery tool, and, on the other hand, it accounts for NLRP3 immunostimulation. One can speculate that antigens that may be contaminated by immunostimulatory agents such as TLR agonists might require only the antigen delivery function of alum to induce efficient adaptive responses. In contrast, alum-induced responses for antigens that are internalized well by antigen-presenting cells but are poorly immunogenic may preferentially rely on NLRP3-dependent alum adjuvanticity.
While these early studies mostly focused on an immune response upon immediate treatment with stimuli activating NLRP3, an increasing number of investigators have begun to study the role of NLRP3 in the induction of adaptive responses in settings that are much more complex. Ghiringhelli et al. reported that activation of the NLRP3 inflammasome within dendritic cells is decisive for the immune response against dying tumor cells upon treatment with oxaliplatin. Anticancer chemotherapy of immune-sensitive thymomas was inefficient in mice deficient for the purinergic receptor P2X7, NLRP3, or caspase-1, and priming of tumor-specific T cells of tumor-draining lymph nodes relied on inflammasome components. In this model, chemotherapeutic drugs induce the release of ATP from dying tumor cells, which in turn activates NLRP3 through engaging the P2X7 receptor [119]. Another study shed light on the down-modulation of inflammasome activation in vivo, which is essential to prevent deleterious effects. NLRP3 activation in macrophages seems to be suppressed by effector and memory T cells by a process that requires cell-to-cell contact [120].
The AIM2 inflammasome
Prokaryotic viral and microbial DNA trigger a number of innate immune pathways that result in the production of type-I interferons and pro-inflammatory cytokines including bioactive IL-1β and IL-18. Unmethylated CpG sequences present in prokaryotic DNA are recognized in endolysosomal compartments via TLR9 [121], but once foreign DNA gains access to the cytosol, additional signaling pathways are engaged. Double-stranded DNA transcribed by RNA polymerase III can be sensed indirectly via RIG-I [122]. In addition, the protein DAI has been reported to recognize cytosolic DNA [123], but other as-yet-unidentified pathways also seem to be operational [124] (for a review see [125]). These signaling pathways lead to NF-κB and interferon regulatory factor (IRF) activation and thus transcription of pro-inflammatory cytokines. Tschopp and colleagues [63] demonstrated that the transfection of bacterial, viral, mammalian, or synthetic DNA additionally triggers the cleavage of caspase-1 and processing of IL-1β in an ASC-dependent manner, yet independent of TLR9, NLRC4, and NLRP3. These results suggested the existence of an additional inflammasome sensor that requires ASC. We and several other groups described the missing DNA sensor to be AIM2, a protein containing the HIN200 and pyrin domains [10–13].
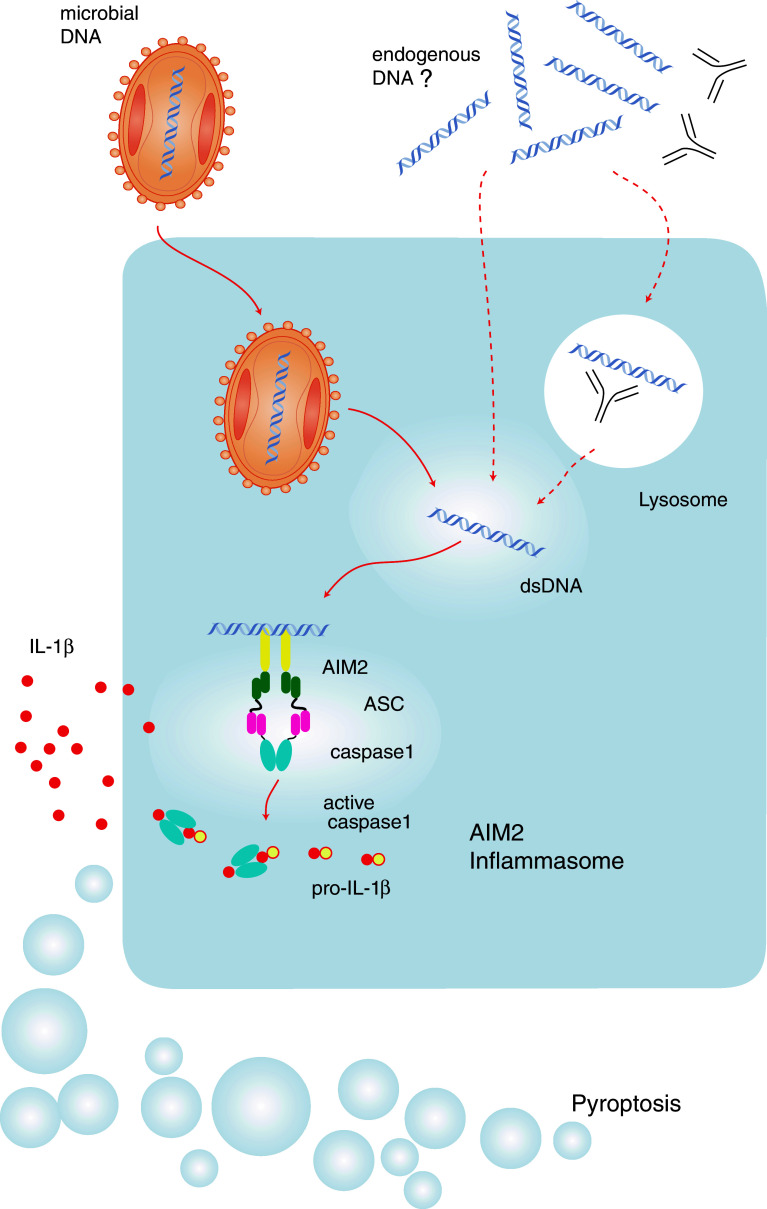
In the murine system, the HIN200 protein family consists of at least nine structurally related members that are all localized to chromosome 1 location 1H3. In contrast, the human system harbors only four homologous genes (MNDA, IFI16, AIM2, and IFIX) that are located on human chromosome 1 location 1q22 [126]. The common denominator of this group of proteins is the so-called HIN200 domain, whose name is an acronym for hematopoietic expression, IFN-inducible, nuclear localization and length of 200 amino acid domains. In the human system, all HIN200 proteins contain an N-terminal pyrin domain, whereas in the murine system, the member p202 lacks a pyrin domain. Grouping the HIN200 proteins due to their sequence similarity in their HIN200 and pyrin domains reveals that AIM2 is special with regards to its pyrin and HIN200 domain sequence [127]. Both in the human and in the murine system, the pyrin and the HIN domains of AIM2 show the most distant homology to other pyrin or HIN200 domains of the PYHIN family. In addition, among the human HIN200 proteins, AIM2 is the only member lacking a nuclear localization domain. Indeed, AIM2 turned out to be the only member of the human HIN200 proteins that localizes to the cytosol, and only the PYD of AIM2 interacts with the PYD of ASC [12]. The HIN200 domain of AIM2 preferentially binds double-stranded DNA, and activated AIM2 recruits the adaptor molecule ASC via PYD–PYD interactions (Fig. 4). ASC in turn induces autocatalytic cleavage of caspase-1 through CARD–CARD interactions [12].
Fig. 4.
The AIM2 inflammasome. AIM2 directly binds to its ligand, which is double-stranded DNA. Following ligand binding, AIM2 recruits ASC and thereby leads to caspase-1 activation. Various exogenous triggers of the AIM2 inflammasome have been identified: vaccinia virus (depicted), mouse cytomegalovirus, L. monocytogenes, and F. tularensis. At the same time, it is tempting to speculate whether DNA of endogenous origin can also lead to AIM2 activation. This could, for example, include DNA antibody complexes found in autoimmune disease or DNA released from phagocytosed apoptotic cells
AIM2 seems to recognize cytosolic dsDNA—independent of species specificity—including DNA from mammalian cells [12, 63]. It is therefore tempting to speculate that AIM2 might participate in autoinflammatory immune responses involving cytosolic DNA. Indeed, HIN200 proteins fall within susceptibility loci for SLE in humans and in mice [128]. Autoinflammatory diseases such as SLE have been linked to missensing of self DNA, and patients with systemic lupus erythematodes develop autoantibodies against double-stranded DNA that form immune complexes. Interestingly, these patients display IFN-α- but also IL-1β-inducible gene signatures, which allows one to speculate that AIM2 is being activated [129].
The relevance of DNA recognition via AIM2 in innate immune responses against pathogens is demonstrated by the fact that inflammasome activation by vaccinia virus mainly relies on AIM2 [12]. Furthermore, AIM2 is also involved in the host response against additional DNA viruses and also bacteria such as Francisella tularensis [130, 131], L. monocytogenes [131, 132], and mouse cytomegalovirus (mCMV) [131] that gain access to the cytosol.
The in vivo function of AIM2 in the recognition of exogenous DNA ligands has been demonstrated by the fact that AIM2 largely contributes to the clearance of F. tularensis and murine cytomegalovirus infections. Whether AIM2 also contributes in the detection of endogenous DNA ligands awaits in-depth characterization in mice deficient for AIM2.
Currently, the AIM2 inflammasome is exceptional among inflammasomes in that a well-defined ligand has been identified and direct ligand binding has been demonstrated [12]. A second aspect making the AIM2 inflammasome unique is that it provides the first example of inflammasome assembly by proteins outside the NLR family. AIM2 lacks a NACHT domain, which is required for oligomerization of NLRs. Homodimerization or multimerization are widely believed to be essential for the recruitment of ASC, so one can speculate that binding of multiple AIM2 proteins to a single molecule of dsDNA could lead to a process mimicking proximity interactions via domains known for dimerization. This hypothesis is further supported by experiments demonstrating a length dependency for DNA ligands that trigger inflammasome assembly. Short double-stranded DNA fails to activate the AIM2 inflammasome [10]. In addition, it was also reported that p202 can self-associate via a sequence stretch that is conserved among the HIN200 family members [133].
The precise role of HIN200 proteins other than AIM2 in inflammatory responses has not been characterized in detail. Roberts et al. [10] described murine p202 as negatively regulating the AIM2 inflammasome, and caspase-1 cleavage was enhanced after RNAi-mediated knock down of p202. While p202 binds to double-stranded DNA and contains two HIN domains, it lacks a PYD domain, and, due to the missing signal transduction domain, it is not able to recruit the adaptor molecule ASC. Whether this potentially inhibiting mechanism is also operational in response to pathogens needs to be determined, as well as its possible presence in humans, since a human HIN200 protein lacking a pyrin domain has not yet been described.
Concluding remarks
Knowledge about how inflammasomes are regulated and etiologically linked with autoinflammatory disorders and metabolic diseases such as gout or pseudogout has already led to a broadened and successful use of IL-1β-targeting drugs. Studies with the interleukin-1 receptor antagonist anakinra have underscored the potential benefit of new therapies that neutralize inflammasome-induced overwhelming innate immune responses [14]. In general, applying the knowledge of the molecular basis of a disease has often led to a more rational and sophisticated drug design. Despite remarkable progress in the NLR field, there are still numerous aspects of inflammasome activation that need to be understood for drug development. In particular, the upstream mechanisms leading to NLRP3 activation are rather unclear. Other challenging open questions include unraveling the role of poorly characterized NLRPs, such as NLRP2 or NLRP4-14 and the PYD-containing protein pyrin (alternatively named MEFV). No activators have been identified for these proteins, but some were shown to assemble inflammasomes [134, 135]. On the other hand, cytosolic sensors other than NLRPs might induce the assembly of inflammasomes. In analogy to the DNA receptor AIM2, the cytosolic helicase RIG-I was recently shown to initiate ASC-dependent caspase-1 activation in response to cytosolic RNA. Whether this MAVS-independent RIG-I signal transduction requires an additional bridging protein to interact with ASC still needs to be determined [93].
Even as the nature of molecular events leading to the activation of established inflammasomes is becoming more and more evident, their precise role in in vivo settings and disease models and their interplay with other pathways such as TLR signaling still need to be resolved in the future.
Acknowledgments
This work was supported by grants from the German Research Foundation (SFB704 and SFB670) and the European Research Council (ERC‐2009‐StG 243046) to Veit Hornung.
Footnotes
F. Bauernfeind and A. Ablasser have contributed equally to this work.
References
- 1.Bella J, Hindle KL, McEwan PA, Lovell SC. The leucine-rich repeat structure. Cell Mol Life Sci. 2008;65:2307–2333. doi: 10.1007/s00018-008-8019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duncan JA, Bergstralh DT, Wang Y, Willingham SB, Ye Z, Zimmermann AG, Ting JP. Cryopyrin/NALP3 binds ATP/dATP, is an ATPase, and requires ATP binding to mediate inflammatory signaling. Proc Natl Acad Sci USA. 2007;104:8041–8046. doi: 10.1073/pnas.0611496104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ting JP, et al. The NLR gene family: a standard nomenclature. Immunity. 2008;28:285–287. doi: 10.1016/j.immuni.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 5.Faustin B, et al. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol Cell. 2007;25:713–724. doi: 10.1016/j.molcel.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 6.Boyden ED, Dietrich WF. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet. 2006;38:240–244. doi: 10.1038/ng1724. [DOI] [PubMed] [Google Scholar]
- 7.Franchi L, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat Immunol. 2006;7:576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- 8.Bauernfeind FG, et al. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J Immunol. 2009;183:787–791. doi: 10.4049/jimmunol.0901363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen IC, et al. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30:556–565. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts TL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 11.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burckstummer T, et al. An orthogonal proteomic–genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 14.Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol. 2009;27:519–550. doi: 10.1146/annurev.immunol.021908.132612. [DOI] [PubMed] [Google Scholar]
- 15.Wilson KP, et al. Structure and mechanism of interleukin-1 beta converting enzyme. Nature. 1994;370:270–275. doi: 10.1038/370270a0. [DOI] [PubMed] [Google Scholar]
- 16.Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: host cell death and inflammation. Nat Rev Microbiol. 2009;7:99–109. doi: 10.1038/nrmicro2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrei C, Margiocco P, Poggi A, Lotti LV, Torrisi MR, Rubartelli A. Phospholipases C and A2 control lysosome-mediated IL-1 beta secretion: implications for inflammatory processes. Proc Natl Acad Sci USA. 2004;101:9745–9750. doi: 10.1073/pnas.0308558101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacKenzie A, Wilson HL, Kiss-Toth E, Dower SK, North RA, Surprenant A. Rapid secretion of interleukin-1beta by microvesicle shedding. Immunity. 2001;15:825–835. doi: 10.1016/s1074-7613(01)00229-1. [DOI] [PubMed] [Google Scholar]
- 19.Qu Y, Franchi L, Nunez G, Dubyak GR. Nonclassical IL-1 beta secretion stimulated by P2X7 receptors is dependent on inflammasome activation and correlated with exosome release in murine macrophages. J Immunol. 2007;179:1913–1925. doi: 10.4049/jimmunol.179.3.1913. [DOI] [PubMed] [Google Scholar]
- 20.Puren AJ, Fantuzzi G, Dinarello CA. Gene expression, synthesis, and secretion of interleukin 18 and interleukin 1beta are differentially regulated in human blood mononuclear cells and mouse spleen cells. Proc Natl Acad Sci USA. 1999;96:2256–2261. doi: 10.1073/pnas.96.5.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okamura H, et al. A novel costimulatory factor for gamma interferon induction found in the livers of mice causes endotoxic shock. Infect Immun. 1995;63:3966–3972. doi: 10.1128/iai.63.10.3966-3972.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmitz J, et al. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity. 2005;23:479–490. doi: 10.1016/j.immuni.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 23.Carriere V, Roussel L, Ortega N, Lacorre DA, Americh L, Aguilar L, Bouche G, Girard JP. IL-33, the IL-1-like cytokine ligand for ST2 receptor, is a chromatin-associated nuclear factor in vivo. Proc Natl Acad Sci USA. 2007;104:282–287. doi: 10.1073/pnas.0606854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cayrol C, Girard JP. The IL-1-like cytokine IL-33 is inactivated after maturation by caspase-1. Proc Natl Acad Sci USA. 2009;106:9021–9026. doi: 10.1073/pnas.0812690106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talabot-Ayer D, Lamacchia C, Gabay C, Palmer G. Interleukin-33 is biologically active independently of caspase-1 cleavage. J Biol Chem. 2009;284:19420–19426. doi: 10.1074/jbc.M901744200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luthi AU, et al. Suppression of interleukin-33 bioactivity through proteolysis by apoptotic caspases. Immunity. 2009;31:84–98. doi: 10.1016/j.immuni.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. 2009;27:229–265. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- 28.Bouchier-Hayes L, Martin SJ. CARDINAL roles in apoptosis and NFkappaB activation. Vitam Horm. 2004;67:133–147. doi: 10.1016/S0083-6729(04)67008-7. [DOI] [PubMed] [Google Scholar]
- 29.Fernandes-Alnemri T, et al. The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 2007;14:1590–1604. doi: 10.1038/sj.cdd.4402194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agostini L, Martinon F, Burns K, McDermott MF, Hawkins PN, Tschopp J. NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity. 2004;20:319–325. doi: 10.1016/s1074-7613(04)00046-9. [DOI] [PubMed] [Google Scholar]
- 31.Faustin B, Chen Y, Zhai D, Le Negrate G, Lartigue L, Satterthwait A, Reed JC. Mechanism of Bcl-2 and Bcl-X(L) inhibition of NLRP1 inflammasome: loop domain-dependent suppression of ATP binding and oligomerization. Proc Natl Acad Sci USA. 2009;106:3935–3940. doi: 10.1073/pnas.0809414106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bruey JM, et al. Bcl-2 and Bcl-XL regulate proinflammatory caspase-1 activation by interaction with NALP1. Cell. 2007;129:45–56. doi: 10.1016/j.cell.2007.01.045. [DOI] [PubMed] [Google Scholar]
- 33.Hsu LC, et al. A NOD2–NALP1 complex mediates caspase-1-dependent IL-1beta secretion in response to Bacillus anthracis infection and muramyl dipeptide. Proc Natl Acad Sci USA. 2008;105:7803–7808. doi: 10.1073/pnas.0802726105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, Philpott DJ, Sansonetti PJ. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278:8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 35.Ferwerda G, et al. Engagement of NOD2 has a dual effect on proIL-1beta mRNA transcription and secretion of bioactive IL-1beta. Eur J Immunol. 2008;38:184–191. doi: 10.1002/eji.200737103. [DOI] [PubMed] [Google Scholar]
- 36.Banks DJ, Ward SC, Bradley KA. New insights into the functions of anthrax toxin. Expert Rev Mol Med. 2006;8:1–18. doi: 10.1017/S1462399406010714. [DOI] [PubMed] [Google Scholar]
- 37.Nour AM, Yeung YG, Santambrogio L, Boyden ED, Stanley ER, Brojatsch J. Anthrax lethal toxin triggers the formation of a membrane-associated inflammasome complex in murine macrophages. Infect Immun. 2009;77:1262–1271. doi: 10.1128/IAI.01032-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Squires RC, Muehlbauer SM, Brojatsch J. Proteasomes control caspase-1 activation in anthrax lethal toxin-mediated cell killing. J Biol Chem. 2007;282:34260–34267. doi: 10.1074/jbc.M705687200. [DOI] [PubMed] [Google Scholar]
- 39.Fink SL, Bergsbaken T, Cookson BT. Anthrax lethal toxin and Salmonella elicit the common cell death pathway of caspase-1-dependent pyroptosis via distinct mechanisms. Proc Natl Acad Sci USA. 2008;105:4312–4317. doi: 10.1073/pnas.0707370105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wickliffe KE, Leppla SH, Moayeri M. Anthrax lethal toxin-induced inflammasome formation and caspase-1 activation are late events dependent on ion fluxes and the proteasome. Cell Microbiol. 2008;10:332–343. doi: 10.1111/j.1462-5822.2007.01044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin Y, Birlea SA, Fain PR, Spritz RA. Genetic variations in NALP1 are associated with generalized vitiligo in a Romanian population. J Invest Dermatol. 2007;127:2558–2562. doi: 10.1038/sj.jid.5700953. [DOI] [PubMed] [Google Scholar]
- 42.Jin Y, Mailloux CM, Gowan K, Riccardi SL, LaBerge G, Bennett DC, Fain PR, Spritz RA. NALP1 in vitiligo-associated multiple autoimmune disease. N Engl J Med. 2007;356:1216–1225. doi: 10.1056/NEJMoa061592. [DOI] [PubMed] [Google Scholar]
- 43.Tu CX, Gu JS, Lin XR. Increased interleukin-6 and granulocyte-macrophage colony stimulating factor levels in the sera of patients with non-segmental vitiligo. J Dermatol Sci. 2003;31:73–78. doi: 10.1016/s0923-1811(02)00151-2. [DOI] [PubMed] [Google Scholar]
- 44.Wagner RN, Proell M, Kufer TA, Schwarzenbacher R. Evaluation of Nod-like receptor (NLR) effector domain interactions. PLoS One. 2009;4:e4931. doi: 10.1371/journal.pone.0004931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poyet JL, Srinivasula SM, Tnani M, Razmara M, Fernandes-Alnemri T, Alnemri ES. Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J Biol Chem. 2001;276:28309–28313. doi: 10.1074/jbc.C100250200. [DOI] [PubMed] [Google Scholar]
- 46.Mariathasan S, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 47.Zamboni DS, et al. The Birc 1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat Immunol. 2006;7:318–325. doi: 10.1038/ni1305. [DOI] [PubMed] [Google Scholar]
- 48.Suzuki T, et al. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 2007;3:e111. doi: 10.1371/journal.ppat.0030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miao EA, Ernst RK, Dors M, Mao DP, Aderem A. Pseudomonas aeruginosa activates caspase 1 through Ipaf. Proc Natl Acad Sci USA. 2008;105:2562–2567. doi: 10.1073/pnas.0712183105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miao EA, Alpuche-Aranda CM, Dors M, Clark AE, Bader MW, Miller SI, Aderem A. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol. 2006;7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 51.Ren T, Zamboni DS, Roy CR, Dietrich WF, Vance RE. Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog. 2006;2:e18. doi: 10.1371/journal.ppat.0020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Warren SE, Mao DP, Rodriguez AE, Miao EA, Aderem A. Multiple Nod-like receptors activate caspase 1 during Listeria monocytogenes infection. J Immunol. 2008;180:7558–7564. doi: 10.4049/jimmunol.180.11.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lightfield KL, et al. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat Immunol. 2008;9:1171–1178. doi: 10.1038/ni.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lamkanfi M, et al. The Nod-like receptor family member Naip5/Birc 1e restricts Legionella pneumophila growth independently of caspase-1 activation. J Immunol. 2007;178:8022–8027. doi: 10.4049/jimmunol.178.12.8022. [DOI] [PubMed] [Google Scholar]
- 55.Sutterwala FS, Mijares LA, Li L, Ogura Y, Kazmierczak BI, Flavell RA. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J Exp Med. 2007;204:3235–3245. doi: 10.1084/jem.20071239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Franchi L, Stoolman J, Kanneganti TD, Verma A, Ramphal R, Nunez G. Critical role for Ipaf in Pseudomonas aeruginosa-induced caspase-1 activation. Eur J Immunol. 2007;37:3030–3039. doi: 10.1002/eji.200737532. [DOI] [PubMed] [Google Scholar]
- 57.Miao EA, Warren SE. Innate immune detection of bacterial virulence factors via the NLRC4 inflammasome. J Clin Immunol. 2010;30:502–506. doi: 10.1007/s10875-010-9386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoffman HM, Mueller JL, Broide DH, Wanderer AA, Kolodner RD. Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nat Genet. 2001;29:301–305. doi: 10.1038/ng756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Manji GA, et al. PYPAF1, a PYRIN-containing Apaf1-like protein that assembles with ASC and regulates activation of NF-kappa B. J Biol Chem. 2002;277:11570–11575. doi: 10.1074/jbc.M112208200. [DOI] [PubMed] [Google Scholar]
- 60.Kanneganti TD, et al. Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J Biol Chem. 2006;281:36560–36568. doi: 10.1074/jbc.M607594200. [DOI] [PubMed] [Google Scholar]
- 61.Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J Exp Med. 2009;206:79–87. doi: 10.1084/jem.20081667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomas PG, et al. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity. 2009;30:566–575. doi: 10.1016/j.immuni.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muruve DA, Petrilli V, Zaiss AK, White LR, Clark SA, Ross PJ, Parks RJ, Tschopp J. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- 64.Mariathasan S, et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 65.Meixenberger K, et al. Listeria monocytogenes-infected human peripheral blood mononuclear cells produce IL-1beta, depending on listeriolysin O and NLRP3. J Immunol. 2010;184:922–930. doi: 10.4049/jimmunol.0901346. [DOI] [PubMed] [Google Scholar]
- 66.Duncan JA, et al. Neisseria gonorrhoeae activates the proteinase cathepsin B to mediate the signaling activities of the NLRP3 and ASC-containing inflammasome. J Immunol. 2009;182:6460–6469. doi: 10.4049/jimmunol.0802696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Joly S, Ma N, Sadler JJ, Soll DR, Cassel SL, Sutterwala FS. Cutting edge: Candida albicans hyphae formation triggers activation of the Nlrp3 inflammasome. J Immunol. 2009;183:3578–3581. doi: 10.4049/jimmunol.0901323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hise AG, Tomalka J, Ganesan S, Patel K, Hall BA, Brown GD, Fitzgerald KA. An essential role for the NLRP3 inflammasome in host defense against the human fungal pathogen Candida albicans . Cell Host Microbe. 2009;5:487–497. doi: 10.1016/j.chom.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gross O, et al. Syk kinase signalling couples to the Nlrp3 inflammasome for anti-fungal host defence. Nature. 2009;459:433–436. doi: 10.1038/nature07965. [DOI] [PubMed] [Google Scholar]
- 70.Ichinohe T, Pang IK, Iwasaki A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat Immunol. 2010;11:404–410. doi: 10.1038/ni.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pelegrin P, Surprenant A. Pannexin-1 mediates large pore formation and interleukin-1beta release by the ATP-gated P2X7 receptor. EMBO J. 2006;25:5071–5082. doi: 10.1038/sj.emboj.7601378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Petrilli V, Papin S, Dostert C, Mayor A, Martinon F, Tschopp J. Activation of the NALP3 inflammasome is triggered by low intracellular potassium concentration. Cell Death Differ. 2007;14:1583–1589. doi: 10.1038/sj.cdd.4402195. [DOI] [PubMed] [Google Scholar]
- 73.Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 74.Chen CJ, Shi Y, Hearn A, Fitzgerald K, Golenbock D, Reed G, Akira S, Rock KL. MyD88-dependent IL-1 receptor signaling is essential for gouty inflammation stimulated by monosodium urate crystals. J Clin Invest. 2006;116:2262–2271. doi: 10.1172/JCI28075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, Fitzgerald KA, Latz E. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dostert C, Petrilli V, Van Bruggen R, Steele C, Mossman BT, Tschopp J. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. 2008;320:674–677. doi: 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cassel SL, et al. The Nalp3 inflammasome is essential for the development of silicosis. Proc Natl Acad Sci USA. 2008;105:9035–9040. doi: 10.1073/pnas.0803933105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Eisenbarth SC, Colegio OR, O′Connor W, Sutterwala FS, Flavell RA. Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature. 2008;453:1122–1126. doi: 10.1038/nature06939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kool M, et al. Cutting edge: alum adjuvant stimulates inflammatory dendritic cells through activation of the NALP3 inflammasome. J Immunol. 2008;181:3755–3759. doi: 10.4049/jimmunol.181.6.3755. [DOI] [PubMed] [Google Scholar]
- 80.Dostert C, et al. Malarial hemozoin is a Nalp3 inflammasome activating danger signal. PLoS One. 2009;4:e6510. doi: 10.1371/journal.pone.0006510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reimer T, et al. Experimental cerebral malaria progresses independently of the Nlrp3 inflammasome. Eur J Immunol. 2010;40(3):764–769. doi: 10.1002/eji.200939996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shio MT, et al. Malarial hemozoin activates the NLRP3 inflammasome through Lyn and Syk kinases. PLoS Pathog. 2009;5:e1000559. doi: 10.1371/journal.ppat.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Griffith JW, Sun T, McIntosh MT, Bucala R. Pure hemozoin is inflammatory in vivo and activates the NALP3 inflammasome via release of uric acid. J Immunol. 2009;183:5208–5220. doi: 10.4049/jimmunol.0713552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Halle A, et al. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol. 2008;9:857–865. doi: 10.1038/ni.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Griffin WS, Stanley LC, Ling C, White L, MacLeod V, Perrot LJ, White CL, 3rd, Araoz C. Brain interleukin 1 and S-100 immunoreactivity are elevated in Down syndrome and Alzheimer disease. Proc Natl Acad Sci USA. 1989;86:7611–7615. doi: 10.1073/pnas.86.19.7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kanneganti TD, Lamkanfi M, Kim YG, Chen G, Park JH, Franchi L, Vandenabeele P, Nunez G. Pannexin-1-mediated recognition of bacterial molecules activates the cryopyrin inflammasome independent of toll-like receptor signaling. Immunity. 2007;26:433–443. doi: 10.1016/j.immuni.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 87.Kanneganti TD, et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006;440:233–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 88.Franchi L, Eigenbrod T, Nunez G. Cutting edge: TNF-alpha mediates sensitization to ATP and silica via the NLRP3 inflammasome in the absence of microbial stimulation. J Immunol. 2009;183:792–796. doi: 10.4049/jimmunol.0900173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Netea MG, et al. Differential requirement for the activation of the inflammasome for processing and release of IL-1beta in monocytes and macrophages. Blood. 2009;113:2324–2335. doi: 10.1182/blood-2008-03-146720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Piccini A, Carta S, Tassi S, Lasiglie D, Fossati G, Rubartelli A. ATP is released by monocytes stimulated with pathogen-sensing receptor ligands and induces IL-1beta and IL-18 secretion in an autocrine way. Proc Natl Acad Sci USA. 2008;105:8067–8072. doi: 10.1073/pnas.0709684105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Perregaux D, Gabel CA. Interleukin-1 beta maturation and release in response to ATP and nigericin. Evidence that potassium depletion mediated by these agents is a necessary and common feature of their activity. J Biol Chem. 1994;269:15195–15203. [PubMed] [Google Scholar]
- 92.Arlehamn CS, Petrilli V, Gross O, Tschopp J, Evans TJ. The role of potassium in inflammasome activation by bacteria. J Biol Chem. 2010;285:10508–10518. doi: 10.1074/jbc.M109.067298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Poeck H, et al. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1 beta production. Nat Immunol. 2010;11:63–69. doi: 10.1038/ni.1824. [DOI] [PubMed] [Google Scholar]
- 94.Fubini B, Hubbard A. Reactive oxygen species (ROS) and reactive nitrogen species (RNS) generation by silica in inflammation and fibrosis. Free Radic Biol Med. 2003;34:1507–1516. doi: 10.1016/s0891-5849(03)00149-7. [DOI] [PubMed] [Google Scholar]
- 95.Cruz CM, Rinna A, Forman HJ, Ventura AL, Persechini PM, Ojcius DM. ATP activates a reactive oxygen species-dependent oxidative stress response and secretion of proinflammatory cytokines in macrophages. J Biol Chem. 2007;282:2871–2879. doi: 10.1074/jbc.M608083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rada B, Leto TL. Oxidative innate immune defenses by Nox/Duox family NADPH oxidases. Contrib Microbiol. 2008;15:164–187. doi: 10.1159/000136357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhou R, Tardivel A, Thorens B, Choi I, Tschopp J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat Immunol. 2010;11:136–140. doi: 10.1038/ni.1831. [DOI] [PubMed] [Google Scholar]
- 98.Pollock JD, et al. Mouse model of X-linked chronic granulomatous disease, an inherited defect in phagocyte superoxide production. Nat Genet. 1995;9:202–209. doi: 10.1038/ng0295-202. [DOI] [PubMed] [Google Scholar]
- 99.Duewell P, et al. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature. 2010;464:1357–1361. doi: 10.1038/nature08938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Masters SL, Simon A, Aksentijevich I, Kastner DL. Horror autoinflammaticus: the molecular pathophysiology of autoinflammatory disease (*) Annu Rev Immunol. 2009;27:621–668. doi: 10.1146/annurev.immunol.25.022106.141627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Feldmann J, et al. Chronic infantile neurological cutaneous and articular syndrome is caused by mutations in CIAS1, a gene highly expressed in polymorphonuclear cells and chondrocytes. Am J Hum Genet. 2002;71:198–203. doi: 10.1086/341357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Neven B, et al. Molecular basis of the spectral expression of CIAS1 mutations associated with phagocytic cell-mediated autoinflammatory disorders CINCA/NOMID, MWS, and FCU. Blood. 2004;103:2809–2815. doi: 10.1182/blood-2003-07-2531. [DOI] [PubMed] [Google Scholar]
- 103.Gattorno M, et al. Pattern of interleukin-1beta secretion in response to lipopolysaccharide and ATP before and after interleukin-1 blockade in patients with CIAS1 mutations. Arthritis Rheum. 2007;56:3138–3148. doi: 10.1002/art.22842. [DOI] [PubMed] [Google Scholar]
- 104.Yu JW, et al. Cryopyrin and pyrin activate caspase-1, but not NF-kappaB, via ASC oligomerization. Cell Death Differ. 2006;13:236–249. doi: 10.1038/sj.cdd.4401734. [DOI] [PubMed] [Google Scholar]
- 105.Anderson JP, Mueller JL, Misaghi A, Anderson S, Sivagnanam M, Kolodner RD, Hoffman HM. Initial description of the human NLRP3 promoter. Genes Immun. 2008;9:721–726. doi: 10.1038/gene.2008.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pontillo A, Brandao LA, Guimaraes RL, Segat L, Athanasakis E, Crovella S. A 3′ UTR SNP in NLRP3 gene is associated with susceptibility to HIV-1 infection. J Acquir Immune Defic Syndr. 2010;54:236–240. doi: 10.1097/QAI.0b013e3181dd17d4. [DOI] [PubMed] [Google Scholar]
- 107.Goldbach-Mansky R, et al. Neonatal-onset multisystem inflammatory disease responsive to interleukin-1beta inhibition. N Engl J Med. 2006;355:581–592. doi: 10.1056/NEJMoa055137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yamamoto M, et al. ASC is essential for LPS-induced activation of procaspase-1 independently of TLR-associated signal adaptor molecules. Genes Cells. 2004;9:1055–1067. doi: 10.1111/j.1365-2443.2004.00789.x. [DOI] [PubMed] [Google Scholar]
- 109.Li P, et al. Mice deficient in IL-1 beta-converting enzyme are defective in production of mature IL-1 beta and resistant to endotoxic shock. Cell. 1995;80:401–411. doi: 10.1016/0092-8674(95)90490-5. [DOI] [PubMed] [Google Scholar]
- 110.Sutterwala FS, et al. Critical role for NALP3/CIAS1/Cryopyrin in innate and adaptive immunity through its regulation of caspase-1. Immunity. 2006;24:317–327. doi: 10.1016/j.immuni.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 111.De Gregorio E, Tritto E, Rappuoli R. Alum adjuvanticity: unraveling a century old mystery. Eur J Immunol. 2008;38:2068–2071. doi: 10.1002/eji.200838648. [DOI] [PubMed] [Google Scholar]
- 112.Morefield GL, Sokolovska A, Jiang D, HogenEsch H, Robinson JP, Hem SL. Role of aluminum-containing adjuvants in antigen internalization by dendritic cells in vitro. Vaccine. 2005;23:1588–1595. doi: 10.1016/j.vaccine.2004.07.050. [DOI] [PubMed] [Google Scholar]
- 113.Jordan MB, Mills DM, Kappler J, Marrack P, Cambier JC. Promotion of B cell immune responses via an alum-induced myeloid cell population. Science. 2004;304:1808–1810. doi: 10.1126/science.1089926. [DOI] [PubMed] [Google Scholar]
- 114.Gavin AL, Hoebe K, Duong B, Ota T, Martin C, Beutler B, Nemazee D. Adjuvant-enhanced antibody responses in the absence of toll-like receptor signaling. Science. 2006;314:1936–1938. doi: 10.1126/science.1135299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nemazee D, Gavin A, Hoebe K, Beutler B (2006) Immunology: toll-like receptors and antibody responses. Nature 441:E4; discussion E4 [DOI] [PMC free article] [PubMed]
- 116.Li H, Willingham SB, Ting JP, Re F. Cutting edge: inflammasome activation by alum and alum’s adjuvant effect are mediated by NLRP3. J Immunol. 2008;181:17–21. doi: 10.4049/jimmunol.181.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McKee AS, Munks MW, MacLeod MK, Fleenor CJ, Van Rooijen N, Kappler JW, Marrack P. Alum induces innate immune responses through macrophage and mast cell sensors, but these sensors are not required for alum to act as an adjuvant for specific immunity. J Immunol. 2009;183:4403–4414. doi: 10.4049/jimmunol.0900164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Franchi L, Nunez G. The Nlrp3 inflammasome is critical for aluminium hydroxide-mediated IL-1beta secretion but dispensable for adjuvant activity. Eur J Immunol. 2008;38:2085–2089. doi: 10.1002/eji.200838549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ghiringhelli F, et al. Activation of the NLRP3 inflammasome in dendritic cells induces IL-1beta-dependent adaptive immunity against tumors. Nat Med. 2009;15:1170–1178. doi: 10.1038/nm.2028. [DOI] [PubMed] [Google Scholar]
- 120.Guarda G, Dostert C, Staehli F, Cabalzar K, Castillo R, Tardivel A, Schneider P, Tschopp J. T cells dampen innate immune responses through inhibition of NLRP1 and NLRP3 inflammasomes. Nature. 2009;460:269–273. doi: 10.1038/nature08100. [DOI] [PubMed] [Google Scholar]
- 121.Hemmi H, et al. A toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 122.Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Takaoka A, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 124.Ishii KJ, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 125.Hornung V, Latz E. Intracellular DNA recognition. Nat Rev Immunol. 2010;10:123–130. doi: 10.1038/nri2690. [DOI] [PubMed] [Google Scholar]
- 126.Landolfo S, Gariglio M, Gribaudo G, Lembo D. The Ifi 200 genes: an emerging family of IFN-inducible genes. Biochimie. 1998;80:721–728. doi: 10.1016/s0300-9084(99)80025-x. [DOI] [PubMed] [Google Scholar]
- 127.Ludlow LE, Johnstone RW, Clarke CJ. The HIN-200 family: more than interferon-inducible genes? Exp Cell Res. 2005;308:1–17. doi: 10.1016/j.yexcr.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 128.Choubey D, Panchanathan R. Interferon-inducible Ifi200-family genes in systemic lupus erythematosus. Immunol Lett. 2008;119:32–41. doi: 10.1016/j.imlet.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Baechler EC, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci USA. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fernandes-Alnemri T, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis . Nat Immunol. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rathinam VA, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kim S, Bauernfeind F, Ablasser A, Hartmann G, Fitzgerald KA, Latz E, Hornung V. Listeria monocytogenes is sensed by the NLRP3 and AIM2 inflammasome. Eur J Immunol. 2010;40:1545–1551. doi: 10.1002/eji.201040425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Koul D, Obeyesekere NU, Gutterman JU, Mills GB, Choubey D. p202 self-associates through a sequence conserved among the members of the 200-family proteins. FEBS Lett. 1998;438:21–24. doi: 10.1016/s0014-5793(98)01263-0. [DOI] [PubMed] [Google Scholar]
- 134.Wang L, et al. PYPAF7, a novel PYRIN-containing Apaf1-like protein that regulates activation of NF-kappa B and caspase-1-dependent cytokine processing. J Biol Chem. 2002;277:29874–29880. doi: 10.1074/jbc.M203915200. [DOI] [PubMed] [Google Scholar]
- 135.Grenier JM, et al. Functional screening of five PYPAF family members identifies PYPAF5 as a novel regulator of NF-kappaB and caspase-1. FEBS Lett. 2002;530:73–78. doi: 10.1016/s0014-5793(02)03416-6. [DOI] [PubMed] [Google Scholar]
- 136.Harder J, Franchi L, Munoz-Planillo R, Park JH, Reimer T, Nunez G. Activation of the Nlrp3 inflammasome by Streptococcus pyogenes requires streptolysin O and NF-kappa B activation but proceeds independently of TLR signaling and P2X7 receptor. J Immunol. 2009;183:5823–5829. doi: 10.4049/jimmunol.0900444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gurcel L, Abrami L, Girardin S, Tschopp J, van der Goot FG. Caspase-1 activation of lipid metabolic pathways in response to bacterial pore-forming toxins promotes cell survival. Cell. 2006;126:1135–1145. doi: 10.1016/j.cell.2006.07.033. [DOI] [PubMed] [Google Scholar]
- 138.Munoz-Planillo R, Franchi L, Miller LS, Nunez G. A critical role for hemolysins and bacterial lipoproteins in Staphylococcus aureus-induced activation of the Nlrp3 inflammasome. J Immunol. 2009;183:3942–3948. doi: 10.4049/jimmunol.0900729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sharp FA, et al. Uptake of particulate vaccine adjuvants by dendritic cells activates the NALP3 inflammasome. Proc Natl Acad Sci USA. 2009;106:870–875. doi: 10.1073/pnas.0804897106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yamasaki K, et al. NLRP3/cryopyrin is necessary for interleukin-1beta (IL-1beta) release in response to hyaluronan, an endogenous trigger of inflammation in response to injury. J Biol Chem. 2009;284:12762–12771. doi: 10.1074/jbc.M806084200. [DOI] [PMC free article] [PubMed] [Google Scholar]