Abstract
MicroRNAs (miRNAs) constitute a novel class of small, non-coding RNAs that act as post-transcriptional regulators of gene expression. Remarkably, it has been shown that these small molecules can coordinately regulate multiple genes coding for proteins with related cellular functions. Previously, we reported that brain-specific miR-338 modulates the axonal expression of cytochrome c oxidase IV (COXIV), a nuclear-encoded mitochondrial protein that plays a key role in oxidative phosphorylation and axonal function. Here, we report that ATP synthase (ATP5G1), like COXIV mRNA, contains a putative miR-338 binding site, and that modulation of miR-338 levels in the axon results in alterations in both COXIV and ATP5G1 expression. Importantly, miR-338 modulation of local COXIV and ATP5G1 expression has a marked effect on axonal ROS levels, as well as axonal growth. These findings point to a mechanism by which miR-338 modulates local energy metabolism through the coordinate regulation of the expression of multiple nuclear-encoded mitochondrial mRNAs in the axon.
Keywords: Axonal protein synthesis, ATP synthesis, Reactive oxygen species, Neurite outgrowth, Sympathetic neurons
Introduction
It is well established that the axon and presynaptic nerve terminal contains large numbers of highly active mitochondria, and that these organelles play a critical role in the function of these remote structural/functional regions of the neuron [1–3]. Mitochondria present in these cellular compartments have been shown to be closely associated with synapses and tethered to vesicle release sites [4, 5]. Synaptic transmission requires mitochondrial ATP generation and control of local [Ca2+]i for neurotransmitter exocytosis, vesicle recruitment, and potentiation of neurotransmitter release and reuptake [6, 7]. In addition, mitochondrial fragmentation has been shown to be one of the hallmarks of neurodegeneration, further supporting the importance of these organelles in axonal maintenance and function [8]. However, little is known about the half-life and biogenesis of synaptically localized mitochondria. Evidence that a portion of neuronal mitochondrial biogenesis occurs in the axon at significant distances from the cell body has been recently provided [3]. Over the past few years, it has also become widely accepted that a distinct subset of nuclear-encoded mitochondrial mRNAs are selectively transported to the distal structural/functional domains of the neuron, including the axon and presynaptic nerve terminal [9–14]. Proteins synthesized from these mRNAs play a key role, not only in mitochondrial function but also in the development of the neuron and the function of the axon and nerve terminal [15–17].
Previous studies have identified microRNAs (miRNAs) as key gene regulators that are abundantly expressed in both the developing and adult mammalian brain [18–20]. MicroRNAs are non-coding gene transcripts that are involved in post-transcriptional regulatory processes, modulating gene expression by binding to the 3′-untranslated region (UTR) of target mRNAs and inhibiting mRNA translation or causing mRNA degradation [21]. Recently, using a compartmentalized neuronal cell culture system [22], we identified 130 miRNAs present in axons of primary sympathetic neurons by microarray analysis and quantitative real-time PCR (qRT-PCR) [23]. These axonally located miRNAs were postulated to play an important regulatory role in the maintenance of synaptic structure and function, as well as in neuronal growth and development [23].
A particularly powerful aspect of miRNA function is the ability of individual miRNAs to coordinately regulate the expression of a multitude of proteins with related cellular functions [24, 25]. Given that we had previously established that the brain-specific miR-338 regulated the levels of COXIV expression in the axon and influenced axonal energy metabolism, as well as function [15], we explored the possibility that miR-338 regulates the local expression of multiple nuclear-encoded mitochondrial mRNAs coding for proteins comprising the oxidative phosphorylation chain, such as COXIV and ATP5G1.
Materials and methods
Primary cell cultures
Primary neuronal cell cultures were prepared from superior cervical ganglion (SCG) obtained from 3-day-old Harlan Sprague–Dawley rats. Ganglia were dissociated, and the cells (e.g., neurons, glia, fibroblasts, and blood cells) were plated in the center compartment of compartmentalized Campenot culture dishes as previously described [11, 16]. Cells were cultured in serum-free Neurobasal medium (Invitrogen) containing 20 mM KCl, NGF (50 ng/ml), and 20 U/ml Penicillin/20 μg/ml Streptomycin (Hyclone) for 3–10 days prior to use. The complete culture media, including NGF, was present in both the central and side compartments throughout the culture period and during all experimental procedures. After 2 days in culture, 5-Fluoro-2′-deoxyuridine (50 μM) was added to the culture medium to inhibit the growth of non-neuronal cells, and remained in the media for the duration of the experiment. The side compartments, which contained the distal axons used in these experiments, were devoid of neuronal cell soma or non-neuronal cells, as judged by phase-contrast microscopy, as well as ethidium bromide and acridine orange staining.
Analyses of mitochondrial mRNA levels
mRNA levels were determined by qRT-PCR, in total RNA samples prepared from SCG axons and parental cell soma using the cells-to-signal lysis buffer (Ambion) and gene-specific primers for COXIV, COXVIc, ATP5G1, and ATP5I (Qiagen). Total RNA was incubated with RNase-free DNaseI to remove trace DNA contamination and qRT-PCR analyses were conducted essentially as previously described [11]. The relative levels of each transcript were normalized to β-actin to provide an internal control for reverse transcription and axonal density. RNA values are expressed relative to control by the comparative threshold (C T) method [26].
Western-blot analysis
For the analysis of ATP5G1 protein levels, distal axons in the side compartments of Campenot chambers were harvested and lysed in 250 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 5 % LDS, 2 % BME, 8 M urea, and complete protease inhibitor cocktail (Sigma). Protein concentration was assessed using the micro BCA Assay Kit (Pierce). Equal amounts of each lysate were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). For SDS-PAGE, axonal homogenates were heated at 70 °C in LDS sample buffer (Invitrogen) and resolved on 12 % mini-gels (Invitrogen). Proteins were transferred to PVDF membranes that were then blocked with ECL Advance Blocking Agent (GE Healthcare), probed with monoclonal antibodies as previously described [16], and developed using the ECL Advance kit (GE Health Care). ATP5G1 and β-actin antibodies were purchased from Sigma.
Transfection of axons with miRNA precursors and siRNAs
The siRNA sense and antisense strands targeting ATP5G1 were purchased from Dharmacon with the following sequences: sense 5′-CCUCAUACCUUUGAUCGUGUU-3′, antisense 5′-CACGAUCAAAGGUAUGAGGUU-3′, and scramble control, sense 5′-UAGCGACUAAACACAUCAA-3′, antisense 5′-UAAGGCUAUGAAGAGAUAC-3′. Two independent siRNAs targeting rat COXIV were used for silencing in SCG neurons. The COXIV siRNA sense and antisense strands were purchased from Dharmacon; the sequences for siRNAs targeting COXIV are provided in [15].
The double-stranded RNA that mimics endogenous rat precursor miRNA-338 (pre-miR-338), and non-targeting miRNA (pre-miR-NT), employed as a negative control, were obtained from Ambion. In addition, the miRNA inhibitor (anti-miR-338) and non-targeting control (anti-miR-NT) were obtained from Ambion. The introduction of small RNAs (miRNAs or siRNAs, each at 25 nM final concentration) into the distal axons located in the side chambers of the culture dishes, or the soma and proximal axons in the center compartment was accomplished by lipofection using siPORT NeoFX (Ambion). For qRT-PCR and Western-blot analyses, 16- and 24-h transfection paradigms were used, respectively. The Campenot multi-chamber culture system used in the present studies contained two lateral compartments that harbored the distal axons. The distal axons present in each of these lateral compartments were transfected independently, and the total RNA obtained from each lateral compartment was tested separately to increase the sample number of the analyses.
ATP and ROS measurements
ATP levels were assessed by luminescence using ViaLight® Plus Kit (Lonza) according to the manufacturer’s instruction. Briefly, SCG neurons were transfected with ATP5G1-siRNA or the NT-siRNA control. After transfection (16–24 h), the distal axons were independently harvested in 50 μl of cell lysis reagent, and AMR Plus assay reagent (100 μl) was added to the cell lysate, and the luminescence was then recorded in a luminometer with an integration time of 1 s per well.
Reactive oxygen species (ROS) were detected using Image-iT™Live Green Reactive Oxygen Species Detection Kit (Invitrogen). After miRNA or siRNA treatment, cultures were incubated in Hank’s balanced salt solution with calcium and magnesium (HBSS/Ca/Mg) containing 5-(and-6)-carboxy-2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA, 25 μM) for 30 min at 37 °C. Cultures were subsequently washed thrice with HBSS/Ca/Mg. Fluorescence was visualized according to the manufacturer’s recommendation, using an inverted fluorescence microscope (Nikon Eclipse TE300). Images were deconvoluted and fluorescence quantitated using the Deconvolution Lab plugin [27] and ImageJ software (NIH).
We used MitoSOX Red (Invitrogen) to determine relative mitochondrial ROS levels. After miRNA or siRNA treatments, cultures were incubated for 10 min at 37 °C in the cell culture medium containing MitoSOX (4 μM). After incubation in MitoSOX, cultures were washed thrice with culture medium and MitoSOX fluorescence was measured according to the manufacturer’s recommendations.
For ROS scavenging experiments, siRNA or miRNA treated cultures were treated with either 100 μM oxypurinol or 10 μM nordihydraguaiaretic acid (NDGA) in culture media at the time of transfection and ROS levels were measured after 16–24 h by MitoSOX and H2DCFDA staining. For the axonal growth rescue experiments, 5 μM NDGA was added to the culture media at the time of transfection of axons with siRNAs, and the axon length was measured 16–24 h after treatment by phase contrast microscopy (see below).
Measurement of axonal growth
Images of axons were obtained using an inverted phase-contrast microscope (Nikon). The length of the distal axons present in the side compartment was measured using the neurite tracing and quantification tool, Neuron J, from ImageJ software (NIH). All axons in each culture dish were measured and a total of at least 35–40 axons for each treatment were used for statistical comparisons.
Bioinformatics and statistical analysis
To investigate putative targets of miR-338, we searched several target databases for conserved, putative rodent targets of miR-338 [28, 29]. The secondary structure prediction analysis of the ATP5G1 and COXIV 3′UTRs was conducted using Mfold [30]. All quantitative data are presented as the mean ± SEM. Student’s t test was used to determine significant differences between two groups. One-way analysis of variance (ANOVA) was employed to analyze differences among multiple groups; p values ≤ 0.05 were considered significant.
Results
As mentioned in the introduction, COXIV mRNA is one of several nuclear-encoded mitochondrial mRNAs that are present in the axons of sympathetic neurons [31]. An Mfold [30] RNA secondary structure prediction analysis of the 3′UTR of COXIV mRNA indicated that a miR-338 targeting site (MTS) was present in a 37-nucleotide hair-pin loop structure in a position that might facilitate miRNA accessibility. The construction of a chimeric luciferase reporter gene containing this stem-loop structure placed the reporter gene expression under the control of miR-338 [15]. Further, in silico analyses for other genes containing the miR-338 MTS, revealed that the nuclear-encoded mitochondrial mRNA encoding ATP5G1, a key component of Complex V of the oxidative phosphorylation chain [32], also contained a putative miR-338 binding site in its 3′UTR (Fig. 1a). The putative MTS in ATP5G1 had a predicted free energy of hybridization with the cognate miRNA of 24.8 kcal/mol. Recently, we reported that ATP5G1 mRNA was present and locally translated in the axons of sympathetic neurons [16].
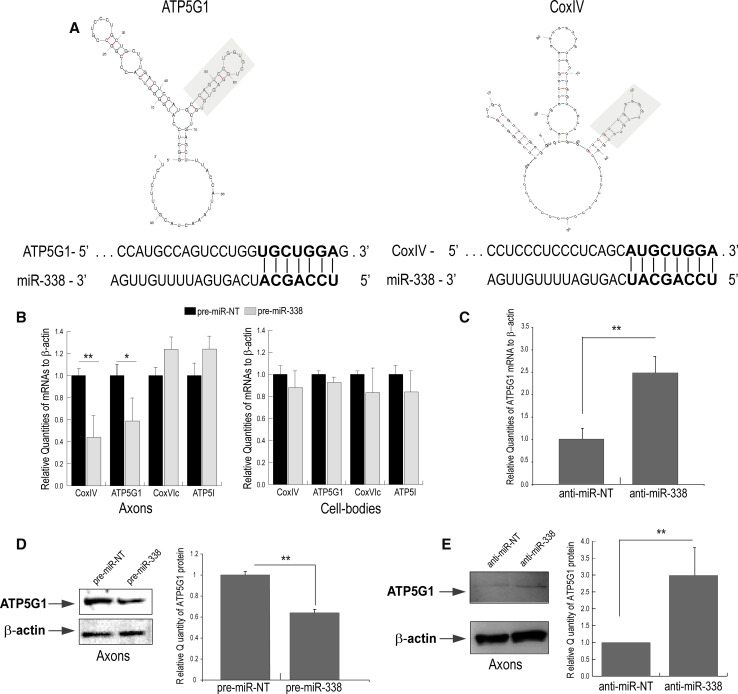
Fig. 1.
miR-338 targets rat ATP5G1. a Putative miR-338 binding site in the rat ATP5G1 and COXIV 3‘UTR. Secondary structure of the ATP5G1 3‘UTR was determined by Mfold secondary structural analysis. The COXIV 3‘UTR secondary structure has been adapted from Aschrafi et al. [15]. The miR-338 target site is indicated in grey. Putative miR-338 binding site sequence in the rat ATP5G1 and COXIV 3‘UTR: The seed sequence of the target site is bolded. b–e miR-338 modulates ATP5G1 expression in the axons of SCG neurons. Quantification of nuclear-encoded mitochondrial mRNA levels in the distal axons and cell soma of SCG neurons transfected with b 25 nM pre-miR-338 or c 25 nM anti-miR-338. mRNA abundance was determined by qRT-PCR 16 h after transfection. All mRNA levels are expressed relative to β-actin mRNA. Error bars represent the SEM for three samples. Student’s t test *p< 0.05, **p<0.01. Western-blot analysis of axonal protein lysates from SCG axons transfected with d pre-miR-338 or e anti-miR-338. β- actin was used as a loading control. NT, non-targeting oligonucleotides. Blots of axonal protein lysates were quantified using ImageJ. Quantification showed significant changes in ATP5G1 protein levels under the influence of miR-338. Values represent the mean ± SEM. Student’s t test ** p< 0.01; *** p<0.001
Regulation of axonal levels of ATP5G1 mRNA and protein by miR-338
To explore whether miR-338 had the capacity to regulate ATP5G1 mRNA levels in the axons of SCG neurons, we quantified ATP5G1 mRNA levels after transfecting the distal axons with the miR-338 precursor (pre-miR-338). In these experiments, COXIV mRNA served as an internal positive control. Similar to earlier results observed with COXIV mRNA, axonal ATP5G1 mRNA levels decreased approximately 40 % when compared to the non-targeting pre-miR-NT 16–24 h post-transfection (Fig. 1b). Transfection of the distal axons with pre-miR-338 had no effect on the levels of COXIV or ATP5G1 in the parental cell soma located in the central compartment of the culture chamber. To evaluate the specificity of these effects, we compared the effects of miR-338 levels on the relative abundance of COXVIc and ATP5I, two nuclear-encoded mitochondrial mRNAs that do not contain a miR-338 binding site in their 3′UTRs. As shown in Fig. 1b, the introduction of pre-miR-338 into the distal axons had no effect on the levels of these mRNAs either in the distal axon or parental cell bodies. On the other hand, lipofection of an inhibitor of endogenous miR-338 activity, anti-miR-338, into the distal axons resulted in a 2.5-fold increase in ATP5G1 mRNA when compared with the non-targeting anti-miRNA control (Fig. 1c).
To assess whether the miR-338-mediated modulation of mRNA also correlated with an alteration in axonal ATP5G1 protein levels, miR-338 was over-expressed in SCG axons by transfection of precursor miR-338, and ATP5G1 protein levels were evaluated by Western analysis. Consistent with the mRNA findings described above, over-expression of miR-338 led to a 40 % decrease in ATP5G1 protein levels in distal axons of SCG neurons (Fig. 1d). Conversely, the inhibition of endogenous miR-338 by transfection of the specific anti-miR-338 into the distal axons led to a 2.5-fold increase in the expression of ATP5G1 protein, while having no effect on the β-actin protein levels in the axons (Fig. 1e). No change in ATP5G1 protein levels was observed in the corresponding cell bodies after treatment of axons with either pre-miR-338 or anti-miR-338 (data not shown).
Modulating local miR-338 levels alters ROS production in the axons of SCG neurons
Recent studies suggest that inhibition of oxidative phosphorylation results in the production of ROS [33, 34]. COXIV and ATP5G1 play rate-limiting roles in the assembly of complexes IV and V, respectively, and dysfunctional COXIV and ATP5G1 resulted in a compromised mitochondrial membrane potential, as well as decreased respiration and ATP levels [15, 16, 35]. Since elevation of miR-338 levels in distal axons lead to a reduction in mitochondrial oxygen consumption and ATP production [15], we investigated whether the miR-338 effects on mitochondrial function resulted in changes in the axonal ROS levels.
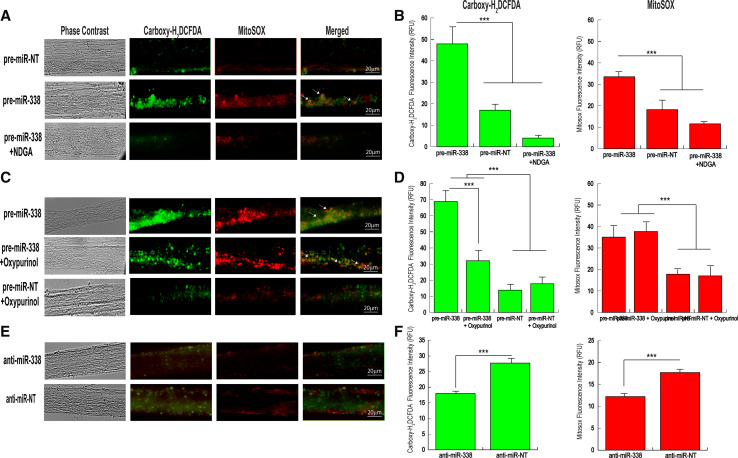
To determine whether alterations in miR-338 levels had the capacity to modulate ROS production in the distal axons, axonal processes located in the side compartments of 7-day-old cultures were transfected with either pre-miR-338 or non-targeting pre-miR-NT, and ROS levels were assessed 16 h later with H2DCFDA and MitoSOX, fluorescent indicators of total and mitochondrial-generated ROS (i.e., superoxide), respectively. We found that over-expression of miR-338 in distal axons resulted in a marked increase in axonal ROS levels: Some of the fluorescent signal appeared punctuate in nature (Fig. 2a, arrows). Quantitation of the fluorescent signal in miR-338-treated axons revealed that total ROS levels increased 2.5-fold (H2DCFDA), whereas mitochondrial-generated ROS was elevated twofold (MitoSOX), as compared to ROS levels visualized in axons transfected with the pre-miR-NT (Fig. 2b). Repetitions of this experiment yielded similar results, with 2.1–2.3-fold elevation in total ROS levels and 1.6–1.9-fold elevation in mitochondrial ROS levels when compared to pre-miR-NT transfected neurons. In both cases, exposure of miR-338-treated axons to NDGA, a powerful ROS scavenger, at the time of transfection returned miRNA-induced increases in ROS levels to control values (Fig. 2a, b). Previous studies indicated that inhibition of ATP production and/or ATP depletion in cultured primary neurons could result in the generation of cytoplasmic ROS via activation of a xanthine oxidase-mediated pathway [36]. To test whether miR-338 induced the generation of mitochondrial ROS, cytosolic ROS, or both, we used oxypurinol to specifically inhibit xanthine oxidase-dependent cytosolic ROS production. In this experiment, miR-338- or miR-NT-treated distal axons (7 DIV) were exposed to oxypurinol at the time of transfection and throughout the duration of the experiment. ROS levels were visualized by both H2DCFDA and MitoSOX 16–24 h later. We observed no effect of oxypurinol on basal levels of ROS in miR-NT-treated control axons (Fig. 2c). In contrast, oxypurinol treatment showed a 2.2-fold reduction in the levels of extra-mitochondrial ROS induced by miR-338, but had little effect on mitochondrial-generated ROS (Fig. 2c, d). Similar results were obtained on repetition of this experiment. No statistically significant change was observed in miR-338-induced mitochondrial ROS levels after oxypurinol treatment, while a 2.3–2.5-fold decrease in cytosolic ROS levels was detected as compared to axons not exposed to oxypurinol. The outcome of this experiment suggested that miR-338-mediated modulation of axonal protein synthesis resulted in a marked increase in mitochondrial ROS.
Fig. 2.
Modulation of axonal ATP5G1 expression alters ROS production. a miR-338-mediated ROS production in distal axons. Distal axons were transfected with pre-miR-338, or non-targeting oligonucleotides (NT). Intra-axonal ROS levels were measured in miRNA-treated axons by fluorescence microscopy using Carboxy-H2DCFDA (green) and Mitosox (red). NDGA, a potent antioxidant, was used to inhibit miR-338-mediated ROS generation in the axon. b Fluorescence intensity was quantified using ImageJ, and fluorescence levels are indicated as relative fluorescence units (RFU). Data are mean ± SEM from the measurement of 35–45 axons. Student’s t test, ***p < 0.0001. c Oxypurinol was employed to inhibit the formation of extra-mitochondrial, xanthine oxidase-generated ROS. e Distal axons were transfected with anti-miR-338, or NT-oligonucleotides. ROS was measured in axons as described above. Scale bar, 20 μm. b, d, f Quantitation of miR-338-mediated induction of ROS levels in the axon. Arrows indicate mitochondrial-associated ROS. Values represent the mean ± SEM. Student’s t test **p < 0.01; ***p < 0.001
Next, we addressed the question as to whether ROS levels could be reduced in distal axons by specifically silencing endogenous miR-338 activity through the transfection of anti-miR-338. Consistent with the increase in the ROS levels observed upon over-expression of miRNA-338, axons transfected with anti-miR-338 displayed a ~50 % reduction in basal axonal ROS levels (Fig. 2e, f). These findings point to the fact that the action of miR-338 on its nuclear-encoded mitochondrial target genes can have significant effects on axonal ROS levels.
Modulation of axonal elongation rates by miR-338
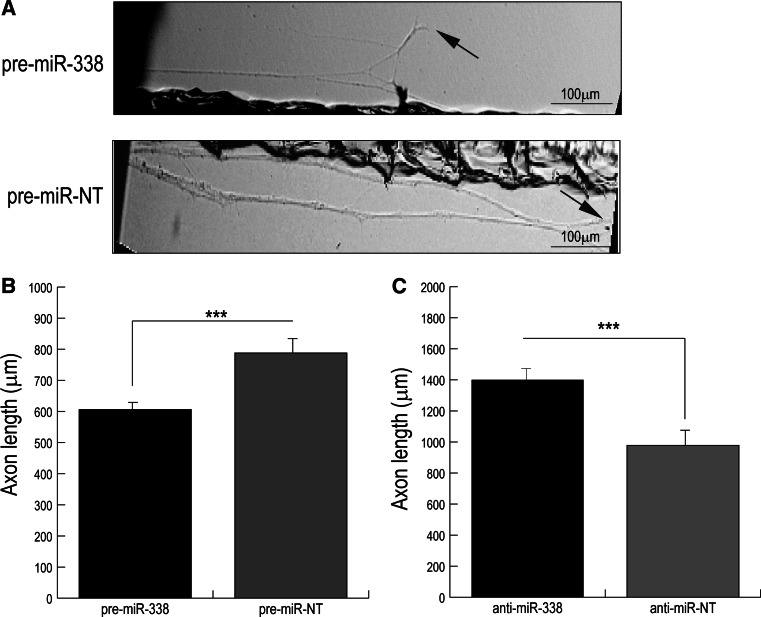
In previous studies, we used 2-day-old SCG cultures, and identified miR-338 as a novel regulator of mitochondrial oxidative phosphorylation and axonal function in the distal axons through local modulation of COXIV levels [15]. The reduction in axonal respiration mediated by this miRNA had pronounced effects on some functions of SCG axons, such as neurotransmitter uptake, yet over-expression or inhibition of miR-338 had only minimal effects on axon morphology and viability, as judged by light microscopy. Therefore, the question arose as to whether the modulation of local mitochondrial protein synthesis by miR-338 would exhibit significant effects on SCG axon growth rate and/or elongation. To investigate the role miR-338 might play in the mediation of axonal growth, we transfected 1-day-old primary SCG neurons with pre-miR-338, and determined the length of SCG axons. Over-expression of miRNA-338 in primary SCG neurons resulted in a significant decrease in axon length in comparison to cells transfected with the non-targeted precursor (Fig. 3a). Conversely, we observed a marked increase in neurite outgrowth of 3-day-old primary neurons that were transfected with a specific inhibitor of endogenous miR-338 (data not shown). As shown in Fig. 3b, the axonal transfection of pre-miR-338 resulted in 20–30 % reduction in axon outgrowth, as compared to axons transfected with a non-targeted precursor. Conversely, inhibition of endogenous miR-338 in axons by transfection with anti-miR-338 resulted in a significant increase in primary neurite outgrowth. The average neurite length for anti-miR-338-treated neurons was approximately 40 % greater than that of the non-targeted anti-miR transfected control axons (Fig. 3c).
Fig. 3.
Elevation of axonal miR-338 levels attenuates axon outgrowth in sympathetic neurons. a Photomicrographs of distal axons of SCG neurons that were cultured for 3 days and axons located in the lateral sides of compartmentalized cultures were transfected with 25 nM pre-miR-338 or 25 nM control pre-miR-NT for 24 h. Arrows indicate location of growth cones. Scale bar: 100 μm. b Distal axons were transfected with pre-miR-338, or NT-oligonucleotides and c anti-miR-338, or NT-control for 24 h. The axon length was measured after transfection. Data are mean ± SEM from the measurement of 35–45 axons. The experiment was repeated three times with similar results. Student’s t test, ***p < 0.0001
Effects of miR-338 on axonal ATP synthesis and ROS production are mediated by local ATP5G1 and COXIV expression
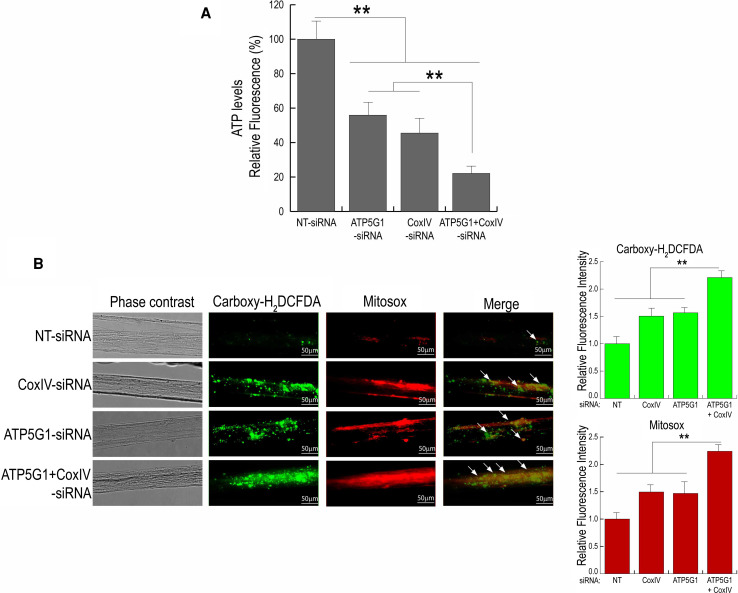
To further evaluate the postulate that the axonal effects of miR-338 are mediated, at least in part, by the local synthesis of ATP5G1 and COXIV, siRNAs targeted against ATP5G1 and COXIV mRNAs were lipofected into the distal axons. We previously reported the siRNA-mediated knock-downs of COXIV and ATP5G1 mRNA and protein in SCG axons. The COXIV siRNA knock-down yielded 70 and 60 % decreases in mRNA and protein levels, respectively, while ATP5G1 siRNA treatment resulted in 70 and 50 % decreases in mRNA and protein levels [15, 16]. Similar to the findings obtained with miR-338, reduction in ATP5G1 and COXIV expression by RNA interference resulted in a 50 and 60 % decrease in axonal ATP levels, respectively, compared to sham controls (Fig. 4a). Additionally, the combined knock-down of ATP5G1 and COXIV resulted in a 70 % reduction in axonal ATP levels (Fig. 4a). Transfection of distal axons with NT- siRNA had no effect on this experimental variable.
Fig. 4.
Knock-down of both ATP5G1 and COXIV levels together leads to a greater increase in ROS and decrease in ATP levels in the axon as compared to knock-down of the individual mRNAs. a SCG neurons (7-DIV) were transfected with siRNA oligonucleotides (25 nM) targeted against ATP5G1 mRNA or COXIV mRNA or both together. ATP levels were assessed using ViaLight® Plus Kit from Lonza. Values were plotted in arbitrary luminescence units. Data represent the mean ± SEM; one-way ANOVA, **p < 0.001. b Knock-down of axonal COXIV and ATP5G1 expression increased ROS levels in the axon. SCG neurons (7 DIV) were transfected with siRNA oligonucleotides (25 nM) targeted against COXIV mRNA or ATP5G1 mRNA or both together. Intra-axonal ROS levels were measured in siRNA-treated axons by fluorescence microscopy using Carboxy-H2DCFDA (green) and Mitosox (red). Values are plotted in arbitrary fluorescence units. Data represent mean ± SEM; one-way ANOVA, **p < 0.001. Scale bars: 50 μm
To test the hypothesis that the miR-338-induced ROS levels were mediated, at least in part, by suppression of axonal ATP5G1 and COXIV levels, siRNAs targeted against ATP5G1 and COXIV mRNAs were introduced individually or in combination into distal axons: ROS levels were assessed 16–24 h post-transfection, as described above. As shown in Fig. 4b, little fluorescence was observed in control, NT-, or siRNA-treated axons with either dye. In contrast, there was a marked elevation in fluorescence in COXIV, as well as ATP5G1, siRNA-treated axons (Fig. 4b, arrows). Quantitation of the fluorescent signal in COXIV or ATP5G1 siRNA-treated axons revealed that total ROS levels increased 1.5-fold, whereas, joint application of COXIV together with the ATP5G1 siRNA resulted in over a twofold elevation in axonal ROS levels.
Silencing of local ATP5G1 and COXIV expression inhibits axonal growth
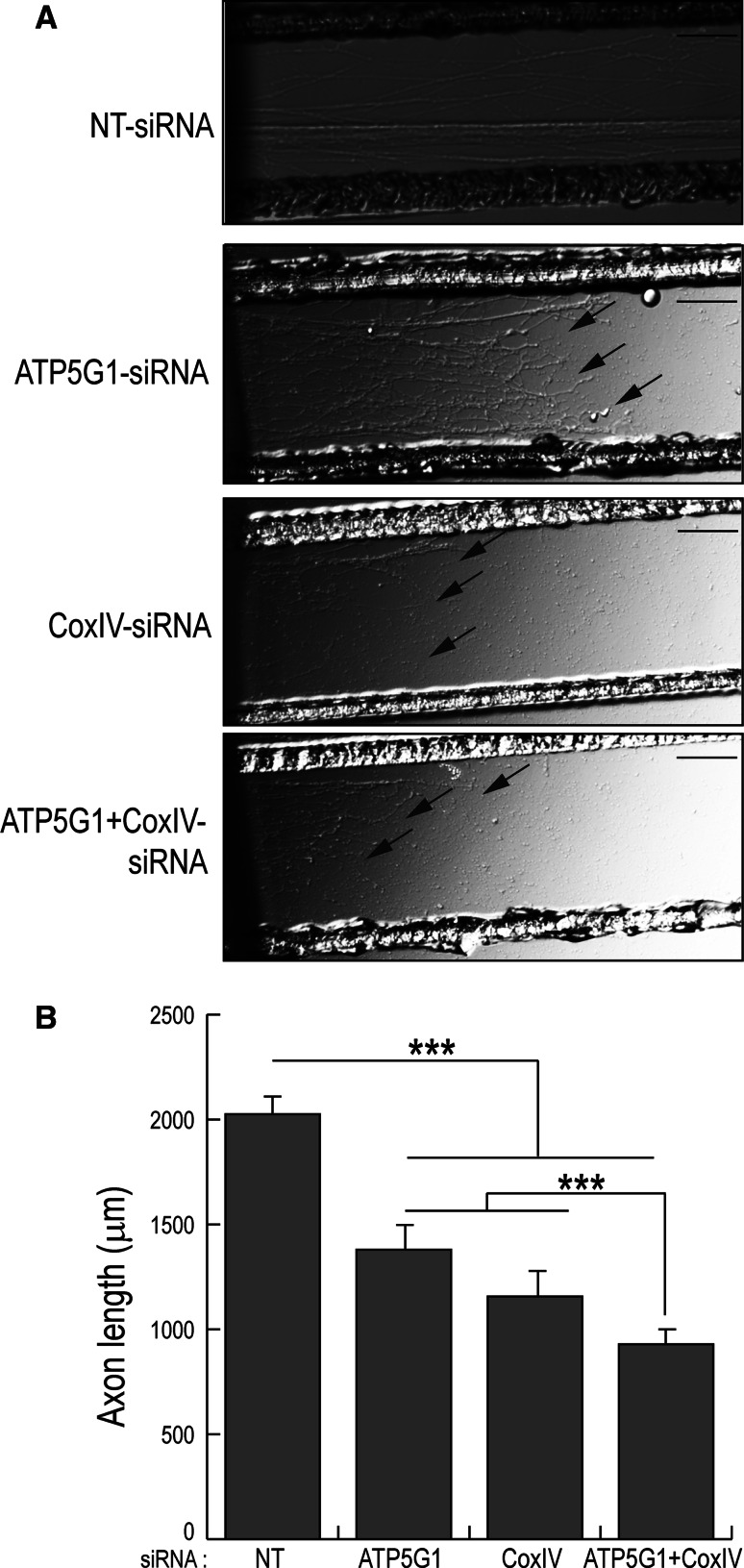
Since knock-down of axonal ATP5G1 and COXIV levels resulted in both a reduction of axonal ATP levels and an increase in axonal ROS production, we tested the hypothesis that the effects of miR-338 on axonal outgrowth were mediated by the co-ordinate regulation of local ATP5G1 and COXIV expression. Toward this end, siRNAs targeted against ATP5G1 and COXIV mRNAs were lipofected into the 3-day-old axons grown in the lateral compartments of Campenot cultures. After 16 h, knock-down of COXIV or ATP5G1 mRNA levels resulted in a 30–40 % reduction in rates of axonal elongation, as compared to control axons. The combined knock-down of COXIV and ATP5G1 mRNAs resulted in a 50 % attenuation of axonal growth (Fig. 5a, b). These data provide evidence that the local translation of ATP5G1 and COXIV mRNAs and their subsequent effects on mitochondrial activity play an important role in regulating outgrowth of the axon, and supports the hypothesis that miR-338 affects axonal growth via the co-ordinate regulation of axonal ATP5G1 and COXIV expression.
Fig. 5.
Knock-down of axonal ATP5G1and COXIV mRNAs shows a greater decrease in axon outgrowth as compared to knock-down of the individual mRNAs. a SCG neurons were cultured for 3 days and the distal axons were subsequently transfected with siRNA oligonucleotides (25 nM) targeted against COXIV mRNA or ATP5G1 mRNA or both together. Arrows indicate location of axon terminal regions. Scale bars: 200 μm. b Axons were measured 16 h post-transfection in the lateral sides of compartmentalized culture. Data are the mean ± SEM from measurement of 30–40 axons. The experiment was repeated three times with similar results. One-way ANOVA, ***p < 0.0001
Discussion
Mitochondria are enriched in subcellular regions of high energy consumption, such as in axons, dendrites, as well as presynaptic nerve terminals [1, 31]. Despite the acknowledged significance of mitochondria in synapse formation and function, there is still a paucity of information regarding the mechanisms that regulate their activity in the distal structural/functional domains of the neuron. In previous studies, we identified miR-338, a brain-specific miRNA, as a local modulator of COXIV expression in the distal axons of SCG neurons. Alteration in the levels of miR-338 in the axon had a profound effect on the activity of the local mitochondrial population and the basal metabolic rate of the axon [15]. That miR-338-mediated modulation of local COXIV expression could have such a profound effect on axonal respiration and function further led us to assess the regulatory impact of miR-338 on axonal mitochondrial activity. We, therefore, conducted a bioinformatics search to identify additional nuclear-encoded mitochondrial mRNAs that contained putative miR-338-binding sites. The 18 additional messages identified represent approximately 1 % of the total mRNAs that encode mitochondrial proteins: 13 of these mRNAs have been identified in the axon by RT-PCR methodology. These mRNAs code for a variety of proteins involved in energy metabolism, membrane transport, DNA replication, mRNA translation, and lipid metabolism (see Table 1, Ref. [15]). One of these 13 axonally localized mRNAs that contained a miR-338 binding site coded for ATP5G1, a key component of Complex V of the oxidative phosphorylation chain [37]. Mitochondrial ATP synthase catalyzes ATP synthesis, utilizing an electrochemical gradient of protons across the inner membrane during oxidative phosphorylation. The present study provides direct evidence to support the hypothesis that miR-338 has the capacity to regulate mitochondrial function by locally regulating the synthesis of ATP5G1 in addition to COXIV in the distal axons of sympathetic neurons. Moreover, our investigation demonstrated that the modulation of axonal levels of ATP5G1 expression by miR-338 has a wide range of functional consequences for local ATP and ROS generation, as well as axonal outgrowth. Taken together, our results also support the notion that these small, non-coding RNAs have the capacity to modulate complex physiological phenotypes by co-ordinately regulating the expression of multiple functionally related genes [38, 39].
Key proteins involved in the miRNA processing and functional pathway, i.e., Dicer and miRISC complexes, can assemble and function in developing axons [40]. In addition, miRNAs and the RISC complex have recently been shown to be present in the axonal compartments where they can actively regulate local mRNA levels [15]. Most recently, we employed the Campenot cell culture system to obtain a pure axonal RNA fraction of superior cervical ganglia (SCG) neurons and determined the composition and miRNA expression levels in this subcellular structural domain [23]. The data revealed the presence of a number of mature miRNAs that were enriched in the axons and presynaptic nerve terminals. Among the 130 miRNAs identified in the axon, miR-15b, miR-16, miR-204, and miR-221 were found to be highly abundant in distal axons relative to their levels in the cell bodies.
The intracellular mechanisms underlying the regulation of axonal growth are not well understood, although it has been demonstrated that extracellular factors, such as NGF, can modulate axon growth rates [41]. Our experiments show that in the presence of NGF, upregulation of both COXIV and ATP5G1 levels by inhibition of endogenous miR-338 activity in the distal axons, enhanced axon growth, while its down-regulation, by transfection of axons with the precursor miR-338, or specific siRNAs, resulted in axon growth attenuation. The findings presented here also establish that local regulation of COXIV and ATP5G1 expression has significant effects on the levels of ATP in the axon. The energy generated by mitochondria is required for a number of pivotal processes during outgrowth, including growth cone motility, and the accumulation of this organelle has been shown to be important for sustained axonal outgrowth. Our studies suggest that regulation of mitochondrial activity by local translation of nuclear-encoded mRNAs contribute to the mitochondria-mediated effects on axonal elongation, and further underscore the importance of local translation in axonal growth.
To initiate investigation into the molecular mechanisms underlying miR-338-mediated mitochondrial dysfunction, which affected axonal growth, we assessed the relative abundance of axonal ROS levels. Earlier studies suggested that enhancement of mitochondrial ROS levels can be generated through decreased complex IV or complex V activity [42–44], and that regulation of ROS levels strongly affected nervous system development, particularly axon pathfinding [45]. The findings presented here demonstrate that significant mitochondrially derived ROS levels are detected as a consequence of suppression of local ATP5G1 and COXIV levels by miR-338 or specific siRNAs. Conversely, the transfection of anti-miR-338 increased ATP5G1 and COXIV levels, and resulted in a significant decrease in axonal ROS production. Taken together, our findings demonstrate that miR-338 modulates the axonal expression of multiple components involved in the mitochondrial oxidative phosphorylation pathway.
Interestingly, an increasing number of studies have indicated that cytotoxic-free radicals, such as superoxides, nitric oxide, and others may contribute to neural damage, including attenuation in axonal growth [46]. ROS has previously been implicated in the pathogenesis of neurodegenerative diseases, as well as in the deficits that occur in normal aging [47]. It has also been reported that ROS production promotes apoptosis of neurons leading to neurodegeneration. Although we have not addressed axonal degeneration in our studies, our results raise the possibility that moderate deficiencies in mitochondrial activity could have detrimental effects on axonal growth as a consequence of excess ROS production, as well as through abnormalities associated with energy deficits, per se (i.e., decreased ATP levels).
Recent studies have proposed a connection between mitochondrial dysfunction and a number of neuropsychiatric disorders, including schizophrenia, bipolar disorder, and neurodegenerative disorders, such as amyotrophic lateral sclerosis, Parkinson’s and Alzheimer’s disease [48, 49]. The outcomes of this investigation support the notion of the significance of local translation of nuclear-encoded mitochondrial mRNAs in the maintenance of mitochondrial function, particularly in neurons with long axons, and propose a novel mechanism for the maintenance of axonal function and viability, which ultimately could underlie neurodegenerative and/or neurodevelopmental disorders. Multiple lines of evidence suggest that altered neuronal plasticity and morphology, as seen in neurodevelopmental disorders, may result from disruption of a common post-translational process that is under tight regulation by miRNAs [50]. Several intellectual disability (ID) syndromes, such as Fragile X syndrome, Rett syndrome, and Down syndrome have been linked to the miRNA pathway [51–53]. Increasing evidence suggests that miRNAs, which act as regulators of neuronal gene circuitries, are associated with complex neuropsychiatric disorders involving abnormalities in synaptic plasticity, as well as neurodegenerative diseases. From a large number of recent studies on neurological diseases, it thus appears that miRNAs play an essential regulatory role in neuronal differentiation, neurite outgrowth, and dendritic spine formation. Thus, the dysregulation of miRNAs could ultimately underlie some of the clinical features observed in these diseases through their effects on the local protein synthetic system(s).
Acknowledgments
This work was supported by the Division of Intramural Research Programs of the National Institute of Mental Health. We thank Ms. Sanah Vohra for her invaluable technical assistance.
Abbreviations
- H2DCFDA
5-(and-6)-Carboxy-2′, 7′-dichlorodihydrofluorescein diacetate
- NDGA
Nordihydraguaiaretic acid
- COX
Cytochorome oxidase
- qRT-PCR
Quantitative real-time reverse transcriptase polymerase chain reaction
- miRNA
MicroRNA
References
- 1.Schuman E, Chan D. Fueling synapses. Cell. 2004;119:738–740. doi: 10.1016/j.cell.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Hollenbeck PJ. Mitochondria and neurotransmission: evacuating the synapse. Neuron. 2005;47:331–333. doi: 10.1016/j.neuron.2005.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amiri M, Hollenbeck PJ. Mitochondrial biogenesis in the axons of vertebrate peripheral neurons. Dev Neurobiol. 2008;68:1348–1361. doi: 10.1002/dneu.20668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang DT, Honick AS, Reynolds IJ. Mitochondrial trafficking to synapses in cultured primary cortical neurons. J Neurosci. 2006;26:7035–7045. doi: 10.1523/JNEUROSCI.1012-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ly CV, Verstreken P. Mitochondria at the synapse. Neuroscientist. 2006;12:291–299. doi: 10.1177/1073858406287661. [DOI] [PubMed] [Google Scholar]
- 6.Bindokas VP, Lee CC, Colmers WF, Miller RJ. Changes in mitochondrial function resulting from synaptic activity in the rat hippocampal slice. J Neurosci. 1998;18:4570–4587. doi: 10.1523/JNEUROSCI.18-12-04570.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zenisek D, Matthews G. The role of mitochondria in presynaptic calcium handling at a ribbon synapse. Neuron. 2000;25:229–237. doi: 10.1016/S0896-6273(00)80885-5. [DOI] [PubMed] [Google Scholar]
- 8.Knott AB, Perkins G, Schwarzenbacher R, Bossy-Wetzel E. Mitochondrial fragmentation in neurodegeneration. Nat Rev Neurosci. 2008;9:505–518. doi: 10.1038/nrn2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gioio AE, Eyman M, Zhang H, Lavina ZS, Giuditta A, Kaplan BB. Local synthesis of nuclear-encoded mitochondrial proteins in the presynaptic nerve terminal. J Neurosci Res. 2001;64:447–453. doi: 10.1002/jnr.1096. [DOI] [PubMed] [Google Scholar]
- 10.Gioio AE, Lavina ZS, Jurkovicova D, Zhang H, Eyman M, Giuditta A, Kaplan BB. Nerve terminals of squid photoreceptor neurons contain a heterogeneous population of mRNAs and translate a transfected reporter mRNA. Eur J Neurosci. 2004;20:865–872. doi: 10.1111/j.1460-9568.2004.03538.x. [DOI] [PubMed] [Google Scholar]
- 11.Hillefors M, Gioio AE, Mameza MG, Kaplan BB. Axon viability and mitochondrial function are dependent on local protein synthesis in sympathetic neurons. Cell Mol Neurobiol. 2007;27:701–716. doi: 10.1007/s10571-007-9148-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang W, van Niekerk E, Willis DE, Twiss JL. RNA transport and localized protein synthesis in neurological disorders and neural repair. Dev Neurobiol. 2007;67:1166–1182. doi: 10.1002/dneu.20511. [DOI] [PubMed] [Google Scholar]
- 13.Willis DE, van Niekerk EA, Sasaki Y, Mesngon M, Merianda TT, Williams GG, Kendall M, Smith DS, Bassell GJ, Twiss JL. Extracellular stimuli specifically regulate localized levels of individual neuronal mRNAs. J Cell Biol. 2007;178:965–980. doi: 10.1083/jcb.200703209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor AM, Berchtold NC, Perreau VM, Tu CH, Li JN, Cotman CW. Axonal mRNA in uninjured and regenerating cortical mammalian axons. J Neurosci. 2009;29:4697–4707. doi: 10.1523/JNEUROSCI.6130-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aschrafi A, Schwechter AD, Mameza MG, Natera-Naranjo O, Gioio AE, Kaplan BB. MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. J Neurosci. 2008;28:12581–12590. doi: 10.1523/JNEUROSCI.3338-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Natera-Naranjo O, Kar AN, Aschrafi A, Gervasi NM, MacGibeny MA, Gioio AE, Kaplan BB. Local translation of ATP synthase subunit 9 mRNA alters ATP levels and the production of ROS in the axon. Mol Cell Neurosci. 2012;49:263–270. doi: 10.1016/j.mcn.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 17.Aschrafi A, Natera-Naranjo O, Gioio AE, Kaplan BB. Regulation of axonal trafficking of cytochrome c oxidase IV mRNA. Mol Cell Neurosci. 2010;43:422–430. doi: 10.1016/j.mcn.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/S0092-8674(01)00616-X. [DOI] [PubMed] [Google Scholar]
- 19.Kim J, Krichevsky A, Grad Y, Hayes GD, Kosik KS, Church GM, Ruvkun G. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci USA. 2004;101:360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, Horvitz HR. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, Biffo S, Merrick WC, Darzynkiewicz E, Pillai RS, Filipowicz W, Duchaine TF, Sonenberg N. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 22.Campenot RB. Local control of neurite development by nerve growth factor. Proc Natl Acad Sci USA. 1977;74:4516–4519. doi: 10.1073/pnas.74.10.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Natera-Naranjo O, Aschrafi A, Gioio AE, Kaplan BB. Identification and quantitative analyses of microRNAs located in the distal axons of sympathetic neurons. RNA. 2010;16:1516–1529. doi: 10.1261/rna.1833310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.An J, Choi KP, Wells CA, Chen YP. Identifying co-regulating microRNA groups. J Bioinform Comput Biol. 2010;8:99–115. doi: 10.1142/S0219720010004574. [DOI] [PubMed] [Google Scholar]
- 25.Lindow M. Prediction of targets for microRNAs. Methods Mol Biol. 2011;703:311–317. doi: 10.1007/978-1-59745-248-9_21. [DOI] [PubMed] [Google Scholar]
- 26.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C (T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 27.Vonesch C, Unser M. A fast thresholded landweber algorithm for wavelet-regularized multidimensional deconvolution. IEEE Trans Image Process. 2008;17:539–549. doi: 10.1109/TIP.2008.917103. [DOI] [PubMed] [Google Scholar]
- 28.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA–target interactions. Nucl Acid Res. 2009;37:D105–D110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaplan BB, Gioio AE, Hillefors M, Aschrafi A. Axonal protein synthesis and the regulation of local mitochondrial function. In: Koenig E, editor. Cell biology of the axon. Berlin: Springer; 2009. pp. 225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vives-Bauza C, Magrané J, Andreu AL, Manfredi G. Novel role of ATPase subunit C targeting peptides beyond mitochondrial protein import. Mol Biol Cell. 2010;21:131–139. doi: 10.1091/mbc.E09-06-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loeb LA, Wallace DC, Martin GM. The mitochondrial theory of aging and its relationship to reactive oxygen species damage and somatic mtDNA mutations. Proc Natl Acad Sci USA. 2005;102:18769–18770. doi: 10.1073/pnas.0509776102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong WK, Han EH, Kim DG, Ahn JY, Park JS, Han BG. Amyloid-beta-peptide reduces the expression level of mitochondrial cytochrome oxidase subunits. Neurochem Res. 2007;32:1483–1488. doi: 10.1007/s11064-007-9336-7. [DOI] [PubMed] [Google Scholar]
- 35.Li Y, Park JS, Deng JH, Bai Y. Cytochrome c oxidase subunit IV is essential for assembly and respiratory function of the enzyme complex. J Bioenerg Biomembr. 2006;38:283–291. doi: 10.1007/s10863-006-9052-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abramov AY, Scorziello A, Duchen MR. Three distinct mechanisms generate oxygen free radicals in neurons and contribute to cell death during anoxia and reoxygenation. J Neurosci. 2007;27:1129–1138. doi: 10.1523/JNEUROSCI.4468-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xing SL, Yan J, Yu ZH, Zhu CQ. Neuronal cell surface ATP synthase mediates synthesis of extracellular ATP and regulation of intracellular pH. Cell Biol Int. 2011;35:81–86. doi: 10.1042/CBI20090441. [DOI] [PubMed] [Google Scholar]
- 38.Backes C, Meese E, Lenhof HP, Keller A. A dictionary on microRNAs and their putative target pathways. Nucleic Acids Res. 2010;38:4476–4486. doi: 10.1093/nar/gkq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 40.Hengst U, Cox LJ, Macosko EZ, Jaffrey SR. Functional and selective RNA interference in developing axons and growth cones. J Neurosci. 2006;26:5727–5732. doi: 10.1523/JNEUROSCI.5229-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Campenot RB. Local control of neurite sprouting in cultured sympathetic neurons by nerve growth factor. Brain Res. 1987;465:293–301. doi: 10.1016/0165-3806(87)90250-1. [DOI] [PubMed] [Google Scholar]
- 42.Yoon YS, Lee JH, Hwang SC, Choi KS, Yoon G. TGF beta1 induces prolonged mitochondrial ROS generation through decreased complex IV activity with senescent arrest in Mv1Lu cells. Oncogene. 2005;24:1895–1903. doi: 10.1038/sj.onc.1208262. [DOI] [PubMed] [Google Scholar]
- 43.Carter BJ, Anklesaria P, Choi S, Engelhardt JF. Redox modifier genes and pathways in amyotrophic lateral sclerosis. Antioxid Redox Signal. 2009;11:1569–1586. doi: 10.1089/ars.2008.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Granata S, Zaza G, Simone S, Villani G, Latorre D, Pontrelli P, Carella M, Schena FP, Grandaliano G, Pertosa G. Mitochondrial dysregulation and oxidative stress in patients with chronic kidney disease. BMC Genomics. 2009;10:388. doi: 10.1186/1471-2164-10-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pocock R, Hobert O. Oxygen levels affect axon guidance and neuronal migration in Caenorhabditis elegans. Nat Neurosci. 2008;11:894–900. doi: 10.1038/nn.2152. [DOI] [PubMed] [Google Scholar]
- 46.Press C, Milbrandt J. Nmnat delays axonal degeneration caused by mitochondrial and oxidative stress. J Neurosci. 2008;28:4861–4871. doi: 10.1523/JNEUROSCI.0525-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 48.Konradi C, Eaton M, MacDonald ML, Walsh J, Benes FM, Heckers S. Molecular evidence for mitochondrial dysfunction in bipolar disorder. Arch Gen Psychiatry. 2004;61:300–308. doi: 10.1001/archpsyc.61.3.300. [DOI] [PubMed] [Google Scholar]
- 49.Shao L, Martin MV, Watson SJ, Schatzberg A, Akil H, Myers RM, Jones EG, Bunney WE, Vawter MP. Mitochondrial involvement in psychiatric disorders. Ann Med. 2008;40:281–295. doi: 10.1080/07853890801923753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hunsberger JG, Austin DR, Chen G, Manji HK. MicroRNAs in mental health: from biological underpinnings to potential therapies. Neuro Mol Med. 2009;11:173–182. doi: 10.1007/s12017-009-8070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, Jongens TA, Nelson DL, Moses K, Warren ST. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nature Neurosci. 2004;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- 52.Kuhn DE, Nuovo GJ, Terry AV, Jr, Martin MM, Malana GE, Sansom SE, Pleister AP, Beck WD, Head E, Feldman DS, Elton TS. Chromosome 21-derived microRNAs provide an etiological basis for aberrant protein expression in human Down syndrome brains. J Biol Chem. 2010;285:1529–1543. doi: 10.1074/jbc.M109.033407. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Szulwach KE, Li X, Smrt RD, Li Y, Luo Y, Lin L, Santistevan NJ, Li W, Zhao X, Jin P. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J Cell Biol. 2010;189:127–141. doi: 10.1083/jcb.200908151. [DOI] [PMC free article] [PubMed] [Google Scholar]