Abstract
Iron homeostasis is maintained at the cellular and systemic levels to assure adequate iron supply while preventing iron overload. The identification of genes mutated in patients with iron-related disorders or animal models with imbalances of iron homeostasis gave insight into the molecular mechanisms underlying processes critical for balancing iron levels, such as iron uptake, storage, export, and monitoring of available iron. MicroRNAs control genes involved in some of these processes adding an additional level of complexity to the regulation of iron metabolism. This review summarizes recent advances how miRNAs regulate iron homeostasis.
Keywords: Iron homeostasis, MicroRNA
Repression of gene expression by microRNAs
MicroRNAs (miRNAs) are approximately 22-nucleotide-long, single-stranded non-coding (nc) RNAs that repress target gene expression. MiRNA genes are frequently localized within intronic regions [1], but are also expressed from non-protein coding transcripts localized in intergenic regions [2]. Most miRNAs are transcribed by RNA polymerase II [3] as long primary transcripts (pri-miRNA) that are processed in two consecutive steps to generate the functional, mature miRNA [4]. Processed miRNAs are inserted into the RNA-induced silencing complex (RISC), to guide the RISC to the target mRNA sequence. MiRNAs recognize miRNA responsive elements (MREs), sequences with partial complementary localized within the 3′ untranslated region (UTR) of mRNAs. Depending on the level of complementarity between miRNA and MRE, this interaction either limits mRNA translation or decreases mRNA stability. To date, more than 1,000 miRNA sequences have been identified in the human genome (miRBase; version 18, November 2011 [5]).
MiRNAs control cellular processes as diverse as metabolism, cancer [6], or neurodegeneration [7]. Vice versa, cellular signals [8, 9] and pathological conditions [10] alter miRNA expression. Molecular networks controlled by miRNAs are highly complex. Computational analyses suggest that miRNAs may repress up to 30 % of the protein-coding genes of each genome [11], whereby each individual mRNA transcript may contain several different miRNA-specific MREs. In addition, it is expected that mRNAs containing identical MREs in their 3′UTR will co-regulate each other’s expression by competing for a limited miRNA pool [12–14]. Likewise, MRE-containing pseudogenes [15] and long non-coding (lnc)RNAs [16] may act as ‘sponges’ for miRNAs. Thus, mRNAs, pseudogenes, and lncRNAs form a network of transcripts named the ‘competing endogenous RNAs (ceRNA)’. These can control the distribution of miRNAs on their target mRNAs [17] despite the fact that the expression of a specific miRNA remains unchanged, hence imposing an additional layer of post-transcriptional control on gene expression.
Heme-mediated control of miRNA biogenesis
miRNA genes are usually transcribed from RNA Polymerase II promoters as long primary transcripts (pri-miRNAs), whereby transcription factor binding sites in miRNA promoters determine tissue- or development-specific miRNA transcription [18]. The approximately 22-nucleotide-long miRNA sequences are located within hairpin loop structures that are recognized by a nuclear protein known as the DiGeorge Syndrome Critical Region 8 (DGCR8) [19]. DGCR8 binds Drosha, the Rnase III that releases the 70 nucleotide precursor-miRNA (pre-miRNA) from the pri-miRNA [20]. The pre-miRNA hairpins are exported to the cytoplasm where they are further processed by the RNase III protein Dicer into 19- to 25-nt miRNA duplex structures [21]. One of the two strands in the duplex is incorporated into a multiple-protein nuclease complex, the RISC, to post-transcriptionally control gene expression. Regulation of miRNA biogenesis has emerged as a critical mechanism to define the spatiotemporal pattern of miRNA expression [22]. Although little is known about how miRNA biogenesis is modulated, one of the control mechanisms seems to depend on the availability of iron-containing heme [23]. In reconstituted pri-miRNA processing assays, heme enhances DGCR8 dimerization, which is required for increased activity of this essential miRNA processing cofactor [24–27]. During this process, heme binds to DGCR8 via its ferric iron co-factor. By contrast, ferrous heme is unable to enhance miRNA processing [27]. It was suggested that heme exerts a regulatory function in miRNA biogenesis, because the heme-binding domain of DGCR8 is not required for processing of pri-miRNAs. Thus, in conditions of heme deficiency, the pri-miRNA cleavage rate is predicted to be slow, which will result in a global decrease of mature miRNA abundance. Further studies will be required to investigate how this mechanism influences miRNA biogenesis in vivo, and whether miRNA biogenesis is impaired in diseases with dysregulated heme synthesis such as porphyrias.
Iron metabolism is controlled at the cellular and systemic level
Iron is an essential micronutrient, which is required as a cofactor for fundamental cellular processes such as oxygen transport, cellular respiration, and DNA synthesis. Continued iron deficiency causes cell death. On the other extreme, free iron excess is also toxic. If not tightly bound to iron-carrier proteins, it will catalyze the formation of reactive oxygen species (ROS) that damage lipid membranes, proteins, and nucleic acids. Therefore, iron levels must be tightly balanced to assure adequate iron supplies while preventing toxicity.
The discovery that systemic iron homeostasis is largely controlled by the antimicrobial peptide hormone hepcidin [28, 29] paved the way to our understanding as to how systemic iron metabolism is regulated. Hepcidin controls systemic iron availability by interacting with its target ferroportin, a transmembrane iron efflux channel highly expressed on cells that release iron, such as enterocytes, macrophages, and hepatocytes. Iron export further requires an extracellular ferroxidase activity that is provided by the multicopper oxidases ceruloplasmin (Cp) and/or hephaestin [30]. Upon hepcidin binding, ferroportin is internalized and degraded [31]. Hepcidin is produced by the liver in response to systemic iron availability, hypoxia, erythroid iron demand, and inflammatory cues. Although hepcidin levels are controlled by systemic iron requirements, iron levels are not directly sensed by hepcidin. Hepcidin expression is balanced by upstream activator proteins, such as Hfe [32, 33], hemojuvelin (Hjv; [34–36]), and Transferrin Receptor 2 (TfR2; [37, 38]), and inhibitors like the transmembrane protease, serine 6 (TMPRSS6; [39]), and Smad family member 7 [40]. Mutations in hepcidin itself or in the genes coding for hepcidin activators cause a metabolic disorder named hereditary hemochromatosis (HH). HH is hallmarked by decreased hepcidin activity, which causes increased iron uptake from the diet and iron release from macrophages. As a consequence, tissue iron overload develops, which leads to a clinical condition that can result in the development of mild to serious pathologies, including abdominal pain, arthritis, heart failure, diabetes, and hepatic cirrhosis.
The hepcidin/ferroportin regulatory system controls the availability of adequate levels of plasma iron, which is tightly bound to the plasma protein transferrin. Transferrin-bound iron is taken up by most cell-types of the body via transferrin receptor (TfR) 1. The expression of genes involved in maintaining cellular iron homeostasis is predominantly controlled post-transcriptionally by the binding of iron regulatory proteins (IRPs) 1 or 2 to cis-regulatory mRNA motifs termed iron responsive elements (IREs). IREs are short, conserved RNA stem-loop structures located in the non-coding sequence (3′- or 5′UTR) of mRNA encoding proteins for iron acquisition [transferrin receptor 1 (TfR1), and the divalent metal transporter 1 (DMT1)], transport [ferritin H (FTH1), and ferritin L (FTL)] utilization [aconitase (ACO2), erythroid 5-Aminolevulinate synthase (ALAS) and hypoxia-inducible factor 2 (HIF2)] and export [ferroportin (FPN1)] [41]. Past and recent evidence suggests that additional mRNAs are regulated by the IRPs [42]. In iron-deficient conditions, IRE/IRP complexes form within an mRNA 5′UTR (e.g., FTH1, FTL, ACO2, and FPN1) to inhibit translation, whereas IRP binding to IREs in the 3′UTR of TfR1 and DMT1 prevents mRNA degradation. Mice lacking both copies of the IRP1 and the IRP2 genes die at the embryonic stage, indicating that the IRE/IRP regulatory network is essential, at least for early development [43]. In addition to the predominant transcriptional control of systemic iron homeostasis via hepcidin, and the post-transcriptional regulation of genes involved in maintaining cellular iron levels, recent studies reveal that miRNAs play a critical role in balancing cellular and tissue iron homeostasis.
Control of cellular iron uptake by miRNAs
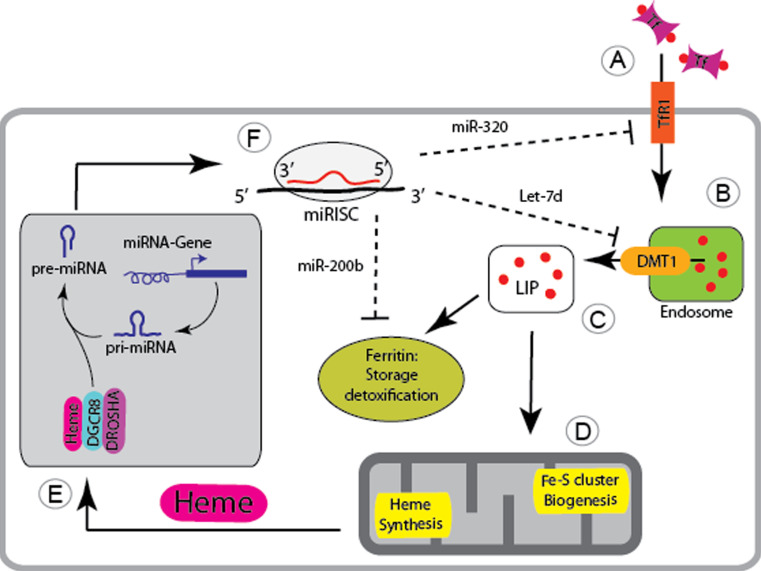
Most mammalian cell types satisfy their iron needs by the uptake of iron-bound transferrin (Tf-Fe2) from plasma. Tf-Fe2 binds to the ubiquitously expressed transferrin receptor 1 (TfR1), and the complex is internalized by clathrin-dependent endocytosis. Subsequent acidification of early endosomes promotes the release of iron from transferrin, which then is reduced to Fe(II) by members of the STEAP family of metalloreductases [44] for transport into the cytoplasm via the endosomally expressed DMT1. TfR1 and apo-transferrin are recycled to the cell surface. Although TfR1 is ubiquitously expressed, the transferrin cycle is most critical for the massive iron demand of erythroid precursor cells. The amount of TfR1 expressed at the cell surface reflects upon cellular iron requirements and is predominantly regulated at the level of transcription and post-transcriptionally by the IRE/IRP regulatory system. Recent data highlight that the transferrin cycle is additionally controlled by miRNAs at two different steps (Fig. 1).
Fig. 1.
microRNA-mediated control of cellular iron metabolism. A The ubiquitously expressed transferrin receptor-1 (TfR1) controls cellular uptake of iron-bound transferrin (Tf, iron indicated by red dots). B The Tf-iron-TfR1 complex is taken up into the cell via endocytosis and the iron is released from the endosome by the nonIRE isoform of Divalent Metal Transporter 1 (DMT1). C In the cytoplasm iron is incorporated within the cellular labile iron pool (LIP), is stored and detoxified by ferritin or D is utilized by mitochondria for heme synthesis and Fe–S cluster biogenesis. E Heme is critical for the processing of miRNA primary transcripts (pri-miRNAs) and binds to the microprocessor complex composed of DGCR8 and DROSHA. F miRNA precursors (pre-miRNA) are exported to the cytoplasm where they are processed to form single stranded miRNAs that are bound by the RISC. The miRISC will target MREs within the 3′ UTR of genes to repress their translation or to trigger mRNA degradation. Several miRNAs control genes involved in maintaining cellular iron homeostasis, whereby miR-320 post-transcriptionally controls TfR1 expression, Let-7d targets the DMT1-nonIRE isoform, and miR-200b regulates the expression of ferritin
It was recognized some time ago that Tfr1 expression is increased in neoplastic cells to satisfy their increased iron requirements for cellular proliferation [45, 46]. Conversely, differentiation of neoplastic cells such as the human leukemia cell line HL-60 by tetradecanoylphorbol acetate (TPA) into the monocytic/macrophage lineage [47] represses TfR1 expression and increases levels of miRNAs predicted to bind to the 3′UTR of TfR1 (miR-22, miR-200a, and miR-320). Of these, miR-320 represses the activity of luciferase reporter vectors fused to sequences of the Tfr1 3′UTR that contain the miR-320-specific MRE. Likewise, enforced miR-320 expression in the lung carcinoma A549 cell line decreases TfR1 surface expression and slows down cell cycle progression and cell growth. Because the additional treatment of cells with a soluble iron salt reverses the growth inhibitory effect, the authors speculate that decreased TfR1 expression in miR-320 overexpressing cells lowered iron availability and inhibited cell proliferation [48]. It is currently unresolved whether miR-320-mediated TfR1 regulation is confined to cancer cells or whether the ubiquitously and highly expressed miRNA (see http://mirnamap.mbc.nctu.edu.tw) is involved in the control of TfR1 expression under physiological conditions.
In addition to miRNA-dependent regulation of TfR1, miRNAs control the transferrin cycle at a second step, the release of iron from the endosome via DMT1. The gene coding for DMT1 (SLC11A2) gives rise to four variant mRNA transcripts that either differ at their 5′ end as a result of alternative promoter usage (DMT1A and 1B protein isoforms) or at the 3′ end due to alternative splicing [49]. While the DMT1A isoform is predominantly expressed in the duodenum, isoform 1B is ubiquitously distributed. The alternative 3′UTR sequences differ as to whether they contain an IRE sequence motif, whereby only the IRE-containing isoforms are controlled in response to cellular iron levels by IRP binding [50]. All different DMT1 isoforms are capable of transporting iron and, except for the 1A isoform, are ubiquitously expressed [51]. Notably, the DMT1 splice variant lacking the IRE sequence (DMT1B-nonIRE) is abundantly expressed in erythroid cells [51], where it is responsible for iron export across the membrane of acidified endosomes into the cytoplasm following iron-bound transferrin uptake [52]. Although the DMT1B-nonIRE isoform plays a fundamental role during erythroid precursor differentiation, the regulatory mechanism underlying its expression remained unclear. A recent study in erythroid cells revealed that the expression of the DMT1B-nonIRE isoform is fine-tuned by the miRNA Let-7d, providing an alternative mode of regulation of this isoform that cannot respond to iron availability through the IRE/IRP regulatory system [53]. In this study, the expression of miRNAs, which were bioinformatically predicted to target the DMT1-nonIRE isoform (miR-15a, miR-15b, miR-223, and Let-7d) were analyzed during erythroid differentiation of CD34+ cells. From these, only Let-7d showed an adequate binding affinity to the DMT1-nonIRE 3′UTR, phylogenetic conservation as well as an opposite expression trend with respect to DMT1-nonIRE mRNA and protein expression, which was upregulated during erythroid differentiation. Transfection of K562 cells with luciferase reporter constructs containing the full-length 3′UTR of human DMT1+IRE and DMT1−IRE mRNA splice variants demonstrated the specificity of Let-7d in repressing the DMT1-nonIRE isoform only. Furthermore, Let-7d overexpression decreases human DMT-1-nonIRE expression at the mRNA and protein level in K562 and HEL cells and leads to the inhibition of erythroid differentiation due to iron accumulation in endosomes of K562 cells. As a consequence of iron retention in endosomes, cytosolic ferritin levels are decreased, and TfR1 mRNA levels are increased, indicating functional cellular iron deficiency. Let-7d is ubiquitously expressed, suggesting that a similar regulatory mechanism might be operational in other cell types, where the DMT1-IRE isoform is expressed. These include cell types with critical DMT-1 functions, such as duodenal enterocytes and neurons where Let-7d-mediated control of DMT1 may play a role. Furthermore, miRNA-controlled DMT1 expression may contribute to the uptake of non-transferrin bound iron (NTBI) in the iron overload disorder Hereditary Hemochromatosis [54]. How miRNA-dependent control of DMT-1 expression is integrated with additional DMT-1 control mechanisms at the transcriptional (by hypoxia-inducible factor (HIF)2a [55, 56]), post-transcriptional (by the IRE/IRP regulatory system [50]), or post-translational (by Ndfips-dependant ubiquitination and proteasomal degradation) level, requires further investigation.
Because the processing of miRNA maturation requires iron in the form of heme [23], the findings that miRNAs control cellular iron uptake open the possibility that a regulatory loop exists in which iron is required for the efficient synthesis of mature miRNAs, while defined mature miRNAs control cellular iron uptake.
Control of cellular iron storage by miR-200b
Ferritin heteropolymers consist of 24 subunits of heavy (FtH1) and light (FtL) chains that bind iron from the cytoplasmic “labile iron pool” (LIP) that is not utilized or exported (see review by Arosio and Levi [77]). Only the FtH1 subunit exerts ferroxidase activity that is necessary for iron deposition into the nanocage, while FtL facilitates iron nucleation and increases the turnover of the ferroxidase site. Ferritin detoxifies excess iron in a redox inactive form to prevent iron-mediated cell and tissue damage; it also constitutes an iron store whose mobilization involves both proteasomal and lysosomal degradation of ferritin.
A recent study showed that human breast cancer cells with an aggressive mesenchymal phenotype (i.e. Hs578T, BT549, and MDA-MB-231) express substantially higher mRNA and protein levels of FtH1 and FtL and lower LIP compared to breast cancer cells with an epithelial and less aggressive phenotype (i.e. MCF7, MDA-MB-361, T47D, and HCC70). A large fraction of FtH1 in these breast cancer cells was associated with the chromatin-bound nuclear fraction, a finding that lacks explanation. Increased FTH1 levels correlated with low expression of miR-200b, a miRNA predicted to bind to the FTH1 3′UTR. A functional role for miR-200b binding to the FTH1 3′UTR was demonstrated by luciferase reporter assays and FTH1 western blotting in the MDA-MB-231 cell line. Unexpectedly, miR-200b transfection also decreased FtL protein levels, although the FtL 3′UTR is not predicted to bind to miR-200b. Transfection of miR-200b further increased sensitivity to doxorubicin, a drug commonly used to treat breast cancer. Whether miR-200b-mediated ferritin regulation is related to previous clinical findings, that patients with higher circulating plasma ferritin levels show worse treatment outcome compared to patients with lower ferritin values, needs to be determined [57]. The authors speculate that down-regulation of miR-200b in human breast cancer could be a contributory factor to cancer aggressiveness. Whether or not this finding may have potential therapeutic implications remains controversial at the moment [58, 59].
Control of systemic iron regulation by miR-122
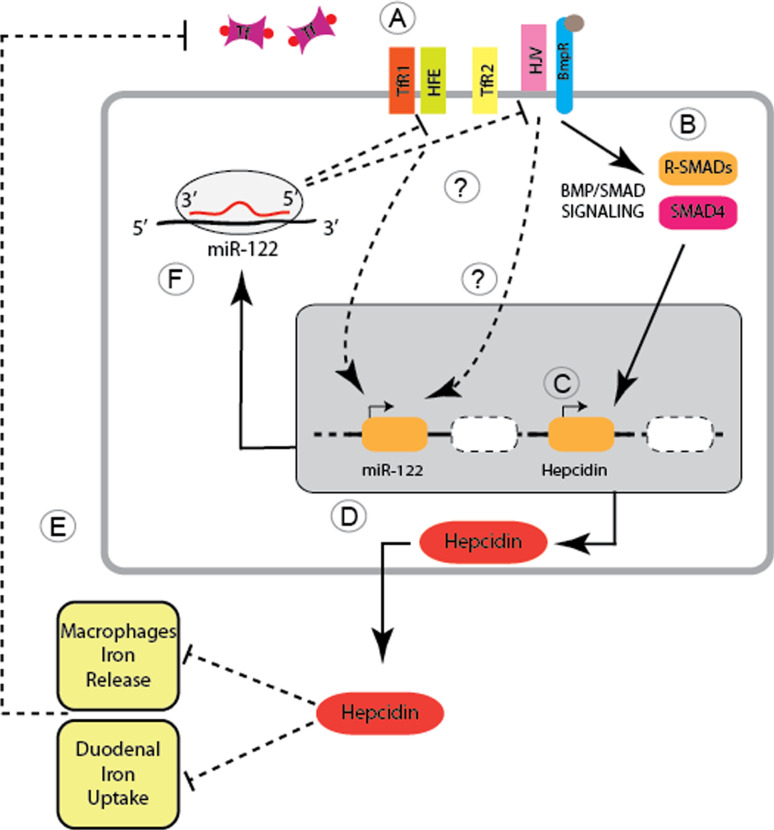
The liver is the major iron storage site and the endocrine organ responsible for the regulation of systemic iron homeostasis. It synthesizes the small peptide hormone hepcidin that controls the amount of circulating iron by interacting with the iron exporter ferroportin. The liver senses systemic iron availability through genes mutated in hereditary hemochromatosis (e.g., Hfe, hemojuvelin, and TfR2), the Bone morphogenetic protein (Bmp)6, and the Smad4 protein, which serve to regulate hepcidin transcription. Inappropriately low hepcidin activity as a result of mutations in Hfe, hemojuvelin, TfR2, or hepcidin itself cause the iron overload disorder hereditary hemochromatosis (HH). MiRNA expression analysis in mouse models of iron overload found that miR-122, a miRNA abundantly and selectively expressed in the liver, is down-regulated in a murine disease model of Hfe-mediated HH as well as in liver biopsies from HH patients with homozygous C282Y mutations. A functional link between miR-122 and iron metabolism was established in wild-type mice subjected to a single, intraperitoneal injection of locked nucleic acid (LNA)-modified antimiR oligonucleotides, which specifically and exclusively inhibited miR-122 in the liver. MiR-122 depleted mice are hallmarked by decreased systemic iron levels, which result in an inadequate iron supply to the erythron that mildly impairs hematopoiesis [60]. Specifically, miR-122 inhibition decreases the mean corpuscular volume (MCV) of erythrocytes, increases reticulocyte counts, and reduces the reticulocyte hemoglobin content. Additionally, the iron content of the liver (site of iron storage), the spleen (site of iron recycling), and the plasma (site of iron transport) were reduced in miR-122-inhibited mice. The decrease in plasma iron levels resulted in reduced transferrin iron binding capacity (TIBC) 3 weeks after antimiR-122 injection. Interestingly, disturbance of systemic iron homeostasis in miR-122-inhibited mice arises from altered mRNA expression levels of those genes that participate in the sensing of systemic iron levels (i.e., Hfe, Hemojuvelin, and Bmpr1a) and that transmit signals at least in part, via the Bmp/Smad signaling pathway to regulate transcription of hepcidin. MRNA expression of Hfe, HJV, Bmpr1a, and hepcidin were increased in miR-122-depleted mice, while mRNA expression of TfR2 or other hepcidin effectors (e.g., Smad7 and Smad4) was not affected. Furthermore, antimiR-122 transfection of murine primary hepatocytes also increased mRNA expression of Hfe, hemojuvelin, and hepcidin. This allowed for the validation of the miR-122 binding sites that were bioinformatically predicted within the 3′UTRs of Hfe (2 MREs), Hemojuvelin (3 MREs) and Hepcidin (a single MRE) mRNAs. Cotransfection of luciferase reporter genes bearing the full length 3′-UTRs of mouse hepcidin, Hfe, and hemojuvelin, together with miR-122 mimics (pre-miR-122) into Hepa 1–6 (mouse) hepatocarcinoma cells, revealed the functionality of the MREs in Hfe and HJV. By contrast, reporter constructs containing the hepcidin 3′UTR did not reveal any specific alterations. These data clearly demonstrate that the 3′-UTRs of Hfe and hemojuvelin are direct targets of miR-122.
In summary, these findings reveal a regulatory loop of miR-122-dependant control of systemic iron homeostasis. The model summarized in Fig. 2 shows that miR-122 inhibition in wild-type mice derepresses the expression of Hfe and hemojuvelin, which then trigger the activation of hepcidin transcription. As a consequence, elevated circulating hepcidin levels enhance the degradation of ferroportin on target cells, thus reducing iron absorption from the diet and iron release from erythrocyte iron recycling macrophages. This will result in plasma and tissue iron deficiency and mild impairment of erythropoiesis. Equally important, the gene Hfe mutated in patients with HH is a critical upstream regulator of miR-122 levels. Of note is that miR-122 levels are not regulated as a consequence of iron that accumulates in the liver of HH patients, but likely as a consequence of the signaling activities impaired by the lack of Hfe. It is known that the lack of Hfe attenuates the BMP/Smad signaling pathway in HH patients and the respective murine disease model [61]. However, an increase in Smad1/5/8 phosphorylation was not detectable in miR-122-depleted mice, suggesting that either BMP/Smad signaling was analyzed in an inappropriate time window following miR-122 depletion or that additional signaling pathways are activated downstream of Hfe.
Fig. 2.
Systemic iron homeostasis is controlled by miR-122. A Levels of iron-bound transferrin (Tf) are monitored by Transferrin Receptor 1 (TfR1) and proteins mutated in different subtypes of hereditary hemochromatosis, the MHC class I-like protein HFE, Transferrin Receptor 2 (TfR2) and the co-receptor for bone morphogenetic proteins (BMPs), Hemojuvelin (HJV). B, C High levels of iron-bound transferrin and BMP6 activate BMP/SMAD signaling and stimulate Hepcidin transcription. D Increased systemic Hepcidin levels inhibit iron release from macrophages and duodenal enterocytes, which will (E) decrease the amount of iron-bound transferrin. F Likewise, HFE and HJV are required to activate the expression of miR-122, while miR-122 specifically represses the HFE and HJV mRNA. Experimental depletion of miR-122 increases HFE and HJV expression, which stimulates hepcidin transcription and triggers systemic iron deficiency. The proposed feedback mechanism will fine tune Hepcidin expression to satisfy systemic iron demands
The finding that miR-122 regulates systemic iron homeostasis adds to a growing list of liver functions that are controlled by miR-122. For example, miR-122 levels are decreased in advanced liver diseases such as cirrhosis [62] and hepatocellular carcinoma [63, 64], pathologies known to be exacerbated by increased liver iron levels [65]. Furthermore, several independent studies have demonstrated that in vivo inhibition of miR-122 reduces systemic cholesterol levels by as yet unidentified molecular mechanisms [66, 67]. Moreover, miR-122 expression is critical for hepatitis C virus (HCV) infection, replication, and the response to interferon therapy [68–70], which likewise involve alterations in iron homeostasis [71].
Concluding remarks
Iron homeostasis is controlled by miRNAs at the level of cellular uptake of iron-bound transferrin, iron storage by ferritin, and the hepatic control of systemic iron levels via hepcidin. Furthermore, tissue iron overload that hallmarks frequent disorders, such as hereditary hemochromatosis, the thalassemias, or Alzheimer’s disease causes oxidative stress that itself has been shown to alter miRNA expression [72, 73]. Similarly, iron deficiency activates the hypoxia-inducible factors (HIF) 1 and 2, critical transcription factors orchestrating adaptations to low oxygen pressure [74]. During hypoxia, Hif1a induces miR-210 [75, 76], which regulates ISCU1 and ISCU2, two proteins facilitating the assembly of FeS-clusters. These findings suggest that miRNAs control large regulatory networks that link microenvironmental stress, such as oxidative stress and hypoxia to the regulation of iron metabolism. As the maintenance of iron homeostasis is critical for many essential cellular functions, we expect to see several more miRNAs that will directly or indirectly control iron-related genes.
References
- 1.Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proc Natl Acad Sci USA. 2007;104:17719–17724. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 5.Griffiths-Jones S, Saini HK, van DS, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun W, Julie Li YS, Huang HD, Shyy JY, Chien S. microRNA: a master regulator of cellular processes for bioengineering systems 1. Annu Rev Biomed Eng. 2010;12:1–27. doi: 10.1146/annurev-bioeng-070909-105314. [DOI] [PubMed] [Google Scholar]
- 7.Bushati N, Cohen SM. MicroRNAs in neurodegeneration 2. Curr Opin Neurobiol. 2008;18:292–296. doi: 10.1016/j.conb.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 8.Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol Cell. 2010;39:373–384. doi: 10.1016/j.molcel.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartley AN, Yao H, Barkoh BA, Ivan C, Mishra BM, Rashid A, Calin GA, Luthra R, Hamilton SR. Complex patterns of altered MicroRNA expression during the adenoma-adenocarcinoma sequence for microsatellite-stable colorectal cancer. Clin Cancer Res. 2011;17:7283–7293. doi: 10.1158/1078-0432.CCR-11-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets 2. Cell. 2003;115:787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 12.Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, DeNicola G, Webster KA, Weiss D, Perez-Mancera PA, et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147:382–395. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di CF, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer A, Llobet-Navas D, Rajbhandari P, Bansal M, Guarnieri P, Silva J, et al. An extensive microRNA-mediated network of RNA–RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–381. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corcoran DL, Pandit KV, Gordon B, Bhattacharjee A, Kaminski N, Benos PV. Features of mammalian microRNA promoters emerge from polymerase II chromatin immunoprecipitation data. PLoS One. 2009;4:e5279. doi: 10.1371/journal.pone.0005279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shiohama A, Sasaki T, Noda S, Minoshima S, Shimizu N. Molecular cloning and expression analysis of a novel gene DGCR8 located in the DiGeorge syndrome chromosomal region. Biochem Biophys Res Commun. 2003;304:184–190. doi: 10.1016/S0006-291X(03)00554-0. [DOI] [PubMed] [Google Scholar]
- 20.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 21.Gregory RI, Chendrimada TP, Shiekhattar R. MicroRNA biogenesis: isolation and characterization of the microprocessor complex. Methods Mol Biol. 2006;342:33–47. doi: 10.1385/1-59745-123-1:33. [DOI] [PubMed] [Google Scholar]
- 22.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 23.Faller M, Matsunaga M, Yin S, Loo JA, Guo F. Heme is involved in microRNA processing. Nat Struct Mol Biol. 2007;14:23–29. doi: 10.1038/nsmb1182. [DOI] [PubMed] [Google Scholar]
- 24.Barr I, Smith AT, Senturia R, Chen Y, Scheidemantle BD, Burstyn JN, Guo F. DiGeorge critical region 8 (DGCR8) is a double-cysteine-ligated heme protein. J Biol Chem. 2011;286:16716–16725. doi: 10.1074/jbc.M110.180844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faller M, Toso D, Matsunaga M, Atanasov I, Senturia R, Chen Y, Zhou ZH, Guo F. DGCR8 recognizes primary transcripts of microRNAs through highly cooperative binding and formation of higher-order structures 1. RNA. 2010;16:1570–1583. doi: 10.1261/rna.2111310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Senturia R, Faller M, Yin S, Loo JA, Cascio D, Sawaya MR, Hwang D, Clubb RT, Guo F. Structure of the dimerization domain of DiGeorge critical region 8 2. Protein Sci. 2010;19:1354–1365. doi: 10.1002/pro.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barr I, Smith AT, Chen Y, Senturia R, Burstyn JN, Guo F. Ferric, not ferrous, heme activates RNA-binding protein DGCR8 for primary microRNA processing. Proc Natl Acad Sci USA. 2012;109:1919–1924. doi: 10.1073/pnas.1114514109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krause A, Neitz S, Magert HJ, Schulz A, Forssmann WG, Schulz-Knappe P, Adermann K. LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS Lett. 2000;480:147–150. doi: 10.1016/S0014-5793(00)01920-7. [DOI] [PubMed] [Google Scholar]
- 29.Park CH, Valore EV, Waring AJ, Ganz T. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J Biol Chem. 2001;276:7806–7810. doi: 10.1074/jbc.M008922200. [DOI] [PubMed] [Google Scholar]
- 30.Ward DM, Kaplan J. Ferroportin-mediated iron transport: expression and regulation. Biochim Biophys Acta. 2012;25:119–131. doi: 10.1016/j.bbamcr.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 32.Muckenthaler M, Roy CN, Custodio AO, Minana B, deGraaf J, Montross LK, Andrews NC, Hentze MW. Regulatory defects in liver and intestine implicate abnormal hepcidin and Cybrd1 expression in mouse hemochromatosis. Nat Genet. 2003;34:102–107. doi: 10.1038/ng1152. [DOI] [PubMed] [Google Scholar]
- 33.Bridle KR, Frazer DM, Wilkins SJ, Dixon JL, Purdie DM, Crawford DH, Subramaniam VN, Powell LW, Anderson GJ, Ramm GA. Disrupted hepcidin regulation in HFE-associated haemochromatosis and the liver as a regulator of body iron homoeostasis 1. Lancet. 2003;361:669–673. doi: 10.1016/S0140-6736(03)12602-5. [DOI] [PubMed] [Google Scholar]
- 34.Papanikolaou G, Samuels ME, Ludwig EH, MacDonald ML, Franchini PL, Dube MP, Andres L, MacFarlane J, Sakellaropoulos N, Politou M, et al. Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis 1. Nat Genet. 2004;36:77–82. doi: 10.1038/ng1274. [DOI] [PubMed] [Google Scholar]
- 35.Huang FW, Pinkus JL, Pinkus GS, Fleming MD, Andrews NC. A mouse model of juvenile hemochromatosis 1. J Clin Invest. 2005;115:2187–2191. doi: 10.1172/JCI25049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niederkofler V, Salie R, Arber S. Hemojuvelin is essential for dietary iron sensing, and its mutation leads to severe iron overload 1. J Clin Invest. 2005;115:2180–2186. doi: 10.1172/JCI25683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Camaschella C, Roetto A, Cali A, De GM, Garozzo G, Carella M, Majorano N, Totaro A, Gasparini P. The gene TFR2 is mutated in a new type of haemochromatosis mapping to 7q22 1. Nat Genet. 2000;25:14–15. doi: 10.1038/75534. [DOI] [PubMed] [Google Scholar]
- 38.Fleming RE, Ahmann JR, Migas MC, Waheed A, Koeffler HP, Kawabata H, Britton RS, Bacon BR, Sly WS. Targeted mutagenesis of the murine transferrin receptor-2 gene produces hemochromatosis 1. Proc Natl Acad Sci USA. 2002;99:10653–10658. doi: 10.1073/pnas.162360699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silvestri L, Pagani A, Nai A, De DI, Kaplan J, Camaschella C. The serine protease matriptase-2 (TMPRSS6) inhibits hepcidin activation by cleaving membrane hemojuvelin 1. Cell Metab. 2008;8:502–511. doi: 10.1016/j.cmet.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mleczko-Sanecka K, Casanovas G, Ragab A, Breitkopf K, Muller A, Boutros M, Dooley S, Hentze MW, Muckenthaler MU. SMAD7 controls iron metabolism as a potent inhibitor of hepcidin expression 1. Blood. 2010;115:2657–2665. doi: 10.1182/blood-2009-09-238105. [DOI] [PubMed] [Google Scholar]
- 41.Muckenthaler MU, Galy B, Hentze MW. Systemic iron homeostasis and the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network. Annu Rev Nutr. 2008;28:197–213. doi: 10.1146/annurev.nutr.28.061807.155521. [DOI] [PubMed] [Google Scholar]
- 42.Sanchez M, Galy B, Schwanhaeusser B, Blake J, Bahr-Ivacevic T, Benes V, Selbach M, Muckenthaler MU, Hentze MW. Iron regulatory protein-1 and -2: transcriptome-wide definition of binding mRNAs and shaping of the cellular proteome by iron regulatory proteins. Blood. 2011;118:e168–e179. doi: 10.1182/blood-2011-04-343541. [DOI] [PubMed] [Google Scholar]
- 43.Smith SR, Cooperman S, Lavaute T, Tresser N, Ghosh M, Meyron-Holtz E, Land W, Ollivierre H, Jortner B, Switzer R, III, et al. Severity of neurodegeneration correlates with compromise of iron metabolism in mice with iron regulatory protein deficiencies 1. Ann NY Acad Sci. 2004;1012:65–83. doi: 10.1196/annals.1306.006. [DOI] [PubMed] [Google Scholar]
- 44.Ohgami RS, Campagna DR, Greer EL, Antiochos B, McDonald A, Chen J, Sharp JJ, Fujiwara Y, Barker JE, Fleming MD. Identification of a ferrireductase required for efficient transferrin-dependent iron uptake in erythroid cells 1. Nat Genet. 2005;37:1264–1269. doi: 10.1038/ng1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Magro G, Cataldo I, Amico P, Torrisi A, Vecchio GM, Parenti R, Asioli S, Recupero D, D’Agata V, Mucignat MT, et al. Aberrant expression of TfR1/CD71 in thyroid carcinomas identifies a novel potential diagnostic marker and therapeutic target 1. Thyroid. 2011;21:267–277. doi: 10.1089/thy.2010.0173. [DOI] [PubMed] [Google Scholar]
- 46.Kwok JC, Richardson DR. The iron metabolism of neoplastic cells: alterations that facilitate proliferation? 1. Crit Rev Oncol Hematol. 2002;42:65–78. doi: 10.1016/S1040-8428(01)00213-X. [DOI] [PubMed] [Google Scholar]
- 47.Rovera G, Santoli D, Damsky C. Human promyelocytic leukemia cells in culture differentiate into macrophage-like cells when treated with a phorbol diester 1. Proc Natl Acad Sci USA. 1979;76:2779–2783. doi: 10.1073/pnas.76.6.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schaar DG, Medina DJ, Moore DF, Strair RK, Ting Y. miR-320 targets transferrin receptor 1 (CD71) and inhibits cell proliferation 1. Exp Hematol. 2009;37:245–255. doi: 10.1016/j.exphem.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 49.Hubert N, Hentze MW. Previously uncharacterized isoforms of divalent metal transporter (DMT)-1: implications for regulation and cellular function 1. Proc Natl Acad Sci USA. 2002;99:12345–12350. doi: 10.1073/pnas.192423399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gunshin H, Allerson CR, Polycarpou-Schwarz M, Rofts A, Rogers JT, Kishi F, Hentze MW, Rouault TA, Andrews NC, Hediger MA. Iron-dependent regulation of the divalent metal ion transporter 1. FEBS Lett. 2001;509:309–316. doi: 10.1016/S0014-5793(01)03189-1. [DOI] [PubMed] [Google Scholar]
- 51.Tchernitchko D, Bourgeois M, Martin ME, Beaumont C. Expression of the two mRNA isoforms of the iron transporter Nramp2/DMTI in mice and function of the iron responsive element 1. Biochem J. 2002;363:449–455. doi: 10.1042/0264-6021:3630449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Canonne-Hergaux F, Zhang AS, Ponka P, Gros P. Characterization of the iron transporter DMT1 (NRAMP2/DCT1) in red blood cells of normal and anemic mk/mk mice 1. Blood. 2001;98:3823–3830. doi: 10.1182/blood.V98.13.3823. [DOI] [PubMed] [Google Scholar]
- 53.Andolfo I, De FL, Asci R, Russo R, Colucci S, Gorrese M, Zollo M, Iolascon A. Regulation of divalent metal transporter 1 (DMT1) non-IRE isoform by the microRNA Let-7d in erythroid cells 1. Haematologica. 2010;95:1244–1252. doi: 10.3324/haematol.2009.020685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chua AC, Olynyk JK, Leedman PJ, Trinder D. Nontransferrin-bound iron uptake by hepatocytes is increased in the Hfe knockout mouse model of hereditary hemochromatosis. Blood. 2004;104:1519–1525. doi: 10.1182/blood-2003-11-3872. [DOI] [PubMed] [Google Scholar]
- 55.Mastrogiannaki M, Matak P, Keith B, Simon MC, Vaulont S, Peyssonnaux C. HIF-2alpha, but not HIF-1alpha, promotes iron absorption in mice 1. J Clin Invest. 2009;119:1159–1166. doi: 10.1172/JCI38499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yeh KY, Yeh M, Polk P, Glass J. Hypoxia-inducible factor-2alpha and iron absorptive gene expression in Belgrade rat intestine 2. Am J Physiol Gastrointest Liver Physiol. 2011;301:G82–G90. doi: 10.1152/ajpgi.00538.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shpyleva SI, Tryndyak VP, Kovalchuk O, Starlard-Davenport A, Chekhun VF, Beland FA, Pogribny IP. Role of ferritin alterations in human breast cancer cells 2. Breast Cancer Res Treat. 2011;126:63–71. doi: 10.1007/s10549-010-0849-4. [DOI] [PubMed] [Google Scholar]
- 58.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells 1. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dykxhoorn DM, Wu Y, Xie H, Yu F, Lal A, Petrocca F, Martinvalet D, Song E, Lim B, Lieberman J. miR-200 enhances mouse breast cancer cell colonization to form distant metastases 1. PLoS One. 2009;4:e7181. doi: 10.1371/journal.pone.0007181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Castoldi M, Vujic SM, Altamura S, Elmen J, Lindow M, Kiss J, Stolte J, Sparla R, D’Alessandro LA, Klingmuller U, et al. The liver-specific microRNA miR-122 controls systemic iron homeostasis in mice. J Clin Invest. 2011;121:1386–1396. doi: 10.1172/JCI44883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bolondi G, Garuti C, Corradini E, Zoller H, Vogel W, Finkenstedt A, Babitt JL, Lin HY, Pietrangelo A. Altered hepatic BMP signaling pathway in human HFE hemochromatosis 1. Blood Cells Mol Dis. 2010;45:308–312. doi: 10.1016/j.bcmd.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Padgett KA, Lan RY, Leung PC, Lleo A, Dawson K, Pfeiff J, Mao TK, Coppel RL, Ansari AA, Gershwin ME. Primary biliary cirrhosis is associated with altered hepatic microRNA expression 1. J Autoimmun. 2009;32:246–253. doi: 10.1016/j.jaut.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kutay H, Bai S, Datta J, Motiwala T, Pogribny I, Frankel W, Jacob ST, Ghoshal K. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas 1. J Cell Biochem. 2006;99:671–678. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 64.Girard M, Jacquemin E, Munnich A, Lyonnet S, Henrion-Caude A. miR-122, a paradigm for the role of microRNAs in the liver 1. J Hepatol. 2008;48:648–656. doi: 10.1016/j.jhep.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 65.Kew MC. Hepatic iron overload and hepatocellular carcinoma. Cancer Lett. 2009;286:38–43. doi: 10.1016/j.canlet.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 66.Elmen J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjarn M, Hansen JB, Hansen HF, Straarup EM, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver 17. Nucleic Acids Res. 2008;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting 18. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 68.Jopling CL, Norman KL, Sarnow P. Positive and negative modulation of viral and cellular mRNAs by liver-specific microRNA miR-122 1. Cold Spring Harb Symp Quant Biol. 2006;71:369–376. doi: 10.1101/sqb.2006.71.022. [DOI] [PubMed] [Google Scholar]
- 69.Jopling CL. Regulation of hepatitis C virus by microRNA-122 1. Biochem Soc Trans. 2008;36:1220–1223. doi: 10.1042/BST0361220. [DOI] [PubMed] [Google Scholar]
- 70.Sarasin-Filipowicz M. Interferon therapy of hepatitis C: molecular insights into success and failure 1. Swiss Med Wkly. 2010;140:3–11. doi: 10.4414/smw.2010.12670. [DOI] [PubMed] [Google Scholar]
- 71.Ryan JD, Altamura S, Devitt E, Mullins S, Lawless MW, Muckenthaler MU, Crowe J. Pegylated interferon-alpha induced hypoferremia is associated with the immediate response to treatment in hepatitis C. Hepatology. 2012 doi: 10.1002/hep.25666. [DOI] [PubMed] [Google Scholar]
- 72.Svasti S, Masaki S, Penglong T, Abe Y, Winichagoon P, Fucharoen S, Umemura T. Expression of microRNA-451 in normal and thalassemic erythropoiesis. Ann Hematol. 2010;89:953–958. doi: 10.1007/s00277-010-0980-7. [DOI] [PubMed] [Google Scholar]
- 73.Geekiyanage H, Chan C. MicroRNA-137/181c regulates serine palmitoyltransferase and in turn amyloid beta, novel targets in sporadic Alzheimer’s disease. J Neurosci. 2011;31:14820–14830. doi: 10.1523/JNEUROSCI.3883-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yoon D, Pastore YD, Divoky V, Liu E, Mlodnicka AE, Rainey K, Ponka P, Semenza GL, Schumacher A, Prchal JT. Hypoxia-inducible factor-1 deficiency results in dysregulated erythropoiesis signaling and iron homeostasis in mouse development 1. J Biol Chem. 2006;281:25703–25711. doi: 10.1074/jbc.M602329200. [DOI] [PubMed] [Google Scholar]
- 75.Favaro E, Ramachandran A, McCormick R, Gee H, Blancher C, Crosby M, Devlin C, Blick C, Buffa F, Li JL, et al. MicroRNA-210 regulates mitochondrial free radical response to hypoxia and krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU 4. PLoS One. 2010;5:e10345. doi: 10.1371/journal.pone.0010345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chan SY, Zhang YY, Hemann C, Mahoney CE, Zweier JL, Loscalzo J. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2 6. Cell Metab. 2009;10:273–284. doi: 10.1016/j.cmet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Arosio P, Levi S. Cytosolic and mitochondrial ferritins in the regulation of cellular iron homeostasis and oxidative damage. Biochim Biophys Acta. 2010;1800:783–792. doi: 10.1016/j.bbagen.2010.02.005. [DOI] [PubMed] [Google Scholar]