Abstract
Centrins are small, highly conserved members of the EF-hand superfamily of calcium-binding proteins that are found throughout eukaryotes. They play a major role in ensuring the duplication and appropriate functioning of the ciliary basal bodies in ciliated cells. They have also been localised to the centrosome, which is the major microtubule organising centre in animal somatic cells. We describe the identification, cloning and characterisation of centrins in multiple eukaryotic species. Although centrins have been implicated in centriole biogenesis, recent results have indicated that centrosome duplication can, in fact, occur in the absence of centrins. We discuss these data and the non-centrosomal functions that are emerging for the centrins. In particular, we discuss the involvement of centrins in nucleotide excision repair, a process that repairs the DNA lesions that are induced primarily by ultraviolet irradiation. We discuss how centrin may be involved in these diverse processes and contribute to nuclear and cytoplasmic events.
Keywords: Centrosome, Centrin, Centriole, Cilia, DNA repair, Centrosome amplification
Introduction
Centrosomes are small organelles that organise an astral microtubule network during interphase and a bipolar spindle during mitosis. They play important roles in controlling various cell cycle transitions. Their ultrastructure is striking: centrosomes contain two distinct cylinders made up of nine microtubule bundles, the centrioles, within a cloud-like proteinaceous matrix, the pericentriolar material (PCM) (reviewed in [1–3]). The centrioles also serve as basal bodies for cilia and flagella. As a stable centrosome number is important in avoiding chromosome segregation problems and aneuploidy [4], centrosome duplication is tightly co-ordinated with the cell cycle [3, 5]. The same cyclin-CDK activities are required for both the chromosome and the centrosome cycles [6–9]. Licensing of centrosomes for duplication is controlled by the same activities that regulate sister chromatid separation at the metaphase-anaphase transition—the protease activity of separase and the kinase activity of Polo-like kinase 1 (Plk1) [10–13]. While the centrosome is thus highly responsive to the state of the chromosome cycle, recent work indicates that the centrosome itself plays a significant part in cell cycle control, acting as a nexus for intracellular signalling, with certain key components of the centrosome playing additional, non-centrosomal roles. An example of a centrosome component with additional functions is the centrins: small, evolutionarily conserved proteins that play an important role in several cellular activities. This review will examine emerging roles for the centrins in DNA repair and discuss how these activities may be related to the centrosome.
DNA damage and centrosome duplication
Centrosome amplification
The implication of centrosome copy number in the control of accurate cell division is a long-established concept, having initially been postulated by Theodor Boveri at the turn of the last century [14]. A key observation in this regard is that cancer cells often carry aberrant numbers of centrosomes, with aneuploidy and chromosomal instability being highly correlated with the appearance of multiple centrosomes [15–19]. Furthermore, centrosome number aberrations in tumour cells are often associated with structural irregularities such as increased size and/or changes in the PCM [20]. Supernumerary or aberrant centrosomes can cause mitotic abnormalities, such as the formation of multipolar spindles. Such spindle abnormalities could, in turn, result in abnormal chromosome segregation and aneuploidy and thus contribute to transformation [21, 22]. However, the massive degree of aneuploidy likely under such a scenario has meant that other mechanisms have been proposed for how multiple centrosomes might drive genome instability. An emerging model involves the multiple centrosomes generating a transient multipolar state before they cluster together to allow a bipolar spindle, which causes a delay during mitosis that allows inappropriate chromosome attachments and chromosome mis-segregation [4]. Although these models describe different effects on mitosis of multiple centrosomes, they both provide mechanisms by which abnormal centrosome numbers can cause aneuploidy. Thus, the appropriate control of centrosome number also regulates genome stability.
Like the chromosomes, centrosomes are normally duplicated in a semi-conservative manner, once per cycle and during S phase. While not templated to the same extent as replicating DNA by the pre-existing copy, centriole assembly occurs at a location that is specified by the existing centriole [5, 23–25]. However, pre-existing centrioles are not sufficient to restrict centriole duplication to a single daughter per mother. Multiple daughter centrioles can form around a single mother after proteasome inhibition, extended S-phase delay or expression of viral oncoprotein [26, 27]. The overexpression of the key kinase, PLK4/SAK, in unfertilised Drosophila embryos leads to de novo centriole formation, providing further evidence that the mother acts more as a scaffold for duplication activities rather than as a direct template for duplication per se [28]. Similarly, overduplication through multiple daughter centrioles occurs after PLK4 overexpession in human cells [29]. Furthermore, experiments demonstrating that the de novo generation of centrioles occurs after centrosome ablation in both transformed and non-transformed human cells showed that the mother centriole controls centriole number rather than being an absolute requirement for centriole biogenesis [30–32].
While extra centrosomes can arise through defects in cytokinesis, genotoxic stress is another driver of centrosome abnormalities. Centrosome amplification has been described after cells have been subjected to a broad range of DNA-damaging insults including ionising radiation [33–35] and DNA replication stress [36, 37]. Overduplication of the centrosome occurs independently of exogenous genotoxic stress in cells with mutations in DNA repair or checkpoint genes [33, 38–44], cells that express mutant forms of telomerase [45] or viral oncogenes [18, 46–48]. How this amplification happens is not yet fully understood. Although multiple centrosomes are seen in cells that fail in cytokinesis due to altered expression of cell cycle regulators such as BRCA2 or Aurora A [37, 49], DNA-damaging treatments do not generally cause tetraploidisation. Therefore, cytokinesis failure is not a sufficient explanation for how centrosome amplification occurs after genotoxic stress. DNA damage signalling appears to impact on centrosome number through the centrosome duplication pathway.
Mechanisms of DNA damage-responsive centrosome duplication
The appropriate control of centrosome duplication involves at least two elements: the regulation of cyclin-dependent kinase activity [50] and the ‘licensing’ of centrosome duplication [51]. CDK2 is a particular activity that links the chromosome and centrosome cycles, and its activity is necessary for centrosome overduplication that occurs during extended S-phase delay in mammalian cells [6–9] or following overexpression of the human papillomavirus (HPV) type 16 E7 oncoprotein [46]. However, Cdk2 is dispensable for normal centrosome duplication in mouse and chicken cells, presumably due to redundancy with other CDKs [46, 52, 53]. The temporally limited licensing of centrosomes for duplication occurs through centriole disengagement, which occurs late in mitosis. Disengagement is mediated by Plk1 and the separase protease, which is normally activated through anaphase promoting complex/cyclosome activity at the metaphase-anaphase transition [10, 11]. The Scc1 cohesin subunit is the key separase target in allowing centriole separation [13]. A final consideration is the requirement for sufficient time to allow assembly of the licensed centriole under conditions of appropriate CDK activity.
There are three major examples in which centrosome amplification occurs through dysregulation of one or more of these controls. An extended S-phase delay is induced by hydroxyurea (HU) treatment and, in many mammalian cell lines, this treatment leads to the appearance of multiple centrosomes in a Cdk-dependent manner, with Cdk2 being a strong candidate [36, 54]. This overduplication fits well with the previously described requirements, as the centrosomes are licensed for duplication at this stage in the cell cycle, and Cdk2 is activated. A second example is the high level of centrosome amplification that is observed in p53-deficient mice and cells [39]. This is believed to arise from abnormal Cdk2 activity in the absence of p53, along with a p53-independent cell cycle arrest to provide sufficient time for amplification [55]. In support of this model, upregulated Cdk2 activity caused by cyclin E overexpression caused centrosome amplification in p53-deficient mouse cells, but not in wild-type rat or mouse fibroblasts [56, 57]. Furthermore, cyclin E overexpression induced centrosome amplification in human tumour cells, but only in the absence of p53 function [58]. The status of the license was not explored in these experiments, however. The third case of centrosome amplification is that arising after induced DNA damage. Ionising radiation induced multiple spindle poles in mammalian cells that subsequently were found to contain centrosomes [35, 59]. Analysis of the underlying mechanism found that the Atm-Chk1-controlled G2-to-M checkpoint was necessary to allow this centrosome amplification [33, 60], providing the requisite time for reduplication. Irradiation of some human and chicken cell lines caused upregulation of Cdk2 activity and, notably, centrosome amplification was dependent on there being Cdk1 or Cdk2 available [61], fulfilling the next of the key control requirements.
DNA damage-induced centrosome amplification is p53-independent, as it was reported to occur with the same frequency in TP53 −/− human HCT116 cells as in controls [34]. Furthermore, the extent to which DNA damage allows centrosome licensing is not yet clear. Centrosome fragmentation has been described in Drosophila and rodent cells following cell cycle progression after incomplete DNA replication [62, 63] and premature centriole splitting, which may reflect disengagement, has been observed after irradiation of human cell lines [64]. Despite the observation that G2 phase centrosomes can acquire a license after irradiation [65], it is not known what effect irradiation has on separase, and several studies have indicated that the other licensing signal, Plk1, is actually inhibited by DNA damage [66–68]. Clearly, the molecular nature of the signal that transmits the report of DNA damage to the centrosome duplication machinery remains to be fully defined.
Many of the key components of the DNA damage response and the cell cycle apparatus localise to the centrosomes (reviewed in [69–71]. For example, the initial activation of the Cdk1-cyclin B complex that permits entry into mitosis occurs at the centrosome [72]. Centrosomal Chk1 kinase has been described as a regulator of the Cdk1-activating phosphatase, Cdc25B, providing a mechanism linking DNA damage-sensitive kinase signalling to the control of the cell cycle [73, 74]. Further regulation of Chk1 occurs through other centrosome components, specifically through Mcph1 and pericentrin [75]. Interestingly, Cdc25B was found to indirectly interact with centrin 2 and to be involved in the centrosomal recruitment of centrins [76]. These activities and the many other DNA damage-responsive proteins that are found at the centrosome provide support for the idea that the centrosome can act as a macromolecular scaffold at which biochemical signals can be amplified, as well as the eventual target of such signalling. Furthermore, centrosomal proteins frequently have additional roles that link nuclear activities with the centrosome, as we will discuss in the case of centrin.
Identification of centrin
Initial identification of centrin
The striated flagellar root is a massive structure in the green alga, Tetraselmis striata. Addition of high concentrations of calcium to cultures of Tetraselmis causes these roots to contract and pull the plasma membrane inward, as well as flagellar loss [77]. Exploration of the mechanism of this calcium-responsive activity revealed that a single protein band comprised >60 % of the total flagellar root protein and that the electrophoretic mobility of this protein changed significantly in the presence of calcium. Antisera raised to this polypeptide of approximately 20 kDa clearly decorated the flagellar roots [77]. These antibodies also recognised a similar protein in the alga Chlamydomonas reinhardtii, which localised to the proximal ends of the basal bodies at the roots of the flagella and to an extended structure that linked the nucleus to the basal bodies [78]. Furthermore, these antisera also detected the basal bodies in the green flagellate Spermatozopsis similis, which radically change their orientation to allow photoreactive changes in swimming direction in a calcium-dependent manner [79]. An extended analysis of 28 green alga taxa confirmed the conservation of this immunolocalisation throughout green algae [80]. The contractile responses of Chlamydomonas flagella are calcium dependent, and the immunolocalisation of this protein to the contractile structures and to the basal body suggested a functional link to such activity [81, 82]. Later work with inhibitory, specific antibodies confirmed that these contractile responses specifically required the protein [83] and detailed immunoelectron microscopy eventually placed this protein throughout an entire filamentous scaffold linking the nucleus and the basal bodies [84]. Purification and analysis of Chlamydomonas basal bodies revealed that the immunolocalisation detected a calmodulin-like protein homologous to the calcium-binding protein from Tetraselmis striated flagellar roots [85], providing a clear indication that this was likely to be an evolutionarily conserved protein.
Microsequencing of the purified Chlamydomonas basal body protein allowed the generation of a probe to screen a Chlamydomonas cDNA library, which led to the cloning of the gene that encodes this calcium-responsive protein [86]. In this study, the protein was dubbed ‘caltractin’ on the basis of it being a component of a calcium-sensitive contractile fibre system. It was found to be highly homologous to calmodulin and to the previously identified product of budding yeast CDC31, mutations of which lead to problems in spindle pole body duplication [87]. However, an alternative designation, ‘centrin’, was advanced by the Salisbury group [88], and has become more established, so we are using this terminology in our review. Centrins are small, highly conserved members of the EF-hand protein superfamily. Members of the EF-hand superfamily carry distinct helix-loop-helix structures that coordinate calcium. This nomenclature arose from the alphabetical labelling of the six alpha-helices of the prototypic carp parvalbumin, the crystal structure of which revealed an orthogonal arrangement of the EF helices around a calcium-binding loop, resembling a pointing forefinger and extended thumb of a right hand [89, 90]. Centrins contain four EF-hand domains, consistent with the calcium-responsive actions of the structures in which the proteins were originally identified. We will discuss the roles of the centrin EF-hands in Sect. “Centrin structure”, below.
Evolutionary conservation of centrins
Given the functional and structural similarities between the basal body and the centrosome, it was of great interest to address the question of whether centrins might be conserved outside the green algae and localise to centrosomes. On the basis of immunological reactivity, a wide distribution amongst eukaryotic species seemed likely [91]. However, although polyclonal antibodies to centrin recognised the centrosome in several mammalian species, it is unclear whether this was actually centrin or some other reactivity, as proteins of different size were predominantly observed in immunoblot analyses [88, 92, 93]. Nevertheless, cloning of centrin orthologues from mouse [94] and human [95, 96] confirmed the evolutionary conservation of centrins and allowed the generation of immunoreagents that were specific for mammalian centrin. These antibodies demonstrated that centrin was indeed a centrosomal protein in mammals [95, 96].
Consistent with evidence from a range of organisms [97], further analysis using monoclonal antibodies raised against Chlamydomonas centrin but with a robust reactivity to mammalian centrins [83] provided clear evidence for centrin being a general feature of centrosomes and basal bodies in animal cells [98–101]. Localisation of the Saccharomyces cerevisiae orthologue, Cdc31p, to the half-bridge of the analogous structure in yeast, the spindle pole body, demonstrated that centrins are found in analogous structures throughout eukaryotes [102]. Focussing on the mammalian centrosome, light and electron microscopy demonstrated that centrin is localised both to the distal ends of the centrioles and to a region between them [101]; a similar localisation to the distal lumen of the basal bodies was seen by electron microscopy of the flagellar apparatus in Chlamydomonas [84]. On the basis of this localisation, the sensitivity of mammalian centrosomes to divalent cations [103] appeared to suggest a mechanical similarity to the centrin-dependent contractile response in algae, although no centrin fibres have been described to date in mammalian cells.
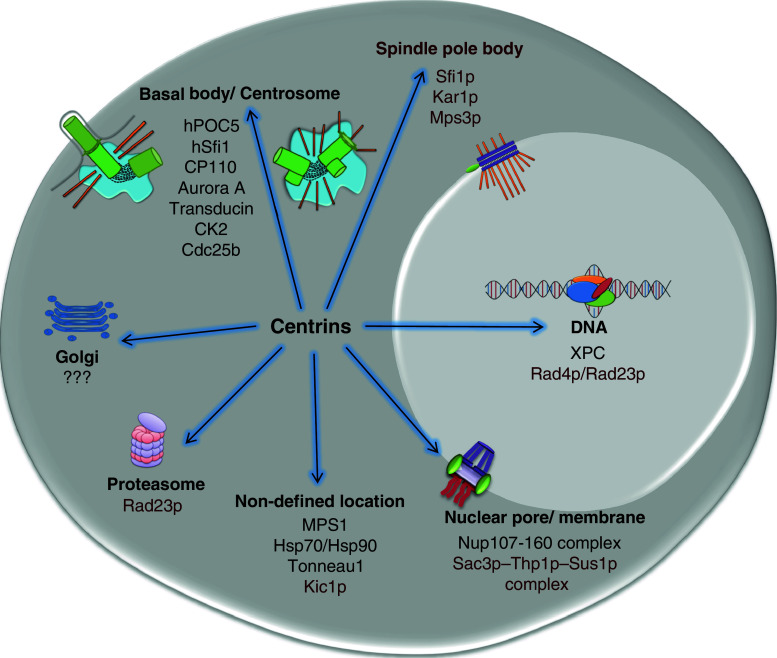
While these data emphasise the centrosomal localisation of centrin, it is important to note that the vast majority of cellular centrin is not incorporated into centrosomes, at least in human lymphoblasts [101]. The observation of a calcium-responsive pericentriolar lattice-like structure in rat kangaroo cells indicated an additional localisation of centrins in mammalian cells [104], although the implication of centrin in nuclear functions, proteasome activities and the nuclear pore complex provides further hints of noncentrosomal roles of centrin [105–108]. The localisations and known interactors of the centrins are shown in Fig. 1.
Fig. 1.
The localisation and interactions of centrins. Cartoon shows centrin interactions in higher eukaryotes and in yeast (in brown), indicating the organelles or structures where centrins were found to associate with their respective partners, as discussed in the text. The references describing these interactions are as follow: hSfi1 [221], hPOC5 [219], CP110 [174], Cdc25b [76], XPC [105], Aurora A [145], MPS1 [115], CK2 [222], Transducin [147], Nup107-160 complex [108], Hsp70/Hsp90 [100], Kic1p [153], Tonneau1 [223], Rad4 [106], Rad23p [106], Sfi1p [142], Kar1p [151], Mps3p [152], Sac3p–Thp1p–Sus1p complex [107], golgi localisation [160]. Note that the elements of the idealised cell shown here are not to scale
The centriolar localisation pattern of centrin has made fluorescently tagged centrins an attractive tool to study centriole duplication in living mammalian cells [31, 109–112]. However, the overexpression of fluorescently tagged centrin can lead to basal body anomalies and the presence of extra non-centrosomal centrin clumps that do not co-localise with γ-tubulin or other centrosomal markers in unicellular organisms [113, 114]. In HeLa and CHO cells, overexpression of centrin 2 caused overduplication of centrioles during extended S-phase arrest, but this effect was not observed in U2OS or RPE1 cells, suggesting some cell-type specificities in the regulation of centriole assembly [115]. Several groups have reported the aggregation of centrin into subcentriolar foci early during centriole overduplication induced by HU treatment or during de novo centrosome formation [31, 110, 116, 117]. However, as discussed below, Plk4- and HU-induced centriole overduplication can occur in the absence of centrin [29, 118], so it is not clear that these centrin aggregates are necessary for centriole duplication.
Centrin genes
The localisation of centrin to centrioles and basal bodies throughout Eukarya and biochemical similarities of the protein from different species suggested that centrin-coding genes would be evolutionarily conserved. Following on from the previously mentioned cloning of the Chlamydomonas centrin and the mammalian orthologues, sufficient data have accumulated on centrins to enable a detailed phylogenetic tree to be established for the family [119]. In fact, a recent evolutionary analysis of centrosomal proteins assigned centrin 2 to a core inventory of 14 centriolar proteins suggested to have been present in the last common ancestor of eukaryotes [120]. Two principal centrin subfamilies have been defined on the basis of comparative sequence analysis, although all are related to calmodulin, as was noted in the initial cloning of centrin from Chlamydomonas [86]. Members of one subfamily are related to budding yeast CDC31, and members of the other are more homologous to the Chlamydomonas centrin [121, 122].
While genome analysis of the unicellular ciliated protozoan Paramecium tetraurelis revealed a centrin family of more than 30 members, several of which are highly divergent from the two principal subfamilies mentioned above [114], only four centrin isoforms have been described to date in mammals. Centrins 1, 2 and 4 are closely related to one another and to the Chlamydomonas centrin [95, 96, 123]. At the amino acid level, human centrins 1 and 2 show 83 % sequence identity in comparison to 52 % observed between centrin 2 and centrin 3. This may suggest some evolutionary constraints on the degree of conservation in the various centrin subdomains.
Cetn2 is ubiquitously expressed, although Cetn1 has been reported to have a more restricted expression pattern, being limited to male germ cells, neurons and ciliated cells [124, 125]. Cetn1 was proposed to have arisen from a retroposition of Cetn2, as it shows certain diagnostic features of a retroposon: Cetn1 lacks introns and internal stop codons, and is bordered by a pair of direct repeats (Hart et al. 1999). Interestingly, Cetn1 is entirely absent from the chicken and zebrafinch genomes, from which it was concluded that its retroposition occurred after the bird-mammal split [118]. Cetn3 is of the CDC31 subfamily and is ubiquitously expressed [121]. Cetn4 is a centrin 2-related gene, initially identified in mouse, with a tissue-restricted expression pattern that has suggested its being limited to ciliated cells [123]. Cetn4 is a pseudogene in human cells and thus is not expressed, while it appears to show unrestricted expression in chicken [118]. Full-length versions of these centrin isoforms all associate with centrioles, but to differing extents that may reflect changing centrin activities during centriole duplication and ciliogenesis [121, 123, 126].
Centrin structure
The involvement of centrin in ion-mediated contractile responses clearly implicates the calcium-binding components of the molecule in regulating its mechanism of action. Our current model for centrin is based on a structure in which its four EF-hand motifs are arranged in pairs of EF-hands, separated by a linker region [127, 128]. Figure 2 illustrates the functional domains of centrins, along with key regulatory residues that are mentioned in more detail below. The mechanistically important interactions of the EF-hand subdomains with calcium and with centrin’s binding partners are complex and appear to vary between species (reviewed in [129]).
Fig. 2.
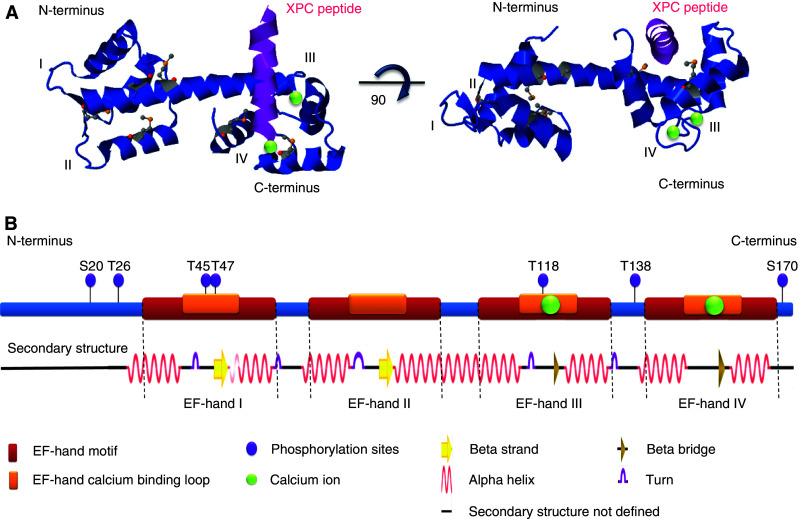
Centrin structure and domains. a Ribbon diagram of centrin 2 based on the structure described for the protein without the first 24 amino acids of the N-terminus [128]. The 3D representation of the structure was assembled and adapted using Jmol version 12.0.41 (Research Collaboratory for Structural Bioinformatics (RCSB); http://home.rcsb.org/). Calcium ions are represented as green spheres and the EF-hands are marked by Roman numerals. b Two-dimensional representation of the principal domains of the 172 amino acid centrin 2 protein, along with the corresponding secondary structure predicted using the algorithm ‘Define Secondary Structure of Proteins’ in the RCSB Protein Data Bank. Regulatory elements that have been described as phosphorylation sites are indicated and were based on the following references: S20, T26 [224]; T45, T47, T118 [115]; T138 [222]; S170 [145, 146]
A general view of how centrins work is that the binding of calcium facilitates target peptide recognition by the protein, with low-affinity peptide binding sites becoming activated in the presence of calcium, as in the case of Scherffelia dubia centrin [130]. In Chlamydomonas centrin, while both the N-terminal and C-terminal domains can bind calcium, the N-terminal EF-hand pair has a greater affinity for calcium and the C-terminal for a Kar1 peptide [127, 131, 132], consistent with a regulatory function in the N-terminal domain [133]. In Leishmania donovani, the C-terminal domain can bind calcium more efficiently than the N-terminal, although both are capable of doing so [134]. Work with the yeast centrin orthologue, Cdc31p, has indicated that the C-terminal EF-hand pair is required for binding of a peptide target, with N- and C-terminal EF-hands implicated in calcium binding [135]. The C-terminal domains of human centrin 1 and centrin 2 carry the high-affinity calcium-binding sites, with only low affinity in the N-terminal EF-hand domain [136–138], while human centrin 3 has three robust calcium binding sites with the one sited in the N-terminal domain also being capable of binding magnesium [139]. The calcium-regulatory and target-binding sites therefore appear to be sited mainly in the C-terminal pair of EF-hands in human centrins, with the N-terminal pair responding to the occupancy status of the C-terminus.
In vitro analysis showed that centrins from algae, yeast and humans form aggregates. The S. dubia centrin, in particular, formed extended filamentous structures in a calcium-dependent manner [140]. Interactors additional to centrin itself described to date include the xeroderma pigmentosum group C protein [105], the yeast spindle pole body proteins Kar1p [141] and Sfi1p [142]. At the very N-terminus of the molecule is a disordered region of some 20 amino acids, which is not well conserved between different centrins, but whose presence distinguishes centrins from closely related EF-hand proteins [86, 128]. This disordered domain is important in the self-aggregation of centrin [138, 143], although not for its association in budding yeast with its partner, Sfi1p, in the assembly of the mitotic apparatus [142, 144].
Centrins are also subject to extensive regulation by post-translational modification. The very initial identification of centrins in Tetraselmis noted that the protein was phosphorylated [77]. Two-dimensional electrophoresis to resolve human centrin allowed the detection of at least ten differently migrating forms, suggesting that centrin undergoes several posttranslational modifications [101]. In addition, centrin was also found to be highly phosphorylated in some human tumours with supernumerary centrosomes [16]. Aurora A and PKA can phosphorylate centrin 2 at serine 170, a modification that regulates the stability of centrin. Additionally, this phosphorylation has been suggested to promote centriole separation in G2 and to be required for Aurora A-induced centrosome amplification [145, 146]. CK2 was also reported to phosphorylate centrins at threonine 138, which regulates their ability to bind to a retinal G-protein complex and ciliary microtubules [147]. Three additional centrin threonine sites (Thr45, Thr47 and Thr118) were described as target sites for the MPS1 kinase in vitro [115]. Each of these sites is necessary for the MPS1-dependent centriole overduplication that results from overexpression of centrin 2 to occur efficiently in HeLa cells. Furthermore, expression of centrin 2 with phosphomimetic mutations of these sites caused more overduplication than the wild-type protein [115]. These results suggested that centrin 2 activity is regulated, to some extent, through these phosphorylation sites. Further regulation of centrin activity is achieved through SUMOylation by SUMO2/3, which determines the subcellular localisation of the protein and which requires the activity of the polycomb protein 2 as an E3 ligase [148].
Centrosomal effects of centrin deficiency
Centrin requirement for basal body/spindle pole body duplication
Mutational analysis and reverse genetics have demonstrated a requirement for centrin in centriolar/basal body functions in a range of organisms. The first such analysis of one major subfamily of centrins identified a Chlamydomonas strain with a mutation in the centrin gene (vfl2), which had increased rates of basal body mis-segregation due to defective nucleus-basal body connections. This mutation also resulted in a variable number of flagella and the absence of both distal striated and stellate fibres [78, 149]. Furthermore, RNAi-mediated depletion of CrCentrin resulted in a strong reduction in the number of basal bodies per cell as well as non-flagellar basal bodies, suggesting a defect in their duplication and/or problems in flagellar assembly or in basal body maturation [113]. Looking at the other major centrin subfamily, CDC31, the prototypic Cetn3 orthologue in Saccharomyces cerevisiae, is an essential gene that is required during the initial duplication of the spindle pole body (SPB), the main microtubule-organising centre in yeast cells that is functionally analogous to the centrosome in animal cells [87, 150]. Cdc31p localises to the half-bridge of the SPB where it forms filaments with Sfi1p and preserves the integrity of this structure [102, 142, 144]. Cdc31p recruitment to the SPB was shown to depend on other SPB proteins, including Kar1p and Mps3p [141, 151, 152], but is required for the activity of its interactor, Kic1p kinase, to ensure proper cell integrity and morphology [153]. Summarising several studies, cdc31 mutations cause defective SPB biogenesis, leading to cell cycle arrest and large-budded cell morphology [87, 153–155]. The fission yeast centrin 3 orthologue, Cdc31p, is also required for SPB function, indicating conservation of centrin functions in yeast [156].
Looking through the evolutionarily diverse range of reverse genetic analyses of centrin, the general impact has been most pronounced in ciliated cells. Centrin knockdown in the water fern, Marsilea vestita, induced spermiogenesis arrest before the assembly of blepharoplasts and basal bodies [157]. However, when cell cycle arrest was induced in this gametophyte by drug treatments (including hydroxyurea), centrin translation and accumulation was not affected, while blepharoplast and the motile apparatus formation was inhibited, indicating that centrin translation was not sufficient for basal body formation de novo [158].
Centrin deletion in the pathogenic trypansosome, Leishmania donovani, resulted in a G2/M cell cycle arrest with defects in basal body duplication and failure in cell division exclusively during one amastigote stage of the parasite’s development, but not during the promastigote [159]. Even though this phenotype was frequently associated with cell death, this observation suggested the possibility of stage- or cell type-specific functions of centrin in Leishmania. Five centrin isoforms have been found in the parasitic protozoan, Trypanosoma brucei [160]. TbCentrin 1 was shown to localise solely to the basal body, whereas TbCentrin 2 and TbCentrin 4 (also described as TbCentrin 1) were also detected at a bilobed structure connected to the Golgi complex [160–162]. RNAi depletion of TbCentrin1 or TbCentrin2 inhibits basal body duplication, with TbCentrin2 ablation also preventing the duplication or segregation of the Golgi complex and other organelles. Since nuclear division, but not cytokinesis, continued in these cells, multi-nucleated cells originated in the absence of cell division [160]. On the other hand, TbCentrin 4 knockdown had no detectable impact on the duplication of basal bodies or other organelles, although it led to the uncoupling of nuclear and cell division and led to the production of anuclear daughter cells, zoids, or enlarged cells with multiple organelles and nuclei [161, 162]. Together, these data indicate discrete roles for individual trypanosome centrins in cell division.
Tetrahymena thermophila centrins localise to basal bodies in the cortical rows, the oral apparatus, and to ciliary rootlets and their accessory structures [163, 164]. Immunoelectron microscopy demonstrated that TtCEN1, which is the closest homologue to human centrin 2 of the centrins described from Tetrahymena, is specifically recruited to the transition zone, to the basal body midzone and to its base [165]. The deletion of TtCEN1 leads to severe deficiencies in basal body stability and maintenance, together with mis-orientated probasal body assembly, and thus is lethal [164, 166]. Rescue experiments with truncated TtCEN1 demonstrated that the N- or C-terminal domains were individually incapable of rescuing viability, but that mutations of the EF-hands within either domain were compatible with survival. This work concluded in a model in which the C-terminal domain of centrin controls its localisation to basal bodies and the N-terminal domain of centrin, the separation and orientation of basal bodies [166].
PtCen2a/b and PtCen3a/b are the Paramecium centrin HsCETN2 and HsCETN3 homologues, respectively [114]. Multiple atypical centrins also exist in Paramecium, and play specialised and nonredundant roles in an extended contractile array, the infraciliary lattice [167, 168]. Both PtCen2ap and PtCen3ap localise to basal bodies, but show distinct locations within these structures, with PtCen2ap in the basal body proper and PtCen3ap on fibrous structures linking basal bodies. Knockdown experiments showed that even though these centrins do not have completely overlapping roles, both are dispensable for basal body assembly but necessary for appropriate basal body orientation and positioning during the duplication cycle [114]. The mis-orientation/positioning of the basal bodies seen in the absence of PtCen2ap and PtCen3ap eventually inhibited basal body duplication [114], suggesting that the inability to position or insert a basal body in the cortex for ciliogenesis may prevent further assembly of these structures, providing an explanation for the severity of the phenotypes seen in the absence of centrin.
Centrin in vertebrate cells
While the extensive work in lower eukaryotes described above provides clear evidence for the importance of centrins in basal body assembly and/or function throughout evolution, their roles in vertebrate cells are less clear. Partial centrin 3 depletion in U2OS cells led to the loss of centrosomal anchorage of microtubules and the disruption of their radial organisation, possibly by affecting the centrosomal recruitment of ninein [169]. A striking phenotype was observed after RNAi depletion of centrin 2 in HeLa cells, namely the inhibition of centriole biogenesis [170]. Cells with at least two centrioles continued to undergo centrosome separation and progress through the cell cycle. Eventually, this resulted in only one or no centrioles being available, so cells assembled abnormal mitotic spindles, cell division was blocked and the number of multinucleated cells increased [170]. However, in contrast to the results seen in this study, depletion of CETN2 using a similar siRNA sequence did not interfere with the recruitment of HsSAS-6 to the base of the mother centriole or the initiation of procentriole biogenesis [171]. In hTERT-RPE1 cells, the individual depletion of several centrosomal proteins led to a p53-dependent cell cycle arrest in G1 phase. CETN2 siRNA resulted in a strong accumulation of G1 cells, suggesting that centrin 2 is required for the normal cell cycle progression of non-transformed human cells [172]. However, centrin 2 and 3 depletion by siRNA in human U2OS cells did not block Plk4-induced procentriole biogenesis, which occurred with normal kinetics. In addition, no defects were detected in the typical recruitment of key duplication components to the nascent procentrioles or in their subsequent behaviour [29]. Another group repeated the siRNA analysis of CETN2 and saw the expected decrease of centrin 2-labelled centrioles, but no major effect on HsSAS-6 recruitment or the number of γ-tubulin foci per cell. However, these authors did note that centrin 2 depletion delayed CP110 incorporation into procentrioles at the beginning of S-phase, without any evidence of an S-phase delay. In addition, although centrin 2-depleted cells were able to assemble multiple centrioles upon the overexpression of a nondegradable form of the centrosomal MPS1 kinase, a considerable fraction of these failed to recruit y-tubulin [115]. Our own analysis used gene targeting in the chicken DT40 cell line to disrupt all centrin genes and observed no impact on centrosome structure, duplication or mitotic functions [118].
An additional deficiency seen with centrin 2 depletion was a marked reduction in primary ciliogenesis capacity after serum starvation of siRNA-treated cells [172, 173]. Centrin localisation at the distal lumen of the centrioles and the preferential association of centrins with the mother centriole [101] are consistent with a requirement for centrins in ciliogenesis, as are the centrin interactions with a known ciliogenesis regulator, CP110 [174]. A recent paper describing the knockdown of Cetn2 in zebrafish embryos using morpholino oligonucleotides noted developmental defects in kidney and the olfactory bulb, as well as other developmental abnormalities that were very similar to phenotypes observed in models of ciliopathy [175]. Analysis of these zebrafish morphants also revealed mitotic delays in the absence of centrin 2, although the precise impact on mitosis was not described, so that it is not clear whether this effect was the consequence of a problem in ciliogenesis, or a cell-autonomous phenotype. Nevertheless, these data offer strong support for a crucial role for the centrins in cilium formation.
Centrins and nucleotide excision repair of DNA damage
It has been noted that the bulk of cellular centrin is not associated with the centrosome, which clearly suggests its having additional roles in the cell [101]. Centrin has been implicated in a major DNA repair process, nucleotide excision repair (NER), which repairs bulky, helix-distorting DNA adducts, particularly those generated by ultraviolet (UV) light. Deficiencies in NER lead to several human diseases, including xeroderma pigmentosum (XP), in which patients have a marked sensitivity to sunlight (UV damage) and UV-mimetic chemicals leading to a high predisposition to skin cancer (reviewed by [176, 177]). Two principal NER subpathways resolve the 6-4 photoproducts and cyclobutane pyrimidine dimers that arise after UV. One is termed transcription-coupled NER, and acts quickly to remove DNA distortions that block transcription and the other is global genome repair, which, while not as rapid, acts genome-wide [178, 179].
Although both methods of NER are similar in their basic mechanism, one major difference exists in the initial recognition of DNA damage. In transcription-coupled NER, the DNA lesion is recognised by the stalling of RNA polymerase II at lesions [180], whereas in global genome NER, the damage is initially detected by an XP group C protein (XPC)-containing complex (Fig. 3) [181]. The initial detection is dependent on the type of distortion. 6-4 Photoproducts are predominantly recognised by XPC but cyclobutane pyrimidine dimers are detected more robustly by the UV-DDB complex before XPC is recruited. The steps subsequent to the initial recognition are common to both modes; DNA is unzipped near the lesion by helicase actions of TFIIH in the presence of XPA, XPG and replication protein A (RPA) [182]. This allows structure-specific endonucleases, namely XPF-ERCC1 and XPG, to excise at both 5′ and 3′ of the damage, removing approximately 25–30 base pairs. The resulting gap is then resolved by DNA polymerase (δ or ε), and the nick is joined by DNA ligase I (reviewed by [176, 177, 183]).
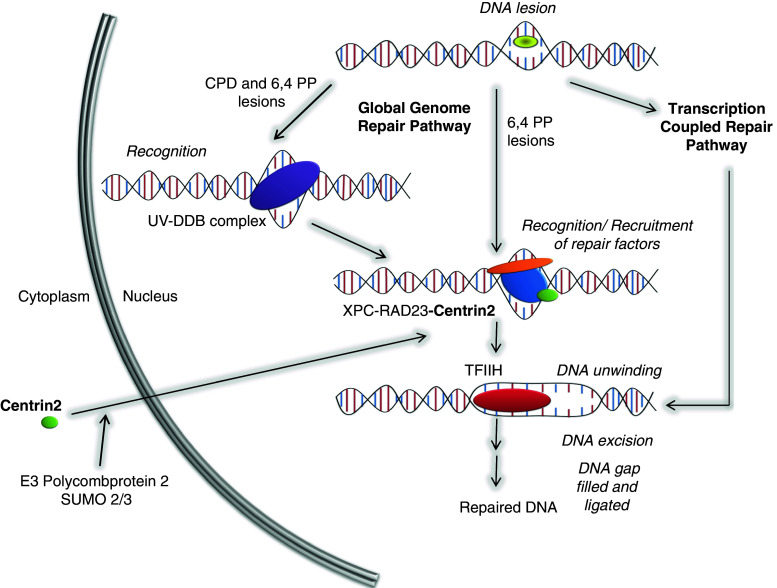
Fig. 3.
Role of centrins in nucleotide excision repair pathways. Diagrammatic representation of the pathways of nucleotide excision repair and the roles of centrin within these pathways. DNA lesions induced by UV irradiation are repaired through one of two pathways, global genome repair (GGR) or transcription coupled repair (TCR). During TCR, the initial recognition of DNA damage is achieved through a block in transcription [180]. However, for regions of DNA not being actively transcribed, this recognition relies on the heterotrimer of XPC-HRAD23-centrin 2, aided by the UV-DDB complex for less helical-distorting lesions, such as cyclobutane pyrimidine dimers (CPD) [199]. Centrin 2 localises to the nucleus through SUMOylation by SUMO 2/3, which involves the E3 ligase Polycomb protein 2 [148]. Once in the nucleus, retention of centrin 2 is dependent on its interaction with XPC, as described in the text. After recognition of the DNA lesion, XPC then recruits TFIIH through interaction with one of its subunits, XPB [225]. Helicase activities of TFIIH unwind DNA, and 25–30 base pairs of DNA are excised. The resulting gap is filled by an appropriate polymerase and the damage repaired
Centrin interaction with XPC in NER
In addition to the description of the bulk of cellular centrin as being non-centrosomal, cell fractionation experiments have positioned it within the nucleus [101, 184]. Protein sequencing identified centrin 2 as a significant component of a chromatographic fraction from HeLa nuclear extracts that was capable of rescuing NER capacity in extracts prepared from human cells with a mutation in XPC [105]. This fraction was shown to contain XPC and its known partner, HRAD23B [185], and centrin 2 was found to bind directly to XPC. Although centrin 2 was dispensable in in vitro NER reactions [186, 187], its presence stabilised XPC in a heat inactivation assay and slightly increased NER activity [105]. A 17 amino acid region in XPC that was predicted on the basis of calmodulin binding site preferences was shown to have a strong affinity to centrin 2 that increased in the presence of calcium [188]. Characterisation of the structure of this peptide complexed to centrin 2 indicated that interaction primarily involved the C-terminal domain of centrin 2 [128]. A mutational analysis of the XPC-centrin 2 interaction confirmed this peptide as the region of XPC required for the centrin interaction, lying in an a-helical region toward the C-terminus (amino acids 847–866 in 940 amino acid human XPC sequence) [189]. Abrogation of the interaction by mutation of this domain led to a significant diminution of NER activity in an in vitro reaction to monitor the excision of a base lesion by a reconstituted XPC cell extract, as well as reduced binding to damaged DNA by the complex [189]. Together, these in vitro observations clearly implicate centrin in NER. Interestingly, the recent biochemical purification and identification of XPC/HRAD23/centrin 2 as a coactivator complex that is selectively required for the synergistic activation of the Nanog gene by the key stem cell-specific transcription factors, Oct4 and Sox2 [190], also link centrins to the control of genome stability and transcriptional regulation in mouse embryonic stem cells.
In yeast, the XPC orthologue Rad4p binds Cdc31p as part of the NEF complex, which also contains Rad23p [106]. Another interactor of Rad4p/Rad23p is the recently described Rad33p, which has been suggested as an alternative to Cdc31p and a possible functional homologue of centrin 2 in NER [191]. Rad23p also functions as a proteasome-targeting vehicle for polyubiquitinated proteins [192, 193]. Temperature-sensitive cdc31 mutants that showed disruption of the Rad4p-Cdc31p interaction were sensitive to UV, in support of a role for centrin in NER. Interestingly, biochemical data showed that Cdc31p was capable of interacting directly with proteasomal components, as well as with polyubiquitinated proteins, thus regulating the stability of its interactors. Cdc31 mutants showed defective protein degradation [106]. As the control of Rad4p stability through Rad23p-mediated proteasome targeting determines UV resistance [194], it is interesting to speculate that Cdc31p may regulate Rad4p through Rad23p. As HRAD23 controls XPC stability [195, 196], a possibility is that centrin 2 might likewise regulate XPC by modulating HRAD23 and thus the proteasome.
In human cells, centrin 2 is found in both the cytoplasm and the nucleus. Overexpression of XPC alters the partitioning of centrin 2, shifting it to the nucleus [197]. This repartitioning requires the XPC interaction, as it is not induced by overexpression of an XPC with a mutation in the centrin interaction sequence [197]. Localisation to the nucleus of centrin 2, but not of XPC, was disrupted by RNAi-mediated depletion of the SUMO E2-conjugating enzyme, UBC9, the E3 ligase, polycomb protein 2, or of SUMO2/3 [148]. This work also showed that centrin 2 is SUMOylated and that centrin’s interaction with XPC was greatly increased by this SUMOylation. XPC, but not XPA, depletion also reduced the level of nuclear centrin 2, indicating the specificity of this interaction in directing nuclear localisation of centrin. In related work, centrin underwent a prominent relocalisation to the nucleus in response to UV irradiation and analogous DNA damage, potentially limiting the level of cytoplasmic centrin [184]. This relocalisation was not observed in XPC null fibroblasts [184] or in HeLa cells after siRNA knockdown of XPC [198]. Conversely, HRAD23 knockdown did not impact on centrin 2 localisation [198], consistent with the idea that nuclear targeting of centrin specifically requires XPC. XPC is regulated by SUMOylation after UV treatment [199–201], but this modification was not correlated with any change in centrin 2 SUMOylation status [148]. Overall, these data indicate that XPC regulates the nuclear activity of centrin 2 by controlling its localisation, although the signals that regulate centrin 2 SUMOylation or nuclear localisation are not yet known. DNA damage-induced phosphorylation might be one candidate, but no major changes in electrophoretic mobility of centrins were observed after UV irradiation [189].
The in vivo involvement of centrins in NER in multicellular organisms is supported by several lines of evidence. MCF-7 breast cancer cells that were partially depleted of centrin 2 by shRNA showed delayed resolution of 6-4 photoproducts [184]. Chicken DT40 cells in which all centrin isoforms had been deleted by gene targeting showed delayed resolution of cyclobutane pyrimidine dimers [118]. These cells showed a marked hypersensitivity to UV (100-fold compared to controls at 15 J/m2), but not IR, in clonogenic survival assays, and rescue experiments indicated NER roles for all centrins. As DNA damage checkpoint responses in the centrin nulls appeared to be normal, it was concluded that they suffered defective repair of UV-induced DNA damage in the absence of centrin [118]. In plants, Arabidopsis thaliana CEN2 mutant plants also showed defective NER, although this was a relatively moderate phenotype, accompanied by a hyperrecombination phenotype whose mechanism is not yet fully understood [202, 203]. Taking all these data together, we can conclude that centrins are required for efficient NER. They are localised appropriately to the sites of DNA damage by XPC and in turn control XPC stability and function.
Other DNA repair pathways and centrosomal proteins
While there are multiple examples of DNA damage checkpoint proteins localising to the centrosome (reviewed in [69–71], there is little evidence for the NER factors acting there, beyond a recent report of the XPB component of TFIIH being observed at the centrosome [204]. No mitotic defects that might reflect centrosomal defects have been reported in NER-deficient mutants without exogenous genotoxic stress [205]. There are several other examples of specific repair pathways whose members localise to centrosomes. Notably, several components of the poly(ADP-ribose) polymerase (PARP) system, which is involved in base excision repair, localise to the centrosome. Light microscopy analysis has indicated that PARP-1 localises to both centrioles and PARP-3 predominantly to the daughter throughout the cell cycle [206, 207]. Tankyrase, a PARP that regulates components of the telomere end-protection complex, has also been localised to mitotic centrosomes [208]. Furthermore, PARG, the glycohydrolase that hydrolyses poly(ADP-ribose) polymers, has been observed at mitotic centrosomes [209]. Poly ADP-ribosylation has been described as a potential regulatory mechanism for centrosomal proteins [210]. Although there have been no clear indications to date that centrosome duplication is aberrant where PARP activities are disrupted, the abnormal DNA damage responses in the absence of fully functional PARP have been implicated in centrosomal numerical abnormalities [211]. Another role for PARP lies in its control of Xrcc1 in single-strand break repair. Xrcc1 and DNA ligase IIIα were both found at centrosomes prior to anaphase and relocated to mitotic chromosomes upon induction of DNA damage [212]. This localisation was abrogated by treatment of cells with PARP inhibitors. Together, these findings provide evidence of robust communication between the nucleus and the centrosome through PARP signalling.
A further link between the centrosome and DNA repair lies in the homologous recombination (HR) apparatus. The Rad51 paralogue, Xrcc2, has been implicated in the control of mitotic centrosome stability [40]. Both Rad51 and Xrcc2 have been described at the centrosome [213], as has BRCA2, which plays important roles in regulating HR [214]. Interestingly, an interaction between Rad51 and γ-tubulin has been described in mammalian nuclei [215]. These observations are consistent with the idea that the centrosome may provide a reservoir for HR factors during mitosis, although this model is still somewhat speculative.
In addition, another distal centriole component has been implicated in both primary ciliogenesis and the DNA damage response. Cep164 is a component of the centriolar distal appendages in human cells that is necessary to allow the formation of primary cilia [173]. Notably, Cep164 interacts with the apical DNA damage response kinases, ATM and ATR, and undergoes ATR-dependent phosphorylation upon UV damage [216]. Cep164 depletion by siRNA greatly reduced the activation of the DNA damage response after both UV and IR treatment, and caused a notable loss of G2-to-M checkpoint delay, leading to the suggestion that it acts as a mediator in the DNA damage response [216]. Cep164 is recruited to UV-induced DNA lesions, but requires XPA for this localisation to occur efficiently after DNA damage. Cep164 knockdown increases cellular sensitivity to UV damage [217]. The activity of Cep164 in centrin-deficient cells is not known, but the intact G2-to-M checkpoint and other DNA damage responses after IR treatment in Cetn nulls argue against Cep164 being a key, centrosomal mediator of centrin activity.
Concluding remarks: key roles for centrin in ciliogenesis and DNA repair?
The impact on centrosome integrity of centrin deficiency in various models has been described in detail above, but it is clear that vertebrate centrioles can at least assemble without centrin [118]. The involvement of centrin in contractile responses of the basal body is well established from many studies in lower eukaryotes. Based on the multiple centrin-interacting sites described in the centrin interactor, Sfi1p [142], it was proposed that the calcium-responsive movement of centrins on a fixed core Sfi1p molecule could allow the contractile response of an extended multimeric filament [218]. hSfi1 knockdown in hTERT-RPE1 cells leads to decreased assembly of primary cilia, a phenotype also observed after centrin 2 depletion [172, 173]. Interestingly, centrin was also shown to interact with a known ciliogenesis regulator, CP110 [174], and with hPOC5, a protein involved in centriole elongation [142, 173, 219].
Combining several observations made in vertebrates with the localisation of centrins at the distal lumen and their enrichment at the mother centriole, a role for centrin in controlling primary ciliogenesis seems reasonable [101]. We have shown that centrin-deficient chicken DT40 cells are able to duplicate their centrosome [118]. Unfortunately, since this is a lymphoid cell line and therefore does not assemble primary cilia [220], we were unable to test centrin's role in ciliogenesis. However, recent work in zebrafish embryos also reported developmental defects in the kidney and olfactory bulb after Cetn2 knockdown. These and other developmental abnormalities were very similar to phenotypes observed in models of ciliopathy, and furthermore, the authors also detected a reduction in cilia numbers [175]. Whether centrin facilitates any dynamic responses in the primary cilium, during its formation or after its establishment, is not known, but a ciliary role appears to be a strong candidate for the major function of centrin in animals [101, 115, 172, 173]. Many of the unicellular organisms in which centrins have been ascribed an essential role all have striking ciliation requirements for their lifestyle. One hypothesis to explain the high level of evolutionary conservation of centrins, as well as the variability of phenotypic outcomes in reverse genetic experiments in vertebrate cells, is that the essential role for centrins lies in control of ciliogenesis, which may indirectly affect centrosome biogenesis or mitotic functioning in certain organisms or cell types.
Aside from the roles of centrin in ciliogenesis and possible functions in centriole activities, there are several emerging questions on its functions in DNA repair. That a core component of the motile apparatus in aquatic organisms might have an additional role in responding to light seems quite reasonable with a view toward phototropism, or in retinal development. How this activity became involved in DNA repair is less obvious. What is centrin’s role in linking centrosomes and DNA repair and is there a mechanistic link, or are these simply independent interactions? Given the quantities of cellular centrin, the centrosome does not seem necessary to serve as a reservoir in which centrin is sequestered for eventual assembly with XPC/ HRAD23 in the nucleus following UV damage. However, the previously mentioned possibility remains that the centrosome acts as a scaffold on which key cellular signals can be generated. With this in mind, is the localisation of centrin at the centrosome permissive or inhibitory of post-translation modification before it can enter the nucleus? Centrin SUMOylation is an attractive candidate for such regulation, but the signals dictating such a modification in response to UV damage are not yet defined. Does centrin, in fact, control other activities in the DNA damage response to UV damage, rather than being a key component in its own right? A further question is at what stage does the NER reaction become impaired in the absence of centrin? A strong hypothesis is that it is at the recognition step that is normally catalysed by XPC/ HRAD23, but an alternative impact on the reaction could change our understanding of the role of centrin in NER. Finally, the role of calcium signalling in cellular responses to UV damage remains unknown. Tackling the questions outlined above will help to define the pressures that have forced the evolutionary conservation of the centrins, despite the diversity of their functions.
Acknowledgments
TJD received a predoctoral fellowship from the Fundação para a Ciência e a Tecnologia (Portugal). This work was supported by Science Foundation Ireland Principal Investigator awards 08/IN.1/B1029 and 10/IN.1/B2972.
References
- 1.Doxsey S, McCollum D, Theurkauf W. Centrosomes in cellular regulation. Annu Rev Cell Dev Biol. 2005;21:411–434. doi: 10.1146/annurev.cellbio.21.122303.120418. [DOI] [PubMed] [Google Scholar]
- 2.Bettencourt-Dias M, Glover DM. Centrosome biogenesis and function: centrosomics brings new understanding. Nat Rev Mol Cell Biol. 2007;8(6):451–463. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- 3.Nigg EA, Stearns T. The centrosome cycle: centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol. 2011;13(10):1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460(7252):278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delattre M, Gonczy P. The arithmetic of centrosome biogenesis. J Cell Sci. 2004;117(Pt 9):1619–1630. doi: 10.1242/jcs.01128. [DOI] [PubMed] [Google Scholar]
- 6.Hinchcliffe EH, Li C, Thompson EA, Maller JL, Sluder G. Requirement of Cdk2-cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science. 1999;283(5403):851–854. doi: 10.1126/science.283.5403.851. [DOI] [PubMed] [Google Scholar]
- 7.Lacey KR, Jackson PK, Stearns T. Cyclin-dependent kinase control of centrosome duplication. Proc Natl Acad Sci USA. 1999;96(6):2817–2822. doi: 10.1073/pnas.96.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsumoto Y, Hayashi K, Nishida E. Cyclin-dependent kinase 2 (Cdk2) is required for centrosome duplication in mammalian cells. Curr Biol. 1999;9(8):429–432. doi: 10.1016/S0960-9822(99)80191-2. [DOI] [PubMed] [Google Scholar]
- 9.Meraldi P, Lukas J, Fry AM, Bartek J, Nigg EA. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat Cell Biol. 1999;1(2):88–93. doi: 10.1038/10054. [DOI] [PubMed] [Google Scholar]
- 10.Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006;442(7105):947–951. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- 11.Tsou MF, Wang WJ, George KA, Uryu K, Stearns T, Jallepalli PV. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev Cell. 2009;17(3):344–354. doi: 10.1016/j.devcel.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakamura A, Arai H, Fujita N. Centrosomal Aki1 and cohesin function in separase-regulated centriole disengagement. J Cell Biol. 2009;187(5):607–614. doi: 10.1083/jcb.200906019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schockel L, Mockel M, Mayer B, Boos D, Stemmann O. Cleavage of cohesin rings coordinates the separation of centrioles and chromatids. Nat Cell Biol. 2011;13(8):966–972. doi: 10.1038/ncb2280. [DOI] [PubMed] [Google Scholar]
- 14.Boveri T. Concerning the origin of malignant tumours by Theodor Boveri. Translated and annotated by Henry Harris. J Cell Sci. 2008;121:1–84. doi: 10.1242/jcs.025742. [DOI] [PubMed] [Google Scholar]
- 15.D’Assoro AB, Lingle WL, Salisbury JL. Centrosome amplification and the development of cancer. Oncogene. 2002;21(40):6146–6153. doi: 10.1038/sj.onc.1205772. [DOI] [PubMed] [Google Scholar]
- 16.Lingle WL, Lutz WH, Ingle JN, Maihle NJ, Salisbury JL. Centrosome hypertrophy in human breast tumors: implications for genomic stability and cell polarity. Proc Natl Acad Sci USA. 1998;95(6):2950–2955. doi: 10.1073/pnas.95.6.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pihan GA, Purohit A, Wallace J, Malhotra R, Liotta L, Doxsey SJ. Centrosome defects can account for cellular and genetic changes that characterize prostate cancer progression. Cancer Res. 2001;61(5):2212–2219. [PubMed] [Google Scholar]
- 18.Duensing S, Lee LY, Duensing A, Basile J, Piboonniyom S, Gonzalez S, Crum CP, Munger K. The human papillomavirus type 16 E6 and E7 oncoproteins cooperate to induce mitotic defects and genomic instability by uncoupling centrosome duplication from the cell division cycle. Proc Natl Acad Sci USA. 2000;97(18):10002–10007. doi: 10.1073/pnas.170093297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nigg EA. Centrosome aberrations: cause or consequence of cancer progression? Nat Rev Cancer. 2002;2(11):815–825. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- 20.Lingle WL, Barrett SL, Negron VC, D’Assoro AB, Boeneman K, Liu W, Whitehead CM, Reynolds C, Salisbury JL. Centrosome amplification drives chromosomal instability in breast tumor development. Proc Natl Acad Sci USA. 2002;99(4):1978–1983. doi: 10.1073/pnas.032479999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brinkley BR. Managing the centrosome numbers game: from chaos to stability in cancer cell division. Trends Cell Biol. 2001;11(1):18–21. doi: 10.1016/S0962-8924(00)01872-9. [DOI] [PubMed] [Google Scholar]
- 22.Sluder G, Nordberg JJ. The good, the bad and the ugly: the practical consequences of centrosome amplification. Curr Opin Cell Biol. 2004;16(1):49–54. doi: 10.1016/j.ceb.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 23.Vorobjev IA, Chentsov YuS. Centrioles in the cell cycle. I. Epithelial cells. J Cell Biol. 1982;93(3):938–949. doi: 10.1083/jcb.93.3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hinchcliffe EH, Sluder G. “It takes two to tango”: understanding how centrosome duplication is regulated throughout the cell cycle. Genes Dev. 2001;15(10):1167–1181. doi: 10.1101/gad.894001. [DOI] [PubMed] [Google Scholar]
- 25.Nigg EA. Centrosome duplication: of rules and licenses. Trends Cell Biol. 2007;17(5):215–221. doi: 10.1016/j.tcb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Guarguaglini G, Duncan PI, Stierhof YD, Holmstrom T, Duensing S, Nigg EA. The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol Biol Cell. 2005;16(3):1095–1107. doi: 10.1091/mbc.E04-10-0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duensing A, Liu Y, Perdreau SA, Kleylein-Sohn J, Nigg EA, Duensing S. Centriole overduplication through the concurrent formation of multiple daughter centrioles at single maternal templates. Oncogene. 2007;26(43):6280–6288. doi: 10.1038/sj.onc.1210456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodrigues-Martins A, Riparbelli M, Callaini G, Glover DM, Bettencourt-Dias M. Revisiting the role of the mother centriole in centriole biogenesis. Science. 2007;316(5827):1046–1050. doi: 10.1126/science.1142950. [DOI] [PubMed] [Google Scholar]
- 29.Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13(2):190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 30.Uetake Y, Loncarek J, Nordberg JJ, English CN, La Terra S, Khodjakov A, Sluder G. Cell cycle progression and de novo centriole assembly after centrosomal removal in untransformed human cells. J Cell Biol. 2007;176(2):173–182. doi: 10.1083/jcb.200607073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.La Terra S, English CN, Hergert P, McEwen BF, Sluder G, Khodjakov A. The de novo centriole assembly pathway in HeLa cells: cell cycle progression and centriole assembly/maturation. J Cell Biol. 2005;168(5):713–722. doi: 10.1083/jcb.200411126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khodjakov A, Rieder CL, Sluder G, Cassels G, Sibon O, Wang CL. De novo formation of centrosomes in vertebrate cells arrested during S phase. J Cell Biol. 2002;158(7):1171–1181. doi: 10.1083/jcb.200205102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dodson H, Bourke E, Jeffers LJ, Vagnarelli P, Sonoda E, Takeda S, Earnshaw WC, Merdes A, Morrison C. Centrosome amplification induced by DNA damage occurs during a prolonged G2 phase and involves ATM. Embo J. 2004;23(19):3864–3873. doi: 10.1038/sj.emboj.7600393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dodson H, Wheatley SP, Morrison CG. Involvement of centrosome amplification in radiation-induced mitotic catastrophe. Cell Cycle. 2007;6(3):364–370. doi: 10.4161/cc.6.3.3834. [DOI] [PubMed] [Google Scholar]
- 35.Sato N, Mizumoto K, Nakamura M, Tanaka M. Radiation-induced centrosome overduplication and multiple mitotic spindles in human tumor cells. Exp Cell Res. 2000;255(2):321–326. doi: 10.1006/excr.1999.4797. [DOI] [PubMed] [Google Scholar]
- 36.Balczon R, Bao L, Zimmer WE, Brown K, Zinkowski RP, Brinkley BR. Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. J Cell Biol. 1995;130(1):105–115. doi: 10.1083/jcb.130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meraldi P, Honda R, Nigg EA. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53−/− cells. Embo J. 2002;21(4):483–492. doi: 10.1093/emboj/21.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bertrand P, Lambert S, Joubert C, Lopez BS. Overexpression of mammalian Rad51 does not stimulate tumorigenesis while a dominant-negative Rad51 affects centrosome fragmentation, ploidy and stimulates tumorigenesis, in p53-defective CHO cells. Oncogene. 2003;22(48):7587–7592. doi: 10.1038/sj.onc.1206998. [DOI] [PubMed] [Google Scholar]
- 39.Fukasawa K, Choi T, Kuriyama R, Rulong S, Vande Woude GF. Abnormal centrosome amplification in the absence of p53. Science. 1996;271(5256):1744–1747. doi: 10.1126/science.271.5256.1744. [DOI] [PubMed] [Google Scholar]
- 40.Griffin CS, Simpson PJ, Wilson CR, Thacker J. Mammalian recombination-repair genes XRCC2 and XRCC3 promote correct chromosome segregation. Nat Cell Biol. 2000;2(10):757–761. doi: 10.1038/35036399. [DOI] [PubMed] [Google Scholar]
- 41.Kraakman-van der Zwet M, Overkamp WJ, van Lange RE, Essers J, van Duijn-Goedhart A, Wiggers I, Swaminathan S, van Buul PP, Errami A, Tan RT, Jaspers NG, Sharan SK, Kanaar R, Zdzienicka MZ. Brca2 (XRCC11) deficiency results in radioresistant DNA synthesis and a higher frequency of spontaneous deletions. Mol Cell Biol. 2002;22(2):669–679. doi: 10.1128/MCB.22.2.669-679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mantel C, Braun SE, Reid S, Henegariu O, Liu L, Hangoc G, Broxmeyer HE. p21(cip-1/waf-1) deficiency causes deformed nuclear architecture, centriole overduplication, polyploidy, and relaxed microtubule damage checkpoints in human hematopoietic cells. Blood. 1999;93(4):1390–1398. [PubMed] [Google Scholar]
- 43.Tutt AN, van Oostrom CT, Ross GM, van Steeg H, Ashworth A. Disruption of Brca2 increases the spontaneous mutation rate in vivo: synergism with ionizing radiation. EMBO Rep. 2002;3(3):255–260. doi: 10.1093/embo-reports/kvf037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamaguchi-Iwai Y, Sonoda E, Sasaki MS, Morrison C, Haraguchi T, Hiraoka Y, Yamashita YM, Yagi T, Takata M, Price C, Kakazu N, Takeda S. Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. Embo J. 1999;18(23):6619–6629. doi: 10.1093/emboj/18.23.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guiducci C, Cerone MA, Bacchetti S. Expression of mutant telomerase in immortal telomerase-negative human cells results in cell cycle deregulation, nuclear and chromosomal abnormalities and rapid loss of viability. Oncogene. 2001;20(6):714–725. doi: 10.1038/sj.onc.1204145. [DOI] [PubMed] [Google Scholar]
- 46.Duensing A, Liu Y, Tseng M, Malumbres M, Barbacid M, Duensing S. Cyclin-dependent kinase 2 is dispensable for normal centrosome duplication but required for oncogene-induced centrosome overduplication. Oncogene. 2006;25(20):2943–2949. doi: 10.1038/sj.onc.1209310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duensing S, Munger K. Human papillomavirus type 16 E7 oncoprotein can induce abnormal centrosome duplication through a mechanism independent of inactivation of retinoblastoma protein family members. J Virol. 2003;77(22):12331–12335. doi: 10.1128/JVI.77.22.12331-12335.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watanabe N, Yamaguchi T, Akimoto Y, Rattner JB, Hirano H, Nakauchi H. Induction of M-phase arrest and apoptosis after HIV-1 Vpr expression through uncoupling of nuclear and centrosomal cycle in HeLa cells. Exp Cell Res. 2000;258(2):261–269. doi: 10.1006/excr.2000.4908. [DOI] [PubMed] [Google Scholar]
- 49.Daniels MJ, Wang Y, Lee M, Venkitaraman AR. Abnormal cytokinesis in cells deficient in the breast cancer susceptibility protein BRCA2. Science. 2004;306(5697):876–879. doi: 10.1126/science.1102574. [DOI] [PubMed] [Google Scholar]
- 50.Hinchcliffe EH, Cassels GO, Rieder CL, Sluder G. The coordination of centrosome reproduction with nuclear events of the cell cycle in the sea urchin zygote. J Cell Biol. 1998;140(6):1417–1426. doi: 10.1083/jcb.140.6.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wong C, Stearns T. Centrosome number is controlled by a centrosome-intrinsic block to reduplication. Nat Cell Biol. 2003;5(6):539–544. doi: 10.1038/ncb993. [DOI] [PubMed] [Google Scholar]
- 52.Hochegger H, Dejsuphong D, Sonoda E, Saberi A, Rajendra E, Kirk J, Hunt T, Takeda S. An essential role for Cdk1 in S phase control is revealed via chemical genetics in vertebrate cells. J Cell Biol. 2007;178(2):257–268. doi: 10.1083/jcb.200702034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adon AM, Zeng X, Harrison MK, Sannem S, Kiyokawa H, Kaldis P, Saavedra HI. Cdk2 and Cdk4 regulate the centrosome cycle and are critical mediators of centrosome amplification in p53-null cells. Mol Cell Biol. 2010;30(3):694–710. doi: 10.1128/MCB.00253-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prosser SL, Straatman KR, Fry AM. Molecular dissection of the centrosome overduplication pathway in S-phase arrested cells. Mol Cell Biol. 2009;29:1760–1773. doi: 10.1128/MCB.01124-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fukasawa K. P53, cyclin-dependent kinase and abnormal amplification of centrosomes. Biochim Biophys Acta. 2008;1786(1):15–23. doi: 10.1016/j.bbcan.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mussman JG, Horn HF, Carroll PE, Okuda M, Tarapore P, Donehower LA, Fukasawa K. Synergistic induction of centrosome hyperamplification by loss of p53 and cyclin E overexpression. Oncogene. 2000;19(13):1635–1646. doi: 10.1038/sj.onc.1203460. [DOI] [PubMed] [Google Scholar]
- 57.Spruck CH, Won KA, Reed SI. Deregulated cyclin E induces chromosome instability. Nature. 1999;401(6750):297–300. doi: 10.1038/45836. [DOI] [PubMed] [Google Scholar]
- 58.Kawamura K, Izumi H, Ma Z, Ikeda R, Moriyama M, Tanaka T, Nojima T, Levin LS, Fujikawa-Yamamoto K, Suzuki K, Fukasawa K. Induction of centrosome amplification and chromosome instability in human bladder cancer cells by p53 mutation and cyclin E overexpression. Cancer Res. 2004;64(14):4800–4809. doi: 10.1158/0008-5472.CAN-03-3908. [DOI] [PubMed] [Google Scholar]
- 59.Sato C, Kuriyama R, Nishizawa K. Microtubule-organizing centers abnormal in number, structure, and nucleating activity in x-irradiated mammalian cells. J Cell Biol. 1983;96(3):776–782. doi: 10.1083/jcb.96.3.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bourke E, Dodson H, Merdes A, Cuffe L, Zachos G, Walker M, Gillespie D, Morrison CG. DNA damage induces Chk1-dependent centrosome amplification. EMBO Rep. 2007;8(6):603–609. doi: 10.1038/sj.embor.7400962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bourke E, Brown JA, Takeda S, Hochegger H, Morrison CG. DNA damage induces Chk1-dependent threonine-160 phosphorylation and activation of Cdk2. Oncogene. 2010;29(4):616–624. doi: 10.1038/onc.2009.340. [DOI] [PubMed] [Google Scholar]
- 62.Sibon OC, Kelkar A, Lemstra W, Theurkauf WE. DNA-replication/DNA-damage-dependent centrosome inactivation in Drosophila embryos. Nat Cell Biol. 2000;2(2):90–95. doi: 10.1038/35000041. [DOI] [PubMed] [Google Scholar]
- 63.Hut HM, Lemstra W, Blaauw EH, Van Cappellen GW, Kampinga HH, Sibon OC. Centrosomes split in the presence of impaired DNA integrity during mitosis. Mol Biol Cell. 2003;14(5):1993–2004. doi: 10.1091/mbc.E02-08-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saladino C, Bourke E, Conroy PC, Morrison CG. Centriole separation in DNA damage-induced centrosome amplification. Environ Mol Mutagen. 2009;50(8):725–732. doi: 10.1002/em.20477. [DOI] [PubMed] [Google Scholar]
- 65.Inanc B, Dodson H, Morrison CG. A centrosome-autonomous signal that involves centriole disengagement permits centrosome duplication in G2 phase after DNA damage. Mol Biol Cell. 2010;21(22):3866–3877. doi: 10.1091/mbc.E10-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smits VA, Klompmaker R, Arnaud L, Rijksen G, Nigg EA, Medema RH. Polo-like kinase-1 is a target of the DNA damage checkpoint. Nat Cell Biol. 2000;2(9):672–676. doi: 10.1038/35023629. [DOI] [PubMed] [Google Scholar]
- 67.van Vugt MA, Smits VA, Klompmaker R, Medema RH. Inhibition of Polo-like kinase-1 by DNA damage occurs in an ATM- or ATR-dependent fashion. J Biol Chem. 2001;276(45):41656–41660. doi: 10.1074/jbc.M101831200. [DOI] [PubMed] [Google Scholar]
- 68.Zhang W, Fletcher L, Muschel RJ. The role of Polo-like kinase 1 in the inhibition of centrosome separation after ionizing radiation. J Biol Chem. 2005;280(52):42994–42999. doi: 10.1074/jbc.M505450200. [DOI] [PubMed] [Google Scholar]
- 69.Loffler H, Lukas J, Bartek J, Kramer A. Structure meets function—centrosomes, genome maintenance and the DNA damage response. Exp Cell Res. 2006;312(14):2633–2640. doi: 10.1016/j.yexcr.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 70.Fukasawa K. Oncogenes and tumour suppressors take on centrosomes. Nat Rev Cancer. 2007;7(12):911–924. doi: 10.1038/nrc2249. [DOI] [PubMed] [Google Scholar]
- 71.Shimada M, Komatsu K. Emerging connection between centrosome and DNA repair machinery. J Radiat Res (Tokyo) 2009;50(4):295–301. doi: 10.1269/jrr.09039. [DOI] [PubMed] [Google Scholar]
- 72.Jackman M, Lindon C, Nigg EA, Pines J. Active cyclin B1-Cdk1 first appears on centrosomes in prophase. Nat Cell Biol. 2003;5(2):143–148. doi: 10.1038/ncb918. [DOI] [PubMed] [Google Scholar]
- 73.Kramer A, Mailand N, Lukas C, Syljuasen RG, Wilkinson CJ, Nigg EA, Bartek J, Lukas J. Centrosome-associated Chk1 prevents premature activation of cyclin-B-Cdk1 kinase. Nat Cell Biol. 2004;6(9):884–891. doi: 10.1038/ncb1165. [DOI] [PubMed] [Google Scholar]
- 74.Loffler H, Bochtler T, Fritz B, Tews B, Ho AD, Lukas J, Bartek J, Kramer A. DNA damage-induced accumulation of centrosomal Chk1 contributes to its checkpoint function. Cell Cycle. 2007;6(20):2541–2548. doi: 10.4161/cc.6.20.4810. [DOI] [PubMed] [Google Scholar]
- 75.Tibelius A, Marhold J, Zentgraf H, Heilig CE, Neitzel H, Ducommun B, Rauch A, Ho AD, Bartek J, Kramer A. Microcephalin and pericentrin regulate mitotic entry via centrosome-associated Chk1. J Cell Biol. 2009;185(7):1149–1157. doi: 10.1083/jcb.200810159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Boutros R, Lorenzo C, Mondesert O, Jauneau A, Oakes V, Dozier C, Gabrielli B, Ducommun B. CDC25B associates with a centrin 2-containing complex and is involved in maintaining centrosome integrity. Biol Cell. 2011;103(2):55–68. doi: 10.1042/BC20100111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Salisbury JL, Baron A, Surek B, Melkonian M. Striated flagellar roots: isolation and partial characterization of a calcium-modulated contractile organelle. J Cell Biol. 1984;99(3):962–970. doi: 10.1083/jcb.99.3.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wright RL, Salisbury J, Jarvik JW. A nucleus-basal body connector in Chlamydomonas reinhardtii that may function in basal body localization or segregation. J Cell Biol. 1985;101(5 Pt 1):1903–1912. doi: 10.1083/jcb.101.5.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McFadden GI, Schulze D, Surek B, Salisbury JL, Melkonian M. Basal body reorientation mediated by a Ca2+-modulated contractile protein. J Cell Biol. 1987;105(2):903–912. doi: 10.1083/jcb.105.2.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schulze D, Robenek H, McFadden GI, Melkonian M. Immunolocalization of a Ca2+-modulated contractile protein in the flagellar apparatus of green algae: the nucleus-basal body connector. Eur J Cell Biol. 1987;45:51–61. [Google Scholar]
- 81.Salisbury JL, Baron AT, Sanders MA. The centrin-based cytoskeleton of Chlamydomonas reinhardtii: distribution in interphase and mitotic cells. J Cell Biol. 1988;107(2):635–641. doi: 10.1083/jcb.107.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Salisbury JL, Sanders MA, Harpst L. Flagellar root contraction and nuclear movement during flagellar regeneration in Chlamydomonas reinhardtii . J Cell Biol. 1987;105(4):1799–1805. doi: 10.1083/jcb.105.4.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sanders MA, Salisbury JL. Centrin plays an essential role in microtubule severing during flagellar excision in Chlamydomonas reinhardtii . J Cell Biol. 1994;124(5):795–805. doi: 10.1083/jcb.124.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Geimer S, Melkonian M. Centrin scaffold in Chlamydomonas reinhardtii revealed by immunoelectron microscopy. Eukaryot Cell. 2005;4(7):1253–1263. doi: 10.1128/EC.4.7.1253-1263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang B, Watterson DM, Lee VD, Schibler MJ. Purification and characterization of a basal body-associated Ca2+-binding protein. J Cell Biol. 1988;107(1):121–131. doi: 10.1083/jcb.107.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang B, Mengersen A, Lee VD. Molecular cloning of cDNA for caltractin, a basal body-associated Ca2+-binding protein: homology in its protein sequence with calmodulin and the yeast CDC31 gene product. J Cell Biol. 1988;107(1):133–140. doi: 10.1083/jcb.107.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Baum P, Furlong C, Byers B. Yeast gene required for spindle pole body duplication: homology of its product with Ca2+-binding proteins. Proc Natl Acad Sci USA. 1986;83(15):5512–5516. doi: 10.1073/pnas.83.15.5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Baron AT, Salisbury JL. Identification and localization of a novel, cytoskeletal, centrosome-associated protein in PtK2 cells. J Cell Biol. 1988;107(6 Pt 2):2669–2678. doi: 10.1083/jcb.107.6.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kretsinger RH, Nockolds CE. Carp muscle calcium-binding protein. II. Structure determination and general description. J Biol Chem. 1973;248(9):3313–3326. [PubMed] [Google Scholar]
- 90.Kretsinger RH. Calcium-binding proteins. Annu Rev Biochem. 1976;45:239–266. doi: 10.1146/annurev.bi.45.070176.001323. [DOI] [PubMed] [Google Scholar]
- 91.Salisbury JL, Baron AT, Coling DE, Martindale VE, Sanders MA. Calcium-modulated contractile proteins associated with the eucaryotic centrosome. Cell Motil Cytoskelet. 1986;6(2):193–197. doi: 10.1002/cm.970060218. [DOI] [PubMed] [Google Scholar]
- 92.Baron AT, Greenwood TM, Bazinet CW, Salisbury JL. Centrin is a component of the pericentriolar lattice. Biol Cell. 1992;76(3):383–388. doi: 10.1016/0248-4900(92)90442-4. [DOI] [PubMed] [Google Scholar]
- 93.Moudjou M, Paintrand M, Vigues B, Bornens M. A human centrosomal protein is immunologically related to basal body-associated proteins from lower eucaryotes and is involved in the nucleation of microtubules. J Cell Biol. 1991;115(1):129–140. doi: 10.1083/jcb.115.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ogawa K, Shimizu T. cDNA sequence for mouse caltractin. Biochim Biophys Acta. 1993;1216(1):126–128. doi: 10.1016/0167-4781(93)90048-i. [DOI] [PubMed] [Google Scholar]
- 95.Errabolu R, Sanders MA, Salisbury JL. Cloning of a cDNA encoding human centrin, an EF-hand protein of centrosomes and mitotic spindle poles. J Cell Sci. 1994;107(Pt 1):9–16. doi: 10.1242/jcs.107.1.9. [DOI] [PubMed] [Google Scholar]
- 96.Lee VD, Huang B. Molecular cloning and centrosomal localization of human caltractin. Proc Natl Acad Sci USA. 1993;90(23):11039–11043. doi: 10.1073/pnas.90.23.11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Levy YY, Lai EY, Remillard SP, Heintzelman MB, Fulton C. Centrin is a conserved protein that forms diverse associations with centrioles and MTOCs in Naegleria and other organisms. Cell Motil Cytoskelet. 1996;33(4):298–323. doi: 10.1002/(SICI)1097-0169(1996)33:4<298::AID-CM6>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 98.Wolfrum U. Cytoskeletal elements in arthropod sensilla and mammalian photoreceptors. Biol Cell. 1992;76(3):373–381. doi: 10.1016/0248-4900(92)90441-3. [DOI] [PubMed] [Google Scholar]
- 99.Stearns T, Kirschner M. In vitro reconstitution of centrosome assembly and function: the central role of gamma-tubulin. Cell. 1994;76(4):623–637. doi: 10.1016/0092-8674(94)90503-7. [DOI] [PubMed] [Google Scholar]
- 100.Uzawa M, Grams J, Madden B, Toft D, Salisbury JL. Identification of a complex between centrin and heat shock proteins in CSF-arrested Xenopus oocytes and dissociation of the complex following oocyte activation. Dev Biol. 1995;171(1):51–59. doi: 10.1006/dbio.1995.1259. [DOI] [PubMed] [Google Scholar]
- 101.Paoletti A, Moudjou M, Paintrand M, Salisbury JL, Bornens M. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J Cell Sci. 1996;109(Pt 13):3089–3102. doi: 10.1242/jcs.109.13.3089. [DOI] [PubMed] [Google Scholar]
- 102.Spang A, Courtney I, Fackler U, Matzner M, Schiebel E. The calcium-binding protein cell division cycle 31 of Saccharomyces cerevisiae is a component of the half bridge of the spindle pole body. J Cell Biol. 1993;123(2):405–416. doi: 10.1083/jcb.123.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Paintrand M, Moudjou M, Delacroix H, Bornens M. Centrosome organization and centriole architecture: their sensitivity to divalent cations. J Struct Biol. 1992;108(2):107–128. doi: 10.1016/1047-8477(92)90011-X. [DOI] [PubMed] [Google Scholar]
- 104.Baron AT, Suman VJ, Nemeth E, Salisbury JL. The pericentriolar lattice of PtK2 cells exhibits temperature and calcium-modulated behavior. J Cell Sci. 1994;107(Pt 11):2993–3003. doi: 10.1242/jcs.107.11.2993. [DOI] [PubMed] [Google Scholar]
- 105.Araki M, Masutani C, Takemura M, Uchida A, Sugasawa K, Kondoh J, Ohkuma Y, Hanaoka F. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J Biol Chem. 2001;276(22):18665–18672. doi: 10.1074/jbc.M100855200. [DOI] [PubMed] [Google Scholar]
- 106.Chen L, Madura K. Centrin/Cdc31 is a novel regulator of protein degradation. Mol Cell Biol. 2008;28(5):1829–1840. doi: 10.1128/MCB.01256-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fischer T, Rodriguez-Navarro S, Pereira G, Racz A, Schiebel E, Hurt E. Yeast centrin Cdc31 is linked to the nuclear mRNA export machinery. Nat Cell Biol. 2004;6(9):840–848. doi: 10.1038/ncb1163. [DOI] [PubMed] [Google Scholar]
- 108.Resendes KK, Rasala BA, Forbes DJ. Centrin 2 localizes to the vertebrate nuclear pore and plays a role in mRNA and protein export. Mol Cell Biol. 2008;28(5):1755–1769. doi: 10.1128/MCB.01697-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Higginbotham H, Bielas S, Tanaka T, Gleeson JG. Transgenic mouse line with green-fluorescent protein-labeled Centrin 2 allows visualization of the centrosome in living cells. Transgenic Res. 2004;13(2):155–164. doi: 10.1023/B:TRAG.0000026071.41735.8e. [DOI] [PubMed] [Google Scholar]
- 110.Kuriyama R, Terada Y, Lee KS, Wang CL. Centrosome replication in hydroxyurea-arrested CHO cells expressing GFP-tagged centrin 2. J Cell Sci. 2007;120(Pt 14):2444–2453. doi: 10.1242/jcs.008938. [DOI] [PubMed] [Google Scholar]
- 111.Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J Cell Biol. 2000;149(2):317–330. doi: 10.1083/jcb.149.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.White RA, Pan Z, Salisbury JL. GFP-centrin as a marker for centriole dynamics in living cells. Microsc Res Tech. 2000;49(5):451–457. doi: 10.1002/(SICI)1097-0029(20000601)49:5<451::AID-JEMT7>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 113.Koblenz B, Schoppmeier J, Grunow A, Lechtreck KF. Centrin deficiency in Chlamydomonas causes defects in basal body replication, segregation and maturation. J Cell Sci. 2003;116(Pt 13):2635–2646. doi: 10.1242/jcs.00497. [DOI] [PubMed] [Google Scholar]
- 114.Ruiz F, Garreau de Loubresse N, Klotz C, Beisson J, Koll F. Centrin deficiency in Paramecium affects the geometry of basal-body duplication. Curr Biol. 2005;15(23):2097–2106. doi: 10.1016/j.cub.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 115.Yang CH, Kasbek C, Majumder S, Yusof AM, Fisk HA. Mps1 phosphorylation sites regulate the function of centrin 2 in centriole assembly. Mol Biol Cell. 2010 doi: 10.1091/mbc.E10-04-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Collins ES, Hornick JE, Durcan TM, Collins NS, Archer W, Karanjeet KB, Vaughan KT, Hinchcliffe EH. Centrosome biogenesis continues in the absence of microtubules during prolonged S-phase arrest. J Cell Physiol. 2010 doi: 10.1002/jcp.22222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Prosser SL, Straatman KR, Fry AM. Molecular dissection of the centrosome overduplication pathway in S-phase-arrested cells. Mol Cell Biol. 2009;29(7):1760–1773. doi: 10.1128/MCB.01124-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dantas TJ, Wang Y, Lalor P, Dockery P, Morrison CG. Defective nucleotide excision repair with normal centrosome structures and functions in the absence of all vertebrate centrins. J Cell Biol. 2011;193(2):307–318. doi: 10.1083/jcb.201012093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Azimzadeh J, Bornens M (2004) The centrosome in evolution. In: Nigg EA (ed) Centrosomes in development and disease. Wiley-VCH, Weinheim, pp 93–122
- 120.Hodges ME, Scheumann N, Wickstead B, Langdale JA, Gull K. Reconstructing the evolutionary history of the centriole from protein components. J Cell Sci. 2010;123(Pt 9):1407–1413. doi: 10.1242/jcs.064873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Middendorp S, Paoletti A, Schiebel E, Bornens M. Identification of a new mammalian centrin gene, more closely related to Saccharomyces cerevisiae CDC31 gene. Proc Natl Acad Sci USA. 1997;94(17):9141–9146. doi: 10.1073/pnas.94.17.9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Middendorp S, Kuntziger T, Abraham Y, Holmes S, Bordes N, Paintrand M, Paoletti A, Bornens M. A role for centrin 3 in centrosome reproduction. J Cell Biol. 2000;148(3):405–416. doi: 10.1083/jcb.148.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gavet O, Alvarez C, Gaspar P, Bornens M. Centrin4p, a novel mammalian centrin specifically expressed in ciliated cells. Mol Biol Cell. 2003;14(5):1818–1834. doi: 10.1091/mbc.E02-11-0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wolfrum U, Salisbury JL. Expression of centrin isoforms in the mammalian retina. Exp Cell Res. 1998;242(1):10–17. doi: 10.1006/excr.1998.4038. [DOI] [PubMed] [Google Scholar]
- 125.Hart PE, Glantz JN, Orth JD, Poynter GM, Salisbury JL. Testis-specific murine centrin, Cetn1: genomic characterization and evidence for retroposition of a gene encoding a centrosome protein. Genomics. 1999;60(2):111–120. doi: 10.1006/geno.1999.5880. [DOI] [PubMed] [Google Scholar]
- 126.Laoukili J, Perret E, Middendorp S, Houcine O, Guennou C, Marano F, Bornens M, Tournier F. Differential expression and cellular distribution of centrin isoforms during human ciliated cell differentiation in vitro. J Cell Sci. 2000;113(Pt 8):1355–1364. doi: 10.1242/jcs.113.8.1355. [DOI] [PubMed] [Google Scholar]
- 127.Veeraraghavan S, Fagan PA, Hu H, Lee V, Harper JF, Huang B, Chazin WJ. Structural independence of the two EF-hand domains of caltractin. J Biol Chem. 2002;277(32):28564–28571. doi: 10.1074/jbc.M112232200. [DOI] [PubMed] [Google Scholar]
- 128.Thompson JR, Ryan ZC, Salisbury JL, Kumar R. The structure of the human centrin 2-xeroderma pigmentosum group C protein complex. J Biol Chem. 2006;281(27):18746–18752. doi: 10.1074/jbc.M513667200. [DOI] [PubMed] [Google Scholar]
- 129.Zhang Y, He CY. Centrins in unicellular organisms: functional diversity and specialization. Protoplasma. 2011 doi: 10.1007/s00709-011-0305-2. [DOI] [PubMed] [Google Scholar]
- 130.Radu L, Durussel I, Assairi L, Blouquit Y, Miron S, Cox JA, Craescu CT. Scherffelia dubia centrin exhibits a specific mechanism for Ca(2+)-controlled target binding. Biochemistry. 2010;49(20):4383–4394. doi: 10.1021/bi901764m. [DOI] [PubMed] [Google Scholar]
- 131.Ortiz M, Sanoguet Z, Hu H, Chazin WJ, McMurray CT, Salisbury JL, Pastrana-Rios B. Dynamics of hydrogen-deuterium exchange in Chlamydomonas centrin. Biochemistry. 2005;44(7):2409–2418. doi: 10.1021/bi0484419. [DOI] [PubMed] [Google Scholar]
- 132.Hu H, Sheehan JH, Chazin WJ. The mode of action of centrin. Binding of Ca2+ and a peptide fragment of Kar1p to the C-terminal domain. J Biol Chem. 2004;279(49):50895–50903. doi: 10.1074/jbc.M404233200. [DOI] [PubMed] [Google Scholar]
- 133.Sheehan JH, Bunick CG, Hu H, Fagan PA, Meyn SM, Chazin WJ. Structure of the N-terminal calcium sensor domain of centrin reveals the biochemical basis for domain-specific function. J Biol Chem. 2006;281(5):2876–2881. doi: 10.1074/jbc.M509886200. [DOI] [PubMed] [Google Scholar]
- 134.Selvapandiyan A, Duncan R, Debrabant A, Bertholet S, Sreenivas G, Negi NS, Salotra P, Nakhasi HL. Expression of a mutant form of Leishmania donovani centrin reduces the growth of the parasite. J Biol Chem. 2001;276(46):43253–43261. doi: 10.1074/jbc.M106806200. [DOI] [PubMed] [Google Scholar]
- 135.Geier BM, Wiech H, Schiebel E. Binding of centrins and yeast calmodulin to synthetic peptides corresponding to binding sites in the spindle pole body components Kar1p and Spc110p. J Biol Chem. 1996;271(45):28366–28374. doi: 10.1074/jbc.271.45.28366. [DOI] [PubMed] [Google Scholar]
- 136.Durussel I, Blouquit Y, Middendorp S, Craescu CT, Cox JA. Cation- and peptide-binding properties of human centrin 2. FEBS Lett. 2000;472(2–3):208–212. doi: 10.1016/S0014-5793(00)01452-6. [DOI] [PubMed] [Google Scholar]
- 137.Matei E, Miron S, Blouquit Y, Duchambon P, Durussel I, Cox JA, Craescu CT. C-terminal half of human centrin 2 behaves like a regulatory EF-hand domain. Biochemistry. 2003;42(6):1439–1450. doi: 10.1021/bi0269714. [DOI] [PubMed] [Google Scholar]
- 138.Yang A, Miron S, Duchambon P, Assairi L, Blouquit Y, Craescu CT. The N-terminal domain of human centrin 2 has a closed structure, binds calcium with a very low affinity, and plays a role in the protein self-assembly. Biochemistry. 2006;45(3):880–889. doi: 10.1021/bi051397s. [DOI] [PubMed] [Google Scholar]
- 139.Cox JA, Tirone F, Durussel I, Firanescu C, Blouquit Y, Duchambon P, Craescu CT. Calcium and magnesium binding to human centrin 3 and interaction with target peptides. Biochemistry. 2005;44(3):840–850. doi: 10.1021/bi048294e. [DOI] [PubMed] [Google Scholar]
- 140.Wiech H, Geier BM, Paschke T, Spang A, Grein K, Steinkotter J, Melkonian M, Schiebel E. Characterization of green alga, yeast, and human centrins. Specific subdomain features determine functional diversity. J Biol Chem. 1996;271(37):22453–22461. doi: 10.1074/jbc.271.37.22453. [DOI] [PubMed] [Google Scholar]
- 141.Spang A, Courtney I, Grein K, Matzner M, Schiebel E. The Cdc31p-binding protein Kar1p is a component of the half bridge of the yeast spindle pole body. J Cell Biol. 1995;128(5):863–877. doi: 10.1083/jcb.128.5.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kilmartin JV. Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J Cell Biol. 2003;162(7):1211–1221. doi: 10.1083/jcb.200307064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tourbez M, Firanescu C, Yang A, Unipan L, Duchambon P, Blouquit Y, Craescu CT. Calcium-dependent self-assembly of human centrin 2. J Biol Chem. 2004;279(46):47672–47680. doi: 10.1074/jbc.M404996200. [DOI] [PubMed] [Google Scholar]
- 144.Li S, Sandercock AM, Conduit P, Robinson CV, Williams RL, Kilmartin JV. Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication. J Cell Biol. 2006;173(6):867–877. doi: 10.1083/jcb.200603153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lukasiewicz KB, Greenwood TM, Negron VC, Bruzek AK, Salisbury JL, Lingle WL. Control of centrin stability by Aurora A. PLoS One. 2011;6(6):e21291. doi: 10.1371/journal.pone.0021291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lutz W, Lingle WL, McCormick D, Greenwood TM, Salisbury JL. Phosphorylation of centrin during the cell cycle and its role in centriole separation preceding centrosome duplication. J Biol Chem. 2001;276(23):20774–20780. doi: 10.1074/jbc.M101324200. [DOI] [PubMed] [Google Scholar]
- 147.Trojan P, Krauss N, Choe HW, Giessl A, Pulvermuller A, Wolfrum U. Centrins in retinal photoreceptor cells: regulators in the connecting cilium. Prog Retin Eye Res. 2008;27(3):237–259. doi: 10.1016/j.preteyeres.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 148.Klein UR, Nigg EA. SUMO-dependent regulation of centrin-2. J Cell Sci. 2009;122(Pt 18):3312–3321. doi: 10.1242/jcs.050245. [DOI] [PubMed] [Google Scholar]
- 149.Taillon BE, Adler SA, Suhan JP, Jarvik JW. Mutational analysis of centrin: an EF-hand protein associated with three distinct contractile fibers in the basal body apparatus of Chlamydomonas. J Cell Biol. 1992;119(6):1613–1624. doi: 10.1083/jcb.119.6.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Schild D, Ananthaswamy HN, Mortimer RK. An endomitotic effect of a cell cycle mutation of Saccharomyces cerevisiae . Genetics. 1981;97(3–4):551–562. doi: 10.1093/genetics/97.3-4.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Biggins S, Rose MD. Direct interaction between yeast spindle pole body components: Kar1p is required for Cdc31p localization to the spindle pole body. J Cell Biol. 1994;125(4):843–852. doi: 10.1083/jcb.125.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jaspersen SL, Giddings TH, Jr, Winey M. Mps3p is a novel component of the yeast spindle pole body that interacts with the yeast centrin homologue Cdc31p. J Cell Biol. 2002;159(6):945–956. doi: 10.1083/jcb.200208169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sullivan DS, Biggins S, Rose MD. The yeast centrin, cdc31p, and the interacting protein kinase, Kic1p, are required for cell integrity. J Cell Biol. 1998;143(3):751–765. doi: 10.1083/jcb.143.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Baum P, Yip C, Goetsch L, Byers B. A yeast gene essential for regulation of spindle pole duplication. Mol Cell Biol. 1988;8(12):5386–5397. doi: 10.1128/mcb.8.12.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ivanovska I, Rose MD. Fine structure analysis of the yeast centrin, Cdc31p, identifies residues specific for cell morphology and spindle pole body duplication. Genetics. 2001;157(2):503–518. doi: 10.1093/genetics/157.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Paoletti A, Bordes N, Haddad R, Schwartz CL, Chang F, Bornens M. Fission yeast cdc31p is a component of the half-bridge and controls SPB duplication. Mol Biol Cell. 2003;14(7):2793–2808. doi: 10.1091/mbc.E02-10-0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Klink VP, Wolniak SM. Centrin is necessary for the formation of the motile apparatus in spermatids of Marsilea. Mol Biol Cell. 2001;12(3):761–776. doi: 10.1091/mbc.12.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Tsai CW, Wolniak SM. Cell cycle arrest allows centrin translation but not basal body formation during spermiogenesis in Marsilea. J Cell Sci. 2001;114(Pt 23):4265–4272. doi: 10.1242/jcs.114.23.4265. [DOI] [PubMed] [Google Scholar]
- 159.Selvapandiyan A, Debrabant A, Duncan R, Muller J, Salotra P, Sreenivas G, Salisbury JL, Nakhasi HL. Centrin gene disruption impairs stage-specific basal body duplication and cell cycle progression in Leishmania. J Biol Chem. 2004;279(24):25703–25710. doi: 10.1074/jbc.M402794200. [DOI] [PubMed] [Google Scholar]
- 160.He CY, Pypaert M, Warren G (2005) Golgi duplication in Trypanosoma brucei requires Centrin 2. Science (New York, NY 310 (5751):1196–1198. doi:10.1126/science.1119969 [DOI] [PubMed]
- 161.Selvapandiyan A, Kumar P, Morris JC, Salisbury JL, Wang CC, Nakhasi HL. Centrin1 is required for organelle segregation and cytokinesis in Trypanosoma brucei . Mol Biol Cell. 2007;18(9):3290–3301. doi: 10.1091/mbc.E07-01-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Shi J, Franklin JB, Yelinek JT, Ebersberger I, Warren G, He CY (2008) Centrin4 coordinates cell and nuclear division in T. brucei. J Cell Sci 121 (Pt 18):3062–3070. doi:10.1242/jcs.030643 [DOI] [PubMed]
- 163.Guerra C, Wada Y, Leick V, Bell A, Satir P. Cloning, localization, and axonemal function of Tetrahymena centrin . Mol Biol Cell. 2003;14(1):251–261. doi: 10.1091/mbc.E02-05-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Stemm-Wolf AJ, Morgan G, Giddings TH, Jr, White EA, Marchione R, McDonald HB, Winey M. Basal body duplication and maintenance require one member of the Tetrahymena thermophila centrin gene family. Mol Biol Cell. 2005;16(8):3606–3619. doi: 10.1091/mbc.E04-10-0919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kilburn CL, Pearson CG, Romijn EP, Meehl JB, Giddings TH, Jr, Culver BP, Yates JR, 3rd, Winey M. New Tetrahymena basal body protein components identify basal body domain structure. J Cell Biol. 2007;178(6):905–912. doi: 10.1083/jcb.200703109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Vonderfecht T, Stemm-Wolf AJ, Hendershott M, Giddings TH, Jr, Meehl JB, Winey M. The two domains of centrin have distinct basal body functions in Tetrahymena. Mol Biol Cell. 2011;22(13):2221–2234. doi: 10.1091/mbc.E11-02-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Madeddu L, Klotz C, Le Caer JP, Beisson J. Characterization of centrin genes in Paramecium. Eur J Biochem. 1996;238(1):121–128. doi: 10.1111/j.1432-1033.1996.0121q.x. [DOI] [PubMed] [Google Scholar]
- 168.Gogendeau D, Klotz C, Arnaiz O, Malinowska A, Dadlez M, de Loubresse NG, Ruiz F, Koll F, Beisson J. Functional diversification of centrins and cell morphological complexity. J Cell Sci. 2008;121(Pt 1):65–74. doi: 10.1242/jcs.019414. [DOI] [PubMed] [Google Scholar]
- 169.Dammermann A, Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol. 2002;159(2):255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Salisbury JL, Suino KM, Busby R, Springett M. Centrin-2 is required for centriole duplication in mammalian cells. Curr Biol. 2002;12(15):1287–1292. doi: 10.1016/S0960-9822(02)01019-9. [DOI] [PubMed] [Google Scholar]
- 171.Strnad P, Leidel S, Vinogradova T, Euteneuer U, Khodjakov A, Gönczy P. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev Cell. 2007;13(2):203–213. doi: 10.1016/j.devcel.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Mikule K, Delaval B, Kaldis P, Jurcyzk A, Hergert P, Doxsey S. Loss of centrosome integrity induces p38-p53-p21-dependent G1-S arrest. Nat Cell Biol. 2007;9(2):160–170. doi: 10.1038/ncb1529. [DOI] [PubMed] [Google Scholar]
- 173.Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA. Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol. 2007;179(2):321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Tsang WY, Spektor A, Luciano DJ, Indjeian VB, Chen Z, Salisbury JL, Sanchez I, Dynlacht BD. CP110 cooperates with two calcium-binding proteins to regulate cytokinesis and genome stability. Mol Biol Cell. 2006;17(8):3423–3434. doi: 10.1091/mbc.E06-04-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Delaval B, Covassin L, Lawson ND, Doxsey S. Centrin depletion causes cyst formation and other ciliopathy-related phenotypes in zebrafish. Cell Cycle. 2011;10:3964–3972. doi: 10.4161/cc.10.22.18150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wood RD. DNA repair in eukaryotes. Annu Rev Biochem. 1996;65:135–167. doi: 10.1146/annurev.bi.65.070196.001031. [DOI] [PubMed] [Google Scholar]
- 177.de Laat WL, Jaspers NG, Hoeijmakers JH. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13(7):768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 178.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40(2):359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 179.Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51(2):241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 180.Selby CP, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260(5104):53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 181.Sugasawa K, Ng JM, Masutani C, Iwai S, van der Spek PJ, Eker AP, Hanaoka F, Bootsma D, Hoeijmakers JH. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol Cell. 1998;2(2):223–232. doi: 10.1016/S1097-2765(00)80132-X. [DOI] [PubMed] [Google Scholar]
- 182.Egly JM (2001) The 14th Datta Lecture. TFIIH: from transcription to clinic. FEBS Lett 498(2–3):124–128 [DOI] [PubMed]
- 183.Sugasawa K. Regulation of damage recognition in mammalian global genomic nucleotide excision repair. Mutat Res. 2010;685(1–2):29–37. doi: 10.1016/j.mrfmmm.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 184.Acu ID, Liu T, Suino-Powell K, Mooney SM, D’Assoro AB, Rowland N, Muotri AR, Correa RG, Niu Y, Kumar R, Salisbury JL. Coordination of centrosome homeostasis and DNA repair is intact in MCF-7 and disrupted in MDA-MB 231 breast cancer cells. Cancer Res. 2010;70(8):3320–3328. doi: 10.1158/0008-5472.CAN-09-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Masutani C, Sugasawa K, Yanagisawa J, Sonoyama T, Ui M, Enomoto T, Takio K, Tanaka K, van der Spek PJ, Bootsma D, et al. Purification and cloning of a nucleotide excision repair complex involving the xeroderma pigmentosum group C protein and a human homologue of yeast RAD23. EMBO J. 1994;13(8):1831–1843. doi: 10.1002/j.1460-2075.1994.tb06452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Araujo SJ, Nigg EA, Wood RD. Strong functional interactions of TFIIH with XPC and XPG in human DNA nucleotide excision repair, without a preassembled repairosome. Mol Cell Biol. 2001;21(7):2281–2291. doi: 10.1128/MCB.21.7.2281-2291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Araki M, Masutani C, Maekawa T, Watanabe Y, Yamada A, Kusumoto R, Sakai D, Sugasawa K, Ohkuma Y, Hanaoka F. Reconstitution of damage DNA excision reaction from SV40 minichromosomes with purified nucleotide excision repair proteins. Mutat Res. 2000;459(2):147–160. doi: 10.1016/S0921-8777(99)00067-1. [DOI] [PubMed] [Google Scholar]
- 188.Popescu A, Miron S, Blouquit Y, Duchambon P, Christova P, Craescu CT. Xeroderma pigmentosum group C protein possesses a high affinity binding site to human centrin 2 and calmodulin. J Biol Chem. 2003;278(41):40252–40261. doi: 10.1074/jbc.M302546200. [DOI] [PubMed] [Google Scholar]
- 189.Nishi R, Okuda Y, Watanabe E, Mori T, Iwai S, Masutani C, Sugasawa K, Hanaoka F. Centrin 2 stimulates nucleotide excision repair by interacting with xeroderma pigmentosum group C protein. Mol Cell Biol. 2005;25(13):5664–5674. doi: 10.1128/MCB.25.13.5664-5674.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Fong YW, Inouye C, Yamaguchi T, Cattoglio C, Grubisic I, Tjian R. A DNA repair complex functions as an Oct4/Sox2 coactivator in embryonic stem cells. Cell. 2011;147(1):120–131. doi: 10.1016/j.cell.2011.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.den Dulk B, van Eijk P, de Ruijter M, Brandsma JA, Brouwer J. The NER protein Rad33 shows functional homology to human Centrin 2 and is involved in modification of Rad4. DNA Repair (Amst) 2008;7(6):858–868. doi: 10.1016/j.dnarep.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 192.Chen L, Madura K. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol Cell Biol. 2002;22(13):4902–4913. doi: 10.1128/MCB.22.13.4902-4913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Rao H, Sastry A. Recognition of specific ubiquitin conjugates is important for the proteolytic functions of the ubiquitin-associated domain proteins Dsk2 and Rad23. J Biol Chem. 2002;277(14):11691–11695. doi: 10.1074/jbc.M200245200. [DOI] [PubMed] [Google Scholar]
- 194.Ortolan TG, Chen L, Tongaonkar P, Madura K. Rad23 stabilizes Rad4 from degradation by the Ub/proteasome pathway. Nucleic Acids Res. 2004;32(22):6490–6500. doi: 10.1093/nar/gkh987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Okuda Y, Nishi R, Ng JM, Vermeulen W, van der Horst GT, Mori T, Hoeijmakers JH, Hanaoka F, Sugasawa K. Relative levels of the two mammalian Rad23 homologs determine composition and stability of the xeroderma pigmentosum group C protein complex. DNA Repair (Amst) 2004;3(10):1285–1295. doi: 10.1016/j.dnarep.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 196.Ng JM, Vermeulen W, van der Horst GT, Bergink S, Sugasawa K, Vrieling H, Hoeijmakers JH. A novel regulation mechanism of DNA repair by damage-induced and RAD23-dependent stabilization of xeroderma pigmentosum group C protein. Genes Dev. 2003;17(13):1630–1645. doi: 10.1101/gad.260003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Charbonnier JB, Renaud E, Miron S, Le Du MH, Blouquit Y, Duchambon P, Christova P, Shosheva A, Rose T, Angulo JF, Craescu CT. Structural, thermodynamic, and cellular characterization of human centrin 2 interaction with xeroderma pigmentosum group C protein. J Mol Biol. 2007;373(4):1032–1046. doi: 10.1016/j.jmb.2007.08.046. [DOI] [PubMed] [Google Scholar]
- 198.Renaud E, Miccoli L, Zacal N, Biard DS, Craescu CT, Rainbow AJ, Angulo JF. Differential contribution of XPC, RAD23A, RAD23B and CENTRIN 2 to the UV-response in human cells. DNA Repair (Amst) 2011;10(8):835–847. doi: 10.1016/j.dnarep.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 199.Sugasawa K, Okuda Y, Saijo M, Nishi R, Matsuda N, Chu G, Mori T, Iwai S, Tanaka K, Hanaoka F. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 2005;121(3):387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 200.Wang QE, Praetorius-Ibba M, Zhu Q, El-Mahdy MA, Wani G, Zhao Q, Qin S, Patnaik S, Wani AA. Ubiquitylation-independent degradation of Xeroderma pigmentosum group C protein is required for efficient nucleotide excision repair. Nucleic Acids Res. 2007;35(16):5338–5350. doi: 10.1093/nar/gkm550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wang QE, Zhu Q, Wani G, El-Mahdy MA, Li J, Wani AA. DNA repair factor XPC is modified by SUMO-1 and ubiquitin following UV irradiation. Nucleic Acids Res. 2005;33(13):4023–4034. doi: 10.1093/nar/gki684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Liang L, Flury S, Kalck V, Hohn B, Molinier J. CENtrin 2 interacts with the Arabidopsis homolog of the human XPC protein (AtRAD4) and contributes to efficient synthesis-dependent repair of bulky DNA lesions. Plant Mol Biol. 2006;61(1–2):345–356. doi: 10.1007/s11103-006-0016-9. [DOI] [PubMed] [Google Scholar]
- 203.Molinier J, Ramos C, Fritsch O, Hohn B. CENtrin 2 modulates homologous recombination and nucleotide excision repair in Arabidopsis. Plant Cell. 2004;16(6):1633–1643. doi: 10.1105/tpc.021378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Weber A, Chung HJ, Springer E, Heitzmann D, Warth R. The TFIIH subunit p89 (XPB) localizes to the centrosome during mitosis. Cell Oncol. 2010;32(1–2):121–130. doi: 10.3233/CLO-2009-0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.de Boer J, Hoeijmakers JH. Cancer from the outside, aging from the inside: mouse models to study the consequences of defective nucleotide excision repair. Biochimie. 1999;81(1–2):127–137. doi: 10.1016/S0300-9084(99)80045-5. [DOI] [PubMed] [Google Scholar]
- 206.Kanai M, Uchida M, Hanai S, Uematsu N, Uchida K, Miwa M. Poly(ADP-ribose) polymerase localizes to the centrosomes and chromosomes. Biochem Biophys Res Commun. 2000;278(2):385–389. doi: 10.1006/bbrc.2000.3801. [DOI] [PubMed] [Google Scholar]
- 207.Augustin A, Spenlehauer C, Dumond H, Menissier-De Murcia J, Piel M, Schmit AC, Apiou F, Vonesch JL, Kock M, Bornens M, De Murcia G. PARP-3 localizes preferentially to the daughter centriole and interferes with the G1/S cell cycle progression. J Cell Sci. 2003;116(Pt 8):1551–1562. doi: 10.1242/jcs.00341. [DOI] [PubMed] [Google Scholar]
- 208.Smith S, de Lange T. Cell cycle dependent localization of the telomeric PARP, tankyrase, to nuclear pore complexes and centrosomes. J Cell Sci. 1999;112(Pt 21):3649–3656. doi: 10.1242/jcs.112.21.3649. [DOI] [PubMed] [Google Scholar]
- 209.Ohashi S, Kanai M, Hanai S, Uchiumi F, Maruta H, Tanuma S, Miwa M. Subcellular localization of poly(ADP-ribose) glycohydrolase in mammalian cells. Biochem Biophys Res Commun. 2003;307(4):915–921. doi: 10.1016/S0006-291X(03)01272-5. [DOI] [PubMed] [Google Scholar]
- 210.Kanai M, Tong WM, Sugihara E, Wang ZQ, Fukasawa K, Miwa M. Involvement of poly(ADP-Ribose) polymerase 1 and poly(ADP-Ribosyl)ation in regulation of centrosome function. Mol Cell Biol. 2003;23(7):2451–2462. doi: 10.1128/MCB.23.7.2451-2462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Kanai M, Tong WM, Wang ZQ, Miwa M. Haploinsufficiency of poly(ADP-ribose) polymerase-1-mediated poly(ADP-ribosyl)ation for centrosome duplication. Biochem Biophys Res Commun. 2007;359(3):426–430. doi: 10.1016/j.bbrc.2007.05.108. [DOI] [PubMed] [Google Scholar]
- 212.Okano S, Lan L, Tomkinson AE, Yasui A. Translocation of XRCC1 and DNA ligase IIIalpha from centrosomes to chromosomes in response to DNA damage in mitotic human cells. Nucleic Acids Res. 2005;33(1):422–429. doi: 10.1093/nar/gki190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Cappelli E, Townsend S, Griffin C, Thacker J. Homologous recombination proteins are associated with centrosomes and are required for mitotic stability. Exp Cell Res. 2011;317(8):1203–1213. doi: 10.1016/j.yexcr.2011.01.021. [DOI] [PubMed] [Google Scholar]
- 214.Nakanishi A, Han X, Saito H, Taguchi K, Ohta Y, Imajoh-Ohmi S, Miki Y. Interference with BRCA2, which localizes to the centrosome during S and early M phase, leads to abnormal nuclear division. Biochem Biophys Res Commun. 2007;355(1):34–40. doi: 10.1016/j.bbrc.2007.01.100. [DOI] [PubMed] [Google Scholar]
- 215.Lesca C, Germanier M, Raynaud-Messina B, Pichereaux C, Etievant C, Emond S, Burlet-Schiltz O, Monsarrat B, Wright M, Defais M. DNA damage induce gamma-tubulin-RAD51 nuclear complexes in mammalian cells. Oncogene. 2005;24:5165–5172. doi: 10.1038/sj.onc.1208723. [DOI] [PubMed] [Google Scholar]
- 216.Sivasubramaniam S, Sun X, Pan YR, Wang S, Lee EY. Cep164 is a mediator protein required for the maintenance of genomic stability through modulation of MDC1, RPA, and CHK1. Genes Dev. 2008;22(5):587–600. doi: 10.1101/gad.1627708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Pan YR, Lee EY. UV-dependent interaction between Cep164 and XPA mediates localization of Cep164 at sites of DNA damage and UV sensitivity. Cell Cycle. 2009;8(4):655–664. doi: 10.4161/cc.8.4.7844. [DOI] [PubMed] [Google Scholar]
- 218.Salisbury JL. Centrosomes: Sfi1p and centrin unravel a structural riddle. Curr Biol. 2004;14(1):R27–R29. doi: 10.1016/j.cub.2003.12.019. [DOI] [PubMed] [Google Scholar]
- 219.Azimzadeh J, Hergert P, Delouvee A, Euteneuer U, Formstecher E, Khodjakov A, Bornens M. hPOC5 is a centrin-binding protein required for assembly of full-length centrioles. J Cell Biol. 2009;185(1):101–114. doi: 10.1083/jcb.200808082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Alieva IB, Vorobjev IA. Vertebrate primary cilia: a sensory part of centrosomal complex in tissue cells, but a “sleeping beauty” in cultured cells? Cell Biol Int. 2004;28(2):139–150. doi: 10.1016/j.cellbi.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 221.Martinez-Sanz J, Yang A, Blouquit Y, Duchambon P, Assairi L, Craescu CT. Binding of human centrin 2 to the centrosomal protein hSfi1. FEBS J. 2006;273(19):4504–4515. doi: 10.1111/j.1742-4658.2006.05456.x. [DOI] [PubMed] [Google Scholar]
- 222.Trojan P, Rausch S, Giessl A, Klemm C, Krause E, Pulvermuller A, Wolfrum U. Light-dependent CK2-mediated phosphorylation of centrins regulates complex formation with visual G-protein. Biochim Biophys Acta. 2008;1783(6):1248–1260. doi: 10.1016/j.bbamcr.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 223.Azimzadeh J, Nacry P, Christodoulidou A, Drevensek S, Camilleri C, Amiour N, Parcy F, Pastuglia M, Bouchez D. Arabidopsis TONNEAU1 proteins are essential for preprophase band formation and interact with centrin. Plant Cell. 2008;20(8):2146–2159. doi: 10.1105/tpc.107.056812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127(3):635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 225.Bernardes de Jesus BM, Bjoras M, Coin F, Egly JM. Dissection of the molecular defects caused by pathogenic mutations in the DNA repair factor XPC. Mol Cell Biol. 2008;28(23):7225–7235. doi: 10.1128/MCB.00781-08. [DOI] [PMC free article] [PubMed] [Google Scholar]