Abstract
The oxygen-sensitive transcription factor hypoxia inducible factor (HIF) is a key regulator of gene expression during adaptation to hypoxia. Crucially, inflamed tissue often displays regions of prominent hypoxia. Recent studies have shown HIF signalling is intricately linked to that of the pro-inflammatory transcription factor nuclear factor kappa B (NFκB) during hypoxic inflammation. We describe the relative temporal contributions of each to hypoxia-induced inflammatory gene expression and investigate the level of crosstalk between the two pathways using a novel Gaussia princeps luciferase (Gluc) reporter system. Under the control of an active promoter, Gluc is expressed and secreted into the cell culture media, where it can be sampled and measured over time. Thus, Gluc constructs under the control of either HIF or NFκB were used to resolve their temporal transcriptional dynamics in response to hypoxia and to cytokine stimuli, respectively. We also investigated the interactions between HIF and NFκB activities using a construct containing the sequence from the promoter of the inflammatory gene cyclooxygenase 2 (COX-2), which includes functionally active binding sites for both HIF and NFκB. Finally, based on our experimental data, we constructed a mathematical model of the binding affinities of HIF and NFκB to their respective response elements to analyse transcriptional crosstalk. Taken together, these data reveal distinct temporal HIF and NFκB transcriptional activities in response to hypoxic inflammation. Furthermore, we demonstrate synergistic activity between these two transcription factors on the regulation of the COX-2 promoter, implicating a co-ordinated role for both HIF and NFκB in the expression of COX-2 in hypoxic inflammation.
Keywords: NFκB, Hypoxia inducible factor, Inflammation, Transcription, Crosstalk, Mathematical modelling
Introduction
Localised hypoxia is a common feature in a variety of biological settings where inflammation is also occurring, including growing tumours and critically inflamed tissues [1–3]. This creates a situation whereby both the hypoxic signalling pathway, through the hypoxia inducible factor (HIF), and the inflammatory signalling pathway, through nuclear factor-κB (NFκB), are activated. Recent studies have shown that hypoxia influences the NFκB pathway and that HIF may play an important role in inflammation [4–8]. However, the relative contributions of HIF and NFκB into creating transcriptional activation profiles leading to a coordinated regulation of hypoxia-induced inflammatory gene expression remain unclear. Developing our understanding of their transcriptional activities and regulation represents a clear goal in this area of systems biology.
HIF is a heterodimeric transcription factor composed of a β-subunit, which is constitutively present in the cell nucleus, and an oxygen-sensitive α-subunit (Hif1α, Hif2α or Hif3α). Although HIF is constitutively synthesised at high levels, it is destabilised in the presence of molecular oxygen as a result of the enzymatic activity of oxygen-sensing enzymes termed prolyl hydroxylases (PHD). In hypoxia, this oxygen-requiring hydroxylation event is inhibited, HIFα escapes degradation and can translocate to the nucleus to form a functional dimer with HIFβ that activates gene expression and triggers the hypoxic response. This transcriptional response allows cellular adaptation to a hypoxic environment, such as a tumour microenvironment [1, 2] or the hematopoietic stem cell niche [9].
NFκB is a family of transcription factors which plays a key role in a wide variety of physiological (such as immunity) and patho-physiological cellular responses (chronic inflammation, diabetes, cancer) [10, 11]. Due to its role as a master regulator of immunity and inflammation, its transcriptional activity and regulatory pathway have been an area of intense research [12]. Substantial evidence now exists that hypoxia can activate NFκB in vivo [8, 13] and in vitro [5, 7, 8, 14]. While the exact mechanism involved in the activation of NFκB remains to be fully elucidated, recent evidence has suggested that the same oxygen-sensing enzymes, which confer oxygen sensitivity to the HIF pathway also play a role in activation of NFκB in response to hypoxia [5, 14].
Crosstalk between the HIF and NFκB pathways has been demonstrated by a number of in vitro and in vivo studies showing that NFκB plays an important role in regulating basal and stimulated HIF-1α expression [7, 15–18]. NFκB can also regulate HIF-2α signalling through an interaction with the NFκB essential modulator (NEMO), which aids in the recruitment of transcriptional co-activators such as CREB binding protein (CBP) and p300, and increases HIF-2α transcriptional activity [19]. Conversely, HIF-1α has been reported to alter NFκB signalling in neutrophils [20]. In addition, a group of pro-inflammatory genes, including cyclooxygenase (COX-2) and inducible nitric oxide synthase (iNOS) contains functional response elements for both HIF and NFκB in their promoter regions. We have previously shown that NFκB both directly but also indirectly, through its regulation of HIF-1α, regulates COX-2 expression in response to hypoxia [8].
In the present study, in order to investigate the crosstalk between HIF and NFκB transcriptional activities in mammalian cells, we generated a novel Gaussia luciferase (Gluc) reporter system. Gluc is derived from the marine copecod Gaussia princeps and belongs to a new class of luciferases that are naturally secreted molecules [21], and has already been used as a sensitive monitor for evaluating promoter activity in algae [22], as well as monitoring tumour growth in vivo [23], NFκB activity in vivo [24] and in vitro [25] and HIF activity in vitro [26]. As Gaussia luciferase is secreted, it is thus possible to monitor temporal transcriptional activity in a single cell population. We designed Gaussia constructs under the control of either HIF or NFκB to study their transcriptional activity under hypoxic or cytokine stimulation. To investigate transcriptional crosstalk, we chose a sequence from the promoter region of the human COX-2 gene which includes functional response elements for both HIF and NFκB. The COX-2 gene encodes for the inducible cyclooxygenase which has been associated with inflammation and cell proliferation [27], and its transcriptional regulation can be through either HIF and/or NFκB activity [4, 8, 28].
Using experimental data and mathematical modelling, we establish it is possible to analyse and dissect the interactions between HIF and NFκB transcriptional activities under hypoxic and inflammatory conditions. From our analysis, we propose that HIF and NFκB bind to the COX-2 promoter independently of each other, but display a synergistic behaviour in the transcriptional regulation under dual hypoxic and inflammatory stimulation.
Materials and methods
Cell lines and cell culture
Human embryonic kidney cell HEK293, human epithelial colorectal adenocarcinoma Caco-2 cells and human cervical cancer Hela cells were obtained from ATCC and cultured in DMEM high-glucose medium supplemented with 10% FCS and 100 U/ml penicillin–streptomycin. Cells were exposed to hypoxia using pre-equilibrated media and maintained in standard normobaric hypoxic conditions (1 or 3% O2, 5% CO2, and 94% N2) in a hypoxia chamber (Coy Laboratories). Normoxic controls were exposed to pre-equilibrated normoxic media and maintained at atmospheric O2 levels (21% O2, 5% CO2) in a tissue culture incubator.
Gaussia constructs
The mammalian expression vector pGluc-TK (NEB) contains the coding sequence for Gaussia luciferase under the control of the Herpes Simplex Virus thymidine kinase (TK) promoter, for constitutive activity. This vector was used to assess the expression and secretion of Gaussia luciferase protein into the culture media in normoxia and hypoxia.
The mammalian expression vector pGluc-Basic (NEB) was modified to include a minimal promoter sequence, and this resultant vector (pGluc-Mp) was used to generate a series of hypoxia-responsive and NFκB-responsive vectors (pGluc-HRE, pGluc-NRE and pGluc-COX2). pGluc-HRE contains four copies of the EPO HREs in the right orientation while pGluc-NRE contains a concatamer of NREs in the left orientation. pGluc-COX2 contains the sequence −4 to −631 of the human COX-2 gene, which includes one HIF response element (HRE) [28] and two NFκB response elements (NRE) [27]. The inserts were amplified by PCR from human genomic DNA using commercially available reagents (Invitrogen), cut using BglII and EcoRI restriction enzymes (Roche) and subcloned into pGluc-Mp. Resulting plasmids were characterised by sequencing (MWG).
Plasmid DNAs were transfected using Lipofectamine 2000 (Invitrogen) at a concentration of 200 ng/40,000 of Caco-2 cells or 200 ng/100,000 of HEK293 cells. Sampling of culture media started 24 h post-transfection. Gaussia luciferase activity was measured using the Biolux Gaussia luciferase Flex Assay kit (NEB) in a plate reader (Synergy HT; Biotek).
The expression and secretion rates of Gaussia luciferase were found to be unchanged during hypoxia, although we have observed lower luciferase activity in the media when the cells were more confluent (data not shown). It is likely that the secretory pathway and mechanism of Gaussia luciferase is dependent on available cellular energy, possibly through ATP binding cassette transporters [29]. Resolving this pathway is beyond the scope of this study, but it is accounted for in all our assays through paired sampling and the use of the constitutively active pGluc-TK construct as internal control in experimental conditions involving hypoxic culture.
Western blot analysis
Whole-cell, nuclear and cytosolic extracts were generated in either normoxia or hypoxia according to previously published protocols [30, 31]. Protein concentration was quantified using a Bradford assay, and samples were normalised accordingly. Samples were separated by SDS-PAGE and immunoblotted as described previously [30, 31] using the following primary Abs and dilutions: HIF-1α (1:250; BD Pharmingen), β-actin (1:10,000; Sigma) and TATA box binding protein (TBP; 1:2,500; Abcam).
Chromatin immunoprecipitation
HEK293 cells were grown on 3 × 145 mm dishes per treatment and exposed to normoxia or hypoxia (1% O2) for 0–24 h. At the end of the time course, cells were removed from the hypoxia chamber or the tissue culture incubator, and medium was aspirated. Cells were immediately fixed (1% formaldehyde and Eagle’s MEM tissue culture media) for 10 min. Fixation was stopped using glycine solution, and cells were scraped in PBS supplemented with PMSF following a PBS wash step. Cells were pelleted by centrifugation and lysed prior to shearing of chromatin by sonication. After precleaning, chromatin was incubated with a specific Ab, and immunocomplexes were subsequently collected using salmon sperm DNA/protein A agarose (Millipore). After a series of washes, immunocomplexes were eluted using an elution buffer (1% SDS and 0.1 M NaHCO3), and cross-links were reversed. DNA was then recovered by phenol/chloroform extraction. Purified DNA (3 μl) was amplified using human COX-2 promoter primers (forward, 5′-GAATTTACCTTTCCCGCCTCTC-3′; reverse, 5′-AAGCCCGGTGGGGGCAGGGTTT-3′) [8] using a thermocycler program (94°C for 3 min; then 36 cycles of 94°C for 20 s, 60°C for 30 s, and 72°C for 30 s; then a hold cycle of 10°C). Samples were run on a 2% agarose gel using ethidium bromide to visualize a 649-bp product.
Reagents
The cell permeable prolyl hydroxylase inhibitor dimethyl-oxaloylglycine (DMOG; Cayman Chemicals) was dissolved in dimethyl sulfoxide (DMSO; Sigma). Tumour necrosis factor-α (TNFα) was from Sigma while interleukin-1β (IL-1β) was obtained from R&D Systems.
Thermo-statistical model
We used a thermo-statistical approach to modelling transcriptional activity as developed in refs. [32, 33]. Briefly, we consider the relative concentrations of transcription factors and the probability of these transcription factors binding to the promoter to initiate transcription. Let [HIF] and [NFκB] denote the concentrations of the transcription factors under consideration. Then, qH = [HIF]/KH and qN = [NFκB]/KN denote the concentrations relative to the effective dissociation constant KH and KN. The latter describe the formation and dissociation of HIF–DNA complexes and NFκB–DNA complexes, respectively. In what follows, we consider only the saturation domain in which [HIF] ≫ KH and [NFκB] ≫ KN holds such that qH ≫ 1 and qN ≫ 1 [33]. Furthermore, let P denote the probability that RNA polymerase occupies the promoter, i.e., the binding probability referred to in the main text, and let P0 denote the basal binding probability. With these definitions at hand, the binding probabilities P(HIF), P(NFκB), and P(dual) for the conditions Ctrl Hypoxia, TNFα Normoxia, and dual activation in the saturation domain are given by [33]
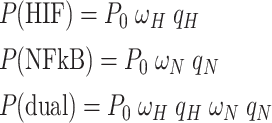 |
1 |
where ωH > 1 and ωN > 1 are certain proportionality factors. Note that the third relationship listed in Eq. (1) holds only under the assumption of independent activation. A key assumption that has been frequently made in the context of thermo-statistical modelling of the transcriptional machinery is that the rate of transcription initiation is proportional to the binding probability P [32–34]. Likewise, the transcription rate of the protein is assumed to be proportional to P. Let rX with X=“0”, “HIF”, “NFκB”, and “dual” denote the transcription rates observed in the respective conditions. Then, the aforementioned assumption implies that we have
 |
2 |
where β > 0 is a proportionality factor. Introducing the rescaled binding probabilities P*X = PX/P0 and the fold changes r*X = rX/r0, from Eq. (2), it follows that they correspond to each other: P*X = r*X (as mentioned in the main text). Furthermore, from Eq. (1) it follows that the fold changes of transcription rates satisfy
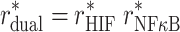 |
3 |
In order to make contact with the experiment, we re-write Eq. (3) in terms of the experimentally measurable variables r0, rHIF, rNFκB and rdual and thus obtain
 |
4 |
For each sample, we substituted the observed values for r0, rHIF, rNFκB in the right-hand side of Eq. (4) in order to obtain the predicted fold changes for dual stimulation. Using sample averages, we compared the predicted fold changes with the measured fold changes, see Fig. 5d.
Fig. 5.
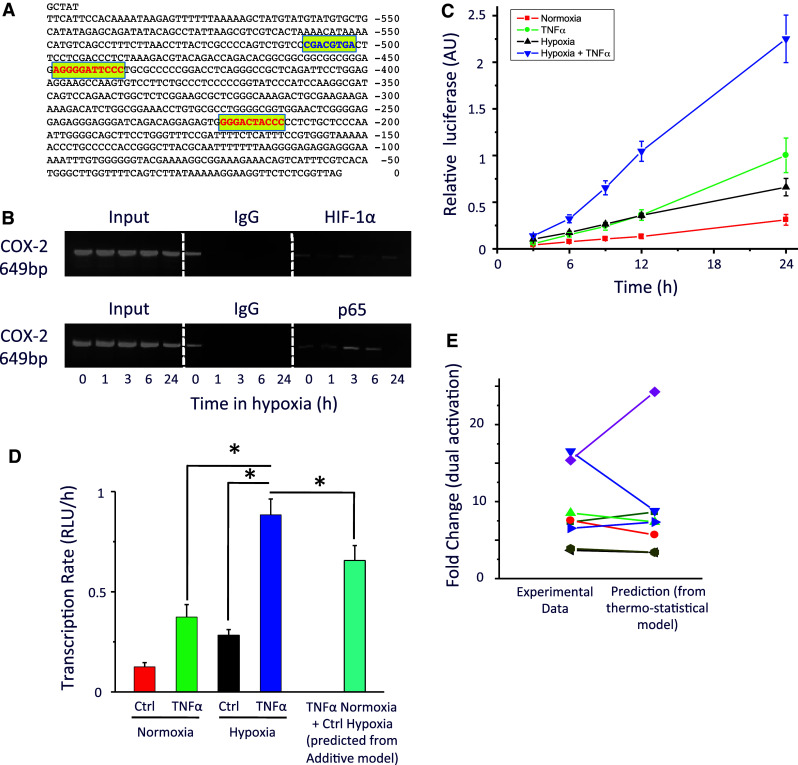
Transcriptional activity of the human COX-2 promoter. a Representation of the human cyclooxygenase-2 (COX-2) promoter region cloned into the pGLuc-Mp vector, showing the binding sites for HIF (blue) and NFκB (red). b Chromatin immunoprecipitation analysis was carried out using an antibody against HIF-1α, p65 or a control antibody as indicated in cells exposed to hypoxia for 0–24 h to assess whether HIF-1α or p65 binds directly to the COX-2 promoter under conditions of hypoxia. (n = 3). c HEK293 cells were transfected with the resultant vector (pGluc-COX2) and cultured under either normoxia (21% O2; filled square), TNFα (1 ng/ml; filled circle), hypoxia (1% O2; filled triangle) or TNFα and hypoxia (filled inverted triangle). d Transcriptional activity under hypoxia, TNFα, dual Hypoxia and TNFα stimulation and predicted transcriptional activity under dual stimulation (additive model). e Fold change under dual activation for each experiment was calculated and compared with the prediction of a thermo-statistical model of transcriptional binding probabilities as described in “Materials and methods”. n = 8. Asterisks significant difference (p < 0.05)
Statistical analysis
All experiments were carried out a minimum of n = 3 independent times unless otherwise indicated and data were expressed as the mean ± SEM. To estimate an indicator of current transcriptional activity from the cumulative luciferase activity data (i.e., time derivatives), where possible we used central difference approximations according to
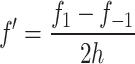 |
5 |
where f′ is the estimated first time derivative at the current time point, f 1 is the cumulative luciferase activity at the next time point, f −1 is the cumulative luciferase activity at the previous time point, and h is the time step. It is only possible to use a central difference approximation when there are two evenly spaced measurements in either direction from the time point of interest, both h time units away. When this was not the case, as for instance with the first data point of the time series, the more widespread and intuitive forward difference approximation was used according to
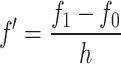 |
6 |
where f 0 denotes the cumulative luciferase activity at the current time point. Central difference approximations were preferred to forward difference approximations because their resulting predictions are less sensitive to measurement noise.
Results
Development of the Gaussia luciferase reporter system
The Gluc reporter system relies on the inherent property of Gaussia luciferase to be secreted from the cell into the culture media, where it can be sampled. To assess whether the rate of secretion is stable over time, we transfected HEK293 cells with the constitutively active vector pGluc-TK which is under the control of the HSV thymidine kinase promoter. The luciferase activity was found to increase linearly over the period measured whereas the activity from HEK293 cells transfected with a Gluc vector driven by a minimal promoter (pGluc-Mp) remained unchanged (Fig. 1a). As Gluc content in the culture media is cummulative, we can estimate the current promoter activity (Fig. 1b) by calculating the time derivative of the curve in Fig. 1a (Eq. 6 of “Materials and methods”). Linear regression of the time derivative data points showed that pGluc-TK had a stable transcriptional activity over the 24-h time course (y = 4 × 10−6 x + 9.2; r 2 = 0.00016). Thus, under the control of a constitutively active promoter, Gaussia luciferase is constitutively produced, expressed and secreted at a constant rate.
Fig. 1.
Development of the Gaussia luciferase assay for non-invasive, reproducible and high temporal resolution of transcriptional activity. a Relative luciferase activity and transcriptional activity from human embryonic kidney cells (HEK293) transfected with a Gluc construct under the control of the Herpes Simplex virus thymidine kinase promoter for constitutive expression (pGluc-TK; filled square) or under the control of a minimal promoter (pGluc-Mp; filled circle). Representative traces are shown for luciferase activity. b Transcriptional activity of pGluc-TK was calculated using primarily central differences as described in “Materials and methods”. Data shown as mean ± SEM
Characterisation of HIF transcriptional activity
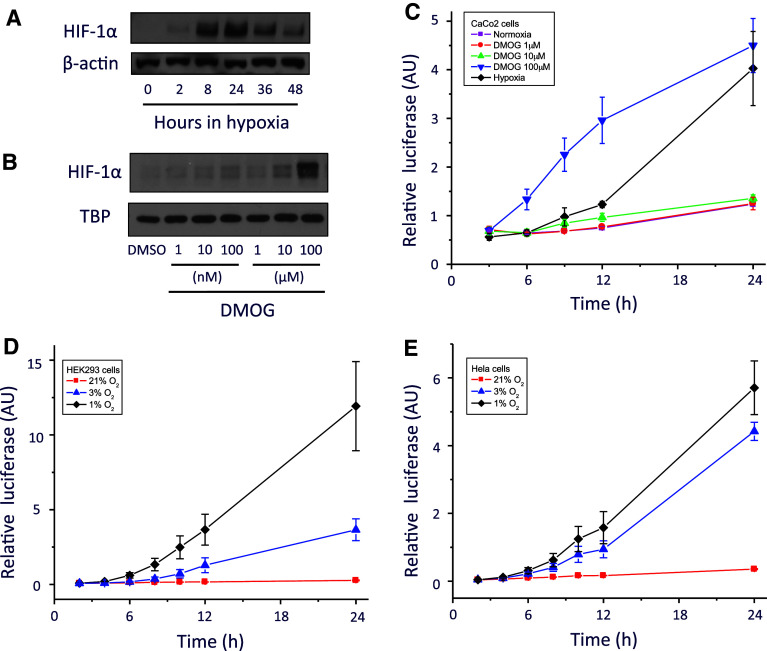
HIF activation has been well characterised in cells exposed to hypoxia [1]. During low oxygen tension, the HIF-1α protein is rapidly stabilised (Fig. 2a). In normoxia, HIF protein degradation is dependent on the activity of the prolyl hydroxylases (PHDs). As such, chemical inhibition of the PHDs by DMOG [35] can also increase HIF stabilisation and localisation in the nucleus (Fig. 2b). We generated a Gaussia luciferase reporter under the control of HIF (pGluc-HRE) to investigate HIF temporal transcriptional activity. Consistent with an increase in HIF protein, we observed a HIF transcriptional activity in Caco-2 cells transfected with pGluc-HRE when exposed to hypoxia (1% O2) or to increasing concentration of DMOG (Fig. 2c). This activity was also observed in pGluc-HRE transfected HEK293 (Fig. 2d) and Hela cells (Fig. 2e) cultured under different degrees of hypoxia (1% O2 or 3% O2). Thus, we show that HIF displays unique temporal transcriptional activity in response to oxygen or to PHD inhibition and demonstrate that pGluc-HRE represents a useful tool to effectively monitor temporal changes in HIF-dependent transcriptional activity.
Fig. 2.
Transcriptional activity of the Hypoxia Inducible Factor (HIF). a Hypoxia induces expression of HIF-1α protein in human epithelial colorectal adenocarcinoma (Caco2) cells. b Nuclear HIF-1α protein is detected after 6 h of chemical inhibition of prolyl-hydroxylases by DMOG. c Caco2 cells transfected with the pGluc-HRE vector and cultured under normoxia (21% O2) under increasing concentration of DMOG or hypoxia (1% O2). HEK293 cells (d) and Hela cells (e) transfected with the pGluc-HRE vector and cultured under different oxygen tension (21% O2, red; 3% O2, purple; 1% O2, black)
Characterisation of NFκB transcriptional activity
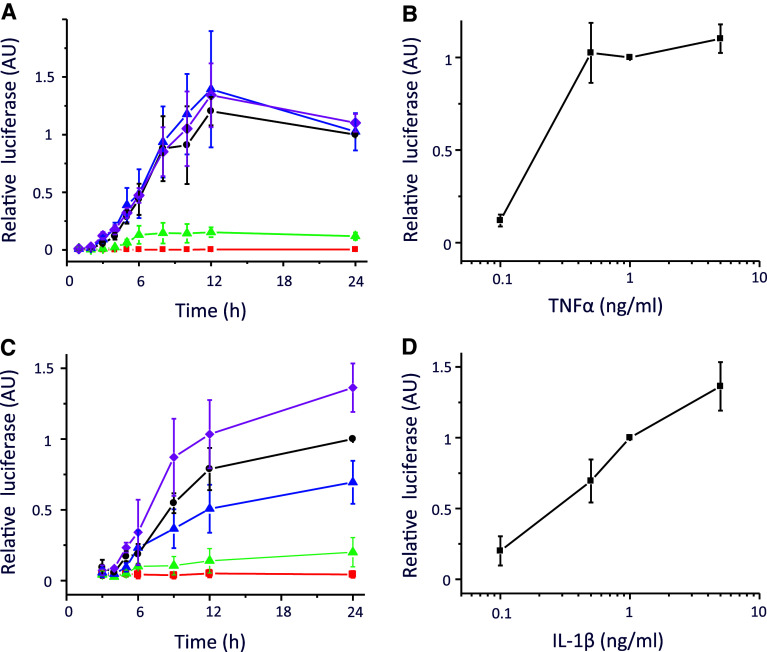
NFκB consists of a family of transcription factors that play critical roles in inflammation, immunity, cell proliferation, differentiation, and survival [10, 11]. A large number of diverse external stimuli can lead to activation of NFκB. We next generated a Gaussia luciferase reporter under the control of NFκB (pGluc-NRE) and used it to investigate the transcriptional response to two different inflammatory stimuli: TNFα (Figs. 3a, b) and IL-1β (Figs. 3c, d). A dose-dependent increase in luciferase activity was observed under both stimuli. Furthermore, consistent with previous studies [5, 7], we observed a hypoxic induction of pGluc-NRE activity, although not as strongly as in pGluc-HRE (Fig. 4a, b). We explored the data further by examining the NFκB transcriptional activity and observed distinct transcriptional responses under increasing concentration of TNFα stimulation (Fig. 4c). These responses were modulated when under dual inflammatory cytokine and hypoxic stimulations, where we observed an elevated response in TNFα-induced NFκB transcriptional activity in a background of hypoxia (Fig. 4d). Thus, we show that stimulation with inflammatory stimuli results in quantitatively and temporally distinct NFκB transcriptional activity, which can be further modulated by hypoxia.
Fig. 3.
Transcriptional activity of the Nuclear Factor Kappa B (NFκB). a Relative luciferase activity from HEK293 transfected with pGluc-NRE under increasing concentration of TNFα (filled square 0; filled triangle 0.1 ng/ml; filled triangle 0.5 ng/ml; filled circle 1 ng/ml and filled diamond 5 ng/ml). b Concentration-dependent luciferase activity after 24 h TNFα stimulus. c Relative luciferase activity from human embryonic kidney cells (HEK293) transfected with pGluc-NRE under increasing concentration of IL-1β (filled square 0; filled triangle 0.1 ng/ml; filled triangle 0.5 ng/ml; filled circle 1 ng/ml and filled diamond 5 ng/ml). d Concentration-dependent luciferase activity after 24 h IL-1β stimulus
Fig. 4.
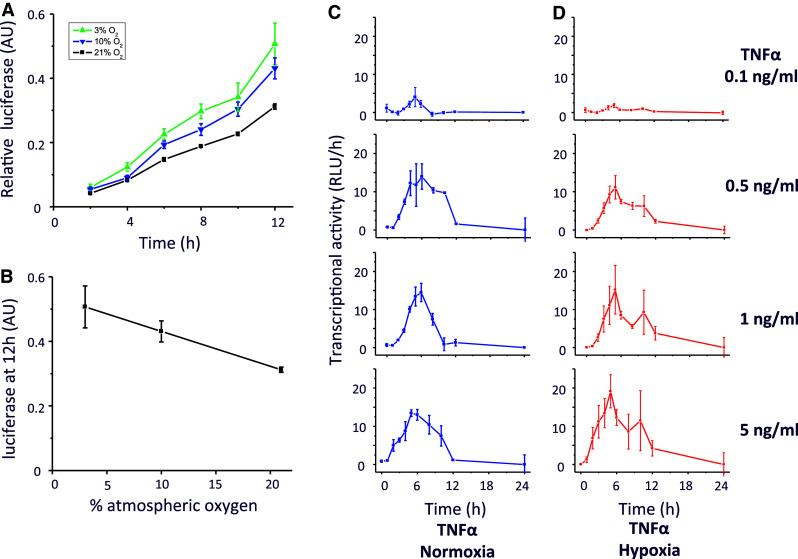
Effect of hypoxia on the transcriptional activity of NFκB. a HEK293 cells transfected with pGluc-NRE were exposed to a range of oxygen tension (21% O2 to 1% O2). b The relative luciferase activity at 12 h is shown as a function of oxygen. c, d The pGluc-NRE responses to TNFα are shown as a function of transcriptional activity per hour is dependent on the concentration of the inflammatory stimulus under normoxia (c) or hypoxia (d). Time derivatives were calculated based on central difference approximation where possible as described in “Materials and methods”
Characterisation of COX-2 promoter activity
We have previously shown there can be crosstalk between HIF and NFκB [8]. A region of the COX-2 promoter sequence (−605 to −5 bp) was reported to include two NFκB binding sites and one HIF binding site (Fig. 5a). We show by chromatin immunoprecipitation that NFκB and HIF can bind to that promoter sequence (Fig. 5b). To investigate this interaction further, we cloned the COX-2 promoter sequence into the pGluc-MP vector. Using the resultant vector pGluc-COX2, we resolved for the first time the time course of the promoter activity of COX-2 under hypoxic, TNFα (1 ng/ml), or dual stimulation (Fig. 5c). In all three conditions, the transcriptional activity was found to be significantly higher than in basal conditions (Fig. 5d; p < 0.05). Thus, our data indicate that TNFα and hypoxic stimuli have similar effect on the transcriptional activity of pGluc-COX2 within 24 h.
Mathematical model of TF binding
Cells would normally receive a wide variety of cellular and environmental signals that are processed in combination to generate a specific genetic response. Hence, stimulation by both hypoxia and inflammation should generate a different transcriptional regulation than from a single stimulus. The data generated from the pGluc-COX2 were used to predict the transcriptional activity under dual hypoxic and inflammatory stimulation using an additive model of cooperative transcriptional activation (i.e., TNFα Normoxia + Ctrl Hypoxia = TNFα Hypoxia; Fig. 5d). The additive transcriptional activity (calculated from the addition of the rate of transcription in the conditions TNFα Normoxia and Ctrl Hypoxia) was found to be significantly lower than the actual transcriptional activity observed (TNFα Hypoxia; p < 0.05). Thus, this ‘greater-than-additive’ transcriptional activity (i.e. violation of the additive model) would demonstrate a synergy between hypoxia and inflammatory stimuli.
Next, we constructed a thermodynamic model of transcriptional regulation (described in “Materials and methods”) to test the probability of RNA polymerase binding to the promoter as a result of transcription factor recruitment [32]. According to the model, under independent dual activation, the binding probability equals the product of the binding probabilities observed during individual stimulation [33] (either TNFα Normoxia or Ctrl Hypoxia but not both). For this relationship to hold, we rescaled the binding probabilities by the basal probabilities such that they actually correspond to fold changes of transcriptional activities (see “Materials and methods”). In line with previous work [33], we exploited the multiplicative relationship in order to test whether the binding of HIF and NFκB to their respective response elements was independent of each other. We did not observe any statistical difference between the experimental and the predicted (‘multiplication rule’) transcriptional activity (n = 8; Fig. 5e), indicating that there is indeed independent activation, i.e. the binding of HIF or NFκB to the promoter is independent of each other.
In short, we show that the dual activation by HIF and NFκB violates the additive model of cooperative activation, but is consistent with the thermo-statistical model for cooperative independent activation that predicts synergistic ‘greater-than-additive’ responses.
Discussion
Reporter assays are useful tools in probing transcriptional activity and regulation. Using the inherent property of the secreted Gaussia luciferase, we here show that we can monitor and measure the temporal transcriptional dynamics of HIF and NFκB activities and analyse the transcriptional crosstalk between the two pathways.
The inclusion of response elements for HIF into the pGluc vector has opened new avenues for investigating HIF-dependent transcriptional activities. Since its discovery in the early 1990s, HIF-1 has rapidly attracted interest for its involvement in fundamental biological processes—such as cardiovascular development [36], tumour metabolism [37] and stem cell differentiation [38]. Its role in regulating the transcriptional response to oxygen deprivation has made it a potential therapeutic target [39]. Using our in-house-generated pGluc-HRE construct, we reveal distinct transcriptional dynamics for HIF in response to graded hypoxia or prolyl hydroxylase inhibition. Interestingly, we observed a gradual increase in HIF transcriptional activity in hypoxia, while pharmacological prolyl hydroxylase inhibition caused a sharp and rapid activation.
The NFκB family of transcription factors plays an important role in the regulation of the immune and inflammatory response, as well as cell division and cell death [10]. Research using real-time single-cell imaging has shown NFκB shuttling in and out of the nucleus under an inflammatory stimulus such as TNFα, matched with firefly-luciferase activity [40]. Here, we show that the data from a Gaussia luciferase reporter under the control of NFκB is a valid measure of the dynamics of transcriptional activity due to NFκB stimulation in a cell population. Using two different inflammatory stimuli, we describe a dose-dependent increase in transcriptional activity, which we found to be very different between TNFα and IL-1β, further reinforcing the view that distinct stimuli may generate quantitatively and temporally distinct genetic responses from the same transcriptional pathway [10, 12]. Indeed, within the range of concentrations tested, we observed an ‘all-or-nothing switch-like’ effect with TNFα stimulation, while the response to IL-1β was ‘analogue-like’. In addition, we confirm that hypoxia enhanced basal NFκB activity and the NFκB response to cytokine stimulation, likely through the activation of the IKK complex and inhibition of prolyl hydroxylase-1 [5, 6], thus providing further evidence for a role of hypoxia in mediating inflammatory response.
Given the growing number of studies demonstrating a high degree of crosstalk between the HIF and NFκB pathways [4–8], we decided to analyse the contribution of each on the cyclooxygenase-2 promoter. The human COX-2 gene was chosen as its regulation can be through either HIF and/or NFκB activity [4, 8, 28]. We constructed a thermodynamic model of transcriptional regulation to test the probability of RNA polymerase binding to the promoter as a result of transcription factor recruitment [32] and found that, while HIF and NFκB were acting independently on their respective response elements, there was also a ‘greater-than-additive’ transcriptional activity under dual stimulation, which would imply synergy. This suggests the capacity for increased recruitment of RNA polymerase arising from the effect of hypoxia on NFκB activity, as shown from our data using the pGluc-NRE construct under cytokine and hypoxic stimulation.
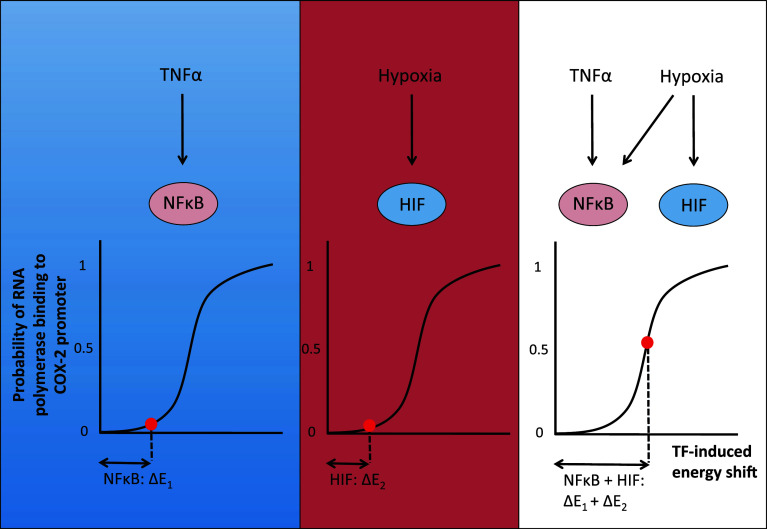
We speculate that the synergy observed under dual cytokine and hypoxic stimulation could be arising at both the signalling network and the promoter levels. Given the thermodynamics model that was used, both transcription factors (HIF and NFκB) bind independently to their respective response element and lower the binding energy for RNA polymerase to bind to the gene (Fig. 6). Due to the non-linearity of the probability of RNA polymerase binding, the addition of the activities of HIF and NFκB generates a higher binding probability than under a single transcription factor [41], which results in a ‘greater-than-additive effect’ on the level of transcription rates. Additionally, given that hypoxia activates the IKK complex and increases the amount of nuclear NFκB [5, 6], it is reasonable to assume that this increased concentration of NFκB would also further lower the binding energy for RNA polymerase, thus enhancing the synergy effect.
Fig. 6.
Simplified scheme illustrating a ‘greater-than-additive’ effect caused by the nonlinearity of the transcriptional machinery as predicted by thermo-statistical approaches. The probability P of RNA polymerase recruitment to the COX-2 promoter as function of the energy shift induced by transcription factors is shown following a Boltzmann distribution law. TNFα stimulation results in NFκB activity which induces a shift in energy ∆E1 required for RNA polymerase to bind to the COX-2 promoter. The binding probability is P(NFκB). In hypoxia, HIF is stabilised, and its activity induces a shift in energy ∆E2 and a binding probability P(HIF). However, under dual TNFα and hypoxia stimulation, the shift in energy results in a greater probability for RNA polymerase to bind to the promoter, i.e. P(NFκB +HIF) > P(NFκB) + P(HIF)
While we have focused on the crosstalk of NFκB and HIF in the promoter regulation of the pro-inflammatory protein COX-2, this crosstalk would probably occur in the regulation of other genes containing both NFκB and HIF response elements in their promoter, including anti-inflammatory genes. For example, the anti-inflammatory protein netrin-1 was shown to be regulated by both NFκB [42] and HIF [43]. We speculate that this duality in the transcriptional crosstalk for regulating both pro- and anti-inflammatory genes might be dependent on other factors in order to resolve inflammation.
In summary, the findings of this study have revealed that the Gaussia luciferase reporter system can be a useful tool in probing the transcriptional dynamics of NFκB and HIF; HIF protein stabilisation from hypoxia or chemical inhibitors elicit distinct transcriptional responses; NFκB transcriptional activity is dependent on the stimulus and can be modulated by hypoxia; and HIF and NFκB act synergistically on the COX-2 promoter under dual hypoxia and cytokine stimulation. This interaction between hypoxia and inflammation underscores the complex crosstalk between the HIF and the NFκB signalling pathways.
Acknowledgments
We thank Drs. Catrióna Johnston and Jens Rauch (Systems Biology Ireland) for discussion. This work was supported by Science Foundation Ireland (Grant No. 06/CE/B1129).
Conflict of interest
None.
Abbreviations
- COX-2
Cyclooxygenase2
- DMOG
Dimethyl-oxaloylglycine
- DMSO
Dimethyl-sulfoxide
- Gluc
Gaussia Luciferase
- HIF
Hypoxia inducible factor
- HRE
Hypoxia response element
- NFκB
Nuclear factor kappa B
- NRE
NFκB response element
- pGluc
Plasmid encoding Gluc
- PHD
Prolyl hydroxylase
Footnotes
U. Bruning, S. F. Fitzpatrick, C. T. Taylor and A. Cheong contributed equally to this work.
References
- 1.Semenza GL. Regulation of physiological responses to continuous and intermittent hypoxia by hypoxia-inducible factor 1. Exp Physiol. 2006;91:803–806. doi: 10.1113/expphysiol.2006.033498. [DOI] [PubMed] [Google Scholar]
- 2.Semenza GL. Regulation of cancer cell metabolism by hypoxia-inducible factor 1. Semin Cancer Biol. 2009;19:12–16. doi: 10.1016/j.semcancer.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 3.Eltzschig HK, Carmeliet P. Hypoxia and inflammation. N Engl J Med. 2011;364:656–665. doi: 10.1056/NEJMra0910283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmedtje JF, Jr, Ji YS, Liu WL, DuBois RN, Runge MS. Hypoxia induces cyclooxygenase-2 via the NF-kappaB p65 transcription factor in human vascular endothelial cells. J Biol Chem. 1997;272:601–608. doi: 10.1074/jbc.272.1.601. [DOI] [PubMed] [Google Scholar]
- 5.Cummins EP, Berra E, Comerford KM, Ginouves A, Fitzgerald KT, et al. Prolyl hydroxylase-1 negatively regulates IkappaB kinase-beta, giving insight into hypoxia-induced NFkappaB activity. Proc Natl Acad Sci USA. 2006;103:18154–18159. doi: 10.1073/pnas.0602235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Culver C, Sundqvist A, Mudie S, Melvin A, Xirodimas D, et al. Mechanism of hypoxia-induced NF-kappaB. Mol Cell Biol. 2010;30:4901–4921. doi: 10.1128/MCB.00409-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koong AC, Chen EY, Giaccia AJ. Hypoxia causes the activation of nuclear factor kappa B through the phosphorylation of I kappa B alpha on tyrosine residues. Cancer Res. 1994;54:1425–1430. [PubMed] [Google Scholar]
- 8.Fitzpatrick SF, Tambuwala MM, Bruning U, Schaible B, Scholz CC, et al. An intact canonical NF-kappaB pathway is required for inflammatory gene expression in response to hypoxia. J Immunol. 2011;186:1091–1096. doi: 10.4049/jimmunol.1002256. [DOI] [PubMed] [Google Scholar]
- 9.Eliasson P, Jonsson JI. The hematopoietic stem cell niche: low in oxygen but a nice place to be. J Cell Physiol. 2010;222:17–22. doi: 10.1002/jcp.21908. [DOI] [PubMed] [Google Scholar]
- 10.Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 11.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 12.Hoffmann A, Natoli G, Ghosh G. Transcriptional regulation via the NF-kappaB signaling module. Oncogene. 2006;25:6706–6716. doi: 10.1038/sj.onc.1209933. [DOI] [PubMed] [Google Scholar]
- 13.Simakajornboon N, Gozal E, Gozal D. Developmental patterns of NF-kappaB activation during acute hypoxia in the caudal brainstem of the rat. Brain Res Dev Brain Res. 2001;127:175–183. doi: 10.1016/S0165-3806(01)00132-8. [DOI] [PubMed] [Google Scholar]
- 14.Cockman ME, Lancaster DE, Stolze IP, Hewitson KS, McDonough MA, et al. Posttranslational hydroxylation of ankyrin repeats in IkappaB proteins by the hypoxia-inducible factor (HIF) asparaginyl hydroxylase, factor inhibiting HIF (FIH) Proc Natl Acad Sci USA. 2006;103:14767–14772. doi: 10.1073/pnas.0606877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belaiba RS, Bonello S, Zahringer C, Schmidt S, Hess J, et al. Hypoxia up-regulates hypoxia-inducible factor-1alpha transcription by involving phosphatidylinositol 3-kinase and nuclear factor kappaB in pulmonary artery smooth muscle cells. Mol Biol Cell. 2007;18:4691–4697. doi: 10.1091/mbc.E07-04-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonello S, Zahringer C, BelAiba RS, Djordjevic T, Hess J, et al. Reactive oxygen species activate the HIF-1alpha promoter via a functional NFkappaB site. Arterioscler Thromb Vasc Biol. 2007;27:755–761. doi: 10.1161/01.ATV.0000258979.92828.bc. [DOI] [PubMed] [Google Scholar]
- 17.van Uden P, Kenneth NS, Rocha S. Regulation of hypoxia-inducible factor-1alpha by NF-kappaB. Biochem J. 2008;412:477–484. doi: 10.1042/BJ20080476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rius J, Guma M, Schachtrup C, Akassoglou K, Zinkernagel AS, et al. NF-kappaB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1alpha. Nature. 2008;453:807–811. doi: 10.1038/nature06905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bracken CP, Whitelaw ML, Peet DJ. Activity of hypoxia-inducible factor 2alpha is regulated by association with the NF-kappaB essential modulator. J Biol Chem. 2005;280:14240–14251. doi: 10.1074/jbc.M409987200. [DOI] [PubMed] [Google Scholar]
- 20.Walmsley SR, Print C, Farahi N, Peyssonnaux C, Johnson RS, et al. Hypoxia-induced neutrophil survival is mediated by HIF-1alpha-dependent NF-kappaB activity. J Exp Med. 2005;201:105–115. doi: 10.1084/jem.20040624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol Ther. 2005;11:435–443. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 22.Ruecker O, Zillner K, Groebner-Ferreira R, Heitzer M. Gaussia-luciferase as a sensitive reporter gene for monitoring promoter activity in the nucleus of the green alga Chlamydomonas reinhardtii . Mol Genet Genomics. 2008;280:153–162. doi: 10.1007/s00438-008-0352-3. [DOI] [PubMed] [Google Scholar]
- 23.Chung E, Yamashita H, Au P, Tannous BA, Fukumura D, et al. Secreted Gaussia luciferase as a biomarker for monitoring tumor progression and treatment response of systemic metastases. PLoS One. 2009;4:e8316. doi: 10.1371/journal.pone.0008316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Badr CE, Niers JM, Morse D, Koelen JA, Vandertop P, et al. Suicidal gene therapy in an NF-kappaB-controlled tumor environment as monitored by a secreted blood reporter. Gene Ther. 2011;18:445–451. doi: 10.1038/gt.2010.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Badr CE, Niers JM, Tjon-Kon-Fat LA, Noske DP, Wurdinger T, et al. Real-time monitoring of nuclear factor kappaB activity in cultured cells and in animal models. Mol Imaging. 2009;8:278–290. [PMC free article] [PubMed] [Google Scholar]
- 26.Bruning U, Cerone L, Neufeld Z, Fitzpatrick SF, Cheong A, et al. MicroRNA-155 promotes resolution of hypoxia-inducible factor 1alpha activity during prolonged hypoxia. Mol Cell Biol. 2011;31:4087–4096. doi: 10.1128/MCB.01276-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Appleby SB, Ristimaki A, Neilson K, Narko K, Hla T. Structure of the human cyclo-oxygenase-2 gene. Biochem J. 1994;302:723–727. doi: 10.1042/bj3020723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaidi A, Qualtrough D, Williams AC, Paraskeva C. Direct transcriptional up-regulation of cyclooxygenase-2 by hypoxia-inducible factor (HIF)-1 promotes colorectal tumor cell survival and enhances HIF-1 transcriptional activity during hypoxia. Cancer Res. 2006;66:6683–6691. doi: 10.1158/0008-5472.CAN-06-0425. [DOI] [PubMed] [Google Scholar]
- 29.Higgins CF. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 30.Cummins EP, Oliver KM, Lenihan CR, Fitzpatrick SF, Bruning U, et al. NF-kappaB links CO2 sensing to innate immunity and inflammation in mammalian cells. J Immunol. 2010;185:4439–4445. doi: 10.4049/jimmunol.1000701. [DOI] [PubMed] [Google Scholar]
- 31.Agbor TA, Cheong A, Comerford KM, Scholz CC, Bruning U, et al. Small ubiquitin-related modifier (SUMO)-1 promotes glycolysis in hypoxia. J Biol Chem. 2011;286:4718–4726. doi: 10.1074/jbc.M110.115931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bintu L, Buchler NE, Garcia HG, Gerland U, Hwa T, et al. Transcriptional regulation by the numbers: models. Curr Opin Genet Dev. 2005;15:116–124. doi: 10.1016/j.gde.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bintu L, Buchler NE, Garcia HG, Gerland U, Hwa T, et al. Transcriptional regulation by the numbers: applications. Curr Opin Genet Dev. 2005;15:125–135. doi: 10.1016/j.gde.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shea MA, Ackers GK. The OR control system of bacteriophage lambda. A physical-chemical model for gene regulation. J Mol Biol. 1985;181:211–230. doi: 10.1016/0022-2836(85)90086-5. [DOI] [PubMed] [Google Scholar]
- 35.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 36.Koong AC, Chen EY, Lee AS, Brown JM, Giaccia AJ. Increased cytotoxicity of chronic hypoxic cells by molecular inhibition of GRP78 induction. Int J Radiat Oncol Biol Phys. 1994;28:661–666. doi: 10.1016/0360-3016(94)90191-0. [DOI] [PubMed] [Google Scholar]
- 37.Pouyssegur J, Dayan F, Mazure NM. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature. 2006;441:437–443. doi: 10.1038/nature04871. [DOI] [PubMed] [Google Scholar]
- 38.Koong AC, Chen EY, Kim CY, Giaccia AJ. Activators of protein kinase C selectively mediate cellular cytotoxicity to hypoxic cells and not aerobic cells. Int J Radiat Oncol Biol Phys. 1994;29:259–265. doi: 10.1016/0360-3016(94)90272-0. [DOI] [PubMed] [Google Scholar]
- 39.Semenza GL. Regulation of vascularization by hypoxia-inducible factor 1. Ann NY Acad Sci. 2009;1177:2–8. doi: 10.1111/j.1749-6632.2009.05032.x. [DOI] [PubMed] [Google Scholar]
- 40.Nelson DE, Ihekwaba AE, Elliott M, Johnson JR, Gibney CA, et al. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science. 2004;306:704–708. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- 41.Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92:5–8. doi: 10.1016/S0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- 42.Paradisi A, Maisse C, Bernet A, Coissieux MM, Maccarrone M, et al. NF-kappaB regulates netrin-1 expression and affects the conditional tumor suppressive activity of the netrin-1 receptors. Gastroenterology. 2008;135:1248–1257. doi: 10.1053/j.gastro.2008.06.080. [DOI] [PubMed] [Google Scholar]
- 43.Rosenberger P, Schwab JM, Mirakaj V, Masekowsky E, Mager A, et al. Hypoxia-inducible factor-dependent induction of netrin-1 dampens inflammation caused by hypoxia. Nature Immunol. 2009;10:195–202. doi: 10.1038/ni.1683. [DOI] [PubMed] [Google Scholar]