Abstract
The Dictyostelium centrosome consists of a layered core structure surrounded by a microtubule-nucleating corona. A tight linkage through the nuclear envelope connects the cytosolic centrosome with the clustered centromeres within the nuclear matrix. At G2/M the corona dissociates, and the core structure duplicates, yielding two spindle poles. CP148 is a novel coiled coil protein of the centrosomal corona. GFP-CP148 exhibited cell cycle-dependent presence and absence at the centrosome, which correlates with dissociation of the corona in prophase and its reformation in late telophase. During telophase, GFP-CP148 formed cytosolic foci, which coalesced and joined the centrosome. This explains the hypertrophic appearance of the corona upon strong overexpression of GFP-CP148. Depletion of CP148 by RNAi caused virtual loss of the corona and disorganization of interphase microtubules. Surprisingly, formation of the mitotic spindle and astral microtubules was unaffected. Thus, microtubule nucleation complexes associate with centrosomal core components through different means during interphase and mitosis. Furthermore, CP148 RNAi caused dispersal of centromeres and altered Sun1 distribution at the nuclear envelope, suggesting a role of CP148 in the linkage between centrosomes and centromeres. Taken together, CP148 is an essential factor for the formation of the centrosomal corona, which in turn is required for centrosome/centromere linkage.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-011-0904-2) contains supplementary material, which is available to authorized users.
Keywords: Dictyostelium, Corona, Microtubules, Centrosome, Nucleus
Introduction
Microtubule organization is of crucial importance not only for formation of the mitotic spindle and chromosome segregation but also for directional transport and positioning of organelles, RNA, and proteins, which in turn direct processes such as cell polarization and cytokinesis. The main microtubule organizing centers are known as centrosomes. Despite their universal functions, centrosomes display remarkably different morphologies in animals, fungi, and lower eukaryotes. All organisms possessing cilia or flagella at least in some cell types have centrosomes with a pair of centrioles. Centrioles consist of a ninefold symmetrical cylindrical arrangement of specialized short microtubules [1]. They are embedded in so-called pericentriolar material (PCM) containing γ complexes and other proteins involved in microtubule nucleation. Since centrosomes participate in the organization of the bipolar mitotic spindle, they have to duplicate once and only once per cell cycle. In case of centriole-containing centrosomes, duplication of the single centriole pair starts at the G1/S transition with the growth of daughter centrioles at the proximal end of the existing mother centrioles. In G2, the cell contains two pairs of centrioles. In late G2, the centriole pairs separate, and are now ready to act as mitotic spindle poles. In late mitosis, the tight orthogonal association of mother and daughter centrioles is abrogated in a process called disengagement. This appears to be the licensing event for the next round of centriole duplication. Proteomic analyses of isolated centrosomes led to the discovery of many new centrosomal and centriolar proteins including several players in centriole duplication (reviewed by Azimzadeh and Marshall (2010) [1]). Centrioles are not the only dynamic part of the centrosome. In late G2, when cells prepare for mitosis, microtubule nucleating activity and the size of the PCM increases. The PCM is formed by recruitment of microtubule-nucleating γ-tubulin ring complexes to scaffolding proteins, which in turn are associated with centrioles. Three such proteins have been characterized in this context, D-PLP/pericentrin, D-Spd-2/Cep192, and Cnn/Cep215/Cdk5Rap2 [2–4]. All of them associate with γ-tubulin ring complexes through NEDD1/GCP-WD [4]. The control of centrosome size has thoroughly been investigated in Drosophila. Here, centrosome size is regulated by Cnn (centrosomin), which is phosphorylated by polo and associates with the centriole components Asl (Asterless, Cep152 in humans) and DSpd-2 (Cep192 in humans) [5, 6].
Except for yeasts, little is known about organization and biogenesis of main microtubule-organizing centers in fungi and lower eukaryotes, where they are often called spindle pole bodies (SPBs) or nucleus-associated bodies (NABs), respectively. Despite the lack of centrioles, the term “centrosome” is used by many researchers also for these organelles, since they all serve common functions and share many conserved proteins. The centrosome of Dictyostelium amoebae is a cytosolic nucleus-associated body (reviewed by Gräf et al. (2004) [7]). It consists of an electron-dense core structure composed of three layered major disks. The core structure is surrounded by a less-structured corona with dense nodules containing γ-tubulin. Thus, the corona is considered the functional equivalent of the pericentriolar material in animal cells. Based on morphological changes, Dictyostelium centrosome duplication is not synchronized with S-phase but with the G2/M transition, similar to the situation in fission yeast [8, 9]. First, the whole organelle increases in size, and in early prophase, the corona disappears and all microtubules are shed from the centrosome and depolymerize. The central disk disappears, and the remaining outer disks insert into the nuclear envelope, which remains intact throughout mitosis. In prometaphase, the disks separate from each other and instantly start to nucleate a central spindle from their former inner surfaces. Upon spindle elongation, the two disks are driven to the poles and act as the mitotic centrosomes. In anaphase, the disks start to fold back onto themselves in a way that the microtubule-nucleating surface is turned outward. In telophase, the folding and maturation process is completed with re-appearance of the central layer and the corona. Elucidation of the molecular processes underlying these substantial morphological events requires information about the proteins involved. Several corona components such as centrin, γ-tubulin complex proteins (γ-tubulin, Spc97, Spc98), the XMAP215-family protein CP224, TACC, EB1, LIS1, and CP248/250 have been thoroughly investigated [10–17]. We were recently able to identify four novel components of the central core structure through a proteomic screen (CP39, CP55, CP75, and CP91) [15, 18]. Among the known corona proteins was no candidate for a function in corona formation and linking its γ-tubulin complexes to the central core component, since they all localized to the corona farther distal from the core structure than γ-tubulin itself. Yet, in our proteomic screen we have also identified a novel protein, CP148, localizing to an intermediate zone, which stained with outer corona markers and core markers. In this work, we provide a functional characterization of CP148, and show that it is required for the integrity and stability of the corona and its association to the components of the centrosomal core structure.
Results
CP148 dissociates from the centrosome in prophase and reappears in telophase
CP148 (dictybase geneID DDB_G0278809) is a 148.7-kDa protein with a length of 1,297 amino acids, about 50% of which are predicted to form coiled coil structures (COILS program, window size 28 aa). In our recently published survey of novel centrosomal proteins, we have shown that CP148 is a genuine centrosomal protein, since the GFP-CP148 fusion protein localized to the centrosomal corona in isolated, microtubule-free centrosomes [15]. In the same study, we also reported that the GFP-fusion protein was absent from centrosomes, i.e., spindle poles in metaphase cells. However, nothing was known about the function of CP148 or the behavior of the endogenous CP148 protein, nor of the time points of its dissociation from the centrosome during mitosis and its re-association. In order to address these gaps of knowledge, we first raised polyclonal antibodies against a C-terminal fragment of 357 amino acids and used these antibodies to localize endogenous CP148 during interphase and all mitotic stages. Figure 1 shows that CP148 is absent from the centrosome already in prophase and reappears only in telophase. This behavior corresponds well to the time frame of dissolution and reformation of the corona during centrosome duplication. However, it is in contrast to the behavior of other corona proteins involved in microtubule nucleation, such as γ-tubulin and the XMAP215-family protein CP224, which are present at centrosomes during the entire cell cycle [10, 14].
Fig. 1.

Endogenous CP148 dissociates from the centrosome in prophase and reappears in late telophase. Dictyostelium AX2 cells were fixed with glutaraldehyde and stained with anti-CP148/anti-rabbit-AlexaFluor 568 (red), anti-tubulin YL1/2 [39]/anti-rat-AlexaFluor 488 (green) and DAPI (blue). Cell cycle stages and stainings are indicated. Bar 3 μm
GFP-CP148 forms cytosolic clusters in telophase that relocate to the centrosome
The centrosomal cell cycle-dependent localization pattern of endogenous CP148 was mimicked by GFP-CP148 (supplementary Fig. S1, Movie S1). In GFP-CP148 cells, we also frequently observed tiny, spot-like clusters of GFP-CP148 in the cytosol, which were never observed in case of endogenous CP148. Upon strong overexpression (~300-fold as judged from Western-blot densitometry, Fig. 3j) of the fusion protein, clusters were more prominent and varied in size and number (Fig. 2a, b). Some but not all of these GFP-CP148 clusters also stained positive for γ-tubulin and CP224 (Fig. 2a, b). Live cell imaging revealed that most of these clusters formed de novo during telophase and migrated in a radial fashion towards the spindle poles where they appeared to fuse (Movie S1). GFP-CP148 clusters were always located in close vicinity to astral microtubules in telophase cells (Fig. 2c). This observation and pattern of movement of GFP-CP148 clusters strongly suggested transport along microtubules through association with a minus end-directed microtubule motor such as dynein.
Fig. 3.

Distribution of centrosomal and nuclear envelope marker proteins upon GFP-CP148 overexpression. Cells in interphase were fixed with methanol (b, e, f, g, h, i) or glutaraldehyde (a, c). GFP-CP148 overexpressing cells were stained with anti-tubulin YL1/2 (a), anti-γ-tubulin (b), anti-Sun1 (c) [22], anti-NE81 (d), anti-CP91 (e), anti-CP55 (f), anti-CP39 (g) [15], and anti-CP224 (h ). The merged images show antibody stainings in red, GFP-CP148 in green, and DAPI in blue. AX2 control cells were stained with anti- CP148 (green) and the corona marker anti-CP224 (red) (I). Bar 3 μm. The extent of GFP-CP148 overexpression is shown on the immunoblot (j). The Western blot of cell extracts was stained with anti-CP148/anti-rabbit-peroxidase and enhanced chemiluminescence visualization (upper panel) and Ponceau S (lower panel) to demonstrate equal protein loading of extracts from AX2 control cells and GFP-CP148 overexpressors. The diagram in k shows the increase in size of the corona measured by the area stained with anti-CP224 in AX2 control cells (n = 96) and GFP-CP148 overexpressors (n = 103). Mean values ± SD are given
Fig. 2.

The corona proteins γ-Tubulin and CP224 partially colocalize at cytosolic GFP-CP148 foci. GFP-CP148 overexpressing cells in telophase were fixed with methanol and stained with anti-γ-tubulin (a) [20], anti-CP224 [40] (b), or anti-tubulin YL1/2 (c). The merged images (a′′′, b′′′, c) show antibody stainings with secondary AlexaFluor 568 conjugates in red, GFP-CP148 in green and DAPI in blue. Bar 3 μm
GFP-CP148 overexpression causes hypertrophy of the centrosomal corona
As a consequence of particle movement and fusion at the centrosome, the corona often appeared enlarged and irregularly shaped. This became obvious by immunofluorescence stainings with antibodies against the corona markers γ-tubulin and CP224 (Fig. 3, Movie S1). If the area stained with anti-CP224 in maximum intensity projections of these specimens was taken as a measure for the size of the corona, we calculated a 2.7-fold increase compared to control cells (Fig. 3k). In contrast, the centrosomal core appeared unaffected by overexpression of GFP-CP148, since it was still visible only as a tiny, spherical dot in immunofluorescence specimens stained with antibodies against the centrosomal core proteins CP39, CP55, and CP91 (Fig. 3). These antibodies only stained the nucleus-associated centrosome, but never any of the cytosolic, non centrosome-associated GFP-CP148 clusters, indicating that these clusters do not represent supernumerary centrosomes. Electron microscopy of GFP-CP148 overexpressing cells more closely revealed the ultrastructural aberrations in centrosomal morphology, and confirmed our conclusions drawn from immunofluorescence microscopy. While the layered core structure appeared more or less unaltered by overexpression, the corona was considerably enlarged. In wild-type cells, it usually consists of only one layer of electron-dense nodules within an amorphous matrix around the core structure (Fig. 4a) [19]. In contrast, in GFP-CP148 overexpressors, the core structure is embedded in a greatly extended amorphous matrix full of nodules (Fig. 4c, c’). Since γ-tubulin has been addressed to the nodules [20], this explains why the γ-tubulin signal was spread all over this enlarged corona structure in immunofluorescence specimens (Fig. 3b).
Fig. 4.
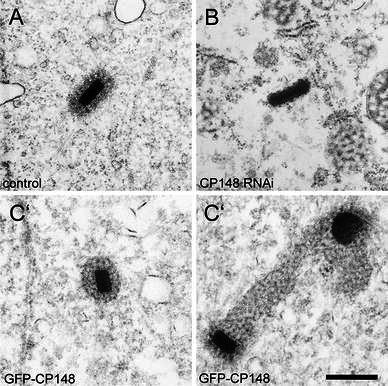
Depletion of CP148 by RNA results in loss of the corona, while overexpression of GFP-CP148 causes hypertrophy of the corona. Ultrathin sections through the centrosome of control cells (a), CP148-RNAi cells (b), and GFP-CP148 overexpressing cells (c, c′) are shown. Bar 0.5 μm
GFP-CP148 shows no significant mobility at the centrosome
In order to get information about the dynamic behavior of CP148 at the centrosome during interphase, we performed fluorescence recovery after photo bleaching (FRAP) experiments. In these experiments, we found no significant recovery of GFP-CP148 fluorescence within a time of 22 min (Fig. 5/Movie S2). Thus, GFP-CP148 behaved clearly differently from other corona components such as CP224, which showed a half time of recovery of only 7.2 s [17]. Taken together with the data of its cell cycle-dependent localization pattern, our FRAP data indicate that CP148 could function as a scaffolding factor for other corona components.
Fig. 5.

GFP-CP148 shows almost no fluorescence recovery after photobleaching. Selected time points of a representative FRAP experiment (Movie S2) with GFP-CP148/mCherry-H2B cells are shown. Upper row, centrosome bleached with a point-focused 473-nm laser pulse at time point 170 s; lower row, non-bleached control cell. Confocal spinning disk microscopy; time lapse acquisition rate was six stacks per minute (at a frame rate of 10 fr/s); maximum intensity projection of seven slices per image stack. The graph shows recovery kinetics from six independent measurements (mean ± SD)
Depletion of CP148 causes disintegration of the corona and interphase microtubule arrays
To prove the idea of CP148 as a scaffolding factor for corona assembly, and to gain more insight into the functions of CP148, we set out to suppress expression of the CP148 protein. Since our attempts to knockout the CP148 gene were not successful, we generated a stable CP148 RNAi strain based on the wild-type-like AX2 strain. Protein expression of CP148 in this strain was suppressed completely as shown by Western blots of Dictyostelium extracts (Fig. 6). CP148 RNAi cells were characterized by a severely disordered appearance of microtubule arrays lacking a detectable MTOC. The corona markers γ-tubulin and CP224 were no longer concentrated at a clearly discernible MTOC (compare Fig. 7a–a′′′ with Fig. 1). In immunofluorescence specimens, γ-tubulin was sometimes still detectable in a few foci, which, however, could not be correlated with an MTOC (Fig. 7b′). CP224 signals were distributed all over the cell with no clear concentration at a point in the nuclear vicinity, where a centrosome would be expected to be localized (Fig. 7b). These results strongly suggested that the complete corona was lost from the centrosome upon CP148 RNAi. This notion was confirmed by the ultrastructural appearance of the centrosomal remnants in CP148-RNAi cells. In ultrathin sections, we detected layered core structures in the cytosol, which were clearly devoid of the usual corona with its electron-dense nodules (Fig. 4b). The size and appearance of these core structures in CP148-RNAi cells was comparable to that of intact centrosomes in control cells (Fig. 4a).
Fig. 6.

CP148 is knocked down almost completely by RNAi. Western blot of cell extracts stained with anti-CP148/anti-rabbit-peroxidase and enhanced chemiluminescence visualization to show the extent of RNAi (left). The right panel shows the corresponding protein staining of the same blot with Ponceau S to demonstrate equal protein loading of extracts from AX2 control cells and CP148-RNAi cells
Fig. 7.

CP148-RNAi disrupts radial microtubule arrays (a) and causes an increased number of microtubule plus ends (c, d). The corona markers CP224 and γ-tubulin can no longer be correlated with an MTOC (b). CP148-RNAi cells (a, b), CP148-RNAi cells expressing the GFP-TACC-domain (c) and GFP-TACC-domain control cells (d) were fixed with glutaraldehyde (a) or methanol (b, c, d) and stained with anti-CP148 and anti-tubulin YL1/2 (a, a′), anti-γ-tubulin (b) and anti-CP224 (b, c, d). The merged images (a′′′–d′′′) show antibody stainings with AlexaFluor 568 conjugates (a–d) in red, GFP fluorescence (c′, d′) and antibody stainings with AlexaFluor 488 conjugates (a′, b′) in green, and DAPI (a″–d″) in blue. Bars 5 μm
The CP224-positive dots within the cytosol visible in Fig. 7b were likely to correspond with microtubule plus ends, indicating that the total number of individual microtubules was increased upon CP148 RNAi. To confirm this, CP148 was also knocked down by RNAi in a strain expressing the TACC-domain fused to GFP. The TACC protein recruits CP224 to microtubule plus ends and the GFP-tagged TACC-domain is the best microtubule plus end marker in Dictyostelium [17]. When these cells with green fluorescent microtubule tips were compared to the GFP-TACCdom control strain (Fig. 7d), it became evident that the number of microtubule plus ends was clearly increased upon CP148 RNAi (Fig. 7c).
Depletion of CP148 causes increased ploidy and disrupts the linkage of the centrosome to clustered centromeres
CP148 RNAi cells were also characterized by an increase in size by a factor of two to five (Figs. 8, 9a). Furthermore, the appearance of nuclei in DAPI stainings suggested that CP148 RNAi frequently caused aneuploidy. In order to investigate this issue further, we knocked down CP148 expression by RNAi also in Dictyostelium GFP-α-tubulin cells. When mixed with AX2 cells, these CP148 RNAi/GFP-α-tubulin cells were easily discernible by their green fluorescent microtubules. Comparison of their DAPI staining intensities with those of AX2 cells in the same sample revealed that the DNA content per nucleus was increased by a factor of 1.67 in CP148 RNAi/GFP-α-tubulin cells (Fig. 9b). When the DNA content of individual cells was plotted by increasing DNA content, it became clear that 42% of all CP148 RNAi/GFP-α-tubulin cells had a DNA content that was more than twofold higher than the average DNA content of control cells, while this occurred in almost none of the control cells (Fig. 9b′, b″), indicating that the increased DNA content was not due to a prolonged time in S-phase. It is rather caused by increased ploidy, and suggests that cells encounter problems in chromosome segregation upon CP148 RNAi. In addition, we observed centromeres distributed over the whole nucleus in interphase (Fig. 8c, d). Since centromeres are usually clustered and linked to the centrosome through the nuclear envelope during the entire cell cycle [21, 22], this observation suggested that CP148 RNAi could also affect the linkage of centrosomes to nuclei. Sun1, a transmembrane protein of the nuclear envelope [22, 23], is a key component of this linkage structure. Therefore, we looked for alterations in the distribution of Sun1 at the nuclear envelope. Indeed, Sun1, which accumulates in the pericentrosomal region in wild-type cells, was evenly distributed over the nuclear envelope in CP148 RNAi cells (Fig. 8a, b). An involvement of CP148 in maintenance of the centrosome/nucleus linkage was also supported by the fact that the centrosomal core components CP55, CP91 and CP39 [15] were visible in single or several immunofluorescent spots, often detached from the nucleus (Fig. 8f′′′, h′′′). The strict colocalization of these core components is in agreement with our ultrastructural data showing that the centrosomal core appeared as an intact structural entity in CP148 RNAi cells (Fig. 4b). The absence of a discernible corona, while core components were frequently detached from the nucleus, indicated a requirement of the corona for centrosome/nucleus attachment. Together with the dispersal of the centromere cluster, and the abrogation of the pericentrosomal localization pattern of Sun1, these observations support our notion that CP148 RNAi is required for maintenance of the centrosome/nucleus linkage.
Fig. 8.

CP148 RNAi causes Sun1 displacement and centromere dispersal while centrosomal core components remain clustered in discrete foci. CP148 was depleted by RNAi in AX2 (b, d) and GFP-CP55 cells (f, h). Corresponding control cells for each antibody staining are shown in a, c, e, g. Cells were fixed with glutaraldehyde (a, b) or methanol (C–H) and stained with anti-Sun1 (a, b), the centromere marker anti-Cenp68 [17] (c, d), anti-CP39 (e, f), anti-CP91 (g, h) and anti-tubulin YL1/2 (a′–d′). The merged images (a′′′–h′′′) show antibody stainings with AlexaFluor 568 conjugates in red, GFP fluorescence (e′–h′) and antibody stainings with AlexaFluor 488 conjugates in green (a′–d′), and DAPI (a″–h″) in blue. Bars 3 μm
Fig. 9.

CP148 RNAi causes an increase in cell size and DNA content. In order to estimate cell size and DNA content within the same specimen, AX2 control cells and GFP-α-tubulin cells were mixed 1:1 and stained with anti-CP148 along with a AlexaFluor 568 labeled secondary antibody and DAPI. RNAi cells were discernible by the lack of green microtubule fluorescence and CP148 labeling. a The area of the cells in phase contrast images was used as a measure for cell size. Mean values ± SD are given; n = 114 (AX2), n = 199 (CP148-RNAi). b–b″ The DNA content is expressed as the product of the area (μm2) and intensity of DAPI staining (16-bit gray scale). Values are normalized to the mean value of AX2 control cells, which was set to 1. Graphs show mean values ± SD (n = 72) (b), and DNA contents of each individual evaluated cell (x-axis) sorted by increasing normalized DNA content (y-axis) (b′, b″). Comparison of both charts (b′, b″) reveals an increased fraction of cells with aberrant ploidy upon CP148-RNAi
Depletion of CP148 does not affect formation of astral microtubules and a central spindle
The strong, aberrant phenotype upon CP148 RNAi, with a completely disorganized microtubule cytoskeleton, raised the question how these cells manage to survive and divide through many passages. Therefore, we investigated the mitotic behavior of CP148 RNAi/GFP-α-tubulin cells by spinning disk confocal microscopy in living cells. Figure 10/Movie S3 shows two cells from G2 through M and cytokinesis. At the G2/M transition, the unordered microtubule cytoskeleton with no discernible MTOC disassembles completely, and a bright spot emerges (upper left cell at time point 540 s and lower left cell at time point 3,165 s). The bright spot duplicates and yields the two poles of a central spindle, showing that the initial bright spot corresponds to the unduplicated prophase centrosome. Later in anaphase, astral microtubules become visible, and the appearance of the microtubule cytoskeleton through mitosis on to cytokinesis was almost indistinguishable from AX2 cells expressing just GFP-α-tubulin (see [24] for an example). However, right after completion of cytokinesis (upper cell at 2,895 s), the radial microtubule cytoskeleton decomposes to the unordered mess of microtubules found in CP148RNAi/GFP-α-tubulin interphase cells. Thus, CP148 is only required for the organization of the radial microtubule cytoskeleton of interphase cells, but not for the organization of the central spindle and astral microtubules.
Fig. 10.

Two mitoses in CP148 RNAi/GFP-α-tubulin cells. Selected time points of live cell imaging (Movie S3) of CP148 RNAi/GFP-α-tubulin cells are shown. The two mitotic cells and their daughter cells are labeled with (*) and (#). Arrowheads point at the site of MTOC formation. After duplication, each mitotic MTOC sits at the pole of a bipolar spindle and organizes astral microtubules. During cytokinesis, the radial arrangement of astral microtubules becomes disrupted again. Confocal spinning disk microscopy; time lapse acquisition rate was four stacks per minute (at a frame rate of 10 fr/s); maximum intensity projection of seven slices per image stack
Discussion
Microtubules are associated with centrosomal core components through different pathways in interphase and mitosis
The corona of the Dictyostelium centrosome is considered the equivalent of the pericentriolar matrix of animal centrosomes. On the ultrastructural level, it consists of electron-dense nodules embedded in an amorphous matrix. The functionally characterized corona proteins so far comprise mainly proteins involved in microtubule nucleation and organization, such as γ-tubulin, which was addressed to the nodules [20], the γ-tubulin complex components Spc97 and Spc98 [10], and the orthologues of XMAP215 (CP224) [25] and TACC [17]. With CP148 we have now characterized the first corona protein required for formation of the corona itself. All the other corona proteins mentioned above also localize to the mitotic spindle poles, which, by contrast to interphase centrosomes, are devoid of a corona [ 9, 26]. In contrast, localization of CP148 at the centrosome correlates exactly with dissociation of the corona in prophase and its reformation in late telophase. The presence and absence of the corona already suggested cell cycle-dependent differences in the recruitment of microtubule nucleation complexes to the centrosome. For the first time we are now able to address this duality to the presence of a specific protein, CP148. Depletion of the protein by RNAi revealed that CP148 is required only for the organization of the radial microtubule cytoskeleton of interphase cells, but not for the organization of the central spindle and astral microtubules. The appearance of normal, mitotic microtubule arrays in CP148-RNAi cells was exactly reciprocal to the presence of CP148 at the centrosome in normal cells. CP148 is clearly not required for nucleation and growth of microtubules, since CP148-RNAi cells contain many microtubules during interphase. Stainings with plus-end markers such as CP224 or TACC (Fig. 7c) suggest even a higher number of microtubules than in control cells. Their minus ends must somehow be protected from depolymerization, perhaps through their association with nucleation complexes, since it is known that the generation of free minus ends, e.g., by laser ablation of the centrosome, causes rapid depolymerization of microtubules [27]. The microtubules of CP148-RNAi cells were arranged in a disordered manner and did not focus at the centrosome, which was no longer discernible as a microtubule-organizing center. In these cells, its position could only be assessed by the localization of the centrosomal core components. In contrast, the formation of ordered, mitotic microtubule arrays was unaffected by the absence of CP148. Yet, assembly of a corona, which appears to be the prerequisite for the organization of the radial microtubule arrays of interphase cells, requires recruitment of CP148 to the new centrosome right after cytokinesis. The amount of CP148 recruited to late telophase centrosomes may be crucial for the size of the corona. Overexpression caused hypertrophic coronas with a greatly enlarged number of electron-dense nodules, while depletion of CP148 by RNAi caused loss of the corona, suggesting that the expression level of CP148 has to be tightly controlled. Core structures in CP148-RNAi cells were largely unaffected by lack of CP148, indicating that the formation of intact core structures of the correct size requires neither CP148 nor other corona proteins.
Taken together, we propose a role of CP148 as a “glue” or scaffolding protein for the organization of corona components at the centrosomal core.
A putative role of astral microtubules and mitotic kinases in corona formation
Our results also shed light on the role of astral microtubules in Dictyostelium. In animal cells, these microtubules, together with cortical dynein, are required for spindle orientation and determination of the plane of cell division, which in epithelial stem cells, e.g., in the developing neocortex, determines cell fate after division [28]. In amoeboid vegetative cells, cell division always yields two identical daughter cells, and spindle orientation is obviously not important to determine cell differentiation. Yet, our results from GFP-CP148 overexpressing cells indicate a role of astral microtubules in corona formation. In early telophase, many GFP-CP148-positive protein clusters arose in the cytosol and were associated with astral microtubules. The fact that these clusters sometimes contained γ-tubulin and moved towards the late mitotic spindle poles in a radial fashion, indicates that microtubule nucleation complexes are preassembled with GFP-CP148 in the cytosol, and then transported along astral microtubules towards the spindle poles to form a centrosome with the layered core structure and a hypertrophic corona. Their direction of migration suggests that dynein is the motor protein driving this movement along astral microtubules. We hypothesize that this preformation of γ-tubulin complexes with CP148 and other proteins also occurs at normal expression levels of CP148, and that these complexes are then transported towards the late mitotic spindle poles employing dynein as a motor. This view is also supported by the fact that both the dynein regulator Lis1 and the dynein heavy chain were localized at the corona of the interphase centrosome [13].
We consider the cell cycle-dependent disassembly and re-formation of the corona a prerequisite for centrosome duplication. Without disassembly of the corona, the two outer layers of the core structure were unable to grow and separate to form the mitotic spindle poles. The dynamic behavior of the corona and the redistribution of the microtubule minus end-associated proteins such as γ-tubulin and CP224 have to be regulated. It is tempting to speculate that this occurs through regulation of CP148 by mitotic kinases. A closer look at the CP148 protein sequence reveals five potential phosphorylation sites for each CDK1 and polo-like kinase (Plk). The known activity of both kinases agrees with the time point of corona disassembly, and disappearance of CP148 from the centrosome. We will investigate the role of these potential phosphorylation sites on CP148 localization in the near future.
The absence of CP148 and a corona at mitotic spindle poles poses the question how mitotic centrosomes organize microtubules. One possibility is that spindle formation and nucleation of mitotic microtubules occurs mainly through the RanGTP-dependent acentrosomal pathway that was first described in Xenopus. Here, formation of RanGTP through a chromatin-associated GEF activates spindle assembly factors including aurora A, motor proteins and microtubule-associated proteins [29, 30]. In this model, kinesins and dynein act together to order microtubules primarily nucleated at chromatin, to form a bipolar spindle. Yet, the presence of astral microtubules in CP148-RNAi cells argues against this idea, since obviously they cannot be formed through the acentrosomal pathway, at least in animal cells [31]. Thus, it is also possible that anchorage of microtubule-nucleation complexes at centrosomal core components employs a different set of proteins during interphase and mitosis.
Similarities of the corona organizer CP148 to known scaffolding proteins of the PCM
Since CP148 behaves like a scaffolding protein for the organization and assembly of corona components around the centrosomal core structure, and the corona can be considered the functional equivalent to the pericentriolar matrix of higher cells, it is interesting to ask how CP148 compares to the known scaffolding factors within the PCM of animal centrosomes. As mentioned above, three large coiled coil proteins have been characterized in this context, D-PLP/pericentrin, D-Spd-2/Cep192, and Cnn/Cep215/Cdk5Rap2. All of them are only weakly conserved with regard to their amino acid sequence similarity among different animals. Not surprisingly, CP148 shows no striking sequence similarity to either of these proteins, except for the general similarity between coiled coil regions. Thus, we cannot judge whether it serves as a functional homologue of one of these proteins. Dictyostelium cells possess a real orthologue of Cep192 [15 ]. Of the remaining two proteins, only Cnn has homologues not only in animals but also in fungi [32]. One similarity of Drosophila Cnn to CP148 is that its absence causes detachment of centrioles from the PCM, resulting in centriole rocketing through the embryo [33]. This phenotype is reminiscent of the detachment of the remaining core structures from the nuclear envelope in CP148 RNAi cells.
A role of CP148 in centrosome nucleus attachment and centromere clustering
Detachment of centrosome cores upon CP148 RNAi also tells us that CP148 is involved in centrosome/nucleus attachment. In line with this observation, the pericentrosomal concentration of Sun1 at the nuclear envelope was abrogated upon CP148 RNAi, and Sun1 was more evenly distributed at the nuclear envelope. Sun1 is a protein of the inner nuclear envelope that binds to KASH-domain proteins in the outer nuclear envelope, which in turn are associated with dynein and other components to tether the centrosome to the nucleus [34]. In Dictyostelium, no KASH domain proteins have been characterized so far, and Sun1 was found concentrated in the pericentrosomal region in both nuclear membranes, where it appears to link the cytosolic centrosome to the clustered centromeres inside the nucleus. Disruption of Sun1 function resulted in detachment of both the centrosome and the centromere cluster from the tethering site [22]. Our results suggest that CP148 links the centrosome to Sun1 either directly, or indirectly through another centrosomal protein, and that this linkage is required to maintain the pericentrosomal concentration of Sun1. The dispersal of the centromere cluster upon CP148 RNAi does not seem to depend on Sun1 displacement alone. We conclude this from our observation that disruption of Sun1 function by overexpression of GFP-Sun1 or a dominant negative fragment resulted only in displacement of the centromere cluster but not in its dispersal [22]. The role of CP148 is most likely an indirect one, since there is no indication for a nuclear population of the protein, or the presence of a nuclear localization signal in the CP148 sequence, which should be expected if the protein played a direct role in centromere clustering. Thus, clarification of the role of CP148 in this process certainly has to await the identification of CP148 interaction partners.
The role of CP148 in the linkage of the centrosome to the centromere cluster may also explain the frequent aneuploidy of CP148 RNAi cells. Both centrosome detachment from the nucleus and dispersal of the centromere cluster may account for mitotic defects, despite the ability of many cells to form normal mitotic spindles upon CP148 RNAi. Both effects may lead to a prolongation of prometaphase, which may cause the cells to switch to interphase with duplicated centrosomes but unsegregated chromosomes. This in turn may cause multipolar spindles and unequal chromosome segregation in the next cell cycle, since the spindle checkpoint of Dictyostelium cells functions less strictly than in other cell types [35]. This became apparent also after treatment with microtubule-depolymerizing drugs. They block mitotic progression in prometaphase only for a few hours, after which the drug-treated cells switch to interphase with duplicated centrosomes but unsegregated chromosomes, resulting in aneuploid cell populations [36].
Conclusions
Taken together, we have shown that the novel Dictyostelium centrosomal protein CP148 is an essential factor for the formation and organization of the microtubule-nucleating centrosomal corona. Moreover, CP148 is required for maintenance of the nuclear envelope-spanning linkage of centrosomes and centromeres. This is also the first indication that centrosome nucleus attachment in Dictyostelium occurs through an interaction of a corona protein, and not a core component, with proteins in the nuclear envelope.
Materials and methods
Vector constructions, and expression of recombinant CP148 for immunization
Cloning of the complete coding sequence of CP148 into the N-terminal GFP fusion vector pIS77 (G418 resistance) was described recently [15]. This vector was used as a basis for all plasmid constructions of this work.
To express a C-terminal fragment of CP148 as an antigen, a DNA fragment encoding amino acids 940–1297 (CP148ΔN) was amplified by PCR employing SalI forward and BamHI reverse linker primers. The resulting DNA fragment was cloned into pMALc2 (NEB, Frankfurt, Germany) and expressed as a maltose-binding fusion protein (MBP-CP148ΔN) in E. coli. CP148ΔN was purified by amylose affinity chromatography according to the manufacturer’s manual (NEB, Frankfurt, Germany) and used for custom immunization of a rabbit (Pab Productions, Hebertshausen, Germany). The antiserum was affinity purified using a column with MBP-CP148ΔN coupled to NHS-activated Sepharose (GE-Healthcare, Munich, Germany).
CP148 RNAi was based on the strategy of Martens et al. (2002) [37], where Dictyostelium cells are transformed with a plasmid containing a long inverted repeat of the sequence of interest downstream from a strong promoter. The CP148 RNAi plasmid was constructed exactly according to Samereier et al. (2011) [17]. Thus, SalI/BamHI linker primers were used for PCR amplification of a CP148 sense fragment corresponding to base positions 1–440 of the coding sequence, and AflII/KpnI linker primers were used to amplify the respective reverse sequence. After cloning of both fragments into pIS193, the resulting vector encoded a long inverted CP148 repeat with a short stuffer fragment derived from mCherry. The CP148-RNAi plasmid was transformed by electroporation into Dictyostelium strains AX2, GFP-TACCdom [17] and GFP-CP55 [15].
Microscopy
Fluorescence light microscopy and image processing of fixed samples was carried out as published recently on a Zeiss CellObserver HS system equipped with PlanApo 1.4/100× a objective, an Axiocam MRm Rev. 3 CCD camera, a piezo stage, and the Axiovision 4.7 fast iterative deconvolution software package [22]. Maximum intensity projections of deconvolved image stacks (focus step size 0.25 μm) were calculated with Axiovision. Live cells were prepared in glass-bottom Petri dishes (Fluorodish, WPI, Berlin, Germany) in LoFlo medium (Formedium, Hunsanton, UK). As indicated in the figures, live cell imaging was either performed on the same wide field system [22] or on a confocal spinning disk system (CellObserver SD, Carl Zeiss Microimaging GmbH, Göttingen, Germany) equipped with two Evolve EM-CCD cameras (Photometrics, Tucson, AZ, U.S.A.). In both cases, the LCI PlanNeofluar 1.3/63× objective was used. The spinning disk system was equipped with a Rapp UGA-40-2L Galvo Scanner (Rapp Optoelectronics, Hamburg, Germany) employing an independent 473-nm laser diode that allows photomanipulation of user-defined regions of interest [24]. For FRAP experiments cell were put under agar overlay [38].
Electron microscopy was performed essentially as described [9]. Cells were fixed on coverslips for 30 min with 1% glutaraldehyde in PHEM buffer (12 mM PIPES, 5 mM HEPES, 1.6 mM EGTA, 1 mM MgCl2) supplemented with 0.5% Triton X-100. After washing the coverslips for 3 × 5 min with 50 mM Na-cacodylate buffer (pH 7.0), they were postfixed for 30 min in cacodylate-buffered 1% OsO4. After dehydration in a graded ethanol series and acetone, specimens were embedded in Spurr’s resin. Ultrathin sections were viewed on a Philips CM100 electron microscope.
Other methods
Cells (Dictyostelium strain AX2) were cultured in HL5c medium (Formedium, Hunsanton, UK) and transformed by electroporation as reported earlier [19, 25]. SDS gel electrophoresis and Western blotting were performed as described [19]. Expression levels of CP148 and GFP-CP148 were estimated by densitometry from immunoblot bands visualized by chemiluminescence on X-ray film. The film was scanned with a transmitted light scanner (Epson V750) and band intensities were measured with the ImageJ program. DAPI fluorescence intensities for estimation of the DNA content were measured in maximum intensity projections of fluorescence microscopy image stacks using ImageJ. Mean areas as a measure for cell and corona sizes were measured with ImageJ.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 2 (MOV 3287 kb). Movie S1. Mitosis in GFP-CP148/mCherry-H2B cells. GFP-CP148 is shown in green, histone-labeled chromosomes are shown in red. The mitotic cell is visible in the middle. Mitotic stages can be judged from the extent of DNA condensation and progression of chromosome segregation. GFP-CP148 disappears from the centrosome in prophase. In telophase, several GFP-CP148 foci appear in the cytosol and migrate in a radial fashion towards the nucleus where they join at the position of the new centrosome. Confocal spinning disk microscopy; time lapse acquisition rate was four stacks per minute (at a frame rate of 10 fr/s); maximum intensity projection of nine slices per image stack
Supplementary material 3 (MOV 1269 kb). Movie S2. GFP-CP148 shows almost no fluorescence recovery after photobleaching in GFP-CP148/cherry-H2B cells. GFP-CP148 is shown in green, histone-labeled chromosomes are shown in red. Left, centrosome bleached with a point-focused 473-nm laser pulse at time point 170 s; right, non-bleached control cell. Confocal spinning disk microscopy; time lapse acquisition rate was six stacks per minute (at a frame rate of 10 fr/s); maximum intensity projection of seven slices per image stack
Supplementary material 4 (MOV 6634 kb). Movie S3. Two mitoses in CP148 RNAi/GFP-α-tubulin cells (upper left, lower left). After duplication each mitotic MTOC sits at the pole of a bipolar spindle and organizes astral microtubules. During cytokinesis, the radial arrangement of astral microtubules becomes disrupted again. Confocal spinning disk microscopy; time lapse acquisition rate was four stacks per minute (at a frame rate of 10 fr/s); maximum intensity projection of eight slices per image stack
Acknowledgments
We would like to thank Belinda Pipke for technical assistance. We also acknowledge Dr. Annette Müller-Taubenberger for providing the mCherry-H2B plasmid and Dr. Alexandra Lepier for critically reading the manuscript. This work was supported by DFG GR1642/3-1 and GR1642/4-1.
References
- 1.Azimzadeh J, Marshall WF. Building the centriole. Curr Biol. 2010;20:R816–R825. doi: 10.1016/j.cub.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dictenberg JB, Zimmerman W, Sparks CA, Young A, Vidair C, Zheng YX, Carrington W, Fay FS, Doxsey SJ. Pericentrin and gamma-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J Cell Biol. 1998;141:163–174. doi: 10.1083/jcb.141.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fong KW, Choi YK, Rattner JB, Qi RZ. CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the {gamma}-tubulin ring complex. Mol Biol Cell. 2008;19:115–125. doi: 10.1091/mbc.E07-04-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haren L, Stearns T, Lüders J. Plk1-dependent recruitment of gamma-tubulin complexes to mitotic centrosomes involves multiple PCM components. PLoS One. 2009;4:e5976. doi: 10.1371/journal.pone.0005976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conduit PT, Brunk K, Dobbelaere J, Dix CI, Lucas EP, Raff JW. Centrioles regulate centrosome size by controlling the rate of Cnn incorporation into the PCM. Curr Biol. 2010;20:2178–2186. doi: 10.1016/j.cub.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 6.Conduit PT, Raff JW. Cnn dynamics drive centrosome size asymmetry to ensure daughter centriole retention in Drosophila neuroblasts. Curr Biol. 2010;20:2187–2192. doi: 10.1016/j.cub.2010.11.055. [DOI] [PubMed] [Google Scholar]
- 7.Gräf R, Daunderer C, Schulz I. Molecular and functional analysis of the dictyostelium centrosome. Int Rev Cytol. 2004;241:155–202. doi: 10.1016/S0074-7696(04)41003-1. [DOI] [PubMed] [Google Scholar]
- 8.Ding R, West RR, Morphew M, Oakley BR, McIntosh JR. The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol Biol Cell. 1997;8:1461–1479. doi: 10.1091/mbc.8.8.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ueda M, Schliwa M, Euteneuer U. Unusual centrosome cycle in Dictyostelium: correlation of dynamic behavior and structural changes. Mol Biol Cell. 1999;10:151–160. doi: 10.1091/mbc.10.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daunderer C, Gräf R. Molecular analysis of the cytosolic Dictyostelium gamma-tubulin complex. Eur J Cell Biol. 2002;81:175–184. doi: 10.1078/0171-9335-00241. [DOI] [PubMed] [Google Scholar]
- 11.Daunderer C, Schliwa M, Gräf R. Dictyostelium centrin-related protein (DdCrp), the most divergent member of the centrin family, possesses only two EF hands and dissociates from the centrosome during mitosis. Eur J Cell Biol. 2001;80:621–630. doi: 10.1078/0171-9335-00198. [DOI] [PubMed] [Google Scholar]
- 12.Rehberg M, Gräf R. Dictyostelium EB1 is a genuine centrosomal component required for proper spindle formation. Mol Biol Cell. 2002;13:2301–2310. doi: 10.1091/mbc.E02-01-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rehberg M, Kleylein-Sohn J, Faix J, Ho TH, Schulz I, Gräf R. Dictyostelium LIS1 is a centrosomal protein required for microtubule/cell cortex interactions, nucleus/centrosome linkage, and actin dynamics. Mol Biol Cell. 2005;16:2759–2771. doi: 10.1091/mbc.E05-01-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gräf R, Euteneuer U, Ho TH, Rehberg M. Regulated expression of the centrosomal protein DdCP224 affects microtubule dynamics and reveals mechanisms for the control of supernumerary centrosome number. Mol Biol Cell. 2003;14:4067–4074. doi: 10.1091/mbc.E03-04-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schulz I, Erle A, Gräf R, Kruger A, Lohmeier H, Putzler S, Samereier M, Weidenthaler S. Identification and cell cycle-dependent localization of nine novel, genuine centrosomal components in Dictyostelium discoideum . Cell Motil Cytoskeleton. 2009;66:915–928. doi: 10.1002/cm.20384. [DOI] [PubMed] [Google Scholar]
- 16.Blau-Wasser R, Euteneuer U, Xiong H, Gassen B, Schleicher M, Noegel AA. CP250, a novel acidic coiled-coil protein of the Dictyostelium centrosome, affects growth, chemotaxis, and the nuclear envelope. Mol Biol Cell. 2009;20:4348–4361. doi: 10.1091/mbc.E09-03-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samereier M, Baumann O, Meyer I, Gräf R. Analysis of Dictyostelium TACC reveals differential interactions with CP224 and unusual dynamics of Dictyostelium microtubules. Cell Mol Life Sci. 2011;68:275–287. doi: 10.1007/s00018-010-0453-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reinders Y, Schulz I, Gräf R, Sickmann A. Identification of novel centrosomal proteins in Dictyostelium discoideum by comparative proteomic approaches. J Proteome Res. 2006;5:589–598. doi: 10.1021/pr050350q. [DOI] [PubMed] [Google Scholar]
- 19.Gräf R, Euteneuer U, Ueda M, Schliwa M. Isolation of nucleation-competent centrosomes from Dictyostelium discoideum . Eur J Cell Biol. 1998;76:167–175. doi: 10.1016/S0171-9335(98)80031-9. [DOI] [PubMed] [Google Scholar]
- 20.Euteneuer U, Gräf R, Kube-Granderath E, Schliwa M. Dictyostelium gamma-tubulin: molecular characterization and ultrastructural localization. J Cell Sci. 1998;111:405–412. doi: 10.1242/jcs.111.3.405. [DOI] [PubMed] [Google Scholar]
- 21.Kaller M, Euteneuer U, Nellen W. Differential effects of heterochromatin protein 1 isoforms on mitotic chromosome distribution and growth in Dictyostelium discoideum . Eukaryot Cell. 2006;5:530–543. doi: 10.1128/EC.5.3.530-543.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schulz I, Baumann O, Samereier M, Zoglmeier C, Gräf R. Dictyostelium Sun1 is a dynamic membrane protein of both nuclear membranes and required for centrosomal association with clustered centromeres. Eur J Cell Biol. 2009;88:621–638. doi: 10.1016/j.ejcb.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 23.Xiong H, Rivero F, Euteneuer U, Mondal S, Mana-Capelli S, Larochelle D, Vogel A, Gassen B, Noegel AA. Dictyostelium Sun-1 connects the centrosome to chromatin and ensures genome stability. Traffic. 2008;9:708–724. doi: 10.1111/j.1600-0854.2008.00721.x. [DOI] [PubMed] [Google Scholar]
- 24.Samereier M, Meyer I, Koonce MP, Gräf R. Live cell-imaging techniques for analyses of microtubules in Dictyostelium . Methods Cell Biol. 2010;97:341–357. doi: 10.1016/S0091-679X(10)97018-0. [DOI] [PubMed] [Google Scholar]
- 25.Gräf R, Daunderer C, Schliwa M. Dictyostelium DdCP224 is a microtubule-associated protein and a permanent centrosomal resident involved in centrosome duplication. J Cell Sci. 2000;113:1747–1758. doi: 10.1242/jcs.113.10.1747. [DOI] [PubMed] [Google Scholar]
- 26.McIntosh JR. Spindle structure and the mechanisms of chromosome movement. Basic Life Sci. 1985;36:197–229. doi: 10.1007/978-1-4613-2127-9_14. [DOI] [PubMed] [Google Scholar]
- 27.Brito DA, Strauss J, Magidson V, Tikhonenko I, Khodjakov A, Koonce MP. Pushing forces drive the comet-like motility of microtubule arrays in Dictyostelium . Mol Biol Cell. 2005;16:3334–3340. doi: 10.1091/mbc.E05-01-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yingling J, Youn YH, Darling D, Toyo-Oka K, Pramparo T, Hirotsune S, Wynshaw-Boris A. Neuroepithelial stem cell proliferation requires LIS1 for precise spindle orientation and symmetric division. Cell. 2008;132:474–486. doi: 10.1016/j.cell.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karsenti E, Vernos I. The mitotic spindle: a self-made machine. Science. 2001;294:543–547. doi: 10.1126/science.1063488. [DOI] [PubMed] [Google Scholar]
- 30.Zheng Y. G protein control of microtubule assembly. Annu Rev Cell Dev Biol. 2004;20:867–894. doi: 10.1146/annurev.cellbio.20.012103.094648. [DOI] [PubMed] [Google Scholar]
- 31.Rieder CL, Faruki S, Khodjakov A. The centrosome in vertebrates: more than a microtubule-organizing center. Trends Cell Biol. 2001;11:413–419. doi: 10.1016/S0962-8924(01)02085-2. [DOI] [PubMed] [Google Scholar]
- 32.Kraemer N, Issa L, Hauck SC, Mani S, Ninnemann O, Kaindl AM. What’s the hype about CDK5RAP2? Cell Mol Life Sci. 2011;68:1719–1736. doi: 10.1007/s00018-011-0635-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lucas EP, Raff JW. Maintaining the proper connection between the centrioles and the pericentriolar matrix requires Drosophila centrosomin. J Cell Biol. 2007;178:725–732. doi: 10.1083/jcb.200704081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Starr DA, Fridolfsson HN (2010). Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol [DOI] [PMC free article] [PubMed]
- 35.Ma S, Triviños Lagos L, Gräf R, Chisholm RL. Dynein intermediate chain mediated dynein-dynactin interaction is required for interphase microtubule organization and centrosome replication and separation in Dictyostelium . J Cell Biol. 1999;147:1261–1274. doi: 10.1083/jcb.147.6.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitanishi T, Shibaoka H, Fukui Y. Disruption of microtubules and retardation of development of Dictyostelium with Ethyl N-phenylcarbamate and thiabendazole. Protoplasma. 1984;120:185–196. doi: 10.1007/BF01282599. [DOI] [Google Scholar]
- 37.Martens H, Novotny J, Oberstrass J, Steck TL, Postlethwait P, Nellen W. RNAi in Dictyostelium: the role of RNA-directed RNA polymerases and double-stranded RNase. Mol Biol Cell. 2002;13:445–453. doi: 10.1091/mbc.01-04-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fukui Y, Yumura S, Yumura TK. Agar-overlay immunofluorescence: high resolution studies of cytoskeletal components and their changes during chemotaxis. Methods Cell Biol. 1987;28:347–356. doi: 10.1016/S0091-679X(08)61655-6. [DOI] [PubMed] [Google Scholar]
- 39.Wehland J, Willingham MC. A rat monoclonal antibody reacting specifically with the tyrosylated form of alpha-tubulin. II. Effects on cell movement, organization of microtubules, and intermediate filaments, and arrangement of Golgi elements. J Cell Biol. 1983;97:1476–1490. doi: 10.1083/jcb.97.5.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gräf R, Daunderer C, Schliwa M. Cell cycle-dependent localization of monoclonal antibodies raised against isolated Dictyostelium centrosomes. Biol Cell. 1999;91:471–477. doi: 10.1111/j.1768-322x.1999.tb01102.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 2 (MOV 3287 kb). Movie S1. Mitosis in GFP-CP148/mCherry-H2B cells. GFP-CP148 is shown in green, histone-labeled chromosomes are shown in red. The mitotic cell is visible in the middle. Mitotic stages can be judged from the extent of DNA condensation and progression of chromosome segregation. GFP-CP148 disappears from the centrosome in prophase. In telophase, several GFP-CP148 foci appear in the cytosol and migrate in a radial fashion towards the nucleus where they join at the position of the new centrosome. Confocal spinning disk microscopy; time lapse acquisition rate was four stacks per minute (at a frame rate of 10 fr/s); maximum intensity projection of nine slices per image stack
Supplementary material 3 (MOV 1269 kb). Movie S2. GFP-CP148 shows almost no fluorescence recovery after photobleaching in GFP-CP148/cherry-H2B cells. GFP-CP148 is shown in green, histone-labeled chromosomes are shown in red. Left, centrosome bleached with a point-focused 473-nm laser pulse at time point 170 s; right, non-bleached control cell. Confocal spinning disk microscopy; time lapse acquisition rate was six stacks per minute (at a frame rate of 10 fr/s); maximum intensity projection of seven slices per image stack
Supplementary material 4 (MOV 6634 kb). Movie S3. Two mitoses in CP148 RNAi/GFP-α-tubulin cells (upper left, lower left). After duplication each mitotic MTOC sits at the pole of a bipolar spindle and organizes astral microtubules. During cytokinesis, the radial arrangement of astral microtubules becomes disrupted again. Confocal spinning disk microscopy; time lapse acquisition rate was four stacks per minute (at a frame rate of 10 fr/s); maximum intensity projection of eight slices per image stack


