Abstract
The establishment and maintenance of rhizobium–legume symbioses require a sequence of highly regulated and coordinated events between the organisms. Although the interaction is mutually beneficial under nitrogen-limited conditions, it can resemble a pathogenic infection at some stages. Some host legumes mount defense reactions, including the production of reactive oxygen species (ROS) and defensin-like antimicrobial compounds. To subvert these host defenses, the infecting rhizobial cells can use measures to passively protect themselves and actively modulate host functions. This review first describes the establishment and maintenance of active nodules, as well as the external and endogenous attack and threat stages. Next, recent studies of ROS scavenging enzymes, the BacA protein originally found in Sinorhizobium meliloti, and the type III/IV secretion systems are discussed, with a focus on two legume–rhizobium model systems.
Keywords: Nitrogen-fixing mutualism, Host defense, Reactive oxygen species, Type III secretion system, Type IV secretion system, BacA protein
Introduction
Bacteria of the family Rhizobiaceae and compatible leguminous plants establish mutualistic relationships to exchange nitrogen and carbon fixed from the atmosphere. The establishment of symbiosis requires multistep reciprocal recognition with exchanges of signal molecules and complex developmental programs, which leads to the formation of nodules on the legume root and the differentiation of rhizobial cells into bacteroids [1]. Although the relationship is beneficial to both participants, it can resemble a pathogenic interaction in that the eukaryotic organism is chronically infected [2]. However, the host plant may suppress its defense mechanisms to maintain a successful symbiotic interaction [3]. Transcriptomic analyses have revealed that many defense- and stress-related genes are up-regulated in the legume host during the early stage of this interaction, but most are subsequently suppressed as the symbiosis proceeds [4, 5]. In addition, the rhizobial infection can be arrested by a mechanism similar to a hypersensitive reaction [6]. Furthermore, established bacteroids can be eliminated in some legume–host pairings by necrosis of the nodules [7, 8]. These indicate that the host plant can attack the infiltrating rhizobial cells. In addition to host threats, nitrogen-fixing bacteroids encounter reactive oxygen species (ROS) that are endogenously produced from ATP-producing respiratory oxidative phosphorylation and harmful to ROS-sensitive nitrogenases.
Rhizobia have developed mechanisms to survive exogenous (host-derived) and endogenous threats during symbiosis, from the initial contact to senescence. Some rhizobia passively protect bacterial cells or functions, whereas others actively interact with the host to reduce attack. This review focuses on proteins involved in ROS scavenging as a passive defense with an emphasis on two legume–rhizobium model systems: Lotus–Mesorhizobium loti, which forms determinate globular nodules without persisting meristems [9], and Medicago–Sinorhizobium meliloti, which forms indeterminate cylindrical nodules with persisting meristems [10]. This review also discusses the significance of the BacA protein, a bacterial factor essential for bacteroid development in Sinorhizobium meliloti [11], and the significance of the type III and type IV secretion systems, which inject proteins into the eukaryotic host [12]. Non-proteinaceous factors including lipochitooligosaccharides (LCOs), lipopolysaccharides (LPSs), and extracellular polysaccharides (EPSs) are also important for protection and evasion [1, 13, 14] but are not discussed in this review.
Stages of legume–rhizobium symbiotic processes
Encounter, attachment, and initial signal exchange
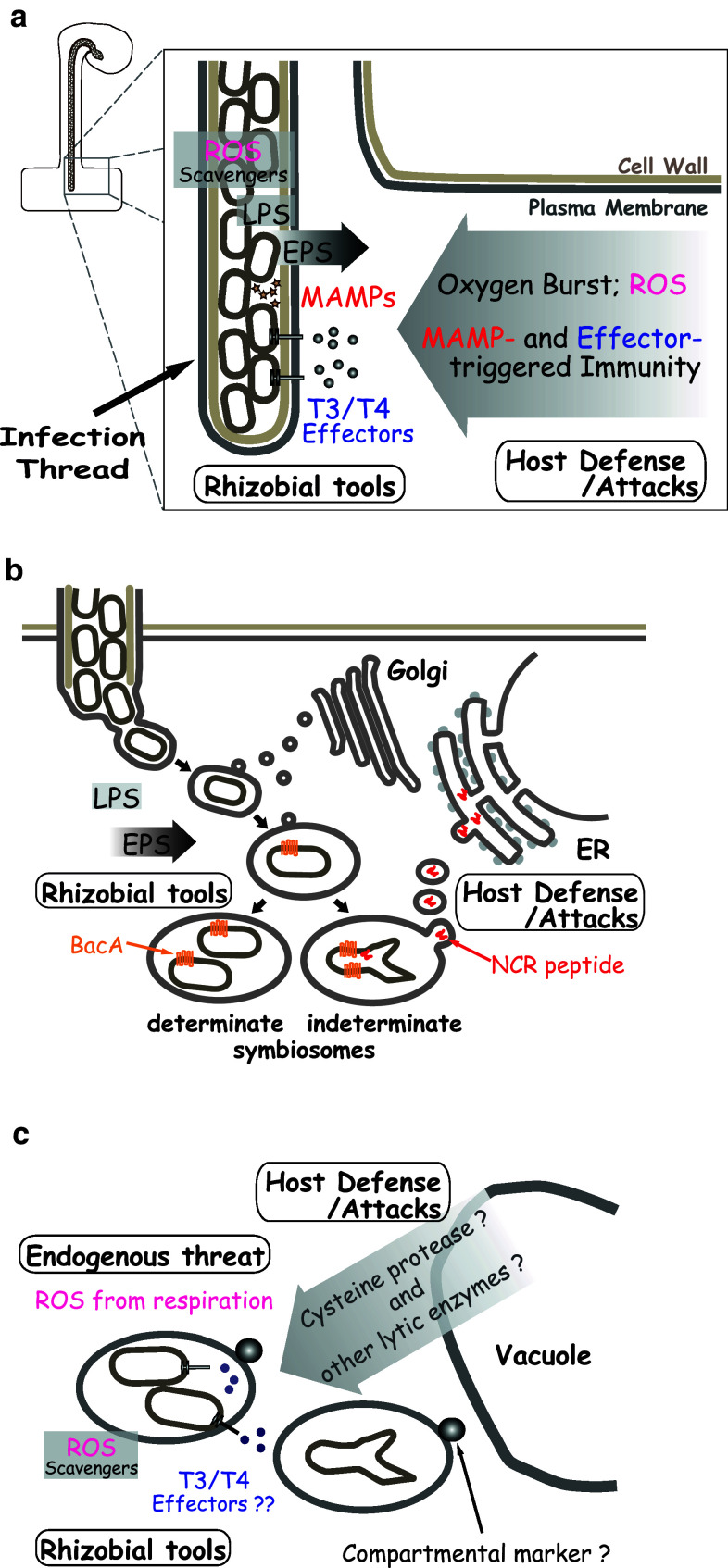
Because rhizobia can exist as saprophytes in the soil, the mutual recognition of two symbionts starts when the rhizobial cells perceive host-specific signal molecules (mostly phenolic flavonoid compounds) exuded to the rhizosphere by the legume roots (Fig. 1a). The signal molecules induce the bacterial regulatory protein NodD to activate transcription of several nod (nodulation) genes that participate in the synthesis of species- or strain-specific signal molecules [LCOs, also known as Nod factors (NFs)] [15]. Rhizobial cells attached to the tip of the emerging root hairs secrete NFs that trigger the nodulation developmental program, including root hair deformation and nodule primordia formation in the cortex (Fig. 1b). Because flavonoid compounds and NFs have specific chemical structures depending on their producers, the combinations of these signaling molecules are the primary determinants of the various host–rhizobium combinations. Hence, most rhizobial species can only establish symbiosis with a few host legumes [16].
Fig. 1.
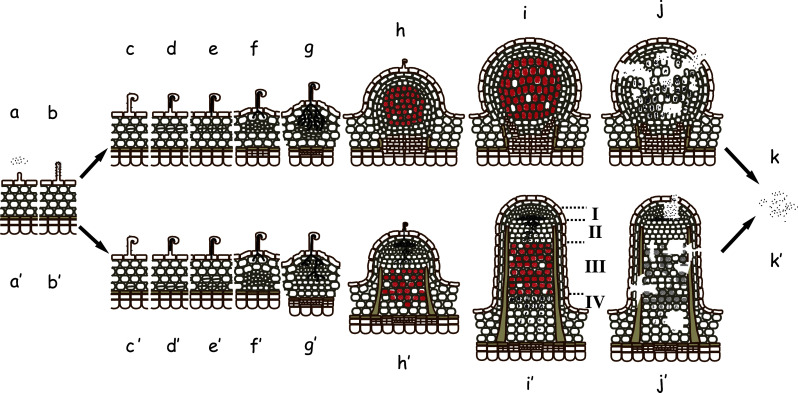
The nodule developmental and decaying process. The processes for determinate (upper) and indeterminate (lower) nodules are shown. Saprophytic rhizobia exist in a rhizosphere (a). The rhizobia attach to root hairs (b), detect flavonoids from legumes and secrete NF to induce root hair deformation (c). The deformed root hair entraps the rhizobia and invaginates to form infection threads, which contain entrapped rhizobial cells (d). The infection thread elongates (e), ramifies, and penetrates the outer or inner cortex cell layers (f, g). The rhizobial cells are then enveloped in a plant-derived membrane and released as droplets into the plant cytosol (h). The released rhizobia differentiate to bacteroids and begin nitrogen fixation; hence, the droplets are called symbiosomes (i). After a period of nitrogen fixation, the nodule cells initiate senescence (j). Most bacteroids in determinate nodules and some undifferentiated rhizobial cells in indeterminate nodules return to a saprophytic lifestyle (k). Zones in mature indeterminate nodule are indicated by I–IV. I Meristematic zone; II invasion zone; III N2-fixing zone; IV senescence zone
Invasion of rhizobia into host cells and establishment of symbiosis
Rhizobial cells on a root hair tip are entrapped in the curled root hair, which is shaped similarly to a shepherd’s crook, and form tubular structures known as infection threads, which contain rhizobial cells. The infection threads elongate inside the root hair, traverse multiple cell layers, ramify and reach the developing nodule primordia (Fig. 1c–f) [17]. The bacterial cells proliferate directionally to the front of the growing infection threads and invade the plant cytoplasm through an endocytosis-like mechanism that enables encapsulation of the engulfed bacteria within the plant plasma membrane. The bacterial cells enlarge, differentiate to bacteroids, and initiate nitrogen fixation in the capsules, which become organelle-like structures known as symbiosomes (Fig. 1g) [18–20]. As the bacteria differentiate, the host genomic DNA replicates in the invaded plant cells without mitosis, and the infected cells become large polyploid cells harboring thousands of symbiosomes [21]. The mature nodules actively fix nitrogen for a length of time that depends on environmental and plant developmental conditions and then enter senescence.
Indeterminate and determinate nodules
The origins of the plant cells that harbor symbiosomes and the fate of the bacteria are considerably different between indeterminate and determinate nodule types (Fig. 1). Indeterminate nodules originate from the nodule primordia formed in the inner root cortex next to the xylem pole (Table 1). Furthermore, these nodules possess a persistent meristem and elongate to become cylindrical so that a meristematic zone forms near the apex and successive zones form for rhizobial invasion, active nitrogen fixation, and senescence (Fig. 1h′–k′). Host cells in the nitrogen-fixation zone contain mature symbiosomes, which typically contain an enlarged, deformed bacteroid with low reproductive viability, while those in the senescence zone have decayed or disintegrating symbiosomes [21–24]. Consequently, an indeterminate nodule accommodates a heterologous population of rhizobial cells in various developmental states in distinct zones. In contrast, determinate nodules originate from the primordia formed in the outer or middle root cortex. In contrast to indeterminate nodules, determinate nodules do not have a persistent meristem and thus become globular (Fig. 1h–j), and the mature symbiosomes contain multiple bacteroids of normal size with high reproductive viability [25–27]. In a determinate nodule, the developmental stages of the host and rhizobial cells are relatively synchronized. Senescence of determinate nodules begins at the center of the nodule and extends to the periphery [28]. Rhizobial cells released from decaying and disintegrating nodules enter a saprophytic life cycle (Fig. 1k).
Table 1.
Representative differences between indeterminate and determinate nodule types
| Property | Indeterminate | Determinate |
|---|---|---|
| Legume examples | Medicago sativa, Pisum sativum, Astragalus sinicus | Glycine max, Phaseolus vulgaris, Lotus japonicus |
| Normal nodule form | Cylindrical/branched | Spherical/globular |
| Site of initial cell division | Inner root cortex | Outer or middle cortex |
| Meristem type | Persistent meristem | No persistent meristem |
| Infection thread | Broad | Narrow |
| Infected cells | Highly vacuolated | Minimal vacuolation |
| Major bacteroid form | Enlarged, branched, one per symbiosome | Normal rod size, high viability, multiple per symbiosome |
| Poly-hydroxybutylate accumulation in bacteroid | Present | Absent |
| Bacteroid reproductivity | Low | High |
Phases of host-derived and endogenous threats to rhizobia
Observations of unsuccessful symbiosis by normal and mutant rhizobia and host legumes indicate that there are at least three major phases of threats to rhizobia [29, 30]. Each phase can be distinguished by nodulation efficiency, nitrogen-fixation capacity, duration and the symbiotic stage in which it takes place.
The initial phase threats appear to be related to the prevention of nodulation (few nodules phenotype). The threats at this phase occur just after rhizobial contact with the root hair or root surface and continue during the elongation of the infection threads (Fig. 2a). Contact between the rhizobial cells and the host root epidermis evokes innate immunity or basal defense responses similar to those that occur with pathogenic infections; however, the responses are transient and regulated during infection with compatible rhizobia, unlike the responses against pathogenic bacteria [4, 5]. The transient responses include the generation of ROS [31–33] and nitric oxide [34] and the expression of gene products similar to pathogen-related (PR) proteins [4, 5, 34, 35]. Some S. meliloti mutants with defective ROS scavenging enzymes have poor nodulation capacity on Medicago sativa [36, 37], indicating that the level of ROS produced by the host is tolerated by wild-type, but not mutant, S. meliloti. Medicago can use ROS and other mechanisms at this phase to interfere with the rhizobial infection process. The accumulation of phenolic compounds and PR proteins has been observed in epidermal cells with aborted infection threads [6]. Therefore, attacks during this phase can be used by the host plant to eliminate excessive nodulation and related to a phenomenon known as autoregulation of nodulation [18].
Fig. 2.
Host-derived and endogenous threats to rhizobia and rhizobial counter measures at the different developmental stages. Host actions to infecting rhizobia and rhizobial counter measures in infection threads (a), in released droplets or young symbiosomes (b), and in mature symbiosomes (c)
The intermediate phase threats involve the reduction of intracellular niche formation by parasitic rhizobia (inactive empty nodules phenotype). The threats at this phase are mainly exerted during and just after the endocytosis-like invasion and are mediated by the drastic change in the bacterial environment within the intracellular encapsulated structure (Fig. 2b). Although the biochemical identities of these threats are not known, some are likely responsible for the aberrant phenotypes of the S. meliloti bacA and lpsB mutants, which are unable to form bacteroids [11, 38]. Intermediate phase threats may also be involved in the phenotypes of S. meliloti EPS mutants, which poorly form bacteria-filled infection threads and small nodules without bacteroids [39, 40]. Empty nodules can be formed by some EPS mutants that escape the initial phase threats but are unable to overcome the intermediate phase threats. While it is difficult to distinguish the passive and positive roles of EPS in the host defense mechanisms, a role for this signaling in the suppression of host defense genes is supported by the transcriptomic analysis of M. truncatula infected with S. meliloti [41].
Late (maintenance) phase threats are divided into two categories: those derived from the host attack of rhizobia established as mature nitrogen-fixing bacteroids and those derived from an endogenous threat from the co-existence of aerobic respiratory energy metabolism and highly oxygen-labile nitrogenase systems (Fig. 2c). An example of the former is the host response of L. japonicus to Rhizobium etli: although nodules formed by Rhizobium etli acquire the capability to fix nitrogen, they lose this capacity within 3 weeks of inoculation and enter early senescence [42]. A L. japonicus mycorrhiza-inducible phosphate transporter knockdown line exhibited nodule necrosis when its normal symbionts M. loti (rhizobium) and Glomus mosseae (mycorrhiza) were inoculated simultaneously, even though the line establishes normal nitrogen-fixing symbiosis when infected with M. loti alone [7]. The senescence process of Medicago truncatula has been studied in detail at the transcriptional level [43, 44] to confirm the involvement of various genes, including some encoding cysteine proteases that likely function in senescence in a number of legumes [28, 44–48]. A recent analysis of compartmental markers has demonstrated that the symbiosomes in M. truncatula change over the course of endocytotic formation to senescence [49]. Such alterations may determine whether each symbiosome persists or enters the lytic pathway, although the mechanism of the alteration is unknown. Although G. max is reported to sanction parasitic rhizobia [50], it has also been reported that some inefficient rhizobia, including Rhizobium sp. NGR234, persist for longer periods in L. japonicus nodules [8]. Thus, it is of interest to elucidate what enables the host to discriminate non-efficient from efficient rhizobia and how marker alterations are triggered.
ROS scavenging enzymes
ROS production occurs during all three phases of threats to rhizobia. Because ROS (especially H2O2) function not only as antimicrobial agents but also as signals for nodule organogenesis, regulation of ROS levels is required for successful symbiosis [51–53]. The ROS level during the initial phase must be low enough to allow the survival of compatible wild-type rhizobium with somewhat redundant ROS scavenging systems. The redundancy often masks some ROS detoxification enzymes, but some mutants with compromised systems have defective symbiotic capacities. S. meliloti has two monofunctional catalases encoded by katA and katC, a bifunctional catalase-peroxidase encoded by katB [36, 54, 55], a superoxide dismutase (SOD) encoded by sodA or sodB [37, 56, 57] and several uncharacterized ROS scavenging enzymes [14, 19]. M. loti has a monofunctional catalase encoded by katE, a bifunctional catalase-peroxidase encoded by katG, a SOD encoded by sodA and other enzymes [58].
Among the ROS scavenging enzymes, catalase genes have been most extensively characterized. Mutation of any of the three kat genes in S. meliloti does not affect H2O2 sensitivity, but a katA katC double mutant and a katB katC double mutant are deficient in nodule formation and nitrogen fixation [36, 55]. The katA katC double mutant can fix nitrogen, but has low nodulation efficiency [55] and sparsely distributed bacteroids, some of which are irregularly shaped [36]. The katB katC mutant has an even lower nodulation efficiency and is unable to form bacteroids and fix nitrogen [36]. Because these double mutants are capable of aerobic growth, S. meliloti likely requires both katB and katC to overcome the initial and intermediate phase attacks from the Medicago host before forming mature symbiosomes. In contrast, an M. loti katE katG double mutant forms a greater number of effective nodules, but they have a slightly lower nitrogen-fixing capacity than wild-type M. loti [59]. Because M. loti katG mutants are at least 30 times more vulnerable to exogenous H2O2 than wild-type M. loti and exhibit slower aerobic growth [59], it is possible that the initial and intermediate attacks from the Lotus host are not strong enough to prevent nodulation and formation of mature symbiosomes. In M. loti, katE contributes to survival during the stationary phase; a katE single mutant has decreased nitrogen-fixation capacity similar to the katE katG double mutant, while the katG single mutant has comparable capacity to wild-type M. loti [59]. This suggests that the monofunctional catalase KatE but not the bifunctional catalase-peroxidase KatG is required for continuing nitrogen fixation or to protect bacteroids from maintenance phase threats. In addition, the S. meliloti katC mutant, which lacks the monofunctional catalase similar to M. loti KatE, forms nodules with slightly decreased nitrogen-fixing capacity [55]. It is unclear why catalase disruption has different effects on nodulation of S. meliloti and M. loti, but the initial and intermediate phase threats to infecting rhizobia appear to be stronger in Medicago than Lotus.
The requirement of rhizobial ROS scavenging proteins to properly maintain nitrogen-fixation capacity is supported by studies examining Rhizobium etli, which nodulates the determinate host Phaseolus vulgaris [60, 61]. R. etli has only one catalase, a catalase-peroxidase encoded by katG, that is responsible for protection from exogenous H2O2 and survival during the stationary phase [60]. Like many rhizobia, this species encodes 2-Cys peroxiredoxin, a non-heme protein that catalyzes the reduction of H2O2 in the presence of thiol [62]. This peroxiredoxin is encoded by prxS and is expressed under symbiotic conditions [61]. katG and prxS single mutants both show a symbiotic phenotype similar to wild-type R. etli, however, a katG prxS double mutant forms nodules with a nitrogen-fixing capacity that is approximately 50% of the capacity of wild-type R. etli [61]. Notably, S. meliloti has another peroxiredoxin encoded by nex1 that is primarily expressed in nodules [63].
The heterologous over-expression of a cyanobacterial flavodoxin in S. meliloti contributes to delayed nodule senescence without significant side effects, although its expression does not result in an increase of plant biomass [64]. As neither S. meliloti nor its host Medicago encodes flavodoxin, the excess of the FMN-containing electron transport protein may have functioned as a sink for ROS. If this strategy to express a ROS scavenging protein is successful, it would be worth to control endogenous enzymes at proper level. Monofunctional catalases, including S. meliloti katC and M. loti katE, might be good candidates for this strategy because they are exceptionally stable [65] and function in a wide pH range [66] to detoxify the membrane-permeable ROS H2O2.
SOD mutants of the same gene in different S. meliloti strains have been reported to have different symbiotic phenotypes. The disruption of sodA in S. meliloti strain Rm5000 results in fewer infection events, poor nodulation on Medicago, failure to differentiate into bacteroids and rapid senescence, all of which result in poor nitrogen fixation [37]. In contrast, disruption of the homologous gene (designated sodB) in S. meliloti strain Rm1021 has minimal effects on symbiosis in terms of plant growth and nitrogenase activity [67]. Because the S. meliloti strains Rm5000 and Rm1021 are derivatives of the same wild-type strain SU47 (synonym of Rm2011) that were selected for spontaneous rifampicin and streptomycin resistance, respectively [68, 69], the phenotypic difference is perplexing. However, SU47 has a slower response to NF than Rm1021 as analyzed by Ca2+ spiking, probably due to the lower expression of nod genes [70]. We have disrupted the sole SOD gene (sodA) in M. loti MAFF303099, and the mutant displays markedly different symbiotic efficiencies depending on the Lotus japonicus cultivar used (Hanyu and Saeki, unpublished data). This suggests that the pairing of rhizobial strains and host cultivars significantly affects the outcome of symbiosis.
BacA protein
bacA was first described in Sinorhizobiummeliloti as a gene essential for bacteroid formation after release into the cytoplasm of the host Medicago sativa [11]. The predicted product of bacA is an integral membrane peptide transporter that has 64% sequence similarity to Escherichia coli SbmA [11]. SbmA sensitizes E. coli to some peptide antibiotics including microcin B17, microcin J25 and bleomycin [71]. S. melilotibacA is also involved in this sensitization and, most likely, in the uptake of peptide antibiotics [72, 73]. Homologues of bacA are found in M. loti MAFF303099 [58] and animal pathogens including Brucella abortus and Mycobacterium tuberculosis [74, 75]. In B. abortus, which induces abortion in chronically infected animals, the bacA homolog is required for effective survival in murine macrophages [74]. In M. tuberculosis, the causative agent of tuberculosis, the lack of the bacA homolog results in the compromised maintenance of persistent infection in mice [75]. Therefore, BacA homologues in symbiotic and pathogenic bacteria may support chronic intracellular survival in their eukaryotic hosts by counteracting host defense reactions [14, 19, 74, 75].
Although many rhizobia have bacA homologues, genetic studies were only recently performed in five rhizobial species other than S. meliloti [76–78]. Gene disruption studies revealed that the bacA homologs of Mesorhizobium huakuii and Rhizobium leguminosarum bv. viciae are essential for symbiosis with Astragalus sinicus (Chinese milk vetch) and Pisum sativum (pea) [76, 77], respectively. In contrast, similar gene disruption studies have revealed that the bacA homologs in Rhizobium leguminosarum bv. phaseoli and Rhizobium etli are dispensable for symbiosis with the host Phaseolus vulgaris (bean) [77] and that the M. loti MAFF303099 bacA homolog is dispensable for symbiosis with L. japonicus [78]. Legume hosts that do not require bacA in compatible rhizobia form determinate nodules, while those that require bacA form indeterminate nodules. The indeterminate hosts Medicago, Pisum and Astragalus belong to the inverted repeat-lacking clade (IRLC), whereas the two determinate hosts, Lotus and Phaseolus, belong to the robinoids and milletioids clades, respectively [79].
Despite their different contributions to the establishment of symbiosis, bacA and its homologues appear to perform similar functions under free-living conditions. BacA-lacking mutants of five rhizobial species have increased resistance to the glycopeptide antibiotic bleomycin and some aminoglycoside antibiotics and increased sensitivity to some membrane-disturbing reagents including detergents [72, 76–78, 80–82]. The M. tuberculosis BacA homologue is reported to be an ABC-transporter, but its ATP-binding cassette is located in an extended carboxyl-terminal portion that is not conserved with rhizobial BacA homologs [75]. This suggests that rhizobial BacA homologs may not function without other synergistic components. The S. meliloti and M. huakuii bacA mutants have abnormal LPS lacking the very-long-chain fatty acid (VLCFA) modification [76, 83, 84]. This is in accordance with the relatively weak but significant structural similarity with the human adrenoleukodystrophy protein (hALDP), and genetic disorders associated with this protein can result in the accumulation of VLCFAs in all body tissues [85]. These properties support a role of BacA homologs as transporters, although there is little direct biochemical evidence.
The rhizobial requirement for BacA is determined by its pairing to a host legume. Rhizobial species absolutely require BacA to establish symbiosis with the IRLC indeterminate legumes Medicago, Pisum and Astragalus, but it is not required for symbiosis with the non-IRLC determinate legumes Phaseolus and Lotus. In nodules of M. truncatula and P. sativum, bacteroids tend to undergo endoreproduction with fragmented DNA and have reduced reproductive viability [21]. A similar fragmentation of bacteroid DNA was also observed in A. sinicus [86]. In contrast, bacteroids in L. japonicus and P. vulgaris maintain quasi-normal sizes and reproductive viability [21]. The two cultivars of R. leguminosarum have essentially identical main chromosomes, which contain bacA and differ only in their symbiotic plasmids [87]; however, their bacA mutants display opposite symbiotic phenotypes. Similarly, although M. loti and M. hukuii are closely related and have similar BacA homologs that differ by only two amino acids, bacA mutants of these two non-IRLC legumes display opposite symbiotic phenotypes [76, 78]. M. loti bacA can partially complement the symbiotic defect of the S. meliloti bacA mutant [78], suggesting that the necessity of BacA to establish symbiosis is solely dependent on the host properties that determine the strength of the intermediate, and possibly maintenance, phase attacks.
IRLC legumes have various nodule-specific cysteine-rich (NCR) peptides that are similar to defensin-type antimicrobial peptides, which may be responsible for the strong attacks (Fig. 2c). The peptides are individually expressed in distinct nodule zones, but neither Lotus nor Phaseolus possesses such peptides [88, 89]. Some NCR peptides have regulatory or bactericidal effects on rhizobial cells and attach to bacteroids in symbiosomes [43]. Notably, NCR peptides attached to bacteroids are delivered via a nodule-specific protein secretion pathway consisting of DNF1, a subunit of the signal peptidase complex [90].
Even if a specific NCR peptide degrades bacA-lacking mutants of S. meliloti and R. leguminosarum bv. viciae, it can be difficult to explain the mechanism underlying the BacA-mediated protection due to the pleiotropy of bacA deletions. All known BacA-lacking mutants have a compromised cell envelope, with the partial loss of VLCFA from LPS, and a decreased sensitivity to some antibiotics. In addition, R. leguminosarum species are deficient in the ability to utilize dicarboxylic acid as a growth substrate [77]. The lack of VLCFA modifications is not directly related to decreased bleomycin sensitivity [82]. To fully understand the protective mechanism of BacA, its biochemical properties, including physiological substrates and components with which it interacts, must be elucidated.
Although the requirement of BacA to establish symbiosis with five legume species has been investigated, it is unclear if the studies in these IRLC and non-IRLC legumes can be extrapolated to indeterminate and determinate legumes. Recently, Oono et al. [91] reported additional classification schemes of bacteroid morphology—swollen (longer than 4 μm or wider than 1.5 μm (for spherical bacteroids) or branched (regardless of size)) and non-swollen (smaller than 2.5 × 1.5 μm)—together with the conventional indeterminate and determinate nodule types. They observed at least four classes of nodules: indeterminate with non-swollen bacteroids, determinate with swollen bacteroids, indeterminate with swollen bacteroids and determinate with non-swollen bacteroids. Based on the distribution of the classes within a legume phylogenetic tree, they proposed multiple evolutionary origins for nodule types and bacterial deformations [91]. It will be of interest to investigate the response of legumes with determinate nodules and swollen bacteroids, as well as legumes with indeterminate nodules and non-swollen bacteroids, to rhizobial cells lacking BacA. Such analyses might help elucidation of factors used to evade host attacks during chronic infection.
Type III and type IV secretion systems and their effectors
The type III and type IV secretion systems (T3SS and T4SS) are important for the virulence of many animal and plant pathogenic bacteria. Bacteria use the T3/T4SS to transfer effector proteins from the bacterial cytoplasm into eukaryotic cells or the external milieu, where they can manipulate host cellular processes to facilitate pathogenicity [92, 93]. Phytopathogenic bacteria often use effector proteins to suppress the host immune response activated by pathogen-associated molecular patterns (PAMPs) [94]. To counteract bacterial effector proteins, host plants use resistance proteins (R proteins) that recognize effector proteins and trigger resistance responses, including the hypersensitive response (HR), which halts pathogen invasion and virulence [95, 96].
T3SS and T4SS are also found in many rhizobia that use effectors to modulate their host specificity and symbiotic efficiency [12, 97, 98]. Rhizobial T3SS genes are often designated nop (nodulation outer protein) [99]. Among the rhizobia of model legumes, the Lotus symbionts M. loti strains MAFF303099 and R7A possess T3SS and T4SS, respectively [58, 100]. The deletion of T3SS in MAFF303099 does not affect its symbiotic performance with its host L. japonicus but modulates its nodulation capacity with other species of the Lotus genus [101]. A negative effector (Mlr6361 protein) against Lotus halophilus possesses a conserved multidomain that is also found in the T3SS genes of several plant pathogens [101]. The absence of T4SS in R7A results in delayed nodulation with L. corniculatus but increases the capacity of productive symbiosis with the tree legume Leucaena leucocephala [102, 103]. Separate insertion mutations in two effectors (Msi059 and Msi061 proteins) have shown that these proteins are at least partially responsible for the positive and negative effects. Notably, it has been demonstrated using the bacteriophage P1 Cre/lox-based CRAft system [104] that these effectors can be transported via the Agrobacterium tumefaciens VirB/D4 system into Arabidopsis thaliana and Saccharomyces cerevisiae [102]. The Mlr6361, Msi059, and Msi061 effectors negatively affect symbiosis with certain hosts and could be recognized as PAMP or virulence factors by the host, whereas those with positive effects might successfully evade host defenses or reinforce symbiotic functions. The Medicago symbiont S. meliloti strain Rm1021 also encodes a T4SS [105], which does not affect its symbiotic capacity with M. sativa or M. truncatula [106, 107]. Rhizobium sp. NGR234, which has a broad host range, can establish symbiosis with L. japonicus [108] and has several T3SS and T4SS [109]; however, experimental studies have focused on one T3SS encoded by a symbiotic plasmid [99, 110–122].
Based on studies in which increased transcription of T3SS genes in Rhizobium sp. NGR234 was observed within 24 h of adding the flavonoid daidzein, T3SS have been assumed to counteract the initial and intermediate phase threats [121, 122]. This hypothesis was supported by the immunocytochemical observation that Sinorhizobium fredii NopX was detected in infection threads, but not within mature nodules, in Glycine max (soybean) and Vigna unguiculata (cowpea) [123] as well as by other omics studies [124, 125]. However, recent experimental data have indicated that rhizobial T3SS function even in mature nodules. Gene-fusion analyses have shown that NopE1 of B. japonicum USDA110 is expressed in mature nodules (4 weeks after infection) and in infection threads [126]. This indicates that some rhizobial effectors are used to subvert the late-phase threats, thereby enabling chronic infection, as is the case in many T3SS effectors of animal pathogens [127, 128].
The presence of B. japonicum NopE1 or its closely related homolog NopE2 positively affects symbiosis with G. max and Macroptilium atropurpureum (Siratro or Purple Bush-Bean) but negatively affects symbiosis with Vigna radiata (mung bean or green gram) [129]. Both NopE1 and NopeE2 are transported into the M. atropurpureum (host) cytoplasm, as demonstrated using an in-frame fusion [129] to the Bordetella pertussis calmodulin-dependent adenylate cyclase (cya gene product), which becomes active only in the eukaryotic host cytoplasm where calmodulin is present [130, 131]. Both NopE1 and NopeE2 become physiologically active only after autonomous cleavage [129]. Modification is also required for the activation of NopL and NopP by phosphorylation by a plant cytosolic kinase in Rhizobium sp. NGR234 [114]. NopP is transported to the cytoplasm of host nodule cells, which was demonstrated using Vigna unguiculata as a host and the fusion of a closely related NopP from S. fredii to the Bordetella adenylate cyclase described above [132]. These studies indicate that some rhizobial T3SS effectors require posttranslational modifications to function in their host.
S. fredii NopP is responsible for suppressing the expression of the host defense-related gene PR1 in G. max [133]. It is plausible that rhizobial T3SS and T4SS effectors can suppress or induce PAMP-triggered host defense responses that might constitute the initial and intermediate phase threats. It is also possible that during the maintenance phase, in addition to defense measures that are common in other non-leguminous plants, legume hosts have nodule-specific attack mechanisms, as exemplified by the expression of NCRs by IRLC legumes. Some T3SS effectors can suppress nodule-specific plant defense measures while others can be activators that increase the expression of nodule-function host genes. Because at least some T3SS effectors are expressed in mature nodules and transported to the host cytoplasm, there can be molecular chaperones that control the timing and order of effector transport, as observed in pathogens [134, 135]. To decipher the function and mode of action for each T3SS or T4SS effector, it is necessary to identify the target molecules in the host plant and to investigate the biochemical properties of the effectors and targets, as well as their molecular chaperones.
Concluding remarks
Rhizobial measures to evade host-derived and endogenous threats do not function equally during the three threat phases (Fig. 2). Furthermore, the measures are not used in the same manner in the model rhizobial species M. loti and S. meliloti. To evade the initial and intermediate phase threats, ROS scavenging enzymes seem to be more critically are required in S. meliloti than in M. loti. It is possible that ROS scavenging subverts the maintenance phase threats in both rhizobial models. The BacA protein is essential to overcome intermediate threats in S. meliloti, but not M. loti. The T3SS and T4SS in M. loti are important for host-specific modulation of symbiotic efficiency, mostly by evading or triggering the host defense mechanisms during the initial and intermediate phases. However, it is possible that some effectors contribute to evade threats during the maintenance phase. The contribution of the T4SS in S. meliloti is currently not known.
Autoregulation of nodulation is necessary for legumes to balance energy expenditure and growth. Significant advances, including the identification of a receptor kinase and signaling peptide, have been made over the last decade (see reviews [18, 136]). However, it is currently not known how host plants arrest infecting rhizobial cells. Several specific questions remain: (1) Do hosts use ROS or other hazardous compounds to eliminate excess rhizobia? (2) Do hosts simply discontinue the organogenesis program as well as the elongation of infection threads? Combined cytological and biochemical analyses are necessary to address these questions.
Because the maintenance of active mature nodules for long periods would be directly beneficial to host legumes and ultimately to agronomy, it is important to investigate the nature of the late-phase threats. It is also important to determine whether they are involved in the plant sanction of inefficient rhizobial cells. This phenomenon has attracted attention because such a selection method for rhizobia would be beneficial for legumes and influence the co-evolution of legume–rhizobia [137]. It has been reported that the legume host G. max supplies less oxygen to nodule cells containing rhizobial B. japonicum cells unable to fix nitrogen than to those with nitrogen-fixing rhizobia [50]. However, whether this phenomenon exists is controversial because there is supporting [138] and opposing [139, 140] evidence. Although it is currently unclear whether the observed sanction is caused by simple metabolic imbalances or by complex surveillance machineries that detect commensal rhizobia, studies on late-phase threats will be useful to breed more efficient rhizobial species.
Acknowledgments
The author thanks Shin Okazaki for many stimulating discussions. This work was supported in part by the Special Coordination Fund for Promoting Science and Technology and KAKENHI (Grant-in-Aid for Scientific Research) on the Priority Area “Comparative Genomics” (17018041) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
It might be noted that cloning of soybean R gene probably to counteract rhizobial T3SS effectors has been reported by Zhu and colleagues [141] during revision process of this review.
References
- 1.Oldroyd GED, Downie JA. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol. 2008;59:519–546. doi: 10.1146/annurev.arplant.59.032607.092839. [DOI] [PubMed] [Google Scholar]
- 2.Parniske M. Intracellular accommodation of microbes by plants: a common developmental program for symbiosis and disease? Curr Opin Plant Biol. 2000;3:320–328. doi: 10.1016/s1369-5266(00)00088-1. [DOI] [PubMed] [Google Scholar]
- 3.Mithöfer A. Suppression of plant defence in rhizobia–legume symbiosis. Trends Plant Sci. 2002;7:440–444. doi: 10.1016/s1360-1385(02)02336-1. [DOI] [PubMed] [Google Scholar]
- 4.El Yahyaoui F, Kuster H, Ben Amor B, Hohnjec N, Puhler A, Becker A, Gouzy J, Vernie T, Gough C, Niebel A, Godiard L, Gamas P. Expression profiling in Medicago truncatula Identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol. 2004;136:3159–3176. doi: 10.1104/pp.104.043612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kouchi H, Shimomura K, Hata S, Hirota A, Wu G-J, Kumagai H, Tajima S, Suganuma N, Suzuki A, Aoki T, Hayashi M, Yokoyama T, Ohyama T, Asamizu E, Kuwata C, Shibata D, Tabata S. Large-scale analysis of gene expression profiles during early stages of root nodule formation in a model legume, Lotus japonicus . DNA Res. 2004;11:263–274. doi: 10.1093/dnares/11.4.263. [DOI] [PubMed] [Google Scholar]
- 6.Vasse J, de Billy F, Truchet G. Abortion of infection during the rhizobium meliloti-alfalfa symbiotic interaction is accompanied by a hypersensitive reaction. Plant J. 1993;4:555–566. [Google Scholar]
- 7.Maeda D, Ashida K, Iguchi K, Chechetka SA, Hijikata A, Okusako Y, Deguchi Y, Izui K, Hata S. Knockdown of an arbuscular Mycorrhiza-inducible phosphate transporter gene of Lotus japonicus suppresses mutualistic symbiosis. Plant Cell Physiol. 2006;47:807–817. doi: 10.1093/pcp/pcj069. [DOI] [PubMed] [Google Scholar]
- 8.Schumpp O, Crevecoeur M, Broughton WJ, Deakin WJ. Delayed maturation of nodules reduces symbiotic effectiveness of the Lotus japonicus-Rhizobium sp. NGR234 interaction. J Exp Bot. 2009;60:581–590. doi: 10.1093/jxb/ern302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Handberg K, Stougaard J. Lotus japonicus, an autogamous, diploid legume species for classical and molecular genetics. Plant J. 1992;2:487–496. [Google Scholar]
- 10.Barker D, Bianchi S, Blondon F, Dattée Y, Duc G, Essad S, Flament P, Gallusci P, Génier G, Guy P, Muel X, Tourneur J, Dénarié J, Huguet T. Medicago truncatula, a model plant for studying the molecular genetics of the rhizobium–legume symbiosis. Plant Mol Biol Rep. 1990;8:40–49. [Google Scholar]
- 11.Glazebrook J, Ichige A, Walker GC. A Rhizobium meliloti homolog of the Escherichia coli peptide-antibiotic transport protein SbmA is essential for bacteroid development. Genes Dev. 1993;7:1485–1497. doi: 10.1101/gad.7.8.1485. [DOI] [PubMed] [Google Scholar]
- 12.Deakin WJ, Broughton WJ. Symbiotic use of pathogenic strategies: rhizobial protein secretion systems. Nat Rev Micro. 2009;7:312–320. doi: 10.1038/nrmicro2091. [DOI] [PubMed] [Google Scholar]
- 13.D’Haeze W, Holsters M. Surface polysaccharides enable bacteria to evade plant immunity. Trends Microbiol. 2004;12:555–561. doi: 10.1016/j.tim.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 14.Gibson KE, Kobayashi H, Walker GC. Molecular determinants of a symbiotic chronic infection. Annu Rev Genet. 2008;42:413–441. doi: 10.1146/annurev.genet.42.110807.091427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spaink HP. The molecular basis of infection and nodulation by rhizobia: the ins and outs of sympathogenesis. Annu Rev Phytopathol. 1995;33:345–368. doi: 10.1146/annurev.py.33.090195.002021. [DOI] [PubMed] [Google Scholar]
- 16.Perret X, Staehelin C, Broughton WJ. Molecular basis of symbiotic promiscuity. Microbiol Mol Biol Rev. 2000;64:180–201. doi: 10.1128/mmbr.64.1.180-201.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gage DJ. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol Mol Biol Rev. 2004;68:280–300. doi: 10.1128/MMBR.68.2.280-300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferguson BJ, Indrasumunar A, Hayashi S, Lin MH, Lin YH, Reid DE, Gresshoff PM. Molecular analysis of legume nodule development and autoregulation. J Integr Plant Biol. 2010;52:61–76. doi: 10.1111/j.1744-7909.2010.00899.x. [DOI] [PubMed] [Google Scholar]
- 19.Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC. How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat Rev Microbiol. 2007;5:619–633. doi: 10.1038/nrmicro1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oldroyd GED, Downie JA. Nuclear calcium changes at the core of symbiosis signalling. Curr Opin Plant Biol. 2006;9:351–357. doi: 10.1016/j.pbi.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 21.Mergaert P, Uchiumi T, Alunni B, Evanno G, Cheron A, Catrice O, Mausset A-E, Barloy-Hubler F, Galibert F, Kondorosi A, Kondorosi E. Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium–legume symbiosis. Proc Natl Acad Sci USA. 2006;103:5230–5235. doi: 10.1073/pnas.0600912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oke V, Long SR. Bacteroid formation in the rhizobium–legume symbiosis. Curr Opin Microbiol. 1999;2:641–646. doi: 10.1016/s1369-5274(99)00035-1. [DOI] [PubMed] [Google Scholar]
- 23.Kijne JW. The fine structure of pea root nodules. 1. Vacuolar changes after endocytotic host cell infection by Rhizobium leguminosarum . Physiol Plant Pathol. 1975;5:75–76. [Google Scholar]
- 24.Kijne JW. The fine structure of pea root nodules. 2. Senescence and disintegration of the bacteroid tissue. Physiol Plant Pathol. 1975;7:17–21. [Google Scholar]
- 25.Gresshoff PM, Rolfe BG. Viability of Rhizobium bacteroids isolated from soybean nodule protoplasts. Planta. 1978;142:329–333. doi: 10.1007/BF00385085. [DOI] [PubMed] [Google Scholar]
- 26.Sutton WD, Paterson AD. Further evidence for a plant host effect on Rhizobium bacteroid viability. Plant Sci Lett. 1983;30:33–41. [Google Scholar]
- 27.Sutton WD, Paterson AD. Effects of the plant host on the detergent sensitivity and viability of Rhizobium bacteroids. Planta. 1980;148:287–292. doi: 10.1007/BF00380040. [DOI] [PubMed] [Google Scholar]
- 28.Puppo A, Groten K, Bastian F, Carzaniga R, Soussi M, Lucas MM, De Felipe MR, Harrison J, Vanacker H, Foyer CH. Legume nodule senescence: roles for redox and hormone signalling in the orchestration of the natural aging process. New Phytol. 2005;165:683–701. doi: 10.1111/j.1469-8137.2004.01285.x. [DOI] [PubMed] [Google Scholar]
- 29.Mitra RM, Shaw SL, Long SR. Six nonnodulating plant mutants defective for Nod factor-induced transcriptional changes associated with the legume–rhizobia symbiosis. Proc Natl Acad Sci USA. 2004;101:10217–10222. doi: 10.1073/pnas.0402186101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitra RM, Long SR. Plant and bacterial symbiotic mutants define three transcriptionally distinct stages in the development of the Medicago truncatula/Sinorhizobium meliloti symbiosis. Plant Physiol. 2004;134:595–604. doi: 10.1104/pp.103.031518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cárdenas L, Martínez A, Sánchez F, Quinto C. Fast, transient and specific intracellular ROS changes in living root hair cells responding to Nod factors (NFs) Plant J. 2008;56:802–813. doi: 10.1111/j.1365-313X.2008.03644.x. [DOI] [PubMed] [Google Scholar]
- 32.Santos R, Hérouart D, Sigaud S, Touati D, Puppo A. Oxidative burst in Alfalfa–Sinorhizobium meliloti symbiotic interaction. Mol Plant Microbe Interact. 2001;14:86–89. doi: 10.1094/MPMI.2001.14.1.86. [DOI] [PubMed] [Google Scholar]
- 33.Shaw SL, Long SR. Nod factor inhibition of reactive oxygen efflux in a host legume. Plant Physiol. 2003;132:2196–2204. doi: 10.1104/pp.103.021113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagata M, Murakami E-i, Shimoda Y, Shimoda-Sasakura F, Kucho K-i, Suzuki A, Abe M, Higashi S, Uchiumi T. Expression of a class 1 hemoglobin gene and production of nitric oxide in response to symbiotic and pathogenic bacteria in Lotus japonicus . Mol Plant Microbe Interact. 2008;21:1175–1183. doi: 10.1094/MPMI-21-9-1175. [DOI] [PubMed] [Google Scholar]
- 35.Gamas P, de Billy F, Truchet G. Symbiosis-specific expression of two Medicago truncatula nodulin genes, MtN1 and MtN13, encoding products homologous to plant defense proteins. Mol Plant Microbe Interact. 1998;11:393–403. doi: 10.1094/MPMI.1998.11.5.393. [DOI] [PubMed] [Google Scholar]
- 36.Jamet A, Sigaud S, Van de Sype G, Puppo A, Hérouart D. Expression of the bacterial catalase genes during Sinorhizobium meliloti–Medicago sativa symbiosis and their crucial role during the infection process. Mol Plant Microbe Interact. 2003;16:217–225. doi: 10.1094/MPMI.2003.16.3.217. [DOI] [PubMed] [Google Scholar]
- 37.Santos R, Hérouart D, Puppo A, Touati D. Critical protective role of bacterial superoxide dismutase in rhizobium–legume symbiosis. Mol Microbiol. 2000;38:750–759. doi: 10.1046/j.1365-2958.2000.02178.x. [DOI] [PubMed] [Google Scholar]
- 38.Campbell GRO, Reuhs BL, Walker GC. Chronic intracellular infection of alfalfa nodules by Sinorhizobium meliloti requires correct lipopolysaccharide core. Proc Natl Acad Sci USA. 2002;99:3938–3943. doi: 10.1073/pnas.062425699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leigh JA, Signer ER, Walker GC. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc Natl Acad Sci USA. 1985;82:6231–6235. doi: 10.1073/pnas.82.18.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leigh JA, Walker GC. Exopolysaccharides of Rhizobium: synthesis, regulation and symbiotic function. Trends Genet. 1994;10:63–67. doi: 10.1016/0168-9525(94)90151-1. [DOI] [PubMed] [Google Scholar]
- 41.Jones KM, Sharopova N, Lohar DP, Zhang JQ, VandenBosch KA, Walker GC. Differential response of the plant Medicago truncatula to its symbiont Sinorhizobium meliloti or an exopolysaccharide-deficient mutant. Proc Natl Acad Sci USA. 2008;105:704–709. doi: 10.1073/pnas.0709338105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Banba M, Siddique A-BM, Kouchi H, Izui K, Hata S. Lotus japonicus forms early senescent root nodules with Rhizobium etli. Mol Plant Microbe Interact. 2001;14:173–180. doi: 10.1094/MPMI.2001.14.2.173. [DOI] [PubMed] [Google Scholar]
- 43.Van de Velde W, Guerra JCP, Keyser AD, De Rycke R, Rombauts S, Maunoury N, Mergaert P, Kondorosi E, Holsters M, Goormachtig S. Aging in legume symbiosis. A molecular view on nodule senescence in Medicago truncatula . Plant Physiol. 2006;141:711–720. doi: 10.1104/pp.106.078691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perez Guerra JC, Coussens G, De Keyser A, De Rycke R, De Bodt S, Van De Velde W, Goormachtig S, Holsters M. Comparison of developmental and stress-induced nodule senescence in Medicago truncatula . Plant Physiol. 2010;152:1574–1584. doi: 10.1104/pp.109.151399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kardailsky IV, Brewin NJ. Expression of cysteine protease genes in pea nodule development and senescence. Mol Plant Microbe Interact. 1996;9:689–695. doi: 10.1094/mpmi-9-0689. [DOI] [PubMed] [Google Scholar]
- 46.Alesandrini F, Mathis R, Van de Sype G, Hérouart D, Puppo A. Possible roles for a cysteine protease and hydrogen peroxide in soybean nodule development and senescence. New Phytol. 2003;158:131–138. [Google Scholar]
- 47.Asp T, Bowra S, Borg S, Holm PB. Cloning and characterisation of three groups of cysteine protease genes expressed in the senescing zone of white clover (Trifolium repens) nodules. Plant Sci. 2004;167:825–837. [Google Scholar]
- 48.Li Y, Zhou L, Chen D, Tan X, Lei L, Zhou J. A nodule-specific plant cysteine proteinase, AsNODF32, is involved in nodule senescence and nitrogen-fixation activity of the green manure legume Astragalus sinicus . New Phytol. 2008;180:185–192. doi: 10.1111/j.1469-8137.2008.02562.x. [DOI] [PubMed] [Google Scholar]
- 49.Limpens E, Ivanov S, van Esse W, Voets G, Fedorova E, Bisseling T. Medicago N2-fixing symbiosomes acquire the endocytic identity marker Rab7 but delay the acquisition of vacuolar identity. Plant Cell. 2009;21:2811–2828. doi: 10.1105/tpc.108.064410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kiers ET, Rousseau RA, West SA, Denison RF. Host sanctions and the legume–rhizobium mutualism. Nature. 2003;425:78–81. doi: 10.1038/nature01931. [DOI] [PubMed] [Google Scholar]
- 51.Matamoros MA, Dalton DA, Ramos J, Clemente MR, Rubio MC, Becana M. Biochemistry and molecular biology of antioxidants in the rhizobia–legume symbiosis. Plant Physiol. 2003;133:499–509. doi: 10.1104/pp.103.025619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Glyan’ko A, Vasil’eva G. Reactive oxygen and nitrogen species in legume–rhizobial symbiosis: a review. Appl Biochem Microbiol. 2010;46:15–22. [PubMed] [Google Scholar]
- 53.Jamet A, Mandon K, Puppo A, Herouart D. H2O2 is required for optimal establishment of the Medicago sativa/Sinorhizobium meliloti symbiosis. J Bacteriol. 2007;189:8741–8745. doi: 10.1128/JB.01130-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Herouart D, Sigaud S, Moreau S, Frendo P, Touati D, Puppo A. Cloning and characterization of the katA gene of Rhizobium meliloti encoding a hydrogen peroxide-inducible catalase. J Bacteriol. 1996;178:6802–6809. doi: 10.1128/jb.178.23.6802-6809.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sigaud S, Becquet V, Frendo P, Puppo A, Herouart D. Differential regulation of two divergent Sinorhizobium meliloti genes for HPII-Like catalases during free-living growth and protective role of both catalases during symbiosis. J Bacteriol. 1999;181:2634–2639. doi: 10.1128/jb.181.8.2634-2639.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Santos R, Bocquet S, Puppo A, Touati D. Characterization of an atypical superoxide dismutase from Sinorhizobium meliloti . J Bacteriol. 1999;181:4509–4516. doi: 10.1128/jb.181.15.4509-4516.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davies BW, Walker GC. Identification of novel Sinorhizobium meliloti mutants compromised for oxidative stress protection and symbiosis. J Bacteriol. 2007;189:2110–2113. doi: 10.1128/JB.01802-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaneko T, Nakamura Y, Sato S, Asamizu E, Kato T, Sasamoto S, Watanabe A, Idesawa K, Ishikawa A, Kawashima K, Kimura T, Kishida Y, Kiyokawa C, Kohara M, Matsumoto M, Matsuno A, Mochizuki Y, Nakayama S, Nakazaki N, Shimpo S, Sugimoto M, Takeuchi C, Yamada M, Tabata S. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti . DNA Res. 2000;7:331–338. doi: 10.1093/dnares/7.6.331. [DOI] [PubMed] [Google Scholar]
- 59.Hanyu M, Fujimoto H, Tejima K, Saeki K. Functional differences of two distinct catalases in Mesorhizobium loti MAFF303099 under free-living and symbiotic conditions. J Bacteriol. 2009;191:1463–1471. doi: 10.1128/JB.01583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.del Carmen Vargas M, Encarnacion S, Davalos A, Reyes-Perez A, Mora Y, Garcia-de los Santos A, Brom S, Mora J. Only one catalase, katG, is detectable in Rhizobium etli, and is encoded along with the regulator OxyR on a plasmid replicon. Microbiology. 2003;149:1165–1176. doi: 10.1099/mic.0.25909-0. [DOI] [PubMed] [Google Scholar]
- 61.Dombrecht B, Heusdens C, Beullens S, Verreth C, Mulkers E, Proost P, Vanderleyden J, Michiels J. Defence of Rhizobium etli bacteroids against oxidative stress involves a complexly regulated atypical 2-Cys peroxiredoxin. Mol Microbiol. 2005;55:1207–1221. doi: 10.1111/j.1365-2958.2005.04457.x. [DOI] [PubMed] [Google Scholar]
- 62.Wood ZA, Schröder E, Robin Harris J, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 63.Oke V, Long SR. Bacterial genes induced within the nodule during the rhizobium–legume symbiosis. Mol Microbiol. 1999;32:837–849. doi: 10.1046/j.1365-2958.1999.01402.x. [DOI] [PubMed] [Google Scholar]
- 64.Redondo FJ, De la Pena TC, Morcillo CN, Lucas MM, Pueyo JJ. Overexpression of flavodoxin in bacteroids induces changes in antioxidant metabolism leading to delayed senescence and starch accumulation in alfalfa root nodules. Plant Physiol. 2009;149:1166–1178. doi: 10.1104/pp.108.129601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chelikani P, Fita I, Loewen PC. Diversity of structures and properties among catalases. Cell Mol Life Sci. 2004;61:192–208. doi: 10.1007/s00018-003-3206-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ardissone S, Frendo P, Laurenti E, Jantschko W, Obinger C, Puppo A, Ferrari RP. Purification and physical-chemical characterization of the three hydroperoxidases from the symbiotic bacterium Sinorhizobium meliloti . Biochemistry. 2004;43:12692–12699. doi: 10.1021/bi048836s. [DOI] [PubMed] [Google Scholar]
- 67.Davies BW, Walker GC. Disruption of sitA compromises Sinorhizobium meliloti for manganese uptake required for protection against oxidative stress. J Bacteriol. 2007;189:2101–2109. doi: 10.1128/JB.01377-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meade HM, Long SR, Ruvkun GB, Brown SE, Ausubel FM. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J Bacteriol. 1982;149:114–122. doi: 10.1128/jb.149.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Finan TM, Hartweig E, LeMieux K, Bergman K, Walker GC, Signer ER. General transduction in Rhizobium meliloti . J Bacteriol. 1984;159:120–124. doi: 10.1128/jb.159.1.120-124.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wais RJ, Wells DH, Long SR. Analysis of differences between Sinorhizobium meliloti 1021 and 2011 strains using the host calcium spiking response. Mol Plant Microbe Interact. 2007;15:1245–1252. doi: 10.1094/MPMI.2002.15.12.1245. [DOI] [PubMed] [Google Scholar]
- 71.Yorgey P, Lee J, Kördel J, Vivas E, Warner P, Jebaratnam D, Kolter R. Posttranslational modifications in microcin B17 define an additional class of DNA gyrase inhibitor. Proc Natl Acad Sci USA. 1994;91:4519–4523. doi: 10.1073/pnas.91.10.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ichige A, Walker GC. Genetic analysis of the Rhizobium meliloti bacA gene: functional interchangeability with the Escherichia coli sbmA gene and phenotypes of mutants. J Bacteriol. 1997;179:209–216. doi: 10.1128/jb.179.1.209-216.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marlow VL, Haag AF, Kobayashi H, Fletcher V, Scocchi M, Walker GC, Ferguson GP. Essential role for the BacA protein in the uptake of a truncated eukaryotic peptide in Sinorhizobium meliloti . J Bacteriol. 2009;191:1519–1527. doi: 10.1128/JB.01661-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.LeVier K, Phillips RW, Grippe VK, Roop RM, II, Walker GC. Similar requirements of a plant symbiont and a mammalian pathogen for prolonged intracellular survival. Science. 2000;287:2492–2493. doi: 10.1126/science.287.5462.2492. [DOI] [PubMed] [Google Scholar]
- 75.Domenech P, Kobayashi H, LeVier K, Walker GC, Barry CE., III BacA, an ABC transporter involved in maintenance of chronic murine infections with Mycobacterium tuberculosis . J Bacteriol. 2009;191:477–485. doi: 10.1128/JB.01132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tan X-J, Cheng Y, Li Y-X, Li Y-G, Zhou J-C. BacA is indispensable for successful Mesorhizobium–Astragalus symbiosis. Appl Microbiol Biotechnol. 2009;84:519–526. doi: 10.1007/s00253-009-1959-y. [DOI] [PubMed] [Google Scholar]
- 77.Karunakaran R, Haag AF, East AK, Ramachandran VK, Prell J, James EK, Scocchi M, Ferguson GP, Poole PS. BacA is essential for bacteroid development in nodules of Galegoid, but not Phaseoloid, legumes. J Bacteriol. 2010;192:2920–2928. doi: 10.1128/JB.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maruya J, Saeki K. The bacA gene homologue, mlr7400, in Mesorhizobium loti MAFF303099 is dispensable for symbiosis with Lotus japonicus but partially capable of supporting the symbiotic function of bacA in Sinorhizobium meliloti . Plant Cell Physiol. 2010;51:1443–1452. doi: 10.1093/pcp/pcq114. [DOI] [PubMed] [Google Scholar]
- 79.Wojciechowski MF, Lavin M, Sanderson MJ. A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Am J Bot. 2004;91:1846–1862. doi: 10.3732/ajb.91.11.1846. [DOI] [PubMed] [Google Scholar]
- 80.LeVier K, Walker GC. Genetic analysis of the Sinorhizobium meliloti BacA protein: differential effects of mutations on phenotypes. J Bacteriol. 2001;183:6444–6453. doi: 10.1128/JB.183.21.6444-6453.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ferguson GP, Roop Ii RM, Walker GC. Deficiency of a Sinorhizobium meliloti bacA mutant in alfalfa symbiosis correlates with alteration of the cell envelope. J Bacteriol. 2002;184:5625–5632. doi: 10.1128/JB.184.20.5625-5632.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ferguson GP, Jansen A, Marlow VL, Walker GC. BacA-mediated bleomycin sensitivity in Sinorhizobium meliloti is independent of the unusual Lipid A modification. J Bacteriol. 2006;188:3143–3148. doi: 10.1128/JB.188.8.3143-3148.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ferguson G, Datta A, Carlson R, Walker G. Importance of unusually modified lipid A in Sinorhizobium stress resistance and legume symbiosis. Mol Microbiol. 2005;56:68–80. doi: 10.1111/j.1365-2958.2005.04536.x. [DOI] [PubMed] [Google Scholar]
- 84.Ferguson GP, Datta A, Baumgartner J, Roop RM, Carlson RW, Walker GC. Similarity to peroxisomal-membrane protein family reveals that Sinorhizobium and Brucella BacA affect lipid-A fatty acids. Proc Natl Acad Sci USA. 2004;101:5012–5017. doi: 10.1073/pnas.0307137101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Valianpour F, Selhorst JJM, van Lint LEM, van Gennip AH, Wanders RJA, Kemp S. Analysis of very long-chain fatty acids using electrospray ionization mass spectrometry. Mol Genet Metab. 2003;79:189–196. doi: 10.1016/s1096-7192(03)00098-2. [DOI] [PubMed] [Google Scholar]
- 86.Kobayashi H, Sunako M, Hayashi M, Murooka Y. DNA synthesis and fragmentation in bacteroids during Astragalus sinicus root nodule development. Biosci Biotechnol Biochem. 2001;65:510–515. doi: 10.1271/bbb.65.510. [DOI] [PubMed] [Google Scholar]
- 87.Young JP, Crossman L, Johnston A, Thomson N, Ghazoui Z, Hull K, Wexler M, Curson A, Todd J, Poole P, Mauchline T, East A, Quail M, Churcher C, Arrowsmith C, Cherevach I, Chillingworth T, Clarke K, Cronin A, Davis P, Fraser A, Hance Z, Hauser H, Jagels K, Moule S, Mungall K, Norbertczak H, Rabbinowitsch E, Sanders M, Simmonds M, Whitehead S, Parkhill J. The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol. 2006;7:R34. doi: 10.1186/gb-2006-7-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mergaert P, Nikovics K, Kelemen Z, Maunoury N, Vaubert D, Kondorosi A, Kondorosi E. A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol. 2003;132:161–173. doi: 10.1104/pp.102.018192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Alunni B, Kevei Z, Redondo-Nieto M, Kondorosi A, Mergaert P, Kondorosi E. Genomic organization and evolutionary insights on GRP and NCR genes, two large nodule-specific gene families in Medicago truncatula . Mol Plant Microbe Interact. 2007;20:1138–1148. doi: 10.1094/MPMI-20-9-1138. [DOI] [PubMed] [Google Scholar]
- 90.Wang D, Griffitts J, Starker C, Fedorova E, Limpens E, Ivanov S, Bisseling T, Long S. A nodule-specific protein secretory pathway required for nitrogen-fixing symbiosis. Science. 2010;327:1126–1129. doi: 10.1126/science.1184096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Oono R, Schmitt I, Sprent JI, Denison RF. Multiple evolutionary origins of legume traits leading to extreme rhizobial differentiation. New Phytol. 2010;187:508–520. doi: 10.1111/j.1469-8137.2010.03261.x. [DOI] [PubMed] [Google Scholar]
- 92.Alvarez-Martinez CE, Christie PJ. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev. 2009;73:775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Block A, Li G, Fu ZQ, Alfano JR. Phytopathogen type III effector weaponry and their plant targets. Curr Opin Plant Biol. 2008;11:396–403. doi: 10.1016/j.jbi.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 95.Abramovitch RB, Anderson JC, Martin GB. Bacterial elicitation and evasion of plant innate immunity. Nat Rev Mol Cell Biol. 2006;7:601–611. doi: 10.1038/nrm1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host–microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 97.Fauvart M, Michiels J. Rhizobial secreted proteins as determinants of host specificity in the rhizobium–legume symbiosis. FEMS Microbiol Lett. 2008;285:1–9. doi: 10.1111/j.1574-6968.2008.01254.x. [DOI] [PubMed] [Google Scholar]
- 98.Downie JA. The roles of extracellular proteins, polysaccharides and signals in the interactions of rhizobia with legume roots. FEMS Microbiol Rev. 2010;34:150–170. doi: 10.1111/j.1574-6976.2009.00205.x. [DOI] [PubMed] [Google Scholar]
- 99.Marie C, Deakin WJ, Viprey V, Kopciñska J, Golinowski W, Krishnan HB, Perret X, Broughton WJ. Characterization of Nops, nodulation outer proteins, secreted via the type III secretion system of NGR234. Mol Plant Microbe Interact. 2003;16:743–751. doi: 10.1094/MPMI.2003.16.9.743. [DOI] [PubMed] [Google Scholar]
- 100.Sullivan JT, Trzebiatowski JR, Cruickshank RW, Gouzy J, Brown SD, Elliot RM, Fleetwood DJ, McCallum NG, Rossbach U, Stuart GS, Weaver JE, Webby RJ, De Bruijn FJ, Ronson CW. Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J Bacteriol. 2002;184:3086–3095. doi: 10.1128/JB.184.11.3086-3095.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Okazaki S, Okabe S, Higashi M, Shimoda Y, Sato S, Tabata S, Hashiguchi M, Akashi R, Göttfert M, Saeki K. Identification and functional analysis of type III effector proteins in Mesorhizobium loti . Mol Plant Microbe Interact. 2010;23:223–234. doi: 10.1094/MPMI-23-2-0223. [DOI] [PubMed] [Google Scholar]
- 102.Hubber A, Vergunst AC, Sullivan JT, Hooykaas PJJ, Ronson CW. Symbiotic phenotypes and translocated effector proteins of the Mesorhizobium loti strain R7A VirB/D4 type IV secretion system. Mol Microbiol. 2004;54:561–574. doi: 10.1111/j.1365-2958.2004.04292.x. [DOI] [PubMed] [Google Scholar]
- 103.Hubber AM, Sullivan JT, Ronson CW. Symbiosis-induced cascade regulation of the Mesorhizobium loti R7A VirB/D4 type IV secretion system. Mol Plant Microbe Interact. 2007;20:255–261. doi: 10.1094/MPMI-20-3-0255. [DOI] [PubMed] [Google Scholar]
- 104.Vergunst AC, Schrammeijer B, den Dulk-Ras A, de Vlaam CMT, Regensburg-Tuink TJG, Hooykaas PJJ. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science. 2000;290:979–982. doi: 10.1126/science.290.5493.979. [DOI] [PubMed] [Google Scholar]
- 105.Galibert F, Finan TM, Long SR, Puhler A, Abola P, Ampe F, Barloy-Hubler F, Barnett MJ, Becker A, Boistard P, Bothe G, Boutry M, Bowser L, Buhrmester J, Cadieu E, Capela D, Chain P, Cowie A, Davis RW, Dreano S, Federspiel NA, Fisher RF, Gloux S, Godrie T, Goffeau A, Golding B, Gouzy J, Gurjal M, Hernandez-Lucas I, Hong A, Huizar L, Hyman RW, Jones T, Kahn D, Kahn ML, Kalman S, Keating DH, Kiss E, Komp C, Lelaure V, Masuy D, Palm C, Peck MC, Pohl TM, Portetelle D, Purnelle B, Ramsperger U, Surzycki R, Thebault P, Vandenbol M, Vorholter FJ, Weidner S, Wells DH, Wong K, Yeh KC, Batut J. The composite genome of the legume symbiont Sinorhizobium meliloti . Science. 2001;293:668–672. doi: 10.1126/science.1060966. [DOI] [PubMed] [Google Scholar]
- 106.Barnett MJ, Fisher RF, Jones T, Komp C, Abola AP, Barloy-Hubler F, Bowser L, Capela D, Galibert F, Gouzy J, Gurjal M, Hong A, Huizar L, Hyman RW, Kahn D, Kahn ML, Kalman S, Keating DH, Palm C, Peck MC, Surzycki R, Wells DH, Yeh K-C, Davis RW, Federspiel NA, Long SR. Nucleotide sequence and predicted functions of the entire Sinorhizobium meliloti pSymA megaplasmid. Proc Natl Acad Sci USA. 2001;98:9883–9888. doi: 10.1073/pnas.161294798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jones KM, Lloret J, Daniele JR, Walker GC. The type IV secretion system of Sinorhizobium meliloti strain 1021 is required for conjugation but not for intracellular symbiosis. J Bacteriol. 2007;189:2133–2138. doi: 10.1128/JB.00116-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hussain AKMA, Jiang Q, Broughton WJ, Gresshoff PM. Lotus japonicus nodulates and fixes nitrogen with the broad host range Rhizobium sp. NGR234. Plant Cell Physiol. 1999;40:894–899. [Google Scholar]
- 109.Schmeisser C, Liesegang H, Krysciak D, Bakkou N, Le Quere A, Wollherr A, Heinemeyer I, Morgenstern B, Pommerening-Roser A, Flores M, Palacios R, Brenner S, Gottschalk G, Schmitz RA, Broughton WJ, Perret X, Strittmatter AW, Streit WR. Rhizobium sp. strain NGR234 possesses a remarkable number of secretion systems. Appl Environ Microbiol. 2009;75:4035–4045. doi: 10.1128/AEM.00515-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kambara K, Ardissone S, Kobayashi H, Saad MM, Schumpp O, Broughton WJ, Deakin WJ. Rhizobia utilize pathogen-like effector proteins during symbiosis. Mol Microbiol. 2009;71:92–106. doi: 10.1111/j.1365-2958.2008.06507.x. [DOI] [PubMed] [Google Scholar]
- 111.Wassem R, Kobayashi H, Kambara K, Le Quéré A, Walker GC, Broughton WJ, Deakin WJ. TtsI regulates symbiotic genes in Rhizobium species NGR234 by binding to tts boxes. Mol Microbiol. 2008;68:736–748. doi: 10.1111/j.1365-2958.2008.06187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Saad MM, Staehelin C, Broughton WJ, Deakin WJ. Protein–protein interactions within type III secretion system-dependent pili of Rhizobium sp. Strain NGR234. J Bacteriol. 2008;190:750–754. doi: 10.1128/JB.01116-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Deakin WJ, Marie C, Saad MM, Krishnan HB, Broughton WJ. NopA is associated with cell surface appendages produced by the type III secretion system of Rhizobium sp. strain NGR234. Mol Plant Microbe Interact. 2007;18:499–507. doi: 10.1094/MPMI-18-0499. [DOI] [PubMed] [Google Scholar]
- 114.Skorpil P, Saad MM, Boukli NM, Kobayashi H, Ares-Orpel F, Broughton WJ, Deakin WJ. NopP, a phosphorylated effector of Rhizobium sp. strain NGR234, is a major determinant of nodulation of the tropical legumes Flemingia congesta and Tephrosia vogelii . Mol Microbiol. 2005;57:1304–1317. doi: 10.1111/j.1365-2958.2005.04768.x. [DOI] [PubMed] [Google Scholar]
- 115.Saad MM, Kobayashi H, Marie C, Brown IR, Mansfield JW, Broughton WJ, Deakin WJ. NopB, a type III secreted protein of Rhizobium sp. strain NGR234, is associated with pilus-like surface appendages. J Bacteriol. 2005;187:1173–1181. doi: 10.1128/JB.187.3.1173-1181.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Marie C, Deakin WJ, Ojanen-Reuhs T, Diallo E, Reuhs B, Broughton WJ, Perret X. TtsI, a key regulator of Rhizobium species NGR234 is required for type III-dependent protein secretion and synthesis of rhamnose-rich polysaccharides. Mol Plant Microbe Interact. 2004;17:958–966. doi: 10.1094/MPMI.2004.17.9.958. [DOI] [PubMed] [Google Scholar]
- 117.Bartsev AV, Deakin WJ, Boukli NM, McAlvin CB, Stacey G, Malnoe P, Broughton WJ, Staehelin C. NopL, an effector protein of Rhizobium sp. NGR234, thwarts activation of plant defense reactions. Plant Physiol. 2004;134:871–879. doi: 10.1104/pp.103.031740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ausmees N, Kobayashi H, Deakin WJ, Marie C, Krishnan HB, Broughton WJ, Perret X. Characterization of NopP, a type III secreted effector of Rhizobium sp. strain NGR234. J Bacteriol. 2004;186:4774–4780. doi: 10.1128/JB.186.14.4774-4780.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bartsev AV, Boukli NM, Deakin WJ, Staehelin C, Broughton WJ. Purification and phosphorylation of the effector protein NopL from Rhizobium sp. NGR234. FEBS Lett. 2003;554:271–274. doi: 10.1016/s0014-5793(03)01145-1. [DOI] [PubMed] [Google Scholar]
- 120.Marie C, Broughton WJ, Deakin WJ. Rhizobium type III secretion systems: legume charmers or alarmers? Curr Opin Plant Biol. 2001;4:336–342. doi: 10.1016/s1369-5266(00)00182-5. [DOI] [PubMed] [Google Scholar]
- 121.Perret X, Freiberg C, Rosenthal A, Broughton WJ, Fellay R. High-resolution transcriptional analysis of the symbiotic plasmid of Rhizobium sp. NGR234. Mol Microbiol. 1999;32:415–425. doi: 10.1046/j.1365-2958.1999.01361.x. [DOI] [PubMed] [Google Scholar]
- 122.Viprey V, Del Greco A, Golinowski W, Broughton WJ, Perret X. Symbiotic implications of type III protein secretion machinery in Rhizobium . Mol Microbiol. 1998;28:1381–1389. doi: 10.1046/j.1365-2958.1998.00920.x. [DOI] [PubMed] [Google Scholar]
- 123.Krishnan HB. NolX of Sinorhizobium fredii USDA257, a type III-secreted protein involved in host range determination, is localized in the infection threads of Cowpea (Vigna unguiculata [L.] Walp) and Soybean (Glycine max [L.] Merr.) nodules. J Bacteriol. 2002;184:831–839. doi: 10.1128/JB.184.3.831-839.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chang W-S, Franck WL, Cytryn E, Jeong S, Joshi T, Emerich DW, Sadowsky MJ, Xu D, Stacey G. An oligonucleotide microarray resource for transcriptional profiling of Bradyrhizobium japonicum . Mol Plant Microbe Interact. 2007;20:1298–1307. doi: 10.1094/MPMI-20-10-1298. [DOI] [PubMed] [Google Scholar]
- 125.Sarma AD, Emerich DW. Global protein expression pattern of Bradyrhizobium japonicum bacteroids: a prelude to functional proteomics. Proteomics. 2005;5:4170–4184. doi: 10.1002/pmic.200401296. [DOI] [PubMed] [Google Scholar]
- 126.Zehner S, Schober G, Wenzel M, Lang K, Göttfert M. Expression of the Bradyrhizobium japonicum type III secretion system in legume nodules and analysis of the associated tts box promoter. Mol Plant Microbe Interact. 2008;21:1087–1093. doi: 10.1094/MPMI-21-8-1087. [DOI] [PubMed] [Google Scholar]
- 127.Ibarra JA, Steele-Mortimer O. Salmonella—the ultimate insider. Salmonella virulence factors that modulate intracellular survival. Cell Microbiol. 2009;11:1579–1586. doi: 10.1111/j.1462-5822.2009.01368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Haraga A, Ohlson MB, Miller SI. Salmonellae interplay with host cells. Nat Rev Microbiol. 2008;6:53–66. doi: 10.1038/nrmicro1788. [DOI] [PubMed] [Google Scholar]
- 129.Wenzel M, Friedrich L, Göttfert M, Zehner S. The type III-secreted protein NopE1 affects symbiosis and exhibits a calcium-dependent autocleavage activity. Mol Plant Microbe Interact. 2010;23:124–129. doi: 10.1094/MPMI-23-1-0124. [DOI] [PubMed] [Google Scholar]
- 130.Sory MP, Cornelis GR. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol Microbiol. 1994;14:583–594. doi: 10.1111/j.1365-2958.1994.tb02191.x. [DOI] [PubMed] [Google Scholar]
- 131.Casper-Lindley C, Dahlbeck D, Clark ET, Staskawicz BJ. Direct biochemical evidence for type III secretion-dependent translocation of the AvrBs2 effector protein into plant cells. Proc Natl Acad Sci USA. 2002;99:8336–8341. doi: 10.1073/pnas.122220299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Schechter LM, Guenther J, Olcay EA, Jang S, Krishnan HB. Translocation of NopP by Sinorhizobium fredii USDA257 into Vigna unguiculata root nodules. Appl Environ Microbiol. 2010;76:3758–3761. doi: 10.1128/AEM.03122-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.López-Baena FJ, Monreal JA, Pérez-Montaño F, Guasch-Vidal B, Bellogín RA, Vinardell JM, Ollero FJ. The absence of Nops secretion in Sinorhizobium fredii HH103 increases GmPR1 expression in Williams Soybean. Mol Plant Microbe Interact. 2009;22:1445–1454. doi: 10.1094/MPMI-22-11-1445. [DOI] [PubMed] [Google Scholar]
- 134.Brutinel ED, Yahr TL. Control of gene expression by type III secretory activity. Curr Opin Microbiol. 2008;11:128–133. doi: 10.1016/j.mib.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Deane J, Abrusci P, Johnson S, Lea S. Timing is everything: the regulation of type III secretion. Cell Mol Life Sci. 2010;67:1065–1075. doi: 10.1007/s00018-009-0230-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Magori S, Kawaguchi M. Long-distance control of nodulation: Molecules and models. Mol Cells. 2009;27:129–134. doi: 10.1007/s10059-009-0016-0. [DOI] [PubMed] [Google Scholar]
- 137.Oono R, Denison FR, Kiers TE. Controlling the reproductive fate of rhizobia: how universal are legume sanctions? New Phytol. 2009;183:967–979. doi: 10.1111/j.1469-8137.2009.02941.x. [DOI] [PubMed] [Google Scholar]
- 138.Sachs JL, Russell JE, Lii YE, Black KC, Lopez G, Patil AS. Host control over infection and proliferation of a cheater symbiont. J Evol Biol. 2010;23:1919–1927. doi: 10.1111/j.1420-9101.2010.02056.x. [DOI] [PubMed] [Google Scholar]
- 139.Marco DE, Pérez-Arnedo R, Hidalgo-Perea Á, Olivares J, Ruiz-Sainz JE, Sanjuán J. A mechanistic molecular test of the plant-sanction hypothesis in legume–rhizobia mutualism. Acta Oecol. 2009;35:664–667. doi: 10.1016/j.jtbi.2009.03.033. [DOI] [PubMed] [Google Scholar]
- 140.Marco DE, Carbajal JP, Cannas S, Pérez-Arnedo R, Hidalgo-Perea Á, Olivares J, Ruiz-Sainz JE, Sanjuán J. An experimental and modelling exploration of the host-sanction hypothesis in legume–rhizobia mutualism. J Theor Biol. 2009;259:423–433. doi: 10.1016/j.jtbi.2009.03.033. [DOI] [PubMed] [Google Scholar]
- 141.Yang S, Tang F, Gao M, Krishnan HB, Zhu H. R gene-controlled host specificity in the legume–rhizobia symbiosis. Proc Natl Acad Sci USA. 2010;107:18735–18740. doi: 10.1073/pnas.1011957107. [DOI] [PMC free article] [PubMed] [Google Scholar]