Abstract
Genome-wide association studies have identified a number of genes associated with human body weight. While some of these genes are large fields within obesity research, such as MC4R, POMC, FTO and BDNF, the majority do not have a clearly defined functional role explaining why they may affect body weight. Here, we searched biological databases and discovered 33 additional genes associated with human obesity (CADM2, GIPR, GPCR5B, LRP1B, NEGR1, NRXN3, SH2B1, FANCL, GNPDA2, HMGCR, MAP2K5, NUDT3, PRKD1, QPCTL, TNNI3K, MTCH2, DNAJC27, SLC39A8, MTIF3, RPL27A, SEC16B, ETV5, HMGA1, TFAP2B, TUB, ZNF608, FAIM2, KCTD15, LINGO2, POC5, PTBP2, TMEM18, TMEM160). We find that the majority have orthologues in distant species, such as D. melanogaster and C. elegans, suggesting that they are important for the biology of most bilateral species. Intriguingly, signalling cascade genes and transcription factors are enriched among these obesity genes, and several of the genes show properties that could be useful for potential drug discovery. In this review, we demonstrate how information from several distant model species, interactomics and signalling pathway analysis represents an important way to better understand the functional diversity of the surprisingly high number of molecules that seem to be important for human obesity.
Keywords: Genome-wide association studies, Signalling cascade genes, Interactomics, Drosophila, C. elegans
Introduction
Tremendous progress has been made in the field of obesity research, with academic and industrial interest growing rapidly. During the 1980s (1981–1990) there were approximately 16,000 publications focused on obesity [1], while in the last year more than 20,000 articles on obesity were published. Interestingly, twin studies showed there is a strong genetic component to obesity [2, 3]. In accordance with this data, Claude Bouchard and colleagues generated a yearly human obesity genetic map between 1996 and 2005 [4], with the last report listing 176 human obesity cases relating to single-gene mutations in 11 different genes, and 50 loci relating to Mendelian syndromes relevant to human obesity that mapped to a specific genomic region. Their mouse obesity gene map identified 248 genes that, when mutated or overexpressed as transgenes in the mouse, resulted in phenotypes affecting body weight or adiposity. Many of these genes are expressed in the hypothalamus, and one of the earliest discovered genes, having a strong genetic association to human obesity, was the hypothalamic MC4 receptor (MC4R). Mc4r is inhibited by the orexigenic peptide Agrp and stimulated by the anorexic agonists α- and β-Msh [5]. The α- and β-Msh peptide hormones originate from the precursor protein Pomc, and humans with a mutated POMC gene have severe obesity [6]. Enormous resources have been put into MC4R research, as it was shown that synthetic and selective agonists and antagonists are very effective in reducing food intake and bodyweight in animal feeding models [7, 8]. To date, this has not resulted in any promising drugs, mainly due to the complexity of the action of the melanocortin system, including effects on the cardiovascular system, creating risks for severe side effects.
Genome-wide association (GWAS) studies have transformed our knowledge of which genes are most important for obesity. The first gene that was strongly associated with obesity was fat mass- and obesity-associated (FTO) [9]. The FTO gene is expressed in many tissues of the body but, interestingly, it is highly expressed in hypothalamic feeding regions [10]. The molecular mechanism of how gene variants of FTO may cause obesity is still unknown, it has been suggested that it affects food intake, as carriers of the risk allele tend to choose high energy and more palatable food [3, 11, 12]. Fto knockout models and mutant mice point towards a role for Fto in energy homeostasis, metabolism and adipogenesis, and mutant mice have clearly decreased body fat mass [13, 14]. Moreover, transgenic mice that overexpress Fto have a higher body weight, and hypothalamic Fto expression is regulated in several animal feeding models [14].
Soon after the discovery of FTO as an obesity gene, six additional loci were found to be associated with obesity [15]. Only one of these genes, SH2B1, had any prior evidence linking it to obesity; knocking out Sh2b1 in mice resulted in obesity, hyperglycemia, insulin resistance, and glucose intolerance. Thorleifsson and collegues identified new sequence variants at seven loci associated with obesity in GWAS of more than 30,000 individuals [16]. In total, 29 variants in 11 chromosomal regions reached a genome-wide significance threshold. These associations included the brain-derived neurotrophic factor (BDNF) gene, which was previously associated with both BMI and eating disorders [17, 18].
Meta-analysis of several GWAS, and other association studies involving almost 250,000 individuals, showed association with 14 known obesity susceptibility loci, while an additional 18 new loci were associated with BMI [19]. It is estimated that, similar to the 37 genes linked to these 32 loci, more than 250 additional common variant loci remain to be discovered that will have effects on BMI. It should be mentioned that GWAS studies identify genomic regions, rather than individual genes, and several of the genes currently linked to obesity by associated SNPs may later prove not be a causative agent in obesity. To date, only 8 of the 32 loci identified by Speliotes et al. [19] are strongly linked to an adjacent common missense SNP, and only 15 of the loci contain genes that can be biologically linked to obesity. This lack of a causative link is a result of the limited understanding of the biology behind obesity and weight regulation, which calls for a comprehensive investigation of these obesity-associated genes.
Among the 32 loci that have emerged as currently the most important ones for BMI [19], MC4R and POMC have clearly understood physiological roles, and their functions are well reviewed. Moreover, the research on BDNF and FTO is very intense, and there are several recent reviews addressing what is known about these genes. Here, we have focused on what is known about a select group of the additional 33 obesity-linked genes (GIPR, GPRC5B, SH2B1, HMGCR, PRKD1, TUB, ZNF608, TFAP2B, KCTD15, SEC16B, and MTCH2) and used the growing number of databases to search for information that can shed light into the functional role of these genes. These genes were chosen because further information beyond the GWAS studies was available, yielding possible evidences for their involvement in obesity. We have in particular taken advantage of model organisms that are distant to humans, as we find that 24 of the 33 newly discovered genes are well conserved in most bilateral species, including D. melanogaster and C. elegans (Table 1).
Table 1.
Evolutionary conservation of obesity-linked genes in major model organisms
| Human | G. gallus | D. rerio | D. melanogaster | C. elegans | S. cerevisiae | D. discoideum |
|---|---|---|---|---|---|---|
| Receptors, ligands and signal transduction | ||||||
| CADM2 | CADM2 | Cadm2a, cadm2b | ||||
| GIPR | GIPR | gipr | ||||
| GPRC5B | GPRC5B | gprc5b | boss | |||
| LRP1B | LRP1B | lrp1b | Lrp1 | lrp-2 | ||
| NEGR1 | NEGR1 | Negr1 | CG11320 | |||
| NRXN3 | NRXN3 | Nrxn3 | Nrx-1 | nrx-1 | ||
| SH2B1 | SH2B1 | sh2b | Lnk | |||
| Enzymes | ||||||
| FANCL | FANCL | fancl | Fancl | fncl | ||
| GNPDA2 | GNPDA2 | gnpda2 | Oscillin | T03F6.3 | gnpda1 | |
| HMGCR | HMGCR | hmgcra | Hmgcr | F08F8.2 | HMG1 | hmgB |
| MAP2K5 | MAP2K5 | map2k5 | ||||
| NUDT3 | NUDT3 | nudt3 | Aps | Y92H12BL.5 | DDP1 | |
| PRKD1 | PRKD1 | prkd1 | PKD | dkf-2 | ||
| QPCTL | QPCTL | qpctl | CG5976 | H27A22.1 | YFR018C | qpct |
| TNNI3K | TNNI3K | tnni3k | C24A1.3 | |||
| Transporters | ||||||
| MTCH2 | MTCH2 | mtch2 | Mtch | F43E2.7 | ||
| DNAJC27 | DNAJC277 | rbj | ||||
| SLC39A8 | SLC39A8 | slc39a8 | ||||
| Protein processes | ||||||
| MTIF3 | MTIF3 | mtif3 | CG13163 | |||
| RPL27A | RPL27A | rpl27a | RpL27A | Y37E3.8 | RPL28 | rpl27a |
| SEC16B | SEC16B | sec16b | Sec16 | F13B9.1 | Sec16 | |
| Transcription factor | ||||||
| ETV5 | ETV5 | etv5 | ETS96B | |||
| HMGA1 | HMGA1 | hmga1 | ||||
| TFAP2B | TFAP2B | tfap2b | AP-2 | aptf-1 | ||
| TUB | TUB | LOC568677 | king-tubby | tub-1 | ||
| ZNF608 | ZNF608 | znf608 | sbb | |||
| Unkown function | ||||||
| FAIM2 | FAIM2 | faim2 | xbx-6 | |||
| KCTD15 | KCTD15 | kctd15 | CG10440 | |||
| LINGO2 | LINGO2 | lingo2 | ||||
| POC5 | C5orf37 | POC5 | ||||
| PTBP2 | PTPB2 | Ptpb2 | heph | ptb-1 | ||
| TMEM18 | TMEM18 | tmem18 | CG30051 | |||
| TMEM160 | tmem160 | |||||
Using NCBI HomoloGene, EMBL-EBI databases (http://www.ebi.ac.uk/) and protein sequence similarity searches (http://hmmer.janelia.org/search/phmmer,UniProtKB), orthologues for the obesity-linked genes in a variety of model organisms were determined. Genes are grouped according to molecular function
A subgroup of obesity-linked genes
In this review, we introduce a select subgroup of obesity-linked genes, including what is known about their domain structure and possible functions in distant model organisms. We also present some possible interactions between obesity-linked genes and how this interaction may regulate homeostasis.
Gastric inhibitory peptide receptor (GIPR): resistance to diet-induced obesity
GIPR is a G protein-coupled receptor (GPCR) belonging to the secretin-family that includes receptors for the peptides GLP1, glucagon, PACAP, VIP and secretin. The GIPR ligand, gastric inhibitory peptide (GIP), is released from intestinal K cells and potentiates glucose-stimulated insulin secretion by elevating cAMP levels, inhibiting the KATP channel and increasing intracellular Ca2+ [20]. GIPR is highly expressed in the pancreas, but is also found in a wide range of peripheral tissues. Studies using Gipr null mice established the importance of Gipr signalling to maintain glucose homeostasis and regulate lipid metabolism [21, 22]. In response to orally administered glucose, mice lacking Gipr signalling exhibited mild glucose intolerance and reduced levels of glucose-stimulated insulin secretion [21, 23]. However, mice given an intraperitoneal glucose challenge displayed normal fasting glucose levels and a normal glycemic index [23]. These findings suggest that GIPR stimulates insulin release, indicating that GIP functions as an incretin hormone. One potential explanation for the mild glucose intolerance observed in mice with a single incretin receptor mutation, either GIPR or the related glucagon-like peptide receptor (GLPR), is that loss of one incretin receptor invokes upregulation of compensatory factors, particularly enhanced activity of the remaining incretin [24, 25]. Support for this hypothesis is derived from observations that Glpr null mice exhibit increased circulating levels of Gip, as well as enhanced sensitivity to the insulinotropic actions of Gip [26].
Gip has no direct effect on food intake or satiety, yet Gipr null mice exhibited resistance to diet-induced obesity, even after months of high-fat feeding, and when crossed with obese (ob/ob) mice, ob/ob diet-induced obesity was attenuated [27]. How could this be explained? It was observed that administration of the GIPR agonist [D-Ala2]GIP increased plasma levels of the plasma adipokine resistin in wild-type mice. This observation confirmed that GIPR signalling is an essential component of the adipocyte response to chronic nutritional excess [27]. Observations linking GIP action to the modulation of resistin and control of energy expenditure explain the divergent effects of GIP action on pancreatic β cells and adipocytes. Whereas loss of GIPR function in β cells impairs the adaptive islet response to metabolic stress, the potential deleterious effects arising from loss of GIPR signalling, and thus impaired insulin secretion, is offset by the persistence of insulin sensitivity, likely arising through a combination of reduced resistin, decreased adipose tissue mass, and increased energy expenditure [28].
Recently, it was observed that the microRNA miR-642 is upregulated during adipogenic differentiation, and in 19 out of 20 obese subjects is highly expressed in fat depots [29, 30]. This is interesting because miR-642 is positioned within intron seven of GIPR, and may share the same promoter [29], leading to the possibility that at least some of the obese phenotypes associated with GIPR may be due to misregulation of miR-642.
G-protein coupled receptor family C group 5 member B (GPRC5B): possible glucose sensor
GPRC5 is a member of the family-C GPCRs, this family includes mGluRs, calcium-sensing receptors (CaRs), type B γ-aminobutyric acid receptors (GABABRs), putative pheromone receptors (V2Rs) and taste receptors (T1Rs), as well as Drosophila bride of sevenless (BOSS) [31, 32]. Within family-C, GPRC5B belongs to the retinoic acid inducible gene (RAIG) subgroup, consisting of at least four orphan receptors [31–35].
Although little is known about GPRC5B signalling in mammals, it has been observed that Gprc5b knockout mice had developmental and behavioural defects. Approximately 30 % of homozygous Gprc5b knockout mice died at postnatal day 0, while a further 50 % died by 4 weeks of age [36]. The 20 % who survived into adulthood had behavioural defects, including reduced activity before light onset, and in the open field test, travelled less distance and spent more time in the centre of the field, compared to control mice [36].
A recent report on Drosophila points to a possible function of GPRC5B in glucose metabolism. The Drosophila GPRC5B homologue, BOSS, was first identified as a ligand for the tyrosine kinase Sevenless, involved in eye differentiation [37]. However, as with other GPRC5B homologues, until recently the physiological function of BOSS as a GPCR was completely unknown. It was observed that boss-deficient flies are reduced in size, suggesting that it might be required for cell growth, cell size and survival [38]. Further analysis determined that BOSS is expressed in a Drosophila nutrient-sensing tissue, the fat body, and is a glucose-responding GPCR required for the homeostatic regulation of glucose and lipids. Boss mutants have downregulated insulin signalling activity, demonstrating that BOSS has a critical function regulating energy homeostasis [38, 39]. This finding represents an example of a glucose-responding GPRC5B homologue in a model organism. Since GPRC5B is conserved from Drosophila to humans, this provides evidence that GPRC5B maybe a nutrient-sensing GPCR.
SH2B adapter protein 1 (SH2B1): signals in the insulin pathway
Mammalian SH2B adapter protein 1 (SH2B1) belongs to a family of adapter proteins known to regulate several different tyrosine kinases, including the receptors for insulin and insulin-like growth factor-1 [40–44]. As a result of these interactions, SH2B proteins are known to function during glucose homeostasis, energy metabolism and reproduction, and in humans, mutations in SH2B1 are associated with metabolic syndrome [45–49]. Furthermore, Sh2b1 deletions in mice produce neonatal growth retardation and infertility possibly due to impaired responses to growth hormone or Igf-1 signalling [50]. Sh2b1 null mice significantly increase their body mass and develop obesity as a result of impaired hypothalamic leptin signalling [47]. Intriguingly, neuronal restoration of Sh2b1 expression rescued leptin resistance, as well as the obesity phenotype, suggesting that Sh2b1 is involved in regulating energy balance and body weight by enhancing leptin sensitivity. In this same study, it was observed that loss of Sh2b1 in peripheral tissues induced insulin sensitivity regardless of body mass [47]. This result indicates that Sh2b1 regulates insulin signalling in peripheral tissues.
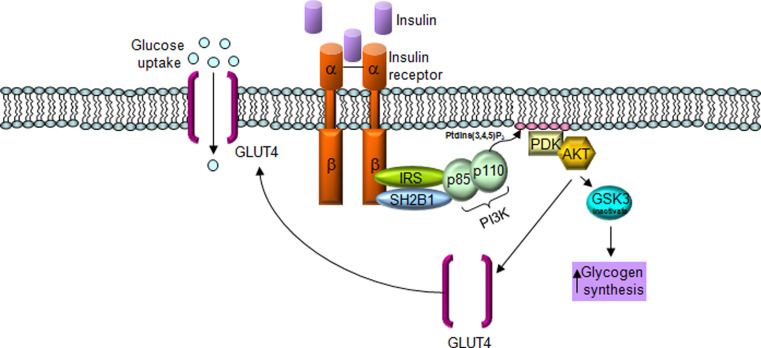
How might SH2B1 function to increase insulin signalling? Recent evidence demonstrated that SH2B1 physically interacts with two components of the insulin-signalling pathway, the insulin receptor (INSR) and insulin receptor substrate 1 (IRS1), and that this interaction is necessary to increase phosphoinositide 3-kinase (PI3K) activation downstream of IRS1 [40]. SH2B1 interaction with IRS1 inhibits dephosphorylation of IRS1 by protein tyrosine phosphotase 1B (PTP1B), leading to increased PI3K activation (Fig. 1). Furthermore, it was shown that SH2B1 interaction with INSR may increase INSR auto-phosphorylation, thus increasing INSR signalling.
Fig. 1.
SH2B1 signals in the insulin pathway. Insulin activates the insulin receptor (INSR), leading to INSR autophosphorylation and activation. Active INSR binds to and phosphorylates insulin receptor substrate-1 (IRS1), leading to activation of the downstream PI3K signalling pathway. Phosphorylated IRS1 is recognized and bound by SH2B1, inhibiting IRS1 dephosphorylation by protein tyrosine phosphatase 1B (PTP1B), thus prolonging insulin signalling pathway activation. Glucose transporter type 4 (GLUT4), Phosphoinositol 3-kinase (PI3K), phosphoinositide-3-kinase, regulatory subunit 1 (p110), phosphoinositide-3-kinase, regulatory subunit 2 (p85), Pyruvate dehydrogenase kinase (PDK), v-akt murine thymoma viral oncogene (AKT), Glycogen synthase kinase 3 (GSK3)
The fact that SH2B1 signals in the insulin pathway was substantiated in Drosophila melanogaster. The Drosophila genome contains a single SH2B homologue, known as Lnk, which shares a similar domain structure to its mammalian counterparts, including the highly conserved c-Cbl binding motif. In Drosophila, loss-of function Lnk mutations produce phenotypes reminiscent of reduced insulin and insulin-like growth factor-1 (IIS) signalling, including growth reduction, developmental delay and female sterility [46, 51]. Furthermore, classical genetic epistasis analysis established that, upon Drosophila insulin receptor (InR) activation during cell growth and division, Lnk signals in parallel with the Drosophila insulin receptor substrate, known as Chico, to activate PI3K [52].
Mutations reducing insulin/IGF-like (IIS) activity in multiple model organisms, including C. elegans, Drosophila and mouse, is known to increase lifespan. In Drosophila, recent studies showed that loss of Lnk increased lifespan, as well as improved survival during oxidative stress and starvation [46, 51, 52]. Furthermore, Lnk loss-of-function results in increased stored energy reserves associated with transcriptional changes in genes involved in both lipid and carbohydrate metabolism. Finally, in Drosophila, genetic analysis indicates that Lnk is itself a direct target for transcriptional regulation by the dFoxo transcription factor, indicating that Lnk transcription is regulated by insulin signalling [51].
3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGCR): regulates cholesterol production
3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGCR) is a transmembrane protein located on the smooth endoplasmic reticulum (SER), necessary to catalyze the production of mevalonate, the rate-limiting step during cholesterol biosynthesis in mammals [53, 54]. Tight control of cholesterol biosynthesis is physiologically critical, as overproduction can induce a predisposition to atherosclerosis [55]. In mammals, HMGCR production is regulated by cholesterol levels in a negative feedback look, while in mammals and Drosophila, HMGCR regulation is linked with levels of insulin, which strongly stimulates its production [56–59]. Insulin regulation of HMGCR involves a family of helix-loop-helix transcription factors, known as sterol response element binding proteins (SREBP) [60–63].
In mammals, mevalonate synthesized by HMGCR is required to produce cholesterol, which is used as a precursor for corticoid production by the adrenal glands or androgen production by the gonads [64, 65]. In insects, cholesterol production is not generated downstream of mevalonate, but in the Drosophila corpus allatum, mevalonate is used to synthesize Juvenile Hormone (JH), in response to insulin signalling from the Pars intercerebralis [59, 66, 67]. In adult flies, JH is necessary to regulate sexual dimorphic behaviour, such as variations in locomotor activity between males and females [59]. HMGCR also regulates adult body size in response to insulin signalling in Drosophila [59, 67].
Protein kinase D1 (PRKD1): regulates insulin secretion
PRKD1 is a member of the serine/threonine-protein kinase family. Along with the kinase domain, PRKD1 is predicted to contain a pleckstrin homology (PH) domain and two C1 domains. PH domains are known to possess multiple functions, including binding to inositol phosphates, as well as being involved in various protein–protein interactions [68–70], while C1 domains bind to the second messenger diacylglycerol (DAG) [71]. Protein kinase D1 is implicated in cancer, possibly through regulation of angiogenesis [72], and has also been shown to be involved in Toll-like receptor signalling in the immune response; specifically, it is necessary for the activity of the adaptor protein Myd88 [73].
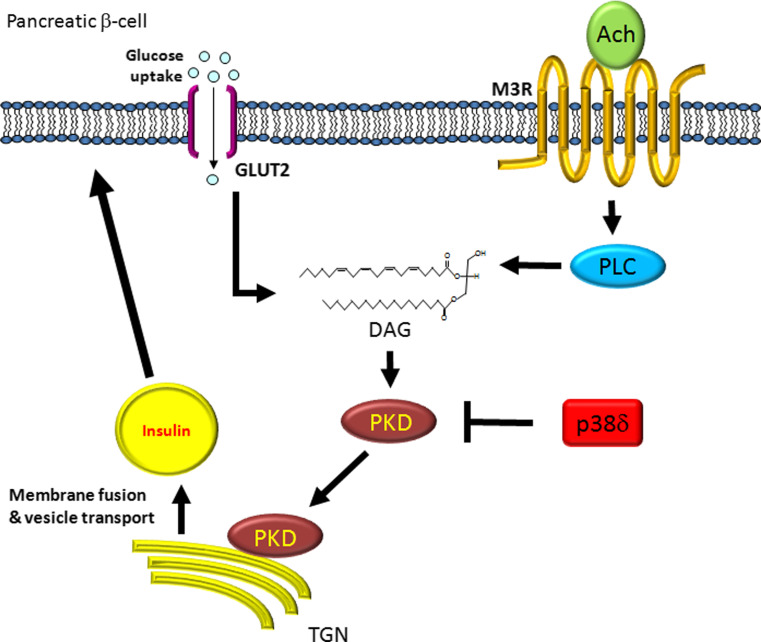
It was previously determined that PKD signals downstream of a Gq protein-coupled receptor (GqPCR) activated by acetylcholine [74]. When acetylcholine binds to the GqPCR M3-muscarinic receptor (M3R), it activates phospholipase C (PLC), leading the production of diacylglycerol (DAG), which in turn activates PKD (Fig. 2) [75]. It has been demonstrated that glucose passing through the SLC2A2 (also known as GLUT2) channel also leads to the production of DAG [76, 77]. DAG-activated PKD translocates to the trans-Golgi network (TGN) where it is necessary for vesicle membrane fusion. It was observed that this was also the case for insulin secretion; blocking PKD in INS-1 cells inhibited insulin secretion, but not insulin production [78]. In these same cells, it has been reported that loss of p38δ lead to increased insulin secretion, and that this increase in insulin secretion was due to hyperactivation of PKD [78]. Interestingly, a link between protein kinase D and insulin signalling was reported in C. elegans. Loss of the C. elegans PRKD1 orthologue, dkf-2, increases the worms lifespan by as much as 40 % over wild-type [79]. This increase in lifespan can be rescued in worms that also lack the C. elegans FOXO orthologue, daf-16. FOXO translocation to the nucleus is inhibited by insulin signalling, and the C. elegans result is further indication that PKD is regulating insulin signalling.
Fig. 2.
Protein kinase D (PKD) is necessary for insuln secretion. Acetylcholine (Ach) and glucose both induce pathways leading to PKD activation. Ach binds to and activates the GPCR M3R, leading to induction of phospholipase C (PLC), or glucose passing through the glucose transporter (GLUT), leading to the production of diacylglycerol (DAG), which in turn activates PKD. Activated PKD interacts with the trans-Golgi network (TGN) to regulate the production and transportation of vesicles, including those which contain insulin, to the plasma membrane. PKD is inhibited by the MAP kinase p38δ. M3R muscarinic acetylcholine receptor M3
Tubby (TUB): affects late-onset obesity
The mouse tubby mutation is the cause of maturity-onset obesity, insulin resistance and sensory deficits [80]. In contrast to the rapid juvenile-onset weight gain seen in diabetes (db) and obese (ob) mice, tubby mice become obese gradually as they age. This phenotype strongly resembles late-onset obesity observed in maturing humans. In the end, in tubby mice, excessive deposition of adipose tissue culminates in a twofold increase of body weight. Although this, along with the insulin resistance and sensory deficit phenotypes, indicate a vital role for tubby proteins, no molecular function has yet been attributed to this family of proteins [81]. TUB belongs to the tubby-like proteins, or TULPs, which are found in both the plant and animal kingdoms.
The N-terminal portion of TULP protein is not very conserved, but mammalian TUB contains a nuclear localisation signal and may have transcriptional-activation activity. The C-terminal residues of TULP family members are highly conserved, containing a cysteine residue that might play an important role in the normal functioning of these proteins. This domain is arranged as a 12-stranded, all anti-parallel, closed beta-barrel that surrounds a central alpha helix that forms most of the hydrophobic core. Structural analyses suggest that TULPs constitute a unique family of bipartite transcription factors [81]. In Drosophila, the TULP orthologue, known as king-tubby, is highly expressed in the developing nervous system [82]. In C. elegans, loss of tub-1, the worm orthologue of TUB, causes an increase in the storage of triglycerides and leads to a significant increase in life span [83]. While the increase in life span was rescued by removing the daf-16, the C. elegans FOXO homologue, the increase in lipid storage required TUB-1 interactions with the Rab GTPase-activating protein RBG-3, the orthologue of human TBC1D5. The results in C. elegans could mean that, similar to PRKD1, TUB is involved in regulating insulin secretion. On the other hand, this same group went on to show that TUB-1 and RBG-3 signal through RAB7 to regulate lipid storage in C. elegans [84]. In mammalian cell lines, TBC1D5 and Rab7 are both involved in regulating autophagy, and recently Rab7 was shown to signal downstream of insulin-like growth factor 1 (IGF1) to regulate autophagy in cultured neuronal cells [85, 86]. Considering the expression pattern of king-tubby in Drosophila, TUB may signal in neuronal cells during development to regulate autophagy.
Zinc finger protein 608 (ZNF608): regulates histone methylation for gene repression
Although nothing is known about possible molecular mechanisms of ZNF608 function, the Drosophila homologue of ZNF608, known as scribbler (sbb), is involved in the resistance to starvation, with sbb mutants being more susceptible to starvation [87]. Also, in Drosophila, Sbb bound directly to Grunge [known as arginine-glutamic acid dipeptide (RE) repeats, RERE, in humans], and together with the nuclear hormone receptor Tailless (Tll), known as TLX or NR2E1 in humans, they repressed the expression of the GAP gene knirps [88]. Finally, in this same study, it was shown that human ZNF608 and a closely related RERE protein, known as Antropin-1, directly interact, showing a conservation of function between the Drosophila and human proteins.
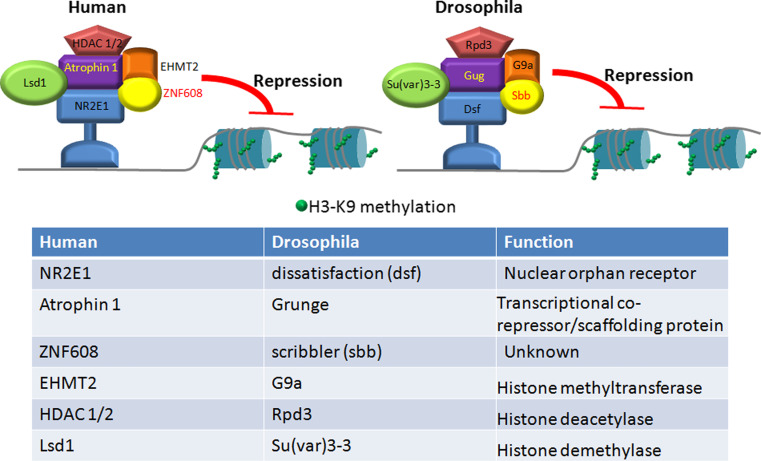
RERE can recruit HDAC1 and HDAC2 and the histone H3-K9 methyltransferase G9a [89] (Fig. 3). By directing the activities of HDAC1/2 and G9a, RERE catalyzes sequential molecular events, first causing deacetylation of H3-K9 and then allowing the deacetylated residue to be methylated by G9a [89]. As a result, chromatin regions where RERE binds are converted to compact structures, favouring gene silencing (Fig. 3). In Drosophila, the Atrophin-Rpd3 (HDAC1/2)-G9a complex represses the EGFR signalling pathway. During metamorphosis, wing vein formation is initiated by activated EGFR [90]. A mutation of Atro, or reduced expression of Atro using RNAi, results in ectopic vein formation in the intervein regions [89, 91, 92]. This ectopic wing vein phenotype is enhanced when G9a or Rpd3 is also mutated [89]. These genetic data indicate that Atro, G9a and Rpd3 act together to repress wing vein formation, perhaps by antagonizing the activities of EGFR.
Fig. 3.
ZNF608 functions in the atrophin transcriptional repression machinery. A model depicting the actions of the Atrophin-protein complex on chromosomes. Atrophin-1, or Grunge in Drosophila, recognizes nuclear hormone receptors and forms a complex with histone deacetylaces (HDACs), histone demethylase (Lsd1 in humans, Su(var)3-3 in Drosophila), histone methyltranserfase (EHMT2 in humans, G9a in Drosophila) and ZNF608 in humans or Scribbler in Drosophila. This complex induces methylation of histone H3 leading to transcriptional repression
Possible interactions between obesity-linked genes
TFAP2B and KCTD15: potential interactions in adipocytes
Transcription factor AP-2beta (TFAP2B) is a member of the AP-2 family of transcription factors, key regulators of various developmental processes [93–95]. AP-2 family members can homo- and heterodimerize through a highly conserved C-terminal helix-span-helix motif and bind to DNA by means of a basic domain immediately N-terminal of the dimerization domain. Potassium channel tetramerisation domain containing 15 (KCTD15) belongs to a family of potassium channel tetramerization domain-containing proteins. Like all KCTD family members, KCTD15 contains a Broad complex, Tramtrack and Bric-a-brac (BTB) domain. To date, only one study on KCTD15 has been performed to elucidate its molecular function. In zebrafish embryos, Kctd15 functions to inhibit the Wnt signalling pathway, in order to restrict neural crest formation, though the exact mechanism of this inhibition is unknown [96]. Interestingly, in Drosophila, the Wnt responsive gene hedgehog plays a role in adipogenesis and in mice is involved in the determination of brown and white adipose tissue [97].
A large-scale yeast two-hybrid screen was carried out in Drosophila, using almost the entire proteome, in an attempt to find all possible protein–protein interactions [98]. In this screen, the Drosophila homologues of TFAP2B (AP-2 in Drosophila) and KCTD15 (CG10440 in Drosophila) had a strong interaction. Furthermore, mRNA in situ analysis in Drosophila embryos demonstrated that AP-2 and CG10440 co-express in the developing brain [99, 100]. These two results, and the fact that both TFAP2B and KCTD15 are linked to obesity in multiple genome-wide association studies, indicate a possible physical interaction in vivo. It must be mentioned that yeast two-hybrid screens are not definitive evidence for an interaction between two proteins, and further experiments need to be performed to ascertain whether or not AP-2 and CG10440 proteins actually interact.
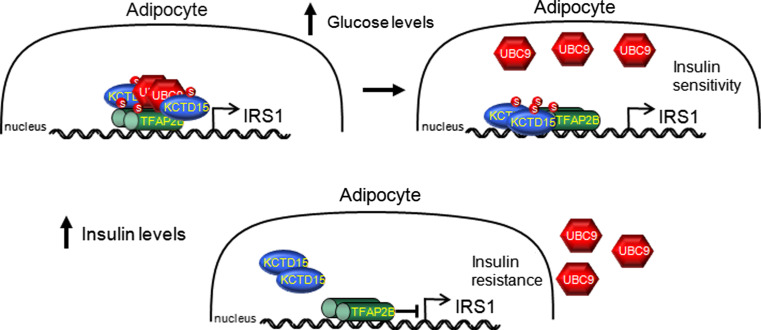
Taking what is known about AP-2 and KCTD family members, we propose the following model for a possible TFAP2B and KDTD15 interaction (Fig. 4). In adipocytes, AP-2β is a negative regulator of insulin receptor substrate-1 (IRS1) expression [93]. IRS1 links signalling from the insulin receptor (INSR) to the phosphoinositol 3-kinase (PI3K) and Akt pathways, and reduced IRS1 expression is one of the key molecular events involved in insulin resistance [101]. Also in adipocytes, Ap2α function was inhibited by sumoylation, requiring the SUMO-conjugating enzyme Ubiquitin carrier protein 9 (UBC9) [102]. In this same study, UBC9 was also shown to sumoylate AP-2β. KCTD5 and 11 interact with cullin E3 ligase (CUL3), and this complex then binds to E2 ubiquitin ligase [103–105]. Though no interaction with the sumoylation complex has been shown, using SUMOsp 2.0 Sumoylation Site Prediction [106, 107], we found a conserved sumoylation site in all KCTD family members. This leads to the possibility that KCTD family members are able to interact with the sumoylation apparatus. Furthermore, in a yeast two-hybrid screen, the Drosophila KCTD15 homolog, CG10440, was shown to interact directly with Lesswright (Lwr), the Drosophila UBC9 homolog [98].
Fig. 4.
Transcription factor AP-2beta (TFAP2B) and potassium channel tetramerization domain containing 15 (KCTD15) may regulate insulin receptor substrate 1 (IRS1) expression. Model predicting a possible interaction between AP-2β and KCTD15 in the regulation of IRS1. High glucose levels would activate the sumylation regulatory protein Ubiquitin carrier protein 9 (UBC9). UBC would then sumoylate KCTD15 and together they would bind to dimers of AP-2β. UBC9 sumoylation of AP-2β would inhibit its function, allowing for increased IRS1 transcription. Increased insulin signalling would feedback to release AP-2β inhibition and reduce IRS1 transcription
In our model (Fig. 4), high glucose levels would potentially induce UBC9 to interact and sumoylate KCTD15, and this complex could then bind to dimers of AP-2β, leading to their sumoylation and inhibition. AP-2β inhibition would allow for increased IRS1 transcription, and thus increased insulin sensitivity. Insulin signalling, in response to increased glucose levels, would inhibit UCB9 activation, leading to AP-2β desumoylation and activation. Activated AP-2β would inhibit IRS1 transcription, thus inducing insulin resistance. It has been published that AP-2α interacts with another KCTD family member, KCTD1. Interestingly, the domain in AP-2α required for interactions with KCTD1 is conserved in AP-2β and in Drosophila AP-2 [108].
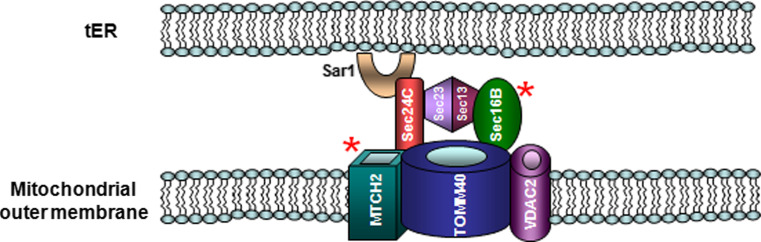
SEC16B and MTCH2 may help to regulate Ca2+ stores
Intracellular Ca2+ is required for the proper regulation of multiple important processes within a cell, and the formation of a precise spatiotemporal Ca2+ signal depends on extensive cellular machinery. Recently, it became evident that there is a complex interplay between the endoplasmic reticulum (ER), a major storehouse for Ca2+, and the mitochondria, to regulate not only cytoplasmic Ca2+ but also ER Ca2+ stores [109].
ER Ca2+ levels are regulated, in part, by the inositol 1,4,5-triphosphate receptor (IP3R) [110]. The amount of Ca2+ transferred from the ER to mitochondria not only depends on the activity of the IP3R but also on the distance between the mouth of the IP3R and voltage-dependent anion channels (VDAC), the major protein family involved in mitochondrial Ca2+ uptake. For this interaction to occur, the distance between the ER and mitochondria must be regulated, too close or too far and Ca2+ transfer is less efficient. This means that the cell needs a mechanism to maintain a stable distance between ER and mitochondrial membranes. Heat shock 70-kDa protein 9 (HSPA9), also known as mortalin, is one protein implicated in bridging this gap. HSPA9 binds to both IP3R and VDAC family members, allowing for better transfer of Ca2+ (Fig. 5) [111, 112]. Although HSPA9 controls interactions between IP3R and VDACs, thus helping to regulate gap distance, it is not the major determinant of the ER–mitochondrial bridge [110, 112].
Fig. 5.
Sec16 homolog B (SEC16B) and mitochondrial carrier 2 (MTCH2) may regulate ER and mitochondria gap distance. Depiction of interactions between the COPII apparatus at the tER and the translocase of the mitochondrial outer membrane 40 (Tom40) complex in the outer mitochondrial membrane. Interactions between the COPII machinery and the Tom40 complex would help regulate protein transit across the mitochondrial membrane, as well as maintain the correct distance between ER and mitochondrial membranes. This would, in turn, control the transfer of Ca2+ between the ER and mitochondria, which is used as a sensor for the induction of apoptosis. SEC24 family, member C (Sec24C), voltage-dependent anion channel 2 (VDAC2)
So what is regulating the gap distance? The ER protein Sec16 homolog B (SEC16B) and the mitochondrial protein mitochondrial carrier 2 (MTCH2) are both linked to obesity. The S. cerevisiae SEC16B homologue, known as Sec16, is a peripheral membrane protein necessary for ER-to-Golgi transport and cell viability [113]. Sec16-family members interact with the COPII machinery, including Sec23, Sec24 and Sec31, and are involved in the recruitment of the GTPase Sar1 to the ER exit sites (tER) (Fig. 5) [113–119].
MTCH2 shares homology with members of the mitochondrial carrier family, but unlike most family members, it is located on the mitochondrial outer membrane, where it co-localizes with a large protein complex, containing the apoptosis regulatory proteins tBID and BCL2-associated X protein (BAX) [120, 121] (Fig. 5). MTCH2 protein is required for tBID recruitment to the mitochondria and plays a essential role tBID-induced cell death [120].
In Drosophila, the COPII assembly proteins Sec16 and Sec24C have been shown to bind to the mitochondrial protein Tom40 [98]. Tom40 is the Drosophila homologue of mammalian translocase of outer mitochondrial membrane 40 (TOMM40), which is a channel-forming protein in the Tom40 complex, involved in mitochondrial protein import [122, 123]. In multiple organisms, the Tom40 complex and VDAC proteins directly interact to regulate each other’s functions [124–127]. It is postulated that MTCH2 may also interact with the Tom40 complex [128].
We suggest the following model for a SEC16B and MTCH2 interaction in regulating cellular homeostasis (Fig. 5). Tomm40 binding to SEC16B and SEC24C at the tER would facilitate mitochondrial protein import. It would also, along with HSPA9, help to anchor mitochondria close the ER, allowing calcium to pass out of the ER, via the IP3R, and into the mitochondria through VDAC. Mitochondrial calcium overload is a major inducer of apoptosis [129, 130], and would trigger MTCH2–BH3 interacting domain death agonist (tBid) interactions. In this way, the cell closely links ER and mitochondrial functions, necessary for the proper regulation of cellular Ca2+ levels. Disruption of this Ca2+ homeostasis would induce apoptosis. Interestingly, in C. elegans, TOMM-40, TOMM-20 and TOMM-22 are necessary for proper insulin secretion [131]. If SEC16B and MTCH2 interact with the TOMM complex in humans, they could also be involved in regulating insulin secretion.
Future perspectives
Current estimates suggest that there could be at least 250 genetic loci in humans important for the regulation of obesity [19], while the number of pathways or networks that these obesity genes participate in is likely to be much lower. Tremendous progress has been made recently in network biology, protein cooperatives, or modules displaying interaction between proteins, i.e. interactome networks. Many of these networks are highly conserved biological pathways throughout evolution. We show here that about half of the human obesity genes are highly evolutionary conserved and likely to be present in most vertebrates, and in some cases most bilateral species.
These obesity genes are not equally distributed through protein classes when compared with the rest of the genome. We see that genes involved in cell signalling, such as receptors (9 genes), ligands (2 genes) and signal transduction molecules (1 gene), corresponding to about one-third of the obesity associated genes, are significantly enriched (p ≤ 0.01; hypergeometric test) when compared to the whole human genome, where about 16 % of the genes are annotated to have comparable functions (source: http://www.uniprot.org, http://www.geneontology.org). Similarly, we also identify a potential enrichment for transcription factors according to UniProt and Gene ontology (p ≤ 0.02; 14 % of the obesity genes, ~4 % of genome). However, when compared with human transcription factors listed in a comprehensive study by Ravasi and colleagues (~10 % of the genome) [132] the enrichment was not validated (p > 0.92) and should be considered as uncertain. The 9 enzymes and 2 transporters found among the obesity genes are not more than expected by chance.
We identified new potentially important protein interactions using expression data from mouse and Drosophila (BioGPS, Flyatlas), and other data obtained from Drosophila (yeast-two hybrid, embryonic mRNA in situ), as well as searching homologies (NCBI homoloGene) and protein interaction maps (Interlog finder, http://www.interlogfinder.org). We show that TFAP2B is linked to KCTD15 and SEC16B is linked to MTCH2. For HMGCR, we see a conservation of expression and function, as it is necessary to make cholesterol-derived products involved in regulating homeostasis, corticoids in the case of mammals and Juvenile hormone (JH) in Drosophila adults. These findings show that studies using model organisms, such as Drosophila melanogaster, can provide a conservation of function that, if exploited, could help in our understanding of how these obesity-linked genes function to regulate the homeostatic system.
Moreover, it is interesting to identify which of the genes in each network are the most suitable against which to develop drugs. One classical feature of drug targets includes the ability of the protein to form a cavity that a small molecule can lock in, and in most cases block the activity of the proteins. Indeed, the 9 receptors (LRP1B, NRXN3, GPRC5B, NEGR1, GIPR, CADM2, MC4R, FAIM2 and LRRN6C) and 9 enzymes (QPCTL, HMGCR, GNPDA2, FANCL, NUDT3, PRKD1, TNNI3 K, MAP2K5 and FTO) that together constitute about half of the obesity genes arguably have this important drug target feature. Also, 13 of the genes (CADM2, FAIM2, GIPR, GPRC5B, HMGCR, LINGO2, LRP1B, MC4R, MTCH2,NRXN3, QPCTL, SLC39A8 and TMEM18) code for transmembrane proteins, a highly used class of proteins in drug development and likely to play key roles in conveying signals over the membrane (another classical feature of drug targets). Furthermore, 14 of the genes (MC4R, GIPR, MAP2K4, PRKD1, TNNI3K, LINGO2, NEGR1, CADM2, KCTD15, SH2B1, QPCTL, GPRC5B, LRP1B and NRXN3) share protein family or protein domains with known drug targets, and HMGCR is targeted by a number of anticholesteremic agents, such as Lovastatin [133]. Hence, there are enormous possibilities that a considerable part of the currently identified obesity-linked genes can be targeted by traditional types of drugs. The recent success of monoclonal antibodies has allowed for additional opportunities for rational target selection and high target specificity. Among the obesity genes, there are two coding for ligands (BDNF and POMC); such gene products are not traditional drug targets, but may become interesting providing that an antibody could reach the intended site of action. This is obviously complicated by the fact that these ligands are expressed in the CNS. In general, while our understanding of the functional roles of obesity-linked genes is currently very vague, no doubt distant model species, interactomics and signalling pathway analysis represent an important way to better understand the functional diversity of the surprisingly high number of molecules seemingly important for human obesity.
Contributor Information
Michael J. Williams, FAX: +46-18-511540, Email: michael.williams@neuro.uu.se
Helgi B. Schiöth, Email: Helgi.Schioth@neuro.uu.se
References
- 1.Bouchard C. How much progress have we made over the last few decades? Int J Obes. 2008;32(Suppl 7):S2–S7. doi: 10.1038/ijo.2008.231. [DOI] [PubMed] [Google Scholar]
- 2.Maes HH, Neale MC, Eaves LJ. Genetic and environmental factors in relative body weight and human adiposity. Behav Genet. 1997;27:325–351. doi: 10.1023/a:1025635913927. [DOI] [PubMed] [Google Scholar]
- 3.Wardle J, Carnell S, Haworth CM, Plomin R. Evidence for a strong genetic influence on childhood adiposity despite the force of the obesogenic environment. Am J Clin Nutr. 2008;87:398–404. doi: 10.1093/ajcn/87.2.398. [DOI] [PubMed] [Google Scholar]
- 4.Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, Perusse L, Bouchard C. The human obesity gene map: the 2005 update. Obesity (Silver Spring) 2006;14:529–644. doi: 10.1038/oby.2006.71. [DOI] [PubMed] [Google Scholar]
- 5.Huszar D, Lynch CA, Fairchild-Huntress V, Dunmore JH, Fang Q, Berkemeier LR, Gu W, Kesterson RA, Boston BA, Cone RD, Smith FJ, Campfield LA, Burn P, Lee F. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell. 1997;88:131–141. doi: 10.1016/s0092-8674(00)81865-6. [DOI] [PubMed] [Google Scholar]
- 6.Krude H, Biebermann H, Luck W, Horn R, Brabant G, Gruters A. Severe early-onset obesity, adrenal insufficiency and red hair pigmentation caused by POMC mutations in humans. Nat Genet. 1998;19:155–157. doi: 10.1038/509. [DOI] [PubMed] [Google Scholar]
- 7.Kask A, Rago L, Korrovits P, Wikberg JE, Schioth HB. Evidence that orexigenic effects of melanocortin 4 receptor antagonist HS014 are mediated by neuropeptide Y. Biochem Biophys Res Commun. 1998;248:245–249. doi: 10.1006/bbrc.1998.8961. [DOI] [PubMed] [Google Scholar]
- 8.Cone RD. Anatomy and regulation of the central melanocortin system. Nat Neurosci. 2005;8:571–578. doi: 10.1038/nn1455. [DOI] [PubMed] [Google Scholar]
- 9.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, Shields B, Harries LW, Barrett JC, Ellard S, Groves CJ, Knight B, Patch AM, Ness AR, Ebrahim S, Lawlor DA, Ring SM, Ben-Shlomo Y, Jarvelin MR, Sovio U, Bennett AJ, Melzer D, Ferrucci L, Loos RJ, Barroso I, Wareham NJ, Karpe F, Owen KR, Cardon LR, Walker M, Hitman GA, Palmer CN, Doney AS, Morris AD, Smith GD, Hattersley AT, McCarthy MI. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fredriksson R, Hagglund M, Olszewski PK, Stephansson O, Jacobsson JA, Olszewska AM, Levine AS, Lindblom J, Schioth HB. The obesity gene, FTO, is of ancient origin, up-regulated during food deprivation and expressed in neurons of feeding-related nuclei of the brain. Endocrinology. 2008;149:2062–2071. doi: 10.1210/en.2007-1457. [DOI] [PubMed] [Google Scholar]
- 11.Speakman JR, Rance KA, Johnstone AM. Polymorphisms of the FTO gene are associated with variation in energy intake, but not energy expenditure. Obesity. 2008;16:1961–1965. doi: 10.1038/oby.2008.318. [DOI] [PubMed] [Google Scholar]
- 12.Tanofsky-Kraff M, Han JC, Anandalingam K, Shomaker LB, Columbo KM, Wolkoff LE, Kozlosky M, Elliott C, Ranzenhofer LM, Roza CA, Yanovski SZ, Yanovski JA. The FTO gene rs9939609 obesity-risk allele and loss of control over eating. Am J Clin Nutr. 2009;90:1483–1488. doi: 10.3945/ajcn.2009.28439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer J, Koch L, Emmerling C, Vierkotten J, Peters T, Bruning JC, Ruther U. Inactivation of the Fto gene protects from obesity. Nature. 2009;458:894–898. doi: 10.1038/nature07848. [DOI] [PubMed] [Google Scholar]
- 14.Church C, Lee S, Bagg EA, McTaggart JS, Deacon R, Gerken T, Lee A, Moir L, Mecinovic J, Quwailid MM, Schofield CJ, Ashcroft FM, Cox RD. A mouse model for the metabolic effects of the human fat mass and obesity associated FTO gene. PLoS Genet. 2009;5:e1000599. doi: 10.1371/journal.pgen.1000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, Berndt SI, Elliott AL, Jackson AU, Lamina C, Lettre G, Lim N, Lyon HN, McCarroll SA, Papadakis K, Qi L, Randall JC, Roccasecca RM, Sanna S, Scheet P, Weedon MN, Wheeler E, Zhao JH, Jacobs LC, Prokopenko I, Soranzo N, Tanaka T, Timpson NJ, Almgren P, Bennett A, Bergman RN, Bingham SA, Bonnycastle LL, Brown M, Burtt NP, Chines P, Coin L, Collins FS, Connell JM, Cooper C, Smith GD, Dennison EM, Deodhar P, Elliott P, Erdos MR, Estrada K, Evans DM, Gianniny L, Gieger C, Gillson CJ, Guiducci C, Hackett R, Hadley D, Hall AS, Havulinna AS, Hebebrand J, Hofman A, Isomaa B, Jacobs KB, Johnson T, Jousilahti P, Jovanovic Z, Khaw KT, Kraft P, Kuokkanen M, Kuusisto J, Laitinen J, Lakatta EG, Luan J, Luben RN, Mangino M, McArdle WL, Meitinger T, Mulas A, Munroe PB, Narisu N, Ness AR, Northstone K, O’Rahilly S, Purmann C, Rees MG, Ridderstrale M, Ring SM, Rivadeneira F, Ruokonen A, Sandhu MS, Saramies J, Scott LJ, Scuteri A, Silander K, Sims MA, Song K, Stephens J, Stevens S, Stringham HM, Tung YC, Valle TT, Van Duijn CM, Vimaleswaran KS, Vollenweider P, Waeber G, Wallace C, Watanabe RM, Waterworth DM, Watkins N, Wellcome Trust Case Control C. Witteman JC, Zeggini E, Zhai G, Zillikens MC, Altshuler D, Caulfield MJ, Chanock SJ, Farooqi IS, Ferrucci L, Guralnik JM, Hattersley AT, Hu FB, Jarvelin MR, Laakso M, Mooser V, Ong KK, Ouwehand WH, Salomaa V, Samani NJ, Spector TD, Tuomi T, Tuomilehto J, Uda M, Uitterlinden AG, Wareham NJ, Deloukas P, Frayling TM, Groop LC, Hayes RB, Hunter DJ, Mohlke KL, Peltonen L, Schlessinger D, Strachan DP, Wichmann HE, McCarthy MI, Boehnke M, Barroso I, Abecasis GR, Hirschhorn JN, Genetic Investigation of ATC Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, Styrkarsdottir U, Gretarsdottir S, Thorlacius S, Jonsdottir I, Jonsdottir T, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Jonsson F, Borch-Johnsen K, Hansen T, Andersen G, Jorgensen T, Lauritzen T, Aben KK, Verbeek AL, Roeleveld N, Kampman E, Yanek LR, Becker LC, Tryggvadottir L, Rafnar T, Becker DM, Gulcher J, Kiemeney LA, Pedersen O, Kong A, Thorsteinsdottir U, Stefansson K. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 17.Gratacos M, Gonzalez JR, Mercader JM, de Cid R, Urretavizcaya M, Estivill X. Brain-derived neurotrophic factor Val66Met and psychiatric disorders: meta-analysis of case-control studies confirm association to substance-related disorders, eating disorders, and schizophrenia. Biol Psychiatry. 2007;61:911–922. doi: 10.1016/j.biopsych.2006.08.025. [DOI] [PubMed] [Google Scholar]
- 18.Gunstad J, Schofield P, Paul RH, Spitznagel MB, Cohen RA, Williams LM, Kohn M, Gordon E. BDNF Val66Met polymorphism is associated with body mass index in healthy adults. Neuropsychobiology. 2006;53:153–156. doi: 10.1159/000093341. [DOI] [PubMed] [Google Scholar]
- 19.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Allen HL, Lindgren CM, Luan J, Magi R, Randall JC, Vedantam S, Winkler TW, Qi L, Workalemahu T, Heid IM, Steinthorsdottir V, Stringham HM, Weedon MN, Wheeler E, Wood AR, Ferreira T, Weyant RJ, Segre AV, Estrada K, Liang L, Nemesh J, Park JH, Gustafsson S, Kilpelainen TO, Yang J, Bouatia-Naji N, Esko T, Feitosa MF, Kutalik Z, Mangino M, Raychaudhuri S, Scherag A, Smith AV, Welch R, Zhao JH, Aben KK, Absher DM, Amin N, Dixon AL, Fisher E, Glazer NL, Goddard ME, Heard-Costa NL, Hoesel V, Hottenga JJ, Johansson A, Johnson T, Ketkar S, Lamina C, Li S, Moffatt MF, Myers RH, Narisu N, Perry JR, Peters MJ, Preuss M, Ripatti S, Rivadeneira F, Sandholt C, Scott LJ, Timpson NJ, Tyrer JP, van Wingerden S, Watanabe RM, White CC, Wiklund F, Barlassina C, Chasman DI, Cooper MN, Jansson JO, Lawrence RW, Pellikka N, Prokopenko I, Shi J, Thiering E, Alavere H, Alibrandi MT, Almgren P, Arnold AM, Aspelund T, Atwood LD, Balkau B, Balmforth AJ, Bennett AJ, Ben-Shlomo Y, Bergman RN, Bergmann S, Biebermann H, Blakemore AI, Boes T, Bonnycastle LL, Bornstein SR, Brown MJ, Buchanan TA, Busonero F, Campbell H, Cappuccio FP, Cavalcanti-Proenca C, Chen YD, Chen CM, Chines PS, Clarke R, Coin L, Connell J, Day IN, Heijer M, Duan J, Ebrahim S, Elliott P, Elosua R, Eiriksdottir G, Erdos MR, Eriksson JG, Facheris MF, Felix SB, Fischer-Posovszky P, Folsom AR, Friedrich N, Freimer NB, Fu M, Gaget S, Gejman PV, Geus EJ, Gieger C, Gjesing AP, Goel A, Goyette P, Grallert H, Grassler J, Greenawalt DM, Groves CJ, Gudnason V, Guiducci C, Hartikainen AL, Hassanali N, Hall AS, Havulinna AS, Hayward C, Heath AC, Hengstenberg C, Hicks AA, Hinney A, Hofman A, Homuth G, Hui J, Igl W, Iribarren C, Isomaa B, Jacobs KB, Jarick I, Jewell E, John U, Jorgensen T, Jousilahti P, Jula A, Kaakinen M, Kajantie E, Kaplan LM, Kathiresan S, Kettunen J, Kinnunen L, Knowles JW, Kolcic I, Konig IR, Koskinen S, Kovacs P, Kuusisto J, Kraft P, Kvaloy K, Laitinen J, Lantieri O, Lanzani C, Launer LJ, Lecoeur C, Lehtimaki T, Lettre G, Liu J, Lokki ML, Lorentzon M, Luben RN, Ludwig B, Magic, Manunta P, Marek D, Marre M, Martin NG, McArdle WL, McCarthy A, McKnight B, Meitinger T, Melander O, Meyre D, Midthjell K, Montgomery GW, Morken MA, Morris AP, Mulic R, Ngwa JS, Nelis M, Neville MJ, Nyholt DR, O’Donnell CJ, O’Rahilly S, Ong KK, Oostra B, Pare G, Parker AN, Perola M, Pichler I, Pietilainen KH, Platou CG, Polasek O, Pouta A, Rafelt S, Raitakari O, Rayner NW, Ridderstrale M, Rief W, Ruokonen A, Robertson NR, Rzehak P, Salomaa V, Sanders AR, Sandhu MS, Sanna S, Saramies J, Savolainen MJ, Scherag S, Schipf S, Schreiber S, Schunkert H, Silander K, Sinisalo J, Siscovick DS, Smit JH, Soranzo N, Sovio U, Stephens J, Surakka I, Swift AJ, Tammesoo ML, Tardif JC, Teder-Laving M, Teslovich TM, Thompson JR, Thomson B, Tonjes A, Tuomi T, van Meurs JB, van Ommen GJ, Vatin V, Viikari J, Visvikis-Siest S, Vitart V, Vogel CI, Voight BF, Waite LL, Wallaschofski H, Walters GB, Widen E, Wiegand S, Wild SH, Willemsen G, Witte DR, Witteman JC, Xu J, Zhang Q, Zgaga L, Ziegler A, Zitting P, Beilby JP, Farooqi IS, Hebebrand J, Huikuri HV, James AL, Kahonen M, Levinson DF, Macciardi F, Nieminen MS, Ohlsson C, Palmer LJ, Ridker PM, Stumvoll M, Beckmann JS, Boeing H, Boerwinkle E, Boomsma DI, Caulfield MJ, Chanock SJ, Collins FS, Cupples LA, Smith GD, Erdmann J, Froguel P, Gronberg H, Gyllensten U, Hall P, Hansen T, Harris TB, Hattersley AT, Hayes RB, Heinrich J, Hu FB, Hveem K, Illig T, Jarvelin MR, Kaprio J, Karpe F, Khaw KT, Kiemeney LA, Krude H, Laakso M, Lawlor DA, Metspalu A, Munroe PB, Ouwehand WH, Pedersen O, Penninx BW, Peters A, Pramstaller PP, Quertermous T, Reinehr T, Rissanen A, Rudan I, Samani NJ, Schwarz PE, Shuldiner AR, Spector TD, Tuomilehto J, Uda M, Uitterlinden A, Valle TT, Wabitsch M, Waeber G, Wareham NJ, Watkins H, Procardis C, Wilson JF, Wright AF, Zillikens MC, Chatterjee N, McCarroll SA, Purcell S, Schadt EE, Visscher PM, Assimes TL, Borecki IB, Deloukas P, Fox CS, Groop LC, Haritunians T, Hunter DJ, Kaplan RC, Mohlke KL, O’Connell JR, Peltonen L, Schlessinger D, Strachan DP, van Duijn CM, Wichmann HE, Frayling TM, Thorsteinsdottir U, Abecasis GR, Barroso I, Boehnke M, Stefansson K, North KE, McCarthy MI, Hirschhorn JN, Ingelsson E, Loos RJ. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tseng CC, Zhang XY. Role of G protein-coupled receptor kinases in glucose-dependent insulinotropic polypeptide receptor signaling. Endocrinology. 2000;141:947–952. doi: 10.1210/endo.141.3.7365. [DOI] [PubMed] [Google Scholar]
- 21.Kim SJ, Nian C, McIntosh CH. Adipocyte expression of the glucose-dependent insulinotropic polypeptide receptor involves gene regulation by PPAR{gamma} and histone acetylation. J Lipid Res. 2011;52:759–770. doi: 10.1194/jlr.M012203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song DH, Getty-Kaushik L, Tseng E, Simon J, Corkey BE, Wolfe MM. Glucose-dependent insulinotropic polypeptide enhances adipocyte development and glucose uptake in part through Akt activation. Gastroenterology. 2007;133:1796–1805. doi: 10.1053/j.gastro.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyawaki K, Yamada Y, Yano H, Niwa H, Ban N, Ihara Y, Kubota A, Fujimoto S, Kajikawa M, Kuroe A, Tsuda K, Hashimoto H, Yamashita T, Jomori T, Tashiro F, Miyazaki J, Seino Y. Glucose intolerance caused by a defect in the entero-insular axis: a study in gastric inhibitory polypeptide receptor knockout mice. Proc Natl Acad Sci USA. 1999;96:14843–14847. doi: 10.1073/pnas.96.26.14843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Preitner F, Ibberson M, Franklin I, Binnert C, Pende M, Gjinovci A, Hansotia T, Drucker DJ, Wollheim C, Burcelin R, Thorens B. Gluco-incretins control insulin secretion at multiple levels as revealed in mice lacking GLP-1 and GIP receptors. J Clinical Invest. 2004;113:635–645. doi: 10.1172/JCI20518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansotia T, Drucker DJ. GIP and GLP-1 as incretin hormones: lessons from single and double incretin receptor knockout mice. Regul Pept. 2005;128(2):125–134. doi: 10.1016/j.regpep.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 26.Pederson RA, Satkunarajah M, McIntosh CH, Scrocchi LA, Flamez D, Schuit F, Drucker DJ, Wheeler MB. Enhanced glucose-dependent insulinotropic polypeptide secretion and insulinotropic action in glucagon-like peptide 1 receptor −/− mice. Diabetes. 1998;47:1046–1052. doi: 10.2337/diabetes.47.7.1046. [DOI] [PubMed] [Google Scholar]
- 27.Miyawaki K, Yamada Y, Ban N, Ihara Y, Tsukiyama K, Zhou H, Fujimoto S, Oku A, Tsuda K, Toyokuni S, Hiai H, Mizunoya W, Fushiki T, Holst JJ, Makino M, Tashita A, Kobara Y, Tsubamoto Y, Jinnouchi T, Jomori T, Seino Y. Inhibition of gastric inhibitory polypeptide signaling prevents obesity. Nat Med. 2002;8:738–742. doi: 10.1038/nm727. [DOI] [PubMed] [Google Scholar]
- 28.Hansotia T, Maida A, Flock G, Yamada Y, Tsukiyama K, Seino Y, Drucker DJ. Extrapancreatic incretin receptors modulate glucose homeostasis, body weight, and energy expenditure. J Clinical Invest. 2007;117:143–152. doi: 10.1172/JCI25483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaragosi LE, Wdziekonski B, Brigand KL, Villageois P, Mari B, Waldmann R, Dani C, Barbry P. Small RNA sequencing reveals miR-642a-3p as a novel adipocyte-specific microRNA and miR-30 as a key regulator of human adipogenesis. Genome Biol. 2011;12:R64. doi: 10.1186/gb-2011-12-7-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinelli R, Nardelli C, Pilone V, Buonomo T, Liguori R, Castano I, Buono P, Masone S, Persico G, Forestieri P, Pastore L, Sacchetti L. miR-519d overexpression is associated with human obesity. Obesity. 2010;18:2170–2176. doi: 10.1038/oby.2009.474. [DOI] [PubMed] [Google Scholar]
- 31.Brauner-Osborne H, Wellendorph P, Jensen AA. Structure, pharmacology and therapeutic prospects of family C G-protein coupled receptors. Curr Drug Targets. 2007;8:169–184. doi: 10.2174/138945007779315614. [DOI] [PubMed] [Google Scholar]
- 32.Urwyler S. Allosteric modulation of family C G-protein-coupled receptors: from molecular insights to therapeutic perspectives. Pharmacol Rev. 2011;63:59–126. doi: 10.1124/pr.109.002501. [DOI] [PubMed] [Google Scholar]
- 33.Pleines I, Elvers M, Strehl A, Pozgajova M, Varga-Szabo D, May F, Chrostek-Grashoff A, Brakebusch C, Nieswandt B. Rac1 is essential for phospholipase C-gamma2 activation in platelets. Pflugers Archiv. 2009;457:1173–1185. doi: 10.1007/s00424-008-0573-7. [DOI] [PubMed] [Google Scholar]
- 34.Rickhag M, Wieloch T, Gido G, Elmer E, Krogh M, Murray J, Lohr S, Bitter H, Chin DJ, von Schack D, Shamloo M, Nikolich K. Comprehensive regional and temporal gene expression profiling of the rat brain during the first 24 h after experimental stroke identifies dynamic ischemia-induced gene expression patterns, and reveals a biphasic activation of genes in surviving tissue. J Neurochem. 2006;96:14–29. doi: 10.1111/j.1471-4159.2005.03508.x. [DOI] [PubMed] [Google Scholar]
- 35.Brauner-Osborne H, Krogsgaard-Larsen P. Sequence and expression pattern of a novel human orphan G-protein-coupled receptor, GPRC5B, a family C receptor with a short amino-terminal domain. Genomics. 2000;65:121–128. doi: 10.1006/geno.2000.6164. [DOI] [PubMed] [Google Scholar]
- 36.Sano T, Kim YJ, Oshima E, Shimizu C, Kiyonari H, Abe T, Higashi H, Yamada K, Hirabayashi Y. Comparative characterization of GPRC5B and GPRC5CLacZ knockin mice; behavioral abnormalities in GPRC5B-deficient mice. Biochem Biophys Res Commun. 2011;412:460–465. doi: 10.1016/j.bbrc.2011.07.118. [DOI] [PubMed] [Google Scholar]
- 37.Simon MA. Signal transduction during the development of the Drosophila R7 photoreceptor. Dev Biol. 1994;166(2):431–442. doi: 10.1006/dbio.1994.1327. [DOI] [PubMed] [Google Scholar]
- 38.Kohyama-Koganeya A, Kim Y-J, Miura M, Hirabayashi Y. A Drosophila orphan G protein-coupled receptor BOSS functions as a glucose-responding receptor: loss of boss causes abnormal energy metabolism. Proc Natl Acad Sci USA. 2008;105:15328–15333. doi: 10.1073/pnas.0807833105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kohyama-Koganeya A, Hirabayashi Y (2010) The Drosophila 7-pass transmembrane glycoprotein BOSS and metabolic regulation: what Drosophila can teach us about human energy metabolism. In: Minoru F (ed) Methods in enzymology, vol 480, Glycobiology. Academic, New York, pp 525–538 [DOI] [PubMed]
- 40.Morris DL, Cho KW, Zhou Y, Rui L. SH2B1 enhances insulin sensitivity by both stimulating the insulin receptor and inhibiting tyrosine dephosphorylation of insulin receptor substrate proteins. Diabetes. 2009;58:2039–2047. doi: 10.2337/db08-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Donatello S, Fiorino A, Degl’Innocenti D, Alberti L, Miranda C, Gorla L, Bongarzone I, Rizzetti MG, Pierotti MA, Borrello MG. SH2B1beta adaptor is a key enhancer of RET tyrosine kinase signaling. Oncogene. 2007;26:6546–6559. doi: 10.1038/sj.onc.1210480. [DOI] [PubMed] [Google Scholar]
- 42.Li M, Li Z, Morris DL, Rui L. Identification of SH2B2beta as an inhibitor for SH2B1- and SH2B2alpha-promoted Janus kinase-2 activation and insulin signaling. Endocrinol. 2007;148:1615–1621. doi: 10.1210/en.2006-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshiga D, Sato N, Torisu T, Mori H, Yoshida R, Nakamura S, Takaesu G, Kobayashi T, Yoshimura A. Adaptor protein SH2-B linking receptor-tyrosine kinase and Akt promotes adipocyte differentiation by regulating peroxisome proliferator-activated receptor gamma messenger ribonucleic acid levels. Mol Endocrinol. 2007;21:1120–1131. doi: 10.1210/me.2006-0413. [DOI] [PubMed] [Google Scholar]
- 44.O’Brien KB, O’Shea JJ, Carter-Su C. SH2-B family members differentially regulate JAK family tyrosine kinases. J Biol Chem. 2002;277:8673–8681. doi: 10.1074/jbc.M109165200. [DOI] [PubMed] [Google Scholar]
- 45.Duan C, Tang C, Liao L, Li C, Su T, Chen Z. Molecular mechanism of SH2B1 in regulating JAK2/IRS2 during obesity development. Zhong nan da xue xue baoYi xue ban. 2010;35:209–214. doi: 10.3969/j.issn.1672-7347.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 46.Song W, Ren D, Li W, Jiang L, Cho KW, Huang P, Fan C, Song Y, Liu Y, Rui L. SH2B regulation of growth, metabolism, and longevity in both insects and mammals. Cell Metab. 2010;11:427–437. doi: 10.1016/j.cmet.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morris DL, Cho KW, Rui L. Critical role of the Src homology 2 (SH2) domain of neuronal SH2B1 in the regulation of body weight and glucose homeostasis in mice. Endocrinology. 2010;151:3643–3651. doi: 10.1210/en.2010-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ren D, Zhou Y, Morris D, Li M, Li Z, Rui L. Neuronal SH2B1 is essential for controlling energy and glucose homeostasis. J Clin Invest. 2007;117:397–406. doi: 10.1172/JCI29417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kubo-Akashi C, Iseki M, Kwon SM, Takizawa H, Takatsu K, Takaki S. Roles of a conserved family of adaptor proteins, Lnk, SH2-B, and APS, for mast cell development, growth, and functions: APS-deficiency causes augmented degranulation and reduced actin assembly. Biochem Biophys Res Commun. 2004;315:356–362. doi: 10.1016/j.bbrc.2004.01.060. [DOI] [PubMed] [Google Scholar]
- 50.Maures TJ, Kurzer JH, Carter-Su C. SH2B1 (SH2-B) and JAK2: a multifunctional adaptor protein and kinase made for each other. Trends Endocrinol Metab. 2007;18:38–45. doi: 10.1016/j.tem.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 51.Slack C, Werz C, Wieser D, Alic N, Foley A, Stocker H, Withers DJ, Thornton JM, Hafen E, Partridge L. Regulation of lifespan, metabolism, and stress responses by the Drosophila SH2B protein. Lnk. PLoS Genet. 2010;6:e1000881. doi: 10.1371/journal.pgen.1000881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Werz C, Kohler K, Hafen E, Stocker H. The Drosophila SH2B family adaptor Lnk acts in parallel to chico in the insulin signaling pathway. PLoS Genet. 2009;5:e1000596. doi: 10.1371/journal.pgen.1000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang J, Yang XF, Liao DF, Wu Q, Liu ZX, He YD. A study of the effects of quercetin on the expression of HMGCR and the cholesterol synthesis of HL-02 cells] Zhonghua gan zang bing za zhi. 2007;15:143–145. [PubMed] [Google Scholar]
- 54.Dietschy JM, Turley SD, Spady DK. Role of liver in the maintenance of cholesterol and low density lipoprotein homeostasis in different animal species, including humans. J Lipid Res. 1993;34:1637–1659. [PubMed] [Google Scholar]
- 55.Reed RM, Iacono A, Defilippis A, Eberlein M, Girgis RE, Jones S. Advanced chronic obstructive pulmonary disease is associated with high levels of high-density lipoprotein cholesterol. J Heart Lung Transplant. 2011;30:674–678. doi: 10.1016/j.healun.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 56.Ness GC, Chambers CM. Feedback and hormonal regulation of hepatic 3-hydroxy-3-methylglutaryl coenzyme A reductase: the concept of cholesterol buffering capacity. Proc Soc Exp Biol Med. 2000;224:8–19. doi: 10.1046/j.1525-1373.2000.22359.x. [DOI] [PubMed] [Google Scholar]
- 57.Easom RA, Zammit VA. Effects of diabetes on the expressed and total activities of 3-hydroxy-3-methylglutaryl-CoA reductase in rat liver in vivo. Reversal by insulin treatment. Biochem J. 1985;230:747–752. doi: 10.1042/bj2300747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stange EF, Fleig WE, Schneider A, Nother-Fleig G, Alavi M, Preclik G, Ditschuneit H. 3-Hydroxy-3-methylglutaryl CoA reductase in cultured hepatocytes. Regulation by heterologous lipoproteins and hormones. Atherosclerosis. 1982;41:67–80. doi: 10.1016/0021-9150(82)90071-5. [DOI] [PubMed] [Google Scholar]
- 59.Belgacem YH, Martin JR. Hmgcr in the corpus allatum controls sexual dimorphism of locomotor activity and body size via the insulin pathway in Drosophila. PLoS ONE. 2007;2:e187. doi: 10.1371/journal.pone.0000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jump DB. Fatty acid regulation of hepatic lipid metabolism. Curr Opin Clin Nutr Metab Care. 2010;14:115–120. doi: 10.1097/MCO.0b013e328342991c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wong RH, Sul HS. Insulin signaling in fatty acid and fat synthesis: a transcriptional perspective. Curr Opin Pharmacol. 2010;10:684–691. doi: 10.1016/j.coph.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kunte AS, Matthews KA, Rawson RB. Fatty acid auxotrophy in Drosophila larvae lacking SREBP. Cell Metab. 2006;3:439–448. doi: 10.1016/j.cmet.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 63.van der Meer DL, Degenhardt T, Vaisanen S, de Groot PJ, Heinaniemi M, de Vries SC, Muller M, Carlberg C, Kersten S. Profiling of promoter occupancy by PPARalpha in human hepatoma cells via ChIP-chip analysis. Nucleic Acids Res. 2010;38(9):2839–2850. doi: 10.1093/nar/gkq012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waterham HR. Inherited disorders of cholesterol biosynthesis. Clin Genet. 2002;61:393–403. doi: 10.1034/j.1399-0004.2002.610601.x. [DOI] [PubMed] [Google Scholar]
- 65.Beg ZH, Brewer HB., Jr Regulation of liver 3-hydroxy-3-methylglutaryl-CoA reductase. Curr Topics Cell Reg. 1981;20:139–184. doi: 10.1016/b978-0-12-152820-1.50008-0. [DOI] [PubMed] [Google Scholar]
- 66.Belles X, Martin D, Piulachs MD. The mevalonate pathway and the synthesis of juvenile hormone in insects. Annu Rev Entomol. 2005;50:181–199. doi: 10.1146/annurev.ento.50.071803.130356. [DOI] [PubMed] [Google Scholar]
- 67.Gruntenko NE, Wen D, Karpova EK, Adonyeva NV, Liu Y, He Q, Faddeeva NV, Fomin AS, Li S, Rauschenbach IY. Altered juvenile hormone metabolism, reproduction and stress response in Drosophila adults with genetic ablation of the corpus allatum cells. Insect Biochem Mol Biol. 2010;40:891–897. doi: 10.1016/j.ibmb.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 68.Haslam RJ, Koide HB, Hemmings BA. Pleckstrin domain homology. Nature. 1993;363:309–310. doi: 10.1038/363309b0. [DOI] [PubMed] [Google Scholar]
- 69.Musacchio A, Gibson T, Rice P, Thompson J, Saraste M. The PH domain: a common piece in the structural patchwork of signalling proteins. Trends Biochem Sci. 1993;18:343–348. doi: 10.1016/0968-0004(93)90071-t. [DOI] [PubMed] [Google Scholar]
- 70.Ingley E, Hemmings BA. Pleckstrin homology (PH) domains in signal transduction. J Cell Biochem. 1994;56:436–443. doi: 10.1002/jcb.240560403. [DOI] [PubMed] [Google Scholar]
- 71.Ono Y, Fujii T, Igarashi K, Kuno T, Tanaka C, Kikkawa U, Nishizuka Y. Phorbol ester binding to protein kinase C requires a cysteine-rich zinc-finger-like sequence. Proc Natl Acad Sci USA. 1989;86:4868–4871. doi: 10.1073/pnas.86.13.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ha CH, Jin ZG. Protein kinase D1, a new molecular player in VEGF signaling and angiogenesis. Mol Cells. 2009;28:1–5. doi: 10.1007/s10059-009-0109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Park JE, Kim YI, Yi AK. Protein kinase D1 is essential for MyD88-dependent TLR signaling pathway. J Immunol. 2009;182:6316–6327. doi: 10.4049/jimmunol.0804239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gilon P, Henquin JC. Mechanisms and physiological significance of the cholinergic control of pancreatic beta-cell function. Endocr Rev. 2001;22:565–604. doi: 10.1210/edrv.22.5.0440. [DOI] [PubMed] [Google Scholar]
- 75.Mitrani P, Srinivasan M, Dodds C, Patel MS. Role of the autonomic nervous system in the development of hyperinsulinemia by high-carbohydrate formula feeding to neonatal rats. Am J Physiol Endocrinol Metab. 2007;292:E1069–E1078. doi: 10.1152/ajpendo.00477.2006. [DOI] [PubMed] [Google Scholar]
- 76.Anichini E, Zamperini A, Chevanne M, Caldini R, Pucci M, Fibbi G, Del Rosso M. Interaction of urokinase-type plasminogen activator with its receptor rapidly induces activation of glucose transporters. Biochemistry. 1997;36:3076–3083. doi: 10.1021/bi9619379. [DOI] [PubMed] [Google Scholar]
- 77.Fibbi G, Caldini R, Chevanne M, Pucci M, Schiavone N, Morbidelli L, Parenti A, Granger HJ, Del Rosso M, Ziche M. Urokinase-dependent angiogenesis in vitro and diacylglycerol production are blocked by antisense oligonucleotides against the urokinase receptor. Lab Invest. 1998;78:1109–1119. [PubMed] [Google Scholar]
- 78.Sumara G, Formentini I, Collins S, Sumara I, Windak R, Bodenmiller B, Ramracheya R, Caille D, Jiang H, Platt KA, Meda P, Aebersold R, Rorsman P, Ricci R. Regulation of PKD by the MAPK p38delta in insulin secretion and glucose homeostasis. Cell. 2009;136:235–248. doi: 10.1016/j.cell.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Feng H, Ren M, Chen L, Rubin CS. Properties, regulation, and in vivo functions of a novel protein kinase D: Caenorhabditis elegans DKF-2 links diacylglycerol second messenger to the regulation of stress responses and life span. J Biol Chem. 2007;282:31273–31288. doi: 10.1074/jbc.M701532200. [DOI] [PubMed] [Google Scholar]
- 80.Kleyn PW, Fan W, Kovats SG, Lee JJ, Pulido JC, Wu Y, Berkemeier LR, Misumi DJ, Holmgren L, Charlat O, Woolf EA, Tayber O, Brody T, Shu P, Hawkins F, Kennedy B, Baldini L, Ebeling C, Alperin GD, Deeds J, Lakey ND, Culpepper J, Chen H, Glucksmann-Kuis MA, Carlson GA, Duyk GM, Moore KJ. Identification and characterization of the mouse obesity gene tubby: a member of a novel gene family. Cell. 1996;85:281–290. doi: 10.1016/s0092-8674(00)81104-6. [DOI] [PubMed] [Google Scholar]
- 81.Boggon TJ, Shan WS, Santagata S, Myers SC, Shapiro L. Implication of tubby proteins as transcription factors by structure-based functional analysis. Science. 1999;286(5447):2119–2125. doi: 10.1126/science.286.5447.2119. [DOI] [PubMed] [Google Scholar]
- 82.Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 83.Mukhopadhyay A, Deplancke B, Walhout AJ, Tissenbaum HA. C. elegans tubby regulates life span and fat storage by two independent mechanisms. Cell Metab. 2005;2:35–42. doi: 10.1016/j.cmet.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 84.Mukhopadhyay A, Pan X, Lambright DG, Tissenbaum HA. An endocytic pathway as a target of tubby for regulation of fat storage. EMBO Rep. 2007;8:931–938. doi: 10.1038/sj.embor.7401055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Popovic D, Akutsu M, Novak I, Harper JW, Behrends C, Dikic I. Rab GTPase-activating proteins in autophagy: regulation of endocytic and autophagy pathways by direct binding to human ATG8 modifiers. Mol Cell Biol. 2012;32:1733–1744. doi: 10.1128/MCB.06717-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bains M, Zaegel V, Mize-Berge J, Heidenreich KA. IGF-I stimulates Rab7-RILP interaction during neuronal autophagy. Neurosci Lett. 2011;488:112–117. doi: 10.1016/j.neulet.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Harbison ST, Yamamoto AH, Fanara JJ, Norga KK, Mackay TF. Quantitative trait loci affecting starvation resistance in Drosophila melanogaster . Genetics. 2004;166(4):1807–1823. doi: 10.1534/genetics.166.4.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Haecker A, Qi D, Lilja T, Moussian B, Andrioli LP, Luschnig S, Mannervik M. Drosophila brakeless interacts with atrophin and is required for tailless-mediated transcriptional repression in early embryos. PLoS Biol. 2007;5:e145. doi: 10.1371/journal.pbio.0050145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang L, Charroux B, Kerridge S, Tsai CC. Atrophin recruits HDAC1/2 and G9a to modify histone H3K9 and to determine cell fates. EMBO Rep. 2008;9:555–562. doi: 10.1038/embor.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Martin-Blanco E, Roch F, Noll E, Baonza A, Duffy JB, Perrimon N. A temporal switch in DER signaling controls the specification and differentiation of veins and interveins in the Drosophila wing. Development. 1999;126:5739–5747. doi: 10.1242/dev.126.24.5739. [DOI] [PubMed] [Google Scholar]
- 91.Charroux B, Freeman M, Kerridge S, Baonza A. Atrophin contributes to the negative regulation of epidermal growth factor receptor signaling in Drosophila. Dev Biol. 2006;291:278–290. doi: 10.1016/j.ydbio.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 92.Kankel MW, Duncan DM, Duncan I. A screen for genes that interact with the Drosophila pair-rule segmentation gene fushi tarazu. Genetics. 2004;168:161–180. doi: 10.1534/genetics.104.027250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Meng X, Kondo M, Morino K, Fuke T, Obata T, Yoshizaki T, Ugi S, Nishio Y, Maeda S, Araki E, Kashiwagi A, Maegawa H. Transcription factor AP-2β: a negative regulator of IRS-1 gene expression. Biochem Biophys Res Commun. 2010;392:526–532. doi: 10.1016/j.bbrc.2010.01.056. [DOI] [PubMed] [Google Scholar]
- 94.Wenke AK, Bosserhoff AK. Roles of AP-2 transcription factors in the regulation of cartilage and skeletal development. FEBS J. 2010;277:894–902. doi: 10.1111/j.1742-4658.2009.07509.x. [DOI] [PubMed] [Google Scholar]
- 95.Eckert D, Buhl S, Weber S, Jager R, Schorle H. The AP-2 family of transcription factors. Genome Biol. 2005;6:246. doi: 10.1186/gb-2005-6-13-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dutta S, Dawid IB. Kctd15 inhibits neural crest formation by attenuating Wnt/β-catenin signaling output. Development. 2010;137:3013–3018. doi: 10.1242/dev.047548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pospisilik JA, Schramek D, Schnidar H, Cronin SJ, Nehme NT, Zhang X, Knauf C, Cani PD, Aumayr K, Todoric J, Bayer M, Haschemi A, Puviindran V, Tar K, Orthofer M, Neely GG, Dietzl G, Manoukian A, Funovics M, Prager G, Wagner O, Ferrandon D, Aberger F, Hui CC, Esterbauer H, Penninger JM. Drosophila genome-wide obesity screen reveals hedgehog as a determinant of brown versus white adipose cell fate. Cell. 2010;140(1):148–160. doi: 10.1016/j.cell.2009.12.027. [DOI] [PubMed] [Google Scholar]
- 98.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M, Burgess S, McDaniel L, Stimpson E, Spriggs F, Williams J, Neurath K, Ioime N, Agee M, Voss E, Furtak K, Renzulli R, Aanensen N, Carrolla S, Bickelhaupt E, Lazovatsky Y, DaSilva A, Zhong J, Stanyon CA, Finley RL, Jr, White KP, Braverman M, Jarvie T, Gold S, Leach M, Knight J, Shimkets RA, McKenna MP, Chant J, Rothberg JM. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 99.Tomancak P, Berman BP, Beaton A, Weiszmann R, Kwan E, Hartenstein V, Celniker SE, Rubin GM. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2007;8:R145. doi: 10.1186/gb-2007-8-7-r145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tomancak P, Beaton A, Weiszmann R, Kwan E, Shu S, Lewis SE, Richards S, Ashburner M, Hartenstein V, Celniker SE, Rubin GM. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2002;3:R88. doi: 10.1186/gb-2002-3-12-research0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ueki K, Yamauchi T, Tamemoto H, Tobe K, Yamamoto-Honda R, Kaburagi Y, Akanuma Y, Yazaki Y, Aizawa S, Nagai R, Kadowaki T. Restored insulin-sensitivity in IRS-1-deficient mice treated by adenovirus-mediated gene therapy. J Clin Invest. 2000;105:1437–1445. doi: 10.1172/JCI7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Eloranta JJ, Hurst HC. Transcription factor AP-2 interacts with the SUMO-conjugating enzyme UBC9 and is sumolated in vivo. J Biol Chem. 2002;277:30798–30804. doi: 10.1074/jbc.M202780200. [DOI] [PubMed] [Google Scholar]
- 103.Bayon Y, Trinidad AG, de la Puerta ML, Del Carmen Rodriguez M, Bogetz J, Rojas A, De Pereda JM, Rahmouni S, Williams S, Matsuzawa S, Reed JC, Crespo MS, Mustelin T, Alonso A. KCTD5, a putative substrate adaptor for cullin3 ubiquitin ligases. FEBS J. 2008;275:3900–3910. doi: 10.1111/j.1742-4658.2008.06537.x. [DOI] [PubMed] [Google Scholar]
- 104.Canettieri G, Di Marcotullio L, Greco A, Coni S, Antonucci L, Infante P, Pietrosanti L, De Smaele E, Ferretti E, Miele E, Pelloni M, De Simone G, Pedone EM, Gallinari P, Giorgi A, Steinkuhler C, Vitagliano L, Pedone C, Schinin ME, Screpanti I, Gulino A. Histone deacetylase and Cullin3-REN(KCTD11) ubiquitin ligase interplay regulates Hedgehog signalling through Gli acetylation. Nat Cell Biol. 2010;12:132–142. doi: 10.1038/ncb2013. [DOI] [PubMed] [Google Scholar]
- 105.Correale S, Pirone L, Di Marcotullio L, De Smaele E, Greco A, Mazza D, Moretti M, Alterio V, Vitagliano L, Di Gaetano S, Gulino A, Pedone EM. Molecular organization of the cullin E3 ligase adaptor KCTD11. Biochimie. 2011;93:715–724. doi: 10.1016/j.biochi.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 106.Ren J, Gao X, Jin C, Zhu M, Wang X, Shaw A, Wen L, Yao X, Xue Y. Systematic study of protein sumoylation: development of a site-specific predictor of SUMOsp 2.0. Proteomics. 2009;9:3409–3412. doi: 10.1002/pmic.200800646. [DOI] [PubMed] [Google Scholar]
- 107.Xue Y, Zhou F, Fu C, Xu Y, Yao X. SUMOsp: a web server for sumoylation site prediction. Nucleic Acids Res. 2006;34:W254–W257. doi: 10.1093/nar/gkl207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ding X, Luo C, Zhou J, Zhong Y, Hu X, Zhou F, Ren K, Gan L, He A, Zhu J, Gao X, Zhang J. The interaction of KCTD1 with transcription factor AP-2alpha inhibits its transactivation. J Cell Biochem. 2009;106:285–295. doi: 10.1002/jcb.22002. [DOI] [PubMed] [Google Scholar]
- 109.Kuba M, Higure Y, Susaki H, Hayato R, Kuba K. Bidirectional Ca2+ coupling of mitochondria with the endoplasmic reticulum and regulation of multimodal Ca2+ entries in rat brown adipocytes. Am J Physiol Cell Physiol. 2007;292:C896–C908. doi: 10.1152/ajpcell.00649.2005. [DOI] [PubMed] [Google Scholar]
- 110.Decuypere J-P, Monaco G, Bultynck G, Missiaen L, De Smedt H, Parys JB. The IP3 receptor–mitochondria connection in apoptosis and autophagy. Biochim Biophys Acta. 2011;1813:1003–1013. doi: 10.1016/j.bbamcr.2010.11.023. [DOI] [PubMed] [Google Scholar]
- 111.Wadhwa R, Taira K, Kaul SC. An Hsp70 family chaperone, mortalin/mthsp70/PBP74/Grp75: what, when, and where? Cell Stress Chaperones. 2002;7:309–316. doi: 10.1379/1466-1268(2002)007<0309:ahfcmm>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Szabadkai G, Bianchi K, Varnai P, De Stefani D, Wieckowski MR, Cavagna D, Nagy AI, Balla T, Rizzuto R. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J Cell Biol. 2006;175:901–911. doi: 10.1083/jcb.200608073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Espenshade P, Gimeno RE, Holzmacher E, Teung P, Kaiser CA. Yeast SEC16 gene encodes a multidomain vesicle coat protein that interacts with Sec23p. J Cell Biol. 1995;131:311–324. doi: 10.1083/jcb.131.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Miller EA, Barlowe C. Regulation of coat assembly—sorting things out at the ER. Curr Opin Cell Biol. 2010;22:447–453. doi: 10.1016/j.ceb.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ivan V, de Voer G, Xanthakis D, Spoorendonk KM, Kondylis V, Rabouille C. Drosophila Sec16 mediates the biogenesis of tER sites upstream of Sar1 through an arginine-rich motif. Mol Biol Cell. 2008;19:4352–4365. doi: 10.1091/mbc.E08-03-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Supek F, Madden DT, Hamamoto S, Orci L, Schekman R. Sec16p potentiates the action of COPII proteins to bud transport vesicles. J Cell Biol. 2002;158:1029–1038. doi: 10.1083/jcb.200207053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gimeno RE, Espenshade P, Kaiser CA. COPII coat subunit interactions: Sec24p and Sec23p bind to adjacent regions of Sec16p. Mol Biol Cell. 1996;7:1815–1823. doi: 10.1091/mbc.7.11.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gimeno RE, Espenshade P, Kaiser CA. SED4 encodes a yeast endoplasmic reticulum protein that binds Sec16p and participates in vesicle formation. J Cell Biol. 1995;131:325–338. doi: 10.1083/jcb.131.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shaywitz DA, Espenshade PJ, Gimeno RE, Kaiser CA. COPII subunit interactions in the assembly of the vesicle coat. J Biol Chem. 1997;272:25413–25416. doi: 10.1074/jbc.272.41.25413. [DOI] [PubMed] [Google Scholar]
- 120.Zaltsman Y, Shachnai L, Yivgi-Ohana N, Schwarz M, Maryanovich M, Houtkooper RH, Vaz FM, De Leonardis F, Fiermonte G, Palmieri F, Gillissen B, Daniel PT, Jimenez E, Walsh S, Koehler CM, Roy SS, Walter L, Hajnoczky G, Gross A. MTCH2/MIMP is a major facilitator of tBID recruitment to mitochondria. Nat Cell Biol. 2010;12:553–562. doi: 10.1038/ncb2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Grinberg M, Schwarz M, Zaltsman Y, Eini T, Niv H, Pietrokovski S, Gross A. Mitochondrial carrier homolog 2 is a target of tBID in cells signaled to die by tumor necrosis factor alpha. Mol Cell Biol. 2005;25:4579–4590. doi: 10.1128/MCB.25.11.4579-4590.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Endo T, Yamano K. Transport of proteins across or into the mitochondrial outer membrane. Biochim Biophys Acta. 2010;1803:706–714. doi: 10.1016/j.bbamcr.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 123.Ryan MT, Wagner R, Pfanner N. The transport machinery for the import of preproteins across the outer mitochondrial membrane. Int J Biochem Cell Biol. 2000;32:13–21. doi: 10.1016/s1357-2725(99)00114-4. [DOI] [PubMed] [Google Scholar]
- 124.Pusnik M, Charriere F, Maser P, Waller RF, Dagley MJ, Lithgow T, Schneider A. The single mitochondrial porin of Trypanosoma brucei is the main metabolite transporter in the outer mitochondrial membrane. Mol Biol Evol. 2009;26:671–680. doi: 10.1093/molbev/msn288. [DOI] [PubMed] [Google Scholar]
- 125.Salinas T, Duchene AM, Delage L, Nilsson S, Glaser E, Zaepfel M, Marechal-Drouard L. The voltage-dependent anion channel, a major component of the tRNA import machinery in plant mitochondria. Proc Natl Acad Sci USA. 2006;103:18362–18367. doi: 10.1073/pnas.0606449103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schleiff E, Silvius JR, Shore GC. Direct membrane insertion of voltage-dependent anion-selective channel protein catalyzed by mitochondrial Tom20. J Cell Biol. 1999;145:973–978. doi: 10.1083/jcb.145.5.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kmita H, Antos N, Wojtkowska M, Hryniewiecka L. Processes underlying the upregulation of Tom proteins in S. cerevisiae mitochondria depleted of the VDAC channel. J Bioenerg Biomembr. 2004;36:187–193. doi: 10.1023/b:jobb.0000023622.66554.a6. [DOI] [PubMed] [Google Scholar]
- 128.Cogliati S, Scorrano L. A BID on mitochondria with MTCH2. Cell Res. 2010;20:863–865. doi: 10.1038/cr.2010.100. [DOI] [PubMed] [Google Scholar]
- 129.Cho SY, Lee JH, Bae HD, Jeong EM, Jang GY, Kim CW, Shin DM, Jeon JH, Kim IG. Transglutaminase 2 inhibits apoptosis induced by calcium-overload through down-regulation of Bax. Expl Mol Med. 2010;42:639–650. doi: 10.3858/emm.2010.42.9.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kaddour-Djebbar I, Choudhary V, Brooks C, Ghazaly T, Lakshmikanthan V, Dong Z, Kumar MV. Specific mitochondrial calcium overload induces mitochondrial fission in prostate cancer cells. Int J Oncol. 2010;36:1437–1444. doi: 10.3892/ijo_00000629. [DOI] [PubMed] [Google Scholar]
- 131.Billing O, Kao G, Naredi P. Mitochondrial function is required for secretion of DAF-28/insulin in C. elegans . PLoS ONE. 2011;6:e14507. doi: 10.1371/journal.pone.0014507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, Akalin A, Schmeier S, Kanamori-Katayama M, Bertin N, Carninci P, Daub CO, Forrest AR, Gough J, Grimmond S, Han JH, Hashimoto T, Hide W, Hofmann O, Kamburov A, Kaur M, Kawaji H, Kubosaki A, Lassmann T, van Nimwegen E, MacPherson CR, Ogawa C, Radovanovic A, Schwartz A, Teasdale RD, Tegnér J, Lenhard B, Teichmann SA, Arakawa T, Ninomiya N, Murakami K, Tagami M, Fukuda S, Imamura K, Kai C, Ishihara R, Kitazume Y, Kawai J, Hume DA, Ideker T, Hayashizaki Y. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rask-Andersen M, Almen MS, Schioth HB. Trends in the exploitation of novel drug targets. Nat Rev Drug Discov. 2011;10:579–590. doi: 10.1038/nrd3478. [DOI] [PubMed] [Google Scholar]