Abstract
An endogenous timing mechanism, the circadian clock, causes rhythmic expression of a considerable fraction of the genome of most organisms to optimally align physiology and behavior with their environment. Circadian clocks are self-sustained oscillators primarily based on transcriptional feedback loops and post-translational modification of clock proteins. It is increasingly becoming clear that regulation at the RNA level strongly impacts the cellular circadian transcriptome and proteome as well as the oscillator mechanism itself. This review focuses on posttranscriptional events, discussing RNA-binding proteins that, by influencing the timing of pre-mRNA splicing, polyadenylation and RNA decay, shape rhythmic expression profiles. Furthermore, recent findings on the contribution of microRNAs to orchestrating circadian rhythms are summarized.
Keywords: Circadian clock, Post-transcriptional, miRNA, RNA-binding proteins, RNA decay, Splicing
Introduction
The periodic environment generated by the rotation of the earth has caused organisms to acquire an awareness of internal time. Endogenous clocks have evolved that govern daily physiological processes such as sleep/wake cycles and metabolic activity in metazoa or growth and expression of photosynthetic genes in plants [1, 2]. These circadian (meaning “about daily”) clocks generate endogenous rhythms with a period of about 24 h.
Circadian oscillators in eukaryotes consist of autoregulatory feedback loops operating at the level of a single cell. Transcription factors (TFs) activate the expression of clock proteins that in turn inhibit the TFs and thus repress transcription of their own genes. To maintain synchrony with the environment, the oscillator components receive input from ambient light and temperature in a process designated entrainment. In turn, they impart rhythmicity to downstream genes to control biochemical and physiological rhythms, the output.
Transcription so far is considered the prime mechanism driving rhythms in gene expression both within the oscillator and in clock output. In addition, post-translational modifications of the clock components regulate their subcellular localization, interaction, activity and turnover, as reviewed in [3–5]. They help to sustain a 24-h period and avoid generation of a steady state. Accumulating evidence points to the importance of post-transcriptional control, a layer between transcription and translation, in the circadian system (Fig. 1a) [6–8].
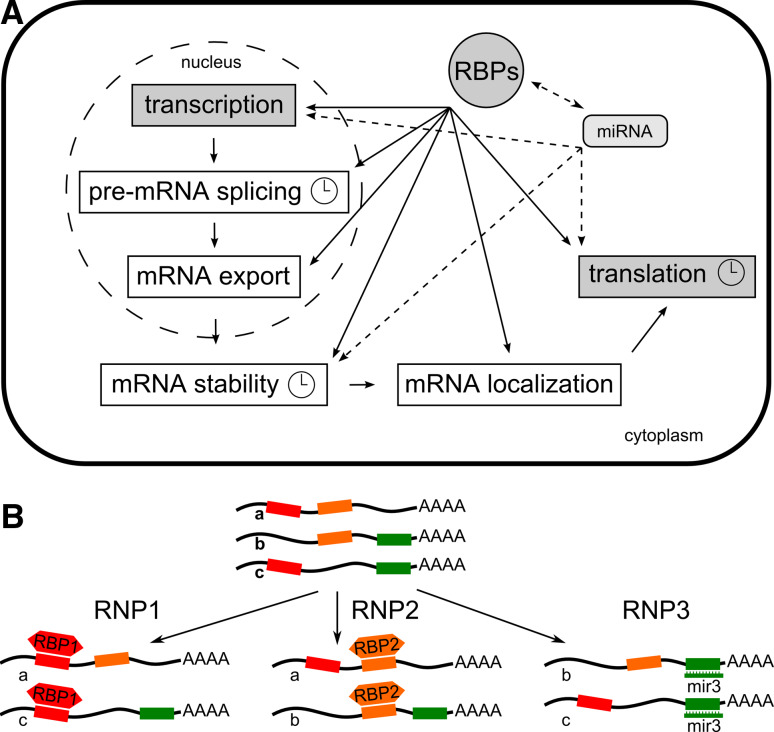
Fig. 1.
Post-transcriptional regulation of gene expression. a The steps of pre-mRNA processing controlled by RNA-binding proteins or microRNAs. Clock symbols denote events known to be associated with circadian regulation. b Post-transcriptional operons defined by RBPs or miRNAs binding to cognate cis-regulatory elements in mRNAs [11]
Throughout their life, mRNAs are bound by a suite of proteins collectively known as heterogenous nuclear ribonucleoproteins (hnRNPs) [9, 10]. A model has been put forward in which ribonucleoprotein complexes organize post-transcriptional regulation of mRNAs [11]. Through interaction with trans-acting RNA-binding proteins (RBPs), subpopulations of mRNAs can be co-ordinately processed, i.e. spliced, exported from the nucleus, turned over or translated (Fig. 1b). Indeed, RBPs can target multiple mRNAs in vivo, suggesting an integrated post-transcriptional regulation [12]. Often, mRNAs targeted by the same RBP code for functionally related gene products. Therefore, in analogy to the organization of coordinately expressed bacterial genes into DNA operons the terms “RNA operons” or “RNA regulons” have been coined [11]. Eukaryotic monocistronic mRNAs are assembled in functional groups much like prokaryotic polycistronic mRNAs, which encode two or more functionally related proteins. With the discovery of a novel cellular regulatory system based on microRNAs, the concept of RBP-driven operons was extended to these small RNAs [13].
One may envisage that the circadian system also relies on organizing clock-regulated genes in post-transcriptional operons [14]. The discovery of RBPs in the circadian transcriptome of several organisms indeed suggests a role in shaping rhythmic transcript profiles at the RNA level [15–17]. Early on, it was observed that nitrate reductase has a constant transcription rate but a rhythmic mRNA accumulation pattern in the model plant Arabidopsis thaliana [18]. Also, the importance of transcriptional control within the core oscillator has been questioned to some degree by the recent observation that mouse fibroblasts treated with RNA polymerase inhibitors retain mRNA rhythms, albeit with a reduced amplitude [19]. Furthermore, half of the proteins that cycle in mouse liver are translated from constitutively expressed mRNAs, pointing to a prominent role for translational control [20].
This review summarizes recent insights into post-transcriptional events in the circadian system of the model organisms Drosophila, mammals, Neurospora, Chlamydomonas and Arabidopsis. After introducing the blueprint of the transcriptional clock feedback loops, we focus on events in circadian regulation that involve cis-active RNA motifs and their trans-acting RBPs or non-coding RNAs. These events include mRNA decay, polyadenylation, pre-mRNA splicing, and translation.
Basic design of transcriptional feedback loops
Generally, in the transcriptional clock circuits positive elements activate the expression of “clock” genes encoding negative elements (Fig. 2a) [21, 22]. The clock proteins physically interact with the positive elements to inhibit their activity. In an additional loop the negative elements can also promote expression of the positive elements. This network structure contributes to stability and robustness.
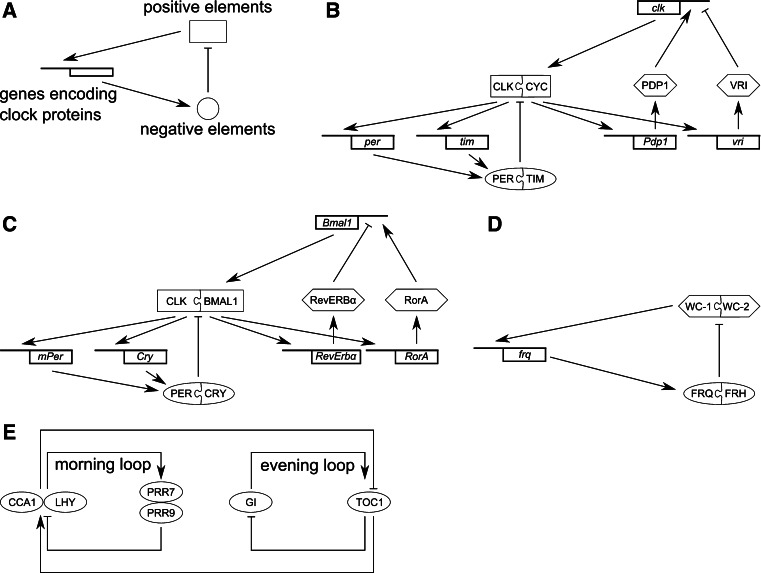
Fig. 2.
Transcriptional clock feedback loops. Common design principle (a) and model of the Drosophila (b), mammalian (c) and Neurospora (d) oscillator. See text for details. e Simplified scheme of the Arabidopsis oscillator consisting of the central CCA1/LHY-TOC1 loop, a morning-phase loop and an evening-phased loop [2, 32]
In the fly Drosophila melanogaster, transcription of the clock genes period (per) and timeless (tim) is activated by the two basic helix- loop-helix (bHLH) TFs CLOCK (CLK) and CYCLE (CYC) (Fig. 2b). After a time delay, PER and TIM proteins enter the nucleus where the association of PER with the CLK/CYC heterodimer inhibits their transcriptional activity. Gradually decreasing levels of TIM and PER relieve autoinhibition and initiate another round of transcription (for review see [23, 24]. CLK/CYC also activate an interdependent feedback loop comprising the activator PAR DOMAIN PROTEIN1 (PDP1) and the repressor VRILLE (VRI) which in turn regulate clk [25].
In mammals, the TFs CLK and BMAL1, the homolog of Drosophila CYC, activate Cryptochrome (Cry1 and Cry2) and Period (mPer1, mPer2 and mPer3) genes (for review, see [26, 27]). The PER/CRY complexes in turn repress BMAL1/CLK-mediated transcription of their own genes [28] (Fig. 2c). Furthermore, BMAL1/CLK activate transcription of the nuclear receptors Rev-erbα and RorA [29]. REV-ERBα represses Bmal1 and RorA activates Bmal1 [30].
In the fungus Neurospora crassa, transcription of the clock gene frequency (frq) is activated by the PAS domain proteins WHITE COLLAR-1 (WC-1) and WC-2 that form the WHITE COLLAR complex (WCC) and bind to the frq promoter (Fig. 2d). FRQ protein then inhibits the activity of WCC and thereby feeds back on its own synthesis (for review, see [22, 31]). The role of the FRQ-interacting helicase FRH is detailed below.
Similar to the Neurospora clock, the circadian clock in the higher plant A. thaliana uses conserved principles of rhythm generation but utilizes different molecular players. Two MYB TFs, CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) inhibit the expression of TIMING OF CAB EXPRESSION 1 (TOC1) by directly binding to its promoter, whereas TOC1 contributes to CCA1 and LHY activation (Fig. 2e). This central loop connects additional morning- and evening-phased loops (for a recent review see [2]). In the morning-phased loop, CCA1 and LHY activate the expression of PSEUDO-RESPONSE REGULATOR 7 (PRR7) and PRR9 that subsequently feed back to repress CCA1 and LHY. In the evening-phased loop, TOC1 expression is activated by GIGANTEA (GI) which in turn is negatively controlled by TOC1. Connected with these loops are additional components whose exact mode of action remains to be defined [2, 32].
In Chlamydomonas reinhardtii, an RBP protein appears to be an important component of the timekeeping system [33], but the molecular makeup of the core oscillator is not yet clear [34]. Notably, in the prokaryotic cyanobacteria, the core oscillator mechanism comprises interactions between the Kai clock proteins and rhythmic KaiC protein phosphorylation [22]. Thus, timekeeping is primarily based on a post-translational oscillator rather than on a transcriptional/translational feedback loop, as described above for the eukaryotic model organisms.
mRNA decay
As described above, periodic transcriptional activity is largely responsible for circadian transcript oscillations. Because rhythmic mRNA profiles show a delayed accumulation and reduced amplitude compared to rhythms in transcriptional activity, these will manifest themselves in high amplitude mRNA oscillations only if a transcript has a sufficiently short half-life [35, 36].
A time-of-day-dependent change in mRNA half-life on top of rhythmic transcription was first discovered for the Drosophila clock gene per. Nuclear run-on assays revealed a delayed per mRNA accumulation relative to per transcription during the rising phase whereas mRNA abundance closely follows transcription during the declining phase [37]. In flies expressing PER from a transgene lacking the authentic promoter, per mRNA cycles with a low amplitude and rescues the behavioral arrhythmicity of a per o mutant [38]. With the help of per-luciferase reporter constructs, it was shown that apart from the promoter an element within the first intron of per is required to faithfully mimic per mRNA oscillations [39]. Direct measurements of run-on transcription uncovered that this intragenic element at least partly operates post-transcriptionally [37].
Generally, mRNA decay is mediated by the interaction between specific cis-acting motifs and cognate trans-acting factors. The two main turnover pathways are both initiated by poly(A) tail removal, followed either by removal of the 5′ 7-methylguanosine cap and 5′ → 3′ exoribonuclease digestion or by 3′ → 5′ digestion [40].
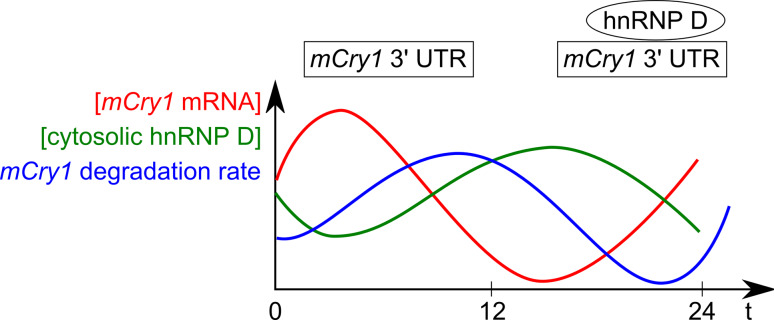
Direct evidence for regulated mRNA decay was obtained for the mammalian clock genes mPer2, mPer3 and mCry1. Mper2 mRNA decays faster during the declining phase than during the rising phase of its oscillation. The degradation is mediated by its 3′UTR and the cognate trans-acting factor was identified as the polypyrimidine tract-binding protein (PTB) [41]. PTB, also termed hnRNP I, preferentially binds pyrimidine-rich regions in introns to regulate alternative splicing but has also been implicated in other post-transcriptional events. When PTB is depleted by RNAi in fibroblasts, mPer2 mRNA is stabilized. Whereas PTB preferentially localizes to the nucleus, increased PTB levels in the cytoplasm and binding of this cytoplasmic PTB to the mPer2 3′UTR are observed at times when the mPer2 mRNA declines [41]. This inverse correlation of PTB in the cytoplasm with mPer2 oscillations suggests that PTB is responsible for the mPer2 degradation during the declining phase. Notably, the mPer2 3′UTR alone is not sufficient to convey circadian oscillations upon a reporter driven by a constitutive promoter, indicating that it cooperates with the transcriptional regulation in shaping the mPer2 profile [41]. Similarly, the mPer3 3′UTR modulates RNA stability but the cognate protein factors have not been identified [42]. The 3′UTR of mCry1 is recognized by hnRNP D that is known to regulate stability or translation of its target mRNAs [43]. Knockdown of hnRNP D stabilizes mCry1 mRNA and delays its circadian phase. As observed for PTB, the amount of hnRNP D in the cytoplasm cycles in antiphase to mCry1, suggesting that regulation by hnRNP D of the degradation rate modulates mCry1 rhythms (Fig. 3).
Fig. 3.
Scheme of the inverse mCry1 mRNA and cytoplasmic hnRNP D profiles. Relative levels of mCry1, hnRNP D protein abundance in the cytoplasm and mCry1 degradation rate across 1 day are shown (adapted from data in [43])
Rhythmic mRNA degradation is also intrinsic to oscillations of serotonin N-acetyltransferase (AANAT), the key enzyme in melatonin synthesis [44]. Melatonin production is activated in the dark, and levels of circulating melatonin increase at night. The AANAT 3′UTR mediates mRNA degradation through interaction with hnRNP R, hnRNP Q and hnRNP L. These accumulate to highest levels around the time of the AANAT peak and thus contribute to its rapid degradation during the declining phase. Notably, hnRNP Q has an additional function in the control of rhythmic AANAT translation [45] (see below).
A cis-acting element widely found in 3′UTRs of short-lived mRNAs in mammals is the AU-rich element (ARE) that targets them for decay. One of the factors interacting with ARE is butyrate response factor1, which destabilizes early response gene transcripts. It undergoes circadian oscillations and has been proposed to impose time-of-day-dependent degradation onto its targets as part of an oscillating post-transcriptional RNA operon [14, 46]. So a picture emerges that RBPs participating in general post-transcriptional regulatory events in the cell are employed by the circadian system to time the half-life of mRNAs both within and downstream of the oscillator.
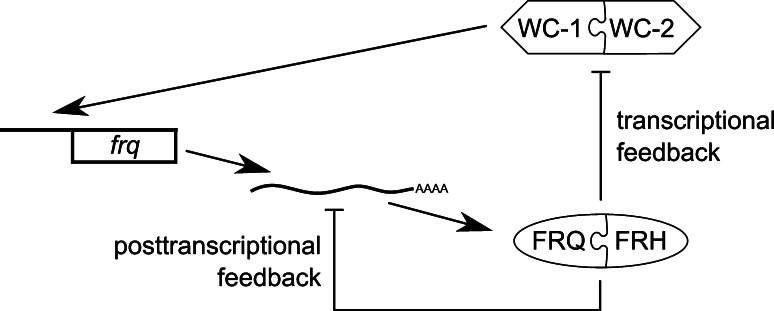
An even more fundamental role for regulated mRNA decay is emerging in Neurospora. The FRQ clock protein forms a complex with FRH (FRQ-interacting helicase) (Fig. 2d), a member of the DExD/H family of RNA helicases that use ATP hydrolysis to unwind double-stranded RNA [47]. Thus, a protein with a predicted function in RNA metabolism is part of the transcriptional feedback loop, repressing frq transcription through interaction with WCC. Recently, it has been found that FRQ and FRH also regulate frq mRNA abundance at the post-transcriptional level [48]. Downregulation of frh increases the stability of frq mRNA that is associated with the FRQ–FRH complex in vivo. Because FRQ itself has no domain predicted to bind nucleic acids, this presumably is through FRH. FRH shows significant homology to Mtr4p, a cofactor of the yeast exosome that is one of the primary machineries for RNA degradation [40]. In Neurospora, FRH interacts with the ortholog of the 3′ → 5′ exonuclease subunit RRP44. Thus, FRH likely targets frq mRNA for degradation when FRQ–FRH levels are high. Indeed, downregulation of RRP44 leads to enhanced frq mRNA stability, higher frq levels and a longer period of frq mRNA oscillations. Thus, FRQ, FRH and the exosome are part of a post-transcriptional negative feedback loop interlocked with the transcriptional feedback loop (Fig. 4) [48]. Liu and coworkers have taken this a step further by showing that the rrp44 transcript itself is clock-controlled. Moreover, the exosome regulates a few other rhythmic transcripts, suggesting that time-of-day-dependent exosome activity may play a wider role in circadian regulation.
Fig. 4.
Transcriptional and post-transcriptional regulation by FRH. In the transcriptional feedback loop, FRQ and FRH inhibit WCC activity leading to repression of frq transcription. In the post-transcriptional feedback loop, FRQ and FRH promote frq decay through the exosome
In Arabidopsis, an approach to globally identify short-lived transcripts has identified a suite of clock-controlled transcripts [49]. For some of them mRNA stability changes over the circadian cycle [50]. Furthermore, conditional instability of the CCA1 transcript in the light has been suggested to contribute to synchronization of the Arabidopsis clock with the environment [51].
Polyadenylation
mRNAs are polyadenylated at their 3′ ends, and poly(A) binding proteins (PABP) associate with this tail, leading to mRNA stabilization. The interaction of PABP with the translation initation factor complex assembled around the 5′ m7G cap brings about circularization of the mRNA and provides the basis for 3′UTR participation in translational control [52].
Daily fluctuations in poly(A) tail size have initially been observed for the mRNA encoding the neuroactive peptide vasopressin that is synthesized within the suprachiasmatic nuclei (SCN) of rats and displays rhythmic concentration changes in the cerebrospinal fluid [53]. Poly(A) tail shortening is catalyzed by deadenylases, Mg2+-dependent 3′ → 5′ exoribonucleases [40]. Mammals contain five deadenylases of which Nocturnin, the homolog of yeast CCR4, is rhythmically expressed [54]. Mice deficient for Nocturnin have defects in lipid homeostasis and response to glucose, suggesting that Nocturnin mediates post-transcriptional regulation of metabolic events by the circadian clock [55]. Direct targets of Nocturnin have not yet been found and thus it is not clear whether it influences degradation or translational activity through the changes in poly(A) tail length. Notably, in Neurospora, the increased stability of frq mRNA upon FRH downregulation (see above) is accompanied by increased poly(A) tail length [48].
Splicing
Drosophilaper comes in two transcript forms that differ by the presence or absence of an intron in the 3′UTR [56]. Cycling of both transcripts is virtually identical; however, transgenic flies that express only the intron-containing transcript show longer activity rhythms and slower accumulation of PER protein. Thus, the retention of the 3′UTR intron may affect per mRNA translation. This splicing event is under clock-control and actually serves to adjust the fly’s activity cycle to seasonal progression [57–59]. Both low temperature and short photoperiod (usually linked in nature) promote splicing of the 3′UTR intron leading to a more rapid increase in per mRNA and PER protein with concomitant earlier evening activity of the flies. In contrast, warm temperatures and long photoperiods decrease the splicing activity and lead to mainly nocturnal activity, thus avoiding desiccation during daytime heat. This temperature-dependent splicing has been attributed to suboptimal splice signals that are less efficiently recognized as the temperature rises [60]. Notably, natural variation of these splice sites has been connected to the adjustment to latitudes.
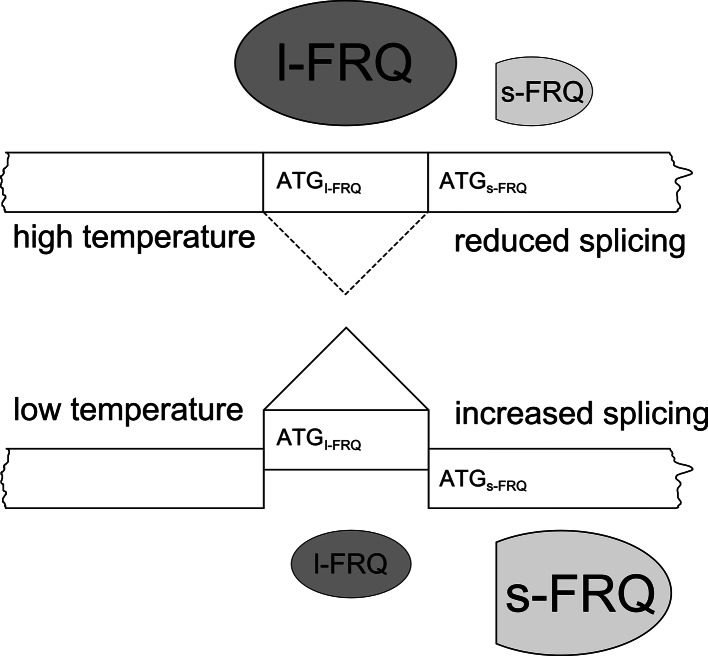
In Neurospora, conditional splicing of an intron containing the FRQ initiation codon in response to ambient temperature is observed. FRQ comes in a large (l) form required to maintain rhythmicity at warm temperatures and a small (s) form required at cold temperatures [61]. Their ratio is controlled by a temperature-dependent choice of start codons that correlates with alternative splicing of an intron encompassing the AUG [62]. The non-canonical splice sites are used with higher efficiency at lower temperatures and, accordingly, the fraction of s-FRQ increases with decreasing temperature (Fig. 5). This regulation of the l- versus s-FRQ ratio in a temperature-dependent fashion allows the fungus to ensure robust rhythmicity over a broad temperature range.
Fig. 5.
Temperature-dependent alternative splicing of an intron encompassing the l-FRQ start codon regulates the ratio of l-FRQ versus s-FRQ. The sizes of l-FRQ and s-FRQ symbols reflect their relative amounts
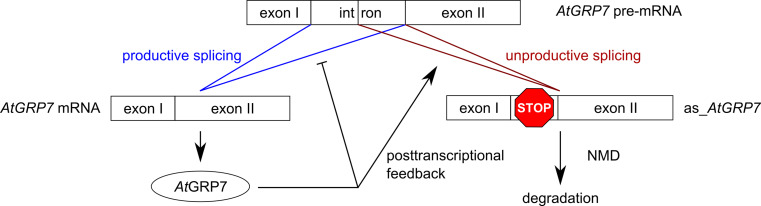
In Arabidopsis, the RRM (RNA recognition motif) proteins AtGRP7 (A. thaliana glycine-rich RBP7) and AtGRP8, also designated CCR2 (COLD AND CIRCADIAN REGULATED2) and CCR1, are clock-controlled at the level of transcription [63–65] and negatively autoregulate through alternative splicing (AS). When the proteins reach a certain threshold, they bind to their own pre-mRNAs and promote the formation of an AS form with a premature termination codon (PTC) in the retained part of the intron [66–69]. The AS form rapidly decays but accumulates in mutants defective in UPF1 or UPF3, the Arabidopsis orthologs of nonsense-mediated decay (NMD) pathway components [70]. NMD is a quality control mechanism that selectively clears the transcriptome of mRNAs harboring PTCs and now emerges as a more global cellular regulator [71]. AtGRP7 and AtGRP8 are the first circadian components proposed to generate a post-transcriptional negative feedback loop by coupling unproductive splicing to decay via UPF1 and UPF3 (Fig. 6). Interestingly, in Neurospora, knockdown of an UPF1 ortholog leads to an altered frq profile and shorter period (A. Mehra and J.C. Dunlap, unpublished).
Fig. 6.
Negative autoregulation of the RNA-binding protein AtGRP7 through alternative splicing and subsequent nonsense-mediated decay (NMD) [70]. In the presence of elevated AtGRP7 protein levels a higher proportion of AtGRP7 pre-mRNA is spliced to a PTC-containing NMD substrate through binding of AtGRP7 to its own intron and subsequent use of a cryptic intronic 5′splice site. The alternative splice form (as_AtGRP7) contains a PTC and is degraded via NMD
The AtGRP7 feedback loop functions in clock output, as AtGRP7 affects several rhythmic transcripts (C. Streitner and D. Staiger, unpublished). Apart from their prominent circadian regulation, AtGRP7 and AtGRP8 distinctly react to biotic and abiotic stresses such as low temperature or oxidative stress and thus may serve to integrate external stimuli with endogenous timing cues [64, 72–74].
Notably, among potentially functional counterparts of the plant glycine-rich RBPs in humans is cold-inducible RNA-binding protein (CIRP). Exposure of mammalian cells to mild cold stress leads to an increase in cold-shock proteins including CIRP that is thought to function as an RNA chaperone [75]. Cirp mRNA oscillates with a peak at the time when body temperature rhythms reach their trough. CIRP has been suggested to affect accumulation or translation of rhythmic transcripts in a temperature-dependent fashion [76, 77]. In addition to the clock in the SCN of the hypothalamus, most organs contain peripheral oscillators of similar composition. Although these are cell-autonomous and self-sustained, they depend on synchronization through systemic cues of the SCN that may comprise rhythms in hormones, metabolites or body temperature. In transgenic mice in which the oscillator in liver is turned off, very few transcripts retain rhythmicity, including CIRP. It is tempting to speculate that the systemically driven CIRP may play a role in entraining peripheral oscillators by temperature rhythms [77].
Indirect hints that the presence or absence of introns relates to circadian patterns come from a timecourse using Arabidopsis whole genome tiling arrays [78]. About 500 transcripts with rhythmic introns are detected. Roughly 40% of the cycling introns are in genes whose exonic sequences are also rhythmic. Some of the introns show a phase difference to the exonic sequences and almost 60% of the cycling introns are in genes with non-rhythmic coding regions [78]. One may assume that intron-containing and intron-less mRNAs are assembled into different mRNPs and thus differentially regulated. Intron retention is frequent in Arabidopsis [79] and obviously can lead to transcripts with PTCs. Such time-of-day-specific appearance of PTCs in otherwise constantly expressed transcripts could cause their disappearance through NMD. Alternatively, gradual appearance of a truncated protein acting as dominant negative inhibitor could terminate the function of the authentic protein, thereby shaping its activity profile. Clearly, more detailed investigations are required to understand the significance of these cycling introns.
Translation
Translation initiation of individual mRNAs is regulated through sequence-specific RBPs or miRNAs [52]. Notably, these interactions occur mostly in the 3′UTR.
In Drosophila, an RBP implicated in circadian translational control is encoded by the lark gene. LARK contains two RRMs and a zinc finger. In lark mutants, the emergence of adult flies from pupal cases shows an earlier phase. This “eclosion” is under clock control with a peak in the morning to avoid water loss of the newly hatched flies [80]. The mutation does not affect the core clock, indicating that LARK operates downstream of the clock to mediate distinct behavioural outputs. Immunoprecipitation of LARK mRNPs with subsequent microarray analysis of complexed RNAs identified RNAs associated with LARK in vivo [81]. Two of these targets give rise to increased protein abundance upon LARK overexpression without an effect on transcript levels consistent with translational control by LARK.
LARK also interacts with dFMRP, the Drosophila orthologue of Fragile X mental retardation protein [82]. Loss of FMRP causes the most common heritable form of mental retardation in mammals and also leads to altered behavioral rhythmicity [83]. FRMP harbors two K-homology (KH) RNA binding domains and an arginine- and glycine-rich domain. It has a widespread role in translation, mRNA stability and trafficking. Flies lacking dFMRP also have disturbed circadian rhythms presumably due to alterations in clock output [82]. dFMRP and LARK appear to function together and, indeed, common target transcripts of dFMRP and LARK regulation have been identified. In mammals, RNA binding motif protein 4 (RBM4) is the putative counterpart of LARK. It binds to the 3′UTR of mPer1, leading to increased mPER1 levels [84]. This increase depends on the cap and poly(A) tail and thus likely affects translation. RBM4 protein levels rise towards the end of the day and could stimulate translation at a time when mPer1 transcript levels decline. Another twist is added by observations that implicate LARK and FMRP in miRNA-mediated regulation both in Drosophila and mammals [82, 85].
A novel type of RBP, CHLAMY1, has been implicated in circadian translational control in the green alga C. reinhardtii. The heteromeric protein binds to UG repeats and consists of the C1 subunit containing three KH motifs and the C3 subunit, a member of the CUG-BP-ETR-3-Like factors (CELF) family with three RRMs [86]. A bioinformatic search for CHLAMY1 targets has identified transcripts with at least seven tandem UGs in their 3′UTR and CHLAMY1 binding has been confirmed for a suite of transcripts associated with CO2- and N-metabolism [87]. Notably, the activity of one of these enzymes, nitrite reductase, increases in the morning when CHLAMY1 binding activity is low, suggesting that CHLAMY1 acts as a translational repressor. The UG repeats of the glutamine synthetase2, argininosuccinate lyase and nitrite reductase1 mRNAs confer rhythmicity upon a reporter gene when inserted in its 3′UTR and determine phase [88]. CHLAMY1 thus can be considered as key component of a post-transcriptional operon coordinating rhythmic translation of enzymes involved in CO2- and N-metabolism. CHLAMY1 has been shown to be crucial for maintaining period, indicating that it may also be part of the core oscillator itself [33].
In rare cases, translation initiation is mediated by internal ribosome entry sites (IRES) that recruit ribosomes cap-independently with the help of trans-acting factors [52]. Oscillations of serotonin N-acetyltransferase in mammals partly rely on rhythmic mRNA degradation mediated by hnRNP R, hnRNP Q and hnRNP L [44] (see above). The circadian pattern of protein accumulation is additionally mediated by the 5′UTR with a region within the AANAT 5′UTR that contains an IRES specifically interacting with hnRNP Q [45]. The level of hnRNP Q increases concurrently with AANAT protein at nighttime, and peak abundance of AANAT protein strongly decreases upon knockdown of hnRNP Q, suggesting that hnRNP Q indeed contributes to AANAT oscillations [45].
MicroRNAs
miRNAs are single-stranded RNA species of ~22 nucleotides. They represent recently discovered post-transcriptional regulators in development, differentiation and response to environmental signals [89, 90]. Their double-stranded precursors (pre-miRNAs) are hairpin loop structures excised from primary transcripts (pri-miRNAs) by DROSHA, assisted by proteins with double-stranded RNA binding domains. Pre-miRNAs are subsequently exported from the nucleus and further processed by DICER to duplexes of ~22 nucleotides which 2-nucleotide overhangs at the 3′ ends. One strand is loaded onto the RNA-induced silencing complex (RISC), which subsequently is directed to the target mRNA. In animals, miRNAs usually have imperfect complementarity to elements in the 3′UTRs of mRNA targets, leading to inhibition of translation and/or stimulation of degradation [91, 92]. As miRNAs control at least a third of mammalian mRNAs and thus represent global regulators of utmost importance [90], it is perhaps not surprising that they appear as novel players in the circadian system.
A microarray experiment was performed in Drosophila to discover rhythmic miRNAs in a circadian timecourse [93]. Dme-miR-263a and -263b were identified that exhibit daily changes in abundance in wild-type flies but not in an arrhythmic mutant defective in CYCLE [93]. Among the predicted targets are several transcripts related to clock function that now need to be validated.
When the miRNA biogenesis pathway in clock neurons in fly heads was downregulated in another study, the amplitude of locomotor activity rhythms was reduced [94]. To identify targets of miRNA regulation, mRNAs associated with RISC have been co-immunoprecipitated from fly head extracts with an antibody against ARGONAUTE1 (AGO1). Rhythmically expressed mRNAs are not significantly enriched but mRNAs for the clock components clk and vri are. Notably, clk association with AGO1 varies across the day and is maximal several hours after its mRNA peak. The clk 3′UTR confers repression upon a luciferase reporter, indicating that indeed it serves as miRNA-binding site. These data suggest that declining CLK levels following the clk mRNA peak partly result from miRNA-mediated repression [94].
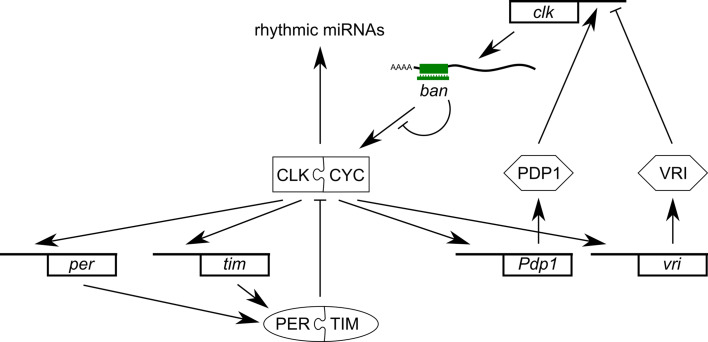
Having thus established a miRNA-based component in the core clock mechanism of Drosophila, an elegant experiment designed to specifically inhibit pri-miRNA maturation in circadian cell types by RNAi against DROSHA and PASHA identifed miRNAs involved. Among the pri-miRNAs accumulating is the precursor of bantam (ban), a miRNA with a prominent role in development [95]. In addition, ban appears to affect the core oscillator through translational regulation of clk conferred by ban binding sites in its 3′UTR (Fig. 7) [94].
Fig. 7.
Involvement of the miRNAs in the Drosophila circadian system. The core oscillator controls rhythmic expression of a number of miRNAs [93]. The bantam miRNA targets the oscillator component CLK through interaction with its 3′UTR [94]
Another strategy was chosen to identify rhythmic miRNAs in mammals, based on the rationale that miRNA oscillations may result from BMAL1/CLK-dependent rhythmic transcription [96]. Chromatin immunoprecipitation revealed that CLK binds to the enhancer region of the miR219-1 gene, and both pre-miR-219 and mature miR-219 indeed display circadian rhythms. Moreover, knockdown by miR-219 antagomirs leads to period lengthening indicating that miR-219 is not only clock-controlled but in turn also influences rhythms. Similarly, the enhancer of the miR-132 precursor is bound by CREB, a light-inducible transcription factor in the SCN, and miR132 modulates light input to the clock [96]. The targets of both miRNAs remain to be determined.
Profiling the miRNome by microarrays has identified several miRNAs with diurnal variation in abundance in mouse retina [97]. Of these, miR-182 and miR-96 target the 3′UTR of Adenylyl cyclase VI mRNA (Adcy6) that oscillates in antiphase with miR-182 and miR-96, suggesting that both miRNAs contribute to Adcy6 cycling. Also among their predicted targets is Clk, adding the possibility for another level of feedback on the core oscillator [97].
In mouse liver, the pri- and pre-miRNA precursors of miR122, the prevalent miRNA in hepatocytes, oscillate about four- to tenfold in abundance, indicating that transcription of the miR-122 locus is under circadian control [98]. However, mature miR-122 levels do not change throughout the day due to their high stability. To unravel whether miR-122 would nevertheless exert circadian control, the global effect of miR-122 downregulation has been analyzed [98]. In these experiments, the steady-state abundance of the target mRNAs served as proxy for their translational repression, since it is often accompanied by reduced mRNA levels. Indeed, transcripts with altered levels in mice injected with miR-122 antisense oligonucleotides were significantly enriched for circadian transcripts. Among them are transcripts for regulators of metabolism suggesting that miR-122 participates in circadian control of hepatic metabolism. Also in line with these results is the finding that the consequence of miR122 depletion on circadian targets depends on the time of the day. Gatfield and colleagues propose that constant repression of basal translation levels by an miRNA can strongly increase the circadian amplitude of a protein synthesized from rhythmic mRNA. Alternatively, the newly loaded RISC complex that depends on processing of rhythmically available pre-miRNA122 could be functionally different from already existing RISC complex [98].
In Arabidopsis, several miRNAs oscillate either diurnally or under clock-control [78, 99]. The functional implications are not yet clear. However, miRNAs have a demonstrated role in clock output processes such as the seasonal timing of floral transition in Arabidopsis [100].
Natural antisense transcripts
Natural antisense transcripts (NATs) have been implicated in clock regulation in Neurospora. The Crosthwaite laboratory has identified a suite of antisense transcripts overlapping the frq gene that cycle in an opposite phase to frq [101]. Like frq itself, these NATs are induced by light and mutants defective in light-induced production of frq NATs show delayed rhythms and enhanced phase shifting by light. Thus, the NATs appear to play a role in light entrainment.
In Arabidopsis, rhythmic NATs are detected for 7% of the protein coding genes using tiling arrays [78]. Among these are the clock-associated proteins LHY, CCA1 and TOC1. In contrast with the frq NATs, these NATs have a phase similar to their sense transcripts [78]. Furthermore, 813 of the rhythmic NATs correspond to sense strands that are arrhythmic. Before any conclusions on the functional significance can be drawn, it needs to be determined to what extent sense and antisense transcripts are co-expressed in the same cell type.
The regulation of Arabidopsis FLOWERING LOCUS C (FLC) by NATs may also be of relevance for the circadian system. Apart from controlling the transition to flowering, FLC lengthens the period of the circadian clock at 27°C and thus has temperature-specific effects on circadian rhythms [102]. An FLC NAT is involved in regulation of FLC abundance through a novel mechanism: alternative 3′end processing of the FLC antisense transcript influences transcription of the FLC sense transcript through chromatin modification [103].
Outlook
The observed regulation at the post-transcriptional level for both circadian output genes and core clock genes enforces the notion that circadian transcription can only partly account for rhythmic gene expression. So far, insights into post-transcriptional events mainly concern one-on-one control: in several instances, RBPs have been identified that, through recognizing cis-active features within the transcripts, shape rhythmic mRNA profiles or time-of-day-dependent translation. Among these are both general RBPs previously identified as mediators of mRNA processing, like hnRNP K or PTB, and RBPs identified on the basis of their circadian function, like CHLAMY1 and AtGRP7. Recently, a genome-wide screen for modifiers of the circadian clock in human cell lines has uncovered several RBPs and splicing factors affecting amplitude or period of clock genes [104].
Large-scale definition of RBP targets will extend the view on post-transcriptional networks. A bioinformatic search based on the known binding site for a circadian RBP has successfully identified transcripts whose rhythmic translation is controlled by CHLAMY1 [87]. However, computational methods to identify conserved RNA binding motifs have their limitations. The structural context of the binding sites is important for target recognition and, thus, programs for RNA sequence alignment have to be informed by structure [105]. Furthermore, microarray profiling of transgenic organisms aberrantly expressing RBPs also reveal indirect targets. Thus, immunoprecipitation of RNPs and identification of the associated RNAs by microarray hybridization or deep sequencing is required to globally define mRNAs associated with RBPs in vivo, as performed for Drosophila LARK. In addition to targeted analysis of circadian RBPs, mining of transcripts associated with non-circadian RBPs will presumably uncover overlaps with the circadian transcriptome in the light of general cellular RBPs influencing clock transcript oscillations [41, 43].
To fully understand the extent of post-transcriptional coordination in the circadian system, appropriate genome-wide screens for post-transcriptional events have to be further developed. Assessment of mRNA turnover by microarray hybridization requires elimination of the transcriptional component through inhibitors of transcription. In this way, oscillating transcripts have been found among unstable transcripts in Arabidopsis [49]. To monitor genome wide time-of-day-specific translational status of mRNAs, their association with polysomes can be assessed [106].
More globally, post-transcriptional processes themselves may be under clock control, as exemplified by circadian regulation of exosome components in Neurospora [48]. The identified miRNA participation in core clock mechanisms adds the possibility of additional layers of feed-back regulation and fine tuning of clock gene expression. Introduction of miRNAs in a theoretical model describing the negative feedback of Drosophila PER on its own RNA has shown that both amplitude and frequency can be affected [107]. The impact of miRNAs on the circadian transcriptome and proteome is likely larger than currently estimated, because both the percentage of genes under control by miRNAs is increasing and novel modes of miRNAs actions are discovered such as a decoy activity interfering with RBPs action on their targets [108].
Acknowledgments
We thank Jay Dunlap for communication of results prior to publication, Stefan Janssen for help in preparing the figures and the anonymous reviewers for their suggestions on the manuscript. Tino Köster is a fellow of the German National Academic Foundation. Work in our laboratory is supported by the German Research Foundation through Grant STA 653/2 and the SFB 613.
Abbreviations
- AANAT
Serotonin N-acetyltransferase
- AGO1
ARGONAUTE1
- ARE
AU-rich element
- AS
Alternative splicing
- bHLH
Basic helix-loop-helix
- CCA1
CIRCADIAN CLOCK ASSOCIATED1
- CIRP
Cold-inducible RNA-binding protein
- CLK
CLOCK
- CRY
CRYPTHOCHROME
- CYC
CYCLE
- FLC
FLOWERING LOCUS C
- FMRP
Fragile X mental retardation protein
- FRH
FRQ-interacting helicase
- FRQ
FREQUENCY
- GI
GIGANTEA
- hnRNP
Heterogenous nuclear ribonucleoprotein
- IRES
Internal ribosome entry sites
- KH
K homology
- LHY
LATE ELONGATED HYPOCOTYL
- NAT
Natural antisense transcripts
- NMD
Nonsense-mediated decay
- PABP
Poly(A) binding protein
- PDP
PAR DOMAIN PROTEIN
- PER
PERIOD
- PRR
PSEUDO RESPONSE REGULATOR
- PTB
Polypyrimidine tract-binding protein
- PTC
Premature termination codon
- RBM
RNA binding motif protein
- RBP
RNA-binding protein
- RRM
RNA recognition motif
- SCN
Suprachiasmatic nuclei
- TF
Transcription factor
- TIM
TIMELESS
- TOC1
TIMING OF CAB EXPRESSION 1
- UTR
Untranslated region
- VRI
VRILLE
- WC
WHITE COLLAR
- WCC
WHITE COLLAR Complex
References
- 1.Gachon F, Nagoshi E, Brown SA, Ripperger J, Schibler U. The mammalian circadian timing system: from gene expression to physiology. Chromosoma. 2004;113:103–112. doi: 10.1007/s00412-004-0296-2. [DOI] [PubMed] [Google Scholar]
- 2.Mas P. Circadian clock function in Arabidopsis thaliana: time beyond transcription. Trends Cell Biol. 2008;18:273–281. doi: 10.1016/j.tcb.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 3.Schöning JC, Staiger D. At the pulse of time: protein interactions determine the pace of circadian clocks. FEBS Lett. 2005;579:3246–3252. doi: 10.1016/j.febslet.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 4.Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 5.Mehra A, Baker CL, Loros JJ, Dunlap JC. Post-translational modifications in circadian rhythms. Trends Biochem Sci. 2009;34:483–490. doi: 10.1016/j.tibs.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edery I. Role of post-transcriptional regulation in circadian clocks: lessons from Drosophila . Chronobiol Int. 1999;16:377–414. doi: 10.3109/07420529908998716. [DOI] [PubMed] [Google Scholar]
- 7.Harms E, Kivimae S, Young MW, Saez L. Post-transcriptional and post-translational regulation of clock genes. J Biol Rhythms. 2004;19:361–373. doi: 10.1177/0748730404268111. [DOI] [PubMed] [Google Scholar]
- 8.Staiger D, Streitner C, Rudolf F, Huang X (2006) Multiple and slave oscillators, In: Hall A, McWatters H (eds) Endogenous plant rhythms. Blackwell, Oxford, pp 57–83
- 9.Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1548. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 11.Keene JD, Tenenbaum SA. Eukaryotic mRNPs may represent post-transcriptional operons. Mol Cell. 2002;9:1161–1167. doi: 10.1016/s1097-2765(02)00559-2. [DOI] [PubMed] [Google Scholar]
- 12.Tenenbaum SA, Carson CC, Lager PJ, Keene JD. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc Natl Acad Sci USA. 2000;97:14085–14090. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 14.Keene JD. Biological clocks and the coordination theory of RNA operons and regulons. Cold Spring Harb Symp Quant Biol. 2007;72:157–165. doi: 10.1101/sqb.2007.72.013. [DOI] [PubMed] [Google Scholar]
- 15.Staiger D. RNA-binding proteins and circadian rhythms in Arabidopsis thaliana . Philos Trans R Soc Lond B. 2001;356:1755–1759. doi: 10.1098/rstb.2001.0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 17.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi H, Hogenesch JB. Coordinated transcription of key pathways in the mouse by circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 18.Pilgrim ML, Caspar T, Quail PH, McClung CR. Circadian and light-regulated expression of nitrate reductase in Arabidopsis . Plant Mol Biol. 1993;23:349–364. doi: 10.1007/BF00029010. [DOI] [PubMed] [Google Scholar]
- 19.Dibner C, Sage D, Unser M, Bauer C, d’Esmond T, Naef F, Schibler U. Circadian gene expression is resilient to large fluctuations in overall transcription rates. EMBO J. 2009;28:123–134. doi: 10.1038/emboj.2008.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reddy AB, Karp NA, Maywood ES, Sage EA, Deery M, O’Neill JS, Wong GK, Chesham J, Odell M, Lilley KS, Kyriacou CP, Hastings MH. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16:1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 21.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 22.Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allada R, Chung BY. Circadian organization of behavior and physiology in Drosophila . Annu Rev Physiol. 2010;72:605–624. doi: 10.1146/annurev-physiol-021909-135815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hardin PE. The circadian timekeeping system of Drosophila . Curr Biol. 2005;15:R714–R722. doi: 10.1016/j.cub.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 25.Cyran SA, Buchsbaum AM, Reddy KL, Lin MC, Glossop NR, Hardin PE, Young MW, Storti RV, Blau J. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112:329–341. doi: 10.1016/s0092-8674(03)00074-6. [DOI] [PubMed] [Google Scholar]
- 26.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 27.Ukai H, Ueda HR. Systems biology of mammalian circadian clocks. Annu Rev Physiol. 2010;72:579–603. doi: 10.1146/annurev-physiol-073109-130051. [DOI] [PubMed] [Google Scholar]
- 28.Sato TK, Yamada RG, Ukai H, Baggs JE, Miraglia LJ, Kobayashi TJ, Welsh DK, Kay SA, Ueda HR, Hogenesch JB. Feedback repression is required for mammalian circadian clock function. Nat Genet. 2006;38:312–319. doi: 10.1038/ng1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 30.Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K-I, Suzuki Y, Sugano S, Masamitsu I, Shigeyoshi Y, Hashimoto S. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 31.Brunner M, Kaldi K. Interlocked feedback loops of the circadian clock of Neurospora crassa. Mol Microbiol. 2008;68:255–262. doi: 10.1111/j.1365-2958.2008.06148.x. [DOI] [PubMed] [Google Scholar]
- 32.Locke JCW, Kozma-Bognar L, Gould PD, Feher B, Kevei E, Nagy F, Turner MS, Hall A, Millar AJ. Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana . Mol Syst Biol. 2006;2:59. doi: 10.1038/msb4100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iliev D, Voytsekh O, Schmidt EM, Fiedler M, Nykytenko A, Mittag M. A heteromeric RNA-binding protein is involved in maintaining acrophase and period of the circadian clock. Plant Physiol. 2006;142:797–806. doi: 10.1104/pp.106.085944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schulze T, Prager K, Dathe H, Kelm J, Kiessling P, Mittag M (2010) How the green alga Chlamydomonas reinhardtii keeps time. Protoplasma. doi:10.1007/s00709-010-0113-0 [DOI] [PubMed]
- 35.Wuarin J, Falvey E, Lavery D, Talbot D, Schmidt E, Ossipow V, Fonjallaz P. The role of the transcriptional activator protein DBP in circadian liver gene expression. J Cell Sci. 1992;16:123–127. doi: 10.1242/jcs.1992.supplement_16.15. [DOI] [PubMed] [Google Scholar]
- 36.Jacobshagen S, Kessler B, Rinehart CA. At least four distinct circadian regulatory mechanisms are required for all phases of rhythms in mRNA amount. J Biol Rhythms. 2008;23:511–524. doi: 10.1177/0748730408325753. [DOI] [PubMed] [Google Scholar]
- 37.So WV, Rosbash M. Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J. 1997;16:7146–7155. doi: 10.1093/emboj/16.23.7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frisch B, Hardin PE, Hamblen-Coyle MJ, Rosbash M, Hall JC. A promoterless period gene mediates behavioral rhythmicity and cyclical per expression in a restricted subset of the Drosophila nervous system. Neuron. 1994;12:555–570. doi: 10.1016/0896-6273(94)90212-7. [DOI] [PubMed] [Google Scholar]
- 39.Stanewsky R, Jamison CF, Plautz JD, Kay SA, Hall JC. Multiple circadian-regulated elements contribute to cycling period gene expression in Drosophila . EMBO J. 1997;16:5006–5018. doi: 10.1093/emboj/16.16.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitchell P, Tollervey D. mRNA stability in eukaryotes. Curr Opin Genet Dev. 2000;10:193–198. doi: 10.1016/s0959-437x(00)00063-0. [DOI] [PubMed] [Google Scholar]
- 41.Woo KC, Kim TD, Lee KH, Kim DY, Kim W, Lee KY, Kim KT. Mouse period 2 mRNA circadian oscillation is modulated by PTB-mediated rhythmic mRNA degradation. Nucleic Acids Res. 2009;37:26–37. doi: 10.1093/nar/gkn893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwak E, Kim TD, Kim KT. Essential role of 3′-untranslated region-mediated mRNA decay in circadian oscillations of mouse Period3 mRNA. J Biol Chem. 2006;281:19100–19106. doi: 10.1074/jbc.M511927200. [DOI] [PubMed] [Google Scholar]
- 43.Woo KC, Ha DC, Lee KH, Kim DY, Kim TD, Kim KT. Circadian amplitude of cryptochrome 1 is modulated by mRNA stability regulation via cytoplasmic hnRNP D oscillation. Mol Cell Biol. 2010;30:197–205. doi: 10.1128/MCB.01154-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim TD, Kim JS, Kim JH, Myung J, Chae HD, Woo KC, Jang SK, Koh DS, Kim KT. Rhythmic serotonin N-acetyltransferase mRNA degradation is essential for the maintenance of its circadian oscillation. Mol Cell Biol. 2005;25:3232–3246. doi: 10.1128/MCB.25.8.3232-3246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim TD, Woo K-C, Cho S, Ha D-C, Jang SK, Kim K-T. Rhythmic control of AANAT translation by hnRNP Q in circadian melatonin production. Genes Dev. 2007;21:797–810. doi: 10.1101/gad.1519507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benjamin D, Schmidlin M, Min L, Gross B, Moroni C. BRF1 protein turnover and mRNA decay activity are regulated by protein kinase B at the same phosphorylation sites. Mol Cell Biol. 2006;26:9497–9507. doi: 10.1128/MCB.01099-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng P, He Q, Wang L, Liu Y. Regulation of the Neurospora circadian clock by an RNA helicase. Genes Dev. 2005;19:234–241. doi: 10.1101/gad.1266805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo J, Cheng P, Yuan H, Liu Y. The exosome regulates circadian gene expression in a post-transcriptional negative feedback loop. Cell. 2009;138:1236–1246. doi: 10.1016/j.cell.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gutierrez RA, Ewing RM, Cherry JM, Green PJ. Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc Natl Acad Sci USA. 2002;99:11513–11518. doi: 10.1073/pnas.152204099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lidder P, Gutierrez RA, Salome PA, McClung CR, Green PJ. Circadian control of messenger RNA stability. Association with a sequence-specific messenger RNA decay pathway. Plant Physiol. 2005;138:2374–2385. doi: 10.1104/pp.105.060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yakir E, Hilman D, Hassidim M, Green RM. CCA1 transcript stability and the entrainment of the circadian clock in Arabidopsis . Plant Physiol. 2007;145:925–932. doi: 10.1104/pp.107.103812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson BG, Frim DM, Schwartz WJ, Majzoub JA. Vasopressin mRNA in the suprachiasmatic nuclei: daily regulation of polyadenylate tail length. Science. 1988;241:342–344. doi: 10.1126/science.3388044. [DOI] [PubMed] [Google Scholar]
- 54.Wang Y, Osterbur DL, Megaw PL, Tosini G, Fukuhara C, Green CB, Besharse JC. Rhythmic expression of Nocturnin mRNA in multiple tissues of the mouse. BMC Dev Biol. 2001;1:9. doi: 10.1186/1471-213X-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Green CB, Douris N, Kojima S, Strayer CA, Fogerty J, Lourim D, Keller SR, Besharse JC. Loss of Nocturnin, a circadian deadenylase, confers resistance to hepatic steatosis and diet-induced obesity. Proc Natl Acad Sci USA. 2007;104:9888–9893. doi: 10.1073/pnas.0702448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng Y, Gvakharia B, Hardin PE. Two alternatively spliced transcripts from the Drosophila period gene rescue rhythms having different molecular and behavioral characteristics. Mol Cell Biol. 1998;18:6505–6514. doi: 10.1128/mcb.18.11.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Collins BH, Rosato E, Kyriacou CP. Seasonal behavior in Drosophila melanogaster requires the photoreceptors, the circadian clock, and phospholipase C. Proc Natl Acad Sci USA. 2004;101:1945–1950. doi: 10.1073/pnas.0308240100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Majercak J, Sidote D, Hardin PE, Edery I. How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron. 1999;24:219–230. doi: 10.1016/s0896-6273(00)80834-x. [DOI] [PubMed] [Google Scholar]
- 59.Majercak J, Chen WF, Edery I. Splicing of the period gene 3′-terminal intron is regulated by light, circadian clock factors, and phospholipase C. Mol Cell Biol. 2004;24:3359–3372. doi: 10.1128/MCB.24.8.3359-3372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Low KH, Lim C, Ko HW, Edery I. Natural variation in the splice site strength of a clock gene and species-specific thermal adaptation. Neuron. 2008;60:1054–1067. doi: 10.1016/j.neuron.2008.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu Y, Garceau NY, Loros JJ, Dunlap JC. Thermally regulated translational control of FRQ mediates aspects of temperature responses in the Neurospora circadian clock. Cell. 1997;89:477–486. doi: 10.1016/s0092-8674(00)80228-7. [DOI] [PubMed] [Google Scholar]
- 62.Colot HV, Loros JJ, Dunlap JC. Temperature-modulated alternative splicing and promoter use in the circadian clock gene frequency. Mol Biol Cell. 2005;16:5563–5571. doi: 10.1091/mbc.E05-08-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heintzen C, Melzer S, Fischer R, Kappeler S, Apel K, Staiger D. A light- and temperature-entrained circadian clock controls expression of transcripts encoding nuclear proteins with homology to RNA-binding proteins in meristematic tissue. Plant J. 1994;5:799–813. doi: 10.1046/j.1365-313x.1994.5060799.x. [DOI] [PubMed] [Google Scholar]
- 64.Carpenter CD, Kreps JA, Simon AE. Genes encoding glycine-rich Arabidopsis thaliana proteins with RNA-binding motifs are influenced by cold treatment and an endogenous circadian rhythm. Plant Physiol. 1994;104:1015–1025. doi: 10.1104/pp.104.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Staiger D, Apel K. Circadian clock-regulated expression of an RNA-binding protein in Arabidopsis: characterisation of a minimal promoter element. Mol Gen Genet. 1999;261:811–819. doi: 10.1007/s004380050025. [DOI] [PubMed] [Google Scholar]
- 66.Heintzen C, Nater M, Apel K, Staiger D. AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1997;94:8515–8520. doi: 10.1073/pnas.94.16.8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Staiger D, Zecca L, Wieczorek Kirk DA, Apel K, Eckstein L. The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA. Plant J. 2003;33:361–371. doi: 10.1046/j.1365-313x.2003.01629.x. [DOI] [PubMed] [Google Scholar]
- 68.Schöning JC, Streitner C, Page DR, Hennig S, Uchida K, Wolf E, Furuya M, Staiger D. Autoregulation of the circadian slave oscillator component AtGRP7 and regulation of its targets is impaired by a single RNA recognition motif point mutation. Plant J. 2007;52:1119–1130. doi: 10.1111/j.1365-313X.2007.03302.x. [DOI] [PubMed] [Google Scholar]
- 69.Schüttpelz M, Schöning JC, Doose S, Neuweiler H, Peters E, Staiger D, Sauer M. Changes of conformational dynamics of mRNA upon AtGRP7 binding studied by fluorescence correlation spectroscopy. J Am Chem Soc. 2008;130:9507–9513. doi: 10.1021/ja801994z. [DOI] [PubMed] [Google Scholar]
- 70.Schöning JC, Streitner C, Meyer IM, Gao Y, Staiger D. Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis . Nucleic Acids Res. 2008;36:6977–6987. doi: 10.1093/nar/gkn847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McGlincy NJ, Smith CW. Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem Sci. 2008;33:385–393. doi: 10.1016/j.tibs.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 72.Fu ZQ, Guo M, Jeong BR, Tian F, Elthon TE, Cerny RL, Staiger D, Alfano JR. A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature. 2007;447:284–288. doi: 10.1038/nature05737. [DOI] [PubMed] [Google Scholar]
- 73.Kim JS, Jung HJ, Lee HJ, Kim KA, Goh C-H, Woo Y, Oh SH, Han YS, Kang H. Glycine-rich RNA-binding protein7 affects abiotic stress responses by regulating stomata opening and closing in Arabidopsis thaliana . Plant J. 2008;55:455–466. doi: 10.1111/j.1365-313X.2008.03518.x. [DOI] [PubMed] [Google Scholar]
- 74.Schmidt F, Marnef A, Cheung M-K, Wilson I, Hancock J, Staiger D, Ladomery M. A proteomic analysis of oligo(dT)-bound mRNP containing oxidative stress-induced Arabidopsis thaliana RNA-binding proteins ATGRP7 and ATGRP8. Mol Biol Rep. 2010;37:839–845. doi: 10.1007/s11033-009-9636-x. [DOI] [PubMed] [Google Scholar]
- 75.Fujita J. Cold shock response in mammalian cells. J Mol Microbiol Biotechnol. 1999;1:243–255. [PubMed] [Google Scholar]
- 76.Nishiyama H, Xue JH, Sato T, Fukuyama H, Mizuno N, Houtani T, Sugimoto T, Fujita J. Diurnal change of the cold-inducible RNA-binding protein (Cirp) expression in mouse brain. Biochem Biophys Res Commun. 1998;245:534–538. doi: 10.1006/bbrc.1998.8482. [DOI] [PubMed] [Google Scholar]
- 77.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hazen SP, Naef F, Quisel T, Gendron JM, Chen H, Ecker JR, Borevitz JO, Kay SA. Exploring the transcriptional landscape of plant circadian rhythms using genome tiling arrays. Genome Biol. 2009;10:R17. doi: 10.1186/gb-2009-10-2-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ner-Gaon H, Halachmi R, Savaldi-Goldstein S, Rubin E, Ophir R, Fluhr R. Intron retention is a major phenomenon in alternative splicing in Arabidopsis . Plant J. 2004;39:877–885. doi: 10.1111/j.1365-313X.2004.02172.x. [DOI] [PubMed] [Google Scholar]
- 80.Newby LM, Jackson FR. Regulation of a specific circadian clock output pathway by lark, a putative RNA-binding protein with repressor activity. J Neurobiol. 1996;31:117–128. doi: 10.1002/(SICI)1097-4695(199609)31:1<117::AID-NEU10>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 81.Huang Y, Genova G, Roberts M, Jackson FR. The LARK RNA-binding protein selectively regulates the circadian eclosion rhythm by controlling E74 protein expression. PLoS ONE. 2007;2:e1107. doi: 10.1371/journal.pone.0001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sofola O, Sundram V, Ng F, Kleyner Y, Morales J, Botas J, Jackson FR, Nelson DL. The Drosophila FMRP and LARK RNA-binding proteins function together to regulate eye development and circadian behavior. J Neurosci. 2008;28:10200–10205. doi: 10.1523/JNEUROSCI.2786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang J, Fang Z, Jud C, Vansteensel MJ, Kaasik K, Lee CC, Albrecht U, Tamanini F, Meijer JH, Oostra BA, Nelson DL. Fragile X-related proteins regulate mammalian circadian behavioral rhythms. Am J Hum Genet. 2008;83:43–52. doi: 10.1016/j.ajhg.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kojima S, Matsumoto K, Hirose M, Shimada M, Nagano M, Shigeyoshi Y, Hoshino S, Ui-Tei K, Saigo K, Green CB, Sakaki Y, Tei H. LARK activates post-transcriptional expression of an essential mammalian clock protein, PERIOD1. Proc Natl Acad Sci USA. 2007;104:1859–1864. doi: 10.1073/pnas.0607567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Höck J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, Urlaub H, Meister G. Proteomic and functional analysis of argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhao B, Schneid C, Iliev D, Schmidt EM, Wagner V, Wollnik F, Mittag M. The circadian RNA-binding protein CHLAMY 1 represents a novel type heteromer of RNA recognition motif and lysine homology domain-containing subunits. Eukaryot Cell. 2004;3:815–825. doi: 10.1128/EC.3.3.815-825.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Waltenberger H, Schneid C, Grosch JO, Bareiss A, Mittag M. Identification of target mRNAs for the clock-controlled RNA-binding protein Chlamy 1 from Chlamydomonas reinhardtii . Mol Genet Genomics. 2001;265:180–188. doi: 10.1007/s004380000406. [DOI] [PubMed] [Google Scholar]
- 88.Kiaulehn S, Voytsekh O, Fuhrmann M, Mittag M. The presence of UG-repeat sequences in the 3′-UTRs of reporter luciferase mRNAs mediates circadian expression and can determine acrophase in Chlamydomonas reinhardtii . J Biol Rhythms. 2007;22:275–277. doi: 10.1177/0748730407301053. [DOI] [PubMed] [Google Scholar]
- 89.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 91.Zdanowicz A, Thermann R, Kowalska J, Jemielity J, Duncan K, Preiss T, Darzynkiewicz E, Hentze MW. Drosophila miR2 primarily targets the m7GpppN cap structure for translational repression. Mol Cell. 2009;35:881–888. doi: 10.1016/j.molcel.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 92.Brodersen P, Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol. 2009;10:141–148. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- 93.Yang M, Lee JE, Padgett RW, Edery I. Circadian regulation of a limited set of conserved microRNAs in Drosophila . BMC Genomics. 2008;9:83. doi: 10.1186/1471-2164-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kadener S, Menet JS, Sugino K, Horwich MD, Weissbein U, Nawathean P, Vagin VV, Zamore PD, Nelson SB, Rosbash M. A role for microRNAs in the Drosophila circadian clock. Genes Dev. 2009;23:2179–2191. doi: 10.1101/gad.1819509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hipfner DR, Weigmann K, Cohen SM. The bantam gene regulates Drosophila growth. Genetics. 2002;161:1527–1537. doi: 10.1093/genetics/161.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, Obrietan K. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem. 2007;282:25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- 98.Gatfield D, Le Martelot G, Vejnar CE, Gerlach D, Schaad O, Fleury-Olela F, Ruskeepaa AL, Oresic M, Esau CC, Zdobnov EM, Schibler U. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev. 2009;23:1313–1326. doi: 10.1101/gad.1781009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sire C, Moreno AB, Garcia-Chapa M, Lopez-Moya JJ, Segundo BS. Diurnal oscillation in the accumulation of Arabidopsis microRNAs, miR167, miR168, miR171 and miR398. FEBS Lett. 2009;583:1039–1044. doi: 10.1016/j.febslet.2009.02.024. [DOI] [PubMed] [Google Scholar]
- 100.Jung JH, Seo YH, Seo PJ, Reyes JL, Yun J, Chua NH, Park CM. The GIGANTEA-regulated microRNA172 mediates photoperiodic Flowering independent of CONSTANS in Arabidopsis . Plant Cell. 2007;19:2736–2748. doi: 10.1105/tpc.107.054528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kramer C, Loros JJ, Dunlap JC, Crosthwaite SK. Role for antisense RNA in regulating circadian clock function in Neurospora crassa . Nature. 2003;421:948–952. doi: 10.1038/nature01427. [DOI] [PubMed] [Google Scholar]
- 102.Edwards KD, Anderson PE, Hall A, Salathia NS, Locke JC, Lynn JR, Straume M, Smith JQ, Millar AJ. FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell. 2006;18:639–650. doi: 10.1105/tpc.105.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu F, Marquardt S, Lister C, Swiezewski S, Dean C. Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science. 2010;327:94–97. doi: 10.1126/science.1180278. [DOI] [PubMed] [Google Scholar]
- 104.Zhang EE, Liu AC, Hirota T, Miraglia LJ, Welch G, Pongsawakul PY, Liu X, Atwood A, Huss JW, 3rd, Janes J, Su AI, Hogenesch JB, Kay SA. A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell. 2009;139:199–210. doi: 10.1016/j.cell.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Brown JW, Birmingham A, Griffiths PE, Jossinet F, Kachouri-Lafond R, Knight R, Lang BF, Leontis N, Steger G, Stombaugh J, Westhof E. The RNA structure alignment ontology. RNA. 2009;15:1623–1631. doi: 10.1261/rna.1601409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zong Q, Schummer M, Hood L, Morris DR. Messenger RNA translation state: the second dimension of high-throughput expression screening. Proc Natl Acad Sci USA. 1999;96:10632–10636. doi: 10.1073/pnas.96.19.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nandi A, Vaz C, Bhattacharya A, Ramaswamy R. miRNA-regulated dynamics in circadian oscillator models. BMC Syst Biol. 2009;3:45. doi: 10.1186/1752-0509-3-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, Liu S, Schwind S, Santhanam R, Hickey CJ, Becker H, Chandler JC, Andino R, Cortes J, Hokland P, Huettner CS, Bhatia R, Roy DC, Liebhaber SA, Caligiuri MA, Marcucci G, Garzon R, Croce CM, Calin GA, Perrotti D. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140:652–665. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]