Abstract
Modification of nuclear and cytosolic proteins by O-linked N-acetylglucosamine (O-GlcNAcylation) is ubiquitous in cells. The in vivo function of the protein O-GlcNAcylation, however, is not well understood. Here, we manipulated the cellular O-GlcNAcylation level in Drosophila and found that it promotes developmental growth by enhancing insulin signaling. This increase in growth is due mainly to cell growth and not to cell proliferation. Our data suggest that the increase in the insulin signaling activity is mediated, at least in part, through O-GlcNAcylation of Akt. These results indicate that O-GlcNAcylation is one of the crucial mechanisms involved in control of insulin signaling during Drosophila development.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-011-0640-7) contains supplementary material, which is available to authorized users.
Keywords: OGT, O-GlcNAcase, Size control, Insulin, Drosophila
Introduction
O-GlcNAcylation is a unique type of protein glycosylation in which a single GlcNAc residue is linked via an O-linked glycosidic bond to either serine or threonine of nuclear or cytoplasmic proteins. It is observed in most multicellular organisms and found on many proteins with diverse functions [1]. A large proportion of these O-GlcNAcylated proteins are also known to be modified by phosphorylation. In fact, O-GlcNAc modification and O-phosphate modification can occur dynamically reciprocal at the same or adjacent sites [1]. The biological functions of O-GlcNAcylation are diverse as is reflected by the large repertoire of target proteins [1].
Enzymes that add or remove O-GlcNAc are the O-linked GlcNAc transferase (OGT) and the O-GlcNAcase, respectively. Their in vivo functions have been investigated in Caenorhabditis elegans and mice. C. elegans null mutants for OGT or O-GlcNAcase are viable and fertile, but display defects in macronutrient storage, suggesting that insulin signaling is impaired [2, 3]. In fact, both mutants interact genetically with a temperature-sensitive allele of daf-2, the gene that encodes an insulin-like receptor, in a way that an elevated cellular O-GlcNAcylation level suppresses insulin signaling. In mice, OGT is an essential gene [4], and studies on conditional knockout animals indicate that loss of OGT causes T-cell apoptosis, neuronal tau hyperphosphorylation, and fibroblast growth arrest [5]. Transgenic mice overexpressing OGT develop insulin resistance and hyperglycemia [6, 7]. Several components of the insulin signaling pathway in mammals are O-GlcNAcylated, such as Akt [8] and FOXO [9]. The mechanism by which O-GlcNAcylation changes their activity is currently not understood. Previous studies of Akt yielded controversial results as to whether O-GlcNAcylation of Akt increases or decreases its phosphorylation and influences its activity [7, 8, 10]. These discrepancies may be attributed to differences in cell type or experimental setup.
The OGT of Drosophila has recently been reported to be a protein encoded by the Polycomb group gene super sex comb (sxc) [11, 12]. The protein is involved in PcG-mediated transcriptional repression. A possible involvement of sxc in insulin signaling, however, is not known. To further analyze the in vivo function of O-GlcNAcylation in this regard, we used Drosophila as a model. We manipulated the cellular O-GlcNAcylation level either pharmacologically or genetically and found that O-GlcNAcylation regulates body growth of Drosophila by modulating insulin signaling. In contrast to findings in worm and mice, we found that O-GlcNAcylation in Drosophila enhances insulin signaling.
Materials and methods
Drosophila stocks
Act-GAL4, en-GAL4, Act5C>CD2>GAL4, hsFLP, and UAS-GFP.nls were obtained from the Bloomington Stock Center (Bloomington, IN). UAS-OGA RNAiX (41823), UAS-OGA RNAiII (41822), UAS-OGT RNAiII (18610), and UAS-OGT RNAiIII (18611) were from Vienna Drosophila RNAi Center (Vienna, Austria), and UAS-PTEN RNAi (5671R-1) and UAS-Akt RNAi (4006R-3) were from the National Institute of Genetics (Shizuoka, Japan).
Antibodies
The following antibodies were used: rabbit anti-dFOXO (1:500) [13], rabbit anti-phospho-histone H3 (1:50; Cell Signaling, 9701), rabbit anti-active caspase 3 (1:50; Cell Signaling, 9661), goat anti-rabbit IgG-FITC (1:200; Jackson ImmunoResearch Labs, 111-096-047), goat anti-rabbit IgG-Texas Red (1:200; Jackson ImmunoResearch Labs, 111-076-047), rabbit anti-β-actin (1:1000; Cell Signaling, 4967), rabbit anti-OGT (1:1000; Santa Cruz Biotechnology, H-300), mouse anti-O-GlcNAc CTD 110.6 (1:1000; Covance, MMS-248R-500), mouse anti-O-GlcNAc RL2 (1:1000; ABR, MA1-072), rabbit anti-Akt (1:1000; Cell Signaling, 9272), rabbit anti-phospho-(Ser505)Akt (1:1000; Cell Signaling, 4054), rabbit anti-phospho-(Ser/Thr) Akt substrate (1:1000; Cell Signaling, 9611), mouse anti-V5 (1:5000; Invitrogen, 46-0705), and anti-FLAG M2 affinity gel (Sigma, A2220).
Immunohistochemistry, co-immunoprecipitation, and immunoblotting
Immunohistochemistry was performed as described previously [14]. For analysis of the fat body tissues, larvae were heat-shocked for 10 min at 34°C 48 h after egg deposition. After staining the tissue with phalloidin-rhodamine (Molecular Probes), the samples were mounted in Prolong Gold plus DAPI (Invitrogen, S36939) and observed with a AxioImager A1 microscope (Carl Zeiss) using a 10 × 0.3 NA Plan Neofluar lens (Carl Zeiss). For image acquisition, AxioCam HRc camera (Carl Zeiss) and AxioVision Rel 4.8 software were used. For imaging wing imaginal discs, samples were mounted in the same anti-fading medium and observed with a LSM 510 META confocal microscope (Carl Zeiss) using a Neo-Fluar 10 × NA 0.3 objective lens at room temperature. Image processing and analysis were performed with Imaris and Adobe Photoshop CS5.
Co-immunoprecipitation and immunoblotting were performed as described previously [15].
Synthesis of 1,2-dideoxy-2′-propyl-α-D-glucopyranoso-[2,1-D]-Δ2′-thiazoline (NButGT)
NButGT (9c compound) was synthesized according Macauley et al. [16].
Results
Reduction of O-GlcNAcase activity increases Drosophila body growth
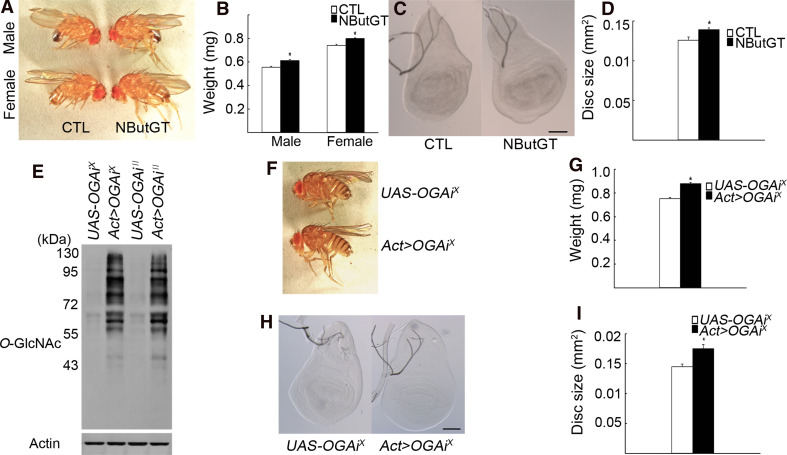
To study the in vivo role of O-GlcNAcylation in fly development, we performed a drug-feeding experiment using the O-GlcNAcase inhibitor NButGT [16] to increase the level of protein O-GlcNAcylation. The optimal amount of NButGT that efficiently elevated O-GlcNAcylation was 200 μl of a 10-mM NButGT solution added on the surface of solidified fly food in a vial (Supplementary Fig. 1a). The body weight of male and female adult flies fed with NButGT from the first to the third instar larval stages was increased by 10.4 and 7.9%, respectively, compared to the mock-fed control groups (Fig. 1a, b). This was due to increased growth of larval imaginal discs since the size of wing imaginal discs was increased by 10.7%, examined in male larvae, compared to control (Fig. 1c, d). This growth-promoting effect of NButGT was not due to its metabolic conversion in an energy source, since the same amount of glucose had no significant effect on growth (Supplementary Fig. 1b). Of note, NButGT-fed larvae eclosed faster, indicative of an accelerated developmental growth rate (Supplementary Fig. 1c).
Fig. 1.
Reduction of O-GlcNAcase activity increases Drosophila body growth. a The size of NButGT-fed flies compared to that of mock-fed control (CTL) flies. b Body weights of control (n = 293 ♂ and n = 297 ♀) and drug-fed (n = 278 ♂ and n = 313 ♀) adult flies. Experiments were done in triplicate with >100 male and >100 female flies in each experiment. c, d The size of wing discs from mock-fed (n = 15) and NButGT-fed (n = 17) male wandering third instar larvae. e Knock-down efficiency of two OGA RNAi transgenes as analyzed by Western blotting using anti-O-GlcNAc RL2 antibody as a measure of cellular O-GlcNAcylation level in larvae of indicated genotypes. β-actin was used as a loading control. f, g The size and weight of Act>OGA RNAi X (n = 277) female flies compared to the UAS-only control flies (n = 259). Since the UAS-OGA RNAi X stock contained the attached X chromosome, only females were analyzed. Experiments were done in triplicate with >100 animals analyzed in each experiment. h, i The sizes of the wing discs of UAS-only (n = 13) and Act>OGA RNAi X (n = 13) female larvae. b, d, g, i Data are mean ± SE *p < 0.001 by Student’s t test. c, h Scale bars represent 100 μm
To show specificity of the NButGT effect, we knocked down O-GlcNAcase using RNAi in vivo. From FlyBase (http://flybase.org), we identified a single Drosophila melanogaster gene, CG5871, that encodes a protein of 1019 amino acids with significant homology to human O-GlcNAcase (35.3% overall identity; 56.3% identity of the two N-terminal halves containing the O-GlcNAcase catalytic domain). No mutant for CG5871 has been reported. Hereafter, we call CG5871 Drosophila OGA. For its knock-down, two RNAi strains were used, UAS-OGA RNAi X and UAS-OGA RNAi II (X and II mark the transgene insertions). Both strains, which were crossed to Actin-GAL4 flies (hereafter named Act>OGA RNAi), were viable and had significantly reduced OGA transcripts (29.8 and 23.2% of the UAS-only controls, respectively; see Supplementary Fig. 1d). Both strains also showed a marked increase in cellular O-GlcNAcylation levels (Fig. 1e). The OGA RNAi X was selected for further studies. The body weight of Act>OGA RNAi X adult females was increased by 16.8% compared to the UAS-only control flies (Fig. 1f, g). The size of the larval wing discs was increased by 21.0% compared to the UAS-only control flies (Fig. 1h, i). Thus, increased O-GlcNAcylation level either due to O-GlcNAcase inhibition or RNAi knock-down results in increased body growth in Drosophila.
RNAi knock-down of OGT decreases Drosophila body growth
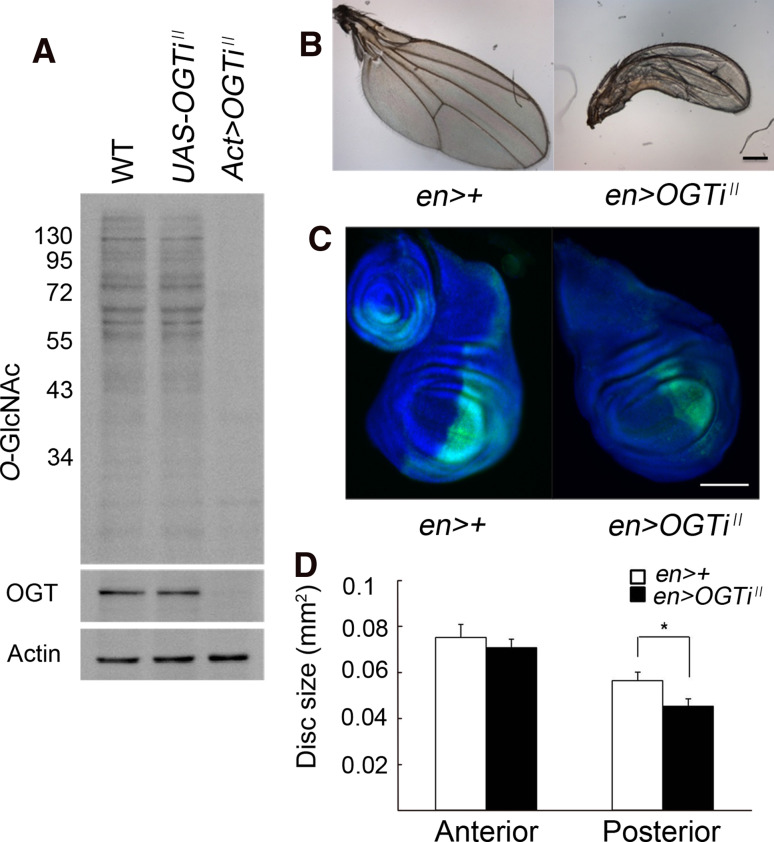
The effect of decreased O-GlcNAcylation on body growth of Drosophila was analyzed by RNAi knock-down of OGT. We tested two different RNAi transgenic strains, UAS-OGT RNAi II and UAS-OGT RNAi III. Only OGT RNAi II, driven by Act-GAL4, resulted in nearly complete loss of OGT protein (Fig. 2a). Act>OGT RNAi II flies were early pupal lethal, similar as reported for the amorphic allele of sxc/OGT [11] and had strongly reduced cellular O-GlcNAcylation level (Fig. 2a). The size of the wing imaginal discs of Act>OGT RNAi II larvae was reduced by 36.5% compared to that of the UAS-only control larvae (Supplementary Fig. 2). Selective expression of OGT RNAi II in the posterior half of the wing discs using engrailed-GAL4 (en-GAL4) not only resulted in growth retardation in this domain, but also in a posteriorly bent wrinkled wing (Fig. 2b–d). Since the OGT RNAi phenotype is the reverse of the OGA RNAi phenotype, and the NButGT-fed flies are a phenocopy of the OGA RNAi flies, this indicates that the RNAi phenotypes of OGA and OGT are gene-specific and not due to an off-target effect. Thus, decreasing the cellular O-GlcNAcylation levels results in decreased body growth of Drosophila.
Fig. 2.
RNAi knock-down of OGT decreases Drosophila body growth. a Knock-down efficiency of the OGT RNAi II transgene was analyzed by Western blotting with anti-O-GlcNAc RL2 antibody. β-actin was used as a loading control. b Selective expression of OGT RNAi II in the posterior compartment of the wing disc using the en-GAL4 driver results in a shrunken, posteriorly bent adult wing. c Growth reduction in the posterior compartment of the wing disc of an en>OGT RNAi II larva. The GFP-positive region is the Engrailed expression domain (dorsal is up, anterior is left). d Measurement of the anterior and the posterior compartments of the wing discs of UAS-only (n = 13) and en>OGT RNAi II (n = 15) male larvae. Data are mean ± SE *p < 0.05 by Student’s t test. b, c Scale bars represent 100 μm
The effects of O-GlcNAcylation on body growth are mainly due to cell growth
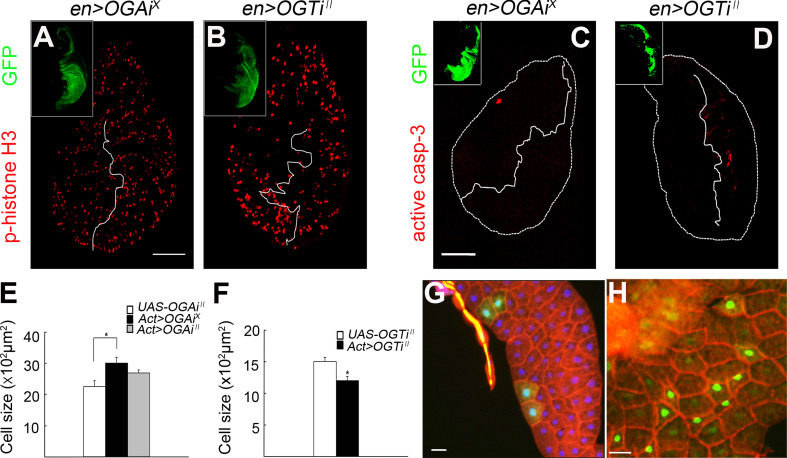
The decrease in body growth in OGT RNAi flies may be due to decreased cell proliferation, decreased cell growth, increased cell death, or any combination of the three. We examined cell proliferation using anti-phospho-histone H3 antibody, which stains dividing cells in M phase [17]. When OGA RNAi X or OGT RNAi II was expressed in the posterior compartment of the wing disc, no significant changes in the phospho-histone H3 staining were detected between RNAi-expressing discs and controls or between the anterior and the posterior compartments of the en>RNAi discs (Fig. 3a, b; data not shown). To determine the apoptotic cell death rate, we analyzed active caspase-3 immunostaining in the wing discs [18]. Only a slight increase in apoptosis in the OGT RNAi II-expressing wing disc domain was found (Fig. 3c, d). Thus, apoptosis may contribute to the altered body size, but it is likely not a major cause (see below). We then assessed cell growth by comparing cell numbers in a defined area of a particular lobe of the fat body from control and RNAi-expressing larvae. The average size of fat body cells of Act>OGA RNAi X larvae was increased by 33.1% compared to the UAS-only control larvae, whereas that of Act>OGT RNAi II larvae was reduced by 20.1% (Fig. 3e, f; also compare Fig. 4h, k to b, e, respectively). The number of wing hairs in a defined area of the Act>OGA RNAi X flies was 87.9% of that in UAS-only control flies (Supplementary Fig. 3A, B), indicating that impairment of cell growth was a major factor for the decreased body size. To validate these findings further, we expressed OGA RNAi X or OGT RNAi II transgenes post-mitotically in a small patch of cells in an otherwise wild-type background using the FLP/FRT-based “flip-out” technique [19]. During the larval stage, most cells, including the fat body cells, grow in size through endoreplication but do not proliferate. By expressing the RNAi transgenes from early to mid-larval stages and simultaneously marking those cells with GFP, we could determine the effect of O-GlcNAcylation exclusively on cell growth. OGA RNAi X-expressing cells were larger than neighboring wild-type cells (Fig. 3g; see Supplementary Fig. 3 for quantification), whereas OGT RNAi II-expressing cells were markedly smaller than neighboring wild-type cells (Fig. 3h). Together, this demonstrates that the effect of O-GlcNAcylation on body growth is mainly through cell growth.
Fig. 3.
The effect of O-GlcNAcylation on body growth is mainly due to cell growth and not proliferation. a, b Demonstration of proliferating cells in the larval wing discs using anti-phospho-histone H3 antibody (red fluorescence). The Engrailed expression domain was labeled with GFP (green in insets). a en>OGA RNAi X. b en>OGT RNAi II. Scale bar represents 100 μm. c, d Demonstration of cells undergoing apoptosis in the larval wing discs using anti-active caspase-3 antibody (red fluorescence). The Engrailed expression domain was labeled with GFP (green in insets). c en>OGA RNAi X. d en>OGT RNAi II. Scale bar represents 100 μm. e, f Fat body cell size of Act>OGA RNAi female and Act>OGT RNAi II male larvae. For a fair comparison, stage-matched third instar larvae were dissected, and the fat body lobe located closest to the gonad was used for size comparison. Cell number was counted in a defined area of the lobe. Data are mean ± SE *p < 0.01. g, h Post-mitotic effects of O-GlcNAcylation on cell growth. OGA or OGT was knocked down using the flip-out method by applying a 10-min heat shock (34°C) 48 h after egg laying. g Genotype: hsFLP 122 /UAS-OGA RNAi X ; +/+; Act5C>CD2>GAL4 UAS-GFP.nls/+ . See Supplementary Fig. 3 for quantification. h Genotype: hsFLP 122 /+ ; UAS-OGT RNAi II /+ ; Act5C>CD2>GAL4 UAS-GFP.nls/+ . OGA or OGT RNAi-expressing cells were labeled with nuclear GFP (green), and the cell boundary was visualized using phalloidin-rhodamine (red). Scale bar represents 50 μm
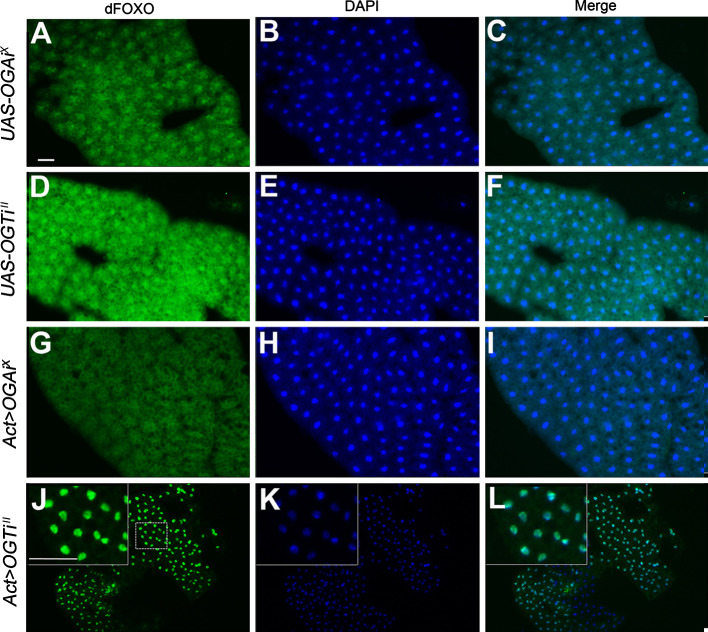
Fig. 4.
O-GlcNAcylation inhibits nuclear localization of dFOXO in the fat bodies. Fat bodies of third instar larvae of UAS-OGA RNAiX only (a–c), UAS-OGT RNAiII only (d–f), Act>OGA RNAiX (g–i), and Act>OGT RNAiII (j–l). a, d, g, and j dFOXO immunostaining (green). b, e, h, and k DAPI staining (blue) for nuclei. c, f, i, and l Merged images. The insets in j–l show the area marked with a dashed line in j at higher magnification. Scale bars represent 50 μm
O-GlcNAcylation enhances insulin signaling activity
The changes in body growth and in eclosion time observed in OGA and OGT RNAi flies are highly reminiscent of the phenotypes of insulin signaling mutants [20]. We reasoned that the O-GlcNAcylation level may influence insulin signaling activity and analyzed it by examining dFOXO in the fat body. dFOXO is a negative regulator of the insulin signaling pathway and is translocated to the nucleus when insulin signaling is inactive [13]. Fat body cells of UAS-OGA RNAi X-only (Fig. 4a–c) or UAS-OGT RNAi II-only (Fig. 4d–f) control larvae showed stronger nuclear to cytosolic staining for dFOXO. In contrast, fat body cells of Act>OGA RNAi X larvae showed nuclear dFOXO staining equal to cytosolic staining (Fig. 4g–i). This indicates enhanced insulin signaling activity in OGA knock-down larvae. Conversely, fat body cells of Act>OGT RNAi II larvae showed a marked increase in nuclear staining and almost no cytosolic staining of dFOXO (Fig. 4j–l; note also the decrease in cell size).
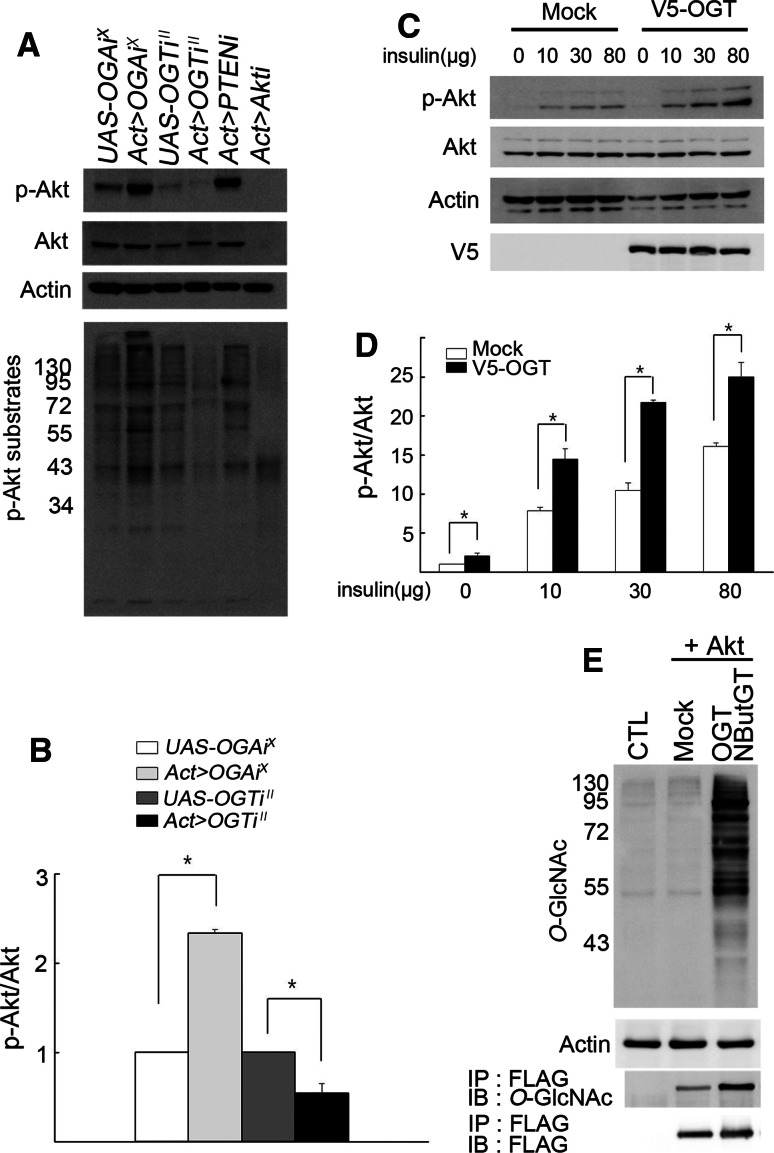
We next analyzed Akt phosphorylation using an antibody that detects phospho-Ser505, which corresponds to phospho-Ser473 of mammalian Akt. Compared to UAS-only controls, Act>OGA RNAi X larvae had an increased ratio of phospho-Akt to total Akt (Fig. 5a, b). Conversely, Act>OGT RNAi II larvae had a substantially decreased ratio of phospho-Akt to total Akt. Thus, the activity of insulin signaling is modulated by the cellular O-GlcNAcylation level.
Fig. 5.
O-GlcNAcylation increases the level of Akt phosphorylation and Akt is O-GlcNAcylated. a Phosphorylation and kinase activity of Akt in OGA RNAiX or OGT RNAiII larvae as analyzed by Western blot. Akt phosphorylation was examined using anti-phospho-(Ser505) Akt and anti-Akt antibodies. β-actin was used as a loading control. To assess Akt kinase activity, phosphorylation state of Akt substrates in larval lysates of the indicated genotypes was analyzed using phospho-Akt substrate antibody. b Quantification of the ratio of phospho-Akt to total Akt. For reason of comparison with Act>OGAiX and Act>OGTiII, the values of the UAS-only controls were arbitrarily set to 1. Experiments were performed in triplicate. Data are mean ± SE *p < 0.025. c The effect of OGT overexpression on insulin-induced Akt phosphorylation in S2 cells. Five milliliters of Drosophila S2 cell culture was treated with 10, 30, or 80 μg of insulin and incubated for 15 min before Western-blot analysis. β-actin was used as a loading control. d Quantification of the results in c. Experiments were done in triplicate. Data are mean ± SE *p < 0.05. eO-GlcNAcylation of Akt. FLAG-tagged Akt was expressed in S2 cells either alone or together with Drosophila OGT and 50 μM of NButGT. Total O-GlcNAcylation level was examined using anti-O-GlcNAc RL2 antibody. β-actin was used as a loading control. To demonstrate O-GlcNAcylation of Akt, Akt was immunoprecipitated using anti-FLAG antibody followed by Western blotting with anti-O-GlcNAc antibody CTD110.6. For control, the same immunoprecipitates were blotted with anti-FLAG antibody
We then examined whether increased cellular O-GlcNAcylation enhances Akt kinase activity as a correlate to the phosphorylation state of Ser505 of Akt. Since no antibody that effectively immunoprecipitates Drosophila Akt was available, we assessed Akt kinase activity using phospho-(Ser/Thr) Akt substrate antibody [21]. Phosphorylation of Akt substrate proteins was markedly increased in Act>OGA RNAi X larvae and substantially decreased in Act>OGT RNAi II larvae (Fig. 5a). To verify that O-GlcNAcylation affects insulin signaling activity in a cell-autonomous manner, we overexpressed V5-tagged OGT in S2 cells. This increased insulin-induced Akt phosphorylation up to 2.2-fold (Fig. 5c, d).
Based on findings in mammals, we predicted Akt a candidate protein for O-GlcNAcylation [7, 8, 10, 22]. Indeed, FLAG-tagged Drosophila Akt was modified with O-GlcNAc, and its O-GlcNAcylation was substantially increased by OGT overexpression and inhibition of O-GlcNAcase (Fig. 5e). Thus, increased O-GlcNAcylation of Akt in vitro correlates not only with its phosphorylation state (Ser505) but also with its kinase activity in vivo, suggesting that modulation of insulin signaling and body growth occurs, at least in part, through O-GlcNAcylation of Akt.
Discussion
Our data demonstrate that O-GlcNAcylation is a key post-translational protein modification that controls insulin signaling in Drosophila, which is in agreement with previous findings in C. elegans and mice [2, 3, 6, 7] and a report published after completion of our manuscript [23]. However, the mechanism of O-GlcNAcylation-mediated modulation of Drosophila insulin signaling is different from the mechanism observed in worm and mice. Whereas O-GlcNAcylation in C. elegans and mice suppresses insulin signaling [2, 7], O-GlcNAcylation in Drosophila enhances insulin signaling, indicating species-specific differences in this evolutionarily conserved mechanism.
How does O-GlcNAcylation affect insulin signaling activity and body growth in Drosophila? Our results suggest that O-GlcNAcylation of Akt and possibly of its upstream signaling components modulates the activity of insulin signaling and consequently of growth in Drosophila during development. This is based on the fact that increased cellular O-GlcNAcylation is paralleled by increased Akt phosphorylation and kinase activity and decreased nuclear localization of dFOXO and vice versa. We found that modulation of cellular O-GlcNAcylation by OGT or OGA RNAi knock-down increases Drosophila body size mainly through cell growth and perhaps apoptosis, but not proliferation. Mutations in different insulin signaling components have been shown to influence growth by different mechanisms. For example, PI3K and the upstream components of the PI3K pathway are necessary for both cell growth and proliferation, whereas components downstream of PI3K are necessary for cell growth, but not for cell proliferation [20, 24]. Thus, regulation of Akt by O-GlcNAcylation appears to be a key component in the insulin signaling pathway of Drosophila.
Protein modification by O-GlcNAc and O-phosphate may have antagonistic effects on each other [1, 25]. However, the two modifications may not be mutually exclusive since they may occur simultaneously at different sites. Treatment with insulin-like growth factor-1 increases both phosphorylation and O-GlcNAcylation of Akt1 in SH-SY5Y neuroblastoma cells [8]. Studies on glutathione peroxidase 1 (GPX1) under hyperglycemic conditions suggest that O-GlcNAcylation on GPX1 increases GPX1 phosphorylation and consequently enhances the activity of the protein [26]. To better understand the significance of Akt O-GlcNAcylation, further investigations, including identification and manipulation of O-GlcNAc modification sites, are warranted.
While our present data suggest Akt O-GlcNAcylation is the cause of the growth change, it is also possible that the growth changes in OGT or OGA RNAi flies result from changes in the production of insulin-like growth factors or larval feeding behavior in a cell non-autonomous manner. However, we believe that a direct modification of the insulin signaling component is the main cause. Namely, en-GAL4-driven UAS-OGT RNAi decreased the growth of the wing imaginal discs specifically in the Engrailed expression domain. Furthermore, expression of OGA or OGT RNAi in small cell patches resulted in cell-size changes in the RNAi-expressing cells only. The degree of increase of cell size in the fat body cells of Act>OGA RNAi animals was comparable to that in the cell-patch experiments and similar results were obtained with OGT RNAi. Last but not least, OGT overexpression increased insulin-induced Akt phosphorylation in S2 cells. Thus, the effects of OGA or OGT RNAi occur largely in a cell-autonomous manner. We also believe that the observed phenotypes are not a secondary consequence of abnormal epigenetic regulation, despite that both OGT and OGA may exert their actions as chromatin remodeling factors [11, 12, 27]. This is related to the known relationship between insulin signaling and growth, and that several components of the insulin signaling pathway are modified by O-GlcNAcylation [7–9, 28].
Our present findings point to the intriguing possibility that O-GlcNAcylation is regulated during Drosophila development, which would add another layer of complexity to the regulation of OGT [1]. Therefore, it will be interesting to investigate the regulation of the protein O-GlcNAcylation machinery, including the hexosamine signaling pathway, during development and in various diseased states.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank K. S. Cho, J. Chung, and K. Yu for fly stocks and DNA clones, and O. Puig and K. Yu for providing the anti-dFOXO antibody. We are also grateful to J. Lee and I. Jang for preliminary results on fly O-GlcNAcylation, other members of the Cho and Choe laboratories for helpful discussions, and E. Y. Kim and K. Yu for critical reading of the manuscript. This work was supported by the National Research Foundation (NRF) funded by the Korean Government (2010-0018923 to J.W.C. and 2010-0027736 to J.R.), World Class University Program (R31-2008-000-10086-0 to J.W.C. and J.R.) and Basic Research Program (2010-0008544 to K.-M.C.) through NRF of Korea funded by the Ministry of Education, Science and Technology and partly by KRF-2006-005-J04502 to J.W.C. Further partial support was received by NRF grant funded by the Korea government through the Center for Bioactive Molecular Hybrids (NO. R11-2003-019-00000-0 to K.S.K.). S.P. and S.-H.P are fellowship awardees of the Brain Korea 21 program. This work was made possible through the use of research facilities in the Yonsei Center for Biotechnology.
Conflict of interest
The authors declare no conflict of interest.
Abbreviations
- GPX1
Glutathione peroxidase 1
- NButGT
1,2-Dideoxy-2′-propyl-α-D-glucopyranoso-[2,1-D]-Δ2′-thiazoline
- O-GlcNAc
O-linked N-acetylglucosamine
- OGT
O-linked GlcNAc transferase
Contributor Information
Jin Won Cho, Email: chojw311@yonsei.ac.kr.
Kwang-Min Choe, Phone: +82-2-21235652, FAX: +82-2-3125657, Email: kmchoe@yonsei.ac.kr.
References
- 1.Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 2.Hanover JA, Forsythe ME, Hennessey PT, Brodigan TM, Love DC, Ashwell G, Krause M. A Caenorhabditis elegans model of insulin resistance: altered macronutrient storage and dauer formation in an OGT-1 knockout. Proc Natl Acad Sci USA. 2005;102:11266–11271. doi: 10.1073/pnas.0408771102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forsythe ME, Love DC, Lazarus BD, Kim EJ, Prinz WA, Ashwell G, Krause MW, Hanover JA. Caenorhabditis elegans ortholog of a diabetes susceptibility locus: oga-1 (O-GlcNAcase) knockout impacts O-GlcNAc cycling, metabolism, and dauer. Proc Natl Acad Sci USA. 2006;103:11952–11957. doi: 10.1073/pnas.0601931103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shafi R, Iyer SP, Ellies LG, O’Donnell N, Marek KW, Chui D, Hart GW, Marth JD. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci USA. 2000;97:5735–5739. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Donnell N, Zachara NE, Hart GW, Marth JD. Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol Cell Biol. 2004;24:1680–1690. doi: 10.1128/MCB.24.4.1680-1690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McClain DA, Lubas WA, Cooksey RC, Hazel M, Parker GJ, Love DC, Hanover JA. Altered glycan-dependent signaling induces insulin resistance and hyperleptinemia. Proc Natl Acad Sci USA. 2002;99:10695–10699. doi: 10.1073/pnas.152346899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang X, Ongusaha PP, Miles PD, Havstad JC, Zhang F, So WV, Kudlow JE, Michell RH, Olefsky JM, Field SJ, Evans RM. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature. 2008;451:964–969. doi: 10.1038/nature06668. [DOI] [PubMed] [Google Scholar]
- 8.Gandy JC, Rountree AE, Bijur GN. Akt1 is dynamically modified with O-GlcNAc following treatments with PUGNAc and insulin-like growth factor-1. FEBS Lett. 2006;580:3051–3058. doi: 10.1016/j.febslet.2006.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuo M, Zilberfarb V, Gangneux N, Christeff N, Issad T. O-glycosylation of FoxO1 increases its transcriptional activity towards the glucose 6-phosphatase gene. FEBS Lett. 2008;582:829–834. doi: 10.1016/j.febslet.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 10.Kang ES, Han D, Park J, Kwak TK, Oh MA, Lee SA, Choi S, Park ZY, Kim Y, Lee JW. O-GlcNAc modulation at Akt1 Ser473 correlates with apoptosis of murine pancreatic beta cells. Exp Cell Res. 2008;314:2238–2248. doi: 10.1016/j.yexcr.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 11.Gambetta MC, Oktaba K, Muller J. Essential role of the glycosyltransferase sxc/Ogt in Polycomb repression. Science. 2009;325:93–96. doi: 10.1126/science.1169727. [DOI] [PubMed] [Google Scholar]
- 12.Sinclair DA, Syrzycka M, Macauley MS, Rastgardani T, Komljenovic I, Vocadlo DJ, Brock HW, Honda BM. Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc) Proc Natl Acad Sci USA. 2009;106:13427–13432. doi: 10.1073/pnas.0904638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puig O, Marr MT, Ruhf ML, Tjian R. Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 2003;17:2006–2020. doi: 10.1101/gad.1098703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee KS, Kwon OY, Lee JH, Kwon K, Min KJ, Jung SA, Kim AK, You KH, Tatar M, Yu K. Drosophila short neuropeptide F signalling regulates growth by ERK-mediated insulin signalling. Nat Cell Biol. 2008;10:468–475. doi: 10.1038/ncb1710. [DOI] [PubMed] [Google Scholar]
- 15.Kang JG, Park SY, Ji S, Jang I, Park S, Kim HS, Kim SM, Yook JI, Park YI, Roth J, Cho JW. O-GlcNAc protein modification in cancer cells increases in response to glucose deprivation through glycogen degradation. J Biol Chem. 2009;284:34777–34784. doi: 10.1074/jbc.M109.026351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macauley MS, Whitworth GE, Debowski AW, Chin D, Vocadlo DJ. O-GlcNAcase uses substrate-assisted catalysis: kinetic analysis and development of highly selective mechanism-inspired inhibitors. J Biol Chem. 2005;280:25313–25322. doi: 10.1074/jbc.M413819200. [DOI] [PubMed] [Google Scholar]
- 17.Marenda DR, Vrailas AD, Rodrigues AB, Cook S, Powers MA, Lorenzen JA, Perkins LA, Moses K. MAP kinase subcellular localization controls both pattern and proliferation in the developing Drosophila wing. Development. 2006;133:43–51. doi: 10.1242/dev.02168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu SY, Yoo SJ, Yang L, Zapata C, Srinivasan A, Hay BA, Baker NE. A pathway of signals regulating effector and initiator caspases in the developing Drosophila eye. Development. 2002;129:3269–3278. doi: 10.1242/dev.129.13.3269. [DOI] [PubMed] [Google Scholar]
- 19.Struhl G, Basler K. Organizing activity of wingless protein in Drosophila . Cell. 1993;72:527–540. doi: 10.1016/0092-8674(93)90072-X. [DOI] [PubMed] [Google Scholar]
- 20.Garofalo RS. Genetic analysis of insulin signaling in Drosophila . Trends Endocrinol Metab. 2002;13:156–162. doi: 10.1016/S1043-2760(01)00548-3. [DOI] [PubMed] [Google Scholar]
- 21.Kane S, Sano H, Liu SC, Asara JM, Lane WS, Garner CC, Lienhard GE. A method to identify serine kinase substrates. Akt phosphorylates a novel adipocyte protein with a Rab GTPase-activating protein (GAP) domain. J Biol Chem. 2002;277:22115–22118. doi: 10.1074/jbc.C200198200. [DOI] [PubMed] [Google Scholar]
- 22.Park SY, Ryu J, Lee W. O-GlcNAc modification on IRS-1 and Akt2 by PUGNAc inhibits their phosphorylation and induces insulin resistance in rat primary adipocytes. Exp Mol Med. 2005;37:220–229. doi: 10.1038/emm.2005.30. [DOI] [PubMed] [Google Scholar]
- 23.Sekine O, Love DC, Rubenstein DS, Hanover JA (2010) Blocking O-GlcNAc cycling in Drosophila insulin-producing cells perturbs glucose-insulin homeostasis. J Biol Chem. doi:10.1074/jbc.M110.155192 [DOI] [PMC free article] [PubMed]
- 24.Verdu J, Buratovich MA, Wilder EL, Birnbaum MJ. Cell-autonomous regulation of cell and organ growth in Drosophila by Akt/PKB. Nat Cell Biol. 1999;1:500–506. doi: 10.1038/70293. [DOI] [PubMed] [Google Scholar]
- 25.Yang WH, Kim JE, Nam HW, Ju JW, Kim HS, Kim YS, Cho JW. Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat Cell Biol. 2006;8:1074–1083. doi: 10.1038/ncb1470. [DOI] [PubMed] [Google Scholar]
- 26.Yang WH, Park SY, Ji S, Kang JG, Kim JE, Song H, Mook-Jung I, Choe KM, Cho JW. O-GlcNAcylation regulates hyperglycemia-induced GPX1 activation. Biochem Biophys Res Commun. 2010;391:756–761. doi: 10.1016/j.bbrc.2009.11.133. [DOI] [PubMed] [Google Scholar]
- 27.Toleman C, Paterson AJ, Whisenhunt TR, Kudlow JE. Characterization of the histone acetyltransferase (HAT) domain of a bifunctional protein with activable O-GlcNAcase and HAT activities. J Biol Chem. 2004;279:53665–53673. doi: 10.1074/jbc.M410406200. [DOI] [PubMed] [Google Scholar]
- 28.Patti ME, Virkamaki A, Landaker EJ, Kahn CR, Yki-Jarvinen H. Activation of the hexosamine pathway by glucosamine in vivo induces insulin resistance of early postreceptor insulin signaling events in skeletal muscle. Diabetes. 1999;48:1562–1571. doi: 10.2337/diabetes.48.8.1562. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.