Abstract
Proteins of the ESCRT (endosomal sorting complex required for transport) complex function in membrane fission processes, such as multivesicular body (MVBs) formation, the terminal stages of cytokinesis, and separation of enveloped viruses from the plasma membrane. In mammalian cells, the machinery consists of a network of more than 20 proteins, organized into three complexes (ESCRT-I, -II, and -III), and other associated proteins such as the ATPase vacuolar protein sorting 4 (Vps4). Early biochemical studies of MVBs biogenesis in yeast support a model of sequential recruitment of ESCRT complexes on membranes. Live-cell imaging of ESCRT protein dynamics during viral budding and cytokinesis now reveal that this long-standing model of sequential assembly and disassembly holds true in mammalian cells.
Keywords: ESCRT machinery, Viral budding, Cytokinesis, Live cell imaging
Introduction
The mammalian ESCRT machinery mediates membrane curvature and fission processes, such as the genesis of multivesicular bodies (MVBs) from late endosomal membranes and budding of enveloped viruses from the plasma membrane. They are also involved in two different stages of cell division: the separation of the two daughter cells during the late steps of cytokinesis and centrosome maintenance [1–3]. Cytokinesis, virus budding, and MVB biogenesis involve membrane separation. At the end of the separation process, the two membranes remain connected by a thin ‘neck’ whose severing depends on ESCRT proteins (Fig. 1). The role played by ESCRT proteins in centrosome duplication remains to be determined.
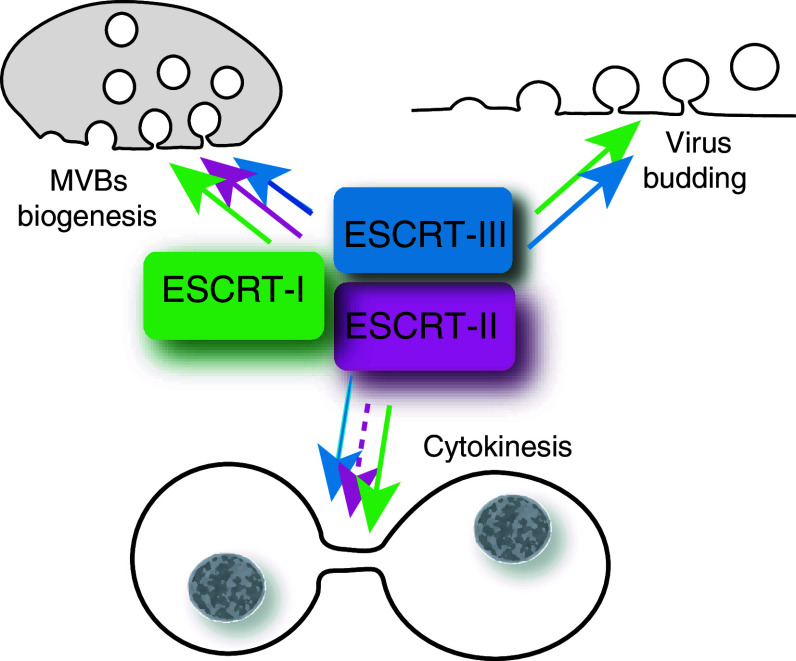
Fig. 1.
Requirement of ESCRT complexes for MVBs biogenesis, viral budding, and cytokinesis. ESCRT proteins mediate membrane fission events such as MVBs biogenesis, viral budding, and late stages of cytokinesis. Despite similar membrane topology, these processes differ in their requirements for the three ESCRT complexes. The ESCRT-II complex is dispensable for HIV-1 budding and its role in cytokinesis is not well defined
Since its discovery in Saccharomyces cerevisiae in 2001 [4], the ESCRT pathway has been intensively studied. Over 500 papers have been published on the structure and function of the different proteins of the network, and these data have been compiled in dozens of excellent reviews (see references [5–8] for recent ones). Biochemical studies of MVB biogenesis in yeast support a model of sequential recruitment of ESCRT complexes at the site of membrane fission. Direct evidence for such dynamic recruitment was lacking until four live-cell imaging studies revealed the kinetic of ESCRT assembly and disassembly during viral budding and cytokinesis [9–12]. These recent studies are the focus of this review.
Overview of the mammalian ESCRT machinery composition and function
The mammalian ESCRT machinery is composed of three multiprotein complexes, termed ESCRT-I, -II, and -III, associated with proteins such as Alix, IST1, the ATPase Vps4 and its cofactor Lip5. The machinery is recruited at sites of function by direct interaction with specific adaptor proteins, such as Hrs for MVB biogenesis and Gag proteins for retroviral budding. The centrosomal protein Cep55 is another of these adaptor proteins and, as for Hrs and Gag, recruits the ESCRT machinery by direct interaction with ESCRT-I proteins.
The mammalian ESCRT-I complex is composed of five proteins: Tsg101, Vps28, Vps37, MVB12, and UBAP1 [13–18]. The soluble form of the complex contains a single copy of each of the different subunits [16, 19]. Tsg101 links membrane-bound proteins, such as Gag and Hrs, to the downstream complexes ESCRT-II and -III. Alix, which is often classified as an ESCRT-I-associated protein, is composed of three distinct domains: an N-terminal Bro1 domain, a coiled-coil V domain, and a C-terminal proline rich domain (PRD) [20–22]. The Bro1 domain, which folds into a banana shape, is a good candidate for induction of the negative curvature required for virus and MVBs budding [20–24]. It interacts with C-terminal residues of the ESCRT-III Chmp4 proteins [25]. The central domain of Alix folds into a V-shaped structure and is involved in interactions with retroviral Gag proteins [21, 22]. The PRD interacts with several cellular factors including Tsg101 and Cep55 [1, 2, 22, 26–28].
Endosomal sorting complex required for transport-II can be isolated as a stable, soluble complex composed of three proteins: EAP30, EAP20, and EAP45. Endosomal sorting complex required for transport-II proteins bind to one another, interact with Tsg101, and with the ESCRT-III Chmp6 [29]. Therefore, they likely function as a linker between ESCRT-I and -III. However, certain ESCRT-mediated processes, such as HIV-1 budding, do not require ESCRT-II [29] (Fig. 1), suggesting that the entire ESCRT-II complex is dispensable in this context and that some other links between ESCRT-I and -III complexes exist. In vitro analyses using purified proteins and giant unilamellar vesicles show that the combination of purified ESCRT-I and -II trigger the formation of MVB-like buds, even at concentrations that are below physiological concentrations in yeast [30]. Endosomal sorting complex required for transport-I and -II colocalize almost exclusively to the necks of such buds and seem to act by stabilizing the bud neck.
The core of the mammalian ESCRT-III complex is composed of 11 charged MVB proteins (Chmp) and of the associated protein IST1. Charged multivesicular body protein proteins are grouped into seven families based on primary sequence similarities (Chmp 1 to 7). They share structural features allowing them to cycle from a resting closed conformation to an activated open one [31, 32]. This conformational change triggers polymerization at the site of membrane fission into membrane detergent-insoluble helical filaments with dome-like caps [10, 33–35]. The assembly of such dome-like filaments would trigger membrane constriction, leading to extreme narrowing of the bud neck at the top of the dome and eventually membrane severing [6, 34]. This dome-like model is particularly appealing since it has been validated by computational approaches [36]. The accessory protein IST1 binds to Vps37, Chmp1, and also to Vsp4 and its cofactor LIP5 [37, 38], possibly consolidating the interaction between the ESCRT-I complex and the downstream ESCRT-III and Vps4 complexes.
Vacuolar protein sorting 4, the only enzyme in the ESCRT machinery, is recruited by ESCRT-III/Chmp1–3 [39, 40]. Vacuolar protein sorting 4 is a type 1 AAA ATPase that assembles into a double-ringed complex with six subunits per ring [41–46]. Early yeast studies showed that expression of Vps4 proteins lacking ATPase activity triggers the formation of enlarged endosomal structures called class E compartments from which ESCRT complexes are unable to dissociate [47, 48], suggesting that Vps4 catalytic activity is required to recycle ESCRT complexes from site of membrane fission. Indeed, disassembly and recycling of the ESCRT machinery by Vps4 have been reconstituted in vitro [30, 33, 49–51]. The Vps4-cofactor Lip5 stimulates subunit assembly and ATPase activity [52, 53]. Structural and cryo-electron microscopy (EM) studies [41–46] suggest a model in which the ESCRT machinery is recycled after being pulled trough the central pore of the Vps4 dodecameric ring, releasing them as soluble molecules into the cytosol. Therefore, Vps4 recycling might participate in membrane scission per se by gradually forcing ESCRT-III filaments through its central pore. Consistent with this model, in the presence of a dominant negative version of Vps4 [47, 48], ESCRT proteins accumulate at the sites of retrovirus budding but are unable to catalyze the fission reaction [11].
Therefore, ESCRT-I and ESCRT-II complexes probably function as linker and stabilizer molecules, but might also be responsible for the membrane curvature leading to neck formation, while ESCRT-III complex functions more directly in membrane fission. In addition to its recycling function, Vps4 might also play an active role in membrane fission. Consistent with a key role for the ESCRT-III and Vps4 subunits in membrane fission per se, an ESCRT machinery, composed of homologues of ESCRT-III and Vps4, is capable of mediating cell division in one phyla of Archaea [54–56]. Moreover, the presence of a functional ESCRT apparatus in such prokaryotic organisms that lack endomembrane structures suggests that the ancestral function of the ESCRT pathway is to mediate cell division, and that the pathway has evolved to diversify in other cellular functions in Eukaryotes.
Yeast biochemical data define the model of sequential recruitment of ESCRT complexes on membranes
Protein sorting along the endocytic pathway requires their sequestration within MVBs. These vesicles are generated by invagination of the limiting membrane of late endosomes and bud into the lumen of the organelle [57]. Mature MVBs subsequently deliver their content to the lumen of lysosomes by fusing with lysosomal membranes. Proteins are then degraded by hydrolytic enzymes [58].
Biochemical and immunofluorescence studies of MVBs biogenesis in yeast have defined the long-standing model of the sequential recruitment of ESCRT complexes on membranes. First, microscopy showed that deletion of the Vps4 gene triggers the formation of aberrant endosomal structures on which ESCRT components are sequestered [47, 48]. Therefore, ESCRT proteins are transiently associated with endosomal membranes and their dissociation from these membranes is dependent on Vps4. Loss of ESCRT-I or ESCRT-II functions impair formation of MVBs, which leads to the accumulation of endosomal cargo in aberrant class E compartments. The ESCRT-I phenotype is partially suppressed when ESCRT-II is overexpressed but the reverse is not true, suggesting that ESCRT-I functions upstream of ESCRT-II. In a Vps4Δ context, immunofluorescence experiments revealed that mutation in ESCRT-II genes dramatically affected the endosomal accumulation of ESCRT-III proteins, suggesting that ESCRT-II regulates the formation of a membranous ESCRT-III complex [47, 48]. Consistent with this, subcellular fractionation assays showed that, in the absence of ESCRT-II, -III proteins remain in a soluble cytoplasmic fraction and do not accumulate into these aberrant endosomal membranes. Based on these early studies, it was proposed that the ESCRT machinery assembles onto membranes in an orderly fashion, with ESCRT-I complex recruited first and ESCRT-III complex recruited last and that the disassembly of the machinery is mediated by Vps4 [4, 47, 48].
More recently, similar biochemical and immunofluorescence microscopy techniques were exploited to determine the sequence of ESCRT-III subunits recruitment on yeast MVBs [59]. Vacuolar protein sorting 20, Snf7, Vps24, and Vps2 form the core of the yeast ESCRT-III complex; they are homologs of the human proteins Chmp6, Chmp4, Chmp3, and Chmp2, respectively. C-terminal GFP fusions of these four ESCRT-III subunits were generated by chromosomal integration. As in mammalian cells [27, 32, 60], ESCRT-III–GFP exhibited a dominant-negative phenotype, triggering the accumulation of the fusion proteins on aberrant endosomal structures. Analysis of the location of these ESCRT-III–GFP subunits by microscopy, in the presence of absence of the other three ESCRT-III core proteins, revealed which ESCRT-III subunits localization on endosomes is dependent on other ESCRT-III subunits presence. For instance, Vps20/Chmp6–GFP localized to the cytoplasm and to endosomes, without the need of the other ESCRT-III subunits, while Snf7/Chmp4–GFP localization on endosomes was dependent on Vps20/Chmp6 [59]. Such analysis demonstrated that the ESCRT-III complex assembles in a sequential manner on endosomal membranes and that Vps20/Chmp6-dependent recruitment of Snf7/Chmp4 may be the first step in the formation of the ESCRT-III complex on membranes. Consistent with this, velocity sedimentation centrifugation and cross-linking experiments on membrane or cytosol fractions revealed that the cytosolic Snf7/Chmp4 sediments in a low molecular weight fraction (M r < 156 kDa) that did not crosslink and that membranous Snf7/Chmp4 sediments in a large fraction (about 450 kDa) that crosslinked into a stable complex [59]. These results suggest the existence of two Snf7/Chmp4 pools: a cytoplasmic one and endosomal one which is part of a 450-kDa complex. Therefore, Vps20/Chmp6 functions as a nucleator, which is required to induce Snf7/Chmp4 oligomerization on endosomes. Using similar approaches, it was shown that recruitment on endosomes of Vps2/Chmp2 and Vps4 depend on Vps24/Chmp3 [59]. Therefore, by contrast to ESCRT-I and -II, which are recruited to membranes as pre-formed complexes, the ESCRT-III components are recruited in a sequential manner to endosomes where they will assemble in a 450-kDa stable complex.
Altogether, these yeast MVB studies suggest that the interaction between ESCRT complexes and endosomal membrane is highly dynamic. However, such studies did not resolve the kinetics of ESCRT assembly and disassembly. Until recently, there was no direct observation available to support the model of sequential recruitment of ESCRT proteins at site of membrane fission.
Dynamics of ESCRT protein recruitment during cytokinesis
As during MVBs biogenesis and viral budding, a thin membrane tubule must be severed from within during the final abscission step of cytokinesis [61]. The center of this thin membrane tubule is a dense protein-rich ring-like structure called the Flemming body. This ring is surrounded on each side by compact microtubule bundles that run parallel to the membranes. The Flemming body and the two microtubule bundles define the midbody. Centrosomal protein 55 is one of many residents of the midbody [62] and is required for midbody organization and for the completion of cytokinesis [63–65].
Dynamic of abscission
Time-lapse imaging of cells expressing fluorescently tagged version of α-tubulin enables to investigate the temporal kinetic of abscission, based on midbody-associated microtubule disassembly [66]. This approach was exploited in Madin–Darby canine kidney cells (MDCK) and in HeLa cells to show that abscission starts with the gradual narrowing of one microtubule bundle and ends with its complete disassembly; a process that lasts approximately 20 min. Disassembly of the first microtubule bundle and membrane severing are well synchronized and occur at the constriction zone, where the intracellular bridge is at its thinnest [9, 10, 67]. The second microtubule bundle disassembles around 20 min later. Abscission consists therefore on two sequential cuts on each side of the midbody [9, 10, 67].
ESCRT protein requirements for cytokinesis
In 2007, yeast two-hybrid assays and proteomic analyses revealed that Alix and Tsg101 bind a series of proteins involved in cytokinesis, including Cep55. Small interfering RNA (siRNA) approaches have shown that both Tsg101 and Alix are required for midbody abscission. Furthermore, overexpression of YFP–Tsg101 and YFP–Alix fusion proteins, which act as dominant-negative proteins, increases the number of multinucleated cells, reflecting a defect in cytokinesis [1, 2]. Endosomal sorting complex required for transport-II subunits localize to centrosome [29, 68], but depletion of the ESCRT-II subunit EAP20 do not lead to significant cytokinesis defects [2]. The role of ESCRT-II in cytokinesis remains therefore to be determined (Fig. 1). Depletion of the 11 different human Chmp proteins inhibited abscission [3], implying non-redundant role for individual ESCRT-III in cytokinesis. Over-expressing fluorescent-tagged version of Chmp4 isoforms identified Chmp4c as the most potent inhibitor of cytokinesis [1, 2, 69]. More evidence supporting the essential role of ESCRT-III in abscission is provided by the cytokinetic defects in cells expressing an Alix mutant lacking the Chmp4-binding site [1, 2, 69]. The ESCRT-III-associated protein IST1 plays a functional role in abscission since its depletion causes daughter cells to remain connected through their midbody for a prolonged period of time before eventually re-uniting into multinucleated cell [37, 38]. Vacuolar protein sorting 4 ATPase activity is also required for efficient cytokinesis since expression of a dominant-negative version of Vps4 results in a dramatic increase in the percentage of cells arrested in telophase with multiple nuclei [2].
Localization of ESCRT proteins at the midbody
Epifluorescence microscopy revealed that the tagged version of Tsg101, Alix, and Cep55 localized to the Flemming body, so is endogenous Alix [1, 2, 63–65]. C-terminally flag-tagged Chmp2A, Chmp4A, and Chmp5 proteins are also recruited to midbodies during cell division but they localized into two distinct rings, one on either side of the Flemming body (Fig. 2). Endogenous Vps4 also forms discrete bands on either side of the midbody. Its recruitment was only observed when the midbodies were particularly thin, suggesting that Vps4 may only be recruited at the final stages of cytokinesis [3]. IST1 localization at the midbody during cytokinesis matches that of Vps4 [37, 38].
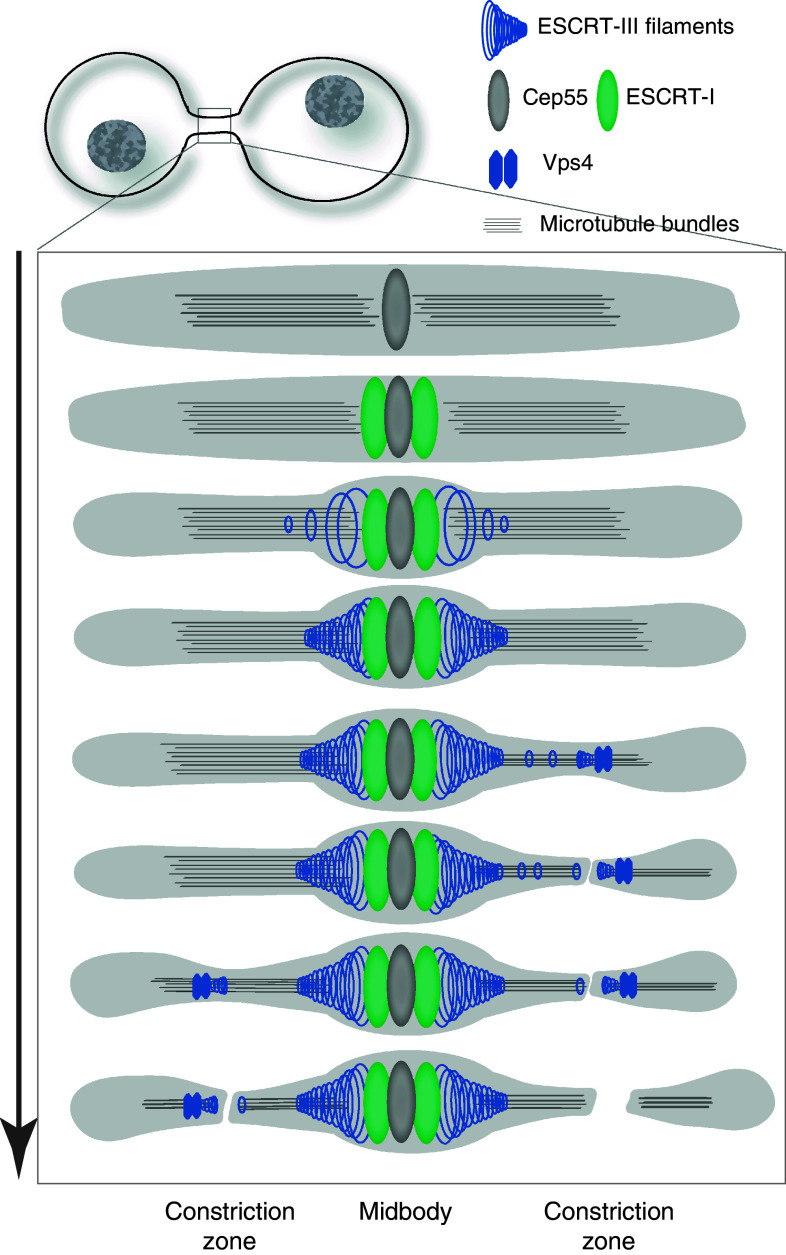
Fig. 2.
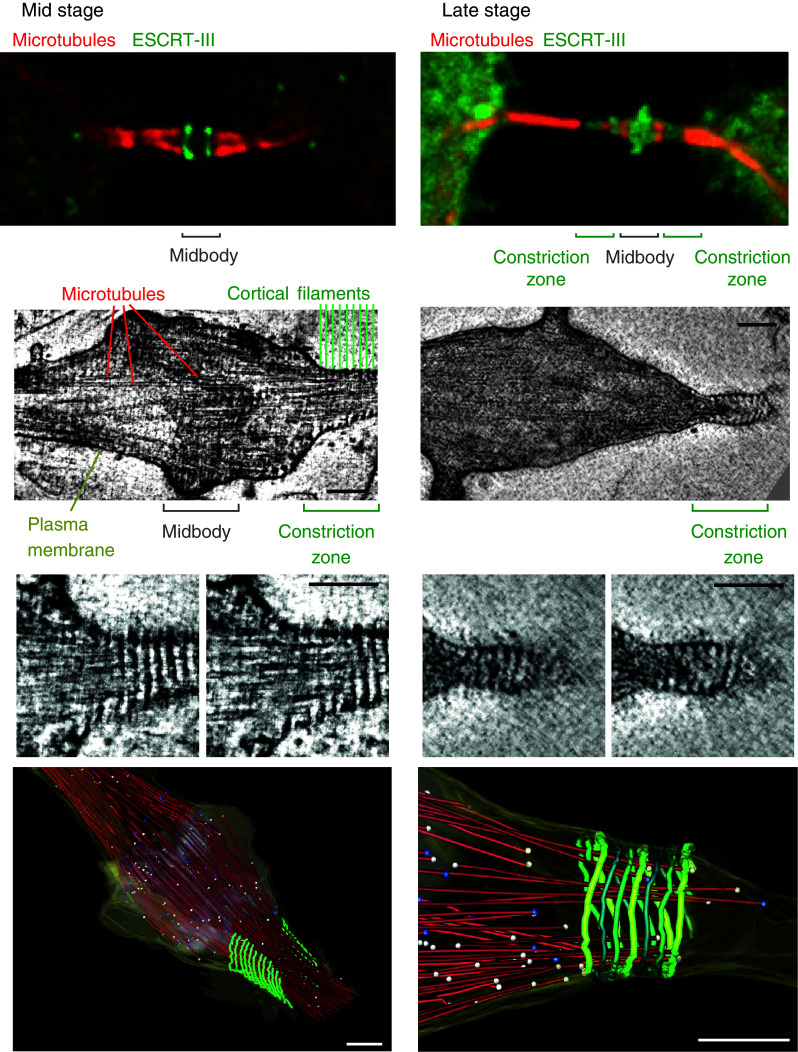
Localization of ESCRT proteins at the midbody. Top panels show the distribution of ESCRT-III proteins at the midbody of dividing HeLa cells stably expressing GFP–Chmp4b (green, mid stage) or GFP–Chmp2a (green, late stage). Samples were stained with anti-α-tubulin antibodies (red) and observed with an epifluorescence microscope. Deconvolved optical sections acquired at the center of the vertical dimension of the cell are shown. Middle panels electron tomography of high-pressure frozen cells reveals cortical filaments at constriction zone of mid- and late-abscission stages. The smaller images are enlarged views of two z-sections of the above corresponding images. Scale bar 100 nm. Bottom panels three-dimensional reconstructions of early and late-stage abscission structures. Red microtubules, green 17-nm-diameter filaments, gray midbody, yellow plasma membrane, white balls open microtubule ends, blue balls closed microtubule ends. Scale bars 200 nm. Middle and bottom panels were reproduced from reference ten with permission
Structured illumination microscopy of cells expressing a low level of fluorescently tagged version of ESCRT proteins revealed the layout of ESCRT proteins at the midbody in finer details [9, 10]. Alix and Cep55 accumulate in the core of the midbody whereas Tsg101 localizes in two distinct, hollow rings located on each side of the midbody centre. In early stage intercellular bridges, Chmp4b, Chmp6, and Chmp2a also localize in two distinct ring-like structures located on each side of the midbody center (Fig. 2), but these were located further away from the core of the midbody. These observations were confirmed for endogeneous Chmp4b and Chmp2b proteins [10]. Such layout suggests an orderly recruitment. In thinner, later-stage intercellular bridges, ESCRT-III extended toward the constriction zone (Fig. 2).
Electron tomography analysis of the intercellular bridge revealed that the constriction zone is spanned by 17-nm-wide helical structures [10] (Fig. 2). These structures do not assemble in the absence of ESCRT-III complex and are therefore probably made of polymerized ESCRT-III proteins. Such helices might generate the contractile force to progressively narrow the membrane tube of the intercellular bridge until abscission occurs.
Dynamics of ESCRT protein recruitment
The dynamic of ESCRT protein recruitment during cytokinesis was recently analyzed by imaging live cells transiently or stably expressing low level of fluorescently tagged version of ESCRT-III and α-tubulin [9, 10]. In MDCK cells, Cep55 and its binding partner MKLP1 were present at the core of the midbody from the start of cytokinesis and remain there during the entire cytokinesis process [9] (Fig. 3). Their levels increased moderately around 90 min before the first microtubule severing and then stabilize. By contrast, Tsg101 is recruited in the mid-phase of cytokinesis, around 60 min before completion of abscission and its level steadily rises until the end of the process [9]. Charged multivesicular body protein 4b is also recruited in the mid-phase of cytokinesis at the midbody, at around the same time as Tsg101, then its level reaches a plateau until a second recruitment step occurs in the late phase of cytokinesis. This second wave of recruitment probably corresponds to the Chmp4b pool that accumulates further away from the core of the midbody, at the constriction site. Charged multivesicular body protein 4b reaches its peak level approximately 15 min before the completion of abscission and is followed by an acute decrease in microtubule diameter that ends in membrane severing. Vacuolar protein sorting 4 recruitment and peak level are occurring closer to the completion of abscission, with an average of 10-min delay compared to Chmp4b recruitment and peak [9].
Fig. 3.
Kinetics of ESCRT protein recruitment during cytokinesis. Cep55 and its binding partners are recruited first at the midbody and remain there during the entire cytokinesis process. ESCRT-I and associated proteins, such as Tsg101 and Alix, are recruited in the mid-phase of cytokinesis, and their level steadily rises until the end of the process. ESCRT-III are also recruited in the mid-phase of cytokinesis at the midbody, at around the same time as Tsg101. In the late phase, their levels increased dramatically and their localization extends toward the first constriction site, where they probably polymerize into helical filaments. ESCRT-III peak level is concomitant with an acute decrease in microtubule diameter and is followed by Vps4 recruitment. ESCRT-III redistribution leads to membrane severing and separation of the two daughter cells
In HeLa cells, Cep55 was recruited to the midbody 60 min before the first microtubule bundles disassembly, and its concentration continues to increase until 10 min prior to abscission [10]. Alix recruitment at the midbody occurs approximately 10 min after that of Cep55 and continues to accumulate until few minutes before abscission. Charged multivesicular body protein 4b was present at very low level within early stage intercellular bridges. Then, 40 min before disassembly of the first microtubule bundle, its levels increased dramatically and its localization extend toward the constriction site [10]. Therefore, as in MDCK cells, Cep55 is recruited to the midbody before Alix and Chmp4b.
These data clearly demonstrate that the ESCRT proteins are sequentially recruited at the midbody (Fig. 3).
Chmp4c controls the abscission checkpoint
Timing is crucial for proper abscission, yet very little is known about the mechanisms that control it. Based on the observation that Chmp4c-depleted cells exhibited a low number of cells connected by midbodies, Carlton and colleagues [70] investigate the possibility that midbodies were resolved faster in cells lacking Chmp4c. Indeed, depletion of Chmp4c, but not Chmp4-a or -b, reduced abscission time by approximately 30 min and over-expression of GFP–Chmp4c resulted in an abscission delay. Comparison of Chmp4 sequences revealed a Chmp4c-specific insertion that resembles canonical phosphorylation sites. Interestingly, the kinase Aurora B, whose activation delays cytokinesis [67] is able to phosphorylate Chmp4c at this specific site, which is also required for Chmp4c localization at the Flemming body [70]. Moreover, Aurora B inhibitor reduced Chmp4c phosphorylation. Altogether, these compelling data suggest that Aurora B-dependent phosphorylation of Chmp4c directs it to the Flemming body, a recruitment that leads to abscission delay, possibly by preventing assembly of a productive abscission complex [70].
Therefore, individual ESCRT-III proteins have specialized into specific function in the late stage of cell division: Chmp2, Chmp3, and Chmp4a/b are probably responsible for the formation, within the intercellular bridge, of helical filaments whose constriction leads to narrowing and severing [10, 33, 51, 71, 72], whereas Chmp4c ensures that the timing of these events are correct [70]. Ordered recruitment of ESCRT-III proteins at site of membrane fission may have facilitate such specialization.
Dynamics of ESCRT protein recruitment during virus budding
Retroviral assembly is a highly regulated process that requires the viral structural components, as well as some key cellular factors, such as ESCRT proteins, to converge to the correct cellular site at the correct time. HIV-1 virions assemble into a ~100-nm-diameter particle containing few thousand Gag proteins [73], one 20th of which are Gag–Pol proteins [74], 8–10 envelope trimers [75] and two copies of the viral genome [76] (Fig. 4).
Fig. 4.
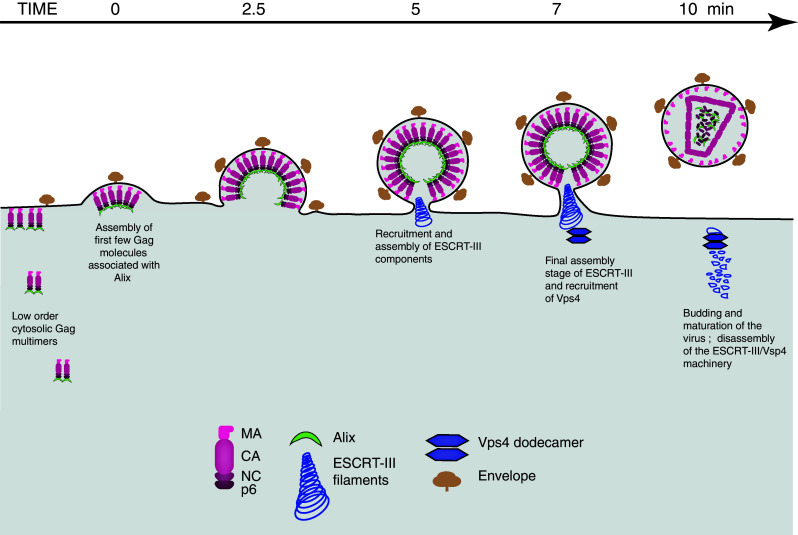
Kinetic of ESCRT protein recruitment during retroviral assembly and budding. Retroviral assembly is a highly regulated process that requires the viral structural components, such as Gag and envelope, as well as cellular factors, such as ESCRT proteins, to converge to the correct site at the correct time. Assembly is initiated at the plasma membrane by few Gag molecules associated to the viral genome (not represented) and is completed by the recruitment of thousand more Gag molecules. The whole assembly process, from the detection of the first few Gag molecules to end of Gag recruitment, takes an average of 10 min. Alix progressively accumulate at viral assembly sites, coincident with the accumulation of Gag, and is not recycled after assembly. In contrast, ESCRT-III and Vps4 are recruited when assembly is completed and are recycled. They stay on average 2 min at site of budding. The four Gag subunits, matrix (MA), capsid (CA), nucleocapsid (NC) and p6 become cleaved from one another upon the action of the viral protease—a process that happens during or shortly after assembly and leads to the formation of mature infectious virions
The assembly process is orchestrated by the polyprotein Gag whose expression is sufficient to generate defective viral-like particles (VLPs) that are morphologically indistinguishable from the immature virions produced by infected cells [77]. Gag contains four domains that become cleaved from each other upon the action of the viral protease; a process that happens during or shortly after assembly and leads to the formation of mature particles [78]. The N-terminal matrix (MA) domain targets Gag to the inner leaflet of the plasma membrane, the capsid (CA) domain drives Gag multimerization, the nucleocapsid (NC) domain packages the dimeric RNA genome and the C-terminal p6 region recruits ESCRT complexes for particle release [78].
The very first stage of assembly occurs in the cytosol with the recognition of the genomic RNA by low-order multimer Gag proteins [79] via direct interaction between NC and highly structured sequences located in the 5′-untranslated region of the genomic RNA [80]. The Gag-RNA complex then traffic to the plasma membrane where its interaction with the membrane triggers higher order Gag multimerization and subsequent particle assembly and release [79, 81–85].
ESCRT proteins requirements for HIV-1 budding
Divergent enveloped viruses, such as Filoviruses, Rhaboviruses, Herpesviruses, Paramyxoviruses, and Arenaviruses exploit the ECRT machinery to bud off the plasma membrane [86–88]. Endosomal sorting complex required for transport proteins are recruited at site of budding via direct interaction with so-called late domain motifs that lie within viral structural proteins. There are three known types of late domain that recruit ESCRT proteins via direct interactions. PTAP and YPDL sequences recognize Tsg101 [89, 90] and Alix [22, 27, 60], respectively, both of which interact with ESCRT-III proteins [27, 91]. PPXY motifs interact with ubiquitin ligases, such as WWP-1 [92], which connect with the class E machinery through ubiquitin chains linked to any viral or cellular substrates within the machinery [93, 94]. Arrestin-related trafficking (ART) proteins might also be involved in PPXY-dependent viral budding, by linking ubiquitin ligases to ESCRT-I component. Alternatively, consistent with a role for ubiquitin in PPXY-dependent viral budding, ubiquitination of ARTs by ubiquitin ligases might trigger the recruitment of the ESCRT machinery on PPXY motifs [95].
The HIV-1 main structural protein, Gag, possesses a PTAP motif that acts as major late domain by recruiting Tsg101 [89, 90, 96]. Two accessory late domains, which recruit Alix, are found in Gag p6 [60] and in the zinc fingers of NC [97–99]. Depletion and over-expression off Tsg101, Vps28, Vps37, and MVB12 inhibit HIV-1 infectivity and induce viral assembly defects, including aberrant virion morphologies and altered viral Gag protein processing [15, 16, 18, 26, 89, 90], demonstrating that these four members of the ESCRT-I complex play a functional role in virus release. By contrast, efficient siRNA depletion of UBAP1 does not alter significantly HIV-1 budding [19]. HIV-1 release and infectivity were not reduced either by depletion of EAP20/ESCRT-II [29], suggesting that ESCRT-II complex is dispensable for HIV-I budding. Therefore, the recruitment of ESCRT-III by HIV-1 remains unclear. Alix could well be the protein bridging ESCRT-I and -III components, since it interacts with both Tsg101 and CHMP4 isoforms [25, 27]. However, Alix clearly plays a minor role in HIV-1 release since its depletion barely affects virion release [27, 100]. Consistent with this, expression of a Chmp4b variant lacking Alix binding site in cells silenced for endogenous Chmp4b has a mild effect on the efficiency of HIV-1 release [101]. However, in an artificial system where HIV-1 Gag Tsg101 binding site is mutated, Alix over-expression potently rescues the release defect of mutants [22, 102], demonstrating that Alix can support HIV-1 budding. This effect of Alix depends on its interaction with the LYPxnL motif in p6, confirming that this motif constitutes a functional late domain. The Bro1 domain of Alix also interacts with NC zinc fingers and mutations that interfere with this interaction lead to a phenotype that resembles that of L domain mutants [97, 99, 103]. In this case, the Alix Bro1 domain is interchangeable with widely divergent Bro1 domains [97]. Therefore, the exact role of Alix in HIV-1 budding remains to be clearly defined, and so is the molecular basis for ESCRT-III recruitment.
Over-expression of most ESCRT-III proteins as fusion proteins drastically inhibits HIV-1 release [27, 60, 91], suggesting a requirement for budding. However, over-expression can lead to sequestration of ESCRT proteins on endosomal membranes and therefore might have an indirect effect on viral release. siRNA approaches have recently allowed a fresh look at the requirement of individual ESCRT-III proteins for budding [101]. Depletion of any individual Chmp protein had a weak effect on the efficiency of viral release, suggesting functional redundancy amongst the Chmp family members. By contrast, when cells were co-depleted of Chmp2 or Chmp4 family members, HIV-1 release was largely inhibited. Moreover, the function of Chmp4 and Chmp2 in release depends on their direct interaction. Co-depletion of other Chmp family members had only minor effects on HIV-1 release. The associated ESCRT-III protein IST1 plays no direct role in HIV-1 budding [37, 38].
Altogether, these studies demonstrate that a subset of ESCRT proteins, including ESCRT-I, Chmp2, and Chmp4 families, constitute the minimal ESCRT requirement for HIV-1 release.
Localization of ESCRT proteins at site of viral budding
In cells stably expressing low level of GFP–Chmp4b, GFP–Chmp4c or Chmp1b–GFP and transiently transfected with HIV-1 Gag and Gag–mCherry, only few percent of fluorescent virions were associated with ESCRT proteins at a giving time. When a catalytically inactive version of Vps4 was added in the system, GFP–ESCRT-III proteins were detectable at more than 75 % of fluorescent virions [11]. These results indicate that the association between the Chmp proteins and Gag are transient, but stabilized when Vps4 activity was inhibited. Immuno-EM analysis of ESCRT protein distribution within the neck of budding virions are not available. However, ring-like striations can be observed within the membrane stalks of budding viruses arrested by dominant-negative Vps4 proteins. These striations probably represent trapped ESCRT machinery, since they are not detectable within the stalks of viruses lacking late-domains [91]. Moreover, EM images of budding virions in Chmp2a/b-depleted cells reveal ring-like electron-dense structures within the membrane neck of virions that fail to separate from the producer cells [101]. Charged multivesicular body protein 4 is known to form circular filaments [71] and these structures probably represent Chmp4 filaments that have not yet been recycled. Therefore, Chmp4 is recruited prior to Chmp2 at site of viral budding and Chmp2 is required for Chmp4 recycling, probably by recruiting Vps4. These EM and epifluorescence analysis suggest that the recruitment of ESCRT-III proteins by Gag is transient and ordered.
Dynamics of HIV-1 and EIAV assembly
The HIV-1 main structural protein, Gag, possesses a PTAP motif that acts as major late domain by recruiting Tsg101 [89, 90, 96], whereas the Gag protein of Equine infectious anemia virus (EIAV), a lentivirus that infects horses [104], encodes a YPDL late domain motif, which binds Alix with high affinity [60]. The assembly of HIV-1 and EIAV VLPs is driven by Gag proteins and takes place at the plasma membrane [83–85]. By using fluorescently tagged derivatives of Gag and total-internal-reflection fluorescent microscopy (TIRFM) techniques, the kinetics of HIV-1 and EIAV assembly have been elucidated [11]. Total-internal-reflection fluorescent microscopy results in a selective excitation of fluorophores within 100 nm of the coverslip [105], therefore minimizing photodamage to the cell and maximizing the signal over background of fluorophores at the cell surface. Total-internal-reflection fluorescent microscopy is a powerful tool to study events that occur at the plasma membrane, including retroviral assembly. TIRFM approaches revealed that the fluorescent intensity generated by Gag during the genesis of VLPs, represented by a punctum, increases steadily over the diffuse Gag background until it reaches its peak of intensity and remains stable (Fig. 5) [11, 81, 106]. Quantification of Gag proteins proximity by FRET demonstrates that Gag become more and more closely packed within virions as assembly proceeds [106]. Fluorescence recovery after photobleaching analysis showed that Gag recruitment into nascent virions becomes irreversible once it reaches a plateau [106], indicating the end of the assembly process. Such techniques have allowed the direct observation of the genesis of thousand of individual virions in real time, from initiation to completion of assembly. Virions appear individually at the plasma membrane, and an average of 9 min is required to complete the assembly of a single HIV-1 VLPs [106]. Importantly, viral particles generated in the context of a full viral genome, in which GFP is inserted into the flexible C-terminus of the matrix region of Gag, assemble with broadly similar kinetics [106, 107]. Similarly, EIAV Gag assembly was completed in a mean of 11.5 min [11].
Fig. 5.
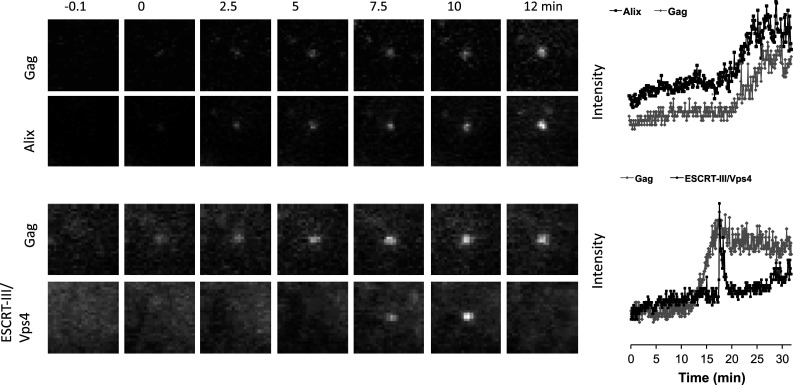
Typical recruitment of Alix and Chmp/Vps4 during equine infectious anemia virus assembly. HeLa cells stably expressing GFP tagged version of ESCRT proteins, such as Alix (top panels) or Chmp/Vps4 (bottom panels) were transfected with HIV-1 Gag/Gag-mCherry and observed under TIR-FM beginning at 6 h post-transfection. Each set of images illustrates the recruitment of ESCRT proteins during the genesis of an individual virion. The time ‘zero’ is set to correspond to the initial Gag detection. Fields are 2.5 × 2.5 μm. The plots represent the fluorescence intensity in arbitrary units emitted by GFP–ESCRT protein (black) and by Gag-mCherry signals (grey) associated with the assembly of an individual virion over time
One caveat into using the very first detectable Gag signal over background as a reference for the start of assembly is that this measurement is dependent on the resolution of the microscope used. Visualization HIV-1 genomes using a technique involving an RNA binding protein, from the bacteriophage MS2, fused to GFP [108] have allowed a better resolution of the very first step of viral assembly [81]. MS2–GFP-bound genomes that subsequently initiate VLP assembly were detectable in the TIR field on average 5 min before detectable levels of Gag accumulated on this Gag-RNA subvirion complex. Therefore, HIV-1 assembly, from genuine initiation (i.e., the docking of a small number of Gag proteins associated with the genome), to end of Gag recruitment is completed on average within 14–15 min [81].
Such dynamic studies of retroviral assembly provide a model system to dissect the recruitment of ESCRT proteins at sites of membrane fission.
Dynamics of ESCRT protein recruitment
Over-expression of fluorescently tagged forms of ESCRT proteins can be deleterious to cell physiology and virion release [18, 27, 60, 109, 110] and was therefore a potential limitation for live-cell analysis of ESCRT-dependent processes. This limitation was bypassed by generating cell lines stably expressing low level of fluorescent version of these proteins [11]. In these cell lines, GFP-tagged ESCRT proteins exhibited similar localization as their endogenous counterpart: they accumulated at midbody during cytokinesis and were recruited to class E compartments induced by expression of a catalytically inactive Vps4. The expression level of GFP-tagged ESCRT proteins in the stable lines was close to the expression level of the native protein and consistent with this, cell division and viral release were not affected. Monitoring the recruitment of ESCRT proteins during the assembly of HIV-1 and EIAV by time-lapse TIRFM microscopy using these cell lines revealed two completely different types of recruitment for ESCRT proteins at the site of viral budding [11].
Alix and EIAV Gag protein signals were perfectly synchronized during assembly, increasing in perfect correlation and reaching a plateau at the same time (Fig. 5). This suggests that the amount of Alix that is recruited at EIAV assembly site is proportional to the number of Gag (Fig. 4). Once viral assembly was completed, Alix did not dissociate from EIAV VLPs, which is consistent with the detection of Alix in purified EIAV and HIV-1 virions [60, 91]. By contrast, the amount of Alix and Tsg101 that are recruited at HIV-1 assembly site is under the detection limit of the TIR microscope, suggesting a different mechanism of ESCRT-I recruitment by HIV-1 Gag. This is agreement with the estimation of the numbers of ESCRT-I components incorporated into highly purified HIV-1 from infected T-cells. On average, 5–25 Vps28 molecules are incorporated into each virus particle [18] and since ESCRT-I complex is a 1:1:1:1 heterotetramer [16], the amount of Tsg101 per HIV-1 particles is in the same order of few molecules.
The recruitment of Chmp and Vps4 proteins differs from that of Alix. Chmp1b, Chmp4a, Chmp4c and Vps4 were only transiently recruited to HIV-1 and EIAV assembly sites around the time that Gag protein accumulation is completed, most often in a single pulse (Fig. 5) [11]. In contrast to Alix, the Chmp and Vps4 proteins behave as if some triggering event concomitant to the completion of viral assembly causes their rapid and transient recruitment. This trigger could be accumulation of a threshold number of Alix molecules, or perhaps a critical degree of membrane curvature, also induced by Alix. Charged multivesicular body proteins and Vps4 remained detectable at the site of budding for an average of 2 min and, in most cases, were completely recycled [11]. Similar transient recruitment of Vps4 at sites of HIV-1 budding was observed in cells overexpressing GFP–Vps4 [12]. However, in this case, Vps4 remained at site of budding for shorter periods of time (35 sec on average).
By averaging the data for all assembly events observed [11], Chmp and Vps4 seemed to be recruited at the same time. However, a small delay between Chmp and Vps4 recruitment is likely because Vps4 is recruited by Chmp proteins [39, 40, 91].
Unlike for cytokinesis studies, the delay between recruitment of ESCRT proteins and membrane fission remains difficult to measure in the context of viral budding. Imaging HIV-1 particles generated with Gag fused to a GFP variant whose fluorescence is diminished at acidic pH [111] has been used to identify which viral particles have pinch off the cell membrane at a given time [106]. Unfortunately, the experimental settings of such approach do not allow dynamic studies. Based on the intuitive assumption that the motility of particles might increase once scission has occurred, increase in particle mobility has been used to measure the time between ESCRT protein recruitment and membrane fission, leading to an estimate of 15 min between the two processes [12]. However, such increase in motility after assembly was rarely observed, which is in agreement with the results obtained with the pH sensitive GFP variant revealing that virions do not move away from the site of assembly, after budding at the interface between the cell ventral surface and the coverslip [106]. This lack of motility after budding is probably due to the action of cell surface adhesion molecules, or to the physical constraints impose by the small space between the cell and the coverslip. The rare cases of virion motility increase observed after assembly [12] might then represent endocytosis events of fully assembled virions. Therefore, the delay between recruitment of ESCRT proteins and viral budding remain unknown. However, since the recruitment of the proteins that are responsible for membrane fission is concomitant with completion of assembly and, because they remain, on average, ~2 min at the site of assembly, one can hypothesize that budding occurs rapidly after completion of assembly.
Altogether, these live cells studies of ESCRT proteins recruitment at viral assembly sites have revealed two completely different behaviors of ESCRT proteins, suggesting a model in which progressive accumulation of early players, such as Alix, to a threshold level triggers the rapid and transient recruitment of ESCRT-III and Vps4 proteins (Figs. 4, 5).
Conclusions
Since the discovery of the ESCRT machinery more than 10 years ago, much indirect evidence for a sequential recruitment of ESCRT proteins at sites of action have been collected trough biochemical and fixed cell microscopic studies of yeast MVB biogenesis, and more recently trough SIM and EM analysis of ESCRT localization at the midbody and at viral assembly sites in mammalian cells. This model is now very well supported by live cell studies of ESCRT assembly and disassembly during retroviral budding and cytokinesis. ESCRT-mediated processes are indeed very dynamic: early players, such as ESCRT-I and Alix, which might be responsible for membrane curvature and membrane neck formation, accumulate progressively, until the process is advanced enough to trigger the rapid and transient recruitment of late players, such as ESCRT-III and Vps4 proteins, which carry out membrane scission and protein recycling.
Finer analysis would come from monitoring fluorescently tagged versions of a couple of ESCRT components in a single cell, which is technically challenging because it requires the generation of cell lines stably expressing few fluorescent proteins and microscopic settings that would separate correctly these signals.
Acknowledgments
I thank the ‘Centre National de la Recherche Scientifique’ for support, Sandy Simon for critical reading of the manuscript, and Juan Martin-Serrano for comments and access to unpublished data.
References
- 1.Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316(5833):1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 2.Morita E, et al. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26(19):4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morita E, et al. Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc Natl Acad Sci USA. 2010;107(29):12889–12894. doi: 10.1073/pnas.1005938107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106(2):145–155. doi: 10.1016/S0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 5.Caballe A, Martin-Serrano J. ESCRT machinery and cytokinesis: the road to daughter cell separation. Traffic. 2011;12(10):1318–1326. doi: 10.1111/j.1600-0854.2011.01244.x. [DOI] [PubMed] [Google Scholar]
- 6.Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it’s all in the neck. Nat Rev Mol Cell Biol. 2010;11(8):556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21(1):77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 8.Guizetti J, Gerlich DW. ESCRT-III polymers in membrane neck constriction. Trends Cell Biol. 2012;22(3):133–140. doi: 10.1016/j.tcb.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Elia N, et al. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc Natl Acad Sci USA. 2011;108(12):4846–4851. doi: 10.1073/pnas.1102714108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guizetti J, et al. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science. 2011;331(6024):1616–1620. doi: 10.1126/science.1201847. [DOI] [PubMed] [Google Scholar]
- 11.Jouvenet N, et al. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat Cell Biol. 2011;13(4):394–401. doi: 10.1038/ncb2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baumgartel V, et al. Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat Cell Biol. 2011;13(4):469–474. doi: 10.1038/ncb2215. [DOI] [PubMed] [Google Scholar]
- 13.Babst M, et al. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic. 2000;1(3):248–258. doi: 10.1034/j.1600-0854.2000.010307.x. [DOI] [PubMed] [Google Scholar]
- 14.Bishop N, Woodman P. TSG101/mammalian VPS23 and mammalian VPS28 interact directly and are recruited to VPS4-induced endosomes. J Biol Chem. 2001;276(15):11735–11742. doi: 10.1074/jbc.M009863200. [DOI] [PubMed] [Google Scholar]
- 15.Eastman SW, et al. Identification of human VPS37C, a component of endosomal sorting complex required for transport-I important for viral budding. J Biol Chem. 2005;280(1):628–636. doi: 10.1074/jbc.M413556200. [DOI] [PubMed] [Google Scholar]
- 16.Morita E, et al. Identification of human MVB12 proteins as ESCRT-I subunits that function in HIV budding. Cell Host Microbe. 2007;2(1):41–53. doi: 10.1016/j.chom.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stefani F, et al. UBAP1 is a component of an endosome-specific ESCRT-I complex that is essential for MVB sorting. Curr Biol. 2011;21(14):1245–1250. doi: 10.1016/j.cub.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 18.Stuchell MD, et al. The human endosomal sorting complex required for transport (ESCRT-I) and its role in HIV-1 budding. J Biol Chem. 2004;279(34):36059–36071. doi: 10.1074/jbc.M405226200. [DOI] [PubMed] [Google Scholar]
- 19.Agromayor M, et al. The UBAP1 Subunit of ESCRT-I Interacts with ubiquitin via a SOUBA domain. Structure. 2012;20:414–418. doi: 10.1016/j.str.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J, et al. Structural basis for endosomal targeting by the Bro1 domain. Dev Cell. 2005;8(6):937–947. doi: 10.1016/j.devcel.2005.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee S, et al. Structural basis for viral late-domain binding to Alix. Nat Struct Mol Biol. 2007;14(3):194–199. doi: 10.1038/nsmb1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fisher RD, et al. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 2007;128(5):841–852. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 23.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438(7068):590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 24.Odorizzi G, et al. Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae . J Cell Sci. 2003;116(Pt 10):1893–1903. doi: 10.1242/jcs.00395. [DOI] [PubMed] [Google Scholar]
- 25.McCullough J, et al. ALIX–CHMP4 interactions in the human ESCRT pathway. Proc Natl Acad Sci USA. 2008;105(22):7687–7691. doi: 10.1073/pnas.0801567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin-Serrano J, Zang T, Bieniasz PD. Role of ESCRT-I in retroviral budding. J Virol. 2003;77(8):4794–4804. doi: 10.1128/JVI.77.8.4794-4804.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin-Serrano J, et al. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc Natl Acad Sci USA. 2003;100(21):12414–12419. doi: 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee HH, et al. Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55. Science. 2008;322(5901):576–580. doi: 10.1126/science.1162042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langelier C, et al. Human ESCRT-II complex and its role in human immunodeficiency virus type 1 release. J Virol. 2006;80(19):9465–9480. doi: 10.1128/JVI.01049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wollert T, et al. Membrane scission by the ESCRT-III complex. Nature. 2009;458(7235):172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shim S, Kimpler LA, Hanson PI. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic. 2007;8(8):1068–1079. doi: 10.1111/j.1600-0854.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- 32.Zamborlini A, et al. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc Natl Acad Sci USA. 2006;103(50):19140–19145. doi: 10.1073/pnas.0603788103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lata S, et al. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321(5894):1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peel S, et al. Divergent pathways lead to ESCRT-III-catalyzed membrane fission. Trends Biochem Sci. 2010;36(4):199–210. doi: 10.1016/j.tibs.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 35.Bodon G, et al. Charged multivesicular body protein 2B (CHMP2B) of the endosomal sorting complex required for transport-III (ESCRT-III) polymerizes into helical structures deforming the plasma membrane. J Biol Chem. 2012;286(46):40276–40286. doi: 10.1074/jbc.M111.283671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fabrikant G, et al. Computational model of membrane fission catalyzed by ESCRT-III. PLoS Comput Biol. 2009;5(11):e1000575. doi: 10.1371/journal.pcbi.1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agromayor M, et al. Essential role of hIST1 in cytokinesis. Mol Biol Cell. 2009;20(5):1374–1387. doi: 10.1091/mbc.E08-05-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bajorek M, et al. Biochemical analyses of human IST1 and its function in cytokinesis. Mol Biol Cell. 2009;20(5):1360–1373. doi: 10.1091/mbc.E08-05-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Obita T, et al. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449(7163):735–739. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- 40.Stuchell-Brereton MD, et al. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449(7163):740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- 41.Scott A, et al. Structural and mechanistic studies of VPS4 proteins. EMBO J. 2005;24(20):3658–3669. doi: 10.1038/sj.emboj.7600818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gonciarz MD, et al. Biochemical and structural studies of yeast Vps4 oligomerization. J Mol Biol. 2008;384(4):878–895. doi: 10.1016/j.jmb.2008.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu Z, et al. Cryo-EM structure of dodecameric Vps4p and its 2:1 complex with Vta1p. J Mol Biol. 2008;377(2):364–377. doi: 10.1016/j.jmb.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hartmann C, et al. Vacuolar protein sorting: two different functional states of the AAA-ATPase Vps4p. J Mol Biol. 2008;377(2):352–363. doi: 10.1016/j.jmb.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 45.Landsberg MJ, et al. Three-dimensional structure of AAA ATPase Vps4: advancing structural insights into the mechanisms of endosomal sorting and enveloped virus budding. Structure. 2009;17(3):427–437. doi: 10.1016/j.str.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 46.Inoue M, et al. Nucleotide-dependent conformational changes and assembly of the AAA ATPase SKD1/VPS4B. Traffic. 2008;9(12):2180–2189. doi: 10.1111/j.1600-0854.2008.00831.x. [DOI] [PubMed] [Google Scholar]
- 47.Babst M, et al. Escrt-III: an endosome-associated hetero oligomeric protein complex required for mvb sorting. Dev Cell. 2002;3(2):271–282. doi: 10.1016/S1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- 48.Babst M, et al. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev Cell. 2002;3(2):283–289. doi: 10.1016/S1534-5807(02)00219-8. [DOI] [PubMed] [Google Scholar]
- 49.Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464(7290):864–869. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saksena S, et al. Functional reconstitution of ESCRT-III assembly and disassembly. Cell. 2009;136(1):97–109. doi: 10.1016/j.cell.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghazi-Tabatabai S, et al. Structure and disassembly of filaments formed by the ESCRT-III subunit Vps24. Structure. 2008;16(9):1345–1356. doi: 10.1016/j.str.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 52.Shim S, Merrill SA, Hanson PI. Novel interactions of ESCRT-III with LIP5 and VPS4 and their implications for ESCRT-III disassembly. Mol Biol Cell. 2008;19(6):2661–2672. doi: 10.1091/mbc.E07-12-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Azmi I, et al. Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta1. J Cell Biol. 2006;172(5):705–717. doi: 10.1083/jcb.200508166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Samson RY, et al. A role for the ESCRT system in cell division in Archaea. Science. 2008;322(5908):1710–1713. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Samson RY, et al. Molecular and structural basis of ESCRT-III recruitment to membranes during archaeal cell division. Mol Cell. 2011;41(2):186–196. doi: 10.1016/j.molcel.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moriscot C, et al. Crenarchaeal CdvA forms double-helical filaments containing DNA and interacts with ESCRT-III-like CdvB. PLoS ONE. 2011;6(7):e21921. doi: 10.1371/journal.pone.0021921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Felder S, et al. Kinase activity controls the sorting of the epidermal growth factor receptor within the multivesicular body. Cell. 1990;61(4):623–634. doi: 10.1016/0092-8674(90)90474-S. [DOI] [PubMed] [Google Scholar]
- 58.Futter CE, et al. Multivesicular endosomes containing internalized EGF–EGF receptor complexes mature and then fuse directly with lysosomes. J Cell Biol. 1996;132(6):1011–1023. doi: 10.1083/jcb.132.6.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Teis D, Saksena S, Emr SD. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev Cell. 2008;15(4):578–589. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 60.Strack B, et al. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114(6):689–699. doi: 10.1016/S0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- 61.Glotzer M. The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat Rev Mol Cell Biol. 2009;10(1):9–20. doi: 10.1038/nrm2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Skop AR, et al. Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science. 2004;305(5680):61–66. doi: 10.1126/science.1097931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fabbro M, et al. Cdk1/Erk2- and Plk1-dependent phosphorylation of a centrosome protein, Cep55, is required for its recruitment to midbody and cytokinesis. Dev Cell. 2005;9(4):477–488. doi: 10.1016/j.devcel.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 64.Martinez-Garay I, et al. The novel centrosomal associated protein CEP55 is present in the spindle midzone and the midbody. Genomics. 2006;87(2):243–253. doi: 10.1016/j.ygeno.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 65.Zhao WM, Seki A, Fang G. Cep55, a microtubule-bundling protein, associates with centralspindlin to control the midbody integrity and cell abscission during cytokinesis. Mol Biol Cell. 2006;17(9):3881–3896. doi: 10.1091/mbc.E06-01-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guizetti J, et al. Correlative time-lapse imaging and electron microscopy to study abscission in HeLa cells. Methods Cell Biol. 2010;96:591–601. doi: 10.1016/S0091-679X(10)96024-X. [DOI] [PubMed] [Google Scholar]
- 67.Steigemann P, et al. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell. 2009;136(3):473–484. doi: 10.1016/j.cell.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 68.Jin Y, et al. The fission yeast homolog of the human transcription factor EAP30 blocks meiotic spindle pole body amplification. Dev Cell. 2005;9(1):63–73. doi: 10.1016/j.devcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 69.Carlton JG, Agromayor M, Martin-Serrano J. Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc Natl Acad Sci USA. 2008;105(30):10541–10546. doi: 10.1073/pnas.0802008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carlton JG, et al. ESCRT-III governs the Aurora B-mediated abscission checkpoint through CHMP4C. Science. 2012;336:220–225. doi: 10.1126/science.1217180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hanson PI, et al. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol. 2008;180(2):389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pires R, et al. A crescent-shaped ALIX dimer targets ESCRT-III CHMP4 filaments. Structure. 2009;17(6):843–856. doi: 10.1016/j.str.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Briggs JA, et al. The stoichiometry of Gag protein in HIV-1. Nat Struct Mol Biol. 2004;11(7):672–675. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]
- 74.Jacks T, et al. Characterization of ribosomal frame shifting in HIV-1 gag-pol expression. Nature. 1988;331(6153):280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 75.Zhu P, et al. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc Natl Acad Sci USA. 2003;100(26):15812–15817. doi: 10.1073/pnas.2634931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen J, et al. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc Natl Acad Sci USA. 2009;106(32):13535–13540. doi: 10.1073/pnas.0906822106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Larson DR, et al. Visualization of retrovirus budding with correlated light and electron microscopy. Proc Natl Acad Sci USA. 2005;102(43):15453–15458. doi: 10.1073/pnas.0504812102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goff SP. Retroviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields virology. Philadelphia: Lippincott Williams and Wilkins; 2001. pp. 1999–2070. [Google Scholar]
- 79.Kutluay SB, Bieniasz PD. Analysis of the initiating events in HIV-1 particle assembly and genome packaging. PLoS Pathog. 2010;6(11):e1001200. doi: 10.1371/journal.ppat.1001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jouvenet N, et al. Cell biology of retroviral RNA packaging. RNA Biol. 2011;8(4):572–580. doi: 10.4161/rna.8.4.16030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jouvenet N, Simon SM, Bieniasz PD. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc Natl Acad Sci USA. 2009;106(45):19114–19119. doi: 10.1073/pnas.0907364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kemler I, Meehan A, Poeschla EM. Live-cell coimaging of the genomic RNAs and Gag proteins of two lentiviruses. J Virol. 2010;84(13):6352–6366. doi: 10.1128/JVI.00363-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jouvenet N, et al. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 2006;4(12):e435. doi: 10.1371/journal.pbio.0040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Finzi A, et al. Productive human immunodeficiency virus type 1 assembly takes place at the plasma membrane. J Virol. 2007;81(14):7476–7490. doi: 10.1128/JVI.00308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Welsch S, et al. HIV-1 buds predominantly at the plasma membrane of primary human macrophages. PLoS Pathog. 2007;3(3):e36. doi: 10.1371/journal.ppat.0030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen BJ, Lamb RA. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology. 2008;372(2):221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bieniasz PD. The cell biology of HIV-1 virion genesis. Cell Host Microbe. 2009;5(6):550–558. doi: 10.1016/j.chom.2009.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Carlton JG, Martin-Serrano J. The ESCRT machinery: new functions in viral and cellular biology. Biochem Soc Trans. 2009;37(Pt 1):195–199. doi: 10.1042/BST0370195. [DOI] [PubMed] [Google Scholar]
- 89.Martin-Serrano J, Zang T, Bieniasz PD. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med. 2001;7(12):1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- 90.Garrus JE, et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107(1):55–65. doi: 10.1016/S0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 91.von Schwedler UK, et al. The protein network of HIV budding. Cell. 2003;114(6):701–713. doi: 10.1016/S0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 92.Martin-Serrano J, et al. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J Cell Biol. 2005;168(1):89–101. doi: 10.1083/jcb.200408155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhadina M, Bieniasz PD. Functional interchangeability of late domains, late domain cofactors and ubiquitin in viral budding. PLoS Pathog. 2010;6(10):e1001153. doi: 10.1371/journal.ppat.1001153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Weiss ER, et al. Rescue of HIV-1 release by targeting widely divergent NEDD4-type ubiquitin ligases and isolated catalytic HECT domains to Gag. PLoS Pathog. 2010;6(9):e1001107. doi: 10.1371/journal.ppat.1001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rauch S, Martin-Serrano J. Multiple interactions between the ESCRT machinery and arrestin-related proteins: implications for PPXY-dependent budding. J Virol. 2011;85(7):3546–3556. doi: 10.1128/JVI.02045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.VerPlank L, et al. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag) Proc Natl Acad Sci USA. 2001;98(14):7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Popov S, et al. Divergent Bro1 domains share the capacity to bind human immunodeficiency virus type 1 nucleocapsid and to enhance virus-like particle production. J Virol. 2009;83(14):7185–7193. doi: 10.1128/JVI.00198-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Popova E, Popov S, Gottlinger HG. Human immunodeficiency virus type 1 nucleocapsid p1 confers ESCRT pathway dependence. J Virol. 2010;84(13):6590–6597. doi: 10.1128/JVI.00035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dussupt V, et al. The nucleocapsid region of HIV-1 Gag cooperates with the PTAP and LYPXnL late domains to recruit the cellular machinery necessary for viral budding. PLoS Pathog. 2009;5(3):e1000339. doi: 10.1371/journal.ppat.1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fujii K, et al. Functional role of Alix in HIV-1 replication. Virology. 2009;391(2):284–292. doi: 10.1016/j.virol.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Morita E, et al. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe. 2011;9(3):235–242. doi: 10.1016/j.chom.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Usami Y, Popov S, Gottlinger HG. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J Virol. 2007;81(12):6614–6622. doi: 10.1128/JVI.00314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dussupt V, et al. Basic residues in the nucleocapsid domain of Gag are critical for late events of HIV-1 budding. J Virol. 2010;85(5):2304–2315. doi: 10.1128/JVI.01562-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Leroux C, Cadore JL, Montelaro RC. Equine infectious anemia virus (EIAV): what has HIV’s country cousin got to tell us? Vet Res. 2004;35(4):485–512. doi: 10.1051/vetres:2004020. [DOI] [PubMed] [Google Scholar]
- 105.Simon SM. Partial internal reflections on total internal reflection fluorescent microscopy. Trends Cell Biol. 2009;19(11):661–668. doi: 10.1016/j.tcb.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jouvenet N, Bieniasz PD, Simon SM. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature. 2008;454(7201):236–240. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ivanchenko S, et al. Dynamics of HIV-1 assembly and release. PLoS Pathog. 2009;5(11):e1000652. doi: 10.1371/journal.ppat.1000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fusco D, et al. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol. 2003;13(2):161–167. doi: 10.1016/S0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lin Y, et al. Interaction of the mammalian endosomal sorting complex required for transport (ESCRT) III protein hSnf7-1 with itself, membranes, and the AAA+ ATPase SKD1. J Biol Chem. 2005;280(13):12799–12809. doi: 10.1074/jbc.M413968200. [DOI] [PubMed] [Google Scholar]
- 110.Howard TL, et al. CHMP1 functions as a member of a newly defined family of vesicle trafficking proteins. J Cell Sci. 2001;114(Pt 13):2395–2404. doi: 10.1242/jcs.114.13.2395. [DOI] [PubMed] [Google Scholar]
- 111.Miesenbock G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 1998;394(6689):192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]