Abstract
The hypoxia-inducible transcription factor (HIF) controls (in an oxygen-dependent manner) the expression of a large number of genes whose products are involved in the response of cells to hypoxia. HIF is an αβ dimer that binds to hypoxia response elements (HREs) in its target genes. Human HIF-α has three isoforms, HIF-1α, HIF-2α and HIF-3α, of which the roles of HIF-3α are largely unknown, although it is usually regarded as a negative regulator of HIF-1α and HIF-2α. The human HIF-3α locus is subject to extensive alternative splicing, leading to at least seven variants. We analyzed here the effects of the long variants and the short variant HIF-3α4 on the hypoxia response. All these variants were found to interact with HIF-β, HIF-1α and HIF-2α. The long HIF-3α variants were localized in the nucleus in hypoxia, while HIF-3α4 was cytoplasmic. Interaction of the HIF-3α variants with HIF-1α inhibited the nuclear translocation of both. None of the long HIF-3α variants was capable of efficient induction of an HRE reporter in overexpression experiments, but instead inhibited the transcriptional activation of the reporter by HIF-1 and HIF-2. Unexpectedly, siRNA knock-down of the endogenous HIF-3α variants led to downregulation of certain HIF target genes, while overexpression of individual long HIF-3α variants upregulated certain HIF target genes in a variant and target gene-specific manner under conditions in which HIF-β was not a limiting factor. These data indicate that the HIF-3α variants may have more versatile and specific roles in the regulation of the hypoxia response than previously anticipated.
Keywords: Hypoxia response, Hypoxia-inducible factor, Hypoxia-inducible factor 3 isoform, Prolyl 4-hydroxylase, Hypoxia response element
Introduction
The hypoxia-inducible transcription factor (HIF) is a master regulator of adaptive cellular responses to decreased oxygen levels, controlling in an oxygen-dependent manner the expression of a large number of genes whose products are involved in functions such as hematopoiesis, angiogenesis, iron transport, glucose utilization, extracellular matrix synthesis, cell proliferation, survival and apoptosis, and tumor progression [1–4]. HIF is a dimer consisting of an unstable α subunit (HIF-α) and a stable β subunit (HIF-β) and binds to specific sequences termed hypoxia response elements (HREs) in its target genes. Hydroxylation of at least one of two conserved prolines in HIF-α mediates its interaction with the von Hippel-Lindau E3 ubiquitin ligase complex, which results in rapid proteasomal degradation of HIF-α in normoxia [5–7]. This hydroxylation is catalyzed in humans by three oxygen-dependent HIF prolyl 4-hydroxylases (HIF-P4Hs 1–3, also known as PHDs 1–3, and EglN2, 1 and 3, respectively) [8–12]. Another hydroxylase, factor inhibiting HIF (FIH), acts on an asparagine in the C-terminal transactivation domain (CTAD) of HIF-α, thereby preventing binding of the coactivator p300 and thus full activity of HIF [13–15]. Hypoxia inhibits both these hydroxylations and leads to stabilization and full transcriptional activity of HIF.
Human HIF-α has three isoforms, HIF-1α, HIF-2α and HIF-3α, of which HIF-1α and HIF-2α have been studied most extensively [1–4]. These two are closely related and have similar domain structures, with an N-terminal basic helix-loop-helix (bHLH) domain followed by two Per-ARNT-Sim (PAS) domains, the oxygen-dependent degradation domain (ODDD) which harbors the two conserved prolines Pro402 and Pro564 in HIF-1α that are hydroxylated by HIF–P4Hs and regulates the stability of the subunit, and N-terminal and C-terminal transactivation domains (TADs), of which the NTAD partially overlaps with the ODDD [16] (see Fig. 1). Analyses of HIF-DNA interactions at various hypoxia-responsive gene loci have identified a core HRE motif RCGTG that is bound by both HIF-1α and HIF-2α. This binding results in efficient activation of HRE-driven reporter genes [17–19], although HIF-1α and HIF-2α show some specificity in their endogenous target genes [16, 20]. The first HIF-3α variants to be identified, mouse HIF-3α and human HIF-3α1, differ from HIF-1α and HIF-2α in that they lack the CTAD [21, 22] and thus cannot bind p300. They also differ from HIF-1α and HIF-2α in that their ODDD contains only one of the two conserved prolines, that corresponding to Pro564 in HIF-1α. The mouse HIF-3α and human HIF-3α1 have been shown to dimerize with HIF-β and to induce expression of an HRE reporter when overexpressed together with HIF-β, but much more weakly than do HIF-1α and HIF-2α, due to the lack of the CTAD [21, 22]. They suppress the activation potential of HIF-1α and HIF-2α under conditions where HIF-β is a limiting factor, however, probably by competing for binding with the latter [21, 22].
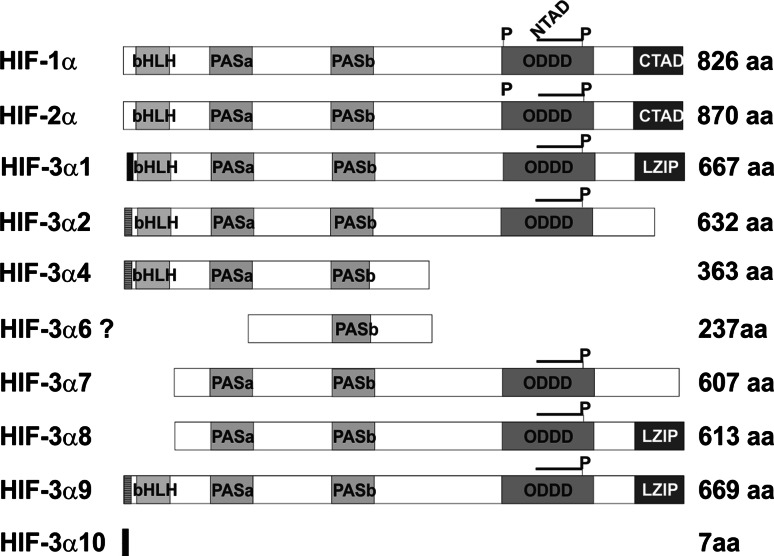
Fig. 1.
Schematic representation of the HIF-1α, HIF-2α, and the HIF-3α variant polypeptides. The structural motifs basic helix-loop-helix (bHLH), Per/Arnt/Sim (PAS), the oxygen-dependent degradation domain (ODDD), the N and C-terminal transactivation domains (NTAD and CTAD), the leucine zipper (LZIP), and the lengths of the polypeptides are indicated. The positions of the prolines hydroxylated by HIF-P4Hs in the ODDD are indicated by P
Three alternatively spliced forms are generated from the mouse HIF-3α locus. The 307-residue mouse inhibitory PAS domain protein (IPAS) differs from the 662-amino-acid mouse HIF-3α in that the first eight amino acids are coded by the IPAS-specific exon 1a instead of exon 1b as used in HIF-3, and in addition to lacking the NTAD and CTAD, it also lacks the ODDD [23, 24]. IPAS does not dimerize with HIF-β, but instead forms an inactive complex with HIF-1α, thus acting as a dominant negative regulator of HIF-1 [23]. A third mouse HIF-3α variant, NEPAS, is a 664-residue polypeptide that differs from mouse HIF-3α only in that exon 1b is replaced by the IPAS exon 1a [25]. Like HIF-3α, NEPAS dimerizes with HIF-β and suppresses HIF-1 and HIF-2 activity [25]. It is expressed mainly during the embryonic and neonatal stages, and its inactivation in mice leads to abnormal heart development and lung remodeling [25].
The human HIF-3α locus is subject to even more extensive alternative splicing. Database analyses first suggested the existence of six splice variants, HIF-3α variants 1–6, utilizing three alternative transcription start sites [26]. We have recently shown that full-length human HIF-3α1, HIF-3α2 and HIF-3α4 are expressed in several human tissues and cell lines (Fig. 1), but we obtained no evidence for the generation of the predicted HIF-3α3 or HIF-3α5 variants [27]. The existence of the predicted HIF-3α6 is also unlikely, as its DNA sequence does not correspond to the predicted domain structure [26] but contains partial duplications of exons 3 and 4 after exon 6 [28]. In addition, we identified four novel human HIF-3α variants, HIF-3α7 to HIF-3α10, that are expressed in many tissues and cell types (Fig. 1), [27]. The short human HIF-3α4 variant is similar to, but not identical with, the mouse IPAS and it has been shown to form a complex with both HIF-1α and HIF-β, and like the mouse IPAS, to inhibit the transcriptional activity of HIF-1 in a dominant negative manner [28, 29].
We set out to study the effects of the long human HIF-3α variants 1, 2, 7, 8 and 9, as well as those of HIF-3α4, on the hypoxia response. We excluded analysis of HIF-3α10 as it consists of only seven amino acids (Fig. 1), [27]. Our data show that all the human HIF-3α variants are able to interact with HIF-β, HIF-1α and HIF-2α. The long HIF-3α variants were localized in the nucleus in hypoxia in overexpression experiments, while the short variant HIF-3α4 was cytoplasmic. Interaction of the HIF-3α variants with HIF-1α inhibited nuclear translocation of both HIF-3α and HIF-1α. None of the long variants was capable of efficient induction of an HRE-reporter in overexpression experiments, but instead inhibited the transcriptional activation of the reporter by HIF-1 and HIF-2 under conditions where HIF-β is likely to be a limiting factor. Unexpectedly, siRNA knock-down of the endogenous HIF-3α variants led to downregulation of certain HIF target genes, while overexpression of individual long HIF-3α variants upregulated HIF target genes in a variant and target gene-specific manner under conditions in which HIF-β was not a limiting factor. These data indicate that the HIF-3α variants may have more versatile and specific roles in regulation of the hypoxia response than previously anticipated.
Materials and methods
Cell culture
Hep3B hepatoma cells were cultured in Eagle’s minimum essential medium (Sigma) and ChoK1 cells in Dulbecco’s minimum essential medium (Biochrom AG) with 0.375% sodium bicarbonate (Sigma). Both culture media were supplemented with 0.1 mM non-essential amino acids (Sigma), 1 mM sodium pyruvate (Sigma), 10% fetal bovine serum (HyClone), 2 mM l-glutamine (Sigma) and 100 U/ml penicillin and 0.1 mg/ml streptomycin (Gibco). In the hypoxia experiments, the cells were incubated in 1% O2, 5% CO2, and 94% N2 in an Invivo2 400 hypoxic workstation (Ruskin Technologies). As the ChoK1 cell line is easily transfectable and allows good expression levels of the transfected constructs, it was selected as a host for the interaction, reporter, and localization studies of the recombinant human HIF-3α polypeptides. Hep3B cells were used in experiments were the effects of the knock-down of endogenous human HIF-3α variants or overexpression of recombinant human HIF-3α variants on the expression of endogenous human hypoxia responsive genes was studied.
Expression plasmids
The HIF-1α-C-ODDD-luciferase reporter plasmid (a gift from FibroGen Inc.) contains the C-terminal region of the HIF-1α ODDD between an NF-κB activation domain and a Gal4 DNA-binding domain (Stratagene) followed by a Gal4-dependent luciferase reporter. The HIF-3α-ODDD-luciferase reporter plasmid was generated in a similar way by cloning cDNA coding for the full-length ODDD (amino acids 411–552) of HIF-3α into the reporter plasmid.
Mammalian expression vectors for full-length human untagged HIF-1α and HIF-2α, and for HIF-2α with a C-terminal V5 tag were gifts from FibroGen Inc, and the expression plasmid for HIF-β with an N-terminal V5 tag was a gift from Dr. Katchinski, University of Göttingen. Cloning of the full-length HIF-3α variants into the pcDNA3.1/Zeo(−) vector has been described previously [27]. To generate expression plasmids for HIF-1α and the HIF-3α variants [27] with C-terminal V5 tags, the full-length cDNAs were amplified by PCR and cloned into the pcDNA3.1–V5–His-A vector (Invitrogen). The HIF-3α2Pro_Ala mutant plasmid in which the codon for the single hydroxylatable Pro was changed to Ala was generated using the QuickChangeXL site-directed mutagenesis kit (Stratagene).
An expression plasmid for untagged HIF-β was generated by amplifying the full-length HIF-β cDNA and cloning it into the pcDNA3.1(+) vector (Invitrogen). For generation of an expression plasmid for HIF-2α with an N-terminal hemagglutinin (HA) tag, the HIF-2α cDNA was amplified by PCR so that the 5′ oligo contained the sequence for the HA-tag followed by the codon for the second HIF-2α amino acid, and the amplified product was then cloned into the pcDNA3.1/Zeo(−) vector (Invitrogen).
The glutathione S-transferase (GST) fusion constructs were generated by amplifying full-length HIF-1α and HIF-2α and HIF-β cDNAs using primers with an artificial BamHI restriction site at the 5′ end and an XbaI or SalI restriction site at the 3′ end to facilitate cloning into a BamHI-XbaI/SalI-digested pGEX-4T1 vector (GE Healthcare Life Sciences) using the Rapid DNA Ligation Kit (Roche).
EGFP-tagged constructs of the full-length human HIF-3α variants 2, 4, 7, and 9 were generated by cloning into an XhoI-HindIII-digested pEGFP-N1 expression vector (BD Biosciences Clontech). The DsRed-monomer-C1-mHIF-1α expression plasmid was a gift from Dr. Teresa Pereira (Karolinska Institutet, Stockholm, Sweden).
All sequences were verified using an automated DNA sequencer (ABI Prism 377, Applied Biosystems).
Analysis of degradation of HIF-3α ODDD and selected full-length long HIF-3α variants by HIF–P4Hs
On 24-well plates, 7 × 104 ChoK1 cells were seeded and cotransfected after 24 h with 300 ng of the HIF-1α-C-ODDD-Luc or HIF-3α-ODDD-Luc plasmids together with 300 and 600 ng of the plasmids coding for HIF-P4H-1, HIF-P4H-2 or HIF-P4H-3 or an empty pcDNA3.1/Zeo(−) vector, and 60 ng of a Renilla luciferase plasmid (Promega) with Lipofectamine™ 2000 (Invitrogen) according to the supplier’s protocol. The cells were cultured for 24 h in normoxia and assayed for luciferase activity using the Dual-luciferase reporter assay system (Promega) according to the manufacturer’s protocol and Luminoskan RS (Labsystems) for measuring the luciferase activity.
In 6-well plates, 3 × 105 ChoK1 cells were seeded and cotransfected after 24 h with 750 ng of the expression plasmids coding for full-length HIF-1α or V5-tagged HIF-3α1, HIF-3α2 or HIF-3α2_ProAla, in which the single hydroxylatable proline in the ODDD had been mutated to alanine, together with increasing amounts (750, 1,000, 1,250, or 1,500 ng) of HIF-P4H-2 with Lipofectamine™ 2000 (Invitrogen) according to the supplier’s protocol. The cells were cultured for 48 h in normoxia and lysed in a buffer containing 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.1% Tween-20, 1% NP-40, 2.5 mM EGTA, and 50 mM Hepes, pH 7.5 [30], supplemented with Complete protease inhibitor cocktail + EDTA (Roche), 1 mM NaVO4, 50 mM NaF and 0.5 mM dithiothreitol upon incubation on ice for 20 min, and centrifuged at 12,000 × g for 20 min. The cell lysates were analyzed by 8% SDS-PAGE under reducing conditions followed by ECL-Western blotting (Amersham Biosciences) with primary antibodies against HIF-1α (BD Biosciences Pharmingen), V5 (Invitrogen) and α-tubulin (Sigma) at 1:1,000, 1:2,000 and 1:10,000 dilutions, respectively.
Protein interaction assays
For the immunoprecipitation analyses, 1.8 × 106 ChoK1 cells were seeded on 10-cm plates and transfected with 5 μg of V5-tagged HIF-3α1, HIF-3α2, HIF-3α4, HIF-3α7, HIF-3α8 or HIF-3α9 together with 2 μg of untagged HIF-1α, HA-tagged HIF-2α or untagged HIF-β using Lipofectamine™ 2000 (Invitrogen) according to the supplier’s protocol. Negative control samples were obtained by transfecting 5 μg of the V5-tagged HIF-3α variants together with 2 μg of an empty pcDNA3.1/Zeo(−) vector (Invitrogen), and positive control samples by transfecting 5 μg of V5-tagged HIF-1α with 2 μg of untagged HIF-β and 5 μg of V5-tagged HIF-β with 2 μg of untagged HIF-1α or HA-tagged HIF-2α. The transfected cells were cultured for 24 h in normoxia followed by a 24 h exposure to hypoxia. The cells were lysed as above. Immunoprecipitation of the lysates was performed with the V5 antibody and a Protein A/G PLUS–Agarose Immunoprecipitation reagent (Santa Cruz) according to the manufacturer’s protocol. The immunoprecipitated samples were analyzed by 8% SDS-PAGE under reducing conditions followed by ECL-Western blotting (Amersham Biosciences) with primary antibodies against V5, HIF-1α and HA (Sigma) at dilutions of 1:2,000, 1:1,000 and 1:1,000, respectively.
For the in vitro binding assays the GST-tagged HIF-1α and HIF-2α expression constructs were transformed into the E. coli BL21 strain. Cells were grown from a single colony at 37°C overnight in 100 ml of Luria–Bertoni medium containing ampicillin (50 μg/ml). The overnight culture was diluted 1:100 and isopropyl-β-D-thiogalatoside was added to a final concentration of 0.1 mM, at which an A600 of 0.6–2 was reached. The cells were harvested by centrifugation 3 h after the induction, resuspended in 10 ml of ice-cold PBS and disrupted by sonication. Triton X-100 was added to a final concentration of 1% and the lysate was centrifuged at 10,000 × g for 5 min at +4°C. The GST-fusion proteins expressed were bound to Protino Glutathione Agarose 4B beads (Macherey–Nagel) according to the manufacturer’s protocol. The ChoK1 cells were transfected with the V5-tagged HIF-3α variants or HIF-β, which served as a positive control. Cells transfected with the empty vector pcDNA3.1/Zeo(−) served as negative controls. The transfected cells were cultured for 24 h in normoxia followed by a 24 h hypoxia treatment. The cells were lysed in a buffer containing 150 mM NaCl, 1% Nonidet P-40, 10% glycerol, 10 mM MgCl2, 1 mM EDTA and 0.25% sodium deoxycholate, 25 mM Hepes, pH 7.5, supplemented with Complete protease inhibitor cocktail + EDTA (Roche), 1 mM NaVO4 and 50 mM NaF. The cell lysates were centrifuged at 12,000 × g for 20 min at +4°C, precleared with mouse IgG together with Protino Glutathione Agarose 4B beads, and incubated with 50 μl of Protino Glutathione Agarose 4B with or without the bound GST-HIF-1α and GST-HIF-2α fusion proteins for 2 h at 4°C. The beads were washed four times with the above buffer, boiled for 5 min in SDS-PAGE loading buffer, centrifuged at 12,000 × g for 2 min at room temperature and analyzed by 8% SDS-PAGE under reducing conditions followed by ECL-Western blotting (Amersham Biosciences) with the primary antibody against V5 at a dilution of 1:2,000.
HIF-3α overexpression experiments
To study the ability of overexpressed HIF-3α variants to activate an HRE reporter, 3 × 105 ChoK1 cells were seeded per well on 6-well plates and transfected 24 h later with a 3xHRE-secreted alkaline phosphates (HRE-SEAP) reporter plasmid (a gift from FibroGen Inc.) together with increasing amounts (500, 1,000, 2,000 ng) of pcDNA.3.1/Zeo(−) (negative control) or expression plasmids for full-length V5-tagged HIF-1α, HIF-2α, HIF-3α1, HIF-3α2, HIF-3α4, HIF-3α7, HIF-3α8 or HIF-3α9 using Lipofectamine™ 2000 (Invitrogen) according to the supplier’s protocol. The cells were cultured for 48 h in normoxia and medium samples were collected and assayed for SEAP activity using the SEAP Reporter Assay Kit (InvivoGen). Absorbance at 405 nm was measured with a Victor™ 1420 Multilabel Counter (Wallac). The cells were lysed in a buffer containing 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.1% Tween-20, 1% NP-40, 2.5 mM EGTA, and 50 mM Hepes, pH 7.5 [30], supplemented with Complete protease inhibitor cocktail + EDTA (Roche), 1 mM NaVO4, 50 mM NaF and 0.5 mM dithiothreitol upon incubation on ice for 20 min, and centrifuged at 12,000 × g for 20 min. Expression of the transfected HIF-α polypeptides was analyzed by 8% SDS-PAGE under reducing conditions followed by ECL-Western blotting (Amersham Biosciences) with the primary antibody against V5 at a dilution of 1:2,000.
To study the capacity of overexpressed HIF-1α and HIF-2α to activate an HRE reporter in the presence of overexpressed HIF-3α variants, 3 × 105 ChoK1 cells were seeded on 6-well plates and transfected after 24 h with 500 ng of the expression plasmids for full-length HIF-1α or HIF-2α together with 500 ng of pcDNA3.1/Zeo(−) or the expression plasmids for the HIF-3α variants together with 750 ng of a 3xHRE-luciferase (HRE-LUC) reporter plasmid (a gift from Dr. Pasi Tavi) and 100 ng of a Renilla luciferase plasmid (Promega) using Lipofectamine™ 2000 (Invitrogen) as above. The cells were cultured for 48 h in normoxia and assayed for luciferase activity as above.
To study the effect of overexpressed human HIF-3α variants on the transactivation capacity of an HRE reporter with respect to endogenous human HIF-1α and HIF-2α, Hep3B cells were transfected as above but without the expression plasmids for human HIF-1α and HIF-2α, cultured for 24 h in normoxia followed by 24 h in hypoxia and assayed for luciferase activity.
To study the effect of overexpressed HIF-3α variants on selected endogenous HIF target genes 3.5 × 106 Hep3B cells were seeded on 6-well plates and transfected 24 h later with 250–1,000 ng of the expression plasmids for the HIF-3α variants together with 1,000 ng of the HIF-β or pcDNA3.1/Zeo(−) plasmid using Lipofectamine™ 2000 (Invitrogen) as above. In the experiments where HIF-β was not limited, i.e., in the presence of the HIF-β plasmid, the cells were cultured in normoxia for 48 h followed by 90 min in hypoxia, while in those where HIF-β should become limiting, i.e., in the presence of the pcDNA3.1/Zeo(−) plasmid, the cells were cultured for 24 h in normoxia followed by 24 h in hypoxia. Total RNA was isolated from the cells using the E.Z.N.A.™ total RNA kit (Omega) and mRNA expression levels of the HIF target genes for erythropoietin (Epo), angiopoietin-related protein 4 (AngPTL4), phosphofructokinase L (Pfkl) and glucose transporter 1 (Glut1) were analyzed by qPCR with normalization to the expression of the TATA-binding protein gene using gene-specific primers (Table 1), iTaq™ SYBR® Green Supermix with ROX (Bio-Rad) or SsoFast EvaGreen® Supermix (Bio-Rad) and the MX3005 (Stratagene) equipment.
Table 1.
Sequences of the oligos used in RNA interference and qPCR analyses of the HIF-3α variants
| Primer | Sequences (5′- 3′) and locations of the oligos | Use |
|---|---|---|
| 3α_a (D-010068-02) | GCAAGAGCAUCCACACCUU, exon 7 and 8 | RNA interference |
| 3α_b (D-010068-05) | CGACAGGAUUGCAGAAGUG, exon 6 and 7 | RNA interference |
| 3α_c (D-010068-21) | UAACAGGGCAGUAUCGCUU, exon 8 | RNA interference |
| TBP_F | GAARARAARCCCAAGCGGTTTG | qPCR |
| TBP_R | ACTTCACATCACAGCTCCCC | qPCR |
| HIF1a_Q_F | CTAGCTTTGCAGAATGCTCAG | qPCR |
| HIF1a_Q_R | GTAGTAGCTGCATGATCGTCTG | qPCR |
| HIF2a_Q_F | CCCAGATCCACCATTACAT | qPCR |
| HIF2a_Q_R | ACTCCAGCTGTCGCTTCA | qPCR |
| Epo_F | CTCCGAACAATCACTGCT | qPCR |
| Epo_R | GGTCATCTGTCCCCTGTCCT | qPCR |
| LOX_QF | ACGGCACTGGCTACTTCCAG | qPCR |
| LOX_QR | GGCCAGACAGTTTTCCTCCG | qPCR |
| Q_GLUT_F | GATTGGCTCCTTCTCTGTGG | qPCR |
| Q_GLUT_R | TCAAAGGACTTGCCCAGTTT | qPCR |
| Q_HK2_F | CAAAGTGACAGTGGGTGTGG | qPCR |
| Q_HK2_R | GCCAGGTCCTTCACTGTCTC | qPCR |
| Q_PFKL_F | CCCGAGGACGGCTGGGAGAA | qPCR |
| Q_PFKL_R | GGGCTTCCCGTTGCGGTCAA | qPCR |
| Q_TRF_F | GTCGCTGGTCAGTTCGTGATT | qPCR |
| Q_TRF_R | AGCAGTTGGCTGTTGTAACCTCTC | qPCR |
| Q_uPAR_F | GGTGACGCCTTCAGCATGA | qPCR |
| Q_uPAR_R | CCCACTGCGGTACTGGACAT | qPCR |
| Q_BNIP_F | GGACAGAGTAGTTCCAGAGGCAGTTC | qPCR |
| Q_BNIP_R | GGTGTGCATTTCCACATCAAACAT | qPCR |
| Q_AngPT4_F | TCTCCGTACCCTTCTCCACT | qPCR |
| Q_AngPT4_R | AGTACTGGCCGTTGAGGTTG | qPCR |
| Q_HO1_F | CTATGTGAAGCGGCTCCACG | qPCR |
| Q_HO1_R | GCTCTGGTCCTTGGTGTCAT | qPCR |
| Q_VEGF_F | AGCCTTGCCTTGCTGCTCTA | qPCR |
| Q_VEGF_R | TCAGGGTACTCCTGGAAGATG | qPCR |
RNA interference
siRNA oligos against HIF-3α (siGENOME Set of 4, MQ-010068-03-0005, Thermo Fisher Scientific) were obtained from Dharmacon, and three siRNA oligos from this set were used either individually (3α_a, b or c) or in combination (3α_a+b+c) in the siRNA experiments. The sequences of the siRNA oligos are given in Table 1. Firefly pGL2 luciferase control duplex siRNA (Eurogentec) was used as a negative siRNA control. All siRNAs were reconstituted under RNase-free conditions according to the manufacturer’s protocol, using the buffers supplied. Hep3B cells were transfected with the siRNAs at a confluence of 80–90% with Lipofectamine ™ 2000 (Invitrogen) in OPTI-MEM®-1 medium (GIBCO®) according to the supplier’s protocol. The final concentration of each individual siRNA oligo in the cell culture medium was 0.08 pmol/μl. The transfection was repeated after 24 h and the cells were cultured for an additional 24 h in hypoxia. Forty-eight hours after the first transfection, the culture medium was removed from the cells and stored in −70°C and total RNA and protein were isolated from the cells with the Paris™ kit (Ambion). No distinct differences in the viability of the HIF-3α and luciferase siRNA treated cells and the mock-transfected cells were detected as the cell cultures were confluent at the time of harvest and the amounts of total RNA and protein isolated from the cells were highly similar. The RNA samples were treated with DNase using the Turbo-DNA-free™ kit (Ambion) and converted to cDNA with the iScript™ cDNA synthesis kit. The protein samples were analyzed by 8% SDS-PAGE under reducing conditions followed by ECL-Western blotting (Amersham Biosciences) with primary antibodies against HIF-1α (BD Biosciences Pharmingen), HIF-2α (Novus Biologicals) and α-tubulin (Sigma) at dilutions of 1:1,000, 1:200, and 1:10,000, respectively. The amount of Epo protein in the medium was analyzed with a Quantikine® IVD® Erythropoietin ELISA kit (R&D Systems Inc.) according to the manufacturer’s protocol. The amount of Epo was measured with a Victor™ 1420 Multilabel Counter (Wallac) using 450 nm as the primary wavelength and 540 nm as the reference wavelength. mRNA expression levels of EPO, ANGPTL4, PFKL, GLUT1, lysyl oxidase (LOX), hexokinase 2 (HK2), transferrin (TRF), urokinase receptor (uPAR), BCL2/adenovirus E1B 19 kDa protein-interacting protein 3 (BNIP3), heme oxygenase 1 (HO1) and vascular endothelial growth factor (VEGF) were analyzed by qPCR as above.
Confocal microscopy
To study the localization of the EGFP-tagged HIF-3α variants in cells, 2.2 × 105 ChoK1 cells were seeded on 35 mm glass-bottomed culture dishes (MatTek Corporation) and transfected 24 h later with 800 ng of the expression plasmids for EGFP-tagged HIF-3α2, HIF-3α4, HIF-3α7 or HIF-3α9 using Lipofectamine™ 2000 (Invitrogen) as above. The cells were incubated for 12 h in normoxia ±1 h in 1% oxygen, fixed with 3% paraformaldehyde for 3 min, washed with PBS and analyzed using an Olympus FluoView FV 1000 confocal laser scanning microscope system. To study the possible colocalization of HIF-3α variants with HIF-1α, cells were seeded as above and transfected with 800 ng of the expression plasmids for each of the EGFP-tagged HIF-3α variants together with DsRed- monomer-C1-mHIF-1α. The cells were cultured for 24 h in normoxia followed by 8 h in 1% oxygen. Cells were fixed and washed as above and stained with DAPI (Sigma) diluted 1:10,000 in PBS for 15 min at room temperature. After staining, the cells were washed with PBS three times and examined as above.
Statistical analysis
The statistical analyses were performed using Student’s two-tailed t test. Data are shown as means ± SD. Values of p < 0.05 were considered statistically significant (*p < 0.05, **p < 0.01, ***p < 0.001).
Results
Regulation of the HIF-3α ODDD by HIF-P4Hs
Synthetic 20–35-residue peptides representing the single hydroxylation site of the human HIF-3α ODDD have been shown to be efficiently hydroxylated in vitro by all three recombinant human HIF-P4Hs [11, 31]. The human HIF-3α ODDD and the full-length human HIF-3α1 have been shown to be targeted for ubiquitylation and proteasomal degradation in a VHL and oxygen-dependent manner, but the full-length HIF-3α1 is ubiquitylated less robustly than the ODDD, suggesting that the full-length protein may have structural elements that inhibit ubiquitylation [26]. It has recently been reported that full-length human HIF-3α2 does not accumulate in cells upon chemical inhibition of HIF-P4Hs, indicating that it is not subject to oxygen-dependent destabilization [32].
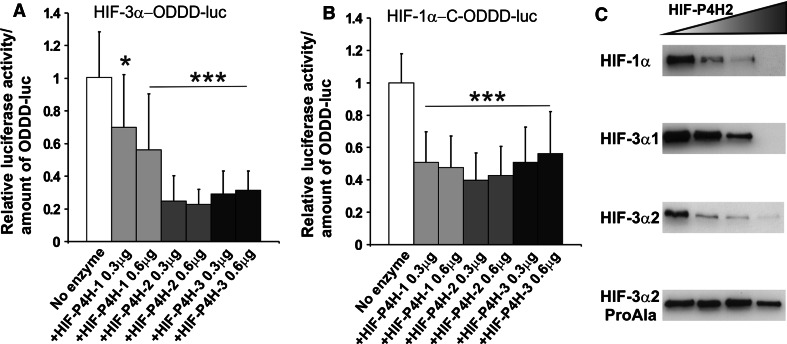
To study this aspect further, we first analyzed the effect of overexpressed HIF-P4Hs on degradation of the HIF-3α ODDD by cotransfecting ChoK1 cells with (a) a constant amount of a luciferase reporter plasmid for the HIF-3α ODDD or the C-terminal region of the HIF-1α ODDD, and (b) increasing amounts of a plasmid encoding HIF-P4H-1, 2 or 3. The cells were cultured for 24 h in normoxia and assayed for luciferase activity. Overexpression of all three HIF-P4Hs led to a 44–60% reduction in the level of the C-terminal region of the HIF-1α ODDD (Fig. 2b). The level of the HIF-3α ODDD was reduced by 30–44% by overexpression of HIF-P4H-1 and by 69–77% with HIF-P4H-2 or 3 (Fig. 2a), indicating that the HIF-3α ODDD is efficiently hydroxylated by all three HIF-P4Hs in cellulo, leading to subsequent VHL-dependent ubiquitylation and proteasomal degradation.
Fig. 2.
Overexpression of HIF-P4H enzymes leads to degradation of a HIF-3α-ODDD reporter and full-length HIF-3 variants. ChoK1 cells were transfected with a HIF-3α-ODDD-luc (a) or HIF-1α-C-ODDD-luc (b) reporter plasmid together with 0.3 or 0.6 μg of HIF-P4H-1, HIF-P4H-2 or HIF-P4H-3 expression plasmids. The transfected cells were cultured for 24 h in normoxia and assayed for luciferase activity. The luciferase activity of the control sample without HIF-P4H overexpression was set as 1. The data represent means ± SD from at least four independent experiments. *p < 0.05, ***p < 0.001. To study the degradation of full-length HIF-α polypeptides, ChoK1 cells were transfected with plasmids encoding full-length HIF-1α, V5-tagged HIF-3α1, HIF-3α2 or HIF-3α2_ProAla together with increasing amounts of HIF-P4H-2 plasmid (c). The cells were cultured for 48 h in normoxia, lysed, and analyzed by 8% SDS-PAGE under reducing conditions followed by Western blotting with HIF-1α, V5 and α-tubulin antibodies. The results of a typical experiment are shown
To study the HIF-P4H-dependent degradation of selected full-length long HIF-3α variants, we cotransfected ChoK1 cells with (a) a constant amount of a plasmid encoding full-length V5-tagged HIF-1α, HIF-3α1, HIF-3α2 or HIF-3α2Pro_Ala mutant polypeptides, and (b) increasing amounts of a plasmid encoding HIF-P4H-2, the main hydroxylase regulating the hypoxia response [33–35]. The cells were cultured for 48 h in normoxia, lysed, and analyzed by Western blotting. The amounts of the HIF-1α, HIF-3α1 and HIF-3α2 polypeptides all decreased in a HIF-P4H-2 dose-dependent manner, while that of the HIF-3α2Pro_Ala mutant polypeptide remained constant (Fig. 2c). These data show that the HIF-3α ODDD also mediates HIF-P4H-dependent degradation in the context of full-length HIF-3α polypeptides, including HIF-3α2.
All human HIF-3α variants interact with HIF-β, HIF-1α, and HIF-2α
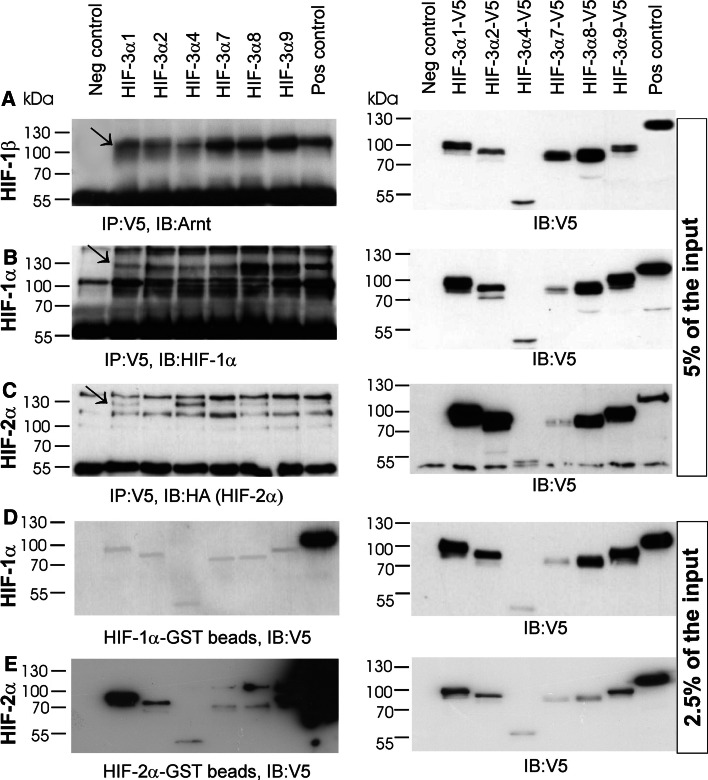
ChoK1 cells were cotransfected with vectors coding for V5-tagged HIF-3α variants together with non-tagged HIF-β, non-tagged HIF-1α or HA-tagged HIF-2α (Fig. 3a–c). Immunoprecipitation with an anti-V5 antibody showed that HIF-β was coprecipitated with all the V5-HIF-3α variants and with the V5-HIF-1α used as a control for positive interaction (Fig. 3a). The interaction of the HIF-3α variants with HIF-β seemed to be just as efficient as that of HIF-1α when the input amounts of the HIF-3α variants and HIF-1α were taken into consideration (Fig. 3a). Likewise, HIF-1α was coimmunoprecipitated with all the V5-HIF-3α variants in the experiments where interaction of HIF-1α with V5-HIF-β served as a positive control (Fig. 3b). HA-HIF-2α was coimmunoprecipitated with all the V5-HIF-3α variants except HIF-3α2 (Fig. 3c). The amount of HA-HIF-2α immunoprecipitated with the V5-tagged HIF-3α variants or HIF-β was in general low, except for V5-HIF-3α4, which pulled down the largest amount of HA-HIF-2α despite the relatively low input amount of V5-HIF-3α4 (Fig. 3c).
Fig. 3.
All HIF-3α variants can bind HIF-1α, HIF-2α and HIF-1β. ChoK1 cells were transfected for immunoprecipitation studies with plasmids for V5-tagged HIF-3α1, HIF-3α2, HIF-3α4, HIF-3α7, HIF-3α8 and HIF-3α9 with or without untagged HIF-1β (a), HIF-1α (b), or HA-tagged HIF-2α (c). In the positive control samples the cells were transfected with plasmids for HIF-1β and V5-tagged HIF1α (a), HIF-1α and V5-tagged HIF-β (b), and HA-tagged HIF-2α andV5-tagged HIF-β (c). The cells were cultured for 24 h in normoxia followed by 24 h in 1% oxygen. The left-hand panels in a–c show the amounts of the HIF-1β, HIF-1α and HIF-2α polypeptides immunoprecipitated with the V5-tagged HIF-3α variants or the positive controls, and the right-hand panels show the amounts of the V5-tagged polypeptides present in the whole cell lysates (5% of the input amount) used in the immunoprecipitation analysis (d–e). For in vitro binding studies HIF-1α and HIF-2α were expressed as GST-fusion proteins in E. coli, purified and bound to glutathione agarose beads. ChoK1 cells were transfected with plasmids for the V5-tagged HIF-3α variants or HIF-β (positive control) or the empty vector pcDNA3.1/Zeo(−) (negative control) and cultured for 24 h in normoxia followed by 24 h in 1% oxygen. The cell lysates were incubated with the GST beads ± GST-HIF-1α or GST-HIF-2α. The left-hand panels show the amounts of the captured V5-tagged HIF-3α variants or positive controls, and the right-hand panels show the amounts of the V5-tagged polypeptides present in the whole cell lysates (2.5% of the input amount) used in the in vitro binding assay
Interaction of the HIF-3α variants with HIF-1α and HIF-2α was studied further by means of an in vitro binding assay. GST-tagged HIF-1α and HIF-2α were produced in E. coli, bound to glutathione beads, and lysates from ChoK1 cells expressing V5-tagged HIF-3α variants or V5-tagged HIF-β were incubated with the beads (Fig. 3d, e). All the HIF-3α variants were bound by the GST-tagged HIF-1α and HIF-2α, but in both cases the interaction of HIF-1α and HIF-2α with HIF-β seemed to be much stronger than with any of the HIF-3α variants (Fig. 3d, e). In the case of GST-HIF-2α the amount of bound HIF-β was so high that it partially masked the neighboring lane representing binding of HIF-3α9, and leakage of HIF-β into the next two lanes representing binding of HIF-3α7 and HIF-3α8 also occurred (Fig. 3e). Bands of the correct size representing bound HIF-3α7, HIF-3α8 and HIF-3α9 were nevertheless seen in these lanes in addition to HIF-β (Fig. 3e).
Overexpression of the human HIF-3α variants does not lead to efficient induction of an HRE-reporter
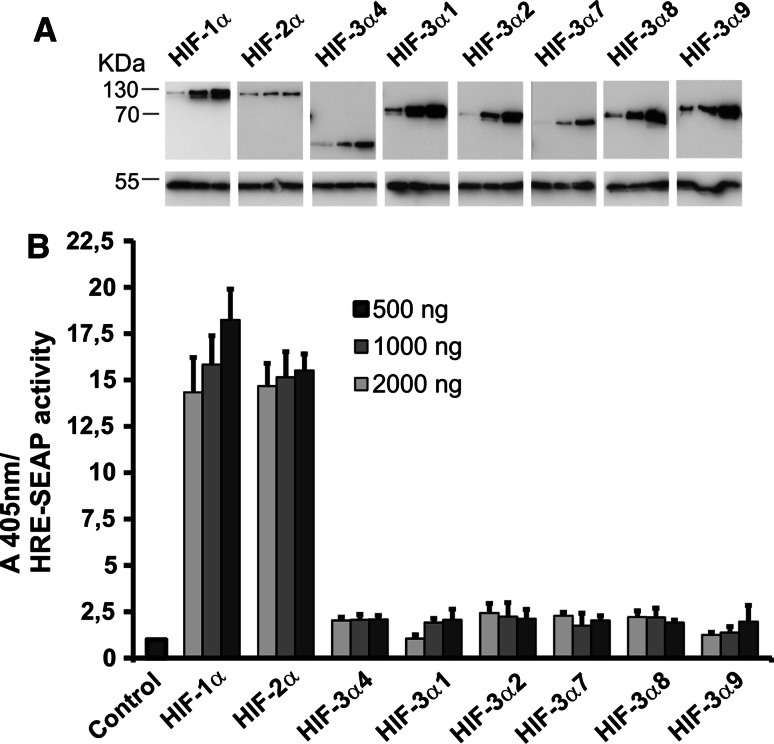
To study the transactivation potential of the HIF-3α variants with respect to a reporter gene driven by an HRE, we transiently transfected ChoK1 cells with a HRE-SEAP reporter plasmid together with increasing amounts of expression plasmids coding for HIF-1α, HIF-2α or the HIF-3α variants. To exclude the effect of endogenous HIF-α subunits, the cells were cultured for 48 h under normoxic conditions after the transfection and assayed for recombinant HIF-α subunit expression and SEAP activity. The amount of overexpressed HIF-α subunits exceeded the hydroxylation capacity of the endogenous HIF-P4Hs, resulting in the accumulation of stable recombinant HIF-α subunits in a dose-dependent manner under normoxic conditions (Fig. 4a), as also reported by others [21–23, 25]. Overexpression of HIF-1α and HIF-2α led to a dose-dependent 14- to 18-fold induction in the HRE-driven SEAP activity, while overexpression of the HIF-3α variants led only to a 2.5-fold induction at most, and this was in most cases independent of the dose (Fig. 4b). It should be noted, however, that overexpression of even the short HIF-3α4 variant, which does not contain any known transactivation domain (Fig. 1), led to about a twofold induction in the SEAP activity (Fig. 4b). This finding may indicate that the 2- to 2.5-fold induction seen with the HIF-3α variants is not a true effect of these variants on HRE but may result from some indirect phenomenon in the experimental setup. This suggestion is supported by the differences in the cellular localization of the short and long HIF-3α variants (see Fig. 8a).
Fig. 4.
Effects of the HIF-3α variants on the activation of a HRE reporter. ChoK1 cells were transfected with an HRE-SEAP reporter plasmid together with increasing amounts (500, 1,000, 2,000 ng) of expression plasmids for HIF-1α, HIF-2α or the HIF-3α variants. The cells were cultured for 48 h in normoxia and analyzed for expression of the HIF-α polypeptides by Western blotting (a) and assayed for SEAP activity (b), which was set to 1 in the control sample without any overexpressed HIF-α polypeptide. The results of a typical experiment are shown in a, and the data in b represent means ± S.D. from at least 4 independent experiments
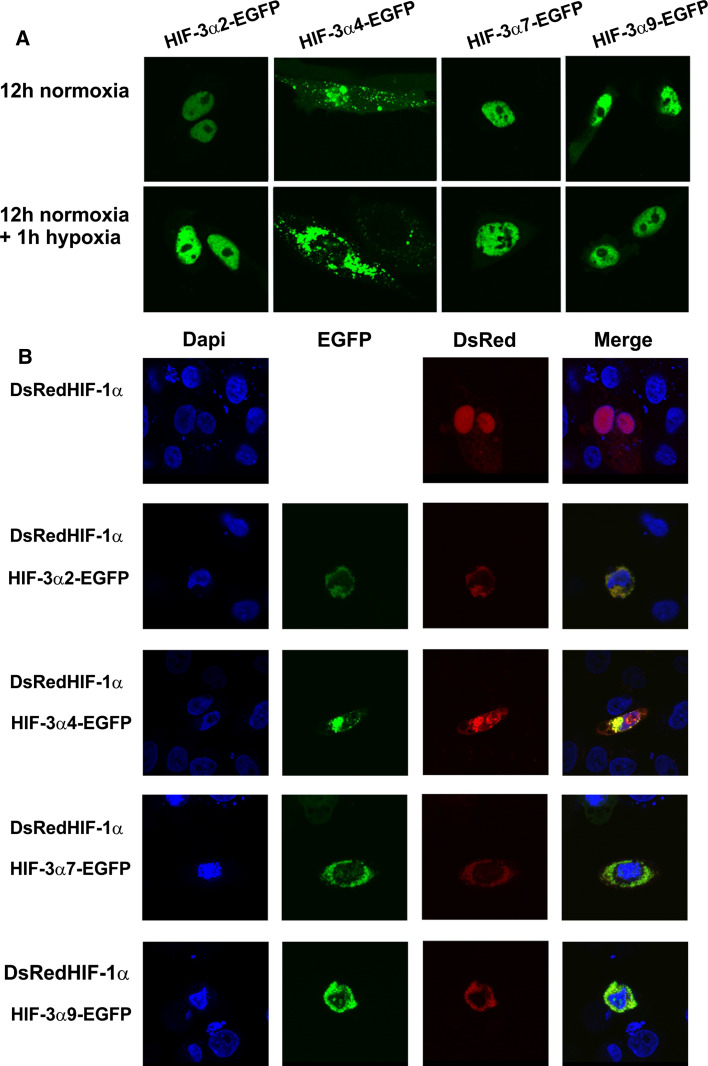
Fig. 8.
Confocal microscopy analysis of the cellular localization of overexpressed HIF-3α variants. a ChoK1 cells were transfected with plasmids encoding EGFP-labeled HIF-3α variants 2, 4, 7, or 9 and cultured in normoxia for 12 h (upper panel) followed by 1 h in 1% oxygen (lower panel). b ChoK1 cells were cotransfected with plasmids encoding EGFP-labeled HIF-3α variants 2, 4, 7, or 9 and DSRed-labeled HIF-1α and cultured for 24 h in normoxia followed by 8 h in 1% oxygen and stained with DAPI after fixation
Overexpression of the human HIF-3α variants inhibits the transcriptional activation of an HRE-reporter by HIF-1α and HIF-2α
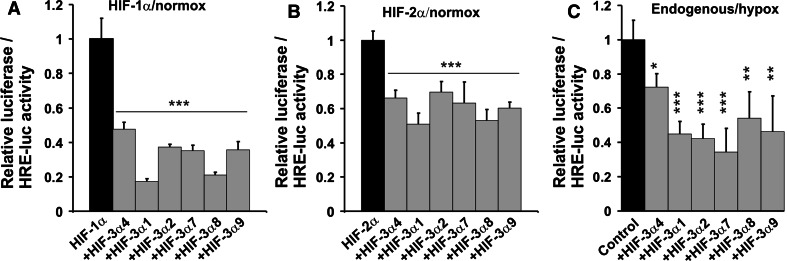
ChoK1 cells were cotransfected with a HRE-LUC reporter plasmid and the HIF-1α or HIF-2α expression plasmids in the presence or absence of an expression plasmid for each of the HIF-3α variants. The cells were cultured for 48 h under normoxic conditions after the transfection and assayed for luciferase activity. Coexpression of all the HIF-3α variants markedly reduced the HRE-LUC induction potential of overexpressed HIF-1α and HIF-2α, the reduction being 52–83% for HIF-1α and 30–50% for HIF-2α (Fig. 5a, b). This suggests that the HIF-3α variants act as dominant negative regulators of HIF-1α and HIF-2α-mediated gene expression, at least under conditions where the amount of HIF-β becomes limiting.
Fig. 5.
HIF-3α variants inhibit the transcriptional activation of an HRE-reporter by HIF-1 and HIF-2. ChoK1 cells were transfected with a 3xHRE-LUC reporter plasmid together with plasmids for HIF-1α (a) or HIF-2α (b) and with or without plasmids for the HIF-3α variants. The cells were cultured for 48 h in normoxia and assayed for luciferase activity. The luciferase activity of cells transfected only with HIF-1α or HIF-2α was set to 1. The data represent means ± SD from at least six independent experiments. ***p < 0.001. Hep3B cells were transfected with a 3xHRE-Luc reporter plasmid together with a empty vector or certain HIF-3α variants (c). Transfected cells were cultured for 24 h in normoxia followed by 24 h in hypoxia and then assayed for luciferase activity. The luciferase activity of the control samples was set to 1
To study the effect of overexpressed HIF-3α variants on the HRE induction potential of endogenous HIF-1α and HIF-2α in hypoxia, Hep3B cells were cotransfected with the HRE-LUC reporter in the presence or absence of an expression plasmid for each of the HIF-3α variants. The cells were cultured for 24 h under normoxic conditions after transfection, followed by a 24 h culture in 1% O2. All the long HIF-3α variants efficiently reduced the hypoxia-induced HRE-LUC activity by 45–66% (Fig. 5c). The inhibitory efficiency of HIF-3α4 was lower, 28% (Fig. 5c), but this may be due to the lower expression level of this variant relative to the long ones (Fig. 4a).
siRNA knock-down of endogenous HIF-3α variants unexpectedly leads to downregulation of many HIF target genes
We next studied the effect of knock-down of the endogenous HIF-3α variants on the expression of a known HIF target gene, Epo, in Hep3B cells. Three individual siRNA oligos targeting all the HIF-3α variants were used either singly or in combination and their efficiency was analyzed by qPCR using oligos that amplify a region from exons 5 and 6 that is common to all the HIF-3α variants. All three HIF-3α siRNAs, either alone or in combination, efficiently downregulated expression of the HIF-3α variants at the mRNA level (80–87% reduction) (Fig. 6a). We also attempted to analyze the siRNA efficiency by Western blotting using a monoclonal HIF-3α antibody or a polyclonal hHIF-3α2 antibody. All the recombinant HIF-3α variants could be identified with these antibodies when overexpressed in Hep3B cells, but no bands of the corresponding sizes representing endogenous HIF-3α variants at the protein level in Hep3B cells were detectable (data not shown). This is most likely due to the generally low expression levels of the HIF-3α variants in various cell lines and tissues [27]. One of the siRNA oligos, 3α_a, led to a non-specific reduction in HIF-1α and HIF-2α mRNA levels by about 13 and 23%, respectively, and the combination of the HIF-3α siRNA oligos also reduced the level of HIF-2α mRNA by about 11.5% (Fig. 6a). These non-specific reductions did not lead to any noticeable reduction in the amounts of stabilized HIF-1α and HIF-2α protein in the hypoxic cells, however (Fig. 6b).
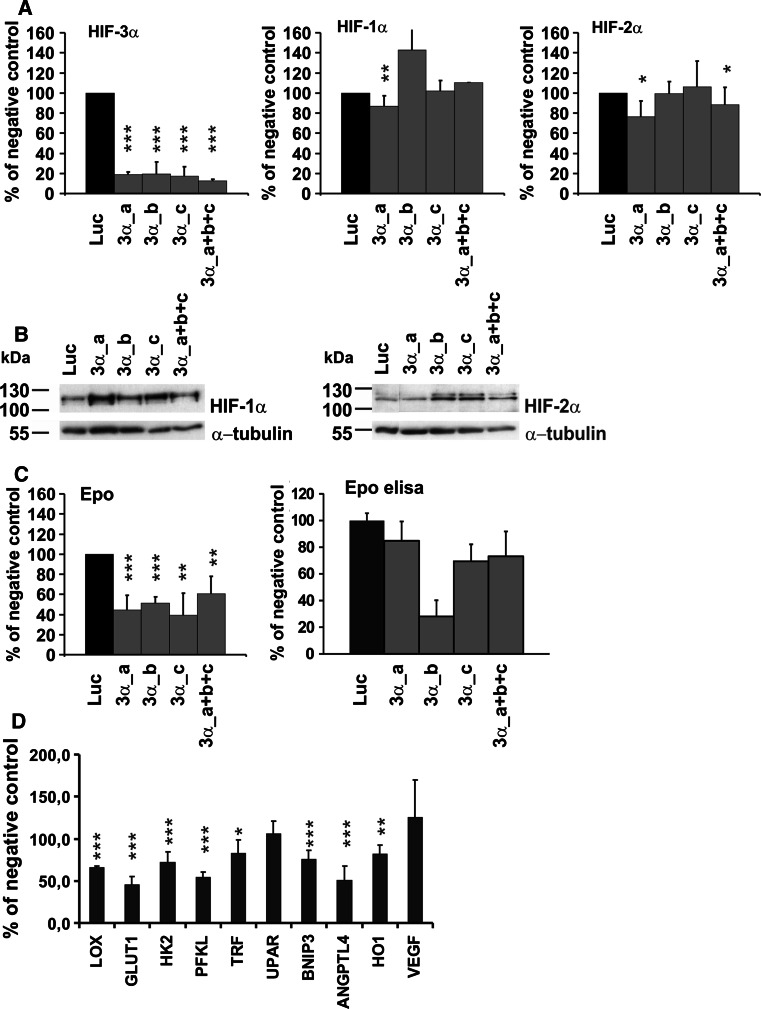
Fig. 6.
siRNA knock-down of endogenous HIF-3α leads to downregulation of many HIF target genes. Hep3B cells were transfected with a negative control siRNA (luc, pGL2 luciferase control duplex siRNA) or with HIF-3α siRNAs (three independent siRNAs a, b, c alone or in combination) twice at an interval of 24 h and cultured in 1% oxygen for 24 h after the second transfection. The expression of HIF-3α (all variants), HIF-1α, HIF-2α (a) and Epo (c) at the mRNA level was analyzed by qPCR and the expression of HIF-1α and HIF-2α (b) and Epo (c) at the protein level by Western blotting and ELISA, respectively. The effect of the HIF-3α siRNA on selected HIF target genes was analyzed by qPCR (d). The data represent means ± SD from at least four independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001. In the case of the Epo ELISA analysis, the data represent the results of a typical experiment
The effect of HIF-3α knock-down on Epo expression was analyzed by qPCR and ELISA. Surprisingly, instead of being upregulated, the Epo mRNA level was reduced by 39–60% and the Epo protein level by 28–73% in the HIF-3α siRNA-treated cells (Fig. 6c). This finding cannot be due to non-specific effects on the HIF-1α and HIF-2α levels, as the reduction was obtained with similar efficiency with the 3α_b and 3α_c siRNA oligos, which had no unspecific effects on HIF-1α or HIF-2α (Fig. 6a). To study this finding further, we analyzed the effect of HIF-3α siRNA on the expression of several other known HIF target genes. The mRNA expression levels of Lox, Glut1, Hk2, Pfkl, Trf, Bnip3, AngPTL4 and HO1 were likewise significantly downregulated by about 18–54%, while those of uPAR and VEGF were not affected (Fig. 6d).
Certain HIF-3α variants upregulate selected HIF target genes when HIF-β is not a limiting factor
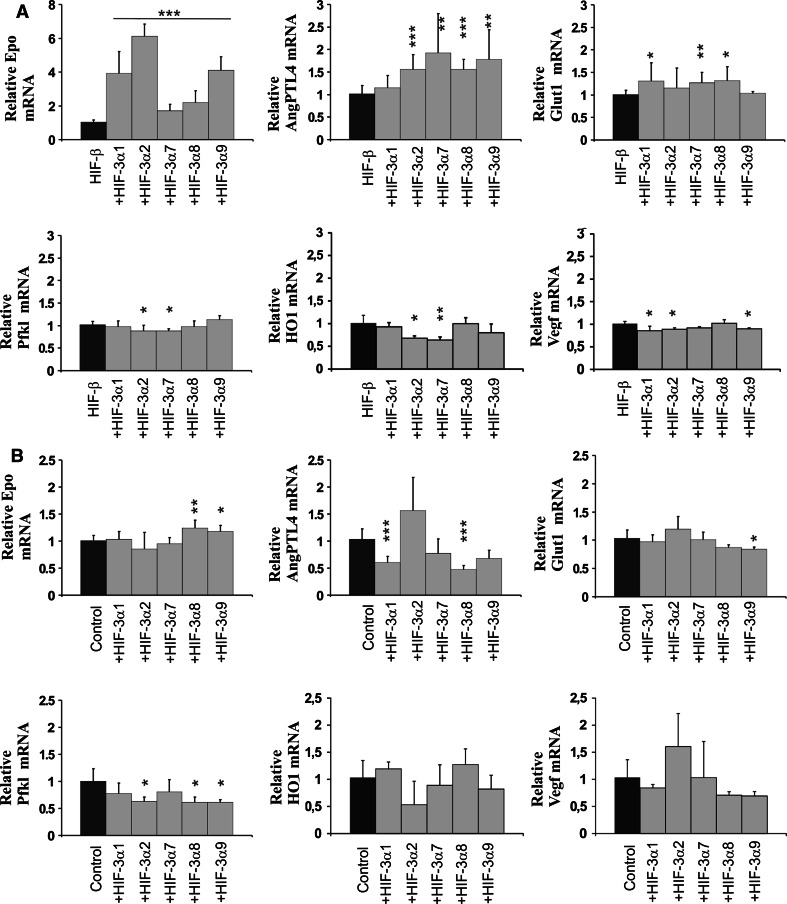
The above data suggest that the long HIF-3α variants may be required for maximal activation of certain HIF target genes in hypoxia. We analyzed this possibility by first cotransfecting Hep3B cells with the expression plasmids for each of the long HIF-3α variants together with the HIF-β plasmid, i.e., under conditions where HIF-β should not be a limiting factor. The cells were cultured for 48 h in normoxia followed by 90 min in hypoxia and the mRNA levels of the HIF target genes Epo, AngPTL4, Pfkl, Glut1, HO-1 and VEGF were determined by qPCR (Fig. 7a). These genes were selected for analysis as knock-down of HIF-3α had the strongest downregulating effect on the expression levels of Epo, AngPTL4, Pfkl and Glut1, an intermediate effect on HO-1 and no effect on VEGF (Fig. 6d). The HIF-3α1, HIF-3α2 and HIF-3α9 variants gave about a four to six fold increase in the hypoxic induction of the Epo mRNA level, while the HIF-3α7 and HIF-3α8 variants gave about a two to three fold increase (Fig. 7a). The AngPTL4 mRNA level was increased by about 1.5- to 2-fold with four of the variants, and the Glut1 mRNA level by up to about 1.4-fold with three variants, while none of the variants had any inductive effect on the Pfkl, HO-1 or VEGF mRNA levels (Fig. 7a).
Fig. 7.
Certain long HIF-3α variants upregulate HIF target genes when HIF-β is not limiting. Hep3B cells were transfected with a plasmid encoding the full-length long HIF-3α variants with (a) or without (b) HIF-β. HIF-3α4 was excluded here as the aim was to study the transactivation potential of HIF-3α variants. The cells were cultured for 48 h in normoxia followed by 90 min in 1% oxygen (a) or for 24 h in normoxia followed by 24 h in 1% oxygen (b). The expression levels of the EPO, ANGPTL4, GLUT1, PFKL, HO-1 and VEGF mRNAs were analyzed by qPCR. The data represent means ± SD from at least three independent experiments. *p < 0.05, **p < 0.01,***p < 0.001
We next studied the effect of the overexpressed HIF-3α variants on these same HIF target genes in the absence of overexpressed HIF-β, i.e., under conditions where the amount of endogenous HIF-β should become limiting. Hep3B cells were transfected with the HIF-3α long variant plasmids, the cells were cultured for 24 h in normoxia followed by 24 h in hypoxia, and the mRNA levels of the above genes were determined by qPCR (Fig. 7b). The expression level of the Epo mRNA was increased by about 1.3-fold with the HIF-3α8 and HIF-3α9 variants and unaffected with the other variants (Fig. 7b). The level of AngPTL4 mRNA was downregulated by about twofold with overexpression of the HIF-3α1, HIF-3α8 and HIF-3α9 variants, and HIF-3α7 also showed a trend of downregulation, while overexpression of HIF-3α2 seemed to have a slight inductive effect (Fig. 7b). The Glut1 mRNA level was in general not affected by the HIF-3α variants, except for HIF-3α9, which caused a small, but statistically significant reduction (Fig. 7b). In the case of the Pfkl mRNA, all the long HIF-3α variants showed a trend for downregulation, about a twofold reduction being obtained with the HIF-3α2, HIF-3α8 and HIF-3α9 variants (Fig. 7b). None of the HIF-3α variants had a statistically significant effect on the HO-1 and VEGF mRNA levels (Fig. 7b).
Cellular localization of overexpressed HIF-3α variants
ChoK1 cells were transfected with EGFP-labeled long HIF-3α variants 2, 7, and 9, and the short variant HIF-3α4 and the cells were cultured for 12 h in normoxia. The long variants accumulated in the nucleus, a similar localization also being observed when culturing of the cells was extended by means of a 1-h exposure to hypoxia (Fig. 8a). In contrast, HIF-3α4 was localized in the cytoplasm and perinuclear regions and did not translocate into the nucleus even in hypoxia (Fig. 8a).
We next analyzed the localization of HIF-1α and the above HIF-3α variants when coexpressed in hypoxic cells. ChoK1 cells were transfected with DsRed-labeled HIF-1α with or without the EGFP-labeled HIF-3α variants 2, 4, 7, and 9, and the cells were cultured for 24 h in normoxia followed by 8 h in 1% O2. When the DsRed-labeled HIF-1α was expressed alone, a nuclear localization was observed after the 8 h hypoxia exposure (Fig. 8b). When coexpressed together with any of the four HIF-3α variants, the localization of the HIF-1α and HIF-3α polypeptides remained cytoplasmic and no accumulation into the nucleus was observed (Fig. 8b). Our interaction studies showed that all the HIF-3α variants can form a complex with HIF-1α (Fig. 3) and the data obtained here indicate that the HIF-1α/HIF-3α complexes formed are not capable of nuclear translocation (Fig. 8b).
Discussion
We set out here to investigate the roles of the human HIF-3α variants in the regulation of the hypoxia response. Our present data demonstrate that HIF-P4H-2 and HIF-P4H-3 hydroxylate the HIF-3α ODDD and target it for proteasomal degradation with an efficiency at least similar to that of the HIF-1α ODDD, if not better, while HIF-P4H-1 seemed to act somewhat less efficiently on the HIF-3α ODDD than on the HIF-1α ODDD. HIF-P4H-1 and HIF-P4H-3 have been shown to hydroxylate a 20-residue synthetic peptide representing the HIF-3α hydroxylation site with identical efficiency [11], and thus the less efficient hydroxylation of the HIF-3α ODDD by HIF-P4H-1 may be attributed to neighboring sequences or conformational effects. Importantly, our results also show that the HIF-3α ODDD is degraded in a HIF-P4H-2-dependent manner when the ODDD is present in a full-length HIF-3α polypeptide, in this case HIF-3α1 or HIF-3α2. In a previous report the full-length human HIF-3α1 as produced in an in vitro transcription-translation system was said to be ubiquitylated less robustly than the HIF-3α ODDD after treatment with a HIF-P4H-enriched extract from rabbit reticulocyte lysate [26], while HIF-3α2 was said not to be regulated in an oxygen-dependent manner in Caki-1 or HeLa cells at all [32]. Our data clearly indicate, however, that the full-length HIF-3α polypeptides, including HIF-3α2, are targeted for proteasomal degradation in a HIF-P4H-dependent and thus also oxygen-dependent manner.
In order to understand the potential effects of the human HIF-3α variants on the hypoxia response, we studied their capacity to form complexes with HIF-β, HIF-1α and HIF-2α. The in vitro binding studies showed that all the HIF-3α variants were able to form a complex with HIF-1α and HIF-2α. Furthermore, the coimmunoprecipitation studies showed that the HIF-3α variants generally associated with HIF-1α and HIF-2α as efficiently as did HIF-β, and with HIF-β as efficiently as did HIF-1α. The only exceptions were the undetectable formation of a HIF-3α2/HIF-2α complex, and the particularly effective formation of a HIF-3α4/HIF-2α complex. The interaction of the HIF-3α variants 2, 4, 7, and 9 with HIF-1α was further verified in confocal microscopy studies, where overexpression of these HIF-3α polypeptides together with HIF-1α was found to inhibit translocation of HIF-1α and the long HIF-3α variants 2, 7, and 9 into the nucleus. The subcellular localization of the short HIF-3α4 variant was cytoplasmic irrespective of the oxygen concentration and this variant also effectively inhibited the nuclear transport of HIF-1α in hypoxia.
Previous analyses of the effects of HIF-3α on the hypoxia response have mainly been carried out by studying the effects on HRE reporters. It has been shown that dimers formed between the mouse HIF-3α or human HIF-3α1 and HIF-β can induce an HRE reporter, but much more weakly than the HIF-1α/HIF-β or HIF-2α/HIF-β dimers [21, 22]. When HIF-β is a limiting factor, however, these HIF-3α variants have been shown to suppress the activation potential of HIF-1 and HIF-2 [21, 22]. In the present study we obtained at most a 2.5-fold induction of an HRE reporter with the long HIF-3α variants under conditions where overexpression of the HIF-1α and HIF-2α polypeptides resulted in a 14- to 18-fold induction. Furthermore, about a twofold induction of the HRE reporter was obtained even with the short HIF-3α4 variant, which does not contain any known transactivation domain, and therefore the inductive potential of the long HIF-3α variants with respect to the HRE reporter must be at least very low, if it exists at all. Our results agree with those of previous studies in that all the HIF-3α variants were found to be effective in inhibiting HIF-1 and HIF-2-mediated HRE-reporter induction in overexpression experiments where HIF-β is likely to be limiting, in addition to which the inhibiting effect was much stronger towards HIF-1α than towards HIF-2α, which agrees with the results of the coimmunoprecipitation studies, which generally showed a stronger interaction of the HIF-3α variants with HIF-1α than with HIF-2α. One exception to this was HIF-3α4, which seemed to interact particularly strongly with HIF-2α in the coimmunoprecipitation experiments but still exerted a stronger inhibitory effect on HIF-1α than on HIF-2α.
In the light of the data obtained in our HRE reporter experiments, the results of the HIF-3α siRNA experiments were highly unexpected. The expectation was that knock-down of the endogenous HIF-3α variants would lead to increased expression of the HIF target genes, but instead it did not have an inductive effect on any of the HIF target genes studied and even significantly reduced the expression levels of certain target genes, especially those of Epo, AngPTL4, Glut1 and Pfkl, where the reduction was 50–60%. Overexpression experiments showed that the HIF-3α1, HIF-3α2 and HIF-3α9 variants induced the Epo mRNA expression level about four to six fold and the HIF-3α7 and HIF-3α8 variants about two to three fold when HIF-β was not a limiting factor. Under the same conditions the inductive effects of the HIF-3α variants on AngPTL4 were weaker, up to about twofold, and those on Glut1 less than 1.5-fold, while no inductive effect was detected on Pfkl. As the siRNA experiments were designed so that all the HIF-3α variants were targeted for knock-down simultaneously, the effects of the individual HIF-3α variants on selected HIF target genes were studied further in the overexpression experiments. Their results indicate that these target genes may have specific response elements of varying strengths for the individual HIF-3α variants. In the case of the Epo gene, overexpression of HIF-3α1, HIF-3α2 and HIF-3α9 alone was enough to exert a strong inductive effect and overexpression of HIF-3α7 and HIF-3α8 alone could exert a somewhat weaker inductive effect, while in the case of the AngPTL4 gene overexpression of HIF-3α2, HIF-3α7, HIF-3α8 and HIF-3α9 alone was enough to give a significant inductive effect and in the case of the Glut1 gene the same was true of the expression of HIF-3α1, HIF-3α7, HIF-3α8 alone. On the other hand, induction of Pfkl and a stronger induction of AngPTL4 and Glut1 is likely to require the concerted action of several HIF-3α variants. Our data also show that the effects of the HIF-3α variants differ in a variant and target gene-dependent manner under conditions where the amount of HIF-β is likely to be limiting. Certain HIF-3α variants had a minor effect in increasing the Epo mRNA level, a reducing effect on the AngPTL4 and Pfkl mRNA levels and little effect on Glut1.
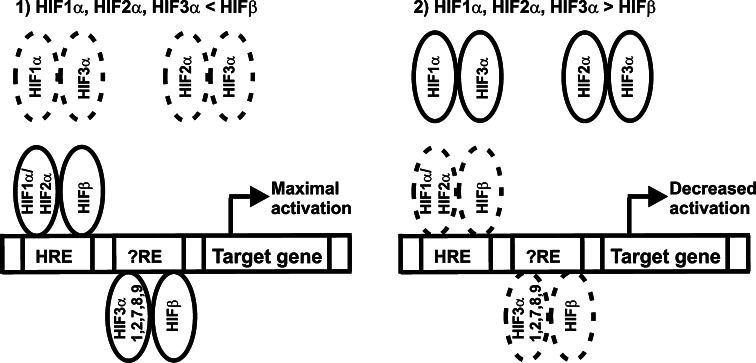
Altogether, our data indicate that the HIF-3α variants are likely to have more versatile and specific roles in the regulation of the hypoxia-responsive genes than has been anticipated previously (Fig. 9). When HIF-β is not limiting, HIF-1α, HIF-2α, and the long HIF-3α variants mainly associate with HIF-β and activate hypoxia-inducible target genes, the long HIF-3α variants being required for maximal activation of a subset of HIF target genes. As the inductive potential of the long HIF-3α variants with respect to an HRE reporter was very low, if it existed at all, the molecular mechanism involved most likely includes binding of the HIF-3α/HIF-β dimer to a response element that is distinct from the canonical HRE element recognized by HIF-1 and HIF-2 and which is only present in a subset of HIF-1 and HIF-2 regulated genes. When the amount of HIF-β is limiting, the long HIF-3α variants associate with HIF-1α and HIF-2α leading to decreased activation of HIF target genes. The short variant HIF-3α4 forms inactive complexes with HIF-1α, HIF-2α and HIF-β and causes downregulation of HIF target genes independent on whether HIF-β is non-limiting or limiting. Formation of these complexes is also dependent on the relative amounts of HIF-3α4, HIF-1α, HIF-2α and HIF-β. As the HIF-3α variants themselves are also HIF-1 targets, and as the expression levels of individual HIF-3α variants vary in different cell types and tissues [27, 32], their contributions to the hypoxia response in cellulo and in vivo will be an important topic for further studies. It is also noteworthy that several other factors can modulate the expression of hypoxia-inducible genes. For example, Bach1 has been shown to downregulate the hypoxic induction of HO-1 in certain cell types, whereas in some other cell types HO-1 is clearly hypoxia-inducible in a HIF-1-dependent manner [37, 38]. Whether HIF-3α can affect the functions of such modulating factors (or vice versa) remains to be studied.
Fig. 9.
Schematic representation of the roles of long HIF-3α variants in the hypoxia response. a When HIF-β is not limiting, HIF-1α, HIF-2α, and the long HIF-3α variants mainly associate with HIF-β and activate hypoxia-inducible target genes. Our data suggest that the long HIF-3α variants are required for maximal activation of certain HIF target genes and are likely to have target elements distinct from the canonical HRE response element. b When the amount of HIF-β is limiting, the long HIF-3α variants associate with HIF-1α and HIF-2α leading to decreased activation of HIF target genes. The short variant HIF-3α4, which is not depicted in the scheme, forms inactive complexes with HIF-1α, HIF-2α and HIF-β and causes downregulation of HIF target genes independent on whether HIF-β is non-limiting or limiting. Formation of these complexes is also dependent on the relative amounts of HIF-3α4, HIF-1α, HIF-2α and HIF-β
Acknowledgments
We thank Anne Kokko and Raija Juntunen for their expert technical assistance. This work was supported by the Academy of Finland (Grants 1114344 and 1211128 to JM and 121789 to MH), the Sigrid Juselius Foundation and FibroGen Inc.
Abbreviations
- HIF
Hypoxia-inducible factor
- ODDD
Oxygen-dependent degradation domain
- P4H
Prolyl 4-hydroxylase
- C-TAD
C-terminal transactivation domain
- bHLH
Basic helix-loop-helix
- PAS
Per-ARNT-Sim
- HRE
Hypoxia response element
- IPAS
Inhibitory PAS domain protein
- LZIP
Leucine zipper
- qPCR
Quantitative real-time RT-PCR
References
- 1.Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Curr Opin Genet Dev. 2010;20:51–56. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 3.Weidemann A, Johnson RS. Biology of HIF-1α. Cell Death Differ. 2008;15:621–627. doi: 10.1038/cdd.2008.12. [DOI] [PubMed] [Google Scholar]
- 4.Myllyharju J. Prolyl 4-hydroxylases, key enzymes in the synthesis of collagens and regulation of the response to hypoxia, and their roles as treatment targets. Ann Med. 2008;40:402–417. doi: 10.1080/07853890801986594. [DOI] [PubMed] [Google Scholar]
- 5.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lanie WS, Kaelin WG., Jr HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 6.Jaakkola P, Mole DR, Tian Y-M, Wilson MI, Gielbert J, Gaskell SJ, Kriegsheim AV, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, Ratcliffe PJ. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 7.Yu F, White SB, Zhao Q, Lee FS. HIF-1α binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc Natl Acad Sci USA. 2001;98:9630–9635. doi: 10.1073/pnas.181341498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 9.Epstein ACR, Gleadle JM, McNeill LA, Hewitson KS, O’Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, Tian Y-M, Masson N, Hamilton DL, Jaakkola P, Barstead R, Hodgkin J, Maxwell PH, Pugh CW, Schofield CJ, Ratcliffe PJ. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/S0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 10.Ivan M, Haberberger T, Gervasi DC, Michelson KS, Günzler V, Kondo K, Yang H, Sorokina I, Conaway RC, Conaway JW, Kaelin WG., Jr Biochemical purification and pharmacological inhibition of a mammalian prolyl hydroxylase acting on hypoxia-inducible factor. Proc Natl Acad Sci USA. 2002;99:13459–13464. doi: 10.1073/pnas.192342099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirsilä M, Koivunen P, Günzler V, Kivirikko KI, Myllyharju J. Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J Biol Chem. 2003;278:30772–30780. doi: 10.1074/jbc.M304982200. [DOI] [PubMed] [Google Scholar]
- 12.Myllyharju J. HIF prolyl 4-hydroxylases and their potential as drug targets. Curr Pharm Des. 2009;15:3878–3885. doi: 10.2174/138161209789649457. [DOI] [PubMed] [Google Scholar]
- 13.Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Asparagine hydroxylation of the HIF transactivation domain: a hypoxic switch. Science. 2002;295:858–861. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- 14.Hewitson KS, McNeill LA, Riordan MV, Tian Y-M, Bullock AN, Welford RW, Elkins JM, Oldham NJ, Bhattacharya S, Gleadle JM, Ratcliffe PJ, Pugh CW, Schofield CJ. Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J Biol Chem. 2002;277:26351–26355. doi: 10.1074/jbc.C200273200. [DOI] [PubMed] [Google Scholar]
- 15.Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, Bruick RK. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel SA, Simon MC. Biology of hypoxia-inducible factor-2α in development and disease. Cell Death Differ. 2008;15:628–634. doi: 10.1038/cdd.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiesener MS, Turley H, Allen WE, Willam C, Eckardt K-U, Talks KL, Wood SM, Gatter KC, Harris AL, Pugh CW, Ratcliffe PJ, Maxwell PH. Induction of endothelial PAS domain protein-1 by hypoxia: characterization and comparison with hypoxia-inducible factor-1α. Blood. 1998;92:2260–2268. [PubMed] [Google Scholar]
- 18.Wenger RH. Cellular adaptation to hypoxia: O2-sensing protein hydroxylases, hypoxia-inducible transcription factors, and O2-regulated gene expression. FASEB J. 2002;16:1151–1162. doi: 10.1096/fj.01-0944rev. [DOI] [PubMed] [Google Scholar]
- 19.Wenger RH, Stiehl DP, Camenisch G. Integration of oxygen signaling at the consensus HRE. Sci. STKE. 2005;306:re12. doi: 10.1126/stke.3062005re12. [DOI] [PubMed] [Google Scholar]
- 20.Mole DR, Blancher C, Copley RR, Pollard PJ, Gleadle JM, Ragoussis J, Ratcliffe PJ. Genome-wide association of hypoxia-inducible factor (HIF)-1α and HIF-2α DNA binding with expression profiling of hypoxia-inducible transcripts. J Biol Chem. 2009;284:16767–16775. doi: 10.1074/jbc.M901790200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu Y-Z, Moran SM, Hogenesch JB, Wartman L, Bradfield CA. Molecular characterization and chromosomal localization of a third alpha-class hypoxia inducible factor subunit, HIF3α. Gene Expr. 1998;7:205–213. [PMC free article] [PubMed] [Google Scholar]
- 22.Hara S, Hamada J, Kobayashi C, Kondo Y, Imura N. Expression and characterization of hypoxia-inducible factor (HIF)-3α in human kidney: suppression of HIF-mediated gene expression by HIF-3α. Biochem Biophys Res Commun. 2001;287:808–813. doi: 10.1006/bbrc.2001.5659. [DOI] [PubMed] [Google Scholar]
- 23.Makino Y, Cao R, Svensson K, Bertilsson G, Åsman M, Tanaka H, Cao Y, Berkenstam A, Poellinger L. Inhibitory PAS domain protein is a negative regulator of hypoxia-inducible gene expression. Nature. 2001;414:550–554. doi: 10.1038/35107085. [DOI] [PubMed] [Google Scholar]
- 24.Makino Y, Kanopka A, Wilson WJ, Tanaka H, Poellinger L. Inhibitory PAS domain protein (IPAS) is a hypoxia-inducible splicing variant of the hypoxia-inducible factor-3α locus. J Biol Chem. 2002;277:32405–32408. doi: 10.1074/jbc.C200328200. [DOI] [PubMed] [Google Scholar]
- 25.Yamashita T, Ohneda O, Nagano M, Iemitsu M, Makino Y, Tanaka H, Miyauchi T, Goto T, Ohneda K, Fujii-Kuriyama Y, Poellinger L, Yamamoto M. Abnormal heart development and lung remodeling in mice lacking the hypoxia-inducible factor-related basic helix-loop-helix PAS protein NEPAS. Mol Cell Biol. 2008;28:1285–1297. doi: 10.1128/MCB.01332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maynard MA, Qi H, Chung J, Lee EHL, Kondo Y, Hara S, Conaway RC, Conaway JW, Ohh M. Multiple splice variants of the human HIF-3α locus are targets of the von Hippel-Lindau E3 ubiquitin ligase complex. J Biol Chem. 2003;278:11032–11040. doi: 10.1074/jbc.M208681200. [DOI] [PubMed] [Google Scholar]
- 27.Pasanen A, Heikkilä M, Rautavuoma K, Hirsilä M, Kivirikko KI, Myllyharju J. Hypoxia-inducible factor (HIF)-3α is subject to extensive alternative splicing in human tissues and cancer cells and is regulated by HIF-1 but not HIF-2. Int J Biochem Cell Biol. 2010;42:1189–1200. doi: 10.1016/j.biocel.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 28.Jang MS, Park JE, Lee JA, Park SG, Myung PK, Lee DH, Park BC, Cho S. Binding and regulation of hypoxia-inducible factor-1 by the inhibitory PAS proteins. Biochem Biophys Res Commun. 2005;337:209–215. doi: 10.1016/j.bbrc.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 29.Maynard MA, Evans AJ, Hosomi T, Hara S, Jewett MAS, Ohh M. Human HIF-3α4 is a dominant-negative regulator of HIF-1 and is down-regulated in renal cell carcinoma. FASEB J. 2005;19:1396–1406. doi: 10.1096/fj.05-3788com. [DOI] [PubMed] [Google Scholar]
- 30.Besson A, Wilson TL, Yong VW. The anchoring protein RACK1 links protein kinase Cε to integrin β chains. Requirements for adhesion and motility. J Biol Chem. 2002;277:22073–22084. doi: 10.1074/jbc.M111644200. [DOI] [PubMed] [Google Scholar]
- 31.Koivunen P, Hirsilä M, Kivirikko KI, Myllyharju J. The length of peptide substrates has a marked effect on hydroxylation by the hypoxia-inducible factor prolyl 4-hydroxylases. J Biol Chem. 2006;281:28712–28720. doi: 10.1074/jbc.M604628200. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka T, Wiesener M, Bernhardt W, Eckardt K-U, Warnecke C. The human HIF (hypoxia-inducible factor)-3α gene is a HIF-1 target gene and may modulate hypoxic gene induction. Biochem J. 2009;424:143–151. doi: 10.1042/BJ20090120. [DOI] [PubMed] [Google Scholar]
- 33.Berra E, Benizri E, Ginouvès A, Volmat V, Roux D, Pouysségur J. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1α in normoxia. EMBO J. 2003;22:4082–4090. doi: 10.1093/emboj/cdg392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Appelhoff RJ, Tian Y-M, Raval RR, Turley H, Harris AL, Pugh CW, Ratcliffe PJ, Gleadle JM. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J Biol Chem. 2004;279:38458–38465. doi: 10.1074/jbc.M406026200. [DOI] [PubMed] [Google Scholar]
- 35.Takeda K, Ho VC, Takeda H, Duan L-J, Nagy A, Fong G-H. Placental but not heart defects are associated with elevated hypoxia-inducible factor alpha levels in mice lacking prolyl hydroxylase domain protein 2. Mol Cell Biol. 2006;26:8336–8346. doi: 10.1128/MCB.00425-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan DA, Sutphin PD, Yen S-E, Giaccia AJ. Coordinate regulation of the oxygen-dependent degradation domains of hypoxia-inducible factor 1α. Mol Cell Biol. 2005;25:6415–6426. doi: 10.1128/MCB.25.15.6415-6426.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee PJ, Jiang B-H, Yoke Chin B, Iyer NV, Alam J, Semenza GL, Choi AMK. Hypoxia-inducible factor-1 mediates transcriptional activation of the heme oxygenase-1 gene in response to hypoxia. J Biol Chem. 1997;272:5375–5381. doi: 10.1074/jbc.272.9.5375. [DOI] [PubMed] [Google Scholar]
- 38.Kitamuro T, Takahashi K, Ogawa K, Udono-Fujimori R, Takeda K, Furuyama K, Nakayama M, Sun J, Fujita H, Hida W, Hattori T, Shirato K, Igarashi K, Shibahara S. Bach1 functions as a hypoxia-inducible repressor for the heme oxygenase-1 gene in human cells. J Biol Chem. 2003;278:9125–9133. doi: 10.1074/jbc.M209939200. [DOI] [PubMed] [Google Scholar]