Abstract
Melanopsin (OPN4) is an opsin photopigment that, in mammals, confers photosensitivity to retinal ganglion cells and regulates circadian entrainment and pupil constriction. In non-mammalian species, two forms of opn4 exist, and are classified into mammalian-like (m) and non-mammalian-like (x) clades. However, far less is understood of the function of this photopigment family. Here we identify in zebrafish five melanopsins (opn4m-1, opn4m-2, opn4m-3, opn4x-1 and opn4x-2), each encoding a full-length opsin G protein. All five genes are expressed in the adult retina in a largely non-overlapping pattern, as revealed by RNA in situ hybridisation and immunocytochemistry, with at least one melanopsin form present in all neuronal cell types, including cone photoreceptors. This raises the possibility that the teleost retina is globally light sensitive. Electrophysiological and spectrophotometric studies demonstrate that all five zebrafish melanopsins encode a functional photopigment with peak spectral sensitivities that range from 470 to 484 nm, with opn4m-1 and opn4m-3 displaying invertebrate-like bistability, where the retinal chromophore interchanges between cis- and trans-isomers in a light-dependent manner and remains within the opsin binding pocket. In contrast, opn4m-2, opn4x-1 and opn4x-2 are monostable and function more like classical vertebrate-like photopigments, where the chromophore is converted from 11-cis to all-trans retinal upon absorption of a photon, hydrolysed and exits from the binding pocket of the opsin. It is thought that all melanopsins exhibit an invertebrate-like bistability biochemistry. Our novel findings, however, reveal the presence of both invertebrate-like and vertebrate-like forms of melanopsin in the teleost retina, and indicate that photopigment bistability is not a universal property of the melanopsin family. The functional diversity of these teleost melanopsins, together with their widespread expression pattern within the retina, suggests that melanopsins confer global photosensitivity to the teleost retina and might allow for direct “fine-tuning” of retinal circuitry and physiology in the dynamic light environments found in aquatic habitats.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-011-0785-4) contains supplementary material, which is available to authorized users.
Keywords: Vertebrate, Zebrafish, Retina, Opsin, Melanopsin, Opn4
Introduction
Light detection is fundamental to the survival of most organisms. The most familiar function of photoreception is to build an image of the world using cone and rod photoreceptors located within the retina. The eye, however, also contains dedicated irradiance detectors, which are used to adjust a broad range of physiological responses to light, including the regulation of sleep, circadian rhythms, melatonin and pupil constriction [1–7]. This second function of the eye has been best studied in mammals and has been shown to be mediated by a small subset of retinal ganglion cells (RGCs) that express the non-ciliary m-class of melanopsin (OPN4), a photopigment that displays characteristic invertebrate-like bistable biochemistry [8–11]. In all mammalian species that have been studied, melanopsin exists only as long and short splice variants of a single gene [12–14]. By contrast, non-mammalian vertebrates possess two evolutionary lineages of the melanopsin gene family, opn4m and opn4x [13, 15], which show varied expression within the retina, including the ganglion cell layer of the eye, retinal pigment epithelium, iris, brain and skin [15–24]. In particular, expression of a subset of opn4m genes was found in the horizontal cells of the zebrafish by reverse-transcription polymerase chain reaction (RT-PCR) and RNA in situ hybridisation (RNA ISH) [15, 21]. However, the specific function of melanopsin in these tissues is largely unknown, and there is no information relating to why the melanopsins exhibit such apparent diversity within a species, as well as across species. For example, in the non-mammalian vertebrates, it is unclear how many different forms of melanopsin are present within a single species; whether variant forms of melanopsin have a tissue-specific pattern of expression; if all are capable of signalling light; and if the signalling pathways of different melanopsins are conserved. Such fundamental questions regarding melanopsin photobiology need to be addressed if the role of this new photopigment system is to be more broadly understood across the vertebrates. To address these issues, we have examined the expression and functional properties of melanopsin photopigments in the zebrafish (Danio rerio). Our results show that zebrafish possess five different forms of melanopsin (encoded by five distinct opn4 genes) that are expressed in different classes of retinal neurons. Furthermore, we show that the functional properties of these melanopsins differ markedly with regards to their chromophore-handling biochemistry. Beyond the surprising finding that all retinal neurons seem to have the potential for intrinsic photosensitivity, these results show that the melanopsin class of photopigments exhibit a much greater level of functional diversity than previously considered, and raise critical questions regarding sensory ecology and the physiological importance of irradiance detection.
Materials and methods
Gene mining and cloning of five melanopsins expressed in zebrafish tissues
The long isoforms of chicken melanopsin, OPN4M (accession number EU124632) and OPN4X (accession number EU124630), were used as bait to search the zebrafish genome (e!Ensembl zebrafish Zv8 assembly, http://www.ensembl.org/Danio_rerio/Info/Index) for opn4-like genes. Zebrafish adult ocular tissues were dissected at 3 h post light onset [zeitgeber 3 (ZT3), where ZT0 is defined as lights on] from animals kept on a constant 12/12 h light/dark cycle and homogenised in TRIzol® Reagent (Invitrogen, UK), and total RNA was isolated according to the manufacturer’s instructions based on the method of Chomczynski and Sacchi [25]. A sample of total RNA (1–2 µg) was primed with 500 ng of oligo-dT12 primer and converted to single-stranded complementary DNA (cDNA) using Superscript™ III reverse transcriptase (Invitrogen, UK), following the protocol provided by the manufacturer. Polymerase chain reaction (PCR) amplification of five full-length zebrafish opn4 sequences was performed using cDNA generated from ocular tissue, with each PCR containing the following components: 33 μl sterile water, 5 μl (10×) KOD DNA polymerase buffer, 2 μl (25 mM) MgSO4, 5 μl (2 mM) dNTP mix, 1 μl (1 U) KOD DNA polymerase, (Novagen, UK), 1.5 μl of each forward and reverse primer (0.3 μM) (Supplementary Material Table 1), and 1 μl template (40 ng). The PCR was conducted under the following thermal cycling conditions: an initial denaturation cycle at 94°C for 2 min and 40 cycles of 94°C for 15 s, 60°C for 30 s and 72°C for 1 min. Amplicons were visualised by staining with ethidium bromide on a 2% (w/v) agarose gel, and single distinct PCR products of the expected size were excised and purified using the QIAquick Gel Extraction kit (Qiagen, UK) by following the manufacturer’s instructions. Each amplicon was subsequently digested overnight at 37°C with EcoRI and SalI restriction enzymes (1 unit of each), subjected to electrophoresis, excised, purified and ligated into the pMT4 mammalian expression vector containing predigested complementary ends for cloning. All cloned inserts were sequenced by Source BioScience (Oxford, UK) in both orientations using the appropriate primers (see Supplementary Material).
Phylogenetic analysis
Five zebrafish melanopsin (opn4m-1, opn4m-2, opn4m-3, opn4x-1, opn4x-2) cDNA sequences were compared with visual and non-visual sequences across a variety of vertebrates (from teleosts to primates) and a Branchiostoma belcheri (lancelet) melanopsin (accession no. AB205400) outgroup to produce a sequence alignment generated by ClustalW [26], which was manually manipulated. Phylogenetic analysis was performed using the MEGA Version 4.0.2 computer package [27] on a codon-matched nucleotide alignment by applying a Kimura 2-parameter substitution matrix [28] to produce a neighbour-joining phylogenetic tree, bootstrapped with 1,000 replicates.
RNase protection assay
Zebrafish adult retinal total RNA was extracted in TRIzol® Reagent (Invitrogen, UK) following the manufacturer’s instructions. Partial sequences of the five zebrafish opn4 cDNAs were cloned into pGEM-T Easy (Promega, UK) using standard molecular biology techniques. Each vector was linearised and used as a template for riboprobe synthesis (Promega, UK) according to the manufacturer’s instructions. RNase protection assays, including tRNA (negative) controls, were carried out as previously described [29].
RNA in situ hybridisation
Whole zebrafish eyes were collected at ZT3 and immediately fixed in 4% paraformaldehyde (PFA) (Pierce, UK) in phosphate-buffered saline (PBS) [140 mM NaCl, 3 mM KCl, 10 mM PO4 buffer) (pH 7.0) (Invitrogen, UK)] for 12 h. Eyes were cryoprotected in 30% sucrose for 48 h prior to embedding in OCT (VWR, UK) and frozen at −80°C. Cryostat sections (18 μm) of zebrafish adult ocular tissue were placed on Superfrost Plus microscope slides (VWR, UK), permeabilised with proteinase K (1 μg/ml) (Sigma, UK) for 5 min at room temperature (RT) and acetylated for 10 min at RT prior to prehybridisation with ULTRAhyb® Ultrasensitive Hybridisation Buffer (Ambion, UK) for at least 1 h at RT. All five full-length zebrafish opn4 cDNA sequences were subcloned into pBluescript II SK plasmid (Stratagene, UK) via EcoRI and SalI restriction enzyme sites and linearised prior to generating riboprobes using the SP6/T7 DIG RNA Labelling Kit (Roche, UK). These digoxigenin (DIG)-labelled riboprobes were mixed in preheated ULTRAhyb® Ultrasensitive Hybridisation Buffer (Ambion, UK) and used to replace the prehybridisation solution present on the pre-treated slides before overnight incubation at 68°C in a humidified chamber. After 12 h, the slides were washed twice in 0.2× saline-sodium citrate (SSC) buffer (Invitrogen, UK) at 72°C prior to blocking in PBS with 10% heat-inactivated goat serum (Sigma, UK) for 1 h at RT. After blocking, the slides were incubated overnight at 4°C with anti-digoxigenin antibody conjugated to alkaline phosphate (AP) (Roche, UK) prior to the detection of hybridised probes using the BCIP/NBT AP Substrate Kit IV (Vector Laboratories, UK) according to the manufacturer’s instructions. Slides were mounted in Glycergel Mounting Medium (Dako, UK) and imaged with a 40× (0.8 NA) water-immersion lens using a Nikon E1000 microscope (Nikon, UK) connected to a digital camera (Jenoptik, Germany) operated by Openlab software (Improvision, UK).
Quantitative PCR of melanopsin mRNA in adult zebrafish tissues
Total RNA was extracted from zebrafish adult eyes (n = 6) and converted to cDNA as described earlier. Removal of the initial RNA strand from the cDNA sample was achieved by digestion with RNase H (New England Biolabs, UK) at 37°C for 15 min and purification through a silica-membrane column using the PCR Purification Kit according to the manufacturer’s instructions (Qiagen, UK). For quantitative PCR (qPCR) analysis, individual melanopsin transcripts were quantified using gene-specific forward and reverse primers (30-mers) designed specifically to amplify individual opn4 mRNAs (Supplementary Material Table 2). In addition, forward and reverse primers were designed to amplify transcripts of two internal control genes, 60S ribosomal protein l13 alpha (rpl13a) and elongation factor 1 alpha (ef1a), where the geometric mean was used to normalise target gene expression and correct for sample-to-sample variation [30]. The specificity of each primer was assessed, and the production and validation of qPCR protocols were performed as previously described (see Supplementary Material). All primer combinations traversed at least one exon-exon boundary and produced amplicons of less than 150 base pairs. All standard curves (semi-log plot of PCR cycle value for above background threshold value against log of input DNA concentration) gave line gradients very close to −3.32 with amplification efficiencies close to 100% when analysed over five orders of magnitude. Assays were performed in triplicate with at least five independent cDNA templates (40 ng), using 1× SYBR Green qPCR Master Mix (Applied Biosystems, UK) and forward and reverse primers at 0.2 μM. A StepOne™ Real-Time PCR Detection system was used to monitor SYBR Green reporter dye fluorescence and included an initial denaturation step at 95°C for 10 min, followed by 40 cycles of 95°C for 10 s (denaturation), 60°C for 20 s (annealing) and 75°C for 20 s (extension), with a plate read at the end of the extension phase for each cycle. All data were analysed offline, using a method adapted from Carleton and Kocher [31], where the relative expression (RE) of melanopsin (REmel) compared to that of an internal control (REint) was calculated as follows:
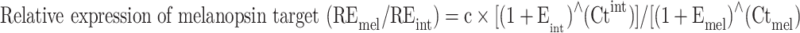 |
where Emel and Eint are the primer efficiencies of each individual melanopsin target and the internal control (geometric mean of rpl13a and ef1a), respectively, and Ctmel and Ctint are the experimentally determined cycle threshold values for each individual melanopsin target and the internal control (geometric mean of rpl13a and ef1a), respectively. In all cases, an arbitrary multiplication constant (c) of 10,000 was used.
Immunocytochemistry of opn4m in the zebrafish adult retina
A rabbit polyclonal antibody (pas350) that recognises a highly conserved region of vertebrate mammalian-like melanopsins (opn4m), including zebrafish opn4m-1, opn4m-2 and opn4m-3, was raised against a 13 amino acid synthetic peptide CVPFPTVDVPDHA conjugated to keyhole limpet hemocyanin (KLH) by Harlan (UK) and characterised according to standard procedures (see Supplementary Material). Whole zebrafish eyes were collected and fixed as described above. Immunocytochemistry was performed using standard techniques as previously described (see Supplementary Material). Briefly, 18 μm cryostat sections were permeabilised in PBS with 0.2% Triton X-100 for 20 min at room temperature (RT) and blocked in PBS with 10% normal donkey serum (Sigma, UK) for 1 h at RT. Rabbit polyclonal anti-melanopsin antibody (pas350) was diluted 1:400 in PBS with 2.5% serum and 0.2% Triton X-100, and incubated for 16 h at 4°C. Alexa Fluor 555 donkey anti-rabbit secondary antibody (Life Technologies, UK) was diluted 1:200 in PBS with 2.5% serum and 0.2% Triton X-100 for 2 h at RT. All wash steps were performed with PBS/0.05% Tween-20. Sections were mounted with Prolong Gold anti-fade media containing 4′,6-diamidino-2-phenylindole (DAPI) (Life Technologies, UK). Images were acquired using Metamorph image acquisition software (Molecular Devices, UK). Excitation and emission filters for DAPI, green and red fluorescence (Chroma, UK) were, 350, 480, 545 and 460, 525 and 600 nm, respectively.
Promoter analysis of all five zebrafish melanopsin genes
As mapping of the transcription start site (TSS) is a prerequisite to promoter identification, the major TSS for each melanopsin transcript was determined by a combination of screening an adult zebrafish ocular 5′-Rapid Amplification of cDNA Ends (5′-RACE) (cDNA) library (Clontech, France) for full-length clones, comparison with zebrafish expressed sequence tag (EST) databases (e.g. http://www.ncbi.nlm.nih.gov/genome/seq/BlastGen/BlastGen.cgi?taxid=7955 and http://www.ensembl.org/Danio_rerio/blastview) and bioinformatics (e.g. http://www.fruitfly.org/seq_tools/promoter.html, with settings of ‘eukaryote’ as organism and a cutoff score value of >0.8). Once identified, the flanking region of the TSS was compared to the consensus initiator (INR) sequence (YYCANWYY), where A represents the TSS [32]. Once mapped, a genomic region of 3 kb upstream of the TSS was subjected to promoter analysis. Initially, CpG islands [33] (http://zeus2.itb.cnr.it/cgi-bin/wwwcpg.pl) and CCCTC-binding factor (CTCF) insulator elements [34, 35] (identified by manual comparison to the consensus sequence of CCGCNNGGNGGCAG [36] and a cutoff score value of >0.7) were mapped as indicators that these putative regions were consistent with being under gene regulation. Proximal core transcription elements within 200 bp upstream of the major TSS, such as TATA boxes, were determined (http://zeus2.itb.cnr.it/~webgene/wwwHC_tata.html) and manually compared with the TATA box consensus sequence (TATAWAWN [37]). Other factors involved in general transcription [38, 39] were also determined within the flanking 200 bp upstream of the TSS and the downstream 5′-untranslated region (5′-UTR): CAAT/CCAAT-box (consensus sequence GGNCAATCT [40]); Downstream Promoter Element (DPE) (consensus sequence RGWCGTG [41]); Motif Ten Element (MTE) (consensus sequence CSARCSSAACG [42]); B Recognition Element (BRE) (consensus sequence SSRCGCC [43]); and GC box (consensus sequence KRGGCGKRRY [44]). Analyses to determine the presence of putative binding sites (cis-elements) for known vertebrate transcription factors [(e.g. those involved in general transcription, such as ap-1 (consensus sequence TGASTCA [45]), as well as those involved in ocular development [46] (e.g. paired box gene 6 (pax-6)) and retinoid signalling [47] (e.g. retinoic acid receptor (rar), retinoid x receptor (rxr))] in the flanking 3 kb promoter region (sense and antisense strands) of each zebrafish melanopsin gene were performed by using the MatrixTM Version 1.0 database (with a cutoff filter to minimize false positives) (http://www.gene-regulation.com/cgi-bin/pub/programs/match/bin/match.cgi) and NSITE: Animal TFD from Ghosh database (http://linux1.softberry.com/berry.phtml?topic=nsite&group=programs&subgroup=promoter). To complement the data obtained from bioinformatics as discussed above, a number of putative binding sites for retinal-specific transcription factors [46, 48] that are not presently included in the NSITE or TRANSFAC transcription factor (http://www.gene-regulation.com/pub/databases/transfac/doc/toc.html) databases were investigated by manual comparison of both sense and antisense genomic regions with binding motif consensus sequences and applying a cutoff score value >0.7. These include: cone photoreceptor regulatory element-1 (cpre-1) (consensus sequence CTGGAGTGWTGGARGCAGGGST [49]); neural retina leucine zipper (nrl) [consensus sequence TGAN6–8GCA [50], where the N6–8 core resembles the binding site for ap-1 (consensus sequence TGASTCA [45]) or a 3′-5′-cyclic adenosine monophosphate (cAMP) response element (TGAGCTCA [51, 52])]; nuclear receptor subfamily 2 group E member 3 (nr2e3) (consensus sequence RAGRTCAAARRTCA [53])]; rar-related orphan receptor beta (rorβ) (consensus sequence WWAWBTAGGTCA [54]) (also employed in circadian gene regulation [55]); cone-rod homeobox (crx) (consensus sequence TAATCA [56]) and crx/orthodenticle homeobox-2 (crx/otx-2) (consensus sequence TYTAATCC [57]); retina-specific region-1/photoreceptor conserved element-1 (ret-1/pce-1) (consensus sequence CAATTAG [58]) and orthodenticle homeobox/bovine AT-rich sequence-1 (otx/bat-1) (consensus sequence TGATTAG [58]); retina-specific region-4 (ret-4) (consensus sequence GCTTAG [59]); glass-like binding motif (consensus sequence ACCCTTGAAATGCC [60]); and Caenorhabditis elegans homeobox-10 (ceh-10) containing homolog (chx10) (consensus sequence YTAATTRR [61]). Similarly, each upstream 3 kb melanopsin promoter region (sense and antisense strands) was investigated for the presence of putative d-box (consensus sequence RTTAYGTAAY [62] with a cutoff score value >0.75) and e-box (consensus sequences CACGTG [62] and CACGTT [62]) motifs known to be important in circadian regulation of rhythmic clock genes.
Transfection of zebrafish opn4 constructs in cell culture
Neuro-2a cells (ECACC) were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) (Sigma, UK), supplemented with 10% foetal bovine serum (FBS) (Life Technologies, UK), 2 mM l-glutamine (Sigma, UK) and 1% (v/v) penicillin/streptomycin (Sigma, UK) in a humidified chamber at 37°C with 5% CO2. Cells were fed fresh media every 48–60 h and passaged prior to reaching confluence. Cells were trypsinised, diluted and cultured at 1 × 105 cells per ml in 90 mm dishes for 24 h before transfection. Transient transfections were performed with 1D4 plasmid vectors containing full-length opn4 sequences, using GeneJuice transfection reagent (Novagen, UK) following standard techniques (see Supplementary Material). In all cases, each melanopsin plasmid was co-transfected with a control pSIREN-DNR-DsRed-Express vector (Clontech, UK) for cell identification prior to electrophysiology. For single transfections, the DNA:GeneJuice ratio was 1 μg DNA per 3 μl GeneJuice. For double transfections, equal amounts of DNA were used. The day after transfection, cell differentiation was induced using 20 μM retinoic acid (Tocris, UK), and all cells were kept in complete darkness until electrophysiological analysis was performed 48–60 h post differentiation.
Whole-cell electrophysiology
The use of 11-cis retinal (the predominantly utilised native chromophore) and 9-cis retinal (commercially available analogue) isomers are interchangeable with regards to the activation of photopigments. However, the peak spectral sensitivity of pigments bound to 9-cis retinal is blue-wavelength shifted compared to those that form stable interactions with native 11-cis retinal. Thus, 9-cis retinal was used for all electrophysiological experiments, where large amounts of chromophore were required, whereas spectral sensitivity was determined by ultraviolet-visible (UV-vis) spectrophotometry with photopigments conjugated to 11-cis retinal.
Transfected cells were identified by mCherry fluorescence using a Zeiss Axioskop FS2 microscope. 20 μM 9-cis retinal (Sigma, UK) or all-trans retinal (Sigma, UK) was added to the perfusion solution (140 mM NaCl, 4 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 5 mM glucose, 10 mM HEPES; pH 7.3–7.4, 24°C), and cells were kept in the dark for at least 1 h prior to recording. Whole-cell patch-clamp recordings were made with pipettes containing 140 mM KCl, 10 mM NaCl, 1 mM MgCl2, 10 mM HEPES and 10 mM EGTA. Osmolarity was adjusted to 285 ± 5 mosmol/l and pH to 7.3–7.4 with KOH. Open pipette resistance was 3.5–5 MΩ, and access resistance during recordings was less than 20 MΩ. Currents were recorded (Axopatch 200B, Axon Instruments, USA) in neurons that were voltage-clamped at holding potentials of −50 mV. The records were filtered at 1 kHz and sampled at 20 kHz. Light stimuli were generated using a Cairn Optoscan Xenon arc source comprising a slit monochromator. All stimuli were 10 s in duration with a 20 nm half-bandwidth. Irradiance was measured using an optical power metre (Macam Photometrics, UK) and converted to photon flux. The magnitude of the responses was defined by the peak sustained current measured using Clampfit (Axon Instruments, USA).
Expression of recombinant opsins
Zebrafish opn4m-2 and opn4x-1 were chosen as interesting representatives of each of the two melanopsin lineages, and both full-length cDNAs were cloned into pMT4 expression vector as previously described (see Supplementary Material). Each plasmid containing zebrafish melanopsin (210 µg DNA) was added to 12 140 mm plates containing human embryonic kidney (HEK293T) cells and transfected with GeneJuice transfection reagent (Novagen, UK) at a DNA:GeneJuice ratio of 3:1. After 48 h post-transfection, the transfected cells were harvested and washed four times with PBS. Independent experiments of both opn4 pigments were regenerated in excess 11-cis retinal (generous gift from Dr. Rosalie Crouch of the Storm Eye Institute, Medical University of South Carolina, USA) or all-trans retinal (Sigma, UK) in the dark. One percent (w/v) dodecyl-maltoside (DDM) and 20 mg/ml phenylmethanesulphonylfluoride (PMSF) was then added before passage over a CNBr-activated Sepharose-binding column coupled to an anti-1D4 monoclonal antibody (see Supplementary Material). Absorbance spectra were recorded in triplicate in the dark using a Shimadzu UV-visible spectrophotometer (UV-2550) (Shimadzu, UK). Pigments were bleached by exposure to light for 1 h before a second spectrum was recorded. The peak sensitivity value, the lambda max (λmax), for each pigment was determined by subtracting the bleached spectrum from the dark spectrum for each pigment to produce a difference spectrum. This was then fitted to a standard Govardovskii rhodopsin A1-template [63] with a bleached retinal curve subtracted using an Excel spread sheet to determine the λmax.
Results
Gene identification and phylogenetic analysis of zebrafish melanopsins
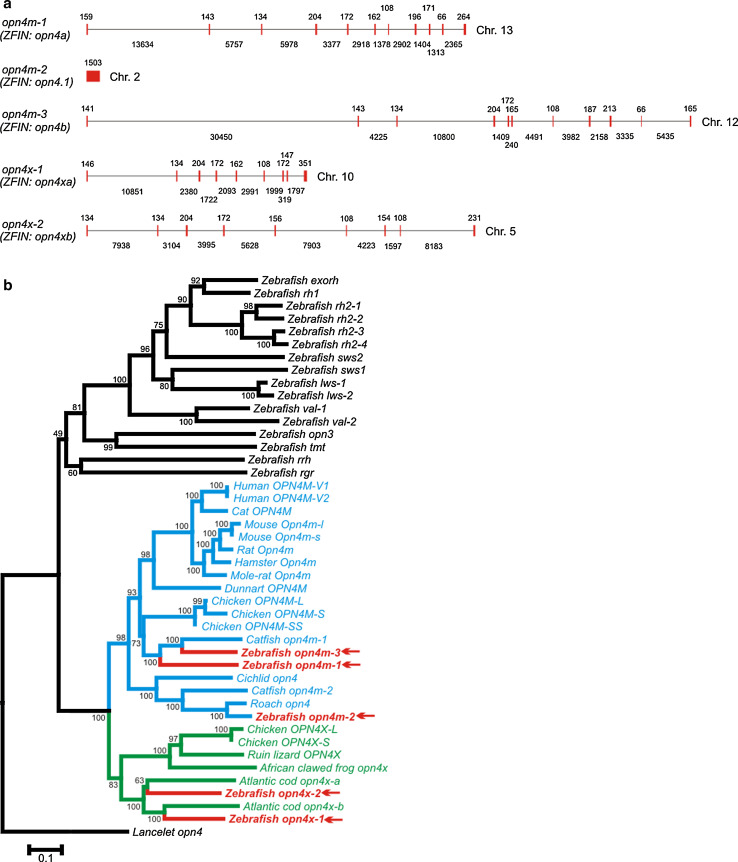
Using a bioinformatics approach and the long isoforms of chicken OPN4M (accession no. EU124632) and OPN4X (accession no. EU124630) as bait, we identified five opn4-like genes (Fig. 1a) in the zebrafish genome, which are consistent with previously predicted partial zebrafish melanopsin-like genes [64]. These melanopsin genes clustered into two groups: those comprising 11 exons (opn4m-like) and those with 9 exons (opn4x-like), with a fifth previously isolated opn4m-like gene completely devoid of introns [15, 21]. This fifth gene may have arisen from an ancient retrotransposon event that reinserted a mature, processed mRNA into the teleost genome, similar to the intronless rod opsin gene of teleosts [65, 66]. Phylogenetic analyses of both nucleotide and amino acid opn4 sequences were consistent and confirmed that three of these melanopsin genes are orthologues of the opn4m class (opn4m-1 on chr. 13, opn4m-2 on chr. 2, and opn4m-3 on chr. 12), and two are orthologues of the opn4x group (opn4x-1 on chr. 10 and opn4x-2 on chr. 6) (Fig. 1b). The full-length sequences for all five opn4 transcripts (GenBank accession nos.: GQ925715-GQ925719) were confirmed by PCR amplification from zebrafish retinal cDNA (Supplementary Material Table 1). These sequences are predicted to produce rhodopsin-like photopigments of the G protein-coupled receptor superfamily based on the presence of seven putative transmembrane domains and all the critical amino acids shown to be important for structural integrity and function, including a conserved lysine at residue 296 for the attachment of the retinal chromophore (Supplementary Material Fig. 1).
Fig. 1.
Zebrafish melanopsin gene structures and phylogeny. a Genomic structure of five zebrafish melanopsin (opn4) genes. b Phylogenetic analyses of visual and non-visual photosensory pigments from representative chordates based on a codon-matched nucleotide alignment. The data show the relative positioning of the five zebrafish melanopsins (red arrow) into the two main clades (m-class and x-class) of melanopsin, with three m-class genes (opn4m-1, opn4m-2, and opn4m-3) and two x-class genes (opn4x-1 and opn4x-2). In the ZFIN database (http://zfin.org), these genes are known as opn4a (opn4m-1), opn4.1 (opn4m-2), opn4b (opn4m-3), opn4xa (opn4x-1) and opn4xb (opn4x-2). However, the nomenclature used throughout this manuscript is consistent with the historical discovery of melanopsin genes in the zebrafish and other teleosts. The degree of support for internal branching is shown as a percentage at the base of each node, and the scale bar indicates the number of nucleotide substitutions per site. The opsin sequences used for generating the tree are as follows: (a) exorhodopsin (exo-rod): zebrafish (Danio rerio), NM131212; (b) rod opsin (rh1): zebrafish (Danio rerio), NM131084; (c) rod opsin-like 2 (rh2): zebrafish (Danio rerio), NM131253 (Rh2-1), NM182891 (Rh2-2), NM182892 (Rh2-3), NM131254 (Rh2-4); (d) short-wavelength-sensitive 2 (sws2): zebrafish (Danio rerio), NM131192; (e) short-wavelength-sensitive 1 (sws1): zebrafish (Danio rerio), NM131319; (f) long-wavelength-sensitive/middle-wavelength-sensitive (lws/mws): zebrafish (Danio rerio), NM131175 (lws-1), NM001002443 (lws-2); (g) vertebrate ancient (va) opsin: zebrafish (Danio rerio), AB035276 (va-1), AY996588 (va-2); (h) panopsin (opn3): zebrafish (Danio rerio), NM001111164; (i) teleost multiple tissue (tmt) opsin: zebrafish (Danio rerio), BC163681; (j) retinal pigment epithelium-specific rhodopsin homolog (rrh) (peropsin): zebrafish (Danio rerio), NM001004654; (k) retinal G protein-coupled receptor (rgr): zebrafish (Danio rerio), NM001017877; (l) mammalian-like melanopsin (opn4m): human (Homo sapiens), NM033282 (OPN4V1), NM001030015 (OPN4V2); cat (Felis catus), AY382594; mouse (Mus musculus), EU303118 (Opn4m-l), EU303117 (Opn4m-s); rat (Rattus norvegicus), NM138860; hamster (Phodopus sungorus), AY726733; mole-rat (Spalax ehrenbergi), AM748539; dunnart (Sminthopsis crassicaudata), DQ383281; chicken (Gallus gallus), EU124632 (OPN4M-L), EU124633 (OPN4M-S), EU124634 (OPN4M-SS); catfish (Ictalurus punctatus), FJ839437 (opn4m-1), FJ839438 (opn4m-2); roach (Rutilus rutilus), AY226847; cichlid (Astatotilapia burtoni), EU523855; zebrafish (Danio rerio), GQ925715 (opn4m-1), GQ925716 (opn4m-2); GQ925717 (opn4m-3); (m) xenopus-like melanopsin (opn4x): chicken (Gallus gallus), EU124630 (OPN4X-L), EU124631 (OPN4X-S); lizard (Podarcis siculus), DQ013043; African clawed frog (Xenopus laevis), AF014797; cod (Gadus morhua), AF385823 (opn4x-1), AY126448 (opn4x-2); zebrafish (Danio rerio), GQ925718 (opn4x-1), GQ925719 (opn4x-2); (n) chordate melanopsin (opn4): lancelet (Branchiostoma belcheri), AB205400 (outgroup). The gene nomenclature used follows the guidelines adopted by the Entrez Gene database (http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene). In brief, the genes of all terrestrial species are in upper case, except for the mouse and rat, where only the first letter is capitalized. The genes of all aquatic species, including amphibians, are in lower case
Expression and location of five zebrafish melanopsins
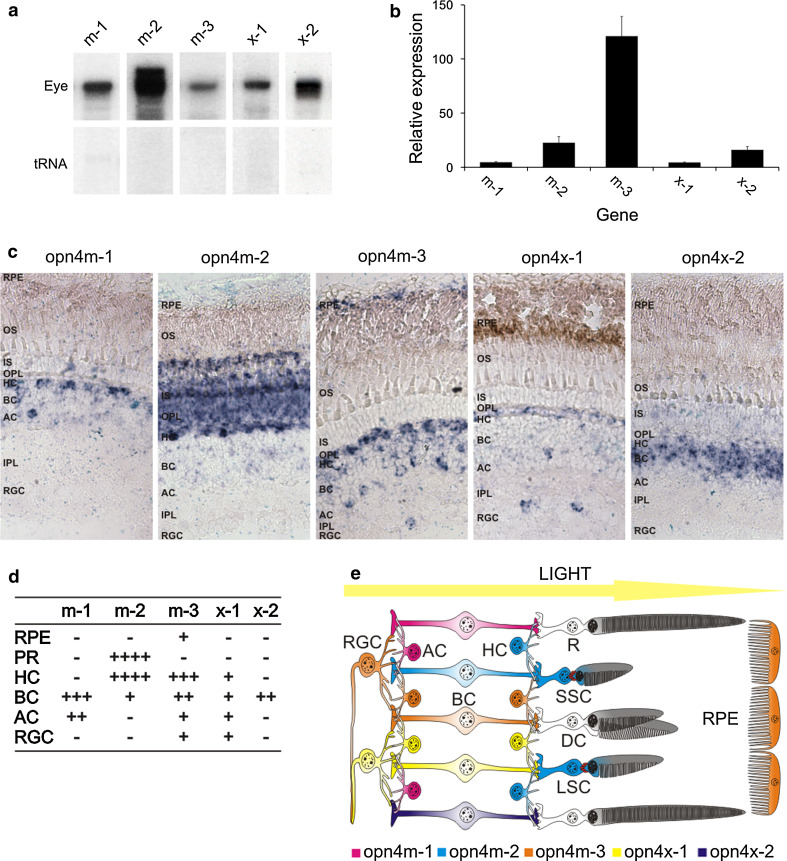
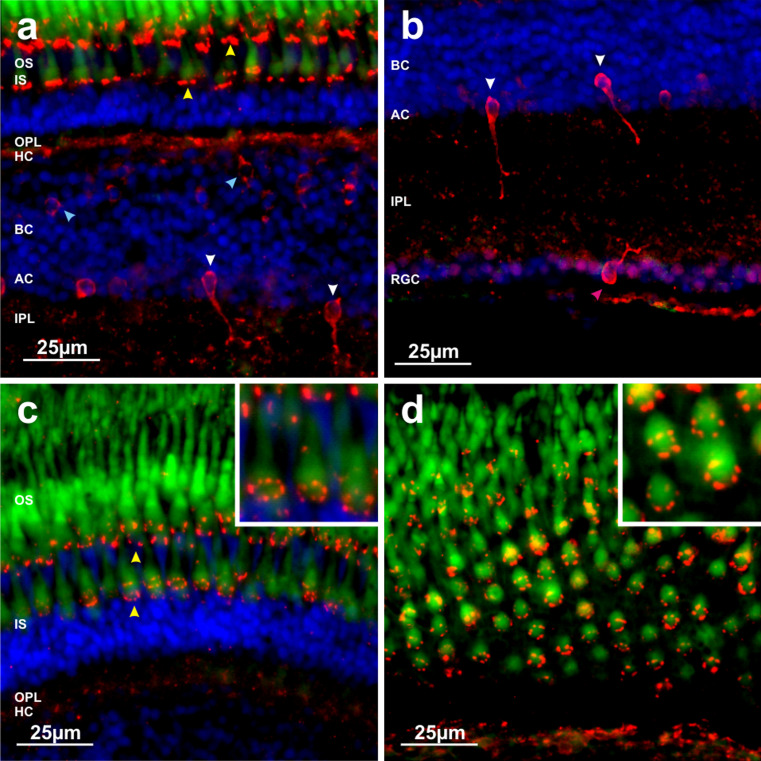
Melanopsin expression was examined by RNase protection assay, quantitative PCR and RNA ISH, where all five opn4 transcripts were shown to be expressed in the adult zebrafish eye (Fig. 2). Overall, opn4m-3 was the most abundant transcript, followed by opn4m-2, opn4x-2, opn4x-1, with opn4m-1 being the least abundant (Fig. 2b). To localise melanopsin expression within the retina, RNA ISH was performed on adult eye sections, where each opn4 transcript exhibited a characteristic, but partially overlapping profile of expression (Fig. 2c, d). All five transcripts were detected in the inner nuclear layer, with horizontal cells expressing predominantly opn4m-2 and to a lesser extent opn4m-3 and opn4x-1, with no mRNA being detected for opn4m-1 and opn4x-2. In bipolar cells, opn4m-1 was strongly expressed, with the other four transcripts being detected to a lesser degree. In amacrine cells, low levels of expression were observed for opn4m-1, opn4m-3 and opn4x-1. In a small subset of retinal ganglion cells (RGCs), opn4x-1 and opn4m-3 were the only melanopsin transcripts to be detected, though it is unclear whether these two transcripts are co-expressed in the same cell. The majority of RGCs did not express any melanopsin isoform. In the retinal pigment epithelium, opn4m-3 was the only melanopsin expressed (Fig. 2c, d). Interestingly, a high level of opn4m-2 expression was also detected in the cell bodies of the photoreceptor layer, demonstrating that melanopsin expression extends across the whole retina and includes all neuronal cell types (Fig. 2e). A previous RNA ISH study using a short riboprobe [21] also showed opn4m-2 expression (relatively low) in horizontal cells, but failed to detect opn4m-2 transcripts in the outer nuclear layer, as shown here. The full-length riboprobe used in the present study is ~3-fold longer and presumably more sensitive, and as such, may account for this discrepancy. Retinal expression of the m-class of melanopsin was examined in further detail by immunocytochemistry (ICC), using an antibody (pas350) that specifically recognises the amino terminus of all three opn4m proteins (Supplementary Material Fig. 2). The cellular distribution of opn4m protein determined by ICC was consistent with the expression profile shown by RNA ISH, confirming that opn4m isoforms were present in all cell types of the inner nuclear layer (horizontal, bipolar and amacrine cells), a subpopulation of RGCs, as well as in cone photoreceptors (Fig. 3 and Supplementary Material Figs. 3–4). Significantly, ICC showed that opn4m protein was expressed in all single short and long cone photoreceptors (that express sws1 and sws2, respectively), but was not detected in double cones (that express lws and rh2). At higher magnification, melanopsin immunolabelling appeared as discrete punctate spots in ring-like structures around the base of the megamitochondrial ellipsoid of the inner segment of both types of single cones. Closer inspection confirmed that the spatial arrangement of opn4m-2 protein across the adult zebrafish retina (Fig. 3d) was consistent with the diamond-like mosaic patterning of both single short and long photoreceptors, but not the hexagonal-like mosaic patterning of double cones [67].
Fig. 2.
Expression of five melanopsin genes (opn4m-1, opn4m-2, opn4m-3, opn4x-1, opn4x-2) in the adult eye of zebrafish. a RNase protection assay and b quantitative PCR (qPCR) analyses showing the presence of all five zebrafish melanopsins in the adult eye. c RNA in situ hybridisation (RNA ISH) of the five melanopsin genes and d summary table showing a global pattern of melanopsin expression across all the main cell types in the adult retina, with each isoform exhibiting a discrete expression profile. e Schematic representation of the zebrafish retina showing the location of melanopsin expression as a function of each opn4 isoform. The presence of opn4m protein located at the base of the ellipsoid of the single cone inner segments is indicated by a ring of red ellipses (see Fig. 2). All qPCR data are shown as mean ± standard error of the mean (SEM) with significance being indicated as p < 0.01 (for the following Student’s t test comparisons: opn4m-1 and opn4m-3; opn4m-2 and opn4m-3; opn4m-3 and opn4x-1; and opn4m-3 and opn4x-2) and p < 0.05 (for the following Student’s t test comparisons: opn4m-1 and opn4m-2; opn4m-1 and opn4x-2; opn4m-2 and opn4x-1; and opn4x-1 and opn4x-2). Retinal pigment epithelium, RPE; rod photoreceptor, R; double cone photoreceptor, DC; long single cone photoreceptor, LSC; short single cone photoreceptor, SSC; outer segment, OS; inner segment, IS; outer plexiform layer, OPL; horizontal cell, HC; bipolar cell, BC; amacrine cell, AC; inner plexiform layer, IPL; retinal ganglion cell, RGC
Fig. 3.
Immunofluorescent labelling of the zebrafish adult retina with an anti-melanopsin (m-form) antibody (pas350). a Image of the outer retina showing melanopsin (m-form) labelling of multiple cell types. Intense punctate spots of labelling are observed that form a distinct and highly organised ring-like pattern that is localised to the base of the megamitochondrial ellipsoid of the inner segment (IS) (probably within the myoid region) of long and short single cone photoreceptors (yellow arrow). Melanopsin immunoreactivity is also observed for horizontal cells (HC), bipolar cells (BC) (blue arrow) in the outer layers of the inner nuclear layer (INL) and also a subset of amacrine cells (AC) (white arrow) located in the INL, close to the border of the inner plexiform layer (IPL). b Image of the inner retina showing melanopsin labelling of amacrine cells located in the INL and also a cell in the retinal ganglion cell (RGC) layer (pink arrow). c Image of the outer retina, showing highly ordered punctuate ring-like staining encircling the cones, with a section of higher magnification shown in the inset. d Image of a retinal section cut almost parallel to the plane of the photoreceptor layer showing a clearly defined mosaic of ring-like staining around individual cone photoreceptors, with a section of higher magnification shown in the inset. Labelling of these ring-like structures was consistently observed for all single cones positioned in these layers with a similar intensity of staining observed across the entire retina. Melanopsin-positive amacrine cells were relatively common, showing a uniform distribution across the retina similar to that shown in a and b. Labelling of cells in the RGC layer was rare with approximately one cell detected in every 3–4 retinal sections examined. Typically, the levels of staining were weaker for bipolar cells and horizontal cells when compared to cones, amacrine cells and ganglion cells. For all images, red is melanopsin immunoreactivity; green is autofluorescence from cone and rod photoreceptors; blue is 4′,6-diamidino-2-phenylindole (DAPI) nuclear counterstain. Outer segment, OS; inner segment, IS; outer plexiform layer, OPL; horizontal cell, HC; bipolar cell, BC; amacrine cell, AC; inner plexiform layer, IPL; retinal ganglion cell, RGC
Bioinformatic analysis of the promoter region for each zebrafish melanopsin gene
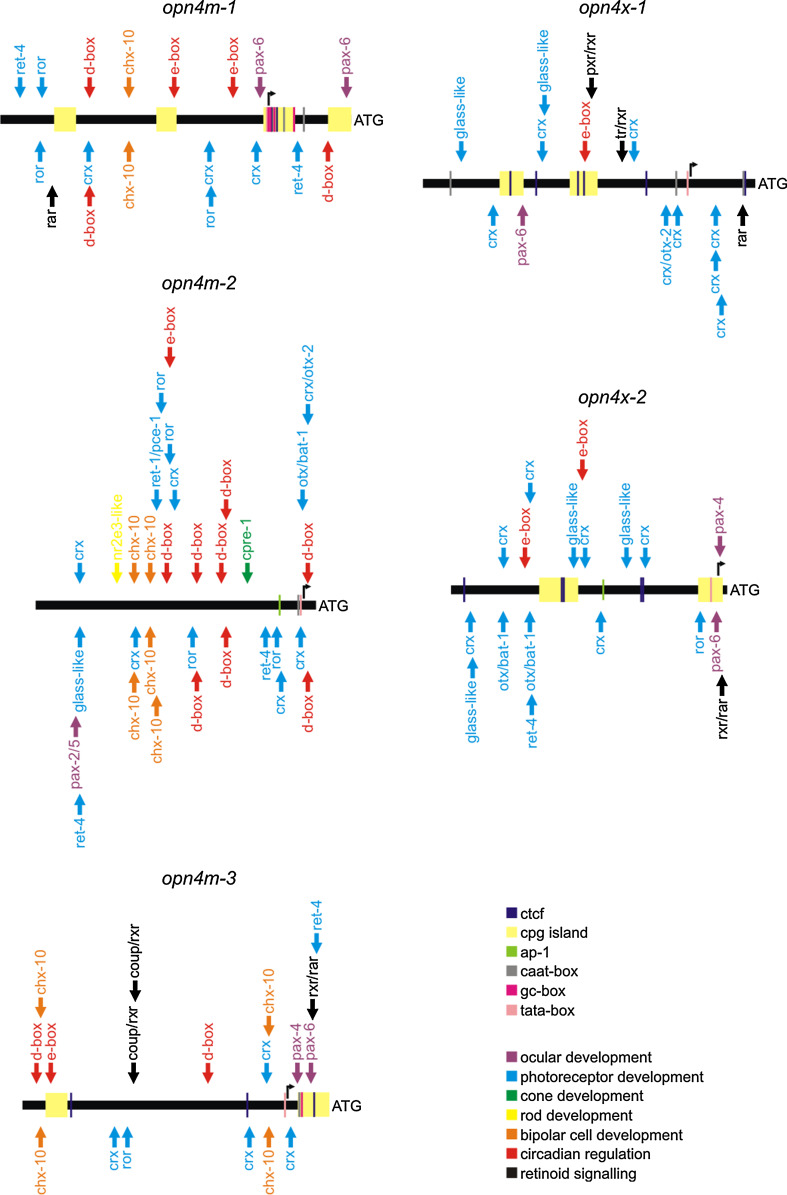
As opn4m-2 is a retrogene, it is possible that this melanopsin gene reintegrated into the zebrafish genome downstream of promoter elements that conferred expression in cone photoreceptors, in addition to cell types of the inner nuclear layer. Indeed, examination of the genomic region upstream of the opn4m-2 transcription start site showed the presence of multiple binding sites for known photoreceptor-specific transcriptional regulators (Fig. 4). Significantly, a cone photoreceptor regulatory element-1 enhancer [49] was found in the proximal upstream region of opn4m-2, with a nuclear receptor subfamily 2 group E member 3-like binding motif [53] in the distal promoter region, which concomitantly may mediate the unique expression of this melanopsin gene in single cones, while repressing double cone expression. Collectively, all five melanopsin promoter regions contained binding sites for transcription factors that are involved at different hierarchical levels of ocular gene regulation, including the master regulator homeobox genes that influence eye development (e.g. paired box gene 6), those that control photoreceptor (e.g. cone-rod homeobox) and bipolar cell [e.g. Caenorhabditis elegans homeobox-10 (ceh-10) containing homolog] differentiation, as well as circadian regulation (e.g. d-box, e-box) and those related to retinoid signalling (e.g. retinoic acid receptor), all of which are consistent with the global expression of melanopsin in the teleost retina. Interesting, opn4m and opn4x transcripts are detected in a zebrafish photoresponsive ZEM-2S cell line [68], where the authors report that both mRNA and protein of opn4x oscillates with an ultradian period of 10–11 h, with peak expression at ZT3 (also the time point at which zebrafish ocular tissues were collected in our study) and ZT12 (to a lesser extent). Closer inspection of the primer sequences and antibody epitopes shows that they were not observing opn4x but opn4m-2 ([15, 21] and this study), specifically. Together, the data suggest that transcription of the zebrafish melanopsin expressed in single cones and horizontal cells (i.e. opn4m-2) may be regulated over a 24 h period. Indeed, the 3 kb upstream promoter region of opn4m-2 contains the highest number (9) of d-box and e-box elements that are known to be involved with circadian regulation [62], suggesting that opn4m-2 may be strongly linked to central or peripheral clocks in the zebrafish. Similarly, the number of d-box and e-box motifs was notable in the promoters of both opn4m-1 (5) and opn4m-3 (3). In contrast, however, their number falls to either 1 or 2 in the promoters of opn4x-1 and opn4x-2, respectively. Thus, it would appear that all zebrafish melanopsins may be circadianally regulated, with opn4m isoforms being more tightly regulated then those of the opn4x lineage.
Fig. 4.
Promoter analysis of the upstream 3 kb region of all five zebrafish melanopsin genes. Using experimental procedures and bioinformatics, the major transcription start site (TSS) for each zebrafish melanopsin gene was mapped to −914 bp for opn4m-1, −129 bp for opn4m-2, −453 bp for opn4m-3, −727 bp for opn4x-1 and −105 bp for opn4x-2, where the methionine of the main AUG translation start site is denoted as +1. For each gene, investigation of putative promoter elements was mapped to a region of 3 kb upstream of the major TSS. Initially, CpG islands and CCCTC-binding factor (CTCF) insulator elements were mapped, and multiple copies were found to be present in the upstream promoter regions of all melanopsin genes except for opn4m-2, which is consistent with this retrogene being randomly reinserted into the zebrafish genome. Overall, each melanopsin promoter contained a putative tata box immediately upstream of the major TSS and binding sites for those transcription factors involved in basal gene regulation (e.g. caat-box, gc-box, ap-1) that are usually found in the proximal core promoters of genes. When analysed for ocular-specific transcription factor binding sites, the melanopsin promoter regions contained putative motifs consistent with gene regulation at different hierarchical levels of ocular development [46], such as the master regulator homeobox genes (e.g. pax-2/5, pax-4, pax-6), factors involved in photoreceptor development and differentiation [46, 48] (e.g. crx, crx/otx-2, ret-1/pce-1, otx/bat-1, ror, ret-4, glass-like) and bipolar cell differentiation [61] (e.g. chx-10), as well as those binding sites important in circadian clock gene regulation [62] (e.g. d-box, e-box) and retinoid signalling [47] (e.g. rar, rxr), all of which are consistent with the global expression of melanopsin in the teleost retina. Significantly, two motifs (cpre-1 and nr2e3-like) were found solely in the promoter of the opn4m-2 retrogene, which concomitantly may play significant roles in the expression of this melanopsin gene in single cones while repressing double cone expression
Functional chacterisation of zebrafish melanopsins
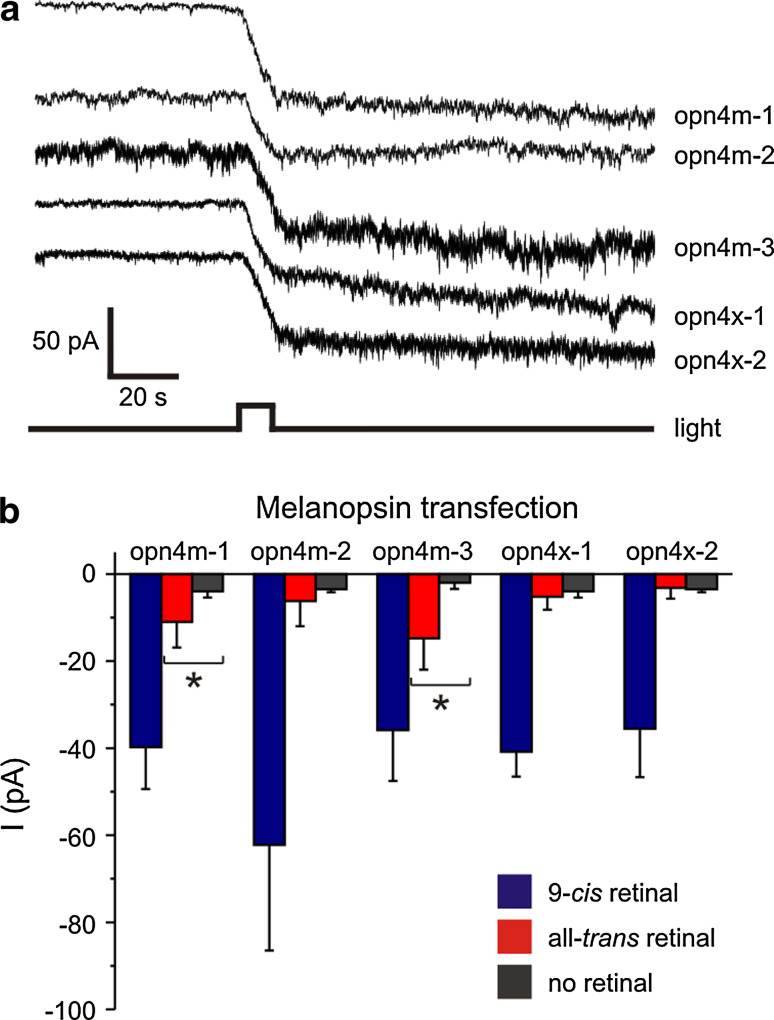
To determine whether all five melanopsin genes encode functional photopigments, a heterologous expression system [8, 69, 70] was used as previously described for human OPN4 [8, 69]. Furthermore, both 9-cis retinal and all-trans retinal chromophores were used to investigate whether these melanopsins function as bistable or monostable photopigments as discussed earlier. Whole-cell patch-clamp recordings from Neuro-2a cells transiently transfected with each full-length cDNA sequence revealed that expression of all five opn4 photopigments resulted in light-dependent inward currents in the presence, but not the absence, of 9-cis retinal chromophore (9-cis retinal: opn4m-1, −40 ± 10 pA; opn4m-2, −62 ± 24 pA; opn4m-3, −36 ± 12 pA; opn4x-1, −41 ± 6 pA; opn4x-2, −36 ± 11 pA) (Fig. 5a). By contrast, when all-trans retinal was used, light-dependent inward currents were only observed in cells transfected with opn4m-1 and opn4m-3, albeit with currents less than those measured with 9-cis-retinal (all-trans retinal: opn4m-1, −11 ± 6 pA; opn4m-2, −6 ± 6 pA; opn4m-3, −14 ± 6 pA; opn4x-1, −5 ± 3 pA; opn4x-2, −3 ± 2 pA) (Fig. 5b). In all cases, the light-dependent responses showed very slow recovery, consistent with the absence of a native deactivation cascade. Bistable photopigments may form stable interactions with multiple chromophores with the same affinity, but this is not always the case. Amphioxus rhodopsin (Branchiostoma belcheri, Amphiop1) is bistable and binds both 11-cis and all-trans retinal isomers, but shows ~50-fold higher affinity for 11-cis retinal compared to all-trans chromophore [71]. Similarly, in our experiments, the increase in light-dependent inward current with 11-cis retinal compared to all-trans retinal was five- and threefold higher for opn4m-1 and opn4m-3, respectively. These functional studies showed that opn4m-1 and opn4m-3 photopigments were able to form stable interactions with both 9-cis and all-trans chromophores, which is consistent with the bistable nature exhibited by chordate melanopsin [8–11] and invertebrate rhabdomeric photopigments [72]. More surprisingly, however, our data indicate that photopigments encoded by opn4m-2, opn4x-1 and opn4x-2 only formed stable interactions with cis-isomers and are therefore monostable, similar to classical vertebrate photopigments. This observation demonstrates for the first time that invertebrate-like and vertebrate-like forms of melanopsin are present within the same species (and in some cases, the same cell types) and reveals that melanopsins are functionally more diverse than previously thought.
Fig. 5.
Light-evoked responses of Neuro-2a cells expressing melanopsin. a Whole-cell recordings from cells expressing melanopsin preincubated with 9-cis retinal and exposed to a 10 s light pulse (420 nm; 8 × 1014 photons cm−2 s−1) revealed that all five zebrafish melanopsin genes encode functional photopigments in vitro. b Quantification of melanopsin-dependent light-evoked currents with 9-cis retinal (n = 5–8 cells) (blue), all-trans retinal (n = 4–6 cells) (red) and no retinal (n = 2 cells) (grey), respectively. Light-evoked currents (mean ± SEM) observed in cells expressing melanopsin preincubated with 9-cis retinal were significantly larger compared with cells with no retinal. Recordings with all-trans retinal showed that light-evoked responses of opn4m-1 and opn4m-3 transfected cells were larger (p < 0.05; Student’s t test) than those with no retinal. All experiments using 9-cis retinal showed larger light-evolved currents compared to no retinal controls (p < 0.01)
Spectral properties of representative zebrafish melanopsins
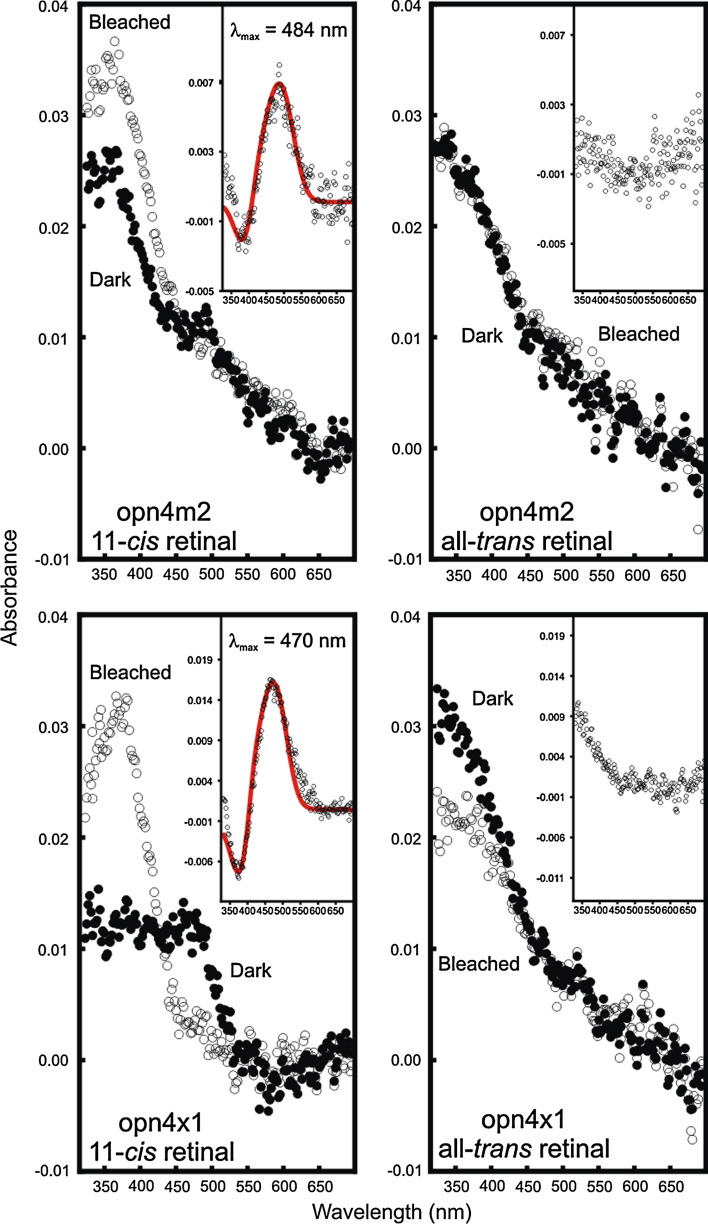
Peak spectral sensitivities of one representative mammalian-like (opn4m-2) and one non-mammalian-like form of melanopsin (opn4x-1) were determined by ultraviolet-visible spectrophotometry. Despite the technical difficulties when working with vertebrate non-cone, non-rod opsins in vitro [13], both opn4m-2 and opn4x-1 photopigments gave dark spectra close to 500 nm when regenerated with 11-cis retinal and spectral peaks of 380 nm when bleached by light, the latter corresponding to the formation of the all-trans retinal oxime (Fig. 6). In both cases, difference (dark minus bleached) spectra were generated that could be fitted to a Govardovskii A1-template (Fig. 6, insets). A spectral peak sensitivity (λmax) of 484 nm was obtained for opn4m-2 and 470 nm for opn4x-1 photopigments, which is broadly consistent with the λmax value for other chordate melanopsins [9, 10, 14]. When performed with all-trans retinal, the spectra for both photopigments failed to show an absorbance peak between 300 and 700 nm (Fig. 6, insets), suggesting that both opn4m-2 and opn4x-1 photopigments were not able to form stable interactions with all-trans retinal. These findings are consistent with these pigments being monostable in the whole-cell patch studies. Despite repeated attempts, opn4m-1, opn4m-3 and opn4x-2 failed to regenerate, although high levels of protein were produced (Supplementary Material Fig. 2), and each was able to generate light evoked, retinal-dependent currents (Fig. 5).
Fig. 6.
Spectral sensitivity analyses of representative zebrafish melanopsins. Absorption spectra of regenerated zebrafish melanopsin photopigments, opn4m-2 (top panels) and opn4x-1 (bottom panels) with 11-cis (left hand panels) and all-trans (right hand panels) retinal chromophores. For all photopigments, dark (closed circles) and bleached (open circles) spectra are shown. Difference spectra (inset) were fitted with a Govardovskii A1-template (red) to determine the λmax value
Examination of monostability and bistability by coexpressing zebrafish melanopsins
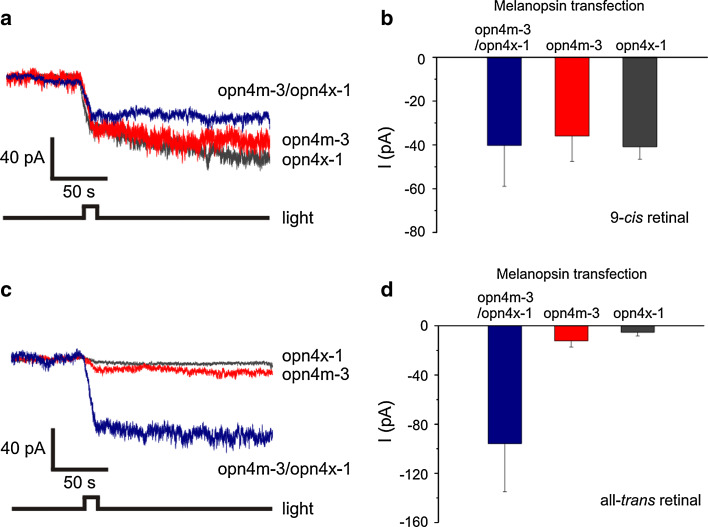
Spectrophotometry and electrophysiology has clearly demonstrated that both opn4m-2 (highly expressed in cone photoreceptors, as well as horizontal cells) and opn4x-1 (expressed in the inner retina and a subset of RGCs) encode photopigments that only form functional complexes with chromophores containing cis-isomers. By contrast, we show that opn4m-1 and opn4m-3 photopigments can form stable interactions with both all-trans retinal and 9-cis retinaldehyde, suggesting that some forms of melanopsin may be capable of endogenous photoisomerase activity (Fig. 5b). We have shown that the RGCs of the inner retina express two forms of melanopsin, namely opn4x-1 and opn4m-3; however, whether they are co-expressed in the same RGC is unclear. Nonetheless, to further investigate the biochemistry of bistability versus monostability in these photopigments and any potential functional interactions between these two opsins, opn4x-1 and opn4m-3 were co-expressed in Neuro-2a cells, and light-evoked currents were studied using whole-cell patch-clamp recording [8]. When opn4x-1 and opn4m-3 were co-expressed, a retinal-dependent, light-evoked current was observed in the presence (but not in the absence) of chromophore (Fig. 7). When 9-cis retinal was used, the light-dependent current for cells co-transfected with opn4x-1 and opn4m-3 (−40 ± 19 pA) were not significantly different from cells that expressed each melanopsin alone (opn4x-1, −41 ± 6 pA; opn4 m-3, −36 ± 12 pA) (Fig. 7). By contrast, the light-dependent current for co-transfected cells (−96 ± 39 pA) was greatly augmented compared to cells transfected only with opn4x-1 (−5 ± 3 pA) or opn4m-3 (−14 ± 6 pA) in the presence of all-trans retinal (Fig. 7). Whilst we cannot exclude complex interactions between heterodimers of zebrafish melanopsins, the most parsimonious conclusion from these findings is that the opn4m-3 photopigment can convert all-trans retinal to a cis-isomer that is, in turn, used by co-expressed melanopsins to generate a more pronounced light-dependent inward current. Similar co-expression experiments were performed with opn4x-1 and opn4m-2; however, no significant enhancement was observed when these melanopsin genes were co-expressed in the presence of either all-trans retinal (opn4m-2/opn4x-1, −11 ± 6 pA; opn4m-2, −6 ± 6 pA; opn4x-1, −5 ± 3 pA) or 9-cis retinal (opn4m-2/opn4x-1, −30 ± 4 pA; opn4m-2, −62 ± 24 pA; opn4x-1, −41 ± 6 pA) (Supplementary Material Fig. 5).
Fig. 7.
Co-expression of opn4m-3 and opn4x-1 in Neuro-2a cells. Representative light-evoked responses (mean ± SEM) from Neuro-2a cells transfected with both opn4m-3 and opn4x-1 (n = 5–8 cells) (blue), or solely with opn4m-3 (n = 5 cells) (red) or opn4x-1 (n = 4–6 cells) (grey), preincubated with (a–b) 9-cis retinal or (c–d) all-trans retinal. Significant differences were found between all permutations of light-evoked responses resulting from opn4m-3 and opn4x-1 experiments when preincubated with all-trans retinal, but not 9-cis retinal (p < 0.01; Student’s t test)
Discussion
Photopigments consist of an opsin protein linked to a photosensitive retinal chromophore based on vitamin A. Classical vertebrate pigments are monostable and form stable associations with only cis-isomers of retinal [73, 74]. By contrast, invertebrate pigments exhibit bistability and are able to form stable associations with both cis- and trans-isomers of retinal chromophore [72]. To date, all the described forms of melanopsin appear to exhibit invertebrate-like bistability, and this has been thought to be indicative of their ancient evolutionary lineage [8–11]. Our data show that opsins encoded by opn4m-2 and opn4x-1 (and presumably opn4x-2) are more akin to classical vertebrate photopigments: they are monostable, without endogenous isomerase activity, and can only form a stable interaction with and respond to light in the presence of cis-isomer chromophores. By contrast, opn4m-1 and opn4m-3 photopigments are bistable, as they form functional interactions with both cis-retinal and trans-isomers, and as such, are similar to both human [8, 69] and mouse [70] melanopsins, as well as melanopsin from cephalochordate lancelets [9]. Furthermore, we provide evidence that suggests these bistable pigments may function as endogenous photoisomerases (most likely converting all-trans retinaldehyde to 11-cis retinal in a light-dependent manner). These data provide the first demonstration to show functional diversity within the melanopsin family of photopigments. Furthermore, it appears that this preferential interaction with particular chromophores (i.e. monostability versus bistability) is not simply restricted to photopigments of either zebrafish opn4 gene lineage (i.e. opn4m versus opn4x), as both states are represented within the same melanopsin group (i.e. m-class) that is phylogenetically related to mammalian OPN4. It has been suggested that the retina of the vertebrate eye arose from cells derived from both protostome/rhabdomeric (invertebrate) and deuterostome/ciliary (vertebrate) lineages [75, 76]. In this way, the cones, rods and bipolar cells originate from a ciliary ancestry and horizontal, amacrine and RGCs derive from a rhabdomeric ancestry [76]. Support for this hypothesis has come from the distribution of opsins in the mammalian retina. The cones and rods possess ciliary opsins typical of the vertebrates, whilst a subset of RGCs express melanopsin, which shows many features of an invertebrate rhabdomeric photopigment [11]. Our finding that melanopsins are expressed within every cell type of the teleost retina suggests the rhabdomeric/ciliary origin of the vertebrate retina may need revision [13].
We have identified and functionally characterised five melanopsin genes in the zebrafish retina. These opsins have specific retinal cellular localisations and discrete functional properties. These data suggest that direct melanopsin photosensitivity in the retinal network may be distributed over a much wider range of cell types than previously appreciated, including RGCs, bipolar, amacrine, horizontal cells (as previously shown in the roach [19] and catfish [22]), and remarkably, cone photoreceptors themselves. Thus, a subset of all the main retinal neuronal cell types appear to express one or more forms of melanopsin protein, with opn4m-2 protein confined to a large subset of the cones. The presence of multiple isoforms of melanopsin within discrete retinal layers may indicate these genes are co-expressed in vivo. If this is the case, monostable pigments (i.e. opn4m-2, opn4x-1 and opn4x-2) would be associated with bistable and presumably chromophore-regenerative melanopsins (i.e. opn4m-1 and opn4m-3) that would provide 11-cis retinal to melanopsins that are distal to the visual cycle in the RPE [77]. The exemption to this observation is the sole expression of monostable opn4m-2 in the photoreceptor layer. However, like the cones and rods of this layer, 11-cis retinal is converted to an all-trans isomer and regenerated in the proximal RPE [77] (which also expresses the bistable opn4m-3 photopigment). Thus, a ready supply of 11-cis retinal would be available to the photoreceptor layer to be utilised by cone and rod opsins and opn4m-2 photopigments, thus removing the requirement for intrinsic sources of cis-isomers of the retinal chromophore. Nonetheless, the function of this melanopsin in cone photoreceptors is unclear and warrants further investigation. One possibility is that opn4m-2 extends the dynamic range of cone photoreceptors. This seems unlikely, however, because melanopsins (unlike cone opsins) do not appear to couple significantly to a cognate α-transducin protein pathway, but instead interact with a Gq/G11-mediated phototransduction cascade [8, 69, 70, 78]. Another possibility is that opn4m-2 is involved in regulatory mechanisms, such as retinomotor movements that occur in teleosts and many other vertebrates. This again seems unlikely because light-driven retinomotor movements appear to be a rod-mediated response [79]. In our view, the most likely role is that opn4m-2 may be involved in light-dependent adaptation that extends the working range of cone photoreceptors. It has been previously shown that light stimulates a transducin-independent increase in cytoplasmic calcium and suppression of current in cones of zebrafish lacking the α-subunit of cone transducin [80]. In view of this striking finding, we suggest that this parallel bright light-mediated response of cone photoreceptors is driven by a melanopsin-dependent cascade. This may be functionally homologous to murine pRGCs, where melanopsin mediates changes in intracellular calcium mobilisation through a Gq/G11-dependent mechanism [81]. How might such a mechanism be important to cone function? Some possibilities include a role in (1) sustaining minimum calcium levels under near saturating bright light and (2) contributing to calcium-dependent adaptation of light responsiveness. The localisation of opn4m-2 close to the inner segment may suggest that the former possibility is the most likely function. We have provided evidence that opn4m-2 is a photopigment that is dependent on cis-retinal, which is functionally consistent with a pigment expressed in cone photoreceptors, where there is an abundance of chromophore.
We have studied the entire set of melanopsin photopigments encoded by five opn4 genes found within the zebrafish genome. Remarkably, these photopigments are distributed across all retinal cell types, including cones. Why should all retinal cell types respond to light? One possibility is that retinal circuitry is constantly being “fine-tuned” in response to a change in environmental light. Zebrafish, like many teleosts, are diurnal animals that rely heavily on their visual sense, but they live in a dynamic light environment. Light levels can change many orders of magnitude as a result of movement through the water column or shading from vegetation or other organisms. Under such circumstances, it would be advantageous to adjust their retinal circuitry to compensate for such dynamic changes and maintain optimal visual processing.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank Dr. Rosalie Crouch and Dr. Jill Cowing for the generous provision of 11-cis retinal and 1D4 antibody; Dr. Stephen Price and Dr. Sofia Godinho for help with RNA in situ hybridisation experiments; Dr. Kara Cerveny for microscopy and photography advice; Dr. Stuart Peirson for his assistance on UV-vis template fitting and useful discussions; and University College London Fish Facility for adult zebrafish. We thank Prof. Shaun Collin for his advice on photoreceptor morphology. This work was supported by a grant awarded by the UK Biotechnology and Biological Research Council (BBSRC) to MWH.
Conflict of interest
The authors declare no competing financial interests.
Footnotes
W. I. L. Davies and L. Zheng contributed equally to this research.
Supplementary material is linked to the online version of the paper at http://www.springer.com/birkhauser/biosciences/journal/18.
References
- 1.Hattar S, Liao HW, Takao M, Berson DM, Yau KW. Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science. 2002;295:1065–1070. doi: 10.1126/science.1069609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rollag MD, Berson DM, Provencio I. Melanopsin, ganglion-cell photoreceptors, and mammalian photoentrainment. J Biol Rhythms. 2003;18:227–234. doi: 10.1177/0748730403018003005. [DOI] [PubMed] [Google Scholar]
- 3.Hankins MW, Peirson SN, Foster RG. Melanopsin: an exciting photopigment. Trends Neurosci. 2008;31:27–36. doi: 10.1016/j.tins.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Bailes HJ, Lucas RJ. Melanopsin and inner retinal photoreception. Cell Mol Life Sci. 2010;67:99–111. doi: 10.1007/s00018-009-0155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller LP, Do MT, Yau KW, He S, Baldridge WH. Tracer coupling of intrinsically photosensitive retinal ganglion cells to amacrine cells in the mouse retina. J Comp Neurol. 2010;518:4813–4824. doi: 10.1002/cne.22490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gamlin PD, McDougal DH, Pokorny J, Smith VC, Yau KW, Dacey DM. Human and macaque pupil responses driven by melanopsin-containing retinal ganglion cells. Vision Res. 2007;47:946–954. doi: 10.1016/j.visres.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hatori M, Panda S. The emerging roles of melanopsin in behavioral adaptation to light. Trends Mol Med. 2010;16:435–446. doi: 10.1016/j.molmed.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melyan Z, Tarttelin EE, Bellingham J, Lucas RJ, Hankins MW. Addition of human melanopsin renders mammalian cells photoresponsive. Nature. 2005;433:741–745. doi: 10.1038/nature03344. [DOI] [PubMed] [Google Scholar]
- 9.Koyanagi M, Kubokawa K, Tsukamoto H, Shichida Y, Terakita A. Cephalochordate melanopsin: evolutionary linkage between invertebrate visual cells and vertebrate photosensitive retinal ganglion cells. Curr Biol. 2005;15:1065–1069. doi: 10.1016/j.cub.2005.04.063. [DOI] [PubMed] [Google Scholar]
- 10.Walker MT, Brown RL, Cronin TW, Robinson PR. Photochemistry of retinal chromophore in mouse melanopsin. Proc Natl Acad Sci USA. 2008;105:8861–8865. doi: 10.1073/pnas.0711397105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mure LS, Cornut PL, Rieux C, Drouyer E, Denis P, Gronfier C, Cooper HM. Melanopsin bistability: a fly’s eye technology in the human retina. PLoS One. 2009;4:e5991. doi: 10.1371/journal.pone.0005991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pires SS, Hughes S, Turton M, Melyan Z, Peirson SN, Zheng L, Kosmaoglou M, Bellingham J, Cheetham ME, Lucas RJ, Foster RG, Hankins MW, Halford S. Differential expression of two distinct functional isoforms of melanopsin (Opn4) in the mammalian retina. J Neurosci. 2009;29:12332–12342. doi: 10.1523/JNEUROSCI.2036-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies WL, Hankins MW, Foster RG. Vertebrate ancient opsin and melanopsin: divergent irradiance detectors. Photochem Photobiol Sci. 2010;9:1444–1457. doi: 10.1039/c0pp00203h. [DOI] [PubMed] [Google Scholar]
- 14.Torii M, Kojima D, Okano T, Nakamura A, Terakita A, Shichida Y, Wada A, Fukada Y. Two isoforms of chicken melanopsins show blue light sensitivity. FEBS Lett. 2007;581:5327–5331. doi: 10.1016/j.febslet.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 15.Bellingham J, Chaurasia SS, Melyan Z, Liu C, Cameron MA, Tarttelin EE, Iuvone PM, Hankins MW, Tosini G, Lucas RJ. Evolution of melanopsin photoreceptors: discovery and characterization of a new melanopsin in nonmammalian vertebrates. PLoS Biol. 2006;4:e254. doi: 10.1371/journal.pbio.0040254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drivenes O, Soviknes AM, Ebbesson LO, Fjose A, Seo HC, Helvik JV. Isolation and characterization of two teleost melanopsin genes and their differential expression within the inner retina and brain. J Comp Neurol. 2003;456:84–93. doi: 10.1002/cne.10523. [DOI] [PubMed] [Google Scholar]
- 17.Bailey MJ, Cassone VM. Melanopsin expression in the chick retina and pineal gland. Brain Res Mol Brain Res. 2005;134:345–348. doi: 10.1016/j.molbrainres.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 18.Chaurasia SS, Rollag MD, Jiang G, Hayes WP, Haque R, Natesan A, Zatz M, Tosini G, Liu C, Korf HW, Iuvone PM, Provencio I. Molecular cloning, localization and circadian expression of chicken melanopsin (Opn4): differential regulation of expression in pineal and retinal cell types. J Neurochem. 2005;92:158–170. doi: 10.1111/j.1471-4159.2004.02874.x. [DOI] [PubMed] [Google Scholar]
- 19.Jenkins A, Munoz M, Tarttelin EE, Bellingham J, Foster RG, Hankins MW. VA opsin, melanopsin, and an inherent light response within retinal interneurons. Curr Biol. 2003;13:1269–1278. doi: 10.1016/S0960-9822(03)00509-8. [DOI] [PubMed] [Google Scholar]
- 20.Provencio I, Jiang G, De Grip WJ, Hayes WP, Rollag MD. Melanopsin: an opsin in melanophores, brain, and eye. Proc Natl Acad Sci USA. 1998;95:340–345. doi: 10.1073/pnas.95.1.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bellingham J, Whitmore D, Philp AR, Wells DJ, Foster RG. Zebrafish melanopsin: isolation, tissue localisation and phylogenetic position. Brain Res Mol Brain Res. 2002;107:128–136. doi: 10.1016/S0169-328X(02)00454-0. [DOI] [PubMed] [Google Scholar]
- 22.Cheng N, Tsunenari T, Yau KW. Intrinsic light response of retinal horizontal cells of teleosts. Nature. 2009;460:899–903. doi: 10.1038/nature08175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grone BP, Sheng Z, Chen CC, Fernald RD. Localization and diurnal expression of melanopsin, vertebrate ancient opsin, and pituitary adenylate cyclase-activating peptide mRNA in a teleost retina. J Biol Rhythms. 2007;22:558–561. doi: 10.1177/0748730407308285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomonari S, Takagi A, Noji S, Ohuchi H. Expression pattern of the melanopsin-like (cOpn4m) and VA opsin-like genes in the developing chicken retina and neural tissues. Gene Expr Patterns. 2007;7:746–753. doi: 10.1016/j.modgep.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 25.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- 26.Higgins DG, Thompson JD, Gibson TJ. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/S0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 27.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 28.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 29.Whitmore D, Foulkes NS, Sassone-Corsi P. Light acts directly on organs and cells in culture to set the vertebrate circadian clock. Nature. 2000;404:87–91. doi: 10.1038/35003589. [DOI] [PubMed] [Google Scholar]
- 30.Vandesompele J, Preter K, Pattyn F, Poppe B, Roy N, Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carleton KL, Kocher TD. Cone opsin genes of African Cichlid fishes: tuning spectral sensitivity by differential gene expression. Mol Biol Evol. 2001;18:1540–1550. doi: 10.1093/oxfordjournals.molbev.a003940. [DOI] [PubMed] [Google Scholar]
- 32.Lo K, Smale ST. Generality of a functional initiator consensus sequence. Gene. 1996;182:13–22. doi: 10.1016/S0378-1119(96)00438-6. [DOI] [PubMed] [Google Scholar]
- 33.Cross SH, Bird AP. CpG islands and genes. Curr Opin Genet Dev. 1995;5:309–314. doi: 10.1016/0959-437X(95)80044-1. [DOI] [PubMed] [Google Scholar]
- 34.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amouyal M. Gene insulation. Part II: natural strategies in vertebrates. Biochem Cell Biol. 2010;88:885–898. doi: 10.1139/o10-111. [DOI] [PubMed] [Google Scholar]
- 36.Ishihara K, Sasaki H. An evolutionarily conserved putative insulator element near the 3′ boundary of the imprinted Igf2/H19 domain. Hum Mol Genet. 2002;11:1627–1636. doi: 10.1093/hmg/11.14.1627. [DOI] [PubMed] [Google Scholar]
- 37.Juo ZS, Chiu TK, Leiberman PM, Baikalov I, Berk AJ, Dickerson RE. How proteins recognize the TATA box. J Mol Biol. 1996;261:239–254. doi: 10.1006/jmbi.1996.0456. [DOI] [PubMed] [Google Scholar]
- 38.Baumann M, Pontiller J, Ernst W. Structure and basal transcription complex of RNA polymerase II core promoters in the mammalian genome: an overview. Mol Biotechnol. 2010;45:241–247. doi: 10.1007/s12033-010-9265-6. [DOI] [PubMed] [Google Scholar]
- 39.Juven-Gershon T, Kadonaga JT. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol. 2010;339:225–229. doi: 10.1016/j.ydbio.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benoist C, O’Hare K, Breathnach R, Chambon P. The ovalbumin gene-sequence of putative control regions. Nucleic Acids Res. 1980;8:127–142. doi: 10.1093/nar/8.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burke TW, Kadonaga JT. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lim CY, Santoso B, Boulay T, Dong E, Ohler U, Kadonaga JT. The MTE, a new core promoter element for transcription by RNA polymerase II. Genes Dev. 2004;18:1606–1617. doi: 10.1101/gad.1193404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lagrange T, Kapanidis AN, Tang H, Reinberg D, Ebright RH. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 1998;12:34–44. doi: 10.1101/gad.12.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Briggs MR, Kadonaga JT, Bell SP, Tjian R. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science. 1986;234:47–52. doi: 10.1126/science.3529394. [DOI] [PubMed] [Google Scholar]
- 45.Lee W, Mitchell P, Tjian R. Purified transcription factor AP-1 interacts with TPA-inducible enhancer elements. Cell. 1987;49:741–752. doi: 10.1016/0092-8674(87)90612-X. [DOI] [PubMed] [Google Scholar]
- 46.Swaroop A, Kim D, Forrest D. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat Rev Neurosci. 2010;11:563–576. doi: 10.1038/nrn2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cvekl A, Wang WL. Retinoic acid signaling in mammalian eye development. Exp Eye Res. 2009;89:280–291. doi: 10.1016/j.exer.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akimoto M. Transcriptional factors involved in photoreceptor differentiation. Semin Ophthalmol. 2005;20:25–30. doi: 10.1080/08820530590921864. [DOI] [PubMed] [Google Scholar]
- 49.Smyth VA, Di Lorenzo D, Kennedy BN. A novel, evolutionarily conserved enhancer of cone photoreceptor-specific expression. J Biol Chem. 2008;283:10881–10891. doi: 10.1074/jbc.M710454200. [DOI] [PubMed] [Google Scholar]
- 50.Rehemtulla A, Warwar R, Kumar R, Ji X, Zack DJ, Swaroop A. The basic motif-leucine zipper transcription factor Nrl can positively regulate rhodopsin gene expression. Proc Natl Acad Sci USA. 1996;93:191–195. doi: 10.1073/pnas.93.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kataoka K, Noda M, Nishizawa M. Maf nuclear oncoprotein recognizes sequences related to an AP-1 site and forms heterodimers with both Fos and Jun. Mol Cell Biol. 1994;14:700–712. doi: 10.1128/mcb.14.1.700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kerppola TK, Curran T. Maf and Nrl can bind to AP-1 sites and form heterodimers with Fos and Jun. Oncogene. 1994;9:675–684. [PubMed] [Google Scholar]
- 53.Chen J, Rattner A, Nathans J. The rod photoreceptor-specific nuclear receptor Nr2e3 represses transcription of multiple cone-specific genes. J Neurosci. 2005;25:118–129. doi: 10.1523/JNEUROSCI.3571-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jetten AM, Kurebayashi S, Ueda E. The ROR nuclear orphan receptor subfamily: critical regulators of multiple biological processes. Prog Nucleic Acid Res Mol Biol. 2001;69:205–247. doi: 10.1016/S0079-6603(01)69048-2. [DOI] [PubMed] [Google Scholar]
- 55.Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 56.Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91:531–541. doi: 10.1016/S0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- 57.Kelley CG, Lavorgna G, Clark ME, Boncinelli E, Mellon PL. The Otx2 homeoprotein regulates expression from the gonadotropin-releasing hormone proximal promoter. Mol Endocrinol. 2000;14:1246–1256. doi: 10.1210/me.14.8.1246. [DOI] [PubMed] [Google Scholar]
- 58.Kimura A, Singh D, Wawrousek EF, Kikuchi M, Nakamura M, Shinohara T. Both PCE-1/RX and OTX/CRX interactions are necessary for photoreceptor-specific gene expression. J Biol Chem. 2000;275:1152–1160. doi: 10.1074/jbc.275.2.1152. [DOI] [PubMed] [Google Scholar]
- 59.Chen S, Zack DJ. Ret 4, a positive acting rhodopsin regulatory element identified using a bovine retina in vitro transcription system. J Biol Chem. 1996;271:28549–28557. doi: 10.1074/jbc.271.45.28549. [DOI] [PubMed] [Google Scholar]
- 60.Moses K, Rubin GM. Glass encodes a site-specific DNA-binding protein that is regulated in response to positional signals in the developing Drosophila eye. Genes Dev. 1991;5:583–593. doi: 10.1101/gad.5.4.583. [DOI] [PubMed] [Google Scholar]
- 61.Dorval KM, Bobechko BP, Fujieda H, Chen S, Zack DJ, Bremner R. CHX10 targets a subset of photoreceptor genes. J Biol Chem. 2006;281:744–751. doi: 10.1074/jbc.M509470200. [DOI] [PubMed] [Google Scholar]
- 62.Zhang J, Dong X, Fujimoto Y, Okamura H. Molecular signals of mammalian circadian clock. Kobe J Med Sci. 2004;50:101–109. [PubMed] [Google Scholar]
- 63.Govardovskii VI, Fyhrquist N, Reuter T, Kuzmin DG, Donner K. In search of the visual pigment template. Vis Neurosci. 2000;17:509–528. doi: 10.1017/S0952523800174036. [DOI] [PubMed] [Google Scholar]
- 64.Shiraki T, Kojima D, Fukada Y. Light-induced body color change in developing zebrafish. Photochem Photobiol Sci. 2010;9:1498–1504. doi: 10.1039/c0pp00199f. [DOI] [PubMed] [Google Scholar]
- 65.Fitzgibbon J, Hope A, Slobodyanyuk SJ, Bellingham J, Bowmaker JK, Hunt DM. The rhodopsin-encoding gene of bony fish lacks introns. Gene. 1995;164:273–277. doi: 10.1016/0378-1119(95)00458-I. [DOI] [PubMed] [Google Scholar]
- 66.Bellingham J, Tarttelin EE, Foster RG, Wells DJ. Structure and evolution of the teleost extraretinal rod-like opsin (errlo) and ocular rod opsin (rho) genes: is teleost rho a retrogene? J Exp Zool B Mol Dev Evol. 2003;297:1–10. doi: 10.1002/jez.b.18. [DOI] [PubMed] [Google Scholar]
- 67.Allison WT, Barthel LK, Skebo KM, Takechi M, Kawamura S, Raymond PA. Ontogeny of cone photoreceptor mosaics in zebrafish. J Comp Neurol. 2010;518:4182–4195. doi: 10.1002/cne.22447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Farhat FP, Martins CB, De Lima LH, Isoldi MC, Castrucci AM. Melanopsin and clock genes: regulation by light and endothelin in the zebrafish ZEM-2S cell line. Chronobiol Int. 2009;26:1090–1119. doi: 10.3109/07420520903249005. [DOI] [PubMed] [Google Scholar]
- 69.Qiu X, Kumbalasiri T, Carlson SM, Wong KY, Krishna V, Provencio I, Berson DM. Induction of photosensitivity by heterologous expression of melanopsin. Nature. 2005;433:745–749. doi: 10.1038/nature03345. [DOI] [PubMed] [Google Scholar]
- 70.Panda S, Nayak SK, Campo B, Walker JR, Hogenesch JB, Jegla T. Illumination of the melanopsin signaling pathway. Science. 2005;307:600–604. doi: 10.1126/science.1105121. [DOI] [PubMed] [Google Scholar]
- 71.Tsukamoto H, Terakita A, Shichida Y. A rhodopsin exhibiting binding ability to agonist all-trans-retinal. Proc Natl Acad Sci USA. 2005;102:6303–6308. doi: 10.1073/pnas.0500378102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hillman P, Hochstein S, Minke B. Transduction in invertebrate photoreceptors: role of pigment bistability. Physiol Rev. 1983;63:668–772. doi: 10.1152/physrev.1983.63.2.668. [DOI] [PubMed] [Google Scholar]
- 73.Wald G. The molecular basis of visual excitation. Nature. 1968;219:800–807. doi: 10.1038/219800a0. [DOI] [PubMed] [Google Scholar]
- 74.Farrens DL, Khorana HG. Structure and function in rhodopsin. Measurement of the rate of metarhodopsin II decay by fluorescence spectroscopy. J Biol Chem. 1995;270:5073–5076. doi: 10.1074/jbc.270.10.5073. [DOI] [PubMed] [Google Scholar]
- 75.Lamb TD. Evolution of vertebrate retinal photoreception. Philos Trans R Soc Lond B Biol Sci. 2009;364:2911–2924. doi: 10.1098/rstb.2009.0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arendt D, Hausen H, Purschke G. The ‘division of labour’ model of eye evolution. Philos Trans R Soc Lond B Biol Sci. 2009;364:2809–2817. doi: 10.1098/rstb.2009.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lamb TD, Pugh EN., Jr Dark adaptation and the retinoid cycle of vision. Prog Retin Eye Res. 2004;23:307–380. doi: 10.1016/j.preteyeres.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 78.Terakita A, Tsukamoto H, Koyanagi M, Sugahara M, Yamashita T, Shichida Y. Expression and comparative characterization of Gq-coupled invertebrate visual pigments and melanopsin. J Neurochem. 2008;105:883–890. doi: 10.1111/j.1471-4159.2007.05184.x. [DOI] [PubMed] [Google Scholar]
- 79.Kirsch M, Wagner HJ, Douglas RH. Rods trigger light adaptive retinomotor movements in all spectral cone types of a teleost fish. Vision Res. 1989;29:389–396. doi: 10.1016/0042-6989(89)90003-5. [DOI] [PubMed] [Google Scholar]
- 80.Brockerhoff SE, Rieke F, Matthews HR, Taylor MR, Kennedy B, Ankoudinova I, Niemi GA, Tucker CL, Xiao M, Cilluffo MC, Fain GL, Hurley JB. Light stimulates a transducin-independent increase of cytoplasmic Ca2+ and suppression of current in cones from the zebrafish mutant nof. J Neurosci. 2003;23:470–480. doi: 10.1523/JNEUROSCI.23-02-00470.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sekaran S, Foster RG, Lucas RJ, Hankins MW. Calcium imaging reveals a network of intrinsically light-sensitive inner-retinal neurons. Curr Biol. 2003;13:1290–1298. doi: 10.1016/S0960-9822(03)00510-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.