Abstract
During the past two decades of research in T cell biology, an increasing number of distinct T cell subsets arising during the transition from naïve to antigen-experienced T cells have been identified. Recently, it has been appreciated that, in different experimental settings, distinct T cell subsets can be generated in parallel within the same immune response. While signals driving a single “lineage” path of T cell differentiation are becoming increasingly clear, it remains largely enigmatic how the phenotypic and functional diversification creating a multi-faceted T cell response is achieved. Here, we review current literature indicating that diversification is a stable trait of CD8+ T cell responses. We showcase novel technologies providing deeper insights into the process of diversification among the descendants of individual T cells, and introduce two models that emphasize either intrinsic noise or extrinsic signals as driving forces behind the diversification of single cell-derived T cell progeny populations in vivo.
Keywords: T cell memory, Subset diversity, Fate, Plasticity, Single T cell-derived progeny, Intrinsic noise, Extrinsic environmental cues
Introduction
A central characteristic of T cell immune responses is their ability to generate a short-lived effector phase, essential for defense against acute infection, as well as a lasting—often life-long—memory towards re-challenge with the same antigen. We know today that this dual capacity of T cell immune responses can be attributed to distinct subsets of antigen-experienced T cells including at least one short-lived subset mounting acute effector functions (such as cytokine secretion or cytotoxicity) and at least one T cell subset harboring the ability to persist over long periods of time, ready to respond to subsequent antigen re-challenge [1]. Historically, it had been difficult to demonstrate the presence of memory- versus effector-prone T cell subsets due to the fact that most known features of memory T cells were either uniformly expressed or uniformly absent from all T cells participating in primary responses to a given antigen [2]. However, further research yielded the identification of markers differentially expressed on responding T cells early after primary antigen contact, and it soon became obvious that, depending on the set of markers examined, T cell subsets differ with regard to their migratory activity, effector function, and future capacity to become memory cells [3].
The advent of major histocompatibility complex (MHC) multimer technology enabled the study of T cell populations of identical peptide–MHC specificity and confirmed aspects of diversification first observed in less rigidly controlled systems. The generation of T cell receptor (TCR)-transgenic mouse lines for use in adoptive transfer experiments increased the homogeneity of responding naïve T cells even further—in this case to clonal TCR identity—and still preserved all aspects of effector versus memory diversification.
The delineation of further subsets among effector and memory T cell populations that originated from phenotypically homogenous naïve T cells is an ongoing process. Current immunology has firmly established the distinction into memory- and effector-prone T cells, but these two subsets seem to encompass many additional diverse paths of differentiation [4]. The stimuli that can direct differentiation down an individual lineage path—while another is obstructed—are currently under intense investigation and are reviewed elsewhere [5]. In this review, we mainly focus on current insights concerning the following challenging questions: How can multiple paths of differentiation evolve simultaneously, coming from the same “source”, i.e., from a pool of naïve T cells sharing the same peptide–MHC specificity or even the same TCR? What is the smallest unit from which the simultaneous generation of distinct T cell subsets may occur? And what are the environmental or cell-intrinsic prerequisites for diversification?
In the first part of our review, we showcase current evidence for antigen-driven diversification in CD8+ T cell responses and discuss models for diversity acquisition within clonal populations. The second part will focus on novel technologies that enable the monitoring of different aspects of diversification, starting out from single T cells. In the last section, we will summarize recent insights into the process of diversification, gained by novel technologies, and will discuss two models for the putative forces driving diversification on the single-cell level.
Diverse capacity for memory and function
During infection or other foreign antigen exposure, naïve T cells recognizing their cognate antigen undergo vigorous proliferation, reaching maximum population sizes at approximately 1 week after priming, and then contract to 5–10 % of their original burst size. As early as day 4–5 after infection with lymphocytic choriomeningitis virus (LCMV), the surface markers Killer cell lectin-like receptor subfamily G member 1 (KLRG1) and interleukin-7 receptor alpha chain, also known as CD127, indicate a first distinction into KLRG1+CD127− short-lived effectors cells (SLECs) and KLRG1−CD127+ memory-prone effector cells (MPECs) [6]. At this time, both subsets contain cells displaying effector characteristics, e.g., cytokine secretion. SLECs, however, are programmed for terminal differentiation and die soon after peak expansion, while the MPEC compartment contains at least a fraction of cells capable of transition to memory. Later during the primary response, around the time of peak expansion, further subdivision of the memory-prone compartment, detected by expression of lymph node homing marker L-Selectin (CD62L), becomes evident [7, 8]. CD62L−CD127+ T cells populate the pool of so-called T effector memory (TEM) cells, while CD62L+CD127+ T cells become T central memory (TCM) cells. The two subsets differ in their migratory behavior as well as their ability for immediate display of effector functions and their proliferative capacity upon re-challenge. While TEM cells home to peripheral organs such as gut or lung and display immediate effector function but poor proliferative activity upon re-challenge, TCM cells re-circulate through secondary lymphatic organs and proliferate vigorously upon antigen re-encounter but display few direct effector functions [9]. More detailed investigation into the paradigm of TCM versus TEM cells in mice clearly confirmed their distinctiveness in terms of proliferative capacity and homing [10] but also provided conflicting data on their commitment to exerting effector functions. In fact, some studies show that TCM and TEM cells are very similar in terms of interferon-γ (IFN-γ) secretion and even cytotoxic capacity [11]. A major functional characteristic that remains a factor of distinction between CD8+ TEF and TCM cells is the ability of the latter to secrete interleukin-2 (IL-2) upon antigen contact [9]; this also seems to be a functional characteristic setting apart memory-prone (IL-2+) and effector-prone (IL-2−) CD8+ T cells early during primary responses [12]. Studies simultaneously evaluating the expression of IFN-γ, IL-2, tumor necrosis factor-α, and macrophage-inflammatory protein-1-β via multi-parameter flow cytometry also show that, even within defined memory subsets, cytokine secretion patterns are far from homogenous: multifunctional T cells (secreting multiple of the above factors) exist in parallel to oligo-functional T cells (secreting, for example, only IFN-γ), creating further subdivisions within established memory subsets [13]. The presence of multi-functional T cells within a given memory compartment seems to be of major importance for the protective quality of T cell memory in mouse infection models [14] and human infections with pathogens such as human immunodeficiency virus [15, 16]. However, the capacity for production of defined cytokines is not irresolvably tied to certain paths of memory; it rather creates another layer of diversity that has to be considered in addition to central or effector memory characteristics [17] (Fig. 1). An optimal T cell immune response to a given pathogen probably does not consist of a single optimal T cell subset. Instead, it is likely to require a division of labor between multiple subsets—long- and short-lived, functional and quiescent—that only through their interplay are able to address the manifold challenges that infection poses. We are only beginning to understand how vaccination strategies can be modulated to preferentially generate a certain T cell subset. However, the truly challenging question might be how to generate a certain diversity of subsets specific to a given antigen. Rational design of vaccines aimed at this goal requires a better understanding of how diversity arises.
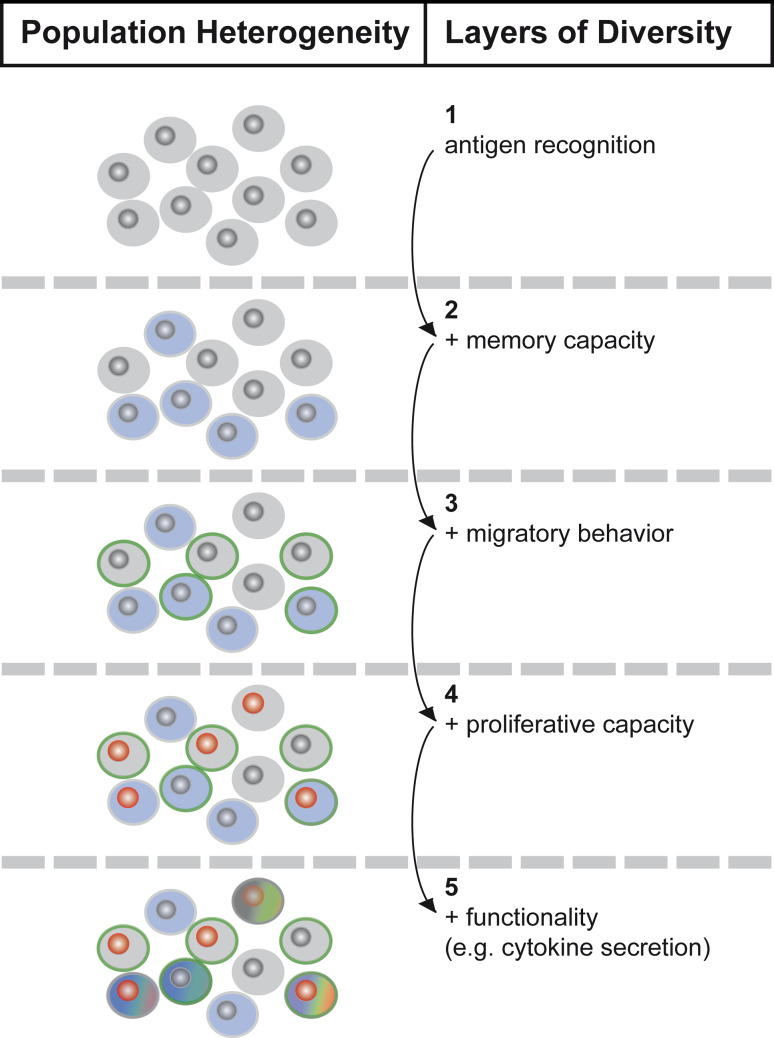
Fig. 1.
Heterogeneity of effector and memory CD8+ T cell subsets. When multiple parameters (1–5) are taken into account, bipolar subdivision into memory and effector T cell subsets does not suffice to describe the phenotypic heterogeneity present within a population of antigen-experienced T cells. Migratory behavior and proliferative capacity further subdivide effector and memory compartments. Within these sub-compartments, distinct cytokine secretion patterns set apart multi- from mono- or non-functional T cells and thus generate further functional diversity
Origin of diversification
Estimates for the number of naïve precursor T cells specific for one peptide-MHC complex range between 50 and 500 cells in mice [18, 19]. The number of T cells expressing identical TCRs has been suggested to range between 10 and 50 cells per mouse [20].
The diversification of naïve T cells can be studied at different resolutions. Either all T cells responsive to a pathogen or only subpopulations specific to a defined antigen or peptide can be analyzed for their capacity to generate memory and effector subsets. Given the conceptual notion that all TCR specificities should potentially be available for recruitment into acute immune responses as well as preservation into memory, the smallest units that should be able undergo sufficient in vivo diversification are clonal TCR-identical populations of naïve T cells (Fig. 2).
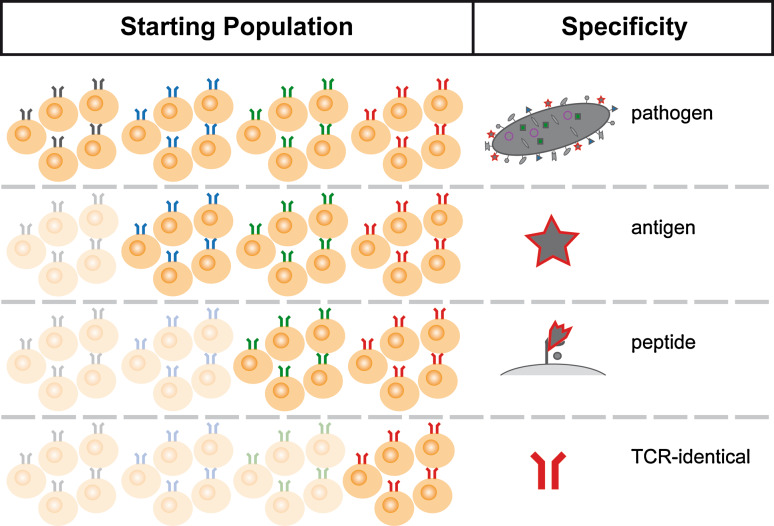
Fig. 2.
Response “units” derived from the naïve T cell repertoire. T cells participating in a fully developed immune response against a complex pathogen are heterogeneous in their clonal descent. The number of clonal ancestral populations contributing to a pathogen-specific response will generally be larger than the number contributing to antigen- or peptide-specific responses. The smallest unit that should be capable of generating both effector and memory T cells is a population of TCR-identical naïve T cells
While MHC multimer staining allows the definition of progeny originating from naïve T cells with identical peptide–MHC specificity [21], even in humans and genetically un-manipulated mice, delineation of clonal progenies in these systems is only possible via TCR repertoire analysis. Two pioneering studies in this field yielded somewhat contradictory information. One study in humans showed TCR repertoires of TCM and TEM cells retrieved from peripheral blood to be largely distinct [22], a discovery interpreted as evidence for the selective recruitment of certain TCR specificities for either central or effector memory differentiation. Another study, however, demonstrated largely overlapping TCR repertoires in TCM and TEM populations derived from various peripheral lymphatic organs in mice [23].
Easy access to TCR-identical populations of T cells is possible through the use of TCR-transgenic mouse lines for adoptive transfer experiments [24]. When the number of naïve TCR-transgenic T cells transferred into wild-type hosts is adapted to physiological precursor frequencies, this approach grants valuable insights into the process of subset diversification [25]. Thus, it could be shown that diverse memory- and effector-prone subsets can originate from small populations of TCR-identical precursors of both CD8+ and CD4+ T cells [26]. These insights have led to different models of how diversification from clonal progenitors might occur.
Current models for the generation of diversity within clonal progeny populations
Different models for the generation of diverse memory and effector subsets have been proposed. The first two models mainly focus on the directionality of differentiation: the linear differentiation model (Fig. 3) was coined based on the initial observation that effector phenotype T cells arise before memory phenotype T cells during the course of the expansion phase of a T cell immune response. This model postulates that after encounter with their cognate antigen, T cells will proliferate and differentiate first into TEF cells, most of which will subsequently die, but a fraction will continue to display TEM characteristics, and a fraction of those will become TCM cells, re-acquiring the potential for re-circulation, homeostatic self renewal, and vigorous re-expansion [27–29]. The passage through a TEF stage, a central part of this model, received recent support from studies showing that large fractions of memory CD8+ and CD4+ T cells had at some point during their ontogeny expressed effector molecules such as Granzyme B and IFN -γ, respectively (since IFN-γ is also secreted by many CD4+ and CD8+ memory T cells, the study using IFN-γ as a reporter provides only limited ontological information) [30, 31]. These studies showed that the acquisition of some effector T cell characteristics is not obstructive to later differentiation towards memory, but they could not establish the passage through an effector phase as obligatory for memory generation.
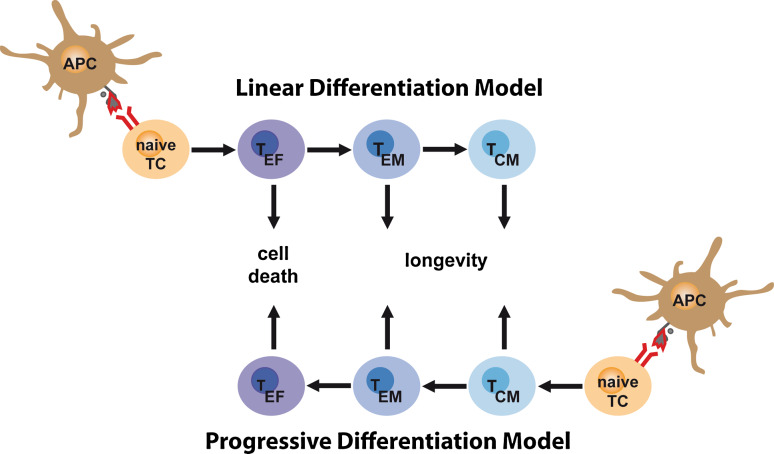
Fig. 3.
Linear versus progressive differentiation model. The ‘linear differentiation model’ proposes that T cells recognizing their cognate antigen on antigen-presenting cells (APCs) and receiving further signals through co-stimulatory molecules and cytokines will first acquire effector function (dark blue cell color); thereafter, they will either go into apoptosis and die or continue to down-regulate effector function (transition from dark to light blue cell color) and become memory T cells (TEM and TCM cells). The ‘progressive differentiation model’ postulates that antigen-experienced T cells will first acquire a status ‘low in effector function’ and prone to TCM development. Stronger activation signals will drive T cells beyond this state and into becoming TEM or even TEF cells. TEF cells in this model are irrevocably destined for cell death
The progressive differentiation model (Fig. 3) proposes the opposite course of events, namely that activated T cells do not differentiate towards a memory phenotype but away from it. Depending on the strength of the signal—integrated out of stimuli received via TCR, costimulatory molecules, cytokines, and other environmental signals—T cells are induced to either become terminally differentiated effector cells (strong signal) or remain outfitted with the naïve-like homing and proliferative characteristics of TCM cells (weak signal). In support of this model, extensive studies show the inability of phenotypically defined effector T cells to re-acquire a memory phenotype upon adoptive transfer and re-exposure to antigen [32]. The progressive differentiation model is also supported by observations showing that short or weak stimulation favors the generation of TCM cells over terminally differentiated TEF cells [33, 34]. Studies showing that late entry into an immune response [35] or the presence of many responding T cells support increased TCM generation (probably due to a decrease in the available stimuli over time or inaccessibility due to competition) [25] point in a similar direction. On the other hand, truly suboptimal signals can also obstruct the road to lasting memory [36].
A synthesis of the two models appears to be the most reasonable representation of physiological events, with a more or less unidirectional flow of differentiation from TCM to TEM to TEF cells in established memory but also a requirement for acquisition of at least some effector functions for memory licensing during the initial expansion phase. In fact, an interesting study by Intlekofer et al. [37] showed that acquisition of effector function and capacity for memory transition seem intrinsically linked. The authors describe that transcription factors T-bet (i.e., T box transcription factor expressed in T cells) and Eomesodermin both induce IFN-γ secretion and make CD8+ T cells simultaneously receptive to the essential homeostatic cytokine IL-15 by inducing expression of the IL-15 receptor beta chain.
Apart from the directionality of differentiation, the time point at which a certain profile of differentiation is fixed and stably transmitted to further progeny is another intensely debated topic. Is it determined on the level of the first cell division, or is it fixed even before the first DC–T cell encounter? Or is differentiation plastic altogether and subjected to multiple alterations during the course of population expansion? Early studies showed that the signal duration required to induce clonal populations of naïve CD8+ T cells to generate diverse progeny is very limited: 2 h of T cell–DC contact are sufficient to induce proliferation, and 24 h are enough to generate both memory and effector phenotype progeny [38, 39]. In addition, modulation of the inflammatory setting very early during infection results in substantially altered distribution into memory-prone and short-lived effector CD8+ T cell populations, modulation later results in only minor effects [40]. These results are generally interpreted as evidence for early programming of differentiation pathways and early determination of T cell memory versus effector fate. However, neither a predetermination before initial antigen contact nor the successive integration of multiple signals received during rapid population expansion can be formally excluded by the data. When following the diversification of T cell populations, it in fact appears impossible to resolve the stalemate between the opposing models favoring early determination or prolonged plasticity of T cells, proliferating and differentiating in response to antigen. Diversity of effector and memory or mono- and multifunctional T cell subsets could arise by determining multiple naïve T cells for divergent paths of differentiation before or during initial priming, thus creating homogenous progeny from one (“one cell–one fate”) but diverse progeny from many naïve ancestors. Alternatively, diversification could be generated by a single naïve T cell whose progeny retains plasticity during clonal expansion and can therefore diversify in response to varying extrinsic or intrinsic stimuli (“one cell–multiple fates”) (Fig. 4).
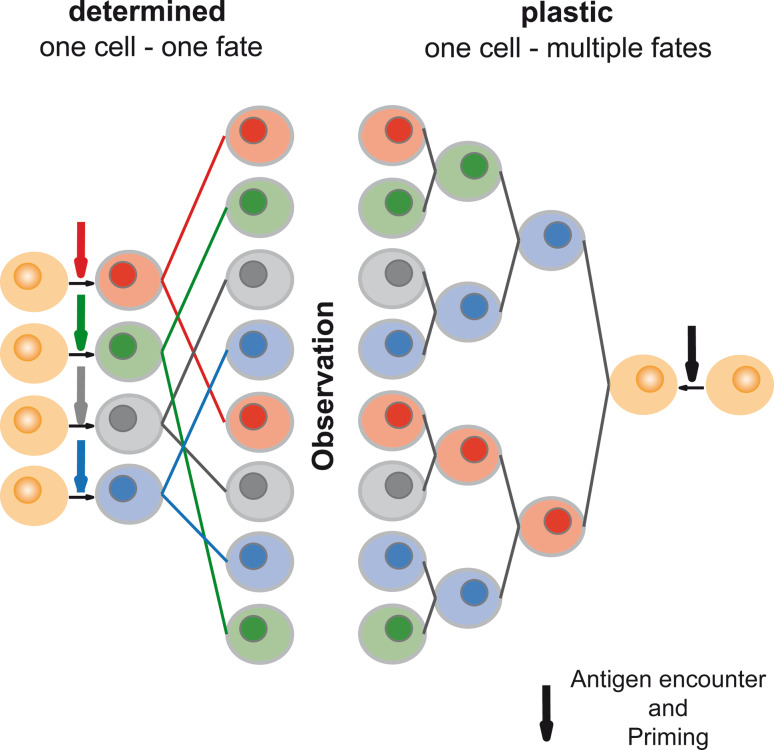
Fig. 4.
One cell–one fate versus one cell–multiple fates models. The ‘one cell–one fate’ model states that the cumulative signal (antigen, co-stimulation and cytokines) received before the first T cell division determines the T cell’s destiny, and all of its progeny adhere to the defined cell fate (e.g., effector T cell differentiation). The ‘one cell–multiple fates’ model proposes that the progeny of a T cell retains plasticity to change direction of differentiation through multiple rounds of division, thus opening the possibility to generate progeny outfitted with multiple distinct fates starting out from a single ancestor. When observing the outcome of differentiation within progeny cells derived from multiple T cells, the result predicted by a deterministic model is indistinguishable from that predicted by a plastic one
Dissecting T cell ontogeny down to the single-cell level
Further understanding of the rules and mechanisms determining T cell diversification into various effector and memory subsets requires techniques enabling the dissection of T cell ontogeny down to the single-cell level. To experimentally establish a direct link between an ancestral T cell and its progeny, two main approaches are conceivable: a single T cell can be continually observed while it is dividing and generating progeny, or a heritable marker specific to an individual ancestral T cell can be introduced, allowing the identification of all marker-positive T cells as being derived from a single ancestor.
The structure of lymphatic organs as well as the distribution and delineation of specific groups of leukocytes has been traditionally investigated using light microscopy. The staining of tissue sections with specific dyes, differentially labeling various populations of leukocytes according to the chemical properties of their sub-cellular components, is a decade-old method still widely applied for basic research and pathological evaluation of clinical specimens. While substantially refined through the use of enzymatically or fluorescently labeled antibodies or reporter systems tying a certain transcriptional activity to the expression of a fluorescent protein, classical immunohistochemistry remains an altogether static method. The requirement for thin sections of fixed tissues restricts it to the display of mere snapshots of the dynamic processes going on in vital organs.
Confocal (single-photon) fluorescence microscopy makes a deeper look into intact organs possible. By focusing a laser beam on a defined point within the tissue and blocking all light emitted from excited fluorophores outside the focal plane, background fluorescence is largely eliminated, allowing thicker tissue sections or even intact organs to be microscopically inspected. In principle, this method enables analysis of vital organs; however, the high light intensities required for sufficient excitation of the tightly defined focal plane lead to rapid photo-damage of analyzed tissues and largely preclude the visualization of dynamic processes [41].
Reduction of photo-damage and increased penetration into vital tissue became possible with the development of two-photon laser scanning microscopy. Here, an infrared laser is used to excite a focal point within a given tissue. The long wavelength of the lasers used guarantees lower light scattering and thus deeper penetration into tissues and decreased photo-damage. Reduction of background fluorescence is achieved due to the lower energy of the laser beam, which induces sufficient fluorophore excitation only when two photons are absorbed simultaneously by a given fluorophore, an event highly unlikely outside the focal point [42]. Using this technique, quite representative tissue volumes of an organ can be observed over periods of up to a few hours while leaving the organ more or less in situ. Not every organ can be analyzed equally well using this technique, and most insights concerning T cell differentiation have been gained by studying lymph nodes draining defined sites of infection [43, 44]. Intravital imaging granted novel insights into the earliest events during antigen-driven T cell responses [45]. Elegant studies could define three phases of CD8+ T cell interactions with dendritic cells (DCs) presenting cognate antigen [46]. Phase 1 is dominated by transient contacts during which upregulation of activation markers such as CD44 or CD69 occurs. Phase 2 is characterized by long-lasting interactions (up to several hours), during which T cell cytokine secretion becomes detectable. Proliferation is initiated in phase 3, and the interactions of DCs with the earliest ensuing progeny again become transient. It could further be shown that DC–T cell ratio as well as peptide–MHC density on presenting DCs and TCR affinity to presented peptides positively correlate with a shortening of phase 1 and rapid establishment of stable interactions [47]. From these discoveries, it was deduced that repetitive contacts during phase 1 allow the generation of a cumulative TCR signal that, when reaching a threshold, enables lasting interaction and induction of proliferation. These data were put into the context of findings from flow cytometrical studies showing that very short prevalence of antigen-presenting DCs or very low-affinity antigen diminishes CD8+ T cell population expansion (possibly by reducing the number of cells reaching phase 2), but leaves differentiation into diverse memory and effector T cell subsets intact [48]. The prevailing conclusion was that defined CD8+ T cell fates are determined before the first cell division, conceptually tying a single cell-derived progeny to a single fate and requiring multiple precursors for generating diversity. A study by Steven Reiner’s group put forward another concept of how different memory and effector subsets might arise. By imaging (in vitro) the first cell division of in vivo-activated CD8+ T cells, they identified a substantially unequal distribution of synaptic signaling components (such as CD8 or leukocyte function-associated antigen-1) onto the two arising daughter cells. This process—reminiscent of asymmetric cell divisions that tissue stem cells undergo—was claimed to generate a proximal effector-prone daughter cell and a distal memory-prone one [49]. However, the inheritance and lasting impact of the initial disequilibrium could not be shown conclusively.
While intravital imaging is an excellent tool for investigating the earliest events of single T cell differentiation within an intact tissue context, it also introduces some unphysiological factors and faces severe restraints in terms of monitoring effector and memory T cell ontogeny in its entirety. First, to allow the visualization of representative numbers of antigen-specific T cells within a tissue volume of approximately 1 mm3 (the volume size examined by standard imaging studies), it is necessary to introduce a very high number of cells (106 or higher) into an experimental animal. This number is approximately 10,000-fold above the threshold established as ensuring physiological conditions for T cell activation and differentiation in adoptive transfer systems [25], thus possibly limiting the physiological relevance of data acquired under such conditions. Second, limited observation time due to accumulating photo-damage and limited observation breadth due to visualization of only a small tissue volume make continuous monitoring of single cell-derived progeny cells through multiple rounds of differentiation currently impossible, precluding further-reaching conclusions regarding T cell ontogeny and kinship [41].
The first experimental approach truly enabling visualization of a complete ontological tree (or at least all of its branches) stemming from a single T cell “root” was conducted by transfer of a single antigen-specific T cell into an otherwise normal wild-type host [50]. Progeny T cells generated from the single ancestor during immunization-driven expansion were followed flow cytometrically via expression of a heritable congenic marker, thus allowing for direct identification of phenotypic and functional traits. Not only did this study show that it is technically possible to recover substantial numbers of single cell-derived progeny cells after transfer of a single T cell but it also demonstrated that progeny derived from a single naïve CD8+ T cell can develop into functionally diverse effector and long-lived memory subsets. These data stand in obvious opposition to the notion that T cell differentiation is stably determined before the first cell division or even pre-determined before antigen contact and emphasize plasticity as a hallmark of T cells proliferating and differentiating in response to antigen contact [51]. While providing the field with very clear insights into the general diversification potential of single naïve T cells, the method of transferring a single congenically marked T cell is intrinsically limited in the volume of data that can be acquired, and rare phenotypes showing, for example, limited subset diversification of single cell-derived progeny cells might be “overlooked”. A solution to this dilemma could lie in an innovative method developed by Schumacher et al. [52], who have introduced multiple unique and heritable genetic labels into an otherwise homogenous population of naïve TCR-transgenic T cells to track the kinship of single cell-derived progeny cells. This procedure—termed “barcoding” by the authors—is carried out by retrovirally transducing a unique short strip of DNA (“barcode”) into a T cell. During T cell proliferation, this barcode is faithfully transmitted to all progeny and can then be detected by subjecting T cells to microarray analysis. A library of over 3,000 different barcodes enables individualization of large populations of naïve T cells and thus provides a wealth of data concerning the shared origin of effector and memory T cells. In essence, studies carried out using the barcoding approach have confirmed that single naïve T cells can generate both effector- and memory-prone progeny and appear to do so very regularly [53]. It is of note that the authors elegantly circumvent the caveat of retroviral transduction, which requires naïve T cell proliferation and activation, by labeling physiologically proliferating thymocytes. However, among the limitations to the “barcoding” approach is the difficulty in ensuring truly unique labeling when transducing bulk T cells with a retroviral library. It must also be mentioned that the authors mainly aimed to resolve qualitative—not quantitative—differences in subset distribution among different single cell-derived progeny populations: two barcodes might, for example, be represented in both memory and effector subsets, but the relative distribution of the two corresponding single cell-derived progeny populations onto the subsets could still differ greatly.
Taken together, novel approaches for tracking single T cells and/or their progeny in vivo have changed our view of how T cell differentiation takes place. Intravital imaging studies support the notion that substantial signal integration occurs during repetitive DC–T cell interactions before the first T cell division. However “barcoding” and single T cell transfer studies show that this cumulative initial signal does not ultimately tie all single cell-derived progeny to a single fate. The observation that multiple subsets arise within a single cell-derived T cell progeny population conclusively shows that proliferating T cells retain differentiation plasticity beyond the first cell division and diversify during the expansion phase. Thus, on the level of multiple progenitors, two phases of diversification can be envisioned: phase 1 lies before the first cell division and generates multiple pre-mitotic T cells outfitted with a different “starting level” of differentiation (e.g., due to micro-anatomical variations in signal strength). In phase 2, the proliferating progeny of these T cells then diversify around the imprinted “starting level” of differentiation. But why does diversification during the expansion phase occur? What are the environmental signals and signal imbalances driving it? Are extrinsic signals required at all, or is T cell subset diversification an altogether stochastic process?
Diversity by default?
Studies monitoring the development of T effector and memory subsets starting out from populations of TCR-transgenic T cells have painted a picture that emphasizes the stunning robustness of diversification. In a study carried out in a CD11c-Diphteria toxin receptor (DTR) transgenic mouse model, where the duration of antigen exposure and inflammation was limited to less than a day by DT-induced depletion of DCs, it was found that, although T cell proliferation was massively diminished, no substantial alteration could be found in the distribution of cells onto T memory and effector subsets compared to non-depleted mice [48]. A study comparing the responses of TCR-transgenic T cells triggered by very low-affinity or very high-affinity antigen expressing L.m.-OVA yielded similar results [54]. Thus, it does not seem to matter what happens after priming: diversity always prevails. These observations could tempt one to speculate that diversification is driven by intrinsic processes leading to stochastically uneven distribution of differentiation signatures among dividing progeny. In fact, the topic of transcriptional noise as a driving force behind probabilistic differentiation of stem cells has drawn substantial attention in recent years [55–57]. A basic concept from this line of research proposes that stem cells shift between different transcriptional states in a probabilistic manner, allowing for extrinsic signal-dependent differentiation of cells in state A, while preserving others in a signal unreceptive state B [58]. While this particular example does not fully abolish the need for extrinsic signals to establish diversity, it shows that these signals do not necessarily have to be diverse themselves but only have to act upon populations of intrinsically state-shifting cells. We can speculate that such diverse states might be created in T cells by, for example, the bi-polar expression of CD25, creating a high or low responder state to autocrine IL-2. In fact, high CD25 expression in a subset of virus-specific CD8+ T cells responding to infection could recently be correlated with an IL-2 dependent bias for effector differentiation and apoptosis [59]. CD25low T cells, although producing higher amounts of IL-2, were shown to preferentially continue towards a memory phenotype.
Diversity due to variable environmental cues?
If it is true that antigen and inflammation do not substantially influence diversification of T cells during the expansion phase, then removing these signals directly after priming should not restrict diversification. While evidence in this direction has been generated on the population level [48], studies investigating this topic with resolution down to the level of single cell-derived progeny cells are largely lacking. However, a recent publication gave a first glimpse of what might really happen when the environment is “turned off”. In a hybrid intravital imaging and flow cytometry-based study, Beuneu and colleagues [60] made use of the Yeti-System, which has a yellow fluorescent protein (YFP) placed under control of the IFN-γ promoter, to examine the early acquisition and heritability of functional characteristics among single CD8+ TCR-transgenic T cells and their ensuing progeny. The authors could show by intravital imaging that substantial differences in IFN-γ promoter activity were detectable before the first cell division. They went on to flow cytometrically sort single YFPhigh and YFPlow pre-mitotic T cells and subjected them to further in vitro expansion in cultures devoid of antigen and supplemented only with IL-2. After approximately 5–6 cell divisions, single cell-derived progeny cells were reexamined for IFN-γ promoter activity via YFP expression and IFN-γ secretion. Surprisingly, both promoter activity and cytokine secretion proved to be remarkably restricted to the ancestors’ phenotype—being either IFN-γ high or IFN-γ low. While this study focused on cytokine production and made no statement concerning effector or memory phenotype, the restrictive cytokine phenotype observed for single cell-derived progeny cells under “environmentally limited” conditions in vitro stands in contrast to diverse cytokine profiles observed after single cell transfer and population expansion in vivo [50, 61]. Thus, although diversification due to stochastic variations in transcriptional activity (i.e., transcriptional noise) is far from being ruled out, environmental influences on diversification need re-investigation. But why should single cell-derived subset diversification be so dependent on environmental cues during expansion phase while population-derived subset diversification is not? The answer to this question might lie in the fact that individual members of a population of T cells could be imprinted with distinct differentiation programs before the first cell division while—quite obviously—a single cell could always only acquire one program before it has divided. Thus, while in the first case diversity already exists before the first cell division, it has to still develop in the second case, making it vulnerable to restriction of environmental cues during the expansion phase.
Conclusion
We have reviewed here our current knowledge of the multiple layers of effector and memory subset diversity that have become evident within clonal populations of CD8+ T cells responding to antigen exposure and have highlighted the capacity of even single naïve CD8+ T cells to generate multi-faceted progeny populations. Major unanswered questions regarding how diversification occurs remain: is diversification driven intrinsically through stochastic processes within every dividing T cell or extrinsically through the heterogeneous exposure of an evolving progeny population to distinct environmental cues? Ultimately, the consequences of environmental modulation at different time points during T cell immune responses will have to be analyzed. Will “turning off” inflammation and/or antigen presentation after initial priming have detrimental effects on the diversity of single cell-derived progeny populations? If not, then the key to diversification lies preferentially within T cells themselves. If so, then extrinsic cues would have to be essential to the process of diversification (Fig. 5), making it well imaginable that not every environment creates optimal conditions for diversification.
Fig. 5.
Driving diversification: intrinsic versus extrinsic factors. The restriction of environmental influences (“cues”) could be a way to experimentally investigate the factors driving subset diversification within single T cell-derived progeny populations. If restriction of antigen exposure and/or inflammatory stimuli during population expansion does not alter the profile and degree of subset diversification, then cell intrinsic processes must drive diversification. If, on the other hand, subset diversification is limited through restriction of environmental input, then diversification must at least partly be dependent on extrinsic signals
Answers to these questions will require monitoring of progeny of single cell origin to unmask distorting effects present in population-based studies. It will additionally require both representative and sensitive methods for monitoring single T cell-derived progeny cells: ‘representative’ by faithfully monitoring many single cell life histories, and ‘sensitive’ by allowing quantitative statements regarding the amount of diversity achieved in a certain experimental setting. Current studies highlight the robustness of diversification. However, this robustness might be the product of a dual process, relying both on the diversification potential of single cell-derived progeny cells under optimal conditions and the efficient recruitment of multiple clonal progenitors [62] to correct for limited single cell-derived diversification under sub-optimal conditions.
Understanding the mechanisms of and the prerequisites for single T cell-derived diversification could have a substantial impact on the development of efficient vaccination strategies. For example, vaccination against target antigens for which only small numbers of naïve precursor T cells pass thymic selection into the periphery (e.g., tumor antigens) will require optimal diversification of responders in order to result in effective immune responses. In the case of chronic infections, there is frequent antigen re-encounter and substantial clonal T cell turnover, and insufficient diversification of progeny populations could lead to the loss of individual T cell clones, which might have substantial implications for the control of (chronic) infection. Therefore, a better understanding of how to guide a single T cell’s progeny towards diversification into short-living effector T cell populations and long-living memory T cells should provide valuable tools to improve current immunotherapies.
Abbreviations
- DCs
Dendritic cells
- DTR
Diphteria toxin receptor
- IFN-γ
Interferon-γ
- IL-2
Interleukin-2
- LCMV
Lymphocytic choriomeningitis virus
- KLRG1
Killer cell lectin-like receptor subfamily G member 1
- MHC
Major histocompatibility complex
- MPECs
Memory-prone effector cells
- SLECs
Short-lived effectors cells
- TCM cells
T central memory cells
- TEM cells
T effector memory cells
- T-bet
T box transcription factor expressed in T cells
- YFP
Yellow fluorescent protein
References
- 1.Williams MA, Bevan MJ. Effector and memory CTL differentiation. Annu Rev Immunol. 2007;25:171–192. doi: 10.1146/annurev.immunol.25.022106.141548. [DOI] [PubMed] [Google Scholar]
- 2.Dutton RW, Bradley LM, Swain SL. T cell memory. Annu Rev Immunol. 1998;16:201–223. doi: 10.1146/annurev.immunol.16.1.201. [DOI] [PubMed] [Google Scholar]
- 3.Huster KM, Stemberger C, Busch DH. Protective immunity towards intracellular pathogens. Curr Opin Immunol. 2006;18(4):458–464. doi: 10.1016/j.coi.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Jameson SC, Masopust D. Diversity in T cell memory: an embarrassment of riches. Immunity. 2009;31(6):859–871. doi: 10.1016/j.immuni.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rutishauser RL, Kaech SM. Generating diversity: transcriptional regulation of effector and memory CD8 T-cell differentiation. Immunol Rev. 2010;235(1):219–233. doi: 10.1111/j.0105-2896.2010.00901.x. [DOI] [PubMed] [Google Scholar]
- 6.Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, Gapin L, Kaech SM. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27(2):281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, Ahmed R. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol. 2003;4(12):1191–1198. doi: 10.1038/ni1009. [DOI] [PubMed] [Google Scholar]
- 8.Huster KM, Busch V, Schiemann M, Linkemann K, Kerksiek KM, Wagner H, Busch DH. Selective expression of IL-7 receptor on memory T cells identifies early CD40L-dependent generation of distinct CD8+ memory T cell subsets. Proc Natl Acad Sci USA. 2004;101(15):5610–5615. doi: 10.1073/pnas.0308054101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401(6754):708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 10.Masopust D, Vezys V, Marzo AL, Lefrançois L. Preferential localization of effector memory cells in nonlymphoid tissue. Science. 2001;291(5512):2413–2417. doi: 10.1126/science.1058867. [DOI] [PubMed] [Google Scholar]
- 11.Unsoeld H, Krautwald S, Voehringer D, Kunzendorf U, Pircher H. Cutting edge: CCR7+ and CCR7− memory T cells do not differ in immediate effector cell function. J Immunol. 2002;169(2):638–641. doi: 10.4049/jimmunol.169.2.638. [DOI] [PubMed] [Google Scholar]
- 12.Sarkar S, Kalia V, Haining WN, Konieczny BT, Subramaniam S, Ahmed R. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J Exp Med. 2008;205(3):625–640. doi: 10.1084/jem.20071641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seder RA, Darrah PA, Roederer M. T-cell quality in memory and protection: implications for vaccine design. Nat Rev Immunol. 2008;8(4):247–258. doi: 10.1038/nri2274. [DOI] [PubMed] [Google Scholar]
- 14.Darrah PA, Patel DT, De Luca PM, Lindsay RW, Davey DF, Flynn BJ, Hoff ST, Andersen P, Reed SG, Morris SL, Roederer M, Seder RA. Multifunctional T(H)1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat Med. 2007;13:843–850. doi: 10.1038/nm1592. [DOI] [PubMed] [Google Scholar]
- 15.Harari A, Petitpierre S, Vallelian F, Pantaleo G. Skewed representation of functionally distinct populations of virus-specific CD4 T cells in HIV-1-infected subjects with progressive disease: changes after antiretroviral therapy. Blood. 2004;103(3):966–972. doi: 10.1182/blood-2003-04-1203. [DOI] [PubMed] [Google Scholar]
- 16.Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, Roederer M, Koup RA. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107(12):4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pantaleo G, Koup RA. Correlates of immune protection in HIV-1 infection: what we know, what we don’t know, what we should know. Nat Med. 2004;10(8):806–810. doi: 10.1038/nm0804-806. [DOI] [PubMed] [Google Scholar]
- 18.Blattman JN, Antia R, Sourdive DJD, Wang X, Kaech SM, Murali-Krishna K, Altman JD, Ahmed R. Estimating the precursor frequency of naive antigen-specific CD8 T cells. J Exp Med. 2002;195(5):657–664. doi: 10.1084/jem.20001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moon J, Chu H, Pepper M, Mcsorley S, Jameson S, Kedl R, Jenkins M. Naive CD4+ T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 2007;27(2):203–213. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casrouge A, Beaudoing E, Dalle S, Pannetier C, Kanellopoulos J, Kourilsky P. Size estimate of the alpha beta TCR repertoire of naive mouse splenocytes. J Immunol. 2000;164(11):5782–5787. doi: 10.4049/jimmunol.164.11.5782. [DOI] [PubMed] [Google Scholar]
- 21.Busch DH, Pilip IM, Vijh S, Pamer EG. Coordinate regulation of complex T cell populations responding to bacterial infection. Immunity. 1998;8(3):353–362. doi: 10.1016/S1074-7613(00)80540-3. [DOI] [PubMed] [Google Scholar]
- 22.Baron V, Bouneaud C, Cumano A, Lim A, Arstila TP, Kourilsky P, Ferradini L, Pannetier C. The repertoires of circulating human CD8(+) central and effector memory T cell subsets are largely distinct. Immunity. 2003;18(2):193–204. doi: 10.1016/S1074-7613(03)00020-7. [DOI] [PubMed] [Google Scholar]
- 23.Bouneaud C, Garcia Z, Kourilsky P, Pannetier C. Lineage relationships, homeostasis, and recall capacities of central- and effector-memory CD8 T cells in vivo. J Exp Med. 2005;201(4):579–590. doi: 10.1084/jem.20040876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy KM, Heimberger AB, Loh DY. Induction by antigen of intrathymic apoptosis of CD4+CD8+TCRlo thymocytes in vivo. Science. 1990;250(4988):1720–1723. doi: 10.1126/science.2125367. [DOI] [PubMed] [Google Scholar]
- 25.Badovinac VP, Haring JS, Harty JT. Initial T cell receptor transgenic cell precursor frequency dictates critical aspects of the CD8(+) T cell response to infection. Immunity. 2007;26(6):827–841. doi: 10.1016/j.immuni.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stemberger C, Neuenhahn M, Buchholz VR, Busch DH. Origin of CD8+ effector and memory T Cell subsets. Cell Mol Immunol. 2007;4(6):399–405. [PubMed] [Google Scholar]
- 27.Opferman JT, Ober BT, Ashton-Rickardt PG. Linear differentiation of cytotoxic effectors into memory T lymphocytes. Science. 1999;283(5408):1745–1748. doi: 10.1126/science.283.5408.1745. [DOI] [PubMed] [Google Scholar]
- 28.Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 2002;111(6):837–851. doi: 10.1016/S0092-8674(02)01139-X. [DOI] [PubMed] [Google Scholar]
- 29.Wherry EJ, Teichgräber V, Becker TC, Masopust D, Kaech SM, Antia R, Von Andrian UH, Ahmed R. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat Immunol. 2003;4(3):225–234. doi: 10.1038/ni889. [DOI] [PubMed] [Google Scholar]
- 30.Harrington LE, Janowski KM, Oliver JR, Zajac AJ, Weaver CT. Memory CD4 T cells emerge from effector T-cell progenitors. Nature. 2008;452(7185):356–360. doi: 10.1038/nature06672. [DOI] [PubMed] [Google Scholar]
- 31.Bannard O, Kraman M, Fearon DT. Secondary replicative function of CD8+ T cells that had developed an effector phenotype. Science. 2009;323(5913):505–509. doi: 10.1126/science.1166831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huster KM, Koffler M, Stemberger C, Schiemann M, Wagner H, Busch DH. Unidirectional development of CD8+ central memory T cells into protective listeria-specific effector memory T cells. Eur J Immunol. 2006;36(6):1453–1464. doi: 10.1002/eji.200635874. [DOI] [PubMed] [Google Scholar]
- 33.Goldrath AW, Bogatzki LY, Bevan MJ. Naive T cells transiently acquire a memory-like phenotype during homeostasis-driven proliferation. J Exp Med. 2000;192(4):557–564. doi: 10.1084/jem.192.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lauvau G, Vijh S, Kong P, Horng T, Kerksiek K, Serbina N, Tuma RA, Pamer EG. Priming of memory but not effector CD8 T cells by a killed bacterial vaccine. Science. 2001;294(5547):1735–1739. doi: 10.1126/science.1064571. [DOI] [PubMed] [Google Scholar]
- 35.D’Souza WN, Hedrick SM. Cutting edge: latecomer CD8 T cells are imprinted with a unique differentiation program. J Immunol. 2006;177(2):777–781. doi: 10.4049/jimmunol.177.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gett AV, Sallusto F, Lanzavecchia A, Geginat J. T cell fitness determined by signal strength. Nat Immunol. 2003;4(4):355–360. doi: 10.1038/ni908. [DOI] [PubMed] [Google Scholar]
- 37.Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, Gapin L, Ryan K, Russ AP, Lindsten T, Orange JS, Goldrath AW, Ahmed R, Reiner SL. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6(12):1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 38.Kaech SM, Ahmed R. Memory CD8+ T cell differentiation: initial antigen encounter triggers a developmental program in naïve cells. Nat Immunol. 2001;2(5):415–422. doi: 10.1038/87720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Stipdonk MJ, Lemmens EE, Schoenberger SP. Naïve CTLs require a single brief period of antigenic stimulation for clonal expansion and differentiation. Nat Immunol. 2001;2(5):423–429. doi: 10.1038/87730. [DOI] [PubMed] [Google Scholar]
- 40.Badovinac VP, Porter BB, Harty JT. CD8+ T cell contraction is controlled by early inflammation. Nat Immunol. 2004;5(8):809–817. doi: 10.1038/ni1098. [DOI] [PubMed] [Google Scholar]
- 41.Germain RN, Miller MJ, Dustin ML, Nussenzweig MC. Dynamic imaging of the immune system: progress, pitfalls and promise. Nat Rev Immunol. 2006;6(7):497–507. doi: 10.1038/nri1884. [DOI] [PubMed] [Google Scholar]
- 42.Cahalan MD, Parker I, Wei SH, Miller MJ. Two-photon tissue imaging: seeing the immune system in a fresh light. Nat Rev Immunol. 2002;2(11):872–880. doi: 10.1038/nri935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller MJ, Wei SH, Parker I, Cahalan MD. Two-photon imaging of lymphocyte motility and antigen response in intact lymph node. Science. 2002;296(5574):1869–1873. doi: 10.1126/science.1070051. [DOI] [PubMed] [Google Scholar]
- 44.Stoll S, Delon J, Brotz TM, Germain RN. Dynamic imaging of T cell-dendritic cell interactions in lymph nodes. Science. 2002;296(5574):1873–1876. doi: 10.1126/science.1071065. [DOI] [PubMed] [Google Scholar]
- 45.Henrickson SE, von Andrian UH. Single-cell dynamics of T-cell priming. Curr Opin Immunol. 2007;19(3):249–258. doi: 10.1016/j.coi.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 46.Mempel TR, Henrickson SE, Von Andrian UH. T-cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature. 2004;427(6970):154–159. doi: 10.1038/nature02238. [DOI] [PubMed] [Google Scholar]
- 47.Henrickson SE, Mempel TR, Mazo IB, Liu B, Artyomov MN, Zheng H, Peixoto A, Flynn MP, Senman B, Junt T, Wong HC, Chakraborty AK, Von Andrian UH. T cell sensing of antigen dose governs interactive behavior with dendritic cells and sets a threshold for T cell activation. Nat Immunol. 2008;9(3):282–291. doi: 10.1038/ni1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prlic M. Duration of the initial TCR stimulus controls the magnitude but not functionality of the CD8+ T cell response. J Exp Med. 2006;203(9):2135–2143. doi: 10.1084/jem.20060928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang JT, Palanivel VR, Kinjyo I, Schambach F, Intlekofer AM, Banerjee A, Longworth SA, Vinup KE, Mrass P, Oliaro J, Killeen N, Orange JS, Russell SM, Weninger W, Reiner SL. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science. 2007;315(5819):1687–1691. doi: 10.1126/science.1139393. [DOI] [PubMed] [Google Scholar]
- 50.Stemberger C, Huster KM, Koffler M, Anderl F, Schiemann M, Wagner H, Busch DH. A single naive CD8 + T cell precursor can develop into diverse effector and memory subsets. Immunity. 2007;27(6):985–997. doi: 10.1016/j.immuni.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 51.Reiner SL, Sallusto F, Lanzavecchia A. Division of labor with a workforce of one: challenges in specifying effector and memory T cell fate. Science. 2007;317(5838):622–625. doi: 10.1126/science.1143775. [DOI] [PubMed] [Google Scholar]
- 52.Schepers K, Swart E, Van Heijst JWJ, Gerlach C, Castrucci M, Sie D, Heimerikx M, Velds A, Kerkhoven RM, Arens R, Schumacher TNM. Dissecting T cell lineage relationships by cellular barcoding. J Exp Med. 2008;205(10):2309–2318. doi: 10.1084/jem.20072462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gerlach C, van Heijst JWJ, Swart E, Sie D, Armstrong N, Kerkhoven RM, Zehn D, Bevan MJ, Schepers K, Schumacher TNM. One naive T cell, multiple fates in CD8+ T cell differentiation. J Exp Med. 2010;207(6):1235–1246. doi: 10.1084/jem.20091175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zehn D, Lee SY, Bevan MJ. Complete but curtailed T-cell response to very low affinity antigen. Nature. 2009;458(7235):211–214. doi: 10.1038/nature07657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morris SA, Teo RTY, Li H, Robson P, Glover DM, Zernicka-Goetz M. Origin and formation of the first two distinct cell types of the inner cell mass in the mouse embryo. Proc Natl Acad Sci USA. 2010;107(14):6364–6369. doi: 10.1073/pnas.0915063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamanaka Y, Lanner F, Rossant J. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development. 2010;137(5):715–724. doi: 10.1242/dev.043471. [DOI] [PubMed] [Google Scholar]
- 57.Chazaud C, Yamanaka Y, Pawson T, Rossant J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev Cell. 2006;10(5):615–624. doi: 10.1016/j.devcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 58.Eldar A, Elowitz MB. Functional roles for noise in genetic circuits. Nature. 2010;467(7312):167–173. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kalia V, Sarkar S, Subramaniam S, Haining WN, Smith KA, Ahmed R. Prolonged interleukin-2Ralpha expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity. 2010;32(1):91–103. doi: 10.1016/j.immuni.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 60.Beuneu H, Lemaître F, Deguine J, Moreau HD, Bouvier I, Garcia Z, Albert ML, Bousso P. Visualizing the functional diversification of CD8+ T cell responses in lymph nodes. Immunity. 2010;33(3):412–423. doi: 10.1016/j.immuni.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 61.Stemberger C, Neuenhahn M, Gebhardt FE, Schiemann M, Buchholz VR, Busch DH. Stem cell-like plasticity of naïve and distinct memory CD8(+) T cell subsets. Semin Immunol. 2009;21:62–68. doi: 10.1016/j.smim.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 62.van Heijst JWJ, Gerlach C, Swart E, Sie D, Nunes-Alves C, Kerkhoven RM, Arens R, Correia-Neves M, Schepers K, Schumacher TNM. Recruitment of antigen-specific CD8+ T cells in response to infection is markedly efficient. Science. 2009;325(5945):1265–1269. doi: 10.1126/science.1175455. [DOI] [PubMed] [Google Scholar]