Abstract
Myocardial stem cell therapies are emerging as novel therapeutic paradigms for myocardial repair, but are hampered by the lack of sources for autologous human cardiomyocytes. An exciting development in the field of cardiovascular regenerative medicine is the ability to reprogram adult somatic cells into pluripotent stem cell lines (induced pluripotent stem cells, iPSCs) and to coax their differentiation into functional cardiomyocytes. This technology holds great promise for the emerging disciplines of personalized and regenerative medicine, because of the ability to derive patient-specific iPSCs that could potentially elude the immune system. The current review describes the latest techniques of generating iPSCs as well as the methods used to direct their differentiation towards the cardiac lineage. We then detail the unique potential as well as the possible hurdles on the road to clinical utilizing of the iPSCs derived cardiomyocytes in the emerging field of cardiovascular regenerative medicine.
Keywords: Induced pluripotent stem cells, Cardiomyocytes, Disease modeling, Drug screening, Cell therapy, Myocardial repair, Heart diseases
Introduction
Heart failure is responsible for significant worldwide morbidity and mortality and represents the leading cause of hospitalizations in the United States [1]. One of the major causes for heart failure is myocardial infarction resulting from acute occlusion of a major coronary artery. The resulting massive cardiac cell loss, scar formation, and ensuing negative structural and functional cardiac remodeling may lead to progressive deterioration in myocardial contractility and eventually to the development of clinical heart failure.
Although it was commonly believed that the adult mammalian heart is devoid of endogenous regenerative capacity and is exclusively a terminally differentiated organ, recent studies have challenged this paradigm by providing evidences for cardiomyocyte turnover in the adult human heart [2, 3]. Two major mechanisms could account for this endogenous regeneration [4]. Cardiomyocytes may re-enter the cell cycle and divide [5]. This process was shown to be robust in the injured mouse heart during the neonatal period [5], but is considered to be limited in the adult heart. The second mechanism may involve proliferation and differentiation of resident populations of cardiac progenitor cells (CPCs). Such cells were recently identified within specific niches in the mouse and human heart [6].
Despite the evidence supporting the presence of endogenous cardiac regeneration, this process is still believed to be insufficient to deal with the massive cardiomyocyte loss that occurs during a large myocardial infarction. With the prevalence of heart failure continuing to grow as the population ages, and with paucity of donors limiting the number of organ transplantation procedures for end-stage heart disease, the search for new therapeutic paradigms for this clinical epidemic has became imperative.
Recent advancements in stem cell biology, cell therapy, and tissue-engineering have paved the way to the introduction of a new discipline in biomedicine; regenerative/reparative medicine. In the heart, strategies for cardiac repair could theoretically target different pathological processes associated with myocardial infarction. These include strategies aiming to improve ischemic tissue salvage (cardio-protective or pro-angiogenic approaches), to modulate inflammation, to attenuate adverse extracellular matrix remodeling and fibrosis, or to induce cardiomyogenesis. The latter could be targeted either by augmenting the endogenous cardiac regeneration processes described above or alternatively by re-populating the scar through the transplantation of exogenous sources of contractile cells [7–10].
The use of stem cells for the generation of large quantities of human cardiomyocytes is a promising approach for such applications. Several stem and progenitor cell types have been proposed initially to differentiate into cardiomyocytes, either in culture or following transplantation, including mesenchymal stem cells (MSCs), skeletal myoblasts, and hematopoietic stem cells [7, 8, 10]. Most of these cells, however, were subsequently shown to have limited or no cardiomyogenic potential despite being able to improve ventricular function in animal models of myocardial infarction. This indicates that non-contractile mechanisms such as alteration of the infarct remodeling process, enhancement of angiogenesis, or augmentation of an endogenous repair mechanism probably underlies most of the benefit seen to date with cell therapy. It is therefore likely that further functional benefit could be achieved with a cell population that is characterized by typical cardiomyocyte properties and can contribute directly to contractility. At present, the most promising candidate cell types, which can potentially give rise to human cardiomyocytes, are adult CPCs [6] and human pluripotent stem cells (hPSCs) such as human embryonic stem cells (hESCs) [11] or human-induced pluripotent stem cells (hiPSCs) [12, 13].
A number of studies in the last decade highlighted the potential of CPCs for myocardial repair. These studies described the ability to isolate different types of resident CPCs from the heart based on the expression of different surface markers (such as c-Kit [14], Sca-1, or Isl-1) or culturing conditions (cardiospheres containing a mixture of cells [15]). The isolated CPCs were shown to be clonogenic and multipotent in vitro and were able to improve myocardial performance following delivery into animal models of cardiac injury. While a detailed description of the biology of CPCs and the approaches used for their isolation, expansion, and eventual myocardial delivery in animals and, also recently in clinical trials, are beyond the scope of this review (which focuses on hiPSCs), the interested reader is referred to a number of reviews on this topic [6, 16, 17].
The first stem cell source that could reliably give rise to cardiomyocytes in vitro was embryonic stem cells. Murine ESCs were first isolated in 1981 [18, 19] and their human counterparts were derived by Thomson et al. [11] in 1998. In both cases, the cellular origin of the ESCs was the inner cell mass of mammalian blastocysts. One of the most important properties of hESCs is their ability to be propagated in the undifferentiated state in vitro and to be coaxed to differentiate into a variety of cell lineages, including into cell types relevant to the cardiovascular system (endothelial cells [20], vascular smooth muscle cells or pericytes [21], and different types of cardiomyocytes [22–28]).
As a result of this cardiomyocyte differentiation capacity, hESCs were shown to serve as a unique tool for modeling human heart development, for drug testing, and as an attractive cell source for cardiac repair [29–33]. Nevertheless, the inability to create patient/disease-specific ESCs from adult individuals, the ethical issues arising from destructive use of human embryos, and the anticipated immune rejection associated with such allogeneic cell transplantation, imposes important hurdles for their clinical utilization.
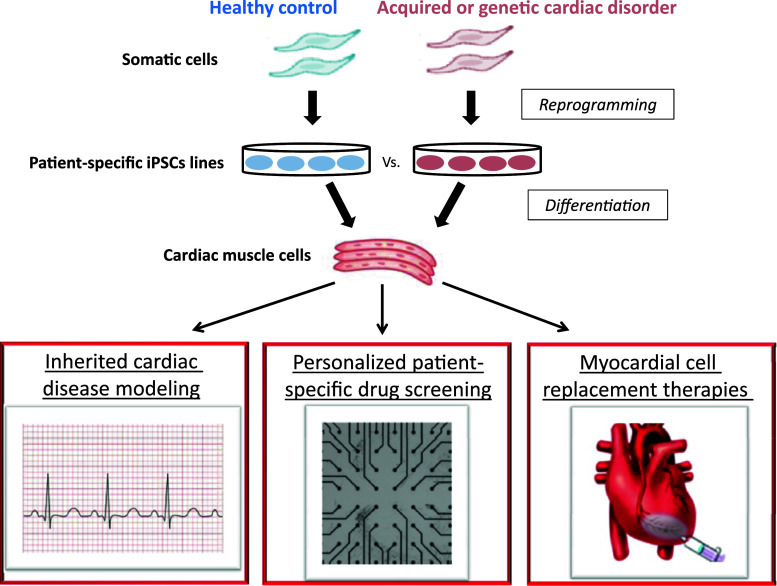
The aforementioned obstacles could be potentially overcome with the introduction of the groundbreaking iPSCs technology [34]. The ability to reprogram adult somatic cells into pluripotency by a set of transcription factors [12, 13, 34] and to differentiate them into cardiomyocytes [35–37] introduces a powerful new research tool, which can potentially be used to develop cell replacement strategies that can evade the immune system, to generate patient/disease-specific models of inherited cardiac disorders [38–40], and to establish screens for drug discovery (Fig. 1). In the current review, we will describe the various techniques used to derive iPSCs and the current progress made in coaxing their differentiation into the cardiac lineage. Particular emphasis will be placed on describing the potential and hurdles associated with the use of human iPSCs-derived cardiomyocytes (hiPSCs-CMs) in the emerging field of cardiovascular regenerative medicine.
Fig. 1.
Potential applications of hiPSCs-derived cardiomyocytes. Somatic cells (fibroblasts) from patients with inherited or acquired cardiac disorders can be reprogrammed to generate hiPSCs by expression of pluripotency-associated genes. These cells can be coaxed to differentiate into cardiac cells that are genetically matched to the patients from whom they were derived. The hiPSCs-CMs can then be used to elucidate basic disease mechanisms, to tailor patient-specific therapies (personalized medicine), for drug discovery and toxicity screening, and for the emerging field of cardiovascular regenerative medicine
Induced pluripotent stem cells
Differentiation, once believed to be a one-way process, has been recently shown to be a dynamic process that can be reversed by transduction of stemness transcription factors into somatic cells [34]. In a landmark study in 2006 [34], Takahashi and Yamanaka transduced mouse fibroblasts with a panel of 24 retroviral vectors, each encoding for a candidate reprogramming gene, and eventually identified Oct4, Sox2, Klf4, and c-Myc as a minimal set of factors capable of yielding cells with characteristics similar to those of mouse ESCs. The reprogrammed mouse pluripotent stem cell lines generated were termed iPSCs. Application of this approach to human somatic cells followed shortly [12], including by using an alternative set of reprogramming factors, in which Klf4 and c-Myc were replaced with Nanog and Lin28 [13]. This resulted in the derivation of hiPSCs, which greatly resembled the previously described hESCs.
While the exact mechanisms underlying the “rewiring” of the epigenetic state of a somatic cell to pluripotency by the ectopic expression of the aforementioned four reprogramming factors are still poorly defined, it seems that it is a stochastic process, which is gradual, slow, and relatively inefficient, and which involves thousands of epigenetic changes [41, 42]. During this process, the reprogrammed cells initially suppress their somatic identity and enter an intermediate partially reprogrammed state. This is followed, in a minority of the cells, by the re-expression of the pluripotent genetic network. Importantly, the reprogramming factors were shown to be required only for the induction of pluripotency, while the stem cell characteristics could be maintained independently of the continuous expression of the ectopic transgenes.
Although fibroblasts were the first target of reprogramming, several other cell types such as keratinocytes [43], neural stem cells, hepatocytes, and gastric epithelial cells [44], adipocytes, B lymphocytes [45], and peripheral blood cells were all successfully reprogrammed. Upon faithful reprogramming, mouse and human iPSCs were shown to have similar (although not identical) properties as to their respective ESC counterparts in term of morphology, gene expression, and genome-wide distribution of epigenetic markers [41, 46]. Furthermore, the reprogrammed cells also share a range of functional pluripotency assays such as differentiation into the three germ layers in vitro and teratoma formation and for mouse iPSCs—contribution to chimera and tetraploid (4N) complementation [41, 46].
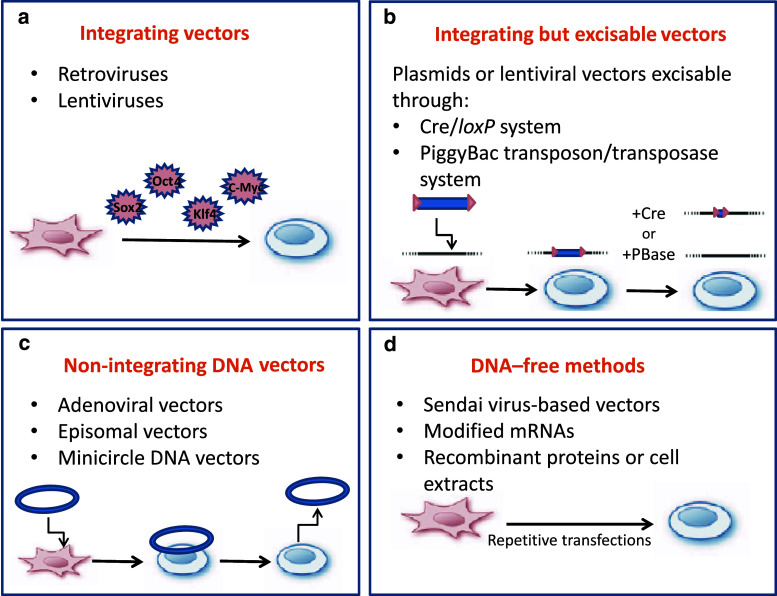
Gene delivery to somatic cells through retroviral or lentiviral vectors provided the initial strategy for ectopic expression of the reprogramming factors, and this turned out to be a particularly useful technique [12, 13, 34]. Retroviruses are highly efficient gene transfer vehicles because they provide prolonged expression of the transgenes after genomic integration and have low immunogenicity. Lentiviruses are a subclass of retroviruses capable of transducing a wide range of both dividing and non-dividing cells. The lentiviral vectors have a less reliable degree of silencing in iPSCs but can be combined with an inducible doxycycline-dependent system [13, 44]. These viral transduction approaches, although highly robust and reproducible, have the problem of multiple insertions into the host genome, which can lead to genetic modifications as well as possible reactivation of the viral vector. This hazard may significantly hamper the potential clinical use of such hiPSCs derivatives.
To overcome this potential limitation, several groups have focused on the development of suitable techniques for generation of integration-free iPSCs. These efforts included, for example, the establishment of integration-dependent vectors flanked with loxP sites that can be subsequently excised upon Cre-recombinase expression in the cells [47]. This system removes the transgenes, but leaves behind small vector element. Complete removal of the transgenes can be achieved with piggyBac transposons, mobile genetic elements that can be introduced into and removed from the host genome by transient expression of transposase [48, 49]. However, the efficiency of both approaches is very low and they also require intensive screening; properties which would make them very challenging to apply in a rapid or high-throughput manner.
The use of non-integrating DNA vectors may represent a more suitable strategy for establishing iPSCs for future cell-based therapies (Fig. 2) [46]. DNA-independent reprogramming is also possible through cell-penetrating reprogramming proteins [50, 51], or by the introduction of modified mRNA molecules encoding the reprogramming factors [52]. Although the aforementioned non-integrating approaches have a lower efficiency when compared to retroviruses/lentiviruses, addition of compounds such as vitamin C, the histone deacetylase inhibitor valproic acid (VPA) [53], or chemical inhibitors of transforming growth factor-β (TGFβ) receptors [54] may increase reprogramming efficiency. Figure 2 summarizes the different methods used for generating iPSCs in this rapidly developing field.
Fig. 2.
Overview of the main strategies to generate iPSCs. a Delivery of the reprogramming factors by retroviruses or lentiviruses. The transgenes are integrated into the cell’s genome. b Excisable methods for generating iPSC through the Cre/loxP or the piggBac transposon/transposase systems. c Methods for generating iPSCs with non-integrating DNA vectors including adenoviral vectors, conventional plasmids, minicircles, and oriP/EBNA1 episomal vectors. d DNA-free methods for generating iPSCs are based on RNA-mediated delivery by Sendai virus-based vectors or by repeated transfection with modified mRNA. An alternative approach focuses on protein delivery using recombinant proteins or cell extracts
Cardiomyocytes differentiation of iPSCs
One of the most important properties of iPSCs is their potential to differentiate in vitro into somatic cells of all tissue types. Cardiomyocyte differentiation of murine iPSCs (miPSCs) was first described by Mauritz et al. [55] who utilized the embryoid body (EB) differentiation system to differentiate miPSCs into cardiomyocytes. In a second study, Narazaki et al. [56] obtained cardiomyocytes by co-culturing induced mesodermal miPSC derivatives (Flk1+ cells) with OP9 mouse stromal cells. In both studies, the resulting miPSC-derived cardiomyocytes were shown to display cardiac-specific molecular, structural, and functional properties.
In the aforementioned studies, cardiomyocytes were differentiated from miPSC lines established using the traditional four-factor reprogramming approach (Oct4, Sox2, Klf4, and c-Myc). In principle, reprogramming without c-Myc may be favorable, given the disregulated gene expression networks and tumorigenic load associated with this oncogenic protein. To this end, recent studies succeeded in establishing cardiomyocyte-differentiating systems from c-Myc-free miPSCs lines [57, 58]. Interestingly, increased cardiomyocyte differentiation capacity was identified in these lines; perhaps due to the potential involvement of the c-Myc pathway in cardiac lineage differentiation and hyperplasia [59].
Cardiomyocyte differentiation of hiPSCs soon followed [35–37]. These studies exploited hiPSCs lines, which were created from different somatic cell sources and using different reprogramming techniques. Zhang et al. [36] utilized hiPSCs generated by lentiviral-mediated transduction of fibroblasts with Oct4, Sox2, Nanog and Lin28; Zwi et al. [37] used a hiPSCs line established by retroviral reprogramming of fibroblasts with Oct4, Sox2, c-Myc and Klf4; and Haase et al. [35] utilized hiPSCs derived from human cord blood. All groups used the spontaneous, EB-based, cardiomyocyte differentiation system. The generated myocytes were demonstrated to display molecular, structural, and functional properties of early-stage human cardiomyocytes [35–37]. Detailed electrophysiological studies revealed the presence of different cardiomyocyte cell types (with ventricular-like, atrial-like, and nodal-like action-potential morphologies) [36] and the expression of a diversity of ionic channel currents [60]. The hiPSCs-derived cardiomyocytes (hiPSCs-CMs) were also demonstrated to respond appropriately to neurohormonal stimulation (with adequate chronotropic response to adrenergic and cholinergic agonists) [37] and to contain functional ryanodine-receptor regulated and caffeine-responsive intracellular calcium stores [61]. Finally, hiPSCs-CMs displayed the development of a functional syncytium with stable pacemaker activity and synchronized action-potential propagation [37].
The aforementioned studies utilized fibroblasts derived from either established cell lines or from healthy or young individuals. More recently, Zwi-Dantsis et al. [62] showed the ability to also establish hiPSCs from patients with advanced heart failure (representing the candidate patient population for future hiPSCs-based cell replacement strategies). Dermal fibroblasts from these patients were reprogrammed to generate hiPSCs by retroviral delivery of Oct4, Sox2, and Klf4 or by using an excisable polycistronic lentiviral vector. Both the resulting transgene-containing and transgene-free hiPSCs could be differentiated into cardiomyocytes portraying similar molecular, structural, and functional properties to those of hiPSCs-CMs derived from healthy control foreskin fibroblasts.
Directed cardiomyocyte differentiation of iPSCs
Although the spontaneous EB-based differentiation system is the traditional method used for cardiomyocyte differentiation of hESCs and hiPSCs, this approach is limited by the relatively low cardiomyocyte yield, by the requirement for serum, and the relative heterogeneous and undefined nature. Achieving a reproducible, defined, animal product-free and efficient cardiomyocyte differentiation system remains one of the most important challenges in the field. Since the cardiomyocyte differentiation processes of both hESCs [63] and hiPSCs [37] seem to parallel those that happen in vivo, efforts have been made to translate lessons learned from developmental biology to enhance cardiomyocyte differentiation in vitro [64, 65]. This developmental biology approach allowed to recapitulate in hESCs and hiPSCs key events that regulate early cardiac-lineage commitment in the embryo, as well as to derive highly enriched cardiovascular progenitor cell-populations [64, 65] (Table 1).
Table 1.
Approaches for cardiomyocyte differentiation of hESC/hiPSC
| Differentiation method | Key steps | Serum-free conditions | Yield of cardiomyocytes | References | |
|---|---|---|---|---|---|
| Spontaneous differentiation | Suspension (EBs) |
hESCs/hiPSCs are detached from the feeder cells and transferred to suspension, where they form three-dimensional aggregates (EBs) After 7–10 days in suspension, EBs are plated on gelatin-coated culture dishes Spontaneous contractions starting 7–8 days after plating |
No | 1–10 % | [22, 23, 27, 36, 37] |
| Guided differentiation | Monolayer |
Undifferentiated hESCs/hiPSCs are treated with bFGF for 6 days Cells are exposed for activin A for 24 h, followed by treatment with BMP4 for another 4 days The cytokines are removed. Beating is observed 12 days after activin treatment |
Yes |
>30 % (>80 % when including Percoll gradient) |
[24] |
| Suspension (EBs) |
hPSCs are guided into the mesodermal lineage by treatment with activin A, BMB4 and bFGF Cardiac induction by treatment with the Wnt inhibitor—DKK1, VEFG and bFGF |
Yes | 40–50 % | [28, 68] | |
| Co-culture |
EBs are continuously cultured in insulin-free medium conditioned with END-2 cells, PGI2 and MAPK inhibitor Beating foci are observed after 9–12 days in culture |
Yes | 5–20 % | [25, 66, 67] | |
| Single cells |
Undifferentiated hESCs/hiPSCs are treated with BMP2 for 4 (hESCs) and 6 (hiPSCs) days, in the presence of SU5402, a FGF receptor inhibitor BMP2-induced SSEA-1+ progenitors are plated in a mix of human cardiac fibroblasts and cardiomyocytes to induce their differentiation into cardiomyocytes |
Yes | 60–80 % | [117] | |
| Small molecules | Suspension (EBs) |
EBs are cultured with 5-aza-deoxycytidine at days 6–8 after initiating of differentiation Percoll gradient separation may be used in order to further enrich the cardiomyocytes population |
No | Two- to threefold induction of cardiac related genes | [27] |
BMP4 bone morphogenic protein-4, BMP2 bone morphogenic protein-2, bFGF basic fibroblast growth factor, DKK1 dickkopf related protein 1, VEGF vascular endothelial growth factor, PGI 2 prostaglandin I2, MAPK MAP kinase
Initial efforts focused on the use of pro-cardiogenic factors released by the visceral endoderm during early development and members of the TGF-β superfamily. For example, co-culture of hESCs with END-2 cells (an endoderm-like cell line) resulted in enhanced cardiomyocyte differentiation [25]. The efficiency of cardiomyocyte differentiation using this approach was further increased (up to ~20 % cardiomyocytes) by serum depletion [66] and by adding a small molecule inhibitor of p38 MAP kinase [67]. Taking an additional step, Laflamme and co-workers [24] were able to establish a directed hESCs cardiomyocyte differentiation system, using a serum-free defined medium supplemented with activin and BMP4, that resulted in cardiomyocyte enrichment to more than 30 %. The third approach, routinely used in the field, was developed by the Keller’s group and involves the initial induction of a primitive streak-like cell population and early cardiac mesoderm by Activin, BMP4, and bFGF followed by canonical Wnt inhibition (using DKK1). This resulted in significant enhancement of cardiomyogenesis (40–50 % cardiomyocytes) [28].
The latter strategy was recently transferred to the mouse iPSCs system, resulting in a differentiated cell population highly enriched with cardiomyocytes [68]. Interestingly, cardiomyocyte yield was less efficient in the hiPSCs system and also required additional pathway inhibitors [68]. In addition, the protocols used for iPSCs cardiomyocyte differentiation had to be optimized individually for each line (perhaps due to variability in the endogenous expression of these factors by the different lines). The authors suggested that monitoring the expression of Flk1/KDR+ or PdgfR-α+ progenitors early during differentiation may be used for the optimization of such protocols [68].
Direct conversion of fibroblasts into cardiomyocytes
Recently, two innovative studies attempted to circumvent the need to reprogram somatic cells into iPSCs before re-directing their differentiation into the cardiac lineage. In the first study, the authors aimed to directly convert fibroblasts into cardiomyocytes by forced expression of cardiac-specific transcription factors. Consequentially, transdifferentiation of postnatal murine fibroblasts into cardiomyocyte-like cells was made possible by ectopic expression of only three cardiac transcription factors: Gata4, Mef2c, and Tbx5 [69].
In the second study, the authors hypothesized that, during the first days following initiation of reprogramming by Yamanka’s factors, the cells enter an initial epigenetic ‘activation phase’. They further reasoned that manipulating the environmental cues to those favoring cardiogenic differentiation may allow the hijacking of conventional reprogramming at this early unstable stage, and specifically shift the outcome towards cardiogenesis [70]. Using this strategy, the authors showed that mouse embryonic fibroblasts could be converted to cardiomyocytes over a brief period of 11–12 days by initial over-expression of Oct4, Sox2, Klf4, and c-Myc followed by short incubation with the reprogramming culture medium, and eventually by changing to a chemically-defined media that includes the cardio-inductive growth factor BMP4.
In both strategies, cardiogenic conversion occurred during a relatively short period, and the possibility that the induced cardiomyocytes originated from an intervening mesodermal or progenitor stage was excluded. In comparison to the iPSCs reprogramming strategy, cardiomyocyte lineage reprogramming seems to be more rapid and efficient and also avoids the presence of residual pluripotent stem cells, thereby lowering the risk of tumor formation.
The most exciting potential, however, lies in the possibility for in vivo use of this technology, where fibroblasts in the infarcted area, for example, may be the target for such cardiogenic reprogramming. Two very recent studies provided proof-of-concept evidence for the validity of this approach. Qian et al. [71] used retroviral delivery of the three cardiogenic transcription factors (Gata4, Mef2c, and Tbx5) to the murine infarct model, while Song et al. [72]. added a fourth factor—hand2. One month after treatment, the reprogrammed cardiomyocyte-like cells comprised up to 6 % of the cardiomyocytes in the infarct border zone in the Song study and as high as 35 % in the Qian report. In both cases, significant improvement in cardiac function was observed as compared to controls. Finally, lineage-tracing studies suggested that the source of these new cardiomyocytes is indeed resident cardiac fibroblasts.
Although highly attractive, these strategies also possess certain limitations. First, the same concepts have not yet been reproduced for human somatic cells. Second, since the induced cardiomyocytes, unlike undifferentiated iPSCs or ESCs, cannot be propagated, this process is less amenable to scaling-up, and the derivation of each batch of new cardiomyocytes would require repeating the entire process of cell transduction and reprogramming. In addition, other safety issues still need to be addressed, including the risk of genomic integration and spontaneous reactivation of the reprogramming factors. Finally, it is also not clear how the induced cardiomyocytes compare with those derived from pluripotent stem cell lines in terms of their cardiomyocyte phenotypic properties and their capacity for cardiac repair.
Myocardial repair using pluripotent stem cells
Myocardial cell therapy with hESCs
The ultimate goal of cardiovascular regenerative medicine is to generate a functional cardiac tissue that will become well integrated with host myocardium and will restore cardiac electro-mechanical properties. The advent of the hESCs cardiomyocyte differentiation system provided a potential sources for human cardiomyocytes for this emerging discipline. Initial studies demonstrated that transplantation of hESCs-CMs into the uninjured hearts of immunocompromised mice, rats, and pigs results in the formation of stable cell grafts [73–77]. The cell grafts expressed human cardiac markers, showed some degree of maturation and vascularization, and continued to proliferate to a certain extent.
More recent studies focused on the impact of hESCs-CMs transplantation in rodent models of myocardial infarction [24, 73, 74, 78–81]. While different strategies were used to differentiate and isolate the hESCs-CMs, to enhance their survival following transplantation, to deliver them into the myocardium, and also in the types of animal models used, all these studies demonstrated the ability of the hESCs-CMs to survive and to form stable cell grafts within the infarcted area. Cell engraftment, in the majority of studies, leads to attenuation of the ventricular remodeling process, delayed heart failure progression, and improved ventricular function when compared to non-myocyte transplantation or vehicle injection alone.
Despite these encouraging results, the underlying mechanisms explaining the observed functional improvement and issues pertaining to its long-term persistence [80] remain a matter of debate. An important open question relates to the ability of the engrafted cardiomyocytes to integrate structurally and functionally with host cardiomyocytes. This issue has significant implications with regards to the mechanism of stem cell therapy (since direct contribution to contractility by the transplanted cells would require electrical coupling with host cardiac cells) as well as to the potential of this approach to be anti-arrhythmic or pro-arrhythmic.
To address these important issues, studies were performed to evaluate the ability of different candidate cell types to integrate with host cardiac tissue. These studies revealed that hESCs-CMs [76] and hiPSCs-CMs [62] can functionally integrate in vitro with primary cultures of neonatal rat ventricular myocytes. Similarly, studies using direct intracellular recordings [82], high-resolution two-photon microscopy calcium imaging [83], and optical mapping using voltage-sensitive dyes [84] all showed that donor cardiomyocytes (derived from ESCs or primary mouse embryonic cardiomyocytes) can also electrically integrate with host cardiac tissue following in vivo engraftment to rodent hearts. In vivo integration was also suggested by the ability of hESCs-CMs to function as “biological pacemakers” and to drive the electrical activity of the entire heart in animal models of slow heart rate, thus also showing the potential of these cells for conduction system repair [76, 85].
Cell-replacement therapy using iPSCs
A major obstacle for the utilization of hESCs derivatives for regenerative medicine is the expected immune rejection associated with such allogenic tissue and the social and religious constraints concerning the use of human embryos. The iPSCs technology may provide a possible solution to these hurdles given the potential to generate isogenic pluripotent cells (that are genetically equivalent to the transplant recipient), which could be propagated, coaxed to differentiate into the desired cell lineage (such as cardiomyocytes), and could potentially evade the immune system following transplantation due to their autologous nature. However, whether iPSCs-derived cardiomyocytes can eventually be used in the desired autologous manner or alternatively as an off-the-shelf allogenic cell product is still unclear. While autologous patient-specific hiPSCs-derived cardiac tissues are assumed to possess immune privilege properties, a recent study challenged this assumption by describing the potential immunogenicity of teratomas generated from undifferentiated mouse iPSCs transplanted in syngeneic animals [86]. Future studies will have to further investigate this important issue in greater detail.
The potential of iPSCs-based cell replacement therapy for the treatment of a variety of non-cardiac disorders was recently demonstrated in a growing number of animal models of disease [87]. For example, Hanna et al. [88] reprogrammed fibroblasts, which were obtained from a humanized transgenic mouse model of sickle cell anemia, into mouse iPSCs. The human sickle hemoglobin allele was then corrected in the established miPSCs by homologous recombination. The resulting ‘healthy’ miPSCs were then induced to differentiate into hematopoietic progenitors, and their ability to rescue the mouse phenotype was demonstrated following transplantation back into the diseased animals.
Given these proof-of-principle studies, and the results of the previous hESCs-CMs transplantation studies in animal infarct models, it seems that immunologically-compatible patient-derived hiPSCs may also bring a unique value for cardiac repair/regeneration. Initial evidence for the potential use of iPSCs in the context of heart disease treatment was reported by Nelson et al. [89]. The authors in this study compared post-ischemic cardiac performance between mice transplanted with parental fibroblasts and mice treated with mouse iPSCs and demonstrated superior functional outcome in the latter group. More recently, the ability of hiPSCs-CMs to engraft, survive and integrate within the healthy rat heart [62], and to alter cardiac remodeling in the infarcted rat heart model, was demonstrated [90].
Prospects and challenges in using iPSCs for myocardial repair
Despite the important progress made in more than a decade since the establishment of the first hESCs lines [11], and in the more than 5 years since the initial description of the iPSCs technology [34], many conceptual, technical, and regulatory obstacles still remain on the road to clinical utilization of hPSCs for myocardial repair [31–33, 91]. These issues are summarized in Table 2 and include: (1) the need to develop efficient protocols for directed cardiomyocyte differentiation; (2) the need to establish strategies for cardiomyocyte selection; (3) the need to develop scaling-up procedures to derive clinically-relevant number of cardiomyocytes using a reproducible, well-defined and animal product-free methodology; (4) the need to address several important regulatory issues such as stem cell line characterization, good manufacturing practice (GMP), and crucial safety issues; and (5) the need to develop cell delivery and cell engineering strategies to enable proper alignment of the grafted tissue and to optimize long-term survival and maturation of the cell grafts. While several of the aforementioned issues that are relevant for both hESCs and hiPSCs have already been detailed above or described elsewhere [31–33, 91], we will further review some of these issues and also outline some of the challenges that are unique to hiPSCs.
Table 2.
Description of the major challenges and potential solutions facing the use of iPSC technology for myocardial repair
| Hurdles | Possible solutions |
|---|---|
| Heterogeneous population of differentiating cells |
Directed cardiac differentiation Specific selection of cardiac cells based on cell surface markers, physical properties, or genetic selection based on cardiac-specific promoters |
| Low cardiomyocyte yield |
Directed cardiac differentiation Upscaling the differentiation process to derive clinically relevant number of cardiomyocytes |
| Survival, integration and maturation of cell-grafts in vivo |
Cell delivery approaches (such as delivery with pro-survival factors, with vascular progenitors, etc.) Tissue engineering strategies Genetic modification to generate cardiomyocytes with enhanced engraftment and survival properties |
| Tumorigenic potential |
Development of suitable techniques for generation of integration-free iPSCs Use of c-Myc-free reprogramming strategies Selection processes which ensure the absence of remaining pluripotent stem cells in the graft |
| Risk of genetic modifications | Genetic screening of the undifferentiated iPSC colonies |
Tumorigenic potential
One of the major concerns associated with hiPSCs-based cell-therapies is the oncogenic risk of such a procedure. This stems from the potential of remaining undifferentiated cells within the cell grafts to form teratomas, from the use of oncogenic reprogramming factors (c-Myc), from the random integration of the viral vectors used (‘insertional-oncogenesis’), and from genetic instability associated with hiPSCs potentially leading to both chromosomal aberrations and mutations [92]. While it is still unclear whether the latter genomic alterations may lead to distorted cellular or oncogenic phenotypes, extensive genomic monitoring should become a standard procedure to ensure hiPSCs phenotypic stability and safety before any clinical use can be expected. Additional measures may include the use of safer reprogramming techniques such as eliminating the use of c-Myc and integrating viral vectors and as discussed above.
Transplantation of undifferentiated mouse and human ESCs (and also in our experience of undifferentiated iPSCMs) into the immunocompromised rodent heart, usually results in the formation of teratomas [73, 93]. This process, which may be cell dose-dependent [94], highlights the need to assure the lack of remaining undifferentiated pluripotent cells within the cell grafts. This task can be achieved either by using a negative selection approach to eliminate cells expressing undifferentiated markers that are responsible for teratoma formation [95] or by positive selection of differentiating cardiomyocyte precursors [75, 96] as discussed below.
Cardiomyocyte selection
Ideally, the directed cardiomyocyte differentiation systems being developed will result in ~100 % cardiomyocyte differentiation efficiency. If this is not achievable, however, then strategies aiming to select only the differentiating cardiomyocytes from the mixed population of differentiating cells may be required. To achieve this goal, a number of approaches have been proposed. The first approach uses manual dissection of the beating areas with the EBs and results in 50–80 % cardiomyocytes [73]. The second approach proposes to isolate cardiomyocytes based on their physical properties; for example, by the use of percoll gradient centrifugation, since these cells have a higher density than most other differentiating cells [24]. A third approach takes advantage of the fact that cardiomyocytes are enriched with mitochondria to select these cells with mitochondria-specific fluorescent dyes [97].
Although the aforementioned approaches can be used to enrich the cardiomyocyte population, the relatively low degree of purity and the inadequate ability for scaling-up may limit their clinical application. An alternative approach involves a transgenic selection strategy utilizing cardiac-specific promoters to drive the expression of selection markers (such as a fluorescent marker for flow cytometry sorting or an antibiotic-resistant gene for antibiotic selection). This method, pioneered by the Field’s group in the murine ESCs system [98], was also recently adapted for hESCs [75, 96, 99] and mouse iPSCs [100] and results in the derivation of highly-purified population of cardiomyocytes. A major drawback of this strategy, however, is the requirement for genetic manipulation with the potential associated risks.
The use of antibodies targeting cardiac-specific surface markers coupled with cell sorting may bypass some of the aforementioned limitations. This approach was hampered until recently by the lack of relevant cardiomyocyte-specific surface markers. The recent identification of such markers including EMILIN2 [101], SIRPA [102], and VCAM [103] has made it possible to isolate highly enriched populations of cardiomyocytes from both hESCs or hiPSCs by FACS or magnetic bead sorting.
Cardiomyocyte maturation
One of the remaining challenges associated with the use of hiPSCs-CMs for cardiac repair as well as for in vitro drug discovery and disease modeling applications is their relative heterogeneous nature and immature phenotype. The cardiomyocytes obtained during hPSCs differentiation represent a mixed population of cells with ‘atrial’, ‘ventricular’, and ‘nodal-like’ action potential properties [22, 36]. Since clinical cell therapy procedures would probably require engraftment of specific cell types (ventricular ‘working’ cardiomyocytes for infarct repair and nodal cells for ‘biological pacemaker’ applications); strategies have to be developed to derive homogenous populations of the desired cell types. This could be achieved either through development of modified differentiation protocols to derive specific cardiomyocyte subtypes (as achieved for pacemaking cells [104]) or by using appropriate selection strategies.
Similarly, although differentiating hiPSCs-CMs has been demonstrated to possess cardiac-like molecular, ultrastructural, electrophysiological (action potentials and diverse ion channels repertoire) [36, 37, 60], and excitation–contraction coupling [61] characteristics; these properties were shown to be relatively immature. From the electrophysiological angle, such early-stage properties includes the presence of spontaneous automaticity, a relatively depolarized resting membrane potential, and increased I f but reduced Ik1 currents.
Optimal regeneration of the infarct would require, in contrast, the generation of pure populations of ‘working’ ventricular myocytes with an adult-like phenotype. Potential strategies to induce hESCs-CMs or hiPSCs-CMs maturation may include prolonged in vitro culturing [105], training the cells with biomechanical forces [106], co-culturing with endothelial cells [106, 107], and potentially also by treatment with certain small molecules. One caveat to this approach is that early-stage cardiomyocytes have been demonstrated to survive significantly better in the in vivo heart than mature adult cells [108]. Hence, achieving significant in vitro maturation prior to cell transplantation may actually hinder cell engraftment and survival. Finally, evidence exists that embryonic mouse cardiomyocytes may actually undergo electrophysiological maturation following in vivo transplantation following the development of functional coupling with host cardiomyocytes [82].
Augmenting cell graft survival, integration, and regeneration
A major hurdle, identified in almost all cardiomyocyte transplantation studies in animal models, is the relatively poor short- and long-term survival of the engrafted cells with the infarcted region. This may stem from several factors including early cell loss due to cell washout, cell death due to the harsh ischemic environment of the infarcted area and the associated inflammatory process, lack of supporting extracellular matrix (anoikis), and lack of supporting signals from neighboring cells [109].
Some of the possible solutions to the challenge of poor cell graft survival may include the use of polymer cell carriers for initial entrapment of the cells, restoration of the missing extracellular matrix within the infarcted area (to prevent anoikis), use of free radical scavengers or anti-inflammatory therapy (to minimize cell death), co-transplantation with non-cardiomyocytes (such as fibroblasts), induction of angiogenesis, and engineering or training the cells to improve their ability to survive (for example, by pre-conditioning the cells with heat-shock or by overexpression of anti-apoptotic proteins) [109].
Using a combination of anti-apoptotic and anti-oncotic factors, Laflamme et al. [24] demonstrated the ability to significantly augment hESCs-CMs survival following transplantation, eventually repopulating up to 11 % of the infarct’s volume with human myocardial tissue. Another recently suggested approach to enhance cell graft survival involves the use of miRNAs. It has been demonstrated that treatment of CPCs with a miRNAs pro-survival cocktail (miR-21, miR-24, and miR-221) significantly improved CPCs engraftment and function in a murine infarct model [110].
An important approach to enhance cell survival following transplantation involves enhancement of vascularization at the cell graft area. Since formation of the heart requires the coordinated functions of cardiac myocytes, smooth muscle cells, endothelial cells, and connective tissue elements, a more complex task would be to attempt to regenerate all these elements that may be lost during myocardial injury. This can be achieved either by attempting to augment host tissue vascularization (for example, by secretion of angiogenic growth factors by the grafted cells) or by co-delivery of vascular precursor cells using novel multicellular cell therapy or tissue engineering approaches [107, 111, 112]. To achieve the latter, a number of investigators selected to co-transplant together with the hPSCs-derived cardiomyocytes and also endothelial cells (alone or together with embryonic fibroblasts or smooth muscle cells). These studies demonstrated the formation of vascular network from donor cells that integrated with the host vasculature and contributed to the perfusion and survival of the graft [111, 112].
Selection of multipotent cardiovascular progenitor cells
An alternative to the multicellular transplantation approach to regenerate different components of cardiac tissue may be the use of earlier cardiovascular progenitor cells. Recent evidence suggests that the different cell types within the heart (cardiomyocytes, endothelial cells, and smooth muscle cells) may arise from common multipotent progenitors [113]. These different cardiovascular progenitors can be recognized and selected during in vitro differentiation of ESCs by the use of different cell surface or genetic markers. These include identifying the expression of the receptor tyrosine kinases KDR (FLK1/VEGFR2) and PDGFRA [68], the expression of cKit and Nkx2.5 [114], or the expression of the second heart field marker Isl-1 [115]. The isolated progenitors could then give rise (either ex vivo or following in vivo engraftment) to cardiomyocytes, endothelial cells, and smooth muscle cells.
This strategy was also recently demonstrated in the context of iPSCs. Moretti et al. [116] showed the ability to isolate Isl1+ cardiovascular progenitors from both mouse and human iPSCs. The mouse Isl1+ progenitors were also shown to differentiate into endothelial, smooth, and cardiac muscle cells in vivo without the formation of teratoma. An alternative strategy for selection of early cardiovascular progenitors was described by Blin et al. [117]. They isolated an early cell population that express OCT4, SSEA1, and MESP1 during the differentiation of various pluripotent stem cells (hESCs, hiPSCs, and Rhesus ESCs). These multipotent progenitors could give rise to cardiomyocytes, smooth muscle cells and endothelial cells. Interestingly, when the Rhesus ESCs-derived SSEA1 + progenitor cells were transplanted into a myocardial infarct model in nonhuman primates, they differentiated into cardiomyocytes and successfully reconstituted the scar tissue.
Tissue engineering strategies
A potential solution to the challenge of increasing the survival and size of the cell grafts and of generating a highly organized three-dimensional anisotropic muscle structure may stem from the emerging field of tissue engineering. Tissue engineering is a multi-disciplinary field that combines functional cells with three-dimensional scaffolds (made from synthetic or biological polymers) to create tissue substitutes. The scaffold serves many purposes, including the delivery of biological signals to control and enhance tissue formation, to provide adequate biomechanical support for the cell graft, to control graft shape and size, to promote angiogenesis, and to protect the cells from physical damage. A wide range of cardiac tissue engineering efforts have been explored to date (with several approaches already using cardiomyocytes derived from pluripotent stem cell sources). Different approaches were used, ranging from the use of hydrogels as in vivo cell carriers [118], to cell-seeded fabricated polymer scaffolds, to decellularized whole organs which are later re-cellularized [119]. While a detailed description of these strategies is beyond the scope of this paper, the interested reader is referred to a number of comprehensive reviews on the subject [33, 120–123].
Summary
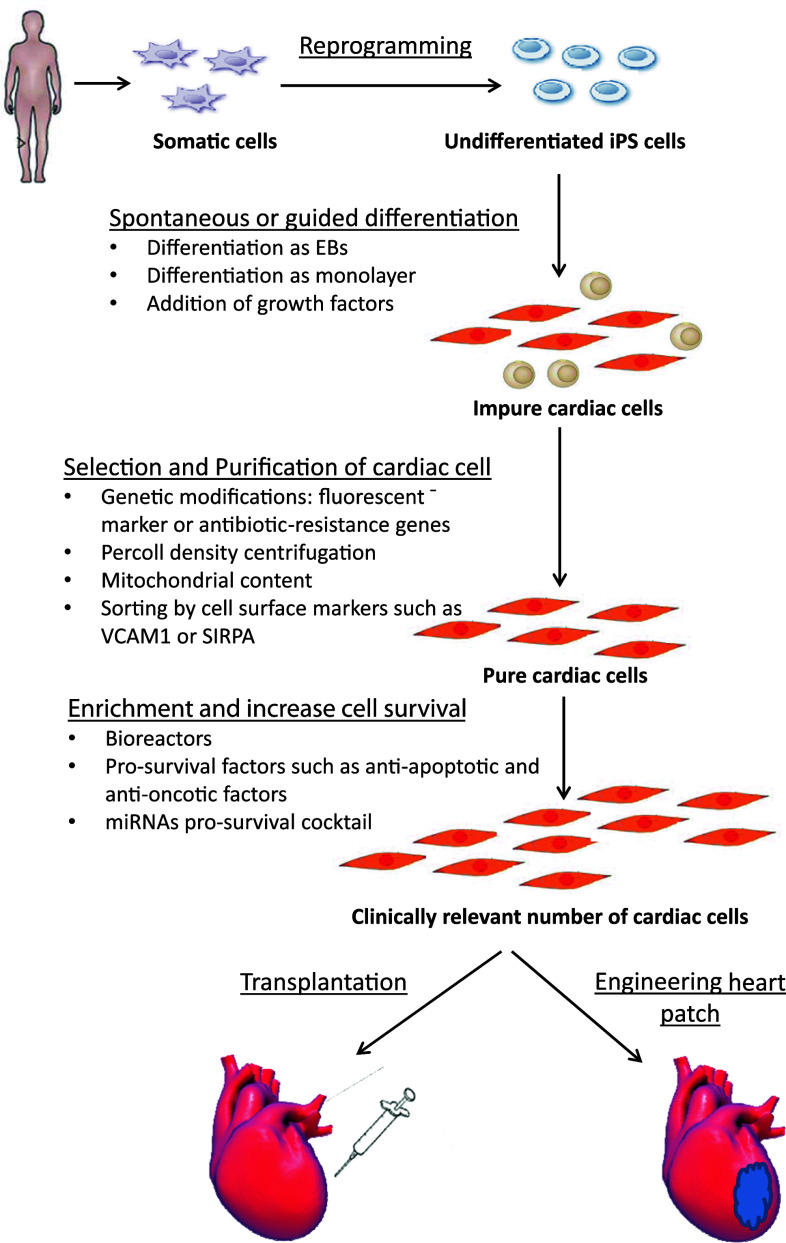
Since the first description of the iPSC technology more than 5 years ago, there has been remarkable progress in this field. When produced from patients with cardiovascular diseases, this technology provides an invaluable source to derive patient-specific cardiomyocytes that can be used to model hereditable cardiovascular diseases and to optimize patient-specific care (personalized medicine). Moreover, hiPSCs-CMs also offer a unique tool for drug development and toxicity testing. Importantly, the ability to generate patient-specific iPSCs and to coax their differentiation into clinically relevant cardiac cells may overcome some of the important barriers in the exciting field of cardiovascular regenerative medicine, tissue engineering, and myocardial repair. Nevertheless, as described above, several milestones have to be achieved (Fig. 3) in order to fully harness the enormous research and clinical potential of this unique technology.
Fig. 3.
Road map for myocardial repair. Somatic cells are obtained from a patient. The cultured cells are reprogrammed to generate iPSCs, and then coaxed to differentiate into patient/disease-specific cardiomyocytes. This procedure is subsequently complemented by various purification methods and selection strategies. The entire process should be scaled-up using bioreactor-related technologies. The generated cardiomyocytes can be engineered or treated with pro-survival factors in order to enhance their survival. Finally, the generated cardiac cells can be transplanted alone or with other cell types directly into the damaged heart (cell therapy) or combined with various scaffolding polymers (tissue engineering approaches) to achieve the desired myocardial repair
Acknowledgments
We apologize to authors whose excellent works were not cited due to space restrictions. This work was supported in part by the Israel Science Foundation [1449/10], by the Israel Science Foundation and Legacy Heritage Foundation (No. 1225/09), by the Lorry Lokey research fund, and by the Nancy and Stephen Grand Philanthropic Fund.
Abbreviations
- CPCs
Cardiac progenitor cells
- EBs
Embryoid bodies
- ESCs
Embryonic stem cells
- hESCs
Human embryonic stem cells
- hiPSCs
Human-induced pluripotent stem cells
- hiPSCs-CMs
Human-induced pluripotent stem cells-derived cardiomyocytes
- hPSCs
Human pluripotent stem cells
- iPSCs
Induced pluripotent stem cells
- miPSCs
Murine-induced pluripotent stem cells
- miRNAs
MicroRNAs
- MSCs
Mesenchymal stem cells
References
- 1.Cohn JN, Bristow MR, Chien KR, Colucci WS, Frazier OH, et al. Report of the National Heart, Lung, and Blood Institute Special Emphasis Panel on Heart Failure Research. Circulation. 1997;95:766–770. doi: 10.1161/01.CIR.95.4.766. [DOI] [PubMed] [Google Scholar]
- 2.Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kajstura J, Gurusamy N, Ogorek B, Goichberg P, Clavo-Rondon C, et al. Myocyte turnover in the aging human heart. Circ Res. 2010;107:1374–1386. doi: 10.1161/CIRCRESAHA.110.231498. [DOI] [PubMed] [Google Scholar]
- 4.Parmacek MS, Epstein JA. Cardiomyocyte renewal. N Engl J Med. 2009;361:86–88. doi: 10.1056/NEJMcibr0903347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leri A, Kajstura J, Anversa P. Role of cardiac stem cells in cardiac pathophysiology: a paradigm shift in human myocardial biology. Circ Res. 2011;109:941–961. doi: 10.1161/CIRCRESAHA.111.243154. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Dimmeler S, Zeiher AM, Schneider MD. Unchain my heart: the scientific foundations of cardiac repair. J Clin Invest. 2005;115:572–583. doi: 10.1172/JCI24283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laflamme MA, Murry CE. Regenerating the heart. Nat Biotechnol. 2005;23:845–856. doi: 10.1038/nbt1117. [DOI] [PubMed] [Google Scholar]
- 9.Passier R, van Laake LW, Mummery CL. Stem-cell-based therapy and lessons from the heart. Nature. 2008;453:322–329. doi: 10.1038/nature07040. [DOI] [PubMed] [Google Scholar]
- 10.Segers VF, Lee RT. Stem-cell therapy for cardiac disease. Nature. 2008;451:937–942. doi: 10.1038/nature06800. [DOI] [PubMed] [Google Scholar]
- 11.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 13.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 14.Beltrami AP, Barlucchi L, Torella D, Baker M, Limana F, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–776. doi: 10.1016/S0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 15.Messina E, De Angelis L, Frati G, Morrone S, Chimenti S, et al. Isolation and expansion of adult cardiac stem cells from human and murine heart. Circ Res. 2004;95:911–921. doi: 10.1161/01.RES.0000147315.71699.51. [DOI] [PubMed] [Google Scholar]
- 16.Barile L, Messina E, Giacomello A, Marban E. Endogenous cardiac stem cells. Prog Cardiovasc Dis. 2007;50:31–48. doi: 10.1016/j.pcad.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 17.Mohsin S, Siddiqi S, Collins B, Sussman MA. Empowering adult stem cells for myocardial regeneration. Circ Res. 2011;109:1415–1428. doi: 10.1161/CIRCRESAHA.111.243071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 19.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levenberg S, Golub JS, Amit M, Itskovitz-Eldor J, Langer R. Endothelial cells derived from human embryonic stem cells. Proc Natl Acad Sci USA. 2002;99:4391–4396. doi: 10.1073/pnas.032074999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dar A, Domev H, Ben-Yosef O, Tzukerman M, Zeevi-Levin N, et al. Multipotent vasculogenic pericytes from human pluripotent stem cells promote recovery of murine ischemic limb. Circulation. 2012;125:87–99. doi: 10.1161/CIRCULATIONAHA.111.048264. [DOI] [PubMed] [Google Scholar]
- 22.He JQ, Ma Y, Lee Y, Thomson JA, Kamp TJ. Human embryonic stem cells develop into multiple types of cardiac myocytes: action potential characterization. Circ Res. 2003;93:32–39. doi: 10.1161/01.RES.0000080317.92718.99. [DOI] [PubMed] [Google Scholar]
- 23.Kehat I, Kenyagin-Karsenti D, Snir M, Segev H, Amit M, et al. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J Clin Invest. 2001;108:407–414. doi: 10.1172/JCI12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, et al. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 2007;25:1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- 25.Mummery C, Ward-van Oostwaard D, Doevendans P, Spijker R, van den Brink S, et al. Differentiation of human embryonic stem cells to cardiomyocytes: role of coculture with visceral endoderm-like cells. Circulation. 2003;107:2733–2740. doi: 10.1161/01.CIR.0000068356.38592.68. [DOI] [PubMed] [Google Scholar]
- 26.Tomescot A, Leschik J, Bellamy V, Dubois G, Messas E, et al. Differentiation in vivo of cardiac committed human embryonic stem cells in postmyocardial infarcted rats. Stem Cells. 2007;25:2200–2205. doi: 10.1634/stemcells.2007-0133. [DOI] [PubMed] [Google Scholar]
- 27.Xu C, Police S, Rao N, Carpenter MK. Characterization and enrichment of cardiomyocytes derived from human embryonic stem cells. Circ Res. 2002;91:501–508. doi: 10.1161/01.RES.0000035254.80718.91. [DOI] [PubMed] [Google Scholar]
- 28.Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 29.Caspi O, Itzhaki I, Kehat I, Gepstein A, Arbel G, et al. In vitro electrophysiological drug testing using human embryonic stem cell derived cardiomyocytes. Stem Cells Dev. 2009;18:161–172. doi: 10.1089/scd.2007.0280. [DOI] [PubMed] [Google Scholar]
- 30.Dick E, Rajamohan D, Ronksley J, Denning C. Evaluating the utility of cardiomyocytes from human pluripotent stem cells for drug screening. Biochem Soc Trans. 2010;38:1037–1045. doi: 10.1042/BST0381037. [DOI] [PubMed] [Google Scholar]
- 31.Gepstein L. Derivation and potential applications of human embryonic stem cells. Circ Res. 2002;91:866–876. doi: 10.1161/01.RES.0000041435.95082.84. [DOI] [PubMed] [Google Scholar]
- 32.Kehat I, Gepstein L. Human embryonic stem cells for myocardial regeneration. Heart Fail Rev. 2003;8:229–236. doi: 10.1023/A:1024709332039. [DOI] [PubMed] [Google Scholar]
- 33.Shiba Y, Hauch KD, Laflamme MA. Cardiac applications for human pluripotent stem cells. Curr Pharm Des. 2009;15:2791–2806. doi: 10.2174/138161209788923804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 35.Haase A, Olmer R, Schwanke K, Wunderlich S, Merkert S, et al. Generation of induced pluripotent stem cells from human cord blood. Cell Stem Cell. 2009;5:434–441. doi: 10.1016/j.stem.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J, Wilson GF, Soerens AG, Koonce CH, Yu J, et al. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ Res. 2009;104:e30–e41. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zwi L, Caspi O, Arbel G, Huber I, Gepstein A, et al. Cardiomyocyte differentiation of human induced pluripotent stem cells. Circulation. 2009;120:1513–1523. doi: 10.1161/CIRCULATIONAHA.109.868885. [DOI] [PubMed] [Google Scholar]
- 38.Carvajal-Vergara X, Sevilla A, D’Souza SL, Ang YS, Schaniel C, et al. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature. 2010;465:808–812. doi: 10.1038/nature09005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Itzhaki I, Maizels L, Huber I, Zwi-Dantsis L, Caspi O, et al. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471:225–229. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- 40.Moretti A, Bellin M, Welling A, Jung CB, Lam JT, et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363:1397–1409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- 41.Amabile G, Meissner A. Induced pluripotent stem cells: current progress and potential for regenerative medicine. Trends Mol Med. 2009;15:59–68. doi: 10.1016/j.molmed.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 42.Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol. 2008;26:1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- 44.Aoi T, Yae K, Nakagawa M, Ichisaka T, Okita K, et al. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008;321:699–702. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- 45.Hanna J, Markoulaki S, Schorderet P, Carey BW, Beard C, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinton DA, Daley GQ. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, et al. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yusa K, Rad R, Takeda J, Bradley A. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat Methods. 2009;6:363–369. doi: 10.1038/nmeth.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim D, Kim CH, Moon JI, Chung YG, Chang MY, et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou H, Wu S, Joo JY, Zhu S, Han DW, et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, et al. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, et al. A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell. 2009;5:491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mauritz C, Schwanke K, Reppel M, Neef S, Katsirntaki K, et al. Generation of functional murine cardiac myocytes from induced pluripotent stem cells. Circulation. 2008;118:507–517. doi: 10.1161/CIRCULATIONAHA.108.778795. [DOI] [PubMed] [Google Scholar]
- 56.Narazaki G, Uosaki H, Teranishi M, Okita K, Kim B, et al. Directed and systematic differentiation of cardiovascular cells from mouse induced pluripotent stem cells. Circulation. 2008;118:498–506. doi: 10.1161/CIRCULATIONAHA.108.769562. [DOI] [PubMed] [Google Scholar]
- 57.Zwi-Dantsis L, Mizrahi I, Arbel G, Gepstein A, Gepstein L. Scalable Production of Cardiomyocytes Derived from c-Myc Free Induced Pluripotent Stem Cells. Tissue Eng Part A. 2011;17:1027–1037. doi: 10.1089/ten.tea.2010.0235. [DOI] [PubMed] [Google Scholar]
- 58.Martinez-Fernandez A, Nelson TJ, Yamada S, Reyes S, Alekseev AE, et al. iPS programmed without c-MYC yield proficient cardiogenesis for functional heart chimerism. Circ Res. 2009;105:648–656. doi: 10.1161/CIRCRESAHA.109.203109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martinez-Fernandez A, Nelson TJ, Ikeda Y, Terzic A. c-MYC independent nuclear reprogramming favors cardiogenic potential of induced pluripotent stem cells. J Cardiovasc Transl Res. 2010;3:13–23. doi: 10.1007/s12265-009-9150-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ma J, Guo L, Fiene SJ, Anson BD, Thomson JA, et al. High purity human-induced pluripotent stem cell-derived cardiomyocytes: electrophysiological properties of action potentials and ionic currents. Am J Physiol Heart Circ Physiol. 2011;301:H2006–H2017. doi: 10.1152/ajpheart.00694.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Itzhaki I, Rapoport S, Huber I, Mizrahi I, Zwi-Dantsis L, et al. Calcium handling in human induced pluripotent stem cell derived cardiomyocytes. PLoS One. 2011;6:e18037. doi: 10.1371/journal.pone.0018037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zwi-Dantsis L, Huber I, Habib M, Winterstern A, Gepstein A, et al. (2012) Derivation and cardiomyocyte differentiation of induced pluripotent stem cells from heart failure patients. Eur Heart J (in press) [DOI] [PubMed]
- 63.Lev S, Kehat I, Gepstein L. Differentiation pathways in human embryonic stem cell-derived cardiomyocytes. Ann NY Acad Sci. 2005;1047:50–65. doi: 10.1196/annals.1341.005. [DOI] [PubMed] [Google Scholar]
- 64.Burridge PW, Keller G, Gold JD, Wu JC. Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell. 2012;10:16–28. doi: 10.1016/j.stem.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 66.Passier R, Oostwaard DW, Snapper J, Kloots J, Hassink RJ, et al. Increased cardiomyocyte differentiation from human embryonic stem cells in serum-free cultures. Stem Cells. 2005;23:772–780. doi: 10.1634/stemcells.2004-0184. [DOI] [PubMed] [Google Scholar]
- 67.Graichen R, Xu X, Braam SR, Balakrishnan T, Norfiza S, et al. Enhanced cardiomyogenesis of human embryonic stem cells by a small molecular inhibitor of p38 MAPK. Differentiation. 2008;76:357–370. doi: 10.1111/j.1432-0436.2007.00236.x. [DOI] [PubMed] [Google Scholar]
- 68.Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 69.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Efe JA, Hilcove S, Kim J, Zhou H, Ouyang K, et al. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat Cell Biol. 2011;13:215–222. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- 71.Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Song K, Nam YJ, Luo X, Qi X, Tan W, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Caspi O, Huber I, Kehat I, Habib M, Arbel G, et al. Transplantation of human embryonic stem cell-derived cardiomyocytes improves myocardial performance in infarcted rat hearts. J Am Coll Cardiol. 2007;50:1884–1893. doi: 10.1016/j.jacc.2007.07.054. [DOI] [PubMed] [Google Scholar]
- 74.Dai W, Field LJ, Rubart M, Reuter S, Hale SL, et al. Survival and maturation of human embryonic stem cell-derived cardiomyocytes in rat hearts. J Mol Cell Cardiol. 2007;43:504–516. doi: 10.1016/j.yjmcc.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huber I, Itzhaki I, Caspi O, Arbel G, Tzukerman M, et al. Identification and selection of cardiomyocytes during human embryonic stem cell differentiation. FASEB J. 2007;21:2551–2563. doi: 10.1096/fj.05-5711com. [DOI] [PubMed] [Google Scholar]
- 76.Kehat I, Khimovich L, Caspi O, Gepstein A, Shofti R, et al. Electromechanical integration of cardiomyocytes derived from human embryonic stem cells. Nat Biotechnol. 2004;22:1282–1289. doi: 10.1038/nbt1014. [DOI] [PubMed] [Google Scholar]
- 77.Laflamme MA, Gold J, Xu C, Hassanipour M, Rosler E, et al. Formation of human myocardium in the rat heart from human embryonic stem cells. Am J Pathol. 2005;167:663–671. doi: 10.1016/S0002-9440(10)62041-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kofidis T, Lebl DR, Swijnenburg RJ, Greeve JM, Klima U, et al. Allopurinol/uricase and ibuprofen enhance engraftment of cardiomyocyte-enriched human embryonic stem cells and improve cardiac function following myocardial injury. Eur J Cardiothorac Surg. 2006;29:50–55. doi: 10.1016/j.ejcts.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 79.Leor J, Gerecht S, Cohen S, Miller L, Holbova R, et al. Human embryonic stem cell transplantation to repair the infarcted myocardium. Heart. 2007;93:1278–1284. doi: 10.1136/hrt.2006.093161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van Laake LW, Passier R, Doevendans PA, Mummery CL. Human embryonic stem cell-derived cardiomyocytes and cardiac repair in rodents. Circ Res. 2008;102:1008–1010. doi: 10.1161/CIRCRESAHA.108.175505. [DOI] [PubMed] [Google Scholar]
- 81.van Laake LW, Passier R, Monshouwer-Kloots J, Nederhoff MG, Ward-van Oostwaard D, et al. Monitoring of cell therapy and assessment of cardiac function using magnetic resonance imaging in a mouse model of myocardial infarction. Nat Protoc. 2007;2:2551–2567. doi: 10.1038/nprot.2007.371. [DOI] [PubMed] [Google Scholar]
- 82.Halbach M, Pfannkuche K, Pillekamp F, Ziomka A, Hannes T, et al. Electrophysiological maturation and integration of murine fetal cardiomyocytes after transplantation. Circ Res. 2007;101:484–492. doi: 10.1161/CIRCRESAHA.107.153643. [DOI] [PubMed] [Google Scholar]
- 83.Rubart M, Pasumarthi KB, Nakajima H, Soonpaa MH, Nakajima HO, et al. Physiological coupling of donor and host cardiomyocytes after cellular transplantation. Circ Res. 2003;92:1217–1224. doi: 10.1161/01.RES.0000075089.39335.8C. [DOI] [PubMed] [Google Scholar]
- 84.Gepstein L, Ding C, Rehemedula D, Wilson EE, Yankelson L, et al. In vivo assessment of the electrophysiological integration and arrhythmogenic risk of myocardial cell transplantation strategies. Stem Cells. 2010;28:2151–2161. doi: 10.1002/stem.545. [DOI] [PubMed] [Google Scholar]
- 85.Xue T, Cho HC, Akar FG, Tsang SY, Jones SP, et al. Functional integration of electrically active cardiac derivatives from genetically engineered human embryonic stem cells with quiescent recipient ventricular cardiomyocytes: insights into the development of cell-based pacemakers. Circulation. 2005;111:11–20. doi: 10.1161/01.CIR.0000151313.18547.A2. [DOI] [PubMed] [Google Scholar]
- 86.Zhao T, Zhang ZN, Rong Z, Xu Y. Immunogenicity of induced pluripotent stem cells. Nature. 2011;474:212–215. doi: 10.1038/nature09971. [DOI] [PubMed] [Google Scholar]
- 87.Wu SM, Hochedlinger K. Harnessing the potential of induced pluripotent stem cells for regenerative medicine. Nat Cell Biol. 2011;13:497–505. doi: 10.1038/ncb0511-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hanna J, Wernig M, Markoulaki S, Sun CW, Meissner A, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- 89.Nelson TJ, Martinez-Fernandez A, Yamada S, Perez-Terzic C, Ikeda Y, et al. Repair of acute myocardial infarction by human stemness factors induced pluripotent stem cells. Circulation. 2009;120:408–416. doi: 10.1161/CIRCULATIONAHA.109.865154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Carpenter L, Carr C, Yang CT, Stuckey DJ, Clarke K, et al. Efficient differentiation of human induced pluripotent stem cells generates cardiac cells that provide protection following myocardial infarction in the rat. Stem Cells Dev. 2012;21:977–986. doi: 10.1089/scd.2011.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yoshida Y, Yamanaka S. Recent stem cell advances: induced pluripotent stem cells for disease modeling and stem cell-based regeneration. Circulation. 2010;122:80–87. doi: 10.1161/CIRCULATIONAHA.109.881433. [DOI] [PubMed] [Google Scholar]
- 92.Ben-David U, Benvenisty N. The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat Rev Cancer. 2011;11:268–277. doi: 10.1038/nrc3034. [DOI] [PubMed] [Google Scholar]
- 93.Nussbaum J, Minami E, Laflamme MA, Virag JA, Ware CB, et al. Transplantation of undifferentiated murine embryonic stem cells in the heart: teratoma formation and immune response. FASEB J. 2007;21:1345–1357. doi: 10.1096/fj.06-6769com. [DOI] [PubMed] [Google Scholar]
- 94.Behfar A, Perez-Terzic C, Faustino RS, Arrell DK, Hodgson DM, et al. Cardiopoietic programming of embryonic stem cells for tumor-free heart repair. J Exp Med. 2007;204:405–420. doi: 10.1084/jem.20061916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tang C, Lee AS, Volkmer JP, Sahoo D, Nag D, et al. An antibody against SSEA-5 glycan on human pluripotent stem cells enables removal of teratoma-forming cells. Nat Biotechnol. 2011;29:829–834. doi: 10.1038/nbt.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Anderson D, Self T, Mellor IR, Goh G, Hill SJ, et al. Transgenic enrichment of cardiomyocytes from human embryonic stem cells. Mol Ther. 2007;15:2027–2036. doi: 10.1038/sj.mt.6300303. [DOI] [PubMed] [Google Scholar]
- 97.Hattori F, Chen H, Yamashita H, Tohyama S, Satoh YS, et al. Nongenetic method for purifying stem cell-derived cardiomyocytes. Nat Methods. 2010;7:61–66. doi: 10.1038/nmeth.1403. [DOI] [PubMed] [Google Scholar]
- 98.Klug MG, Soonpaa MH, Koh GY, Field LJ. Genetically selected cardiomyocytes from differentiating embronic stem cells form stable intracardiac grafts. J Clin Invest. 1996;98:216–224. doi: 10.1172/JCI118769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Elliott DA, Braam SR, Koutsis K, Ng ES, Jenny R, et al. NKX2-5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat Methods. 2011;8:1037–1040. doi: 10.1038/nmeth.1740. [DOI] [PubMed] [Google Scholar]
- 100.van Laake LW, Qian L, Cheng P, Huang Y, Hsiao EC, et al. Reporter-based isolation of induced pluripotent stem cell- and embryonic stem cell-derived cardiac progenitors reveals limited gene expression variance. Circ Res. 2010;107:340–347. doi: 10.1161/CIRCRESAHA.109.215434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Van Hoof D, Dormeyer W, Braam SR, Passier R, Monshouwer-Kloots J, et al. Identification of cell surface proteins for antibody-based selection of human embryonic stem cell-derived cardiomyocytes. J Proteome Res. 2010;9:1610–1618. doi: 10.1021/pr901138a. [DOI] [PubMed] [Google Scholar]
- 102.Dubois NC, Craft AM, Sharma P, Elliott DA, Stanley EG, et al. SIRPA is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nat Biotechnol. 2011;29:1011–1018. doi: 10.1038/nbt.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Uosaki H, Fukushima H, Takeuchi A, Matsuoka S, Nakatsuji N, et al. Efficient and scalable purification of cardiomyocytes from human embryonic and induced pluripotent stem cells by VCAM1 surface expression. PLoS One. 2011;6:e23657. doi: 10.1371/journal.pone.0023657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhu WZ, Xie Y, Moyes KW, Gold JD, Askari B, et al. Neuregulin/ErbB signaling regulates cardiac subtype specification in differentiating human embryonic stem cells. Circ Res. 2010;107:776–786. doi: 10.1161/CIRCRESAHA.110.223917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sartiani L, Bettiol E, Stillitano F, Mugelli A, Cerbai E, et al. Developmental changes in cardiomyocytes differentiated from human embryonic stem cells: a molecular and electrophysiological approach. Stem Cells. 2007;25:1136–1144. doi: 10.1634/stemcells.2006-0466. [DOI] [PubMed] [Google Scholar]
- 106.Tulloch NL, Muskheli V, Razumova MV, Korte FS, Regnier M, et al. Growth of engineered human myocardium with mechanical loading and vascular coculture. Circ Res. 2011;109:47–59. doi: 10.1161/CIRCRESAHA.110.237206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Caspi O, Lesman A, Basevitch Y, Gepstein A, Arbel G, et al. Tissue engineering of vascularized cardiac muscle from human embryonic stem cells. Circ Res. 2007;100:263–272. doi: 10.1161/01.RES.0000257776.05673.ff. [DOI] [PubMed] [Google Scholar]
- 108.Reinecke H, Zhang M, Bartosek T, Murry CE. Survival, integration, and differentiation of cardiomyocyte grafts: a study in normal and injured rat hearts. Circulation. 1999;100:193–202. doi: 10.1161/01.CIR.100.2.193. [DOI] [PubMed] [Google Scholar]
- 109.Robey TE, Saiget MK, Reinecke H, Murry CE. Systems approaches to preventing transplanted cell death in cardiac repair. J Mol Cell Cardiol. 2008;45:567–581. doi: 10.1016/j.yjmcc.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hu S, Huang M, Nguyen PK, Gong Y, Li Z, et al. Novel microRNA prosurvival cocktail for improving engraftment and function of cardiac progenitor cell transplantation. Circulation. 2011;124:S27–S34. doi: 10.1161/CIRCULATIONAHA.111.017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lesman A, Habib M, Caspi O, Gepstein A, Arbel G, et al. Transplantation of a tissue-engineered human vascularized cardiac muscle. Tissue Eng Part A. 2010;16:115–125. doi: 10.1089/ten.TEA.2009.0130. [DOI] [PubMed] [Google Scholar]
- 112.Stevens KR, Kreutziger KL, Dupras SK, Korte FS, Regnier M, et al. Physiological function and transplantation of scaffold-free and vascularized human cardiac muscle tissue. Proc Natl Acad Sci USA. 2009;106:16568–16573. doi: 10.1073/pnas.0908381106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wu SM, Chien KR, Mummery C. Origins and fates of cardiovascular progenitor cells. Cell. 2008;132:537–543. doi: 10.1016/j.cell.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wu SM, Fujiwara Y, Cibulsky SM, Clapham DE, Lien CL, et al. Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell. 2006;127:1137–1150. doi: 10.1016/j.cell.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 115.Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 116.Moretti A, Bellin M, Jung CB, Thies TM, Takashima Y, et al. Mouse and human induced pluripotent stem cells as a source for multipotent Isl1+ cardiovascular progenitors. FASEB J. 2010;24:700–711. doi: 10.1096/fj.09-139477. [DOI] [PubMed] [Google Scholar]
- 117.Blin G, Nury D, Stefanovic S, Neri T, Guillevic O, et al. A purified population of multipotent cardiovascular progenitors derived from primate pluripotent stem cells engrafts in postmyocardial infarcted nonhuman primates. J Clin Invest. 2010;120:1125–1139. doi: 10.1172/JCI40120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Habib M, Shapira-Schweitzer K, Caspi O, Gepstein A, Arbel G, et al. A combined cell therapy and in situ tissue-engineering approach for myocardial repair. Biomaterials. 2011;32:7514–7523. doi: 10.1016/j.biomaterials.2011.06.049. [DOI] [PubMed] [Google Scholar]
- 119.Ott HC, Matthiesen TS, Goh SK, Black LD, Kren SM, et al. Perfusion-decellularized matrix: using nature’s platform to engineer a bioartificial heart. Nat Med. 2008;14:213–221. doi: 10.1038/nm1684. [DOI] [PubMed] [Google Scholar]
- 120.Jawad H, Lyon AR, Harding SE, Ali NN, Boccaccini AR. Myocardial tissue engineering. Br Med Bull. 2008;87:31–47. doi: 10.1093/bmb/ldn026. [DOI] [PubMed] [Google Scholar]
- 121.Masuda S, Shimizu T, Yamato M, Okano T. Cell sheet engineering for heart tissue repair. Adv Drug Deliv Rev. 2008;60:277–285. doi: 10.1016/j.addr.2007.08.031. [DOI] [PubMed] [Google Scholar]
- 122.Segers VF, Lee RT. Biomaterials to enhance stem cell function in the heart. Circ Res. 2011;109:910–922. doi: 10.1161/CIRCRESAHA.111.249052. [DOI] [PubMed] [Google Scholar]
- 123.Zimmermann WH. Embryonic and embryonic-like stem cells in heart muscle engineering. J Mol Cell Cardiol. 2011;50:320–326. doi: 10.1016/j.yjmcc.2010.10.027. [DOI] [PubMed] [Google Scholar]