Abstract
By being the “integration” center of transcriptional control as they move and target transcription factors, corepressors fine-tune the epigenetic status of the nucleus. Many of them utilize enzymatic activities to modulate chromatin through histone modification or chromatin remodeling. The clinical and etiological relevance of the corepressors to neoplastic growth is increasingly being recognized. Aberrant expression or function (both loss and gain of) of corepressors has been associated with malignancy and contribute to the generation of transcriptional “inflexibility” manifested as distorted signaling along certain axes. Understanding and predicting the consequences of corepressor alterations in tumor cells has diagnostic and prognostic value, and also have the capacity to be targeted through selective epigenetic regimens. Here, we evaluate corepressors with the most promising therapeutic potential based on their physiological roles and involvement in malignant development, and also highlight areas that can be exploited for molecular targeting of a large proportion of clinical cancers and their complications.
Keywords: Corepressors, HDAC, Inhibitor, Cancer, Therapy
Introduction
The new era of translational medicine directs the future applications in every aspect of medicine, from bench to bedside. Cancer, a top-notch player in this perception, is primarily manifested through a broad spectrum of genetic and epigenetic aberrations in signaling pathways, highlighting the gigantic perspective of targeting the transcriptional apparatus [1]. Transcription of protein-coding genes requires the assembly of the basal transcriptional machinery along with gene-specific transcription factors and coregulators (corepressors/coactivators), creating an intricate cross-talk network [2].
Transcription corepressors along with coactivators orchestrate gene transcription and maintain cellular homeostasis by controlling nuclear epigenetic status. Corepressors form multi-protein complexes that generally facilitate gene repression through interactions with different transcription factors, such as the nuclear receptors, activator protein 1 (AP-1), and nuclear factor kappa B (NF-κΒ) [3, 4]. Their role is to coordinate the assembly of a wide gamut of proteins that gather around them. Corepressor complexes utilize diverse mechanisms for the enzymatic repression of transcription, which involve mainly histone-tail modifications, namely deacetylation, methylation, deimination/citrullination, and ubiquitylation, as well as ATP-dependent nucleosome modulations [3, 5]. A plethora of corepressor complexes have been reported up to date, including nuclear receptor corepressor 1 (NCoR1), silencing mediator of retinoid and thyroid hormone receptor (SMRT), RE1-silencing transcription factor corepressor (CoREST), nucleosome remodeling and histone deacetylase (NURD) and Swi-independent 3 (SIN3).
Nuclear receptors are transcription factors that convey signals from steroid, thyroid, retinoid, and other lipophilic hormones implicated in physiological processes and the pathogenesis of various diseases, including cancer [2, 6, 7]. Additional reciprocal relations with coregulators modify their transcriptional potential [2]. The prevailing model suggests that coactivators bind to liganded nuclear receptors, while corepressors bind to unliganded ones [6]. Ligands function as an on/off switch, permitting when absent, a nuclear receptor–corepressor interaction through their ligand-binding domain (LBD) and corepressor nuclear receptor (CoRNR) box motif, respectively [5, 6]. The absence of the ligand leaves an open conformation in the receptor allowing interaction with the corepressor. NCoR1/SMRT complex, SIN3 complex, the corepressor Alien and also orphan nuclear receptors are included in this category, serving as constitutive repressors [6]. On the other hand, the existence of a circuitry where corepressors and coactivators are involved in constant turnover has been proposed. Active transcription, which marks a new cycle, requires the release of a corepressor and the binding of a coactivator. Analogous models are implicated in the transcription of NF-κΒ, peroxisome proliferator-activated receptor gamma (PPARγ) and Wnt target genes [5]. Recent experimental data from genome-wide profiling of histone acetyltransferases (HATs) and histone deacetylases (HDACs) binding on chromatin, however, reveal a novel three-phase gene status. An active state is associated with high levels of both HATs and HDACs. HDACs remove any signs of modification required for the re-establishment of the pattern. A second category includes genes that are not yet active but have entered an alert state. A cyclical transient addition of acetyl-groups followed by their removal keeps genes ready when the incoming signal arrives. Finally, low numbers of both HATs and HDACs are detected in silent genes [8]. The typical view of corepressors binding to unliganded receptors is questioned by the presence of a unique category of corepressors, such as receptor-interacting protein 140 (RIP140) and ligand-dependent corepressor (LCoR), which manifest agonist/antagonist-bound-dependent corepression [6, 9]. Interestingly, although corepressor complexes that contain NCoR, SMRT, and HDACs can interact directly with promoter regions of inflammation-related genes controlled by NF-κΒ and AP-1, they are also involved in agonist-dependent transrepression of these genes through their recruitment by PPAR, liver X receptor (LXR) and glucocorticoid receptor (GR) [5, 6].
Naturally, in order to secure smooth cellular procedures, cells control corepressor complexes in a multi-level manner. Ligand-binding conformational changes lead to the dissociation of corepressors from their receptors [5]. Another level of regulation is achieved through direct phosphorylation, followed by nuclear export and/or degradation of the corepressor. In this vein, acetylation, sumoylation, ubiquitylation, and methylation may also function as a derepression signal [5, 10].
Transcription corepressors in carcinogenesis
Transcription corepressors exert a fundamental role in the epigenetic control of cancer-related pathways responsible for proliferation, resistance to apoptosis, and migration, encouraging the endeavor to employ them as rational targets of novel epigenetic therapies (Table 1).
Table 1.
Co-repressor complexes in various types of cancer
| Complex | Members | Function | Interactions | Cancer | References |
|---|---|---|---|---|---|
| NCoR/SMRT | NCoR/SMRT, HDAC1,3,7, TBL1, GP52 TAK1, CORO2A | Histone deacetylase | Nuclear receptors, AP-1, NF-κB, ETO, MYOD, MAD/MXI, CBF, TFIIB | Prostate, breast, bladder, colorectal, endometrial, astrocytoma, leukemia | [5, 11, 13] |
| CoREST | CoREST, HDAC1/2, BHC80, BRAF35, LSD1 | Histone deacetylase, histone demethylase | REST, ZNF217, SWI/SNF, CTBP | Prostate, breast, colorectal | [11, 27, 32] |
| CTBP | CTBP, HDAC1/2, LSD1, Polycomb group proteins, NADH dehydrogenase | Histone deacetylase, histone demethylase | INK4a, INK4b, E-cadherin, PTEN, PERP, BAX, p21, Noxa, CoREST | Colorectal, breast, hepatocellular | [11, 33, 34] |
| SIN3A,B | SIN3A,B, HDAC1/2, BRMS1, RbAp4, RbAp7, SAP18, SAP30, ING1/2, SD53 | Histone deacetylase |
SIN3A: SMRT, MeCP2 SIN3B: CIITA, E2F-Rb Both: Mad1, KLF repressors, REST, ESET |
Breast, NSCLC | [37, 38, 41] |
| SWI/SNF | BRM, BRG1, BAF47, BAF155, BAF170, BAF250A (ARID1A), BAF250B (ARID1B), BAF180, BAF200, BRD7 | ATP-dependent chromatin remodeling, histone deacetylase recruitment | AP-1, steroid receptors, CoREST, MYC, p53, Rb, OCT4, SOX2, INK4a, EZH2, RHOA, ROCK1, Hedgehog, IFNB, CDKN1A (p21), SMAD3 | Breast, colorectal, pancreatic, melanoma, bladder, renal, lung, ovarian clear cell and endometrioid, uterine endometrioid | [11, 43, 45–47] |
| NURD |
CHD3, CHD4, HDAC1, HDAC2, LSD1, MBD2, MDB3, MTA1, MTA2, MTA3, RBBP4, RBBP7, GATAD2A, GATAD2B |
ATP-dependent chromatin remodeling, histone deacetylase, histone demethylase | ER, TWIST, SNAIL, BCL, JUN, PML–RAR, INK4, Rb, BRCA, MYC, HER2, HIF1, p53, PAX | Breast, colorectal, gastric, esophageal, endometrial, pancreatic, ovarian, lung, leukemia, lymphoma | [49–52] |
| PRAME | Unknown | Recruitment of EZH2 | RAR in the presence of RA | Melanoma, leukemia, multiple myeloma, lymphoma, head and neck, breast | [9] |
| PRC | PRC1: RING1a, RING1, CBX2, CBX4, CBX7, CBX8, BMI1, MEL18, BLR, NSPC1, PHC1, PHC2, PHC3 PRC2: EZH2, EZH1, EED, RBBP4, BBP7, PCL1, PCL2, PCL3, JARID2 |
PRC1: histone monoubiquitylation PRC2: histone methylation |
RNA Pol II, DNA methyltransferases, PML–RAR, PZLF–RAR, ARF, INK4b, INK4a, Hedgehog, Wnt, Notch |
Bladder, breast, prostate, squamous cell carcinomas, leukemia, neuroblastoma, colorectal | [56] |
NCoR1/SMRT
NCoR1 and SMRT (NCoR2), the first identified corepressors, are large regulatory proteins that bind to unliganded nuclear receptors [i.e., androgen receptor (AR), estrogen receptor (ER), thyroid hormone receptor (TR), GR, vitamin D receptor (VDR), retinoid receptor (RAR)] and serve as scaffolds for the formation of multi-protein complexes, which facilitate repression of nuclear receptor-target genes through chromatin deacetylation [10]. They share many similarities in both the protein level and the structure of their complexes, which contain HDAC1,3,7, transducin beta-like 1 (TBL1), GP52 and a set of other proteins, such as transforming growth factor-activated kinase 1 (TAK1) and coronin 2A (CORO2A). Most likely, however, HDAC3 is responsible for deacetylase activity [5]. Alternative splicing of SMRT may generate a group of isoforms with diverse nuclear receptor-binding capacities [11]. Recent data depict the recruitment of NCoR1/SMRT by activated transcription factors, such as PPARγ, to mediate transrepression [5, 11]. In the long list of their different targets, AP-1, NF-κB, eight-twenty-one (ETO) nuclear corepressor, myogenic differentiation (MyoD) protein, core-binding factor (CBF), and transcription factor IIB (TFIIB) are also included [5, 7, 10, 11]. The involvement of NCoR1/SMRT in the complex cross-talk transcriptional network highlights the maintenance of cellular integrity. Indeed, gene-knockout has been linked with embryonic lethality, adipogenesis and myocardial development [5, 7, 11]. Regarding tumorigenesis, NCoR1/SMRT is often overexpressed, resulting in the silencing of genes engaged in tumor suppression, as in the case of acute promyelocytic leukemia (APML). In APML, a fusion between RARα and PML or promyelocytic leukemia zinc finger (PLZF) results in the constant activation of NCoR1 and blockage of the RAR-regulated hematopoietic differentiation genes. Administration of retinoic acid (RA) combined with HDAC inhibitors (HDACi) restores differentiation [11, 12]. NCoR1/SMRT assembly in acute myeloid leukemia (AML) is the result of AML1/ETO fusion [11, 13]. It seems that the myeloid-Nervy-DEAF-1 (MYND) domain of AML1/ETO mediates the interaction with NCoR1/SMRT [13].
In solid tumors, however, the state and localization of corepressors is context-dependent. Their expression may change during disease progression and additional alterations may occur due to intervening transcription factors [11]. Prostate (androgen-independent), bladder and breast cancer cells often present with upregulation of NCoR1 and SMRT, a phenomenon that leads to epigenetic silencing of VDR, RAR, and PPARγ and their tumor-suppressive target genes [14–16]. It has also been postulated that ER promotes the proteosomal degradation of NCoR1 and consequently the loss of antiproliferative effect of VDR [11]. A combination of receptor ligands (natural or chemical) with HDACi may reverse the disrupted gene expression and provide an additional targeted therapy. Moreover, tamoxifen combined with HDACi has proven to be beneficial in cases of advanced breast cancer unresponsive to hormonal therapy. Co-administration of HDACi with an aromatase inhibitor is also effective for the treatment of ERα-negative and endocrine-resistant breast cancers [17]. Interestingly, ER facilitates breast cancer progression through the transcription of estrogen response element (ERE)-containing target genes and blockade of p53 antiproliferative actions via NCoR1/SMRT. These actions are inverted by antiestrogens [18]. Inhibition of mitogen-activated protein kinase (MAPK) signaling pathway promotes the recruitment of NCoR1/SMRT to tamoxifen-bound ERs [19]. ARs are important in prostate cancer. Activators and repressors compete for AR binding. Eventually, cancer becomes androgen-independent and resistant to corepressors [20]. NCoR1/SMRT bind to both agonist- and antagonist-bound ARs, which lessens the receptor’s tumor-promoting ability [21, 22]. Activated MAPK cascade may attenuate the corepressor recruitment [23]. Additionally, certain regions on the AR may influence the degree of the complex binding and concomitantly favoring coactivators [24]. Aberrant expression of NCoR1/SMRT has been observed in glioma specimens. Their presence may be associated with tumor proliferation, differentiation, and cancer stem cells. Pharmacologic inhibition via administration of RA and the protein phosphatase 1 (PP1) inhibitor okadaic acid leads to disease remission [5, 25]. Mutated nuclear receptors may alter the corepressor release in response to signals, as has been detected in cases of renal, hepatocellular, and thyroid carcinoma [26].
Finally, nuclear export of NCoR1/SMRT due to post-translational modifications may have an impact on the development of various cancers, including colorectal and endometrial [5, 11].
CoREST
CoREST is a transcription corepressor that was initially identified as an interacting factor with the REST repressor in neurogenesis. It functions as a docking station for the assembly of HDAC1/2, BHC80, BRAF35, and the lysine-specific histone demethylase 1 (LSD1) [27]. LSD1 is upregulated in various solid tumors, including breast [28], neuroblastoma [29], and prostate [30] cancer and has been correlated with a poor prognosis. Additionally, experimental data from colorectal cancer cell lines demonstrated the abnormal epigenetic silencing of tumor-suppressor genes by LSD1 [31]. LSD1 inhibition, in combination with other compounds (i.e., DNA methyltransferase inhibitors), may restore the expression of silenced genes [28–31]. CoREST can also be fused with ZNF217 in breast cancer, an event associated with the loss of transforming growth factor-beta (TGF-β) responsiveness and the suppression of tumor-suppressor genes, such as p15ink4b [32]. CoREST is also required in carcinogenesis, in cooperation with SWItch/Sucrose Non-Fermentable (SWI/SNF) and C-terminal-binding protein (CTBP) [11].
CTBP
CTBP 1 and 2, two evolutionary conserved and resembling corepressors that have been implicated in various steps of tumorigenesis and cancer progression, are considered as attractive targets of personalized medicine [33, 34]. CTBP inhibits transcription via interactions with transcription factors and the recruitment of chromatin-remodeling proteins, including HDAC1/2, LSD1, and Polycomb group proteins. The protein bears an intrinsic redox-sensing capacity through a dehydrogenase, and its activity is enhanced by nicotinamide adenine dinucleotide hydrogen (NADH) [33, 35]. Hypoxia and elevated extracellular glucose, a common tumor microenvironment, increases NADH and subsequently CTBP dimerization and activity [33, 34]. CTBP promotes proliferation, anti-apoptosis, epithelial-mesenchymal transition (EMT), and invasion by suppressing members of INK4 cell-cycle control family pro-apoptotic genes, such as Bax, E-cadherin and phosphatase and tensin homologue (PTEN) [34]. Tumor suppressors target and inactivate CTBP. Involvement in cancer, however, is context-dependent. In colorectal cancer, CTBP elevation is correlated with adenomatous polyposis coli (APC) and alternative reading frame (ARF) loss. Familiar adenomatous polyposis (FAP) patients present high levels of CTBP. Similarly, in breast and hepatocellular cancer, it suppresses ER and INK4 target genes, respectively. On the contrary, low CTBP levels in melanoma permit upregulation of T-cell factor/lymphoid-enhancing factor (TCF/LEF)-related genes, which facilitate invasiveness [11]. The compound 4-methylthio-2-oxobutyric acid (MTOB) has been utilized in both in vitro and in vivo experiments for the treatment of colorectal cancer. MTOB binding to CTBP triggers conformational changes that lead to its dislocation from the promoter of the targeted gene [33]. An alternative therapeutic method is the reduction of NADH levels via antioxidants, which disarray CTBP from its interacting proteins [36].
SIN3
SIN3 is a large protein scaffold that may function as both a coactivator and a corepressor. With its many protein-interacting domains, it can attract a wide variety of proteins and form a complex that, apart from its main enzymatic catalysts HDAC1/2, also contains breast cancer metastasis suppressor 1 (BRMS1), retinoblastoma-binding protein (RBBP) 4, RBBP7, Sin3A-associated protein (SAP)18, SAP30, ING1/2, and SD53 [37, 38]. SIN3 may directly interact with DNA-binding transcription factors as well as with other coregulators, such as NCoR1/SMRT. Post-translational modifications intervene in the complex’s activity. The broad repertoire of SIN3 actions includes control of cell cycle, DNA methylation, DNA damage repair and gene activation. Decreased levels of SIN3 have been reported in non-small cell lung cancer (NSCLC) and in clear cell renal carcinoma [37]. In ERα-positive breast cancer cells, however, SIN3 potentiates tumor proliferation by blocking pro-apoptotic genes [39]. Administration of a small-molecule inhibitor of the binding domain of SIN3 with its partner proteins hinders cell growth, forces the expression of silenced genes, and restores responsiveness to estradiol and retinoids in triple-negative breast cancer [40]. BRMS1 blocks several steps in the metastasis cascade by either recruiting chromatin-remodeling complexes or inhibiting NF-κB. BRMS1 is often lost in breast, melanoma, ovarian, and NSCLC and correlated with a poor outcome [41]. Recently, HDACi contributed to the deconstruction of the SIN3 complex and reactivation of silenced antiproliferative genes, such as p21 [42]. This notion suggests an alternative role apart from the profound inhibition of the catalytic center of corepressor complexes.
SWI/SNF
SWI/SNF is an ATP-dependent chromatin-remodeling complex. SWI/SNFs coordinate, by forming a dynamic equilibrium (activation/repression), a wide range of pathways involved in differentiation, self-renewal, and proliferation [43]. They are related to a diverse population of transcription factors, including AP-1, steroid receptors, Myc, p53, and Rb among others. The complexes contain the ATPase subunits Brahma (BRM) or Brahma-related gene 1 (BRG1), exclusively for each complex, accompanied by a set of regulatory proteins collectively termed BRM- or BRG1-associated factors (BAFs) [44]. Remodeling is achieved through mobilization of nucleosomes that involves both sliding and the insertion of histone octamers and HDAC recruitment [45]. Inactivating mutations of the SWI/SNF members, in addition to post-translational silencing modifications and protein stability issues, often occur in malignant tissues (breast, colon, ovaries, pancreas, melanoma, bladder), validating the tumor-suppressive role of the complex [43, 45]. Furthermore, the hypothesis that SWI/SNF is a bona fide tumor suppressor is supported via the involvement of ARID1A (BAF250A) subunit in a wide variety of cancers. ARID1A is frequently mutated in ovarian clear cell (46 %) and endometrioid (30 %) cancers and in uterine endometrioid tumors [46, 47]. ARID1A, in cooperation with p53, controls the expression of cyclin-dependent kinase inhibitor 1A (CDKN1A), which encodes p21, and SMAD3 and concomitantly cell proliferation in gynecologic cancers [46]. The ARID1A subunit is also aberrantly expressed in medulloblastomas, in breast and renal cancer, and in lung carcinoma cell lines [45]. Therapeutic approaches may incorporate selective HDACi and DNA/histone methyltransferase inhibitors, restoring the epigenetic silencing of the complex [43]. Surprisingly, tumors arising from loss of complementary proteins, leading to the dissociation of the complex, may depend on the residual ATPase activity of BRG1. Thus, small-molecule ATPase inhibitors represent an alternative, yet rational, strategy [48].
NURD
The NURD complex is an ATP-dependent chromatin-remodeling complex that can also recruit HDAC1,2 and LSD1. Additionally, the complex contains the RBBP4,7 and GATAD2A,2B structural subunits and the auxiliary proteins metastasis tumor antigen (MTA)1,2,3 and methyl-CpG-binding domain (MBD), which help the complex to interact with methylated DNA and transcription factors [44]. NURD is implicated in the preservation of DNA integrity. It may function, depending on the context, as a tumor promoter or repressor [49–51] and is involved in multiple stages of tumor initiation, progression, and migration. NURD complexes inhibit p53 through deacetylation interactions with SNAIL and TWIST in the process of EMT in many cancers. MTA1 is upregulated by the oncogene myc, which correlates with an invasive phenotype and poor clinical outcome in many cancers, including breast, colorectal, gastric, esophageal, endometrial, pancreatic, ovarian, NSCL, and prostate cancer [49, 50]. In breast cancer, the HER2 pathway upregulates MTA1 and both MTA1,2 block estrogen actions. On the other hand, MTA3 competes with MTA1, is regulated by estrogens, hampers EMT, and attracts LSD1-containing NURD complexes, which block tumor-promoting pathways, including TGF-β and MAPK [49, 50, 52]. Moreover, in APML, NURD is recruited by the fusion protein PML–RAR, impairing cell differentiation [51]. Experimental data suggest that MBD proteins recruit repressor complexes in hypermethylated promoters of tumor-suppressor genes to reinforce silencing. Putative therapeutic options are inhibition of the enzymatic region, e.g., HDACi, interventions in the associated proteins/pathways, and also alterations in the post-translational control [49]. The natural compound resveratrol exhibited p53-activating properties in prostate cancer cells through destabilization of NURD, caused by downregulation of MTA. HDACi enhanced this phenomenon [53]. The use of a low-molecular-mass compound that mimics the function of an MTA1 splice variant, which regulates estrogens’ nuclear localization, has proven to improve breast cancer in an in vivo model [26].
Other corepressor complexes
The number of corepressors engaged in tumorigenesis is constantly growing. Corepressors of agonist-bound nuclear receptors, such as LCoR and RIP140, may function as tumor suppressors in prostate and endometrial cancer, respectively [54, 55]. Preferentially expressed antigen in melanoma (PRAME) is found overexpressed and correlates with poor prognosis in several malignancies, including leukemia, multiple myeloma, lymphoma, and breast cancer. Its presence is related to HDAC-independent RA unresponsiveness [9]. The Polycomb-repressive complexes (PRC) 1 and 2 are considered putative tumor-promoting genes. PRC1 possesses histone-ubiquitylation abilities, while PRC2 recruits the methyltransferase EZH2. The complexes promote tumorigenesis through the inhibition of differentiation and the promotion of self-renewal. Augmented levels have been detected in breast, prostate, and colorectal cancer and leukemias. Their expression has been related to the presence of cancer stem cells [56]. Other newly introduced cancer-related corepressors are the Groucho/Transducin-Like Enhancer of split (TLE) proteins and the Alien [57, 58].
Therapeutic potential
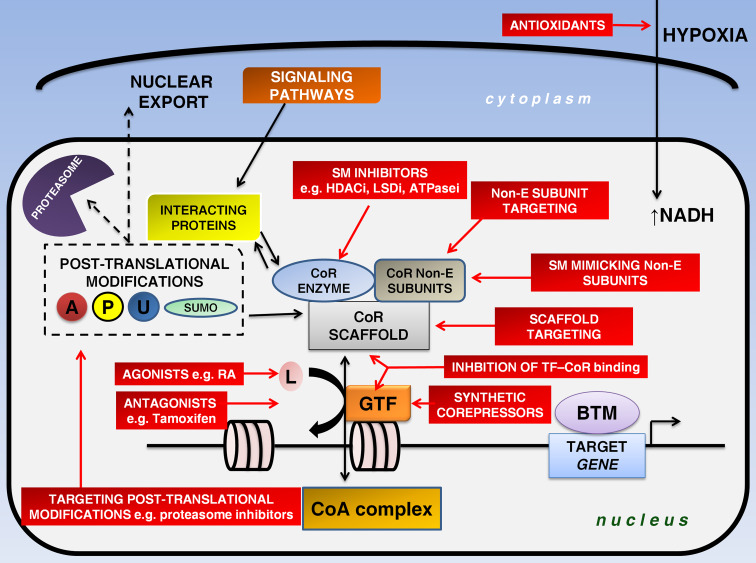
Epigenetic modifications, in contrast to gene mutations, represent a reversible process that has the potential to be altered through the administration of small molecules in combination with conventional tumor therapeutic measures, bearing in mind the contextual nature of corepressors [5, 59]. A broad spectrum of targets for the design of small-molecule drugs exists within a corepressor complex, namely (i) enzymatic region, (ii) complex dissociation, (iii) transcription factor-interacting area, (iv) derepression, (v) corepressor-related pathways and (vi) artificial corepressors with high specificity [11, 26, 33, 40, 45, 49, 53, 60] (Fig. 1).
Fig. 1.
Rationale behind corepressor targeting. Corepressors fine-tune epigenetic signaling in cooperation with gene-specific transcription factors and other interacting proteins. Ligand binding regulates the competition between coregulators (repressors and activators). Post-translational modifications (acetylation, phosphorylation, ubiquitylation, sumoylation) may alter the activity of a corepressor. Red rectangles represent putative therapeutic targets. The enzymatic component of the complex is a prime target of small-molecule inhibitors. An alternative approach entails destabilization of the complex that can be achieved through targeting of the complex subunits or the use of small molecules that mimic normal subunits. Additionally, the interaction between corepressors and transcription factors may also be blocked. Recently, the generation of a synthetic corepressor was presented. Corepressor therapies seem to be more beneficial when combined with agonist or antagonist administration. A acetylation, ATPasei adenosine triphosphatase inhibitor, BTM basal transcriptional machinery, CoA coactivator, CoR corepressor, GTF gene-specific transcription factor, HDACi histone deacetylase inhibitor, L ligand, LSDi lysine-specific histone demethylase inhibitor, Non-E non-enzymatic, P phosphorylation, RA retinoic acid, SM small-molecule, SUMO sumoylation, TF transcription factor, U ubiquitylation
The family of HDACs comprises 11 members and it is subdivided into three classes: I (1–3, 8), II (4–7, 9, 10), and III (11) [5]. Acetylation of histone tails results in chromatin loosening and hence enhances transcription, whereas removal of acetyl-groups has the exact opposite effect [12]. Class I members catalyze chromatin modifications for most corepressor complexes [5]. A volume of data indicates the link between abnormal HDAC expression and carcinogenesis [12]. Reasonably, pharmacological HDAC inhibition has become an appealing target [59, 61]. Natural and synthetic compounds have been utilized in this direction. Vorinostat and romidepsin gained approval for the treatment of cutaneous T-cell lymphoma [61]. It seems that additional benefit occurs from the combination of HDACi with agonists or antagonists of related receptors, as in the case of APML and breast cancer [11, 53, 62]. Recently, it was reported that the production of specific HDACi may be feasible [63]. Similarly, histone methyltransferase and demethylase inhibitors may function alone or in combination with compounds like RA and tamoxifen [12, 62].
Concluding remarks
Transcription factor corepressors represent a class of epigenetic silencers, which although not the leading actors in the transcriptional scenery, they mediate a crucial “behind-the-scene” role through the manipulation of various transcription factors and their target genes. Corepressors function as docking stations and facilitate the assembly of histone modifying enzymes along with auxiliary proteins. Aberrant expression has been observed in various types of malignancies, including leukemias, breast, and prostate cancer among others. Elevated expression, as in the case of NCoR1 and SMRT, may lead to inhibition of tumor-suppressive genes. By contrast, corepressor silencing, such as BRMS1, may enhance the expression of tumor-promoting genes. Corepressors have become rational targets in the pursuit of higher therapeutic efficacy combined with less side effects. Evidently, the era of translational medicine opens a new horizon and highlights the need of renovating our therapeutic arsenal towards individuality.
References
- 1.Grivas PD, Kiaris H, Papavassiliou AG. Tackling transcription factors: challenges in antitumor therapy. Trends Mol Med. 2011;17:537–538. doi: 10.1016/j.molmed.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 2.Lonard DM, O’Malley BW. Nuclear receptor coregulators: judges, juries, and executioners of cellular regulation. Mol Cell. 2007;27:691–700. doi: 10.1016/j.molcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 3.Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 4.Payankaulam S, Li LM, Arnosti DN. Transcriptional repression: conserved and evolved features. Curr Biol. 2010;20:R764–R771. doi: 10.1016/j.cub.2010.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nat Rev Gen. 2010;11:109–123. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- 6.Stewart MD, Wong J. Nuclear receptor repression: regulatory mechanisms and physiological implications. Prog Mol Biol Transl Sci. 2009;87:235–259. doi: 10.1016/S1877-1173(09)87007-5. [DOI] [PubMed] [Google Scholar]
- 7.Lonard DM, Lanz RB, O’Malley BW. Nuclear receptor coregulators and human disease. Endocr Rev. 2007;28:575–587. doi: 10.1210/er.2007-0012. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gurevich I, Flores AM, Aneskievich BJ. Corepressors of agonist-bound nuclear receptors. Toxicol Appl Pharmacol. 2007;223:288–298. doi: 10.1016/j.taap.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watson PJ, Fairall L, Schwabe JW. Nuclear hormone receptor co-repressors: structure and function. Mol Cell Endocrinol. 2012;348:440–449. doi: 10.1016/j.mce.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Battaglia S, Maguire O, Campbell MJ. Transcription factor co-repressors in cancer biology: roles and targeting. Int J Cancer. 2010;126:2511–2519. doi: 10.1002/ijc.25181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biancotto C, Frige G, Minucci S. Histone modification therapy of cancer. Adv Genet. 2010;70:341–386. doi: 10.1016/B978-0-12-380866-0.60013-7. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Chen W, Gaudet J, Cheney MD, Roudaia L, Cierpicki T, Klet RC, Hartman K, Laue TM, Speck NA, Bushweller JH. Structural basis for recognition of SMRT/N-CoR by the MYND domain and its contribution to AML1/ETO’s activity. Cancer Cell. 2007;11:483–497. doi: 10.1016/j.ccr.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abedin SA, Banwell CM, Colston KW, Carlberg C, Campbell MJ. Epigenetic corruption of VDR signalling in malignancy. Anticancer Res. 2006;26:2557–2566. [PubMed] [Google Scholar]
- 15.Abedin SA, Thorne JL, Battaglia S, Maguire O, Hornung LB, Doherty AP, Mills IG, Campbell MJ. Elevated NCOR1 disrupts a network of dietary-sensing nuclear receptors in bladder cancer cells. Carcinogenesis. 2009;30:449–456. doi: 10.1093/carcin/bgp005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Battaglia S, Maguire O, Thorne JL, Hornung LB, Doig CL, Liu S, Sucheston LE, Bianchi A, Khanim FL, Gommersall LM, Coulter HS, Rakha S, Giddings I, O’Neill LP, Cooper CS, McCabe CJ, Bunce CM, Campbell MJ. Elevated NCOR1 disrupts PPARalpha/gamma signaling in prostate cancer and forms a targetable epigenetic lesion. Carcinogenesis. 2010;31:1650–1660. doi: 10.1093/carcin/bgq086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mann M, Cortez V, Vadlamudi RK. Epigenetics of estrogen receptor signaling: role in hormonal cancer progression and therapy. Cancers. 2011;3:1691–1707. doi: 10.3390/cancers3021691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konduri SD, Medisetty R, Liu W, Kaipparettu BA, Srivastava P, Brauch H, Fritz P, Swetzig WM, Gardner AE, Khan SA, Das GM. Mechanisms of estrogen receptor antagonism toward p53 and its implications in breast cancer therapeutic response and stem cell regulation. Proc Natl Acad Sci USA. 2010;107:15081–15086. doi: 10.1073/pnas.1009575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong W, Chen L, Li J, Yao Z. Inhibition of MAP kinase promotes the recruitment of corepressor SMRT by tamoxifen-bound estrogen receptor alpha and potentiates tamoxifen action in MCF-7 cells. Biochem Biophys Res Commun. 2010;396:299–303. doi: 10.1016/j.bbrc.2010.04.085. [DOI] [PubMed] [Google Scholar]
- 20.Reeb CA, Gerlach C, Heinssmann M, Prade I, Ceraline J, Roediger J, Roell D, Baniahmad A. A designed cell-permeable aptamer-based corepressor peptide is highly specific for the androgen receptor and inhibits prostate cancer cell growth in a vector-free mode. Endocrinology. 2011;152:2174–2183. doi: 10.1210/en.2011-0149. [DOI] [PubMed] [Google Scholar]
- 21.Chmelar R, Buchanan G, Need EF, Tilley W, Greenberg NM. Androgen receptor coregulators and their involvement in the development and progression of prostate cancer. Int J Cancer. 2007;120:719–733. doi: 10.1002/ijc.22365. [DOI] [PubMed] [Google Scholar]
- 22.van de Wijngaart DJ, Dubbink HJ, van Royen ME, Trapman J, Jenster G. Androgen receptor coregulators: recruitment via the coactivator binding groove. Mol Cell Endocrinol. 2012;352:57–69. doi: 10.1016/j.mce.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Eisold M, Asim M, Eskelinen H, Linke T, Baniahmad A. Inhibition of MAPK-signaling pathway promotes the interaction of the corepressor SMRT with the human androgen receptor and mediates repression of prostate cancer cell growth in the presence of antiandrogens. J Mol Endocrinol. 2009;42:429–435. doi: 10.1677/JME-08-0084. [DOI] [PubMed] [Google Scholar]
- 24.Buchanan G, Need EF, Barrett JM, Bianco-Miotto T, Thompson VC, Butler LM, Marshall VR, Tilley WD, Coetzee GA. Corepressor effect on androgen receptor activity varies with the length of the CAG encoded polyglutamine repeat and is dependent on receptor/corepressor ratio in prostate cancer cells. Mol Cell Endocrinol. 2011;342:20–31. doi: 10.1016/j.mce.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campos B, Bermejo JL, Han L, Felsberg J, Ahmadi R, Grabe N, Reifenberger G, Unterberg A, Herold-Mende C. Expression of nuclear receptor corepressors and class I histone deacetylases in astrocytic gliomas. Cancer Sci. 2011;102:387–392. doi: 10.1111/j.1349-7006.2010.01792.x. [DOI] [PubMed] [Google Scholar]
- 26.Hsia EY, Goodson ML, Zou JX, Privalsky ML, Chen HW. Nuclear receptor coregulators as a new paradigm for therapeutic targeting. Adv Drug Deliv Rev. 2010;62:1227–1237. doi: 10.1016/j.addr.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lakowski B, Roelens I, Jacob S. CoREST-like complexes regulate chromatin modification and neuronal gene expression. J Mol Neurosci. 2006;29:227–239. doi: 10.1385/JMN:29:3:227. [DOI] [PubMed] [Google Scholar]
- 28.Lim S, Janzer A, Becker A, Zimmer A, Schule R, Buettner R, Kirfel J. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis. 2010;31:512–520. doi: 10.1093/carcin/bgp324. [DOI] [PubMed] [Google Scholar]
- 29.Schulte JH, Lim S, Schramm A, Friedrichs N, Koster J, Versteeg R, Ora I, Pajtler K, Klein-Hitpass L, Kuhfittig-Kulle S, Metzger E, Schule R, Eggert A, Buettner R, Kirfel J. Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy. Cancer Res. 2009;69:2065–2071. doi: 10.1158/0008-5472.CAN-08-1735. [DOI] [PubMed] [Google Scholar]
- 30.Kahl P, Gullotti L, Heukamp LC, Wolf S, Friedrichs N, Vorreuther R, Solleder G, Bastian PJ, Ellinger J, Metzger E, Schule R, Buettner R. Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence. Cancer Res. 2006;66:11341–11347. doi: 10.1158/0008-5472.CAN-06-1570. [DOI] [PubMed] [Google Scholar]
- 31.Huang Y, Stewart TM, Wu Y, Baylin SB, Marton LJ, Perkins B, Jones RJ, Woster PM, Casero RA., Jr Novel oligoamine analogues inhibit lysine-specific demethylase 1 and induce reexpression of epigenetically silenced genes. Clin Cancer Res. 2009;15:7217–7228. doi: 10.1158/1078-0432.CCR-09-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thillainadesan G, Isovic M, Loney E, Andrews J, Tini M, Torchia J. Genome analysis identifies the p15ink4b tumor suppressor as a direct target of the ZNF217/CoREST complex. Mol Cell Biol. 2008;28:6066–6077. doi: 10.1128/MCB.00246-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Straza MW, Paliwal S, Kovi RC, Rajeshkumar B, Trenh P, Parker D, Whalen GF, Lyle S, Schiffer CA, Grossman SR. Therapeutic targeting of C-terminal binding protein in human cancer. Cell Cycle. 2010;9:3740–3750. doi: 10.4161/cc.9.18.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chinnadurai G. The transcriptional corepressor CtBP: a foe of multiple tumor suppressors. Cancer Res. 2009;69:731–734. doi: 10.1158/0008-5472.CAN-08-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao LZ, Chinnadurai G. Incapacitating CtBP to kill cancer. Cell Cycle. 2010;9:3645–3646. doi: 10.4161/cc.9.18.13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng Y, Liu J, Han G, Lu SL, Wang SY, Malkoski S, Tan AC, Deng C, Wang XJ, Zhang Q. Redox-dependent Brca1 transcriptional regulation by an NADH-sensor CtBP1. Oncogene. 2010;29:6603–6608. doi: 10.1038/onc.2010.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grzenda A, Lomberk G, Zhang JS, Urrutia R. Sin3: master scaffold and transcriptional corepressor. Biochim Biophys Acta. 2009;1789:443–450. doi: 10.1016/j.bbagrm.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silverstein RA, Ekwall K. Sin3: a flexible regulator of global gene expression and genome stability. Curr Genet. 2005;47:1–17. doi: 10.1007/s00294-004-0541-5. [DOI] [PubMed] [Google Scholar]
- 39.Ellison-Zelski SJ, Alarid ET. Maximum growth and survival of estrogen receptor-alpha positive breast cancer cells requires the Sin3A transcriptional repressor. Mol Cancer. 2010;9:263. doi: 10.1186/1476-4598-9-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farias EF, Petrie K, Leibovitch B, Murtagh J, Chornet MB, Schenk T, Zelent A, Waxman S. Interference with Sin3 function induces epigenetic reprogramming and differentiation in breast cancer cells. Proc Natl Acad Sci USA. 2010;107:11811–11816. doi: 10.1073/pnas.1006737107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hurst DR, Welch DR. Unraveling the enigmatic complexities of BRMS1-mediated metastasis suppression. FEBS Lett. 2011;585:3185–3190. doi: 10.1016/j.febslet.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith KT, Martin-Brown SA, Florens L, Washburn MP, Workman JL. Deacetylase inhibitors dissociate the histone-targeting ING2 subunit from the Sin3 complex. Chem Biol. 2010;17:65–74. doi: 10.1016/j.chembiol.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene. 2009;28:1653–1668. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- 44.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 46.Guan B, Wang TL, Shih IeM. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res. 2011;71:6718–6727. doi: 10.1158/0008-5472.CAN-11-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, Yang W, Heravi-Moussavi A, Giuliany R, Chow C, Fee J, Zayed A, Prentice L, Melnyk N, Turashvili G, Delaney AD, Madore J, Yip S, McPherson AW, Ha G, Bell L, Fereday S, Tam A, Galletta L, Tonin PN, Provencher D, Miller D, Jones SJ, Moore RA, Morin GB, Oloumi A, Boyd N, Aparicio SA, Shih IeM, Mes-Masson AM, Bowtell DD, Hirst M, Gilks B, Marra MA, Huntsman DG. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X, Sansam CG, Thom CS, Metzger D, Evans JA, Nguyen PT, Roberts CW. Oncogenesis caused by loss of the SNF5 tumor suppressor is dependent on activity of BRG1, the ATPase of the SWI/SNF chromatin remodeling complex. Cancer Res. 2009;69:8094–8101. doi: 10.1158/0008-5472.CAN-09-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lai AY, Wade PA. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat Rev Cancer. 2011;11:588–596. doi: 10.1038/nrc3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manavathi B, Singh K, Kumar R. MTA family of coregulators in nuclear receptor biology and pathology. Nucl Recept Signal. 2007;5:e010. doi: 10.1621/nrs.05010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramirez J, Hagman J. The Mi-2/NuRD complex: a critical epigenetic regulator of hematopoietic development, differentiation and cancer. Epigenetics. 2009;4:532–536. doi: 10.4161/epi.4.8.10108. [DOI] [PubMed] [Google Scholar]
- 52.Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, Liang J, Sun L, Yang X, Shi L, Li R, Li Y, Zhang Y, Li Q, Yi X, Shang Y. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009;138:660–672. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- 53.Kai L, Samuel SK, Levenson AS. Resveratrol enhances p53 acetylation and apoptosis in prostate cancer by inhibiting MTA1/NuRD complex. Int J Cancer. 2010;126:1538–1548. doi: 10.1002/ijc.24928. [DOI] [PubMed] [Google Scholar]
- 54.Asim M, Hafeez BB, Siddiqui IA, Gerlach C, Patz M, Mukhtar H, Baniahmad A. Ligand-dependent corepressor acts as a novel androgen receptor corepressor, inhibits prostate cancer growth, and is functionally inactivated by the Src protein kinase. J Biol Chem. 2011;286:37108–37117. doi: 10.1074/jbc.M111.292771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng YH, Utsunomiya H, Pavone ME, Yin P, Bulun SE. Retinoic acid inhibits endometrial cancer cell growth via multiple genomic mechanisms. J Mol Endocrinol. 2011;46:139–153. doi: 10.1530/JME-10-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Richly H, Aloia L, Di Croce L. Roles of the Polycomb group proteins in stem cells and cancer. Cell Death Dis. 2011;2:e204. doi: 10.1038/cddis.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jennings BH, Ish-Horowicz D. The Groucho/TLE/Grg family of transcriptional co-repressors. Genome Biol. 2008;9:205. doi: 10.1186/gb-2008-9-1-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Papaioannou M, Melle C, Baniahmad A. The coregulator alien. Nucl Recept Signal. 2007;5:e008. doi: 10.1621/nrs.05008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Graham JS, Kaye SB, Brown R. The promises and pitfalls of epigenetic therapies in solid tumours. Eur J Cancer. 2009;45:1129–1136. doi: 10.1016/j.ejca.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 60.Cerchietti LC, Ghetu AF, Zhu X, Da Silva GF, Zhong S, Matthews M, Bunting KL, Polo JM, Farès C, Arrowsmith CH, Yang SN, Garcia M, Coop A, Mackerell AD, Jr, Privé GG, Melnick A. A small-molecule inhibitor of BCL6 kills DLBCL cells in vitro and in vivo. Cancer Cell. 2010;17:400–411. doi: 10.1016/j.ccr.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dworkin AM, Huang TH, Toland AE. Epigenetic alterations in the breast: implications for breast cancer detection, prognosis and treatment. Semin Cancer Biol. 2009;19:165–171. doi: 10.1016/j.semcancer.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bantscheff M, Hopf C, Savitski MM, Dittmann A, Grandi P, Michon AM, Schlegl J, Abraham Y, Becher I, Bergamini G, Boesche M, Delling M, Dumpelfeld B, Eberhard D, Huthmacher C, Mathieson T, Poeckel D, Reader V, Strunk K, Sweetman G, Kruse U, Neubauer G, Ramsden NG, Drewes G. Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat Biotechnol. 2011;29:255–265. doi: 10.1038/nbt.1759. [DOI] [PubMed] [Google Scholar]