Abstract
Krueppel-like factor 4 (Klf4) belongs to the Sp/Klf family of zinc-finger transcription factors and is indispensable for terminal maturation of epithelial tissues. Furthermore, it is part of a small set of proteins that are used to generate pluripotent embryonic stem cells from differentiated tissues. Herein, we describe that a Klf4 zinc-finger domain mutant induces self-renewal and block of maturation, while wild-type Klf4 induces terminal macrophage differentiation. Moreover, we present the crystal structure of the zinc-finger domain of Klf4 bound to its target DNA, revealing that primarily the two C-terminal zinc-finger motifs are required for site specificity. Lack of those two zinc fingers leads to deficiency of Klf4 to induce macrophage differentiation. The first zinc finger, on the other hand, inhibits the otherwise cryptic self-renewal and block of differentiation activity of Klf4. Our data show that impairing the DNA binding could potentially contribute to a monocytic leukemia.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-010-0618-x) contains supplementary material, which is available to authorized users.
Keywords: Zinc finger, Crystal structure, Self-renewal, Macrophage differentiation
Introduction
Klf4 (Gklf, gut-enriched Krueppel-like factor) is a transcription factor belonging to the specificity protein/Krueppel-like factor (Sp/Klf) superfamily of gene regulatory proteins [1]. Sp/Klf proteins are characterized by three highly conserved zinc-finger motifs, which bind to GC/GT-rich regions of DNA to mediate activation and/or repression of transcription [2, 3]. Proteins of the Klf subfamily are implicated in many biological processes, including cell proliferation, apoptosis, differentiation, and development [2] and are also potential targets for cancer therapy [4].
To date, 17 members of the Klf family have been identified in mammals. They are referred to as Klf1 to Klf17 [3, 5]. The founding member of this group in mammals, Klf1 (Eklf, erythroid Krueppel-like factor), was shown to be critical for red blood cell maturation [6, 7]. Klf2 (Lklf, lung Krueppel-like factor) is involved in maintaining quiescence of individual positive T-lymphocytes [8] and in T-lymphocyte trafficking [9]. Ectopic expression of Klf4 has recently been shown to induce macrophage maturation, and bone marrow chimera lacking Klf4 showed reduced numbers of Ly-6Chigh monocytes in the peripheral blood [10, 11]. In a somewhat surprising contrast to the induction of terminal differentiation, Klf4 is part of a group of proteins (together with Oct4, Sox2, and c-Myc) that permit reprogramming of terminally differentiated cells into pluripotent stem cells [12].
Next to its C-terminal zinc-finger domain, Klf4 comprises an N-terminal poly proline and acidic amino acid stretch, an inhibitory region, and a conserved nuclear localization signal sequence adjacent to the zinc finger [13]. Little is known about the cellular function of the different protein domains of Klf4. Moreover, no structural information about the DNA-binding properties of the zinc-finger domain of Klf proteins is available. To date, only crystal structures of related C2H2-type zinc-finger proteins such as Zif268 [14], Wilms tumor suppressor protein [15], GLI oncogene product [16], and TFIIIA [17] are known. In addition, solution structures of isolated zinc-finger domains of Sp1 [18], Klf3 [19], Klf10 (PDB-ID 2epa, unpublished), Klf15 (PDB-ID 2ent, unpublished), and uncomplexed Klf5 (PDB-ID 2ebt, unpublished) have been reported (http://www.pdb.org). To investigate the function of the different protein domains of Klf4, we determined the crystal structure of the zinc-finger domain in complex with target DNA and furthermore investigated the function of Klf4 in primary hematopoietic cells. Herein, we present for the first time the structural basis for DNA recognition of a protein from the Klf subfamily. We demonstrate that individual structural contribution of the different zinc fingers to DNA-binding links to the function of the protein to induce terminal differentiation. We further report that Klf4 lacking the DNA-binding domain induces block of differentiation and self-renewal in hematopoietic cells.
Materials and methods
Cloning, expression, and purification
DNA encoding the zinc-finger domain of murine Klf4 (amino acids 396–483) was subcloned into the pQLinkH vector [20]. The gene encoding the N-terminal His7-tagged protein was over-expressed at 20°C in E. coli Rosetta (DE3). The purification procedure comprises an affinity chromatography on a 5 ml HisTrap FF crude column (GE Healthcare), charged with Zn2+, and a size-exclusion chromatography on a Superdex 75 prep grade column (26 × 60, GE Healthcare). The His7-tag was cleaved with tobacco etch virus protease prior to the gel-filtration step.
Protein crystallization
The purified protein (in 20 mM HEPES-NaOH pH 7.5, 0.2 M NaCl, 5 mM DTT, 10 μM ZnSO4) was complexed with 1.5-fold molar excess of DNA (7mer: 5′ GAG GCG C 3′; 10mer: 5′ GAG GCG TGG C 3′, [1]) and co-concentrated to 4–7 mg/ml. The Klf4 zinc-finger domain was crystallized using the vapor diffusion method at 20°C by mixing equal volumes of protein and reservoir solution. The complex of Klf4 with heptameric DNA was crystallized using 10% (w/v) PEG 6000 and 0.1 M HEPES-NaOH pH 7.0 as reservoir buffer. The reservoir buffer for the crystallization of the Klf4-decamer DNA complex contained 27% (w/v) PEG 3350, 0.2 M sodium acetate, and 0.1 M Tris-HCl pH 8.5. Before flash-freezing in liquid nitrogen, the crystal was transformed in a cryoprotectant consisting of reservoir solution supplemented with 24% glycerol.
Data collection, structure determination, and refinement
X-ray diffraction data were collected at beamlines BL14.1 and BL14.2 [21] of the Helmholtz-Zentrum Berlin für Materialien und Energie and Freie Universität Berlin at BESSY. Data reduction was performed using Xds and Xscale. Data collection statistics are reported in Table 1 and Supplementary Table 1. For the Klf4-heptamer DNA complex, the zinc substructure was solved using ShelXD [22], and the identified three zinc positions were used for phasing and density modification as implemented in Sharp/autoSharp [23]. The resulting experimental electron density map was used for automatic chain tracing using Arp/wArp [24]. The heptameric DNA fragment was placed manually into density using Coot [25]. This model was transferred to the high-resolution native data by using rigid-body refinement with Refmac [26] and refined (Supplementary Table 2). The structure of the Klf4-decamer DNA complex was solved by molecular replacement using the program Phaser [27] and the Klf4-heptamer DNA complex structure as search model. The structure was refined using simulated annealing in Phenix followed by multiple rounds of refinement in Refmac [26]. The graphics program Coot was used for model building and visualization [25]. Statistics on structure refinement are summarized in Table 1. Figures were created with Pymol [28]. The coordinates and structure factors have been deposited in the Protein Data Bank, http://www.pdb.org (PDB ID codes 2wbs, 2wbu).
Table 1.
Data collection and refinement statistics for the Klf4:DNA-10 complex
| Klf4:DNA-10 | |
|---|---|
| Data collection | |
| Wavelength (Å) | 0.91841 |
| Resolution (Å) | 46.93–2.50 (2.565–2.50) |
| Space group | P43212 |
| Unit-cell parameters (Å) | a = 50.28, c = 131.03 |
| Measured reflections | 51,838 (3,731) |
| Unique reflections | 6,277 (446) |
| R meas (%) | 8.2 (64.1) |
| Completeness (%) | 99.5 (100.0) |
| <I>/<σ(I)> | 20.4 (4.2) |
| Wilson B-factor (Å2) | 47.8 |
| Refinement | |
| Resolution range (Å) | 46.93–2.50 |
| R cryst | 0.215 |
| R free (test set of 8%) | 0.278 |
| No. of non-protein molecules | |
| DNA | 1 |
| Zinc | 3 |
| Water | 21 |
| Average isotropic B-factor (Å2) | |
| Overall | 23.1 |
| DNA | 34.1 |
| Zinc | 35.9 |
| Water | 25.1 |
| RMSD for bond lengths (Å) | 0.016 |
| RMSD for bond angles (°) | 1.355 |
| Ramachandran regions | |
| Most favored (%) | 92.8 |
| Allowed (%) | 7.2 |
| Outliers (%) | – |
Numbers in parentheses are for the highest resolution shell
Mice
Mice (129/sv × C57BL/6 or pure C57BL/6 background) were bred under pathogen-free conditions in accordance with the German animal protection law.
The retroviral constructs
The pMIEG3 plasmid (provided by D.A. Williams, Children’s Hospital Medical Center, Cincinnati, OH, USA) is a bicistronic murine stem cell virus (MSCV)-based retroviral vector, enabling co-expression of the marker eGFP along with the gene of interest. The ERT2 fragment (estrogen-receptor ligand-binding domain) was obtained from the Cre-ERT2 plasmid (provided by Daniel Metzger, INSERM, Strasbourg, France) [29]. The Klf4 (NM_010637.3) fragments were obtained by PCR amplification using bone-marrow macrophage-derived cDNA as template and sequence specific primers (available upon request) and included amino acids 63–400 (ΔN62Klf4ΔC), 117–400 (ΔN116Klf4ΔC), 1–400 (Klf4ΔC), and Klf4ΔZf2/3 (1–425, the two C-terminal zinc fingers deleted). Amplified sequences were verified by sequencing.
Retroviral transfection of progenitor-enriched bone-marrow cells and stimulation
The progenitor-enriched bone-marrow cells were obtained 72 h after injection of 150 mg/ml 5-FU into the peritoneal cavity and plated in the stimulation medium (IMDM, supplemented with 10% FCS, 2 mM glutamine, 0.1 mM 2-mercaptoethanol, 2% pen/strep, and recombinant cytokines: rrSCF, rhu G-CSF (a kind gift from AMGEN, Thousand Oaks, CA, USA), and rhTPO (a kind gift from Kirin Brewery, Tokyo, Japan). After 48 h of stimulation, cells were loaded on fibronectin-coated plates (Retronectin™, Takara, Japan) and infected by sequentially adding 2 ml of undiluted virus supernatant (30 min at 37°C), 2 ml of fresh supernatant diluted with 2 ml of stimulation medium (incubated over night), and virus-free stimulation medium (incubated for 6 h). The infection procedure was repeated twice. After the second infection, cells were incubated for another 48 h in the stimulation medium, detached from the Retronectin™ by using cold Cell Dissociation buffer (PAA Laboratories, Pasching, Germany), and sorted for eGFP expression on a MoFlo High Speed cell sorter (DakoCytomation, Hamburg, Germany). After sorting, cells were cultivated for another 3–8 days using the same cytokines and stimulated with either 1 μM tamoxifen (4-OHT; Sigma, Deisenhofen, Germany) or control diluent (ethanol). For determination of RNA expression, RNA was extracted using the peqGOLD MicroSpin Total RNA Kit (peqLab, Erlangen, Germany).
Fluorescence-assisted cell sorting (FACS)
Cell samples of 1–5 × 105 cells were resuspended (2% FCS, 2 mM EDTA, 0.1% NaN3 in PBS) and labeled with fluorochrome-coupled antibodies: CD11b, Gr1(Ly-6G/C; clone RB6-8C5), (BD Biosciences PharMingen, San Diego, CA, USA), and F4/80 (Serotec, Düsseldorf, Germany). Analysis was performed on the flow cytometer FACSCalibur, with CellQuest software (BD).
CFU assay
After FACS sorting, 1 × 105 cells were plated into methylcellulose medium (MethoCult M3134, Stem Cell Technologies, Vancouver, Canada) supplemented with 30% FCS, 2 mM glutamine, 2% P/S, and 0.1 mM β-mercaptoethanol in replica (first culture). The methylcellulose medium was supplemented with cytokines as follows: 100 ng/ml rhuG-CSF, 100 ng/ml rhuTPO, 100 ng/ml rmSCF, and 1 μM 4-OHT. In some instances, rmGM-CSF (5 U/ml) or rhu M-CSF (100 ng/ml) was used. The CFU colonies (composed of a minimum of 50 single cells) were counted on day 7 post-plating. After counting, the cells were rinsed from the plates by washing several times with PBS. These cells were used for replating experiments. Again 1 × 105 cells were plated per methylcellulose plate in replica. The same cytokine stimulation was maintained as for the primary culture; one half of the secondary culture was stimulated with tamoxifen, the other half with diluent (ethanol) only. The colony number was determined again on day 10. These cells were again washed from the plates and replated as stated above. Remaining cells from the first and the secondary culture were investigated by flow-cytometric analysis. Cytospins were prepared, May-Gruenwald-Giemsa stained and the cell morphology was analyzed.
Results
Crystal structure of the zinc-finger domain of Klf4
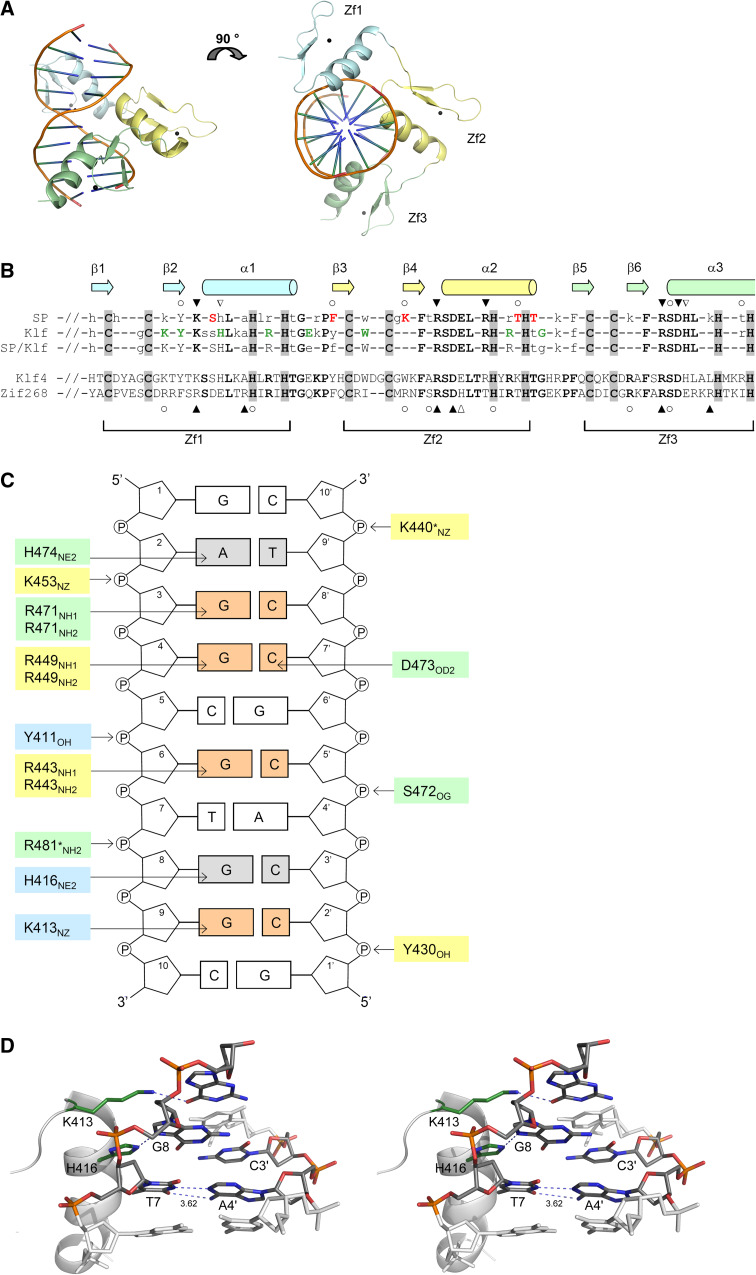
Klf4 contains three C2H2-type zinc-finger motifs at its C-terminus, representing the DNA-binding site of the transcription factor. By base-specific mutagenesis, a double-stranded DNA with the sequence 5′-G/A G/A GG C/T G C/T-3′ was found to contain the minimal essential binding site for Klf4 [1]. To investigate the structural basis for these preferences, we aimed to analyze the structure of Klf4 in complex with DNA. We first determined the crystal structure of Klf4 bound to this heptameric target DNA at 1.7 Å resolution by using intrinsic zinc for MAD phasing, revealing that the first zinc finger was not involved in oligonucleotide binding (Supplementary Fig. 1). We therefore chose a 3′-extended decameric DNA fragment for further co-crystallization experiments, resembling a DNA sequence that is present in the basic transcription element of the CYP1A1 gene, which interacts with Sp1 and several Sp1-like transcription factors [1]. The overall structure of the Klf4-decamer DNA complex is represented in Fig. 1a. The final model includes amino-acid residues 399–483 of Klf4 and a 10-bp DNA duplex (statistics in Table 1). Each zinc finger has a conserved ββα structure and binds one zinc ion that is sandwiched between the two-stranded antiparallel β-sheet and the α-helix (Fig. 1a). Each zinc ion tetrahedrally coordinates to the side chains of two cysteines at the end of the second β-sheet and two histidines in the C-terminal span of the α-helix (Fig. 1a), residues that are highly conserved within the entire Sp/Klf family (Fig. 1b).
Fig. 1.
Crystal structure of the zinc-finger domain of Klf4 in complex with DNA. The zinc-finger motifs are highlighted in blue (motif 1), yellow (motif 2), and green (motif 3), respectively. a The overall fold of the Klf4 monomer in complex with a decameric double-stranded DNA molecule is presented as a cartoon model. The right model corresponds to a 90° rotation of the complex around the x-axis. The N- and C-termini are marked with N or C. Zinc ions are shown as gray spheres. b Consensus sequences for the zinc-finger domains of Sp and Klf factors and the entire Sp/Klf family (modified after [3]). Bold capital letters indicate residues that are 100% conserved between all family members (black), between all Klf proteins (green), or between all Sp factors (red). Capitalized residues indicate >90% conservation, lower-case letters >75% conservation. The cysteine and histidines residues involved in zinc coordination are highlighted in gray. The secondary structure elements shown above the alignment correspond to murine Klf4 (this work). Klf4 residues involved in specific base contacts are marked with filled inverted triangle, additional base interactions with inverted triangle, and unspecific DNA-phosphate-backbone interactions with open circle. Direct DNA interactions of Zif268 (taken from [31]) are indicated below the alignment of murine Klf4 and Zif268. c Schematic representation of protein-DNA interactions. DNA bases that are specifically recognized by hydrogen bonding to Klf4 side chains are highlighted in red, other base interactions in gray. Residues marked with an asterisk originate from a symmetry-related molecule. d Stereo representation of the interaction of Klf4 with its target DNA. Residues involved in direct DNA-base contacts are shown as sticks. A stacking interaction between His416 and the T7 base is observed, altering the stacking geometry between T7-A4′ and G8-C3′ and the interbase hydrogen bonds between T7 and A4′. Blue dashed lines indicate hydrogen bonds
The related C2H2-zinc-finger protein Zif268 is the closest structural homolog of Klf4 as revealed by the SSM program [30]. The Cα atoms in the structures of Klf4 and Zif268 (1aay, [31]) superimpose with a root-mean-square deviation (RMSD) of 1.2 Å (over 83 Cα-atoms). Other structural homologs are the Wilms tumor suppressor protein (2prt, [15]), the Ying-Yang 1 protein (YY1, 1ubd, [32]), the GLI oncogene product (2gli, [16]), and the transcription factor TFIIIA (1tf6, [17]) that superimpose onto Klf4 with RMSD values of 1.7, 2.0, 2.7, and 2.2 Å, respectively (over 82, 80, 85, and 85 Cα atoms). While secondary structure elements superimpose well, structural changes are observed in the loop regions connecting the first and second β-sheet of each zinc finger (Supplementary Fig. 2), regions that are least conserved within the Sp/Klf superfamily (Fig. 1b).
In Klf4, binding of the identified minimal essential binding site [1] is exclusively mediated by the second and third zinc-finger motifs (Supplementary Fig. 1). Residues that determine DNA-binding specificity are highly conserved within the entire Sp/Klf superfamily (Fig. 1b), accounting for the recognition of related DNA promoter sequences by the family members. The recognition pattern between Klf4 and DNA is displayed in Fig. 1c. Three arginine residues of the two C-terminal zinc fingers form bidentate hydrogen bonding interactions involving the guanidinium group of the arginine side chain and the N7 and O6 atoms of the guanine base in positions G3, G4, and G6, in this way conferring most of the DNA-binding specificity. Two of the arginine residues (R443 and R471, helix position −1) are stabilized by a salt bridge with a neighboring aspartate side chain (D445 and D473, helix position 2). The third arginine residue (R449, helix position 6) forms a salt bridge to a conserved glutamate residue (E446, helix position 3). Interestingly, in Klf1, a critical erythroid regulatory transcription factor, this conserved glutamate residue within the second zinc finger (Fig. 1b) is mutated in the semi-dominant mouse mutation Nan (neonatal anemia) (E339D in Klf1), altering the DNA-binding specificity of Klf1 so that it no longer binds promoters of a subset of its DNA targets [33]. Although this glutamate (E446, Klf4 numbering) does not make any direct DNA interactions, it forms two water-mediated contacts to the DNA and most importantly is salt-bridged to the side chain of R449, which it orients for proper DNA binding, explaining the profound pathological changes in the Nan mouse mutation of Klf1.
Whilst the arginine and aspartate residues that confer sequence specificity are conserved within the entire Sp/Klf superfamily, sequence conservation is not observed for most of the residues that are involved in DNA-phosphate contacts (Fig. 1b). These residues are likely to be important in the energetics but not the specificity of zinc finger-DNA recognition [34].
In the Klf4-decamer DNA complex structure, adjacent symmetry-related segments form a pseudo-continuous double helix throughout the crystal with the α-helix of each zinc finger contacting the major groove. Curves 5.3 analysis [35] shows that the DNA in the crystal is bent by 8° and resembles B-form DNA with a mean twist of 32.9° and rise of 3.26 Å per base-pair step. Major deviations from B-DNA parameters occur in the vicinity of the first zinc finger. Base-pair T7-A4′ is characterized by a 14° opening, accompanied by a loosened hydrogen contact between T7 O4 and A4′ N6 and a shear of −0.6 Å. These perturbations are linked to the stacking interaction of T7 with the side chain of His416, which is hydrogen bonded to and in plane with the G8-C3′ base pair, forming a super base pair (Fig. 1c, d). Notably, this histidine residue is strictly conserved only within the Klfs of the Sp/Klf super-family (Fig. 1b).
The perturbation of DNA conformation induced by His416 occurs at a CpA/TpG base-pair step. These steps have been known for their deformability for some time [36]. In crystal structures of B-DNA fragments, they were frequently found to adopt a usual geometry with greatly increased helix twist and slide [37, 38]. Recently, this slide displacement at CpA/TpG steps was implicated in nucleosome positioning [39]. While the formation of the His416-G8-C3′ base triple in the Klf4-DNA complex induces a different conformational perturbation into the DNA, it agrees well with the previously described structural malleability of the CpA/TpG step.
In Zif268-type C2H2 zinc-finger proteins, each zinc-finger motif partakes in specific recognition of three adjacent base pairs in one strand, and a single fourth base in the complementary strand of the DNA. The binding specificity is conferred by the amino-acid residues within the α-helix at positions −1, 2, 3, and 6 relative to the start of the α-helix, with the position 2 contacting the base in the opposite strand [34]. Generally, this canonical recognition mode is also seen in Klf4. However, due to changes in the amino acid sequence, fewer interactions conferring binding specificity are observed (Fig. 1b). In Klf4 the amino-acid residues within the α-helix at positions −1 and 3 of the first zinc finger, in the second zinc-finger residues at positions −1 and 6 of the recognition helix, and in the third zinc-finger residues at the helical positions −1, 2, and 3 partake in DNA-base interactions. Moreover, the contribution of the first zinc finger to DNA binding of Klf4 is smaller than that of the two C-terminal zinc fingers (Fig. 1b, c). This reduced DNA binding of the first zinc finger of Klf4 has also been reported in the solution structure of the isolated first zinc-finger motif of the specificity protein Sp1 [18], indicating that this might be a common property within the entire Sp/Klf superfamily of transcription factors.
Klf4 induces macrophage differentiation in hematopoietic cells
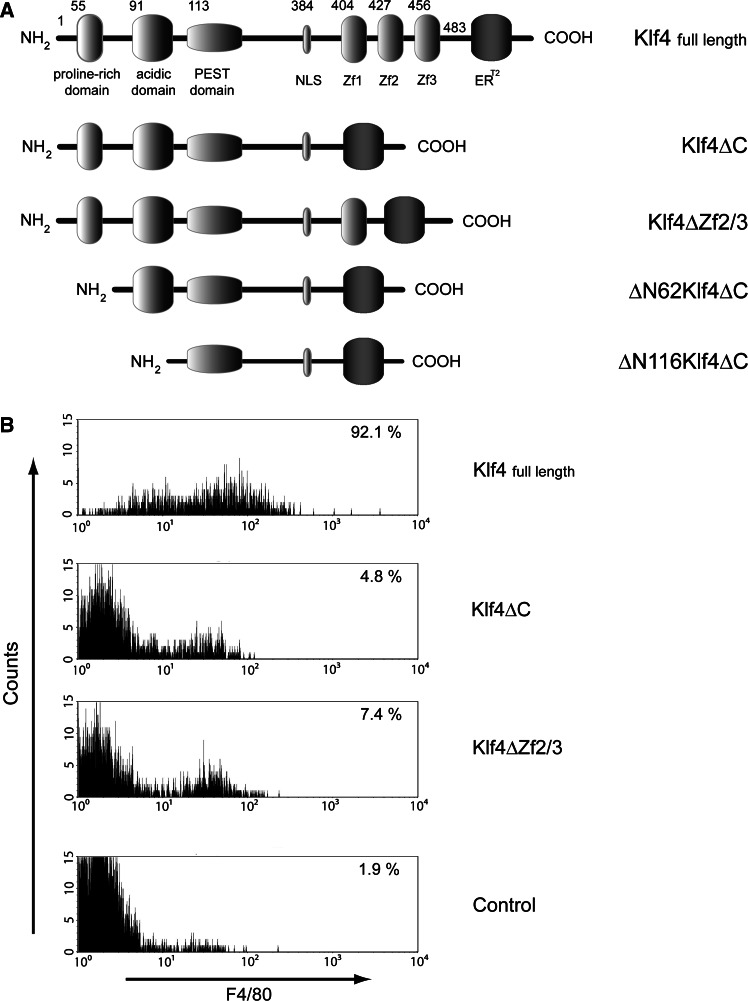
To investigate the cellular function of the various Klf4 domains, we expressed different mutants of Klf4 via retroviral gene transfer in primary murine hematopoietic cells. We fused the modified estrogen receptor ERT2 [29, 40] at the C-terminus of wild-type Klf4 and Klf4 mutants, allowing temporal control of activation. The following mutants were analyzed: Klf4ΔC (lacking the entire zinc-finger domain), Klf4ΔZf2/3 (lacking the second and third zinc fingers), ΔN62Klf4ΔC (lacking the three zinc fingers and the poly proline region [41], and ΔN116Klf4ΔC (additionally lacking the acidic region; Fig. 2a). Primary murine cells from wild-type mice were infected with the retrovirus carrying the cDNAs for the different Klf4-ERT2 fusion proteins, co-expressing the green fluorescent protein (eGFP). GFP-positive cells were isolated via high-speed flow cytometry. Isolated cells were cultivated in CFU assays with cytokines instructive for development into the granulocytic lineage, and the expression of the fusion protein was induced by tamoxifen. Full-length Klf4 fused to ERT2 induced macrophage differentiation (see below).
Fig. 2.
Klf4 induces macrophage differentiation. a Klf4 mutants: full-length Klf4 (Klf4; NM_010637.3), fused in frame to the ERT2 fragment, was cloned into a retroviral vector co-expressing eGFP from an internal ribosomal entry site. The mutants lacking the entire zinc-finger domain (Klf4ΔC) or the last two zinc fingers (Klf4ΔZf2/3), a mutant lacking the first 62 amino acids and the zinc-finger domain (ΔN62Klf4ΔC), and a mutant lacking the first 116 amino acids (including both the poly proline stretch [40] and the acidic transactivation region [42]) and the zinc-finger domain (ΔN116Klf4ΔC) were also fused to the ERT2 fragment. Numbers indicate the position of the respective amino acid. b FACS histograms indicate differentiation as surface expression of the macrophage differentiation marker F4/80 on primary myeloid cells, stably transduced with either full-length Klf4, Klf4ΔC, Klf4ΔZf2/3, or empty control vector (Mieg 3). Percentages of cells expressing F4/80 are indicated. Cells were grown in a cytokine cocktail promoting granulocyte differentiation. While expression of full-length Klf4 induced macrophage differentiation, expression of a Klf4 mutant lacking at least the two C-terminal zinc-finger motifs failed to do so
Our structural analysis demonstrated that specific DNA binding is mediated primarily by the second and third zinc-finger motifs. To test the functional relevance of this finding, we compared the expression of a variant lacking the two C-terminal zinc fingers (Klf4ΔZf2/3) with the expression of the wild-type protein. As a functional readout, we tested for the induction of macrophage differentiation, measured by the expression of the cell surface protein F4/80 [42]. As shown in Fig. 2b, full-length Klf4 showed strong induction of F4/80 expression in comparison to control vector. Expression of the Klf4ΔZf2/3 mutant, in contrast, did not significantly alter the differentiation of transduced cells. Thus, the variant retaining only the N-terminal zinc finger is functionally inactive with respect to the capacity of full-length Klf4 to induce macrophage differentiation. These functional data are in line with the structural findings, since specific DNA binding was shown to be mainly accomplished by the second and third zinc fingers. Moreover, expression of a variant lacking the entire zinc-finger domain did not significantly induce F4/80 expression compared to wild-type Klf4 (Fig. 2b). Thus, the Klf4 variant lacking the entire zinc-finger domain behaved similarly to the variant where only the two C-terminal zinc fingers have been deleted. Taken together, our data indicate that macrophage differentiation is linked with the specific DNA-binding capacity of the zinc-finger domain of Klf4.
Deletion of the zinc-finger domain of Klf4 results in block of monocytic differentiation and induces self-renewal
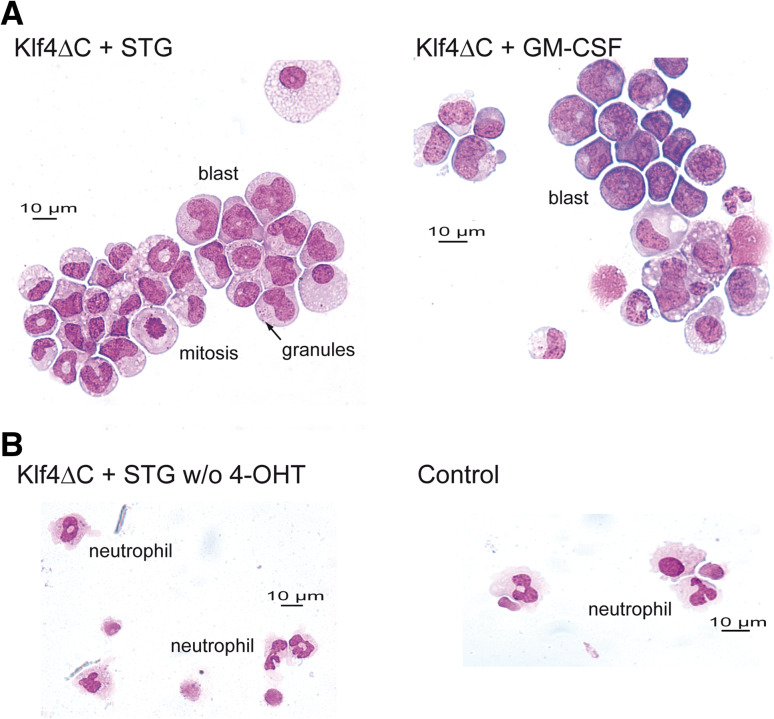
We next studied the function of the Klf4 variant with a deletion of the entire DNA-binding zinc-finger domain (Klf4ΔC) in more detail. We observed that expression of this mutant in hematopoietic cells resulted in colony formation with a very distinctive morphology. Numbers of colonies were higher, and colonies showed a dense accumulation of small cells (not shown). Morphological analysis of individual cells from cytospins of those colonies showed cells with a high nucleus-to-cytoplasm ratio. Cells tended to form clusters and had a blast-like appearance. Nuclei were convoluted, and in several instances prominent cytoplasmic granules were present (Fig. 3a). Morphological analysis of cells washed off from whole plates and cytospun showed little granulocytic differentiation in comparison to control cultures (Fig. 3b). Mitotic figures were frequently found, and cells appeared like monoblasts. Thus, expression of Klf4ΔC blocks monocytic differentiation with cells arresting at a blast-like stage.
Fig. 3.
Klf4 induces block of terminal macrophage differentiation in hematopoietic cells. Cytospins of myeloid cells from cultures grown in CFU assays supplemented with either rhuSCF, rhuTPO, rhuG-CSF (STG), or rmGM-CSF were stained with May-Gruenwald-Giemsa. Photographs were taken at 400× magnification. a Morphology of cells expressing Klf4ΔC. A majority of cells expressing the Klf4ΔC mutant are of blast-like morphology with the appearance of monocytic differentiation (monoblasts) and show cytoplasmic granules. b Morphology of cells in control cultures. Klf4ΔC was either not induced (w/o 4-OHT, without tamoxifen) or transduced with a control vector (Mieg3) in the presence of tamoxifen. As shown, a significant proportion of cells differentiate into the neutrophil granulocytic lineage. Abnormal granulocytic morphology is typical for the unphysiologically high cytokine concentrations in culture
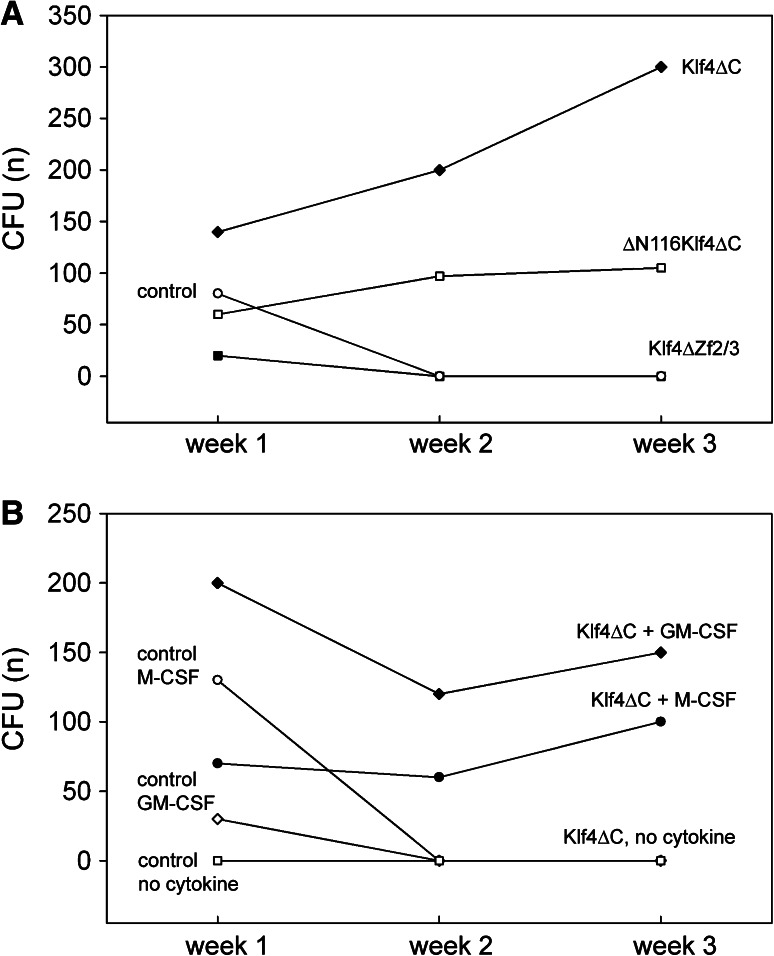
Since the cells resembled leukemic monoblastic cells and unlimited self-renewal is an inherent property of cancer cells, we next investigated whether those cells were capable of self-renewal. For this purpose, a defined number of cells were serially replated in the same cytokine cocktail in the presence or absence of tamoxifen. As shown in Fig. 4a, colonies formed in both secondary cultures. In CFU assays induced with tamoxifen, colonies with similar morphology developed as in the primary cultures, while colonies were large and spread in the absence of tamoxifen (not shown). Nevertheless, the number of observed colonies was lower in CFU assays without tamoxifen, i.e., when Klf4ΔC was not induced, and the resulting morphology was significantly different. Cytomorphological analysis identified the cells from those colonies as macrophages (not shown). In contrast, cells from the colonies induced with tamoxifen again showed blast-like appearance (not shown), indicating that Klf4ΔC indeed was able to induce self-renewal.
Fig. 4.
Klf4 induces self-renewal in hematopoietic cells. Myeloid cells stably expressing either specific Klf4 mutants or control vector were grown in CFU assays. After 1 week of culturing, colonies were counted and replated in the same cytokine condition. Colonies forming after 2 weeks were again counted and replated to test for self-renewal activity. Colonies were again counted on week 3. a CFU assays were supplemented with tamoxifen, rhuSCF, rhuTPO, and rhuG-CSF (STG). While cells expressing Klf4ΔC showed continuous self-renewal, cells expressing control vector did not. Cells expressing ΔN116Klf4ΔC (lacking the N-terminal poly proline and acidic transactivation regions and the zinc-finger domain) still showed continuous self-renewal, while cells expressing Klf4ΔZf2/3 (lacking the two C-terminal zinc fingers) did not. b CFU assays were supplemented with tamoxifen and rmGM-CSF, rhu M-CSF, or no cytokines, respectively. While cells expressing Klf4ΔC showed continuous self-renewal independent of the cytokine used to promote culture, cells expressing Klf4ΔC not stimulated with cytokines did not grow
We also determined whether Klf4ΔC could induce cytokine-independent growth and whether other cytokines would allow for self-renewal. As a result, without addition of cytokines, no colonies were produced, while colonies with self-renewal activity grew in the presence of GM-CSF and M-CSF (Fig. 4b).
Other Klf4 domains required for blockade of differentiation and self-renewal
The N-terminal part of Klf4 bears two polypeptide regions with distinct proposed functions. A polypeptide comprising the poly proline stretch has been proposed to mediate binding to SH3 domains [41]. Another region containing acidic amino acids has been shown to repress transcription of a Klf4-Gal4 fusion protein [43]. Indeed, this is the only part N-terminal of the zinc-finger domain and the nuclear localization sequence for which a specific function has been demonstrated. Furthermore, for human KLF4, it has been shown that this acidic amino-acid region binds to p300/CREB proteins and that this binding can be inhibited by competition with the adenovirus protein E1A [43]. Therefore, this domain is potentially of functional importance for transactivation of genes regulated by Klf4. To test whether these regions mediate part of the observed induction of self-renewal and differentiation block, we expressed mutants lacking (1) both the zinc-finger and the poly proline region (ΔN62Klf4ΔC), (2) additionally lacking the acidic amino acid stretch (ΔN116Klf4ΔC), and (3) the Klf4ΔZf2/3 mutant lacking the two C-terminal zinc fingers. Cells expressing the Klf4 mutant lacking both the zinc-finger domain and the poly proline region (ΔN62Klf4ΔC) induced self-renewal similarly compared to cells expressing Klf4ΔC lacking the zinc-finger domain only (not shown). In contrast, cells expressing the Klf4 mutant where the acidic domain is additionally deleted (ΔN116Klf4ΔC) showed a largely reduced capacity to produce colonies in the primary (week 1) CFU assay (Fig. 4a). Nevertheless, those cells were capable of self-renewal (Fig. 4a). In contrast, cells expressing Klf4 retaining only the first zinc finger (Klf4ΔZf2/3) failed to show continuous self-renewal (Fig. 4a). From those experiments we conclude that the proline-rich stretch of amino acids influences neither transactivation nor repression. Unexpectedly, self-renewal and block of differentiation also occured in the absence of the acidic transactivation region, indicating that this region is dispensable for this function. Furthermore, induction of self-renewal seems to require loss of specific DNA binding. In the presence of the first zinc finger only, neither induction of self-renewal nor terminal macrophage differentiation was observed.
Discussion
In this work we investigated the structure of the zinc-finger domain of Klf4 and aimed to resolve the function of this and other Klf4 protein domains. The crystal structure obtained from the zinc-finger domain of Klf4 is the first DNA complex structure of a Klf protein. We demonstrate that primarily the two C-terminal zinc fingers contact Klf4-specific sequences within the promoters of target genes, while the first zinc finger binds outside the consensus sequence and contributes less to specific DNA binding. Nevertheless, the first zinc finger is of functional relevance. Complete lack of all three zinc fingers, resulting in a Klf4 protein completely lacking DNA binding, induces a maturation arrest and self-renewal in hematopoietic cells. This is prevented when the first zinc finger is present. In particular, this mutant (Klf4ΔZf2/3) neither induces macrophage differentiation such as full-length Klf4 nor self-renewal as does Klf4 lacking the entire zinc-finger domain (Klf4ΔC). Formally, we cannot distinguish whether the function of the first zinc finger is mediated by protein-protein interactions and/or by weak DNA binding.
A similar structure-function analysis of Klf4 has been reported in iPS cell induction [44], where mutants lacking the entire zinc-finger domain or parts thereof did not show an autonomous function with respect to induction of pluripotency. Indeed, all analyzed mutants comprising part of the zinc-finger domain inhibited the intrinsic function of wild-type Klf4. To our knowledge, induction of self-renewal and pluripotency has always been attributed to full-length Klf4. In this respect, the function of the mutants lacking the zinc-finger domain described here is entirely novel as they induce self-renewal in hematopoietic cells under circumstances where full-length Klf4 induces terminal differentiation. Furthermore, a mutant lacking the two C-terminal zinc fingers is the only functionally inactive mutant with respect to pluripotency induction we describe in our study.
How oncogenes block differentiation and induce self-renewal during the process of oncogenesis is only partially understood today. Furthermore, for most myeloid leukemias, it is largely unclear how partial differentiation into a particular myeloid lineage is achieved as reflected by the current WHO classification of myeloid neoplasms [45]. Our data show that impairing of the DNA binding, either by a mutation within the Klf4 gene or via an interacting protein, could potentially induce a monocytic leukemia. We furthermore observed that Klf4 is expressed in murine GMP as well as in several cells of the monocytic lineage (M.M., J.K., D.C.; unpublished observations and NCBI GEO accession number: GSM27216). The function of Klf4 for monocytic development is currently under investigation in our laboratory.
Sequence analyses of the Klf4 protein predict the N-terminus to be unfolded. One-dimensional 1H-NMR data showed that the N-terminal acidic polypeptide stretch (amino acids 63–116) of Klf4 is indeed unstructured (A.S., Peter Schmieder, FMP Berlin, D.C.; unpublished). Deletion of this region, however, does not impair the observed block of differentiation and induction of self-renewal. It therefore remains unclear which N-terminal part of the remaining Klf4 protein is responsible for the observed induction of self-renewal and block of differentiation.
In conclusion, we analyzed the structural basis for specific DNA binding of Klf4 and relate the structural findings to the cellular function of Klf4. Specific DNA binding mediated by the two C-terminal zinc fingers is required for the induction of terminal macrophage differentiation, while the first zinc finger inhibits an otherwise cryptic self-renewal and block of differentiation activity of Klf4. Our findings (1) significantly advance the understanding of the molecular basis of DNA binding of proteins of the Sp/Klf family in general and its member Klf4 in particular. They (2) advance our understanding of how Klf4 induces pluripotency in differentiating cells, and the described Klf4 mutants may be suitable tools in this respect. Furthermore, they (3) yield important insights into the functional anatomy of the Klf4 protein and into the development of myeloid malignancies and are of significant interest to stem-cell research, as the technology to induce pluripotency through expression of defined factors, one of which is Klf4 [12], is rapidly advancing.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft to D.C. (CA 306/1-1; 1-2) and the “Deutsche Krebshilfe” to D.C. The Protein Sample Production Facility at the Max Delbrück Center is funded by the Helmholtz Association of German Research Centres. We thank Janett Tischer, Silke Kurths, Ingrid Berger, and Tracy Dornblut for excellent technical assistance. We are also thankful to the kind staff of the cell sorting facility of the Deutsche Rheumaforschungszentrum and acknowledge Uwe Müller and the beamline support from the staff of Helmholtz-Zentrum Berlin für Materialien und Energie at BESSY.
Abbreviation
- Klf4
Krueppel-like factor 4
References
- 1.Shields JM, Yang VW. Identification of the DNA sequence that interacts with the gut-enriched Krüppel-like factor. Nucleic Acids Res. 1998;26:796–802. doi: 10.1093/nar/26.3.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pearson R, Fleetwood J, Eaton S, Crossley M, Bao S. Krüppel-like transcription factors: a functional family. Int J Biochem Cell Biol. 2008;40:1996–2001. doi: 10.1016/j.biocel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 3.Suske G, Bruford E, Philipsen S. Mammalian SP/KLF transcription factors: bring in the family. Genomics. 2005;85:551–556. doi: 10.1016/j.ygeno.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Safe S, Abdelrahim M. Sp transcription factor family and its role in cancer. Eur J Cancer. 2005;41:2438–2448. doi: 10.1016/j.ejca.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Kaczynski J, Cook T, Urrutia R. Sp1- and Krüppel-like transcription factors. Genome Biol. 2003;4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature. 1995;375:316–318. doi: 10.1038/375316a0. [DOI] [PubMed] [Google Scholar]
- 7.Perkins AC, Sharpe AH, Orkin SH. Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature. 1995;375:318–322. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- 8.Kuo CT, Veselits ML, Leiden JM. LKLF: a transcriptional regulator of single-positive T cell quiescence and survival. Science. 1997;277:1986–1990. doi: 10.1126/science.277.5334.1986. [DOI] [PubMed] [Google Scholar]
- 9.Carlson CM, Endrizzi BT, Wu J, Ding X, Weinreich MA, Walsh ER, Wani MA, Lingrel JB, Hogquist KA, Jameson SC. Krüppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442:299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- 10.Alder JK, Georgantas RW, 3rd, Hildreth RL, Kaplan IM, Morisot S, Yu X, McDevitt M, Civin CI. Krüppel-like factor 4 is essential for inflammatory monocyte differentiation in vivo. J Immunol. 2008;180:5645–5652. doi: 10.4049/jimmunol.180.8.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feinberg MW, Wara AK, Cao Z, Lebedeva MA, Rosenbauer F, Iwasaki H, Hirai H, Katz JP, Haspel RL, Gray S, Akashi K, Segre J, Kaestner KH, Tenen DG, Jain MK. The Krüppel-like factor KLF4 is a critical regulator of monocyte differentiation. EMBO J. 2007;26:4138–4148. doi: 10.1038/sj.emboj.7601824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zaehres H, Scholer HR. Induction of pluripotency: from mouse to human. Cell. 2007;131:834–835. doi: 10.1016/j.cell.2007.11.020. [DOI] [PubMed] [Google Scholar]
- 13.Yet SF, McA’Nulty MM, Folta SC, Yen HW, Yoshizumi M, Hsieh CM, Layne MD, Chin MT, Wang H, Perrella MA, Jain MK, Lee ME. Human EZF, a Krüppel-like zinc finger protein, is expressed in vascular endothelial cells and contains transcriptional activation and repression domains. J Biol Chem. 1998;273:1026–1031. doi: 10.1074/jbc.273.2.1026. [DOI] [PubMed] [Google Scholar]
- 14.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 Å. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 15.Stoll R, Lee BM, Debler EW, Laity JH, Wilson IA, Dyson HJ, Wright PE. Structure of the Wilms tumor suppressor protein zinc finger domain bound to DNA. J Mol Biol. 2007;372:1227–1245. doi: 10.1016/j.jmb.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 16.Pavletich NP, Pabo CO. Crystal structure of a five-finger GLI-DNA complex: new perspectives on zinc fingers. Science. 1993;261:1701–1707. doi: 10.1126/science.8378770. [DOI] [PubMed] [Google Scholar]
- 17.Nolte RT, Conlin RM, Harrison SC, Brown RS. Differing roles for zinc fingers in DNA recognition: structure of a six-finger transcription factor IIIA complex. Proc Natl Acad Sci USA. 1998;95:2938–2943. doi: 10.1073/pnas.95.6.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oka S, Shiraishi Y, Yoshida T, Ohkubo T, Sugiura Y, Kobayashi Y. NMR structure of transcription factor Sp1 DNA binding domain. Biochemistry. 2004;43:16027–16035. doi: 10.1021/bi048438p. [DOI] [PubMed] [Google Scholar]
- 19.Simpson RJ, Cram ED, Czolij R, Matthews JM, Crossley M, Mackay JP. CCHX zinc finger derivatives retain the ability to bind Zn(II) and mediate protein–DNA interactions. J Biol Chem. 2003;278:28011–28018. doi: 10.1074/jbc.M211146200. [DOI] [PubMed] [Google Scholar]
- 20.Scheich C, Kümmel D, Soumailakakis D, Heinemann U, Büssow K. Vectors for co-expression of an unrestricted number of proteins. Nucleic Acids Res. 2007;35:e43. doi: 10.1093/nar/gkm067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinemann U, Büssow K, Mueller U, Umbach P. Facilities and methods for the high-throughput crystal structural analysis of human proteins. Acc Chem Res. 2003;36:157–163. doi: 10.1021/ar010129t. [DOI] [PubMed] [Google Scholar]
- 22.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D. 2002;58:1772–1779. doi: 10.1107/S0907444902011678. [DOI] [PubMed] [Google Scholar]
- 23.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 24.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 25.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 27.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 28.DeLano WL (2010) The PyMOL molecular graphics system. DeLano Scientific, San Carlos. http://www.pymol.org
- 29.Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- 30.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 31.Elrod-Erickson M, Rould MA, Nekludova L, Pabo CO. Zif268 protein-DNA complex refined at 1.6 Å: a model system for understanding zinc finger–DNA interactions. Structure. 1996;4:1171–1180. doi: 10.1016/S0969-2126(96)00125-6. [DOI] [PubMed] [Google Scholar]
- 32.Houbaviy HB, Usheva A, Shenk T, Burley SK. Cocrystal structure of YY1 bound to the adeno-associated virus P5 initiator. Proc Natl Acad Sci USA. 1996;93:13577–13582. doi: 10.1073/pnas.93.24.13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siatecka M, Sahr KE, Andersen SG, Mezei M, Bieker JJ, Peters LL. Severe anemia in the Nan mutant mouse caused by sequence-selective disruption of erythroid Kruppel-like factor. Proc Natl Acad Sci USA. 2010;107:15151–15156. doi: 10.1073/pnas.1004996107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 35.Lavery R, Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J Biomol Struct Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 36.Heinemann U, Alings C, Hahn M. Crystallographic studies of DNA helix structure. Biophys Chem. 1994;50:157–167. doi: 10.1016/0301-4622(94)85028-3. [DOI] [PubMed] [Google Scholar]
- 37.Heinemann U, Alings C. Crystallographic study of one turn of G/C-rich B-DNA. J Mol Biol. 1989;210:369–381. doi: 10.1016/0022-2836(89)90337-9. [DOI] [PubMed] [Google Scholar]
- 38.Heinemann U, Alings C. The conformation of a B-DNA decamer is mainly determined by its sequence and not by crystal environment. EMBO J. 1991;10:35–43. doi: 10.1002/j.1460-2075.1991.tb07918.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tolstorukov MY, Colasanti AV, McCandlish DM, Olson WK, Zhurkin VB. A novel roll-and-slide mechanism of DNA folding in chromatin: implications for nucleosome positioning. J Mol Biol. 2007;371:725–738. doi: 10.1016/j.jmb.2007.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shields JM, Christy RJ, Yang VW. Identification and characterization of a gene encoding a gut-enriched Krüppel-like factor expressed during growth arrest. J Biol Chem. 1996;271:20009–20017. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schaller E, Macfarlane AJ, Rupec RA, Gordon S, McKnight AJ, Pfeffer K. Inactivation of the F4/80 glycoprotein in the mouse germ line. Mol Cell Biol. 2002;22:8035–8043. doi: 10.1128/MCB.22.22.8035-8043.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geiman DE, Ton-That H, Johnson JM, Yang VW. Transactivation and growth suppression by the gut-enriched Krüppel-like factor (Krüppel-like factor 4) are dependent on acidic amino acid residues and protein–protein interaction. Nucleic Acids Res. 2000;28:1106–1113. doi: 10.1093/nar/28.5.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei Z, Yang Y, Zhang P, Andrianakos R, Hasegawa K, Lyu J, Chen X, Bai G, Liu C, Pera M, Lu W. Klf4 interacts directly with Oct4 and Sox2 to promote reprogramming. Stem Cells. 2009;27:2969–2978. doi: 10.1634/stemcells.2008-0333. [DOI] [PubMed] [Google Scholar]
- 45.Vardiman JW, Harris NL, Brunning RD. The World Health Organization (WHO) classification of the myeloid neoplasms. Blood. 2002;100:2292–2302. doi: 10.1182/blood-2002-04-1199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.