Abstract
Intracellular deposits of aggregated alpha-synuclein are a hallmark of Parkinson’s disease. Protein–protein interactions are critical in the regulation of cell proteostasis. Synphilin-1 interacts both in vitro and in vivo with alpha-synuclein promoting its aggregation. We report here that synphilin-1 specifically inhibits the degradation of alpha-synuclein wild-type and its missense mutants by the 20S proteasome due at least in part by the interaction of the ankyrin and coiled-coil domains of synphilin-1 (amino acids 331–555) with the N-terminal region (amino acids 1–60) of alpha-synuclein. Co-expression of synphilin-1 and alpha-synuclein wild-type in HeLa and N2A cells produces a specific increase in the half-life of alpha-synuclein, as degradation of unstable fluorescent reporters is not affected. Synphilin-1 inhibition can be relieved by co-expression of Siah-1 that targets synphilin-1 to degradation. Synphilin-1 inhibition of the proteasomal pathway of degradation of alpha-synuclein may help to understand the pathophysiological changes occurring in PD and other synucleinopathies.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-010-0592-3) contains supplementary material, which is available to authorized users.
Keywords: Synphilin, Synuclein, Proteasome, Proteolysis, Degradation, Parkinson, Synucleinopathies
Introduction
Parkinson’s disease (PD), and other synucleinopathies, are characterized pathologically by proteinaceous inclusions commonly referred to as Lewy pathology in postmortem brain tissue samples [1]. Alpha-synuclein is the major protein constituent of Lewy bodies [2]. Three missense point mutations (Ala53Thr, Ala30Pro, Glu46Lys) in the alpha-synuclein gene have been linked to dominant early onset familial PD [3–5]. Locus duplication [6, 7] and triplication [8] have been described in certain forms of familial PD and hypomethylation of alpha-synuclein intron 1 is found in sporadic PD patients [9], indicating that increased expression of alpha-synuclein is also directly involved in the pathogenesis of Lewy bodies diseases and neurodegeneration, as exemplified in several animal models [10].
Alpha-synuclein is a small acidic protein of 140 amino acids. The N-terminal part of the protein contains seven imperfect repeats with the consensus core sequence Lys-Thr-Lys-Glu-Gly-Val. The C-terminal portion has no recognized motifs except for a strong negative charge. The precise function of alpha-synuclein is not known. Alpha-synuclein is a natively unfolded protein with the ability of self-aggregation in a nucleation-dependent process to both non-fibrillar oligomers, protofibrils, and fibrillar aggregates with amyloid properties that are potentially cytotoxic [11].
In globular proteins, binding promiscuity is favored through unstructured protein regions. In fact, both the intrinsic disorder content and the linear motif content of a protein are predictive of dosage sensitivity in yeast, because promiscuous (“off-target”) low-affinity interactions are favored by over-expression due to simple mass action [12]. Naturally unfolded proteins, like alpha-synuclein, are likely to be promiscuous in the cell, and both gain-of-function mutations and over expression, alpha-synuclein gene duplication, triplication, and hypomethylation of alpha-synuclein intron 1 (see references above), could be deleterious for the cell [13]. Alpha-synuclein fits well into this theoretical framework. Alpha-synuclein has been described to associate with many protein partners in the cell [14]. Among those partners it has been described that alpha-synuclein can associate with: Tau [15], 14.3.3 proteins [16–18], synphilin [19, 20], tubulin [21], and phospholipase D2 promoting its inhibition [22, 23] inhibitory effects not replicated in a recent publication [24]. The natural tendency of alpha-synuclein to aggregate may be the reason that several chaperones also interact with alpha-synuclein. In fact, αB-crystallin [25, 26] and Hsp90 [27] have been shown to be associated with synuclein aggregates in the brain. Hsp70 over-expression, cooperating with Hsp40, has been shown to prevent alpha-synuclein aggregation and toxicity in cell and Drosophila models that over-expressed alpha-synuclein [28–31], while no beneficial effect has been recently reported in A53T transgenic mice [32].
One way to study the strength of alpha-synuclein interaction with its potential protein partners is to get an in vitro read-out that can score the “strength” of the interactions. A general experimental approach to study protein–protein interactions uses the sensitivity of protein partners to proteases as an indirect measurement of the affinity and the mass action of the equilibrium of the protein–protein complex. In this approach, two proteins in solution may form a heterodimer, A + B ↔ AB, and the relative concentrations of each molecular species is dependent on both the amount of both protein partners and the affinity constant. When a protease is added to the mixture, A, B, and the complex AB can be degraded, either partially or totally with different rates, or not degraded. Partial or limited degradation of both partners has also been used to map protein regions of A and B that are resistant to proteolysis due to AB complex formation.
The rational of our work was to approach the relevance of alpha-synuclein protein–protein interactions by in vitro studies of the degradation of alpha-synuclein by the proteasome, as it has been shown that alpha-synuclein is directly degraded by the 20S proteasome [33–35] and may also participate in alpha-synuclein degradation in situ [36, 37].
We performed initial studies with tubulin, the results obtained showed that tubulin is not degraded, and does not significantly affect the degradation of alpha-synuclein, by the 20S proteasome. As a consequence, we direct our attention to synphilin-1 because it has been shown to interact in vivo with alpha-synuclein [19]. Synphilin-1 promotes inclusion formation in cells [19, 38] and is also present in Lewy bodies [39]. Synphilin-1 has been shown to interact through its central domain amino acids 349–555 with the N-terminal (1–65) region of alpha-synuclein by yeast two-hybrid analysis [20] and more recently Xie et al. [40] reported a detailed in vitro study of the interaction of alpha-synuclein and synphilin-1 confirming and extending those previous results.
Here we show that synphilin-1 specifically inhibits in a dose-dependent manner the degradation of alpha-synuclein by the 20S proteasome, this inhibition is due, at least in part, to the interaction of synphilin-1 central region 331–555 with the N-terminal region 1–60 of alpha-synuclein. Furthermore, over-expression of synphilin-1 by cell transfection increases the half-life of alpha-synuclein. These results demonstrate for the first time that synphilin-1 acts on alpha-synuclein turn-over preventing its degradation by the proteasome and this inhibition of alpha-synuclein degradation can be reversed by targeting of synphilin-1 to degradation by ubiquitylation mediated by Siah-1 over-expression.
Materials and methods
Plasmid constructs
Human synphilin-1 331–919 and 331–555 were obtained by PCR with the appropriate oligonucleotides (forward 331 NdeI, 5′-CGGCTACATATGATCTCTCTCCTGCCACACC-3′; reverse 555 SalI, 5′-GCGAGCGTCGACTTACTTGCCCTCTGATTTCTGGGC-3′; and reverse 919 SalI, 5′-CGCGACGTCGACTTATGCTGCCTTATTCTTTCCTTT-3′) using pcDNA 3.1 hSynphilin-1 construct was kindly provided by Mark R. Cookson (National Institutes of Health, Bethesda, MD, USA) [41] and subcloned into the NdeI/XhoI sites of pET15B vector. Human wild-type alpha-synuclein cDNA was obtained by PCR with appropriate oligonucleotides from a human placental cDNA library and cloned into pT7-7 and pcDNA 3.1 vectors, pT7-7 Alpha-synuclein A30P, A53T and E46K were obtained by PCR with QuikChange™ Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA), as described [42].
His-tag full length alpha-synuclein, His-tag alpha-synuclein 1–60 and 61–140 synuclein fragments were obtained by PCR from pcDNA construct with the appropriate oligonucleotides forward BglII alpha-synuclein full length, 5′-GGCGGCGAGATCTATGGATGTATTCATGAAAGG-3′; forward for synuclein 61–140, 5′-GGCCACATCTATGGAGCAAGTGACAAATGTTGGAGG-3′; reverse for full length and 61–140 construct SalI, 5′-AGAGTCGACTTAGGCTTCAGGTTCG-3′; reverse for 1–60 construct, SalI, 5′-GGGCCGTCGACCTATTTGGTCTTCTCAGCCACT-3′ and cloned into the BamH1/XhoI sites of pRSET-A vector. Myc-synphilin-1, HA-Siah-1 tagged cDNAs were kindly provided by Dr. Simone Engelender (Technion-Israel Institute of Technology, Haifa, Israel).
Protein purification
Rat liver 20S proteasome was purified as described [43]. Recombinant His-synphilin-1 331–919 and 331–555, His-alpha-synuclein full length, His-alpha-synuclein 1–60 and 61–140 were obtained from induced bacterial cultures and purified using Ni–NTA agarose (Qiagen, Hilden, Germany), according to the manufacturer’s instructions, and dialyzed against 25 mM Tris–Cl, pH 7.5, 25 mM NaCl, insoluble proteins were removed by centrifugation at 100,000 × g for 30 min. Wild-type, A30P, A53T, and E46K α-synucleins were purified as described [42]. Tubulin dimers were purified from rat brain after three cycles of polymerization as described [44].
In vitro degradation assays
Degradation reactions were incubated at 37°C for the times indicated and contained in a final volume of 20 μl: 20 mM HEPES, pH 7.4, 2 mM EDTA, 1 mM EGTA, 2 μg of alpha-synuclein (7 μM, molecular mass: 14,000 Da), and 0.25–2.5 μg of purified rat liver proteasome (17.36–173.6 nM, molecular mass: 720,000 Da). To study the effect of synphilin-1 in the degradation of alpha-synuclein by the 20S proteasome, 1 μg His-synphilin-1 331–919 (0.75 μM, calculated monomeric molecular mass: 66,433 Da), 0.5 μg His-synphilin-1 331–555 (0.92 μΜ, calculated monomeric molecular mass: 27,062 Da), were preincubated with alpha-synuclein for 15 min at RT with occasional shaking, prior its addition to the degradation mixture containing the 20S proteasome. Reactions were stopped at different times with concentrated SDS-PAGE sample buffer and after boiling for 5 min were loaded onto 14% SDS-PAGE. Gels were stained (Coomassie Blue), destained and dried under vacuum. Transfer and immunoblots were performed with anti-alpha-synuclein antibodies (see below) or anti-His-tag antibodies (1:1,000, Novagen, USA). Quantification was performed using the Quantity-one software (Bio-Rad SA, Madrid, Spain).
Cell degradation assays
HeLa and N2A cell lines were cultured in Dulbecco's modified Eagle’s media containing 10% fetal bovine serum. Cells were plated in 35-mm dishes and transiently transfected with the indicated DNAs using Lipofectamine 2000 reagent (Invitrogen SA, Barcelona, Spain). Typically 300,000 cells were transfected with 200 ng of pcDNA alpha-synuclein expression vectors. Where indicated, 400 ng of Myc-synphilin-1 and/or 400 ng of HA-Siah-1 were used. Total transfected DNA was kept constant to 1 μg by adding corresponding amounts of empty pcDNA vector as carrier. Treatments with cycloheximide (25 μg/ml) and lactacystin (10 μM) (both from Sigma-Aldrich, St. Louis, MO, USA) were performed 48 h after transfection, and the culture media were replaced every 12 h during the course of the experiments. Cells were collected at the indicated times, washed by centrifugation at 4°C with cold PBS and directly resuspended in concentrated SDS-PAGE sample buffer, all time points were done by duplicate and cell viability was assessed by Trypan Blue exclusion, >90% cells were viable. Protein expression was analyzed by Western immunoblotting with the following antibodies: anti-synuclein (1:2,000, Syn-1, BD Biosciences, Franklin Lakes, NJ, USA), anti-Myc tag (1:5,000, clone 9B11, Cell Signalling, Danvers, MA, USA) and anti-HA tag (1:1,000, clone 3F10, Roche, Mannheim, Germany). Anti-α-tubulin monoclonal antibody (1:5,000, Sigma) was used as the control of protein loading. Quantification was performed by chemiluminescence using DNR MF-ChemiBIS 3.2 and Totallab TL100 (DNR Bio-Imaging Systems Ltd, Jerusalem, Israel). Similar experiments were performed with unstable fluorescent protein constructs EGFPd2 and EGFPu and analyzed by immunoblotting as described previously [45]. Results presented are expressed as mean ± SEM for three independent experiments.
Results
Wild-type synuclein and the missense mutants are degraded by the 20S proteasome
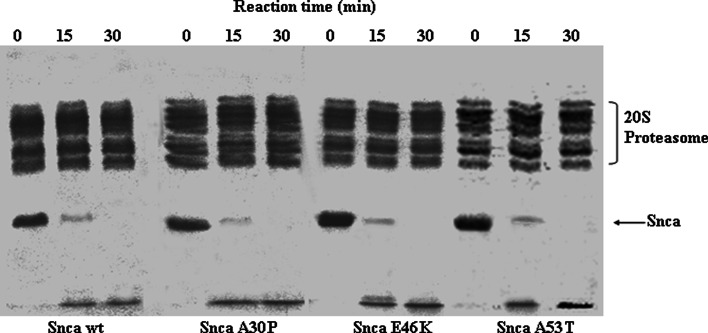
Wild-type alpha-synuclein is directly degraded by the 20S proteasome [33–35]. We extend here those observations demonstrating that alpha-synuclein wild-type and its missense mutants (A30P, E46K, and A53T) are also efficiently degraded by the 20S proteasome as shown in Fig. 1, and the degradation was completely prevented by the presence of proteasome inhibitors (see Supplementary Fig. 1). Furthermore, there is no significant difference in the kinetic parameters of the degradation of the different alpha-synuclein proteins by the proteasome; apparent half-saturation at 5 ± 1 μM and V max of 3.4 ± 0.4 nmoles of alpha-synuclein degraded per minute per nmole of 20S proteasome, which corresponds to a turnover number of 0.056 ± 0.006 s−1.
Fig. 1.
Time course of the degradation of alpha-synuclein by 20S proteasome. Purified alpha-synuclein (Snca) wild-type, A30P, E46K, and A53T were incubated with purified 20S proteasome as indicated in the figure. Results show representative Coomassie Blue-stained gels using 1 μg (3.57 μM) of the respective alpha-synuclein and 2.5 μg (173 nM) of 20S proteasome in the reaction mixtures
Synphilin inhibits the degradation of synuclein by 20S proteasome
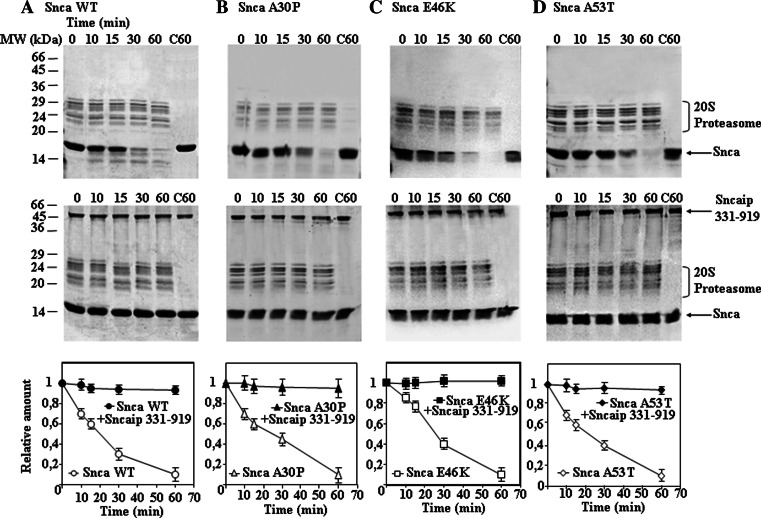
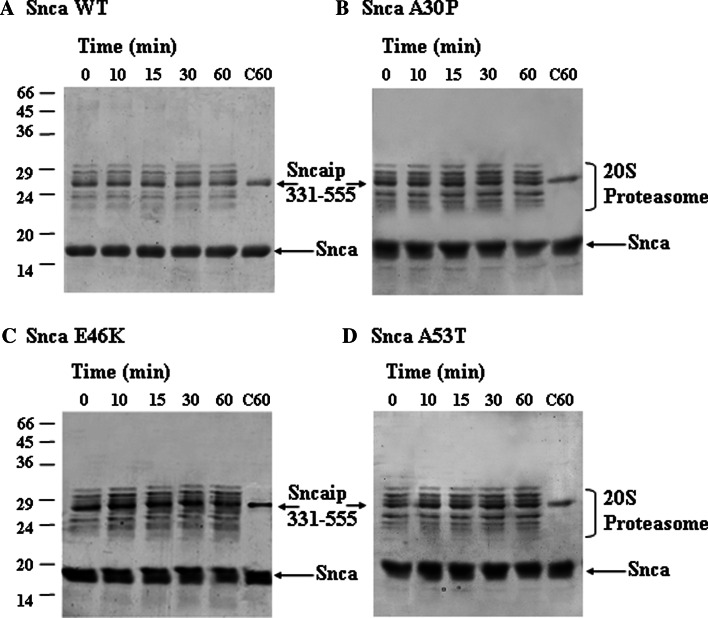
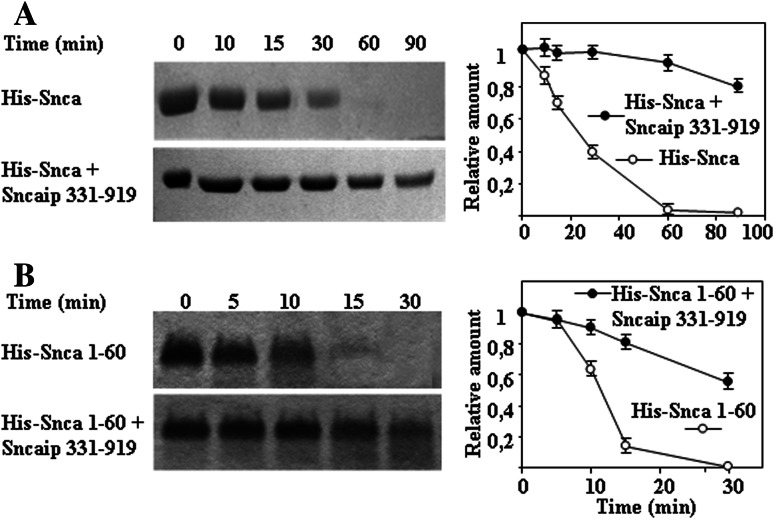
Initially, we studied the possible effect of tubulin on alpha-synuclein degradation and found that tubulin (5 μM) is not degraded by the proteasome, and has no effect on the time course of degradation of alpha-synuclein (3 μM) by the 20S proteasome (69.4 nM). As a consequence, we decided to study if synphilin-1 interaction with alpha-synuclein could affect synuclein degradation by the 20S proteasome. We were unable to obtain bacterial expression of the full-length synphilin-1 in sufficient amounts due to its aggregation into inclusion bodies and the need of 8 M urea for its solubilization casting concerns about getting the corrected folded structure of the synphilin-1 after removal of urea. Guided by the previous work of Neystat et al. [20] the His-tag version of synphilin-1 from amino acids 331 to 919 was obtained and effectively produced in bacteria in a soluble form and purified with enough good yields to perform further experiments. Results presented in Fig. 2 show that synphilin-1 331–919 was able to inhibit the degradation of alpha-synuclein wild-type and the missense mutants (A30P, E46K, and A53T). Synphilin-1 331–919 alone (see Supplementary Fig. 2) or in the presence of alpha-synuclein (see also Fig. 2) is not significantly degraded by the 20S proteasome. Results presented in Fig. 2 were performed with 7 μM alpha-synuclein and 0.75 μM synphilin-1 331–919 (based on monomeric molecular mass) in the presence of 69.4 nM 20S proteasome, and similar results were obtained if analysis was performed by Western immunoblotting with anti-alpha-synuclein antibodies. Following the report of Neystat et al. [20] that maps the region of interaction of synphilin-1 with alpha-synuclein to the ankyrin-like and coiled-coil domain of synphilin-1, a new synphilin-1 construct was made comprising amino acids 331–555, and assayed for its possible effect under the same assay conditions used above with 0.9 μM synphilin-1 331–555 (calculation of concentration based on monomeric molecular mass of synphilin-1 331–555). Synphilin-1 331–555 (see Fig. 3) also inhibits the degradation of alpha-synuclein wild-type and its missense mutants (A30P, E46K, and A53T) by the 20S proteasome, similar results were obtained when analysis was performed by Western immunoblotting with anti-alpha-synuclein antibodies. Note again that synphilin-1 331–555 alone (see Supplementary Fig. 2) or in the presence of alpha-synuclein (Fig. 3) is not significantly degraded by the 20S proteasome. To determine the region of alpha-synuclein that interacts with synphilin-1, and guided again by the work of Neystat et al. [20] that map the alpha-synuclein region of interaction to the N-terminal region of alpha-synuclein, a His-tag version of full-length wild-type alpha-synuclein, 1–60 and 61–140 were obtained and purified. The addition of a His-tag to the N-terminus of full-length alpha-synuclein did not affect its degradation by the proteasome (Fig. 4). Furthermore, the His-tag N-terminal (1–60) alpha-synuclein was also efficiently degraded by the proteasome (see also Fig. 4). The presence of synphilin-1 (Fig. 4) strongly inhibits the degradation of His-tag alpha-synuclein full-length and the 1–60 N-terminal. The His-tag synuclein 61–140 was not significantly degraded by the 20S proteasome and as a consequence synphillin addition had no effect. Taken together, in all the above results it can be concluded that synphilin-1 inhibits the degradation of alpha-synuclein by the 20S proteasome, at least in part, through interaction of its ankyrin-like and coiled-coil domains with the N-terminal region 1–60 of alpha-synuclein.
Fig. 2.
Effect of synphilin-1 313–919 on the degradation of alpha-synuclein wild-type and its missense mutants by 20S proteasome. The panels show representative Coomassie Blue-stained gels of the degradation reactions performed in the absence (upper panels) and in the presence of synphilin-1 (Sncaip, lower panels) for alpha-synuclein (Snca) wild-type and the missense mutants (A30P, E46K, and A53T) as indicated. The quantifications for each of the reaction sets are shown in the corresponding graphical representations below each set of experiments. Reaction conditions were: 69.4 nM proteasome, 7 μM alpha-synuclein, and 0.75 μM synphilin-1 (331–919). Control reactions with synphilin-1and alpha-synuclein for 60 min in the absence of proteasome are also shown
Fig. 3.
Effect of ankyrin and coiled-coil region of synphilin-1 on the degradation of alpha-synuclein by the 20S proteasome. The panels show representative Coomassie Blue-stained gels of the degradations reactions of alpha-synuclein (Snca) wild-type and its missense mutants by 20S proteasome in the presence of synphilin-1 (Sncaip, amino acids 331–555). Reaction conditions: 69.4 nM proteasome, 7 μM alpha-synuclein, and 0.92 μM synphilin-1 (331–515). Control reactions with synphilin-1 and alpha-synuclein for 60 min in the absence of proteasome are also shown
Fig. 4.
Synphilin-1 inhibits the degradation of alpha-synuclein by interacting with the N-terminal region of alpha-synuclein. a Representative experiments analyzed by SDS-PAGE and Coomassie Blue staining and quantification of the degradation of N-terminal His-tag alpha-synuclein (His-Snca wt, 7 μM) full length by 20S proteasome (69.4 nM) in the absence and in the presence of synphilin-1 (Sncaip 331–919). b Similar experiments and quantification using N-terminal His-tag alpha-synuclein 1–50 (His-Snca 1–60, 7 μM)
Synphilin-1 specificity and dose-dependent inhibition of alpha-synuclein degradation by 20S proteasome
The data presented above show that synphilin-1 is able to inhibit alpha-synuclein degradation, but formally, even if it is not a substrate degraded by the proteasome, synphilin-1 could bind and behave as a direct inhibitor of proteasome activity, independently of its specific interaction with alpha-synuclein. We have shown that myelin basic protein (MBP) is an excellent substrate for the 20S proteasome [46]. If synphilin-1 effects are due, at least in part, to a direct effect on proteasome activity, it should also affect the degradation of MBP by the 20S proteasome. As shown in Supplementary Fig. 3, 0.75 μM synphilin-1 331–919 or 0.9 μM synphilin-1 331–555 were unable to inhibit the degradation of MBP by the 20S proteasome at different concentrations of proteasome and MBP. These results discarded that synphilin-1 by itself, or a contaminant that could be present in the purified recombinant protein, directly inhibited in a specific, or a non-specific way, the activity of the 20S proteasome. Furthermore, from the kinetic point of view, it is unlikely that synphilin-1 inhibition can be explained by formation of a direct inactive complex with the proteasome or by binding to the substrate binding site on the proteasome, as the formation of those complexes should also inhibits the degradation of other protein substrates, and this is not the case as shown above.
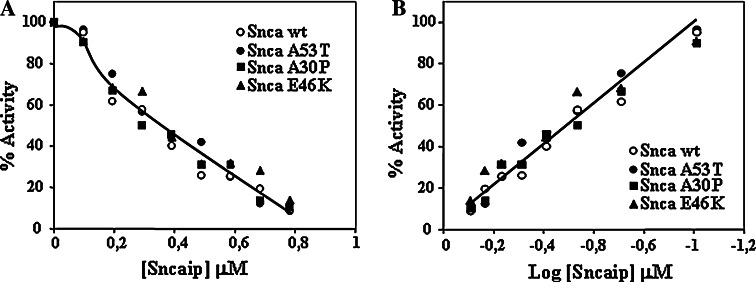
At micromolar concentrations of synphilin-1, we observed almost complete inhibition (70–100%) of the rate of alpha-synuclein degradation; as a consequence it was important to determine the actual IC0.5 of synphilin-1 for the inhibition of alpha-synuclein degradation by the proteasome. To that end, synphilin-1 dose-dependence studies were performed using 3.57 μM alpha-synuclein and 69.4 nM 20S proteasome. The dose-curves for all four forms of alpha-synuclein are presented in Fig. 5. The apparent IC0.5 for the inhibition of degradation of alpha-synuclein by synphilin-1 (331–919) was estimated to be 250 nM (based on monomeric synphilin-1 molecular mass) and no significant difference was observed for the different alpha-synucleins, wild-type, and missense mutants. Accordingly, the inhibition of degradation of alpha-synuclein by synphillin cannot be pure competition by direct one to one binding (sequestering) of alpha-synuclein to synphilin-1 [40].
Fig. 5.
Dose-dependence of the synphilin-1 inhibition of alpha-synuclein degradation by the proteasome. Different amounts of synphilin-1 (Sncaip 313–919) were incubated with a fixed amount (3.57 μM) of the different types of alpha-synuclein (Snca wild-type, A30P, E46K, and A53T) and degradation was performed with 69.4 nM 20S proteasome. The amount of alpha-synuclein remaining after incubation of the reactions for 30–45 min was quantitated after analysis by SDS-PAGE and Coomassie Blue staining. Left graph shows the direct plot of % activity vs. synphilin-1 concentration in the assay, taking 100% as the activity obtained in the absence of synphilin-1. Right graph shows the log transformation of the direct data to calculate the IC0.5, estimated to be 0.25 μM under this reaction conditions. Results are averaged from three different experiments with deviations below 10%
Synphilin-1 effect on alpha-synuclein degradation in cells
The above experiments show that synphilin-1 behaves in vitro as a specific inhibitor of alpha-synuclein degradation by the proteasome. While interesting mechanistically, it may be less relevant in situ. In cells, the unstructured alpha-synuclein will have many possible protein–protein interactions and the 20S proteasome have also many protein substrates and interacts with other protein complexes, most notably with the PA700/19S particle forming the 26S proteasome complex. Accordingly, it was mandatory to test the natural prediction from the above in vitro experiments that synphilin-1 should inhibit the degradation of alpha-synuclein in cells. Using antibodies against synphilin-1 we were unable to detect any significant expression of synphilin-1 in HeLa or N2A cells. Therefore, the degradation of alpha-synuclein after protein synthesis inhibition with cycloheximide (CHX) in HeLa and N2A cells was studied in the absence or in the presence of transfected synphilin-1. Transfected alpha-synuclein has a long half-life in both cell lines and its degradation rate is markedly decreased when synphilin-1 is co-transfected and the degradation of alpha-synuclein is completely prevented by treatment of cells with proteasome inhibitors (Fig. 6). Again, it was important to determine if the effect of synphilin-1 is specific or could affect in a general form the ubiquitin–proteasome pathway. To that end, we have used two unstable fluorescent reporters: EGFPu [47], a 26S ubiquitin-dependent substrate and EGFPd2 [48], a 26S ubiquitin-independent substrate. As shown in Fig. 7, synphilin-1 expression does not affect the rate of degradation of any of the two reporter fluorescent proteins, indicating that the effect of synphilin-1 on alpha-synuclein degradation is specific and not due to a substantial non-specific inhibition of the ubiquitin–proteasome pathway by synphilin-1.
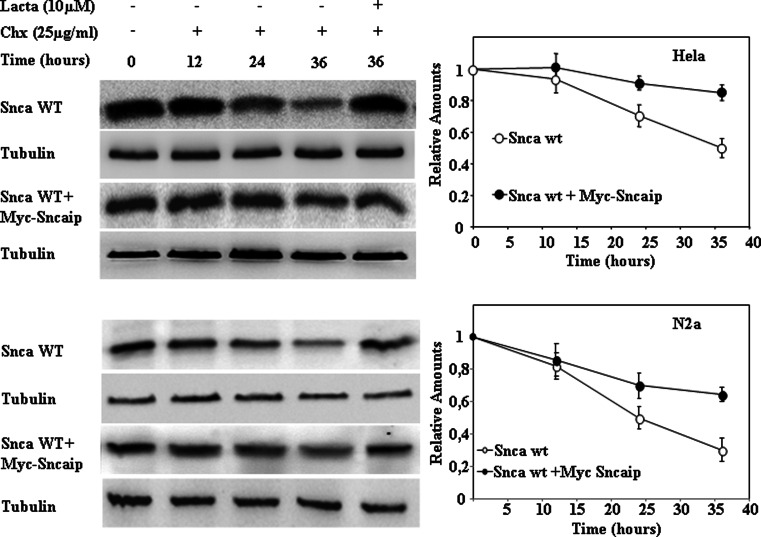
Fig. 6.
Synphilin-1 inhibits the degradation of alpha-synuclein in HeLa and N2a cells. HeLa and N2a cells were transfected with alpha-synuclein (Snca wt) or co-transfected with alpha-synuclein and synphilin-1 (Sncaip). Cells 48 h after transfection were treated with cycloheximide (CHX) either in the absence or in the presence of 10 μM lactacystin for the times indicated. The panels show representative immunoblots with anti-alpha-synuclein antibodies, and anti-tubulin antibodies used as protein loading controls. Quantifications are shown in the right graphs as means ± SEM. from three different experiments. Upper panels, HeLa cells; lower panels, N2A cells
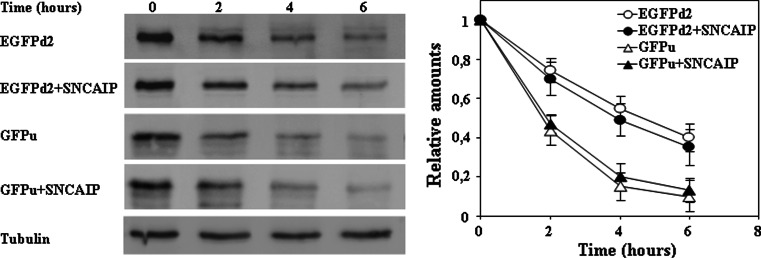
Fig. 7.
Effect of synphilin-1 on the degradation of unstable fluorescent proteins. HeLa cells were transfected with EGFPu or EGFPd2 and co-transfected with synphilin-1 (Sncaip), as indicated. Cells 48 h after transfection were treated with cycloheximide (CHX). The panels show representative immunoblots with anti-EGFP antibodies, and anti-tubulin antibodies used as protein loading controls. Quantifications are shown in the right graph as means ± SEM from three different experiments
If the interpretation of the above experiments is correct, a prediction can be made. Any treatment of cells co-expressing alpha-synuclein and synphilin-1 that significantly reduces synphilin-1 expression levels, should release the inhibition of alpha-synuclein degradation produced by synphilin-1. Synphilin-1 has been shown to be ubiquitylated by Siah-1, producing its fast degradation by the proteasome [49, 50]. As reported, co-transfection of synphilin-1 and Siah-1 in HeLa cells effectively diminished the levels of steady-state expression of synphilin-1 during the co-transfection period (Fig. 8a). Then, we studied the rate of degradation of synphilin-1 and Siah-1 by co-transfection experiments in HeLa cells transfected with alpha-synuclein. Inhibition of protein synthesis by treatment of the transfected (48 h after transfection) cells with CHX (Fig. 8b) produced a slow rate decline of synphilin-1 (40–50% reduction) after 36 h treatment with CHX. When synphilin-1 is co-transfected with Siah-1, first the initial steady-state levels of expression of synphilin-1(as shown in Fig. 8a) was already very low compared to the control and declined to undetectable levels after treatment of cells with CHX for 12 h (Fig. 8b). In contrast, Siah-1 levels did not change significantly after 36 h of CHX treatment of the cells. Identical results are obtained when alpha-synuclein is omitted in the co-transfection experiments (data not shown). The results presented in Fig. 8b are in full agreement with those reported previously [49] performed also in the absence of transfected alpha-synuclein. When the time course of alpha-synuclein degradation was studied in the presence of co-transfected Siah-1, there was no significant change with respect to the control, while co-transfection with synphilin-1 clearly inhibited alpha-synuclein degradation (Fig. 8c), as already described above (Fig. 6). In contrast, triple co-transfections with alpha-synuclein, synphilin-1, and Siah-1 showed a time course of degradation of alpha-synuclein identical to the controls where alpha-synuclein was transfected alone. These results indicate that Siah-1 very efficiently targets synphilin-1 to degradation, and, as a consequence, the inhibitory effect of synphilin-1 on alpha-synuclein degradation is relieved.
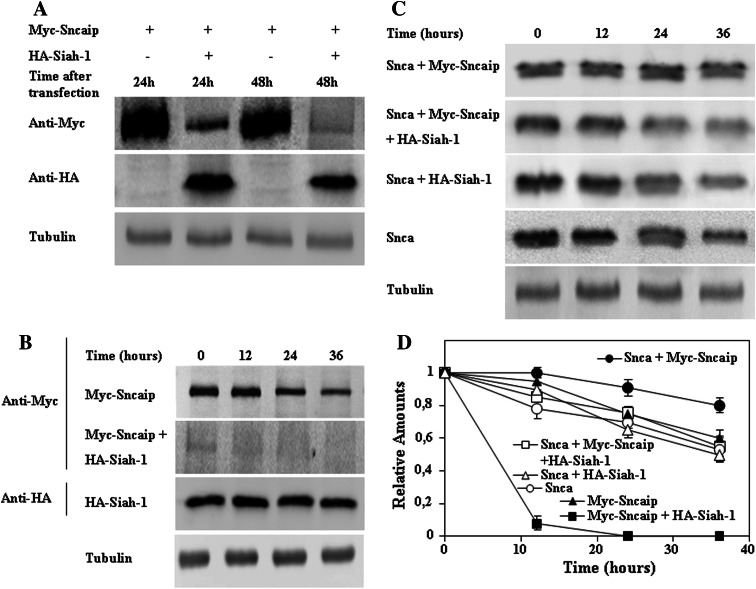
Fig. 8.
Siah-1 effects on synphilin-1 and alpha-synuclein degradation. HeLa cells were single, double, or triple transfected with different plasmids. a Representative immunoblots of the steady-state expression of synphilin-1 (Sncaip) after transfection in the absence or in the presence of Siah-1, as indicated. The blots have been over-exposed in order to show synphilin-1 expression in the presence of transfected Siah-1. b HeLa cells 48 h after transfection with the indicated plasmids were treated with cycloheximide (CHX). Representative immunoblots with anti-Myc (synphilin-1, Sncaip) and anti-HA (Siah-1) are presented and data quantification are shown in d. c HeLa cells 48 h after transfection with the indicated plasmids were treated with CHX. Representative immunoblots with anti-alpha-synuclein (Snca) antibodies are presented and data quantification are shown in d. d Graph showing the quantification of data presented in b and c expressed as means ± SEM from three different experiments. Anti-tubulin antibodies were used as protein-loading controls
Discussion
In this study, we described that synphilin-1 specifically inhibits the degradation of alpha-synuclein by the proteasome, at least in part, through interaction of its ankyrin and coiled-coil region with the N-terminal region of alpha-synuclein, other regions of alpha-synuclein and synphilin-1 may also participate and can not be formally excluded. The unfolded native state of alpha-synuclein allows its tendency to interact with many cellular protein partners. The in vitro and in situ studies presented here demonstrate that alpha-synuclein interaction with synphilin-1 is effective in preventing alpha-synuclein degradation by the proteasome. The specificity of this inhibition was also experimentally demonstrated both by in vitro and in situ experiments, as synphilin-1 did not affect the degradation of MBP in vitro, nor the degradation of two unstable reporter proteins, EGFPd2 or EGFPu, in HeLa cells. These last results, based on kinetic data of the degradation of the proteasome reporters in the absence of protein synthesis discard that synphilin-1 has a pronounced general inhibitory effect on the ubiquitin–proteasome pathway, but does not rule out that synphilin-1 may also inhibit the degradation of other proteasome substrates that may also interact with synphilin-1. A previous report describes an increase in the steady-state levels of GFPu in the presence of transfected synphilin-1 [51], but those experiments do not demonstrate that the proteasomal degradation rate is affected as the results presented can also be explained by direct or indirect effects of synphilin-1 expression on transcription and translation of GFPu, something that could not occur in our experimental setting where actual kinetics of degradation of the reporter proteins were studied in the presence of CHX.
The long half-life of alpha-synuclein in cells, shown here in HeLa and N2a, but also reported in HEK293, PC12, SH-SY5Y, and post-mitotic neurons [37, 52–54] is unusual for a naturally unfolded protein that is a direct substrate of the 20S proteasome in vitro. Ubiquitin-independent and proteasomal dependent degradation is apparently responsible of the degradation of an increasing number of proteins, that at least in vitro can be accomplished by the 20S proteasome, or the 26S proteasome, and they have in general much shorter half-lives than alpha-synuclein [55]. Alpha-synuclein has also been shown to be degraded in vitro by the 26S proteasome [35] and to interact with S6’ (Rpt5, PSMC3, Tbp1) subunit of the 19S proteasome cap complex [56, 57]. Synphilin-1 is reported [58] to interact with the 26S proteasome through binding to Tbp7 (Rpt3, PSMC4, S6). The results presented here in transfected cells clearly indicate that synphilin-1 inhibits alpha-synuclein degradation, similarly to what is obtained in vitro using 20S proteasome. Accordingly, irrespective of the proteasome complex degrading alpha-synuclein in the cell (20S or 26S proteasome) and of the interactions that may have alpha-synuclein or synphilin-1with the 19S complex, the final outcome is that synphilin-1 expression inhibits alpha-synuclein proteasomal degradation in the cell, and synphilin-1 does not behave as a significant non-specific inhibitor of the ubiquitin–proteasome pathway. Alpha-synuclein degradation in the cell can be achieved by the proteasome pathway and by the autophagic pathways [36, 37, 54]. The proteasome role in alpha-synuclein degradation in the cell is also demonstrated here by its degradation in the presence of CHX, a known inhibitor of the autophagic pathways [59] and its inhibition by specific proteasomal inhibitors.
Synphilin-1 is, like alpha-synuclein, a protein with a long half-life in the cell, which likely explains why we were able to find its inhibitory effect on alpha-synuclein degradation in the presence of CHX. A prediction from those results is that any treatment that reduces synphilin-1 levels should release the inhibition of alpha-synuclein degradation by the proteasome. In relation to these experimental results, it is noteworthy that the apparent opposite conclusion that aynphilin-1 expression should increase the steady-state levels of alpha-synuclein can not be deduced from the experiments presented, because the steady-state levels of alpha-synuclein are due to the balance of alpha-synuclein protein synthesis and degradation by the proteasome and autophagic pathways. Synphilin-1 is known to be ubiquitylated by different E3 ubiquitin-ligases, including parkin [60], dorfin [61, 62], and Siah-1 [49, 50]. As shown here, Siah-1 effectively targets synphilin-1 to fast degradation, confirming previous published results [49], and as predicted Siah-1 expression suppresses the inhibitory effect of synphilin-1 on the degradation of alpha-synuclein by the proteasomal pathway. Again, Siah-1 is a protein with a long half-life (Fig. 8) in the cell, also reported previously [49]. While Siah-1, and much more efficiently Siah-2, has been reported to interact with alpha-synuclein resulting in its monoubiquitylation [50], under our assays conditions we were not able to detect any alpha-synuclein modification following Siah-1 expression. The apparent conflicting results can be explained by the fact that monoubiquitylation of alpha-synuclein is quantitatively a minor modification [50, 63] that could not be readily detected by direct immunoblotting of whole cell extracts, and in any case Siah-1 expression did not significantly affect alpha-synuclein degradation by the proteasome pathway in the absence of synphilin-1 (Fig. 8).
Taken together, with the results presented here and those published previously, we can advance a central regulatory mechanism: a long half-life protein, synphilin-1, prevents the degradation of another long half-life protein, alpha-synuclein, by the proteasome pathway. Naturally, this regulatory mechanism could only operate significantly in cells expressing both proteins, for example brain, heart, and skeletal muscle [19]. Accordingly with this central hypothesis, the prediction that reducing the synphilin-1 expression levels should relieve inhibition of alpha-synuclein degradation by the proteasome has been demonstrated by co-expression of another long-life protein, Siah-1, that is also expressed at enough levels in brain, heart, and skeletal muscle [49]. This rational could explain in part the apparent selectivity of tissues damaged in PD and other synucleinopathies (diffuse Lewy bodies disease, multiple system atrophy, etc.) in an age-dependent process that would be exacerbated in familiar forms of PD with over-expression of alpha-synuclein due to locus duplication [6, 7], triplication [8] and hypomethylation of alpha-synuclein intron 1[9] by simple mass-action.
The connection between alpha-synuclein, synphilin-1, and proteasome is also illustrated by the formation of alpha-synuclein–synphilin-1 aggregates upon proteasomal inhibition in cells, forming aggresome-like structures [19, 38, 40, 50, 64–67]. The recent work of Xie et al. [40] also demonstrates that synphilin-1 can not recruit the N-terminal truncated alpha-synuclein to form aggregates and inclusions, so the N-terminal part of alpha-synuclein is essential for both interaction with synphilin-1 and for recognition by the proteasome (as shown here). Accordingly, synphilin-1 by the same molecular mechanism, interaction with the N-terminal region of alpha-synuclein, prevents alpha-synuclein degradation by the proteasome and promotes its aggregation and inclusion formation. Proteasome inhibition also promotes the formation of inclusions that contains polyubiquitylated synphilin-1 bound to Siah-1 [50]. In addition, synphilin-1 is present together with alpha-synuclein in Lewy bodies of PD and diffuse Lewy bodies disease [39, 65, 68] and Siah-1 is also found in Lewy bodies [50]. Finally, in mice, over-expression of wild-type synphilin-1 or a missense synphilin-1 mutant R621C driven by a PrP-promoter in transgenic mice [69] or by adenoviral infection of dopaminergic neurons [70] produce alpha-synuclein aggregates in neurons promoting cell degeneration. In contrast to these two reports, a recent publication indicates that synphilin-1 overexpression may be cytoprotective; delaying, while not totally reverting, the pathological consequence of overexpression of A53T alpha-synuclein in mice by activation of the autophagic pathway [71].
Studies in vitro and in cultured cells of alpha-synuclein-synphilin-1 interaction have limitations respect to the interpretation to the in vivo situation. At present, the in vitro and in situ (in cells) results presented here and published before are very coherent with two of the in vivo reports in transgenic mice, but failed to be reconciled with another report in mice (see above). Further experiments become necessary to clarify the possible role of synphilin-1 on autophagic clearance of alpha-synuclein in view of the fact that loss of basal autophagy without affecting proteasome function promotes neurodegeneration in mice [72, 73]. Those studies would help to a further understanding of the regulation by synphilin-1 of alpha-synuclein degradation by the proteasome and autophagic pathways that are involved in alpha-synuclein proteostasis.
Normal Snca proteostasis in the cell makes it a long half-life protein by balance of its synthesis and degradation through proteasomal and autophagic pathways, even in the absence of synphilin-1 expression. In those cells where synphilin-1 and alpha-synuclein are co-expressed, synphilin-1 could effectively slow-down alpha-synuclein degradation by the proteasomal pathway as demonstrated here, but this does not necessarily imply that alpha-synuclein steady-state levels should be increased. Synphilin-1 inhibition of the proteasomal pathway of degradation of Snca may contribute to its aggregation, as is thoroughly documented in the literature, but it remains to be studied regarding its possible effect on autophagic alpha-synuclein clearance in order to have a complete picture of the regulation of alpha-synuclein degradation by Sncaip and the pathophysiological changes occurring in PD and other synucleinopathies.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1 Alpha-synuclein degradation by the proteasome is prevented by proteasome inhibitors. Panel shows a representative Coomassie Blue-stained gel of the time-course of degradation by 138 nM 20S proteasome of alpha-synuclein wild-type (Snca wt, 3.57 μM). Last lane shows reaction for 30 min in the presence of 10 μM lactacystin (Lacta). (TIFF 133 kb)
Supplementary Fig. 2 Synphilin is not degraded by the 20S proteasome. The panels show representative experiments of synphilin-1 protein constructs (Sncaip 313-919 and 331-555) degradation reactions in the presence of 69 nM 20S proteasome analyzed; a by Coomassie Blue-stained gel and b by Western immunoblots with anti-His-tag antibodies (overexposed to demonstrate that there is not significant degradation of synphilin protein constructs) (TIFF 190 kb)
Supplementary Fig. 3 Synphilin-1 does not affect the degradation of Myelin Basic Protein (MBP) by the proteasome. The panels show representative Coomassie Blue-stained gels of the degradation by 69.4 nM 20S proteasome of MBP (2 μM); a in the absence; b, in the presence of 0.75 μM Synphilin-1 (Sncaip 331-919), and c in the presence of 0.92 μM synphilin-1 (Sncaip 331-555). d Time course of degradation of MBP (10 μM) in the absence or in the presence of 0.75 μM Synphilin-1 (Sncaip 331-919) with 17.36 nM 20S proteasome. At both doses of MBP and proteasome, there is no effect of synphilin-1 on the degradation of MBP by the proteasome. (TIFF 203 kb)
Acknowledgments
We want to express our thanks to Dr. Mark R. Cookson (National Institutes of Health, Bethesda, MD, USA) and Dr. Simone Engelender (Technion-Israel Institute of Technology, Haifa, Israel) for providing us synphilin-1 and Siah-1 expression constructs. This work was supported by grants from SAF-2008-00766, CM SAL-0202, & CIBERNED to J. G. C.
References
- 1.Goedert M. Parkinson’s disease and other alpha-synucleinopathies. Clin Chem Lab Med. 2001;39:308–312. doi: 10.1515/CCLM.2001.047. [DOI] [PubMed] [Google Scholar]
- 2.Spillantini MG, Crowther RA, Jakes R, Hasegawa M, Goedert M. Alpha-synuclein in filamentous inclusions of Lewy bodies from Parkinson’s disease and dementia with lewy bodies. Proc Natl Acad Sci USA. 1998;95:6469–6473. doi: 10.1073/pnas.95.11.6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, Di Iorio G, Golbe LI, Nussbaum RL. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 4.Kruger R, Kuhn W, Muller T, Woitalla D, Graeber M, Kosel S, Przuntek H, Epplen JT, Schols L, Riess O. Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson’s disease. Nat Genet. 1998;18:106–108. doi: 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- 5.Zarranz JJ, Alegre J, Gomez-Esteban JC, Lezcano E, Ros R, Ampuero I, Vidal L, Hoenicka J, Rodriguez O, Atares B, Llorens V, Gomez TE, Del Ser T, Munoz DG, de Yebenes JG. The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann Neurol. 2004;55:164–173. doi: 10.1002/ana.10795. [DOI] [PubMed] [Google Scholar]
- 6.Chartier-Harlin MC, Kachergus J, Roumier C, Mouroux V, Douay X, Lincoln S, Levecque C, Larvor L, Andrieux J, Hulihan M, Waucquier N, Defebvre L, Amouyel P, Farrer M, Destee A. Alpha-synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet. 2004;364:1167–1169. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- 7.Ibanez P, Bonnet AM, Debarges B, Lohmann E, Tison F, Pollak P, Agid Y, Durr A, Brice A. Causal relation between alpha-synuclein gene duplication and familial Parkinson’s disease. Lancet. 2004;364:1169–1171. doi: 10.1016/S0140-6736(04)17104-3. [DOI] [PubMed] [Google Scholar]
- 8.Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, Lincoln S, Crawley A, Hanson M, Maraganore D, Adler C, Cookson MR, Muenter M, Baptista M, Miller D, Blancato J, Hardy J, Gwinn-Hardy K. Alpha-synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 9.Jowaed A, Schmitt I, Kaut O, Wullner U. Methylation regulates alpha-synuclein expression and is decreased in Parkinson’s disease patients’ brains. J Neurosci. 2010;30:6355–6359. doi: 10.1523/JNEUROSCI.6119-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maries E, Dass B, Collier TJ, Kordower JH, Steece-Collier K. The role of alpha-synuclein in Parkinson’s disease: insights from animal models. Nat Rev Neurosci. 2003;4:727–738. doi: 10.1038/nrn1199. [DOI] [PubMed] [Google Scholar]
- 11.Cookson MR. The biochemistry of Parkinson’s disease. Annu Rev Biochem. 2005;74:29–52. doi: 10.1146/annurev.biochem.74.082803.133400. [DOI] [PubMed] [Google Scholar]
- 12.Vavouri T, Semple JI, Garcia-Verdugo R, Lehner B. Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity. Cell. 2009;138:198–208. doi: 10.1016/j.cell.2009.04.029. [DOI] [PubMed] [Google Scholar]
- 13.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Y, Gu G, Goodlett DR, Zhang T, Pan C, Montine TJ, Montine KS, Aebersold RH, Zhang J. Analysis of alpha-synuclein-associated proteins by quantitative proteomics. J Biol Chem. 2004;279:39155–39164. doi: 10.1074/jbc.M405456200. [DOI] [PubMed] [Google Scholar]
- 15.Jensen PH, Hager H, Nielsen MS, Hojrup P, Gliemann J, Jakes R. alpha-synuclein binds to Tau and stimulates the protein kinase A-catalyzed tau phosphorylation of serine residues 262 and 356. J Biol Chem. 1999;274:25481–25489. doi: 10.1074/jbc.274.36.25481. [DOI] [PubMed] [Google Scholar]
- 16.Ostrerova N, Petrucelli L, Farrer M, Mehta N, Choi P, Hardy J, Wolozin B. alpha-synuclein shares physical and functional homology with 14-3-3 proteins. J Neurosci. 1999;19:5782–5791. doi: 10.1523/JNEUROSCI.19-14-05782.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu J, Kao SY, Lee FJ, Song W, Jin LW, Yankner BA. Dopamine-dependent neurotoxicity of alpha-synuclein: a mechanism for selective neurodegeneration in Parkinson disease. Nat Med. 2002;8:600–606. doi: 10.1038/nm0602-600. [DOI] [PubMed] [Google Scholar]
- 18.Sato S, Chiba T, Sakata E, Kato K, Mizuno Y, Hattori N, Tanaka K. 14–3-3eta is a novel regulator of parkin ubiquitin ligase. EMBO J. 2006;25:211–221. doi: 10.1038/sj.emboj.7600774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engelender S, Kaminsky Z, Guo X, Sharp AH, Amaravi RK, Kleiderlein JJ, Margolis RL, Troncoso JC, Lanahan AA, Worley PF, Dawson VL, Dawson TM, Ross CA. Synphilin-1 associates with alpha-synuclein and promotes the formation of cytosolic inclusions. Nat Genet. 1999;22:110–114. doi: 10.1038/8820. [DOI] [PubMed] [Google Scholar]
- 20.Neystat M, Rzhetskaya M, Kholodilov N, Burke RE. Analysis of synphilin-1 and synuclein interactions by yeast two-hybrid beta-galactosidase liquid assay. Neurosci Lett. 2002;325:119–123. doi: 10.1016/S0304-3940(02)00253-7. [DOI] [PubMed] [Google Scholar]
- 21.Payton JE, Perrin RJ, Clayton DF, George JM. Protein–protein interactions of alpha-synuclein in brain homogenates and transfected cells. Brain Res Mol Brain Res. 2001;95:138–145. doi: 10.1016/S0169-328X(01)00257-1. [DOI] [PubMed] [Google Scholar]
- 22.Jenco JM, Rawlingson A, Daniels B, Morris AJ. Regulation of phospholipase D2: selective inhibition of mammalian phospholipase D isoenzymes by alpha- and beta-synucleins. Biochemistry. 1998;37:4901–4909. doi: 10.1021/bi972776r. [DOI] [PubMed] [Google Scholar]
- 23.Payton JE, Perrin RJ, Woods WS, George JM. Structural determinants of PLD2 inhibition by alpha-synuclein. J Mol Biol. 2004;337:1001–1009. doi: 10.1016/j.jmb.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 24.Rappley I, Gitler AD, Selvy PE, LaVoie MJ, Levy BD, Brown HA, Lindquist S, Selkoe DJ. Evidence that alpha-synuclein does not inhibit phospholipase D. Biochemistry. 2009;48:1077–1083. doi: 10.1021/bi801871h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murayama S, Arima K, Nakazato Y, Satoh J, Oda M, Inose T. Immunocytochemical and ultrastructural studies of neuronal and oligodendroglial cytoplasmic inclusions in multiple system atrophy. 2. Oligodendroglial cytoplasmic inclusions. Acta Neuropathol. 1992;84:32–38. doi: 10.1007/BF00427212. [DOI] [PubMed] [Google Scholar]
- 26.Pountney DL, Treweek TM, Chataway T, Huang Y, Chegini F, Blumbergs PC, Raftery MJ, Gai WP. Alpha B-crystallin is a major component of glial cytoplasmic inclusions in multiple system atrophy. Neurotox Res. 2005;7:77–85. doi: 10.1007/BF03033778. [DOI] [PubMed] [Google Scholar]
- 27.Uryu K, Richter-Landsberg C, Welch W, Sun E, Goldbaum O, Norris EH, Pham CT, Yazawa I, Hilburger K, Micsenyi M, Giasson BI, Bonini NM, Lee VM, Trojanowski JQ. Convergence of heat shock protein 90 with ubiquitin in filamentous alpha-synuclein inclusions of alpha-synucleinopathies. Am J Pathol. 2006;168:947–961. doi: 10.2353/ajpath.2006.050770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Auluck PK, Chan HY, Trojanowski JQ, Lee VM, Bonini NM. Chaperone suppression of alpha-synuclein toxicity in a Drosophila model for Parkinson’s disease. Science. 2002;295:865–868. doi: 10.1126/science.1067389. [DOI] [PubMed] [Google Scholar]
- 29.McLean PJ, Klucken J, Shin Y, Hyman BT. Geldanamycin induces Hsp70 and prevents alpha-synuclein aggregation and toxicity in vitro. Biochem Biophys Res Commun. 2004;321:665–669. doi: 10.1016/j.bbrc.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 30.Klucken J, Shin Y, Hyman BT, McLean PJ. A single amino acid substitution differentiates Hsp70-dependent effects on alpha-synuclein degradation and toxicity. Biochem Biophys Res Commun. 2004;325:367–373. doi: 10.1016/j.bbrc.2004.10.037. [DOI] [PubMed] [Google Scholar]
- 31.Dedmon MM, Christodoulou J, Wilson MR, Dobson CM. Heat shock protein 70 inhibits alpha-synuclein fibril formation via preferential binding to prefibrillar species. J Biol Chem. 2005;280:14733–14740. doi: 10.1074/jbc.M413024200. [DOI] [PubMed] [Google Scholar]
- 32.Shimshek DR, Mueller M, Wiessner C, Schweizer T, van der Putten PH. The HSP70 molecular chaperone is not beneficial in a mouse model of alpha-synucleinopathy. PLoS ONE. 2010;5:e10014. doi: 10.1371/journal.pone.0010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayo I, Rodriguez-Vilariño S, Castaño J. G. 20S proteasome degrades alpha-synuclein. Cold Spring Harbor Meeting on Proteolysis and Biological Control, p. 92
- 34.Tofaris GK, Layfield R, Spillantini MG. Alpha-synuclein metabolism and aggregation is linked to ubiquitin-independent degradation by the proteasome. FEBS Lett. 2001;509:22–26. doi: 10.1016/S0014-5793(01)03115-5. [DOI] [PubMed] [Google Scholar]
- 35.Liu CW, Corboy MJ, Demartino GN, Thomas PJ. Endoproteolytic activity of the proteasome. Science. 2003;299:408–411. doi: 10.1126/science.1079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bennett MC, Bishop JF, Leng Y, Chock PB, Chase TN, Mouradian MM. Degradation of alpha-synuclein by proteasome. J Biol Chem. 1999;274:33855–33858. doi: 10.1074/jbc.274.48.33855. [DOI] [PubMed] [Google Scholar]
- 37.Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. Alpha-synuclein is degraded by both autophagy and the proteasome. J Biol Chem. 2003;278:25009–25013. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- 38.O’Farrell C, Murphy DD, Petrucelli L, Singleton AB, Hussey J, Farrer M, Hardy J, Dickson DW, Cookson MR. Transfected synphilin-1 forms cytoplasmic inclusions in HEK293 cells. Brain Res Mol Brain Res. 2001;97:94–102. doi: 10.1016/S0169-328X(01)00292-3. [DOI] [PubMed] [Google Scholar]
- 39.Wakabayashi K, Engelender S, Yoshimoto M, Tsuji S, Ross CA, Takahashi H. Synphilin-1 is present in Lewy bodies in Parkinson’s disease. Ann Neurol. 2000;47:521–523. doi: 10.1002/1531-8249(200004)47:4<521::AID-ANA18>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 40.Xie YY, Zhou CJ, Zhou ZR, Hong J, Che MX, Fu QS, Song AX, Lin DH, Hu HY. Interaction with synphilin-1 promotes inclusion formation of {alpha}-synuclein: mechanistic insights and pathological implication. FASEB J. 2009;24:196–295. doi: 10.1096/fj.09-133082. [DOI] [PubMed] [Google Scholar]
- 41.O’Farrell C, Pickford F, Vink L, McGowan E, Cookson MR. Sequence conservation between mouse and human synphilin-1. Neurosci Lett. 2002;322:9–12. doi: 10.1016/S0304-3940(02)00068-X. [DOI] [PubMed] [Google Scholar]
- 42.Martin-Clemente B, Alvarez-Castelao B, Mayo I, Sierra AB, Diaz V, Milan M, Farinas I, Gomez-Isla T, Ferrer I, Castaño JG. Alpha-synuclein expression levels do not significantly affect proteasome function and expression in mice and stably transfected PC12 cell lines. J Biol Chem. 2004;279:52984–52990. doi: 10.1074/jbc.M409028200. [DOI] [PubMed] [Google Scholar]
- 43.Arribas J, Castaño JG. Kinetic studies of the differential effect of detergents on the peptidase activities of the multicatalytic proteinase from rat liver. J Biol Chem. 1990;265:13969–13973. [PubMed] [Google Scholar]
- 44.Wandosell F, Serrano L, Hernandez MA, Avila J. Phosphorylation of tubulin by a calmodulin-dependent protein kinase. J Biol Chem. 1986;261:10332–10339. [PubMed] [Google Scholar]
- 45.Alvarez-Castelao B, Martin-Guerrero I, Garcia-Orad A, Castaño JG. CMV promoter up-regulation is the major cause of increased protein levels of unstable reporter proteins after treatment of living cells with proteasome inhibitors. J Biol Chem. 2009;284:28253–28262. doi: 10.1074/jbc.M109.004101. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Arribas J, Arizti P, Castaño JG. Antibodies against the C2 COOH-terminal region discriminate the active and latent forms of the multicatalytic proteinase complex. J Biol Chem. 1994;269:12858–12864. [PubMed] [Google Scholar]
- 47.Gilon T, Chomsky O, Kulka RG. Degradation signals for ubiquitin system proteolysis in Saccharomyces cerevisiae . EMBO J. 1998;17:2759–2766. doi: 10.1093/emboj/17.10.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Corish P, Tyler-Smith C. Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng. 1999;12:1035–1040. doi: 10.1093/protein/12.12.1035. [DOI] [PubMed] [Google Scholar]
- 49.Nagano Y, Yamashita H, Takahashi T, Kishida S, Nakamura T, Iseki E, Hattori N, Mizuno Y, Kikuchi A, Matsumoto M. Siah-1 facilitates ubiquitination and degradation of synphilin-1. J Biol Chem. 2003;278:51504–51514. doi: 10.1074/jbc.M306347200. [DOI] [PubMed] [Google Scholar]
- 50.Liani E, Eyal A, Avraham E, Shemer R, Szargel R, Berg D, Bornemann A, Riess O, Ross CA, Rott R, Engelender S. Ubiquitylation of synphilin-1 and alpha-synuclein by SIAH and its presence in cellular inclusions and Lewy bodies imply a role in Parkinson’s disease. Proc Natl Acad Sci USA. 2004;101:5500–5505. doi: 10.1073/pnas.0401081101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Avraham E, Szargel R, Eyal A, Rott R, Engelender S. Glycogen synthase kinase 3beta modulates synphilin-1 ubiquitylation and cellular inclusion formation by SIAH: implications for proteasomal function and Lewy body formation. J Biol Chem. 2005;280:42877–42886. doi: 10.1074/jbc.M505608200. [DOI] [PubMed] [Google Scholar]
- 52.Okochi M, Walter J, Koyama A, Nakajo S, Baba M, Iwatsubo T, Meijer L, Kahle PJ, Haass C. Constitutive phosphorylation of the Parkinson’s disease associated alpha-synuclein. J Biol Chem. 2000;275:390–397. doi: 10.1074/jbc.275.1.390. [DOI] [PubMed] [Google Scholar]
- 53.Lee HJ, Khoshaghideh F, Patel S, Lee SJ. Clearance of alpha-synuclein oligomeric intermediates via the lysosomal degradation pathway. J Neurosci. 2004;24:1888–1896. doi: 10.1523/JNEUROSCI.3809-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 55.Jariel-Encontre I, Bossis G, Piechaczyk M. Ubiquitin-independent degradation of proteins by the proteasome. Biochim Biophys Acta. 2008;1786:153–177. doi: 10.1016/j.bbcan.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 56.Ghee M, Fournier A, Mallet J. Rat alpha-synuclein interacts with Tat binding protein 1, a component of the 26S proteasomal complex. J Neurochem. 2000;75:2221–2224. doi: 10.1046/j.1471-4159.2000.0752221.x. [DOI] [PubMed] [Google Scholar]
- 57.Snyder H, Mensah K, Theisler C, Lee J, Matouschek A, Wolozin B. Aggregated and monomeric alpha-synuclein bind to the S6’ proteasomal protein and inhibit proteasomal function. J Biol Chem. 2003;278:11753–11759. doi: 10.1074/jbc.M208641200. [DOI] [PubMed] [Google Scholar]
- 58.Marx FP, Soehn AS, Berg D, Melle C, Schiesling C, Lang M, Kautzmann S, Strauss KM, Franck T, Engelender S, Pahnke J, Dawson S, von Eggeling F, Schulz JB, Riess O, Kruger R. The proteasomal subunit S6 ATPase is a novel synphilin-1 interacting protein-implications for Parkinson’s disease. FASEB J. 2007;21:1759–1767. doi: 10.1096/fj.06-6734com. [DOI] [PubMed] [Google Scholar]
- 59.Finn PF, Mesires NT, Vine M, Dice JF. Effects of small molecules on chaperone-mediated autophagy. Autophagy. 2005;1:141–145. doi: 10.4161/auto.1.3.2000. [DOI] [PubMed] [Google Scholar]
- 60.Chung KK, Zhang Y, Lim KL, Tanaka Y, Huang H, Gao J, Ross CA, Dawson VL, Dawson TM. Parkin ubiquitinates the alpha-synuclein-interacting protein, synphilin-1: implications for Lewy-body formation in Parkinson disease. Nat Med. 2001;7:1144–1150. doi: 10.1038/nm1001-1144. [DOI] [PubMed] [Google Scholar]
- 61.Hishikawa N, Niwa J, Doyu M, Ito T, Ishigaki S, Hashizume Y, Sobue G. Dorfin localizes to the ubiquitylated inclusions in Parkinson’s disease, dementia with Lewy bodies, multiple system atrophy, and amyotrophic lateral sclerosis. Am J Pathol. 2003;163:609–619. doi: 10.1016/S0002-9440(10)63688-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ito T, Niwa J, Hishikawa N, Ishigaki S, Doyu M, Sobue G. Dorfin localizes to Lewy bodies and ubiquitylates synphilin-1. J Biol Chem. 2003;278:29106–29114. doi: 10.1074/jbc.M302763200. [DOI] [PubMed] [Google Scholar]
- 63.Szargel R, Rott R, Eyal A, Haskin J, Shani V, Balan L, Wolosker H, Engelender S. Synphilin-1A inhibits SIAH and modulates alpha-synuclein monoubiquitylation and inclusion formation. J Biol Chem. 2009;284:11706–11716. doi: 10.1074/jbc.M805990200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee G, Junn E, Tanaka M, Kim YM, Mouradian MM. Synphilin-1 degradation by the ubiquitin–proteasome pathway and effects on cell survival. J Neurochem. 2002;83:346–352. doi: 10.1046/j.1471-4159.2002.01136.x. [DOI] [PubMed] [Google Scholar]
- 65.Bandopadhyay R, Kingsbury AE, Muqit MM, Harvey K, Reid AR, Kilford L, Engelender S, Schlossmacher MG, Wood NW, Latchman DS, Harvey RJ, Lees AJ. Synphilin-1 and parkin show overlapping expression patterns in human brain and form aggresomes in response to proteasomal inhibition. Neurobiol Dis. 2005;20:401–411. doi: 10.1016/j.nbd.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 66.Wong ES, Tan JM, Soong WE, Hussein K, Nukina N, Dawson VL, Dawson TM, Cuervo AM, Lim KL. Autophagy-mediated clearance of aggresomes is not a universal phenomenon. Hum Mol Genet. 2008;17:2570–2582. doi: 10.1093/hmg/ddn157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zaarur N, Meriin AB, Gabai VL, Sherman MY. Triggering aggresome formation. Dissecting aggresome-targeting and aggregation signals in synphilin 1. J Biol Chem. 2008;283:27575–27584. doi: 10.1074/jbc.M802216200. [DOI] [PubMed] [Google Scholar]
- 68.Wakabayashi K, Hayashi S, Yoshimoto M, Kudo H, Takahashi H. NACP/alpha-synuclein-positive filamentous inclusions in astrocytes and oligodendrocytes of Parkinson’s disease brains. Acta Neuropathol. 2000;99:14–20. doi: 10.1007/PL00007400. [DOI] [PubMed] [Google Scholar]
- 69.Nuber S, Franck T, Wolburg H, Schumann U, Casadei N, Fischer K, Calaminus C, Pichler BJ, Chanarat S, Teismann P, Schulz JB, Luft AR, Tomiuk J, Wilbertz J, Bornemann A, Kruger R, Riess O. Transgenic overexpression of the alpha-synuclein interacting protein synphilin-1 leads to behavioral and neuropathological alterations in mice. Neurogenetics. 2009;11:107–120. doi: 10.1007/s10048-009-0212-2. [DOI] [PubMed] [Google Scholar]
- 70.Krenz A, Falkenburger BH, Gerhardt E, Drinkut A, Schulz JB. Aggregate formation and toxicity by wild-type and R621C synphilin-1 in the nigrostriatal system of mice using adenoviral vectors. J Neurochem. 2009;108:139–146. doi: 10.1111/j.1471-4159.2008.05755.x. [DOI] [PubMed] [Google Scholar]
- 71.Smith WW, Liu Z, Liang Y, Masuda N, Swing DA, Jenkins NA, Copeland NG, Troncoso JC, Pletnikov M, Dawson TM, Martin LJ, Moran TH, Lee MK, Borchelt DR, Ross CA. Synphilin-1 attenuates neuronal degeneration in the A53T {alpha}-synuclein transgenic mouse model. Hum Mol Genet. 2010;19:2087–2098. doi: 10.1093/hmg/ddq086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 73.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1 Alpha-synuclein degradation by the proteasome is prevented by proteasome inhibitors. Panel shows a representative Coomassie Blue-stained gel of the time-course of degradation by 138 nM 20S proteasome of alpha-synuclein wild-type (Snca wt, 3.57 μM). Last lane shows reaction for 30 min in the presence of 10 μM lactacystin (Lacta). (TIFF 133 kb)
Supplementary Fig. 2 Synphilin is not degraded by the 20S proteasome. The panels show representative experiments of synphilin-1 protein constructs (Sncaip 313-919 and 331-555) degradation reactions in the presence of 69 nM 20S proteasome analyzed; a by Coomassie Blue-stained gel and b by Western immunoblots with anti-His-tag antibodies (overexposed to demonstrate that there is not significant degradation of synphilin protein constructs) (TIFF 190 kb)
Supplementary Fig. 3 Synphilin-1 does not affect the degradation of Myelin Basic Protein (MBP) by the proteasome. The panels show representative Coomassie Blue-stained gels of the degradation by 69.4 nM 20S proteasome of MBP (2 μM); a in the absence; b, in the presence of 0.75 μM Synphilin-1 (Sncaip 331-919), and c in the presence of 0.92 μM synphilin-1 (Sncaip 331-555). d Time course of degradation of MBP (10 μM) in the absence or in the presence of 0.75 μM Synphilin-1 (Sncaip 331-919) with 17.36 nM 20S proteasome. At both doses of MBP and proteasome, there is no effect of synphilin-1 on the degradation of MBP by the proteasome. (TIFF 203 kb)