Abstract
Neural stem (NS) cells are a self-renewing population of symmetrically dividing multipotent radial glia-like stem cells, characterized by homogeneous expansion in monolayer. Here we report that fetal NS cells isolated from different regions of the developing mouse nervous system behave in a similar manner with respect to self-renewal and neuropotency, but exhibit distinct positional identities. For example, NS cells from the neocortex maintain the expression of anterior transcription factors, including Otx2 and Foxg1, while Hoxb4 and Hoxb9 are uniquely found in spinal cord-derived NS cells. This molecular signature was stable for over 20 passages and was strictly linked to the developmental stage of the donor, because only NS cells derived from E14.5 cortex, and not those derived from E12.5 cortex, carried a consistent transcription factor profile. We also showed that traits of this positional code are maintained during neuronal differentiation, leading to the generation of electrophysiologically active neurons, even if they do not acquire a complete neurochemical identity.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-010-0548-7) contains supplementary material, which is available to authorized users.
Keywords: Neural stem cells, Neural development, Positional identity, Neuronal differentiation, Transcription factors
Introduction
Neural induction is the initial step in the generation of the nervous system in vertebrates. Soon after, neural cells acquire distinct identities and different fates depending on their positions along the rostrocaudal (R-C) and dorsoventral (D-V) axes of the neural tube. Patterning thus largely depends on the ability of the cells to acquire specific positional information and then a consistent molecular identity [1].
Early during central nervous system (CNS) development, morphologically distinct structures appear along the R-C axis that constitute the prosencephalon, mesencephalon, rhombencephalon, and spinal cord. A unique combination of transcription factors is expressed over time in each progenitor domain, specifying distinct CNS regions [2]. Hox genes are particularly important in establishing compartment identity within the rhombencephalon and spinal cord [3]. More rostrally, Otx2 and Gbx2 orchestrate the prosencephalon and mesencephalon/metencephalon territories [4], while Foxg1, Pax6, Emx1 and Emx2 are involved in telencephalon specification [5]. D-V patterning of the neural tube provides a further element of anatomical specification of the CNS. For instance, in the spinal cord, Pax7 and Pax6 are expressed dorsally, whereas Nkx2-2 and Nkx6-1 occur ventrally [6]. On the other hand, the telencephalon can be subdivided into ventral, lateral, and dorsal compartments on the basis of the expression of Pax6 (dorsal), Gsx2 (ventral, in particular in the medial and lateral ganglionic eminences, MGE/LGE) and Nkx2-1 (MGE) [7].
The recent generation of protocols for extracting and propagating stem/progenitor cells from the developing nervous system has allowed establishment of a controlled biological system suitable for molecular and cellular investigations. Radial glia-like neural stem (NS) cells represent one such system because they are a self-renewing population of tripotent neural stem cells, with properties of neurogenic radial glia [8–10]. NS cells can be clonally derived and undergo long-term and homogeneous expansion in adherent culture [8]. They can be derived from embryonic stem cells (ESCs), as well as from fetal and adult mouse brain, and remain highly neurogenic even after long-term expansion in vitro (>100 passages). Interestingly, NS cells derived from the adult subventricular zone (SVZ), exposed to an optimized differentiation protocol, consistently and reproducibly differentiate into mature GABAergic neurons, most of which can fire action potentials and express functionally active neurotransmitter receptors [11]. Furthermore, ESC-derived NS cells, subjected to a newly developed neuronal differentiation protocol, exhibit an 85% yield of molecularly and electrophysiologically mature neurons in the presence of limited cell death [22]. NS cells can also be efficiently derived from induced pluripotent stem cells obtained from the somatic reprogramming of fibroblasts [12] or NS cells [13]. More primitive and multipotent NS cells (called long-term neuroepithelial stem cells) have also been derived from human ESCs [14] and induced pluripotent stem cells (Falk et al., unpublished work).
Little is known about the positional identity of in vitro expanded neural stem cells isolated from the developing brain. For example, bona fide neural stem cells grown as neurospheres (heterogeneous aggregates of neural progenitors, mixed with neuronal precursors and astrocytes) show region-specific differences, such as Emx1 expression in cortex-derived neurospheres, Dlx2 in MGE neurospheres, and Hoxb1 in neurospheres derived from the hindbrain [15]. However, this gene expression profile has been confirmed only in the very initial phases of cell propagation, i.e. after one or two passages, as their prolonged culture (six passages) in vitro impairs the expression of some regional markers [15]. Accordingly, others have found that in vivo positional identity gene expression code is not preserved in neural stem cells grown in neurospheres, as shown by Foxg1 expression in neurospheres derived from posterior regions, and lack of expression of Hox genes in derivatives of spinal cord [16]. On the other hand, Zappone et al. [17] demonstrated the expression of Foxg1 (Bf1), Otx1, Tbr2 and Emx2 in passage 12 of cortex-derived neurospheres, but not in neurospheres derived from spinal cord. Moreover, only one study has investigated the molecular code found in neurospheres derived from adult SVZ versus different embryonic regions, finding a general downregulation of regional transcription factors in neurospheres, mainly concerning the D-V axis [18].
In this study, we characterized the molecular and biological properties of a number of fetal mouse CNS-derived NS cell lines isolated from different brain regions and grown in monolayer and serum-free medium according to the method of Conti et al. [8]. Importantly, we demonstrated that NS cells, isolated from different germinal zone regions along the R-C and D-V axes of the developing CNS, display region-specific positional identities that mainly concern the R-C identity. We also showed that different NS cell lines maintain a region-specific pattern of transcription factors that is stable for over 20 passages in culture. On the other hand, the D-V identity is affected by in vitro culture conditions, thus confirming FGF-2 ventralizing activity [18–20]. Furthermore, we provide evidence that the donor developmental stage is critical for the derivation of committed NS cells that maintain an in vitro positional identity consistent with that of the region of derivation. We also showed that fetus-derived NS cells promptly differentiate into mature, electrophysiologically functional neurons that express a set of cell determinants consistent with their region of derivation, even if they do not acquire the full molecular spectrum of markers that characterize a complete and specific neurochemical identity.
Materials and methods
NS cell derivation and culture
Fetus-derived NS cells were generated from different areas of developing CNS of “green mice” [21] in which eGFP is under the control of chicken β-actin and CMV promoter. In particular, NS cells were generated from five germinative regions: NS12CB, from prospective cerebellum; NS12CX and NS14CX from E12.5 and E14.5 developing neocortex, respectively; NS12Mes, from ventral mesencephalon; NS12SC, from developing spinal cord; and NS12ST, from ganglionic eminences (the striatum anlage). The standard dissection procedure was as follows: carefully dissected areas of interest from E12.5 and E14.5 mouse embryo were placed into a 1.5-ml Eppendorf tube and held in ice-cold PBS. Tissue pieces were spun at 800 rpm for 3 min, followed by removal of the supernatant and placement of the tube on ice for 10 min. Every 2–3 min, the bottom of the tube was “flicked” to encourage tissue dissociation. By using tissue at these stages, any enzymatic dissociation could be avoided simply by leaving the tissue on ice. Tissue fragments were delicately resuspended in 1 ml PBS. The cell suspension was centrifuged in a total volume of 1.5 ml PBS at 1,000 rpm for 3 min. Once the supernatant had been removed, the pellet was resuspended in NS medium to 5 × 105 cells/ml. Cells were then plated onto a laminin-coated (3 μg/ml laminin in cold PBS) six-well plastic tissue culture plate (IWAKI, Bibby Scientific) at a density of 1 × 106 cells per well and grown for 1–2 days at 37°C. The day after, cells adhering to the plastic and others still in suspension were visible. The medium was changed, with retention of the adherent cells. Cells in suspension were resuspended and spun at 1,000 rpm for 3 min. The pellet was resuspended in NS medium and plated again onto a laminin-coated (3 μg/ml laminin in cold PBS) six-well plastic tissue culture plate (IWAKI) at a density of 1 × 106 cells per well. After 3–4 days, growing cells were dissociated into single cells by treatment with Accutase solution diluted 1:2 in PBS, replated, and propagated in NS medium onto laminin-coated plastic (2 μg/ml laminin in cold PBS).
NS 46C CAG and NS R1 cell lines were generated as previously described by Conti et al. [8] and by Spiliotopoulos et al. [22] from ES 46C CAG and ES R1 cell lines, respectively. Cells were plated onto an uncoated T-25 flask (IWAKI) in Euromed-N medium (Euroclone, Milan, Italy), supplemented with N2 and 20 ng/ml of both EGF (Peprotech, Tebu-Bio) and FGF-2 (Peprotech, Tebu-bio), the standard NS cell proliferating medium.
The adult SVZ-derived NS cell line aNS-1 was derived from the SVZ of a 2-month-old wild-type (CD1 strain) mouse, as described by Goffredo et al. [11]. For routine passaging, all NS cells were dissociated with Accutase (Sigma) and split 1:4 or 1:5 every 2–3 days. All the described NS cell lines can be requested from the cell repository at Biorep S.r.l. (http://www.biorep.it/eng/).
Neuronal differentiation of fetus-derived NS12ST and NS12SC
NS12ST and NS12SC were differentiated according to an optimized protocol in serum-free conditions. The neuronal differentiation procedure is one step, based on six progressive changes of medium. Differentiating conditions require Euromed-N (M1–M4 medium), Neurobasal/DMEM/F12 in specific ratios (M4*), and defined supplements. Briefly, NS12ST and NS12SC cells were collected following Accutase treatment and then plated (plating density 6 × 104–7 × 104 cells/cm2) on a laminin-coated (Gibco, Invitrogen) T-25 flask or 35-mm dishes (Greiner Bio-One) in Euromed-N medium, supplemented with N2 and 20 ng/ml of both EGF and FGF-2. After 12 h, the proliferation medium was removed and the cells were incubated in a predifferentiation medium, M1 consisting of Euromed-N medium containing 0.5% N2 (Invitrogen) and 1% B27 supplements (Invitrogen) and supplemented with 10 ng/ml of FGF-2. After 3 days, the cultures were shifted to M2 medium composed of Euromed-N containing 0.5% N2, 1% B27 supplements, 5 ng/ml FGF-2, and 20 ng/ml of BDNF (Peprotech, Tebu-Bio). After 3 days of culture in M2 medium, the cells were exposed for another 3 days to M3 medium, identical to M2 except for augmentation of BDNF from 20 to 30 ng/ml. Subsequently, the fetal NS cells were shifted to a new medium, M4, identical to M3 except for a decrease in FGF-2 from 5 to 3 ng/ml and incubated for another 3 days for differentiation. For terminal differentiation, the cells were incubated in M4* medium composed of a 3:1 mixture of Neurobasal and DMEM/F12 medium containing 0.5% N2 and 1% B27 supplements and containing 3 ng/ml FGF-2 and 30 ng/ml BDNF. The cells were maintained under these conditions for an additional 7–15 days, and the M4* medium was partially changed every 3 days.
Immunofluorescence
Cell cultures were fixed with 4% paraformaldehyde for 15 min at room temperature. For labelling intracellular antigens, cells were permeabilized in PBS containing 0.5% Triton X-100. After washing in PBS, cells were blocked with 5% fetal calf serum in PBS for 1 h and incubated overnight in 3% fetal calf serum in PBS with the following primary antibodies: nestin (1:200, Becton Dickinson), SOX2 (1:100, Chemicon), vimentin (1:100, Developmental Studies Hybridoma Bank), PAX6 (1:100, MBL Eppendorf), βIII-tubulin (1:1000, Promega), MAP2ab (1:500, Becton Dickinson), GFAP (1:800, Dako), O4 (1:50, R&D Systems), synapsin (1:200, Synaptic System), GABA (1:500, Sigma), RC2 (1:50, DSHB), and GAP-43 (1:500, Chemicon). After two washes in PBS, appropriate secondary antibodies conjugated to Alexa fluorophores 350 or 568 (Molecular Probes, Invitrogen) were diluted at 1:500 in blocking solution and mixed with Hoechst 33258 (5 μg/ml; Molecular Probes, Invitrogen) to counterstain the nuclei. The cells were kept in the blocking solution for 1 h at room temperature and were then washed twice in PBS buffer. Images were acquired with a Leica DMI 6000B microscope (×20 objective) and LAS-AF imaging software and then processed using Adobe Photoshop.
RT-PCR analysis
Total RNA was extracted from NS cells with Trizol reagent (Invitrogen) according to the manufacturer’s recommendations. Before reverse transcription, RNA was treated with DNaseI (Qiagen) and purified using the RNeasy kit (Qiagen). The cDNA was generated with Superscript III reverse transcriptase (Invitrogen) according to the manufacturer’s recommendations. The cDNA was produced from 1 µg total RNA. RT-PCR was performed for 30 cycles for all markers, except Hb9 (35 cycles), as follows: denaturing for 45 s at 94°C; annealing for 30 s; and extension at 72°C. PCR products were resolved on a 2% agarose gel. Primer sequences, annealing temperatures and product sizes are listed in Table 1.
Table 1.
RT-PCR primer sequences, related conditions and amplicons
| Gene | Forward primer | Reverse primer | Annealing temperature (°C) | Product size (bp) |
|---|---|---|---|---|
| Arpp21 | ATGTCTGAGCAAGGAGGACTG | TCTCTTGGTTCTGAGCCGC | 58 | 154 |
| Ascl1 | CGTCCTCTCCGGAACTGAT | CGTCCTCTCCGGAACTGAT | 56 | 482 |
| Ascl1 (RT-qPCR) | CACGAGAAGAGTGACTGGTGT | GGCTAAGACAGCTCTGACAGA | 58 | 194 |
| β-Actin | GGCCCAGAGCAAGAGAGGTATCC | ACGCACGATTTCCCTCTCAGC | 58 | 460 |
| β-Actin (RT-qPCR) | AGTGTGACGTTGACATCCGTA | GCCAGAGCAGTAATCTCCTTCT | 60 | 112 |
| Blbp | GGGTAAGACCCGAGTTCCTC | ATCACCACTTTGCCACCTTC | 56 | 213 |
| Chrm4 | TCCTCACCTGGACACCCTAC | ACGTAGCAGAGCCAGTAGCC | 60 | 103 |
| Dlx5 | AGCCCCTACCACCAGTACG | TGTGTTTGCGTCAGTCCTAGAGAG | 58 | 245 |
| Dlx6 | TCAATACCTGGCCCTTCCCG | GGTGTCCTGGTGTGGTGAG | 58 | 292 |
| Drd1 | GAGCGTAGTCTCCCAGATCG | GGATGCTGCCTCTTCTTCTG | 58 | 104 |
| Drd2 | CAAGCGTAGCAGCCGAGCTT | CATTCTCCGCCTGTTCACTG | 60 | 136 |
| Emx2 | ACGCTTTTGAGAAGAACCATTACGT | GTCTGAGGTCACATCTATTTCCTCC | 60 | 242 |
| Foxg1 | CTGAGTGTGGACCGGCTG | CGTGCTGGTCTGCGAAGTC | 58 | 213 |
| Foxp1 | TGACAAGCAACCAGCTCTTCAG | ATGTTGCTGTTGGAGAAGTTGAAGC | 58 | 226 |
| Foxp2 | TCACTTATGCAACACTCATAAGGCAG | CGCTTCTGGTATTCGACTTCGTC | 58 | 243 |
| Gabbr1 | AGGTTGATCACTCGAGGTGAA | CATCTGTTTCCCAGAGTACTGA | 56 | 476 |
| Gfap | GCTCCAAGATGAAACCAACCTGAG | CTTAAGACTGGATCTCCTCCTCCA | 56 | 554 |
| Glast | CCAAAAGCAACGGAGAAGAG | ACCTCCCGGTAGCTCATTTT | 56 | 229 |
| Glra1 | GAAGCCTGACTTGTTCTTTGCC | GTTGCATGATACACGTCTGTAC | 58 | 185 |
| Glra2 | CGTACAGTGAGTACCCAGATG | CAACTTGCACTGGACCATCAC | 58 | 315 |
| Gsx2 | CACTACCTACAACATGTCGGACC | CCTTCTTGTGCTTCACGCGAC | 58 | 254 |
| Hb9 | TCGAACCTCTTGGGGAAGTG | AATCTTCACCTGAGTCTCGGT | 61 | 159 |
| Hoxa4 | TGTGCCCTACTCATCTCCTG | GCTGTCTGGCTGACTCAAAG | 53 | 260 |
| Hoxb4 | TGCAAAGAGCCCGTCGTC | GGAACCAGATCTTGATCTGGCG | 60 | 185 |
| Hoxb9 | TCCATTTCTGGGACGCTTAG | CCCTCCTGAGGTACATATTG | 60 | 230 |
| Irx3 | GACGAGGAGGGCAATGCTTAT | AAATGGGTCCCAGGCCGAAGT | 60 | 259 |
| Islet-1 | CGTCTGATTTCCCTGTGTGTTGG | AAGTCGTTCTTGCTGAAGCCTATG | 58 | 239 |
| Lim3 | ACAGCGACGCCGAAGTCT | ACTGCTCTGGACCCGTCAAG | 60 | 168 |
| Meis2 | GAGACGTCTGTTCCTCTGACTC | TCTCATCAATCACGAGGTCAATGG | 58 | 243 |
| Nkx2-1 | ACCGGGTTCAGACTCAGTTC | ATCGACATGATTCGGCGTCGG | 60 | 221 |
| Nkx6-1 | TACTTGGCAGGACCAGAGAG | CGCTGGATTTGTGCTTTTTC | 58 | 270 |
| NMDAR1 | TTGGCCGATTTAAGGTGAAC | AAAACCACATGGCAGAGGAC | 58 | 72 |
| Olig2 | GGCGGTGGCTTCAAGTCATC | TAGTTTCGCGCCAGCAGCAG | 60 | 260 |
| Otx1 | GGCGCTGTTCGCAAAGACTC | AGTGGCTCTGGCACCGATAC | 60 | 429 |
| Otx1 (RT-qPCR) | GTCCAGAGTCCAGGTTTGGTTC | GACGAAGAAGCAGAGCTAGATACG | 58 | 180 |
| Otx2 | TACCTCAGTCCCAACCATTG | TCTGAGAGCATCGTTCCATC | 58 | 572 |
| Pax6 | AGTCACAGCGGAGTGAATCAGC | AGCCAGGTTGCGAAGAACTCTG | 58 | 426 |
| Pax7 | GTCACTAAGCATGGGTAGATG | GCTACCAGTACAGCCAGTATG | 56 | 328 |
| Pax7 (RT-qPCR) | CATGAACCCTGTCAGCAATGG | CTGTAGCCAGTGGTGCTGTA | 58 | 230 |
| Rarβ1 | CTTCTGTGTGATTCTGGACCTGTAG | CCACTGGGAGGTGGGTG | 58 | 219 |
| Shh | TCTGTGATGAACCAGTGGCC | GCCACGGAGTTCTCTGCTTT | 60 | 242 |
| Six3 | CAGCAGAGTCACCGTCCAC | TGGAGGTTACCGAGAGGATCG | 58 | 100 |
| Sox2 endo | TAGAGCTAGACTCCGGGCGATGA | TTGCCTTAAACAAGACCACGAAA | 56 | 297 |
| Tlx | TGTGGTGACCGCAGCTC | TCGACACGCCCTGCATTG | 58 | 168 |
| Vglut2 | TCGGCTTCTGCATATCCTTC | CCTGGAATCTGGGTGATGAT | 56 | 190 |
Quantitative RT-PCR analysis
Quantitative RT-PCR (RT-qPCR) analyses were done in triplicate for the analysed gene. An iCycler thermal cycler with a multicolour teal-time PCR detection system (Bio-Rad) was used. All reactions were performed in a total volume of 25 μl containing 50 ng cDNA, 50 mM KCl, 20 mM Tris-HCl, pH 8.4, 0.2 mM dNTPs, 25 U/ml iTaq DNA polymerase, 3 mM MgCl2, SYBR Green I, 10 nM fluorescein, stabilizers (iQ SYBR Green Supermix; BioRad), and 0.2 μM forward and reverse primers. Amplification cycles consisted of an initial denaturing cycle at 95°C for 3 min, followed by 45 cycles of 30 s at 95°C, 30 s at 60°C, and 30 s at 72°C. Fluorescence was quantified during the 60°C annealing step, and product formation was confirmed by a melting curve analysis (55–94°C). Amounts of target gene mRNA were normalized to a reference gene, β-actin. Primer sequences, annealing temperature, and product sizes are listed in Table 1. Statistical analysis was performed by means of one-way ANOVA and the Tukey-Kramer multiple comparison test.
Whole-cell patch-clamp recordings
The electrophysiological experiments were performed using the patch-clamp technique in whole-cell configuration. Seals between the electrode and the cell were established in a bath solution containing 155 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 3 mM KCl and 10 mM HEPES/NaOH (pH 7.4). Patch pipettes were filled with 128 mM KCl, 10 mM NaCl, 11 mM EGTA and 10 mM HEPES/KOH (pH 7.4). Ligand-gated currents elicited following application of GABA (20 μM), dopamine (100 μM), glutamate (20 and 100 μM), glycine (100 and 300 μM), acetylcholine (30 μM), nicotine (50 μM) and carbachol (20 μM) were recorded under voltage-clamp (holding potential −80 mV) during application of neurotransmitters or their agonists to the cell using a perfusion system positioned near the recorded cell. Receptor currents were derived with the following external solution: 145 mM NaCl, 3 mM KCl, 1 mM MgCl2, 1 mM CaCl2, 5 mM TEA-Cl, 5 mM 4-AP and 10 mM HEPES/NaOH (pH 7.4). The patch pipette was filled with 145 mM CsCl, 1.1 mM EGTA, 2 mM MgCl2, 0.1 mM CaCl2, 20 mM TEA-Cl and 5 mM HEPES/CsOH (pH 7.3). Patch pipettes were made of borosilicate glass tubing (Hilgenberg, Malsfeld, Germany) and fire-polished to a final resistance of 3.0–5.0 MΩ when filled with internal solution. All of the experiments were performed at room temperature (22–24°C).
Whole-cell currents and potentials were recorded using an Axopatch 200B amplifier (Axon Instruments, Burlingame, CA), digitized at sampling intervals of 10–50 μs using a DigiData 1322A AD/DA converter (Axon Instruments). Stimulation, acquisition, and data analysis were carried out using pCLAMP (Axon Instruments) and ORIGIN software (Microcal Software, Northampton, MA). Fast capacitive transients were reduced online using analogic circuitry, and residual capacitive and leak currents were removed by P/4 subtraction. The currents were filtered at 5 kHz. All data are presented as the means ± SE of n observations.
Results
Fetus-derived NS cells have features of neurogenic radial glia
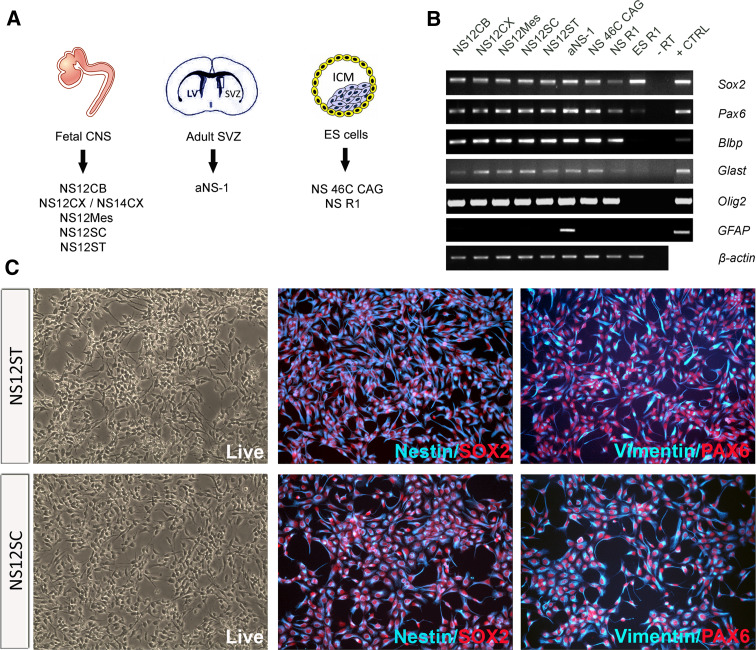
Mouse NS cell lines were generated from five germinative areas of the fetal CNS (NS12CB, from prospective cerebellum; NS12CX and NS14CX from E12.5 and E14.5 developing neocortex, respectively; NS12Mes, from ventral mesencephalon; NS12SC, from spinal cord; NS12ST, from striatum anlage). Fetal NS cells were compared to NS cells derived from the SVZ of adult brain (aNS-1) [9, 11] and to ESC-derived NS 46C CAG and NS R1 cell lines [8, 22] (Fig. 1a).
Fig. 1.
Phenotypic and genetic characterization of self-renewing mouse NS cells. a NS cell lines were generated from the germinative areas of the fetal CNS (CB cerebellum, CX neocortex, MES mesencephalon, SC spinal cord, ST striatum) at different embryonic stages (E12.5 and E14.5), from the SVZ (aNS-1) of the adult brain, and from ESCs (NS 46C CAG, NSR1). b RT-PCR analysis of a set of markers diagnostic of neural progenitors/radial glia. NS cells express Sox2, Pax6, Blbp, Glast and Olig2. GFAP expressed only in aNS-1 cells, according to the presence of GFAP in stem cells of the SVZ. Mouse fetal brain was used as positive control (+CTRL); −RT negative control of retrotranscription. c Self-renewing NS cells homogeneously show double immunoreactivity for nestin/vimentin and SOX2/PAX6
All NS cell lines tested underwent homogeneous expansion in fully defined adherent culture conditions in medium containing EGF and FGF-2. RT-PCR showed that they all expressed typical neural progenitor markers, including Sox2, Pax6, Blbp, Glast and Olig2, a set of markers considered diagnostic for NS cells with radial glia properties (Fig. 1b) [8]. As expected, the astrocytic marker GFAP was not expressed in the self-renewal condition in fetus- and ESC-derived NS cells (Fig. 1b), but was specifically present only in the aNS-1 line (Fig. 1b), a feature resembling GFAP expression in SVZ stem cells in vivo [23]. Interestingly, in the expansion phase, NS cell lines expressed Id1, Id2, and Id3, which act as passive repressors of proneural gene activity [24] (Fig. S1). Hes1 and Hes3, essential effectors of notch signalling which regulates the maintenance of undifferentiated cells [25], were expressed at high levels in all analysed NS cells lines (Fig. S1).
Three fetus-derived NS cell lines, NS12ST, NS12SC and NS14CX, were further analysed by immunofluorescence. All NS12ST and NS12SC cells were immunopositive for nestin/SOX2 and vimentin/PAX6 (Fig. 1c, S2), demonstrating that fetal NS cells consist of a homogeneous population of neural stem cells. The same homogeneous expression of neural stem cell-associated markers was found in clonal populations derived from NS12CX and NS12SC (Fig. S3).
Fetal NS cells show a conserved positional identity along the R-C axis
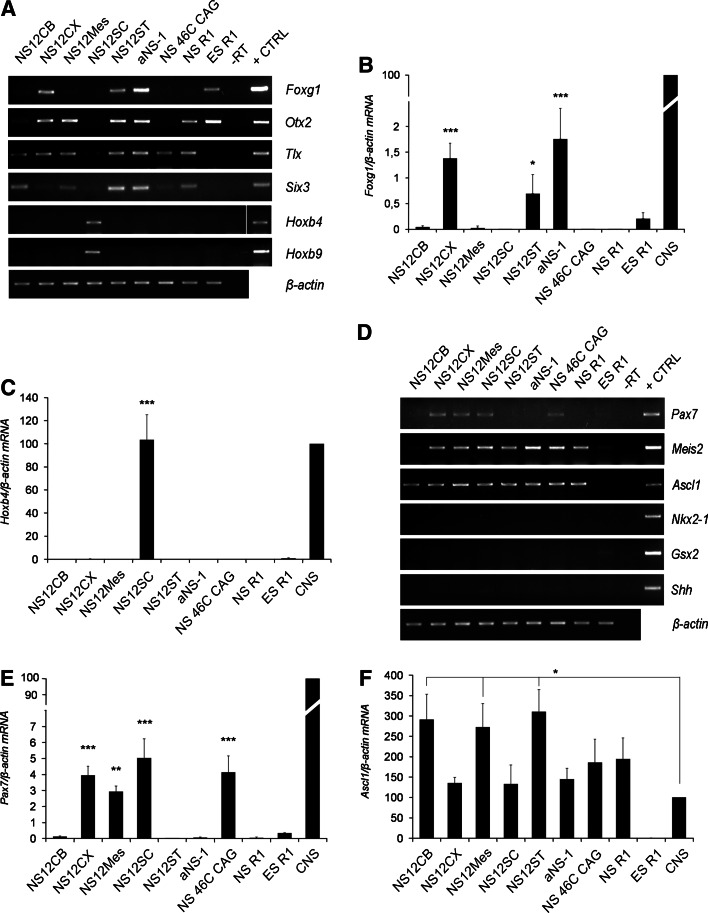
Fetal NS cells derived from the five histogenic compartments along the developing neuraxis were expanded for at least 20 passages. To determine whether these different NS cell lines could show a unique regional identity in culture corresponding to the region of origin, we investigated the expression of a subset of transcription factors that establish positional identity along the CNS R-C and D-V axes. As shown in Fig. 2a, of the several fetal NS cell lines, Foxg1, a winged helix transcriptional repressor specifically found in the telencephalon [26], was expressed only in telencephalon-derived NS cells, i.e. NS12CX and NS12ST. Foxg1 was also strongly expressed in SVZ-derived aNS-1 cells (Fig. 2a), consistent with their region of derivation. RT-qPCR further confirmed significant Foxg1 expression in NS12CX (p < 0.001), NS12ST (p < 0.05), and aNS-1 cells (p < 0.001) in comparison to the other fetal NS cell lines (Fig. 2b).
Fig. 2.
Analysis of the in vitro positional identity in NS cell lines. a RT-PCR analysis of a subset of R-C markers shows that fetus-derived NS cells possess a region-specific transcription factor pattern: Foxg1 is expressed in telencephalic NS lines, Otx2 and Six3 in anterior CNS-derived NS cells, and Hoxb4 and Hoxb9 in NS12SC cells. Mouse fetal brain was used as positive control (+CTRL); -RT was the negative control b, c RT-qPCR analysis showing the expression levels of two relevant R-C markers, Foxg1 (b) and Hoxb4 (c). Data are expressed as percentages (100% was assigned to fetal CNS-E13 brain and spinal cord). d RT-PCR analysis of some indicative D-V markers shows constitutive expression of important ventral markers, including Meis2 and Ascl1. Mouse fetal brain was used as positive control (+CTRL); -RT was the negative control. e, f RT-qPCR analysis showing the expression levels of two relevant D-V markers, Pax7 (e) and Ascl1 (f). Data are expressed as percentages (100% was assigned to fetal CNS). *p < 0.05, **p < 0.01, ***p < 0.001, as calculated by ANOVA using the Tukey-Kramer post-test
Similarly, the prosencephalic/mesencephalic homeobox gene, Otx2, was expressed in the anterior CNS-derived NS cells (NS12CX, NS12ST, NS12Mes and aNS-1) (Fig. 2a). Tlx, a forebrain marker [27], distinguishes NS12CX, NS12ST, NS12Mes and, to a lesser extent, NS12CB (Fig. 2a). Tlx expression was also found in aNS-1 cells (Fig. 2a), in agreement with its in vivo expression in adult SVZ [28]. Anterior CNS-derived NS cell lines and those from cerebellum anlage (i.e. NS12ST, NS12Mes, NS12CB and aNS-1) showed a strong Six3 expression, while a lower expression was detectable in NS12CX cells (Fig. 2a).
On the other hand, the hindbrain/spinal cord homeobox genes, Hoxb4 and Hoxb9, were exclusively expressed in spinal cord-derived NS12SC (Fig. 2a), at a level comparable to the in vivo expression in E13 spinal cord, as shown by RT-qPCR (p > 0.05; Fig. 2c). These results suggested that fetal NS cells retain in vitro their R-C transcriptional code according to the region of derivation, like the Otx2, Foxg1 and Tlx expression in forebrain-derived NS and Hox genes in spinal cord NS cells.
ESC-derived NS cells also showed a specific regional identity. For example, NS 46C CAG cells did not express anterior markers (except for Tlx) (Fig. 2a–c) but were positive for HoxA4 (13) (data not shown), thus demonstrating a posterior identity. On the other hand, NSR1 cells expressed Otx2, Tlx and Six3, but were negative for posterior markers (Fig. 2a–c). These discrepancies between the NS cells derived from different ESCs may be the result of either different exposure times to neural induction protocols or different capacities for responding to induction stimuli.
Partial deregulation of D-V patterning in NS cells
We next investigated a subset of D-V markers. Meis2 and Ascl1, which distinguish a ventral regional identity, were expressed in all NS cell lines (Meis2 was absent only in NS12CB) (Fig. 2d), demonstrating a constitutive ventral specification.
Ascl1 was strongly expressed in all NS cell lines, at similar or higher levels than in vivo expression (p < 0.05; Fig. 2f). More interestingly, Pax7, a dorsal marker of mesencephalon and spinal cord, was present in NS12Mes and NS12SC cells, demonstrating an expression pattern consistent with their region of derivation (p < 0.05 and p < 0.001 for NS12Mes and NS12SC, respectively, vs. NS12CB, NS12ST, aNS-1 and NSR1; Fig. 2d, e). Surprisingly, we noted Pax7 expression in NS12CX cells (Fig. 2d, e), a result that did not mirror the normal gene expression pattern found in the developing neocortex (see below). On the other hand, the ventral forebrain transcription factor-coding genes Nkx2-1 and Gsx2 were silent in all NS cell lines tested (Fig. 2d). These findings demonstrate that D-V patterning in NS cells is partially deregulated. All NS cells expressed ventral markers, even in the absence of endogenous Sonic hedgehog (Shh) expression (Fig. 2d; Calabrese et al., unpublished observations), probably because of the exposure to the ventralizing action of FGF-2 during culture [19].
Developmental stage relevance for derivation of committed NS cells and temporal stability of positional identity in culture
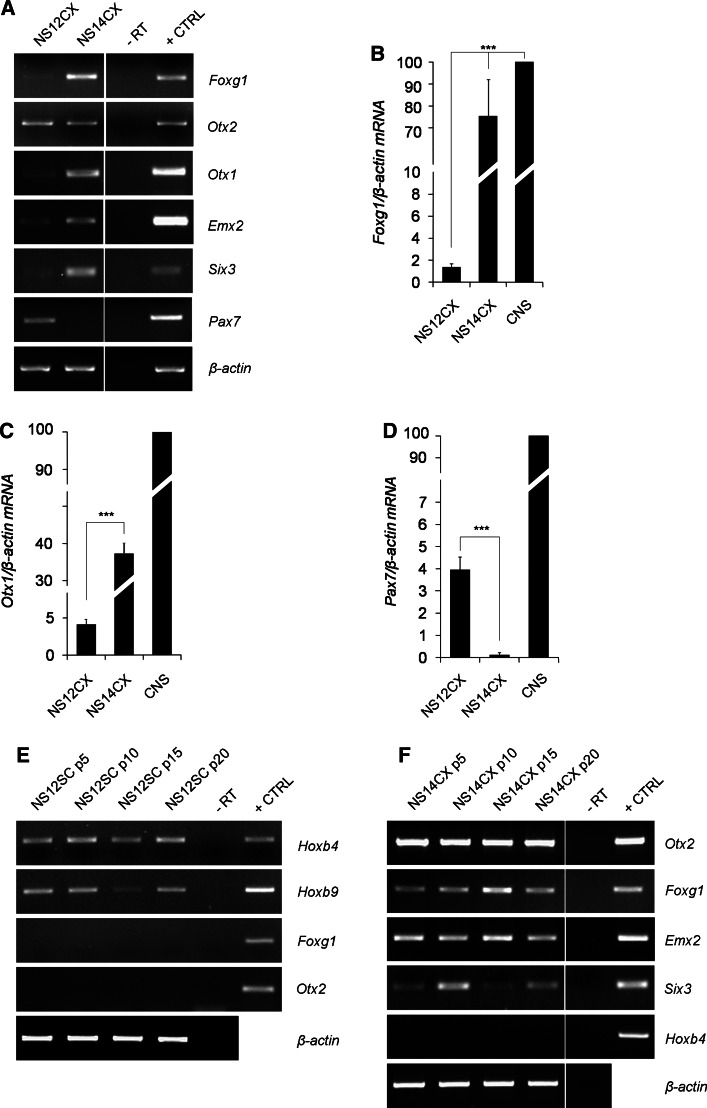
NS12CX cells are characterized by low expression of Six3 and the misexpression of Pax7. We reasoned that, in vivo, developmental timing orchestrates the specification and determination of the neural progenitors within the neuraxis, with a C-R polarity. At E12.5, spinal cord precursors are specified and start to differentiate, while cortical progenitors are still proliferating. We therefore tested whether the stage of derivation of NS cells could be important for establishing an NS cell line that consistently mirrors the gene expression hallmarks of the in vivo compartment of derivation. We compared the features of NS14CX cells (derived from E14.5 neocortex, at the peak of corticogenesis [29]) with those of NS12CX cells. RT-PCR showed that Foxg1, Six3 and Emx2 were more expressed in NS14CX cells (Fig. 3a).
Fig. 3.
Gene expression profiles of NS12CX and NS14CX and temporal stability of gene expression. a NS14CX exhibits high expression of Foxg1, Otx1, Six3 and Emx2 and no evidence of Pax7 expression, in contrast to NS12CX. These data suggest that NS14CX, derived from a later developmental stage, better characterized neocortical progenitor identity. b-d RT-qPCR analysis for Foxg1, Otx1 and Pax7 mRNA levels in NS cells. Data are expressed as percentages (100% was assigned to fetal CNS-E13 brain and spinal cord). e, f Stable expression of regional markers in NS12SC and NS14CX, considered every five passages. Mouse fetal brain was used as positive control (+CTRL); -RT was the negative control. ***p < 0.001, as calculated by ANOVA using the Tukey-Kramer post-test
Moreover, Otx1, a homeobox gene involved in corticogenesis, was also significantly upregulated in NS14CX cells (p < 0.001; Fig. 3a, c). RT-qPCR for Foxg1 showed a 55-fold increase in NS14CX cells compared to NS12CX cells (p < 0.001; Fig. 3b). Interestingly, Foxg1 expression level was comparable to the in vivo expression in E13 mouse fetal brain (p > 0.05; Fig. 3b). Conversely, NS14CX cells no longer expressed Pax7, in contrast to NS12CX cells (p < 0.001; Fig. 3a, d). These results indicate that NS14CX cells are more committed to a dorsal-telencephalic identity, demonstrating that the stage of derivation is important for establishing an NS cell line that can retain a specific regional molecular identity in vitro.
We next investigated whether the positional identity of NS cells is retained in vitro by analysing NS12SC and NS14CX cells every five passages for 20 passages. NS12SC cells showed stable expression of Hoxb4 and Hoxb9 for over 20 passages and did not express forebrain markers (Fig. 3e). Similarly, NS14CX cells stably expressed Otx2, Foxg1, Six3 and Emx2, while hindbrain/spinal cord markers, like Hoxb4, were never detectable (Fig. 3f).
Lineage-specific neurogenic potential of fetus-derived NS cells
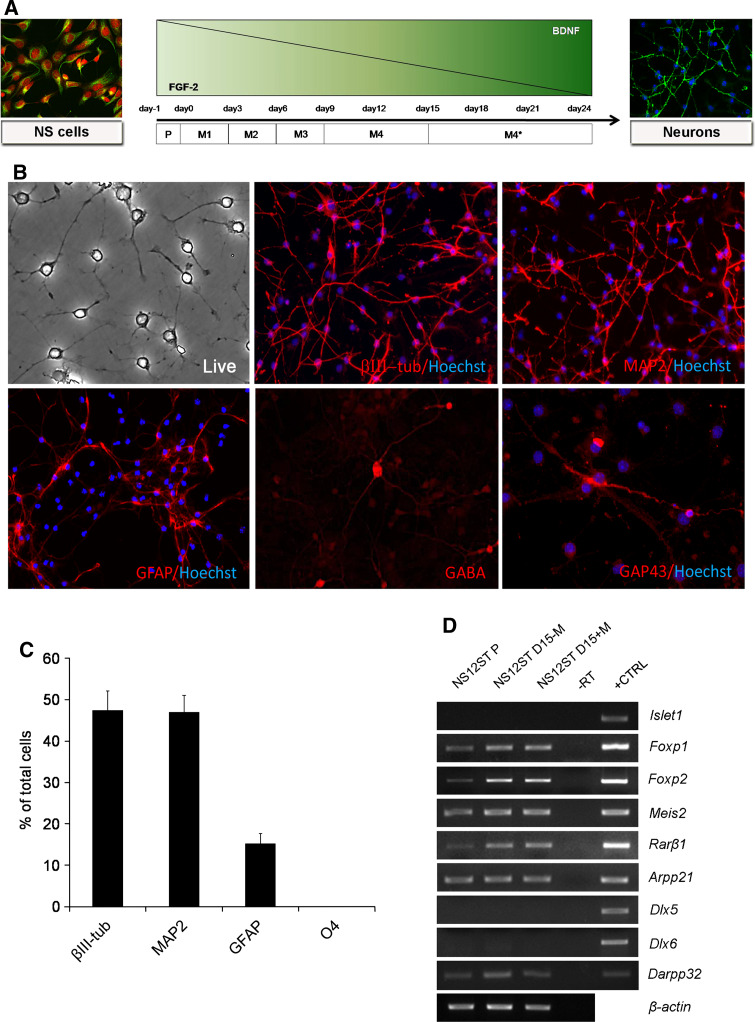
NS cells are multipotent and give rise to neurons, oligodendrocytes and astrocytes, the three primary lineages of the CNS [8]. To test the neural differentiation ability of fetal NS cells, we set up a new one-step protocol (Fig. 4a) because of the limited differentiation and reduced survival using the protocol developed for the ESC-derived NS cells [22] or aNS-1 [11] (not shown). The protocol involved EGF withdrawal and a gradual decrease in FGF-2, with a simultaneous increase in BDNF in the differentiation medium up to a final concentration of 30 ng/ml (Fig. 4a). After 3–5 days of differentiation, morphological maturation of the cells and acquisition of a neuronal phenotype were remarkable. With differentiation, the neuronal precursors began to express the neuronal markers βIII-tubulin and MAP2ab, and expression increased in the following days.
Fig. 4.
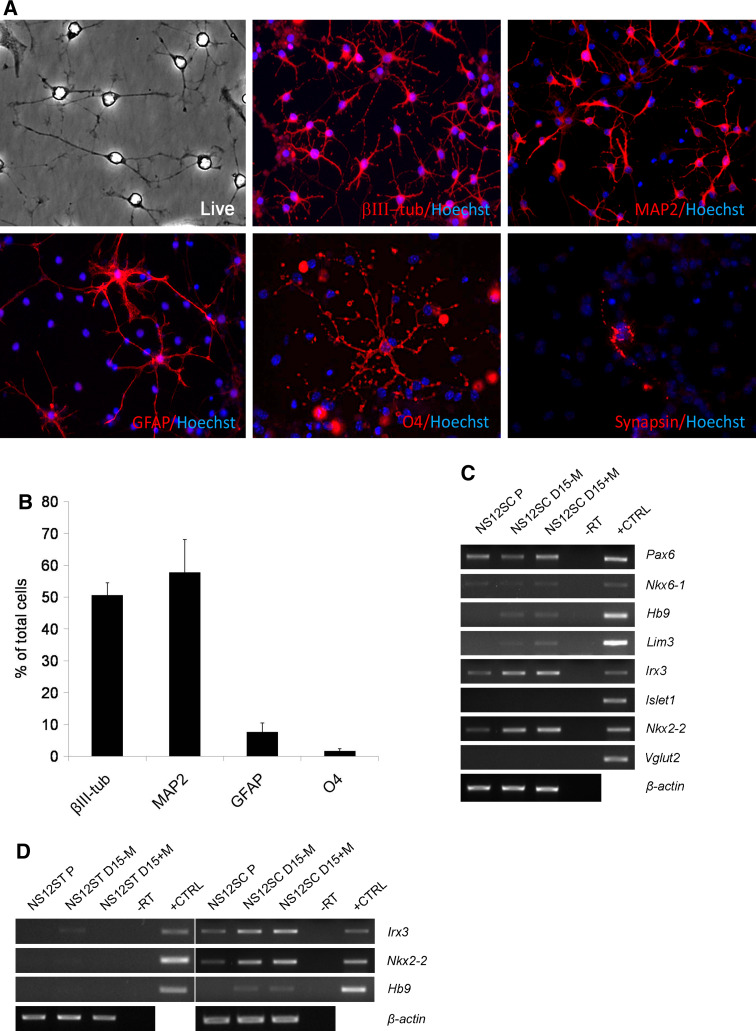
Neuronal differentiation of NS12ST cells. a Scheme showing the neural differentiation protocol applied to fetus-derived NS cells. Growth factors were withdrawn, while BDNF concentration was augmented. b After 23 days of differentiation in vitro, NS12ST cells gave rise to βIII-tubulin-, MAP2-, and GFAP-positive cells. GABA- and GAP-43-positive cells were present (×4, magnified after acquisition). c At the end of differentiation, 47.4 ± 4.6% of cells were immunopositive for βIII-tubulin, 47.0 ± 4.0% for MAP2, and 15.2 ± 2.4% for GFAP, while there were no O4-positive cells (n = 1,600 cells for βIII-tubulin; n = 1,163 cells for MAP2 and GFAP; n = 1,585 cells for O4) (columns represent averages, error bars standard deviations). d Gene expression analysis by RT-PCR on NS12ST cells in proliferation (P) and after 15 days of differentiation in the absence (D15-M) or in the presence (D15+M) of morphogens. Mouse fetal brain was used as positive control (+CTRL); -RT was the negative control
The NS12ST and NS12SC cell lines were subjected to more thorough investigations. After 23 days of differentiation, NS12ST cells exhibited thin, elongated processes typical of mature neurons (Fig. 4b) and gave rise to βIII-tubulin-, MAP2ab- and GFAP-positive cells (Fig. 4b). GABA-positive neurons were present at the end of differentiation (Fig. 4b), in accordance with the typical GABAergic phenotype of striatal neurons and confirming the tendency of NS cell to differentiate towards the GABAergic fate [11, 12, 22]. We noted the expression of GAP-43 in some neuronal cells, demonstrating their active neurite growth and plasticity (Fig. 4b).
Quantitative analysis of neural cell frequency after the NS12ST differentiation protocol demonstrated that 47.4 ± 4.6% of cells were immunopositive for βIII-tubulin, 47 ± 4.0% for MAP2ab, and 15.2 ± 2.4% for GFAP, while there were no O4-positive cells (Fig. 4c). These results are comparable to those obtained for most NS cell lines, which have a neurogenic potential ranging between 50% and 85% [11, 12, 22].
We next investigated the lineage-specific differentiation of NS12ST cells by analysing the expression of a panel of neuronal markers typical of striatal neurons (Fig. 4d). We exposed NS12ST cells to retinoic acid (1 μM), a morphogen known to act in determining the fate of the LGE and in striatal neuron differentiation, and to valproic acid (1 mM), employed for GABAergic neuronal differentiation [30, 31]. Candidate gene expression analysis was performed on NS12ST cells in proliferation and after 15 days of differentiation in the absence or presence of retinoic acid (Fig. 4d). Dlx5 is a gene primarily expressed in the SVZ and the mantle while Dlx6 is expressed primarily in the mantle in the LGE, labelling striatal precursors. As shown in Fig. 4d, we observed no Dlx5 or Dlx6 expression in differentiated NS12ST cells. Also, Isl1, a LIM-homeobox gene expressed in the SVZ of the LGE and developing mantle and expressed in early postmitotic striatal precursors [32], was not expressed in differentiating NS12ST cells (Fig. 4d). Foxp1, Foxp2, Meis2 and Rarβ1, markers of striatal precursors and medium spiny neurons, which represent the vast majority (95%) of striatal neurons, were not differentially expressed in the absence or presence of retinoic acid and valproic acid (Fig. 4d). Darpp-32 was barely detectable (immunostaining did not reveal any positive cells, data not shown) while Arpp-21 showed a constitutive expression under self-renewal and differentiation conditions (Fig. 4d). These results suggest that although NS12ST cells express some striatum-specific genes during differentiation, they do not acquire a full panel of markers typical of a terminally differentiated striatal neuron.
The described protocol for neuronal differentiation was also applied to NS12SC cells. After 23 days of differentiation, NS12SC cells gave rise to βIII-tubulin-positive cells (50.5 ± 3.9%), MAP2ab-positive cells (57.7 ± 10.3%), and a few GFAP-positive cells (7.5 ± 2.8%) (Fig. 5a). Also, 1.5 ± 0.7% O4-positive oligodendrocytes were present (Fig. 5a). Synapsin-positive and GAP-43 neurons were also present (Figs. 5a, S4).
Fig. 5.
Neuronal differentiation of NS12SC cells. a After 23 days of differentiation in vitro, NS12SC cells gave rise to βIII-tubulin-, MAP2-, GFAP-, and O4-positive cells. A representative synapsin-positive cell is shown after 21 days of differentiation (×4, magnified after acquisition). b At the end of differentiation, 50.5 ± 3.9% of cells were immunopositive for βIII-tubulin, 57.7 ± 10.3% for MAP2, 7.5 ± 2.8% for GFAP, and 1.5 ± 0.7% for O4 (n = 1,068 cells for βIII-tubulin; n = 1,184 cells for MAP2 and GFAP; n = 1,645 cells for O4) (columns represent averages, error bars standard deviations). c Gene expression analysis by RT-PCR on NS12SC cells in proliferation (P) and after 15 days differentiation in the absence (D15-M) or in the presence (D15+M) of morphogens. d Spinal marker expression was restricted in NS12SC and not detectable in NS12ST cells. Mouse fetal brain was used as positive control (+CTRL); -RT was the negative control
We investigated the expression of markers of spinal neurons in NS12SC cells exposed to the differentiation protocol (Fig. 5c). In particular, we exposed NS12SC cells to retinoic acid (1 μM) and SHH (100 ng/ml), two morphogens known to be active in determining the fate of motorneuron (MN) cells. Moreover, we added NT-3 (10 ng/ml) and GDNF (10 ng/ml), two neurotrophins that support MN differentiation [33]. RT-PCR analysis was next performed on NS12SC cells in proliferation and after 15 days of differentiation in the absence and presence of morphogens (Fig. 5c). Different types of interneurons and MNs are generated in stereotypical locations in specified progenitor domains defined in part by a homeodomain protein code [34]. Progenitor cells in the pMN domain are characterized by the expression of two homeobox genes, Pax6 and Nkx6-1 [35, 36]. Moreover, their combined activities drive MN progenitors and the direct expression of downstream transcription factors, notably Lim3 and Hb9, that consolidate the identity of postmitotic MNs [33]. We found that differentiated NS12SC cells expressed Pax6, Nkx6-1 and, at a low level, Lim3 and Hb9, thus demonstrating their commitment and terminal differentiation towards spinal MNs (Fig. 5c). On the other hand, NS12SC cells expressed at a high level Irx3, a marker of spinal interneurons [37] that together with Nkx6-1 defines V2 interneurons, thus demonstrating a propensity of NS12SC cells to differentiate towards this lineage (Fig. 5c). MNs and interneuron markers are expressed by NS12SC-derived neurons whether or not the morphogens are added (Fig. 5c).
During ventral spinal cord differentiation, after MN production is completed, the domains of NKX2-2 and Olig2 expression, which previously were mutually exclusive, overlap and oligodendrocytes appear [38]. We found that differentiated NS12SC cells also express Nkx2-2, supporting the evidence of oligodendrocyte differentiation (Fig. 5a, c). On the other hand, we found no expression of lslet1 (Fig. 5c), a gene expressed in MNs and dorsal interneurons. Finally, we verified that the expression of the analysed spinal cord-specific markers was restricted to the NS12SC cell line. In fact, NS12ST cells never expressed Irx3, Nkx2-2 or Hb9, demonstrating that NS12SC cells show a restricted gene expression pattern (Fig. 5d).
Development of sodium currents, regenerative voltage response, and functional GABA receptor expression in neuronal progeny from fetal NS cells
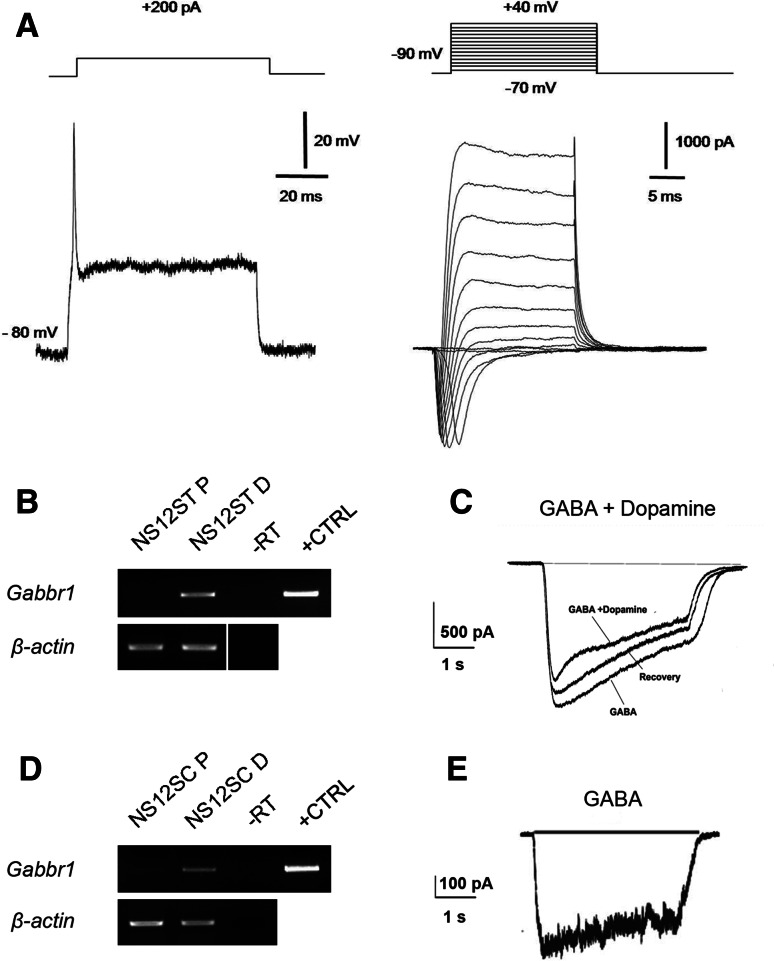
We used the expression of voltage-gated tetrodotoxin-sensitive Na+ current and the ability to fire action potentials as landmarks to assess the neuronal differentiation of NS12ST and NS12SC cells towards a mature phenotype. To monitor the electrophysiological extent of the neuronal conversion, we used the whole-cell patch-clamp technique both in current- and voltage-clamp mode. Following exposure to the differentiating protocol, about 30% of the cells were able to elicit a voltage-gated fast Na+ current and delayed rectifier K+ current. Figure 6a shows sample records of the total current derived from the NS12SC cells and evoked at a test potential of −10 mV from a holding voltage of −90 mV. Overshooting action potentials were obtained from NS12SC cells (Fig. 6a) and NS12ST cells (not shown).
Fig. 6.
Differentiated fetal NS cells produce action potentials and exhibit ligand-gated channel currents. a Sample records of the total current derived from the NS12SC cell demonstrate the presence of a tetrodotoxin-sensitive Na+ current (right). A representative overshooting action potential, evoked on the same cell, is shown on the left. b RT-PCR performed on NS12ST cells in proliferation (P) and in differentiation for the expression of the GABAB receptor-coding gene (Gabbr1). c Sample traces showing the effects of GABAergic agonists on differentiated NS12ST cells. Coapplication of dopamine (100 μM) significantly reduced the inward current evoked by GABA (20 μM) in differentiated NS12ST cells. This effect was partially reversible. d RT-PCR performed on NS12SC cells in proliferation (P) and in differentiation (D). e Sample trace showing the current evoked by GABAergic agonist on differentiated NS12SC cells
Furthermore, to explore the potential ability of these cells to develop a functional synaptic network, we assessed whether they carried ligand-gated channels that inhibitory or excitatory neurotransmitters could activate. We observed that a large percentage of cells responded to the application of GABAergic agonists. RT-PCR performed at the end of differentiation demonstrated that these cells expressed GABA-B receptors (Fig. 6b, d). To verify whether or not the differentiated NS12ST and NS12SC cells expressed functional GABAergic receptors, we also applied GABA locally to differentiated cells. Figure 6e shows the current traces elicited in differentiated NS12SC cells during application of 20 μM GABA from a holding potential of −80 mV. The GABAergic response occurred in 70% of NS12ST cells (n = 7/10) and 64% of NS12SC cells (n = 9/14) (data not shown). Differentiated NS12ST and NS12SC cells responding to GABA application showed an average ionic current of 221 ± 65 pA (mean ± SE) and 156 ± 69 pA, respectively. These data confirm the presence of functional GABA receptors on neurons from fetal NS cells and their ability to generate a functional GABAergic network in vitro.
We next evaluated the presence and functionality of receptors that have been described in striatal and spinal cord neurons in vivo. More precisely, we recorded the membrane currents during application of glutamate (20 and 100 μM), glycine (100 and 300 μM), acetylcholine (30 μM), nicotine (50 μM), carbachol (20 μM; a selective agonist of muscarinic cholinergic receptors), and dopamine (100 μM). No responses could be elicited in NS12SC and NS12ST cells after more than 2 weeks of differentiation in vitro following application of any of these compounds (data not shown). However, RT-PCR data demonstrated that NS12ST cells consistently and significantly expressed dopaminergic D2 receptors (Fig. S5). Following this indication and results in the literature [39], we decided to evaluate a possible modulatory effect produced by dopamine on GABA-evoked currents. As shown in Fig. 6c, dopamine (100 μM) significantly reduced (23 ± 5%; n = 7 cells) the inward current evoked by GABA (20 μM) in NS12ST cells differentiated for more than 2 weeks. This effect was partially reversible. We also found that about 64% of the striatal NS cells (n = 7/11) responded to dopamine. Altogether these data indicate that although a great percentage of NS12ST and NS12SC cells have functional GABAergic receptors on their membranes, they do not express functional glutamatergic, glycinergic, or cholinergic receptors, even if the muscarinic receptor (Chrm4) is detectable by RT-PCR (Fig. S5). In addition, only differentiated NS12ST cells expressed functional dopaminergic receptors, which can act as neuromodulators by decreasing the inhibitory effect elicited by GABA application.
Discussion
The possibility of keeping neural progenitors ready for further differentiation is appealing; however, unlimited self-renewal has not been observed in neural progenitors in vivo [40]. Indeed, long-term self-renewal of neural progenitors is usually achieved with extracellular cues such as FGF-2 and EGF, which alter aspects of cellular identity and the potential of neural progenitors and stem cells [14, 19]. This question is relevant because the isolation of multipotent cells from the adult CNS has fuelled hope for the reconstitution of degenerating neurons and neuronal networks in neuropathological conditions.
Positional information is a general feature that organizes cellular identities on the basis of their anatomic location in multicellular organisms. Comparison of global gene expression patterns of different cell types (e.g. fibroblasts and bones) derived from different anatomic sites demonstrate that cells retain stereotypic differences in their expression patterns based on their sites of origin. Many of these differences are retained with ex vivo passaging, suggesting a “transcriptional memory” of positional information [41].
Little information is available about the positional identity of cultured neural stem cells, mainly inferred from the neurosphere system. Region-specific differences among neurosphere-containing neural stem cells originating from different regions have been reported by Hitoshi et al. [15] even though a correct molecular profile was present only in neurospheres propagated for one or two passages, which could be considered more close to primary cultures and not a homogeneous population of stem cells. Accordingly, other reports have identified significant changes in region-specific features of neurospheres maintained in culture [17–19] regarding both R-C and D-V regional markers.
In this study, we demonstrated that NS cells isolated from different germinal zone regions of the developing CNS behave in a similar manner regarding self-renewal and neuropotency and display region-specific positional identities. Our candidate transcription factor analysis in NS cell lines derived from cerebellum, striatum, neocortex, mesencephalon, and spinal cord reveals that a clear R-C identity is maintained in culture, as demonstrated by the stable (>20 passages) expression of Foxg1, Otx2 and Tlx in anterior-derived NS cells and by Hoxb4 and Hoxb9 specifically expressed only in spinal cord-derived NS cells. These findings are not in accordance with those of a previous study that demonstrated the loss of Hoxb9 expression in spinal cord-derived neurospheres and widespread expression of Bf1 (Foxg1) in diencephalic and mesencephalic neurospheres [17]. However, neurospheres consist of heterogeneous cultures of neural progenitors and differentiating progenies where stem cells represent only a small percentage [42]. In this regard, Sun et al. [43] demonstrated that NS cells derived from human fetal CNS maintain the expression of regional markers in culture, such as OTX2 in brain-derived NS and HOXB9 in spinal cord-derived NS cells.
We investigated also the D-V identity that results altered under in vitro culture conditions, thus confirming the FGF-2 ventralizing activity [18–20]. We found that NS cells show a general expression of intermediate and ventral genes, such as Olig2, Meis2 and Ascl1 (although a neocortical-VZ expression is present for the latter [29]). Surprisingly, this ventral identity is maintained as a default activation of the Shh pathway, even in the absence of Shh expression, probably because of the presence of a low Ihh expression (Calabrese et al., unpublished observations).
By comparing the expression profile of neocortical NS cells, derived at E12.5 and E14.5, respectively, we also found evidence that the developmental stage is critical for the derivation of committed NS cells that maintain an in vitro positional identity consistent with that of the region of derivation. The change in commitment level is a known phenomenon occurring during cortical development, as demonstrated by some elegant experiments involving Mash1 (Ascl1) misexpression, which causes respecification and ventralization of early (E12.5) cortical progenitors with expression of Dlx1, Dlx6 and Gad1 [29]. On the other hand, respecification by Mash1 misexpression was unsuccessful at mid-corticogenesis (E14.5), when cortical progenitors are already irreversibly committed [29].
We also showed that neuronal differentiation of NS cells leads to electrophysiologically active neurons expressing a set of cell determinants consistent with their positional identity, even if they did not acquire a full molecular spectrum in terms of gene expression and specific neurochemical phenotype. For example, striatal NS cells differentiated into GABAergic neurons responded to a modulatory effect of dopamine but did not express Darpp-32. On the other hand, spinal cord NS cells exhibited some molecular evidence of MN cell fate acquisition (such as Hb9 expression) but did not show cholinergic markers. Spatial pattern formation and specification of positional information in general precedes and is independent of molecular differentiation [1]. So far, NS cells, even though they possess a correct positional identity, do not develop a full biochemical phenotype, thus confirming that there is no strict correlation between a cell state—determined by a transcription factor code—and the future behaviour. This consideration can be applied also to many other neural stem cell propagation systems that are even less plastic or regionally specified than the NS cells.
In conclusion, these findings shed light on the biology of cultured NS cells and delineate both the power and the limits. Further strategies for neuronal differentiation should be considered to push in vitro-generated precursor cells towards acquisition of the complete features of a neuron.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1. RT–PCR analysis of selected neural progenitor/stem cell markers (JPEG 750 kb)
Figure S2. Immunofluorescence characterization of E14.5 neocortex-derived NSCs. Self-renewing NS14CX cells homogeneously showed double immunoreactivity for Nestin/Vimentin and SOX2/PAX6 (JPEG 1804 kb)
Figure S3. Immunofluorescence characterization of 3 different clones of NS cells derived from E12.5 neocortex (NS12CX) and 3 clones of NS cells derived from E12.5 spinal cord (NS12SC). Self-renewing NS cell clones homogeneously express the typical NS cell markers Nestin, BLBP, RC2, Vimentin, and OLIG2 (JPEG 2075 kb)
Figure S4. Neuronal differentiation of NS12SC. This image shows a GAP-43-positive neuron derived from NS12SC cells differentiated for 23 d (JPEG 646 kb)
Figure S5. RT–PCR analysis of a subset of neurotransmitter receptors, described in striatal and spinal cord neurons in vivo. Neither proliferating (P) nor differentiating (D) NS cells expressed functional glutamatergic, glycinergic, or cholinergic receptors, even if the muscarinic receptor (Chrm4) was detectable (JPEG 579 kb)
Acknowledgments
This research was supported by initial funding from Fondazione Cariplo (Italy) in the context of the awarded N.O.B.E.L. (Operational Network for Biomedicine par Excellence in Lombardy) project entitled “A genetic toolkit for the analyses of neural stem cells—acronym: Mouse NS-toolkit”. This work is also supported by EuroSystem (FP7, European Union Health-F4-2008-200720), Neuroscreen (FP6, European Union LSHB-CT-2007-037766), and NeuroStemcell (FP7, European Union HEALTH-2008-B-222943). The laboratory also acknowledges the contribution of Unicredit Banca S.p.A. (Italy) and of Tavola Valdese (Italy).
Abbreviations
- BDNF
Brain-derived neurotrophic factor
- CNS
Central nervous system
- D-V
Dorsoventral
- EGF
Epidermal growth factor
- ESC
Embryonic stem cell
- FGF-2
Fibroblast growth factor 2
- GABA
γ-Aminobutyric acid
- GAP-43
Growth-associated protein 43
- GFAP
Glial fibrillary acid protein
- LGE
Lateral ganglionic eminence
- MAP2
Microtubule-associated protein 2
- MGE
Medial ganglionic eminence
- MN
Motorneuron
- NS cells
Radial glia-like neural stem cells
- R-C
Rostrocaudal
- Shh
Sonic hedgehog
- SVZ
Subventricular zone
Footnotes
An erratum to this article is available at http://dx.doi.org/10.1007/s00018-016-2307-x.
References
- 1.Wolpert L. Positional information and the spatial pattern of cellular differentiation. J Theor Biol. 1969;25:1–47. doi: 10.1016/S0022-5193(69)80016-0. [DOI] [PubMed] [Google Scholar]
- 2.Lupo G, Harris WA, Lewis KE. Mechanisms of ventral patterning in the vertebrate nervous system. Nat Rev Neurosci. 2006;7:103–114. doi: 10.1038/nrn1843. [DOI] [PubMed] [Google Scholar]
- 3.Krumlauf R. Hox genes and pattern formation in the branchial region of the vertebrate head. Trends Genet. 1993;9:106–112. doi: 10.1016/0168-9525(93)90203-T. [DOI] [PubMed] [Google Scholar]
- 4.Wassef M, Joyner AL. Early mesencephalon/metencephalon patterning and development of the cerebellum. Perspect Dev Neurobiol. 1997;5:3–16. [PubMed] [Google Scholar]
- 5.Rubenstein JL, Beachy PA. Patterning of the embryonic forebrain. Curr Opin Neurobiol. 1998;8:18–26. doi: 10.1016/S0959-4388(98)80004-4. [DOI] [PubMed] [Google Scholar]
- 6.McMahon AP. Neural patterning: the role of Nkx genes in the ventral spinal cord. Genes Dev. 2000;14:2261–2264. doi: 10.1101/gad.840800. [DOI] [PubMed] [Google Scholar]
- 7.Campbell K. Dorsal-ventral patterning in the mammalian telencephalon. Curr Opin Neurobiol. 2003;13:50–56. doi: 10.1016/S0959-4388(03)00009-6. [DOI] [PubMed] [Google Scholar]
- 8.Conti L, Pollard SM, Gorba T, Reitano E, Toselli M, Biella G, Sun Y, Sanzone S, Ying QL, Cattaneo E, Smith A. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pollard SM, Conti L, Sun Y, Goffredo D, Smith A. Adherent neural stem (NS) cells from fetal and adult forebrain. Cereb Cortex. 2006;16(Suppl 1):i112–i120. doi: 10.1093/cercor/bhj167. [DOI] [PubMed] [Google Scholar]
- 10.Glaser T, Pollard SM, Smith A, Brustle O. Tripotential differentiation of adherently expandable neural stem (NS) cells. PLoS One. 2007;2:e298. doi: 10.1371/journal.pone.0000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goffredo D, Conti L, Di Febo F, Biella G, Tosoni A, Vago G, Biunno I, Moiana A, Bolognini D, Toselli M, Cattaneo E. Setting the conditions for efficient, robust and reproducible generation of functionally active neurons from adult subventricular zone-derived neural stem cells. Cell Death Differ. 2008;15:1847–1856. doi: 10.1038/cdd.2008.118. [DOI] [PubMed] [Google Scholar]
- 12.Onorati M, Camnasio S, Binetti M, Jung CB, Moretti A, Cattaneo E. Neuropotent self-renewing neural stem (NS) cells derived from mouse induced pluripotent stem (iPS) cells. Mol Cell Neurosci. 2010;43:287–295. doi: 10.1016/j.mcn.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Kim JB, Zaehres H, Arauzo-Bravo MJ, Scholer HR. Generation of induced pluripotent stem cells from neural stem cells. Nat Protoc. 2009;4:1464–1470. doi: 10.1038/nprot.2009.173. [DOI] [PubMed] [Google Scholar]
- 14.Koch P, Opitz T, Steinbeck JA, Ladewig J, Brustle O. A rosette-type, self-renewing human ES cell-derived neural stem cell with potential for in vitro instruction and synaptic integration. Proc Natl Acad Sci U S A. 2009;106:3225–3230. doi: 10.1073/pnas.0808387106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hitoshi S, Tropepe V, Ekker M, van der Kooy D. Neural stem cell lineages are regionally specified, but not committed, within distinct compartments of the developing brain. Development. 2002;129:233–244. doi: 10.1242/dev.129.1.233. [DOI] [PubMed] [Google Scholar]
- 16.Santa-Olalla J, Baizabal JM, Fregoso M, del Carmen Cardenas M, Covarrubias L. The in vivo positional identity gene expression code is not preserved in neural stem cells grown in culture. Eur J Neurosci. 2003;18:1073–1084. doi: 10.1046/j.1460-9568.2003.02824.x. [DOI] [PubMed] [Google Scholar]
- 17.Zappone MV, Galli R, Catena R, Meani N, De Biasi S, Mattei E, Tiveron C, Vescovi AL, Lovell-Badge R, Ottolenghi S, Nicolis SK. Sox2 regulatory sequences direct expression of a (beta)-geo transgene to telencephalic neural stem cells and precursors of the mouse embryo, revealing regionalization of gene expression in CNS stem cells. Development. 2000;127:2367–2382. doi: 10.1242/dev.127.11.2367. [DOI] [PubMed] [Google Scholar]
- 18.Hack MA, Sugimori M, Lundberg C, Nakafuku M, Gotz M. Regionalization and fate specification in neurospheres: the role of Olig2 and Pax6. Mol Cell Neurosci. 2004;25:664–678. doi: 10.1016/j.mcn.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 19.Gabay L, Lowell S, Rubin LL, Anderson DJ. Deregulation of dorsoventral patterning by FGF confers trilineage differentiation capacity on CNS stem cells in vitro. Neuron. 2003;40:485–499. doi: 10.1016/S0896-6273(03)00637-8. [DOI] [PubMed] [Google Scholar]
- 20.Bithell A, Finch SE, Hornby MF, Williams BP. Fibroblast growth factor 2 maintains the neurogenic capacity of embryonic neural progenitor cells in vitro but changes their neuronal subtype specification. Stem Cells. 2008;26:1565–1574. doi: 10.1634/stemcells.2007-0832. [DOI] [PubMed] [Google Scholar]
- 21.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–319. doi: 10.1016/S0014-5793(97)00313-X. [DOI] [PubMed] [Google Scholar]
- 22.Spiliotopoulos D, Goffredo D, Conti L, Di Febo F, Biella G, Toselli M, Cattaneo E. An optimized experimental strategy for efficient conversion of embryonic stem (ES)-derived mouse neural stem (NS) cells into a nearly homogeneous mature neuronal population. Neurobiol Dis. 2009;34:320–331. doi: 10.1016/j.nbd.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 23.Doetsch F, Garcia-Verdugo JM, Alvarez-Buylla A. Cellular composition and three-dimensional organization of the subventricular germinal zone in the adult mammalian brain. J Neurosci. 1997;17:5046–5061. doi: 10.1523/JNEUROSCI.17-13-05046.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- 25.Selkoe D, Kopan R. Notch and Presenilin: regulated intramembrane proteolysis links development and degeneration. Annu Rev Neurosci. 2003;26:565–597. doi: 10.1146/annurev.neuro.26.041002.131334. [DOI] [PubMed] [Google Scholar]
- 26.Tao W, Lai E. Telencephalon-restricted expression of BF-1, a new member of the HNF-3/fork head gene family, in the developing rat brain. Neuron. 1992;8:957–966. doi: 10.1016/0896-6273(92)90210-5. [DOI] [PubMed] [Google Scholar]
- 27.Monaghan AP, Grau E, Bock D, Schutz G. The mouse homolog of the orphan nuclear receptor tailless is expressed in the developing forebrain. Development. 1995;121:839–853. doi: 10.1242/dev.121.3.839. [DOI] [PubMed] [Google Scholar]
- 28.Shi Y, Chichung Lie D, Taupin P, Nakashima K, Ray J, Yu RT, Gage FH, Evans RM. Expression and function of orphan nuclear receptor TLX in adult neural stem cells. Nature. 2004;427:78–83. doi: 10.1038/nature02211. [DOI] [PubMed] [Google Scholar]
- 29.Britz O, Mattar P, Nguyen L, Langevin LM, Zimmer C, Alam S, Guillemot F, Schuurmans C. A role for proneural genes in the maturation of cortical progenitor cells. Cereb Cortex. 2006;16(Suppl 1):i138–i151. doi: 10.1093/cercor/bhj168. [DOI] [PubMed] [Google Scholar]
- 30.Laeng P, Pitts RL, Lemire AL, Drabik CE, Weiner A, Tang H, Thyagarajan R, Mallon BS, Altar CA. The mood stabilizer valproic acid stimulates GABA neurogenesis from rat forebrain stem cells. J Neurochem. 2004;91:238–251. doi: 10.1111/j.1471-4159.2004.02725.x. [DOI] [PubMed] [Google Scholar]
- 31.Aubry L, Bugi A, Lefort N, Rousseau F, Peschanski M, Perrier AL. Striatal progenitors derived from human ES cells mature into DARPP32 neurons in vitro and in quinolinic acid-lesioned rats. Proc Natl Acad Sci U S A. 2008;105:16707–16712. doi: 10.1073/pnas.0808488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stenman JM, Wang B, Campbell K. Tlx controls proliferation and patterning of lateral telencephalic progenitor domains. J Neurosci. 2003;23:10568–10576. doi: 10.1523/JNEUROSCI.23-33-10568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wichterle H, Lieberam I, Porter JA, Jessell TM. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/S0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 34.Stone D, Rosenthal A. Achieving neuronal patterning by repression. Nat Neurosci. 2000;3:967–969. doi: 10.1038/79894. [DOI] [PubMed] [Google Scholar]
- 35.Ericson J, Briscoe J, Rashbass P, van Heyningen V, Jessell TM. Graded sonic hedgehog signaling and the specification of cell fate in the ventral neural tube. Cold Spring Harb Symp Quant Biol. 1997;62:451–466. [PubMed] [Google Scholar]
- 36.Sander M, Sussel L, Conners J, Scheel D, Kalamaras J, Dela Cruz F, Schwitzgebel V, Hayes-Jordan A, German M. Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of beta-cell formation in the pancreas. Development. 2000;127:5533–5540. doi: 10.1242/dev.127.24.5533. [DOI] [PubMed] [Google Scholar]
- 37.Jacob J, Briscoe J. Gli proteins and the control of spinal-cord patterning. EMBO Rep. 2003;4:761–765. doi: 10.1038/sj.embor.embor896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kessaris N, Pringle N, Richardson WD. Ventral neurogenesis and the neuron-glial switch. Neuron. 2001;31:677–680. doi: 10.1016/S0896-6273(01)00430-5. [DOI] [PubMed] [Google Scholar]
- 39.Ariano MA, Cepeda C, Calvert CR, Flores-Hernandez J, Hernandez-Echeagaray E, Klapstein GJ, Chandler SH, Aronin N, DiFiglia M, Levine MS. Striatal potassium channel dysfunction in Huntington’s disease transgenic mice. J Neurophysiol. 2005;93:2565–2574. doi: 10.1152/jn.00791.2004. [DOI] [PubMed] [Google Scholar]
- 40.Gaspard N, Gaillard A, Vanderhaeghen P. Making cortex in a dish: in vitro corticopoiesis from embryonic stem cells. Cell Cycle. 2009;8:2491–2496. doi: 10.4161/cc.8.16.9276. [DOI] [PubMed] [Google Scholar]
- 41.Chang HY. Anatomic demarcation of cells: genes to patterns. Science. 2009;326:1206–1207. doi: 10.1126/science.1175686. [DOI] [PubMed] [Google Scholar]
- 42.Conti L, Cattaneo E. Neural stem cell systems: physiological players or in vitro entities? Nat Rev Neurosci. 2010;11:176–187. doi: 10.1038/nrn2938. [DOI] [PubMed] [Google Scholar]
- 43.Sun Y, Pollard S, Conti L, Toselli M, Biella G, Parkin G, Willatt L, Falk A, Cattaneo E, Smith A. Long-term tripotent differentiation capacity of human neural stem (NS) cells in adherent culture. Mol Cell Neurosci. 2008;38:245–258. doi: 10.1016/j.mcn.2008.02.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. RT–PCR analysis of selected neural progenitor/stem cell markers (JPEG 750 kb)
Figure S2. Immunofluorescence characterization of E14.5 neocortex-derived NSCs. Self-renewing NS14CX cells homogeneously showed double immunoreactivity for Nestin/Vimentin and SOX2/PAX6 (JPEG 1804 kb)
Figure S3. Immunofluorescence characterization of 3 different clones of NS cells derived from E12.5 neocortex (NS12CX) and 3 clones of NS cells derived from E12.5 spinal cord (NS12SC). Self-renewing NS cell clones homogeneously express the typical NS cell markers Nestin, BLBP, RC2, Vimentin, and OLIG2 (JPEG 2075 kb)
Figure S4. Neuronal differentiation of NS12SC. This image shows a GAP-43-positive neuron derived from NS12SC cells differentiated for 23 d (JPEG 646 kb)
Figure S5. RT–PCR analysis of a subset of neurotransmitter receptors, described in striatal and spinal cord neurons in vivo. Neither proliferating (P) nor differentiating (D) NS cells expressed functional glutamatergic, glycinergic, or cholinergic receptors, even if the muscarinic receptor (Chrm4) was detectable (JPEG 579 kb)