Abstract
Alzheimer’s disease (AD) is a neurodegenerative disorder occurring in the elderly. It is widely accepted that the amyloid beta peptide (Aβ) aggregation and especially the oligomeric states rather than fibrils are involved in AD onset. We used infrared spectroscopy to provide structural information on the entire aggregation pathway of Aβ(1–40), starting from monomeric Aβ to the end of the process, fibrils. Our structural study suggests that conversion of oligomers into fibrils results from a transition from antiparallel to parallel β-sheet. These structural changes are described in terms of H-bonding rupture/formation, β-strands reorientation and β-sheet elongation. As antiparallel β-sheet structure is also observed for other amyloidogenic proteins forming oligomers, reorganization of the β-sheet implicating a reorientation of β-strands could be a generic mechanism determining the kinetics of protein misfolding. Elucidation of the process driving aggregation, including structural transitions, could be essential in a search for therapies inhibiting aggregation or disrupting aggregates.
Keywords: Amyloid-beta, Alzheimer’s disease, Aggregation, Secondary structure changes, Infrared spectroscopy
Introduction
The accumulation of aggregated protein is a typical hallmark of many human degenerative disorders, among which Alzheimer’s, Parkinson’s and Creutzfeldt–Jakob’s diseases are the most known. Misfolding of amyloidogenic proteins gives rise to self-assembled aggregates [1]. Whereas monomeric forms are not toxic, these aggregates have been identified as toxic species responsible for the disease. Because a wide range of amyloidogenic proteins which do not share sequence homology at all, show similar biophysical characteristics after aggregation, it has been assumed that diverse amyloidoses may share a common mechanism of pathogenesis [2].
Alzheimer’s disease (AD) is characterized by a progressive decline of cognitive abilities as a consequence of synaptic and neuronal loss. The principal neuropathological feature of this brain disease consists in polymorphous extracellular deposits called “senile plaques”. The amyloid-beta peptide (Aβ), the primary component of these plaques, was suggested to be the key factor in the development of AD pathology [2, 3]. This peptide is produced by the proteolytic cleavage of the amyloid precursor protein by the β- and γ-secretases to predominantly form 40–42 amino acid peptides [4]. In its native soluble state, the peptide adopts both helical and random-coil conformations [5]. Assembly of Aβ monomers leads to soluble aggregates, namely oligomers, presenting β-structure [6]. Many types of Aβ oligomers have been described and linked to AD etiology: dimers [7], trimers, tetramers, dodecamers (Aβ*56) [8], Aβ-derived diffusible ligands (ADDLs), protofibrils and also annular oligomers [9]. Further aggregation gives rise to insoluble fibrils causing extraneuronal deposits in the brain.
In the first “amyloid cascade hypothesis” [10], Aβ fibrils were supposed to be responsible for the initiation of AD. However, many publications over the last decade rather blamed oligomers. They were indeed shown to be responsible for the formation of tau protein tangles [11], neuronal oxidative stress [12], prolonged long-term depression [13] and inhibition of long-term potentiation [14]. Other groups linked oligomers to abnormal dendritic spine morphology, spine loss, change in synaptic receptor composition [15] and finally cell death. Oligomers have thus replaced fibrils as the cause of the disorder in the “new amyloid cascade hypothesis” [16].
Despite the general interest concerning oligomers toxicity compared to fibrils, few structural data are available [17, 18] and so far, no consensus model has been proposed. Nevertheless, infrared spectroscopy on Aβ(1–40) [19] and (1–42) [20, 21] oligomers shows an antiparallel β-sheet organization.
However, the mechanisms leading to accumulation of misfolded peptide and fibrils formation still remain a matter of debate. Fibrils formation has been shown to occur by a nucleation-dependent kinetics, whereas the rate-determining key step is the formation of oligomers, after which fibrillization rapidly takes place [22]. Subsequent experiments have proved that aggregation of Aβ and its self-assembly into mature fibrils do not necessarily occur in a linear way [23]. Many aggregation intermediates or oligomers can either give rise to fibrils or remain as such, without further aggregating [24].
Biophysical studies describe fibrils as cross-β-structure filaments [25–27]. However, structural studies of oligomers and the conformational transitions taking place during the aggregation process are limited by the size range and transitory nature of these aggregates [28]. Elucidation of the mechanisms driving the aggregation, including structural transitions in the peptide, could be essential in a search for therapies inhibiting aggregation or disrupting aggregates [29]. Our group already demonstrated that oligomers and fibrils of Aβ(1–42) do not share the same β-sheet organization. Indeed, infrared spectroscopy performed in attenuated total reflection mode (ATR-FTIR) displays an antiparallel organization of the β-sheets for oligomers while fibrils are composed of parallel β-sheets [20]. This study led to a putative model of Aβ(1–42) oligomers tridimensional organization. We suggested that the antiparallel organization of the β-sheets in oligomers could lead to the formation of a β-barrel, particularly suited to insert into lipidic bilayers.
Our main objective here is to use spectroscopic techniques for which the insoluble nature of aggregates does not represent an issue [20], in order to provide a structural description of the entire aggregation process of Aβ(1–40), from the very beginning with Aβ in its monomeric state to the last step of the process when Aβ turns into insoluble fibrils. As Aβ(1–40) is known to have a different folding, aggregation process and toxicity compared to the Aβ(1–42), it is not evident whether this peptide formed oligomers with the same conformation or have the same structural changes when it aggregates. Species formed along the aggregation pathway were characterized by atomic force microscopy (AFM), SDS-PAGE and accessibility to monoclonal antibodies.
Materials and methods
Reagents and chemicals
Aβ(1–40) was purchased from American Peptide Co. (Sunnyvale, CA, USA). Dimethylsulfoxide (DMSO 99.9% purity) and Thioflavin T (ThT) were obtained from Sigma-Aldrich (St. Louis, MO, USA). The antibodies used (6E10, 4G8, 11A50-B10) were from Covance (Emeryville, CA, USA). The conformational A11 antibody was kindly provided by Dr C. Glabe (UC, Irvine). Horseradish peroxidase-conjugated anti-mouse and anti-rabbit antibodies were, respectively, purchased from Millipore (Billerica, MA, USA) and Cell Signaling Technology (Danvers, MA, USA). Supersignal West Pico Chemiluminescent Substrate and ECL plus western blot detection system were, respectively obtained from Pierce (Biotechnology, Rockford, IL, USA) and GE Healthcare (Piscataway, NJ, USA).
Aβ(1–40) preparation
The peptide was dissolved in cold milliQ water at a 2 mg/ml concentration and then aliquoted. MilliQ water was evaporated using a Speed Vac (Thermo Savant). The resulting Aβ(1–40) film was stored at −20°C until further manipulation. Before any incubation, we use DMSO for initial resuspension of the peptide (5 mM final concentration) to avoid pre-existing structures in the sample.
Aβ(1–40) aggregation
Aggregation of Aβ(1–40) was carried out at 37°C by incubating the peptide in 20 mM Tris–HCl, pH 7.4, 100 mM NaCl (Tris-buffered saline, TBS). The final Aβ concentration is 100 μM. Samples were taken during the time course of peptide aggregation to perform the experiments described below.
Western blot analysis of SDS-PAGE
Samples were diluted in SDS-PAGE sample buffer (Laemmli sample buffer) and separated on a 12% bis–Tris gel using MES as running buffer. The separated bands were transferred onto a nitrocellulose membrane and detected with the mouse monoclonal anti-Aβ antibody 6E10 (dilution 1:3,000). Detection was carried out using horseradish peroxidase-conjugated anti-mouse antibody (1:2,000) and the Supersignal West Pico Chemiluminescent Substrate. Pictures were recorded and analyzed using the ImageQuant 400 gel imager and ImageQuant TL software (GE Healthcare). Molecular weights were estimated using a prestained protein ladder from Fermentas (St. Leon-Rot, Germany).
Atomic force microscopy
AFM characterization of Aβ(1–40) samples has been handled on a Nanoscope IIIa, as reported in Cerf et al. [20]. All images were taken at 21°C in tapping mode on Aβ samples diluted to 20 μM and plated on freshly cleaved mica.
Thioflavin T (ThT) fluorescence
To perform ThT fluorescence assay on our samples, we followed the well-established protocol of H. Levine III [30]. Five micromolar of ThT were freshly dissolved in 50 mM glycine-NaOH pH 8.5 buffer. For each measurement, we diluted 4.5 μg of Aβ(1–40) in the ThT buffer. Temperature was kept constant at 25°C; emission wavelength: 482 nm; excitation wavelength: 450 nm.
IR spectroscopy
IR spectra were recorded on an Equinox 55 infrared spectrophotometer (Bruker Optics, Ettlingen, Germany) placed in a thermoregulated room (21°C). For more details see [20]. Each spectrum represents the mean of 128 repetitions, taken at a resolution of 2 cm−1. One microgram of 100 μM Aβ(1–40) samples [prepared as described in (“Aβ(1–40) species morphology along aggregation”) and (“Accessibility of the different Aβ(1–40) entities to specific monoclonal antibodies”)], was used for each measurement and spread on the surface of the internal reflection element made of a diamond crystal. Excess water was removed under nitrogen flow. For salt elimination, the peptide solution was washed three times with excess milliQ water at the surface of the internal reflection element. Finally, excess water was removed under nitrogen flow. The curve fitting analysis has been obtained according to [31]. Briefly, Fourier self-deconvolution was applied to increase the resolution of the spectra in the amide I region, which is the most sensitive to the secondary structure of proteins. The self-deconvolution was carried out using a Lorentzian line shape for the deconvolution and a Gaussian line shape for the apodization. To quantify the area of the different components of amide I revealed by the self-deconvolution, a least square iterative curve fitting was performed to fit a mixture of Gaussian and Lorentzian line shapes to the spectrum between 1,700 and 1,600 cm−1. Prior to curve fitting, a straight baseline passing through the ordinates at 1,700 and 1,600 cm−1 was subtracted. To avoid introducing artifacts due to the self-deconvolution procedure, the fitting was performed on the non-deconvoluted spectrum. The proportion of a particular structure is computed to be the sum of the area of all the fitted bands having their maximum in the frequency region where that structure occurs divided by the area of all the Lorentzian bands having their maximum between 1,700 and 1,600 cm−1 [31, 32].
Dot blot analysis
Each dot consists in 1 μg of Aβ(1–40) sample. The blocking, washing and antibody incubation protocols of the nitrocellulose membrane were already detailed in [20]. We detected Aβ(1–40) dots using either 6E10, 4G8, 11A50-B10 (mouse monoclonal antibodies) or A11 (rabbit monoclonal antibody), all diluted 3,000 times. The secondary antibodies were horseradish peroxidase-conjugated: anti-mouse IgG for 6E10, 4G8 and 11A50-B10 and anti-rabbit IgG for A11. Detection and quantification of the dots intensities have been handled as for western blot.
Results
Aβ(1–40) species distribution during aggregation
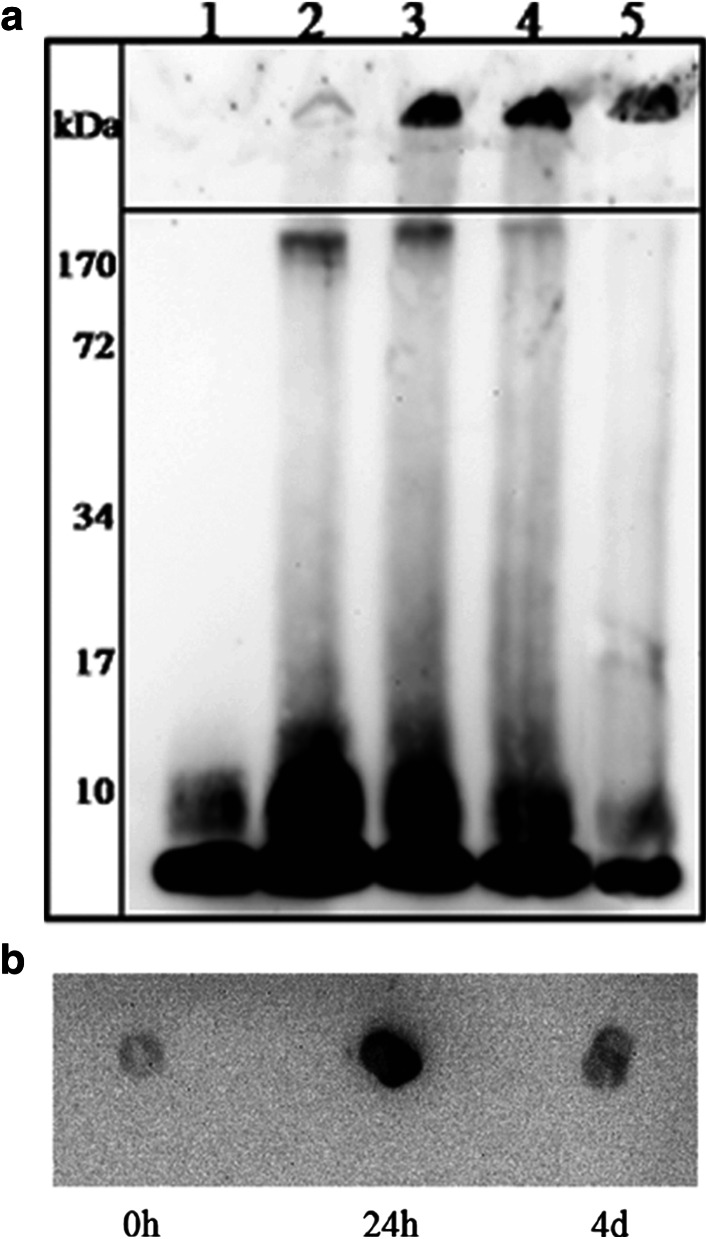
Samples of the peptide in TBS buffer at 37°C were collected at different aggregation time points and characterized by bis–Tris SDS-PAGE followed by 6E10 monoclonal antibody recognition (western blot). At the very beginning of the aggregation process, Aβ(1–40) is mainly present as monomers (84% as calculated with ImageQuant TL software) (Fig. 1a, lane 1). Dimers (8.1 kDa) are already detected but in lower proportions (16%). Further aggregation then gives rise to different types of oligomers: dimers (mainly), trimers, a slight amount of tetramers and higher molecular weight oligomers (~170 kDa) (Fig. 1a, lane 2). These 24 h-entities are recognized by A11, an oligomeric conformation-specific antibody (Fig. 1b). Continued aggregation leads to extremely high molecular weight species formation, remaining in the stacking gel and corresponding to fibrils (Fig. 1a, lanes 3, 4, 5). At the same time, we observe a concomitant and progressive decrease of the oligomeric content with incubation, most probably transforming into fibrils. Four-days samples become weakly reactive to A11 (Fig. 1b). After 2 weeks of incubation, the main species showing up on the gel are fibrils (Fig. 1a, lane 5). Therefore, the aggregation process involves a size distribution of aggregates which evolve to become, for most of them, the final entities of the aggregation pathway.
Fig. 1.
Visualization of Aβ(1–40) species during aggregation. a Western blot analysis of Aβ(1–40) samples, separated on a 12% bis–Tris SDS-PAGE and probed with 6E10 monoclonal antibody. Aβ(1–40) was incubated in TBS, pH 7.4 for 0 h (lane 1), 24 h (lane 2), 48 h (lane 3), 72 h (lane 4) and 16 days (lane 5). b Reactivity of Aβ(1–40) aggregates to A11 antibody. Dot blot analysis: Aβ(1–40) after 0 h, 24 h and 4 days in TBS, pH 7.4
Although, SDS-PAGE is a common technique used to study amyloidogenic proteins, it is not devoid of potential drawbacks. This is why we further analyzed our samples with complementary techniques.
Aβ(1–40) species morphology along aggregation
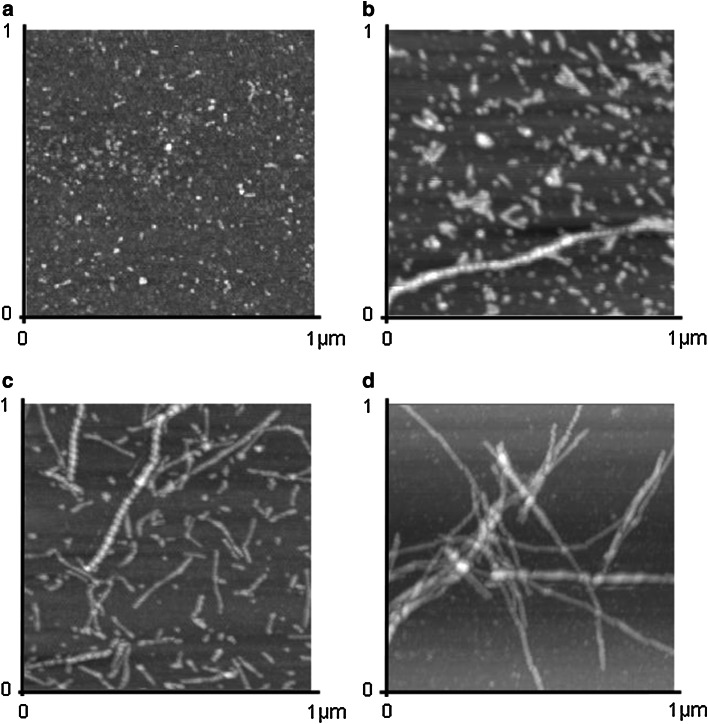
To further depict the different species formed during the aggregation process, we used AFM as an imaging technique. At the starting point, Aβ(1–40) forms small spherical entities, which are in agreement with previously described oligomers [20, 33] (Fig. 2a). Oligomers particles show an average height and diameter of ~1.5 nm and 60 nm, respectively. No fibrillar assemblies were detected at this time point. After several hours of aggregation (4 and 24 h; Fig. 2b, c), Aβ(1–40) forms large oligomers (also known as ADDLs), with a height and a diameter around 9 nm and 75 nm, respectively. Several thread-like structures, which are most likely protofibrils, are simultaneously seen. These two pictures display the most heterogeneity in terms of size and shape of the species observed. Continuing sample incubation leads to elongated forms of Aβ(1–40), corresponding to fibrils (Fig. 2d) weakly reactive to A11 (see Fig. 1b). At higher magnification, fibrils display a periodicity along the axis (result not shown). The morphology of the samples is not modified as a consequence of the interaction with the mica plate. This was investigated earlier [20].
Fig. 2.
AFM images of Aβ(1–40). AFM images were recorded in 1 × 1 μm tapping mode. Aβ(1–40) was incubated in TBS, pH 7.4 after a 0 h (total z range = 15 nm), b 4 h (total z range = 20 nm), c 24 h (total z range = 15 nm) and d 1 week (total z range = 30 nm)
Accessibility of the different Aβ(1–40) entities to specific monoclonal antibodies
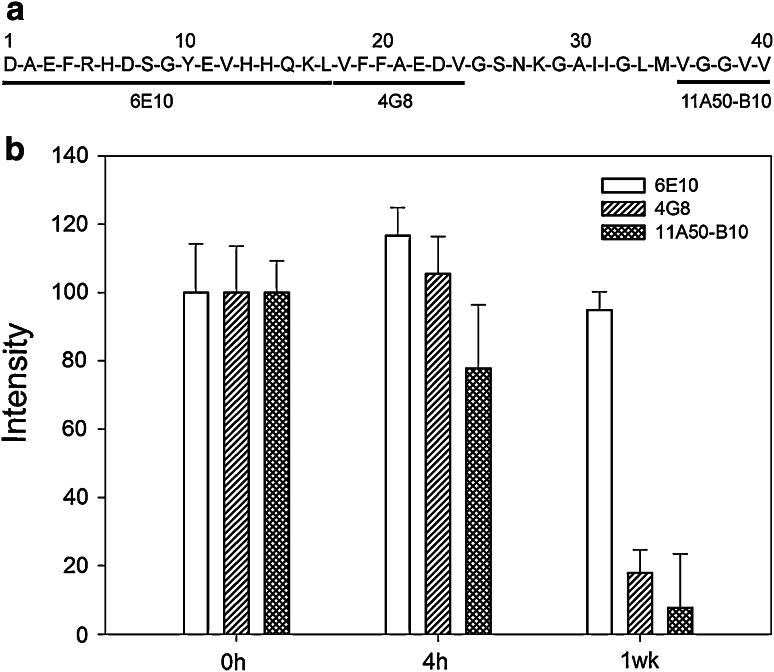
To investigate which regions of Aβ(1–40) are accessible during the time course of aggregation, a dot blot analysis was performed using three monoclonal antibodies raised against the N-terminal (6E10), central (4G8) and C-terminal (11A50-B10) parts of Aβ (Fig. 3a). Dots intensities were further quantified with ImageQuant TL software (GE Healthcare). Aβ(1–40) oligomers obtained in the early stages of aggregation show a slightly decreased (~20%) accessibility of the C-terminal end (aa 36–40) compared to the freshly resuspended peptide. Interestingly, the central (aa 17–24) and C-terminal regions become almost completely inaccessible in the fibrillar forms of the peptide (decrease in accessibility of ~80 and 90%, respectively) (Fig. 3b).
Fig. 3.
Accessibility of the aggregates to region-specific monoclonal antibodies. a The three monoclonal targets in Aβ(1–40) sequence. b Dot blot intensities for 6E10 (white), 4G8 (single hatched) and 11A50-B10 (double hatched). The intensities were analyzed using ImageQuant TL software
Secondary structure changes occurring during aggregation
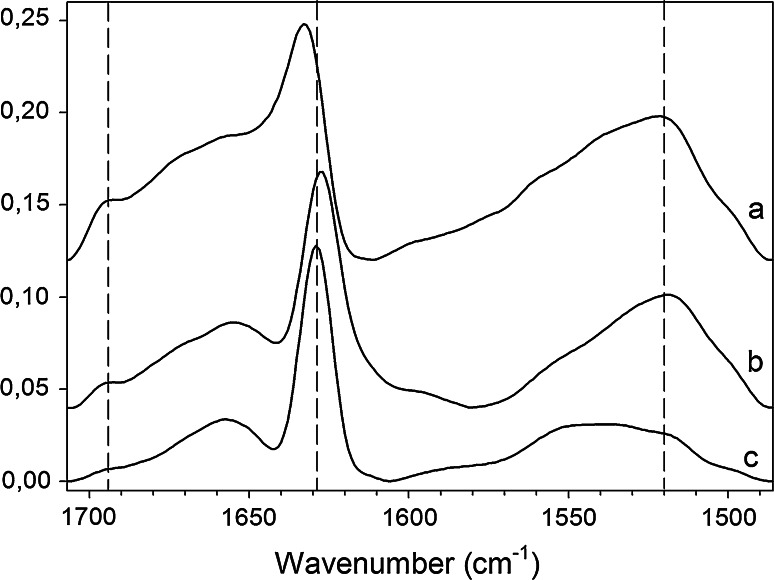
Based on the amide I band analysis (1,700–1,600 cm−1), Aβ(1–40) mainly displays β-sheet structure during the aggregation process with a main peak around ~1,630 cm−1. Interestingly, the amide I of the 0 h and 24 h spectra shows an additional minor peak around 1,695 cm−1 (Fig. 4a, b). The presence of both components is typical of an antiparallel arrangement of the β-strands [34–36] and is attributed to oligomers [20]. When the peptide is incubated longer until it turns into fibrils, the 1,695 cm−1 peak vanishes, meaning most likely that the peptide reorganizes its β-strands from an antiparallel to a parallel β-sheet (Fig. 4c). The progressive disappearance of the 1,695 cm−1 peak perfectly correlates with oligomers loss as observed by SDS-PAGE, A11 Dot blot and AFM (Figs. 1, 2).
Fig. 4.
ATR-FTIR spectra of Aβ(1–40) in the amide I and II regions. Aβ(1–40) in TBS, pH 7.4, a after 0 h, b after 24 h, and c after 96 h. Spectral intensities were normalized to the intensity of the 1,630 cm−1 peaks and were shifted for better visualization. Spectra were deconvoluted using a Lorentzian deconvolution factor with a full width at half height of 20 cm−1 and a Gaussian apodization factor with a full width at half height of 13.33 cm−1 to obtain a resolution enhancement factor K = 1.5. Spectra are representative of at least three independent experiments
The amide II band (1,600–1,500 cm−1), which is more complex than the amide I but also sensitive to secondary structure, shows a maximum of absorbance that evolves from 1,520 cm−1 to higher wavenumbers upon peptide aggregation. This initial position specifically corresponds to antiparallel β-sheet [37] and its shift to higher wavenumbers is in agreement with the progressive decrease of the 1,695 cm−1 peak upon aggregation.
During aggregation, the peak around 1,630 cm−1 seems to evolve to a narrower peak, shifted to smaller wavenumbers, from 1,632 to 1,627 cm−1. This shift is interesting because it indicates an increase of the number of β-strands involved the β-sheet and/or the formation of stronger H-bonding (i.e. shorter H-bonds) [38, 39] as expected in extremely stable structure such as fibrils.
A quantitative analysis of the amide I region by deconvolution followed by curve fitting confirms the gradual evolution in the secondary structure content. The β-sheet structure is the most abundant secondary structure (1,630 and 1,695 cm−1 peaks), which increases from 42 to 57% while the peptide aggregates. Additionally, a loss in random coil and helical contribution is observed from 40 to 20% (1,657 cm−1 peak). The latter could be attributable to the lowering amount of monomers along aggregation as observed on SDS-PAGE (from 86 to 39%) (Fig. 1a, lanes 1 and 5). The β-turns content represents 15–20% for all aggregated forms. The secondary structure determination within Aβ(1–40) fibrils is in excellent agreement with previous studies [25].
Thioflavin T and FTIR: complementary tools for aggregates identification
The relative intensity ratio of the 1,695 and the 1,630 cm−1 peaks has been suggested to be proportional to the percentage of antiparallel arrangement of the strands in a β-sheet [34–36]. Whereas the 1,695/1,630 cm−1 intensity ratio describes the amount of oligomers during the time course of aggregation, Thioflavin T fluorescence was used to probe the formation of fibrils (Fig. 5). As expected, the freshly resuspended peptide has a low ThT binding capacity. In contrast, 48 h samples, in which high molecular weight aggregates become the main entities (Figs. 1, 2), reach a plateau whereas the 1,695 cm−1 peak becomes almost undetectable. The evolution of the 1,695/1,630 cm−1 intensity ratios along Aβ aggregation correlates nicely with the increase of ThT fluorescence.
Fig. 5.
ThT fluorescence emission intensity and 1,695/1,630 cm−1 ratios of Aβ during the aggregation process. Comparison of the average ThT fluorescence emission intensity (circles) with the 1,695/1,630 cm−1 intensity ratios (triangles) of Aβ(1–40) samples along the aggregation process. The 1,695/1,630 cm−1 ratios were calculated on the basis of deconvoluted spectra. Means and error bars have been calculated on basis of three independent repetitions
Because of the drying process, ATR-FTIR may perturb the system by enhancing the aggregation. However, this comparison between ATR-FTIR and Thioflavin T fluorescence spectroscopy data provides evidence that there is an obvious correlation between the same samples observed either on a surface or in solution.
Discussion
The initial and final states of Aβ fibrillogenesis have been well established and characterized by biophysical methods [26, 27, 40, 41], but conversely, the dynamic process leading to aggregation and to the formation of oligomeric intermediates is much less understood and still under investigation [24, 42]. Tycko’s group provided evidence for a parallel β-sheet organization in the fibrils using solid state NMR implicating residues 12–40 with a turn located around residues 21–30 [25, 26, 41]. This strand-turn-strand motif is packed by the strands side chains and the turn is called β-arc. Other structural organizations have been suggested for fibrils based on cryo electron microscopy images [27]. Nevertheless, in all these models, the β-sheets are described as parallel and therefore should give rise to similar infrared spectral features. On the other hand, monomeric states are harder to study than fibrils because of their transitory nature. Moreover, their secondary structure seems to depend on the environmental conditions. In its native state, Aβ is supposed to adopt an α-helix structure embedded in the membrane, just after APP cleavage by secretases [43]. Lately, a high resolution structure of Aβ monomers in solution stabilized by a phage-display selected Affibody® protein showed a β-hairpin structure implicating two short β-strands (aa 17–23 and aa 30–36) in an antiparallel organization [40]. The mechanism leading to fibrils formation thus somehow implicates a structural transition, which has been much less investigated because of the transitory and heterogeneous nature of aggregation intermediates currently considered as key factors in AD development. The structural transition from native protein to cross-β-sheet is nevertheless accepted to be essential for fiber formation. It is believed that these conformational changes are due to competition between hydrophobicity and hydrogen bonding [44, 45]. As the knowledge concerning the neurotoxicity of Aβ aggregates has evolved and now points out the higher toxicity and the crucial role of oligomers in the cognitive symptoms associated with AD [46], we found it important to gain a deeper understanding of the aggregation process. Even though Aβ oligomers are privileged theoretical targets in therapies efforts [47], current structural data are still imprecise and/or missing. In a previous work, we specifically formed either the oligomeric or fibrillar species of Aβ(1–42) peptide in order to characterize their secondary structures using infrared spectroscopy. This study led to a putative model of Aβ(1–42) oligomers tridimensional organization [20]. Our main objective here is to provide a structural description of the entire aggregation process of Aβ(1–40), starting from the very beginning with Aβ in its monomeric state, to the last step of the process, when Aβ turns into insoluble fibrils.
There is an excellent correlation between ATR-FTIR and Thioflavin T fluorescence spectroscopy data concerning Aβ aggregation level (Fig. 5). While the 1,695/1,630 cm−1 FTIR peaks intensity ratio is representative of the oligomeric content, ThT probes the growth and/or the presence of amyloid fibrils. From a structural point of view, ThT curve corresponds to the disappearance of the 1,695 cm−1 peak in the amide I band in agreement with the reorganization of the β-strands from antiparallel to parallel β-sheets (Fig. 5).
As neither the structural diversity nor the size range or insoluble nature of Aβ aggregates represent an obstacle for ATR-FTIR spectroscopy, this technique can be used to follow Aβ aggregation as well as the ThT probe assay.
Aβ aggregation is known to be complex and involves more than a simple conversion from monomers to fibers. Aggregation intermediates could be on-pathway or off-pathway to give rise to fibrils [9, 24]. Moreover, it has been suggested that oligomerization and fibrillization could be independent processes which may be selectively inhibited [23, 48, 49]. On the other hand, fibrils formation could be directly initiated by monomers and/or oligomers. Our in vitro data tend to show a disappearance of oligomers during the aggregation process suggesting that oligomers are intermediates in this process (Figs. 1 and 5). The role of oligomers as compulsory intermediates in this conversion is still to be solved and it is not our purpose to determine whether they are or not. Nevertheless, the aggregation process implicates a distribution of aggregates with different sizes and results in a dynamic equilibrium between those species. At the beginning of the aggregation, FTIR spectrum is representative of antiparallel β-sheet (42%) and α-helix and random coil structures (40%) (Fig. 4a). These data suggest that monomers already adopt a partially β-sheet folded structure showing high accessibility to region specific monoclonal antibodies (Fig. 3b) compared to oligomers and fibrils. The dimers formation and their quite constant amount (25–30%) (Fig. 1a, lanes 2, 3, 4) during the process are in agreement with data suggesting that Aβ(1–40) dimers are particularly stable at low concentration [50]. Their simultaneous presence with higher molecular weight oligomers suggests that dimers may be required in Aβ(1–40) nucleation (Fig. 1a, lanes 2, 3, 4). Our experiments show that monomers and dimers seem to have an important role in the aggregation process as they are present in the sample along the entire aggregation process. Dynamic equilibrium between the different species and especially between fibrils and monomers/dimers during the elongation phase may explain their contribution on SDS-PAGE (Fig. 1a) and FTIR spectra (Fig. 4) [48].
Our observations suggest that self-assembling of Aβ(1–40) from oligomers to fibrils involves the central (aa 17–24) and C-terminal parts (aa 30–40) of the peptide (Fig. 3). In the oligomeric state, shorter β-strands are supposed to be formed within these residues as attested by the initial position of the ~1,630 cm−1 peak and the lower β-sheet content (42%) calculated by curve fitting. Concomitant loss of helical and random coil structures and band shift (1,632–1,627 cm−1) observed during aggregation suggest the formation of β-sheets involving an increasing number of strands as observed by the increasing sizes of species on the AFM images. Aβ(1–40) forms linear entities with a length of ~50 nm (Fig. 2c) which evolve into mature ~1-μm-long fibrils (Fig. 2d) that display a parallel β-sheet structure. Formation of this structure supposes that intra-sheet H-bonding stabilizing the oligomers in antiparallel β-sheets becomes weaker than the increasing inter-sheet interactions therefore leading to a parallel β-sheet. According to [40], the conformational change from antiparallel to parallel β-sheet could be explained by a reorientation of the strands by 90°. In this case, oligomers represent a possible intermediate conformation on the fibrils pathway. Their formation as a transitory reservoir for fibrils and particularly the reorientation of the β-strands in the β-hairpin could determine the nucleation step which corresponds to the low ThT intensity sample (Fig. 5). This is in agreement with studies reporting that β-hairpin formation is responsible for the aggregation and toxicity [51].
If the presence of the two characteristic peaks (1,630 and 1,695 cm−1) in the infrared spectrum, which is the hallmark of an antiparallel β-sheet organization, was jeopardized in some specific cases [52], nevertheless the results observed here are in very good agreement with such an assignment. Moreover, the antiparallel β-sheet organization of Aβ(1–40) oligomers has been previously suggested in Habicht et al. [19] and is in agreement with previous structural studies carried out on Aβ(1–42) oligomers [20, 21] as well as on other proteins forming oligomers like β2-microglobulin [53] and PrP (prion related protein) peptide [PrP-(82–146)] [54]. Additionally, Aβ(1–40) oligomers are recognized by the A11 antibody. Kayed and co-workers [2] showed that this conformational antibody also reacts with oligomers of other amyloidogenic proteins linked to well-known diseases such as α-synuclein, islet amyloid polypeptide (IAPP), polyglutamine, lysosyme and human insulin. These results suggest a common structure for those oligomers implicating a common mechanism of pathogenesis. Recently, Götz and co-workers [55] found that Aβ(1–42) and human islet amyloid polypeptide (hIAPP) share a common toxicity mechanism via mitochondrial dysfunction.
In conclusion, we demonstrate that ATR-FTIR can be used to follow Aβ aggregation as well as the ThT probe assay. The different spectral features of oligomers and fibrils are used to determine the oligomeric content over aging time. This is, in our opinion, essential as the cytotoxicity was found to depend on the conformational organization of oligomers [56]. One could speculate that toxicity of oligomers may be related to their structural order, their morphologies and their ability to impair cellular processes by interacting with membranes as suggested by [6, 57].
As antiparallel β-sheet structure is also observed for other proteins forming oligomers [53, 54], the reorganization of β-sheet implicating a reorientation of β-strands could be a generic mechanism determining the kinetics and controlling the protein misfolding to form inert fibrils.
Acknowledgments
R.S. is Research Fellow for the Fund for Research in the Industry and Agriculture (Belgium), E.C. is Research Fellow for the National Fund for Scientific Research (Belgium), Y.F.D. and V.R. are Senior Research Associate and E.G. is Research Director for the National Fund for Scientific Research (Belgium). We thank Dr. C. Glabe for kindly providing A11 antibody, Dr. P. Boussard and Dr. Y. Looze for access to the fluorimeter.
Abbreviations
- Aβ
Amyloid-beta
- AD
Alzheimer’s disease
- ADDLs
Aβ-derived diffusible ligands
- AFM
Atomic force microscopy
- ATR
Attenuated total reflection
- DMSO
Dimethylsulfoxide
- FTIR
Fourier-transform infrared spectroscopy
- TBS
Tris-buffered saline solution
- ThT
Thioflavin T
Footnotes
R. Sarroukh and E. Cerf have equally contributed to this work.
References
- 1.Stefani M, Dobson CM. Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J Mol Med. 2003;81:678–699. doi: 10.1007/s00109-003-0464-5. [DOI] [PubMed] [Google Scholar]
- 2.Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 3.Vigo-Pelfrey C, Lee D, Keim P, Lieberburg I, Schenk DB. Characterization of beta-amyloid peptide from human cerebrospinal fluid. J Neurochem. 1993;61:1965–1968. doi: 10.1111/j.1471-4159.1993.tb09841.x. [DOI] [PubMed] [Google Scholar]
- 4.Wilquet V, De Strooper B. Amyloid-beta precursor protein processing in neurodegeneration. Curr Opin Neurobiol. 2004;14:582–588. doi: 10.1016/j.conb.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Xu Y, Shen J, Luo X, Zhu W, Chen K, Ma J, Jiang H. Conformational transition of amyloid beta-peptide. Proc Natl Acad Sci USA. 2005;102:5403–5407. doi: 10.1073/pnas.0501218102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simmons LK, May PC, Tomaselli KJ, Rydel RE, Fuson KS, Brigham EF, Wright S, Lieberburg I, Becker GW, Brems DN. Secondary structure of amyloid beta peptide correlates with neurotoxic activity in vitro. Mol Pharmacol. 1994;45:373–379. [PubMed] [Google Scholar]
- 7.Shankar GM, Li S, Mehta TH, Garcia-Munoz A, Shepardson NE, Smith I, Brett FM, Farrell MA, Rowan MJ, Lemere CA, Regan CM, Walsh DM, Sabatini BL, Selkoe DJ. Amyloid-beta protein dimers isolated directly from Alzheimer’s brains impair synaptic plasticity and memory. Nat Med. 2008;14:837–842. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lesne S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, Gallagher M, Ashe KH. A specific amyloid-beta protein assembly in the brain impairs memory. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 9.Kayed R, Pensalfini A, Margol L, Sokolov Y, Sarsoza F, Head E, Hall J, Glabe C. Annular protofibrils are a structurally and functionally distinct type of amyloid oligomer. J Biol Chem. 2009;284:4230–4237. doi: 10.1074/jbc.M808591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hardy JA, Higgins GA. Alzheimer’s disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 11.De Felice FG, Wu D, Lambert MP, Fernandez SJ, Velasco PT, Lacor PN, Bigio EH, Jerecic J, Acton PJ, Shughrue PJ, Chen-Dodson E, Kinney GG, Klein WL. Alzheimer’s disease-type neuronal tau hyperphosphorylation induced by Aβ oligomers. Neurobiol Aging. 2008;29:1334–1347. doi: 10.1016/j.neurobiolaging.2007.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Felice FG, Velasco PT, Lambert MP, Viola K, Fernandez SJ, Ferreira ST, Klein WL. Abeta oligomers induce neuronal oxidative stress through an N-methyl-d-aspartate receptor-dependent mechanism that is blocked by the Alzheimer drug memantine. J Biol Chem. 2007;282:11590–11601. doi: 10.1074/jbc.M607483200. [DOI] [PubMed] [Google Scholar]
- 13.Wang HW, Pasternak JF, Kuo H, Ristic H, Lambert MP, Chromy B, Viola KL, Klein WL, Stine WB, Krafft GA, Trommer BL. Soluble oligomers of beta amyloid (1–42) inhibit long-term potentiation but not long-term depression in rat dentate gyrus. Brain Res. 2002;924:133–140. doi: 10.1016/S0006-8993(01)03058-X. [DOI] [PubMed] [Google Scholar]
- 14.Lambert MP, Barlow AK, Chromy BA, Edwards C, Freed R, Liosatos M, Morgan TE, Rozovsky I, Trommer B, Viola KL, Wals P, Zhang C, Finch CE, Krafft GA, Klein WL. Diffusible, nonfibrillar ligands derived from Abeta1–42 are potent central nervous system neurotoxins. Proc Natl Acad Sci USA. 1998;95:6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lacor PN, Buniel MC, Furlow PW, Clemente AS, Velasco PT, Wood M, Viola KL, Klein WL. Abeta oligomer-induced aberrations in synapse composition, shape, and density provide a molecular basis for loss of connectivity in Alzheimer’s disease. J Neurosci. 2007;27:796–807. doi: 10.1523/JNEUROSCI.3501-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 17.Ahmed M, Davis J, Aucoin D, Sato T, Ahuja S, Aimoto S, Elliott JI, Van Nostrand WE, Smith SO. Structural conversion of neurotoxic amyloid-beta(1–42) oligomers to fibrils. Nat Struct Mol Biol. 2010;17:561–567. doi: 10.1038/nsmb.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu L, Edalji R, Harlan JE, Holzman TF, Lopez AP, Labkovsky B, Hillen H, Barghorn S, Ebert U, Richardson PL, Miesbauer L, Solomon L, Bartley D, Walter K, Johnson RW, Hajduk PJ, Olejniczak ET. Structural characterization of a soluble amyloid beta-peptide oligomer. Biochemistry. 2009;48:1870–1877. doi: 10.1021/bi802046n. [DOI] [PubMed] [Google Scholar]
- 19.Habicht G, Haupt C, Friedrich RP, Hortschansky P, Sachse C, Meinhardt J, Wieligmann K, Gellermann GP, Brodhun M, Gotz J, Halbhuber KJ, Rocken C, Horn U, Fandrich M. Directed selection of a conformational antibody domain that prevents mature amyloid fibril formation by stabilizing Abeta protofibrils. Proc Natl Acad Sci USA. 2007;104:19232–19237. doi: 10.1073/pnas.0703793104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cerf E, Sarroukh R, Tamamizu-Kato S, Breydo L, Derclaye S, Dufrene YF, Narayanaswami V, Goormaghtigh E, Ruysschaert JM, Raussens V. Antiparallel beta-sheet: a signature structure of the oligomeric amyloid beta-peptide. Biochem J. 2009;421:415–423. doi: 10.1042/BJ20090379. [DOI] [PubMed] [Google Scholar]
- 21.Eckert A, Hauptmann S, Scherping I, Meinhardt J, Rhein V, Drose S, Brandt U, Fandrich M, Muller WE, Gotz J. Oligomeric and fibrillar species of beta-amyloid (Aβ 42) both impair mitochondrial function in P301L tau transgenic mice. J Mol Med. 2008;86:1255–1267. doi: 10.1007/s00109-008-0391-6. [DOI] [PubMed] [Google Scholar]
- 22.Come JH, Fraser PE, Lansbury PT., Jr A kinetic model for amyloid formation in the prion diseases: importance of seeding. Proc Natl Acad Sci USA. 1993;90:5959–5963. doi: 10.1073/pnas.90.13.5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Necula M, Kayed R, Milton S, Glabe CG. Small molecule inhibitors of aggregation indicate that amyloid beta oligomerization and fibrillization pathways are independent and distinct. J Biol Chem. 2007;282:10311–10324. doi: 10.1074/jbc.M608207200. [DOI] [PubMed] [Google Scholar]
- 24.Wetzel R. Kinetics and thermodynamics of amyloid fibril assembly. Acc Chem Res. 2006;39:671–679. doi: 10.1021/ar050069h. [DOI] [PubMed] [Google Scholar]
- 25.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. A structural model for Alzheimer’s beta-amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci USA. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balbach JJ, Petkova AT, Oyler NA, Antzutkin ON, Gordon DJ, Meredith SC, Tycko R. Supramolecular structure in full-length Alzheimer’s beta-amyloid fibrils: evidence for a parallel beta-sheet organization from solid-state nuclear magnetic resonance. Biophys J. 2002;83:1205–1216. doi: 10.1016/S0006-3495(02)75244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt M, Sachse C, Richter W, Xu C, Fandrich M, Grigorieff N. Comparison of Alzheimer Abeta(1–40) and Abeta(1–42) amyloid fibrils reveals similar protofilament structures. Proc Natl Acad Sci USA. 2009;106:19813–19818. doi: 10.1073/pnas.0905007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teplow DB, Lazo ND, Bitan G, Bernstein S, Wyttenbach T, Bowers MT, Baumketner A, Shea JE, Urbanc B, Cruz L, Borreguero J, Stanley HE. Elucidating amyloid beta-protein folding and assembly: a multidisciplinary approach. Acc Chem Res. 2006;39:635–645. doi: 10.1021/ar050063s. [DOI] [PubMed] [Google Scholar]
- 29.Chen D, Martin ZS, Soto C, Schein CH. Computational selection of inhibitors of Abeta aggregation and neuronal toxicity. Bioorg Med Chem. 2009;17:5189–5197. doi: 10.1016/j.bmc.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LeVine H., III Quantification of beta-sheet amyloid fibril structures with thioflavin T. Methods Enzymol. 1999;309:274–284. doi: 10.1016/S0076-6879(99)09020-5. [DOI] [PubMed] [Google Scholar]
- 31.Goormaghtigh E, Raussens V, Ruysschaert JM. Attenuated total reflection infrared spectroscopy of proteins and lipids in biological membranes. Biochim Biophys Acta. 1999;1422:105–185. doi: 10.1016/s0304-4157(99)00004-0. [DOI] [PubMed] [Google Scholar]
- 32.Goormaghtigh E, Cabiaux V, Ruysschaert JM. Secondary structure and dosage of soluble and membrane proteins by attenuated total reflection Fourier-transform infrared spectroscopy on hydrated films. Eur J Biochem. 1990;193:409–420. doi: 10.1111/j.1432-1033.1990.tb19354.x. [DOI] [PubMed] [Google Scholar]
- 33.Mastrangelo IA, Ahmed M, Sato T, Liu W, Wang C, Hough P, Smith SO. High-resolution atomic force microscopy of soluble Abeta42 oligomers. J Mol Biol. 2006;358:106–119. doi: 10.1016/j.jmb.2006.01.042. [DOI] [PubMed] [Google Scholar]
- 34.Chirgadze YN, Nevskaya NA. Infrared spectra and resonance interaction of amide-I vibration of the antiparallel-chain pleated sheet. Biopolymers. 1976;15:607–625. doi: 10.1002/bip.1976.360150402. [DOI] [PubMed] [Google Scholar]
- 35.Krimm S, Bandekar J. Vibrational spectroscopy and conformation of peptides, polypeptides, and proteins. Adv Protein Chem. 1986;38:181–364. doi: 10.1016/S0065-3233(08)60528-8. [DOI] [PubMed] [Google Scholar]
- 36.Miyazawa T, Blout ER. The Infrared spectra of polypeptides in various conformations: amide I and II bands. J Am Chem Soc. 1961;83(3):712–719. doi: 10.1021/ja01464a042. [DOI] [Google Scholar]
- 37.Oberg KA, Ruysschaert JM, Goormaghtigh E. The optimization of protein secondary structure determination with infrared and circular dichroism spectra. Eur J Biochem. 2004;271:2937–2948. doi: 10.1111/j.1432-1033.2004.04220.x. [DOI] [PubMed] [Google Scholar]
- 38.Kubelka J, Keiderling TA. Differentiation of beta-sheet-forming structures: ab initio-based simulations of IR absorption and vibrational CD for model peptide and protein beta-sheets. J Am Chem Soc. 2001;123:12048–12058. doi: 10.1021/ja0116627. [DOI] [PubMed] [Google Scholar]
- 39.Zandomeneghi G, Krebs MR, McCammon MG, Fandrich M. FTIR reveals structural differences between native beta-sheet proteins and amyloid fibrils. Protein Sci. 2004;13:3314–3321. doi: 10.1110/ps.041024904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoyer W, Gronwall C, Jonsson A, Stahl S, Hard T. Stabilization of a beta-hairpin in monomeric Alzheimer’s amyloid-beta peptide inhibits amyloid formation. Proc Natl Acad Sci USA. 2008;105:5099–5104. doi: 10.1073/pnas.0711731105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer’s beta-amyloid fibrils. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bitan G, Fradinger EA, Spring SM, Teplow DB. Neurotoxic protein oligomers—what you see is not always what you get. Amyloid. 2005;12:88–95. doi: 10.1080/13506120500106958. [DOI] [PubMed] [Google Scholar]
- 43.Tomaselli S, Esposito V, Vangone P, van Nuland NA, Bonvin AM, Guerrini R, Tancredi T, Temussi PA, Picone D. The alpha-to-beta conformational transition of Alzheimer’s Abeta-(1–42) peptide in aqueous media is reversible: a step by step conformational analysis suggests the location of beta conformation seeding. Chembiochem. 2006;7:257–267. doi: 10.1002/cbic.200500223. [DOI] [PubMed] [Google Scholar]
- 44.Fraser PE, Nguyen JT, Surewicz WK, Kirschner DA. pH-dependent structural transitions of Alzheimer amyloid peptides. Biophys J. 1991;60:1190–1201. doi: 10.1016/S0006-3495(91)82154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peralvarez-Marin A, Mateos L, Zhang C, Singh S, Cedazo-Minguez A, Visa N, Morozova-Roche L, Graslund A, Barth A. Influence of residue 22 on the folding, aggregation profile, and toxicity of the Alzheimer’s amyloid beta peptide. Biophys J. 2009;97:277–285. doi: 10.1016/j.bpj.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walsh DM, Selkoe DJ. Aβ oligomers—a decade of discovery. J Neurochem. 2007;101:1172–1184. doi: 10.1111/j.1471-4159.2006.04426.x. [DOI] [PubMed] [Google Scholar]
- 47.Dasilva KA, Shaw JE, McLaurin J. Amyloid-beta fibrillogenesis: structural insight and therapeutic intervention. Exp Neurol. 2009;223(2):311–321. doi: 10.1016/j.expneurol.2009.08.032. [DOI] [PubMed] [Google Scholar]
- 48.Necula M, Breydo L, Milton S, Kayed R, van der Veer WE, Tone P, Glabe CG. Methylene blue inhibits amyloid Abeta oligomerization by promoting fibrillization. Biochemistry. 2007;46:8850–8860. doi: 10.1021/bi700411k. [DOI] [PubMed] [Google Scholar]
- 49.Ferrao-Gonzales AD, Robbs BK, Moreau VH, Ferreira A, Juliano L, Valente AP, Almeida FC, Silva JL, Foguel D. Controlling {beta}-amyloid oligomerization by the use of naphthalene sulfonates: trapping low molecular weight oligomeric species. J Biol Chem. 2005;280:34747–34754. doi: 10.1074/jbc.M501651200. [DOI] [PubMed] [Google Scholar]
- 50.Garzon-Rodriguez W, Sepulveda-Becerra M, Milton S, Glabe CG. Soluble amyloid Abeta-(1–40) exists as a stable dimer at low concentrations. J Biol Chem. 1997;272:21037–21044. doi: 10.1074/jbc.272.34.21037. [DOI] [PubMed] [Google Scholar]
- 51.Liao MQ, Tzeng YJ, Chang LY, Huang HB, Lin TH, Chyan CL, Chen YC. The correlation between neurotoxicity, aggregative ability and secondary structure studied by sequence truncated Abeta peptides. FEBS Lett. 2007;581:1161–1165. doi: 10.1016/j.febslet.2007.02.026. [DOI] [PubMed] [Google Scholar]
- 52.Khurana R, Fink AL. Do parallel beta-helix proteins have a unique fourier transform infrared spectrum? Biophys J. 2000;78:994–1000. doi: 10.1016/S0006-3495(00)76657-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fabian H, Gast K, Laue M, Misselwitz R, Uchanska-Ziegler B, Ziegler A, Naumann D. Early stages of misfolding and association of beta2-microglobulin: insights from infrared spectroscopy and dynamic light scattering. Biochemistry. 2008;47:6895–6906. doi: 10.1021/bi800279y. [DOI] [PubMed] [Google Scholar]
- 54.Natalello A, Prokorov VV, Tagliavini F, Morbin M, Forloni G, Beeg M, Manzoni C, Colombo L, Gobbi M, Salmona M, Doglia SM. Conformational plasticity of the Gerstmann–Straussler–Scheinker disease peptide as indicated by its multiple aggregation pathways. J Mol Biol. 2008;381:1349–1361. doi: 10.1016/j.jmb.2008.06.063. [DOI] [PubMed] [Google Scholar]
- 55.Lim YA, Rhein V, Baysang G, Meier F, Poljak A, Raftery MJ, Guilhaus M, Ittner LM, Eckert A, Gotz J. Abeta and human amylin share a common toxicity pathway via mitochondrial dysfunction. Proteomics. 2010;10:1621–1633. doi: 10.1002/pmic.200900651. [DOI] [PubMed] [Google Scholar]
- 56.Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 57.Ravault S, Soubias O, Saurel O, Thomas A, Brasseur R, Milon A. Fusogenic Alzheimer’s peptide fragment Abeta (29–42) in interaction with lipid bilayers: secondary structure, dynamics, and specific interaction with phosphatidyl ethanolamine polar heads as revealed by solid-state NMR. Protein Sci. 2005;14:1181–1189. doi: 10.1110/ps.041291405. [DOI] [PMC free article] [PubMed] [Google Scholar]