Abstract
Regulation of apoptosis at various stages of differentiation plays an important role in spermatogenesis. Therefore, the identification and characterisation of highly expressed genes in the testis that are involved in apoptosis is of great value to delineate the mechanism of spermatogenesis. Here, we reported that Fank1, a novel gene highly expressed in testis, functioned as an anti-apoptotic protein that activated the activator protein 1 (AP-1) pathway. We found that Jab1 (Jun activation domain-binding protein 1), a co-activator of AP-1, specifically interacted with Fank1. Reporter analyses showed that Fank1 activated AP-1 pathway in a Jab1-dependent manner. Fank1 overexpression also increased the expression and activation of endogenous c-Jun. Further study showed that Fank1 inhibited cell apoptosis by upregulating and activating endogenous c-Jun and its downstream target, Bcl-3. This process was shown to be Jab1 dependent. Taken together, our results indicated that by interacting with Jab1, Fank1 could suppress cell apoptosis by activating the AP-1-induced anti-apoptotic pathway.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-010-0559-4) contains supplementary material, which is available to authorized users.
Keywords: Fank1, Jab1, AP-1, Bcl-3, Apoptosis
Introduction
Spermatogenesis is a complex developmental process that includes mitosis of spermatogonial stem cells, meiosis of spermatocytes and spermiogenesis [1]. During spermatogenic differentiation, more than half of the differentiating spermatogenic cells die by apoptosis before they mature into spermatozoa [2]. Therefore, regulation of apoptosis at various stages of differentiation plays an important role in spermatogenesis. Studies show that many genes highly expressed in testis are involved in the regulation of apoptosis during spermatogenesis. Therefore, the identification and characterisation of these genes are of great value to delineating the mechanism of spermatogenesis.
Fibronectin type III and ankyrin repeat domains 1 (Fank1) is a nuclear protein expressed during the transition from the meiotic phase to the haploid phase of spermatogenesis [3]. While little is known about the function of Fank1 during spermatogenesis, gene ontology analysis suggests that it may be a transcription factor. We explored the role of Fank1 using the yeast two-hybrid system to screen for cellular proteins that may interact with Fank1. In this screen, we identified the Jun activation domain-binding protein 1 (Jab1) as a potential Fank1-interacting protein.
Jab1 is the fifth component of the COP9 signalosome (CSN) complex [4], and it was initially identified as a co-activator of activator protein 1 (AP-1) [5]. Jab1 enhances binding of the AP-1 complex, which contains c-Jun, to its DNA consensus site and increases transactivation of an AP-1-dependent promoter. The AP-1 transcription factor is a dimeric complex composed of members that belong to the Jun, Fos, ATF (activating transcription factor) and MAF (musculoaponeurotic fibrosarcoma) protein families. AP-1 has been implicated in the regulation of cell proliferation, differentiation and apoptosis. Recent findings have shown that AP-1 has a dual function in the regulation of apoptosis: it induces apoptosis in some cell types but is required for cell survival in others. For instance, in fibroblasts, c-Jun binds to a variant AP-1 site in the p53 promoter, thereby downregulating the transcription of p53 and stimulating apoptosis [6]. In melanoma cells, c-Jun inhibits Fas transcription, resulting in apoptosis [7]. However, c-Jun blocks apoptosis in IL-4-deprived T cells by upregulating the expression of Bcl-3, an anti-apoptotic protein [8]. The different apoptotic functions of AP-1 in different cell types could be explained by the regulation of different downstream target genes.
In the present study, we provide evidence for an interaction between Fank1 and Jab1 in vivo. In addition, we investigate the biological effect of this interaction on the modulation of the anti-apoptotic effect of AP-1. Our observations indicate that Fank1 can function as an anti-apoptotic protein. These results will allow us to gain further insight into the biological and molecular functions of Fank1 in spermatogenesis.
Materials and methods
Cell culture and transfection
HEK 293T, HeLa, MCF-7 and CHO cells were purchased from ATCC and cultured in fresh Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% foetal bovine serum in 5% CO2 incubators at 37°C. Adherent cells were passaged every 2–3 days with 0.5 mg/ml trypsin (1:250) and 0.53 mM ethylenediaminetetraacetic acid (EDTA). Transfections of plasmid DNA and Jab1 siRNA (siRNA1: 5′-GGACUAAGGAUCACCAUUACU-3′, siRNA2: 5′-AAGCUCAGAGUAUCGAUGAAA-3′ or scrambled siRNA: 5′-UUCUCCGAACGUGUCACGU-3′) were performed using Lipofectamin™ 2000 (Invitrogen, CA). Briefly, HEK 293T cells were plated in 3.5 cm dishes at 40–50% confluence 1 day before transfection. After 16–18 h, transfections were performed using 2 µg plasmid DNA and 100 nmol siRNA. Cells were harvested for further analysis after 2 days incubation.
Co-immunoprecipitation
Cells were transiently transfected with p3×Flag-CMV-14, p3×Flag-CMV-14-Fank1, p3×Flag-CMV-14-FN3, p3×Flag-CMV-14-ANK and pCDNA6/V5-hisB-Jab1 as indicated. After a 48 h incubation, cells were harvested and lysed in lysis buffer [50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% NP-40, 1 mM EDTA and 10% glycerol] containing protease inhibitor cocktails (Roche). Flag-Fank1 protein complexes were isolated using an anti-Flag M2 affinity gel (Sigma). The bound protein complexes were solubilised in 1.25× SDS loading buffer and analysed by Western blot.
GST pull-down
In vivo GST pull-down assays were performed using 400 μg of HEK 293T cell lysates [50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% NP-40, 1 mM EDTA and protease inhibitor cocktails] mixed with 5 μg of purified GST-Fank1, GST-FN3 or GST, immobilized on glutathione-Sepharose beads for 2 h at 4°C and washed three times with 1× PBS. Next, the GST pull-down products were run in a 12% SDS-PAGE gel and immunoblotted with anti-Jab1 and anti-GST antibodies.
Western blot
Proteins were extracted with RIPA buffer (Sigma) and quantified using the BCA protein assay reagent (Pierce). Extracts were loaded in a 12% SDS-PAGE gel, separated and then electrophoretically transferred to a PVDF membrane (GE Healthcare). The membrane was blocked in 5% skim milk for 1 h at room temperature and then incubated overnight with the indicated antibodies at 4°C. The membrane was incubated with anti-rabbit or anti-mouse HRP-IgG (Santa Cruz) for 1 h at room temperature. Chemiluminescence was detected using an ECL blot detection system (Santa Cruz). The following primary antibodies were used (1:1,000 unless otherwise indicated): Bcl-3 (Santa Cruz, sc-185); GAPDH (Santa Cruz, sc-25778, 1:5,000); GFP (Santa Cruz, sc-9996, 1:5,000); phospho-c-Jun(Ser63) (Cell Signaling Technology, 9261S); phospho-c-Jun(Ser73) (Cell Signaling Technology, 9164S); JNK (Cell Signaling Technology, 9252); phospho-JNK (Cell Signaling Technology, 9251S); PARP (Cell Signaling Technology, 9542); BCL-2 (Cell Signaling Technology, 2876); BCL-xL (Cell Signaling Technology, 2764); c-Jun (BD Pharmingen, 610326); FLAG (Sigma, F2555); V5 (Sigma, V8137); Jab1 (Abcam, ab495); LaminA/C (Abcam, ab4789, 1:50); GST (MBL International, JM-3997-100); caspase 7 (MBL International, M053-3).
Transcription factor profiling (Mercury system detection)
For transcription factor profiling, a stable cell line that expressed GFP-Fank1 recombinant protein (CHO/GFP-Fank1) was generated, and a stable cell line that expressed GFP (CHO/GFP) was used as a control. The stable cell lines were transfected with different SEAP constructs (Mercury™ pathway profiling system from CLONTECH Laboratories) using Lipofectamine™ 2000. After 24 h, SEAP activity was measured using the SEAP reporter assay kit (CLONTECH Laboratories, Inc.) according to the manufacturer’s instructions.
Luciferase assays
The pAP1-Luc plasmid (Stratagene) was transfected into HEK 293T cells using Lipofectamine™ 2000. In addition, 293T cells were cotransfected with a Renilla luciferase control vector (Promega) to monitor the transfection rates. Firefly and Renilla luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega). Briefly, the cells were washed with PBS and lysed with passive lysis buffer. Cell lysates were mixed with Luciferase Assay Reagent II, and the firefly luminescence was measured using a luminometer (TD-20/20, Turner BioSystems, USA). Next, samples were mixed with the stop reagent and Renilla luciferase activity was measured as an internal control. Relative luciferase activity was calculated as the ratio of firefly luciferase activity to Renilla luciferase activity.
Nuclear protein extraction
HEK 293T cells were transfected with p3×FLAG-CMV-14 or p3×FLAG-CMV-14 -Fank1 plasmid independently. For the Jab1 RNA interference assay, HEK 293T cells were first transfected with siRNA-NC or siRNA-Jab1, and 48 h later, the cells were transfected with p3×FLAG-CMV-14-Fank1 plasmid. After 24 h, the cells were collected, and nuclear protein was prepared using a NE-PER® Nuclear and Cytoplasmic Extraction kit (Thermo Scientific) according to the manufacturer’s instructions.
Generation of stable cell lines
To generate CHO/GFP and CHO/GFP-Fank1 stable cell lines, a pEGFP-N1-Fank1 plasmid was constructed. CHO cells were then transfected with either the pEGFP-N1 or the pEGFP-N1-Fank1 plasmid. After 48 h, 0.5 μg/ml G418 was added to the cell medium to screen for stable cells. After 2 weeks, all non-transfected cells were killed, and a stable population of cells was selected.
To generate stable HeLa/Con and HeLa/Flag-Fank1 cell lines, a pMSCVpuro-Fank1-Flag plasmid was constructed. HeLa cells were then transfected with either pMSCVpuro or pMSCVpuro-Fank1-Flag plasmid. After 48 h, 2 μg/ml puromycin was added to the cell medium to screen for stable cells. After 1 week, all non-transfected cells were killed, and a stable population of cells was selected.
Flow cytometric analysis
Cells were exposed to 40 J/m2 UV or 10 μM Camptothecin (CPT) to induce apoptosis. At indicated times, cells were incubated with 0.25% trypsin-EDTA at 37°C for 3–5 min, collected, and fixed with 75% ethanol at 4°C overnight. Cells were stained with 50 μg/ml propidium iodide (PI) and 50 μg/ml RNase A in PBS at 37°C for 20 min. The DNA content of 10,000 cells was analysed using a COULTER flow cytometer (EPICS-XL) with EXPO32-ADC software. The percentage of apoptotic cells (% of total cells) was analysed by the program EXPO32-ADC, and the results are shown as a bar chart (Microsoft Excel). Double staining with FITC-Annexin V and PI was carried out using the FITC-Annexin V kit (NeoBioscience) according to the manufacturer’s recommendations and then analysed using the Accuri C6 flow cytometer system.
Electrophoresis mobility shift assay
AP-1 activation was measured using LightShift Chemiluminescent EMSA Kit according to the manufacturer’s instructions (Thermo Scientific). Briefly, nuclear extracts (4 μg) was incubated with the labelled probe in binding buffer (10 mM Tris, 50 mM potassium chloride, 1 mM dithiothreitol, 1 μg poly-dI·dC, 0.1 mM EDTA, 2.5% glycerol, 5 mM magnesium chloride, total volume of 20 μl) for 20 min at room temperature. Competition was carried out with unlabeled probe for 20 min prior to addition of biotinylated probes. The samples were separated in a 6% nondenaturing polyacrylamide gel and blotted onto a Biodyne B (0.45 μm) positively charged nylon membrane (Pall Schweiz, Basel, Switzerland). The biotinylated nucleotides were detected using streptavidin conjugated to HRP. The membrane was then incubated with the chemiluminescent reagent ECL and exposed to X-ray film. The sequences of consensus AP-1 probe were 5′-cgcttgatgactcagccggaa-3′. The sequences of double-strand oligonucleotide fragments containing the AP-1-binding site (–1,171 to –1,165 bp) in the human Bcl-3 promoter were 5′- cccatctccagcctgagtcatgcccccaacccg-3′.
Chromatin immunoprecipitation (ChIP) assay
HEK 293T cells were transfected with p3×FLAG-CMV-14-Fank1 plasmid. After 24 h, formaldehyde (1%) was added for crosslinking at room temperature for 15 min. The cells were collected and resuspended in 0.5 ml of lysis buffer. Sheared DNA was diluted in dilution buffer. Two percent of each sample was stored as control input DNA, and the remaining lysates were cleared with 20 μl of mouse IgG (Santa Cruz, sc-2025) and 50 μl of protein G-Sepharose (Roche, 11243233001) for 4 h at 4°C. Immunoprecipitation (IP) was done with specific anti-FLAG, anti-Jab1, anti-c-Jun antibodies or anti-mouse IgG overnight at 4°C, followed by incubation with protein G-agarose beads (50 μl) for 4 h at 4°C. The beads were washed, and precipitated chromatin complexes were extracted. Extracts were reverse crosslinked at 65°C overnight. DNA fragments were purified and analysed by PCR. A 209 bp fragment containing AP-1 binding site was amplified by using primers of 5′-gtgactcagtgacccggact-3′ (–1,338 to –1,318) and 5′-gttgatgggtggggaaca-3′ (–1,141 to –1,123) in human Bcl-3 promoter. Relative levels of DNA were estimated with respect to input DNA.
Results
Identification of Jab1 as a Fank1-interacting protein
Fank1 encodes a protein that is composed of one fibronectin type III domain and six ankyrin repeats (Fig. 1a). To identify Fank1-interacting proteins, we performed a yeast two-hybrid screen using the full-length cDNA of Fank1 as bait. We found that one of the clones that interacted with Fank1 encoded Jab1 (data not shown).
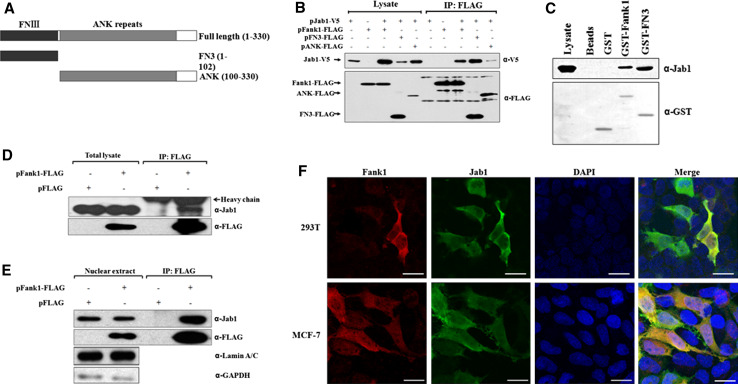
Fig. 1a–f.
Interaction of Fank1 with Jab1 in vivo. a Fank1 deletion constructs. b V5-tagged Jab1 was transfected into HEK 293T cells with or without Flag-tagged full-length Fank1 or indicated deletion mutants. Whole cell lysates were immunoprecipitated with anti-Flag M2 affinity gel and analysed by Western blot with the indicated antibodies. c HEK 293T cell lysates were incubated with GST, GST-Fank1 or GST-FN3 fusion proteins. Bound proteins were analysed by immunoblotting with the indicated antibodies. d Flag-tagged Fank1 was transfected into HEK293T cells. Cell lysates were immunoprecipitated with an anti-Flag M2 affinity gel and analysed with the indicated antibodies. e Flag-tagged Fank1 was transfected into HEK293T cells. Nuclear proteins were extracted with NE-PER reagents (Thermo Scientific) and were immunoprecipitated with an anti-Flag M2 affinity gel and analysed with the indicated antibodies. f HEK 293T and MCF-7 cells were co-transfected with Flag-tagged Fank1 and Myc-tagged Jab1 expression vectors. Subcellular localisation of Fank1 and Jab1 was analysed by immunofluorescence with anti-Flag and anti-Myc antibodies. Nuclei were stained with DAPI. Cells were visualised by confocal microscopy (Leica). Scale bars 15 μm
To verify the interaction between Fank1 and Jab1, we performed a co-immunoprecipitation experiment in vivo. We co-transfected HEK 293T cells with a V5-Jab1 vector and either a control or Flag-Fank1 vector. When co-expressed, Fank1 associated with Jab1 (Fig. 1b). To further characterise the Fank1-binding domain involved in the interaction with Jab1, we generated N-terminal (FN3: 1-102AA) and C-terminal (ANK: 100-330AA) deletion mutants of Fank1 (Fig. 1a). The ability of each mutant to bind to Jab1 was then examined. As shown in Fig. 1b, the N-terminal deletion mutant of Fank1 strongly interacted with Jab1, while the C-terminal deletion mutant only exhibited a slight interaction with Jab1. This suggests that the FN3 domain of Fank1 is important for the interaction between Fank1 and Jab1. To further confirm the interaction, we purified GST-Fank1 and GST-FN3 fusion proteins and then performed a GST pull-down assay. As shown in Fig. 1c, both GST-Fank1 and GST-FN3 bound to endogenous Jab1. We also confirmed the interaction of Flag-Fank1 with endogenous Jab1 by co-immunoprecipitation (Fig. 1d). Jab1 interacted with c-Jun in the nucleus and increased transactivation of an AP-1-dependent promoter [5]. We then asked whether Fank1 and Jab1 formed a complex in the nucleus. HEK 293T cells were transfected with Flag-tagged Fank1 or with a control expression vector. After 24 h, cytoplasmic and nuclear proteins were isolated and analysed by Western blot. As shown in Fig. 1e, Flag-tagged Fank1 could specifically co-immunoprecipitate with endogenous Jab1 in the nucleus. Next, we assessed the subcellular localisation of Fank1 and Jab1 using confocal immunofluorescence microscopy. Flag-tagged Fank1 and Myc-tagged Jab1 expression vectors were co-transfected into either HEK 293T or MCF-7 cells, and the nuclei were stained with DAPI. As shown in Fig. 1f, Fank1 and Jab1 co-localised to both the cytoplasm and nucleus in two cell lines. Taken together, these results consistently demonstrate a specific interaction between Fank1 and Jab1 in both the cytoplasm and nucleus.
Regulation of AP-1 activity by the physical interaction of Fank1 with Jab1
To study the function of Fank1, we generated a cell line that stably expressed GFP-Fank1 recombinant protein (CHO/GFP-Fank1) and a cell line that stably expressed GFP (CHO/GFP) as a control (Fig. 2a–d). Expression of the GFP-Fank1 recombinant protein was demonstrated by Western blot using a GFP antibody (Fig. 2e). To obtain preliminary evidence regarding the role of Fank1 in the activation of key signalling pathways, we performed reporter gene analysis in CHO/GFP-Fank1 and CHO/GFP using CLONTECH’s Mercury pathway profiling SEAP system. As shown in Fig. 2f, Fank1 overexpression markedly activated the HSE and AP-1 signalling pathways. This suggests that Fank1 exerts its function via the HSE and AP-1 signalling pathways. Because Jab1 has been reported to activate an AP-1-dependent promoter [5] and our results showed that Fank1 could interact with Jab1 in vivo, we focused our study on the effect of Fank1 on AP-1 transcriptional activity. HEK 293T cells were transfected with an AP-1 promoter-driven luciferase reporter gene in the presence of c-Jun, Jab1 and Fank1. The cells were then assayed for luciferase activity. The results indicate that Fank1 overexpression increased the relative activation levels of the AP-1 reporter and Fank1 enhanced AP-1 activity in a concentration-dependent manner (Fig. 2g). In addition, we found that either the FN3 domain or ankyrin repeats of Fank1 could increase AP-1 activity, but full-length Fank1 enhanced this activity more than either domain alone (Fig. 2h). To investigate the role of Jab1 in Fank1-dependent AP-1 activation further, we knocked down endogenous Jab1 using Jab1 siRNA and performed luciferase assays. Two independent siRNAs against Jab1 were employed in order to avoid non-specific and off-target effects. As shown in Fig. 2i, transfection of either Jab1 siRNA1 or siRNA2 significantly reduced endogenous Jab1 expression, and scrambled siRNA did not affect Jab1 expression. Luciferase analysis showed that knockdown of Jab1 inhibited Fank1-dependent activation of the AP-1 reporter gene. Taken together, these results suggest that Fank1 could activate the AP-1 pathway and that Jab1 is involved in Fank1-mediated activation of AP-1.
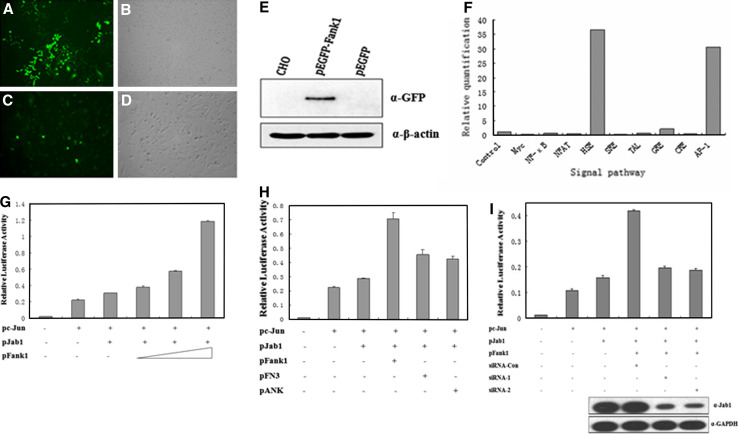
Fig. 2a–i.
Regulation of AP-1 activity by the interaction of Fank1 with Jab1. Stable CHO/GFP (a, b) and CHO/GFP-Fank1 (c, d) cell lines were generated. e Expression of Fank1 in the CHO/GFP-Fank1 stable cell line was confirmed by Western blot. f CHO/GFP-Fank1 and CHO/GFP stable cells were analysed with the Mercury system for transcription factor profiling. The SEAP activity was measured and is shown here as a bar graph. g HEK 293T cells were co-transfected with an AP-1-driven luciferase reporter, c-Jun, Jab1 and Fank1. Relative luciferase activity was measured. h HEK 293T cells were co-transfected with an AP-1-driven luciferase reporter, c-Jun, Jab1 and Fank1 (full-length or deletion mutants). Relative luciferase activity was measured. i HEK 293T cells were co-transfected with an AP-1-driven luciferase reporter, c-Jun, Jab1, Fank1 and the indicated siRNA. The lower panel indicates the knockdown of Jab1 by Jab1 siRNA. GAPDH was analysed as a loading control. The luciferase activity of each condition was normalised to a co-transfected pRL-TK vector. A representative example of at least three independent experiments, performed in triplicate, is shown. The error bars indicate standard deviation
The upregulation and activation of endogenous c-Jun by Fank1 are Jab1 dependent
To further define the mechanism of AP-1 activation by Fank1, the distribution and phosphorylation status of endogenous c-Jun were examined. HEK 293T cells were transfected with Flag-tagged Fank1 or with a control expression vector. After 24 h, the cells were harvested. Cytoplasmic and nuclear proteins were isolated and analysed by Western blot. As shown in Fig. 3a, Fank1 increased c-Jun expression in the nucleus. Phosphorylation of both Ser-63 and Ser-73 of c-Jun was elevated when Fank1 was over-expressed. Because JNK has been reported to bind to the c-Jun transactivation domain and phosphorylate Ser-63 and Ser-73 [9], we studied the effect of Fank1 on JNK activation. The results showed that JNK was activated in both the control and Fank1 groups. However, Fank1 overexpression did not alter JNK activation level (Fig. 3a). To further confirm the effect of Fank1 on c-Jun activation, we performed EMSA analysis by using biotin-labelled consensus AP-1 probe. As shown in Fig. 3b, Fank1 overexpression increased activated c-Jun binding. To investigate the role of Jab1 in endogenous c-Jun activation by Fank1, we suppressed the expression of endogenous Jab1 by RNA interference methods and isolated cytoplasmic and nuclear proteins for Western blot. The increase in c-Jun expression and activation by Fank1 was visibly inhibited when Jab1 was knocked down (Fig. 3c). Taken together, these results suggest that Fank1 could enhance endogenous c-Jun activity and that this regulation is Jab1 dependent.
Fig. 3a–c.
Activation of endogenous c-Jun by Fank1 is Jab1 dependent. a HEK 293T cells were transfected with Flag-tagged Fank1 or control expression vector. Cytoplasmic and nuclear proteins were extracted for Western blot using the indicated antibodies. b HEK 293T cells were transfected with Flag-tagged Fank1 or control expression vector. Nuclear proteins were extracted and incubated with biotin-labelled probe or competed with 100× or 200× unlabelled cold competitor for EMSA analysis. The specific complexes are indicated by arrows. The protein levels of c-Jun were demonstrated by Western blot. Lamin A/C was analysed as a loading control. c HEK 293T cells were transfected with Flag-tagged Fank1 expression vector and siRNA (Jab1 siRNA2 or control siRNA). Samples were analysed using the methods described in a. The band detected with the anti-Flag antibody in the Western blot assay indicated Fank1 expression
Fank1 inhibits cell apoptosis by upregulating Bcl-3 expression
AP-1 transcription factors are involved in both the induction and prevention of apoptosis [6–8]. To observe the effect of Fank1 on cell apoptosis, we generated a cell line that stably expressed the Flag-Fank1 recombinant protein (HeLa/Flag-Fank1) and a control stable cell line (HeLa/Con). Expression of the constructs was confirmed by reverse transcriptase-PCR and Western blot (Fig. 4a). We measured apoptosis by FACS analysis and observed that apoptosis decreased significantly at indicated times in stable HeLa/Flag-Fank1 cells in comparison to control stable cells after UV exposure (Fig. 4b). In general, activation of the caspase cascade plays an important role in apoptosis. Therefore, to evaluate the inhibition of apoptosis in stable HeLa/Flag-Fank1 cells after UV stimulation, we measured the cleavage of caspase 7 and poly(ADP-ribose) polymerase (PARP). When compared with the control stable cells, the level of cleaved caspase 7 and PARP decreased in stable HeLa/Flag-Fank1 cells (Fig. 4c), suggesting that Fank1 has an anti-apoptotic effect when cells are exposed to UV, which usually induces cell apoptosis.
Fig. 4a–e.
Fank1 inhibits UV-induced cell apoptosis, and this anti-apoptotic effect of Fank1 is Jab1 dependent. a Confirmation of Fank1 expression in the HeLa/Flag-Fank1 stable cell line by RT-PCR and Western blot. b PI incorporation FACS assay to analyse apoptosis in stable HeLa/Flag-Fank1 and HeLa/Con cell lines after UV irradiation. The percentage of apoptotic cells (% of total cells) was determined by program EXPO32-ADC, and the results are shown as a bar graph (*p < 0.05). The data are representative of three different experiments, and error bars represent the standard deviations of triplicate samples. c Stable cells were UV-irradiated. The cell lysates were analysed by immunoblotting with the indicated antibodies. GAPDH was used as a loading control. d Annexin V/PI FACS assay to analyse apoptosis in stable cells transfected with the indicated siRNA (Jab1 siRNA2 or control siRNA). The percentage of apoptotic cells (% of total cells), including early apoptotic cells (Annexin V+, PI−) and late apoptotic cells (Annexin V+, PI+), is shown as a bar graph (*p < 0.05). The data are representative of three different experiments, and error bars represent the standard deviations of triplicate sample. e Stable cells were transfected with the indicated siRNA (Jab1 siRNA2 or control siRNA). The cell lysates were analysed by immunoblotting with the same methods described in c. The band detected with the anti-Flag antibody in the Western blot indicated Fank1 expression
Our following results are in accordance with a report showing that UV exposure resulted in rapid and vigorous JNK activation [9], leading to phosphorylation of c-Jun, which enhanced its transcriptional capacity [10, 11]. Furthermore, we found that overexpressed Fank1 could enhance the total expression and phosphorylation levels of c-Jun when compared with the control group after UV stimulation (Fig. 4c). However, Fank1 did not affect the total expression and phosphorylation levels of JNK (Fig. 4c). The AP-1 inhibition of apoptosis is probably due to differential regulation of pro-apoptotic and anti-apoptotic target genes such as BCL-2, BCL-xL and Bcl-3 [8, 12, 13]. Therefore, we observed the effect of Fank1 overexpression on these AP-1-regulated target genes. Compared with the control group, Fank1 significantly upregulated Bcl-3 expression after UV exposure, but it did not affect the expression of BCL-xL and BCL-2 (Fig. 4c).
In addition, camptothecin (CPT) was used as another apoptosis inducer to detect Fank1’s anti-apoptotic role. When cells were challenged with CPT, we also observed that apoptosis was inhibited in stable HeLa/Flag-Fank1 cells as compared with control stable cells by FACS analysis (Supp. Fig. 1). The level of cleaved PARP decreased in stable HeLa/Flag-Fank1 cells compared to control stable cells after CPT stimulation (Supp. Fig. 2). Likewise, we found that overexpression of Fank1 upregulated c-Jun and Bcl-3 expression after CPT stimulation, but it did not affect the expression of JNK, BCL-xL or BCL-2 (Supp. Fig. 2). Taken together, the above findings suggested that Fank1 exerted its anti-apoptotic effect by activating AP-1 activity, which then upregulated the expression of Bcl-3.
Anti-apoptotic effect of Fank1 is Jab1 dependent
The above results indicate that knockdown of Jab1 inhibited Fank1-dependent AP-1 activation. Therefore, to investigate the functional linkage between Fank1 and Jab1 in the regulation of apoptosis, we silenced Jab1 in HeLa/Flag-Fank1 cells and examined apoptosis by FACS analysis. In all groups in which Jab1 was silenced, we found that Fank1 could not exert its anti-apoptotic effect after UV exposure (Fig. 4d). Compared with control HeLa/Flag-Fank1 cells which were transfected with scrambled siRNA, the level of cleaved PARP increased in Jab1-depleted HeLa/Flag-Fank1 cells after UV stimulation. Meanwhile, the upregulation of c-Jun phosphorylation and Bcl-3 expression by Fank1 was significantly suppressed when Jab1 was knocked down (Fig. 4e). Taken together, these results indicate that the inhibition of apoptosis by Fank1 is Jab1 dependent.
Fank1/Jab1/c-Jun complex binds to Bcl-3 promoter
To substantiate our model that Fank1 inhibits apoptosis by upregulating Bcl-3 expression, we asked whether Fank1/Jab1/c-Jun complex bound to Bcl-3 gene regulatory region. We searched the nucleotide sequence of this gene regulatory region for AP-1 binding sites, using ECR Browser online (http://ecrbrowser.dcode.org). We identified the sequence TGAGTCA, which fully matches the AP-1 consensus motif, in intron 1, located 1,171 bp upstream of the translation start codon in the second exon (Fig. 5a). To verify physical interaction between the Fank1/Jab1/c-Jun complex and the AP-1 binding site in the human Bcl-3 gene regulatory region in vitro and in vivo, we performed EMSA and ChIP assays. As shown in Fig. 5b, incubation of biotin-labelled oligonucleotides containing the newly identified AP-1 binding site in Bcl-3 promoter with nuclear proteins yielded a slowly migrating band, indicated as a protein-DNA complex. This specific band formed by the protein-DNA complex can be completed with an excess amount of unlabelled oligonucleotides containing AP-1 binding site in Bcl-3 promoter. Next, we performed ChIP assays to determine whether Fank1/Jab1/c-Jun complex was able to bind to the endogenous Bcl-3 promoter. As shown in Fig. 5c, Fank1, Jab1 and c-Jun were found in the same protein complex on the Bcl-3 promoter, suggesting that Fank1/Jab1/c-Jun complex bound to the regulatory portion of the Bcl-3 gene. Taken together, both EMSA and ChIP assays support the idea that Bcl-3 is targeted by the Fank1/Jab1/c-Jun complex.
Fig. 5a, b.
Fank1/Jab1/c-Jun complex binds to human Bcl-3 promoter region. a A schematic diagram of the Bcl-3 promoter that contains the identified AP-1 binding site. b Nuclear extract from HEK 293T cells transfected with Flag-Fank1 vector was incubated with biotin-labelled probe or competed with 50× or 200× unlabelled cold competitor for EMSA analysis. The specific complexes are indicated by arrows. c HEK 293T cells were transfected with Flag-Fank1 vector and were cross-linked with formaldehyde (final 1%) for 15 min at room temperature and then subjected to chromatin immunoprecipitation using specific anti-FLAG, anti-Jab1, anti-c-Jun antibodies or IgG (negative control) and the indicated primers. Reaction products were resolved by electrophoresis
Discussion
Fank1 is a protein that is highly expressed in testis. The function of Fank1 remains undefined. To gain insight into the potential function of Fank1, we searched for Fank1 interacting proteins using a yeast two-hybrid strategy. This assay identified Jab1, a critical component of the CSN complex. In mammalian cells, the CSN complex is composed of eight subunits that exhibit significant similarity to the eight subunits of the lid of the 26S proteasome and is involved in protein degradation [14, 15]. The precise biochemical functions of the CSN complex are not clear. However, each subunit of the CSN has been independently characterised. Jab1, the most studied subunit, has been shown to interact with a variety of proteins and to control their functions, including the regulation of gene transcription and the cell cycle [16]. For instance, Jab1 has been shown to play a critical role in the nuclear export and degradation of several tumour suppressors, including p27Kip1 [17], p53 [18] and Smad4 [19]. In the present study, we showed that Fank1 interacted with Jab1 in vivo by performing co-immunoprecipitation and GST pull-down assays. Fank1 encodes a protein that contains an FNIII domain and six ankyrin repeats. Deletion analysis revealed that the FNIII domain of Fank1 was required for the Fank1 interaction with Jab1. Tandem repeats of the FNIII domain have been shown to contain binding sites for DNA, heparin and the cell surface [20]. In addition, the ankyrin repeat is one of the most common protein–protein interaction motifs in nature [21]. Therefore, we speculate that the ankyrin repeats of Fank1 are required for interaction with other proteins. Our further analyses demonstrated the formation of a Fank1-Jab1 complex in the nucleus and the recruitment of this complex to the promoter-proximal region to regulate gene transcription.
Fank1 is supposed to be a transcription factor that regulates gene transcription and expression by activating specific signalling pathways. Our reporter gene analysis showed that Fank1 significantly activated both the HSE and AP-1 signalling pathways. Because we found that Fank1 interacted with Jab1 and it has been previously reported that Jab1 could bind to c-Jun and activate an AP-1-dependent promoter, we studied the effect of Fank1 on AP-1 transcriptional activity by luciferase analysis. While we found that full-length Fank1 induced the greatest enhancement of AP-1 activity, the FNIII domain or ankyrin repeats of Fank1 alone could partially increase AP-1 activity. Therefore, we conclude that maximum activation of the AP-1 reporter gene requires the combined action of both domains of Fank1. Due to its association with Jab1, it is easy to understand how the FNIII domain of Fank1 could partially activate the AP-1-driven luciferase reporter gene. However, it remains unclear from our experimental results how the ankyrin repeats of Fank1 are able to partially enhance AP-1 activity. We propose two possible mechanisms. First, the slight interaction observed between the ankyrin repeats of Fank1 and Jab1 is enough to activate the AP-1 reporter gene. Second, the ankyrin repeats of Fank1 increase AP-1 activity by associating with other proteins that could activate the AP-1 reporter gene. Finally, we observed the role of Jab1 in the Fank1-dependent mediation of AP-1 activation. These results showed that knockdown of Jab1 inhibited the activation of the AP-1 reporter gene by Fank1. This conclusion excludes the possibility that Fank1 could activate the AP-1 pathway independently of Jab1.
AP-1 activation is induced, in part, by the phosphorylation of c-Jun [22, 23]. Phosphorylation of Ser-63 and Ser-73, located within the trans-activation domain of c-Jun, potentiates the ability of c-Jun to activate transcription [24–26]. Previous studies have shown that induction of c-Jun transcription in response to TPA is mediated by Jun/AP-1 binding to a high-affinity AP-1 binding site (–1,056 to –1,049 bp: TGACATCA) in human c-Jun promoter region, suggesting that c-Jun is positively regulated by its own product, Jun/AP-1 [27]. Consistent with these reports, our experiments showed that Fank1 overexpression significantly increased the total expression and phosphorylation levels of endogenous c-Jun in the nucleus. JNK has been reported to be responsible for both the phosphorylation and activation of c-Jun [9]. In our experiment, Fank1 overexpression did not enhance JNK activation compared with the control group, excluding the possibility that the increased phosphorylation level of endogenous c-Jun induced by Fank1 is due to Fank1-mediated JNK activation. RNA interference experiments demonstrated that activation of endogenous c-Jun by Fank1 was Jab1 dependent. Therefore, we hypothesise that a more stable complex will be formed from Fank1, Jab1 and phosphorylated c-Jun that further enhances the activation of c-Jun by Fank1.
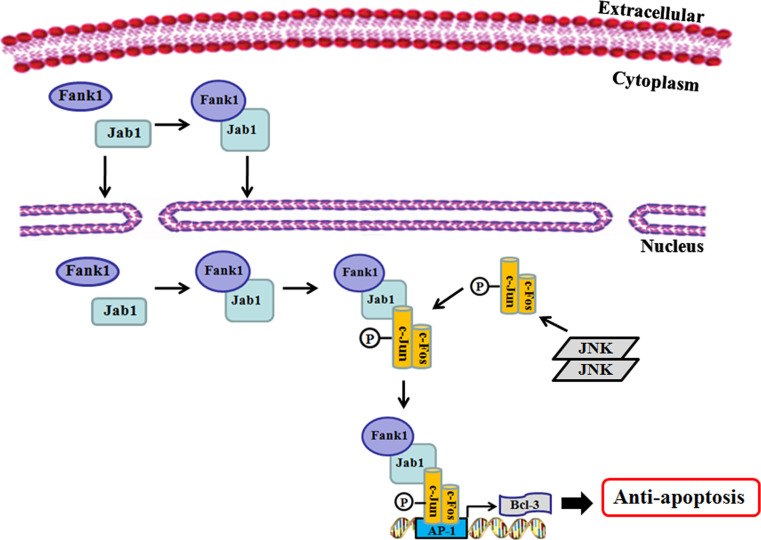
The AP-1 transcription factor regulates cell apoptosis in many cases [28–30]. In the current study, we found that Fank1 upregulated c-Jun expression and activated the AP-1 pathway, which allows us to speculate on the role of Fank1 in the regulation of apoptosis. After UV stimulation, we performed FACS analysis and observed a clear Fank1-dependent anti-apoptotic effect. Previous reports have demonstrated that c-Jun is the major component of the AP-1 complex induced after UV irradiation [31, 32]. Consistent with these reports, we found that Fank1 significantly enhanced the activation of c-Jun after UV stimulation, indicating that Fank1 exerts its anti-apoptotic effect by activating c-Jun. c-Jun has been reported to protect cells from apoptosis induced by UV irradiation [33], but the molecular biological evidence for such a role is lacking. In our experiments, we detected the expression of apoptosis-associated genes regulated by c-Jun. We found that Bcl-3, an anti-apoptotic protein regulated by c-Jun, was upregulated after UV stimulation, and Fank1 could further enhance Bcl-3 expression. To further support our model, we chose another apoptosis inducer, CPT, to repeat our experiments. After CPT stimulation, we observed that overexpression of Fank1 inhibited apoptosis and meanwhile upregulated c-Jun and Bcl-3 expression, which is consistent with our findings obtained from UV stimulation. Our subsequent experiments suggested that Jab1 is necessary for the further activation of c-Jun and Bcl-3 by Fank1. Finally, we identified a new AP-1 binding site in human Bcl-3 promoter, and both EMSA and ChIP assays supported that Bcl-3 was targeted by the Fank1/Jab1/c-Jun complex. Based on the experimental data provided herein and the previously published results, we are now able to propose a model for the anti-apoptotic mechanism of Fank1. First, a complex containing Fank1 and Jab1 will be formed in the nucleus. Then, the complex will bind to phosphorylated c-Jun through Jab1, forming a stable transcription complex that constantly binds to the AP-1 binding site in the Bcl-3 promoters. Finally, the transcription and expression of Bcl-3 will be visibly enhanced (Fig. 6). Note that this model forms a positive regulatory loop: Fank1 enhances the expression and activation of c-Jun through Jab1 binding; then, activated c-Jun participates in the formation of an activated transcription complex, which further upregulates the anti-apoptotic protein Bcl-3.
Fig. 6.
Model depicting the anti-apoptotic mechanism of Fank1
In summary, these findings demonstrate that Fank1 is an anti-apoptotic protein that inhibits apoptosis of cells by activating the AP-1 pathway. It is interesting to note that both Fank1 and c-Jun are expressed at peak levels in the late stage of spermatogenesis [3, 34]. Taken together, we speculate that Fank1 has an anti-apoptotic effect in spermatogenesis, although further study is still needed to confirm this speculation.
Electronic supplementary material
Below is the link to the electronic supplementary material.
PI incorporation FACS analysis of Fank1 inhibition of CPT induced apoptosis. Stable HeLa/Flag-Fank1 and HeLa/Con cell lines were exposed to 10 µM CPT. At indicated times, cells were treated as described in ‘Materials and Methods’ section and analysed by FACS. The percentage of apoptotic cells (% of total cells) was determined by program EXPO32-ADC, and the results were shown as a bar graph (*p<0.05). The data were representative of three different experiments and error bars represented the standard deviations of triplicate samples. (TIFF 19882 kb)
Fank1 inhibited CPT induced apoptosis by upregulating c-Jun and Bcl-3 expression. Stable HeLa/Flag-Fank1 and HeLa/Con cell lines were exposed to 10 µM CPT. The cell lysates were analysed by immunoblotting with the indicated antibodies. GAPDH was used as a loading control. (TIFF 24484 kb)
Annexin V/PI FACS assay to analyse apoptosis in stable cells transfected with the indicated siRNA (Jab1 siRNA2 or control siRNA). Stable cells were treated as described in ‘Materials and Methods’. After staining with FITC-Annexin V and PI, cells were analysed by the Accuri C6 flow cytometer system. Inset numbers represent the percentage of each population in the quadrants. Early apoptotic cells (Annexin V+/PI-, Q1-LR), late apoptotic cells (Annexin V+/PI+, Q1-UR) and dead cells (Annexin V-/PI+, Q1-UL). Data were from one of three independent experiments which yielded similar results. (TIFF 25386 kb)
Acknowledgments
We thank Dr. Dong-Yan Jin for constructive suggestions and critical reading of manuscript. This work was supported by grants from the National Program for Important Research Plans (2006CB944002) and the National Program for Key Basic Research Projects (2006CB504001) from the Ministry of Science and Technology of China, a grant for Creative Research Group (No. 30721063) from the National Natural Science Foundation of China, the National Laboratory Special Fund (2060204), and the Institute Fund for Young Scientist (2009PY10).
Contributor Information
Wei Song, Email: Roy_sw0925@yahoo.com.cn.
Linfang Wang, FAX: +86-010-65240529, Email: wang.linfang@imicams.ac.cn.
References
- 1.Kierszenbaum AL. Mammalian spermatogenesis in vivo and in vitro: a partnership of spermatogenic and somatic cell lineages. Endocr Rev. 1994;15:116–134. doi: 10.1210/edrv-15-1-116. [DOI] [PubMed] [Google Scholar]
- 2.Sinha Hikim AP, Swerdloff RS. Hormonal and genetic control of germ cell apoptosis in the testis. Rev Reprod. 1999;4:38–47. doi: 10.1530/ror.0.0040038. [DOI] [PubMed] [Google Scholar]
- 3.Zheng Z, Zheng H, Yan W. Fank1 is a testis-specific gene encoding a nuclear protein exclusively expressed during the transition from the meiotic to the haploid phase of spermatogenesis. Gene Expr Patterns. 2007;7:777–783. doi: 10.1016/j.modgep.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Wei N, Tsuge T, Serino G, Dohmae N, Takio K, Matsui M, Deng XW. The COP9 complex is conserved between plants and mammals and is related to the 26S proteasome regulatory complex. Curr Biol. 1998;8:919–922. doi: 10.1016/S0960-9822(07)00372-7. [DOI] [PubMed] [Google Scholar]
- 5.Claret FX, Hibi M, Dhut S, Toda T, Karin M. A new group of conserved coactivators that increase the specificity of AP-1 transcription factors. Nature. 1996;383:453–457. doi: 10.1038/383453a0. [DOI] [PubMed] [Google Scholar]
- 6.Schreiber M, Kolbus A, Piu F, Szabowski A, Mohle-Steinlein U, Tian J, Karin M, Angel P, Wagner EF. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev. 1999;13:607–619. doi: 10.1101/gad.13.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivanov VN, Bhoumik A, Krasilnikov M, Raz R, Owen-Schaub LB, Levy D, Horvath CM, Ronai Z. Cooperation between STAT3 and c-jun suppresses Fas transcription. Mol Cell. 2001;7:517–528. doi: 10.1016/S1097-2765(01)00199-X. [DOI] [PubMed] [Google Scholar]
- 8.Rebollo A, Dumoutier L, Renauld JC, Zaballos A, Ayllon V, Martinez AC. Bcl-3 expression promotes cell survival following interleukin-4 deprivation and is controlled by AP1 and AP1-like transcription factors. Mol Cell Biol. 2000;20:3407–3416. doi: 10.1128/MCB.20.10.3407-3416.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hibi M, Lin A, Smeal T, Minden A, Karin M. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 10.Gupta S, Campbell D, Derijard B, Davis RJ. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 11.Karin M. The regulation of AP-1 activity by mitogen-activated protein kinases. J Biol Chem. 1995;270:16483–16486. doi: 10.1074/jbc.270.28.16483. [DOI] [PubMed] [Google Scholar]
- 12.Passegue E, Jochum W, Schorpp-Kistner M, Mohle-Steinlein U, Wagner EF. Chronic myeloid leukemia with increased granulocyte progenitors in mice lacking junB expression in the myeloid lineage. Cell. 2001;104:21–32. doi: 10.1016/S0092-8674(01)00188-X. [DOI] [PubMed] [Google Scholar]
- 13.Kashatus D, Cogswell P, Baldwin AS. Expression of the Bcl-3 proto-oncogene suppresses p53 activation. Genes Dev. 2006;20:225–235. doi: 10.1101/gad.1352206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwechheimer C. The COP9 signalosome (CSN): an evolutionary conserved proteolysis regulator in eukaryotic development. Biochim Biophys Acta. 2004;1695:45–54. doi: 10.1016/j.bbamcr.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 15.Tomoda K, Kubota Y, Arata Y, Mori S, Maeda M, Tanaka T, Yoshida M, Yoneda-Kato N, Kato JY. The cytoplasmic shuttling and subsequent degradation of p27Kip1 mediated by Jab1/CSN5 and the COP9 signalosome complex. J Biol Chem. 2002;277:2302–2310. doi: 10.1074/jbc.M104431200. [DOI] [PubMed] [Google Scholar]
- 16.Chamovitz DA, Segal D. Jab1/CSN5 and the COP9 signalosome. a complex situation. EMBO Rep. 2001;2:96–101. doi: 10.1093/embo-reports/kve028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomoda K, Kubota Y, Kato J. Degradation of the cyclin-dependent-kinase inhibitor p27Kip1 is instigated by Jab1. Nature. 1999;398:160–165. doi: 10.1038/18230. [DOI] [PubMed] [Google Scholar]
- 18.Bech-Otschir D, Kraft R, Huang X, Henklein P, Kapelari B, Pollmann C, Dubiel W. COP9 signalosome-specific phosphorylation targets p53 to degradation by the ubiquitin system. EMBO J. 2001;20:1630–1639. doi: 10.1093/emboj/20.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wan M, Cao X, Wu Y, Bai S, Wu L, Shi X, Wang N, Cao X. Jab1 antagonizes TGF-beta signaling by inducing Smad4 degradation. EMBO Rep. 2002;3:171–176. doi: 10.1093/embo-reports/kvf024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skorstengaard K, Jensen MS, Sahl P, Petersen TE, Magnusson S. Complete primary structure of bovine plasma fibronectin. Eur J Biochem. 1986;161:441–453. doi: 10.1111/j.1432-1033.1986.tb10464.x. [DOI] [PubMed] [Google Scholar]
- 21.Mosavi LK, Minor DL, Jr, Peng ZY. Consensus-derived structural determinants of the ankyrin repeat motif. Proc Natl Acad Sci USA. 2002;99:16029–16034. doi: 10.1073/pnas.252537899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:131–136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 23.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 24.Binetruy B, Smeal T, Karin M. Ha-Ras augments c-Jun activity and stimulates phosphorylation of its activation domain. Nature. 1991;351:122–127. doi: 10.1038/351122a0. [DOI] [PubMed] [Google Scholar]
- 25.Smeal T, Binetruy B, Mercola DA, Birrer M, Karin M. Oncogenic and transcriptional cooperation with Ha-Ras requires phosphorylation of c-Jun on serines 63 and 73. Nature. 1991;354:494–496. doi: 10.1038/354494a0. [DOI] [PubMed] [Google Scholar]
- 26.Smeal T, Binetruy B, Mercola D, Grover-Bardwick A, Heidecker G, Rapp UR, Karin M. Oncoprotein-mediated signalling cascade stimulates c-Jun activity by phosphorylation of serines 63 and 73. Mol Cell Biol. 1992;12:3507–3513. doi: 10.1128/mcb.12.8.3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Angel P, Hattori K, Smeal T, Karin M. The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell. 1988;55:875–885. doi: 10.1016/0092-8674(88)90143-2. [DOI] [PubMed] [Google Scholar]
- 28.Ham J, Babij C, Whitfield J, Pfarr CM, Lallemand D, Yaniv M, Rubin LL. A c-Jun dominant negative mutant protects sympathetic neurons against programmed cell death. Neuron. 1995;14:927–939. doi: 10.1016/0896-6273(95)90331-3. [DOI] [PubMed] [Google Scholar]
- 29.Estus S, Zaks WJ, Freeman RS, Gruda M, Bravo R, Johnson EM., Jr Altered gene expression in neurons during programmed cell death: identification of c-jun as necessary for neuronal apoptosis. J Cell Biol. 1994;127:1717–1727. doi: 10.1083/jcb.127.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bossy-Wetzel E, Bakiri L, Yaniv M. Induction of apoptosis by the transcription factor c-Jun. EMBO J. 1997;16:1695–1709. doi: 10.1093/emboj/16.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Angel P, Baumann I, Stein B, Delius H, Rahmsdorf HJ, Herrlich P. 12-O-tetradecanoyl-phorbol-13-acetate induction of the human collagenase gene is mediated by an inducible enhancer element located in the 5′-flanking region. Mol Cell Biol. 1987;7:2256–2266. doi: 10.1128/mcb.7.6.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Angel P, Allegretto EA, Okino ST, Hattori K, Boyle WJ, Hunter T, Karin M. Oncogene jun encodes a sequence-specific trans-activator similar to AP-1. Nature. 1988;332:166–171. doi: 10.1038/332166a0. [DOI] [PubMed] [Google Scholar]
- 33.Wisdom R, Johnson RS, Moore C. c-Jun regulates cell cycle progression and apoptosis by distinct mechanisms. EMBO J. 1999;18:188–197. doi: 10.1093/emboj/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shalini S, Bansal MP. Role of selenium in spermatogenesis: differential expression of cjun and cfos in tubular cells of mice testis. Mol Cell Biochem. 2006;292:27–38. doi: 10.1007/s11010-006-9168-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PI incorporation FACS analysis of Fank1 inhibition of CPT induced apoptosis. Stable HeLa/Flag-Fank1 and HeLa/Con cell lines were exposed to 10 µM CPT. At indicated times, cells were treated as described in ‘Materials and Methods’ section and analysed by FACS. The percentage of apoptotic cells (% of total cells) was determined by program EXPO32-ADC, and the results were shown as a bar graph (*p<0.05). The data were representative of three different experiments and error bars represented the standard deviations of triplicate samples. (TIFF 19882 kb)
Fank1 inhibited CPT induced apoptosis by upregulating c-Jun and Bcl-3 expression. Stable HeLa/Flag-Fank1 and HeLa/Con cell lines were exposed to 10 µM CPT. The cell lysates were analysed by immunoblotting with the indicated antibodies. GAPDH was used as a loading control. (TIFF 24484 kb)
Annexin V/PI FACS assay to analyse apoptosis in stable cells transfected with the indicated siRNA (Jab1 siRNA2 or control siRNA). Stable cells were treated as described in ‘Materials and Methods’. After staining with FITC-Annexin V and PI, cells were analysed by the Accuri C6 flow cytometer system. Inset numbers represent the percentage of each population in the quadrants. Early apoptotic cells (Annexin V+/PI-, Q1-LR), late apoptotic cells (Annexin V+/PI+, Q1-UR) and dead cells (Annexin V-/PI+, Q1-UL). Data were from one of three independent experiments which yielded similar results. (TIFF 25386 kb)