Abstract
Poly(ADP-ribosyl)ation is the covalent attachment of ADP-ribose subunits from NAD+ to target proteins and was first described in plants in the 1970s. This post-translational modification is mediated by poly(ADP-ribose) polymerases (PARPs) and removed by poly(ADP-ribose) glycohydrolases (PARGs). PARPs have important functions in many biological processes including DNA repair, epigenetic regulation and transcription. However, these roles are not always associated with enzymatic activity. The PARP superfamily has been well studied in animals, but remains under-investigated in plants. Although plants lack the variety of PARP superfamily members found in mammals, they do encode three different types of PARP superfamily proteins, including a group of PARP-like proteins, the SRO family, that are plant specific. In plants, members of the PARP family and/or poly(ADP-ribosyl)ation have been linked to DNA repair, mitosis, innate immunity and stress responses. In addition, members of the SRO family have been shown to be necessary for normal sporophytic development. In this review, we summarize the current state of plant research into poly(ADP-ribosyl)ation and the PARP superfamily in plants.
Keywords: ADP-ribose, PARP, mART, Post-translational modification, Stress, Cell proliferation
Introduction
Post-translational modifications of proteins, such as phosphorylation, ubiquitination and acetylation, allow dynamic and reversible changes to function. The attachment of multiple ADP-ribose moieties to proteins, poly(ADP-ribosyl)ation, is a post-translational modification mediated by poly(ADP-ribose) polymerases (PARPs) and was first discovered in the 1960s [26, 27, 42, 50, 95, 114]. Although originally described in mammals, members of the PARP superfamily have now been identified through sequence similarity in all major groups of eukaryotes [32]. The PARP catalytic site, called the PARP signature, which consists of a ß-alpha-loop-B-alpha NAD+ fold [101, 120], characterizes members of this superfamily. These proteins are best studied in humans, where 17 members are found, which combine diverse functional domains with the PARP catalytic domain [9, 60, 125]. True PARPs attach ADP-ribose subunits to target proteins; depending on the specific PARP involved, several to hundreds of ADP-ribose units can be attached to the target [78]. This process uses nicotinamide adenine dinucleotide (NAD+), releasing nicotinamide as a reaction byproduct. However, not all proteins with PARP signatures actually function in poly(ADP-ribosyl)ation. For example, while both HstiPARP and HsPARP10 have non-conserved residues instead of an important catalytic glutamic acid [3, 88, 148], Hsti-PARP has PARP activity [88] while HsPARP10 acts as a mono(ADP-ribose) transferase (mART) [31, 79, 148]. In addition, two other human proteins that have replaced catalytic residues, HsPARP9 and HsPARP13, are enzymatically inactive [79, 136]. This suggests that the functions of the PARP superfamily extends beyond poly(ADP-ribosyl)ation.
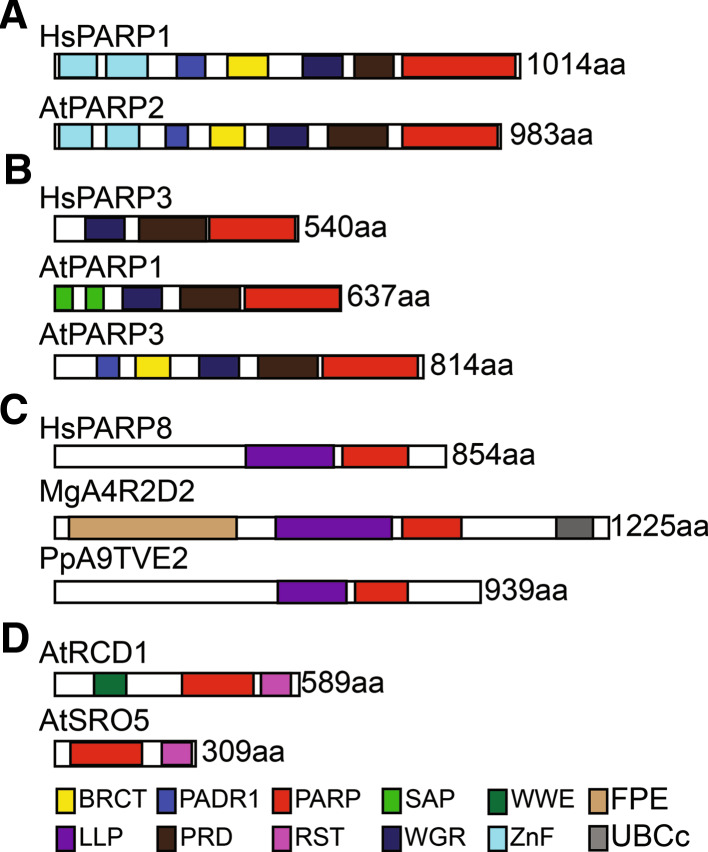
PARP superfamily members are involved in a broad range of functions, including DNA damage repair, cell death pathways, transcription and chromatin modification/remodeling (reviewed in [60, 78, 125]) and are targets for the development of anti-cancer drugs [61]. The first identified member of the PARP superfamily, human PARP1 (HsPARP1), remains the best studied. The PARP1 encoding gene is expressed nearly ubiquitously in mouse and other mammals [124]. The protein contains N-terminal DNA-binding zinc fingers, necessary to bind to double-strand breaks (Fig. 1a). In addition, the protein has a PADR1 domain, a domain of unknown function found in PARP1 and its orthologs [128] as well as a BRCA1 C-Terminus domain (BRCT), found predominantly in proteins involved in cell cycle checkpoints [21]. In addition, HsPARP1 contains a WGR domain of unknown function, although postulated to bind nucleic acids, and a PARP regulatory domain (PRD) [120, 127], before the C-terminal catalytic domain. HsPARP1 was originally found to function in association with DNA damage and repair pathways. In this context, it is activated upon binding to DNA strand breaks and autopoly(ADP-ribosyl)ates [58, 60]. The negatively charged ADP-ribose residues interfere with protein–protein and protein–DNA interactions, including the association of histones with DNA. In fact, histones bind poly(ADP-ribose) (PAR) with high affinity and this stimulates the removal of histones from DNA [7]. It is thought that this change in nucleosome occupancy frees DNA at the site of damage for efficient DNA repair. Histones themselves are also targets of mono- and poly(ADP-ribosyl)ation in mammals and in plants [145, 146], including on lysines [53, 91, 140]. The functional significance of this histone modification remains to be determined.
Fig. 1.
Schematic representation of domains found in PARP superfamily proteins. Protein domains are illustrated by colored boxes and are defined according to Pfam 25.0 [34], unless otherwise noted. Proteins are shown to scale with their lengths in amino acids indicated. a Land plants contain proteins similar to HsPARP1. b Land plants contain proteins similar to HsPARP3. c Plants and other eukaryotes contain HsPARP8 orthologs. d The SRO family is land plant specific. BRCT BRCA-1 C-terminus domain (PF00533), FPE fungal PARP E2-associated domain [32], LLP domain of unknown function found in the PARP8 subfamily of the PARP superfamily (Citarelli, Lee and Lamb, unpublished data), PADR1 domain of unknown function found in PARPs (PF08063), PRD PARP regulatory domain, PARP PARP catalytic domain (PF00644), RST RCD-SRO-TAF4 domain (PF12174), SAP presumed nucleic acid binding domain (PF02037), UBCc ubiquitin E2 catalytic domain (PF00179), WGR domain defined by conserved tryptophan, glycine and arginine residues (PF05406), WWE presumed protein-protein interaction domain characterized by tryptophan and glutamic acid residues (PF02825), ZnF DNA binding zinc finger domain (PF00645). At Arabidopsis thaliana, Hs Homo sapiens, Mg Magnaporthe grisea, Pp Physcomitrella patens
HsPARP1 and it orthologs in animal systems have been implicated in numerous other functions. Enzymatically silent PARP1 acts as a structural protein in chromatin and inhibits transcription by contributing to the condensation of chromatin [82, 97, 102, 139, 143]. However, when activated by developmental or environmental signals, PARP1 auto-modifies itself and other chromatin-associated proteins, opening chromatin to facilitate gene expression [59, 82, 138]. PARP1 has also been shown to have more direct roles in transcriptional regulation (reviewed in [83]). The broad range of roles for PARP1 suggests that other members of the PARP superfamily may also have many context-dependent roles.
In addition to HsPARP1, two other PARP superfamily members, HsPARP2 and HsPARP3, which are found in the same subgroup of the family as HsPARP1 [32], have been implicated in DNA repair in humans and other mammals. PARP2 is expressed in a broad range of tissues and cell types [124]; however, PARP3 expression shows tissue specificity in mouse [141]. It is thought that PARP2 and PARP3 function in conjunction with PARP1 during DNA repair and other processes such as gene regulation. Consistent with this hypothesis, while single mutant animals appear normal, parp1/parp2 knockout mice die during embryogenesis [40], while parp1/parp3 mice are very sensitive to DNA-damaging agents [19]. Both HsPARP2 and HsPARP3 have been shown to act in poly(ADP-ribosyl)ation with activity stimulated by DNA fragments [8, 19, 121]. However, HsPARP3 has also been reported to act as a mART toward histone H1 and to not be stimulated by DNA [87], suggesting that its enzymatic function maybe context dependent.
Removal of ADP-ribose from proteins
Poly(ADP-ribosyl)ation is a reversible modification, and poly(ADP-ribose) glycohydrolase (PARG) enzymes catalyze the hydrolysis of the glycosidic linkages of ADP-ribose polymers to produce free ADP-ribose. Thus, PARG reverses or counteracts the function of PARPs. In contrast to the PARP superfamily, only one or a handful of genes encode PARGs, although alternative splicing can produce multiple isoforms in mammals. PARG has been reported to be present in several subcellular compartments, including the nucleus and cytoplasm as well as mitochondria [20]. PARGs have not been extensively analyzed in plants. In the plant Arabidopsis thaliana, two genes encode putative PARGs, AtPARG1/TEJ (At2g31870) and AtPARG2 (At2g31865), which resulted from gene duplication [23].
Free ADP-ribose is known to be toxic, and it has been speculated that the lethality of mutations in PARG genes in animals is at least partially due to buildup of this metabolite [57, 80]. Nudix (nucleoside diphosphates linked to moiety X) hydrolases catalyze the hydrolysis of a variety of nucleoside diphosphate derivatives [18]. Three members of this family in Arabidopsis, AtNUDT2, AtNUDT6 and AtNUDT7, have been shown to hydrolyze both ADP-ribose (to AMP and ribose-5-phosphate) and NADH in vitro [52, 99] and these seem to be physiological substrates [64, 98]. Overexpression of AtNUDT2 has been shown to protect plants from depletion of NAD+ and ATP under oxidative stress conditions, suggesting its involvement in recycling the ADP-ribose generated by ADP-ribosylation [98]. Similarly, inhibition of removal of ADP-ribose through loss of function in AtNUDT7 has been shown to decrease the level of poly(ADP-ribose) in Arabidopsis, correlating with an increase in ADP-ribose [64], suggesting that PARP activity is inhibited in this background.
Distribution of PARP superfamily proteins in plants
Plants contain at least three types of PARP superfamily members
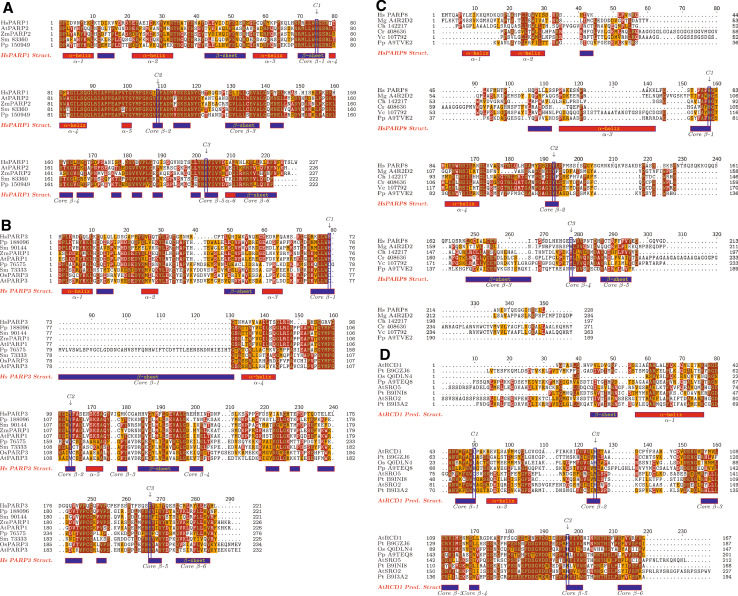
PARP activity in plants was first described in the 1970s, when it was demonstrated that a biochemical activity in the nuclei of wheat [145] and Nicotiana tabacum [146] was poly(ADP-ribosyl)ating histones. Since then, PARP superfamily members have been identified in plants, both by sequence similarity to animal proteins [13, 85] and through genetics [5]. Compared to humans, where this protein family contains 17 members and there are at least five different types of PARP subfamilies [32, 60], plants contain relatively few such proteins. All land plants contain orthologs of HsPARP1 and others that seem to be more related to HsPARP3 based on the sequence of their catalytic domains (Fig. 2 and Table 1; [32]). The HsPARP1 orthologs, typified by Arabidopsis thaliana PARP2 (AtPARP2), share a conserved domain structure with this protein (Fig. 1a). In common with other proteins that act in poly(ADP-ribosyl)ation, the plant HsPARP1 orthologs contain a so-called catalytic triad consisting of histidine-tyrosine-glutamic acid (HYE) residues within the PARP signature (Fig. 2a). The first two residues of this triad are necessary for NAD+ binding [116] while the third is important for polymer formation [90]. Although none of the plant proteins in these groups have been demonstrated to have poly(ADP-ribosyl)ation activity to date, it is very likely that they do in fact have this activity, based on the high identity of the plant catalytic domains to those of HsPARP1 (Fig. 2a). AtPARP2 is broadly expressed (Genevestigator; [151]), consistent with other HsPARP1 orthologs.
Fig. 2.
Multiple alignments of the PARP catalytic domains of land plant PARP proteins. These alignments only show the conserved PARP catalytic domain and the numbers indicate amino acids within these domains. Dots indicated gaps introduced to optimize the alignment. Identical amino acids are indicated by red shading and similar amino acids by orange shading. The amino acids present in the catalytic triad are boxed in blue and labeled C1, C2 and C3. The alignments were generated using the MUSCLE3.8.31 multiple alignment tool, using default settings [45]. Structures were obtained from the RCSB Protein Data Bank [117], unless otherwise noted. a Multiple alignment of HsPARP1 and its land plant orthologs. The structural elements present in HsPARP1 are shown at the bottom of the alignment. b Multiple alignment of HsPARP3 and its land plant orthologs. The structural elements present in HsPARP3 are shown at the bottom of the alignment. c Multiple alignment of HsPARP8, MgA4R2D2 and their green algal and moss orthologs. The structural elements present in HsPARP8 are shown below the alignment. d Multiple alignment of members of the SRO family. The predicted structural elements present in AtRCD1 are shown below the alignment. The structural prediction was done using Phyre [76]. Hs Homo sapiens, At Arabidopsis thaliana, Zm Zea mays, Sm Selaginella moellendorfii, Pp Physcomitrella patens, Os Oryza sativa, Mg Magnaporthe grisea, Ch Chlorella sp 142271, Cr Chlamydomonas reinhardtii, Vc Volvox carteri, Pt Populus trichocarpa
Table 1.
Poly(ADP-ribose) polymerases in Arabidopsis thaliana
| Arabidopsis thaliana gene | Locus ID | Human orthologsa | Selected plant orthologsa | Expression patternb | Enzyme activity | References |
|---|---|---|---|---|---|---|
| AtPARP1 | At4g02390 | HsPARP3 | ZmPARP1 | Expressed in all organs | Yes [13] | [85] |
| OsPARP1 | ||||||
| VvA5AIW8 | ||||||
| Pp188096 | ||||||
| Sm90144 | ||||||
| AtPARP2 | At2g31320 | HsPARP1 | ZmPARP2 | Expressed in all organs | ND | [13] |
| OsPARP2 | ||||||
| VvD7U2A8 | ||||||
| Pp150949 | ||||||
| Sm83360 | ||||||
| Sm76668 | ||||||
| AtPARP3 | At5g22470 | HsPARP3 | OsPARP3 | Seeds | ND | [16] |
| PARP3 | ||||||
| VvA5AUF8 | ||||||
| VvD7TCW5 | ||||||
| MtPARP3 | ||||||
| Pp76575 | ||||||
| Sm73333 | ||||||
| AtRCD1 | At1g32230 | NA | OsQ0DLN4 | Expressed in all organs | No [74] | [17] |
| OsQ336N3 | ||||||
| OsQ0J949 | ||||||
| OsQ654Q5 | ||||||
| VvA7PC35 | ||||||
| VvA5BDE5 | ||||||
| PtB9MU68 | ||||||
| PtB9GZJ6 | ||||||
| AtSRO1 | At2g35510 | NA | See AtRCD1 c | Expressed in all organs | ND | [17] |
| AtSRO2 | At1g23550 | NA | PtB9INI8 | Expressed in all organs | ND | [17] |
| PtB9HDP9 | ||||||
| PtB9HDP8 | ||||||
| PtB9HDP5 | ||||||
| AtSRO3 | At1g70440 | NA | See AtSRO2 c | ND | ND | [17] |
| AtSRO4 | At3g47720 | NA | VvA5BFU2 | ND | ND | [17] |
| PtB9I3A2 | ||||||
| PtB9IES0 | ||||||
| AtSRO5 | At5g62520 | NA | See AtSRO4 c | Expressed in all organs | ND | [5] |
The PARP superfamily members that are more similar to HsPARP3 within their catalytic domains are also found throughout land plants and are split into two distinct groups [32]. The first group, which contains the first plant PARP cloned, AtPARP1/APP [85], has an apparently plant-specific domain structure, containing two SAP domains in the N-terminus (Fig. 1b). SAP domains have been shown to bind to nucleic acids [100] and have also been demonstrated to function in localizing proteins to the kinetochore during mitosis [122]. In addition to the SAP domains, these proteins contain a WGR domain, a PRD domain and catalytic domain at the very C-terminus. These PARP superfamily members have the HYE catalytic triad within their PARP signature (Fig. 2b), and both AtPARP1 (Table 1; [13]) and Zea mays PARP1 (ZmPARP1; [89]) have been shown to have poly(ADP-ribosyl)ation activity, suggesting that all members of this group likely function in poly(ADP-ribosyl)ation. The second group of proteins similar to HsPARP3, typified by AtPARP3, do not contain SAP domains. They more closely resemble HsPARP2 in domain structure (Fig. 1b). Interestingly, these proteins have acquired changes in the catalytic domain such that they no longer contain an intact HYE catalytic triad (Fig. 2b). All members of this group contain a cysteine instead of the histidine at the first position and retain the glutamic acid at the third position. In angiosperms, the second position contains a valine instead of a tyrosine while seedless plants retain the tyrosine (Fig. 2b; [32]). The impact of these changes on catalytic activity of these proteins is unclear, although they would be predicted to eliminate NAD+ binding and therefore enzymatic activity. In Arabidopsis thaliana, AtPARP3 is expressed in developing seeds [16], suggesting a function during this part of the plant lifecycle.
Physcomitrella patens, a moss found in the basal land plant group the bryophytes, contains PARP superfamily members that are orthologous to HsPARP6, 8 and 16 (the PARP8 clade; [32]). Similar proteins are also found in some green algae, including Chlamydomonas reinhardtii and Volvox carteri, as well as many fungi and Trichomonas vaginalis within the Excavata [32]. However, this group of proteins appears to have been lost from vascular plants. No member of this clade, including those found in humans, have been functionally characterized. Interestingly, no green algae with sequenced genomes have any PARP superfamily members other than the PARP8-type. Other eukaryotic lineages have also lost most or all PARP-encoding genes from their genomes [32]. The moss protein in this group, A9TVE2, contains a long N-terminus with no known functional domains but containing an LLP domain, a domain of unknown function found only in the PARP8 clade of the PARP superfamily (Fig. 1c; Citarelli et al., in preparation). This protein contains a C-terminal extension with no apparent functional motifs. This domain structure is similar to that seen in the human PARP8 protein (Fig. 1c); interestingly, the fungal members of this clade contain a C-terminal UBCc domain (Fig. 1c; [32]). UBCc domains are the catalytic domains found in E2 Ub-conjugating enzymes, which carry Ub and transfer it either directly to a substrate or to an E3 ligase [35]. The presence of this domain in some PARP8 clade proteins suggests a connection between ADP-ribosylation and ubiquitination. In common with its orthologs in other groups of eukaryotes, the catalytic domain of PpA9TVE2 contains changes suggesting that it functions in mono(ADP-ribosyl)ation rather than as a bona fide PARP, retaining the first two residues of the catalytic triad, but replacing the third residue (Fig. 2c). However, since no members of the PARP8 clade have been characterized either functionally or biochemically, it is not clear what function these proteins may be playing.
Finally, land plants have acquired a novel group of PARP-like proteins, the SRO family. These proteins are found throughout land plants and consist of two subgroups [32, 74]. The first is ubiquitous in land plants and contains a WWE protein-protein interaction domain [11] in the N-terminus and a C-terminal extension past the PARP catalytic domain (Fig. 1d). This extension contains an RST domain [73]. The second subgroup is found only in the eudicot group of flowering plants and contains proteins that have lost the N-terminal region and retain only the catalytic domain and the RST domain (Fig. 1d). These proteins contain variant PARP signatures and may not act enzymatically (Fig. 2d). In fact, AtRCD1, the first member of this group identified, appears to be inactive (Table 1; [74]). However, the catalytic triads within this group vary, with some members, such as PpA9TEQ8, retaining the NAD+ binding residues (Fig. 2d; [32]). This suggests that activity must be assayed for multiple members before any conclusions on biochemical function can be formed for the family. Two members of this group from Arabidopsis, RCD1 and SRO1, have been shown to bind to transcription factors in yeast two-hybrid assays [17, 72]. The RST domain characteristic of the SRO family is also found in the transcription initiation complex component TAF4 [73]. These facts, along with the known roles of HsPARP1 in regulation of transcription and chromatin structure in absence of enzymatic activity, suggest that the SRO family may function in gene regulation at the transcriptional and/or chromatin level.
Functions of PARP superfamily members in plants
PARP proteins in DNA repair and the cell cycle
Bona fide PARPs were first discovered in association with DNA repair pathways and this is still the best-known and studied role of this class of enzymes (reviewed in [61]). HsPARP1 orthologs have been shown to be involved or implicated in DNA repair in animals [8, 118, 137], fungi [81, 126], Trypanosoma cruzi [46] and Dictyostelium discoideum [112]. It is likely that a role in DNA repair is shared by most orthologs of HsPARP1, HsPARP2 and HsPARP3. Although this has not been extensively studied in plants, there is some evidence supporting the involvement of PARPs in DNA repair in this group of organisms as well. In Arabidopsis, AtPARP1 and AtPARP2 expression is higher in genetic backgrounds that have increased DNA damage or replication stress [13, 113, 150] and in rapidly dividing tissues and stem cells in which the genome needs to be protected [93]. Their expression is induced by radiation [36, 43], genotoxic stress [30] and Gemini virus infection, presumably due to the presence of nicked viral replication intermediates [12]. AtPARP2 has been shown to bind DNA breaks [43]. PARP inhibitor studies suggest that PARP enzymes may participate in control of DNA recombination [110]. However, the studies done with PARP inhibitors need to be interpreted carefully. In plants, two inhibitors have been used: 3-aminobenzamide (3AB) and nicotinamide. 3AB is a competitive inhibitor of the poly(ADP-ribosyl)ation reaction [44] and is thought to act primarily on HsPARP1 and HsPARP2-like enzymes and to a lesser extent on HsPARP3-like enzymes, while nicotinamide, as one of the products of the ADP-ribosylation reaction, therefore inhibits it and would be expected to inhibit any enzyme with ADP-ribosylation activity or that produces nicotinamide (reviewed in [47]). However, both these compounds have suboptimal inhibitory potencies [44]. In addition, they are capable of inhibiting other enzymes in addition to PARPs. In particular, both 3AB (to a minimal extent) and nicotinamide inhibit sirtuins [55], histone deacetylases that couple lysine deacetylation to NAD+ hydrolysis [54]. These enzymes are found throughout plants [54], and their activities are likely to be altered by application of nicotinamide and perhaps 3AB. Arabidopsis encodes two sirtuins, AtSRT1 and AtSRT2 [106]. Therefore, phenotypes seen upon 3AB and nicotinamide application should be examined closely, as it is unclear which PARP superfamily members are inhibited, to what extent and if sirtuins are also impacted. In addition, inhibitor studies offer no insights into any non-enzymatic functions PARP proteins might have. Despite the limitations of PARP inhibitor studies done to date and the lack of a direct demonstration of activity in DNA repair, it is very likely that plant PARPs similar to AtPARP1 and AtPARP3 are involved in this process.
In addition to roles in DNA damage response and repair, various members of the PARP superfamily have been shown to be involved in cell cycle control and mitosis. PAR accumulates along spindles in animals, suggesting a role in spindle formation or regulation [29]. Tankyrase, a type of PARP confined to animals [32], is necessary for sister chromatid separation [63] and spindle function [28]. In plants, some studies also link poly(ADP-ribosyl)ation to cell cycle activity. PARP activity increases during exponential growth of Arabidopsis cell cultures and expression of AtPARP1 and AtPARP2 peaks during this time. This peak of PARP activity correlates with that of glutathione reductase and with an increase in the NAD+/NADH pool size and ratio [48, 107]. A more direct link between plant PARPs and mitosis was found by examining the localization of AtPARP1 and AtPARP2 in mitotic cells. GFP-labeled proteins expressed in tobacco cells localized with chromosomes and the spindle during mitosis, while AtPARP1 also associated with these structures in Arabidopsis roots [14]. AtPARP1’s association with chromosomes required the N-terminal SAP domains, while the localization of AtPARP2 required the two zinc fingers and associated regions. Based on these observations, it is likely that poly(ADP-ribosyl)ation and/or PARP proteins are likely to function during the cell cycle and mitosis in plant cells.
PARP superfamily proteins influence abiotic stress responses in plants
Poly(ADP-ribosyl)ation has been implicated in stress response in eukaryotes since at least the 1970s. In particular, HsPARP1 and other similar PARPs are known to be important in the balance between cell survival and death in response to a number of stresses. Overactivation of HsPARP1 can result in massive necrotic cell death and has been implicated in disease (reviewed in [37, 58–60, 68, 125, 130]). However, it is required for genomic integrity and to protect proliferating cells and some non-proliferating cell types against cell death induced by oxidative stress [39, 40, 66]. During apoptosis, HsPARP1 is proteolytically inactivated by cleavage into characteristic fragments by executioner caspases [25, 94, 96, 134, 149]; this inactivation is hypothesized to prevent energy depletion through NAD+ consumption. The cleavage fragments have been shown to act in a dominant negative manner to prevent full-length HsPARP1 activity [149]. Therefore, HsPARP1’s role as either a survival factor or a death signal is context dependent. In plants, the involvement of PARPs and poly(ADP-ribosyl)ation in abiotic stress response has been the best-studied function for the protein family.
The land plant PARPs similar to both HsPARP1 and HsPARP3 have been implicated in cell death. In soybean cells, upon induction of oxidative stress, PARPs are activated and cellular NAD+ levels are reduced. This is followed by programmed cell death (PCD). This PCD could be inhibited by PARP inhibitors and/or by downregulation of PARP [10]. Consistent with this, overexpression of AtPARP1 in soybean improved the resistance of soybean cells to mild oxidative stress [10]. The cleavage of PARP during cell death may also be shared with animals. Cultured tobacco cells undergo cell death upon heat shock. An antibody raised against HsPARP1 recognized a tobacco protein cleaved to approximately 89 KDa upon heat shock, a size consistent with the known cleavage product produced during apoptotic death in animal systems [135]. Application of 3AB and nicotinamide reduced the amount of death induced by heat shock, suggesting that PARP activity may be involved in the cell death.
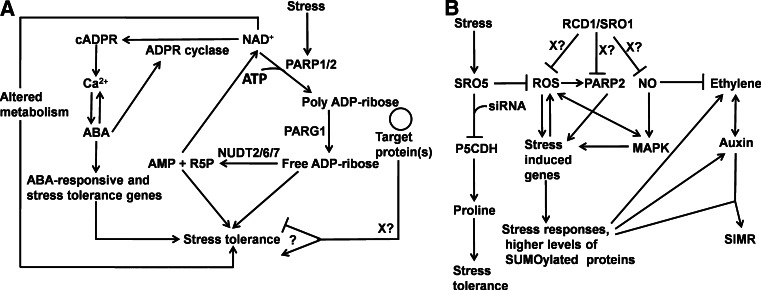
Most of the work connecting PARP superfamily members with stress response in plants has been done in Arabidopsis (Fig. 3). Expression studies in this plant have implicated both AtPARP1 and AtPARP2 in response to abiotic stress. In particular, AtPARP2 is upregulated by oxidative stress and salinity [30, 43, 98]. AtPARP3, although under normal conditions only expressed in seeds, can be upregulated by paraquat, salinity, high light intensity and drought [98]. Functionally, AtPARP1 and AtPARP2 have been implicated in abiotic stress response. Induction of PARP activity under oxidative stress conditions results in depletion of NAD+ and ATP levels, and PARP inhibition can enhance the tolerance to this stress, suggesting that PARP activity is detrimental to plants under stress conditions (Fig. 3a; [98]). Consistent with this, downregulation of AtPARP1 and AtPARP2 by RNAi reduced stress-induced poly(ADP-ribosyl)ation and NAD+ consumption, preventing ATP depletion. This prevented accumulation of reactive oxygen species and increased stress tolerance [38]. However, the consistent upregulation of these genes under abiotic stress conditions suggests that these genes must have a positive impact on survival at a certain level, similar to the situation seen in animals. As no true knockouts have been examined, this remains to be established. Another alternative hypothesis for the increased stress tolerance seen in parp-deficient plants, involves the potential increased levels of the cyclic nucleotide ADP-ribose (cADPR), which is synthesized from NAD+ [142]. The plant hormone abscisic acid (ABA), important in abiotic stress response, signals through Ca2+, and cADPR has been shown to act as a second messenger in this pathway before changes in gene expression [147]. Increased levels of cADPR levels in Arabidopsis can induce more than 100 ABA-responsive genes [123]. parp-deficient plants consume less NAD+, increasing the availability of this metabolite and would be expected to facilitate increased production of cADPR. This increased cADPR could cause production of ABA-regulated stress response genes, conferring tolerance to the parp-deficient plants (Fig. 3a).
Fig. 3.
PARP superfamily members are important for stress responses in Arabidopsis thaliana. a The activities of the bona fide PARPs, AtPARP1 and AtPARP2, must be balanced by PARGs and nudix hydrolases in order to maximize stress tolerance. The expression and/or activities of these proteins are induced under stress conditions, where they function to modify proteins that influence stress response. In order to avoid depletion of NAD+, PARG and nudix activities release AMP and R5P, which can be recycled. In the absence of PARP activity, accumulated NAD+ can be used to produce cADPR to activate ABA-responsive genes. b SRO family members modulate stress responses by repression of ROS and nitric acid. ABA absisic acid, AMP adenosine monophosphate, ATP adenosine triphosphate, cADPR cyclic nucleotide ADP-ribose, MAPK mitogen-activated protein kinase, NAD + nicotinamide adenine dinucleotide, NO nitric oxide, P5CDH Δ1-pyrroline-5-carboxylate dehydrogenase, R5P ribose-5-phosphate, ROS reactive oxygen species, SIMR stress-induced morphogenetic response. References: [1, 4–6, 22, 33, 38, 49, 64, 65, 70, 71, 98, 104, 109, 115, 123, 131, 133, 142, 147]
In Arabidopsis, several plant-specific SRO family members of PARP-like proteins have been implicated in stress response. RCD1, the founding member of this group, was originally identified as a stress response gene [104] and encodes an SRO family member with a WWE domain in its N-terminus [5, 6, 104]. rcd1 mutants are hypersensitive to ozone and other sources of apoplastic reactive oxygen species (ROS) [103, 131] as well as salt [131]. However, rcd mutants are resistant to UV-B and the herbicide paraquat, which generates reactive oxygen species in the plastid [5, 49]. RCD1’s paralog, SRO1, is also involved in stress response. Unlike rcd1 plants, sro1-1 plants are not resistant to chloroplastic ROS induced by paraquat and are resistant to apoplastic ROS. sro1-1 plants also display opposite responses to rcd1 mutants under salt stress, being resistant. Loss of either RCD1 or SRO1 confers resistance to osmotic stress [131]. Even under normal conditions rcd1-3; sro1-1 plants show expression of stress response genes and accumulation of SUMOylated proteins (which have been shown to accumulate under multiple stresses [84, 92]), suggesting that RCD1 and SRO1 may function as inhibitors of stress responses [133]. Consistent with this interpretation is the fact that rcd1 single mutants have been shown to accumulate reactive oxygen species (ROS; [104]) and nitric oxide [4] under non-stress conditions. In fact, many aspects of the developmental phenotype of rcd1-3; sro1-1 plants (described below) resemble those of plants exposed to chronic low-level abiotic stress. The common stress-associated phenotypes seen under various abiotic stresses have been termed stress-induced morphogenetic response (SIMR; [109]). If RCD1 and SRO1 act as inhibitors of abiotic stress response, particularly accumulation of ROS (Fig. 3b), the growth phenotypes of their mutants may share pathways with SIMR. Interestingly, expression of AtPARP2 increases in rcd1-3; sro1-1 plants [133], implying that the SRO family of PARP-like proteins may regulate more traditional PARP-encoding genes, directly or indirectly.
Arabidopsis contains four members of the eudicot-specific group of short SRO family members, SRO2-5. Very little work has been done on these genes. SRO2 is upregulated in response to high light in chloroplastic ascorbate peroxidase mutants [75]. SRO5 is the best-explored member of this group. SRO5 expression is very low under normal conditions but its expression has been shown to be induced by salt treatment [22] and repressed by high light [77]. sro5 plants were more sensitive to H2O2-mediated oxidative stress and to salt stress [22]. SRO5 has also been implicated in regulation of proline metabolism at the small RNA level [22]. Δ1-pyroline-5-carboxylate (P5C) is an intermediate in proline synthesis and catabolism. It has been shown to promote ROS production, reduce growth and induce the expression of stress genes [41, 62, 67]. SRO5 and the gene-encoding Δ1-pyrroline-5-carboxylate dehydrogenase (P5CDH), an enzyme that catabolizes P5C, overlap each other in the antisense orientation. This causes the formation of siRNAs from both loci. When SRO5 is induced by salt stress, a 24-nt SRO5/P5CDH siRNA is produced, eventually leading to the downregulation of P5CDH and the accumulation of proline important for salt tolerance (Fig. 3b). Downregulation of P5CDH will also lead to ROS accumulation; SRO5 counteracts this ROS accumulation, implying it can act to inhibit some stress responses. It has not been shown that this regulatory relationship between SRO5 and P5CDH is found in any other plant species; however, the role of SRO5 in reducing ROS accumulation could be a core function of SRO family members.
Not surprisingly, since ADP-ribosylation has been implicated in response to environmental conditions, PARG genes are also implicated in stress response. AtPARG1 is upregulated in response to gamma irradiation [36] and AtPARG2 is upregulated by treatment with paraquat, salinity, high light and drought [98]. parg1 mutants have lower tolerance to osmotic, drought and oxidative stress. Consistent with this, parg1 plants had lower expression of two oxidative stress defense genes, AtALTERNATIVE OXIDASE 1 and AtASCORBATE PEROXIDASE 2 [86]. Clearly, there is a complicated relationship between poly(ADP-ribosylation) and abiotic stress tolerance.
The PARP superfamily and responses to biotic stress
Poly(ADP-ribosyl)ation and PARP superfamily proteins were first implicated in pathogen response indirectly. AtNUDT7 was identified through gene expression studies to act downstream of Arabidopsis thaliana ENHANCED DISEASE SUSCEPTIBILITY1 (AtEDS1), an important regulator of both basal and receptor-triggered immunity [15]. atnudt7 mutants had enhanced basal resistance, increased levels of salicylic acid (SA), a crucial signaling molecule required for expression of plant defense responses (reviewed in [56]), and spontaneous lesions on leaves, suggesting that defense responses were constitutive in these plants. The same year, this gene was identified by mutational analysis as having constitutive resistance against the biotrophic pathogenic bacteria Pseudomonas syringae [69]. Subsequent work has implicated AtNUDT7 in EDS-driven cell death and both SA-dependent and SA-independent defense pathways, perhaps by modulating redox balance or PARP activity [51, 52, 70, 129]
More direct evidence implicating PARPs and poly(ADP-ribosyl)ation in plant defense has been provided by examination of Arabidopsis thaliana–Pseudomonas syringae plant disease resistance gene (R)–bacterial avirulence (Avr) gene interactions. AtNUDT7, discussed above, and AtPARG2 were induced in multiple such plant–pathogen interactions and also by the elicitor flg22 (flagellin epitope). PARP activity, as measured by amount of poly(ADP-ribose) accumulation, is increased after exposure of Arabidopsis to avirulent Pseudomonas syringae [2]. Consistent with an involvement of PARPs in the innate immune response in plants, the PARP inhibitor 3AB perturbs some responses to microbe-associated molecular patterns including callose deposition and other cell wall modifications [1, 2]. However, as mentioned above, 3AB can inhibit sirtuins. AtSRT2 has been implicated in biotic defense and has been shown to negatively regulate plant basal defense against Pseudomonas syringae [144], again suggesting that results with inhibitors need be interpreted carefully. Plants mutant for both of the genes encoding PARGs in the Arabidopsis genome, AtPARG1/TEJ and AtPARG2, have accelerated necrotic disease symptoms when infected with Botrytis [1], supporting roles in defense against both biotropic and necrotrophic pathogens. Taken together, these results suggest that PARPs and poly(ADP-ribosyl)ation are involved in pathogen response in plants.
PARP superfamily members and plant development
Although PARPs have been extensively studied for their roles in genome integrity, stress response and apoptosis, relatively little work has been done on their role in development, although there is some evidence for their involvement. Mouse parp1/parp2 mutants die embryonically, demonstrating that these genes are essential for development [40]. This is also true in Drosophila melanogaster, where loss of the single PARP gene found in this organism causes larval lethality [139]. Recently, DrPARP3 was shown to act in ectodermal specification and neural crest development in zebrafish [119]. Taken together, this suggests that in animals at least PARP1, 2 and 3 are necessary for proper development. Downregulation of PARPs in the amoebae Dictyostelium discoideum has been shown to arrest development at the slug stage [111], suggesting that in Amoebozoa, sister group to the Opithokonts (animals, fungi and choanoflagalletes), development is also regulated by these enzymes.
In land plants, little is known about the roles in development of the PARPs orthologous to those found in other eukaryotes. Using in vitro induction of tracheary elements in both Helianthus tuberosus and Pisum sativum, 3AB treatment was shown to inhibit this process, suggesting that PARPs or sirtuins or both could be involved in this process [108]. Knockdown of AtPARP1 and/or AtPARP2 was not found to alter Arabidopsis development [38], although development was not closely examined. Silencing of the AtPARP2 ortholog in oilseed rape (Brassica napus) did not alter its development either [142]. However, it is likely that the knockdown lines used in these studies retain some gene function; it is conceivable that total loss of these PARPs would be lethal in plants as has been seen in animals. As mentioned above, no functional work has been down on AtPARP3 or its orthologs in other plants. Poly(ADP-ribosyl)ation has been implicated in establishment of circadian period in Arabidopsis. AtPARG1/TEJ was originally identified through a genetic screen for altered period length. tej mutants have a longer period and this can be rescued by 3AB treatment, suggesting that the phenotype is caused by the buildup of poly(ADP-ribosyl)ated proteins, although it is unclear what these proteins are [105]. It is likely that AtPARP1 and/or AtPARP2 are the proteins responsible for the enzymatic activity regulating the circadian clock.
In contrast to the canonical PARP genes in plants, the SRO family members RCD1 and SRO1 appear essential for proper development in Arabidopsis. Loss of RCD1 causes dramatic developmental defects including reduced stature, malformed leaves, early flowering, abnormal phyllotaxy, small and deformed floral organs, increased lateral root number and length, and shorter primary roots [5, 104, 131]. sro1 plants display some subtle developmental defects similar to the root and flower phenotypes of rcd1-3. However, plant height and leaf shape and size appear normal [131]. The majority of rcd1-3; sro1-1 individuals die during embryogenesis [131], demonstrating that the SRO family may be necessary for land plant development. rcd1-3; sro1-1 plants are very small; at least some of this decrease in height is caused by a decrease in cell elongation [131]. However, double mutant plants also make fewer cells [132]. Roots of these plants are shorter than wild-type and have a reduction in the size of the division zone. The division zone has fewer total cells and fewer mitotic cells. Morphology of the roots cells and planes of cell division are abnormal in addition to the defect in the number of cells. The shoot of rcd1-3; sro1-1 plants also contains fewer cells. Although the division zone is small and disorganized in the root of rcd1-3; sro1-1 plants, cell fate is generally correctly laid down as assayed by reporter gene expression. This suggests that cell fate decisions are not globally disrupted in this mutant background. Consistent with retention of proper cell fate decisions in the root, maxima of the plant hormone auxin are formed at the root pole and tips of forming cotyledons in the embryo and at the root tip postembryonically in double mutant plants and expression and polarization of components of the polar auxin transport system are largely normal. However, cell differentiation is disrupted in double mutant plants. Cells fail to elongate properly and specialized cell walls are not well formed [131, 132]. These results suggest that RCD1 and SRO1 are necessary for cell division and cell differentiation and, by extension, suggest that members of the SRO family in other plants are likely to be necessary for similar processes. As mentioned above, some of these growth phenotypes resemble those of SIMR, suggesting that accumulation or misregulation of ROS may be responsible for some of the phenotypes seen in rcd1 and/or sro1 plants, consistent with a role for SRO family members in redox regulation.
Conclusions
The PARP superfamily and poly(ADP-ribosyl)ation is clearly important for a variety of biological functions in plants, including DNA recombination and stress responses. In particular, the role of PARPs in abiotic stress responses is especially well established. In addition, a land plant-specific group of PARP-like proteins, the SRO family, is necessary for normal plant development. The PARP superfamily within plants contains several unique subfamilies, making plants an excellent system to examine interplays between different subfamilies of this large family. At least three subfamilies of PARPs appear to have arisen at the base of the land plants. These include the AtPARP1 group, which contains SAP domains, unique for PARP superfamily members, the AtPARP3 group, with unique substitutions within the catalytic domain and the SRO family. It is tempting, given the association of poly(ADP-ribosyl)ation and the SRO family with stress response and control of ROS, to speculate that these proteins evolved to help plant contend with the rigors of life on land, including increased exposure to UV-B light and desiccation stress. However, there is no information on gene content for the sister group to land plants, the streptophyte green algae. Until it is determined when these PARP subfamilies emerged, this question will remain open.
Fundamental questions remain in the field. First, only two proteins (AtPARP1 and ZmPARP1), which are similar to HsPARP3, have been demonstrated to have poly(ADP-ribosyl)ation activity while one member of the SRO family is enzymatically inactive. Given the variation within the catalytic domains of the plant PARP proteins, in particular the AtPARP3 subfamily and the SRO family, it is essential to determine which proteins have poly(ADP-ribosyl)ation or mono(ADP-ribose) transferase activity. Second, given the results that indicate overlap in function between HsPARP1 and HsPARP3, it is necessary to determine the extent to which the land plant PARPs similar to these proteins have redundant functions. This necessitates the isolation of null alleles in these genes and careful analysis of single, double and triple mutants. This will also reveal if PARPs are essential in plants as they are in animals. Third, no targets of ADP-ribosylation other than histones have been identified in plants. Even for histones, it is unclear which PARP(s) in plants is responsible for the activity that has been detected in the nucleus. It will be essential to identify targets of the various PARP enzymes and determine the effect of poly(ADP-ribosyl)ation on these proteins. Identification of these targets will provide insight into plant immunity and stress responses, as well as development. We are now in a position to develop a better understanding of the roles of PARPs and poly(ADP-ribosyl)ation in plants and the diversification of the proteins involved in this process in eukaryotes.
Acknowledgments
We thank Dr. Patrice Hamel, Dr. David Mackey and Dr. Iris Meier (Ohio State University), members of the Lamb laboratory and an anonymous reviewer for helpful advice on the manuscript. We apologize to colleagues whose work we could not include due to space limitations.
Footnotes
A recent review on this subject covering similar but not identical data was published during review of this manuscript [24].
References
- 1.Adams-Phillips L, Briggs AG, Bent AF. Disruption of poly(ADP-ribosyl)ation mechanisms alters responses of Arabidopsis to biotic stress. Plant Physiol. 2010;152:267–280. doi: 10.1104/pp.109.148049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams-Phillips L, Wan J, Tan X, Dunning FM, Meyers BC, Michelmore RW, Bent AF. Discovery of ADP-ribosylation and other plant defense pathway elements through expression profiling of four different Arabidopsis-Pseudomonas R-avr interactions. Mol Plant Microbe Interact. 2008;21:646–657. doi: 10.1094/MPMI-21-5-0646. [DOI] [PubMed] [Google Scholar]
- 3.Aguiar RC, Yakushijin Y, Kharbanda S, Salgia R, Fletcher JA, Shipp MA. BAL is a novel risk-related gene in diffuse large B-cell lymphomas that enhances cellular migration. Blood. 2000;96:4328–4334. [PubMed] [Google Scholar]
- 4.Ahlfors R, Brosche M, Kollist H, Kangasjarvi J. Nitric oxide modulates ozone-induced cell death, hormone biosynthesis and gene expression in Arabidopsis thaliana. Plant J. 2008;58:1–12. doi: 10.1111/j.1365-313X.2008.03756.x. [DOI] [PubMed] [Google Scholar]
- 5.Ahlfors R, Lang S, Overmyer K, Jaspers P, Brosche M, Tauriainen A, Kollist H, Tuominen H, Belles-Boix E, Piippo M, Inze D, Palva ET, Kangasjarvi J. Arabidopsis RADICAL-INDUCED CELL DEATH1 belongs to the WWE protein-protein interaction domain protein family and modulates abscisic acid, ethylene, and methyl jasmonate responses. Plant Cell. 2004;16:1925–1937. doi: 10.1105/tpc.021832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahlfors R, Macioszek V, Rudd J, Brosche M, Schlichting R, Scheel D, Kangasjarvi J. Stress hormone-independent activation and nuclear translocation of mitogen-activated protein kinases in Arabidopsis thaliana during ozone exposure. Plant J. 2004;40:512–522. doi: 10.1111/j.1365-313X.2004.02229.x. [DOI] [PubMed] [Google Scholar]
- 7.Althaus FR, Hofferer L, Kleczkowska HE, Malanga M, Naegeli H, Panzeter PL, Realini CA. Histone shuttling by poly ADP-ribosylation. Mol Cell Biochem. 1994;138:53–59. doi: 10.1007/BF00928443. [DOI] [PubMed] [Google Scholar]
- 8.Ame JC, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, Muller S, Hoger T, Menissier-de Murcia J, de Murcia G. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J Biol Chem. 1999;274:17860–17868. doi: 10.1074/jbc.274.25.17860. [DOI] [PubMed] [Google Scholar]
- 9.Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 10.Amor Y, Babiychuk E, Inze D, Levine A. The involvement of poly(ADP-ribose) polymerase in the oxidative stress responses in plants. FEBS Lett. 1998;440:1–7. doi: 10.1016/S0014-5793(98)01408-2. [DOI] [PubMed] [Google Scholar]
- 11.Aravind L. The WWE domain: a common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem Sci. 2001;26:273–275. doi: 10.1016/S0968-0004(01)01787-X. [DOI] [PubMed] [Google Scholar]
- 12.Ascencio-Ibanez JT, Sozzani R, Lee TJ, Chu TM, Wolfinger RD, Cella R, Hanley-Bowdoin L. Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 2008;148:436–454. doi: 10.1104/pp.108.121038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Babiychuk E, Cottrill PB, Storozhenko S, Fuangthong M, Chen Y, O’Farrell MK, Van Montagu M, Inze D, Kushnir S. Higher plants possess two structurally different poly(ADP-ribose) polymerases. Plant J. 1998;15:635–645. doi: 10.1046/j.1365-313x.1998.00240.x. [DOI] [PubMed] [Google Scholar]
- 14.Babiychuk E, Van Montagu M, Kushnir S. N-terminal domains of plant poly(ADP-ribose) polymerases define their association with mitotic chromosomes. Plant J. 2001;28:245–255. doi: 10.1046/j.1365-313X.2001.01143.x. [DOI] [PubMed] [Google Scholar]
- 15.Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE. Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell. 2006;18:1038–1051. doi: 10.1105/tpc.105.039982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Becerra C, Puigdomenech P, Vicient CM. Computational and experimental analysis identifies Arabidopsis genes specifically expressed during early seed development. BMC Genomics. 2006;7:38. doi: 10.1186/1471-2164-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belles-Boix E, Babiychuk E, Van Montagu M, Inze D, Kushnir S. CEO1, a new protein from Arabidopsis thaliana, protects yeast against oxidative damage. FEBS Lett. 2000;482:19–24. doi: 10.1016/S0014-5793(00)02016-0. [DOI] [PubMed] [Google Scholar]
- 18.Bessman MJ, Frick DN, O’Handley SF. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J Biol Chem. 1996;271:25059–25062. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- 19.Boehler C, Gauthier LR, Mortusewicz O, Biard DS, Saliou JM, Bresson A, Sanglier-Cianferani S, Smith S, Schreiber V, Boussin F, Dantzer F. Poly(ADP-ribose) polymerase 3 (PARP3), a newcomer in cellular response to DNA damage and mitotic progression. Proc Natl Acad Sci USA. 2011;108:2783–2788. doi: 10.1073/pnas.1016574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonicalzi ME, Haince JF, Droit A, Poirier GG. Regulation of poly(ADP-ribose) metabolism by poly(ADP-ribose) glycohydrolase: where and when? Cell Mol Life Sci. 2005;62:739–750. doi: 10.1007/s00018-004-4505-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- 22.Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis . Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bowers JE, Chapman BA, Rong J, Paterson AH. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- 24.Briggs AG, Bent AF. Poly(ADP-ribosyl)ation in plants. Trends Plant Sci. 2011;16:372–380. doi: 10.1016/j.tplants.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 25.Casciola-Rosen L, Nicholson DW, Chong T, Rowan KR, Thornberry NA, Miller DK, Rosen A. Apopain/CPP32 cleaves proteins that are essential for cellular repair: a fundamental principle of apoptotic death. J Exp Med. 1996;183:1957–1964. doi: 10.1084/jem.183.5.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chambon P, Weil JD, Doly J, Strosser MT, Mandel P. On the formation of a novel adenylic compound by enzymatic extracts of liver nuclei. Biochem Biophys Res Commun. 1966;25:638–643. doi: 10.1016/0006-291X(66)90502-X. [DOI] [Google Scholar]
- 27.Chambon P, Weill JD, Mandel P. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem Biophys Res Commun. 1963;11:39–43. doi: 10.1016/0006-291X(63)90024-X. [DOI] [PubMed] [Google Scholar]
- 28.Chang P, Coughlin M, Mitchison TJ. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat Cell Biol. 2005;7:1133–1139. doi: 10.1038/ncb1322. [DOI] [PubMed] [Google Scholar]
- 29.Chang P, Jacobson MK, Mitchison TJ. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432:645–649. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- 30.Chen IP, Haehnel U, Altschmied L, Schubert I, Puchta H. The transcriptional response of Arabidopsis to genotoxic stress—a high-density colony array study (HDCA) Plant J. 2003;35:771–786. doi: 10.1046/j.1365-313X.2003.01847.x. [DOI] [PubMed] [Google Scholar]
- 31.Chou HY, Chou HT, Lee SC. CDK-dependent activation of poly(ADP-ribose) polymerase member 10 (PARP10) J Biol Chem. 2006;281:15201–15207. doi: 10.1074/jbc.M506745200. [DOI] [PubMed] [Google Scholar]
- 32.Citarelli M, Teotia S, Lamb RS. Evolutionary history of the poly(ADP-ribose) polymerase gene family in eukaryotes. BMC Evol Biol. 2010;10:308. doi: 10.1186/1471-2148-10-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clarke A, Desikan R, Hurst RD, Hancock JT, Neill SJ. NO way back: nitric oxide and programmed cell death in Arabidopsis thaliana suspension cultures. Plant J. 2000;24:667–677. doi: 10.1046/j.1365-313x.2000.00911.x. [DOI] [PubMed] [Google Scholar]
- 34.Coggill P, Finn RD, Bateman A (2008) Identifying protein domains with the Pfam database. Curr Protoc Bioinformatics Chapter 2: Unit 2 5 [DOI] [PubMed]
- 35.Cook WJ, Jeffrey LC, Sullivan ML, Vierstra RD. Three-dimensional structure of a ubiquitin-conjugating enzyme (E2) J Biol Chem. 1992;267:15116–15121. doi: 10.2210/pdb1aak/pdb. [DOI] [PubMed] [Google Scholar]
- 36.Culligan KM, Robertson CE, Foreman J, Doerner P, Britt AB. ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 2006;48:947–961. doi: 10.1111/j.1365-313X.2006.02931.x. [DOI] [PubMed] [Google Scholar]
- 37.D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342:249–268. doi: 10.1042/0264-6021:3420249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Block M, Verduyn C, De Brouwer D, Cornelissen M. Poly(ADP-ribose) polymerase in plants affects energy homeostasis, cell death and stress tolerance. Plant J. 2005;41:95–106. doi: 10.1111/j.1365-313X.2004.02277.x. [DOI] [PubMed] [Google Scholar]
- 39.de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, LeMeur M, Walztinger C, Chambon P, de Murcia G. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci USA. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Murcia JM, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, Schreiber V, Ame J-C, Dierich A, LeMeur M, Sabatier L, Chambon P, de Murcia G. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 2003;22:2255–2263. doi: 10.1093/emboj/cdg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deuschle K, Funck D, Forlani G, Stransky H, Biehl A, Leister D, van der Graaff E, Kunze R, Frommer WB. The role of [Delta]1-pyrroline-5-carboxylate dehydrogenase in proline degradation. Plant Cell. 2004;16:3413–3425. doi: 10.1105/tpc.104.023622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doly J, Petek F. Etude de la structure d’un compose “poly(ADP-ribose)” synthetise par des extraits nucleaires de foie de poulet. CR Hebd Scanc Acad Sci Ser D Sci Nat. 1966;263:1341–1344. [Google Scholar]
- 43.Doucet-Chabeaud G, Godon C, Brutesco C, de Murcia G, Kazmaier M. Ionising radiation induces the expression of PARP-1 and PARP-2 genes in Arabidopsis . Mol Genet Genomics. 2001;265:954–963. doi: 10.1007/s004380100506. [DOI] [PubMed] [Google Scholar]
- 44.Durkacz BW, Omidiji O, Gray DA, Shall S. (ADP-ribose)n participates in DNA excision repair. Nature. 1980;283:593–596. doi: 10.1038/283593a0. [DOI] [PubMed] [Google Scholar]
- 45.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fernandez Villamil SH, Baltanas R, Alonso GD, Vilchez Larrea SC, Torres HN, Flawia MM. TcPARP: a DNA damage-dependent poly(ADP-ribose) polymerase from Trypanosoma cruzi . Int J Parasitol. 2008;38:277–287. doi: 10.1016/j.ijpara.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 47.Ferraris DV. Evolution of poly(ADP-ribose) polymerase-1 (PARP-1) inhibitors. From concept to clinic. J Med Chem. 2010;53:4561–4584. doi: 10.1021/jm100012m. [DOI] [PubMed] [Google Scholar]
- 48.Foyer CH, Pellny TK, Locato V, De Gara L. Analysis of redox relationships in the plant cell cycle: determinations of ascorbate, glutathione and poly (ADPribose)polymerase (PARP) in plant cell cultures. Methods Mol Biol. 2008;476:199–215. doi: 10.1007/978-1-59745-129-1_14. [DOI] [PubMed] [Google Scholar]
- 49.Fujibe T, Saji H, Arakawa K, Yabe N, Takeuchi Y, Yamamoto KT. A methyl viologen-resistant mutant of Arabidopsis, which is allelic to ozone-sensitive rcd1, is tolerant to supplemental ultraviolet-B irradiation. Plant Physiol. 2004;134:275–285. doi: 10.1104/pp.103.033480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fujimura S, Hasegawa S, Shimizu Y, Sugimura T. Polymerization of the adenosine 5’-diphosphate-ribose moiety of nicotinamide-adenine dinucleotide by nuclear enzyme. I. Enzymatic reactions. Biochim Biophys Acta. 1967;145:247–259. doi: 10.1016/0005-2787(67)90043-3. [DOI] [PubMed] [Google Scholar]
- 51.Ge X, Li GJ, Wang SB, Zhu H, Zhu T, Wang X, Xia Y. AtNUDT7, a negative regulator of basal immunity in Arabidopsis, modulates two distinct defense response pathways and is involved in maintaining redox homeostasis. Plant Physiol. 2007;145:204–215. doi: 10.1104/pp.107.103374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ge X, Xia Y. The role of AtNUDT7, a Nudix hydrolase, in the plant defense response. Plant Signal Behav. 2008;3:119–120. doi: 10.4161/psb.3.2.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Golderer G, Grobner P. ADP-ribosylation of core histones and their acetylated subspecies. Biochem J. 1991;277:607–610. doi: 10.1042/bj2770607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Greiss S, Gartner A. Sirtuin/Sir2 phylogeny, evolutionary considerations and structural conservation. Mol Cells. 2009;28:407–415. doi: 10.1007/s10059-009-0169-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grozinger CM, Chao ED, Blackwell HE, Moazed D, Schreiber SL. Identification of a class of small molecule inhibitors of the sirtuin family of NAD-dependent deacetylases by phenotypic screening. J Biol Chem. 2001;276:38837–38843. doi: 10.1074/jbc.M106779200. [DOI] [PubMed] [Google Scholar]
- 56.Halim VA, Vess A, Scheel D, Rosahl S. The role of salicylic acid and jasmonic acid in pathogen defence. Plant Biol (Stuttg) 2006;8:307–313. doi: 10.1055/s-2006-924025. [DOI] [PubMed] [Google Scholar]
- 57.Hanai S, Kanai M, Ohashi S, Okamoto K, Yamada M, Takahashi H, Miwa M. Loss of poly(ADP-ribose) glycohydrolase causes progressive neurodegeneration in Drosophila melanogaster. Proc Natl Acad Sci USA. 2004;101:82–86. doi: 10.1073/pnas.2237114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biol Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hassa PO, Hottiger MO. The functional role of poly(ADP-ribose)polymerase 1 as novel coactivator of NF-kappaB in inflammatory disorders. Cell Mol Life Sci. 2002;59:1534–1553. doi: 10.1007/s00018-002-8527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci. 2008;13:3046–3082. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- 61.Heitz F, Harter P, Ewald-Riegler N, Papsdorf M, Kommoss S, du Bois A. Poly(ADP-ribosyl)ation polymerases: mechanism and new target of anticancer therapy. Expert Rev Anticancer Ther. 2010;10:1125–1136. doi: 10.1586/era.10.53. [DOI] [PubMed] [Google Scholar]
- 62.Hellmann H, Funck D, Rentsch D, Frommer WB. Hypersensitivity of an Arabidopsis sugar signaling mutant toward exogenous proline application. Plant Physiol. 2000;123:779–789. doi: 10.1104/pp.123.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hsiao SJ, Smith S. Sister telomeres rendered dysfunctional by persistent cohesion are fused by NHEJ. J Cell Biol. 2009;184:515–526. doi: 10.1083/jcb.200810132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ishikawa K, Yoshimura K, Harada K, Fukusaki E, Ogawa T, Tamoi M, Shigeoka S. AtNUDX6, an ADP-ribose/NADH pyrophosphohydrolase in Arabidopsis, positively regulates NPR1-dependent salicylic acid signaling. Plant Physiol. 2010;152:2000–2012. doi: 10.1104/pp.110.153569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ishikawa K, Yoshimura K, Ogawa T, Shigeoka S. Distinct regulation of Arabidopsis ADP-ribose/NADH pyrophosphohydrolases, AtNUDX6 and 7, in biotic and abiotic stress responses. Plant Signal Behav. 2010;5:839–841. doi: 10.4161/psb.5.7.11820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ishizuka S, Martin K, Booth C, Potten CS, de Murcia G, Burkle A, Kirkwood TB. Poly(ADP-ribose) polymerase-1 is a survival factor for radiation-exposed intestinal epithelial stem cells in vivo. Nucleic Acids Res. 2003;31:6198–6205. doi: 10.1093/nar/gkg840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iyer S, Caplan A. Products of proline catabolism can induce osmotically regulated genes in rice. Plant Physiol. 1998;116:203–211. doi: 10.1104/pp.116.1.203. [DOI] [Google Scholar]
- 68.Jagtap P, Szabo C. Poly(ADP-ribose) polymerase and the therapeutic effects of its inhibitors. Nat Rev Drug Discov. 2005;4:421–440. doi: 10.1038/nrd1718. [DOI] [PubMed] [Google Scholar]
- 69.Jambunathan N, Mahalingam R. Analysis of Arabidopsis growth factor gene 1 (GFG1) encoding a nudix hydrolase during oxidative signaling. Planta. 2006;224:1–11. doi: 10.1007/s00425-005-0183-y. [DOI] [PubMed] [Google Scholar]
- 70.Jambunathan N, Penaganti A, Tang Y, Mahalingam R. Modulation of redox homeostasis under suboptimal conditions by Arabidopsis nudix hydrolase 7. BMC Plant Biol. 2010;10:173. doi: 10.1186/1471-2229-10-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jammes F, Song C, Shin D, Munemasa S, Takeda K, Gu D, Cho D, Lee S, Giordo R, Sritubtim S, Leonhardt N, Ellis BE, Murata Y, Kwak JM. MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc Natl Acad Sci USA. 2009;106:20520–20525. doi: 10.1073/pnas.0907205106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jaspers P, Blomster T, Brosche M, Salojarvi J, Ahlfors R, Vainonen JP, Reddy RA, Immink R, Angenent G, Turck F, Overmyer K, Kangasjarvi J. Unequally redundant RCD1 and SRO1 mediate stress and developmental responses and interact with transcription factors. Plant. 2009;J:268–279. doi: 10.1111/j.1365-313X.2009.03951.x. [DOI] [PubMed] [Google Scholar]
- 73.Jaspers P, Brosche M, Overmyer K, Kangasjarvi J. The transcription factor interacting protein RCD1 contains a novel conserved domain. Plant Signal Behav. 2010;5:78–80. doi: 10.4161/psb.5.1.10293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jaspers P, Overmyer K, Wrzaczek M, Vainonen JP, Blomster T, Salojarvi J, Reddy RA, Kangasjarvi J. The RST and PARP-like domain containing SRO protein family: analysis of protein structure, function and conservation in land plants. BMC Genomics. 2010;11:170. doi: 10.1186/1471-2164-11-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kangasjarvi S, Lepisto A, Hannikainen K, Piippo M, Luomala EM, Aro EM, Rintamaki E. Diverse roles for chloroplast stromal and thylakoid-bound ascorbate peroxidases in plant stress responses. Biochem J. 2008;412:275–285. doi: 10.1042/BJ20080030. [DOI] [PubMed] [Google Scholar]
- 76.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 77.Khandelwal A, Elvitigala T, Ghosh B, Quatrano RS. Arabidopsis transcriptome reveals control circuits regulating redox homeostasis and the role of an AP2 transcription factor. Plant Physiol. 2008;148:2050–2058. doi: 10.1104/pp.108.128488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 2005;19:1951–1967. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- 79.Kleine H, Poreba E, Lesniewicz K, Hassa PO, Hottiger MO, Litchfield DW, Shilton BH, Luscher B. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell. 2008;32:57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 80.Koh DW, Lawler AM, Poitras MF, Sasaki M, Wattler S, Nehls MC, Stoger T, Poirier GG, Dawson VL, Dawson TM. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc Natl Acad Sci USA. 2004;101:17699–17704. doi: 10.1073/pnas.0406182101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kothe GO, Kitamura M, Masutani M, Selker EU, Inoue H. PARP is involved in replicative aging in Neurospora crassa . Fungal Genet Biol. 2010;47:297–309. doi: 10.1016/j.fgb.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kotova E, Jarnik M, Tulin AV. Uncoupling of the transactivation and transrepression functions of PARP1 protein. Proc Natl Acad Sci USA. 2010;107:6406–6411. doi: 10.1073/pnas.0914152107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kraus WL. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr Opin Cell Biol. 2008;20:294–302. doi: 10.1016/j.ceb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kurepa J, Walker JM, Smalle J, Gosink MM, Davis SJ, Durham TL, Sung DY, Vierstra RD. The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J Biol Chem. 2003;278:6862–6872. doi: 10.1074/jbc.M209694200. [DOI] [PubMed] [Google Scholar]
- 85.Lepiniec L, Babiychuk E, Kushnir S, Van Montagu M, Inze D. Characterization of an Arabidopsis thaliana cDNA homologue to animal poly(ADP-ribose) polymerase. FEBS Lett. 1995;364:103–108. doi: 10.1016/0014-5793(95)00335-7. [DOI] [PubMed] [Google Scholar]
- 86.Li G, Nasar V, Yang Y, Li W, Liu B, Sun L, Li D, Song F. Arabidopsis poly(ADP-ribose) glycohydrolase 1 is required for drought, osmotic and oxidative stress responses. Plant Sci. 2011;180:283–291. doi: 10.1016/j.plantsci.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 87.Loseva O, Jemth AS, Bryant HE, Schuler H, Lehtio L, Karlberg T, Helleday T. Poly(ADP-ribose) polymerase-3 (PARP-3) is a mono-ADP ribosylase that activates PARP-1 in absence of DNA. J Biol Chem. 2010;285:8054–8060. doi: 10.1074/jbc.M109.077834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ma Q, Baldwin KT, Renzelli AJ, McDaniel A, Dong L. TCDD-inducible poly(ADP-ribose) polymerase: a novel response to 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin. Biochem Biophys Res Commun. 2001;289:499–506. doi: 10.1006/bbrc.2001.5987. [DOI] [PubMed] [Google Scholar]
- 89.Mahajan PB, Zuo Z. Purification and cDNA cloning of maize Poly(ADP)-ribose polymerase. Plant Physiol. 1998;118:895–905. doi: 10.1104/pp.118.3.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Marsischky GT, Wilson BA, Collier RJ. Role of glutamic acid 988 of human poly-ADP-ribose polymerase in polymer formation. Evidence for active site similarities to the ADP-ribosylating toxins. J Biol Chem. 1995;270:3247–3254. doi: 10.1074/jbc.270.7.3247. [DOI] [PubMed] [Google Scholar]
- 91.Messner S, Altmeyer M, Zhao H, Pozivil A, Roschitzki B, Gehrig P, Rutishauser D, Huang D, Caflisch A, Hottiger MO. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 2010;38:6350–6362. doi: 10.1093/nar/gkq463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Muthuswamy S, Meier I. Genetic and environmental changes in SUMO homeostasis lead to nuclear mRNA retention in plants. Planta. 2011;233:201–208. doi: 10.1007/s00425-010-1278-7. [DOI] [PubMed] [Google Scholar]
- 93.Nawy T, Lee JY, Colinas J, Wang JY, Thongrod SC, Malamy JE, Birnbaum K, Benfey PN. Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell. 2005;17:1908–1925. doi: 10.1105/tpc.105.031724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nicholson DW, Ali A, Thornberry NA, Vaillancourt JP, Ding CK, Gallant M, Gareau Y, Griffin PR, Labelle M, Lazebnik YA, et al. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature. 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 95.Nishizuka Y, Ueda K, Nakazawa K, Hayaishi O. Studies on the polymer of adenosine diphosphate ribose. I. Enzymic formation from nicotinamide adenine dinuclotide in mammalian nuclei. J Biol Chem. 1967;242:3164–3171. [PubMed] [Google Scholar]
- 96.Nosseri C, Coppola S, Ghibelli L. Possible involvement of poly(ADP-ribosyl) polymerase in triggering stress-induced apoptosis. Exp Cell Res. 1994;212:367–373. doi: 10.1006/excr.1994.1156. [DOI] [PubMed] [Google Scholar]
- 97.Nusinow DA, Hernandez-Munoz I, Fazzio TG, Shah GM, Kraus WL, Panning B. Poly(ADP-ribose) polymerase 1 is inhibited by a histone H2A variant, MacroH2A, and contributes to silencing of the inactive X chromosome. J Biol Chem. 2007;282:12851–12859. doi: 10.1074/jbc.M610502200. [DOI] [PubMed] [Google Scholar]
- 98.Ogawa T, Ishikawa K, Harada K, Fukusaki E, Yoshimura K, Shigeoka S. Overexpression of an ADP-ribose pyrophosphatase, AtNUDX2, confers enhanced tolerance to oxidative stress in Arabidopsis plants. Plant J. 2009;57:289–301. doi: 10.1111/j.1365-313X.2008.03686.x. [DOI] [PubMed] [Google Scholar]
- 99.Ogawa T, Ueda Y, Yoshimura K, Shigeoka S. Comprehensive analysis of cytosolic Nudix hydrolases in Arabidopsis thaliana . J Biol Chem. 2005;280:25277–25283. doi: 10.1074/jbc.M503536200. [DOI] [PubMed] [Google Scholar]
- 100.Okubo S, Hara F, Tsuchida Y, Shimotakahara S, Suzuki S, Hatanaka H, Yokoyama S, Tanaka H, Yasuda H, Shindo H. NMR structure of the N-terminal domain of SUMO ligase PIAS1 and its interaction with tumor suppressor p53 and A/T-rich DNA oligomers. J Biol Chem. 2004;279:31455–31461. doi: 10.1074/jbc.M403561200. [DOI] [PubMed] [Google Scholar]
- 101.Oliver AW, Ame JC, Roe SM, Good V, de Murcia G, Pearl LH. Crystal structure of the catalytic fragment of murine poly(ADP-ribose) polymerase-2. Nucleic Acids Res. 2004;32:456–464. doi: 10.1093/nar/gkh215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ouararhni K, Hadj-Slimane R, Ait-Si-Ali S, Robin P, Mietton F, Harel-Bellan A, Dimitrov S, Hamiche A. The histone variant mH2A1.1 interferes with transcription by down-regulating PARP-1 enzymatic activity. Genes Dev. 2006;20:3324–3336. doi: 10.1101/gad.396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Overmyer K, Brosche M, Pellinen R, Kuittinen T, Tuominen H, Ahlfors R, Keinanen M, Saarma M, Scheel D, Kangasjarvi J. Ozone-induced programmed cell death in the Arabidopsis radical-induced cell death1 mutant. Plant Physiol. 2005;137:1092–1104. doi: 10.1104/pp.104.055681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Overmyer K, Tuominen H, Kettunen R, Betz C, Langebartels C, Sandermann H, Jr, Kangasjarvi J. Ozone-sensitive arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. Plant Cell. 2000;12:1849–1862. doi: 10.1105/tpc.12.10.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Panda S, Poirier GG, Kay SA. tej defines a role for poly(ADP-ribosyl)ation in establishing period length of the Arabidopsis circadian oscillator. Dev Cell. 2002;3:51–61. doi: 10.1016/S1534-5807(02)00200-9. [DOI] [PubMed] [Google Scholar]
- 106.Pandey R, Muller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, Bender J, Mount DW, Jorgensen RA. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002;30:5036–5055. doi: 10.1093/nar/gkf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pellny TK, Locato V, Vivancos PD, Markovic J, De Gara L, Pallardo FV, Foyer CH. Pyridine nucleotide cycling and control of intracellular redox state in relation to poly (ADP-ribose) polymerase activity and nuclear localization of glutathione during exponential growth of Arabidopsis cells in culture. Mol Plant. 2009;2:442–456. doi: 10.1093/mp/ssp008. [DOI] [PubMed] [Google Scholar]
- 108.Phillips RHSW. Characteristics of the inhibition of induced tracheary element differentiation by 3-aminobenzamide and related compounds. J Exp Bot. 1985;36:119–128. doi: 10.1093/jxb/36.1.119. [DOI] [Google Scholar]
- 109.Potters G, Pasternak TP, Guisez Y, Palme KJ, Jansen MA. Stress-induced morphogenic responses: growing out of trouble? Trends Plant Sci. 2007;12:98–105. doi: 10.1016/j.tplants.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 110.Puchta H, Swoboda P, Gal S, Blot M, Hohn B. Somatic intrachromosomal homologous recombination events in populations of plant siblings. Plant Mol Biol. 1995;28:281–292. doi: 10.1007/BF00020247. [DOI] [PubMed] [Google Scholar]
- 111.Rajawat J, Mir H, Begum R. Differential role of Poly(ADP-ribose) polymerase in D. discoideum growth and development. BMC Dev Biol. 2011;11:14. doi: 10.1186/1471-213X-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rajawat J, Vohra I, Mir HA, Gohel D, Begum R. Effect of oxidative stress and involvement of poly(ADP-ribose) polymerase (PARP) in Dictyostelium discoideum development. FEBS J. 2007;274:5611–5618. doi: 10.1111/j.1742-4658.2007.06083.x. [DOI] [PubMed] [Google Scholar]
- 113.Ramirez-Parra E, Gutierrez C. E2F regulates FASCIATA1, a chromatin assembly gene whose loss switches on the endocycle and activates gene expression by changing the epigenetic status. Plant Physiol. 2007;144:105–120. doi: 10.1104/pp.106.094979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Reeder RH, Ueda K, Honjo T, Nishizuka Y, Hayaishi O. Studies on the polymer of adenosine diphosphate ribose. II. Characterization of the polymer. J Biol Chem. 1967;242:3172–3179. [PubMed] [Google Scholar]
- 115.Rodriguez MC, Petersen M, Mundy J. Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol. 2010;61:621–649. doi: 10.1146/annurev-arplant-042809-112252. [DOI] [PubMed] [Google Scholar]
- 116.Rolli V, O’Farrell M, Menissier-de Murcia J, de Murcia G. Random mutagenesis of the poly(ADP-ribose) polymerase catalytic domain reveals amino acids involved in polymer branching. Biochemistry. 1997;36:12147–12154. doi: 10.1021/bi971055p. [DOI] [PubMed] [Google Scholar]
- 117.Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlic A, Quesada M, Quinn GB, Westbrook JD, Young J, Yukich B, Zardecki C, Berman HM, Bourne PE. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39:D392–D401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rouleau M, McDonald D, Gagne P, Ouellet ME, Droit A, Hunter JM, Dutertre S, Prigent C, Hendzel MJ, Poirier GG. PARP-3 associates with polycomb group bodies and with components of the DNA damage repair machinery. J Cell Biochem. 2007;100:385–401. doi: 10.1002/jcb.21051. [DOI] [PubMed] [Google Scholar]
- 119.Rouleau M, Saxena V, Rodrigue A, Paquet ER, Gagnon A, Hendzel MJ, Masson JY, Ekker M, Poirier GG. A key role for poly(ADP-ribose) polymerase 3 in ectodermal specification and neural crest development. PLoS One. 2011;6:e15834. doi: 10.1371/journal.pone.0015834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ruf A, Mennissier de Murcia J, de Murcia G, Schulz GE. Structure of the catalytic fragment of poly(AD-ribose) polymerase from chicken. Proc Natl Acad Sci USA. 1996;93:7481–7485. doi: 10.1073/pnas.93.15.7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rulten SL, Fisher AE, Robert I, Zuma MC, Rouleau M, Ju L, Poirier G, Reina-San-Martin B, Caldecott KW. PARP-3 and APLF function together to accelerate nonhomologous end-joining. Mol Cell. 2011;41:33–45. doi: 10.1016/j.molcel.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 122.Ryu H, Azuma Y. Rod/Zw10 complex is required for PIASy-dependent centromeric SUMOylation. J Biol Chem. 2010;285:32576–32585. doi: 10.1074/jbc.M110.153817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sanchez JP, Duque P, Chua NH. ABA activates ADPR cyclase and cADPR induces a subset of ABA-responsive genes in Arabidopsis . Plant J. 2004;38:381–395. doi: 10.1111/j.1365-313X.2004.02055.x. [DOI] [PubMed] [Google Scholar]
- 124.Schreiber V, Ame JC, Dolle P, Schultz I, Rinaldi B, Fraulob V, Menissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J Biol Chem. 2002;277:23028–23036. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]
- 125.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 126.Semighini CP, Savoldi M, Goldman GH, Harris SD. Functional characterization of the putative Aspergillus nidulans poly(ADP-ribose) polymerase homolog PrpA. Genetics. 2006;173:87–98. doi: 10.1534/genetics.105.053199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Simonin F, Hofferer L, Panzeter PL, Muller S, de Murcia G, Althaus FR. The carboxyl-terminal domain of human poly(ADP-ribose) polymerase. Overproduction in Escherichia coli, large scale purification, and characterization. J Biol Chem. 1993;268:13454–13461. [PubMed] [Google Scholar]
- 128.Staub E, Fiziev P, Rosenthal A, Hinzmann B. Insights into the evolution of the nucleolus by an analysis of its protein domain repertoire. Bioessays. 2004;26:567–581. doi: 10.1002/bies.20032. [DOI] [PubMed] [Google Scholar]
- 129.Straus MR, Rietz S, Loren Ver, van Themaat E, Bartsch M, Parker JE. Salicylic acid antagonism of EDS1-driven cell death is important for immune and oxidative stress responses in Arabidopsis . Plant J. 2010;62:628–640. doi: 10.1111/j.1365-313X.2010.04178.x. [DOI] [PubMed] [Google Scholar]
- 130.Szabo C, Dawson VL. Role of poly(ADP-ribose) synthetase in inflammation and ischaemia-reperfusion. Trends Pharmacol Sci. 1998;19:287–298. doi: 10.1016/S0165-6147(98)01193-6. [DOI] [PubMed] [Google Scholar]
- 131.Teotia S, Lamb RS. The paralogous genes RADICAL-INDUCED CELL DEATH1 and SIMILAR TO RCD ONE1 have partially redundant functions during Arabidopsis development. Plant Physiol. 2009;151:180–198. doi: 10.1104/pp.109.142786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Teotia S, Lamb RS. RCD1 and SRO1 are necessary to maintain meristematic fate in Arabidopsis thaliana . J Exp Bot. 2011;62:1271–1284. doi: 10.1093/jxb/erq363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Teotia S, Muthuswamy S, Lamb RS. RADICAL-INDUCED CELL DEATH1 and SIMILAR TO RCD ONE1 and the Stress-Induced Morphogenetic Response. Plant Signal Behav. 2010;5:143–145. doi: 10.4161/psb.5.2.10400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tewari M, Quan LT, O’Rourke K, Desnoyers S, Zeng Z, Beidler DR, Poirier GG, Salvesen GS, Dixit VM. Yama/CPP32 beta, a mammalian homolog of CED-3, is a CrmA-inhibitable protease that cleaves the death substrate poly(ADP-ribose) polymerase. Cell. 1995;81:801–809. doi: 10.1016/0092-8674(95)90541-3. [DOI] [PubMed] [Google Scholar]
- 135.Tian R, Zhang GY, Yan CH, Dai YR. Involvement of poly(ADP-ribose) polymerase and activation of caspase-3-like protease in heat shock-induced apoptosis in tobacco suspension cells. FEBS Lett. 2000;474:11–15. doi: 10.1016/S0014-5793(00)01561-1. [DOI] [PubMed] [Google Scholar]
- 136.Till S, Diamantara K, Ladurner AG. PARP: a transferase by any other name. Nat Struct Mol Biol. 2008;15:1243–1244. doi: 10.1038/nsmb1208-1243. [DOI] [PubMed] [Google Scholar]
- 137.Trucco C, Oliver FJ, de Murcia G, Menissier-de Murcia J. DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res. 1998;26:2644–2649. doi: 10.1093/nar/26.11.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tulin A, Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299:560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- 139.Tulin A, Stewart D, Spradling AC. The Drosophila heterochromatic gene encoding poly(ADP-ribose) polymerase (PARP) is required to modulate chromatin structure during development. Genes Dev. 2002;16:2108–2119. doi: 10.1101/gad.1003902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ueda K, Omachi A, Kawaichi M, Hayaishi O. Natural occurrence of poly(ADP-ribosyl) histones in rat liver. Proc Natl Acad Sci USA. 1975;72:205–209. doi: 10.1073/pnas.72.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Urbanek P, Paces J, Kralova J, Dvorak M, Paces V. Cloning and expression of PARP-3 (Adprt3) and U3-55 k, two genes closely linked on mouse chromosome 9. Folia Biol (Praha) 2002;48:182–191. doi: 10.14712/fb2002048050182. [DOI] [PubMed] [Google Scholar]
- 142.Vanderauwera S, De Block M, Van de Steene N, van de Cotte B, Metzlaff M, Van Breusegem F. Silencing of poly(ADP-ribose) polymerase in plants alters abiotic stress signal transduction. Proc Natl Acad Sci USA. 2007;104:15150–15155. doi: 10.1073/pnas.0706668104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wacker DA, Ruhl DD, Balagamwala EH, Hope KM, Zhang T, Kraus WL. The DNA binding and catalytic domains of poly(ADP-ribose) polymerase 1 cooperate in the regulation of chromatin structure and transcription. Mol Cell Biol. 2007;27:7475–7485. doi: 10.1128/MCB.01314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang C, Gao F, Wu J, Dai J, Wei C, Li Y. Arabidopsis putative deacetylase AtSRT2 regulates basal defense by suppressing PAD4, EDS5 and SID2 expression. Plant Cell Physiol. 2010;51:1291–1299. doi: 10.1093/pcp/pcq087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Whitby AJ, Stone PR, Whish WJ. Effect of polyamines and Mg++ on poly(ADP-ribose) synthesis and ADP-ribosylation of histones in wheat. Biochem Biophys Res Commun. 1979;90:1295–1304. doi: 10.1016/0006-291X(79)91177-X. [DOI] [PubMed] [Google Scholar]
- 146.Willmitzer L. Demonstration of in vitro covalent modification of chromosomal proteins by poly(ADP) ribosylation in plant nuclei. FEBS Lett. 1979;108:13–16. doi: 10.1016/0014-5793(79)81167-9. [DOI] [PubMed] [Google Scholar]
- 147.Wu Y, Kuzma J, Marechal E, Graeff R, Lee HC, Foster R, Chua NH. Abscisic acid signaling through cyclic ADP-ribose in plants. Science. 1997;278:2126–2130. doi: 10.1126/science.278.5346.2126. [DOI] [PubMed] [Google Scholar]
- 148.Yu M, Schreek S, Cerni C, Schamberger C, Lesniewicz K, Poreba E, Vervoorts J, Walsemann G, Grotzinger J, Kremmer E, Mehraein Y, Mertsching J, Kraft R, Austen M, Luscher-Firzlaff J, Luscher B. PARP-10, a novel Myc-interacting protein with poly(ADP-ribose) polymerase activity, inhibits transformation. Oncogene. 2005;24:1982–1993. doi: 10.1038/sj.onc.1208410. [DOI] [PubMed] [Google Scholar]
- 149.Zhu P, Martinvalet D, Chowdhury D, Zhang D, Schlesinger A, Lieberman J. The cytotoxic T lymphocyte protease granzyme A cleaves and inactivates poly(adenosine 5’-diphosphate-ribose) polymerase-1. Blood. 2009;114:1205–1216. doi: 10.1182/blood-2008-12-195768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhu Y, Weng M, Yang Y, Zhang C, Li Z, Shen WH, Dong A. Arabidopsis homologues of the histone chaperone ASF1 are crucial for chromatin replication and cell proliferation in plant development. Plant. 2011;J:443–455. doi: 10.1111/j.1365-313X.2011.04504.x. [DOI] [PubMed] [Google Scholar]
- 151.Zimmermann P, Hennig L, Gruissem W. Gene-expression analysis and network discovery using Genevestigator. Trends Plant Sci. 2005;10:407–409. doi: 10.1016/j.tplants.2005.07.003. [DOI] [PubMed] [Google Scholar]